- 1Plant Breeding & Genetics Division, Marker Assisted Breeding Group (MABG), Nuclear Institute for Agriculture and Biology (NIAB), Faisalabad, Pakistan
- 2State Key Laboratory of Crop Gene Resources and Breeding, Institute of Crop Science, Chinese Academy of Agricultural Sciences, Beijing, China
- 3National Institute for Genomics and Advanced Biotechnology (NIGAB), Islamabad, Pakistan
- 4Nuclear Institute for Agriculture and Biology College, Pakistan Institute of Engineering and Applied Sciences (NIAB-C, PIEAS), Faisalabad, Pakistan
- 5Department of Botany and Plant Sciences, University of California, Riverside, Riverside, CA, United States
Low phytate level is a desirable trait because it promotes mineral bioavailability and thus offers a solution to tackle mineral deficiencies. The objectives of the present study were to characterize low phytate (lpa) Basmati rice mutants for the identification of novel mutations in target gene(s) and to develop a PCR-based CAPS (cleaved amplified polymorphic sequence) marker for low phytate Basmati rice. For this purpose, cultivar Super Basmati (Q4) was irradiated with gamma rays (60Co source) and three mutants named Q1 (lpa-5-9), Q2 (lpa-9-13), and Q3 (lpa-59-14) were isolated. Four genes previously been reported for the low phytic acid trait in rice were sequenced in these mutants and no mutation was observed in Q1 and Q2. However, in Q3 (lpa14) mutant a novel mutation in OsMRP5 gene (LOC_Os03g04920`) was detected. Sequence analysis displayed a substitution in the first exon of OsMRP5 at position 1142 bp resulting in the amino acid change from glycine (Gly) to alanine (Ala) at position 381a.a. To facilitate low-phytate breeding program, CAPS marker was developed to confirm this mutation site using the restriction digestion by AluI restriction enzyme. After enzyme digestion, Q3 produces four bands (32, 220, 154, and 32 bp) while Q4 (parent cultivar Super Basmati) produces only 3 bands (32, 374, and 32 bp). These results showed that this CAPS marker is 100% linked with this mutation and can be used for future breeding programs. Present findings provided insights in molecular basis of low phytate trait in rice paving the way for developing low-phytate rice varieties through marker-assisted breeding.
1 Introduction
In cereals, the major form of phosphorus (P) is myo-inositol-1, 2, 3, 4, 5, 6-hexakisphosphate. This was the first P-containing compound found in seeds and hence known as phytic acid. In humans, due to chelation ability, it is known as an anti-nutrient because it reduces the bioavailability of minerals i.e., Zn2+ and Fe2+ (Abid et al., 2017; Akhter et al., 2012; Ibrahim et al., 2022). This prevents the absorption of micro-nutrients and leads to malnutrition. Moreover, in monogastric animals, the digestibility of phytate is less and it is excreted as such from the body which causes environmental pollution (Abdel-Megeed and Tahir, 2015; Cosgrove and Irving, 1980; Kumar et al., 2010; Perera et al., 2018; Pramitha et al., 2021).
Rice (Oryza sativa) is a staple food for nearly half of the human population and is also rich in magnesium, phosphorus, manganese, selenium, iron, folic acid, thiamin, and niacin (Ravichanthiran et al., 2018; Ahmad et al., 2022). In terms of global production, it is grown in more than 100 countries with Asia accounting for 90% of the total production (Fukagawa and Ziska, 2019). In Pakistan, it is the 2nd most-produced cereal in the country with 7.2 million metric tons (MMT) of production, a 0.6% share in Pakistan’s GDP, and a US$ 2 billion export value (Anonymous, 2019). Almost 10% of Pakistan’s cultivated land area is covered by this crop (Shakoor et al., 2015). Phytic acid (also known as inositol hexaphosphate and IP6) is the storage compound of phosphorus in crops like rice, maize, barley, wheat, etc (Tong et al., 2017). It is an anti-nutritional factor in the seeds of cereals and legumes (Ibrahim et al., 2022). It is a negatively charged molecule that binds positively charged mineral elements, such as Ca, Mg, P, Mn, and Zn; the process is known as chelation (Jacob et al., 2024). These chelates hamper the bioavailability of mineral elements in the body of non-ruminants; the situation is termed hidden hunger or mineral malnutrition which leads to mineral deficiencies induced physiological disorders such as anemia and osteoporosis (Tan et al., 2013). Developed countries of the world have evolved strategies like food fortification, medication, and food supplementation to combat the above-mentioned physiological disorders; however, these approaches are not affordable in developing countries of the world due to high costs. For developing countries, bio-fortification i.e., enhanced bioavailability of the mineral elements, through genetic modification for low phytate crops, is the most economical and sustainable solution.
Eutrophication of lakes and rivers is another drawback of high phytate crops (Gupta et al., 2015; Dwivedi and Ortiz, 2021). It results in poor bioavailability of micronutrients in the body of ruminants and non-ruminants resulting in mineral malnutrition. It is a proven fact that an increase in inorganic phosphorus is inversely proportional to phytic acid phosphates and testing for an increased inorganic phosphate as compared with phytic acid is a simple, inexpensive, and fast approach to select the low phytate genotypes in various cereals and grains (Cichy and Raboy, 2009). Low phytate/biofortified genotypes/cultivars have been discovered and isolated in rice (Oryzae sativa L.; Larson et al., 2000; Qamar et al., 2019; Kumar et al., 2021; Utami et al., 2024), wheat (Triticum aestivum L.; Guttieri et al., 2004; Frittelli et al., 2023), soybean (Glycine max L.; Hitz et al., 2002; Lin et al., 2024), barley (Hordeum vulgare L.; Larson et al., 1998; Rasmussen and Hatzack, 1998; Huo et al., 2023), maize (Raboy and Gerbasi, 1996; Raboy et al., 2000; Liu et al., 2007; Yathish et al., 2023), and pea cultivars (Pisum sativum L.; Chigwedere et al., 2023; Subasi et al., 2024; Warkentin et al., 2012).
Low phytate mutants have enhanced bioavailability of mineral elements as compared to wild type parental genotypes (Pramitha et al., 2021). Developed countries of the world have evolved strategies like food fortification, medication, and food supplementation to combat mineral deficiencies-induced physiological disorders like iron deficiency-induced anemia during pregnancy, osteoporosis, etc. However, these approaches are not affordable in developing countries of the world due to high costs (Qamar et al., 2019; Perera et al., 2018). As rice is the staple food for the majority of the world population, the aim of producing low phytate mutants rice has great significance to enhance the bioavailability of essential micronutrients (Tan et al., 2013). The development of low-phytate maize, rice, soybean, wheat, and barley by United States Department of Agriculture are important examples to address mineral deficiencies through bio-fortification intervention (Raboy, 2002; Dorsch et al., 2003). Numerous genes that may be responsible for the metabolism of inositol phosphates and PA synthesis have been recognized in rice including, 2-phosphoglycerate kinase (2-PGK), OsMRP5, OsSULTR3;3, inositol monophosphates (IMP), inositol-pentakisphosphate 2-kinase 1 (IPK1), 1D–Myo-inositol 3-phosphate synthase (MIPS), inositol polyphosphate 2-kinase 2 (IPK2), inositol 1,3,4-trisphosphate 5/6-kinase (ITP5/6 K) and myoinositol kinase (MIK) (Suzuki et al., 2007; Kim et al., 2008a, b; Zhao et al., 2016; Li et al., 2014).
