- National Institute of Agricultural Sciences, Rural Development Administration, Jeonju, Republic of Korea
The environmental conditions play a crucial role in determining crop yield, which is essential for ensuring food and nutritional security. However, rapid climate change is exacerbating global environmental stress, leading to severe biotic pressures on crops. Therefore, enhancing crop resilience to pathogens has become one of the most pressing challenges for humanity. Large-scale mutant library screening is the most efficient strategy for identifying numerous genes associated with specific traits. The revolutionary CRISPR/Cas9 system has ushered in a new era in the construction of mutant library. However, its application in crop plants has been relatively scarce compared to mammals, largely due to challenges in accessibility. Fortunately, several research groups have recently developed CRISPR/Cas9-based mutant libraries, successfully identifying a variety of genes involved in crop immunity. In this review, we present an overview and discussion of studies that have generated significant results through the use of CRISPR/Cas9 library screening to identify novel genes associated with resistance to biotic stresses within the field of plant research.
Introduction
Rapid climate change, driven by environmental degradation and the excessive use of fossil fuels, presents an existential threat to humanity. The detrimental effects of climate change extend to agriculture, significantly jeopardizing crop production, which serves as a critical source of energy and materials for the global population (Rezaei et al., 2023). Crop yields are severely compromised by various pathogens and pest insects. Additionally, climate change tends to promote pathogen proliferation and negatively impacts plant immunity, further intensifying the damage caused by diseases (Velasquez et al., 2018; Subedi et al., 2023; Roussin-Leveillee et al., 2024). Therefore, developing stress-resilient crop varieties and identifying novel genes that enhance stress resilience are crucial for ensuring human survival.
For decades, botanists have used mutant lines to identify novel genes associated with phenotypic traits and elucidate their functions. Large-scale mutant libraries represent important materials for functional genomics and plant breeding (Wang et al., 2013). Mutant libraries have been constructed using traditional methods such as physical mutagenesis (e.g., X-rays, gamma-rays, fast neutrons, and ultraviolet-C radiation), chemical mutagenesis (e.g., ethyl methanesulfonate, ethyl nitrosourea, 1,2:3,4-diepoxybutane, and N-nitroso-N-methylurea), and insertional mutagenesis (e.g., transposons and T-DNA) (Cheng et al., 2017; Ram et al., 2019; Salava et al., 2021). Although mutant libraries generated via traditional methods are valuable, these methods require a lot of time and labor because they generate random mutations and because identifying targeted mutations is difficult (Viana et al., 2019). The development of the revolutionary clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated nuclease 9 (Cas9)-mediated genome editing technique marked a new era in plant breeding (Son and Park, 2022a). Indeed, some mutant libraries have been constructed using CRISPR/Cas9 tools in crops such as cotton (Gossypium hirsutum), maize (Zea mays), rice (Oryza sativa), soybean (Glycine max), and tomato (Solanum lycopersicum) (Jacobs et al., 2017; Lu et al., 2017; Meng et al., 2017; Bai et al., 2020; Liu et al., 2020; Ramadan et al., 2021; Chen et al., 2022; Son et al., 2024a; Sun et al., 2024; Wang et al., 2024).
In mammals, genomic approaches utilizing CRISPR library screening to explore target genes and methods for alleviating disease symptoms have been extensively carried out (Srivastava and Pandit, 2023; Chen et al., 2024). However, these efforts have not been actively performed in plants. Although Jacobs et al. generated several tomato leucine-rich repeat subfamily XII gene mutant lines and demonstrated that S. lycopersicum FLAGELLIN-SENSITIVE 2.1 (SlFLS2.1) plays a crucial role in the 22–amino acid region of bacterial flagellin (flg22)-induced pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI), this was not based on a library screening (Jacobs et al., 2017). Recently, CRISPR/Cas9-mediated mutant library screening approach was utilized in cotton and rice (Chen et al., 2022; Son et al., 2024a; Sun et al., 2024; Wang et al., 2024), leading to the identification of novel genes associated with resistance to biotic stresses (Figure 1).
