
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 18 September 2024
Sec. Plant Pathogen Interactions
Volume 15 - 2024 | https://doi.org/10.3389/fpls.2024.1444271
This article is part of the Research Topic Innovative Strategies for Enhancing Plant Resilience to Phytopathogenic Microbes View all 9 articles
 Tania Chancellor1†
Tania Chancellor1† Daniel P. Smith1†
Daniel P. Smith1† Wanxin Chen1
Wanxin Chen1 Suzanne J. Clark1
Suzanne J. Clark1 Eudri Venter1†
Eudri Venter1† Kirstie Halsey1
Kirstie Halsey1 Esther Carrera2
Esther Carrera2 Vanessa McMillan1†
Vanessa McMillan1† Gail Canning1
Gail Canning1 Victoria J. Armer1
Victoria J. Armer1 Kim E. Hammond-Kosack1*
Kim E. Hammond-Kosack1* Javier Palma-Guerrero1†
Javier Palma-Guerrero1†Take-all disease, caused by the Ascomycete fungus Gaeumannomyces tritici, is one of the most important root diseases of wheat worldwide. The fungus invades the roots and destroys the vascular tissue, hindering the uptake of water and nutrients. Closely related non-pathogenic species in the Magnaporthaceae family, such as Gaeumannomyces hyphopodioides, occur naturally in arable and grassland soils and have previously been reported to reduce take-all disease in field studies. However, the mechanism of take-all protection has remained unknown. Here, we demonstrate that take-all control is achieved via local but not systemic host changes in response to prior G. hyphopodioides root colonisation. A time-course wheat RNA sequencing analysis revealed extensive transcriptional reprogramming in G. hyphopodioides–colonised tissues, characterised by a striking downregulation of key cell wall–related genes, including genes encoding cellulose synthases (CESA), and xyloglucan endotransglucosylase/hydrolases (XTH). In addition, we characterise the root infection biologies of G. tritici and G. hyphopodioides in wheat. We investigate the ultrastructure of previously described “subepidermal vesicles” (SEVs), dark swollen fungal cells produced in wheat roots by non-pathogenic G. hyphopodioides, but not by pathogenic G. tritici. We show that G. hyphopodioides SEVs share key characteristics of fungal resting structures, containing a greater number of putative lipid bodies and a significantly thickened cell wall compared to infection hyphae. We hypothesise that SEVs are fungal resting structures formed due to halted hyphal growth in the root cortex, perhaps as a stress response to locally induced wheat defence responses. In the absence of take-all resistant wheat cultivars or non-virulent G. tritici strains, studying closely related non-pathogenic G. hyphopodioides provides a much needed avenue to elucidate take-all resistance mechanisms in wheat.
Wheat (Triticum aestivum) is one of the most important cereal crops worldwide, providing around 20% of human caloric intake globally. Sustaining excellent root health is critical for the acquisition of water and essential nutrients. As global temperatures continue to rise, root health is predicted to face increasing threats from various soil-borne fungal pathogens (Delgado-Baquerizo et al., 2020). The necrotrophic fungal pathogen Gaeumannomyces tritici, belonging to the Magnaporthaceae family, is responsible for take-all disease, one of the most important root problems of wheat crops worldwide (Freeman and Ward, 2004; Palma-Guerrero et al., 2021). The disease drastically diminishes grain yields during heavy infection episodes. However, due to the genetic intractability of G. tritici, both the pathogen and the cereal-pathosystem remain understudied by the molecular plant–microbe interaction community. Root-confined vascular infection by G. tritici results in the development of characteristic necrotic lesions originating from the stele, which severely disrupt root functions, causing premature crop ripening and reduced grain yield/quality (Asher and Shipton, 1981; Huang et al., 2001). Take-all fungal inoculum builds up in the soil following consecutive wheat crops, and, although recent surveys of take-all disease levels are lacking, yield losses of up to 60% have been reported in the UK (McMillan et al., 2011). At present, take-all resistant wheat cultivars are not commercially available, and current fungicide seed treatments do not provide complete protection (Freeman et al., 2005). For a comprehensive review on our current understanding of the G. tritici–wheat pathogen system, including detailed spatial and temporal descriptions of the G. tritici life cycle and infection process, please see the work of Palma-Guerrero et al. (2021).
Understanding root immunity is essential for the development of take-all resistant cultivars. However, the classical model of immunity, characterised by the concerted effect of pathogen-associated molecular pattern (PAMP)–triggered immune responses and effector-triggered immune responses, is predominantly based on foliar pathogens (Boller and Felix, 2009; Jones and Dangl, 2006; Pok et al., 2022). Roots must constantly interact with a diverse soil microbiome and distinguish pathogenic microbes from, sometimes closely related, non-pathogenic endophytes or beneficial symbionts (Thoms et al., 2021). How plants engage with beneficial microorganisms while restricting damaging pathogens is regarded as one of the top 10 unanswered questions by the molecular plant–microbe interaction (MPMI) research community (Harris et al., 2020). The selective response of plants to microbes with different lifestyles can be partly explained by the compartmentalisation of localised immune responses in roots (Zhou et al., 2020), and the recognition of microbe-associated molecular patterns (MAMPs), damage-associated molecular patterns (DAMPs), and PAMPs by multiple receptors (Thoms et al., 2021). However, further comparative studies into endophytic and pathogenic plant infecting microbes are sorely needed.
Several members of the Magnaporthaceae family are classified within the Gaeumannomyces-Phialophora complex (Hernández-Restrepo et al., 2016). Phialophora species, such as Gaeumannomyces hyphopodioides, occur naturally in grasslands and arable field sites although do not cause disease symptoms in arable crops (Deacon, 1973; Ulrich et al., 2000; Ward and Bateman, 1999). For this reason, such species have been described as “non-pathogenic.” Wheat root colonisation by non-pathogenic Magnaporthaceae species can be easily distinguished from wheat root infection by pathogenic G. tritici due to the inability of non-pathogenic species to infect the vascular tissues and due to the production of dark swollen fungal cells by non-pathogenic species in the root cortex. The swollen cells measure between 12 µm and 30 µm in diameter, depending on the fungal species, and may be formed following growth cessation of a hyphal apex (Deacon, 1976a). These enigmatic structures have been previously described as pigmented cells (Holden, 1976), growth cessation structures (Deacon, 1976a), or subepidermal vesicles (SEVs) (Osborne et al., 2018). The closely related rice leaf blast pathogen Magnaporthe oryzae is also reported to form SEV-like structures in cereal roots. M. oryzae can infect rice root tissues, producing brown spherical structures resembling SEVs in epidermal and cortical cells (Dufresne and Osbourn, 2001; Marcel et al., 2010). SEV-like structures are also common among dark septate endophytes (DSEs), a diverse group of root colonising ascomycete fungi with elusive functions, often found inhabiting stressful environments. The DSEs are morphologically characterised by the production of highly melanised septate hyphae which grow intraradically, producing structures referred to as “microsclerotia,” but which bear striking similarities to the SEVs of the Magnaporthaceae species (Ban et al., 2012; Knapp et al., 2018). The function of these “microsclerotia” in the fungal life cycle of the DSE remains unknown, and their ultrastructure has yet to be investigated. Thus, SEVs appear to be specific to root colonising/infecting fungi, although little is known about SEV formation, SEV ultrastructure, or the involvement of SEVs in root infection processes.
Prior colonisation by certain non-pathogenic Magnaporthaceae species is reported to provide protection against take-all disease in field studies (Wong et al., 1996). Furthermore, Osborne et al. (2018) demonstrated that certain elite winter wheat varieties have an improved ability to promote G. hyphopodioides populations in field soils, suggesting that careful cultivar choice during wheat rotations could provide a natural level of biocontrol. However, as far as we are aware, disease protection by non-pathogenic Magnaporthaceae species has not been reported in any recent publications, and the mechanisms underlying disease control remain unknown.
Root endophytes can provide protection against plant pathogens directly or indirectly. Direct antagonism may be achieved by mycoparasitism, antibiosis, or competition for nutrients, whereas indirect antagonism can be achieved via host-mediated defence responses. Importantly, the exact mechanisms underpinning host-mediated defence can differ between the endophyte species and the host plant in question. The two most well characterised forms of induced resistance, induced systemic resistance (ISR) and systemic acquired resistance (SAR), can be distinguished by the type of elicitors involved and the signalling pathways that are induced (Pieterse et al., 2014). SAR can be triggered by plant detection of both MAMPs and DAMPs and is dependent on salicylic acid (SA) signalling. Conversely, ISR is generally induced via the plant detection of MAMPs and results in the priming of jasmonic acid (JA)/ethylene (ET)–responsive genes, resulting in a faster and stronger defence response against subsequent pathogen attacks (Choudhary et al., 2007). However, induced resistance mechanisms are not restricted to SAR and ISR. Non-pathogenic strains in the Fusarium oxysporum species complex are known to provide protection against Fusarium wilt disease, a major disease caused by pathogenic F. oxysporum strains (de Lamo and Takken, 2020). This phenomenon, termed by the authors as endophyte-mediated resistance (EMR), is reportedly independent of JA, ET, and SA signalling, although little else is known about the mechanisms responsible for EMR (Constantin et al., 2019). Serendipita indica (= Pirisformospora indica), an endophyte with a diverse host range, is known to provide disease resistance against a large number of fungal pathogens via multiple defence pathways (Qiang et al., 2012). In wheat, pre-treatment with S. indica provided effective protection against root rot disease caused by Fusarium graminearum (Li et al., 2022). Although the mechanism of protection is likely to be complex, transcriptomic analyses suggested that the activation of mitogen-activated protein kinase (MAPK) cascades involved in immunity, and the upregulation of phenylpropanoid biosynthesis genes may play a role in the observed disease resistance (Li et al., 2022). In summary, EMR mechanisms are species-specific and highly complex. Detailed in vitro, in planta, and transcriptomic analyses are needed to determine the mode(s) of EMR in specific plant species.
In this study, we set out to elucidate the mechanism of take-all disease control by G. hyphopodioides in wheat using in vitro and in planta assays combined with detailed host transcriptome profiling. In addition, we aimed to better characterise the infection/colonisation processes of G. tritici and G. hyphopodioides. Finally, we aimed to study the ultrastructure of fungal SEVs produced by G. hyphopodioides. We provide the first comparative analysis of wheat transcriptional responses to G. tritici and G. hyphopodioides across key stages of early fungal infection. We characterise the different fungal structures produced and some of the wheat cell-wall changes occurring during root infection. Our findings shed light on the distinct plant responses to these two closely related root infecting fungi with contrasting lifestyles and help to pinpoint localised mechanisms for the control of take-all disease by G. hyphopodioides. Together, our findings contribute to an improved understanding of wheat root resistance against take-all disease.
G. hyphopodioides (taxon id: 1940676) and G. tritici (taxon id: 36779) strains were isolated from field soils at Rothamsted Farm using the soil baiting method (McMillan et al., 2011; Osborne et al., 2018). Fungal isolates (see Table 1) were maintained on potato dextrose agar (PDA) plates at 21°C in the dark.
For seedling pot experiments, plastic pots (7.5-cm wide × 11-cm tall) were filled with damp horticultural sand, and 10 untreated wheat seeds cv. Hereward were sown on the surface. Hereward was chosen due to its strong susceptibility to take-all disease and its ability to build high levels of take-all inoculum (McMillan et al., 2011). Seeds were covered with a thin layer of grit and pots were placed in a controlled environment growth room for 2 weeks (16-h day, light intensity of 250 µmols, 15°C day, 10°C night). G. tritici (isolate 16.NZ.1d) and G. hyphopodioides (isolate NZ.129.2C.17) inocula were prepared by placing 10 fungal plugs (7-mm diameter) taken from the leading edge of each colony into a 1-L flask containing 400 mL of potato dextrose broth. Flasks were placed in an orbital incubator for 7 days at 25°C, 120 revolutions per minute (RPM). Liquid cultures were homogenised by passing through a 2.8-mm sterile sieve. Homogenised cultures were diluted with sterile distilled water in a 2:3 ratio. The first inoculum treatment was added into the pots after 2 weeks of plant growth. Inoculum (50 mL) was poured directly onto the root system using a funnel inserted into the sand. All seedlings were harvested 3 weeks after the final inoculum addition to allow take-all disease symptoms to develop (see Supplementary Figure S1A). Five replicates were prepared per treatment, and the experiment was repeated twice. An additional experiment using wheat cv. Chinese Spring was also carried out due to the availability of an annotated genome (IWGSC RefSeq v2.1) (Zhu et al., 2021).
