- 1College of Life Sciences, Zhejiang Normal University, Jinhua, China
- 2National Engineering Research Center for Vegetables, Beijing Vegetable Research Center, Beijing Academy of Agriculture and Forestry Science, Beijing, China
The regulation of gene expression is crucial for biological plant growth and development, with transcription factors (TFs) serving as key switches in this regulatory mechanism. GOLDEN2-LIKE (GLK) TFs are a class of functionally partially redundant nuclear TFs belonging to the GARP superfamily of MYB TFs that play a key role in regulating genes related to photosynthesis and chloroplast biogenesis. Here, we summarized the current knowledge of the pleiotropic roles of GLKs in plants. In addition to their primary functions of controlling chloroplast biogenesis and function maintenance, GLKs have been proven to regulate the photomorphogenesis of seedlings, metabolite synthesis, flowering time, leaf senescence, and response to biotic and abiotic stress, ultimately contributing to crop yield. This review will provide a comprehensive understanding of the biological functions of GLKs and serve as a reference for future theoretical and applied studies of GLKs.
Introduction
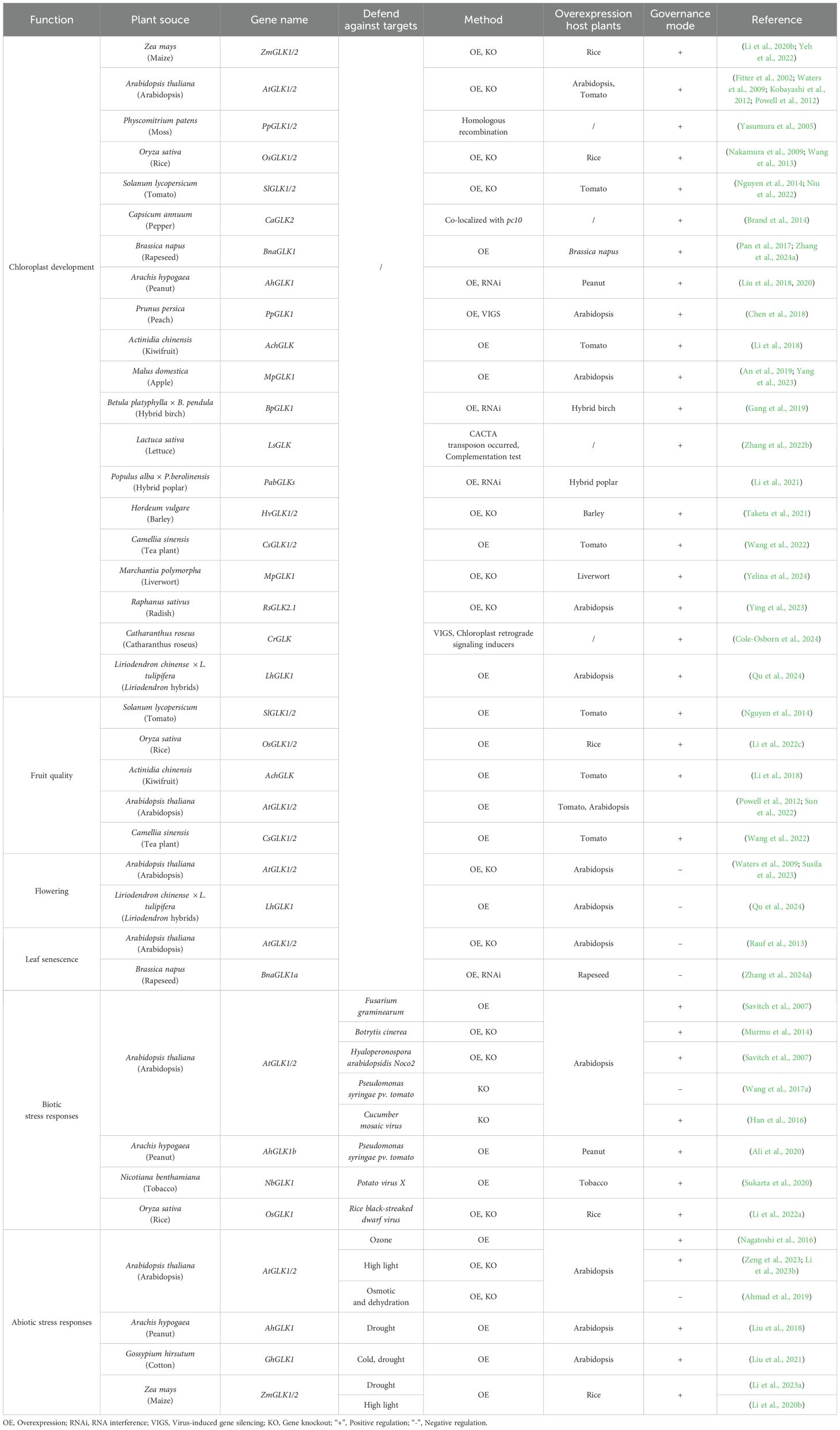
GOLDEN2-LIKEs (GLKs) are plant-specific transcription factors (TFs) involved in multiple biological processes in plants (Chen et al., 2016; Lambret-Frotte et al., 2023). GLKs are members of the GARP superfamily, containing a nuclear localization signal, a DNA-binding domain (DBD), a proline-rich domain and a GLK/C-terminal (GCT) box (Riechmann et al., 2000; Safi et al., 2017). The DBD consists of three α-helices followed by a highly conserved motif of AREAEAA, which confers specific characteristics to GLKs and distinguishes GLKs from other GAPR members (Fitter et al., 2002). To date, GLKs are widespread in land plants, and the last common ancestor of GLKs might be from Embryophyta (Wang et al., 2013; Hernández-Verdeja and Lundgren, 2023). GLKs are demonstrated to be the key regulators for chloroplast biogenesis from lower plants to higher plants (Table 1; Figure 1). Additionally, mounting evidence shows that the GLKs also function in multiple aspects through the entire lifetime of plants, including seedling photomorphogenesis, hormone signalling, leaf senescence, flowering, fruit nutrition and bio- or abiotic stress responses (Table 1; Figures 1, 2). GLKs might be a node of signaling networks in plants, which are valuable to research for crop improvement in molecular breeding.
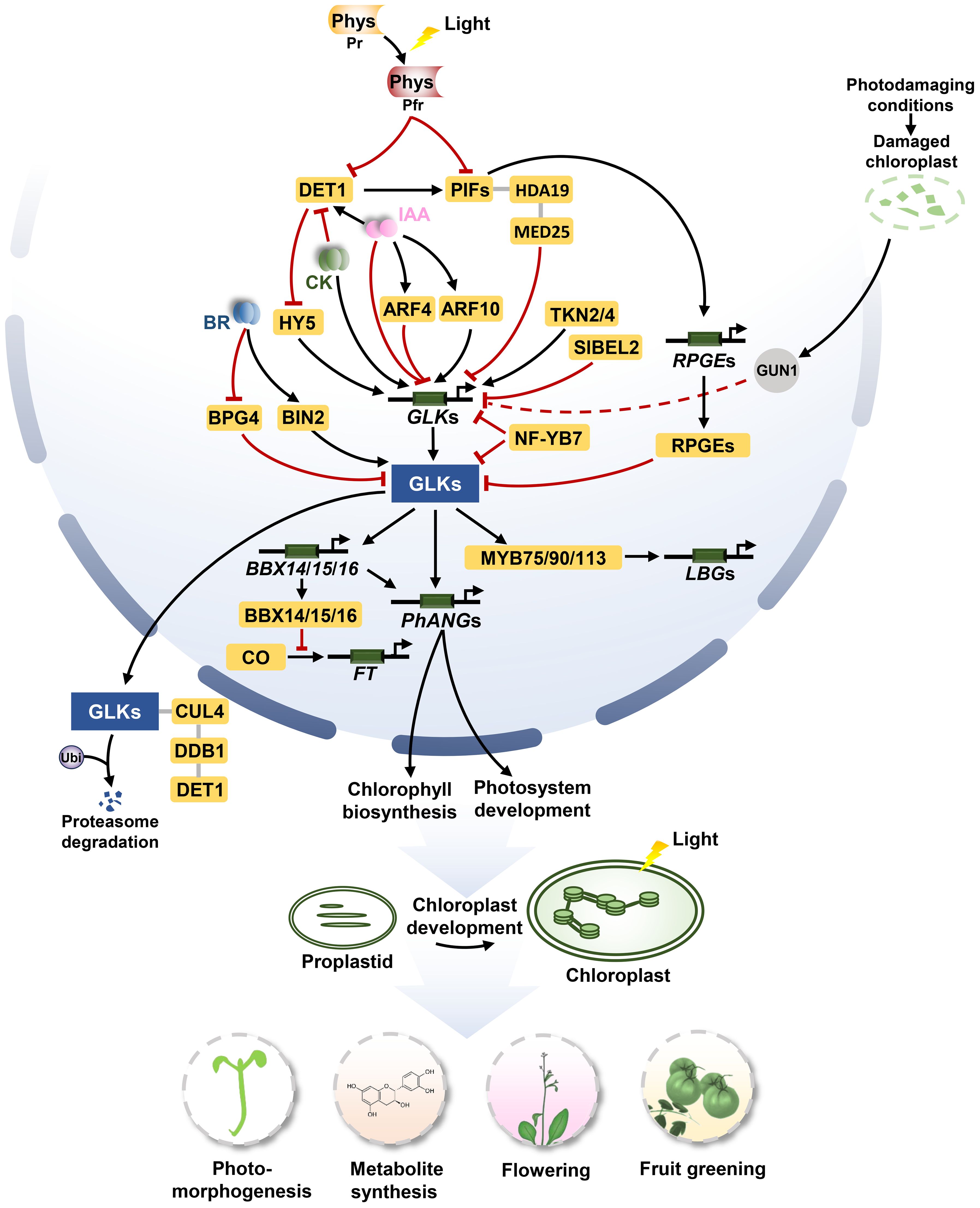
Figure 1. The signaling pathways of GLKs in regulating chloroplast biosynthesis, photomorphogenesis, flowering, and metabolite synthesis. For chloroplast biogenesis, GLKs activate the expression of PhANGs to promote the development of chloroplast. TKN2 and TKN4 activate the expression of GLK2, while BEL2 negatively regulates the expression of GLK2 to promote the establishment of the ‘green shoulder’ in tomato fruits. ARF10 directly induces the expression of GLK1 and ARF4 inhibits the transcription of GLK1. For