- 1State Key Laboratory for Managing Biotic and Chemical Threats to the Quality and Safety of Agro Products, Key Laboratory of Agricultural Microbiome of MARA and Zhejiang Province, Key Laboratory of Biotechnology in Plant Protection of MARA and Zhejiang Province, Institute of Plant Protection and Microbiology, Zhejiang Academy of Agricultural Sciences, Hangzhou, China
- 2State Key Laboratory of Rice Biology and Breeding, Key Laboratory of Molecular Biology of Crop Pathogens and Insects of MARA, Key Laboratory of Biology of Crop Pathogens and Insects of Zhejiang Province, Institute of Biotechnology, Zhejiang University, Hangzhou, China
The rice blast disease, caused by the fungus Magnaporthe oryzae, is a significant agricultural problem that adversely impacts rice production and food security. Understanding the precise molecular pathways involved in the interaction between the pathogen and its host is crucial for developing effective disease management strategies. This study examines the crucial function of the nucleolin MoNsr1 in regulating M. oryzae physiological functions. ΔMoNsr1 deletion mutants showed reduced fungal growth, asexual sporulation, and pathogenicity compared to the wild-type. Mutants exhibited impaired conidial germination and appressoria formation, reducing infection progression. Additionally, ΔMoNsr1 deletion mutant had less turgor pressure, confirming that MoNsr1 is essential for cell wall biogenesis and resistant to external stresses. Furthermore, ΔMoNsr1 deletion mutant showed enhanced sensitivity to oxidative stress, reactive oxygen species, and cold tolerance. Our results offer a thorough understanding of the function of MoNsr1 in the virulence and stress-resilient capability in M. oryzae. These findings provide insights into the novel targets and contribute to the emergence of innovative approaches for managing rice blast disease.
1 Introduction
Magnaporthe oryzae, a disease that can reduce crop yields, is a global priority due to its economic importance as a staple meal (Dean et al., 2012; Hossain, 2022). Over the past decade, M. oryzae has been a key model pathogen, conducting various molecular research on fungal-plant interactions (Ebbole, 2007; Ra, 2005). So far, numerous functional genes involved in the pathogenic process of rice blast have been cloned and analyzed, which are involved in spore production, germination, appressoria formation, pathogen penetration, and post-invasion expansion (Ebbole, 2007; Galhano and Talbot, 2011; Wilson and Talbot, 2009). Several important signaling pathways involved in growth and development have been shown to affect appressorium differentiation and pathogenesis, including the G protein signaling pathway, cAMP signaling pathway, MAPK signaling pathway, and calcium ion signaling pathway etc (Li et al., 2007; Liu et al., 2022; Mehrabi et al., 2009). Up to the present, all potential genes within the rice blast fungus genome and their encoded proteins have been predicted (Li et al., 2024). This advancement facilitates comprehensive and profound research into several key fundamental life processes of the rice blast fungus.
In eukaryotic cells, the ribosomes are the factory for protein synthesis. The ribosomal synthesis starts with pre-rRNA transcription and processing, followed by ribosomal subunit assembly and nucleocytoplasmic transport (Fromont-Racine et al., 2003; Venema and Tollervey, 1999). Nucleolin is a highly abundant protein and is involved in the process of ribosomal synthesis. Nucleolin influences rDNA transcription, rRNA processing, ribosome assembly, and nucleocytoplasmic transport of ribosome particles (Cong et al., 2011; Ginisty et al., 1999; Tajrishi et al., 2011). Nucleolin usually contains three distinct structural domains: an acidic region at the N terminus and the RNA-binding domain in the middle portion. The glycine-arginine-rich (GAR) domain is found in the C-terminal region (Ginisty et al., 1999). The GAR domain plays a role in interacting with ribosomal proteins and has been reported to influence ribosomal assembly and transport (Tuteja and Tuteja, 1998).
Studying the actions of nucleolin is difficult since it performs a wide range of mechanisms that impact DNA and RNA metabolism, and it is found in various regions in subcellular locations (Mongelard and Bouvet, 2007). Nucleolin not only binds to RNA/DNA and aids in adequately folding pre-rRNA, but it also interacts with numerous proteins during the ribosome assembly process. Additionally, it functions in regulating RNA polymerase-I-based transcription (Kojima et al., 2007).
Although nucleolin was identified in the early 1980s, its function in fungi has been poorly reported. Functional studies of nucleolin have only been reported in yeast, which is primarily involved in pre-RNA processing (Gulli et al., 1995; Kondo and Inouye, 1992; Lee et al., 1992). In yeast, deletion of the nucleolin gene ScNsr1 causes the accumulation of 35S pre-rRNA and subsequent reduction in mature 18S rRNA in the mutants and leads to the severe growth of mutant (Gulli et al., 1995; Kondo and Inouye, 1992; Lee et al., 1992), thereby M. oryzae MoNsr1 is likely to share a similar functions in ribosome biogenesis, involving in the processing and maturation of 35S pre-rRNA into the small subunit (18S rRNA) and the large subunit (5.8S and 25S rRNAs).
This study investigated the role of MoNsr1 homologs in M. oryzae. The nucleolin MoNsr1 is located in the nucleus and is essential for various processes, including vegetative growth, conidia germination, appressorium maturity, and full virulence. Additionally, the MoNsr1 gene is crucial for maintaining cell wall integrity, and its deletion leads to increased sensitivity to reactive oxygen species (ROS) and cold.
2 Materials and methods
2.1 Fungal strains and culture conditions
In this study, the wild-type strain Guy11 of rice blast fungus was used to investigate the functions of the MoNsr1. Phenotypic analysis of vegetative growth, chemical sensitivity, and asexual sporulation of rice blast fungus mutants was performed in a complete medium (CM) (Talbot et al., 1993). After the rice blast fungus was inoculated on CM plates and grown at 28 °C with 12-hour light-dark alternation for 9 days, conidia were obtained by washing the colonies with 10 ml sterile water and filtering them through 4 layers of sterile lens paper. After the rice blast fungus was inoculated into liquid CM medium and cultured in the dark at 28°C for 3 days, the culture was collected, and genomic DNA was recovered using EZNA® Fungal DNA Mini Kit (Omega Bio-tek, Inc., Norcross, GA).
2.2 Subcellular localization of MoNsr1
In order to investigate the subcellular localization of the MoNsr1 protein, an integrated expression plasmid for MoNsr1 and GFP was constructed using plasmid p1300BAR as the basic skeleton (Li et al., 2014). The whole MoNsr1 gene fragment (containing the 1.5kb promoter and MoNsr1 without a stop codon), as well as the GFP gene fragment, will be inserted between the EcoRI and XbaI restriction enzyme cleavage sites of plasmid p1300BAR. After the obtained plasmid pMoNsr1-GFP was sequenced and confirmed to be correct, the wild-type strain was introduced using the Agrobacterium tumefaciens-mediated transformation (AtMT) method described by (Rho et al., 2001). Green fluorescent transformants were obtained for subcellular localization investigation of MoNsr1 using CM plate screening containing 200 μg/ml glufosinate ammonium and fluorescence microscopy (ZEISS Imager. A2). The nuclear localization of rice blast fungus was performed using the DAPI staining method (Odenbach et al., 2009), and the subcellular localization of MoNsr1 was observed and recorded under a laser confocal microscope (ZEISS LSM780).
2.3 Targeted gene replacement and complementation
MoNsr1 gene-deletion strains were created according to the principle of homologous recombination. In short, the upstream and downstream fragments of the MoNsr1 gene were first amplified using a wild-type genomic DNA template (approximately 1.0 kb each) and cloned into the plasmid p1300-KO (Li et al., 2014), respectively, between the restriction enzyme cleavage sites EcoRI and SmaI, and between BamHI and XbaI. After obtaining the plasmid pKO-Nsr1 and confirming its correctness through sequencing, it was used for the AtMT transformation of wild-type strains. Gene deletion mutants were obtained through CM plate screening and PCR validation containing 200 μg/ml hygromycin B. The primers used for PCR validation are shown in Supplementary Table S1. The MoNsr1 gene, to obtain a complementary mutant, was amplified using the wild-type genomic DNA as a template, including the promoter region (located 1.5 kb upstream of the start codon), and cloned between the EcoRI and XbaI restriction endonuclease sites of plasmid p1300BAR. The obtained complement plasmid was introduced into a MoNsr1 gene deletion mutant, and the complemented mutant was obtained through CM plate screening and PCR validation containing 200 μg/ml glufosinate-ammonium.
