- 1College of Forest Resources and Environmental Science, Michigan Technological University, Houghton, MI, United States
- 2Department of Biological Sciences, Eastern Illinois University, Charleston, IL, United States
Woody biomass serves as a renewable resource for various industries, including pulp and paper production, construction, biofuels, and electricity generation. However, the molecular mechanisms behind biomass traits are poorly understood, which significantly curtails the speed and efficiency of their improvement. We used activation tagging to discover genes that can positively affect tree biomass-associated traits. We generated and screened under greenhouse conditions a population of 2,700 independent activation tagging lines. A total of 761 lines, which had significantly and positively affected at least one biomass-associated trait, were discovered. The tag was positioned in the genome for forty lines which were affected in multiple traits and activation of proximal genes validated for a subset. For two lines we fully recapitulated the phenotype of the original lines through overexpression. Moreover, the overexpression led to more pronounced and additional improvements, not observed in the original lines. Importantly, the overexpression of a Fasciclin-like gene (PtaFLA10) and a Patatin-like gene (PtaPAT) was found to substantially improve biomass, with a 40% increase in dry-stem weight, and enhance drought tolerance, respectively. Additionally, PtaPAT overexpression increased cellulose content, which is crucial for biofuel production. Our work shows that the activation tagging approach applied even on a non-genome saturation scale in a poplar tree can be successfully used for the discovery of genes positively modify biomass productivity. Such dominant forward genetics approaches can aid in biotechnological manipulation of woody biomass traits and help unravel the functions and mechanisms of individual genes, gene families, and regulatory modules.
Introduction
Dedicated bioenergy crops like poplar, willow, and others are projected to displace 30 % of current US petroleum consumption (Perlack et al., 2005). In addition, woody biomass provides a renewable resource for production of pulp and paper, structural construction timber and multiple other products (de Vries et al., 2021; Skog, 2008). Despite the economic and ecological importance of woody biomass, the underlying molecular mechanisms of biomass-related traits remain poorly understood, and this significantly curtails the speed and efficiency of their improvement.
Biomass is a complex trait resulting from the integration of numerous processes, encompassing molecular, cellular, developmental, physiological, and metabolic levels (Carpita and McCann, 2020; Li et al., 2024; Zhu and Li, 2023). There has been substantial and long-standing interest in understanding biomass-related traits from both improvement and fundamental perspective (Groover, 2005; Li et al., 2024; Zhu and Li, 2023). However, dissecting these traits is challenging due to their complexity and the long generation cycle of trees, which makes traditional genetic, and mutagenesis approaches impractical (Busov et al., 2005a).
The value of forward genetics approaches involving insertional and other forms of mutagenesis is well established (Alonso et al., 2003). However, these methods are difficult to use in trees, largely because of their long generation cycles. Only dominant approaches like activation tagging and full-length overexpression (FOX) approaches are feasible because they can generate mutations in the first generation (Busov et al., 2005a; Rauschendorfer et al., 2020). Activation tagging uses a T-DNA vector with strong constitutive enhancer elements positioned near its left or right border. Insertion of the T-DNA into the genome typically leads to the up-regulation of a proximal flanking gene, resulting in a gain-of-function, dominant mutation (Deng et al., 2020). Dissecting gene functions through loss-of-function mutations is challenging because many genes exist in multiple copies, often organized in large gene families with partially redundant functions. Activation tagging, which creates gain-of-function mutations, offers an alternative for functional characterization of gene families (Deng et al., 2020; Nakazawa et al., 2003). The presence of such gene families is a particular problem in poplar, which has undergone whole-genome duplication events (Tuskan et al., 2006). Indeed, many of the genes we identified through activation tagging in Populus belong to large gene families such as Gibberellin (GA) 2-oxidase, AP2/ERF transcription factor (TF), AT-hook domain TF, and Lateral Organ Boundary (LBD) TF (Azeez et al., 2021; Busov et al., 2003; Trupiano et al., 2013; Yordanov et al., 2010, Yordanov et al., 2014). Activation tagging also offers several other advantages: easy characterization of the insertion site using the tag sequence (Liu et al., 1995); preferential insertion in gene-rich genomic regions (Kim and Veena, 2007); and discovery of poorly annotated or non-protein coding loci (Palatnik et al., 2003).
Here, we demonstrate the successful application of activation tagging in poplar to identify genes that affect biomass traits. Notably, we were able to identify a large number of mutations that positively affect one or several biomass-associated traits. For a subset of these mutants, we mapped the tag within the genome, identified the proximal genes, confirmed their upregulation, and recapitulated the mutant phenotypes via overexpression of the putative causative genes. Our findings demonstrate the efficacy of a dominant forward tagging approach applied on a non-genome saturation scale for uncovering genes that positively impact biomass traits in a tree.
Results
Phenotypic screens for mutations affecting biomass-associated traits
To identify genes influencing traits linked to biomass growth in Populus, we generated and screened under controlled greenhouse conditions a population of 2,700 activation-tagged lines (see Material and Methods). We focused on traits that are linked to biomass growth (Figure 1A). A total of 761 lines exhibited significant effect on at least one trait, categorized as phenotypic mutant (Figure 1B). The various mutations had an approximately evenly distributed impact over the 10 studied traits. The number of internodes showing the greatest impact among all traits (Figure 1A). In contrast, traits such as diameter at leaf plastochron index (LPI) 20, green density, dry weight at the base and dry leaf weight were least affected (Figure 1A). More than half of the mutants (402) were affected in only one trait (Figure 1B). In many cases, mutations impacted simultaneously as many as 5-8 traits (Figure 1B).
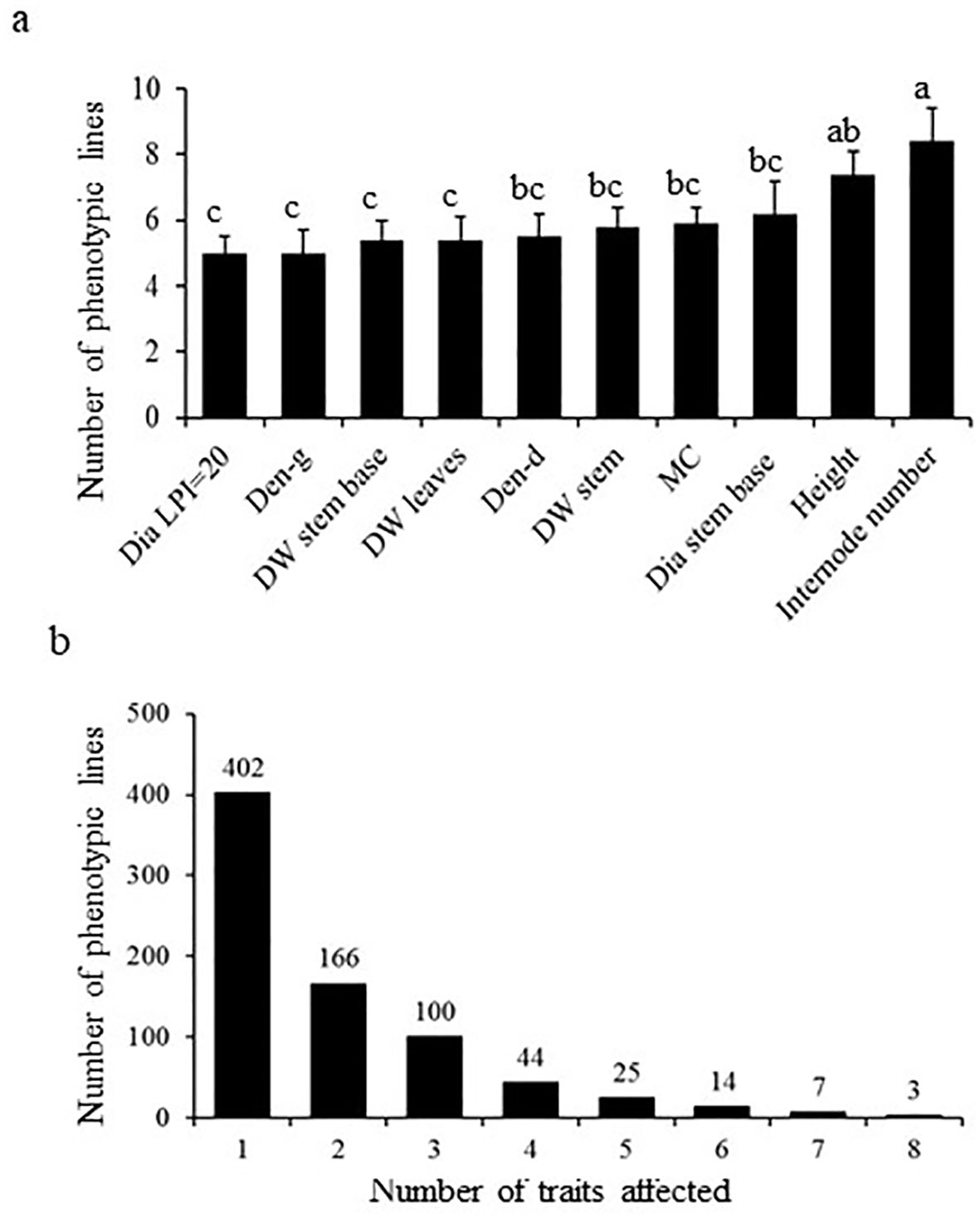
Figure 1. Summary of mutants and biomass traits affected in the poplar activation tagging screen. (A) Number of mutants identified to be positively and significantly affected in the various measured biomass-associated traits. Different letters indicate significance as determined by a one-way ANOVA followed by Fisher’s test (p < 0.05). (B) Illustrate the number of mutants affected in single versus multiple biomass traits.
