
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 25 October 2024
Sec. Functional and Applied Plant Genomics
Volume 15 - 2024 | https://doi.org/10.3389/fpls.2024.1463147
 Diksha Kumari1†
Diksha Kumari1† Bishun Deo Prasad2*†
Bishun Deo Prasad2*† Padmanabh Dwivedi1*
Padmanabh Dwivedi1* Sangita Sahni3
Sangita Sahni3 Mankesh Kumar4
Mankesh Kumar4 Saud Alamri5
Saud Alamri5 Muhammad Faheem Adil6
Muhammad Faheem Adil6 Khaled A. Alakeel7*
Khaled A. Alakeel7*Pathogenesis-related (PR) proteins, found in plants, play a crucial role in responding to both biotic and abiotic stresses and are categorized into 17 distinct families based on their properties and functions. We have conducted a phylogenetic analysis of OsPR1 genes (rice PR1 genes) in conjunction with 58 putative PR1 genes identified in Brachypodium distachyon, Hordeum vulgare, Brassica rapa, and Zea mays through BLASTP predictions. We extensively investigated the responses of the remaining 11 rice PR1 genes, using OsPR1a as a reference, under various stress conditions, including phytohormone treatments (salicylic acid and brassinosteroid [BR]), wounding, and heat stress (HS). In rice, of the 32 predicted OsPR1 genes, 12 have been well-characterized for their roles in disease resistance, while the functions of the remaining genes have not been studied extensively. In our study, we selected an additional 11 OsPR1 genes for further analysis and constructed a phylogenetic tree based on the presence of a 10-amino-acid-long conserved motif within these proteins. The phylogenetic analysis revealed that both OsPR1a from earlier studies and OsPR1-74 from our current study belong to the same clade. These genes consistently exhibit upregulation in response to diverse stress treatments such as biotic stress and abiotic stresses such as heat, drought, and salinity, indicating their potential roles in enhancing stress tolerance in rice. Significantly, this study delves into the previously unexplored role of OsPR1 genes in responding to Brassinosteroid (BR) and heat stress (HS) treatments, confirming their involvement in stress responses through qRT-PCR analysis. We found that seven genes were upregulated by EBR treatment. During heat stress (HS), six and seven genes were upregulated at 1hand 4h HS, respectively. The remaining genes OsPR1-22 and OsPR1-75 were upregulated at 1h but downregulated at 4h HS and under EBR treatment. In contrast, OsPR1-76 was upregulated at both 1h and 4h HS, but downregulated under EBR treatment. Promoters of PR1 genes in rice and other crops are rich in transcription factor binding sites (TFBSs) and feature a conserved Cysteine-rich secretory protein (SCP or CAP) motif. This study advances our understanding of PR1 gene regulation and its potential to enhance stress tolerance in rice.
Plants, despite their immobility, constantly face the threat of numerous invading phytopathogens and abiotic stresses. However, their ability to successfully survive these stresses demonstrates their resilience to environmental challenges (Mazumder et al., 2013; Akbudak et al., 2020; Anuradha et al., 2022). Several studies have shown that complex defense signaling pathways are activated in plants to protect them against these environmental stresses (Kundu et al., 2013; Mazumder et al., 2013; Sahni et al., 2016). The interactions between hosts and pathogens or the environment, which ultimately determine the resistance or susceptibility of host plants, have been studied for decades (Timilsina et al., 2020). These interactions, from the early recognition of stress stimuli to the activation of defense responses, follow a well-organized sequence (Prasad et al., 2009; Kundu et al., 2013; Sahni et al., 2016). Defense responses are triggered by various factors, including the generation of reactive oxygen species (ROS), the induction of the hypersensitive response (HR), the interplay of phytohormones, and the synthesis of pathogenesis-related (PR) proteins (Kundu et al., 2013; Mazumder et al., 2013). PR proteins serve as key modulators in plants, accumulating in response to both biotic and abiotic stresses. Low molecular weight PR proteins encompass a diverse array of functions, including roles as transcription factors (TFs) and metabolism-promoting enzymes (Van Loon and Van Strien, 1999; Graham et al., 2003; Hegde and Keshgond, 2013; Jiang et al., 2015). PR1a was the first isolated protein from TMV-infected tobacco leaves (Van Loon and Van Strien, 1999). The expression of PR1 is induced by a variety of biotic and abiotic stress conditions in Musa species and Duram wheat (Akbudak et al., 2020; Anuradha et al., 2022; Zribi et al., 2023) and it serves as an important marker for systemic acquired resistance (SAR). In rice, out of 32 predicted OsPR1 genes (van Loon et al., 2006), 12 are well characterized for disease resistance in rice (Mitsuhara et al., 2008; Prasad et al., 2009). The functional characteristics of these 12 OsPR1 genes were performed after treatment with Xanthomonas oryzae, Maganaporthe oryzae, wounding, SA, JA, and ethylene (Mitsuhara et al., 2008). However, the role of brassinosteroids (BRs) in regulating the 12 OsPR1 genes has not been studied, despite the fact that BRs are known to regulate numerous genes involved in stress resistance and PR1-mediated resistance in Arabidopsis (Zhang et al., 2015). Brassinosteroid (BR) play a significant role in enhancing both abiotic and biotic stress tolerance in various crops and have crosstalk with other defense hormones (Divi et al., 2010; Sahni et al., 2016). BRs have been shown to enhance SA-mediated defense responses. For instance, BRs can amplify the expression of SA-responsive genes and improve resistance against biotrophic pathogens (Divi et al., 2010). PR1 genes are involved in addressing various stressors, such as salinity and water deficiency, in rice (Kothari et al., 2016) and Arabidopsis (Liu et al., 2013). However, there is a surprising lack of research on the effects of heat stress on rice, despite its significant impact on growth, development, yield, and susceptibility to pathogens (Ali et al., 2018a).
Despite significant interest over several decades, the precise roles and functions of all the PR1 genes in plant defense remain poorly understood. To address this knowledge gap, we selected 11 OsPR1 proteins based on the presence of a 10-amino-acid-long conserved sequence (WCCHGCCCYP), which is unique to OsPR1 proteins (Mitsuhara et al., 2008). These 11 OsPR1 genes, whose responses to both biotic and abiotic stresses have not been explored to date, were chosen for functional characterization. For this purpose, we conducted treatments on rice plants involving wounding, heat stress (HS), as well as the application of two defense hormones: salicylic acid (SA) and brassinosteroid (BR). An important characteristic of pathogen-inducible promoters is their rapid activation in response to multiple phytopathogens. As a result of their interaction with signaling molecules of defense such as SA, jasmonic acid (JA), and ethylene (ET), pathogen-inducible promoters typically contain many potential cis-regulatory elements (Mazarei et al., 2008). Despite this, little is known about the molecular mechanisms governing PR gene expression. According to Lodhi et al. (2008), there is a relationship between tobacco PR1 expression and promoter sequence architecture, nucleosome positioning, and targeting of transcription factors. To elucidate plant defense mechanisms, it is crucial to examine the regulation of the PR1 gene. Plant defense responses are modulated by transcription factors (Liu et al., 2022) interacting with cis-acting regulatory elements (5–20 bp) specific to certain genes (Kaur and Pati, 2016). Therefore, we conducted a comprehensive in silico study of the promoter sequence of OsPR1 genes to gain insights into the regulation of gene expression by transcription factor binding sites (TFBSs). Furthermore, we employed a similar methodology to identify PR1 genes in Brachypodium distachyon, Hordeum vulgare, Brassica rapa, and Zea mays, focusing on the presence of a 10-amino-acid-long conserved sequence. Recent advancements in techniques such as RNAi and RNA-Seq analysis have made it possible to identify and explore the promoter regions of target genes, though these methods are often cost-intensive. Consequently, computational methods are increasingly utilized to discover cis-elements in various promoter regions that govern gene expression (Kaur et al., 2017). The PlantCARE software used for examine the cis-acting regulatory elements (CAREs) within the promoter sequences of PR genes, along with an analysis of their effects, will enhance our comprehension of PR gene regulation. A deeper understanding of trans-acting elements can also improve our ability to manipulate gene expression in a desired manner, offering new avenues for applying plant genetic engineering to safeguard crops against various environmental stresses.
In this study, rice seeds (Oryza sativa cv. Rajendra Kasturi) were obtained from Bihar Agricultural University, Sabour, Bhagalpur. Plants were grown up to the four-leaf stage and used for salicylic acid (SA) and wounding treatments under greenhouse conditions. We have taken three biological replicates for both mock and treatment conditions. Each replicate consisted of four individual plants. For 2, 4-epi-brassinolide (EBR) and heat stress (HS) treatments, surface-sterilized seeds (Prasad et al., 2016) were grown on Murashige and Skoog (MS) medium (HiMedia) in magenta boxes (Tarson, India) for 15 days at 25°C under a 14/10 h light/dark cycle in a growth chamber. Rajendra Kasturi is a short-grained, high-yielding aromatic cultivar but is very prone to several abiotic and biotic stresses, reducing its productivity (Kumar et al., 2020).
Twenty-one-day-old rice plants were treated with 3 mM sodium salicylate. To prepare the working solution, a 1 M stock solution of sodium salicylate was initially prepared by dissolving the required amount of sodium salicylate in absolute ethanol. The volume was then adjusted to the desired final volume with distilled water. To prepare the 2, 4-epi-brassinolide (EBR) stock solution, a 100 mM/mL EBR solution was dissolved in absolute ethanol, and a 100 µM/mL working solution was prepared from this stock. For the experiment, a final concentration of 1 µM EBR was used, and for the mock treatment, 0.02% ethanol (the solvent used for EBR) was added to the MS media in test tubes. For the preparation of MS media, MS salts and vitamins (HiMedia) were added at a concentration of 4.42 g/L. Fe-EDTA (200X) was prepared by dissolving 557 mg FeSO4•7H2O and 745 mg Na2EDTA, and 5 mL/L of this solution was added to the MS media. Additionally, 7.5 g/L of Clarigel (HiMedia) was included, and the pH of the media was adjusted to 5.8.
X. oryzae pv. oryzae (NCBI GenBank: MH986180) was used for the experiment, following the leaf clipping method as previously described (Kumari et al., 2020, 2022). The 21 day old rice leaves were clipped with scissors dipped in a bacterial suspension (1 × 108-9 cfu/mL) in saline (0.9%) with 0.05% Triton-X-100, and mock treatments used sterile water with 0.05% Triton-X-100 (Kumari et al., 2020, 2022).
For HS treatment, 15-day-old rice seedlings grown on MS media were exposed to 42°C for 1 hour and 4 hours, then returned to the growth chamber set to 20°C. Untreated seedlings served as controls.
To comprehensively investigate the presence of PR1 proteins in B. distachyon, H. vulgare, B. rapa, Z. mays, and O. sativa, we conducted BLASTP and HMMER (HMMER 3.1b2; http://hmmer.org/; accessed on 29 April 2020) searches as described previously (Prakash et al., 2017). All the twelve OsPR1 protein sequences were individually subjected for similarity searches against B. distachyon, H. vulgare, B. rapa, and Z. mays using BLASTP searches with these sequences as queries (https://www.ncbi.nlm.nih.gov/) (Agarwala et al., 2018) (accessed on 12 May 2020). The top ten hits from each plant species were selected and combined. Through manual curation, redundant, incomplete, and closely similar entries in terms of amino acid sequences were eliminated. Additionally, we conducted multiple sequence alignments (MSA) of all the newly identified putative PR proteins along with OsPR1s to verify the presence of a conserved 10-amino-acid-long sequence [W(10x)C(8x)C(x)H(12x)GC(4x)C(9x)C(1x)Y(10x)P]. The conservation of the 10 amino acids is a typical characteristic of PR1 proteins (Mitsuhara et al., 2008). Finally, eighty one putative PR1 sequences were used for phylogenetic tree construction. using the neighbor-joining (NJ) method with bootstrapping (1000 replicates) in the MEGA 11 Program (http://www.megasoftware.net; Analysis version 1.01; accessed on 20 August 2020) (Tamura et al., 2021).
All PR1 proteins were subjected for domain and subcellular localization studies. The domain prediction was performed using the Pfam tool (Pfam 37.0; http://pfam.xfam.org/search/sequence; accessed on 10 September 2021) (Mistry et al., 2021) and subcellular localization was predicted using the Balanced Subcellular Localization Predictor tool (BaCelLo) (http://gpcr.biocomp.unibo.it/bacello/info.htm; accessed on 10 September 2021) (Pierleoni et al., 2006). Chromosomal positions of PR1 genes were studied using Phytozome v12.1 database (https://phytozome.jgi.doe.gov/pz/portal.html; Analysis version v12.1; accessed on 20 August 2020). Chromosome maps of O. sativa PR1 gene were constructed using Chromosome Map Tools Oryza base (http://viewer.shigen.info/oryzavw/maptool/MapTool.do; accessed on 27 August 2022).
The Rice Annotation Project Database (RAP-DB), (http://rapdb.dna.affrc.go.jp/tools/dump; accessed on 21 January 2021) (Sakai et al., 2013) was utilized to obtain 1000 base pair upstream sequences of 23 OsPR1 genes. These retrieved sequences were subsequently analyzed using PlantPan3, (http://plantpan3.itps.ncku.edu.tw/; accessed on 21 January 2021) (Chow et al., 2016), for the identification of transcription factor binding sites (TFBS), and PlantCARE, (http://bioinformatics.psb.ugent.be/webtools/plantcare/html; accessed on 21 January 2021) (Lescot, 2002), for the analysis of cis-regulatory DNA elements in plants.
In addition, we retrieved 1000 base pair upstream sequences for newly identified PR1 genes in various plant species using Phytozome 12 (https://phytozome.jgi.doe.gov/pz/portal.html; Analysis version v12.1; accessed on 30 January 2021) (Goodstein et al., 2012). TFBSs and cis-regulatory DNA elements within these sequences were analyzed using the same approach as described above. A detailed flow chart for the identification of cis-regulatory sequences in rice and other plants is depicted in Supplementary Figure 1.
The relative expression of 11 OsPR1 genes, including the reference gene OsPR1a, in rice seedlings treated with salicylic acid (SA), epibrassinolide (EBR), mechanical wounding, and heat stress (HS) was measured by qRT-PCR analysis. Total RNA was extracted from frozen plant tissues using Promega’s SV Total RNA Isolation System following the manufacturer’s protocol (Promega, Madison, WI). For cDNA synthesis, 1µg of total RNA was reverse transcribed using random hexamer primers according to manufacturer’s protocol (Promega, Madison, WI). The resulting cDNA was diluted in nucleases free water (1:5) and used for qRT-PCR (Kumari et al., 2022; Chaudhary et al., 2024). The qRT-PCR was carried out in a Light Cycler system (Applied Biosystems) using SYBR Green dye. Each reaction mixture (10 µL) contained 5 µL of SYBR Green dye (2×) (Promega, Madison, WI), 0.5 µL of forward/reverse gene-specific primers (10 µM), and 1 µL of diluted cDNA.