For split-root experiments, roots from 2-week-old wheat seedlings (cv. Chinese Spring) were split across two pots (pot A and pot B) joined at one side. Pots were filled with sand and covered with grit. Roots in pot A received G. hyphopodioides liquid inoculum (isolate NZ.129.2C.17), using the method described above. Plants were left to grow for one week before inoculating with G. tritici liquid inoculum (isolate 17LH(4)19d1). To investigate whether G. hyphopodiodes provides local control against take-all disease, G. tritici inoculum was added to G. hyphopodioides–colonised roots in pot A. To investigate whether G. hyphopodiodes provides systemic control against take-all disease, G. tritici inoculum was added to uninoculated roots in pot B (see Supplementary Figure S1B). Plants were harvested 3 weeks later. Five replicates were prepared per treatment, and the experiment was repeated three times.
Visual disease assessments were carried out as previously described (McMillan et al., 2011), and qPCR quantification of G. tritici fungal biomass was performed by targeting a 105-bp partial DNA sequence of the translation elongation factor 1-alpha (EF1-α) gene, using primers GtEFF1 (5′-CCCTGCAAGCTCTTCCTCTTAG-3′) and GtEFR1 (5′-GCATGCGAGGTCCCAAAA-3′) with the TaqMan probe (5′-6FAM-ACTGCACAGACCATC-MGB-3′) (Thermo Scientific™, USA) (Keenan et al., 2015).
A precision inoculation method was developed to enable the investigation of local plant responses to fungal infection (see Supplementary Figure S2). Wheat seeds cv. Chinese Spring were surface sterilised with 5% (v/v) sodium hypochlorite for 5 min and pre-germinated in a controlled environment growth chamber cabinet (20°C day, 16°C night, 16-h light cycle) for 2 days. Three pre-germinated seeds were transplanted onto a square petri dish plate (12 cm × 12 cm) containing 1.5% (w/v) water agar. Five replicates were prepared for each treatment. Plates were placed vertically in the growth cabinet. After 4 days, one root from each plant was inoculated with a fungal plug (4 cm × 0.5 cm) cut from the leading edge of a 2-week-old fungal colony growing on 1.5% water agar. Inoculated roots were sampled daily from 2 to 6 days postinoculation (dpi). Briefly, two 1-cm root samples were harvested from the inoculated area on each root and snap-frozen in liquid nitrogen for RNA extraction. To determine the stage of fungal colonisation in these harvested samples, 2 cm × 0.5 cm root pieces were sampled from the areas directly above and below each sample. Root pieces were stored in 50% ethanol for subsequent assessment by confocal microscopy.
To assess colonisation in whole root pieces, samples were cleared in 10% w/v potassium hydroxide for 5 min at 70°C, before staining with propidium iodide (PI) (10 µg/mL) and wheat germ agglutinin (WGA), Alexa Fluor™ 488 Conjugate (WGA) (10 µg/mL). To visualise vascular infection by G. tritici, transversal root sections were cut by hand using a fine edged razor blade under a dissecting microscope. Confocal microscopy was performed using a ZEISS 780 Confocal Laser Scanning Microscope (ZEISS, Germany). WGA fluorescence was excited at 495 nm and detected at 519 nm. PI fluorescence was excited at 535 nm and detected at 617 nm.
Following confocal assessment (see above), root pieces (1 cm each) at the same stage of fungal colonisation were pooled together to create a single sample for RNA extraction. Total RNA was extracted from frozen root material using the E.Z.N.A.® Plant RNA Kit with the associated RNase-free DNase I Set (Omega-Biotek, USA), following the standard protocol provided. RNA quality was assessed based on the RNA integrity number (RIN), measured using the Bioanalyser 2100 with the corresponding RNA 6000 Nano Kit (Agilent, USA), as per manufacturer’s instructions.
mRNA library preparation was carried out by Novogene (China) using the Novogene RNA Library Prep Set (PT042) for polyA enrichment. Libraries were sequenced by Illumina NovaSeq to generate 150-bp paired-end reads, with a target of 40 million paired-end reads per sample.
Quality control of reads was performed using MultiQC (https://multiqc.info/). Sequence trimming of recognised adaptors was performed using Trimmomatic where appropriate (Bolger et al., 2014). Reads were mapped to the Chinese Spring (IWGSC RefSeq v2.1) (Zhu et al., 2021) using HiSat2 (Kim et al., 2019). To ensure that fungal biomass was consistent among replicates of the same treatment, reads were also mapped to the G. tritici genome (Okagaki et al., 2015). Three samples were identified as outliers based on standardised residuals of the percentage of reads mapped to G. tritici. Outliers were subsequently excluded from further analyses (Supplementary Table S10). All treatments contained at least four biological replicates, with the majority containing five biological replicates. Reads were not aligned to G. hyphopodioides due to the lack of a high-quality genome. Count determination was performed using FeatureCounts (Liao et al., 2014) on the R Bioconductor platform (https://bioconductor.org/).
Library normalisation and differential expression (DE) calling was carried out using the Bioconductor package DESeq2 (Love et al., 2014) in R studio. Gene expression levels were compared between G. tritici–infected and G. hyphopodioides–colonised samples and the uninoculated control samples for each time point individually. DE genes were identified by applying a log2 fold change filter of ≥1 or ≤−1. The DESeq2 implementation of Benjamini–Hochberg (Benjamini and Hochberg, 1995) was used to control for multiple testing (q < 0.05). Gene Ontology (GO) enrichment analysis was performed for significantly up- and downregulated wheat genes separately via http://www.geneontology.org, using the Panther classification system.
Statistical analyses were done using Genstat 20th Edition (VSN International Ltd, Hemel Hempstead, UK). Percentage disease data were analysed using a Generalised Linear Regression Model (GLM) with a binomial distribution and LOGIT link function. Analyses were adjusted for over-dispersion and treatment effects tested using deviance ratios (F-statistics) when the residual mean deviance was greater than 1. Data were back-transformed from the LOGIT scale [using the equation exp(x)/(1+exp(x))] for graphical presentation. For continuous outcome variables, namely fungal colony diameter, plant biomass, G. tritici fungal biomass and phytohormone concentration, data were analysed by analysis of variance (ANOVA). Prior to analysis, data were checked for normality by assessing histograms of residuals. G. tritici fungal biomass data were square root transformed prior to ANOVA testing, and data were back-transformed for graphical presentation. Tukey’s multiple comparisons test was carried out when more than one interaction was of interest.
To investigate the role of direct hyphal interaction in take-all control, a series of fungal confrontation assays were conducted on PDA plates. Prior to hyphal contact, the individual growth rates of G. tritici and G. hyphopodioides colonies did not significantly differ from the dual colony controls (Supplementary Table S1). The same was true when the two species were grown in a “sandwich” plate set-up (Figure 1A, Supplementary Table S2), suggesting that, prior to hyphal contact, G. hyphopodioides does not produce diffusible antifungal compounds or volatile organic compounds capable of inhibiting G. tritici. When hyphae of the two fungal species interacted in confrontation assays, a dark barrage was observed in the interaction zone (Figure 1A). The observed barrage formed 1–2 days following hyphal interaction, perhaps suggesting that direct interaction causes hyphal stress in at least one of the interacting species. A dark barrage was not observed when isolates of the same species were confronted (Supplementary Figure S3). Despite in vitro barrage formation, fungal antibiosis is unlikely to be the mechanism responsible for take-all control.
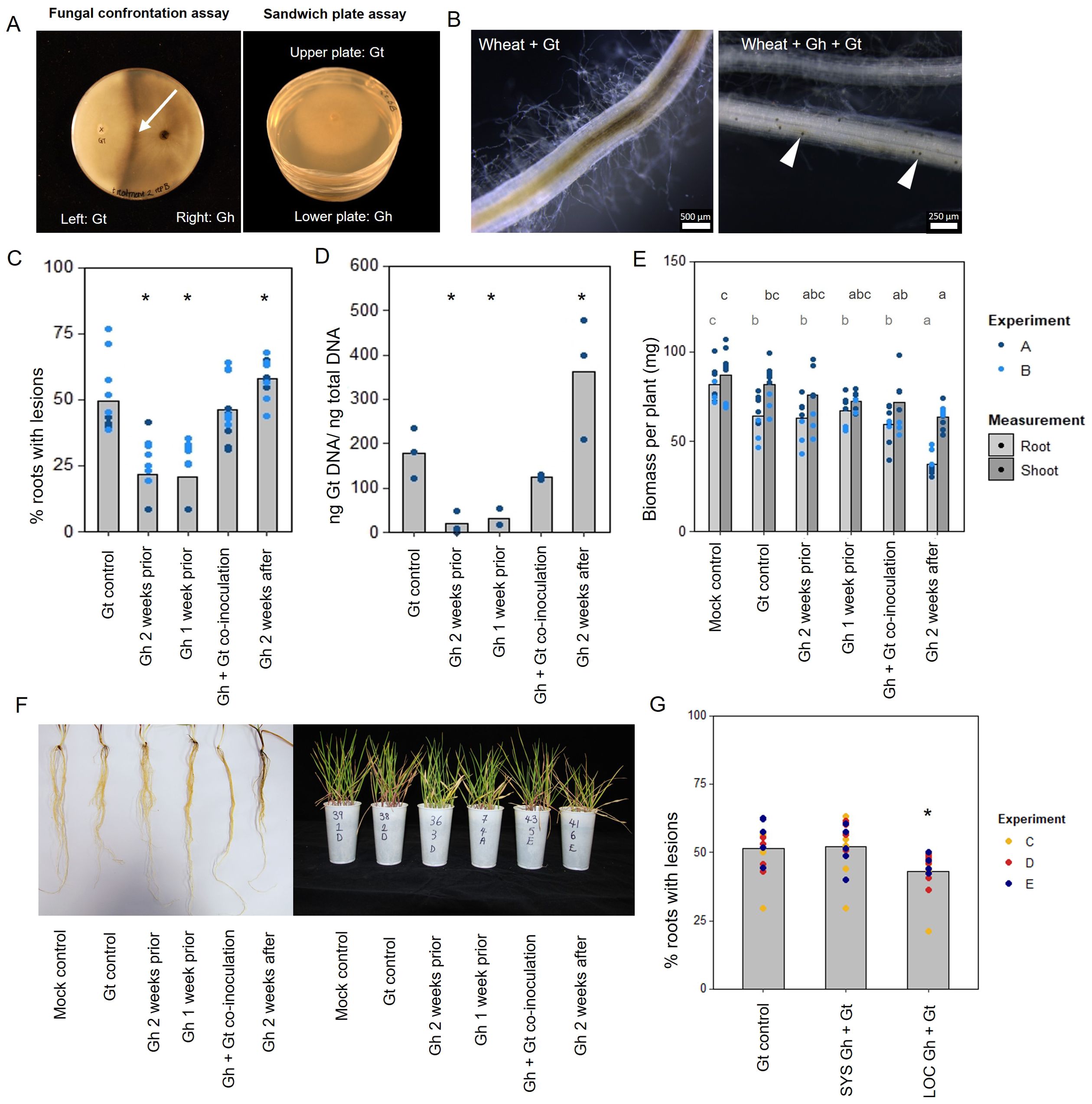
Figure 1. In vitro and in planta interaction studies involving endophytic G. hyphopodioides and pathogenic G. tritici. (A) In vitro fungal interaction assays on PDA plates. Fungal confrontation assays imaged 2 days following hyphal interaction, and colonies in sandwich plate assays imaged 6 days after establishment. Arrow indicates a dark barrage in the interaction zone. (B) Stereomicroscope images of wheat roots infected with G. tritici only or co-inoculated with G. hyphopodioides. Arrowheads indicate G. hyphopodioides sub-epidermal vesicles (SEVs). (C) Percentage of wheat roots (cv. Hereward) with take-all root lesions in co-inoculation experiments with G. hyphopodioides (GLM: F = 25.99, d.f. 4, 49, p < 0.001). (D) G. tritici fungal biomass (ng G. tritici DNA/ng total DNA) in co-inoculation experiments with G. hyphopodioides, as quantified by qPCR (F = 61.10, d.f. 4, 38, p < 0.001). (E) Shoot and root dry biomass (mg) in G. tritici co-inoculation experiments with G. hyphopodioides (F = 6.49, d.f. 5, 36, p < 0.001; F = 4.50, d.f. 5, 36, p < 0.01, respectively). (F) Representative images of wheat roots (left) and shoots (right) in co-inoculation experiments. (G) The percentage of wheat roots (cv. Chinese Spring) with take-all lesions in split root co-inoculation experiments with G. hyphopodioides (F = 29.44, d.f. 2, 36, p = 0.007). Asterisks indicate a significant difference to the G. tritici control as calculated by Dunnett’s post-hoc test (p < 0.05). Letters indicate significant differences as calculated by Tukey’s multiple comparisons test (p < 0.05). Gt, G. tritici; Gh, G. hyphopodioides; SYS, systemic; LOC, local.
