- 1Research Center of Deciduous Oaks, Beijing Forestry University, Beijing, China
- 2Deciduous Oak Improvement and Regeneration Innovation Team of State Forestry and Grassland Administration, Beijing Forestry University, Beijing, China
- 3Key Laboratory for Silviculture and Conservation, Ministry of Education, Beijing Forestry University, Beijing, China
Hydrolyzable tannins (HTs) have garnered significant attention due to their proven beneficial effects in the clinical treatment of various diseases. The cupule of Chinese cork oak (Quercus variabilis Blume) has been used as raw material of traditional medicine for centuries for its high content of HTs. Previous studies have identified UGT84A13 as a key enzyme in the HT biosynthesis pathway in Q. variabilis, but the transcriptional regulation network of UGT84A13 remains obscure. Here, we performed a comprehensive genome-wide identification of the TCP transcription factors in Q. variabilis, elucidating their molecular evolution and gene structure. Gene expression analysis showed that TCP3 from the CIN subfamily and TCP6 from the PCF subfamily were co-expressed with UGT84A13 in cupule. Further functional characterization using dual-luciferase assays confirmed that TCP3, rather than TCP6, played a role in the transcriptional regulation of UGT84A13, thus promoting HT biosynthesis in the cupule of Q. variabilis. Our work identified TCP family members in Q. variabilis for the first time, and provided novel insights into the transcriptional regulatory network of UGT84A13 and HT biosynthesis in Q. variabilis, explaining the reason why the cupule enriches HTs that could be used for traditional medicine.
1 Introduction
Tannins are water-soluble polyphenolic compounds synthesized by plants through secondary metabolism. They are produced via the biological inheritance pathway from galloyl esters or low polymerized proanthocyanidins (Lewis and Yamamoto, 1989). As plant defense chemicals, tannins protect plants from fungi, pathogens, and herbivores (Gallardo et al., 2019; Xie et al., 2019; Das et al., 2020). Based on their phenolic group connections, tannins are categorized as hydrolyzable tannins (HTs) and condensed tannins (CTs). For human health, HTs exhibit antioxidant and antimicrobial effects, showing potential as effective natural antibiotics (Zhou et al., 2019). Additionally, HTs can relax vascular smooth muscle and are effective against arterial hypertension (Marrone et al., 2023). HTs can treat diabetic complications by acting on the hexosamine pathway (Laddha and Kulkarni, 2019). HTs can alleviate stress-induced depression by modulating cortisol and monoamine neurotransmitter levels, reducing oxidative stress, and potentially serving as a complementary treatment for depression (Chandrasekhar et al., 2017). Interest in utilizing natural HTs as potential nutritional supplements or adjunct therapies is increasing (Melo et al., 2023). Plants such as pomegranate, chestnut, and oak species are rich sources of HTs (Deng, 2018; Habashi et al., 2019; Sun et al., 2021; Yang et al., 2023).
Due to the widespread application of HTs, there is significant interest in their biosynthetic pathway. However, model plants including Arabidopsis, rice, and poplar do not contain abundant levels of HTs, leading to uncertainties in the regulatory pathways of HT biosynthesis. Previous studies have indicated that HT synthesis relies on the catalysis of dehydroquinate dehydratase/shikimate dehydrogenase (DQD/SDH) and UDP-dependent glycosyltransferase (UGT). DQD/SDHs have been identified as enzymes involved in the biosynthesis of gallic acid, the precursor to HT production. In Vitis vinifera, VvSDH3 and VvSDH4 are involved in gallic acid biosynthesis (Bontpart et al., 2016). In Eucalyptus camaldulensis, EcDQD/SDH2 and EcDQD/SDH3 are involved in gallate formation, dehydroquinate dehydratase activity, and shikimate dehydrogenase activity, resulting in the accumulation of HTs which significantly enhances aluminum tolerance (Tahara et al., 2021). In Q. variabilis, UGT, rather than SDH, exhibits tissue-specific expression (Yang et al., 2024). β-glucogallin, as a biomarker of HTs in oak cupule, is formed by the combination of UDP-glucose and gallic acid, with UGT being the critical enzyme in this process (Niemetz and Gross, 2005; Tahara et al., 2021; Yang et al., 2024). Expression patterns and correlation analyses in Canarium album L. have revealed that the highly expressed CaUGT84A77 can catalyze the production of β-glucogallin from gallic acid (Ye et al., 2021). UGT84A13 is a pivotal enzyme in the biosynthesis of β-glucogallin in oak species, further influencing HT synthesis, as confirmed in Q. robur and Q. variabilis (Mittasch et al., 2014; Yang et al., 2024). However, the transcriptional regulation of these UGTs remains largely unknown. Our previous research found that Q. variabilis WRKY32/59 and HB26 can activate the expression of UGT84A13 (Yang et al., 2023, 2024). However, other transcription factors such as ERFs, TCP, and bHLH may also be regulatory factors in HT biosynthesis, and the molecular regulatory network involving UGT has yet to be established.
TCP family is a class of transcription factors widely present in plants, named after their prototypical members Teosinte branched (TB1), Cycloidea (CYC), and Proliferating Cell Factor (PCF) (Cubas et al., 1999). The TCP family contains an atypical basic-helix-loop-helix (bHLH) secondary structure composed of 55-60 amino acid residues, known as the TCP conserved domain. This domain can bind to DNA and interact with other proteins to regulate the transcription of target genes (Aggarwal et al., 2010). Based on differences in the conserved domain, the TCP family is divided into Class I (PCF subfamily) and Class II (CIN and CYC/TB1 subfamilies) (Lin et al., 2016). TCPs are broadly involved in various physiological processes of plant growth and development, including seed germination (Tatematsu et al., 2008), bud dormancy (Wang et al., 2020), fruit development (Xie et al., 2020), response to abiotic stress (Xu et al., 2021), and secondary metabolism. Overexpression of AtTCP4 can promote the expression of the LOX2 gene, leading to the accumulation of jasmonic acid and accelerating leaf senescence (Li et al., 2024). PeTCP10 from Phyllostachys edulis, when transformed into Arabidopsis, significantly enhances the salt tolerance of transgenic Arabidopsis during the vegetative growth phase by promoting catalase activity to improve antioxidant capacity (Xu et al., 2021). miRNA319-mediated silencing of the transcription factor OsTCP21 and its target genes blocks JA signal transduction to manipulate rice immune responses (Zhang et al., 2018). However, it remains unclear whether TCP transcription factors are involved in the biosynthesis of HTs.
Chinese cork oak (Quercus variabilis) is a dominant and resource-worthy species in East Asian (Cao and Chen, 2015; Hu et al., 2016). The cupule of Q. variabilis has been used as raw material for traditional medicine for centuries. Traditionally, its branches and cupules are used in Chinese medicine to treat malignant sores and diarrhea (Jia et al., 2013). The ethanolic extract of Q. variabilis cupules exhibits significant inhibitory effects on Salmonella paratyphi A and Staphylococcus aureus, surpassing the activity of pomegranate peel ethanolic extract and ellagic acid (Zhou, 2016). Modern research indicates that the main active components of Q. variabilis extracts are HTs (Yang et al., 2023). However, the transcriptional regulation of HT biosynthesis still remains largely unknown.
In this study, we comprehensively analyzed TCP family genes in Q. variabilis genome. Combined with transcriptome analysis of different tissues and organs of Q. variabilis, we explored the regulatory function of the TCP family in HTs biosynthesis. Dual-luciferase assays were conducted to investigate the regulatory functions of the TCP gene family in HTs production in Q. variabilis. To our knowledge, this research is the first comprehensive analysis of TCP genes in Q. variabilis. It has also refined the molecular regulatory network regulating HT biosynthesis in Q. variabilis involving UGT84A13, explaining the reason why the cupule enriches HTs that could be used for traditional medicine.
