- 1The United Graduate School of Agricultural Sciences, Tottori University, Tottori, Japan
- 2Faculty of Agriculture, Tottori University, Tottori, Japan
- 3Functional Genomics Facility, National Institute for Basic Biology Core Research Facilities, Okazaki, Japan
- 4Unused Bioresource Utilization Center, Tottori University, Tottori, Japan
Orchids and arbuscular mycorrhiza (AM) plants evolved independently and have different structures and fungal partners, but they both facilitate nutrient uptake. Orchid mycorrhiza (OM) supports orchid seed germination, but unlike AM, its role in disease resistance of mature plants is largely unknown. Here, we examined whether OM induces systemic disease resistance against a necrotrophic pathogen in a similar fashion to AM. We investigated the priming effect of mycorrhizal fungi inoculation on resistance of a terrestrial orchid, Bletilla striata, to soft rot caused by Dickeya fangzhongdai. We found that root colonization by a compatible OM fungus primed B. striata seedlings and induced systemic resistance against the infection. Transcriptome analysis showed that priming was mediated by the downregulation of jasmonate and ethylene pathways and that these pathways are upregulated once infection occurs. Comparison with the reported transcriptome of AM fungus–colonized rice leaves revealed similar mechanisms in B. striata and in rice. These findings highlight a novel aspect of commonality between OM and AM plants in terms of induced systemic resistance.
Highlights
● Unlike arbuscular mycorrhizal (AM) plants, our knowledge of orchid mycorrhizal fungus ability to induce systemic resistance is limited. Orchid mycorrhiza evolves independently from AM plants but retains common components for the regulation of symbiosis at molecular levels (Miura et al., Mol. Plant Microbe Interact., 2018). Thus, we are aiming to explore this commonality between the two in terms of their impact on the host plant defense response against pathogens. This study elaborates the positive correlation between Bletilla striata root colonization by mycorrhizal fungus and induced systemic resistance against a necrotrophic pathogen, Dickeya fangzhongdai (Pectobacteriaceae). The magnitude of systemic resistance is determined by the compatibility with mycorrhizal fungus itself. Also, comparative transcriptomic results shows that priming by T. calospora involves upregulation of jasmonate, and ethylene, which is commonly known to occur in AM plants. Our findings contribute to a better understanding of mycorrhiza contribution to induced systemic resistance in orchids as well as its molecular basis. This work is important because not only orchid mycorrhiza is understudied but also the understanding of induced systemic resistance in orchids may give clues to the mechanisms of the evolution of orchid mycorrhiza and its impact on survival against diseases.
Introduction
Mycorrhiza is one of the oldest symbioses, in which plant roots are colonized by fungi forming a specialized structure inside or outside root cells where nutrient uptake and transfer occur (Smith and Read, 2008). The most common type of mycorrhiza, arbuscular mycorrhiza (AM), is found in almost 80% of flowering plants (Brundrett and Tedersoo, 2018) and appeared in the rooting system at the latest during the Carboniferous (Strullu-Derrien et al., 2018). During mycorrhizal colonization, AM fungi transfer nitrogen, phosphate and water to the host plants (Parniske, 2008; Kakouridis et al., 2022). In return, plants provide carbon compounds to the fungi. The extent of each of these physiological functions often differs among mycorrhizal types and plant species. On the basis of these differences, several other mycorrhizal types are distinguished, including ectomycorrhiza, orchid mycorrhiza (OM), and ericoid mycorrhiza. OM is unique in that the fungal structures formed inside orchid cells are transient and are gradually degraded; this phenomenon is most prominent during early seed germination (Selosse et al., 2017). Mycorrhizal associations in orchids are obligatory during early germination, although in green, leafy species their extent decreases with time due to the plants’ autotrophic nature (Cameron et al., 2008; Girlanda et al., 2011; Wang et al., 2023).
Orchidaceae have evolved endomycorrhizas with Basidiomycota or occasionally Ascomycota (Fracchia et al., 2016). The dominant group, Rhizoctonia-like fungi, includes three main genera—Ceratobasidium, Sebacina/Serendipita, and Tulasnella (Smith and Read, 2008)—while some lower taxa of subfamily Epidendroideae are sometimes associated with ectomycorrhizal fungi (Bidartondo et al., 2004; McKendrick et al., 2004; Zahn et al., 2023) or even wood-rotting, mushroom-forming fungi (Umata et al., 2013). Both OM and AM are involved in nitrogen, carbon, and phosphate transfer (Cameron et al., 2007, 2008; Kuga et al., 2014; Fochi et al., 2016) and have common integral molecular mechanisms (Miura et al., 2018). It was recently discovered that gibberellin inhibits both OM and AM colonization, but it inhibits seed germination only in orchids (Miura et al., 2024). These findings further encourage the exploration of the commonalities between OM and AM, including ecophysiological aspects such as disease resistance.
Two distinct defense responses occur upon pathogen infection: an early, local response at the infection site and a systemic response at distal sites (David et al., 2019); the latter includes systemic acquired resistance (SAR) (Ross, 1966) and induced systemic resistance (ISR) (van Peer et al., 1990; Wei et al., 1991). Systemic acquired resistance occurs when pathogens directly infect leaves and involves mainly salicylic acid, whereas ISR is induced by beneficial microbes (bacteria, endophytic fungi, and mycorrhiza) interacting with roots and involves jasmonate and ethylene (Vlot et al., 2021). During ISR, the plant enters an alert state called priming, allowing enhanced resistance once infection occurs, mainly against necrotrophic pathogens (Pieterse et al., 2014). Primed plants often show no visible changes before infection; thus, ISR is best studied in plants challenged with pathogens, which allows changes at the cellular level between non-primed and primed plants to be compared. Due to the diversity of microbes interacting with roots, the molecular mechanisms of ISR are more diverse than those of SAR because different microbe species induce the accumulation of different signaling compounds (Haney et al., 2018; Dreischhoff et al., 2020). ISR induced by an AM fungus in Medicago truncatula increases the defense response against Xanthomonas campestris (Liu et al., 2007), whereas Solanum lycopersicum has a similar response against Fusarium oxysporum (Wang et al., 2022a). Almost all types of pathogens attack orchids, including bacteria (Keith et al., 2005; Suharjo et al., 2014; Khamtham and Akarapisan, 2019), fungi (Silva and Pereira, 2007; Lopes et al., 2009; Srivastava et al., 2018; Suwannarach et al., 2018), and viruses (Fogell et al., 2019; Tsai et al., 2022). Very little is known about ISR in orchids; to the best of our knowledge, only two studies have reported the occurrence of ISR and its role in alleviating soft rot (Wu et al., 2011; Ye et al., 2019). Thus, enhancing our knowledge on ISR in orchids will give clues on the consequences of evolving mycorrhizal association.
On the basis of the arguments that colonization by AM fungi primes host defense responses against pathogens (Marquez et al., 2018) and major molecular components of AM symbiosis signaling are also present in OM (Miura et al., 2018), here we hypothesized that colonization by OM fungi (OMF) also causes ISR in orchids just as in AM plants. We used a necrotrophic, pectinolytic Gram-negative bacterium known to cause leaf soft rot in orchids (Cating and Palmateer, 2011; Joko et al., 2014; Wei et al., 2021). As recommended by Eck and co-workers (Eck et al., 2022), we also investigated the relationship between mycorrhizal colonization rate and ISR. We focused on a generalist orchid species, Bletilla striata (Orchidaceae, tribe Arethuseae), because it is relatively easy to grow and it associates with OMF from different genera (Zettler et al., 2001; Yamamoto et al., 2017; Miura et al., 2019). Using two OMF species previously reported to associate with B. striata (Miura et al., 2019; Fuji et al., 2020) we evaluated ISR and its molecular regulation in leaves through transcriptomic analysis.
Materials and methods
Plant and fungal materials
Bletilla striata Rchb.f. ‘Murasakishikibu’ plants were purchased from a local nursery in Japan (Yamamoto et al., 2017). For seed production, flowers were self-pollinated and allowed to set seeds for 6 months (until dehiscence). Seeds were randomly picked from capsules harvested between 2017 and 2020. In a pathogen inoculation assay, aside from B. striata, Arabidopsis thaliana (L.) Heynh. ecotype Columbia (Col-0) and Nicotiana benthamiana were also used.
Our earlier studies showed that several strains of Tulasnella and Serendipita vermifera form OM associations with B. striata during seed germination (Yamamoto et al., 2017; Miura et al., 2019; Fuji et al., 2020). Among those fungi, we chose T. calospora (Boud.) Juel MAFF305805 (coded as T.cal05) and S. vermifera (Oberw.) P. Roberts MAFF305830 (coded as S.ver30).
Symbiotic seed germination
In symbiotic seed germination, S. vermifera and T. calospora were cultured in agar (1.5 g/L) containing 2.5 g/L oatmeal (Becton Dickinson, Sparks, MD, USA) until maximum hyphal growth. Bletilla striata seeds were surface-sterilized in 3 mL of 5% NaOCl containing 5 µL Tween-80 for 2 min and were sown directly on top of the mycelia. The plates were incubated at 25°C for 2 weeks.
Asymbiotic seed germination and direct root inoculation of OMF
In asymbiotic seed germination, B. striata seeds were surface-sterilized as above and sown on solid half-strength Murashige–Skoog (MS) medium supplemented with 20 g/L sucrose, 20 g/L banana homogenate, 1 g/L tryptone, 3 g/L activated charcoal, MS vitamin mixture (pH 5.8), and 0.8 g/mL agar. Seedlings were grown for 8 weeks until formation of true leaves and roots. For the first subculture, the plantlets were transferred into half-strength P668 medium (Phytotech Labs, Lenexa, KS, USA) supplemented with 20 g/L sucrose, 0.1 g/L tryptone, 0.2 mg/L naphthalene acetic acid, 0.5 mg/L 6-benzylaminopurine, MS vitamin mixture (pH 5.8), and 0.8 g/mL agar for another 6–8 weeks. Seedlings with at least two true leaves and new roots longer than 2 cm were transferred into small pots filled with a mixture of akadama soil, kanuma soil, and vermiculite (4:4:1 volume ratio) under a 16 h light/8 h dark photoperiod at 25°C for another 2 weeks for hardening (Supplementary Figure 1). S. vermifera and T. calospora were cultured in YEPG liquid medium containing 3 g/L yeast extract, 3 g/L peptone, and 20 g/L glucose for 4 weeks until sufficient mycelial growth. The mycelial mass was harvested and homogenized in distilled water (4 mL/g mycelial mass) using a homogenizer (Nissei, Osaka, Japan). A 2-mL aliquot of the mycelial suspension was inoculated beneath each seedling.
Visualization and quantification of OMF colonization in protocorms and roots
Root colonization is defined as the presence of pelotons, both intact and degraded (Yamamoto et al., 2017). Protocorms and roots were fixed in 70% ethanol and cleared in 5% KOH at 90°C for 1 h (Peterson et al., 2004). To evaluate colonization, the protocorms were stained with 5% black ink (Sheaffer, Fort Madison, IA, USA) in 5% acetic acid for 10 min (Vierheilig et al., 1998). Protocorms were photographed under a light microscope (BX53; Olympus, Tokyo, Japan) equipped with a digital camera (DP27; Olympus). By using ImageJ v.1.53a, pelotons present in protocorms were counted with the multipoint tool and root colonization was measured with the segmented line tool. Root colonization was quantified as described: each piece of root was divided into imaginary 1-mm segments. Each segment was further divided horizontally into four subsegments, especially the middle part by the vascular bundle. The ratio of cortical colonization within each segment was scored as 0 (no OMF colonization on either side), 0.25 (one-quarter of the segment is colonized), 0.5 (only one side is fully colonized), 0.75 (three-quarters of the segment is colonized), or 1 (both sides are colonized). The colonization rate (CR) for a single root was calculated through the colonization index (CI) using the following equations:
where i is a positive integer from 0 to 4, ni/4 is the number of segments colonized at a particular level, and N is the total number of segments in one root. This index can vary from 0 to 1. The final CR of a seedling was then expressed as the arithmetic average value of CR obtained from all roots. The detailed method is available at Zenodo (https://doi.org/10.5281/zenodo.10464582).