Correlations between affected phenotypic traits
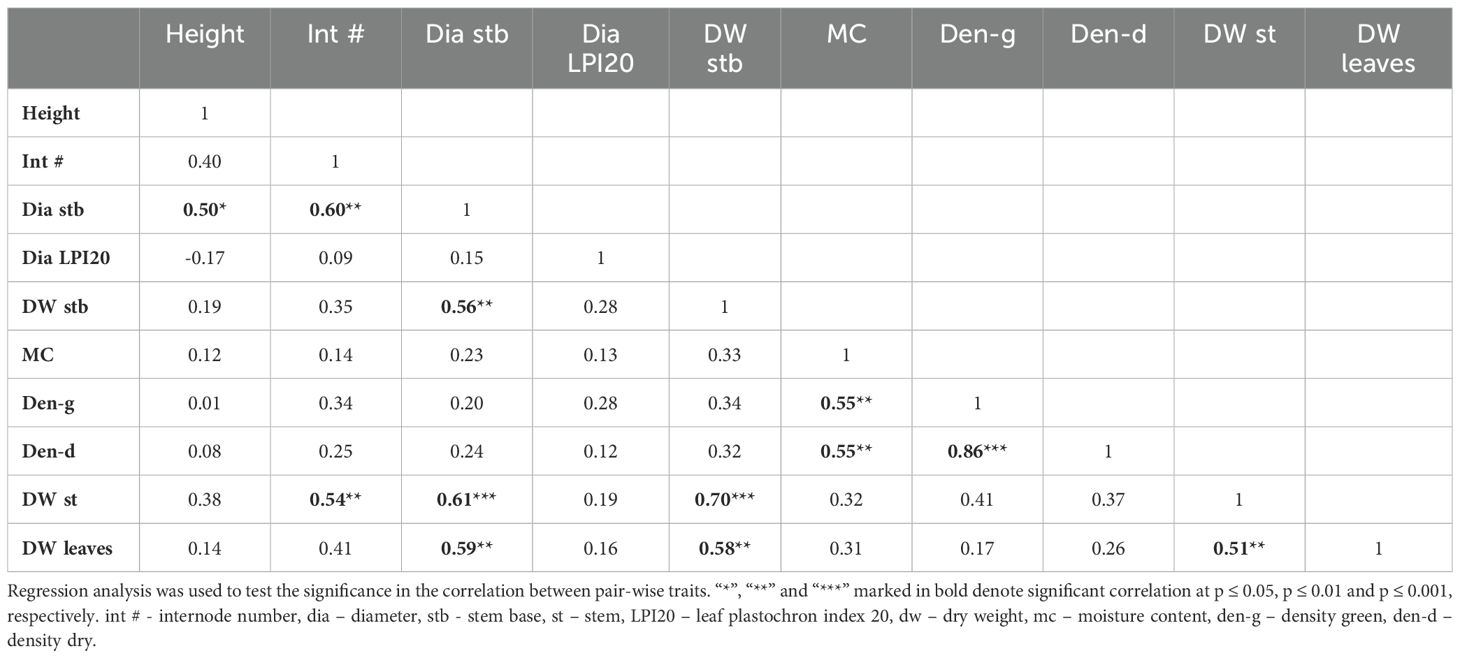
To explore potential correlation between traits, pair-wise phenotypic correlations were calculated across the entire experiment (Table 1). As expected, strong correlations were evident between closely related traits, such as green and dry density, and between total dry biomass of the whole stem and the stem base. Interestingly, less intuitive correlations were also observed. For instance, dry leaf biomass showed significant and positive correlation with the stem base diameter and dry stem biomass. Additionally, dry stem biomass correlated positively with internode number and the stem base diameter. These correlations may suggest underlaying morphological or physiological connections that are not apparent and require further investigations.
The tag insertions are proximal to genes
We characterized T-DNA insertions (Supplementary Table S1) in 40 mutant lines (Table 2). Chromosome 1 had the highest number of insertions (7), followed by 6 insertions on chromosome 6, and 5 insertions on each chromosome 10 and 14 (Figure 2A). A significant correlation between chromosome size and the number of insertions suggests random insertion of the tag (Figure 2B). Most (37.5%/15) of the insertions were located within the 10 Kbp 5′/3′ regions proximal to genes, while 17.5 % (7) in exons and 12.5 % (5) in introns (Figure 2C). Additionally, 22.5 % (9) were positioned in the 5’- or 3’-untranslated gene regions (UTR) (Figure 2C).
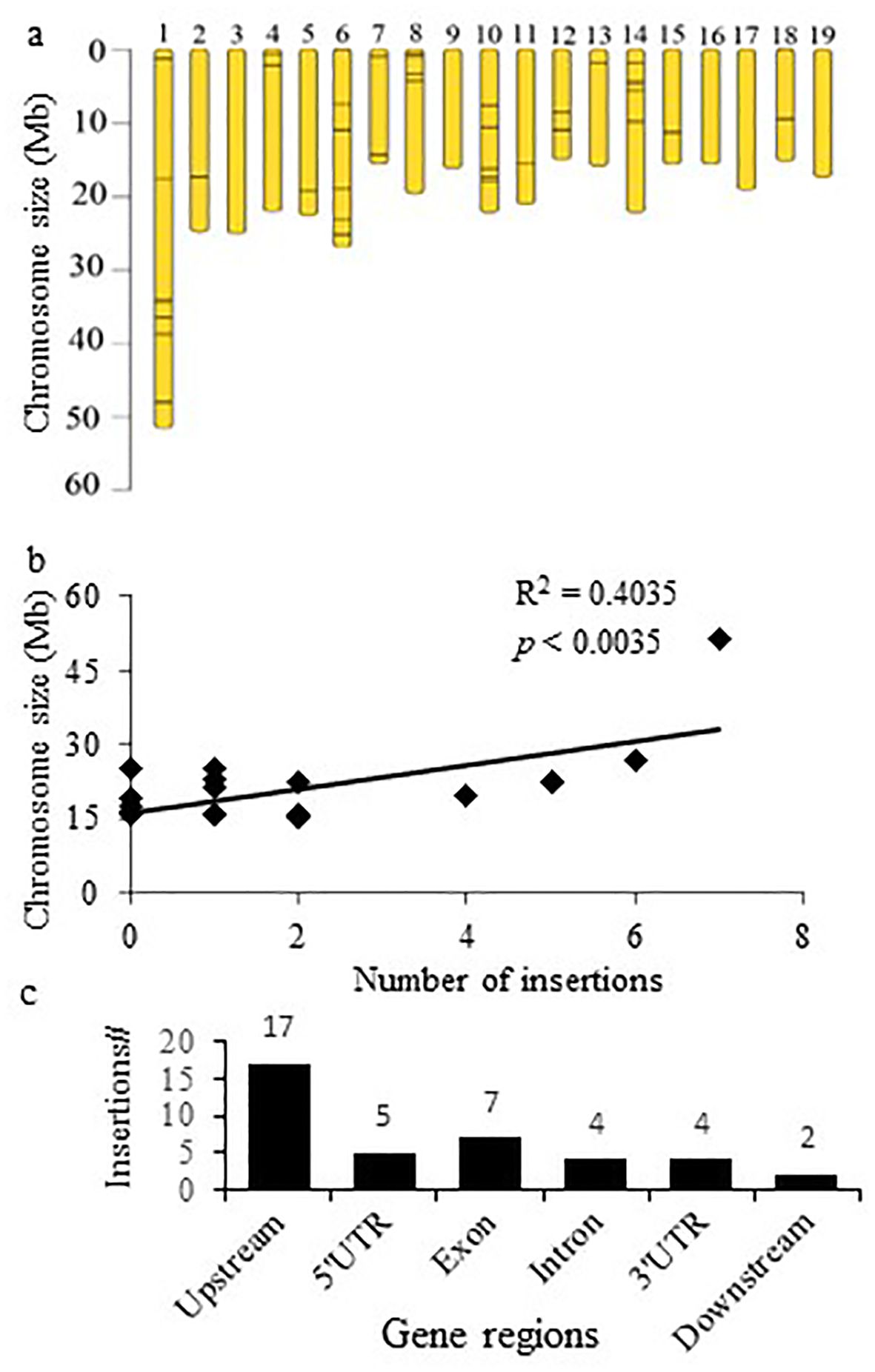
Figure 2. Characterization of the activation tag integration in the Populus genome. (A) Distribution of the insertions within the chromosomes. (B) Correlation between chromosome size and number of insertions per chromosome. (C) Predominant identification of T-DNA insertions upstream from the gene coding region.
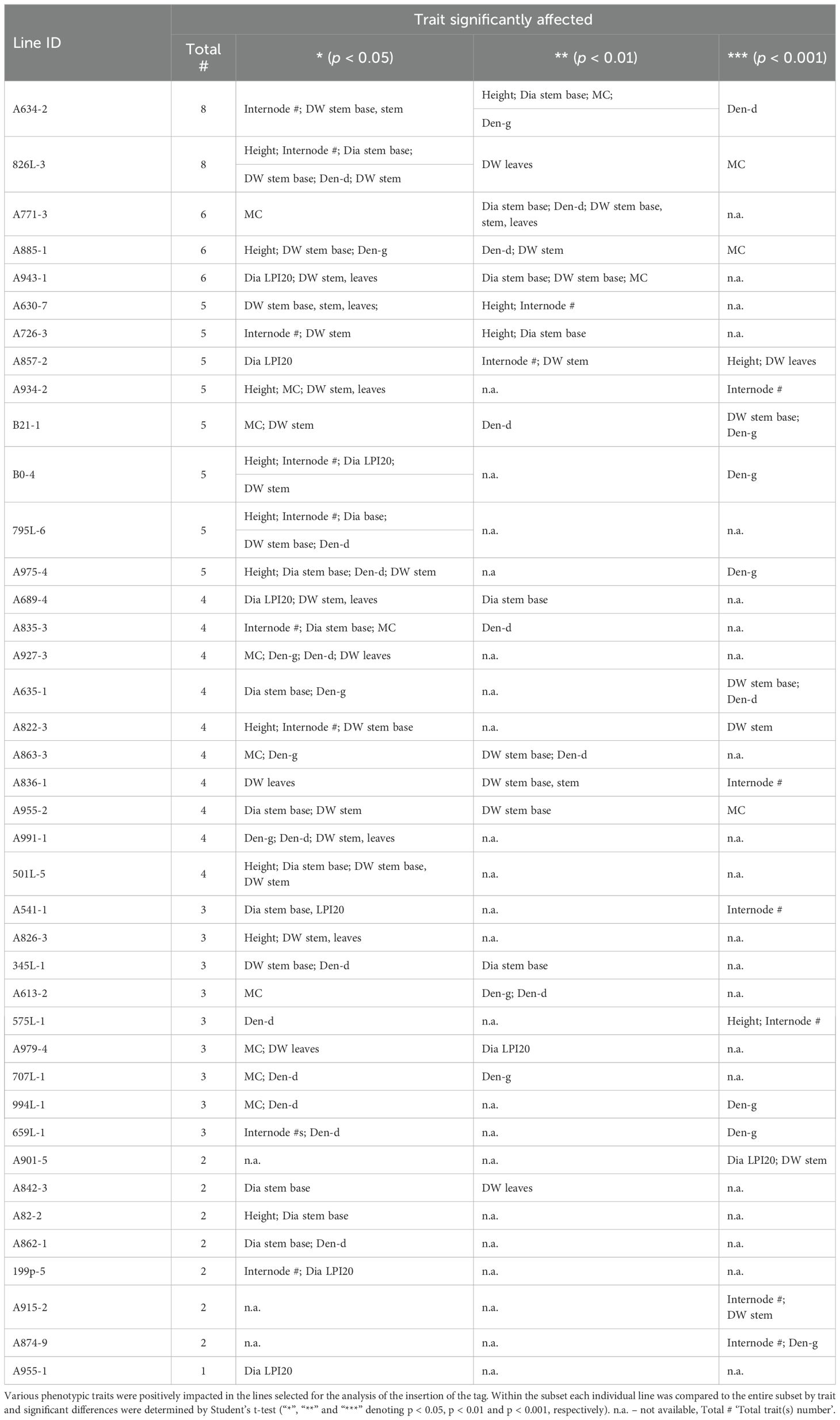
Tagged genes are activated
For nine mutants, we proceeded with further characterization of the expression of the genes near to the insertion sites (Table 3). These mutants were selected based on the magnitude of their impact on the affected trait, the number of traits affected, and the novelty of the gene function (Table 3). Since the tag is often inserted near multiple genes, and thus potentially impacts more than one, we studied the expression of all potential candidates. In most cases (8 out of 9 lines), we found only one of the nearby genes activated (Table 3). However, in only one line (A630-7), both genes proximal to the tag showed activation.
