Introduction
The improvement of crop yield and quality is an eternal theme to face climate change and population growth. The key to improving crop varieties lies in precisely manipulating gene expression. Recent advancements in CRISPR/Cas9 technology have made gene knockout increasingly straightforward, yet for genes related to important agronomic traits, it is crucial to regulate their expression levels appropriately. Complete knockout often results in defects in other aspects. In addition, many agronomic traits require upregulation of target gene expression for their improvement. Therefore, the development of novel methods for precise upregulation or downregulation of gene expression, without altering gene protein sequences or introducing new genome fragments, will significantly bolster the technical foundation for crop genetic improvement.
N6-methyladenosine (m6A) is the most abundant and reversible internal chemical modification in eukaryotic mRNA, which is installed, removed, and recognized by methyltransferases (writers), demethylases (erasers), and m6A-binding proteins (readers), respectively (Tang et al., 2023). Currently, two types of m6A methyltransferases have been identified in plants: multiprotein complexes and a single protein. The multiprotein complex includes MTA, MTB, FIP37, VIRILIZER (VIR), HAKAI, and HIZ2 (HAKAI interacting zinc finger protein 2), which catalyze the majority of m6A modifications in mRNA (Parker et al., 2021; Ruzicka et al., 2017; Shen et al., 2016; Zhang et al., 2022; Zhong et al., 2008). The single protein FIONA1 also exhibits methyltransferase activity in Arabidopsis (Wang et al., 2022; Xu et al., 2022), catalyzing approximately 10% of m6A modifications in mRNA. Several m6A demethylases, which belong to the Fe (II)/α - kg dependent dioxygenase superfamily, have been identified in plants, including Arabidopsis AtALKBH10B and AtALKBH9B (Martinez-Perez et al., 2017), rice OsALKBH9 (Tang et al., 2024), and tomato SlALKBH2 (Zhou et al., 2019). m6A is recognized by m6A-binding proteins, such as ECTs in Arabidopsis, which contains the YTH domain. In plants, the ratio of m6A/A in poly A+ RNA varied among different tissues, with a range of 0.36–0.75% in Arabidopsis and 0.52–0.67% in rice, suggesting its high abundance (Wang et al., 2024). At the transcriptome level, m6A sites are primarily enriched within the 3′-untranslated region (3′ UTR), followed by the coding DNA sequence (CDS) and 5′-untranslated region (5′ UTR). Recent studies have demonstrated the crucial roles of m6A in regulating gene expression in plants, primarily by influencing mRNA stability, translation, and 3′ UTR processing (Tang et al., 2023). Among them, mRNA stability regulation is one of the primary functions of m6A, which involves two aspects: acceleration of RNA decay or preservation of RNA stability, depending on the specific m6A-binding proteins. Recent studies combining proteomics and m6A analysis have shown that m6A in the untranslated regions is negatively correlated with protein abundance, suggesting that m6A in UTR is likely to inhibit protein abundance in plants (Li et al., 2024). Therefore, manipulating m6A could lead to an increase in protein abundance. m6A modifications have also been found to play crucial roles in plant biology, such as embryo development, floral transition, stem cell fate determination, pollen development, fruit ripening, photomorphogenesis, circadian clock, nitrate signaling, and responses to biotic and abiotic stress (Tang et al., 2023). Given the high abundance and crucial roles of m6A modifications in gene expression regulating, altering m6A modifications in genes holds promise as a strategy for enhancing crop agronomic traits.
Manipulating m6A modification to improve crop agronomic traits
In mammals, the m6A demethylase FTO, known as an obesity gene, plays a crucial role in regulating body weight. Researchers have genetically engineered rice and potato to express FTO. In field experiments, the yield and biomass of genetically engineered rice and potato increased by approximately 50% (Yu et al., 2021). Further research indicates that the expression of FTO in rice promotes root growth, tiller bud formation, photosynthetic efficiency, and drought resistance, and these phenotypes are dependent on the m6A demethylase activity of FTO (Yu et al., 2021). In strawberries, inhibiting the expression of m6A methyltransferase genes FveMTA or FveMTB can delay fruit ripening, while upregulating the expression of either gene can accelerate fruit ripening, suggesting that manipulating m6A modification can regulate the ripening time of strawberries (Zhou et al., 2021). The above research suggests that manipulating the m6A modification level overall can enhance the agronomic traits of crops.