2 Materials and methods
2.1 Identification of TCP gene family members
The Hidden Markov Model (HMM) file was downloaded from the Pfam database (https://pfam.xfam.org/) using the protein accession number PF03634. We then used the protein sequences of the Arabidopsis TCP gene family for sequence alignment to identify and preliminarily screen the TCP gene family. Employing HMMER3 software with the HMM domain of TCP as the query, we searched the entire protein sequence database of the Q. variabilis genome (Yang et al., 2024). Additionally, we downloaded amino acid sequences of Arabidopsis TCP gene family members from TAIR11 (https://www.arabidopsis.org/) and used them as query sequences in BLAST comparisons within TBtools. We utilized the Q. variabilis whole-genome protein sequences as the reference library, setting an E-value threshold of 1e-5. The sequences obtained from both methods were integrated, leading to the identification of 22 TCP family genes. We used the Gene Location plugin in TBtools along with the annotation data of the Q. variabilis genome and the ID numbers of the target genes to determine the relative chromosomal positions and gene density on each chromosome. We then visualized the chromosomal distribution. The physicochemical properties of the TCP gene family were assessed using the Protein Parameter Calc plugin in TBtools. The amino acid sequences of the identified TCP gene family were then modeled using the SWISS-MODEL online platform (https://swissmodel.expasy.org/). Upon reviewing multiple results, the optimal protein structure model was selected based on the GMQE score (ranging from 0 to 1, with higher values indicating greater reliability) and the QMEAN score (ranging from -4 to 0, with values closer to 0 indicating a higher degree of similarity between the target and template proteins).
2.2 Evolutionary analysis of the TCP gene family
We constructed an evolutionary tree using the full-length protein sequences from 24 Arabidopsis thaliana TCP gene family members, 13 Quercus robur TCP gene family members, and 22 TCP gene family members. The analysis was performed using MEGA 7.0 software (Kumar et al., 2016), with settings including the Neighbor-Joining (NJ) method, a bootstrap value of 1000, the Poisson model, and pairwise deletion. Subfamily classification of the TCP gene family members was based on established methods used for the Arabidopsis TCP gene family (Yao et al., 2007). The evolutionary tree was visually refined using the Chiplot online tool (https://www.chiplot.online/#) (Xie et al., 2023).
2.3 Motif analysis and gene structure of the TCP gene family
Motif analysis was conducted using the MEME online tool (http://memesuite.org/tools/meme), specifying the identification of 6 motifs. The results were then imported into TBtools software for visualization of conserved protein motifs. The annotation file for the Q. variabilis genome, along with the IDs of its TCP gene family members, was uploaded into TBtools (Chen et al., 2023) to generate a visual representation of their gene structures.
2.4 Promoter cis-element analysis of the TCP gene family
The TBtools software was used to extract the 2000 bp upstream sequence of the TCP genes’ CDS regions. Cis-elements within the TCP promoters were identified using the PlantCARE database (https://bioinformatics.psb.ugent).Visualization of the data was performed using TBtools software (Chen et al., 2023).
2.5 Intraspecific collinearity analysis of the TCP gene family
We used the MCScanX (Wang et al., 2012) plugin in TBtools (Chen et al., 2023) to calculate and analyze collinearity within the Q. variabilis genome, and between A. thaliana and Q. variabilis, using default parameter settings. The collinearity analysis results were visualized using the Dual Synteny Plot plugin within TBtools (Chen et al., 2023).
2.6 Expression pattern analysis of TCP genes in different organs
To identify TCP genes potentially involved in the transcriptional regulatory network of UGT84A13 and its regulation of HTs, we analyzed our published transcriptome data from various tissues of Q. variabilis (Yang et al., 2023). This analysis provided expression profiles of TCP genes across different tissues. We integrated the expression levels of UGT84A13 and the content data of HTs in various tissues generated in our pervious study (Yang et al., 2023). We also analyzed the candidate TCP genes, along with the previously reported HB26, WRKY32, and WRKY59 associated with HT biosynthesis (Yang et al., 2023, 2024), to investigate potential correlations in their expression. Normalization and row standardization were performed using the HeatMap plugin in TBtools software, and the results were visualized (Chen et al., 2023).
2.7 Bioinformatics platform predicts TCP transcription factors
We utilized JASPAR 2024 (https://jaspar.elixir.no/analysis) (Rauluseviciute et al., 2024) to make predictions with the TCP template in the database. The promoter region of UGT84A13 was inputted into the database for predicting TCP transcription factor binding sites, with the relative profile score threshold set at 80%. Potential binding sites were selected based on their scores.
2.8 Dual luciferase assay
To further elucidate the mechanism of direct interaction between TCP and UGT84A13, a dual-luciferase reporter assay was conducted as previously described (Yang et al., 2020). The complete CDSs of TCP3 and TCP6 were cloned into the pGreen II 0029 62-SK vector, while the UGT84A13 promoter, encompassing 2000 bp upstream, was cloned into the pGreen II 0800-LUC vector. These constructs were introduced into Agrobacterium tumefaciens strain GV3101 cells containing the pSoup vector. Following the method outlined by Yang et al. (2020), the optical density (OD) of the SK and LUC bacterial suspensions was adjusted to 1.0, mixed in a 10:1 ratio, and the mixture was used to infect tobacco leaves. After 56 hours of infection, the dual-LUC assays were performed using the Dual Luciferase Reporter Assay Kit (Vazyme, Nanjing, China) as instructed. Fluorescent signals were measured using a Glomax Discover and Explorer (Promega, USA). The experiment was conducted with six biological replicates.
3 Results
3.1 Genome-wide identification of TCP genes
By using BLAST and HMMER search, we identified a total of 22 TCP genes in the genome of Q. variabilis for further analysis. Chromosomal localization analysis (Figure 1A) revealed that these genes are distributed across chromosomes 1, 2, 3, 5, 6, 7, 8, 9, 10, and 12. The 22 TCP genes were sequentially renamed from TCP1 to TCP22 based on their chromosomal positions.
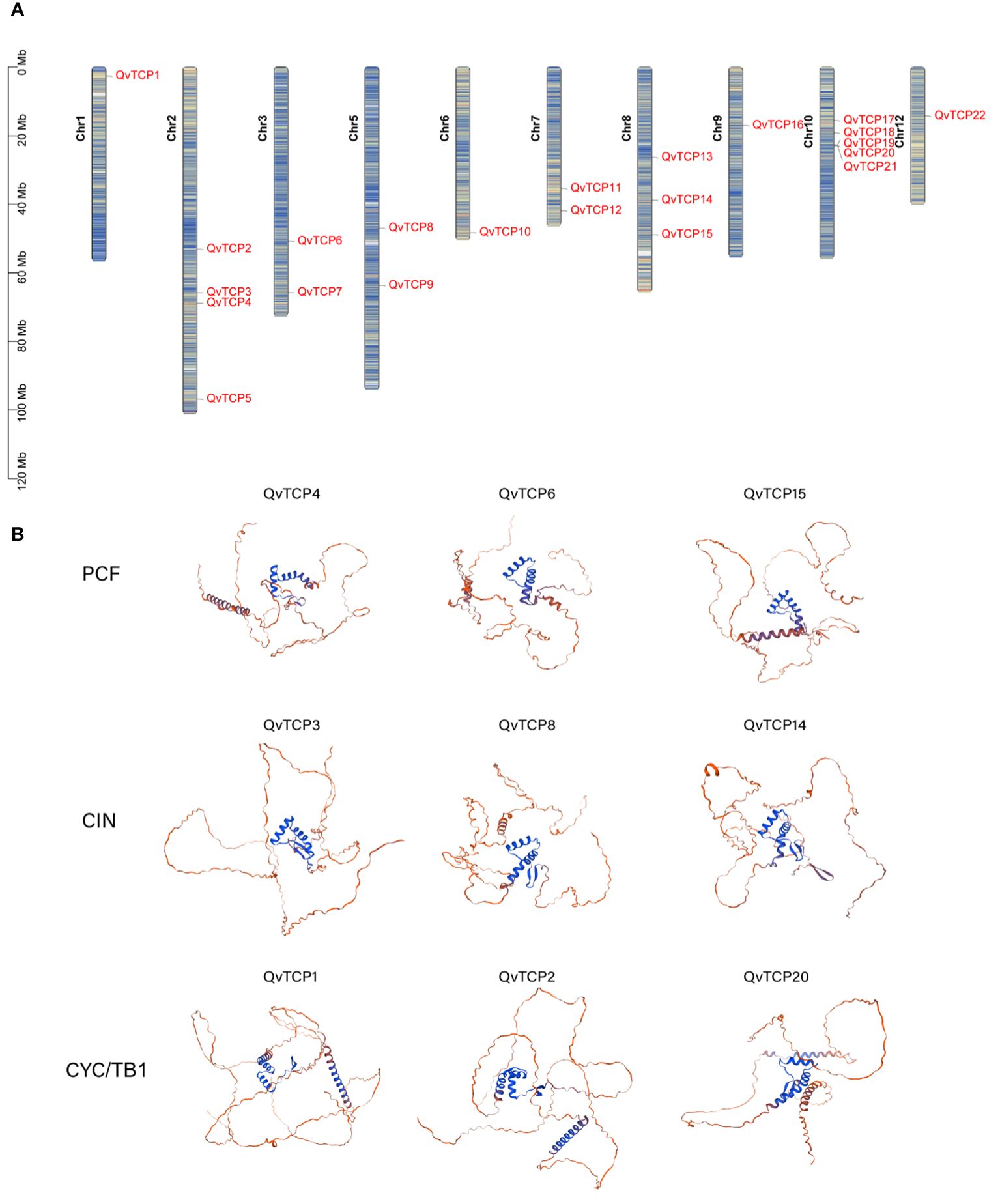
Figure 1 (A) Shows the distribution of the TCP gene on the chromosome in Q. variabilis. The color blocks, ranging from blue to red, indicate a gradual increase in gene density. (B) Predicted protein structure models of TCP gene family in Q. variabilis. In this figure, three members of each subfamily were randomly selected for display, and the complete TCP family structure prediction is shown in Supplementary Figure S1. The predicted protein structure models of the TCP family in Q. variabilis are displayed. The blue color signifies regions where the prediction results are considered highly reliable, while the red color signifies regions with lower reliability in the prediction results.