photomorphogenesis, activated phytochromes (Phys) repress PIF and DET1 under light conditions. DET1 promotes the stability of PIF1 proteins, meanwhile, it mediates the proteasome degradation of GLK by interacting with CUL4 and DDB1 to form a ubiquitin ligase complex. The PIF1/PIF3-HDA19-MED25 complex reduces transcriptional repression of GLK1 under light conditions. Activated BIN2 phosphorylates and thus stabilizes GLKs under light conditions. BPG4 suppress the transcription activity of GLKs via inhibition to their DNA-binding ability. HY5 binds the promoter of GLKs, inducing their activities to promote chloroplast development. Under dark conditions, PIFs can directly bind to the GLK1 promoter to repress the expression of GLK1. Moreover, PIFs activate the expression of RPGEs. RPGEs interact with GLKs to disrupt the DNA-binding activity of GLKs. In photodamaging conditions, the activity of GUN1 appears to down-regulate the expression of GLK1 when plastids are dysfunctional. For flowering, GLKs directly activate the expression of BBX14, BBX15 and BBX16, and the BBX proteins physically interact with the circadian clock regulator protein CO in the nucleus, which prevents CO-mediated FT transcription from repressing flowering. For metabolite synthesis, GLK1 interacts with the MBW complexes MYB75/90/113 and activates the transcriptional activity to enhance the expression of genes related to anthocyanin-specific biosynthetics including LBGs. Arrows and lines with end lines indicate positive regulation and negative regulation, respectively. Grey lines indicate interaction. Dashed arrow represents indirect effects through unknown intermediate factors.
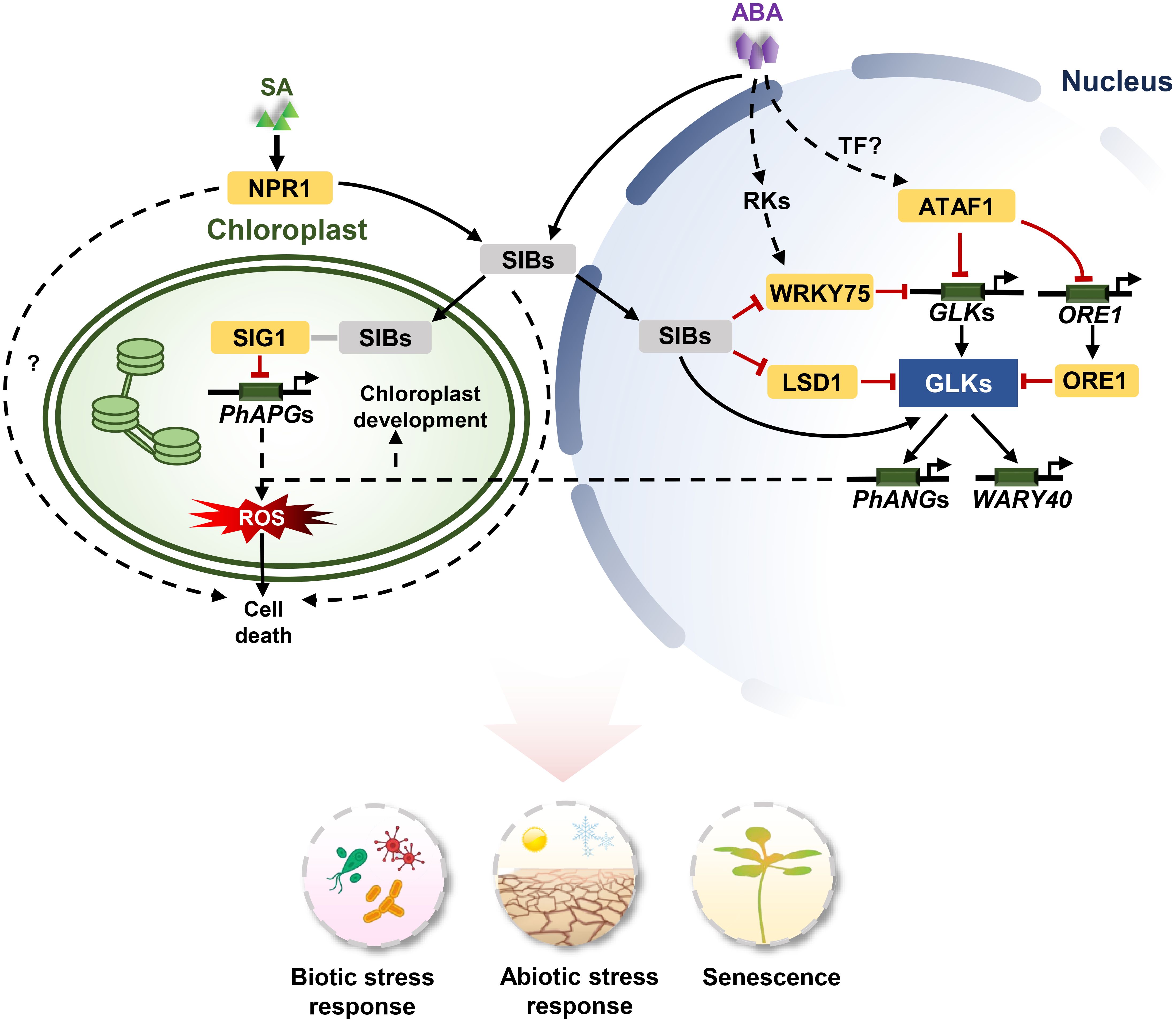
Figure 2. The signaling pathways of GLKs in stress response and senescence. For biotic stresses, SA-mediated NPR1 activation leads to the expression of SIB1. SIB1 proteins are targeted to both the nucleus and chloroplasts. SIB1 interacts with SIG1 to inhibit PhAPGs expression in chloroplasts, and SIB1 activate GLKs to induce the expression of PhANGs in the nucleus. The uncoupled expression of PhANGs and PhAPGs leads to an increase of 1O2 and PQH2 levels in chloroplasts. The reactive oxygen species (ROS) burst contributes to cell death. LSD1 antagonistically regulate the expression of GLKs with SIB1 and functions in cell death. For abiotic stress, For abiotic stress, SIBs are induced by ABA and interact with WRKY75 to inhibit its transcriptional function. WRKY75 directly binds to the promoters of GLKs to repress their expression. ATAF1 responds to ABA and suppresses the expression of GLK1 by directly binding to the promoters of GLK1 and ORE1. ORE1 interacts with GLK1 to inhibit its transcriptional activity. ATAF1 expression is regulated by unknown upstream TFs. ABA activates GLKs via core ABA signalling components PYL/PYRs-PP2Cs-SnRKs, and subsequently GLKs induce the expression of WRKY40. Arrows and lines with an end line indicate positive regulation and negative regulation, respectively. Grey lines indicate interaction. Dashed arrows represent indirect effects through unknown intermediate factors.
