- Division of Agricultural Microbiology, National Institute of Agricultural Science, Rural Development Administration, Wanju, Republic of Korea
Induced systemic resistance (ISR) is a crucial concept in modern agriculture, explaining plant defense mechanisms primed by rhizosphere stimuli and activated by subsequent infections. Biological factors contributing to ISR generally include plant growth-promoting microbes3 (PGPM). Bacillus spp., Pseudomonas spp., and Trichoderma spp. have been extensively studied for their plant growth-promoting characteristics and ISR effect against above-ground pathogens and insect infestations. These phenomena elucidate the bottom-up effects of how beneficial rhizosphere microbes help plants resist above-ground attacks. Conversely, soil microbiome analysis in the rhizosphere of plants infected by above-ground pathogens has shown increased beneficial microbes in the soil, a phenomenon termed 'soil legacy effects'. This represents the top-down effects of above-ground attackers on plants' rhizosphere environments. Interestingly, recent studies have shown that above-ground stimuli not only recruit PGPM in the rhizosphere but also that these PGPM influence plant defense responses against subsequent pathogen infections. This can be seen as a four-step plant defense mechanism involving above-ground attackers, host plants, rhizosphere microbes, and subsequent attacks. This represents an active defense mechanism that overcomes the limitations of sessile plants. This review summarizes plant ISR mechanisms in terms of triple inter-organism interactions and provides molecular evidence for each step.
Introduction
Similar to the T memory cells in animal immune systems that enhance defense resistance against secondary infections, plants possess a comparable immune strategy known as 'defense priming’. Defense priming is a plant-specific immune strategy for faster and stronger responses to secondary attacks (Hönig et al., 2023). To initiate plant defense priming, primary stimuli such as biological (pathogenic and non-pathogenic microbes), chemical or physical stimuli are required (Jung et al., 2018, 2020). Once the plant's defense mechanism is primed, it develops systemic resistance, which is divided into systemic acquired resistance (SAR) and induced systemic resistance (ISR). SAR and ISR are generally known to have defense responses dependent on the salicylic acid (SA) pathway and the jasmonic acid (JA)/ethylene (ET) pathways, respectively (Choudhary et al., 2007). However, current studies demonstrate that both SA and JA/ET signaling are involved in inducing ISR (Yu et al., 2022).
ISR is primarily primed by non-pathogenic microbes in the rhizosphere, which are regarded as plant growth-promoting microbes (PGPM) (Kloepper, 1978; Hyakumachi, 1994). Numerous studies have shown that PGPM such as Bacillus spp. and Pseudomonas spp. promote plant growth and trigger ISR, focusing on bottom-up effects and elucidating the influence of below-ground microbes on above-ground plant immune enhancement (Valenzuela-Soto et al., 2010; Beneduzi et al., 2012; Xie et al., 2018).
Furthermore, ongoing microbiome analyses over recent decades have improved our understanding of the interactions between plants and PGPM. Interestingly, above-ground stimuli in plants result in microbiome reshaping in their rhizosphere, recruiting beneficial microbes into their root environment (Lee et al., 2012; Kong et al., 2016; Wang et al., 2021). For example, above-ground whitefly infestation in pepper has been shown to increase colonization of the rhizosphere with beneficial microbes such as Pseudomonas spp. Indeed, Pseudomonas spp. isolated from pepper roots after above-ground whitefly-infestation showed higher insect-killing activity in Galleria mellonella during an in vitro mortality assay (Kong et al., 2016). Additionally, aphid infestation in potato (Solanum tuberosum L.) leaves leads to lower hatching rates of the endoparasitic nematode Globodera pallida in the soil (Hoysted et al., 2018). These phenomena could be regarded as top-down effects, highlighting the influence from the above-ground to the below-ground environment.
Considering both top-down and bottom-up effects, plants actively recruit beneficial microbes into their rhizosphere and utilize them to acquire ISR and resist further pathogen attacks (Wang et al., 2021; Zhu et al., 2021). This can be considered a sessile plant's active defense strategy employing other organisms for their uses. Therefore, in this review, we discuss how plants recruit beneficial microbes in their rhizosphere and use them for defense resistance development, considering both top-down and bottom-up effects.
Top-down effect: above-ground stimulation alters below-ground microbes
Plants are exposed to both abiotic and biotic stimuli, occurring both above and below ground. How do plant roots perceive signals from above-ground external stimuli, and how do these signals influence below-ground interactions? Recent studies have clearly demonstrated that top-down effects from above-ground bacterial infection or pest infestation can alter the below-ground microbiome (Kong et al., 2016; Wang et al., 2021).
Above-ground signal perception and long-distance signal transduction
Plants have conserved immune signaling initiated by the recognition of microbe-associated molecular patterns (MAMPs) from external microbes (Muthamilarasan and Prasad, 2013).
Pattern recognition receptors (PRRs) localized on the plant cell surface can recognize MAMPs of pathogenic or non-pathogenic microbes, triggering MAMP-triggered immunity (MTI) in plants (Boller and Felix, 2009; Muthamilarasan and Prasad, 2013). Once PRRs recognize MAMPs, they activate a mitogen-activated protein kinase (MAPK) cascade, which eventually activates WRKY family transcription factors (Pandey and Somssich, 2009; Muthamilarasan and Prasad, 2013). WRKY transcription factors induce the biosynthesis of defense-related phytohormones, such as salicylic acid (SA), jasmonic acid (JA), and ethylene (ET), and promote the production and secretion of antimicrobial compounds into vacuoles (Muthamilarasan and Prasad, 2013).
