
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 30 October 2024
Sec. Plant Symbiotic Interactions
Volume 15 - 2024 | https://doi.org/10.3389/fpls.2024.1468812
This article is part of the Research Topic Highlights of iMMM2023 - International Molecular Mycorrhiza Meeting View all 8 articles
Introduction: Plants strictly control root endosymbioses with nutrient-scavenging arbuscular endomycorrhizal fungi or nodule inducing diazotrophic bacteria. The GRAS-type transcriptional regulator NODULATION SIGNALING PATHWAY 2 (NSP2) is a conserved hub in this process. The NSP2-regulated transcriptional network is instrumental in balancing nutrient homeostasis with symbiotic interactions. NSP2 activity is modulated post-transcriptionally by a specific microRNA. Overriding this control mechanism by ectopic expression of a miRNA-resistant NSP2 transgene enhances the symbiotic permissiveness to arbuscular endomycorrhizal fungi. Such engineered plants may possess enhanced capacities for nutrient uptake. However, the trade-off of this strategy on plant development or other symbiotic interactions, like nodulation, is yet to be fully understood.
Method: We used the nodulating Cannabaceae species Parasponia andersonii as an experimental system to study the effect of ectopic NSP2 expression. Parasponia and legumes (Fabaceae) diverged 100 million years ago, providing a unique comparative system to dissect the nodulation trait.
Results: Six independent transgenic Parasponia lines were generated that differed in the level of NSP2 expression in the root from 6 to 95-fold higher when compared to the empty vector control plants. Analysis of these plants revealed a positive correlation between mycorrhization and the NSP2 expression level, as well as with the expression of the symbiosis transcription factor CYCLOPS and the rate-limiting enzyme in the carotenoid biosynthetic pathway PHYTOENE SYNTHASE1 (PSY1). Yet ectopic expression of NSP2 affected plant architecture and root nodule organogenesis.
Discussion: This indicates a significant trade-off when leveraging NSP2 over-expression to enhance endomycorrhization.
Plants explore mutualistic relationships with soil microorganisms to enhance access to essential nutrients. The endosymbiotic interactions with arbuscular mycorrhizal fungi and nodulating nitrogen-fixing bacteria like rhizobia are the most advanced examples of such ecosystem services. As these microbes are engulfed in plant cells, they largely depend on the plant’s supply of carbon sources. These intimate endosymbiotic associations necessitate stringent regulation to prevent the microbes from exploiting the plant, especially when excess exogenous nutrients are in the soil.
The GRAS-type transcriptional regulator NODULATION SIGNALING PATHWAY2 (NSP2) acts as a central hub integrating the plant’s nutrient homeostasis and symbiotic engagement to orchestrate adaptive responses (Quilbé and Arrighi, 2021). NSP2 is first identified in legumes, where it is essential for rhizobium-induced nodule formation (Oldroyd and Long, 2003; Kaló et al., 2005; Heckmann et al., 2006; Ali et al., 2014; Shtark et al., 2016; Peng et al., 2021). Subsequent genome studies revealed the gene is present in many plant species and predates the evolution of angiosperms (Lauressergues et al., 2012; Cenci and Rouard, 2017). NSP2 of different species is, to a large extent, functionally conserved, as was shown in trans-complementation studies of a Lotus japonicus nsp2 nodulation mutant using OsNSP2 of rice (Oryza sativa) (Heckmann et al., 2006). In legumes and non-legumes, NSP2 also controls mycorrhizal infection levels (Lauressergues et al., 2012; Shtark et al., 2016; Li et al., 2022). Studies in barley (Hordeum vulgare) suggest that NSP2 may do so by regulating the expression of several common symbiosis signaling genes, including the transcription factor HvCYCLOPS (Li et al., 2022).
NSP2 also acts as a transcriptional regulator of the strigolactone biosynthesis pathway in a nutrient status-dependent manner (Liu et al., 2011; Li et al., 2022). Strigolactones act in planta and ex planta. In planta, strigolactones function as developmental hormones and control axillary bud and lateral root outgrowth (Ruyter-Spira et al., 2011; Brewer et al., 2015; Marzec and Melzer, 2018). Upon phosphate starvation, plants exude strigolactones into their rhizosphere, affecting the microbial community composition, including the infectiveness of arbuscular mycorrhizal fungi (Schlemper et al., 2017; Kobae et al., 2018; Carvalhais et al., 2019). Together, these findings highlight the complex regulatory role of NSP2 as a signaling hub.
NSP2 interacts with a suite of transcription factors to control the expression of target genes. Studies in heterologous systems showed it interacts with other GRAS-type transcriptional regulators like NSP1, DELLA, REQUIRED FOR ARBUSCULAR MYCORRHIZATION1 (RAM1), and REQUIRED FOR ARBUSCULAR DEVELOPMENT1 (RAD1), and the MYB-type transcription factor INTERACTING PROTEIN OF NSP2 (IPN2) to control nodulation and/or mycorrhization (Hirsch et al., 2009; Gobbato et al., 2012; Park et al., 2015; Xue et al., 2015; Fonouni-Farde et al., 2016; Heck et al., 2016; Jin et al., 2016; Pimprikar et al., 2016; Xiao et al., 2020). These transcriptional modules are regulated in a complex manner. Studies in rice showed that NSP2 expression is controlled by the MYB transcription factor PHOSPHATE STARVATION RESPONSE 2 (OsPHR2), in response to phosphate starvation (Das et al., 2022; Yuan et al., 2023). Additionally, NSP2 translation is controlled post-transcriptionally by microRNA miR171h (Lauressergues et al., 2012; Hofferek et al., 2014). Transcript levels of miR171h correlate with available phosphate concentration, suggesting that NSP2 is controlled transcriptionally and post-transcriptionally in a nutrient-dependent manner (Hofferek et al., 2014).
To enhance sustainable agricultural productivity, reducing reliance on inorganic fertilizers is essential. This can be achieved by better utilizing the arbuscular mycorrhizal symbiosis. The ectopic expression of NSP2 improves mycorrhization under exogenous phosphate concentrations that generally inhibit the symbiosis, suggesting it represents a potential biotechnological strategy to enhance nutrient uptake by the plant (Li et al., 2022; Isidra-Arellano et al., 2024; Yuan et al., 2023). However, at this stage, it remains elusive what the trade-off of such a strategy will be on plant development or other symbiotic interactions, like nodulation.
Studies in the legume models showed that NSP2 overexpression had no effect on nodulation (Medicago truncatula) or resulted in clustering of nodules on the root though not affecting the total number of nodules (L. japonicus) (Lauressergues et al., 2012; Murakami et al., 2013). We used the nodulating species Parasponia andersonii (Parasponia) as a comparative experimental system complementary to legumes to study the effect of ectopic NSP2 expression. The nodulation trait in Parasponia (Cannabaceae) and legumes (Fabaceae) share an evolutionary origin but diverged soon after (~100 million years ago) (van Velzen et al., 2019). Parasponia nodules have a different ontogeny when compared to legumes (Behm et al., 2014) Nevertheless, the same core set of symbiotic genes is used to establish nitrogen-fixing root nodules, including NSP2 (van Velzen et al., 2018; van Zeijl et al., 2018; Bu et al., 2020; Rutten et al., 2020; Alhusayni et al., 2024). Parasponia is amenable for efficient Agrobacterium tumefaciens-mediated transformation, giving T0 transgenic plantlets within ~3 months that can subsequently be propagated vegetatively (Wardhani et al., 2019). We generated independent transgenic Parasponia lines that differ in the level of NSP2 expression from 6 up to 95-fold compared to the empty vector control plants. Analysis of these plants revealed a correlation between mycorrhization and the level of expression of NSP2, the transcription factor CYCLOPS, and all genes in the biosynthetic pathways required for the formation of strigolactones. Yet ectopic expression of NSP2 affected plant shoot development and root nodule organogenesis.
Parasponia PanNSP2 was studied previously, revealing it represents a single-copy gene essential for rhizobium-induced nodulation (van Zeijl et al., 2018). To enable ectopic expression of a stable PanNSP2 transcript, we first identified the putative miR171h target site (Lauressergues et al., 2012). Such a putative site was identified in the coding region and subsequently removed by introducing synonymous DNA mutations (Supplementary Figure S1A). An additional alteration was included in the PanNSP2 coding region, which may enhance expression stability; the insertion of an intron (Supplementary Figure S1B) (Feike et al., 2019). Ultimately, two miR171h resistant PanNSP2 versions (mNSP2) were used for Parasponia transformation, a construct with and one without an engineered intron, both driven by the constitutive L. japonicus UBIQUITIN1 promoter (pLjUBQ1) (Supplementary Figure S1B) (Maekawa et al., 2008).
Six transgenic Parasponia lines were selected, displaying a gradient level of PanNSP2 ectopic expression ranging from a 6 to 95-fold increase in root tissue (Figure 1A). In the shoots of these transgenic lines, mNSP2 was also expressed; in tissues where PanNSP2 transcripts are normally not detected (Supplementary Figure S2A). Noteworthy, lines possessing the mNSP2intron construct generally have a lower transgene expression when compared to constructs possessing mNSP2 with only an adapted miR171h target site.
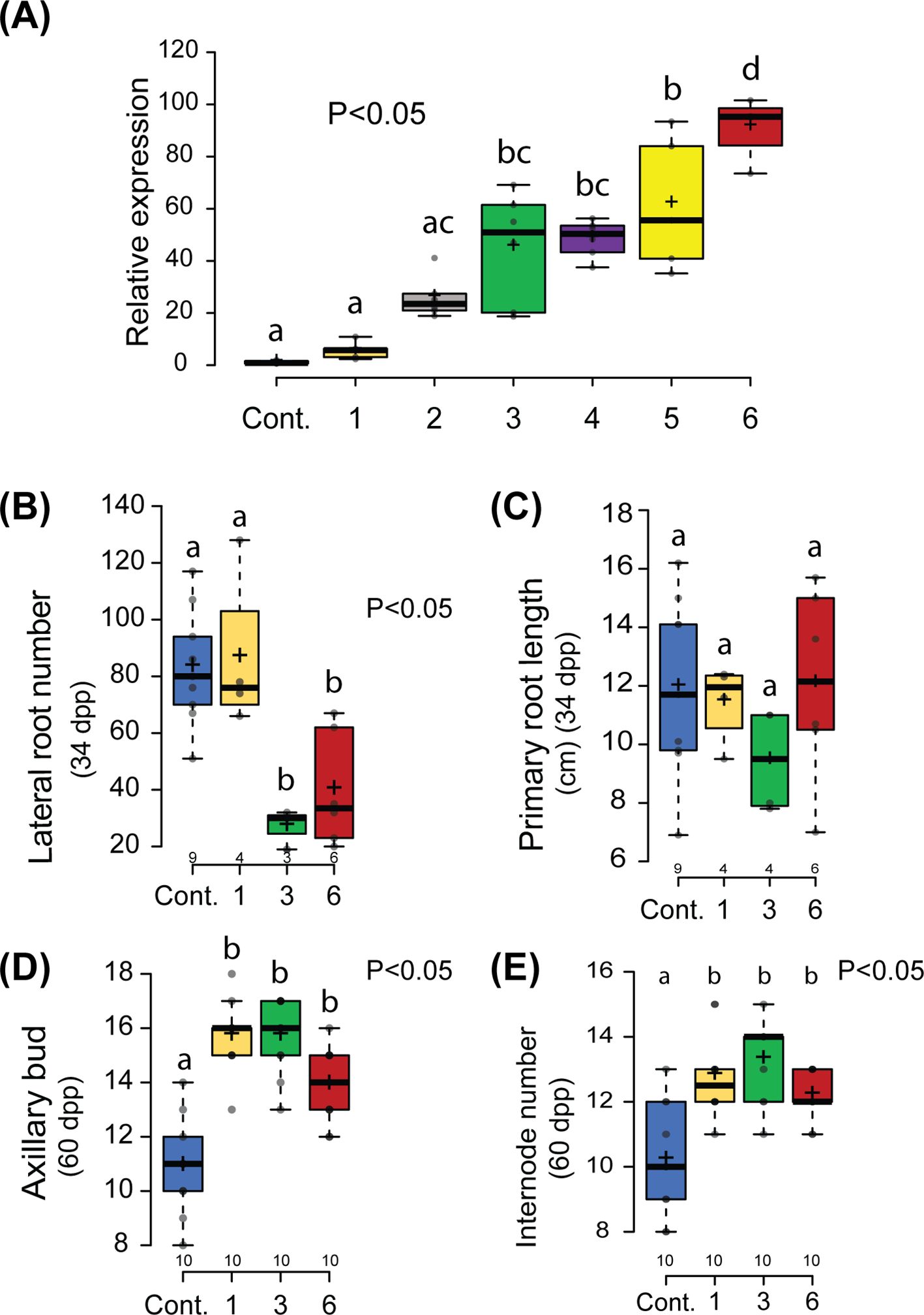
Figure 1. Non-symbiotic phenotype of Parasponia mNSP2ox lines. (A) Relative expression of NSP2 in root tissue of 6 independent lines compared to an empty vector control line (Cont.). mNSP2ox lines 1 to 3 contain the transgene construct pLjUBQ1:mPanNSP2intron, whereas lines 4 to 6 carry pLjUBQ1:mNSP2. Expression was measured using qPCR (n=3). (B, C) Quantification of the number of lateral roots (B) and primary root length (C) of control and transgenic lines 1,3, and 6 in vitro grown 34 days post planting (dpp). (D, E) Quantification of number of lateral shoot branching (D) and internode (E) of mNSP2ox lines 1, 3, and 6 at 60 days post planting (dpp). Different letters indicate significant differences (p<0.05) between these lines as determined by One-way ANOVA in combination with Tukey’s post hoc test). All data are displayed in box plots, showing data points, the median, and the interquartile range (IQR).
