- 1Yunnan Provincial Engineering and Research Center for Sustainable Utilization of Honey Bee Resources, Eastern Bee Research Institute, College of Animal Science and Technology, Yunnan Agricultural University, Kunming, China
- 2Institute of Sericulture and Apiculture, Yunnan Academy of Agricultural Sciences, Mengzi, China
- 3College of Food Science and Technology, Yunnan Agricultural University, Kunming, China
Endophytic fungi are extensive in plant tissues and involved in the defense against stress from harmful microbes. The interaction between pedicel endophytic fungi and nectar yeasts is critical for maintaining nectar homeostasis. This study used Camellia reticulata as the research subject. High-throughput sequencing revealed that the community composition of endophytic fungi in the pedicel is dominated by Ascomycota and Basidiomycota. Their abundance varies at different taxonomic levels, showing sample variability. In total, 27 endophytic fungal isolates were isolated and screened from the pedicel under laboratory conditions. They exhibited antagonistic effects against three nectar yeasts (Metschnikowia reukaufii, Cryptococcus laurentii, and Rhodotorula glutinis) and displayed morphological and physiological diversity. The isolates were classified into the phylum Ascomycota and further categorized into the genera Alternaria, Trichoderma, Fusarium, and Dactylaria. The endophytic fungus D23, which effectively antagonizes nectar yeasts, was identified as Alternaria alternata. This fungus produces various secondary metabolites, including antibiotics such as penicillin G, grandiomycin, and cephalosporin C. The metabolic pathways involved include the biosynthesis of plant secondary metabolites, phenylpropanoids, amino acids, nucleotides, and antibiotics. The endophytic fungal community in C. reticulata pedicel is rich and diverse, making it a valuable material for screening antagonistic strains. This study provides a theoretical basis for the antagonistic effects of endophytic fungal metabolites from the pedicel of C. reticulata against nectar yeasts, highlighting their significance in maintaining nectar stability and reproductive fitness in cross-pollinated plants.
1 Introduction
Nectar is an energy reward from plants for pollinators. Yeast commonly inhabits the nectar of most plants. Increased pollinator visits enhance yeast density in the nectar. This increase in density and subsequent metabolic activity alters the chemical composition of the nectar, particularly affecting sugar and amino acid levels (Canto and Herrera, 2012; Peay et al., 2012). However, such changes may weaken the nectar’s appeal to specific pollinators and reduce visitation rates (Canto and Herrera, 2012; Schaeffer et al., 2017). Yeasts in plant nectar, primarily classified under Ascomycota and Basidiomycota, include genera such as Metschnikowia, Cryptococcus, Rhodotorula, Saccharomyces, and Sporobolomyces (Herrera et al., 2009; Pozo et al., 2012; Jacquemyn et al., 2013). Without effective antimicrobial mechanisms, nutrient-rich nectar likely supports high microorganism proliferation, diminishing its value for plant reproduction. Researchers have investigated the chemical components and defensive functions of nectar worldwide. They found that factors like pH, high osmotic pressure, and various secondary metabolites or proteins contribute to the antimicrobial properties of the nectar (Pusey, 1999; Brysch-Herzberg, 2004; Herrera et al., 2011; Álvarez-Pérez et al., 2013). However, research into the antimicrobial substances in nectar and their functions remains exploratory, and the role of endophytes is also under examination.
Endophytic fungi are crucial for the plant microenvironment, establishing a dynamic balance through colonization and operational mechanisms. They form communities with significant ecological functions (Philippot et al., 2013). These fungi enhance plant stress resistance, promote growth (Compant et al., 2005; Zhang et al., 2006), and suppress pathogen populations by spatial localization. They produce various antimicrobial metabolites that help plants resist microbial stress (Zhang et al., 2006; Pieterse et al., 2014). For instance, Stinson et al. (2003) isolated Gliocladium sp. from Eucryphia cordifolia. This fungus produces volatile organic compounds, such as cyclooctatetraene and 3-methyl-1-butanol, with strong antimicrobial effects against plant pathogens like Pythium ultimum and Verticillium dahliae. Richardson et al. (2015) isolated two Massarinaceae species from Pinus strobus, producing phenolic compounds that inhibit Bacillus subtilis and Microbotryum violaceum. Hiremani et al. (2020) isolated two species of Nigrospora sphaerica from Gossypium arboreum, producing volatile organic compounds effective against Corynespora cassiicola. Recent research has found that endophyte colonization can alter the composition of volatile metabolites in flowers, attract more pollinators, and enhance the reproductive adaptability of plants, providing new insights into the resistance mechanism of endophyte-assisted nectar against yeast (González-Mas et al., 2023). However, studies on the antagonistic effects of endophytic fungi against nectar yeasts are rare.
Camellia reticulata has been cultivated in Yunnan for a long time and is a rare plant protected in China (Feng, 1980). Its seed-extracted oil is rich in unsaturated fatty acids and antioxidants and has nutritional value comparable to olive oil, which makes it a geographical indication product in China (Qin et al., 2023). Due to self-incompatibility, C. reticulata heavily depends on pollinators for fruit set and seed production. The plant has a long flowering period, produces abundant nectar, and faces significant microbial contamination during pollination. Preliminary studies in our laboratory found that nectar from flowers blooming for many days maintains low microbial concentration. However, yeast abundance significantly increases during ex vivo fermentation, deteriorating nectar’s physicochemical properties (unpublished data). Importantly, the antimicrobial mechanism within C. reticulata nectar remains unclear. The bacteriostatic effects of endophytic fungi colonizing plant tissues may provide new explanations for their antimicrobial mechanisms. Pedicels, which are plant tissues connecting to flowers, play a crucial role in spatial distribution and serve as important conduits for nutrient supply (Zhou, 2015). The endophytic fungi colonize the pedicel near the flowers for extended periods, making them valuable microbial resources that can antagonize nectar yeasts in an auxiliary role. However, there is limited understanding of the community composition and functions of endophytic fungi colonizing C. reticulata pedicels. Accordingly, this study focuses on C. reticulata pedicels to analyze its endophytic fungal communities, screen isolates inhibiting nectar yeast, and analyze their metabolic products. Specific research objectives include (1) high-throughput sequencing to analyze endophytic fungi diversity in C. reticulata pedicels; (2) isolation of endophytic fungi from pedicels; (3) screening of endophytic fungi with antagonistic activity against nectar yeasts and evaluating these isolates’ inhibitory effects; (4) morphological and molecular biological identification of these isolates; (5) evaluation of minimum inhibitory concentration (MIC) and antibacterial potential of ethyl acetate extract from efficient isolate D23; and (6) non-targeted metabolomics analysis of metabolic products and antimicrobial substances from efficient isolate D23. This study analyzed endophytic fungi diversity in C. reticulata pedicels and screened for fungi antagonizing nectar yeast. Our results provide insights into mechanisms by which cross-pollinated plants resist yeast stress, regulate nectar stability, and maintain reproductive fitness.
2 Materials and methods
2.1 Sampling information and plant sample collection
In January and February 2024, C. reticulata plant samples were collected from Shaba Forestry Farm (98°5’86’’ E, 24°9’57’’ N) in Tengchong, Yunnan Province. This site has a tropical monsoon climate, with an average annual temperature of 14.6°C, approximately 2,167 h of sunshine per year, an average annual precipitation of 1,531 mm, and an average altitude of 2,149 m. During sampling, healthy C. reticulata plants were selected. Flower buds and the lower 6 cm of stems were cut using sterilized scissors (sterilized with 75% alcohol) as plant samples. These samples were placed in sterile bags and transported to the laboratory at 4°C for further processing.
2.2 Sample surface sterilization
Plant samples were washed in flowing tap water. Under sterile conditions, flower buds and stems were trimmed to leave only pedicels. Following the method of Cardoso et al. (2020), pedicels were subjected to stringent surface sterilization. They were first rinsed with sterile distilled water and then immersed in 75% ethanol for 1 min. Pedicels were washed four times with sterile distilled water and dried with sterile filter paper. Next, they were soaked in 5% sodium hypochlorite solution for 2 min, rinsed four times with sterile distilled water, and dried again with sterile filter paper. To verify sterilization effectiveness, 100 µL of the sterile water from the final wash was plated on Luria broth (LB) and tryptic soy agar (TSA) media and incubated at 30°C for 72 h. The absence of microorganism growth confirmed that sterilization was successful and that pedicels were suitable for subsequent endophytic fungal screening and identification.