GLKs control chloroplast biogenesis and function maintenance
Chloroplast is an important place for photosynthesis in plants (Jarvis and López-Juez, 2014). Solid evidence indicated that GLKs control chloroplast biogenesis by transcriptionally targeting photosynthesis-related nuclear genes (PhANGs), including chlorophyll biosynthesis and photosynthesis-related genes (Waters et al., 2009; Martín et al., 2016). Constitutive expression of GLKs could increase chloroplast numbers and chlorophyll content in photosynthetic tissues, such as leaves or fruits (Nguyen et al., 2014), and even in non-photosynthetic tissues such as roots and callus in Arabidopsis (Arabidopsis thaliana) (Nakamura et al., 2009; Kobayashi et al., 2012). In tomato (Solanum lycopersicum), the expression of GLK2 gradiently reduced from the shoulder to the base in fruit, which influences a gradient of chloroplast development of fruit forming the ‘green shoulder’ fruits (Powell et al., 2012; Nguyen et al., 2014). The TFs KNOTTED1-like Homeobox (KNOX) TKN2 and TKN4 activate the expression of GLK2 to promote the establishment of ‘green shoulder’ fruit in tomato (Nadakuduti et al., 2014). However, BEL1-like HOMEODOMAIN 2 (BEL2) affects the formation of ‘green shoulder’ in tomato fruits by negatively regulating the gradient expression of GLK2 (Niu et al., 2022). In addition, GLKs were affected by AUXIN RESPONSE FACTORs (ARFs) in regulating chlorophyll accumulation in tomato fruit (Sagar et al., 2013; Yuan et al., 2018). In rice (Oryza sativa), a member of the nuclear factor Y (NF-Y) TF family, OsNF-YB7, inactivates the transactivation activity of GLK1 at multiple regulatory layers to inhibit chlorophyll accumulation in the embryo of rice (Yang et al., 2024). In radish (Raphanus sativus), GLK2 interacts with NUCLEAR FACTOR Y, SUBUNIT A 9a (NF-YA9a) to increase the expression of the chlorophyll biosynthesis gene, RsHEMA2, which improves the chloroplast development (Figure 1; Ying et al., 2023).
Interestingly, GLKs are functionally redundant in C3 plants. In Arabidopsis and rice, the glk1 or glk2 single mutant has no phenotypic difference from the wild type (WT), and the glk1/glk2 double mutant displayed pale green leaves and abnormal chloroplast structure (Fitter et al., 2002; Wang et al., 2013). However, the functional redundancy of GLKs does not exist in the C4 plant. For instance, maize (Zea mays) glk2 single mutant showed yellow leaves with abnormal chloroplast structure (Rossini et al., 2001). It is well known that the chloroplasts become different between the C3 and the C4 plants, the former has only one type of chloroplast in mesophyll cells (MC), while the latter has two types of chloroplasts in the bundle sheath cells (BSC) and the MC, respectively (Majeran et al., 2009). The development of chloroplasts in the BSC provides an anatomical basis for efficient photosynthesis in C4 plants (Miyake, 2016). In C4 plants such as maize and sorghum (Sorghum bicolor), GLK1 expressed much more in MC than that in BSC, while GLK2 expressed more in BSC contrarily (Wang et al., 2013; John et al., 2014). In addition, the tissue-expression pattern of GLK1 and GLK2 are almost similar in Arabidopsis (Supplementaryray Figure S1), but different in maize (Supplementary Figure S2). It was considered that both GLK orthologs retained the ability to induce chloroplast biogenesis and play important roles in regulating the differentiation of chloroplast development in C4 plants (Rossini et al., 2001), but recent studies showed that GLK2 adopted a more prominent developmental role, particularly in relation to chloroplast activation in BSC (Lambret-Frotte et al., 2023).
To maintain the functional stability of chloroplasts in plants, the chloroplast-to-nucleus retrograde signalling (RS) is essential for coordinating the expression of PhANGs and photosynthesis-associated plastid genes (PhAPGs; Pogson et al., 2008). Defective chloroplasts in mutants of plastid protein emphasize coordination between chloroplastic protein processing and nuclear transcription (Chan et al., 2016). GENOMES UNCOUPLED1 (GUN1), a chloroplast-localized pentatricopeptide-repeat protein, is a central integrator participating in multiple RS pathways. In photodamaging conditions, the activity of GUN1 appears to down-regulate the expression of GLK1 when plastids are dysfunctional (Kakizaki et al., 2010); GUN1/GLK1 module represses the expression of B-box structural domain PROTEIN16 (BBX16) to regulate the well-established expression of PhANGs (Figure 1; Veciana et al., 2022). However, aside from the GUN1/GLK1 module, studies also showed that the ubiquitin-proteasome system participates in the degradation of Arabidopsis GLK1 in response to plastid signals in a GUN1-independent manner (Tokumaru et al., 2017).
GLKs modulate the photomorphogenesis of seedlings
Seedling photomorphogenesis is coordinately processed as inhibition of hypocotyl elongation, the opening of cotyledon, and chloroplast development when exposed to light. In Arabidopsis, GLKs are induced by light (Fitter et al., 2002). The glk1/glk2 double mutant displayed decreased chlorophyll content, longer hypocotyls and less separated cotyledons (Martín et al., 2016; Alem et al., 2022). PHYTOCHROME-INTERACTING FACTORs (PIFs) are central regulators of photomorphogenesis in plants (Leivar and Monte, 2014). PIFs can form a complex with the histone deacetylase HDA19 and the Mediator subunit MED25, thus attenuating the transcriptional repression of GLK1 by binding to the PBE motif (CACATG) on GLK1 promoter in darkness (Martín et al., 2016; Guo et al., 2023), while light-activated phytochrome reverses this activity, thereby inducing GLKs expression (Martín et al., 2016). Interestingly, PIFs can also induce the expression of the REPRESSOR OF PHOTOSYNTHETIC GENES 1 (RPGE1) and RPGE2 in darkness, and then the RPGEs inhibit the DNA-binding activity of GLK1 by disrupting its dimerization, revealing another mechanism of PIF-mediated GLK repression (Kim et al., 2023). Besides, rice Phytochrome-Interacting Factor-Like1 (OsPIL1), a basic helix-loop-helix transcription factor, is also involved in the promotion of chlorophyll biosynthesis (Sakuraba et al., 2017). Moreover, DEETIOLATED 1 (DET1), a repressor of light-induced photomorphogenesis, not only promotes the protein stability of PIF1 (Shi et al., 2015), but also interacts with GLKs and promotes the degradation of GLK proteins by ubiquitination (Tang et al., 2016; Zhang et al., 2024b). Another regulator of photomorphogenesis, ELONGATED HYPOCOTYL5 (HY5) not only directly activates the expression of GLKs, but also interacts with the GLK proteins, suggesting that HY5 might first activates the expression of GLKs promote chlorophyll biosynthesis and photosystem formation, and then interacts with GLK proteins to inhibit hypocotyl elongation (Zhang et al., 2024b). Furthermore, indole-3-acetic acid (IAA) and cytokinin (CK) regulate GLK2 in the opposing directions at the transcriptional level in a HY5-dependent manner to regulate chlorophyll biosynthesis in Arabidopsis roots (Kobayashi et al., 2012).
Additionally, the transcription factor, TEOSINTE BRANCHED 1, CYCLOIDEA, and PROLIFERATING CELL FACTOR 15 (TCP15), participates in the expression of PhANGs and binds to the same promoter regions of target genes as GLK1. It is postulated that GLK1 helps to recruit TCP15 for coordinating the expression of cell expansion genes with that of genes involved in the development of the photosynthetic apparatus (Alem et al., 2022). A regulator involved in BR signalling, BRASSINOSTEROID INSENSITIVE2 (BIN2), regulates physically interacts with and phosphorylates GLKs, and this phosphorylation stabilizes and activates GLKs to promote chloroplast development and photomorphogenesis (Zhang et al., 2021). Conversely, BRZINSENSITIVE-PALE GREEN 4 (BPG4) inhibits the transcriptional activity of GLKs by interacting with the GCT-box of GLKs and plays an inhibitory role in regulating chloroplast development and homeostasis (Figure 1; Tachibana et al., 2024).
GLKs participate in the synthesis of metabolites
Photosynthetic products of chloroplasts generally contribute to the accumulation of carbohydrates, lycopene, carotenoids or other nutrient related substances in fruits (Klee and Giovannoni, 2011; Jia et al., 2020). Interestingly, GLKs can interact with the G-box Binding Factor (GBF) and activate the transcription of PHYTOENE SYNTHASE (PSY), promoting the biosynthesis of carotenoids (Sun et al., 2022). Overexpression of the exogenous GLKs increases the contents of carbohydrates, carotenoids, and tocopherol (vitamin E) in fruits of tomato (Powell et al., 2012; Nguyen et al., 2014; Lupi et al., 2019). Endosperm-specific overexpression of rice GLK1 promotes the biosynthesis of carotenoids in the endosperm (Li et al., 2022c). Ectopic overexpression of the GLK homolog from pepper (Capsicum annuum), kiwifruit (Actinidia chinensis), and tea (Camellia sinensis) in tomato resulted in higher levels of carotenoids and sugar in the ripened fruits (Brand et al., 2014; Li et al., 2018; Wang et al., 2022). In addition, GLKs induce the biosynthesis of secondary metabolites including catechin and anthocyanin. CsGLKs are also involved in light-regulated catechin accumulation in tea plants by regulating the expression of CsMYB5b (Wang et al., 2022). In Arabidopsis, GLK1 interacts with the WD40-BHLH-MYB (MBW) complexes MYB75/90/113 and activates the transcriptional activity to enhance the expression of genes related to anthocyanin-specific biosynthetic including late biosynthesis genes (LBGs) (Li et al., 2023b). Meanwhile, GLK2 activates the expression of LBGs and TRANSPARENT TESTA GLABRA 1 (TTG1) through AtHY5-mediated light signalling and positively regulates anthocyanin biosynthesis in Arabidopsis (Figure 1; Liu et al., 2022; Zeng et al., 2023).
