- State Key Laboratory for Quality Ensurance and Sustainable Use of Dao-di Herbs, Institute of Medicinal Plant Development, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China
The rhizosphere, as the “frontline” of plant life, connects plant roots, rhizosphere microorganisms, and surrounding soil, plays a crucial role in plant growth and health, particularly in sustainable agriculture. Despite the well-established contribution of plant-microbe interactions to plant health, the specific molecular mechanisms remain insufficiently understood. This review aims to summarize the physiological adjustments and signal modulation that both plants and microorganisms undergo within this unique ecological niche to ensure successful colonization. By analyzing key processes such as chemotaxis, root attachment, immune evasion, and biofilm formation, we uncover how plants precisely modulate root exudates to either recruit or repel specific microorganisms, thereby shaping their colonization patterns. These findings provide new insights into the complexity of plant-microbe interactions and suggest potential directions for future research in sustainable agriculture.
Introduction
The rhizosphere is the nutrient-rich zone of soil surrounding plant roots, which is characterized by intense microbial activity and diversity. It is regarded as one of the most intricate ecosystems on Earth, mediating subterranean interactions between plants and microorganisms (Goswami and Deka, 2022). These interactions, ranging from mutualism to parasitism, can be classified as forms of symbiosis (Plett and Martin, 2018; Du et al., 2021a). In mutualistic relationships, beneficial microorganisms such as rhizobia, mycorrhizae, endophytes (including plant growth-promoting microorganisms, PGPMs), and epiphytes establish positive interactions with plants, providing protection against stresses, enhancing growth, nutrient uptake, and improving soil conditions (Kiers et al., 2011; Zamioudis and Pieterse, 2012; Jin et al., 2019; Ding et al., 2022; Koprivova and Kopriva, 2022). In contrast, parasitism causes diseases that negatively affect plant growth (Venturi and Fuqua, 2013; Zhang et al., 2023).
“Rhizosphere colonization” refers to the process through which microorganisms establish themselves in the rhizosphere soil or on the root surface, thereby forming stable communities (Liu et al., 2024). Among these microorganisms, bacteria are particularly significant due to their widespread presence and profound influence on plant health and soil ecosystems (Liu et al., 2024). The root surface serves as a critical interface for direct interactions between bacteria and plants, making it essential to understand the mechanisms of bacterial colonization in this area to elucidate plant-microbe interactions. Consequently, this review focuses primarily on the process of bacterial colonization on root surfaces. The colonization process typically follows four key steps (Figure 1): (i) chemotactic signal recognition; (ii) attachment to the root surface; (iii) evasion of plant immune defenses; and (iv) biofilm formation on the root surface. Upon completing these steps, endophytic bacteria may subsequently enter plant internal tissues, facilitating more direct interactions with the plant (Dudeja et al., 2021; Mushtaq et al., 2023). Throughout the colonization process, both plants and bacteria adjust their physiological states, guided by molecular signals, which include chemical molecules and effector proteins. These signals, whether specific or non-specific, are perceived through receptors and signaling pathways, influencing bacterial colonization behavior (Badri et al., 2009). This “ molecular dialogue” is crucial for the successful colonization and the establishment of symbiotic relationships in the rhizosphere.
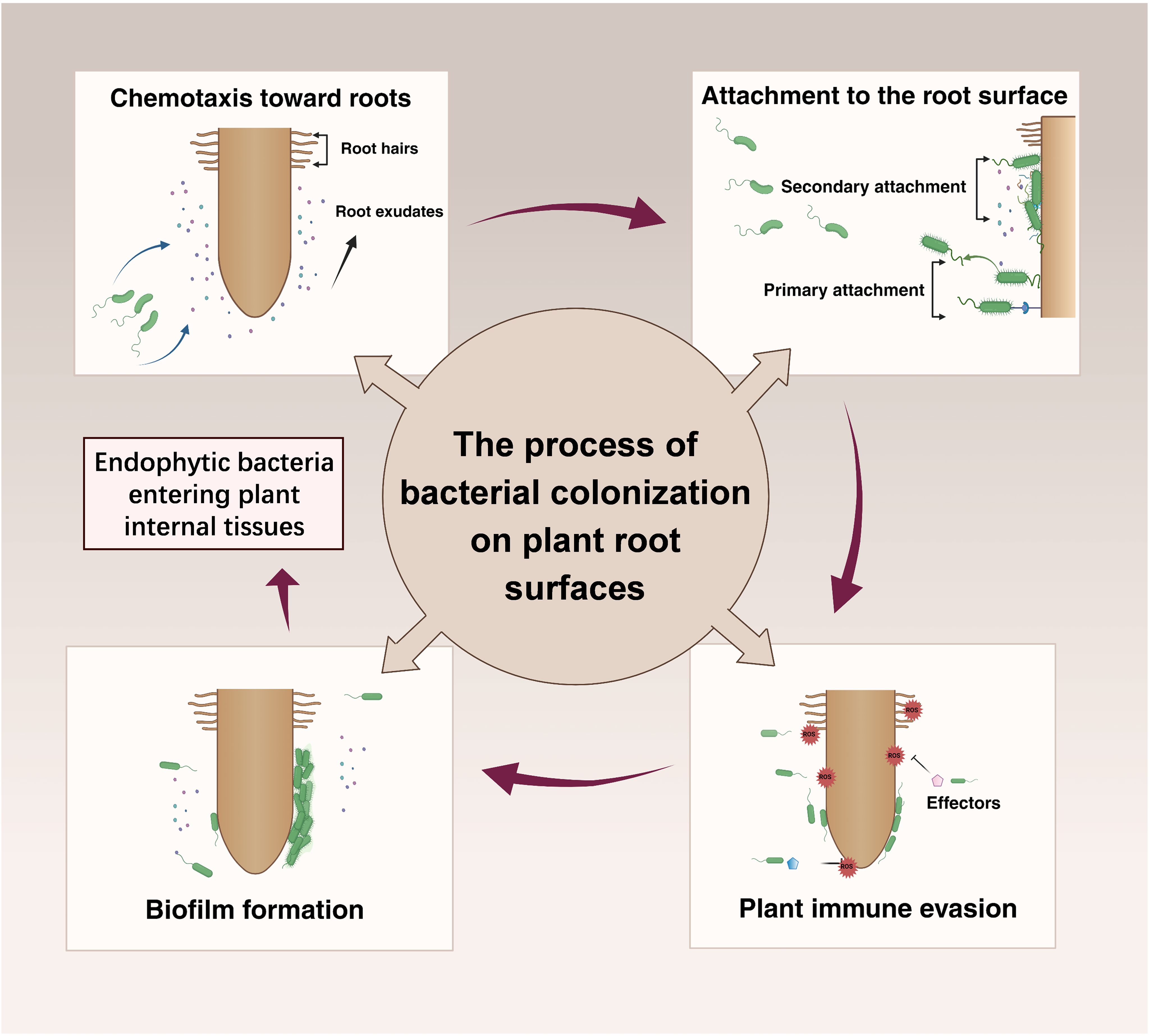
Figure 1. The colonization process of most bacteria can generally be divided into the following steps: chemotaxis, attachment, growth on the root surface, and biofilm formation. The chemotaxis and motility of microorganisms not only determine their movement toward the rhizosphere but also influence their initial positioning and migration of colonization sites. Following attachment to the root surface, microorganisms must overcome plant immune responses. Biofilm formation is essential for most bacteria colonizing the rhizosphere soil and root surfaces, and during this process, the bacteria utilize root exudates as a carbon source, which is a prerequisite for biofilm formation. Root exudates play a crucial role in influencing microbial colonization, and throughout the colonization process, a series of molecular dialogues occur between the interacting partners, indicating complex plant-microbe interactions.
In this review, we comprehensively summarize the process of rhizosphere microbial colonization and its interactions with plants. By conducting an in-depth analysis of regulatory signals, such as chemical molecules and effector proteins, we delve into the “dialogue” mechanisms existing between plants and microorganisms. This review aims to provide a reference for future research and to advance the application of microorganisms in modern agriculture.
Chemotactic signals emitted by plants roots
The chemotactic movement of soil microorganisms constitutes the fundamental basis of plant-microbe interactions, where root exudates frequently serve as communication molecules within this process (Chagas et al., 2018). Root exudates consist of significant amounts of carbon generated by plant photosynthesis, including low molecular weight compounds such as sugars, organic acids, and secondary metabolites (e.g., flavonoids, glucosinolates, and coumarins). These compounds make the rhizosphere a more active site for microbial colonization compared to bulk soil (Bais et al., 2006). Additionally, there are high molecular weight compounds like mucilage and proteins, despite their lower variability, they constitute the principal components of exudates (Chen and Liu, 2024). It is worth noting that due to the differential utilization and metabolic potential of various substrates among soil microorganisms, the chemical valence of rhizosphere exudates can drive positive or negative chemotactic movements in microbes (Feng et al., 2021; Knights et al., 2021).The following are detailed examples further illustrating the specific roles of root exudates in microbial chemotaxis:
1. Primary metabolites, such as sugars, amino acids, and organic acids, play essential roles in the rhizosphere by serving as carbon sources, chemoattractants, and chemical signals, thereby shaping microbial community structure and facilitating root colonization. Sugars are important carbon sources and colonization signals for rhizosphere microorganisms, influencing microbial community structure. In barley, key carbon compounds in root exudates help beneficial Pseudomonas adapt. A decrease in exudate diversity and higher glucose and fructose levels lead to more Pseudomonas (Pacheco-Moreno et al., 2024). Sugar molecules also positively influence the colonization of beneficial Gram-positive bacteria, particularly Bacillus. Additionally, sucrose, a widely present disaccharide, selectively shapes Bacillus in soil microbial communities. When Bacillus subtilis encounters sucrose exuded by Arabidopsis roots, it initiates the “levan detour” signaling cascade, activating solid surface motility (SSM), thereby enhancing its movement and colonization ability in the root environment (Tian et al., 2021).
