- 1Key Laboratory of Southern Rice Innovation & Improvement, Ministry of Agriculture and Rural Affairs, Yuan Longping High-Tech Agriculture Co., Ltd., Changsha, China
- 2State Key Laboratory of Hybrid Rice, Hunan Hybrid Rice Research Center, Changsha, Hunan, China
- 3Yuelushan Laboratory, Changsha, China
- 4College of Life Sciences, Hunan Normal University, Changsha, Hunan, China
- 5State Key Laboratory for Biology of Plant Diseases and Insect Pests, Institute of Plant Protection, Chinese Academy of Agricultural Sciences, Beijing, China
Phytopathogens represent an ongoing threat to crop production and a significant impediment to global food security. During the infection process, these pathogens spatiotemporally deploy a large array of effectors to sabotage host defense machinery and/or manipulate cellular pathways, thereby facilitating colonization and infection. However, besides their pivotal roles in pathogenesis, certain effectors, known as avirulence (AVR) effectors, can be directly or indirectly perceived by plant resistance (R) proteins, leading to race-specific resistance. An in-depth understanding of the intricate AVR-R interactions is instrumental for genetic improvement of crops and safeguarding them from diseases. Magnaporthe oryzae (M. oryzae), the causative agent of rice blast disease, is an exceptionally virulent and devastating fungal pathogen that induces blast disease on over 50 monocot plant species, including economically important crops. Rice-M. oryzae pathosystem serves as a prime model for functional dissection of AVR effectors and their interactions with R proteins and other target proteins in rice due to its scientific advantages and economic importance. Significant progress has been made in elucidating the potential roles of AVR effectors in the interaction between rice and M. oryzae over the past two decades. This review comprehensively discusses recent advancements in the field of M. oryzae AVR effectors, with a specific focus on their multifaceted roles through interactions with corresponding R/target proteins in rice during infection. Furthermore, we deliberated on the emerging strategies for engineering R proteins by leveraging the structural insights gained from M. oryzae AVR effectors.
Introduction
Plants, being sessile, are constantly besieged by a plethora of phytopathogens such as fungi, bacteria, viruses, oomycetes and nematodes, which are capable of causing extensive damage to agrosystems, ecosystems, and human livelihoods (Milgroom, 2015). Unlike animals, plants are devoid of an adaptive immune system and specialized mobile immune cells to fend off the numerous potential threats posed by these pathogens (Spoel and Dong, 2012). Instead, they have evolved a sophisticated two-tiered innate immune machinery, composed of pathogen/microbe-associated molecular patterns (PAMPs/MAMPs)-triggered immunity (PTI) and effector-triggered immunity (ETI), which is fundamental for their survival in nature (Jones and Dangl, 2006; Boller and Felix, 2009; Macho and Zipfel, 2014). Highly conserved PAMPs/MAMPs, such as bacterial flagellin, peptidoglycan (PGN), lipopolysaccharide (LPS) and fungal chitin, are recognized by plant cell surface-localized pattern-recognition receptors (PRRs). This recognition triggers basal immune responses known as PTI, which serves as the first tier of plant immunity thwarting pathogen proliferation (Nürnberger et al., 2004; Jones and Dangl, 2006; Block et al., 2008). Plant PRRs are classified as either transmembrane receptor-like kinases (RLKs) or receptor-like proteins (RLPs), which possess highly variable ectodomains for the detection of a wide range of ligands (Ngou et al., 2021). To circumvent PTI, adapted phytopathogens deliver an arsenal of virulence factors known as effectors into plant apoplast or cytoplasm, where they suppress immune responses and create a favorable niche for pathogenesis, leading to effector-triggered susceptibility (ETS) (Boller and He, 2009). As a counter response, plants employ intracellular immune receptors, called resistance (R) proteins, to detect certain pathogen effectors, referred to as avirulence (AVR) effectors, either through direct interactions or indirect interactions. This recognition triggers effector-triggered immunity (ETI), which represents the second tier of plant immunity (Dodds et al., 2018). Among the diverse types of R proteins, nucleotide-binding, leucine rich-repeat receptors (NLRs) represent the largest group and they share a multi‐domain architecture typically composed of a central nucleotide‐binding (NB‐ARC) domain, a C‐terminal leucine-rich repeat (LRR) domain, and either a coiled‐coil (CC) domain, RPW8-like CC domain, or a Toll/interleukin‐1 receptor (TIR) at N-terminus, and are thus called CNLs, RNLs or TNLs, respectively (Takken and Goverse, 2012). CNLs and RNLs are found in both dicot and monocot plant species, while TNLs are absent in monocots (Shao et al., 2016; Liu et al., 2021a). Notably, RNLs as helpers act downstream of sensor NLRs, transducing immune signals rather than sensing AVR effectors (Jubic et al., 2019). Consistent with their specific roles in immunity, RNLs are usually characterized by a relatively low copy number in plant genomes (Zhong and Cheng, 2016). Furthermore, many NLRs contain additional noncanonical domains called the integrated domains (ID), such as the heavy metal-associated (HMA) domain, BED domain, RIN4/NOI domain or WRKY domain, which serve as baits to trap AVR effectors or monitor their activities (Cesari et al., 2014; Wu et al., 2015a; Duxbury et al., 2016; Kroj et al., 2016; De la Concepcion et al., 2022; Shimizu et al., 2022). Direct binding of AVRs or AVR-host target complexes to these IDs results in NLR activation and initiation of immune responses (Cesari et al., 2014; Fujisaki et al., 2015, 2017; Cesari, 2018; De la Concepcion et al., 2022). NLRs can function as single entities, in pairs, or within intricate networks (Adachi et al., 2019). In comparison to PTI, ETI elicits stronger defense responses, and is usually associated with localized plant cell death, termed the hypersensitive response (HR), to limit the spread of phytopathogens into neighboring uninfected cells (Jones and Dangl, 2006; Duxbury et al., 2021). To counteract ETI, phytopathogens are subject to either loss of function or production of the altered forms of their AVR effectors under selective forces. These adaptations allow them to evade recognition by R proteins or the target proteins (Stergiopoulos and de Wit, 2009). The ‘zig-zag-zig’ model, which depicts the relationship between PTI, ETS, and ETI, is the most widely used and concise model to date (Jones and Dangl, 2006). However, this model is increasingly being challenged. Firstly, AVR effectors are not always detected by NLRs, but can also be recognized by PRRs (Thomas et al., 1997). Secondly, PTI and ETI were initially considered as two separate and sequential branches of the plant immune system mediated by different receptors with distinct ligands perceived and activation modes in the model, but they actually share many downstream immune responses, such as mitogen-activated protein kinase (MAPK) cascades activation, Ca2+ flux, reactive oxygen species (ROS) burst and phytoalexins production (Jones and Dangl, 2006; Tsuda and Katagiri, 2010; Yu et al., 2017; Lolle et al., 2020; Lu and Tsuda, 2021; Liu et al., 2023b). Recently, accumulating evidence has revealed crosstalk between ETI and PTI, indicating that these two branches of the immune system are not entirely independent. Instead, they can synergistically enhance each other, thereby eliciting more robust immune responses against pathogen infections (Ngou et al., 2021; Pruitt et al., 2021; Tian et al., 2021; Yuan et al., 2021). The findings imply a much more intricate and interconnected nature of plant immune responses than previously hypothesized.
Given the pivotal role of AVR effectors in adapted phytopathogens, a profound comprehension of their mode of action is potentially conductive to conceptualize novel strategies for sustainable management of plant diseases. In this review, taking the phytopathogenic fungus Magnaporthe oryzae (M. oryzae), the causal agent of globally important rice blast disease, as an example, we elaborate the dual nature of functions of AVR effectors in rice blast resistance/susceptibility. We present updated findings on the molecular interactions between M. oryzae AVR effectors and rice R/target proteins, as well as the underlying structural basis. We also present recent progress in genetic engineering of R proteins to produce robust resistance in rice based on the structural knowledge.
M. oryzae and rice blast disease
Rice (Oryza sativa L.) is a staple cereal food crop for over 3.5 billion people around the world and sustainable rice production is crucial in ensuring global food security (Khush, 2013; Muthayya et al., 2014). Besides, rice cultivation is the major source of income and employment for more than 200 million smallholder farmers in rice-growing regions (Tonini and Cabrera, 2011). Over decades, rice production has witnessed a remarkable surge, attributed to the adoption of innovative agro-technologies including exploitation of semi-dwarf gene, utilization of heterosis and improvements in farming management practices (Ma and Yuan, 2015). In 2022, world rice production was approximately 776.5 million tons, marking a significant increase of 3.6 times compared to the production levels in 1961 (FAO, https://www.fao.org/faostat/en/). However, it is insufficient to meet the projected demands of continuously increasing global population, which is expected to reach 9.7 billion by 2050 (Hu et al., 2022a). This challenge is further exacerbated by the shrinkage of arable land and escalating influence of various biotic (pests, weeds, diseases, etc.) and abiotic factors (drought, cold, acidity, heat, salinity, etc.) (Sandhu et al., 2020). Among the biotic constraints, diseases caused by phytopathogens accounting for extensive yield losses represent a significant threat to rice production. A wide array of rice diseases caused by fungi, bacteria, viruses and nematodes have been recorded (Slough, 1985). Notably, blast disease, caused by the filamentous ascomycete fungus Magnaporthe oryzae B.C. Couch (anamorph: Pyricularia oryzae Cavara), is undoubtedly the most devastating disease of rice (Wang et al., 2014). It is also known as an ancient disease with records dating back to the 17th century in China (Couch et al., 2005). Nowadays, this notorious disease has a widespread distribution across rice-growing regions globally (Kato, 2001; Skamnioti and Gurr, 2009). Rice blast disease is responsible for average rice yield losses of about 10% to 30% per year, which could fulfill the annual rice consumption of 60 million people (Dean et al., 2005). Under favorable conditions, its regional epidemics can be more destructive, leading to yield loss up to 100% (Dean et al., 2012). In a survey from phytopathologists worldwide, M. oryzae was ranked first in the Top 10 scientifically and economically fungal pathogen list (Dean et al., 2012).
M. oryzae is a complex species with a broad host range. It is capable of plaguing more than 50 Poaceae and Cyperaceae species, including agriculturally important crop species such as rice (Oryza sativa), wheat (Triticum aestivum), maize (Zea maydis), barley (Hordeum vulgare), foxtail millet (Setaria italica) and finger millet (Eleusine coracana), as well as wild grasses such as weeping lovegrass (Eragrostis curvula), ryegrass (Lolium perenne) and goosegrass (Eleusine indica) (Khang et al., 2010; Hossain, 2022). Phylogenetic analyses have shown that M. oryzae’s wide host range is associated with intraspecific diversity (Couch et al., 2005; Chiapello et al., 2015; Yoshida et al., 2016; Inoue et al., 2017; Gladieux et al., 2018b). M. oryzae can be divided into several, genetically differentiated lineages that are associated with a specific or limited number of hosts (Gladieux et al., 2018a). All rice-infecting isolates (Oryza lineage) belong to a genetic lineage which is closely related to isolates infecting foxtail millet (Setaria lineage) (Couch et al., 2005; Ceresini et al., 2018; Gladieux et al., 2018a). Rice blast disease was thus inferred to emerge as a result of a host shift from foxtail millet in the Middle Yangtze Valley of China approximately 2,500 to 7,500 years ago (Couch et al., 2005). The globally distributed rice-infecting isolates can be further subdivided into four main lineages with one recombining lineage and three clonal lineages, which were estimated to have diverged around 1,000 years ago (Zhong et al., 2018; Gladieux et al., 2018a, b; Latorre et al., 2020; Thierry et al., 2022).
Almost all rice plant tissues at any growth stage can be attacked by this pathogen (Wilson and Talbot, 2009; Fisher et al., 2012). M. oryzae invades rice aerial tissues in a hemi-biotrophic manner, but it adopts a biotrophic strategy to infect roots (Marcel et al., 2010). During the infection of rice arial tissues, M. oryzae initially grows in living host cells as a biotroph to suppress the host immunity (Yan and Talbot, 2016). Subsequently, the invasive hyphae spread into neighboring cells through plasmodesmata and the fungus switches to a necrotrophic lifestyle. The initially infected host cells are destroyed, enabling the fungus to utilize nutrients released from the dead cells and sporulate from necrotic disease lesions on the leaf surface (Yan et al., 2023). The newly formed conidia are dispersed by wind or rain splashes, re-infecting healthy tissues and plants in the vicinity.
