
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 23 October 2024
Sec. Plant Pathogen Interactions
Volume 15 - 2024 | https://doi.org/10.3389/fpls.2024.1481650
This article is part of the Research Topic Insights In Plant-Pathogen Interactions: 2023 View all 19 articles
Cell-cell communication is essential for both unicellular and multicellular organisms. Secreted peptides that act as diffusive ligands are utilized by eukaryotic organisms to transduce information between cells to coordinate developmental and physiological processes. In plants, The CLAVATA3/EMBRYO SURROUNDING REGION-RELATED (CLE) genes encode a family of secreted small peptides which play pivotal roles in stem cell homeostasis in various types of meristems. Accumulated evidence has revealed that CLE peptides mediate trans-kingdom interactions between plants and microbes, including pathogens and symbionts. This review highlights the emerging roles of CLE peptide signaling in plant-microbe interactions, focusing on their involvement in nodulation, immunity, and symbiosis with arbuscular mycorrhizal fungi. Understanding these interactions provides insights into the sophisticated regulatory networks to balance plant growth and defense, enhancing our knowledge of plant biology and potential agricultural applications.
Cell-to-cell communication is essential for multicellular organisms to coordinate their growth and development. Plants transmit information between cells through phytohormones, proteins/peptides, small RNAs, and metabolites. Secreted peptides acting as ligands are defined as extracellular proteins (less than 100 amino acids in length) derived from precursor proteins called prepropeptides (Tavormina et al., 2015). Peptide ligands are typically recognized by receptor kinases localized at the plasma membrane, thereby provoking an intracellular signal transduction cascade that changes the activity of downstream genes to modify cellular programs. The CLAVATA3 (CLV3)/EMBRYO SURROUNDING REGION-RELATED (CLE) gene family is one of the largest signaling peptide families in plants, with model plant Arabidopsis thaliana (hereafter Arabidopsis) genome containing 32 CLE genes. Canonical CLE prepropeptides possess a signal peptide at the N-terminus, a highly conserved motif at the C-terminus called the CLE domain, and a variable domain between the signal peptide and the CLE domain (Betsuyaku et al., 2011) (Figure 1). It is thought that CLE prepropeptides are cleaved by an endoplasmic reticulum-localized signal peptide peptidase to remove their signal peptide, resulting in a propeptide (Figure 1A). Propeptides are processed by post-translational modifications and are proteolytically cleaved at the N- and C- termini of the CLE domain by subtilases to generate a mature peptide in length of 12-14 amino acids (Betsuyaku et al., 2011; Stührwohldt et al., 2020). For instance, the mature CLV3 peptide has been identified as two distinct forms; (1) a 12 amino acids peptide with hydroxyproline (Hyp) residues in the 4th and 7th positions; (2) a 13 amino acids peptide that, with Hyp at the 4th position and tri-arabinosylated Hyp at the 7th position, contains an additional histidine residue at the 13th position (Figure 1B), those two forms can directly bind to extracellular domain of the plasma membrane-localized leucine-rich repeat receptor-like kinase (LRR-RLK) AtCLV1 (Kondo et al., 2006; Ohyama et al., 2009). The tri-arabinosylation is catalyzed by the Hyp O-arabinosyltransferases (HPATs) localized at Golgi (Ogawa-Ohnishi et al., 2013). This modification is critical for the binding affinity with CLV1, and therefore important for the bioactivity of the CLV3 peptide.
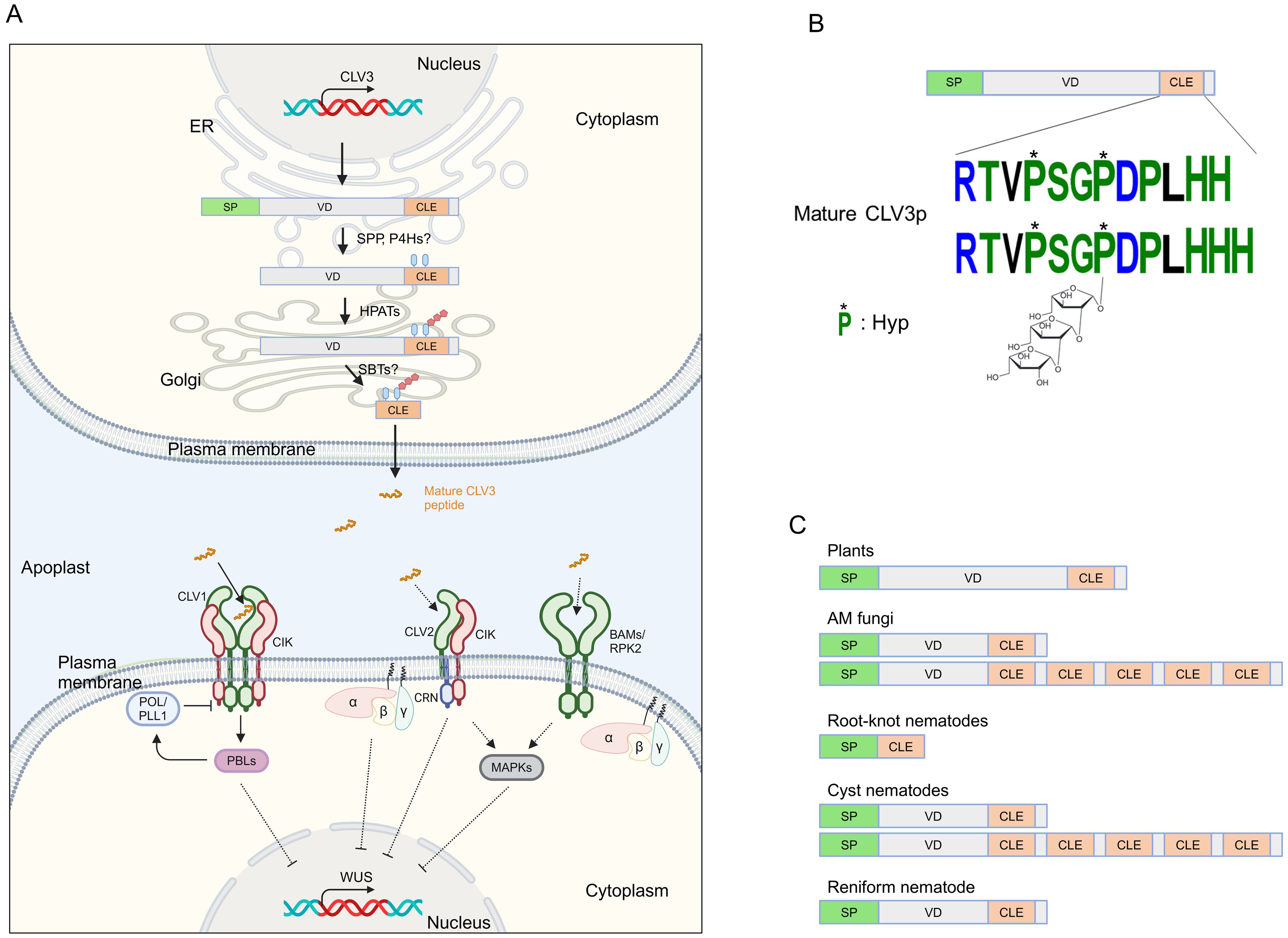
Figure 1. Illustrating signaling components of the CLAVATA pathway. (A) CLV3 signaling in the shoot apical meristem. The CLV3 prepropeptide is processed by hydroxyproline (Hyp) O-arabinosyltransferases (HPATs), and presumably by a signal peptide peptidase (SPP), prolyl-4-hydroxylases (P4Hs) and subtilases (SBTs) in ER-Golgi, and then mature CLV3 peptide is secreted to the apoplast. The CLV3 peptide is recognized by plasma membrane-associated receptor complexes, which in turn, WUSCHEL (WUS) is repressed through heterotrimeric G proteins, receptor-like cytoplasmic kinases (RLCKs) PBS1-LIKEs (PBLs), phosphatases POLTERGEIST (POL) and POL-LIE 1 (PLL1), and MAPKs. Dashed lines indicate putative or indirect pathways. (B) A model of the CLV3 prepropeptide and CLV3 peptide. The CLV3 prepropeptide consists of the N-terminal signal peptide (SP), the C-terminal CLE domain, and the variable domain (VD) between SP and CLE. (C) Representative structures of CLE prepropeptides produced by plants and microbes. Figure adapted from images created with BioRender.com.
Secreted mature CLE peptides are perceived by LRR-RLKs and coreceptors such as CLV1 and CLV3 INSENSITIVE RECEPTOR KINASEs (CIKs), LRR receptor-like protein (LRR-RLP) CLV2, and RLK CORYNE (CRN), which lacks the extracellular domain and acts with CLV2 and CIKs (Hu et al., 2018; Fletcher, 2020; Jones et al., 2021) (Figure 1A). CLV-type receptor complexes have been well characterized for their role in shoot apical meristem (SAM) maintenance, where CLE peptides are recognized by their cognate receptors, reading to the activation of an intracellular signaling cascade such as the receptor-like cytoplasmic kinases (RLCKs) PBS1-LIKEs (PBLs), the protein phosphatases POLTERGEIST (POL) and POLTERGEIST-LIKE1 (PLL1), MAP kinases, and the heterotrimeric GTP binding proteins (Yamaguchi et al., 2016; Wang et al., 2021c). In the SAM, the expression of the homeodomain transcription factor WUSCHEL (WUS) that promotes stem cell activity is suppressed by the CLV3-receptors module, thereby maintaining stem cell homeostasis. CLE-receptor modules also regulate stem cell homeostasis in inflorescence meristem, root apical meristem, and vascular meristem, and some physiological responses (Fletcher, 2020; Bashyal et al., 2023).
In nature, plants are associated with various microbes including bacteria, fungi, oomycetes, viruses, and nematodes. Plant-associated microbes affect host developmental processes and physiological responses as infection outcomes; in turn, host plants regulate microbial associations and behaviors to shape optimal interactions between plants and microbes. Accumulating evidence has depicted the emerging roles of CLE signaling in plant-microbe interactions. In this review, we describe CLE signaling pathways participating in plant-microbe interaction regulation and how CLE peptides and their receptors may respond to biotic and abiotic stimuli and culminate in changes to downstream signal transduction.
The relationship between legume plants (members of the family Fabaceae) and nitrogen-fixing bacteria, so-called rhizobia, is one of the most successful symbioses in nature. Endosymbiotic rhizobia inhabiting nodules which are lateral organs formed on the legume roots supply ammonia converted from atmospheric nitrogen to the host, while the host supplies malate converted from sucrose as the primary source of carbon (Udvardi and Poole, 2013). Excessive nodule formation can be detrimental to the host growth since plants lose carbon sources assimilated by photosynthesis. Therefore, legume plants have evolved a sophisticated regulatory mechanism called autoregulation of nodulation (AON) to prevent excess nodulation (Chaulagain and Frugoli, 2021). AON is a complex mechanism modulated through long-distance signaling between roots and shoots and is highly conserved across legume plants (Chaulagain and Frugoli, 2021; Li et al., 2022). Here, we focus on the role of signaling components constituted by CLE peptides and their cognate receptors mainly in Lotus japonicus and Medicago truncatula (Figure 2A).