Plant hormones like SA, JA, ABA, ET, and cytokinin play roles in plant defense resistance. Notably, JA and cytokinin are reported to be mobile among leaves, from shoot to root, and root to shoot (Figure 1A) (Soler et al., 2013). Although SA does not move among leaves, Methyl salicylate (MeSA) moves from cell to cell among leaves or through phloem and is converted to SA in distal leaves (Park et al., 2007) (Figure 1A). Among other hormones, ABA is mobile among leaves and from root to shoot, while ET has been reported to move from root to shoot (Soler et al., 2013) (Figure 1A). As ET is gaseous hormone, it can diffuse throughout the plant. However, this diffusion refer to general movement rather than directional transport with specific orientation (Binder, 2020). These hormones could potentially induce local defense responses again upon reaching a new location.
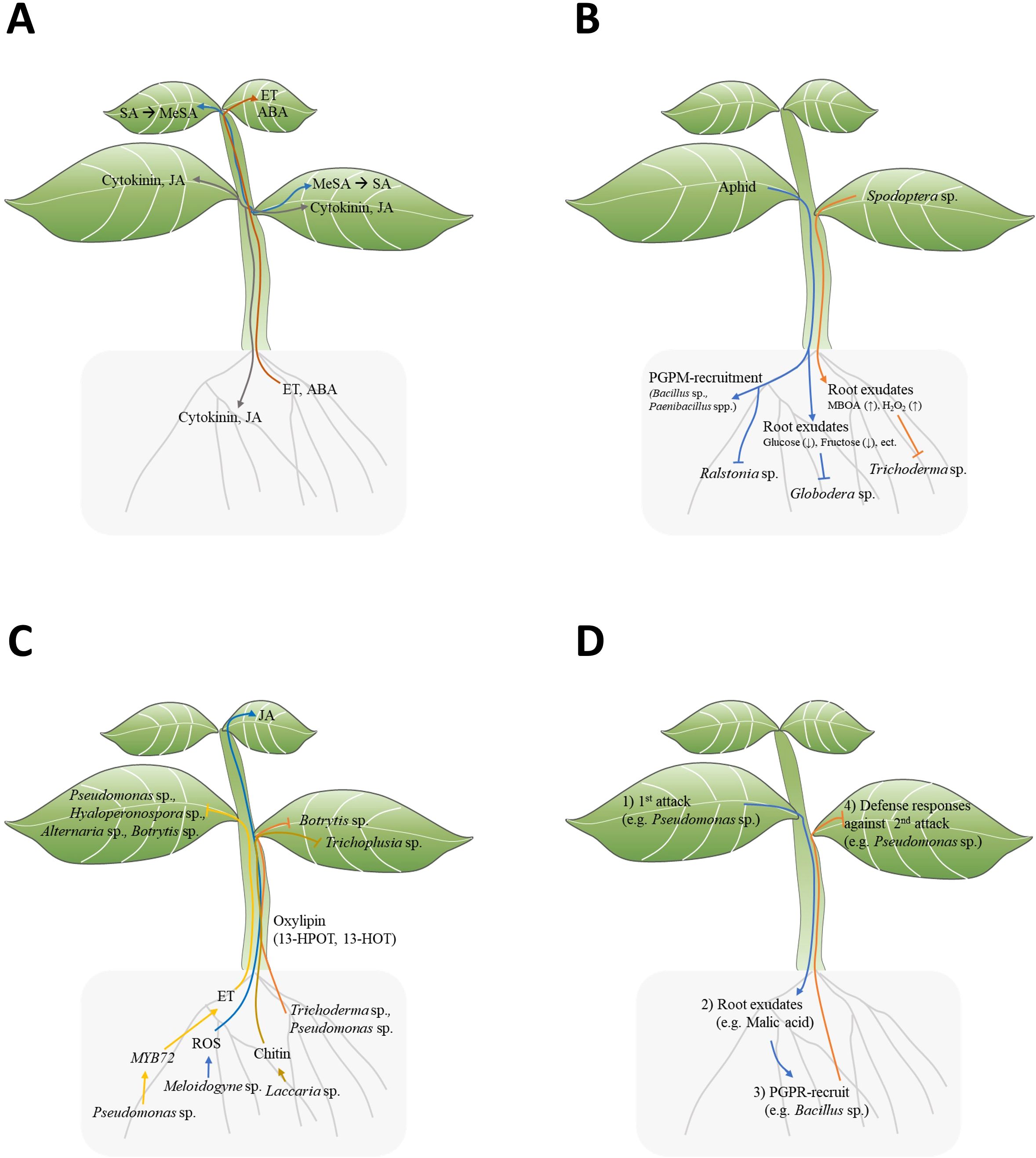
Figure 1. Signal transduction between plant above-ground and below-ground. (A) Mobile signal transduction of plant hormones: SA, MeJA, JA, ET, ABA, Cytokinin. (SA, salicylic acid; MeJA, methyl jasmonate; JA, jasmonic acid; ET, ethylene; ABA, abscisic acid). (B) Top-down effect of insect infestation on leaves affecting the below-ground environment. (C) Bottom-up effect of PGPM on above-ground plant defense responses. (D) Triple interactions among above-ground herbivores/pathogens (1st attack), host plants, beneficial rhizosphere microbes, and herbivores/pathogens (2nd attack) in the context of ISR.
