- Department of Entomology and Plant Pathology, University of Arkansas, Fayetteville, AR, United States
Introduction
Plants are constantly exposed to a diverse spectrum of environmental stressors that typically occur concurrently as multifactorial challenges, both spatially and temporally (Joshi et al., 2024). These stressors include abiotic stresses, such as drought, flooding, salinity, nutrient deficiencies, metal toxicity and extreme temperatures, as well as biotic pressures such as herbivory, pathogen attacks, and inter and intraspecific competition (Porter et al., 2020; González Guzmán et al., 2022). Drought and herbivory are arguably among the most critical factors that limit crop productivity in agricultural settings (Gautam et al., 2024), making it imperative to understand plant responses to their combined effects. While conventional research has tended to examine plant responses to these stressors individually, in both natural and agricultural settings, plants frequently encounter multiple stresses either simultaneously or sequentially, leading to complex interactions with consequences that cascade to multiple trophic levels.
Drought, a major abiotic stress, induces a range of morphophysiological changes in plants, such as reduced leaf area, stomatal closure, and enhanced root growth characterized by increased root biomass, surface area, and root volume (Farooq et al., 2009a). Beyond these immediate responses, drought stress can also prime plants for future stress events, influencing their susceptibility or resistance to subsequent biotic and abiotic challenges (Bilichak et al., 2015; Li et al., 2019; Sallam et al., 2019). Drought stress induced priming can also lead to epigenetic changes in gene expression, creating transcriptional memory that has been found to enhance both plant survival and response to subsequent drought events (Avramova, 2019; Nguyen et al., 2022; Kambona et al., 2023). For instance, in Arabidopsis, abiotic stress induces histone demethylation at the promoter of the proline biosynthetic gene Δ1-pyrroline-5-carboxylate synthetase (P5CS), leading to increased P5CS expression and proline accumulation, which assists in tolerating stress. This epigenetic modification remains even after the stress removal, indicating the formation of stress memory that improves tolerance to future stress (Banerjee and Roychoudhury, 2015). Similarly, Sani et al. (2013) (Sani et al., 2013) discovered that sodium ion pre-treated Arabidopsis plants gained enhanced drought tolerance through histone modification changes, specifically reduced H3K27me3 levels, which activated the HKT1 gene responsible for salt stress responses (Banerjee and Roychoudhury, 2015). This highlights the critical role of H3K27me3 in somatic stress memory (Kim et al., 2015). Although, drought stress memory, characterized by epigenetic changes in gene expression, has garnered recent attention, the mechanisms through which drought primes plants for future stressors are not well understood (Godwin and Farrona, 2020; Tian et al., 2024).
Similarly, herbivory imposes significant biotic stress on plants, often leading to changes in direct and indirect plant defenses, ecophysiology, defense-fitness trade-offs, production of defensive metabolites, and activation of defense genes, and consequently- affecting trophic interactions (Howe and Jander, 2008; Kariyat et al., 2013; Kersten et al., 2013; Müller et al., 2019; Huang and Huang, 2023). Herbivore-induced plant volatiles (HIPVs), released in response to insect damage or herbivore-cues such as saliva, frass or eggs, serve as signals for predators and parasitoids to locate herbivores at different spatial locations (Kariyat et al., 2012; Ali et al., 2023). Notably, herbivory stress responses can also prime plants to subsequent attacks. For example, feeding by caterpillars on Arabidopsis and tomato increased resistance in the subsequent progeny, indicating the presence of transgenerational memory (Van Hulten et al., 2006; Rasmann et al., 2012; Holeski et al., 2012). While existing research on herbivory primarily focuses on plant defenses and their impact on herbivore growth and development (Kariyat et al., 2017, 2018; Tayal et al., 2020; Kaur and Kariyat, 2023), a comprehensive examination in this area should also include physiological responses, such as immune response in herbivores, identification of molecular markers for faster diagnosis, and potential epigenetic modifications that may influence growth and yield traits in subsequent generations (Schmitz et al., 2019).
Despite significant advancements in molecular biology, there remains a lack of a comprehensive molecular understanding of plant responses to abiotic and biotic stresses, when they arrive in tandem. It should be noted that significant progress has been made with techniques like genome sequencing, quantitative trait locus (QTL) mapping which has enabled the identification of key genes involved in drought and herbivory tolerance (Pfalz et al., 2007; Yesudas et al., 2010; Gahlaut et al., 2017; Hassan et al., 2023). And genome wide association studies (GWAS) to identify stress-related traits (Davila Olivas et al., 2017; Thoen et al., 2017; Huang et al., 2024), clustered regularly interspaced short palindromic repeats (CRISPR) for precise genome editing to enhance stress tolerance (Liu et al., 2020; Ghosh and Dey, 2022; Sun et al., 2024), and RNA interference (RNAi) for targeted gene silencing (Li et al., 2024; Zhang et al., 2024), there remains a crucial gap in integrating these molecular tools with ecological interactions. While various biotic and abiotic stressors exist, this manuscript focuses on drought and herbivory as a model due to their synergistic effects, which often intensify plant stress responses (Gautam et al., 2024). By focusing on drought and herbivory, we aim to elucidate how plants manage multiple stresses and propose integrated approaches for studying these complex interactions. This integrated perspective will help bridge the gap between molecular biology and ecological interactions, offering new insights into plant resilience and stress management in natural and agricultural ecosystems.
Eco-physiological and biochemical approaches for studying drought and herbivory in agricultural crops
Drought and herbivory bring significant changes to the crop eco-physiological responses including root architecture, and physiological parameters (Makbul et al., 2011; Bansal et al., 2013; Kunert et al., 2016; Xu et al., 2018). Reduced stomatal conductance, impaired photosynthesis, and decreased chlorophyll content have been consistently observed under drought stress (de Freitas Bueno et al., 2009; Farooq et al., 2009b). Contrary to the consistent effects under drought, herbivory has differential impacts on plant performance; some studies show negative impact on crop performance while others report positive compensatory effects on photosynthesis and crop growth (Kucharik et al., 2016; Dong et al., 2019; Zheng et al., 2021) with an overall negative impact on crop yield (de Freitas Bueno et al., 2009; Grinnan et al., 2013; Owen et al., 2013; Musser et al., 2022). The eco-physiology of the crops under drought and herbivory are mostly resulted by the biochemical changes in the host crops (Abid et al., 2018; Chávez-Arias et al., 2021; Pati et al., 2023). For example, Pati et al. (2023) reported that the action of several defense related enzymes such as peroxidase, polyphenol oxidase, and catalase was higher in resistant genotypes of rice against brown plant hopper (Nilaparvata lugens, Stal). At the same time, Chávez-Arias et al. (2021) also summarized those changes in osmolytes such as proline, glycine, soluble sugars, and ions (K+, Na+) aid in alleviating the impacts of drought stress in crops like maize and rice.
