- 1College of Life Science and Engineering, Henan University of Urban Construction, Pingdingshan, China
- 2Department of Plant Sciences, College of Life Sciences, Wuhan University, Wuhan, China
The Jacalin-related lectins (JRLs) gene family play a crucial role in regulating plant development and responding to environmental stress. However, a systematic bioinformatics analysis of the JRL gene family in Gramineae plants has been lacking. In this study, we identified 101 JRL proteins from five Gramineae species and classified them into eight distinct clades. Most of the AtJRL proteins clustered in the same group and were differentiated from the Gramineae JRL proteins. The analysis of protein motifs, gene structures and protein domain revealed that the JRL genes play diverse functions in Gramineae plants. Duplication events indicated that tandem duplication significantly contributed to the expansion of the JRL family, with most JRL members underwent purifying selection. Tissue expression profile analysis showed that most OsJRL genes were highly expressed in the roots, while ZmJRL genes exhibited high expression in inflorescences. Furthermore, the expression level of OsJRL and ZmJRL genes were influenced by drought, cold, heat and salt stresses, respectively, implying that these genes play important roles in response to various abiotic stresses. RT-qPCR results demonstrated that OsJRL4 was up-regulated under PEG6000 and NaCl stresses, while OsJRL12 and OsJRL26 were down-regulated under PEG6000. These findings provide comprehensive insights into the JRL gene family in Gramineae plants and will facilitate further functional characterization of JRLs.
1 Introduction
The Poaceae family comprises the most widely distributed monocotyledonous plants, and is also the most economically significant large family. Gramineous plants serve as the primary source of human food and livestock feed, functioning as essential raw materials for the production of starch, sugar, wine, paper, textiles, and construction materials. In recent years, the rapid advancement of genomics has led to the sequencing for an increasing number of Gramineous plant genomes (Mockler et al., 2010; Tao et al., 2021; Shang et al., 2023; He et al., 2024). The successful completion of whole-genome sequencing for these species established a robust foundation for further exploration of genomic information, evolutionary conduction and genetic studies. The genomes of Gramineous plants have primarily evolved through a process of gene duplication and loss (Shuai et al., 2002; Lin et al., 2003). Following these events, duplicated genes may experience either purifying or positive selection thus facilitating adaptive evolution. Although these genomes exhibit certain degrees of collinearity, the genomic information varies significantly among species. Notably, many gene families within Gramineae plants display species-specific characteristics due to gene duplication or loss across different species (Gossmann et al., 2010; Duan et al., 2023; Liu et al., 2024).
Plant lectins, exhibits unique molecular structure, are being classified into three types of ‘merolectins’, ‘hololectins’, and ‘chimerolectins’ (Tsaneva and Van Damme, 2020). JRL proteins, known as new plant lectins, play crucial roles in mediating stress signaling pathways across various plant species (Subramanyam et al., 2008; De Coninck and Van Damme, 2021; Marothia et al., 2023). Early studies have highlighted JRL isoforms, Orysata1 and Orysata2, in responding to NaCl stress (Zhang et al., 2000). Recent studies indicate that the PeDJ01 from moso bamboo is up-regulated in response to salt or cold stress, highlighting its significant role in stress regulation (Ma et al., 2021). Over-expression of OsJAC1 in rice enhances resistance to oomycetes and fungi, with its JRL domain being crucial for binding to penetration site of powdery mildew (Weidenbach et al., 2016). Furthermore, OsJAC1 and wheat VER2 play a important role in lignan formation (Burlat et al., 2001; Jiang et al., 2006; Wang and Ma, 2005; Yong et al., 2003).
The expression levels of JRL genes are responsive to various plant hormones. Notably, OsJAC demonstrates high transcription levels in leaves and is inducible by jasmonate (JA) treatment (Jiang et al., 2006). The JRL-like protein TaJRLL1, which contains two jacalin-like lectin domains, is involved in the salicylic acid (SA)/JA signaling pathway (Xiang et al., 2011). In Helianthus tuberosus callus, the level of HTA gene were notably up-regulated by MeJA treatment (Nakagawa et al., 2000). Although, the expression of LEM2 was rapidly induced by SA, it was decreased when responding to drought, dehydration and abscisic acid (ABA) treatments (Abebe et al., 2005).
The sequencing of plant genomes has enabled the identification of JRL genes in Arabidopsis, rice, and wheat (Nagano et al., 2008; Jiang et al., 2010; Song et al., 2013). However, comprehensive bioinformatic analysis of JRL genes in Gramineous plants is lacking. In this study, we screened and identified JRL family members in the genomes of 5 Gramineae plants and examined their evolutionary relationships, cis-acting elements, gene collinearity, and duplication events. RNA-seq and RT-qPCR results were utilized to explore the expression patterns of JRL genes under diverse abiotic stresses. This research serves as a valuable resource for further researches on the biological functions and stress resistance mechanisms involving JRL genes in plants.
2 Results
2.1 Identification and phylogenetic analysis of JRL family members
In this study, 27, 18, 17, 16, and 23 JRL protein members were identified from Oryza sativa (O. sativa), Brachypodium distachyon (B. distachyon), Zea mays (Z. mays), Sorghum bicolor (S. bicolor), and Setaria italica (S. italica), respectively (Figure 1; Supplementary Table 1). We found that O. sativa has more JRL genes than S. italica, S. bicolor, B. distachyon and Z. mays, implying a gene expansion of the JRL gene family among different Gramineae species. Further, we named these JRL genes according to their location on the chromosomes: OsJRL1-OsJRL27; BdJRL1-BdJRL18; ZmJRL1-ZmJRL17; SbJRL1-SbJRL16 and SiJRL1-SiJRL23 (Figure 1). The JRL genes of O. sativa are distributed on 9 chromosomes (Figure 1A). BdJRL genes (Figure 1B) and AtJRL genes (Figure 1F) are focusly distributed on 4 chromosomes. While ZmJRL genes and SiJRL genes are distributed on 7 chromosomes, respevtively (Figures 1C, E), SbJRL genes are distributed on chromosomes 1, 2, 3, 5, and 9 (Figure 1D). And these JRL genes are often distributed together in clusters (Figure 1).
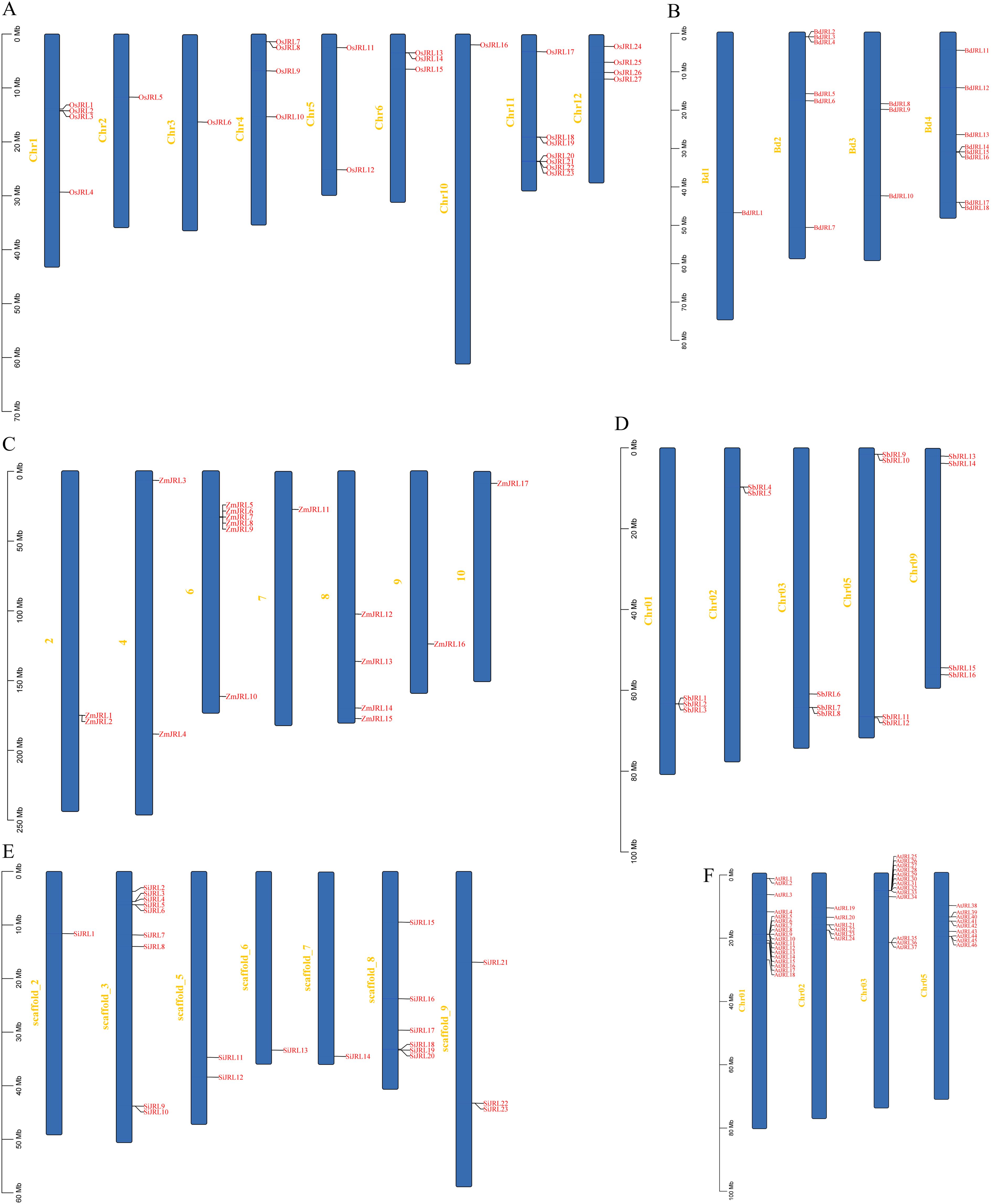
Figure 1. The chromosomal location of the JRL genes in six species, including (A) O. sativa, (B) B. distachyon, (C) Z. mays, (D) S. bicolor, (E) S. italica and (F) A. thaliana.