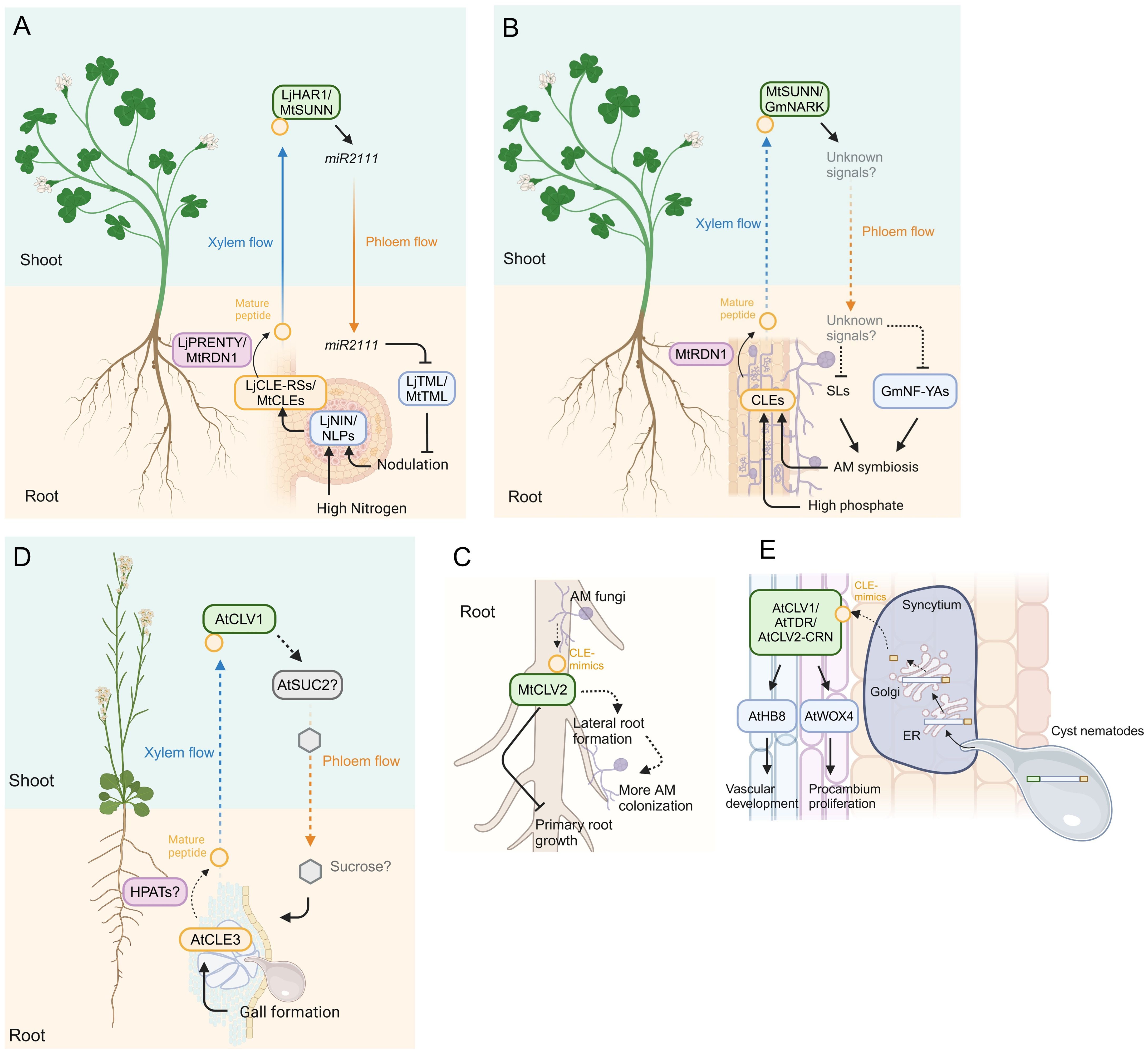
Figure 2. Model of CLE signaling in plant-microbe interactions. (A) Model of systemic signals mediated by CLE signaling in legume-rhizobium interactions. The transcription factors NODULE INCEPTION (NIN) and NIN-LIKE PROTEINs (NLPs) induce CLE gene expression in response to nodulation and high nitrogen conditions, respectively. Mature CLE peptides are post-translationally modified by HPATs LjPRENTY or MtRDN1 and translocate to the shoot via xylem, where the cognate receptors recognize the mature peptides. The miR2111 translocates from the shoot to the root via phloem and suppresses excessive nodulation by downregulating TOO MUCH LOVEs (TMLs) expression. (B) Model of systemic signals mediated by CLE signaling in plant-AM fungi interactions. AM colonization and high phosphate conditions induce the accumulation of CLE peptides, which are post-translationally modified by RDN1 and translocated to the shoot, where the cognate receptors recognize the mature CLE peptides. Strigolactones (SLs) and NUCLEAR FACTOR-YA transcription factors (NF-YAs) accumulation is suppressed through yet unknown long-distance signals translocating from the shoot to the root downstream of the CLE peptides perception, thereby suppressing additional AM symbiosis. (C) Model of local signal mediated by CLE peptide mimics in M. truncatula. AM fungi produce CLE mimics that inhibit primary root growth partially dependent on CLV2. The AM-CLE mimics also induce more AM colonization probably due to their induction effect of lateral root formation. (D) Model of systemic signal mediated by CLE signaling in plant and root-knot nematode interaction in Arabidopsis. The CLE3, which is upregulated in root-knot nematode-induced galls, regulates gall formation in a shoot-expressed CLV1-dependent manner, presumably via SUCROSE-PROTON SYMTORTER 2 (SUC2)-regulated sucrose translocation from the shoot to the root. (E) Model of local signal mediated by CLE peptide mimics in plant-cyst nematode interaction in Arabidopsis. Mature CLE mimics are secreted to the apoplast via the host ER-Golgi network. The host receptors recognize the mimics and induce expression of WUSCHEL-RELATED HOMEOBOX 4 (WOX4) and HOMEOBOX GENE 8 (AtHB8) which regulate procambium activity and vascular formation, thereby supporting cynsytium formation. Dashed lines indicate putative or indirect pathways. Figure adapted from images created with BioRender.com.
CLE peptides act as root-derived mobile signals in AON. In L. japonicus, a model legume plant, the expression of LjCLE-ROOT SIGNAL 1 (LjCLE-RS1), LjCLE-RS2, and LjCLE-RS3 is induced in the roots where rhizobia infect (Okamoto et al., 2009; Nishida et al., 2016). Experiments using transgenic hairy roots revealed that constitutive expression of these genes suppressed nodulation on not only transgenic hairy roots but also untransformed roots (Okamoto et al., 2009; Nishida et al., 2016). The mature LjCLE-RS2 peptide was detected in xylem sap collected at the hypocotyl of the plants that have transgenic hairy roots transformed with LjCLE-RS2, indicating that LjCLE-RS2 peptide is a root-derived mobile signal (Okamoto et al., 2013). As well as the mature form of AtCLV3 peptide, mature LjCLE-RS2 peptide is a 13 amino acids peptide that is derived from the C-terminal region of the precursor polypeptide and the 7th Hyp residue of this peptide is modified with three residues of arabinose (Ohyama et al., 2009; Okamoto et al., 2013). These CLE genes that act as a negative regulator on AON are evolutionarily conserved in other legumes, such as M. truncatula, Glycine max (soybean), Pisum sativum (pea), and Phaseolus vulgaris (common bean). It has been shown that CLE genes of M. truncatula (MtCLE12, MtCLE13, MtCLE34, and MtCLE35), G. max (RHIZOBIA-INDUCED CLE 1: GmRIC1 and GmRIC2), P. sativum (PsCLE12, PsCLE13, and other ten PsCLEs) and P. vulgaris (PvRIC1 and PvRIC2) are upregulated by rhizobial infection (Mortier et al., 2010; Lim et al., 2011; Reid et al., 2011; Ferguson et al., 2014; Alves‐Carvalho et al., 2015; Kassaw et al., 2017; Samorodova et al., 2018; Lebedeva et al., 2020; Mens et al., 2021). Constitutive expression of these genes in transgenic hairy roots leads to suppression of nodulation on systemic roots. However, the mature forms of most of these CLE peptides have yet to be elucidated. In L. japonicus, M. truncatula, and G. max, upregulation of CLE genes (LjCLE-RS1, LjCLE-RS2, MtCLE13, GmRIC1, and GmRIC2) in AON requires NODULE INCEPTION (NIN), which is an indispensable transcription factor for multiple aspects of rhizobial symbioses and is consistently lost in non-nodulating species (Soyano et al., 2014; Wang et al., 2019; Laffont et al., 2020; Shen and Feng, 2024). NIN has evolved from NIN-LIKE PROTEINs (NLPs) which control the expression of nitrate-responsive genes (Konishi and Yanagisawa, 2013; Marchive et al., 2013), suggesting that the regulation of CLE gene expression on AON may have evolved from the mechanism for fine-tuning in planta nitrogen status.
Post-translational modification of CLE peptides is critical for AON. An M. truncatula mutant lacking ROOT DETERMINED NODULATION 1 (MtRDN1) gene that encodes HPAT exhibits hyper nodulation phenotype (Schnabel et al., 2011; Kassaw et al., 2017). Through a combination of biochemical and genetic analyses to test functional implications of arabinosylation on AON, Imin and colleagues demonstrated that chemically synthesized MtCLE12 and MtCLE13 peptides with tri-arabinosylation at the 7th Hyp residue exerted nodulation inhibition in the wild-type and rdn1 mutant when they were applied to the roots or the cotyledons, but these oligopeptides without this modification no longer inhibited nodulation (Imin et al., 2018). Consistent with this, tri-arabinosylated LjCLE-RS1/2/3 and GmRIC1/2 peptides can weaken the hyper nodulation phenotype of mutants lacking LjPLENTY and PsNOD3, which are orthologs to MtRDN1 (Hastwell et al., 2019; Yoro et al., 2019). The MtRDN1 and LjPLENTY are localized to the Golgi (Kassaw et al., 2017; Yoro et al., 2019). Indeed, tri-arabinosylated LjCLE-RS2 peptide was detected in shoot xylem sap collected from soybean plants that developed transformed hairy roots having the genomic region of LjCLE-RS2; its modification significantly impacts binding affinity to the receptor mentioned below (Okamoto et al., 2013). These observations indicate that HPATs-mediated tri-arabinosylation of CLE peptides plays a key role in AON. As Kassaw and coauthors have provided evidence that MtRDN1 is required for the function of MtCLE12, but not MtCLE13 (Kassaw et al., 2017), further studies would reveal substrate preferences/specificities of HPATs for individual CLE peptides in each legume.
CLE-mediated AON requires shoot-acting cognate receptors. Grafting experiments and genetic assays showed that the LjCLE-RS peptides were recognized by shoot-acting LRR-RLKs HYPERNODULATION ABERRANT ROOT FORMATION 1 (LjHAR1) and KLAVIER (LjKLV) and LRR-RLP LjCLV2, which have high similarity to AtCLV1, RECEPTOR-LIKE PROTEIN KINASE 2 (AtRPK2) and AtCLV2, respectively (Wopereis et al., 2000; Krusell et al., 2002, 2011; Nishimura et al., 2002; Oka-Kira et al., 2005; Okamoto et al., 2009, 2013; Miyazawa et al., 2010; Nishida et al., 2016). Indeed, synthetic tri-arabinosylated LjCLE-RS1 and -RS2 peptides directly bind to the extracellular domain of LjHAR1 (Okamoto et al., 2013). Akin to AtCLV1, LjHAR1, LjKLV, and LjCLV2 can form isoforms, with homo- and hetero-dimerization (Miyazawa et al., 2010; Krusell et al., 2011). Since AtCLV2 acts with RLK AtCRN through a protein-protein interaction manner, LjCLV2 may also heterodimerize with an unidentified L. japonicus CRN orthologue (Bleckmann et al., 2009; Krusell et al., 2011). AtCLV1 and AtCLV2 like receptors have been characterized as negative regulators of nodulation in other legume plants: SUPER NUMERIC NODULES (MtSUNN, an AtCLV1 paralog) and MtCLV2 in M. truncatula, NODULE AUTOREGULATION RECEPTOR KINASE (GmNARK/PvNARK, an AtCLV1 paralog) in G. max and P. vulgaris, and PsSYM29 and PsSYM28 (AtCLV1 and AtCLV2 paralog, respectively) in P. sativum (Krusell et al., 2002, 2011; Searle et al., 2003; Schnabel et al., 2005; Ferguson et al., 2014). In M. truncatula, MtCLV2 and MtCRN can form heteromers, in heterologous expression system in tobacco leaf cells, and they act as a negative regulator on AON downstream of MtCLE12 and MtCLE13 peptides (Crook et al., 2016; Nowak et al., 2019). Interestingly, unlike AtCLV1, a mutant lacking LjHAR1 does not show any shoot phenotypes, whereas klv mutant of L. japonicus shows clv1-like shoot phenotypes such as fasciated stems, suggesting that AtCLV1 orthologs of legumes have evolved to control nodulation in shoots (Oka-Kira et al., 2005; Miyazawa et al., 2010).
CLE-mediated regulation of nodule formation is exerted by not only rhizobial infection but also several abiotic conditions. For instance, LjCLE-RS2, LjCLE-RS3, LjCLE40, MtCLE34, MtCLE35, and NITRATE-INDUCED CLE (GmNIC1) are induced in response to high nitrogen conditions, and among them, overexpression of LjCLE-RS2, LjCLE-RS3, MtCLE35, and GmNIC1 suppresses nodulation through their cognate receptors (Okamoto et al., 2009; Reid et al., 2011; Nishida et al., 2016; Lebedeva et al., 2020; Lebedeva et al. 2022; Mens et al., 2021; Moreau et al., 2021). Recent studies have shown that LjNLP1, LjNLP4, MtNLP1, GmNLP1, and GmNLP4, which are homologs of NIN transcription factors, are required to induce these nitrogen-responsive CLEs by directly binding to their promoter regions (Luo et al., 2021; Moreau et al., 2021; Nishida et al., 2021; Fu et al., 2024). Phosphate is another abiotic factor that controls nodulation mediated by CLE signaling. PvRIC1 and PvRIC2 are upregulated under low phosphate conditions as well as rhizobial infection (Isidra-Arellano et al., 2018; Isidra‐Arellano et al., 2020). Suppression of nodulation by low phosphate is not observed in the NARK mutants of G. max and P. vulgaris, suggesting that the peptides-receptors module required for phosphate-responsive nodule regulation is shared with AON pathway.
As described above, root-derived CLE peptides travel to the shoot where they are perceived by shoot-acting receptor complexes, and, in turn, downstream signals suppress excessive nodule formation in roots. Therefore, there should be the signaling molecule(s) that, downstream of CLE perception, translocates from shoots to roots. To regulate nodule numbers during rhizobial infection, two shoot-to-root mobile signals have been proposed by studies of L. japonicus and M. truncatula to date: (1) phytohormone cytokinin and (2) micro-RNA miR2111 (Sasaki et al., 2014; Tsikou et al., 2018; Gautrat et al., 2020; Okuma et al., 2020; Zhang et al., 2021). Rhizobial infection activates the cytokinin production in the shoot by inducing a cytokinin biosynthesis gene LjIPT3 in a LjHAR1-dependent manner, which can suppress nodule formation (Sasaki et al., 2014). An experiment using isotope-labeled cytokinin demonstrated that cytokinin fed to leaves was transported to roots in L. japonicus, probably via phloem, suggesting cytokinin is a shoot-to-root mobile signal downstream of LjHAR1 on AON. On the contrary to cytokinin, an miR2111 has been characterized as a positive regulator of nodulation. Mature miR2111s were highly accumulated in uninfected plants to repress nodule suppressor TOO MUCH LOVE (TML) in roots. In contrast, the expression level of miR2111s in leaf phloem was downregulated depending on the signaling module constituted by LjCLE-RS peptides and LjHAR1 in infected plants, thereby derepressing TML to inhibit excessive nodulation (Tsikou et al., 2018; Okuma et al., 2020). The miR2111 expression in shoots was required for TML repression in roots, suggesting miR2111 acts as a shoot-to-root mobile signal on AON. This miR2111-mediated systemic AON through the CLE signaling is conserved in M. truncatula and G. max as well (Gautrat et al., 2020; Moreau et al., 2021; Zhang et al., 2021).