Three independent biological replicates as well as two technical repeats were conducted for each qRT-PCR analysis. The qRT-PCR conditions included an initial denaturation at 95°C for 2 minutes, followed by 40 cycles of amplification consisting of denaturation, annealing, and extension at 95°C, 53°C, and 72°C for 20 s, 30 s, and 30 s, respectively. The specificity of amplification was confirmed by melting curve analysis. ACTIN was used to normalize the expression data of OsPR1 genes. The fold-change in expression levels of genes was estimated using the 2-ΔΔCt method as described previously (Livak and Schmittgen, 2001).
Gene expression data was statistically analyzed using the computer software SPSS. Significance of differences were analyzed by one-way analysis of variance (ANOVA).
The BLASTP analysis yielded 82 PR1 proteins using OsPR1 as the query sequence, and the HMMER search results identified 145 PR1 proteins. To ensure accuracy, we manually excluded PR1 proteins that were common between the BLASTP and HMMER searches. The initial extensive list of 227 PR1 proteins was refined to 81 proteins based on the presence of a conserved 10-amino-acid sequence [W(10x)C(8x)C(x)H(12x)GC(4x)C(9x)C(1x)Y(10x)P]. Among the 81 PR1 proteins analyzed, 23 were identified in Oryza sativa, while putative orthologs were found in Brachypodium distachyon, Hordeum vulgare, Brassica rapa, and Zea mays, totaling 58 proteins across these species. In this study, 23 OsPR1 genes were selected for phylogenetic analysis, with a subset of 11 OsPR1 genes chosen specifically for expression analysis experiments.
Each gene sequence was designated according to its chromosomal position and gene order (Tables 1, 2). For example, OsPR1-21 (LOC_OS02G54530.1) is located on chromosome 2, while OsPR1-61 (LOC_OS06G24290.1) is situated on chromosome 6. Notably, chromosome 7 houses a cluster of eight PR1 genes, namely OsPR1-71, -72, -73, -74, -75, -76, -77, and -78. This cluster is organized in a consecutive arrangement from the 5’ end to the 3’ end, all aligned in the sense orientation, as described by Mitsuhara et al. (2008).
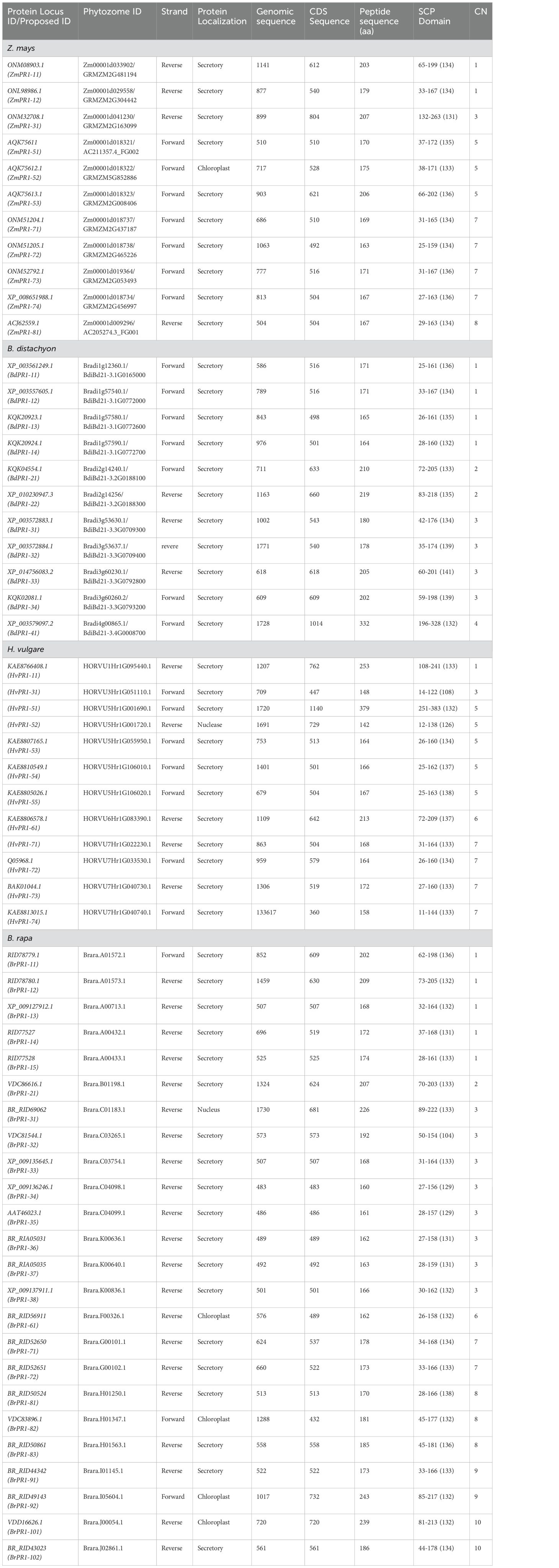
Table 2. Description of pathogenesis related (PR1) genes in Z. mays, Brachypodium distachyon, Hordeum vulgare, and Brassica rapa.
The neighbor-joining (N-J) method of MEGA 11 Program (Tamura et al., 2021) was used to create a phylogenetic tree to compare OsPR1 and its potential homologs in both monocot and dicot plants. The analysis resulted in the formation of three main clusters: Cluster I, II, and III (Figure 1). Further, these clusters were divided into groups (a-f) based on clade division.
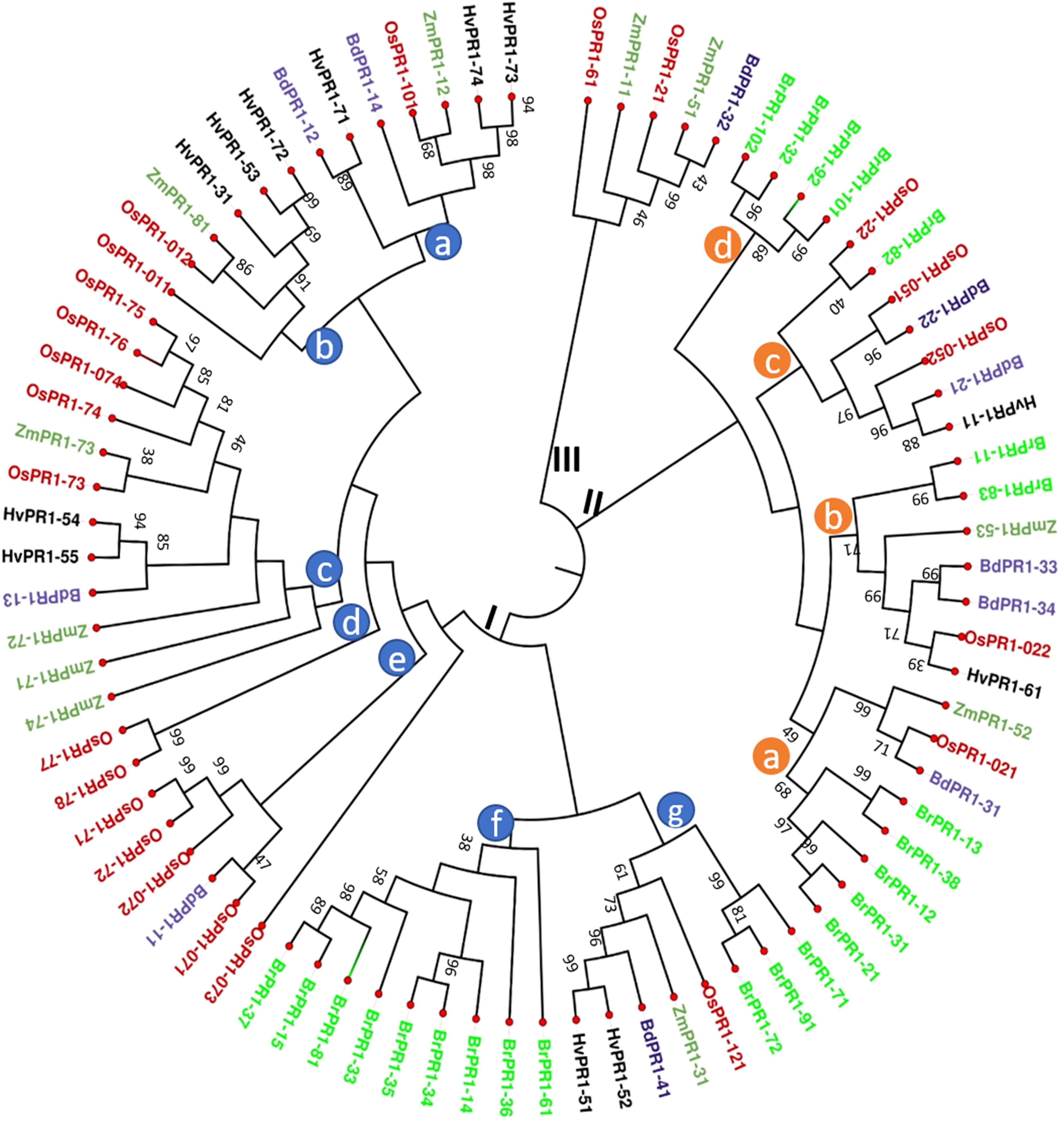
Figure 1. Phylogenetic tree analysis of PR1 protein families. The phylogenetic relationship of OsPR1 proteins with homologs from other important plant species was constructed using the MEGA 11 program, after aligning the protein sequences with MUSCLE. The protein sequences were obtained from Phytozome12. Different PR1 proteins are highlighted in various color codes for easy identification: OsPR1 (Oryza sativa) in red, ZmPR1 (Zea mays) in green, BdPR1 (Brachypodium distachyon) in blue, HvPR1 (Hordeum vulgare) in black, and BrPR1 (Brassica rapa) in fluorescent green. The phylogenetic tree analysis revealed distinct clusters, denoted as Cluster I, Cluster II, and Cluster III. Cluster I can be further divided into several subgroups, represented as clade groups a, b, c, d, e, f, and g, which are identified with blue circles on the phylogenetic tree. Notably, Cluster I contains the highest number of PR1 proteins, totaling 50. Cluster II consists of subgroups a, b, c, and d, which are indicated by brown circles on the phylogenetic tree.
Cluster I contains the largest number (50) of PR1 proteins, including Group C proteins such as OsPR1a and OsPR1-74. Previous reports have used OsPR1a as a versatile marker for both biotic and abiotic studies in rice (Yan et al., 2022). Within Cluster I, OsPR1-011 (OsPR1b) is positioned closer to OsPR1-75. Notably, OsPR1-011 demonstrates upregulation after salicylic acid (SA) treatment but downregulation following wounding (Mitsuhara et al., 2008). Similarly, our study observed the same trend for OsPR1-75. In Cluster I, the Group a, b, and c proteins are clustered together, containing ZmPR1-12 and OsPR1-101, ZmPR1-81 and OsPR1-012, and ZmPR1-73 and OsPR1-73 within the same clade BdPR1-14 (Bradi1g57590) and OsPR1-101. Notably, both of these genes were observed to be upregulated following SA treatment (Mitsuhara et al., 2008; Kakei et al., 2015). Cluster II represents the second largest group of PR1 proteins (Figure 1). In Groups B and C of Cluster II, HvPR1-61 and BrPR1-82 along with OsPR1-022 and OsPR1-22, are grouped within the same clade (Figure 1).
All the identified PR1 proteins contain a conserved motif belonging to the cysteine-rich secretory protein (SCP or CAP) domain, which ranges from 104 to 145 amino acids (Tables 1, 2). Subcellular localization predictions indicated that the majority of PR1 proteins are secretory in nature (Tables 1, 2). However, some proteins, such as OsPR1-76, HvPR1-52, HvPR1-53, and HvPR1-73, were predicted to localize in the nucleus. ZmPR1-52 was predicted to localize in the chloroplast.
Our findings indicate that BdPR1, HvPR1, BrPR1, ZmPR1, and OsPR1 genes are situated on distinct chromosomes (Figure 2). Notably, chromosome 7 exhibited the highest concentration of PR1 genes, comprising 25.5% of the total. Chromosomes 1 and 3 collectively harbored 16% of the PR1 genes, while the remaining genes were distributed across other chromosomes (Tables 1, 2; Figure 2).