In addition to altering the overall m6A modification level, researchers have also endeavored to edit m6A modification levels on individual genes. In plants, a precise editing system for plant mRNA m6A has been successfully developed by fusing dCas13a with the plant m6A methyltransferase core complex MTA-MTB or the mammalian demethylase ALKBH5 (Shi et al., 2024). By specifically editing the m6A modification on the SHR transcript, which is a key gene for root development, it was found that an increase in the m6A modification level of the SHR transcript can promote the enlargement of the aboveground and root parts of plants, increase leaf area, plant height, biomass, and grain yield, thereby promoting plant growth (Shi et al., 2024). In cotton, similar m6A editing tools have also been developed, combining CRISPR/dCas13(Rx) with the methyltransferase GhMTA (Targeted RNA Methylation Editor, TME) or the demethyltransferase GhALKBH10 (Targeted RNA Demethylation Editor, TDE) (Yu et al., 2024). Using TME editor, the m6A level of GhDi19 transcript increased, and the plants with increased m6A levels results in a significant increase in root length and enhanced drought resistance. Both works indicate that manipulating the m6A modification level of key genes can regulate plant phenotype and improve agronomic traits.
Strategies for improving crop traits through m6A modification
According to current research, three strategies have been proposed for enhancing crop agronomic traits through m6A modification. (1) Altering RNA m6A modification regulatory proteins (RMRPs) (methyltransferases, demethylases, and recognition proteins) and their interacting proteins. Given that m6A regulatory proteins typically regulate a multitude of substrate genes, this strategy exhibits a high degree of randomness and uncertainty in improving agronomic traits, and may frequently result in other phenotypic abnormalities; (2) Conducting m6A editing by fusing Cas13 (dCas13) with RNA-modified methyltransferase or demethylase in specific mRNA. However, the RNA-based m6A editing strategy poses limitations in crop breeding applications: the m6A editing vector must be maintained in the offspring, otherwise the editing effect of m6A cannot be preserved. Currently, they are not suitable for practical breeding applications; (3) Editing m6A motif of specific gene to remove m6A modifications. For example, using a base editor to accurately replace m6A-modified adenine on DNA. Regarding the m6A modification site on the UTR, the CRISPR/Cas9 system can be employed to disrupt the m6A modification motif on the UTR, thereby eliminating the m6A modification. This strategy involves directly altering m6A motifs on DNA, enabling the production of m6A editing materials without transgenic vectors in offspring, which can be utilized in breeding applications.
Steps for m6A editing
Steps 1: drawing single-base resolution m6A modification map
For the purpose of m6A editing, accurate m6A modification maps at single-base resolution must be generated initially. With the advancements in m6A detection technology, various single base resolution m6A sequencing methods have been reported, among them, m6A-selective allyl chemical labeling and sequencing (m6A-SAC-seq) (Hu et al., 2022), Nanopore direct RNA sequencing (DRS) (Zheng et al., 2020) and Glyoxal and nitrite-mediated deamination of unmethylated adenosine and sequencing (GLORI-seq) (Liu et al., 2022) are three methods that have broad application prospects (Figure 1). Recent research has reported the transcriptome-wide m6A maps at single-base resolution in various tissues of rice (plumule dark, plumule light, seedling at 8 days, seedling at 2 weeks, panicle, flag leaf at 10 days after anthesis, endosperm at 10 days after anthesis, and embryo at 10 days after anthesis) and Arabidopsis (seedling, shoot, root, rosette leaf, cauline leaf, stem, flower, silique, and seed) using m6A-SAC-seq, identifying a total of 205,691 m6A sites distributed across 22,574 genes in rice, and 188,282 m6A sites across 19,984 genes in Arabidopsis, offering comprehensive resources for investigating single-base resolution m6A sites and their functions in plants (Wang et al., 2024). Given the advancements in SAC-seq, DRS, and GLORI-seq, there are no technical limitations to m6A single base resolution sequencing, and single-base resolution maps for other crops can be constructed rapidly.