To analyze whether morphogenic phenotypes are associated with mNSP2 ectopic expression in Parasponia, we selected lines 1, 3, and 6 that possess a 6-, 51-, and 95-fold enhanced NSP2 expression in root tissue compared to empty vector control plants. First, the root systems of 34-day-old plantlets grown in vitro on EKM medium containing low levels of ammonium nitrate (0.375 mM) and high levels of phosphate (3 mM) were analyzed. Control plants transformed with an empty vector showed a highly branched root system with up to 80 secondary lateral roots. This contrasts with what was observed in mNSP2ox lines 3 and 6, in which the root system was less branched, with only ~30 secondary lateral roots, whereas the primary root length is unaffected (Figures 1B, C). Such root phenotype was not observed on mNSP2ox line 1. As this line has only moderately levels of transgene expression compared to lines 3 and 6, it suggests that repression of lateral root growth has a threshold of PanNSP2 expression. Next, we analyzed the shoot architecture of 60-day-old plantlets grown on potting soil. In the case of shoot branching, control plants transformed with an empty vector exhibited an average of 11 branches. In contrast, mNSP2ox lines showed increased shoot branching, with an average of 14 to 16 branches, respectively (Figure 1D). This increased number of lateral branches is associated with an increased number of internodes (Figure 1E). This phenotype is also observed in mNSP2ox line 1, indicating that NSP2-controlled developmental effects on plant architecture in root and shoot might have a different threshold. Quantification of internode diameter suggests that ectopic PanNSP2 expression may also increase stem thickness, even though the trees were all similar in size (Supplementary Figure S3). Taken together, the data show that ectopic mNSP2 expression affects shoot and root development in Parasponia.
Over-expression of an NSP2 miR171h resistant allele in M. truncatula roots enhances arbuscular mycorrhizal colonization under phosphate-limited conditions (Lauressergues et al., 2012). Furthermore, a more recent study showed that in M. truncatula and barley miR171h resistant NSP2 over-expression promotes arbuscular mycorrhizal root colonization even under exogenous phosphate conditions that are less amenable for the mycorrhizal symbiosis. This response is associated with the up-regulation of the strigolactone biosynthesis pathway (Li et al., 2022). We questioned whether Parasponia mNSP2ox lines also possess enhanced mycorrhization colonization levels. We tested two phosphate conditions; 20 μM and 3 mM exogenous Pi, and inoculated all six mNSP2ox lines with Rhizophagus irregularis DOAM197198 spores. Mycorrhization levels were scored 6 weeks post inoculation. Empty vector control plants can be colonized under both conditions, though the high exogenous phosphate concentration affects arbuscular mycorrhizal infection levels, most notably the number of vesicles formed. In the mNSP2ox lines, the number of hyphae, arbuscules, and vesicles was significantly increased under both phosphate conditions when compared to the empty vector control line (Figure 2). At high exogenous phosphate conditions, the level of mycorrhizal colonization is associated with the level of PanNSP2 expression. Thus, we conclude that ectopic expression of PanNSP2 in Parasponia promotes mycorrhization, which is consistent with reported data in M. truncatula and barley.
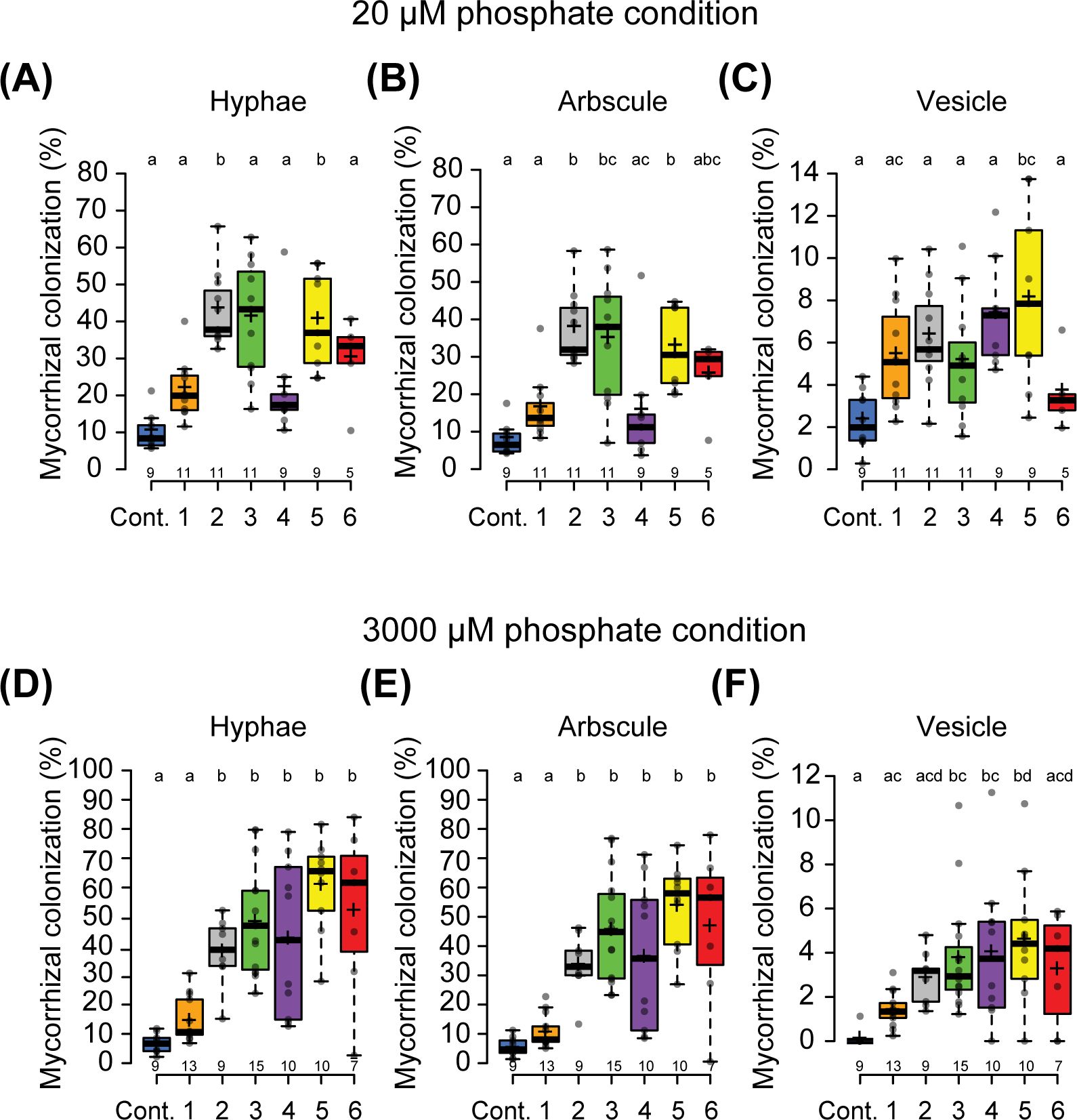
Figure 2. Ectopic expression of PanNSP2 in Parasponia enhances mycorrhizal colonization. Mycorrhizal colonization grown at low exogenous phosphate condition (20 µM PO43-) (A–C) and high exogenous phosphate condition (3000 µM PO43-) (D–F). Parasponia lines 1 to 3 are transformed with pLjUBQ1:mPanNSP2intron construct, whereas lines 4 to 6 NSP2 contain pLjUBQ1:mPanNSP2. Different letters indicate significant differences (p<0.05) between these lines as determined by One-way ANOVA followed by a Tukey’s post hoc test. Plants were harvested 6 weeks post-inoculation (6 wpi) with 300 spores/plant. All data are displayed in box plots, showing data points, the median, and the interquartile range (IQR).
To identify genes that are potentially directly or indirectly regulated by NSP2 in Parasponia, we investigated the transcriptional effect of increased mNSP2 expression under two distinct nutrient conditions. First plants were grown for 6 weeks on nutrient-rich medium containing 24.72 mM NO₃⁻, 2.6 mM NH₄+, and 2.6 mM PO43- and subsequently transferred for 3 weeks to nutrient starved medium (no nitrogen (N) or phosphate (P) source). This treatment will enhance the mycorrhizal responsiveness of the plants. In the second treatment, the plants were transferred from the nutrient-rich medium to nodulation medium, which contains high phosphate levels (0.375 mM NH4NO3 and 3 mM PO43-) (see Materials & Methods). The Pearson correlation test revealed that under nodulation permissive conditions, 2.73% (873 genes) exhibited a positive correlation with NSP2 expression, while only 0.95% (305 genes) showed a negative correlation (p-value, BH < 0.05). Conversely, under nutrient starved conditions, 1.70% (525 genes) were positively correlated with NSP2, and 1.74% (536 genes) were negatively correlated (Supplementary Table S1). Most genes displayed a non-significant correlation with NSP2 under either condition (Figure 3A).
