- 1Rice Pathology Department, Plant Pathology Research Institute, Agricultural Research Center, Sakha, Kafrelsheikh, Egypt
- 2Business Administration Department, Community College, King Khalid University, Guraiger, Abha, Saudi Arabia
- 3Rice Research Department, Field Crops Research Institute, Agricultural Research Center, Sakha, Kafrelsheikh, Egypt
- 4Department of Biology, College of Science, United Arab Emirates University, Al-ain, Abu-Dhabi, United Arab Emirates
- 5Department of Agricultural Botany, Faculty of Agriculture, Tanta University, Tanta, Egypt
Sheath blight, caused by Rhizoctonia solani AG1 IA, is a challenging disease of rice worldwide. In the current study, nine R. solani isolates, within the anastomosis group AG-1 IA, were isolated, characterized based on their macroscopic and microscopic features, as well as their ability to produce cell wall degrading enzymes (CWDEs), and further molecularly identified via ITS sequencing. Although all isolates were pathogenic and produced typical sheath blight symptoms the susceptible rice cultivar, Sakha 101, R. solani AG1 IA -isolate SHBP9 was the most aggressive isolate. The virulence of isolate SHBP9 was correlated with its overproduction of CWDEs, where it had the highest pectinase, amylase, and cellulase activity in vitro. R. solani AG1 IA -isolate SHBP9 was able to infect 12 common rice-associated weeds from the family Poaceae, as well as over 25 economic crops from different families, except chickpea (Cicer arietinum) from Fabaceae, Rocket (Eruca sativa) from Brassicaceae, and the four crops from Solanaceae. Additionally, rice genotype-based resistance was evaluated using 11 rice genotypes for their response to R. solani isolates, morphological traits, yield components, and using 12 SSR markers linked to sheath blight resistance. Briefly, the tested 11 rice genotypes were divided into three groups; Cluster “I” included only two resistant genotypes (Egyptian Yasmine and Giza 182), Cluster “II” included four moderately resistant genotypes (Egyptian hybrid 1, Giza 178, 181, and 183), whereas Cluster “III” included five susceptible (Sakha 104, 101, 108, Super 300 and Giza 177). Correspondingly, only surface-mycelium growth was microscopically noticed on the resistant cultivar Egyptian Yasmine, as well as the moderately resistant Egyptian hybrid 1, however, on the susceptible Sakha 104, the observed mycelium was branched, shrunk, and formed sclerotia. Accordingly, Indica and Indica/Japonica rice genotypes showed more resistance to R. solani than Japonica genotypes. These findings provide insights into its pathogenicity mechanisms and identify potential targets for disease control which ultimately contributes to the development of sustainable eco-friendly disease management strategies. Moreover, our findings might pave the way for developing resistant rice varieties by using more reliable resistance sources of non-host plants, as well as, rice genotype-based resistance as a genetic resource.
1 Introduction
Rice (Oryza sativa L.; family Poaceae) is a monocotyledonous, semi-aquatic annual cereal crop that is cultivated worldwide across a wide range of land areas and in diverse environmental conditions, from wetlands to uplands and temperate to tropical conditions (Hussain et al., 2020; Nawaz et al., 2022; Zahra et al., 2022). Globally, rice is grown over 165.04 million hectares with a production of 776.46 million tons (FAOSTAT, 2023). In Egypt, rice is cultivated over 646.32 thousand hectares, mainly in the northern and Delta areas, representing only 3.91% of the rice area harvested in Africa (16.52 million hectares). However, this small-harvested area in Egypt produces approximately 5.80 million tons, accounting for 14.54% of the estimated African rice production (39.87 million tons) (FAOSTAT, 2023). This might be due to the high productivity of rice per land unit in Egypt. The Egyptian estimated yield (8.97 tons per hectare) is almost double the global yield per land unit (4.70 tons per hectare) and 3.7-fold of the estimated African yield (2.41 tons per hectare) (FAOSTAT, 2023). Although rice consumption increases annually, the spread of various rice diseases, disorders, and pests is also increasing to pose greater challenges to rice production worldwide. Unfortunately, rice plants are susceptible to numerous fungal, bacterial, viral, and other diseases caused by phytoplasmas and nematodes (Mew et al., 2004; Gnanamanickam and Gnanamanickam, 2009; Cartwright et al., 2018). Fungal diseases of rice include, but are not limited to, rice blast (Magnaporthe oryzae; Anamorph: Pyricularia oryzae), brown spot (Bipolaris oryzae; Anamorph: Helminthosporium oryzae), sheath blight (Rhizoctonia solani), bakanae disease (Fusarium fujikuroi), false smut (Ustilaginoidea virens), narrow brown leaf spot (Cercospora janseana), kernel smut (Tilletia barclayana), and foot rot (F. moniliforme) (Mew et al., 2004; Gnanamanickam and Gnanamanickam, 2009; Cartwright et al., 2018).
Among these diseases, Sheath blight (ShB) is a soil-borne disease caused by fungal pathogens (Rhizoctonia solani); it is recorded as one of the most important diseases (Wamishe et al., 2007). The fungus responsible for rice sheath blight disease, R. solani Kühn teleomorph (Thanatephorus cucumeris) anastomosis group (AG) AG-1 IA, was initially identified in Japan at the start of the early twentieth century (Lee and Rush, 1983), then it has been observed throughout the world’s major rice-growing regions (Ou, 1985). In Egypt, the rice sheath blight was recorded as a serious disease caused by the phytopathogenic fungus R. solani AG1 IA (Hassan, 2021). Distinctive mycelium properties of the Rhizoctonia fungus as, at the point of branching, typical right angles are usually seen at hyphal constrictions, the mycelium consists of hyphae partitioned into individual cells by dolipore septa, the sclerotia appeared after three days of inoculation on PDA media (Hassan, 2021). The symptoms of the disease include greenish-gray elliptical or oval-shaped spots with yellow margins mostly found on leaf sheaths, but, at times, leaf blades are also infected (Vidhyasekaran et al., 1997).
Plant cell walls are primarily composed of cellulose, hemicellulose, and lignin, with cellulose being the most prevalent component (Han et al., 2003; Keegstra, 2010). Cell wall degradation enzymes (CWDEs) play an important role in the fungus growth by releasing polysaccharides and nutrients, further facilitating hyphal penetration, and increasing branching around the host (De Lorenzo et al., 1997). There is a correlation between the infection severity and activity of CWDEs (Gawade et al., 2017), in the case of R. solani IG1 IA isolates, a high ability to produce CWDEs is linked to the rapid appearance of symptoms related to rice sheath blight disease (Hassan, 2021). In the process of fungal infection in plants, CWDEs, particularly pectolytic enzymes, are essential because they break down pectin, which gives the pathogen a carbon source, and expose cell wall components to other enzymes like cellulase and hemicellulase, which further break down the cell wall (Jia et al., 2009). In addition to pectolytic and cellulolytic enzymes, lipases and proteases are key enzymes in pathogenesis that attack the plasmalemma following the breakdown of the cell wall by proteases (Hameed et al., 1994).
R. solani has a wide host range and high pathogenicity; the main obstacle in managing sheath blight disease is the lack of an identified germplasm with an adequate level of resistance for use in the resistance breeding program (Poonguzhali et al., 2022), because of the lack of identified resistant donors in cultivated varieties (Bonman et al., 1992) and, until now not determined sources of stable resistance (Molla et al., 2020). About 250 different host plant species, including those of the Poaceae, Fabaceae, Solanaceae, Amaranthaceae, Brassicaceae, Rubiaceae, Malvaceae, Asteraceae, Araceae, Moraceae, and Linaceae families, are susceptible to R. solani (Chahal et al., 2003). R. solani has a total of 14 distinct anastomosis groups (AG1 to AG13 and AGBI), all of which show significant variation in colony morphology, host range, aggressiveness, and nutritional requirements (Guillemaut et al., 2003; Ahvenniemi et al., 2009; Ajayi-Oyetunde and Bradley, 2018). The three subgroups of R. solani AG1 isolates (IA, IB, and IC) are separated based on sclerotia size and morphology as well as sequence homology (Sneh et al., 1991), this all results in rice ShB, with AG1-IA being identified as the causative agent most frequently (Bernardes-De-Assis et al., 2009; Gonzalez et al., 2011). As a result of climatic change, the excessive usage of fungicides to control sheath blight disease is unfriendly to the environment and the general health (Goswami et al., 2019). Moreover, the development of resistant rice genotypes to sheath blight was the most eco-friendly environmental strategy to stop the disease from spreading in the soil (Liu et al., 2009). However, the available breeding strategies are time-consuming (Sherif et al., 2023).
The sheath blight pathogen can produce sclerotia, which may remain dormant over the years in the soil, and stubble and re-infect healthy rice plants in the subsequent rice season (Dey et al., 2016). Long-term and broad-spectrum resistance have been linked to quantitative resistance, which makes it an important tool for rice breeding. Furthermore, the disease resistance is controlled by multiple genes, which confer quantitative resistance (Molla et al., 2020). The revolution of next-generation sequencing and genome-wide association analysis (GWAS) helps in the desiccation of the molecular basis for disease resistance and reduces the breeding cycle for sheath blight resistance (Liu et al., 2021). Recently, two major loci controlling the disease resistance were identified on chromosomes 9 and 11 (Liu et al., 2009; Channamallikarjuna et al., 2010), and then a dozen QTLs were reported on chromosomes 1, 2, 3, 5, 7, and 8 (Chen et al., 2023). Using resistance-linked molecular markers would save time during the breeding programs.
Sheath blight is an emerging disease in Egypt and our knowledge about sources of rice genotypes resistance to sheath blight pathogen is still limited (Hassan, 2021; Kalboush et al., 2024). The current study aims (i) to isolate and characterize the causal agent of sheath blight on rice and to determine the relationship between the pathogen’s virulence and its ability to produce CWDEs which could provide insights into its pathogenicity mechanisms and identify potential targets for disease control. (ii) To determine the host range of the R. solani AG1 IA and short-list the non-host plants which could lead to the development of resistant rice varieties using non-host plants as a genetic resource. And (iii) to evaluate the rice genotype-based resistance against R. solani, to find more reliable resistance sources that could contribute to the rice breeding programs. The findings of this study could provide a comprehensive understanding of the pathogen, its interaction with its host (s), and the potential for developing resistant rice varieties, ultimately contributing to the development of better sustainable and eco-friendly disease management strategies.
2 Materials and methods
2.1 Plant materials
Eleven commercial rice cultivars with the pedigree and type; Giza 177 (Giza 171/Yomji No.1/PiNo.4- Japonica), Giza 178 (Giza 175/Milyang 49 - Indica/Japonica), Giza 181 (IR23/IR22- Indica), Egyptian Hybrid 1 (IR69625A/Giza 178 R- Indica/Japonica), Giza 182 (Giza181/IR39422-161-1-3//Giza 181- Indica), Giza 183 (Giza 178/SKC23893- Indica/Japonica), Egyptian Yasmine (IR262/KDML-105- Indica), Sakha 101 (Giza 176/Milyang 79- Japonica), Sakha 104 (GZ 4096-8-1/GZ 4100-9-1- Japonica), Sakha 108 (Sakha 101/HR5824/Sakha 101- Japonica) and Sakha super 300 (PTGMS38/China2- Indica/Japonica) was used as experimental plant materials throughout this study. Seeds were kindly provided by the Rice Research Department, Field Crops Research Institute (FCRI), Agricultural Research Center (ARC), Egypt. Moreover, seeds of 12 common rice-associated weeds (Table 1) were obtained from the Weed Research Central Laboratory (WRCL), ARC, Egypt. Likewise, seeds/grains of 31 economic crops (Table 2) were generously provided by the Department of Food Legumes Research, FCRI-ARC, Egypt. All seeds were healthy, uniform, and homologous in size and color.
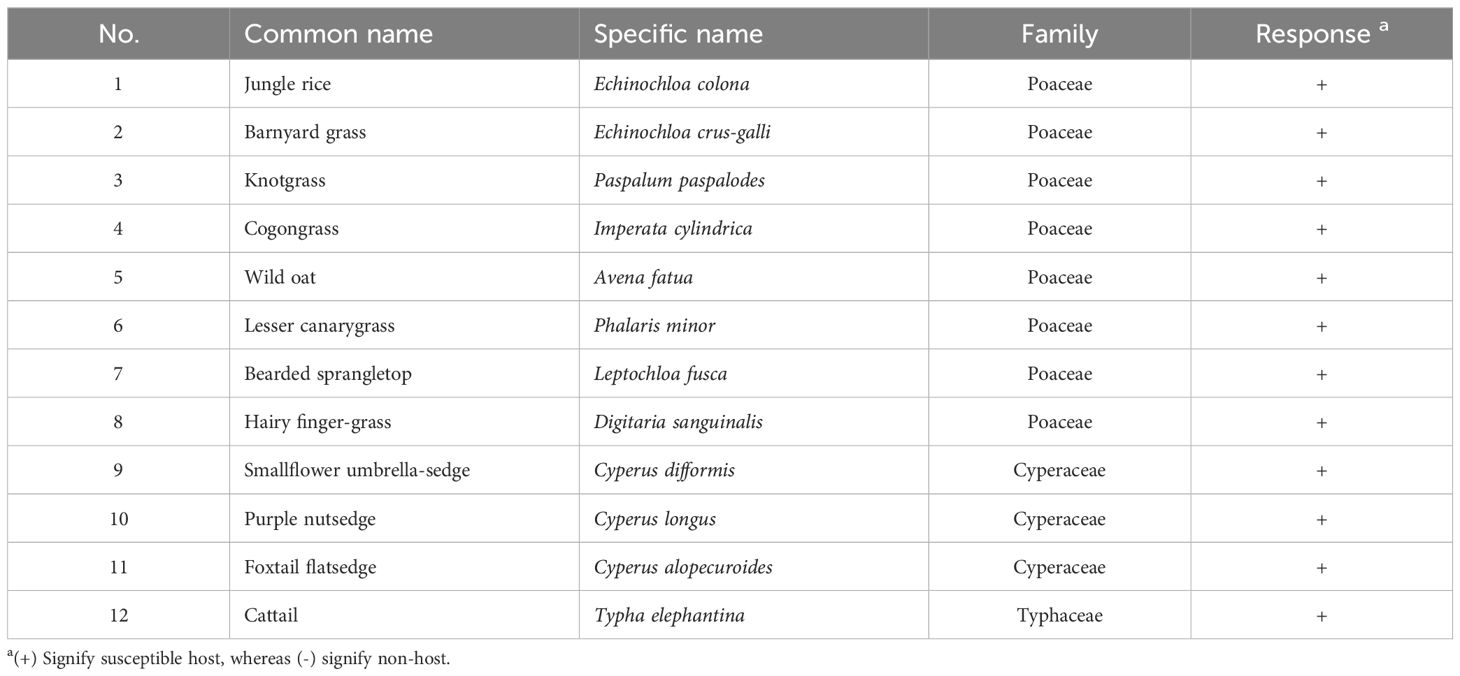
Table 1. List of common rice-associated weeds tested for host range assessment of Rhizoctonia solani under greenhouse conditions.