Pathogen inoculation
Erwinia chrysanthemi MAFF311045, which was isolated from Phalaenopsis (Suharjo et al., 2014), was cultured in Luria–Bertani (LB) medium overnight. Bacterial suspension at several concentrations and OD600 = 0.0025 was mixed with 0.01% Tween-20 (10:1 volume ratio). Two leaves per seedling of B. striata, A. thaliana, and N. benthamiana were used. For each leaf, one small drop (3 µL) was placed on the syringe-wounded adaxial surface. During infection, high humidity was maintained by flooding the planting tray with water and covering the plantlets with a moist plastic lid. Symptoms were observed and leaf samples were collected 3 days after inoculation. Infected leaves (diameter 4 mm) were excised at the site of initial infection and macerated in 1 mL liquid selective enhancement medium containing 3.75 g/L MgSO4·7H2O, 1 g/L (NH4)2SO4, 1 g/L K2HPO4, 0.2 mL/L 5N NaOH, and 1.7 g/L pectin at pH 7.2 (Wako Pure Chemicals, Osaka, Japan) for 24 h at 28°C (Hélias et al., 2012). Because of the ability of this pathogen to produce indigoidine (Lee and Yu, 2006; Alič et al., 2019; Wei et al., 2021), we used a selective nutrient agar medium containing crystal violet, 70 mg/mL glutamine, and 4 mg/mL MnCl2·2H2O at pH 6.5–6.8. Plates were incubated at 28°C for 1 day and bluish colonies were selected for counting.
Leaf symptoms, indicators of photosynthetic damage, and peroxide content
To visualize the necrotic area, leaves were immediately placed into 0.05% trypan blue, stained at 37°C overnight, and destained in absolute ethanol for another night. To visualize peroxide distribution around the infected area, infected leaf samples were directly incubated in 1 mg/mL diaminobenzidine at 37°C overnight in the dark. The leaves were fully decolorized in several changes of 70% ethanol at 70°C for 1 h and stored in 30% glycerol until observation.
Photosynthetic quantum yield was measured with a miniPPM-300 photosynthesis meter (EARS, Wageningen, Netherlands) after dark preconditioning for at least 30 min. Subsequently, individual leaves were weighed, and chlorophyll was extracted with 1 mL of dimethyl sulfoxide per leaf overnight. Quantification of total chlorophyll content was based on extract absorbance at 649 and 665 nm and an equation suggested by (Wellburn, 1994). The values expressed in µg/mL solvent were converted into µg/g leaf fresh weight.
The total peroxide content of leaves was measured by luminol chemiluminescence catalyzed by Co2+ (Pérez and Rubio, 2006). Samples were frozen in liquid nitrogen, ground in 0.5 mL 5% trichloroacetic acid, and centrifuged at 13,000 rpm and 4°C for 10 min. The supernatant was supplemented with 5% polyvinylpyrrolidone and diluted 100-fold with water. An aliquot of the diluted extract (20 µL) was added to 1 mL of diluted luminol–cobalt mixture (1 g/L luminol and 0.7 g/L CoCl2·6H2O in carbonate buffer pH 10.2, incubated for at least 1 h, and diluted 10-fold before use). Chemiluminescence was measured 5 s after the addition of luminol in a Gene Light GL-220 luminometer (Microtec Nition, Chiba, Japan). The values were expressed as log10 (relative light unit)/g fresh weight.
RNA extraction and RNA-sequencing
Aside from control, only T. calospora-colonized seedlings, regardless of their infection status, were used for RNA extraction and sequencing. Each replicate contained materials collected from at least five individuals. Total leaf RNA was extracted using a Total RNA Extraction Kit (Plant) (RBC Bioscience, Taipei, Taiwan) according to the manufacturer’s protocol. Residual genomic DNA was removed with RNase-free DNase I (Toyobo, Osaka, Japan). The quality and quantity of the total RNA were checked using a Qubit RNA HS Assay Kit and Qubit 2.0 Fluorometer (Thermo Fisher Scientific, Waltham, MA, USA). RNA-seq library construction from the total RNA using an MGIEasy RNA Directional Library Prep Set (MGI, Shenzhen, China) and sequencing with strand-specific and paired-end reads (150 bp) using the DNBSEQ-T7RS sequencing platform were performed by Genome-Lead Co. (Takamatsu, Kagawa, Japan).
Transcriptome analysis
The obtained raw reads were filtered using Fastp v.0.23.2 (Chen et al., 2018) to remove low-quality reads (<Q30) and adapter sequences. Reads were mapped to de novo assembled transcript sequences (Miura et al., 2018) using Bowtie2 v.2.5.2 (Langmead and Salzberg, 2012) (Supplementary Table 1). Differentially expressed genes (DEGs) were analyzed using the edgeR package. Library size was corrected using the trimmed mean of M value method and fitted using the glmTreat (filtered at logFC 1.5) (Smyth et al., 2018). Gene Ontology (GO) enrichment was analyzed in the topGO package (Alexa and Rahnenfuhrer, 2023) using the elim parameter and Fisher exact test, and the p-values were adjusted with the Benjamini–Hochberg method. The false discovery rate (FDR) threshold was set at 0.05. Genes of B. striata orthologous to rice genes were identified using SonicParanoid (Cosentino and Iwasaki, 2019) with default parameters. The rice genome assembly (Os-Nipponbare-Reference-IRGSP-1.0; Kawahara et al., 2013) were retrieved from Ensembl Plants (https://plants.ensembl.org/index.html). Additional genes involved in jasmonate, ethylene biosynthesis, and phosphate metabolism were retrieved from rice transcriptomic data (Campo and San Segundo, 2020) and used as queries for local TBLASTX against B. striata genome assembly. Only B. striata genes with an E-value less than 10−5 were selected. The whole TBLASTX process was conducted in Genetyx v.15 software (Genetyx, Tokyo, Japan). For visualization, a heatmap was generated in the RStudio v.4.1.2 package Complexheatmap (Gu et al., 2016), and an UpSet plot was generated manually using CorelDRAW 2021 v.23.1.0.389 (Alludo, Ottawa, ON, Canada).
Statistical analysis
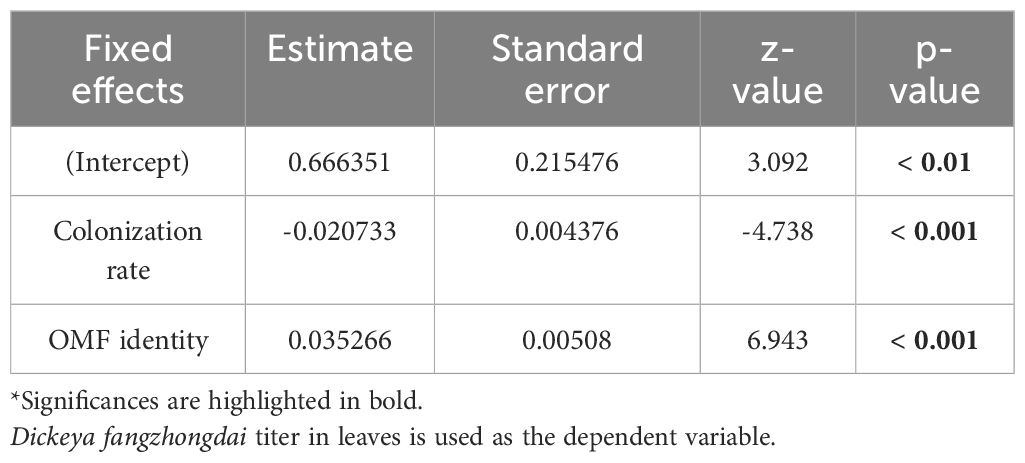
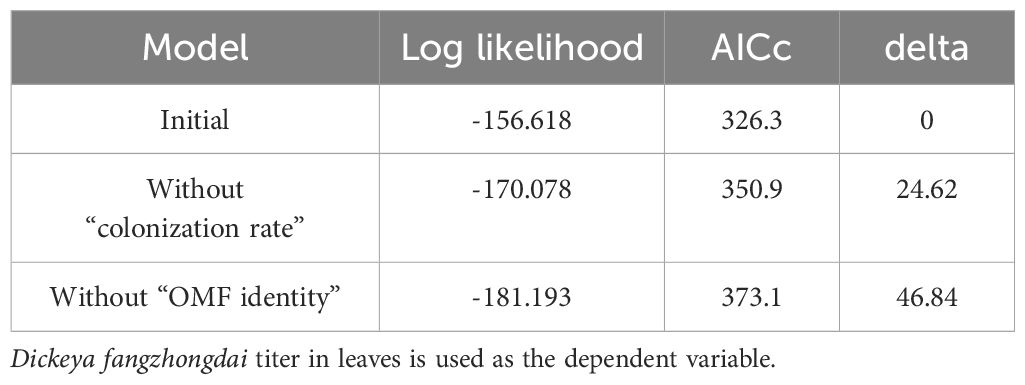
For protocorm symbiotic cell count, root colonization rate, D. fangzhongdai titer, photosynthetic quantum yield, chlorophyll content, and peroxide content data, whenever the data did not meet the normality assumption, logarithmic data transformation was used to enable analyses with parametric tests according to the corresponding data skewness. The statistical analyses were conducted in RStudio v.4.1.2 with Student’s t-test, ANOVA, or a Kruskal–Wallis test. To investigate whether the relationship between D. fangzhongdai titer and root colonization rate among individual seedlings with leaf soft rot is linked to OMF identity, we used a generalized linear mixed model (GLMM) for negative binomial implemented in the lmerTest package (Kuznetsova et al., 2017). Considering the possibility of different responses among individuals and infection occurrence, experimental batches and individual seedlings were used as a random factor and OMF identity and colonization rate were used as fixed factors. The final syntax was constructed without the interaction between OMF colonization rate and identity to produce, the most maximal yet parsimonious model: glmer.nb (colony-forming unit [CFU] count ~ colonization rate + OMF identity + [1| experimental batch/treatment]). To determine the importance of OMF colonization rate and OMF identity in determining CFU count, the model.sel function of the MuMIn package (Bartoń, 2023) was applied to the constructed GLMM. The omitted factor with the largest change in the corrected Akaike information criterion value was regarded as the most important fixed factor.
Results
Tulasnella calospora and S. vermifera colonize B. striata roots with similar symbiotic affinity in protocorms
We established an ex vitro growing system for B. striata seedlings in a common potting mix and cultured OMF in liquid medium to obtain maximum mycelial growth and facilitate direct root inoculation. We tested the compatibility of B. striata with OMF candidates by direct root inoculation described in the current study, and by a symbiotic germination assay established previously (Fuji et al., 2020). Within 2 weeks, we confirmed the presence of pelotons in protocorms and roots, which is the primary sign of OM establishment (Figure 1A, Supplementary Figure 2). In protocorms, OMF colonization started from the basal part and was restricted to the lower half until the formation of shoot primordium (Supplementary Figure 2), as reported earlier (Miura et al., 2019). In roots, OMF colonization occurred everywhere, except the quiescent center. The colonization rate was higher for T. calospora than for S. vermifera in protocorms (Figure 1B) and in roots (Figure 1C).