GLKs negatively regulate flowering time and leaf senescence
The flowering time of plants is tightly controlled by endogenous or exogenous signals (Bouché, et al., 2016). It was reported that chloroplasts RS regulated flowering mediated by the floral repressor FLOWERING LOCUS C (FLC) in Arabidopsis (Feng et al., 2016). GLK1 and GLK2 act as downstream components of the chloroplast RS pathway that negatively regulates flowering time. The glk1/glk2 double mutant of Arabidopsis displays early flowering, and overexpression of AtGLK1, AtGLK2 or LhGLK1 in Arabidopsis delayed flowering time (Waters et al., 2009; Qu et al., 2024). GLKs directly activate the expression of BBX14, BBX15 and BBX16, and these BBXs proteins physically interact with the circadian clock regulatory CONSTANS (CO) in the nucleus, which prevent CO-mediated FLOWERING LOCUST (FT) transcription and repress flowering (Figure 1; Susila et al., 2023).
The chloroplast displays early signs of senescence symptoms, including a decrease in chlorophyll and a decline in photosynthetic efficiency (Soudry et al., 2005). PIF3, 4, and 5 are up-regulated during age-triggered and dark-induced leaf senescence, and the accumulation of PIFs protein inhibits the expression of GLKs to impair chloroplast development and chlorophyll biosynthesis, leading to leaf senescence (Song et al., 2014). In addition, GLKs also respond to abscisic acid (ABA) in regulating plant senescence. The ABA pathway generally promotes leaf senescence, while GLKs negatively modulate ABA-mediated leaf senescence. Both SIBs and WRKY75 are upregulated during leaf senescence and induced by ABA. SIBs interact with WRKY75 and thereby repress its transcriptional function, thus negatively regulating ABA-induced leaf senescence in a WRKY75-dependent manner. In contrast, WRKY75 positively modulates ABA-mediated leaf senescence in a GLK-dependent manner by directly binding to the W-box (T/CTGACC/T) in the GLKs promotor and inhibits their expressions (Zhang et al., 2022a; Lee et al., 2023). In addition, ABA can activate a NAC transcription factor ATAF1, which activates ORESARA1 (ORE1) and represses GLK1 expression by directly binding to the promoters of both genes. ORE1 also interacts with GLKs to inhibit the transcriptional activity of GLK1, resulting in impairing the expression of GLK target genes and leaf senescence (Figure 2; Rauf et al., 2013; Garapati et al., 2015). In Brassica napus, GLK1a has also been shown to directly influence the ABA signalling pathway. Overexpressing BnGLK1a delayed the leaf senescence upon ABA treatment (Zhang et al., 2024a).
GLKs are involved in biotic and abiotic stress response
Current studies have shown that GLKs participate in the defence response of plants. The glk1/glk2 double mutant of Arabidopsis showed enhanced resistance to Pseudomonas syringae pv. tomato and Hyaloperonospora arabidopsidis (Wang et al., 2017a). However, overexpression of AtGLK1 contributes to inducing the expression of pathogenesis-related (PR) genes, which in turn confers resistance to Fusarium graminearum (Savitch et al., 2007). Additionally, overexpression of AtGLK1 enhances the resistance to Botrytis cinerea in a jasmonic acid (JA)-independent manner, while increasing the susceptibility to Hyaloperonospora arabidopsidis Noco2 in a JA-dependant manner (Savitch et al., 2007; Murmu et al., 2014). GLKs play positive roles in resistance to cucumber mosaic virus (CMV), the Potato virus X (PVX), the rice black-streaked dwarf virus (RBSDV) and the maize rough dwarf disease (MRDD) (Han et al., 2016; Sukarta et al., 2020; Li et al., 2022b; Xu et al., 2023). Nevertheless, the virulence protein P69 of Turnip yellow mosaic virus (TYMV) interacts with GLKs and suppresses GLKs transcriptional activity, affecting the normal growth of plants and causing disease symptoms (Ni et al., 2017). Salicylic acid (SA) is an important hormone that regulates the defence responses to environmental stresses and against pathogens in plants (Kunkel and Brooks, 2002). LESION-SIMULATING DISEASE 1 (LSD1) is an SA-induced cell death regulator and a negative regulator that inhibits the DNA-binding activity of GLK1 towards its target promoters, and SIB1 proteins appeared to interrupt the LSD1-GLK interaction, and the subsequent SIB1-GLK interaction activated EX1-mediated singlet oxygen (1O2) signalling, leading to cell death and stress response in plants (Li et al., 2022a).
In addition, GLKs actively participate in the response to abiotic stresses. AhGLK1 upregulates the expression of AhPORA during recovery from drought in peanuts (Arachis hypogaea), stimulating chlorophyll biosynthesis and photosynthesis to increase the survival rate from drought (Liu et al., 2018). Virus-induced silencing of GhGLK1 in cotton (Gossypium hirsutum) leads to a great impact on growth and yield under drought and cold stress, and GhGLK1 helps to increase the adaptability of Arabidopsis in drought and cold stress (Liu et al., 2021). Overexpression of maize GLK genes in rice improves light harvesting efficiency via Photosystem II (PSII), thus buffering the adverse effects of photoinhibition under high or fluctuating light conditions (Li et al., 2023a). In addition, GLKs affect ABA sensitivity and ion channel activity of plants to regulate stomatal movements under stresses. The ABA-responsive genes WRKY40 is regulated by GLKs to increase the sensitivity of seedlings to osmotic stress, and the core ABA signalling components, PYL/PYRs-PP2Cs-SnRKs, possibly act as the intermediary in GLKs-induced WRKY40 expression (Ahmad et al., 2019). In Arabidopsis, the chimeric repressors for GLKs (GLKs-SRDX) downregulate the genes for inwardly rectifying K+in channels and K+in channel activity to close the stomata to enhance the tolerance to ozone (Nagatoshi et al., 2016). Recently, the role of GLKs in various abiotic stress responses has been predicted in multiple species through genome-wide analysis, including soybean (Glycine max), millet (Setaria italica), bamboo (Phyllostachys edulis), orange (Citrus sinensis) and western balsam poplar (Populus trichocarpa) (Alam et al., 2022; Chen et al., 2022; Wu et al., 2022; Xiong et al., 2022; Wu et al., 2023). These facts indicate a broad and conserved function in the abiotic stress response of GLKs in plants, which awaits further validation.
Molecular breeding application of GLKs in crops
Improving plant photosynthesis efficiency is an effective strategy for high-yield breeding in crops. Mounting evidence indicates that manipulation of GLKs achieves yield improvement in plants. In Arabidopsis, leaf-specific and silique wall-specific promoters were used to drive high expression of AtGLK1, resulting in enhanced leaf and silique wall photosynthesis and increased seed oil content by 2.88% and 10.75%, respectively (Zhu et al., 2018). In B. napus, overexpression of BnGLK1a resulted in a 10% increase in the thousand-seed weight of rapeseed (Zhang et al., 2024a). These results suggest that GLKs are promising tools for improving seed yield and oil production in oilseed crops.
Since the photosynthesis efficiency of C4 plants is much higher than that of C3 plants (von Caemmerer et al., 2012), the ectopic expression of maize (C4 plant) ZmGLKs was carried in rice (C3 plant) to improve its yield. The engineering rice plants induced chloroplast development in BSC accompanied by the accumulation of photosynthetic enzymes and intercellular connections (Wang et al., 2017b; Yeh et al., 2022). Overexpression of the ZmGLK1 and ZmGLK2 in rice increased the yield by 30% to 40% (Li et al., 2020b), while expression of ZmGLKs driven by its native promoter in rice increased the yield by 47% to 70% (Yeh et al., 2022).