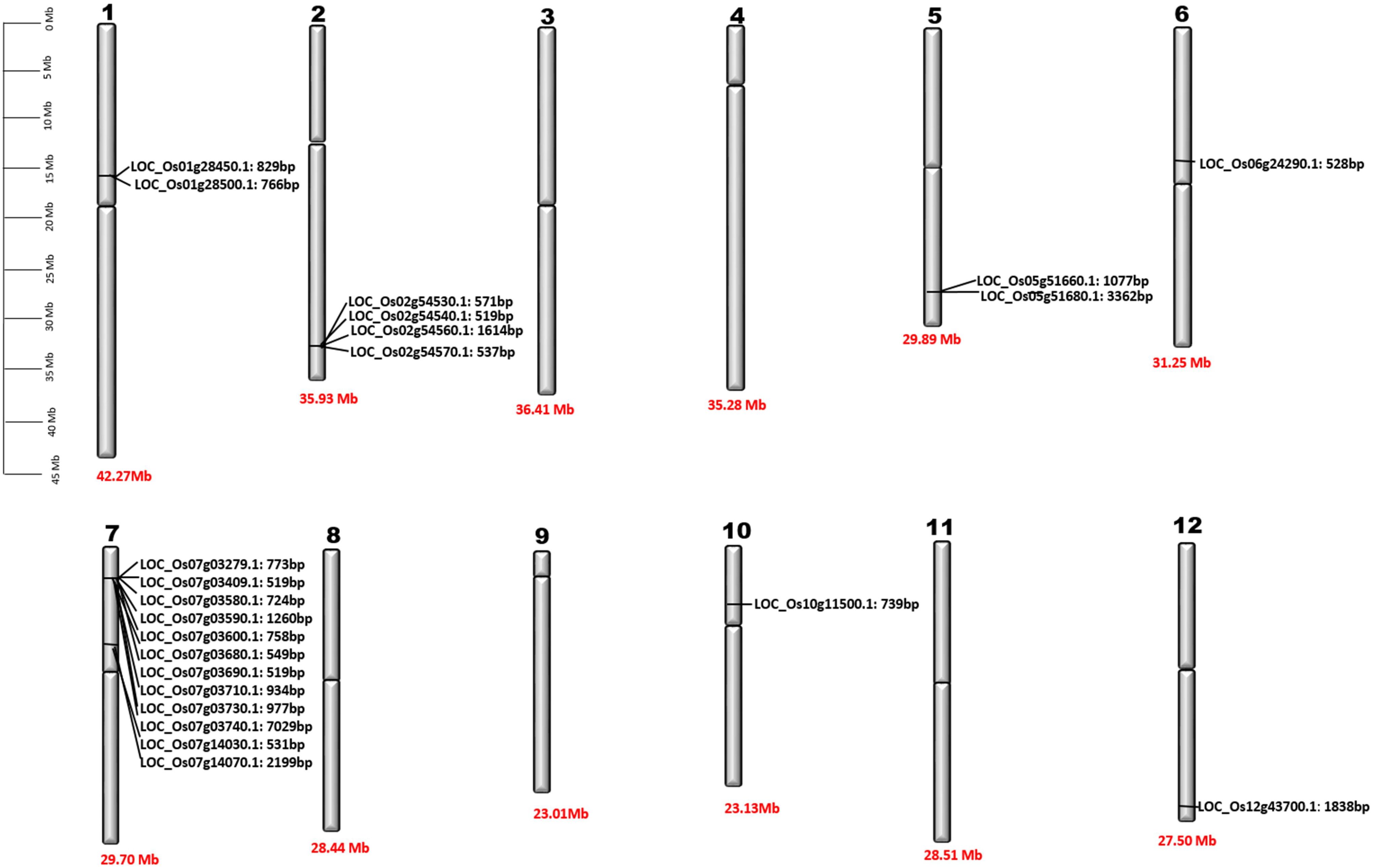
Figure 2. Chromosomal location of OsPR1 genes. Detailed physical maps of individual chromosomes have been meticulously generated, clearly indicating the specific positions of individual OsPR1 genes in base pair (bp) units. Furthermore, the length of each chromosome is precisely denoted at the bottom of the graphical representations, expressed in Megabase (Mb) measurements. It highlights an enrichment pattern of OsPR1 genes on chromosomes 7, 2, 5, 1, 6, 10, and 12, indicating a higher density of these genes in these genomic regions. Interestingly, no OsPR1 genes were found on chromosomes 3, 4, 8, 9, and 11.
Plant hormones, salicylic acid (SA) and brassinosteroids (BR), have been identified as key regulators of the plant defense system (Kumari et al., 2022). While the role of SA in regulating PR1 genes during plant defense is well-established (Takatsuji et al., 2010), the precise impact of BR on PR1 gene expression is largely unknown. In the present investigation, a total of 11 OsPR1 genes, whose significant roles had not been previously studied, were meticulously selected for comprehensive expression analysis, along with the OsPR1a marker gene included for reference.
Upon subjecting the rice seedlings to SA treatment, eight genes OsPR1-21(~21-fold induction), OsPR1-22 (~37 fold induction), OsPR1-61 (~6-fold induction), OsPR1-71(~1.5-fold induction), OsPR1-72 (~45-fold induction), OsPR1-74 (~9.5-fold induction), OsPR1-75 (~5.6-fold induction), and OsPR1-77 (~3.1-fold induction)) showed upregulation, while OsPR1-76 displayed downregulation. Notably, OsPR1-73 and OsPR1-78 were not amplified in uniform manner in treated or mock samples (Table 3, Figure 3A).
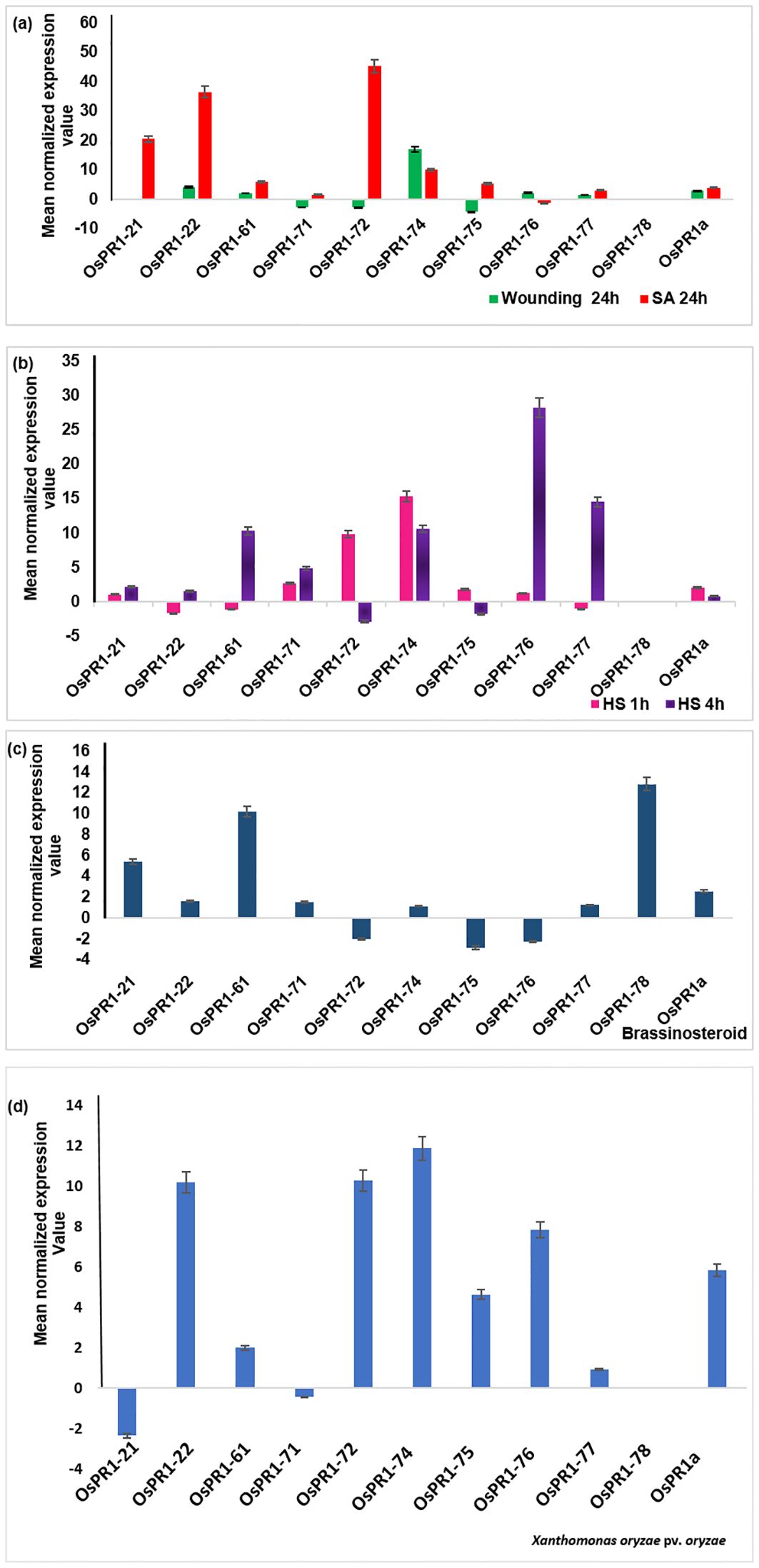
Figure 3. Transcriptional response of 11 OsPR1 genes to selected treatments salicylic acid, wounding, heat, and 2, 4-epibrassinolide treatments were studied (A). Transcriptional response of 11 OsPR1 genes to with salicylic acid. Rice seedlings were treated with 3mM salicylic acid and wounded in criss-cross manner to analyze the transcript levels by qRT-PCR at 24h post treatment. (B). Transcriptional response of 11 OsPR1 genes treated with heat stress. Rice seedling grown in half MS media placed in incubator at 42 °C. (C). Transcriptional response of 11 OsPR1 genes in rice seedlings treated with 2, 4-epibrassinolide. Rice seedlings were grown for 15 days on MS medium supplemented with EBR. (D) Rice seedlings were infected with Xanthomonas. oryzae pv. oryzae, and the transcript levels of OsPR1 genes were analyzed using qRT-PCR at 24h post-infection. Relative transcript abundance is expressed as mean normalized expression value relative to the mock treatment. OsPR1a expression served as a positive control, indicating successful infection of rice seedlings by salicylic acid. Results are representative of three independent experiments. Error bars represent standard error (SE) of mean for three replicates.
Following EBR-treated, seven OsPR1 genes OsPR1-21, OsPR1-22, OsPR1-61, OsPR1-71, OsPR1-74, OsPR1-77, and OsPR1-78 were found to be upregulated whereas three genes OsPR1-72, OsPR1-75, and OsPR1-76 were downregulated. The maximum induction was observed in OsPR1-78 (~12- fold induction), followed by OsPR1-61 (~10-fold induction) and OsPR1-21 (~5-fold induction) (Figure 3C).
Upon 4h of HS treatment, OsPR1-76 showed the highest upregulation (~28-fold induction) while OsPR1-77 showed the lowest upregulation (~14 -fold induction). Both OsPR1-74 and OsPR1-61 showed upregulated to the same level (~10-fold induction) upon 4h of HS. OsPR1-72 and OsPR1-75 genes were downregulated ~2– ~3-fold after 4 h of HS, whereas OsPR1-22, OsPR1-61, and OsPR1-77 were downregulated (~1.5-fold) after 1 h of HS treatment. OsPR1-74 showed the highest upregulation (~15-fold induction) after 1 h of HS-treatment, followed by OsPR1-72 (~10-fold induction), while the remaining upregulated genes showed upregulation in range of ~ 1-3-fold. Following exposure to HS, OsPR1-73 and OsPR1-78 were unamplified (Figure 3B).
The leaves of 21 day old rice seedlings were subjected to precise and effective cross-cross wounding employing sterile scissors as described previously (Mitsuhara et al., 2008). Our result revealed that the maximum expression was observed in OsPR1-74 gene (~17-fold induction), followed by the OsPR1-22 gene (~4-fold induction). Three genes-OsPR1-75 (~4-fold), OsPR1-71 (~2.7-fold), and OsPR1-72 (~4.3-fold) were found to be downregulated (Figure 3A). The responses of all 11 OsPR1 genes to different treatments are summarized in Table 3.
Genes that were highly upregulated at 24 hours post-infection with X. oryzae pv. oryzae included OsPR1-74 (~12-fold), followed by OsPR1-72 and OsPR1-22 (~10-fold), OsPR1-76 (~7.86-fold), OsPR1-75 (~4.64-fold), and OsPR1-61 (~2-fold). In contrast, OsPR1-73 and OsPR1-78 did not show consistent amplification across all biological replicates (Figure 3D).
The promoter sequences located up to 1 kilobase (kb) upstream from the translation start site of each PR gene in Oryza sativa (rice) were meticulously analyzed using PlantCARE and PlantPAN 3 programs. The primary goal was to identify potential plant cis-acting regulatory elements (PCAREs) and transcription factor binding sites (TFBSs) associated with defense responses. A diverse set of important defense-responsive TFBSs, such as bHLH, bZIP, C2H2, EIN3, GATA, LEA, MYB, Myb/SANT, NAC, NAM, and WRKY, were successfully identified in both the positive and negative strands of the promoter sequences of OsPR1 genes (Figures 4, 5). The analysis further focused on determining the enrichment and distribution of all TFBSs within proximal (<500 bp) and distal (>500 bp) regions of all OsPR1 genes. Remarkably, it was observed that the majority of these TFBSs were significantly enriched in the proximal upstream regions of most OsPR1 genes. This finding highlights the potential significance of these regulatory elements in modulating the expression of OsPR1 genes during defense responses in rice (Figures 4, 5). In our experiment, the highest number of WRKY TFBSs was found in OsPR1-73 (93); however, this gene was not expressed under any of the treatments. In contrast, a significant enrichment of WRKY TFBSs was observed in OsPR1-72 (88), which was upregulated 45-fold and 10-fold following SA and heat stress treatments, respectively (Supplementary Table 2). This suggests that WRKY TFBSs play a major role in PR1-mediated biotic and abiotic responses. OsPR1-76 has the highest number of AP2 (175) and bZIP (61) TFBSs (Supplementary Table 2) and shows maximum expression during 4 hours of heat stress (28-fold), as well as 1 hour of heat stress and wounding (3-fold). The presence of a large number of AP2 and bZIP TFBSs may play a significant role in regulating both biotic and abiotic stress responses.