In addition to sugars, amino acids in root exudates serve as key chemoattractants for various rhizosphere bacteria. For example, in Sesbania rostrata, the amino acids histidine, arginine, and aspartate act as chemoattractants for Azorhizobium caulinodans, not only facilitating bacterial chemotaxis but also upregulating the expression of genes involved in chemotaxis and motility. This dual role of amino acids underscores their importance in biofilm formation and root colonization (Liu et al., 2019). Moreover, in Pseudomonas fluorescens Pf0-1, the chemotaxis sensory proteins CtaA, CtaB, and CtaC have been identified as critical for amino acid-driven root colonization. Mutants lacking these proteins demonstrate reduced competitiveness, highlighting the pivotal role of amino acids as chemoattractants in the colonization process (Oku et al., 2012).
Organic acids further contribute to the rhizosphere’s ecological dynamics by regulating soil pH and increasing mineral solubility, thereby promoting nutrient uptake by plants. They also act as chemical signals during the chemotactic colonization of rhizosphere bacteria. Studies have shown that the high release of citric, pyruvic, succinic, and fumaric acids may account for the enrichment of Comamonadaceae (Wen et al., 2020). When Arabidopsis thaliana leaves are infected by the tomato pathogen Pseudomonas syringae pv. tomato DC3000, roots secrete L-malic acid (MA) as a signal to recruit beneficial rhizosphere bacteria Bacillus subtilis FB17 (Rudrappa et al., 2008).
2. Secondary metabolites can serve as attractants for specific microbial strains, promoting beneficial interactions within the rhizosphere. For instance, the secretion of coumarins by Arabidopsis thaliana under iron deficiency attracts beneficial microbes, which enhance plant growth by improving iron uptake (Harbort et al., 2020). On the other hand, secondary metabolites often act as negative chemotactic agents, playing a crucial role in defending plants against pathogens. For instance, glucosinolates (GSLs), secondary metabolites unique to cruciferous plants, are hydrolyzed to produce glucosinolate aglycones, which rearrange into isothiocyanates with potent antimicrobial activity (Wittstock and Burow, 2010; Siebers et al., 2018). Moreover, studies on flavonoids provide important insights into another mechanism by which secondary metabolites resist pathogens. In high-density microbial populations, intercellular communication triggers the production of virulence factors through a process known as quorum sensing (QS) (Fuqua et al., 2001). While quorum sensing is a mechanism of communication that facilitates cooperative behavior in many microbial communities, it may not function as a virulence strategy in all microbes. Research has demonstrated that catechins significantly reduce the expression of key QS regulatory genes lasI, lasR, rhlI, and rhlR in Pseudomonas aeruginosa, leading to decreased production of QS factors and forming a line of defense against pathogen attacks (Vandeputte et al., 2010). This suggests that secondary metabolites can not only directly kill pathogens but also inhibit pathogen virulence by disrupting intercellular bacterial communication, thereby exerting negative chemotactic effects on pathogenic bacteria.
The regulatory role of secondary metabolites in the rhizosphere extends beyond direct interactions with pathogens. For instance, the decomposition product of benzoxazinoids, 6-methoxy-benzoxazolin-2-one (MBOA), indirectly determines the composition of the next generation of rhizosphere microorganisms by altering the associated microbial community around the roots, thereby extending its regulatory effects into the soil (Hu et al., 2018). Benzoxazinoids can also influence soil microbial communities by regulating the release of secondary metabolites from rhizosphere plants, particularly flavonoids (Cotton et al., 2019). These findings further demonstrate that secondary metabolites, as signaling molecules, play a crucial role not only in facilitating communication between plants and microorganisms, but also in influencing interactions among microorganisms themselves. Additionally, these compounds significantly impact the soil environment within the rhizosphere. The regulatory effects of secondary metabolites are highly complex, depending on both the diversity of species that respond to these signals and the specific environmental conditions under which they function.
3. Volatile organic compounds (VOCs) primarily consist of terpenes, aromatic compounds, nitrogen-containing compounds, and fatty acid derivatives, as well as volatile plant hormones like methyl jasmonate and methyl salicylate as low molecular weight substances released by plant roots (Holopainen, 2004). VOCs, such as essential oils, have long been recognized for their broad antimicrobial activity, potentially playing a crucial role in mediating negative chemotaxis of pathogens. However, recent evidence indicates that VOCs can also serve as energy sources or signaling molecules, promoting the enrichment of beneficial bacteria by regulating bacterial growth, chemotaxis, and competitive ability, thereby exerting positive chemotactic effects (de la Porte et al., 2020; Sharifi et al., 2022). Due to their physicochemical properties, VOCs are more likely to diffuse throughout the soil layer, attracting bacteria at distances ranging from a few millimeters to as far as 12 centimeters from the roots, thereby regulating the structure of a broad microbial community (Schulz-Bohm et al., 2018). Schulz-Bohm et al. (2018) utilized an olfactometer system to verify that VOCs released by the roots of Carex arenaria could induce long-distance migration of soil bacteria toward the roots, explaining the crucial role of plant VOCs in facilitating long-distance plant-microbe interactions. Therefore, VOCs play a key role in long-distance interactions in the soil rhizosphere.
4. Complex polymers such as polysaccharides, proteins, and enzymes, known as high molecular weight (HMW) compounds, are also present in root exudates, although they are not easily utilized by soil microorganisms (Goswami and Deka, 2022). However, mucilage (polysaccharides) can form protective barriers, promote soil particle adhesion, and assist in the colonization of rhizosphere microorganisms (Read and Gregory, 1997). Additionally, proteins in root exudates, particularly those with enzymatic activity, enhance plant defense mechanisms through their constitutive release. For example, peroxidases can break down pathogen cell walls, induce defense signaling, and trigger plant immune responses, effectively curbing pathogen invasion. These enzymatic proteins can also recognize and recruit beneficial microorganisms, promoting the formation of symbiotic relationships and further enhancing plant health and stress resistance (Wen et al., 2007; De-la-Peña et al., 2010).
Chemotactic signal reception and response by microorganisms
Upon perceiving chemotactic signals from plants, soil microorganisms move along a chemical gradient towards (attraction) or away from (repulsion) the signal source, engaging in chemotactic movement (Kearns, 2010). The chemotaxis system in bacteria is one of the most complex signal transduction systems in prokaryotes, with signaling and regulatory mechanisms relatively conserved across all bacteria. Specifically, bacterial chemoreceptor sensor proteins (MCPs) serve as receptors for chemotactic agents, sensing rhizosphere chemical effectors (Allard-Massicotte et al., 2016; Compton et al., 2018). Typically, MCPs are transmembrane proteins that form a ternary complex with histidine kinase CheA and coupling protein CheW (Sampedro et al., 2015). MCPs are located in the ligand-binding domain (LBD) of the cell membrane, which exhibits high structural variability to sense a wide range of chemical signals. For instance, the PGPR strain Bacillus velezensis SQR9 can sense various substances, including organic acids, sugars, amino acids, and sugar alcohols, primarily due to the pivotal role of its eight MCPs in mediating host interactions (Corral-Lugo et al., 2016; Feng et al., 2019; Tohidifar et al., 2020). When MCPs selectively recognize and bind specific root exudates, they trigger subsequent signal transduction and execution modules (Sampedro et al., 2015). Specifically, signal transduction regulates the autophosphorylation rate of histidine kinase CheA in a CheW-dependent manner, and phosphorylated CheA further influences the transphosphorylation of the CheY response regulator (McEvoy et al., 1999; Lacal et al., 2010). Ultimately, phosphorylated CheY interacts with motility proteins, mediating bacterial movement (Sampedro et al., 2015). The motility of rhizosphere bacteria manifests in various phenotypic forms, including swarming motility, swimming motility, gliding motility, and twitching motility (Kearns, 2010). These motility forms are driven primarily by flagellar rotation of individual bacterial cells, coordinated flagellar complex movement within the population, an extension of polar type IV pili, or passive surface spreading (Mattick, 2002; Mignot, 2007; Kearns, 2010). This sophisticated chemotaxis system enables bacteria to keenly sense the concentration gradient of extracellular signaling molecules, allowing them to adapt to changes in their surrounding environment. However, current research primarily focuses on bacterial responses to single chemotactic signals. In reality, both root exudates and bacterial MCPs exhibit high diversity, indicating that responses to composite signals may differ significantly from responses to single signals (Figure 2). Therefore, future research should focus more on bacterial responses to composite signals to gain a more comprehensive understanding of their behavior in complex environments.
