- 1State Key Laboratory of Subtropical Silviculture, Zhejiang A&F University, Hangzhou, China
- 2Plant Physiology Laboratory, Department of Botany, Chaudhary Mahadeo Prasad (C.M.P.) Degree College, University of Allahabad, Prayagraj, India
Auxins play significant roles in plant growth and development. The transporter inhibitor response1/auxin signaling F-box (TIR1/AFB) gene family encodes the auxin receptor proteins and plays an essential role in the auxin signaling pathway. Here we identified and characterized the TIR1/AFB family in Carya cathayensis (Cc) plants (named as CcTIR1/AFB). Seven CcTIR1/AFBs were identified and further confirmed by cloning. All proteins encoded by these genes conservatively contained two domains, the F-box and leucine-rich repeat (LRR) domains. The CcTIR1/AFBs were located in the nucleus. Phylogenetic analysis suggested that CcTIR1/AFBs were evenly scattered in four different subgroups. The cis-acting element analysis indicates that CcTIR1/AFBs might be activated by auxin. The spatial and temporal expression of CcTIR1/AFBs during grafting suggested that both CcAFB1 and CcAFB2 in scions and CcAFB4 in the rootstocks were significantly upregulated at 3 days after grafting, which indicated the specialization of three CcAFBs during grafting. The Y2H assay indicated that three CcAFBs were capable of interacting with CcIAA16, CcIAA27b, and CcIAA29a, among which CcAFB4 interacted strongly with CcIAA1 and CcIAA16. Our study provides the opportunity to understand the potential role of not only CcTIR1/AFBs but also special CcAFBs (CcAFB1, CcAFB2, and CcAFB4), which is a great aspect to further explore the molecular mechanism during the grafting process.
1 Introduction
The auxins play a significant role in the processes of plant development and growth, including embryogenesis, vascular formation, and flower development (Zhao, 2010). The core components of auxin signaling pathway come from three protein families: the commonly acknowledged auxin receptor protein, TIR1/AFB (transporter inhibitor response1/auxin signaling F-box); the transcriptional repressor, Aux/IAA (auxin/indol-3-acetic acid); and the transcription factor, ARF (auxin response factor) (Lavy and Estelle, 2016). TIR1/AFB is one of the key elements of SCFTIR1/AFB complex which serves as the ubiquitin–protein ligase catalyzing the transfer of ubiquitin from E2 (ubiquitin-conjugating enzyme) to the target protein (Dharmasiri et al., 2005; Parry et al., 2009). Aux/IAAs play the role of a transcriptional repressor via forming the heterodimers with ARFs and preventing the interaction between ARFs and DNA sequences (Luo et al., 2018). The ARFs, as the transcription factor, are able to form the homodimer which bind the specific cis-element AuxRE (auxin response element) and activate the expression of genes downstream from the AuxRE (Chandler, 2016). The IAA with high concentration in the nucleus contributes to the formation of TIR/AFB–IAA–Aux/IAA complex and triggers the Aux/IAA degradation, which results in the release of ARFs from ARF–Aux/IAA heterodimer and the activation of gene expression regulated by ARFs. On the contrary, the function of ARFs is repressed at a low concentration of IAA because of the failed formation of the Aux/IAA degradation complex (Guo et al., 2021). Jogawat et al. (2021) have done an in-depth analysis of auxin crosstalk with other hormones along with TIR1 gene function in plants.
There are six members of the TIR1/AFB protein family which are involved in a variety of development processes and display extensive biological functions in Arabidopisis. TIR1 and AFB2, with the ability to interact much stronger with Aux/IAA proteins than AFB1 and AFB3 in vitro, are the principal auxin receptors in roots (Parry et al., 2009). AFB3 is involved in auxin signaling modulated by nitrate and participates in the alteration of the root system architecture (Vidal et al., 2010). AFB4 has a significant role in seedlings in response to the elevated temperature, and AFB5 is the principal target of picloram family (Greenham et al., 2011; Prigge et al., 2016). The TIR1/AFB protein contains two critical conserved domains, the F-box domain and the transporter inhibitor response 1 domain (also called leucine-rich repeat (LRR) domain) (Hayashi et al., 2008). The former is capable of interacting with the SPK subunit in the SCF complex, and the latter displayed as a pocket to bind auxin and to interact with Aux/IAA (Kepinski, 2007). However, the reports related to the function of TIR1/AFB protein in the other species are limited.
Plant grafting, an ancient vegetative propagation technique, which connects two cut parts (known as scion and rootstock) originated from either the same or different plant species, is widely used in agriculture (Wang et al., 2017). For plants, grafting contributes to many optimized attributes, like the improvement of tolerance against biotic and abiotic stresses, augmentation of yield, and elongation of the harvest phase (Lee et al., 2010). In Arabidopsis, a successful graft union formation starts with necrotic layer development, followed by callus tissue formation and new vascular tissue differentiation within the scion and finished with vascular reconnection between scion and rootstock (Flaishman et al., 2008). Plant hormones such as cytokinin, ethylene, jasmonic acid, and auxin are critical regulators in response to wounding and grafting (Asahina et al., 2011; Iwase et al., 2011). Particularly, auxin plays an important role in vascular development via the TIR1/AFB signaling pathway (Mazur et al., 2020). In Arabidopsis, iaa18 mutant and tir1afb2afb3 triple mutant are severely incapable of reconnecting the phloem after grafting (Melnyk et al., 2015). In rice, Ostir1Osafb2 double mutant is no longer capable of generating calli even when being treated with 10 µM of 2,4-dichlorophenoxyacetic acid (Guo et al., 2021). Besides this, PtrFBL1 and PtrFBL7, the homologs of TIR1/AFBs in Populus, are mainly expressed in cambial and vascular tissues (Shu et al., 2015).
Chinese hickory (Carya cathayensis Sarg.) is a well-known economic tree and popular in China for its extremely nutritious nuts, containing high amounts of polyunsaturated fatty acids which aid in the brain development (Huang et al., 2022). However, the long juvenile phase of Chinese hickory seriously represses the enlargement of its industry and the increment of fruit growers’ wealth. Grafting, as an asexual reproduction technology in plant, is capable of shortening the juvenile phase and facilitates the development of the hickory industry efficiently. It is the vascular reconnection between the scion and the rootstock that plays a critical role in avoiding the failure of grafting as reported in previous research (Loupit et al., 2023; Melnyk et al., 2015). A majority of evidence indicates that auxin plays a central role in the progress of plant vascular formation (Sharma and Zheng, 2019). However, knowledge about the involvement of auxin in grafting is still limited. In the current investigation, the CcTIR1/AFB gene family, as the auxin receptor, was identified and characterized. Additionally, the potential function of this family during the grafting process in Chinese hickory was further explored in detail. Results of the current investigation may be helpful in further understanding the molecular mechanisms regulated by the CcTIR1/AFB gene family during the grafting process.
2 Materials and methods
2.1 Plant materials
Regarding the grafting union, the rootstock is derived from 2-year-old Chinese hickory trees cultivated for 5 months before grafting in the greenhouse of Zhejiang Agriculture and Forestry University (temperature 25 ± 1°C, 12 h day/12 h night photoperiod, 60%–70% relative humidity, watered once a week), and the scions were collected from the stem of 1-year-old Chinese hickory trees (Yuan et al., 2018).
The conjunction parts of the grafted individuals were sampled at 0, 3, 7, and 14 days (Yang et al., 2022) after grafting, and the sampling time points were determined based on previous research about the changing of morphology during the process of the grafted Chinese hickory’s being successfully reconnected between scions and rootstocks and being alive. The 0-day-after-grafting unions which were sampled just after grafting were taken as the control.
2.2 Isolation and cloning of TIR1/AFB gene family
The seven TIR1/AFB genes were screened and identified from published Chinese hickory genome (Huang et al., 2019). The total RNA as the template for the reverse transcriptase in the subsequence experiments was extracted from the tissue of hickory using TaKaRa MiniBEST Universal RNA Extraction Kit (Takara Bio Inc., Kusatsu, Japan). The cDNA synthesis reaction was performed with the PrimeScript™ II 1st Strand cDNA Synthesis Kit and conducted according to its manufacturer’s protocol. For the first step of that reaction, 1 μg total RNA extracted previously was mixed firstly with 1 μL dNTP mixture (10 Mm each), 1 μL Oligo dT Primer (50 Mm), and the RNase Free dH2O up to the volume of 10 μL. Then, the mixture was kept at the condition of 65°C for 5 min and cooled by burying in ice. For the second step, the 10-μL mixture from step one was infused with 4 μL 5×PrimeScript II Buffer, 0.5 μL RNase Inhibitor (40 U/μL), 1 μL PrimeScript II Rtase (200 U/μL), and RNase Free dH2O up to the volume of 20 μL in the PCR tube. The PCR tube was put into the thermal cycler and operated with the program as follows: heating at 42°C for 60 min and 70°C for 15 min. The TIR1/AFB genes were cloned as per the next several procedures. cDNA was taken as the template for the amplification of target genes with PrimeSTAR Max Premix (2 ×) (Takara Bio Inc., Kusatsu, Japan). The PCR product was purified with FastPure Gel DNA Extraction Mini Kit (Vazyme, Nanjing, China). The purified products were ligated to the cloning vector, pEASY-Blunt Zero, from pEASY-Blunt Zero Cloning Kit (TransGen Biotech, Beijing, China) and transformed into DH5α competent cells of Escherichia coli (TransGen Biotech, Beijing, China). The clones grown on selected medium were picked and sent to Sangon Biotech (Shanghai) Co., Ltd. (China) for sequencing.
2.3 Sequence analysis of TIR1/AFB family genes derived from Chinese hickory
The TIR1/AFB family protein sequences of three other species (Oryza sativa, Populus, and Arabidopsis) were downloaded from Phytozome V13 database (https://phytozome-next.jgi.doe.gov/). Coupled with the seven CcTIR1/AFB protein sequences, multiple sequence alignment was performed with ClustalW, and then the phylogenetic tree of the TIR1/AFB family of the four species was constructed based on the neighbor-joining method associated with a bootstrap of 1,000 replicates on the MEGA X software.