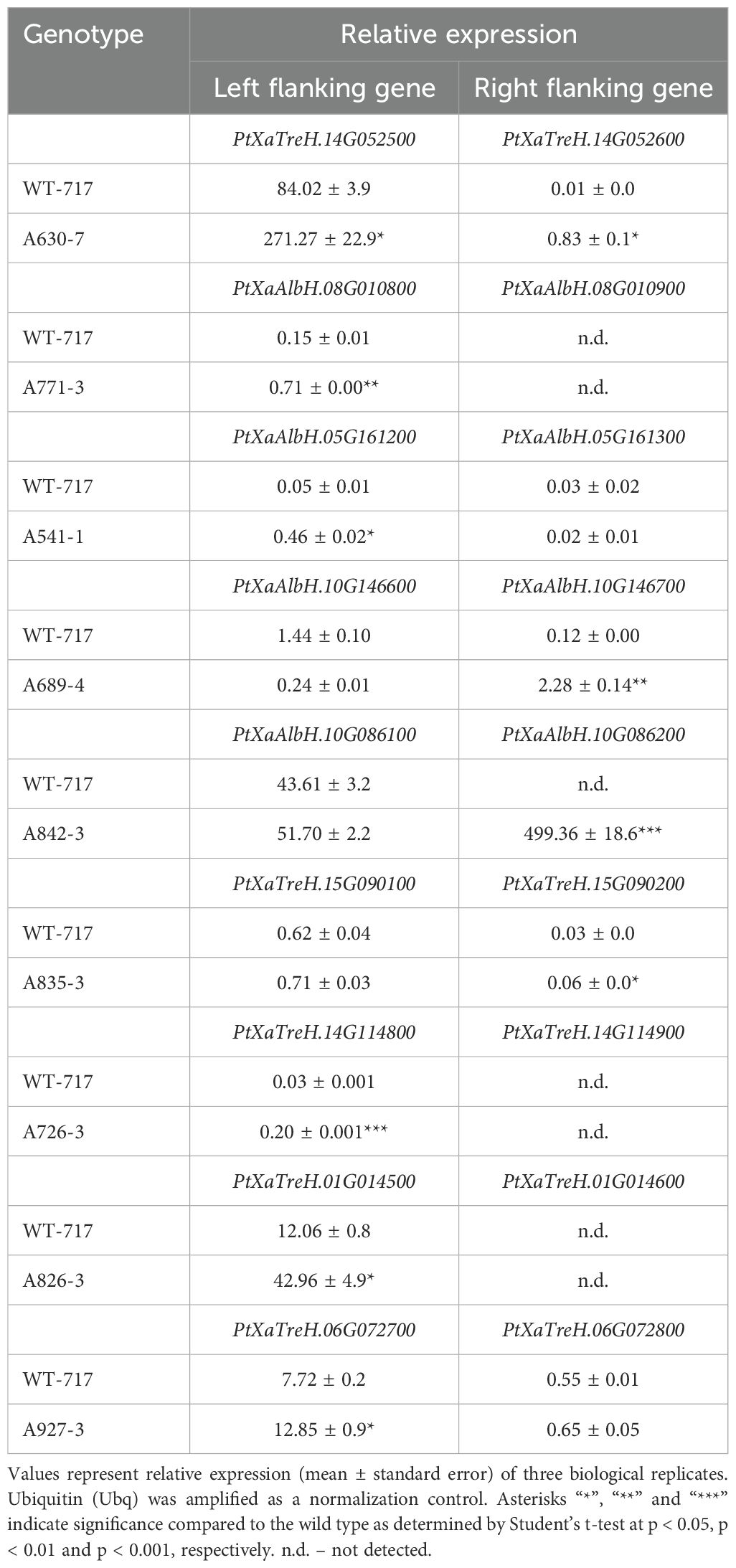
Table 3. Real-time quantitative RT-PCR analysis of proximal genes flanking the T-DNA insertion site.
Tagged genes show diverse and tissue-specific native expression patterns
We characterized the expression of the activated genes in various organs, including the apex, leaf, stem, and root of wild type (WT-717) plants (Figure 3). These genes exhibited diverse tissue specific expression patterns, suggesting varied influence on biomass traits. For example, PtXaTreH.14G052500, PtXaTreH.14G052600 and PtXaAlbH.08G010800 were predominantly expressed in the apex. PtXaAlbH.10G086200 and PtXaTreH.15G090200 were abundant in the root, whereas PtXaAlbH.05G161200 was mainly expressed in both the apex and roots. PtXaTreH.06G072700 was expressed in all studied tissues.
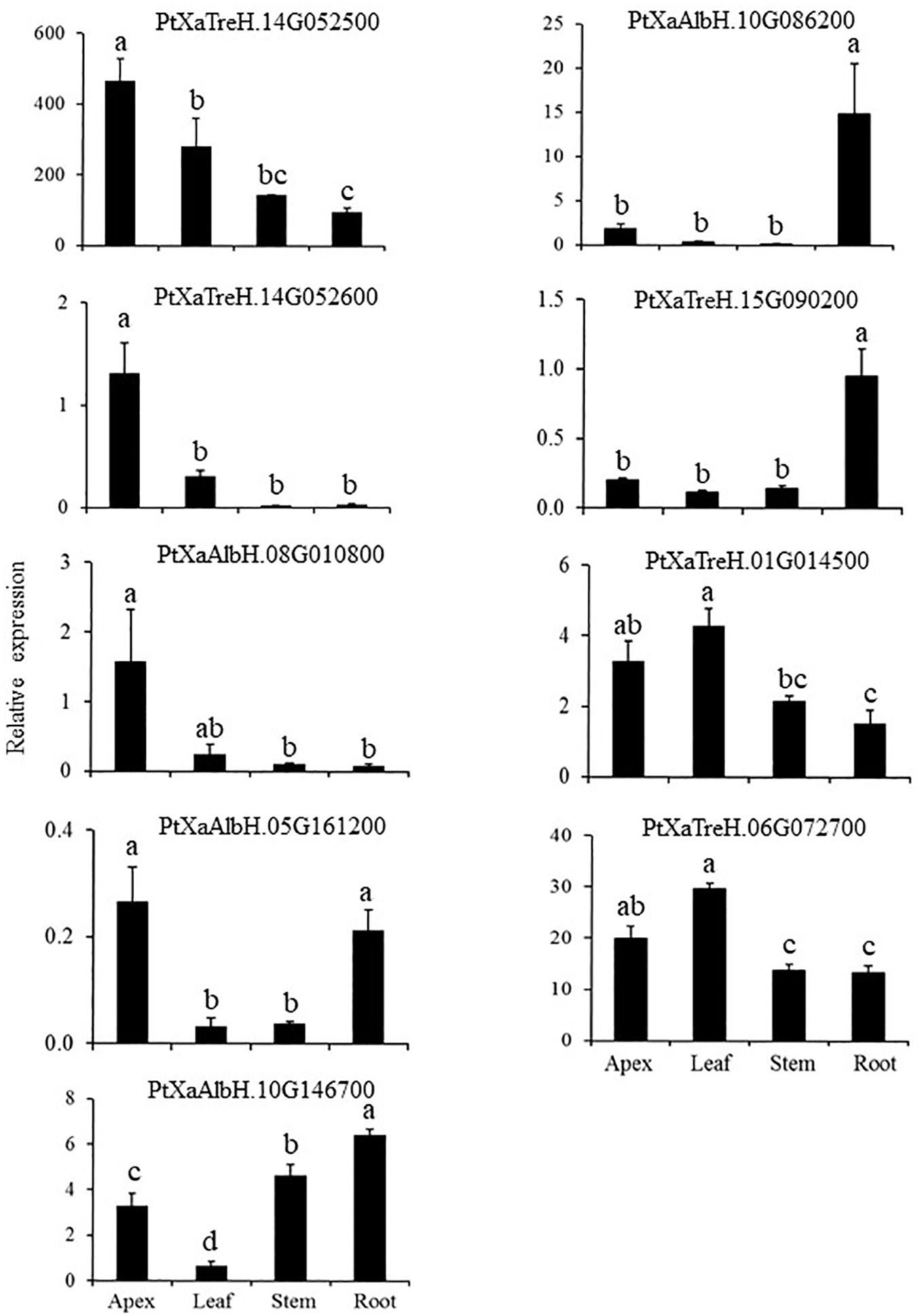
Figure 3. Expression of the tagged genes in different organs/tissues. Bars represent means of four biological replicates ± standard error. Different letters indicate significant differences as determined by a one-way ANOVA followed by Fisher’s test (p < 0.05).