Physicochemical property analysis (Supplementary Table S1) indicated that the amino acid sequences encoded by the TCP genes range from 211 amino acids (TCP12) to 566 amino acids (TCP5). The isoelectric points span from 6.05 (TCP21) to 9.72 (TCP10), and the molecular weights range from 22.44 kDa (TCP12) to 60.16 kDa (TCP5). The instability index of TCP proteins varies between 34.72 (TCP20) and 67.46 (TCP17). Additionally, the aliphatic index ranges from 54.08 (TCP5) to 76.52 (TCP2), and the average hydrophilicity coefficient ranges from -0.930 (TCP21) to -0.226 (TCP11), suggesting that TCP proteins are all hydrophilic.
Prediction of the tertiary structures of the TCP proteins (Figure 1B) showed that their structures mainly consist of random coils and an atypical bHLH domain. The bHLH domain forms a reverse parallel conformation with two helices, and the predicted TCP protein structure indicates that the angle between the helices is nearly perpendicular, similar to the helical topology of the RHH domain. These structural characteristics are consistent with those observed in TCP proteins from other species such as rice and Arabidopsis (Sun et al., 2020).
3.2 Evolutionary analysis of TCP genes
To further elucidate the evolutionary relationships among TCP genes in Q. variabilis and other species, we constructed an evolutionary tree using 22 identified TCP protein sequences from Q. variabilis, 24 from A. thaliana, and 13 from Quercus robur (Figure 2). Based on the classification of the Arabidopsis TCP gene family, TCP family members were categorized into three subfamilies: CYC/TB1, CIN, and PCF (Yao et al., 2007). The CYC/TB1 subfamily contains 5 members, CIN includes 6 members, and PCF comprises the largest number with 11 members. In Q. robur, the CYC/TB1 subfamily has 2 members, CIN has 4 members, and PCF, the largest subfamily, has 7 members. In A. thaliana, CYC/TB1 contains 8 members, CIN has 3 members, and PCF, with the most members, has 13. In the evolutionary tree, shorter distances between genes indicate higher similarity in their motifs, suggesting they may share similar biological functions.
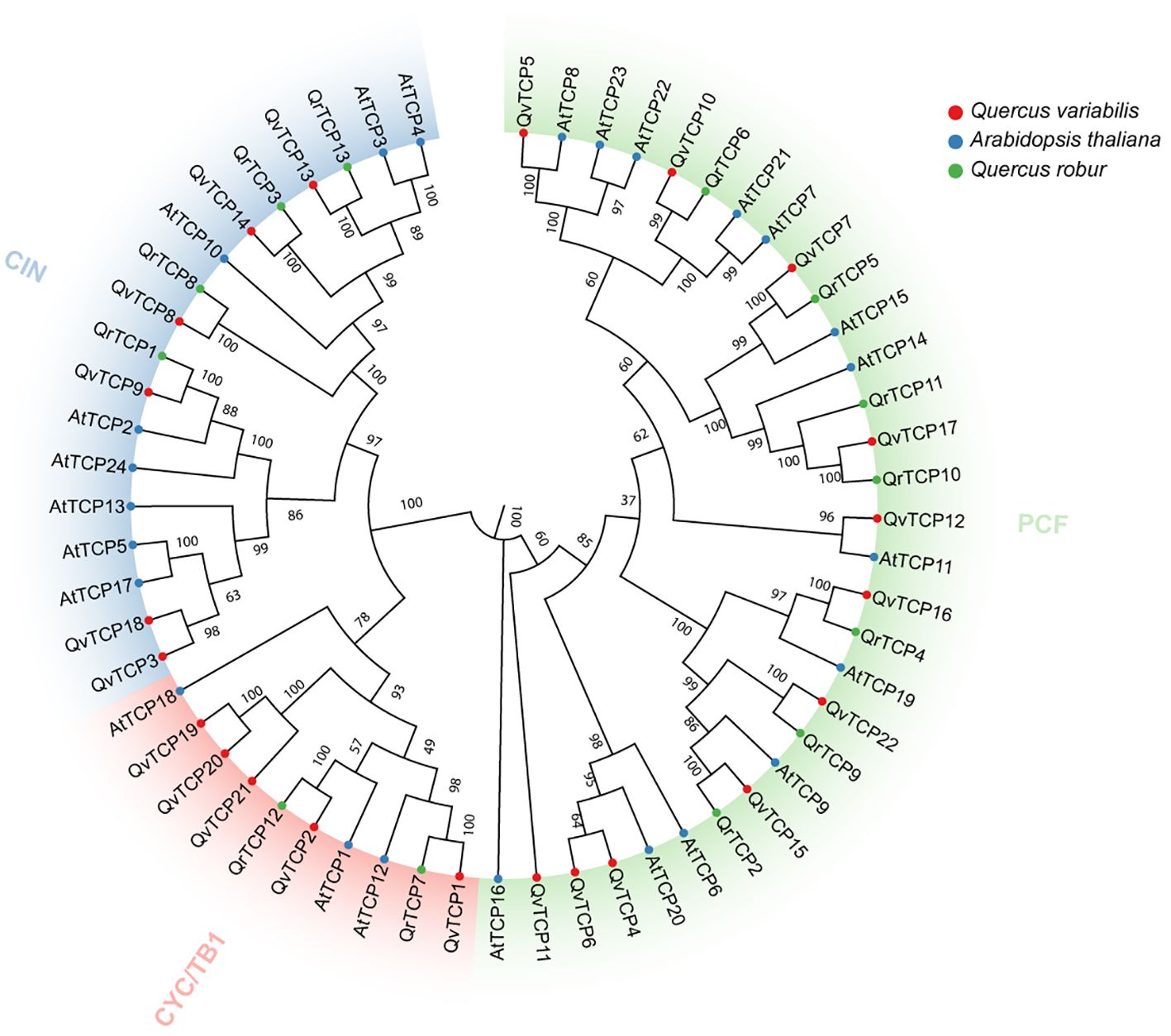
Figure 2 Evolutionary analysis of 59 TCP family genes from Q. variabilis, A. thaliana and Q. robur based on their protein alignment. The evolutionary tree was constructed using the NJ method, with 1000 bootstrap replicates in MEGA. The TCP genes were clustered into three clades (CYC/TB1, CIN and PCF), each one colored differently. The less distance between genes, the higher the similarity in the motifs they contain, suggesting potential shared biological functions.
3.3 Motif analysis and gene structure of TCPs
To analyze the conservation of the TCP family protein sequences, we visualized the gene structure using TBtools software. Based on MEME software, six conserved motifs were identified within the protein sequences. Motif 1 is present in all TCPs and is similarly positioned (Figures 3A, B). Upon alignment, it was found that the sequence of Motif 1 has a high degree of consistency with the TCP conserved domain sequence. Protein sequences within the same subfamily are more similar. In the Class I subfamily, Motif 1 and Motif 2 appear exclusively in combination, while in the Class II subfamily, Motif 1 and Motif 4 are exclusively combined. Motif 5 and Motif 6 are only present in the CYC/TB1 subgroup of Class II and are found only in proteins TCP19, TCP20, and TCP21.From this, it can be deduced that each subfamily has its unique combination of motifs, and the conserved motifs within the same subfamily are very similar or identical. This analysis is consistent with our evolutionary tree analysis, indicating significant differences in conserved motifs among TCP proteins from different subfamilies, suggesting distinct functional roles among members of different subfamilies.