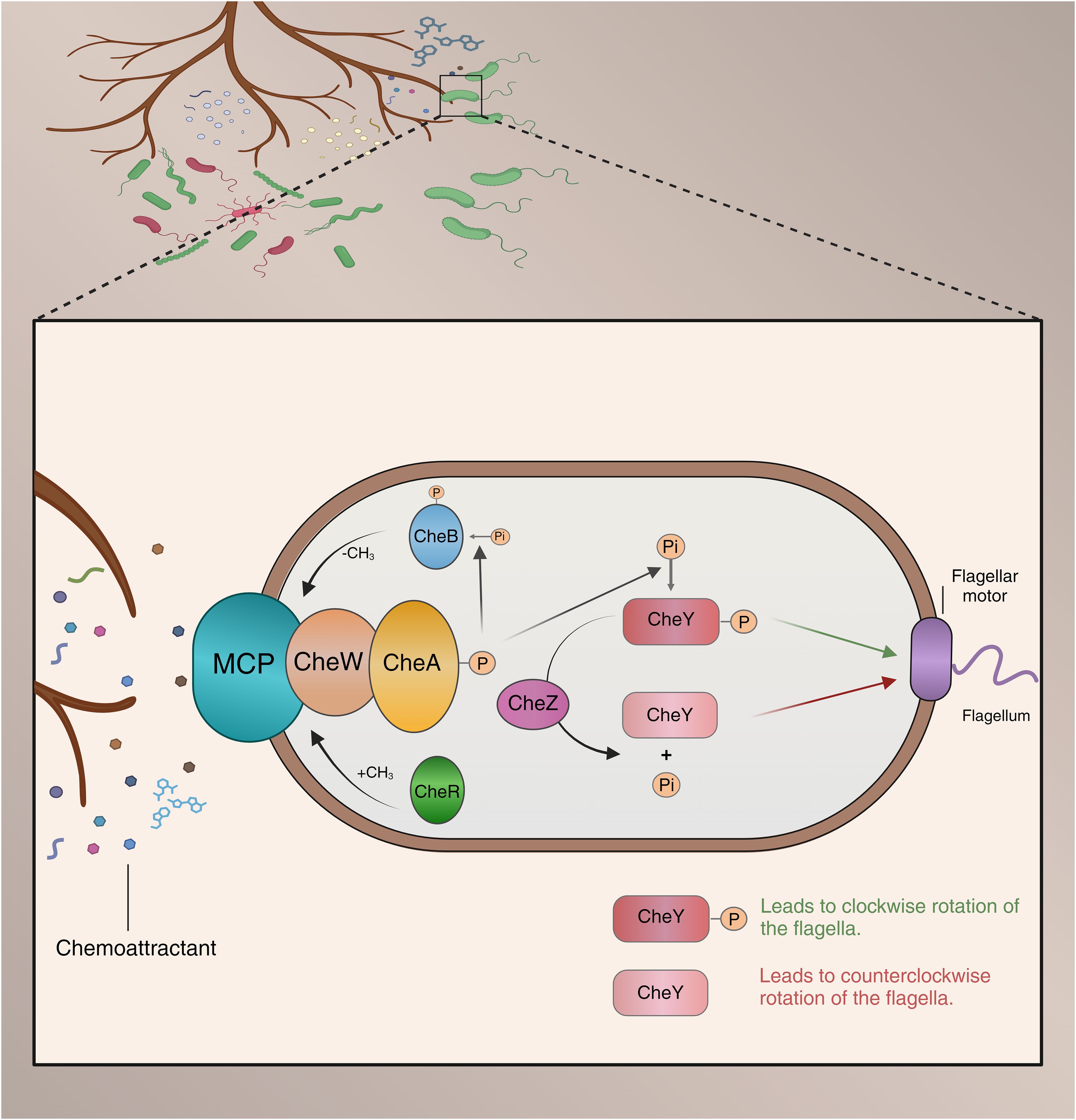
Figure 2. Model of microbial chemotaxis in the rhizosphere Chemical compounds secreted by plant roots attract beneficial bacteria while repelling harmful ones. These compounds are detected by transmembrane chemoreceptors on the bacteria, which regulate the autophosphorylation of CheA through the adaptor protein CheW. Once phosphorylated, CheA (CheA-P) phosphorylates the response regulator CheY. The phosphorylated CheY (CheY-P) binds to the flagellar motor, causing a switch in the direction of flagellar rotation from counterclockwise to clockwise, thereby altering the bacterial movement direction. The phosphatase CheZ dephosphorylates CheY-P, rapidly terminating the signal. CheB-P regulates methylation levels, working in conjunction with CheR to mediate the adaptive regulation of the chemoreceptors.
Bacterial attachment to root surfaces
Microorganisms move along a positive chemotactic signal gradient towards the host roots, selecting regions of the root with high concentrations of exudates as their colonization sites. Interestingly, microbial chemotaxis and motility play crucial roles in the selection of colonization sites on the roots. For instance, organic acids are strong attractants for Azospirillum brasilense, and A. brasilense mutants lacking chemotaxis fail to preferentially colonize the surface of the root maturation and elongation zones, which produce organic acids (O’Neal and Alexandre, 2020). The study also found that A. brasilense avoids the root tip region, which may produce toxic reactive oxygen species (ROS). This is because the root tip, being an area of active growth, generates hydrogen peroxide, which has a repellent effect on bacteria. Similarly, microorganisms must continuously migrate to avoid the immunologically active sites that change as the root develops (Tsai et al., 2023). These findings suggest that the host plays a significant regulatory role in bacterial selection of attachment sites on the roots.
Subsequently, microorganisms cease movement and adhere to the root surface, a critical step in achieving root colonization. Most agricultural microorganisms follow a common biphasic model for root attachment, which includes an initial attachment phase and a secondary attachment phase. During the initial attachment phase, rhizosphere bacterial cells form weak and reversible bonds with the root surface; in the secondary attachment phase, the bacteria’s attachment to the root surface becomes more secure, irreversible, and specific (Wheatley and Poole, 2018). Throughout the attachment process, microorganisms alter their physiological structures and secrete adhesive substances to successfully adhere. For example, bacteria may swing their flagella or pili to overcome electrostatic repulsion and secrete porins, outer membrane proteins, and lipopolysaccharides (LPS) as root surface adhesins (Berne et al., 2015; Knights et al., 2021). Additionally, bacteria secrete the signaling molecule di-AMP as an extracellular signal to regulate root attachment and biofilm formation, with Bacillus subtilis being a typical example (Townsley et al., 2018). This attachment is not merely a passive process; it involves active communication between the microbe and the plant. Plant surface molecules, such as lectins and arabinogalactan proteins, play a role in recognizing and binding microbial cells. This interaction is often species-specific, ensuring that only compatible microbes establish a symbiotic relationship (Imberty and Varrot, 2008). During this process, both the host and microorganisms change their lifestyles, achieving a balanced state through mutual regulation via molecular and physiological mechanisms.
After Pathogen-Associated Molecular Pattern (PAMP) recognition, PRRs located on the cell surface recruit the co-receptor BRI1-Associated Kinase 1 (BAK1), forming a receptor complex that activates downstream Receptor-Like Cytoplasmic Kinase (RLCK)-VII family members, such as Botrytis-Induced Kinase 1 (BIK1). Subsequently, RLCKs phosphorylate downstream targets, including Respiratory Burst Oxidase Homolog D (RBOHD) and Mitogen-Activated Protein Kinase Kinase Kinases (MAPKKKs), triggering a series of defense responses, such as ROS burst, calcium influx, Mitogen-Activated Protein Kinase (MAPK) activation, transcriptional reprogramming, and the production of plant hormones.
The figure also highlights the role of Type III secreted bacterial effectors (T3Es) in modulating plant defense mechanisms. Since some effectors interfere with multiple plant targets, they are represented multiple times in the figure.
Bacterial immune recognition evasion
Once microorganisms attach to the root surface, overcoming the plant immune system becomes one of the crucial challenges for successful rhizosphere colonization. At this stage, microorganisms deploy a series of molecular strategies to adapt to and modify the plant environment, thereby alleviating the immune attacks they face during colonization (Jones and Dangl, 2006; Khavkin, 2021; Kong and Yang, 2023). The following chapter summarizes three strategies evolved by colonizing microorganisms (particularly pathogens and symbionts) in response to the evolutionary selection pressures exerted by plant pattern recognition receptors (PRRs) that activate host immunity (Figure 3).
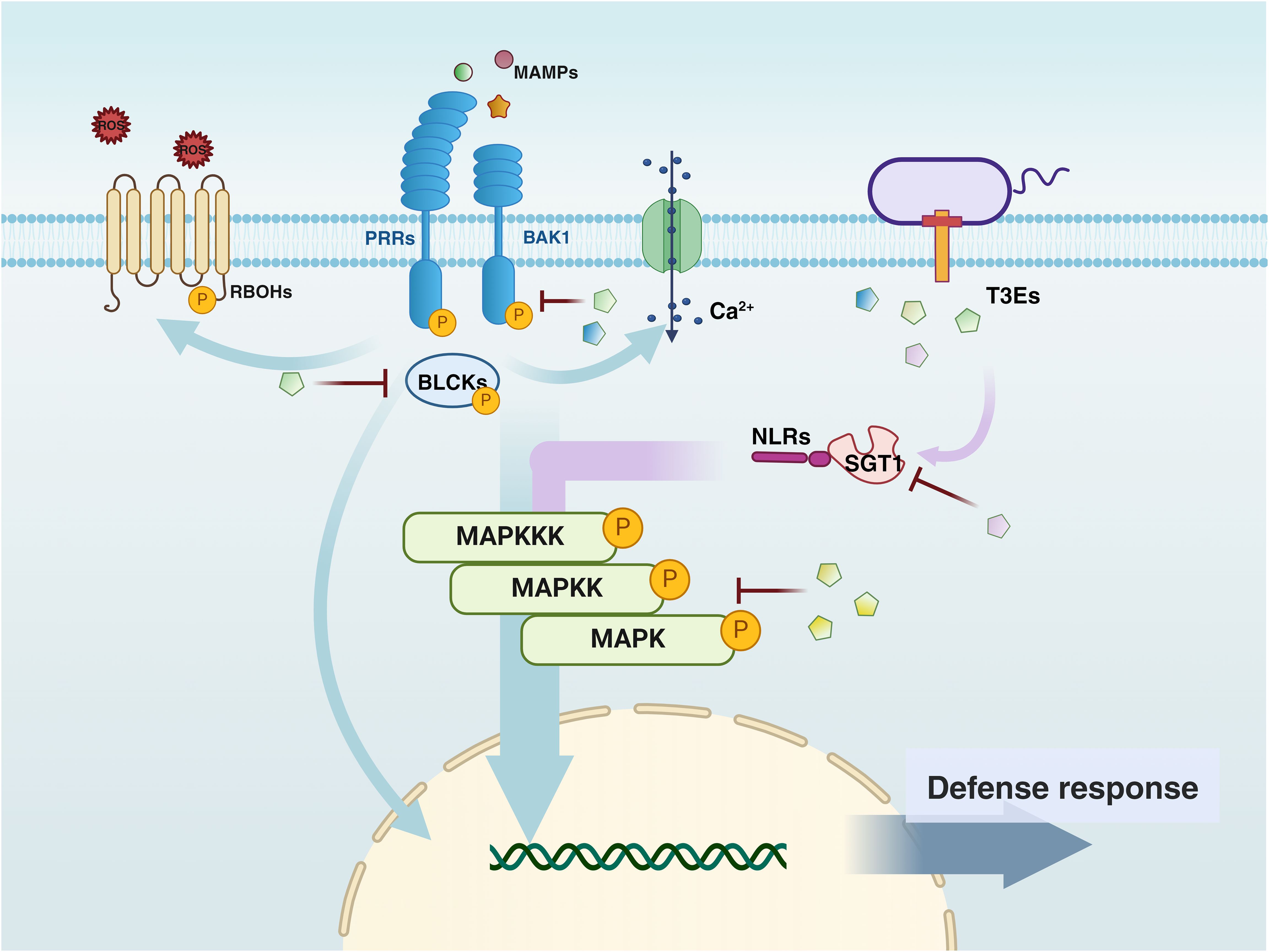
Figure 3. The figure illustrates the key components of PAMP-Triggered Immunity (PTI) and Effector-Triggered Immunity (ETI) signaling pathways in plant defense against bacterial pathogens, as well as the interconnections between these pathways. Arrows indicate the flow of defense signals, with light blue representing Pattern Recognition Receptor (PRR)-dependent signals and purple representing Nucleotide-binding Leucine-rich Repeat (NLR)-dependent signals. The convergence of these two signaling pathways is depicted by arrows that blend purple and light blue. PRR and NLR signals jointly regulate the plant immune response and interact at multiple critical points.