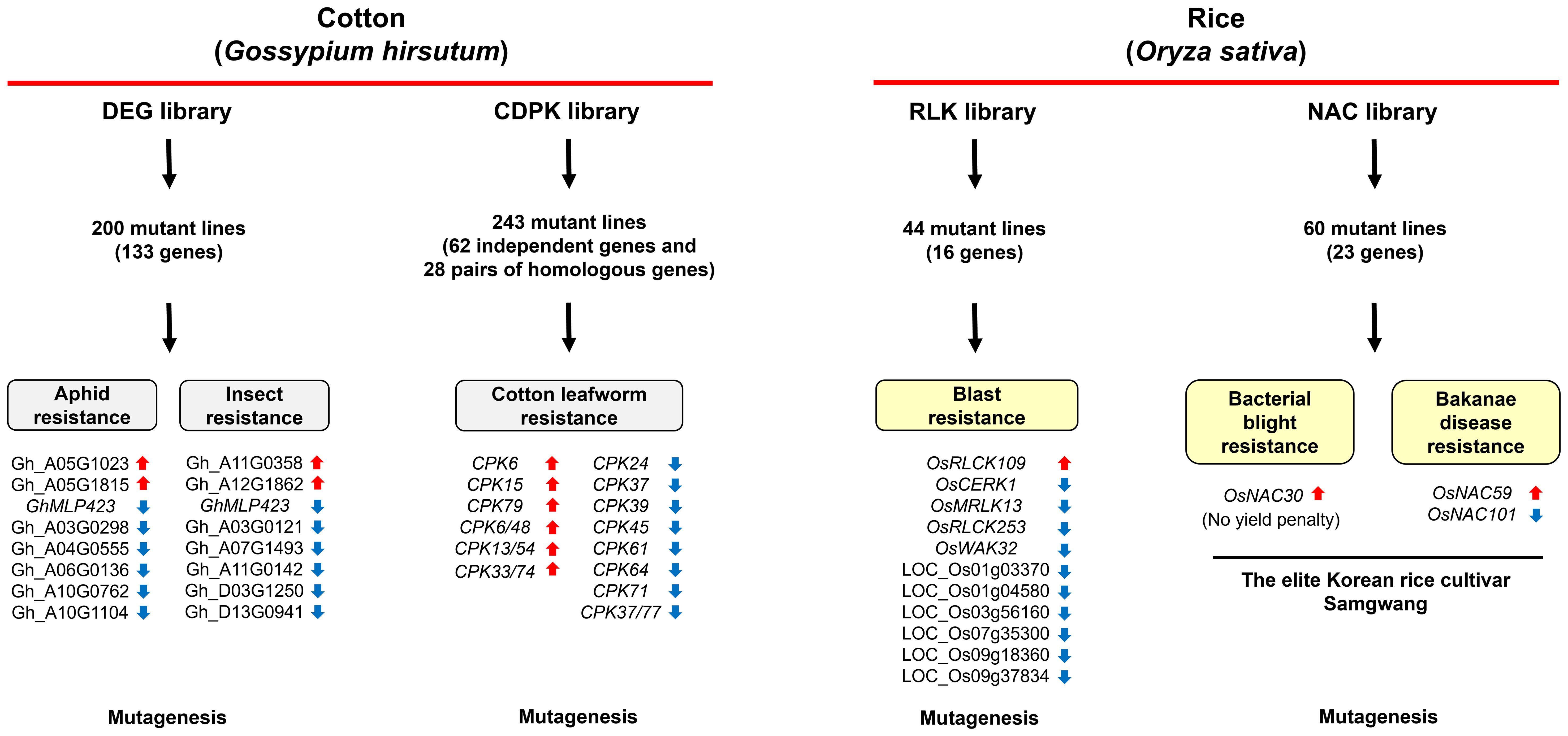
Figure 1. CRISPR/Cas9-mediated mutant library screening for the identification of genes associated with innate immunity in cotton and rice. Using a high-throughput CRISPR/Cas9 system, the differentially expressed gene (DEG) mutant library was constructed for cotton, leading to the identification of 15 genes that significantly influence resistance to various insect pests such as aphids (Sun et al., 2024). Another study utilized CRISPR/Cas9 to develop a cotton calcium-dependent protein kinase (CDPK/CPK) mutant library, identifying six mutant lines with enhanced resistance and eight mutant lines with reduced resistance to Spodoptera litura larvae (Wang et al., 2024). In rice, a receptor-like kinase (RLK) mutant library was generated using the FLASH (PCR fragment-length markers for distinguishing gRNA) pipeline for arrayed CRISPR library construction. This approach revealed that mutagenesis of RECEPTOR-LIKE CYTOPLASMIC KINASE 109 (OsRLCK109) heightened rice resistance to Magnaporthe oryzae, while mutations in 10 other RLKs led to reduced immunity (Chen et al., 2022). Additionally, targeted mutagenesis of rice NAC (no apical meristem, arabidopsis transcription activation factor, and cup-shaped cotyledon) transcription factor genes using CRISPR/Cas9 revealed that OsNAC30 mutations in the elite rice cultivar Samgwang enhanced resistance to Xanthomonas oryzae pv. oryzae without yield penalty, whereas OsNAC59 mutations increased resistance to Fusarium fujikuroi and OsNAC101 mutations reduced it (Son et al., 2024a). These studies underscore the utility of CRISPR/Cas9-mediated mutant libraries in identifying key genes governing both biotic stress resistance in cotton and rice.