Secretion of M. oryzae effectors during host invasion
During the process of host invasion, M. oryzae undergoes several morphogenetic transitions. Initially, the three-celled conidium germinates to form a germ tube and differentiates into a dome-shaped infection structure called appressoria after perceiving physical and chemical cues on the leaf surface (Bourett and Howard, 1990). Subsequently, a penetration peg emerges from an appressorium for puncturing the host epidermal cell with huge turgor pressure and it then differentiates into the narrow tubular primary invasive hyphae (IH) and the bulbous secondary IH (Dagdas et al., 2012). The IH are enclosed by a host-derived plasma membrane termed the extra-invasive hyphal membrane (EIHM) (Kankanala et al., 2007). Once filled with the bulbous IH, the colonized host cells die. Meanwhile, the fungus protrudes into the neighboring host cells through pit field sites containing plasmodesmata, resulting in typical lesion formation and transition of the fungus from biotrophic to the necrotrophic phase (Martin-Urdiroz et al., 2016). During the biotrophic phase, M. oryzae express and secretes a set of effectors around or into host cells to modulate its cellular and metabolic processes, thereby favoring successful invasion and proliferation within plant tissues (Zhai et al., 2022). These effectors can thus be broadly categorized into apoplastic effectors and cytoplastic effectors based on their subcellular localizations in the host. Their deliveries are dependent on different secretion pathways (Mentlak et al., 2012; Yan and Talbot, 2016). Apoplastic effectors are delivered into the space between the fungal cell wall and host plasma membrane via a classical Golgi-dependent secretion pathway that can be blocked by the pharmacological drug brefeldin A (BFA) (Giraldo et al., 2013; Rocafort et al., 2020). Cytoplasmic effectors are secreted and accumulate in an extended dome-shaped interfacial region known as the biotrophic interfacial complex (BIC) near the tip of the first bulbous cell (Khang et al., 2010; Giraldo et al., 2013; Oliveira-Garcia et al., 2023). The cytoplasmic effectors within BIC are further packaged in dynamic vesicle-like membranous effector compartments (MECs), which are bounded by the host plasma membrane and CLATHRIN LIGHT CHAIN 1, a component of clathrin-mediated endocytosis (CME) (Oliveira-Garcia et al., 2023). Inhibition of CME by gene silencing or chemical treatments prevents MEC formation and pathogenicity, which indicates that CME facilitates the internalization of cytoplasmic effectors into host cells (Oliveira-Garcia et al., 2023). The emergence of the BIC structure is a feature of successful infection, but it is not observed during incompatible reactions (Mosquera et al., 2009; Khang et al., 2010; Jones et al., 2016; Shipman et al., 2017). Once internalized, these cytoplasmic effectors execute function in the cytoplasm and/or organelles of infected host cells, and even migrate to the adjacent cells (Khang et al., 2010).
AVR effectors in M. oryzae
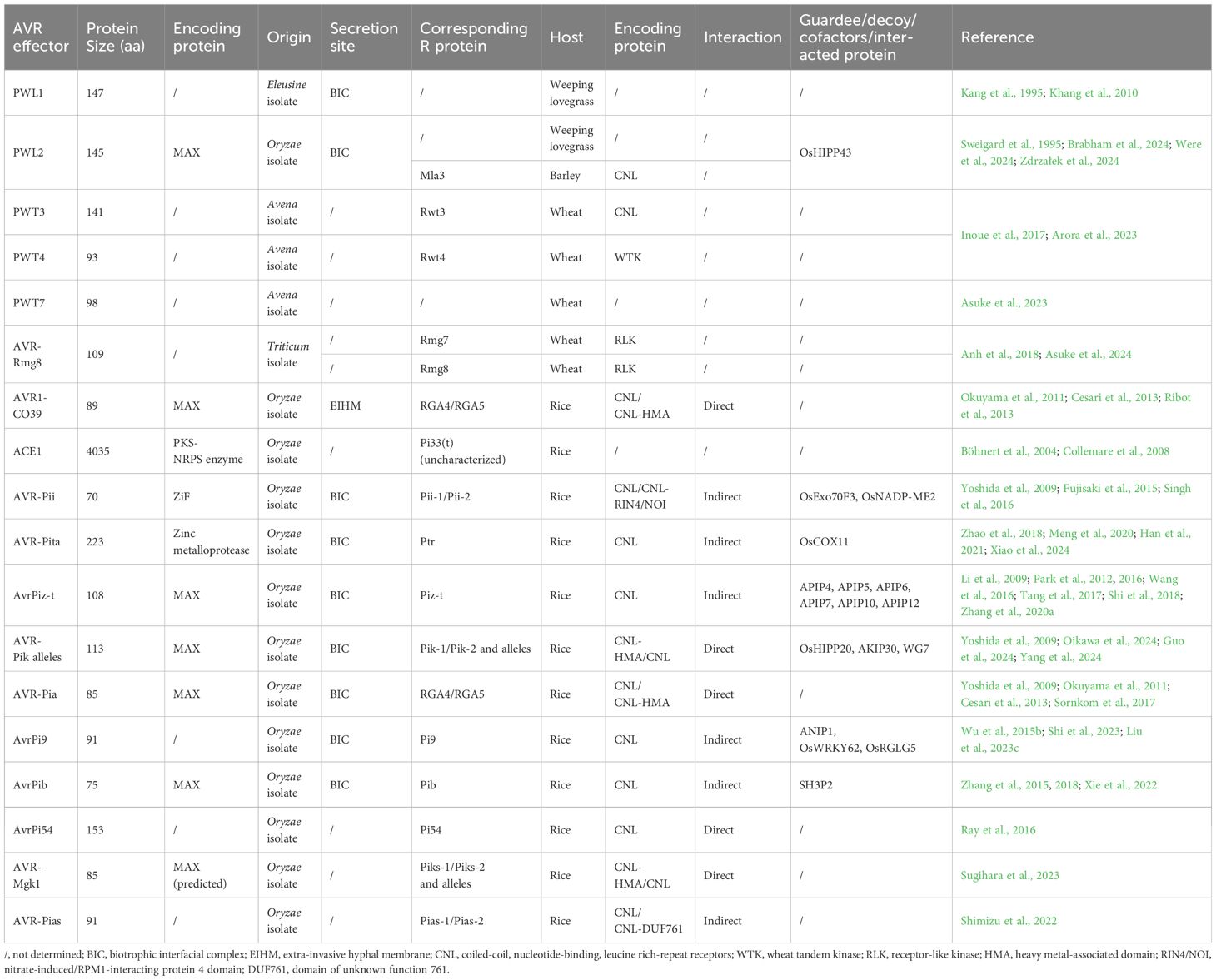
Molecular characterization of effectors stands as a fundamental step for understanding pathogen pathogenesis and plant immunity. Through genomic and transcriptomic analysis, researchers have pinpointed hundreds of potential effector candidates in M. oryzae (Dean et al., 2005; Soanes et al., 2008; Yoshida et al., 2009; Choi et al., 2010; Chen et al., 2013; Dong et al., 2015; Yan et al., 2023; Liu et al., 2023a). More than forty AVR genes have been genetically identified and 18 have been molecularly characterized thus far, including PWL1, PWL2, PWT3, PWT4, PWT7, AVR-Rmg8, AVR-Pita, ACE1, AVR-Pia, AvrPii, AvrPiz‐t, Avr1‐CO39, AvrPib, AvrPi9, AvrPi54, AVR-Pias, AVR-Mgk1 and Avr-Pik (Kang et al., 1995; Sweigard et al., 1995; Orbach et al., 2000; Böhnert et al., 2004; Li et al., 2009; Yoshida et al., 2009; Ribot et al., 2013; Zhang et al., 2015; Wu et al., 2015b; Ray et al., 2016; Inoue et al., 2017; Anh et al., 2018; Zhang et al., 2020b; Shimizu et al., 2022; Asuke et al., 2023; Sugihara et al., 2023) (Table 1). PWL2 encoding a glycine-rich, hydrophilic protein, is the first isolated M. oryzae AVR gene from Oryzae isolates (Sweigard et al., 1995). It belongs to a gene family with three other PWL (pathogenicity toward weeping lovegrass) genes (Kang et al., 1995). Both PWL1 and PWL2 are two host-specificity determinants conferring avirulence on weeping lovegrass but not on rice. However, PWL3 and PWL4 are nonfunctional. PWL2 is a core effector of the blast fungus, since it is ubiquitous in M. oryzae and has undergone substantial copy number expansion (Zdrzałek et al., 2024). PWT3, PWT4 and AVR-Rmg8 conditioning avirulence of M. oryzae isolates from different hosts on wheat, are able to trigger defense responses in wheat cultivars containing R proteins Rwt3, Rwt4 and Rmg7/Rmg8, respectively (Takabayashi et al., 2002; Vy et al., 2014; Inoue et al., 2017; Anh et al., 2018; Arora et al., 2023). PWT3 homologs were found widely distributed across both Triticum and non-Triticum isolates, while PWT4 homologs showed limited distribution in some isolates. Wheat cultivars without Rwt3, introduced to Brazil in the early 1980s, served as springboards for host jumps of Lolium isolates containing PWT3 to wheat, followed by loss of function of PWT3 due to the imposed selection by cultivars with Rwt3 and wheat blast epidemics in South America, Asia as well as Africa (Inoue et al., 2017). PWT7 from an Avena isolate confers avirulence on wheat only at the seedling stage (Asuke et al., 2023). AVR-Rmg8, identified from a Triticum isolate, was found to be recognized by either Rmg7 in tetraploid wheat or Rmg8 in hexaploid wheat, conferring resistance at both the seedling and heading stage (Anh et al., 2018). Among the 12 other AVR genes displaying avirulence toward rice, ten code for small proteins less than 200 amino acids (aa) with N-terminal signal peptides and share low sequence similarity to other proteins of known function in public databases. ACE1 and AVR-Pita are the two exceptions, which encode larger proteins and contain known-function domains or motifs. ACE1 is a secondary metabolism (SM) gene encoding a non-secreted PKS-NRPS hybrid (polyketide synthase and non-ribosomal peptide synthetase) enzyme (Böhnert et al., 2004). AVR-Pita encodes a putative neutral zinc metalloprotease (Orbach et al., 2000). The genetic instability of AVR genes in M. oryzae is considered to be a common mechanism in gaining virulence and causing rapid resistance erosion of their cognate R genes (Huang et al., 2014). Different mechanisms including insertion, point mutation, and deletion, as well as sexual mating and parasexual recombination are responsible for the loss of avirulence function of AVR genes (Noguchi et al., 2006; Tsujimoto Noguchi, 2011; Hu et al., 2022b). Among the cloned AVR genes of M. oryzae, AVR-Pita has been widely studied due to its relatively high variability. For example, AVR-Pita was found to be almost or completely absent in over half of the blast isolates in the Sichuan Basin, China, and five haplotypes with avirulent function were identified (Hu et al., 2022b). In an investigation of M. oryzae isolates from Thailand, AVR-Pita was detected in only around one third of them and six haplotypes of were identified with one deletion and 12 amino acid substitutions (Sutthiphai et al., 2022). Additionally, 40 AVR-Pita haplotypes were identified in avirulent isolates collected from Southern US (Zhang et al., 2020b). In contrast, AVR-Pi9 is much more stable and it can be detected in all the M. oryzae samples in Sichuan and Yunan province, China, as well as Thailand (Hu et al., 2022b; Sutthiphai et al., 2022; Lu et al., 2023). Sequence analysis indicated that AVR-Pi9 had a relatively low genetic diversity (Sutthiphai et al., 2022; Lu et al., 2023).