The conserved motif distribution was predicted with protein sequences using the MEME Suite set with eight motifs. The exon–intron structure information was acquired from the Chinese hickory genome. Both the motif distribution and the gene structure were visualized with Tbtools (Chen et al., 2020). The protein sequences of TIR1/AFBs were taken to conduct the conserved domain prediction with CDD (conserved domain database) in NCBI and then visualized with R package, ggplot2.
2.4 cis-acting element analysis of CcTIR1/AFB promoters
The upstream 2,000-bp sequence of CcTIR1/AFB genes, as the promoter of each gene, was acquired from Carya cathayensis genome. cis-acting regulatory element analysis was performed for element prediction after submitting the promoter sequences to the Plant-CARE website (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/). The elements related with light, stress, and phytohormone were selected for further analysis. The results were visualized with R package, ggplot2.
2.5 Subcellular localization
The TIR/AFB family genes ligated to pEASY-Blunt Zero vector were amplified by PCR and purified. The pCambia1300-GFP vector was cut and linearized with two restriction endonucleases, KpnI and XbaI (Takara Bio Inc., Kusatsu, Japan). The amplified fragments were inserted into the pCambia1300-GFP vector by homologous recombination with ClonExpress II One Step Cloning Kit (Vazyme, Nanjing, China). The constructed vector for subcellular localization was transformed into DH5α competent cells of Escherichia coli (TransGen Biotech, Beijing, China). The clones were picked and sent to Sangon Biotech (Shanghai) Co., Ltd. (China) for sequencing. The recombinant plasmids were extracted from DH5α and transformed into GV3101 chemically competent cells of Agrobacterium tumefaciens (Weidi Bio., Shanghai, China). The tobaccos were planted and cultivated for some 6 weeks based on previous research (Yang et al., 2021). The target genes having been integrated into GV3101 are transiently co-transformed into the tobacco leaves associated with nuclear-localized marker. After cultivation in the dark for 48 h, the tobacco leaves were cut and observed to detect fluorescent signals using a confocal microscope with argon lasers at the wavelength of 488 and 594 nm.
2.6 RNA extraction and quantitative RT-PCR
In order to uncover the function of TIR1/AFB in the process of growing grafted Chinese hickory, qRT-PCR was performed to study the target gene expression in that process. The scions and the rootstocks of the graft union were separated gently. The conjunction parts of both the scions and rootstocks at each time point (0, 3, 7, and 14 days after grafting) were sampled separately for RNA extraction. In addition, the RNA originated from the scions and rootstocks were extracted with TaKaRa MiniBEST Universal RNA Extraction Kit (Takara Bio Inc., Kusatsu, Japan) and based on its manufacturer’s protocol. Total RNA was extracted. The 0 day after grafting was taken as the control. cDNA synthesis for qRT-PCR was performed using PrimeScript™ RT Master Mix (Perfect Real Time) (Takara Bio Inc., Kusatsu, Japan).
The primers designed for qRT-PCR are designed and listed in Supplementary Table S1. cDNA was diluted into 50 ng/μL and used as the template for qRT-PCR amplification. CcActin, the Actin gene in Chinese hickory, was taken as the reference gene functioning in calculating the relative fold change of CcTIR1/AFBs from both the different parts of the graft union and the different time points after grafting. The amplification system was set according to TB Green® Premix Ex Taq™ (Tli RnaseH Plus) (Takara Bio Inc., Kusatsu, Japan). The qRT-PCR procedures were set as follows: 95°C for 3 min, 40 cycles of 95°C for 10 s, and 60°C for 30 s. All of the expression analyses were repeated four times. The relative gene expression of CcTIR1/AFBs was calculated based on the 2−ΔΔCt method.
2.7 Protein interaction prediction and Y2H assay
The coding sequences of CcTIR1/AFBs and CcIAAs were acquired from NCBI. The coding sequences were translated into protein sequences and then submitted to the STRING database website (https://cn.string-db.org/). The homologs of those proteins in Arabidopsis were matched and selected based on the identity score to construct the predicted protein interaction network.
Y2H assay was performed to test the interaction between CcTIR1/AFBs and CcIAAs. The primer was designed based on the genome data of Chinese hickory. The coding sequences of both CcTIR1/AFBs and CcIAAs were amplified from the cDNA of Chinese hickory and were inserted into the bait vector pGADT7 and the prey vector pGBKT7, respectively. The two vectors were linearized with EcoRI and BamHI during plasmid construction. The bait (CcTIR1/AFB) and prey (CcIAA) constructs were co-transformed into the yeast strain Y2Hgold (Weidi Bio., Shanghai, China) using the lithium acetate method according to the manufacturer’s instructions (Clontech, Mountain View, CA, USA). The transformants were selected on the (DDO) SD-Trp/-Leu medium (Coolaber, Beijing, China) and were further confirmed by being spotted on the DDO medium for at least 2 days at 30°C. To examine the interactions between CcTIR1/AFBs and CcIAAs and to uncover the role of IAA in the interaction, the transformants were transferred from SD-Trp/-Leu medium to SD-Trp/-Leu/-Ade/-His (Coolaber, Beijing, China) using a series of concentrations of IAA ranging from 0 to 100 mM and were cultured at 30°C (Prigge et al., 2010).
2.8 Statistical analysis
One-way analysis of variance was performed, and multiple comparison was conducted with the Duncan method at the significant level of 0.05 by the R package, agricolae.
3 Results
3.1 Identification and cloning of TIR1/AFB family genes from Chinese hickory
Seven TIR/AFB sequences were identified totally based on the Chinese genome data. To confirm the accuracy of the sequences, seven pairs of primers used for sequence cloning were designed according to the genome data. The sequencing and alignment results indicated that the sequences cloned from the cDNA of Chinese hickory are identical with those searched from the Chinese hickory genome. The seven CcTIR1/AFB genes are named compared to their putative orthologs in Arabidopsis based on the phylogenetic relationships (Figure 1). The basic information about the CcTIR1/AFB sequences, including gene name, gene id, CDS length, and properties of the predicted peptides, is listed in Table 1. The CDS lengths of CcTIR1/AFB range from 1,716 base pairs (CcAFB3) to 1,896 base pairs (CcAFB4); the peptide lengths range from 571 amino acids (CcAFB3) to 631 amino acids (CcAFB4) and the molecular weights range from 64.19 to 70.61 kDa. The predicted isoelectric points of CcTIR1/AFBs have a range from 5.42 (CcAFB4) to 6.67 (CcAFB3). As predicted, all the family members of CcTIR1/AFB are localized in the nucleus.
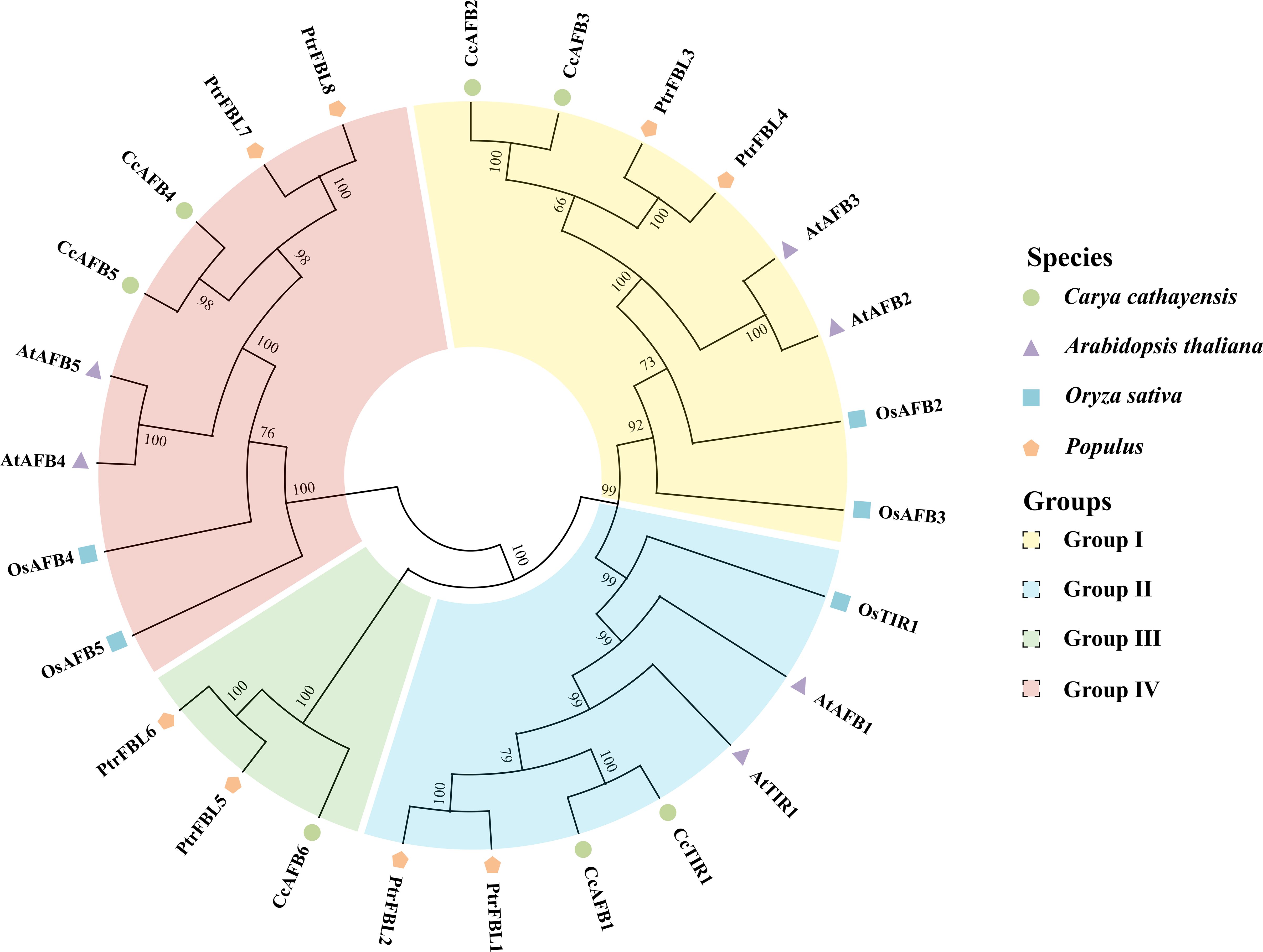
Figure 1. Phylogenetic tree of TIR1/AFB derived from Carya cathayensis, Oryza sativa, Populus, and Arabidopsis. The phylogenetic tree was created by MEGA X using neighbor-joining method with 1,000 bootstrap replicates. The numbers located in each node indicate bootstrap values. The TIR1/AFB family members were divided into four groups. Different species and groups were distinguished with the colored dots or polygons and the colored blocks, respectively. TIR1/AFB, transporter inhibitor response1/auxin signaling F-box. CcTIR1/AFB, AtTIR1/AFB, OsTIR1/AFB, and PtrFBL6 represent the TIR1/AFB family in Carya cathayensis, Arabidopsis, Oryza sativa, and Populus, respectively.