Despite highly efficient, fast, and low-cost colorimetric assays that are readily available, the DNA markers such as cleaved amplified polymorphic sequences (CAPS) are being used for the molecular characterization of crops in the world’s major breeding programs (Thiel et al., 2003; Varshney, 2009; Venegas et al., 2022). DNA markers have become a substitute for labor-intensive phenotypic assays (Thiel et al., 2003). CAPS markers are used because these are efficient in detecting polymorphism and are co-dominant with simple interpretation. Marker-assisted selection has been made easier using CAPS markers (Alavi et al., 2009; Song et al., 2023). The CAP1/Hinc II and CAP3/Hpy 99I are recommended and will be useful for the improvement of blast resistance, especially for the early-season indica rice (Lestari and Koh, 2013; Wang et al., 2012; Fan et al., 2023). Four markers have been derived from mutations in four genes that will be useful for enhancing/regulating SPC in pigeon pea crop improvement (Obala et al., 2019). In wheat, various CAPS markers have been developed for stripe rust resistance gene (Rani et al., 2019), leaf rust resistance gene, Lr-1 (Liu et al., 2022a) and yield related QTLs (Duan et al., 2020). In barley, CAPS markers were identified for the spot blotch susceptibility gene (Leng et al., 2018, 2020) and leaf rust resistance gene Rph28 (Mehnaz et al., 2021). Similarly, in maize, CAPS markers have been developed for cytoplasmic male sterility restorer gene (Liu et al., 2022b), drought resistant genes (Liu et al., 2015) and using SNPs for sugarcane mosaic virus resistance gene (Quint et al., 2002).
Through induced mutagenisis several mutants has been developed with number of useful mutations for biotic and abiotic stress tolerance in coarse and Basmati rice (Hameed et al., 2023; Ashraf et al., 2021; Amjad et al., 2022; Zafar et al., 2020; Qamar et al., 2012; Ashraf et al., 2007a, b; Qamar et al., 2005; Cheema et al., 2004; Ashraf et al., 2003; Rashid et al., 2003; Ashraf et al., 2021; Mumtaz et al., 2020, 2018). For the development of novel traits, induced mutation is a biologically safe and non-transgenic approach (Qamar et al., 2024; Rizwan et al., 2019). In our previous reports regarding development of low phytate rice mutants, three homozygous lpa mutants lpa-5-9, lpa-9-13, and lpa-59-14 were developed through induced mutations (gamma rays 60Co) and identified by colorimetric and High-performance Liquid Chromatography (HPLC) analysis (Cheema et al., 2008; Qamar et al., 2019). In these mutants, phytic acid content reduced to 54-63% but with poor germination and yield. Previously reported Lpa1-CAPS and Lpa1-InDel and functional molecular markers (Tan et al., 2013) were applied but indicated the absence of the Z9B-Lpa allele and XS-Lpa mutation in the OsMRP5 gene in these mutants (Qamar et al., 2019). Based on these previous studies, we proposed that there must be new mutations or novel alleles in our developed low phytate mutants that needs to be identified. Further attempts were made to improve germination and yield traits through hybridization, and backcross breeding.
In this view, present study was conducted for molecular characterization of low phytate basmati mutants with improved germination. Efforts were made to characterize low phytate (lpa) Basmati rice mutants for the identification of novel mutations in target gene(s) and to develop a PCR-based CAPS (cleaved amplified polymorphic sequence) marker for new mutation in low phytate Basmati rice. In this study, a CAPS marker for a new mutation in OsMRP5 gene in low phytate Basmati rice mutant was developed.
2 Materials and methods
2.1 Experimental material
The experimental material consisted of three low phytate mutant lines Q1 (lpa-5-9), Q2 (lpa-9-13), and Q3 (lpa-59-14) and non-low phytate Super Basmati with optimum germination and yield. These low phytate mutants (~58% reduction in phytic acid) were developed using gamma radiations (60Co source) at 250Gy dose under an IAEA project RAS7/014. Other materials include different breeding populations developed through the hybridization of LPA mutant lines with Super Basmati and backcross breeding.
2.2 Confirmation of lpa mutants using a colorimetric assay
Colorimetric Assay was performed according to the method previously described by (Chen et al., 1956; Qamar et al., 2019). Briefly, healthy seeds of each genotype were collected and the husk was removed by hand separately. Seeds were cut with the help of a sharp blade into two parts. One part containing the embryo was stored for germination and another half part was grounded. Grinding was performed by using a machine “Geno-Grinder”, for this purpose, half seed was kept in a 2mL Eppendorf tube (EP) and one metal ball was added into the EP tube. EP tubes were dipped in liquid nitrogen for one minute. The machine’s speed was set at 1450 rpm for 40 seconds. After grinding into a fine powder, 200 µL of 0.4 M HCl was added to each sample. Sample tubes were shacked gently by the hand and kept at 4°C for overnight (incubation). The next morning, 10 µL of the extracted sample was mixed with 90 µL of ddH2O, and 100 µL of colorimetric reagent was added. Incubation at room temperature for 1 hour was performed.
2.2.1 Colorimetric reagent
Mix 1 volume (1 mL) of 6 N sulfuric acid (Dilute 18 mL of commercial-grade sulfuric acid to 108 mL of ddH2O. For this purpose, 90 mL of ddH2O was taken and 18 mL of H2SO4 was added slowly) with 2 volumes (2 mL) of ddH2O and 1 volume (1 mL) of 2.5% ammonium molybdate (2.5 g of ammonium molybdate (NH4)6 Mo7O24.4H2O was dissolved in 70 mL ddH2O and adjusted the final volume up to 100 mL), then add 1 volume (1 mL) of 10% ascorbic acid (10 g of ascorbic acid was dissolved in ddH2O and the final volume was adjusted up to 100 mL). The solution was stored at 2-4°C for use. It can be stored for up to 7 weeks but better to prepare fewer amounts and mix well. On each day it was prepared freshly. All these operations were carried out in the laminar air hood and special care was taken during handling with acids.
2.3 Germination test
For further confirmation of low phytate mutants, we sow the already screened seeds from colorimetric assay on a half-strength MS medium in glass bottles (diameter = 6 cm, and 18 cm in height). These bottles were placed in the growth chamber at 23-26 °C for 16 hours of light and 8 hours of dark, while the relative humidity was kept at 70%. Subsequently, these seeds were confirmed through genotyping (sequencing).
2.4 Molecular characterization of low phytic mutants for reported mutations in genes
In present study, three low phytate (lpa) mutants named as Q1 (lpa-5-9), Q2 (lpa-9-13), and Q3 (lpa-59-14), along with parent cultivar Q4 (Super Basmati) were used for different evaluations. Four low phytic acid mutant genes (Table 1) were selected to find any mutation in our lpa mutants. Primers for these selected genes were designed (Table 2). Some walking primers according to the PCR product size were also designed for complete sequencing. For this purpose, 8 days old seedlings were used for DNA isolation following the CTAB method. After PCR amplification, the PCR product was sent to BGI Genomics, Beijing Company for sequencing.
2.5 Development of CAPS marker to detect the Q3 (lpa-59-14) mutant site in the OsMRP5
For the confirmation of the mutation site, we designed a CAPS (cleaved amplified polymorphic sequence) marker (Table 3). For this purpose, we performed a PCR, and then the PCR product was digested with the AluI (AGCT) restriction enzyme. 1 µl of AluI with 3 µl of cut-smart buffer was added to 30 µl of PCR product. This mixture was kept at 37°C for 90 minutes. Then it was run on the 1% agarose gel. After the electrophoresis, the size of the band was measured under UV light using Gel Doc Quantity One software (Version 4.5.1, BIO-RAD®).