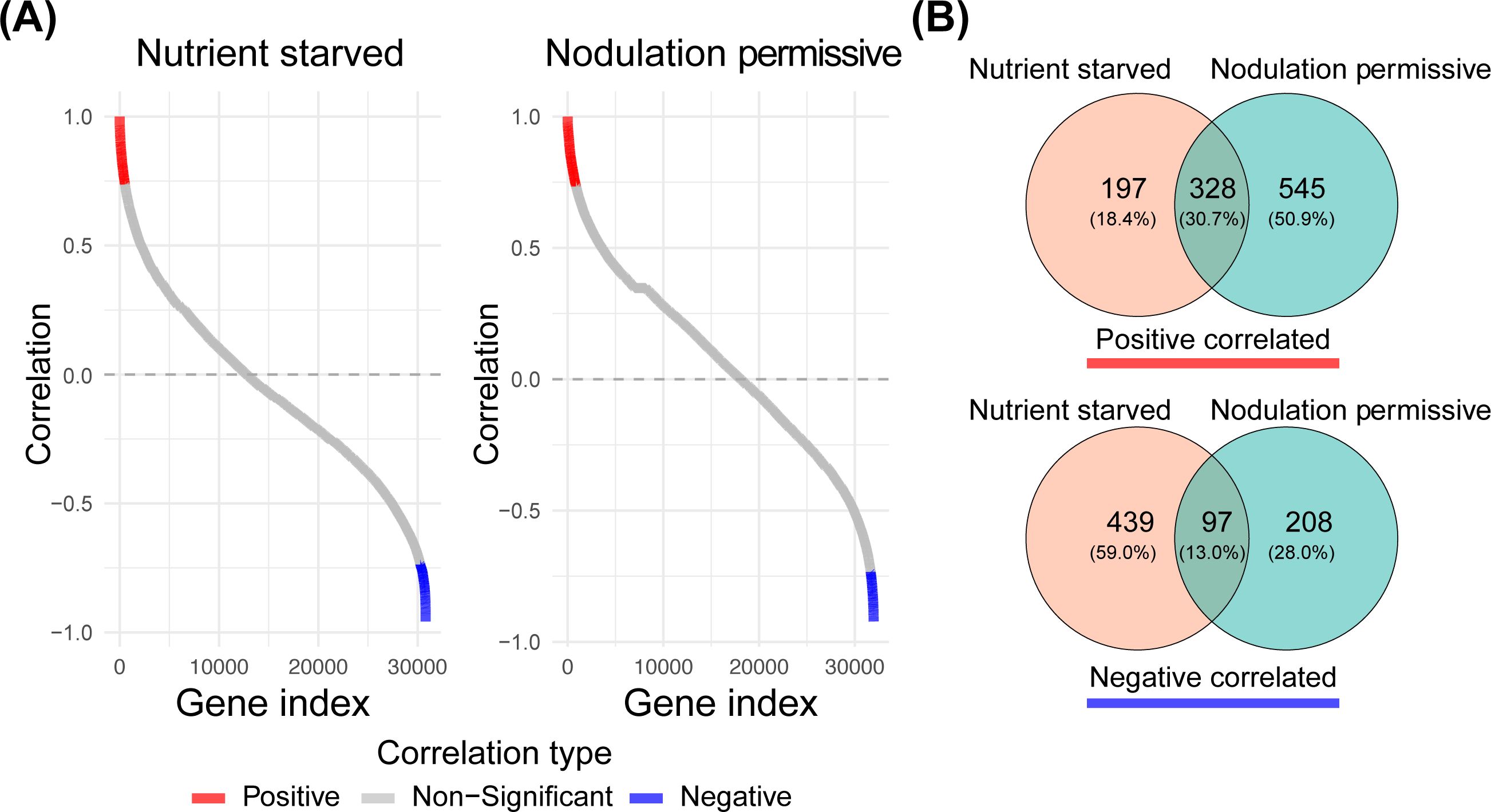
Figure 3. Pearson correlation of all Parasponia genes versus NSP2 profiles under arbuscular mycorrhizal and nodulation permissive nutrient conditions. (A) Pearson correlation coefficients are sorted in ascending order. Genes that are significantly positively correlated are colored in red, while negatively correlated genes are colored in blue. (Benjamini-Hochberg adjusted p-value, BH < 0.05) (B) Venn diagrams illustrate the overlap between genes that positively and negatively correlate with NSP2 expression for the two nutrient conditions.
We performed a hypergeometric test on the overlap between the positive and negative correlated gene sets under both nutrient conditions. The results indicated a p-value of 0 for the overlap in positively correlated genes and a p-value of 1.250571e-96 for the negatively correlated gene set. These findings led us to conclude that NSP2 consistently regulates a core set of genes, regardless of nutrient conditions, with a subset of genes showing differential regulation attributable to the nutrient environment (Figure 3B).
Studies in M. truncatula, rice, and barley suggest that ectopic expression of NSP2 enhances the colonization of plant roots by arbuscular mycorrhizal fungi, potentially through the modulation of the strigolactone biosynthetic pathway (Liu et al., 2011; Li et al., 2022). We investigated the activation of the methylerythritol phosphate (MEP), carotenoid, strigolactone, zaxinone, and abscisic acid (ABA) biosynthetic pathways in roots of Parasponia mNSP2ox lines (Figure 4A; Supplementary Table S1). This revealed a positive correlation between the expression of NSP2 and all genes encoding enzymes in the carotenoid, strigolactone, and zaxinone pathways (Figure 4B; Supplementary Table S1). The encoded enzymes of these genes convert geranylgeranyl pyrophosphate at the start of the carotenoid pathway into strigolactones (Okada et al., 2000; Ablazov et al., 2023). In addition, the expression levels of genes required for the biosynthesis of the apocarotenoid zaxinone correlate with the NSP2 expression. Zaxinone may act in a positive feedforward loop towards strigolactone biosynthesis, as was shown in rice (Votta et al., 2022). Taken together it is most probable that NSP2 ectopic expression leads to an increased biosynthesis of strigolactones in Parasponia root tissue.
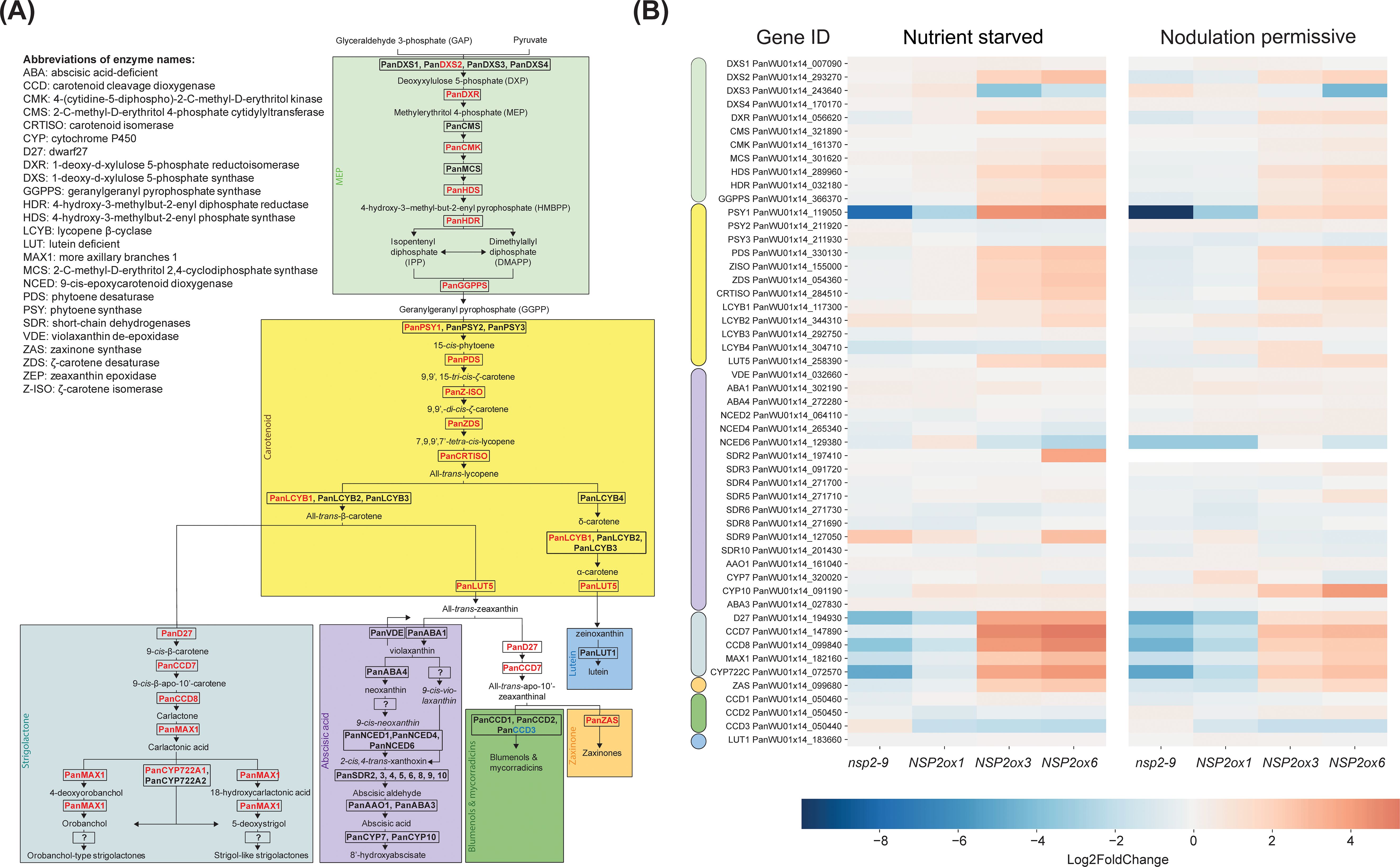
Figure 4. Schematic representation of the carotenoid biosynthetic pathway in Parasponia and heatmap of differential gene expression in root tissue in Parasponia under nutrient starved mycorrhiza and nodulation permissive conditions. (A) Parasponia genes found to be positively correlated with NSP2 over-expression level are colored in red, while non correlating genes are colored in black. The MEP biosynthetic pathway is shaded with green, the carotenoid biosynthetic pathway is shaded with yellow, the strigolactones biosynthetic pathway is shaded cyan, the ABA biosynthesis pathway is shaded with magenta, the zaxinone biosynthesis is shaded orange, the blumenol and Mycorradicins biosynthesis is shaded green, and the Lutein biosynthesis is shaded light blue. Enzyme names are based on annotations in Arabidopsis thaliana and Parasponia andersonii. Biosynthetic pathway is based on Wakabayashi et al., 2019; Wakabayashi et al., 2020; and Li et al., 2022. (B) Heatmap of gene expression in the carotenoid pathway across different lines: Pannsp2-9 knockout mutant, mNSP2ox1, mNSP2ox3, and mNSP2ox6 ectopic expression lines. Expression levels are indicated by color intensity, with the scale representing log2fold changes. Each row represents a gene from the MEP (green), carotenoid (yellow), ABA (magenta), strigolactone (cyan), zaxinone (orange), blumenols (green, and lutein (blue).
Next, we questioned whether the transcriptional activation of the strigolactone biosynthetic pathway also occurs in the shoot of mNSP2ox lines. To test this, we used real-time polymerase chain reaction (qRT-PCR) to quantify the expression of three Parasponia genes, DWARF27 (PanD27), CAROTENOID CLEAVAGE DIOXYGENASE7 (PanCCD7), and PanCCD8, in shoots of lines 1, 3, and 6 under both nutrient conditions (Supplementary Figure S2, Supplementary Table S4). This revealed that NSP2 can also activate the expression of these genes in shoot tissue.
Together, these observations lead us to conclude that enhanced NSP2 expression in Parasponia markedly enhances the expression of genes essential for strigolactone biosynthesis and the apocarotenoid zaxinone across varying nutrient conditions.
We also analyzed the expression dataset for symbiosis genes of which the expression correlates with mNSP2 expression. This revealed that the expression of only a limited subset of the symbiosis-related genes positively correlated with mNSP2 expression (Figure 5; Supplementary Table S1). Among the positively correlated genes, the transcription factor PanCYCLOPS, the two transporters STUNTED ARBUSCULE2 (PanSTR2) and YELLOW STRIPE-LIKE 1 (PanYSL1), and ANNEXIN 1 (PanANN1) were shared between both growth conditions. Intriguingly, the genes encoding a nuclear envelope localized cation channel CASTOR and the VAMP-associated protein VAPYRIN (PanVPY) show only positive correlation with NSP2 expression under nodulation permissive conditions, whereas studies in legumes indicated they are also essential for mycorrhizal symbiosis (Imaizumi-Anraku et al., 2005; Pumplin et al., 2010; Murray et al., 2011). Conversely, under nutrient starved conditions, a negative correlation with NSP2 expression was observed with several transcription factor genes, including NIN-LIKE PROTEIN1 (PanNLP1) and several NUCLEAR FACTOR Y genes: PanNFYA5, PanNFYA6, PanNFYA7, PanNFYB6, PanNFYB8, and PanNFYC4 (Supplementary Table S3). This indicates a complex regulatory network where enhanced NSP2 expression differentially influences the expression of a few symbiosis-related genes depending on the nutrient status of the plant. This observation suggests a nuanced role of NSP2 in the regulatory network governing symbiotic permissiveness.
