- Department of Crop Genetics, John Innes Centre, Norwich, United Kingdom
Winter annual crops are sown in late summer or autumn and require chilling to promote flowering the following spring. Floral initiation begins in autumn and winter, and in winter oilseed rape (OSR), continued chilling during flower development is necessary for high yield potential. This can be a problem in areas where chilling is not guaranteed, or as a result of changing climates. Here, we used chilling disruption and low chilling to identify loci with the potential to increase chilling efficiency in winter OSR. We report that time to flowering and yield potential under low chill conditions are affected by variation at the PLANT HOMOLOGOUS TO PARAFIBROMIN gene, a component of the plant PAF1c complex. We show that increases in winter chilling given to developing flowers can improve seed yields and that loss of function of BnaPHP.A05 leads to early flowering in B. rapa and B. napus and an increase in seed set where chilling is limited. Because PHP is known to specifically target the FLOWERING LOCUS C (FLC) gene in Arabidopsis, we propose that variation at PHP is useful for breeding modifications to chilling responses in polyploid crops with multiple copies of the FLC gene.
Introduction
Winter annual crops are widely grown in Europe for their high yields. In the Brassica family, winter annual habit is conferred by high expression of orthologs of Arabidopsis FLOWERING LOCUS C (FLC), which prevent flowering during the longer days of early autumn but are silenced epigenetically in response to prolonged chilling (Tadege et al., 2001; Hou et al., 2012; Yin et al., 2020; Calderwood et al., 2021). FLC genes also play a role in the modulation of flowering time in rapid-cycling Brassica such as B. rapa (Wu et al., 2012). In winter annual Brassica napus and in Arabidopsis thaliana, vernalization occurs during the lower temperatures of mid-autumn rather than the cold temperatures of winter, followed by floral initiation, which takes place under chilling conditions in the short days of late autumn or early winter (Hepworth et al., 2018; O’Neill et al., 2019; Matar et al., 2021). Silencing of FLC genes requires the recruitment of POLYCOMB REPRESSIVE COMPLEX 2 (PRC2) to intron 1 of FLC and facultative heterochromatin formation (Questa et al., 2016). On the other hand, active transcription by RNA polymerase II attracts deposition of active epigenetic marks via the activity of the PAF1c complex, which recruits histone methyltransferases to sites of POLII transcription, including FLC (Park et al., 2010). For unclear reasons, one subunit of the eukaryotic PAF1c complex known variously as CDC73 or PARAFIBROMIN appears to have a more specialized role in plants and is specifically required for active FLC and FLC-related gene transcription (Nasim et al., 2022).
After floral initiation, at least two isoforms of FLC remain expressed in developing inflorescence meristems in winter oilseed rape (OSR). Further chilling during winter silences these FLCs, accelerates flowering, and is also necessary for normal inflorescence development (Lu et al., 2022). In contrast, warming during flower development in winter resulted in a delay to flower bud growth and increased abscisic acid levels indicative of the induction of bud dormancy. Partial fulfillment of the winter chilling requirement is associated with yield declines in a wide range of temperate crops (Atkinson et al., 2013). In the United Kingdom and in China, chilling during early winter is unreliable and reduced chilling correlates with low OSR yields (He et al., 2017; Brown et al., 2019). Climate change is likely to result in a decrease in available winter chill in temperate western Europe and OSR yields globally are vulnerable to changing weather between growing seasons (Rondanini et al., 2012; Campoy et al., 2019), meaning that plant breeding is required to adapt winter OSR to warmer winter environments without compromising yield. Adaptation to lower chill environments requires genotypes that can delay flower initiation until the short days of winter while being reliably chilled at a wider range of chilling temperatures.
Previously, we described a variety trial designed to investigate the genetic control of flowering time in B. napus (Lu et al., 2022). In this experiment, a panel of winter, spring, and semi-winter OSR and swedes were grown with sequential sowing such that floral initiation began at the start of winter in Norwich UK in each crop type. The panel was then given control cold winters or a winter warming treatment during flower development, and the effect on flowering time was observed. Our data showed that interrupting flower bud chilling during development initiated a type of winter bud dormancy in lines with high FLC expression in reproductive tissues. Here, we use association transcriptomics to further dissect the control of bud dormancy and flowering time in B. napus and reveal a role for PLANT HOMOLOGOUS TO PARAFIBROMIN (PHP) in determining the efficiency of winter chilling in B. napus. Because of the known relationship between PHP and FLC gene function during chilling in Arabidopsis (Nasim et al., 2022), we further explored its role in bud development in B. napus and show that weak PHP alleles are associated with increased seed production in laboratory experiments where chilling is limited, suggesting that weak alleles of PHP may be useful for crop adaptation to warming winters.
Results
Identification of an association between BnaPHP.A05 genotype and flower bud responses to chilling
Previous work suggested that effects of winter temperature variation on flower bud development depend on the crop type and are correlated with the FLC genotype (Lu et al., 2022). To further understand how temperature variation affects winter OSR flowering behavior, we used Associative Transcriptomics (Havlickova et al., 2018) to uncover SNP variation genetically linked to the effects of flower bud warming on the timing of flowering (Figure 1A). We used mean time from floral initiation to BBCH60 in control and warmed treatments plus the ratio between the two measures to uncover loci important on the control of OSR flower bud behavior in response to temperature. From this work, we found several loci with putative associations with flowering time in either the control or warmed treatments, or the effect of temperature on flowering (Figure 1). This included one SNP close to BnaAGL24.A01 on chromosome 1. Exome capture data (Williams et al., 2023) revealed that BnaAGL24.A01 (BnaA01g13920) has deletions primarily in a small number of Chinese semi-winter lines or unusual spring OSR with fast flowering (Supplementary Table S1), but further analysis is complicated by the small number of variant varieties in each crop type in our dataset. A second locus on chromosome A05 had a single SNP associated with the effect of warming on degree days to BBCH60 (Figure 1A). Further analysis of the gene implicated on chromosome A05 revealed that this single-nucleotide polymorphism (SNP) is in one of two orthologs of Arabidopsis PHP, encoding a component of the Polymerase-Associated Factor 1 complex (PAF1C). PAF1C catalyzes transcription elongation and the interaction of RNA polymerase II with complexes that deposit the active epigenetic marks H3K4 and H3K36 at target gene chromatin, and in so doing promotes active transcription. Loss of PAF1C in Arabidopsis leads to a highly pleiotropic syndrome consistent with widespread disruption in transcriptional control (Nasim et al., 2022). However, PHP is unusual among PAF1C complex proteins in that loss-of-function mutations appear only to reduce FLC transcription, causing early flowering in both rapid-cycling and vernalization-requiring Arabidopsis accessions (Yu and Michaels, 2010; Park et al., 2010). Brassica napus has two orthologs of the plant PHP gene with a second gene found on chromosome C05, but only the A05 gene was implicated in flowering time control. The time from floral initiation to flowering in the control treatment was further associated with two genes with likely roles in plastid function based on homology with Arabidopsis genes, whereas time to flowering in the warmed bud treatment was associated with a gene of unknown function on chromosome A06. These were not followed up in more detail in this study.