Molecular interactions between M. oryzae AVR effectors and rice R/target proteins
The detection of AVR effectors by cognate R proteins occurs via either direct or indirect interactions in Rice-M. oryae and other pathosystems (Stergiopoulos and de Wit, 2009). Direct recognition depends on physical binding of AVR effectors to the R proteins and indirect recognition involves the perception of effector-induced modifications of other host targets (usually termed guardees or decoys) by R proteins. It is considered that guardees play certain roles in plant immunity, while decoys specialize in trapping effectors without immune function (Van der Hoorn and Kamoun, 2008; Khan et al., 2016; Ao and Li, 2022). NLRs, as the most prevalent group of characterized R proteins in rice, function as singletons or pairs (Xi et al., 2022). Among over 40 cloned rice R genes, only Pid-2, pi21, Ptr, Pi65 and Pb4 encode non-NLR proteins (Devanna et al., 2022; Shimizu et al., 2022; Xiao et al., 2023; Fan et al., 2024). Most of the rice NLR pairs are genetically linked in a head-to-head orientation. One containing a noncanonical ID acts as a sensor NLR (sNLR) to directly detect the presence of the AVR effector(s), whereas the other one is a canonical NLR acting as a helper (hNLR) to transduce signals to activate immunity (Lüdke et al., 2022; Cadiou et al., 2023; Contreras et al., 2023). Singleton NLRs are capable of mediating both AVR effectors perception and downstream defense signaling initiation without relying on partner NLRs. Recent studies have revealed an extremely complex picture of M. oryzae AVR effectors and rice R/target proteins (Figure 1).
AVR effector vs. singleton NLR
Pita-AVR-Pita is the earliest studied pair of R-AVR in rice-M. oryzae pathosystem and has long been accepted as a classic example of direct AVR effector binding by NLR (Bryan et al., 2000; Jia et al., 2000; Orbach et al., 2000). The mature form of AVR-Pita containing 176 aa at the C-terminus was found to bind specifically to the LRR region of Pita (Orbach et al., 2000). Single amino acid substitutions in either the LRR region of Pita or protease motif of AVR-Pita disrupt the physical interaction, resulting in failure of defense response initiation. Ptr/Pita2 was later found to be not only closely linked to Pita but also involved in Pita-mediated resistance (Zhao et al., 2018; Meng et al., 2020). Ptr/Pita2 encodes an atypical R protein with four Armadillo (ARM) repeats and its disruption leads to a loss of resistance to some AVR-Pita containing isolates, suggesting that it is required for the complete function of Pita. However, a very recent study indicated that Pita is involved in neither Pita resistance nor AVR-Pita detection (Xiao et al., 2024). It also has no role in Ptr/Pita2-mediated resistance. The Pita resistance is indeed provided by one of Ptr alleles, designated PtrB, which recognizes a restricted set of AVR-Pita alleles through an indirect way. PtrA can detect all natural AVR-Pita alleles and confers Pita2 resistance. Additionally, AVR-Pita was found to target OsCOX11, a cytochrome C oxidase (COX) assembly protein, in rice mitochondria (Han et al., 2021). OsCOX11 participates in ROS metabolism and plays a negative role in rice resistance. The AVR-Pita-OsCOX11 interaction increases the COX activity in ROS metabolism, thereby inhibiting ROS accumulation and suppressing rice innate immunity. Pi54-AvrPi54 is currently the only case of direct interaction between AVR effector and singleton NLR (Ray et al., 2016). Pi54 physically interacts with AvrPi54 at the host plasma membrane, which restricts the movement of AvrPi54 into nucleus for its virulence function (Saklani et al., 2023).
Three other singleton NLRs Pib, Pi9 and Piz-t recognize their cognate AVR effectors AvrPib, AvrPi9 and AvrPiz-t, respectively, via indirect way. SH3P2, an SH3 domain-containing protein mediates indirect AvrPib-Pib recognition (Xie et al., 2022). SH3P2 functions as a ‘‘protector’’ to associate with Pib mainly at clathrin-coated vesicles (CCV) in rice cells, which is an important coated vesicle responsible for endocytosis and many post-Golgi trafficking processes. The SH3P2-Pib association interferes with the Pib homodimerization by disrupting CC domain self-association, thus maintaining Pib in static state under normal growth conditions. Since SH3P2 associates with CCVs, which suggests SH3P2 may possess conserved intracellular trafficking functions and transfer Pib cargo to vacuoles for degradation, thereby maintaining a low abundance of Pib in the absence of blast fungus infection. Interestingly, it was found that SH3P2 also associates with the CC domains of Pi2 and Pita, but it is unclear whether SH3P2 affects resistance mediated by these two R proteins. SH3P2 can also bind to AvrPib in CCVs with higher affinity. Upon invasion of M. oryzae isolates containing AvrPib, the competitive binding of AvrPib to SH3P2 releases Pib from the OsSH3P2-Pib complex and alleviates the inhibition of Pib homodimerization, thus eventually activates Pib-mediated resistance. ANIP1-OsWRKY62 module was recently found to be targeted by AvrPi9 and regulates rice immunity in the presence/absence of Pi9 in distinct ways (Shi et al., 2023). ANIP1 is a rice ubiquitin-like domain-containing protein (UDP) subjected to 26S proteasome-mediated degradation. Both AvrPi9 and Pi9 can directly interact with and stabilize ANIP1 through disturbing its degradation. Moreover, ANIP1 physically interacts with the rice WRKY transcription factor OsWRKY62 and affects its abundance by promoting its degradation. OsWRKY62 was also found to interact with AvrPi9 and Pi9. In the absence of Pi9, lower abundance of ANIP1 leads to OsWRKY62 accumulation in rice plants and enhanced immunity during infection by M. oryzae isolates without AvrPi9. When infected by M. oryzae isolates with AvrPi9, ANIP1 is stabilized by AvrPi9 that more efficiently promotes the degradation of OsWRKY62, thus decreasing the immune response. In the presence of Pi9, it binds to and stabilize ANIP1-OsWRKY62 module. They form a complex with unknown adaptor(s) to maintain Pi9 in its inactive state under normal growth conditions. Under invasion by non-AvrPi9 M. oryzae isolates, the forming complex decreases plant immunity. Under invasion by M. oryzae isolates carrying AvrPi9, AvrPi9 promotes the dissociation of ANIP1 from Pi9, which further activate Pi9 and downstream immune responses. In contrast to ANIP as a negative regulator of rice immunity, the AvrPi9 interacting proteins OsRGLG5 and PICI1 were found to positively regulate rice defense (Zhai et al., 2022; Liu et al., 2023c). Both of these two proteins can be targeted for degradation. OsRGLG5, encoding a functional RING-type E3 ubiquitin ligase, functions as a positive regulator of basal resistance, but it is not required for Pi9-mediated blast resistance and no physical interaction between OsRGLG5 and Pi9 was observed, which suggested that OsRGLG5 may not be a guardee in the Pi9-AvrPi9 interaction (Liu et al., 2023c). In response, OsRGLG5 ubiquitinates and subsequently degrades AvrPi9 through the 26S proteasome pathway. The deubiquitinase PICI1, acts as an immune hub for both PTI and ETI through the methionine-ethylene cascade (Zhai et al., 2022). AvrPi9 was found to promote PICI1 degradation in a proteasome-independent manner, which in turns promotes methionine synthases OsMETS1 and OsMETS2 ubiquitination and degradation, leading to reduced methionine and ethylene biosynthesis, as well as comprised PTI. While NLRs, such as PigmR, Pi9 and Pizt, protect PICI1 from AvrPi9 binding in a competitive manner to reboot the methionine-ethylene-mediated immunity. AvrPiz-t was reported to target 12 APIPs (AvrPiz-t interacting proteins) in rice and the immune functions of several APIPs including APIP4 (Bowman-Birk trypsin inhibitor protein), APIP5 (bZIP transcription factor), APIP6 (Ring type E3 ubiquitin ligase), APIP7 (plasma membrane potassium channel), APIP10 (RING-type E3 ubiquitin ligases) and APIP12 (homologue of nucleoporin protein Nup98) have been well characterized (Park et al., 2012, 2016; Wang et al., 2016; Tang et al., 2017; Shi et al., 2018; Zhang et al., 2020a). AvrPiz-t can block the E3 ligase activity of APIP6 and APIP10 to suppress rice PTI (Park et al., 2012, 2016). In return, these two E3 ligase ubiquitinate and degrade AvrPiz-t to reduce its suppressive effects on rice PTI. APIP10 also promotes degradation of the Piz-t protein through the 26S proteasome system, although no direct interaction between these two proteins were observed. During M. oryzae infection, AvrPiz-t interferes the negative regulation of APIP10 on Piz-t, which leads to rapid accumulation of Piz-t protein and initiation of ETI (Park et al., 2016). The transcription factors OsVOZ1 and OsVOZ2 were found to bridge the connection between APIP10 and Piz-t (Wang et al., 2021). They function synergistically to negatively regulate basal defense but positively regulate Piz-t-mediated immunity. ROD1 is a C2 domain Ca2+ sensor, which recruits catalase CatB (OsCATB) to increase its activity for ROS elimination. AvrPiz-t structurally mimics ROD1 and executes similar ROS-scavenging-mediated immune suppression (Gao et al., 2021). But both ROD1 and AvrPiz-t can be targeted for ubiquitin-mediated degradation by APIP6 and the other E3 ligase RIP1. Besides, APIP6 can also ubiquitinate the catalase OsCATC, the peroxisomal receptor protein OsPEX5, and OsELF3-2, an ortholog of the Arabidopsis ELF3, and promotes their degradation via the 26S proteasome pathway to positively regulate basal defense against M. oryzae (You et al., 2022, 2023). OsPEX5 was further found to stabilize the aldehyde dehydrogenase OsALDH2B1 to enhance its repression of the defense-related gene OsAOS2. The Bowman-Birk trypsin inhibitor (BBTI) APIP4 functions as a positive regulator of rice blast resistance (Zhang et al., 2020a). The interaction between AvrPiz-t and APIP4 suppress its trypsin inhibitor activity, while the binding of APIP4 with Piz-t potentially promotes the activity of APIP4, resulting in enhanced rice immunity. Like APIP4, APIP5 is the target of both AvrPiz-t and Piz-t. It plays a critical role in preventing effector-triggered necrosis (ETN) during the necrotrophic stage of M. oryzae infection (Wang et al., 2016; Zhang et al., 2022). APIP5 directly targets the cell wall-associated kinase gene OsWAK5 and the cytochrome P450 gene CYP72A1 as a transcription factor to inhibit their expression, resulting in less lignin, ROS and defense compounds accumulation. Besides, APIP5 regulates mRNA turnover of the cell death- and defense-related genes OsLSD1 and OsRac1 as an RNA-binding protein. AvrPiz-t attenuates the transcriptional activity and protein accumulation of APIP5, leading to ETN at the necrotrophic stage. Piz-t can stabilize APIP5 and reduce the AvrPiz-t-mediated APIP5 turnover to prevent ETN. In turn, APIP5 is essential for the accumulation of Piz-t for the activation of ETI. A recent work showed that APIP5 directly suppresses the transcription of APIP4 and its homolog OsBBTI5, thereby attenuating their trypsin inhibitor activity to weaken the disease resistance (Zhang et al., 2024a). APIP4 and OsBBTI5 were further proved to associate and stabilize the defense-related protein OsPR1aL, which positively regulates rice blast resistance. APIP7 (OsAKT1) forms a complex with OsCBL1 and OsCIPK23, modulating K+ signal transduction for plant growth and development, as well as immunity (Shi et al., 2018). AvrPiz-t suppresses the activity of APIP7 and/or interferes with the APIP7-OsCIPK23 complex to subvert inward K+ currents in favor of M. oryzae pathogenesis. APIP12, targeted by both AvrPiz-t and APIP6, is involved in the basal resistance but not the Piz-t mediated resistance (Tang et al., 2017). AvrPiz-t was also found to interact with OsRac1 to suppress ROS generation (Bai et al., 2019).
