- Zhejiang Key Laboratory of Intelligent Food Logistic and Processing, College of Biological and Environmental Sciences, Zhejiang Wanli University, Ningbo, China
The hypersensitive-induced reaction (HIR) gene family is associated with the hypersensitive response (HR) in plant defense against pathogens. Although rice (Oryza sativa L.) is a crucial food crop, studies on its HIR genes are limited. This study identified six HIR members, categorized into three phylogenetic clades. The analyses of phylogenetics, gene structures, and collinear relationships revealed a high conservation of these genes, featuring the stomatin/prohibitin/flotillin/HflK/C domain. OsHIR genes were regulated by cis-acting elements, including ARE, SARE, DRE, LTR, and GARE. OsHIRs were widely expressed in multiple plant organs, including roots, stems, and leaves. These genes respond to various abiotic stresses (like drought and low temperature) and hormone treatments (including ABA, SA, GA, and MeJA) with overlapping yet distinct expression patterns. Our results indicate that OsHIRs are involved in abiotic stresses and hormone responses, which provides a basis for further functional analysis of OsHIRs in rice crop plants.
1 Introduction
Plants suffer various kinds of biotic and abiotic stresses during growth. They developed effector-triggered immunity (ETI) and PAMP-triggered immunity (PTI) as defense system against pathogens (Jones and Dangl, 2006). ETI can trigger a hypersensitive response (HR), which restricts pathogen spread via programmed cell death (Block and Alfano, 2011; Greenberg and Yao, 2004; Jones and Dangl, 2006). Hypersensitive-induced reaction (HIR) proteins, homologs of Nicotiana tabacum’s NG1, cause HR-like lesions upon overexpressed (Karrer et al., 1998). Three proteins from maize (Zea mays), namely ZmHIR1, ZmHIR2, and ZmHIR3, were discovered as the first NG1 homologs (Nadimpalli et al., 2000). Many additional HIR genes have since been discovered according to DNA and amino acid similarities (Chen et al., 2007; Hakozaki et al., 2004; Rostoks et al., 2003; Yu et al., 2008, 2013; Chen et al., 2017; Zhao et al., 2019; Li et al., 2022).
The HIR protein belongs to the proliferation, ionization, and death (PID) superfamily and features the stomatin/prohibitin/flotillin/HIK (SPFH) structural domain, essential for ion channels regulation, cell proliferation, and apoptosis. Numerous PID family members are known to be associated with lipid rafts, forming membrane microdomains (Nadimpalli et al., 2000). Predominantly, HIR proteins localize to the plasma membrane (Choi et al., 2011; Duan et al., 2013; Qi et al., 2011; Wang et al., 2024; Li et al., 2019; Zhao et al., 2019; Zhou et al., 2010), with a minor portion found in the nucleus, extracellular matrix, tonoplast, nuclear membrane, and mitochondria, as reported by various studies (Li et al., 2022, 2019; Zhao et al., 2019; Choi et al., 2011; Zhou et al., 2010).
HIR proteins are vital for plant stress responses. In wheat (Triticum aestivum L.), the expression levels of TaHIR1 and TaHIR3 were reduced when exposed to temperature, drought, and high salt stresses, as well as with exogenous abscisic acid (ABA) and ethrel treatments (Duan et al., 2013). Studies have demonstrated that methyl jasmonate (MeJA) treatment elevated MdHIR4 transcript level in apple (Zhao et al., 2019). The identification of phytohormone response elements, specifically the abscisic acid response element (ABRE), within the promoter region of the BnHIR gene indicates its involvement in ABA response (Li et al., 2022). Additionally, HIR genes have been implicated in the plant’s defense mechanisms against a range of pathogens, including fungi (Yu et al., 2008; Wang et al., 2024; Sun et al., 2021), bacteria (Zhou et al., 2010; Choi et al., 2011; Zhao et al., 2019), and viruses (Mei et al., 2020; Li et al., 2019).
Rice is a crucial economic crop in the world with a rich history of farming and consumption. However, our knowledge of the OsHIR family remains limited. In this research, we identified six OsHIR-encoding genes within the Oryza sativa genome and conducted bioinformatics analyses, expression profiling, and assessments of their responses to abiotic stresses and hormone treatments. Additionally, we performed subcellular localization analysis to confirm OsHIR protein functions. Our findings contribute crucial insights into the functions of OsHIR protein in rice.
2 Materials and methods
2.1 Sequence retrieval and date sources
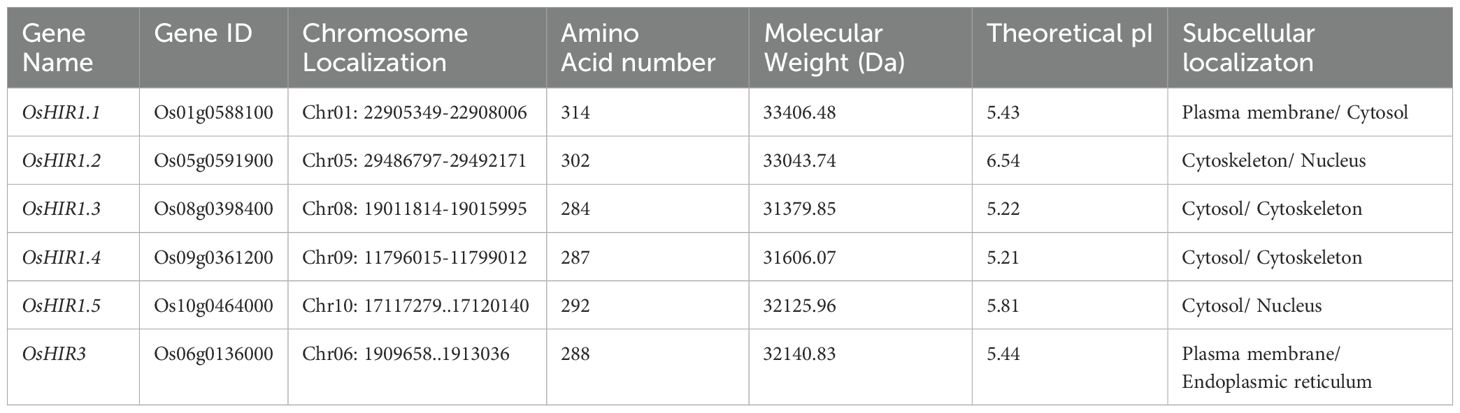
The HIR amino acid sequences of Aabidopsis thaliana, Triticum aestivum, Hordeum vulgare, and Zea mays were obtained from the NCBI website. These sequences were then utilized as queries in a protein BLAST search against rice (Oryza sativa spp. japonica) RefSeq Protein (FASTA). The OsHIR genes were identified based on their chromosomal location in rice and by a BLAST comparison using the Arabdopsis Information Resource (https://www.arabidopsis.org/). The amino acid numbers, isoelectric points (theoretical pI), molecular weights, and subcellular localizations of the identified OsHIR protein candidates were predicted by ProtParam (https://www.expasy.org/resources/protparam) and GenScript WoLF PSORT tools (https://www.genscript.com/wolf-psort.html/).
2.2 Phylogenetic and synteny analyses
The phylogenetic analysis of HIR proteins were conducted in software MEGA 11 using the Maximum Likelihood method, following the approach described by Yoshida and Nei (2016). Synteny relationship analysis was performed using Advanced Circos integrated with TBtools for handling and visualizing large-scale datasets across the whole genome (Chen et al., 2023).
2.3 Structural characterization analysis
The OsHIR gene structures were assessed through CDD-search, and the conserved motifs of OsHIR proteins were found via the MEME tool (https://meme-suite.org/meme/). Results from these analyses were shown by TBtools.