Collectively, these conserved peptide-receptor pairs of legumes function in AON during rhizobial colonization in the same context. Interestingly, some parts of these signaling components are often utilized for regulation of lateral organ formations such as lateral root formation in both leguminous and non-leguminous plants (Araya et al., 2014; Huault et al., 2014; Soyano et al., 2019; Zhang et al., 2021; Hayashi-Tsugane and Kawaguchi, 2022; Kang et al., 2022; Lebedeva et al., 2023; Nakagami et al., 2023a; Sexauer et al., 2023). Therefore, it seems that organogenesis-regulating molecular network mediated by CLE signaling shares with the lateral organ developmental pathway, which is regulated in response to changing nutritional status. Nevertheless, our knowledge of CLE signaling components on AON is still insufficient, necessitating further investigation.
The interaction between plants and Arbuscular Mycorrhizal (AM) fungi that belong to the phylum Glomeromycotina is often recognized as mutualistic because host plants receive phosphorus and some other micronutrients from AM fungi, with the hosts providing carbohydrates and lipids to the fungus (Shi et al., 2023). The host plants suppress excessive AM colonization to prevent excess loss of the photosynthetic products. Similar to AON, the hosts exert a long-distance negative feedback regulation that controls AM symbiosis in already colonized roots, called autoregulation of mycorrhization (AOM). Here, we introduce a molecular mechanism involved in AOM mediated by the CLE signaling pathway (Figure 2B).
AM colonization induces CLE genes in roots of L. japonicus (LjCLE7 and other 5 LjCLEs), M. truncatula (MtCLE53), Solanum lycopersicum (tomato) (SlCLE11), and a monocotyledon Brachypodium distachyon (Bd1g49027 and Bd2g50170) (Handa et al., 2015; Müller et al., 2019; Karlo et al., 2020; Wulf et al., 2023). M. truncatula plants ectopically overexpressing MtCLE53 in the roots showed low colonization levels of the AM fungus, while a mutant lacking MtCLE53 was more colonized than the wild-type plant (Müller et al., 2019; Karlo et al., 2020). Overexpression of MtCLE33, which is not induced by AM colonization but phosphate-inducible, also reduced AM colonization, suggesting the CLE signaling negatively controls AM symbiosis responding to both AM colonization and the nutritional status (Müller et al., 2019). In S. lycopersicum plants, loss- and gain-of-function studies showed that SlCLE11 repressed AM symbiosis by responding to AM infection (Wulf et al., 2023). Akin to the function of the CLE peptides on AON, genes encoding HPATs are required for the function of the CLEs involving AOM. The mutant lacking MtRDN1 showed high levels of AM colonization, with overexpression of MtCLE53 in the roots showing no change in AM colonization in the rdn1, suggesting that MtCLE53 requires HPAT for AOM (Karlo et al., 2020). Corresponding with the effect of HPAT on the CLE function in M. truncatula, a defect of FASCIATED INFLORESCENCE (SlFIN), which encodes HPAT in S. lycopersicum, caused an increase in AM colonization levels and lost the low colonization phenotype caused by SlCLE11 overexpression (Wang et al., 2021a; Wulf et al., 2023). Although genetic studies have revealed that the HPATs are required for MtCLE53 and SlCLE11 function, whether mature forms of these peptides are indeed arabinosylated has not been identified yet. Furthermore, how CLE genes are induced during AM symbioses is still elusive, which necessitates further investigation.
Recognition of CLE peptides involved in AOM requires its cognate receptors, and accumulating evidence indicates that orthologs of AtCLV1 or AtCLV2 are signaling components of AOM. It has been shown that mutants defective in LjHAR1, MtSUNN, GmNARK, PsNARK, SlCLV2, FASCIATED AND BRANCHED (SlFAB), and FLORAL ORGAN NUMBER 1 (BdFON1) exhibit more colonization phenotypes (Morandi et al., 2000; Zakaria Solaiman et al., 2000; Meixner et al., 2005; Müller et al., 2019; Karlo et al., 2020; Wang et al., 2021a). The suppressive effect of MtCLE53 overexpression on AM symbiosis is MtSUNN dependent, suggesting MtSUNN is a cognate receptor for MtCLE53p (Karlo et al., 2020); however, a receptor required for MtCLE33 peptide recognition in AOM has not been characterized. Considering the observation that MtCLE53 and MtCLE33 are expressed in the root vascular tissues and MtSUNN is expressed in the vasculatures of both the root and shoot (Schnabel et al., 2012; Müller et al., 2019), the MtCLEs-MtSUNN module could act locally and/or systemically on AOM. Further investigations using grafting and split-root system would be valuable in distinguishing between these possibilities. Grafting experiments showed that shoot-expressed GmNARK controlled AM colonization systemically (Sakamoto and Nohara, 2009). Genetic studies revealed that SlCLE11 required neither SlFAB nor SlCLV2 (Wulf et al., 2023), suggesting the possibility that there are other CLE peptides involved in AOM and other receptor(s) that can perceive SlCLE11. Grafting experiments showed that SlFAB acts only in roots, while SlCLV2 acts in both roots and shoots (Wang et al., 2021a); whether regulation of AOM mediated by distinct CLE-receptor modules is exerted locally or systemically remains unclear.
Several nutritional conditions also trigger CLE-mediated regulation of AM symbiosis. As described above, MtCLE33 is not induced by AM colonization but phosphate-inducible, with MtCLE33 overexpression reducing AM colonization in a MtSUNN-dependent manner (Müller et al., 2019). Similarly, SlCLE10 is upregulated in responding to high nitrogen and phosphate, and SlCLE10 overexpression suppresses AM colonization (Wulf et al., 2023); however, its cognate receptor has not been identified yet. Although other CLE genes (two LjCLEs, four MtCLEs, three SlCLEs, and three BdCLEs) that respond to phosphate and some other nutrition status have been characterized in several plant species (Funayama-Noguchi et al., 2011; Handa et al., 2015; Müller et al., 2019; Karlo et al., 2020; Wulf et al., 2023), there is a lack of evidence as to whether these CLEs control AM symbioses.
Following CLE perception by their receptor modules, its downstream signaling regulates AM colonization in roots. Gene expression analyses revealed that biosynthesis genes of phytohormone strigolactones that promote AM colonization were downregulated both in MtCLE53 and MtCLE33 overexpressing roots in an MtSUNN-dependent manner, in consequence, strigolactones content was reduced in the roots (Müller et al., 2019). Exogenous treatment of a strigolactone analog GR24 to the MtCLEs overexpressing roots rendered AM colonization levels, indicating that strigolactone signaling is downstream of the MtCLE-MtSUNN module on AM symbiosis. Other factors downstream of the CLE-receptor module are transcription factor NUCLEAR FACTOR-Y (NF-Y) genes. Gene expression studies using split-root experiments showed that GmNF-YA1a and GmNF-YA1b were systemically downregulated in non-infected roots GmNARK-dependently, with knocking down of these two genes weakening hyper AM colonization phenotype of nark mutants (Schaarschmidt et al., 2013). Considering that the CLE peptides are perceived by their cognate receptors in the shoot during AM symbiosis, a shoot-derived descending signal is required for the downstream factors acting in the roots. Further investigation would identify yet unknown shoot-to-root factors. Previous studies have identified two shoot-to-root mobile factors involved in AM symbiosis regulation, ELONGATED HYPOCOTYL 5 (HY5) transcription factor that regulates strigolactone levels in roots, and micro RNAs such as miR399 that induces phosphate transporter genes in response to phosphate starvation (Wu et al., 2013; Müller and Harrison, 2019; Ge et al., 2022). It would be interesting to investigate whether these mobile factors are downstream of CLE signaling.
Plant-associated microbes also produce phytohormones or their mimics that manipulate phytohormone signaling networks in plants, thereby supporting their colonization of the hosts (Nakano et al., 2022). Le Marquer et al. (2019) identified CLE-like genes in the genome of five AM species. The prepropeptides of four Rhizophagus species were comprised of a predicted signal peptide at the N-terminus, a relatively shorter variable domain, and a CLE domain at the C-terminus as well as that of plants, while that of Gigaspora rosea possessed five CLE domains (Figure 1C). Expression of the fungal CLE-like genes of Rhizophagus irregularis and G. rosea (RiCLE1 and GrCLE1) was induced during symbiotic conditions. Exogenous application of synthetic RiCLE1 peptide on roots of M. truncatula, P. sativum and A. thaliana reduced primary root growth (Le Marquer et al., 2019). Consistent with a previous study in AtCLE peptides, this effect was partially dependent on CLV2 type receptor but not CLV1 (Fiers et al., 2005; Le Marquer et al., 2019), suggesting that RiCLE1 peptide is perceived by the host receptors. In addition, RiCLE1 peptide application to M. truncatula roots increased lateral root branching and AM colonization (Le Marquer et al., 2019). Since AM colonization often leads to an increase in lateral root formation and lateral roots are preferentially colonized by AM fungi (Sukumar et al., 2013), these observations raise the possibility that their increased abundance mediated by the fungal CLE peptides may lead to an enlarged interface for the plant-AM fungi interaction (Figure 2C). However, there is the possibility that increasing lateral root growth may merely be caused by the inhibition of primary root growth (Potters et al., 2009), further studies should investigate the biological implications of the fungal CLE peptides on AM symbiosis.
Phytoparasitic nematodes are obligate parasitic animals, which are recognized to cause severe economic losses in agriculture worldwide (Jones et al., 2013). Accumulating evidence has shown that the CLE signaling pathway is utilized for successful infection. Here, we introduce the roles of CLE signaling in plant-nematode interactions.
Root-knot nematodes (RKN; Meloidogyne spp.) parasite most vascular plants and are distributed worldwide. Infective juveniles move toward plant roots in the soil and invade through the root meristematic zone. After invasion, they inject an effector cocktail into the host procambial cells that suppresses host defense responses and manipulates host developmental pathways, thereby triggering the formation of their feeding sites called galls or root-knots on the host root by modulating the host auxin pathway (Favery et al., 2016; Olmo et al., 2020; Suzuki et al., 2022; Abril-Urias et al., 2023; Noureddine et al., 2023). Gene expression analyses in M. incognita-infected Arabidopsis showed that AtCLE1, AtCLE3, AtCLE4, and AtCLE7, which are orthologs of the symbiosis-induced CLEs described above, were upregulated in galls (Yamaguchi et al., 2017; Nakagami et al., 2023b). Single mutants lacking AtCLE1, AtCLE3, or AtCLE7 exhibited reduced gall numbers, and the higher-order mutant of AtCLE1 to AtCLE7 (cle1~7) showed an additive effect on the reduction of gall formation, whereas CLE overexpression led to increased gall formation, showing that these CLE genes positively regulate gall formation (Nakagami et al., 2023b). Grafting and split-root experiments revealed that the CLE-mediated regulation of gall formation required shoot-expressed AtCLV1, with synthetic AtCLE3 peptide directly binding to the extracellular domain of AtCLV1, indicating that AtCLE1~7-AtCLV1 module is the systemic pathway. Consistent with this, MtSUNN positively controls gall formation during M. javanica infection (Costa et al., 2020). These results raise the question of how shoot-expressed AtCLV1 regulates gall formation on roots after AtCLE peptides perception. A recent study showed that AtCLE2 and AtCLE3 were induced in roots when the roots were exposed to sucrose starvation (Okamoto et al., 2022). Root-expressed AtCLE1~AtCLE7 maintained SUCROSE-PROTON SYMTORTER 2 (SUC2) expression in the leaves, thereby balancing the root sucrose levels and growth. Indeed, exogenous sucrose application to the M. incognita-infected roots compensated the gall-reducing phenotype of the cle1~7 and clv1 mutants (Nakagami et al., 2023b), suggesting that maintenance of sucrose levels in roots mediated by leaf-expressed SUC2 is a possible downstream mechanism of AtCLE peptides/CLV1 on RKN infection (Figure 2D). Therefore, M. incognita may manipulate host sucrose homeostasis to acquire sucrose as a nutrient efficiently and/or allocate sucrose as an energy source for gall formation.
Our knowledge of AtCLE/CLV1 on plant-nematode interactions is still fragmented. For instance, there is a lack of explanation for how M. incognita induces these AtCLEs in the host roots, whether these AtCLEs require arabinosylation for their function, and if so, which HPAT arabinosylates these AtCLEs. In another aspect of CLE signaling in plant-RKN interactions, like cyst nematodes (see below), M. incognita possesses a gene harboring a ligand-like motif somewhat similar to the CLE motif, named 16D10, that is required for RKN parasitism, and the 16D10 peptide can interact with host transcription factors, not receptor-like proteins, making it unique among secreted peptides (Huang et al., 2006b, 2006a; Yang et al., 2013). In addition, RKNs harbor CLE-like genes constituted by only a signal peptide domain and a CLE domain, but their function is unknown (Mitchum and Liu, 2022) (Figure 1C). However, several studies provide evidence that RKNs possess functional peptide mimics to facilitate their successful infection (Kim et al., 2018; Zhang et al., 2020; Mishra et al., 2023; Yimer et al., 2023). Investigating the functions of CLE-like genes of RKN will underscore the ecological significance of CLE signaling in plant-RKN interactions. Moreover, the ways which RKN and rhizobia infect host plants share similarities even though RKNs and rhizobia are parasites and symbionts, respectively (Costa et al., 2021). Advancing our knowledge about plant-rhizobia interactions may help to reveal the mechanisms of plant-RKN interactions.