2.4 Pathogenicity assays
Referring to previous method (Liu et al., 2016a), pathogenicity testing was conducted on the susceptible rice (cv. CO-39) seedlings and 7-day-old barley (cv. ZJ-8) plants at 3-4 true leaf stage. The rice plants were inoculated with 0.25% gelatin suspension with a concentration of 1 × 105 conidia/ml by spray. After inoculation, the rice plants were moved into a dark room with a humidity of 25 °C and above 95% for dark cultivation for 24 hours and then continued to grow alternately in light and dark for 16/8 hours at the same temperature until the wild-type inoculated plants showed typical symptoms. The disease severity of the leaves of inoculated rice plants was investigated and recorded to determine the pathogenicity of all tested strains. For the detached barley leaf inoculation assay, 20 µL of gradient diluted conidia suspension was spot inoculated on the barley leaves. The leaves were cultured in the dark at 25°C for 24 h and then illuminated until the wild-type inoculation treatment showed typical symptoms. The disease severity of each strain was compared and recorded. The experiment was carried out at least three times, with three replicates each time, to obtain similar results.
2.5 Analysis of infection-related morphogenesis
20 μL of 5 × 104 conidia/ml suspension was inoculated onto a plastic coverslip (Thermo Fisher Scientific, Waltham, MA, USA) and cultured in the dark at 25°C. Conidia germination and appressorium differentiation were observed and recorded under a microscope for 2h, 4h, 8h, 12h and 24h after inoculation. The appressorium turgor pressure of rice blast fungus was determined according to the previous method (Dixon et al., 1999). The appressorium formed by induction on the hydrophobic surface for 24h was treated with 1-4 M glycerol for 10 minutes, and the collapse rate of mutant and wild-type appressorium after glycerol treatment was counted under a microscope. The test was repeated three times, and the number of conidia or appressorium observed for each strain was no less than 200. For the rice blast infection rate test, barley leaves grown for 7 days were selected and sprayed with 1 × 10-5/mL rice blast conidia suspension. After 36 h or 48 h of inoculation, the barley leaves were decolorized with methanol, and the number of infected appressorium was counted under a light microscope. The experiment was repeated 3 times, and no less than 50 appressorium was observed for each strain.
2.6 Phosphorylation assays
The mycelium of all tested strains grown in liquid CM was used to extract the proteins. Western blotting was used to identify the amounts of phosphorylated Pmk1 and Mps1. To detect phosphor-Mps1 and phospho-Pmk1, a primary anti-phospho-p44/42 MAPK antibody (Cell Signaling Technology, USA) was employed. Two antibodies were applied to detect Mps1 and Pmk1, anti-ERK1/2 MAPK antibody (Cell Signaling Technology, USA) for Mps1 and anti-p44/42 MAPK antibody (Santa Cruz, USA) for Pmk1. A monoclonal anti-GAPDH antibody (HUABIO, China) was also used to detect the samples. Using the ImageJ program, the band intensity was calculated.
2.7 Statistical analysis
The relevant experimental data were processed with Excel 2016, and one-way ANOVA was analyzed using DPS (Data Processing System) v20.05. Significance levels were determined using Duncan’s new multiple-range test at p < 0.05.
3 Results
3.1 Identification of MoNsr1
The homologous genes of Saccharomyces cerevisiae ScNsr1 were searched in the EsdmblFungi (https://fungi.ensembl.org) database using the BLASTp program. The resultant MoNsr1 (MGG_01268) was obtained and comprised 486 amino acids. The MoNsr1-encoding gene comprises 1592 bp, with 2 introns and an exon. After amino acid sequence alignment, MoNsr1 showed 54% homology with ScNsr1 (Figure 1A), including 4 conserved domains. The N-terminus includes an acidic domain composed of serine and acidic residues. This structure may be related to the interaction between nucleolin and ribosomal proteins, histone H1, U3 snoRNP, etc (Erard et al., 1988; Ginisty et al., 1998; Sicard et al., 1998). The middle part contains two RRMs (RNA recognition motifs), which allow the protein to bind to RNA specifically, and the C-terminal GAR domain. The GAR domain is often found in nucleolar proteins, and the GAR domain in nucleolin interacts with ribosomal proteins (Bouvet et al., 1998).
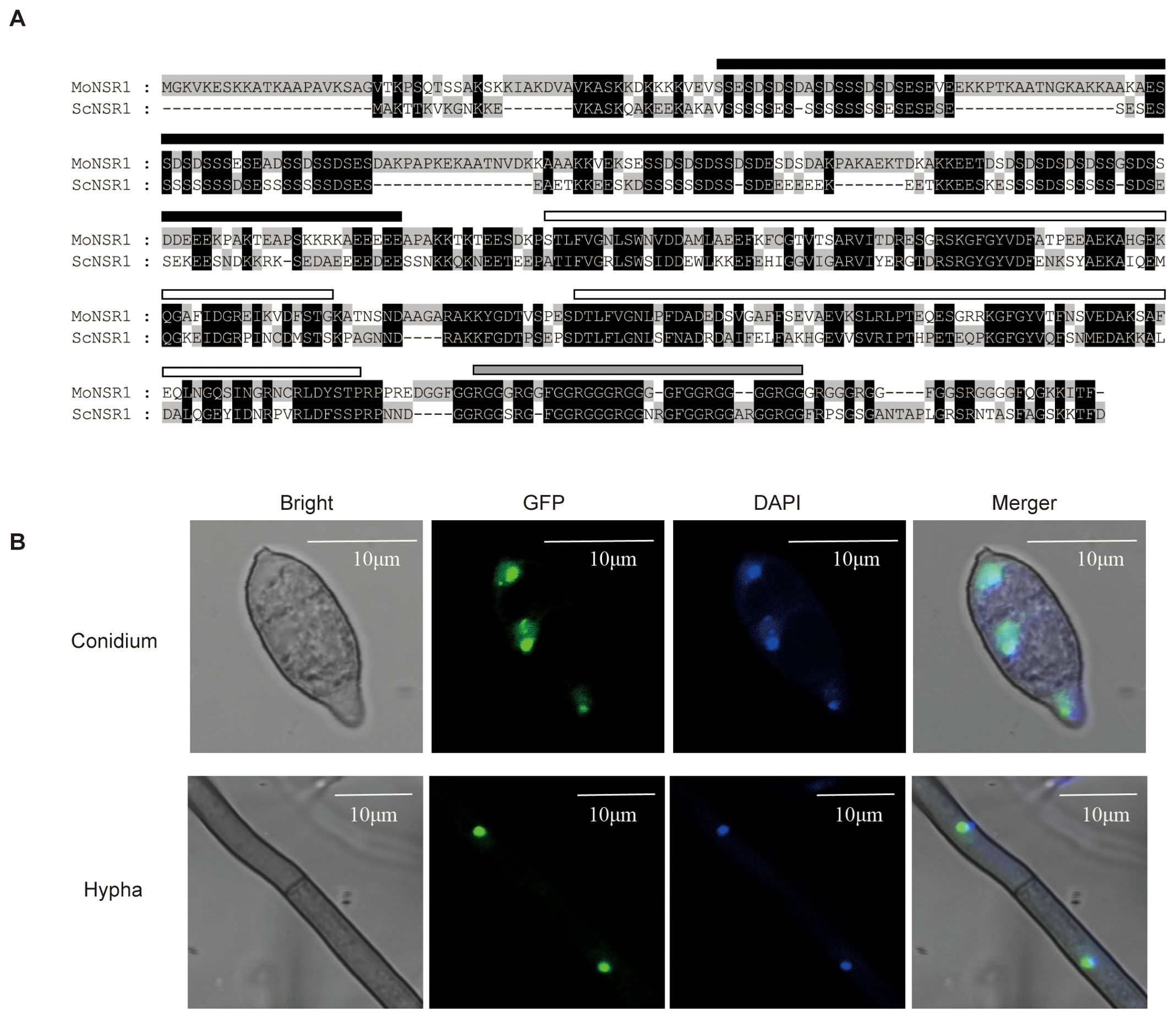
Figure 1. Primary structure and subcellular localization of MoNsr1. (A) Comparison of amino acid sequence of MoNsr1 and ScNsr1. The conserved amino acids are black-shaded. The black rectangle represents the acidic N-terminal domain, the white rectangles show the two RRM domains and the gray rectangle represents the GAR domain. (B) The MoNsr1-GFP protein was co-localized with DAPI-stained DNA in M. oryzae. Bar = 10 μm.