2.4 Prediction of cis-acting elements in the promoters
The promoter sequences (2.0 kb) of OsHIR genes were obtained from the rice gene annotation file. Regulatory elements within the promoter regions were examined via PlantCARE (Rombauts et al., 1999; Lescot et al., 2002), and the results were displayed using the Simple BioSequence Viewer in TBtools.
2.5 Subcellular localization analysis
OsHIR cDNA was amplified through RT-PCR and inserted into pCAMBIA1301-GFP plasmid using specific primers listed in Supplementary Table S1. Agrobacterium cells containing 35S::OsHIRs-GFP or 35S::GUS-GFP (control) were injected into tobacco leaves. GFP fluorescence was captured in the FITC (EGFP) channel via a confocal laser microscope system (Nikon A1+, Tokyo, Japan) 60 hours post-infiltration.
2.6 Plant material, growth conditions and treatments
‘N1P’ rice (Oryza sativa spp. japonica) seedlings were cultivated in a glasshouse with a 16-h light/8-h dark photoperiod at 28-30°C and 60% humidity. Roots, stems, and leaves from four-week-old plants were rapidly frozen in liquid nitrogen and stored at -80°C for RT-qPCR analysis.
The rice seedlings experienced low temperature stress at 4°C and drought stress with 10% PEG6000. For phytohormone treatments, seedlings were sprayed with 100 μM abscisic acid (ABA), 2 mM salicylic acid (SA), 100 μM gibberellin (GA), or 100 μM methyl jasmonate (MeJA) with 0.02% (v/v) Tween 20, while control seedlings were treated with Tween 20 only. Leaf samples were collected at designated time points and stored at -80°C.
2.7 RT-qPCR analysis
RNA extraction, reverse transcription, and quantitative PCR (qPCR) were conducted as previously detailed (Li et al., 2019). The gene-specific primers used in this study was listed in Supplementary Table S1.
2.8 Statistical analysis
Statistical analyses were performed using SPSS 22.0 software (SPSS Inc., Chicago, IL, USA). Results were presented as mean ± standard error and evaluated using Student’s t-test (* p≤ 0.05, ** p≤ 0.01, *** p≤ 0.001).
3 Results
3.1 Genome-wide identification and naming of OsHIR members
To identify the HIR genes in rice (Oryza sativa L.), the amino acid sequences of three Arabidopsis HIR proteins (AtHIR1.1, AtHIR1.2, and AtHIR3) were served as queries for Protein BLAST against the Oryza sativa L. Genome Database. Six HIR-encoding genes, namely OsHIR1.1, OsHIR1.2, OsHIR1.3, OsHIR1.4, OsHIR1.5, and OsHIR3, were identified. These OsHIR genes were located on distinct chromosomes, as shown in Table 1; Supplementary Figure S1. The OsHIR proteins ranged from 284 (OsHIR1.3) to 314 (OsHIR1.1) amino acids, with molecular weights (MW) spanning 31379.85 (OsHIR1.3) to 33406.48 (OsHIR1.1) Da and isoelectric points (pI) from 5.21 (OsHIR1.4) to 6.54 (OsHIR1.2) (Table 1). The subcellular localization prediction analysis indicated that most OsHIR proteins were localized to the plasma membrane and cytoplasm, except OsHIR3, which localized to the plasma membrane, and OsHIR1.2 localized to both the nucleus and cytoskeleton (Table 1). To validate these predictions, four OsHIR proteins were transiently expressed in tobacco (N. benthamiana) leaf cells. The GFP signals of OsHIR1.2 and OsHIR3 proteins were distinctly visible in the plasma membrane, whereas the OsHIR1.4-GFP fusion protein signal was observed in both the plasma membrane and cytoplasmic aggregates (Figure 1).
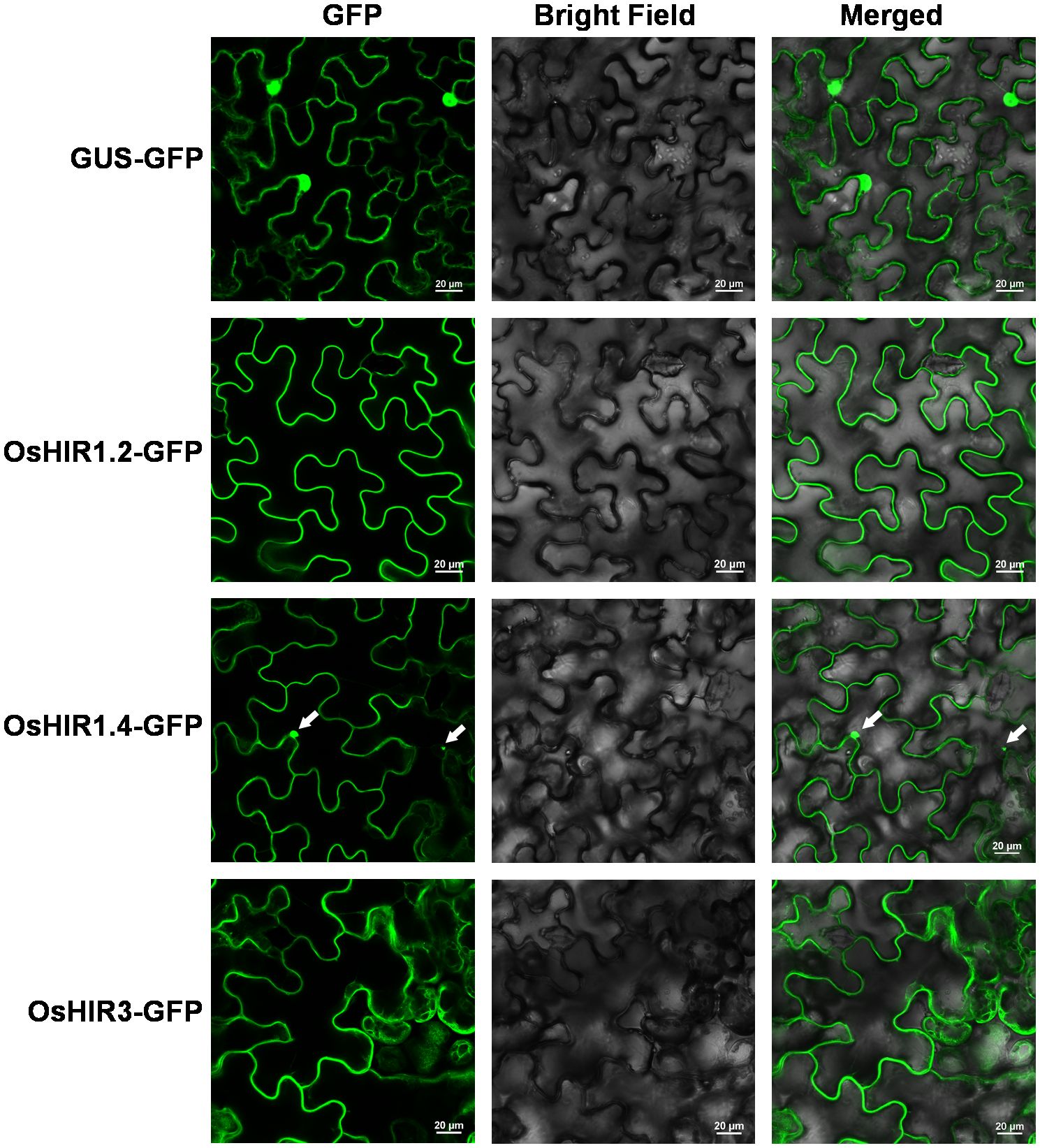
Figure 1. Subcellular localization of OsHIR proteins in Nicotiana benthamiana leaf epidermal cells. Arrow indicates the cytoplasmic aggregates. Scale bar: 20 μm.
