- 1Institute of Grape Wine, College of Pharmacy, Binzhou Medical University, Yantai, Shandong, China
- 2Department of Plant Science, University of Cambridge, Cambridge, United Kingdom
- 3The Engineering Research Institute of Agriculture and Forestry, College of Horticulture, Ludong University, Yantai, Shandong, China
In wine grape, the multi-functional cytochrome P450 enzyme VvCYP76F14 sequentially catalyzes the formation of linalool-derived compounds, including (E)-8-hydroxylinalool, (E)-8-oxolinalool, and (E)-8-carboxylinalool, which are crucial precursors for the wine bouquet. However, molecular basis towards VvCYP76F14 in regulating the wine bouquet precursor production remain unknown. In this study, both wine bouquet precursor contents and catalytic activities of VvCYP76F14s varied among the three different wine bouquet type varieties. Subcellular localization analysis revealed that VvCYP76F14s are predominantly localized in the endoplasmic reticulum. Notably, a maltose-binding protein (MBP) fusion-tag was added to each of the three VvCYP76F14 proteins in the Escherichia coli expression system, significantly induced the concentration of the MBP-VvCYP76F14 fusion proteins. Site-directed mutation of 4 amino acid residues (I120L, L298V, E378G, and T389A) in VvCYP76F14 resulted in a significant decrease in VvCYP76F14 enzymatic activities, respectively. Furthermore, the transient expression of VvCYP76F14 cloned from ‘Yanniang No.2’ significantly increased the levels of (E)-8-hydroxylinalool, 8-oxolinalool, and (E)-8-carboxylinalool compounds in the transformed ‘Yanniang No.2’, ‘Italian Riesling’, and ‘Marselan’ berries, respectively. In conclusion, VvCYP76F14 dominates the production of wine bouquet precursors and could be a fingerprint marker for screening superior hybrid offspring with desired levels of wine bouquet precursors.
1 Introduction
In wine grape (Vitis vinifera L.), aroma serves as a pivotal sensory indicator of wine quality, captivating enthusiasts and connoisseurs alike (Thomas-Danguin et al., 2011; Robinson et al., 2014; Alegre et al., 2020; Riffle et al., 2022; Zhai et al., 2023). The aroma of wine principally composed of primary aromas and wine bouquet (or secondary aromas). In particular, flavor compounds within the berries contribute to primary aromas, embodied in herbal, sweet, floral, and fruity notes, while wine bouquet is formed during fermentation and ageing through biochemical processes, transforming flavor precursors into a diverse array of aromatic compounds (Lukic et al., 2017; Wang et al., 2017; Alegre et al., 2020; Riffle et al., 2022; Zhai et al., 2023). As wine ages, the primary aromas derived from the wine grape tend to diminish, while the wine bouquet becomes more prominent, destroying the typical characteristics associated with the wine’s aroma. The evolving bouquet is implicated in dominating the unique aromatic profile as the wine matures and develops (Thomas-Danguin et al., 2011; Parker et al., 2017). Notably, Picard et al. (2015) revealed a limited correlation between wine bouquet and the berry aroma directly derived from wine grape berries.
To date, a plethora of compounds contributing to the complex and diverse wine bouquet have been identified (Ghaste et al., 2015; Lin et al., 2019; Alegre et al., 2020). However, only a relatively limited amounts of compounds have been unveiled in shaping the characteristical wine bouquet (Alegre et al., 2020). Particularly, bicyclic monoterpene lactones play pivotal roles in controlling wine bouquet formation. Notably, these bicyclic monoterpene lactones are derived from the crucial precursor (E)-8-carboxylinalool during wine ageing (Giaccio et al., 2011; Ilc et al., 2017). The biosynthesis of (E)-8-carboxylinalool involves multiple steps (hydroxylation, dehydrogenation, and carboxylation), which was catalyzed by a cytochrome P450 enzyme VvCYP76F14 (Ilc et al., 2017; Lin et al., 2019; Peng et al., 2024). Unlike most of the monofunctional P450s that only catalyze a single substrate in plants, the CYP76 family enzymes are multifunctional monooxygenases capable of catalyzing multiple substrates (Paine et al., 2005). In Arabidopsis thaliana, the CYP76 family enzyme AtCYP76C1 can catalyze the conversion of linalool into 8-hydroxylinalool, 8-oxolinalool, and 8-carboxyllinalool as well as lilac aldehydes and lilac alcohols (Boachon et al., 2015; Kunert et al., 2023). However, the molecular basis underlying the regulation of wine bouquet precursor production by VvCYP76F14 in wine grapes are largely unknown.
In accordance with previous studies, grape varieties are typically described as ‘Neutral’ (low bouquet density), ‘Aromatic’ (middle bouquet density), and ‘Full-Bodied’ (high bouquet density), contributing differently to the wine bouquet (Giaccio et al., 2011; Thomas-Danguin et al., 2011; Yang et al., 2019; Alegre et al., 2020; Peng et al., 2024). In this study, we selected three distinct wine bouquet types (‘Italian Riesling’, Neutral; ‘Marselan’, Aromatic; ‘Yanniang No.2’, Full-bodied) to investigate sequence differences and enzymatic activities among VvCYP76F14s derived from these varieties. Subsequently, site-directed mutagenesis and in vitro and in vivo functional characterization were conducted to identify the key amino acid residues of VvCYP76F14s responsible for the three catalytic reaction processes. This study provides molecular insights for investigating the physiological function of VvCYP76F14 in wine grape and revealing the application of VvCYP76F14 as a fingerprint marker for selecting wine grape varieties with desired amounts of wine bouquet precursors.
2 Materials and methods
2.1 Chemicals
According to the description of Peng et al. (2024), (E)-8-hydroxylinalool, (E)-8-oxolinalool, and (E)-8-carboxylinalool were synthesized and purified in Accela ChemBio Co. Ltd. (Shanghai, China). The primary precursor of linalool was purchased from J and K Scientific Co. Ltd. (Shanghai, China). The other chemicals used in this study were purchased from China National Pharmaceutical Group Chemical Reagents Shanghai Co., Ltd (Shanghai, China).
2.2 Wine grape varieties and grape wine samples
V. vinifera cv. Italian Riesling, V. vinifera cv. Marselan, and V. vinifera × V. labrusca cv. Yanniang No.2 berries were collected from the National Grape Germplasm Repository in Yantai, China at 90, 100, and 110 DAFB (day after full bloom), respectively, exhibiting similar maturity stages as outlined in the Grape Grower’s Handbook (Goldammer, 2018). Berry samples were promptly frozen in liquid nitrogen for subsequent analysis. Three biological replicates were carried out, each with 40 individual wine grapes.
2.3 Determination of linalool-derived compounds in berries
The levels of linalool, (E)-8-hydroxylinalool, (E)-8-oxolinalool, and (E)-8-carboxylinalool in the wine grape berries were assessed using High-Performance Liquid Chromatography combined with High-Resolution Mass Spectrometry (HPLC-HRMS) (Waters, Milford, MA, USA) by Shanghai Bioprofile Technology Co. Ltd. (Shanghai, China).
2.4 Cloning and sequence analysis of VvCYP76F14
The coding sequences (CDSs) of VvCYP76F14 were cloned and sequenced from three wine bouquet type varieties (‘Italian Riesling’, ‘Marselan’, and ‘Yanniang No.2’). Total RNA was extracted using the MiniBEST Plant RNA Extraction Kit (TaKaRa, Dalian, China), and the remaining DNA contamination was removed with RNase-free Recombinant DNase I (TaKaRa, Dalian, China). The quantity and quality of the extracted RNA were determined by Invitrogen Qubit Flex Fluorometer (Thermo Fisher Scientific, Waltham, USA). Then, the first-strand cDNA was synthesized using the PrimeScript II First Strand cDNA Synthesis Kit (TaKaRa, Dalian, China).
The CDSs of VvCYP76F14 were cloned using the Prime STAR™ HS DNA polymerase (TaKaRa, Dalian, China). Specific primers of VvCYP76F14 (Forward: 5’-ATGGAGTTGTTGAGTTGTCTG-3’; Reverse: 5’-TCAAACCCGTACAGGTAGAGCTTGCAG-3’) were synthesized in Shenggong Bioengineering Co., Ltd. (Shanghai, China). The PCR fragments were then cloned into pMD 18-T (TaKaRa, Dalian, China) and sequenced by Shenggong Bioengineering Co., Ltd. (Shanghai, China).