Biochemical approaches are usually indicated by decline in membrane stability, increased reactive oxygen species (ROS) production, lipid peroxidation and injury to the membranes (Abid et al., 2018) under drought stress. Higher accumulation of proline, free amino acids, and soluble sugars have also been identified as mechanisms of plants for osmotic adjustments under drought stress (Oh and Komatsu, 2015; Mwenye et al., 2016; Du et al., 2020; Chávez-Arias et al., 2021). Similarly, based on the types of crops and herbivores, crops may respond differentially with varying quantities of phenols, soluble proteins, and activities of defense enzymes including peroxidases, and catalases (Pati et al., 2023). Interestingly, production of secondary metabolites in crops such as flavonoids, terpenoids, alkaloids, and glucosinolates are triggered in response to both drought and herbivory (Lin et al., 2023). Besides secondary metabolites, drought and herbivory also alter the volatiles (VOCs) emitted by the crops. For instance, herbivory by Spodoptera exigua in potato (Solanum tuberosum) under lower water availability induced lower VOCs than under well-watered plants (Vázquez-González et al., 2022). Most importantly, volatile compounds and secondary metabolites induced under drought and herbivory leads to signaling cascades in crops triggering the production of systemic phytohormones (Jogawat et al., 2021).
Phytohormones such as jasmonic acid (JA), salicylic acid (SA), and abscisic acid (ABA) have been studied to explore the possible signaling mechanism for crop responses under drought and herbivory. A decreasing trend was observed for JA and methyl jasmonate (Me-JA) under drought and herbivory stress in soybeans compared to herbivory only treatment, but SA showed a reduction only under drought but not due to insect infestation (Faustino et al., 2021). However, compared to control treatments (no drought or herbivory), JA and Me-JA levels were still upregulated indicating a defensive response under drought and herbivory interaction. Interestingly, under severe drought and simulated herbivory (exogenous application of MeJA), several volatile metabolites including methyl salicylate is strongly induced potentially due to priming under drought stress (Scott et al., 2019). At the same time, under drought stress, ABA has been found to synergize with JA to provide resistance against herbivores (Nguyen et al., 2016). Jogawat et al. (2021) mentioned that changes in level of ethylene (ET) affected growth responses in wheat and drought induced the genes encoding for ET receptors in rice, Arabidopsis thaliana and tobacco. Moreover, the expression levels of lipoxygenases (LOX) genes that are involved in the first step of jasmonic acid (JA) biosynthesis were upregulated under drought but was repressed when followed by herbivory in soybeans (Faustino et al., 2021). In a meta-analysis, transcriptomics of major crops such as wheat, maize, and tomato revealed that ET pathway and ABA pathway-related transcription factors are upregulated under drought (Benny et al., 2019). In addition to eco-physiology, biochemical and phytohormonal assessment, examination of several omics and epigenetics approaches can potentially extrapolate the mechanistic understanding of drought and herbivory interaction in crops. The current approaches focused on phenotypic effects, therefore, are mostly limited to few model species, pairwise interactions and lack an integrated approach. Omics approaches allow us to expand our understanding on the underlying mechanisms of drought and herbivory interactions that we still lack and ask more landscape level questions, which is what we propose, an integration of different approaches.
Redefining drought and herbivory interaction from omics approach
Omics approaches help in elucidating the interaction between the environment and genes, characterization and identification of biomarkers and phenotyping (Chávez-Arias et al., 2021). Innovative approaches that combine data from multiple omics layers, such as transcriptomics metabolomics, proteomics, epigenomics, and metagenomics—collectively referred to as panomics - are some of the approaches under ‘Omics’ which have recently become critical for understanding the pathways, genetic and biochemical basis of drought and herbivory interaction in agricultural crops (Figure 1).