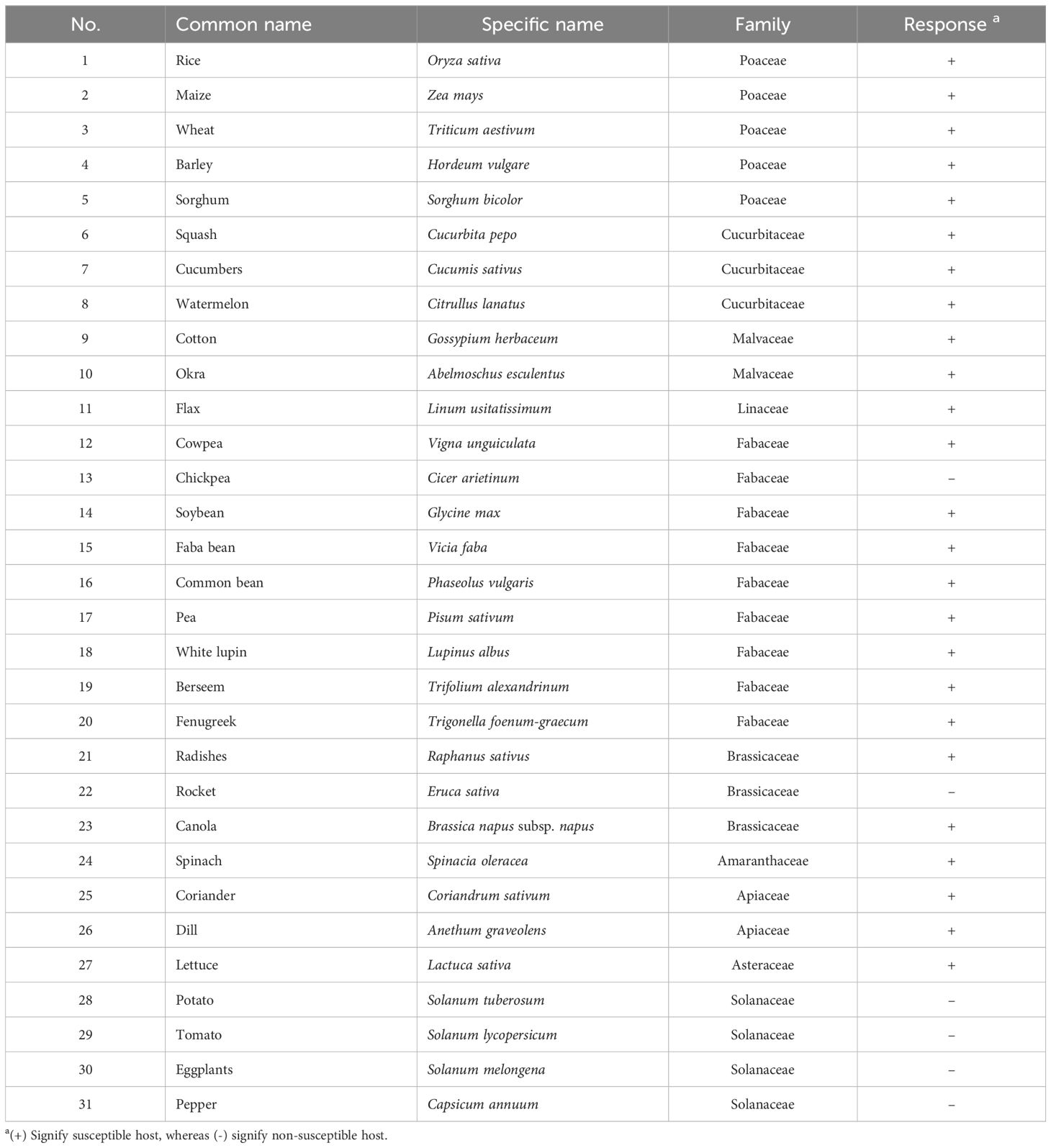
Table 2. List of major economic crops tested for host range assessment of Rhizoctonia solani under greenhouse conditions.
All experiments were conducted at the Rice Pathology Department, Rice Research and Training Center (RRTC), ARC, Egypt. Seeds were initially sterilized for 3-4 minutes using sodium hypochlorite (NaClO) solution (1%) and then washed twice with sterile water, then planted in plastic pots (30 cm inner diameter and 45 cm in depth) filled with sterilized clay soils. All pots were maintained under greenhouse conditions at 27 ± 2°C, >75% RH, and 8D:16L light cycle. Plants were irrigated daily, or earlier if needed, and fertilized with macro- and micronutrients according to the recommendations of the Egyptian Ministry of Agriculture and Land Reclamation for these cultivars/species.
2.2 Isolation, characterization, and molecular identification of R. solani isolates
2.2.1 Pathogen isolation
Samples of rice plants showing typical symptoms of sheath blight disease (disease severity: 20-40%) were collected from commercial farms within three different governorates (Beheira, Dakahlia, and Kafrelsheikh) (Table 3). Initially, collected infected materials were surface sterilized for 3-4 min using 1% NaOCl, then washed twice using sterilized distilled water, and dried between sterilized filter papers to remove the extra water. The fungal causal agents were isolated as described by Hassan et al. (2023). Briefly, the sterilized parts were placed on a water agar medium (WA), and then incubated at 26 ± 2°C for 24 h. moreover, the hyphal tips method (Brown, 1924) was used to purify the fungal isolates from the mixed or contaminated cultures. Briefly, the terminal of the growing hypha was cut off and transferred to a new potato dextrose agar (PDA) then incubated at 26 ± 2°C until the whole surface of the plate was covered with mycelium.
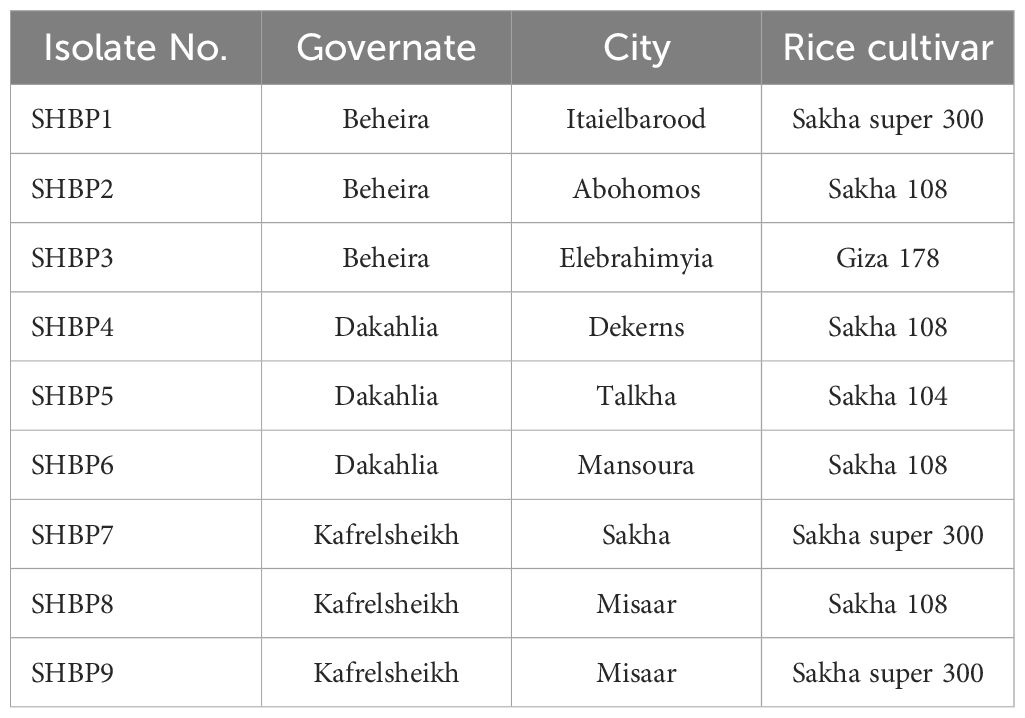
Table 3. Sources and plant materials used to isolate the causal agent of rice sheath blight disease used in this study.
2.2.2 Macroscopic and microscopic characterization of R. solani isolates
All fungal isolates were initially identified based on their macroscopic features (cultural morphology), and microscopic characteristics (mycelium morphology, and other observed structures) (Barnett and Hunter, 1972). Briefly, 9-cm Petri dishes containing PDA media were inoculated at the center with a 0.5-cm plug of three-day-old cultures of the fungal isolates and incubated for seven days as described above. All tested isolates were examined for morphological characteristics according to Moni et al. (2016). Each isolate was maintained with five biological replicates and repeated twice (two technical replicates). The purified fungal isolates were first identified based on their cultural morphology characteristics and then the identification was confirmed using microscopic features as R. solani.
2.2.3 Molecular identification of R. solani isolates
Moreover, the identification of the nine isolates was further confirmed based on the sequencing of the Internal transcribed spacer (ITS) region. Briefly, the isolates were grown on PD broth and incubated at 26 ± 2°C for 5-7 days. Then, the fungal mycelium was harvested, filtered through cheesecloth, washed twice with sterile water, and dried using sterile filter paper. The genomic DNA was extracted according to Kuramae (1997) with slight modifications as described by Atallah et al. (2022, 2024). Subsequently, the 18S rDNA fragment was amplified using the ITS1 (5’ TCCGTAGGTGAACCTGCGG 3’) and ITS4 (5’ CCTCCGCTTATTGATATGC 3’) universal primers (Usyk et al., 2017). Molecular analysis was done through the Sigma Company, Germany. The query sequence was compared to the recent available data in Gen Bank, the National Center for Biotechnology Information website (NCBI, http://www.ncbi.nlm.nih.gov/gene/; access) using the Nucleotide–Nucleotide Basic Local Alignment Search Tool (BLASTn) (Altschul et al., 1997). Due to the variation in the length of ITS-based sequence accessions, sequences were initially aligned using the ClustalW algorism, then trimmed (~480 bp) while used for phylogenetic analysis. The Molecular Evolutionary Genetics Analysis software (MEGA11) was used to align the query sequence with other 40 Rhizoctonia strains/isolates from different anastomosis groups (AGs) retrieved from the recently available data in NCBI GenBank (Table 4). The Neighbor-Joining method was used to infer the phylogenetic tree for the identified isolates (Saitou and Nei, 1987). Furthermore, the phylogeny test was done using the Bootstrap method.
Table 4. Sequences of internal transcribed spacer (ITS) region from Rhizoctonia strains/isolates from different anastomosis groups (AGs) used for phylogenetic analysis.
2.2.4 Pathogenicity of R. solani isolates
To fulfill Koch’s postulates, the pathogenicity of the nine R. solani isolates was assessed on the commercial susceptible rice cultivar ‘Sakha 101’ as described by Park et al. (2008) with slight modifications. Briefly, rice seeds were sterilized for 3-4 minutes using sodium hypochlorite (NaClO) solution (1%) and then washed twice with sterile water. The sterilized seeds were planted in sterile soil until the seedlings reached about twenty-one days of age. Afterward, the seedlings were transferred to new sterilized soil as individual plants in each pot (30 cm in diameter) had two seedlings. The seedlings were maintained and fertilized well (1 g 46.5% N/pot) until they reached 55 days from planting and produced the maximum number of branches. The isolates were prepared and inoculated on the sheaths of rice plants using a 5-mm plug of three-day-old cultures of each isolate separately. Fungal plugs were placed on the sheaths, starting from the base of the plants, and the pieces were wrapped in aluminum foil. After 3-4 days of inoculation, the aluminum foil was removed. Plants were monitored at 30 ± 2°C with a relative humidity of 80-100% until symptoms appeared. This experiment was completely randomized with five replicates, each with four pots. The experiment was repeated twice under the same conditions. Disease scoring was calculated as described by the International Rice Research Institute’s standard evaluation system (IRRI, 2013) on a scale of 0 to 9. The disease severity was taken after one month of symptoms appeared, as relative lesion height (RLH) using the formula described by Sharma and Teng (1990).
2.2.5 in vitro enzymatic activity of cell wall degradation enzymes of R. solani isolates
The screening of cell wall degradation enzymes (CWDE) including cellulase, pectinase, and amylase activity was assessed by transferring a 5-mm mycelial plug of a 7-day-old culture of each R. solani isolate on the media containing carboxymethyl cellulose (CMC), pectin, or starch, respectively (Leopold and SamsˇinˇÁková, 1970; Hankin and Anagnostakis, 1975; Jalgaonwala and Mahajan, 2011), then incubated at 26 ± 2°C for 3-7 days. To visualize the CWDE activities, CMC and pectin media with the R. solani were flooded with 0.3% Congo red solution for 15min and plates were destained with 0.1% NaCl for another 15 minutes, whereas the plates with starch agar medium were flooded with iodine solution, kept for one minute, and then poured off. The plates were visualized for the halo/clear zone, indicating cellulase, pectinase, and amylase production (Anand et al., 2010). Data were expressed as Fungal linear growth (cm) as an indicator for better utilization of CMC, pectin, or starch from the media.
2.2.6 Expression of cell wall degradation genes of R. solani isolates using real-time PCR
Fungal mycelia of R. solani were collected after 3 days of incubation on the media containing pectin to assess the gene expression of some CWD genes including pectate lyase B (RsPLB; AG1IA_00690) with sequencing F (GTCGGGACGGTAAGCATAAA) and R (GCTAGCTTCAGAGGCGAATAA), putative pectate lyase C (RsPLC; AG1IA_10120) with sequencing F (TATTCGACCATGTCAGCGTATC) and R (GAATGTCTCATCTGCCTTCCA), and polysaccharide lyase family 1 protein (RsPSL; AG1IA_06890) with sequencing F (AGGACTACTACGATGGGTTACT) and R (TCTTCGTCTTCGTTGCTATCC), using qRT-PCR. Briefly, total fungal RNA was isolated using a Simply P Total RNA Extraction Kit (catalog number BSC52S1), according to the manufacturer’s procedure. Then, cDNA was synthesized using a TOP script™ cDNA Synthesis Kit as described in the manufacturer’s protocol. The primer sequences for the three genes (RsPLB, RsPLC, and RsPSL) are previously described by (Bhaskar Rao et al., 2020). β-Tubulin was used as a housekeeping gene, and the 2−ΔΔCT method was used for the calculation of relative gene expression (Livak and Schmittgen, 2001). The expression level of these genes in R. solani grown on a pectin-free medium was used as a reference point for comparison.
2.3 Greenhouse experiments
2.3.1 Host range assessment of R. solani AG1 IA isolate SHBP9 under greenhouse conditions
Seeds of 12 common rice-associated weeds (Table 1), and 31 economic crops (Table 2) were initially sterilized using NaClO (1%) for 3-4 min, washed twice with sterile water, and then transferred into sterilized conical flasks. A 5-mm mycelial plug of a 7-day-old culture from R. solani AG1 IA isolate SHBP9 was transferred separately into each flask with a mixture of sterilized corn, rice husk, and sand (1:2:1). The flasks were incubated at 26 ± 2°C for 15 days until the fungus produced the sclerotia. The pots (30 cm) containing sterile soil were inoculated with sandy media containing R. solani AG1 IA isolate SHBP9 and mixed well, then irrigated and maintained under greenhouse conditions for about 24h. The seeds were sown in the inoculated pots, covered with sterile soil, irrigated again, then maintained at 26 ± 2°C under greenhouse conditions. After 21-28 days of sowing for different crops, the disease symptoms were developed and recorded as (+) susceptible host and (-) for non-host. On the other hand, weed plants were left to 60 days post-sowing (dps), and the disease score was recorded. A complete randomized design with three replicates was used for the experiment; each replicate had four pots. The whole experiment was repeated twice to ensure the reliability and validity of the results.
2.3.2 Evaluation of rice genotypes under artificial inoculation with R. solani AG1 IA (SHBP9 isolate) at greenhouse conditions
Eleven rice genotypes were used in this experiment. The seeds of rice genotypes were initially sterilized and sown as described above. Subsequently, rice seedlings were thinned as 2 seedlings per pot and fertilized as recommended (1 g 46.5% N/pot) at 21 dps and maintained under greenhouse conditions as described above. At 60 dps, rice plants were inoculated with R. solani AG1 IA isolate SHBP9 as described by Park et al. (2008). The inoculated genotypes were monitored at 26 ± 2°C with a relative humidity of 80-100% until symptoms appeared. The plants were observed for symptom expression after 30 days post-inoculation (dpi). The rice genotypes inoculated with PDA plugs without any fungal mycelia served as negative control (Mock) plants and were used for comparison of the vegetative and yield parameters in R. solani-inoculated and non-inoculated (Mock) rice genotypes. The whole experiment was repeated twice to ensure the reliability and validity of the results.
2.3.2.1 Assessment of sheath blight disease
Disease scoring for rice genotypes was calculated as described by IRRI (2013) on a scale of 0 to 9. The disease severity was taken after 30 days of symptoms appearance, as Relative lesion height (RLH) and the Percent disease index (PDI) were calculated using the formula provided by Sharma and Teng (1990) and Naveenkumar et al. (2022), respectively.
2.3.2.2 Assessment of vegetative and yield parameters
The vegetative traits including plant height (cm) at the maturing stage, number of tillers per plant, and stem diameter (mm) at the maximum tiller stage were recorded. Moreover, flag leaf area (FLA; cm2) was calculated using the formula FLA = K (L × W) of Palaniswamy and Gomez (1974), where L is the maximum leaf length, W is the maximum width of the leaf and K=0.75. Likewise, the yield components including panicle length (cm), panicle weight (g),1000-grain weight (g), and the number of discolored grains were recorded for R. solani-infected and non-infected rice genotype plants at the harvest stage. All parameters were collected according to the guidelines described in IRRI (2002).
2.3.2.3 Preparation of rice sheath for microscopy
Infected materials were collected from the tested eleven rice genotypes at 7 dpi. Subsequently, each infected sheath was chopped into small pieces (2-5 mm), submerged in trypan blue solution (0.05%) for 10 min, then washed with lactophenol three times. After washing, stained samples were transferred onto glass slides and examined using a compound microscope (Basu et al., 2016).