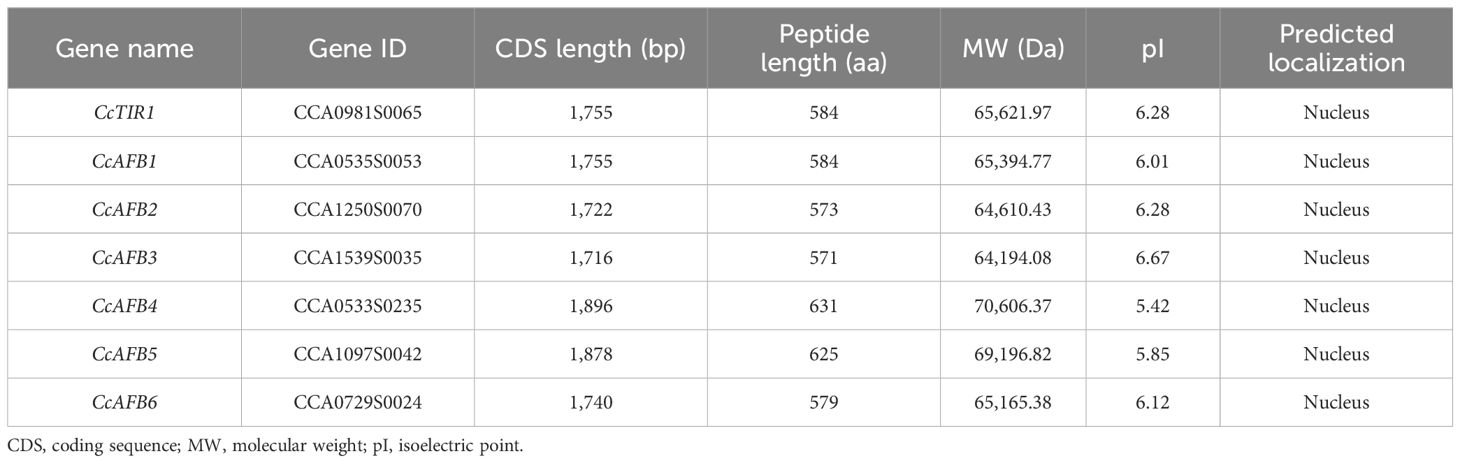
Table 1. Sequence information about Carya cathayensis transporter inhibitor response1/auxin signaling F-box (CcTIR1/AFB) gene family.
To explore the localization on which the CcTIR1/AFBs might perform their functions, some of the CcTIR1/AFBs were selected randomly and transiently transformed into tobacco leaves. The results showed that the CcTIR1/AFB proteins were located in the cellular nucleus (Figure 2).
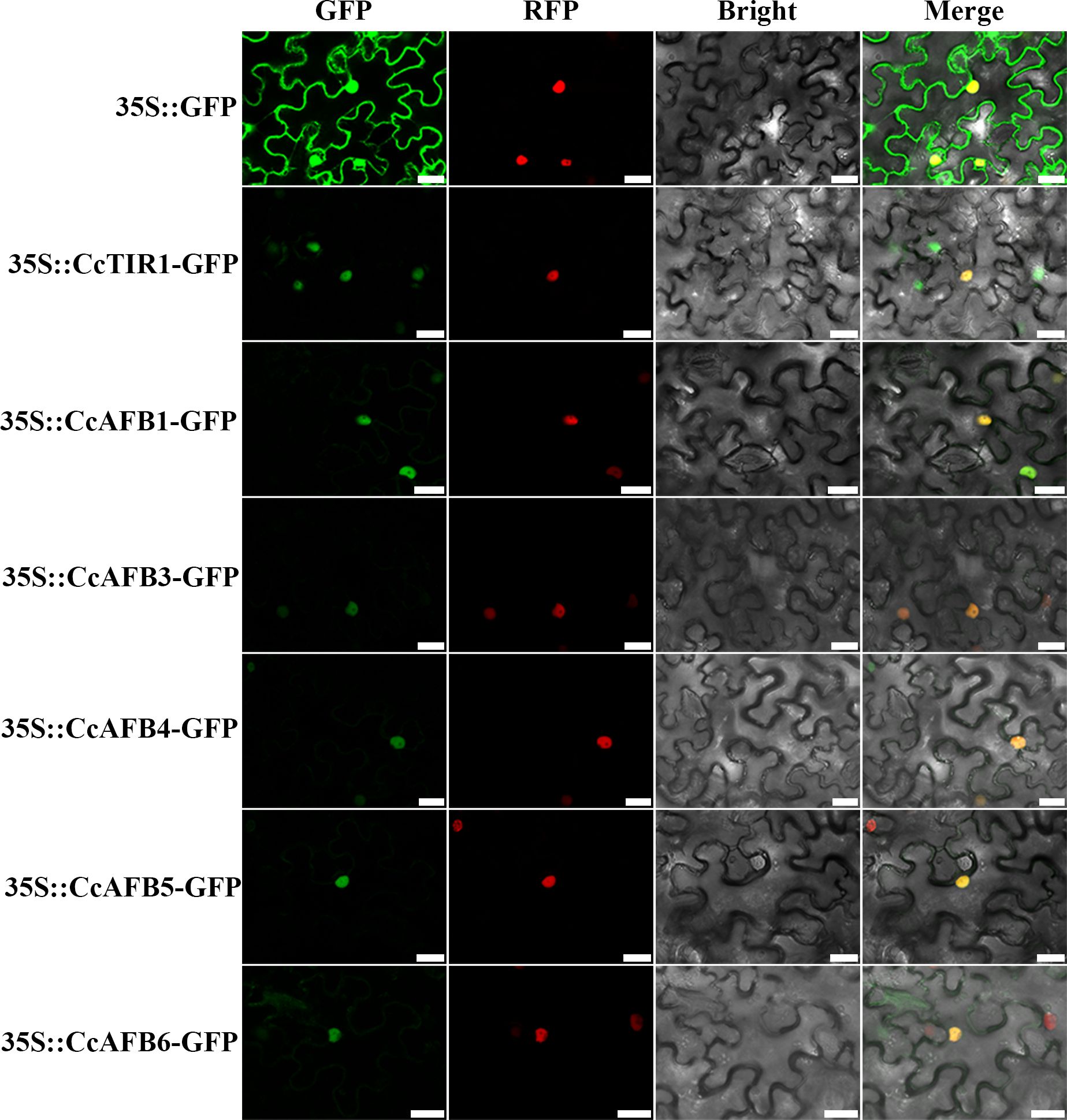
Figure 2. Subcellular localization of CcTIR1/AFB in Nicotiana tabacum leaves. The CDS sequences of CcTIR1/AFBs are inserted into the pCambia 1300-GFP vector and are under the control of 35S promoter. GFP, GFP fluorescence; RFP, RFP fluorescence; Bright, bright field; Merge, GFP/RFP/bright field overlay. The bar indicates 25 μm. CcTIR1/AFB, Carya cathayensis transporter inhibitor response1/auxin signaling F-box; GFP, green florescent protein; RFP, red florescent protein; 35S, CaMV35S promoter.
3.2 Phylogenetic relationship of TIR1/AFB proteins derived from Carya cathayensis, Oryza sativa, Populus, and Arabidopsis
In order to uncover the phylogenetic organization of CcTIR1/AFB and to reveal the phylogenetic relationship of TIR1/AFB of Carya cathayensis and of other different species, the TIR1/AFB family members from three species, Oryza sativa, Populus, and Arabidopsis, were chosen for the phylogenetic tree construction using neighbor-joining method with MEGA X (Figure 1). According to the phylogenetic relationships, we divided the TIR1/AFB family members into four groups and successively named those as clades I, II, III, and IV. The members of each class were varied from three to eight. The TIR1/AFBs from Carya cathayensis were scattered in all four groups and always clustered stronger with those from Populus. CcTIR1/AFB was distributed almost evenly in each class. The number of CcTIR1/AFB in clades I, II, III, and IV were two, two, one, and two, respectively. It was noteworthy that only group III contained the family genes from Carya cathayensis and Populus, but the other groups contained those family genes from all four species.