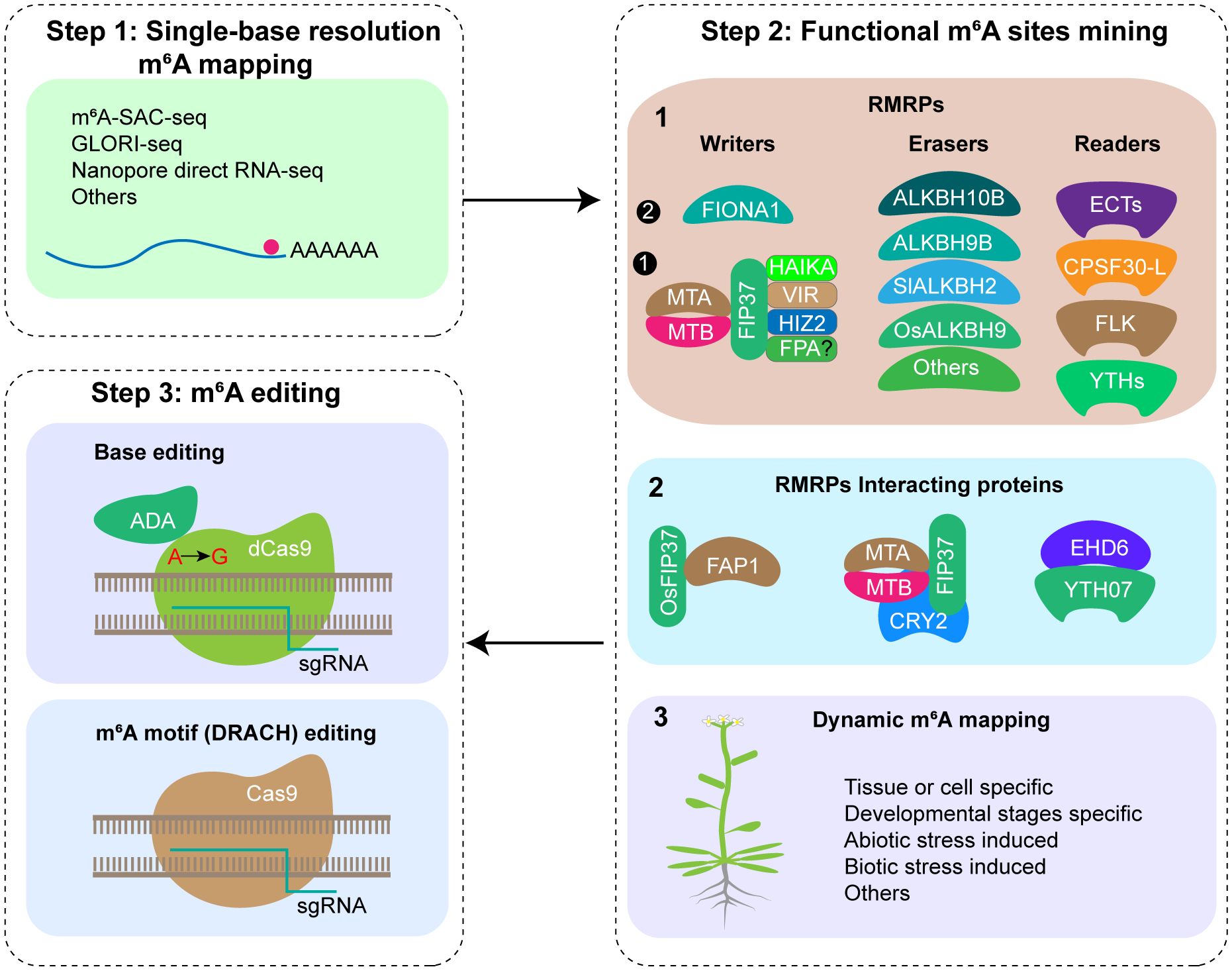
Figure 1. Putative steps of m6A editing in crop traits improvement. Step 1: Mapping mRNA m6A modification at single-base resolution; Step 2: Functional m6A sites mining by functional study of RMRPs, RMRPs interacting proteins, and dynamic m6A modification mapping; Step 3: m6A editing using CRISPR/Cas9 system or base editor.
