- 1College of Forestry, Shanxi Agricultural University, Taigu, Shanxi, China
- 2Integrative Omics and Molecular Modeling Laboratory, Department of Bioinformatics and Biotechnology, Government College University Faisalabad (GCUF), Faisalabad, Pakistan
C-repeat binding factor (CBF) transcription factors can activate the expression of a series of cold regulation-related genes, thereby improving the cold resistance of plants. However, no detailed information is known about the biological functions of CBF proteins in yellowhorn (Xanthoceras sorbifolium). In this study, a total of 59 CBF gene family members were identified in five yellowhorn cultivars (WF18, Zhongshi 4, Jinguanxipei 2021, Zhong Guan NO.2, and XsoG11), revealing their intraspecific structural and functional diversity, with 8 core genes present in all cultivars. Phylogenetic and motif analyses highlighted conserved features and species-specific adaptations. Gene duplication events revealed that tandem duplicates are major factors involved in the expansion of this gene family in yellowhorn. Expression profiling under stress conditions demonstrated the involvement of these genes in stress responses. Of particular interest was Xg11_CBF11, which showed strong induction by low-temperature stress. Overexpression of Xg11_CBF11 in Arabidopsis thaliana was performed to validate its cold resistance function. The wild-type and T2 transgenic A. thaliana plants were subjected to low-temperature stress at 4°C for 0, 24, and 48 h, and physiological indexes related to antioxidant enzyme activity, photosynthesis, and cell membrane permeability were determined by comparative test. The results were as follows: the POD and SOD activities of transgenic lines were significantly higher than those of wild-type lines, indicating Xg11_CBF11 improved the adaptability of A. thaliana to low-temperature; The increase of relative conductivity and malondialdehyde, the decrease of chlorophyll content in transgenic lines were smaller than those of wild-type lines, indicating Xg11_CBF11 enhanced the resistance of A. thaliana to low-temperature stress. These results implied that Xg11_CBF11 has a positive regulatory effect on A. thaliana ‘s response to low-temperature stress.
1 Introduction
AP2/ERF is a widely known gene family that regulates responses to environmental stresses in higher plants. Several reports have shown that plants with these overexpressed factors show better tolerance to environmental stresses (Feng et al., 2020). Over time it has evolved into a signal transduction pathway, the ICE1-CBF-COR pathway. When plants are subjected to abiotic stress such as cold, the ICE1, which is the inducer of CBF expression binds to the MYC recognition site. The ICE1 encodes MYC-like bHLH transcriptional activator which binds to the sequence in MYC. This MYC recognition sequence is present in the promoter sequence of the CBF genes and helps activate transcription. The overexpression of CBF proteins activates the COR (cold-responsive) genes and helps plants tolerate cold stress (Chinnusamy et al., 2010; Verma et al., 2020).
The AP2/ERF genes contain AP2/ERF DNA binding domains which are 60-70 amino acids long. These domains contain RAYD and YRG motifs. Based on similarity among amino acid sequences and the domains, the AP2/ERF genes can be classified into four categories. The sequences belonging to AP2 family contain two AP2 domains. The members of EREBP family contain one AP2 domain (Kagaya et al., 1999). The EREBP family is further subdivided into DREB and ERF subfamilies. The RAV family members contain B3 and AP2 domains. These RAV family members are regulated by brassinosteroid or ethylene and regulate responses to environmental stresses (Hu et al., 2022). The Soloist family members contain single AP2 domain and participate in disease regulatory mechanisms (Giri et al., 2014). Thus, these TFs are involved in various defense-related mechanisms and induce responses to various stimuli including salt, drought, and heat stress. The CBF genes contain two AP2 domains which contain the transcriptional activator. This transcriptional activator binds to the CCGAC, the low-temperature-responsive element, thereby, inducing the expression of some genes related to cold stress responsiveness (Lv et al., 2020).
In A. thaliana, three CBF genes; CBF1, CBF2, and CBF3 have been reported to show cold tolerance (Lv et al., 2020). Other plant species that exhibit cold tolerance contain signature sequences in CBF-like proteins. CBF orthologs express rapidly in Brassica napus (Gao et al., 2002), barley (J. Ding et al., 2021), tomato (Pontaroli et al., 2009), and rice (Dubouzet et al., 2003). An O. sativa AP2/ERF gene, OsDREBL was cloned and showed responsiveness to cold stress (Chen et al., 2003). In wild soybean, the member of ERF family is involved in alkaline stress tolerance. The GsERF6 gene was introduced in A. thaliana and induced tolerance to alkaline stress (Yu et al., 2016).
Yellowhorn, belonging to the genus Xanthoceras, is a deciduous shrub or small tree, as well as a unique woody oil tree in China. The oil content of the seed kernel can reach about 60%, which can be used as high-quality raw material for advanced edible oil and biodiesel, and has high economic use value (Li et al., 2022). As an important oil tree species, the main research content focuses on oil metabolism, processing and extraction, and seedling cultivation. Cold stress influences the plant’s survival by causing cellular dehydration in tissues. Plants can respond to cold stress through a wide range of transcriptional changes to maintain cellular homeostasis and improve cold tolerance. Still, there are few studies on the internal mechanism of cold resistance in yellowhorn. The cold resistance function of CBF transcription factor genes has been demonstrated in various plants. Under freezing or chilling temperature conditions, CBFs induce several cold-stress-responsive genes that provide tolerance against this stress. However, the roles of CBFs in yellowhorn still remains unclear.
The identification studies of yellowhorn CBF genes are important as CBF genes play vital roles in stress resistance and have not been identified in yellowhorn. Traditional approaches for gene family identification studies utilize a single reference genome and, thus, cannot identify the members of gene families present in other genomes but absent in the reference genome (Ding et al., 2024). Wang et al. reported the assembly as well as comparative pan-genome-wide gene family studies among yellowhorn and other species genomes, providing an important resource for further functional studies (Wang et al., 2023, 2024; Hu et al., 2024; Jia et al., 2023).
The current study reports the pan-genome-wide identification of CBF genes from five yellowhorn cultivars including WF18 (Xwf8), Zhongshi 4 (Xzs4), Jinguanxipei 2021 (Xjg), Zhoug Guan NO.2 (Xzg2), and XsoG11 (Xg11). These genes were further investigated for intraspecies diversity and functional conservation. Further, the expression level of these genes was checked in seed tissues and under abiotic stress. We focused on Xg11_CBF11, which showed strong induction under low-temperature stress. Based on the cloning of the Xg11_CBF11 gene, expression vector was constructed and was transformed into A. thaliana. The transgenic plants were subjected to cold stress to verify the cold resistance function of the Xg11_CBF11 gene, by comparing the changes in antioxidant enzyme activity, chlorophyll content, relative conductivity, and malondialdehyde content. The analysis of the physiological indicators of cold resistance in Xg11_CBF11-overexpressing A. thaliana is the expansion and deepening of the molecular mechanism of cold resistance in yellowhorn.
2 Materials and methods
2.1 Pan-genome-wide identification of CBF gene family members in five yellowhorn cultivars and analysis of their physicochemical properties
Protein sequences of CBF from A. thaliana, O. sativa, and Triticum aestivum were obtained from the NCBI Protein database (https://www.ncbi.nlm.nih.gov/protein/) (Geer et al., 2010). These sequences were used as queries for a local BLASTp search (Altschul et al., 1997) against five yellowhorn cultivars (Xzs4, Xwf8, Xjg, Xg11, and Xzg2). To confirm the presence of the full AP2 domain, the candidate sequences were analyzed using the Pfam (https://pfam.xfam.org/) (Mistry et al., 2021), CDD (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) (Marchler-Bauer et al., 2015), and SMART (http://smart.embl-heidelberg.de/) (Schultz et al., 2000) databases. Sequences that contained the complete AP2 domain were finalized for subsequent analysis and named according to their chromosomal order. The physicochemical properties of the CBF proteins were predicted using the ExPASy ProtParam tool (https://web.expasy.org/protparam/) (Gasteiger et al., 2005). Subcellular localization predictions were made using the WoLF PSORT tool (https://wolfpsort.hgc.jp/) (Horton et al., 2007).
2.2 Phylogenetic analysis of yellowhorn CBFs
Using the default settings of ClustalW, CBF protein sequences from A. thaliana, O. sativa, Lolium perenne, Hordeum vulgare, T. aestivum, Secale cereale, and five yellowhorn cultivars were aligned (Kumar et al., 2016; Zameer et al., 2022). A phylogenetic tree was then constructed using the Maximum Likelihood (ML) method with 1000 bootstrap replicates in the IQ-TREE Web Server, and JTT+F+I+G4 was selected as the best-fit substitution model based on the BIC scores (http://iqtree.cibiv.univie.ac.at/) (Trifinopoulos et al., 2016). The resulting phylogenetic tree was edited and visualized using iTOL: Interactive Tree of Life v6 (https://itol.embl.de/) (Sadaqat et al., 2024; Letunic and Bork, 2021).
2.3 Exon-Intron representation and assessment of conserved motif
The gene structures of CBFs were analyzed using the Gene Structure View (advanced) tool in Tbtools, with the annotation file as input (Chen et al., 2020). Conserved motifs of CBFs were identified using the amino acid sequences as input in the MEME website (https://meme-suite.org/) (Bailey et al., 2015), with the maximum number of motifs set to 20.
2.4 Chromosomal localization, Ka/Ks ratios, gene duplication analysis
The chromosomal positions of all CBF genes in the five yellowhorn cultivars were obtained from their annotation files. The positional localization of the CBF genes was performed and visualized using TBtools software (Chen et al., 2018). Gene duplication events for CBF genes were identified based on the criteria that the shorter gene covered at least 70% of the length of the longer gene and that the similarity between the two aligned genes was at least 70% (Tahir and Sadaqat, 2023). Tandem and segmental duplications were reported as the two main mechanisms underlying gene family expansion. The synonymous substitution rate (Ks), nonsynonymous substitution rate (Ka), and the Ka/Ks ratio were used to assess the selection history and duplication events (Fatima et al., 2023). The Ks and Ka values of duplicated CBF genes were computed using DnaSP v6 (Rozas et al., 2017). The following formula was used to calculate the duplication time: T= Ks/(2 × 1.5 × 10−8)*10−6 million years ago (Mya) (Ma et al., 2024; Yang et al., 2024).
2.5 Protein-Protein interaction and gene ontology enrichment analysis
The amino acid sequences of the yellowhorn Xg11 variety were used to analyze the interactions among CBF proteins using STRING database (Mering et al., 2003). The top 10 interactions were kept and 0.4 was selected as the interaction threshold. Cytoscape software (Shannon et al., 2003) was used to visualize the interactions. The components of GO enrichment were predicted using PANNZER database (Törönen et al., 2018).