3.3 Gene structure, motif, and domain analysis of CcTIR1/AFB proteins in Chinese hickory
The investigation of the gene structure, the conserved domains, and the motifs of CcTIR1/AFB showed that the protein sequences of CcTIR1/AFBs were similar and conserved. The construction of phylogenetic tree of CcTIR1/AFBs was performed with MEGA X based on neighbor-joining method (Figure 3A). Their exon–intron structure was obtained from the Chinese hickory genome. Motif analysis was conducted using the MEME Suite with eight conserved motifs. All eight motifs were detected in every CcTIR1/AFB protein (Figure 3B). Except for CcAFB4 which contained two exons and one intron, all of the other six CcTIR1/AFBs contained three exons and two introns (Figure 3C). Conserved domain analysis was performed using CCD and visualized with ggplot2. The results showed that all seven CcTIR1/AFBs contained both the F-box domain and the LRR domain (also called transporter inhibitor response 1 domain) (Hayashi et al., 2008; Sheard et al., 2010). Besides that, all the CcTIR1/AFBs contained one to two complete or incomplete AMN1 (antagonist of mitotic exit network protein 1) superfamily domain which was composed of a number of leucine-rich repeats (Figures 3D, E) (Wang et al., 2003).
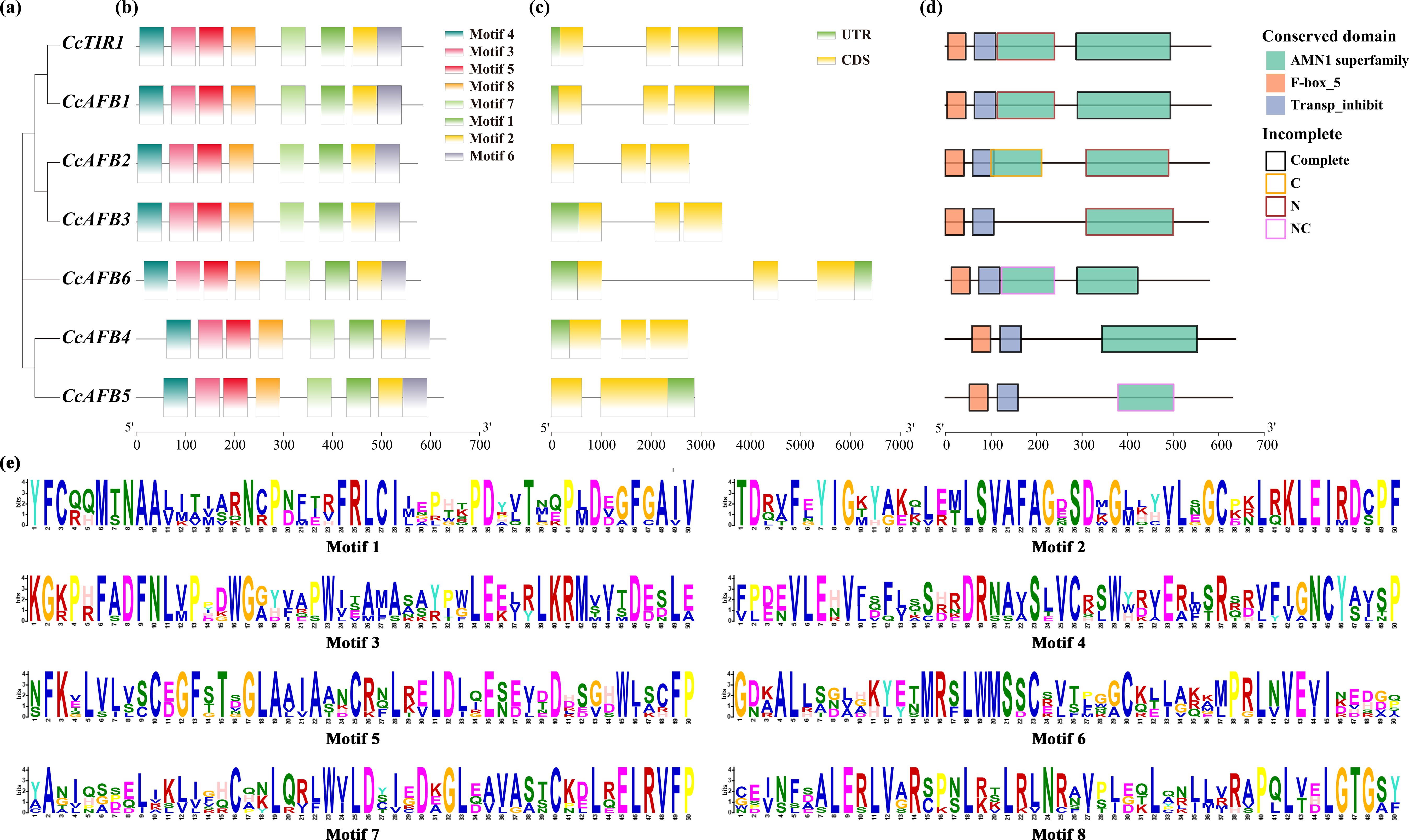
Figure 3. Gene structure, conserved motifs, and conserved domains of CcTIR1/AFB. (A) Phylogenetic tree of CcTIR1/AFBs being constructed with the protein sequences. (B) Motif distributions of the CcTIR1/AFB proteins generated in MEME Suite. Eight motifs were represented with eight blocks with different colors. (C) Exon–intron structure of CcTIR1/AFBs. The UTRs (untranslated region), exons, and introns were represented with green blocks, yellow blocks, and gray lines, respectively. (D) Conserved domain of CcTIR1/AFBs. The red, blue, and green blocks indicate the TIR1 protein domain, the F-box domain, and the AMN1 superfamily domain, respectively. The outline of the blocks represented the incomplete degree of the AMN1 superfamily domains. The description of the legends is as follows: Complete, the domain was complete; C, the domain was incomplete in C terminal; N, incomplete in N terminal; NC, incomplete in both N and C terminals. (E) Information of the discovered motifs from CcTIR1/AFBs. The size of the letters suggests the conservation degree of the amino acids. CcTIR1/AFB, Carya cathayensis transporter inhibitor response1/auxin signaling F-box; UTR, untranslated region; CDS, coding sequence; AMN1, antagonist of mitotic exit network protein 1; Trans_inhibit, transporter inhibitor response 1 domain.
3.4 cis-acting element analysis of CcTIR1/AFB promoters
It is extremely vital to explore the CcTIR1/AFBs transcriptional regulation and to infer their biological function. Therefore, prediction of the cis-acting elements was conducted by plant-CARE after submitting the upstream 2,000 base pairs of CcTIR1/AFBs. All CcTIR1/AFB promoters contained six kinds of cis-acting element of interest, including light response element, temperature response element, stress response element, and three kinds of phytohormone response elements, including auxin response elements, abscisic acid response elements, and MeJA response elements. Each element class contained 12, one, one, two, one, and two kinds of cis-acting elements, respectively.
The light response elements, as the majority of the cis-acting types of CcTIR1/AFB promoters, are scattered evenly on the promoters, suggesting that CcTIR1/AFB might play a significant role in the response to light. The stress and temperature response elements were set upstream and downstream of the promoter, respectively. As for the phytohormone response element, the auxin response elements mostly occupy the downstream part of the promoters, but both the abscisic acid response and the MeJA response elements are mostly concentrated on the upstream part (Figure 4A).
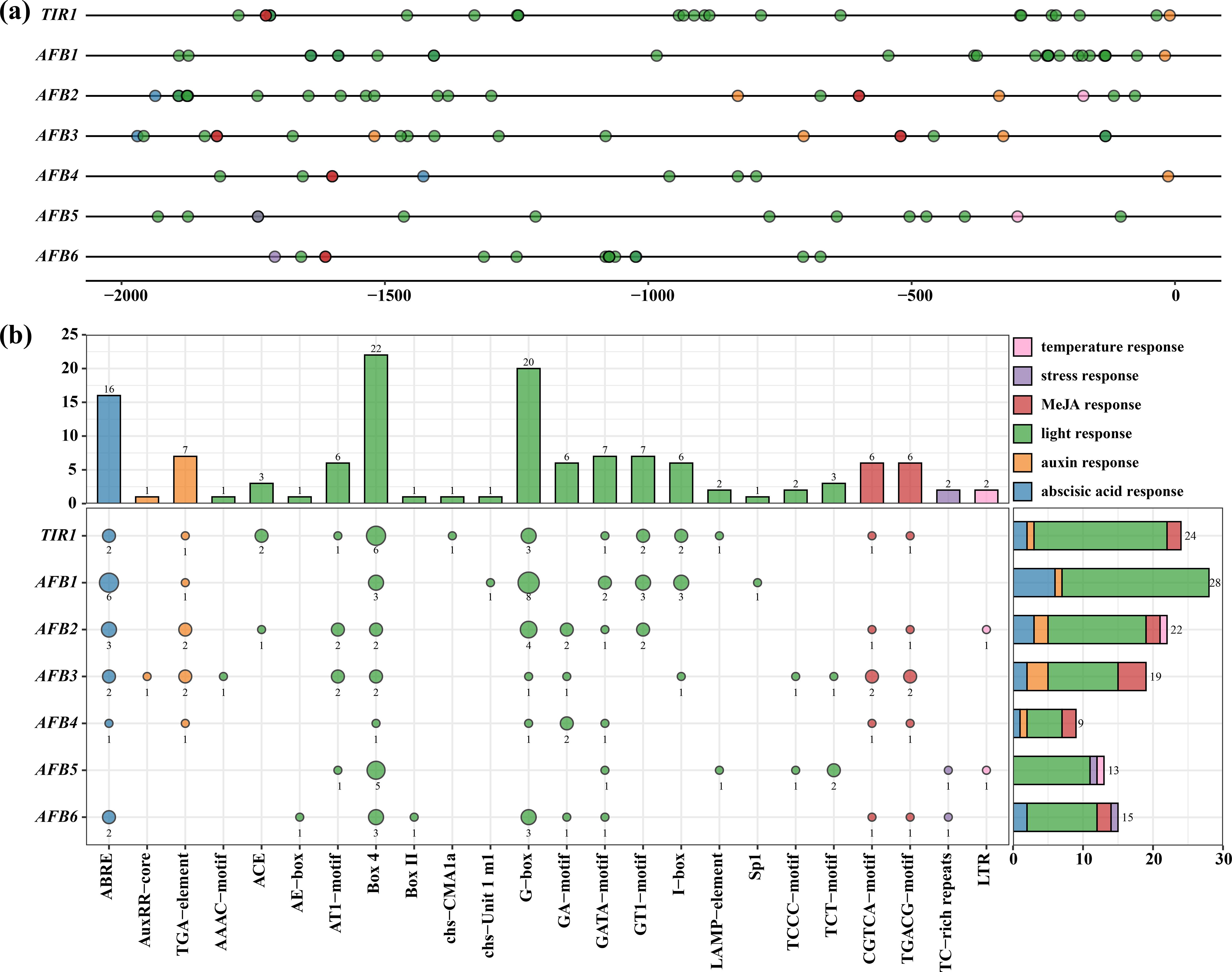
Figure 4. cis-acting element analysis of CcTIR1/AFB promoters. The upstream 2,000-bp sequences of CcTIR1/AFB genes were identified as the promoters. (A) Distribution of cis-acting elements on the promoter of CcTIR1/AFB. (B) The amounts of cis-acting elements belonging to different response classes on the promoter of each gene. The color of each bubble indicates each responsiveness class. The size of the bubble on the left plot suggests the quantity of each cis-acting element type, and the exact numbers are shown below the bubbles. The bar plot on the right illustrates the relative amounts of each response element on the promoter of each gene. TIR1/AFB, Carya cathayensis transporter inhibitor response1/auxin signaling F-box. On the x-axis, ABRE to LTR represent different cis-acting elements.