2.5 Real-time quantitative PCR
The qPCR was carried out using the LightCycler® 480 system (Roche, Inc., Basel, Switzerland) and SYBR Green qPCR Master Mix (TaKaRa, Dalian, China). The positive recombinant VvCYP76F14-PMD 18-T plasmids were extracted using the Plasmid Miniprep Kit (Tiangen, Beijing, China), followed by the manufacturer’s instructions. The concentration of the extracted plasmids was assessed using the Qubit Flex Fluorometer (Thermo Fisher Scientific, Waltham, USA). Absolute quantification was conducted using the specific primers (Forward: 5’-TGTTATCCAACACCATAT-3’; Reverse: 5’-TCCCAGCTTCCTCCATCACA-3’) and the first-strand cDNA template. To generate a standard curve, each recombinant plasmid was diluted and used as a template for qPCR.
2.6 Subcellular localization of VvCYP76F14
To investigate the subcellular localization of VvCYP76F14, the CDSs of VvCYP76F14 from ‘Italian Riesling’, ‘Marselan’, and ‘Yanniang No.2’ were cloned into the pBWA(V)HS-ccdb-GLosgfp vector (RiORUN, Wuhan, China) using BsaI and Eco31I restriction sites, resulting in the generation of pBWA(V)HS-CYP76F14-GLosgfp constructs. The pBWA(V)HS-sper-LK-mKATE was used as an endoplasmic reticulum marker (Mravec et al., 2009). The Agrobacterium GV3101 strain, containing either pBWA(V)HS-CYP76F14-GLosgfp or the marker vector, was independently infiltrated into Nicotiana benthamiana leaves. Two days after infiltration, selected leaves were excised for confocal observations using an LSM880 microscope (Carl Zeiss, Oberkochen, Germany). The GFP fluorescence was observed using excitation/emission wavelengths of 488/510 nm, the Chlorophyll autofluorescence was observed using excitation/emission wavelengths of 640/660 nm, while the mKATE fluorescence was observed using excitation/emission wavelengths of 561/580 nm.
2.7 Site-directed mutagenesis
To unveil the catalytic activities of key amino acid residues in VvCYP76F14 from ‘Yanniang No.2’, alanine-scanning method was employed to generate alanine-substituted VvCYP76F14-SMs (Song et al., 2021; Peng et al., 2024). Sequence codons were optimized and synthesized at GenScript Co. Ltd. (Nanjing, China). Each of the candidate amino acid residues (N46S, T107I, N111K, I120L, R175Q, L222V, M264I, S286N, L298V, K325T, E378G, T380A, E383D, and T386A) was substituted with a corresponding amino acid with opposite polarity.
2.8 Heterologous expression of VvCYP76F14 in Escherichia coli
To enhance production and facilitate the folding of recombinant VvCYP76F14 proteins, the pMAL-c6T vector (New England Biolabs, Beijing, China) containing a maltose-binding protein (MBP) tag was used for heterologous expression (Lebendiker and Danieli, 2011). The complete CDSs of the VvCYP76F14s and VvCYP76F14-SMs, with AlwN I and Sbf I sites added at the 5’ and 3’ ends, respectively, were synthesized by GenScript Co. Ltd. (Nanjing, China). The correctness of the pMAL-c6T-VvCYP76F14s constructs was validated through sequencing (Biomarker Co. Ltd. Beijing, China), using the same primer pairs (Forward: 5’-ATGGAGTTGTTGAGTTGTCTG-3’; Reverse: 5’-TCAAACCCGTACAGGTAGAGCTTGCAG-3’). These validated constructs were then expressed in the Escherichia coli BL21(DE3) strain (TaKaRa, Dalian, China). The NEBExpress® MBP Fusion and Purification System (New England Biolabs, Hitchin, UK) was employed to facilitate the purification of MBP-VvCYP76F14s, using the affinity between MBP and amylose resin. Subsequently, the MBP-VvCYP76F14s were cleaved from the MBP-tag using TEV Protease (New England Biolabs, Hitchin, UK) and purified using sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) assay. The confirmation of the recombinant proteins was performed using HPLC-HRMS (Waters, Milford, MA, USA).
2.9 In vitro enzymatic activity assay
In vitro enzyme activities of recombinant VvCYP76F14 from ‘Yanniang No.2’ (Full-Bodied) and VvCYP76F14-SMs were determined using linalool, (E)-8-hydroxylinalool, and (E)-8-oxolinalool as the substrate, respectively. Preliminary assays were conducted to determine the optimum reaction system. The Arabidopsis NADPH cytochrome P450 reductase (ATR1, Werck-Reichhart and Feyereisen, 2000; Jensen and Møller, 2010; Lin et al., 2019; Kunert et al., 2023) was chosen as the electron transport redox partner of VvCYP76F14. The enzyme activity assays were conducted in a final volume of 5 mL, supplied with 100 mM Na+/K+ phosphate buffer (pH 5.0), 1 mM NADPH, and adjusted ratio of VvCYP76F14 to ATR1 (2:1). The reactions were conducted at 25 ± 1°C for 1 h with agitation, and the resulting product was gathered and determined using HPLC-HRMS (Waters, Milford, MA, USA). Boiled protein (nonfunctional) was taken as the control. For the determination of kinetic parameters, substrate reduction was both qualitatively and quantitatively determined. The turnover number (kcat) and affinity (km) were individually calculated. All assays were conducted in sextuplicate.
2.10 Overexpression of full-bodied variety VvCYP76F14 in neutral variety berries
To assess the enzymatic activity of VvCYP76F14 in berries, the CDS of VvCYP76F14 was isolated from the Full-Bodied variety ‘Yanniang No.2’ and synthesized by GenScript Co. Ltd. (Nanjing, China), and further cloned into the pMDC32-HPB vector (Addgene: 32078) to generate the recombinant plasmid pMDC32-HPB-VvCYP76F14. Subsequently, the recombinant plasmid and empty vector were transferred into the Agrobacterium GV3101 strain (WEIDI, Shanghai, China), respectively. In detail, 1 mL of the Agrobacterium GV3101 suspension with an OD600 of 0.8 was injected into the berries of ‘Yanniang No.2’ at 110 days after full bloom (DAFB), ‘Marselan’ at 100 DAFB, or ‘Italian Riesling’ at 90 DAFB, in the early morning (approximately 24-26 °C) at four points individually around the diameter of the wine grape surface. After injection for 3 days, the contents of linalool, (E)-8-hydroxylinalool, (E)-8-oxolinalool, and (E)-8-carboxylinalool in the grape berries were determined by HPLC-HRMS (Waters, Milford, MA, USA).
2.11 Statistical analysis
The significant differences were conducted in IBM SPSS Statistics 23 (Armonk, New York, USA), using ANOVA followed by Fisher’s LSD test method.
3 Results
3.1 Different wine bouquet precursor contents among neutral, aromatic, and full-bodied wine grape varieties
The core collection of the National Grape Germplasm Repository in Yantai, China has proven to be a valuable resource for studying and evaluating the contributions of various wine grape varieties to wine bouquet (Xiao et al., 2020; Peng et al., 2024). An analysis of wine bouquet precursor contents revealed that there was no difference in the levels of linalool among ‘Italian Riesling’ (Neutral), ‘Marselan’ (Aromatic), and ‘Yanniang No.2’ (Full-bodied) berries (Figure 1; Supplementary Table 1). However, the levels of (E)-8-hydroxylinalool, 8-oxolinalool, and (E)-8-carboxylinalool varied significantly among the three wine bouquet type varieties, respectively (Figure 1). Notably, the maximum amounts of (E)-8-hydroxylinalool, 8-oxolinalool, and (E)-8-carboxylinalool were observed in ‘Yanniang No.2’ berries, followed by Marselan’ and ‘Italian Riesling’ berries (Figure 1).