AVR effector vs. paired NLRs
The genetically and molecularly co-acting NLR pairs are prevalent in rice and other plant genomes (Duxbury et al., 2021; Xi et al., 2022). The Pia pair RGA4/RGA5 recognize two sequence-unrelated AVR effectors, AVR-Pia and AVR1-CO39 (Cesari et al., 2013). Both these two AVR effectors bind to the HMA ID integrated into the sNLR, RGA5. RGA4, as the hNLR, is autoactive and its function is repressed by RGA5 in the absence of pathogen (Cesari et al., 2014). This repression is relieved upon direct interaction of AVR-Pia or AVR1-CO39 with the HMA domain in RGA5, leading to activation of ETI (Cesari et al., 2013; Ortiz et al., 2017). Pias pair Pias-1/Pias-2, which is allelic Pia pair, detects the AVR effector AVR-Pias. Interestingly, the sNLR Pias-2 carries a different ID, DUF761, and no direct binding between AVR-Pias and DUF761 of Pias-2 was observed (Shimizu et al., 2022). For the Pik pair Pik-1/Pik-2 and its cognate AVR effector Avr-Pik, both of them exist in an allelic series in rice and M. oryzae, respectively (Yoshida et al., 2009; Kanzaki et al., 2012; Wu et al., 2014; De la Concepcion et al., 2018). At least 7 Pik alleles (Pi1, Pik, Pikm, Pikp, Piks, Pikh and Pike) and 6 AVR-Pik variants (A-F) have been reported. The Pik alleles showing different recognition specificities to AVR-Pik variants. Pik-1 recognition of AVR-Pik is mediated by direct binding of the AVR effector to a HMA domain, integrated into between the CC and NB‐ARC domains. In contrast to RGA4, the hNLR Pik-2 does not show autoimmunity in an ectopic expression system, and both NLRs are required to trigger an immune response upon perceiving the matching AVR effector (Maqbool et al., 2015). Besides, AVR-Pik variants interact with a subset of small HMA‐containing (sHMA) protein, which belong to heavy metal-associated plant proteins (HPPs) and heavy metal-associated isoprenylated plant proteins (HIPPs) (Maidment et al., 2021; Oikawa et al., 2024). AVR-PikD binds and stabilizes OsHIPP19 and OsHIPP20 in plant cells. The binding affects the subcellular distribution of the OsHIPP19 and OsHIPP20. Knockout of OsHIPP20 conferred enhanced resistance to infection by the blast pathogen, suggesting OsHIPP20 is a susceptibility gene. Therefore, it is hypothesized that AVR-Pik-mediated stabilization of sHMA proteins suppresses rice defenses. Additionally, AVR-Mgk1, an effector sharing no sequence similarity to known AVR-Pik family, is found on a mini-chromosome and detected by Piks as well as other multiple Pik alleles (Sugihara et al., 2023). Recent studies reported that Avr-PikD interacts with the zinc finger−type transcription factor WG7 and the LSD1-like transcription factor AKIP30. WG7 negatively regulates immunity through SA signaling pathway (Yang et al., 2024). Avr-PikD suppresses rice immunity by targeting WG7 in nucleus and promoting its transcriptional activity. By contrast, AKIP30 is also a positive regulator of rice immunity. Avr-PikD interferes with the expression, subcellular localization and transcriptional activity of AKIP30, thereby facilitating ETS (Guo et al., 2024). AVR-Pii interacts with two members of rice Exo70 family, OsExo70F2 and OsExo70F3, suggesting that the pathogen may target exocyst-mediated trafficking as a virulence-associated mechanism (Fujisaki et al., 2015). Exo70, a component of the exocyst complex, plays crucial roles in tethering and fusion of the vesicles and plasma membrane at the site of polarized exocytosis (Munson and Novick, 2006). It was revealed that OsExo70F3 is specifically involved in Pii-dependent resistance (Fujisaki et al., 2015). The association of AVR-Pii with OsExo70F3 is monitored by Pii through an unconventional RIN4/NOI domain integrated in the sNLR Pii-2 (Fujisaki et al., 2017). AVR-Pii also targets OsNADP-ME2, a rice nicotinamide adenine dinucleotide phosphate-malic enzyme, and inhibit its activity to limit ROS accumulation and suppress basal resistance (Singh et al., 2016).
AVR effector vs. uncharacterized R protein
Even though PWL2 is capable of being recognized by the NLR protein Mla3 in barely which confers resistance to Blumeria graminis and M. oryzae, its corresponding R protein in rice has not yet been identified (Brabham et al., 2024). More recently, it was reported PWL2 specifically binds to HIPP43 in rice and its orthologs from other grass species (Were et al., 2024; Zdrzałek et al., 2024). HIPP43 is a susceptibility factor for infection, since overexpression of HIPP43 suppresses PAMP-induced ROS in transgenic plants. PWL2 targets HIPP43 to stabilize and alter plasmodesmata localization of HIPP43, thus enhancing susceptibility (Were et al., 2024). ACE1, coding for a PKS-NRPS hybrid, is the only non-secreted AVR effector in M. oryzae to date (Böhnert et al., 2004). It is located in a secondary metabolite gene cluster exclusively expressed during fungal appressorium-mediated penetration (Collemare et al., 2008). The AVR signal detected by the R protein Pi33(t) is not the ACE1, but the secondary metabolite synthesized by it. However, the expression of ACE1 is under strict temporal and cell type-specific regulation and its produced secondary metabolite is extremely difficult to isolate. Ectopic expression of ACE1 indicated that the metabolite is likely to be a tyrosine-derived cytochalasan compound (Song et al., 2015). But the exact AVR molecule remains to be determined.
Structural overview of M. oryzae AVR effectors and their interactions with rice R/target proteins
In natural pathosystems, AVR effectors are under strong selection pressure to adapt to specific or new hosts and evade immunity, which has driven their rapid expansion and diversification (Fouché et al., 2018). The majority of fungal AVR effectors share low sequence similarity with each other or with other proteins of known function (Ellis et al., 2009). Therefore, the prediction on their function is challenging. A protein’s three-dimensional structure can provide key insights into function and evolution. As such, structural determination has become an avenue pursued to understand roles of the effectors in the infection process. Till now, the 3-dimensional structures of seven AVR effectors including AvrPiz-t, AVR-Pia, AVR1-CO39, AVR-Pik variants, AvrPib, AVR-Pii and PWL2 have been experimentally solved (Zhang et al., 2013; De Guillen et al., 2015; Maqbool et al., 2015; Ose et al., 2015; Zhang et al., 2018; De la Concepcion et al., 2022). All these AVR effectors except AVR-Pii belong to the MAX (Magnaporthe AVRs and ToxB-like) effector family, which accounts for 5-10% of the effector repertoire in M. oryzae (De Guillen et al., 2015; Kotsaridis et al., 2023). The crystal structure of AVR-Pii/OsExo70F2 complex revealed a fold for AVR-Pii based on a zinc-finger (ZiF) motif sustained by four residues coordinating a Zn2+ atom and the structure has not been previously reported for other phytopathogen effectors. AVR-Pii binds to Exo70 via a conserved hydrophobic pocket (De la Concepcion et al., 2022). MAX effectors share a common fold with six-stranded β-sheet sandwich and the fold is stabilized by at least one disulfide bond between conserved cysteins connecting β1 and β5 (De Guillen et al., 2015). Even containing the similar structure, distinct shapes and surface properties due to the varying orientation and length of β-strands and loops constitute the basis of diversity in their functions. For example, AvrPib and AvrPiz-t contain the shorter β-strand β6 at the C-terminus, while the shorter one of AVR1-CO39, AVR-Pia and AVR-Pik is β5 (Zhang et al., 2018). AVR1-CO39, AvrPiz-t and AvrPib have dominant charge patch(es) on the surfaces, but AVR-Pia and AVR-Pik have only hydrophobic patch with multiple charged residues distributed separately on the surfaces.
Bioengineering of rice NLRs guided by structural knowledge of NLR-AVR interactions
Management of rice blast disease is cumbersome, even though rice R genes have been extensively used in breeding (Wang and Valent, 2017; Younas et al., 2023). The recognition spectra of R proteins tend to be specific and M. oryzae may delete AVR effectors from their genome or evolve novel AVR variants that evade detection by the R proteins to re-establish infection (Maekawa et al., 2011). With increasing mechanistic and structural insights into the NLR-ID-AVR interactions, bioengineering of NLR’s ID has emerged as a promising approach to expand its recognition specificities. Recent studies have reported that HMA domain engineering is an effective way to generate new resistance specificities. A binding interface was grafted from Pikm-1-HMA onto Pikp-1-HMA by mutating two residues in Pikp-1 and the engineered variant gained an expanded recognition profile to AVR-Pik variants previously unrecognized by Pikp in N. benthamiana (De la Concepcion et al., 2019). Introduction of the HMA or three specific residues in the interface of OsHIPP19 into Pikp-1-HMA creates Pikp-1 variants that recognize all known AVR-Pik alleles including AVR-PikC and AVR-PikF, which are not detected by naturally occurring Pik-1, not only in N. benthamiana but also in rice (Maidment et al., 2021). Integration of the HMA of OsHIPP43 into the Pikm-1 switches recognition from AVR-Pik to PWL2, as well as PWL1 and PWL3 in N. benthamiana (Zdrzałek et al., 2024). By combining the AVR-PikD binding residues of Pikp-1-HMA into RGA5-HMA, a variant gained an extended resistance specificity in N. benthamiana but not in transgenic rice (Cesari et al., 2022). The modified sites may affect NLR activation or additional interactions with RGA5 outside the ID might be important for recognition. In another two studies, RGA5-HMA was engineered by comparing the structures of AVR1-CO39 and the noncorresponding AVR-Pib and AVR-PikD for predicting their potential interface. The engineered RGA5 confers specific resistance to M. oryzae strains expressing AvrPib or AVR-PikD in transgenic rice (Liu et al., 2021b; Zhang et al., 2024b). More recently, a groundbreaking approach for molecular engineering of Pikm-1 by replacing HMA ID with camelid-derived nanobodies of fluorescent proteins (FP) was reported (Kourelis et al., 2023). The synthetic Pikm-1s with nanobodies trigger HR in the presence of Pikm-2 and the corresponding fluorescent proteins in N. benthamiana and confer resistance against plant viruses expressing FPs. These studies collectively demonstrated the potential for engineering IDs to alter the recognition profiles of the NLR proteins.
Conclusion and future perspectives
Over the past three decades, despite our understanding the roles of AVR effectors of M. oryzae in establishing interactions with rice and other hosts is increasing, many issues and challenges (listed below) remain to be resolved:
(1) The AVR effectors corresponding to the majority of known R proteins, particularly those with broad-spectrum resistance, such as Pigm, Pi2, etc., have not yet been isolated. These AVR effectors may be highly conserved and prevalent across M. oryzae population in the field. The loss of them likely imposes fitness penalties on the pathogen (Leach et al., 2001; Bart et al., 2012).
(2) Since wheat is currently threatened by the expanding blast pandemic, research efforts are urgent to isolate more AVR effector and R protein pairs. It will enable the study of their molecular interactions and the potential for engineering resistance against the Triticum pathotype of M. oryzae.
(3) What is the final product synthetized by ACE1 and how does Pi33 detect the AVR signal?
(4) The structural mechanism underlying the transformation of NLRs from their static to activated states upon recognition of AVR effectors needs further investigation.
(5) What are the detailed molecular events downstream once R protein is activated by AVR effectors?
(6) Little is known about the detailed mechanism by which the cytoplasmic AVR effectors are internalized and transported into plant cells. Once entering into the cytoplasm, how these AVR effectors move into the cellular organelles for virulence and avirulence functions remains to be addressed.
Future research in these fields will undoubtedly reveal novel strategies of M. oryzae AVR effectors participating in rice resistance/susceptibility that can be exploited to control blast disease with high efficiency and durability.
Author contributions
XL: Writing – original draft, Writing – review & editing. XH: Writing – original draft, Writing – review & editing. ZT: Writing – review & editing, Writing – original draft. ZS: Writing – review & editing. PQ: Funding acquisition, Writing – review & editing. YL: Writing – review & editing. XC: Writing – review & editing. ZL: Supervision, Writing – review & editing. NJ: Writing – original draft, Writing – review & editing, Supervision. YY: Funding acquisition, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by grants from the STI 2030-Major Projects (2022ZD0400202).
Conflict of interest
Authors XL, XH, ZT, ZS, PQ, YL, XC, NJ, and YY were employed by Yuan Longping High-Tech Agriculture Co., Ltd.