Parasitic cyst nematodes (CN; Heterodera and Globodera spp.) are obligate parasites to vascular plants including economically important crops (Jones et al., 2013). They penetrate their host roots and inject various effectors into the host procambial cells to establish their feeding site called syncytium in the root (Davis et al., 2008). Like AM fungi, CNs possess functional CLE-like genes in their genome as effectors. The prepropeptides of Hterodera species were similar to that of plants, while that of Globodera species possessed five CLE domains in addition to a signal peptide and a variable domain (Mitchum and Liu, 2022) (Figure 1C). Several studies in H. glycines, and G. rostochiensis showed that transcript of GrCLE1 and GrCLE4 was accumulated in dorsal gland secretory cells of the infective and parasitic juveniles, while immunolocalization experiments revealed that HgCLE peptides were localized in dorsal gland cells of parasitic juveniles (Wang et al., 2005, 2010; Bakhetia et al., 2007; Lu et al., 2009). Neither HsCLEs nor HgCLEs were expressed in pre-parasitic juveniles, implying that CN CLE-like genes act only in parasitic stages (Wang et al., 2010; Fosu-Nyarko et al., 2016). Heterologous expression of CN CLE-like genes (HsCLE1, HsCLE2, HgCLE1, HgCLE2, GrCLE1, and GrCLE4) that have high similarities of AtCLE1~AtCLE7 mimicked AtCLE function; exogenous application of the 12 amino acids form of them inhibited primary root growth of Arabidopsis as well as AtCLE peptides (Wang et al., 2005, 2010, 2011; Lu et al., 2009; Replogle et al., 2011, 2013; Chen et al., 2015). Moreover, in planta-expressed GrCLE1 was post-translationally processed, producing a 12 amino acids triarabinosylated GrCLE1 peptide which had high structural similarity to mature plant-CLE peptides, and this can directly bind to CLV2 of potato (Chen et al., 2015). The effect of CLE-like genes and peptides depends on the host receptor complexes such as AtCLV1 and AtCLV2 (Replogle et al., 2011, 2013; Chen et al., 2015; Guo et al., 2015, 2017), suggesting that the CN CLE-like peptides are recognized by receptors of the host plants. So, how do CN-CLE peptides injected into the host cells translocate to the apoplast to be perceived by the extracellular receptors? It has been shown that the variable domain of the CN CLE-like proteins has the function of a translocation signal that enables the CN CLE-like peptides to be delivered from the cytoplasm of syncytial cells to the apoplast through the host endoplasmic reticulum (Wang et al., 2010, 2021b).
The host receptor complexes also play a role in syncytium formation and CN fecundity. Mutants of AtCLV1, AtCLV2/AtCRN, AtRPK2, GmCLV2, and GmRPK2 exhibit defects of syncytium formation and low levels of CN fecundity (Replogle et al., 2011, 2013; Guo et al., 2015). In addition, exogenous treatment of HsCLEB, which is similar to TRACHEARY ELEMENT DIFFERENTIATION INHIBITORY FACTOR (AtTDIF), induces the expression of WUSCHEL-RELATED HOMEOBOX 4 (WOX4) transcription factor, which promotes procambial cell proliferation, through TDIF-RECEPTOR (TDR) in roots, thereby facilitating syncytium formation (Guo et al., 2017). Gene expression profiles and genetic studies revealed that HOMEOBOX GENE 8 (AtHB8), which controls vascular cell differentiation as well as WOX4, is a downstream component of CLE signaling during CN infection (Smetana et al., 2019; Liu and Mitchum, 2024). Also, direct binding of several CN CLE-like peptide/plant receptor pairs has been shown (Guo et al., 2011, 2015; Chen et al., 2015). Thus, it has been proposed that CNs co-opt the host developmental programs through CLAVATA pathways to support their infection (Figure 2E). However, whether CN CLE-like peptides injected into the host cells are processed during infection is not yet clear (Frei dit Frey and Favery, 2021).
Reniform nematodes (RN) also possess CLE-like genes. An RN, Rotylenchulus reniformis, is an obligate semi-endoparasite of more than 300 plant species. Three RrCLE genes were identified in the R. reniformis genome and consisted of a signal peptide at the N-terminus, a cryptic signal peptide within a variable domain, and a CLE domain at the C-terminus (Wubben et al., 2015) (Figure 1C). The RrCLE transcripts were detected in parasitic juveniles and localized in their dorsal gland. Another study in G. max infected by R. reniformis showed that several GmCLE genes were upregulated in the roots similarly to in the RKN-induced galls (Redding et al., 2018; Nakagami et al., 2023b). Therefore, RNs may use their CLE mimics and/or host-endogenous CLE peptides for their infection.
Some phytoparasitic insects induce gall formation on their host tissues where they acquire nutrients and are protected from enemies and environments (Favery et al., 2020). Phyloxera (Daktulosphaira vitifoliae), which parasitizes wild grapevine (Vitis riparia), induces the formation of flower-like gall on the host leaf. A transcriptomic analysis has found that the CLE signaling pathway in regulating cambial cell homeostasis is activated in D. vitifoliae-inducing galls (Schultz et al., 2019). For instance, VrCLE44, VrTDR, and VrWOX4 are up-regulated in insect-inducing galls, as well as in RKN- and CN-inducing galls/syncytia (Yamaguchi et al., 2017; Schultz et al., 2019). Gall-forming insects may therefore use the mechanism for the cambial cell maintenance of their hosts.
Plants employ RKs and RLPs as pattern recognition receptors (PRRs) to recognize pathogen-associated molecular patterns (PAMPs), damage-associated molecular patterns (DAMPs), and phytocytokines (Zhang et al., 2023b). PRRs induce pattern-triggered immunity (PTI) by recognizing PAMPs. One of the best-studied PRRs is FLAGELLIN SENSITIVE 2 (FLS2) which recognizes bacterial flagellin with the co-receptor BAK1 via a 22 amino acids epitope (flg22) (DeFalco and Zipfel, 2021). Emerging evidence suggests that CLE signaling is involved in plant immune responses and signaling by CLEs-CLV1/BAMs in development and flg22-FLS2 in immunity uses similar downstream factors (DeFalco et al., 2022) (Figure 3A). Here we discuss the current knowledge on the contribution of CLE signaling to plant immunity.
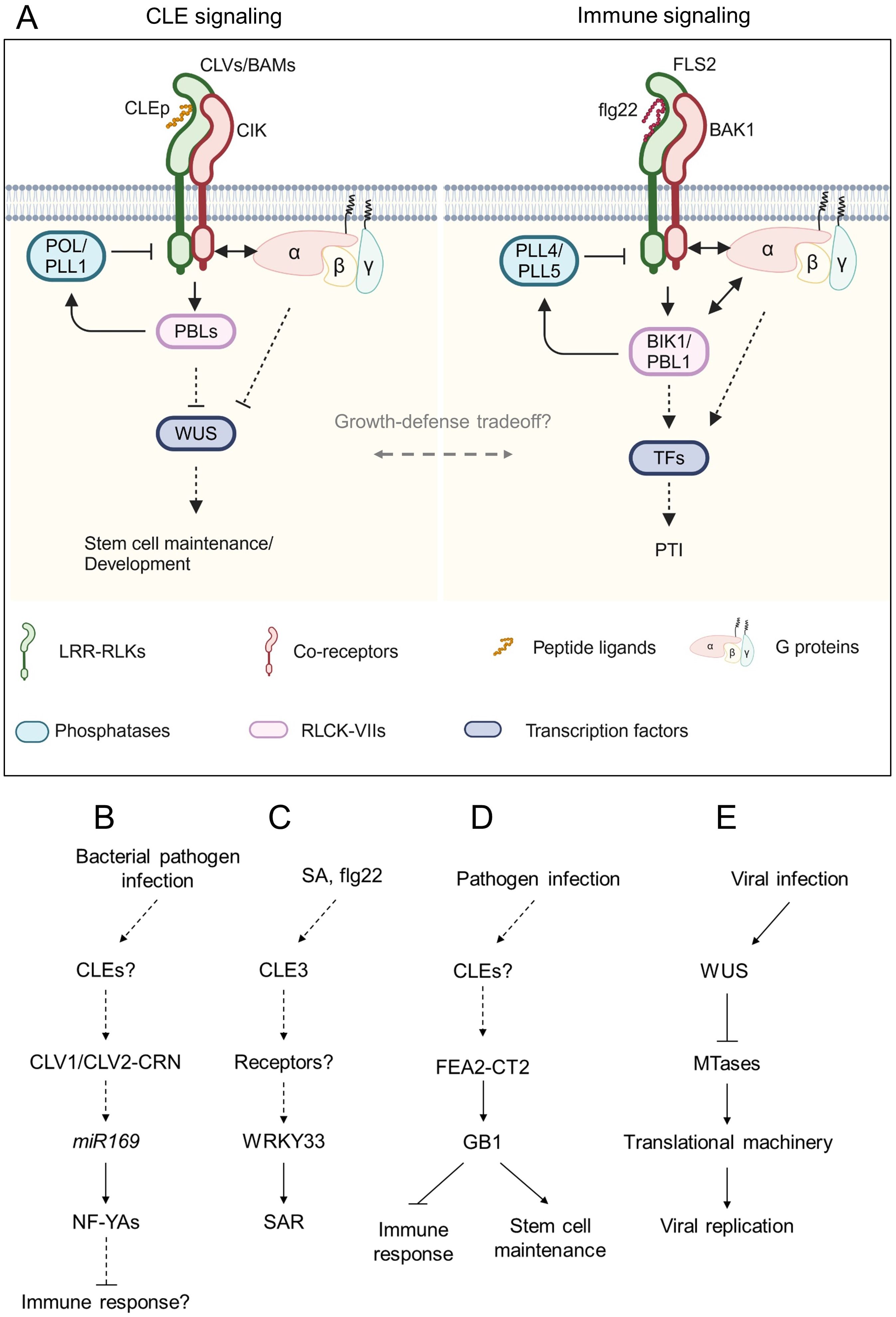
Figure 3. Schematic model of CLE signaling involved in immune response. (A) Representative model of the conserved signaling pathway between CLE signaling and immune signaling. Peptide ligands are recognized by leucine-rich repeat receptor-like kinases (LRR-RLKs) with their co-receptors. The signals are transduced via receptor-like cytoplasmic kinases (RLCKs) and G proteins and dampened by phosphatases to regulate downstream transcription factors. (B-E) Schematic flowcharts of CLE signaling involving plant-pathogen interactions. (B-D) Involvement of CLE signaling in plant-bacterial pathogen interactions in Arabidopsis (B, C) and maize (D). (E) Involvement of WUS in plant-virus interaction in Arabidopsis. Dashed lines indicate putative or indirect pathways. Figure adapted from images created with BioRender.com.
AtCLV1 and AtCLV2 participate in susceptibility to bacterial, fungal, and oomycete pathogens (Figure 3B). The clv1 and clv2 mutants exhibited increased resistance against the bacterial pathogen Ralstonia solanacearum and the oomycete pathogen Hyaloperonospora arabidopsidis compared to the wild-type plant (Hanemian et al., 2016). Conversely, clv1 was more susceptible to the bacterial pathogen Pseudomonas syringae pv. tomato DC3000 and the fungal pathogens Plectosphaerella cucumerina and Botrytis cinerea. Multiple NF-YA genes were upregulated in clv1 and clv2 mutants infected with R. solanacearum in an miR169-dependent manner. The overexpression of miR169 that suppresses the NF-YA genes facilitated susceptibility of clv1 and clv2 to R. solanacearum (Sorin et al., 2014; Hanemian et al., 2016). In contrast to the more susceptible phenotype of clv1 to B. cinerea, the mutant lacking AtACR4, which recognizes CLE40p coordinately with AtCLV1, was more resistant to this fungal pathogen (E-Zereen and Ingram, 2012; Czyzewicz et al., 2016). On the one hand, work by Lee et al. (2011) has proposed that AtCLV3 peptide is recognized by AtFLS2 leading to activation of immune response in the SAM. On the other hand, work by Segonzac et al. (2012) has provided experimental evidence that AtFLS2 does not recognize AtCLV3 peptide and that the immunity of the SAM to DC3000 is independent of AtCLV3 peptide perception. Another study focused on gene expression analyses of AtCLE genes has revealed that AtCLE3 expression is induced in the roots by exogenous treatments of flg22, Pep2, which is a 23 amino acids peptide known as a DAMP, and the defense phytohormone salicylic acid (SA) (Ma et al., 2022). Root-expressing AtCLE3 is required for upregulation of shoot-expressed WRKY33 that contributes to systemic acquired resistance (Wang et al., 2018; Ma et al., 2022), suggesting that the AtCLE3 peptide may act as a systemic signal to transduce SA signaling from roots to shoots (Figure 3C). However, it has not been tested yet whether AtCLE3 peptide contributes to immune response, particularly affecting pathogen growth in planta. Therefore, careful investigation of whether and how the CLE signaling is involved in plant immunity is necessary.
Recent studies have also provided evidence that downstream components of the CLE-receptor modules are involved in plant immunity. Genetic studies have shown that Arabidopsis G proteins AtAGB1 (Gβ), AtAGG1, and AtAGG2 (Gγ) are required for FLS2-mediated immune response (Liu et al., 2013). Furthermore, AtAGB1 and EXTRA-LARGE GTP BINDING PROTEIN 2 (AtXLG2), a noncanonical Gα, directly interact with FLS2 and BOTRYTIS-INDUCED KINASE 1 (BIK1), conferring stability on the receptor complex (Liang et al., 2016). AtAGB1 also interacts with AtRPK2, which maintains meristem activity in the SAM (Ishida et al., 2014), suggesting that G proteins control both meristem development and immune response. A recent study found that the Zea mays Gβ subunit 1 (ZmGB1) mutants showed seedling-lethal phenotype due to autoimmunity (Wu et al., 2020b). A viable ZmGB1 mutant fea*183 was identified from ethyl methanesulfonate-mutagenesis screen and showed striking inflorescence defects, reminiscent of mutants lacking ZmCT2 (Gα) (Bommert et al., 2013; Wu et al., 2020b). ZmFEA2, an ortholog of AtCLV2, and ZmCT2 were epistatic to ZmGB1, suggesting that the CLE-CLV-G protein signaling circuit may balance the tradeoff between growth and defense (Figure 3D). The SAM-specific transcription factor WUS protects the SAM from infection by cucumber mosaic virus (CMV) (Wu et al., 2020a; Lopes et al., 2021). WUS inhibits transcript levels of S-adenosyl-Lmethionine-dependent methyltransferases (MTases), which are involved in rRNA processing and ribosome stability, by responding to CMV infection, thereby sabotaging CMV replication and its invasion ability to the SAM (Wu et al., 2020a) (Figure 3E).