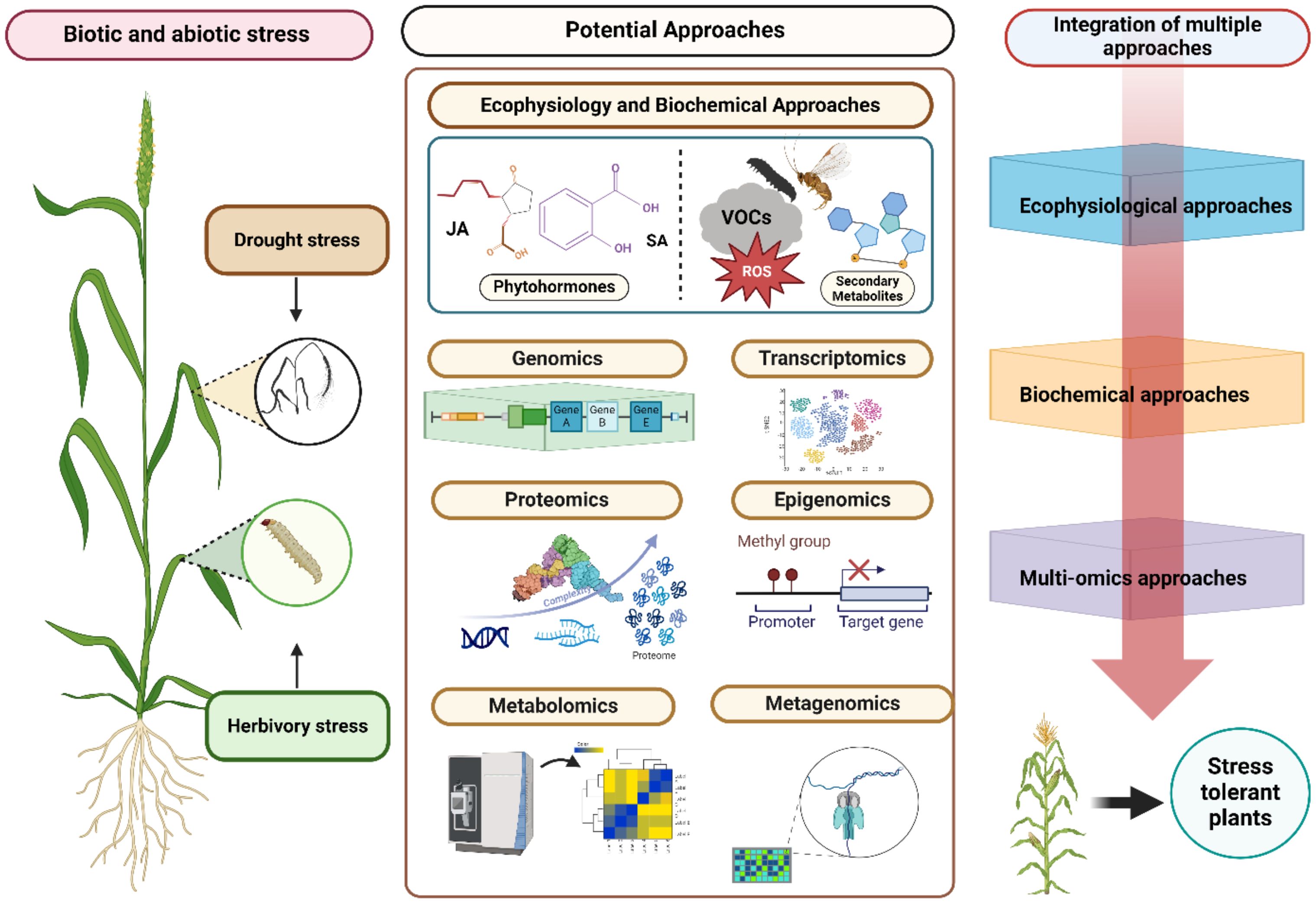
Figure 1. Schematic representation of integration of eco-physiological, biochemical and omics approaches to understand plant environment interactions under biotic and abiotic stress. Eco-physiological approaches involve assessing plant growth, water use efficiency, and stress tolerance, providing valuable insights into how plants adapt to changing environments. Biochemical approaches help in identifying and quantifying stress-related metabolites, enzymes, and hormones, which help elucidate the molecular mechanisms driving stress responses. Omics approaches, including genomics, transcriptomics, proteomics, and metabolomics, generate extensive datasets that uncover the complex regulatory networks and pathways involved in stress adaptation.
Differential expression of genes under drought and herbivory
Transcriptome profiles under simultaneous drought and herbivory stress have been widely used to elucidate their interactive effects on several plants. (Coolen et al., 2016) used RNAseq to explore the transcriptome of Arabidopsis under herbivory when previously exposed to drought stress and found that the transcriptional changes due to caterpillars masked the drought-induced changes that occurred previously. Using exogenous coronatine (COR), (Attaran et al., 2014) found that JA-induced signaling repressed photosynthetic genes but defense genes were induced in Arabidopsis. Through a comprehensive RNAseq analysis, it was clear that JA-induced signaling (stress response) redirected the photosynthetic metabolism towards defense responses. Similarly, the transcriptional responses were detected with a significant level of upregulation of GmCDPK genes suggesting their integral roles in soybean-drought-insect interactions (Hettenhausen et al., 2016). In rice, 6,885 transcripts and 238 lncRNAs were detected to be contributing for drought stress response by (Li et al., 2019). Coolen et al. (2016) (Coolen et al., 2016) also reported several unique DEGs expressed under drought compared to herbivory in Arabidopsis thaliana. In soybean, the transcriptomic profiles have not been explored much in response to drought and herbivory stress either in sequence or in tandem. It is important to note that majority of such studies have been focused on model species such as Arabidopsis and it is imperative to drive future works towards exploring drought and herbivory interaction in non-model agricultural crops that vary in their degree of drought or herbivory tolerance.
Drought and herbivory interactions drives the changes in metabolome
Analysis of metabolites accumulated in plants under drought stress and herbivory provide a more holistic view on the plant responses towards both stressors. (He et al., 2022) showed that drought stress induced the production of Daidzin which also plays an anti-herbivore role in poplars. An untargeted metabolomics using ultra-performance liquid chromatography-mass spectrometry (UPLC-MS) on the metabolites of wheat under drought revealed increased accumulation of phenolics, thymine and pyrimidine in wheat seedlings which provided further resistance against drought stress (Guo et al., 2020). Likewise, metabolomics study in maize has allowed us to identify indole, terpenoids, and green leaf volatiles [(Z)-3-hexanal and (Z)-3-hexen1-ol] as primary secondary metabolites involved in defense against herbivores (Chávez-Arias et al., 2021). Compared to other ‘Omics’ tools, metabolomics seems to be more effectively utilized by researchers when exploring drought and herbivory. However, comprehensive understanding of drought and herbivory interaction in agricultural crops using metabolomics by itself is difficult due to the involvement of myriads of biotic and abiotic factors other than drought and herbivores themselves.
Overlapping drought and herbivory stressors can alter protein expression
Proteomic approaches elucidate the role of genes related to a specific protein associated with stress, providing a snapshot of the proteome of crop under biotic or abiotic stress (Bhadauria et al., 2010; Ahmed et al., 2024). Although many studies have focused on the individual impacts of drought or herbivory on the proteomic composition of different crops, very few studies have addressed the combined effects of these stressors on crop proteomes (Zhang et al., 2010; Li et al., 2014; Gu et al., 2020). Verdugo et al. (2015) (Verdugo et al., 2015) found that Myzus persicae; (Green peach aphids) on drought-stressed susceptible plants showed higher protein expression levels compared to those on well-watered susceptible plants. A study by Dworak et al. (2016) (Dworak et al., 2016) highlights the maize leaf proteome responses to soil drought and two-spotted spider mite (Tetranychus urticae) stresses applied separately and concurrently. The findings revealed that the protein carbonylation level, a key marker of oxidative damage, increased with both soil drought and mite feeding when applied separately. However, when these stressors occurred simultaneously, there was a decrease in the protein carbonylation level, indicative of a unique response when the stresses overlap. This underscores the complexity of the interplay between different stressors and their impact on the proteomic profiles of crops, emphasizing the need for thorough research to understand the underlying mechanisms governing these interactions.