Cotton mutant library screening for insect pest resistance
A variety of pests cause substantial reductions in crop yields, resulting in considerable economic losses and jeopardizing global food production (Subedi et al., 2023). Conventional pest control strategies, including the use of chemical pesticides, are associated with environmental risks and can drive the development of resistance in pest populations over time. To address these limitations, Sue et al. present a significant advancement in generating a mutant library of insect-resistant host plants using a high-throughput CRISPR/Cas9 system (Sun et al., 2024). They not only examine the efficacy of this method but also identify a resistance genes, which has the potential to facilitate the development of crops with durable protection against diverse insect species.
To identify cotton genes conferring insect resistance, Sun et al. identified 502 differentially expressed genes (DEGs) and attempted to generate a CRISPR/Cas9-mediated mutant library targeting these genes (Sun et al., 2024). Since these DEGs are implicated in the host plant’s response to insect attacks, they could provide valuable information into the molecular mechanisms governing plant-insect interactions, enabling the identification of potential genetic targets for enhancing pest resistance. Indeed, Sun et al. obtained over 2,000 T0 mutant lines and randomly selected 200 independent T1 mutant lines, harboring mutations in 133 genes, to assess their insect resistance. The mutagenesis of eight genes (i.e., Gh_A03G1240, Gh_A03G0298, Gh_A04G0555, Gh_A05G1023, Gh_A05G1815, Gh_A06G0136, Gh_A10G0762, and Gh_A10G1104) resulted in altered resistance to aphids, while the mutagenesis of the genes (i.e., Gh_A03G1240, Gh_A03G0121, Gh_A07G1493, Gh_A11G0142, Gh_A11G0358, Gh_A12G1862, Gh_D03G1250, and Gh_D13G0941) led to changes in resistance to chewing pests (Sun et al., 2024). The mutations in Gh_A03G1240, corresponding to MAJOR LATEX-LIKE PROTEIN 423 (GhMLP423), compromised immune responses to both aphids and chewing pests, while GhMLP423-overexpressing cotton plants showed enhanced broad-spectrum insect resistance genes (Sun et al., 2024). In subsequent studies, Sun et al. demonstrated that GhMLP423 interacts with the calcium-binding protein (CBP) EPIDERMAL GROWTH FACTOR RECEPTOR SUBSTRATE 15 (GhEPS15) to induce calcium (Ca²+) flux, leading to hydrogen peroxide (H2O2) accumulation, which activates systemic acquired resistance (SAR) (Sun et al., 2024). Therefore, this study establishes a foundational framework for developing cotton with enhanced resilience to a variety of insect pests through the integration of DEG analysis and CRISPR/Cas9-mediated genome editing.
A recent study also reported the development of a calcium-dependent protein kinase (CDPK/CPK) mutant library in cotton. CDPKs play a pivotal role in plant immunity by acting as key mediators of calcium signaling, which is essential for activating various defense responses (Boudsocq and Sheen, 2013). Upon exposure to stress from pathogens or insect pests, plants experience an elevation in intracellular calcium concentrations, which activates CDPKs to initiate a cascade of protective responses. These responses involve the upregulation of defense-associated genes, synthesis of antimicrobial metabolites, and fortification of cell walls to inhibit pathogen penetration (Bacete et al., 2018; Wang and Luan, 2024). By translating calcium signals into downstream immune reactions, CDPKs orchestrate a robust defense, positioning them as key regulators in the plant’s resistance to biotic stresses. Elucidating the specific functions of CDPKs in plant immunity is crucial for advancing strategies aimed at enhancing crop resilience and promoting sustainable agriculture. Therefore, to address the challenge of increased dependence on insect pests and reduce the need for harmful pesticides, Wang et al. generated 518 T0 mutant lines using the CRISPR/Cas9 system and subsequently analyzed 243 T1 and/or T2 mutant plants, involving editing of 62 independent GhCPKs and 28 pairs of homologous GhCPKs, to assess resistance to larvae of Spodoptera litura (Wang et al., 2024). Six mutant lines (i.e., cpk6, cpk15, cpk79, cpk6/48, cpk13/54, and cpk33/74) exhibited enhanced resistance to S. litura, whereas eight mutant lines (i.e., cpk24, cpk37, cpk39, cpk45, cpk61, cpk64, cpk71, and cpk37/77) showed reduced resistance to the pest. Notably, while the cpk33/74 double mutant line exhibited the highest level of insect resistance, neither the cpk33 nor the cpk74 single mutation lines had a significant effect on resistance. Moreover, cotton plants overexpressing GhCPK33 or GhCPK74 exhibited increased susceptibility to S. litura, indicating that GhCPK33 and GhCPK74 have redundant functions in downregulating the immune response against this insect (Wang et al., 2024). Indeed, GhCPK33 and GhCPK74 inhibited Ca²+ flux and jasmonic acid (JA) synthesis, thereby impairing cotton immunity against S. litura (Wang et al., 2024). These GhCPKs also interacted with both S-ADENOSYLMETHIONINE SYNTHETASE 1 (GhSAMS1) and GhSAMS2 which are positive regulators of resistance to S. litura (Wang et al., 2024). However, the regulatory mechanism of GhCPK33 and GhCPK74 remains to be elucidated.