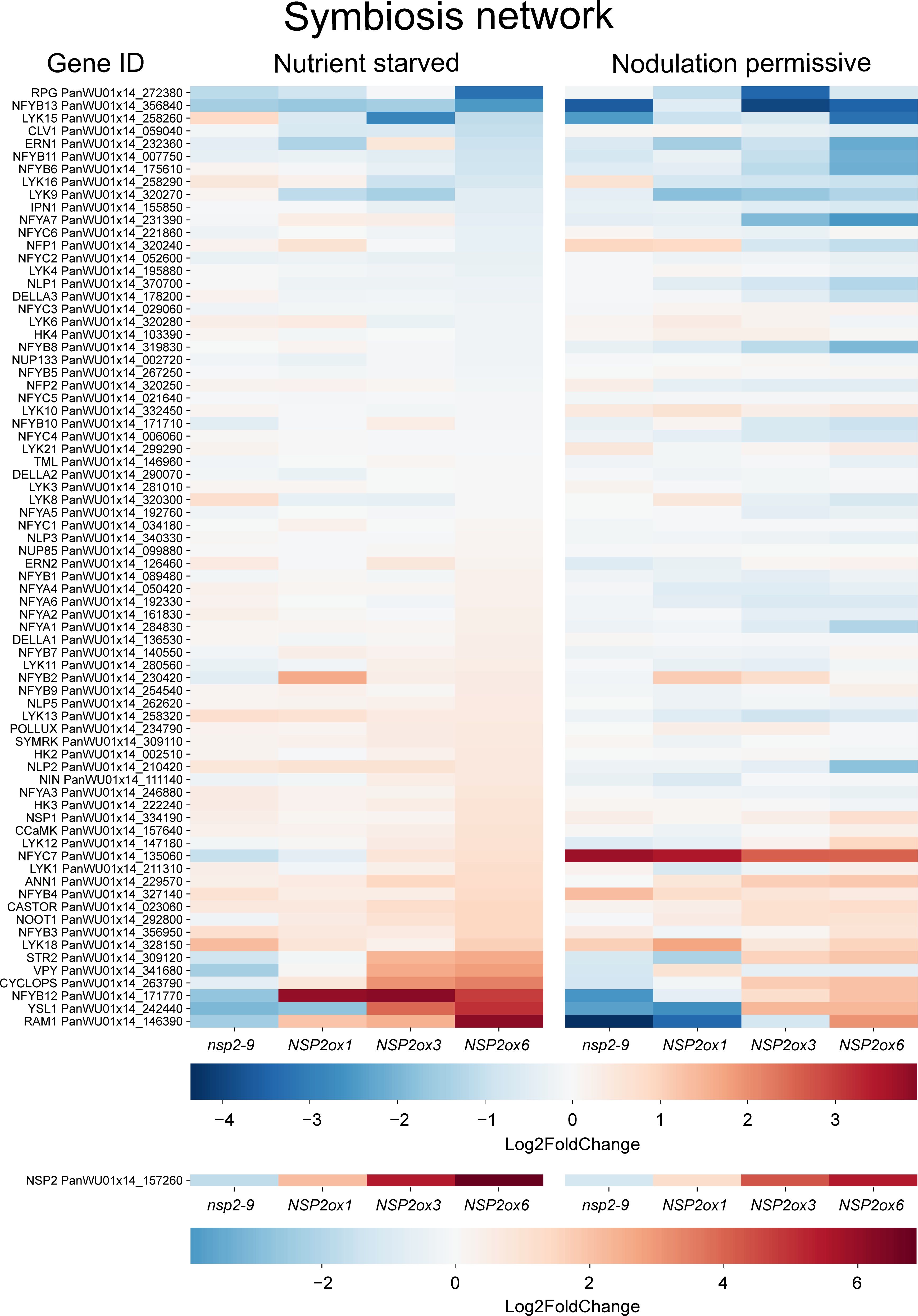
Figure 5. Heatmap of genes involved in the symbiosis network across different lines: Pannsp2-9 knockout mutant, mNSP2ox1, mNSP2ox3, and mNSP2ox6 ectopic expression lines. Expression levels are indicated by color intensity, with the scale representing Log2fold changes. Each row represents a gene from the symbiosis pathway.
To investigate the gene expression patterns during mycorrhization and nodulation in Parasponia and their interplay with NSP2 expression, we plotted the Log2fold changes of genes in mycorrhizal roots and young developing nodules (Supplementary Table S3) against their Pearson correlation coefficients with PanNSP2 expression under both conditions (Figure 6). This revealed that PanSTR2 not only exhibits upregulation in mycorrhizal roots, but also is transcriptionally induced young developing Parasponia nodules. When focusing on the carotenoid and strigolactone biosynthetic pathways, only a single gene stood out; namely PHYTOENE SYNTHASE1 (PanPSY1). Parasponia possesses three gene copies encoding phytoene synthases (Supplementary Figure S4), of which only one correlates with NSP2 expression. PanPSY1 is transcriptionally enhanced in mycorrhizal roots (0.58 Log2fold (1.5 fold) upregulated) and young nodules (6.47 Log2fold (89.7 fold) upregulated) (Supplementary Table S3). Phytoene synthase catalyzed the first step in carotenoid biosynthesis and is a major rate-limiting enzyme (Zhou et al., 2022). This suggests that NSP2-controlled of PanPSY1 expression represent a critical step in the biosynthesis of downstream products, including strigolactones in Parasponia roots that associate with arbuscular mycorrhizal fungi or nitrogen-fixing rhizobia.
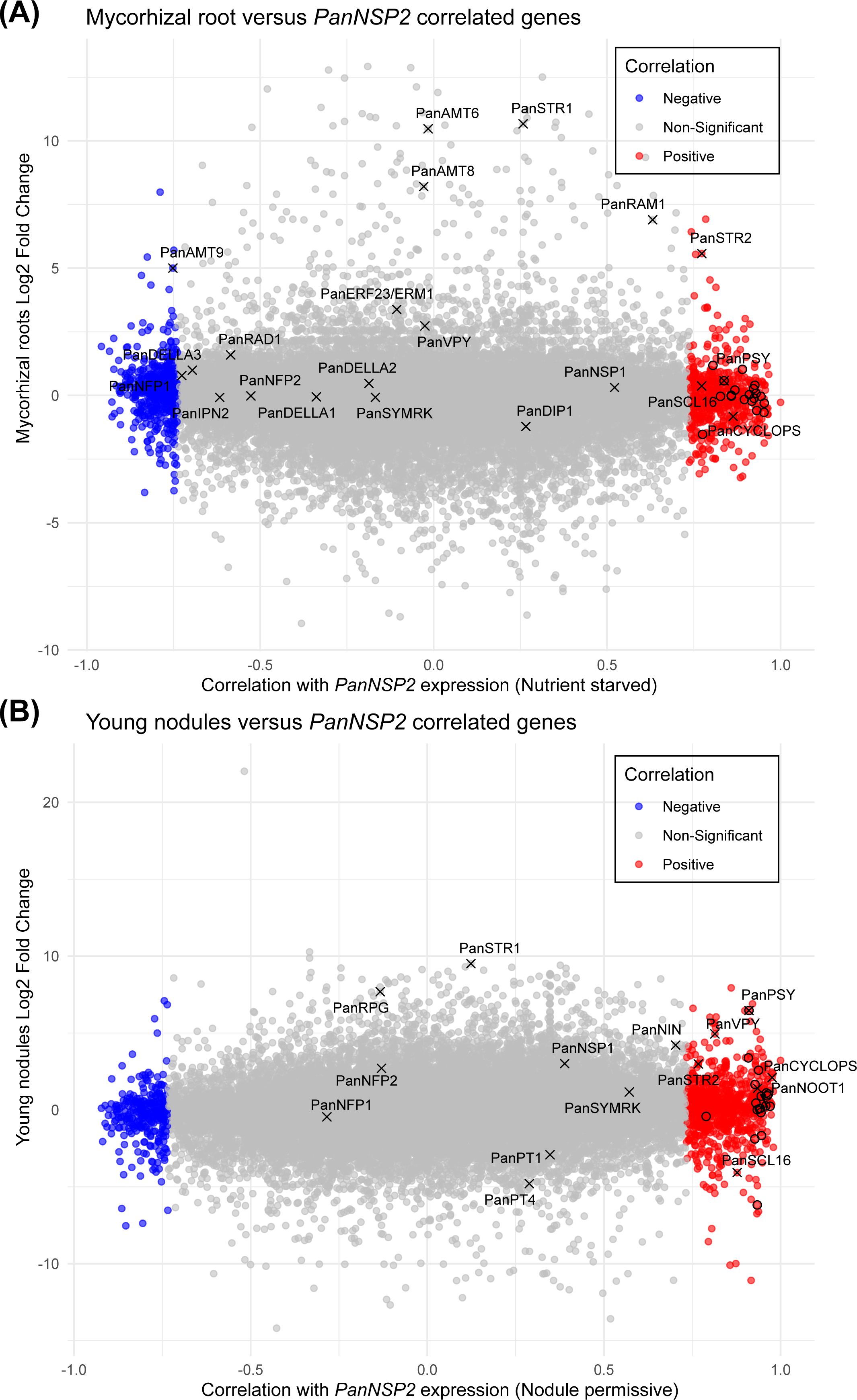
Figure 6. Scatterplot displaying the expression correlation between mNSP2 and mycorrhizal-induced and nodulation-induced genes. (A) Scatterplot representing the Log2fold change in gene expression of wild type mycorrhizal Parasponia roots against the corresponding Pearson correlation (Supplementary Table S1) with mNSP2 overexpression under nutrient starved conditions. (B) Scatterplot representing the Log2fold change of gene expression in young wild type Parasponia nodules against the corresponding Pearson correlation (Supplementary Table S1) with mNSP2 expression in nodulation permissive conditions. Each point indicates a Parasponia gene; blue points denote genes negatively correlated with mNSP2 expression, red points represent genes with a positive correlation, and gray points signify non-significant correlations. Several marker genes of mycorrhization and/or nodulation are indicated. Genes representing the carotenoid pathway are indicated by a circle.
Given that NSP2 is critical for both mycorrhization and nodulation, we investigated whether overexpression of mNSP2 could enhance symbiotic interactions with rhizobia. To test that, we grew all six mNSP2ox lines on nodulation medium (0.375 mM NH4NO3, 3 mM PO43-) and scored the number of nodules at 8 weeks post inoculation with Mesorhizobium plurifarium BOR2. We observed fewer nodules on lines with the highest mNSP2 expression (Figure 7A). We examined the cytoarchitecture of these nodules, revealing a wild-type phenotype where infection threads and fixation threads were formed (Figures 7B–D). A surprising observation was the occurrence of abnormal cell divisions on roots of plants highly expressing mNSP2. To determine if this cell proliferation was spontaneous due to the level of PanNSP2 expression or a response to rhizobium application, we grew plantlets of three PanNSP2 over-expressor lines with fold change in gene expression of 6, 51, and 95 on nodulation medium either non-inoculated or inoculated with M. plurifarium BOR2. Under the non-inoculated condition, all mNSP2ox lines showed no such epidermal cell divisions, whereas under inoculated conditions, mNSP2ox lines with 51 and 95-fold increase in mNSP2 expression, but not 6-fold, exhibited foci of massive cell divisions (Figures 7E–G). This shows that high levels of NSP2 promote cell proliferation in response to rhizobium inoculation. Sections of these aberrant structures revealed mitotic activity in epidermal, cortical and endodermal cell layers, (Figures 7H, I). We concluded that increased NSP2 expression in Parasponia negatively regulates nodule formation and causes abnormal proliferation of root tissue in response to rhizobium inoculation.