The remaining author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Adachi, H., Derevnina, L., Kamoun, S. (2019). NLR singletons, pairs, and networks: evolution, assembly, and regulation of the intracellular immunoreceptor circuitry of plants. Curr. Opin. Plant Biol. 50, 121–131. doi: 10.1016/j.pbi.2019.04.007
Anh, V. L., Inoue, Y., Asuke, S., Vy, T. T. P., Anh, N. T., Wang, S., et al. (2018). Rmg8 and Rmg7, wheat genes for resistance to the wheat blast fungus, recognize the same avirulence gene AVR-Rmg8. Mol. Plant Pathol. 19, 1252–1256. doi: 10.1111/mpp.12609
Ao, K., Li, X. (2022). Indirect recognition of pathogen effectors by NLRs. Essays Biochem. 66, 485–500. doi: 10.1042/ebc20210097
Arora, S., Steed, A., Goddard, R., Gaurav, K., O’Hara, T., Schoen, A., et al. (2023). A wheat kinase and immune receptor form host-specificity barriers against the blast fungus. Nat. Plants 9, 385–392. doi: 10.1038/s41477-023-01357-5
Asuke, S., Horie, A., Komatsu, K., Mori, R., Vy, T. T. P., Inoue, Y., et al. (2023). Loss of PWT7, located on a supernumerary chromosome, is associated with parasitic specialization of Pyricularia oryzae on wheat. Mol. Plant Microbe Interact. 36, 716–725. doi: 10.1094/mpmi-06-23-0078-r
Asuke, S., Morita, K., Shimizu, M., Abe, F., Terauchi, R., Nago, C., et al. (2024). Evolution of wheat blast resistance gene Rmg8 accompanied by differentiation of variants recognizing the powdery mildew fungus. Nat. Plants 10, 971–983. doi: 10.1038/s41477-024-01711-1
Bai, P., Park, C. H., Shirsekar, G., Songkumarn, P., Bellizzi, M., Wang, G. L. (2019). Role of lysine residues of the Magnaporthe oryzae effector AvrPiz-t in effector- and PAMP-triggered immunity. Mol. Plant Pathol. 20, 599–608. doi: 10.1111/mpp.12779
Bart, R., Cohn, M., Kassen, A., McCallum, E. J., Shybut, M., Petriello, A., et al. (2012). High-throughput genomic sequencing of cassava bacterial blight strains identifies conserved effectors to target for durable resistance. Proc. Natl. Acad. Sci. U.S.A. 109, E1972–E1979. doi: 10.1073/pnas.1208003109
Block, A., Li, G., Fu, Z. Q., Alfano, J. R. (2008). Phytopathogen type III effector weaponry and their plant targets. Curr. Opin. Plant Biol. 11, 396–403. doi: 10.1016/j.pbi.2008.06.007
Böhnert, H. U., Fudal, I., Dioh, W., Tharreau, D., Notteghem, J. L., Lebrun, M. H. (2004). A putative polyketide synthase/peptide synthetase from Magnaporthe grisea signals pathogen attack to resistant rice. Plant Cell 16, 2499–2513. doi: 10.1105/tpc.104.022715
Boller, T., Felix, G. (2009). A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60, 379–406. doi: 10.1146/annurev.arplant.57.032905.105346
Boller, T., He, S. Y. (2009). Innate immunity in plants: an arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science 324, 742–744. doi: 10.1126/science.1171647
Bourett, T. M., Howard, R. J. (1990). In vitro development of penetration structures in the rice blast fungus Magnaporthe grisea. Can. J. Bot. 68, 329–342. doi: 10.1139/b90-044
Brabham, H. J., Gómez de la Cruz, D., Were, V., Shimizu, M., Saitoh, H., Hernández-Pinzón, I., et al. (2024). Barley MLA3 recognizes the host-specificity effector Pwl2 from Magnaporthe oryzae. Plant Cell 36 (2), 447–470. doi: 10.1093/plcell/koad266
Bryan, G. T., Wu, K. S., Farrall, L., Jia, Y., Hershey, H. P., McAdams, S. A., et al. (2000). A single amino acid difference distinguishes resistant and susceptible alleles of the rice blast resistance gene Pi-ta. Plant Cell 12, 2033–2046. doi: 10.1105/tpc.12.11.2033
Cadiou, L., Brunisholz, F., Cesari, S., Kroj, T. (2023). Molecular engineering of plant immune receptors for tailored crop disease resistance. Curr. Opin. Plant Biol. 74, 102381. doi: 10.1016/j.pbi.2023.102381
Ceresini, P. C., Castroagudín, V. L., Rodrigues, F., Rios, J. A., Eduardo Aucique-Pérez, C., Moreira, S. I., et al. (2018). Wheat Blast: past,present, and future. Annu. Rev. Phytopathol. 56, 427–456. doi: 10.1146/annurev-phyto-080417-050036
Cesari, S. (2018). Multiple strategies for pathogen perception by plant immune receptors. New Phytol. 219, 17–24. doi: 10.1111/nph.14877
Cesari, S., Bernoux, M., Moncuquet, P., Kroj, T., Dodds, P. N. (2014). A novel conserved mechanism for plant NLR protein pairs: the “integrated decoy” hypothesis. Front. Plant Sci. 5. doi: 10.3389/fpls.2014.00606
Cesari, S., Thilliez, G., Ribot, C., Chalvon, V., Michel, C., Jauneau, A., et al. (2013). The rice resistance protein pair RGA4/RGA5 recognizes the Magnaporthe oryzae effectors AVR-Pia and AVR1-CO39 by direct binding. Plant Cell 25, 1463–1481. doi: 10.1105/tpc.112.107201
Cesari, S., Xi, Y., Declerck, N., Chalvon, V., Mammri, L., Pugnière, M., et al. (2022). New recognition specificity in a plant immune receptor by molecular engineering of its integrated domain. Nat. Commun. 13, 1524. doi: 10.1038/s41467-022-29196-6
Chen, S., Songkumarn, P., Venu, R. C., Gowda, M., Bellizzi, M., Hu, J., et al. (2013). Identification and characterization of in planta-expressed secreted effector proteins from Magnaporthe oryzae that induce cell death in rice. Mol. Plant Microbe Interact. 26, 191–202. doi: 10.1094/mpmi-05-12-0117-r
Chiapello, H., Mallet, L., Guérin, C., Aguileta, G., Amselem, J., Kroj, T., et al. (2015). Deciphering genome content and evolutionary relationships of isolates from the fungus Magnaporthe oryzae attacking different host plants. Genome Biol. Evol. 7, 2896–2912. doi: 10.1093/gbe/evv187
Choi, J., Park, J., Kim, D., Jung, K., Kang, S., Lee, Y. H. (2010). Fungal secretome database: integrated platform for annotation of fungal secretomes. BMC Genom. 11, 105. doi: 10.1186/1471-2164-11-105
Collemare, J., Pianfetti, M., Houlle, A. E., Morin, D., Camborde, L., Gagey, M. J., et al. (2008). Magnaporthe grisea avirulence gene ACE1 belongs to an infection-specific gene cluster involved in secondary metabolism. New Phytol. 179, 196–208. doi: 10.1111/j.1469-8137.2008.02459.x
Contreras, M. P., Lüdke, D., Pai, H., Toghani, A., Kamoun, S. (2023). NLR receptors in plant immunity: making sense of the alphabet soup. EMBO Rep. 24, e57495. doi: 10.15252/embr.202357495
Couch, B. C., Fudal, I., Lebrun, M. H., Tharreau, D., Valent, B., van Kim, P., et al. (2005). Origins of host-specific populations of the blast pathogen Magnaporthe oryzae in crop domestication with subsequent expansion of pandemic clones on rice and weeds of rice. Genetics 170, 613–630. doi: 10.1534/genetics.105.041780
Dagdas, Y. F., Yoshino, K., Dagdas, G., Ryder, L. S., Bielska, E., Steinberg, G., et al. (2012). Septin-mediated plant cell invasion by the rice blast fungus, Magnaporthe oryzae. Science 336, 1590–1595. doi: 10.1126/science.1222934
Dean, R. A., Talbot, N. J., Ebbole, D. J., Farman, M. L., Mitchell, T. K., Orbach, M. J., et al. (2005). The genome sequence of the rice blast fungus Magnaporthe grisea. Nature 434, 980–986. doi: 10.1038/nature03449
Dean, R., Van Kan, J. A., Pretorius, Z. A., Hammond-Kosack, K. E., Di Pietro, A., Spanu, P. D., et al. (2012). The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 13, 414–430. doi: 10.1111/j.1364-3703.2011.00783.x
De Guillen, K., Ortiz-Vallejo, D., Gracy, J., Fournier, E., Kroj, T., Padilla, A. (2015). Structure analysis uncovers a highly diverse but structurally conserved effector family in phytopathogenic fungi. PloS Pathog. 11, e1005228. doi: 10.1371/journal.ppat.1005228
De la Concepcion, J. C., Franceschetti, M., MacLean, D., Terauchi, R., Kamoun, S., Banfield, M. J. (2019). Protein engineering expands the effector recognition profile of a rice NLR immune receptor. Elife. 8, e47713. doi: 10.7554/eLife.47713
De la Concepcion, J. C., Franceschetti, M., Maqbool, A., Saitoh, H., Terauchi, R., Kamoun, S., et al. (2018). Polymorphic residues in rice NLRs expand binding and response to effectors of the blast pathogen. Nat. Plants 4, 576–585. doi: 10.1038/s41477-018-0194-x
De la Concepcion, J. C., Fujisaki, K., Bentham, A. R., Cruz Mireles, N., Sanchez de Medina Hernandez, V., Shimizu, M., et al. (2022). A blast fungus zinc-finger fold effector binds to a hydrophobic pocket in host Exo70 proteins to modulate immune recognition in rice. Proc. Natl. Acad. Sci. U.S.A. 119, e2210559119. doi: 10.1073/pnas.2210559119
Devanna, B. N., Jain, P., Solanke, A. U., Das, A., Thakur, S., Singh, P. K., et al. (2022). Understanding the dynamics of blast resistance in rice-Magnaporthe oryzae interactions. J. Fungi(Basel) 8 (6), 584. doi: 10.3390/jof8060584
Dodds, P. N., Lawrence, G. J., Pryor, A., Ellis, J. G., Dickinson, M., Beynon, J. (2018). “Genetic analysis and evolution of plant disease resistance genes,” in Annual Plant Reviews online (UK: Wiley), 92–112.