3.2 MoNsr1 is localized to the nucleus
In yeast, Nsr1 is localized in the nucleus and binds to the (TG1-3)n telomeric DNA (Lee et al., 1991; Lin and Zakian, 1994). The MoNsr1-GFP fusion expression vector constructed in this study was transformed into the rice blast fungus strain Guy11 by AtMT, and 6 transformants with detectable green fluorescence were obtained. There was no significant difference in the obtained transformants growth and sporulation phenotypes compared with the wild-type. The hyphae and spores of the GFP-labeled transformants were stained with 0.8 μg/ml DAPI solution and observed under a fluorescence microscope. The blue nuclei labeled with DAPI fluorescence in the cells co-localized with the green fluorescence of the MoNsr1-GFP fusion expression protein (Figure 1B). The results showed that MoNsr1, like ScNsr1, is localized in the nucleus.
3.3 Deletion of MoNsr1 affected the vegetative growth and asexual reproduction in M. oryzae
We performed gene knockout and complementation to investigate the function of the MoNsr1 gene. Inoculate the wild-type strain, gene deletion mutant ΔMoNsr1, and the complementary strain MoNsr1c onto CM medium with alternate light and dark cultivation at 28 °C for 12 hours for 6 days. The results showed that the average colony diameter of the ΔMoNsr1 mutant was significantly smaller than that of the wild-type (p<0.05) (Figures 2A, C). Rectangular mycelium blocks grown on CM medium for 4 days were cut and placed on a glass slide. The conidia induction experiment was carried out by alternating light and dark at 28 °C for 12 and 48 hours. Microscopic observation confirmed that compared with the wild-type and complemented strains, the ΔMoNsr1 mutant had fewer conidiophores and produced sparse conidia (Figure 2B). Conidia grown on CM medium for 6 days were collected for statistics. The results showed that the yield of conidia per unit area of the ΔMoNsr1 mutant was only 35% of that of the wild-type (Figure 2D). The above results indicate that the loss of the MoNsr1 gene is involved in the vegetative growth and asexual reproduction of rice blast fungus.
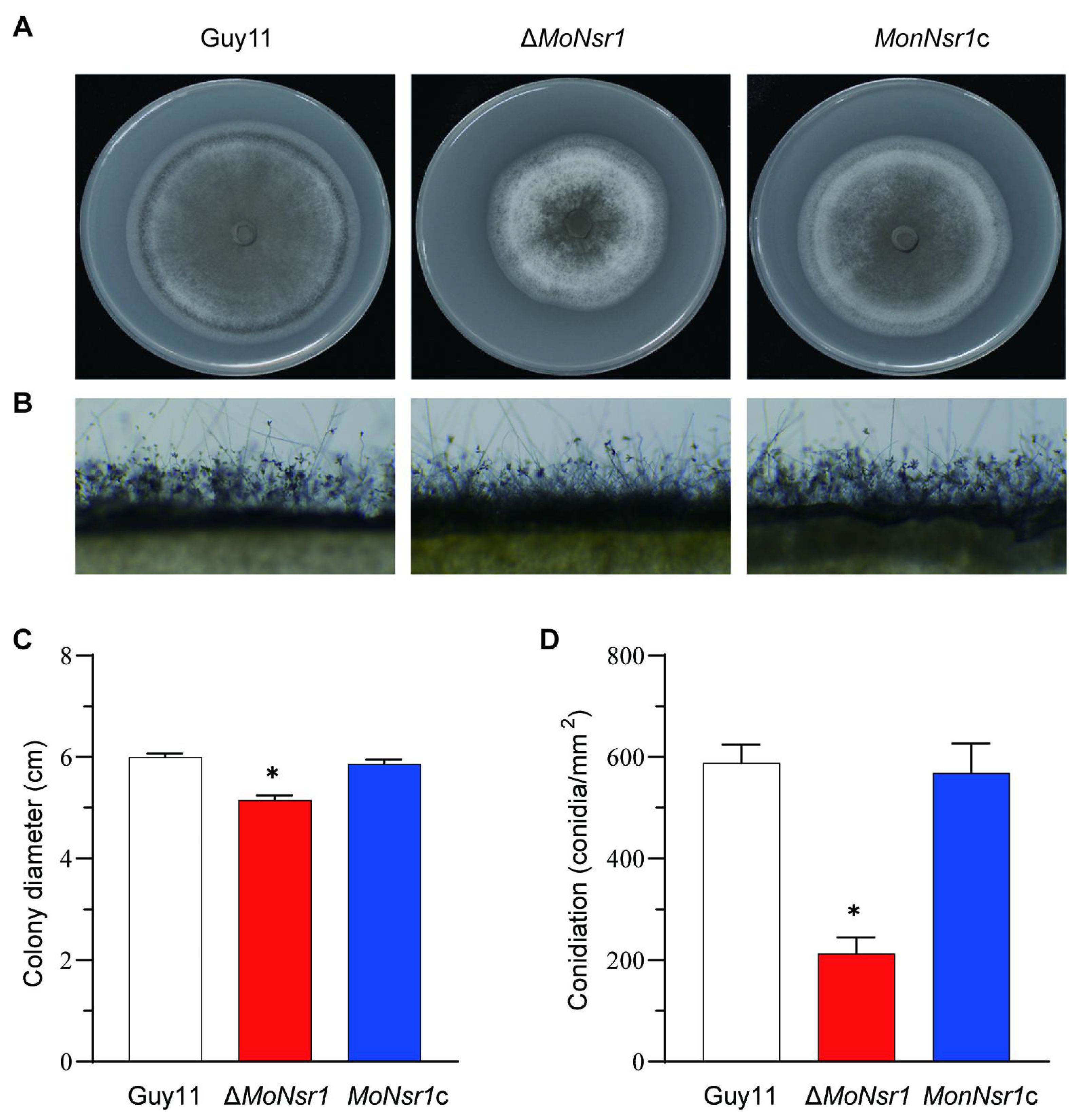
Figure 2. Vegetative growth and conidiation of M. oryzae strains in CM medium. (A) Colony morphology on CM plates for 6 days at 28°C with 12 h light and dark alternation. (B) Development of conidia on conidiophores on CM. (C) Statistical analysis of the vegetative growth rate of the tested strain on CM plates. (D) Statistical analysis of the number of conidia on CM plates. Asterisks in each data column indicate significant differences at p < 0.05.
3.4 MoNsr1 is indispensable for full virulence of M. oryzae
In order to clarify the role of the MoNsr1 in the pathogenesis of rice blast fungus, the wild-type strain Guy11, gene deletion mutant ΔMoNsr1, and compensatory strain MoNsr1c were used to inoculate barley leaves and 14-day-old susceptible rice seedlings (CO-39). The pathogenicity results of detached barley leaves showed that the pathogenicity of the ΔMoNsr1 mutant was lower than that of wild-type and complement treated with the same concentration of spore solution. Pathogenicity weakening became more pronounced with decreased spore concentration (Figure 3A). The results of inoculation of rice seedlings with 1 x 105 spores/ml by spray showed that the mutant ΔMoNsr1 produced fewer disease spots, and the disease severity significantly decreased compared with that of the wild-type strain (Figures 3B, C).
