- 1State Key Laboratory of Crop Gene Exploration and Utilization in Southwest China, Sichuan Agricultural University, Chengdu, China
- 2Triticeae Research Institute, Sichuan Agricultural University, Chengdu, China
- 3Key Laboratory of Wheat Biology and Genetic Improvement on Southwestern China (Ministry of Agriculture and Rural Affairs), Crop Research Institute, Sichuan Academy of Agricultural Sciences, Chengdu, China
- 4Environment-Friendly Crop Germplasm Innovation and Genetic Improvement Key Laboratory of Sichuan Province, Crop Research Institute, Sichuan Academy of Agricultural Sciences, Chengdu, China
Anthocyanins are plant secondary metabolites belonging to the polyphenol class of natural water-soluble phytopigments. The accumulation of anthocyanins in different plant tissues can improve plant survival under adverse conditions. In addition, plants with the resulting colorful morphology can be utilized as landscape plants. Triticum boeoticum (syn. Triticum monococcum ssp. aegilopoides, 2n=2x=14, AbAb) serves as a valuable genetic resource for the improvement of its close relative common wheat in terms of enhancing resilience to various biotic and abiotic stresses. In our previous study, the EMS-mutagenized mutant Z2921 with a red glume, stem, and rachis was generated from T. boeoticum G52, which has a green glume, stem, and rachis. In this study, the F1, F2, and F2:3 generations of a cross between mutant-type Z2921 and wild-type G52 were developed. A single recessive gene, tentatively designated RgM4G52, was identified in Z2921 via genetic analysis. Using bulked segregant exome capture sequencing (BSE-Seq) analysis, RgM4G52 was mapped to chromosome 6AL and was flanked by the markers KASP-58 and KASP-26 within a 3.40-cM genetic interval corresponding to 1.71-Mb and 1.61-Mb physical regions in the Chinese Spring (IWGSC RefSeq v1.1) and Triticum boeoticum (TA299) reference genomes, respectively, in which seven and four genes related to anthocyanin synthesis development were annotated. Unlike previously reported color morphology-related genes, RgM4G52 is a recessive gene that can simultaneously control the color of glumes, stems, and rachis in wild einkorn. In addition, a synthetic Triticum dicoccum–T. boeoticum amphiploid Syn-ABAb-34, derived from the colchicine treatment of F1 hybrids between tetraploid wheat PI 352367 (T. dicoccum, AABB) and Z2921, expressed the red stems of Z2921. The flanking markers of RgM4G52 developed in this study could be useful for developing additional common wheat lines with red stems, laying the foundation for marker-assisted breeding and the fine mapping of RgM4G52.
Introduction
Anthocyanins are flavonoid pigments that are important for plant adaptation to biotic and abiotic stress conditions (Laikova et al., 2005). In common bread wheat (Triticum aestivum L.), pigmentation caused by anthocyanins can occur on leaves, stems, auricles, glumes, pericarp aleurone, coleoptiles, and anthers. Pigment accumulation in crop plants can not only be used as a morphological marker to assist in breeding and research related to impurity removal, gene functions, pigment synthesis pathways, photosynthesis, and other related theories but also provide agroecological tourism value for humans (Song et al., 2020). The anthocyanin pigmentation of different parts of plants is related to their adaptation to environmental stress conditions. In wheat, purple coleoptiles, stems, and anthers are reportedly related to resistance to bunt (Bogdanova et al., 2002). It was shown that the lines with dark-purple grains and coleoptiles demonstrated a higher seedling drought tolerance than plants with uncolored pericarp and light purple coleoptiles (Shoeva et al., 2017). The relationship between accumulation of anthocyanins in wheat coleoptiles and cold treatment has been shown (Gordeeva et al., 2013). Furthermore, the purple-grained NILs had better viability after accelerated aging compared with the recurrent parent lacking anthocyanins (Gordeeva and Khlestkina, 2013). The knowledge about specific features of anthocyanin biosynthesis regulation in wheat can be useful for improvement of its adaptation to biotic and abiotic stress conditions. In addition, anthocyanins are important for maintaining human health and preventing cardiovascular diseases, carcinogenesis, inflammation, and many other human pathological conditions (Lila, 2004).
Anthocyanin accumulation occurs naturally in some species. However, it has also been reported that some changes in pigment accumulation occur in mutant plant populations induced by chemical mutagens. Chemical mutagens, among which ethyl methane sulfonate (EMS) is the most widely used and effective in crop mutagenesis breeding, can induce plants to produce heritable mutants (Li et al., 2017; Greene et al., 2003; Henry et al., 2014). There is a high frequency of EMS-induced point mutagenesis, primarily involving G–C to A–T conversion, with relatively few chromosomal aberrations. Most of the mutations are easy to screen (Greco et al., 2001) and can be used to induce variation in plants, construct mutant libraries, and provide rich basic genetic material for the study of plant gene function. Liu et al. (2009) obtained a rice (Oryza sativa L.) YGL4 mutant with a yellow–green leaf color through the EMS mutagenesis of Jinhui 10 cultivar. EMS-induced mutant germplasm can be effectively used to mine new genes, facilitate functional genomics research, and accelerate breeding programs (Hohmann et al., 2005). Mutagenetic breeding techniques make it possible to overcome the disadvantages of the long conventional breeding cycle, which is slow and comes with difficulties in producing mutation variation. Moreover, mutagenic breeding techniques can bring about breakthroughs in the creation of new crop cultivars, germplasms, and genetic materials and can enable some special problems in breeding work to be solved.
Triticum boeoticum (Syn. Triticum monococcum ssp. aegilopoides, 2n=2x=14, AbAb), the wild progenitor of Triticum monococcum ssp. monococcum (2n=2x=14, AmAm), represents a valuable genetic resource for improving the ability of its close relative common wheat in terms of coping with various biotic and abiotic stresses (Budak et al., 2013; Ahmed et al., 2023; Wang et al., 2023). In addition, T. boeoticum is one of the sources of the blue grain trait controlled by blue aleurone layer 2 (Ba2) (Zeller et al., 1991; Zeven, 1991; Dubcovsky et al., 1996; Singh et al., 2007; Yu et al., 2017; Liu et al., 2021). To date, the salt tolerance genes Nax2 and Nax1; the powdery mildew resistance genes Pm25, PmTb7A.1, and PmTb7A.2; the leaf rust resistance gene Sr22; the stripe rust resistance locus QYrtb.pau-5A; and the stripe rust resistance gene YrZ15-1370 have been successfully introduced into common wheat (Paull et al., 1994; Shi et al., 1998; Chhuneja et al., 2008; Munns et al., 2012; Tounsi et al., 2016; Elkot et al., 2015; Zhang et al., 2021).
Synthetic amphiploids play an important role in wheat breeding and evolutionary studies (Gill et al., 1988; Megyeri et al., 2011; Ahmed et al., 2014; Badaeva et al., 2016; Li et al., 2018; Liu et al., 2022, 2023). Many studies have aimed to generate amphidiploids from hybrids of wheat with related species from the genera Aegilops, Secale, Thinopyrum, and Triticum (Nemeth et al., 2015). Recently, 18 synthetic Triticum turgidum–Triticum boeoticum amphiploids were identified using cytological methods, and their nutritional compositions were evaluated (Liu et al., 2023). Artificial amphidiploids are regarded as a source of genetic variation for improving wheat (Kroupin et al., 2019).
Since the first mention of the expression of purple color traits in wheat, studies on the inheritance of these characteristics have made great progress in revealing the molecular-genetic mechanisms of anthocyanin pigment biosynthesis and its regulation in wheat (Shoeva and Khlestkina, 2015). In our previous study, a mutant, Z2921, with a red glume, stem, and rachis was produced from an EMS-mutagenized population of T. boeoticum G52 in M4 generation. In addition, a synthetic T. dicoccum–T. boeoticum amphiploid, Syn-ABAb-34, derived from the colchicine treatment of F1 hybrids of T. dicoccum PI 352367 and Z2921, expressed the red stem from Z2921. The objectives of the present study were (1) to identify and map the genes conferring resistance to the red glume, stem, and rachis in Z2921 by using bulked segregant exome capture sequencing (BSE-Seq) analysis and (2) to characterize the new synthetic T. dicoccum–T. boeoticum amphiploid Syn-ABAb-34 via cytological methods.
Materials and methods
Plant materials
T. boeoticum G52, the Z2921 mutant, T. dicoccum PI352367, and the synthetic T. dicoccum–T. boeoticum amphiploid Syn-ABAb-34 were used in this study. The M4 generation of Z2921, which originated from the 0.4% EMS treatment of 3,000 seeds of the T. boeoticum accession G52, was used. Syn-ABAb-34 was derived from the colchicine treatment of F1 hybrids from PI 352367×Z2921. G52 was kindly provided by George Fedak at the Ottawa Research and Development Centre for Agriculture and Agri-Food (Ottawa, ON, Canada) in 2012. PI 352367 was kindly provided by the USDA-ARS germplasm bank (http://www.ars-grin.gov). All materials used in this study were kept at the Triticeae Research Institute of Sichuan Agricultural University.