Microorganisms possess widely conserved molecular patterns (MAMPs), such as flg22, elongation factor Tu (EF-Tu), cold shock proteins (CSP), lipopolysaccharides (LPS), chitin, phospholipids, and Nep1-like proteins, which can be recognized by different pattern recognition receptors (PRRs) in plants. The activation of immune signaling events, known as MAMP-triggered immunity (MTI), plays a crucial role in eliminating potential pathogenic infections and forms the first barrier to microbial colonization (Zipfel and Oldroyd, 2017). Microbial pathogens have evolved complex strategies to evade plant immunity, allowing them to effectively colonize roots. A key strategy involves avoiding detection by PRRs, which primarily includes altering the structure of MAMPs, degrading MAMP precursors, inhibiting their biosynthesis, or preventing MAMP release. For example, Pseudomonas syringae secretes the protease ArpA to degrade flagellin monomers, preventing the release of the immunogenic epitope flg22 and thus evading immune detection during root colonization (Pel et al., 2014).
Beneficial microorganisms also employ similar strategies to evade recognition by the plant immune system. However, unlike pathogenic microorganisms, which rapidly adjust the properties of MAMPs, the immune evasion of beneficial microorganisms is more often achieved through the positive selection of PRRs over long-term evolution. For instance, variations in the flagellin protein sequence of the nitrogen-fixing symbiont Sinorhizobium meliloti result in its inability to activate immune responses in Arabidopsis (Felix et al., 1999). Similarly, the FLS2 receptor in grapevines has a weaker recognition of the flg22 epitope derived from beneficial Bacillus subtilis compared to that from pathogenic Pseudomonas syringae and Xanthomonas campestris, leading to a significantly reduced immune response (Trdá et al., 2014). Beneficial symbiotic microorganisms can also avoid detection by creating environments unfavorable to immune recognition. For example, Pseudomonas capeferrum WCS358 lowers environmental pH by secreting organic acids, thereby inhibiting plant recognition of flg22 (Yu et al., 2019). However, current research on immune evasion primarily focuses on flg22, and future studies should extend to other MAMPs to uncover the mechanisms by which rhizosphere microorganisms evade immune recognition during colonization. In addition to MAMPs, mutualistic symbiotic microorganisms also secrete symbiosis-related molecules, which are recognized by host symbiotic receptors and initiate signal transduction (Zipfel and Oldroyd, 2017). For example, Nod factors from rhizobia are lipochitooligosaccharides similar in structure to MAMPs like chitin and peptidoglycan, and they initiate the symbiotic process of rhizobia (Oldroyd, 2013). Interestingly, many symbiotic molecules from beneficial microorganisms seem to suppress the local immune responses triggered by MAMPs in roots (Gourion et al., 2015). For instance, Nod factors from rhizobia significantly suppress MAMP-induced immune responses in both leguminous and non-leguminous plants like Arabidopsis, leading to a significant reduction in the levels of homologous PRR proteins on the cell membrane (Liang et al., 2013; Gourion et al., 2015).
Although the mechanisms by which plant immunity regulates pathogenic and beneficial microorganisms have been elucidated (Yang et al., 2022), microorganisms in the rhizosphere often exist in community forms, which means plants need to sense the presence of entire communities. The FLS2 receptor in plants plays a crucial role in this process, acting as a community sensor to detect the relative proportions of danger signals (canonical peptides) from pathogens and signals (evading and modulating peptides) from safe microorganisms. This sensing mechanism influences the assembly of specific pathogenic and symbiotic microbial communities, enabling the plant to distinguish between friends and foes (Colaianni et al., 2021). This discovery challenges the traditional view that the flg22-FLS2 complex only drives immune signal transduction, showcasing the complexity and subtlety of plant regulation in immunity and symbiosis.
Bacterial effectors interfere with immune signaling
Bacteria can secrete effectors, primarily composed of effector proteins, into the apoplast or deliver them into plant cells through specialized secretion systems (Jones and Dangl, 2006). These effectors bypass plant MTI by targeting immune signal transduction components. Research into the mechanisms of pathogen effectors provides important insights into how bacterial effectors suppress plant immune responses by interfering with key immune signaling molecules. Specifically, BRI1-Associated Kinase 1 (BAK1) and Bacterial-Like Cytoplasmic Kinases (BLCKs) are major targets for various pathogen effectors (Wang et al., 2022). BAK1 acts as a co-receptor for multiple pattern recognition receptors (PRRs) and plays a critical role in immune signal transduction induced by various MAMPs (Yasuda et al., 2017). For example, the pathogen Phytophthora sojae effector PsAvh110 specifically binds to the soybean heterochromatin protein GmLHP1-2, disrupting its assembly with the transcriptional complex GmPHD6, thereby inhibiting the expression of a set of immunity-related genes, including the BRI1-associated receptor kinase GmBAK1-3 (Qiu et al., 2023). Botrytis-induced Kinase 1 (BIK1), a member of the BLCK-VII subfamily, is a core regulator of plant immunity, located downstream of PRR and BAK1 signaling pathways, and is responsible for transmitting signals to downstream MAPK cascades, initiating a series of cellular defense responses, and coordinating the plant’s defensive actions (Ma et al., 2020). The type III secretion effector RipAC from the bacterium Ralstonia solanacearum targets the plant E3 ubiquitin ligase PUB4, suppressing pattern-triggered immunity (PTI) and leading to the degradation of the critical immune kinase BIK1 (Yu et al., 2022). Additionally, within plant cells, Nucleotide-binding Leucine-rich Repeat-like receptors (NLRs) recognize effectors either directly or indirectly, activating effector-triggered immunity (ETI), which triggers a more robust immune response (Gourion et al., 2015). Pathogen effectors also disrupt multiple stages of ETI signal transduction. As mentioned earlier, the Ralstonia solanacearum effector RipAC not only drives BIK1 degradation but also interferes with the phosphorylation of SGT1 (a regulator of NLR accumulation or activation) and its interaction with MAPKs (Yu et al., 2020). The multifaceted mechanisms by which RipAC regulates plant immune responses suggest that the targets of effectors are often not singular, reflecting the complex regulatory methods and further emphasizing the multi-layered strategies pathogens employ to evade plant immunity. It is noteworthy that pathogens can evade NLR receptor recognition by regulating effector gene expression, such as altering promoter regions or applying epigenetic modifications. However, as this involves more epigenetic aspects, it will not be detailed in this article, but relevant content can be found in other literature (Wang et al., 2022).
Activation of the MAPK cascade is an early signal transduction event shared by both PTI and ETI and plays a key role in regulating immune outputs such as callose deposition, hormone production, and transcriptional reprogramming (Tzipilevich et al., 2021). This critical node is also often targeted by pathogens to weaken host immunity. For instance, the P. syringae T3Es effectors HopA1 and HopF2 inactivate MAPKs and their upstream kinases through different mechanisms. HopA1 acts as a phosphothreonine lyase, physically inactivating MPK3 and/or MPK6, while HopF2 inactivates MAPK kinase 5 (MKK5) through ADP-ribosylation (Zhang et al., 2007; Wang et al., 2010). Additionally, the P. syringae effector AvrB interferes with plant hormone signaling by activating MAPK MPK4, thereby negatively regulating pathogen defense (Cui et al., 2010). These findings demonstrate that the regulation by effector proteins is often not singular; they frequently activate different mechanisms to enhance their effects. The Phytophthora infestans RXLR effector PITG20303 further emphasizes this conclusion, targeting the stabilization of the potato MAPK cascade protein StMKK1, suppressing PTI, promoting pathogen colonization, and evading recognition by the resistance protein Rpi-blb2, thereby negatively regulating the plant’s PTI response (Du et al., 2021b).
Similar to plant pathogens, symbiotic rhizobia in legumes use a type III protein secretion system (T3SS) to deliver toxin-like type III effectors (T3Es) into plant cells, utilizing multiple effectors to suppress plant immune activation. These T3Es not only inhibit the immune response in legumes but also stimulate the production of nodulation signals, which are crucial for establishing symbiosis (Ferguson et al., 2010; Oldroyd, 2013). The NopL effector secreted by Sinorhizobium sp. strain NGR234 is a typical example; its serine-proline motif is essential for its effector activity during symbiosis, and over-phosphorylated NopL mutants show significantly reduced activity during symbiosis (Ge et al., 2016). The mechanism by which NopL suppresses immune responses by interfering with the plant MAPK cascade is gradually being uncovered. For example, NopL forms a complex with the tobacco MAP kinase SIPK and is phosphorylated by SIPK in vitro, thereby suppressing MAP kinase signal-mediated defense responses. Additionally, NopL mimics MAPK substrates and interferes with their signal transduction, inhibiting host cell death and premature nodule senescence (Zhang et al., 2011). Thus, it can be inferred that NopL suppresses plant defense responses by interfering with the MAPK signaling process or its downstream events. Recent studies have shown that, unlike the strategy of pathogen and rhizobial symbionts that rely on T3SS to inject highly specific effector proteins, beneficial rhizosphere symbionts primarily interfere with MTI signal transduction through non-specific extracellular mechanisms. The synthetic communities (SynComs) constructed from diverse MTI-suppressing strains in Arabidopsis roots primarily inhibit MTI by modulating specific and conserved parts of the host immune system. Gene screening further revealed that although Dyella japonica MF79 carries T3SS genes, its potent MTI-suppressing function mainly relies on the type II secretion system (T2SS), while T3SS is not essential for its inhibitory activity (Ökmen et al., 2013).