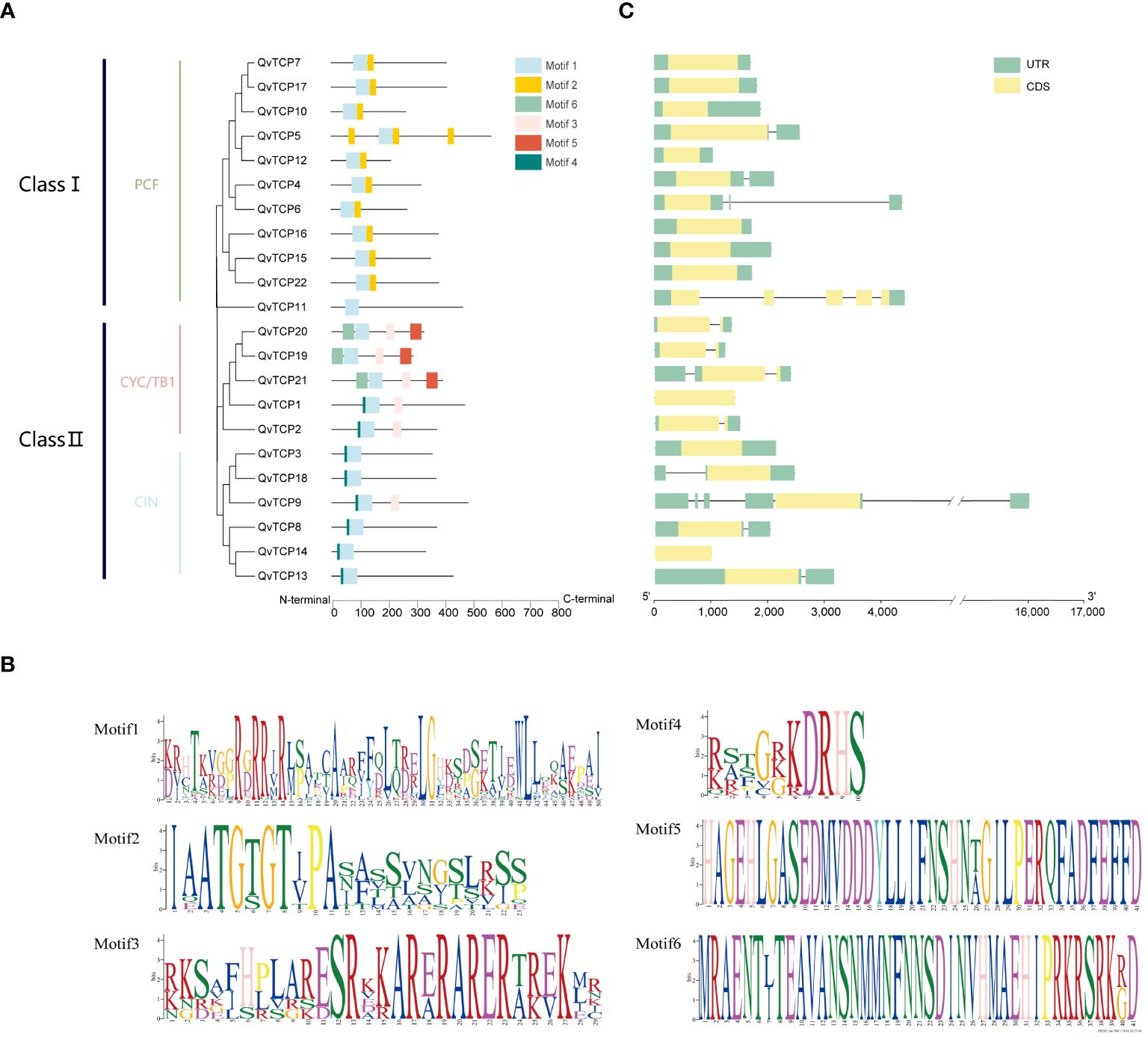
Figure 3 (A) Protein motif and gene structure analysis of TCP family genes identified in Q. variabilis. The evolutionary tree of TCP protein sequences is shown, with conserved protein motifs colored differently. (B) Exon-intron distribution analysis of TCP genes. The green boxes represent coding sequences (CDS), the green boxes represent untranslated regions (UTR), and the black lines represent intron positions, respectively. (C) The conserved motifs of the TCP family members in Q. variabilis.
To explore the diversity of TCP gene structures, we conducted a gene structure analysis. The results (Figure 3C) show significant variation in the number of exons and introns among the 22 TCP genes. Generally, genes that cluster together have similar gene structures, and homologous genes within the same branch exhibit consistent structures, suggesting that members of the same subfamily may have similar biological functions. All TCP gene regions contain introns, with the number of exons ranging from 1 to 6 and introns from 1 to 5. Most TCP genes have relatively simple structures, though a few genes, such as TCP9, have intron lengths significantly different from others. The consistent structures of homologous genes within the same branch further suggest that members of the same subfamily may have similar biological functions.
3.4 Cis-acting elements analysis of TCP genes
Promoters play a regulatory role in the transcription and expression of genes. To better understand the biological functions and mechanisms of TCP genes, this study analyzed the genomic sequences of 2,000 bp upstream of the coding regions of the TCP genes, as potential promoter regions. Ten important cis-acting elements were identified (Figure 4). These elements are primarily classified into hormone responses, environmental responses, abiotic stress responses, and stress responses. Hormone response elements, such as those for gibberellins, abscisic acid, methyl jasmonate, salicylic acid, auxin, and zeatin, are widely present in the promoters of the TCP gene family, indicating that TCP family genes are closely related to hormone signal regulation. Among the stress-related elements, some TCPs contain cis-acting elements responsive to cold stress, defense, and stress, suggesting that these TCPs play roles in resistance to and response to cold stress and adversity. Additionally, light-responsive elements are widely distributed across the TCP family, suggesting that the TCP gene family may be regulated by light signals. All 22 TCP genes contain one or more types of cis-elements involved in various functions, implying that TCP family genes may participate in multiple regulatory aspects during plant growth and development. Further research revealed that members of both Class I and Class II subfamilies are generally involved in the aforementioned cis-elements, although there are some differences in the number of gene members associated with each type of cis-element.