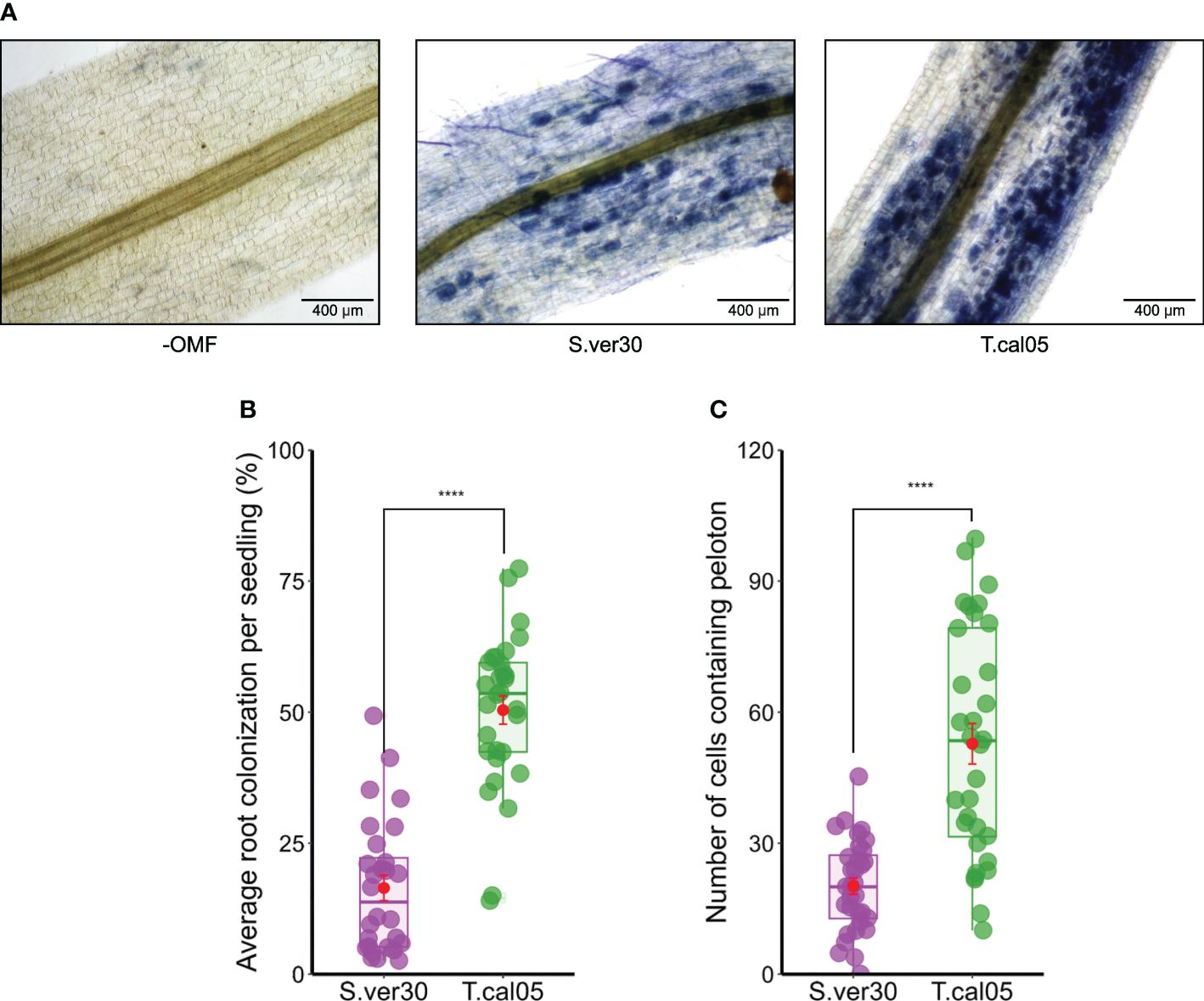
Figure 1 Colonization of Bletilla striata by orchid mycorrhizal fungi (OMF). Seedlings were inoculated with the homogenized mycelial suspension of OMF into soil and incubated for 2 weeks. (A) Root colonization of OMF was visualized by ink staining. Pelotons are visible inside the root cortex as blue dots. Bar = 400 µm. (B) Average root colonization rate per individual seedling. Symbiotic seed germination was conducted by co-culture with OMF for 2 weeks. (C) Protocorms were subjected to ink staining, and the number of symbiotic cells per protocorm was counted. Red dots indicate mean ± standard error. ****p< 0.0001, Student’s t-test. S.ver30, Serendipita vermifera; T.cal05, Tulasnella calospora.
Erwinia chrysanthemi MAFF311045 is synonymous to Dickeya fangzhongdai
Our pathosystem for B. striata was based on a study on orchid infection by Erwinia species (Wu et al., 2011; Ye et al., 2019). We chose Erwinia chrysanthemi because of its ability to infect orchids. The Erwinia genus is rather complex (Samson et al., 2005). Our phylogenetic tree based on 16S ribosomal RNA sequences of various species within Pectobacteriaceae assigned the E. chrysanthemi MAFF311045 to Dickeya fangzhongdai (Supplementary Figure 3). Leaf inoculation with this pathogen resulted in a lesion and bacterial spread throughout the leaf, often causing the leaf to drop, in B. striata, A. thaliana, and N. benthamiana (Supplementary Figures 4A–C).
Induced systemic resistance is linked to the degree of OMF colonization
The effects of OMF colonization on ISR against pathogens were examined in the B. striata–D. fangzhongdai pathosystem. Both S. vermifera and T. calospora were able to colonize B. striata roots (Figure 1). Upon D. fangzhongdai infection, lesions on detached leaves were wider in the presence of OMF colonization than in its absence (Figure 2A). T. calospora colonization induced the defense response, as indicated by a reduced D. fangzhongdai titer (Figure 2B). On the other hand, S. vermifera colonization did not affect resistance since D. fangzhongdai titer was not reduced as in uncolonized seedlings.
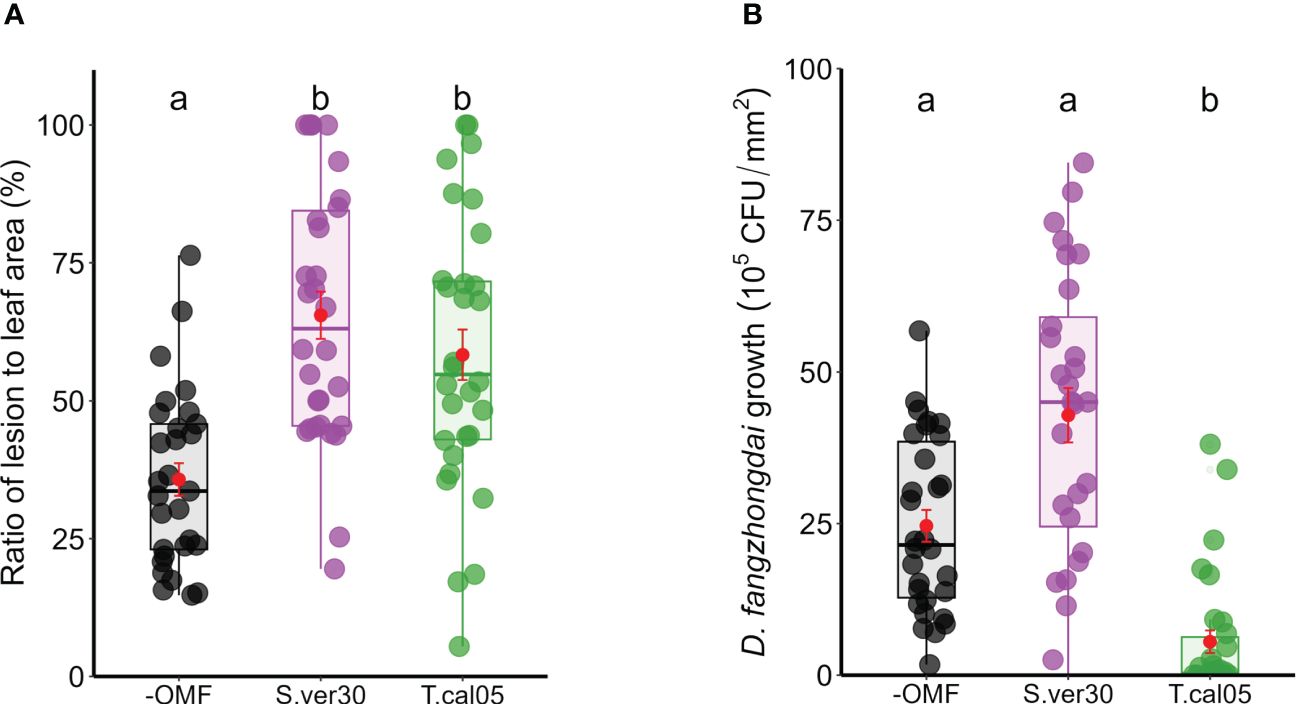
Figure 2 Induced systemic resistance on Bletilla striata leaves upon Dickeya fangzhongdai infection. Leaves of OMF-colonized B striata seedlings were inoculated with bacterial suspension of D fangzhongdai, and (A) ratio of lesion area to total leaf area and (B) colony-forming unit (CFU) count were measured 3 days after inoculation. Red dots indicate mean ± standard error. The different letters show that the values are significantly different at p< 0.05 based on Kruskal–Wallis test. −OMF, non-colonized control; S.ver30, Serendipita vermifera; T.cal05, Tulasnella calospora.
To further test the physiological changes associated with the induction of disease resistance, we assessed the indicators of photosynthetic damage and peroxide production. Photosynthetic damage is a general proxy for disease resistance in leaves, and reactive oxygen species are general inducers of disease resistance (Lu and Yao, 2018; Li and Kim, 2022). Regardless of lesion size, the necrotic area was relatively constant, as revealed by trypan blue staining (Supplementary Figures 4D, E). The photosynthetic quantum yield was reduced in all infected leaves, but the photosynthetic damage caused by the infection was significantly smaller when T. calospora colonized the roots (Figure 3A). Similarly, the decrease in total chlorophyll content of seedlings caused by D. fangzhongdai infection was lowest in the presence of T. calospora (Figure 3B). Hydrogen peroxide was also accumulated in infected leaves (Supplementary Figure 5A), and its content was significantly higher in OMF-colonized seedlings (Supplementary Figure 5B). Overall, the ability of OMF to systemically induce disease resistance in orchids is not necessarily related to symptom appearance, e.g., lesion size, but rather to reduced pathogen proliferation. This is also supported by the data that both OMF identity and colonization rate strongly affected ISR (n = 88 plants; p< 0.001; Tables 1, 2, Supplementary Figure 6).