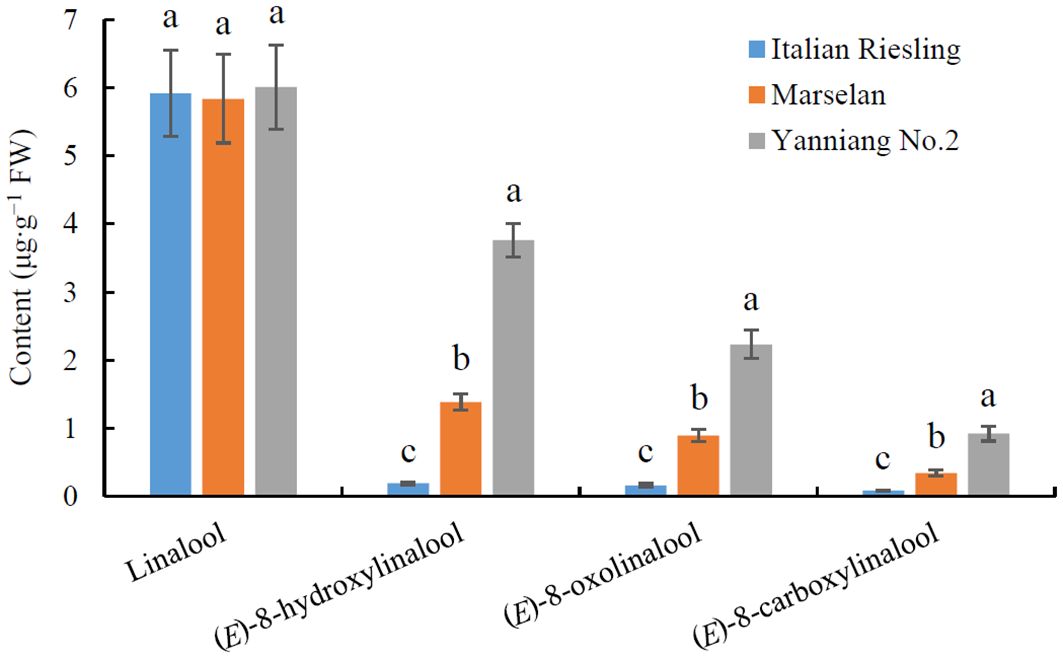
Figure 1. Determination of wine bouquet precursor contents from three typical wine bouquet types of grape varieties. Vitis vinifera cv. Italian Riesling, V. vinifera cv. Marselan, and V. vinifera × V. labrusca cv. Yanniang No.2 berries were collected at 90, 100, and 110 DAFB (day after full bloom), respectively. The contents of linalool, (E)-8-hydroxylinalool, (E)-8-oxolinalool and (E)-8-carboxylinalool in the grape berries were determined using HPLC-HRMS. Data are presented as means ± SEs (n = 3). Letters represent significant differences among three wine bouquet type varieties at a significance level of p ≤ 0.05, as determined using ANOVA followed by Fisher’s LSD test.
3.2 Identification of amino acid residue variations in VvCYP76F14s
To investigate sequence differences among these three wine grape varieties, we isolated the CDSs of VvCYP76F14s from ‘Italian Riesling’ (Neutral), ‘Marselan’ (Aromatic), and ‘Yanniang No.2’ (Full-bodied) berries. Sequencing results revealed that all three VvCYP76F14s possessed the identical gene structure that contained with a long 5’-UTR region, two exons, and one intron, respectively (Figure 2A). In addition, all three VvCYP76F14s encoded a predicted polypeptide consisting of 499 amino acids (Figure 2B). In particular, sequence variations were observed among these three wine grape varieties. Compared to ‘Yanniang No.2’, 14 site variations (N46S, T107I, N111K, I120L, R175Q, L222V, M264I, S286N, L298V, K325T, E378G, T380A, E383D, and T386A) were found in ‘Italian Riesling’ and 12 site variations (N46S, N111K, R175Q, L222V, M264I, S286N, L298V, K325T, E378G, T380A, E383D, and T386A) were observed in ‘Marselan’ (Figure 2B). Furthermore, the expression levels of VvCYP76F14 exhibited no differences in berries of these three varieties (Supplementary Figure 1).

Figure 2. Gene structure and amino acid sequence alignment analysis. (A) Gene structures of VvCYP76F14s from three typical wine bouquet types of grape varieties. (B) Amino acid sequence alignment revealed amino acid residue variations in VvCYP76F14s derived from Neutral and Aromatic wine grape varieties. Multiple sequence alignment was performed on the amino acid sequences of VvCYP76F14s from wine grape varieties representing three types of wine bouquet (V. vinifera cv. Italian Riesling, V. vinifera cv. Marselan, and V. vinifera × V. labrusca cv. Yanniang No.2). Putative key amino acid residues in the Neutral variety (Italian Riesling) were indicated by brown frames and stars. Conserved regions are highlighted in red frames. The alignment analysis was conducted using the ClustalW program within the MEGA 13.0 software.
3.3 VvCYP76F14 is localized in the endoplasmic reticulum
In this study, the coding sequence of VvCYP76F14s derived from ‘Italian Riesling’ (Neutral), ‘Marselan’ (Aromatic), or ‘Yanniang No.2’ (Full-bodied) was cloned into the pBWA(V)HS-ccdb-GLosgfp vector (RiORUN, Wuhan, China) and expressed in N. benthamiana leaves, respectively. The pBWA(V)HS-sper-LK-mKATE was used as an endoplasmic reticulum marker. Confocal observations revealed that all VvCYP76F14s derived from ‘Italian Riesling’ (Neutral), ‘Marselan’ (Aromatic), and ‘Yanniang No.2’ (Full-bodied) were predominantly localized in the endoplasmic reticulum (Figure 3).
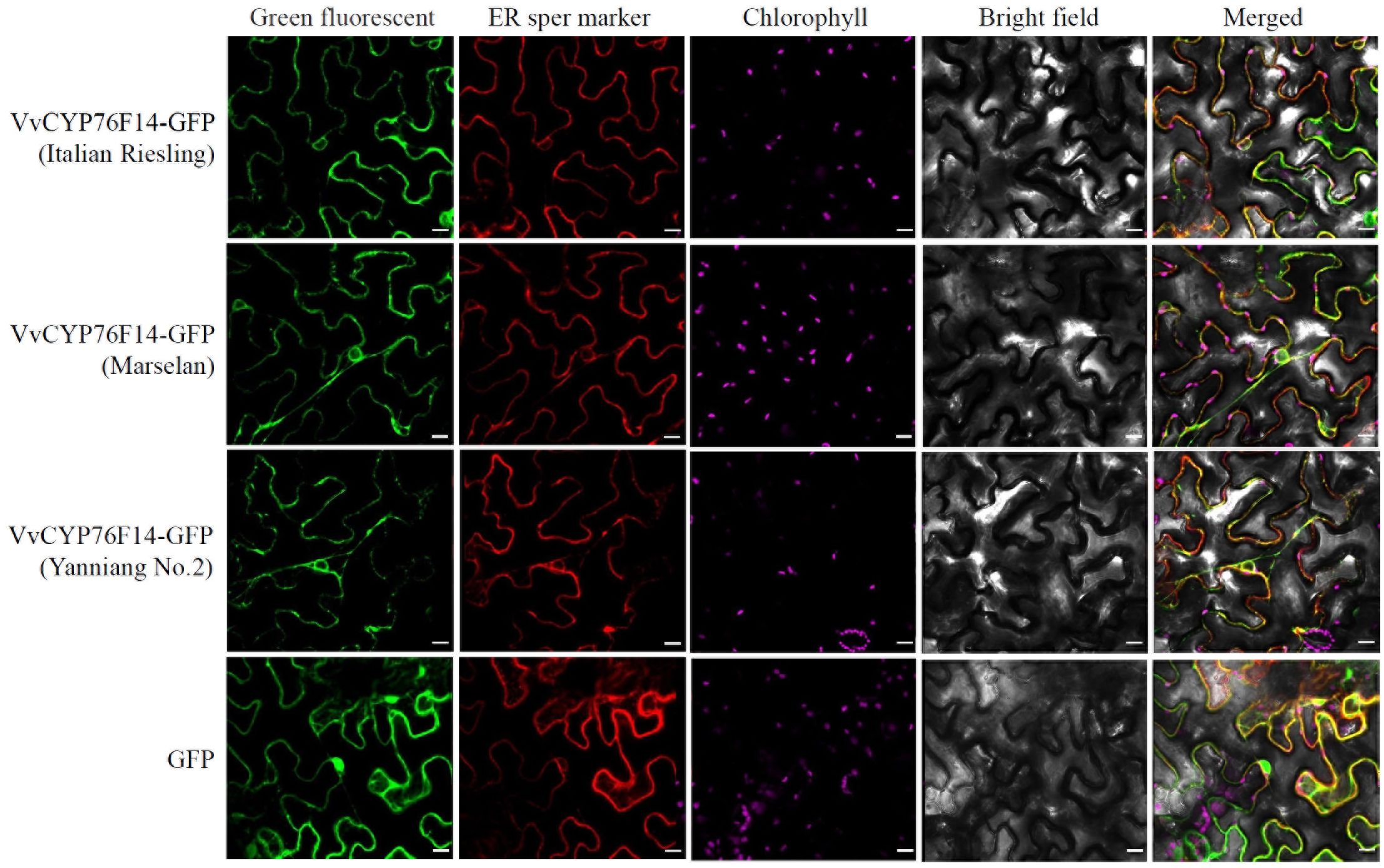
Figure 3. Subcellular localization analysis of VvCYP76F14s from three typical wine bouquet type varieties. The coding sequence of VvCYP76F14s derived from ‘Italian Riesling’ (Neutral), ‘Marselan’ (Aromatic), or ‘Yanniang No.2’ (Full-bodied) was cloned into the pBWA(V)HS-ccdb-GLosgfp vector (RiORUN, Wuhan, China). The pBWA(V)HS-sper-LK-mKATE was used as an endoplasmic reticulum marker. The Agrobacterium GV3101 strain containing either the pBWA(V)HS-CYP76F14-GLosgfp or the marker vector was independently infiltrated into Nicotiana benthamiana leaves. Two days after infiltration, selected leaves were excised for confocal observations using a LSM880 microscope. The GFP fluorescence was observed using excitation/emission wavelengths of 488/510 nm, the Chlorophyll autofluorescence was observed using excitation/emission wavelengths of 640/660 nm, while the sper mKATE fluorescence was observed using excitation/emission wavelengths of 561/580 nm. Scale bar = 10 μm.