2.3 Diversity analysis of endophytic fungi
Surface-sterilized pedicels were ground in liquid nitrogen, and total DNA from the pedicel microbiota was extracted using the E.Z.N.A.® Soil DNA Kit (Omega Bio-tek, Norcross, GA, U.S.). DNA concentration and purity were measured with a NanoDrop 2000 machine, and DNA quality was assessed using 1% agarose gel electrophoresis. PCR amplification of the ITS1-ITS2 region of genomic DNA was performed using endophytic fungal primers ITS1F (5’-CTTGGTCATTTAGAGGAAGTAA-3’) and ITS2R (5’-GCTGCGTTCTTCATCGATGC-3’) (Gardes and Bruns, 1993). PCR products were recovered using the AxyPrep DNA Gel Recovery Kit, and the target fragment size was confirmed with 2% agarose gel electrophoresis. Quantification was conducted using the QuantiFluor™-ST blue fluorescence quantification system. Purified PCR products were sent to Shanghai Majorbio Biomedical Technology Co., Ltd. for Illumina high-throughput sequencing. Sequences from raw data were optimized, and OTU clustering analysis of non-redundant sequences was performed using the Usearch software platform (similarity: 97%). Diversity analysis was conducted using the I-Sanger bioinformatics cloud platform.
2.4 Isolation, purification, and preservation of endophytic fungi
Sterilized pedicels were cut into small pieces using a sterile scalpel and transferred to the potato dextrose agar (PDA) medium. They were incubated at 28°C in an incubator for 3-7 days. Once fungal mycelium grew at the edges of the plant tissue, hyphae were isolated from the medium based on colony characteristics using a scalpel and transferred to a fresh PDA medium for purification. After several rounds of purification to achieve uniform colony morphology, colonies were transferred to a PDA slant and stored at 4°C for preservation, with subculturing every 30 days to prevent isolate degradation. Additionally, surface mycelium was scraped using an inoculation spatula, placed in a 50% glycerol cryopreservation tube, mixed thoroughly, and frozen at -80°C for long-term storage.
2.5 Screening of isolates antagonizing nectar yeasts
Three types of nectar yeasts provided by the China General Microbiological Culture Collection Center were used for testing: Metschnikowia reukaufii, Cryptococcus laurentii, and Rhodotorula glutinis. The yeast strains were first revived using the malt extract broth (MEB) medium. After 24 h, 1 mL of each strain suspension was taken and diluted with sterile phosphate buffered saline (PBS) to a 0.5 McFarland standard (approximately 108 CFU/mL). The prepared yeast suspension was evenly spread onto a PDA medium using a sterile cotton swab for testing. Additionally, 5 mm diameter agar discs medium covered with fungal mycelium were punched from the PDA medium. After overnight incubation at 28°C, isolates exhibiting inhibition zones were initially selected. The endophytic fungi initially screened with antagonistic activity against nectar yeast were then inoculated into potato dextrose broth (PDB) medium and cultured at 135 rpm and 28°C for 5 days in a shaking incubator. Afterward, each endophytic fungal culture (10 mL) was centrifuged at 12,000 rpm for 15 min to obtain the supernatant, which was then filtered through a 0.22 μm membrane to produce a sterile fermentation extract for each isolate. Oxford cups with a diameter of 5 mm were placed on the medium surface, and 100 μL of the fermentation extract was added to each cup. PDB medium served as the blank control (CK), and Amphotericin B was used as the positive control (+). All media were wrapped with sealing film and incubated at 28°C for 24 h. Finally, the inhibition zone diameters for each isolate were measured.
2.6 Identification of endophytic fungi
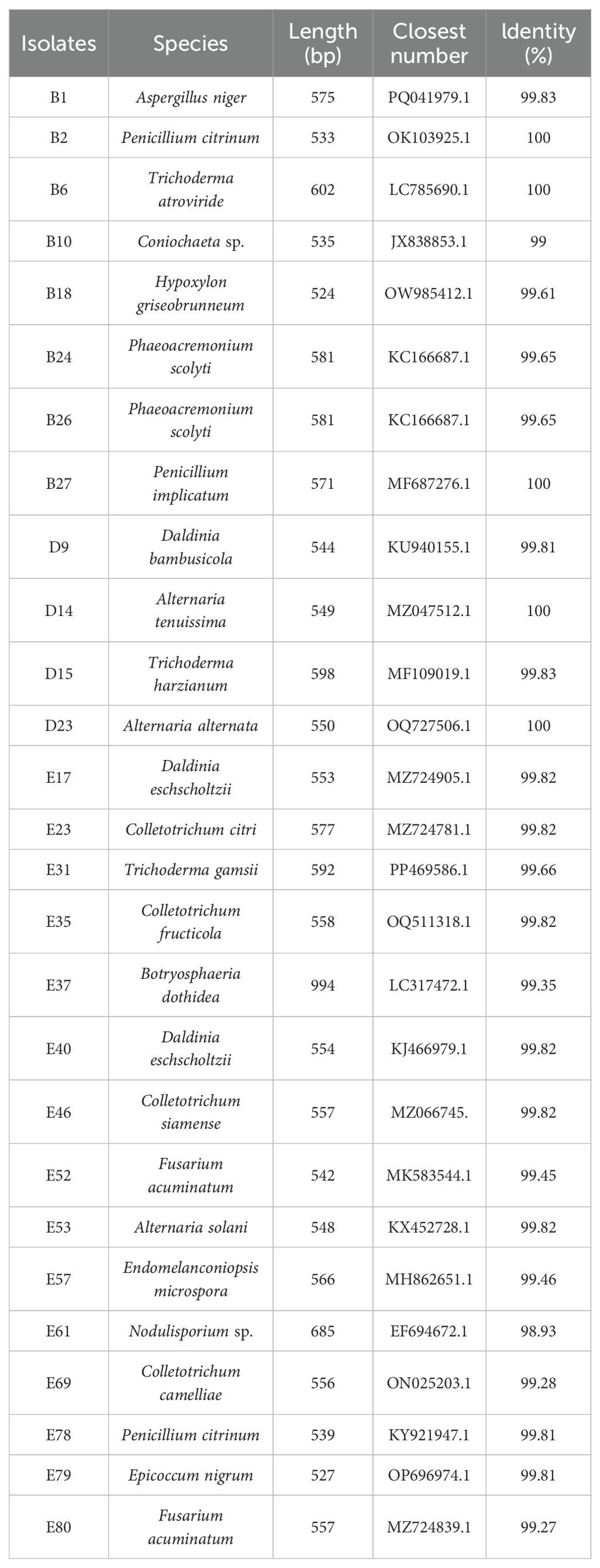
Firstly, morphological characteristics of endophytic fungi exhibiting antagonistic activity against nectar yeast were recorded, including colony color, size, surface features, and pigment production. DNA was extracted from selected endophytic fungi using the E.Z.N.A.® Soil DNA Kit, following the manufacturer’s instructions. PCR amplification was performed with fungal primers ITS1 (5’-TCCGTAGGTGAACCTGCGG-3’) and ITS4 (5’-TCCTCCGCTTATTGAATGC-3’) (White et al., 1990). Amplification results were analyzed by 1% agarose gel electrophoresis (150V/100mA for 20 min). Target DNA fragments were recovered using a QIAquick agarose gel extraction kit. The purified PCR products were sequenced using the ABI3730XL system. Sequencing data were compared to the NCBI database using BLAST. The accession number and percent identity for the strain that showed the highest alignment score with the isolate were obtained.
2.7 Analysis of isolate metabolites showing effective antagonism against nectar yeasts
2.7.1 Extraction of secondary metabolites of isolate
First, the endophytic fungus D23 was activated and rejuvenated. The fungus was transferred to PDA medium and incubated at 28°C for 4 days. After spores and mycelium matured, 5 mm mycelial discs were prepared and inoculated into a PDB medium. The culture was incubated at 28°C and 135 rpm for 3 days to obtain a fungal liquid culture. Next, 200 g of rice and 250 mL of MEB medium were added to each 2L wide-mouthed bottle. After autoclaving and cooling, 100 mL of fungal liquid culture was inoculated, mixed well, and incubated at 28°C for 30 days. Once the rice was fully colonized by mycelium, it was transferred to a sterilized tray, dried completely at 45°C, and then crushed. To extract the compounds, 100 g of the powdered rice was mixed with 500 mL of ethyl acetate. The mixture was extracted in a 45°C water bath for 3 h, then filtered, and the organic phase was collected. The extraction was repeated with ethyl acetate for each batch of powder a total of three times. The combined organic phases were transferred to a rotary evaporator to evaporate the solvent at 45°C. After evaporation, the organic solvent was recovered, and the crude extract was dissolved in methanol, dried in a ventilated cabinet, and stored at 4°C for future use.
2.7.2 MIC estimation of isolate metabolites against nectar yeasts
MIC values were determined using the broth dilution method following Clinical and Laboratory Standards Institute (CLSI) guidelines, with Amphotericin B as the positive control (Wang et al., 2023). In the first row of a 96-well plate, 180 µL of yeast suspension (1.25 × 106 CFU/mL) was added, while 100 µL was added to the remaining wells. A crude extract solution of D23 in DMSO (10 mg/mL) was prepared, and 20 µL was added to the first row of the wells. A two-fold serial dilution was then performed across the plate, and turbidity in each well was observed to determine the lowest D23 concentration required to inhibit yeast growth.