3.2 Phylogenetic analysis
To explore the evolutionary relationship of OsHIR proteins, the amino acid sequences of 24 HIR proteins from Arabidopsis, Triticum aestivum (wheat), Zea mays (maize), Hordeum vulgare (barley), and Oryza sativa (rice) were analyzed to build a phylogenetic tree. These proteins were grouped into three clades: clade I, which included two OsHIR proteins (OsHIR1.3 and OsHIR1.4); clade II, which consisted of three OsHIR proteins (OsHIR1.1, OsHIR1.2, and OsHIR1.5); and clade III, which had a single OsHIR3 protein (Figure 2). Furthermore, synteny relationship analysis of OsHIR genes revealed collinearity among four out of the six OsHIRs (Supplementary Figure S1).
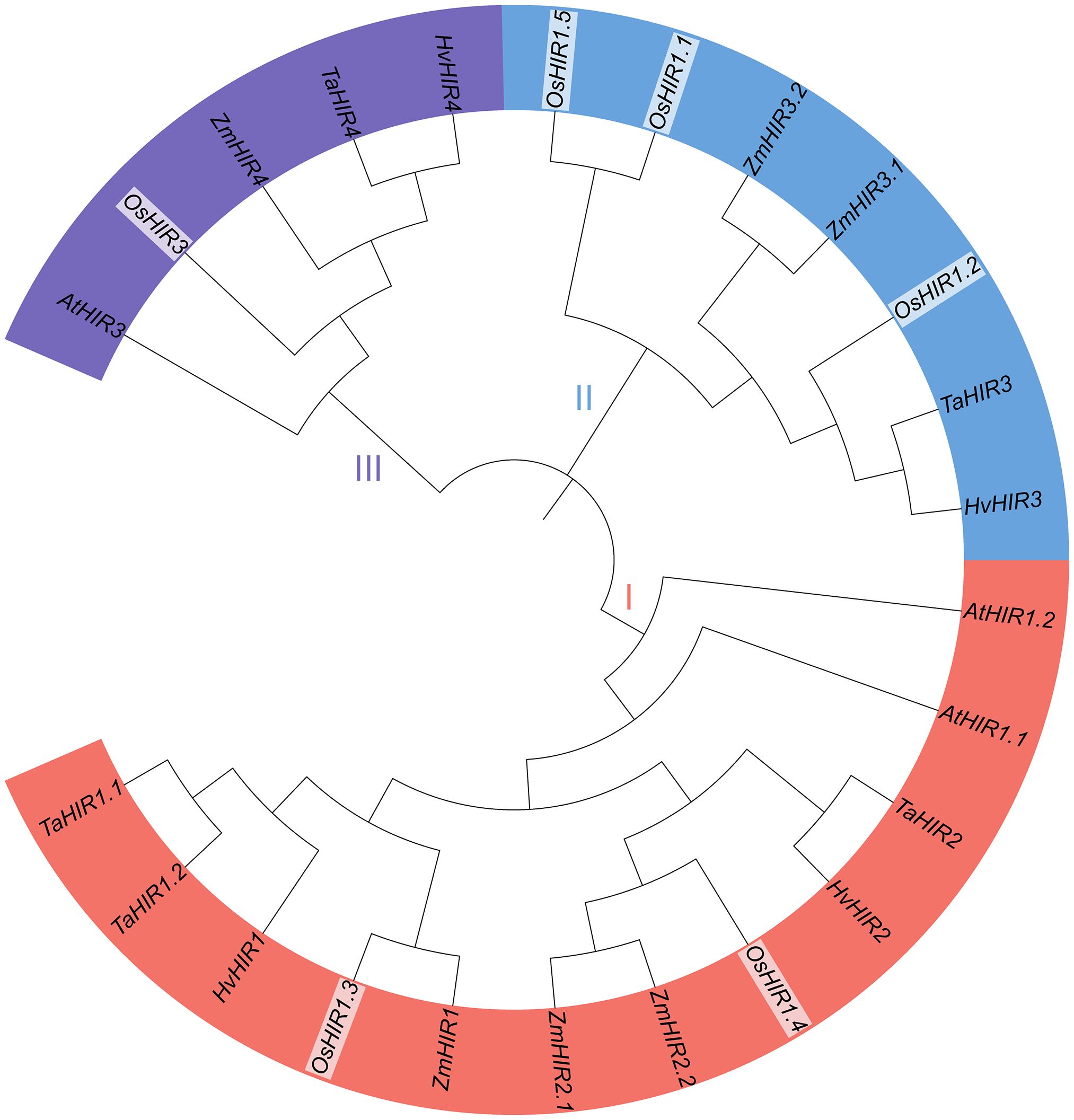
Figure 2. Phylogenetic analysis of HIR proteins among five plant species. The numbers on branches are bootstrap values calculated from 1000 replicates. AtHIRs, HIRs of Arabidopsis; OsHIRs, HIRs of Oryza sativa (rice); ZmHIRs, HIRs of Zea mays (maize); TaHIRs, HIRs of Triticum aestivum (wheat); HvHIRs, HIRs of Hordeum vulgare (barley). Clade I (red), II (blue), and III (purple) present the three phylogenetic groups.
To further clarify their evolutionary relationships, syntenic maps between Oryza sativa and Zea mays, Triticum aestivum, and Hordeum vulgare were constructed. 11, 8, and 4 homologous HIR gene pairs were shown between O. sativa and T. aestivum, Zea mays, and H. vulgare, respectively (Figure 3), whereas no pairs were found between A. thaliana and O. sativa (Supplementary Figure S2). These results indicated a closer evolutionary link between O. sativa and T. aestivum and Zea mays.
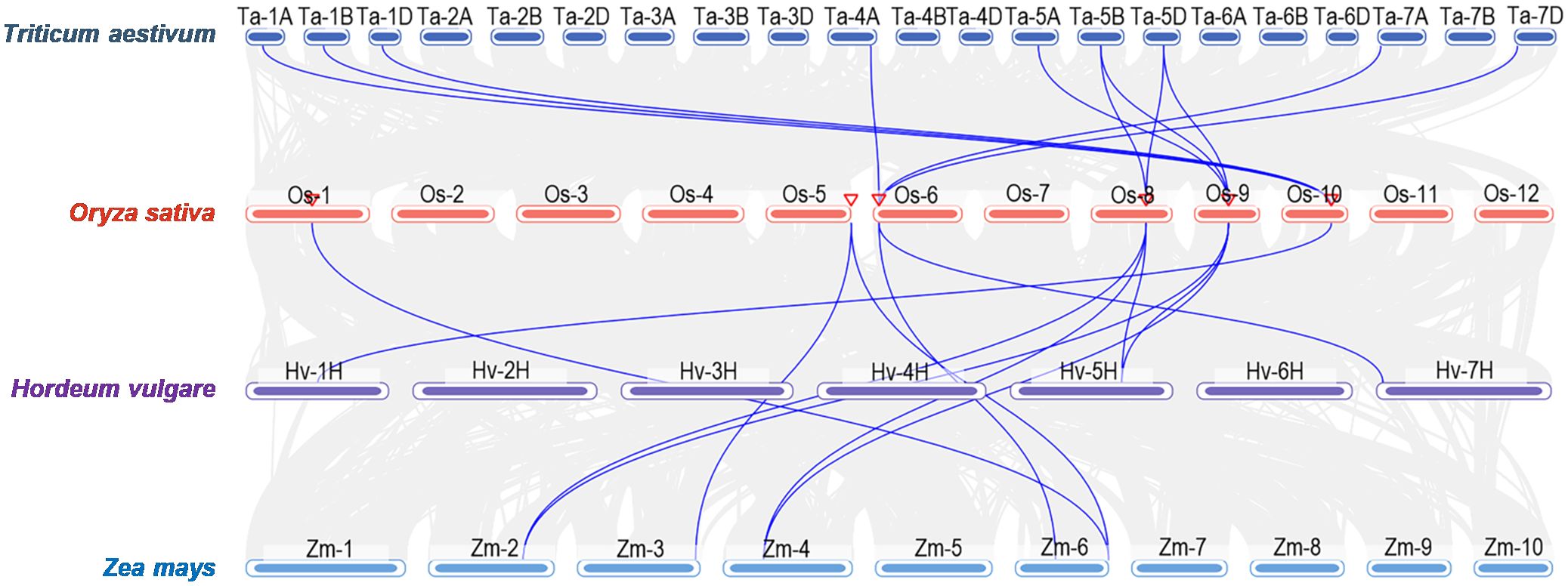
Figure 3. Syntenic analysis of HIR genes between Oryza sativa (rice) and other plant species including Zea mays (maize), Triticum aestivum (wheat), and Hordeum vulgare (barley).