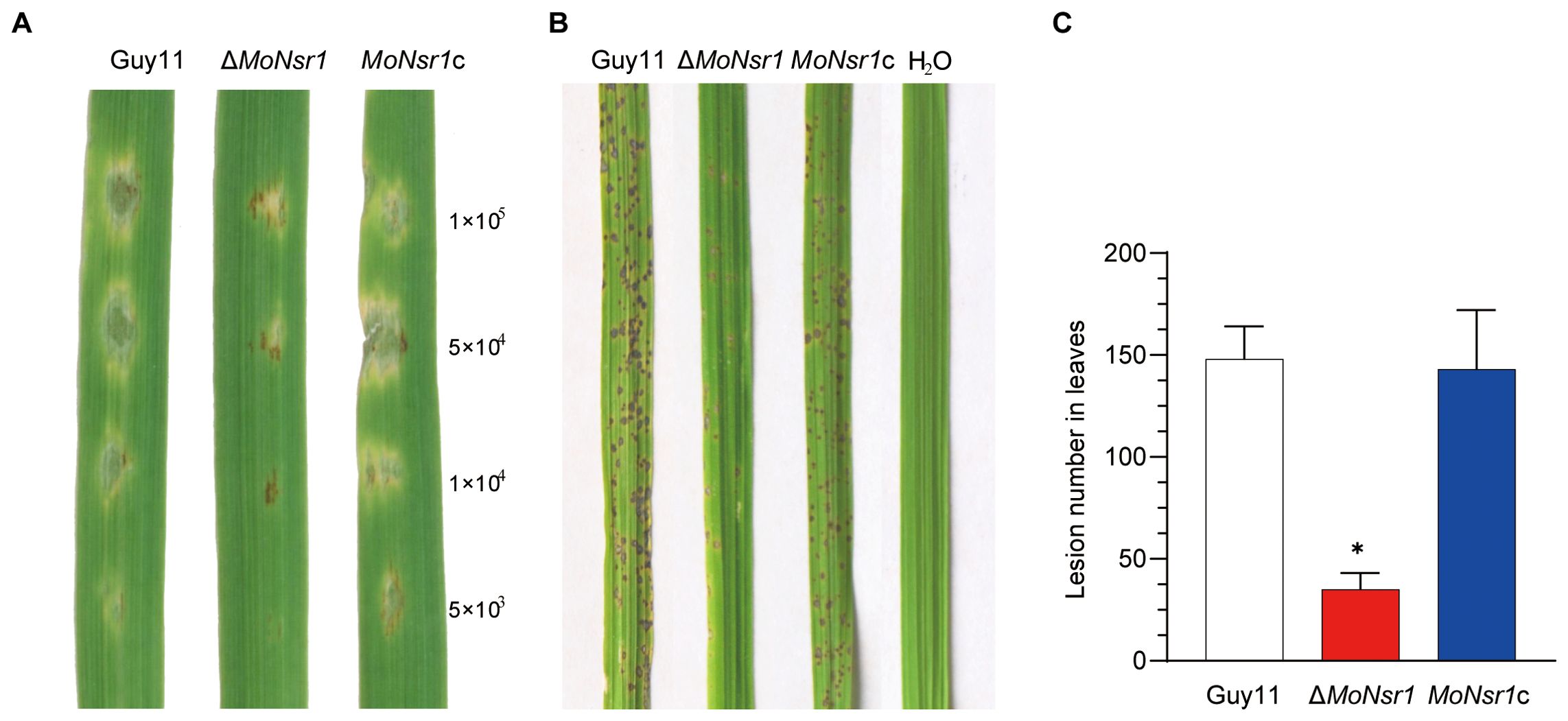
Figure 3. Pathogenicity of the tested M. oryzae strains. (A) Drop inoculation with 20 μL serial dilutions of conidia suspension on barley leaf segments. (B) Spray inoculation with 14-day-old rice seedlings. Representative leaves were photographed 6 days post-inoculation. (C) Lesion number in rice leaves. Asterisks in each data column indicate significant differences at p < 0.05.
3.5 MoNsr1 is involved in the germination of conidia and maturation of appressorium
In order to investigate the impact of MoNsr1 gene deletion on the pathogenicity of rice blast fungus, we analyzed the effects of gene deletion mutants on conidia germination, appressorium differentiation, and host epidermal penetration ability. The results of the conidial germination experiment showed that the conidia of the ΔMoNsr1 mutant could germinate and form an appressorium. Compared with the wild-type, the conidial germination rate and appressorium formation rate of the ΔMoNsr1 mutant were significantly reduced (Figures 4A, B). When the appressorium induced for 24 hours was treated with 1-4 M glycerol, the collapse rate of the appressorium in the ΔMoNsr1 mutant was significantly higher than that in the wild-type (Figure 4C). These results indicated that MoNsr1 is involved in conidial germination, appressorium differentiation, and maturation.
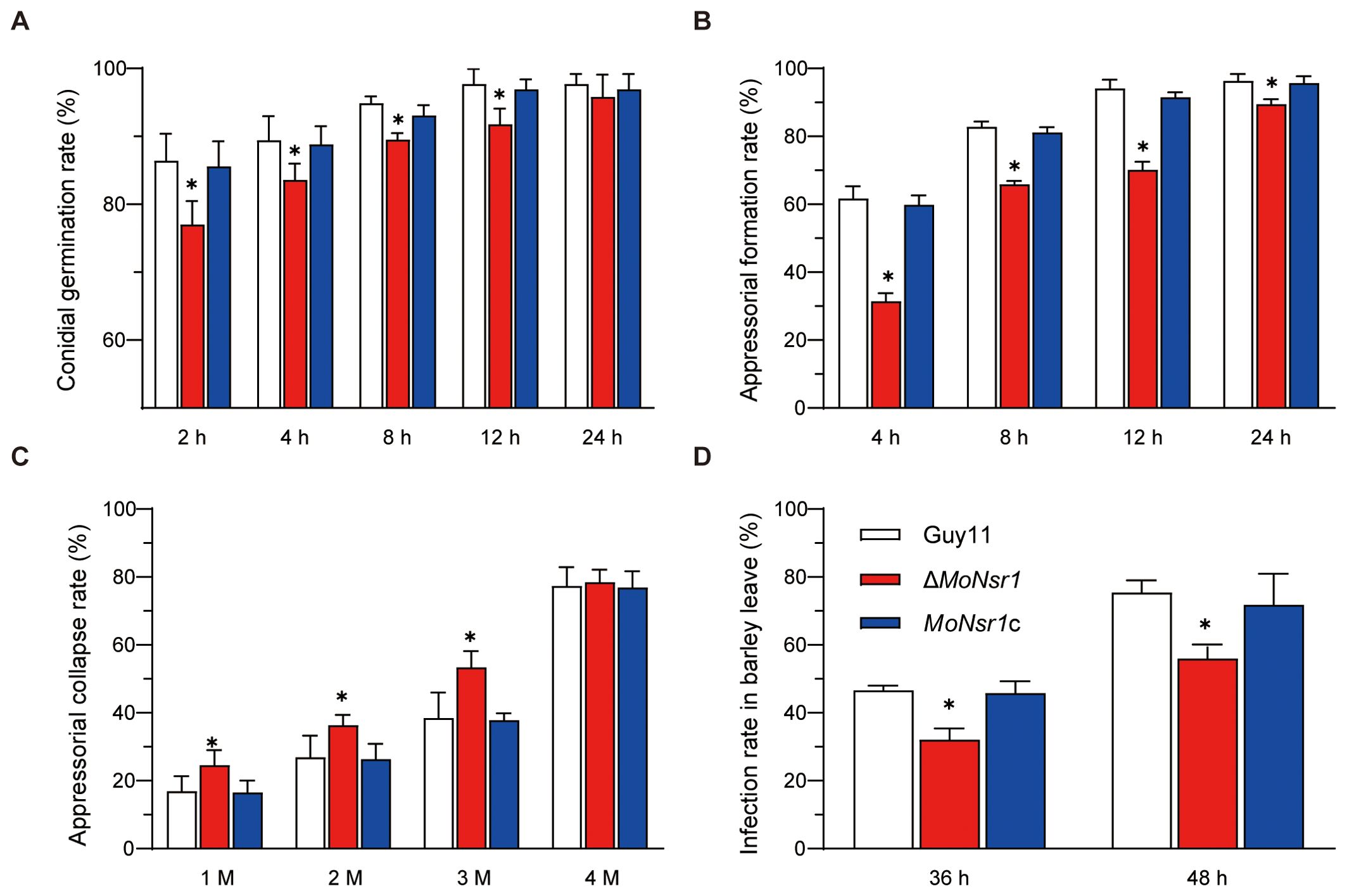
Figure 4. Analysis of infection-related morphogenesis of ΔMoNsr1 deletion mutant. (A) conidial germination rate; (B) appressorial formation rate; (C) appressorial collapse rate; (D) Infection rate in barley leaf cells. Asterisks in each data column indicate significant differences at p < 0.05.
We analyzed the infection rate in barley leaves to determine whether the decrease in turgor pressure of the ΔMoNsr1 mutant caused the decrease in host pathogenicity. The results showed that after inoculating barley leaves with a suspension of 5 × 104 spores/ml in vitro for 36 and 48 hours, the infection rates of the ΔMoNsr1 mutant on barley epidermal cells decreased by 31% and 26%, respectively, compared to that of the wild-type under a microscope (Figure 4D). The above indicates that the absence of the MoNsr1 leads to a decrease in mutant infection rate, resulting in a decrease in mutant pathogenicity. This result is consistent with the decrease in appressorial turgor pressure in the ΔMoNsr1 mutant.