Discussion
GLK is a key regulator of chloroplast development. Knockout of GLKs lead to abnormal chloroplast structure but not complete distortion of chloroplast biogenesis (Fitter et al., 2002; Wang et al., 2013), suggesting the existence of other genes which can partly compensate for GLKs function in chloroplast development. Besides, though GLKs are considered to play important roles in regulating the differentiation of chloroplast development in C4 plants (Rossini et al., 2001), the molecular mechanism remains unclear. Recently, it was shown that the pleiotropic role of GLKs beyond chloroplast regulation, including photomorphogenesis, synthesis of secondary metabolites, flowering, senescence and response to biotic and abiotic stresses (Table 1). Regarding GLKs being functionally redundant in chloroplast development in C3 plants, it’s natural to think whether GLKs are also redundant in regulating other aspects of life. Clarifying these questions would be helpful in understanding the bio-function of GLK in plants.
As core regulators in plant, GLKs are involved in multiple molecular modes of action including response to upstream genes, binding to downstream target genes and protein-protein interactions. However, so far, some studies only proved the interaction relationship between GLK and target proteins. The specific binding elements still await further research. The expression of GLK can be regulated by the upstream regulators by binding to specific cis-elements in the promoter, such as T/CTGACC/T (W-box), CACGTG (G-box) or CACATG (E-box) (Zhang et al., 2022a; Sakuraba et al., 2017). Besides, GLK can also bind to the promotor of target genes downstream to regulate their expression. The highly conserved motif CCAATC is considered a widely shared cis-acting element for downstream targets of GLKs (Waters et al., 2009). Comparative cross-species analyses of GLKs have shown that most of the binding sites of GLKs were species-specific (Tu et al., 2022), providing support for further exploration of binding sites rich in downstream targets of GLKs in the future. Furthermore, the DNA-binding domain and GCT-box of GLK proteins are specific binding domains for most regulatory factors. Interestingly, a few proteins also bind to proline-rich regions of GLK proteins, such as LSD1 (Li et al., 2022a). As for the degradation, SlGLK2 is proven a substrate of the CULLIN4 (CUL4) - UV-DAMAGED DNA BINDING PROTEIN 1 (DDB1) - DET1 ubiquitin ligase complex for the proteasome degradation (Tang et al., 2016). However, the ubiquitin-proteasome system is also shown to participate in the degradation of Arabidopsis GLK1 in response to plastid signals (Tokumaru et al., 2017). Would it also be a part of the ‘CUL4-DDB1-DET1 degradation pathway’? Further research is needed to clarify their relationship.
In addition, GLKs have shown a rosy application prospect. By regulating the gene expression of GLKs, not only can the photosynthetic efficiency of crops be increased which in turn improves crop yields, but leaf morphogenesis can also be changed. It makes GLKs potentially applicable to agronomic trait improvement, horticultural plant breeding and ornamental plant improvement. However, overexpression of GLKs has certain negative effects. For example, transgenic rice of ZmG1 drived by the constitutive promoter resulted in reduced seed size and no increase in yield (Yeh et al., 2022). Overexpression of OsGLK1 in rice causes abnormal tapetum development and low seed setting rates, and also increased endosperm chalkiness of rice grains (Zheng et al., 2022; Li et al., 2022c). To mitigate the potential negative effects, the expression level of GLKs may be tightly regulated by selecting appropriate promoters, or ‘Knock-up’ by gene-editing techniques (Lu et al., 2021; Wang et al., 2024). Accurate regulation of the expression of GLKs will help improve crop overall quality and bring breakthroughs in agricultural production.
Author contributions
MZ: Writing – original draft, Writing – review & editing. XW: Writing – original draft, Writing – review & editing. JL: Writing – original draft, Writing – review & editing. BM: Writing – review & editing. DL: Project administration, Supervision, Writing – review & editing. XC: Project administration, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was funded by the National Natural Science Foundation of China (32272096), Innovation and Development Program of the Beijing Vegetable Research Center (KYCX202304) and the Natural Science Foundation of Zhejiang Province (LZ23C130004 and Z24C130016).
Acknowledgments
We thank Dr. Tianhua He (Murdoch University, Australia) for critical reading of this manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1445875/full#supplementary-material
References
Ahmad, R., Liu, Y., Wang, T. J., Meng, Q., Yin, H., Wang, X., et al. (2019). GOLDEN2-LIKE transcription factors regulate WRKY40 expression in response to abscisic acid. Plant Physiol. 179, 1844–1860. doi: 10.1104/pp.18.01466
Alam, I., Manghwar, H., Zhang, H., Yu, Q., Ge, L. (2022). Identification of GOLDEN2-like transcription factor genes in soybeans and their role in regulating plant development and metal ion stresses. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1052659
Alem, A. L., Ariel, F. D., Cho, Y., Hong, J. C., Gonzalez, D. H., Viola, I. L. (2022). TCP15 interacts with GOLDEN2-LIKE 1 to control cotyledon opening in Arabidopsis. Plant J. 110, 748–763. doi: 10.1111/tpj.15701
Ali, N., Chen, H., Zhang, C., Khan, S. A., Gandeka, M., Xie, D., et al. (2020). Ectopic expression of AhGLK1b (GOLDEN2-like transcription factor) in Arabidopsis confers dual resistance to fungal and bacterial pathogens. Genes 11, 343. doi: 10.3390/genes11030343
An, X. H., Tian, Y., Chen, Y. H., Li, E. M., Li, M., Cheng, C. G. (2019). Functional identification of apple MdGLK1 which regulates chlorophyll biosynthesis in Arabidopsis. J. Plant Growth Regul. 38, 778–787. doi: 10.1007/s00344–018-9889–5
Bouché, F., Lobet, G., Tocquin, P., Perilleux, C. (2016). FLOR-ID: an interactive database of flowering-time gene networks in Arabidopsis thaliana. Nucleic Acids Res. 44, 1167–1171. doi: 10.1093/nar/gkv1054
Brand, A., Borovsky, Y., Hill, T., Rahman, K. A., Bellalou, A., Van Deynze, A., et al. (2014). CaGLK2 regulates natural variation of chlorophyll content and fruit color in pepper fruit. Theor. Appl. Genet. 127, 2139–2148. doi: 10.1007/s00122–014-2367-y
Chan, K. X., Phua, S. Y., Crisp, P., Mcquinn, R., Pogson, B. J. (2016). Learning the languages of the chloroplast: retrograde signaling and beyond. Annu. Rev. Plant Biol. 67, 25–53. doi: 10.1146/annurev-arplant-043015–111854
Chen, M., Ji, M., Wen, B., Liu, L., Li, S., Chen, X., et al. (2016). GOLDEN 2-LIKE transcription factors of plants. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01509
Chen, M., Liu, X., Jiang, S., Wen, B., Yang, C., Xiao, W., et al. (2018). Transcriptomic and functional analyses reveal that PpGLK1 regulates chloroplast development in peach (Prunus persica). Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00034
Chen, H., Qin, L., Tian, J., Wang, X. (2022). Identification and evolutionary analysis of the GOLDEN2-LIKE gene family in Foxtail Millet. Trop. Plant Biol. 15, 301–318. doi: 10.1007/s12042–022-09324–8
Cole-Osborn, L. F., Mccallan, S. A., Prifti, O., Abu, R., Sjoelund, V., Lee-Parsons, C. W. T. (2024). The role of the Golden2-like (GLK) transcription factor in regulating terpenoid indole alkaloid biosynthesis in Catharanthus roseus. Plant Cell Rep. 43, 141. doi: 10.1007/s00299–024-03208–9