To investigate the evolutionary mechanism of JRL proteins in Gramineous, we used the Neighbor-Joining (N-J) method to construct an evolutionary tree including 46 AtJRL, 27 OsJRL, 23 SiJRL, 16 SbJRL, 18 BdJRL and 17 ZmJRL proteins. As shown in Figure 2, these JRL proteins are divided into 8 subgroups. The JRL-IV group contains the most members, including 12 OsJRL, 1 AtJRL, 10 SiJRL, 12 ZmJRL, 8 SbJRL, and 2 BdJRL members. In contrast, JRL-I contains the fewest JRL members. Interestingly, the JRL-III and JRL-VI groups only include the Gramineous JRL proteins. And JRL-VIII is unique to Arabidopsis, containing 33 AtJRL proteins (71.7%). In addition, we found that the ZmJRL is lost in the group of JRL-III and JRL-V. Meanwhile, both JRL-IV and JRL-VII contain members of Gramineae JRL and Arabidopsis JRL, suggesting that JRL members in these two groups may function conserved between Arabidopsis and Gramineae (Figure 2).
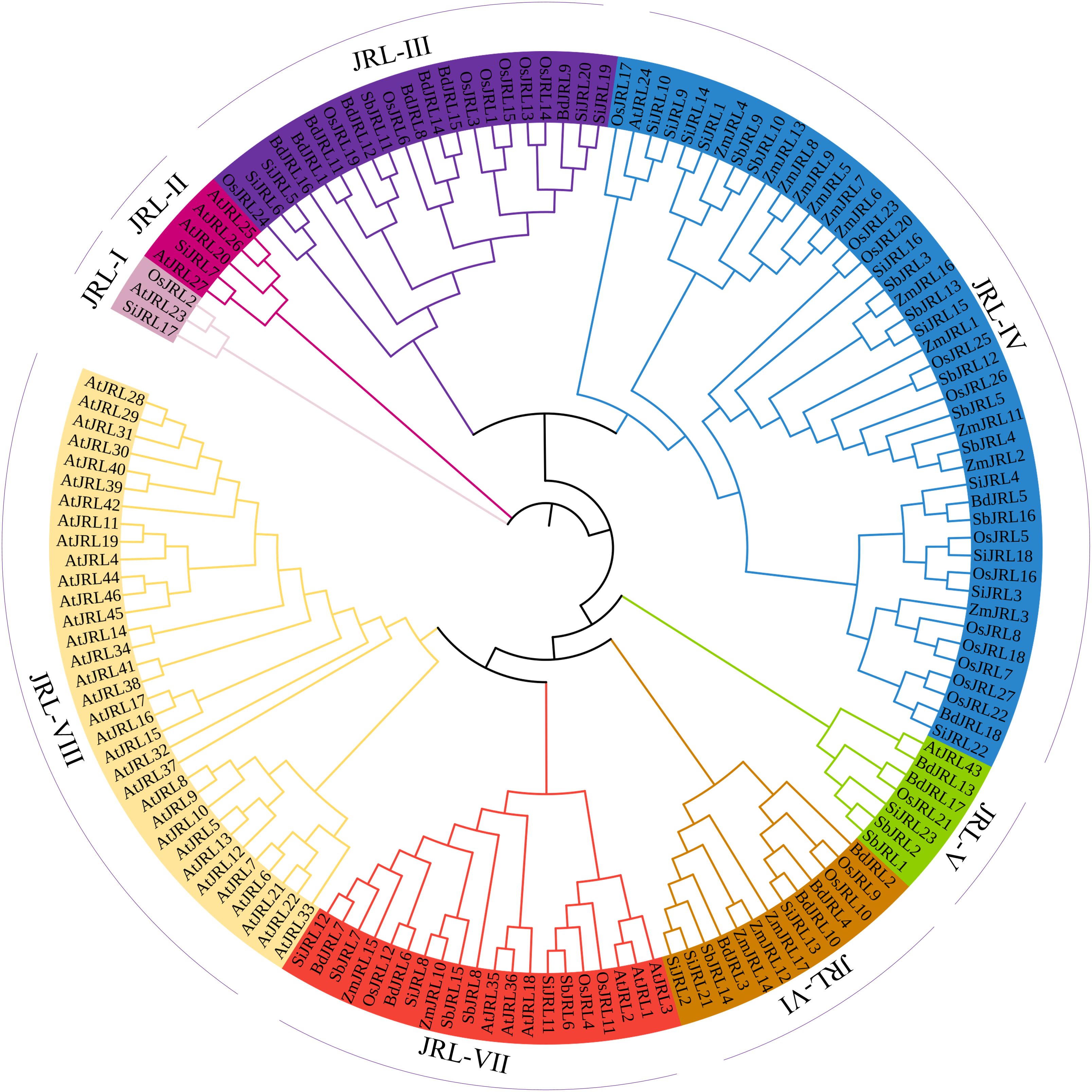
Figure 2. The evolutionary analysis of the 147 identified JRLs proteins sequences from O. sativa, Z. mays, B.distachyon, S. bicolor, S. italica, and Arabidopsis. 147 JRLs were divided into eight subgroups of JRL-I, JRL-II, JRL-III, JRL-IV, JRL-V,JRL-VI, JRL-VII and JRL-VIII respectively. The evolutionary tree was analyzed by MEGA 7 (method: neighbor-joining (N-J); bootstrap values: 1000 replicates).
2.2 Gene structure and protein motif analysis
The diversity of gene structure can reflect the evolutionary processes of gene family. We analyzed the JRL gene structure, protein motif and conserved domain in O. sativa, B. distachyon, Z. mays, S. bicolor, S. italica and A. thaliana. The protein motif analysis showed that nearly all JRL proteins contain motif 3, motif 4, motif 8, motif 9 and motif 10, indicating that these motifs may be conserved for the functions roles of JRL proteins. We observed that JRL members of the same subgroup exhibit similar conserved motifs (Figures 3A, B). In contrast to the other six subgroups, members of the JRL-VII and JRL-VIII subgroups contain two or three Motif 1 and Motif 2 in tandem. While JRL-VIII comprises only Arabidopsis members, JRL-VII includes both Gramineae JRL genes and AtJRL genes. These suggests that the members of these two subgroups may possess unique functions in the evolutionary process, warranting further investigation.
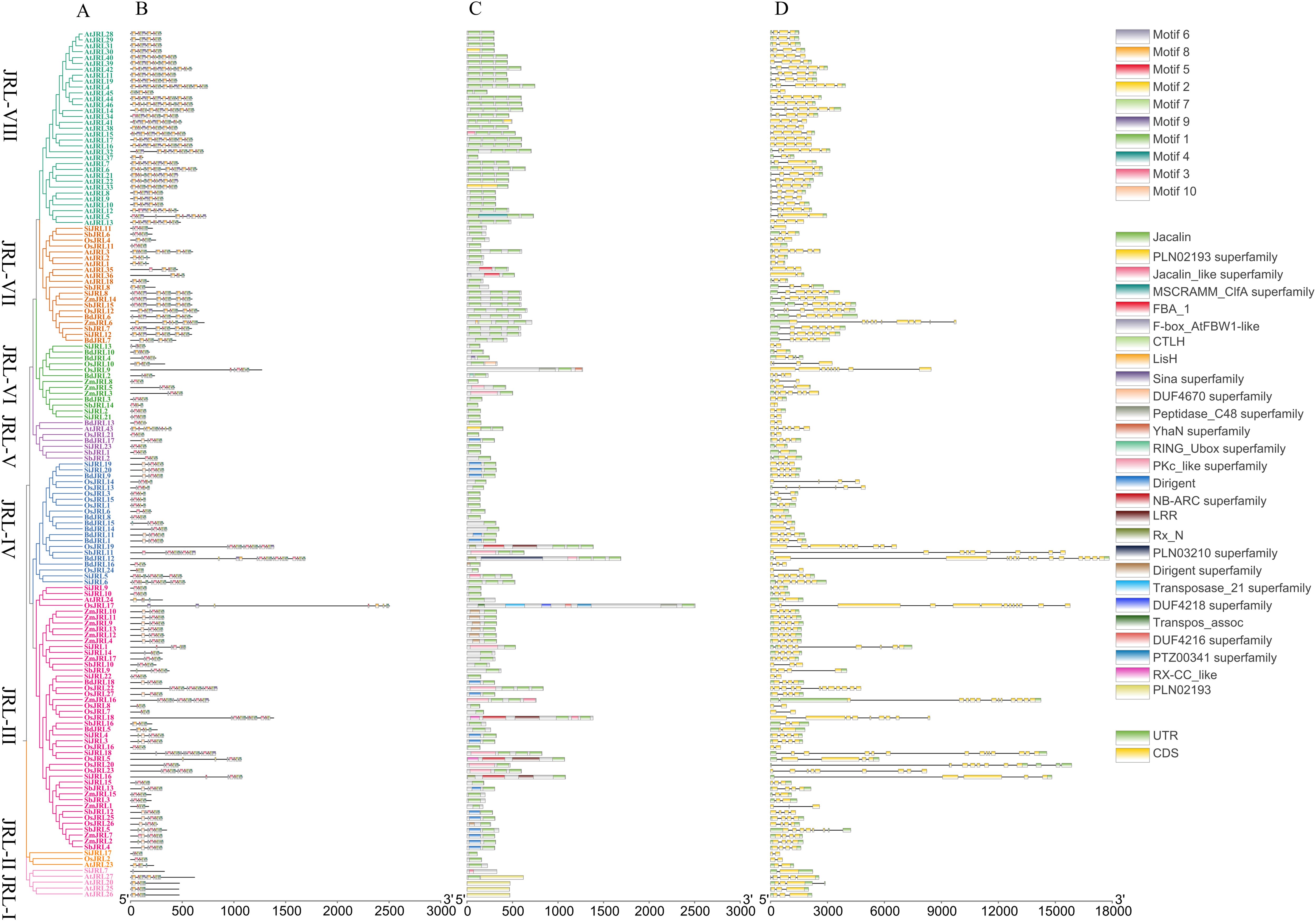
Figure 3. Phylogenetic relationship, conserved motif, conserved domain and gene structure analysis of JRLs in Gramineae and Arabidopsis. (A) Phylogenetic tree of JRL proteins in Gramineae and Arabidopsis. The phylogenetic tree was constructed with MEGA 7 through neighbor-joining method using the full-length amino acid sequences of JRL proteins, and the bootstrap test replicate was set as 1000. (B) Distributions of conserved motifs in JRL proteins in Gramineae and Arabidopsis. Ten putative motifs are indicated in different colored boxes. (C) The conserved domain of JRL full-length amino acid sequences in Gramineae and Arabidopsis. (D) Exon/intron organization of JRL genes in Gramineae and Arabidopsis. Yellow boxes represent exons, gray lines represent introns and green boxes represent UTR.