2.3.3 Molecular analysis using simple sequence repeat markers
Young leaves of eleven rice genotypes were collected for DNA extraction according to Murray and Thompson (1980). Furthermore, the purity of the extracted DNA was checked by Agarose gel (0.8%) and stored at -20°C for further investigations. Twelve SSR markers (Table 5) were selected from the previous studies on sheath blight. The polymerase chain reaction was conducted in 20 µl reaction volume containing 2 µl DNA templet (10 ng), 10 ul master mix (2X Taq Plus Master Mix with dye), 0.5 ul of each forward and reversed primers (10 pM), and 7 µl of d2H2O. In addition, the gel electrophoresis was done using Agarose gel of 3%, and the band size was calculated by GelAnalyzer 23.1.1 (www.gelanalyzer.com). The cluster analysis and its associated dendrogram were prepared by Paleontological Statistics (Past 4.13) software (Hammer and Harper, 2001).
2.4 Data analysis
All in vitro experiments were performed in a completely randomized design with five biological replicates and two technical replicates. Likewise, all in vivo experiments were performed in a completely randomized design, unless otherwise stated, with three replicates, each replicate had four pots, and all experiment was repeated twice. All data were statistically analyzed according to the analysis of variance technique (ANOVA), followed by Tukey’s Honestly Significant Difference (HSD) Test as a post hoc analysis. On the other hand, the experiment of evaluation of rice genotypes against R. solani infection was laid out using a split-plot design comprising R. solani infection in the main plots (infection) and eleven rice genotypes in subplots (genotypes). Data were statistically analyzed using ANOVA, followed by the HSD Test as a post hoc analysis based on the p-value of infection (p infection< 0.05), genotypes (p genotypes< 0.05), and their interaction (p infection × genotypes< 0.05). Principal component analysis (PCA) was performed and its associated scatter and loading plots were generated using the response of the 11 tested rice genotypes to the 9 isolates of R. solani as expressed by relative lesion height (RLH; %). Moreover, two-way Hierarchical Cluster Analysis (HCA) was performed using the average of each RLH in each treatment. Distance and linkage were done using the Ward method (Ward Jr, 1963) and RLH similarities were presented as a heat map.
3 Results
3.1 Isolation and Identification of R. solani isolates
Nine isolates (named SHBP 1- 9) were isolated from diseased rice cultivars showing typical symptoms of sheath blight disease (four isolates from Sakha 108, three isolates from Sakha super 300, and one isolate from Giza 178 and Sakha 104 each) collected from three Egyptian governorates included Beheira (Itaielbarood, Abohomos, Elebrahimyia), Dakahlia (Dekerns, Talkha, Mansoura), and Kafrelsheikh (Sakha, Misaar, Kellan) (Table 3). Isolates SHBP 1-9 showed typical morphological characteristics, colony texture, and abundance of asexual reproductive structures (mycelium and sclerotia) with R. solani when grown on a PDA medium (Figure 1A). The mycelium growth showed two types: moderate speed with flat shape in five isolates (SHBP1, 3, 4, 7, and 9) and rapid with flat/arial in four isolates (SHBP2, 5, 6, and 8) (Figure 1A; Table 6).
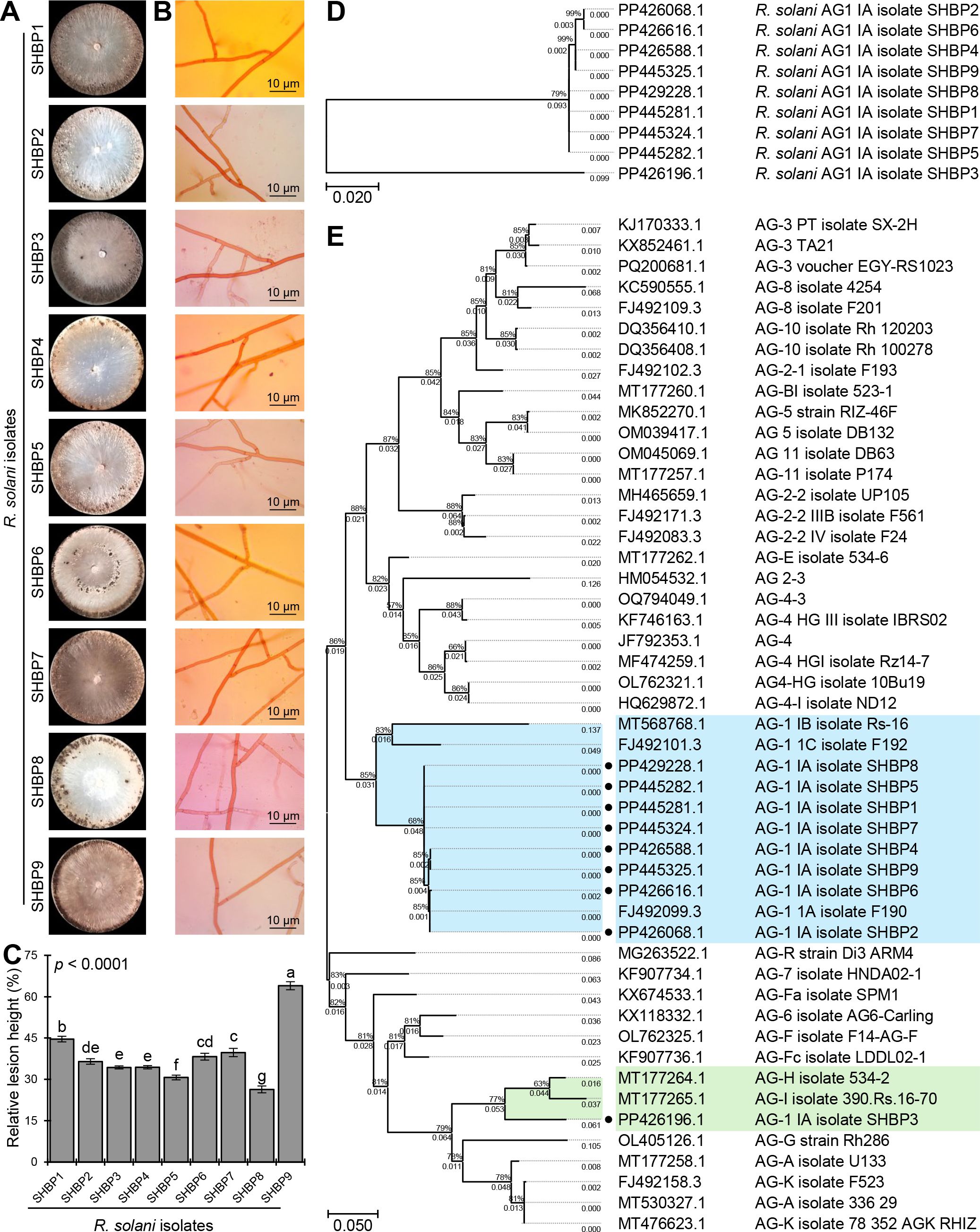
Figure 1. The macroscopic and microscopic features, pathogenicity, and molecular identification of the phytopathogenic fungus Rhizoctonia solani, the causal agent of sheath blight disease on rice. (A) The growth and macroscopic features of the nine isolates of R. solani on potato dextrose agar (PDA) media after 10 days of incubation at 26 ± 2°C. (B) Microscopic characteristics and mycelium features of the nine isolates of R. solani. (C) The relative lesion height (%) of nine isolates of R. solani on susceptible rice cultivar Sakha 101 under greenhouse conditions. Bars denote the means ± standard deviations (means ± SD) of five biological replicates. Different letters signify statistically significant differences among isolates using the Tukey HSD test (p< 0.05). (D, E) The evolutionary analysis of only R. solani AG1 IA isolates SHBP 1-9 by themselves and in comparison with other 40 R. solani strains/isolates from different anastomosis groups (AGs) retrieved from the recent available data in National Center for Biotechnology Information (NCBI) GenBank (https://www.ncbi.nlm.nih.gov/). The evolutionary history was inferred using the Maximum Likelihood method and the Tamura-Nei model. The tree with the highest log likelihood ( (-1242.36 and -6738.22, respectively) is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches. The proportion of sites where at least 1 unambiguous base is present in at least 1 sequence for each descendent clade is shown next to each internal node in the tree. Analysis in panel D involved 9 nucleotide sequences with a total of 608 positions in the final dataset, whereas analysis in panel E involved 49 nucleotide sequences with a total of 694 positions in the final dataset. There were Evolutionary analyses were conducted in MEGA11.
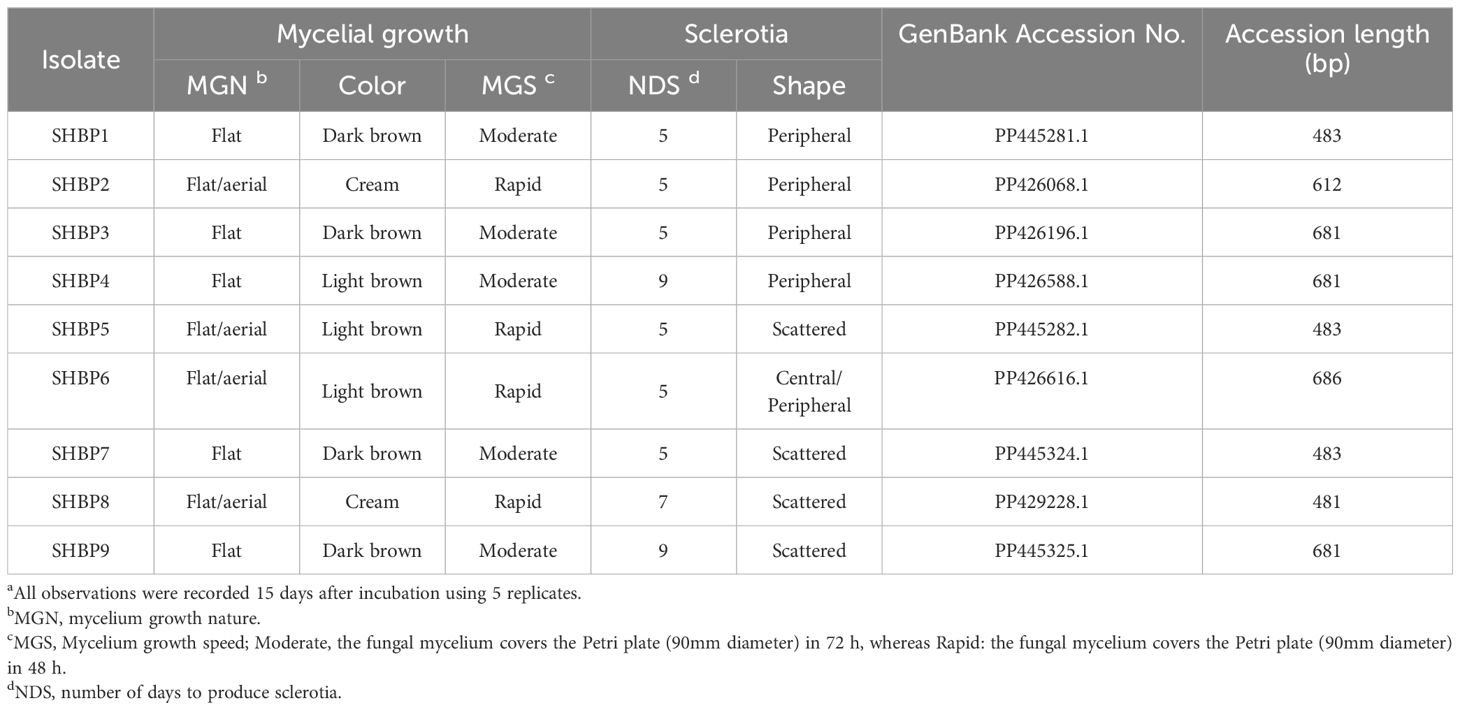
Table 6. Morphological characterization of Rhizoctonia solani isolates, the causal fungus of rice sheath blight disease a.
Furthermore, we noticed four color patterns: cream (two isolates), light, and dark brown (three isolates each), and a unique isolate, SHBP3, with a black-brown color (Figure 1A). Furthermore, the sclerotia appeared in two shapes: peripheral in four isolates (SHBP1, 2, 3, and 4) and scattered in four isolates (SHBP5, 7, 8, and 9); meanwhile, SHBP6 showed a central peripheral shape. In addition, SHBP 1, 2, 5, 6, and 7 recorded the least number of days to produce sclerotia (5 days), and SHBP4 and 9 recorded the greatest number of days to produce sclerotia (9 days) (Figure 1A and Table 6).
In situ microscopic observation revealed that the mycelium is generally dark or hyaline, its cells are lengthy, and its branch septa typically branch off from the main hyphae. Normal right angles were typically seen at the hyphal constrictions at the point of branching (Figure 1B). The mycelium was made up of hyphae that have been divided into separate cells by dolipore septa, which have extensive taxonomic characteristics.
3.2 R. solani AG1 IA - isolate SHBP9 is the most aggressive isolate
Although the nine isolates were pathogenic and produced typical symptoms of sheath blight disease on the susceptible rice cultivar (Sakha 101) under greenhouse conditions, SHBP 9 was the most aggressive one and resulted in the highest relative lesion height % (63.98± 1.45%) on infected rice plants (Figure 1C). On the other hand, SHBP8 showed the lowest performance (26.36± 1.23%) compared with all the isolates.
3.3 Molecular identification of R. solani isolates
The identification of the nine R. solani isolates (SHBP1 to 9) was further confirmed based on the sequencing of the Internal transcribed spacer (ITS) region. Briefly, nine PCR products ranged from 481 to 686 bp. The ITS sequence of each isolate was aligned on NCBI for molecular identification, and then each sequence was submitted to the GenBank sequence database of the National Center for Biotechnology Information (NCBI; https://www.ncbi.nlm.nih.gov/). Finally, the isolates accession numbers were as fellow; PP445281, PP426068, PP426196, PP426588, PP445282, PP426616, PP445324, PP429228 and PP445325, for SHBP1 to 9 respectively. The phylogenetic analysis between the nine R. solani isolates showed high similarity (about 99%) between isolates except R. solani AG1 IA isolate SHBP3 (PP426196.1; 79%) which was clustered separately at the bottom of the dendrogram (Figure 1D). it is worth noting that the most aggressive isolate SHBP9 (PP445325.1) was more similar to R. solani AG1 IA isolate SHBP4 (PP426588.1) than other isolates.
Moreover, to better understand the relationship between the R. solani AG1 IA and other anastomosis groups (AGs), the evolutionary analysis of R. solani AG1 IA (isolates SHBP 1-9) in comparison with other 40 R. solani strains/isolates from different anastomosis groups (AGs) (Figure 1E). Interestingly, eight of the nine R. solani isolates (except SHBP3) showed high similarity with the Japanese isolate of R. solani AG-1 1A isolate F190 (GenBank accession No. FJ492099.3, 644 bp) isolated from sugar beet. Moreover, the same eight isolates were also clustered with the Turkish isolate AG-1 IB isolate Rs-16 (GenBank accession No. MT568768.1, 692 bp) isolated from Brassica oleracea and the Japanese one AG-1 1C isolate F192 (GenBank accession No. FJ492101.3, 636 bp) isolated from sugar beet (Figure 1E). On the other hand, R. solani AG1 IA isolate SHBP3 was clustered separately from other isolates and shared similar evolutionary behavior with two Canadian isolates including R. solani AG-H isolate 534-2 (GenBank accession No. MT177264.1; 458 bp) isolated from Triticum sp. and R. solani AG-I isolate 390.Rs.16-70 (GenBank accession No. MT177265.1; 665 bp) from Glycine max (Figure 1E).
3.4 R. solani isolates differentially produce cell wall degradation enzymes in vitro
The nine isolates showed a wide response for the cell wall degradation enzymes (CWDE) production when cultured on a minimal inductive medium supplemented with plant-derived polysaccharides (e.g., pectin, starch, or carboxymethyl cellulose [CMC]) (Figure 2A). It is well known that fungal linear growth rates on CWDE-inductive media are correlated with enzyme production. Faster growth on inductive media indicates robust CWDE production. The fungal linear growth of R. solani isolates on pectin-containing medium ranged from 3.2 to 7.0 cm (Figure 2B), whereas it ranged from 5.5 to 8.9 cm on starch-containing medium (Figure 2C) and ranged from 3.9-6.13 cm on CMC-containing medium (Figure 2D). R. solani AG1 IA isolate SHBP9 recorded the highest linear growth on pectin-, starch-, and CMC- containing media (7.00 ± 0.20, 8.90 ± 0.20, and 6.13 ± 0.13 cm, respectively).