Drought and herbivory induce epigenetic modifications
Epigenetics plays a pivotal role in deciphering how plants respond to biotic and abiotic stresses, offering unique insights into the molecular mechanisms underlying stress adaptation (Mladenov et al., 2021). Epigenetic changes induced by biotic stressors like bacteria, fungi, or insect herbivores can be passed down to offspring, resulting in transgenerational priming (Holeski et al., 2012; Harris et al., 2023). Recent studies have shown that inherited epigenetic modifications can lead to acquired resistance patterns that persist for multiple generations following exposure to biotic stressors (Stassen et al., 2018). Specifically, alterations in DNA methylation, particularly in the CG context, have been observed in response to stress (Huang and Jin, 2022). Some other studies on Thale cress (Arabidopsis sp.) and rice (Oryza sp.) reveal that long-term stresses like drought-induced changes in DNA methylation can be transferred together with stress memory and improved stress tolerance to multiple generations (Wang et al., 2011; Zheng et al., 2017). Using epigenetic recombinant inbred lines (epiRILs) by (Furci et al., 2019) from a ddm1 mutant crossed with wild-type plants, a screening for downy mildew pathogen Hyaloperonospora arabidopsidis (Hpa) -resistant Arabidopsis revealed enhanced defense responses and no growth impairments post-infection, indicating defense priming. Transcriptome and DNA methylome analysis showed that priming defense genes across the genome provides lasting, inheritable disease resistance, indicating a crucial role for epigenetic responses in transgenerational acquired resistance in Arabidopsis. Hence, by studying the epigenetic landscape of plants under biotic stress conditions, researchers can identify key epigenetic signatures associated with defense responses and priming mechanisms.
Meta-genomics reveal microbial diversity under drought and herbivory
Drought and herbivory have a broader impact on multi-tropic level interactions in agricultural settings— microbial communities that could potentially directly/indirectly impact the host crop and herbivore performance are often overlooked in most of the studies. Through metagenomics analysis, (Dai et al., 2019) found that several bacteria such as Actinobacteria, and Acidobacteria significantly increased in peanut rhizosphere during drought stress. On the other hand, metagenomics along with metabolomic profile analysis in oilseed rape (Brassica napus) under root herbivory by cabbage root fly (Delia radicum) revealed significant increment in the abundances of bacterial genera like Bacillus, Pseudomonas, and Stenotrophomonas (Ourry et al., 2018). It is evident that meta-genomics in combination with other ‘Omics’ approaches can be effective in obtaining a holistic understanding of drought and herbivory interaction in crops. However, to the date, no metagenomics analysis has been conducted to unravel the interaction of these two stressors in terms of both host plants and herbivores and therefore require extensive work in the future.
Conclusion and future directions
The integration of multi-omics approaches in plant research presents a transformative opportunity to deeply explore the intricate cellular responses that underpin stress tolerance mechanisms. By integrating data from diverse omics pipelines, such as transcriptomics, proteomics, epigenomics and metabolomics, researchers gain a comprehensive understanding of how plants respond to and manage drought and herbivory. This integration allows for the identification of key regulatory networks, biomarkers, and candidate genes that differentiate between stress-tolerant and sensitive plants, providing valuable insights for breeding resilient crop varieties. Moreover, incorporating physiological studies and phytohormone signaling pathways into this multi-omics framework will further enhance our understanding of the complex interactions between plants and their environment. These approaches will elucidate how changes in physiology and hormonal signaling contribute to plant resilience under combined stress conditions. Thus, the integration of multi-omics data in plant research, augmented by physiological and phytohormonal insights, will not only deepen our understanding of stress responses and adaptive mechanisms but also pave the way for the development of resilient crop varieties that can thrive in changing environmental conditions. By merging advanced technologies with traditional breeding methods, researchers can fully harness the potential of omics-assisted breeding for ensuring food security and sustainability in agriculture.
Author contributions
IS: Writing – review & editing, Writing – original draft, Visualization, Validation, Supervision, Resources, Project administration, Methodology, Investigation, Conceptualization. MG: Writing – review & editing, Visualization, Validation, Resources, Methodology, Conceptualization. RK: Writing – review & editing, Visualization, Resources, Investigation, Conceptualization.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abid, M., Ali, S., Qi, L. K., Zahoor, R., Tian, Z., Jiang, D., et al. (2018). Physiological and biochemical changes during drought and recovery periods at tillering and jointing stages in wheat (Triticum aestivum L.). Sci. Rep. 8, 1–15. doi: 10.1038/s41598-018-21441-7
Ahmed, S., Khan, M. S. S., Xue, S., Islam, F., Ikram, A. U., Abdullah, M., et al. (2024). A comprehensive overview of omics-based approaches to enhance biotic and abiotic stress tolerance in sweet potato. Hortic. Res. 11, 1–17. doi: 10.1093/hr/uhae014
Ali, M. Y., Naseem, T., Holopainen, J. K., Liu, T., Zhang, J., Zhang, F. (2023). Tritrophic interactions among arthropod natural enemies, herbivores and plants considering volatile blends at different scale levels. Cells 12, 251. doi: 10.3390/cells12020251
Attaran, E., Major, I. T., Cruz, J. A., Rosa, B. A., Koo, A. J. K., Chen, J., et al. (2014). Temporal dynamics of growth and photosynthesis suppression in response to jasmonate signaling. Plant Physiol. 165, 1302–1314. doi: 10.1104/pp.114.239004
Avramova, Z. (2019). Defence-related priming and responses to recurring drought: Two manifestations of plant transcriptional memory mediated by the ABA and JA signalling pathways. Plant Cell Environ. 42, 983–997. doi: 10.1111/pce.13458