2.7.3 LC-MS/MS analysis of metabolites composition in the isolate
Non-targeted metabolomics LC-MS/MS was conducted to analyze active components in the ethyl acetate extract of D23. The MSConvert tool in the Proteowizard software package (v3.0.8789) was used to convert the original mass spectrometry downloaded file into mzXML format. Peak detection, filtering, and alignment were performed using the R XCMS package (v3.12.0), resulting in a quantitative metabolite list. Systematic errors were corrected using support vector regression based on QC samples, and substances with relative standard deviation (RSD) > 30% in QC samples were filtered out. Substance identification was conducted using public databases such as HMDB, MassBank, LipidMaps, mzCloud, and KEGG, with parameters set to < 30 ppm. Metabolite identification was conducted at Shanghai Majorbio Biomedical Technology Co., Ltd.
3 Results
3.1 Community structure of endophytic fungi in the pedicel of C. reticulata
3.1.1 OTU analysis of endophytic fungi in pedicel of C. reticulata
After the final rinse of the pedicel, plating on LB and TSA media followed by 72 h of incubation revealed no colony growth, indicating successful tissue sterilization. Consequently, the isolated fungi were derived from the endophytic colonization of the pedicel and not from the environment.
OTUs of endophytic fungi from the pedicel were classified based on 97% similarity, resulting in an average of 72, 061 effective sequences with a mean length of 238 bp. Sequence lengths primarily ranged between 201 and 280 bp, aligning with expected amplification lengths and demonstrating good concentration (Figures 1A, B). A total of 1,034 OTUs was obtained, distributed across 4 phyla, 24 classes, 77 orders, 161 families, and 267 genera. This indicates that the endophytic fungi in the pedicel of C. reticulata exhibit rich community diversity at various taxonomic levels.
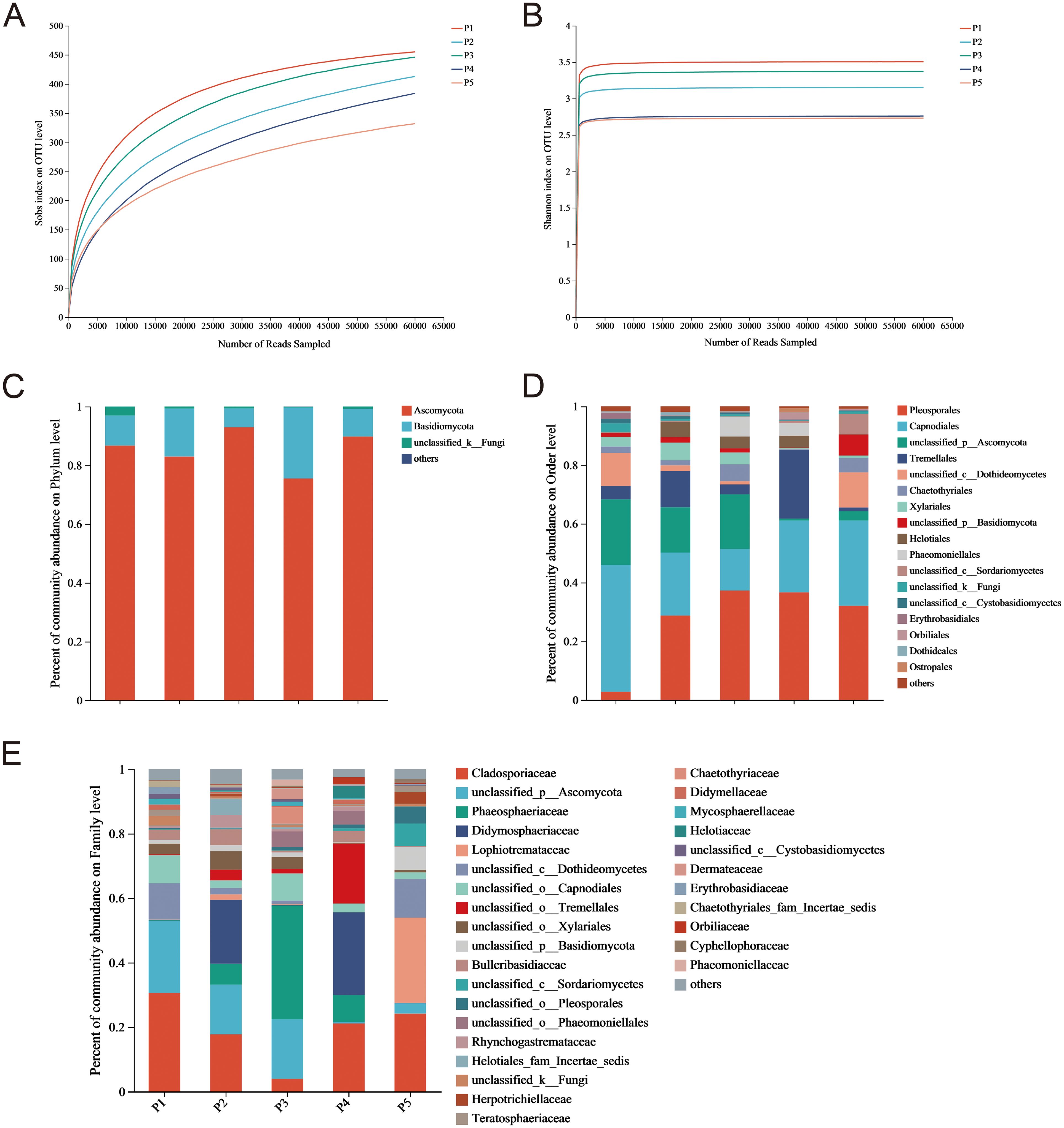
Figure 1. Rarefaction curves for (A) Sobs and (B) Shannon Indices, and relative abundance of endophytic fungi in C. reticulata pedicels at the (C) phylum, (D) order, and (E) family levels.
3.1.2 Abundance analysis of endophytic fungi in C. reticulata pedicel
The endophytic fungal communities detected in C. reticulata pedicels included Ascomycota, Basidiomycota, and unclassified_k_Fungi (Figure 1C). Ascomycota and Basidiomycota together accounted for 90% of the abundance. The proportion of Ascomycetes in each group reached 60%, indicating it is a dominant phylum. These endophytic fungi were distributed across 15 orders, with varying proportions and strain uniformity in each sample. Pleosporales, Capnodiales, unclassified_p_Ascomycota, Tremellales, unclassified_c_Dothideomycetes, Xylariales, and Chaetothyriales were the dominant orders, representing over 80% of each sample (Figure 1D). These fungi were also distributed across 30 families, but dominant families differed among samples (Figure 1E). In some samples, dominant families accounted for over 20%, including Cladosporiaceae, unclassified_p_Ascomycota, Phaeosphaeriaceae, Didymosphaeriaceae, and Lophiotremataceae. However, only some endophytic fungi were classified to the family level, with others classified only to the order, class, phylum, or kingdom level. This suggests that C. reticulata pedicels harbor a diverse range of fungal species, warranting further exploration.
3.2 Screening of endophytic fungi antagonizing nectar yeasts in C. reticulata pedicels
After rigorous surface sterilization, 118 endophytic fungal isolates were isolated from C. reticulata pedicels based on different colony characteristics. Of these, 27 isolates were preliminarily screened for antagonistic activity against the three nectar yeasts, indicated by inhibition zones. Among these, 20 isolates were inhibitory to M. reukaufii, 25 to C. laurentii, and 22 to R. glutinis. Isolates B1, B2, B10, B18, B27, D14, D23, E23, E52, E53, E57, E78, E79, and E80 exhibited antagonistic activity against all three nectar yeasts (Table 1).