To investigate the hypothesis that non-pathogenic G. hyphopodioides provides protection against take-all disease by inducing wheat resistance, seedling co-inoculation experiments were carried out under controlled environment conditions. G. hyphopodioides inoculum was added to wheat seedlings (cv. Hereward) growing in pots 1 week prior, 2 weeks prior, at the same time as, and 2 weeks after inoculation with pathogenic G. tritici. Characteristic black necrotic root lesions were observed in G. tritici–infected control plants. SEVs were observed in plants co-inoculated with G. hyphopodioides (Figure 1B). The data revealed a significant reduction in both take-all disease levels and G. tritici fungal biomass in plants pre-treated with G. hyphopodioides 2 weeks prior or 1 week prior to G. tritici inoculation (Figures 1C, D). Hence, even very early colonisation by G. hyphopodioides is sufficient for take-all control. These findings were consistent with additional experiments involving a different G. tritici isolate (Gt 17LH(4)19d1) and wheat cultivar (cv. Chinese Spring) (Supplementary Figure S4).
Seedlings co-inoculated with G. hyphopodioides and G. tritici at the same time had no effect on take-all disease levels or G. tritici fungal biomass. Importantly, adding G. hyphopodioides after G. tritici resulted in increased levels of take-all disease and G. tritici fungal biomass (Figures 1C, D). Furthermore, the shoot and root dry biomass of plants in this latter treatment were significantly reduced, indicating that seedling health is negatively affected when G. hyphopodioides infections occur in addition to G. tritici infection (Figures 1E, F). These findings suggest that the earlier G. tritici infections somehow lead to the suppression of critical local defence responses, and this now permits G. hyphopodioides to colonise throughout the wheat root system. These findings should be taken into careful consideration when evaluating the potential of G. hyphopodioides as a biocontrol agent.
Split-root experiments were carried out to determine whether G. hyphopodioides provides local or systemic protection against take-all disease. Significant disease reduction was achieved only in roots which had been directly inoculated with G. hyphopodioides (LOC) and not in systemic roots (SYS), which had not been directly inoculated with G. hyphopodioides (Figure 1G). Taken together, we demonstrate that local induced wheat resistance plays a crucial role in the control of take-all disease by G. hyphopodioides, and this response is consistent across both winter and spring wheat types.
To study fungal infection processes during early root colonisation, wheat seedlings (cv. Chinese Spring) were root inoculated with either G. hyphopodioides (NZ.129.2C.17) or G. tritici (Gt 17LH(4)19d1) in an agar plate system. Plants were harvested at 2, 4, and 5 dpi to capture key stages of fungal infection for later RNA-seq analysis. At 2 dpi, very few hyaline runner hyphae were detected on the root surface of plants inoculated with either G. tritici or G. hyphopodioides, and hyphae had not yet penetrated the epidermal cells in either interaction (Figures 2A, B). By 4 dpi, hyaline runner hyphae covered a greater area of the root surface (Supplementary Figure S5) and hyphae were detected in epidermal and cortical cells of both G. tritici– and G. hyphopodioides–inoculated roots (Figures 2A, B). At 5 dpi, hyaline runner hyphae were detected across a large area of the root surface (Supplementary Figure S5) in both fungal treatments. G. tritici hyphae infected the stele, whereas G. hyphopodioides hyphal growth was arrested in the cortex (Figure 2B). G. hyphopodioides hyphae were detected in cortical cells, from which SEVs were formed (Figure 2A).
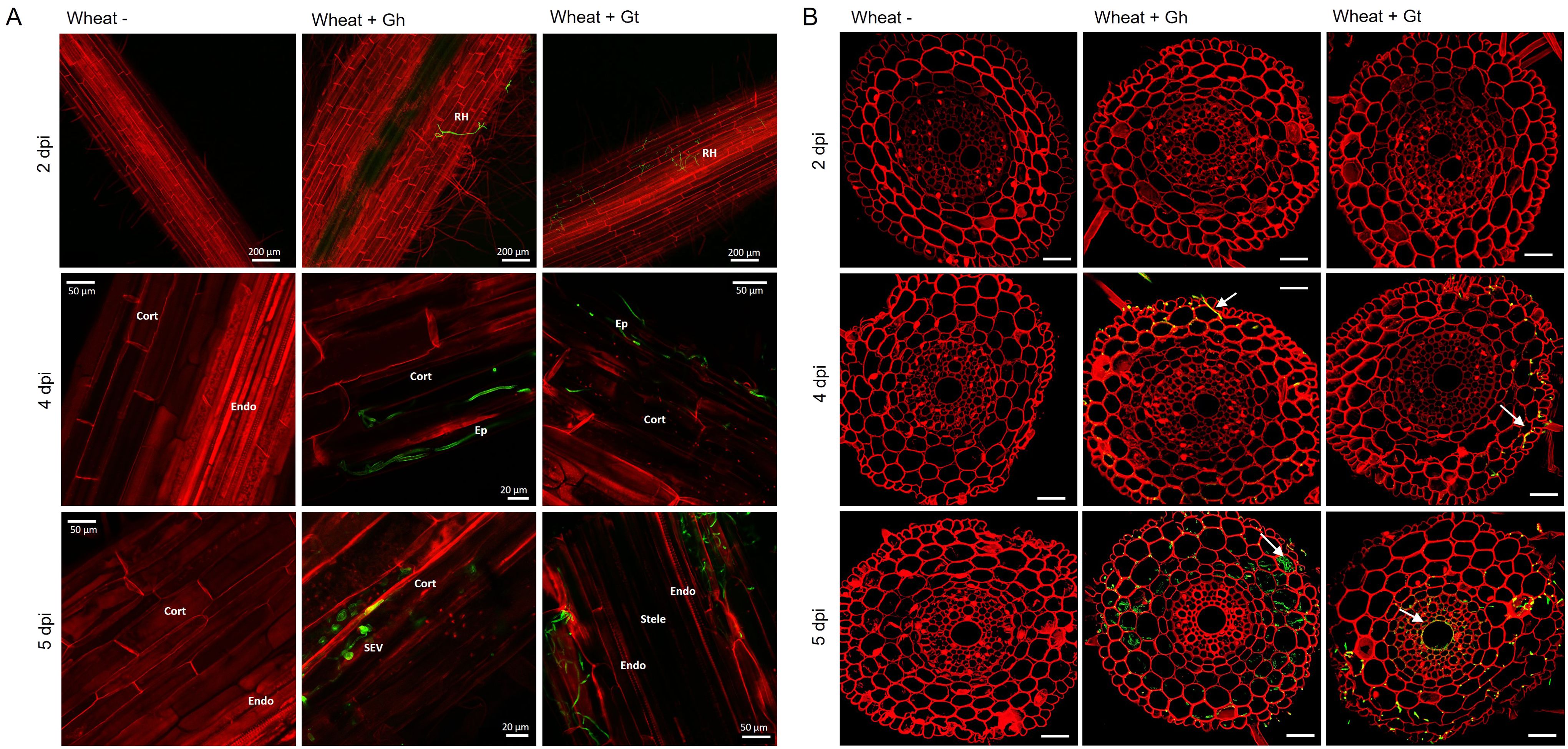
Figure 2. Fluorescence images obtained by confocal microscopy of mock inoculated, G. hyphopodioides–colonised, or G. tritici–infected wheat roots. (A) Confocal micrographs of whole root pieces highlighting fungal infection structures. (B) Z-stack images of transversal sections showing colonisation of different root cell layers across time points. Gt, G. tritici; Gh, G. hyphopodioides; RH, runner hyphae; Ep, epidermal cell; Cort, cortical cell; SEV, subepidermal vesicle; Endo, endodermal barrier. Fungal hyphae (green) are stained with WGA-AF488, and plant cell walls (red) are stained with propidium iodide. White arrows in panel (B) indicate fungal hyphae. Scale bars in panel (B) represent 50 µm.
Although the role of fungal SEVs in the infection process of G. hyphopodioides is unknown, the presence of these characteristic structures serves as visual confirmation of G. hyphopodioides colonisation in the root cortex (Osborne et al., 2018). Newly formed SEVs could be visualised by WGA staining, whereas mature SEVs, which were darker in colour, could not be visualised by WGA staining (Supplementary Figure S6). G. tritici did not produce SEVs in wheat roots at any time point, and G. hyphopodioides hyphae were not observed in the stele at any time point (Supplementary Figure S5). To investigate the structure of mature SEVs, wheat plants (cv. Hereward) were inoculated with G. hyphopodioides (NZ.129.2C.17) in a seedling pot infection assay. Colonised plants were harvested at 5 weeks postinoculation and imaged by transmission electron microscopy (TEM). Comparative analysis of intraradical fungal hyphae and SEVs revealed that SEVs contain a greater number of putative lipid bodies and a significantly thickened cell wall, comprising two to three layers of differing densities (Figures 3A, B). Multiple SEVs were often observed in a single plant cell (Figure 3C) and SEVs were often found appressed to the plant cell wall (Figures 3D, E).
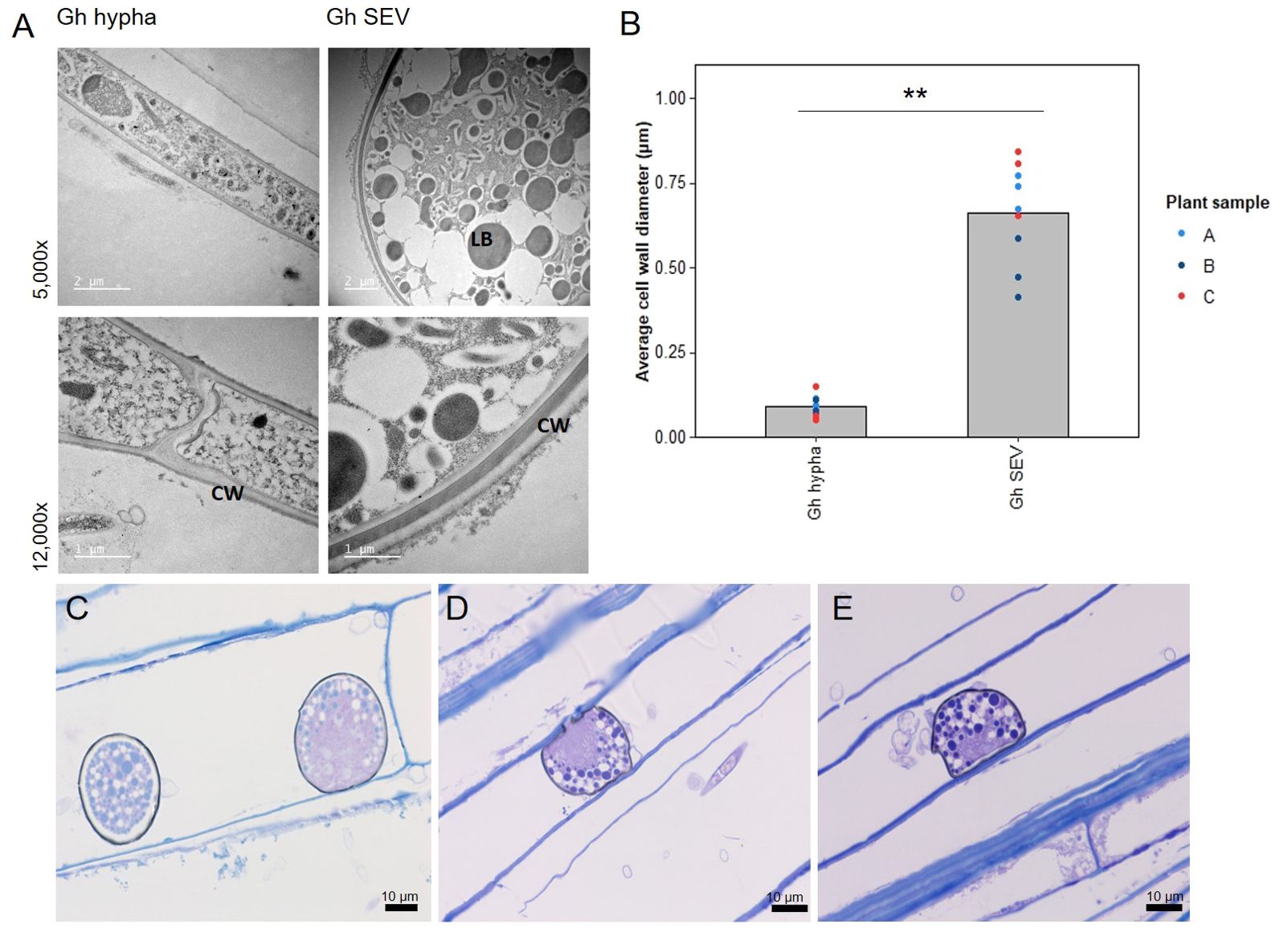
Figure 3. Subepidermal vesicles (SEVs) produced by G. hyphopodioides (Gh) in wheat roots. (A) Transmission electron micrographs (TEM) of Gh hyphae (left) and SEVs (right). (B) Average cell wall diameter (µm) of Gh fungal structures (F = 8.3, d.f. 2, 12, p <0.01). (C–E) Light micrographs of Gh SEVs in semi-thin sections, stained with toluidine blue. CW, plant cell wall; LB, putative lipid body. **=p<0.01.