We found that AtJRL28, AtJRL29, SiJRL5, and BdJRL7 contain two “Jacalin domain”, while some JRL proteins like SiJRL8, ZmJRL14, OsJRL12, and SbJRL15 possess three “Jacalin domain”. Interestingly, we found the members in JRL-VII and JRL-VIII subgroups contain 1-5 Jacalin domains. However, the conserved domain of S. bicolor is relatively simple, comprising only three types of domains: Jacalin, Dirigent and PKc_like superfamily. In contrast, the OsJRL members contain more complex conserved domains (Figures 3A, C). These suggests that there are both functional conservation and functional differentiation among the Gramineae JRL family members. Moreover, most AtJRL proteins that only contained Jacalin domain and no other domains (Figure 3). Notably, AtJRL proteins are more likely to have multiple tandem Jacalin domain repeats. For example, AtJRL4 is composed of 5 tandem Jacalin domains (Figures 3A, C). It is noteworthy that a dirigent domain, which is the key domain of DIR proteins (Gong et al., 2023), was found at the N-terminal of the Gramineous JRL proteins, but was not present in Arabidopsis.
The number of exons in JRL genes ranged from 2 to 14 (Figures 3A, C, D). In O. sativa, the number of introns varied from 1 to 12. However, AtJRL genes exhibited a narrower intron range of 1-6 (Figure 3D). Furthermore, we observed that members of the same subgroup tend to exhibit similar gene structures. Previous studies have indicated that in the same gene family, genes with multiple introns play significant roles in stress resistance compared to those without introns (Shaul, 2017). The considerable variation in gene structure among Gramineous plants suggests potential substantial differences in gene function.
2.3 Analysis of duplication event of JRL genes
During evolution, tandem duplication (TD) and whole genome duplication (WGD) have played crucial roles in the expansion of gene families. In this study, we analyzed the duplication events of the JRL gene family members in O. sativa, Z. mays, S. italica, S. bicolor, B. distachyon and Arabidopsis. As shown in Figure 4; Supplementary Table 2, we identified 61 TD and 18 WGD genes. Specifically, we found 6, 7, 6, 7, 8, and 27 TD genes, and 0, 4, 2, 4, 4 and 3 WGD genes in O. sativa, S. bicolor, B. distachyon, Z. mays, S. italica, and Arabidopsis, respectively. The TD genes of AtJRL6-13 (Figure 4) are predominantly located in Group VIII of Figure 2. Moreover, some TD genes of O. sativa, Z. mays, S. italica, S. bicolor and B. distachyon were grouped into the same subfamily (Figure 2), suggesting that the expansion of JRL gene families in Z. mays, S. italica, S. bicolor and B. distachyon is primarily driven by the significant expansion of a specific subfamily. It can be inferred that both TD and WGD events jointly promote the expansion of JRL gene families in O. sativa, Z. mays, S. italica, S. bicolor, B. distachyon and Arabidopsis, with TD playing a a major dominant role.
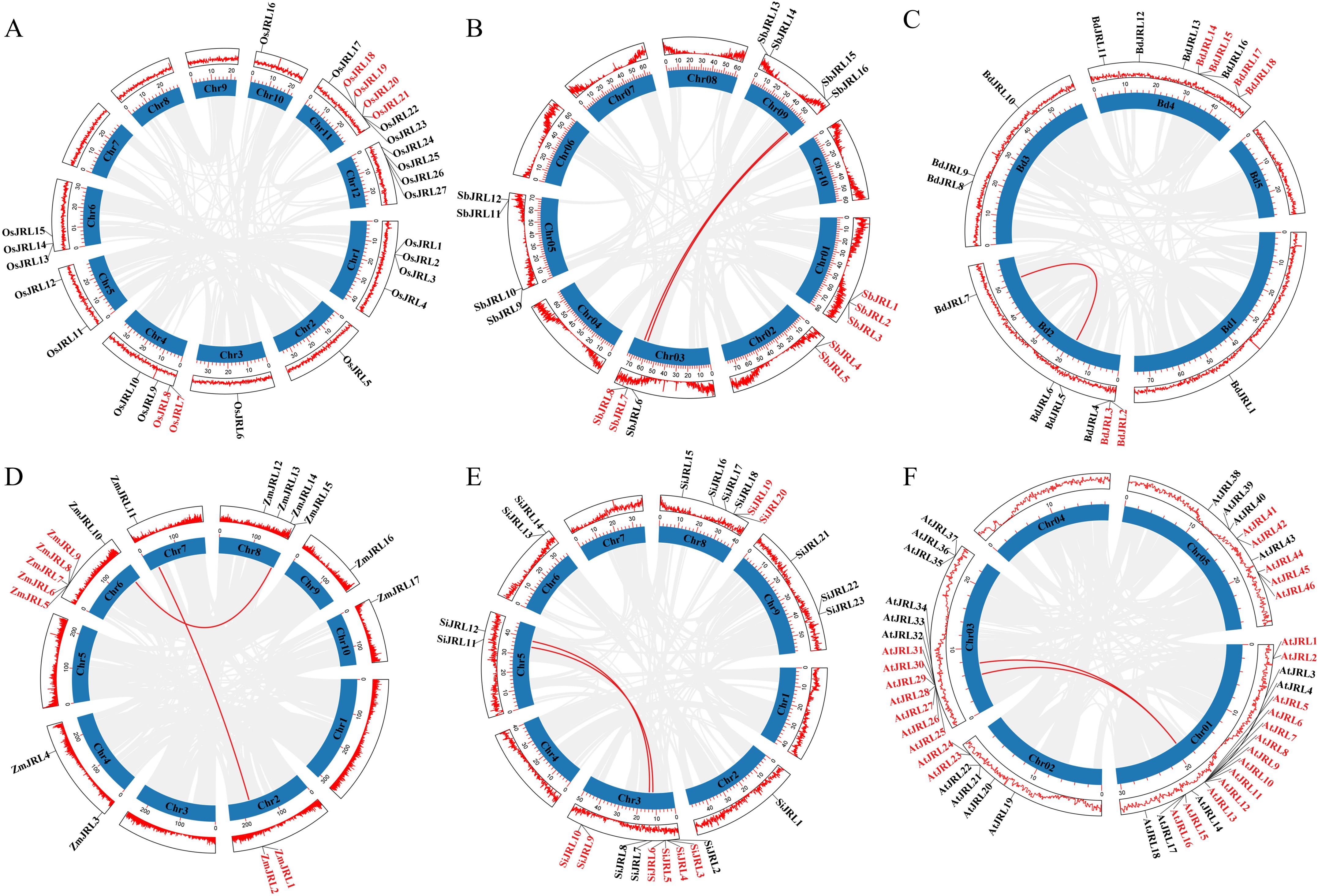
Figure 4. The duplication events analysis of JRL genes in Gramineae and Arabidopsis, including (A) O. sativa, (B) S. bicolor, (C) B. distachyon, (D) Z. mays, (E) S. italica and (F) Arabidopsis. The name of TD genes are indicated in red color. The WGD genes are indicated in red lines.
Further, we calculated the ratio of Ka/Ks on TD and WGD gene pairs. And except for one gene pair in rice was larger than 1, the ratio of Ka/Ks of all other replicated gene pairs were smaller than 1 (Figure 5A). Additionally, we evaluated the appearance time of these TD and WGD genes. And the divergence time of TD genes in the ZmJRL, BdJRL, SiJRL, OsJRL, SbJRL and AtJRL ranged from 2.77 to 62.68, while WGD genes in these species ranged from 31.15 to 80.44 (Figure 5B). These results hint that most duplicate genes of the JRL family were under purification selection, and TD duplication events might contributed chiefly to their evolution in O. sativa, A. thaliana, S. italica, B.distachyon, S.bicolor and Z. may.