Although the mechanisms of action for individual bacterial effectors have been extensively studied, the synergistic effects of multiple effectors within the same pathogen and the temporal and spatial regulation by different pathogens in secreting effectors at various infection stages to evade host defense systems remain unclear. These complex molecular dialogues and regulatory networks require further research to elucidate their synergistic mechanisms and interactions in plant immunity.
Microbial tolerance to immune responses
Activated immune signaling cascades ultimately trigger a series of powerful immune responses, including the secretion of proteases and inhibitors, the release of antimicrobial molecules, and bursts of ROS. Plants with activated defense responses synthesize and secrete various defense molecules and proteases, generating resistance to pathogenic microorganisms through different mechanisms, thereby altering the rhizosphere environment (Jones and Dangl, 2006; Wang et al., 2022). Meanwhile, symbiotic microorganisms cleverly evade these immune responses and even modify the environment to successfully promote their survival and proliferation. To successfully infect host plants and overcome their defense mechanisms, rhizosphere microorganisms have developed various strategies, which can be categorized into active neutralization and passive adaptation.
Active Neutralization Strategies include directly intervening in and regulating host plant immune responses by inhibiting the production of plant defense substances and secreting specific inhibitors. For example, the effectors AVRblb2 from Phytophthora infestans and Avh240 from Phytophthora sojae target plant PLCP C14 and aspartic protease AP1, preventing the secretion and activation of these defense proteins (Bozkurt et al., 2011; Guo et al., 2019). Similarly, the Kazal-like protease inhibitor EPI1 secreted by P. infestans can inhibit the plant resistance protease P69B, thereby hindering the maturation of immunogenic peptides (Paulus et al., 2020; Wang et al., 2021). In addition, pathogenic microorganisms suppress ROS production or interfere with their transport in host plants as a key strategy to actively neutralize host defense mechanisms. Host plants primarily produce ROS through peroxidases and membrane-bound NADPH oxidases. To suppress ROS, pathogenic microorganisms secrete various effectors. For example, Ustilago maydis effector Pep1 directly inhibits maize peroxidase POX12 (Hemetsberger et al., 2012), while Phytophthora parasitica effector PpE18 weakens the immunity of Nicotiana benthamiana by inhibiting the ROS-scavenging function of NbAPX3-1 and interfering with its interaction with NbANKr2 (Cao et al., 2024). Similarly, the Phytophthora sojae effectors Avr3b and CRN78 reduce ROS accumulation and transport by disrupting NADH availability and phosphorylating aquaporin PIP2;2 (Ai et al., 2021).
Passive Adaptation Strategies involve setting up biological barriers and altering physiological structures to adapt to plant defenses. Pathogenic microorganisms evade host immune responses through post-translational modifications of effector molecules. For instance, the virulence factor XEG1 produced by Phytophthora sojae undergoes glycosylation modifications to evade degradation by the host protease AP5 (Xia et al., 2020). Furthermore, pathogenic microorganisms can detoxify antimicrobial compounds synthesized by plants, such as the conversion of toxic α-tomatine into less toxic derivatives by the enzyme secreted by Cladosporium fulvum (Ökmen et al., 2013). Microbial pathogens have also developed various strategies to evade oxidative stress during infection. A typical example is that bacterial pathogens can produce extracellular polysaccharides to form protective layers, shielding themselves from oxidative stress induced by the host (Fones and Preston, 2012).
Similar to pathogenic microorganisms, beneficial microorganisms employ both active and passive adaptation strategies to cope with ROS bursts in plants, allowing them to successfully colonize the rhizosphere. Enhancing tolerance is a key strategy for responding to activated root immune responses, such as ROS bursts. For example, the colonization of beneficial bacterium Bacillus velezensis triggers plant immune responses and ROS production, which in turn stimulate bacterial auxin production, reducing ROS toxicity and playing a crucial role in root colonization (Tzipilevich et al., 2021). Bacillus subtilis SQR9 induces oxidative bursts in cucumber and Arabidopsis through its flg22 homologs, demonstrating high H2O2 tolerance and the ability to suppress oxidative bursts, with the ResD-ResE signal transduction system in SQR9 playing a key role in tolerating plant oxidative stress and root colonization (Zhang et al., 2021). Additionally, recently revealed spatial adaptation mechanisms indicate that plant cortical, endodermal, and root hair cells exhibit different defense capabilities, and beneficial microorganism Pseudomonas simiae WCS417 selects colonization sites with lower stress, demonstrating a spatially host-immune-driven adaptation strategy (Verbon et al., 2023).
Active Regulation: For example, the NopM effector from rhizobium NGR234 significantly suppresses flg22-induced ROS bursts when expressed in Nicotiana benthamiana, blocking ROS-related defense responses (Xin et al., 2012). This demonstrates that beneficial microorganisms can suppress transient ROS bursts by secreting effectors, cleverly interfering with plant immune responses, and showcasing their strategies and potential in actively regulating plant immunity.
Through the detailed analysis of the strategies employed by beneficial and pathogenic microorganisms in coping with plant immune systems, we can see that both types of microorganisms use many similar strategies in immune recognition evasion, immune signal interference, and immune response suppression. However, their ultimate goals are entirely different: pathogenic microorganisms aim to overcome plant immune systems to cause infection, while beneficial microorganisms regulate plant immune systems to establish symbiotic relationships.
Biofilm formation and stable colonization
Biofilm formation is a key strategy employed by microbes to ensure stable colonization on the root surface. Biofilms are complex communities of microorganisms embedded in a self-produced extracellular matrix, which provides protection against environmental stresses and host immune responses (Flemming et al., 2023). This matrix mainly consists of polysaccharides, proteins, amyloids, lipids, extracellular DNA (eDNA), as well as membrane vesicles and humic-like microbially derived refractory substances (Flemming et al., 2023). The properties of these polymers confer resistance or antimicrobial tolerance to biofilms under certain adverse conditions. Beneficial rhizosphere bacteria first halt their motility and then form biofilms, a process generally controlled by one or more of their own transcriptional regulators (Arnaouteli et al., 2021; Nie et al., 2022; Ivanova et al., 2023). However, increasing evidence suggests that this process is also co-regulated by global transcriptional regulators mediated by environmental factors such as root exudates. For example, Bacillus subtilis colonizes Arabidopsis roots by forming biofilms dependent on extracellular matrix genes, a process triggered by plant polysaccharides and transmitted through the regulation of the phosphorylation state of the master regulator Spo0A (Beauregard et al., 2013). Interestingly, these polysaccharides not only serve as signals but also as sugar sources for synthesizing the extracellular polysaccharides (ESP) in the matrix, demonstrating the critical role of external regulation in bacterial root colonization (Beauregard et al., 2013). Similarly, phenolic compounds in root exudates attract and promote endophyte biofilm formation on the host root surface, facilitating colonization. After colonization, endophytes enhance the host’s intrinsic defense mechanisms by increasing the levels of phenolic antimicrobial substances in the rhizosphere exudates, significantly boosting the plant’s resistance to pathogen attacks (Ray et al., 2018). Root exudates also influence biofilm formation in pathogenic bacteria. Pseudomonas aeruginosa strains PAO1 and PA14 can infect the roots of Arabidopsis and sweet basil, forming biofilms and causing plant death. Inducing the secretion of rosmarinic acid (RA) from the host roots before infection or the exogenous addition of RA effectively counters Pseudomonas aeruginosa (Walker et al., 2004). Notably, although chemotaxis and biofilm formation are two distinct processes in rhizosphere colonization, the same molecule secreted by the host can regulate chemotaxis or biofilm formation in rhizosphere microorganisms depending on its concentration (Zhang et al., 2014; López-Farfán et al., 2019). This dose-dependent signaling is very common in the regulation of biofilm formation and chemotaxis in rhizosphere bacteria, highlighting the critical role of concentration gradients of environmental signals in microbial behavior.
Summary and future perspectives
Previous research on microbial colonization processes has predominantly focused on isolating beneficial strains from rhizosphere bacteria. However, future studies should prioritize the regulation of these colonization processes to effectively mitigate pathogen invasion and enhance the efficiency of beneficial microbes in field applications. Potential regulatory strategies may involve manipulating the composition of root exudates, modulating microbial signaling pathways, or employing gene-editing technologies to optimize the colonization potential of beneficial microorganisms. By elucidating the interaction mechanisms between various microbial functional groups in the rhizosphere, we can enhance microbial symbiosis and competitive dynamics under conditions that closely mimic natural environments, thereby promoting plant health and advancing agricultural sustainability.
Future research should focus on investigating microbial colonization mechanisms within complex ecological contexts, examining the roles of soil physicochemical properties, microbial community structure dynamics, and interspecies interactions in shaping plant health. A multi-layered, systems-based approach that integrates the soil microenvironment and the ecological dynamics of microbial communities will provide a more precise theoretical foundation for agricultural management practices. This, in turn, will enhance crop resilience, optimize yield, and contribute robustly to sustainable agricultural development.
Author contributions
LY: Formal analysis, Investigation, Visualization, Writing – original draft. XQ: Investigation, Conceptualization, Writing – review & editing. ZZ: Investigation, Writing – original draft. YW: Investigation, Writing – original draft. GD: Methodology, Supervision, Writing – review & editing. XX: Methodology, Supervision, Writing – review & editing, Funding acquisition.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (grant no. 32170013), National Key Research and Development Program of China (grant no.2023YFD2201802), and CAMS Initiative for Innovative Medicine (grant no. 2021-I2M-1-031).