3.3 Conserved motifs, gene structure, and domains analyses
To elucidate the evolution of OsHIR members, MEME analysis identified 20 conserved motifs. OsHIR1.2 and OsHIR1.5 contain all 20 predicted motifs, while OsHIR1.3 and OsHIR1.4 possess 19 motifs except motif 20. OsHIR1.1 contains 16 motifs, and OsHIR3 contains 17 motifs, excluding motif 13, 18, and 20 (Figure 4A). The gene structures of OsHIR are largely conserved, typically featuring 3-5 introns and 2-3 exons (Figure 4B). Notably, all OsHIR proteins contained the typical conserved SPFH domain, categorized under the PID superfamily (Supplementary Figure S3), indicating evolutionary conservation of functional domains among OsHIR proteins.
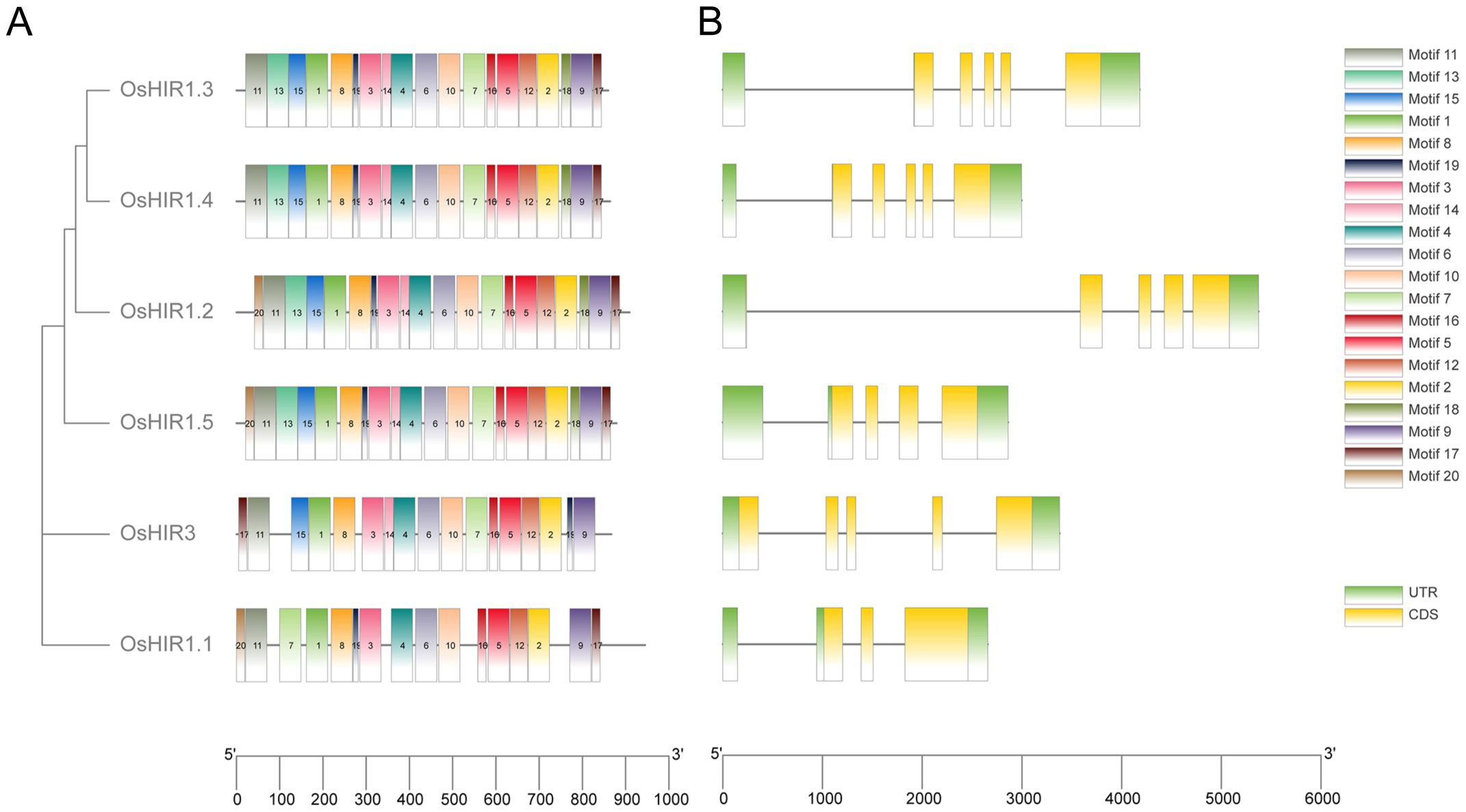
Figure 4. Analysis of conserved motifs and gene structure of Oryza sativa HIR genes. (A) Motif distribution of OsHIRs. Motifs from 1 to 20 are marked by different colors. (B) Gene structure analysis of OsHIRs. Untranslated regions, exons, and introns are shown as light green boxes, yellow boxes and horizontal lines, respectively.
3.4 Prediction of cis-acting elements in promoters
To investigate the roles of OsHIR genes, the promoter analysis was conducted. A total of 244 cis-acting elements, including 12 significant ones, with 48% being light responsive, were identified (Figure 5). Furthermore, 5 of these elements were linked to plant hormones, while 3 were related to abiotic stress responses, highlighting the importance of OsHIR genes in hormone and stress responses in plants (Figure 5).