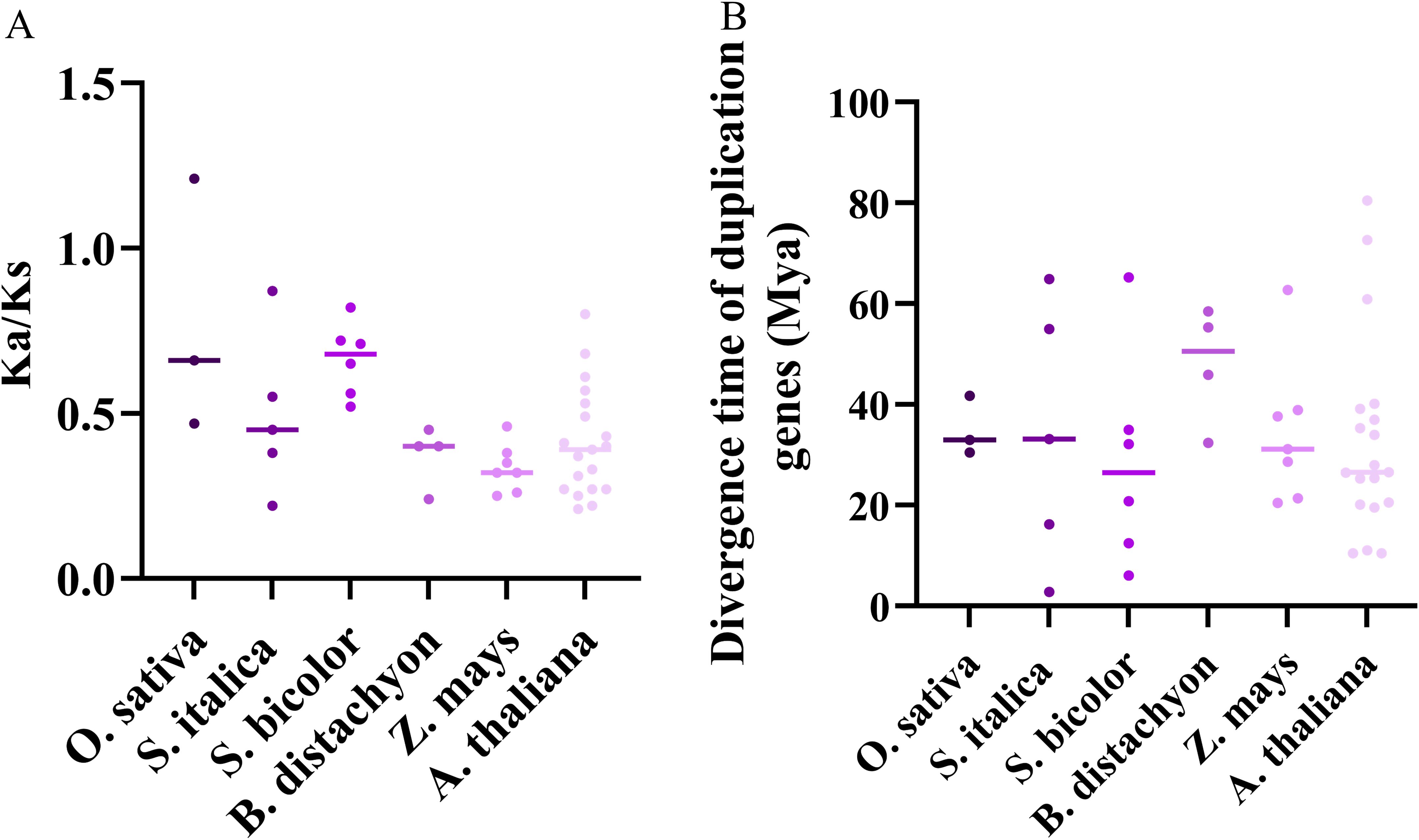
Figure 5. Selection pressure and divergence time analysis of JRL gene pairs in O. sativa, A. thaliana, S. italica, B.distachyon, S.bicolor and Z. may. (A) Ka/Ks ratio calculation of the JRL gene pairs. (B) The divergence time prediction of JRL gene pairs.
2.4 Collinearity analysis
To further investigate the evolutionary mechanism of the JRL gene, we conducted a collinearity analysis across O. sativa, B. distachyon, S. bicolor, S. italica, Z. mays, and A. thaliana. As shown in Figure 6, the number of collinear gene pairs between O. sativa and the other species are follows: 9 with B. distachyon, 8 with S.bicolor, 8 with S. italica, 6 with Z. may and 0 with A. thaliana. It is noteworthy that some OsJRL genes exhibit collinear relationships with at least two genes. For example, OsJRL4 is collinear with SbJRL6 and SbJRL16; OsJRL12 is collinear with SbJRL7, SbJRL15, ZmJRL10 and ZmJRL15.. These findings suggest that these genes may represent significant genetic resources. Notably, the genetic relationship between O. sativa and B. distachyon is closer than that observed among S. italica, Z. may, and S.bicolor. However, no co-linear gene pairs between O. sativa and A. thaliana was found, which demonstrates the function differences between monocotyledonous and dicotyledonous plants, to some extent.
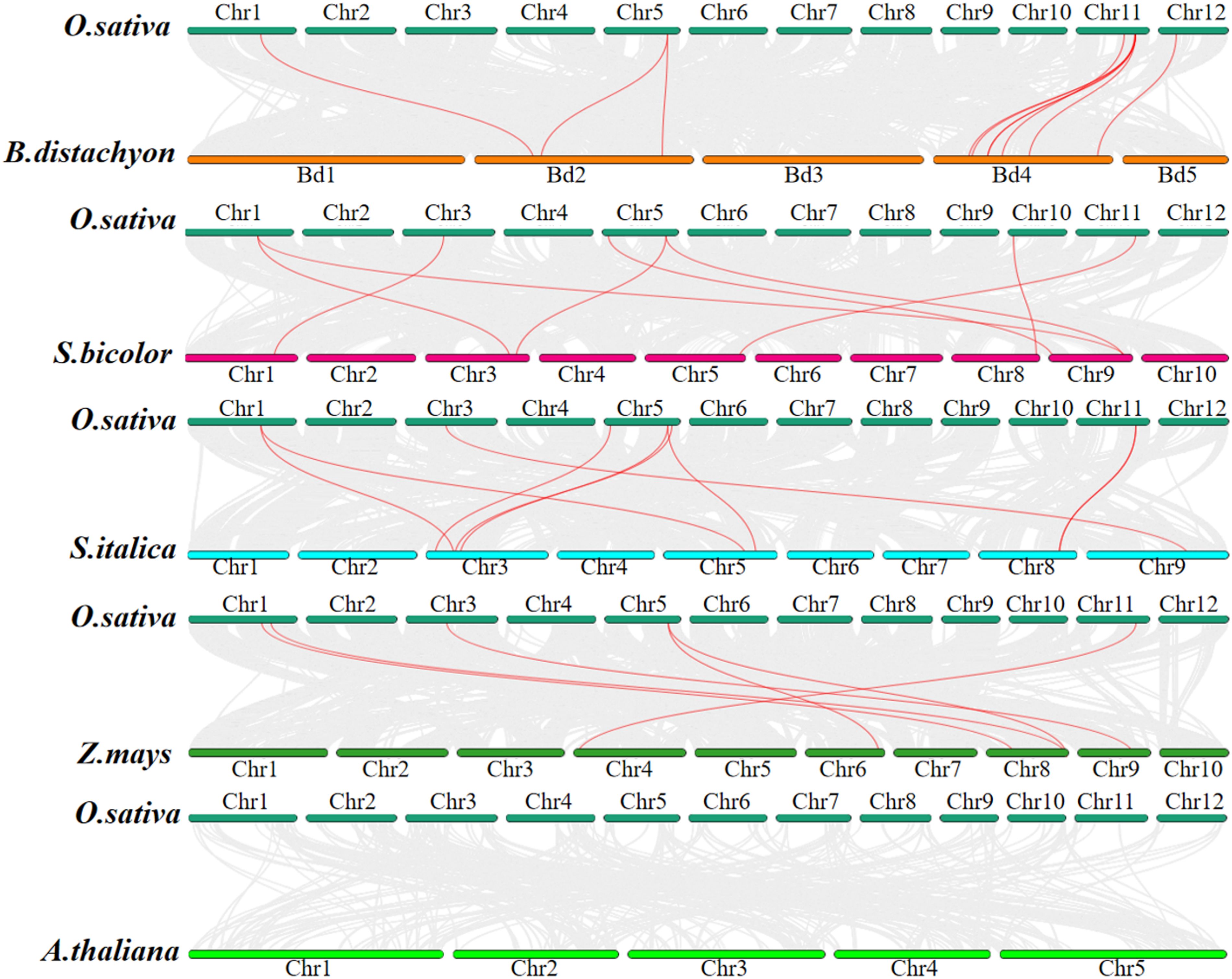
Figure 6. Synteny analysis of O. sativa between and B. distachyon, S. bicolor, S. italica, Z. mays and A. thaliana, respectively. The collinear blocks between O.sativa and other plants were shown in gray lines. The syntenic JRL gene pairs between O.sativa and other plants were highlighted in red lines.
2.5 Analysis of promoter elements
To better understand the function, we analyzed the cis-elements of 147 JRL genes using their 2k-length promoter sequences. Our results revealed a strong connection between the JRL gene and various processes, such as meristem expression, circadian rhythm, root-specific and seed-specific regulatory elements. In addition, the JRL gene displayed a wide array of responsiveness to plant hormones, indicating its involvement in a complex hormone signaling network. And the most prevalent observed elements included abscisic acid response elements (ABRE) followed by jasmonic acid response elements (CGTCA-motif and TGACG-motif), implying the significant role of the JRL gene in coping with abiotic stress. The existence of light-responsive elements such as G-box and I-box implies that JRL genes might play a role in photosynthesis. The discovery of transcription factor binding sites, including W-box, indicates that multiple transcription factors may regulate the JRL genes’ transcription. Additionally, we observed that the cis-elements of MBSI associated with flavonoid biosynthesis was found in A. thaliana, O.sativa, and Z. mays, but not in B. distachyon, S. italica and S. bicolor. On the contrary, the cis-elements of motif I associated with root specific was found in S. italica and S. bicolor, but not in other four species (Figure 7; Supplementary Table 3). Moreover, members of the same subgroup have similar cis-acting elements (Figure 7). These results indicate that the JRL genes may simultaneously have functional consistency and differences.