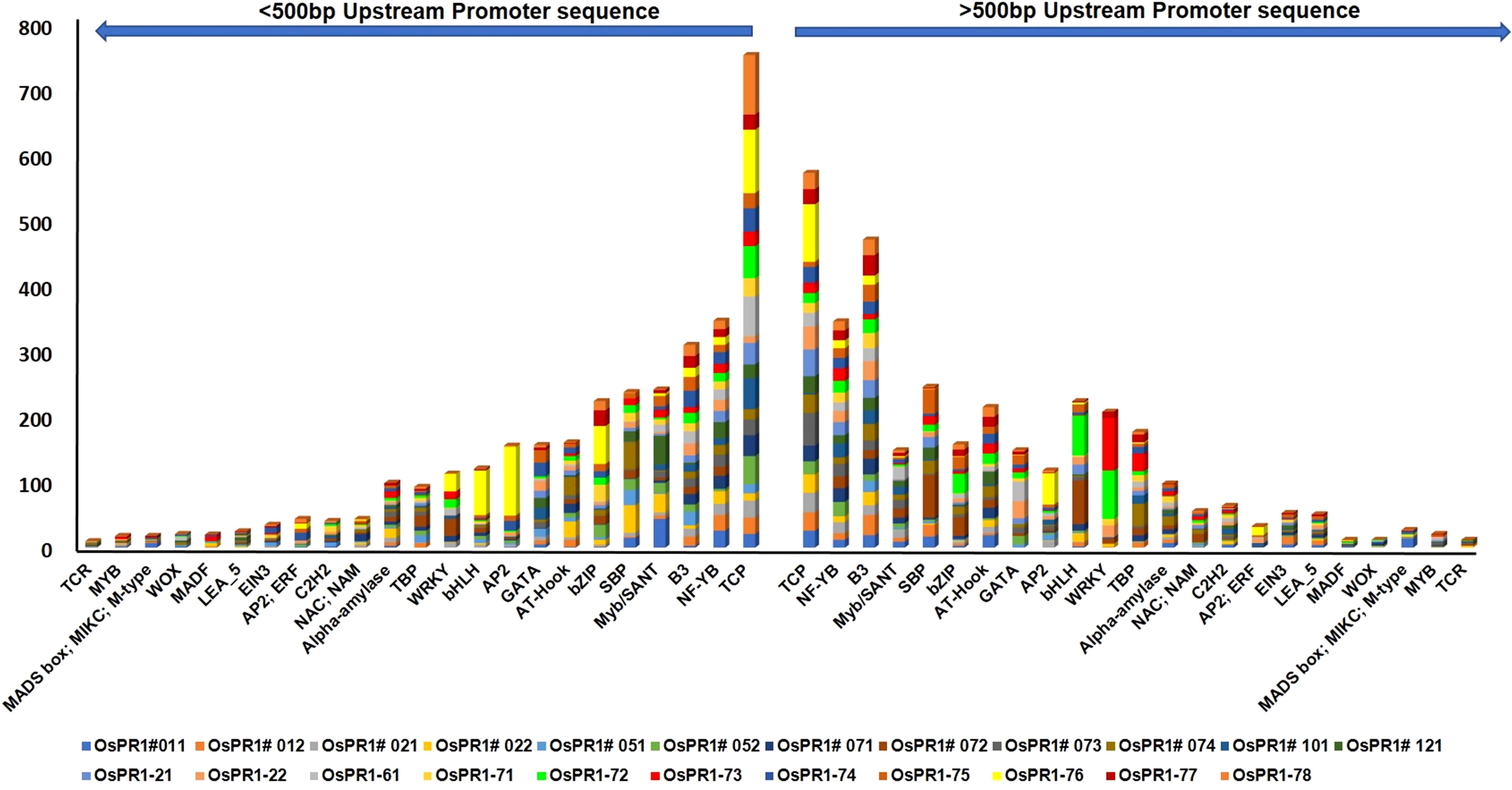
Figure 4. Enrichment of transcription factor binding sites (TFBSs) in OsPR1 promoter. The abundance of stress and developmental related TFBSs shown in OsPR1 promoter proximal (<500 bp) and distal (>500 bp) region. TFs is key component in transcription regulation of gene expression during multiple stress.
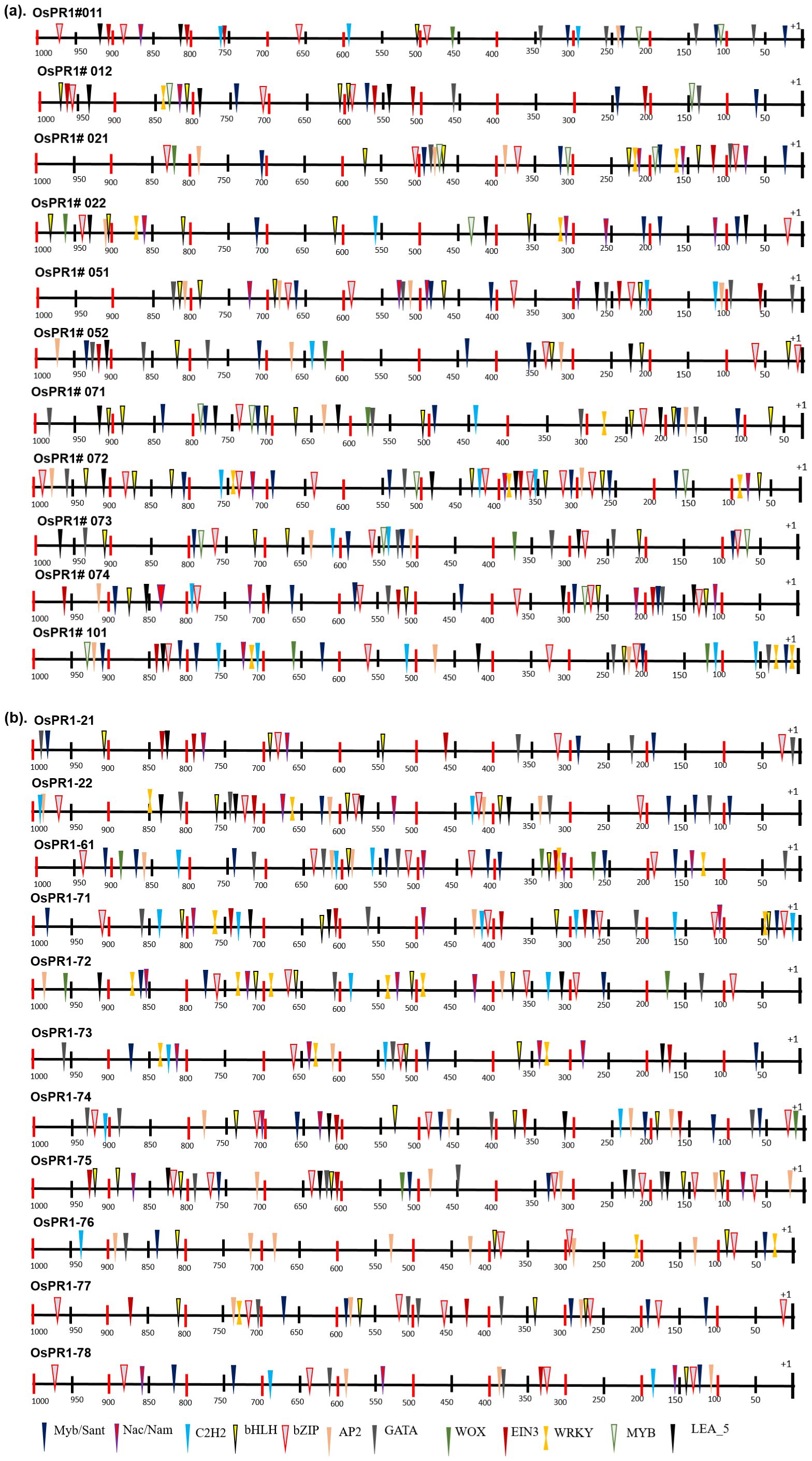
Figure 5. Distribution of transcription factor binding sites (TFBSs) in 1000 bp promoter region of rice PR1 genes. Different TFBSs denoted by different shapes and color code combination as identified by PlantPan3. Promoter prediction tool. (A) Distribution of transcription factor binding sites (TFBSs) of previously reported OsPR1 (Mitsuhara et al., 2008). (B) Distribution of transcription factor binding sites (TFBSs) in currently studied OsPR1.
The study conducted on Oryza sativa (rice) identified a total of 57 cis-acting regulatory elements (CAREs) in its promoter regions. These cis-elements varied in length from 4 to 10 base pairs (bp), with the majority being 6 to 7 bp long. The cis-elements were further categorized into different functional groups, revealing that a significant proportion (63%) were associated with stress responsiveness, with light-responsive elements constituting 34% of the stress-responsive group. The occurrence frequency of different cis-elements was nearly identical in both the forward and reverse strands of the 1-kilobase promoter region. Interestingly, the distribution analysis revealed that most cis-elements were clustered between 16 to 493 bp on the forward strand (+) and 546 to 842 bp on the reverse strand (−) (Figure 5).
Furthermore, specific cis-elements such as TATA, CAAT box (not shown in Supplementary Table S3), Box 4, and G-box were found to be more abundant in the OsPR1 genes compared to other cis-elements. These findings provide valuable insights into the regulatory mechanisms governing the expression of OsPR1 genes and highlight their responsiveness to various stress conditions in rice. The distribution of CAREs was unequal throughout the upstream region of the genes, with nearly equal numbers found in the proximal (<500 bp) and distal (>500 bp) regions. Gene expression is correlated with the presence of PCAREs and the number of transcription factor binding sites (TFBSs). Following SA, EBR, wounding, and heat stress treatment (HST), the OsPR1-71 and OsPR1-74 genes were upregulated and enriched with the greatest number of PCAREs. OsPR1-74 was found to be enriched with defense-responsive TFBSs such as WRKY (32), AP2 (74), and developmentally related TFBSs such as TCP (186) and α-amylase (10) (Supplementary Table 2). OsPR1-71 and OsPR1-74 were enriched with W-box (TTGACC), SA-responsive element TCA (CCATCTTTTT), and EBR-binding element MBS (CAACTG), suggesting SA-mediated EBR expression of the genes (Supplementary Table 3). In addition, the OsPR1-71, OsPR1-74, OsPR1-76, and OsPR1-21 genes, which were upregulated after 1 h and 4 h of heat stress treatment (HST), contain the highest number of hormone-responsive PCAREs. These genes were enriched with the maximum number of methyl jasmonate-responsive TGACG-motif and CGTCA-motif, suggesting their role in basal thermotolerance. Previous research in Arabidopsis has shown that jasmonic acid (JA) can mitigate the effects of high light and high temperature conditions, which is crucial for plants to thrive in various agroclimatic environments. Furthermore, OsPR1-72 and OsPR1-76 contain the GC-motif (CCCCCG), which is associated with anoxic-specific inducibility (Supplementary Table 3). This finding suggests that the OsPR1 genes may be involved in submergence regulation.
The promoter sequences of BdPR1, HvPR1, BrPR1, and ZmPR1 were studied to retrieve important stress-responsive TFBSs and CAREs. Comparatively, within the promoter sequences of all PR1 genes, stress-responsive TFBSs exhibit a higher prevalence compared to those associated with developmental processes. In BdPR1, HvPR1, and BrPR1, the number of total CAREs varies from 52 to 53, with 61% of them being stress-responsive. In contrast, ZmPR1 has a total of 49 CAREs, with 70% being stress-responsive (Supplementary Table 4). The numbers of developmental-related CAREs in BrPR1 and ZmPR1 are compared to those in BdPR1 and HvPR1 (Supplementary Table 4, Figure 6). All the PR1 genes were enriched with TATA-box, CAAT-box, G-box, MYC, ARE, Myb, abscisic acid (CGTACGTGCA), and methyl-jasmonate (TGACG, CGTCA) responsive PCAREs (Supplementary Table 4). The maximum number of TFBSs was found in BdPR1-11 [bHLH (252), bZIP (140), WRKY (109), NAC;NAM (102), BES1 (44), Myb-related (31), AP2;ERF (93)), followed by BdPR1-13 (bZIP (239), EIN3 (11)] (Figure 7). In ZmPR1, the maximum enrichment of defense-responsive TFBSs was found in ZmPR1-51 and ZmPR1-31, though the total enrichment was lower compared to others. In contrast, the enrichment of TFBSs in members of HvPR1 and BrPR1 was the highest (Figure 7).
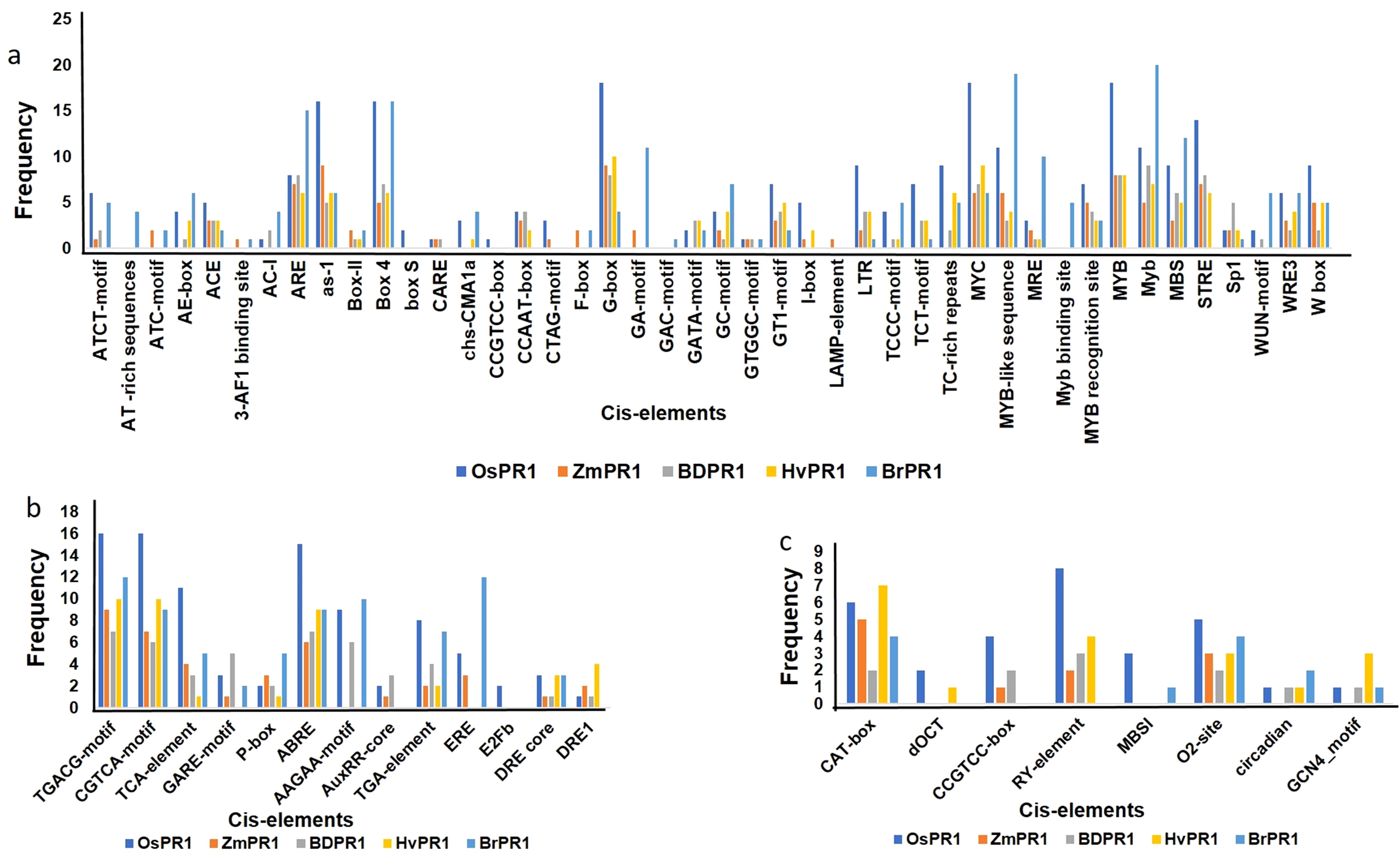
Figure 6. Frequencies of cis-regulatory motifs identified in the 1000 bp upstream promoter regions of Pathogenesis-Related 1 (PR1) genes in different plant species using PlantCARE. The figure presents the distribution of different categories of motifs identified in the promoter regions of PR1 genes in Oryza sativa (OsPR1), Zea mays (ZmPR1), Brachypodium distachyon (BdPR1), Hordeum vulgare (HvPR1), and Brassica rapa (BrPR1). (A) Stress-related motifs, such as W-box, TC-rich repeats, and LTR, are shown in the promoter regions across species. (B) Hormone-responsive motifs, including ABRE, TGA-element, and GARE-motif, indicating regulation by abscisic acid, salicylic acid, and gibberellins. (C) Tissue-specific motifs, such as CAT-box, RY-element, and O2-site, are compared across the PR1 genes of the different species, showing variability in their potential roles in developmental expression patterns.