In addition to hormones, several long-distance mobile signals have been identified as key players in SAR. These include glycolipids, which are lipid-based molecules acting as signaling compounds in plant defense responses (Chaturvedi et al., 2008). Another important signal is azelaic acid, a dicarboxylic acid that primes plants for enhanced defense activation and contributes to SAR signal generation (Jung et al., 2009). Glycerol-3-phosphate (G3P) also plays a crucial role in SAR as a metabolite synthesized in the cytosol and chloroplasts (Chanda et al., 2011; Shine et al., 2019). Pipecolic acid, a non-protein amino acid, acts as a critical regulator of plant systemic immunity (Návarová et al., 2012). It accumulates in both local and distal tissues following pathogen infection and is essential for SAR activation and priming of plant defenses (Návarová et al., 2012). These diverse molecules work in concert to establish and maintain SAR, enhancing the plant's ability to defend against subsequent pathogen attacks.
The above examples explain mobile phytohormones and small molecules that transfer and function in distal locations. Signals that migrate to the roots could potentially alter the composition of root exudates. These studies support that plant root exudates function to reshape the rhizosphere microbiome, thereby directly affecting belowground evaders or enabling the utilization of surrounding beneficial substances.
Above-ground stimuli alter root exudation and reshape the rhizosphere microbiome
Above-ground stimuli in plants change below-ground root exudation components, which directly and indirectly lead to the reshaping of the rhizosphere, providing feedback to the plants (Lee et al., 2012; Kim et al., 2016; Hoysted et al., 2018; Friman et al., 2021). Plants with above-ground stimulation may selectively attract specific microorganisms into their rhizosphere. Interestingly, phloem-feeding aphid infestation in pepper (Capsicum annuum) leaves attracts beneficial bacteria, such as Bacillus subtilis and Paenibacillus spp., but not pathogenic bacteria like Ralstonia solanacearum, to their roots (Figure 1B; Table 1) (Lee et al., 2012; Kim et al., 2016). These results suggest that plants actively and selectively reshape the below-ground microbiome in response to above-ground stimuli. How do plants selectively recruit beneficial microbes to the rhizosphere? For example, Pst DC3000 infection in Arabidopsis leaves led to the secretion of malic acid from their roots (Table 1) (Rudrappa et al., 2008). Interestingly, B. subtilis strain FB17 exhibits malic acid chemotaxis, suggesting that malic acid may be a key factor in recruiting B. subtilis strain FB17 (Rudrappa et al., 2008). Finally, the B. subtilis strain FB17 induced systemic resistance against Pst DC3000, demonstrating how sessile plants actively trigger defense responses using surrounding beneficial bacteria (Figure 1D) (Rudrappa et al., 2008).
Above-ground attacks on plant leaves may influence plant root exudate. Recent studies have shown that root exudate components change following above-ground infections (Doornbos et al., 2012; Kim et al., 2016). Root exudation profiles in Pseudomonas syringae pv. tomato (Pst)-infected Arabidopsis exhibited significantly higher levels of amino acids, nucleotides, and long-chain organic acids (LCOAs) (C > 6) and lower levels of sugars, alcohols, and short-chain organic acids (SCOAs) (C ≤ 6) (Table 1) (Yuan et al., 2018). Interestingly, since exogenous amino acids and LCOA application to plants showed disease-suppressive responses, the pst-induced root exudation may function as plant’s defense enhancement (Yuan et al., 2018). Similarly, aphid-infested potato leaves also showed reduced sugar contents in root exudates (Hoysted et al., 2018). Root exudates from aphid-infested potato (Solanum tuberosum L.) showed reduced glucose and fructose contents, resulting in lower hatching rates of below-ground endoparasitic nematode Globodera pallida (Table 1; Figure 1B) (Hoysted et al., 2018). However, sugar supplementation did not recover the hatching potential of G. pallida, indicating that root exudation may involve other important factors for egg hatching (Figure 1B) (Hoysted et al., 2018). In the case of chewing insect stimuli, leaf area and biomass decrease by Spodoptera frugiperda-infested maize leaves correlated with changes in root exudate compounds (Adame-Garnica et al., 2023). The degree of S. frugiperda infestation modulated the root exudation contents, non-structural carbohydrates (NSCs), 6-methoxy-2-benzoxaxolin-2-one (MBOA), and the reactive oxygen species H2O2, leading to the growth inhibition of the rhizosphere fungus Trichoderma atroviride (Table 1; Figure 1B) (Adame-Garnica et al., 2023). This result clearly demonstrates the top-down effect of above-ground insect attacks, showing how they influence root exudation and, consequently, effect on rhizosphere fungus.
Molecular evidence of root exudate-mediated rhizosphere microbiota shaping and disease resistance in plants
Recent studies have provided molecular evidence of changes in root exudates. For example, JA is a critical hormone involved in plant defense responses, has been shown to influence root exudation. Root exudates from Arabidopsis JA mutants, such as myc2 and med25, showed reduced levels of asparagine, ornithine, and tryptophan (Carvalhais et al., 2015). Additionally, these myc2 mutants had an increased abundance of Streptomyces, Bacillus, and Lysinibacillus taxa in the rhizosphere, suggesting that JA-dependent root exudates alterations affect microbiome communities (Carvalhais et al., 2015).