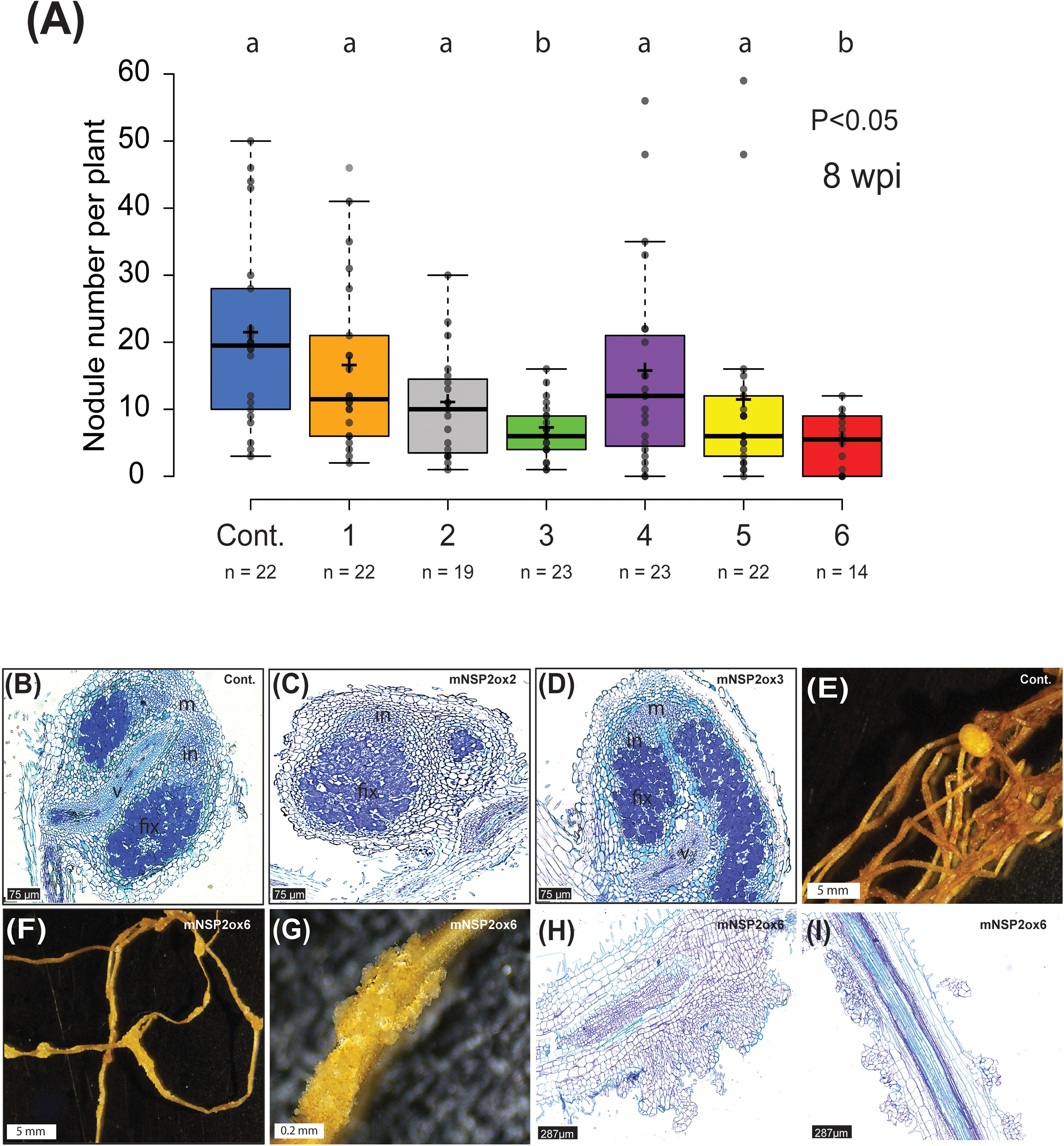
Figure 7. Rhizobium-induced nodulation on Parasponia NSP2ox lines. (A) Nodule number per plant of empty vector control (Cont.) and mNSP2ox lines 1 to 6. 8 weeks post-inoculation (wpi) with M. plurifarium BOR2. Different letters indicate significant differences (p<0.05) between these lines as determined by One-way ANOVA followed by a Tukey’s post hoc test. post hoc Data is displayed in a box plot, showing data points, the median, and the interquartile range (IQR) (B–D) Representative image of a section through a mature nodule formed on an empty vector control line (cont.) (B), mNSP2ox lines 2 (C), and mNSP2ox lines 3 (D) (8 wpi). Scale bar is 75 µm. m, nodule meristem; in, infection zone; fix, fixation zone; v, nodule vasculature (E–G) Bright-field image of nodulated root of the empty vector control line (cont.) (E), and the root of mNSP2ox line 6 showing rhizobium-induced irregular cell divisions (F, G) (10 wpi). Scale bar in (E, F) is 5 mm and in (G) is 0.2 mm. (H, I) Longitudinal sections of rhizobium-induced irregular cell divisions on the root of mNSP2ox line 6 (10 wpi). Scale bar is 287µm.
The GRAS-type transcriptional regulator NSP2 serves as a conserved hub that regulates the expression of symbiotic genes and genes essential for strigolactone biosynthesis. As such, NSP2 performs a dual function. The gene is essential for rhizobium-induced root nodule formation in legumes and the Cannabaceae species Parasponia (Kaló et al., 2005; Heckmann et al., 2006; van Zeijl et al., 2018; Peng et al., 2021). Additionally, NSP2-controlled strigolactones are exuded into the rhizosphere, serving as stimulants for arbuscular mycorrhizal fungi (Liu et al., 2011). Remarkably, ectopic expression of NSP2 in barley and rice has been shown to increase arbuscular mycorrhizal colonization rates under various nutrient conditions, while the reported trade-offs in plant developmental traits are relatively minor (Li et al., 2022; Yuan et al., 2023). This opens up opportunities to investigate the effects of enhanced NSP2 expression on plant development and endosymbioses in different species (Isidra-Arellano et al., 2024). We focused on Parasponia, the only non-legume known to form nitrogen-fixing root nodules with rhizobium and revealed novel phenotypes associated with increased NSP2 expression.We generated six independent Parasponia NSP2 ectopic expression lines that differ in NSP2 over-expression from 6 to 95-fold compared to the root tissue of control plants. These transgenic lines allowed us to correlate phenotypic responses to NSP2 expression levels. This revealed that the mycorrhizal infection levels positively correlate with NSP2 expression.
We also explored the ectopic expression lines to identify genes of which the expression correlates positively with NSP2 expression. Thereby we focused on two gene sets; the pathways that lead to strigolactone biosynthesis and a set of (putative) symbiosis genes. In the case of symbiosis genes, only a few genes showed a correlation with NSP2 expression (Supplementary Table S1). Of these PanCYCLOPS is most relevant. CYCLOPS is a transcription factor that functions in the common symbiosis signaling pathway as the last shared hub of arbuscular mycorrhizal- and rhizobium-induced signaling (Capoen and Oldroyd, 2008). Studies in a range of plant species showed that CYCLOPS is critical for both symbioses (Chen et al., 2008; Yano et al., 2008; Horváth et al., 2011; Das et al., 2019; Jin et al., 2018; Prihatna et al., 2018). CYCLOPS and NSP2 can form a complex having DELLA as an intermediate (Jin et al., 2016). Studies in M. truncatula showed that the CYCLOPS orthologs INTERACTING PROTEIN OF DMI3 (MtIPD3) and IPD3-LIKE (MtIPD3L) are critical for mycorrhizal-induced signaling when plants are grown at higher exogenous phosphate levels (Lindsay et al., 2019). Furthermore, constitutive expression of auto active variants of MtIPD3 and MtIPD3L induces the expression of several arbuscular mycorrhizal symbiosis genes, including MtVPY (Lindsay et al., 2019; Lindsay et al., 2022). We found PanVPY to be associated with mNSP2ox expression in Parasponia when grown at nodulation permissive conditions, which contains a relatively high phosphate concentration, but not under nutrient starved (mycorrhizal permissive) conditions. This led us to conclude that NSP2-controlled PanVPY expression is phosphate-dependent in Parasponia, a response that may require CYCLOPS-DELLA-NSP2 complex formation.
Analyzing the strigolactone biosynthetic pathways revealed that the expression of all genes encoding essential enzymes for the biosynthesis of carotenoids, strigolactones, and zaxinone positively correlates with NSP2 expression. In contrast, side branches leading to abscisic acid and lutein do not show such a response. A critical gene in the carotenoid pathway, namely PanPSY1, is also enhanced upon rhizobium and mycorrhizal inoculation. PSY commits the first step in the carotenoid pathway converting geranylgeranyl pyrophosphate into 15-cis-phytoene and is generally considered the rate-limiting step in carotenoid biosynthesis (Zhou et al., 2022). PSY functioning is controlled by several mechanisms, in which transcriptional regulation plays an important role (Stanley and Yuan, 2019). Enhancing PSY expression has been utilized extensively in plant biotech approaches to enhance carotenoid biosynthesis (Zhou et al., 2022). We hypothesize that in Parasponia the symbiotic regulation of PanPSY1 in response to arbuscular mycorrhizal infection or rhizobium-induced nodulation is a critical factor ultimately in the biosynthesis of strigolactones.
High expression levels of NSP2 also cause pleiotropic phenotypes. Some of these, like reduced axillary bud outgrowth, increased secondary thickening of the stem, and reduced lateral root formation are known strigolactone -controlled responses (Waters et al., 2017). In addition, we observed irregular cell divisions upon rhizobium-inoculation in roots of some Parasponia mNSP2ox lines. The structures formed resemble enlarged pre-nodules, which can serve as infection pockets to facilitate rhizobial crack entry (Lancelle and Torrey, 1984). A function of NSP2 in the formation of rhizobium infection pockets is also known in the legume Aeschynomene avenia. Like in Parasponia, rhizobium penetrates A. avenia by crack entry. Under nodulation permissive conditions, but in absence of rhizobium, A. avenia forms clusters of multicellular auxiliary hair-like cells at the base of newly formed lateral roots. These structures can be explored by rhizobium as an infection pocket that guides bacterial entry (Bonaldi et al., 2011). A. avenia nsp2 mutants do not form such infection pockets (Quilbé et al., 2022). Together, these experiments suggest the importance of a balanced NSP2 expression in the formation of pockets that ultimately can be explored by rhizobium as a starting point of intracellular infection.
Ectopic expression of NSP2 also affects plant architecture. mNSP2ox Parasponia trees have a reduced number of lateral roots but more lateral shoot branches. Studies in rice revealed similar developmental phenotypes, where OsNSP2ox results in increased tiller formation and a reduced number of lateral roots (Liu et al., 2011; Yuan et al., 2023). These phenotypes question whether exploring NSP2 ectopic expression in a biotech approach aiming to improve crop yield in low phosphate environments is a realistic strategy (Isidra-Arellano et al., 2024). Our findings advocate using tissue-specific promoters to obtain enhanced mycorrhization while minimizing pleiotropic phenotypes in plant architecture. Studies in rice indicate that using native promoter elements to enhance expression of GRAS-type transcriptional regulators might be the way forward to improve agronomic traits under low and medium-phosphorus conditions (Yuan et al., 2023).
The tissue culture propagation and maintenance of Parasponia plants were done on agar plates supplemented with Parasponia propagation medium. This medium contains 3.2 g/L SH-basal salt, 1 g/L SH-vitamin mixture, 10 g/L sucrose, 1 mL/L BAP (1 mg/mL), 100 μL/L IBA (1 mg/mL), 3 mL/L MES (1 M, pH 5.8), and 8 g/L Daishin agar (van Zeijl et al., 2018; Wardhani et al., 2019). Performance of nodulation and mycorrhization assays was carried out on platelets vegetatively grown and rooted following previously published procedures (van Zeijl et al., 2018; Wardhani et al., 2019).
Rooted plantlets were grown in 1 liter crystal-clear polypropylene pots (OS14BOX, Duchefa Biochemie, Netherlands) half-full, with a mixture (1:2 weight ratio) of river sand and agri perlite (Massmond-Westland, Netherlands). The mixture was watered with modified EKM medium [15 μM Fe-Citrate, 3 mM MES (pH 6.6), 6.6 μM MnSO4, 4.1 μM Na2MoO4, 2.08 mM MgSO4, 0.70 mM Na2SO4, 1.4 mM CaCl2, 0.375 mM NH4NO3, 1.5 μM ZnSO4, 0.88 mM KH2PO4, 1.6 μM CuSO4, 2.07 mM K2HPO4, and 4 μM H3BO3] (Becking, 1983) and inoculated with Mesorhizobium plurifarium BOR2 (OD600 = 0.025). Inoculated plants were then incubated in a controlled climate room at 85% humidity, under a 16/8 h day/night growth condition for eight weeks. Plants were removed from pots and washed under running tap water followed by quantification of total nodule number and harvest of some nodules for sectioning. Subsequently, harvested nodules were fixed in 0.1 M phosphate buffer (pH 7.2) containing 0.5% glutaraldehyde followed by vacuum application for 60 minutes. Next, nodules were embedded in infiltration plastic, Technovit 7100 (Heraeus-Kulzer, Germany), following the protocol provided by the manufacturer. A Leica RJ2035 microtome was used to make 4.5 μm plastic sections which were stained with 0.5% toluidine blue. A Leica DFC425c camera was used to make high resolution images of the cytoarchitecture of the fixed nodules.