Rice RLK mutant library screening for blast resistance
The receptor-like kinase (RLK) superfamily plays important roles in the detection of pathogens and the subsequent activation of defense responses. Although some RLKs, such as receptor-like cytoplasmic kinases (RLCKs), exhibit different subcellular localizations or structural variations, RLKs are typically membrane proteins with extracellular receptor domains (Liu et al., 2024). These membrane-localized proteins function as sensors, recognizing specific PAMPs or damage-associated molecular patterns and triggering a cascade of signaling pathways that enhance the plant’s ability to resist biotic stress (Wu and Zhou, 2013). Upon activation, RLKs initiate various immune responses, including the production of reactive oxygen species, the expression of defense-related genes, and the reinforcement of cell walls, all of which contribute to a robust defense against pathogens (Huang and Joosten, 2024). The variety of RLKs in plants allows for responses to a broad spectrum of pathogens, including bacteria, fungi, and nematodes. Comprehending the specific functions of various RLKs in plant immunity is crucial for deciphering the intricate signaling networks that govern plant defense (Tang et al., 2017; Huang and Joosten, 2024). Moreover, characterizing these receptors can yield critical insights for devising strategies aimed at improving disease resistance in crops, thereby supporting sustainable agricultural practices and enhancing food security. As researchers further explore the roles of RLKs, their importance in plant immunity becomes increasingly evident, underscoring the necessity for innovative strategies to exploit these kinases in crop enhancement.
In rice, with over 1,000 RLK genes whose functions are largely unknown, Chen et al. generated a RLK mutant library using CRISPR/Cas9 technology to identify novel genes involved in immune responses (Chen et al., 2022). To increase efficiency, they introduced the FLASH (PCR fragment-length markers for distinguishing gRNA) gene editing pipeline to generate an arrayed CRISPR/Cas9 library. As a result, they successfully obtained a rice RLK gene mutant library covering 936 RLK genes out of a total of 1,072 rice RLK members. Blast disease, caused by the filamentous fungus Magnaporthe oryzae, is among the most devastating diseases affecting rice crops (Wilson, 2021). To identify RLK genes involved in defense responses to M. oryzae, the causal agent of rice blast, the mutant library was screened. For the initial screening, RLK expression levels were assessed, and 14 RLK genes exhibiting over 4-fold induction at 72 hours after M. oryzae inoculation were selected (Chen et al., 2022). Among these, 9 T1 RLK mutant lines (LOC_Os01g03370, LOC_Os01g04580, LOC_Os03g56160, LOC_Os04g24220 [WALL-ASSOCIATED KINASE GENE 32, OsWAK32], LOC_Os04g22470 [MALECTIN/MALECTIN-LIKE RECEPTOR-LIKE KINASE 13, OsMRLK13], LOC_Os07g35300, LOC_Os08g28710 [OsRLCK253], LOC_Os09g18360, and LOC_Os09g37834) demonstrated a diminished immune response to M. oryzae (Chen et al., 2022). Therefore, this study not only provides a streamlined method for large-scale gene function analysis but also lays the groundwork for identifying critical RLKs that can be targeted to enhance rice resilience and productivity. The findings have significant implications for global food security and the sustainable improvement of rice, a staple crop for millions.
Rice NAC mutant library screening for resistance to bacterial and fungal pathogens
In plants, numerous transcription factor families exist, with six major superfamilies—NAC (no apical meristem, arabidopsis transcription activation factor, and cup-shaped cotyledon), APETALA2/ethylene response factor, basic helix-loop-helix, basic leucine zipper, myeloblastosis, and WRKY DNA-binding protein—playing essential roles in mediating innate immunity against a diverse array of pathogens (Tsuda and Somssich, 2015). Especially, the NAC transcription factor superfamily, which plays important roles in plant responses to biotic stress, is associated with multiple stress responses and crop yields. Therefore, the NAC transcription factors are consequently regarded as significant targets in plant breeding (Singh et al., 2021). In rice, there are approximately 146 NAC transcription factors, and previous reports have shown that OsNACs act as both positive and negative regulators of rice immunity against various pathogens. Bacterial blight, caused by the bacterial pathogen Xanthomonas oryzae pv. oryzae (Xoo), and bakanae disease, caused by the fungus Fusarium fujikuroi, result in rice yield losses of up to 80% and 50%, respectively, with the potential for more severe damage anticipated due to climate change (Son et al., 2024b). Therefore, identifying the novel genes involved in resistance to these pathogens is crucial for rice breeding. Among the 146 genes, four OsNAC genes associated with bacterial blight have been identified, whereas no genes related to bakanae disease had been reported until recently. OsNAC58, OsNAC66, and OsNAC96 act as positive regulators of resistance to Xoo (Park et al., 2017; Liu et al., 2018; Wang et al., 2021; Yuan et al., 2021), while OsNAC2, which impairs salicylic acid (SA) synthesis and OsEREBP1, negatively regulate rice immunity to Xoo (Zhong et al., 2024). The rice online expression profiles array database and the rice transcription factor phylogenomics database showed almost 35% of OsNAC genes responded to Xoo; 20 OsNAC genes were upregulated and 30 OsNAC genes were downregulated in response to this treatment (Chandran et al., 2019), suggesting many OsNACs involved in immune responses. However, OsNAC genes regulating rice immunity against Xoo and F. fujikuroi are largely unknown.