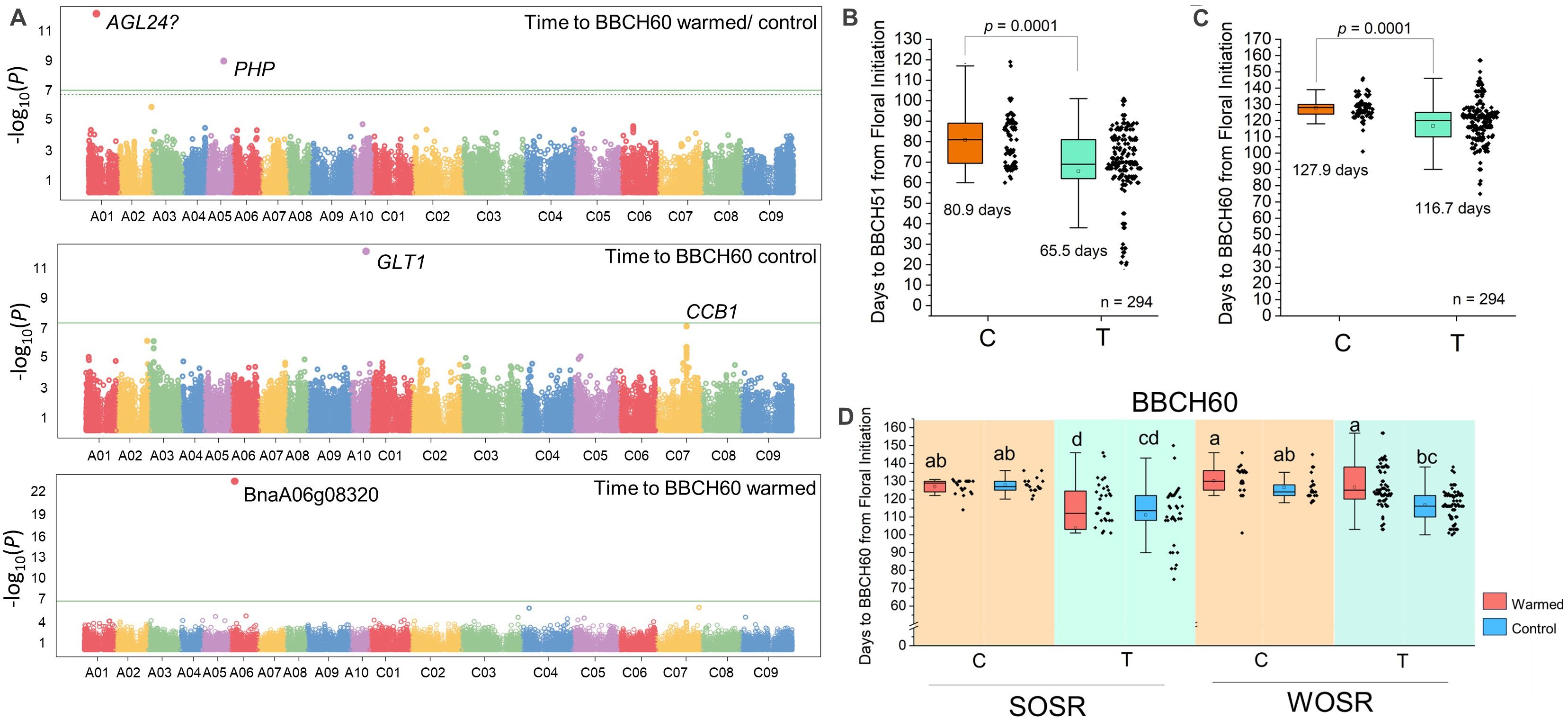
Figure 1. Identification of variation at BnaPHP.A05 that correlates with the effect of winter temperature variation applied to developing flower buds on flowering time in OSR. (A) Manhattan plots reveal loci at which variation correlates with time to BBCH60 (first flower) from floral initiation in either the control winter, warm winter, or the ratio of the two treatments (see Materials and Methods). Significant SNP associations after Bonferroni correction are shown above the horizontal green line. In addition to BnaPHP.A05, other loci identified but not analyzed further were orthologs of A. thaliana AGAMOUS-LIKE 24 (AGL24), GLUCOSE TRANSPORTER 1 (GLT1), COFACTOR OF COMPLEX ASSEMBLY 1 (CCB1), and the unknown protein encoded by BnaA06g08320D. (B, C) The association between the two identified variants of SNP Cab041204.2:750 (Havlickova et al., 2018) at the BnaPHP.A05 locus and time from floral initiation to the flowering stages BBCH51 (buds visible; B) and BBCH60 (first flowering; C) combining flowering data from all the varieties and treatments. Mean times to flowering stages for varieties with each BnaPHP.A05 SNP call are shown on the charts. (D) Time to BBCH60 vs. Cab041204.2:750 SNP call with data from spring and winter OSR in both control and warmed treatments. Significant differences at p < 0.05 are indicated using the Tukey post-hoc test.
Further analysis of BnaPHP.A05 variation revealed that across the diversity set, lines with the T allele reached BBCH51 a mean of 15.4 ± 2.3 days earlier and first flowering 11.2 ± 2.6 days earlier than lines with the C SNP call (Figures 1B, C). The allelic effect showed a significant interaction with crop type, and a much weaker interaction with the effect of warming on different crop types (Supplementary Table S1). Because only spring and winter OSR types contained greater than five varieties with both allelic variants (Supplementary Table S2), we further analyzed the effect of SNP variation at BnaPHP.A05 on spring and winter OSR separately. In spring OSR, we found that, on average, varieties with a T SNP flowered substantially earlier than those with a C SNP call, and this was true for both control and warmed plants (Figure 1D). Notably, we observed wide variation in flowering time in spring OSR varieties with a T SNP call (Figure 1D). In winter OSR, only varieties carrying the T allele were significantly delayed to flowering by the warming treatment applied to developing flower buds, and again, large variation in flowering times was observed in varieties carrying the T allele.
Haplotype variation at BnaPHP.A05 and association with time to flowering
The change from C to T in BnaPHP.A05 is synonymous and not predicted to affect protein function. To understand how sequence diversity at the BnaPHP.A05 locus might affect flowering, we analyzed transcriptome sequence data for 48 varieties with either allele including both spring and winter OSR (Havolikova et al., 2019). All varieties with the C PHP allele had a BnaPHP.A05 sequence (HAP1) that matched the Darmor-bzh reference sequence (Chalhoub et al., 2014; Figure 2A). In contrast, varieties with the T allele showed substantial variation that we broadly categorized into two additional haplotypes. The most frequent, HAP2, contained a large deletion of the putative 5′ untranslated region (UTR): this was up to and included the predicted ATG start codon, suggesting disruption to the PHP protein sequence (we cannot rule out translation of protein variants using alternative start codons). Additional BnaPHP.A05 variants were found only in spring OSR and combined variable deletions of the 5′ UTR with additional deletions in either the first or the first and second exons (Figure 2A). These were grouped into a third heterogeneous haplotype (HAP3) that appears likely to cause loss of function. A small number of SNPs in BnaPHP.A05 HAP3 genes (Figure 2A) mainly led to synonymous amino acid substitutions in the protein sequence; thus, we concluded that deletions were the most likely pertinent variation for BnaPHP.A05 HAP3. Because the 5′ deletion found in HAP2 and HAP3 could affect promoter activity or transcript stability, we analyzed RNAseq data for PHP in the third leaf of the 48 varieties (Supplementary Figure S1). HAP2 varieties had a modest 1.5-fold increase in BnaPHP.A05 expression compared to HAP1 or HAP3 varieties. However, because HAP2 plants share an early flowering phenotype with Arabidopsis php mutants, we concluded that changes to the upstream sequences likely do not cause PHP gain of function by changing promoter activity or mRNA stability. To understand the contribution of the three haplotypes to flowering time variation, we first compared time from floral initiation to flowering in the control winter and warm winter treatments (Figure 2; Supplementary Table S1). In winter OSR, HAP1 varieties flowered at a similar time in control and warmed treatments (Supplementary Table S2). However, warming caused a significant mean delay of 10 calendar days to flowering in HAP2 varieties. HAP2 varieties were earlier flowering than HAP1 varieties primarily in the control treatment (Figure 2B). This suggests that BnaPHP.A05 HAP2 leads to an enhanced response to chilling in winter OSR. For spring OSR the biggest differences were observed in HAP3 plants containing 5′ UTR and exon deletions. These were earlier flowering than HAP1 and HAP2 regardless of treatment (Figure 2B). There was no significant difference in the mean flowering time of HAP1 and HAP2 varieties in spring OSR, unlike in winter OSR. This early flowering phenotype of BnaPHP.A05 disruption plants resembles that of php mutants in rapid-cycling Arabidopsis accessions (Park et al., 2010), raising the prospect that BnaPHP.A05 loss of function causes early flowering in B. napus. Taken together, our analysis shows that HAP2 leads to a greater flowering time sensitivity to chilling than HAP1 in winter OSR.