Step 2: identification of functional m6A sites
Since there are more than ten thousands m6A sites present in the plant transcriptome, which ones have regulatory functions remains a key question to be clarified. Currently, three strategies are employed for identifying functional m6A sites (Figure 1). (1) Functional study of RMRPs. By utilizing mutants, RNA interference, or overexpression techniques for RMRPs, analyzing the phenotypes of relevant plant materials at different growth and development stages and in response to various biotic and abiotic stresses. Next, m6A sequencing and corresponding molecular biology methods are employed to ultimately confirm the m6A modification sites associated with the observed phenotypes. Utilizing RMRPs, several m6A modification sites with crucial functions have been identified, such as m6A in STM and WUS mRNA regulating shoot stem cell fate in Arabidopsis (Shen et al., 2016), FT, SPL3, and SPL9 mRNA regulating floral transition in Arabidopsis (Duan et al., 2017), and OsYUCCA3, TDR, and GAMYB mRNA regulating rice pollen development (Cheng et al., 2022; Tang et al., 2024), etc. In the future, we should enhance the application of genetic materials, comprehensively study their functions under various growth and development stages, environmental stress and stimulation, and different pest and disease stress in the field, and explore important functional m6A sites. (2) Functional study of proteins interacting with RMRPs. Interaction proteins of RMRPs might be involved in the processes of installing, removing, or recognizing m6A modifications at specific sites on specific mRNAs. Several studies have demonstrated the selection of specific m6A sites by interacting proteins of RMRPs. In Arabidopsis, CRY2 recruits the m6A writer complex (MTA/MTB/FIP37) to the photobody under blue light, thereby promoting m6A modification of circadian clock associated 1 (CCA1) and enhancing its mRNA stability (Wang et al., 2021). In rice, OsFIP37 is recruited by the RNA-binding protein OsFAP1 to deposit the m6A modification on OsYUCCA3 mRNA, thereby regulating male germ cell miosis (Cheng et al., 2022). EARLY HEADING DATE6 (EHD6) recruits the m6A reader YTH07 and sequesters OsCOL4 mRNA into phase-separated ribonucleoprotein condensates, thereby promoting rice flowering (Cui et al., 2024). (3) Exploring key m6A modification sites through dynamic m6A modification mapping. By mapping the changes in m6A modification maps under various tissues, developmental stages of the same tissue, and stress treatments of crops, m6A modification sites specific to corresponding processes are expected to be identified, which may have relevant regulatory functions.
Step 3: m6A editing using CRISPR/Cas9 system or base editor
After generating a single-base resolution m6A modification map and identifying functional m6A modification sites, m6A motif of specific gene can be edited using the CRISPR/Cas9 system or base editor (Figure 1). After gene editing, screening transgenic positive seedlings, identifying mutations, and confirming m6A modification changes, m6A-edited plants were obtained. Subsequently, phenotype analysis will be conducted and transgenic free plants will be screened for subsequent breeding applications.
Discussion
Fine-tuning gene expression is of great significance for crop improvement. m6A, the most abundant modification in mRNAs, plays crucial roles in regulating gene expression, positioning it as a potential tool for manipulating gene expression. With the advent of new m6A sequencing methods, mapping m6A at single base resolution is no longer an obstacle. The primary limitation of m6A application is the insufficient mining of functional m6A sites. The most effective approach to explore functional m6A sites in crops involves studying the biological functions of m6A methyltransferases, demethylases, or m6A binding proteins in crop development, biotic and abiotic stress response, and other processes. However, fewer RMRPs have been identified in crops so far, and future research should strengthen the identification and functional research of crop RMRPs. In summary, because of the vital roles of m6A in regulating gene expression post-transcriptionally, the m6A editing strategy holds significant potential for crop improvement.