3.4 Full-bodied type VvCYP76F14 exhibited higher enzymatic activity in vitro
Considering the previously observed low concentration of VvCYP76F14 produced (too low to quantify) in a heterologous expression system (Boachon et al., 2015; Ilc et al., 2017), a maltose-binding protein (MBP) fusion-tag was added to each of the three VvCYP76F14 proteins in the E. coli expression system. Taking the Full-Bodied type VvCYP76F14 (‘Yanniang No.2’) for example, this modification resulted in a significantly higher concentration of the MBP-VvCYP76F14 fusion protein (Figure 4). Subsequently, the MBP-VvCYP76F fusion was cleaved using TEV protease, resulting in the isolation of VvCYP76F (Figure 4).
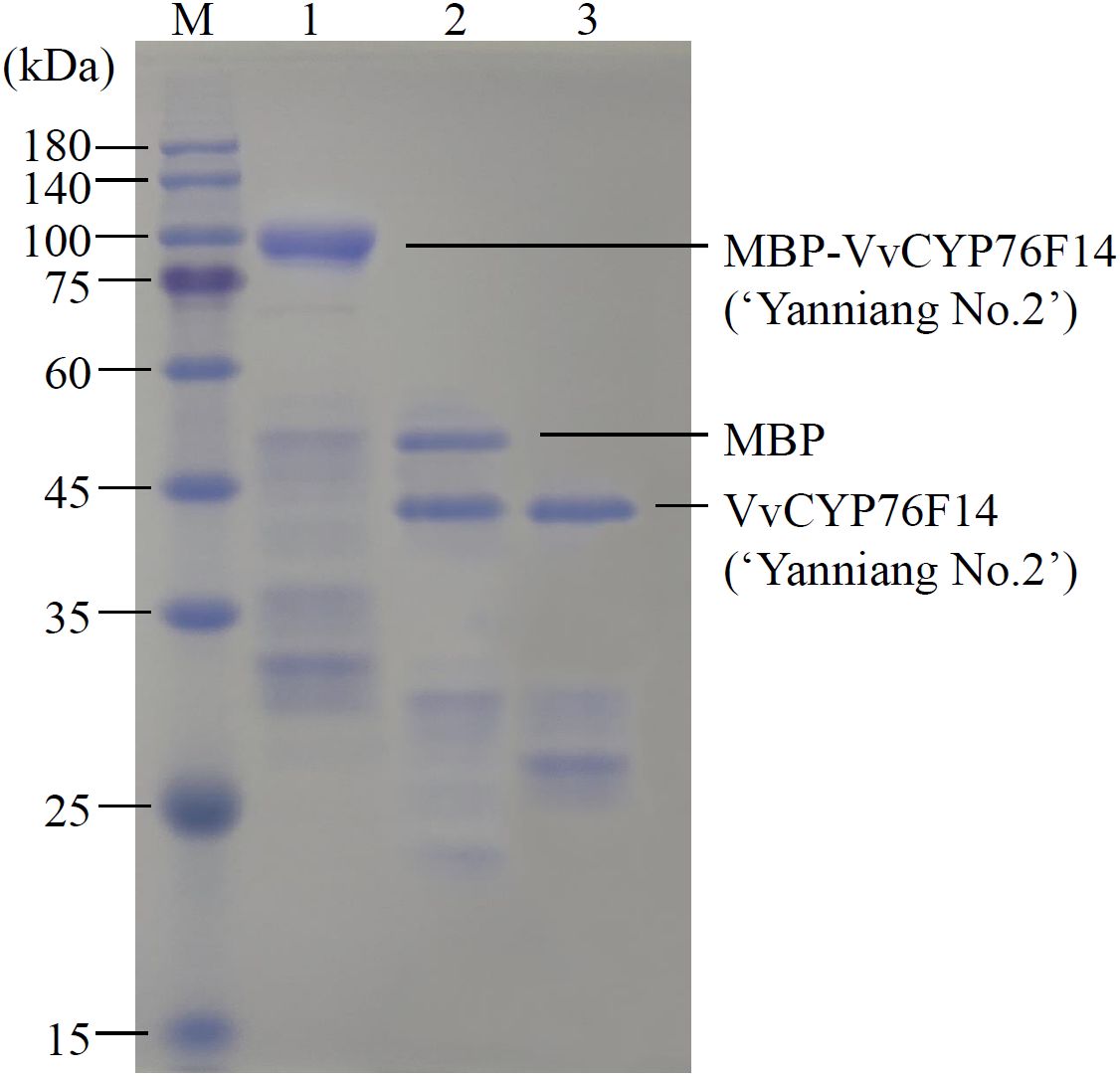
Figure 4. SDS-PAGE of MBP fusion analysis of VvCYP76F14s. SDS-PAGE analysis of the recombinant VvCYP76F14 from the Full-bodied variety of ‘Yanniang No.2’ in E. coli. The pMAL-c6T-VvCYP76F14s (Biomarker Co., Ltd, Beijing, China) constructs were expressed in the E. coli BL21(DE3) strain. The protein standard marker (M) was loaded on the gel. Lane 1 represents purified MBP-VvCYP76F14 eluted from amylose column with maltose. Lane 2 represents purified VvCYP76F14 after TEV protease cleavage. Lane 3 represents isolated and partially purified VvCYP76F14.
The subsequent enzyme activity analysis involved monitoring the decrease in the substrate concentration. All three VvCYP76F14s demonstrated catalytic activity in reactions utilizing linalool (hydroxylation), (E)-8-hydroxylinalool (dehydrogenation), and (E)-8-oxolinalool (carboxylation) as substrates (Figure 5; Supplementary Table 2). In detail, the Full-Bodied type VvCYP76F14 exhibited a significantly higher depletion of linalool and (E)-8-hydroxylinalool substrates, compared to the other two types, reflects the fact that the catalytic activity of ‘Yanniang No.2’ VvCYP76F14 was higher than those of ‘Italian Riesling’ and ‘Marselan’ VvCYP76F14s. However, there were no noticeable differences in catalytic efficiencies among the three VvCYP76F14s when using (E)-8-oxolinalool as a substrate, means that no significant difference in the catalytic activity of three VvCYP76F14s. Furthermore, there were no significant changes in the catalytic activities utilizing linalool, (E)-8-hydroxylinalool, or (E)-8-oxolinalool as substrate, respectively, for the empty controls without recombinant VvCYP76F14s (Figure 5).