2.6 Multiple sequence alignment and phylogenetic analysis
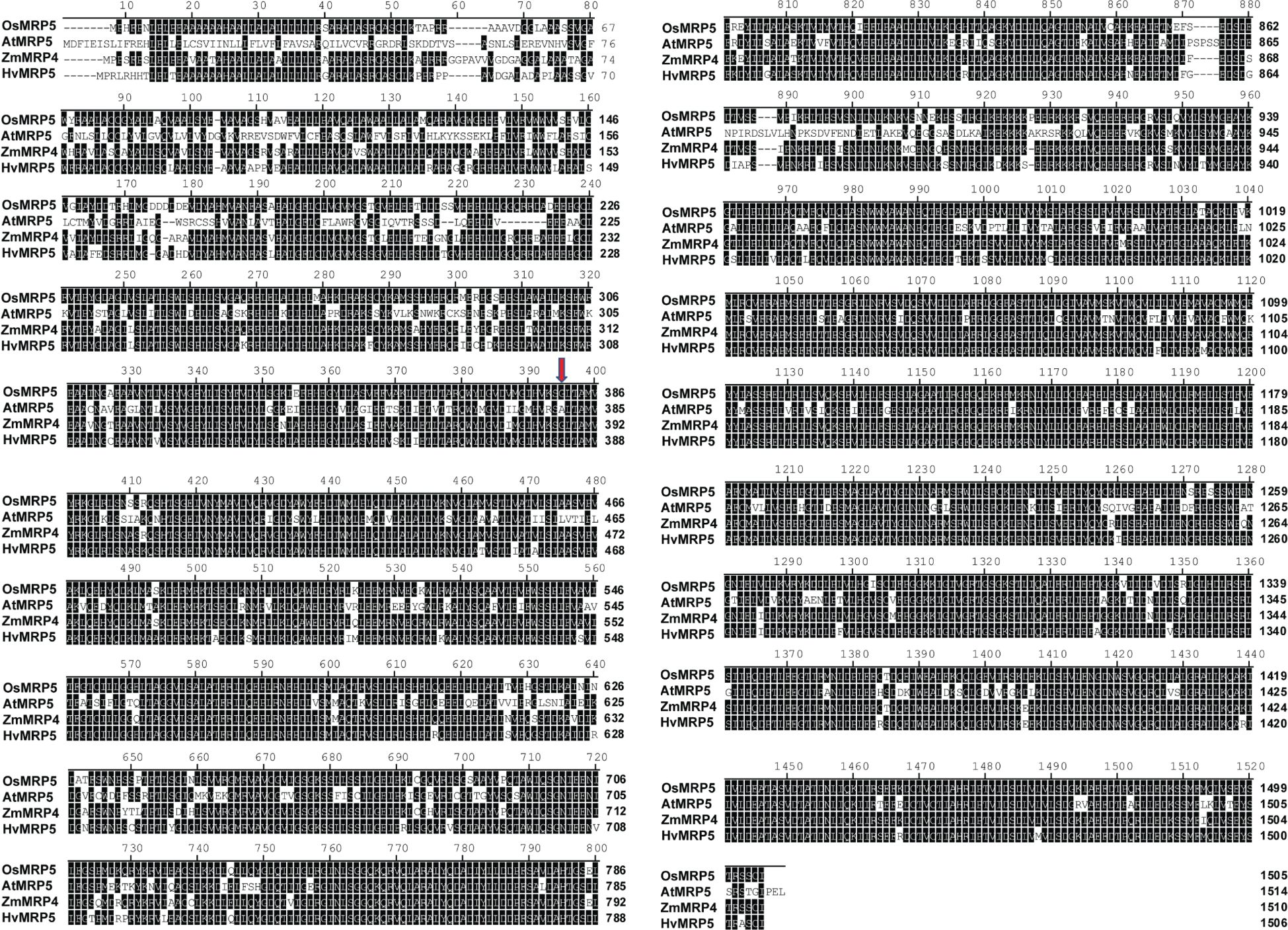
The amino acid sequences of MRP5 and their orthologue genes from different species such as rice, maize, sorghum, and Arabidopsis were retrieved from Ensembl plants (https://plants.ensembl.org/index.html). The BioEdit tool was used to align the sequences by using ClustalW multiple alignments. MEGA7 software was utilized for the construction of a phylogenetic tree. For this purpose, a neighbor-joining method with 1000 bootstraps was used.
3 Results and discussion
3.1 Confirmation of the lpa mutants through colorimetric assay
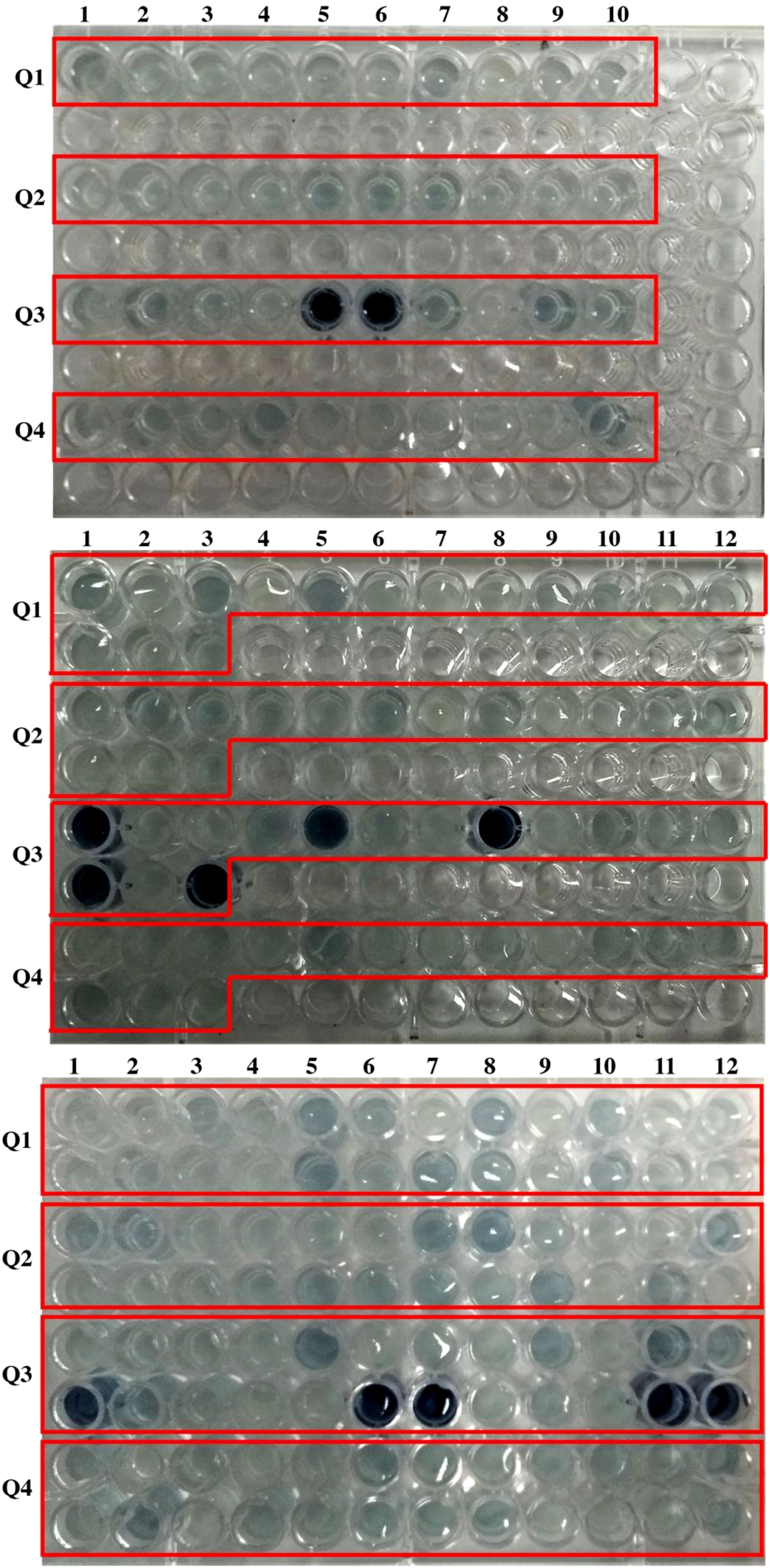
Colorimetric assays are a standard approach for detecting and quantifying phytate levels in plant samples. Plant types that have been genetically engineered or chosen to generate seeds or grains with lower amounts of phytate are known as low-phytate mutants. Plant breeders and farmers are interested in developing low phytate mutants because phytate may bind to key minerals such as iron, zinc, and calcium, rendering them less accessible for absorption by the human body. In these tests, a reagent is added to a plant sample, which interacts with inorganic P to cause a color change. Decreases in seed phytate increase seed inorganic P (the “High Inorganic P” phenotype) and since there is low inorganic P in wild-type seeds, this increase is easy to detect with this colorimetric assay. During this study, 49 seeds of each genotype (Q1-Q4) were used for colorimetric assay (Figure 1). Individual seed extracts were visually observed. There was no detection of blue color in the Q1 and Q2 genotypes. While in the Q3 genotype, 12 out of 49 seeds showed dark blue color which indicates 3:1 segregation ratio (Figure 1).