2.6 Prediction of cis-regulatory elements and expression profiling of Xg11 CBFs
The 2kb upstream regions of yellowhorn CBFs genes were retrieved and searched for cis-elements using the PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Rombauts et al., 1999; Zia et al., 2024). The RNA-seq data of yellowhorn Xg11 cultivar was analyzed to check the expression profiles of CBF genes in low and high-temperature stress; drought stress (BioProject: PRJNA974867); salt and alkali stress; and tissues (BioProject: PRJNA923394). The transcriptomic data was obtained from SRA-NCBI database (https://www.ncbi.nlm.nih.gov/sra) and the quality of reads was evaluated using FastQC tool (Wingett and Andrews, 2018). The genome was indexed and the clean-reads were mapped onto it using HISAT (Kim et al., 2019). StringTie (Pertea et al., 2016) was further used to estimate the expression in fragments per kilobase of transcript per million mapped reads (FPKM) values (Sadaqat et al., 2023).
2.7 Plant materials and treatments
The study utilized yellowhorn seedlings and wild-type A. thaliana plants. The Xg11_CBF11 gene was introduced into A. thaliana, resulting in T2 transgenic lines (L1, L5, L6) with high expression. “G11” (superior line of yellowhorn) seeds were collected from Shanxi Agricultural University, sown, and grown until seedlings reached about 15 cm. Leaves of yellowhorn seedlings treated at 4°C for 12 hours were harvested for cloning Xg11_CBF11. Wild-type A. thaliana seeds were grown in a light incubator with a relative humidity of 60%-70%, a photoperiod of 16/8h, and a temperature of 25°C. Both wild-type and transgenic A. thaliana lines were treated with 4°C cold stress and sampled at 0, 24, and 48h. Various physiological indicators, including antioxidant enzyme activity (POD, SOD), chlorophyll content, relative conductivity, and malondialdehyde content, were measured.
2.8 Gene cloning and transformation
For gene cloning and transformation, RNA was extracted from yellowhorn seedlings subjected to cold stress using the RNAprep pure Plant Kit (Tiangen, Beijing, China). cDNA was synthesized using the Fastking reverse transcription kit, TaKaRa, Dalian, China, and the Xg11_CBF11 gene was amplified using specific primers designed from the full-length CDS sequence. The specific primer pairs (Xg11_CBF11-F/R) of the selected genes were designed using Primer 3.0 software (https://bioinfo.ut.ee/primer3-0.4.0/) (Supplementary Table S1) and synthesized by Tsingke Biotech. The amplified product was inserted into the pMD19-T vector and transformed into E. coli DH5α for cloning.
The Xg11_CBF11 gene was then inserted into the pEGOEP35S-H vector with Xg11_CBF11-e-F/R primers (Supplementary Table S1), and Agrobacterium tumefaciens containing the recombinant plasmid was used to infect A. thaliana plants through the floral-dip method. Transgenic seeds (T1) were selected on a hygromycin-containing medium and further confirmed by PCR (Ding et al., 2021, 2023).
To precisely quantify the expression levels of Xg11_CBF11 in transgenic A. thaliana, a quantitative real-time polymerase chain reaction (qRT-PCR) was conducted employing specifically designed Xg11_CBF11-q-F/R primers (Supplementary Table S1). For normalization and accurate quantification, AtActin2, a constitutively expressed gene from A. thaliana, was utilized as the internal reference gene.
2.9 Determination of physiological indicators and data analysis
To determine physiological indicators, fresh leaves (0.2 g) were ground, and enzyme activities were measured using specific kits (Solarbio, Beijing, China). Absorbance values were recorded at 470 nm and 560 nm for POD and SOD, respectively. Chloroplast pigments were extracted with 95% ethanol, and absorbance values were measured at 665 nm and 649 nm to calculate chlorophyll a and chlorophyll b content. Leaf strips were soaked in deionized water for 12 h, and conductivity was measured before and after boiling to assess cell membrane permeability. Malondialdehyde (MDA) was determined using thiobarbituric acid, with absorbance measured to calculate content, indicating lipid peroxidation levels. The physiological data were analyzed using ANOVA in SPSS software. Mean and standard error values were plotted using Origin software to visualize the trends and differences between treatments.
3 Results
3.1 Pan-genome−wide Identification of CBF genes from five yellowhorn cultivars
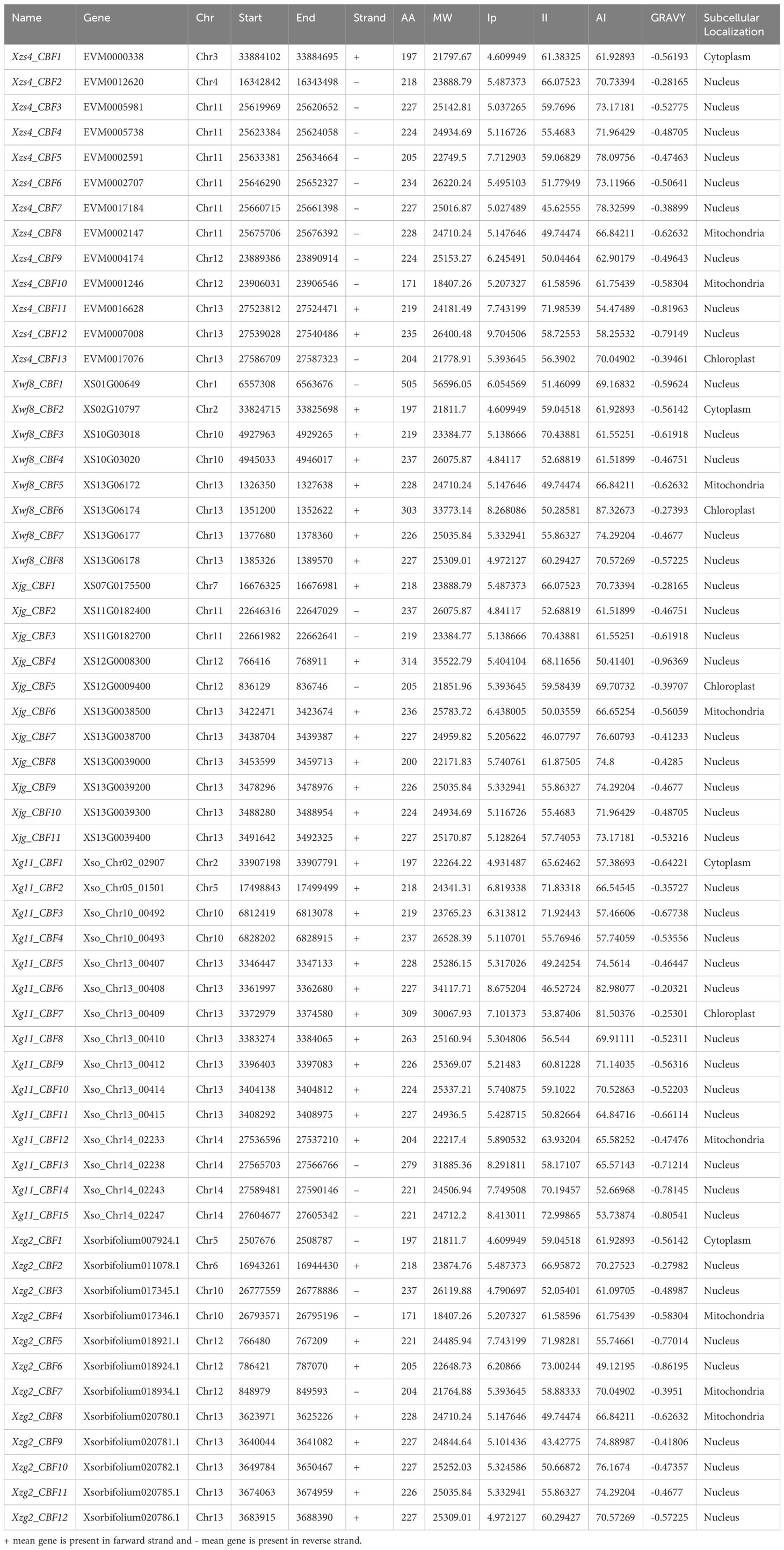
A total of 13, 8, 11, 15, and 12 CBF genes were identified from the genomes of Xzs4, Xwf8, Xjg, Xg11, and Xzg2, respectively. CBF1-8 genes were present in all five genomes. CBF9-CBF13 were dispensable genes present in genomes except Xwf8. Two unique cultivar-specific genes CBF14 and CBF15 were identified that were present only in the Xg11 genome.
All the sequences contain the conserved AP2 domain. The physical and chemical properties of all CBF proteins were analyzed. There were no significant differences in amino acid residue number (AA), molecular weight (MW), isoelectric point (pI), instability index (II), aliphatic index (AI), and GRAVY among the five cultivars. The protein length ranged from 171-505 aa, MW ranged from 18.41-56.60 kDa, pI ranged from 4.61-9.70, II ranged from 43-73, AI ranged from 49.12-87.33, and GRAVY value ranged from -0.203 to -0.964. Most of the CBFs were localized in the nucleus (Table 1).
3.2 Phylogenetic relationships of CBF family members from five yellowhorn cultivars
To analyze the possible evolutionary characteristics of the CBF gene family in yellowhorn, we constructed a phylogenetic tree based on 129 CBF amino acid sequences including 4 from A. thaliana, 12 from L. perenne, 12 from H. vulgare, 17 from O. sativa, 16 from T. aestivum, 9 from S. cereale, and 59 from five yellowhorn cultivars. All CBF proteins were clustered into five groups. Group V contained the most CBF gene family members, followed by group I, II, III, and Group IV had the fewest members. Group I and II had the same number of CBF genes (Figure 1).