3.6 MoNsr1 is involved in cell wall integrity
We investigated the mechanism effects of the MoNsr1 on the cell wall integrity of the gene deletion mutant and the sensitivity of the ΔMoNsr1 mutant to cell wall disruptors. The results showed that the growth of the ΔMoNsr1 mutant was inhibited on CM plates containing 25 μg/mL Congo red (CR) and 12.5 μg/mL Calcolflour white (CFW), with inhibition rates of 24.6% and 12.1%, respectively, which were significantly higher than those of the wild-type (Figures 5A, B). The results indicate that gene deletion leads to defects in the integrity of the ΔMoNsr1 mutant cell wall. The cell wall integrity signaling pathway is a key mechanism for sensing and adapting to various environmental conditions, and the Mps1-mediated MAPK signaling pathway is its core component (Lu et al., 2023). The phosphorylation level of Mps1 in the ΔMoNsr1 mutant was analyzed, and the results showed that compared with the wild-type, the Mpsl phosphorylation level of the ΔMoNsr1 mutant was lower (Figure 5C). The above results indicate that MoNsr1 may respond to the cell integrity signaling pathway by regulating Mpsl phosphorylation. Surprisingly, the phosphorylation level of Pmkl in the ΔMoNsr1 mutant was also lower than that of the wild-type. Pmk1 is an important signaling pathway that regulates appressorium differentiation and infection in rice blast fungus (Zhang et al., 2011). Therefore, MoNsr1 may also participate in the differentiation of appressorium by regulating the phosphorylation of Pmk1.
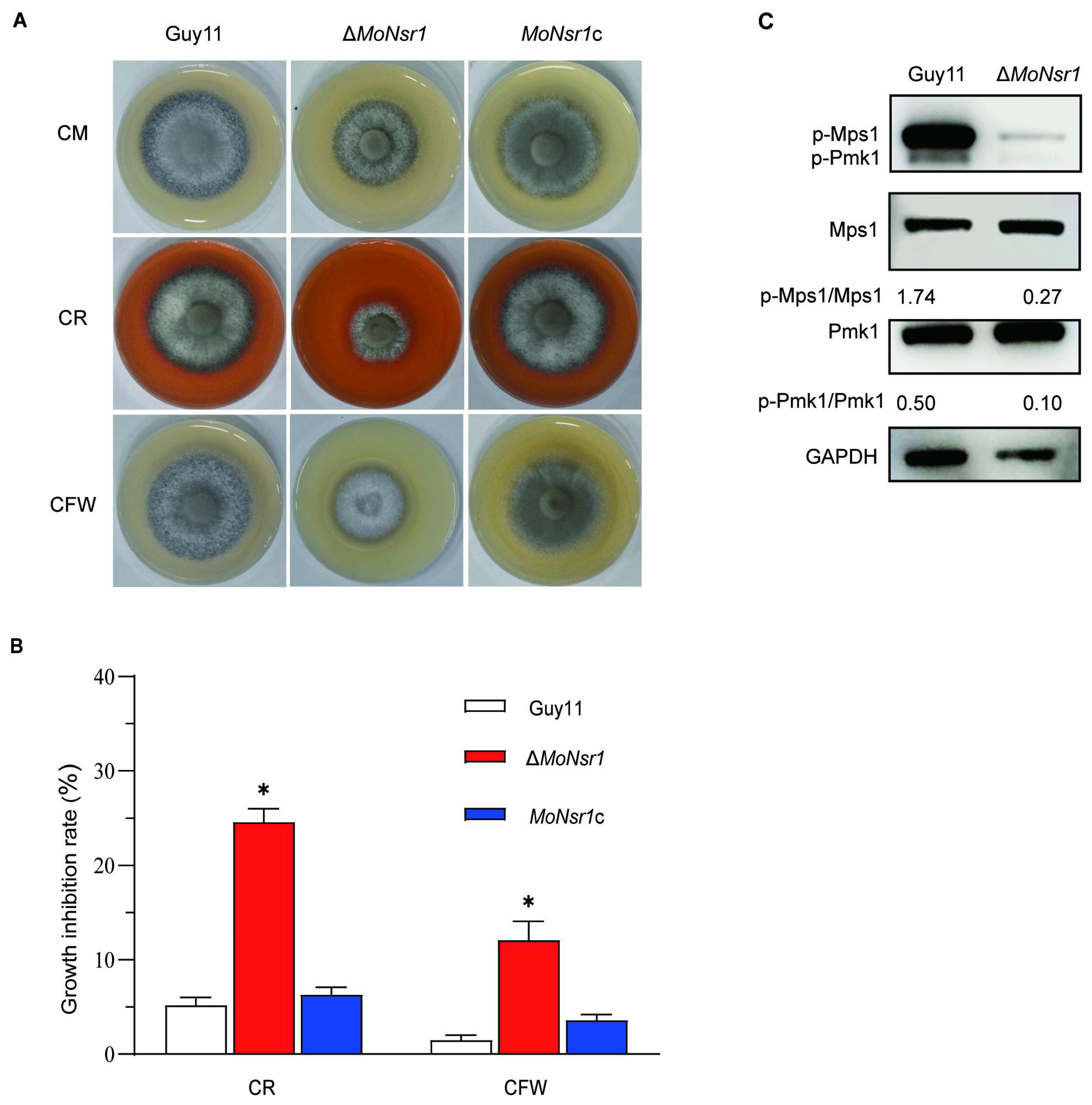
Figure 5. Effect of cell wall disruptors on vegetative growth of the tested strains. (A) Colony morphology of the tested strains grown on CM containing Congo red (25 μg/mL) or Calcolflour white (12.5 μg/mL) at 28°C for 6 days; (B) Relative inhibition rate of the tested strains under Congo red or Calcolflour White stress; (C) The phosphorylation level of Mps1 and Pmk1 in the tested strains. Asterisks in each data column indicate significant differences at p < 0.05.
3.7 MoNsr1 deletion results in sensitivity to ROS and cold
In addition, we also found that the ΔMoNsr1 mutant is more sensitive to oxidants and cold stress. The relative growth inhibition rates of mutants on CM plates containing 2 mM methyl viologen (MV) or 7.35 mM H2O2 were 42% and 43%, respectively, significantly lower than that of the wild-type (Figures 6A, B). The relative growth inhibition rate of mutants cultured at 20 °C was 49%, significantly higher than that of the wild-type (40%) (Figures 7A, B). These findings indicate that MoNsr1 plays a positive role in resisting oxidative and low-temperature stress.
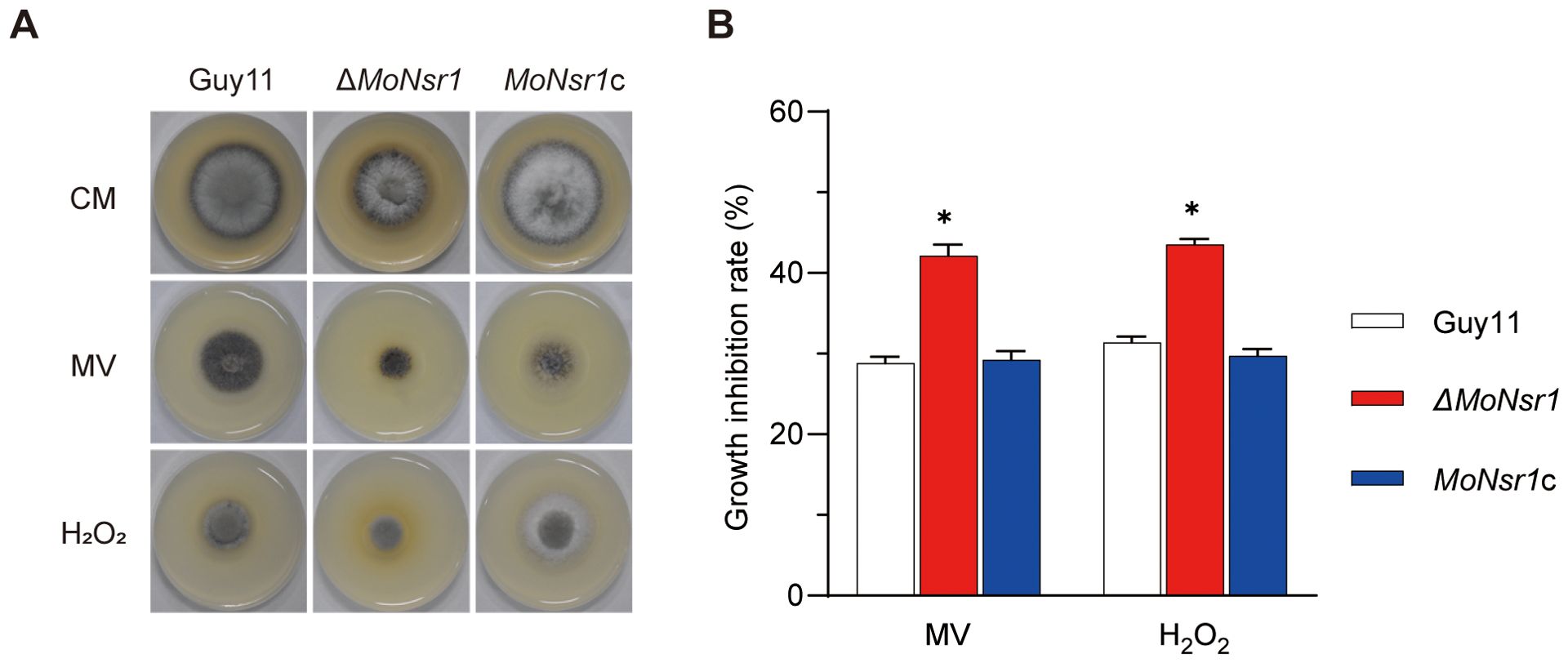
Figure 6. Effect of oxidants on vegetative growth of the tested M. oryzae strains. (A) Colony morphology of the tested strains grown on CM containing methyl viologen (MV, 2.0 mM) and H2O2 (7.35 mM) at 28°C for 6 days; (B) Relative inhibition rate under ROS stress. Asterisks in each data column indicate significant differences at p < 0.05.