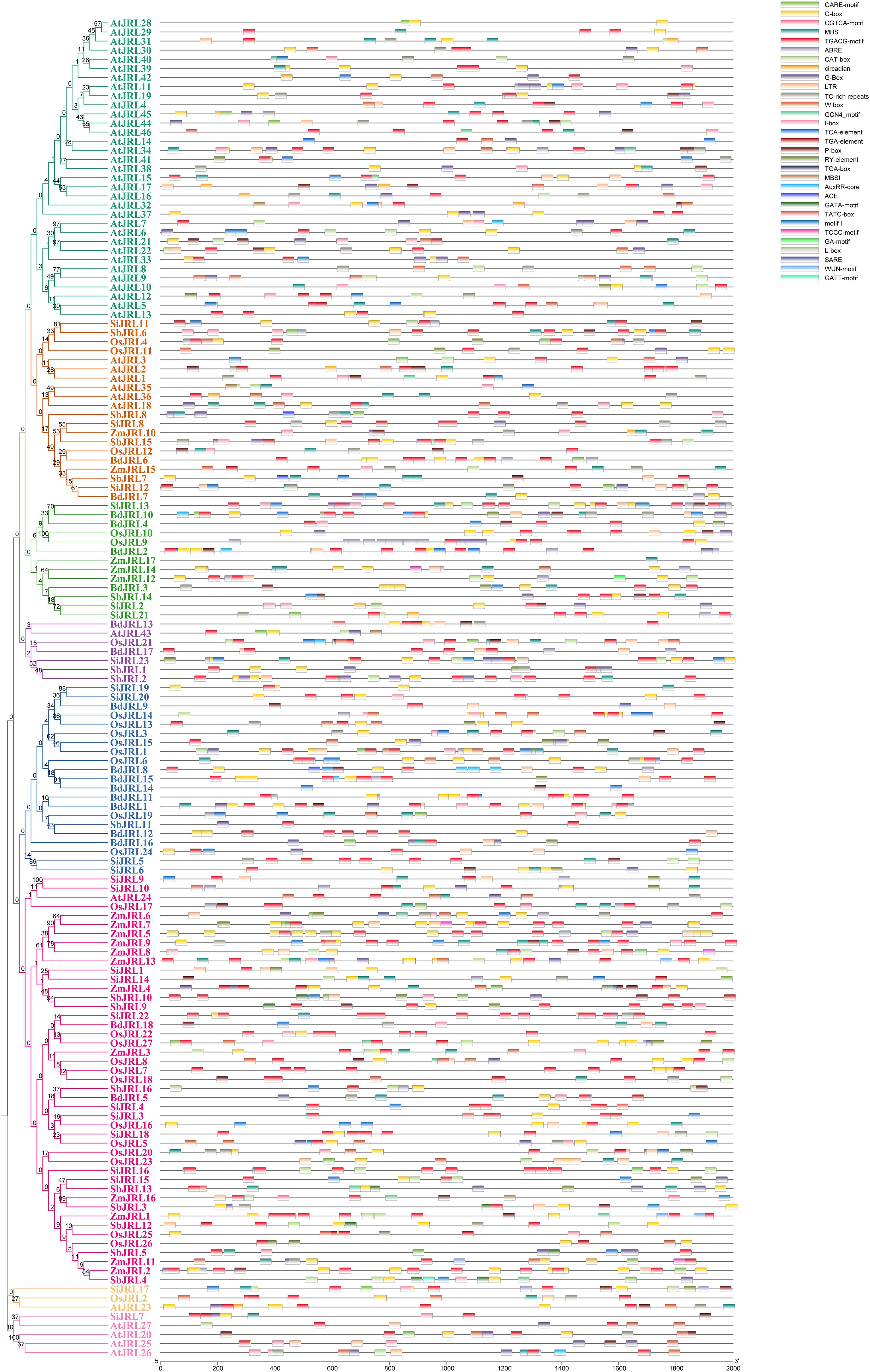
Figure 7. Distribution of cis-acting elements of JRL gene family. The cis-elements were predicted in the 2000 bp promoter sequences of JRL genes. The Cis-elements was analyzed by plantcare.
2.6 Tissues expression pattern analysis of OsJRL and ZmJRL genes
To enhance our understanding about the potential function of the JRL genes, we analyzed the tissue expression patterns of OsJRL and ZmJRL genes. Our findings revealed that OsJRL2/8/9/17/21/22 and ZmJRL1/6/13/17 were not expressed in any of the four examined tissues (Figure 8; Supplementary Table 4). And 44.44% of OsJRL genes and 17.65% of ZmJRL genes exhibited high expression levels in roots, while 11.11% of OsJRL genes and 29.41% of ZmJRL genes were highly expressed in flowers. OsJRL3/13/14/15/23/26 and ZmJRL5/12 were exclusively expressed in roots, whereas OsJRL7 and ZmJRL7 were exclusively expressed in flowers. Additionally, OsJRL4/5/12/16 and ZmJRL10/15 were highly expressed in stems; OsJRL10/24 and ZmJRL4/11 are highly expressed in leaf (Figure 8). Interestingly, the collinear gene pairs OsJRL6/ZmJRL16 were both highly expressed in their inflorescence. And the collinear genes OsJRL12, ZmJRL10 and ZmJRL15 demonstrated high expression levels in stems (Figure 8). However, the tissue expression patterns of the collinear gene pairs OsJRL20/ZmJRL3 exhibited differently: OsJRL20 was highly expressed in roots, while ZmJRL3 showed broad expression across root, flower, stem, and leaf.
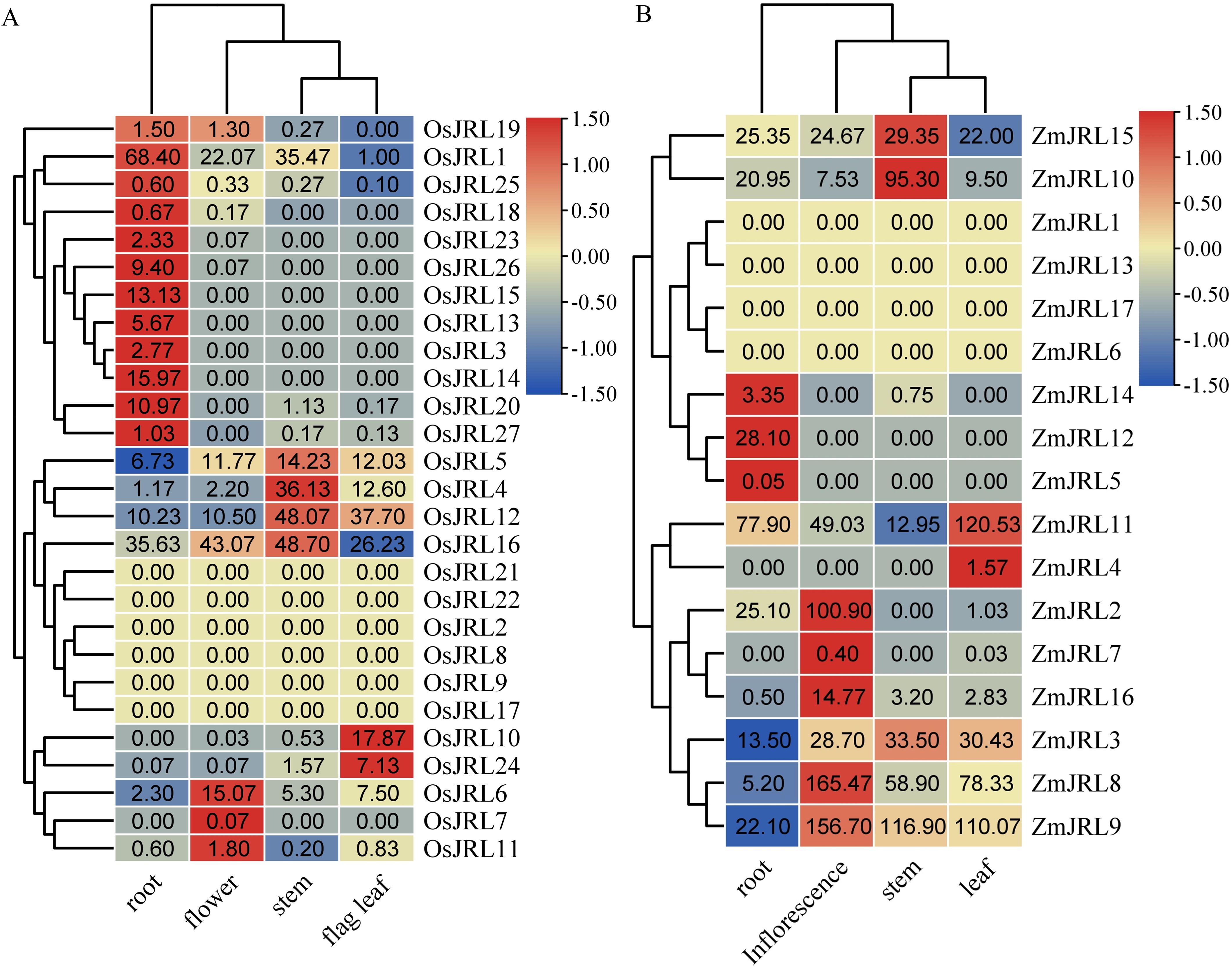
Figure 8. Tissue expression pattern analysis of OsJRL and ZmJRL genes in different tissues. (A). The expression heat map of OsJRL genes in 4 tissues (root, flower, stem, and flag leaf). (B). The expression heat map of ZmJRL genes in 4 tissues (root, inflorescence, stem, and leaf). The data are gene expression that caculated by row-scaled FPKM values, and the heatmap is constructed by TBtools software. Red and blue boxes indicate high and low expression levels of OsJRL and ZmJRL genes, respectively.
2.7 Expression pattern analysis under abiotic stress treatments
For further investigation, we explored the expression levels of ZmJRLs and OsJRLs under abiotic stress by mining transcriptome data. As shown in Figure 9; Supplementary Table 5, the expression levels of OsJRL1/4/27 genes were significantly up-regulated, while OsJRL6/9/17/18/20/24 genes were down-regulated under drought stress. Under salt, heat and cold stresses, OsJRL5/14 genes showed similar up-regulated expression patterns. Interestingly, some OsJRL genes displayed opposing expression patterns under different stress treatments. For instance, the expression levels of OsJRL15/18/20/23 were up-regulated under heat stress, but down-regulated under drought, salt and cold stresses (Figure 9A). In addition, we observed that the expression levels of ZmJRL6/9/11/15 genes were up-regulated under salt stress, with ZmJRL6 showing an extremely significant up-regulation to approximately 51-fold. The expression level of the ZmJRL3 gene was slightly up-regulated under cold stress, but it did not respond to drought, salt and heat stress (Figure 9B). Moreover, we note a descending trend in the expression levels of ZmJRL1 and ZmJRl10 under salt stress.