Most recently, Son et al. developed CRISPR/Cas9 plasmids specifically designed to target 146 OsNAC genes, which were introduced into the elite rice cultivar Samgwang via Agrobacterium-mediated transformation (Son et al., 2024a). This resulted in the generation of 60 T1 homozygous mutant lines harboring mutations in 23 OsNAC genes. Given that bacterial blight and bakanae disease are destructive pathogens affecting rice production (Liu and Wang, 2016), disease resistance assays were conducted using homozygous mutant lines to identify the OsNAC transcription factors involved in innate immunity against these pathogens (Son et al., 2024a). The mutagenesis of OsNAC30 resulted in enhanced resistance to Xoo without any yield penalty. Additionally, along with an elevated SA-mediated defense response, which is crucial for plant immunity against Xoo, the transcription levels of SUGAR WILL EVENTUALLY BE EXPORTED TRANSPORTER 13 (OsSWEET13) and OsSWEET14, which are induced by Xoo to promote susceptibility, were reduced in the osnac30 mutant lines (Son et al., 2024a).
Moreover, OsNAC59 and OsNAC101 were identified as a regulator of innate immunity against bakanae disease. The osnac59 mutant lines exhibited enhanced resistance to F. fujikuroi, indicating that OsNAC59 functions as a negative regulator of resistance to this pathogen (Son et al., 2024a). In osnac59 mutants, the expression levels of both gibberellin (GA)-related genes, which promote susceptibility to F. fujikuroi, and jasmonate ZIM-domain (JAZ) genes, encoding proteins that inhibit the JA-mediated defense response conferring resistance to F. fujikuroi, were reduced (Son et al., 2024a). Conversely, the osnac101 mutant lines displayed reduced resistance to F. fujikuroi, indicating that OsNAC101 positively regulates resistance to the pathogen (Son et al., 2024a). Therefore, through targeted mutagenesis, Son et al. identified specific NAC transcription factors implicated in rice-pathogen interactions and elucidated how individual NAC genes influence the plant’s innate immune responses. Furthermore, this research showed that the CRISPR/Cas9 system is a powerful tool for generating elite rice cultivars that confer enhanced disease resistance through genome editing.
Discussion
Traditional methods of mutagenesis, while useful, present challenges due to the time and labor required to generate and identify useful mutations. However, new plant breeding technologies (i.e., site-directed nucleases, oligonucleotide-directed mutagenesis, cisgenesis, intragenesis, RNA-dependent DNA methylation, grafting, reverse breeding, Agrobacterium-mediated infiltration, and synthetic genomics) have initiated a revolution in the field of plant breeding (Son and Park, 2022b). Especially, the amazing genome editing technology CRISPR is widely regarded as one of the most significant advancements in the history of biological science and technology. Recently studies using CRISPR/Cas9-mediated mutant libraries have led to significant advancements in crop breeding. DEG and CDPK mutant libraries have revealed key genes involved in insect pest resistance in cotton, particularly those related to calcium signaling pathways and protein kinases essential for immune responses (Figure 1). In rice, RLK and NAC mutant libraries have identified critical genes associated with bacterial and fungal pathogen resistance, providing insights into pathogen recognition and transcriptome reprogramming (Figure 1). These findings could potentially provide how crops are bred for biotic stress resilience.
While CRISPR/Cas9 technology has proven highly effective, there are still several challenges that need addressing (Mao et al., 2019; Son and Park, 2022a). One key challenge is the restrictive regulatory framework governing genetically modified organisms (GMOs), which hinders large-scale implementation, especially in regions where legal restrictions and public opposition to GMO crops persist. Therefore, further advancements in DNA-free genome editing techniques are essential to overcome these regulatory hurdles and facilitate broader adoption. Moreover, to date, there are many limitations on the application of this technique to various crops. Therefore, ongoing technological development is necessary to address these challenges. The advancement of genome-editing and transformation technologies and the screening of diverse mutant libraries will pave the way for a significant leap forward in crop development in the future.