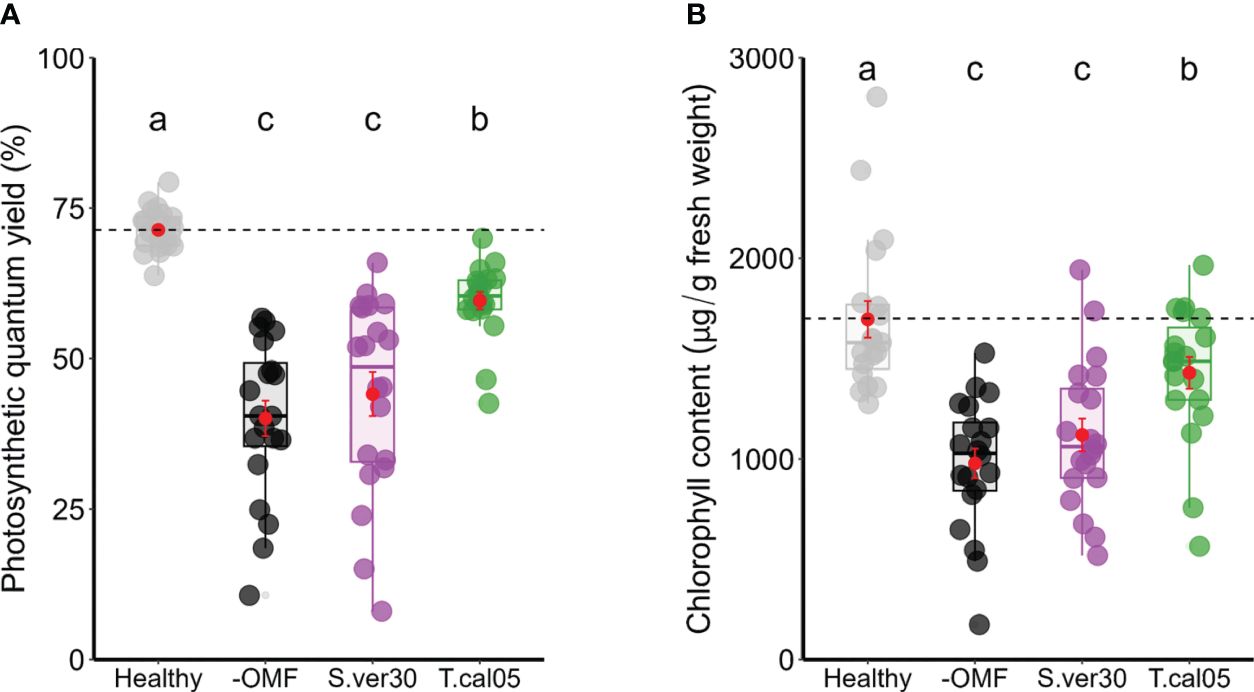
Figure 3 Photosynthetic damage caused by Dickeya fangzhongdai infection. (A) Photosynthetic quantum yield and (B) total chlorophyll contents were measured in D fangzhongdai-infected leaves of OMF-colonized Bletilla striata seedlings 3 days after infection and those of healthy uninoculated seedlings. Red dots indicate mean ± standard error. The different letters show that the values are significantly different at p< 0.05 based on Kruskal–Wallis test. −OMF, non-colonized control; S.ver30, Serendipita vermifera; T.cal05, Tulasnella calospora.
Transcriptome analysis of leaves of B. striata seedlings inoculated with OMF and pathogen
The lowest D. fangzhongdai titer and photosynthetic damage in T. calospora–infected leaves led us to hypothesize that T. calospora colonization enhances disease resistance by systemically inducing changes in gene transcription. We analyzed transcriptomes of leaves upon root inoculation with T. calospora (T treatment), upon leaf infection by D. fangzhongdai (P treatment), or both (PT treatment).
Up- and downregulated DEGs were identified by comparisons among the three treatments (Supplementary Tables 2–4). A heatmap of DEG clustering showed distinct expression patterns among the treatments (Figure 4A). A total of 2660 genes in P, 1391 in T, and 1628 in PT seedlings were upregulated, and 2987 in P, 2319 in T, and 857 in PT seedlings were downregulated. Among them, 6 DEGs (4 upregulated and 2 downregulated) were shared among all treatments, 1641 DEGs (397 upregulated and 1244 downregulated) between P and T, 63 DEGs (45 upregulated and 18 downregulated) between T and PT, and 5 upregulated DEGs between P and PT (Figure 4A). The transcriptional profile was less similar between T and PT seedlings than between P and PT seedlings, as indicated by MDS plot (Supplementary Figure 7). These data demonstrated that OM symbiosis greatly affects the leaf transcriptome. Since both T and PT treatments involved T. calospora colonization, it systemically and conspicuously altered leaf transcriptional changes before and after D. fangzhongdai infection.
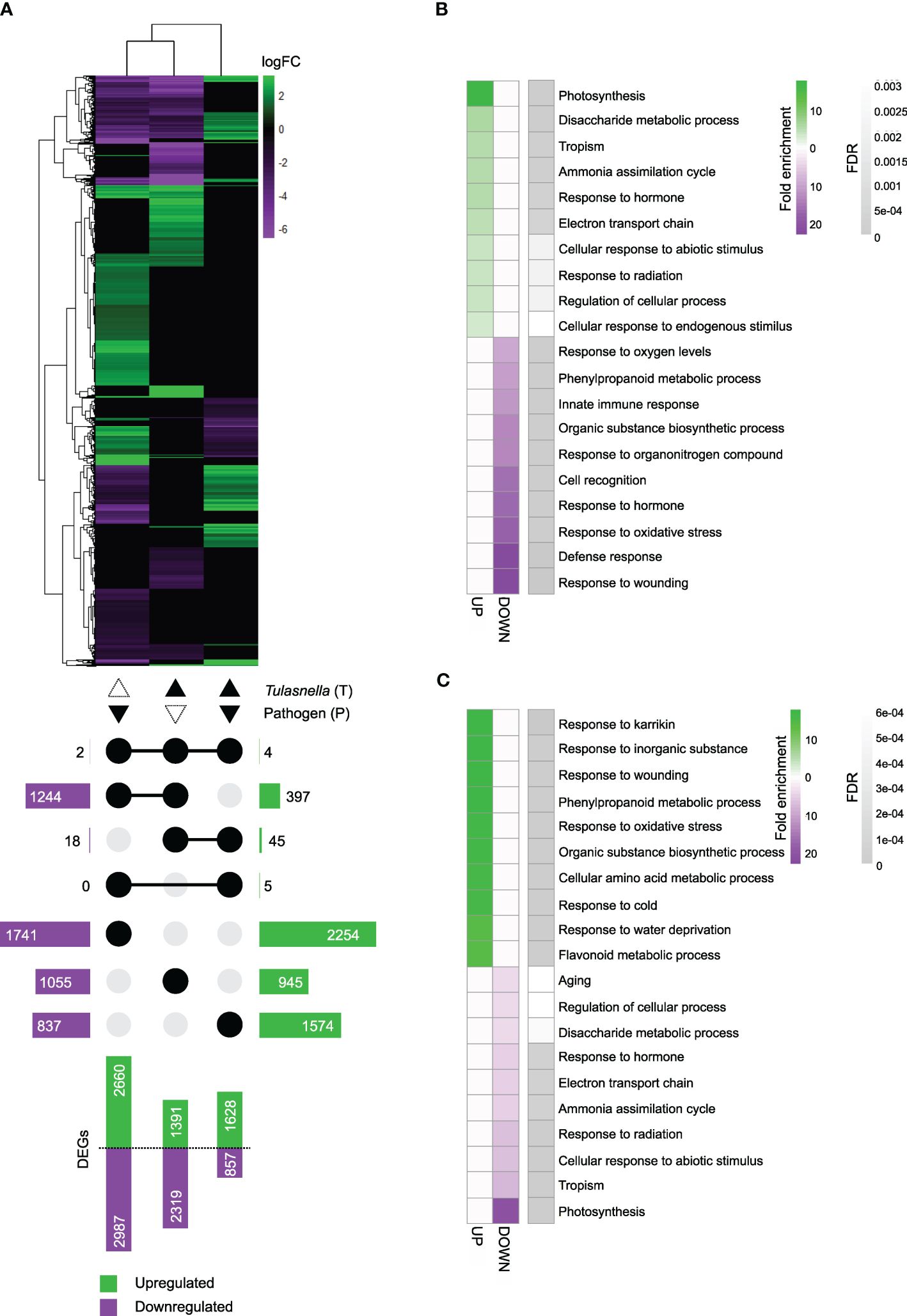
Figure 4 RNA-seq analysis. Root colonization of Tulasnella calospora in Bletilla striata seedlings was conducted by inoculating the homogenized mycelial suspension into soil, and then seedlings were cultured for 2 weeks. Leaves of seedlings were infected with Dickeya fangzhongdai by drop inoculation and harvested 3 days after inoculation. Three biological replicates (each replicate contains leaves collected from different plants) were used for RNA extraction and sequencing. (A) Heatmap showing fold changes of differentially expressed genes (DEGs) in leaves of seedlings treated with the pathogen D. fangzhongdai only (P); T. calospora only (T); or both (PT), each compared with non-colonized and uninoculated healthy control (without T. calospora and D. fangzhongdai). An UpSet plot below the heatmap shows the number of upregulated and downregulated DEGs attached to bar plots. Connected dots show the treatment sets that share the respective DEGs. The log fold change (logFC) was calculated as log2[treatment/uncolonized] > |1.5| with FDR< 0.05. (B, C) Heatmaps of the top 10 enriched biological process gene ontology (GO) terms for upregulated and downregulated DEGs in (B) T treatment and (C) PT treatment. FDR, false discovery rate. All GO terms are listed in Supplementary Tables 5, 6.
GO enrichment analysis of biological processes revealed that terms related to photosynthesis were overrepresented among the upregulated DEGs in T-treated seedling leaves. The categories of disaccharide metabolic process, tropism, ammonia assimilation cycle, and innate immune response were also enriched. Among downregulated DEGs, response to wounding, defense response, and response to oxidative stress were among the overrepresented terms (Figure 4B, Supplementary Table 5). In PT-treated seedlings, terms related to responses to karrikin, inorganic substance, and wounding were overrepresented among the upregulated DEGs, whereas photosynthesis, tropism, and ammonia assimilation cycle were among the overrepresented terms among downregulated DEGs (Figure 4C, Supplementary Table 6). These results implied that upon T. calospora colonization probably by increasing primary metabolism, e.g., photosynthesis and sugar metabolism, and then resistance was increased upon pathogen infection. Thus, it is worth noting that there is a potential of growth-defense tradeoff.
Comparisons of expression levels of phosphate and jasmonate/ethylene metabolism-related genes in rice and B. striata
We hypothesized that transcriptional patterns are conserved across different mycorrhizal types, especially in comparison with those in AM plants. To test this hypothesis, we compared our data with the published data on leaf transcriptional changes in rice (Oryza sativa ssp. japonica cv. ‘Loto’) inoculated with AM fungus Funneliformis mosseae (Campo and San Segundo, 2020). The most noticeable transcriptional changes in rice leaves were in phosphate- and jasmonate/ethylene metabolism-related genes. Jasmonate and ethylene are involved in resistance against necrotrophic pathogens and in ISR (Shoresh et al., 2005; Zhu, 2014). To identify the orthologous genes by SonicParanoid, we used rice genes as queries against B. striata de novo assembly (Supplementary Table 7). We also used local TBLASTX (Supplementary Table 8). The expression of the identified B. striata orthologs of phosphate metabolism–related genes did not necessarily mirror changes in the expression of rice genes (Figure 5A, Supplementary Table 8), but the identified genes involved in the first steps of jasmonate and ethylene biosynthesis were similarly downregulated in rice and B. striata, such as phospholipase A1 (PLA1), lipoxygenase (LOX), allene oxide synthase (AOS), oxophytodienoic acid reductase (OPR), and S-adenosylmethionine synthase (SAMS) (Figure 5B, Supplementary Table 8). The expression of these genes was suppressed in T and P seedlings, but increased in PT seedlings.