Cytological observations
Sc-GISH was conducted using T. boeoticum genomic DNA as a probe according to previously published methods (Wang et al., 2019). Total genomic DNA from G52 was labeled with fluorescein-12-dUTP (Roche Diagnostics Australia, Castle Hill, NSW) using nick translation. The chromosomes were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) and pseudocolored red. Hybridization signals were visualized and captured using an Olympus BX-63 epifluorescence microscope equipped with a Photometric SenSys DP70 CCD camera (Olympus, Tokyo, Japan). The raw images were processed using Photoshop v.7.1 (Adobe Systems Inc., San Jose, CA, USA).
Chromosome pairing observation in pollen mother cells (PMCs) was performed as described previously (Zhang et al., 2007). For meiotic analysis, at least 50 PMCs were observed for Z5471. Ring bivalents (ring II) and rod bivalents (rod II) were counted, and their average numbers were calculated.
Phenotypic investigation
Field evaluations of glumes, stems, and rachides in G52, Z2921, PI 352367, Syn-ABAb-34, G52×Z2921 F1, and G52×Z2921 F2 individuals as well as their corresponding F2:3 families were performed at the experimental field of the Triticeae Research Institute, Sichuan Agricultural University, Wenjiang. The colors of the glumes, stems, and rachides were recorded as red or green from the jointing stage to the mature stage during the growth cycle. Each plant was 10 cm apart within rows and 30 cm apart between rows and was 1.5 m in length. The color phenotypes of all the materials in each generation were investigated.
Bulked segregant exome capture sequencing
Genomic DNA was extracted via the CTAB method (Chatterjee et al., 2002). Phenotypically contrasting F2:3 families with different glume/stem/rachis colorations in the field were used to construct red and green glume/stem/rachis DNA pools for BSE-Seq. Equal amounts of DNA from 20 homozygous red-phenotype families and 20 homozygous green-phenotype families were pooled for bulked segregant exome capture sequencing (Ji et al., 2023). The DNA samples were subjected to exome capture sequencing, a technology developed by Chengdu Tcuni Technology (Chengdu, China). Sequence quality was controlled using Trimmomatic v0.36 software (Bolger et al., 2014). DNA reads of the wild-type and mutant bulks were aligned to the reference genome sequence of Chinese Spring v1.1 (IWGSC, 2018) using STARv2.5.1b software (Dobin et al., 2013). The unique and high-confidence alignments were applied to call SNP variants using GATK v3.6 software (McKenna et al., 2010). SNP variants with Fisher’s exact test (FET) P values < 1e−8 and allele frequency difference (AFD) >0.6 were considered associated with the red glume/stem/rachis phenotype and were then used as templates to develop SNP markers (Li et al., 2020).
Kompetitive allele−specific PCR assays
The red-phenotype-related SNPs and the 500-bp flanking sequences were used to design the Kompetitive allele−specific PCR (KASP) primers and test polymorphisms in the parental lines and the wild-type and mutant DNA bulks. Polymorphic markers that could be reliably scored were genotyped in the F2 segregation population of G52×Z2921. For each KASP assay, a 10-µl reaction volume containing 5 µl of 2 KASP master mix (Biosearch Technologies), 1.4 µl of primer mix (a mixture of 0.168 µM each forward A1 and A2 primer and 0.42 µM of reverse primer), 100 ng of genomic DNA, and 2.6 µl of ddH2O was prepared. The CFX96 Touch™ real-time PCR detection system (Bio-Rad, USA) was used for amplification under the following conditions: 15 min at 94°C, 10 touchdown cycles of 20 s at 94°C, 60 s at 65°C–57°C (decreasing by 0.8°C per cycle), and 32 cycles of 20 s at 94°C, 60 s at 57°C.
Data analysis
Chi-square (χ2) tests were used to determine the goodness of fit for the observed segregation and expected ratios of the F2 and F2:3 populations. Linkage analysis was performed using MAPMAKER/EXP v3.0b (Lander et al., 1987). The Kosambi function was used to convert recombination values to genetic distances (Kosambi, 1943). A logarithmic odds (LOD) ratio of 3.0 and a maximum distance of 50.0 cM were set as the thresholds for the declaration of linkage. The genetic linkage map was drawn using MapDraw v2.1 software (Liu and Meng, 2003).
Candidate gene analysis
The corresponding sequences of the markers KASP-58 and KASP-26 linked to RgM4G52 were subjected to BLAST searches against the genomes of common wheat cv. Chinese Spring v1.1 (Appels et al., 2018) and the genome of Triticum boeoticum TA299 (Ahmed et al., 2023). Gene annotations between the flanking markers of the two genomes were retrieved from the databases Ensembl Plants (http://plants.ensembl.org/index.html) and Swiss-Prot (http://www.gpm-aw.com/html/swi-ss-prot.html). Collinearity analysis was performed on the DEGs related to the function of anthocyanin synthesis among the parents and mixed pools.
Results
Genetic analysis of genes related to red glumes, stems, and rachides
From the jointing stage to the mature stage, the EMS mutant Z2921 exhibited red glumes, stems and rachides (Figures 1A–D), and G52 exhibited green glumes, stems, and rachides. Z2921 was crossed with G52 to develop F1, F2, and F2:3 populations for genetic analysis of the genes conferring red glumes, stems, and rachides in Z2921. The glume, stem, and rachis coloration of all the F1 plants were similar to those of the parent G52 plants, with green glumes, stems, and rachides (Figures 1E–H). The F2 population segregated into 46 green-phenotype and 18 red-phenotype strains (Figures 1I–L), fitting a 1G:3R ratio (χ2 = 0.333, p=0.564) (Table 1), indicating that the red phenotype was conferred by a single recessive gene, tentatively designated RgM4G52. The segregation rate of the F2:3 population composed of 64 families was 14 (homozygous green type):30 (heterozygous):17 (homozygous red type) (χ1:2:12 = 0.097, p = 0.953), which is consistent with the segregation results of the F2 population (Table 1).
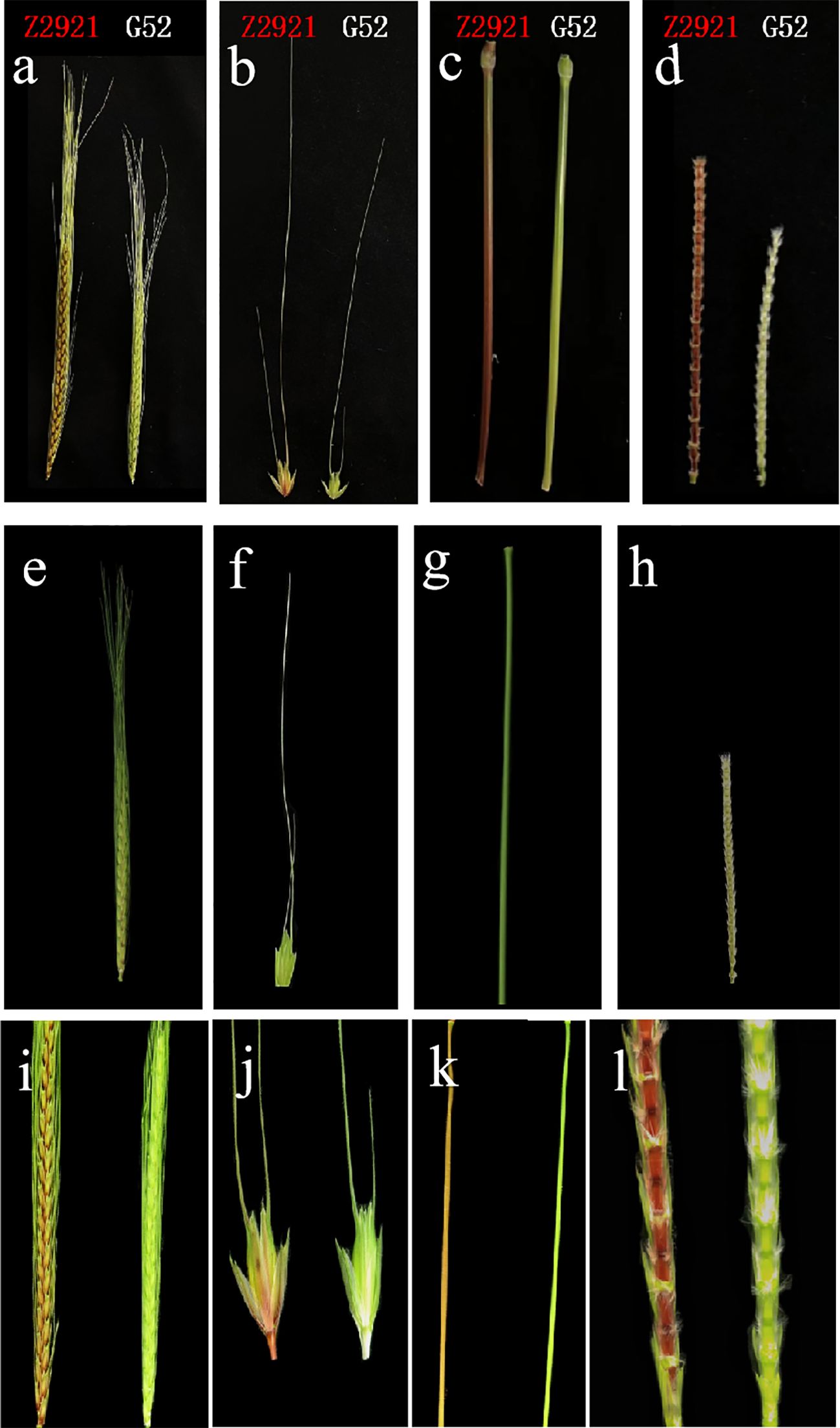
Figure 1. The coloration of spikes (A), spikelets (glumes) (B), stems (C) and rachides (D) of Z2921 and G52; The coloration of spikes (E), spikelets (glumes) (F), stems (G), and rachides (H) of G52×Z2921 F1; The coloration of spikes (I), spikelets (glumes) (J), stems (K), and rachides (L) of G52×Z2921 F2 individual plants.