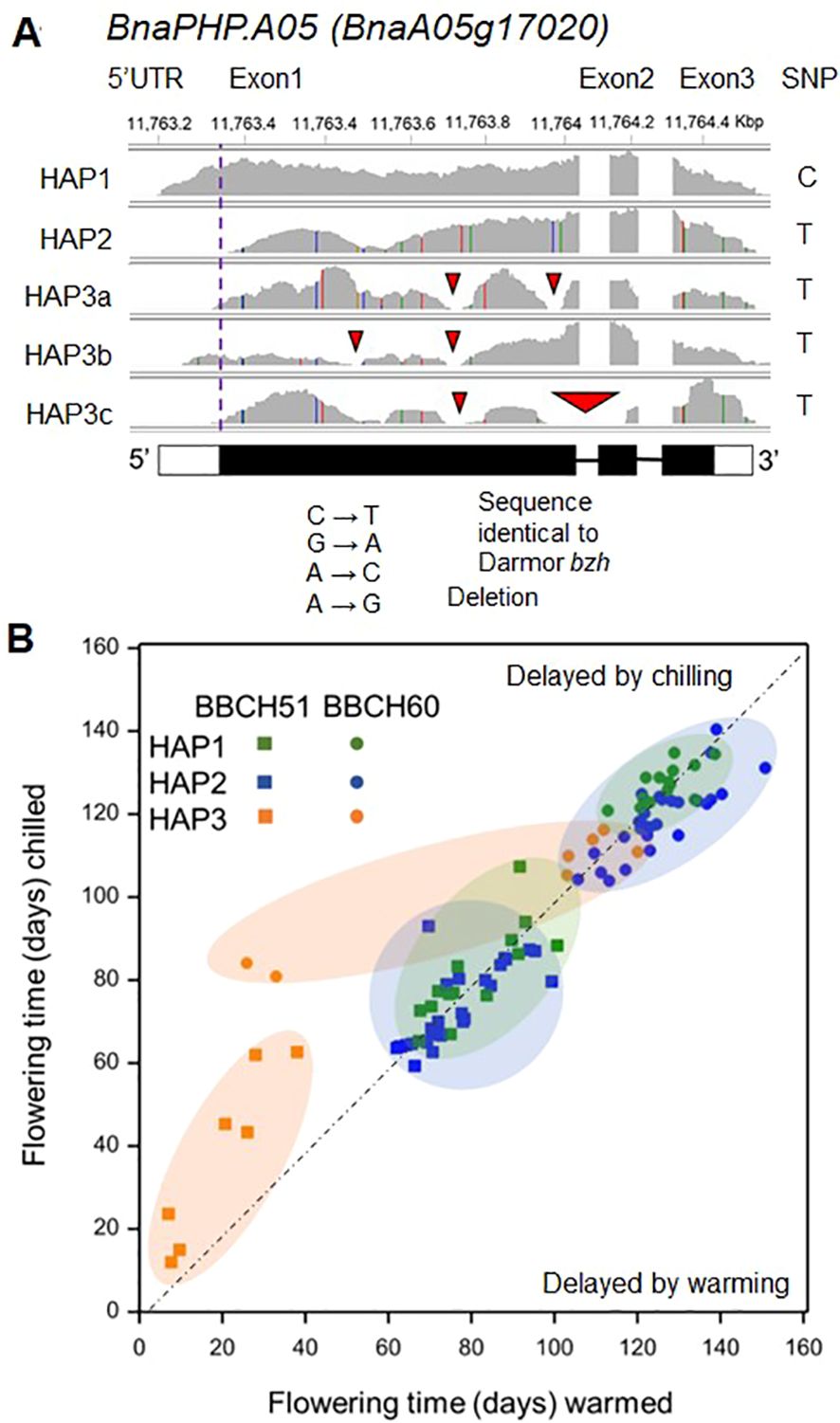
Figure 2. Haplotype variation at BnaPHP.A05 and its relationship with flowering time in B. napus. (A) Sequence analysis of BnaPHP.A05 haplotypes aligned to Darmor-bzh reference genome revealed that varieties with the Cab041204.2:750:CSNP call resembled Darmor-bzh, whereas Cab041204.2:750:T varieties were divided into two categories, HAP2 with a predicted 5′ UTR deletion and HAP3 varieties, which also contained a variety of predicted exon deletions. (B) Scatter plot to show the effect of the BnaPHP.A05 haplotype on mean time to buds visible (BBCH51) or first flowering (BBCH60). Data from Lu et al. (2022). Dashed line divides those varieties accelerated by flower bud warming and those delayed from flower bud warming. An accompanying statistical analysis of the data is shown in Supplementary Table S2.
In a second experiment, we tested whether variation in vernalization temperature between 5°C and 15°C affects time to flowering in a manner that correlates with BnaPHP.A05 allelic variation (see Materials and Methods). These temperatures are below the upper temperature limit for vernalization but may promote flowering with differing efficiencies (Tommey and Evans, 1991; Wollenberg and Amasino, 2012). In spring OSR, increasing the vernalization temperature from 5°C to 10°C had no significant effect on flowering time, but at 15°C, BnaPHP.A05 HAP1 and HAP2 plants showed a delay in flowering that varied substantially between varieties (Figure 3A). There was no significant difference in flowering time between HAP1- and HAP2-containing varieties, supporting our previous conclusion that HAP2 does not affect flowering time in spring OSR (Figure 2). HAP3 varieties flowered earlier than HAP1 and HAP2 varieties in all treatments, and in contrast to HAP1 and HAP2 plants, HAP3 plants flowered earlier at warmer vernalization temperatures. This again suggests that HAP3 confers earlier flowering.