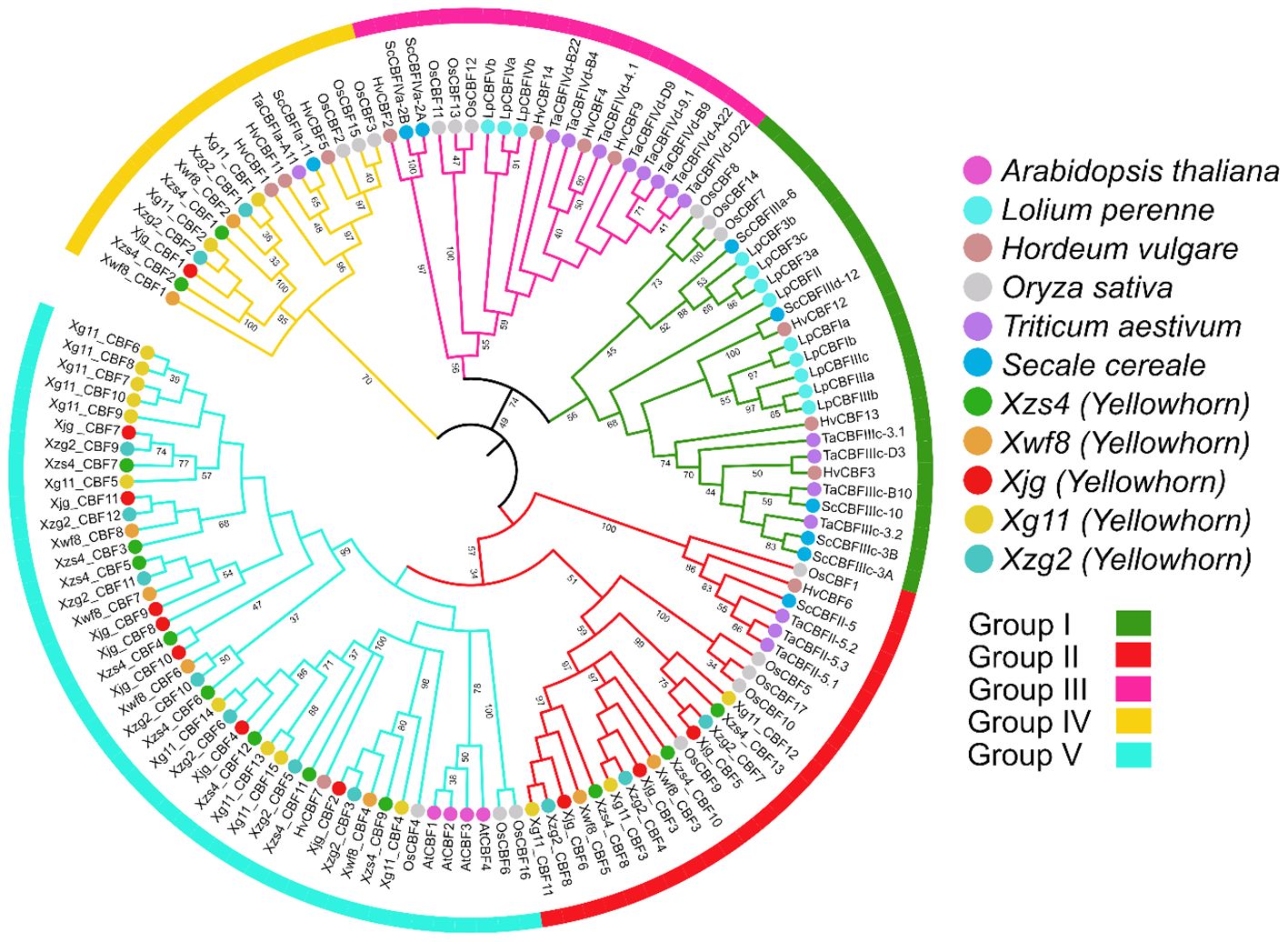
Figure 1. An ML phylogenetic tree (circular tree) constructed with full-length amino acid sequences of the 129 CBFs, 1000 bootstrap replicates, and set JTT+F+I+G4 best-fit substitution model through IQ Tree software. The tree was viewed in the iTOL software and divided into five groups: pink, yellow, green, cyan, and red.
3.3 Exon-Intron and conserved motif distribution analysis of the CBF family
To gain insight into potential functions and diversification among CBFs, the exon–intron organizations and encoded conserved motifs were compared. As expected, most phylogenetically closely related STPs shared similar motifs and structures (Figure 2). The exon/intron structures exhibited a highly conserved organization in CBF genes. Most of the CBFs presented one exon while few members have more than one exon. Maximum number of exons observed for CBF2 was 4. Twenty predicted motifs were identified throughout the CBF protein sequences. Motifs 1 and 5 were present in all analyzed CBFs. Motifs 2, 3, and 4 were also present in most of the members. Motif 7 was only conserved among all the members of Xg11 and motif 6 was only conserved among all the members of Xzg2 cultivars (Supplementary Figure S1-S4).
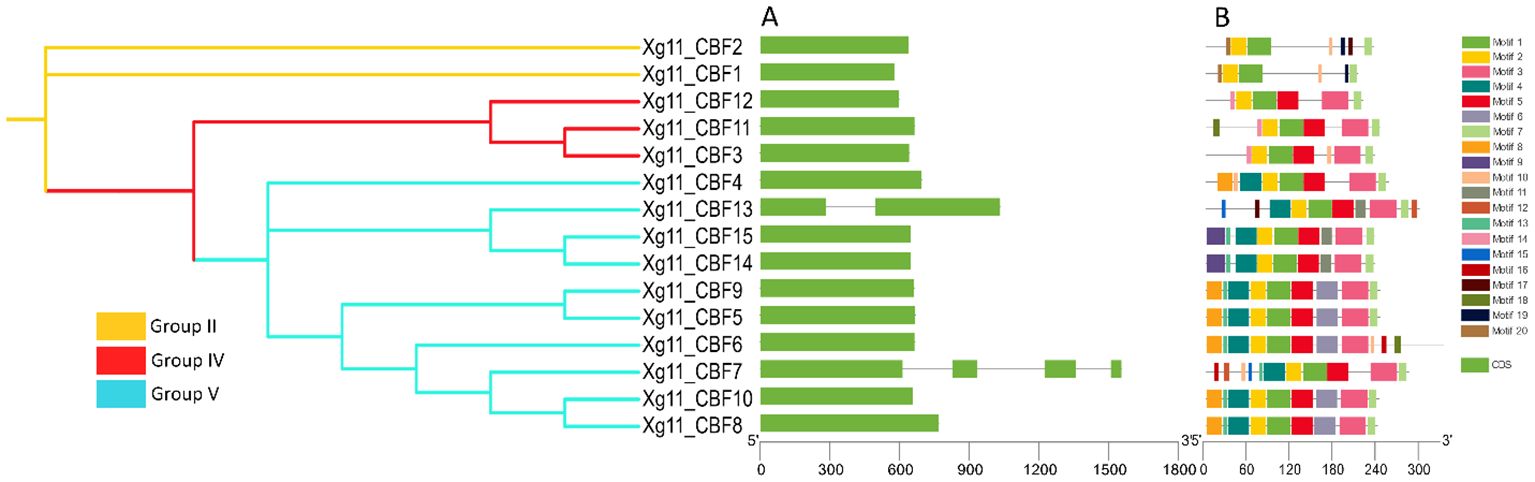
Figure 2. Structural and motif analyses of Xg11_CBFs. (A) Exon/intron structures of CBFs, (B) Schematic representation of the conserved motif compositions.
3.4 Chromosomal mapping and duplication events
All CBF genes of five yellowhorn cultivars were located on 15 chromosomes. The CBFs were mapped on their corresponding position on chromosomes. In Xg11, most of the CBF genes were located on Chr13 followed by Chr14. Chr 2, 5, and 10 contained 1,1, and 2 CBF genes, respectively (Figure 3D). The other four cultivars contained most of the genes on Chr13. In all five yellowhorn cultivars Chr8 and 15 contained no CBF genes localized on them (Figures 3A–C, E).
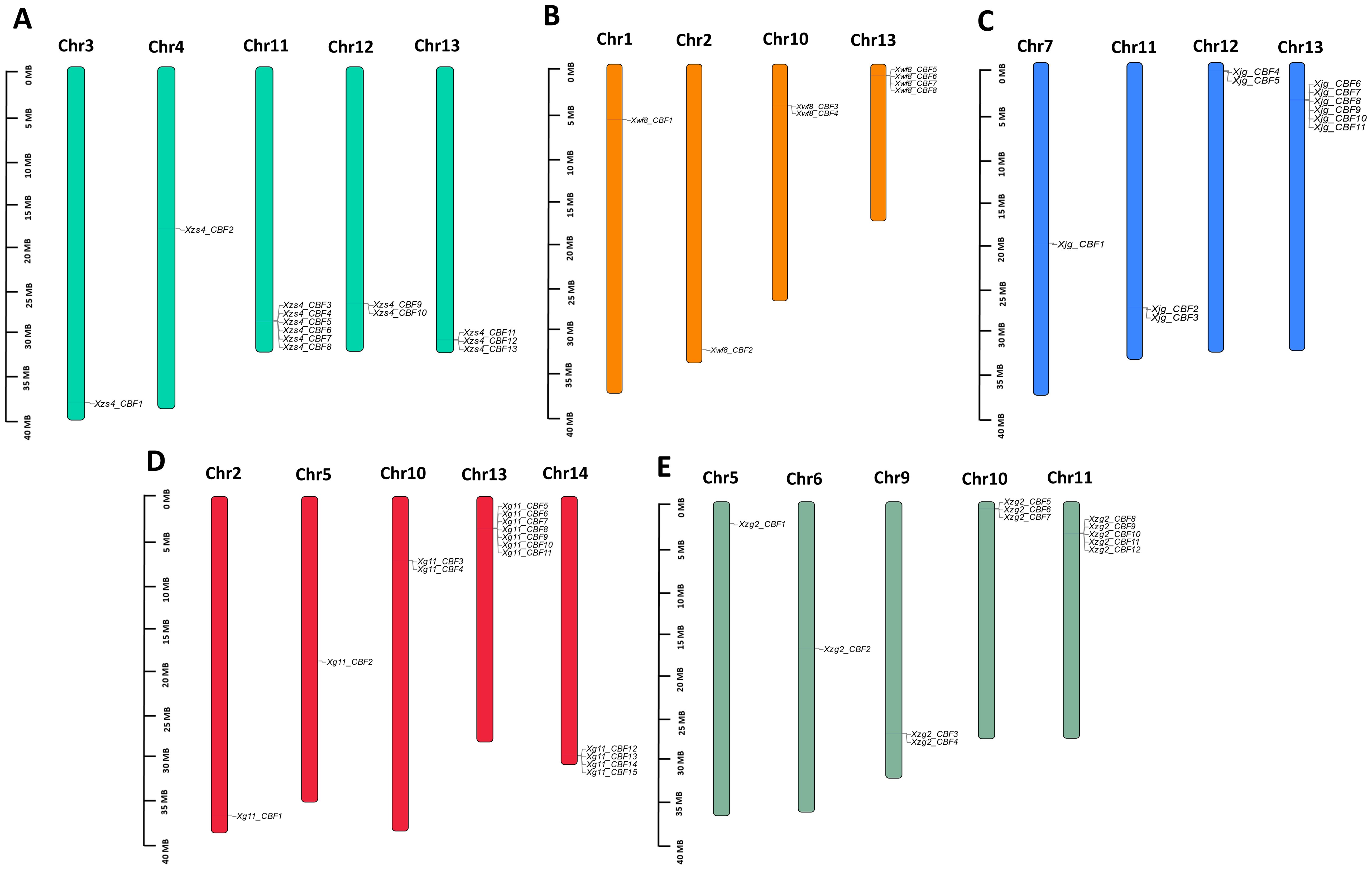
Figure 3. Localization of CBF genes on chromosomes. (A) Xzs4_CBF (B) Xwf8_CBF, (C) Xjg_CBF, (D) Xg11_CBF, and (E) Xzg2_CBF. Each chromosome representation displays the chromosomal number at the top.