Banerjee, A., Roychoudhury, A. (2015). WRKY proteins: Signaling and regulation of expression during abiotic stress responses. Sci. World J. 2015 (1), 807560. doi: 10.1155/2015/807560
Bansal, S., Hallsby, G., Löfvenius, M. O., Nilsson, M. C. (2013). Synergistic, additive and antagonistic impacts of drought and herbivory on Pinus sylvestris: Leaf, tissue and whole-plant responses and recovery. Tree Physiol. 33, 451–463. doi: 10.1093/treephys/tpt019
Benny, J., Pisciotta, A., Caruso, T., Martinelli, F. (2019). Identification of key genes and its chromosome regions linked to drought responses in leaves across different crops through meta-analysis of RNA-Seq data. BMC Plant Biol. 19, 1–18. doi: 10.1186/s12870-019-1794-y
Bhadauria, V., Banniza, S., Wang, L. X., Wei, Y. D., Peng, Y. L. (2010). Proteomic studies of phytopathogenic fungi, oomycetes and their interactions with hosts. Eur. J. Plant Pathol. 126, 81–95. doi: 10.1007/s10658-009-9521-4
Bilichak, A., Ilnytskyy, Y., Woycicki, R., Kepeshchuk, N., Fogen, D., Kovalchuk, I. (2015). The elucidation of stress memory inheritance in Brassica rapa plants. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00005
Chávez-Arias, C. C., Ligarreto-Moreno, G. A., Ramírez-Godoy, A., Restrepo-Díaz, H. (2021). Maize responses challenged by drought, elevated daytime temperature and arthropod herbivory stresses: A physiological, biochemical and molecular view. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.702841
Coolen, S., Proietti, S., Hickman, R., Davila Olivas, N. H., Huang, P. P., Van Verk, M. C., et al. (2016). Transcriptome dynamics of Arabidopsis during sequential biotic and abiotic stresses. Plant J. 86, 249–267. doi: 10.1111/tpj.13167
Dai, L., Zhang, G., Yu, Z., Ding, H., Xu, Y., Zhang, Z. (2019). Effect of drought stress and developmental stages on microbial community structure and diversity in peanut rhizosphere soil. Int. J. Mol. Sci. 20, 1–17. doi: 10.3390/ijms20092265
Davila Olivas, N. H., Kruijer, W., Gort, G., Wijnen, C. L., van Loon, J. J., Dicke, M. (2017). Genome-wide association analysis reveals distinct genetic architectures for single and combined stress responses in Arabidopsis thaliana. New Phytol. 213, 838–851. doi: 10.1111/nph.2017.213.issue-2
de Freitas Bueno, A., de Freitas Bueno, R. C. O., Nabity, P. D., Higley, L. G., Fernandes, O. A. (2009). Photosynthetic response of soybean to twospotted spider mite (Acari: Tetranychydae) injury. Braz. Arch. Biol. Technol. 52, 825–834. doi: 10.1590/S1516-89132009000400005
Dong, S., Jiang, Y., Dong, Y., Wang, L., Wang, W., Ma, Z., et al. (2019). A study on soybean responses to drought stress and rehydration. Saudi J. Biol. Sci. 26, 2006–2017. doi: 10.1016/j.sjbs.2019.08.005
Du, Y., Zhao, Q., Chen, L., Yao, X., Zhang, W., Zhang, B., et al. (2020). Effect of drought stress on sugar metabolism in leaves and roots of soybean seedlings. Plant Physiol. Biochem. 146, 1–12. doi: 10.1016/j.plaphy.2019.11.003
Dworak, A., Nykiel, M., Walczak, B., Miazek, A., Szworst-Łupina, D., Zagdańska, B., et al. (2016). Maize proteomic responses to separate or overlapping soil drought and two-spotted spider mite stresses. Planta 244, 939–960. doi: 10.1007/s00425-016-2559-6
Farooq, M., Wahid, A., Kobayashi, N., Fujita, D. B. (2009b). Plant drought stress : effects, mechanisms and management To cite this version : Review article. Agron. Sustain. Dev. 29, 185–212. doi: 10.1051/agro:2008021
Farooq, M., Wahid, A., Kobayashi, N., Fujita, D., Basra, S. M. A. (2009a). Plant drought stress : effects, mechanisms and management. Agron. Sustain. Dev. 29, 185–212. doi: 10.1051/agro:2008021
Faustino, V. A., Gouveia, A. d. S., Coutinho, F. S., da Silva Júnior, N. R., Barros, R. d. A., Meriño Cabrera, Y., et al. (2021). Soybean plants under simultaneous signals of drought and Anticarsia gemmatalis herbivory trigger gene expression and metabolic pathways reducing larval survival. Environ. Exp. Bot. 190, 1-14. doi: 10.1016/j.envexpbot.2021.104594
Furci, L., Jain, R., Stassen, J., Berkowitz, O., Whelan, J., Roquis, D., et al. (2019). Identification and characterisation of hypomethylated DNA loci controlling quantitative resistance in Arabidopsis. Elife 8, 1–23. doi: 10.7554/eLife.40655
Gahlaut, V., Jaiswal, V., Tyagi, B. S., Singh, G., Sareen, S., Balyan, H. S., et al. (2017). QTL mapping for nine drought-responsive agronomic traits in bread wheat under irrigated and rain-fed environments. PloS One 12, e0182857. doi: 10.1371/journal.pone.0182857
Gautam, M., Shafi, I., Kariyat, R. (2024). Compensation of physiological traits under simulated drought and herbivory has functional consequences for fitness in soybean (Glycine max (L.) Merrill). Environ. Exp. Bot. 226, 105944. doi: 10.1016/j.envexpbot.2024.105944
Ghosh, S., Dey, G. (2022). Biotic and abiotic stress tolerance through CRISPR-Cas mediated genome editing. J. Plant Biochem. Biotechnol. 31, 227–238. doi: 10.1007/s13562-021-00746-1
Godwin, J., Farrona, S. (2020). Plant epigenetic stress memory induced by drought: a physiological and molecular perspective. Plant Epigenet. epigenom. Methods Protoc. 2093, 243–259. doi: 10.1007/978-1-0716-0179-2_17
González Guzmán, M., Cellini, F., Fotopoulos, V., Balestrini, R., Arbona, V. (2022). New approaches to improve crop tolerance to biotic and abiotic stresses. Physiol. Plant 174, 1-18. doi: 10.1111/ppl.13547
Grinnan, R., Carter, T. E., Johnson, M. T. J. (2013). The effects of drought and herbivory on plant-herbivore interactions across 16 soybean genotypes in a field experiment. Ecol. Entomol. 38, 290–302. doi: 10.1111/een.12017
Gu, H., Wang, Y., Xie, H., Qiu, C., Zhang, S., Xiao, J., et al. (2020). Drought stress triggers proteomic changes involving lignin, flavonoids and fatty acids in tea plants. Sci. Rep. 10, 1–11. doi: 10.1038/s41598-020-72596-1
Guo, X., Xin, Z., Yang, T., Ma, X., Zhang, Y., Wang, Z., et al. (2020). Metabolomics response for drought stress tolerance. Plants 9, 520. doi: 10.3390/plants9040520
Harris, C. J., Amtmann, A., Ton, J. (2023). Epigenetic processes in plant stress priming: Open questions and new approaches. Curr. Opin. Plant Biol. 75, 102432. doi: 10.1016/j.pbi.2023.102432
Hassan, M. A., Dahu, N., Hongning, T., Qian, Z., Yueming, Y., Yiru, L., et al. (2023). Drought stress in rice: morpho-physiological and molecular responses and marker-assisted breeding. Front. Plant Sci. 14, 1215731.