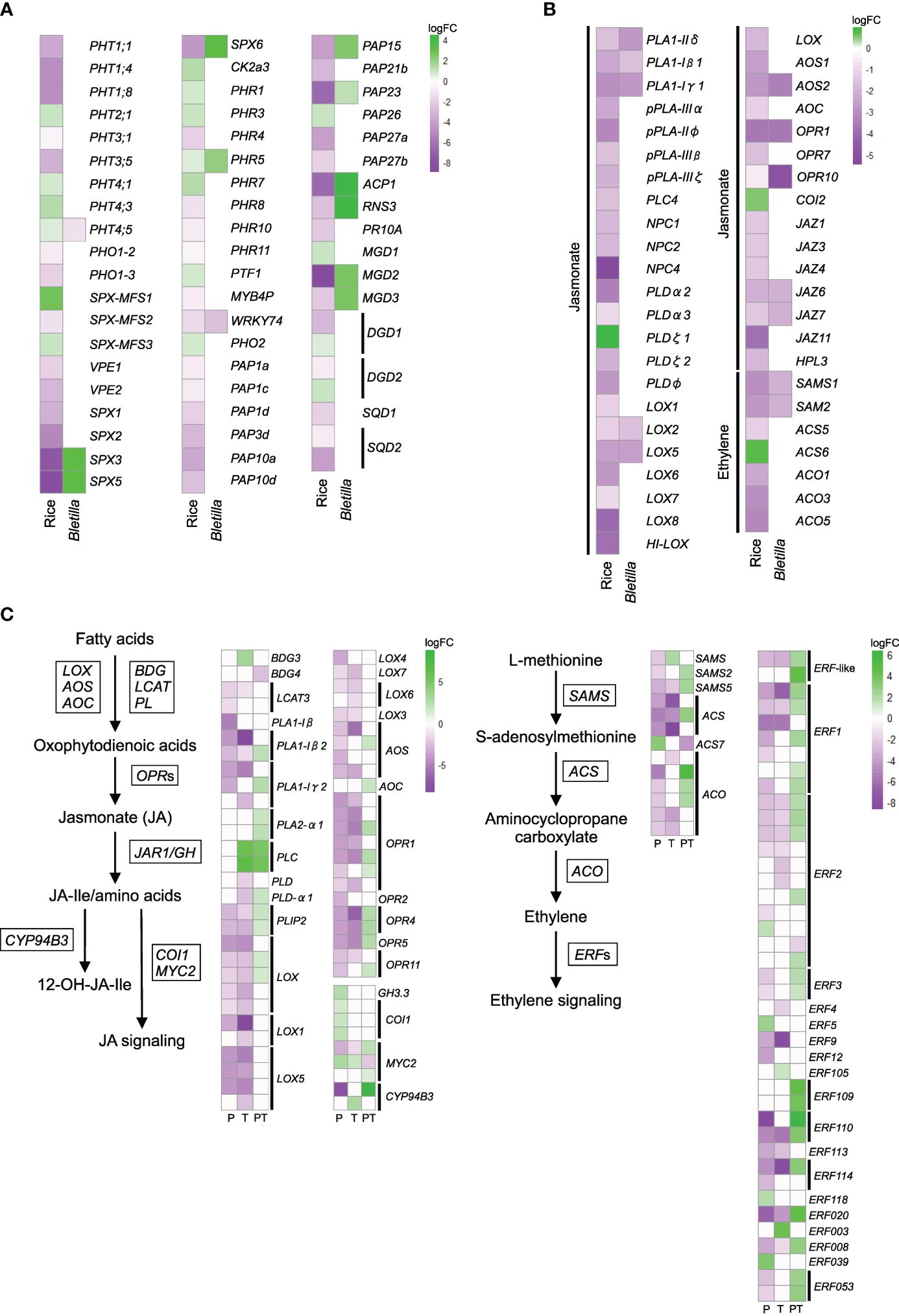
Figure 5 Comparative analysis of the expression patterns of phosphate-acquisition and defense hormone–related genes in leaves of arbuscular mycorrhizal rice (Oryza sativa spp. japonica, cv. Loto; Campo and San Segundo, 2020) and Bletilla striata seedlings treated with the pathogen Dickeya fangzhongdai only (P); Tulasnella calospora only (T); or both (PT). (A) Heatmap showing fold changes of DEGs involved in phosphate metabolism and (B) jasmonate and ethylene signaling/metabolism in T. The orthologous genes of B striata were identified by SonicParanoid and TBLASTX. (C) The expression of jasmonate- and ethylene-related genes of B striata was determined from transcriptomic analysis. The log fold change (logFC) was calculated as log2[treatment/uncolonized] > |1.5| with FDR< 0.05. See Supplementary Tables 8, 9 for details.
We further checked the expression patterns of homologous jasmonate and ethylene biosynthesis–related genes extracted from DEG lists (FDR< 0.05) and obtained similar results. The overall results showed that priming suppresses the jasmonate and ethylene signaling pathways and that leaf infection by pathogen activates them, confirming the occurrence of ISR in B. striata (Figure 5C, Supplementary Table 9).
Discussion
In this study, we demonstrated that a compatible OMF colonization, T. calospora, systemically induces resistance against a necrotrophic pathogen as well as its possible role in defense priming in orchids (Figure 6). We also showed partial commonalities between AM and OM plants in aboveground transcriptomic changes during root colonization.
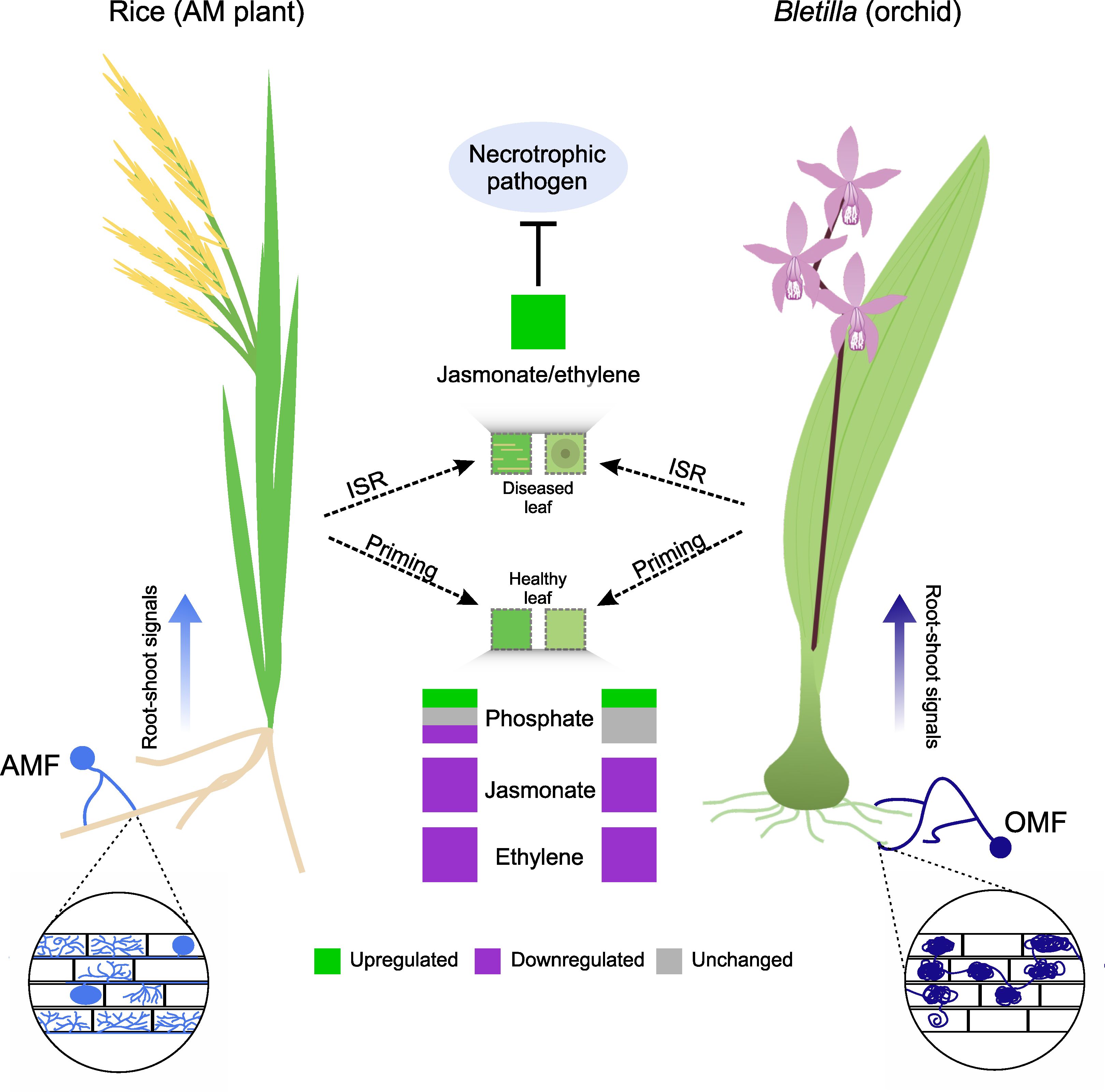
Figure 6 Commonalities in induced systemic resistance (ISR) primed by mycorrhizal fungus colonization in an AM plant (rice) and orchid (Bletilla striata). Despite structural differences, colonization by AMF and OMF causes priming by regulating the expression of genes related to primary metabolism (e.g., phosphate metabolism). This is more variable in AM plants than in orchids. However, the expression of genes related to the jasmonate and ethylene signaling pathways is downregulated in both species. When a necrotrophic pathogen infects the plants, AMF or OMF colonization triggers ISR in leaves, which increases the expression of genes involved in jasmonate and ethylene signaling pathways and in turn inhibits pathogen proliferation.
Mycorrhizal fungi improve host fitness, which varies among AM fungus–plant pairings (Säle et al., 2021; Cope et al., 2022; Guigard et al., 2023). The driving factors for the functional diversity are largely unknown, but plants may simply choose mycorrhizal fungi that provide the highest growth with minimal metabolic costs, which is important for survival. The same applies to orchids, which underwent rapid diversification in their growth habits during the Cenozoic (Dearnaley et al., 2012; Chomicki et al., 2015; Xing et al., 2019). Orchids with the highest demand for mycorrhizal association during early seed germination prioritize different OMF species as their partners (Otero et al., 2005; Meng et al., 2019; Pujasatria et al., 2022). Previously, we showed that B. striata associates with multiple strains of Tulasnella during seed germination (Yamamoto et al., 2017; Fuji et al., 2020). It could be assumed that B. striata remains compatible with an OMF from seed germination until seedling development and quantified OMF colonization in seedlings with well-developed roots. Colonization during seed germination and root colonization was higher for T. calospora than for S. vermifera. Other Bletilla sister taxa, the members of tribe Arethuseae (e.g., Arundina, Pleione, and Coelogyne), are also associated with Tulasnella, as suggested by metagenomic studies and in vitro assays (Yukawa et al., 2009; Sathiyadash et al., 2014; Meng et al., 2019; Qin et al., 2019). These suggest that the convergent evolution of OM still involves mycorrhizal partner selection, in addition to molecular-level mechanisms shared with AM plants, such as the roles of phytohormones and transcriptional regulations (Miura et al., 2018; Miura et al., 2024).
Dickeya fangzhongdai, as well as Erwinia sensu lato, infects orchids (Fu et al., 2012; Alič et al., 2019; Ye et al., 2019; Zhou et al., 2021), Arabidopsis (Kraepiel et al., 2011), and tobacco (Sobiczewski et al., 2017), resulting in brownish soft necrotic lesions. The lesions, however, should not be used as the sole proxy of resistance; pathogen titer should be added as another criterion, as shown in this study. In Phalaenopsis, lesions are smaller upon OMF colonization (Wu et al., 2011). In this study, the lesions were visually larger upon priming, but D. fangzhongdai titer was significantly lower. Whether these phenomena were due to the physiological difference between these orchid species is unknown. However, it is worth mentioning that leaf soft rot is usually unstoppable, causing the preventive way is by cutting the whole infected leaf because the symptom spreads quickly within days. This phenomenon is commonly observed in cultivation of various orchid species (Chi et al., 2022). Regardless, in this study, disease resistance against necrotrophic pathogens were systemically induced in orchids colonized by mycorrhizal fungi.