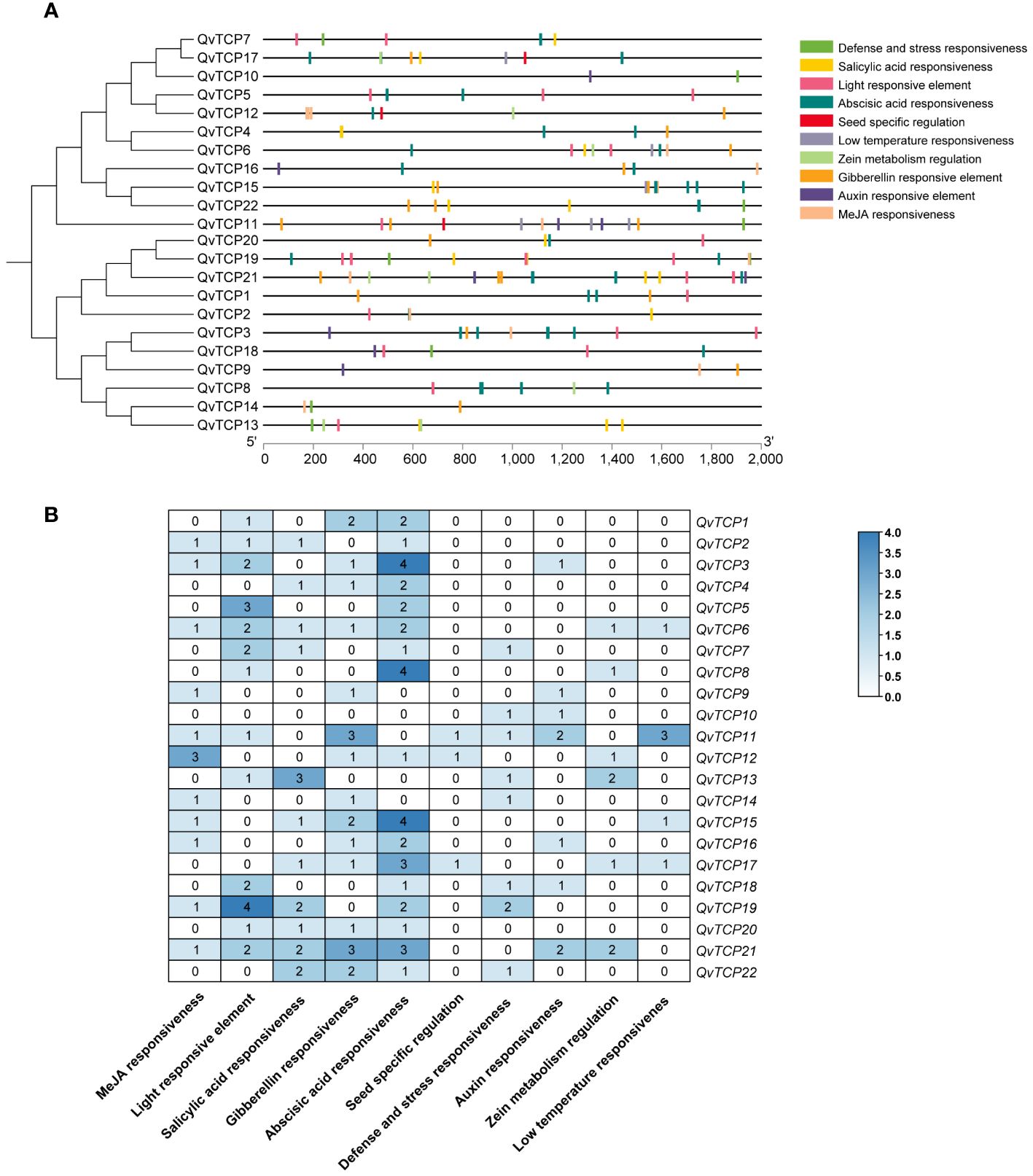
Figure 4 Cis-acting element distribution of TCP gene family in Q. variabilis. (A) Analysis of cis-regulatory elements within TCP gene promoters. (B) The numerical values and colors represent the number of cis-elements identified within each TCP gene promoter.
3.5 Evolutionary analysis of the TCP family
To detect instances of gene duplication within the TCP family, we conducted a syntenic analysis across the Q. variabilis genome, focusing on the TCP family. This is illustrated by the purple lines representing syntenic relationships (Figure 5A). These collinear gene pairs may have originated from the single ancient whole-genome duplication (Yang et al., 2024). We identified a total of eight syntenic gene pairs (Supplementary Table S3) and eight syntenic blocks and, the Ka/Ks values for all eight syntenic gene pairs ranged from 0.1 to 0.4 (Supplementary Table S4; Figure 5B), indicating that the expansion of the TCP family has likely been subject to purifying selection, which eliminates deleterious non-synonymous mutations during the natural selection process in species evolution.
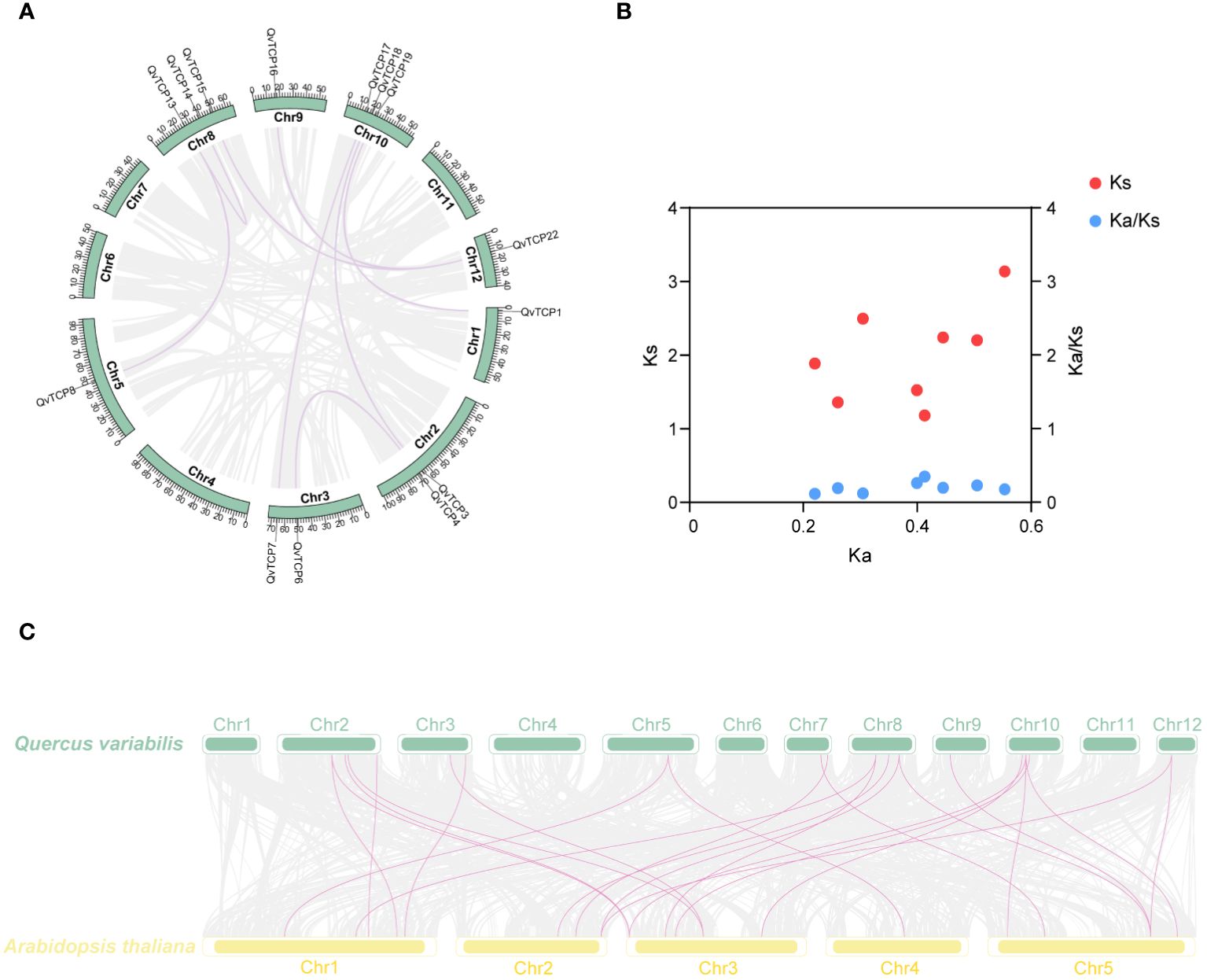
Figure 5 (A) Shows a schematic diagram of the motif and gene structure of TCP family members. The black vertical lines on the chromosomes indicate gene localization, while the inner lines represent collinearity. Gray lines indicate genome-wide collinearity, and purple lines represent the colinear region of TCPs. (B) Ka, Ks, Ka/Ks ratio calculation of the paralog pairs of TCPs in Q. variabilis. (C) Collinear analysis of TCP gene family between A. thaliana and Q. variabilis.
To examine the evolution of the Q. variabilis TCP gene family, we performed a syntenic analysis between the TCP gene families of Q. variabilis and A. thaliana, identifying 24 syntenic gene pairs (Figure 5C). With 22 members within the TCP gene family, this result suggested that the majority of the family members were highly conserved throughout the evolutionary process.