Fasciclin-like gene enhances multiple biomass traits
One of the lines selected for recapitulation experiments was A630-7, chosen for its simultaneous impacts on multiple traits (five in total, as shown in Table 2). Most importantly, the mutation significantly enhanced stem dry weight, a crucial aspect of biomass production. Additionally, the line exhibited increased leaf dry biomass, which, as previously mentioned, is positively correlated with stem dry weight (Table 1). This prompted our interest in identifying the gene responsible for these changes. Position and expression analyses indicated the upregulation of two genes near the tag insertion site (Table 3). One gene showed the highest homology to Arabidopsis's Fasciclin 10 (FLA10), which we named PtaFLA10. The other gene exhibited the highest sequence homology to Growth Regulating Factor 9 (GRF9) transcription factor from Arabidopsis, which was called PtaGRF9. It was unclear which of the two genes was responsible for the phenotypic changes. We thus produced overexpression constructs for both genes and transformed them into transgenic plants. Numerous independent events were regenerated with the PtaFLA overexpression construct (oe-PtaFLA10). However, despite several transformations attempts, we could only recover four transgenic plants with PtaGRF9 overexpression construct (oe-PtaGRF9), suggesting that PtaGRF interferes with the regeneration process.
PtaFLA10 overexpression positively affected several phenotypic traits linked to biomass growth compared to WT-717 plants (Figure 4). For instance, both the original mutant A630-7 line and oe-PtaFLA lines were about 9 % taller than the wild type. Additionally, the number of internodes increased by 15% in the A630-7, while the oe-PtaFLA lines exhibited an average increase of 20 % (Figure 4). The oe-PtaFLA lines also displayed changes not observed in the original mutant, likely due to the strong overexpression. For example, there was a significant increase in the diameter at the 20th internode and at the stem base (Figure 4). Most notably, a 40 % increase in the dry stem biomass was observed in both the original A630-7 mutant line and oe-PtaFLA transgenics. Consistent with the strong correlation between leaf and stem dry weight (Table 1), leaf dry weight was also increased in both the mutant and recapitulation transgenics.
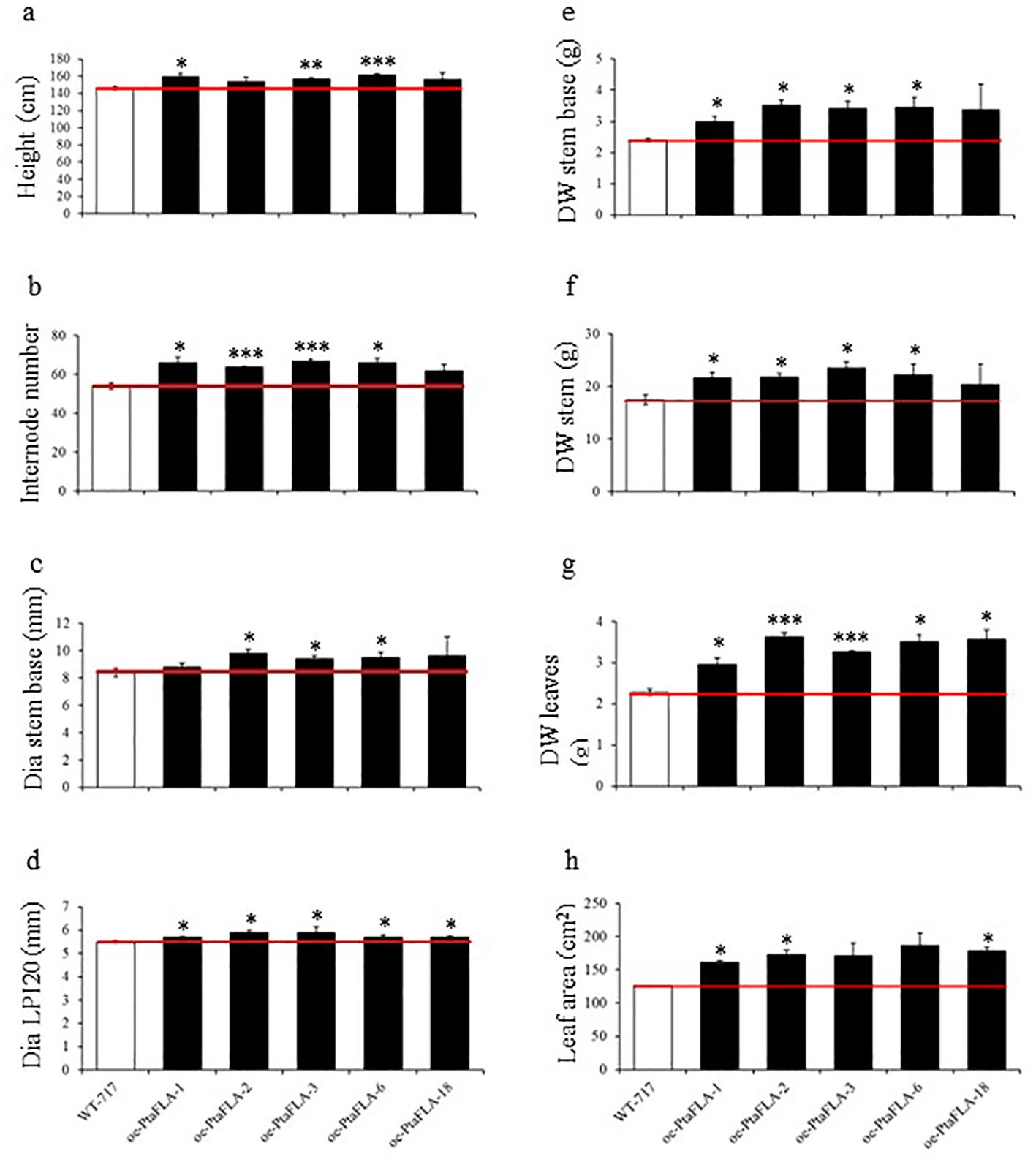
Figure 4. Recapitulation of A630-7 phenotype. (A–H) Changes of traits in mutant and oe-PtaFLA lines. Values are reported as mean ± SE (n = 4). White bars represent wild type and black bars – PtaFLA OE=overexpression lines. Asterisks indicate significant differences between transgenics and wild type plants as determined by Student’s t-test (*, ** and *** denoting p < 0.05, p < 0.01 and p < 0.001, respectively). Red line corresponds to the wild types’ threshold.
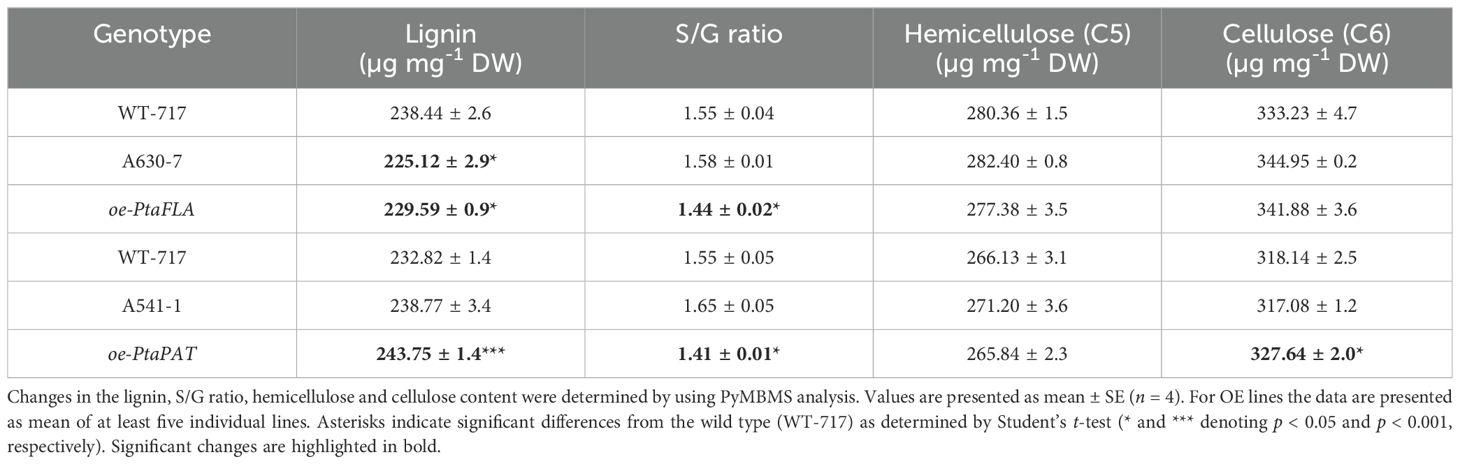
In addition to enhanced biomass growth, both the original line and the overexpression transgenics showed a significant decrease in lignin content (Table 4). The overexpression transgenics also exhibited a reduced S/G ratio, a change not observed on original line (Table 4). No significant changes were measured in cell wall carbohydrates.
Overexpression of the other gene, PtaGRF9, did not result in significant phenotypic changes, suggesting that the observed mutant phenotypic characteristics are due to the upregulation of PtaFLA10 gene.
A patatin-like gene affects biomass growth, leaf development and response to drought
We also further characterized the A541-1 mutant line through recapitulation experiments. The activated gene in this line encoded a protein with high similarity to patatin, and thus named PtaPAT. PtaPAT overexpression led to an increase in stem diameter, but only in stems undergoing primary growth, specifically 5th and 10th internodes (Figures 5A, B; Table 2). Consistent with the original A541-1 mutant line, the oe-PtaPAT lines also exhibited an increased number of internodes (Figure 5C; Table 2). Additionally, overexpression of PtaPAT resulted in leaves with an uneven adaxial leaf surface, reminiscent of potato leaves (Figures 6A–C).