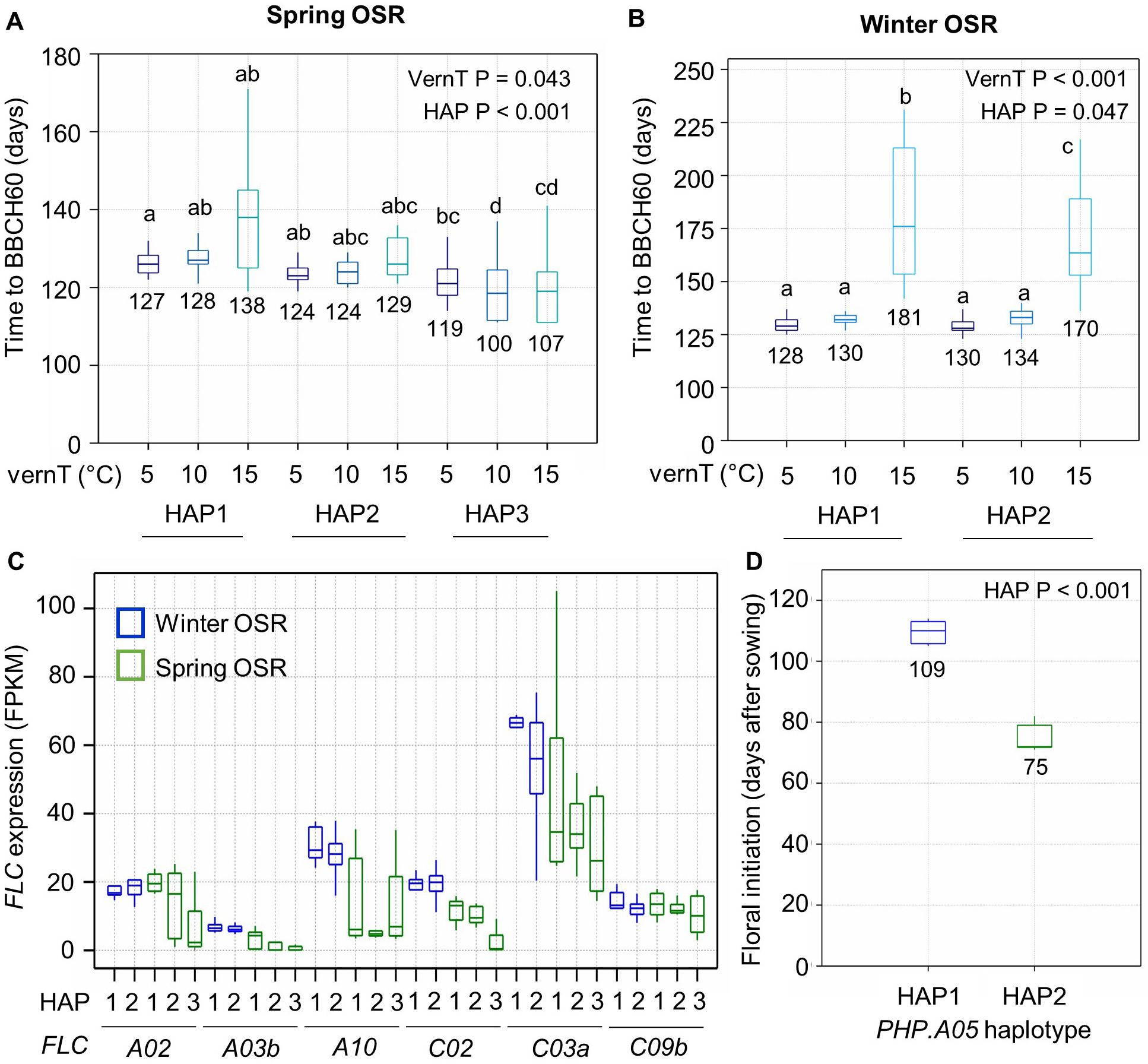
Figure 3. BnaPHP.A05 variation correlates with vernalization responses and FLC expression in B. napus. (A) Boxplot to show the flowering time of spring OSR with three main BnaPHP.A05 haplotypes after vernalization at three temperatures. p-values from two-way ANOVA using vernalization temperature (vernT) and haplotype (HAP) as factors are shown. Significant differences between groups are shown (p < 0.05). (B) As for A but for winter OSR and the two identified haplotypes of BnaPHP.A05 in winter oilseed rape varieties. (C) Expression of the six highest expressed B. napus FLC orthologs in the third leaf prior to vernalization. Spring OSR HAP1, n = 7; spring OSR HAP2, n = 5; and spring OSR HAP3, n = 7. Winter OSR HAP1, n = 6; winter OSR HAP2, n = 21. (D) Time for completion of the floral transition from autumn sowing in winter OSR with either BnaPHP.A05 HAP1 or HAP2. Data indicate the mean and standard error of five plants per genotype. Differences between haplotypes generated by one-way ANOVA.
In winter OSR, increasing the vernalization temperature from 5°C to 10°C again had no significant effect on flowering time and HAP1 and HAP2 varieties showed no significant difference in flowering time. However, 15°C vernalization temperatures conferred a strong delay to flowering that was 11 days longer in HAP1 plants than HAP2 plants, suggesting that in winter OSR, HAP2 confers a more effective chilling response at warmer chilling temperatures (Figure 3B). This result supports our previous conclusion that HAP2 is more responsive in flowering time to lower levels of chill.
Vernalization dynamics of OSR depend on the FLC genotype, starting FLC expression level, and rate of silencing in the falling temperatures of autumn (Schiessl et al., 2019; O’Neill et al., 2019; Calderwood et al., 2021). Because we observed an effect on time to flowering after chilling, we checked for a relationship between FLC expression prior to vernalization and PHP haplotype, focusing on the most highly expressed FLC genes (Figure 3C). In spring OSR, while some FLC genes were absent or rarely expressed (Schiessl et al., 2019), we found that several orthologs were lower expressed in lines with putative weaker alleles of PHP, especially HAP3 plants. This could be because HAP3 causes lower starting FLC levels, but we cannot rule out that the very early flowering of HAP3 lines results in part because of allelic variation at FLC in addition to PHP. In winter OSR, only FLC.C03a had a clear difference between HAP1 and HAP2 plants. To further test whether BnaPHP.A05variation affects the speed at which vernalization occurs, we used ecotilling (Woodhouse et al., 2021) to identify varieties of winter OSR with identical FLC genotypes but different haplotypes of BnaPHP.A05 (Figure 3D). We found five lines with identical FLC genotypes, and a sixth (Castille) with variation in only one FLC haplotype at BnaFLC.C02: three of these had the putative weak HAP2 and three contained the A. thaliana-like HAP1 genotype. Sown in late summer, the HAP2 lines underwent floral initiation a full 34 days prior to the HAP1 lines on average, despite near-identical FLC genotypes (Figure 3D). While we cannot rule out the presence of other genetic variation that contributes to this affect, this experiment supports the other findings reported here that putative weak alleles of PHP have reduced chilling requirements in winter OSR. These observations are consistent with previous observations in A. thaliana, which showed that effects of PAF1C disruption on flowering time were stronger in plants subjected to lower temperatures than those maintained at warm temperatures (Yu and Michaels, 2010; Park et al., 2010; Nasim et al., 2022).
Chilling and weak BnaPHP.A05 alleles promote gains in seed set
Next, we tested whether changing the vernalization temperature or BnaPHP.A05 genotype had a relationship with plant yield in winter and spring OSR, measuring seed weight per pod on the primary raceme in plants vernalized at different temperatures (see Materials and Methods). In spring OSR, the vernalization temperature had no discernible effect on seed weight per pod (Figure 4). This is not surprising given that spring varieties are known to have a minimal vernalization response. However, HAP3 plants in general had a lower yield than HAP1 and HAP2 plants regardless of the vernalization temperature, perhaps because they are earlier to flower (Figure 2). In winter OSR, the picture was strikingly different: firstly, increasing the vernalization temperature resulted in a clear decline in seed weight per pod, regardless of the BnaPHP.A05 genotype. This shows that in addition to well-known effects of chilling on flowering time and branching, increased chilling also promotes changes to reproductive development that improve seed yield within individual pods. Considering BnaPHP.A05 variation in winter OSR, HAP2 plants had a 10% higher seed weight per pod (p = 0.007) at all tested vernalization intensities, consistent with the enhanced flowering time response to chilling observed in HAP2 varieties. Furthermore, variation at BnaPHP.A05 can enhance the effect of chilling on seed yield. Thus, we concluded that chilling promotes seed yield in winter OSR and that BnaPHP.A05 HAP2 increases the yield response to applied chilling within the range tested.