Apart from the CLE signaling pathway, plant endogenous signaling peptides play roles in plant immunity, abiotic stress response, and growth regulation (Fitrianti et al., 2022; Liu et al., 2022; Rzemieniewski and Stegmann, 2022; Rzemieniewski et al., 2022; Taleski et al., 2024). In addition, a recent study has revealed that a RALF-FERONIA signaling module that modulates the formation of FLS2-BAK1 receptor complex affects the rhizosphere microbiome (Song et al., 2021). This suggests that peptide-receptor modules are central regulators of diverse aspects of plant physiology, and their roles in immunity, development, and beyond are mechanistically coupled. Yet, how the CLE signaling harmonizes multiple aspects of plant immunity, development, and physiology is still elusive.
Peptide ligand and its receptor pairs play important roles in plant development and plant-microbe interactions. The CLE gene family is one of the largest among those that encode peptide ligands, and they are conserved across green algae to higher plants (Olsson et al., 2019). Plants have evolved the CLE signaling to develop more complex multicellular bodies and be adapted to environmental nutritional status that is constantly changing. Both developing de novo organs induced by plant-associated microbes and coping with the nutritional status of their host are pivotal for microbial colonization. Thus, plant-associated microbes may utilize the host-CLE signaling to achieve those.
Secreted peptides are perceived by distinct receptors in the apoplast, in consequence, their signals are transduced into the cell. In this process, post-translational modifications to form mature peptides are required. However, the mature form of most CLE peptides involved in plant-microbe interactions remains uncharacterized. In addition, most of the knowledge about peptide-receptor pairs has been brought by genetic studies, it is still largely unknown whether distinct peptides bind their cognate receptors. Interestingly, it has been shown that AI-based prediction to identify peptide ligand and receptor pairs is a powerful tool for structure-function analysis of peptide-receptor pairs (Snoeck et al., 2024). Applying this in silico method to CLE peptides and their postulated receptors holds the potential to identify bona fide peptide-receptor pairs more and distinguish functions of different peptides.
While there are increasing studies on plant-microbe interactions, such efforts still lag behind those on plant development and physiology due to more complex phenomena and technical obstacles. Recently, several studies have tackled unraveling molecular mechanisms underlying plant-microbe interactions by using the co-transcriptomics of plants inoculated with microbes and the microbes in planta (Roux et al., 2014; Hacquard et al., 2016; Nobori et al., 2018, 2022; Sauviac et al., 2022; Siddique et al., 2022; Zhang et al., 2023a), spacial transcriptomics (Frank et al., 2023; Nobori et al., 2023; Schnabel et al., 2023; Tang et al., 2023; Verbon et al., 2023; Zhu et al., 2023), and the technique that is combined with both (Serrano et al., 2024a, 2024b), which will identify yet unknown upstream and downstream components of the CLE-receptor modules, and accelerate our understanding of ecological implications of the plant-microbe interactions.
SN: Conceptualization, Funding acquisition, Visualization, Writing – original draft, Writing – review & editing. TK: Writing – original draft, Writing – review & editing. KT: Supervision, Writing – review & editing. SS: Conceptualization, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. Research on this topic is supported by Human Resources Department of Huazhong Agricultural University (Fellowship ID: TH9009350) to SN and KAKENHI (Grant Numbers: 20H00422, 20KK0135, 21K19273, and 23H04748) to SS.
SN thanks members of the Kenichi Tsuda and Xiaowei Han laboratories (Huazhong Agricultural University) and members of the Shinichiro Sawa laboratory (Kumamoto University) for useful discussions.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abril-Urias, P., Ruiz-Ferrer, V., Cabrera, J., Olmo, R., Silva, A. C., Díaz-Manzano, F. E., et al. (2023). Divergent regulation of auxin responsive genes in root-knot and cyst nematodes feeding sites formed in Arabidopsis. Front. Plant Sci 14. doi: 10.3389/fpls.2023.1024815
Alves-Carvalho, S., Aubert, G., Carrère, S., Cruaud, C., Brochot, A., Jacquin, F., et al. (2015). Full-length de novo assembly of RNA-seq data in pea ( P isum sativum L.) provides a gene expression atlas and gives insights into root nodulation in this species. Plant J 84, 1–19. doi: 10.1111/tpj.12967
Araya, T., Miyamoto, M., Wibowo, J., Suzuki, A., Kojima, S., Tsuchiya, Y. N., et al. (2014). CLE-CLAVATA1 peptide-receptor signaling module regulates the expansion of plant root systems in a nitrogen-dependent manner. Proc. Natl. Acad. Sci 111, 2029–2034. doi: 10.1073/pnas.1319953111
Bakhetia, M., Urwin, P. E., Atkinson, H. J. (2007). QPCR analysis and RNAi define pharyngeal gland cell-expressed genes of Heterodera glycines required for initial interactions with the host. Mol. Plant-Microbe Interact 20, 306–312. doi: 10.1094/MPMI-20-3-0306
Bashyal, S., Gautam, C. K., Müller, L. M. (2023). CLAVATA signaling in plant-environment interactions. Plant Physiol 194, 1336–1357. doi: 10.1093/plphys/kiad591
Betsuyaku, S., Sawa, S., Yamada, M. (2011). The function of the CLE peptides in plant development and plant-microbe interactions. Arabidopsis Book 9, e0149. doi: 10.1199/tab.0149
Bleckmann, A., Weidtkamp-Peters, S., Seidel, C. A. M., Simon, R. (2009). Stem cell signaling in arabidopsis requires CRN to localize CLV2 to the plasma membrane. Plant Physiol 152, 166–176. doi: 10.1104/pp.109.149930
Bommert, P., Je, B. I., Goldshmidt, A., Jackson, D. (2013). The maize Gα gene COMPACT PLANT2 functions in CLAVATA signalling to control shoot meristem size. Nature 502, 555–558. doi: 10.1038/nature12583
Chaulagain, D., Frugoli, J. (2021). The regulation of nodule number in legumes is a balance of three signal transduction pathways. Int. J. Mol. Sci 22, 1117. doi: 10.3390/ijms22031117
Chen, S., Lang, P., Chronis, D., Zhang, S., De Jong, W. S., Mitchum, M. G., et al. (2015). In planta processing and glycosylation of a nematode CLAVATA3/ENDOSPERM SURROUNDING REGION-Like effector and its interaction with a host CLAVATA2-Like receptor to promote parasitism. Plant Physiol 167, 262–272. doi: 10.1104/pp.114.251637
Costa, S. R., Chin, S., Mathesius, U. (2020). Infection of medicago truncatula by the root-knot nematode meloidogyne javanica does not require early nodulation genes. Front. Plant Sci 11. doi: 10.3389/fpls.2020.01050
Costa, S. R., Ng, J. L. P., Mathesius, U. (2021). Interaction of symbiotic rhizobia and parasitic root-knot nematodes in legume roots: From molecular regulation to field application. MPMI 34, 470–490. doi: 10.1094/MPMI-12-20-0350-FI
Crook, A. D., Schnabel, E. L., Frugoli, J. A. (2016). The systemic nodule number regulation kinase SUNN in Medicago truncatula interacts with MtCLV2 and MtCRN. Plant J 88, 108–119. doi: 10.1111/tpj.13234
Czyzewicz, N., Nikonorova, N., Meyer, M. R., Sandal, P., Shah, S., Vu, L. D., et al. (2016). The growing story of (ARABIDOPSIS) CRINKLY 4. J. Exp. Bot 67, 4835–4847. doi: 10.1093/jxb/erw192
Davis, E. L., Hussey, R. S., Mitchum, M. G., Baum, T. J. (2008). Parasitism proteins in nematode–plant interactions. Curr. Opin. Plant Biol 11, 360–366. doi: 10.1016/j.pbi.2008.04.003
DeFalco, T. A., Anne, P., James, S. R., Willoughby, A. C., Schwanke, F., Johanndrees, O., et al. (2022). A conserved module regulates receptor kinase signalling in immunity and development. Nat. Plants 8, 356–365. doi: 10.1038/s41477-022-01134-w
DeFalco, T. A., Zipfel, C. (2021). Molecular mechanisms of early plant pattern-triggered immune signaling. Mol. Cell 81, 3449–3467. doi: 10.1016/j.molcel.2021.07.029
E-Zereen, J., Ingram, G. (2012). A possible involvement of ACR4, a receptor like kinase, in plant defence mechanism. Bangladesh Pharm. J 15, 127–130. doi: 10.3329/bpj.v15i2.12576
Favery, B., Dubreuil, G., Chen, M.-S., Giron, D., Abad, P. (2020). Gall-inducing parasites: convergent and conserved strategies of plant manipulation by insects and nematodes. Annu. Rev. Phytopathol 58, 1–22. doi: 10.1146/annurev-phyto-010820-012722
Favery, B., Quentin, M., Jaubert-Possamai, S., Abad, P. (2016). Gall-forming root-knot nematodes hijack key plant cellular functions to induce multinucleate and hypertrophied feeding cells. J. Insect Physiol 84, 60–69. doi: 10.1016/j.jinsphys.2015.07.013
Ferguson, B. J., Li, D., Hastwell, A. H., Reid, D. E., Li, Y., Jackson, S. A., et al. (2014). The soybean (Glycine max) nodulation-suppressive CLE peptide, GmRIC1, functions interspecifically in common white bean (Phaseolus vulgaris), but not in a supernodulating line mutated in the receptor PvNARK. Plant Biotechnol. J 12, 1085–1097. doi: 10.1111/pbi.12216
Fiers, M., Golemiec, E., Xu, J., van der Geest, L., Heidstra, R., Stiekema, W., et al. (2005). The 14-amino acid CLV3, CLE19, and CLE40 peptides trigger consumption of the root meristem in Arabidopsis through a CLAVATA2-dependent pathway. Plant Cell 17, 2542–2553. doi: 10.1105/tpc.105.034009
Fitrianti, A. N., Mai, T. L., Phuong, L. T., Monden, H., Shiiba, N., Matsui, H., et al. (2022). CEP peptide induces susceptibility of Arabidopsis thaliana to non-adapted pathogens. J. Gen. Plant Pathol 88, 287–292. doi: 10.1007/s10327-022-01077-2
Fletcher, J. C. (2020). Recent advances in arabidopsis CLE peptide signaling. Trends Plant Sci 25, 1005–1016. doi: 10.1016/j.tplants.2020.04.014
Fosu-Nyarko, J., Nicol, P., Naz, F., Gill, R., Jones, M. G. K. (2016). Analysis of the transcriptome of the infective stage of the beet cyst nematode, H. schachtii. PloS One 11, e0147511. doi: 10.1371/journal.pone.0147511
Frank, M., Fechete, L. I., Tedeschi, F., Nadzieja, M., Nørgaard, M. M. M., Montiel, J., et al. (2023). Single-cell analysis identifies genes facilitating rhizobium infection in lotus japonicus. Nat. Commun 14, 7171. doi: 10.1038/s41467-023-42911-1
Frei dit Frey, N., Favery, B. (2021). Plant-parasitic nematode secreted peptides hijack a plant secretory pathway. New Phytol 229, 11–13. doi: 10.1111/nph.16842
Fu, M., Yao, X., Li, X., Liu, J., Bai, M., Fang, Z., et al. (2024). GmNLP1 and GmNLP4 activate nitrate-induced CLE peptides NIC1a/b to mediate nitrate-regulated root nodulation. Plant J 119, 783–795. doi: 10.1111/tpj.16795
Funayama-Noguchi, S., Noguchi, K., Yoshida, C., Kawaguchi, M. (2011). Two CLE genes are induced by phosphate in roots of Lotus japonicus. J. Plant Res 124, 155–163. doi: 10.1007/s10265-010-0342-5
Gautrat, P., Laffont, C., Frugier, F. (2020). Compact Root Architecture 2 Promotes Root Competence for Nodulation through the miR2111 Systemic Effector. Curr. Biol 30, 1339–1345.e3. doi: 10.1016/j.cub.2020.01.084
Ge, S., He, L., Jin, L., Xia, X., Li, L., Ahammed, G. J., et al. (2022). Light-dependent activation of HY5 promotes mycorrhizal symbiosis in tomato by systemically regulating strigolactone biosynthesis. New Phytol 233, 1900–1914. doi: 10.1111/nph.17883
Guo, X., Chronis, D., de la Torre, C. M., Smeda, J., Wang, X., Mitchum, M. G. (2015). Enhanced resistance to soybean cyst nematode Heterodera glycines in transgenic soybean by silencing putative CLE receptors. Plant Biotechnol. J 13, 801–810. doi: 10.1111/pbi.12313