Dong, Y., Li, Y., Zhao, M., Jing, M., Liu, X., Liu, M., et al. (2015). Global genome and transcriptome analyses of Magnaporthe oryzae epidemic isolate 98-06 uncover novel effectors and pathogenicity-related genes, revealing gene gain and lose dynamics in genome evolution. PloS Pathog. 11, e1004801. doi: 10.1371/journal.ppat.1004801
Duxbury, Z., Ma, Y., Furzer, O. J., Huh, S. U., Cevik, V., Jones, J. D., et al. (2016). Pathogen perception by NLRs in plants and animals: Parallel worlds. Bioessays 38, 769–781. doi: 10.1002/bies.201600046
Duxbury, Z., Wu, C. H., Ding, P. (2021). A comparative overview of the intracellular guardians of plants and animals: NLRs in innate immunity and beyond. Annu. Rev. Plant Biol. 72, 155–184. doi: 10.1146/annurev-arplant-080620-104948
Ellis, J. G., Rafiqi, M., Gan, P., Chakrabarti, A., Dodds, P. N. (2009). Recent progress in discovery and functional analysis of effector proteins of fungal and oomycete plant pathogens. Curr. Opin. Plant Biol. 12, 399–405. doi: 10.1016/j.pbi.2009.05.004
Fan, Y., Ma, L., Pan, X., Tian, P., Wang, W., Liu, K., et al. (2024). Genome-wide association study identifies rice panicle blast-resistant gene Pb4 encoding a wall-associated kinase. Int. J. Mol. Sci. 25 (2), 830. doi: 10.3390/ijms25020830
Fisher, M. C., Henk, D. A., Briggs, C. J., Brownstein, J. S., Madoff, L. C., McCraw, S. L., et al. (2012). Emerging fungal threats to animal, plant and ecosystem health. Nature 484, 186–194. doi: 10.1038/nature10947
Fouché, S., Plissonneau, C., Croll, D. (2018). The birth and death of effectors in rapidly evolving filamentous pathogen genomes. Curr. Opin. Microbiol. 46, 34–42. doi: 10.1016/j.mib.2018.01.020
Fujisaki, K., Abe, Y., Ito, A., Saitoh, H., Yoshida, K., Kanzaki, H., et al. (2015). Rice Exo70 interacts with a fungal effector, AVR-Pii, and is required for AVR-Pii-triggered immunity. Plant J. 83, 875–887. doi: 10.1111/tpj.12934
Fujisaki, K., Abe, Y., Kanzaki, E., Ito, K., Utsushi, H., Saitoh, H., et al. (2017). An unconventional NOI/RIN4 domain of a rice NLR protein binds host EXO70 protein to confer fungal immunity. bioRxiv. doi: 10.1101/239400
Gao, M., He, Y., Yin, X., Zhong, X., Yan, B., Wu, Y., et al. (2021). Ca2+ sensor-mediated ROS scavenging suppresses rice immunity and is exploited by a fungal effector. Cell 184, 5391–5404 e5317. doi: 10.1016/j.cell.2021.09.009
Giraldo, M. C., Dagdas, Y. F., Gupta, Y. K., Mentlak, T. A., Yi, M., Martinez-Rocha, A. L., et al. (2013). Two distinct secretion systems facilitate tissue invasion by the rice blast fungus Magnaporthe oryzae. Nat. Commun. 4, 1996. doi: 10.1038/ncomms2996
Gladieux, P., Condon, B., Ravel, S., Soanes, D., Maciel, J. L. N., Nhani, A., Jr., et al. (2018a). Gene flow between divergent cereal-and grass-specific lineages of the rice blast fungus Magnaporthe oryzae. mBio 9 (1), e01219–17. doi: 10.1128/mBio.01219-17
Gladieux, P., Ravel, S., Rieux, A., Cros-Arteil, S., Adreit, H., Milazzo, J., et al. (2018b). Coexistence of multiple endemic and pandemic lineages of the rice blast pathogen. mBio 9 (2), e01806–17. doi: 10.1128/mBio.01806-17
Guo, J., Wu, Y., Huang, J., Yu, K., Chen, M., Han, Y., et al. (2024). The Magnaporthe oryzae effector Avr-PikD suppresses rice immunity by inhibiting an LSD1-like transcriptional activator. Crop J. 12, 482–492. doi: 10.1016/j.cj.2024.01.011
Han, J., Wang, X., Wang, F., Zhao, Z., Li, G., Zhu, X., et al. (2021). The fungal effector Avr-pita suppresses innate immunity by increasing COX activity in rice mitochondria. Rice(N Y) 14, 12. doi: 10.1186/s12284-021-00453-4
Hossain, M. M. (2022). Wheat blast: A review from a genetic and genomic perspective. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.983243
Hu, H. W., Chen, Q. L., He, J. Z. (2022a). The end of hunger: fertilizers, microbes and plant productivity. Microb. Biotechnol. 15, 1050–1054. doi: 10.1111/1751-7915.13973
Hu, Z. J., Huang, Y. Y., Lin, X. Y., Feng, H., Zhou, S. X., Xie, Y., et al. (2022b). Loss and natural variations of blast fungal avirulence genes breakdown rice resistance genes in the Sichuan basin of China. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.788876
Huang, J., Si, W., Deng, Q., Li, P., Yang, S. (2014). Rapid evolution of avirulence genes in rice blast fungus Magnaporthe oryzae. BMC Genet. 15, 45. doi: 10.1186/1471-2156-15-45
Inoue, Y., Vy, T. T. P., Yoshida, K., Asano, H., Mitsuoka, C., Asuke, S., et al. (2017). Evolution of the wheat blast fungus through functional losses in a host specificity determinant. Science 357, 80–83. doi: 10.1126/science.aam9654
Jia, Y., McAdams, S. A., Bryan, G. T., Hershey, H. P., Valent, B. (2000). Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J. 19, 4004–4014. doi: 10.1093/emboj/19.15.4004
Jones, J. D., Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Jones, K., Kim, D. W., Park, J. S., Khang, C. H. (2016). Live-cell fluorescence imaging to investigate the dynamics of plant cell death during infection by the rice blast fungus Magnaporthe oryzae. BMC Plant Biol. 16, 69. doi: 10.1186/s12870-016-0756-x
Jubic, L. M., Saile, S., Furzer, O. J., El Kasmi, F., Dangl, J. L. (2019). Help wanted: helper NLRs and plant immune responses. Curr. Opin. Plant Biol. 50, 82–94. doi: 10.1016/j.pbi.2019.03.013
Kang, S., Sweigard, J. A., Valent, B. (1995). The PWL host specificity gene family in the blast fungus Magnaporthe grisea. Mol. Plant Microbe Interact. 8, 939–948. doi: 10.1094/mpmi-8-0939
Kankanala, P., Czymmek, K., Valent, B. (2007). Roles for rice membrane dynamics and plasmodesmata during biotrophic invasion by the blast fungus. Plant Cell 19, 706–724. doi: 10.1105/tpc.106.046300
Kanzaki, H., Yoshida, K., Saitoh, H., Fujisaki, K., Hirabuchi, A., Alaux, L., et al. (2012). Arms race co-evolution of Magnaporthe oryzae AVR-Pik and rice Pik genes driven by their physical interactions. Plant J. 72, 894–907. doi: 10.1111/j.1365-313X.2012.05110.x
Khan, M., Subramaniam, R., Desveaux, D. (2016). Of guards, decoys, baits and traps: pathogen perception in plants by type III effector sensors. Curr. Opin. Microbiol. 29, 49–55. doi: 10.1016/j.mib.2015.10.006
Khang, C. H., Berruyer, R., Giraldo, M. C., Kankanala, P., Park, S. Y., Czymmek, K., et al. (2010). Translocation of Magnaporthe oryzae effectors into rice cells and their subsequent cell-to-cell movement. Plant Cell 22, 1388–1403. doi: 10.1105/tpc.109.069666
Khush, G. S. (2013). Strategies for increasing the yield potential of cereals: case of rice as an example. Plant Breed. 132, 433–436. doi: 10.1111/pbr.1991
Kotsaridis, K., Tsakiri, D., Sarris, P. F. (2023). Understanding enemy’s weapons to an effective prevention: common virulence effects across microbial phytopathogens kingdoms. Crit. Rev. Microbiol. 49, 528–542. doi: 10.1080/1040841x.2022.2083939
Kourelis, J., Marchal, C., Posbeyikian, A., Harant, A., Kamoun, S. (2023). NLR immune receptor-nanobody fusions confer plant disease resistance. Science 379, 934–939. doi: 10.1126/science.abn4116
Kroj, T., Chanclud, E., Michel-Romiti, C., Grand, X., Morel, J. B. (2016). Integration of decoy domains derived from protein targets of pathogen effectors into plant immune receptors is widespread. New Phytol. 210, 618–626. doi: 10.1111/nph.13869
Latorre, S. M., Reyes-Avila, C. S., Malmgren, A., Win, J., Kamoun, S., Burbano, H. A. (2020). Differential loss of effector genes in three recently expanded pandemic clonal lineages of the rice blast fungus. BMC Biol. 18, 88. doi: 10.1186/s12915-020-00818-z
Leach, J. E., Vera Cruz, C. M., Bai, J., Leung, H. (2001). Pathogen fitness penalty as a predictor of durability of disease resistance genes. Annu. Rev. Phytopathol. 39, 187–224. doi: 10.1146/annurev.phyto.39.1.187
Li, W., Wang, B., Wu, J., Lu, G., Hu, Y., Zhang, X., et al. (2009). The Magnaporthe oryzae avirulence gene AvrPiz-t encodes a predicted secreted protein that triggers the immunity in rice mediated by the blast resistance gene Piz-t. Mol. Plant Microbe Interact. 22, 411–420. doi: 10.1094/mpmi-22-4-0411
Liu, D., Lun, Z., Liu, N., Yuan, G., Wang, X., Li, S., et al. (2023a). Identification and characterization of novel candidate effector proteins from Magnaporthe oryzae. J. Fungi(Basel) 9 (5), 574. doi: 10.3390/jof9050574
Liu, Z., Qiu, J., Shen, Z., Wang, C., Jiang, N., Shi, H., et al. (2023c). The E3 ubiquitin ligase OsRGLG5 targeted by the Magnaporthe oryzae effector AvrPi9 confers basal resistance against rice blast. Plant Commun. 4, 100626. doi: 10.1016/j.xplc.2023.100626
Liu, Y., Zeng, Z., Zhang, Y. M., Li, Q., Jiang, X. M., Jiang, Z., et al. (2021a). An angiosperm NLR Atlas reveals that NLR gene reduction is associated with ecological specialization and signal transduction component deletion. Mol. Plant 14, 2015–2031. doi: 10.1016/j.molp.2021.08.001
Liu, Y., Zhang, Y. M., Tang, Y., Chen, J. Q., Shao, Z. Q. (2023b). The evolution of plant NLR immune receptors and downstream signal components. Curr. Opin. Plant Biol. 73, 102363. doi: 10.1016/j.pbi.2023.102363
Liu, Y., Zhang, X., Yuan, G., Wang, D., Zheng, Y., Ma, M., et al. (2021b). A designer rice NLR immune receptor confers resistance to the rice blast fungus carrying noncorresponding avirulence effectors. Proc. Natl. Acad. Sci. U.S.A. 118 (44), e2110751118. doi: 10.1073/pnas.2110751118
Lolle, S., Stevens, D., Coaker, G. (2020). Plant NLR-triggered immunity: from receptor activation to downstream signaling. Curr. Opin. Immunol. 62, 99–105. doi: 10.1016/j.coi.2019.12.007
Lu, Y., Tsuda, K. (2021). Intimate association of PRR-and NLR-mediated signaling in plant immunity. Mol. Plant Microbe Interact. 34, 3–14. doi: 10.1094/mpmi-08-20-0239-ia
Lu, L., Wang, Q., Shi, Z., Li, C., Guo, Z., Li, J. (2023). Emergence of rice blast AVR-Pi9 resistance breaking haplotypes in Yunnan province, China. Life (Basel) 13 (6), 1320. doi: 10.3390/life13061320
Lüdke, D., Yan, Q., Rohmann, P. F. W., Wiermer, M. (2022). NLR we there yet? Nucleocytoplasmic coordination of NLR-mediated immunity. New Phytol. 236, 24–42. doi: 10.1111/nph.18359
Ma, G.-h., Yuan, L.-p. (2015). Hybrid rice achievements, development and prospect in China. J. Integr. Agric. 14, 197–205. doi: 10.1016/S2095-3119(14)60922-9
Macho, A. P., Zipfel, C. (2014). Plant PRRs and the activation of innate immune signaling. Mol. Cell. 54, 263–272. doi: 10.1016/j.molcel.2014.03.028
Maekawa, T., Kufer, T. A., Schulze-Lefert, P. (2011). NLR functions in plant and animal immune systems: so far and yet so close. Nat. Immunol. 12, 817–826. doi: 10.1038/ni.2083
Maidment, J. H. R., Franceschetti, M., Maqbool, A., Saitoh, H., Jantasuriyarat, C., Kamoun, S., et al. (2021). Multiple variants of the fungal effector AVR-Pik bind the HMA domain of the rice protein OsHIPP19, providing a foundation to engineer plant defense. J. Biol. Chem. 296, 100371. doi: 10.1016/j.jbc.2021.100371
Maqbool, A., Saitoh, H., Franceschetti, M., Stevenson, C. E., Uemura, A., Kanzaki, H., et al. (2015). Structural basis of pathogen recognition by an integrated HMA domain in a plant NLR immune receptor. Elife 4, e08709. doi: 10.7554/eLife.08709
Marcel, S., Sawers, R., Oakeley, E., Angliker, H., Paszkowski, U. (2010). Tissue-adapted invasion strategies of the rice blast fungus Magnaporthe oryzae. Plant Cell 22, 3177–3187. doi: 10.1105/tpc.110.078048
Martin-Urdiroz, M., Oses-Ruiz, M., Ryder, L. S., Talbot, N. J. (2016). Investigating the biology of plant infection by the rice blast fungus Magnaporthe oryzae. Fungal. Genet. Biol. 90, 61–68. doi: 10.1016/j.fgb.2015.12.009
Meng, X., Xiao, G., Telebanco-Yanoria, M. J., Siazon, P. M., Padilla, J., Opulencia, R., et al. (2020). The broad-spectrum rice blast resistance (R) gene Pita2 encodes a novel R protein unique from Pita. Rice(N Y) 13, 19. doi: 10.1186/s12284-020-00377-5
Mentlak, T. A., Kombrink, A., Shinya, T., Ryder, L. S., Otomo, I., Saitoh, H., et al. (2012). Effector-mediated suppression of chitin-triggered immunity by Magnaporthe oryzae is necessary for rice blast disease. Plant Cell 24, 322–335. doi: 10.1105/tpc.111.092957
Milgroom, M. G. (2015). Population biology of plant pathogens: genetics, ecology, and evolution (USA: APS Press).