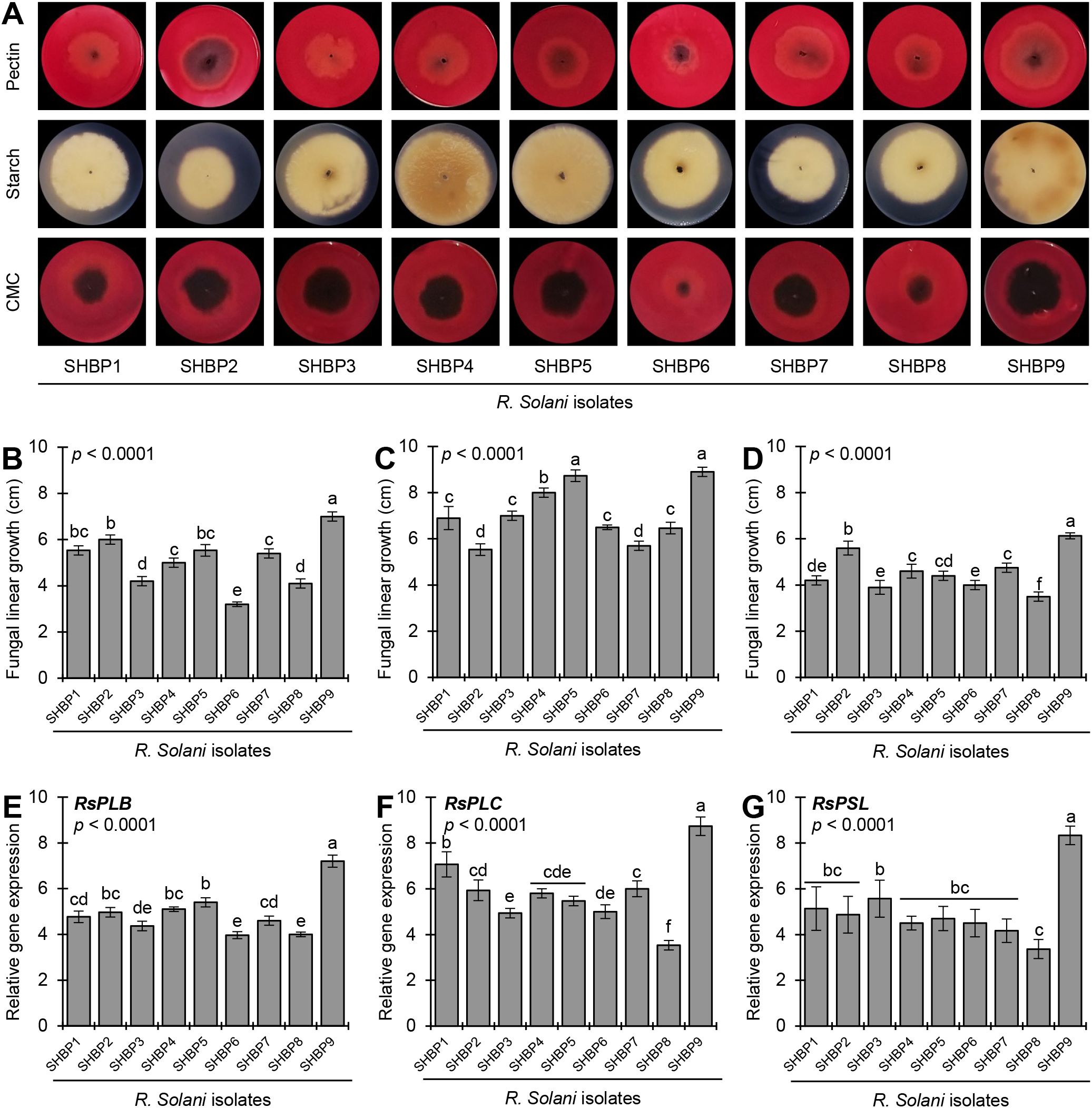
Figure 2. In vitro production of cell wall degradation enzymes (CWDEs) by the phytopathogenic fungus Rhizoctonia solani, the causal agent of sheath blight disease on rice. (A) In-situ visualization of CWDEs production of the nine isolates of R. solani growing on a minimal inductive medium supplemented with plant-derived polysaccharides (e.g., pectin, starch, or carboxymethyl cellulose [CMC]) for 7 days at 26 ± 2°C. Congo red solution (0.3%) was used to visualize the degradation of pectin and CMC as a halo zone, whereas iodine solution was used for the degradation of starch. (B-D) Fungal linear growth (cm) of nine isolates of R. solani growing on a minimal inductive medium supplemented with pectin, starch, or CMC, respectively. (E-G) Relative gene expression of pectate lyase B (RsPLB), putative pectate lyase C (RsPLC), and polysaccharide lyase family 1 protein (RsPSL), respectively, of nine isolates of R. solani growing on inductive pectin-containing medium for 4 days at 26 ± 2°C. Bars represent the average of five biological replicates (n = 5), whereas whiskers represent the standard deviation (means ± SD). Different letters signify statistically significant differences among isolates using the Tukey HSD test (p< 0.05).
Likewise, the gene expression of pectate lyase B (RsPLB, Figure 2E), putative pectate lyase C (RsPLC, Figure 2F), and polysaccharide lyase family 1 protein (RsPSL, Figure 2G) supported the enzymatic activity analysis. The studied genes were upregulated in all R. solani isolates when grown on a pectin-containing medium compared with those grown on a pectin-free medium. For instance, R. solani AG1 IA isolate SHBP9 had the highest transcript levels of RsPLB (up to 7.20-folds), RsPLC (8.73-folds), and RsPSL (8.33-folds). On the other hand, R. solani AG1 IA isolate SHBP8 had the lowest transcript levels of RsPLB, RsPLC, and RsPSL (4.00-, 3.53-, and 3.37-folds, respectively) when grown on pectin-containing medium compared with those grown on pectin-free medium (Figures 2E–G).
3.5 Pathogenicity of R. solani isolates on different Egyptian rice genotypes
All nine isolates were pathogenic and produced typical symptoms of sheath blight disease on 11 different Egyptian rice genotypes under greenhouse conditions. The percentage of relative lesion height ranged from 10.93% (isolate SHBP4 on Egyptian Yasmine) to 68.70% (Isolate SHBP9 on Sakha 104) (Figure 3A). Moreover, two-way Hierarchical Cluster Analysis (HCA) and its associated heatmap were performed using the individual response (RLH) of each genotype to different R. solani isolates (Figure 3A). Interestingly, the HCA-associated dendrogram among rice genotypes showed that all tested genotypes were clustered into three distinct clusters. Cluster “I” included three resistant genotypes (Egyptian Hybrid 1, Giza 182, and Egyptian Yasmine), Cluster “II” included four moderately resistant genotypes (Giza 177, Giza 178, Giza 183, and Giza 181), whereas Cluster “II” included four susceptible genotypes (Sakha 101, Sakha 104, Sakha 108, and Sakha Super-300) (Figure 3A). It is worth noting that while Egyptian Yasmine showed the highest resistance to all isolates, Sakha 104 had almost the lowest resistance to all isolates. On the other hand, the HCA-associated dendrogram among R. solani isolates clustered both SHBP8 and SHBP9 separately from other isolates. R. solani AG1 IA isolate SHBP9 was the most aggressive isolate and caused the highest relative lesion height percentage on Sakha 104 (68.70 ± 1.22%), Sakha 101 (64.32 ± 1.14%), Sakha Super 300 (62.53 ± 1.04%), and Sakha-108 (59.40 ± 0.95%) (Figure 3A). On the other hand, Egyptian Yasmine was the lowest susceptible rice genotype and recorded the lowest relative lesion height percentage when infected with isolates SHPB4 (10.93 ± 0.51%), SHPB8 (11.02 ± 0.51%), SHPB1 (11.14 ± 0.42%), and SHPB5 (11.30 ± 0.72%) (Figure 3A).
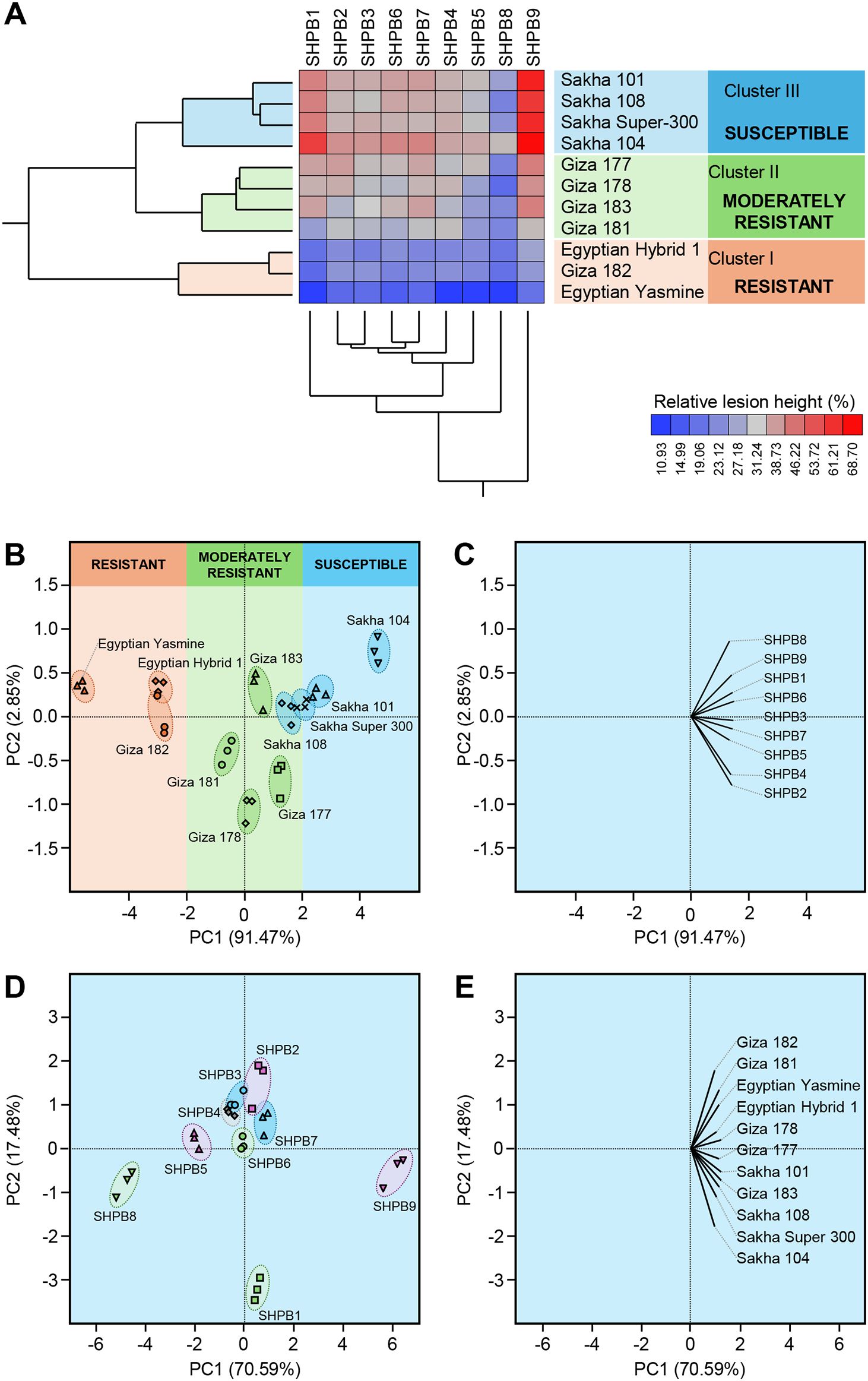
Figure 3. Two-way hierarchical cluster analysis (HCA) and Principal component analysis (PCA) of relative lesion height (%) of 11 commercial rice cultivars after the infection with individual Rhizoctonia solani AG1 IA Isolates (SHBP 1-9). (A) HCA-associated eat map and cluster dendrograms of relative lesion height (%) of 11 commercial rice cultivars after the infection with individual R. solani AG1 IA Isolates (SHBP 1-9). The differences in relative lesion height (%) are visualized in the heat map diagram. Isolates are presented in columns and rice cultivars are presented in rows. Isolates and cultivars are organized using two-way hierarchical cluster analysis based on similarities in auto-scaled values and correlations, respectively. Low relative lesion height (%) is colored blue, whereas higher relative lesion height (%) is colored red (see the scale at the right bottom corner of the graph). (B, D) PCA-associated scatter plots of rice cultivars and R. solani isolates, respectively, (C, E) PCA-associated loading plots of rice cultivars and R. solani isolates, respectively.
Likewise, principal component analysis (PCA) revealed the differences between rice genotypes as well as R. solani isolates (Figures 3B–E). Briefly, The PCA-associated scatter plot showed a clear separation among all studied rice genotypes concerning the total variation of PC1 (91.47%) and PC2 (2.85%) (Figures 3A, B). in agreement with HCA, Egyptian Hybrid 1, Giza 182, and Egyptian Yasmine were clustered together at the left side of the scatter plot and separately from other genotypes. Moderately resistant genotypes (Giza 177, Giza 178, Giza 183, and Giza 181) were grouped in the center of the scatter plot, whereas susceptible rice genotypes (Sakha 101, Sakha 104, Sakha 108, and Sakha Super-300) were grouped into the right side of the scatter plot (Figure 3B). Moreover, the PCA-associated loading plot showed that all R. solani isolates were positively correlated with the susceptible rice genotypes (Figure 3C).
Similarly, the PCA-associated scatter plot of R. solani isolates showed a clear separation among all isolates concerning the total variation of PC1 (70.59%) and PC2 (17.48%) (Figures 3D, E). The most aggressive isolate, R. solani isolate AG1 IA SHBP9, and the lowest aggressive isolate, R. solani isolate AG1 IA SHBP8 were grouped far away from each other (Figure 3D). Furthermore, the PCA-associated loading plot showed that all rice genotypes were positively correlated with the virulence of R. solani AG1 IA isolate SHBP9 (Figure 3E).
3.6 Assessment of R. solani AG1 IA (isolate SHBP9) host range at greenhouse condition
The virulence of R. solani isolate AG1 IA SHBP9 was tested against 12 common rice-associated weeds (Table 1), and 31 economic crops (Table 2) belonging to 12 families (3 Monocotyledons and 9 Dicotyledons) to assess the host range of R. solani AG1 IA. All the Monocotyledon plants, 13 of Poaceae, 3 of Cyperaceae, and 1 of Typeaceae, were susceptible to R. solani AG1 IA isolate SHBP9 (Figures 4, 5). Meanwhile, 6 plant species belonging to Dicotyledons were resistant to the SHBP9 including chickpea (Cicer arietinum) from Fabaceae, Rocket (Eruca sativa) from Brassicaceae, and the four studied crops from Solanaceae (potato, tomato, eggplant, and pepper) (Table 2). In addition, the remaining 20 Dicotyledons plant species: 3 of Cucurbitaceae, 2 of Malvaceae, 1 of Linaceae, 8 of Fabaceae, 2 of Brassicaceae, 1 of Amaranthaceae, 2 of Apiaceae, and 1 of Asteraceae, were susceptible to R. solani isolate AG1 IA isolate SHBP9 (Figure 5; Table 2).

Figure 4. Symptoms’ development of sheath blight disease induced by Rhizoctonia solani AG1 IA - isolate SHBP9 on 12 common rice-associated weeds from Poaceae under greenhouse conditions.

Figure 5. Symptoms’ development of sheath blight disease and similar symptoms induced by Rhizoctonia solani AG1 IA - isolate SHBP9 on economic crops from different families under greenhouse conditions.
3.7 Evaluation of rice genotypes under artificial inoculation with R. solani AG1 IA (isolate SHBP9) under greenhouse conditions
Eleven rice genotypes (five Japonica, three Indica-Japonica, and three Indica) were used to evaluate their resistance against the sheath blight disease caused by R. solani AG1 IA (isolate SHBP 9). Greenhouse findings showed that the typical symptoms of sheath blight disease were observed on the eleven tested rice genotypes at least 30 dpi, (Figure 6). Rice genotypes differed in their susceptibility to sheath blight (Figures 6, 7). Sakha 104 recorded the highest RLH (69.63%; Figure 7A), disease score (9; Figure 7B), disease index (67.31%; Figure 7C), and sheath lesion area (9.75 cm2; Figure 7D), followed by Sakha 101, Sakha super 300, Sakha 108 and Giza 177 in terms of RLH (64.22, 62.52, 59.90 and 45.16%), disease score (7), disease index (62.08, 60.44, 57.93 and 43.65%), and lesion sheath area (7.18, 5.13, 2.96 and 1.53 cm2), respectively. Meanwhile, the lowest disease traits were observed by indica type and indica japonica; particularly Egyptian Yasmine with RLH (19.10%), disease score (1), disease index (18.4%), and Lesion sheath area (1.24 cm), followed by Egyptian hybrid 1 and Giza 182 RLH (26.99 and 28.23%), disease score (3), disease index (26.09 and 26.95%) and Lesion sheath area (1.60 and 1.77 cm, respectively). In addition, indica type, i.e., Giza 181, Giza 182, Giza 178, and Giza 183, recorded moderate disease traits ranging from 32- 44.75% for RLH and 5 for disease score (Figures 7A, B).