Guo, Y., Ni, J., Denver, R., Wang, X., Clark, S. E. (2011). Mechanisms of molecular mimicry of plant CLE peptide ligands by the parasitic nematode Globodera rostochiensis. Plant Physiol 157, 476–484. doi: 10.1104/pp.111.180554
Guo, X., Wang, J., Gardner, M., Fukuda, H., Kondo, Y., Etchells, J. P., et al. (2017). Identification of cyst nematode B-type CLE peptides and modulation of the vascular stem cell pathway for feeding cell formation. PloS Pathog 13, e1006142. doi: 10.1371/journal.ppat.1006142
Hacquard, S., Kracher, B., Hiruma, K., Münch, P. C., Garrido-Oter, R., Thon, M. R., et al. (2016). Survival trade-offs in plant roots during colonization by closely related beneficial and pathogenic fungi. Nat. Commun 7, 11362. doi: 10.1038/ncomms11362
Handa, Y., Nishide, H., Takeda, N., Suzuki, Y., Kawaguchi, M., Saito, K. (2015). RNA-seq transcriptional profiling of an arbuscular mycorrhiza provides insights into regulated and coordinated gene expression in Lotus japonicus and Rhizophagus irregularis. Plant Cell Physiol 56, 1490–1511. doi: 10.1093/pcp/pcv071
Hanemian, M., Barlet, X., Sorin, C., Yadeta, K. A., Keller, H., Favery, B., et al. (2016). Arabidopsis CLAVATA1 and CLAVATA2 receptors contribute to Ralstonia solanacearum pathogenicity through a miR169-dependent pathway. New Phytol 211, 502–515. doi: 10.1111/nph.13913
Hastwell, A. H., Corcilius, L., Williams, J. T., Gresshoff, P. M., Payne, R. J., Ferguson, B. J. (2019). Triarabinosylation is required for nodulation-suppressive CLE peptides to systemically inhibit nodulation in Pisum sativum: Tri-arabinosylated CLE peptides in nodulation. Plant Cell Environ 42, 188–197. doi: 10.1111/pce.13325
Hayashi-Tsugane, M., Kawaguchi, M. (2022). Lotus japonicus HAR1 regulates root morphology locally and systemically under a moderate nitrate condition in the absence of rhizobia. Planta 255, 95. doi: 10.1007/s00425-022-03873-8
Hu, C., Zhu, Y., Cui, Y., Cheng, K., Liang, W., Wei, Z., et al. (2018). A group of receptor kinases are essential for CLAVATA signalling to maintain stem cell homeostasis. Nat. Plants 4, 205–211. doi: 10.1038/s41477-018-0123-z
Huang, G., Allen, R., Davis, E. L., Baum, T. J., Hussey, R. S. (2006a). Engineering broad root-knot resistance in transgenic plants by RNAi silencing of a conserved and essential root-knot nematode parasitism gene. Proc. Natl. Acad. Sci 103, 14302–14306. doi: 10.1073/pnas.0604698103
Huang, G., Dong, R., Allen, R., Davis, E. L., Baum, T. J., Hussey, R. S. (2006b). A root-knot nematode secretory peptide functions as a ligand for a plant transcription factor. MPMI 19, 463–470. doi: 10.1094/MPMI-19-0463
Huault, E., Laffont, C., Wen, J., Mysore, K. S., Ratet, P., Duc, G., et al. (2014). Local and systemic regulation of plant root system architecture and symbiotic nodulation by a receptor-like kinase. PloS Genet 10, e1004891. doi: 10.1371/journal.pgen.1004891
Imin, N., Patel, N., Corcilius, L., Payne, R. J., Djordjevic, M. A. (2018). CLE peptide tri-arabinosylation and peptide domain sequence composition are essential for SUNN-Dependent autoregulation of nodulation in Medicago Truncatula. New Phytol 218, 73–80. doi: 10.1111/nph.15019
Ishida, T., Tabata, R., Yamada, M., Aida, M., Mitsumasu, K., Fujiwara, M., et al. (2014). Heterotrimeric G proteins control stem cell proliferation through CLAVATA signaling in Arabidopsis. EMBO Rep 15, 1202–1209. doi: 10.15252/embr.201438660
Isidra-Arellano, M. C., Pozas-Rodríguez, E. A., Del Rocío Reyero-Saavedra, M., Arroyo-Canales, J., Ferrer-Orgaz, S., Del Socorro Sánchez-Correa, M., et al. (2020). Inhibition of legume nodulation by Pi deficiency is dependent on the autoregulation of nodulation (AON) pathway. Plant J 103, 1125–1139. doi: 10.1111/tpj.14789
Isidra-Arellano, M., Reyero-Saavedra, M., Sánchez-Correa, M., Pingault, L., Sen, S., Joshi, T., et al. (2018). Phosphate deficiency negatively affects early steps of the symbiosis between common bean and rhizobia. Genes 9, 498. doi: 10.3390/genes9100498
Jones, J. T., Haegeman, A., Danchin, E. G. J., Gaur, H. S., Helder, J., Jones, M. G. K., et al. (2013). Top 10 plant-parasitic nematodes in molecular plant pathology: Top 10 plant-parasitic nematodes. Mol. Plant Pathol 14, 946–961. doi: 10.1111/mpp.12057
Jones, D. S., John, A., VanDerMolen, K. R., Nimchuk, Z. L. (2021). CLAVATA signaling ensures reproductive development in plants across thermal environments. Curr. Biol 31, 220–227.e5. doi: 10.1016/j.cub.2020.10.008
Kang, J., Wang, X., Ishida, T., Grienenberger, E., Zheng, Q., Wang, J., et al. (2022). A group of CLE peptides regulates de novo shoot regeneration in Arabidopsis thaliana. New Phytol 235, 2300–2312. doi: 10.1111/nph.18291
Karlo, M., Boschiero, C., Landerslev, K. G., Blanco, G. S., Wen, J., Mysore, K. S., et al. (2020). The CLE53–SUNN genetic pathway negatively regulates arbuscular mycorrhiza root colonization in medicago truncatula. J. Exp. Bot 71, 4972–4984. doi: 10.1093/jxb/eraa193
Kassaw, T., Nowak, S., Schnabel, E., Frugoli, J. (2017). ROOT DETERMINED NODULATION1 Is Required for M. truncatula CLE12, But Not CLE13, Peptide Signaling through the SUNN Receptor Kinase. Plant Physiol 174, 2445–2456. doi: 10.1104/pp.17.00278
Kim, J., Yang, R., Chang, C., Park, Y., Tucker, M. L. (2018). The root-knot nematode meloidogyne incognita produces a functional mimic of the arabidopsis INFLORESCENCE DEFICIENT IN ABSCISSION signaling peptide. J. Exp. Bot 69, 3009–3021. doi: 10.1093/jxb/ery135
Kondo, T., Sawa, S., Kinoshita, A., Mizuno, S., Kakimoto, T., Fukuda, H., et al. (2006). A plant peptide encoded by CLV3 identified by in situ MALDI-TOF MS analysis. Science 313, 845–848. doi: 10.1126/science.1128439
Konishi, M., Yanagisawa, S. (2013). Arabidopsis NIN-like transcription factors have a central role in nitrate signalling. Nat. Commun 4, 1617. doi: 10.1038/ncomms2621
Krusell, L., Madsen, L. H., Sato, S., Aubert, G., Genua, A., Szczyglowski, K., et al. (2002). Shoot control of root development and nodulation is mediated by a receptor-like kinase. Nature 420, 422–426. doi: 10.1038/nature01207
Krusell, L., Sato, N., Fukuhara, I., Koch, B. E. V., Grossmann, C., Okamoto, S., et al. (2011). The Clavata2 genes of pea and Lotus japonicus affect autoregulation of nodulation. Plant J 65, 861–871. doi: 10.1111/j.1365-313X.2010.04474.x
Laffont, C., Ivanovici, A., Gautrat, P., Brault, M., Djordjevic, M. A., Frugier, F. (2020). The NIN transcription factor coordinates CEP and CLE signaling peptides that regulate nodulation antagonistically. Nat. Commun 11, 3167. doi: 10.1038/s41467-020-16968-1
Lebedeva, M., Azarakhsh, M., Yashenkova, Y., Lutova, L. (2020). Nitrate-induced CLE peptide systemically inhibits nodulation in medicago truncatula. Plants 9, 1456. doi: 10.3390/plants9111456
Lebedeva, M. A., Dobychkina, D. A., Yashenkova, Y., Romanyuk, D. A., Lutova, L. A. (2023). Local and systemic targets of the MtCLE35-SUNN pathway in the roots of Medicago truncatula. J. Plant Physiol 281, 153922. doi: 10.1016/j.jplph.2023.153922
Lebedeva, M., Dvornikova, K., Lutova, L. (2022). Nitrate-induced mtCLE34 gene lacks the ability to reduce symbiotic nodule number and carries nonsense mutation in a few accessions of medicago truncatula. Agronomy 12, 842. doi: 10.3390/agronomy12040842
Lee, H., Chah, O.-K., Sheen, J. (2011). Stem-cell-triggered immunity through CLV3p–FLS2 signalling. Nature 473, 376–379. doi: 10.1038/nature09958
Le Marquer, M., Bécard, G., Frei dit Frey, N. (2019). Arbuscular mycorrhizal fungi possess a CLAVATA3/embryo surrounding region-related gene that positively regulates symbiosis. New Phytol 222, 1030–1042. doi: 10.1111/nph.15643
Li, Y., Pei, Y., Shen, Y., Zhang, R., Kang, M., Ma, Y., et al. (2022). Progress in the self-regulation system in legume nodule development-AON (Autoregulation of nodulation). Int. J. Mol. Sci 23, 6676. doi: 10.3390/ijms23126676
Liang, X., Ding, P., Lian, K., Wang, J., Ma, M., Li, L., et al. (2016). Arabidopsis heterotrimeric G proteins regulate immunity by directly coupling to the FLS2 receptor. eLife 5, e13568. doi: 10.7554/eLife.13568
Lim, C. W., Lee, Y. W., Hwang, C. H. (2011). Soybean nodule-enhanced CLE peptides in roots act as signals in GmNARK-Mediated nodulation suppression. Plant Cell Physiol 52, 1613–1627. doi: 10.1093/pcp/pcr091
Liu, X., Mitchum, M. (2024). A major role of class III HD-ZIPs in promoting sugar beet cyst nematode syncytium formation in Arabidopsis. bioRxiv. doi: 10.1101/2024.02.15.580549
Liu, J., Ding, P., Sun, T., Nitta, Y., Dong, O., Huang, X., et al. (2013). Heterotrimeric G proteins serve as a converging point in plant defense signaling activated by multiple receptor-like kinases. Plant Physiol 161, 2146–2158. doi: 10.1104/pp.112.212431
Liu, L., Song, W., Huang, S., Jiang, K., Moriwaki, Y., Wang, Y., et al. (2022). Extracellular ph sensing by plant cell-surface peptide-receptor complexes. Cell 185, 3341–3355.e13. doi: 10.1016/j.cell.2022.07.012
Lopes, F. L., Galvan-Ampudia, C., Landrein, B. (2021). WUSCHEL in the shoot apical meristem: old player, new tricks. J. Exp. Bot 72, 1527–1535. doi: 10.1093/jxb/eraa572
Lu, S.-W., Chen, S., Wang, J., Yu, H., Chronis, D., Mitchum, M. G., et al. (2009). Structural and functional diversity of CLAVATA3/ESR (CLE)-like genes from the potato cyst nematode Globodera rostochiensis. Mol. Plant-Microbe Interact 22, 1128–1142. doi: 10.1094/MPMI-22-9-1128
Luo, Z., Lin, J., Zhu, Y., Fu, M., Li, X., Xie, F. (2021). NLP1 reciprocally regulates nitrate inhibition of nodulation through SUNN-CRA2 signaling in medicago truncatula. Plant Commun 2, 100183. doi: 10.1016/j.xplc.2021.100183
Ma, D., Endo, S., Betsuyaku, E., Fujiwara, T., Betsuyaku, S., Fukuda, H. (2022). Root-specific CLE3 expression is required for WRKY33 activation in arabidopsis shoots. Plant Mol. Biol. 108, 225–239. doi: 10.1007/s11103-021-01234-9
Marchive, C., Roudier, F., Castaings, L., Bréhaut, V., Blondet, E., Colot, V., et al. (2013). Nuclear retention of the transcription factor NLP7 orchestrates the early response to nitrate in plants. Nat. Commun 4, 1713. doi: 10.1038/ncomms2650
Meixner, C., Ludwig-Müller, J., Miersch, O., Gresshoff, P., Staehelin, C., Vierheilig, H. (2005). Lack of mycorrhizal autoregulation and phytohormonal changes in the supernodulating soybean mutant nts1007. Planta 222, 709–715. doi: 10.1007/s00425-005-0003-4
Mens, C., Hastwell, A. H., Su, H., Gresshoff, P. M., Mathesius, U., Ferguson, B. J. (2021). Characterisation of Medicago truncatula CLE34 and CLE35 in nitrate and rhizobia regulation of nodulation. New Phytol 229, 2525–2534. doi: 10.1111/nph.17010
Mishra, S., Hu, W., DiGennaro, P. (2023). Root-knot-nematode-encoded CEPs increase nitrogen assimilation. Life 13, 2020. doi: 10.3390/life13102020
Mitchum, M. G., Liu, X. (2022). Peptide effectors in phytonematode parasitism and beyond. Annu. Rev. Phytopathol 60, 97–119. doi: 10.1146/annurev-phyto-021621-115932
Miyazawa, H., Oka-Kira, E., Sato, N., Takahashi, H., Wu, G.-J., Sato, S., et al. (2010). The receptor-like kinase KLAVIER mediates systemic regulation of nodulation and non-symbiotic shoot development in lotus japonicus. Development 137, 4317–4325. doi: 10.1242/dev.058891