The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE242417 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE242417). Three time points (2, 4, and 5 dpi) were selected for RNA-seq analysis based on the stage of fungal infection (Supplementary Figure S7). Principal component analysis (PCA) of sample distances demonstrated a good level of clustering between biological replicates, although G. tritici–infected samples exhibited comparatively higher levels of variation (Figure 4A). Gene expression levels were compared between G. tritici–infected or G. hyphopodioides–colonised plants and the uninoculated control plants at each time point individually. Full lists of the differentially expressed genes (DEGs) can be found in Supplementary Table S3. As expected, the number of wheat DEGs between the uninoculated control and G. tritici–infected or G. hyphopodioides–colonised plants was low at 2 dpi (77 and 62, respectively). By 4 dpi, G. tritici infection and G. hyphopodioides colonisation resulted in the DE of 1,061 and 1,635 wheat genes, respectively. At 5 dpi, a striking number of wheat genes were DE in response to G. hyphopodioides colonisation (7,532), whereas the number of DEGs in response to G. tritici infection (1,074) showed little change compared to 4 dpi (Figure 4B).
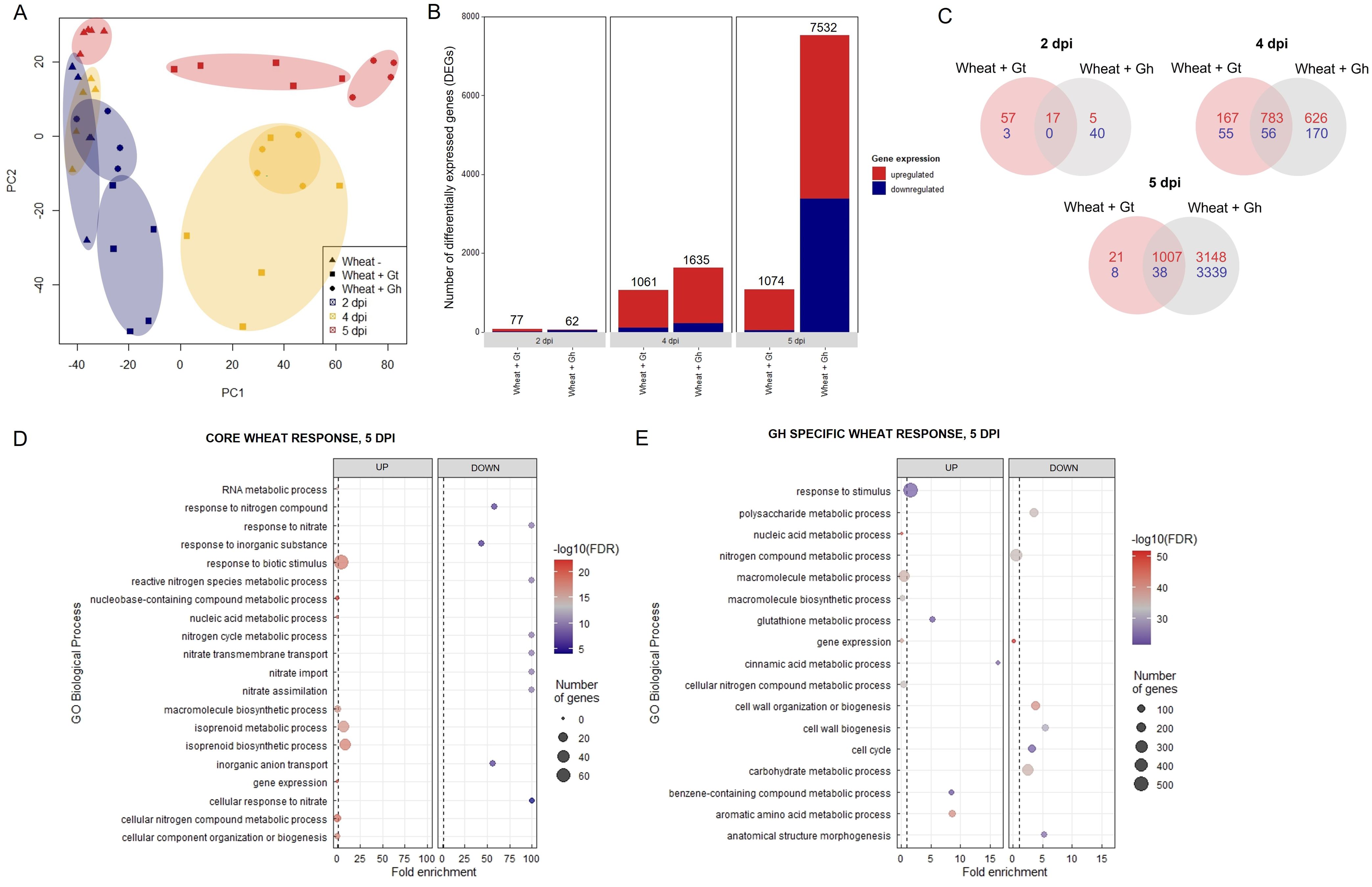
Figure 4. Transcriptional profiling of G. hyphopodioides–colonised or G. tritici–infected wheat roots. (A) Principal component analysis (PCA) plot of sample distances based on transformed (variance stabilising transformation) gene count data. Data points have been categorised by shape and colour to denote treatment and time point, respectively. (B) The number of differentially expressed genes (DEGs) in wheat colonised by G. hyphopodioides or G. tritici compared to uninoculated control samples. (C) Venn diagram highlighting the number of unique and shared wheat DEGs in G. tritici–infected or G. hyphopodioides–colonised samples compared to the uninoculated control samples. (D) Top 10 enriched biological process GO terms among DEGs in the shared wheat response to both G. tritici and G. hyphopodioides at 5 dpi. (E) Top 10 enriched biological process GO terms unique to the wheat response to G. hyphopodioides colonisation at 5 dpi. The top 10 GO terms were determined by false discovery rate (FDR).
To investigate wheat transcriptional changes during the infection progression of G. hyphopodioides compared to G. tritici, GO enrichment analyses were carried out on the sets of DEGs described above. At 2 dpi, genes involved in the diterpenoid biosynthetic process/metabolic process and gibberellin biosynthesis process were upregulated in G. tritici–inoculated roots. A large number of broad-spectrum phytoalexins in wheat are diterpenoids, requiring the biosynthesis of gibberellins (GAs) (Polturak et al., 2022). Thus, early responses to G. tritici may involve the production of antimicrobial compounds. Meanwhile, genes involved in the biosynthetic process/metabolic process were downregulated in G. hyphopodioides–inoculated roots (Supplementary Figures S8A, B). At 4 dpi, genes involved in the cinnamic acid biosynthetic/metabolic process and the L-phenylalanine metabolic/catabolic process were upregulated in G. hyphopodioides–colonised wheat roots, suggesting that lignin biosynthesis is important at this time point. However, these GO terms were not significantly enriched until 5 dpi in G. tritici–infected roots, suggesting that lignin biosynthesis is also involved in the defence response to G. tritici, although at a later stage than G. hyphopodioides. Other enriched terms in G. hyphopodioides–colonised plants at 5 dpi included response to wounding, regulation of defence responses and regulation of the JA signalling pathway. Downregulated terms included gene expression and RNA metabolic process, suggesting fine-tuning of wheat expression patterns in response to G. hyphopodioides. In addition, plant-type cell-wall organisation or biogenesis was downregulated, suggesting that the cell wall may play a role in the local wheat response to G. hyphopodioides (Supplementary Figures S8A, B).
Next, we compared the unique and shared wheat transcriptional responses to the two fungal species. At 5 dpi, 97% of the genes which were DE in response to G. tritici infection were also DE in response to G. hyphopodioides colonisation (Figure 4C). Within this core set of genes at 5 dpi, highly enriched GO biological process terms included plant response to biotic stimulus and isoprenoid biosynthetic/metabolic process (Figure 4D). The plant response to biotic stimulus term comprised 42 DE genes, 14 of which encoded proteins containing small cysteine-rich protein domains, often associated with pathogenesis-related proteins. Six genes encoded chitinases, two encoded wound-induced proteins (WIN) and a further four encoded protein kinase domain-containing proteins, thus indicating a clear defence response to both fungi (Supplementary Table S4). Enriched biological process GO terms among shared downregulated genes included response to nitrate, nitrate transmembrane transport, and nitrate assimilation (Figure 4D). Highly enriched molecular function GO terms among upregulated genes included manganese ion binding, oxidoreductase activity, and heme binding. Highly enriched molecular function GO terms among downregulated genes included nitrate transmembrane transporter activity and oxygen binding (Supplementary Table S5A). Highly enriched cellular component GO terms among upregulated genes included extracellular region (Supplementary Table S5B).
GO enrichment analysis was repeated on the subset of genes that were DE in response to G. hyphopodioides, but not G. tritici, at 5 dpi. Genes in this subset were of particular interest due to their potential role in G. hypophodioides–mediated take-all control. Cinnamic acid metabolic process and glutathione metabolic process were among the top 10 upregulated biological process GO terms. Phenolic compounds such as cinnamic acid are known to have antifungal properties and increased production of cinnamic acid can inhibit pathogen growth (Shalaby and Horwitz, 2015). Furthermore, glutathione plays diverse roles in plant immunity, both acting as an antioxidant and as a modulator of signalling processes associated with disease resistance (Zechmann, 2020). Further investigation is required to assess the role of plant phenolics and glutathione in the G. hyphopodioides–wheat interaction. Cell-wall organisation or biogenesis, cell cycle, and anatomical structure morphogenesis were among the top 10 downregulated biological process GO terms (Figure 4E). Highly enriched molecular function GO terms among upregulated genes included phenylalanine ammonia lyase (PAL) activity, glutathione transferase activity, and ion binding. In contrast, structural constituents of chromatin, tubulin binding and nucleosome binding were highly enriched among the downregulated genes (Supplementary Table S6A). Highly enriched cellular component function GO terms among downregulated genes included microtubule cytoskeleton, nucleosome, and protein–DNA complex (Supplementary Table S6B). The plant cytoskeleton plays a crucial and dynamic role in plant–microbe interactions (Wang et al., 2022). In addition, histone modification and chromatin remodelling are known to contribute to plant disease resistance (Kang et al., 2022). Indeed, there is evidence that pathogen modulated microtubule dynamics can be regulated by histone modifications (Hu et al., 2014). Thus, it is tempting to suggest that similar mechanisms could be at play in the G. hyphopodioides–wheat interaction.
Regulation of the JA signalling pathway was identified as a newly upregulated GO term at 5 dpi in G. hyphopodioides–colonised roots (see above). The GO term comprised 26 DEGs (out of a total of 77 known genes in wheat), all of which encoded TIFY transcriptions factors (TIFY TFs). In contrast, just three genes encoding TIFY TFs (TIFY10C-like_TraesCS5D02G219300, TIFY10C-like _TraesCS5B02G211000, and TIFY11E-like_ TraesCS7D02G204700) were DE in G. tritici–infected plants compared to the control. Although not identified by GO enrichment analysis, we also investigated the expression of JA biosynthesis genes. In total, 23 JA biosynthesis–related genes were DE (17 up/6 down) in response to G. hyphopodioides at 5 dpi. The list included lipoxygenase (LOX), allene oxide synthase (AOS) and AOS-like, 12-oxophytodienoate reductase (OPR) and OPR-like, and 3-ketoacyl-CoA thiolase (KAT-like) genes. Of these genes, only one (LOX8_TraesCS7B02G145200) was differentially expressed in response to G. tritici at 5 dpi (Figure 5A, Supplementary Table S7).