Figure 6. Symptoms’ development of sheath blight disease induced by Rhizoctonia solani AG1 IA - isolate SHBP9 on eleven rice genotypes under greenhouse conditions.
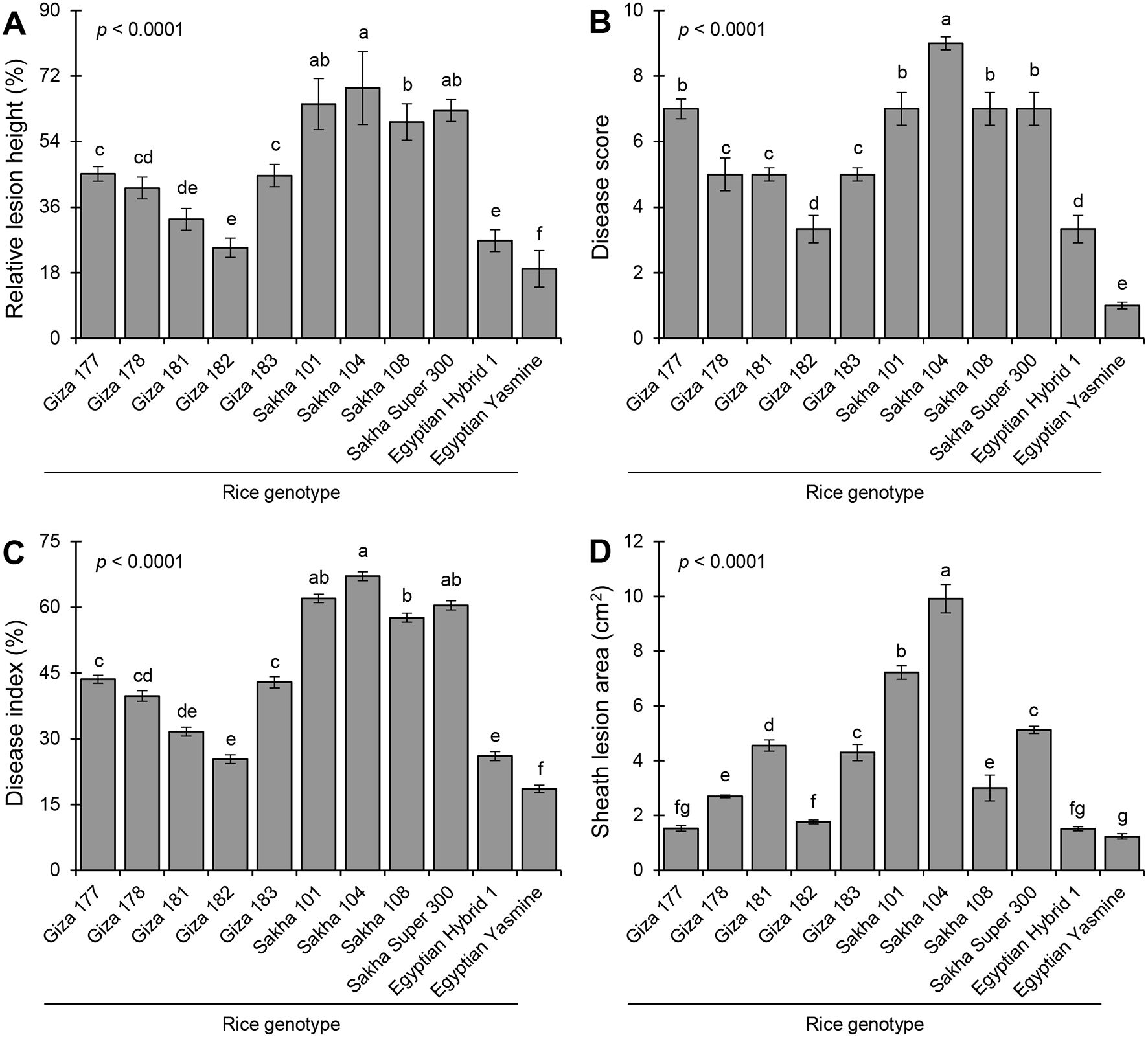
Figure 7. Sheath blight disease assessment of 11 rice genotypes at 30 days post inoculation (dpi) with Rhizoctonia solani under greenhouse conditions. (A) Relative lesion height (%), (B) Disease score, (C) percent disease index, and (D) sheath lesion area (cm2). Bars represent the average of three biological replicates (n = 3), whereas whiskers represent the standard deviation (means ± SD). Different letters signify statistically significant differences among rice cultivars using the Tukey HSD test (p< 0.05).
3.8 Morphological traits of rice genotypes inoculated with R. solani AG1 IA (isolate SHBP9) under greenhouse conditions
The morphological traits of healthy and R. solani-infected rice genotypes differed significantly (Figure 8). Infection with R. solani significantly altered plant height (p Infection< 0.0001; Figure 8A), stem diameter (p Infection = 0.0003; Figure 8C), and flag leaf area (p Infection = 0.0234; Figure 8D), but not the number of tillers per plant (p Infection = 0.1001; Figure 8B). Mock-inoculated (healthy) Sakha Super 300 had the plant height (125.3 ± 3.00 cm), whereas R. solani-infected Sakha 108 showed the lowest plant height (85.60 ± 6.00 cm) (Figure 8A). Likewise, healthy Giza 182 recorded the highest number of tillers per plant (15.60 ± 0.40) which was similar to the R. solani-infected Sakha 108 (15.60 ± 0.40) with no significant differences between them (Figure 8B). However, Giza 177, either infected or not, had the lowest number of tillers per plant (6.90 ± 0.30). Meanwhile, healthy Sakha Super 300, healthy Egyptian Yasmine, and R. solani-infected Egyptian Yasmine had the highest stem diameter (5.67 ± 0.20 5.60 ± 0.40, and 5.58 ± 0.20 mm, respectively) without significant differences between them. Nevertheless, infected Giza 177 and Sakha 101 recorded the thinnest stem diameter (3.93 ± 0.20 and 3.92 ± 0.40 mm, respectively) without significant differences between them (Figure 8C). Finally, the flag leaf area did not differ significantly among most of the rice genotypes, either infected or not. However, healthy Egyptian Yasmine had the highest flag leaf area (58.30 ± 4.0 cm2), whereas R. solani-infected Sakha 104 had the lowest flag leaf area (30.04 ± 3.89 cm2) (Figure 8D).
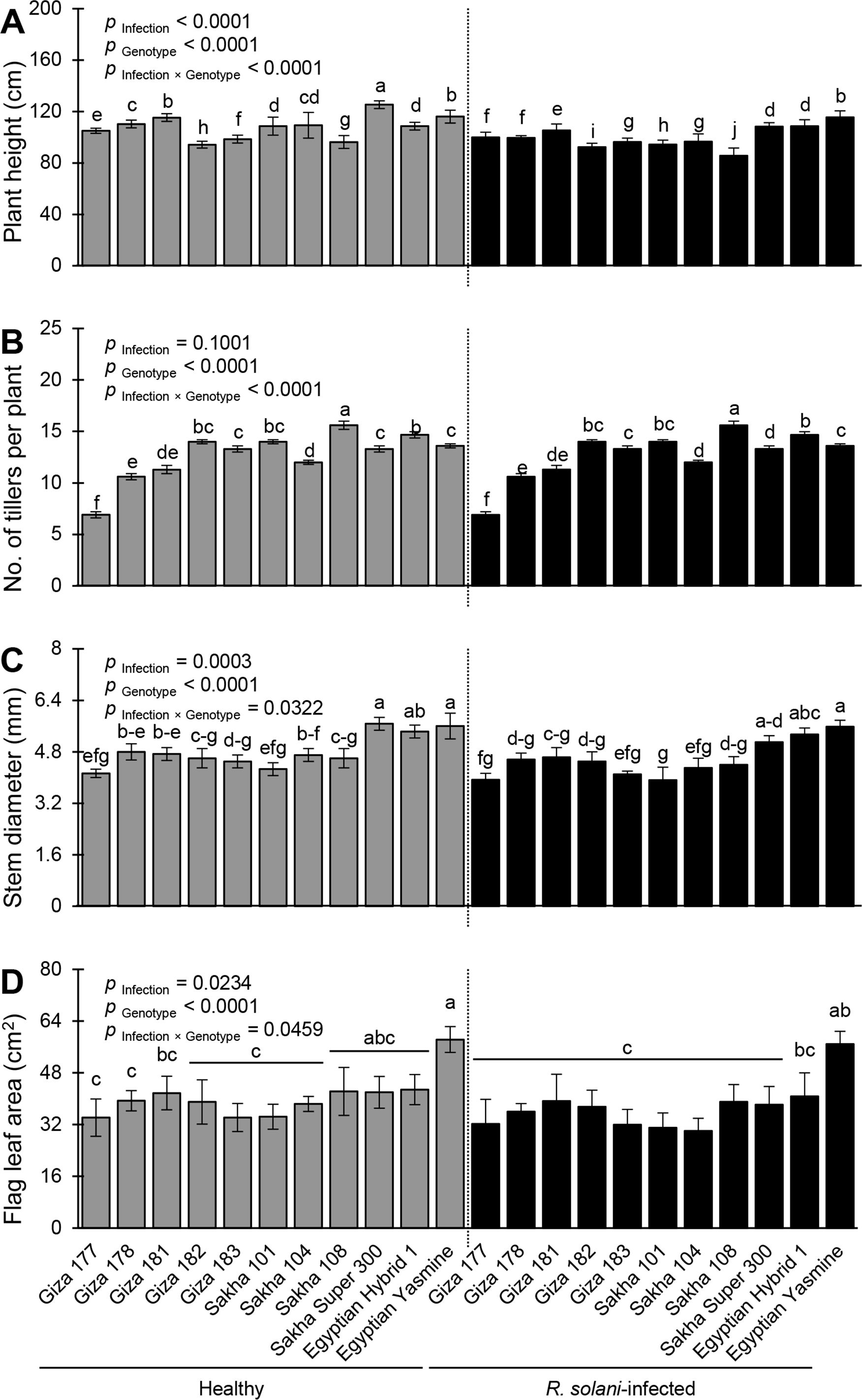
Figure 8. Performance and morphological traits of 11 rice genotypes with or without the infection with Rhizoctonia solani AG1 IA – isolate SHBP9 under greenhouse conditions. (A) plant height (cm), (B) number of tillers per plant, (C) stem diameter (mm), and (D) Flag leaf area (cm2). Bars represent the average of three biological replicates (n = 3), whereas whiskers represent the standard deviation (means ± SD). Different letters signify statistically significant differences among cultivars using the Tukey HSD test (p Infection × Genotype< 0.05).
3.9 Yield components of rice genotypes inoculated with R. solani AG1 IA (isolate SHBP9) under greenhouse conditions
Infection with R. solani significantly altered all studied yield components including panicle length (p Infection< 0.0001; Figure 9A), panicle weight (p Infection = 0.0019; Figure 9B), 1000-grain weight (p Infection< 0.0001; Figure 9C), and number of discolored grains (p Infection< 0.0001; Figure 9D). Mock-inoculated Giza 181 recorded the highest panicle length (27.67 ± 0.30 cm), whereas infected Sakha Super 300 had the lowest panicle length (17.20 ± 0.20cm) (p Infection × Genotype< 0.0001; Figure 9A). Likewise, the highest performance of panicle weight was recorded by the healthy Sakha Super 300 (4.70 ± 0.20 g) while the lowest performance of panicle weight was recorded by healthy Giza 178 (1.56 ± 0.20 g) and R. solani-infected Giza 178 (1.50 ± 0.20 cm) with no significant differences between them (p Infection × Genotype = 0.0473; Figure 9B). In terms of 1000-grain weight, non-infected Sakha 101 had the highest 1000-grain weight (28.2 ± 0.20 g) whereas infected Giza 178 had the lowest value (15.00 ± 0.20 g) (p Infection × Genotype< 0.0001; Figure 9C). Finally, R. solani-infected Giza 181 recorded the highest number of discolored grains (28.00 ± 0.20) whereas Giza 177 had the lowest number (6.00 ± 0.20) (p Infection × Genotype< 0.0001; Figure 9D).
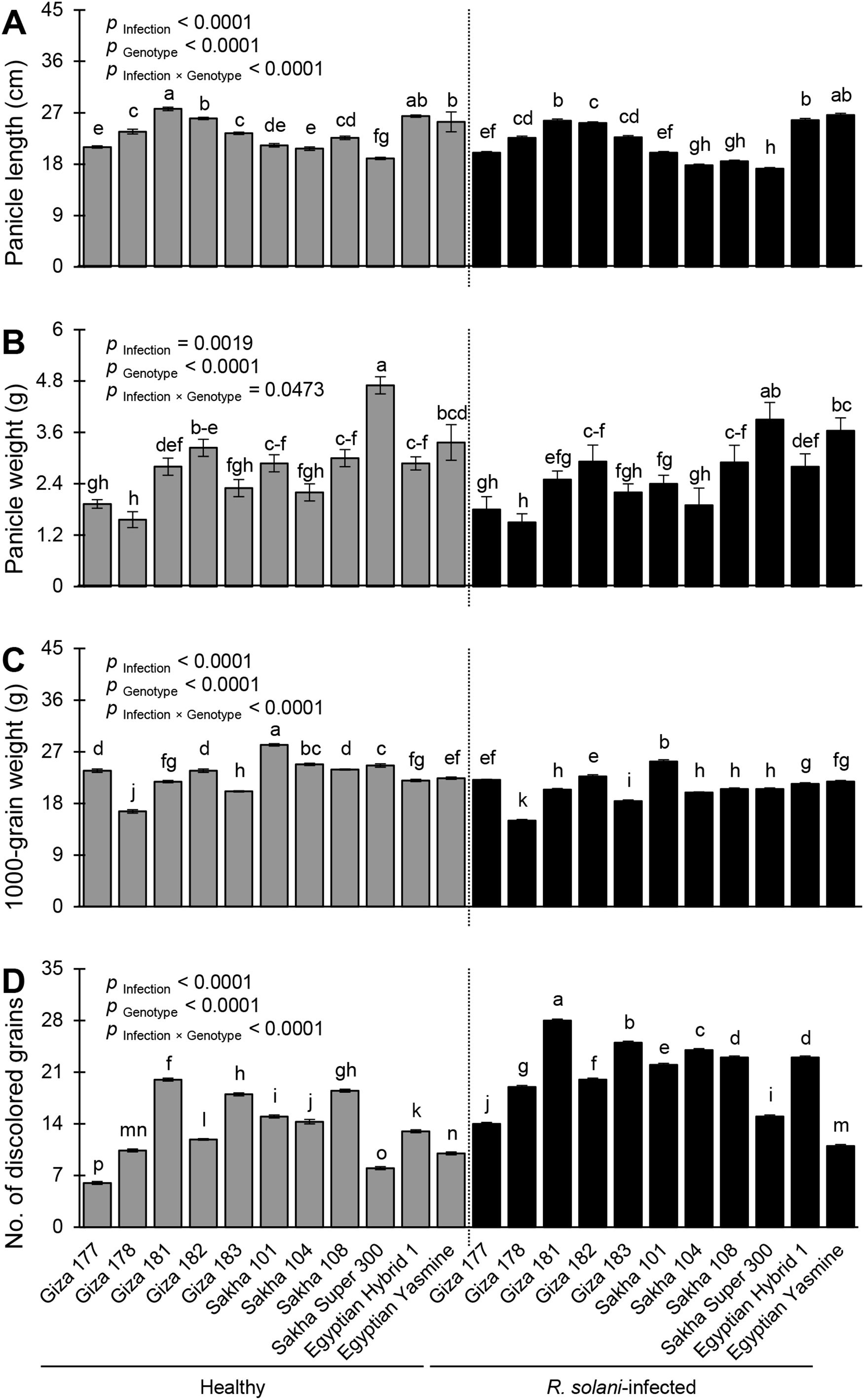
Figure 9. Yield components of 11 rice genotypes with or without the infection with Rhizoctonia solani AG1 IA – isolate SHBP9 under greenhouse conditions. (A) Panicle length (cm), (B) Panicle weight (g), (C) 1000-grain weight (g), and (D) number of discolored grains. Bars represent the average of three biological replicates (n = 3), whereas whiskers represent the standard deviation (means ± SD). Different letters signify statistically significant differences among cultivars using the Tukey HSD test (p Infection × Genotype< 0.05).