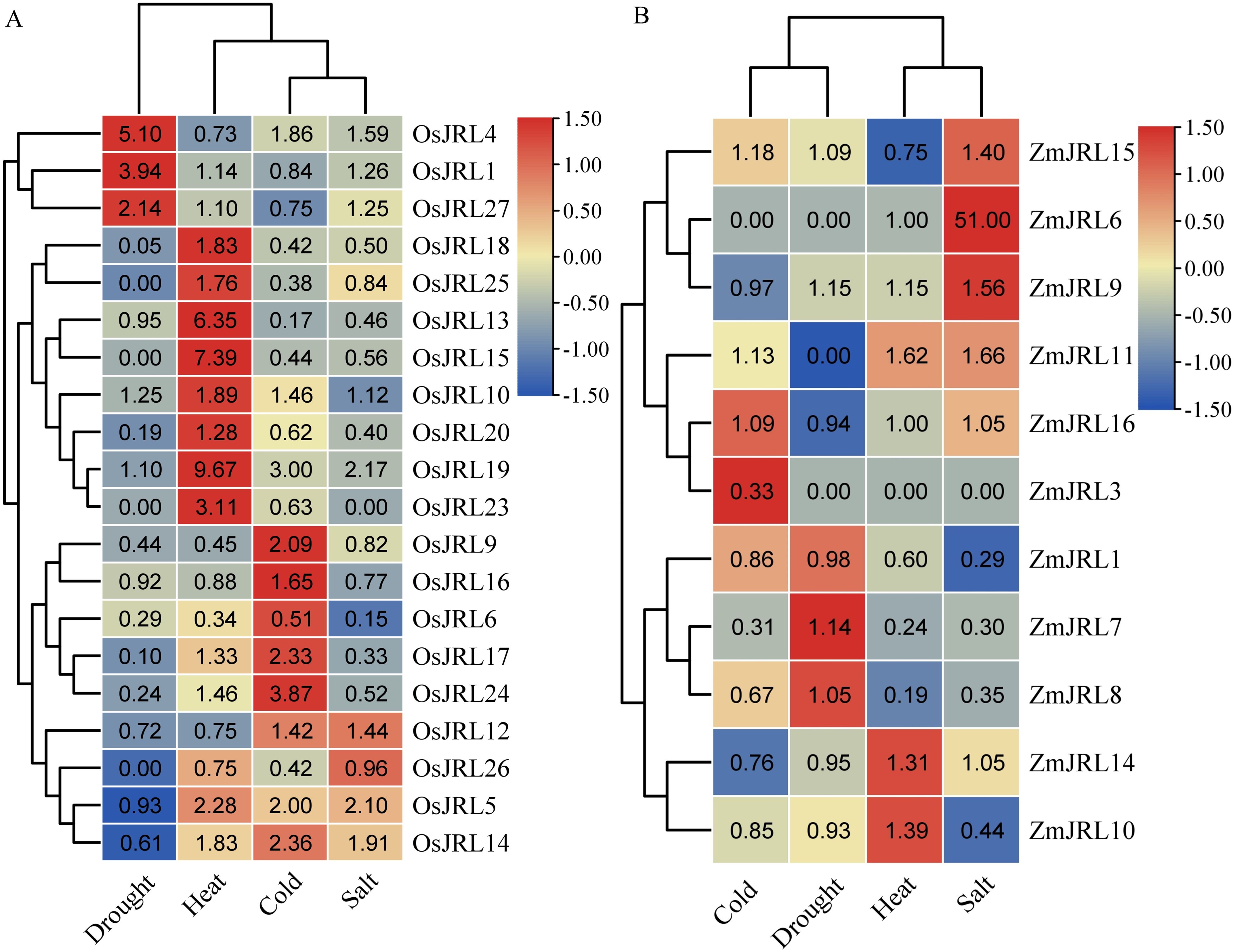
Figure 9. Expression profiles of the OsJRL and ZmJRL genes. (A) The heat map exhibit the ratio of the expression levels of OsJRL genes under abiotic stresses treatment (drought, heat, cold and salt). (B) The heat map exhibit the ratio of the expression levels of ZmJRL genes under abiotic stresses treatment (cold drought, heat, and salt). The data are gene expression that caculated by row-scaled FPKM values, and the heatmap is constructed by TBtools software. Red and blue boxes indicate high and low expression levels of OsJRL and ZmJRL genes, respectively.
To further explore the potential function of the JRL gene, we randomly selected six OsJRL genes that were expressed in roots and responsive to abiotic stress. The mRNA levels of these six genes were assessed using RT-qPCR following treatment with 30% PEG6000 and 150 mM NaCl, respectively (Figure 10). The transcription level of OsJRL4 was significantly up-regulated under PEG6000 treatment and peaking at 24 h, which was similar to its response under salt stress (Figures 10A, G). The expression levels of OsJRL12 and OsJRL26 were down-regulated under 30% PEG6000 treatment (Figures 10B, F), while OsJRL15 exhibited a significant up-regulation (Figure 10D). Under salt stress, the expression levels of OsJRL12, OsJRL14 and OsJRL26 were significantly up-regulated (Figures 10H, I, L), whereas OsJRL15 was inhibited (Figure 10J). Interestingly, the expression level of OsJRL26 was initially increased and then decreased under salt stress (Figure 10L).
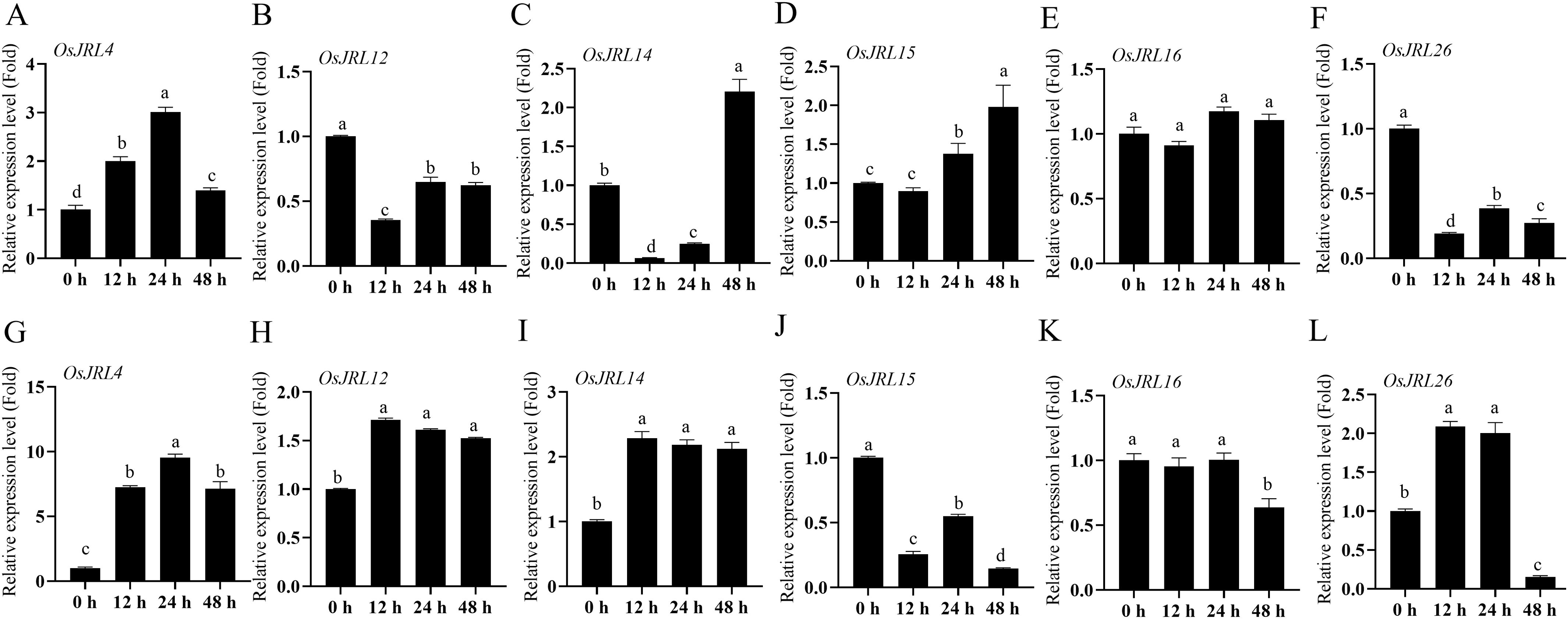
Figure 10. Expression patterns of OsJRL genes under abiotic stress by RT-qPCR. (A–F) The expression level of OsJRLs gene under 30% PEG6000 treatment. (G-L) The expression level of OsJRLs under 150 mM NaCl treatment. The expression levels were calculated using 2-△△Ct methods. Means ± SDs (n = 3). The expression of 0 h were regarded as “1”. Different lowercase letters indicate significant differences among means (P<0.01). qRT-PCR analysis were performed using the rice OsActin as an internal control.
3 Discussion
In this study, a comprehensive analysis of 101 JRL proteins from five Gramineous plants have been carried out. And we then focused on rice and maize to investigate their expression patterns in different tissues, as well as their expression characteristics in response to various stresses.
Here, we identified 27, 17, 23, 16, and 18 JRL genes from O. sativa, Z. mays, S. italica, S. bicolor and B. distachyon, respectively (Figure 1; Supplementary Table 1). With the exception of O. sativa and S. italica, the number of JRL gene in the Gramineous family is relatively similar. Gramineous plants diverged approximately 50-70 million years ago (Paterson et al., 2003) and may have expanded at a comparable rate. Monocotyledonous and dicotyledonous plants diverged around 140 to 150 million years ago (Kumar et al., 2017). And the number of JRL genes in Arabidopsis (46) (Nagano et al., 2008) is approximately twice that of Gramineous plants, indicating a faster expansion rate of JRL genes in Arabidopsis.