3.2 Confirmation of lpa through germination
Previously it is reported that low phytate mutants have poor germination (Zhao et al., 2008; Qamar et al., 2019). In the current study, we also checked the germination of pre-selected seeds (Figure 2; Table 4). The Q1, Q2 and Q4 genotypes showed similar germination 92%, 92% and 84%, respectively. Seeds from Q3 were divided into two parts (blue color Q3C and without color Q3 WC) and sown separately. The germination rate of seeds showing blue color was lower as compared with Q3 WC and Q4. The seedling growth rate of Q3C is also very slow as compared with Q4 (Figure 2B). This indicates that these are the possible true low phytate mutant seeds.
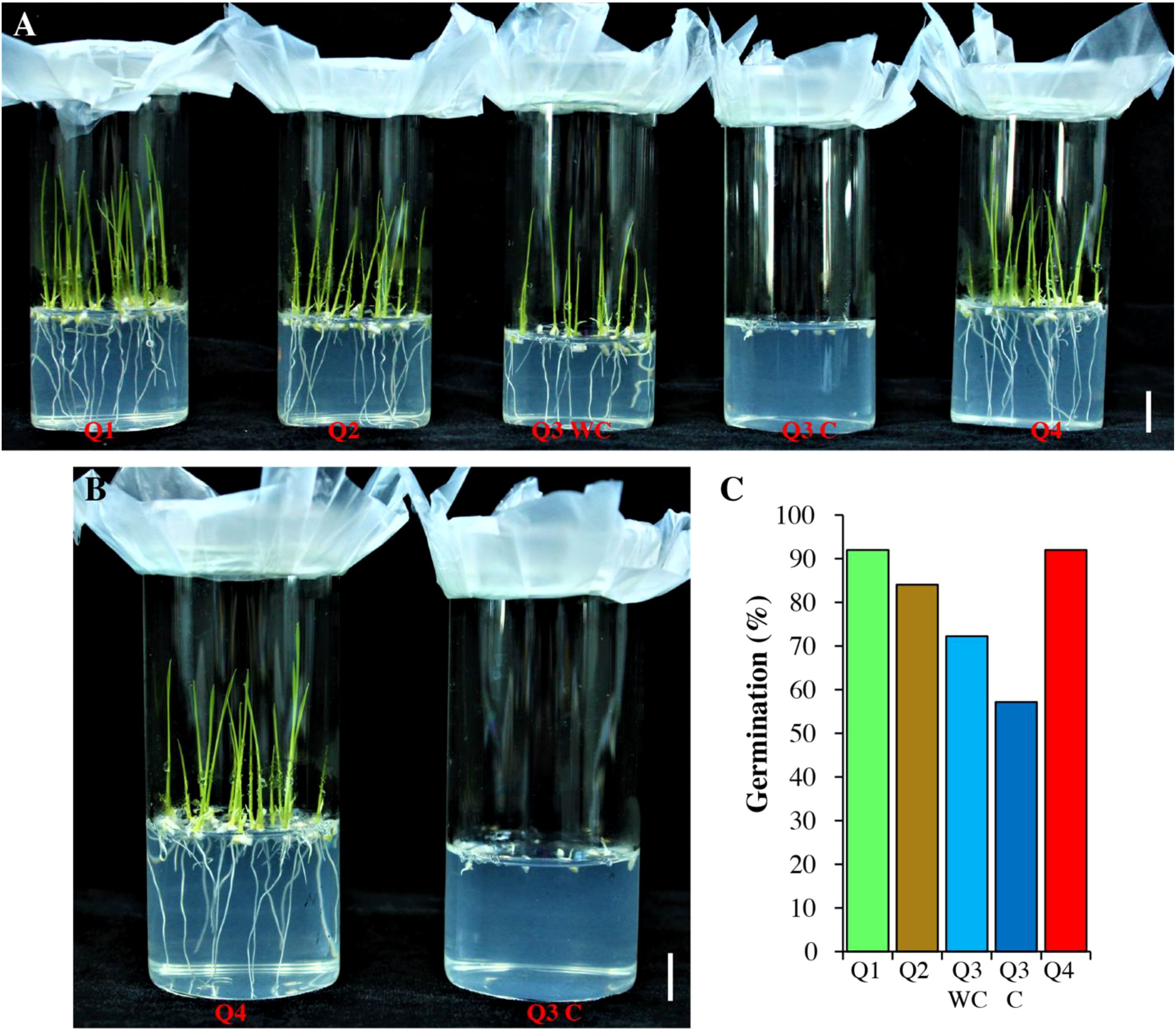
Figure 2. Germination test of low phytate genotypes. (A) Four genotypes Q1-Q4 were grown on ½ MS medium. Pictures were captured 5 days after sowing. WC, without color; C, colored. (B) Comparison of Q3 C with Q4. Scale bar = 1 cm. (C) Quantification of germination (%).
3.3 Genotyping of low phytate mutants
Twelve seeds (already screened through colorimetric assay) of each genotype (Q1-Q4) were used for genotyping (Table 5). Q1 and Q2 both showed WT (9/12, AA) and heterozygous (3/12, Aa) plants, while Q4 have all (12/12, AA) WT plants. We also used 9 plants of Q3 WC (no color in colorimetric assay), all the plants in this genotype were also WT (AA). These results further support the findings (seeds are not pure) of the colorimetric assay and germination test.
3.4 Identification of mutation sites in the lpa-59-14 mutant
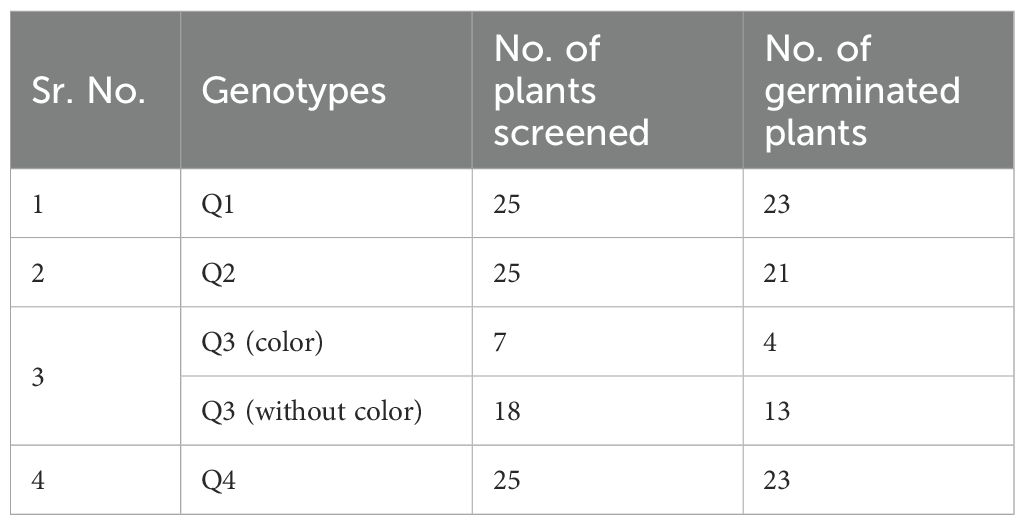
The results of sequencing of the PCR product revealed that we did not find any mutation sites in Q1 and Q2 genotypes in the four reported low phytate mutant genes in rice (Table 1). However, we did find a mutation site in Q3 (lpa-59-14) in the OsMRP5 gene (LOC_Os03g04920). There is a substitution in the first exon at position 1142 bp (G to A). Due to this substitution, the amino acid sequence also changed from Gly to Ala at position 381a.a (Figure 3A). This was also confirmed through sequencing (Figure 3B). There was a single peak in the mutant which further confirmed the G to A substitution.