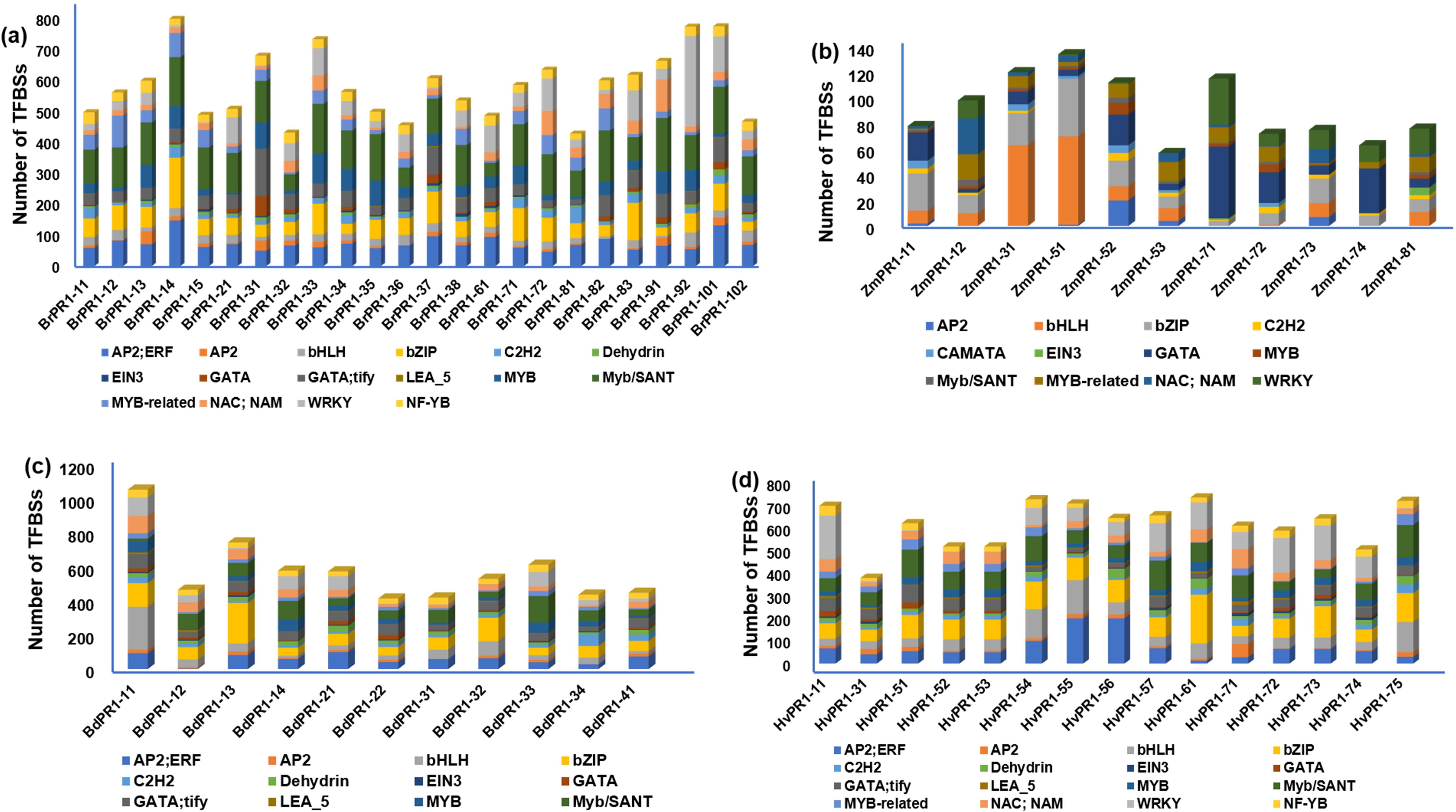
Figure 7. Distribution of transcription factor binding sites (TFBSs) in the 1000 bp upstream promoter regions of Pathogenesis-Related 1 (PR1) genes in different plant species. The graphs display the presence of various TFBS categories identified in PR1 gene promoter regions of (A) Brassica rapa (BrPR1), (B) Zea mays (ZmPR1), (C) Brachypodium distachyon (BdPR1), and (D) Hordeum vulgare (HvPR1). The color-coded bars represent the number of TFBSs associated with different transcription factor families, including AP2/ERF, AP2, bHLH, bZIP, C2H2, EIN3, GATA, MYB, WRKY, and others, indicating the complexity and variability of transcriptional regulation across different species.
Plant pathogens exhibit a remarkable range of ecological adaptations and can inflict substantial damage on plant growth, often surpassing the impact of many other organisms. While plants have not naturally evolved resistance to all diseases, researchers have implemented various strategies to enhance plant resistance. These approaches include modifying signaling pathways, gene pyramiding (stacking multiple resistance genes), and overexpressing disease-responsive genes, such as PR genes (Pathogenesis-Related genes) (Grover and Gowthaman, 2003). The studied PR1 genes belong to the CAP (Cysteine-rich Antifungal Protein) or SCP (Secretory Cysteine-rich Protein) family, which is known for its roles in the defense system, including ion binding, antifungal activity, cell wall degradation, sterol binding and transport, and reproduction (Schneiter and Di Pietro, 2013; Chen et al., 2014). The selection of these specific PR1 genes was based on the presence of ten highly conserved amino acid residues in their C-terminal region. This conservation was observed not only in the PR1 genes chosen for this study but also in all 36 PR1-type proteins identified across a diverse range of 14 plant species (Mitsuhara et al., 2008; Chen et al., 2014). PR1 proteins are thought to function in plants by utilizing regions in their C-terminus. In this study, out of the 11 OsPR1 genes selected for expression analysis, 9 exhibited upregulation following SA treatment. In numerous studies, PR1 has been shown to play a significant role in plant defense by acting as a signaling marker for SA pathways. Under optimal environmental conditions, healthy and young rice leaves contain high amounts of salicylic acid (SA), with levels reaching up to 10 µg per gram of fresh leaf. In contrast, tobacco and Arabidopsis leaves contain much lower amounts of SA, approximately 20 ng and 30 ng, respectively (Silverman et al., 1995; Park et al., 2007; Seo et al., 2007). Rice leaves are known to possess remarkably high levels of SA, ranging from 50 to 500 times greater than those found in tobacco or Arabidopsis leaves. These SA levels in rice are comparable to the levels induced by Tobacco mosaic virus (TMV) infection, which typically leads to hypersensitive response (HR) lesion formation. Given the elevated SA levels in rice, it is conceivable that the expression of OsPR1 genes could be significantly influenced by treatments with SA, EBR, heat stress (HS), and wounding (Mitsuhara et al., 2008). To compare the nature of 11 OsPR1 genes, the results are summarized in Supplementary Table 1. The phylogenetic analysis reveals that OsPR1#074 (OsPR1a) and OsPR1-74 fall into the same clade, exhibiting similar characteristics and forming transcripts after all treatments. OsPR1-74 could act as a common marker for multiple stresses. OsPR1#011 (OsPR1b) was found to be upregulated after SA treatment but downregulated after wounding. Similar trends were observed in OsPR1-75, which is located close to OsPR #011 in the phylogenetic tree. The process of wounding is known to activate numerous genes, including PR1 genes, as demonstrated in studies by Mitsuhara et al. (2008) and Ali et al. (2018b). Investigating the expression pattern of OsPR1 genes under wounding treatment in rice holds a significant promise in uncovering the detailed role of these genes under various stress conditions. Investigating the expression pattern of OsPR1 genes under wounding treatment in rice holds significant promise for uncovering the detailed roles of these genes under various stress conditions. OsPR1 genes are typically induced by biotic stresses, such as pathogen infection. Upon infection with X. oryzae pv. oryzae (Xoo), OsPR1 expression is significantly upregulated, which correlates with an enhanced defense response in rice plants (Mitsuhara et al., 2008). Among these PR1 genes, OsPR1a is a well-characterized member known to be involved in rice defense mechanisms (Yan et al., 2022). Investigating the expression of other OsPR1 genes were upregulated after treatment.
By understanding how OsPR1 responds to wounding stress, we can gain valuable insights into its broader functions during different types of stress in rice. In the present study, 5 OsPR1 genes were upregulated after wounding, 24 hours post-treatment, indicating a late wounding response. Both early and late inducible wounding-responsive PR1 genes have been reported in tomato and B. napus (Scranton et al., 2013; Ali et al., 2018). Additionally, it has been reported that wounding and the defense response utilize similar signaling pathways, including SA, JA, and ET (Maleck, 1999). The expression profile of OsPR1 genes was found to be similar to that observed in Mitsuhara et al. (2008) study.
Brassinosteroids (BRs) mediated response in multiple stress is extensively studied but PR1 mediated response need to be verified in rice (Li and Chory, 1999; Sreeramulu et al., 2013). Previous studies, conducted by Nakashita et al. (2003), Divi et al. (2010), and Sahni et al. (2016), have demonstrated that the application of exogenous brassinosteroids (BR) enhances overall plant resistance against pathogens. These findings highlight the positive impact of BR treatment on bolstering plant defenses against various pathogens. Similarly, we have studied the OsPR1 response after exogenous treatment of BR and found that six genes along with OsPR1a upregulated after treatment. Further, we found that 4 OsPR1 genes (OsPR1-22, OsPR1-61, OsPR1-74, OsPR1-77) were upregulated after all the treatments. All the PR1 genes that were upregulated in heat stress follow the same trend in BR treatment. However, the observed downregulation of OsPR1-76 after BR treatment might suggest that OsPR1-76 is not a BR-responsive gene. In brassica, several overlapping findings came in light of how BR modulated the expression of WRKY, PR, HSP, protein synthesis, calcium signaling, and ROS-associated genes (Sahni et al., 2016) as well as multiple stress responsive genes. In Arabidopsis cpd mutants, which are deficient in BR biosynthesis, a decrease in the PR1, PR2, and PR5, whereas the CPD over-expressed transgenic line shows higher expression (Szekeres et al., 1996), suggesting that BL induces PR gene expression. Contrary to its effects in rice and other plants, exogenous application of brassinosteroids (BR) did not result in changes in the expression of PR genes in tobacco, as evidenced by the study conducted by Nakashita et al. (2003). This observation suggests that NPR1, a key regulator of SA-mediated defense genes, may play a crucial role in the enhancement of thermotolerance and salt tolerance triggered by BR. However, NPR1 is not necessary for the BR-mediated induction of PR-1 gene expression in tobacco. This finding indicates that BR can exert its anti-stress effects independently, and it may also interact with other hormonal pathways to elicit its effects, as reported by Divi et al. (2010). Thus, BR appears to modulate stress responses through diverse mechanisms, and its interactions with other hormones contribute to its multifaceted effects on plant defense and stress tolerance. In this experiment, OsPR1-78 gene showed enhanced expression (~12.8-fold) after EBR treatment, whereas no change in expression was observed after SA, HS, or wounding treatment. So, in future it is need to study critically the link between BR and SA pathway.
Moreover, PR1 gene has role in multiple type of abiotic stress, but PR1 gene expression after abiotic stress has not been studied in rice. For the first time, we studied the 11 OsPR1 gene expression after heat stress treatment. In this study, we found that four OsPR1 genes along with OsPR1a gene were upregulated after 1 h and 4 h of heat stress treatment, whereas 6 and 7 OsPR1 individually upregulated in 1 h and 4 h of heat stress treatment, respectively. OsPR1-76 expressed maximally after 4 h of HS treatment indicates that OsPR1 have a role in amelioration effect on plants against heat stress. It is also believed that high temperature is responsible for rice’s susceptibility to pathogens (Dossa et al., 2017). PR1 gene expression in abiotic stressed condition studied in many crops like wheat, Arabidopsis, tomato, grape (Liu et al., 2013; Wang et al., 2019; Akbudak et al., 2020; Li et al., 2021) help to study the regulation of gene. TaPR-1-1 gene expression studied in wheat after osmotic stress, freezing, and salinity treatment and overexpression of TaPR-1-1 leads to stress tolerance response in Arabidopsis and yeast (Wang et al., 2019). In Arabidopsis, the Di19 (drought-induced) protein has been shown to positively regulate the expression of pathogenesis-related genes, specifically PR-1, PR-2, and PR-5 (Liu et al., 2013). 13 SlPR-1 reported in tomato, each gene leads to upregulation (as high as 50-fold) after drought stress treatment (Akbudak et al., 2020). Additionally, the genes PR1a2 and PR1b1 in tomato were consistently upregulated after 12- and 24-hour heat stress treatments, and heat shock element motifs were detected in their promoters (Arofatullah et al., 2018). In grape, the expression of VvHSP24, a class B heat shock protein has been found to be regulated by PR1. In transgenic Arabidopsis thaliana plants expressing VvNPR1 (NPR1 from Vitis vinifera) along with several salicylic acid (SA)-inducible genes, including PR1, PR2, and PR5, a notable decrease in resistance to Botrytis cinerea (a fungal pathogen) was observed compared to wildtype plants. The overexpression of VvNPR1 and these SA-inducible genes seemingly compromised the plant’s defense response, leading to reduced resistance against B. cinerea infection when compared to the natural or wildtype counterparts (Li et al., 2021). It has been reported that HuPR-1 is up-regulated by heat stress treatment, and it can also be induced by salt stress (Nong et al., 2019) and overexpressing of HuPR-1 in Arabidopsis results in tolerance to heat and salt stress. Pepper PR-1 protein shows both biotic and abiotic tolerance (Sarowar et al., 2005). These results provide new insights into heat stress regulation that could lead to a better understanding of how PR-1 genes are regulated in response to abiotic stresses in future studies (Jiao et al., 2021). These studies indicated that PR genes function not only in responses to biotic stress, but also in response to abiotic stress in rice.