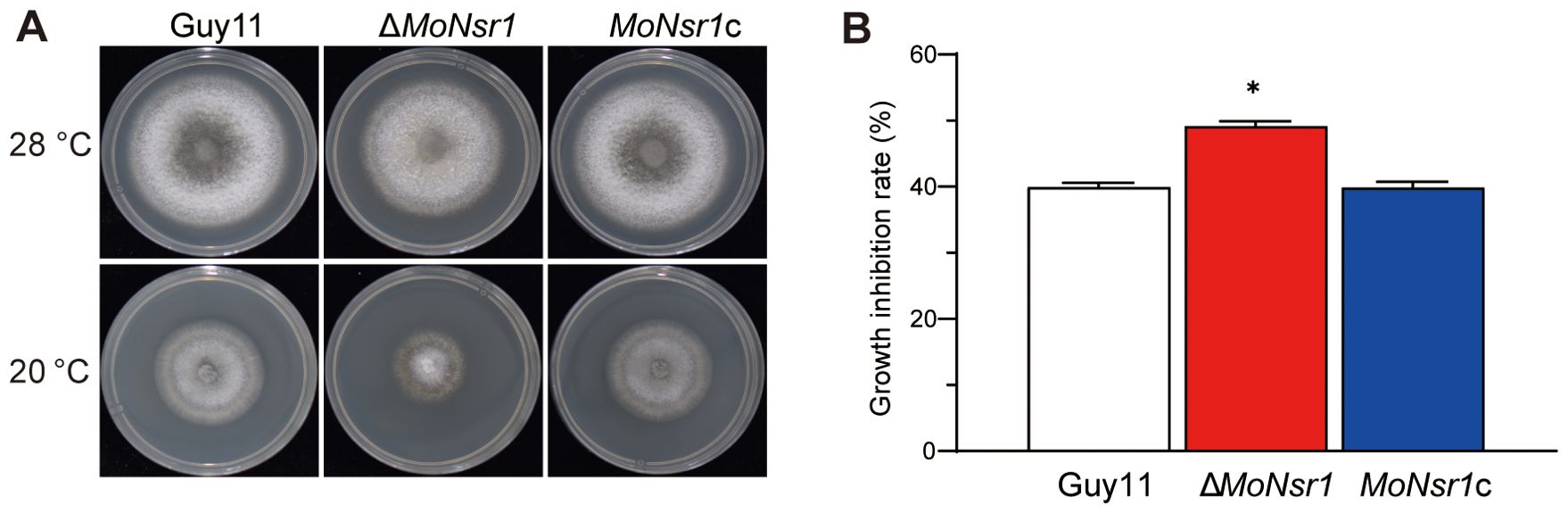
Figure 7. Effect of cold on vegetative growth of the tested M. oryzae strains. (A) Colony morphology of the tested strains grown on CM at 28°C and 20°C for 6 days; (B) Relative inhibition rate under cold stress. Asterisks in each data column indicate significant differences at p < 0.05.
4 Discussion
Nucleolin is a multifunctional non-ribosomal protein, which directly interacts with the pre-rRNA by its RRM domains to participate in the cleavages of pre-rRNA A1 and A2 and interact with ribosomal proteins to participate in pre-ribosomal particle assembly (Ginisty et al., 1998). Several potential CK2 phosphorylation sites were found in the N-terminal acidic region. The phosphorylated nucleolin regulates cell growth, proliferation, apoptosis and DNA damage response (Xiao et al., 2014; Zhang et al., 2018). In the present study, a yeast nucleolin ScNsr1 homolog in M. oryzae, MoNsr1, was identified. MoNsr1 has a similar protein structure to ScNsr1 and is located in the nucleus. The homology in nuclear distribution and protein structure between MoNsr1 and ScNsr1 implies that both proteins likely have comparable functions, such as being involved with ribosomal RNA processing, as evidenced by the presence of RRM and GAR domains in their sequences. Due to defects of growth, spore production, and pathogenicity of gene deletion mutants, it was implied that nucleolin MoNsr1 plays a pivotal role in crucial cellular processes required for growth.
The identification of MoNsr1 functions in the processing of ribosomal RNA and its influence on cellular activities including growth and proliferation improves comprehension of the complex molecular mechanisms present in M. oryzae. Ribosomes in eukaryotic cells are responsible for protein translation, which biosynthesis is a complex process involving the coordination of rDNA, rRNA, RNA polymerase I, and a lot of the non-ribosomal or ribosomal proteins. Nucleolin is involved in pre-rRNA cleavage to produce mature 18S rRNA (Gulli et al., 1995). The loss function of nucleolin affects the accumulation of 40S ribosomal subunits. In M. oryzae, it was documented that ribosome biosynthesis plays an important role in growth and pathogenicity (Cao et al., 2016; Li et al., 2019; Liu et al., 2016b, 2018). For example, ScYvh1 and ScRei1 are involved in 60S ribosomal subunit biogenesis (Meyer et al., 2010), and ScFap7 is a ribosome assembly factor involved in 40S ribosomal subunit biogenesis (Kemmler et al., 2009).
The phenotype of ΔMoNsr1 mutants in vegetative growth, asexual reproduction, pathogenicity, cell wall, and oxidative stress were similar to that of the mutants of the above three genes. Given that nucleolin plays an early role in ribosome synthesis, the evidence suggests that MoNsr1 might participate in the early stage of ribosomal biosynthesis. This participation could subsequently influence the development and pathogenicity of the rice blast fungus, making further studies focusing on this hypothesis essential for understanding its impact on crop health. The correlation between MoNsr1 and ribosome biosynthesis has significant effects on concentrating on crucial individuals involved in the life cycle of the pathogen, like MoNsr1, we can interrupt the progression of the disease and promote the development of robust crops.
Mitogen-activated protein kinase (MAPK) cascade responds to host and environmental signals in M. oryzae (Mehrabi et al., 2009). The Mps1-mediated MAPK signaling pathway regulates penetration, conidiation, and cell wall integrity in M. oryzae. The ΔMps1 mutant is reduced in aerial hyphal growth and conidiation and is hypersensitive to cell wall disruptors (Xu et al., 1998). The suppression of the ΔMoNsr1 mutant growth on media containing CR and CFW provides evidence to support the concept that MoNsr1 plays a role in maintaining the cell wall. This data, supported by additional evidence, shows a reduction in the phosphorylation level of Mps1 in the ΔMoNsr1. Significantly, they also discovered a decreased degree of phosphorylation of Pmk1 in the ΔMoNsr1 mutant, which is a crucial regulator of appressorium differentiation and infection (Xu and Hamer, 1996; Sakulkoo et al., 2018; Zhang et al., 2011). The complex regulatory network that controls the pathogen’s interaction with the host is indicated by the influence of MoNsr1 on MAPK signaling pathways and the association with cell wall integrity and stress responses. Develop measures that can lessen the adverse impacts of M. oryzae and other associated diseases on crop health, these pathways must be understood.