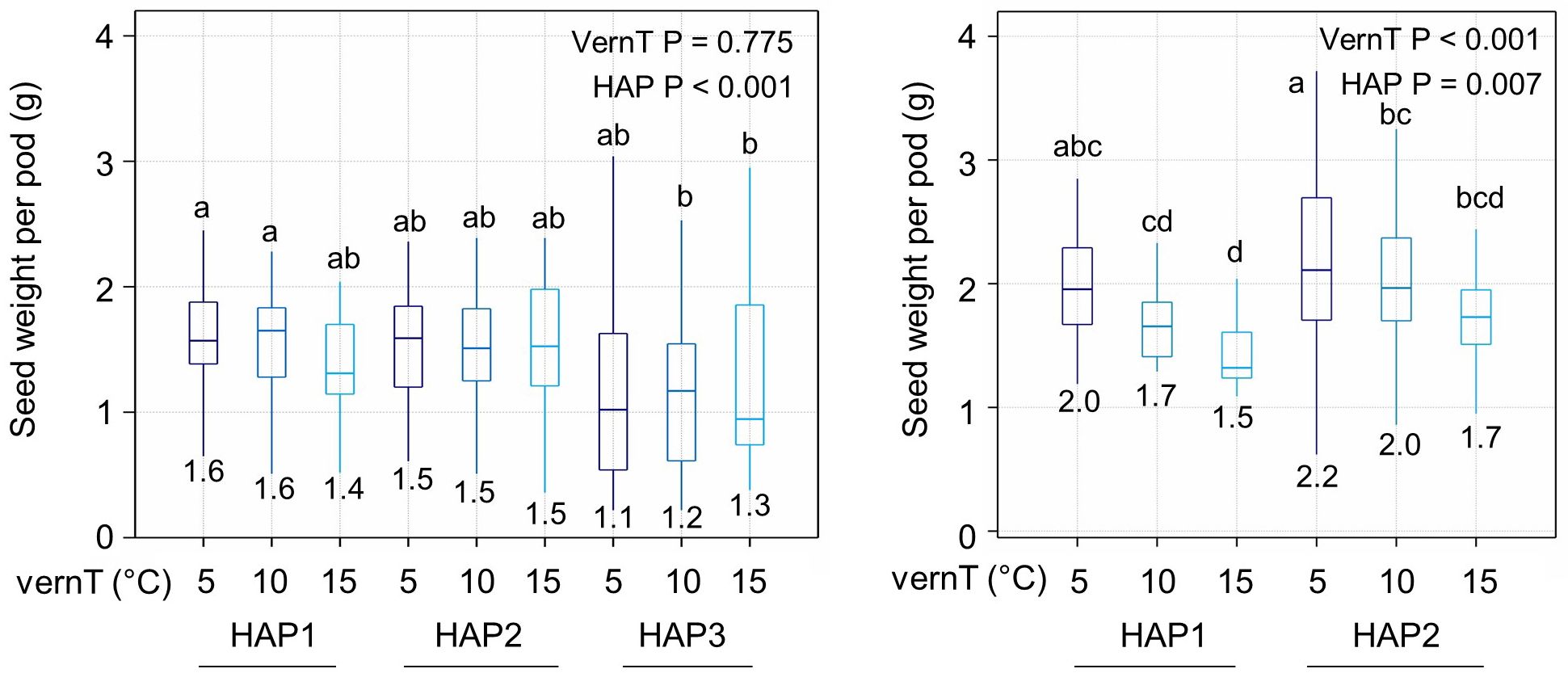
Figure 4. OSR seed yield parameters vary with the BnaPHP.A05 haplotype. Mean seed weight per pod is shown for lines with each haplotype in each vernalization temperature. p-values were calculated by two-way ANOVA with vernalization temperature (vernT) and haplotype (HAP) as factors. Differences between groups are shown at p < 0.05. Left plot, spring OSR. Right plot, winter OSR. Mean values for each treatment are indicated on plots. Significant differences are indicated by letters (P < 0.05).
PHP.A05 loss of function in Brassica rapa causes early flowering
Based on the mutant phenotype reported previously in A. thaliana, we assumed that Brassica loss-of-function PHP alleles would flower early compared to wild type whether in rapid-cycling or vernalization-requiring backgrounds (Yu and Michaels, 2010; Park et al., 2010). To directly test whether variation at BnaPHP.A05 can cause differences in flowering time, we used a Brassica rapa R-o-18 TILLING population (Stephenson et al., 2010) to identify putative loss-of-function alleles of the BraPHP.A05 gene. Brassica rapa shares the Brassica A genome with B. napus, allowing us to examine the consequences of PHP.A05 disruption without the complication of tetraploidy. R-o-18 phenology resembles that of rapid-cycling Arabidopsis accessions in that there is no requirement for vernalization for flowering, although there are up to five functional FLC orthologs in the species that nevertheless affect time to flowering, as in rapid-cycling Arabidopsis accessions (Wu et al., 2012; Takada et al., 2019; Akter et al., 2023). We found one line, ji41194-b, carrying a C-to-T mutation that converted a codon for a glutamine residue at position 186 to a stop codon, a change that should result in premature termination of translation midway through the first exon (Figure 5A). From a segregating population, we isolated plants homozygous for the Braphp.A05 mutant and wild-type alleles and compared their time to flowering. We found that the Braphp.A05 mutant reached BBCH51 an average of 8 days earlier than wild type and BBCH60 an average of 5 days earlier than wild type in a heated long-day glasshouse (Figures 5B–E). Because these differences closely resemble those we found between B. napus PHP haplotypes, we concluded that PHP.A05 loss of function can accelerate flowering across Brassica species and crop types.