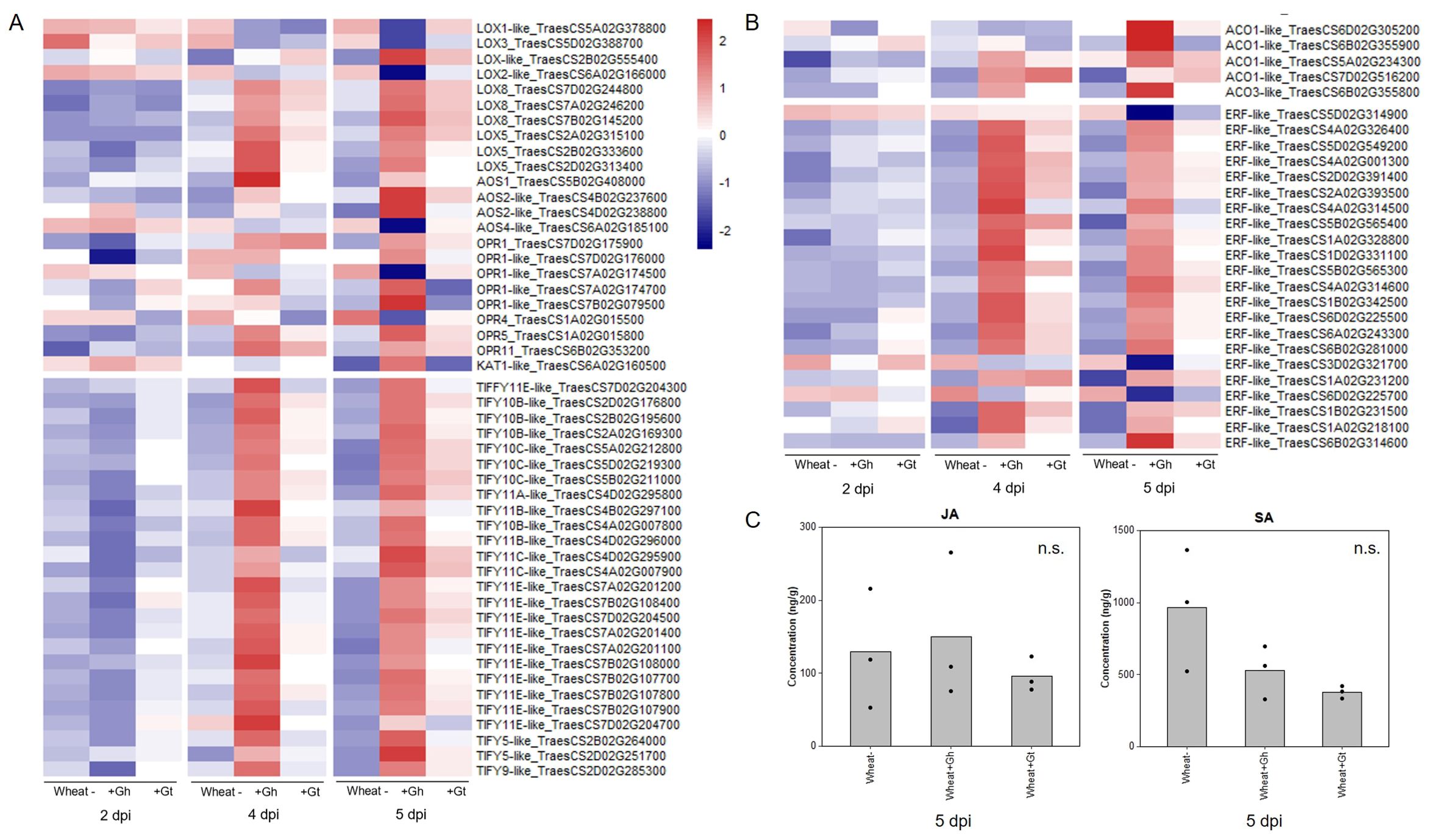
Figure 5. Wheat phytohormone-associated gene responses and JA quantification in response to G. hyphopodioides (Gh) colonisation or G. tritici (Gt) infection. (A) Expression of genes involved in the biosynthesis of JA and the regulation of the JA-mediated signalling pathway. (B) Expression of genes involved in ET biosynthesis and downstream ET signalling pathways. Heatmap data represent LOG transformed normalised genes counts. (C) Phytohormone quantification of JA and SA in roots harvested at 5 dpi (F = 0.30, d.f. 2, 6, p = 0.75; F = 4.19, d.f. 2, 6, p = 0.07, respectively). n.s., not significant. Data have been back-transformed from a square root scale. ACO, 1-aminocyclopropane-1-carboxylic acid oxidase; AOC, allene oxide cyclase; AOS, allene oxide synthase; ERF, ethylene responsive transcription factor; KAT, 3-ketoacyl-CoA thiolase; LOX, lipoxygenase; OPR, 12-oxophytodienoate reductase; TIFY, TIFY-domain containing transcription factor.
The JA and ET signalling are often closely linked. Therefore, we investigated genes involved in ET biosynthesis and signalling. Five ACC-oxidase (ACO-like) genes were upregulated in response to G. hyphopodioides at 5 dpi. In addition, 22 ET-responsive transcription factor-like (ERF-like) genes, key integrators of downstream ET and JA signal transduction pathways (Lorenzo et al., 2003), were DE (19 up/3 down) in response to G. hyphopodioides by 5 dpi. In contrast, four ERF-like genes (TraesCS4A02G001300, TraesCS5B02G565400, TraesCS1A02G231200, and TraesCS1B02G231500) were upregulated at 4 dpi and two (TraesCS5B02G565400 and TraesCS1A02G231200) were upregulated at 5 dpi in G. tritici–infected roots compared to the control (Figure 5B, Supplementary Table S7). SA is another key phytohormone involved in the plant response to pathogen invasion. SA signalling was not identified as a significantly enriched GO term in response to G. hyphopodioides or G. tritici at any time point.
To investigate whether the local transcriptional changes described above resulted in altered hormone levels, hormone quantifications of JA and SA were carried out in G. hyphopodioides–colonised, G. tritici–infected, and uninoculated control roots at 5 dpi. We found no significant difference in the levels of JA or SA between any treatments (Figure 5C).
PAL activity, essential for the lignin biosynthesis pathway, was identified as a significantly enriched molecular function GO term in the unique wheat response to G. hyphopodioides at 5 dpi (see Supplementary Table S6A). To investigate root lignification in response to G. hyphopodioides and G. tritici, we explored the expression of key genes involved in the lignin biosynthesis pathway in wheat (Figure 6A). G. hyphopodioides colonisation resulted in the earlier upregulation of lignin biosynthesis genes compared to G. tritici, with key genes such as arogenate dehydratase (ADT), phenylalanine ammonia-lyase (PAL), cinnamate 4-hydroxylase (4CL), and cinnamoyl-CoAreductase (CCR) significantly upregulated at 4 dpi (Figure 6B). However, two of eight caffeic acid O-methyltransferase (COMT) genes detected were already significantly upregulated in response to G. tritici at 2 dpi (TraesCS5D02G488800 and TraesCS5D02G488900). Interestingly, the remaining COMT genes (TraesCS2B02G066100, TraesCSU02G024300, TraesCS3B02G612000, TraesCS7D02G539100, TraesCS6D02G008200, and TraesCS7D02G538900) were strongly downregulated in response to G. hyphopodioides by 5 dpi, suggesting a decrease in the proportion of syringyl (S)-lignin. Most striking, however, was the significant upregulation of 37 PAL genes in response to G. hyphopodioides, compared to the upregulation of just 12 PAL genes in response to G. tritici at 5 dpi (Figure 6B, Supplementary Table S8).
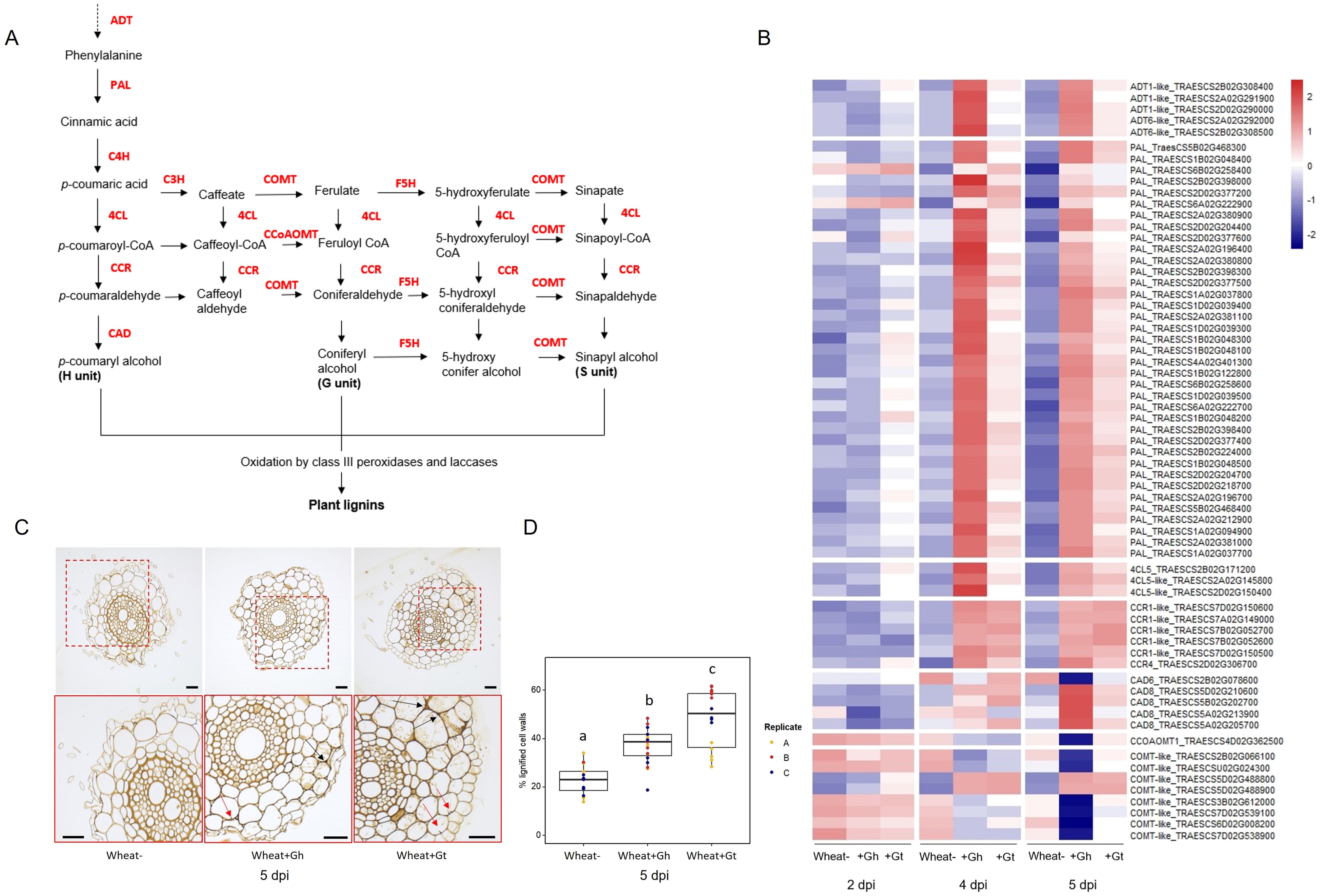
Figure 6. Lignin biosynthesis pathway and the lignin responses to G. hyphopodioides colonisation or G tritici infection. (A) Schematic of the lignin biosynthesis pathway in plants [adapted from Nguyen et al. (2016)]. (B) Expression of key genes involved in the lignin biosynthesis pathway, based on LOG transformed normalised gene counts. (C) Micrographs of transversal root sections stained with potassium permanganate for the visualisation of cell wall lignification in response to fungal infection at 5 dpi. Black arrows indicate lignified cell wall thickenings, and red arrows indicate plant lignitubers. Scale bars represent 50 µm. (D) Mean percentage of total cell wall area stained within dark parameters, indicating relative cell wall lignification (F = 34.61, d.f. 2, 50, p < 0.001). Lowercase letters indicate Tukey post-hoc groupings. ADT, arogenate dehydratase; PAL, phenylalanine ammonia-lyase; C4H, cinnamate 4-hydroxylase; 4CL, 4-coumarate CoA ligase; HCT, quinateshikimate p-hydroxycinnamoyltransferase; C3H, p-coumaroylshikimate 3′-hydroxylase; CCoAOMT, caffeoyl-CoA O-methyltransferase; CCR, cinnamoyl-CoAreductase; F5H, ferulate 5-hydroxylase; CAD, cinnamyl alcohol dehydrogenase; COMT, caffeic acid O-methyltransferase.
To visualise lignification of infected root tissues, potassium permanganate staining was performed on transverse sections of samples harvested at 5 dpi (Figure 6C). The percentage of total cell-wall area with dark potassium permanganate staining (measured in ImageJ) was used to quantify relative cell-wall lignification. Based on these measurements, G. tritici–infected roots exhibited the highest levels of cell-wall lignification, although both G. hyphopodioides and G. tritici infection resulted in increased cell-wall lignin levels compared to uninoculated control roots at 5 dpi (Figure 6D). Plant lignitubers, lignified callose deposits surrounding hyphal tips (Bradshaw et al., 2020; Huang et al., 2001; Park et al., 2022), were often detected in cells containing fungal hyphae, although these structures were more common in G. tritici–infected samples.