BSE-Seq analysis
The DNA samples of the red bulk and the green bulk were subjected to BSE-Seq analysis, which generated 224,290,036 and 177,194,890 raw reads, respectively. After quality control, 224,266,906 and 177,176,312 high-quality reads from the red bulk and the green bulk, respectively, were uniquely mapped to the Chinese Spring genome (IWGSC RefSeq v1.1). A total of 5651 SNPs (p< 1e-8 and |AFD|>0.6) were identified from these reads using GATK v4.0 software (Figure 2B). A total of 348 SNPs were located within a 5-Mb genomic interval (555 Mb–560 Mb) on the long arm of chromosome 6A (Figure 2A) in the Chinese Spring reference genome; these SNPs were regarded as candidate SNPs linked to RgM4G52.
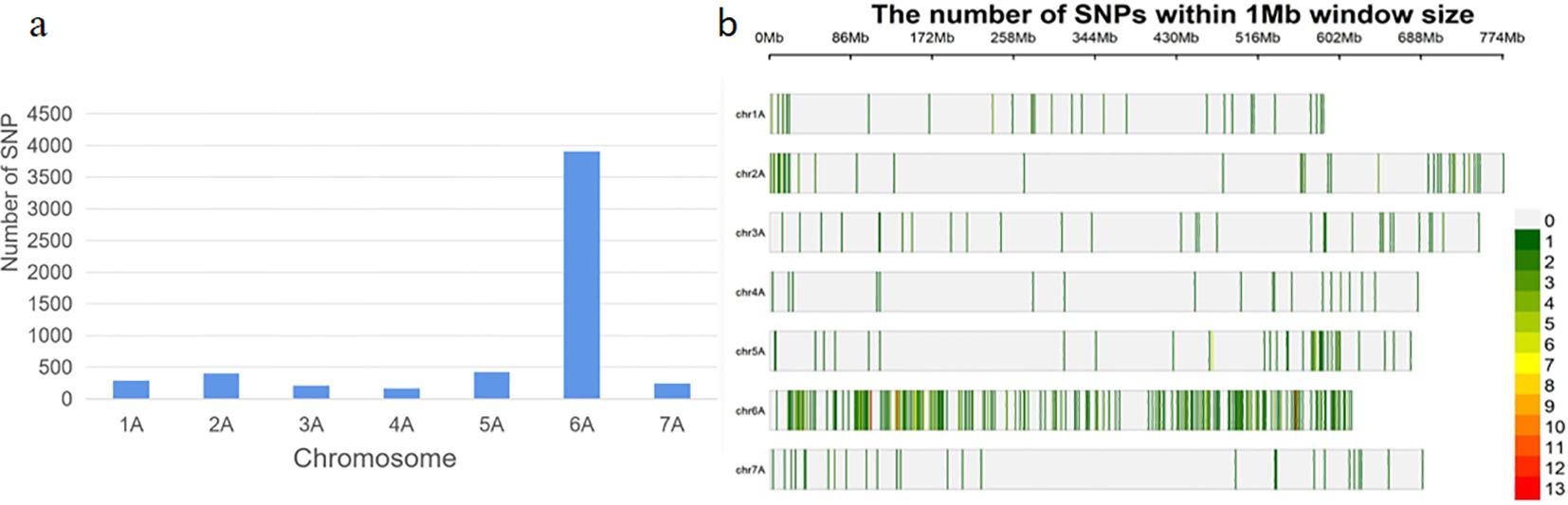
Figure 2. Distribution of SNPs from DNA bulks. (A) Number of SNPs (|AFD| > 0.6, P value <1e−8) distributed on different wheat chromosomes; (B) enrichment of SNPs within a 1-Mb window size on wheat chromosomes.
Molecular mapping of RgM4G52
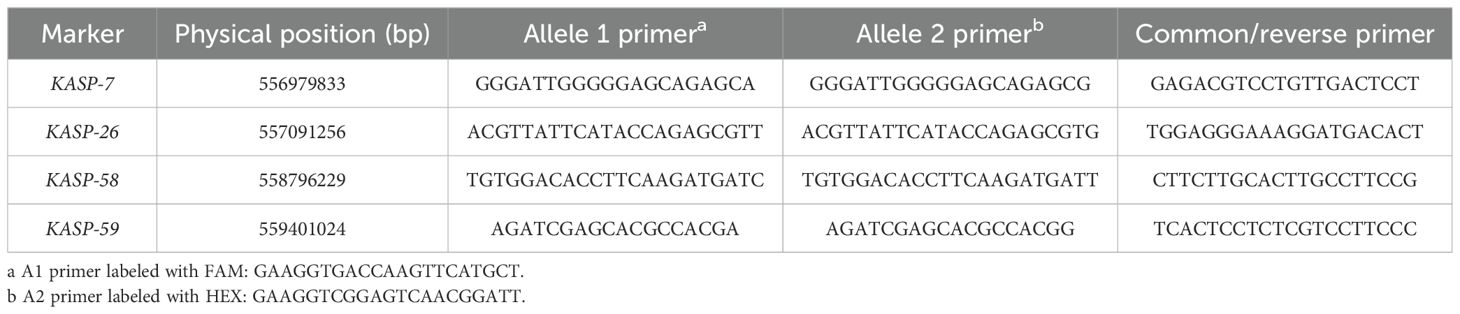
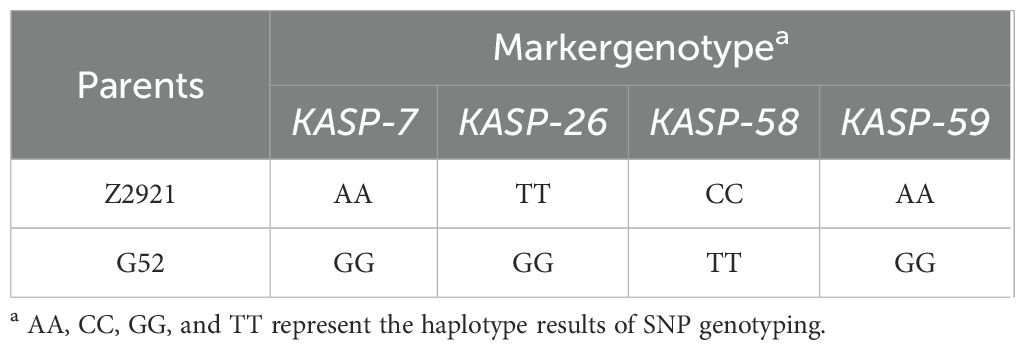
There were 59 of the 171 clustered SNPs on 6AL 555–560 Mb (p<1e-8 and |AFD|>0.6) chosen for the development of KASP markers. Four of them were successfully converted into KASP markers (KASP-7, KASP-26, KASP-58, KASP-59) (Table 2) and scored reliably on the parents as well as the red and green bulks (Table 3). These KASP markers were subsequently used to genotype 64 F2 plants derived from a cross between green-type G52 and red-type Z2921. Linkage analysis indicated that KASP-58 was mapped 2.73 cM distal and that KASP-26 was located 0.67 cM proximal to RgM4G52 (Figures 3A–C).