Rooted plantlets were placed in pots half-filled with a mixture (5:1 weight ration) of river sand and commercial potting soil and watered with half-strength Hoagland’s medium containing 20 μM phosphate (low P condition) or 3 mM (high P condition). Plants then were incubated in a controlled climate room at 85% humidity, under a 16/8 h day/night regime for two weeks followed by inoculation with 300 spores of Rhizophagus irregularis fungi (Agronutrion-DAOM197198, Carbonne, France) and subsequent incubation for 6 weeks. Afterwards, plants’ roots were harvested and treated with 10% (w/v) KOH and boiled for 30 minutes at 90°C and then stained in a staining solution contains 5% acetic acid and 3% ink (Waterman Vulpen, Zwolle, the Netherland) for 15 minutes at 90 °C. Next, fungal colonization and formation of arbuscules and vesicles were quantified, using the gridline intersect method (Giovannetti and Mosse, 1980), and high-resolution images were taken, using a Leica CTR66000 microscope.
All vectors used in this study were propagated in Escherichia coli strain DH5α and assembled, using Golden Gate Cloning (GGC) as was previously described (Engler et al., 2009). Level one and two acceptors and binary vectors used for the assembly were obtained from the GGC toolbox (Engler et al., 2014). Parasponia clone of NSP2 coding sequence (CDS) with and without insertion of intron 10 of Arabidopsis thaliana UBQ10 gene (intron) in the first putative splicing site in NSP2 CDS, including silent mutation in microR171h putative site and golden gate BsaI or BpiI restriction sites was synthesized as Level zero vectors. Subsequently, level zero vectors of NSP2, Lotus japonicus UBQ1 (LjUBQ1) promoter, C-terminal Myc tag, and Agrobacterium tumefaciens nopaline synthase terminator (T-NOS) were recombined into level one acceptor. Finally, level one vectors of NSP2 with or without (intron), including Kanamycin resistant Level one vectors were recombined into Level 2 binary vectors. All binary GGC vectors were validated by restriction enzymes and sequencing before transformation.
Level 2 binary vectors were transformed into Agrobacterium tumefaciens AGL1 strain, using Eppendorf Eporator (Eppendorf SE, Hamburg Germany). Transformed AGL1 were then grown on lysogeny broth (LB) agar plate supplemented with appropriate antibiotics and incubated at 28°C for 2 days. Parasponia plant transformation was carried out as previously described (van Zeijl et al., 2018; Wardhani et al., 2019), so young stems and petioles were collected from Parasponia trees grown in controlled greenhouse conditions at 85% humidity and under a 16/8 h day/night regime. The surface of the stems and petioles was sterilized with 2.5% bleach with a few drops of Tween 20, and subsequently stems and petioles washed six times with sterile water followed by a co-cultivation with AGL1 strains carrying constructs of interest for 2 days. Afterwards, transformed stems and petioles were transferred to callus induction medium supplemented with cefotaxime (300μg/ml) and kanamycin (50μg/ml) antibiotics for one week followed by transferring to plant propagation medium supplemented with cefotaxime (300μg/ml) and kanamycin (50μg/ml) antibiotics, refreshed every 1-2 weeks until transgenic shoots were formed. Genotyping was performed using a pair of primers designed to anneal to the L. japonicus UBIQUITIN 1 promoter (pLjUBQ1) and the C-terminal Myc tag, with amplification carried out using the Phire Plant Direct PCR kit (Thermo Fisher, F130WH), following the manufacturer’s protocol.
Parasponia plant lines, consisting of three NSP2 overexpression mutants (mNSP2ox1, mNSP2ox3, and mNSP2ox6), a nsp2 knockout (nsp2-9), and control line 46 with an empty vector were selected. The NSP2 overexpression mutants displayed NSP2 expression levels of 6, 51, and 95-fold higher than empty vector control. Selected plants were cultivated for 6 weeks on nutrient rich medium containing 24.72 mM NO₃⁻, 2.6 mM NH₄+ and 2.6 mM PO43-. Next, cultivated plants were transferred for 3 weeks to either nutrient starved medium (no N and P resource), or the nodulation permissive medium (0.375 mM NH4NO3 and 3 mM PO43-).
Subsequently, healthy plantlets were transferred to pots containing a 2:1 weight ratio of perlite to sand. Each pot was supplemented with 150 mL of EKM medium, with or without nitrogen and phosphorus, referred to as nodulation permissive (low nitrate, high phosphate) and nutrient starved (low nitrate and low phosphate) treatments, respectively. Pots were populated with three plantlets of a single genotype and grown under a 16-hour light/8-hour dark photoperiod for 21 days. Following this period, roots from each pot were pooled for RNA extraction.
For each treatment, five root samples weighing 50 mg each were collected. Tissue lysis was performed using a QIAGEN TissueLyser LT, followed by sequential phenol:chloroform and chloroform extractions. Nucleic acid precipitation was performed using sodium acetate, followed by sequential washing steps with isopropanol and ethanol to purify the samples. Subsequently, the samples were treated with DNase to eliminate genomic DNA contamination. A second round of phenol:chloroform and chloroform extractions was conducted. The RNA was then re-precipitated in ethanol and re-suspended in nuclease-free water. The concentration of the isolated RNA was quantified using both a Biochrom SimpliNano™ Spectrophotometer and a Qubit 2.0 Fluorometer, utilizing Broad Range Assay Kits. RNA integrity was confirmed through gel electrophoresis. To validate NSP2 expression levels, quantitative polymerase chain reaction (qPCR) was employed. ACTIN and ELONGATION FACTOR1 ALPHA (EF1α) served as the housekeeping genes for normalization.
For each harvested sample, mRNA was isolated from total RNA utilizing poly-T oligo-attached magnetic beads. The resulting mRNA served as the template for first-strand cDNA synthesis, which was initiated with random hexamer primers, and subsequently followed by second-strand cDNA synthesis. Sample quality and nucleic acid concentration were quantitatively assessed using Qubit and qPCR assays, using EF1α and ACTIN for normalization genes. Size distribution of the samples was further confirmed via bioanalyzer analysis. Three biological replicates for each sample exhibiting the best characteristics (quantity and quality) for sequencing were selected for library preparation and sequencing, conducted at Novogene UK (Cambridge, United Kingdom). The resulting libraries were pooled and subjected to paired-end sequencing on an Illumina NovaSeq 6000 platform. Each biological replicate generated approximately 25 million reads, with each read consisting of 150 base pairs.
The quality of the sequenced reads was assessed using FastQC and subsequently visually inspected through MultiQC. Remaining adapters and low-quality bases were trimmed using Trimmomatic with settings TRAILING:3, SLIDINGWINDOW:4:15, MINLEN:50, HEADCROP:12. Transcript abundances were quantified with Kallisto, and the resulting files were loaded into R Studio version 2022.07.2 using R version 4.1.1. Differential gene expression analysis was conducted using DESeq2 for each condition (Nodulation permissive and nutrient starved) separately (Supplementary Table S2).
Pearson correlation analysis was performed for each nutrient condition (nodulation permissive and nutrient starved) to assess the expression correlation with NSP2. The analysis was conducted in R, calculating the correlation, p-value, and False Discovery Rate (FDR, or adjusted p-value). An FDR cutoff of 0.05 was applied and used to categorize the correlation as positive, non-significant, or negative. The Pearson correlation graphs were plotted using the ggplot2 package. Overlapping genes between nutrient conditions were visualized using the VennDiagram package in R.
Prominent pathways featuring upregulated genes under NSP2 overexpression conditions, as well as genes of interest related to nodulation and mycorrhization in Parasponia, were selected for detailed investigation. The first set, named the “Symbiosis Network,” consists of a gene set in Parasponia previously annotated (Supplementary Table S1). The second set includes genes involved in the carotenoid biosynthesis pathway in Parasponia. Orthologous genes in Parasponia involved in carotenoid biosynthesis were identified using gene IDs from Arabidopsis thaliana, as reported in the study by (Li et al., 2022) (Supplementary Table S1). Corresponding gene IDs for Parasponia were identified through a reciprocal best BLAST hit approach, using the Arabidopsis gene IDs as references. BLASTP was performed with the following parameters: E-value cutoff of 1e-5, BLOSUM62 scoring matrix, gap penalties of 11 for existence and 1 for extension, and a word size of 3. The top BLAST hits in Parasponia were then used for a reverse search against the TAIR Araport 11 protein set to verify orthologous relationships.
Parasponia wild type young nodule data were previously published under PRJNA272473. Additionally, an experiment was conducted on Parasponia plants inoculated with Rhizophagus irregularis (MYC+) or left uninoculated (MYC-) [(van Velzen et al., 2018; van Zeijl et al., 2018; Bu et al., 2020; Rutten et al., 2020; Alhusayni et al., 2023)]. Parasponia plants were grown under controlled greenhouse conditions with a 16-hour light/8-hour dark photoperiod at 28°C and 85% humidity. The growth medium consisted of a mixture (5:1 ratio) of river sand and commercial potting soil and watered with half-strength Hoagland solution containing 20 μM PO₄3- every other week. After placing the plants in pots, they were allowed to recover for 2 weeks before being inoculated with 300 R. irregularis spores. Root samples were harvested 42 days after inoculation.
Total RNA was extracted from collected samples using the RNeasy Plant Mini Kit (Qiagen). mRNA was isolated from total RNA using poly-T oligo-attached magnetic beads. The isolated mRNA served as the template for first-strand cDNA synthesis, initiated with random hexamer primers, followed by second-strand cDNA synthesis. Sample quality and nucleic acid concentration were assessed using Qubit and qPCR assays. Size distribution of the samples was confirmed via bioanalyzer analysis. Three biological replicates per sample exhibiting the best characteristics (quantity and quality) were selected for library preparation and sequencing.
Sequencing was conducted at Novogene UK (Cambridge, United Kingdom) using an Illumina NovaSeq 6000 platform. Each biological replicate generated approximately 25 million paired end reads of 150 base pairs. The quality of the sequenced reads was assessed using FastQC and subsequently visually inspected through MultiQC.
To analyze the RNA-seq dataset from nodule stages 1, 2, and 3 versus roots, as well as mycorrhization data of Parasponia wild type MYC- and MYC+ data, the following procedures were performed using the R programming language. Transcript abundances were quantified using Kallisto. The resulting files were imported into R Studio version 2022.07.2 with R version 4.1.1 for subsequent analysis using DESeq2 as previously described (Supplementary Table S2). The differential expression results were integrated with existing annotated datasets for nodulation permissive and nutrient starved conditions. Ggplot2 was used to plot Log2fold change versus the NSP2 correlation (Supplementary Table S3).
Box plot graphs were generated using BoxPlotR, an online tool developed by (Spitzer et al., 2014). Bar charts were created in Microsoft Excel for Mac (Version 16.89). The Kruskal–Wallis test, followed by Tukey’s post-hoc test, was employed for statistical analysis. Statistical significance was established at a threshold of p < 0.05. Additionally, the quantification of internode diameter was assessed using a Student’s t-test, with statistical significance also defined as p < 0.05.
The RNA-seq datasets analyzed in this study are available in the NCBI SRA repository under BioProject number PRJNA1029573. Plant material and seeds used in this study can be obtained upon request from the authors. Scripts used in the differential gene expression and visualizationvisualisation of the heatmaps can be found on GitHub: https://github.com/kleinjoel/NSP2_overexpression_analysis. Young nodules data were previously published under BioProject number PRJNA272473.
SA: Validation, Writing – review & editing, Writing – original draft, Visualization, Supervision, Resources, Methodology, Investigation, Formal analysis, Conceptualization. NK: Writing – review & editing, Writing – original draft, Visualization, Software, Investigation, Formal analysis, Data curation. RH: Writing – review & editing, Writing – original draft, Validation, Investigation, Formal analysis. RG: Writing – review & editing, Writing – original draft, Visualization, Validation, Supervision, Resources, Project administration, Methodology, Investigation, Funding acquisition, Data curation, Conceptualization. JK: Validation, Project administration, Data curation, Writing – review & editing, Writing – original draft, Visualization, Supervision, Software, Methodology, Investigation, Formal analysis, Conceptualization.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The project was funded by the ENSA project funded by the Bill & Melinda Gates Foundation to the University of Cambridge (to RG) [OPP1172165], and the Ministry of Education, King Faisal University, Saudi Arabia (grant no. 10598 to SA), RH is funded by Dutch Science Organization (Nederlandse Organisatie voor Wetenschappelijk Onderzoek VI.Veni.212.132), and JK by UK Research and Innovation (UKRI) under the UK government’s MCSA Postdoctoral Fellowship funding guarantee [grant number EP/X023672/1].