Morandi, D., Sagan, M., Prado-Vivant, E., Duc, G. (2000). Influence of genes determining supernodulation on root colonization by the mycorrhizal fungus Glomus mosseae in Pisum sativum and Medicago truncatula mutants. Mycorrhiza 10, 37–42. doi: 10.1007/s005720050285
Moreau, C., Gautrat, P., Frugier, F. (2021). Nitrate-induced CLE35 signaling peptides inhibit nodulation through the SUNN receptor and miR2111 repression. Plant Physiol 185, 1216–1228. doi: 10.1093/plphys/kiaa094
Mortier, V., Den Herder, G., Whitford, R., Van de Velde, W., Rombauts, S., D’haeseleer, K., et al. (2010). CLE peptides control Medicago truncatula nodulation locally and systemically. Plant Physiol 153, 222–237. doi: 10.1104/pp.110.153718
Müller, L. M., Flokova, K., Schnabel, E., Sun, X., Fei, Z., Frugoli, J., et al. (2019). A CLE–SUNN module regulates strigolactone content and fungal colonization in arbuscular mycorrhiza. Nat. Plants 5, 933–939. doi: 10.1038/s41477-019-0501-1
Müller, L. M., Harrison, M. J. (2019). Phytohormones, miRNAs, and peptide signals integrate plant phosphorus status with arbuscular mycorrhizal symbiosis. Curr. Opin. Plant Biol 50, 132–139. doi: 10.1016/j.pbi.2019.05.004
Nakagami, S., Aoyama, T., Sato, Y., Kajiwara, T., Ishida, T., Sawa, S. (2023a). CLE3 and its homologs share overlapping functions in the modulation of lateral root formation through CLV1 and BAM1 in Arabidopsis thaliana. Plant J 113, 1176–1191. doi: 10.1111/tpj.16103
Nakagami, S., Notaguchi, M., Kondo, T., Okamoto, S., Ida, T., Sato, Y., et al. (2023b). Root-knot nematode modulates plant CLE3-CLV1 signaling as a long-distance signal for successful infection. Sci. Adv. 9, eadf4803. doi: 10.1126/sciadv.adf4803
Nakano, M., Omae, N., Tsuda, K. (2022). Inter-organismal phytohormone networks in plant-microbe interactions. Curr. Opin. Plant Biol 68, 102258. doi: 10.1016/j.pbi.2022.102258
Nishida, H., Handa, Y., Tanaka, S., Suzaki, T., Kawaguchi, M. (2016). Expression of the CLE-RS3 gene suppresses root nodulation in Lotus japonicus. J. Plant Res 129, 909–919. doi: 10.1007/s10265-016-0842-z
Nishida, H., Nosaki, S., Suzuki, T., Ito, M., Miyakawa, T., Nomoto, M., et al. (2021). Different DNA-binding specificities of NLP and NIN transcription factors underlie nitrate-induced control of root nodulation. Plant Cell 33, 2340–2359. doi: 10.1093/plcell/koab103
Nishimura, R., Hayashi, M., Wu, G.-J., Kouchi, H., Imaizumi-Anraku, H., Murakami, Y., et al. (2002). HAR1 mediates systemic regulation of symbiotic organ development. Nature 420, 426–429. doi: 10.1038/nature01231
Nobori, T., Cao, Y., Entila, F., Dahms, E., Tsuda, Y., Garrido-Oter, R., et al. (2022). Dissecting the cotranscriptome landscape of plants and their microbiota. EMBO Rep 23, e55380. doi: 10.15252/embr.202255380
Nobori, T., Monell, A., Lee, T. A., Zhou, J., Nery, J. R., Ecker, J. R. (2023). Time-resolved single-cell and spatial gene regulatory atlas of plants under pathogen attack. bioRxiv. doi: 10.1101/2023.04.10.536170
Nobori, T., Velásquez, A. C., Wu, J., Kvitko, B. H., Kremer, J. M., Wang, Y., et al. (2018). Transcriptome landscape of a bacterial pathogen under plant immunity. Proc. Natl. Acad. Sci. U.S.A 115, E3055–E3064. doi: 10.1073/pnas.1800529115
Noureddine, Y., Da Rocha, M., An, J., Médina, C., Mejias, J., Mulet, K., et al. (2023). AUXIN RESPONSIVE FACTOR8 regulates development of the feeding site induced by root-knot nematodes in tomato. J. Exp. Bot 74, 5752–5766. doi: 10.1093/jxb/erad208
Nowak, S., Schnabel, E., Frugoli, J. (2019). The Medicago truncatula CLAVATA3-LIKE CLE12/13 signaling peptides regulate nodule number depending on the CORYNE but not the COMPACT ROOT ARCHITECTURE2 receptor. Plant Signaling Behav 14, 1598730. doi: 10.1080/15592324.2019.1598730
Ogawa-Ohnishi, M., Matsushita, W., Matsubayashi, Y. (2013). Identification of three hydroxyproline O-arabinosyltransferases in arabidopsis thaliana. Nat. Chem. Biol 9, 726–730. doi: 10.1038/nchembio.1351
Ohyama, K., Shinohara, H., Ogawa-Ohnishi, M., Matsubayashi, Y. (2009). A glycopeptide regulating stem cell fate in arabidopsis thaliana. Nat. Chem. Biol 5, 578–580. doi: 10.1038/nchembio.182
Oka-Kira, E., Tateno, K., Miura, K., Haga, T., Hayashi, M., Harada, K., et al. (2005). Klavier (klv), a novel hypernodulation mutant of lotus japonicus affected in vascular tissue organization and floral induction: Klavier, a novel hypernodulation mutant of lotus. Plant J 44, 505–515. doi: 10.1111/j.1365-313X.2005.02543.x
Okamoto, S., Kawasaki, A., Makino, Y., Ishida, T., Sawa, S. (2022). Long-distance translocation of CLAVATA3/ESR-related 2 peptide and its positive effect on roots sucrose status. Plant Physiol 189, 2357–2367. doi: 10.1093/plphys/kiac227
Okamoto, S., Ohnishi, E., Sato, S., Takahashi, H., Nakazono, M., Tabata, S., et al. (2009). Nod Factor/Nitrate-Induced CLE genes that drive HAR1-Mediated systemic regulation of nodulation. Plant Cell Physiol 50, 67–77. doi: 10.1093/pcp/pcn194
Okamoto, S., Shinohara, H., Mori, T., Matsubayashi, Y., Kawaguchi, M. (2013). Root-derived CLE glycopeptides control nodulation by direct binding to HAR1 receptor kinase. Nat. Commun 4, 2191. doi: 10.1038/ncomms3191
Okuma, N., Soyano, T., Suzaki, T., Kawaguchi, M. (2020). MIR2111-5 locus and shoot-accumulated mature miR2111 systemically enhance nodulation depending on HAR1 in Lotus japonicus. Nat. Commun 11, 5192. doi: 10.1038/s41467-020-19037-9
Olmo, R., Cabrera, J., Díaz-Manzano, F. E., Ruiz-Ferrer, V., Barcala, M., Ishida, T., et al. (2020). Root-knot nematodes induce gall formation by recruiting developmental pathways of post-embryonic organogenesis and regeneration to promote transient pluripotency. New Phytol 227, 200–215. doi: 10.1111/nph.16521
Olsson, V., Joos, L., Zhu, S., Gevaert, K., Butenko, M. A., De Smet, I. (2019). Look closely, the beautiful may be small: precursor-derived peptides in plants. Annu. Rev. Plant Biol 70, 153–186. doi: 10.1146/annurev-arplant-042817-040413
Potters, G., Pasternak, T. P., Guisez, Y., Jansen, M. A. K. (2009). Different stresses, similar morphogenic responses: integrating a plethora of pathways. Plant Cell Environ 32, 158–169. doi: 10.1111/j.1365-3040.2008.01908.x
Redding, N. W., Agudelo, P., Wells, C. E. (2018). Multiple nodulation genes are up-regulated during establishment of reniform nematode feeding sites in soybean. Phytopathology 108, 275–291. doi: 10.1094/PHYTO-04-17-0154-R
Reid, D. E., Ferguson, B. J., Gresshoff, P. M. (2011). Inoculation- and nitrate-induced CLE peptides of soybean control NARK-Dependent nodule formation. MPMI 24, 606–618. doi: 10.1094/MPMI-09-10-0207
Replogle, A., Wang, J., Bleckmann, A., Hussey, R. S., Baum, T. J., Sawa, S., et al. (2011). Nematode CLE signaling in arabidopsis requires CLAVATA2 and CORYNE: Nematode CLE receptors. Plant J 65, 430–440. doi: 10.1111/j.1365-313X.2010.04433.x
Replogle, A., Wang, J., Paolillo, V., Smeda, J., Kinoshita, A., Durbak, A., et al. (2013). Synergistic interaction of CLAVATA1, CLAVATA2, and RECEPTOR-LIKE PROTEIN KINASE 2 in cyst nematode parasitism of Arabidopsis. Mol. Plant-Microbe Interact 26, 87–96. doi: 10.1094/MPMI-05-12-0118-FI
Roux, B., Rodde, N., Jardinaud, M.-F., Timmers, T., Sauviac, L., Cottret, L., et al. (2014). An integrated analysis of plant and bacterial gene expression in symbiotic root nodules using laser-capture microdissection coupled to RNA sequencing. Plant J 77, 817–837. doi: 10.1111/tpj.12442
Rzemieniewski, J., Leicher, H., Lee, H. K., Broyart, C., Nayem, S., Wiese, C., et al. (2022). CEP signaling coordinates plant immunity with nitrogen status. Plant Biol. doi: 10.1101/2022.12.20.521212
Rzemieniewski, J., Stegmann, M. (2022). Regulation of pattern-triggered immunity and growth by phytocytokines. Curr. Opin. Plant Biol 68, 102230. doi: 10.1016/j.pbi.2022.102230
Sakamoto, K., Nohara, Y. (2009). Soybean (Glycine max [L.] Merr.) shoots systemically control arbuscule formation in mycorrhizal symbiosis. Soil Sci. Plant Nutr 55, 252–257. doi: 10.1111/j.1747-0765.2009.00358.x
Samorodova, A. P., Tvorogova, V. E., Tkachenko, A. A., Potsenkovskaya, E. A., Lebedeva, М.А., Tikhonovich, I. A., et al. (2018). Agrobacterial tumors interfere with nodulation and demonstrate the expression of nodulation-induced CLE genes in pea. J. Plant Physiol 221, 94–100. doi: 10.1016/j.jplph.2017.12.005
Sasaki, T., Suzaki, T., Soyano, T., Kojima, M., Sakakibara, H., Kawaguchi, M. (2014). Shoot-derived cytokinins systemically regulate root nodulation. Nat. Commun 5, 4983. doi: 10.1038/ncomms5983
Sauviac, L., Rémy, A., Huault, E., Dalmasso, M., Kazmierczak, T., Jardinaud, M., et al. (2022). A dual legume-rhizobium transcriptome of symbiotic nodule senescence reveals coordinated plant and bacterial responses. Plant Cell Environ 45, 3100–3121. doi: 10.1111/pce.14389
Schaarschmidt, S., Gresshoff, P. M., Hause, B. (2013). Analyzing the soybean transcriptome during autoregulation of mycorrhization identifies the transcription factors GmNF-YA1a/b as positive regulators of arbuscular mycorrhization. Genome Biol 14, R62. doi: 10.1186/gb-2013-14-6-r62
Schnabel, E., Journet, E.-P., De Carvalho-Niebel, F., Duc, G., Frugoli, J. (2005). The Medicago truncatula SUNN gene encodes a CLV1-like leucine-rich repeat receptor kinase that regulates nodule number and root length. Plant Mol. Biol 58, 809–822. doi: 10.1007/s11103-005-8102-y
Schnabel, E., Karve, A., Kassaw, T., Mukherjee, A., Zhou, X., Hall, T., et al. (2012). The M. truncatula SUNN gene is expressed in vascular tissue, similarly to RDN1, consistent with the role of these nodulation regulation genes in long distance signaling. Plant Signaling Behav 7, 4–6. doi: 10.4161/psb.7.1.18491
Schnabel, E. L., Kassaw, T. K., Smith, L. S., Marsh, J. F., Oldroyd, G. E., Long, S. R., et al. (2011). The ROOT DETERMINED NODULATION1 gene regulates nodule number in roots of Medicago truncatula and defines a highly conserved, uncharacterized plant gene family. Plant Physiol 157, 328–340. doi: 10.1104/pp.111.178756
Schnabel, E., Thomas, J., El-Hawaz, R., Gao, Y., Poehlman, W. L., Chavan, S., et al. (2023). Laser capture microdissection transcriptome reveals spatiotemporal tissue gene expression patterns of medicago truncatula roots responding to rhizobia. Mol. Plant-Microbe Interact 36, 805–820. doi: 10.1094/MPMI-03-23-0029-R
Schultz, J. C., Edger, P. P., Body, M. J. A., Appel, H. M. (2019). A galling insect activates plant reproductive programs during gall development. Sci. Rep 9, 1833. doi: 10.1038/s41598-018-38475-6
Searle, I. R., Men, A. E., Laniya, T. S., Buzas, D. M., Iturbe-Ormaetxe, I., Carroll, B. J., et al. (2003). Long-distance signaling in nodulation directed by a CLAVATA1-like receptor kinase. Science 299, 109–112. doi: 10.1126/science.1077937
Segonzac, C., Nimchuk, Z. L., Beck, M., Tarr, P. T., Robatzek, S., Meyerowitz, E. M., et al. (2012). The shoot apical meristem regulatory peptide CLV3 does not activate innate immunity. Plant Cell 24, 3186–3192. doi: 10.1105/tpc.111.091264
Serrano, K., Bezrutczyk, M., Goudeau, D., Dao, T., O’Malley, R., Malmstrom, R. R., et al. (2024a). Spatial co-transcriptomics reveals discrete stages of the arbuscular mycorrhizal symbiosis. Nat. Plants 10, 673–688. doi: 10.1038/s41477-024-01666-3
Serrano, K., Tedeschi, F., Andersen, S. U., Scheller, H. V. (2024b). Unraveling plant–microbe symbioses using single-cell and spatial transcriptomics. Trends Plant Sci 10, 1360–1385. doi: 10.1016/j.tplants.2024.06.008