The specific binding of transcription factors (TFs) to their respective transcription factor binding sites (TFBSs) is a crucial mechanism for mediating the transcriptional regulation of target genes. This process allows TFs to exert precise control over gene expression by interacting with specific DNA sequences in the regulatory regions of their target genes (Narlikar and Ovcharenko, 2009; Jayaram et al., 2016). The evidence of high throughput experimental methods accelerate the identification of TFBSs (Jayaram et al., 2016). However, in silico identification of TFBSs was still a method of choice to study various defense makers of plants (Boeva, 2016; Kumari et al., 2018). PR1 genes were acting as defense markers through activating a series of elements including transcription factors, MAPK and ROS. PR1 is a family of proteins whose expression regulates different functions during stress. The in silico analysis of the promoter regions of PR1 genes revealed the presence of multiple cis-regulatory elements, providing strong evidence for their involvement in multiple stress responses.
WRKY (W box) has an extensive role in biotic and abiotic stress along with a wide range of development process (Choi et al., 2015; Rinerson et al., 2015). Different PR1 genes of this study enriched with conserved consensus sequence TTGAC(C/T/A/G) (Supplementary Table 3). WRKY proteins play a crucial and diverse role in plant responses to various stresses and are involved in essential processes related to plant development, embryogenesis, dormancy, as well as responses to abiotic stresses like drought, salt, and heat in rice (Kalde et al., 2003; Jiang and Deyholos, 2009; Chen et al., 2012; Rushton et al., 2012). In addition, OsWRKY03 regulates a cascade of Jasmonic acid (JA) and salicylic acid signaling pathways to protect plants against bacterial and fungal infections (Liu et al., 2005). WRKY have been shown to exert a positive regulatory effect on the heat tolerance of plants. For example, in pepper plants, CaWRKY40 plays a vital role in the response to high-temperature stresses, contributing to enhanced heat tolerance. Similarly, in rice, OsWRKY11 is involved in the plant’s resistance to heat stress, further emphasizing the significance of WRKY in facilitating heat stress adaptation in different plant species (Wu et al., 2009; Cai et al., 2015). In Arabidopsis, it was observed that PR1 genes expression levels were notably higher in plants overexpressing WRKY39 compared to wild-type plants following heat treatment. This finding led researchers to speculate that WRKY39 likely acts as an upstream regulator of PR1, influencing its transcription and subsequently contributing to the plant’s response to heat stress (Li et al., 2010).
The basic region/leucine zipper motif (bZIP) plays a crucial role in regulating diverse stress and developmental responses in plants. These bZIP proteins exert their regulatory functions by binding to specific transcription factor binding sites (TFBSs) on DNA. Notably, plant bZIP proteins have a preference for recognizing the ACGT core in DNA sequences, particularly in regions known as the A-box, C-box, and G-box (Jakoby et al., 2002), CCAAT-box, TGA-element, NON, ABRE (Holdsworth et al., 1995; Jakoby et al., 2002; Liao et al., 2008) AS-1 (Després et al., 2000), TATCCAT/C-motif (Kong et al., 2018). G-box element imparts response to abscisic acid ABRE (ABA-responsive element), methyl-jasmonate, Anaerobiosis, defense responses ethylene induction as well as in seed specific expression found in most of studied PR1 genes (Kong et al., 2018). The TATCCAT/C-motif (Kong et al., 2018) and TGACGTCA, commonly referred to as the G-box, is abundantly present in the promoter regions of many stress-inducible genes, including the PR1 promoter. This interplay between SA, the TGA subfamily, and NPR1 represents a crucial mechanism underlying the activation of promoters involved in the plant’s defense against pathogens (Zander et al., 2014). In a previous report were found that AS-1 cis element is an oxidative stress-responsive element and activated by SA by binding TGA cis element (Zander et al., 2014). bZIP proteins have impart their major role in activating a number of defense genes (Alves et al., 2013).
AP2 is one of the biggest plant TF super families which play a significant role in regulating plant growth and stress responses (Najafi et al., 2018). The AP2 (APETALA2)/ERF family is categorized into three distinct subfamilies based on the number of AP2/ERF domains they possess. These subfamilies are known as AP2, ethylene responsive factor (ERF), and RAV. Each subfamily is distinguished by its unique arrangement of AP2/ERF domains (Nakano et al., 2014). The ERF subfamily plays a crucial role in regulating a wide array of genes associated with both biotic and abiotic stresses in rice. These stresses include challenging environmental conditions like drought, high salinity, and cold temperatures (Pandey et al., 2014). The AP2 subfamily is known to be involved in regulations of developmental process such as flower development, ovule development, seed set and regulating organ-specific growth while RAV subfamily genes response to expression induced by ethylene, brassinosteroids, and biotic and abiotic stresses (Xu et al., 2013). ERF subfamily members bind to the conserved nucleotide sequences AGCCGCC (Xu et al., 2013) and TACCGACAT (GCC-box) (Pandey et al., 2014) in the upstream regions of genes while the RAV binds to CAACA and CACCTG motif (Song et al., 2013). AP2/ERF transcription factors have been identified as regulators that can trigger the synthesis of ethylene (ET), salicylic acid (SA), and jasmonic acid (JA). These signaling molecules play a significant role in enhancing the expression of pathogenesis-related (PR) genes during instances of pathogen infestation (Singh, 2002; Brown et al., 2003). Moreover, the presence of AP2/ERF transcription factor binding sites (TFBSs) in defense-responsive genes has been found to strengthen resistance against specific biotic and abiotic stresses (Berrocal-Lobo et al., 2002). The ERF family of TFs was shown to regulate abiotic and biotic stresses (Pandey et al., 2014) as well as involved in ethylene mediated PR genes expression which confer tolerance against cold and dehydration stresses (Liu et al., 2007). In the AP2/EREBP family, a subset called DREBs is classified under the EREBP subfamily and plays a pivotal role in plants’ response to various abiotic stresses. Notably, specific DREB members, such as rice OsDREB2B and maize ZmDREB2A, are known to be induced and expressed in response to high-temperature stress. This induction of DREBs assists plants in coping with the challenges posed by elevated temperature conditions (Qin et al., 2007; Matsukura et al., 2010). CG-1;CAMTAs is comprised of core sequence CGTG (Silva, 1994) which associated with ABRE cis- element. In our study of PR1 genes we found the same consensus sequence and another cis-element chs-CMA1a (TTACTTAA) were also present. Calmodulin binding transcription activators (CAMTAs) are proteins that exhibit responsiveness to a diverse range of external signals, including cold, wounding, and drought. Additionally, they are influenced by hormonal signals like ethylene and ABA (Reddy et al., 2000; Yang and Poovaiah, 2003). Our research findings indicate that Calmodulin signaling responsive genes are predominantly linked to ABRE (CGTG) cis-elements.
This may be possible that PR1 signaling response is linked to ABA hormone signaling. bHLH can recognize the MYC (CATTTG), G-box, E-box, WRE3 (CACCT) (Miyamoto et al., 2012). bHLH transcription factors (TFs) play a crucial role in various aspects of plant biology, including SA and JA mediated stress responses, light-induced hormone signaling, wound and drought stress management, shoot branching, as well as fruit and flower development, root development, and other pathways (Carretero-Paulet et al., 2010). Moreover, bHLH TFs are also involved in inducing ABA-dependent signaling to cope with cold stress, specifically in rice (Chinnusamy et al., 2003). MYB transcription factors (TFs) are universally present in eukaryotes and are associated with specific TFBS sequences, such as TAACTG, CAACTG, AACGG, and C/TAACNA/G, as reported by Xu et al. (2008) and Qin et al. (2012). These MYBs play essential roles in various biological processes, including plant development, metabolism, and stress responses. In rice, MYB TFs have been demonstrated to be crucial in managing abiotic stresses like dehydration, salt, and cold stresses (Abe et al., 1997). They are also involved in the ABA and SA-signaling pathways, which confer responses to both biotic and abiotic stresses (Ambawat et al., 2013). One specific member of the MYB family, OsMYB55, has been shown to enhance the tolerance of rice plants to high temperatures. This is achieved by increasing the expression of downstream genes like OsGS1;2, GAT1, and GAD3, which are involved in amino acid metabolism (El-Kereamy et al., 2015).
TCP proteins possess a distinctive TCP domain, and their binding sites exhibit variation, including sequences like GGNCCCAC, GTGGNCCC, and TGGGCC, as identified by Giraud et al. (2011). These TCP genes play a crucial role in influencing the development of axillary structures, petals, and stamens in plants (Cubas et al., 1999). The SBP protein has been shown to interact with the GTAC core sequence, as demonstrated by Wang et al. (2015). In plants, the SBP protein is actively involved in various developmental processes, including flower and fruit development, architecture formation, responses to copper and fungal toxins, as well as regulation of GA (gibberellic acid) levels (Wang et al., 2009). B3 cis -elements are combining with core sequences of CATGC (RY-element) verified in all PR1 genes. The B3 family of proteins plays significant roles in regulating flowering time, organ growth, and organ polarity in plants. These B3 members are distributed in various plant tissues, including pistils, stamens, germinated seeds, and roots (Swaminathan et al., 2008). Together with these TFBSs, many more found in PR1 genes, which regulate the multiple stress along with developmental process.
This study provides valuable insights into the multifaceted nature of PR1 proteins. While these proteins play a crucial role in connecting biotic and abiotic stress responses, certain aspects of their characteristics remain unexplored and require further investigation. There is a requirement for further expansions in the classification of PR-proteins, extending the existing 17 families. The considerable expansion of the PR proteins and the differential expression of several OsPR1 genes in response to abiotic and biotic stresses supports the importance of further investigating PR1 genes as targets for increasing stress tolerance in crop plants. Several PR1 proteins, which may play a role in plant defense mechanisms, have been uncovered from our study. We detected PR1 homologues in different crops by performing computational prediction analysis and querying the PR1 protein dataset from OsPR1. These 58 putative PR1 proteins shared sequence, subcellular localization, SCP domain, similarities with the query PR1 proteins from OsPR1. The expression characteristic studied in eleven OsPR1 genes after SA, wounding, BR, HS treatment, which proves its predominant role as a marker of biotic and abiotic stress. The expression profiling reveals the evident role in the SA-dependent pathway. Additionally, critical promoter analysis of 1000 bp was performed to find the role in TFBSs in response to multiple stress. In short, the findings of this study may provide new insight into the regulatory mechanism of OsPR1 in response to BR and HS that could aid future study to examine the role of OsPR1 genes in response to different abiotic stress. The in-silico promoter analysis of O. sativa, Z. mays, B. distachyon, H. vulgare, B. rapa further can provide plant genetic engineering technology for protection of crops against biotic and abiotic stress.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
DK: Writing – review & editing, Writing – original draft, Visualization, Supervision, Methodology, Funding acquisition, Conceptualization. BP: Writing – review & editing, Conceptualization. PD: Writing – review & editing, Investigation, Conceptualization. SS: Writing – review & editing, Supervision. MK: Writing – review & editing, Visualization, Supervision. SA: Writing – review & editing. MA: Writing – review & editing. KA: Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The authors extend their appreciation to the Researchers Supporting Project number (RSP2024R194), King Saud University, Riyadh, Saudi Arabia.
DK kindly acknowledges the BHU-UGC fellowship. The authors extend their appreciation to the Researchers Supporting Project number (RSP2024R194), King Saud University, Riyadh, Saudi Arabia.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1463147/full#supplementary-material
Supplementary Figure 1 | Workflow of promoter analysis of PR1 genes in different crops. A complete analysis of promoter region involves various steps which start from downloading naïve protein sequencing. The major steps shown in screenshot of regions which link the steps. The users may change the species name by selecting species name in Phytozome12. Tool. Downloaded protein sequence cross was verified in Phytozome12. Tool afterwards promoter sequence was retrieved from genomic region.
Supplementary Table 1 | List of OsPR1 Primers used in gene expression analysis.
Supplementary Table 2 | Characterization of Transcription Factor Binding Sites in Promoter Regions of Rice Pathogenesis-Related (OsPR1) Genes.
Supplementary Table 3 | Identification and Functional Categorization of Cis-Elements in the 1000 bp Upstream Promoter Region of Pathogenesis-Related 1 (PR1) Genes in (Oryza sativa).
Supplementary Table 4 | Comparative Analysis and Categorization of Cis-Elements in the 1000 bp Upstream Promoter Regions of Pathogenesis-Related 1 (PR1) Genes Across Zea mays, Brachypodium distachyon, Hordeum vulgare, and Brassica rapa.
Supplementary Table 5 | List of Genes taken for Search BLASTP search of other genes.
Supplementary Table 6 | List of Genes obtained after BLASTP search.
Supplementary Table 7 | List of Genes obtained after HMM search.