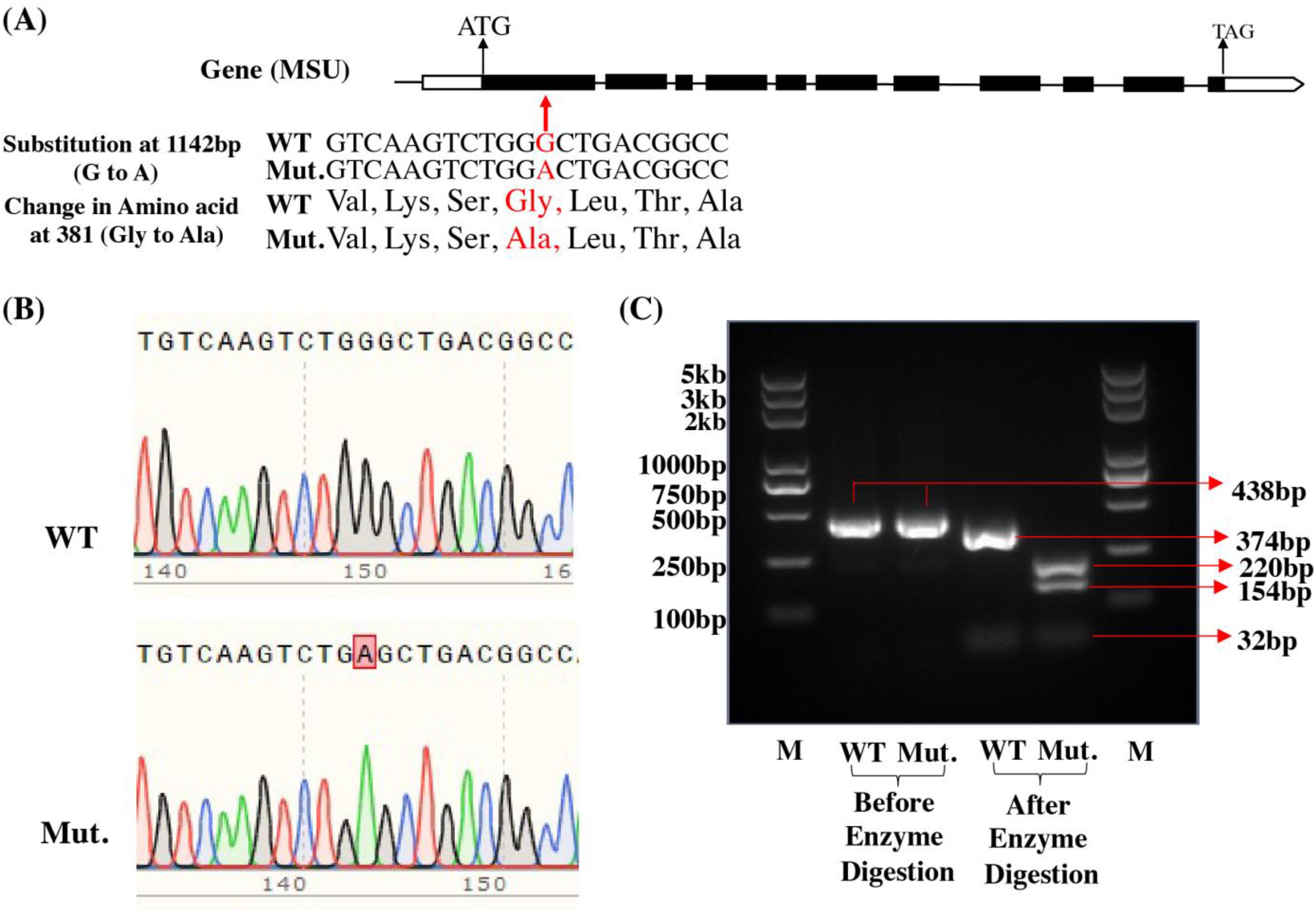
Figure 3. Confirmation of LPA3 gene. (A) Gene structure and mutation site. (B) Sequencing results of WT and mutant (Mut). (C) Confirmation of mutation through Alu1 enzyme digestion. PCR product size is 432bp. After digestion with the restriction enzyme AluI (recognition site: AGCT), WT(Qamar4) produced 3 bands: 32bp, 374bp, 32bp; the lpa-59-14 mutant (Qamar3) produced 4 bands: 32, 154, 220, 32bp.
3.5 Use of a CAPS marker to detect the lpa-59-14 mutant site in the OsMRP5
To further confirm this mutation site, we designed a CAPS (cleaved amplified polymorphic sequence) marker. For this purpose, we performed a PCR, and then the PCR product was digested with the AluI restriction enzyme. After enzyme digestion, Q3 produces 4 bands (32, 220, 154, and 32 bp) while Q4 produces only 3 (32, 374, and 32 bp) bands (Figure 3C). This marker is 100% linked with this mutation and can be used for future breeding programs. Previously, CAPS markers for low phytate grain trait were developed for maize kernels (Abhijith et al., 2020) and Rice (Tan et al., 2013) for different genes but this is the first study for the development of functional molecular marker based on MRP5 gene which adds further diversity into our pool of molecular markers.
3.6 Multiple sequences alignment and phylogenetic analysis
To scan the sequence of MRP5 gene across different cereals versus model plant Arabidopsis, we did a sequence alignment of amino acid sequences of MRP5 (Figure 4). A phylogenetic tree (Figure 5) was constructed using orthologue genes from different species such as rice, maize, barley, and Arabidopsis. Results indicated that all these genes are closely related to each other’ but OsMRP5 is more closely related to maize MRP4 which suggest their more recent evolution from a common ancestor.
4 Conclusion
In conclusion, we were able to detect a novel mutation in OsMRP5 gene (LOC_ Os03g04920) in Q3 (lpa-59-14) mutant. Sequence analysis displayed a substitution in the first exon of OsMRP5 at position 1142 bp resulting in the amino acid change from glycine (Gly) to alanine (Ala) at position 381a.a.a. A breeder friendly CAPS marker for a new mutation in OsMRP5 gene in low phytate Basmati rice mutant was developed. This CAPS marker provided 100% confirmation of new low phytic acid mutation and can be used for future marker assisted breeding programs.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author contributions
Z-U-Q: Visualization, Methodology, Investigation, Conceptualization, Writing – review & editing, Writing – original draft. MU: Validation, Writing – review & editing, Formal analysis. AH: Writing – review & editing, Validation. SZ: Writing – review & editing, Methodology. XL: Writing – review & editing, Validation, Supervision, Resources.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. Studies carried out in China were supported by the Agricultural Variety Improvement Project of Shandong Province (2021LZGC020). Studies carried out in Pakistan, were supported by International Atomic Energy agency, (IAEA project No. RAS 7/014) and Pakistan Science Foundation Project No. PSF/NSL/P-NIAB/149.