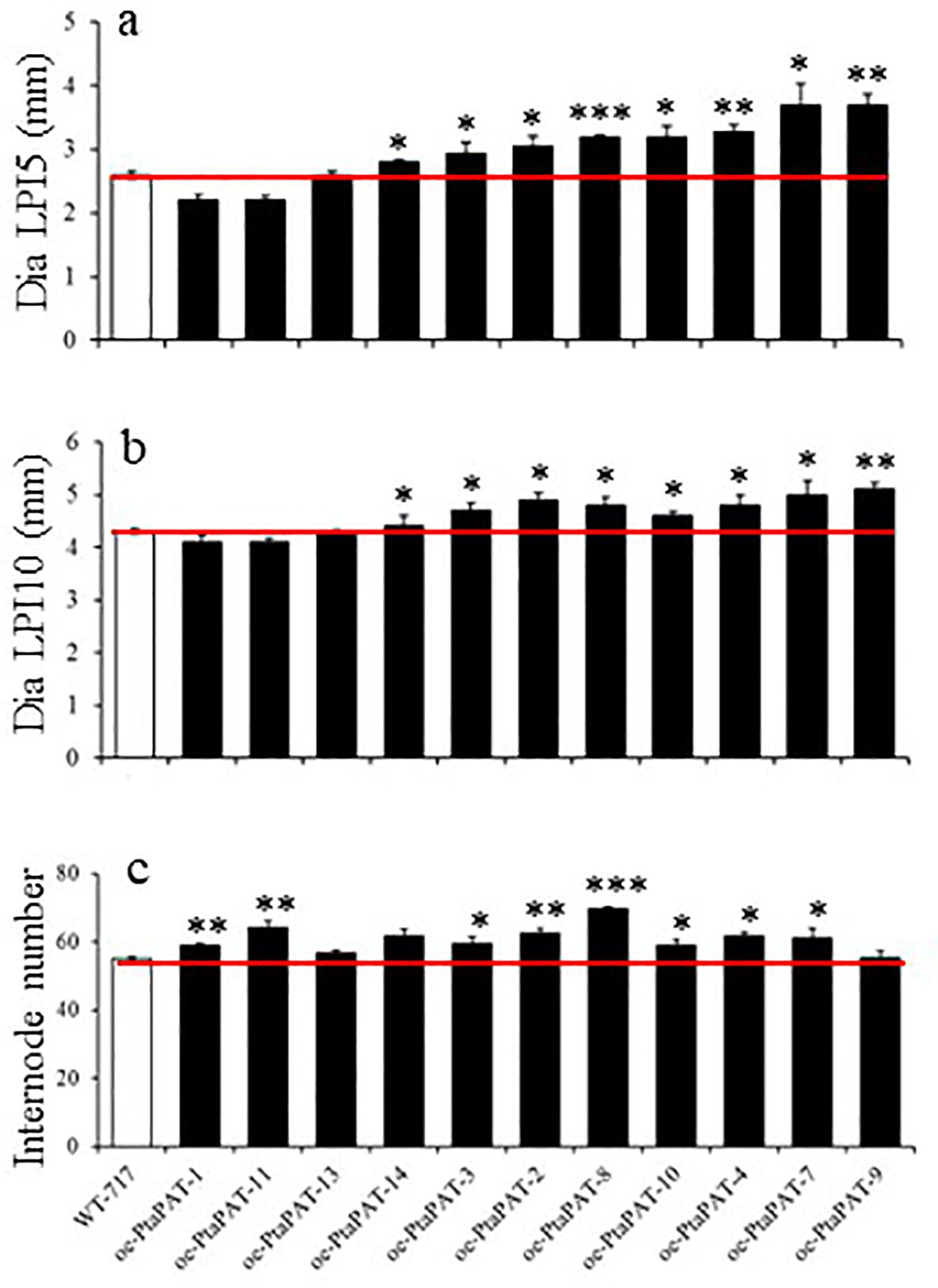
Figure 5. Recapitulation of A541-1 phenotype. (A–C) Changes in traits of oe-PtaPAT lines. Bars represent mean of four biological replicates ± SE (n = 4). White bars represent wild type and black bars represent Patatin OE lines. Asterisks indicate significant differences between transgenic and WT plants as determined by Student’s t-test (‘*’, ‘**’ and ‘***’ denoting p < 0.05, p < 0.01 and p < 0.001, respectively).
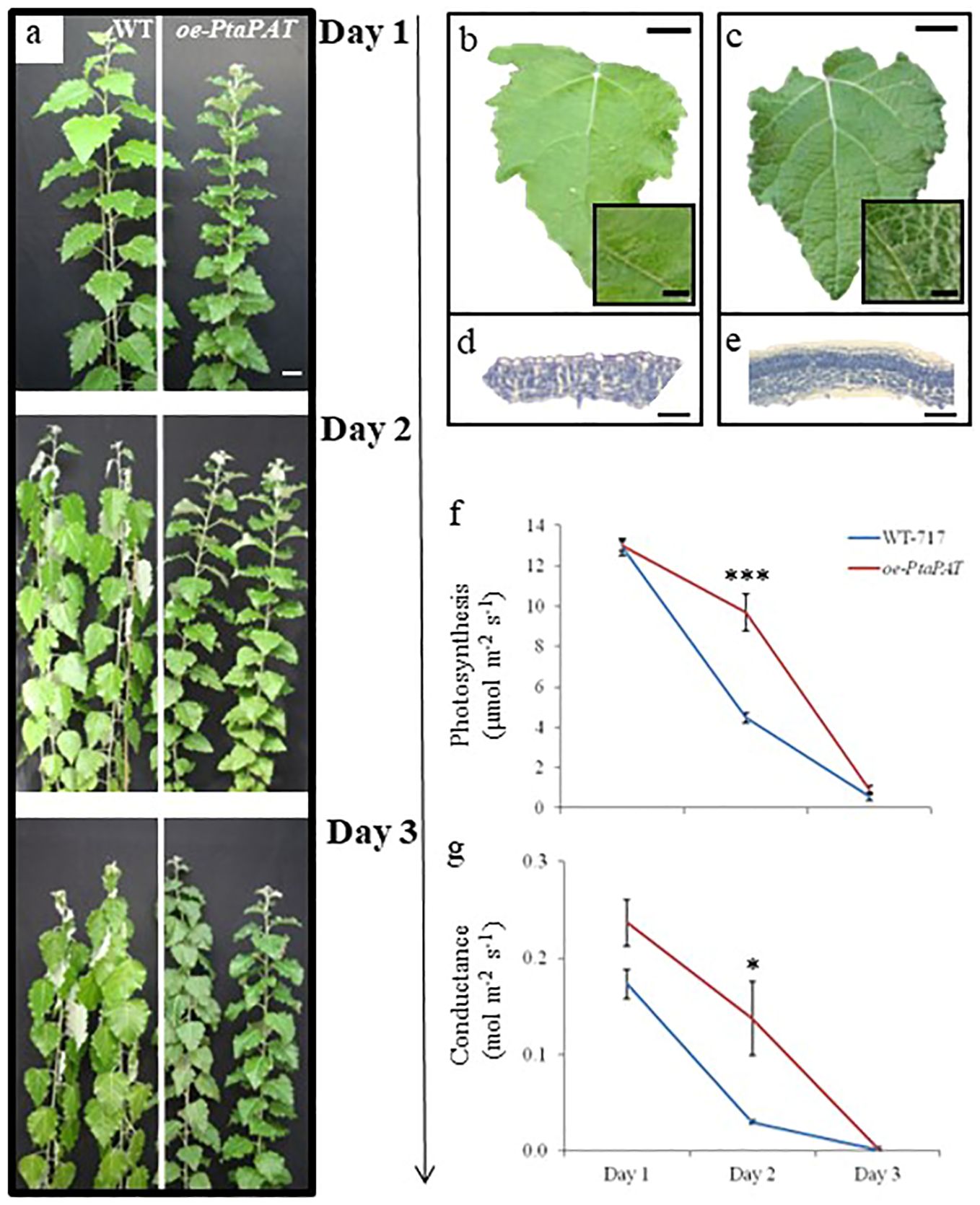
Figure 6. Poplar patatin OE lines showed better performance under drought conditions. (A) Drought experiment including wild type (WT-717) and oe-PtaPAT lines. Plants were maintained without water for 3 days. Pictures were taken daily and show representative appearance of multiple ramets and lines. Leaf texture and cross-section of WT-717 (B, D) and oe-PtaPAT (C, E) control plants (n=3 lines with 4 ramets per line). Analysis of photosynthetic activity and (F) stomatal conductance (G) during drought stress. Data were collected every day during the treatment period. Asterisks indicate significant differences between transgenic and WT-717 plants as determined by Student’s t-test (n=4, * and *** denoting p< 0.05 and p < 0.001, respectively). Scale bars (A) = 5 cm, (B, C) = 2 cm, (D, E) = 50 µm.
Given the distinct leaf surface observed in the oe-PtaPAT lines, we performed histological analysis (Figures 6 D, E). The oe-PtaPAT lines displayed a much denser palisade mesophyll cell layer, with a larger number of closely spaced cells compared to the WT plants (Figures 6 D, E).
Because of these unique changes in leaf structure, we wanted to study the performance of the oe-PtaPAT lines under drought conditions (Figures 6A, F, G). Wild-type and oe-PtaPAT lines were subjected to drought stress for 3 days and CO2 assimilation and stomatal conductance were monitored during this period. On the 2nd day of the stress treatment oe-PtaPAT lines were not wilted (Figure 6A) and showed higher photosynthetic activity (Figure 6F) and stomatal conductance (Figure 6G) compared to the wild-type plants. These results indicate a possible and yet unknown role of the patatin gene in the response to the drought and create new opportunities for further gene manipulations to improve drought tolerance in plants.
The oe-PtaPAT lines also showed increases in lignin, cellulose, and S/G ratio, changes not observed in the original mutant line and likely attributable to the much higher level of overexpression (Table 4).
Discussion
In this study, we applied a moderate, non-genome saturation size of poplar activation tagging population to discover genes that positively affect biomass. Despite the moderate population size and apparent randomness of the tag insertion, we identified a significant number of ‘productive’ mutations. Several pieces of evidence suggest that these mutations result from genuine activation tagging. These include the tag’s insertion near genes, the upregulation of the adjacent genes, and the successful recapitulation of the phenotypic changes through overexpression. The high success of activation tagging in poplar is attributed to the preferential insertion of the T-DNA near genes, likely due to Agrobacterium's tendency to target transcriptionally active, open chromatin regions, a phenomenon also observed in Arabidopsis and rice (Alonso et al., 2003; An et al., 2003; Chen et al., 2003; Forsbach et al., 2003; Rosso et al., 2003; Sallaud et al., 2004; Szabados et al., 2002). The bias toward open chromatin enhances the likelihood of successful activation and productive mutation, while our analysis revealed a random insertion pattern correlated with chromosomal size. Majority (37.5%) of the positioned tags were found with the 10 kb upstream or downstream of the genes, confirming our previous finding (Busov et al., 2010) with a much smaller population. Finally, the large size of poplar also facilitates the detection of the phenotypic changes. As a perennial species, these phenotypic changes could be further accentuated if the screening were performed under multi-year field conditions, where the changes would accumulate over time. Unfortunately, due to stringent regulatory regimes, conducting such field trials remains logistically challenging.