The domains of JRL proteins exhibit considerable variability, with significant differences observed between Gramineous species and Arabidopsis. Gramineous JRL proteins can contain up to 3 Jacalin domains, while AtJRLs may possess as many as 5 tandem Jacalin domains (Figure 3), indicating that only one complete domain is required for the normal functioning of JRL proteins. And the formation of multiple domains may be necessary for the conformation of a specific protein structure, or it may be necessary for plants to adapt to complex environmental changes. Also, these differences in JRL protein domains may suggest that monocotyledonous and dicotyledonous plants have evolved distinct strategies. Some JRL protein domains are connected to the dirigent domains (Figure 3), which may be related to their specific functions and this will be the focus of our future research. Interestingly, the members in JRL-III subfamily have more introns than other subfamilies. It has been reported that intron-rich genes have several advantages for species evolution, including increased gene length and recombination frequency between genes. Additionally, introns often contain regulatory elements such as enhancers, and differential splicing of introns allows a single gene to produce multiple protein isoforms (Back and Walther, 2021). Therefore, the increase of introns in the JRL-III subfamily may facilitate an adaptation to the changing environment.
TD and WGD play indispensable roles during the evolution of gene family (Ohno, 1971; Semple and Wolfe, 1999; Xiang et al., 2011; Zhang et al., 2022). Our results suggest that tandem replication events contribute more to the evolution of Gramineous JRL genes (Figure 4). The TD is also a process of new gene generation and functional diversification, which helps plants adapt to environmental changes. Therefore, the TD events analysis hints that these JRL proteins might play specific functions in plant environmental adaptation. Purification selection pressure targets to gradually remove harmful and detrimental genetic variants out through evolution, thus beneficial variants may confer advantages (Chang et al., 2015). Ka/Ks values indicates that the evolution of JRL genes in Arabidopsis and Gramineous may be predominantly influenced by purification selection (Figure 5), and this could promote their better adaptation to complex environmental changes. And the extensive involvement of JRL genes in both biological and abiotic stresses (Figures 8-10) further support this hypothesis.
The expression profiles indicated that OsJRL and ZmJRL genes were expressed in different tissues. Specifically, OsJRL genes exhibited high expression levels in the roots, while ZmJRL genes were predominantly expressed in inflorescences (Figure 8). As sessile organisms, Gramineous crops have evolved the ability to adapt to various unfavorable environments, such as high salt, drought, cold and heat to some extent. And we investigated and found that OsJRL genes respond diversely to drought, cold, salt, and heat stresses, which aligns with the reported essential roles of JRL genes (Abebe et al., 2005; Ma et al., 2021; Gao et al., 2023). Interestingly, the expression levels of the orthologous genes OsJRL12/ZmJRL15 are up-regulated under salt stress (Figure 9), while the orthologous genes OsJRL6/ZmJRL16 exhibit opposing expression patterns under salt stress. Though the mechanisms by which JRL genes respond to abiotic stresses require further investigation, these results indicate that JRL gene is not only functionally differentiated but also functionally conserved across different species. Collectively, these findings illuminate that JRL genes can effectively respond to complex and adverse environmental conditions.
4 Materials and methods
4.1 Plant materials and treatments
The plant material utilized in this study was the rice variety Zhonghua 11 (ZH11). Rice seeds were initially soaked and incubated in purified water overnight in a dark incubator set at 28°C. After two days, uniform seedlings were selected and transferred to be grown in Kimura nutrient solutions, and they were grown under a photoperiod of a 10-hour light/14-hour darkness, with a relative humidity of 50%. Following a 10-day cultivation period, various stress treatments were administered. Drought stress was simulated by application of 30% polyethylene glycol (PEG6000), while salt stress was treated using 150 mM NaCl. Plant roots were collected from both the control and experimental groups at 0, 12, 24, and 48 hours post-treatment. The samples were rapidly frozen in liquid nitrogen, and stored at –80°C for future analysis. The experiment was conducted with three biological replicates.
4.2 Sources of genomic data for different species
The species selected for this study include Oryza sativa (O. sativa), Brachypodium distachyon (B. distachyon), Sorghum bicolor (S. bicolor), Zea mays (Z. mays), Setaria italica (S. italica) and Arabidopsis thaliana (A. thaliana). The genome files and gene structure annotation files of the above-mentioned species were downloaded from the Phytozome database (https://phytozome-next.jgi.doe.gov/) (Goodstein et al., 2012). The sequences of the JRL protein family in A. thaliana were downloaded from TAIR (https://www.Arabidopsis.org/).
4.3 Identification of JRL family proteins in different species
The protein sequences of AtJRLs were used as reference sequences for bidirectional BLAST alignment (E-value < 1e-5) to identify potential candidate JRL proteins across various organisms. The unique domain file (PF01419) specific to the JRL protein family was obtained from the Pfam website (http://pfam-legacy.xfam.org/) (Mistry et al., 2021). Subsequently, HMMER 3.0 software was employed to screen for potential candidate JRL family members within the five species under investigation (Potter et al., 2018). The presence of the conserved JRL domain in the candidate proteins was confirmed using NCBI-CDD (https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi) and SMART (http://smart.embl-heidelberg.de/). Proteins that lacked the JRL domain were excluded from the candidate list based on this analysis.
4.4 Evolution analysis of JRL proteins
The JRL protein sequences that we have identified from Gramineae plants were used to construct an evolutionary tree by MEGA7.0 software and neighbor joining (NJ) method (Kumar et al., 2016), with parameter settings of poisson model, pair wise deletion, and 1000 bootstrap replicates.
4.5 Analysis of chromosome localization, gene replication, collinearity, and Ka/Ks ratio
The genome location of the JRL genes in each species was obtained from their gene structure annotation files (GFF3), named according to their order on chromosomes, and visualized by TBtools. Gene replication events were explored by the Multiple Collinearity Scan Toolkit (MCScan), with parameters set to default (Wang et al., 2012). Using rice as a reference, collinearity analysis was conducted with A. thaliana, B. distachyon, S. bicolor, Z. mays and S. italica. The results were visualized by TBtools software (Chen et al., 2020). The ratio of Ka/Ks for tandem replication and segmental replication gene pairs were calculated through TBtools software (Chen et al., 2020).
4.6 Cluster heat map analysis
Gene expression data from different tissues of rice and maize under abiotic stress were obtained from the Plant Public RNA-seq Database (PPRD) (http://ipf.sustech.edu.cn/pub/plantrna/) (Yu et al., 2022). The gene expression was calculated using FPKM values. To highlight gene expression changes, we use Log2 converted values with a multiple change of 1+FPKM (treatment group)/1+FPKM (control group). Heat maps were generated by TBtools software.
4.7 RNA isolation and quantitative real-time PCR
The primer 5.0 software was used to design specific primers for JRL genes in this study (Supplementary Table 6). Total RNA was extracted using the KKFast Plant RNApure Kit (ZOMANBIO, ZP405K-2). The cDNA was synthesized by EX RT kit (gDNA remover) (ZOMANBIO, ZR108-3). The quantitative real-time PCR (RT-qPCR) system program was performed according to the previous research (Duan et al., 2023). The gene expression was analyzed by the 2−ΔΔCT method as used in the previous study (Xue et al., 2024).
5 Conclusions
In this study, the JRL gene families in five Gramineous plants were comprehensively analyzed, resulting in the identification of 101 JRL genes, which were categorized into eight subgroups. The gene structures of Gramineae JRLs exhibit considerable variability. Duplication events reveal that TD possibly play a major role in the expansion of the gramineous JRL gene family. And the expression profile analysis demonstrates that ZmJRLs and OsJRLs respond differently to drought, cold, salt and heat stress. These findings illustrate that JRL genes may function not only conservatively but also divergently across species. Although the biological functions and regulatory mechanisms of JRL proteins remain unclear, our explorations provide a foundation for further analysis of the biological roles of JRL genes in Gramineous crops.
Data availability statement
Gene expression data were obtained from the Plant Public RNA-seq Database (PPRD) (http://ipf.sustech.edu.cn/pub/plantrna/) (Yu et al., 2022).