Acknowledgments
The authors would like to express their sincere appreciation to all institutions and individuals involved in establishing and supporting the Memorandum of Understanding (MOUs) between ICS-CAAS through XL and P.B.G. D. NIAB through AH and Z-U-Q. These MOUs have fostered a productive collaboration resulting in enhancing the research and exchange of ideas leading to this exceptional research work.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abdel-Megeed, A., Tahir, A. (2015). Reduction of phosphorus pollution from broilers waste through supplementation of wheat based broilers feed with phytase. J. @ Chem. 1, 867014. doi: 10.1155/2015/867014
Abhijith, K. P., Muthusamy, V., Chhabra, R., Dosad, S., Bhatt, V., Chand, G., et al. (2020). Development and validation of breeder-friendly gene-based markers for lpa1-1 and lpa2-1 genes conferring low phytic acid in maize kernel. 3 Biotech. 10, 1–12. doi: 10.1007/s13205-020-2113-x
Abid, N., Khatoon, A., Maqbool, A., Irfan, M., Bashir, A., Asif, I., et al. (2017). Transgenic expression of phytase in wheat endosperm increases bioavailability of iron and zinc in grains. Transg. Res. 26, 109–122. doi: 10.1007/s11248-016-9983-z
Ahmad, H., Zafar, S. A., Naeem, M. K., Shokat, S., Inam, S., Rehman, M. A. U., et al. (2022). Impact of pre-anthesis drought stress on physiology, yield-related traits, and drought-responsive genes in green super rice. Front. In Genet. 13, 832542. doi: 10.3389/fgene.2022.832542
Akhter, S., Saeed, A., Irfan, M., Malik, K. A. (2012). In vitro dephytinization and bioavailability of essential minerals in several wheat varieties. J. Cereal Sci. 56, 741–746. doi: 10.1016/j.jcs.2012.08.017
Alavi, M., Ahmadikhah, A., Kamkar, B., Kalateh, M. (2009). Mapping Rf3 locus in rice by SSR and CAPS markers. Intl. J. Genet. Mol. Biol. 7, 121–126.
Amjad, I., Kashif, M., Akhtar, S., Qamar, Z. U. (2022). Evaluation of the tolerance of rice (oryza sativa) for submergence and drought using various yield related stress indices. Biol. Clin. Sci. Res. J. 1). doi: 10.54112/bcsrj.v2022i1.106
Anonymous (2019). Economic survey of Pakistan (Islamabad, Pakistan: Government of Pakistan, Finance Division, Economic Advisor’s Wing).
Ashraf, M., Cheema, A. A., Qamar, Z.-u., Rashid, M. (2007a). Detection of polymorphism in rice germplasm using RAPD markers. Pak. J. Bot. 39, 2483–2493.
Ashraf, M., Cheema, A. A., Rashid, M., Qamar, Z.-u. (2003). Effect of gamma rays on M1 generation in Basmati rice. Pak. J. Bot. 35, 791–795.
Ashraf, M., Cheema, A. A., Rashid, M., Qamar, Z.-u. (2007b). Induction and breeding of long grain basmati rice. Biologia 53, 83–88.
Ashraf, M., Qamar, Z. U., Hameed, A., Sarsu, F. (2021). Correlation of yield with early seedling performance and physio-biochemical traits in basmati rice mutants subjected to heat stress. Aust.J. Crop Sci. 15, 78–89. doi: 10.3316/informit.176411304308955
Cheema, A. A., Qamar, Z.-u., Rashid, M., Ashraf, M. (2008). “Induction and evaluation of low phytic acid mutants in basmati rice (Abstract),” in Proceedings of International symposium on induced mutations in plants (ISIM), International Atomic Energy Agency (IAEA), Joint FAO/IAEA Division of Nuclear Techniques in Food and Agriculture, Plant Breeding Section, (edits), Vienna (Austria), 12-15, Aug 2008, Vienna, Austria. IAEA-CN-167-410. 64.
Cheema, A. A., Rashid, M., Ashraf, M., Qamar, Z.-u. (2004). Genetic divergence in rice collections. Pak. J. Bot. 36, 557–566.
Chen, P. S., Toribara, T. Y., Warner, H. (1956). Microdeterminations of phosphorous. Anal. Chem. 28, 1756–1758. doi: 10.1021/ac60119a033
Chigwedere, C. M., Stone, A., Konieczny, D., Lindsay, D., Huang, S., Glahn, R., et al. (2023). Examination of the functional properties, protein quality, and iron bioavailability of low-phytate pea protein ingredients. Eur. Food Res. Technol. 249, 1517–1529. doi: 10.1007/s00217-023-04232-x
Cichy, K., Raboy, V. (2009). “Evaluation and Development of Low-Phytate Crops,” in Modi. of Seed Comp. Prom. Health Nutri, vol. 51. , 177–200.
Cosgrove, D. J., Irving, G. C. J. (1980). Inositol phosphates: their chemistry, biochemistry, and physiology (Amsterdam: Elsevier), 191.
Dorsch, J. A., Cook, A., Young, K. A., Anderson, J. M., Bauman, A. T., Volkmann, C. J., et al. (2003). Seed phosphorus and inositol phosphate phenotype of barley low phytic acid genotypes. Phytochem 62, 691–706. doi: 10.1016/S0031-9422(02)00610-6
Duan, X., Yu, H., Ma, W., Sun, J., Zhao, Y., Yang, R., et al. (2020). A major and stable QTL controlling wheat thousand grain weight: Identification, characterization, and CAPS marker development. Mol. Br. 40, 68. doi: 10.1007/s11032-020-01147-3
Dwivedi, S. L., Ortiz, R. (2021). Food crops low in seed-phytate improve human and other non-ruminant animals’ Health. Crop Br. Genet. Genom. 3, e210009. doi: 10.20900/cbgg20210009
Fan, Y., Zhang, Z., Huang, D., Huang, T., Wang, H., Zhuang, J., et al. (2023). A novel PCR-based functional marker of rice blast resistance gene pi25. Agric 13, 1926. doi: 10.3390/agriculture13101926
Frittelli, A., Botticella, E., Palombieri, S., Masci, S., Celletti, S., Fontanella, M. C., et al. (2023). The suppression of TdMRP3 genes reduces the phytic acid and increases the nutrient accumulation in durum wheat grain. Front. Plant Sci. 14, 1079559. doi: 10.3389/fpls.2023.1079559
Fukagawa, N. K., Ziska, L. H. (2019). Rice: importance for global nutrition. J.Nutr. Sci.Vitaminol. 65, S2–S3. doi: 10.3177/jnsv.65.S2
Gupta, R. K., Gangoliya, S. S., Singh, N. K. (2015). Reduction of phytic acid and enhancement of bioavailable micronutrients in food grains. J.Food Sci.Tech. 52, 676–684. doi: 10.1007/s13197-013-0978-y
Guttieri, M., Bowen, D., Dorsch, J. A., Raboy, V., Souza, E. (2004). Identification and characterization of a low phytic acid wheat. Crop Sci. 44, 418–424. doi: 10.2135/cropsci2004.4180
Hameed, K., Habib, M., Awan, F. S., Sadia, B., Hussain, S., Qamar, Z. U., et al. (2023). Detection of genetic divergence among putative ethyl methane sulfonate mutants of super basmati using microsatellite markers. Mol. Biol.Rep. 50, 8799–8808. doi: 10.1007/s11033-023-08425-1
Hitz, W. D., Carlson, T. J., Kerr, P. S., Sebastian, S. A. (2002). Biochemical and molecular characterization of a mutation that confers a decreased raffinosaccharide and phytic acid phenotype on soybean seeds. Available online at.