Acknowledgments
The text elements of this review were generated using ChatGPT with the GPT-4 model and were thoroughly reviewed by the author prior to integration into the document.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ai, G., Xia, Q., Song, T., Li, T., Zhu, H., Peng, H., et al. (2021). A Phytophthora sojae CRN effector mediates phosphorylation and degradation of plant aquaporin proteins to suppress host immune signaling. PloS Pathog. 17, e1009388. doi: 10.1371/journal.ppat.1009388
Allard-Massicotte, R., Tessier, L., Lécuyer, F., Lakshmanan, V., Lucier, J.-F., Garneau, D., et al. (2016). Bacillus subtilis early colonization of Arabidopsis thaliana roots involves multiple chemotaxis receptors. mBio 7, e01664–e01616. doi: 10.1128/mBio.01664-16
Arnaouteli, S., Bamford, N. C., Stanley-Wall, N. R., Kovács, Á. T. (2021). Bacillus subtilis biofilm formation and social interactions. Nat. Rev. Microbiol. 19, 600–614. doi: 10.1038/s41579-021-00540-9
Badri, D. V., Weir, T. L., van der Lelie, D., Vivanco, J. M. (2009). Rhizosphere chemical dialogues: plant–microbe interactions. Curr. Opin. Biotechnol. 20, 642–650. doi: 10.1016/j.copbio.2009.09.014
Bais, H. P., Weir, T. L., Perry, L. G., Gilroy, S., Vivanco, J. M. (2006). The role of root exudates in rhizosphere interactions with plants and other organisms. Annu. Rev. Plant Biol. 57, 233–266. doi: 10.1146/annurev.arplant.57.032905.105159
Beauregard, P. B., Chai, Y., Vlamakis, H., Losick, R., Kolter, R. (2013). Bacillus subtilis biofilm induction by plant polysaccharides. Proc. Natl. Acad. Sci. U. S. A. 110, E1621–E1630. doi: 10.1073/pnas.1218984110
Berne, C., Ducret, A., Hardy, G. G., Brun, Y. V. (2015). Adhesins involved in attachment to abiotic surfaces by gram-negative bacteria. Microb. Biofilms, 163–199. doi: 10.1128/9781555817466.ch9
Bozkurt, T. O., Schornack, S., Win, J., Shindo, T., Ilyas, M., Oliva, R., et al. (2011). Phytophthora infestans effector AVRblb2 prevents secretion of a plant immune protease at the haustorial interface. Proc. Natl. Acad. Sci. U. S. A. 108, 20832–20837. doi: 10.1073/pnas.1112708109
Cao, Y., Zhang, Q., Liu, Y., Yan, T., Ding, L., Yang, Y., et al. (2024). The RXLR effector PpE18 of Phytophthora parasitica is a virulence factor and suppresses peroxisome membrane-associated ascorbate peroxidase NbAPX3-1-mediated plant immunity. New Phytol. 243, 1472-1489. doi: 10.1111/nph.19902
Chagas, F. O., Pessotti, R., de, C., Caraballo-Rodríguez, A. M., Pupo, M. T. (2018). Chemical signaling involved in plant–microbe interactions. Chem. Soc Rev. 47, 1652–1704. doi: 10.1039/C7CS00343A
Chen, L., Liu, Y. (2024). The function of root exudates in the root colonization by beneficial soil rhizobacteria. Biology 13, 95. doi: 10.3390/biology13020095
Colaianni, N. R., Parys, K., Lee, H.-S., Conway, J. M., Kim, N. H., Edelbacher, N., et al. (2021). A complex immune response to flagellin epitope variation in commensal communities. Cell Host Microbe 29, 635-649.e9. doi: 10.1016/j.chom.2021.02.006
Compton, K. K., Hildreth, S. B., Helm, R. F., Scharf, B. E. (2018). Sinorhizobium meliloti chemoreceptor McpV senses short-chain carboxylates via direct binding. J. Bacteriol. 200, e00519–e00518. doi: 10.1128/jb.00519-18
Corral-Lugo, A., de la Torre, J., Matilla, M. A., Fernández, M., Morel, B., Espinosa-Urgel, M., et al. (2016). Assessment of the contribution of chemoreceptor-based signalling to biofilm formation. Environ. Microbiol. 18, 3355–3372. doi: 10.1111/1462-2920.13170
Cotton, T. E. A., Pétriacq, P., Cameron, D. D., Meselmani, M. A., Schwarzenbacher, R., Rolfe, S. A., et al. (2019). Metabolic regulation of the maize rhizobiome by benzoxazinoids. ISME J. 13, 1647–1658. doi: 10.1038/s41396-019-0375-2
Cui, H., Wang, Y., Xue, L., Chu, J., Yan, C., Fu, J., et al. (2010). Pseudomonas syringae effector protein AvrB perturbs Arabidopsis hormone signaling by activating MAP kinase 4. Cell Host Microbe 7, 164–175. doi: 10.1016/j.chom.2010.01.009
De-la-Peña, C., Badri, D. V., Lei, Z., Watson, B. S., Brandão, M. M., Silva-Filho, M. C., et al. (2010). Root secretion of defense-related proteins is development-dependent and correlated with flowering time. J. Biol. Chem. 285, 30654–30665. doi: 10.1074/jbc.M110.119040
de la Porte, A., Schmidt, R., Yergeau, É., Constant, P. (2020). A gaseous milieu: extending the boundaries of the rhizosphere. Trends Microbiol. 28, 536–542. doi: 10.1016/j.tim.2020.02.016
Ding, H., Liu, T., Hu, Q., Liu, M., Cai, M., Jiang, Y., et al. (2022). Effect of microbial community structures and metabolite profile on greenhouse gas emissions in rice varieties. Environ. pollut. 306, 119365. doi: 10.1016/j.envpol.2022.119365
Du, Y., Chen, X., Guo, Y., Zhang, X., Zhang, H., Li, F., et al. (2021b). Phytophthora infestans RXLR effector PITG20303 targets a potato MKK1 protein to suppress plant immunity. New Phytol. 229, 501-515. doi: 10.1111/nph.16861
Du, J. X., Li, Y., Ur-Rehman, S., Mukhtar, I., Yin, Z., Dong, H., et al. (2021a). Synergistically promoting plant health by harnessing synthetic microbial communities and prebiotics. iScience 24, 102918. doi: 10.1016/j.isci.2021.102918
Dudeja, S. S., Suneja-Madan, P., Paul, M., Maheswari, R., Kothe, E. (2021). Bacterial endophytes: Molecular interactions with their hosts. J. Basic Microbiol. 61, 475–505. doi: 10.1002/jobm.202000657
Felix, G., Duran, J. D., Volko, S., Boller, T. (1999). Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 18, 265–276. doi: 10.1046/j.1365-313x.1999.00265.x
Feng, H., Fu, R., Hou, X., Lv, Y., Zhang, N., Liu, Y., et al. (2021). Chemotaxis of beneficial rhizobacteria to root exudates: The first step towards root–microbe rhizosphere interactions. Int. J. Mol. Sci. 22, 6655. doi: 10.3390/ijms22136655
Feng, H., Zhang, N., Fu, R., Liu, Y., Krell, T., Du, W., et al. (2019). Recognition of dominant attractants by key chemoreceptors mediates recruitment of plant growth-promoting rhizobacteria. Environ. Microbiol. 21, 402–415. doi: 10.1111/1462-2920.14472
Ferguson, B. J., Indrasumunar, A., Hayashi, S., Lin, M.-H., Lin, Y.-H., Reid, D. E., et al. (2010). Molecular analysis of legume nodule development and autoregulation. J. Integr. Plant Biol. 52, 61–76. doi: 10.1111/j.1744-7909.2010.00899.x
Flemming, H.-C., van Hullebusch, E. D., Neu, T. R., Nielsen, P. H., Seviour, T., Stoodley, P., et al. (2023). The biofilm matrix: multitasking in a shared space. Nat. Rev. Microbiol. 21, 70–86. doi: 10.1038/s41579-022-00791-0
Fones, H., Preston, G. M. (2012). Reactive oxygen and oxidative stress tolerance in plant pathogenic Pseudomonas. FEMS Microbiol. Lett. 327, 1–8. doi: 10.1111/j.1574-6968.2011.02449.x
Fuqua, C., Parsek, M. R., Greenberg, E. P. (2001). Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Annu. Rev. Genet. 35, 439–468. doi: 10.1146/annurev.genet.35.102401.090913
Ge, Y.-Y., Xiang, Q.-W., Wagner, C., Zhang, D., Xie, Z.-P., Staehelin, C. (2016). The type 3 effector NopL of Sinorhizobium sp. strain NGR234 is a mitogen-activated protein kinase substrate. J. Exp. Bot. 67, 2483–2494. doi: 10.1093/jxb/erw065
Goswami, M., Deka, S. (2022). “Rhizodeposits: An essential component for microbial interactions in rhizosphere,” in Re-visiting the Rhizosphere Eco-system for Agricultural Sustainability. Eds. Singh, U. B., Rai, J. P., Sharma, A. K. (Springer Nature, Singapore), 129–151. doi: 10.1007/978-981-19-4101-6_7
Gourion, B., Berrabah, F., Ratet, P., Stacey, G. (2015). Rhizobium–legume symbioses: the crucial role of plant immunity. Trends Plant Sci. 20, 186–194. doi: 10.1016/j.tplants.2014.11.008
Guo, B., Wang, H., Yang, B., Jiang, W., Jing, M., Li, H., et al. (2019). Phytophthora sojae effector PsAvh240 inhibits host aspartic protease secretion to promote infection. Mol. Plant 12, 552–564. doi: 10.1016/j.molp.2019.01.017
Harbort, C. J., Hashimoto, M., Inoue, H., Niu, Y., Guan, R., Rombolà, A. D., et al. (2020). Root-secreted coumarins and the microbiota interact to improve iron nutrition in Arabidopsis. Cell Host Microbe 28, 825–837.e6. doi: 10.1016/j.chom.2020.09.006
Hemetsberger, C., Herrberger, C., Zechmann, B., Hillmer, M., Doehlemann, G. (2012). The Ustilago maydis effector Pep1 suppresses plant immunity by inhibition of host peroxidase activity. PloS Pathog. 8, e1002684. doi: 10.1371/journal.ppat.1002684