A recent study indicated that the ripening-inhibitor (RIN) transcription factor plays a role in assembling disease-suppressive rhizosphere microbiota by altering root exudates. RIN tomato mutant exhibited lower concentration of 3-hydroxyflavone and riboflavin in their root exudates, leading to reduction in pathogen-suppressing Actinobacteria (Streptomyces) in the rhizosphere (Yang et al., 2023). The disease suppressiveness of the rin plant microbiome was restored by complementing with 3-hydroxyflavone and riboflavin (Yang et al., 2023).
Interestingly, plant root exudates also changed in response to fusaric acid (FA), produced by Fusarium oxysporum (Jin et al., 2024). FA differently altered the root exudate components of two tomato cultivars; F. oxysporum f. sp. lycopersici resistant Z19 and susceptible D72. FA enhanced colonization of disease-suppressive bacteria, Sphingomonas sp., in the resistant cultivar Z19 (Jin et al., 2024). These microbiome changes provided feedback, leading to defense resistance to F. oxysporum in the resistant cultivar Z19 (Jin et al., 2024).
Bottom-up effect: signal recognition and transduction from root to leaf
For a mutually beneficial relationship between plants and microbes, plants need to recognize certain microbes as symbiotic friends rather than enemies to fight. However, MTI is induced regardless of whether microbes are beneficial or harmful. Beneficial microbes, therefore, need to suppress or evade plant immune responses to establish a symbiotic relationship (Yu et al., 2019b). How do beneficial microbes colonize the rhizosphere while suppressing the plant immune system? Some evidence suggests that beneficial microbes suppress plant immunity in roots, facilitating their colonization in the rhizosphere.
Symbiotic relationship by MTI suppression
Flg22 is highly conserved domain of bacterial flagellin, recognized as a MAMP by the plant FLS2 receptor, resulting in MTI (Felix et al., 1999). Pseudomonas capeferrum WCS358, a well-known PGPM (Meziane et al., 2005), suppresses flg22-mediated Arabidopsis root immunity by producing gluconic acid and shows enhanced colonization in Arabidopsis roots compared to gluconic acid production mutants (pqqF::Tn5 and cyoB::Tn5) (Table 1) (Yu et al., 2019a). Another study, using a high-throughput transposon (Tn-Seq) screening system in Pseudomonas sp. WCS365, found that morA and spuC mutants induced MTI, enhanced biofilms formation, and inhibited the growth of Arabidopsis compared to the wild type (Table 1) (Liu et al., 2018). MorA (encoding phosphodiesterase) and SpuC (encoding putrescine aminotransferase) are related to the suppression of biofilm formation. This biofilm inhibition may be one of the microbes' strategies for evading MTI and fitting within the rhizosphere.
Considering that ISR occurs through the recognition of microbial-derived factors by the plant, the symbiosis between plants and microbes is crucial (Zamioudis and Pieterse, 2012). However, current studies suggest that some ISR responses might not require a symbiotic association (Vishwanathan et al., 2020). For example, the ectomycorrhizal fungus Laccaria bicolor can trigger ISR against the insect Trichoplusia ni in Arabidopsis, a nonmycorrhizal plant (Vishwanathan et al., 2020). Therefore, the crucial factor may lie more in how effectively the plant responds to ISR elicitors secreted by microbes, rather than the duration of the plant-microbes symbiotic relationship.
Microbe elicitor-plant recognition and signal transduction
Plants may recognize signals from rhizosphere microbes for inducing ISR. Then, what happens in plant roots for initiating ISR signaling?
Pseudomonas fluorescens WCS417r is known for inducing ISR against a broad spectrum of pathogens in Arabidopsis. However, P. fluorescence WCS417r showed impaired ISR in myb72-1 and myb72-2 Arabidopsis mutant against Pseudomonas syringae pv tomato, Hyaloperonospora parasitica, Alternaria brassicicola, and Botrytis cinerea (Table 1; Figure 1C) (Van der Ent et al., 2008). Indeed, the root-specific transcription factor MYB72 appears to be a convergence node, as it is essential for early signaling in ISR induction by P. fluorescence WCS417r and Trichoderma asperellum T34 (Van der Ent et al., 2008; Segarra et al., 2009). The author suggest MYB72 is upstream of ET signaling, as WCS417r activated MYB72 in ethylene-insensitive ein2-1 plants, and exogeneous ethylene precursor 1-aminocyclopropane-1-carboxylate (ACC) induced ISR responses in myb72-1 mutants (Figure 1C) (Van der Ent et al., 2008).
Chitin, one of the MAMPs of microbes, has been shown to induce ISR (Vishwanathan et al., 2020; Takagi et al., 2022). L.bicolor triggers ISR in Arabidopsis against Trichoplusia ni (Figure 1C) (Vishwanathan et al., 2020). Molecular evidence suggest that chitin derived from L.bicolor is an ISR-inducing molecules, as heat-killed L. bicolor or chitin also trigger ISR, but the chitin receptor mutant cerk1-2 could not trigger ISR by L.bicolor or chitin (Table 1) (Vishwanathan et al., 2020). Another study showed that chitin-induced ISR against Bipolaris oryzae in rice is related to down-regulated cytokinin signaling, resulting in alterations of cell-wall components (Table 1) (Takagi et al., 2022). Therefore, chitin-induced ISR is mediated by perturbation in cell-wall biogenesis in leaves.