Interestingly, we found that the mutants were susceptible to cold stress. The cold stress response in yeast is transduced by the Hog1-mediated MAPK signaling pathway (Panadero et al., 2006). These findings indicate that MoNsr1 might have a broader function in the infection process and cold stress, potentially playing a role in MAPK signaling pathways. The increased vulnerability of MoNsr1-deficient mutants to cold stress emphasizes the protein’s wider involvement in M. oryzae ability to adjust to different environmental circumstances.
Based on the growth of the ΔMoNsr1 mutant on the medium containing H2O2, it was speculated that the growth of the infected hyphae of the mutant could be blocked within the host, indicating a potential limitation in its pathogenicity. Previous investigations have demonstrated that MoYvh1 can increase ribosome production, which promotes the synthesis and secretion of ROS-scavenging extracellular proteins, allowing the infected mycelium to grow in host tissue under oxidative stress (Liu et al., 2018). Thus, we speculate that ribosome biosynthesis in the ΔMoNsr1 mutant may also be affected, leading to removing the ROS extracellular protein synthesis and increasing the sensitivity to ROS.
Our results revealed that nucleolin MoNsr1 plays a crucial significance in the biology of this destructive rice blast. MoNsr1 is a multifunctional protein required for vegetative growth, conidiogenesis, pathogenicity, and the MAPK pathways. Discovering the various activities of MoNsr1 offers potential for future research, promoting greater exploration of the complex interplay between fungal genes and their impact on disease-causing capability and environmental adaption.
5 Conclusions
The results of this study provide significant insights into the role of the nucleolin gene, MoNsr1, in the pathogenicity and stress adaptation of the rice blast fungus, M. oryzae. The findings demonstrate that MoNsr1 is crucial for various physiological processes, including vegetative growth, asexual reproduction, and full virulence of the pathogen. The deletion of MoNsr1 led to reduced fungal growth, impaired conidial germination, and defective appressorium formation, all of which are critical factors in the infection process. Furthermore, the ΔMoNsr1 deletion mutant exhibited decreased turgor pressure, confirming the essential role of MoNsr1 in maintaining cell wall integrity. The increased sensitivity to oxidative stress and cold tolerance in the mutants highlights the importance of MoNsr1 in stress response mechanisms. Overall, these findings underline the pivotal function of MoNsr1 in the virulence and stress adaptability of M. oryzae, offering potential novel targets for developing strategies to control rice blast disease.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
ZZ: Conceptualization, Formal Analysis, Investigation, Writing – original draft. MSI: Conceptualization, Investigation, Writing – review & editing. JX: Investigation, Writing – review & editing. XF: Investigation, Writing – review & editing. MN: Writing – review & editing. JW: Data curation, Formal Analysis, Writing – review & editing. ZH: Writing – review & editing. HQ: Resources, Writing – review & editing. RC: Resources, Writing – review & editing. YC: Writing – review & editing. YW: Writing – review & editing. JyW: Conceptualization, Formal Analysis, Funding acquisition, Project administration, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was supported by the “Pioneer” and “Leading Goose” R&D Program of Zhejiang (Grant no. 2022C02047 and 2023C02018).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1482934/full#supplementary-material
References
Bouvet, P., Diaz, J. J., Kindbeiter, K., Madjar, J. J., Amalric, F. (1998). Nucleolin interacts with several ribosomal proteins through its RGG domain. J. Biol. Chem. 273, 19025–19029. doi: 10.1074/jbc.273.30.19025
Cao, H., Huang, P., Zhang, L., Shi, Y., Sun, D., Yan, Y., et al. (2016). Characterization of 47 Cys2-His2 zinc finger proteins required for the development and pathogenicity of the rice blast fungus Magnaporthe oryzae. New Phytol. 211, 1035–1051. doi: 10.1111/nph.2016.211.issue-3
Cong, R., Das, S., Bouvet, P. (2011). “The multiple properties and functions of nucleolin,” in The Nucleolus (Springer New York, New York, NY), 185–212.
Dean, R., Van Kan, J. A. L., Pretorius, Z. A., Hammond-Kosack, K. E., Di Pietro, A., Spanu, P. D., et al. (2012). The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 13, 414–430. doi: 10.1111/j.1364-3703.2011.00783.x
Dixon, K. P., Xu, J. R., Smirnoff, N., Talbot, N. J. (1999). Independent signaling pathways regulate cellular turgor during hyperosmotic stress and appressorium-mediated plant infection by Magnaporthe grisea. Plant Cell 11, 2045–2058. doi: 10.1105/tpc.11.10.2045
Ebbole, D. J. (2007). Magnaporthe as a model for understanding host-pathogen interactions. Annu. Rev. Phytopathol. 45, 437–456. doi: 10.1146/annurev.phyto.45.062806.094346
Erard, M. S., Belenguer, P., Caizergues-Ferrer, M., Pantaloni, A., Amalric, F. (1988). A major nucleolar protein, nucleolin, induces chromatin decondensation by binding to histone Hl. Eur. J. Biochem. 175, 525–530. doi: 10.1111/j.1432-1033.1988.tb14224.x
Fromont-Racine, M., Senger, B., Saveanu, C., Fasiolo, F. (2003). Ribosome assembly in eukaryotes. Gene 313, 17–42. doi: 10.1016/S0378-1119(03)00629-2
Galhano, R., Talbot, N. J. (2011). The biology of blast: Understanding how Magnaporthe oryzae invades rice plants. Fungal Biol. Rev. 25, 61–67. doi: 10.1016/j.fbr.2011.01.006
Ginisty, H., Amalric, F., Bouvet, P. (1998). Nucleolin functions in the first step of ribosomal RNA processing. EMBO J. 17, 1476–1486. doi: 10.1093/emboj/17.5.1476
Ginisty, H., Sicard, H., Roger, B., Bouvet, P. (1999). Structure and functions of nucleolin. J. Cell Sci. 112, 761–772. doi: 10.1242/jcs.112.6.761
Gulli, M. P., Girard, J. P., Zabetakis, D., Lapeyre, B., Melese, T., Caizergues-Ferrer, M. (1995). Gar2 is a nucleolar protein from Schizosaccharomyces pombe required for 18S rRNA and 40S ribosomal subunit accumulation. Nucleic Acids Res. 23, 1912–1918. doi: 10.1093/nar/23.11.1912
Hossain, M. M. (2022). Wheat blast: A review from a genetic and genomic perspective. Front. Microbiol. 13, 983243. doi: 10.3389/fmicb.2022.983243
Kemmler, S., Occhipinti, L., Veisu, M., Panse, V. G. (2009). Yvh1 is required for a late maturation step in the 60S biogenesis pathway. J. Cell Biol. 186, 863–880. doi: 10.1083/jcb.200904111
Kojima, H., Suzuki, T., Kato, T., Enomoto, K., Sato, S., Kato, T., et al. (2007). Sugar-inducible expression of the nucleolin-1 gene of Arabidopsis thaliana and its role in ribosome synthesis, growth and development. Plant J. 49, 1053–1063. doi: 10.1111/j.1365-313X.2006.03016.x
Kondo, K., Inouye, M. (1992). Yeast NSR1 protein that has structural similarity to mammalian nucleolin is involved in pre-rRNA processing. J. Biol. Chem. 267, 16252–16258. doi: 10.1016/S0021-9258(18)41993-X
Lee, W. C., Xue, Z., Mélèse, T. (1991). The NSR1 gene encodes a protein that specifically binds nuclear localization sequences and has two RNA recognition motifs. J. Cell Biol. 113, 1–12. doi: 10.1083/jcb.113.1.1
Lee, W. C., Zabetakis, D., Mélèse, T. (1992). NSR1 is required for pre-rRNA processing and for the proper maintenance of steady-state levels of ribosomal subunits. Mol. Cell. Biol. 12, 3865–3871. doi: 10.1128/mcb.12.9.3865-3871.1992