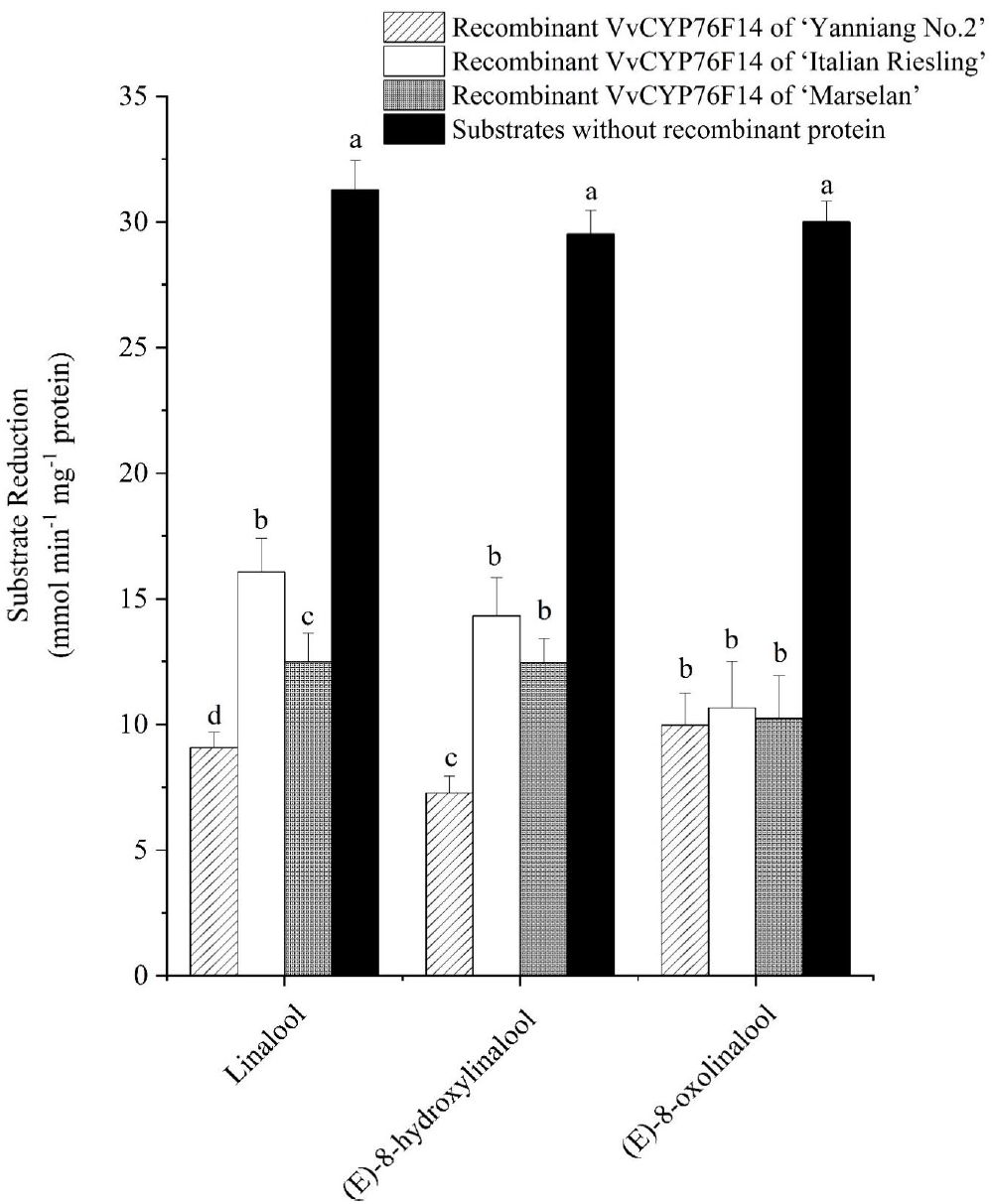
Figure 5. The in vitro specific activity of the recombinant VvCYP76F14s from E. coli. The in vitro specific activity of three wine bouquet type VvCYP76F14s was assessed by measuring the residual levels of substrates. Data are presented as means ± SE (n =3). Letters indicate significant differences among VvCYP76F14s from three wine bouquet type varieties at a significance level of p ≤ 0.05, as determined using ANOVA followed by Fisher’s LSD test.
3.5 Substitution of key amino acid residues diminishes activity of VvCYP76F14 in vitro
In particular, 14 site variations and 12 site variations were observed in VvCYP76F14 of ‘Italian Riesling’ and ‘Marselan’, respectively, compared to ‘Yanniang No.2’ (Figure 2B). To estimate the individual contribution of each candidate amino acid residue to the reactions of VvCYP76F14 from ‘Yanniang No.2’, their functions were examined using site-directed mutagenesis (SM). VvCYP76F14 and VvCYP76F14-SMs were heterologously expressed in E. coli as above and further identified by HPLC-HRMS.
Based on the enzyme kinetic analyses, the substitution of I120L, L298V, E378G, and T389A lead to a significant reduction in the kcat/km ratios, correspondingly, implying decreased activities in the respective VvCYP76F14-SMs (Figure 6; Supplementary Table 3). However, there is no differences in enzymatic activity between the remaining 10 VvCYP76F14-SMs (N46S, T107I, N111K, R175Q, L222V, M264I, S286N, K325T, E383D, and T386A) and the wild type VvCYP76F14, respectively (Figure 6).
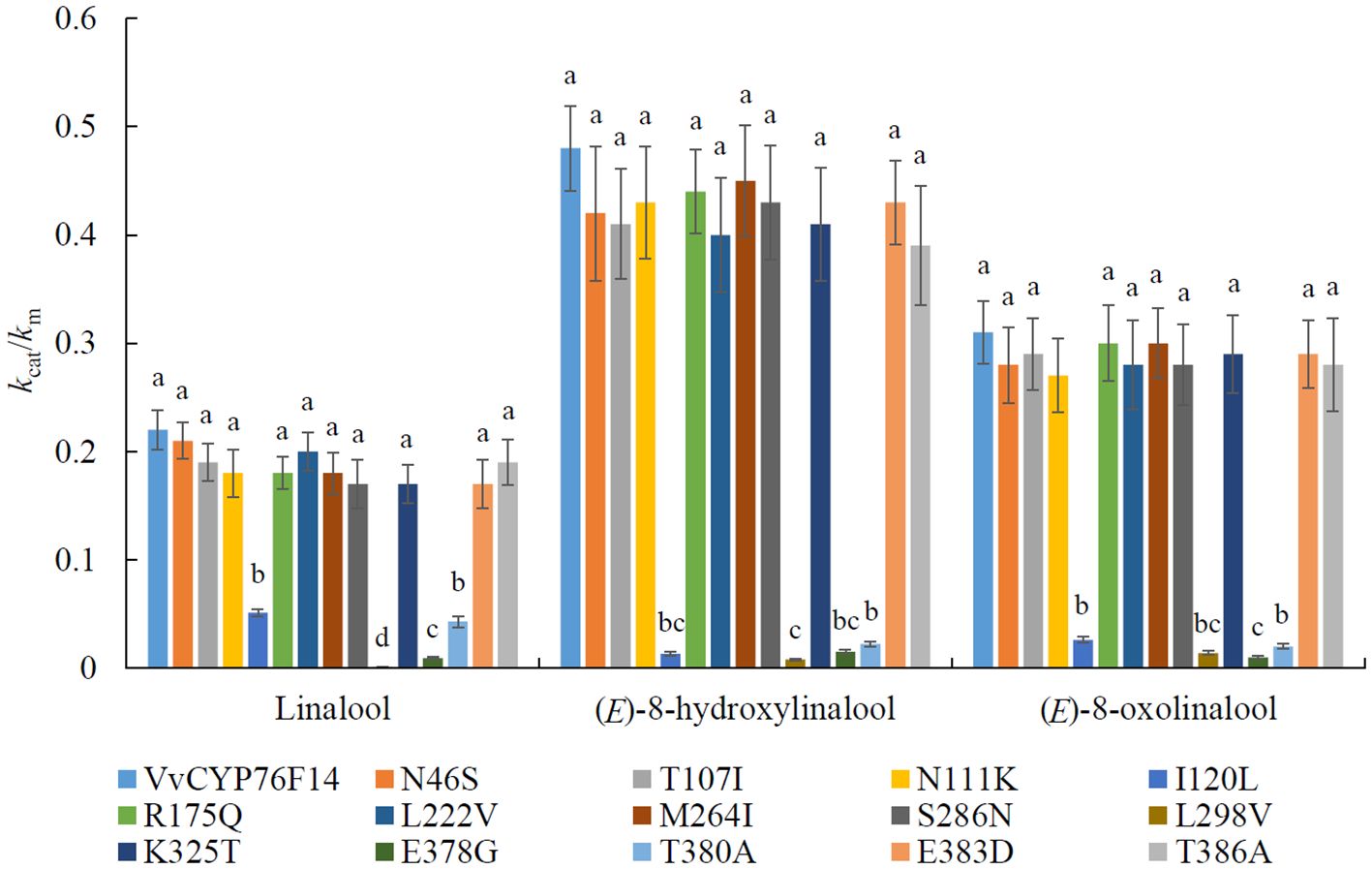
Figure 6. Enzyme kinetics of VvCYP76F14 and its site-directed mutant proteins (VvCYP76F14-SMs) using linalool, (E)-8-hydroxylinalool and (E)-8-oxolinalool as substrate, respectively. VvCYP76F14 was isolated from the Full-Bodied variety ‘Yanniang No.2’. The reaction product was collected and analyzed using HPLC-HRMS (Waters, Milford, MA, USA). Boiled protein (nonfunctional) served as a control. For the determination of kinetic parameters, substrate reduction was both qualitatively and quantitatively determined by HPLC-HRMS (Waters, Milford, MA, USA). The turnover number (kcat) and affinity (km) were individually calculated. Data were presented as the means ± SE (n = 3). Letters represent significant differences among VvCYP76F14 and VvCYP76F14-SMs at a significance level of p ≤ 0.05, as determined using ANOVA followed by Fisher’s LSD test.