3.6 Expression profiles of TCP genes and identification of candidates for HT biosynthesis
Gene expression patterns are often closely linked to their functions, making the analysis of these patterns crucial for understanding gene function. To explore the expression of TCP genes across various tissues and to further comprehend the impact of the TCP gene family on HT content and related mechanisms in Q. variabilis tissues, we analyzed the expression patterns of TCP family genes using heatmaps and transcriptional data (Transcripts Per Million, TPM). The transcriptional data were log-transformed and row-normalized to generate heatmaps and perform clustering (Figure 6A). Given that the expression variation of UGT84A13 is a primary factor contributing to the differential accumulation of β-glucogallin and HTs (Yang et al., 2024), we screened for TCP transcription factors that are specifically highly expressed in cupule and have high TPM values in the cupule. Preliminary screening identified five transcription factors: TCP3, TCP4, TCP6, TCP15, and TCP19. Additionally, we conducted a correlation analysis between the expression levels of TCP and UGT84A13, as well as the abundance of HTs measured in our previous study (Yang et al., 2023). The results indicated that the expression levels of two candidate genes (Figure 6B), TCP3 and TCP6, were significantly correlated (p < 0.001) with the expression level of UGT84A13. Expression patterns of genes associated with HT biosynthesis in different organs of Q. variabilis (Figure 6C) revealed that these genes were highly expressed in the cupule. We conducted correlation analyses between the expression levels of TCP3/6 and genes associated with HT biosynthesis (Figure 6D). The results indicated a significant correlation between the expression levels of TCP3 and TCP6 and those of HB26 and WRKY32/59 (p < 0.001). Therefore, we identified TCP3 and TCP6 as potential candidate transcription factors that may regulate the biosynthesis of HTs in the cupule.
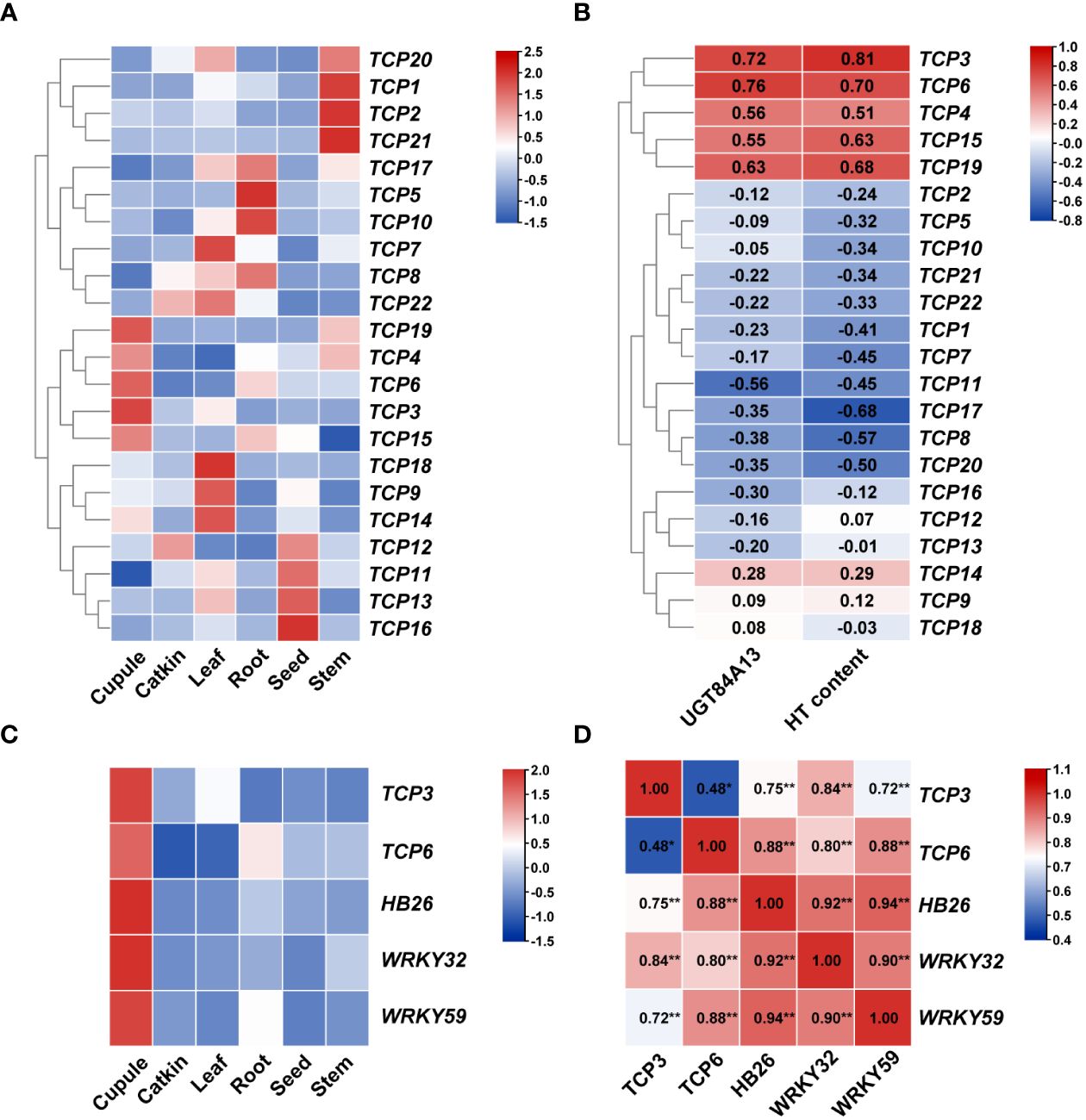
Figure 6 (A) The expression patterns of TCP family members in various organs of Q. variabilis. Red blocks indicate a relatively high level of TPM values, while blue blocks indicate a relatively low level of TPM values after normalization. The red gene name indicates the TCP genes that is highly expressed specifically in the cupule. (B) Correlation of TCP genes with UGT84A13 expression and HT content. (C) The expression patterns of genes associated with HT biosynthesis in various organs of Q. variabilis. (D) Correlation of genes associated with HT biosynthesis expression. *Significant correlation at the 0.05 level; **Significant correlation at the 0.01 level.
3.7 Dual luciferase assay
JASPAR was used to predict a number of potential binding sites on the UGT84A13 promoter where the TCP transcription factors could bind (Figure 7A). To verify the role of TCP3 and TCP6 in regulating UGT84A13, we cloned the full-length CDS (coding sequence) of these two genes into SK vector (Figure 7B). The co-expression of proUGT84A13-LUC and QvTCP3-SK significantly increased luciferase activity (Figure 7C); However, there was no significant change with QvTCP6-SK (Figure 7C). These results confirmed that TCP3, rather than TCP6, interacted with the promoter of UGT84A13 and activated its expression, suggesting the potential role of TCP transcription factors in HT biosynthesis.
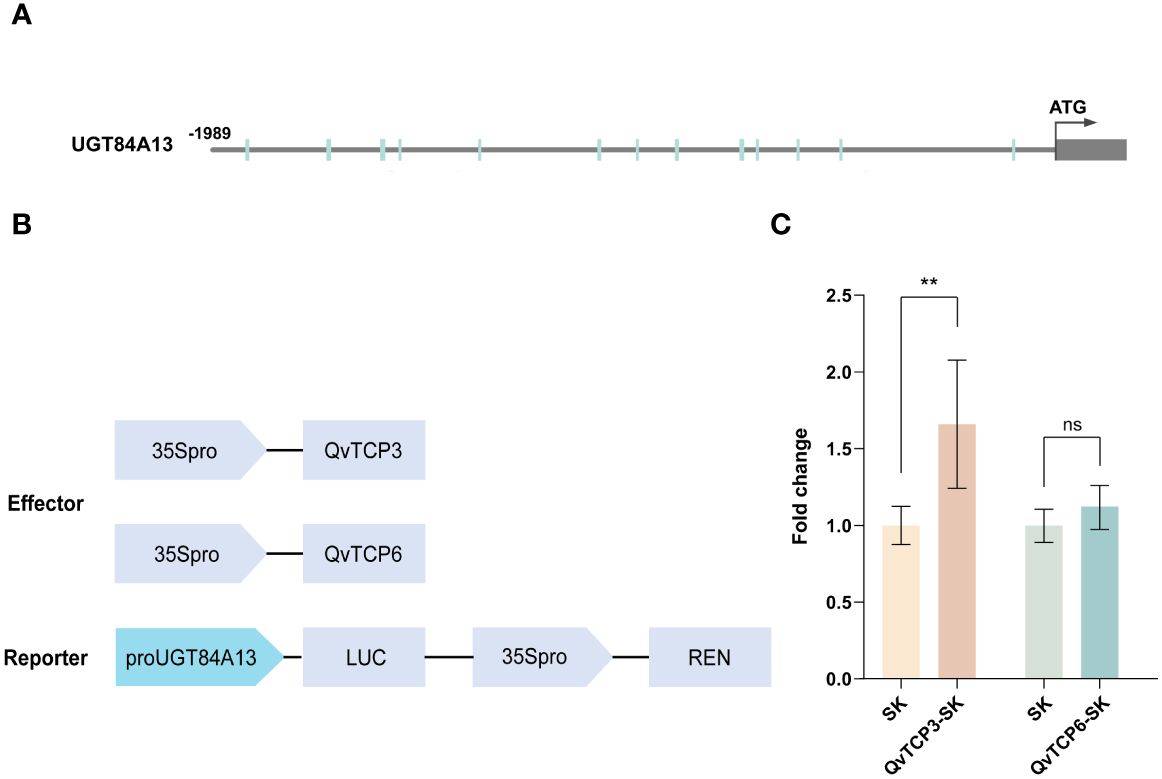
Figure 7 (A) JASPAR predicts the potential binding sites of TCP transcription factors in the UGT84A13 promoter. (B) Sketch map of the vectors used in dual-luciferase assays. (C) The dual luciferase assay for QvTCP3 and QvTCP6. Error bars represent the standard deviation of 6 biological replicates. Asterisks indicate a significant difference between the experimental group and the control group, while ns denotes no significant difference (Student’s t-test; **, p < 0.01).