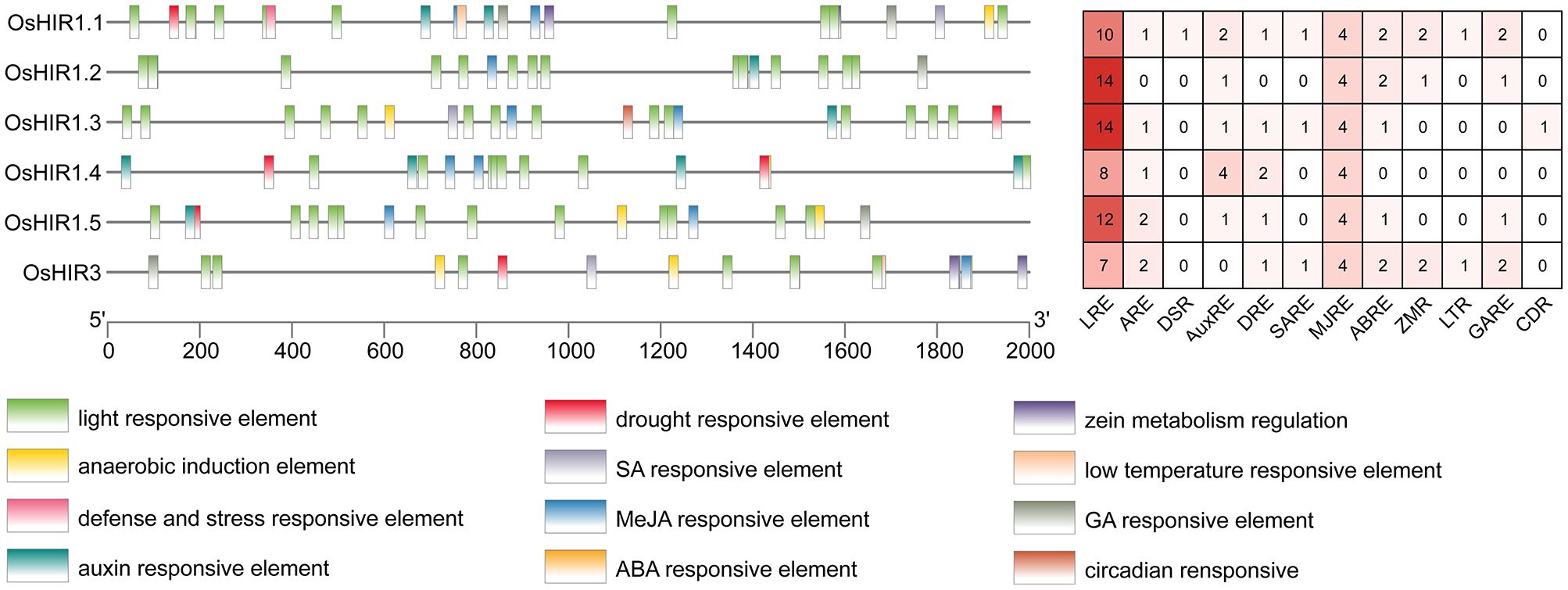
Figure 5. Cis-acting elements in the promoter regions of OsHIR genes. Different cis-acting elements in the promoters of OsHIRs were presented with differently colored boxes. The numbers of cis-acting elements was shown in the right. LRE, light responsive element; ARE, anaerobic induction element; DSR, defense and stress responsive element; AuxRE, auxin responsive element; DRE, drought responsive element; SARE, SA responsive element; MJRE, MeJA responsive element; ABRE, ABA responsive element; ZMR, zein metabolism regulation; LTR, low temperature responsive element; GARE, GA responsive element, CDR, circadian responsive.
3.5 Expression patterns of OsHIR genes in various tissues
The expression pattern of six OsHIR members across various tissues (root, stem, leaf) was analyzed using RT-qPCR. OsHIR3, OsHIR1.2, and OsHIR1.3 were expressed highly in the leaf, while the expression of OsHIR1.1, OsHIR1.4, and OsHIR1.5 was very low or undetectable in the stem and leaf (Figure 6).
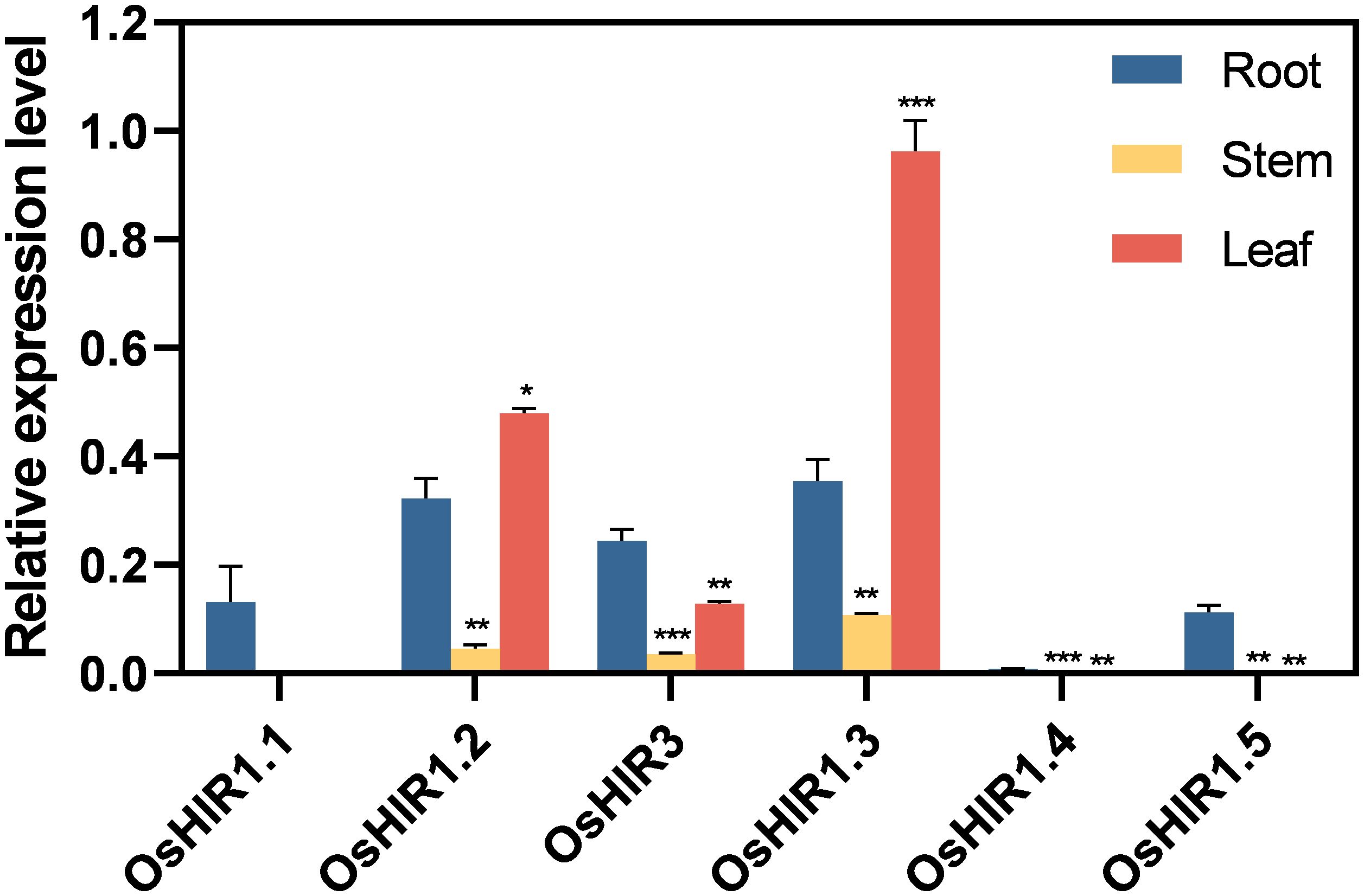
Figure 6. Expression profiles of OsHIR genes in different tissues. Data are mean ± SE (n = 3). The housekeeping gene OsActin was used as the reference transcript, which was set up as ‘1’. Asterisk represents a significant difference between the root and other tissues (*p< 0.05, **p< 0.01, and ***p< 0.001). Three independent experiments were conducted for experimental reproducibility.
3.6 Expression profiles under abiotic stress and hormonal treatment
Plants activate stress response genes when exposed to abiotic stress. Here, the transcription levels of four representative OsHIR members in rice treated with PEG6000, low temperature, ABA, GA, SA, or MeJA over varying durations were analyzed. Following PEG treatment, OsHIR3 expression level increased twofold at 6 h, while OsHIR1.2 and OsHIR1.3 were down-regulated at 3, 6, and 12 h. OsHIR1.2 was first down-regulated then up-regulated under cold stress, whereas OsHIR3 and OsHIR1.3 remained down-regulated. In contrast, OsHIR1.4 exhibited initial up-regulation followed by down-regulation under drought and cold stress (Figure 7).