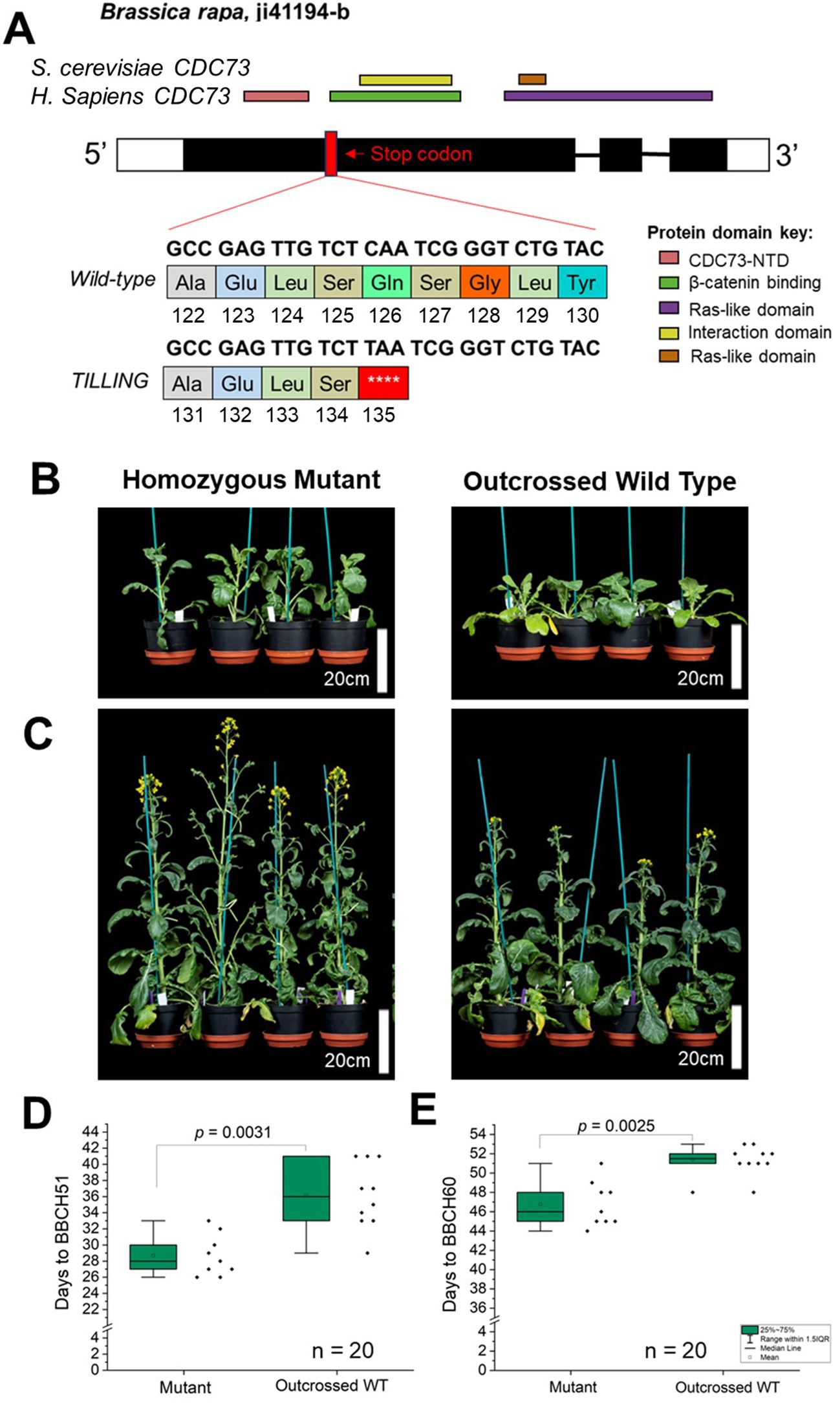
Figure 5. Loss of BraPHP.A05 function in Brassica rapa leads to early flowering. (A) Figure to show the structure of the B. rapa PHP.A05 locus and the predicted effect of the tilling mutant ji41194-b on PHP function. (B, C) Images to show the flowering time of the php.a05 mutant and WT segregant. (D, E) Plots to show the time to BBCH51 and BBCH60 of WT and php.a05, n = 10 plants per genotype. Significant differences indicated by one-way ANOVA are shown.
Discussion
In Arabidopsis, loss of PHP causes early flowering in vernalization-requiring and vernalization non-requiring backgrounds. In rapid-cycling Arabidopsis, the largest effects of PHP on flowering are observed at lower temperatures, approximately 10°C, although PHP affects flowering time at all tested temperatures in Col-0 (Nasim et al., 2022). Our data show that this role appears to be highly conserved in Brassica species. We observed that incomplete haplotypes of BnaPHP.A05 are, in general, associated with either early flowering in spring OSR or an enhanced flowering time response to chilling in winter OSR. We also show that loss of function of the identical gene in B. rapa causes a clear early flowering phenotype (Figure 5), confirming that in Brassica sp., PHP acts to delay flowering as previously observed in Arabidopsis. The observed enhanced response to chilling in weak php alleles observed in B. napus is also consistent with the known role of PAF1c in delaying flowering via effects on FLC chromatin state (Yu and Michaels, 2010).
However, here, we additionally reveal that, in B. napus, increases in chilling intensity have additional post-flowering effects on yield components, increasing seed yield within individual pods (Figure 4). This yield-promoting effect of chilling is consistent with our earlier observation that interruption to chilling in winter OSR affects crop yields (Brown et al., 2019; Lu et al., 2022). The precise mechanism by which chilling affects yield on a per-pod basis remains to be clarified. Overall, this suggests that in the absence of other considerations, yield gains can be achieved in winter OSR by reducing the vernalization requirement as long as winter annual habit can be maintained. Interestingly, weak BnaPHP.A05 alleles appear to act additively with chilling, increasing seed yield per pod at all chilling intensities tested (Figure 4). PAF1c couples transcription elongation to the deposition of active genetic marks (Krogan et al., 2003; Obermeyer et al., 2022), with this positive feedback important for maintaining the active chromatin state. Our data show that disrupting this feedback can increase the effectiveness of chilling treatments, leading to earlier flowering and higher yields, specifically in winter OSR.
In general, the PAF1c complex appears a poor choice for variation for crop improvement because of its central role in plant chromatin dynamics (Yu and Michaels, 2010). However, PHP appears to be an exception because, in Arabidopsis, its function appears to be limited to the regulation of flowering time via FLC expression, with detectable but apparently phenotypically irrelevant roles in the expression efficiency of long transcripts (Park et al., 2010; Nasim et al., 2022; Obermeyer et al., 2022). Because php mutants lack the pleiotropic effects of other PAF1c mutants, PHP alleles are well suited for adapting OSR crops to lower winter chill or increasing seed yield per pod for which a strong chilling response is advantageous (Figure 4). This makes PHP an interesting target for adapting winter OSR to warmer climates, especially maritime Europe where winter chilling is already low in some growing seasons. Fixing weaker BnaPHP.A05 haplotypes may be useful in winter OSR breeding, especially in adapting the crop to warmer winter environments in maritime Western Europe.
Materials and methods
Plant materials
The B. napus diversity fixed foundation set (DFFS) has been described previously and was used for the genome-wide association study. This set contains a mixture of winter OSR, spring OSR, Chinese semi-winter OSR, fodder rapes, and swede crop types. Brassica rapa R-o-18 and tilling mutant ji41194-b were obtained from the Germplasm Resources Unit at the John Innes Centre, Norwich, UK (https://www.seedstor.ac.uk/).
Genotyping of B. napus and B. rapa mutants
The presence or absence of the ji41194-b C-to-T mutation in the first exon of Brassica rapa PHP.A05 that converts glutamine at position 126 to a stop codon was confirmed by PCR from genomic DNA purified using a Macherey-Nagel™ NucleoSpin™ Gel and PCR Clean-up Kit according to the manufacturer’s instructions. BrPHP.A05 was amplified from B. rapa using the forward primer 5′-GTAATCAACGTCCTCTCC-3′ and the reverse primer 5′-GTTGGACCGAATCAGCATAATG-3′, and the presence or absence of the mutation was confirmed by sanger sequencing. The B. napus DFFS was genotyped for the SNP at BnaPHP.A05 with the highest p-value (Cab041204.2:750; Havlickova et al., 2018) in BnaPHP.A05 by PCR amplification from genomic DNA using the forward primer 5′-ATGGATCCGTTATCGGTGCTCAAGG-3′ and the reverse primer 5′-GAACAATTTAACTCACCAGTACTGG-3′ followed by sanger sequencing of the resulting fragment.
Data
Genotype (SNP) and expression level datasets (Havlickova et al., 2018) were downloaded from York Knowledgebase (http://yorknowledgebase.info) and refined to include only the lines used within this study. Sequence variation in BnaAGL24.A01 was determined from exome capture data (Williams et al., 2023), NCBI accession number PRJEB57649. BnaPHP.A05 sequence variation, BnaPHP.A05, and B. napus FLC expression data were extracted from pre-existing leaf transcriptome data from the Sequence Read Archive accession number PRJNA309367.