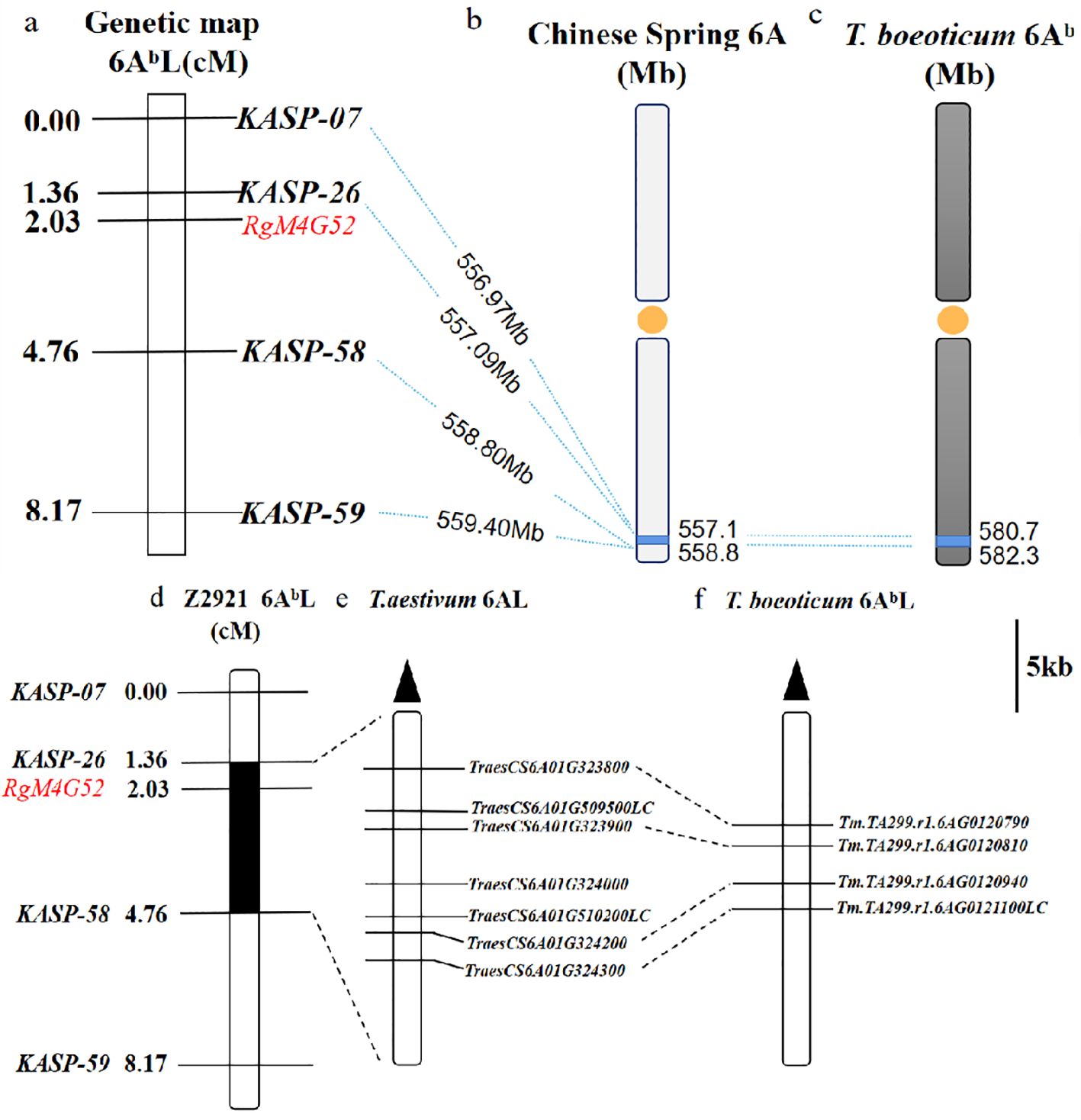
Figure 3. Genetic linkage map of the RgM4G52 gene on chromosome 6AL showing the physical location of RgM4G52. (A) Linkage map of RgM4G52; (B) physical interval (blue) where the four KASP markers are linked to RgM4G52 anchored in Chinese Spring, with orange dots representing centromeres and dotted lines indicating the physical positions of each marker; (C) physical intervals anchored by markers linked to RgM4G52 in T. boeoticum. Collinearity relationship of candidate genes in the RgM4G52 gene mapping interval between T. aestivum Chinese Spring and T. boeoticum TA299. (D) Genetic linkage map of RgM4G52; (E, F) physical mapping of candidate genes in Chinese Spring and TA299.
Gene analysis of the RgM4G52 genomic region
The sequences of the closely linked markers KASP-26 and KASP-58 were subjected to BLAST searches against the genome of Chinese Spring and that of T. boeoticum TA299 to determine their physical positions. RgM4G52 was physically mapped to a 1.71-Mb region between the 557.09-Mb and 558.97-Mb regions of the Chinese Spring 6AL chromosome (IWGSC RefSeq v1.1) and between the 580.72-Mb and 582.33-Mb regions (1.61 Mb) of the TA299 6AbL chromosome (T. boeoticum) (Figures 3A–C). There were 40 and 42 predicted genes in the target physical regions in Chinese Spring and T. boeoticum TA299, respectively (IWGSC RefSeq v1.1; T. boeoticum TA299, Supplementary Tables S1, S2). In the Chinese Spring genome, seven genes may be associated with the anthocyanin biosynthesis pathway, including four cytochrome P450 family protein-related genes (Tanaka et al., 2009; Yang et al., 2004) (TraesCS6A02G509500LC, TraesCS6A02G323900, TraesCS6A02G324000, TraesCS6A02G510200LC), one universal stress protein family gene (Song et al., 2023a, 2023b; Gonzalez et al., 2008; Baudry et al., 2004; Stracke et al., 2010). (TraesCS6A02G323800), one peroxidase gene (TraesCS6A02G324200) (Grommeck and Markakis, 1964; Zapata et al., 1995; Kader et al., 2002; Zhang et al., 2005), and one F-box family protein-encoding gene (Zhang et al., 2017; Feder et al., 2015) (TraesCS6A02G324300). Four genes, namely, one cytochrome P450 protein-related gene (Tm.TA299.r1.6AG0120810), one peroxidase gene (Tm.TA299.r1.6AG0120940), a universal stress protein family gene, Tm.TA299.r1.6AG0120790, and an F-box family protein-encoding gene, Tm.TA299.r1.6AG0121100LC, were found in the T. boeoticum (TA299) genome and had a good collinear relationship with those of Chinese Spring (Figures 3D–F).
Phenotypic and cytological molecular characterization of the amphiploid Syn-ABAb-34
An investigation of the number of chromosomes in the root tip showed that seven plants had 42 chromosomes and 2 plants had 41 chromosomes in the nine plants tested from amphiploid Syn-ABAb-34. Plants with 42 chromosomes were used for mc-GISH identification and chromosome pairing observation. Mc-GISH revealed 28 A-genome chromosomes among the amphiploid Syn-ABAb-34 chromosomes in the A-genome of Z5471 (Figure 4A). Chromosome pairing in PMCs (observed PMCs >50) at meiotic metaphase I was 2.58 rod II+17.92 ring II+0.16 I+0.16 III+0.08 IV in these plants with 42 chromosomes (Figure 4B), indicating the cytological stability of amphiploid Syn-ABAb-34. The amphiploid Syn-ABAb-34 was detected using the KASP markers KASP-26 and KASP-58 linked to RgM4G52; the results indicated that Syn-ABAb-34 carried the gene RgM4G52 (Figures 4C, D). Field evaluation revealed that the color of stem changes from green to purple over time and finally changes from purple to red. Syn-ABAb-34 exhibited stem pigment accumulation similar to the diploid parent Z2921, whereas no pigment accumulation was detected in the tetraploid parent PI352367 (Figures 5A–C). Additionally, it was observed that the average length and width of 10 grains of Syn-ABAb-34 were greater than those of the parents Z2921 and PI352367 (Figures 5D, E).