3.10 Behavior and development of the fungal mycelium on the sheath surface of rice genotypes inoculated with R. solani AG1 IA (isolate SHBP9) under greenhouse conditions
The behavior of mycelium growth on different genotypes varies due to the genetic background of each genotype (Figures 10A–L). Only growing mycelium was noticed on Egyptian hybrid 1, Giza 182, and Egyptian Yasmine (Figures 10D, E, G). The growing mycelium started to branch outside the sheath, as in Giza 177 (Figure 10A). The branching mycelium penetrated the stem and branched inside it as in Giza 178 and Giza 183, (Figures 10B, F), then the mycelium shrank as in Giza 181 and Sakha 108 and Super 300 (Figures 10C, K, L respectively), and converted to mycelium cushion as in Sakha 101 (Figure 10H), finally formed sclerotia as in Sakha 104 (Figure 10I). So, the fungus can invade the sheath of Sakha 104 faster than the other genotypes. It was observed that there was more branching and growth density of the fungus on the Japanese types, especially the Sakha 104, where there was a significant development in the growth of the mycelium and its transformation into a cushion, as well as the formation of sclerotia on the sheath’s surface (Figure 10J).
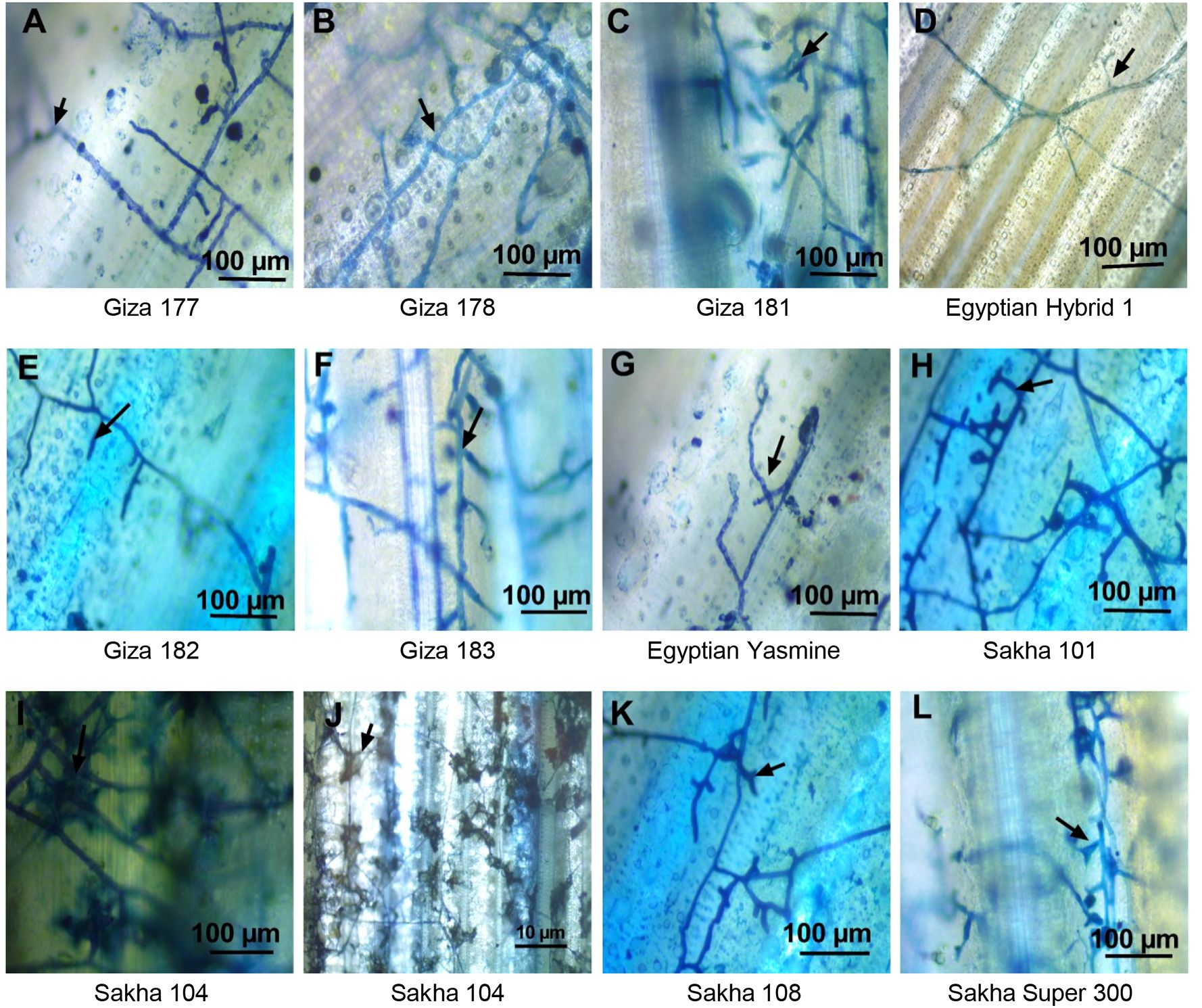
Figure 10. Light microscopy analysis of the behavior and development of the mycelium of Rhizoctonia solani AG1 IA – isolate SHBP9 on the surface of infected sheaths of 11 rice genotypes with different degrees of susceptibility. (A) Typical mycelium of R. solani on the sheath surface of Giza 177 rice cultivar with flat mycelium growth and distribution with more branches, (B) Giza 178 cv increases the mycelium growth in the sheath surface and clamped with trichome, (C) Giza 181 more mycelium branching and formed to mycelium cushion, (D) Egyptian hybrid 1 only mycelium grows on the surface without branching, (E) Giza 182 only mycelium grows on the surface without branching, (F) Giza 183 more mycelium branching, (G) Egyptian Yasmine with aerial mycelium growth no branch, (H) Sakha 101 branching mycelium penetrated the stem and branched inside, then the mycelium shrank, (I, J) Sakha 104 converted to mycelium cushion and formed sclerotia, (K) Sakha 108 branching mycelium penetrated the stem and branched inside, then the mycelium shrank, (L) Sakha super 300 branching mycelium penetrated the stem and branched inside, then the mycelium shrank. Arrows indicate the mycelium branching.
3.11 SSR marker associated with morphological traits and sheath blight resistance
Out of the 12 tested SSR markers (Figure 11A), 11 SSR markers were polymorphic, and among them, only three markers differentiated between the resistance and susceptible genotypes. Furthermore, these markers, RM202, RM426, and RM6971, showed a specific band (~300, ~144, and ~152bp), respectively, for the resistance genotypes (Egyptian Yasmine and Giza 182) (Figure 11A). The SSR—based phylogenetic tree was divided into three main clades (Figure 11B). Clade I contained only two resistant genotypes (Giza 182 and Egyptian Yasmine), Clade II contained four moderately resistant genotypes (Giza 181, Giza 183, Giza 178, Egyptian Hybrid 1), and Clade III contained five susceptible genotypes (Sakha Super 300, Giza 177, Sakha 101, Sakha 104, Sakha 108). Finally, these three SSR markers, RM202, RM426, and RM6971, could be used in a breeding program for sheath blight resistance. Collectively, these findings suggest the high specificity of RM202, RM426, and RM6971 markers, as well as their potential utilization in breeding programs since they uniquely differentiate between resistant and susceptible rice genotypes.
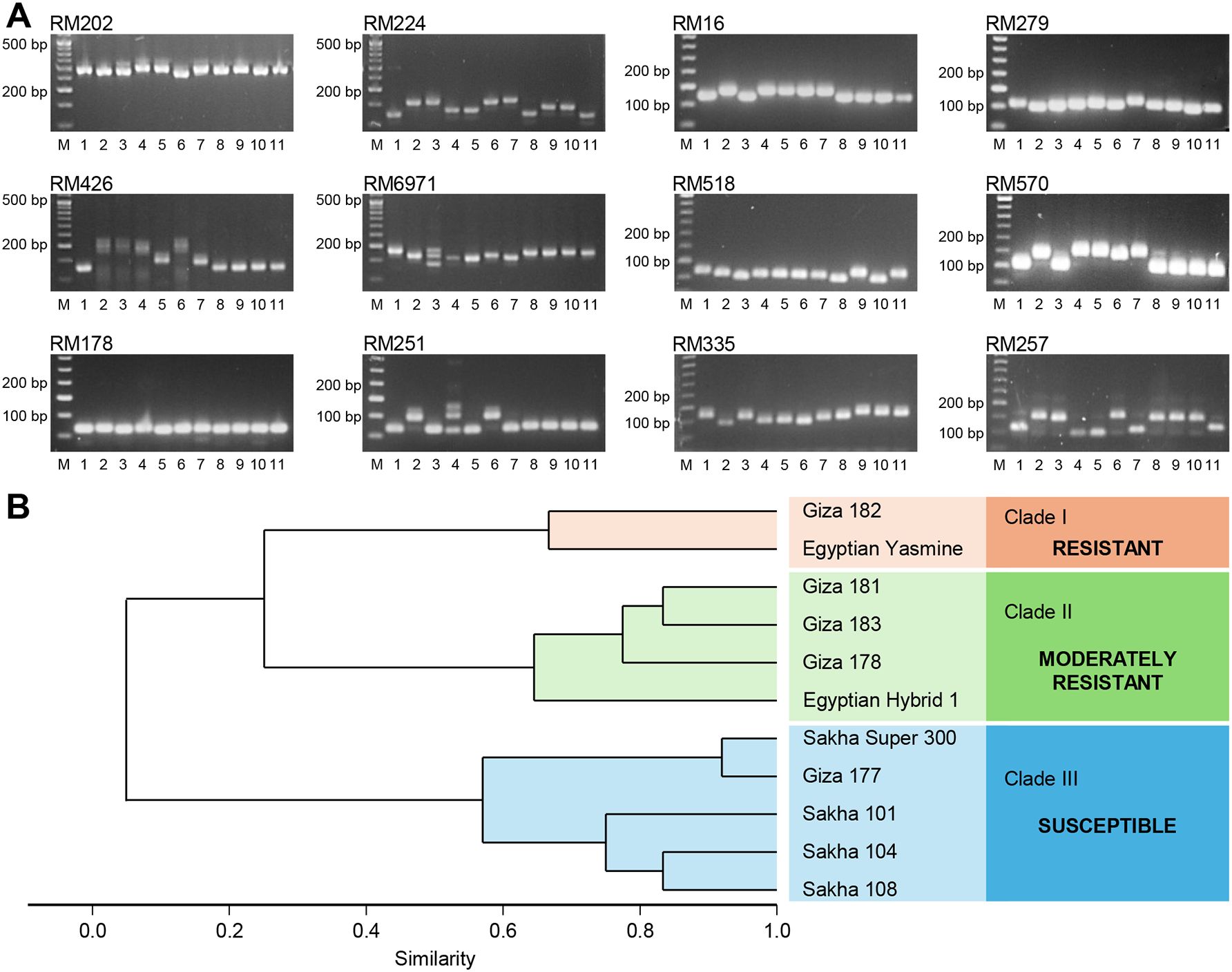
Figure 11. Diversity Analysis of 11 rice genotypes with different degrees of susceptibility based on 12 SSR Markers. (A) Representative gel images and banding patterns of 12 SSR markers (RM202, RM224, RM16, RM279, RM426, RM6971, RM518, RM570, RM178, RM251, RM335, and RM257) linked to sheath blight resistance in rice. Lane M represents the standard DNA ladder, whereas lanes 1–11 represent rice genotypes (Giza177, Giza178, Egyptian Hybrid 1, Giza181, Giza182, Giza183, Egyptian Yasmine, Sakha101, Sakha104, Sakha108, and Sakha Super 300, respectively). (B) Dendrogram derived from UPGMA cluster analysis of 11 rice genotypes.
4 Discussion
Rhizoctonia solani is a common fungus found in soil and infects a large number of families (Chahal et al., 2003). Given the importance of this fungus, its pathogenic ability to infect rice plants was recently discovered in Egypt, causing sheath blight disease, especially the strain belonging to the AG1 IA group (Hassan, 2021), and rice worldwide (Wamishe et al., 2007). Isolates differ in their morphological and pathogenic characteristics depending on geographical locations. It was found that the nine isolates of R. solani AG1 IA were different in morphological characteristics, their ability to produce some cell wall degrading enzymes, and their pathogenic behavior on rice plants. Many researchers (Moni et al., 2016; Mughal et al., 2017) agree that R. solani isolates differ in their morphological and pathological characteristics. This difference may be due to the occurrence of anastomosis compatibility (Mughal et al., 2017) or parasexual cycle (Tinline and MacNeill, 1969; Qu et al., 2008) between Rhizoctonia isolates, which leads to the production of new, different strains.
Many pathogenic fungi produce enzymes that degrade the cell wall (Shieh et al., 1997; Have et al., 1998). It was found that R. solani can produce these enzymes. The high or low ability to produce these enzymes is due to the virulence of the isolates (Mondal et al., 2013). The fungi benefit from the substances resulting from the process of cell wall decomposition to provide a source of food for the fungus and also facilitate the process of penetration and spread within the host cells (De Lorenzo et al., 1997). Our results showed that R. solani AG1 IA isolate SHBP9 excelled in producing the enzymes associated with cell wall degradation and its pathogenic ability to cause infection in rice plants through artificial inoculation under greenhouse conditions. The appearance of symptoms and their severity were positively associated with an increase in the amount of cell wall-degrading enzymes in the susceptible rice cultivar under conditions of infection with R. solani (Hassan, 2021).
The necrotrophic fungus R. solani is soilborne and is one of the fungi that can grow on the appropriate host to cause sheath blight of rice and cause losses (Senapati et al., 2022). It can grow on some other hosts as a parasite and also cause different symptoms such as root rot, crown, damping off, and black peeling, but it does not cause losses (Nagaraj et al., 2017; Shekhawat et al., 2023). Species of R. solani are classified into multinucleate and binucleate isolates based on the number of nuclei inside each cell and each of these groups is divided into anastomosis groups (AGs) based on the ability of hyphae to fuse (Lee et al., 2021). To date, 21 AGs (named AG-A to AG-U) were reported within the binucleate group, whereas only 13 AGs (named AG-1 to AG-13) were reported within the multinucleate group (Muzhinji et al., 2015; Muzhinji and Lekota, 2024). In the current study, all nine tested R. solani isolates were classified within the multinucleate group AG1-IA. Moreover, the most aggressive isolate R. solani AG1 IA isolate SHBP9 was virulent on 12 common rice-associated weeds, and 25 economic crops belonging to 12 families, however, it did not show any symptoms on chickpea from Fabaceae, rocket from Brassicaceae, and the four crops from Solanaceae including potato, tomato, eggplant, and pepper. It was reported previously that seven AGs were associated with wet root rot in chickpea including AG1, AG2-2, AG2-2LP, AG2-3, AG3, AG4, and AG5 (Ganeshamoorthi and Dubey, 2013). Likewise, rocket (Eruca sativa) seedlings were susceptible to R. solani – AG-4 and exhibited strong damping-off symptoms (Pane et al., 2020). Nevertheless, in the current study, R. solani AG1 IA isolate SHBP9 did not show any significant symptoms either on chickpea or rocket plants under greenhouse conditions.
Likewise, several previous studies experimentally reported the susceptibility of potato and other solanaceous plants to R. solani – AG-3 (Woodhall et al., 2007; Tsror, 2010; Yarmeeva et al., 2021; Naqvi et al., 2024), particularly AG-3 PT (Zrenner et al., 2020; Hejazi et al., 2022; Iradukunda et al., 2022; Monazzah et al., 2022). The most updated AG distribution in potato worldwide confirms the association of some other AGs rather than only AG-3 PT such as AG2-1 (Woodhall et al., 2007), AG-5, and AG-K (Yarmeeva et al., 2021), AG-2, AG-4 (HGI and HGII), AG-6, AG-7, and AG-9 (López-Corrales et al., 2024) which markedly reflects the population dynamics of the pathogen. Recently, several R. solani-AG-1 (AG-1 IA, AG-1 IB, and AG-1 IC) were reported to be associated with potato diseases in Northern Sinaloa, Mexico (López-Corrales et al., 2024). However, the aggressive Egyptian R. solani AG1 IA isolate SHBP9 (isolated from rice) did not show any symptoms on potato plants under greenhouse conditions. These findings suggest potential differences in host specificity and pathogenicity among R. solani isolates, even within the same anastomosis group (AG-1). Collectively, these findings highlight the complexity of host-pathogen interactions within R. solani.