All individuals who contributed to this manuscript are included as authors.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1468812/full#supplementary-material
Supplementary Figure S1 | Construct design to ectopically express Parasponia NSP2. (A) Alignment of putative miR171h target sites in Medicago truncatula (Lauressergues et al., 2012) and Parasponia NSP2 as well as mNSP2 a modified version of Parasponia NSP2 in which the target site of miR171hR is silenced. Matching nucleotides at the miR171h binding site are shown in red, mismatching nucleotides in black, and silencing nucleotides in green. (B) A schematic representation of the transformation constructs to achieve ectopic expression of PanNSP2 in Parasponia.
Supplementary Figure S2 | Ectopic PanNSP2 expression enhances the expression of strigolactone biosynthesis genes in shoot tissue. (A–D) qRT-PCR-based expression of PanNSP2, PanCCD7, PanCDD8, and PanD27 in shoot of plants grown under nodulation permissive condition (n=3), and (E–H) nutrient starved condition (n=3).
Supplementary Figure S3 | Shoot phenotype of Parasponia NSP2ox lines. post hoc quantification of internode diameter of the empty vector control line (cont.) and mNSP2ox lines 1,3 and 6. Asterisk indicates significant difference relative to cont. (p < 0.05) as determined by Student’s t-test (*p < 0.05). Stem diameters were measured at the middle of internodes (IN) for the first 12 internodes, from the bottom to the top of 60 days old plants (n=10).
Supplementary Figure S4 | Phylogenetic relation of Parasponia PSY proteins. Phylogenetic relation is reconstructed based on an alignment of protein sequences from the following species: Parasponia andersonii, Trema orientalis, Arabidopsis thaliana, Medicago truncatula, Glycine max, Fragaria vesca, Eucalyptus grandis, and Manihot esculenta. Parasponia and Arabidopsis proteins are in bold letter-type. Three orthogroups are recognized and marked in distinct colors. Node labels indicate posterior probability.
Supplementary Table S1 | RNA-seq analysis of mNSP2 overexpression in Parasponia. Pearson correlation tests and a quantitative analysis of gene correlations under mycorrhizal and nodulation permissive conditions. The table also details genes in carotenoid biosynthesis and symbiosis networks, highlighting their correlation values in relation to PanNSP2 overexpression.
Supplementary Table S2 | Integrated RNA-seq Analysis of PanNSP2 overexpression in Parasponia, Log2fold change (LFC) values for all Parasponia genes under both nodulation permissive and nutrient starved conditions, detailing results from mNSP2 overexpressing and knockout lines.
Supplementary Table S3 | RNA-seq analysis of Parasponia genes under arbuscular mycorrhiza permissive conditions. This table includes base means, Log2fold changes between non-mycorrhized and mycorrhized roots, and adjusted p-values (padj). It also provides annotations, Pearson correlation values for NSP2 under nutrient starved and nodulation permissive conditions, along with their respective p-values, false discovery rates (FDR), and correlation types.
Supplementary Table S4 | Primers used in this study. List of primers, their purpose, and their sequence that were used in various applications in this study.
Ablazov, A., Votta, C., Fiorilli, V., Wang, J. Y., Aljedaani, F., Jamil, M., et al. (2023). ZAXINONE SYNTHASE 2 regulates growth and arbuscular mycorrhizal symbiosis in rice. Plant physiology 191, 328–399.
Alhusayni, S., Roswanjaya, Y. P., Rutten, L., Huisman, R., Bertram, S., Sharma, T., et al. (2023). A rare non-canonical splice site in Trema orientalis SYMRK does not affect its dual symbiotic functioning in endomycorrhiza and rhizobium nodulation. BMC Plant Biol. 23, 587. doi: 10.21203/rs.3.rs-2757645/v1
Ali, L., Madrid, E., Varshney, R. K., Azam, S., Millan, T., Rubio, J., et al. (2014). Mapping and identification of a Cicer arietinum NSP2 gene involved in nodulation pathway. TAG. Theor. Appl. Genet. Theoretische und angewandte Genetik 127, 481–488. doi: 10.1007/s00122-013-2233-3
Becking, J. H. (1983). TheParasponia parviflora—Rhizobium symbiosis. Host specificity, growth and nitrogen fixation under various conditions. Plant Soil 75, 309–342. doi: 10.1007/BF02369969
Behm, J.E., Geurts, R., Kiers, E. T. (2014). Parasponia: a novel system for studying mutualism stability. Trends Plant Sci. 19, 757–763.
Bonaldi, K., Gargani, D., Prin, Y., Fardoux, J., Gully, D., Nouwen, N., et al. (2011). Nodulation of Aeschynomene afraspera and A. indica by photosynthetic Bradyrhizobium Sp. strain ORS285: the nod-dependent versus the nod-independent symbiotic interaction. Mol. Plant-Microbe interactions: MPMI 24, 1359–1371. doi: 10.1094/MPMI-04-11-0093
Brewer, P. B., Dun, E. A., Gui, R., Mason, M. G., Beveridge, C. A. (2015). Strigolactone inhibition of branching independent of polar auxin transport. Plant Physiol. 168, 1820–1829. doi: 10.1104/pp.15.00014
Bu, F., Rutten, L., Roswanjaya, Y. P., Kulikova, O., Rodriguez-Franco, M., Ott, T., et al. (2020). Mutant analysis in the nonlegume Parasponia andersonii identifies NIN and NF-YA1 transcription factors as a core genetic network in nitrogen-fixing nodule symbioses. New Phytol. 226, 541–554. doi: 10.1111/nph.v226.2
Capoen, W., Oldroyd, G. (2008). How CYCLOPS keeps an eye on plant symbiosis. Proc. Natl. Acad. Sci. U. S. A. 105, 20053–20054. doi: 10.1073pnas.0811417106
Carvalhais, L. C., Rincon-Florez, V. A., Brewer, P. B., Beveridge, C. A., Dennis, P. G., Schenk, P. M., et al. (2019). The ability of plants to produce strigolactones affects rhizosphere community composition of fungi but not bacteria. Rhizosphere 9, 18–26. doi: 10.1016/j.rhisph.2018.10.002
Cenci, A., Rouard, M. (2017). Evolutionary analyses of GRAS transcription factors in angiosperms. Front. Plant Sci. 8, 273. doi: 10.3389/fpls.2017.00273
Chen, C., Ané, J.-M., Zhu, H. (2008). OsIPD3, an ortholog of the Medicago truncatula DMI3 interacting protein IPD3, is required for mycorrhizal symbiosis in rice. New Phytol. 180, 311–315. doi: 10.1111/j.1469-8137.2008.02612.x
Das, D. R., Horváth, B., Kundu, A., Kaló, P., DasGupta, M., et al. (2019). Functional conservation of CYCLOPS in crack entry legume Arachis hypogaea. Plant Sci. 281, 232–241. doi: 10.1016/j.plantsci.2018.12.003
Das, D., Paries, M., Hobecker, K., Gigl, M., Dawid, C., Lam, H. M., et al. (2022). PHOSPHATE STARVATION RESPONSE transcription factors enable arbuscular mycorrhiza symbiosis. Nat. Commun. 13, 477. doi: 10.1038/s41467-022-27976-8
Engler, C., Gruetzner, R., Kandzia, R., Marillonnet, S. (2009). Golden gate shuffling: a one-pot DNA shuffling method based on type IIs restriction enzymes. PloS One 4, e5553. doi: 10.1371/journal.pone.0005553
Engler, C., Youles, M., Gruetzner, R., Ehnert, T. M., Werner, S., Jones, J. D., et al. (2014). A golden gate modular cloning toolbox for plants. ACS synthetic Biol. 3, 839–843. doi: 10.1021/sb4001504
Feike, D., Korolev, A.V., Soumpourou, E., Murakami, E., Reid, D., Breakspear, A., et al. (2019). Characterizing standard genetic parts and establishing common principles for engineering legume and cereal roots. Plant Biotechnol. J. 17, 2234–2245. doi: 10.1111/pbi.13135. Preprint.
Fonouni-Farde, C., Tan, S., Baudin, M., Brault, M., Wen, J., Mysore, K. S., et al. (2016). DELLA-mediated gibberellin signalling regulates Nod factor signalling and rhizobial infection. Nat. Commun. 7, 12636. doi: 10.1038/ncomms12636
Giovannetti, M., Mosse, B. (1980). An evaluation of techniques for measuring vesicular arbuscular mycorrhizal infection in roots. New Phytol. 84, 489–500. doi: 10.1111/j.1469-8137.1980.tb04556.x
Gobbato, E., Marsh, J. F., Vernié, T., Wang, E., Maillet, F., Kim, J., et al. (2012). A GRAS-type transcription factor with a specific function in mycorrhizal signaling. Curr. Biol. 22, 2236–2241. doi: 10.1016/j.cub.2012.09.044
Heck, C., Kuhn, H., Heidt, S., Walter, S., Rieger, N., Requena, N., et al. (2016). Symbiotic fungi control plant root cortex development through the novel GRAS transcription factor MIG1. Curr. Biol. 26, 2770–2778. doi: 10.1016/j.cub.2016.07.059
Heckmann, A. B., Lombardo, F., Miwa, H., Perry, J. A., Bunnewell, S., Parniske, M., et al. (2006). Lotus japonicus nodulation requires two GRAS domain regulators, one of which is functionally conserved in a non-legume. Plant Physiol. 142, 1739–1750. doi: 10.1104/pp.106.089508
Hirsch, S., Kim, J., Muñoz, A., Heckmann, A. B., Downie, J. A., Oldroyd, G. E., et al. (2009). GRAS proteins form a DNA binding complex to induce gene expression during nodulation signaling in Medicago truncatula. Plant Cell 21, 545–557. doi: 10.1105/tpc.108.064501
Hofferek, V., Mendrinna, A., Gaude, N., Krajinski, F., Devers, E. A. (2014). MiR171h restricts root symbioses and shows like its target NSP2 a complex transcriptional regulation in Medicago truncatula. BMC Plant Biol. 14, 199. doi: 10.1186/s12870-014-0199-1
Horváth, B., Yeun, L. H., Domonkos, A., Halász, G., Gobbato, E., Ayaydin, F., et al. (2011). Medicago truncatula IPD3 is a member of the common symbiotic signaling pathway required for rhizobial and mycorrhizal symbioses. Mol. Plant-Microbe interactions: MPMI 24, 1345–1358. doi: 10.1094/MPMI-01-11-0015
Imaizumi-Anraku, H., Takeda, N., Charpentier, M., Perry, J., Miwa, H., Umehara, Y., et al. (2005). Plastid proteins crucial for symbiotic fungal and bacterial entry into plant roots. Nature 433, 527–531. doi: 10.1038/nature03237
Isidra-Arellano, M. C., Singh, J., Valdés-López, O. (2024). Unraveling the potential of the strigolactones-NSP1/NSP2 friendship in crop improvement. Trends Plant Sci. 29, 501–503. doi: 10.1016/j.tplants.2023.12.012. Preprint.