There is substantial evidence indicating significant interdependence between the regulators of biomass production and cell wall thickening (Maleki et al., 2020). Similar findings were observed in Arabidopsis (Hu et al., 2018). For instances, enzymes involved in cell wall loosening result significant increases in biomass accumulation (Park et al., 2003; Shani et al., 2004), highlighting the intricate link between cell wall biology and growth processes in plants. Overexpression of a cellulose synthase led to increased biomass production in poplar (Maleki et al., 2020). Therefore, it is not surprising that both genes we recapitulated (e.g., patatin and fasciclin) are associated with cell wall metabolism.
Fasciclins have long been known to be involved in secondary wall thickening, though their exact role remains elusive (Dharmawardhana et al., 2010; Huang et al., 2013; Janz et al., 2012; Lafarguette et al., 2004; Ma et al., 2022, Ma et al., 2023; MacMillan et al., 2010; Wang et al., 2015a, Wang et al., 2017, Wang et al., 2015a). Fasciclin-like arabinogalactan proteins (FLAs) contain a characteristic fasciclin-like domain, which plays a crucial role in cell-cell and cell-matrix interactions, as well as in cell expansion, in animal cells. Recent studies have identified approximately 50 FLA proteins in Populus trichocarpa (poplar), with most of these proteins being highly expressed in developing xylem. Notably, group A FLAs are specifically associated with lignified internodes, highlighting their potential role in wood formation and structural integrity (Zhen et al., 2023). Indeed, the PtrFLA40/45 mutant exhibited a significant increase in lignin content, which was accompanied by the upregulation of six lignin biosynthetic genes (Zhen et al., 2023). Interestingly, several other FLA genes in poplar have been implicated in the formation of tension wood, operating through a pathway associated with Gibberellin A3 signaling (Wang et al., 2017; Lafarguette et al., 2004). Since secondary cell wall constitutes the bulk of lignocellulosic biomass, it is not surprising that the modification in fasciclin expression leads to changes in biomass yield and properties. The fasciclin family is large and complex with a significant variation in the number and types of domains (Dharmawardhana et al., 2010). The gene identified in our study is of particular interest for several reasons, First, it increases biomass by 40 % on a dry biomass basis. Additionally, the cell wall shows a significant decrease in lignin content. These changes are highly consistent with the effects observed with other members of the fasciclin family (Ma et al., 2022, Ma et al., 2023; Wang et al., 2015a).
Patatins, a nonspecific lipid acyl hydrolase reported to play a role in plant signaling were only recently linked to cell wall metabolism, specifically lignin and cellulose biosynthesis (Huang et al., 2001; Jang and Lee, 2020; Li et al., 2011; Simiyu et al., 2023). These lipolytic enzymes, primarily known for their role in lipid metabolism, have an yet unclear link to lignin biosynthesis. However, substantial experimental evidence, including in poplar (Jang and Lee, 2020), is pointing to a connection between lipid metabolism and lignin biosynthesis pathway (Huang et al., 2001; Jang and Lee, 2020; Li et al., 2011; Simiyu et al., 2023). One compelling explanation presented by Ali et al. is that patatins play a pivotal role in regulating central carbon flux during cell wall biosynthesis (Ali et al., 2022). Interestingly, one of the first patatins identified was through activation tagging in Arabidopsis (Huang et al., 2001). The patatin gene was named STURDY, after the mutant’s tougher stems, resulting from changes in lignin biosynthesis (Huang et al., 2001).The mutant also displayed increased stem girth due to enhanced cell proliferation.
Additionally, our study uncovered a fascinating link between patatins and drought stress response, which was not observed in the original mutant but only when the gene was highly overexpressed. It is unclear if this is a pleiotropic effect of the ectopic expression or a result of the much higher expression levels than in the WT plants. Nevertheless, the evidence is pointing to a highly positive effect of patatin overexpression on drought resistance. We are still unclear of the underpinnings of this phenotype. One potential factor is the denser leaf structure, which may reduce transpiration levels, as suggested by our measurements of photosynthesis and stomatal conductance. Given that the patatins are lipolytic enzymes, their role in drought resistance might involve a wide array of changes, such as membrane organization, mobilization of storage reserves, and modification of the cell’s osmotic potential (Li et al., 2020; Scherer et al., 2010). Patatins are also induced by abiotic stress, suggesting they likely play a significant yet unknown role in stress responses (Li et al., 2020; Matos et al., 2001, Matos et al., 2008). Further investigations into this underlying mechanism could lead to new strategies for engineering stress response. Most excitingly, our work indicates a simultaneous increase in biomass yield and drought resistance, unlike many other strategies for engineering drought resistance that often result in growth penalties (Hwang et al., 2010; Schluepmann et al., 2012; Sreenivasulu et al., 2012).
Fascinatingly, the leaf surface of the patatin overexpression transgenics resembles the leaf morphology of potato leaves, where patatin is highly expressed, particularly in the tubers (Andrews et al., 1988; Rosahl et al., 1986). The change in leaf morphology and tissue organization are reminiscent of the mutant phenotypes associated with disruption in hormone metabolism and/or signaling, leading to imbalances in cell division, proliferation and differentiation. Some studies linked patatins to auxin, which could explain the significant growth/developmental phenotypes observed in our study (Dong et al., 2014; Labusch et al., 2013).
Our work demonstrates the feasibility and efficacy of activation tagging for discovering genes that positively influence biomass-associated traits in poplar. Recapitulation experiments indicate that greater improvements can be achieved through overexpression, with additional enhancements such as drought resistance identified, suggesting potential for simultaneously improving plant growth and resilience.
Methods
Plant transformation and validation
A hybrid aspen clone, Populus tremula x Populus alba INRA 717-IB4 (referred to as WT or 717), was used in all experiments, including the transgenic manipulations. The activation tagging population was generated using a binary vector pSKI074 via an Agrobacterium-mediated procedure (Han et al., 2000). All putative transformants were PCR verified for the presence of neomycin phosphotransferase II (NPT), as a selectable marker (Yordanov et al., 2010). Only the NPT-positive transformant were used in further experiments. These verified transformant were propagated and maintained in vitro on ½ MS media with 20 g/l sucrose (Caisson), 0.1mg/l IBA (Sigma-Aldrich), vitamins (Han et al., 2000) solidified with 2.5 g/l Gelrite (Sigma) and 4 g/l Phytablend agar (Caisson), at 16/8 h day/night photoperiod (20 µmol m-2s-1).
Plant growth conditions
For the greenhouse experiments, plants were first propagated and grown in vitro for four weeks on ½ MS solid media (as describe above). The rooted plantlets were then transferred to soil and gradually acclimated to greenhouse conditions. Once acclimated, uniformly developed plants were transplanted and grown in greenhouse for approximately 3 months as previously described (Rauschendorfer et al., 2020). The experiment was conducted in a completely randomized design with three replications.
Biometric measurements and harvesting
Measurements of the height and basal diameter of the stem, counting of the internodes of each plant were performed regularly to analyze the growth characteristics of poplar plants. A slide digital caliper was used to determine the basal diameter of each plant above the pot surface. For harvest the above-ground part of each plant was separated into leaves and stems. The fifth leaf from the top of the plant was used to determine the leaf area. Digital images were taken with Nicon Coolpix camera. Leaves and stems were air dried, and their dry weight was measured until was unchanged. We measured green wood density (Den-g, g/cm3), basic/dry wood density (Den-d, g/cm3), and moisture content (MC, %) of wood samples collected at the base (15 cm from the soil surface) of the stem. For each sample, we determine the green disc mass and green volume, using water displacement. The stem sections were kiln-dried at 105°C and again weighed. Den-g and Den-d were estimated for each tree as sample mass (g) / disc green volume (cm3).MC will be estimated as [(Den-g – Den-d) / Den-g] x 100.
Measurements of photosynthesis
Two-month-old greenhouse-grown plants subjected to drought stress, were used to measure photosynthesis. Net photosynthetic rate was measured using LI-6400XT portable photosynthesis system (Li-Cor Inc., Lincoln, NE, USA). The measurements were conducted during mid- and late- morning (usually 09:00 – 11:30am) on uniformly sunny days. Leaves of each genotype (four biological replicates/genotype) were measured under the following conditions: 400 μmol s -1 flow rate, 400 μmol mol -1 reference CO 2 concentration and photosynthetic photon flux density (PPFD) of 1500 μmol m -2 s -1, provided by a red–blue light source (6400-02B). Relative humidity was maintained between 50 % and 75 % inside the chamber (RH_S_%). The control temperature was set at 30°C.
Tag mapping and validation of gene activation
Recovery of sequence flanking the insertion site of the activation tag was performed as previously described (Yordanov et al., 2010). The isolated DNA fragments were positioned in the poplar 717 genome using BLAST searches in the Phythozome v13 (http://www.phythozome.net/poplar.php) database and proximal genes to the insertion site identified. Expression of the flanking genes was studied using RT-PCR with gene specific primers (Supplementary Table S2) and Ubiquitin (Ubq) gene (Wei et al., 2013) as a loading control.
Generation of constructs and transformation
The open reading frame of candidate genes were amplified using the following primers (Supplementary Table S3). The amplified product was then cloned into pDONR221 vector (ThermoFisher Scientific) using the BP Gateway system (Invitrogen), sequence validated and transferred into the overexpression pK7WG2 vector (Karimi et al., 2002) using the LR Gateway system (Invitrogen). The binary vectors were transformed into Agrobacterium strain AGL1 (Lazo et al., 1991) and transformed into the hybrid aspen clone, Populus tremula x Populus alba INRA 717-IB4 as previously described (Han et al., 2000).
Real time RT-PCR
Total RNA was extracted as previously described (Busov et al., 2003). Reverse transcription was performed on 1 μg of DNAase I-treated total RNA in a final reaction volume of 20 μl using an MMuLV (Moloney Murine Leukemia Virus) reverse transcriptase (ThermoFisher Scientific) following the manufacturer's protocol. Quantitative RT-PCR (qRT-PCR) was performed using the StepOnePlus Real Time System (Applied Biosystems) with the Maxima SYBR Green detection system (Thermo Fisher). Each PCR reaction contained 1× Maxima SYBR Green qPCR master mix, 0.1 μM of each forward and reverse primer (Eurofins MWG Operon), 1 μl of 10 × diluted cDNA solution and nuclease-free water. The final volume of each PCR reaction was 20 μl. The qRT-PCR cycling stages consisted of initial denaturation at 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min, and a final melting curve stage of 95°C for 15 s, 60°C for 1 min and 95°C for 15 s. qRT-PCR was performed with three biological and two technical replicates for each sample. Relative gene expression was calculated as previously described (Livak and Schmittgen, 2001; Schmittgen and Livak, 2008; Tsai et al., 2006; Yordanov et al., 2017). The Ubq gene expression was used as a loading control (Yordanov et al., 2010). All primers used in the gene expression analyses are shown in Supplementary Table S2. Primers were designed using the Primer-BLAST web resource at NCBI (National Center for Biotechnology Information; http://www.ncbi.nlm.nih.gov/BLAST).