According to the defined criteria, the analysis of gene duplication events showed that there were 44 pairs of tandem duplication genes in the five yellowhorn cultivars including 11 duplicated pairs in Xzs4, 3 in Xwf8, 9 in Xjg, 14 in Xg11, and 7 in Xzg2. No segmental duplicated pair was observed in all of the five cultivars. The unique genes Xg11_CBF13 and Xg11_CBF14 were found to exhibit tandem duplication. The Ka/Ks ratio ranged from 0.24-1.93 and the duplication time was observed from 0.35-5.07 Mya (Table 2).
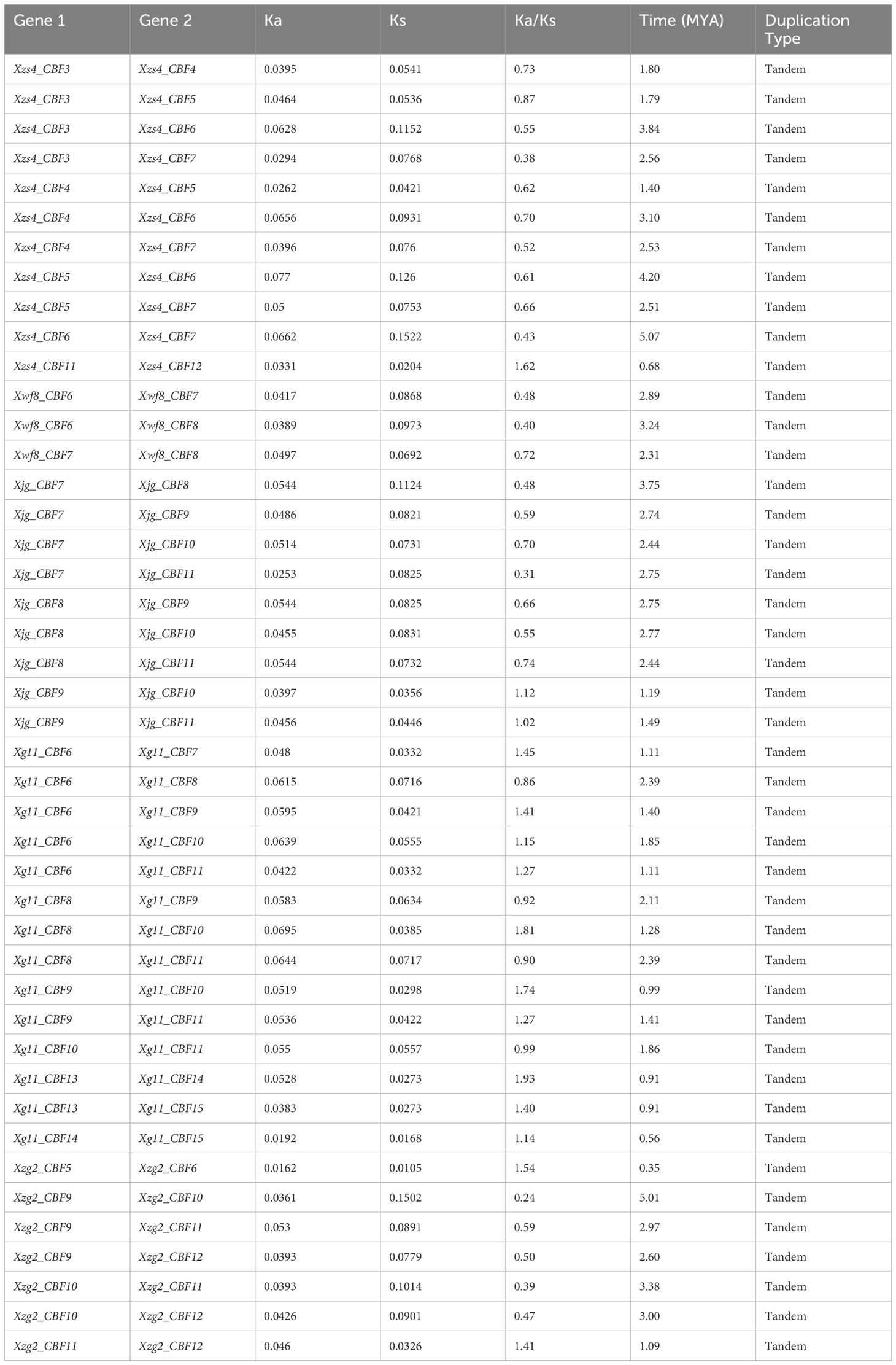
Table 2. Duplicated pairs of five yellowhorn genes, synonymous and non-synonymous mutations, duplication time, and type of duplication between the genes.
3.5 PPI and GO enrichment analysis
A PPI network of Xg11_CBFs was generated to perform functional evaluation. The seven Xg11_CBFs including Xg11_CBF1, Xg11_CBF3, Xg11_CBF4, Xg11_CBF10, Xg11_CBF11, Xg11_CBF12, and Xg11_CBF15 showed interactions with other homologous proteins of A. thaliana. These homologous proteins were found to have higher expression in various abiotic stresses such as drought, salt, cold, and osmotic stresses, suggesting the potential roles of Xzs4_CBFs in abiotic stress regulation pathways (Figure 4A).
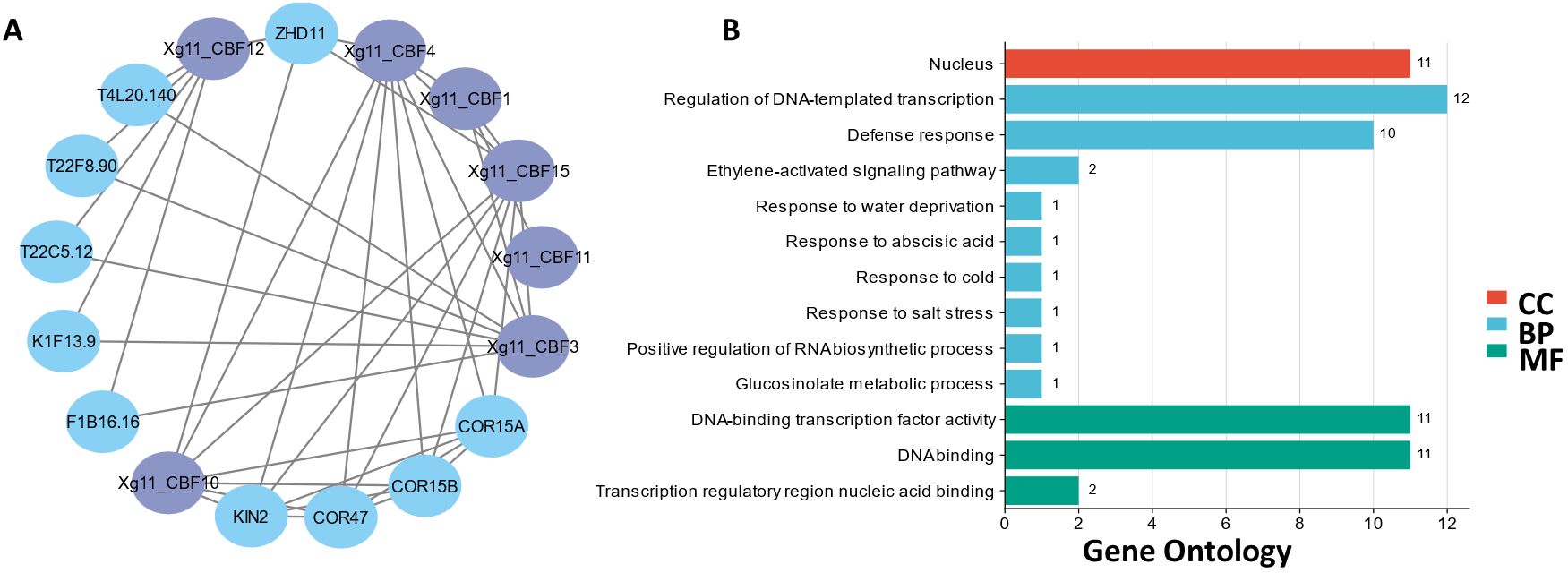
Figure 4. (A) Interactions among Xg11_CBF proteins and other homologous proteins. (B) Predicted biological processes (BP), cellular components (CC), and molecular functions (MF) associated with Xg11_CBFs.
The GO enrichment analysis showed that these proteins are found in the nucleus component of the cell. Further, the Xg11_CBFs proteins are found to be involved in responses to water deprivation, cold, and salt stress. Moreover, these are also involved in the ethylene-activated signaling pathway and regulation of RNA biosynthetic pathway. Further, these are also involved in several MFs including DNA-binding transcription factor activity, and transcription regulatory region for nucleic acid binding (Figure 4B).