Sheath blight is a disease that is widespread in most rice crops in the world. Due to the limited existing sources of resistance to this disease, we always resort to using alternative treatments to stop the spread of the disease and limit its losses (Molla et al., 2020). However, varieties that have partial resistance to this disease are considered among the best environmentally friendly and safe methods (Hassan, 2021). Therefore, searching for sources of resistance in Egyptian cultivated varieties is one of the important means of resisting the disease. The economic losses for the sheath blight disease vary among genotypes; thus, the identification of resistance sources is very important. Rice cultivars (Indica type) showed resistance to sheath blight under artificial infection conditions with the R. solani, with less loss in 1000-grain weight compared to Japonica-type varieties, which were more susceptible to the fungus and also the greatest loss in weight per thousand grains, under the same conditions.
Map populations produced from indica or japonica rice have been used to identify several quantitative trait loci (QTL) for sheath blight resistance. Different rice cultivars exhibiting partial resistance have mapped numerous QTLs, including Tetep, Teqing, jasmine, and additional germplasm lines (Bal et al., 2020). It has been found that cultivars from the indica subspecies are more resistant to sheath blight than cultivars from the japonica subspecies (Jia et al., 2012). The study measured the morphological traits of the infection and non-infected varieties, and the loss was determined. It was found that the Indica type (Egyptian Yasmine and Giza 182) is the least affected by the infection. The lesser susceptibility to infection may be due to the difference in morphological traits in the Indica-type cultivars from the Japanese ones. It was found that the Egyptian Yasmine variety has a higher height than other varieties, as well as the area of the flag leaf, so the fungus may take longer to reach the panicle and cause loss compared to the Japanese types. Hossain et al. (2016) found that morphological traits of rice genotypes were associated with resistance to sheath blight disease. The morphological characteristics of the cultivars may be positively related to the environmental conditions (Groth and Nowick, 1992) that lead to reducing the severity of the infection. Plant growth factors such as plant height, and flag leaf area indirectly affect rice sheath blight resistance (Zou et al., 2000; Pinson et al., 2005). It was found that the area of the flag leaf in the Egyptian Yasmine was large compared to other varieties. Perhaps the larger leaf width increases the area exposed to solar radiation and thus reduces the disease.
Rice sheath blight is a disease controlled by polygenes and is a quantitative trait linked to morphological traits (Hossain et al., 2016). Twelve SSR markers divided rice genotypes into three groups: susceptible genotypes (japonica type), moderately resistant genotypes (indica/japonica and indica type), and resistance genotypes (indica type). Many researchers (Willocquet et al., 2012; Wen et al., 2015) have agreed that Indica-type genotypes are more resistant than japonica types. Although most tested SSR markers in this study showed polymorphism among tested genotypes without a clear indication of resistance among rice genotypes, three markers RM202, RM426, and RM6971 demonstrated a specific band (~300, ~144, and ~152bp, respectively) only in resistant genotypes like Egyptian Yasmine and Giza 182. The high specificity of these three markers suggests their potential as a reliable genetic source for identifying resistance traits at the molecular level, vital for selecting disease-resistant rice genotypes. It is well known that traditional breeding for disease resistance in cereal crops, particularly rice, is time-consuming and significantly affected by environmental variations. We believe that RM202, RM426, and RM6971 could be used as accurate molecular indicators for sheath blight resistance during the marker-assisted selection (MAS) breeding programs to save time and resources. Briefly, screening rice seedlings for these markers at early stages might help breeders identify resistant materials without waiting for field-based disease assessments which will accelerate the development of resistant rice varieties and augment the reliability of breeding outcomes.
5 Conclusion
Sheath blight, caused by R. solani, is a serious emerging disease of rice in Egypt and worldwide. Although all characterized R. solani isolates fall within the anastomosis group anastomosis group (AG) AG-1 IA, virulence variability was noticed due to the variation in their ability to produce CWDEs, as well as geographical distribution. R. solani isolate AG1 IA SHBP9 (PP445325.1) was the most aggressive isolate and was able to infect 12 common rice-associated weeds from Poaceae and several hosts from different families, except some hosts of the family Solanaceae and Fabaceae. Indica rice genotypes (such as Giza 182 and Egyptian Yasmine) and Indica/Japonica rice genotypes (such as Giza 178 and Giza 183) were more resistant to R. solani than Japonica rice genotypes (Sakha 101, Sakha 104, and Sakha 108). Accordingly, we encourage the adoption and cultivation of resistant genotypes such as Egyptian Hybrid 1, Giza 182, and Egyptian Yasmine, rather than the susceptible genotypes such as Sakha 101, 104, 108, and Super-300 under Egyptian conditions. However, further studies are required to confirm the resistance behavior of commercial rice cultivars over the years, particularly the moderately resistant genotypes such as Giza 177, 178, 181, and 183 which are threatened to be more susceptible to R. solani under the current climate change and the invasion of new isolates. While we acknowledge the limited number of isolates and genotypes in the current study, we believe that future studies with expanded sample sizes could provide additional insights into this emerging disease.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
ZK: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Resources, Supervision, Validation, Writing – original draft, Writing – review & editing. YM: Data curation, Funding acquisition, Methodology, Project administration, Resources, Writing – original draft, Writing – review & editing. AH: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Resources, Validation, Writing – original draft, Writing – review & editing. AS: Formal analysis, Investigation, Methodology, Validation, Writing – original draft, Writing – review & editing. WG: Formal analysis, Investigation, Methodology, Validation, Writing – original draft, Writing – review & editing. QA: Data curation, Formal analysis, Funding acquisition, Resources, Software, Writing – original draft, Writing – review & editing. YN: Conceptualization, Data curation, Formal analysis, Methodology, Project administration, Resources, Software, Supervision, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The work was funded by the Deanship of Research and Graduate Studies at King Khalid University for funding this work through a Large Research Project under grant number RGP2/249/45.
Acknowledgments
The authors extend their appreciation to the Deanship of Research and Graduate Studies at King Khalid University for funding this work through a Large Research Project under grant number RGP2/249/45. We also gratefully acknowledge the Rice Pathology Department, Plant Pathology Research Institute (PPRI), Agricultural Research Center (ARC), Egypt. Additionally, YN would like to extend his appreciation to the Graduate Student and Research Affairs Sector of Tanta University, Egypt. QA thanks the United Arab Emirates University for providing a postdoctoral grant on climate action under grant number 12S140. We just used the free version of "Grammarly" for English check and "Mendeley" for reference management.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ahvenniemi, P., Wolf, M., Lehtonen, M. J., Wilson, P., German-Kinnari, M., Valkonen, J. P. (2009). Evolutionary diversification indicated by compensatory base changes in ITS2 secondary structures in a complex fungal species, Rhizoctonia solani. J. Mol. Evol. 69, 150–163. doi: 10.1007/s00239-009-9260-3
Ajayi-Oyetunde, O. O., Bradley, C. A. (2018). Rhizoctonia solani: taxonomy, population biology and management of Rhizoctonia seedling disease of soybean. Plant Pathol. 67, 3–17. doi: 10.1111/ppa.12733
Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Anand, P., Alwin, A., Vennison, S. J., Sankar, S. G., Gilwax Prabhu, D. I., Vasan, P. T., et al. (2010). Isolation and characterization of bacteria from the gut of Bombyx mori that degrade cellulose, xylan, pectin and starch and their impact on digestion. J. Insect Sci. 10, 107. doi: 10.1673/031.010.10701
Atallah, O. O., Hassanin, A., Yassin, S. M., Aloufi, A. S., Almanzalawi, E. A., Abdelkhalek, A., et al. (2024). Pathological characterization and management of Lasiodiplodia theobromae, a hemi-biotroph with an interkingdom host range. Plant Dis. 108, 3243–3257. doi: 10.1094/PDIS-03-24-0713-RE
Atallah, O. O., Mazrou, Y. S., Atia, M. M., Nehela, Y., Abdelrhim, A. S., Nader, M. M. (2022). Polyphasic characterization of four Aspergillus species as potential biocontrol agents for white mold disease of bean. J. Fungi 8, 626. doi: 10.3390/jof8060626
Bal, A., Samal, P., Chakraborti, M., Mukherjee, A. K., Ray, S., Molla, K. A., et al. (2020). Stable quantitative trait locus (QTL) for sheath blight resistance from rice cultivar CR 1014. Euphytica 216, 1–19. doi: 10.1007/s10681-020-02702-x
Barnett, H., Hunter, B. (1972). Illustrated genera of imperfect fungi. 3rd edn (Minneapolis: Burgess).
Basu, A., Chowdhury, S., Ray Chaudhuri, T., Kundu, S. (2016). Differential behavior of sheath blight pathogen Rhizoctonia solani in tolerant and susceptible rice varieties before and during infection. Plant Pathol. 65, 1333–1346. doi: 10.1111/ppa.2016.65.issue-8
Bernardes-De-Assis, J., Storari, M., Zala, M., Wang, W., Jiang, D., Shidong, L., et al. (2009). Genetic structure of populations of the rice-infecting pathogen Rhizoctonia solani AG-1 IA from China. Phytopathology 99, 1090–1099. doi: 10.1094/PHYTO-99-9-1090
Bhaskar Rao, T., Chopperla, R., Prathi, N. B., Balakrishnan, M., Prakasam, V., Laha, G. S., et al. (2020). A Comprehensive Gene Expression Profile of Pectin Degradation Enzymes Reveals the Molecular Events during Cell Wall Degradation and Pathogenesis of Rice Sheath Blight Pathogen Rhizoctonia solani AG1-IA. J. Fungi 6, 71. doi: 10.3390/jof6020071
Bonman, J. M., Khush, G. S., Nelson, R. J. (1992). Breeding rice for resistance to pests. Annu. Rev. Phytopathol. 30, 507–528. doi: 10.1146/annurev.py.30.090192.002451
Brown, W. (1924). II. A method of isolating single strains of fungi by cutting out a hyphal tip. Ann. Bot. 38, 402–404.
Cartwright, R. D., Groth, D. E., Wamishe, Y. A., Greer, C. A., Calvert, L. A., Vera Cruz, C. M., et al. (2018). Compendium of rice diseases and pests. (St. Paul, Minnesota, U.S.A: The American Phytopathological Society - APS PRESS), 158-pp.
Chahal, K. S., Sokhi, S., Rattan, G. (2003). Investigations on sheath blight of rice in Punjab. Indian Phytopathol. 56, 22–26.
Channamallikarjuna, V., Sonah, H., Prasad, M., Rao, G. J. N., Chand, S., Upreti, H. C., et al. (2010). Identification of major quantitative trait loci qSBR11-1 for sheath blight resistance in rice. Mol. Breed. 25, 155–166. doi: 10.1007/s11032-009-9316-5
Chen, J., Xuan, Y., Yi, J., Xiao, G., Yuan, D. P., Li, D. (2023). Progress in rice sheath blight resistance research. Front. Plant Sci. 14, 1–13. doi: 10.3389/fpls.2023.1141697
Chen, X., Temnykh, S., Xu, Y., Cho, Y., Mccouch, S. (1997). Development of a microsatellite framework map providing genome-wide coverage in rice (Oryza sativa L.). Theor. Appl. Genet. 95, 553–567. doi: 10.1007/s001220050596
De Lorenzo, G., Castoria, R., Bellincampi, D., Cervone, F. (1997). “Fungal invasion enzymes and their inhibition,” in Plant Relationships: Part A. Eds. Carroll, G. C., Tudzynski, P. (Springer Berlin Heidelberg, Berlin, Heidelberg), 61–83.
Dey, S., Badri, J., Prakasam, V., Bhadana, V. P., Eswari, K. B., Laha, G. S., et al. (2016). Identification and agro-morphological characterization of rice genotypes resistant to sheath blight. Australas. Plant Pathol. 45, 145–153. doi: 10.1007/s13313-016-0404-9
FAOSTAT (2023). Crops and livestock products (Rice). Crop Livest Prod. Available online at: https://www.fao.org/faostat/en/#data/QCL/visualize (accessed August 6, 2024).
Ganeshamoorthi, P., Dubey, S. C. (2013). Anastomosis grouping and genetic diversity analysis of Rhizoctonia solani isolates causing wet root rot in chickpea. Afr. J. Biotechnol. 12, 6159–6169. doi: 10.5897/AJB2013.12439
Gawade, D. B., Perane, R. R., Suryawanshi, A. P., Deokar, C. D. (2017). Extracellular enzymes activity determining the virulence of Rhizoctonia bataticola, causing root rot in soybean. Physiol. Mol. Plant Pathol. 100, 49–56. doi: 10.1016/j.pmpp.2017.06.003
Gnanamanickam, S. S., Gnanamanickam, S. S. (2009). “Major diseases of rice,” in Gnanamanickam S.S. Biological control of rice diseases (Dordrecht, Netherlands: Springer), 13–42. doi: 10.1007/978-90-481-2465-7
Gonzalez, M., Pujol, M., Metraux, J.-P., Gonzalez-Garcia, V., Bolton, M. D., Borrás-Hidalgo, O. (2011). Tobacco leaf spot and root rot caused by Rhizoctonia solani Kühn. Mol. Plant Pathol. 12, 209–216. doi: 10.1111/j.1364-3703.2010.00664.x
Goswami, S. K., Singh, V., Kashyap, P. L., Singh, P. K. (2019). Morphological characterization and screening for sheath blight resistance using Indian isolates of Rhizoctonia solani AG1IA. Indian Phytopathol. 72, 107–124. doi: 10.1007/s42360-018-0103-2
Groth, D., Nowick, E. (1992). Selection for resistance to rice sheath blight through number of infection cushions and lesion type. Plant Dis. 76, 721–723. doi: 10.1094/PD-76-0721
Guillemaut, C., Edel-Hermann, V., Camporota, P., Alabouvette, C., Richard-Molard, M., Steinberg, C. (2003). Typing of anastomosis groups of Rhizoctonia solani by restriction analysis of ribosomal DNA. Can. J. Microbiol. 49, 556–568. doi: 10.1139/w03-066
Hameed, M. A., Natt, M. J., Iqbal, J., P. (1994). The role of protease and lipase in plant pathogenesis. Phytopathology 6, 13–16.
Hammer, Ø., Harper, D. A. (2001). Past: paleontological statistics software package for education and data analysis. Palaeontol. Electronica 4, 1–9.
Han, S. O., Yukawa, H., Inui, M., Doi, R. H. (2003). Regulation of expression of cellulosomal cellulase and hemicellulase genes in Clostridium cellulovorans. J. Bacteriol. 185, 6067–6075. doi: 10.1128/JB.185.20.6067-6075.2003
Hankin, L., Anagnostakis, S. L. (1975). The use of solid media for detection of enzyme production by fungi. Mycologia 67, 597–607. doi: 10.1080/00275514.1975.12019782
Hassan, A. A. (2021). Molecular, physiological and infection behavior studies of Rhizoctonia solani causing the Rice Sheath Blight disease. Egyptian J. Agric. Res. 99, 248–261. doi: 10.21608/ejar.2021.89516.1130
Hassan, A. A., Gabr, W. E., Kalboush, Z. A. (2023). Biogenic silica: A promoter of cell wall strengthening and induced resistance against sheath blight disease in rice via salicylic acid signaling and phenolic metabolism. Physiol. Mol. Plant Pathol. 128, 1–12. doi: 10.1016/j.pmpp.2023.102152
Have, T., Rimko, Hartmans, S., Teunissen, P. J., Field, J. A. (1998). Purification and characterization of two lignin peroxidase isozymes produced by Bjerkandera sp. strain BOS55. FEBS Lett. 422, 391–394. doi: 10.1016/S0014-5793(98)00044-1
Hejazi, R., Esfahani, M. N., Maleki, M., Sedaghatfar, E. (2022). Susceptibility assessment and genetic population structure associated with Rhizoctonia solani AG3-PT-Potato stem canker disease. Physiol. Mol. Plant Pathol. 119, 101835. doi: 10.1016/j.pmpp.2022.101835
Hossain, M. K., Jena, K. K., Bhuiyan, M. A., Wickneswari, R. (2016). Association between QTLs and morphological traits toward sheath blight resistance in rice (Oryza sativa L.). Breed. Sci. 66, 613–626. doi: 10.1270/jsbbs.15154
Hussain, S., Huang, J., Huang, J., Ahmad, S., Nanda, S., Anwar, S., et al. (2020). “Rice production under climate change: adaptations and mitigating strategies,” in Environment, climate, plant and vegetation growth, eds. Fahad, S., (Cham, Switzerland: Springer), 686. doi: 10.1007/978-3-030-49732-3_26
Iradukunda, L., Wang, Y.-P., Nkurikiyimfura, O., Wang, T., Yang, L.-N., Zhan, J. (2022). Establishment and application of a multiplex PCR assay for the rapid detection of Rhizoctonia solani anastomosis group (AG)-3PT, the pathogen causing potato black scurf and stem canker. Pathogens 11, 627. doi: 10.3390/pathogens11060627
IRRI (2002). Standard Evaluation Systems for Rice (Manila, Philippines: International Rice Research Institute), 19.