Associative transcriptomics
Associative transcriptomics was performed using the GAGA (GEM and GWAS Automation version 1.0) pipeline (available at https://github.com/bsnichols/GAGA) in R version 4.3.1 for Windows. Association was performed using GAPIT3 and the BLINK multilocus model (Wang and Zhang, 2021; Huang et al., 2019). BLINK runs two fixed-effect models iteratively. The first tests each marker with associated markers fitted as covariates to control for population stratification. The second selects those associated covariate markers and controls for spurious associations that arise in place of kinship (Liu et al., 2016). Loci were selected for further analysis based on significant Bonferroni-corrected p-values (Havlickova et al., 2018) and sorting for genes with known roles in flower development. For the further analysis of BnaPHP.A05, SNP calls extracted from Havlickova et al. (2018) were tested for a significant correlation with time from floral initiation to flowering using two-way ANOVA. Differences between groups were identified using a Bonferroni post-hoc test at p < 0.05.
Vernalization experiments
The B. napus DFFS was grown in a heated and lit glasshouse with a daylength of 16 h, a day temperature of 20°C, a night temperature of 15°C for 21 days, and then vernalized for 12 weeks at 5°C, 10°C, or 15°C in controlled environment chambers otherwise matched for light level (80–140 µm m−2 sec−1) in 8-hour days. After vernalization, plants were potted into 1-L pots and maintained in the long-day glasshouse with a temperature of 18°C for flowering and seed set. Flowering time was scored using the BBCH scale (Weber and Bleiholder, 1990). Yield per plant was estimated by analyzing seed number, size, and weight from 20 filled pods from the primary raceme, as described previously (Miller et al., 2019). Mean flowering time for each variety was used for statistical association with the BnaPHPA05 haplotype using two-way factorial ANOVA. Differences between groups were identified using a Bonferroni post-hoc test at p < 0.05.
In a second experiment to monitor the timing of floral initiation after vernalization, six varieties of winter OSR were grown, Rafal, Dippes, and Eurol with PHP.A05 HAP1, and Castille, Temple, and Rameses with PHP.A05 HAP2, in an unheated unlit glasshouse with seeds sown at the beginning of autumn (September 2022). These lines had identical FLC haplotypes, except for Castille, which has a homoeologous exchange of FLC.C02 for an extra copy of FLC.A02. FLC haplotype data have been described previously (Lu et al., 2022). Plants were monitored for the completion of vernalization by the timing of floral initiation using manual dissection under a Leica SP6 dissecting microscope.
Winter warming experiment
The B. napus DFFS was sown in an unheated unlit polytunnel, with timing of sowing staggered such that all varieties initiated flowers in November in Norwich, UK. Plants were maintained in the polytunnel over winter or moved 1 week after flowering to an unlit glasshouse at 20°C for 4 weeks. Precise details of temperature treatments were monitored using Gemini Tinytag Plus 2 TGP-44017 temperature and humidity loggers and are shown in Supplementary Figure S2. More details have been given previously (Lu et al., 2022). These conditions approximately match known field conditions in the UK in high- and low-yielding growing seasons (Supplementary Figure S3). After 4 weeks, all plants were returned to the unheated polytunnel, randomized, and maintained until flowering. Flowering time was scored on the BBCH scale.
Data availability statement
Genotype (SNP) and expression level datasets (Havlickova et al., 2018) were downloaded from York Knowledgebase (http://yorknowledgebase.info) and refined to include only the lines used within this study. Sequence variation in BnaAGL24.A01 was determined from exome capture data (Williams et al., 2023), NCBI accession number PRJEB57649. BnaPHP.A05 sequence variation, BnaPHP.A05 and B. napus FLC expression data were extracted from pre-existing leaf transcriptome data from the Sequence Read Archive accession number PRJNA309367.
Author contributions
SP: Conceptualization, Formal analysis, Funding acquisition, Supervision, Writing – original draft, Writing – review & editing. SW: Data curation, Formal analysis, Investigation, Writing – review & editing. CO: Conceptualization, Methodology, Supervision, Writing – review & editing. RD: Investigation, Writing – review & editing. RW: Conceptualization, Data curation, Formal analysis, Funding acquisition, Project administration, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was funded by the Biotechnology and Biological Sciences Research Council grants BB/W000415/1 to SP and BBSRC grants BB/X01102X/1 and BB/P003095/1 to SP and RW.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1481282/full#supplementary-material
References
Akter, A., Kakizaki, T., Itabashi, E., Kunita, K., Shimizu, M., Akter, M. A., et al. (2023). Characterization of FLOWERING LOCUS C 5 in brassica rapa L. Mol. Breed. 43, 58. doi: 10.1007/s11032-023-01405-0
Brown, J. K. M., Beeby, R., Penfield, S. (2019). Yield instability of winter oilseed rape modulated by early winter temperature. Sci. Rep. 9, 6953. doi: 10.1038/s41598-019-43461-7
Calderwood, A., Lloyd, A., Hepworth, J., Tudor, E. H., Jones, D. M., Woodhouse, S., et al. (2021). Total FLC transcript dynamics from divergent paralogue expression explains flowering diversity in Brassica napus. New Phytol. 229, 3534–3548. doi: 10.1111/nph.v229.6
Campoy, J. A., Darbyshire, R., Dirlewanger, E., Quero-García, J., Wenden, B. (2019). Yield potential definition of the chilling requirement reveals likely underestimation of the risk of climate change on winter chill accumulation. Int. J. Biometeorol. 63, 183–192. doi: 10.1007/s00484-018-1649-5
Chalhoub, B., Denoeud, F., Liu, S., Parkin, I. A. P., Tang, H., Wang, X., et al. (2014). Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345, 950–953. doi: 10.1126/science.1253435
Havlickova, L., He, Z., Wang, L., Langer, S., Harper, A. L., Kaur, H., et al. (2018). Validation of an updated associative transcriptomics platform for the polyploid crop species Brassica napus by dissection of the genetic architecture of erucic acid and tocopherol isoform variation in seeds. Plant J. 93, 181–192. doi: 10.1111/tpj.2018.93.issue-1
He, Y., Revell, B. J., Leng, B., Feng, Z. (2017). The effects of weather on oilseed rape (OSR) yield in China: future implications of climate change. Sustainability 9, 418. doi: 10.3390/su9030418
Hepworth, J., Antoniou-Kourounioti, R. L., Bloomer, R. H., Selga, C., Berggren, K., Cox, D., et al. Absence of warmth permits epigenetic memory of winter in Arabidopsis. Nat Commun. (2018) 9 (1), 639. doi: 10.1038/s41467-018-03065-7