As mentioned previously, biological process GO term “cell-wall organisation and biogenesis” was identified as being unique to the wheat response to G. hyphopodioides at 5 dpi (see Figure 4E). In total, 124 genes involved in cell-wall organisation and biogenesis (out of 1,122 total known genes involved in cell-wall organisation and biogenesis in wheat) were downregulated in response to G. hyphopodioides at 5 dpi (Supplementary Table S9). In contrast, G. tritici infection did not lead to the DE of any genes within the cell-wall organisation and biogenesis GO term. Focusing on the top 30 DE genes within this GO term in G. hyphopodioides–colonised plants at 5 dpi, we identified six xyloglucan endotransglycosylases/hydrolases (XTH) genes, three cellulose synthase CESA–like A-like (CSLA) genes, one CESA-like F (CSLF) gene, and three fasciclin-like arabinogalactan (FLA) genes (Figure 7A). Although just one CESA-like gene was present in the list of top 30 DEGs, a total of 10 CESA genes were downregulated at 5 dpi (Supplementary Table S9). To validate gene expression in the RNA-seq dataset, we identified key cell wall–related genes where all three wheat homoeologs were downregulated in G. hyphopodioides–colonised plant roots relative to the mock-inoculated controls (Figure 7B). RT-qPCR analyses of the selected targets revealed that, as expected, cell wall–related genes CESA7-like, COBL-5D, and FLA11 were significantly downregulated in G. hyphopodioides–colonised plants compared to the mock-inoculated controls (Figure 7C).
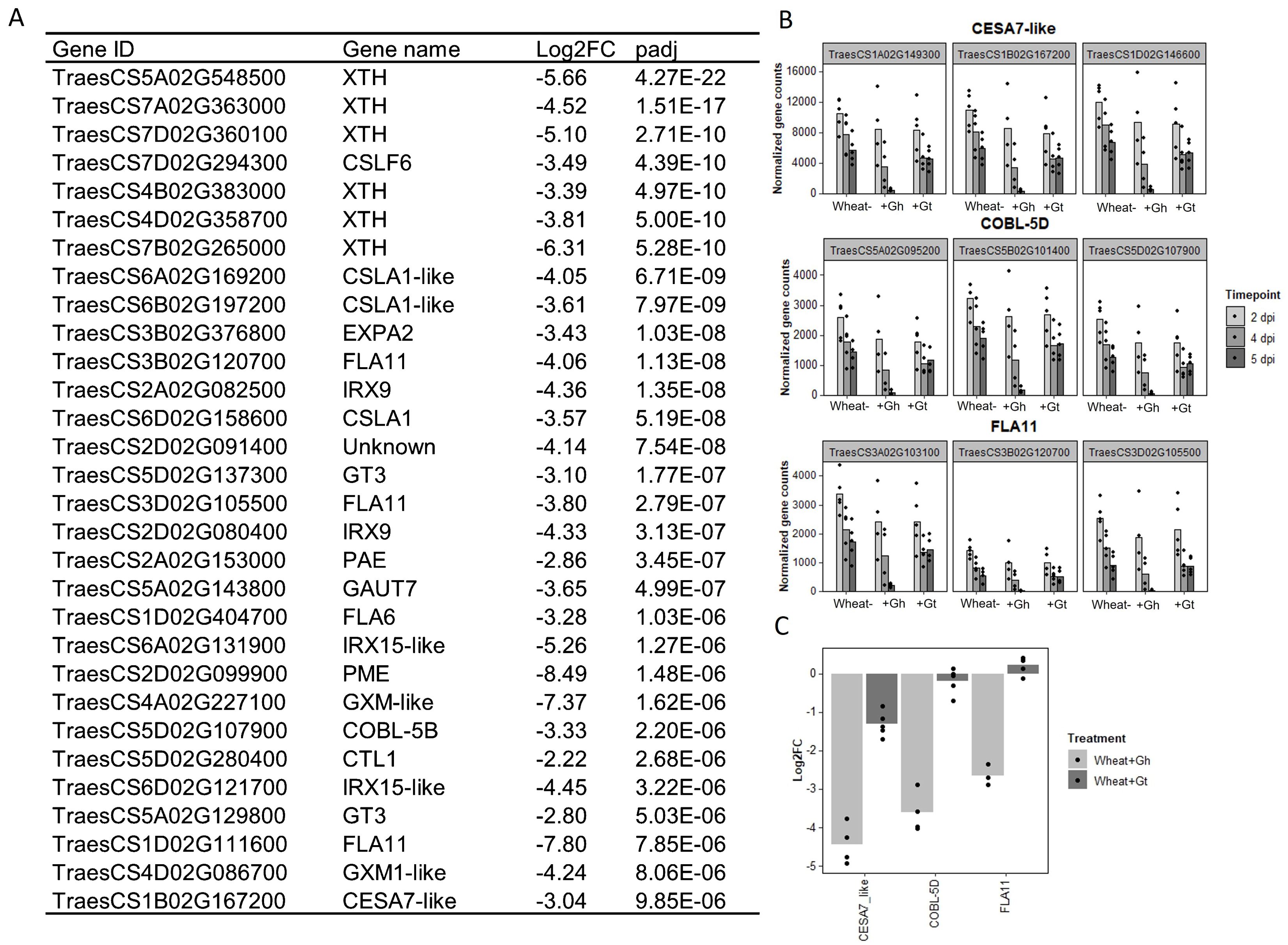
Figure 7. Colonisation by G. hyphopodioides results in local downregulation of cell wall–related genes. (A) The top 30 most differentially expressed genes relating to cell wall organisation and biogenesis in G. hyphopodioides–colonised plants at 5 dpi. Ordered by significance (padj). (B) Gene counts of selected genes across time points. (C) qPCR expression analysis of CESA7-like, COBL-5D and FLA11 in G. hyphopodioides–colonised or G. tritici–infected plants at 5 dpi (Log2FC relative to the mock-inoculated control).
Collectively, a wide and diverse range of transcriptional changes occurs in wheat roots in response to G. hyphopodioides colonisation, creating a mutually antagonistic structural, physiological, and chemical environment that limits G. tritici hyphal progress into the deeper root tissues. There is a possibility that G. hyphopodioides downregulates the wheat genes involved in cell-wall organisation and biogenesis in an attempt to maintain hyphal access to locally available water and nutrient sources, but, where these changes fail to occur, hyphal growth rates slow and these abiotic stresses trigger SEV formation.
Although transcriptional studies into the G. tritici–wheat interaction have been carried out previously for both host (Kang et al., 2019a; Yang et al., 2015; Zhang et al., 2020) and pathogen (Gazengel et al., 2020; Kang et al., 2019b), this is not the case for the G. hyphopodioides–wheat interaction. To investigate early wheat responses to G. tritici infection and uncover the local wheat defence mechanisms responsible for G. hyphopodiodes–induced disease control, we performed comparative transcriptome profiling of G. hyphopodioides–colonised and G. tritici–infected wheat using a precision inoculation method. Through detailed screening of infected root material by confocal microscopy, we were able to characterise infection progression across several time points. In support of early studies into non-pathogenic Magnaporthaceae species (Holden, 1976; Speakman and Lewis, 1978), we observed that, whereas pathogenic G. tritici grew into the vascular tissues of wheat at 5 dpi, growth of endophytic G. hyphopodioides was always limited to the inner cortex. In addition, we observed the formation of G. hyphopodioides SEVs in cortical cells at 5 dpi, whereas SEVs were not observed in G. tritici–infected roots at any time point. Interestingly, the formation of G. hyphopodioides SEVs at 5 dpi was concomitant with a dramatic increase in the number of wheat DEGs. In contrast, the number of wheat DEGs in G. tritici–infected roots showed minimal increase between 4 dpi and 5 dpi.
TEM analysis of mature G. hyphopodioides SEVs revealed that SEVs share key similarities with fungal resting structures such as chlamydospores, both being characterised by a significantly thickened, multi-layered cell wall and an increased number of putative lipid bodies (Francisco et al., 2019). Therefore, we hypothesise that G. hyphopodioides SEVs are fungal resting structures where growth has ceased and SEVs may be produced as a stress response to locally induced host defences, as indicated by extensive transcriptional reprogramming at 5 dpi. Further investigations are required to test this hypothesis and to determine what function, if any, SEVs may play in fungal root infection. In contrast, G. tritici infections resulted in far fewer DEGs at 5 dpi (1,074), the majority of which were upregulated. Interestingly, almost all DEGs identified in response to G. tritici infection were also shared with the wheat response to G. hyphopodioides colonisation. Despite triggering a significant wheat defence response, G. tritici successfully causes disease, suggesting an ability to either suppress or overcome the local wheat defences triggered. Therefore, future studies should focus on the elucidation of G. tritici effectors, enzymes, and secondary metabolites, which no doubt contribute to G. tritici pathogenicity. One such effector, the ortholog of the barley powdery mildew effector gene BEC1019, has already been associated with G. tritici virulence in wheat (Zhang et al., 2019).
Strikingly, 11% of all known cell-wall organisation/biogenesis related genes in wheat were downregulated in G. hyphopodioides–colonised plants at 5 dpi, whereas none were significantly downregulated in response to G. tritici. Impairment of cell-wall integrity (CWI) by pathogen invasion triggers the release of antimicrobial compounds and DAMPs, the latter inducing plant innate immune responses upon recognition by plant pattern recognition receptors (Miedes et al., 2014). In this study, G. hyphopodioides colonisation triggered the downregulation of 13 FLA genes and 18 XTH genes. FLA proteins contain a putative cell adhesion domain, which may link the cell membrane and the cell wall. FLA mutants in Arabidopsis exhibit a range of phenotypes including reduced cellulose content, altered secondary cell-wall deposition, and reduced tensile strength (Ashagre et al., 2021). XTH genes are also involved in the maintenance of CWI; these genes encode xyloglucan modifying enzymes that cleave xyloglucan chains to enable cell-wall expansion or alter cell-wall strength (Cosgrove, 2005). In addition, we detected the downregulation of 10 CESA genes. Although the exact mechanism is not yet known, a number of studies in Arabidopsis indicate a link between CESA expression, CWI, and disease resistance. Mutations in the CESA4, CESA7, and CESA8 genes, required for secondary cell-wall formation in Arabidopsis, confer enhanced resistance to the necrotrophic fungus Plectrosphaerella cucumerina and the biotrophic bacterium Ralstonia solanacearum (Hernández-Blanco et al., 2007). In addition, pathogenic Fusarium oxysporum root infection of Arabidopsis results in the downregulation of various CESAs, causing an alteration in primary cell-wall cellulose and contributing to disease resistance (Menna et al., 2021). Furthermore, mutations in CESA genes in Arabidopsis trigger the activation of defence responses and the biosynthesis of lignin, regulated at least in part, by the JA and ET signalling pathways (Caño-Delgado et al., 2003). A link between JA/ET signalling and reduced cellulose levels has also previously been reported by Ellis et al. (2002). Thus, our finding that G. hyphopodioides colonisation results in the upregulation of lignin biosynthesis genes and JA/ET signalling genes is pertinent.
Lignin polymers are a major component of the secondary cell wall of vascular plants, and lignin biosynthesis is known to be upregulated in response to pathogen attack by functioning as a physical barrier. Increased lignification is well-known to cause a physical barrier to microbial invasion (Miedes et al., 2014). Increased levels of lignin are present in cell-wall appositions such as papillae, which form one of the first layers of plant immunity, and in lignitubers, which surround invading hyphae. Both responses slow pathogen growth (Łaźniewska et al., 2012). Increased lignification has been documented in response to colonisation by beneficial fungi such as Trichoderma species, potentially playing a role in induced resistance against plant pathogens (Basińska-Barczak et al., 2020). Indeed, previous studies have reported higher levels of cell-wall lignification in response to colonisation by several non-pathogenic Magnaporthaceae species (Huang et al., 2001; Speakman and Lewis, 1978). In the present study, we detected earlier and higher expression of lignin biosynthesis genes in G. hyphopodioides–colonised tissues compared to G. tritici–infected tissues. In contrast, local cell-wall lignification (as determined by potassium permanganate staining) was more prominent in G. tritici–infected roots at 5 dpi. However, the downregulation of several COMT genes in response to G. hyphopodioides is noteworthy. COMT genes are involved in the synthesis of the S unit of lignin, and downregulation of these genes has a minimal effect on total lignin content (Nguyen et al., 2016). Such changes in lignin composition can drastically alter the outcome of plant-pathogen interactions (Höch et al., 2021; Ma et al., 2018; Quentin et al., 2009). Therefore, despite contrasting results, cell-wall lignification could play an important role in G. hyphopodioides–mediated take-all control.
In our dataset, 26 genes encoding TIFY TFs, involved in the cross-talk between JA and other phytohormones (Singh and Mukhopadhyay, 2021) were upregulated in response to G. hyphopodioides at 5 dpi. Just three TIFY TFs were significantly upregulated in response to G. tritici. In addition, G. hyphopodioides colonisation resulted in the upregulation of a greater number of ERF-like genes, known to integrate ET and JA signal transduction pathways (Lorenzo et al., 2003). Phytohormone quantifications using ultrahigh-performance liquid chromatography yielded highly variable results, and we were unable to detect significant differences in JA or SA levels between treatments. Although a somewhat surprising finding, this finding could potentially be explained by the early stage of infection captured, at which point take-all root lesions had not yet developed. The high levels of variability may also be a result of the transient nature of phytohormone signalling, particularly in the case of JA (Ruan et al., 2019). Future studies should probably quantify phytohormone levels across different time points, from early infection through to disease symptom development. Thus, unlike in EMR by non-pathogenic Fusarium species, G. hyphopodioides–induced resistance is potentially mediated, at least to some extent, by the JA/ET signalling pathway. Further investigation is required to determine whether the disruption of CWI mechanisms is directly responsible for the activation of JA/ET-mediated defence pathway and the lignin biosynthesis pathway. In addition, future studies should investigate plant and fungal gene expression during G. tritici infection of roots already colonised by G. hyphopodioides.