Huo, Y., Wang, J., Xu, Y., Hu, D., Zhang, K., Chen, B., et al. (2023). The impact of various organic phosphorus carriers on the uptake and use efficiency in barley. Intl. J. Mol.Sci. 24, 17191. doi: 10.3390/ijms242417191
Ibrahim, S., Saleem, B., Rehman, N., Zafar, S. A., Naeem, M. K., Khan, M. R. (2022). CRISPR/Cas9 mediated disruption of Inositol Pentakisphosphate 2-Kinase 1 (TaIPK1) reduces phytic acid and improves iron and zinc accumulation in wheat grains. J. Adv. Res. 37, 33–41. doi: 10.1016/j.jare.2021.07.006
Jacob, R. H., Afify, A. S., Shanab, S. M., Shalaby, E. A. (2024). Chelated amino acids: biomass sources, preparation, properties, and biological activities. Biom. Conver. Bioref. 14, 2907–2921. doi: 10.1007/s13399-022-02333-3
Kim, S. I., Andaya, C. B., Goyal, S. S., Tai, T. H. (2008a). The rice OsLpa1 gene encodes a novel protein involved in phytic acid metabolism. Theor. Appl. Genet. 117, 769–779. doi: 10.1007/s00122-008-0818-z
Kim, S. I., Andaya, C. B., Newman, J. W., Goyal, S. S., Tai, T. H. (2008b). Isolation and characterization of a low phytic acid rice mutant reveals a mutation in the rice orthologue of maize MIK. Theor. Appl. Genet. 117, 1291–1301. doi: 10.1007/s00122-008-0863-7
Kumar, A., Nayak, S., Ngangkham, U., Sah, R. P., Lal, M. K., TP, A., et al. (2021). A single nucleotide substitution in the SPDT transporter gene reduced phytic acid and increased mineral bioavailability from Rice grain (Oryza sativa L.). J. Food. Chem. 45, e13822. doi: 10.1111/jfbc.13822
Kumar, V., Sinha, A. K., Makkar, H. P., Becker, K. (2010). Dietary roles of phytate and phytase in human nutrition: A review. Food Chem. 120, 945–959. doi: 10.1016/j.foodchem.2009.11.052
Larson, S. R., Rutger, J. N., Young, K. A., Raboy, V. (2000). Isolation and genetic mapping of a non-lethal rice (Oryza sativa L.) low phytic acid mutation. Crop Sci. 40, 1397–1405.
Larson, S. R., Young, K. A., Cook, A., Blake, T. K., Raboy, V. (1998). Linkage mapping of two mutations that reduce phytic acid content of barley grain. Theor. Appl. Genet. 97, 141–146.
Leng, Y., Zhao, M., Fiedler, J., Dreiseitl, A., Chao, S., Li, X., et al. (2020). Molecular mapping of loci conferring susceptibility to spot blotch and resistance to powdery mildew in barley using the sequencing-based genotyping approach. Phytopathology 110, 440–446. doi: 10.1094/PHYTO-08-19-0292-R
Leng, Y., Zhao, M., Wang, R., Steffenson, B. J., Brueggeman, R. S., Zhong, S. (2018). The gene conferring susceptibility to spot blotch caused by Cochliobolus sativus is located at the Mla locus in barley cultivar Bowman. Theor.Appl. Genet. 131, 1531–1539. doi: 10.1007/s00122-018-3095-5
Lestari, P., Koh, H. J. (2013). Development of new CAPS/dCAPS and SNAP markers for rice eating quality. HAYATI J. Biosci. 20, 15–23. doi: 10.4308/hjb.20.1.15
Li, W. X., Zhao, H. J., Pang, W. Q., Cui, H. R., Poirier, Y., Shu, Q. Y. (2014). Seed-specific silencing of OsMRP5 reduces seed phytic acid and weight in rice. Transg. Res. 23, 585–599. doi: 10.1007/s11248-014-9792-1
Lin, W., Bai, M., Peng, C., Kuang, H., Kong, F., Guan, Y. (2024). Genome editing toward biofortified soybean with minimal trade-off between low phytic acid and yield. aBIOTECH 5 (2), 196–201. doi: 10.1007/s42994-024-00158-4
Liu, Q. L., Xu, X. H., Ren, X. L., Fu, H. W., Wu, D. X., Shu, Q. Y. (2007). Generation and characterization of low phytic acid germplasm in rice (Oryza sativa L.). Theo.Appl. Genet. 114, 803–814. doi: 10.1007/s00122-006-0478-9
Liu, S., Hao, Z., Weng, J., Li, M., Zhang, D., Pan, G., et al. (2015). Identification of two functional markers associated with drought resistance in maize. Mol.Breeding 35, 1–10. doi: 10.1007/s11032-015-0231-7
Liu, X. J., Liu, X. C., Sun, H. Y., Hao, C. Y., Wang, X. X., Rong, Z. J., et al. (2022a). Validation of CAPS marker WR003 for the leaf rust resistance gene Lr1 and the molecular evolution of Lr1 in wheat. Czech J. Genet. Plant Br. 58, 223–232. doi: 10.17221/119/2021-CJGPB
Liu, Y., Zhang, L., Liu, X., Zhang, P., Zhao, Z., Yi, H., et al. (2022b). Identification of maize rf4-restorer lines and development of a CAPS marker for rf4. Agron 12, 1506. doi: 10.3390/agronomy12071506
Mehnaz, M., Dracatos, P., Pham, A., March, T., Maurer, A., Pillen, K., et al. (2021). Discovery and fine mapping of Rph28: A new gene conferring resistance to Puccinia hordei from wild barley. Theo. Appl. Genet. 134, 2167–2179. doi: 10.1007/s00122-021-03814-1
Mumtaz, M. Z., Saqib, M., Akhtar, J., Abbas, G., Qamar, Z.-u. (2018). Field evaluation of physiology, growth and yield of different rice genotypes under salt affected field conditions. J. Plant Nutr. 41, 233–242. doi: 10.1080/01904167.2017.1385796
Mumtaz, M. Z., Saqib, M., Akhtar, J., Abbas, G., Qamar, Z.-u. (2020). Drought stress impairs grain yield and quality of different rice genotypes under field conditions by impaired photosynthetic attributes and K nutrition. Rice Sci. 27, 5–9. doi: 10.1016/j.rsci.2019.12.001
Obala, J., Saxena, R. K., Singh, V. K., Saxena, K., Kumar, C. V. S., Kumar, A., et al. (2019). Genetic analysis of seed protein content and its association with seed weight and yield in pigeonpea. J. Food. Legumes 32, 65–69. doi: 10.59797/jfl.v32i2.671
Perera, I., Seneweera, S., Hirotsu, N. (2018). Manipulating the phytic acid content of rice grain toward improving micronutrient bioavailability. Rice 11, 1–13. doi: 10.1186/s12284-018-0200-y
Pramitha, J. L., Rana, S., Aggarwal, P. R., Ravikesavan, R., Joel, A. J., Muthamilarasan, M. (2021). Diverse role of phytic acid in plants and approaches to develop low-phytate grains to enhance bioavailability of micronutrients. Adv.Genet 107, 89–120. doi: 10.1016/bs.adgen.2020.11.003
Qamar, Z. U., Akhtar, J., Ashraf, M., Akram, M., Hameed, A. (2012). A multivariate analysis of rice genetic resources. Pak. J.Bot. 44, 1335–1340.
Qamar, Z. U., Cheema, A. A., Ashraf, M., Rashid, M., Tahir, G. R. (2005). Association analysis of some yield influencing traits in aromatic and non-aromatic rice. Pak. J.Bot. 37, 613–627.
Qamar, Z. U., Hameed, A., Ashraf, M., Rizwan, M., Akhtar, M. (2019). Development and molecular characterization of low phytate basmati rice through induced mutagenesis, hybridization, backcross, and marker assisted breeding. Front.Plant Sci. 10, 1525. doi: 10.3389/fpls.2019.01525
Qamar, Z., Rizwan, M., Rani, R., Shahzad, R., Jamil, S., Faheem, M., et al. (2024). Trends towards the production of biologically safe marker free transgenic plants. Appl.Ecol.Env. Res. 22, 221–247. doi: 10.15666/aeer/2201_221247
Quint, M., Mihaljevic, R., Dussle, C., Xu, M., Melchinger, A., Lübberstedt, T. (2002). Development of RGA-CAPS markers and genetic mapping of candidate genes for sugarcane mosaic virus resistance in maize. Theo.Appl.Genet 105, 355–363. doi: 10.1007/s00122-002-0953-x
Raboy, V. (2002). Progress in breeding low phytate crops. J.Plant.Nut 132, 503S–505S. doi: 10.1093/jn/132.3.503S
Raboy, V., Gerbasi, P. (1996). “Genetics of myo-inositol phosphate synthesis and accumulation,” myo-Inositol phosphates, phosphoinositides, and signal transduction. Biswas, B.B., Biswas, S. Eds. (New York: Plenum Press), 257–285.