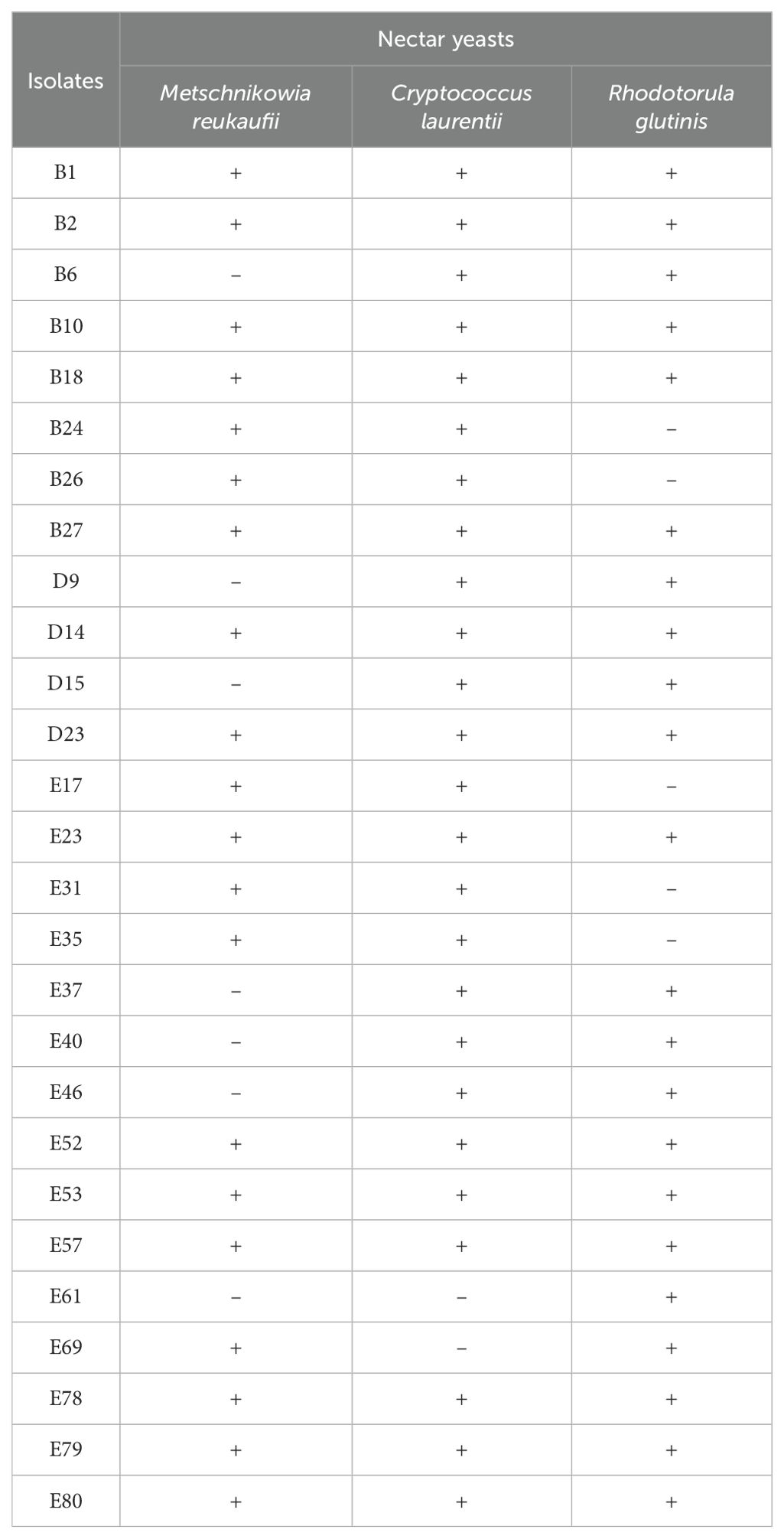
Table 1. Preliminary screening results of endophytic fungi with antagonistic activity (indicated by inhibition zone) against nectar yeasts in C. reticulata pedicels.
In addition, endophytic fungi with antagonistic activity against nectar yeast were again screened using fermentation substances. Our results showed that fermentation substances from isolates with preliminary antagonistic activity also exhibited antagonistic activity against yeasts (Table 2). Inhibition zone measurements revealed that endophytic fungus D23 had the strongest inhibitory effect on all three yeasts, with inhibition zone diameters of 16.37 ± 0.09 mm for M. reukaufii, 15.03 ± 0.65 mm for C. laurentii, and 13.97 ± 0.46 mm for R. glutinis.
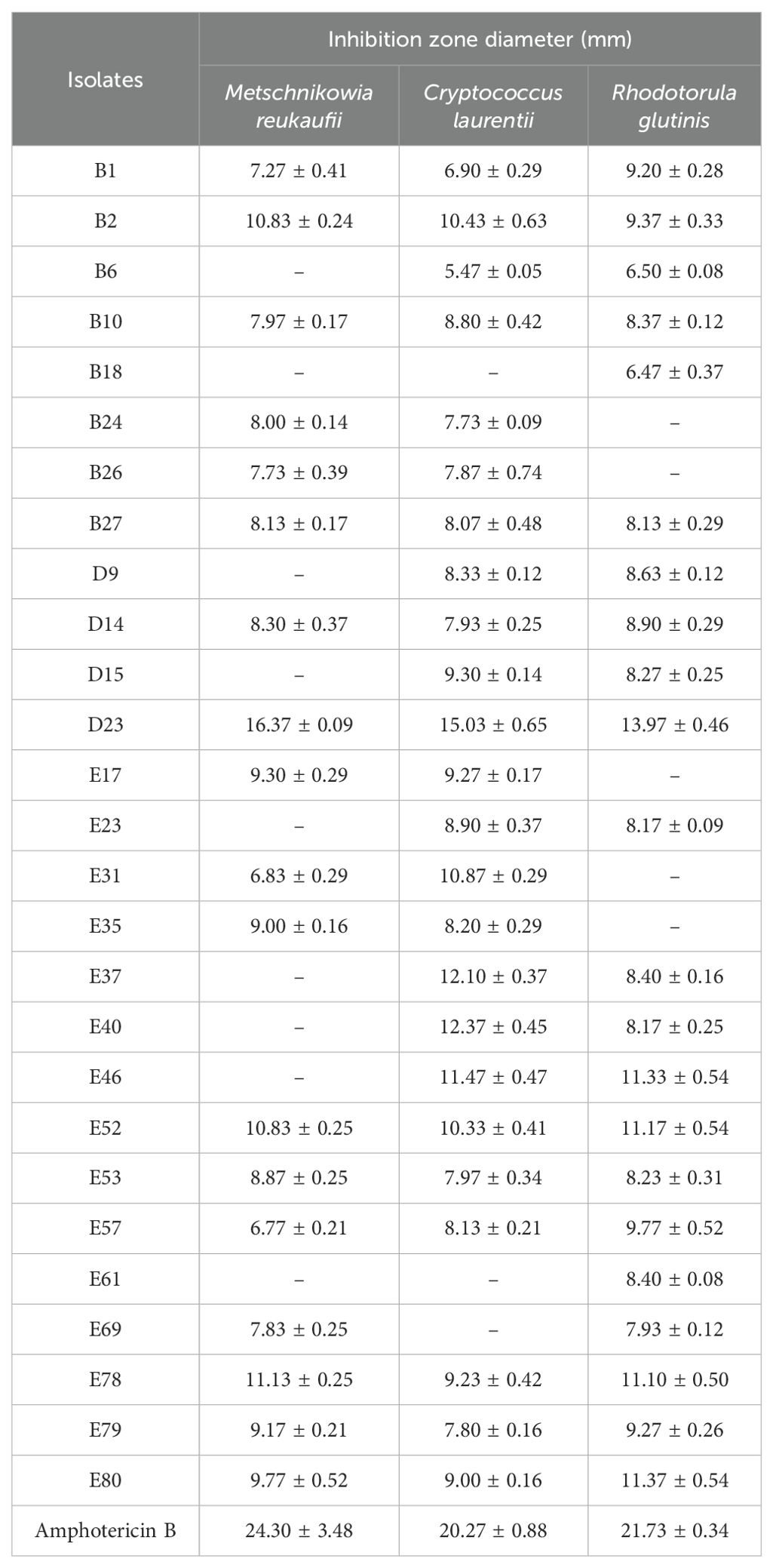
Table 2. Inhibition zone diameters of endophytic fungi with antagonistic activity against nectar yeasts (mean ± S.E., unit: mm).
3.3 Identification of endophytic fungi antagonizing nectar yeasts
3.3.1 Morphological characterization of the isolates
Isolates isolated from the C. reticulata pedicel showed diverse colony features, including color and hyphal characteristics. In early growth stages, colonies were mainly white or gray, with some light yellow or light pink. As colonies grew, colors gradually changed to gray-green, gray-brown, black, or orange. Some produced pigments altering the medium’s color to gray-brown, gray-yellow, light yellow, brown, reddish-brown, brown-yellow, and black (Figure 2).

Figure 2. Colony morphology of endophytic fungi within the C. reticulata pedicel under cultivation conditions of 28°C on PDA medium.
3.3.2 Molecular identification of isolates
Endophytic fungi isolated from the C. reticulata pedicel were all Ascomycota, classified into the Eurotiomycetes, Sordariomycetes, and Dothideomycetes classes. They were further categorized into genera such as Trichoderma, Fusarium, Daldinia, Nodulisporium, Hypoxylon, Colletotrichum, Coniochaeta, Epicoccum, Alternaria, Endomelanconiopsis, and Botryosphaeria (Table 3). Among them, fungus D23, with efficient antagonism against nectar yeast, was identified as Alternaria alternata.
3.4 Antagonistic activity of A. alternata against nectar yeasts
3.4.1 Determination of A. alternata MIC against nectar yeasts
The MIC of A. alternata was below 500 µg/mL for all three tested nectar yeasts, showing significant potential to inhibit them, similar to Amphotericin B. Specifically, A. alternata exhibited the highest antimicrobial activity against M. reukaufii, with a MIC of 125 µg/mL, followed by a MIC of 250 µg/mL against C. laurentii and 500 µg/mL against R. glutinis. For Amphotericin B, MIC values for the three nectar yeasts were 1.95, 3.91, and 3.91 µg/mL, respectively.