We investigated the potential similarity in ISR between orchids and AM plants through comparative transcriptomics. Upon root colonization by mycorrhizal fungi, the host plants alter various metabolic pathways (Fontana et al., 2009; Jacott et al., 2017; Goddard et al., 2021; Orine et al., 2022; Li et al., 2023); host plants increase their primary metabolic activity (nutrient uptake and remobilization) and temporarily lower the expression of defense-related genes (Schoenherr et al., 2019; Jing et al., 2022). In AM plants such as rice, genes associated with phosphate metabolism and phospholipid biosynthesis are upregulated, whereas non-phosphate lipids, which are later used in jasmonate biosynthesis, are downregulated (Campo and San Segundo, 2020). In Bletilla, the annotations related to phosphate metabolism are insufficient to conclude whether phosphate metabolism is also aligned with that of AM plants. Although it is unknown whether Bletilla does not prioritize phosphate metabolism or its phosphate-related genes are not differentially expressed, most studies on nutrient acquisition through OM show that orchids primarily focus on carbon and nitrogen from OMF (Stöckel et al., 2014; Fochi et al., 2016; Dearnaley and Cameron, 2017; Zahn et al., 2023). Inorganic phosphate is also taken up through OMF hyphae (Cameron et al., 2007; Davis et al., 2022) and its content is determined by trophic mode (Minasiewicz et al., 2023): mycoheterotrophic species have the highest phosphorus content, followed by partial mycoheterotrophic and autotrophic ones. Assuming that B. striata seedlings used in this study were already fully autotrophic, it is possible that most phosphate metabolism–related genes were not dominantly expressed even after plant colonization by OMF, unlike in AM plants.
On the other hand, after infection, the defense response was stronger after priming than before it. This is often indicated by an increase in defense-related gene expression and content of phytohormones, notably jasmonate and ethylene, which are involved in ISR and play a major role in defense against necrotrophic pathogens (Shoresh et al., 2005; Wasternack and Feussner, 2018). Mycorrhiza-colonized plants typically prioritize growth and thus allocate less resources to defense responses, including jasmonate biosynthesis (Huot et al., 2014). Once the leaf is attacked by a pathogen, jasmonate is readily increased for defense (Galis et al., 2009). Again, our comparative transcriptome analysis was still too inconclusive due to the low number of genes matched with those of rice. However, it was hinted that the expression pattern could be similar since the genes involved in the first steps of jasmonate/ethylene biosynthesis were also downregulated during priming and then upregulated after D. fangzhongdai infection onset. In turn, D. fangzhongdai titer was reduced, similar to that in other pathogen infections of AM plants. Our transcriptional evidence further supports the presence of defense priming in orchids.
In summary, our study demonstrated that the association between orchid and mycorrhizal fungi leads to ISR, thus providing a new clue for commonalities between AM and OM. As in AM plants, OMF compatibility also reflects the best colonization. The presence of a particular OMF in a substrate is a key factor in orchid seedling establishment (Gowland et al., 2013; Pujasatria et al., 2022) to provide the protocorms with nutrients and to protect against stress. We also propose that the ISR in B. striata involves a complex mechanism in which regulation of metabolites, defense responses, photosynthesis, and oxidative stress responses is synchronized to reduce D. fangzhongdai proliferation during leaf soft rot onset. Most AM studies have focused on leaves (Fujita et al., 2022; Wang et al., 2022b), but priming could be more variable in orchids than in other plants because orchid diseases also occur on pseudobulbs, rhizomes, and even flowers (Ito and Aragaki, 1977; Swett and Uchida, 2015; Li et al., 2022). Thus, the priming effects in orchids are still largely unknown if one takes into account pathogen trophic modes, latency, and the attacked organs. Further studies are needed to clarify how the defense responses were activated at the molecular level, allowing to reveal more commonalities among AM plants and orchids as well as various types of pathogens.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
GP: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Visualization, Writing – original draft, Writing – review & editing. CM: Conceptualization, Data curation, Methodology, Supervision, Writing – original draft, Writing – review & editing. KY: Data curation, Resources, Writing – review & editing. SS: Data curation, Resources, Writing – review & editing. HK: Conceptualization, Project administration, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Acknowledgments
This work was supported by the National Institute for Basic Biology (NIBB) Cooperative Research Programs (Next-generation DNA Sequencing Initiative: 22NIBB403, 23NIBB401). We are grateful to the Japanese Ministry of Education, Culture, Sports, Science, and Technology (MEXT) for a scholarship to GP, and to the Japan Society for the Promotion of Science (JSPS) for Research Fellowships for Young Scientists to CM. We also thank the Data Integration and Analysis Facility, NIBB for supporting the RNA sequencing and providing computational resources.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1447050/full#supplementary-material
References
Alexa, A., Rahnenfuhrer, J. (2023). topGO: Enrichment Analysis for Gene Ontology. Available online at: https://bioconductor.org/packages/topGO.
Alič, Š., Pédron, J., Dreo, T., Van Gijsegem, F. (2019). Genomic characterisation of the new Dickeya fangzhongdai species regrouping plant pathogens and environmental isolates. BMC Genom. 20, 1–18. doi: 10.1186/s12864-018-5332-3
Bidartondo, M. I., Burghardt, B., Gebauer, G., Bruns, T. D., Read, D. J. (2004). Changing partners in the dark: isotopic and molecular evidence of ectomycorrhizal liaisons between forest orchids and trees. Proc. R. Soc Lond. B. Biol. Sci. 271, 1799–1806. doi: 10.1098/rspb.2004.2807
Brundrett, M. C., Tedersoo, L. (2018). Evolutionary history of mycorrhizal symbioses and global host plant diversity. New Phytol. 220, 1108–1115. doi: 10.1111/nph.14976
Cameron, D. D., Johnson, I., Leake, J. R., Read, D. J. (2007). Mycorrhizal acquisition of inorganic phosphorus by the green-leaved terrestrial orchid Goodyera repens. Ann. Bot. 99, 831–834. doi: 10.1093/aob/mcm018
Cameron, D. D., Johnson, I., Read, D. J., Leake, J. R. (2008). Giving and receiving: Measuring the carbon cost of mycorrhizas in the green orchid, Goodyera repens. New Phytol. 180, 176–184. doi: 10.1111/j.1469-8137.2008.02533.x
Campo, S., San Segundo, B. (2020). Systemic induction of phosphatidylinositol-based signaling in leaves of arbuscular mycorrhizal rice plants. Sci. Rep. 10, 1–17. doi: 10.1038/s41598-020-72985-6
Cating, R. A., Palmateer, A. J. (2011). Bacterial Soft Rot of Oncidium Orchids Caused by a Dickeya sp. (Pectobacterium chrysanthemi) in Florida. Plant Dis. 95, 74. doi: 10.1094/PDIS-07-10-0523
Chen, S., Zhou, Y., Chen, Y., Gu, J., Gu, Z., Eils, R., et al. (2018). Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, 2847–2849. doi: 10.1093/bioinformatics/bty560
Chi, N. M., Anh, D. T. K., Hung, T. X., Nhung, N. P., Bao, H. Q., Toan, D. V., et al. (2022). Soft rot caused by Dickeya fangzhongdai in epiphytic orchids in Vietnam. Can. J. Plant Path. 44, 386–399. doi: 10.1080/07060661.2021.1998226
Chomicki, G., Bidel, L. P. R., Ming, F., Coiro, M., Zhang, X., Wang, Y., et al. (2015). The velamen protects photosynthetic orchid roots against UV-B damage, and a large dated phylogeny implies multiple gains and losses of this function during the Cenozoic. New Phytol. 205, 1330–1341. doi: 10.1111/nph.13106
Cope, K. R., Kafle, A., Yakha, J. K., Pfeffer, P. E., Strahan, G. D., Garcia, K., et al. (2022). Physiological and transcriptomic response of Medicago truncatula to colonization by high- or low-benefit arbuscular mycorrhizal fungi. Mycorrhiza 32, 281–303. doi: 10.1007/s00572-022-01077-2
Cosentino, S., Iwasaki, W. (2019). SonicParanoid: Fast, accurate and easy orthology inference. Bioinformatics 35, 149–151. doi: 10.1093/bioinformatics/bty631
David, L., Harmon, A. C., Chen, S. (2019). Plant immune responses - from guard cells and local responses to systemic defense against bacterial pathogens. Plant Signal Behav. 14, 1–9. doi: 10.1080/15592324.2019.1588667
Davis, B., Lim, W. H., Lambers, H., Dixon, K. W., Read, D. J. (2022). Inorganic phosphorus nutrition in green-leaved terrestrial orchid seedlings. Ann. Bot. 129, 669–678. doi: 10.1093/aob/mcac030
Dearnaley, J. D. W., Cameron, D. D. (2017). Nitrogen transport in the orchid mycorrhizal symbiosis – further evidence for a mutualistic association. New Phytol. 213, 10–12. doi: 10.1111/nph.14357
Dearnaley, J. D. W., Martos, F., Selosse, M. A. (2012). “Orchid mycorrhizas: Molecular ecology, physiology, evolution and conservation aspects,” in Fungal Associations. Ed. Hock, B. (Springer Berlin, Heidelberg), 207–230. doi: 10.1007/978-3-642-30826-0_12
Dreischhoff, S., Das, I. S., Jakobi, M., Kasper, K., Polle, A. (2020). Local responses and systemic induced resistance mediated by ectomycorrhizal fungi. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.590063
Eck, J. L., Kytöviita, M. M., Laine, A. L. (2022). Arbuscular mycorrhizal fungi influence host infection during epidemics in a wild plant pathosystem. New Phytol. 236, 1922–1935. doi: 10.1111/nph.18481
Fochi, V., Chitarra, W., Kohler, A., Voyron, S., Singan, V. R., Lindquist, E. A., et al. (2016). Fungal and plant gene expression in the Tulasnella calospora - Serapias vomeracea symbiosis provides clues about nitrogen pathways in orchid mycorrhizas. New Phytol. 213, 365–379. doi: 10.1111/nph.14279
Fogell, D. J., Kundu, S., Roberts, D. L. (2019). Genetic homogenisation of two major orchid viruses through global trade-based dispersal of their hosts. Plants People Planet 1, 356–362. doi: 10.1002/ppp3.46
Fontana, A., Reichelt, M., Hempel, S., Gershenzon, J., Unsicker, S. B. (2009). The effects of arbuscular mycorrhizal fungi on direct and indirect defense metabolites of Plantago lanceolata L. J. Chem. Ecol. 35, 833–843. doi: 10.1007/s10886-009-9654-0
Fracchia, S., Aranda-Rickert, A., Rothen, C., Sede, S. (2016). Associated fungi, symbiotic germination and in vitro seedling development of the rare Andean terrestrial orchid. Chloraea riojana. Flora 224, 106–111. doi: 10.1016/j.flora.2016.07.008