Author contributions
JT: Writing – original draft, Writing – review & editing. XW: Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the earmarked fund for China Agriculture Research System (CARS-34) and the National Crop Germplasm Resources Center (NCGRC-63).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Cheng, P., Bao, S., Li, C., Tong, J., Shen, L., Yu, H. (2022). RNA N6-methyladenosine modification promotes auxin biosynthesis required for male meiosis in rice. Dev. Cell. 57, 246–259.e4. doi: 10.1016/j.devcel.2021.12.014
Cui, S., Song, P., Wang, C., Chen, S., Hao, B., Xu, Z., et al. (2024). The RNA binding protein EHD6 recruits the m6A reader YTH07 and sequesters OsCOL4 mRNA into phase-separated ribonucleoprotein condensates to promote rice flowering. Mol. Plant 17, 935–954. doi: 10.1016/j.molp.2024.05.002
Duan, H. C., Wei, L. H., Zhang, C., Wang, Y., Chen, L., Lu, Z., et al. (2017). ALKBH10B is an RNA N6-methyladenosine demethylase affecting arabidopsis floral transition. Plant Cell 29, 2995–3011. doi: 10.1105/tpc.16.00912
Hu, L., Liu, S., Peng, Y., Ge, R., Su, R., Senevirathne, C., et al. (2022). m6A RNA modifications are measured at single-base resolution across the mammalian transcriptome. Nat. Biotechnol. 40, 1210–1219. doi: 10.1038/s41587-022-01243-z
Li, S.-T., Ke, Y., Zhu, Y., Zhu, T.-Y., Huang, H., Li, L., et al. (2024). Mass spectrometry-based proteomic landscape of rice reveals a post-transcriptional regulatory role of N6-methyladenosine. Nat. Plants. 10, 1201–1214. doi: 10.1038/s41477-024-01745-5
Liu, C., Sun, H., Yi, Y., Shen, W., Li, K., Xiao, Y., et al. (2022). Absolute quantification of single-base m6A methylation in the mammalian transcriptome using GLORI. Nat. Biotechnol. 41, 355–366. doi: 10.1038/s41587-022-01487-9
Martinez-Perez, M., Aparicio, F., Lopez-Gresa, M. P., Belles, J. M., Sanchez-Navarro, J. A., Pallas, V. (2017). Arabidopsis m6A demethylase activity modulates viral infection of a plant virus and the m6A abundance in its genomic RNAs. Proc. Natl. Acad. Sci. U.S.A. 114, 10755–10760. doi: 10.1073/pnas.1703139114
Parker, M. T., Knop, K., Zacharaki, V., Sherwood, A. V., Tome, D., Yu, X., et al. (2021). Widespread premature transcription termination of arabidopsis thaliana NLR genes by the spen protein FPA. Elife 10, e65537. doi: 10.7554/eLife.65537
Ruzicka, K., Zhang, M., Campilho, A., Bodi, Z., Kashif, M., Saleh, M., et al. (2017). Identification of factors required for m6A mRNA methylation in arabidopsis reveals a role for the conserved E3 ubiquitin ligase HAKAI. New Phytol. 215, 157–172. doi: 10.1111/nph.14586
Shen, L., Liang, Z., Gu, X., Chen, Y., Teo, Z. W., Hou, X., et al. (2016). N6-methyladenosine RNA modification regulates shoot stem cell fate in arabidopsis. Dev. Cell 38, 186–200. doi: 10.1016/j.devcel.2016.06.008
Shi, C., Zou, W., Liu, X., Zhang, H., Li, X., Fu, G., et al. (2024). Programmable RNA N6-methyladenosine editing with CRISPR/dCas13a in plants. Plant Biotechnol. J. 22, 1867–1880. doi: 10.1111/pbi.14307