3.6 Transient expression of the full-bodied type VvCYP76F14 resulted in the restoration of wine bouquet precursor levels in the each of the three wine bouquet varieties
To further investigate the physiological function of VvCYP76F14 in vivo, pMDC32-HPB-VvCYP76F14 (containing the CDS of VvCYP76F14 isolated from the Full-Bodied variety ‘Yanniang No.2’) was introduced into ‘Yanniang No.2’, ‘Marselan’, and ‘Italian Riesling’ berries, respectively. Results showed that the levels of (E)-8-hydroxylinalool, 8-oxolinalool, and (E)-8-carboxylinalool compounds in the transformed ‘Yanniang No.2’ (Figure 7A), ‘Marselan’ (Figure 7B), and ‘Italian Riesling’ (Figure 7C) berries carrying pMDC32-HPB-VvCYP76F14 were significantly higher than that of wild-type berries and berries transformed with the empty vector, whereas the levels of linalool were significantly reduced in all tested VvCYP76F14 overexpressing berries (Figure 7; Supplementary Table 4).
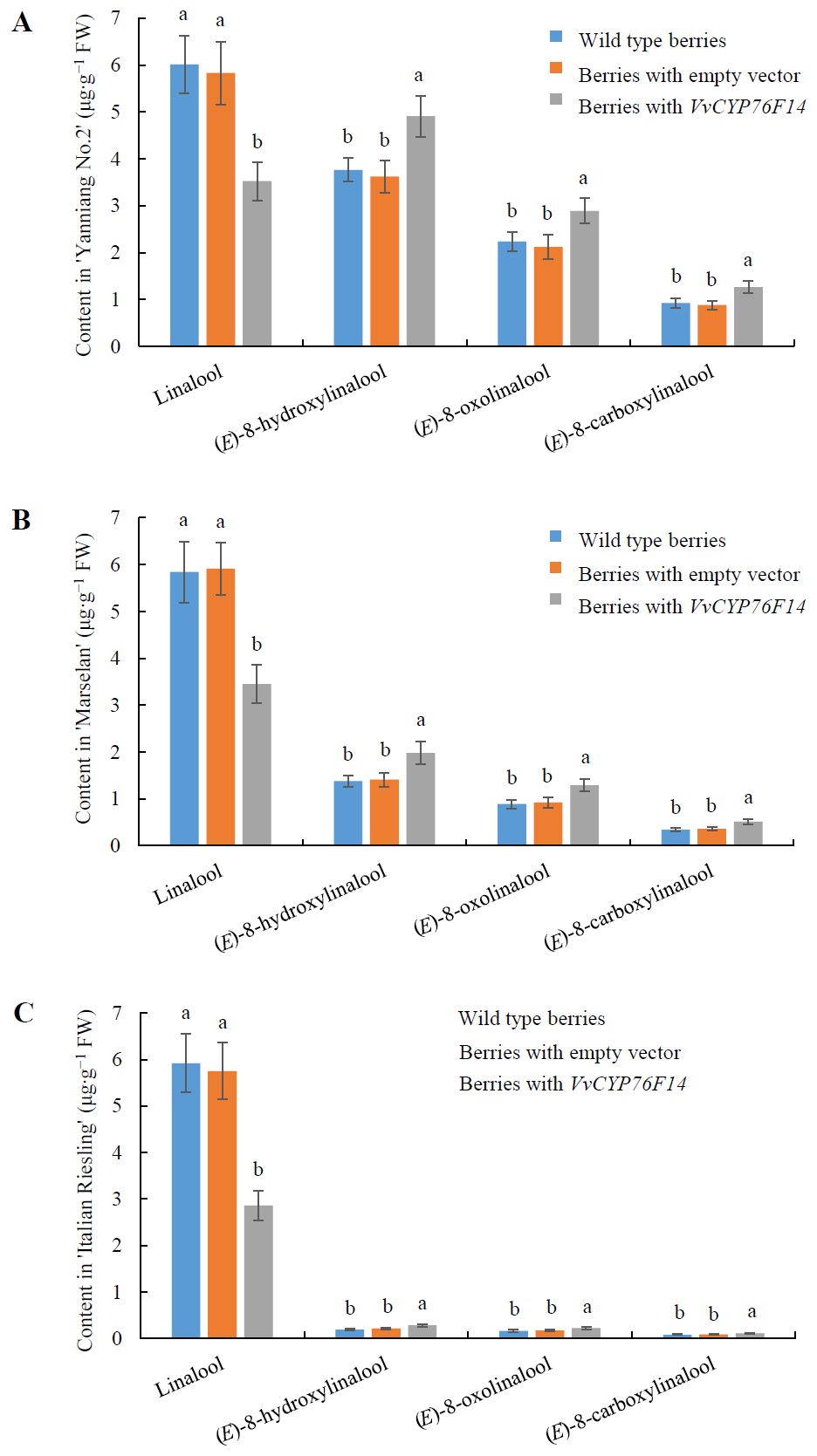
Figure 7. Transient expression of Full-Bodied type VvCYP76F14 in three wine bouquet type varieties. The CDS of VvCYP76F14 was isolated from the Full-Bodied variety ‘Yanniang No.2’ and further cloned into the pMDC32-HPB (Addgene: 32078) to generate the recombinant plasmid pMDC32-HPB-CYP76F14. The pMDC32-HPB-VvCYP76F14 overexpression vector and empty vector were transferred into Agrobacterium GV3101 (WEIDI, Shanghai, China) strain, respectively. The Agrobacterium GV3101 suspension with an OD600 of 0.8 was injected into the berries of ‘Yanniang No.2’ at 110 DAFB (A), ‘Marselan’ at 100 DAFB (B), or ‘Italian Riesling’ at 90 DAFB (C), respectively. After injection for 3 days, the contents of linalool, (E)-8-hydroxylinalool, (E)-8-oxolinalool and (E)-8-carboxylinalool in the grape berries were determined using HPLC-HRMS. Data are presented as means ± SEs (n = 3). Letters represent significant differences among wild type berries, berries transformed with empty vector, and berries transformed with VvCYP76F14 at a significance level of p ≤ 0.05, as determined using ANOVA followed by Fisher’s LSD test. FW means fresh weight.
4 Discussion
In wine grapes, the cytochrome P450 VvCYP76F14 can catalyze trisubstrate-triproduct reaction processes (hydroxylation, dehydrogenation and carboxylation) to produce (E)-8-carboxylinalool, which is an important wine bouquet precursor during wine making and wine ageing processes (Ilc et al., 2017; Yang et al., 2019; Zhai et al., 2023). However, physiological function of wine grape VvCYP76F14 in regulating the production of wine bouquet precursors remains unknown.
Picard et al. (2015) revealed that there is limited correlation between wine bouquet and the berry aroma derived directly from wine grape berries. In contrast, we found significant variations in catalytic activity among different wine grape varieties (mutants) of the VvCYP76F14 enzyme. Simultaneously, numerous genetic differences have also been observed in 1-deoxy-dxylulose 5-phosphate synthase (Battilana et al., 2009), terpene synthase (Martin et al., 2010), and glycosyl transferase (Schwab et al., 2014; Bönisch et al., 2014) among different varieties that have the potential to enhance wine bouquet quality (Lin et al., 2019).