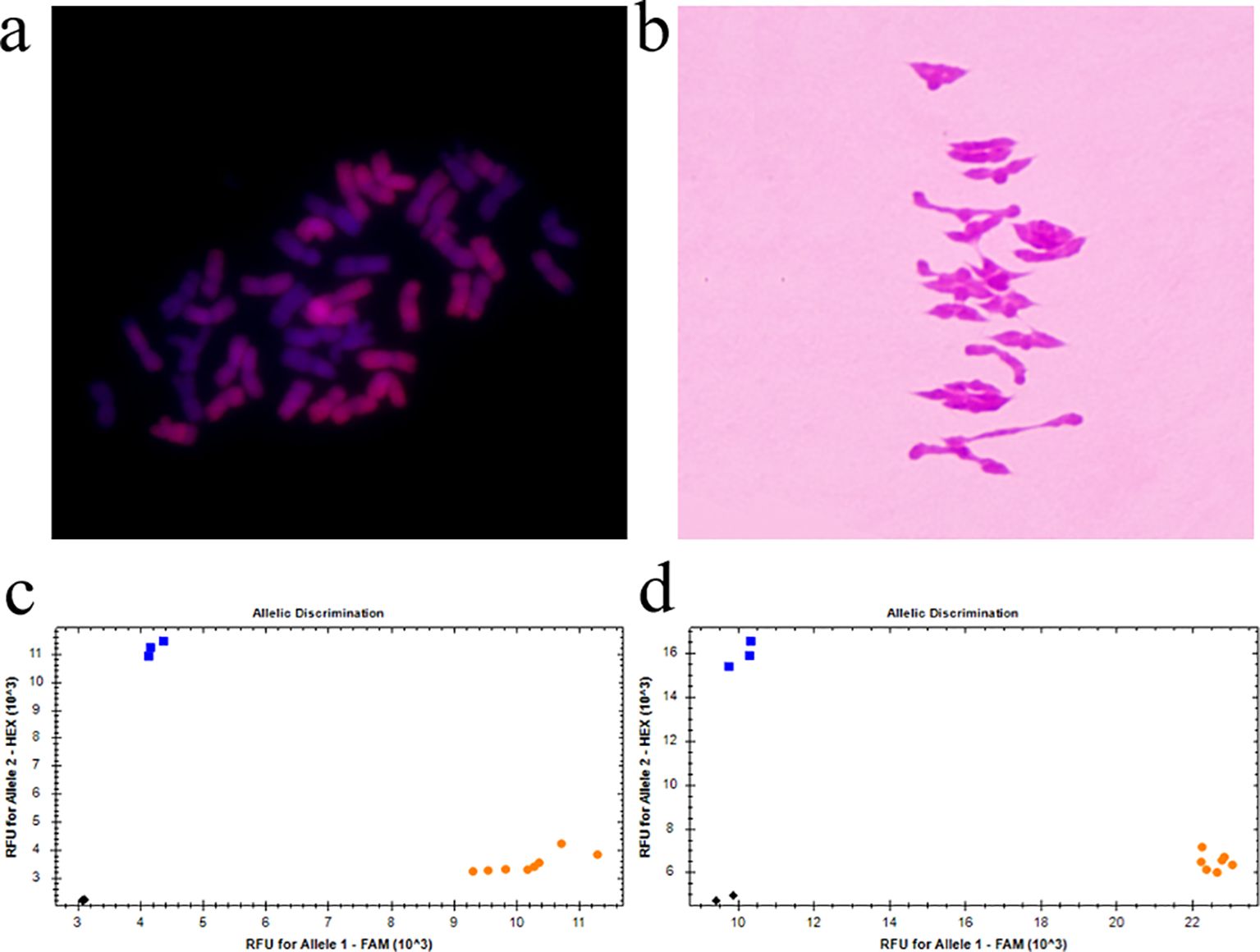
Figure 4. Cytological observations of the T. dicoccum–T. boeoticum amphiploid Syn-ABAb-34 and molecular detection using KASP markers linked to RgM4G52. (A) GISH identification of Syn-ABAb-34 using G52 genomic DNA as a probe; (B) chromosome pairing of PMCs in Syn-ABAb-34 with 21 bivalents; (C) molecular marker detection of Syn-ABAb-34 and its parents G52 and Z2921 using the KASP marker KASP-26, which is linked to RgM4G52. G52 in blue, Z2921 and Syn-ABAb-34 in yellow; (D) molecular marker detection of Syn-ABAb-34 and its parents G52 and Z2921 using the KASP marker KASP-58 linked to RgM4G52. G52, blue; Z2921 and Syn-ABAb-34, yellow.

Figure 5. Phenotype of the T. dicoccum–T. boeoticum amphiploid Syn-ABAb-34 and its parents PI 352367 and Z2921. (A) The coloration of different stems from T. dicoccum–T. boeoticum amphiploid Syn-ABAb-34; (B) the stem coloration of Syn-ABAb-34 and its parents Z2921 and PI352367; (C) the plants of Syn-ABAb-34 and its parents Z2921 and PI352367; (D) the seed length of Syn-ABAb-34 and its parents Z2921 and PI352367; (E) the seed width of Syn-ABAb-34 and its parents Z2921 and PI352367.
Discussion
Anthocyanins cause pigmentation in plant tissues and enhance plant resistance to biotic and abiotic stresses (Laikova et al., 2005). To date, there have been some reports of genes associated with pigmentation on leaves, glume shells, culms, seeds, etc., in wheat. Rg genes control glume shell color in diploid, tetraploid, and hexaploid wheat and are located on chromosomes 1A, 1B, 1D, and 2A (Khlestkina et al., 2010; McIntosh et al., 2013). Among these genes, Rg-A1b and Rg-A1c control the red and black (dark brown) glumes of diploid, tetraploid, and hexaploid wheat, respectively (Blanco et al., 1998; Salina et al., 2006) whereas Rg-D1b and Rg-D1c control the red (brown) and gray glumes, respectively, of hexaploid wheat (Khlestkina and Salina, 2006, 2009b, 2002a, 2002b). Three homoeologous genes for purple stems (Pc-A1, Pc-B1, Pc-D1), three homoeologs for purple leaf sheaths (Pls-A1, Pls-B1, Pls-D1), and three homoeologs for purple leaf blades (Plb-A1, Plb-B1, Plb-D1) have been mapped in close linkage with the red coleoptile genes Rc-A1, Rc-B1, and Rc-D1 in wheat (Khlestkina et al., 2009a, 2010). Two genes responsible for purple anthers (Pan-A1 and Pan-D1) have been mapped on chromosomes 7A (Blanco et al., 1998) and 7D (Khlestkina et al., 2009a) at short distances from Rc-A1 and Rc-D1, respectively.
However, there have only been a few reports of pigmentation in glumes, culms, and rachides being simultaneously controlled by a single gene. In this study, a new pigmentation gene, RgM4G52, conferring red glumes, stems, and rachides, was identified in the T. boeoticum mutant Z2921 and mapped on chromosome arm 6AL flanked by the markers KASP-26 and KASP-58 within a 3.40-cM genetic interval corresponding to a 1.71-Mb physical region in the Chinese Spring genome (IWGSC RefSeq v1.1).
RgM4G52 was physically mapped to a 1.61-Mb region between 580.72 Mb and 582.33 Mb on the TA299 6AbL chromosome arm (T. boeoticum TA299) (Figures 3A–C). Based on the gene functional annotation, there were four protein-coding genes, Tm.TA299.r1.6AG0120790, Tm.TA299.r1.6AG0120810, Tm.TA299.r1.6AG0120940, and Tm.TA299.r1.6AG0121100LC, in the target physical regions of the T. boeoticum genome (Supplementary Tables S1, S2; Figures 3D, E).
Tm.TA299.r1.6AG0120790 was not annotated, and the homologous gene of Tm.TA299.r1.6AG0120790 in Chinese Spring was TraesCS6A02G323800. Its functional annotation was that of a universal stress protein family gene. The universal stress protein family is involved in UV-B-induced flavonoid biosynthesis, and VcUSPs are coexpressed mainly with transcription factors from the MYB, AP2, zinc finger, and bHLH families (Song et al., 2023a, 2023b; Gonzalez et al., 2008; Baudry et al., 2004; Stracke et al., 2010). Tm.TA299.r1.6AG0121100LC was not annotated, and the homologous gene of Tm.TA299.r1.6AG0121100LC in the Chinese Spring is TraesCS6A02G324300. Its functional annotation was that of an F-box family protein. Members of the F-box serve as crucial negative regulators by mediating CHS and PAL degradation, which coordinately controls flavonoid biosynthesis (Zhang et al., 2017; Feder et al., 2015).
Tm.TA299.r1.6AG0120810 was annotated as a desmethyl-deoxy-podophyllotoxin synthase, and the homologous gene of Tm.TA299.r1.6AG0120810 in Chinese Spring was TraesCS6A02G323900. Its functional annotation was that of a cytochrome P450 protein-related gene. The enzymes flavonoid 3′-hydroxylase (F3′H) and flavonoid 3′,5′-hydroxylase (F3′5′H) play important roles in the anthocyanin biosynthesis pathway, and the genes encoding flavonoid 3′5′-hydroxylase (F3′5′h) and flavonoid 3′-hydroxylase (F3′h) belong to the cytochrome P450 monooxygenase gene family (Tanaka et al., 2009). There are no data on the cloning and/or mapping of these genes in wheat, except regarding one partial nucleotide sequence, F3′5′h (Yang et al., 2004). In the present study, Tm.TA299.r1.6AG0120940 was annotated as a peroxidase gene, and the gene homologous to Tm.TA299.r1.6AG0120940 in Chinese Spring was TraesCS6A02G324200. Its functional annotation was the same as that for Tm.TA299.r1.6AG0120940. All flavonoids have shown great binding affinity to peroxidase, and peroxidase can be degraded by anthocyanins as direct crop plant substrates or in the presence of H2O2 with anthocyanidins as substrates to oxidize and decolorize the anthocyanidins. The activity of peroxidase is negatively correlated with that of anthocyanins (Grommeck and Markakis, 1964; Zapata et al., 1995; Kader et al., 2002; Zhang et al., 2005). Now, the work is ongoing to clone and sequence these four genes, namely, Tm.TA299.r1.6AG0120790, Tm.TA299.r1.6AG0120810, Tm.TA299.r1.6AG0120940, and Tm.TA299.r1.6AG0121100LC. Then, candidate genes will be screened by sequence alignment. Finally, the function of candidate genes will be verified by the transgenic technique.