IRRI (2013). Standard evaluation system for rice (SES) (Manila: International Rice Research Institute).
Jalgaonwala, R. E., Mahajan, R. T. (2011). Evaluation of hydrolytic enzyme activities of endophytes from some indigenous medicinal plants. Int. J. Agric. Technol. 7, 1733–1741.
Jia, Y. J., Feng, B. Z., Sun, W. X., Zhang, X. G. (2009). Polygalacturonase, Pectate Lyase and Pectin Methylesterase Activity in Pathogenic Strains of Phytophthora capsici Incubated under Different Conditions. J. Phytopathol. 157, 585–591. doi: 10.1111/j.1439-0434.2008.01533.x
Jia, L., Yan, W., Zhu, C., Agrama, H. A., Jackson, A., Yeater, K., et al. (2012). Allelic analysis of sheath blight resistance with association mapping in rice. PloS One 7, e32703. doi: 10.1371/journal.pone.0032703
Kalboush, Z. A., Hassan, A. A., Sherif, A., Gabr, W. E. (2024). Endophytic bacteria nano-formula: A promotor for plant growth and systemic acquired resistance against sheath blight disease on rice. Physiol. Mol. Plant Pathol. 131, 1–12. doi: 10.1016/j.pmpp.2024.102295
Kuramae, E. (1997). A rapid, easy and high yield protocol for total genomic DNA isolation from Colletotrichum gloeosporioides and Fusarium oxysporum for RAPD. Rev. UNIMAR 19, 683–689.
Lee, D.-Y., Jeon, J., Kim, K.-T., Cheong, K., Song, H., Choi, G., et al. (2021). Comparative genome analyses of four rice-infecting Rhizoctonia solani isolates reveal extensive enrichment of homogalacturonan modification genes. BMC Genomics 22, 1–15. doi: 10.1186/s12864-021-07549-7
Leopold, J., SamsˇinˇÁková, A. (1970). Quantitative estimation of chitinase and several other enzymes in the fungus Beauveria bassiana. J. Invertebrate Pathol. 15, 34–42. doi: 10.1016/0022-2011(70)90095-9
Liu, G., Jia, Y., Correa-Victoria, F. J., Prado, G. A., Yeater, K. M., Mcclung, A., et al. (2009). Mapping quantitative trait Loci responsible for resistance to sheath blight in rice. Phytopathology 99, 1078–1084. doi: 10.1094/PHYTO-99-9-1078
Liu, Z., Zhu, Y., Shi, H., Qiu, J., Ding, X., Kou, Y. (2021). Recent progress in rice broad-spectrum disease resistance. Int. J. Agric. Technol. 22, 11658–11674. doi: 10.3390/ijms222111658
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2 (-Delta Delta C (T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
López-Corrales, R., Michereff, S. J., García-Estrada, R. S., Correia, K. C., Mora-Romero, G. A., León-Félix, J., et al. (2024). Diversity and pathogenicity of anastomosis groups of Rhizoctonia solani isolates associated with potato diseases in northern Sinaloa, Mexico. Potato Res. 67, 1011–1026. doi: 10.1007/s11540-023-09676-y
McCouch, S. R., Teytelman, L., Xu, Y., Lobos, K. B., Clare, K., Walton, M., et al. (2002). Development and mapping of 2240 new SSR markers for rice (Oryza sativa L.). DNA Res. 9, 199–207. doi: 10.1093/dnares/9.6.199
Mew, T. W., Leung, H., Savary, S., Vera Cruz, C. M., Leach, J. E. (2004). Looking ahead in rice disease research and management. Crit. Rev. Plant Sci. 23, 103–127. doi: 10.1080/07352680490433231
Molla, K. A., Karmakar, S., Molla, J., Bajaj, P., Varshney, R. K., Datta, S. K., et al. (2020). Understanding sheath blight resistance in rice: the road behind and the road ahead. Plant Biotechnol. J. 18, 895–915. doi: 10.1111/pbi.13312
Monazzah, M., Esfahani, M. N., Enferadi, S. T. (2022). Genetic structure and proteomic analysis associated in potato to Rhizoctonia solani AG-3PT-stem canker and black scurf. Physiol. Mol. Plant Pathol. 122, 101905. doi: 10.1016/j.pmpp.2022.101905
Mondal, A., Dutta, S., Kuiry, S., Chakraborty, D., Nandi, S., Das, S., et al. (2013). The biochemical constituents and pectinase activities associated with the virulence of Rhizoctonia solani isolates in rice in West Bengal, India. Afr. J. Agric. Res. 8, 3029–3035.
Moni, Z. R., Ali, M. A., Alam, M. S., Rahman, M. A., Bhuiyan, M. R., Mian, M. S., et al. (2016). Morphological and genetical variability among Rhizoctonia solani isolates causing sheath blight disease of rice. Rice Sci. 23, 42–50. doi: 10.1016/j.rsci.2016.01.005
Mughal, M. N., Bashir, S., Bhat, N. A., Bhat, K. (2017). Cultural and morphological variability and identification of anastomosis group of Rhizoctonia solani (Thanatephorus cucumeris) causing sheath blight of rice in Kashmir. Int. J. Curr. Microbiol. Appl. Sci. 6, 3787–3794. doi: 10.20546/ijcmas.2017.611.443
Murray, M., Thompson, W. (1980). Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8, 4321–4326. doi: 10.1093/nar/8.19.4321
Muzhinji, N., Lekota, M. (2024). Binucleate Rhizoctonia on potato: Geographic distribution, identification, taxonomy, genomics, host range and disease management. J. Phytopathol. 172, e13364. doi: 10.1111/jph.v172.4
Muzhinji, N., Truter, M., Woodhall, J., van der Waals, J. E. (2015). Anastomosis groups and pathogenicity of Rhizoctonia solani and binucleate Rhizoctonia from potato in South Africa. Plant Dis. 99, 1790–1802. doi: 10.1094/PDIS-02-15-0236-RE
Nagaraj, B., Sunkad, G., Pramesh, D., Naik, M., Patil, M. (2017). Host range studies of rice sheath blight fungus Rhizoctonia solani (Kuhn). Int. J. Curr. Microbiol. Appl. Sci. 6, 3856–3864. doi: 10.20546/ijcmas.2017.611.452
Naqvi, S., Abbas, A., Farhan, M., Kiran, R., Hassan, Z., Mehmood, Y., et al. (2024). Unveiling the Genetic Tapestry: Exploring Rhizoctonia solani AG-3 Anastomosis Groups in Potato Crops across Borders. Plants 13, 715. doi: 10.3390/plants13050715
Naveenkumar, R., Anandan, A., Singh, V., Prabhukarthikeyan, S. R., Parameswaran, C., Sangeetha, G., et al. (2022). Deciphering environmental factors and defense response of rice genotypes against sheath blight disease. Physiol. Mol. Plant Pathol. 122, 1–16. doi: 10.1016/j.pmpp.2022.101916
Nawaz, A., Rehman, A. U., Rehman, A., Ahmad, S., Siddique, K. H., Farooq, M. (2022). Increasing sustainability for rice production systems. J. Cereal Sci. 103, 103400. doi: 10.1016/j.jcs.2021.103400
Palaniswamy, K., Gomez, K. (1974). Length-width method for estimating leaf area of rice 1. Agron. J. 66, 430–433. doi: 10.2134/agronj1974.00021962006600030027x
Panaud, O., Chen, X., Mccouch, S. R. (1996). Development of microsatellite markers and characterization of simple sequence length polymorphism (SSLP) in rice (Oryza sativa L.). Mol. Gen. Genet. 252, 597–607. doi: 10.1007/BF02172406
Pane, C., Caputo, M., Francese, G., Manganiello, G., Lo Scalzo, R., Mennella, G., et al. (2020). Managing Rhizoctonia damping-off of rocket (Eruca sativa) seedlings by drench application of bioactive potato leaf phytochemical extracts. Biology 9, 270. doi: 10.3390/biology9090270
Park, D. S., Sayler, R. J., Hong, Y. G., Nam, M. H., Yang, Y. (2008). A method for inoculation and evaluation of rice sheath blight disease. Plant Dis. 92, 25–29. doi: 10.1094/PDIS-92-1-0025
Pinson, S. R., Capdevielle, F. M., Oard, J. H. (2005). Confirming QTLs and finding additional loci conditioning sheath blight resistance in rice using recombinant inbred lines. Crop Sci. 45, 503–510. doi: 10.2135/cropsci2005.0503
Poonguzhali, P., Chauhan, A., Kar, A., Lavale, S., Nayak, S. N., Prashanthi, S. K. (2022). New sources of resistance and identification of DNA marker loci for sheath blight disease caused by Rhizoctonia solani Kuhn, in rice. Plant Pathol. J. 38, 572–582. doi: 10.5423/PPJ.OA.04.2022.0054
Qu, P., Aratani, A., Syoji, T., Toda, T., Kubota, M., Hyakumachi, M. (2008). Use of single-protoplast isolates in the study of the mating phenomena of Rhizoctonia solani (Thanatephorus cucumeris) AG-1 IC and IA. Mycoscience 49, 132–137. doi: 10.1007/S10267-007-0400-6
Saitou, N., Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. doi: 10.1093/oxfordjournals.molbev.a040454
Senapati, M., Tiwari, A., Sharma, N., Chandra, P., Bashyal, B. M., Ellur, R. K., et al. (2022). Rhizoctonia solani Kühn pathophysiology: status and prospects of sheath blight disease management in rice. Front. Plant Sci. 13, 881116–888138. doi: 10.3389/fpls.2022.881116
Sharma, N., Teng, P. (1990). Comparison of rice sheath blight (ShB) assessment methods. Int. Rice Res. Newslett. 15, 20–21.
Shekhawat, D. S., Bagri, R., Yadav, A., Bhati, P., Yadav, B. B., Kumawat, S. (2023). Studies on Different Host Range of Root Rot (Rhizoctonia solani Kühn) under Pot House. Int. J. Plant Soil Sci. 35, 393–397. doi: 10.9734/ijpss/2023/v35i224147
Sherif, A., Zhang, B., Wu, B., Hu, Y., Li, S., Zhou, X., et al. (2023). A Pseudo-near isogenic F2 population strategy for rapid QTL cloning. Mol. Breed. 43, 61. doi: 10.1007/s11032-023-01408-x
Shieh, M.-T., Brown, R. L., Whitehead, M. P., Cary, J. W., Cotty, P. J., Cleveland, T. E., et al. (1997). Molecular genetic evidence for the involvement of a specific polygalacturonase, P2c, in the invasion and spread of Aspergillus flavus in cotton bolls. Appl. Environ. Microbiol. 63, 3548–3552. doi: 10.1128/aem.63.9.3548-3552.1997
Sneh, B., Burpee, L., Ogoshi, A. (1991). Identification of Rhizoctonia species (St. Paul, Minnesota, U.S.A.: APS press), 133.
Temnykh, S., Declerck, G., Lukashova, A., Lipovich, L., Cartinhour, S., Mccouch, S. (2001). Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): frequency, length variation, transposon associations, and genetic marker potential. Genome Res. 11, 1441–1452. doi: 10.1101/gr.184001
Temnykh, S., Park, W. D., Ayres, N., Cartinhour, S., Hauck, N., Lipovich, L., et al. (2000). Mapping and genome organization of microsatellite sequences in rice (Oryza sativa L.). Theor. Appl. Genet. 100, 697–712. doi: 10.1007/s001220051342
Tinline, R., MacNeill, B. (1969). Parasexuality in plant pathogenic fungi. Annu. Rev. Phytopathol. 7, 147–168. doi: 10.1146/annurev.py.07.090169.001051
Tsror, L. (2010). Biology, epidemiology and management of Rhizoctonia solani on potato. J. Phytopathol. 158, 649–658. doi: 10.1111/j.1439-0434.2010.01671.x
Usyk, M., Zolnik, C. P., Patel, H., Levi, M. H., Burk, R. D. (2017). Novel ITS1 fungal primers for characterization of the mycobiome. mSphere 2, e00488–17. doi: 10.1128/mSphere.00488-17
Vidhyasekaran, P., Ponmalar, T. R., Samiyappan, R., Velazhahan, R., Vimala, R., Ramanathan, A., et al. (1997). Host-specific toxin production by Rhizoctonia solani, the rice sheath blight pathogen. Phytopathology 87, 1258–1263. doi: 10.1094/PHYTO.1997.87.12.1258
Wamishe, Y. A., Yulin, J., Singh, P., Cartwright, R. D. (2007). Identification of field isolates of Rhizoctonia solani to detect quantitative resistance in rice under greenhouse conditions. Front. Agric. China 1, 361–367. doi: 10.1007/s11703-007-0061-4
Ward, J. H., Jr (1963). Hierarchical grouping to optimize an objective function. J. Am. Stat. Assoc. 58, 236–244. doi: 10.1080/01621459.1963.10500845
Wen, Z., Zeng, Y., Ji, Z., Yang, C. (2015). Mapping quantitative trait loci for sheath blight disease resistance in Yangdao 4 rice. Genet. Mol. Res. 14, 1636–1649. doi: 10.4238/2015.March.6.10
Willocquet, L., Noel, M., Sackville Hamilton, R., Savary, S. (2012). Susceptibility of rice to sheath blight: an assessment of the diversity of rice germplasm according to genetic groups and morphological traits. Euphytica 183, 227–241. doi: 10.1007/s10681-011-0451-9
Woodhall, J., Lees, A., Edwards, S., Jenkinson, P. (2007). Characterization of Rhizoctonia solani from potato in Great Britain. Plant Pathol. 56, 286–295. doi: 10.1111/j.1365-3059.2006.01545.x
Yarmeeva, M. M., Kokaeva, L. Y., Chudinova, E. M., Kah, M. O., Kurchaev, M. L., Zeyruk, V. N., et al. (2021). Anastomosis groups and sensitivity to fungicides of Rhizoctonia solani strains isolated from potato in Russia. J. Plant Dis. Prot. 128, 1253–1261. doi: 10.1007/s41348-021-00490-7
Zahra, N., Hafeez, M. B., Nawaz, A., Farooq, M. (2022). Rice production systems and grain quality. J. Cereal Sci. 105, 103463. doi: 10.1016/j.jcs.2022.103463
Zou, J., Pan, X., Chen, Z., Xu, J., Lu, J., Zhai, W., et al. (2000). Mapping quantitative trait loci controlling sheath blight resistance in two rice cultivars (Oryza sativa L.). Theor. Appl. Genet. 101, 569–573. doi: 10.1007/s001220051517
Keywords: sheath blight, rhizoctonia, host range, rice genotype, SSR marker, weeds, cell wall
Citation: Kalboush ZA, Mazrou YSA, Hassan AA, Sherif A, Gabr WE, Ali Q and Nehela Y (2024) Revisiting the emerging pathosystem of rice sheath blight: deciphering the Rhizoctonia solani virulence, host range, and rice genotype-based resistance. Front. Plant Sci. 15:1499785. doi: 10.3389/fpls.2024.1499785
Received: 21 September 2024; Accepted: 02 December 2024;
Published: 19 December 2024.
Edited by:
Omer Frenkel, Agricultural Research Organization (ARO), IsraelReviewed by:
Dinesh Pandey, G. B. Pant University of Agriculture and Technology, IndiaBishnu Maya Bashyal, Indian Agricultural Research Institute (ICAR), India
Copyright © 2024 Kalboush, Mazrou, Hassan, Sherif, Gabr, Ali and Nehela. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Amr A. Hassan, YW1yX2FiXzIwMjhAeWFob28uY29t; Qurban Ali, cmF0dGFycXVyYmFuQHVhZXUuYWMuYWU=; Yasser Nehela, eWFzc2VyLm5laGVsYUBhZ3IudGFudGEuZWR1LmVn
 Zeinab A. Kalboush1
Zeinab A. Kalboush1 Amr A. Hassan
Amr A. Hassan Ahmed Sherif
Ahmed Sherif Qurban Ali
Qurban Ali Yasser Nehela
Yasser Nehela