4 Discussion
Tannins, also known as plant polyphenols, are widely found in the leaves, bark, roots, and fruits of plants (Shirmohammadli et al., 2018). They constitute an important class of secondary metabolites in plants, providing protection against herbivory and enhancing resistance to pests and diseases (Tondi et al., 2013). Traditionally, tannins are classified into two main types: condensed tannins and hydrolyzable tannins (Freudenberg, 1960). HTs possess anti-inflammatory and immunomodulatory functions, can lower blood sugar and lipids, have antioxidant capabilities, and exhibit DNA repair and anti-cancer effects (Smeriglio et al., 2017). Additionally, the ester bonds of HTs can be hydrolyzed by human digestive enzymes, making them easier to break down into smaller molecules after ingestion. These molecules are then metabolized by intestinal microbiota and absorbed in the intestines (Smeriglio et al., 2017), making HTs an ideal health supplement and nutritional aid. HTs are mainly found in oak, grape seeds, pomegranate peels, and other sources that are not easily supplemented through a daily diet (Fischer et al., 2011). Therefore, a plant rich in HTs is needed, and methods to extract HTs from it must be devised to meet demand. Studies have found that the tannins in oak species are mainly HTs (Yin et al., 2019). Oaks have garnered attention due to their high tannin content in tissues, large reserves, and easy access to raw materials. Among these, Q. variabilis has a higher HT content than other oak species such as Q. aliena and Q. dentata (Yang et al., 2024), indicating that Q. variabilis is an ideal material for extracting HTs.
Compared to the well-studied CTs (Saigo et al., 2020), the biosynthetic regulatory pathways of HTs remain incompletely understood. Current research indicates that the expression of the key enzyme, UDP-glucose transferase (UGT), determines the synthesis of HTs (Mittasch et al., 2014). Additionally, UGT is an essential enzyme for the biosynthesis of HTs in plants such as pomegranates (Yuan et al., 2018). UGT84A13s are pivotal enzymes in the HT synthesis pathway of oak genus plants (Mittasch et al., 2014), capable of catalyzing gallic acid into β-glucogallin. β-glucogallin, a precursor of HT synthesis, possesses numerous pharmacological activities, including antioxidant and anti-inflammatory effects, anti-diabetic properties, prevention of diabetic retinopathy complications, protection against eye diseases, and defense against ultraviolet radiation (Khan et al., 2022; Rohilla et al., 2023). However, the regulation of UGT84A13 in the HT biosynthetic pathway of oaks has not been fully elucidated, impeding further improvement in HT production efficiency. Our preliminary research identified 14 transcription factors that co-express significantly with UGT84A13, among which the role of TCP genes in the HT synthesis pathway remains unknown (Yang et al., 2024). In a study on the de-astringency of persimmon fruits, it was found that the expression of 2 TCP transcription factors was significantly suppressed during the reduction of HT content following maturation (Deng, 2018). In this study, we identified the entire genome of the TCP gene family and selected 2 members, TCP3 and TCP6, which may regulate UGT84A13. The identification of the TCP gene family will provide more information for understanding the characteristics and potential capabilities of this plant-specific transcription factor.
TCP transcription factors are an ancient, conserved family of proteins widely distributed across various plant species, such as Arabidopsis, rice, and tomato (Cubas et al., 1999; Takeda et al., 2003; Parapunova et al., 2014). TCPs regulate plant morphogenesis and environmental adaptation by participating in multiple growth and developmental processes. In Q. variabilis, we identified a total of 22 TCP gene family members, which is fewer than the number of TCP genes identified in other higher plants, such as 24 in Arabidopsis, 52 in apple, 35 in peony, and 38 in cotton (Yao et al., 2007; Ma et al., 2014; Li et al., 2022; Tabarelli et al., 2022). This may suggest a contraction of the TCP gene family in Q. variabilis. Our study examined the characteristics of the TCP protein family, including the number of exons and introns, isoelectric points, and molecular weights. The TCP domain, consisting of about 60 amino acids and characterized by an atypical bHLH secondary structure, is a hallmark of TCP proteins that we analyzed. In addition to the conserved TCP domain, members of the TCP transcription factor family also possess other domains, such as the ECE domain (Howarth and Donoghue, 2006), which is found exclusively in the Class II CYC subgroup and whose function remains unclear and requires further investigation. Further research indicates that most genes within the same evolutionary branch have similar exon-intron compositions, suggesting a correlation between the genetic makeup of the TCP domain and its evolutionary history.The distribution of TCP genes is irregular, with the 22 TCP genes unevenly distributed across 10 Q. variabilis chromosomes (Figure 1A), and no copies of TCP genes were found on chromosomes 4 and 11. Based on the evolutionary analysis of Arabidopsis TCP genes, we divided the TCP family members into three subfamilies: CYC/TB1, CIN, and PCF, and constructed an evolutionary tree of TCP protein sequences in Q. variabilis (Figure 2).This allows us to further understand the potential functions of the TCP gene family. Evolutionarily, we found that the structure of TCP genes is closely related to their phylogeny. Members within the same subfamily have identical motif compositions, indicating that they have similar functions at the protein level.
According to reports, proteins from the CIN and PCF subfamilies of the TCP gene family are involved in responses to abiotic stresses (Xu et al., 2019; Hao et al., 2023). This aligns with the stress-resistant function of hydrolyzable tannins. It was found that TCP3 belongs to the CIN subfamily, while TCP6 belongs to the PCF subfamily (Figure 2). Studies have shown that defense-related plant hormones, including jasmonic acid (JA) and abscisic acid (ABA), regulate hydrolyzable tannins (HTs) in response to biotic and abiotic stresses (Iqbal and Poór, 2024). The application of exogenous ABA can increase the content of ellagic acid in the pericarp of Vitis rotundifolia (Sandhu et al., 2011). In studies on pomegranate, treatment with MeJA led to the accumulation of β-glucogallin in the leaves (Chang et al., 2021), and MeJA treatment increased the expression of PgUGT84A23 and PgUGT84A24, affecting the content of HTs and confirming the key role of MeJA in HT production (Chang et al., 2021). Cis-element analysis of the TCP family indicates that the promoters of TCP genes contain various plant-specific binding elements, including those related to abiotic stress, such as ABA, MeJA, and SA (Figure 4). Numerous studies have shown that the TCP family responds to these plant hormones. In Arabidopsis, research confirmed that TCP2 (CIN subfamily) positively regulates LOX2 promoter activity, directly regulating LOX2, an enzyme catalyzing a key step in JA biosynthesis (Schommer et al., 2008). In Ginkgo biloba, exogenous MeJA treatment significantly upregulated the expression of GbTCP6 (CIN subfamily), GbTCP11 (CIN subfamily), and GbTCP13 (PCF subfamily) (Yu et al., 2022). In Vitis vinifera L., many members are strongly regulated by ABA treatment. Among them, VvTCP14 (PCF subfamily) showed higher transcriptional expression levels after exogenous ABA treatment (Jiu et al., 2019). However, whether ABA or MeJA treatment regulates members of the TCP family and further increases the content of HTs in the cupule of Q. variabilis remains to be studied (Figure 6A). The correlation between TCP6 and other TFs identified in our previous studies was higher than that between TCP3 and the other TFs, suggesting that TCP6 might indirectly activate UGT84A13 through other TFs, which need further validation. However, the expression of TCP3 in the cupules was much higher than that of TCP6. Considering the direct activation of TCP3 on UGT84A13 promoter, we concluded that TCP3 is a key TF regulating the expression of UGT84A13 and HT biosynthesis (Figure 7).