The flanking markers KASP-26 and KASP-58 developed in this study could be used as molecular markers to screen recombinant heterozygous plants, construct secondary F2 populations and develop markers, and further narrow the location interval to finely map and clone RgM4G52. There have been rare reports of pigmentation in glumes, culms, and rachides being simultaneously controlled by a single gene. RgM4G52 may be a new recessive pigmentation gene. According to previous reports on the relationship between anthocyanin accumulation and plants, the red pigment accumulation in wheat with RgM4G52 may be related to their adaptation to environmental stress conditions. It may be used as a morphological marker to assist in breeding and research related to gene functions and pigment synthesis pathways. Furthermore, it can also be used as a landscape crop for agro-ecological popular science tourism.
The expression of superior genes has been found to decrease or be completely inhibited when foreign genes have been transferred from a lower ploidy level (Ma et al., 1997; Ahmed et al., 2014). In this study, the amphiploid Syn-ABAb-34 had red stems at the jointing stage but had green glumes and rachides, indicating that the expression of RgM4G52 was suppressed in the glumes and rachides. According to Hao et al. (2019), the amphiploid Syn-ABAb-34 can serve as a bridge to hybridize with elite wheat varieties, transferring this trait to common wheat and providing new germplasm resources for wheat breeding. Combined with molecular marker-assisted selection, the transfer of RgM4G52 from diploid wheat to common wheat cultivars are ongoing using the amphiploid Syn-ABAb-34 as a bridge. The breeding of new wheat cultivars with red glumes/stems/rachides will provide new materials for the breeding of widely adaptable wheat varieties.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
LC: Data curation, Investigation, Methodology, Software, Writing – original draft. JZ: Investigation, Methodology, Software, Writing – original draft. PM: Investigation, Software, Writing – original draft. YM: Investigation, Software, Writing – review & editing. LW: Investigation, Software, Writing – review & editing. KZ: Investigation, Software, Writing – review & editing. JY: Investigation, Software, Writing – review & editing. MZ: Investigation, Software, Writing – review & editing. XL: Investigation, Software, Writing – review & editing. BJ: Software, Supervision, Writing – review & editing. MH: Investigation, Software, Writing – review & editing. LH: Investigation, Supervision, Writing – review & editing. SN: Investigation, Supervision, Writing – review & editing. XJC: Data curation, Investigation, Writing – review & editing. XC: Investigation, Software, Writing – review & editing. DL: Funding acquisition, Methodology, Supervision, Writing – review & editing. HW: Methodology, Supervision, Writing – review & editing. LZ: Conceptualization, Methodology, Project administration, Supervision, Validation, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was supported by the Key Research and Development Program of Sichuan Province (2021YFYZ0002), Sichuan Science and Technology Program (2022ZDZX0016; 2023NSFSC1925), the Open Fund of the Key Laboratory of Wheat Biology and Genetic Improvement in Southwestern China (ZWS2024001), and the Sichuan Provincial Finance Department (1 + 9KJGG001; YSCX2035-001).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1459505/full#supplementary-material
References
Ahmed, H. I., Heuberger, M., Schoen, A., Koo, D. H., Quiroz-Chavez, J., Adhikari, L., et al. (2023). Einkorn genomics sheds light on history of the oldest domesticated wheat. Nature 620, 830–838. doi: 10.1038/s41586-023-06389-7
Ahmed, S., Bux, H., Rasheed, A., Gul Kazi, A., Rauf, A., Mahmood, T., et al. (2014). Stripe rust resistance in Triticum durum–T. monococcum and T. durum–T. urartu amphiploids. Australas. Plant Pathol. 43, 109–113. doi: 10.1007/s13313-013-0237-8
Appels, R., Eversole, K., Stein, N., Feuillet, C., Keller, B., Jane, R., et al. (2018). Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361, eaar7191. doi: 10.1126/science.aar7191
Badaeva, E. D., Ruban, A. S., Zoshchuk, S. A., Surzhikov, S. A., Knüpffer, H., Kilian, B. (2016). Molecular cytogenetic characterization of Triticum timopheevii chromosomes provides new insight on genome evolution of T. zhukovskyi. Plant Syst. Evol. 302, 943–956. doi: 10.1007/s00606-016-1309-3
Baudry, A., Heim, M. A., Dubreucq, B., Caboche, M., Weisshaar, B., Lepiniec, L. (2004). TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 39, 366–380. doi: 10.1111/j.1365-313X.2004.02138.x
Blanco, A., Bellomo, M. P., Cenci, A., De Giovanni, C., D’ovidio, R., Iacono, E., et al. (1998). A genetic linkage map of durum wheat. Theor. Appl. Genet. 97, 721–728. doi: 10.1007/s001220050948
Bogdanova, E. D., Sarbaev, A. T., Makhmudova, K. K. (2002). “Resistance of common wheat to bunt,” in Proceedings of the research conference on genetics, Moscow, Russia: Research Conference on Genetics. 43–44.
Bolger, A. M., Lohse, M., Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Budak, H., Kantar, M., Yucebilgili Kurtoglu, K. (2013). Drought tolerance in modern and wild wheat. Sci. World J. 4, 66–75. doi: 10.1155/2013/548246
Chatterjee, A., Moulik, S. P., Majhi, P. R., Sanyal, S. K. (2002). Studies on surfactant–biopolymer interaction. I. Microcalorimetric investigation on the interaction of cetyltrimethylammonium bromide (CTAB) and sodium dodecylsulfate (SDS) with gelatin (Gn), lysozyme (Lz) and deoxyribonucleic acid (DNA). Biophys. Chem. 98, 313–327. doi: 10.1016/S0301-4622(02)00107-2
Chhuneja, P., Kaur, S., Garg, T., Ghai, M., Kaur, S., Prashar, M., et al. (2008). Mapping of adult plant stripe rust resistance genes in diploid A genome wheat species and their transfer to bread wheat. Theor. Appl. Genet. 116, 313–324. doi: 10.1007/s00122-007-0668-0
Dobin, A., Davis, C. A., Schlesinger, F., Drenkow, J., Zaleski, C., Jha, S., et al. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. doi: 10.1093/bioinformatics/bts635
Dubcovsky, J., Luo, M. C., Zhong, G. Y., Bransteitter, R., Desai, A., Kilian, A., et al. (1996). Genetic map of diploid wheat, Triticum monococcum L., and its comparison with maps of Hordeum vulgare L. Genetics 143, 983–999. doi: 10.1093/genetics/143.2.983
Elkot, A. F. A., Chhuneja, P., Kaur, S., Saluja, M., Keller, B., Singh, K. (2015). Marker assisted transfer of two powdery mildew resistance genes PmTb7A.1 and PmTb7A.2 from Triticum boeoticum (Boiss.) to Triticum aestivum (L.). PloS One 10, e0128297. doi: 10.1371/journal.pone.0128297
Feder, A., Burger, J., Gao, S., Lewinsohn, E., Katzir, N., Schaffer, A. A., et al. (2015). A Kelch domain-containing F-Box coding gene negatively regulates flavonoid accumulation in muskmelon. Plant Physio 169, 1714–1726. doi: 10.1104/pp.15.01008
Gill, R. S., Dhaliwal, H. S., Multani, D. S. (1988). Synthesis and evaluation of Triticum durum-T. monococcum amphiploids. Theor. Appl. Genet. 75, 912–916. doi: 10.1007/BF00258053
Gonzalez, A., Zhao, M., Leavitt, J. M., Lloyd, A. M. (2008). Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 53, 814–827. doi: 10.1111/j.1365-313x.2007.03373.x
Gordeeva, E. I., Khlestkina, E. K. (2013). “Relationship between accumulation of anthocyanins in wheat pericarp and response to artificial ageing of seeds,” in Abstracts of conference free radicals and antioxidants in chemistry, biology and medicine, Novosibirsk, Russian.