3.4.2 Compositional analysis of the A. alternata metabolites
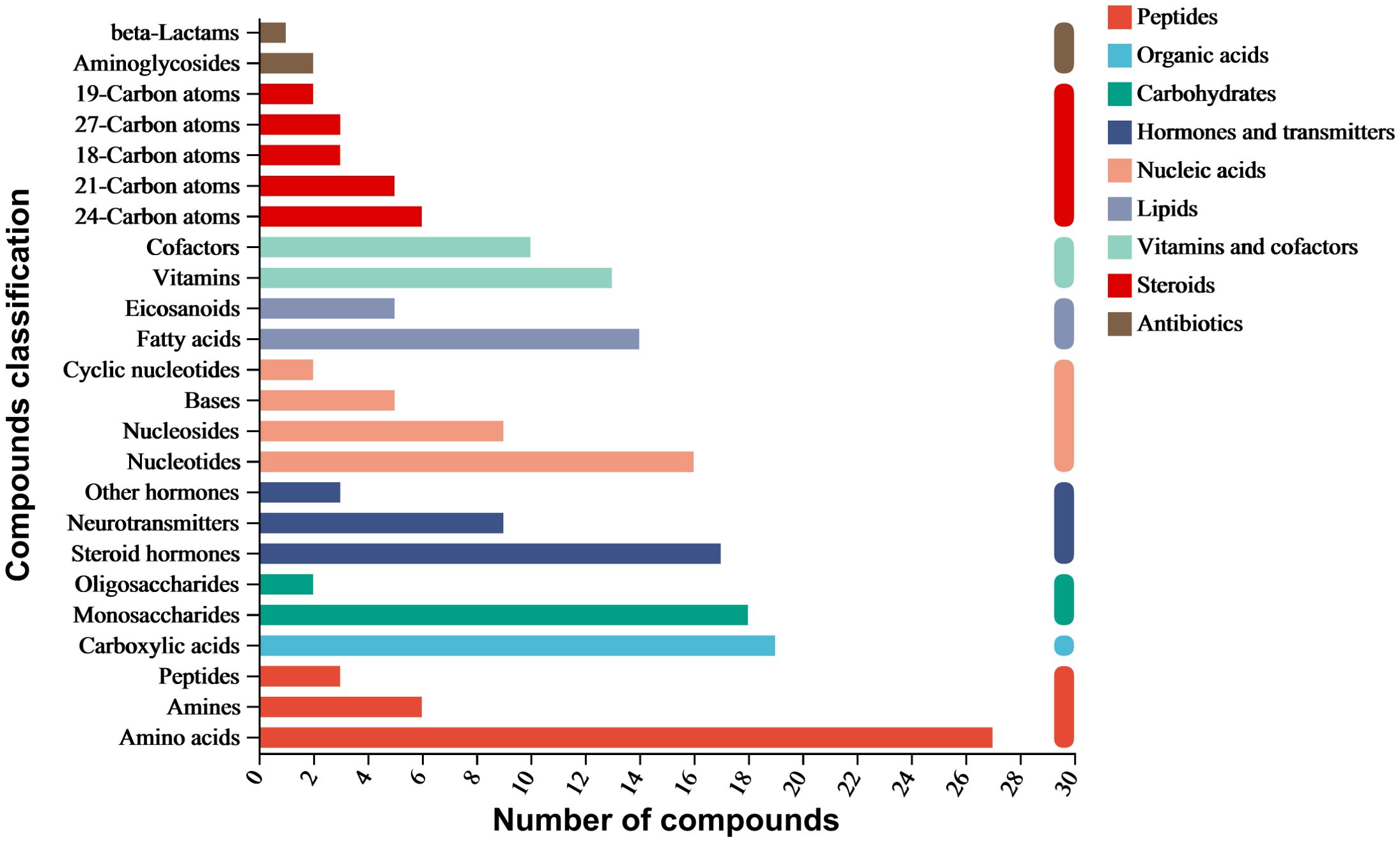
The ethyl acetate extract of A. alternata was analyzed using LC-MS/MS, identifying 4036 metabolites categorized into nine classes: organic acids, lipids, carbohydrates, nucleic acids, peptides, vitamins and cofactors, steroids, hormones and transmitters, and antibiotics. Amino acids had the highest proportion, followed by carboxylic acids, monosaccharides, steroid hormones, nucleotides, fatty acids, vitamins, cofactors, neurotransmitters, nucleosides, and eicosanoids. In addition, aminoglycosides, beta-lactams, quinolones, and other antibiotics were also present (Figure 3).
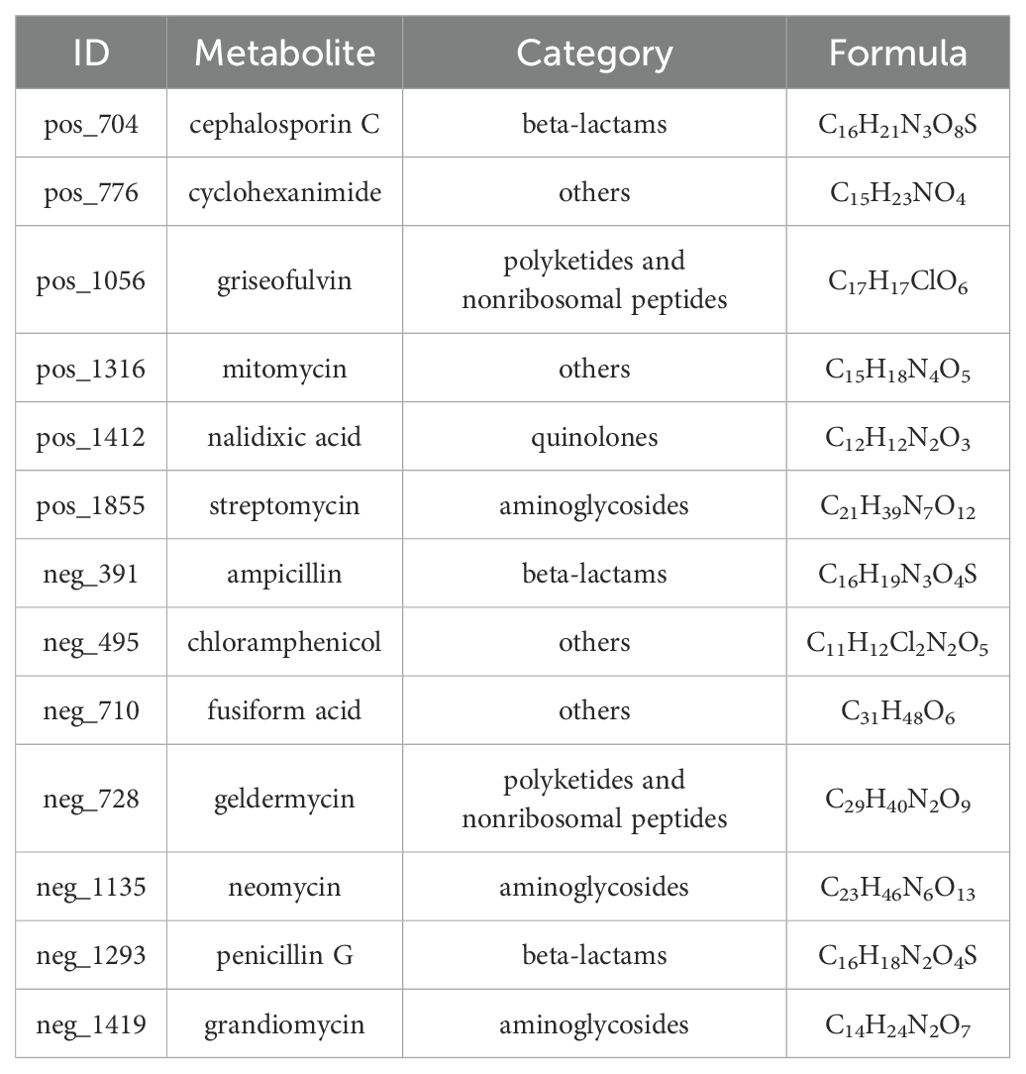
Six antibiotics were detected in positive ion mode: cephalosporin C, cyclohexanimide, griseofulvin, mitomycin, nalidixic acid, and streptomycin. Additionally, seven antibiotics related to antimicrobial activity were detected in negative ion mode: ampicillin, chloramphenicol, fusiform acid, geldermycin, neomycin, penicillin G, and grandiomycin. These antibiotics mainly belong to beta-lactams, polyketides and nonribosomal peptides, quinolones, aminoglycosides, and other protein families (Table 4).