Abe, H., Yamaguchi-Shinozaki, K., Urao, T., Iwasaki, T., Hosokawa, D., Shinozaki, K. (1997). Role of arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell 9, 1859–1868. doi: 10.1105/tpc.9.10.1859
Agarwala, R., Barrett, T., Beck, J., Benson, D. A., Bollin, C., Bolton, E., et al. (2018). Database resources of the National Center for biotechnology information. Nucleic Acids Res. 46, D8–D13. doi: 10.1093/nar/gkx1095
Akbudak, M. A., Yildiz, S., Filiz, E. (2020). Pathogenesis related protein-1 (PR-1) genes in tomato (Solanum lycopersicum L.): Bioinformatics analyses and expression profiles in response to drought stress. Genomics 112, 4089–4099. doi: 10.1016/j.ygeno.2020.07.004
Ali, S., Ganai, B. A., Kamili, A. N., Bhat, A. A., Mir, Z. A., Bhat, J. A., et al. (2018a). Pathogenesis-relted proteins and peptides as promising tools for engineering plants with multiple stress tolerance. Microbiol. Res. 212–213, 29–37. doi: 10.1016/j.micres.2018.04.008
Ali, S., Mir, Z. A., Bhat, J. A., Tyagi, A., Chandrashekar, N., Yadav, P., et al. (2018b). Isolation and characterization of systemic acquired resistance marker gene PR1 and its promoter from Brassica juncea. 3 Biotech. 8, 10. doi: 10.1007/s13205-017-1027-8
Alves, M., Dadalto, S., Gonçalves, A., De Souza, G., Barros, V., Fietto, L. (2013). Plant bZIP transcription factors responsive to pathogens: A review. Int. J. Mol. Sci. 14, 7815–7828. doi: 10.3390/ijms14047815
Ambawat, S., Sharma, P., Yadav, N. R., Yadav, R. C. (2013). MYB transcription factor genes as regulators for plant responses: an overview. Physiol. Mol. Biol. Plants 19, 307–321. doi: 10.1007/s12298-013-0179-1
Anuradha, C., Chandrasekar, A., Backiyarani, S., Thangavelu, R., Giribabu, P., Uma, S. (2022). Genome-wide analysis of pathogenesis-related protein 1 (PR-1) gene family from Musa spp. and its role in defense response during stresses. Gene 821, 146334. doi: 10.1016/j.gene.2022.146334
Arofatullah, N. A., Hasegawa, M., Tanabata, S., Ogiwara, I., Sato, T. (2018). Heat Shock-Induced Resistance Against Pseudomonas syringae pv. tomato (Okabe) Young et al. via Heat Shock Transcription Factors in Tomato. Agronomy 9, 2. doi: 10.3390/agronomy9010002
Berrocal-Lobo, M., Molina, A., Solano, R. (2002). Constitutive expression of ETHYLENE-RESPONSE-FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi. Plant J. 29, 23–32. doi: 10.1046/j.1365-313x.2002.01191.x
Boeva, V. (2016). Analysis of genomic sequence motifs for deciphering transcription factor binding and transcriptional regulation in eukaryotic cells. Front. Genet. 7. doi: 10.3389/fgene.2016.00024
Brown, R. L., Kazan, K., McGrath, K. C., Maclean, D. J., Manners, J. M. (2003). A role for the GCC-box in Jasmonate-mediated activation of the PDF1.2 gene of arabidopsis. Plant Physiol. 132, 1020–1032. doi: 10.1104/pp.102.017814
Cai, H., Yang, S., Yan, Y., Xiao, Z., Cheng, J., Wu, J., et al. (2015). CaWRKY6 transcriptionally activates CaWRKY40, regulates Ralstonia solanacearum resistance, and confers high-temperature and high-humidity tolerance in pepper. J. Exp. Bot. 66, 3163–3174. doi: 10.1093/jxb/erv125
Carretero-Paulet, L., Galstyan, A., Roig-Villanova, I., Martínez-García, J. F., Bilbao-Castro, J. R., Robertson, D. L. (2010). Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae. Plant Physiol. 153, 1398–1412. doi: 10.1104/pp.110.153593
Chaudhary, B., Prasad, B. D., Sahni, S., Kumari, D., Sharma, V. K., Dwivedi, P. (2024). Brassinosteroid mediated modulation in the gene expression analysis of genes associated with arsenic uptake and transport in rice (Oryza sativa L.) for effective mitigation of arsenic. J. Plant Growth Regul. 43, 3159–3170. doi: 10.1007/s00344-023-11073-1
Chen, L., Song, Y., Li, S., Zhang, L., Zou, C., Yu, D. (2012). The role of WRKY transcription factors in plant abiotic stresses. Biochim. Biophys. Acta - Gene Regul. Mech. 1819, 120–128. doi: 10.1016/j.bbagrm.2011.09.002
Chen, Y.-L., Lee, C.-Y., Cheng, K.-T., Chang, W.-H., Huang, R.-N., Nam, H. G., et al. (2014). Quantitative peptidomics study reveals that a wound-induced peptide from PR-1 regulates immune signaling in tomato. Plant Cell 26, 4135–4148. doi: 10.1105/tpc.114.131185
Chinnusamy, V., Ohta, M., Kanrar, S., Lee, B.-H., Hong, X., Agarwal, M., et al. (2003). ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 17, 1043–1054. doi: 10.1101/gad.1077503
Choi, C., Hwang, S., Fang, I. R., Kwon, S., II, Park, S. R., Ahn, I., et al. (2015). Molecular characterization of Oryza sativa WRKY 6, which binds to W-box-like element 1 of the Oryza sativa pathogenesis-related (PR) 10a promoter and confers reduced susceptibility to pathogens. New Phytol. 208, 846–859. doi: 10.1111/nph.13516
Chow, C.-N., Zheng, H.-Q., Wu, N.-Y., Chien, C.-H., Huang, H.-D., Lee, T.-Y., et al. (2016). PlantPAN 2.0: an update of plant promoter analysis navigator for reconstructing transcriptional regulatory networks in plants. Nucleic Acids Res. 44, D1154–D1160. doi: 10.1093/nar/gkv1035
Cubas, P., Lauter, N., Doebley, J., Coen, E. (1999). The TCP domain: a motif found in proteins regulating plant growth and development. Plant J. 18, 215–222. doi: 10.1046/j.1365-313X.1999.00444.x
Després, C., DeLong, C., Glaze, S., Liu, E., Fobert, P. R. (2000). The Arabidopsis NPR1/NIM1 protein enhances the DNA binding activity of a subgroup of the TGA family of bZIP transcription factors. Plant Cell 12, 279–290. Available at: http://www.ncbi.nlm.nih.gov/pubmed/10662863.
Divi, U. K., Rahman, T., Krishna, P. (2010). Brassinosteroid-mediated stress tolerance in Arabidopsis shows interactions with abscisic acid, ethylene and salicylic acid pathways. BMC Plant Biol. 10, 151. doi: 10.1186/1471-2229-10-151
Dossa, G. S., Torres, R., Henry, A., Oliva, R., Maiss, E., Cruz, C. V., et al. (2017). Rice response to simultaneous bacterial blight and drought stress during compatible and incompatible interactions. Eur. J. Plant Pathol. 147, 115–127. doi: 10.1007/s10658-016-0985-8
El-Kereamy, A., Bi, Y.-M., Mahmood, K., Ranathunge, K., Yaish, M. W., Nambara, E., et al. (2015). Overexpression of the CC-type glutaredoxin, OsGRX6 affects hormone and nitrogen status in rice plants. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00934
Giraud, E., Ng, S., Carrie, C., Duncan, O., Low, J., Lee, C. P., et al. (2011). TCP transcription factors link the regulation of genes encoding mitochondrial proteins with the circadian clock in arabidopsis thaliana. Plant Cell 22, 3921–3934. doi: 10.1105/tpc.110.074518
Goodstein, D. M., Shu, S., Howson, R., Neupane, R., Hayes, R. D., Fazo, J., et al. (2012). Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178–D1186. doi: 10.1093/nar/gkr944
Graham, M., Weidner, J., Wheeler, K., Pelow, M., Graham, T. (2003). Induced expression of pathogenesis-related protein genes in soybean by wounding and the Phytophthora sojae cell wall glucan elicitor. Physiol. Mol. Plant Pathol. 63, 141–149. doi: 10.1016/j.pmpp.2003.11.002
Grover, A., Gowthaman, R. (2003). Strategies for development of fungus-resistant transgenic plants. Curr. Sci. 84, 330–340.
Hegde, Y. R., Keshgond, R. S. (2013). Role of pathogenesis-related proteins in plant disease management -a review. Agric. Rev. 34, 145–151.
Holdsworth, M. J., Muñoz-Blanco, J., Hammond-Kosack, M., Colot, V., Schuch, W., Bevan, M. W. (1995). The maize transcription factor Opaque-2 activates a wheat glutenin promoter in plant and yeast cells. Plant Mol. Biol. 29, 711–720. doi: 10.1007/BF00041162
Jakoby, M., Weisshaar, B., Dröge-Laser, W., Vicente-Carbajosa, J., Tiedemann, J., Kroj, T., et al. (2002). bZIP transcription factors in Arabidopsis. Trends Plant Sci. 7, 106–111. doi: 10.1016/S1360-1385(01)02223-3
Jayaram, N., Usvyat, D., Martin, A. C. R. (2016). Evaluating tools for transcription factor binding site prediction. BMC Bioinf. 17, 547. doi: 10.1186/s12859-016-1298-9
Jiang, L., Wu, J., Fan, S., Li, W., Dong, L., Cheng, Q., et al. (2015). Isolation and Characterization of a Novel Pathogenesis-Related Protein Gene (GmPRP) with Induced Expression in Soybean (Glycine max) during Infection with Phytophthora sojae. PloS One 10, e0129932. doi: 10.1371/journal.pone.0129932
Jiang, Y., Deyholos, M. K. (2009). Functional characterization of Arabidopsis NaCl-inducible WRKY25 and WRKY33 transcription factors in abiotic stresses. Plant Mol. Biol. 69, 91–105. doi: 10.1007/s11103-008-9408-3
Jiao, Z., Xu, W., Nong, Q., Zhang, M., Jian, S., Lu, H., et al. (2021). An integrative transcriptomic and metabolomic analysis of red pitaya (Hylocereus polyrhizus) seedlings in response to heat stress. Genes (Basel) 12, 1714. doi: 10.3390/genes12111714
Kakei, Y., Mochida, K., Sakurai, T., Yoshida, T., Shinozaki, K., Shimada, Y. (2015). Transcriptome analysis of hormone-induced gene expression in Brachypodium distachyon. Sci. Rep. 5, 14476. doi: 10.1038/srep14476
Kalde, M., Barth, M., Somssich, I. E., Lippok, B. (2003). Members of the arabidopsis WRKY group III transcription factors are part of different plant defense signaling pathways. Mol. Plant-Microbe Interact. 16, 295–305. doi: 10.1094/MPMI.2003.16.4.295
Kaur, A., Pati, P. K., Pati, A. M., Nagpal, A. K. (2017). In-silico analysis of cis-acting regulatory elements of pathogenesis-related proteins of Arabidopsis thaliana and Oryza sativa. PloS One 12, e0184523. doi: 10.1371/journal.pone.0184523
Kaur, G., Pati, P. K. (2016). Analysis of cis-acting regulatory elements of Respiratory burst oxidase homolog (Rboh) gene families in Arabidopsis and rice provides clues for their diverse functions. Comput. Biol. Chem. 62, 104–118. doi: 10.1016/j.compbiolchem.2016.04.002
Kong, W., Ding, L., Cheng, J., Wang, B. (2018). Identification and expression analysis of genes with pathogen-inducible cis-regulatory elements in the promoter regions in Oryza sativa. Rice 11, 52. doi: 10.1186/s12284-018-0243-0
Kothari, K. S., Dansana, P. K., Giri, J., Tyagi, A. K. (2016). Rice stress associated protein 1 (OsSAP1) interacts with aminotransferase (OsAMTR1) and pathogenesis-related 1a protein (OsSCP) and regulates abiotic stress responses. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01057
Kumar, A., Singh, J. K., Upadhyay, P. K. (2020). Effect of INM in aromatic rice (Oryza sativa) under SRI and its residual effect on wheat (Triticum aestivum). Indian J. Agric. Sci. 90, 1258–1266. doi: 10.56093/ijas.v90i7.105576
Kumari, D., Parasad, B. D., Sahni, S., Ghatak, A. (2020). Identification and Functional Characterization of Xanthomonas oryzae Pv. oryzae Isolates. Curr. J. Appl. Sci. Technol. 78–84. doi: 10.9734/cjast/2020/v39i430533
Kumari, D., Parasad, B. D., Singh, A. K., Solanki, I. S. (2018). In silico comparative analysis of transcriptional factor binding sites in rice and arabidopsis calmodulin binding protein 60s genes. Curr. J. Appl. Sci. Technol. 31, 1–9. doi: 10.9734/cjast/2018/45903
Kumari, D., Prasad, B. D., Sahni, S., Nonhebel, H. M., Krishna, P. (2022). The Expanded and Diversified Calmodulin-Binding Protein 60 (CBP60) Family in Rice (Oryza sativa L.) Is Conserved in Defense Responses against Pathogens. Agronomy 12, 3060. doi: 10.3390/agronomy12123060
Kundu, S., Chakraborty, D., Kundu, A., Pal, A. (2013). Proteomics approach combined with biochemical attributes to elucidate compatible and incompatible plant-virus interactions between Vigna mungo and Mungbean Yellow Mosaic India Virus. Proteome Sci. 11, 15. doi: 10.1186/1477-5956-11-15
Lescot, M. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Li, C., Cao, S., Wang, K., Lei, C., Ji, N., Xu, F., et al. (2021). Heat shock protein HSP24 is involved in the BABA-induced resistance to fungal pathogen in postharvest grapes underlying an NPR1-dependent manner. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.646147
Li, J., Chory, J. (1999). Brassinosteroid actions in plants. J. Exp. Bot. 50, 275–282. doi: 10.1093/jxb/50.332.275
Li, S., Zhou, X., Chen, L., Huang, W., Yu, D. (2010). Functional characterization of Arabidopsis thaliana WRKY39 in heat stress. Mol. Cells 29, 475–483. doi: 10.1007/s10059-010-0059-2
Liao, Y., Zou, H.-F., Wei, W., Hao, Y.-J., Tian, A.-G., Huang, J., et al. (2008). Soybean GmbZIP44, GmbZIP62 and GmbZIP78 genes function as negative regulator of ABA signaling and confer salt and freezing tolerance in transgenic Arabidopsis. Planta 228, 225–240. doi: 10.1007/s00425-008-0731-3
Liu, X. Q., Bai, X. Q., Qian, Q., Wang, X. J., Chen, M. S., Chu, C. C. (2005). OsWRKY03, a rice transcriptional activator that functions in defense signaling pathway upstream of OsNPR1. Cell Res. 15, 593–603. doi: 10.1038/sj.cr.7290329
Liu, T., Chen, T., Kan, J., Yao, Y., Guo, D., Yang, Y., et al. (2022). The GhMYB36 transcription factor confers resistance to biotic and abiotic stress by enhancing PR1 gene expression in plants. Plant Biotechnol. J. 20, 722–735. doi: 10.1111/pbi.13751
Liu, J.-X., Srivastava, R., Che, P., Howell, S. H. (2007). Salt stress responses in Arabidopsis utilize a signal transduction pathway related to endoplasmic reticulum stress signaling. Plant J. 51, 897–909. doi: 10.1111/j.1365-313X.2007.03195.x
Liu, W.-X., Zhang, F.-C., Zhang, W.-Z., Song, L.-F., Wu, W.-H., Chen, Y.-F. (2013). Arabidopsis di19 functions as a transcription factor and modulates PR1, PR2, and PR5 expression in response to drought stress. Mol. Plant 6, 1487–1502. doi: 10.1093/mp/sst031
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lodhi, N., Ranjan, A., Singh, M., Srivastava, R., Singh, S., Chaturvedi, C., et al. (2008). Interactions between upstream and core promoter sequences determine gene expression and nucleosome positioning in tobacco PR-1a promoter. Biochim. Biophys. Acta - Gene Regul. Mech. 1779, 634–644. doi: 10.1016/j.bbagrm.2008.07.010
Maleck, K. (1999). Defense on multiple fronts: how do plants cope with diverse enemies? Trends Plant Sci. 4, 215–219. doi: 10.1016/S1360-1385(99)01415-6
Matsukura, S., Mizoi, J., Yoshida, T., Todaka, D., Ito, Y., Maruyama, K., et al. (2010). Comprehensive analysis of rice DREB2-type genes that encode transcription factors involved in the expression of abiotic stress-responsive genes. Mol. Genet. Genomics 283, 185–196. doi: 10.1007/s00438-009-0506-y
Mazarei, M., Teplova, I., Hajimorad, M., Stewart, C. (2008). Pathogen phytosensing: plants to report plant pathogens. Sensors 8, 2628–2641. doi: 10.3390/s8042628
Mazumder, M., Das, S., Saha, U., Chatterjee, M., Bannerjee, K., Basu, D. (2013). Salicylic acid-mediated establishment of the compatibility between Alternaria brassicicola and Brassica juncea is mitigated by abscisic acid in Sinapis alba. Plant Physiol. Biochem. 70, 43–51. doi: 10.1016/j.plaphy.2013.04.025
Mistry, J., Chuguransky, S., Williams, L., Qureshi, M., Salazar, G. A., Sonnhammer, E. L. L., et al. (2021). Pfam: The protein families database in 2021. Nucleic Acids Res. 49, D412–D419. doi: 10.1093/nar/gkaa913
Mitsuhara, I., Iwai, T., Seo, S., Yanagawa, Y., Kawahigasi, H., Hirose, S., et al. (2008). Characteristic expression of twelve rice PR1 family genes in response to pathogen infection, wounding, and defense-related signal compounds (121/180). Mol. Genet. Genomics 279, 415–427. doi: 10.1007/s00438-008-0322-9
Miyamoto, K., Shimizu, T., Lin, F., Sainsbury, F., Thuenemann, E., Lomonossoff, G., et al. (2012). Identification of an E-box motif responsible for the expression of jasmonic acid-induced chitinase gene OsChia4a in rice. J. Plant Physiol. 169, 621–627. doi: 10.1016/j.jplph.2011.12.008
Najafi, S., Sorkheh, K., Nasernakhaei, F. (2018). Characterization of the APETALA2/Ethylene-responsive factor (AP2/ERF) transcription factor family in sunflower. Sci. Rep. 8, 11576. doi: 10.1038/s41598-018-29526-z
Nakano, T., Fujisawa, M., Shima, Y., Ito, Y. (2014). The AP2/ERF transcription factor SlERF52 functions in flower pedicel abscission in tomato. J. Exp. Bot. 65, 3111–3119. doi: 10.1093/jxb/eru154
Nakashita, H., Yasuda, M., Nitta, T., Asami, T., Fujioka, S., Arai, Y., et al. (2003). Brassinosteroid functions in a broad range of disease resistance in tobacco and rice. Plant J. 33, 887–898. doi: 10.1046/j.1365-313X.2003.01675.x
Narlikar, L., Ovcharenko, I. (2009). Identifying regulatory elements in eukaryotic genomes. Briefings Funct. Genomics Proteomics 8, 215–230. doi: 10.1093/bfgp/elp014
Nong, Q., Yang, Y., Zhang, M., Zhang, M., Chen, J., Jian, S., et al. (2019). RNA-seq-based selection of reference genes for RT-qPCR analysis of pitaya. FEBS Open Bio 9, 1403–1412. doi: 10.1002/2211-5463.12678
Pandey, B., Sharma, P., Saini, M., Pandey, D. M., Sharma, I. (2014). Isolation and characterization of dehydration-responsive element-binding factor 2: (DREB2) from Indian wheat (Triticum aestivum L.) cultivars. Aust. J. Crop Sci. 8, 44–54.