The promoter of CcTIR1/AFBs, from CcTIR1 to CcAFB6, contained 24, 28, 22, 19, nine, 12, and 15 cis-acting elements of interesting, respectively. The CcTIR1 promoter contains four classes of cis-acting elements of interest, except for the temperature response and the stress response. CcAFB1 contains the most abscisic acid response elements (ABRE) and light response elements. CcAFB2 possesses five classes of response types of cis-acting elements except for the stress response. According to the prediction, the temperature and stress response elements only exist in the promoter of CcAFB2, CcAFB5, and CcAFB6 and occupy a small part of all CcTIR1/AFB cis-acting elements. As for the cis-acting elements of the promoters, Box-4 and G-box are the two most common cis-acting elements belonging to the light response, the number of which are 22 and 20, respectively. ABRE (abscisic acid response elements) follows with a total number of 16. The element classes with the minimum numbers are the temperature response class and the stress response class, which only contained two LTR and two TC-rich repeats, respectively (Figure 4B).
3.5 The interactions between CcTIR1/AFBs and CcIAAs are regulated by different concentrations of IAA
The TIR1/AFB genes play a critical role in the auxin signaling pathway. The interactions of TIR1/AFB proteins with Aux/IAA proteins contribute to the degradation of Aux/IAA, resulting in the release of ARFs which, with the presence of IAA, activate the expression genes involved in a series of biological processes. Based on that, it is necessary to explore the interactions between CcTIR1/AFBs and Cc/IAAs.
The amino acid sequences of CcTIR1/AFB and of CcIAAs acquired from NCBI were selected to perform the interaction prediction with the online database, STRING. Each Chinese hickory sequence was mapped to the homologs in Arabidopsis based on sequence similarity. The Arabidopsis homologs were taken to construct the protein interaction network. The most similar Chinese hickory sequences were labeled next to the Arabidopsis homologs. The edge boldness indicated the interaction confidence. The predicted interaction network was composed of 14 nodes and 66 edges. There were a variety of strong interactions not only between two gene families but also in the same gene family (Figure 5A).
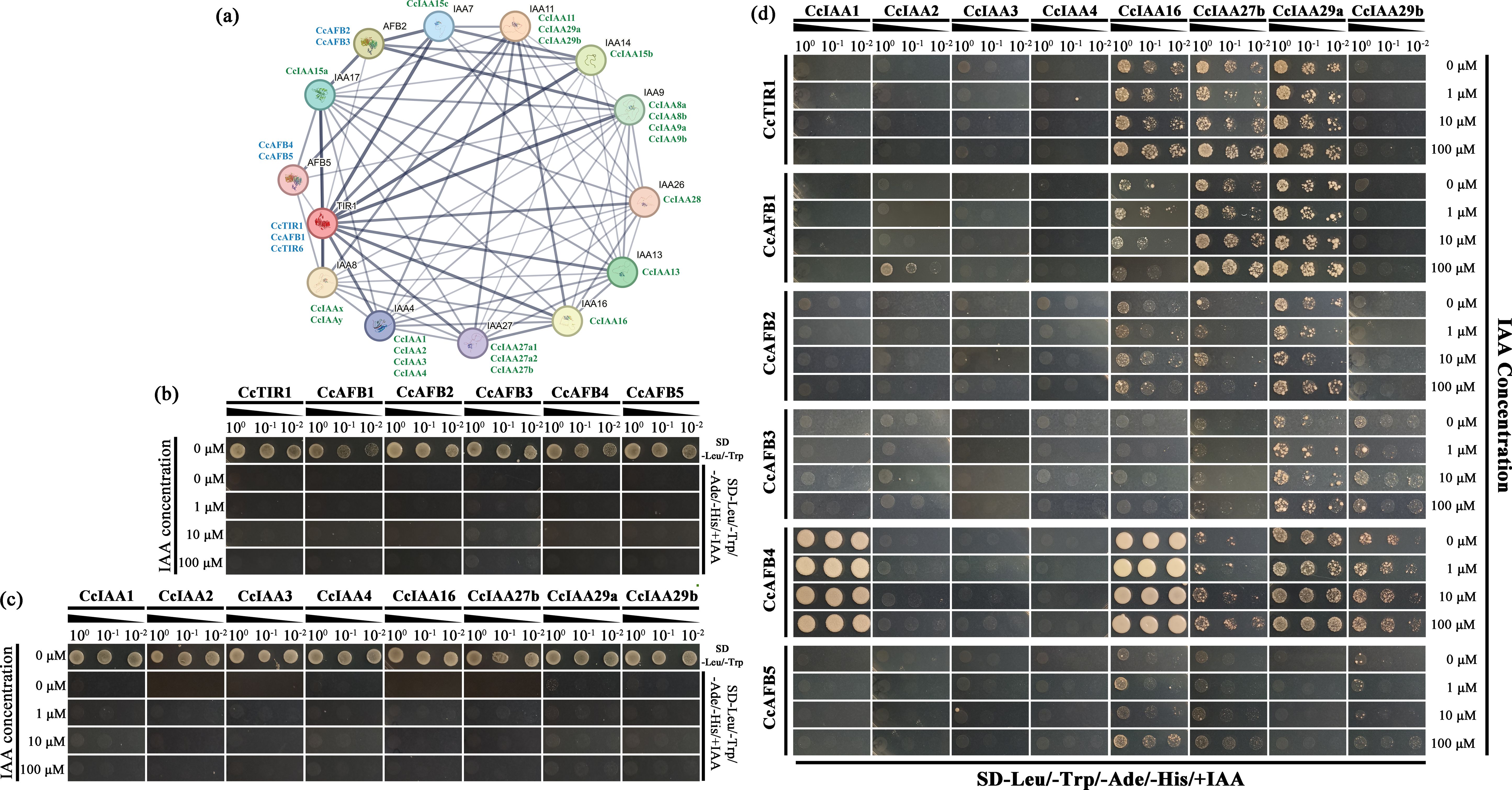
Figure 5. Interactions between CcTIR/AFBs and CcIAAs. (A) Prediction of the interaction network of CcTIR/AFBs and CcIAAs. Both the TIR1/AFBs and IAAs from Arabidopsis thaliana are shown with black names. CcTIR1/AFBs and CcIAAs are shown with blue and green names, respectively, and were labeled near to the counterpart in Arabidopsis based on the sequence similarity. The boldness of the edge lines in the network suggests the confidence of the interactions. (B) Identification of the co-transformed yeast (CcTIR1/AFBs in pGADT7 and pGBKT7 empty vector) with DDO (SD-Leu/-Trp) medium and the self-activation test with the QDO associated with different IAA concentrations (SD-Leu/-Trp/-Ade-/His/+IAA). (C) Identification of the co-transformed yeast (CcIAAs in pGBKT7 and pGADT7 empty vector) with DDO medium and the self-activation test with the QDO associated with different IAA concentrations. (D) Yeast two-hybrid experiment used to identify the 6 CcTIR1/AFBs’ interactions with eight CcIAAs. The CcTIR1/AFBs and CcIAAs were inserted into pGADT7 and pGBKT7, respectively. Diploids containing GAL4-AD-CcTIR1/AFBs and GAL4-BD-CcIAAs were generated by the co-transforming method and were spotted on the QDO selective medium with increasing concentrations of IAA. CcTIR1/AFB, Carya cathayensis transporter inhibitor response1/auxin signaling F-box; CcIAA, Carya cathayensis auxin/indole-3-acetic acid; IAA, indole acetic acid; SD, synthetic defined medium; Leu, leucine; Trp, tryptophan; Ade, adenine; His, histidine; DDO, double dropout medium; QDO, quadruple dropout medium.