Regarding PGPM interactions with plant roots, the root hair is the first part of the plant to interact with microbes. Root hair-specific syntaxin gene SYP123 is suggested to be necessary for PGPM-triggered ISR, as ISR marker genes (PR1, MYC2, and PDF1.2) did not increase in syp123 Arabidopsis mutants in response to beneficial Pseudomonas species (Rodriguez-Furlán et al., 2016; Zhu et al., 2022).
In the case of root-to-shoot communications, the root-knot nematode (RNK) Meloidogyne incognita induces the transition of ROS signals from root to leaves of tomato, resulting in JA accumulation in leaves (Figure 1C) (Wang et al., 2019). The M. incognita-induced signal transduction involves RBOH1, GLR3.5, and MPK1/2-dependent JA accumulation, as JA accumulation in leaves was abolished in grafting experiments with scion of the mutant GLUTAMATE RECEPTOR-LIKE 3.5 (GLR3.5), RESPIRATORY BURST OXIDASE HOMOLOG1 (RBOH1), and plant silenced for mitogen-activated kinases1 (MPK1) or MPK2 (Wang et al., 2019). The RNK M. incognita induced-JA accumulation in leaves transfers to the roots, triggering resistance to RNK (Table 1) (Wang et al., 2019).
Oxylipins are oxidized lipid signals that regulate plant physiological responses to abiotic and biotic stress, including defense responses against pathogens and insects (Wang et al., 2019). Several non-jasmonate oxylipins have been identified as crucial regulators for ISR in various plant species. In maize, two xylem-mobile oxylipins, 12-oxo-phytodienoic acid (12-OPDA) and an γ-ketol of octadecadienoic acid (KODA), play important roles in ISR (Wang et al., 2019; Carella, 2020). Additionally, two γ-ketols, 12-Oxo-9-hydroxy-10(E)-octadecenoic acid (9,12-KOMA) and 12-Oxo-9-hydroxy-10(E),15(Z)-octadecadienoic acid (9,12-KODA), have been identified as ISR priming agents in maize (Wang et al., 2020). The small secreted protein Sm1 from PGPM virens triggers ISR regulating 12-OPDA and KODA biosynthesis in maize plants (Table 1) (Wang et al., 2019). Similarly, Pseudomonas putida BTP1 induced ISR against Botrytis cinerea by accumulating two antifungal oxylipin, free 13-hydroperoxy-octadecatrienoic (13-HPOT) and 13-hydroxy-octadecatrienoic acids (13-HOT), in tomato (Figure 1C; Table 1) (Mariutto et al., 2011).
Recently, microRNAs have also been suggested as mobile signals for plant-microbe interactions (Xie et al., 2019). Bacillus velezensis FZB42, which is reported to induce ISR, showed that four miRNAs, zma-miR169a-5p, zma-miR169c-5p, zma-miR169i-5p, and zma-miR395b-5p, belonging to the miR169 family, are associated with triggering ISR by regulating nuclear factor Y transcription factor in maize (Table 1) (Xie et al., 2019).
Perspectives
This study explored the triple interaction among above-ground herbivores/pathogen (1st attack), host plants, beneficial rhizosphere microbes PGPM, and herbivores/pathogen (2nd attack) in the context of ISR. ISR is triggered by beneficial microbes interacting with plant roots, enhancing the plant's defenses against pathogens and pests. In this review, we focused on how PGPM enhance systemic resistance in plants through ISR and activate defense mechanisms against herbivores and pathogens. Many previous studies have shown that plants can actively assemble beneficial microbes in the rhizosphere according to their needs. Even in the absence of a symbiotic associations, these microbes can induce ISR in plants.
There are naturally occurring complex interactions: (1) plant leaves are exposed to pathogens and insects (above ground microbe’s first attack-plant interaction), and (2) plant roots interact with various microbes in their rhizosphere (below ground microbes-plant interaction). We propose a concept of 'triple interactions' by adding the plant's active immune response, specifically ISR, for a second above-ground attack. This raises the question: How can we apply these complex plant immune systems to agriculture?
Since plants can actively gather the microbes they need in the rhizosphere, it is possible to study the types of beneficial microbes that can control pests and pathogens that are currently difficult to manage. For instance, by inoculating plants with target pests or pathogens and then observing the changes in the microbial community over time, it may be possible to identify microbes that the plant recruits to combat the target pest/pathogen. Alternatively, since root exudates released under specific stress conditions ultimately gather beneficial microbes, identifying the root exudate components that attract PGPM would also be beneficial for future agricultural applications.
Unlike animals, plants do not have T-cells and cannot be vaccinated in the traditional sense. However, by actively recruiting beneficial microbes to the rhizosphere after an attack, plants can enhance their systemic resistance, functioning similarly to a vaccine. Of course, there are also cases where microbial communities harmful to the plant assemble in response to an above-ground attack. Therefore, future studies could investigate whether there is a threshold beyond which a stressed plant's ability to recruit beneficial microbes becomes ineffective. If this threshold is exceeded, it could potentially neutralize the plant's capacity to assemble beneficial microbial communities. Moreover, it is important to acknowledge that not all plants can acquire immunity against every pest or pathogen they encounter. This limitation means that the concept under discussion may not apply universally. Further research is needed to understand the boundaries of a plant's ability to recruit beneficial microbes under various stress conditions and how these limitations impact overall plant health and resistance.