Tang, J., Chen, S., Jia, G. (2023). Detection, regulation, and functions of RNA N6-methyladenosine modification in plants. Plant Commun. 4, 100546. doi: 10.1016/j.xplc.2023.100546
Tang, J., Lei, D., Yang, J., Chen, S., Wang, X., Huang, X., et al. (2024). OsALKBH9-mediated m6A demethylation regulates tapetal PCD and pollen exine accumulation in rice. Plant Biotechnol. J. 22, 2410–2423. doi: 10.1111/pbi.14354
Wang, C., Yang, J., Song, P., Zhang, W., Lu, Q., Yu, Q., et al. (2022). FIONA1 is an RNA N6-methyladenosine methyltransferase affecting Arabidopsis photomorphogenesis and flowering. Genome Biol. 23, 40. doi: 10.1186/s13059-022-02612-2
Wang, G., Li, H., Ye, C., He, K., Liu, S., Jiang, B., et al. (2024). Quantitative profiling of m6A at single base resolution across the life cycle of rice and Arabidopsis. Nat. Commun. 15, 4881. doi: 10.1038/s41467-024-48941-7
Wang, X., Jiang, B., Gu, L., Chen, Y., Mora, M., Zhu, M., et al. (2021). A photoregulatory mechanism of the circadian clock in Arabidopsis. Nat. Plants 7, 1397–1408. doi: 10.1038/s41477-021-01002-z
Xu, T., Wu, X., Wong, C. E., Fan, S., Zhang, Y., Zhang, S., et al. (2022). FIONA1-mediated m6A modification regulates the floral transition in arabidopsis. Adv. Sci. (Weinh), e2103628. doi: 10.1002/advs.202103628
Yu, L., Alariqi, M., Li, B., Hussain, A., Zhou, H., Wang, Q., et al. (2024). CRISPR/dCas13(Rx) derived RNA N6-methyladenosine (m6A) dynamic modification in plant. Advanced Sci. e2401118. doi: 10.1002/advs.202401118
Yu, Q., Liu, S., Yu, L., Xiao, Y., Zhang, S., Wang, X., et al. (2021). RNA demethylation increases the yield and biomass of rice and potato plants in field trials. Nat. Biotechnol. 39, 1581–1588. doi: 10.1038/s41587-021-00982-9
Zhang, M., Bodi, Z., Mackinnon, K., Zhong, S., Archer, N., Mongan, N. P., et al. (2022). Two zinc finger proteins with functions in m6A writing interact with HAKAI. Nat. Commun. 13, 1127. doi: 10.1038/s41467-022-28753-3
Zheng, H. X., Zhang, X. S., Sui, N. (2020). Advances in the profiling of N6-methyladenosine (m6A) modifications. Biotechnol. Adv. 45, 107656. doi: 10.1016/j.biotechadv.2020.107656
Zhong, S., Li, H., Bodi, Z., Button, J., Vespa, L., Herzog, M., et al. (2008). MTA is an arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant Cell 20, 1278–1288. doi: 10.1105/tpc.108.058883
Zhou, L., Tang, R., Li, X., Tian, S., Li, B., Qin, G. (2021). N6-methyladenosine RNA modification regulates strawberry fruit ripening in an ABA-dependent manner. Genome Biol. 22, 168. doi: 10.1186/s13059-021-02385-0
Keywords: RNA modification, gene expression, m6A modification, m6A application, crop improvement, m6A editing
Citation: Tang J and Wang X (2024) Strategies for RNA m6A modification application in crop improvement. Front. Plant Sci. 15:1477240. doi: 10.3389/fpls.2024.1477240
Received: 07 August 2024; Accepted: 30 September 2024;
Published: 15 October 2024.
Edited by:
Xuming Li, Hugo Biotechnologies Co., Ltd., ChinaReviewed by:
Ji Shi, Peking University, ChinaCopyright © 2024 Tang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jun Tang, dGFuZ2p1bkBjYWFzLmNu; Xuemin Wang, d2FuZ3h1ZW1pbkBjYWFzLmNu
 Jun Tang
Jun Tang Xuemin Wang
Xuemin Wang