3.6 Cis-regulatory element analysis of yellowhorn CBFs
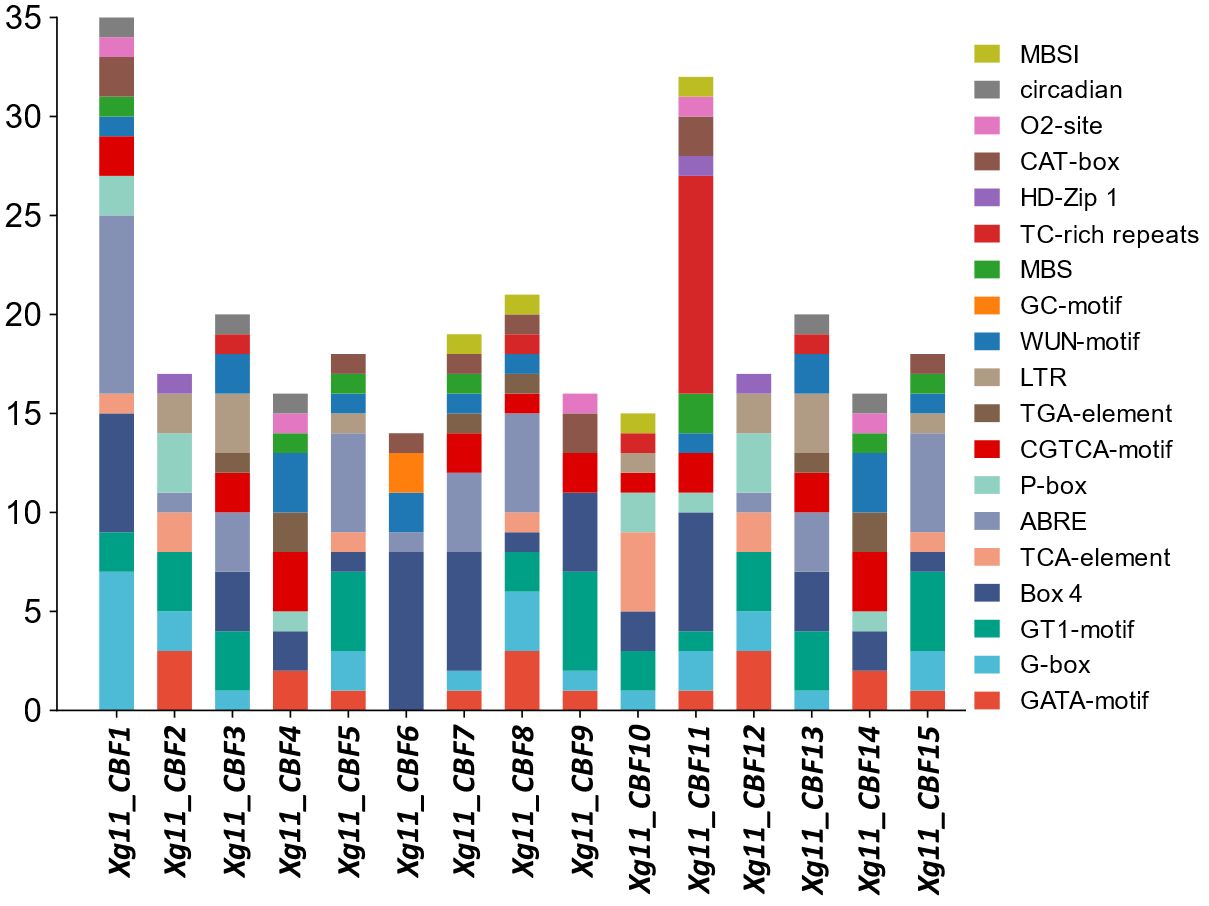
To further evaluate the stress-responsive behaviors of yellowhorn CBFs, their promoter regions were analyzed to find stress-related cis-regulatory elements. The elements such as G-box, GATA-motif, Box 4, and GT1-motif were found in almost all identified members. These elements are associated with light stress regulation. Five elements including TGA-element, P-box, TCA-element, CGTCA-motif, and ABRE were found which are linked to hormone responsiveness. The stress-responsive elements; LTR, GC-motif, MBS, and TC-rich repeats were also found. Development-related elements which include CAT-box, MBSI, circadian, HD-Zip 1, and o2-site were found. All these elements show the potential involvement of yellowhorn CBFs genes in light stress, abiotic stresses, and development-related mechanisms (Figure 5, Supplementary Figure S5).
3.7 Expression profiling of Xg11 CBFs in tissues and abiotic stress
The expression profiling of Xg11 CBF genes was analyzed under low and high-temperature stress conditions (Figure 6A). Genes including Xg11_CBF1, Xg11_CBF4, Xg11_CBF8, Xg11_CBF10, and Xg11_CBF11 showed change in expression levels under control, low-temperature, and high-temperature conditions. Xg11_CBF11 was highly expressed during low-temperature stages and showed no change in expression under high-temperature. For drought stress, Xg11_CBF1 showed a minimum change in expression and Xg11_CBF4 contained an increase in expression (Figure 6B). Under salt and alkali stress conditions, Xg11_CBF1 contained a fluctuated expression. Xg11_CBF3 was highly expressed under salt treatment and contained a minimum change in expression in alkali conditions. Similarly, Xg11_CBF11 also showed a change in expression pattern (Figure 6C). Xg11_CBF1 was highly expressed in seed coat with minimal expression in seed kernel. Xg11_CBF2 and Xg11_CBF4 exhibited a change in expression pattern in seed kernel and coat (Figure 6D).
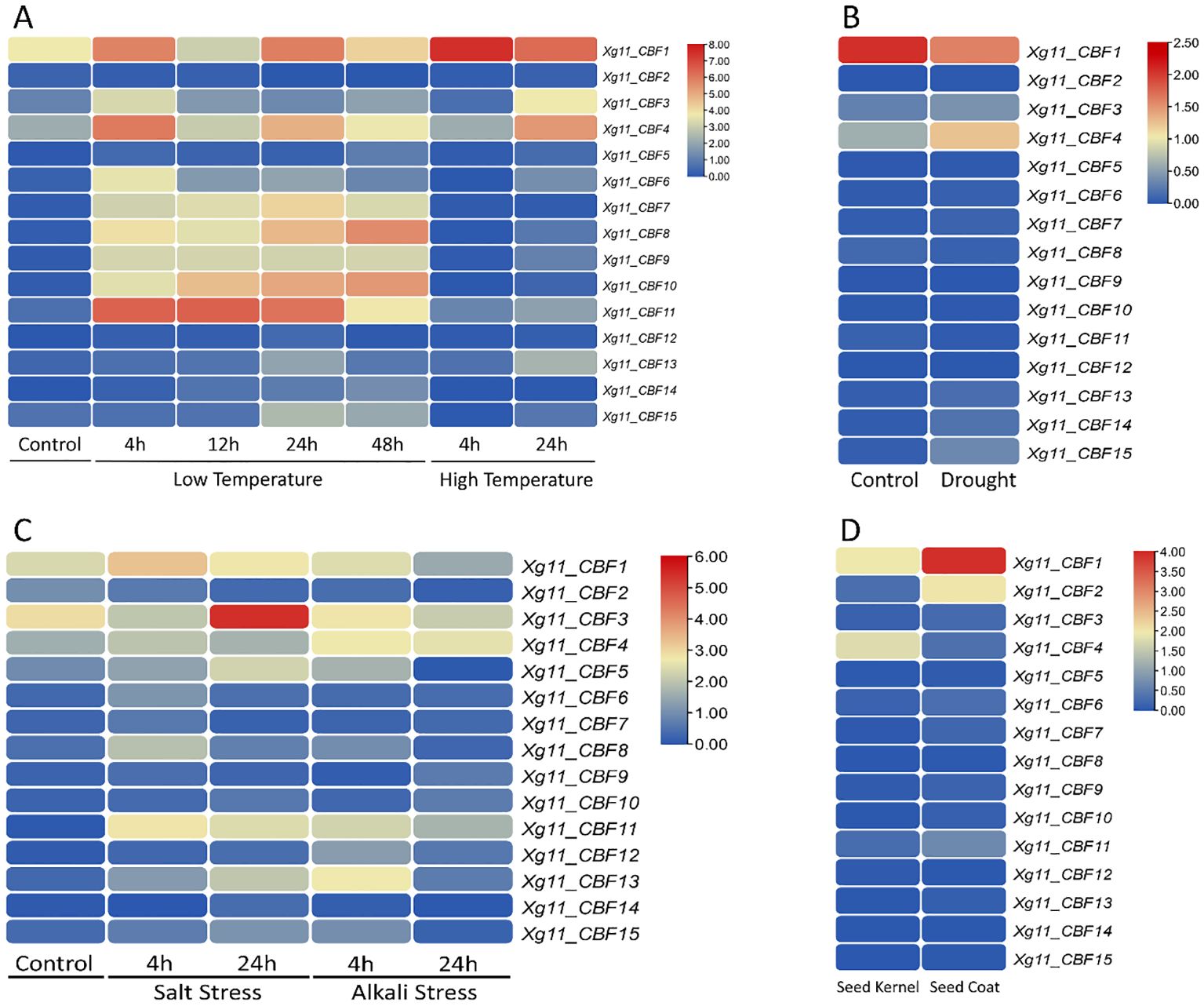
Figure 6. The expression profiles of Xg11 CBF genes in (A) low and high-temperature stress conditions, (B) drought stress, (C) salt, and alkali stress, and (D) in seed kernel and seed coat.
3.8 Phenotype, physical, and chemical characteristics of the transgenes under low-temperature stress
3.8.1 Changes in growth phenotype of A. thaliana seedlings
The expression vector pEGOEP35S-H-Xg11_CBF11 was transformed into A. thaliana plants with the floral-dip method. DNA was extracted and PCR amplified for eight resistant A. thaliana lines, of which 6 lines had a targeted band (Supplementary Figure S6). The expression levels of Xg11_CBF11 in the transgenic plants were checked using qRT-PCR, the expression levels of transgenic L1, L5, L6 lines were higher than other lines (Supplementary Table S2), which were selected for the follow experiment.
The growth of transgenic lines showed a non-significant difference compared to wild type before low-temperature stress. After low-temperature stress, all lines were affected to different degrees, and their leaves turned yellow and withered. At 24 h of cold stress, there was no significant change in the growth of each line. At 48 h of cold stress, the wilting degree of transgenic lines was light than that of wild type, indicating that transgenic A. thaliana lines were more cold-tolerance (Figure 7A).