Author contributions
JJ: Writing – original draft, Writing – review & editing. SA: Writing – review & editing. D-HK: Writing – review & editing. MR: Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was supported by grants from the Agenda Project (Project No. PJ01727502) of the Rural Development Administration (RDA), Republic of Korea.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Adame-Garnica, S. G., Contreras-Cornejo, H. A., Luna-Cruz, A., García-Pineda, E., Macías-Rodríguez, L. (2023). Root exudates from maize plants with high levels of foliar herbivory alters beneficial plant fungus growth. Rhizosphere 26, 100703. doi: 10.1016/j.rhisph.2023.100703
Beneduzi, A., Ambrosini, A., Passaglia, L. M. (2012). Plant growth-promoting rhizobacteria (PGPR): Their potential as antagonists and biocontrol agents. Genet. Mol. Biol. 35(4, 1044–1051. doi: 10.1590/S1415-47572012000600020
Binder, B. M. (2020). Ethylene signaling in plants. J. Biol. Chem. 295, 7710–7725. doi: 10.1074/jbc.REV120.010854
Boller, T., Felix, G. (2009). A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60, 379–406. doi: 10.1146/annurev.arplant.57.032905.105346
Carella, P. (2020). Xylem-mobile oxylipins are critical regulators of induced systemic resistance in maize. Plant Cell 32, 13–14. doi: 10.1105/tpc.19.00924
Carvalhais, L. C., Dennis, P. G., Badri, D. V., Kidd, B. N., Vivanco, J. M., Schenk, P. M. (2015). Linking jasmonic acid signaling, root exudates, and rhizosphere microbiomes. Mol. Plant Microbe Interact. 28, 1049–1058. doi: 10.1094/MPMI-01-15-0016-R
Chanda, B., Xia, Y., Mandal, M. K., Yu, K., Sekine, K. T., Gao, Q. M., et al. (2011). Glycerol-3-phosphate is a critical mobile inducer of systemic immunity in plants. Nat. Genet. 43, 421–427. doi: 10.1038/ng.798
Chaturvedi, R., Krothapalli, K., Makandar, R., Nandi, A., Sparks, A. A., Roth, M. R., et al. (2008). Plastid omega3-fatty acid desaturase-dependent accumulation of a systemic acquired resistance inducing activity in petiole exudates of Arabidopsis thaliana is independent of jasmonic acid. Plant J. 54, 106–117. doi: 10.1111/j.1365-313X.2007.03400.x
Choudhary, D. K., Prakash, A., Johri, B. N. (2007). Induced systemic resistance (ISR) in plants: mechanism of action. Indian J. Microbiol. 47, 289–297. doi: 10.1007/s12088-007-0054-2
Doornbos, R. F., van Loon, L. C., Bakker, P. A. H. M. (2012). Impact of root exudates and plant defense signaling on bacterial communities in the rhizosphere. A review. Agron. Sustain. Dev. 32, 227–243. doi: 10.1007/s13593-011-0028-y
Felix, G., Duran, J. D., Volko, S., Boller, T. (1999). Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 18, 265–276. doi: 10.1046/j.1365-313X.1999.00265.x
Friman, J., Karssemeijer, P. N., Haller, J., de Kreek, K., van Loon, J. J. A., Dicke, M. (2021). Shoot and root insect herbivory change the plant rhizosphere microbiome and affects cabbage-insect interactions through plant-soil feedback. New Phytol. 232, 2475–2490. doi: 10.1111/nph.v232.6
Hönig, M., Roeber, V. M., Schmülling, T., Cortleven, A. (2023). Chemical priming of plant defense responses to pathogen attacks. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1146577
Hoysted, G. A., Bell, C. A., Lilley, C. J., Urwin, P. E. (2018). Aphid colonization affects potato root exudate composition and the hatching of a soil borne pathogen. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01278
Hyakumachi, M. (1994). Plant-growth-promoting fungi from turfgrass rhizosphere with potential for disease suppression. Soil Microorg. 44, 53–68. doi: 10.18946/JSSM.44.0_53
Jin, X., Jia, H., Ran, L., Wu, F., Liu, J., Schlaeppi, K., et al. (2024). Fusaric acid mediates the assembly of disease-suppressive rhizosphere microbiota via induced shifts in plant root exudates. Nat. Commun. 15, 5125. doi: 10.1038/s41467-024-49218-9
Jung, H. W., Tschaplinski, T. J., Wang, L., Glazebrook, J., Greenberg, J. T. (2009). Priming in systemic plant immunity. Science 324, 89–91. doi: 10.1126/science.1170025
Jung, J., Kim, S.-K., Jung, S.-H., Jeong, M.-J., Ryu, C.-M. (2020). Sound vibration-triggered epigenetic modulation induces plant root immunity against ralstonia solanacearum. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.01978
Jung, J., Kim, S. K., Kim, J. Y., Jeong, M. J., Ryu, C. M. (2018). Beyond chemical triggers: evidence for sound-evoked physiological reactions in plants. Front. Plant Sci. 9, 25. doi: 10.3389/fpls.2018.00025
Kim, B., Song, G. C., Ryu, C. M. (2016). Root exudation by aphid leaf infestation recruits root-associated paenibacillus spp. to lead plant insect susceptibility. J. Microbiol. Biotechnol. 26, 549–557. doi: 10.4014/jmb.1511.11058
Kloepper, J. W. (1978). “Plant growth-promoting rhizobacteria on radishes,” in Proc. of the 4th Internet. Conf. on Plant Pathogenic Bacter, Station de Pathologie Vegetale et Phytobacteriologie, Angers, France (INRA).