Author contributions
LG: Writing – original draft, Writing – review & editing. YL: Writing – review & editing. YW: Writing – review & editing. FH: Writing – review & editing. TZ: Writing – review & editing. BX: Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by Doctoral Research Start up Fund from Henan University of Urban Construction, grant no. K-Q2023022, Science and Technology Research Projects of Henan province in China, grant no. 242102110310.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1501975/full#supplementary-material
References
Abebe, T., Skadsen, R. W., Kaeppler, H. F. (2005). A proximal upstream sequence controls tissue-specific expression of Lem2, a salicylate-inducible barley lectin-like gene. Planta 221, 170–183. doi: 10.1007/s00425-004-1429-9
Back, G., Walther, D. (2021). Identification of cis-regulatory motifs in first introns and the prediction of intron-mediated enhancement of gene expression in Arabidopsis thaliana. BMC Genomics 22, 390. doi: 10.1186/s12864-021-07711-1
Burlat, V., Kwon, M., Davin, L. B., Lewis, N. G. (2001). Dirigent proteins and dirigent sites in lignifying tissues. Phytochemistry 57, 883–897. doi: 10.1016/S0031-9422(01)00117-0
Chang, F., Zou, W. C., Gao, F. L., Shen, J. G., Zhan, J. S. (2015). Comparative analysis of population genetic structure of Potato virus Y from different hosts. Yi. Chuan. 37, 292–301. doi: 10.16289/j.yczz.14-396
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
De Coninck, T., Van Damme, E. J. M. (2021). Review: The multiple roles of plant lectins. Plant Sci. 313, 111096. doi: 10.1016/j.plantsci.2021.111096
Duan, W., Xue, B. P., He, Y. Q., Liao, S. H., Li, X. M., Li, X. Y., et al. (2023). Genome-wide identification and expression pattern analysis of dirigent members in the Genus Oryza. Int. J. Mol. Sci. 24, 7189. doi: 10.3390/ijms24087189
Gao, Q., Yin, X., Wang, F., Zhang, C., Xiao, F., Wang, H., et al. (2023). Jacalin-related lectin 45 (OsJRL45) isolated from 'sea rice 86' enhances rice salt tolerance at the seedling and reproductive stages. BMC. Plant Biol. 23, 553. doi: 10.1186/s12870-023-04533-z
Gong, L., Li, B., Zhu, T., Xue, B. (2023). Genome-wide identification and expression profiling analysis of DIR gene family in Setaria italica. Front. Plant Sci. 14, 1243806. doi: 10.3389/fpls.2023.1243806
Goodstein, D. M., Shu, S., Howson, R., Neupane, R., Hayes, R. D., Fazo, J., et al. (2012). Phytozome: a comparative platform for green plant genomics. Nucleic. Acids Res. 40, D1178–D1186. doi: 10.1093/nar/gkr944
Gossmann, T. I., Song, B. H., Windsor, A. J., Mitchell-Olds, T., Dixon, C. J., Kapralov, M. V., et al. (2010). Genome wide analyses reveal little evidence for adaptive evolution in many plant species. Mol. Biol. Evol. 27, 1822–1832. doi: 10.1093/molbev/msq079
He, Q., Wang, C., He, Q., Zhang, J., Liang, H., Lu, Z., et al. (2024). A complete reference genome assembly for foxtail millet and Setaria-db, a comprehensive database for Setaria. Mol. Plant 17, 219–222. doi: 10.1016/j.molp.2023.12.017
Jiang, J. F., Han, Y., Xing, L. J., Xu, Y. Y., Xu, Z. H., Chong, K. (2006). Cloning and expression of a novel cDNA encoding a mannose-specific jacalin-related lectin from Oryza sativa. Toxicon 47, 133–139. doi: 10.1016/j.toxicon.2005.10.010
Jiang, S. Y., Ma, Z., Ramachandran, S. (2010). Evolutionary history and stress regulation of the lectin superfamily in higher plants. BMC. Evol. Biol. 10, 79. doi: 10.1186/1471-2148-10-79
Kumar, S., Stecher, G., Suleski, M., Hedges, S. B. (2017). TimeTree: A resource for timelines, timetrees, and divergence times. Mol. Biol. Evol. 4, 1812–1819. doi: 10.1093/molbev/msx116
Kumar, S., Stecher, G., Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lin, W. C., Shuai, B., Springer, P. S. (2003). The Arabidopsis LATERAL ORGAN BOUNDARIES-domain gene ASYMMETRIC LEAVES2 functions in the repression of KNOX gene expression and in adaxial-abaxial patterning. Plant Cell. 15, 2241–2252. doi: 10.1105/tpc.014969
Liu, H., Darya, K., Aamir, A. K., Zhang, Y., Wei, L., Babar, U., et al. (2024). Characterization of MADS-box gene family and its unique response to drought and nickel stresses with melatonin-mediated tolerance in dragon fruit (Selenicereus undatus L.). Plant Stress. 12, 10049. doi: 10.1016/j.stress.2024.100492
Ma, R. F., Huang, B., Chen, J. L., Huang, Z. N., Yu, P. Y., Ruan, S. Y., et al. (2021). Genome-wide identification and expression analysis of dirigent-jacalin genes from plant chimeric lectins in moso bamboo (Phyllostachys edulis). PLoS. One 16, e0248318. doi: 10.1371/journal.pone.0248318
Marothia, D., Kaur, N., Jhamat, C., Sharma, I., Pati, P. K. (2023). Plant lectins: Classical molecules with emerging roles in stress tolerance. Int. J. Biol. Macromol. 244, 125272. doi: 10.1016/j.ijbiomac.2023.125272
Mistry, J., Chuguransky, S., Williams, L., Qureshi, M., Salazar, G. A., Sonnhammer, E. L. L., et al. (2021). Pfam: The protein families database in 2021. Nucleic. Acids Res. 49, D412–D419. doi: 10.1093/nar/gkaa913
Mockler, T. C., Schmutz, J., Rokhsar, D., Bevan, M. W., Barry, K., Lucas, S., et al. (2010). Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463, 763–768. doi: 10.1038/nature08747
Nagano, A. J., Fukao, Y., Fujiwara, M., Nishimura, M., Hara-Nishimura (2008). Antagonistic jacalin-related lectins regulate the size of ER bodytype beta-glucosidase complexes in Arabidopsis thaliana. Plant Cell. Physiol. 49, 969–980. doi: 10.1093/pcp/pcn075
Nakagawa, R., Yasokawa, D., Okumura, Y., Nagashima, K. (2000). Cloning and sequence analysis of cDNA coding for a lectin from Helianthus tuberosus callus and its jasmonate-induced expression. Biosci. Biotechnol. Biochem. 64, 1247–1254. doi: 10.1271/bbb.64.1247
Ohno, S. (1971). Evolution by gene and chromosome duplication. Lancet 2, 1299–1300. doi: 10.1016/S0140-6736(71)90612-X
Paterson, A. H., Bowers, J. E., Peterson, D. G., Estill, J. C., Chapman, B. A. (2003). Structure and evolution of cereal genomes. Curr. Opin. Genet. Dev. 13, 644–650. doi: 10.1016/j.gde.2003.10.002
Potter, S. C., Luciani, A., Eddy, S. R., Park, Y., Lopez, R., Finn, R. D. (2018). HMMER web server: 2018 update. Nucleic. Acids Res. 46, W200–W204. doi: 10.1093/nar/gky448
Semple, C., Wolfe, K. H. (1999). Gene duplication and gene conversion in the Caenorhabditis elegans genome. J. Mol. Evol. 48, 555–564. doi: 10.1007/PL00006498
Shang, L., He, W., Wang, T., Yang, Y., Xu, Q., Zhao, X., et al. (2023). A complete assembly of the rice Nipponbare reference genome. Mol. Plant 16, 1232–1236. doi: 10.1016/j.molp.2023.08.003
Shaul, O. (2017). How introns enhance gene expression. Int. J. Biochem. Cell. Biol. 91, 145–155. doi: 10.1016/j.biocel.2017.06.016
Shuai, B., Reynaga-Peña, C. G., Springer, P. S. (2002). The lateral organ boundaries gene defines a novel, plant-specific gene family. Plant Physiol. 129, 747–761. doi: 10.1104/pp.010926
Song, M., Xu, W., Xiang, Y., Jia, H., Zhang, L., Ma, Z. (2013). Association of jacalin-related lectins with wheat responses to stresses revealed by transcriptional profiling. Plant Mol. Biol. 84, 95–110. doi: 10.1007/s11103-013-0121-5
Subramanyam, S., Smith, D. F., Clemens, J. C., Webb, M. A., Sardesai, N., Williams, C. E. (2008). Functional characterization of HFR1, a high-mannose N-glycan-specific wheat lectin induced by Hessian fly larvae. Plant Physiol. 147, 1412–1426. doi: 10.1104/pp.108.116145
Tao, Y., Luo, H., Xu, J., Cruickshank, A., Zhao, X., Teng, F., et al. (2021). Extensive variation within the pan-genome of cultivated and wild sorghum. Nat. Plants. 7, 766–773. doi: 10.1038/s41477-021-00925-x
Tsaneva, M., Van Damme, E. J. M. (2020). 130 years of plant lectin research. Glycoconj. J. 37, 533–551. doi: 10.1007/s10719-020-09942-y
Wang, X. M., Ma, Q. H. (2005). Characterization of a jasmonate-regulated wheat protein related to a beta-glucosidase-aggregating factor. Plant Physiol. Biochem. 43, 185–192. doi: 10.1016/j.plaphy.2005.01.018
Wang, Y., Tang, H., DeBarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic. Acids Res. 40, e49. doi: 10.1093/nar/gkr1293
Weidenbach, D., Esch, L., Möller, C., Hensel, G., Kumlehn, J., Höfle, C., et al. (2016). Polarized defense against fungal pathogens is mediated by the jacalin-related lectin domain of modular poaceae-specific proteins. Mol. Plant 9, 514–527. doi: 10.1016/j.molp.2015.12.009
Xiang, Y., Song, M., Wei, Z., Tong, J., Zhang, L., Xiao, L., et al. (2011). A jacalin-related lectin-like gene in wheat is a component of the plant defence system. J. Exp. Bot. 62, 5471–5483. doi: 10.1093/jxb/err226
Xue, B., Duan, W., Gong, L., Zhu, D., Li, X., Li, X., et al. (2024). The OsDIR55 gene increases salt tolerance by altering the root diffusion barrier. Plant J. 118, 1550–1568. doi: 10.1111/tpj.v118.5
Yong, W. D., Xu, Y. Y., Xu, W. Z., Wang, X., Li, N., Wu, J. S., et al. (2003). Vernalization-induced flowering in wheat is mediated by a lectin-like gene VER2. Planta 217, 261–270. doi: 10.1007/s00425-003-0994-7
Yu, Y., Zhang, H., Long, Y., Shu, Y., Zhai, J. (2022). Plant Public RNA-seq Database: a comprehensive online database for expression analysis of ~45 000 plant public RNA-Seq libraries. Plant Biotechnol. J. 20, 806–808. doi: 10.1111/pbi.13798
Zhang, W., Peumans, W. J., Barre, A., Astoul, C. H., Rovira, P., Rougé, P., et al. (2000). Isolation and characterization of a jacalin-related mannose-binding lectin from salt-stressed rice (Oryza sativa) plants. Planta 210, 970–978. doi: 10.1007/s004250050705
Keywords: JRL gene family, evolution, expression pattern, stress response, gramineous plants
Citation: Gong L, Lu Y, Wang Y, He F, Zhu T and Xue B (2024) Comparative analysis of the JRL gene family in the whole-genome of five gramineous plants. Front. Plant Sci. 15:1501975. doi: 10.3389/fpls.2024.1501975
Received: 26 September 2024; Accepted: 20 November 2024;
Published: 24 December 2024.
Edited by:
Meng Jiang, Zhejiang University, ChinaReviewed by:
Fengli Zhao, China National Rice Research Institute, ChinaZengxu An, Zhejiang University, China
Elly Poretsky, Agricultural Research Service (USDA), United States
Xiaohong Tong, Chinese Academy of Agricultural Sciences, China
Copyright © 2024 Gong, Lu, Wang, He, Zhu and Xue. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Baoping Xue, eHVlYmFvcGluZ0B3aHUuZWR1LmNu
 Luping Gong
Luping Gong Yitong Lu1
Yitong Lu1