Raboy, V., Gerbasi, P. F., Young, K. A., Stoneberg, S. D., Pickett, S. G., Bauman, A. T., et al. (2000). Origin and seed phenotype of maize low phytic acid 1 1 and low phytic acid 2 1. Plant Physiol. 124, 355–368.
Rani, R., Singh, R., Yadav, N. R. (2019). Evaluating stripe rust resistance in Indian wheat genotypes and breeding lines using molecular markers. Comptes. Rendus. Biologies 342, 154–174. doi: 10.1016/j.crvi.2019.04.002
Rashid, M., Cheema, A. A., Ashraf, M., Zia-ul-Qamar and Mahmood, Z. (2003). Development of basmati Rice varieties through the use of induced mutations and related techniques in rice. Pak. J. Bot. 35, 811–818.
Rasmussen, S. K., Hatzack, F. (1998). Identification of two low-phytate barley (Hordeum vulgare L.) grain mutants by TLC and genetic analysis. Hereditas 129, 107–112. doi: 10.1111/j.1601-5223.1998.00107.x
Ravichanthiran, K., Ma, Z. F., Zhang, H., Cao, Y., Wang, C. W., Muhammad, S., et al. (2018). Phytochemical profile of brown rice and its nutrigenomic implications. Antioxidants 7, 71. doi: 10.3390/antiox7060071
Rizwan, M., Hussain, M., Shimelis, H., Hameed, M. U., Atif, R., Azhar, R. M., et al. (2019). Gene flow from major genetically modified crops and strategies for containment and mitigation of transgene escape: a review. Appl. Ecol. Env. Res. 17, 11191–11208. doi: 10.15666/aeer/1705_1119111208
Shakoor, U., Saboor, A., Baig, I., Afzal, A., Rahman, A. (2015). Climate variability impacts on rice crop production in Pakistan. Pak. J. Agric. Res. 28, 19–27.
Song, L., Wang, R., Yang, X., Zhang, A., Liu, D. (2023). Molecular markers and their applications in marker-assisted selection (MAS) in bread wheat (Triticum aestivum L.). Agric 13, 642. doi: 10.3390/agriculture13030642
Subasi, B. G., Forghani, B., Abdollahi, M. (2024). Exploring Swedish pea varieties suitable for protein isolation, focusing on antinutrients and off-flavors. J. Food Comp. Analy 128, 105988. doi: 10.1016/j.jfca.2024.105988
Suzuki, M., Tanaka, K., Kuwano, M., Yoshida, K. T. (2007). Expression pattern of inositol phosphate-related enzymes in rice (Oryza sativa L.): implications for the phytic acid biosynthetic pathway. Gene 405, 55–64. doi: 10.1016/j.gene.2007.09.006
Tan, Y. Y., Fu, H. W., Zhao, H. J., Lu, S., Fu, J. J., Li, Y. F., et al. (2013). Functional molecular markers and high-resolution melting curve analysis of low phytic acid mutations for marker-assisted selection in rice. Mol. Br. 31, 517–528. doi: 10.1007/s11032-012-9809-5
Thiel, T., Michalek, W., Varshney, R., Graner, A. (2003). Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Theo. Appl. Genet. 106, 411–422. doi: 10.1007/s00122-002-1031-0
Tong, C., Chen, Y., Tan, Y., Liu, L., Waters, D. L., Rose, T. J., et al. (2017). Analysis of lyso phospho lipid content in low phytate rice mutants. J. Agric. Food. Chem. 65, 5435–5441. doi: 10.1021/acs.jafc.7b01576
Utami, D. W., Lestari, P., Yuriyah, S., Prangdimurti, E. (2024). “Molecular breeding of biofortified rice and its bioaccessibility on cooked rice,” in INTERNATIONAL CONFERENCE ON ORGANIC AND APPLIED CHEMISTRY (ICOAC) 2022.
Varshney, R. K. (2009). “Gene-based marker systems in plants: high throughput approaches for marker discovery and genotyping,” in Mol. Techniq. Crop Improvement, 2nd Edition (Netherlands: Springer), 119–142.
Venegas, J., Guttieri, M. J., Boehm, J. D., Jr., Graybosch, R., Bai, G., St. Amand, P. C., et al. (2022). Genetic architecture of the high inorganic phosphate phenotype derived from a low phytate mutant in winter wheat. Crop Sci. 62, 1228–1241. doi: 10.1002/csc2.20738
Wang, H. M., Chen, J., Shi, Y. F., Pan, G., Shen, H. C., Wu, J. L. (2012). Development and validation of CAPS markers for marker-assisted selection of rice blast resistance gene Pi25. Acta Agronomica Sin. 38, 1960–1968. doi: 10.3724/SP.J.1006.2012.01960
Warkentin, T. D., Delgerjav, O., Arganosa, G., Rehman, A. U., Bett, K. E., Anbessa, Y., et al. (2012). Development and characterization of low-phytate pea. Crop Sci. 52, pp.74–pp.78. doi: 10.2135/cropsci2011.05.0285
Xu, X.-H., Zhao, H.-J., Liu, Q.-L., Frank, T., Engel, K.-H., An, G., et al. (2009). Mutations of the multi-drug resistance-associated protein ABC transporter gene 5 result in reduction of phytic acid in rice seeds. Theor. Appl. Genet. 119, 75–83.
Yathish, K. R., Karjagi, C. G., Gangoliya, S. S., Gadag, R. N., Mallikarjuna, M. G., Sekhar, J. C., et al. (2023). Development of low-phytate maize inbred lines through marker-assisted introgression of lpa1. Crop Pasture Sci. 74, 843–855. doi: 10.1071/CP22238
Zafar, S. A., Hameed, A., Ashraf, M., Khan, A. S., Qamar, Z. U., Li, X., et al. (2020). Agronomic, physiological and molecular characterisation of rice mutants revealed the key role of reactive oxygen species and catalase in high-temperature stress tolerance. Funct. Plant Biol. 47, 440–453. doi: 10.1071/FP19246
Zhao, H., Frank, T., Tan, Y., Zhou, C., Jabnoune, M., Arpat, A. B., et al. (2016). Disruption of OsSULTR3; 3 reduces phytate and phosphorus concentrations and alters the metabolite profile in rice grains. New Phytol. 211, 926–939. doi: 10.1111/nph.13969
Keywords: gamma rays, bio fortification, mineral deficiency, Oryza sativa, phytic acid
Citation: Qamar Z-u-, Uzair M, Hameed A, Zafar SA and Li X (2024) Identification of a novel mutation in the OsMRP5 gene in low phytate Basmati rice mutant and development of CAPS marker for marker-assisted breeding. Front. Plant Sci. 15:1455219. doi: 10.3389/fpls.2024.1455219
Received: 26 June 2024; Accepted: 25 November 2024;
Published: 17 December 2024.
Edited by:
Francesco Sestili, University of Tuscia, ItalyReviewed by:
Victor Raboy, Independent Researcher, Portland, OR, United StatesGayacharan, Indian Council of Agricultural Research (ICAR), India
Copyright © 2024 Qamar, Uzair, Hameed, Zafar and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zia-ul- Qamar, emlhX3VsX3FhbWFyMjAwM0B5YWhvby5jb20=; Xueyong Li, bGl4dWV5b25nQGNhYXMuY24=; Amjad Hameed, YW1qYWQ0NnBrQHlhaG9vLmNvbQ==
†These authors have contributed equally to this work
 Zia-ul- Qamar
Zia-ul- Qamar Muhammad Uzair
Muhammad Uzair Amjad Hameed
Amjad Hameed Syed Adeel Zafar
Syed Adeel Zafar Xueyong Li
Xueyong Li