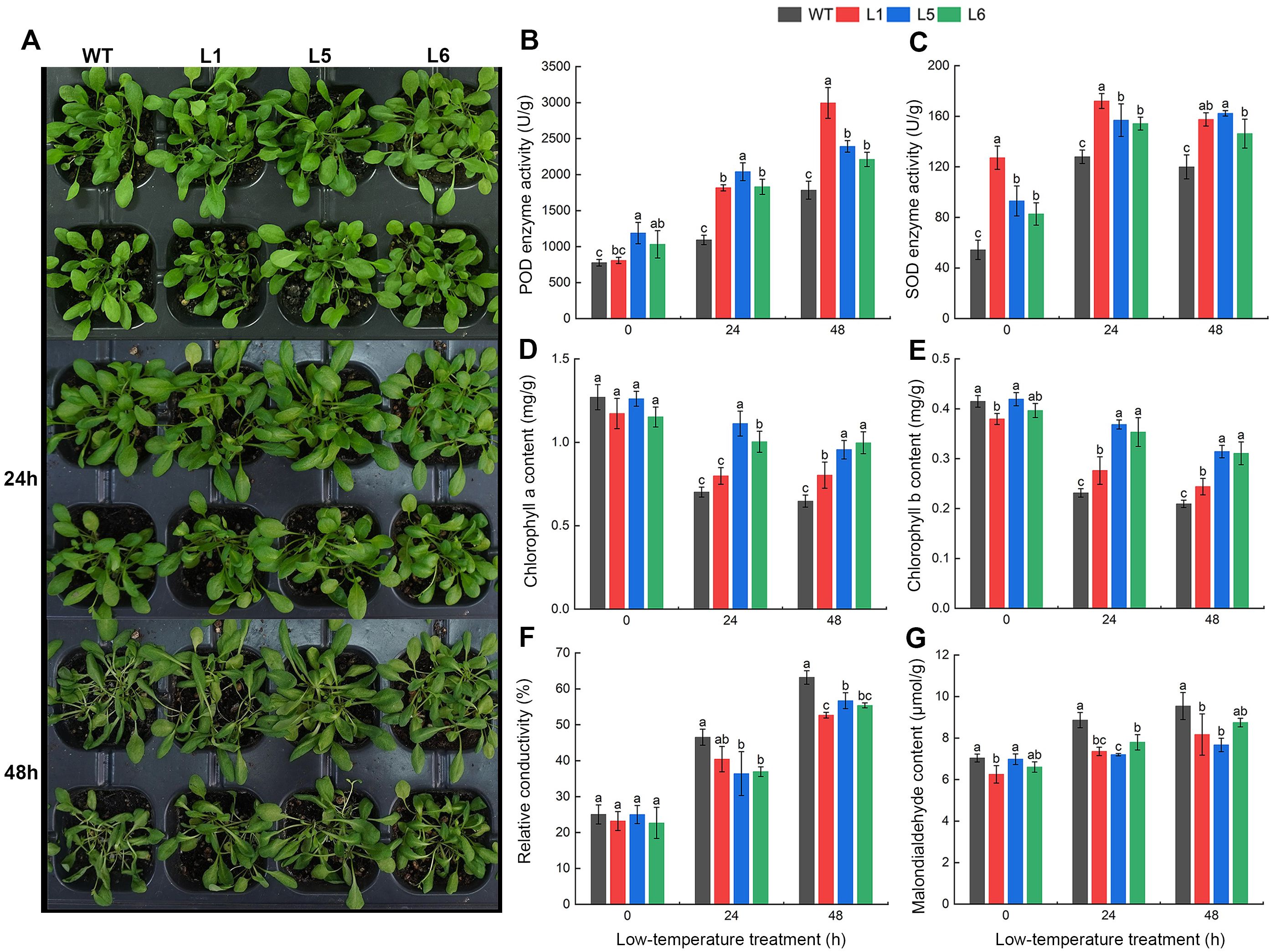
Figure 7. Growth phenotype and physiological indexes in wild-type (WT) and transgenic A. thaliana lines (L1, L5, L6) under 4°C cold stress at 0, 24, and 48h. (A) Growth phenotype. (B) POD enzyme activity. (C) SOD enzyme activity. (D) Chlorophyll a content. (E) Chlorophyll b content. (F) Relative conductivity. (G) Malondialdehyde (MDA) content. Different lowercase letters in the figure indicate significant differences (p < 0.05).
3.8.2 Changes in antioxidant enzyme activity of A. thaliana leaves
Continuous low-temperature stress increased the POD activity in all A. thaliana lines. The POD activity of the three transgenic lines were higher compared to wild type at every time point of low-temperature treatment. After 48 h of cold treatment, WT, L1, L5 and L6 increased by 1007.49 U/g, 2187.45 U/g, 1203.74 U/g and 1179.32 U/g, respectively, suggesting that transgenic lines had a greater increase than that of wild-type lines (Figure 7B). Transgenic lines showed significantly higher POD activity, indicating Xg11_CBF11 enhanced antioxidant defense mechanisms compared to wild-type lines.
The SOD activity of L5 showed an upward trend while the other three lines increased first and then decreased. Meanwhile, the SOD activity of all transgenic lines was significantly higher than that of WT at any treatment time. At 0 h and 24 h of cold treatment, L1 had the highest SOD activity, which was 2.34 times and 1.34 times that of WT, respectively. At 48 h of cold treatment, L5 was the highest at 162.36 U/g, which was 1.35 times higher than that of WT (Figure 7C). Transgenic lines exhibited significantly higher SOD activity, further supporting enhanced antioxidant defenses compared to wild-type plants.
3.8.3 Changes in chlorophyll contents of A. thaliana leaves
The chlorophyll a and chlorophyll b content decreased in all A. thaliana lines with increasing stress time, while the wild-type lines decreased more than the transgenic lines. Under normal culture conditions (0 h), there was no significant difference in chlorophyll indexes of the three transgenic lines compared with the wild-type lines. At 24 h and 48 h of cold treatment, the chlorophyll index of the wild-type lines was significantly lower than that of the transgenic lines (Figures 7D, E). Transgenic lines showed higher chlorophyll content compared to wild-type, indicating better preservation of photosynthetic pigments and maintenance of photosynthesis under stress.
3.8.4 Changes in the membrane permeability of A. thaliana leaves
The relative conductivity of each A. thaliana line was constantly increased during the continuous low-temperature treatment. Under untreated conditions (0 h), the relative conductivity of seedling leaves was low, ranging from 20 to 30%, and the differences among different lines were not obvious. After 48 h of low-temperature stress, the relative conductivity of all lines exceeded 50%, while the wild-type lines were significantly higher than the transgenic lines (Figure 7F). Transgenic lines exhibited lower increases in relative conductivity, indicating reduced cell membrane damage compared to wild-type plants.
With the prolongation of cold stress time, the malondialdehyde (MDA) content in all A. thaliana lines increased continuously. After 48 h of low-temperature treatment, the MDA content of WT, L1, L5, and L6 lines increased by 35.6%, 30.7%, 9.8%, and 32.4%, respectively, compared to before low-temperature treatment (Figure 7G). Transgenic lines exhibited lower MDA content, suggesting reduced lipid peroxidation and cell membrane damage compared to wild-type plants.
4 Discussion
CBF gene families have been identified in several plant cultivars and sequences have also been analyzed in A. thaliana (Novillo et al., 2012), rice (O. sativa) (Cao et al., 2008), ryegrass (L. perenne) (D. Wang et al., 2023), rye (S. cereal) (Jung and Seo, 2018), wheat (T. aestivum) (Mohseni et al., 2012), cotton (G. hirsutum) (Ma et al., 2014), and barley (H. Vulgare) (Choi et al., 2002). In this study, we identified 13, 8, 11, 15, and 12 CBF genes from the five yellowhorn cultivars (Xzs4, Xwf8, Xjg, Xg11, and Xzg2). Only eight genes were present in all yellowhorn varieties. Five genes were dispensable genes present in different cultivars but not in all. Two unique genes were present in only one variety, Xg11. A similar phenomenon was observed in other cultivars also. For example, in a recent maize pan-genome-wide study, 20 out of 30 genes were present in all varieties (Sun et al., 2023). Similarly, the rice TPS gene family analysis showed that one gene was absent in the reference genome (Sun et al., 2022). Phylogenetic analysis revealed that among the cultivars under study, yellowhorn CBF sequences are more closely related to those of T. aestivum, O. sativa, and H. vulgare as compared to others. The protein length of all five yellowhorn cultivars is almost equal to the length of reported CBF genes in other plants. Most of the CBFs were localized in the nucleus and few localized in the cytoplasm, and mitochondria, while in S. cereale the CBFs were localized in the same subcellular localization. Furthermore, the phylogenetic tree revealed that all the yellowhorn CBFs were present in three groups. The same trend was observed in L. perenne. (D. Wang et al., 2023). The gene structure and motif patterns were almost conserved in all CBFs. Motif 7 was only conserved in the Xg11 and motif 6 was only conserved in the Xzg2 cultivars. The motif conservation pattern of all five yellowhorn cultivars is similar to the L. perenne (D. Wang et al., 2023) and S. cereale (Jung and Seo, 2018). CBFs were distributed unevenly on chromosomes for all five yellowhorn cultivars. Tandem duplication is the major factor behind the duplication of CBF genes and these are duplicated about 0.35-5.07 Mya ago. This trend of chromosomal distribution and duplication was also similar in O. sativa (Cao et al., 2008), S. cereal (Jung and Seo, 2018), T. aestivum (Mohseni et al., 2012), G. hirsutum (Ma et al., 2014), and H. Vulgare (Choi et al., 2002). The cis-regulatory elements in the promoter region, the PPI, and GO insisted on the potential involvement of these CBF genes in abiotic stress responsiveness. Previously breakthroughs have been made in the mechanism of cold tolerance in plants, among which ICE-CBF-COR signaling cascade pathway is one of the main cold tolerance pathways in higher plants and plays a crucial role under cold stress (Feng et al., 2020). The expression profiling of these genes in low and high temperature; drought; salt and alkali; and in seed kernel and coat showed a similar expression pattern of these genes as shown in previous studies. It has been reported that AtCBF2 negatively regulated AtCBF3 and AtCBF1, while AtCBF4 functioned in drought stress tolerance (Haake et al., 2002). Similarly, AcerApseCBFs, AcyanCBF2/4, AtruCBF5 genes are involved in drought inducibility (Zhao et al., 2023). Moreover, the core genes present in all yellowhorn varieties were showing a change in expression pattern, and potential involvement of these genes in stress responsiveness. While, for dispensable genes, a higher expression of Xg11_CBF7-11 was observed which showed these genes are potentially involved in temperature stress responsiveness in Xg11 variety.
In nature, plants often encounter various abiotic stresses, among which low-temperature is a very important environmental stress factor, which is an important obstacle in agricultural and forestry production in China. Compared with animals, plants themselves cannot move freely, so a system of antioxidant protective enzymes has evolved to reduce stress damage. POD is one of the key enzymes of the enzymatic defense system in plants under stress conditions, and the enzyme activity rises when encountering stress. The study found that the POD enzyme activity was significantly higher than that of nontransgenic tobacco, which improved the cold resistance of tobacco (Gu et al., 2021). In this study, the POD enzyme activity in the transgenic A. thaliana lines was continuously increased under cold stress, and the increase was significantly higher than that in the wild-type lines. However, the change trend of SOD enzyme is complex and does not always rise under stress. The SOD enzyme activity may decline continuously, fall first and then rise, or remain unchanged. In this study, the SOD enzyme activity in the L5 lines rose linearly with the duration of low-temperature stress, while the WT, L1, and L6 lines increased first and then decreased. Low-temperature stress affects the normal growth and development of plants, reducing the effective photosynthetic area, resulting in blocked chlorophyll synthesis and reduced content (Song et al., 2021). After silencing CBF1 in cucumber, the chlorophyll content in gene-silenced plants decreased more than in wild-type plants, indicating that the CBF1 gene plays an important role in cold stress (Gupta et al., 2012). In this study, the chlorophyll content of both transgenic and wild-type A. thaliana lines was gradually decreased under continuous low-temperature stress, indicating that low-temperature inhibited the normal photosynthesis of A. thaliana; at 24 h and 48 h, the chlorophyll content of the transgenic lines was higher than that of the wild-type line, with a smaller reduction. Therefore, it can be speculated that the Xg11_CBF11 gene has a protective effect on chlorophyll to ensure the chlorophyll content.