Mosquera, G., Giraldo, M. C., Khang, C. H., Coughlan, S., Valent, B. (2009). Interaction transcriptome analysis identifies Magnaporthe oryzae BAS1-4 as biotrophy-associated secreted proteins in rice blast disease. Plant Cell 21, 1273–1290. doi: 10.1105/tpc.107.055228
Munson, M., Novick, P. (2006). The exocyst defrocked, a framework of rods revealed. Nat. Struct. Mol. Biol. 13, 577–581. doi: 10.1038/nsmb1097
Muthayya, S., Sugimoto, J. D., Montgomery, S., Maberly, G. F. (2014). An overview of global rice production, supply, trade, and consumption. Ann. N. Y. Acad. Sci. 1324, 7–14. doi: 10.1111/nyas.12540
Ngou, B. P. M., Ahn, H. K., Ding, P., Jones, J. D. G. (2021). Mutual potentiation of plant immunity by cell-surface and intracellular receptors. Nature 592, 110–115. doi: 10.1038/s41586-021-03315-7
Noguchi, M. T., Yasuda, N., Fujita, Y. (2006). Evidence of genetic exchange by parasexual recombination and genetic analysis of pathogenicity and mating type of parasexual recombinants in rice blast fungus, Magnaporthe oryzae. Phytopathology 96, 746–750. doi: 10.1094/phyto-96-0746
Nürnberger, T., Brunner, F., Kemmerling, B., Piater, L. (2004). Innate immunity in plants and animals: striking similarities and obvious differences. Immunol. Rev. 198, 249–266. doi: 10.1111/j.0105-2896.2004.0119.x
Oikawa, K., Fujisaki, K., Shimizu, M., Takeda, T., Nemoto, K., Saitoh, H., et al. (2024). The blast pathogen effector AVR-Pik binds and stabilizes rice heavy metal-associated (HMA) proteins to co-opt their function in immunity. bioRxiv. doi: 10.1101/2020.12.01.406389
Okuyama, Y., Kanzaki, H., Abe, A., Yoshida, K., Tamiru, M., Saitoh, H., et al. (2011). A multifaceted genomics approach allows the isolation of the rice Pia-blast resistance gene consisting of two adjacent NBS-LRR protein genes. Plant J. 66, 467–479. doi: 10.1111/j.1365-313X.2011.04502.x
Oliveira-Garcia, E., Tamang, T. M., Park, J., Dalby, M., Martin-Urdiroz, M., Rodriguez Herrero, C., et al. (2023). Clathrin-mediated endocytosis facilitates the internalization of Magnaporthe oryzae effectors into rice cells. Plant Cell 35, 2527–2551. doi: 10.1093/plcell/koad094
Orbach, M. J., Farrall, L., Sweigard, J. A., Chumley, F. G., Valent, B. (2000). A telomeric avirulence gene determines efficacy for the rice blast resistance gene Pi-ta. Plant Cell 12, 2019–2032. doi: 10.1105/tpc.12.11.2019
Ortiz, D., de Guillen, K., Cesari, S., Chalvon, V., Gracy, J., Padilla, A., et al. (2017). Recognition of the Magnaporthe oryzae effector AVR-Pia by the decoy domain of the rice NLR immune receptor RGA5. Plant Cell 29, 156–168. doi: 10.1105/tpc.16.00435
Ose, T., Oikawa, A., Nakamura, Y., Maenaka, K., Higuchi, Y., Satoh, Y., et al. (2015). Solution structure of an avirulence protein, AVR-Pia, from Magnaporthe oryzae. J. Biomol. NMR 63, 229–235. doi: 10.1007/s10858-015-9979-7
Park, C. H., Chen, S., Shirsekar, G., Zhou, B., Khang, C. H., Songkumarn, P., et al. (2012). The Magnaporthe oryzae effector AvrPiz-t targets the RING E3 ubiquitin ligase APIP6 to suppress pathogen-associated molecular pattern-triggered immunity in rice. Plant Cell 24, 4748–4762. doi: 10.1105/tpc.112.105429
Park, C. H., Shirsekar, G., Bellizzi, M., Chen, S., Songkumarn, P., Xie, X., et al. (2016). The E3 ligase APIP10 connects the effector AvrPiz-t to the NLR receptor Piz-t in rice. PloS Pathog. 12, e1005529. doi: 10.1371/journal.ppat.1005529
Pruitt, R. N., Locci, F., Wanke, F., Zhang, L., Saile, S. C., Joe, A., et al. (2021). The EDS1-PAD4-ADR1 node mediates Arabidopsis pattern-triggered immunity. Nature 598, 495–499. doi: 10.1038/s41586-021-03829-0
Ray, S., Singh, P. K., Gupta, D. K., Mahato, A. K., Sarkar, C., Rathour, R., et al. (2016). Analysis of Magnaporthe oryzae genome reveals a fungal effector, which is able to induce resistance response in transgenic rice line containing resistance gene, Pi54. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01140
Ribot, C., Césari, S., Abidi, I., Chalvon, V., Bournaud, C., Vallet, J., et al. (2013). The Magnaporthe oryzae effector AVR1-CO39 is translocated into rice cells independently of a fungal-derived machinery. Plant J. 74, 1–12. doi: 10.1111/tpj.12099
Rocafort, M., Fudal, I., Mesarich, C. H. (2020). Apoplastic effector proteins of plant-associated fungi and oomycetes. Curr. Opin. Plant Biol. 56, 9–19. doi: 10.1016/j.pbi.2020.02.004
Saklani, B. K., Ray, S., Arora, K., Asthana, R. K., Sharma, T. R. (2023). Magnaporthe oryzae encoded effector protein AvrPi54 interacts in vivo with rice encoded cognate resistance protein Pi54 at the host plasma membrane. J. Plant Biochem. Biot. 32, 274–283. doi: 10.1007/s13562-022-00803-3
Sandhu, N., Yadav, S., Kumar, A. (2020). “Advances in developing multigene abiotic and biotic stress-tolerant rice varieties,” in Abiotic Stress in Plants. Ed. Fahad., S. (IntechOpen, London).
Shao, Z. Q., Xue, J. Y., Wu, P., Zhang, Y. M., Wu, Y., Hang, Y. Y., et al. (2016). Large-scale analyses of angiosperm nucleotide-binding site-leucine-rich repeat genes reveal three anciently diverged classes with distinct evolutionary patterns. Plant Physiol. 170, 2095–2109. doi: 10.1104/pp.15.01487
Shi, X., Long, Y., He, F., Zhang, C., Wang, R., Zhang, T., et al. (2018). The fungal pathogen Magnaporthe oryzae suppresses innate immunity by modulating a host potassium channel. PloS Pathog. 14, e1006878. doi: 10.1371/journal.ppat.1006878
Shi, X., Xiong, Y., Zhang, K., Zhang, Y., Zhang, J., Zhang, L., et al. (2023). The ANIP1-OsWRKY62 module regulates both basal defense and Pi9-mediated immunity against Magnaporthe oryzae in rice. Mol. Plant 16, 739–755. doi: 10.1016/j.molp.2023.03.001
Shimizu, M., Hirabuchi, A., Sugihara, Y., Abe, A., Takeda, T., Kobayashi, M., et al. (2022). A genetically linked pair of NLR immune receptors shows contrasting patterns of evolution. Proc. Natl. Acad. Sci. U.S.A. 119, e2116896119. doi: 10.1073/pnas.2116896119
Shipman, E. N., Jones, K., Jenkinson, C. B., Kim, D. W., Zhu, J., Khang, C. H. (2017). Nuclear and structural dynamics during the establishment of a specialized effector-secreting cell by Magnaporthe oryzae in living rice cells. BMC Cell Biol. 18, 11. doi: 10.1186/s12860-017-0126-z
Singh, R., Dangol, S., Chen, Y., Choi, J., Cho, Y. S., Lee, J. E., et al. (2016). Magnaporthe oryzae effector AVR-Pii helps to establish compatibility by inhibition of the rice NADP-malic enzyme resulting in disruption of oxidative burst and host innate immunity. Mol. Cell 39, 426–438. doi: 10.14348/molcells.2016.0094
Skamnioti, P., Gurr, S. J. (2009). Against the grain: safeguarding rice from rice blast disease. Trends Biotechnol. 27, 141–150. doi: 10.1016/j.tibtech.2008.12.002
Soanes, D. M., Alam, I., Cornell, M., Wong, H. M., Hedeler, C., Paton, N. W., et al. (2008). Comparative genome analysis of filamentous fungi reveals gene family expansions associated with fungal pathogenesis. PloS One 3, e2300. doi: 10.1371/journal.pone.0002300
Song, Z., Bakeer, W., Marshall, J. W., Yakasai, A. A., Khalid, R. M., Collemare, J., et al. (2015). Heterologous expression of the avirulence gene ACE1 from the fungal rice pathogen Magnaporthe oryzae. Chem. Sci. 6, 4837–4845. doi: 10.1039/c4sc03707c
Sornkom, W., Miki, S., Takeuchi, S., Abe, A., Asano, K., Sone, T. (2017). Fluorescent reporter analysis revealed the timing and localization of AVR-Pia expression, an avirulence effector of Magnaporthe oryzae. Mol. Plant Pathol. 18, 1138–1149. doi: 10.1111/mpp.12468
Spoel, S. H., Dong, X. (2012). How do plants achieve immunity? Defence without specialized immune cells. Nat. Rev. Immunol. 12, 89–100. doi: 10.1038/nri3141
Stergiopoulos, I., de Wit, P. J. (2009). Fungal effector proteins. Annu. Rev. Phytopathol. 47, 233–263. doi: 10.1146/annurev.phyto.112408.132637
Sugihara, Y., Abe, Y., Takagi, H., Abe, A., Shimizu, M., Ito, K., et al. (2023). Disentangling the complex gene interaction networks between rice and the blast fungus identifies a new pathogen effector. PloS Biol. 21, e3001945. doi: 10.1371/journal.pbio.3001945
Sutthiphai, T., Damchuay, K., Neupane, R. C., Longya, A., Sriwongchai, T., Songkumarn, P., et al. (2022). Genetic variation of avirulence genes (AVR-Pi9, AVR-Pik, AVR-Pita1) and genetic diversity of rice blast fungus, Pyricularia oryzae, in Thailand. Plant Pathol. 71, 322–333. doi: 10.1111/ppa.13460
Sweigard, J. A., Carroll, A. M., Kang, S., Farrall, L., Chumley, F. G., Valent, B. (1995). Identification, cloning, and characterization of PWL2, a gene for host species specificity in the rice blast fungus. Plant Cell 7, 1221–1233. doi: 10.1105/tpc.7.8.1221
Takabayashi, N., Tosa, Y., Oh, H. S., Mayama, S. (2002). A gene-for-gene relationship underlying the species-specific parasitism of Avena/Triticum isolates of Magnaporthe grisea on wheat cultivars. Phytopathology 92, 1182–1188. doi: 10.1094/phyto.2002.92.11.1182
Takken, F. L., Goverse, A. (2012). How to build a pathogen detector: structural basis of NB-LRR function. Curr. Opin. Plant Biol. 15, 375–384. doi: 10.1016/j.pbi.2012.05.001
Tang, M., Ning, Y., Shu, X., Dong, B., Zhang, H., Wu, D., et al. (2017). The Nup98 homolog APIP12 targeted by the effector AvrPiz-t is involved in rice basal resistance against Magnaporthe oryzae. Rice(N Y) 10, 5. doi: 10.1186/s12284-017-0144-7
Thierry, M., Charriat, F., Milazzo, J., Adreit, H., Ravel, S., Cros-Arteil, S., et al. (2022). Maintenance of divergent lineages of the Rice Blast Fungus Pyricularia oryzae through niche separation, loss of sex and post-mating genetic incompatibilities. PloS Pathog. 18, e1010687. doi: 10.1371/journal.ppat.1010687
Thomas, C. M., Jones, D. A., Parniske, M., Harrison, K., Balint-Kurti, P. J., Hatzixanthis, K., et al. (1997). Characterization of the tomato Cf-4 gene for resistance to Cladosporium fulvum identifies sequences that determine recognitional specificity in Cf-4 and Cf-9. Plant Cell 9, 2209–2224. doi: 10.1105/tpc.9.12.2209
Tian, H., Wu, Z., Chen, S., Ao, K., Huang, W., Yaghmaiean, H., et al. (2021). Activation of TIR signalling boosts pattern-triggered immunity. Nature 598, 500–503. doi: 10.1038/s41586-021-03987-1
Tonini, A., Cabrera, E. (2011). Opportunities for global rice research in a changing world 31. doi: 10.22004/ag.econ.287644
Tsuda, K., Katagiri, F. (2010). Comparing signaling mechanisms engaged in pattern-triggered and effector-triggered immunity. Curr. Opin. Plant Biol. 13, 459–465. doi: 10.1016/j.pbi.2010.04.006
Tsujimoto Noguchi, M. (2011). Parasexual recombination in Magnaporthe oryzae. JARQ-JPN AGR Res. Q 45, 39–45. doi: 10.6090/jarq.45.39
Van der Hoorn, R. A., Kamoun, S. (2008). From guard to decoy: a new model for perception of plant pathogen effectors. Plant Cell 20, 2009–2017. doi: 10.1105/tpc.108.060194
Vy, T. T. P., Hyon, G.-S., Nga, N. T. T., Inoue, Y., Chuma, I., Tosa, Y. (2014). Genetic analysis of host–pathogen incompatibility between Lolium isolates of Pyricularia oryzae and wheat. J. Gen. Plant Pathol. 80, 59–65. doi: 10.1007/s10327-013-0478-y
Wang, X., Lee, S., Wang, J., Ma, J., Bianco, T., Jia, Y. (2014). “Current advances on genetic resistance to rice blast disease,” in Rice Germplasm, Genetics and Improvement. Eds. Wengui, Y., Jinsong, B. (IntechOpen, Rijeka).