CRISPR/Cas9 libraries have been established in a limited number of crop plants (Jacobs et al., 2017; Lu et al., 2017; Meng et al., 2017; Bai et al., 2020; Liu et al., 2020; Ramadan et al., 2021; Chen et al., 2022; Son et al., 2024a; Sun et al., 2024; Wang et al., 2024). Furthermore, the application of CRISPR/Cas9 library screening for the identification of immune-related genes has been realized only recently (Chen et al., 2022; Son et al., 2024a; Sun et al., 2024; Wang et al., 2024). These mutant libraries have been analyzed only in the context of specific pathogens. Therefore, further screening of these libraries across a range of biotic and abiotic stresses will reveal a wealth of genes involved in regulating stress responses. Broad-spectrum resistance is a highly valuable trait to incorporate into crop plants (Li et al., 2020; Koseoglou et al., 2022; Zhao et al., 2024). Continuous accumulation of results from CRISPR library screening for resistance to various pathogens will provide crucial insights into achieving broad-spectrum resistance in crops. Another challenge lies in understanding the complex molecular pathways governing plant immunity. While several key genes have been identified, the interplay between different genes and proteins is not always fully understood. Potential solutions include further research into plant-pathogen interactions and expanding the scope of CRISPR libraries to include genes with unknown functions, as demonstrated in cotton and rice studies.
The development of crops with enhanced resilience to biotic and abiotic stresses through CRISPR-based technologies presents significant potential for addressing global food security issues. As climate change increasingly affects agriculture, the capacity to engineer crops capable of withstanding extreme environmental conditions and pathogen pressures will be critical for sustaining agricultural productivity. Expanding CRISPR research to more plant species and traits, such as stress resilience or nutrient efficiency, could significantly contribute to the development of more sustainable agricultural practices. Moreover, advancements in CRISPR technology, such as the development of more efficient gene-editing pipelines will improve the speed and precision of gene identification and editing. This will enable more targeted breeding programs, resulting in crops that not only exhibit enhanced plant immunity but are also more nutritious and better adapted to evolving climate conditions.
Here, we presented an examination and analysis of a range of studies that have produced notable findings through CRISPR library screening, specifically aimed at the identification of novel genes implicated in enhancing crop immunity, thereby contributing to advancements within the broader scope of plant biology research. New gene-editing technologies are continuously being developed and advanced based on this remarkable technology. Therefore, the CRISPR library screening approach is an influential strategy for identifying new genes and advancing plant improvement. Despite challenges such as scaling the technology and navigating regulatory hurdles, the future prospects for CRISPR-mediated plant breeding are promising and have the potential to transform global agriculture.
Author contributions
SRP: Writing – original draft, Writing – review & editing. SS: Conceptualization, Supervision, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by the Research Program for Agricultural Science and Technology Development (Project No. PJ01570601) and was supported by the Fellowship Program (Project No. PJ01661001) of the National Institute of Agricultural Sciences, Rural Development Administration, Republic of Korea.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Bacete, L., Melida, H., Miedes, E., Molina, A. (2018). Plant cell wall-mediated immunity: cell wall changes trigger disease resistance responses. Plant J. 93, 614–636. doi: 10.1111/tpj.13807
Bai, M., Yuan, J., Kuang, H., Gong, P., Li, S., Zhang, Z., et al. (2020). Generation of a multiplex mutagenesis population via pooled CRISPR-Cas9 in soya bean. Plant Biotechnol. J. 18, 721–731. doi: 10.1111/pbi.13239
Boudsocq, M., Sheen, J. (2013). CDPKs in immune and stress signaling. Trends Plant Sci. 18, 30–40. doi: 10.1016/j.tplants.2012.08.008
Chandran, A. K. N., Moon, S., Yoo, Y. H., Gho, Y. S., Cao, P., Sharma, R., et al. (2019). A web-based tool for the prediction of rice transcription factor function. Database (Oxford) 2019, baz061. doi: 10.1093/database/baz061