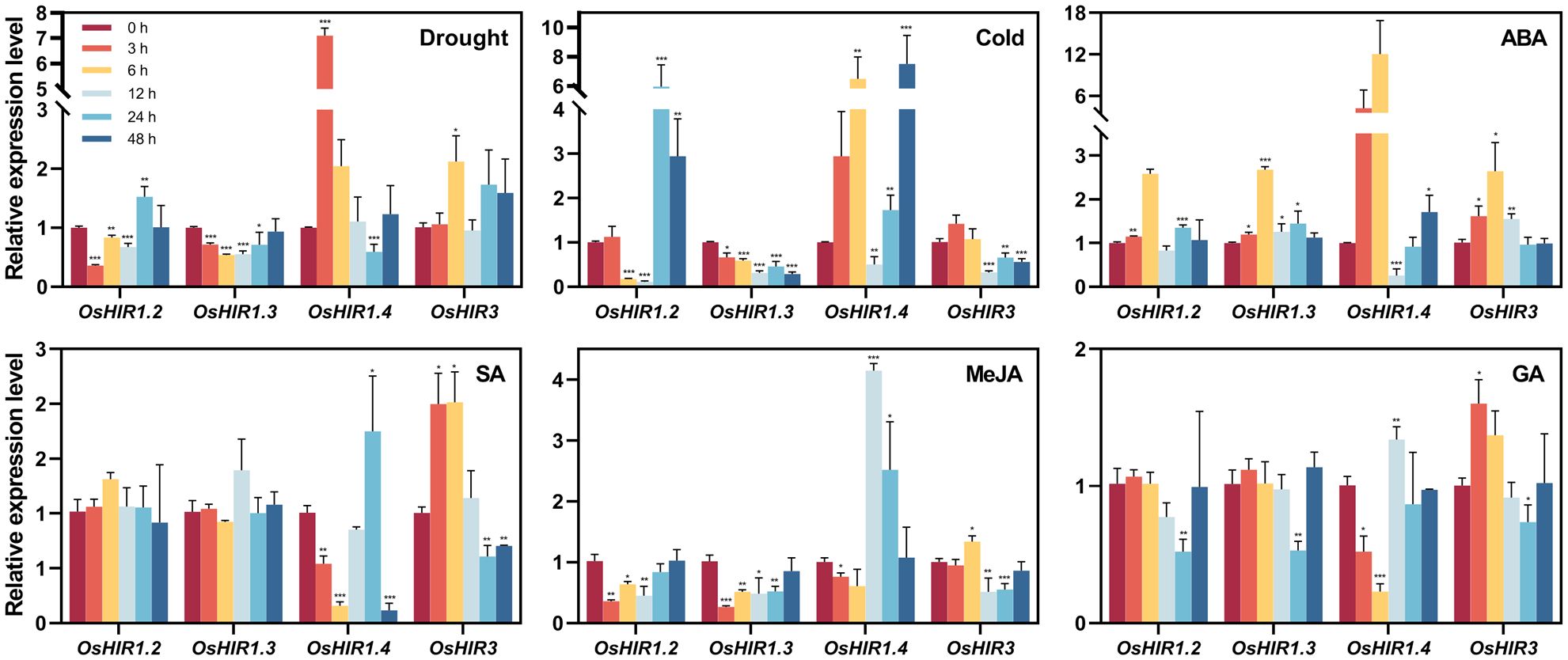
Figure 7. Expression profiles of OsHIR genes in response to different abiotic stress and plant hormone treatments. Four-week-old rice seedling were sprayed with 10% PEG6000, low temperature, 100 μM ABA, 100 μM GA, 100 μM MeJA or 2 mM SA with 0.02%(v/v) Tween 20. Plants sprayed with 0.02%(v/v) Tween 20 were used as mock control. After treatments, leaves were collected at 0, 3, 6, 12, 24, and 48h. Gene expression was normalized to control unstressed expression level, which was set up as ‘1’. The housekeeping gene OsActin was used as the reference transcript. Data are mean ± SE (n = 3). Asterisk represents a significant difference between the control and different treatment (*p< 0.05, **p< 0.01, and ***p< 0.001). Three independent experiments were conducted for experimental reproducibility.
Most OsHIR genes were up-regulated by ABA but down-regulated by GA and MeJA. OsHIR1.4 expression was inhibited at 12 h post-ABA treatment, showing an initial down-regulation followed by up-regulation under GA and MeJA. Notably, OsHIR3 expression doubled at 3 and 6 h following SA treatment, remained stable at 12 h, and fell at 24 and 48 h, while OsHIR1.2 and OsHIR1.3 remained unchanged. OsHIR1.4 showed significant inhibition at 3, 6, and 48 h, while it was up-regulated at 24 h (Figure 7). These findings reveal that OsHIR gene expression varies in response to abiotic stress and hormonal treatments.
4 Discussion
Hypersensitive-induced reaction (HIR) proteins, part of the PID superfamily, have been shown to be involved in plant defense responses (Nadimpalli et al., 2000). The number of HIR genes varies among species, reflecting the evolution of this gene family. Currently, researchers have identified four HIR genes in Arabidopsis (Qi et al., 2011), three in maize (Zea mays) (Nadimpalli et al., 2000), four in barley (Hordeum vulgare) (Rostoks et al., 2003), five in wheat (Triticum aestivum) (Yu et al., 2008), three in soybean (Glycine max L. Merr.) (Luan et al., 2019), as well as additional HIR genes in legume (Lotus japonicus), cucumber (Cucumis sativus), pepper (Capsicum annuum), apple (Malus domestica), rapeseed (Brassica napus L.) and rice (Li et al., 2022; Hakozaki et al., 2004; Kim et al., 2004; Zhao et al., 2019; Jung and Hwang, 2007; Chen et al., 2007). The identification of six HIR genes in the Oryza sativa genome suggested a diversification of the functions of OsHIR (Table 1).
Previous researches shown that most HIR proteins were localized to the plasma membrane (PM) (Choi et al., 2011; Duan et al., 2013; Qi et al., 2011; Wang et al., 2024; Li et al., 2019; Zhao et al., 2019; Zhou et al., 2010). Our findings corroborate this, as we observed that OsHIR proteins were also PM-localized and exhibited the characteristic SPFH domains typical of HIR proteins (Table 1, Figure 1; Supplementary Figure S3). Additonally, the phylogenetic analysis of HIR proteins from Arabidopsis, maize, wheat, barley, and rice demonstrated their division into three distinct groups. OsHIR1.3 and OsHIR1.4 were grouped with AtHIR1.1, AtHIR1.2, HvHIR1, HvHIR2, ZmHIR1, and TaHIR2 proteins. On the one hand, OsHIR1.1, OsHIR1.2, OsHIR1.5, ZmHIR3.1, ZmHIR3.2, TaHIR3, and HvHIR3 grouped together as a cluster. On the other hand, OsHIR3, AtHIR3, ZmHIR4, TaHIR4, and HvHIR4 formed another cluster (Figure 2). These results indicate a significant conservation level among HIR proteins across different plant species. Our analysis revealed that four out of the six OsHIR genes exhibited collinearity (Supplementary Figure S1), indicating the notion of evolutionary conservation within this gene family. Furthermore, the lack of collinearity between OsHIR3 and OsHIR1.5 with HIRs suggests possible evolutionary novelty (Supplementary Figure S1). Consistent with an earlier rice genome study (Huang et al., 2012), syntenic maps revealed that O. sativa shares a closer evolutionary relationship with T. aestivum and Zea mays (Figure 3). Collectively, these findings indicate that OsHIR proteins may have various activities in rice.