Sexauer, M., Bhasin, H., Schön, M., Roitsch, E., Wall, C., Herzog, U., et al. (2023). A micro RNA mediates shoot control of root branching. Nat. Commun 14, 8083. doi: 10.1038/s41467-023-43738-6
Shen, L., Feng, J. (2024). NIN—at the heart of NItrogen-fixing Nodule symbiosis. Front. Plant Sci 14. doi: 10.3389/fpls.2023.1284720
Shi, J., Wang, X., Wang, E. (2023). Mycorrhizal symbiosis in plant growth and stress adaptation: from genes to ecosystems. Annu. Rev. Plant Biol 74, 569–607. doi: 10.1146/annurev-arplant-061722-090342
Siddique, S., Radakovic, Z. S., Hiltl, C., Pellegrin, C., Baum, T. J., Beasley, H., et al. (2022). The genome and lifestage-specific transcriptomes of a plant-parasitic nematode and its host reveal susceptibility genes involved in trans-kingdom synthesis of vitamin B5. Nat. Commun 13, 6190. doi: 10.1038/s41467-022-33769-w
Smetana, O., Mäkilä, R., Lyu, M., Amiryousefi, A., Sánchez Rodríguez, F., Wu, M.-F., et al. (2019). High levels of auxin signalling define the stem-cell organizer of the vascular cambium. Nature 565, 485–489. doi: 10.1038/s41586-018-0837-0
Snoeck, S., Lee, H. K., Schmid, M. W., Bender, K. W., Neeracher, M. J., Fernández-Fernández, A. D., et al. (2024). Leveraging coevolutionary insights and AI-based structural modeling to unravel receptor-peptide ligand-binding mechanisms. Proc. Natl. Acad. Sci 121, e2400862121. doi: 10.1073/pnas.2400862121
Song, Y., Wilson, A. J., Zhang, X.-C., Thoms, D., Sohrabi, R., Song, S., et al. (2021). FERONIA restricts pseudomonas in the rhizosphere microbiome via regulation of reactive oxygen species. Nat. Plants 7, 644–654. doi: 10.1038/s41477-021-00914-0
Sorin, C., Declerck, M., Christ, A., Blein, T., Ma, L., Lelandais-Brière, C., et al. (2014). A mi R 169 isoform regulates specific NF - YA targets and root architecture in A rabidopsis. New Phytol 202, 1197–1211. doi: 10.1111/nph.12735
Soyano, T., Hirakawa, H., Sato, S., Hayashi, M., Kawaguchi, M. (2014). NODULE INCEPTION creates a long-distance negative feedback loop involved in homeostatic regulation of nodule organ production. Proc. Natl. Acad. Sci 111, 14607–14612. doi: 10.1073/pnas.1412716111
Soyano, T., Shimoda, Y., Kawaguchi, M., Hayashi, M. (2019). A shared gene drives lateral root development and root nodule symbiosis pathways in lotus. Science 366, 1021–1023. doi: 10.1126/science.aax2153
Stührwohldt, N., Ehinger, A., Thellmann, K., Schaller, A. (2020). Processing and formation of bioactive CLE40 peptide are controlled by posttranslational proline hydroxylation. Plant Physiol 184, 1573–1584. doi: 10.1104/pp.20.00528
Sukumar, P., Legué, V., Vayssières, A., Martin, F., Tuskan, G. A., Kalluri, U. C. (2013). Involvement of auxin pathways in modulating root architecture during beneficial plant–microorganism interactions. Plant Cell Environ 36, 909–919. doi: 10.1111/pce.12036
Suzuki, R., Kanno, Y., Abril-Urias, P., Seo, M., Escobar, C., Tsai, A. Y.-L., et al. (2022). Local auxin synthesis mediated by YUCCA4 induced during root-knot nematode infection positively regulates gall growth and nematode development. Front. Plant Sci 13. doi: 10.3389/fpls.2022.1019427
Taleski, M., Jin, M., Chapman, K., Taylor, K., Winning, C., Frank, M., et al. (2024). CEP hormones at the nexus of nutrient acquisition and allocation, root development, and plant–microbe interactions. J. Exp. Bot 75, 538–552. doi: 10.1093/jxb/erad444
Tang, B., Feng, L., Hulin, M. T., Ding, P., Ma, W. (2023). Cell-type-specific responses to fungal infection in plants revealed by single-cell transcriptomics. Cell Host Microbe 31, 1732–1747. doi: 10.1016/j.chom.2023.08.019
Tavormina, P., De Coninck, B., Nikonorova, N., De Smet, I., Cammue, B. P. A. (2015). The plant peptidome: an expanding repertoire of structural features and biological functions. Plant Cell 27, 2095–2118. doi: 10.1105/tpc.15.00440
Tsikou, D., Yan, Z., Holt, D. B., Abel, N. B., Reid, D. E., Madsen, L. H., et al. (2018). Systemic control of legume susceptibility to rhizobial infection by a mobile microRNA. Science 362, 233–236. doi: 10.1126/science.aat6907
Udvardi, M., Poole, P. S. (2013). Transport and metabolism in legume-rhizobia symbioses. Annu. Rev. Plant Biol 64, 781–805. doi: 10.1146/annurev-arplant-050312-120235
Verbon, E. H., Liberman, L. M., Zhou, J., Yin, J., Pieterse, C. M. J., Benfey, P. N., et al. (2023). Cell type-specific transcriptomics reveals that root hairs and endodermal barriers play important roles in beneficial plant-rhizobacterium-interactions. Mol. Plant 16, 1160–1177. doi: 10.1016/j.molp.2023.06.001
Wang, J., Dhroso, A., Liu, X., Baum, T. J., Hussey, R. S., Davis, E. L., et al. (2021b). Phytonematode peptide effectors exploit a host post-translational trafficking mechanism to the ER using a novel translocation signal. New Phytol 229, 563–574. doi: 10.1111/nph.16765
Wang, W., Hu, C., Li, X., Zhu, Y., Tao, L., Cui, Y., et al. (2021c). Receptor-like cytoplasmic kinases PBL34/35/36 are required for CLE peptide-mediated signaling to maintain shoot apical meristem and root apical meristem homeostasis in arabidopsis. Plant Cell 34, 1289–1307. doi: 10.1093/plcell/koab315
Wang, J., Lee, C., Replogle, A., Joshi, S., Korkin, D., Hussey, R., et al. (2010). Dual roles for the variable domain in protein trafficking and host-specific recognition of Heterodera glycines CLE effector proteins. New Phytol 187, 1003–1017. doi: 10.1111/j.1469-8137.2010.03300.x
Wang, X., Mitchum, M. G., Gao, B., Li, C., Diab, H., Baum, T. J., et al. (2005). A parasitism gene from a plant-parasitic nematode with function similar to CLAVATA3/ESR (CLE) of Arabidopsis thaliana. Mol. Plant Pathol 6, 187–191. doi: 10.1111/j.1364-3703.2005.00270.x
Wang, J., Replogle, A., Hussey, R., Baum, T., Wang, X., Davis, E. L., et al. (2011). Identification of potential host plant mimics of CLAVATA3/ESR (CLE)-like peptides from the plant-parasitic nematode heterodera schachtii: Beet cyst nematode CLEs. Mol. Plant Pathol 12, 177–186. doi: 10.1111/j.1364-3703.2010.00660.x
Wang, Y., Schuck, S., Wu, J., Yang, P., Döring, A.-C., Zeier, J., et al. (2018). A MPK3/6-WRKY33-ALD1-Pipecolic acid regulatory loop contributes to systemic acquired resistance. Plant Cell 30, 2480–2494. doi: 10.1105/tpc.18.00547
Wang, L., Sun, Z., Su, C., Wang, Y., Yan, Q., Chen, J., et al. (2019). A gmNINa-miR172c-NNC1 regulatory network coordinates the nodulation and autoregulation of nodulation pathways in soybean. Mol. Plant 12, 1211–1226. doi: 10.1016/j.molp.2019.06.002
Wang, C., Velandia, K., Kwon, C.-T., Wulf, K. E., Nichols, D. S., Reid, J. B., et al. (2021a). The role of CLAVATA signalling in the negative regulation of mycorrhizal colonization and nitrogen response of tomato. J. Exp. Bot 72, 1702–1713. doi: 10.1093/jxb/eraa539
Wopereis, J., Pajuelo, E., Dazzo, F. B., Jiang, Q., Gresshoff, P. M., de Bruijn, F. J., et al. (2000). Short root mutant of lotus japonicus with a dramatically altered symbiotic phenotype. Plant J 23, 97–114. doi: 10.1046/j.1365-313x.2000.00799.x
Wu, H., Qu, X., Dong, Z., Luo, L., Shao, C., Forner, J., et al. (2020a). WUSCHEL triggers innate antiviral immunity in plant stem cells. Science 370, 227–231. doi: 10.1126/science.abb7360
Wu, P., Shou, H., Xu, G., Lian, X. (2013). Improvement of phosphorus efficiency in rice on the basis of understanding phosphate signaling and homeostasis. Curr. Opin. Plant Biol 16, 205–212. doi: 10.1016/j.pbi.2013.03.002
Wu, Q., Xu, F., Liu, L., Char, S. N., Ding, Y., Je, B. I., et al. (2020b). The maize heterotrimeric G protein β subunit controls shoot meristem development and immune responses. Proc. Natl. Acad. Sci 117, 1799–1805. doi: 10.1073/pnas.1917577116
Wubben, M. J., Gavilano, L., Baum, T. J., Davis, E. L. (2015). Sequence and spatiotemporal expression analysis of CLE-Motif containing genes from the reniform nematode (rotylenchulus reniformis linford & oliveira). J. Nematol 47, 159–165.
Wulf, K., Sun, J., Wang, C., Ho-Plagaro, T., Kwon, C.-T., Velandia, K., et al. (2023). The role of CLE peptides in the suppression of mycorrhizal colonization of tomato. Plant And Cell Physiol 65, 107–119. doi: 10.1093/pcp/pcad124
Yamaguchi, Y. L., Ishida, T., Sawa, S. (2016). CLE peptides and their signaling pathways in plant development. J. Exp. Bot 67, 4813–4826. doi: 10.1093/jxb/erw208
Yamaguchi, Y. L., Suzuki, R., Cabrera, J., Nakagami, S., Sagara, T., Ejima, C., et al. (2017). Root-knot and cyst nematodes activate procambium-associated genes in arabidopsis roots. Front. Plant Sci 8. doi: 10.3389/fpls.2017.01195
Yang, Y., Jittayasothorn, Y., Chronis, D., Wang, X., Cousins, P., Zhong, G.-Y. (2013). Molecular characteristics and efficacy of 16D10 siRNAs in inhibiting root-knot nematode infection in transgenic grape hairy roots. PloS One 8, e69463. doi: 10.1371/journal.pone.0069463
Yimer, H. Z., Luu, D. D., Coomer Blundell, A., Ercoli, M. F., Vieira, P., Williamson, V. M., et al. (2023). Root-knot nematodes produce functional mimics of tyrosine-sulfated plant peptides. Proc. Natl. Acad. Sci 120, e2304612120. doi: 10.1073/pnas.2304612120
Yoro, E., Nishida, H., Ogawa-Ohnishi, M., Yoshida, C., Suzaki, T., Matsubayashi, Y., et al. (2019). PLENTY, a hydroxyproline O-arabinosyltransferase, negatively regulates root nodule symbiosis in Lotus japonicus. J. Exp. Bot 70, 507–517. doi: 10.1093/jxb/ery364
Zakaria Solaiman, M., Senoo, K., Kawaguchi, M., Imaizumi-Anraku, H., Akao, S., Tanaka, A., et al. (2000). Characterization of Mycorrhizas Formed by Glomus sp. on Roots of Hypernodulating Mutants of Lotus japonicus. J. Plant Res 113, 443–448. doi: 10.1007/PL00013953
Zhang, L., Hua, C., Janocha, D., Fliegmann, J., Nürnberger, T. (2023b). Plant cell surface immune receptors-novel insights into function and evolution. Curr. Opin. Plant Biol 74, 102384. doi: 10.1016/j.pbi.2023.102384
Zhang, X., Peng, H., Zhu, S., Xing, J., Li, X., Zhu, Z., et al. (2020). Nematode-encoded RALF peptide mimics facilitate parasitism of plants through the FERONIA receptor kinase. Mol. Plant 13, 1434–1454. doi: 10.1016/j.molp.2020.08.014
Zhang, M., Su, H., Gresshoff, P. M., Ferguson, B. J. (2021). Shoot-derived miR2111 controls legume root and nodule development. Plant Cell Environ 44, 1627–1641. doi: 10.1111/pce.13992
Zhang, D., Wu, Q., Zhao, Y., Yan, Z., Xiao, A., Yu, H., et al. (2023a). Dual RNA-Seq Analysis Pinpoints a Balanced Regulation between Symbiosis and Immunity in Medicago truncatula-Sinorhizobium meliloti Symbiotic Nodules. Int. J. Mol. Sci 24, 16178. doi: 10.3390/ijms242216178
Keywords: peptide, plant-microbe interaction, systemic signaling, plant immunity, nodulation, phytoparasitic nematode, AM symbiosis
Citation: Nakagami S, Kajiwara T, Tsuda K and Sawa S (2024) CLE peptide signaling in plant-microbe interactions. Front. Plant Sci. 15:1481650. doi: 10.3389/fpls.2024.1481650
Received: 16 August 2024; Accepted: 02 October 2024;
Published: 23 October 2024.
Edited by:
Choong-Min Ryu, Korea Research Institute of Bioscience and Biotechnology (KRIBB), Republic of KoreaReviewed by:
Gabriel Castrillo, University of Nottingham, United KingdomCopyright © 2024 Nakagami, Kajiwara, Tsuda and Sawa. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Satoru Nakagami, c25ha2FnYW1pQG1haWwuaHphdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.