Kong, H. G., Kim, B. K., Song, G. C., Lee, S., Ryu, C. M. (2016). Aboveground whitefly infestation-mediated reshaping of the root microbiota. Front. Microbiol. 7, 1314. doi: 10.3389/fmicb.2016.01314
Lee, B., Lee, S., Ryu, C.-M. (2012). Foliar aphid feeding recruits rhizosphere bacteria and primes plant immunity against pathogenic and non-pathogenic bacteria in pepper. Ann. Bot. 110, 281–290. doi: 10.1093/aob/mcs055
Liu, Z., Beskrovnaya, P., Melnyk, R. A., Hossain, S. S., Khorasani, S., O'Sullivan, L. R., et al. (2018). A genome-wide screen identifies genes in rhizosphere-associated pseudomonas required to evade plant defenses. mBio 9. doi: 10.1128/mBio.00433-18
Mariutto, M., Duby, F., Adam, A., Bureau, C., Fauconnier, M. L., Ongena, M., et al. (2011). The elicitation of a systemic resistance by Pseudomonas putida BTP1 in tomato involves the stimulation of two lipoxygenase isoforms. BMC Plant Biol. 11, 29. doi: 10.1186/1471-2229-11-29
Meziane, H., Van Der Sluis, I., Van Loon, L. C., Höfte, M., Bakker, P. A. (2005). Determinants of Pseudomonas putida WCS358 involved in inducing systemic resistance in plants. Mol. Plant Pathol. 6, 177–185. doi: 10.1111/j.1364-3703.2005.00276.x
Muthamilarasan, M., Prasad, M. (2013). Plant innate immunity: an updated insight into defense mechanism. J. Biosci. 38, 433–449. doi: 10.1007/s12038-013-9302-2
Návarová, H., Bernsdorff, F., Döring, A. C., Zeier, J. (2012). Pipecolic acid, an endogenous mediator of defense amplification and priming, is a critical regulator of inducible plant immunity. Plant Cell 24, 5123–5141. doi: 10.1105/tpc.112.103564
Pandey, S. P., Somssich, I. E. (2009). The role of WRKY transcription factors in plant immunity. Plant Physiol. 150, 1648–1655. doi: 10.1104/pp.109.138990
Park, S.-W., Kaimoyo, E., Kumar, D., Mosher, S., Klessig, D. F. (2007). Methyl salicylate is a critical mobile signal for plant systemic acquired resistance. Science 318, 113–116. doi: 10.1126/science.1147113
Rodriguez-Furlán, C., Salinas-Grenet, H., Sandoval, O., Recabarren, C., Arraño-Salinas, P., Soto-Alvear, S., et al. (2016). The root hair specific SYP123 regulates the localization of cell wall components and contributes to rizhobacterial priming of induced systemic resistance. Front. Plant Sci. 7, 1081. doi: 10.3389/fpls.2016.01081
Rudrappa, T., Czymmek, K. J., Paré, P. W., Bais, H. P. (2008). Root-secreted Malic acid recruits beneficial soil bacteria. Plant Physiol. 148, 1547–1556. doi: 10.1104/pp.108.127613
Segarra, G., van der Ent, S., Trillas, I., Pieterse, C. M. J. (2009). MYB72, a node of convergence in induced systemic resistance triggered by a fungal and a bacterial beneficial microbe. Plant Biol. 11, 90–96. doi: 10.1111/j.1438-8677.2008.00162.x
Shine, M. B., Gao, Q.-m., Chowda-Reddy, R. V., Singh, A. K., Kachroo, P., Kachroo, A. (2019). Glycerol-3-phosphate mediates rhizobia-induced systemic signaling in soybean. Nat. Commun. 10, 5303. doi: 10.1038/s41467-019-13318-8
Soler, R., Erb, M., Kaplan, I. (2013). Long distance root–shoot signalling in plant–insect community interactions. Trends Plant Sci. 18, 149–156. doi: 10.1016/j.tplants.2012.08.010
Takagi, M., Hotamori, K., Naito, K., Matsukawa, S., Egusa, M., Nishizawa, Y., et al. (2022). Chitin-induced systemic disease resistance in rice requires both OsCERK1 and OsCEBiP and is mediated via perturbation of cell-wall biogenesis in leaves. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1064628
Valenzuela-Soto, J. H., Estrada-Hernández, M. G., Ibarra-Laclette, E., Délano-Frier, J. P. (2010). Inoculation of tomato plants (Solanum lycopersicum) with growth-promoting Bacillus subtilis retards whitefly Bemisia tabaci development. Planta 231, 397–410. doi: 10.1007/s00425-009-1061-9
Van der Ent, S., Verhagen, B. W., Van Doorn, R., Bakker, D., Verlaan, M. G., Pel, M. J., et al. (2008). MYB72 is required in early signaling steps of rhizobacteria-induced systemic resistance in Arabidopsis. Plant Physiol. 146, 1293–1304. doi: 10.1104/pp.107.113829