Park, S.-W., Kaimoyo, E., Kumar, D., Mosher, S., Klessig, D. F. (2007). Methyl salicylate is a critical mobile signal for plant systemic acquired resistance. Sci. (80-.) 318, 113–116. doi: 10.1126/science.1147113
Pierleoni, A., Martelli, P. L., Fariselli, P., Casadio, R. (2006). BaCelLo: a balanced subcellular localization predictor. Bioinformatics 22, e408–e416. doi: 10.1093/bioinformatics/btl222
Prakash, A., Jeffryes, M., Bateman, A., Finn, R. D. (2017). The HMMER web server for protein sequence similarity search. Curr. Protoc. Bioinforma 60, 3–15. doi: 10.1002/cpbi.40
Prasad, B. D., Creissen, G., Lamb, C., Chattoo, B. B. (2009). Overexpression of rice (Oryza sativa L.) osCDR1 leads to constitutive activation of defense responses in rice and arabidopsis. Mol. Plant-Microbe Interact. 22, 1635–1644. doi: 10.1094/MPMI-22-12-1635
Prasad, B. D., Kumar, P., Sahni, S., Kumar, V., Kumari, S., Kumar, P., et al. (2016). An improved protocol for agrobacterium-mediated genetic transformation and regeneration of indica rice (Oryza sativa L. Var. Rajendra kasturi) 16, 5597–5606.
Qin, F., Kakimoto, M., Sakuma, Y., Maruyama, K., Osakabe, Y., Tran, L.-S. P., et al. (2007). Regulation and functional analysis of ZmDREB2A in response to drought and heat stresses in Zea mays L. Plant J. 50, 54–69. doi: 10.1111/j.1365-313X.2007.03034.x
Qin, Y., Wang, M., Tian, Y., He, W., Han, L., Xia, G. (2012). Over-expression of TaMYB33 encoding a novel wheat MYB transcription factor increases salt and drought tolerance in Arabidopsis. Mol. Biol. Rep. 39, 7183–7192. doi: 10.1007/s11033-012-1550-y
Reddy, A. S.N., Reddy, V. S., Golovkin, M. (2000). A calmodulin binding protein from arabidopsis is induced by Ethylene and contains a DNA-binding motif. Biochem. Biophys. Res. Commun. 279, 762–769. doi: 10.1006/bbrc.2000.4032
Rinerson, C. I., Scully, E. D., Palmer, N. A., Donze-Reiner, T., Rabara, R. C., Tripathi, P., et al. (2015). The WRKY transcription factor family and senescence in switchgrass. BMC Genomics 16, 912. doi: 10.1186/s12864-015-2057-4
Rushton, D. L., Tripathi, P., Rabara, R. C., Lin, J., Ringler, P., Boken, A. K., et al. (2012). WRKY transcription factors: key components in abscisic acid signalling. Plant Biotechnol. J. 10, 2–11. doi: 10.1111/j.1467-7652.2011.00634.x
Sahni, S., Prasad, B. D., Liu, Q., Grbic, V., Sharpe, A., Singh, S. P., et al. (2016). Overexpression of the brassinosteroid biosynthetic gene DWF4 in Brassica napus simultaneously increases seed yield and stress tolerance. Sci. Rep. 6, 28298. doi: 10.1038/srep28298
Sakai, H., Lee, S. S., Tanaka, T., Numa, H., Kim, J., Kawahara, Y., et al. (2013). Rice annotation project database (RAP-DB): an integrative and interactive database for rice genomics. Plant Cell Physiol. 54, e6–e6. doi: 10.1093/pcp/pcs183
Sarowar, S., Kim, Y. J., Kim, E. N., Kim, K. D., Hwang, B. K., Islam, R., et al. (2005). Overexpression of a pepper basic pathogenesis-related protein 1 gene in tobacco plants enhances resistance to heavy metal and pathogen stresses. Plant Cell Rep. 24, 216–224. doi: 10.1007/s00299-005-0928-x
Schneiter, R., Di Pietro, A. (2013). The CAP protein superfamily: function in sterol export and fungal virulence. Biomol. Concepts 4, 519–525. doi: 10.1515/bmc-2013-0021
Scranton, M. A., Fowler, J. H., Girke, T., Walling, L. L. (2013). Microarray analysis of tomato’s early and late wound response reveals new regulatory targets for leucine aminopeptidase A. PloS One 8, e77889. doi: 10.1371/journal.pone.0077889
Seo, S., Katou, S., Seto, H., Gomi, K., Ohashi, Y. (2007). The mitogen-activated protein kinases WIPK and SIPK regulate the levels of jasmonic and salicylic acids in wounded tobacco plants. Plant J. 49, 899–909. doi: 10.1111/j.1365-313X.2006.03003.x
Silva, O.da C.e (1994). CG-1, a parsley light-induced DNA-binding protein. Plant Mol. Biol. 25, 921–924. doi: 10.1007/BF00028887
Silverman, P., Seskar, M., Kanter, D., Schweizer, P., Metraux, J. P., Raskin, I. (1995). Salicylic acid in rice (Biosynthesis, conjugation, and possible role). Plant Physiol. 108, 633–639. doi: 10.1104/pp.108.2.633
Singh, K. (2002). Transcription factors in plant defense and stress responses. Curr. Opin. Plant Biol. 5, 430–436. doi: 10.1016/S1369-5266(02)00289-3
Song, X., Li, Y., Hou, X. (2013). Genome-wide analysis of the AP2/ERF transcription factor superfamily in Chinese cabbage (Brassica rapa ssp. pekinensis). BMC Genomics 14, 573. doi: 10.1186/1471-2164-14-573
Sreeramulu, S., Mostizky, Y., Sunitha, S., Shani, E., Nahum, H., Salomon, D., et al. (2013). BSKs are partially redundant positive regulators of brassinosteroid signaling in Arabidopsis. Plant J. 74, 905–919. doi: 10.1111/tpj.12175
Swaminathan, K., Peterson, K., Jack, T. (2008). The plant B3 superfamily. Trends Plant Sci. 13, 647–655. doi: 10.1016/j.tplants.2008.09.006
Szekeres, M., Németh, K., Koncz-Kálmán, Z., Mathur, J., Kauschmann, A., Altmann, T., et al. (1996). Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in arabidopsis. Cell 85, 171–182. doi: 10.1016/S0092-8674(00)81094-6
Takatsuji, H., Jiang, C.-J., Sugano, S. (2010). Salicylic acid signaling pathway in rice and the potential applications of its regulators. Japan Agric. Res. Q. JARQ 44, 217–223. doi: 10.6090/jarq.44.217
Tamura, K., Stecher, G., Kumar, S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Timilsina, S., Potnis, N., Newberry, E. A., Liyanapathiranage, P., Iruegas-Bocardo, F., White, F. F., et al. (2020). Xanthomonas diversity, virulence and plant–pathogen interactions. Nat. Rev. Microbiol. 18, 415–427. doi: 10.1038/s41579-020-0361-8
van Loon, L. C., Rep, M., Pieterse, C. M. J. (2006). Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 44, 135–162. doi: 10.1146/annurev.phyto.44.070505.143425
Van Loon, L. C., Van Strien, E. A. (1999). The families of pathogenesis-related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiol. Mol. Plant Pathol. 55, 85–97. doi: 10.1006/pmpp.1999.0213
Wang, J., Mao, X., Wang, R., Li, A., Zhao, G., Zhao, J., et al. (2019). Identification of wheat stress-responding genes and TaPR-1-1 function by screening a cDNA yeast library prepared following abiotic stress. Sci. Rep. 9, 141. doi: 10.1038/s41598-018-37859-y
Wang, Y., Hu, Z., Yang, Y., Chen, X., Chen, G. (2009). Function annotation of an SBP-box gene in arabidopsis based on analysis of co-expression networks and promoters. Int. J. Mol. Sci. 10, 116–132. doi: 10.3390/ijms10010116
Wang, X., Gao, J., Zhu, Z., Dong, X., Wang, X., Ren, G., et al. (2015). TCP transcription factors are critical for the coordinated regulation of ISOCHORISMATE SYNTHASE 1 expression in Arabidopsis thaliana. Plant J. 82, 151–162. doi: 10.1111/tpj.12803
Wu, X., Shiroto, Y., Kishitani, S., Ito, Y., Toriyama, K. (2009). Enhanced heat and drought tolerance in transgenic rice seedlings overexpressing OsWRKY11 under the control of HSP101 promoter. Plant Cell Rep. 28, 21–30. doi: 10.1007/s00299-008-0614-x
Xu, W., Li, F., Ling, L., Liu, A. (2013). Genome-wide survey and expression profiles of the AP2/ERF family in castor bean (Ricinus communis L.). BMC Genomics 14, 785. doi: 10.1186/1471-2164-14-785
Xu, Z.-S., Ni, Z.-Y., Liu, L., Nie, L.-N., Li, L.-C., Chen, M., et al. (2008). Characterization of the TaAIDFa gene encoding a CRT/DRE-binding factor responsive to drought, high-salt, and cold stress in wheat. Mol. Genet. Genomics 280, 497–508. doi: 10.1007/s00438-008-0382-x
Yan, G., Liu, Y., Lan, J., Zhang, T., Wang, T., Li, L., et al. (2022). The rapid induction of OsPR1A protein is crucial in Xa21-mediated rice bacterial blight resistance. J. Plant Pathol. 104, 969–978. doi: 10.1007/s42161-022-01105-2
Yang, T., Poovaiah, B. (2003). Calcium/calmodulin-mediated signal network in plants. Trends Plant Sci. 8, 505–512. doi: 10.1016/j.tplants.2003.09.004
Zander, M., Thurow, C., Gatz, C. (2014). TGA transcription factors activate the salicylic acid-suppressible branch of the ethylene-induced defense program by regulating ORA59 expression. Plant Physiol. 165, 1671–1683. doi: 10.1104/pp.114.243360
Zhang, D.-W., Deng, X.-G., Fu, F.-Q., Lin, H.-H. (2015). Induction of plant virus defense response by brassinosteroids and brassinosteroid signaling in Arabidopsis thaliana. Planta 241, 875–885. doi: 10.1007/s00425-014-2218-8
Keywords: Pathogenesis-related protein1 (PR1), Brassinosteroids (BR), Heat stress (HS), Promoter, Transcription Factor Binding Sites (TFBSs), Cysteine-rich secretory protein (CAP)
Citation: Kumari D, Prasad BD, Dwivedi P, Sahni S, Kumar M, Alamri S, Adil MF and Alakeel KA (2024) Comprehensive analysis of transcription factor binding sites and expression profiling of rice pathogenesis related genes (OsPR1). Front. Plant Sci. 15:1463147. doi: 10.3389/fpls.2024.1463147
Received: 11 July 2024; Accepted: 10 September 2024;
Published: 25 October 2024.
Edited by:
Syed Adeel Zafar, University of California, Riverside, United StatesReviewed by:
Muhammad Ali Mumtaz, Cornell University, United StatesCopyright © 2024 Kumari, Prasad, Dwivedi, Sahni, Kumar, Alamri, Adil and Alakeel. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bishun Deo Prasad, YmRwcmFzYWRiYXVAZ21haWwuY29t; YmRwcmFzYWRAcnBjYXUuYWMuaW4=; Padmanabh Dwivedi, cGR3aXZlZGkyNUByZWRpZmZtYWlsLmNvbQ==; Khaled A. Alakeel, a2FhbGFrZWVsQGthY3N0Lmdvdi5zYQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.