Holopainen, J. K. (2004). Multiple functions of inducible plant volatiles. Trends Plant Sci. 9, 529–533. doi: 10.1016/j.tplants.2004.09.006
Hu, L., Robert, C. A. M., Cadot, S., Zhang, X., Ye, M., Li, B., et al. (2018). Root exudate metabolites drive plant-soil feedbacks on growth and defense by shaping the rhizosphere microbiota. Nat. Commun. 9, 2738. doi: 10.1038/s41467-018-05122-7
Imberty, A., Varrot, A. (2008). Microbial recognition of human cell surface glycoconjugates. Curr. Opin. Struct. Biol. 18, 567–576. doi: 10.1016/j.sbi.2008.08.001
Ivanova, L. A., Egorov, V. V., Zabrodskaya, Y. A., Shaldzhyan, A. A., Baranchikov, A. Y., Tsvigun, N. V., et al. (2023). Matrix is everywhere: extracellular DNA is a link between biofilm and mineralization in Bacillus cereus planktonic lifestyle. NPJ Biofilms Microbiomes 9, 1–10. doi: 10.1038/s41522-023-00377-5
Jin, Y., Zhu, H., Luo, S., Yang, W., Zhang, L., Li, S., et al. (2019). Role of maize root exudates in promotion of colonization of Bacillus velezensis strain S3-1 in rhizosphere soil and root tissue. Curr. Microbiol. 76, 855–862. doi: 10.1007/s00284-019-01699-4
Jones, J. D. G., Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Kearns, D. B. (2010). A field guide to bacterial swarming motility. Nat. Rev. Microbiol. 8, 634–644. doi: 10.1038/nrmicro2405
Khavkin, E. E. (2021). Plant–Pathogen molecular dialogue: Evolution, mechanisms and agricultural implementation. Russ. J. Plant Physiol. 68, 197–211. doi: 10.1134/S1021443721020072
Kiers, E. T., Duhamel, M., Beesetty, Y., Mensah, J. A., Franken, O., Verbruggen, E., et al. (2011). Reciprocal rewards stabilize cooperation in the mycorrhizal symbiosis. Science 333, 880–882. doi: 10.1126/science.1208473
Knights, H. E., Jorrin, B., Haskett, T. L., Poole, P. S. (2021). Deciphering bacterial mechanisms of root colonization. Environ. Microbiol. Rep. 13, 428–444. doi: 10.1111/1758-2229.12934
Kong, F., Yang, L. (2023). Pathogen-triggered changes in plant development: Virulence strategies or host defense mechanism? Front. Microbiol. 14. doi: 10.3389/fmicb.2023.1122947
Koprivova, A., Kopriva, S. (2022). Plant secondary metabolites altering root microbiome composition and function. Curr. Opin. Plant Biol. 67, 102227. doi: 10.1016/j.pbi.2022.102227
Lacal, J., García-Fontana, C., Muñoz-Martínez, F., Ramos, J.-L., Krell, T. (2010). Sensing of environmental signals: classification of chemoreceptors according to the size of their ligand binding regions. Environ. Microbiol. 12, 2873–2884. doi: 10.1111/j.1462-2920.2010.02325.x
Liang, Y., Cao, Y., Tanaka, K., Thibivilliers, S., Wan, J., Choi, J., et al. (2013). Nonlegumes respond to rhizobial Nod factors by suppressing the innate immune response. Science 341, 1384–1387. doi: 10.1126/science.1242736
Liu, X., Xie, Z., Wang, Y., Sun, Y., Dang, X., Sun, H. (2019). A dual role of amino acids from Sesbania rostrata seed exudates in the chemotaxis response of Azorhizobium caulinodans ORS571. Mol. Plant Microbe Interact. 32, 1134–1147. doi: 10.1094/MPMI-03-19-0059-R
Liu, Y., Xu, Z., Chen, L., Xun, W., Shu, X., Chen, Y., et al. (2024). Root colonization by beneficial rhizobacteria. FEMS Microbiol. Rev. 48, fuad066. doi: 10.1093/femsre/fuad066
López-Farfán, D., Reyes-Darias, J. A., Matilla, M. A., Krell, T. (2019). Concentration dependent effect of plant root exudates on the chemosensory systems of Pseudomonas putida KT2440. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.00078
Ma, X., Claus, L. A. N., Leslie, M. E., Tao, K., Wu, Z., Liu, J., et al. (2020). Ligand-induced monoubiquitination of BIK1 regulates plant immunity. Nature 581, 199–203. doi: 10.1038/s41586-020-2210-3
Mattick, J. S. (2002). Type IV pili and twitching motility. Annu. Rev. Microbiol. 56, 289–314. doi: 10.1146/annurev.micro.56.012302.160938
McEvoy, M. M., Bren, A., Eisenbach, M., Dahlquist, F. W. (1999). Identification of the binding interfaces on CheY for two of its targets the phosphatase CheZ and the flagellar switch protein FliM1. J. Mol. Biol. 289, 1423–1433. doi: 10.1006/jmbi.1999.2830
Mignot, T. (2007). The elusive engine in Myxococcus xanthus gliding motility. Cell. Mol. Life Sci. 64, 2733–2745. doi: 10.1007/s00018-007-7176-x
Mushtaq, S., Shafiq, M., Tariq, M. R., Sami, A., Nawaz-ul-Rehman, M. S., Bhatti, M. H. T., et al. (2023). Interaction between bacterial endophytes and host plants. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1092105
Nie, H., Xiao, Y., Song, M., Wu, N., Peng, Q., Duan, W., et al. (2022). Wsp system oppositely modulates antibacterial activity and biofilm formation via FleQ-FleN complex in Pseudomonas putida. Environ. Microbiol. 24, 1543–1559. doi: 10.1111/1462-2920.15905
O’Neal, L., Alexandre, G. (2020). Specific root exudate compounds sensed by dedicated chemoreceptors shape Azospirillum brasilense chemotaxis in the rhizosphere. Appl. Environ. Microbiol. 86, e01026–e01020. doi: 10.1128/AEM.01026-20
Ökmen, B., Etalo, D. W., Joosten, M. H. A. J., Bouwmeester, H. J., de Vos, R. C. H., Collemare, J., et al. (2013). Detoxification of α-tomatine by Cladosporium fulvum is required for full virulence on tomato. New Phytol. 198, 1203–1214. doi: 10.1111/nph.12208
Oku, S., Komatsu, A., Tajima, T., Nakashimada, Y., Kato, J. (2012). Identification of chemotaxis sensory proteins for amino acids in Pseudomonas fluorescens Pf0-1 and their involvement in chemotaxis to tomato root exudate and root colonization. Microbes Environ. 27, 462–469. doi: 10.1264/jsme2.ME12005
Oldroyd, G. E. D. (2013). Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nat. Rev. Microbiol. 11, 252–263. doi: 10.1038/nrmicro2990
Pacheco-Moreno, A., Bollmann-Giolai, A., Chandra, G., Brett, P., Davies, J., Thornton, O., et al. (2024). The genotype of barley cultivars influences multiple aspects of their associated microbiota via differential root exudate secretion. PloS Biol. 22, e3002232. doi: 10.1371/journal.pbio.3002232
Paulus, J. K., Kourelis, J., Ramasubramanian, S., Homma, F., Godson, A., Hörger, A. C., et al. (2020). Extracellular proteolytic cascade in tomato activates immune protease Rcr3. Proc. Natl. Acad. Sci. U.S.A. 117, 17409–17417. doi: 10.1073/pnas.1921101117
Pel, M. J. C., van Dijken, A. J. H., Bardoel, B. W., Seidl, M. F., van der Ent, S., van Strijp, J. A. G., et al. (2014). Pseudomonas syringae evades host immunity by degrading flagellin monomers with alkaline protease AprA. Mol. Plant Microbe Interact. 27, 603–610. doi: 10.1094/MPMI-02-14-0032-R
Plett, J. M., Martin, F. M. (2018). Know your enemy, embrace your friend: using omics to understand how plants respond differently to pathogenic and mutualistic microorganisms. Plant J. 93, 729–746. doi: 10.1111/tpj.13802
Qiu, X., Kong, L., Chen, H., Lin, Y., Tu, S., Wang, L., et al. (2023). The Phytophthora sojae nuclear effector PsAvh110 targets a host transcriptional complex to modulate plant immunity. Plant Cell 35, 574–597. doi: 10.1093/plcell/koac300
Ray, S., Mishra, S., Bisen, K., Singh, S., Sarma, B. K., Singh, H. B. (2018). Modulation in phenolic root exudate profile of Abelmoschus esculentus expressing activation of defense pathway. Microbiol. Res. 207, 100–107. doi: 10.1016/j.micres.2017.11.011
Read, D. B., Gregory, P. J. (1997). Surface tension and viscosity of axenic maize and lupin root mucilages. New Phytol. 137, 623–628. doi: 10.1046/j.1469-8137.1997.00859.x
Rudrappa, T., Czymmek, K. J., Paré, P. W., Bais, H. P. (2008). Root-secreted Malic acid recruits beneficial soil bacteria. Plant Physiol. 148, 1547–1556. doi: 10.1104/pp.108.127613
Sampedro, I., Parales, R. E., Krell, T., Hill, J. E. (2015). Pseudomonas chemotaxis. FEMS Microbiol. Rev. 39, 17–46. doi: 10.1111/1574-6976.12081
Schulz-Bohm, K., Gerards, S., Hundscheid, M., Melenhorst, J., de Boer, W., Garbeva, P. (2018). Calling from distance: attraction of soil bacteria by plant root volatiles. ISME J. 12, 1252–1262. doi: 10.1038/s41396-017-0035-3
Sharifi, R., Jeon, J.-S., Ryu, C.-M. (2022). Belowground plant–microbe communications via volatile compounds. J. Exp. Bot. 73, 463–486. doi: 10.1093/jxb/erab465