Vishwanathan, K., Zienkiewicz, K., Liu, Y., Janz, D., Feussner, I., Polle, A., et al. (2020). Ectomycorrhizal fungi induce systemic resistance against insects on a nonmycorrhizal plant in a CERK1-dependent manner. New Phytol. 228, 728–740. doi: 10.1111/nph.v228.2
Wang, G., Hu, C., Zhou, J., Liu, Y., Cai, J., Pan, C., et al. (2019). Systemic root-shoot signaling drives jasmonate-based root defense against nematodes. Curr. Biol. 29, 3430–3438.e3434. doi: 10.1016/j.cub.2019.08.049
Wang, K.-D., Borrego, E. J., Kenerley, C. M., Kolomiets, M. V. (2019). Oxylipins other than jasmonic acid are xylem-resident signals regulating systemic resistance induced by trichoderma virens in maize. Plant Cell 32, 166–185. doi: 10.1105/tpc.19.00487
Wang, K. D., Gorman, Z., Huang, P. C., Kenerley, C. M., Kolomiets, M. V. (2020). Trichoderma virens colonization of maize roots triggers rapid accumulation of 12-oxophytodienoate and two γ-ketols in leaves as priming agents of induced systemic resistance. Plant Signal Behav. 15, 1792187. doi: 10.1080/15592324.2020.1792187
Wang, Q., Yang, R., Peng, W., Yang, Y., Ma, X., Zhang, W., et al. (2021). Tea plants with gray blight have altered root exudates that recruit a beneficial rhizosphere microbiome to prime immunity against aboveground pathogen infection. Front. Microbiol. 12, 774438. doi: 10.3389/fmicb.2021.774438
Xie, S., Jiang, H., Ding, T., Xu, Q., Chai, W., Cheng, B. (2018). Bacillus amyloliquefaciens FZB42 represses plant miR846 to induce systemic resistance via a jasmonic acid-dependent signalling pathway. Mol. Plant Pathol. 19, 1612–1623. doi: 10.1111/mpp.2018.19.issue-7
Xie, S., Yu, H., Li, E., Wang, Y., Liu, J., Jiang, H. (2019). Identification of miRNAs Involved in Bacillus velezensis FZB42-Activated Induced Systemic Resistance in Maize. Int. J. Mol. Sci. 20, 5057. doi: 10.3390/ijms20205057
Yang, K., Fu, R., Feng, H., Jiang, G., Finkel, O., Sun, T., et al. (2023). RIN enhances plant disease resistance via root exudate-mediated assembly of disease-suppressive rhizosphere microbiota. Mol. Plant 16, 1379–1395. doi: 10.1016/j.molp.2023.08.004
Yu, K., Liu, Y., Tichelaar, R., Savant, N., Lagendijk, E., van Kuijk, S. J. L., et al. (2019a). Rhizosphere-associated pseudomonas suppress local root immune responses by gluconic acid-mediated lowering of environmental pH. Curr. Biol. 29, 3913–3920.e3914. doi: 10.1016/j.cub.2019.09.015
Yu, K., Pieterse, C. M. J., Bakker, P. A. H. M., Berendsen, R. L. (2019b). Beneficial microbes going underground of root immunity. Plant Cell Environ. 42, 2860–2870. doi: 10.1111/pce.v42.10
Yu, Y., Gui, Y., Li, Z., Jiang, C., Guo, J., Niu, D. (2022). Induced systemic resistance for improving plant immunity by beneficial microbes. Plants 11, 386. doi: 10.3390/plants11030386
Yuan, J., Zhao, J., Wen, T., Zhao, M., Li, R., Goossens, P., et al. (2018). Root exudates drive the soil-borne legacy of aboveground pathogen infection. Microbiome 6, 156. doi: 10.1186/s40168-018-0537-x
Zamioudis, C., Pieterse, C. M. (2012). Modulation of host immunity by beneficial microbes. Mol. Plant Microbe Interact. 25, 139–150. doi: 10.1094/MPMI-06-11-0179
Zhu, L., Huang, J., Lu, X., Zhou, C. (2022). Development of plant systemic resistance by beneficial rhizobacteria: Recognition, initiation, elicitation and regulation. Front. Plant Sci. 13, 952397. doi: 10.3389/fpls.2022.952397
Keywords: induced systemic resistance, plant defense mechanism, insect, pathogen, root exudates
Citation: Jung J, Ahn S, Kim D-H and Riu M (2024) Triple interactions for induced systemic resistance in plants. Front. Plant Sci. 15:1464710. doi: 10.3389/fpls.2024.1464710
Received: 15 July 2024; Accepted: 05 November 2024;
Published: 22 November 2024.
Edited by:
Michael V. Kolomiets, Texas A and M University, United StatesReviewed by:
Eli James Borrego, Rochester Institute of Technology (RIT), United StatesCopyright © 2024 Jung, Ahn, Kim and Riu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jihye Jung, amhqdW5nMTIyNEBrb3JlYS5yZS5rcg==
 Jihye Jung
Jihye Jung Seongho Ahn
Seongho Ahn