Cis-acting elements are particular sequences of DNA that interact with specific transcription factors (TFs) to regulate the expression of downstream genes. This study identified numerous cis-acting elements linked to responses to light, anaerobic, hormonal, and stress, located 2.0 kb upstream of OsHIR genes (Figure 5). In wheat (Triticum aestivum L.), HIR genes have been proven to respond to environmental stimuli like drought, high salinity, and low temperature (Duan et al., 2013). Here, light responsive elements (LREs), drought responsive elements (DREs), and low temperature responsive elements (LTRs) were also observed in OsHIR genes promoters, indicating their roles in abiotic stress response (Figure 5). In accordance with previous studies on wheat, apple, and rapeseed (Duan et al., 2013; Zhao et al., 2019; Li et al., 2022), diverse phytohormone response elements, such as the abscisic acid responsive element (ABRE), methyl jasmonate responsive element (MJRE), salicylic acid responsive element (SARE), and gibberellin responsive element (GARE), were found in all OsHIR genes promoters (Figure 5). These findings suggest that OsHIR genes play roles in the signal transduction network for stress response regulation in Oryza sativa.
Researches on functional diversity of HIR genes in various plant species have shown their roles in stress regulation, such as drought and cold stress responses in wheat (Duan et al., 2013), ABA response in apple (Zhao et al., 2019), and pathogen defense in Arabidopsis and other species (Mei et al., 2020; Sun et al., 2021; Wang et al., 2024; Zhao et al., 2019). All OsHIR genes exhibited consistent expression under normal conditions (Figure 6), highlighting their essential roles in rice. Consistent with the expression of HvHIRs in barley and BnHIRs in rapeseed (Rostoks et al., 2003; Li et al., 2022), OsHIR3, OsHIR1.2, and OsHIR1.3 genes were primarily expressed in leaves, with lower levels in stems (Figure 6). Resistance is one of the physiological basis for high crop yield, with leaves serving as primary organs for plant responses. The expression levels of four representative OsHIR members (OsHIR3, OsHIR1.2, OsHIR1.3, and OsHIR1.4) from three clades under both normal and stress conditions were evaluated to explore their functions in rice. Gene expression level was normalized to unstressed controls, which was set up as ‘1’. The expression patterns of OsHIR genes shifted under various abiotic stresses and hormone treatments. In wheat, TaHIR1 and TaHIR3 levels dropped during low temperature, drought, and salinity (Duan et al., 2013), a trend we observed in our study (Figure 7). PEG treatment down-regulated OsHIR1.2 and OsHIR1.3 at 3, 6, and 12 h, while OsHIR3 was up-regulated twofold at 6 h. Cold stress down-regulated OsHIR3 and OsHIR1.3, whereas OsHIR1.2 initially decreased then increased. Notably, OsHIR1.4 increased then decreased under drought and cold stress (Figure 7). Previous research has shown raised SA levels in OsHIR3-overexpressed rice and CaHIR1-silenced plants during X. campestris pv. Vesicatoria infection, suggesting a potential link between SA and HIRs (Li et al., 2019; Choi et al., 2011). The qPCR analysis showed that OsHIR3 and OsHIR1.4 were sensitive to all hormonal treatments. Specifically, OsHIR3 peaked at 3 or 6 h, followed by a decline at 24 h after treatments with ABA, GA, SA, and MeJA. Conversely, OsHIR1.4 was first down-regulated, then up-regulated by those hormonal treatments (Figure 7). It can be speculated that OsHIR3 and OsHIR1.4 may have significant roles in response to both abiotic and hormonal stresses. In contrast to reports that ABA down-regulated TaHIR1 and TaHIR3, and MeJA increased MdHIR4 (Duan et al., 2013; Zhao et al., 2019), we noted that the other OsHIR members (OsHIR1.2 and OsHIR1.3) was up-regulated by ABA, down-regulated by GA and MeJA, while unchanged by SA treatment. These observations indicated diverse HIR responses to plant hormones, particularly the defense hormones SA and MeJA. Our findings highlighted that OsHIRs from different clades might play varied and dominant roles in maintaining a balance of hormones in plants at different stages of resistance response.
5 Conclusion
In conclusion, we identified a total number of six OsHIR genes from the Oryza sativa genome. Analyses of physicochemical properties, phylogenetics, conserved motifs, gene structure, cis-acting elements in promoters, and expression patterns were conducted to understand the functions of OsHIR genes. The findings indicate that all four OsHIR members tested responded to PEG6000, low temperatures, ABA, SA, MeJA, and GA, highlighting the significant roles of O. sativa HIR genes in mediating plant reactions to different abiotic stimuli. This genome-wide identification and characterization of the HIR family in O. sativa serves as a foundation for further functional studies and for enhancing breeding and genetic improvement in Oryza sativa.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
JL: Methodology, Visualization, Writing – original draft. LShao: Data curation, Validation, Writing – original draft, Writing – review & editing. QW: Formal analysis, Investigation, Writing – original draft. LShi: Validation, Writing – original draft. WW: Methodology, Writing – original draft. WC: Supervision, Writing – original draft. ZY: Resources, Writing – review & editing. SL: Resources, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was supported by the National Natural Science Foundation of China (Young Program) (32102304), and the Zhejiang Provincial Top Discipline of Biological Engineering (ZS2022005, Level A, Zhejiang Wanli University, Ningbo 315100, China).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1492026/full#supplementary-material
Supplementary Figure 1 | Chromosomal distribution and synteny analysis of OsHIR genes from Oryza sativa. The blue boxes with different lengths labeled as Chr1 to Chr12 stood for the different chromosomes of O. sativa. The locations of each OsHIR gene from O. sativa on the chromosomes were displayed in black. The synteny relationships between different OsHIRs were indicated with red lines.
Supplementary Figure 2 | Syntenic analysis of HIR genes between Oryza sativa (rice) and Arabidopsis thaliana.
Supplementary Figure 3 | Domain prediction of Oryza sativa HIR proteins. Green boxes: SPFH domains.