Low-temperature stress will cause peroxidation of cell membrane lipids, increase membrane permeability, and lead to an increase in relative conductivity and malondialdehyde content. Therefore, the degree of cell membrane damage is usually measured by the relative conductivity and malondialdehyde content (Dong et al., 2020). The species-specific CBF genes in yellowhorn cultivars may contribute to physiological responses such as improved antioxidant enzyme activity, reduced membrane damage, and higher chlorophyll retention under low temperatures. The phenotypic differences observed among the five yellowhorn cultivars may enhance the cold tolerance through species-specific motifs and expression profiles. Thus, the species-specific genes not only contribute to cold tolerance but may also play a role in defining the unique adaptive traits of each cultivar. The overexpression of A. thaliana CBF1 gene in the transgenic lines changed the relative conductivity and malondialdehyde content less than that of the non-transgenic control verifying the cold resistance function of the A. thaliana CBF1 gene (Akhtar et al., 2012). Similarly, the A. thaliana CBF1 gene was transformed into grape and tobacco (Park et al., 2001; Siddiqua and Nassuth, 2011), and the relative conductivity of the transgenic lines was also lower than the non-transgenic lines after cold stress, which announced the cold resistance of grape and tobacco. Similar results were obtained in this study, and the relative conductivity and malondialdehyde in transgenic lines were less elevated than those in wild-type lines, indicating that the Xg11_CBF11 gene protected the cell membrane of A. thaliana and reduced the degree of damage.
5 Conclusion
This study has identified and characterized the CBF gene family across five yellowhorn cultivars, providing new insights into their structural and functional diversity. Phylogenetic and motif analyses highlighted both conserved features and species-specific adaptations among these cultivars, with gene duplication, especially tandem duplications, playing a significant role in the expansion of this gene family. Expression profiling revealed the involvement of these genes in abiotic stress responses, particularly under cold conditions. Functional validation of the Xg11_CBF11 gene in transgenic A. thaliana demonstrated its positive role in enhancing cold tolerance, as evidenced by increased antioxidant enzyme activity, maintained chlorophyll levels, and reduced cellular damage. These findings enrich our understanding of the molecular mechanisms behind cold stress tolerance in yellowhorn and provide potential targets for further genetic improvement in this species. This research contributes to the broader effort of improving plant resilience to environmental stress, with practical implications for agricultural and forestry applications.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
JW: Writing – original draft. XL: Writing – review & editing. WZ: Writing – review & editing. AK: Writing – review & editing. YW: Writing – review & editing. SL: Writing – review & editing. MQ: Writing – review & editing, Validation, Conceptualization. XW: Writing – review & editing. JG: Writing – review & editing, Validation, Supervision, Software, Resources, Project administration, Methodology, Funding acquisition, Conceptualization.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was funded by Basic Research Program of Shanxi Province (202103021224168); Shanxi Province doctoral graduates and postdoctoral researchers come to work in Shanxi Province to reward the fund scientific research project (SXBYKY2021061); Shanxi Agricultural University Doctoral Research Initiation Project (2021BQ17).
Acknowledgments
The authors would like to acknowledge Shanxi Agricultural University for providing necessary facilities for this research.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1481358/full#supplementary-material
References
Akhtar, M., Jaiswal, A., Taj, G., Jaiswal, J. P., Qureshi, M. I., Singh, N. K. (2012). DREB1/CBF transcription factors: their structure, function and role in abiotic stress tolerance in plants. J. Genet. 91, 385–395. doi: 10.1007/s12041-012-0201-3
Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Bailey, T. L., Johnson, J., Grant, C. E., Noble, W. S. (2015). The MEME suite. Nucleic Acids Res. 43, W39–W49. doi: 10.1093/nar/gkv416
Cao, Y., Wang, J., Guo, L., Xiao, K. (2008). Identification, characterization and expression analysis of transcription factor (CBF) genes in rice (Oryza sativa L.). Front. Agric. China 2, 253–261. doi: 10.1007/s11703-008-0052-0
Chen, C., Rui, X., Hao, C., He, Y. (2018). TBtools, a Toolkit for Biologists integrating various HTS-data handling tools with a user-friendly interface. TBtools Toolkit Biologists integrating various HTS-data handling Tools user-friendly Interface 13, 289660. doi: 10.1101/289660
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, J.-Q., Dong, Y., Wang, Y., Liu, Q., Zhang, J., Chen, S., et al. (2003). An AP2/EREBP-type transcription-factor gene from rice is cold-inducible and encodes a nuclear-localized protein. TAG. Theor. Appl. Genet. Theoretische und angewandte Genetik 107, 972–979. doi: 10.1007/s00122-003-1346-5
Chinnusamy, V., Zhu, J.-K., Sunkar, R. (2010). Gene regulation during cold stress acclimation in plants. Methods Mol. Biol. (Clifton N.J.) 639, 39–55. doi: 10.1007/978-1-60761-702-0_3
Choi, D. W., Rodriguez, E. M., Close, T. J. (2002). Barley CBF3 gene identification, expression pattern, and map location. Plant Physiol. 129, 1781–1787. doi: 10.1104/pp.003046
Ding, B., Liu, T., Hu, Y., Song, Y., Hao, R., Feng, X., et al. (2021). Comparative analysis of transcriptomic profiling to identify genes involved in the bulged surface of pear fruit (Pyrus bretschneideri Rehd. cv. Yuluxiangli). Physiol. Mol. Biol. Plants 27, 69–80. doi: 10.1007/s12298-021-00929-z
Ding, J., Karim, H., Li, Y., Harwood, W., Guzmán, C., Lin, N., et al. (2021). Re-examination of the APETALA2/Ethylene-Responsive factor gene family in Barley (Hordeum vulgare L.) indicates a role in the regulation of starch synthesis. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.791584
Ding, B., Hu, H., Cao, Y., Xu, R., Lin, Y., Muhammad, T. U. Q., et al. (2024). Pear genomes display significant genetic diversity and provide novel insights into the fruit quality traits diferentiation. Hortic Plant J. 10, 1274–1290. doi: 10.1016/j.hpj.2024.05.005
Dong, X., Yan, Y., Jiang, B., Shi, Y., Jia, Y., Cheng, J., et al. (2020). The cold response regulator CBF1 promotes Arabidopsis hypocotyl growth at ambient temperatures. EMBO J. 39, e103630. doi: 10.15252/embj.2019103630
Dubouzet, J. G., Dubouzet, J., Sakuma, Y., Ito, Y., Kasuga, M., Dubouzet, E. G., et al. (2003). OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant journal : Cell Mol. Biol. 33, 751–763. doi: 10.1046/j.1365-313x.2003.01661.x
Fatima, K., Sadaqat, M., Azeem, F., Rao, M. G., Alshammari, A., Tahir, M. (2023). Integrated omics and machine learning-assisted profiling of cysteine-rich-receptor-like kinases from three peanut spp. revealed their role multiple stresses. Front. Genet. 14. doi: 10.3389/fgene.2023.1252020
Feng, K., Hou, X., Xing, G., Liu, J., Duan, A., Xu, Z., et al. (2020). Critical reviews in biotechnology advances in AP2 / ERF super-family transcription factors in plant. Crit. Rev. Biotechnol. 40, 1–27. doi: 10.1080/07388551.2020.1768509
Gao, M.J., Allard, G., Byass, L., Flanagan, A., Singh, J. (2002). Regulation and characterization of four CBF transcription factors from Brassica napus. Plant Mol. Biol. 49, 459–471. doi: 10.1023/a:1015570308704
Gasteiger, E., Hoogland, C., Gattiker, A., Duvaud, S., Wilkins, M. R., Appel, R. D., et al. (2005). Protein identification and analysis tools on the ExPASy server. Proteomics Protoc. Handb., 571–607. doi: 10.1385/1-59259-890-0:571
Geer, L. Y., Marchler-Bauer, A., Geer, R. C., Han, L., JHe, J., He, S., et al. (2010). The NCBI biosystems database. Nucleic Acids Res. 38, D492–D496. doi: 10.1093/nar/gkp858
Giri, M. K., Swain, S., Gautam, J. K., Singh., S., Singh., N., Bhattacharjee, L., et al. (2014). The Arabidopsis thaliana At4g13040 gene, a unique member of the AP2/EREBP family, is a positive regulator for salicylic acid accumulation and basal defense against bacterial pathogens. J. Plant Physiol. 171, 860–867. doi: 10.1016/j.jplph.2013.12.015
Gu, K., Hou, S, Chen, J., Guo, J., Wang, F., He, C., et al. (2021). The physiological response of different tobacco varieties to chilling stress during the vigorous growing period. Sci. Rep. 11, 22136. doi: 10.1038/s41598-021-01703-7
Gupta, N., Rathore, M., Goyary, D., Khare, N., Anandhan, S., Pande, V., et al. (2012). Marker-free transgenic cucumber expressing Arabidopsis CBF1 gene confers chilling stress tolerance. Biol. Plantarum 56, 57–63. doi: 10.1007/s10535-012-0016-3
Haake, V., Cook, D., Riechmann, J., Pineda, O., Thomashow, M. F., Zhang, J. Z. (2002). Transcription factor CBF4 is a regulator of drought adaptation in Arabidopsis. Plant Physiol. 130, 639–648. doi: 10.1104/pp.006478
Horton, P., Park, K. J., Obayashi, T., Fujita, N., Harada, H., Adams-Collier, C. J., et al. (2007). WoLF PSORT: protein localization predictor. Nucleic Acids Res. 35, W585–W587. doi: 10.1093/nar/gkm259
Hu, H., Li, R., Zhao, J., Batley, J., Edwards, D. (2024). Technological development and advances for constructing and analyzing plant pangenomes. Genome Biol. Evol. 16, evae081. doi: 10.1093/gbe/evae081
Hu, F., Zhang, Y., Guo, J. (2022). Identification and characterization of AP2/ERF transcription factors in Yellow Horn. Int. J. Mol. Sci. 23, 14991. doi: 10.3390/ijms232314991
Jia, Y., Xu, M., Hu, H., Chapman, B., Watt, C. B., Buerte, B., et al. (2023). Comparative gene retention analysis in barley, wild emmer, and bread wheat pangenome lines reveals factors affecting gene retention following gene duplication. BMC Biol. 21, 25. doi: 10.1186/s12915-022-01503-z