Chen, B., Deng, Y., Ren, X., Zhao, J., Jiang, C. (2024). CRISPR/Cas9 screening: unraveling cancer immunotherapy’s ‘Rosetta Stone’. Trends Mol. Med. 30, 736–749. doi: 10.1016/j.molmed.2024.04.014
Chen, K., Ke, R., Du, M., Yi, Y., Chen, Y., Wang, X., et al. (2022). A FLASH pipeline for arrayed CRISPR library construction and the gene function discovery of rice receptor-like kinases. Mol. Plant 15, 243–257. doi: 10.1016/j.molp.2021.09.015
Cheng, X., Liu, G., Ke, W., Zhao, L., Lv, B., Ma, X., et al. (2017). Building a multipurpose insertional mutant library for forward and reverse genetics in Chlamydomonas. Plant Methods 13, 36. doi: 10.1186/s13007-017-0183-5
Huang, W. R. H., Joosten, M. (2024). Immune signaling: receptor-like proteins make the difference. Trends Plant Sci. [In press]. doi: 10.1016/j.tplants.2024.03.012
Jacobs, T. B., Zhang, N., Patel, D., Martin, G. B. (2017). Generation of a collection of mutant tomato lines using pooled CRISPR libraries. Plant Physiol. 174, 2023–2037. doi: 10.1104/pp.17.00489
Koseoglou, E., van der Wolf, J. M., Visser, R. G. F., Bai, Y. (2022). Susceptibility reversed: modified plant susceptibility genes for resistance to bacteria. Trends Plant Sci. 27, 69–79. doi: 10.1016/j.tplants.2021.07.018
Li, W., Deng, Y., Ning, Y., He, Z., Wang, G. L. (2020). Exploiting broad-spectrum disease resistance in crops: from molecular dissection to breeding. Annu. Rev. Plant Biol. 71, 575–603. doi: 10.1146/annurev-arplant-010720-022215
Liu, H. J., Jian, L., Xu, J., Zhang, Q., Zhang, M., Jin, M., et al. (2020). High-throughput CRISPR/cas9 mutagenesis streamlines trait gene identification in maize. Plant Cell 32, 1397–1413. doi: 10.1105/tpc.19.00934
Liu, J., Li, W., Wu, G., Ali, K. (2024). An update on evolutionary, structural, and functional studies of receptor-like kinases in plants. Front. Plant Sci. 15. doi: 10.3389/fpls.2024.1305599
Liu, Q., Yan, S., Huang, W., Yang, J., Dong, J., Zhang, S., et al. (2018). NAC transcription factor ONAC066 positively regulates disease resistance by suppressing the ABA signaling pathway in rice. Plant Mol. Biol. 98, 289–302. doi: 10.1007/s11103-018-0768-z
Liu, W., Wang, G.-L. (2016). Plant innate immunity in rice: a defense against pathogen infection. Natl. Sci. Rev. 3, 295–308. doi: 10.1093/nsr/nww015
Lu, Y., Ye, X., Guo, R., Huang, J., Wang, W., Tang, J., et al. (2017). Genome-wide targeted mutagenesis in rice using the CRISPR/Cas9 system. Mol. Plant 10, 1242–1245. doi: 10.1016/j.molp.2017.06.007
Mao, Y., Botella, J. R., Liu, Y., Zhu, J. K. (2019). Gene editing in plants: progress and challenges. Natl. Sci. Rev. 6, 421–437. doi: 10.1093/nsr/nwz005
Meng, X., Yu, H., Zhang, Y., Zhuang, F., Song, X., Gao, S., et al. (2017). Construction of a genome-wide mutant library in rice using CRISPR/Cas9. Mol. Plant 10, 1238–1241. doi: 10.1016/j.molp.2017.06.006
Park, S. R., Kim, H. S., Lee, K. S., Hwang, D.-J., Bae, S.-C., Ahn, I.-P., et al. (2017). Overexpression of rice NAC transcription factor OsNAC58 on increased resistance to bacterial leaf blight. J. Plant Biotechnol. 44, 149–155. doi: 10.5010/JPB.2017.44.2.149
Ram, H., Soni, P., Salvi, P., Gandass, N., Sharma, A., Kaur, A., et al. (2019). Insertional mutagenesis approaches and their use in rice for functional genomics. Plants (Basel) 8, 310. doi: 10.3390/plants8090310
Ramadan, M., Alariqi, M., Ma, Y., Li, Y., Liu, Z., Zhang, R., et al. (2021). Efficient CRISPR/Cas9 mediated Pooled-sgRNAs assembly accelerates targeting multiple genes related to male sterility in cotton. Plant Methods 17, 16. doi: 10.1186/s13007-021-00712-x
Rezaei, E. E., Webber, H., Asseng, S., Boote, K., Durand, J. L., Ewert, F., et al. (2023). Climate change impacts on crop yields. Nat. Rev. Earth Environ. 4, 831–846. doi: 10.1038/s43017-023-00491-0
Roussin-Leveillee, C., Rossi, C. A. M., Castroverde, C. D. M., Moffett, P. (2024). The plant disease triangle facing climate change: a molecular perspective. Trends Plant Sci. 29, 895–914. doi: 10.1016/j.tplants.2024.03.004
Salava, H., Thula, S., Mohan, V., Kumar, R., Maghuly, F. (2021). Application of genome editing in tomato breeding: mechanisms, advances, and prospects. Int. J. Mol. Sci. 22, 682. doi: 10.3390/ijms22020682
Singh, S., Koyama, H., Bhati, K. K., Alok, A. (2021). The biotechnological importance of the plant-specific NAC transcription factor family in crop improvement. J. Plant Res. 134, 475–495. doi: 10.1007/s10265-021-01270-y