Wang, R., Ning, Y., Shi, X., He, F., Zhang, C., Fan, J., et al. (2016). Immunity to rice blast disease by suppression of effector-triggered necrosis. Curr. Biol. 26, 2399–2411. doi: 10.1016/j.cub.2016.06.072
Wang, G. L., Valent, B. (2017). Durable resistance to rice blast. Science 355, 906–907. doi: 10.1126/science.aam9517
Wang, J., Wang, R., Fang, H., Zhang, C., Zhang, F., Hao, Z., et al. (2021). Two VOZ transcription factors link an E3 ligase and an NLR immune receptor to modulate immunity in rice. Mol. Plant 14, 253–266. doi: 10.1016/j.molp.2020.11.005
Were, V., Yan, X., Foster, A. J., Sklenar, J., Langner, T., Bentham, A., et al. (2024). The blast effector Pwl2 is a virulence factor that modifies the cellular localisation of host protein HIPP43 to suppress immunity. bioRxiv. doi: 10.1101/2024.01.20.576406
Wilson, R. A., Talbot, N. J. (2009). Under pressure: investigating the biology of plant infection by Magnaporthe oryzae. Nat. Rev. Microbiol. 7, 185–195. doi: 10.1038/nrmicro2032
Wu, J., Kou, Y., Bao, J., Li, Y., Tang, M., Zhu, X., et al. (2015b). Comparative genomics identifies the Magnaporthe oryzae avirulence effector AvrPi9 that triggers Pi9-mediated blast resistance in rice. New Phytol. 206, 1463–1475. doi: 10.1111/nph.13310
Wu, C. H., Krasileva, K. V., Banfield, M. J., Terauchi, R., Kamoun, S. (2015a). The “sensor domains” of plant NLR proteins: more than decoys? Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00134
Wu, W., Wang, L., Zhang, S., Li, Z., Zhang, Y., Lin, F., et al. (2014). Stepwise arms race between AvrPik and Pik alleles in the rice blast pathosystem. Mol. Plant Microbe Interact. 27, 759–769. doi: 10.1094/mpmi-02-14-0046-r
Xi, Y., Cesari, S., Kroj, T. (2022). Insight into the structure and molecular mode of action of plant paired NLR immune receptors. Essays Biochem. 66, 513–526. doi: 10.1042/ebc20210079
Xiao, G., Laksanavilat, N., Cesari, S., Lambou, K., Baudin, M., Jalilian, A., et al. (2024). The unconventional resistance protein PTR recognizes the Magnaporthe oryzae effector AVR-Pita in an allele-specific manner. Nat. Plants 10, 994–1004. doi: 10.1038/s41477-024-01694-z
Xiao, N., Wu, Y., Zhang, X., Hao, Z., Chen, Z., Yang, Z., et al. (2023). Pijx confers broad-spectrum seedling and panicle blast resistance by promoting the degradation of ATP β subunit and OsRbohC-mediated ROS burst in rice. Mol. Plant 16, 1832–1846. doi: 10.1016/j.molp.2023.10.001
Xie, Y., Wang, Y., Yu, X., Lin, Y., Zhu, Y., Chen, J., et al. (2022). SH3P2, an SH3 domain-containing protein that interacts with both Pib and AvrPib, suppresses effector-triggered, Pib-mediated immunity in rice. Mol. Plant 15, 1931–1946. doi: 10.1016/j.molp.2022.10.022
Yan, X., Talbot, N. J. (2016). Investigating the cell biology of plant infection by the rice blast fungus Magnaporthe oryzae. Curr. Opin. Microbiol. 34, 147–153. doi: 10.1016/j.mib.2016.10.001
Yan, X., Tang, B., Ryder, L. S., MacLean, D., Were, V. M., Eseola, A. B., et al. (2023). The transcriptional landscape of plant infection by the rice blast fungus Magnaporthe oryzae reveals distinct families of temporally co-regulated and structurally conserved effectors. Plant Cell 35, 1360–1385. doi: 10.1093/plcell/koad036
Yang, T., Song, L., Hu, J., Qiao, L., Yu, Q., Wang, Z., et al. (2024). Magnaporthe oryzae effector AvrPik-D targets a transcription factor WG7 to suppress rice immunity. Rice(N Y) 17, 14. doi: 10.1186/s12284-024-00693-0
Yoshida, K., Saitoh, H., Fujisawa, S., Kanzaki, H., Matsumura, H., Yoshida, K., et al. (2009). Association genetics reveals three novel avirulence genes from the rice blast fungal pathogen Magnaporthe oryzae. Plant Cell 21, 1573–1591. doi: 10.1105/tpc.109.066324
Yoshida, K., Saunders, D. G., Mitsuoka, C., Natsume, S., Kosugi, S., Saitoh, H., et al. (2016). Host specialization of the blast fungus Magnaporthe oryzae is associated with dynamic gain and loss of genes linked to transposable elements. BMC Genomics 17, 370. doi: 10.1186/s12864-016-2690-6
You, X., Zhang, F., Liu, Z., Wang, M., Xu, X., He, F., et al. (2022). Rice catalase OsCATC is degraded by E3 ligase APIP6 to negatively regulate immunity. Plant Physiol. 190, 1095–1099. doi: 10.1093/plphys/kiac317
You, X., Zhu, S., Sheng, H., Liu, Z., Wang, D., Wang, M., et al. (2023). The rice peroxisomal receptor PEX5 negatively regulates resistance to rice blast fungus Magnaporthe oryzae. Cell Rep. 42, 113315. doi: 10.1016/j.celrep.2023.113315
Younas, M. U., Wang, G., Du, H., Zhang, Y., Ahmad, I., Rajput, N., et al. (2023). Approaches to reduce rice blast disease using knowledge from host resistance and pathogen pathogenicity. Int. J. Mol. Sci. 24 (5), 4985. doi: 10.3390/ijms24054985
Yu, X., Feng, B., He, P., Shan, L. (2017). From chaos to harmony: responses and signaling upon microbial pattern recognition. Annu. Rev. Phytopathol. 55, 109–137. doi: 10.1146/annurev-phyto-080516-035649
Yuan, M., Jiang, Z., Bi, G., Nomura, K., Liu, M., Wang, Y., et al. (2021). Pattern-recognition receptors are required for NLR-mediated plant immunity. Nature 592, 105–109. doi: 10.1038/s41586-021-03316-6
Zdrzałek, R., Xi, Y., Langner, T., Bentham, A. R., Petit-Houdenot, Y., de la Concepcion, J. C., et al. (2024). Bioengineering a plant NLR immune receptor with a robust binding interface toward a conserved fungal pathogen effector. Proc. Natl. Acad. Sci. U.S.A. 121, e2402872121. doi: 10.1073/pnas.2402872121
Zhai, K., Liang, D., Li, H., Jiao, F., Yan, B., Liu, J., et al. (2022). NLRs guard metabolism to coordinate pattern- and effector-triggered immunity. Nature 601, 245–251. doi: 10.1038/s41586-021-04219-2
Zhang, C., Fang, H., Shi, X., He, F., Wang, R., Fan, J., et al. (2020a). A fungal effector and a rice NLR protein have antagonistic effects on a Bowman-Birk trypsin inhibitor. Plant Biotechnol. J. 18, 2354–2363. doi: 10.1111/pbi.13400
Zhang, F., Fang, H., Wang, M., He, F., Tao, H., Wang, R., et al. (2022). APIP5 functions as a transcription factor and an RNA-binding protein to modulate cell death and immunity in rice. Nucleic Acids Res. 50, 5064–5079. doi: 10.1093/nar/gkac316
Zhang, C., Fang, H., Wang, J., Tao, H., Wang, D., Qin, M., et al. (2024a). The rice E3 ubiquitin ligase-transcription factor module targets two trypsin inhibitors to enhance broad-spectrum disease resistance. Dev. Cell 59 (15), 2017–2033.e5. doi: 10.1016/j.devcel.2024.05.003
Zhang, X., He, D., Zhao, Y., Cheng, X., Zhao, W., Taylor, I. A., et al. (2018). A positive-charged patch and stabilized hydrophobic core are essential for avirulence function of AvrPib in the rice blast fungus. Plant J. 96, 133–146. doi: 10.1111/tpj.14023
Zhang, Z., Jia, Y., Wang, Y., Sun, G. (2020b). A rapid survey of avirulence genes in field isolates of Magnaporthe oryzae. Plant Dis. 104, 717–723. doi: 10.1094/pdis-08-19-1688-re
Zhang, X., Liu, Y., Yuan, G., Wang, S., Wang, D., Zhu, T., et al. (2024b). The synthetic NLR RGA5HMA5 requires multiple interfaces within and outside the integrated domain for effector recognition. Nat. Commun. 15, 1104. doi: 10.1038/s41467-024-45380-2
Zhang, S., Wang, L., Wu, W., He, L., Yang, X., Pan, Q. (2015). Function and evolution of Magnaporthe oryzae avirulence gene AvrPib responding to the rice blast resistance gene Pib. Sci. Rep. 5, 11642. doi: 10.1038/srep11642
Zhang, Z. M., Zhang, X., Zhou, Z. R., Hu, H. Y., Liu, M., Zhou, B., et al. (2013). Solution structure of the Magnaporthe oryzae avirulence protein AvrPiz-t. J. Biomol. NMR 55, 219–223. doi: 10.1007/s10858-012-9695-5
Zhao, H., Wang, X., Jia, Y., Minkenberg, B., Wheatley, M., Fan, J., et al. (2018). The rice blast resistance gene Ptr encodes an atypical protein required for broad-spectrum disease resistance. Nat. Commun. 9, 2039. doi: 10.1038/s41467-018-04369-4
Zhong, Z., Chen, M., Lin, L., Han, Y., Bao, J., Tang, W., et al. (2018). Population genomic analysis of the rice blast fungus reveals specific events associated with expansion of three main clades. ISME J. 12, 1867–1878. doi: 10.1038/s41396-018-0100-6
Keywords: rice, blast disease, Magnaporthe oryzae, AVR effector, R protein, resistance
Citation: Liu X, Hu X, Tu Z, Sun Z, Qin P, Liu Y, Chen X, Li Z, Jiang N and Yang Y (2024) The roles of Magnaporthe oryzae avirulence effectors involved in blast resistance/susceptibility. Front. Plant Sci. 15:1478159. doi: 10.3389/fpls.2024.1478159
Received: 09 August 2024; Accepted: 19 September 2024;
Published: 09 October 2024.
Edited by:
Wen-Ming Wang, Sichuan Agricultural University, ChinaReviewed by:
Jinling Liu, Hunan Agricultural University, ChinaYan-Yan Huang, Sichuan Agricultural University, China
Copyright © 2024 Liu, Hu, Tu, Sun, Qin, Liu, Chen, Li, Jiang and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhiqiang Li, emhpcWlhbmdkb183NzFAMTYzLmNvbQ==; Nan Jiang, SmlhbmduYW4xOTg0NzMxQDEyNi5jb20=; Yuanzhu Yang, eXpodXlhaEAxNjMuY29t
†These authors have contributed equally to this work
 Xin Liu
Xin Liu Xiaochun Hu1,2,3†
Xiaochun Hu1,2,3† Zhiqiang Li
Zhiqiang Li Nan Jiang
Nan Jiang Yuanzhu Yang
Yuanzhu Yang