Cell wall analyses
Wood samples were milled to a 20-mesh using a Wiley mill. Approximately 4 mg of milled wood sample was measured and loaded into metal cups, which were then placed into an auto-sampler tray. The cell wall composition was analyzed by studying pyrolysis vapors produced using a commercially available molecular beam mass spectrometer (PyMBMS) designed specifically for biomass analysis as previously described (Sykes et al., 2009).
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
TG: Conceptualization, Formal analysis, Investigation, Methodology, Writing – review & editing. YY: Formal analysis, Investigation, Methodology, Writing – review & editing. EY: Formal analysis, Investigation, Resources, Validation, Writing – review & editing. MK: Formal analysis, Validation, Visualization, Writing – review & editing. KL: Formal analysis, Writing – review & editing, Validation. VB: Conceptualization, Formal analysis, Funding acquisition, Project administration, Resources, Supervision, Validation, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was supported by grants from the Plant Feedstock Genomics for Bioenergy: a Joint Research Program of USDA and DOE (2009-65504-05767 and DE-SC0008462) and USDA McIntire Stennis Fund (Grant 1001498).
Acknowledgments
We thank Naomi Ojala and Galina Agapova for their technical help.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1468905/full#supplementary-material
References
Ali, U., Lu, S., Fadlalla, F., Iqbal, S., Yue, H., Yang, B., et al. (2022). The functions of phospholipases and their hydrolysis products in plant growth, development and stress responses. Prog. Lipid Res. 86, 101158. doi: 10.1016/j.plipres.2022.101158
Alonso, J. M., Stepanova, A. N., Leisse, T. J., Kim, C. J., Chen, H., Shinn, P., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657. doi: 10.1126/science.1086391
An, S., Park, S., Jeong, D. H., Lee, D. Y., Kang, H. G., Yu, J. H., et al. (2003). Generation and analysis of end sequence database for T-DNA tagging lines in rice. Plant Physiol. 133, 2040–2047. doi: 10.1104/pp.103.030478
Andrews, D. L., Beames, B., Summers, M. D., Park, W. D. (1988). Characterization of the lipid acyl hydrolase activity of the major potato (Solanum tuberosum) tuber protein, patatin, by cloning and abundant expression in a baculovirus vector. Biochem. J. 252, 199–206. doi: 10.1042/bj2520199
Azeez, A., Zhao, Y. C., Singh, R. K., Yordanov, Y. S., Dash, M., Miskolczi, P., et al. (2021). EARLY BUD-BREAK 1 and EARLY BUD-BREAK 3 control resumption of poplar growth after winter dormancy. Nat. Commun. 12, 1123. doi: 10.1038/s41467-021-21449-0
Busov, V., Fladung, M., Groover, A., Strauss, S. (2005a). Insertional mutagenesis in Populus: relevance and feasibility. Tree Genet. Genomes 1, 135–142. doi: 10.1007/s11295-005-0019-8
Busov, V., Yordanov, Y., Gou, J., Meilan, R., Ma, C., Regan, S., et al. (2010). Activation tagging is an effective gene tagging system in Populus Tree. Gen. Genom. doi: 10.1007/s11295-010-0317-7
Busov, V. B., Meilan, R., Pearce, D. W., Ma, C., Rood, S. B., Strauss, S. H. (2003). Activation tagging of a dominant gibberellin catabolism gene (GA 2-oxidase) from poplar that regulates tree stature. Plant Physiol. 132, 1283–1291. doi: 10.1104/pp.103.020354
Carpita, N. C., McCann, M. C. (2020). Redesigning plant cell walls for the biomass-based bioeconomy. J. Biol. Chem. 295, 15144–15157. doi: 10.1074/jbc.REV120.014561
Chen, S., Jin, W., Wang, M., Zhang, F., Zhou, J., Jia, Q., et al. (2003). Distribution and characterization of over 1000 T-DNA tags in rice genome. Plant journal: Cell Mol. Biol. 36, 105–113. doi: 10.1046/j.1365-313X.2003.01860.x
Deng, M., Wang, Y., Kuzma, M., Chalifoux, M., Tremblay, L., Yang, S., et al. (2020). Activation tagging identifies Arabidopsis transcription factor AtMYB68 for heat and drought tolerance at yield determining reproductive stages. Plant journal: Cell Mol. Biol. 104, 1535–1550. doi: 10.1111/tpj.15019
de Vries, L., Guevara-Rozo, S., Cho, M., Liu, L. Y., Renneckar, S., Mansfield, S. D. (2021). Tailoring renewable materials via plant biotechnology. Biotechnol. Biofuels 14, 167. doi: 10.1186/s13068-021-02010-z
Dharmawardhana, P., Brunner, A. M., Strauss, S. H. (2010). Genome-wide transcriptome analysis of the transition from primary to secondary stem development in Populus trichocarpa. BMC Genomics 11. doi: 10.1186/1471-2164-11-150
Dong, Y., Li, M., Zhang, P., Wang, X., Fan, C., Zhou, Y. (2014). Patatin-related phospholipase pPLAIIIdelta influences auxin-responsive cell morphology and organ size in Arabidopsis and Brassica napus. BMC Plant Biol. 14, 332. doi: 10.1186/s12870-014-0332-1
Forsbach, A., Schubert, D., Lechtenberg, B., Gils, M., Schmidt, R. (2003). A comprehensive characterization of single-copy T-DNA insertions in the Arabidopsis thaliana genome. Plant Mol. Biol. 52, 161–176. doi: 10.1023/A:1023929630687
Han, K. H., Meilan, R., Ma, C., Strauss, S. H. (2000). An Agrobacterium tumefaciens transformation protocol effective on a variety of cottonwood hybrids (genus Populus). Plant Cell Rep. 19, 315–320. doi: 10.1007/s002990050019
Hu, H., Zhang, R., Tao, Z., Li, X., Li, Y., Huang, J., et al. (2018). Cellulose synthase mutants distinctively affect cell growth and cell wall integrity for plant biomass production in arabidopsis. Plant Cell Physiol. 59, 1144–1157. doi: 10.1093/pcp/pcy050
Huang, S., Cerny, R. E., Bhat, D. S., Brown, S. M. (2001). Cloning of an Arabidopsis patatin-like gene, STURDY, by activation T-DNA tagging. Plant Physiol. 125, 573–584. doi: 10.1104/pp.125.2.573
Huang, G. Q., Gong, S. Y., Xu, W. L., Li, W., Li, P., Zhang, C. J., et al. (2013). A fasciclin-like arabinogalactan protein, GhFLA1, is involved in fiber initiation and elongation of cotton. Plant Physiol. 161, 1278–1290. doi: 10.1104/pp.112.203760
Hwang, S.-G., Chen, H.-C., Huang, W.-Y., Chu, Y.-C., Shii, C.-T., Cheng, W.-H. (2010). Ectopic expression of rice OsNCED3 in Arabidopsis increases ABA level and alters leaf morphology. Plant Sci. 178, 12–22. doi: 10.1016/j.plantsci.2009.09.014
Jang, J. H., Lee, O. R. (2020). Patatin-related phospholipase atpPLAIIIalpha affects lignification of xylem in arabidopsis and hybrid poplars. Plants (Basel) 9.
Janz, D., Lautner, S., Wildhagen, H., Behnke, K., Schnitzler, J. P., Rennenberg, H., et al. (2012). Salt stress induces the formation of a novel type of pressure wood’ in two Populus species. New Phytol. 194, 129–141. doi: 10.1111/j.1469-8137.2011.03975.x
Karimi, M., Inze, D., Depicker, A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7, 193–195. doi: 10.1016/S1360-1385(02)02251-3
Kim, S. I., Veena, S. B. (2007). Genome-wide analysis of Agrobacterium T-DNA integration sites in the Arabidopsis genome generated under non-selective conditions. Plant J. 51, 779–791. doi: 10.1111/j.1365-313X.2007.03183.x
Labusch, C., Shishova, M., Effendi, Y., Li, M., Wang, X., Scherer, G. F. (2013). Patterns and timing in expression of early auxin-induced genes imply involvement of phospholipases A (pPLAs) in the regulation of auxin responses. Mol. Plant 6, 1473–1486. doi: 10.1093/mp/sst053
Lafarguette, F., Leple, J. C., Dejardin, A., Laurans, F., Costa, G., Lesage-Descauses, M. C., et al. (2004). Poplar genes encoding fasciclin-like arabinogalactan proteins are highly expressed in tension wood. New Phytol. 164, 107–121. doi: 10.1111/j.1469-8137.2004.01175.x
Lazo, G. R., Stein, P. A., Ludwig, R. A. (1991). A dna transformation-competent arabidopsis genomic library in agrobacterium. Bio-Technol 9, 963–967. doi: 10.1038/nbt1091-963
Li, M., Bahn, S. C., Guo, L., Musgrave, W., Berg, H., Welti, R., et al. (2011). Patatin-related phospholipase pPLAIIIbeta-induced changes in lipid metabolism alter cellulose content and cell elongation in Arabidopsis. Plant Cell 23, 1107–1123. doi: 10.1105/tpc.110.081240
Li, J., Li, M., Yao, S., Cai, G., Wang, X. (2020). Patatin-related phospholipase pPLAIIIgamma involved in osmotic and salt tolerance in arabidopsis. Plants (Basel) 9.