Jin, Y., Liu, H., Luo, D., Yu, N., Dong, W., Wang, C., et al. (2016). DELLA proteins are common components of symbiotic rhizobial and mycorrhizal signalling pathways. Nat. Commun. 7, 12433. doi: 10.1038/ncomms12433
Jin, Y., Chen, Z., Yang, J., Mysore, K. S., Wen, J., Huang, J., et al. (2018). IPD3 and IPD3L function redundantly in rhizobial and mycorrhizal symbioses. Front. Plant Sci. 9, 267. doi: 10.3389/fpls.2018.00267
Kaló, P., Gleason, C., Edwards, A., Marsh, J., Mitra, R.M., Hirsch, S., et al. (2005). Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science 308, 1786–1789. doi: 10.1126/science.1110951
Kobae, Y., Kameoka, H., Sugimura, Y., Saito, K., Ohtomo, R., Fujiwara, T., et al. (2018). Strigolactone biosynthesis genes of rice are required for the punctual entry of arbuscular mycorrhizal fungi into the roots. Plant Cell Physiol. 59, 544–553. doi: 10.1093/pcp/pcy001
Lancelle, S. A., Torrey, J. G. (1984). Early development of Rhizobium-induced root nodules of Parasponia rigida. I. Infection and early nodule initiation. Protoplasma 123, 26–37. doi: 10.1007/BF01283179
Lauressergues, D., Delaux, P. M., Formey, D., Lelandais-Brière, C., Fort, S., Cottaz, S., et al. (2012). The microRNA miR171h modulates arbuscular mycorrhizal colonization of Medicago truncatula by targeting NSP2. Plant J. 72, 512–522. doi: 10.1111/j.1365-313X.2012.05099.x
Li, X.-R., Sun, J., Albinsky, D., Zarrabian, D., Hull, R., Lee, T., et al. (2022). Nutrient regulation of lipochitooligosaccharide recognition in plants via NSP1 and NSP2. Nat. Commun. 13, 6421. doi: 10.1038/s41467-022-33908-3
Lindsay, P. L., Williams, B. N., MacLean, A., Harrison, M. J. (2019). A phosphate-dependent requirement for transcription factors IPD3 and IPD3L during arbuscular mycorrhizal symbiosis in medicago truncatula. Mol. Plant Microbe Interact. 32, 1277–1290. doi: 10.1094/MPMI-01-19-0006-R
Lindsay, P. L., Ivanov, S., Pumplin, N., Zhang, X., Harrison, M. J., et al. (2022). Distinct ankyrin repeat subdomains control VAPYRIN locations and intracellular accommodation functions during arbuscular mycorrhizal symbiosis. Nat Commun. 13, 5228.
Liu, W., Kohlen, W., Lillo, A., Op den Camp, R., Ivanov, S., Hartog, M., et al. (2011). Strigolactone biosynthesis in Medicago truncatula and rice requires the symbiotic GRAS-type transcription factors NSP1 and NSP2. Plant Cell 23, 3853–3865. doi: 10.1105/tpc.111.089771
Maekawa, T., Kusakabe, M., Shimoda, Y., Sato, S., Tabata, S., Murooka, Y., et al. (2008). Polyubiquitin promoter-based binary vectors for overexpression and gene silencing in Lotus japonicus. Mol. Plant-Microbe interactions: MPMI 21, 375–382. doi: 10.1094/MPMI-21-4-0375
Marzec, M., Melzer, M. (2018). Regulation of root development and architecture by strigolactones under optimal and nutrient deficiency conditions. Int. J. Mol. Sci. 19. doi: 10.3390/ijms19071887
Murakami, Y., Yokoyama, H., Fukui, R., Kawaguchi, M.. (2013). Down-Regulation of NSP2 Expression in Developmentally Young Regions of Lotus japonicus Roots in Response to Rhizobial Inoculation. Plant Cell Physiol. 54, 518–27.
Murray, J. D., Muni, R.R., Torres-Jerez, I., Tang, Y., Allen, S., Andriankaja, M., et al. (2011). Vapyrin, a gene essential for intracellular progression of arbuscular mycorrhizal symbiosis, is also essential for infection by rhizobia in the nodule symbiosis of Medicago truncatula. Plant journal: Cell Mol. Biol. 65, 244–252. doi: 10.1111/j.1365-313X.2010.04415.x
Okada, K., Saito, T., Nakagawa, T., Kawamukai, M., Kamiya, Y., et al. (2000). Five geranylgeranyl diphosphate synthases expressed in different organs are localized into three subcellular compartments in Arabidopsis. Plant Physiol. 122, 1045–1056.
Oldroyd, G. E., Long, S. R. (2003). Identification and characterization of nodulation-signaling pathway 2, a gene of Medicago truncatula involved in Nod factor signaling. Plant Physiol. 131, 1027–1032. doi: 10.1104/pp.102.010710
Park, H.-J., Floss, D. S., Levesque-Tremblay, V., Bravo, A., Harrison, M. J. (2015). Hyphal branching during arbuscule development requires reduced arbuscular mycorrhiza1. Plant Physiol. 169, 2774–2788. doi: 10.1104/pp.15.01155
Peng, Z., Chen, H., Tan, L., Shu, H., Varshney, R. K., Zhou, Z., et al. (2021). Natural polymorphisms in a pair of NSP2 homoeologs can cause loss of nodulation in peanut. J. Exp. Bot. 72, 1104–1118. doi: 10.1093/jxb/eraa505
Pimprikar, P., Carbonnel, S., Paries, M., Katzer, K., Klingl, V., Bohmer, M.J., et al. (2016). A CCaMK-CYCLOPS-DELLA complex activates transcription of RAM1 to regulate arbuscule branching. Curr. Biol. 26, 987–998. doi: 10.1016/j.cub.2016.01.069
Prihatna, C., Larkan, N. J., Barbetti, M. J., Barker, S. J. (2018). Tomato CYCLOPS/IPD3 is required for mycorrhizal symbiosis but not tolerance to Fusarium wilt in mycorrhiza-deficient tomato mutant rmc. Mycorrhiza 28, 495–507. doi: 10.1007/s00572-018-0842-z
Pumplin, N., Mondo, S. J., Topp, S., Starker, C. G., Gantt, J. S., Harrison, M. J., et al. (2010). Medicago truncatula Vapyrin is a novel protein required for arbuscular mycorrhizal symbiosis. Plant J. 61, 482–494. doi: 10.1111/j.1365-313X.2009.04072.x
Quilbé, J., Arrighi, J.-F. (2021). NSP2, a key symbiotic regulator in the spotlight. J. Exp. Bot. 72, 959–963. doi: 10.1093/jxb/eraa540
Quilbé, J., Nouwen, N., Pervent, M., Guyonnet, R., Cullimore, J., Gressent, F., et al. (2022). A mutant-based analysis of the establishment of Nod-independent symbiosis in the legume Aeschynomene evenia. Plant Physiol. 190, 1400–1417. doi: 10.1093/plphys/kiac325. Preprint.
Rutten, L., Miyata, K., Roswanjaya, Y. P., Huisman, R., Bu, F., Hartog, M., et al. (2020). Duplication of symbiotic lysin motif receptors predates the evolution of nitrogen-fixing nodule symbiosis. Plant Physiol. 184, 1004–1023. doi: 10.1104/pp.19.01420
Ruyter-Spira, C., Kohlen, W., Charnikhova, T., van Zeijl, A., van Bezouwen, L., de Ruijter, N., et al. (2011). Physiological effects of the synthetic strigolactone analog GR24 on root system architecture in Arabidopsis: another belowground role for strigolactones? Plant Physiol. 155, 721–734. doi: 10.1104/pp.110.166645
Schlemper, T. R., Leite, M. F. A., Lucheta, A. R., Shimels, M., Bouwmeester, H. J., van Veen, J. A., et al. (2017). Rhizobacterial community structure differences among sorghum cultivars in different growth stages and soils. FEMS Microbiol. Ecol. 93. doi: 10.1093/femsec/fix096
Shtark, O. Y., Sulima, A.S., Zhernakov, A.I., Kliukova, M.S., Fedorina, J.V., Pinaev, A.G., et al. (2016). Arbuscular mycorrhiza development in pea (Pisum sativum L.) mutants impaired in five early nodulation genes including putative orthologs of NSP1 and NSP2. Symbiosis 68, 129–144. doi: 10.1007/s13199-016-0382-2
Spitzer, M., Wildenhain, J., Rappsilber, J., Tyers, M., et al. (2014). BoxPlotR: a web tool for generation of box plots. Nature methods 11, 121–122.
Stanley, L., Yuan, Y.-W. (2019). Transcriptional regulation of carotenoid biosynthesis in plants: So many regulators, so little consensus. Front. Plant Sci. 10, 1017. doi: 10.3389/fpls.2019.01017
van Velzen, R., Doyle, J. J., Geurts, R. (2019). A resurrected scenario: single gain and massive loss of nitrogen-fixing nodulation. Trends Plant Sci. 24, 49–57. doi: 10.1016/j.tplants.2018.10.005
van Velzen, R., Holmer, R., Bu, F., Rutten, L., van Zeijl, A., Liu, W., et al. (2018). Comparative genomics of the nonlegume Parasponia reveals insights into evolution of nitrogen-fixing rhizobium symbioses. Proc. Natl. Acad. Sci. U. S. A. 115, E4700–E4709. doi: 10.1073/pnas.1721395115
van Zeijl, A., Wardhani, T. A., Seifi Kalhor, M., Rutten, L., Bu, F., Hartog, M., et al. (2018). CRISPR/cas9-mediated mutagenesis of four putative symbiosis genes of the tropical tree parasponia andersonii reveals novel phenotypes. Front. Plant Sci. 9, 284. doi: 10.3389/fpls.2018.00284
Votta, C., Fiorilli, V., Haider, I., Wang, J. Y., Balestrini, R., Petřík, I., et al. (2022). Zaxinone synthase controls arbuscular mycorrhizal colonization level in rice. Plant journal: Cell Mol. Biol. 111, 1688–1700. doi: 10.1111/tpj.v111.6
Wakabayashi, T., Hamana, M., Mori, A., Akiyama, R., Ueno, K., Osakabe, K., et al. (2019). Direct conversion of carlactonoic acid to orobanchol by cytochrome P450 CYP722C in strigolactone biosynthesis. Science advances 5, eaax9067.
Wakabayashi, T., Shida, K., Kitano, Y., Takikawa, H., Mizutani, M., Sugimoto, Y., et al. (2020). CYP722C from Gossypium arboreum catalyzes the conversion of carlactonoic acid to 5-deoxystrigol. Planta 251, 97.
Wardhani, T. A. K., Roswanjaya, Y.P., Dupin, S., Li, H., Linders, S., Hartog, M., et al. (2019). Transforming, genome editing and phenotyping the nitrogen-fixing tropical cannabaceae tree parasponia andersonii. J. Vis. Exp. 150, e59971. doi: 10.3791/59971
Waters, M. T., Gutjahr, C., Bennett, T., Nelson, D. C. (2017). Strigolactone signaling and evolution. Annual review of plant biology 68, 291–322.
Xiao, A., Yu, H., Fan, Y., Kang, H., Ren, Y., Huang, X., et al. (2020). Transcriptional regulation of NIN expression by IPN2 is required for root nodule symbiosis in Lotus japonicus. New Phytol. 227, 513–528. doi: 10.1111/nph.v227.2
Xue, L., Cui, H., Buer, B., Vijayakumar, V., Delaux, PM., Junkermann, S., et al. (2015). Network of GRAS transcription factors involved in the control of arbuscule development in Lotus japonicus. Plant Physiol. 167, 854–871. doi: 10.1104/pp.114.255430
Yano, K., Yoshida, S., Müller, J., Singh, S., Banba, M., Vickers, K., et al. (2008). CYCLOPS, a mediator of symbiotic intracellular accommodation. Proc. Natl. Acad. Sci. United States America 105, 20540–20545. doi: 10.1073/pnas.0806858105
Yuan, K., Zhang, H., Yu, C., Luo, N., Yan, J., Zheng, S., et al. (2023). Low phosphorus promotes NSP1-NSP2 heterodimerization to enhance strigolactone biosynthesis and regulate shoot and root architecture in rice. Mol. Plant 16, 1811–1831. doi: 10.1016/j.molp.2023.09.022
Keywords: arbuscular mycorrhiza, nodulation, Parasponia, NSP2, CYCLOPS, phytoene synthase, carotenoids
Citation: Alhusayni S, Kersten N, Huisman R, Geurts R and Klein J (2024) Ectopic expression of the GRAS-type transcriptional regulator NSP2 in Parasponia triggers contrasting effects on symbioses. Front. Plant Sci. 15:1468812. doi: 10.3389/fpls.2024.1468812
Received: 22 July 2024; Accepted: 03 October 2024;
Published: 30 October 2024.
Edited by:
Andrea Genre, University of Turin, ItalyReviewed by:
Nicolas Frei Dit Frey, Université de Toulouse, FranceCopyright © 2024 Alhusayni, Kersten, Huisman, Geurts and Klein. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rene Geurts, cmVuZS5nZXVydHNAd3VyLm5s; Joël Klein, am9lbC5rbGVpbmpvZWxAZ21haWwuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.