The biocontrol potential of several non-pathogenic Magnaporthaceae species has been reported since the 1970s (Deacon, 1973, 1976b; Wong and Southwell, 1980). However, the precise mechanisms of control and the molecular pathways underpinning these interactions have remained underexplored. In this study, we show that induced wheat resistance mechanisms play a key role in G. hyphopodioides–mediated disease reduction. Furthermore, we demonstrate that these resistance mechanisms operate at a local scale, with effective disease protection conferred in roots pre-treated with G. hyphopodioides. However, adding G. hyphopodioides after G. tritici resulted in increased take-all disease levels; hence, the application of G. hyphopodioides to wheat fields already harbouring significant levels of G. tritici inoculum is unlikely to provide disease control. Such a strategy may even be detrimental to disease control efforts. Therefore, the potential for G. hyphopodioides to become pathogenic in wheat and/or other cereal crops requires careful investigation. Future studies should also investigate the potential cost of G. hyphopodioides–mediated resistance on plant health and yield. Though G. hyphopodioides–mediated resistance operates at a local scale, the induction of diverse defence pathways may create a trade-off with plant growth (He et al., 2022). In addition, cell-wall reinforcement with lignins may limit root growth, potentially having downstream effects on soil anchorage and water/nutrient uptake pathways. Nevertheless, farmers may carefully exploit the disease suppression ability of G. hyphopodioides by growing wheat cultivars known to support natural G. hyphopodioides populations, particularly when placed early in wheat rotations prior to the buildup of G. tritici inoculum (Osborne et al., 2018).
In summary, we demonstrate rapid and extensive transcriptional reprogramming in G. hyphopodioides–colonised wheat roots, characterised by the strong local induction of diverse plant defence mechanisms. We propose that the collective effects of these local defence mechanisms, particularly relating to cell wall–related resistance, are responsible for G. hyphopodioides–mediated take-all control. Due to the lack of high-quality annotated G. tritici and G. hyphopodioides genomes, comparative analysis of fungal gene expression during G. hyphopodioides colonisation and G. tritici infection was not possible in this study. When combined with the RNA-seq dataset presented here, future genome sequencing projects will no doubt facilitate the investigation of novel G. tritici pathogenicity factors. In addition, further analysis of non-pathogenic and pathogenic fungi within the diverse Magnaporthaceae family may help to address wider questions relating to pathogen organ specificity, conserved fungal root infection strategies, and the determinants of fungal pathogenicity.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
TC: Writing – review & editing, Writing – original draft, Visualization, Validation, Resources, Project administration, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. DS: Writing – review & editing, Writing – original draft, Methodology, Formal analysis, Data curation. WC: Writing – review & editing, Investigation, Formal analysis. SC: Writing – review & editing, Methodology, Formal analysis. EV: Writing – review & editing, Methodology, Investigation. KH: Writing – review & editing, Methodology, Investigation. EC: Writing – review & editing, Methodology, Investigation. VM: Writing – review & editing, Supervision, Methodology, Funding acquisition, Conceptualization. GC: Writing – review & editing, Methodology, Investigation. VA: Writing – review & editing, Methodology, Investigation. KH-K: Writing – review & editing, Writing – original draft, Supervision, Resources, Methodology, Funding acquisition, Conceptualization. JP-G: Writing – review & editing, Writing – original draft, Supervision, Resources, Project administration, Methodology, Investigation, Funding acquisition, Conceptualization.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. TC was supported by the Biotechnology and Biological Sciences Research Council (BBSRC) funded University of Nottingham Doctoral Training Programme (BB/M008770/1). JP-G, DS, and KH-K were supported by the BBSRC Institute Strategic Programme (ISP) Grant, Designing Future Wheat (BBS/E/C/000I0250). In addition, DS is supported by the BBSRC ISP Grant (BB/CCG2280/1) and KH-K by the BBSRC ISP Grant, Delivering Sustainable Wheat (BB/X011003/1 and BBS/E/RH/230001B). GC is supported by the DEFRA funded Wheat Genetic Improvement Network, WGIN (CH0109) and the BBSRC ISP Growing Health (BBS/E/RH/230003A). VA is supported by the BBSRC funded South West Biosciences Doctoral Training Partnership (BB/T008741/1).
We thank Smita Kurup and Hannah Walpole (Plant Sciences for the Bioeconomy, Rothamsted Research) for advice and assistance with fluorescent staining and confocal microscopy of wheat roots; Jess Hammond (Protecting Crops and the Environment, Rothamsted Research) for help with fungal isolations and plant care; and Matthew Dickinson (University of Nottingham) for his feedback on the project.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1444271/full#supplementary-material
Ashagre, H., Zaltzman, D., Idan-Molakandov, A., Romano, H., Tzfadia, O., Harpaz-Saad, S. (2021). FASCICLIN-LIKE 18 is a new player regulating root elongation in Arabidopsis thaliana. Front. Plant Sci. 12. doi: 10.3389/FPLS.2021.645286/BIBTEX
Asher, M. J. C., Shipton, P. J. (1981). Biology and control of take-all. Academic Press, London, United Kingdom.
Ban, Y., Tang, M., Chen, H., Xu, Z., Zhang, H., Yang, Y. (2012). The response of dark septate endophytes (DSE) to heavy metals in pure culture. PLoS ONE 7 (10), e47968. doi: 10.1371/journal.pone.0047968
Basińska-Barczak, A., Błaszczyk, L., Szentner, K. (2020). Plant cell wall changes in common wheat roots as a result of their interaction with beneficial fungi of Trichoderma. Cells 9 (10), 2319. doi: 10.3390/cells9102319
Benjamini, Y., Hochberg, Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Society: Ser. B (Methodological) 57 (10), 289–300. doi: 10.1111/J.2517-6161.1995.TB02031.X
Bolger, A. M., Lohse, M., Usadel, B. (2014). Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Boller, T., Felix, G. (2009). A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60, 379–406. doi: 10.1146/annurev.arplant.57.032905.105346
Bradshaw, R. E., Bellgard, S. E., Black, A., Burns, B. R., Gerth, M. L., McDougal, R. L., et al. (2020). Phytophthora agathidicida: research progress, cultural perspectives and knowledge gaps in the control and management of kauri dieback in New Zealand. Plant Pathol. 69, 3–16. doi: 10.1111/ppa.13104
Caño-Delgado, A., Penfield, S., Smith, C., Catley, M., Bevan, M. (2003). Reduced cellulose synthesis invokes lignification and defense responses in Arabidopsis thaliana. Plant J. 34, 351–362. doi: 10.1046/j.1365-313X.2003.01729.x
Choudhary, D. K., Prakash, A., Johri, B. N. (2007). Induced systemic resistance (ISR) in plants: Mechanism of action. Indian J. Microbiol. 47, 289–297. doi: 10.1007/s12088-007-0054-2
Constantin, M. E., de Lamo, F. J., Vlieger, B. V., Rep, M., Takken, F. L. W. (2019). Endophyte-mediated resistance in tomato to fusarium oxysporum is independent of ET, JA, and SA. Front Plant Sci. 10, 1–14. doi: 10.3389/fpls.2019.00979
Cosgrove, D. J. (2005). Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 6, 850–861. doi: 10.1038/nrm1746
Deacon, J. W. (1973). Phialophora radicicola and Gaeumannomyces graminis on roots of grasses and cereals. Trans. Br. Mycological Soc. 61, 471–485. doi: 10.1016/s0007-1536(73)80117-2
Deacon, J. W. (1976a). Biology of the Gaeumannomyces graminis Arx & Olivier/Phialophora radicicola Cain Complex on Roots of the Gramineae. EPPO Bull. 6, 349–363. doi: 10.1111/j.1365-2338.1976.tb02033.x
Deacon, J. W. (1976b). Biological control of the take-all fungus, Gaeumannomyces graminis, by Phialophora radicicola and similar fungi. Soil Biol. Biochem. 8, 275–283. doi: 10.1016/0038-0717(76)90057-2
de Lamo, F. J., Takken, F. L. W. (2020). Biocontrol by Fusarium oxysporum using endophyte-mediated resistance. Front. Plant Sci. 11. doi: 10.3389/FPLS.2020.00037/BIBTEX
Delgado-Baquerizo, M., Guerra, C. A., Cano-Díaz, C., Egidi, E., Wang, J. T., Eisenhauer, N., et al. (2020). The proportion of soil-borne pathogens increases with warming at the global scale. Nat. Climate Change 10, 550–554. doi: 10.1038/s41558-020-0759-3
Dufresne, M., Osbourn, A. E. (2001). Definition of tissue-specific and general requirements for plant infection in a phytopathogenic fungus. Mol. Plant-Microbe Interact. 14, 300–307. doi: 10.1094/MPMI.2001.14.3.300
Ellis, C., Karafyllidis, I., Turner, J. G. (2002). Constitutive activation of jasmonate signaling in an Arabidopsis mutant correlates with enhanced resistance to Erysiphe cichoracearum, Pseudomonas syringae, and Myzus persicae. Mol. Plant-Microbe Interact. 15, 1025–1030. doi: 10.1094/MPMI.2002.15.10.1025
Francisco, C. S., Ma, X., Zwyssig, M. M., McDonald, B. A., Palma-Guerrero, J. (2019). Morphological changes in response to environmental stresses in the fungal plant pathogen Zymoseptoria tritici. Sci. Rep. 9, 1–18. doi: 10.1038/s41598-019-45994-3
Freeman, J., Ward, E. (2004). Gaeumannomyces graminis, the take-all fungus and its relatives. Mol. Plant Pathol. 5, 235–252. doi: 10.1111/j.1364-3703.2004.00226.x
Freeman, J., Ward, E., Gutteridge, R. J., Bateman, G. L. (2005). Methods for studying population structure, including sensitivity to the fungicide silthiofam, of the cereal take-all fungus, Gaeumannomyces graminis var. tritici. Plant Pathol. 54, 686–698. doi: 10.1111/j.1365-3059.2005.01252.x
Gazengel, K., Lebreton, L., Lapalu, N., Amselem, J., Guillerm-Erckelboudt, A. Y., Tagu, D., et al. (2020). pH effect on strain-specific transcriptomes of the take-all fungus. PloS One 15 (7), e0236429. doi: 10.1371/journal.pone.0236429
Harris, J. M., Balint-Kurti, P., Bede, J. C., Day, B., Gold, S., Goss, E. M., et al. (2020). What are the top 10 unanswered questions in molecular plant-microbe interactions? Mol. Plant-Microbe Interact. 33, 1354–1365. doi: 10.1094/MPMI-08-20-0229-CR
He, Z., Webster, S., He, S. Y. (2022). Growth–defense trade-offs in plants. Curr. Biol. 32, 634–639. doi: 10.1016/j.cub.2022.04.070
Hernández-Blanco, C., Feng, D. X., Hu, J., Sánchez-Vallet, A., Deslandes, L., Llorente, F., et al. (2007). Impairment of cellulose synthases required for Arabidopsis secondary cell wall formation enhances disease resistance. Plant Cell 19, 890–903. doi: 10.1105/tpc.106.048058
Hernández-Restrepo, M., Groenewald, J. Z., Elliott, M. L., Canning, G., McMillan, V. E., Crous, P. W. (2016). Take-all or nothing. Stud. Mycology 83, 19–48. doi: 10.1016/j.simyco.2016.06.002
Höch, K., Koopmann, B., von Tiedemann, A. (2021). Lignin composition and timing of cell wall lignification are involved in Brassica napus resistance to stem rot caused by Sclerotinia sclerotiorum. Phytopathology 111 (8), 1438–1448. doi: 10.1094/PHYTO-09-20-0425-R
Holden, J. (1976). Infection of wheat seminal roots by varieties of Phialophora radicicola and Gaeumannomyces graminis. Soil Biol. Biochem. 8, 109–119. doi: 10.1016/0038-0717(76)90074-2
Hu, M., Pei, B. L., Zhang, L. F., Li, Y. Z. (2014). Histone H2B monoubiquitination is involved in regulating the dynamics of microtubules during the defense response to Verticillium dahliae toxins in arabidopsis. Plant Physiol. 164, 1857–1865. doi: 10.1104/pp.113.234567
Huang, L., Kang, Z., Buchenauer, H. (2001). Comparison of infection of wheat roots by Phialophora graminicola and Gaeumannomyces graminis var. tritici by ultrastructural and cytochemical studies. J. Plant Dis. Prot. 108, 593–607.