In vitro enzymatic assays have been commonly used to characterize CYP450 family members in plants, including those from Solanum tuberosum (Grausem et al., 2014), A. thaliana (Boachon et al., 2015), and Salvia miltiorrhiza (Li et al., 2021). It is worthy noticing that sequence differences of VvCYP76F14 between two wine grape varieties were also identified in the study of Ilc et al. (2017), but no qualitative differences were found in vitro between the two VvCYP76F14 enzymes. This could be due to the limited amount of recombinase obtained for quantitative analysis and the presence of nonsense mutations in many of the identified variation sites. In this present study, we investigated the key amino acid residues of VvCYP76F14 obtained from a Full-Bodied variety and elucidated their contribution to the enzyme activity through enzymatic assays via the help of MBP fusion-tag expression system in E. coli. All VvCYP76F14s from three bouquet types could catalyze the monoterpenol reaction using linalool, (E)-8-hydroxylinalool, and (E)-8-oxolinalool as substrates, and the Full-Bodied type VvCYP76F14 definitely exhibited the strongest enzyme activity, which was in line with the findings of Peng et al. (2024).
To be honest, we tried and failed to obtain the crystal structure of VvCYP76F14 in this present study. According to the reported crystal structure of S. miltiorrhiza CYP76 homologs (Protein Data Bank no. 5YLW, Li et al., 2021), their active center is A (G) G (A) XD (E) T, which is consistent with the conserved active domain of three wine grape VvCYP76F14s (Figure 2B). This directly proves that mutation of key amino acid residues in VvCYP76F14s is similar to S. miltiorrhiza CYP76 homologs, and the functional amino acid residues are located around the active center (Li et al., 2021). Mutating key enzymes to select cultivars with desired characteristics has been well documented in various crops (Karunarathna et al., 2020; Zhang et al., 2022). Among distinct fruit crop cultivars, substitutions in key enzymes may dominant the phenotypic variability in aromatic component contents (Dunemann et al., 2012; Peng et al., 2020; Song et al., 2021). In this present study, key amino acid variations (I120L, L298V, E378G, and T389A) in VvCYP76F14 were observed in ‘Italian Riesling’ (Neutral) and ‘Marselan’ (Aromatic) berries, which might be implicated in all three reactions that significantly decreased the enzymatic activities of VvCYP76F14-SMs in vitro. These findings indicate that these substitutions are responsible for decreased levels of wine bouquet precursors. Furthermore, the overexpression of the Full-Bodied VvCYP76F14 in all the three wine bouquet type varieties validated the role of VvCYP76F14 in linalool-derivative production in vivo, which are in line with the in vitro catalytic capabilities of VvCYP76F14, implying a close relationship among key amino acid substitutions, VvCYP76F14 activity, and linalool-derivative production.
Favorably, VvCYP76F14 may be a selective marker to screen VvCYP76F14 variants in grape varieties that contribute differently to wine bouquet. Nonetheless, the wine grape VvCYP76F14 is implicated in dominating the production of wine bouquet precursors.
5 Conclusion
The present study underscores the identification of key amino acid substitutions in VvCYP76F14s derived from different wine grape cultivars. Site-directed mutation of 4 amino acid residues (I120L, L298V, E378G, and T389A) in VvCYP76F14 resulted in a significant decrease in enzyme activities. Transient expression of VvCYP76F14 cloned from high wine bouquet ‘Yanniang No.2’ significantly increased the levels of (E)-8-hydroxylinalool, 8-oxolinalool, and (E)-8-carboxylinalool compounds in the transformed ‘Yanniang No.2’, ‘Italian Riesling’, and ‘Marselan’ berries. This study provides an opportunity to use VvCYP76F14 as a fingerprint marker for screening hybrid offspring with desired levels of linalool-derivatives.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author contributions
GX: Funding acquisition, Investigation, Methodology, Validation, Visualization, Writing – review & editing, Formal analysis, Resources. MS: Formal analysis, Methodology, Validation, Writing – review & editing, Conceptualization, Data curation. WX: Formal analysis, Investigation, Methodology, Resources, Validation, Visualization, Writing – original draft. AD: Methodology, Validation, Writing – review & editing, Investigation, Supervision, Visualization, Writing – original draft. ZS: Investigation, Methodology, Validation, Visualization, Writing – original draft, Writing – review & editing, Funding acquisition, Project administration.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by grants from the Major Project of Science and Technology of Shandong Province (2022CXGC010605), Basic Research on Wine Industry Technology in Yantai Regions (50012305073), Fund of China Scholarship Council (202208370080), and UKRI BBSRC (X008843/1).
Acknowledgments
The authors are grateful to Professor Julia M. Davies, Department of Plant Sciences, University of Cambridge for critical reading and valuable suggestions.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1450251/full#supplementary-material
Supplementary Figure 1 | Expression levels of VvCYP76F14s from three wine bouquet types of grape varieties. Data are presented as means ± SEs (n = 3). Letters represent significant differences at a significance level of p ≤ 0.05, as determined using ANOVA followed by Fisher’s LSD test.
Abbreviations
CDS, coding sequences; CPR, cytochrome P450 reductase; MBP, maltose-binding protein; MM-GBSA, molecular mechanics-generalized born surface area; PCA, principal coordinate analysis; qPCR, real-time quantitative PCR; SDS-PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis; SM, site-directed mutagenesis; HPLC-HRMS, high-performance liquid chromatography combined with high-resolution mass spectrometry.
References
Alegre, Y., Saenz-Navajas, M. P., Hernandez-Orte, P., Ferreira, V. (2020). Sensory, olfactometric and chemical characterization of the aroma potential of Garnacha and Tempranillo winemaking grapes. Food Chem. 331, 127207. doi: 10.1016/j.foodchem.2020.127207
Battilana, J., Costantini, L., Emanuelli, F., Sevini, F., Segala, C., Moser, S., et al. (2009). The 1-deoxy-d-xylulose 5-phosphate synthase gene colocalizes with a major QTL affecting monoterpene content in grapevine. Theor. Appl. Genet. 118, 653–669. doi: 10.1007/s00122-008-0927-8
Boachon, B., Junker, R. R., Miesch, L., Bassard, J. E., Hofer, R., Caillieaudeaux, R., et al. (2015). CYP76C1 (Cytochrome P450)-mediated linalool metabolism and the formation of volatile and soluble linalool oxides in Arabidopsis flowers: A strategy for defense against floral antagonists. Plant Cell 27, 2972–2990. doi: 10.1105/tpc.15.00399
Bönisch, F., Frotscher, J., Stanitzek, S., Rühl, L., Wüst, M., Bitz, O., et al. (2014). A UDP-glucose: monoterpenol glucosyltransferase adds to the chemical diversity of the grapevine metabolome. Plant Physiol. 165, 561–581. doi: 10.1104/pp.113.232470
Dunemann, F., Ulrich, D., Malysheva-Otto, L., Weber, W. E., Longhi, S., Velasco, R., et al. (2012). Functional allelic diversity of the apple alcohol acyl-transferase gene MdAAT1 associated with fruit ester volatile contents in apple cultivars. Mol. Breed. 29, 609–625. doi: 10.1007/s11032-011-9577-7
Ghaste, M., Narduzzi, L., Carlin, S., Vrhovsek, U., Shulaev, V., Mattivi, F. (2015). Chemical composition of volatile aroma metabolites and their glycosylated precursors that can uniquely differentiate individual grape cultivars. Food Chem. 188, 309–319. doi: 10.1016/j.foodchem.2015.04.056
Giaccio, J., Capone, D. L., Håkansson, A. E., Smyth, H. E., Elsey, G. M., Sefton, M. A., et al. (2011). The formation of wine lactone from grape-derived secondary metabolites. J. Agric. Food. Chem. 59, 660–664. doi: 10.1021/jf1038162
Goldammer, T. (2018). Grape Grower’s Handbook: A Guide To Viticulture for Wine Production (U.S.A: Apex Publishers).