Feng, P., Guo, H., Chi, W., Chai, X., Sun, X. W., Xu, X. M., et al. (2016). Chloroplast retrograde signal regulates flowering. PANS 113, 10708–10713. doi: 10.1073/pnas.1521599113
Fitter, D. W., Martin, D. J., Copley, M. J., Scotland, R. W., Langdale, J. A. (2002). GLK gene pairs regulate chloroplast development in diverse plant species. Plant J. 31, 713–727. doi: 10.1046/j.1365–313x.2002.01390.x
Gang, H., Li, R., Zhao, Y., Liu, G., Chen, S., Jiang, J. (2019). Loss of GLK1 transcription factor function reveals new insights in chlorophyll biosynthesis and chloroplast development. J. Exp. Bot. 70, 3125–3138. doi: 10.1093/jxb/erz128
Garapati, P., Xue, G. P., Munné-Bosch, S., Balazadeh, S. (2015). Transcription factor ATAF1 in Arabidopsis promotes senescence by direct regulation of key chloroplast maintenance and senescence transcriptional cascades. Plant Physiol. 168, 1122–1139. doi: 10.1104/pp.15.00567
Guo, Q., Jing, Y. J., Gao, Y., Liu, Y. T., Fang, X. F., Lin, R. C. (2023). The PIF1/PIF3-MED25-HDA19 transcriptional repression complex regulates phytochrome signaling in Arabidopsis. New Phytol. 240, 1097–1115. doi: 10.1111/nph.19205
Han, X. Y., Li, P. X., Zou, L. J., Tan, W. R., Zheng, T., Zhang, D. W., et al. (2016). GOLDEN2-LIKE transcription factors coordinate the tolerance to Cucumber mosaic virus in Arabidopsis. Biochem. Biophys. Res. Commun. 477, 626–632. doi: 10.1016/j.bbrc.2016.06.110
Hernández-Verdeja, T., Lundgren, M. R. (2023). GOLDEN2-LIKE transcription factors: A golden ticket to improve crops? Plants People Planet. 6, 79–93. doi: 10.1002/ppp3.10412
Jarvis, P., López-Juez, E. (2014). Biogenesis and homeostasis of chloroplasts and other plastids. Nat. Rev. Mol. Cell Biol. 14, 787–802. doi: 10.1038/nrm3702
Jia, T., Cheng, Y., Khan, I., Zhao, X., Gu, T., Hu, X. (2020). Progress on understanding transcriptional regulation of chloroplast development in fleshy fruit. Int. J. Mol. Sci. 21, 6951. doi: 10.3390/ijms21186951
John, C. R., Smith-Unna, R. D., Woodfield, H., Covshoff, S., Hibberd, J. M. (2014). Evolutionary convergence of cell-specific gene expression in independent lineages of C4 grasses. Plant Physiol. 165, 62–75. doi: 10.1104/pp.114.238667
Kakizaki, T., Matsumura, H., Nakayama, K., Che, F. S., Terauchi, R., Inaba, T. (2010). Coordination of plastid protein import and nuclear gene expression by plastid-to-nucleus retrograde signaling. PlantPhysiol 151, 1339–1353. doi: 10.1104/pp.109.145987
Kim, N., Jeong, J., Kim, J., Oh, J., Choi, G. (2023). Shade represses photosynthetic genes by disrupting the DNA binding of GOLDEN2-LIKE1. Plant Physiol. 191, 2334–2352. doi: 10.1093/plphys/kiad029
Klee, H. J., Giovannoni, J. J. (2011). Genetics and control of tomato fruit ripening and quality attributes. Annu. Rev. Genet. 45, 41–59. doi: 10.1146/annurev-genet-110410–132507
Kobayashi, K., Baba, S., Obayashi, T., Sato, M., Toyooka, K., Keränen, M., et al. (2012). Regulation of root greening by light and auxin/cytokinin signaling in Arabidopsis. Plant Cell. 24, 1081–1095. doi: 10.1105/tpc.111.092254
Kunkel, B. N., Brooks, D. M. (2002). Cross talk between signaling pathways in pathogen defense. Curr. Opin. Plant Biol. 5, 325–331. doi: 10.1016/s1369–5266(02)00275–3
Lambret-Frotte, J., Smith, G., Langdale, J. A. (2023). GOLDEN2-like 1 is sufficient but not necessary for chloroplast biogenesis in mesophyll cells of C4 grasses. Plant J. 117, 416–431. doi: 10.1007/s00299–024-03208–9
Lee, K. P., Li, M., Li, M., Liu, K., Medina-Puche, L., Qi, S., et al. (2023). Hierarchical regulatory module GENOMES UNCOUPLED1-GOLDEN2-LIKE1/2-WRKY18/40 modulates salicylic acid signaling. Plant Physiol. 192, 3120–3133. doi: 10.1093/plphys/kiad251
Leivar, P., Monte, E. (2014). PIFs: systems integrators in plant development. Plant Cel. 26, 56–78. doi: 10.1105/tpc.113.120857
Li, G., Chen, D., Tang, X., Liu, Y. (2018). Heterologous expression of kiwifruit (Actinidia chinensis) GOLDEN2-LIKE homolog elevates chloroplast level and nutritional quality in tomato (Solanum lycopersicum). Planta 247, 1351–1362. doi: 10.1007/s00425–018-2853–6
Li, Z., Gao, J., Wang, B., Xu, J., Fu, X., Han, H., et al. (2022c). Rice carotenoid biofortification and yield improvement conferred by endosperm-specific overexpression of OsGLK1. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.951605
Li, Y., Gu, C., Gang, H., Zheng, Y., Liu, G., Jiang, J. (2021). Generation of a golden leaf triploid poplar by repressing the expression of GLK genes. Forest. Res. 1. doi: 10.3389/fpls.2022.952877
Li, M., Lee, K. P., Liu, T., Vivek, D., Duan, J., Li, M., et al. (2022a). Antagonistic modules, SIB1 and LSD1, regulate photosynthesis-associated nuclear genes via GOLDEN2-LIKE transcription factors in Arabidopsis. Plant Physiol. 188, 2308–2324. doi: 10.1093/plphys/kiab600
Li, Y., Lei, W., Zhou, Z., Li, Y., Zhang, D., Lin, H. (2023b). Transcription factor GLK1 promotes anthocyanin biosynthesis via an MBW complex-dependent pathway in Arabidopsis thaliana. J. Integr. Plant Biol. 65, 1521–1535. doi: 10.1111/jipb.13471
Li, X., Li, J., Wei, S., Gao, Y., Pei, H., Geng, R., et al. (2023a). Maize GOLDEN2-LIKE proteins enhance drought tolerance in rice by promoting stomatal closure. Plant Physiol. 194, 774–786. doi: 10.1093/plphys/kiad561
Li, X., Lin, F., Li, C., Du, L., Liu, Z., Shi, W., et al. (2022b). Golden 2-like transcription factor contributes to the major QTL against rice black-streaked dwarf virus disease. Theor. Appl. Genet. 135, 4233–4243. doi: 10.1007/s00122–022-04214–9
Li, X., Wang, P., Li, J., Wei, S., Yan, Y., Yang, J., et al. (2020b). Maize GOLDEN2-LIKE genes enhance biomass and grain yields in rice by improving photosynthesis and reducing photoinhibition. Commun. Biol. 3, 151. doi: 10.1038/s42003–020-0887–3
Liu, X., Li, L., Li, M., Su, L., Lian, S., Zhang, B., et al. (2018). AhGLK1 affects chlorophyll biosynthesis and photosynthesis in peanut leaves during recovery from drought. Sci. Rep. 8, 2250. doi: 10.1038/s41598–018-20542–7
Liu, X., Li, L., Zhang, B., Zeng, L., Li, L. (2020). AhHDA1-mediated AhGLK1 promoted chlorophyll synthesis and photosynthesis regulates recovery growth of peanut leaves after water stress. Plant Sci. 294, 110461. doi: 10.1016/j.plantsci.2020.110461
Liu, J., Mehari, T. G., Xu, Y., Umer, M. J., Hou, Y., Wang, Y., et al. (2021). GhGLK1 a key candidate gene from GARP family enhances cold and drought stress tolerance in cotton. Front. Plant Sci. 12, 759312. doi: 10.3389/fpls.2021.759312
Liu, D., Zhao, D., Li, X., Zeng, Y. (2022). AtGLK2, an Arabidopsis GOLDEN2-LIKE transcription factor, positively regulates anthocyanin biosynthesis via AtHY5-mediated light signaling. Plant Growth Regul. 96, 79–90. doi: 10.1007/s10725–021-00759–9
Lu, Y., Wang, J. Y., Chen, B., Mo, S. D., Lian, L., Luo, Y. M., et al. (2021). A donor-DNA-free CRISPR/Cas-based approach to gene knock-up in rice. Nat. Plants. 7, 1445–1452. doi: 10.1038/s41477–021-01019–4
Lupi, A. C. D., Lira, B. S., Gramegna, G., Trench, B., Alves, F. R. R., Demarco, D., et al. (2019). Solanum lycopersicum GOLDEN 2-LIKE 2 transcription factor affects fruit quality in a light- and auxin-dependent manner. PloS One 14, 212224. doi: 10.1371/journal.pone.0212224
Majeran, W., Van Wijk, K. J. (2009). Cell-type-specific differentiation of chloroplasts in C4 plants. Trends Plant Sci. 14, 100–109. doi: 10.1016/j.tplants.2008.11.006