Son, S., Park, S. R. (2022a). Challenges facing CRISPR/Cas9-based genome editing in plants. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.902413
Son, S., Park, S. R. (2022b). Climate change impedes plant immunity mechanisms. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1032820
Son, S., Song, G., Nam, S., Lee, J., Hwang, D.-J., Suh, E.-J., et al. (2024b). OsWRKY65 enhances immunity against fungal and bacterial pathogens in rice. Crop J. 12, 470–481. doi: 10.1016/j.cj.2024.01.007
Son, S., Song, G., Nam, S., Lee, G., Im, J., Lee, K. S., et al. (2024a). CRISPR/Cas9-mediated mutagenesis of rice NAC transcription factor genes results in altered innate immunity. Plant Physiol. 195, 1138–1142. doi: 10.1093/plphys/kiae084
Srivastava, K., Pandit, B. (2023). Genome-wide CRISPR screens and their applications in infectious disease. Front. Genome Editing 5. doi: 10.3389/fgeed.2023.1243731
Subedi, B., Poudel, A., Aryal, S. (2023). The impact of climate change on insect pest biology and ecology: Implications for pest management strategies, crop production, and food security. J. Agric. Food Res. 14, 100733. doi: 10.1016/j.jafr.2023.100733
Sun, L., Alariqi, M., Wang, Y., Wang, Q., Xu, Z., Zafar, M. N., et al. (2024). Construction of host plant insect-resistance mutant library by high-throughput CRISPR/Cas9 system and identification of A broad-spectrum insect resistance gene. Adv. Sci. 11, e2306157. doi: 10.1002/advs.202306157
Tang, D., Wang, G., Zhou, J. M. (2017). Receptor kinases in plant-pathogen interactions: more than pattern recognition. Plant Cell 29, 618–637. doi: 10.1105/tpc.16.00891
Tsuda, K., Somssich, I. E. (2015). Transcriptional networks in plant immunity. New Phytol. 206, 932–947. doi: 10.1111/nph.13286
Velasquez, A. C., Castroverde, C. D. M., He, S. Y. (2018). Plant-pathogen warfare under changing climate conditions. Curr. Biol. 28, R619–R634. doi: 10.1016/j.cub.2018.03.054
Viana, V. E., Pegoraro, C., Busanello, C., Costa de Oliveira, A. (2019). Mutagenesis in rice: the basis for breeding a new super plant. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.01326
Wang, H., Bi, Y., Gao, Y., Yan, Y., Yuan, X., Xiong, X., et al. (2021). A Pathogen-Inducible Rice NAC Transcription Factor ONAC096 Contributes to Immunity Against Magnaprothe oryzae and Xanthomonas oryzae pv. oryzae by Direct Binding to the Promoters of OsRap2.6, OsWRKY62, and OsPAL1. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.802758
Wang, F., Liang, S., Wang, G., Hu, T., Fu, C., Wang, Q., et al. (2024). CRISPR-Cas9-mediated construction of a cotton CDPK mutant library for identification of insect-resistance genes. Plant Commun. 5, 101047. doi: 10.1016/j.xplc.2024.101047
Wang, N., Long, T., Yao, W., Xiong, L., Zhang, Q., Wu, C. (2013). Mutant resources for the functional analysis of the rice genome. Mol. Plant 6, 596–604. doi: 10.1093/mp/sss142
Wang, C., Luan, S. (2024). Calcium homeostasis and signaling in plant immunity. Curr. Opin. Plant Biol. 77, 102485. doi: 10.1016/j.pbi.2023.102485
Wilson, R. A. (2021). Magnaporthe oryzae. Trends Microbiol. 29, 663–664. doi: 10.1016/j.tim.2021.03.019
Wu, Y., Zhou, J. M. (2013). Receptor-like kinases in plant innate immunity. J. Integr. Plant Biol. 55, 1271–1286. doi: 10.1111/jipb.12123
Yuan, X., Wang, H., Bi, Y., Yan, Y., Gao, Y., Xiong, X., et al. (2021). ONAC066, A stress-responsive NAC transcription activator, positively contributes to rice immunity against magnaprothe oryzae through modulating expression of osWRKY62 and three cytochrome P450 genes. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.749186
Zhao, S., Li, M., Ren, X., Wang, C., Sun, X., Sun, M., et al. (2024). Enhancement of broad-spectrum disease resistance in wheat through key genes involved in systemic acquired resistance. Front. Plant Sci. 15. doi: 10.3389/fpls.2024.1355178
Keywords: CRISPR/Cas9, crop, genetic resources, mutant library screening, new breeding technology, plant immunity
Citation: Park SR and Son S (2024) CRISPR/Cas9-based mutant library screening: the discovery of novel genes regulating immune responses in cotton and rice. Front. Plant Sci. 15:1501092. doi: 10.3389/fpls.2024.1501092
Received: 24 September 2024; Accepted: 28 October 2024;
Published: 14 November 2024.
Edited by:
Hyun Uk Kim, Sejong University, Republic of KoreaReviewed by:
Sareena Sahab, Victoria State Government, AustraliaCopyright © 2024 Park and Son. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Seungmin Son, bGluZXdpbmRAa29yZWEua3I=
 Sang Ryeol Park
Sang Ryeol Park Seungmin Son
Seungmin Son