Besides that, six CcTIR1/AFBs (CcTIR1 and CcAFB1–5) and eight CcIAAs (CcIAA1–4, CcIAA16, CcIAA27b, CcIAA29a, and CcIAA29b) were selected randomly to confirm the interactions between CcTIR1/AFBs and Aux/IAA via the Y2H assay. The diploids were generated and confirmed with DDO medium (Figure 5B). The diploids transformed only with CcTIR1/AFBs or only with CcIAAs were not able to grow ordinarily on the DDO medium, which suggested that both of them could not activate the reporter genes independently (Figure 5C). From CcTIR1/AFBs’ perspective, each of the three CcTIR1/AFBs (CcTIR1, CcAFB1, and CcAFB2) was able to interact with CcIAA16, CcIAA27b, and CcIAA29a. Besides that, CcAFB3 shows the ability to interact with CcIAA27, CcIAA29a, and CcIAA29b. There also existed interactions between CcAFB5 and three CcIAAs (CcIAA16, CcIAA27b, and CcIAA29b). Particularly, CcAFB4 interacted with five CcIAAs (CcIAA1, CcIAA16, CcIAA27b, CcIAA29a and CcIAA29b). The interaction strength between CcAFB4 and both CcIAA1 and CcIAA16 was stronger than the other tested interaction combinations. Meanwhile, from the CcIAAs’ perspective, the results showed that CcIAA2, CcIAA3, and CcIAA4 were incapable of interacting with any of the six TIR1/AFBs. On the contrary, CcIAA16, CcIAA27b, and CcIAA29a could interact with the majority of the six CcTIR1/AFBs. CcIAA29b interacted with half of the selected CcTIR1/AFBs (CcAFB3, CcAFB4, and CcAFB5). CcIAA1 interacted only with CcAFB4, which was associated with strong interaction strength. As the concentrations of IAA were increased, the yeast grew more quickly in the QDO medium, which indicated that the IAA molecule could strengthen the interactions between CcTIR1/AFBs and CcIAAs (Figure 5D).
3.6 Expression profiles of CcTIR1/AFB genes during grafting of Chinese hickory
To examine the response of CcTIR1/AFB genes during the grafting process of Chinese hickory, the relative expression of CcTIR1/AFBs, both at different stages during grafting (0, 3, 7, and 14 days after grafting) and at different parts of the grafting units (the scion and rootstock of Chinese hickory), was quantified using RT-qPCR. It was shown that the expression of most CcTIR1/AFBs (CcTIR1, CcAFB3, CcAFB5, and CcAFB6), with the highest expression at 0 days after grafting (DAG), was downregulated after grafting both in the scion and the rootstock. Those CcTIR1/AFBs were mostly expressed higher in the scion than in the rootstock at the same stage after grafting. Besides that, CcAFB1 and CcAFB2 show a similar expression pattern: the expression of CcAFB1/CcAFB2 is lower in the scion than in the rootstock at 0 DAG. The expression of CcAFB1/CcAFB2 in the scion was significantly upregulated at 3 DAG and gradually downregulated until 14 DAG and that in rootstock are downregulated from 0 DAG to 14 DAG. The expressions of CcAFB1/CcAFB2 in both scion and rootstock were decreased to nearly the same low at 14 DAG. Finally, CcAFB4 displayed a distinct manner compared with the others. The expression of CcAFB4 was downregulated in the scion but upregulated firstly, downregulated secondly, and upregulated finally in the rootstock. In general, the results suggested that CcTIR1, CcAFB3, CcAFB5, and CcAFB6 might function similarly and CcAFB1, CcAFB2, and CcAFB4 might play a critical role in the Chinese hickory grafting process (Figure 6).
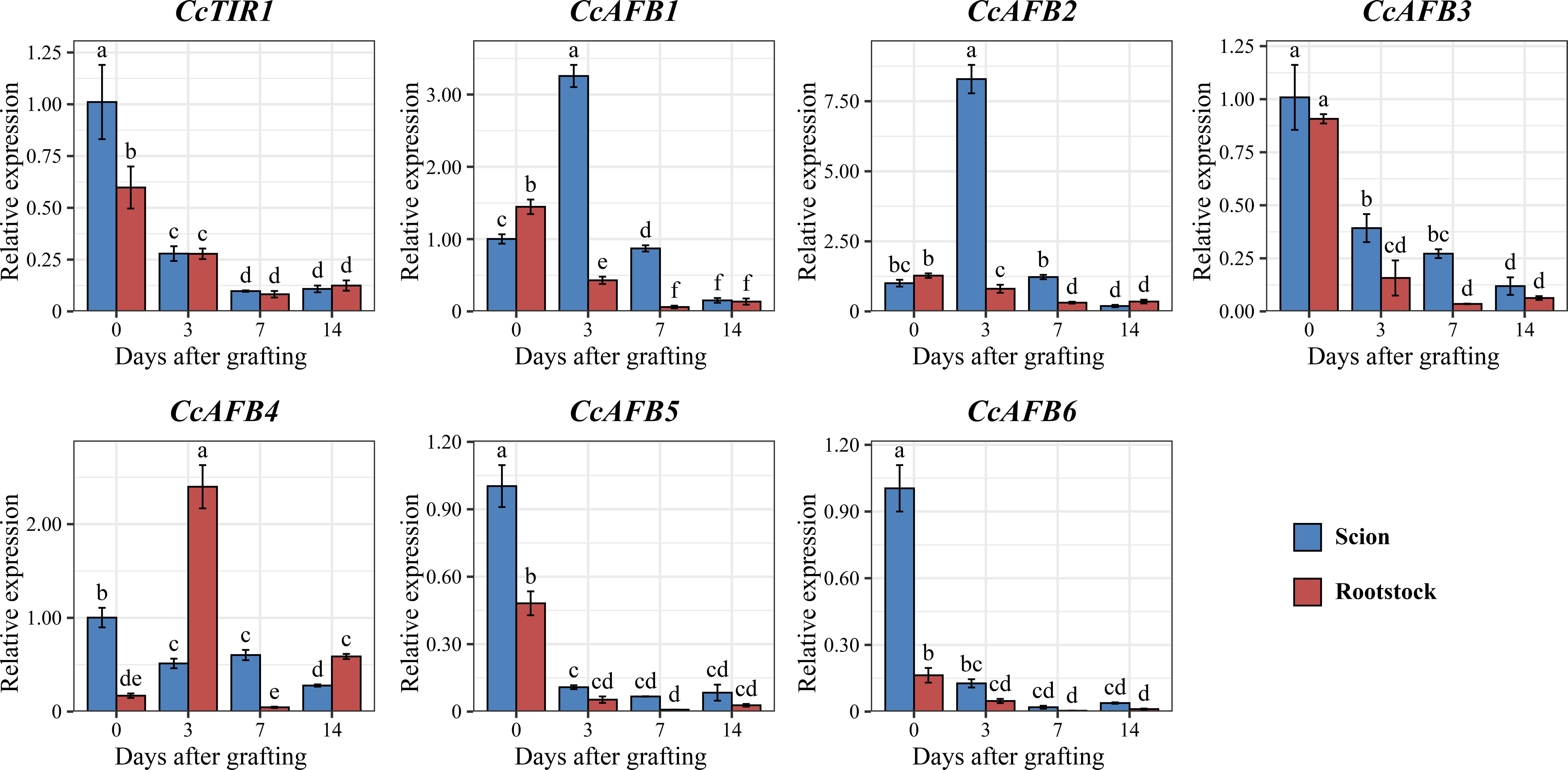
Figure 6. Expression pattern of CcTIR1/AFB genes both in scion and in rootstock at different stages (0, 3, 7, and 14 days after grafting). The letters above the error bar display the significant differences between each sample group and are determined by Duncan’s test (p < 0.05). CcTIR1/AFB, Carya cathayensis transporter inhibitor response1/auxin signaling F-box).
4 Discussion
Auxins are involved in a series of plant growth and development processes via the auxin signaling pathway (Carrillo et al., 2023). The TIR1/AFB family works as the auxin molecule receptor and plays a vital role in that signaling pathway (Ang and Østergaard, 2023). So far, the TIR1/AFB families containing all members have been identified in a few plant species, including six members in Arabidopsis (Parry et al., 2009), five members in rice (Guo et al., 2021), eight members in Populus (Shu et al., 2015), and 18 members in Brassica juncea (Cai et al., 2019), which indicates that the TIR1/AFB gene family is small in most plant species. The Chinese hickory (Carya cathayensis), classified in the Juglandaceae family, is known for its nutritious nuts and economic value (Huang et al., 2022). In our study, seven CcTIR1/AFBs were identified based on the genome data and were cloned for further confirmation. The TIR1/AFB gene family in Chinese hickory is larger than in Arabidopsis and in rice but smaller than in Populus and Brassica juncea.
The TIR1/AFB gene was firstly reported in 1998 and called transport inhibitor response 1 because of the deficiency of auxin transport in the Arabidopsis mutant (tir1) (Ruegger et al., 1998). Then, TIR1 and its homologies (AFBs) are identified as the auxin receptor (Dharmasiri et al., 2005; Greenham et al., 2011). The TIR1/AFB proteins mainly contained two conserved domains, F-box domain and LRR domain. The former domain is required for TIR1/AFBs to interact with the SKP protein, which facilitates the formation of SCF complex, and the latter containing a number of leucine-rich repeats is required for them to bind auxin and to interact with Aux/IAAs (Yu et al., 2015). In our study, all seven CcTIR1/AFBs contain one F-box domain and one LRR domain on the N terminus of the protein, which is consistent with BjuTIR1/AFBs as described in Brassica juncea (Cai et al., 2019).