He, F., Wu, Z., Zhao, Z., Chen, G., Wang, X., Cui, X., et al. (2022). Drought stress drives sex-specific differences in plant resistance against herbivores between male and female poplars through changes in transcriptional and metabolic profiles. Sci. Total Environ. 845, 157171. doi: 10.1016/j.scitotenv.2022.157171
Hettenhausen, C., Sun, G., He, Y., Zhuang, H., Sun, T., Qi, J., et al. (2016). Genome-wide identification of calcium-dependent protein kinases in soybean and analyses of their transcriptional responses to insect herbivory and drought stress. Sci. Rep. 6, 1–14. doi: 10.1038/srep18973
Holeski, L. M., Jander, G., Agrawal, A. A. (2012). Transgenerational defense induction and epigenetic inheritance in plants. Trends Ecol. Evol. 27, 618–626. doi: 10.1016/j.tree.2012.07.011
Howe, G. A., Jander, G. (2008). Plant immunity to insect herbivores. Annu. Rev. Plant Biol. 59, 41–66. doi: 10.1146/annurev.arplant.59.032607.092825
Huang, P., El-Soda, M., Wolinska, K. W., Zhao, K., Davila Olivas, N. H., van Loon, J. J., et al. (2024). Genome-wide association analysis reveals genes controlling an antagonistic effect of biotic and osmotic stress on Arabidopsis thaliana growth. Mol. Plant Pathol. 25, e13436. doi: 10.1111/mpp.13436
Huang, Y., Huang, J. (2023). Analysis of plant expression profiles revealed that aphid attack triggered dynamic defense responses in sorghum plant. Front. Genet. 14. doi: 10.3389/fgene.2023.1194273
Huang, C. Y., Jin, H. (2022). Coordinated epigenetic regulation in plants: A potent managerial tool to conquer biotic stress. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.795274
Jogawat, A., Yadav, B., Chhaya, Lakra, N., Singh, A. K., Narayan, O. P. (2021). Crosstalk between phytohormones and secondary metabolites in the drought stress tolerance of crop plants: A review. Physiol. Plant 172, 1106–1132. doi: 10.1111/ppl.13328
Joshi, S., Patil, S., Shaikh, A., Jamla, M., Kumar, V. (2024). Modern omics toolbox for producing combined and multifactorial abiotic stress tolerant plants. Plant Stress 11, 100301. doi: 10.1016/j.stress.2023.100301
Kambona, C. M., Koua, P. A., Léon, J., Ballvora, A. (2023). Stress memory and its regulation in plants experiencing recurrent drought conditions. Theor. Appl. Genet. 136, 1–21. doi: 10.1007/s00122-023-04313-1
Kariyat, R. R., Balogh, C. M., Moraski, R. P., de Moraes, C. M., Mescher, M. C., Stephenson, A. G. (2013). Constitutive and herbivore-induced structural defenses are compromised by inbreeding in Solanum Carolinense (Solanaceae). Am. J. Bot. 100, 1014–1021. doi: 10.3732/ajb.1200612
Kariyat, R. R., Hardison, S. B., De Moraes, C. M., Mescher, M. C. (2017). Plant spines deter herbivory by restricting caterpillar movement. Biol. Lett. 13, 1–5. doi: 10.1098/rsbl.2017.0176
Kariyat, R. R., Hardison, S. B., Ryan, A. B., Stephenson, A. G., De Moraes, C. M., Mescher, M. C. (2018). Leaf trichomes affect caterpillar feeding in an instar-specific manner. Commun. Integr. Biol. 11, 1–5. doi: 10.1080/19420889.2018.1486653
Kariyat, R. R., Mauck, K. E., De Moraes, C. M., Stephenson, A. G., Mescher, M. C. (2012). Inbreeding alters volatile signalling phenotypes and influences tri-trophic interactions in horsenettle (Solanum carolinense L.). Ecol. Lett. 15, 301–309. doi: 10.1111/j.1461-0248.2011.01738.x
Kaur, I., Kariyat, R. (2023). Trichomes mediate plant–herbivore interactions in two Cucurbitaceae species through pre- and post-ingestive ways. J. Pest Sci. 96, 1077–1089. doi: 10.1007/s10340-023-01611-x
Kersten, B., Ghirardo, A., Schnitzler, J. P., Kanawati, B., Schmitt-Kopplin, P., Fladung, M., et al. (2013). Integrated transcriptomics and metabolomics decipher differences in the resistance of pedunculate oak to the herbivore Tortrix viridana L. BMC Genomics 14, 1–21. doi: 10.1186/1471-2164-14-737
Kim, J. M., Sasaki, T., Ueda, M., Sako, K., Seki, M. (2015). Chromatin changes in response to drought, salinity, heat, and cold stresses in plants. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00114
Kucharik, C. J., Mork, A. C., Meehan, T. D., Serbin, S. P., Singh, A., Townsend, P. A., et al. (2016). Evidence for compensatory photosynthetic and yield response of soybeans to aphid herbivory. J. Econ. Entomol. 109, 1177–1187. doi: 10.1093/jee/tow066
Kunert, K. J., Vorster, B. J., Fenta, B. A., Kibido, T., Dionisio, G., Foyer, C. H. (2016). Drought stress responses in soybean roots and nodules. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01015