Jung, W. J., Seo, Y. W. (2018). Identification of novel C-repeat binding factor (CBF) genes in rye (Secale cereale L.) and expression studies. Gene 684, 82–94. doi: 10.1016/j.gene.2018.10.055
Kagaya, Y., Ohmiya, K., Hattori, T. (1999). RAV1, a novel DNA-binding protein, binds to bipartite recognition sequence through two distinct DNA-binding domains uniquely found in higher plants. Nucleic Acids Res. 27, 470–478. doi: 10.1093/nar/27.2.470
Kim, D., Paggi, J. M., Park, C., Bennett, C., Salzberg, S. L. (2019). Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915. doi: 10.1038/s41587-019-0201-4
Kumar, S., Stecher, G., Tamura, K. (2016). MEGA7 : Molecular evolutionary genetics analysis version 7. 0 for bigger datasets brief communication. Mol Biol Evol. 33, 7, 1870–1874. doi: 10.1093/molbev/msw054
Letunic, I., Bork, P. (2021). Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296. doi: 10.1093/nar/gkab301
Li, X. Z., Chen, C. C., Kang, X. (2022). Research on the cultivation of sustainable development ability of higher vocational students by creative thinking teaching method. Front. Psychol. 13. doi: 10.3389/fpsyg.2022.979913
Lv, K., Li, J., Zhao, K., Chen, S., Nie, J., Zhang, W., et al. (2020). Overexpression of an AP2/ERF family gene, BpERF13, in birch enhances cold tolerance through upregulating CBF genes and mitigating reactive oxygen species. Plant Sci. 292, 110375. doi: 10.1016/j.plantsci.2019.110375
Ma, L. F., Zhang, J., Huang, G., Li, Y., Li, X., Zheng, Y., et al. (2014). Molecular characterization of cotton C-repeat/dehydration-responsive element binding factor genes that are involved in response to cold stress. Mol. Biol. Rep. 41, 4369–4379. doi: 10.1007/s11033-014-3308-1
Ma, Y., Sun, J., Zhang, X., Sadaqat, M., Tahir, M., Liu, T., et al. (2024). Comparative genomics analysis of pheophorbide a oxygenase (PAO) genes in eight pyrus genomes and their regulatory role in multiple stress responses in Chinese pear (Pyrus bretschneideri). Front. Genet. 15. doi: 10.3389/fgene.2024.1396744
Marchler-Bauer, A., Derbyshire, M. K., Gonzales, N. R., Lu, S., Chitsaz, F., Geer, L. Y., et al. (2015). CDD: NCBI’s conserved domain database. Nucleic Acids Res. 43, D222–D226. doi: 10.1093/nar/gku1221
Mering, C., Huynen, M., Jaeggi, D., Schmidt, S., Bork, P., Snel, B., et al. (2003). STRING: a database of predicted functional associations between proteins. Nucleic Acids Res. 31, 258–261. doi: 10.1093/nar/gkg034
Mistry, J., Chuguransky, S., Williams, L., Qureshi, M., Salazar, G. A., Sonnhammer, E. L. L., et al. (2021). Pfam: The protein families database in 2021. Nucleic Acids Res. 49, D412–D419. doi: 10.1093/nar/gkaa913
Mohseni, S., Che, H., Djillali, Z., Dumont, E., Nankeu, J., Danyluk, J. (2012). Wheat CBF gene family: identification of polymorphisms in the CBF coding sequence. Genome 55, 865–881. doi: 10.1139/gen-2012-0112
Novillo, F., Medina, J., Rodríguez-Franco, M., Neuhaus, G., Salinas, J. (2012). Genetic analysis reveals a complex regulatory network modulating CBF gene expression and Arabidopsis response to abiotic stress. J. Exp. Bot. 63, 293–304. doi: 10.1093/jxb/err279
Park, J. M., Park, C. J., Lee, S. B., Ham, B. K., Shin, R., Paek, K. H. (2001). Overexpression of the tobacco Tsi1 gene encoding an EREBP/AP2-type transcription factor enhances resistance against pathogen attack and osmotic stress in tobacco. Plant Cell 13, 1035–1046. doi: 10.1105/tpc.13.5.1035
Pertea, M., Kim, D., Pertea, G. M., Leek, J. T., Salzberg, S. L. (2016). Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 11, 1650–1667. doi: 10.1038/nprot.2016.095
Pontaroli, A. C., Rogers, R. L., Zhang, Q., Shields, M. E., Davis, T. M., Folta, K. M., et al. (2009). Gene content and distribution in the nuclear genome of Fragaria vesca. Plant Genome 2, 93–101. doi: 10.3835/plantgenome2008.09.0007
Rombauts, S., Déhais, P., Montagu, M. V., Rouzé, P. (1999). PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Res. 27, 295–296. doi: 10.1093/nar/27.1.295
Rozas, J., Ferrer-Mata, A., Sánchez-DelBarrio, J. C., Guirao-Rico, S., Librado, P., Ramos-Onsins, S. E., et al. (2017). DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 34, 3299–3302. doi: 10.1093/molbev/msx248
Sadaqat, M., Umer, B., Attia, K. A., Abdelkhalik, A. F., Azeem, F., Javed, M. R., et al. (2023). Genome-wide identification and expression profiling of two-component system (TCS) genes in Brassica oleracea in response to shade stress. Front. Genet. 14, 1142544. doi: 10.3389/fgene.2023.1142544
Sadaqat, M., Fatima, K., Azeem, F., Shaheen, T., Rahman, M., Ali, T., et al. (2024). Computational analysis and expression profiling of Two-component system (TCS) gene family members in Mango (Mangifera indica) indicated their roles in stress response. Funct. Plant Biol. 51. doi: 10.1071/FP24055
Schultz, J., Copley, R. R., Doerks, T., Ponting, C. P., Bork, P. (2000). SMART: A web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 28, 231–234. doi: 10.1093/nar/28.1.231
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Siddiqua, M., Nassuth, A. (2011). Vitis CBF1 and Vitis CBF4 differ in their effect on Arabidopsis abiotic stress tolerance, development and gene expression. Plant Cell Environ. 34, 1345–1359. doi: 10.1111/j.1365-3040.2011.02334.x
Song, Y., Zhang, X., Li, M., Yang, H., Fu, D., Lv, J., et al. (2021). The direct targets of CBFs: In cold stress response and beyond. J. Integr. Plant Biol. 63, 1874–1887. doi: 10.1111/jipb.13161
Sun, Y., Zhang, P., Kou, D., Han, Y., Fang, J., Ni, J., et al. (2022). Terpene synthases in rice pan-genome and their responses to Chilo suppressalis Larvae Infesting. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.905982
Sun, Y., Xiao, W., Wang, Q., Wang, J., Kong, X., Ma, W., et al. (2023). Multiple variation patterns of terpene synthases in 26 maize genomes. BMC Genomics 24, 1–10. doi: 10.1186/s12864-023-09137-3
Tahir, M., Sadaqat, M., Zhu, X., Li, H., Huang, X., Fatima, K., et al. (2023). Comparative genomics profiling revealed multi-stress responsive roles of the CC-NBS-LRR genes in three mango cultivars. Front Plant Sci 14, 1–20. doi: 10.3389/fpls.2023.1285547
Törönen, P., Medlar, A., Holm, L. (2018). PANNZER2: A rapid functional annotation web server. Nucleic Acids Res. 46, W84–W88. doi: 10.1093/nar/gky350
Trifinopoulos, J., Nguyen, L. T., Haeseler, A. V., Minh, B. Q. (2016). W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44, W232–W235. doi: 10.1093/nar/gkw256
Verma, R. K., Santosh, V. V., Yadav, S. K., Kumar, T. S., Rao, M. V., Chinnusamy, V., et al. (2020). Overexpression of Arabidopsis ICE1 enhances yield and multiple abiotic stress tolerance in indica rice. Plant Signaling Behav. 15, 1814547. doi: 10.1080/15592324.2020.1814547
Wang, D., Cui, B., Guo, H., Liu, Y., Nie, S. (2023). Genome-wide identification and expression analysis of the CBF transcription factor family in Lolium perenne under abiotic stress. Plant Signaling Behav. 18, 2086733. doi: 10.1080/15592324.2022.2086733
Wang, J., Hu, H., Liang, X., Tahir, M., Zhang, Y., Zhao, J., et al. (2023). High-quality genome assembly and comparative genomic profiling of yellowhorn (Xanthoceras sorbifolia) revealed environmental adaptation footprints and seed oil contents variations. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1147946
Wang, J., Hu, H., Jiang, X., Zhang, S., Yang, W., Dong, J., et al. (2024). Pangenome-wide association study and transcriptome analysis reveal a novel QTL and candidate genes controlling both panicle and leaf blast resistance in rice. Rice 17, 27. doi: 10.1186/s12284-024-00707-x
Wingett, S. W., Andrews, S. (2018). FastQ Screen: A tool for multi-genome mapping and quality control. F1000Research 7, 1338. doi: 10.12688/f1000research.15931.2
Yang, S., Yu, X., Gao, X., Fatima, K., Tahir, M. (2024). Comparative genomic profiling of transport inhibitor Response1/Auxin signaling F-box (TIR1/AFB) genes in eight Pyrus genomes revealed the intraspecies diversity and stress responsiveness patterns. Front. Genet. 15. doi: 10.3389/fgene.2024.1393487
Yu, Y., Liu, A., Duan, X., Wang, S., Sun, X., Duanmu, H., et al. (2016). GsERF6, an ethylene-responsive factor from Glycine soja, mediates the regulation of plant bicarbonate tolerance in Arabidopsis. Planta 244, 681–698. doi: 10.1007/s00425-016-2532-4
Zameer, R., Fatima, K., Azeem, F., Algwaiz, H. I. M., Sadaqat, M., Rasheed, A., et al. (2022). Genome-wide characterization of superoxide dismutase (SOD) genes in Daucus carota: novel insights into structure, expression, and binding interaction with hydrogen peroxide (H2O2) under abiotic stress condition. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.870241
Zhao, Q., Han, R., Cai, K., Yan, H., Li, Y., Qu, G., et al. (2023). Identification and analysis of the CBF gene family in three species of acer under cold stress. Int. J. Mol. Sci. 24, 2088. doi: 10.3390/ijms24032088
Keywords: Xanthoceras sorbifolium, pangenome-wide, gene ontology, low-temperature stress, physiological response, transgene
Citation: Wang J, Liang X, Zhang W, Khalil A, Wu Y, Liu S, Tahir ul Qamar M, Wang X and Guo J (2024) Comparative genomic profiling of CBFs pan-gene family in five yellowhorn cultivars and functional identification of Xg11_CBF11. Front. Plant Sci. 15:1481358. doi: 10.3389/fpls.2024.1481358
Received: 15 August 2024; Accepted: 10 October 2024;
Published: 19 November 2024.
Edited by:
Yong Jia, Murdoch University, AustraliaReviewed by:
Muhammad Amjad Nawaz, Tomsk State University, RussiaXiangshu Dong, Yunnan University, China
Copyright © 2024 Wang, Liang, Zhang, Khalil, Wu, Liu, Tahir ul Qamar, Wang and Guo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinping Guo, amlucGd1b0AxMjYuY29t; Muhammad Tahir ul Qamar, dGFoaXJ1bHFhbWFyQGdjdWYuZWR1LnBr
 Juan Wang1
Juan Wang1 Xizhen Liang
Xizhen Liang Asma Khalil
Asma Khalil Muhammad Tahir ul Qamar
Muhammad Tahir ul Qamar Jinping Guo
Jinping Guo