The ability of TIR1/AFBs to interact with Aux/IAAs, whose strength is enhanced along with the increment of auxin concentration, has been reported in other species with the yeast two-hybrid assay. In Arabidopsis, TIR1, AFB1, and AFB2 are able to interact with IAA3, IAA5, IAA7, and IAA8, respectively (Calderón et al., 2012). Besides that, the TIR1 can also interact with IAA1 (Gilkerson et al., 2015) and IAA17 (Nan et al., 2014). In peas, PsTIR1a, PsTIR1b, PsAFB4, and PsAFB6 are able to interact with PsIAA6 (Ozga et al., 2022). In Brassica napus, BnTIR1 is capable of interacting with BnIAA7 (Ping et al., 2022). Both the TIR1/AFB family proteins and the Aux/IAA family proteins are mostly located in the cell nucleus and influence the transcription response directly in many other species, such as Arabidopsis (Prigge et al., 2020), rice (Guo et al., 2021), and rapeseed (Ping et al., 2022). In our study, it is firstly predicted with Cell-PLoc and further confirmed with the subcellular localization assay that the CcTIR1/AFBs of Chinese hickory are located in the nucleus, which increased the possibilities of spatial interactions between CcTIR1/AFBs and CcIAAs. Given that, the yeast two-hybrid assay associated with different auxin concentrations was conducted. The results show that each of the CcTIR1/AFBs being tested is capable of interacting with some CcIAAs. As the auxin concentration is increased, the interaction of each combination is strengthened. The results also suggest that the CcTIR1/AFBs interact with CcIAAs in an auxin-dependent way in Chinese hickory, which has also been reported in Arabidopsis (Yu et al., 2013). CcTIR1, CcAFB1, and CcAFB2 are able to interact with CcIAA16, CcIAA27b, and CcIAA29a, respectively. It is suggested that CcTIR1, CcAFB1, and CcAFB2 might be similar in function based on the Y2H assay. Interestingly, CcAFB4 shows an interaction pattern distinct from the other CcTIR1/AFBs. CcAFB4 shows a strong ability to interact with CcIAA1 and CcIAA16. It is indicated that CcAFB4 might show a different manner in the biological process from the other CcTIR1/AFBs.
Both the scions and the rootstocks are essentially the plant segments cut from different plant individuals (Wang et al., 2017). A successful grafting mostly depends on the adhesion of scions and rootstocks, a process called graft union formation (Gautier et al., 2019). The vascular tissues from both the scion and the rootstock are regenerated and reconnected in that process, which determines the plant survival after being grafted (Melnyk et al., 2018). Auxin and auxin signaling pathway play a critical role in vascular regeneration after grafting—for instance, auxin alters PIN polarity via the Aux/IAA-ARF-dependent pathway to facilitate the regeneration of vascular tissue at the wound (Sauer et al., 2006). ARF6 and ARF8 are involved in the regulation of tissue reunion in the Arabidopsis stems (Weerasak et al., 2014). The TIR1/AFB gene family plays as the auxin receptor and the necessary element in the auxin signaling pathway (Salehin et al., 2015). The TIR1/AFB signaling pathway is essential for vascular regeneration after wounding (Mazur et al., 2020). In order to understand the role of CcTIR1/AFBs during Chinese hickory grafting, their expression patterns in both the scion and the rootstock of Chinese hickory at different time points were analyzed. The four time points were determined according to the morphological changes based on the previous study, such as isolation layer formation at 3 DAG, callus formation at 7 DAG, and vascular reconnection at 14 DAG (Yang et al., 2022). Particularly, both CcAFB1 and CcAFB2 in the scion and CcAFB4 in the rootstock are significantly upregulated at 3 DAG. The special expression pattern indicates the special role of CcAFB1, CcAFB2, and CcAFB4 in Chinese hickory grafting. In Arabidopsis, phloem reconnection is severely affected at 4 DAG in the triple mutant, tir1afb2afb3, which suggests that TIR1, AFB2, and AFB3 are indispensable for vascular regeneration (Melnyk et al., 2015). TIR1/AFBs are capable of interacting with Aux/IAAs and facilitate the degradation of the latter. Aux/IAA might prevent the vascular tissue from generation—for instance, the downregulation of SlIAA15 contributes to the increasing number of xylem cells in tomato (Deng et al., 2014). In our study, CcIAA16, CcIAA27b, and CcIAA29a are able to interact with CcAFB1, CcAFB2, and CcAFB4. CcIAA1 and CcIAA16 interact with CcAFB4 strongly. The cis-acting element analysis showed that CcAFB1, CcAFB2, and CcAFB4 might be involved in the auxin response process. These results indicate that they are worthy of being studied further in Chinese hickory grafting.
5 Conclusion
Chinese hickory is an economic nut tree famous for its nutrient value and is broadly planted in east of China. In our study, seven CcTIR1/AFB family genes were identified, cloned, and characterized. The expression pattern analysis and the Y2H assay further indicate the potential role of the CcTIR1/AFB family and provide the special CcAFBs worthy of further study to explore the molecular mechanism during the grafting process.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
JM: Data curation, Formal Analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. XT: Formal Analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. YG: Investigation, Writing – original draft, Writing – review & editing. HL: Investigation, Writing – original draft, Writing – review & editing. YY: Writing – original draft, Writing – review & editing. QS: Writing – original draft, Writing – review & editing. LY: Writing – original draft, Writing – review & editing. BL: Writing – original draft, Writing – review & editing. JZ: Writing – original draft, Writing – review & editing. VS: Writing – original draft, Writing – review & editing. AS: Conceptualization, Validation, Writing – original draft, Writing – review & editing. HY: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Writing – original draft, Writing – review & editing. BZ: Conceptualization, Funding acquisition, Project administration, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by the National Natural Science Foundation of China (32071807, 31901346, and 31971695), Key Agricultural New Varieties Breeding Projects founded by Zhejiang Province Science and Technology Department (2021C02066-12), Open Foundation of State Key Laboratory of Subtropical Silviculture, Zhejiang A&F University (SKLSS-KF2023-07, KF202007, and KF201903), The Talent Foundation of Zhejiang A&F University (2022LFR001), and Overseas Expertise Introduction Project for Discipline Innovation (111 Project D18008).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1494579/full#supplementary-material
References
Ang, A. C. H., Østergaard, L. (2023). Save your TIRs-more to auxin than meets the eye. New Phytol. 238, 971–976. doi: 10.1111/nph.18783
Asahina, M., Azuma, K., Pitaksaringkarn, W., Yamazaki, T., Mitsuda, N., Ohme-Takagi, M., et al. (2011). Spatially selective hormonal control of RAP2.6L and ANAC071 transcription factors involved in tissue reunion in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 108, 16128–16132. doi: 10.1073/pnas.1110443108
Cai, Z., Zeng, D., Liao, J., Cheng, C., Sahito, Z. A., Xiang, M., et al. (2019). Genome-wide analysis of auxin receptor family genes in Brassica juncea var. tumida. Genes 10, 165. doi: 10.3390/genes10020165
Calderón, V. I. A., Lee, S., Oliveira, C. D., Ivetac, A., Brandt, W., Armitage, L., et al. (2012). A combinatorial TIR1/AFB-Aux/IAA co-receptor system for differential sensing of auxin. Nat. Chem. Biol. 8, 477–485. doi: 10.1038/nchembio.926
Carrillo, C. V. P., Hernandez, G. J., Mutte, S. K., Weijers, D. (2023). The birth of a giant: evolutionary insights into the origin of auxin responses in plants. EMBO J. 42, e113018. doi: 10.15252/embj.2022113018
Chandler, J. W. (2016). Auxin response factors. Plant Cell Environ. 39, 1014–1028. doi: 10.1111/pce.12662
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Deng, W., Yan, F., Liu, M., Wang, X., Li, Z. (2014). Down-regulation of SlIAA15 in tomato altered stem xylem development and production of volatile compounds in leaf exudates. Plant Signal Behav. 7, 911–913. doi: 10.4161/psb.20723
Dharmasiri, N., Dharmasiri, S., Estelle, M. (2005). The F-box protein TIR1 is an auxin receptor. Nature 435, 441–445. doi: 10.1038/nature03543
Flaishman, M. A., Loginovsky, K., Golobowich, S., Lev-Yadun, S. (2008). Arabidopsis thaliana as a model system for graft union development in homografts and heterografts. J. Plant Growth Regul. 27, 231–239. doi: 10.1007/s00344-008-9050-y
Gautier, A. T., Chambaud, C., Brocard, L., Ollat, N., Gambetta, G. A., Delrot, S., et al. (2019). Merging genotypes: graft union formation and scion-rootstock interactions. J. Exp. Bot. 70, 747–755. doi: 10.1093/jxb/ery422
Gilkerson, J., Kelley, D. R., Tam, R., Estelle, M., Callis, J. (2015). Lysine residues are not required for proteasome-mediated proteolysis of the auxin/indole acidic acid protein IAA11. Plant Physiol. 168, 708–720. doi: 10.1104/pp.15.00402
Greenham, K., Santner, A., Castillejo, C., Mooney, S., Sairanen, I., Ljung, K., et al. (2011). The AFB4 auxin receptor is a negative regulator of auxin signaling in seedlings. Curr. Biol. 21, 520–525. doi: 10.1016/j.cub.2011.02.029