Li, P., Song, W., Zhang, Y., Yang, Y., Li, S., Zhang, J. (2024). Drought stress enhances plastid-mediated RNA interference for efficient the willow leaf beetle management. Pesticide Biochem. Physiol. 204, 106037. doi: 10.1016/j.pestbp.2024.106037
Li, X., Yang, Y., Sun, X., Lin, H., Chen, J., Ren, J., et al. (2014). Comparative physiological and proteomic analyses of poplar (Populus yunnanensis) plantlets exposed to high temperature and drought. PloS One 9, 1–15. doi: 10.1371/journal.pone.0107605
Li, P., Yang, H., Wang, L., Liu, H., Huo, H., Zhang, C., et al. (2019). Physiological and transcriptome analyses reveal short-term responses and formation of memory under drought stress in rice. Front. Genet. 10. doi: 10.3389/fgene.2019.00055
Lin, P. A., Kansman, J., Chuang, W. P., Robert, C., Erb, M., Felton, G. W. (2023). Water availability and plant–herbivore interactions. J. Exp. Bot. 74 (9), 2811–2828. doi: 10.1093/jxb/erac481
Liu, L., Zhang, J., Xu, J., Li, Y., Guo, L., Wang, Z., et al. (2020). CRISPR/Cas9 targeted mutagenesis of SlLBD40, a lateral organ boundaries domain transcription factor, enhances drought tolerance in tomato. Plant Sci. 301, 110683. doi: 10.1016/j.plantsci.2020.110683
Makbul, S., Saruhan Güler, N., Durmuş, N., Güven, S. (2011). Changes in anatomical and physiological parameters of soybean under drought stress. Turk. J. Bot. 35, 369–377. doi: 10.3906/bot-1002-7
Mladenov, V., Fotopoulos, V., Kaiserli, E., Karalija, E., Maury, S., Baranek, M., et al. (2021). Deciphering the epigenetic alphabet involved in transgenerational stress memory in crops. Int. J. Mol. Sci. 22, 1–20. doi: 10.3390/ijms22137118
Müller, N. A., Kersten, B., Fladung, M., Schroeder, H. (2019). RNA-seq of eight different poplar clones reveals conserved up-regulation of gene expression in response to insect herbivory. BMC Genomics 20, 1–10. doi: 10.1186/s12864-019-6048-8
Musser, F. R., Catchot, A. L., Davis, J. A., Difonzo, C., Graham, S. H., Greene, J. K., et al. (2022). 2021 Soybean insect losses in the United States. Midsouth Entomol. 15, 39–63.
Mwenye, O. J., van Rensburg, L., van Biljon, A., van der Merwe, R. (2016). The role of proline and root traits on selection for drought-stress tolerance in soybeans: a review. South Afr. J. Plant Soil 33, 245–256. doi: 10.1080/02571862.2016.1148786
Nguyen, D., D’Agostino, N., Tytgat, T. O. G., Sun, P., Lortzing, T., Visser, E. J. W., et al. (2016). Drought and flooding have distinct effects on herbivore-induced responses and resistance in Solanum dulcamara. Plant Cell Environ. 39, 1485–1499. doi: 10.1111/pce.12708
Nguyen, N. H., Vu, N. T., Cheong, J. J. (2022). Transcriptional stress memory and transgenerational inheritance of drought tolerance in plants. Int. J. Mol. Sci. 23, 1–25. doi: 10.3390/ijms232112918
Oh, M. W., Komatsu, S. (2015). Characterization of proteins in soybean roots under flooding and drought stresses. J. Proteomics 114, 161–181. doi: 10.1016/j.jprot.2014.11.008
Ourry, M., Lebreton, L., Chaminade, V., Guillerm-Erckelboudt, A. Y., Hervé, M., Linglin, J., et al. (2018). Influence of belowground herbivory on the dynamics of root and rhizosphere microbial communities. Front. Ecol. Evol. 6. doi: 10.3389/fevo.2018.00091
Owen, L. N., Catchot, A. L., Musser, F. R., Gore, J., Cook, D. C., Jackson, R., et al. (2013). Impact of defoliation on yield of group IV soybeans in Mississippi. Crop Prot. 54, 206–212. doi: 10.1016/j.cropro.2013.08.007
Pati, P., Jena, M., Bhattacharya, S., Behera, K. S., Pal, S., Shivappa, R., et al. (2023). Biochemical defense responses in red rice genotypes. Insect 14, 632. doi: 10.3390/insects14070632
Pfalz, M., Vogel, H., Mitchell-Olds, T., Kroymann, J. (2007). Mapping of QTL for resistance against the crucifer specialist herbivore Pieris brassicae in a new Arabidopsis inbred line population, Da (1)-12× Ei-2. PloS One 2, e578. doi: 10.1371/journal.pone.0000578
Porter, S. S., Bantay, R., Friel, C. A., Garoutte, A., Gdanetz, K., Ibarreta, K., et al. (2020). Beneficial microbes ameliorate abiotic and biotic sources of stress on plants. Funct. Ecol. 34, 2075–2086. doi: 10.1111/1365-2435.13499
Rasmann, S., De Vos, M., Casteel, C. L., Tian, D., Halitschke, R., Sun, J. Y., et al. (2012). Herbivory in the previous generation primes plants for enhanced insect resistance. Plant Physiol. 158 (2), 854–863.