Jones, J. D. G., Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Kang, H., Fan, T., Wu, J., Zhu, Y., Shen, W. H. (2022). Histone modification and chromatin remodeling in plant response to pathogens. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.986940
Kang, X., Guo, Y., Leng, S., Xiao, L., Wang, L., Xue, Y., et al. (2019a). Comparative Transcriptome Profiling of Gaeumannomyces graminis var. tritici in Wheat Roots in the Absence and Presence of Biocontrol Bacillus velezensis CC09. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.01474
Kang, X., Wang, L., Guo, Y., Zain Ul Arifeen, M., Cai, X., Xue, Y., et al. (2019b). A Comparative Transcriptomic and Proteomic Analysis of Hexaploid Wheat’s Responses to Colonization by Bacillus velezensis and Gaeumannomyces graminis, Both Separately and Combined. Mol. Plant-Microbe Interact. 32. doi: 10.1094/MPMI-03-19-0066-R
Keenan, S., Cromey, M. G., Harrow, S. A., Bithell, S. L., Butler, R. C., Beard, S. S., et al. (2015). Quantitative PCR to detect Gaeumannomyces graminis var. tritici in symptomatic and non-symptomatic wheat roots. Australas. Plant Pathol. 44, 591–597. doi: 10.1007/s13313-015-0379-y
Kim, D., Paggi, J. M., Park, C., Bennett, C., Salzberg, S. L. (2019). Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915. doi: 10.1038/s41587-019-0201-4
Knapp, D. G., Németh, J. B., Barry, K., Hainaut, M., Henrissat, B., Johnson, J., et al. (2018). Comparative genomics provides insights into the lifestyle and reveals functional heterogeneity of dark septate endophytic fungi. Sci. Rep. 8, 1–13. doi: 10.1038/s41598-018-24686-4
Łaźniewska, J., Macioszek, V. K., Kononowicz, A. K. (2012). Plant-fungus interface: The role of surface structures in plant resistance and susceptibility to pathogenic fungi. Physiol. Mol. Plant Pathol. 78, 24–30. doi: 10.1016/j.pmpp.2012.01.004
Li, L., Guo, N., Feng, Y., Duan, M., Li, C. (2022). Effect of Piriformospora indica-Induced Systemic Resistance and Basal Immunity Against Rhizoctonia cerealis and Fusarium graminearum in Wheat. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.836940
Liao, Y., Smyth, G. K., & Shi, W. (2014). FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. doi: 10.1093/bioinformatics/btt656
Lorenzo, O., Piqueras, R., Sánchez-Serrano, J. J., Solano, R. (2003). ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 15, 165–178. doi: 10.1105/TPC.007468
Love, M. I., Huber, W., Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. doi: 10.1186/s13059-014-0550-8
Marcel, S., Paszkowski, U., Sawers, R., Oakeley, E., Angliker, H. (2010). Tissue-adapted invasion strategies of the rice blast fungus Magnaporthe oryzae. Plant Cell 22, 3177–3187. doi: 10.1105/tpc.110.078048
Ma, Q. H., Zhu, H. H., Qiao, M. Y. (2018). Contribution of both lignin content and sinapyl monomer to disease resistance in tobacco. Plant Pathology 67 (3), 642–650. doi: 10.1111/PPA.12767
McMillan, V. E., Hammond-Kosack, K. E., Gutteridge, R. J. (2011). Evidence that wheat cultivars differ in their ability to build up inoculum of the take-all fungus, Gaeumannomyces graminis var. tritici, under a first wheat crop. Plant Pathol. 60, 200–206. doi: 10.1111/j.1365-3059.2010.02375.x
Menna, A., Dora, S., Sancho-Andrés, G., Kashyap, A., Meena, M. K., Sklodowski, K., et al. (2021). A primary cell wall cellulose-dependent defense mechanism against vascular pathogens revealed by time-resolved dual transcriptomics. BMC Biol. 19, 1–20. doi: 10.1186/s12915-021-01100-6
Miedes, E., Vanholme, R., Boerjan, W., Molina, A. (2014). The role of the secondary cell wall in plant resistance to pathogens. Front. Plant Sci. 5. doi: 10.3389/fpls.2014.00358
Nguyen, T. N., Son, S. H., Jordan, M. C., Levin, D. B., Ayele, B. T. (2016). Lignin biosynthesis in wheat (Triticum aestivum L.): Its response to waterlogging and association with hormonal levels. BMC Plant Biol. 16. doi: 10.1186/s12870-016-0717-4
Okagaki, L. H., Nunes, C. C., Sailsbery, J., Clay, B., Brown, D., John, T., et al. (2015). Genome sequences of three phytopathogenic species of the magnaporthaceae family of fungi. Genes, Genomes, Genetics 5. doi: 10.1534/g3.115.020057
Osborne, S. J., McMillan, V. E., White, R., Hammond-Kosack, K. E. (2018). Elite UK winter wheat cultivars differ in their ability to support the colonization of beneficial root-infecting fungi. J. Exp. Bot. 69, 3103–3115. doi: 10.1093/jxb/ery136
Palma-Guerrero, J., Chancellor, T., Spong, J., Canning, G., Hammond, J., McMillan, V. E., et al. (2021). Take-all disease: new insights into an important wheat root pathogen. Trends Plant Sci. 26, 836–848. doi: 10.1016/j.tplants.2021.02.009
Park, J. R., Kim, E. G., Jang, Y. H., Yun, B. J., Kim, K. M. (2022). Selection strategy for damping-off resistance gene by biotechnology in rice plant. Plant Soil 474, 277–296. doi: 10.1007/s11104-022-05334-3
Pieterse, C. M. J., Zamioudis, C., Berendsen, R. L., Weller, D. M., Van Wees, S. C. M., Bakker, P. A. H. M. (2014). Induced systemic resistance by beneficial microbes. Annu. Rev. Phytopathol. 52, 347–375. doi: 10.1146/annurev-phyto-082712-102340
Pok, B., Ngou, M., Ding, P., Jones, J. D. G. (2022). Thirty years of resistance: Zig-zag through the plant immune system. Plant Cell 34, 1447–1478. doi: 10.1093/PLCELL/KOAC041
Polturak, G., Dippe, M., Stephenson, M. J., Misra, R. C., Owen, C., Ramirez-Gonzalez, R. H., et al. (2022). Pathogen-induced biosynthetic pathways encode defense-related molecules in bread wheat. PNAS 119 (116), e2123299119. doi: 10.1073/pnas.2123299119
Qiang, X., Weiss, M., Kogel, K.-H., Schäfer, P. (2012). Piriformospora indica-a mutualistic basidiomycete with an exceptionally large plant host range: mutualistic root symbiosis. Mol. Plant Pathol. 13, 508–518. doi: 10.1111/j.1364-3703.2011.00764.x
Quentin, M., Allasia, V., Pegard, A., Allais, F., Ducrot, P. H., Favery, B., et al. (2009). Imbalanced lignin biosynthesis promotes the sexual reproduction of homothallic oomycete pathogens. PLoS Pathogens 5 (1), 1–9. doi: 10.1371/journal.ppat.1000264
Ruan, J., Zhou, Y., Zhou, M., Yan, J., Khurshid, M., Weng, W., et al. (2019). Jasmonic acid signaling pathway in plants. Int. J. Mol. Sci. 20, 2479. doi: 10.3390/ijms20102479
Shalaby, S., Horwitz, B. A. (2015). Plant phenolic compounds and oxidative stress: integrated signals in fungal–plant interactions. Curr. Genet. 61, 347–357. doi: 10.1007/s00294-014-0458-6
Singh, P., Mukhopadhyay, K. (2021). Comprehensive molecular dissection of TIFY Transcription factors reveal their dynamic responses to biotic and abiotic stress in wheat (Triticum aestivum L.). Sci. Rep. 11, 1–17. doi: 10.1038/s41598-021-87722-w
Speakman, J. B., Lewis, B. G. (1978). Limitation of Gaeumannomyces graminis by wheat root responses to Phialophora radicicola. New Phytol. 80, 373–380. doi: 10.1111/j.1469-8137.1978.tb01571.x
Thoms, D., Liang, Y., Haney, C. H. (2021). Maintaining symbiotic homeostasis: How do plants engage with beneficial microorganisms while at the same time restricting pathogens? Mol. Plant-Microbe Interact. 34, 462–469. doi: 10.1094/MPMI-11-20-0318-FI
Ulrich, K., Augustin, C., Werner, A. (2000). Identification and characterization of a new group of root-colonizing fungi within the Gaeumannomyces-Phialophora complex. New Phytol. 145, 127–135. doi: 10.1046/j.1469-8137.2000.00553.x
Wang, J., Lian, N., Zhang, Y., Man, Y., Chen, L., Yang, H., et al. (2022). The cytoskeleton in plant immunity: dynamics, regulation, and function. Int. J. Mol. Sci. 23, 15553. doi: 10.3390/ijms232415553
Ward, E., Bateman, G. L. (1999). Comparison of Gaeumannomyces- and Phialophora-like fungal pathogens from maize and other plants using DNA methods. New Phytol. 141, 323–331. doi: 10.1046/j.1469-8137.1999.00346.x
Wong, P. T. W., Mead, J. A., Holley, M. P. (1996). Enhanced field control of wheat take-all using cold tolerant isolates of Gaeumannomyces graminis var. graminis and Phialophora sp. (lobed hyphopodia). Plant Pathol. 45, 285–293. doi: 10.1046/j.1365-3059.1996.d01-132.x
Wong, P. T. W., Southwell, R. J. (1980). Field control of take-all of wheat by avirulent fungi. Ann. Appl. Biol. 94, 41–49. doi: 10.1111/j.1744-7348.1980.tb03894.x
Yang, L., Xie, L., Xue, B., Goodwin, P. H., Quan, X., Zheng, C., et al. (2015). Comparative Transcriptome Profiling of the Early Infection of Wheat Roots by Gaeumannomyces graminis var. tritici. PloS One 10, e0120691. doi: 10.1371/journal.pone.0120691
Zechmann, B. (2020). Subcellular roles of glutathione in mediating plant defense during biotic stress. Plants 9, 1–21. doi: 10.3390/plants9091067
Zhang, Y., Xu, K., Yu, D., Liu, Z., Peng, C., Li, X., et al. (2019). The highly conserved barley powdery mildew effector BEC1019 confers susceptibility to biotrophic and necrotrophic pathogens in wheat. Int. J. Mol. Sci. 20, 1–16. doi: 10.3390/ijms20184376
Zhang, J., Yan, H., Xia, M., Han, X., Xie, L., Goodwin, P. H., et al. (2020). Wheat root transcriptional responses against Gaeumannomyces graminis var. tritici. Phytopathol. Res. 2. doi: 10.1186/s42483-020-00066-7
Zhou, F., Emonet, A., Dénervaud Tendon, V., Marhavy, P., Wu, D., Lahaye, T., et al. (2020). Co-incidence of damage and microbial patterns controls localized immune responses in roots. Cell 180, 440–453. doi: 10.1016/j.cell.2020.01.013
Keywords: wheat root transcriptome, root endophyte, root pathogen, wheat defences, cell-wall modifications, fungal biocontrol
Citation: Chancellor T, Smith DP, Chen W, Clark SJ, Venter E, Halsey K, Carrera E, McMillan V, Canning G, Armer VJ, Hammond-Kosack KE and Palma-Guerrero J (2024) A fungal endophyte induces local cell wall–mediated resistance in wheat roots against take-all disease. Front. Plant Sci. 15:1444271. doi: 10.3389/fpls.2024.1444271
Received: 05 June 2024; Accepted: 06 August 2024;
Published: 18 September 2024.
Edited by:
Yasser Nehela, University of Florida, United StatesReviewed by:
Ruchi Agarrwal, National Research Center on Pomegranate (ICAR), IndiaCopyright © 2024 Chancellor, Smith, Chen, Clark, Venter, Halsey, Carrera, McMillan, Canning, Armer, Hammond-Kosack and Palma-Guerrero. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kim E. Hammond-Kosack, a2ltLmhhbW1vbmQta29zYWNrQHJvdGhhbXN0ZWQuYWMudWs=
†Present addresses: Tania Chancellor, Crop Science Centre, Department of Plant Sciences, University of Cambridge, Cambridge, United Kingdom
Daniel P. Smith, John Innes Centre, Department of Computational and Systems Biology, Norwich John Innes Centre, Norwich, United Kingdom
Eudri Venter, JEOL (UK) Ltd., Welwyn Garden City, Hertfordshire, United Kingdom
Vanessa McMillan, National Institute of Agricultural Botany (NIAB), Cambridge, United Kingdom
Javier Palma-Guerrero, Department of Crop Sciences, Research Institute of Organic Agriculture (FIBL), Frick, Switzerland
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.