Gordeeva, E. I., Shoeva, O. Y., Khlestkina, E. K. (2013). Cold stress response of wheat genotypes having different Rc alleles. Cereal Res. Commun. 41, 519–526. doi: 10.1556/CRC.2013.0029
Greco, R., Ouwerkerk, P. B., Taal, A. J., Favalli, C., Beguiristain, T., Puigdomenech, P., et al. (2001). Early and multiple Ac transpositions in rice suitable for efficient insertional mutagenesis. Plant Mol. Biol. 46, 215–227. doi: 10.1023/A:1010607318694
Greene, E. A., Codomo, C. A., Taylor, N. E., Henikoff, J. G., Till, B. J., Reynolds, S. H., et al. (2003). Spectrum of chemically induced mutations from a large-scale reverse-genetic screen in Arabidopsis. Genetics 164, 731–740. doi: 10.1093/genetics/164.2.731
Grommeck, R., Markakis, P. (1964). The effect of peroxidase on anthocyanin pigments a. J. Food Sci. 29, 53–57. doi: 10.1111/j.1365-2621.1964.tb01693.x
Hao, M., Zhang, L., Zhao, L., Dai, S., Li, A., Yang, W., et al. (2019). A breeding strategy targeting the secondary gene pool of bread wheat: introgression from a synthetic hexaploid wheat. Theor. Appl. Genet. 132, 2285–2294. doi: 10.1007/s00122-019-03354-9
Henry, I. M., Nagalakshmi, U., Lieberman, M. C., Ngo, K. J., Krasileva, K. V., Vasquez-Gross, H., et al. (2014). Efficient genome-wide detection and cataloging of EMS-induced mutations using exome capture and next-generation sequencing. Plant Cell 26, 1382–1397. doi: 10.1105/TPC.113.121590
Hohmann, U., Jacobs, G., Jung, C. (2005). An EMS mutagenesis protocol for sugar beet and isolation of non-bolting mutants. Plant Breed 124, 317–321. doi: 10.1111/j.1439-0523.2005.01126.x
IWGSC, Bellec, A., Berges, H., Appels, R., Eversole, K., Stein, N., et al. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science, 2018, 361:6403. doi: 10.1126/science.aar7191
Ji, G., Xu, Z., Fan, X., Zhou, Q., Chen, L., Yu, Q., et al. (2023). Identification and validation of major QTL for grain size and weight in bread wheat (Triticum aestivum L.). Crop J. 11, 564–572. doi: 10.1016/j.cj.2022.06.014
Kader, F., Irmouli, M., Nicolas, J. P., Metche, M. (2002). Involvement of blueberry peroxidase in the mechanisms of anthocyanin degradation in blueberry juice. J. Food Sci. 67, 910–915. doi: 10.1111/j.1365-2621.2002.tb09427.x
Khlestkina, E. K., Pestsova, E. G., Röder, M. S., Börner, A. (2002a). Molecular mapping, phenotypic expression and geographical distribution of genes determining anthocyanin pigmentation of coleoptiles in wheat (Triticum aestivum L.). Theor. Appl. Genet. 104, 632–637. doi: 10.1007/s00122-001-0788-x
Khlestkina, E. K., Pestsova, E. G., Salina, E., Roder, M. S., Arbuzova, V. S., Koval, S. F., et al. (2002b). Genetic mapping and tagging of wheat genes using RAPD, STS and SSR markers. Cell. Mol. Biol. Let 7, 795–802. doi: 10.1007/s00018-002-8414-x
Khlestkina, E. K., Pshenichnikova, T. A., Röder, M. S., Börner, A. (2009a). Clustering anthocyanin pigmentation genes in wheat group 7 chromosomes. Cereal. Res. Commun. 37, 391–398. doi: 10.1556/crc.37.2009.3.8
Khlestkina, E. K., Röder, M. S., Börner, A. (2010). Mapping genes controlling anthocyanin pigmentation on the glume and pericarp in tetraploid wheat (Triticum durum L.). Euphytica 171, 65–69. doi: 10.1007/s10681-009-9994-4
Khlestkina, E. K., Salina, E. A. (2006). SNP markers: methods of analysis, ways of development, and comparison on an example of common wheat. Russian. J. Genet. 42, 585–594. doi: 10.1134/s1022795406060019
Khlestkina, E. J., Salina, E. A., Pshenichnikova, T. A., Röder, M. S., Börner, A. (2009b). Glume coloration in wheat: allelism test, consensus mapping and its association with specific microsatellite allele. Cereal Res. Commun. 37, 37–43. doi: 10.1556/CRC.37.2009.1.5
Kosambi, D. D. (1943). The estimation of map distances from recombination values. Ann. Eugen. 12, 172–175. doi: 10.1111/j.1469-1809.1943.tb02321.x
Kroupin, P. Y., Divashuk, M., Karlov, G. (2019). Gene resources of perennial wild cereals involved in breeding to improve wheat crop. Selskokhoziaistvennaia Biol. 54, 409–425. doi: 10.15389/agrobiology.2019.3.409eng
Laikova, L. I., Arbuzova, V. S., Efremova, T. T., Popova, O. M. (2005). Genetic analysis of anthocyanin of the anthers and culm pigmentation in common wheat. Genetika 41, 1428–1433. doi: 10.1007/s11177-005-0216-4
Lander, E. S., Green, P., Abrahamson, J., Barlow, A., Daly, M. J., Lincoln, S. E., et al. (1987). MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1, 174–181. doi: 10.1016/j.ygeno.2008.12.003
Li, H. Y., Liu, X. J., Zhang, M. H., Feng, Z., Liu, D. C., Ayliffe, M., et al. (2018). Development and identification of new synthetic T. turgidum–T. monococcum amphiploids. Plant Genet. Resour 16, 555–563. doi: 10.1017/S1479262118000175
Li, W., Guo, H., Wang, Y., Xie, Y., Zhao, L., Gu, J., et al. (2017). Identification of novel alleles induced by EMS-mutagenesis in key genes of kernel hardness and starch biosynthesis in wheat by TILLING. Genes Genom 39, 387–395. doi: 10.1007/s13258-016-0504-5
Li, Y., Shi, X., Hu, J., Wu, P., Qiu, D., Qu, Y., et al. (2020). Identification of a recessive gene PmQ conferring resistance to powdery mildew in wheat landrace Qingxinmai using BSR-Seq analysis. Plant Dis. 104, 743–751. doi: 10.1094/PDIS-08-19-1745-RE
Lila, M. A. (2004). Anthocyanins and human health: an in vitro investigative approach. J. BioMed. Biotechnol. 2004, 306–313. doi: 10.1155/S111072430440401X
Liu, M., Sang, X., Ling, Y., Zhao, F., Yang, Z., He, G. (2009). Genetic analysis and molecular mapping of a yellow green leaf gene (YGL4) in rice (Oryza sativa L.). Acta Agro. Sin. 35, 1405–1409. doi: 10.3724/SP.J.1006.2009.01405
Liu, R. H., Meng, J. (2003). MapDraw: a microsoft excel macro for drawing genetic linkage maps based on given genetic linkage data. Hereditas 25, 317–321. doi: 10.3321/j.issn:0253-9772.2003.03.019
Liu, X., Jiang, X., Zhang, J., Ye, H., Shen, M., Wu, L., et al. (2023). Molecular cytogenetic identification and nutritional composition evaluation of newly synthesized Triticum turgidum-Triticum boeoticum amphiploids (AABBAbAb). Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1285847
Liu, X. J., Yang, H., Zhang, M. H., Liu, X., Peng, T., Hao, M., et al. (2022). Molecular cytogenetic identification of a new synthetic amphiploid (Triticum timococcum, AtAtGGAmAm) with a seed setting rate comparable with that of natural Triticum zhukovskyi. Plant Breed 141, 558–565. doi: 10.1111/pbr.13030
Liu, X., Zhang, M., Jiang, X., Li, H., Jia, Z., Hao, M., et al. (2021). TbMYC4A is a candidate gene controlling the blue aleurone trait in a wheat-Triticum boeoticum substitution line. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.762265
Ma, H., Singh, R. P., Mujeeb-Kazi, A. (1997). Resistance to stripe rust in durum wheats, A-genome diploids, and their amphiploids. Euphytica 94, 279–286. doi: 10.1023/A:1002979706378
McIntosh, R. A., Yamazaki, Y., Dubcovsky, J., Rogers, J., Morris, C., Appels, R., et al. (2013). “Catalogue of gene symbols for wheat,” in Proceedings of the 12th international wheat genetics symposium, Yokohama Japan. 8–13.