Metabolic pathways involving A. alternata metabolites were mainly related to ABC transporters, biosynthesis of phenylpropanoids, alkaloids derived from the shikimate pathway, nucleotide metabolism, carbon metabolism, tyrosine metabolism, pyrimidine metabolism, and degradation of aromatic compounds (Figure 4).
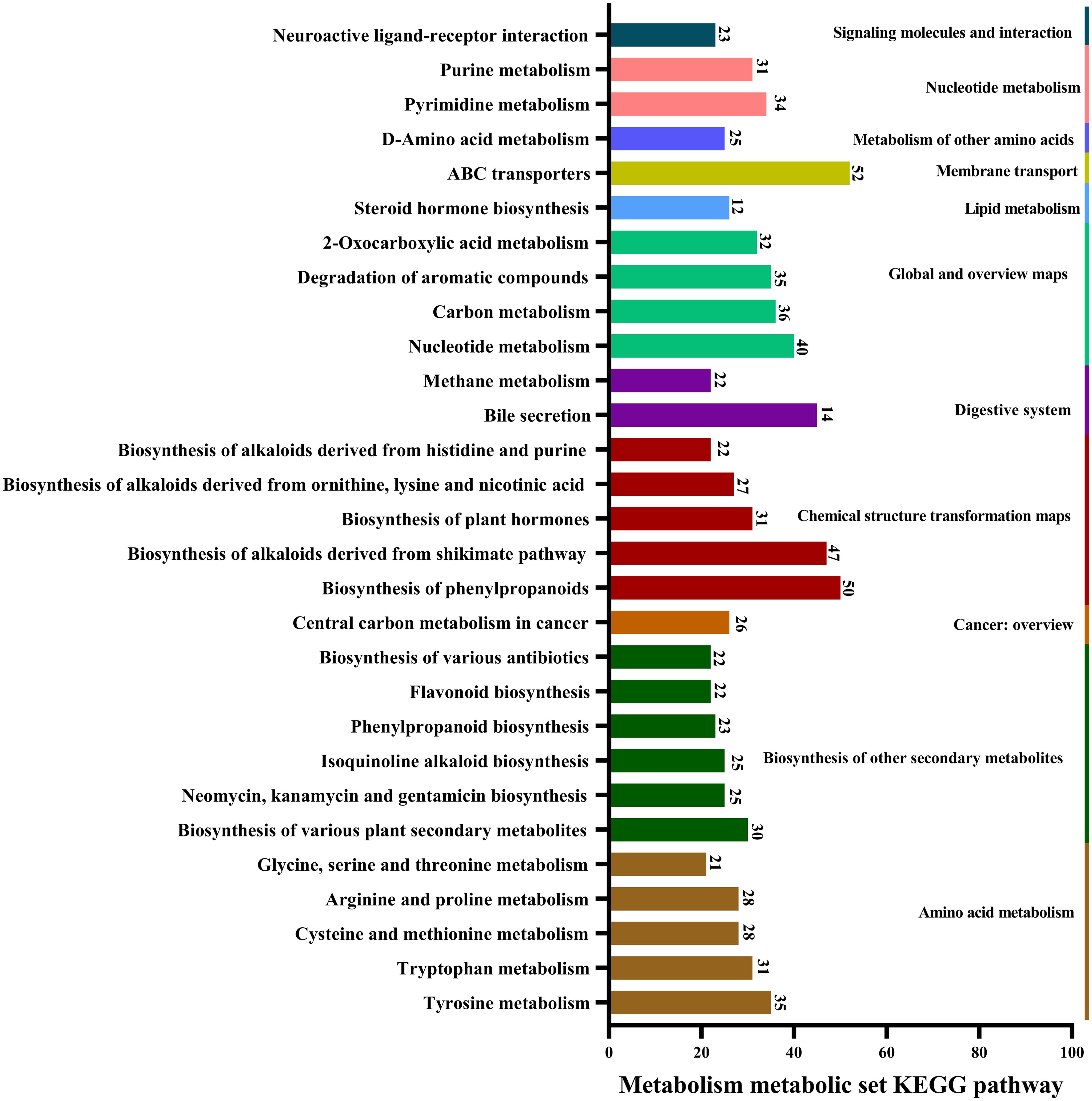
Figure 4. Major pathways involved in the metabolism of A. alternata, revealed by KEGG database annotations.
4 Discussion
Ascomycota and Basidiomycota were the dominant phyla of endophytic fungi found in the pedicel of C. reticulata. Previous studies have shown that in C. oleifera, Ascomycota is the most abundant phylum across all tissues, followed by Basidiomycota (Ercolani, 1991). Similarly, most plants also have Ascomycota as the dominant endophytic phylum, with Basidiomycota as the second most common, such as in Aechmea, Fagopyrum tataricum, and Lunasia amara (Rajamanikyam et al., 2017; Istikorini and Hartoyo, 2019; Leroy et al., 2019; Chen et al., 2024). However, most fungal species in C. reticulata pedicel were only classified to the order level, indicating the potential for discovering new species. Additionally, significant differences in the abundance of endophytic fungi at the family level indicate a diverse distribution of species in C. reticulata. No clear patterns were found in the dominant fungal families in C. reticulata pedicel compared to other Camellia species, possibly due to differences in the plant’s growth environment, which significantly influences endophytic fungal diversity.
Endophytic fungi are widely existed in plant tissues and an increasing number of these fungi have been isolated and identified (Hardoim et al., 2015). Their metabolites can effectively control diseases in agricultural and forestry crop (Abbasi et al., 2002). For instance, Penicillium vinaceum, isolated from Crocus sativus, secretes quinazoline alkaloids and exhibits significant antimicrobial activity against various pathogens (Zheng et al., 2012). Similarly, Phomopsis cassiae, isolated from Cassia spectabilis, produces five different sesquiterpenes and demonstrates strong antimicrobial activity against two plant-pathogenic bacteria (Silva et al., 2006). Nectar is rich in various nutrients, and yeast is commonly found within it. Yeast can significantly alter the composition and concentration of sugars and amino acids in nectar, accelerate flower aging, reduce attractiveness to pollinators, and impact the reproductive fitness of cross-pollinated plants (Herrera et al., 2008; Canto and Herrera, 2012). Given the antibacterial properties of endophytic fungi, they may assist nectar in antagonizing yeast proliferation. In this study, numerous endophytic fungi with inhibitory effects on yeasts have been screened from the C. reticulata pedicel, opening avenues for further research into the role of these endophytes in maintaining nectar stability. However, although research has shown that endophytes can change their colonization sites through rhizosphere competence and motility (Hardoim et al., 2015), the hypothesis that endophytic fungi within the pedicels could migrate to the nectar and influence its composition remains unconfirmed.
Endophytic fungi with antagonistic activity against nectar yeasts, isolated from the pedicel of C. reticulata under laboratory conditions, belong to Ascomycota phylum. Numerous species from the genera Trichoderma, Penicillium, and Fusarium are recognized as effective biocontrol agents. Fungi with antagonistic properties identified in previous studies predominantly belong to the Ascomycota, aligning with the findings of this study (Arnold and Lutzoni, 2007). The genus Trichoderma is a highly competitive biocontrol agent, with extensive research and applications documented (Peng et al., 2021). Trichoderma sp. protects several crops from diseases (Youssef et al., 2016; Swain et al., 2018). Penicillium, another endophytic genus, produces various antimicrobial compounds that have been used in crop disease and nematode control (Chen et al., 2022). Su et al. (2010) reported antibacterial activity in the culture filtrate of Aspergillus endophytes from tea trees, effective against Camellia brown spot disease. Furthermore, this study identified two antagonistic endophytic fungi only at the genus level, highlighting the potential for new species discovery in C. reticulata.
The identified A. alternata exhibited superior inhibition of nectar yeasts, affecting M. reukaufii, C. laurentii, and R. glutinis. Secondary metabolites from the genus Alternaria include pyrones, quinones, alkaloids, steroids, and terpenes, all of which demonstrate significant antimicrobial activity (Tantry et al., 2018; Lee et al., 2019; Chen et al., 2020). Specifically, capsaicin from A. alternata has been shown to exert antimicrobial effects against Staphylococcus aureus, Escherichia coli, and Aspergillus niger (Devari et al., 2014). Additionally, naphthalene and pyrone compounds from marine-derived A. alternata inhibit Candida albicans and Fusarium oxysporum (Shaaban et al., 2012). Alternaramide, a cyclic peptide from marine-derived A. alternata, is effective against Bacillus subtilis and Staphylococcus aureus (Kim et al., 2009). The identified metabolites in this study include antibiotics with antimicrobial properties, such as chloramphenicol, penicillin G, grandiomycin, and cephalosporin C, thus expanding the known data on A. alternata metabolites. Moreover, key metabolic pathways in A. alternata encompass the biosynthesis of plant secondary metabolites, phenylpropanoids, amino acids, antibiotics, and alkaloids. This analysis provides a basis for further research into its applications and mechanisms.