Fu, S. F., Tsai, T. M., Chen, Y. R., Liu, C. P., Haiso, L. J., Syue, L. H., et al. (2012). Characterization of the early response of the orchid, Phalaenopsis amabilis, to Erwinia chrysanthemi infection using expression profiling. Physiol. Plant 145, 406–425. doi: 10.1111/j.1399-3054.2012.01582.x
Fuji, M., Miura, C., Yamamoto, T., Komiyama, S., Suetsugu, K., Yagame, T., et al. (2020). Relative effectiveness of Tulasnella fungal strains in orchid mycorrhizal symbioses between germination and subsequent seedling growth. Symbiosis 81, 53–63. doi: 10.1007/s13199-020-00681-0
Fujita, M., Kusajima, M., Fukagawa, M., Okumura, Y., Nakajima, M., Akiyama, K., et al. (2022). Response of tomatoes primed by mycorrhizal colonization to virulent and avirulent bacterial pathogens. Sci. Rep. 12, 1–12. doi: 10.1038/s41598-022-08395-7
Galis, I., Gaquerel, E., Pandey, S. P., Baldwin, I. T. (2009). Molecular mechanisms underlying plant memory in JA-mediated defence responses. Plant Cell Environ. 32, 617–627. doi: 10.1111/j.1365-3040.2008.01862.x
Girlanda, M., Segreto, R., Cafasso, D., Liebel, H. T., Rodda, M., Ercole, E., et al. (2011). Photosynthetic Mediterranean meadow orchids feature partial mycoheterotrophy and specific mycorrhizal associations1. Am. J. Bot. 98, 1148–1163. doi: 10.3732/ajb.1000486
Goddard, M. L., Belval, L., Martin, I. R., Roth, L., Laloue, H., Deglène-Benbrahim, L., et al. (2021). Arbuscular mycorrhizal symbiosis triggers major changes in primary metabolism together with modification of defense responses and signaling in both roots and leaves of Vitis vinifera. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.721614
Gowland, K. M., van der Merwe, M. M., Linde, C. C., Clements, M. A., Nicotra, A. B. (2013). The host bias of three epiphytic Aeridinae orchid species is reflected, but not explained, by mycorrhizal fungal associations. Am. J. Bot. 100, 764–777. doi: 10.3732/ajb.1200411
Gu, Z., Eils, R., Schlesner, M. (2016). Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847–2849. doi: 10.1093/bioinformatics/btw313
Guigard, L., Jobert, L., Busset, N., Moulin, L., Czernic, P. (2023). Symbiotic compatibility between rice cultivars and arbuscular mycorrhizal fungi genotypes affects rice growth and mycorrhiza-induced resistance. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1278990
Haney, C. H., Wiesmann, C. L., Shapiro, L. R., Melnyk, R. A., O’Sullivan, L. R., Khorasani, S., et al. (2018). Rhizosphere-associated Pseudomonas induce systemic resistance to herbivores at the cost of susceptibility to bacterial pathogens. Mol. Ecol. 27, 1833–1847. doi: 10.1111/mec.14400
Hélias, V., Hamon, P., Huchet, E., Wolf, J., Andrivon, D. (2012). Two new effective semiselective crystal violet pectate media for isolation of Pectobacterium and Dickeya. Plant Pathol. 61, 339–345. doi: 10.1111/j.1365-3059.2011.02508.x
Huot, B., Yao, J., Montgomery, B. L., He, S. Y. (2014). Growth-defense tradeoffs in plants: A balancing act to optimize fitness. Mol. Plant 7, 1267–1287. doi: 10.1093/mp/ssu049
Ito, J. S., Aragaki, M. (1977). Botrytis blossom blight of Dendrobium. Phytopathology 67, 820–824. doi: 10.1094/Phyto-67-820
Jacott, C. N., Murray, J. D., Ridout, C. J. (2017). Trade-offs in arbuscular mycorrhizal symbiosis: Disease resistance, growth responses and perspectives for crop breeding. Agronomy 7, 1–18. doi: 10.3390/agronomy7040075
Jing, S., Li, Y., Zhu, L., Su, J., Yang, T., Liu, B., et al. (2022). Transcriptomics and metabolomics reveal effect of arbuscular mycorrhizal fungi on growth and development of apple plants. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1052464
Joko, T., Subandi, A., Kusumandari, N., Wibowo, A., Priyatmojo, A. (2014). Activities of plant cell wall-degrading enzymes by bacterial soft rot of orchid. Arch. Phytopathol. Plant Prot. 47, 1239–1250. doi: 10.1080/03235408.2013.838374
Kakouridis, A., Hagen, J. A., Kan, M. P., Mambelli, S., Feldman, L. J., Herman, D. J., et al. (2022). Routes to roots: direct evidence of water transport by arbuscular mycorrhizal fungi to host plants. New Phytol. 236, 210–221. doi: 10.1111/nph.18281
Kawahara, Y., de la Bastide, M., Hamilton, P., Kanamori, H., McCombie, W. R., Ouyang, S. (2013). Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice. 6, 1–10. doi: 10.1186/1939-8433-6-4
Keith, L. M., Sewake, K. T., Zee, F. T. (2005). Isolation and characterization of Burkholderia gladioli from orchids in Hawaii. Plant Dis. 89, 1273–1278. doi: 10.1094/PD-89-1273
Khamtham, J., Akarapisan, A. (2019). Acidovorax avenae subsp. cattleyae causes bacterial brown spot disease on terrestrial orchid Habenaria lindleyana in Thailand. J. Plant Pathol. 101, 31–37. doi: 10.1007/s42161-018-0135-6
Kraepiel, Y., Pédron, J., Patrit, O., Simond-Côte, E., Hermand, V., van Gijsegem, F. (2011). Analysis of the plant Bos1 mutant highlights necrosis as an efficient defence mechanism during D. dadantii/Arabidopsis thaliana interaction. PloS One 6, 1–10. doi: 10.1371/journal.pone.0018991
Kuga, Y., Sakamoto, N., Yurimoto, H. (2014). Stable isotope cellular imaging reveals that both live and degenerating fungal pelotons transfer carbon and nitrogen to orchid protocorms. New Phytol. 202, 594–605. doi: 10.1111/nph.12700
Kuznetsova, A., Brockhoff, P. B., Christensen, R. H. B. (2017). lmerTest Package: Tests in Linear Mixed Effects Models. J Stat Softw. 82: 1–26. doi: 10.18637/jss.v082.i13
Langmead, B., Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Method. 9, 357–359. doi: 10.1038/nmeth.1923
Lee, Y. A., Yu, C. P. (2006). A differential medium for the isolation and rapid identification of a plant soft rot pathogen, Erwinia chrysanthemi. J. Microbiol. Method. 64, 200–206. doi: 10.1016/j.mimet.2005.04.031
Li, M., Kim, C. (2022). Chloroplast ROS and stress signaling. Plant Commun. 3, 100264. doi: 10.1016/j.xplc.2021.100264
Li, Y., Nan, Z., Matthew, C., Wang, Y., Duan, T. (2023). Arbuscular mycorrhizal fungus changes alfalfa (Medicago sativa) metabolites in response to leaf spot (Phoma medicaginis) infection, with subsequent effects on pea aphid (Acyrthosiphon pisum) behavior. New Phytol. 239, 286–300. doi: 10.1111/nph.18924
Li, J., Zhang, M., Yang, Z., Li, C. (2022). Botrytis cinerea causes flower gray mold in Gastrodia elata in China. Crop Prot. 155, 1–4. doi: 10.1016/j.cropro.2022.105923
Liu, J., Maldonado-Mendoza, I., Lopez-Meyer, M., Cheung, F., Town, C. D., Harrison, M. J. (2007). Arbuscular mycorrhizal symbiosis is accompanied by local and systemic alterations in gene expression and an increase in disease resistance in the shoots. Plant J. 50, 529–544. doi: 10.1111/j.1365-313X.2007.03069.x
Lopes, U. P., Zambolim, L., Pereira, O. L. (2009). First report of Lasiodiplodia theobromae causing leaf blight on the orchid Catasetum fimbriatum in Brazil. Australas. Plant Dis. Note. 4, 64–65. doi: 10.1071/DN09027
Lu, Y., Yao, J. (2018). Chloroplasts at the crossroad of photosynthesis, pathogen infection and plant defense. Int. J. Mol. Sci. 19, 1–37. doi: 10.3390/ijms19123900
Marquez, N., Giachero, M. L., Gallou, A., Debat, H. J., Cranenbrouck, S., di Rienzo, J. A., et al. (2018). Transcriptional changes in mycorrhizal and nonmycorrhizal soybean plants upon infection with the fungal pathogen Macrophomina phaseolina. Mol. Plant-Microbe Interact. 31, 842–855. doi: 10.1094/MPMI-11-17-0282-R
McKendrick, S. L., Leake, J. R., Lee Taylor, D., Read, D. J. (2004). Symbiotic germination and development of the myco-heterotrophic orchid Neottia nidus-avis in nature and its requirement for locally distributed Sebacina spp. New Phytol. 163, 405–423. doi: 10.1111/j.1469-8137.2004.01115.x
Meng, Y. Y., Zhang, W. L., Selosse, M. A., Gao, J. Y. (2019). Are fungi from adult orchid roots the best symbionts at germination? A case study. Mycorrhiza 29, 541–547. doi: 10.1007/s00572-019-00907-0
Minasiewicz, J., Zwolicki, A., Figura, T., Novotná, A., Bocayuva, M. F., Jersáková, J., et al. (2023). Stoichiometry of carbon, nitrogen and phosphorus is closely linked to trophic modes in orchids. BMC Plant Biol. 23, 1–11. doi: 10.1186/s12870-023-04436-z
Miura, C., Furui, Y., Yamamoto, T., Kanno, Y., Honjo, M., Yamaguchi, K., et al. (2024). Autoactivation of mycorrhizal symbiosis signaling through gibberellin deactivation in orchid seed germination. Plant Physiol. 194, 1–18. doi: 10.1093/plphys/kiad517
Miura, C., Saisho, M., Yagame, T., Yamato, M., Kaminaka, H. (2019). Bletilla striata (Orchidaceae) seed coat restricts the invasion of fungal hyphae at the initial stage of fungal colonization. Plants 8, 1–11. doi: 10.3390/plants8080280
Miura, C., Yamaguchi, K., Miyahara, R., Yamamoto, T., Fuji, M., Yagame, T., et al. (2018). The mycoheterotrophic symbiosis between orchids and mycorrhizal fungi possesses major components shared with mutualistic plant-mycorrhizal symbioses. Mol. Plant Microbe Interact. 31, 1032–1047. doi: 10.1094/MPMI-01-18-0029-R
Orine, D., Defossez, E., Vergara, F., Uthe, H., van Dam, N. M., Rasmann, S. (2022). Arbuscular mycorrhizal fungi prevent the negative effect of drought and modulate the growth-defence trade-off in tomato plants. J. Sustain. Agric. Environ. 1, 177–190. doi: 10.1002/sae2.12018
Otero, J. T., Bayman, P., Ackerman, J. D. (2005). Variation in mycorrhizal performance in the epiphytic orchid Tolumnia variegata in vitro: The potential for natural selection. Evol. Ecol. 19, 29–43. doi: 10.1007/s10682-004-5441-0
Parniske, M. (2008). Arbuscular mycorrhiza: the mother of plant root endosymbioses. Nat. Rev. Microbiol. 6, 763–775. doi: 10.1038/nrmicro1987
Pérez, F. J., Rubio, S. (2006). An improved chemiluminescence method for hydrogen peroxide determination in plant tissues. Plant Growth Regul. 48, 89–95. doi: 10.1007/s10725-005-5089-y
Peterson, R. L., Massicote, H. B., Melville, L. H. (2004). Mycorrhizas: Anatomy and Cell Biology (Ottawa: NRC Research Press). doi: 10.1017/s0269-915x(05)21306-x
Pieterse, C. M. J., Zamioudis, C., Berendsen, R. L., Weller, D. M., Van Wees, S. C. M., Bakker, P. A. H. M. (2014). Induced systemic resistance by beneficial microbes. Annu. Rev. Phytopathol. 52, 347–375. doi: 10.1146/annurev-phyto-082712-102340
Pujasatria, G. C., Nishiguchi, I., Miura, C., Yamato, M., Kaminaka, H. (2022). Orchid mycorrhizal fungi and ascomycetous fungi in epiphytic Vanda falcata roots occupy different niches during growth and development. Mycorrhiza 32, 481–495. doi: 10.1007/s00572-022-01089-y
Qin, J., Zhang, W., Ge, Z. W., Zhang, S. B. (2019). Molecular identifications uncover diverse fungal symbionts of Pleione (Orchidaceae). Fungal Ecol. 37, 19–29. doi: 10.1016/j.funeco.2018.10.003
Ross, A. F. (1966). “Systemic effects of local lesion formation,” in Viruses of Plants: Their Isolation, Purification, and Characterization: the Mechanism of Plant Virus Infection, Synthesis of Viral Protein and Viral Nucleic Acid, and Plant Reactions Evoked by Viruses. Eds. Beemster, A. B. R., Dijkstra, J. (North-Holland Publishing Company, Wageningen), 127–150.