McKenna, A., Hanna, M., Banks, E., Sivachenko, A., Cibulskis, K., Kernytsky, A., et al. (2010). The genome analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome. Res. 20, 1297–1303. doi: 10.1101/gr.107524.110
Megyeri, M., Miko′, P., Molna′r, I., Kova′cs, G. (2011). Development of synthetic amphiploids based of Triticum turgidum × T. monococcum crosses to improve the adaptability of cereals. Acta Agron. Hung 59, 267–274. doi: 10.1556/AAgr.59.2011.3.11
Munns, R., James, R. A., Xu, B., Athman, A., Conn, S. J., Jordans, C., et al. (2012). Wheat grain yield on saline soils is improved by an ancestral Na+ transporter gene. Nat. Biotechnol. 30, 360–364. doi: 10.1038/nbt.2120
Nemeth, C., Yang, C., Kasprzak, P., Hubbart, S., Scholefield, D., Mehra, S., et al. (2015). Generation of amphidiploids from hybrids of wheat and related species from the genera Aegilops, Secale, Thinopyrum, and Triticum as a source of genetic variation for wheat improvement. Genome 58, 71–79. doi: 10.1139/gen-2015-0002
Paull, J. G., Pallotta, M. A., Langridge, P., The, T. T. (1994). RFLP markers associated with Sr22 and recombination between chromosome 7A of bread wheat and the diploid species Triticum boeoticum. Theor. Appl. Genet. 89, 1039–1045. doi: 10.1007/BF00224536
Salina, E. A., Leonova, I. N., Efremova, T. T., Röder, M. S. (2006). Wheat genome structure: translocations during the course of polyploidization. Funct. Integr. Genomic 6, 71–80. doi: 10.1007/s10142-005-0001-4
Shi, A. N., Leath, S., Murphy, J. P. (1998). A major gene for powdery mildew resistance transferred to common wheat from wild einkorn wheat. Phytopathology 88, 144–147. doi: 10.1094/PHYTO.1998.88.2.144
Shoeva, O. Y., Gordeeva, E. I., Arbuzova, V. S., Khlestkina, E. K. (2017). Anthocyanins participate in protection of wheat seedlings from osmotic stress. Cereal Res. Commun. 45, 47–56. doi: 10.1556/0806.44.2016.044
Shoeva, O. Y., Khlestkina, E. K. (2015). “The specific features of anthocyanin biosynthesis regulation in wheat,” in Advances in wheat genetics: from genome to field: Proceedings of the 12th International wheat genetics symposium (Springer, Japan), 147–157.
Singh, K., Ghai, M., Garg, M., Chhuneja, P., Kaur, P., Schnurbusch, T., et al. (2007). An integrated molecular linkage map of diploid wheat based on a Triticum boeoticum × T. monococcum RIL population. Theor. Appl. Genet. 115, 301–312. doi: 10.1007/s00122-007-0543-z
Song, Y., Ma, B., Feng, X., Guo, Q., Zhou, L., Zhang, X., et al. (2023a). Genome-wide analysis of the universal stress protein gene family in Blueberry and their transcriptional responses to UV-B irradiation and abscisic acid. Int. J. Mol. Sci. 24, 16819. doi: 10.3390/ijms242316819
Song, Y., Ma, B., Guo, Q., Zhou, L., Zhou, X., Ming, Z., et al. (2023b). MYB pathways that regulate UV-B-induced anthocyanin biosynthesis in blueberry (Vaccinium corymbosum). Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1125382
Song, W. J., Zhong, M., Yu, L., Xia, W. H., Shu, X. L., Wu, D. X., et al. (2020). Current status on research and utilization of colored rice. Chin. J. @ Rice Sci. 34, 191. doi: 10.16819/j.1001-7216.2020.9137
Stracke, R., Jahns, O., Keck, M., Tohge, T., Niehaus, K., Fernie, A. R., et al. (2010). Analysis of PRODUCTION OF FLAVONOL GLYCOSIDES-dependent flavonol glycoside accumulation in Arabidopsis thaliana plants reveals MYB11-, MYB12-and MYB111-independent flavonol glycoside accumulation. New Phytol. 188, 985–1000. doi: 10.1111/j.1469-8137.2010.03421.x
Tanaka, Y., Brugliera, F., Chandler, S. (2009). Recent progress of flower colour modification by biotechnology. Int. J. Mol. Sci. 10, 5350–5369. doi: 10.3390/ijms10125350
Tounsi, S., Amar, S. B., Masmoudi, H., Sentenac, H., Brini, F., Very, A.-A. (2016). Characterization of two HKT1;4 transporters from Triticum monococcum to elucidate the determinants of the wheat salt tolerance Nax1 QTL. Plant Cell Physiol. 57, 2047–2057. doi: 10.1093/pcp/pcw123
Wang, J., Liu, C., Guo, X., Wang, K., Du, L., Lin, Z., et al. (2019). Development and genetic analysis of wheat double substitution lines carrying Hordeum vulgare 2H and Thinopyrum intermedium 2Ai# 2 chromosomes. Crop J. 7, 163–175. doi: 10.1016/j.cj.2018.11.003
Wang, X., Li, H., Shen, T., Wang, X., Yi, S., Meng, T., et al. (2023). A near-complete genome sequence of einkorn wheat provides insight into the evolution of wheat A subgenomes. Plant Commun. 5 (5), 100768. doi: 10.1016/j.xplc.2023.100768
Yang, G. H., Li, B., Gao, J. W., Liu, J. Z., Zhao, X. Q., Zheng, Q., et al. (2004). Cloning and expression of two chalcone synthase and a flavonoid 3’5’-hydroxylase 3’-end cDNAs from developing seeds of blue-grained wheat involved in anthocyanin biosynthetic pathway. Acta Bot. Sin. 46, 588–594. doi: 1672-6650(2004)46:5>588:CAEOTC>2.0.TX;2-1
Yu, K., Liu, D., Wu, W., Yang, W., Sun, J., Li, X., et al. (2017). Development of an integrated linkage map of einkorn wheat and its application for QTL mapping and genome sequence anchoring. Theor. Appl. Genet. 130, 53–70. doi: 10.1007/s00122-016-2791-2
Zapata, J. M., Calderon, A. A., Ros Barcelo, A. (1995). Actual browning and peroxidase level are not correlated in red and white berries from grapevine (Vitis vinifera) cultivars. Fruit Variet J. 49, 82–84.
Zeven, A. (1991). Wheats with purple and blue grains: a review. Euphytica 56, 243–258. doi: 10.1007/BF00042371
Zeller, F. J., Cermeño, M. C., Miller, T. E (1991). Cytological analysis on the distribution and origin of the alien chromosome pair conferring blue aleurone color in several European common wheat (Triticum aestivum L.) strains. Theor Appl Genet, 81(4): 551–558. doi: 10.1007/BF00219448
Zhang, L. Q., Yen, Y., Zheng, Y. L., Liu, D. C. (2007). Meiotic restriction in emmer wheat is controlled by one or more nuclear genes that continue to function in derived lines. Sex Plant Reprod. 20, 159–166. doi: 10.1007/s00497-007-0052-x
Zhang, M. H., Liu, X., Peng, T., Wang, D. H., Liang, D. Y., Li, H. Y., et al. (2021). Identification of a recessive gene YrZ15-1370 conferring adult plant resistance to stripe rust in wheat-Triticum boeoticum introgression line. Theor. Appl. Genet. 134, 2891–2900. doi: 10.1007/s00122-021-03866-3
Zhang, Y., Liu, Z., Liu, J., Lin, S., Wang, J., Lin, W., et al. (2017). GA-DELLA pathway is involved in regulation of nitrogen deficiency-induced anthocyanin accumulation. Plant Cell Rep. 36, 557–569. doi: 10.1007/s00299-017-2102-7
Keywords: Triticum boeoticum, synthetic amphiploid, red glume, gene mapping, recessive gene, BSE-Seq
Citation: Chen L, Zhang J, Ma P, Miao Y, Wu L, Zhou K, Yang J, Zhang M, Liu X, Jiang B, Hao M, Huang L, Ning S, Chen X, Chen X, Liu D, Wan H and Zhang L (2024) Identification of a recessive gene RgM4G52 conferring red glume, stem, and rachis in a Triticum boeoticum mutant. Front. Plant Sci. 15:1459505. doi: 10.3389/fpls.2024.1459505
Received: 04 July 2024; Accepted: 01 August 2024;
Published: 26 August 2024.
Edited by:
Chongmei Dong, The University of Sydney, AustraliaReviewed by:
Xingguo Ye, Chinese Academy of Agricultural Sciences, ChinaJindong Liu, Chinese Academy of Agricultural Sciences, China
Peter Sharp, The University of Sydney, Australia
Copyright © 2024 Chen, Zhang, Ma, Miao, Wu, Zhou, Yang, Zhang, Liu, Jiang, Hao, Huang, Ning, Chen, Chen, Liu, Wan and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongshen Wan, d2FuaG9uZ3NoZW5AMTI2LmNvbQ==; Lianquan Zhang, emhhbmdsaWFucXVhbjE5NzdAMTI2LmNvbQ==
†ORCID: Hongshen Wan, orcid.org/0000-0002-3212-3561
Lianquan Zhang, orcid.org/0000-0001-9005-8705
 Longyu Chen1,2
Longyu Chen1,2 Yongping Miao
Yongping Miao Ming Hao
Ming Hao Lin Huang
Lin Huang Shunzong Ning
Shunzong Ning Dengcai Liu
Dengcai Liu Hongshen Wan
Hongshen Wan Lianquan Zhang
Lianquan Zhang