Importantly, nectar generally contains antimicrobial substances that are transported through the phloem and xylem during the nectar secretion process, enhancing pollination services and safeguarding rewards for flowers and/or pollinators (Adler, 2000; Stevenson et al., 2017). The nectar of C. reticulata is abundant and it depends on pollinators for fertilization. Endophytic fungi colonize the pedicels and produce a wealth of secondary metabolites, including various antibiotics. Under laboratory conditions, some of these fungi have demonstrated promising inhibitory effects against nectar yeasts. Our research findings provide insights into potential strategies that cross-pollinated plants may employ to cope with yeast stress, regulate nectar stability, and maintain reproductive fitness. However, further research is needed to determine whether metabolites secreted by endophytic fungi are transported from the phloem sieve tubes to the nectar to exert their effects, along with the underlying mechanisms and potential applications in antagonistic nectar yeasts.
5 Conclusion
Endophytic fungal diversity in pedicels of C. reticulata was high, primarily comprising Ascomycota and Basidiomycota. Twenty-seven isolates of endophytic fungi that exhibited antagonistic activity against three types of nectar yeasts were isolated and cultured. These isolates exhibited diverse morphologies and were identified into genera such as Trichoderma, Fusarium, and Alternaria. Among them, isolate D23, identified as A. alternata, demonstrated the highest antimicrobial potential. Its metabolites included antibiotics like penicillin G, grandiomycin, and cephalosporin C, and it participated in metabolic pathways related to phenylpropanoid and alkaloid biosynthesis. This research provided insights into the antagonistic effects of endophytic fungal metabolites against nectar yeasts in C. reticulata pedicels, emphasizing the importance of endophytes in assisting nectar maintain stability and support reproductive fitness in cross-pollinating plants.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.
Author contributions
RH: Formal analysis, Software, Visualization, Writing – original draft, Writing – review & editing. QM: Software, Visualization, Writing – original draft, Writing – review & editing. LX: Formal analysis, Writing – original draft. XW: Methodology, Writing – original draft. DY: Formal analysis, Writing – original draft. WZ: Methodology, Writing – original draft. XD: Conceptualization, Writing – original draft. XG: Writing – review & editing. KD: Data curation, Funding acquisition, Project administration, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by National Natural Science Foundation of China (32060241 and 31572339), the China Agriculture Research System of the Ministry of Finance and the Ministry of Agriculture and Rural Affairs (CARS-44-KXJ13), and the Reserve Talents Training Program for Young and Middle-Aged Academic and Technical Leaders in Yunnan (2018HB041).
Acknowledgments
We gratefully acknowledge the support of the Forestry Bureau of Tengchong, Yunnan Province, and the Tengchong Camellia Resource Bank at Shaba Forestry Farm for their contributions to this project, as well as MJEditor (www.mjeditor.com) for their English editing services during the preparation of this manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abbasi, P. A., Al-Dahmani, J., Sahin, F., Hoitink, H. A. J., Miller, S. A. (2002). Effect of compost amendments on disease severity and yield of tomato in conventional and organic production systems. Plant Dis. 86, 156–161. doi: 10.1094/pdis.2002.86.2.156
Adler, L. S. (2000). The ecological significance of toxic nectar. Oikos 91, 409–420. doi: 10.1034/j.1600-0706.2000.910301.x
Álvarez-Pérez, S., Lievens, B., Jacquemyn, H., Herrera, C. M. (2013). Acinetobacter nectaris sp. nov. and Acinetobacter boissieri sp. nov., isolated from floral nectar of wild Mediterranean insect-pollinated plants. Int. J. Systematic Evolutionary Microbiol. 63, 1532–1539. doi: 10.1099/ijs.0.043489-0
Arnold, A. E., Lutzoni, F. (2007). Diversity and host range of foliar fungal endophytes: are tropical leaves biodiversity hotspots? Ecology 88, 541–549. doi: 10.1890/05-1459
Brysch-Herzberg, M. (2004). Ecology of yeasts in plant–bumblebee mutualism in Central Europe. FEMS Microbiol. Ecol. 50, 87–100. doi: 10.1016/j.femsec.2004.06.003
Canto, A., Herrera, C. M. (2012). Micro-organisms behind the pollination scenes: microbial imprint on floral nectar sugar variation in a tropical plant community. Ann. Bot. 110, 1173–1183. doi: 10.1093/aob/mcs183
Cardoso, V. M., Campos, F. F., Santos, A. R. O., Ottoni, M. H. F., Rosa, C. A., Almeida, V. G., et al. (2020). Biotechnological applications of the medicinal plant Pseudobrickellia brasiliensis and its isolated endophytic bacteria. J. Appl. Microbiol. 129, 926–934. doi: 10.1111/jam.14666
Chen, S., Deng, Y., Yan, C., Wu, Z., Guo, H., Liu, L., et al. (2020). Secondary metabolites with nitric oxide inhibition from marine-derived fungus Alternaria sp. 5102. Mar. Drugs 18, 426. doi: 10.3390/md18080426
Chen, M., Ding, Z., Zhou, M., Shang, Y., Li, C., Li, Q., et al. (2024). The diversity of endophytic fungi in Tartary buckwheat (Fagopyrum tataricum) and its correlation with flavonoids and phenotypic traits. Front. Microbiol. 15. doi: 10.3389/fmicb.2024.1360988
Chen, S., Tian, D., Wei, J., Li, C., Ma, Y., Gou, X., et al. (2022). Citrinin derivatives from Penicillium Citrinum Y34 that inhibit α-Glucosidase and ATP-Citrate lyase. Front. Mar. Sci. 9. doi: 10.3389/fmars.2022.961356
Compant, S., Duffy, B., Nowak, J., Clément, C., Barka, E. A. (2005). Use of plant growth-promoting bacteria for biocontrol of plant diseases: principles, mechanisms of action, and future prospects. Appl. Environ. Microbiol. 71, 4951–4959. doi: 10.1128/AEM.71.9.4951-4959.2005
Devari, S., Jaglan, S., Kumar, M., Deshidi, R., Guru, S., Bhushan, S., et al. (2014). Capsaicin production by Alternaria alternata, an endophytic fungus from Capsicum annum; LC–ESI–MS/MS analysis. Phytochemistry 98, 183–189. doi: 10.1016/j.phytochem.2013.12.001
Ercolani, G. L. (1991). Distribution of epiphytic bacteria on olive leaves and the influence of leaf age and sampling time. Microbial Ecol. 21, 35–48. doi: 10.1007/BF02539143
Feng, G. (1980). Species and Utilization of Camellia in Yunnan (Kunming: Yunnan people’s Publishing House).