Sallam, A., Alqudah, A. M., Dawood, M. F. A., Baenziger, P. S., Börner, A. (2019). Drought stress tolerance in wheat and barley: Advances in physiology, breeding and genetics research. Int. J. Mol. Sci. 20, 1–36. doi: 10.3390/ijms20133137
Sani, E., Herzyk, P., Perrella, G., Colot, V., Amtmann, A. (2013). Hyperosmotic priming of Arabidopsis seedlings establishes a long-term somatic memory accompanied by specific changes of the epigenome. Genome Biol. 14, 1–23. doi: 10.1186/gb-2013-14-6-r59
Schmitz, R. J., Lewis, Z. A., Goll, M. G. (2019). DNA methylation: shared and divergent features across eukaryotes. Trends Genet. 35, 818–827. doi: 10.1016/j.tig.2019.07.007
Scott, E. R., Li, X., Kfoury, N., Morimoto, J., Han, W. Y., Ahmed, S., et al. (2019). Interactive effects of drought severity and simulated herbivory on tea (Camellia sinensis) volatile and non-volatile metabolites. Environ. Exp. Bot. 157, 283–292. doi: 10.1016/j.envexpbot.2018.10.025
Stassen, J. H. M., López, A., Jain, R., Pascual-Pardo, D., Luna, E., Smith, L. M., et al. (2018). The relationship between transgenerational acquired resistance and global DNA methylation in Arabidopsis. Sci. Rep. 8, 1–13. doi: 10.1038/s41598-018-32448-5
Sun, L., Alariqi, M., Wang, Y., Wang, Q., Xu, Z., Zafar, M. N., et al. (2024). Construction of host plant insect-resistance mutant library by high-throughput CRISPR/cas9 system and identification of A broad-spectrum insect resistance gene. Adv. Sci. 11, 2306157. doi: 10.1002/advs.202306157
Tayal, M., Somavat, P., Rodriguez, I., Thomas, T., Christoffersen, B., Kariyat, R. (2020). Polyphenol-rich purple corn pericarp extract adversely impacts herbivore growth and development. Insects 11, 1–20. doi: 10.3390/insects11020098
Thoen, M. P., Davila Olivas, N. H., Kloth, K. J., Coolen, S., Huang, P. P., Aarts, M. G., et al. (2017). Genetic architecture of plant stress resistance: multi-trait genome-wide association mapping. New Phytol. 213, 1346–1362. doi: 10.1111/nph.2017.213.issue-3
Tian, Z., Li, K., Sun, Y., Chen, B., Pan, Z., Wang, Z., et al. (2024). Physiological and transcriptional analyses reveal formation of memory under recurring drought stresses in seedlings of cotton (Gossypium hirsutum). Plant Sci. 338, 111920. doi: 10.1016/j.plantsci.2023.111920
Van Hulten, M., Pelser, M., Van Loon, L. C., Pieterse, C. M., Ton, J. (2006). Costs and benefits of priming for defense in Arabidopsis. Proc. Nat. Acad. Sci. 103 (14), 5602–5607.
Vázquez-González, C., Pombo-Salinas, L., Martín-Cacheda, L., Rasmann, S., Röder, G., Abdala-Roberts, L., et al. (2022). Effect of water availability on volatile-mediated communication between potato plants in response to insect herbivory. Funct. Ecol. 36, 2763–2773. doi: 10.1111/1365-2435.14159
Verdugo, J. A., Sauge, M. H., Lacroze, J. P., Francis, F., Ramirez, C. C. (2015). Drought-stress and plant resistance affect herbivore performance and proteome: The case of the green peach aphid Myzus persicae (Hemiptera: Aphididae). Physiol. Entomol. 40, 265–276. doi: 10.1111/phen.12111
Wang, W. S., Pan, Y. J., Zhao, X. Q., Dwivedi, D., Zhu, L. H., Ali, J., et al. (2011). Drought-induced site-specific DNA methylation and its association with drought tolerance in rice (Oryza sativa L.). J. Exp. Bot. 62, 1951–1960. doi: 10.1093/jxb/erq391
Xu, C., Xia, C., Xia, Z., Zhou, X., Huang, J., Huang, Z., et al. (2018). Physiological and transcriptomic responses of reproductive stage soybean to drought stress. Plant Cell Rep. 37, 1611–1624. doi: 10.1007/s00299-018-2332-3
Yesudas, C. R., Sharma, H., Lightfoot, D. A. (2010). Identification of QTL in soybean underlying resistance to herbivory by Japanese beetles (Popillia japonica, Newman). Theor. Appl. Genet. 121, 353–362. doi: 10.1007/s00122-010-1314-9
Zhang, J. H., Sun, L. W., Liu, L. L., Lian, J., An, S. L., Wang, X., et al. (2010). Proteomic analysis of interactions between the generalist herbivore Spodoptera exigua (Lepidoptera: Noctuidae) and Arabidopsis thaliana. Plant Mol. Biol. Rep. 28, 324–333. doi: 10.1007/s11105-009-0156-6
Zhang, Y., Zhong, J., Munawar, A., Cai, Y., He, W., Zhang, Y., et al. (2024). Knocking down a DNA demethylase gene affects potato plant defense against a specialist insect herbivore. J. Exp. Bot. 75, 483–499. doi: 10.1093/jxb/erad387
Zheng, X., Chen, L., Xia, H., Wei, H., Lou, Q., Li, M., et al. (2017). Transgenerational epimutations induced by multi-generation drought imposition mediate rice plant’s adaptation to drought condition. Sci. Rep. 7, 1–13. doi: 10.1038/srep39843
Keywords: omics integration, water-stress, herbivore-pressure, resilience, stress priming
Citation: Shafi I, Gautam M and Kariyat R (2024) Integrating ecophysiology and omics to unlock crop response to drought and herbivory stress. Front. Plant Sci. 15:1500773. doi: 10.3389/fpls.2024.1500773
Received: 23 September 2024; Accepted: 14 October 2024;
Published: 04 November 2024.
Edited by:
Raul Antonio Sperotto, Federal University of Pelotas, BrazilReviewed by:
Marcelo Lattarulo Campos, Federal University of Mato Grosso, BrazilCopyright © 2024 Shafi, Gautam and Kariyat. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rupesh Kariyat, cmthcml5YXRAdWFyay5lZHU=
 Insha Shafi
Insha Shafi Manish Gautam
Manish Gautam Rupesh Kariyat
Rupesh Kariyat