Martín, G., Leivar, P., Ludevid, D., Tepperman, J. M., Quail, P. H., Monte, E. (2016). Phytochrome and retrograde signalling pathways converge to antagonistically regulate a light-induced transcriptional network. Nat. Commun. 7, 11431. doi: 10.1038/ncomms11431
Miyake, H. (2016). Starch accumulation in the bundle sheaths of C3 plants: A possible pre-condition for C4 photosynthesis. Plant Cell Physiol. 57, 890–896. doi: 10.1093/pcp/pcw046
Murmu, J., Wilton, M., Allard, G., Pandeya, R., Desveaux, D., Singh, J., et al. (2014). Arabidopsis GOLDEN2-LIKE (GLK) transcription factors activate jasmonic acid (JA)-dependent disease susceptibility to the biotrophic pathogen Hyaloperonospora arabidopsidis, as well as JA-independent plant immunity against the necrotrophic pathogen Botrytis cinerea. Mol. Plant Pathol. 15, 174–184. doi: 10.1111/mpp.12077
Nadakuduti, S. S., Holdsworth, W. L., Klein, C. L., Barry, C. S. (2014). KNOX genes influence a gradient of fruit chloroplast development through regulation of GOLDEN2-LIKE expression in tomato. Plant J. 78, 1022–1033. doi: 10.1111/tpj.12529
Nagatoshi, Y., Mitsuda, N., Hayashi, M., Inoue, S., Okuma, E., Kubo, A., et al. (2016). GOLDEN2-LIKE transcription factors for chloroplast development affect ozone tolerance through the regulation of stomatal movement. Proc. Natl. Acad. Sci. U.S.A. 113, 4218–4223. doi: 10.1073/pnas.1513093113
Nakamura, H., Muramatsu, M., Hakata, M., Ueno, O., Nagamura, Y., Hirochika, H., et al. (2009). Ectopic overexpression of the transcription factor OsGLK1 induces chloroplast development in non-green rice cells. Plant Cell Physiol. 50, 1933–1949. doi: 10.1093/pcp/pcp138
Nguyen, C. V., Vrebalov, J. T., Gapper, N. E., Zheng, Y., Zhong, S., Fei, Z., et al. (2014). Tomato GOLDEN2-LIKE transcription factors reveal molecular gradients that function during fruit development and ripening. Plant Cell. 26, 585–601. doi: 10.1105/tpc.113.118794
Ni, F., Wu, L., Wang, Q., Hong, J., Qi, Y., Zhou, X. (2017). Turnip Yellow Mosaic Virus P69 interacts with and suppresses GLK transcription factors to cause Pale-Green symptoms in Arabidopsis. Mol. Plant 10, 764–766. doi: 10.1016/j.molp.2016.12.003
Niu, X. L., Li, H. L., Li, R., Liu, G. S., Peng, Z. Z., Jia, W., et al. (2022). Transcription factor SlBEL2 interferes with GOLDEN2-LIKE and influences green shoulder formation in tomato fruits. Plant J. 112, 982–997. doi: 10.1111/tpj.15989
Pan, Y. L., Pan, Y., Qu, C. M., Su, C. G., Li, J. H., Zhang, X. G. (2017). Identification and cloning of GOLDEN2-LIKE1 (GLK1), a transcription factor associated with chloroplast development in Brassica napus L. Genet. Mol. Res. 16. doi: 10.4238/gmr16018942
Pogson, B. J., Woo, N. S., Forster, B., Small, I. D. (2008). Plastid signalling to the nucleus and beyond. Trends Plant Sci. 13, 602–609. doi: 10.1016/j.tplants.2008.08.008
Powell, A. L., Nguyen, C. V., Hill, T., Cheng, K. L., Figueroa-Balderas, R., Aktas, H., et al. (2012). Uniform ripening encodes a Golden2-like transcription factor regulating tomato fruit chloroplast development. Science 336, 1711–1715. doi: 10.1126/science.1222218
Qu, H. X., Liang, S., Hu, L. F., Yu, L., Liang, P. X., Hao, Z. D., et al. (2024). Overexpression of Liriodendron Hybrid LhGLK1 in Arabidopsis leads to excessive chlorophyll synthesis and improved growth. Int. J. Mol. Sci. 25, 6869. doi: 10.3390/ijms25136968
Rauf, M., Arif, M., Dortay, H., Matallana-Ramírez, L. P., Waters, M. T., Gil Nam, H., et al. (2013). ORE1 balances leaf senescence against maintenance by antagonizing G2-like-mediated transcription. EMBO Rep. 14, 382–388. doi: 10.1038/embor.2013.24
Riechmann, J. L., Heard, J., Martin, G., Reuber, L., Jiang, C., Keddie, J., et al. (2000). Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290, 2105–2110. doi: 10.1126/science.290.5499.2105
Rossini, L., Cribb, L., Martin, D. J., Langdale, J. A. (2001). The maize golden 2 gene defines a novel class of transcriptional regulators in plants. Plant Cell. 13, 1231–1244. doi: 10.1105/tpc.13.5.1231
Safi, A., Medici, A., Szponarski, W., Ruffel, S., Lacombe, B., Krouk, G. (2017). The world according to GARP transcription factors. Curr. Opin. Plant Biol. 39, 159–167. doi: 10.1016/j.pbi.2017.07.006
Sagar, M., Chervin, C., Mila, I., Hao, Y., Roustan, J. P., Benichou, M., et al. (2013). SlARF4, an auxin response factor involved in the control of sugar metabolism during tomato fruit development. Plant Physiol. 161, 1362–1374. doi: 10.1104/pp.113.213843
Sakuraba, Y., Kim, E. Y., Han, S. H., Piao, W., An, G., Todaka, D., et al. (2017). Rice Phytochrome-Interacting Factor-Like1 (OsPIL1) is involved in the promotion of chlorophyll biosynthesis through feed-forward regulatory loops. J. Exp. Bot. 68, 4103–4114. doi: 10.1093/jxb/erx231
Savitch, L. V., Subramaniam, R., Allard, G. C., Singh, J. (2007). The GLK1 ‘regulon’ encodes disease defense related proteins and confers resistance to Fusarium graminearum in Arabidopsis. Biochem. Biophys. Res. Commun. 359, 234–238. doi: 10.1016/j.bbrc.2007.05.084
Shi, H., Wang, X., Mo, X., Tang, C., Zhong, S., Deng, X. W. (2015). Arabidopsis DET1 degrades HFR1 but stabilizes PIF1 to precisely regulate seed germination. Proc. Natl. Acad. Sci. U S A. 112, 3817–3822. doi: 10.1073/pnas.1502405112
Song, Y., Yang, C., Gao, S., Zhang, W., Li, L., Kuai, B. (2014). Age-triggered and dark-induced leaf senescence require the bHLH transcription factors PIF3, 4, and 5. Mol. Plant 7, 1776–1787. doi: 10.1093/mp/ssu109
Soudry, E., Ulitzur, S., Gepstein, S. (2005). Accumulation and remobilization of amino acids during senescence of detached and attached leaves: in planta analysis of tryptophan levels by recombinant luminescent bacteria. J Exp Bot. 56, 695–702. doi: 10.1093/jxb/eri054
Sukarta, O. C. A., Townsend, P. D., Llewelyn, A., Dixon, C. H., Slootweg, E. J., Pålsson, L. O., et al. (2020). A DNA-Binding Bromodomain-containing protein interacts with and reduces Rx1-mediated immune response to potato virus X. Plant Commun. 1, 100086. doi: 10.1016/j.xplc.2020.100086
Sun, T., Zeng, S., Wang, X., Owens, L. A., Fe, Z., Zhao, Y., et al. (2022). GLKs directly regulate carotenoid biosynthesis via interacting with GBFs in nuclear condensates in plants. bioRxiv. doi: 10.1101/2022.09.09.507346
Susila, H., Nasim, Z., Gawarecka, K., Jung, J. Y., Jin, S., Youn, G., et al. (2023). Chloroplasts prevent precocious flowering through a GOLDEN2-LIKE-B-BOX DOMAIN PROTEIN module. Plant Commun. 4, 100515. doi: 10.1016/j.xplc.2023.100515
Tachibana, R., Abe, S., Marugami, M., Yamagami, A., Akema, R., Ohashi, T., et al. (2024). BPG4 regulates chloroplast development and homeostasis by suppressing GLK transcription factors and involving light and brassinosteroid signaling. Nat. Commun. 15, 370. doi: 10.1038/s41467–023-44492–5
Taketa, S., Hattori, M., Takami, T., Himi, E., Sakamoto, W. (2021). Mutations in a Golden2-Like Gene Cause Reduced Seed Weight in Barley albino lemma 1 Mutants. Plant Cell Physiol. 62, 447–457. doi: 10.1093/pcp/pcab001
Tang, X., Miao, M., Niu, X., Zhang, D., Cao, X., Jin, X., et al. (2016). Ubiquitin-conjugated degradation of golden 2-like transcription factor is mediated by CUL4-DDB1-based E3 ligase complex in tomato. New Phytol. 209, 1028–1039. doi: 10.1111/nph.13635
Tokumaru, M., Adachi, F., Toda, M., Ito-Inaba, Y., Yazu, F., Hirosawa, Y., et al. (2017). Ubiquitinproteasome dependent regulation of the golden 2-like 1 transcription factor in response to plastid signals. Plant Physiol. 173, 524–535. doi: 10.1104/pp.16.01546