Li, W., Lin, Y. J., Chen, Y. L., Zhou, C., Li, S., De Ridder, N., et al. (2024). Woody plant cell walls: Fundamentals and utilization. Mol. Plant 17, 112–140. doi: 10.1016/j.molp.2023.12.008
Liu, Y. G., Mitsukawa, N., Oosumi, T., Whittier, R. F. (1995). Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J. 8, 457–463. doi: 10.1046/j.1365-313X.1995.08030457.x
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Ma, Y., MacMillan, C. P., de Vries, L., Mansfield, S. D., Hao, P., Ratcliffe, J., et al. (2022). FLA11 and FLA12 glycoproteins fine-tune stem secondary wall properties in response to mechanical stresses. New Phytol. 233, 1750–1767. doi: 10.1111/nph.17898
Ma, Y., Shafee, T., Mudiyanselage, A. M., Ratcliffe, J., MacMillan, C. P., Mansfield, S. D., et al. (2023). Distinct functions of FASCILIN-LIKE ARABINOGALACTAN PROTEINS relate to domain structure. Plant Physiol. 192, 119–132. doi: 10.1093/plphys/kiad097
MacMillan, C. P., Mansfield, S. D., Stachurski, Z. H., Evans, R., Southerton, S. G. (2010). Fasciclin-like arabinogalactan proteins: specialization for stem biomechanics and cell wall architecture in Arabidopsis and Eucalyptus. Plant journal: Cell Mol. Biol. 62, 689–703. doi: 10.1111/tpj.2010.62.issue-4
Maleki, S. S., Mohammadi, K., Movahedi, A., Wu, F., Ji, K. S. (2020). Increase in cell wall thickening and biomass production by overexpression of pmCesA2 in poplar. Front. Plant Sci. 11, 110. doi: 10.3389/fpls.2020.00110
Matos, A. R., d’Arcy-Lameta, A., Franca, M., Petres, S., Edelman, L., Kader, J., et al. (2001). A novel patatin-like gene stimulated by drought stress encodes a galactolipid acyl hydrolase. FEBS Lett. 491, 188–192. doi: 10.1016/S0014-5793(01)02194-9
Matos, A. R., Gigon, A., Laffray, D., Petres, S., Zuily-Fodil, Y., Pham-Thi, A. T. (2008). Effects of progressive drought stress on the expression of patatin-like lipid acyl hydrolase genes in Arabidopsis leaves. Physiol. Plant 134, 110–120. doi: 10.1111/j.1399-3054.2008.01123.x
Nakazawa, M., Ichikawa, T., Ishikawa, A., Kobayashi, H., Tsuhara, Y., Kawashima, M., et al. (2003). Activation tagging, a novel tool to dissect the functions of a gene family. Plant J. 34, 741–750. doi: 10.1046/j.1365-313X.2003.01758.x
Palatnik, J. F., Allen, E., Wu, X., Schommer, C., Schwab, R., Carrington, J. C., et al. (2003). Control of leaf morphogenesis by microRNAs. Nature 425, 257–263. doi: 10.1038/nature01958
Park, Y. W., Tominaga, R., Sugiyama, J., Furuta, Y., Tanimoto, E., Samejima, M., et al. (2003). Enhancement of growth by expression of poplar cellulase in Arabidopsis thaliana. Plant J. 33, 1099–1106. doi: 10.1046/j.1365-313X.2003.01696.x
Perlack, R. D., Wright, L. L., Turhollow, A. F., Graham, R. L. (2005). Biomass as feedstock for bioenergy and bioproducts industry: the technical feasbility of a billion-ton supply (Oak Ridge, TN: Oak Ridge National Laboratory).
Rauschendorfer, J., Yordanov, Y., Dobrev, P., Vankova, R., Sykes, R., Kulheim, C., et al. (2020). Overexpression of a developing xylem cDNA library in transgenic poplar generates high mutation rate specific to wood formation. Plant Biotechnol. J. 18, 1434–1443. doi: 10.1111/pbi.13309
Rosahl, S., Schmidt, R., Schell, J., Willmitzer, L. (1986). Isolation and characterization of a gene from solanum-tuberosum encoding patatin, the major storage protein of potato-tubers. Mol. Gen. Genet. 203, 214–220. doi: 10.1007/BF00333957
Rosso, M. G., Li, Y., Strizhov, N., Reiss, B., Dekker, K., Weisshaar, B. (2003). An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol. Biol. 53, 247–259. doi: 10.1023/B:PLAN.0000009297.37235.4a
Sallaud, C., Gay, C., Larmande, P., Bes, M., Piffanelli, P., Piegu, B., et al. (2004). High throughput T-DNA insertion mutagenesis in rice: a first step towards in silico reverse genetics. Plant journal: Cell Mol. Biol. 39, 450–464. doi: 10.1111/j.1365-313X.2004.02145.x
Scherer, G. F., Ryu, S. B., Wang, X., Matos, A. R., Heitz, T. (2010). Patatin-related phospholipase A: nomenclature, subfamilies and functions in plants. Trends Plant Sci. 15, 693–700. doi: 10.1016/j.tplants.2010.09.005
Schluepmann, H., Berke, L., Sanchez-Perez, G. F. (2012). Metabolism control over growth: a case for trehalose-6-phosphate in plants. J. Exp. Bot. 63, 3379–3390. doi: 10.1093/jxb/err311
Schmittgen, T. D., Livak, K. J. (2008). Analyzing real-time PCR data by the comparative C-T method. Nat. Protoc. 3, 1101–1108. doi: 10.1038/nprot.2008.73
Shani, Z., Dekel, M., Tsabary, G., Goren, R., Shoseyov, O. (2004). Growth enhancement of transgenic poplar plants by overexpression of Arabidopsis thaliana endo-1,4-0-glucanase (cel1). Mol. Breed. 14, 321–330. doi: 10.1023/B:MOLB.0000049213.15952.8a
Simiyu, D. C., Jang, J. H., Lee, O. R. (2023). A group III patatin-like phospholipase gene pPLAIIIδ regulates lignin biosynthesis and influences the rate of seed germination in Arabidopsis thaliana. Front. Plant Sci. 14, 1212979. doi: 10.3389/fpls.2023.1212979
Skog, K. E. (2008). Sequestration of carbon in harvested wood products for the United States. For. Prod J. 58, 56–72.
Sreenivasulu, N., Harshavardhan, V. T., Govind, G., Seiler, C., Kohli, A. (2012). Contrapuntal role of ABA: does it mediate stress tolerance or plant growth retardation under long-term drought stress? Gene 506, 265–273. doi: 10.1016/j.gene.2012.06.076
Sykes, R., Yung, M., Novaes, E., Kirst, M., Peter, G., Davis, M. (2009). High-throughput screening of plant cell-wall composition using pyrolysis molecular beam mass spectroscopy. Methods Mol. Biol. 581, 169–183.
Szabados, L., Kovacs, I., Oberschall, A., Abraham, E., Kerekes, I., Zsigmond, L., et al. (2002). Distribution of 1000 sequenced T-DNA tags in the Arabidopsis genome. Plant J. 32, 233–242. doi: 10.1046/j.1365-313X.2002.01417.x
Trupiano, D., Yordanov, Y., Regan, S., Meilan, R., Tschaplinski, T., Scippa, G. S., et al. (2013). Identification, characterization of an AP2/ERF transcription factor that promotes adventitious, lateral root formation in Populus. Planta 238, 271–282. doi: 10.1007/s00425-013-1890-4
Tsai, C. J., Harding, S. A., Tschaplinski, T. J., Lindroth, R. L., Yuan, Y. N. (2006). Genome-wide analysis of the structural genes regulating defense phenylpropanoid metabolism in Populus. New Phytol. 172, 47–62. doi: 10.1111/j.1469-8137.2006.01798.x
Tuskan, G. A., Difazio, S., Jansson, S., Bohlmann, J., Grigoriev, I., Hellsten, U., et al. (2006). The genome of black cottonwood, Populus trichocarpa (Torr. Gray). Science. 313, 1596–1604. doi: 10.1126/science.1128691
Wang, H., Jiang, C., Wang, C., Yang, Y., Yang, L., Gao, X., et al. (2015a). Antisense expression of the fasciclin-like arabinogalactan protein FLA6 gene in Populus inhibits expression of its homologous genes and alters stem biomechanics and cell wall composition in transgenic trees. J. Exp. Bot. 66, 1291–1302. doi: 10.1093/jxb/eru479
Wang, H., Jin, Y., Wang, C., Li, B., Jiang, C., Sun, Z., et al. (2017). Fasciclin-like arabinogalactan proteins, PtFLAs, play important roles in GA-mediated tension wood formation in Populus. Sci. Rep. 7, 6182. doi: 10.1038/s41598-017-06473-9
Wei, H. R., Yordanov, Y. S., Georgieva, T., Li, X., Busov, V. (2013). Nitrogen deprivation promotes Populus root growth through global transcriptome reprogramming and activation of hierarchical genetic networks. New Phytol. 200, 483–497. doi: 10.1111/nph.12375
Yordanov, Y. S., Ma, C., Strauss, S. H., Busov, V. B. (2014). EARLY BUD-BREAK 1 (EBB1) is a regulator of release from seasonal dormancy in poplar trees. Proc. Natl. Acad. Sci. United States America 111, 10001–10006.
Yordanov, Y. S., Ma, C., Yordanova, E., Meilan, R., Strauss, S. H., Busov, V. B. (2017). BIG LEAF is a regulator of organ size and adventitious root formation in poplar. PloS One 12, e0180527. doi: 10.1371/journal.pone.0180527
Yordanov, Y., Regan, S., Busov, V. (2010). Members of the lateral organ boundaries domain (LBD) transcription factors family are involved in regulation of secondary growth in populus the plant cell 22, 3662–3677. doi: 10.1105/tpc.110.078634
Zhen, C., Hua, X., Jiang, X., Tong, G., Li, C., Yang, C., et al. (2023). Cas9/gRNA-mediated mutations in ptrFLA40 and ptrFLA45 reveal redundant roles in modulating wood cell size and SCW synthesis in poplar. Int. J. Mol. Sci. 24, 427. doi: 10.3390/ijms24010427
Keywords: activation tag, poplar, fasciclin like gene, patatin, drought stress, gene discovery tools
Citation: Georgieva T, Yordanov Y, Yordanova E, Khan MRI, Lyu K and Busov V (2024) Discovery of genes that positively affect biomass and stress associated traits in poplar. Front. Plant Sci. 15:1468905. doi: 10.3389/fpls.2024.1468905
Received: 22 July 2024; Accepted: 30 September 2024;
Published: 18 October 2024.
Edited by:
Shun Sakuma, Tottori University, JapanReviewed by:
Glenn Thorlby, New Zealand Forest Research Institute Limited (Scion), New ZealandHeather D. Coleman, Syracuse University, United States
Joseph Colbert, Syracuse University, United States, in collaboration with reviewer HC, Syracuse
Copyright © 2024 Georgieva, Yordanov, Yordanova, Khan, Lyu and Busov. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Victor Busov, dmJ1c292QG10dS5lZHU=
 Tatyana Georgieva1
Tatyana Georgieva1 Md Rezaul Islam Khan
Md Rezaul Islam Khan Victor Busov
Victor Busov