Säle, V., Palenzuela, J., Azcón-Aguilar, C., Sánchez-Castro, I., da Silva, G. A., Seitz, B., et al. (2021). Ancient lineages of arbuscular mycorrhizal fungi provide little plant benefit. Mycorrhiza 31, 559–576. doi: 10.1007/s00572-021-01042-5
Samson, R., Legendre, J. B., Christen, R., Fischer-Le Saux, M., Achouak, W., Gardan, L. (2005). Transfer of Pectobacterium chrysanthemi (Burkholder et al. 1953) Brenner et al. 1973 and Brenneria paradisiaca to the genus Dickeya gen. nov. as Dickeya chrysanthemi comb. nov. and Dickeya paradisiaca comb. nov. and delineation of four novel species, Dick. Int. J. Syst. Evol. Microbiol. 55, 1415–1427. doi: 10.1099/ijs.0.02791-0
Sathiyadash, K., Muthukumar, T., Murugan, S. B., Sathishkumar, R., Pandey, R. R. (2014). In vitro symbiotic seed germination of South Indian endemic orchid Coelogyne nervosa. Mycoscience 55, 183–189. doi: 10.1016/j.myc.2013.08.005
Schoenherr, A. P., Rizzo, E., Jackson, N., Manosalva, P., Gomez, S. K. (2019). Mycorrhiza-induced resistance in potato involves priming of defense responses against cabbage looper (Noctuidae: Lepidoptera). Environ. Entomol. 48, 370–381. doi: 10.1093/ee/nvy195
Selosse, M. A., Minasiewicz, J., Boullard, B. (2017). An annotated translation of Noël Bernard’s 1899 article ‘On the germination of Neottia nidus-avis.’. Mycorrhiza 27, 611–618. doi: 10.1007/s00572-017-0774-z
Shoresh, M., Yedidia, I., Chet, I. (2005). Involvement of jasmonic acid/ethylene signaling pathway in the systemic resistance induced in cucumber by Trichoderma asperellum T203. Phytopathology 95, 76–84. doi: 10.1094/PHYTO-95-0076
Silva, M., Pereira, O. L. (2007). First report of Guignardia endophyllicola leaf blight on Cymbidium (Orchidaceae) in Brazil. Australas. Plant Dis. Note. 2, 31–32. doi: 10.1071/DN07015
Smyth, G. K., Ritchie, M. E., Law, C. W., Alhamdoosh, M., Su, S., Dong, X., et al. (2018). RNA-seq analysis is easy as 1-2-3 with limma, Glimma and edgeR. F1000Res 5, 1–30. doi: 10.12688/f1000research.9005.3
Sobiczewski, P., Iakimova, E. T., Mikiciński, A., Węgrzynowicz-Lesiak, E., Dyki, B. (2017). Necrotrophic behaviour of Erwinia amylovora in apple and tobacco leaf tissue. Plant Pathol. 66, 842–855. doi: 10.1111/ppa.12631
Srivastava, S., Kadooka, C., Uchida, J. Y. (2018). Fusarium species as pathogen on orchids. Microbiol. Res. 207, 188–195. doi: 10.1016/j.micres.2017.12.002
Stöckel, M., Těšitelová, T., Jersáková, J., Bidartondo, M. I., Gebauer, G. (2014). Carbon and nitrogen gain during the growth of orchid seedlings in nature. New Phytol. 202, 606–615. doi: 10.1111/nph.12688
Strullu-Derrien, C., Selosse, M. A., Kenrick, P., Martin, F. M. (2018). The origin and evolution of mycorrhizal symbioses: from palaeomycology to phylogenomics. New Phytol. 220, 1012–1030. doi: 10.1111/nph.15076
Suharjo, R., Sawada, H., Takikawa, Y. (2014). Phylogenetic study of Japanese Dickeya spp. and development of new rapid identification methods using PCR-RFLP. J. Gen. Plant Pathol. 80, 237–254. doi: 10.1007/s10327-014-0511-9
Suwannarach, N., Kumla, J., Lumyong, S. (2018). Leaf spot on Cattleya orchid caused by Neoscytalidium orchidacearum in Thailand. Can. J. Plant Pathol. 40, 109–114. doi: 10.1080/07060661.2017.1414882
Swett, C. S., Uchida, J. Y. (2015). Characterization of Fusarium diseases on commercially grown orchids in Hawaii. Plant Pathol. 64, 648–654. doi: 10.1111/ppa.12290
Tsai, C. F., Huang, C. H., Wu, F. H., Lin, C. H., Lee, C. H., Yu, S. S., et al. (2022). Intelligent image analysis recognizes important orchid viral diseases. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1051348
Umata, H., Ota, Y., Yamada, M., Watanabe, Y., Gale, S. W. (2013). Germination of the fully myco-heterotrophic orchid Cyrtosia septentrionalis is characterized by low fungal specificity and does not require direct seed-mycobiont contact. Mycoscience 54, 343–352. doi: 10.1016/j.myc.2012.12.003
van Peer, R., Niemann, G. J., Schippers, B. (1990). Induced resistance and phytoalexin accumulation in biological control of fusarium wilt of carnation by pseudomonas sp. Strain WCS417r. Phytopathology 81, 728–734. doi: 10.1094/Phyto-81-728
Vierheilig, H., Coughlan, A. P., Wyss, U., Piche, Y. (1998). Ink and vinegar, a simple staining technique for arbuscular-mycorrhizal fungi. Appl. Environ. Microbiol. 64, 5004–5007. doi: 10.1128/AEM.64.12.5004-5007.1998
Vlot, A. C., Sales, J. H., Lenk, M., Bauer, K., Brambilla, A., Sommer, A., et al. (2021). Systemic propagation of immunity in plants. New Phytol. 229, 1234–1250. doi: 10.1111/nph.16953
Wang, D., Gebauer, G., Jacquemyn, H., Zahn, F. E., Gomes, S. I. F., Lorenz, J., et al. (2023). Variation in mycorrhizal communities and the level of mycoheterotrophy in grassland and Forest populations of Neottia ovata (Orchidaceae). Funct. Ecol. 37, 1948–1961. doi: 10.1111/1365-2435.14354
Wang, H., Hao, Z., Zhang, X., Xie, W., Chen, B. (2022a). Arbuscular mycorrhizal fungi induced plant resistance against fusarium wilt in jasmonate biosynthesis defective mutant and wild type of tomato. J. Fungi 8, 1–14. doi: 10.3390/jof8050422
Wang, M., Tang, W., Xiang, L., Chen, X., Shen, X., Yin, C., et al. (2022b). Involvement of MdWRKY40 in the defense of mycorrhizal apple against Fusarium solani. BMC Plant Biol. 22, 1–15. doi: 10.1186/s12870-022-03753-z
Wasternack, C., Feussner, I. (2018). The oxylipin pathways: biochemistry and function. Annu. Rev. Plant Biol. 69, 363–386. doi: 10.1146/annurev-arplant-042817-040440
Wei, X. Y., Deng, W. L., Chu, C. C. (2021). Phylogenetic and phenotypic analyses on Dickeya spp. isolated from different host plants in Taiwan. J. Phytopathol. 169, 678–691. doi: 10.1111/jph.13038
Wei, G., Kloepper, J. W., Tuzun, S. (1991). Induction of systemic resistance of cucumber to colletotrichum orbiculare by select strains of plant growth-promoting rhizobacteria. Phytopathology 81, 1508–1512. doi: 10.1094/Phyto-81-1508
Wellburn, A. R. (1994). The Spectral Determination of Chlorophylls a and b, as well as Total Carotenoids, Using Various Solvents with Spectrophotometers of Different Resolution. J. Plant Physiol. 144, 307–313. doi: 10.1016/S0176-1617(11)81192-2
Wu, P. H., Huang, D. D., Chang, D. C. N. (2011). Mycorrhizal symbiosis enhances Phalaenopsis orchid’s growth and resistance to Erwinia chrysanthemi. Afr. J. Biotechnol. 10, 10095–10100. doi: 10.5897/AJB11.1310
Xing, X., Jacquemyn, H., Gai, X., Gao, Y., Liu, Q., Zhao, Z., et al. (2019). The impact of life form on the architecture of orchid mycorrhizal networks in tropical forest. Oikos 128, 1254–1264. doi: 10.1111/oik.06363
Yamamoto, T., Miura, C., Fuji, M., Nagata, S., Otani, Y., Yagame, T., et al. (2017). Quantitative evaluation of protocorm growth and fungal colonization in Bletilla striata (Orchidaceae) reveals less-productive symbiosis with a non-native symbiotic fungus. BMC Plant Biol. 17, 1–10. doi: 10.1186/s12870-017-1002-x
Ye, W., Jiang, J., Lin, Y., Yeh, K. W., Lai, Z., Xu, X., et al. (2019). Colonisation of Oncidium orchid roots by the endophyte Piriformospora indica restricts Erwinia chrysanthemi infection, stimulates accumulation of NBS-LRR resistance gene transcripts and represses their targeting micro-RNAs in leaves. BMC Plant Biol. 19, 1–16. doi: 10.1186/s12870-019-2105-3
Yukawa, T., Ogura-Tsujita, Y., Shefferson, R. P., Yokoyama, J. (2009). Mycorrhizal diversity in Apostasia (Orchidaceae) indicates the origin and evolution of orchid mycorrhiza. Am. J. Bot. 96, 1997–2009. doi: 10.3732/ajb.0900101
Zahn, F. E., Söll, E., Chapin, T. K., Wang, D., Gomes, S. I. F., Hynson, N. A., et al. (2023). Novel insights into orchid mycorrhiza functioning from stable isotope signatures of fungal pelotons. New Phytol. 239, 1449–1463. doi: 10.1111/nph.18991
Zettler, L. W., Stewart, S. L., Bowles, M. L., Jacobs, K. A. (2001). Mycorrhizal fungi and cold-assisted symbiotic germination of the federally threatened eastern prairie fringed orchid, platanthera leucophaea (Nuttall) lindley. Am. Midl. Nat. 145, 168–175. doi: 10.1674/0003-0031(2001)145[0168:mfacas]2.0.co;2
Zhou, A., Nie, J., Tian, Y., Chuan, J., Hu, B., Zou, J., et al. (2021). First report of dickeya fangzhongdai causing soft rot in orchid in Canada. Plant Dis. 105, 4149. doi: 10.1094/PDIS-04-21-0771-PDN
Keywords: Bletilla striata, defense priming, Dickeya fangzhongdai, induced systemic resistance (ISR), mycorrhizal fungi, necrotrophic pathogen, orchids
Citation: Pujasatria GC, Miura C, Yamaguchi K, Shigenobu S and Kaminaka H (2024) Colonization by orchid mycorrhizal fungi primes induced systemic resistance against necrotrophic pathogen. Front. Plant Sci. 15:1447050. doi: 10.3389/fpls.2024.1447050
Received: 11 June 2024; Accepted: 16 July 2024;
Published: 31 July 2024.
Edited by:
Hiromu Kameoka, Chinese Academy of Sciences (CAS), ChinaCopyright © 2024 Pujasatria, Miura, Yamaguchi, Shigenobu and Kaminaka. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hironori Kaminaka, a2FtaW5ha2FAdG90dG9yaS11LmFjLmpw
†ORCID: Galih Chersy Pujasatria, orcid.org/0000-0002-2768-8250
Chihiro Miura, orcid.org/0000-0001-8460-1791
Katsushi Yamaguchi, orcid.org/0000-0001-6871-7882
Shuji Shigenobu, orcid.org/0000-0003-4640-2323
Hironori Kaminaka, orcid.org/0000-0002-3685-8688
 Galih Chersy Pujasatria1†
Galih Chersy Pujasatria1† Chihiro Miura
Chihiro Miura Katsushi Yamaguchi
Katsushi Yamaguchi Shuji Shigenobu
Shuji Shigenobu Hironori Kaminaka
Hironori Kaminaka