Tu, X., Ren, S., Shen, W., Li, J., Li, Y., Li, C., et al. (2022). Limited conservation in cross-species comparison of GLK transcription factor binding suggested wide-spread cistrome divergence. Nat. Commun. 13, 7632. doi: 10.1038/s41467–022-35438–4
Veciana, N., Martín, G., Leivar, P., Monte, E. (2022). BBX16 mediates the repression of seedling photomorphogenesis downstream of the GUN1/GLK1 module during retrograde signalling. New Phytol. 234, 93–106. doi: 10.1111/nph.17975
von Caemmerer, S., Quick, W. P., Furbank, R. T. (2012). The development of C4 rice: current progress and future challenges. Science 336, 1671–1672. doi: 10.1126/science.1220177
Wang, P., Fouracre, J., Kelly, S., Karki, S., Gowik, U., Aubry, S., et al. (2013). Evolution of GOLDEN2-LIKE gene function in C3 and C4 plants. Planta 237, 481–495. doi: 10.1007/s00425–012-1754–3
Wang, P., Khoshravesh, R., Karki, S., Tapia, R., Balahadia, C. P., Bandyopadhyay, A., et al. (2017b). Re-creation of a key step in the evolutionary switch from C3 to C4 leaf anatomy. Curr. Biol. 27, 3278–3287. doi: 10.1016/j.cub.2017.09.040
Wang, H., Seo, J. K., Gao, S., Cui, X., Jin, H. (2017a). Silencing of AtRAP, a target gene of a bacteria-induced small RNA, triggers antibacterial defense responses through activation of LSU2 and down-regulation of GLK1. New Phytol. 215, 1144–1155. doi: 10.1111/nph.14654
Wang, L., Tang, X., Zhang, S., Xie, X., Li, M., Liu, Y., et al. (2022). Tea GOLDEN2-LIKE genes enhance catechin biosynthesis through activating R2R3-MYB transcription factor. Hortic. Res. 9, 117. doi: 10.1093/hr/uhac117
Wang, H., Zhang, D., Chen, M., Meng, X., Bai, S., Xin, P., et al. (2024). Genome editing of 3’ UTR-embedded inhibitory region enables generation of gene knock-up alleles in plants. Plant Commun. 5, 100745. doi: 10.1016/j.xplc.2023.100745
Waters, M. T., Wang, P., Korkaric, M., Capper, R. G., Saunders, N. J., Langdale, J. A. (2009). GLK transcription factors coordinate expression of the photosynthetic apparatus in Arabidopsis. Plant Cell. 21, 1109–1128. doi: 10.1105/tpc.108.065250
Wu, R., Guo, L., Guo, Y., Ma, L., Xu, K., Zhang, B., et al. (2023). The G2-Like gene family in Populus trichocarpa: identification, evolution and expression profiles. BMC Genom. Data. 24, 37. doi: 10.1186/s12863–023-01138–1
Wu, R., Guo, L., Wang, R., Zhang, Q., Yao, H. (2022). Genome-wide identification and characterization of G2-Like transcription factor genes in moso bamboo (Phyllostachys edulis). Molecules 27, 5491. doi: 10.3390/molecules27175491
Xiong, B., Gong, Y., Li, Q., Li, L., Mao, H., Liao, L., et al. (2022). Genome-wide analysis of the GLK gene family and the expression under different growth stages and dark stress in sweet orange (Citrus sinensis). Horticulturae 8, 1076. doi: 10.3390/horticulturae8111076
Xu, Z., Zhou, Z., Cheng, Z., Zhou, Y., Wang, F., Li, M., et al. (2023). A transcription factor ZmGLK36 confers broad resistance to maize rough dwarf disease in cereal crops. Nat. Plants. 9, 1720–1733. doi: 10.1038/s41477–023-01514-w
Yang, Z., Bai, T., Zhiguo, E., Niu, B., Chen, C. (2024). OsNF-YB7 inactivates OsGLK1 to inhibit chlorophyll biosynthesis in rice embryo. bioRxiv. doi: 10.1101/2024.02.05.578907
Yang, S., Wang, X., Yan, W., Zhang, Y., Song, P., Guo, Y., et al. (2023). Melon yellow-green plant (Cmygp) encodes a Golden2-like transcription factor regulating chlorophyll synthesis and chloroplast development. Theor. Appl. Genet. 136, 66. doi: 10.1007/s00122–023-04343–9
Yasumura, Y., Moylan, E. C., Langdale, J. A. (2005). A conserved transcription factor mediates nuclear control of organelle biogenesis in anciently diverged land plants. Plant Cell. 17, 1894–1907. doi: 10.1105/tpc.105.033191
Yeh, S. Y., Lin, H. H., Chang, Y. M., Chang, Y. L., Chang, C. K., Huang, Y. C., et al. (2022). Maize Golden2-like transcription factors boost rice chloroplast development, photosynthesis, and grain yield. Plant Physiol. 188, 442–459. doi: 10.1093/plphys/kiab511
Yelina, N. E., Frangedakis, E., Wang, Z., Schreier, T. B., Rever, J., Tomaselli, M., et al. (2024). Streamlined regulation of chloroplast development in the liverwort Marchantia polymorpha. bioRxiv. doi: 10.1101/2023.01.23.525199
Ying, J., Wang, Y., Xu, L., Yao, S., Wang, K., Dong, J., et al. (2023). RsGLK2.1-RsNF-YA9a module positively regulates the chlorophyll biosynthesis by activating RsHEMA2 in green taproot of radish. Plant Sci. 334, 111768. doi: 10.1016/j.plantsci.2023.111768
Yuan, Y., Mei, L., Wu, M., Wei, W., Shan, W., Gong, Z., et al. (2018). SlARF10, an auxin response factor, is involved in chlorophyll and sugar accumulation during tomato fruit development. J. Exp. Bot. 69, 5507–5518. doi: 10.1093/jxb/ery328
Zeng, X., Ye, L., Zhang, R., Wang, P. (2023). Transcription factor GLK2 regulates key steps of anthocyanin biosynthesis to antagonize photo-oxidative stress during greening of Arabidopsis seedlings. bioRxiv. doi: 10.1101/2023.03.10.532066
Zhang, H., Ji, Y., Jing, Y., Li, L., Chen, Y., Wang, R., et al. (2022a). Arabidopsis SIGMA FACTOR BINDING PROTEIN1 (SIB1) and SIB2 inhibit WRKY75 function in abscisic acid-mediated leaf senescence and seed germination. J. Exp. Bot. 73, 182–196. doi: 10.1093/jxb/erab391
Zhang, Q. W., Mao, Y. Y., Zhao, Z. K., Hu, X., Hu, R., Yin, N. W., et al. (2024a). A Golden2-like transcription factor, BnGLK1a, improves chloroplast development, photosynthesis, and seed weight in rapeseed. J. Integr. Agric. 23, 1481–1493. doi: 10.1016/j.jia.2023.06.020
Zhang, L., Qian, J., Han, Y., Jia, Y., Kuang, H., Chen, J. (2022b). Alternative splicing triggered by the insertion of a CACTA transposon attenuates LsGLK and leads to the development of pale-green leaves in lettuce. Plant J. 109, 182–195. doi: 10.1111/tpj.15563
Zhang, D., Tan, W., Yang, F., Han, Q., Deng, X., Guo, H., et al. (2021). A BIN2-GLK1 signaling module integrates brassinosteroid and light signaling to repress chloroplast development in the dark. Dev. Cell. 56, 310–324. doi: 10.1016/j.devcel.2020.12.001
Zhang, T., Zhang, R., Zeng, X. Y., Lee, S., Ye, L. H., Tian, S. L., et al. (2024b). GLK transcription factors accompany ELONGATED HYPOCOTYL5 to orchestrate light-induced seedling development in Arabidopsis. Plant Physiol. 194, 2400–2421. doi: 10.1093/plphys/kiae002
Zheng, S. Y., Dong, J. F., Lu, J. Q., Li, J., Jiang, D. G., Yu, H. P., et al. (2022). A cytosolic pentatricopeptide repeat protein is essential for tapetal plastid development by regulating OsGLK1 transcript levels in rice. New Phytol. 234, 1678–1695. doi: 10.1111/nph.18105
Keywords: GOLDEN2-LIKEs (GLKs), transcription factor, function, signalling pathway, in plants
Citation: Zheng M, Wang X, Luo J, Ma B, Li D and Chen X (2024) The pleiotropic functions of GOLDEN2-LIKE transcription factors in plants. Front. Plant Sci. 15:1445875. doi: 10.3389/fpls.2024.1445875
Received: 08 June 2024; Accepted: 31 July 2024;
Published: 19 August 2024.
Edited by:
Huihui Li, Chinese Academy of Agricultural Sciences, ChinaReviewed by:
Jean-David Rochaix, University of Geneva, SwitzerlandYanjun Jing, Chinese Academy of Sciences (CAS), China
Copyright © 2024 Zheng, Wang, Luo, Ma, Li and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dayong Li, bGlkYXlvbmdAbmVyY3Yub3Jn; Xifeng Chen, eGZjaGVuQHpqbnUuY24=
†These authors share first authorship
‡ORCID: Xinyu Wang, orcid.org/0009-0009-7314-8915
 Mengyi Zheng
Mengyi Zheng Xinyu Wang
Xinyu Wang Jie Luo1
Jie Luo1 Dayong Li
Dayong Li Xifeng Chen
Xifeng Chen