References
Block, A., Alfano, J. R. (2011). Plant targets for Pseudomonas syringae type III effectors: virulence targets or guarded decoys? Curr. Opin. Microbiol. 14, 39–46. doi: 10.1016/j.mib.2010.12.011
Chen, C., Wu, Y., Li, J., Wang, X., Zeng, Z., Xu, J., et al. (2023). TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 16, 1733–1742. doi: 10.1016/j.molp.2023.09.010
Chen, F., Yuan, Y., Li, Q., He, Z. (2007). Proteomic analysis of rice plasma membrane reveals proteins involved in early defense response to bacterial blight. Proteomics 7, 1529–1539. doi: 10.1002/pmic.200500765
Chen, K. Q., Zhao, X. Y., An, X. H., Tian, Y., Liu, D. D., You, C. X., et al. (2017). MdHIR proteins repress anthocyanin accumulation by interacting with the MdJAZ2 protein to inhibit its degradation in apples. Sci. Rep. 7, 44484. doi: 10.1038/srep44484
Choi, H. W., Kim, Y. J., Hwang, B. K. (2011). The hypersensitive induced reaction and leucine-rich repeat proteins regulate plant cell death associated with disease and plant immunity. Mol. Plant Microbe Interact. 24, 68–78. doi: 10.1094/MPMI-02-10-0030
Duan, Y. H., Guo, J., Shi, X. X., Guan, X. N., Liu, F. R., Bai, P. F., et al. (2013). Wheat hypersensitive-induced reaction genes TaHIR1 and TaHIR3 are involved in response to stripe rust fungus infection and abiotic stresses. Plant Cell Rep. 32, 273–283. doi: 10.1007/s00299-012-1361-6
Greenberg, J. T., Yao, N. (2004). The role and regulation of programmed cell death in plant-pathogen interactions. Cell Microbiol. 6, 201–211. doi: 10.1111/j.1462-5822.2004.00361.x
Hakozaki, H., Endo, M., Masuko, H., Park, J. I., Ito, H., Uchida, M., et al. (2004). Cloning and expression pattern of a novel microspore-specific gene encoding hypersensitive-induced response protein (LjHIR1) from the model legume, Lotus japonicus. Genes Genet. Syst. 79, 307–310. doi: 10.1266/ggs.79.307
Huang, X., Kurata, N., Wei, X., Wang, Z. X., Wang, A., Zhao, Q., et al. (2012). A map of rice genome variation reveals the origin of cultivated rice. Nature 490, 497–501. doi: 10.1038/nature11532
Jones, J. D., Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Jung, H. W., Hwang, B. K. (2007). The leucine-rich repeat (LRR) protein, CaLRR1, interacts with the hypersensitive induced reaction (HIR) protein, CaHIR1, and suppresses cell death induced by the CaHIR1 protein. Mol. Plant Pathol. 8, 503–514. doi: 10.1111/j.1364-3703.2007.00410.x
Karrer, E. E., Beachy, R. N., Holt, C. A. (1998). Cloning of tobacco genes that elicit the hypersensitive response. Plant Mol. Biol. 36, 681–690. doi: 10.1023/a:1005949304445
Kim, M. S., Kim, Y. C., Cho, B. H. (2004). Gene expression analysis in cucumber leaves primed by root colonization with Pseudomonas chlororaphis O6 upon challenge-inoculation with Corynespora cassiicola. Plant Biol. (Stuttg) 6, 105–108. doi: 10.1055/s-2004-817803
Lescot, M., Dehais, P., Thijs, G., Marchal, K., Moreau, Y., Van De Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Li, M. Q., Tang, Y. Q., Yu, M. N., Fan, Y. H., Khan, S. U., Chang, W., et al. (2022). Systematic Characterization of Brassica napus HIR Gene Family Reveals a Positive Role of BnHIR2.7 in Sclerotinia sclerotiorum Resistance. Horticulturae 8, 874. doi: 10.3390/horticulturae8100874
Li, S., Zhao, J., Zhai, Y., Yuan, Q., Zhang, H., Wu, X., et al. (2019). The hypersensitive induced reaction 3 (HIR3) gene contributes to plant basal resistance via an EDS1 and salicylic acid-dependent pathway. Plant J. 98, 783–797. doi: 10.1111/tpj.14271
Luan, H., Liao, W., Niu, H., Cui, X., Chen, X., Zhi, H. (2019). Comprehensive analysis of soybean mosaic virus P3 protein interactors and hypersensitive response-like lesion-inducing protein function. Int. J. Mol. Sci. 20, 3388. doi: 10.3390/ijms20143388
Mei, Y., Ma, Z., Wang, Y., Zhou, X. (2020). Geminivirus C4 antagonizes the HIR1-mediated hypersensitive response by inhibiting the HIR1 self-interaction and promoting degradation of the protein. New Phytol. 225, 1311–1326. doi: 10.1111/nph.16208
Nadimpalli, R., Yalpani, N., Johal, G. S., Simmons, C. R. (2000). Prohibitins, stomatins, and plant disease response genes compose a protein superfamily that controls cell proliferation, ion channel regulation, and death. J. Biol. Chem. 275, 29579–29586. doi: 10.1074/jbc.M002339200
Qi, Y., Tsuda, K., Nguyen Le, V., Wang, X., Lin, J., Murphy, A. S., et al. (2011). Physical association of Arabidopsis hypersensitive induced reaction proteins (HIRs) with the immune receptor RPS2. J. Biol. Chem. 286, 31297–31307. doi: 10.1074/jbc.M110.211615
Rombauts, S., Dehais, P., Van Montagu, M., Rouze, P. (1999). PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Res. 27, 295–296. doi: 10.1093/nar/27.1.295
Rostoks, N., Schmierer, D., Kudrna, D., Kleinhofs, A. (2003). Barley putative hypersensitive induced reaction genes: genetic mapping, sequence analyses and differential expression in disease lesion mimic mutants. Theor. Appl. Genet. 107, 1094–1101. doi: 10.1007/s00122-003-1351-8
Sun, Y., Ruan, X., Wang, Q., Zhou, Y., Wang, F., Ma, L., et al. (2021). Integrated gene co-expression analysis and metabolites profiling highlight the important role of zmHIR3 in maize resistance to gibberella stalk rot. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.664733
Wang, P., Wang, Y., Hu, Y., Chen, Z., Han, L., Zhu, W., et al. (2024). Plant hypersensitive induced reaction protein facilitates cell death induced by secreted xylanase associated with the pathogenicity of Sclerotinia sclerotiorum. Plant J. 118, 90–105. doi: 10.1111/tpj.16593
Yoshida, R., Nei, M. (2016). Efficiencies of the NJp, maximum likelihood, and bayesian methods of phylogenetic construction for compositional and noncompositional genes. Mol. Biol. Evol. 33, 1618–1624. doi: 10.1093/molbev/msw042
Yu, X. M., Yu, X. D., Qu, Z. P., Huang, X. J., Guo, J., Han, Q. M., et al. (2008). Cloning of a putative hypersensitive induced reaction gene from wheat infected by stripe rust fungus. Gene 407, 193–198. doi: 10.1016/j.gene.2007.10.010
Yu, X. M., Zhao, W. Q., Yang, W. X., Liu, F., Chen, J. P., Goyer, C., et al. (2013). Characterization of a hypersensitive response-induced gene taHIR3 from wheat leaves infected with leaf rust. Plant Mol. Biol. Rep. 31, 314–322. doi: 10.1007/s11105-012-0504-9
Zhao, X. Y., Qi, C. H., Jiang, H., Zheng, P. F., Zhong, M. S., Zhao, Q., et al. (2019). Functional identification of apple on MdHIR4 in biotic stress. Plant Sci. 283, 396–406. doi: 10.1016/j.plantsci.2018.10.023
Keywords: rice, HIR, gene family, expression profiles, abiotic stress
Citation: Li J, Shao L, Wang Q, Shi L, Wu W, Chen W, Yang Z and Li S (2024) Genome-wide identification and expression profile of HIR gene family members in Oryza sativa L. Front. Plant Sci. 15:1492026. doi: 10.3389/fpls.2024.1492026
Received: 06 September 2024; Accepted: 31 October 2024;
Published: 19 November 2024.
Edited by:
Meng Jiang, Zhejiang University, ChinaReviewed by:
Gaojie Hong, Zhejiang Academy of Agricultural Sciences, ChinaLi Zhu, Chinese Academy of Agricultural Sciences, China
Tianye Zhang, Ningbo University, China
Copyright © 2024 Li, Shao, Wang, Shi, Wu, Chen, Yang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhenfeng Yang, eWFuZ3pmQHp3dS5lZHUuY24=; Saisai Li, c3NsaTEyMTJAend1LmVkdS5jbg==
 Jiahao Li
Jiahao Li Lingyu Shao
Lingyu Shao Liyu Shi
Liyu Shi Wei Wu
Wei Wu Zhenfeng Yang
Zhenfeng Yang Saisai Li
Saisai Li