Guo, F., Huang, Y., Qi, P., Lian, G., Hu, X., Han, N., et al. (2021). Functional analysis of auxin receptor OsTIR1/OsAFB family members in rice grain yield, tillering, plant height, root system, germination, and auxinic herbicide resistance. New Phytol. 229, 2676–2692. doi: 10.1111/nph.17061
Hayashi, K., Tan, X., Zheng, N., Hatate, T., Kimura, Y., Kepinski, S., et al. (2008). Small-molecule agonists and antagonists of F-box protein-substrate interactions in auxin perception and signaling. Proc. Natl. Acad. Sci. U S A. 105, 5632–5637. doi: 10.1073/pnas.0711146105
Huang, C., Li, Y., Wang, K., Xi, J., Xu, Y., Si, X., et al. (2022). Analysis of lipidomics profile of Carya cathayensis nuts and lipid dynamic changes during embryonic development. Food Chem. 370, 130975. doi: 10.1016/j.foodchem.2021.130975
Huang, Y., Xiao, L., Zhang, Z., Zhang, R., Wang, Z., Huang, C., et al. (2019). The genomes of pecan and Chinese hickory provide insights into Carya evolution and nut nutrition. GigaScience 8, giz036. doi: 10.1093/gigascience/giz036
Iwase, A., Mitsuda, N., Koyama, T., Hiratsu, K., Kojima, M., Arai, T., et al. (2011). The AP2/ERF transcription factor WIND1 controls cell dedifferentiation in Arabidopsis. Curr. Biol. 21, 508–514. doi: 10.1016/j.cub.2011.02.020
Jogawat, A., Yadav, B., Lakra, N., Singh, A. K., Narayan, O. P. (2021). Crosstalk between phytohormones and secondary metabolites in the drought stress tolerance of crop plants: a review. Physiologia Plantarum 172, 1106–1132. doi: 10.1111/ppl.v172.2
Kepinski, S. (2007). The anatomy of auxin perception. Bioessays 29, 953–956. doi: 10.1002/bies.20657
Lavy, M., Estelle, M. (2016). Mechanisms of auxin signaling. Development 143, 3226–3229. doi: 10.1242/dev.131870
Lee, J., Kubota, C., Tsao, S. J., Bie, Z., Echevarria, P. H., Morra, L., et al. (2010). Current status of vegetable grafting: Diffusion, grafting techniques, automation. Sci. Hortic. 127, 93–105. doi: 10.1016/j.scienta.2010.08.003
Loupit, G., Brocard, L., Ollat, N., Cookson, S. J., Lunn, J. (2023). Grafting in plants: recent discoveries and new applications. J. Exp. Bot. 74, 2433–2447. doi: 10.1093/jxb/erad061
Luo, J., Zhou, J., Zhang, J. (2018). Aux/IAA gene family in plants: molecular structure, regulation, and function. Int. J. Mol. Sci. 19, 259. doi: 10.3390/ijms19010259
Mazur, E., Kulik, I., Hajný, J., Friml, J. (2020). Auxin canalization and vascular tissue formation by TIR1/AFB-mediated auxin signaling in Arabidopsis. New Phytol. 226, 1375–1383. doi: 10.1111/nph.16446
Melnyk, C. W., Gabel, A., Hardcastle, T. J., Robinson, S., Miyashima, S., Grosse, I., et al. (2018). Transcriptome dynamics at Arabidopsis graft junctions reveal an intertissue recognition mechanism that activates vascular regeneration. Proc. Natl. Acad. Sci. U S A. 115, E2447–E2456. doi: 10.1073/pnas.1718263115
Melnyk, C. W., Schuster, C., Leyser, O., Meyerowitz, E. M. (2015). A developmental framework for graft formation and vascular reconnection in Arabidopsis thaliana. Curr. Biol. 25, 1306–1318. doi: 10.1016/j.cub.2015.03.032
Nan, W., Wang, X., Yang, L., Hu, Y., Wei, Y., Liang, X., et al. (2014). Cyclic GMP is involved in auxin signalling during Arabidopsis root growth and development. J. Exp. Bot. 65, 1571–1583. doi: 10.1093/jxb/eru019
Ozga, J. A., Jayasinghege, C. P. A., Kaur, H., Gao, L., Nadeau, C. D., Reinecke, D. M. (2022). Auxin receptors as integrators of developmental and hormonal signals during reproductive development in pea. J. Exp. Bot. 73, 4094–4112. doi: 10.1093/jxb/erac152
Parry, G., Calderon-Villalobos, L. I., Prigge, M., Peret, B., Dharmasiri, S., Itoh, H., et al. (2009). Complex regulation of the TIR1/AFB family of auxin receptors. Proc. Natl. Acad. Sci. U S A. 106, 22540–22545. doi: 10.1073/pnas.0911967106
Ping, X., Ye, Q., Yan, M., Zeng, J., Yan, X., Li, H., et al. (2022). Integrated genetic mapping and transcriptome analysis reveal the BnaA03.IAA7 protein regulates plant architecture and gibberellin signaling in Brassica napus L. Theor. Appl. Genet. 135, 3497–3510. doi: 10.1007/s00122-022-04196-8
Prigge, M. J., Greenham, K., Zhang, Y., Santner, A., Castillejo, C., Mutka, A. M., et al. (2016). The Arabidopsis auxin receptor F-box proteins AFB4 and AFB5 are required for response to the synthetic auxin picloram. G3 (Bethesda). 6, 1383–1390. doi: 10.1534/g3.115.025585
Prigge, M. J., Lavy, M., Ashton, N. W., Estelle, M. (2010). Physcomitrella patens auxin-resistant mutants affect conserved elements of an auxin-signaling pathway. Curr. Biol. 20, 1907–1912. doi: 10.1016/j.cub.2010.08.050
Prigge, M. J., Platre, M., Kadakia, N., Zhang, Y., Greenham, K., Szutu, W., et al. (2020). Genetic analysis of the Arabidopsis TIR1/AFB auxin receptors reveals both overlapping and specialized functions. Elife 9, e54740. doi: 10.7554/eLife.54740
Ruegger, M., Dewey, E., Gray, W. M., Hobbie, L., Turner, J., Estelle, M. (1998). The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast grr1p. Genes Dev. 12, 198–207. doi: 10.1101/gad.12.2.198
Salehin, M., Bagchi, R., Estelle, M. (2015). SCFTIR1/AFB-based auxin perception: mechanism and role in plant growth and development. Plant Cell. 27, 9–19. doi: 10.1105/tpc.114.133744
Sauer, M., Balla, J., Luschnig, C., Wisniewska, J., Reinöhl, V., Friml, J., et al. (2006). Canalization of auxin flow by Aux/IAA-ARF-dependent feedback regulation of PIN polarity. Genes Dev. 20, 2902–2911. doi: 10.1101/gad.390806
Sharma, A., Zheng, B. (2019). Molecular Responses during plant grafting and its regulation by auxins, cytokinins, and gibberellins. Biomolecules 9, 397. doi: 10.3390/biom9090397
Sheard, L. B., Tan, X., Mao, H., Withers, J., Ben-Nissan, G., Hinds, T. R., et al. (2010). Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468, 400–405. doi: 10.1038/nature09430
Shu, W., Liu, Y., Guo, Y., Zhou, H., Zhang, J., Zhao, S., et al. (2015). A Populus TIR1 gene family survey reveals differential expression patterns and responses to 1-naphthaleneacetic acid and stress treatments. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00719
Vidal, E. A., Araus, V., Lu, C., Parry, G., Green, P. J., Coruzzi, G. M., et al. (2010). Nitrate-responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U S A. 107, 4477–4482. doi: 10.1073/pnas.0909571107
Wang, J., Jiang, L., Wu, R. (2017). Plant grafting: how genetic exchange promotes vascular reconnection. New Phytol. 214, 56–65. doi: 10.1111/nph.14383
Wang, Y., Shirogane, T., Liu, D., Harper, J. W., Elledge, S. J. (2003). Exit from exit: resetting the cell cycle through AMN1 inhibition of G protein signaling. Cell 112, 697–709. doi: 10.1016/S0092-8674(03)00121-1
Weerasak, P., Sumie, I., Masashi, A., Shinobu, S. (2014). ARF6 and ARF8 contribute to tissue reunion in incised Arabidopsis inflorescence stems. Plant Biotechnol. 31, 49–53. doi: 10.5511/plantbiotechnology.13.1028b
Yang, Y., Huang, Q., Wang, X., Mei, J., Sharma, A., Tripathi, D. K., et al. (2021). Genome-wide identification and expression profiles of ABCB gene family in Chinese hickory (Carya cathayensis Sarg.) during grafting. Plant Physiol. Biochem. 168, 477–487. doi: 10.1016/j.plaphy.2021.10.029
Yang, Y., Mei, J., Chen, J., Yang, Y., Gu, Y., Tang, X., et al. (2022). Expression analysis of PIN family genes in Chinese hickory reveals their potential roles during grafting and salt stress. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.999990
Yu, H., Moss, B. L., Jang, S. S., Prigge, M., Klavins, E., Nemhauser, J. L., et al. (2013). Mutations in the TIR1 auxin receptor that increase affinity for auxin/indole-3-acetic acid proteins result in auxin hypersensitivity. Plant Physiol. 162, 295–303. doi: 10.1104/pp.113.215582
Yu, H., Zhang, Y., Moss, B. L., Bargmann, B. O., Wang, R., Prigge, M., et al. (2015). Untethering the TIR1 auxin receptor from the SCF complex increases its stability and inhibits auxin response. Nat. Plants.1 3), 14030. doi: 10.1038/nplants.2014.30
Yuan, H., Zhao, L., Chen, J., Yang, Y., Zheng, B. (2018). Identification and expression profiling of the aux/iaa gene family in chinese hickory (Carya cathayensis sarg.) during the grafting process. Plant Physiol. Bioch. 127, 55–63. doi: 10.1016/j.plaphy.2018.03.010
Keywords: auxin, TIR1/AFB, Chinese hickory, grafting, Aux/IAA
Citation: Mei J, Tang X, Gu Y, Lu H, Yang Y, Shen Q, Yang L, Li B, Zuo J, Singh VP, Sharma A, Yuan H and Zheng B (2024) Role of TIR1/AFB family genes during grafting in Carya cathayensis. Front. Plant Sci. 15:1494579. doi: 10.3389/fpls.2024.1494579
Received: 11 September 2024; Accepted: 25 October 2024;
Published: 22 November 2024.
Edited by:
Tanveer Alam Khan, Leibniz Institute of Plant Genetics and Crop Plant Research (IPK), GermanyReviewed by:
Bindu Yadav, Jawaharlal Nehru University, IndiaMuhammad Ahsan Altaf, Hainan University, China
Copyright © 2024 Mei, Tang, Gu, Lu, Yang, Shen, Yang, Li, Zuo, Singh, Sharma, Yuan and Zheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anket Sharma, YW5rZXRzaGFybWFAZ21haWwuY29t; Huwei Yuan, aHd5dWFuQHphZnUuZWR1LmNu; Bingsong Zheng, YnN6aGVuZ0B6YWZ1LmVkdS5jbg==
†These authors have contributed equally to this work
 Jiaqi Mei1†
Jiaqi Mei1† Vijay Pratap Singh
Vijay Pratap Singh Anket Sharma
Anket Sharma Huwei Yuan
Huwei Yuan Bingsong Zheng
Bingsong Zheng