Grausem, B., Widemann, E., Verdier, G., Nosbusch, D., Aubert, Y., Beisson, F., et al. (2014). CYP77A19 and CYP77A20 characterized from Solanum tuberosum oxidize fatty acids in vitro and partially restore the wild phenotype in an Arabidopsis thaliana cutin mutant. Plant Cell Environ. 37, 2102–2115. doi: 10.1111/pce.2014.37.issue-9
Ilc, T., Halter, D., Miesch, L., Lauvoisard, F., Kriegshauser, L., Ilg, A., et al. (2017). A grapevine cytochrome P450 generates the precursor of wine lactone, a key odorant in wine. New Phytol. 213, 264–274. doi: 10.1111/nph.2017.213.issue-1
Jensen, K., Møller, B. L. (2010). Plant NADPH-cytochrome P450 oxidoreductases. Phytochemistry 71, 132–141. doi: 10.1016/j.phytochem.2009.10.017
Karunarathna, N. L., Wang, H., Harloff, H. J., Jiang, L., Jung, C. (2020). Elevating seed oil content in a polyploid crop by induced mutations in SEED FATTY ACID REDUCER genes. Plant Biotechnol. J. 18, 2251–2266. doi: 10.1111/pbi.v18.11
Kunert, M., Langley, C., Lucier, R., Ploss, K., López, C. Z. R., Guerrero, A. D. S., et al. (2023). Promiscuous CYP87A enzyme activity initiates cardenolide biosynthesis in plants. Nat. Plants 9, 1607–1617. doi: 10.1038/s41477-023-01515-9
Lebendiker, M., Danieli, T. (2011). Purification of proteins fused to maltose-binding protein. Methods Mol.Biol. 681, 281–293. doi: 10.1007/978-1-60761-913-0_15
Li, B., Li, J., Chai, Y., Huang, Y., Li, L., Wang, D., et al. (2021). Targeted mutagenesis of CYP76AK2 and CYP76AK3 in Salvia miltiorrhiza reveals their roles in tanshinones biosynthetic pathway. Int. J. Biol. Macromol. 189, 455–463. doi: 10.1016/j.ijbiomac.2021.08.112
Lin, J., Massonnet, M., Cantu, D. (2019). The genetic basis of grape and wine aroma. Horti Res. 6, 81. doi: 10.1038/s41438-019-0163-1
Lukic, I., Lotti, C., Vrhovsek, U. (2017). Evolution of free and bound volatile aroma compounds and phenols during fermentation of Muscat blanc grape juice with and without skins. Food Chem. 232, 25–35. doi: 10.1016/j.foodchem.2017.03.166
Martin, D. M., Aubourg, S., Schouwey, M. B., Daviet, L., Schalk, M., Toub, O., et al. (2010). Functional annotation, genome organization and phylogeny of the grapevine (Vitis vinifera) terpene synthase gene family based on genome assembly, FLcDNA cloning, and enzyme assays. BMC Plant Biol. 10, 226. doi: 10.1186/1471-2229-10-226
Mravec, J., Skůpa, P., Bailly, A., Hoyerová, K., Křeček, P., Bielach, A., et al. (2009). Subcellular homeostasis of phytohormone auxin is mediated by the ER-localized PIN5 transporter. Nature 459, 1136–1140. doi: 10.1038/nature08066
Paine, M. J. I., Scrutton, N. S., Munro, A. W., Gutierrez, A., Roberts, G. C. K., Wolf, C. R. (2005). “Electron transfer partners of cytochrome P450,” in cytochrome P450: structure, mechanism, and biochemistry. Ed. Ortiz de Montellano, P. R. (Springer US, Boston, MA), 115–148.
Parker, M., Capone, D. L., Francis, I. L., Herderich, M. J. (2017). Aroma precursors in grapes and wine: flavor release during wine production and consumption. J. Agric. Food Chem. 66, 2281–2286. doi: 10.1021/acs.jafc.6b05255
Peng, B., Ran, J. G., Li, Y. Y., Tang, M. L., Xiao, H. L., Shi, S. P., et al. (2024). Site-directed mutagenesis of VvCYP76F14 (cytochrome P450) unveils its potential for selection in wine grape varieties linked to the development of wine bouquet. J. Agric. Food Chem. 72, 3683–3694. doi: 10.1021/acs.jafc.3c09083
Peng, B., Yu, M. L., Zhang, B. B., Xu, J. L., Ma, R. J. (2020). Differences in PpAAT1 activity in high- and low-aroma peach varieties affect γ-decalactone production. Plant Physiol. 182, 2065–2080. doi: 10.1104/pp.19.00964
Picard, M., Tempere, S., de Revel, G., Marchand, S. (2015). A sensory study of the ageing bouquet of red Bordeaux wines: A three-step approach for exploring a complex olfactory concept. Food Qual. Prefer. 42, 110–122. doi: 10.1016/j.foodqual.2015.01.014
Riffle, V. L., Arredondo, J. A., LoMonaco, I., Appel, C., Catania, A. A., Peterson, J. C. D., et al. (2022). Vine age affects vine performance, grape and wine chemical and sensory composition of cv. Zinfandel from California. Am. J. Eno. Viticul. 73, 277–293. doi: 10.5344/ajev.2022.22014
Robinson, A. L., Boss, P. K., Solomon, P. S., Trengove, R. D., Heymann, H., Ebeler, S. E. (2014). Origins of grape and wine aroma. Part 2. Chemical and sensory analysis. Am. J. Eno. Viticul. 65, 25–42. doi: 10.5344/ajev.2013.13106
Schwab, W., Fischer, T. C., Giri, A., Wüst, M. (2014). Potential applications of glucosyltransferases in terpene glucoside production: impacts on the use of aroma and fragrance. Appl. Microbiol. Biotechnol. 99, 165–174. doi: 10.1007/s00253-014-6229-y
Song, Z. Z., Peng, B., Gu, Z. X., Tang, M. L., Li, B., Liang, M. X., et al. (2021). Site-directed mutagenesis identified the key active site residues of alcohol acyltransferase PpAAT1 responsible for aroma biosynthesis in peach fruits. Horti. Res. 8, 32. doi: 10.1038/s41438-021-00461-x
Thomas-Danguin, T., Ishii-Foret, A., Atanasova, B., Etievant, P. (2011). “Wine bouquet: the perceptual integration of chemical complexity,” in Wine active compounds Conference object (Oenoplurimédia, Beaune, France Chaintré (France).
Wang, X. J., Tao, Y. S., Wu, Y., An, R. Y., Yue, Z. Y. (2017). Aroma compounds and characteristics of noble-rot wines of Chardonnay grapes artificially botrytized in the vineyard. Food Chem. 226, 41–50. doi: 10.1016/j.foodchem.2017.01.007
Werck-Reichhart, D., Feyereisen, R. (2000). Cytochromes P450: a success story. Genome Biol. 1, REVIEWS3003. doi: 10.1186/gb-2000-1-6-reviews3003
Xiao, H. L., Li, J. W., Liu, W. H., Lu, J. S., Wang, J. P., Zheng, Q. L., et al. (2020). Effect of spraying abscisic acid to grape fruit on the quality of Heihuxiang wine. China Brewing 39, 100–105 (in Chinese).
Yang, Y., Jin, G. J., Wang, X. J., Kong, C. L., Liu, J., Tao, Y. S. (2019). Chemical profiles and aroma contribution of terpene compounds in Meili (Vitis vinifera L.) grape and wine. Food Chem. 284, 155–161. doi: 10.1016/j.foodchem.2019.01.106
Zhai, H. Y., Li, S. Y., Zhao, X., Lan, Y. B., Zhang, X. K., Shi, Y., et al. (2023). The compositional characteristics, influencing factors, effects on wine quality and relevant analytical methods of wine polysaccharides: A review. Food Chem. 403, 134467. doi: 10.1016/j.foodchem.2022.134467
Keywords: wine grape, wine bouquet, cytochrome P450 enzyme, sequence variation, site-directed mutagenesis
Citation: Xia G, Shi M, Xu W, Dark A and Song Z (2024) Cytochrome P450 VvCYP76F14 dominates the production of wine bouquet precursors in wine grapes. Front. Plant Sci. 15:1450251. doi: 10.3389/fpls.2024.1450251
Received: 17 June 2024; Accepted: 25 September 2024;
Published: 11 October 2024.
Edited by:
Mohammad Irfan, Cornell University, United StatesReviewed by:
Pawan Kumar, National Institute of Plant Genome Research (NIPGR), IndiaDharanidaran Jayachandran, Buckman Laboratories, United States
Ved Prakash, Kansas State University, United States
Copyright © 2024 Xia, Shi, Xu, Dark and Song. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhizhong Song, enMzMTZAY2FtLmFjLnVr
 Guangli Xia1
Guangli Xia1 Zhizhong Song
Zhizhong Song