Siebers, M., Rohr, T., Ventura, M., Schütz, V., Thies, S., Kovacic, F., et al. (2018). Disruption of microbial community composition and identification of plant growth promoting microorganisms after exposure of soil to rapeseed-derived glucosinolates. PloS One 13, e0200160. doi: 10.1371/journal.pone.0200160
Tian, T., Sun, B., Shi, H., Gao, T., He, Y., Li, Y., et al. (2021). Sucrose triggers a novel signaling cascade promoting Bacillus subtilis rhizosphere colonization. ISME J. 15, 2723–2737. doi: 10.1038/s41396-021-00966-2
Tohidifar, P., Bodhankar, G. A., Pei, S., Cassidy, C. K., Walukiewicz, H. E., Ordal, G. W., et al. (2020). The unconventional cytoplasmic sensing mechanism for ethanol chemotaxis in Bacillus subtilis. mBio 11, e02177-20. doi: 10.1128/mbio.02177-20
Townsley, L., Yannarell, S. M., Huynh, T. N., Woodward, J. J., Shank, E. A. (2018). Cyclic di-AMP acts as an extracellular signal that impacts Bacillus subtilis biofilm formation and plant attachment. mBio. 9, e00341-18. doi: 10.1128/mbio.00341-18
Trdá, L., Fernandez, O., Boutrot, F., Héloir, M.-C., Kelloniemi, J., Daire, X., et al. (2014). The grapevine flagellin receptor VvFLS2 differentially recognizes flagellin-derived epitopes from the endophytic growth-promoting bacterium Burkholderia phytofirmans and plant pathogenic bacteria. New Phytol. 201, 1371–1384. doi: 10.1111/nph.12592
Tsai, H.-H., Wang, J., Geldner, N., Zhou, F. (2023). Spatiotemporal control of root immune responses during microbial colonization. Curr. Opin. Plant Biol. 74, 102369. doi: 10.1016/j.pbi.2023.102369
Tzipilevich, E., Russ, D., Dangl, J. L., Benfey, P. N. (2021). Plant immune system activation is necessary for efficient root colonization by auxin-secreting beneficial bacteria. Cell Host Microbe 29, 1507–1520.e4. doi: 10.1016/j.chom.2021.09.005
Vandeputte, O. M., Kiendrebeogo, M., Rajaonson, S., Diallo, B., Mol, A., El Jaziri, M., et al. (2010). Identification of catechin as one of the flavonoids from Combretum albiflorum bark extract that reduces the production of quorum-sensing-controlled virulence factors in Pseudomonas aeruginosa PAO1. Appl. Environ. Microbiol. 76, 243–253. doi: 10.1128/AEM.01059-09
Venturi, V., Fuqua, C. (2013). Chemical signaling between plants and plant-pathogenic bacteria. Annu. Rev. Phytopathol. 51, 17–37. doi: 10.1146/annurev-phyto-082712-102239
Verbon, E. H., Liberman, L. M., Zhou, J., Yin, J., Pieterse, C. M. J., Benfey, P. N., et al. (2023). Cell-type-specific transcriptomics reveals that root hairs and endodermal barriers play important roles in beneficial plant-rhizobacterium interactions. Mol. Plant 16, 1160–1177. doi: 10.1016/j.molp.2023.06.001
Walker, T. S., Bais, H. P., Déziel, E., Schweizer, H. P., Rahme, L. G., Fall, R., et al. (2004). Pseudomonas aeruginosa-plant root interactions: pathogenicity, biofilm formation, and root exudation. Plant Physiol. 134, 320–331. doi: 10.1104/pp.103.027888
Wang, Y., Li, J., Hou, S., Wang, X., Li, Y., Ren, D., et al. (2010). A Pseudomonas syringae ADP-ribosyltransferase inhibits Arabidopsis mitogen-activated protein kinase kinases. Plant Cell 22, 2033–2044. doi: 10.1105/tpc.110.075697
Wang, Y., Pruitt, R. N., Nürnberger, T., Wang, Y. (2022). Evasion of plant immunity by microbial pathogens. Nat. Rev. Microbiol. 20, 449–464. doi: 10.1038/s41579-022-00710-3
Wang, S., Xing, R., Wang, Y., Shu, H., Fu, S., Huang, J., et al. (2021). Cleavage of a pathogen apoplastic protein by plant subtilases activates host immunity. New Phytol. 229, 3424–3439. doi: 10.1111/nph.17120
Wen, F., VanEtten, H. D., Tsaprailis, G., Hawes, M. C. (2007). Extracellular proteins in pea root tip and border cell exudates. Plant Physiol. 143, 773–783. doi: 10.1104/pp.106.091637
Wen, T., Yuan, J., He, X., Lin, Y., Huang, Q., Shen, Q. (2020). Enrichment of beneficial cucumber rhizosphere microbes mediated by organic acid secretion. Hortic. Res. 7, 154. doi: 10.1038/s41438-020-00380-3
Wheatley, R. M., Poole, P. S. (2018). Mechanisms of bacterial attachment to roots. FEMS Microbiol. Rev. 42, 448–461. doi: 10.1093/femsre/fuy014
Wittstock, U., Burow, M. (2010). Glucosinolate breakdown in Arabidopsis: Mechanism, regulation and biological significance. Arabidopsis Book 8, e0134. doi: 10.1199/tab.0134
Xia, Y., Ma, Z., Qiu, M., Guo, B., Zhang, Q., Jiang, H., et al. (2020). N-glycosylation shields Phytophthora sojae apoplastic effector PsXEG1 from a specific host aspartic protease. Proc. Natl. Acad. Sci. U.S.A. 117, 27685–27693. doi: 10.1073/pnas.2012149117
Xin, D.-W., Liao, S., Xie, Z.-P., Hann, D. R., Steinle, L., Boller, T., et al. (2012). Functional analysis of NopM, a novel E3 ubiquitin ligase (NEL) domain effector of Rhizobium sp. strain NGR234. PloS Pathog. 8, e1002707. doi: 10.1371/journal.ppat.1002707
Yang, J., Lan, L., Jin, Y., Yu, N., Wang, D., Wang, E. (2022). Mechanisms underlying legume-rhizobium symbioses. J. Integr. Plant Biol. 64, 244–267. doi: 10.1111/jipb.13207
Yasuda, S., Okada, K., Saijo, Y. (2017). A look at plant immunity through the window of the multitasking coreceptor BAK1. Curr. Opin. Plant Biol. 38, 10–18. doi: 10.1016/j.pbi.2017.04.007
Yu, G., Derkacheva, M., Rufian, J. S., Brillada, C., Kowarschik, K., Jiang, S., et al. (2022). The Arabidopsis E3 ubiquitin ligase PUB4 regulates BIK1 and is targeted by a bacterial type-III effector. EMBO J. doi: 10.15252/embj.2020107257
Yu, K., Liu, Y., Tichelaar, R., Savant, N., Lagendijk, E., van Kuijk, S. J. L., et al. (2019). Rhizosphere-associated Pseudomonas suppress local root immune responses by gluconic acid-mediated lowering of environmental pH. Curr. Biol. 29, 3913–3920.e4. doi: 10.1016/j.cub.2019.09.015
Yu, G., Xian, L., Xue, H., Yu, W., Rufian, J. S., Sang, Y., et al. (2020). A bacterial effector protein prevents MAPK-mediated phosphorylation of SGT1 to suppress plant immunity. PloS Pathog. 16, e1008933. doi: 10.1371/journal.ppat.1008933
Zamioudis, C., Pieterse, C. M. J. (2012). Modulation of host immunity by beneficial microbes. Mol. Plant-Microbe Interact. 25, 139–150. doi: 10.1094/MPMI-06-11-0179
Zhang, L., Chen, X.-J., Lu, H.-B., Xie, Z.-P., Staehelin, C. (2011). Functional analysis of the type 3 effector nodulation outer protein L (NopL) from Rhizobium sp. NGR234: symbiotic effects, phosphorylation, and interference with mitogen-activated protein kinase signaling. J. Biol. Chem. 286, 32178–32187. doi: 10.1074/jbc.M111.265942
Zhang, S., Kan, J., Liu, X., Wu, Y., Zhang, M., Ou, J., et al. (2023). Phytopathogenic bacteria utilize host glucose as a signal to stimulate virulence through LuxR homologues. Mol. Plant Pathol. 24, 359–373. doi: 10.1111/mpp.13302
Zhang, H., Liu, Y., Wu, G., Dong, X., Xiong, Q., Chen, L., et al. (2021). Bacillus velezensis tolerance to the induced oxidative stress in root colonization contributed by the two-component regulatory system sensor ResE. Plant Cell Environ. 44, 3094–3102. doi: 10.1111/pce.14068
Zhang, J., Shao, F., Li, Y., Cui, H., Chen, L., Li, H., et al. (2007). A Pseudomonas syringae effector inactivates MAPKs to suppress PAMP-induced immunity in plants. Cell Host Microbe 1, 175–185. doi: 10.1016/j.chom.2007.03.006
Zhang, N., Wang, D., Liu, Y., Li, S., Shen, Q., Zhang, R. (2014). Effects of different plant root exudates and their organic acid components on chemotaxis, biofilm formation and colonization by beneficial rhizosphere-associated bacterial strains. Plant Soil 374, 689–700. doi: 10.1007/s11104-013-1915-6
Keywords: rhizosphere, root colonization, molecular dialogue, root exudates, plant-microbe interactions
Citation: Yang L, Qian X, Zhao Z, Wang Y, Ding G and Xing X (2024) Mechanisms of rhizosphere plant-microbe interactions: molecular insights into microbial colonization. Front. Plant Sci. 15:1491495. doi: 10.3389/fpls.2024.1491495
Received: 05 September 2024; Accepted: 17 October 2024;
Published: 06 November 2024.
Edited by:
Marzena Sujkowska-Rybkowska, Warsaw University of Life Sciences, PolandReviewed by:
Anindita Seal, University of Calcutta, IndiaKarin E. Groten, Max Planck Institute for Chemical Ecology, Germany
Copyright © 2024 Yang, Qian, Zhao, Wang, Ding and Xing. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gang Ding, Z2RpbmdAaW1wbGFkLmFjLmNu; Xiaoke Xing, eGt4aW5nMjAwOUBob3RtYWlsLmNvbQ==
†These authors share first authorship
 Luna Yang
Luna Yang Xin Qian†
Xin Qian† Zeyu Zhao
Zeyu Zhao Gang Ding
Gang Ding Xiaoke Xing
Xiaoke Xing