Gardes, M., Bruns, T. D. (1993). ITS primers with enhanced specificity for basidiomycetes - application to the identification of mycorrhizae and rusts. Mol. Ecol. 2, 113–118. doi: 10.1111/j.1365-294X.1993.tb00005.x
González-Mas, N., Cuenca-Medina, M., García-Mozo, H., Muñoz-Redondo, J. M., Moreno-Rojas, J. M., Padilla-Álvarez, F., et al. (2023). Endophytic Beauveria bassiana modifies flowering phenology, floral volatile profile and pollinator behaviour in melon. Entomologia Generalis 43, 961–969. doi: 10.1127/entomologia/2023/1991
Hardoim, P. R., Overbeek, L. S. V., Berg, G., Pirttilä, A. M., Compant, S., Campisano, A., et al. (2015). The hidden world within plants: Ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol. Mol. Biol. Rev. 79, 293–320. doi: 10.1128/MMBR.00050-14
Herrera, C. M., de Vega, C., Canto, A., Pozo, M. I. (2009). Yeasts in floral nectar: a quantitative survey. Ann. Bot. 103, 1415–1423. doi: 10.1093/aob/mcp026
Herrera, C. M., García, I. M., Pérez, R. (2008). Invisible floral larcenies: microbial communities degrade floral nectar of bumble bee-pollinated plants. Ecology 89, 2369–2376. doi: 10.1890/08-0241.1
Herrera, C. M., Pozo, M. I., Bazaga, P. (2011). Clonality, genetic diversity and support for the diversifying selection hypothesis in natural populations of a flower-living yeast. Mol. Ecol. 20, 4395–4407. doi: 10.1111/j.1365-294X.2011.05217.x
Hiremani, N. S., Verma, P., Gawande, S. P., Sain, S. K., Nagrale, D. T., Salunkhe, V. N., et al. (2020). Antagonistic potential and phylogeny of culturable endophytic fungi isolated from desi cotton (Gossypium arboreum L.). South Afr. J. Bot. 134, 329–335. doi: 10.1016/j.sajb.2020.03.008
Istikorini, Y., Hartoyo, A. P. P. (2019). The diversity of endophytic fungi in kemaitan (Lunasia amara blanco). IOP Conf. Series: Earth Environ. Sci. 394, 12016. doi: 10.1088/1755-1315/394/1/012016
Jacquemyn, H., Lenaerts, M., Tyteca, D., Lievens, B. (2013). Microbial diversity in the floral nectar of seven Epipactis (Orchidaceae) species. MicrobiologyOpen 2, 644–658. doi: 10.1002/mbo3.103
Kim, M. Y., Sohn, J. H., Ahn, J. S., Oh, H. (2009). Alternaramide, a cyclic depsipeptide from the marine-derived fungus Alternaria sp. SF-5016. J. Natural Products 72, 2065–2068. doi: 10.1021/np900464p
Lee, C., Li, W., Bang, S., Lee, S. J., Kang, N., Kim, S., et al. (2019). Secondary metabolites of the endophytic fungus Alternaria alternata JS0515 isolated from Vitex rotundifolia and their effects on pyruvate dehydrogenase activity. Molecules. 24, 4450. doi: 10.3390/molecules24244450
Leroy, C., Maes, A. Q., Louisanna, E., Séjalon-Delmas, N. (2019). How significant are endophytic fungi in bromeliad seeds and seedlings? Effects on germination, survival and performance of two epiphytic plant species. Fungal Ecol. 39, 296–306. doi: 10.1016/j.funeco.2019.01.004
Peay, K. G., Belisle, M., Fukami, T. (2012). Phylogenetic relatedness predicts priority effects in nectar yeast communities. Proc. R. Soc. B: Biol. Sci. 279, 749–758. doi: 10.1098/rspb.2011.1230
Peng, K. C., Lin, C. C., Liao, C. F., Yu, H. C., Lo, C. T., Yang, H. H., et al. (2021). Expression of L-amino acid oxidase of Trichoderma harzianum in tobacco confers resistance to Sclerotinia sclerotiorum and Botrytis cinerea. Plant Sci. 303, 110772. doi: 10.1016/j.plantsci.2020.110772
Philippot, L., Raaijmakers, J. M., Lemanceau, P., van der Putten, W. H. (2013). Going back to the roots: the microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 11, 789–799. doi: 10.1038/nrmicro3109
Pieterse, C. M. J., Zamioudis, C., Berendsen, R. L., Weller, D. M., Wees, S. C. M. V., Bakker, P. A. H. M. (2014). Induced systemic resistance by beneficial microbes. Annu. Rev. Phytopathol. 52, 347–375. doi: 10.1146/annurev-phyto-082712-102340
Pozo, M. I., Lachance, M. A., Herrera, C. M. (2012). Nectar yeasts of two southern Spanish plants: the roles of immigration and physiological traits in community assembly. FEMS Microbiol. Ecol. 80, 281–293. doi: 10.1111/j.1574-6941.2011.01286.x
Pusey, P. L. (1999). Effect of nectar on microbial antagonists evaluated for use in control of fire blight of pome fruits. Phytopathology 89, 39–46. doi: 10.1094/phyto.1999.89.1.39
Qin, S., Chen, K., Zhang, W., Xiang, X., Zuo, Z., Guo, C., et al. (2023). Phylogenomic insights into the reticulate evolution of Camellia sect. Paracamellia Sealy (Theaceae). J. Systematics Evol. 62, 38–54. doi: 10.1111/jse.12948
Rajamanikyam, M., Vadlapudi, V., Upadhyayula, S. M. (2017). Endophytic fungi as novel resources of natural therapeutics. Braz. Arch. Biol. Technol. 60, e17160542. doi: 10.1590/1678-4324-2017160542
Richardson, S. N., Nsiama, T. K., Walker, A. K., McMullin, D. R., Miller, J. D. (2015). Antimicrobial dihydrobenzofurans and xanthenes from a foliar endophyte of Pinus strobus. Phytochemistry 117, 436–443. doi: 10.1016/j.phytochem.2015.07.009
Schaeffer, R. N., Mei, Y. Z., Andicoechea, J., Manson, J. S., Irwin, R. E. (2017). Consequences of a nectar yeast for pollinator preference and performance. Funct. Ecol. 31, 613–621. doi: 10.1111/1365-2435.12762
Shaaban, M., Shaaban, K. A., Abdel-Aziz, M. S. (2012). Seven naphtho-γ-pyrones from the marine-derived fungus Alternaria alternata: structure elucidation and biological properties. Organic medicinal Chem. Lett. 2, 1–8. doi: 10.1186/2191-2858-2-6
Silva, G. H., Teles, H. L., Zanardi, L. M., Marx Young, M. C., Eberlin, M. N., Hadad, R., et al. (2006). Cadinane sesquiterpenoids of Phomopsis cassiae, an endophytic fungus associated with Cassia spectabilis (Leguminosae). Phytochemistry 67, 1964–1969. doi: 10.1016/j.phytochem.2006.06.004
Stevenson, P. C., Nicolson, S. W., Wright, G. A. (2017). Plant secondary metabolites in nectar: impacts on pollinators and ecological functions. Funct. Ecol. 31, 65–75. doi: 10.1111/1365-2435.12761
Stinson, M., Ezra, D., Hess, W. M., Sears, J., Strobel, G. (2003). An endophytic Gliocladium sp. of Eucryphia cordifolia producing selective volatile antimicrobial compounds. Plant Sci. 165, 913–922. doi: 10.1016/S0168-9452(03)00299-1
Su, J., Wang, G., Yang, M. (2010). Mixed culture of endophytic fungi isolated from Camellia sinensis enhancing the antagonistic on plant pathogenic fungi. Mycosystema 29, 753–759. doi: 10.13346/j.mycosystema.2010.05.013
Swain, H., Adak, T., Mukherjee, A. K., Mukherjee, P. K., Bhattacharyya, P., Behera, S., et al. (2018). Novel Trichoderma strains isolated from tree barks as potential biocontrol agents and biofertilizers for direct seeded rice. Microbiological Res. 214, 83–90. doi: 10.1016/j.micres.2018.05.015
Tantry, M. A., Idris, A. S., Williamson, J. S., Shafi, T., Dar, J. S., Malik, T. A., et al. (2018). Perylenequinones from an endophytic Alternaria sp. of Pinus ponderosa. Heliyon 4, e01046. doi: 10.1016/j.heliyon.2018.e01046
Wang, H., Liu, Z., Duan, F., Chen, Y., Qiu, K., Xiong, Q., et al. (2023). Isolation, identification, and antibacterial evaluation of endophytic fungi from Gannan navel orange. Front. Microbiol. 14, 1172629. doi: 10.3389/fmicb.2023.1172629
White, T. J., Bruns, T. D., Lee, S. B., Taylor, J. W. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR Protocols: a guide to methods and applications. Eds. Innis, M. A., Gelfand, D. H., Sninsky, J. J., White, T. J. (Academic Press, San Diego), 315–322.
Youssef, S. A., Tartoura, K. A., Abdelraouf, G. A. (2016). Evaluation of Trichoderma harzianum and Serratia proteamaculans effect on disease suppression, stimulation of ROS-scavenging enzymes and improving tomato growth infected by Rhizoctonia solani. Biol. Control 100, 79–86. doi: 10.1016/j.biocontrol.2016.06.001
Zhang, H., Song, Y., Tan, R. (2006). Biology and chemistry of endophytes. Natural Product Rep. 23, 753–771. doi: 10.1039/B609472B
Zheng, C., Li, L., Zou, J., Han, T., Qin, L. (2012). Identification of a quinazoline alkaloid produced by Penicillium vinaceum, an endophytic fungus from Crocus sativus. Pharm. Biol. 50, 129–133. doi: 10.3109/13880209.2011.569726
Keywords: Camellia reticulata, endophytic fungi, nectar yeasts, antagonistic activity, antimicrobial mechanism, Alternaria alternata
Citation: Huang R, Meng Q, Xun L, Wu X, Yue D, Zhao W, Dong X, Gong X and Dong K (2024) Antagonistic effects of endophytic fungi from Camellia reticulata pedicels on yeasts: implications for antimicrobial mechanism of nectar. Front. Plant Sci. 15:1494855. doi: 10.3389/fpls.2024.1494855
Received: 11 September 2024; Accepted: 30 October 2024;
Published: 15 November 2024.
Edited by:
Marie-Joelle Virolle, Centre National de la Recherche Scientifique (CNRS), FranceReviewed by:
Ann C. Lawrie, RMIT University, AustraliaHany H. A. El-Sharkawy, Agricultural Research Center, Egypt
Copyright © 2024 Huang, Meng, Xun, Wu, Yue, Zhao, Dong, Gong and Dong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xueyang Gong, eHVleWFuZ2cxMUAxMjYuY29t; Kun Dong, ZG9uZ2t1bjE5NzIyMDA0QGFsaXl1bi5jb20=
†These authors have contributed equally to this work and share first authorship
 Rong Huang
Rong Huang Qingxin Meng
Qingxin Meng Lijie Xun1,2
Lijie Xun1,2 Kun Dong
Kun Dong