Hou, J., Long, Y., Raman, H., Zou, X., Wang, J., Dai, S., et al. (2012). A Tourist-like MITE insertion in the upstream region of the BnFLC.A10 gene is associated with vernalization requirement in rapeseed (Brassica napus L.). BMC Plant Biol. 12, 238. doi: 10.1186/1471-2229-12-238
Huang, M., Liu, X., Zhou, Y., Summers, R. M., Zhang, Z. (2019). BLINK: a package for the next level of genome-wide association studies with both individuals and markers in the millions. Gigascience 8, giy154. doi: 10.1093/gigascience/giy154
Krogan, N. J., Dover, J., Wood, A., Schneider, J., Heidt, J., Boateng, M. A., et al. (2003). The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: Linking transcriptional elongation to histone methylation. Mol. Cell 11, 721–729. doi: 10.1016/S1097-2765(03)00091-1
Liu, X., Huang, M., Fan, B., Buckler, E. S., Zhang, Z. (2016). Iterative usage of fixed and random effect models for powerful and efficient genome-wide association studies. PloS Genet. 12, e1005767. doi: 10.1371/journal.pgen.1005767
Lu, X., O'Neill, C. M., Warner, S., Xiong, Q., Chen, X., Wells, R., et al. (2022). Winter warming post floral initiation delays flowering via bud dormancy activation and affects yield in a winter annual crop. Proc. Natl. Acad. Sci. U.S.A. 119, e2204355119. doi: 10.1073/pnas.2204355119
Matar, S., Kumar, A., Holtgräwe, D., Weisshaar, B., Melzer, S. (2021). The transition to flowering in winter rapeseed during vernalization. Plant Cell Env. 44, 506–518. doi: 10.1111/pce.13946
Miller, C., Wells, R., McKenzie, N., Trick, M., Ball, J., Fatihi, A., et al. (2019). Variation in expression of the HECT E3 ligase UPL3 modulates LEC2 levels, seed size, and crop yields in brassica napus. Plant Cell 31, 2370–2385. doi: 10.1105/tpc.18.00577
Nasim, Z., Susila, H., Jin, S., Youn, G., Ahn, J. H. (2022). Polymerase II-associated factor 1 complex-regulated FLOWERING LOCUS C-clade genes repress flowering in response to chilling. Front. Plant Sci. 13, 817356. doi: 10.3389/fpls.2022.817356
O’Neill, C. M., Lu, X., Calderwood, A., Tudor, E. H., Robinson, P., Wells, R., et al. (2019). Vernalization and floral transition in autumn drive winter annual life history in oilseed rape. Curr. Biol. 29, 4300–4306. doi: 10.1016/j.cub.2019.10.051
Obermeyer, S., Stöckl, R., Schneckenburger, T., Moehle, C., Schwartz, U., Grasser, K. D. (2022). Distinct role of subunits of the Arabidopsis RNA polymerase II elongation factor PAF1C in transcriptional reprogramming. Front. Plant Sci. 13, 974625. doi: 10.3389/fpls.2022.974625
Park, S., Oh, S., Ek-Ramos, J., van Nocker, S. (2010). PLANT HOMOLOGOUS TO PARAFIBROMIN is a component of the PAF1 complex and assists in regulating expression of genes within H3K27ME3-enriched chromatin. Plant Physiol. 153, 821–831. doi: 10.1104/pp.110.155838
Qüesta, J. I., Song, J., Geraldo, N., An, H., Dean, C. Arabidopsis transcriptional repressor VAL1 triggers Polycomb silencing at FLC during vernalization. Science. (2016) 353 (6298), 485–8. doi: 10.1126/science.aaf7354
Rondanini, D. P., Gomez, N. V., Belen Agosti, M., Mirallas, D. J. (2012). Global trends of rapeseed grain yield stability and rapeseed-to-wheat yield ratio in the last four decades. Eur. J. Agron. 37, 56–65. doi: 10.1016/j.eja.2011.10.005
Schiessl, S. V., Quezada-Martinez, D., Tebartz, E., Snowdon, R. J., Qian, L. (2019). The vernalisation regulator FLOWERING LOCUS C is differentially expressed in biennial and annual Brassica napus. Sci. Rep. 9, 14911. doi: 10.1038/s41598-019-51212-x
Stephenson, P., Baker, D., Girin, T., Perez, A., Amoah, S., King, G. J., et al. (2010). A rich TILLING resource for studying gene function in Brassica rapa. BMC Plant Biol. 10, 62. doi: 10.1186/1471-2229-10-62
Tadege, M., Sheldon, C. C., Helliwell, C. A., Stoutjesdijk, P., Dennis, E. S., Peacock, W. J. (2001). Control of flowering time by FLC orthologues in Brassica napus. Plant J. 28, 545–553. doi: 10.1046/j.1365-313X.2001.01182.x
Takada, S., Akter, A., Itabashi, E., Nishida, N., Shea, D. J., Miyaji, N., et al. (2019). The role of FRIGIDA and FLOWERING LOCUS C genes in flowering time of Brassica rapa leafy vegetables. Sci. Rep. 9, 13843. doi: 10.1038/s41598-019-50122-2
Tommey, A. M., Evans, E. J. (1991). Temperature and daylength control of flower initiation in winter oilseed rape (Brassica napus L.). Ann. Appl. Biol. 118, 201–208. doi: 10.1111/j.1744-7348.1991.tb06098.x
Wang, J., Zhang, Z. (2021). GAPIT Version 3: Boosting power and accuracy for genomic association and prediction. Genomics Proteomics Bioinf. 19, 629–640. doi: 10.1016/j.gpb.2021.08.005
Weber, E., Bleiholder, H. (1990). Erläuterungen zu den BBCH-dezimal-codes für die entwicklungsstadien von mais, raps, fabaBohne, sonnenblume und erbse-mit abbildungen. Gesunde Pflanz 42, 308–321.
Williams, K., Hepworth, J., Nichols, B. S., Corke, F., Woolfenden, H., Paajanen, P., et al. (2023). Integrated Phenomics and Genomics reveals genetic loci associated with inflorescence growth in Brassica napus. bioRxiv. doi: 10.1101/2023.03.31.535149
Wollenberg, A. C., Amasino, R. M. (2012). Natural variation in the temperature range permissive for vernalization in accessions of Arabidopsis thaliana. Plant Cell Env. 35, 2181–2191. doi: 10.1111/j.1365-3040.2012.02548.x
Woodhouse, S., He, Z., Woolfenden, H., Steuernagel, B., Haerty, W., Bancroft, I., et al. (2021). Validation of a novel associative transcriptomics pipeline in Brassica oleracea: identifying candidates for vernalisation response. BMC Genomics 22, 539. doi: 10.1186/s12864-021-07805-w
Wu, J., Wei, K., Cheng, F., Li, S., Wang, Q., Zhao, J., et al. (2012). A naturally occurring InDel variation in BraA.FLC.b (BrFLC2) associated with flowering time variation in Brassica rapa. BMC Plant Biol. 12, 151. doi: 10.1186/1471-2229-12-151
Yin, S., Wan, M., Guo, C., Wang, B., Li, H., Li, G., et al. (2020). Transposon insertions within alleles of BnaFLC.A10 and BnaFLC.A2 are associated with seasonal crop type in rapeseed. J. Exp. Bot. 71, 4729–4741. doi: 10.1093/jxb/eraa237
Keywords: chilling, vernalization, Brassica, temperature, flowering, oilseed rape, climate change
Citation: Warner S, O’Neill CM, Doherty R, Wells R and Penfield S (2024) Adaptation to reductions in chilling availability using variation in PLANT HOMOLOGOUS TO PARAFIBROMIN in Brassica napus. Front. Plant Sci. 15:1481282. doi: 10.3389/fpls.2024.1481282
Received: 15 August 2024; Accepted: 23 September 2024;
Published: 22 October 2024.
Edited by:
Zhanwu Dai, Chinese Academy of Sciences (CAS), ChinaReviewed by:
Fuqiang Cui, Zhejiang Agriculture and Forestry University, ChinaYuanyuan Zhang, Chinese Academy of Agricultural Sciences, China
Copyright © 2024 Warner, O’Neill, Doherty, Wells and Penfield. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Steven Penfield, c3RldmVuLnBlbmZpZWxkQGppYy5hYy51aw==
 Samuel Warner
Samuel Warner Carmel M. O’Neill
Carmel M. O’Neill Rachel Wells
Rachel Wells Steven Penfield
Steven Penfield