Li, L., Wang, J., Zhang, Z., Wang, Y., Liu, M., Jiang, H., et al. (2014). MoPex19, which is essential for maintenance of peroxisomal structure and woronin bodies, is required for metabolism and development in the rice blast fungus. PLoS One 9, e85252. doi: 10.1371/journal.pone.0085252
Li, L., Wright, S. J., Krystofova, S., Park, G., Borkovich, K. A. (2007). Heterotrimeric G protein signaling in filamentous fungi. Annu. Rev. Microbiol. 61, 423–452. doi: 10.1146/annurev.micro.61.080706.093432
Li, L., Zhu, X. M., Shi, H. B., Feng, X. X., Liu, X. H., Lin, F. C. (2019). MoFap7, a ribosome assembly factor, is required for fungal development and plant colonization of Magnaporthe oryzae. Virulence 10, 1047–1063. doi: 10.1080/21505594.2019.1697123
Li, Z., Yang, J., Ji, X., Liu, J., Yin, C., Bhadauria, V., et al. (2024). First telomere-to-telomere gapless assembly of the rice blast fungus Pyricularia oryzae. Sci. Data 11, 380. doi: 10.1038/s41597-024-03209-z
Lin, J. J., Zakian, V. A. (1994). Isolation and characterization of two Saccharomyces cerevisiae genes that encode proteins that bind to (TG1–3) n single strand telomeric DNA in vitro. Nucleic Acids Res. 22, 4906–4913. doi: 10.1093/nar/22.23.4906
Liu, C., Liu, T., Lv, Z., Qin, M., Qu, Z., Zhang, Z., et al. (2022). A calcineurin regulator MoRCN1 Is Important for asexual development, stress response, and plant infection of Magnaporthe oryzae. Front. Plant Sci. 13, 925645. doi: 10.3389/fpls.2022.925645
Liu, X., Cai, Y., Zhang, X., Zhang, H., Zheng, X., Zhang, Z. (2016a). Carbamoyl phosphate synthetase subunit MoCpa2 affects development and pathogenicity by modulating arginine biosynthesis in Magnaporthe oryzae. Front. Microbiol. 7, 2023. doi: 10.3389/fmicb.2016.02023
Liu, X., Qian, B., Gao, C., Huang, S., Cai, Y., Zhang, H., et al. (2016b). The putative protein phosphatase MoYvh1 functions upstream of MoPdeH to regulate the development and pathogenicity in Magnaporthe oryzae. Mol. Plant Microbe Interact. 29, 496–507. doi: 10.1094/MPMI-11-15-0259-R
Liu, X., Yang, J., Qian, B., Cai, Y., Zou, X., Zhang, H., et al. (2018). MoYvh1 subverts rice defense through functions of ribosomal protein MoMrt4 in Magnaporthe oryzae. PLoS Pathog. 14, e1007016. doi: 10.1371/journal.ppat.1007016
Lu, K., Chen, R., Yang, Y., Xu, H., Jiang, J., Li, L. (2023). Involvement of the cell wall–integrity pathway in signal recognition, cell-wall biosynthesis, and virulence in Magnaporthe oryzae. Mol. Plant Microbe Interact. 36, 608–622. doi: 10.1094/MPMI-11-22-0231-CR
Mehrabi, R., Zhao, X., Kim, Y., Xu, J. R. (2009). “The cAMP signaling and map kinase pathways in plant pathogenic fungi,” in Plant Relationships (Springer Berlin Heidelberg, Berlin, Heidelberg), 157–172.
Meyer, A. E., Hoover, L. A., Craig, E. A. (2010). The Cytosolic J-protein, Jjj1, and Rei1 function in the removal of the Pre-60 S subunit factor Arx1. J. Biol. Chem. 285, 961–968. doi: 10.1074/jbc.M109.038349
Mongelard, F., Bouvet, P. (2007). Nucleolin: A multifaceted protein. Trends. Cell Biol. 17, 80–86. doi: 10.1016/j.tcb.2006.11.010
Odenbach, D., Thines, E., Anke, H., Foster, A. J. (2009). The Magnaporthe grisea class VII chitin synthase is required for normal appressorial development and function. Mol. Plant Pathol. 10, 81–94. doi: 10.1111/j.1364-3703.2008.00515.x
Panadero, J., Pallotti, C., Rodríguez-Vargas, S., Randez-Gil, F., Prieto, J. A. (2006). A downshift in temperature activates the high osmolarity glycerol (HOG) pathway, which determines freeze tolerance in Saccharomyces cerevisiae. J. Biol. Chem. 281, 4638–4645. doi: 10.1074/jbc.M512736200
Ra, D. (2005). The genome sequence of the rice blast fungus Magnaporthe grisea. Nature 434, 980–986. doi: 10.1038/nature03449
Rho, H.-S., Kang, S., Lee, Y. H. (2001). Agrobacterium tumefaciens-mediated transformation of the plant pathogenic fungus, Magnaporthe grisea. Mol. Cell 12, 407–411. doi: 10.1016/S1016-8478(23)17116-0
Sakulkoo, W., Osés-Ruiz, M., Oliveira Garcia, E., Soanes, D. M., Littlejohn, G. R., Hacker, C., et al. (2018). A single fungal MAP kinase controls plant cell-to-cell invasion by the rice blast fungus. Science 359, 1399–1403. doi: 10.1126/science.aaq0892
Sicard, H., Faubladier, M., Noaillac-Depeyre, J., Léger-Silvestre, I., Gas, N., Caizergues-Ferrer, M. (1998). The role of the Schizosaccharomyces pombe gar2 protein in nucleolar structure and function depends on the concerted action of its highly charged N terminus and its RNA-binding domains. Mol. Biol. Cell. 9, 2011–2023. doi: 10.1091/mbc.9.8.2011
Tajrishi, M. M., Tuteja, R., Tuteja, N. (2011). Nucleolin. Commun. Integr. Biol. 4, 267–275. doi: 10.4161/cib.4.3.14884
Talbot, N. J., Ebbole, D. J., Hamer, J. E. (1993). Identification and characterization of MPG1, a gene involved in pathogenicity from the rice blast fungus Magnaporthe grisea. Plant Cell 5, 1575–1590. doi: 10.1105/tpc.5.11.1575
Tuteja, R., Tuteja, N. (1998). Nucleolin: A multifunctional major nucleolar phosphoprotein. Crit. Rev. Biochem. Mol. Biol. 33, 407–436. doi: 10.1080/10409239891204260
Venema, J., Tollervey, D. (1999). Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 33, 261–311. doi: 10.1146/annurev.genet.33.1.261
Wilson, R. A., Talbot, N. J. (2009). Under pressure: investigating the biology of plant infection by Magnaporthe oryzae. Nat. Rev. Microbiol. 7, 185–195. doi: 10.1038/nrmicro2032
Xiao, S., Caglar, E., Maldonado, P., Das, D., Nadeem, Z., Chi, A., et al. (2014). Induced expression of nucleolin phosphorylation-deficient mutant confers dominant-negative effect on cell proliferation. PLoS One 9, e109858. doi: 10.1371/journal.pone.0109858
Xu, J. R., Hamer, J. E. (1996). MAP kinase and cAMP signaling regulate infection structure formation and pathogenic growth in the rice blast fungus Magnaporthe grisea. Genes. Dev. 10, 2696–2706. doi: 10.1101/gad.10.21.2696
Xu, J. R., Staiger, C. J., Hamer, J. E. (1998). Inactivation of the mitogen-activated protein kinase Mps1 from the rice blast fungus prevents penetration of host cells but allows activation of plant defense responses. Proc. Natl. Acad. Sci. 95, 12713–12718. doi: 10.1073/pnas.95.21.12713
Zhang, X., Xiao, S., Rameau, R. D., Devany, E., Nadeem, Z., Caglar, E., et al. (2018). Nucleolin phosphorylation regulates PARN deadenylase activity during cellular stress response. RNA Biol. 15, 251–260. doi: 10.1080/15476286.2017.1408764
Keywords: cell wall integrity, Magnaporthe oryzae, MoNsr1, nucleolin, pathogenicity, stress response
Citation: Zhang Z, Islam MS, Xia J, Feng X, Noman M, Wang J, Hao Z, Qiu H, Chai R, Cai Y, Wang Y and Wang J (2024) The nucleolin MoNsr1 plays pleiotropic roles in the pathogenicity and stress adaptation in the rice blast fungus Magnaporthe oryzae. Front. Plant Sci. 15:1482934. doi: 10.3389/fpls.2024.1482934
Received: 22 August 2024; Accepted: 23 September 2024;
Published: 15 October 2024.
Edited by:
Qinhu Wang, Northwest A&F University, ChinaReviewed by:
Huawei Zheng, Minjiang University, ChinaMuxing Liu, Nanjing Agricultural University, China
Copyright © 2024 Zhang, Islam, Xia, Feng, Noman, Wang, Hao, Qiu, Chai, Cai, Wang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiaoyu Wang, d2FuZ2ppYW95dTc4QHNpbmEuY29t
†These authors have contributed equally to this work
 Zhen Zhang1†
Zhen Zhang1† Jiuzhi Xia
Jiuzhi Xia Muhammad Noman
Muhammad Noman Jiaoyu Wang
Jiaoyu Wang