5 Conclusions
In summary, this study presents the first comprehensive genome-wide analysis of the TCP gene family in Q. variabilis. A total of 22 TCP genes were identified. Tissue-specific expression analysis and whole-genome identification of TCP genes led to the selection of TCP3 and TCP6 as candidate genes for regulating UGT84A13 expression and HTs biosynthesis. A dual-luciferase assay confirmed that TCP3, but not TCP6, is involved in the transcriptional regulation of UGT84A13, consequently affecting HTs synthesis in Q. variabilis. This study provides new insights into the transcriptional regulation of UGT84A13 and HTs biosynthesis, enhancing the understanding for the reason why the cupule of Q. variabilis could be used in medicine.
Data availability statement
The RNA-Seq raw data can be obtained from CNGB Sequence Archive (CNSA) of China National GeneBank DataBase (CNGBdb, https://db.cngb.org/) with accession nos.:CNP0003737. Genome of Q. variabilis can be obtained from CNGBdb with accession number. Genome annotations of Q. variabilis are available at FigShare. (10.6084/m9.figshare.23910282). Other data that support the findings of this study are available from the corresponding authors upon reasonable request.
Author contributions
YW: Conceptualization, Data curation, Formal analysis, Investigation, Validation, Visualization, Writing – original draft, Writing – review & editing. JL: Conceptualization, Data curation, Investigation, Visualization, Writing – review & editing. YC: Data curation, Formal analysis, Investigation, Visualization, Writing – review & editing. ZY: Data curation, Formal analysis, Resources, Visualization, Writing – review & editing. PL: Investigation, Writing – review & editing. GL: Project administration, Resources, Supervision, Writing – review & editing. QY: Conceptualization, Funding acquisition, Methodology, Project administration, Resources, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Key R&D Program of China (2021YFD2200302), the China Postdoctoral Science Foundation (2021M700451) and the 5·5 Engineering Research & Innovation Team Project at the Beijing Forestry University (BLRC2023B08).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1444081/full#supplementary-material
References
Aggarwal, P., Das Gupta, M., Joseph, A. P., Chatterjee, N., Srinivasan, N., Nath, U. (2010). Identification of specific DNA binding residues in the TCP family of transcription factors in arabidopsis. Plant Cell 22, 1174–1189. doi: 10.1105/tpc.109.066647
Bontpart, T., Marlin, T., Vialet, S., Guiraud, J. L., Pinasseau, L., Meudec, E., et al. (2016). Two shikimate dehydrogenases, VvSDH3 and VvSDH4, are involved in gallic acid biosynthesis in grapevine. J. Exp. Bot. 67, 3537–3550. doi: 10.1093/jxb/erw184
Cao, Y., Chen, Y. (2015). Biomass, carbon and nutrient storage in a 30-year-old Chinese cork oak (Quercus variabilis) forest on the south slope of the Qinling mountains, China. Forests 6, 1239–1255. doi: 10.3390/f6041239
Chandrasekhar, Y., Ramya, E. M., Navya, K., Phani Kumar, G., Anilakumar, K. R. (2017). Antidepressant-like effects of hydrolysable tannins of Terminalia catappa leaf extract via modulation of hippocampal plasticity and regulation of monoamine neurotransmitters subjected to chronic mild stress (CMS). Biomed. Pharmacother. 86, 414–425. doi: 10.1016/j.biopha.2016.12.031
Chang, L., Wu, S., Tian, L. (2021). Methyl jasmonate elicits distinctive hydrolyzable tannin, flavonoid, and phyto-oxylipin responses in pomegranate (Punica granatum L.) leaves. Planta 254, 89. doi: 10.1007/s00425-021-03735-9
Chen, C., Wu, Y., Li, J., Wang, X., Zeng, Z., Xu, J., et al. (2023). TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 16, 1733–1742. doi: 10.1016/j.molp.2023.09.010
Cubas, P., Lauter, N., Doebley, J., Coen, E. (1999). The TCP domain: a motif found in proteins regulating plant growth and development. Plant J. 18, 215–222. doi: 10.1046/j.1365-313X.1999.00444.x
Das, A. K., Islam, M. N., Faruk, M. O., Ashaduzzaman, M., Dungani, R., Sveriges, L. (2020). Review on tannins: Extraction processes, applications and possibilities. South Afr. J. Bot. 135, 58–70. doi: 10.1016/j.sajb.2020.08.008
Deng, C. (2018). Isolation and characterization of transcription inhibitors in persimmon fruit postharvest deastringency. Hangzhou (Zhejiang): Zhejiang University.
Fischer, U. A., Dettmann, J. S., Carle, R., Kammerer, D. R. (2011). Impact of processing and storage on the phenolic profiles and contents of pomegranate (Punica granatum L.) juices. Eur. Food Res. Technol. 233, 797–816. doi: 10.1007/s00217-011-1560-3
Freudenberg, K. (1960). Catechine und Hydroxy-flavandiole als Gerbstoffbildner. Experientia 16, 101–105. doi: 10.1007/BF02158087
Gallardo, A., Morcuende, D., Solla, A., Moreno, G., Pulido, F., Quesada, A. (2019). Regulation by biotic stress of tannins biosynthesis in Quercus ilex: Crosstalk between defoliation and Phytophthora cinnamomi infection. Physiol. Plant. 165, 319–329. doi: 10.1111/ppl.12848
Habashi, R., Hacham, Y., Dhakarey, R., Matityahu, I., Holland, D., Tian, L., et al. (2019). Elucidating the role of shikimate dehydrogenase in controlling the production of anthocyanins and hydrolysable tannins in the outer peels of pomegranate. BMC Plant Biol. 19, 476. doi: 10.1186/s12870-019-2042-1
Hao, J., Zheng, L., Han, Y., Zhang, H., Hou, K., Liang, X., et al. (2023). Genome-wide identification and expression analysis of TCP family genes in Catharanthus roseus. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1161534
Howarth, D. G., Donoghue, M. J. (2006). Phylogenetic analysis of the “ECE” (CYC/TB1) clade reveals duplications predating the core eudicots. Proc. Natl. Acad. Sci. United States America 103, 9101–9106. doi: 10.1073/pnas.0602827103
Hu, X., Zhang, W., Zhou, J. (2016). Plastic responses in tree architecture to different light intensity habitats: A case of Chinese cork oak. Polish J. Ecol. 64, 500–508. doi: 10.3161/15052249PJE2016.64.4.005
Iqbal, N., Poór, P. (2024). Plant protection by tannins depends on defence-related phytohormones. J. Plant Growth Regul. doi: 10.1007/s00344-024-11291-1
Jia, L. Y., Wang, J., Lv, C. N., Xu, T. Y., Jia, B. Z., Lu, J. C. (2013). A new cycloartane nortriterpenoid from stem and leaf of Quercus variabilis. J. Asian Natural Prod. Res. 15, 1050–1054. doi: 10.1080/10286020.2012.762911
Jiu, S., Xu, Y., Wang, J., Wang, L., Wang, S., Ma, C., et al. (2019). Genome-Wide identification, characterization, and transcript analysis of the TCP transcription factors in Vitis vinifera. Front. Genet. 10. doi: 10.3389/fgene.2019.01276
Khan, A. N., Singh, R., Bhattacharya, A., Chakravarti, R., Roy, S., Ravichandiran, V., et al. (2022). A short review on glucogallin and its pharmacological activities. Mini-Reviews Med. Chem. 22, 2820–2830. doi: 10.2174/1389557522666220513150907
Kumar, S., Stecher, G., Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Laddha, A. P., Kulkarni, Y. A. (2019). Tannins and vascular complications of Diabetes: An update. Phytomedicine 56, 229–245. doi: 10.1016/j.phymed.2018.10.026
Lewis, N. G., Yamamoto, E. (1989). Tannins — their place in plant metabolism. Chem. Signific. Condensed Tannins, 23–46. doi: 10.1007/978-1-4684-7511-1_2
Li, C., Du, J., Xu, H., Feng, Z., Chater, C. C. C., Duan, Y., et al. (2024). UVR8-TCP4-LOX2 module regulates UV-B tolerance in Arabidopsis. J. Integr. Plant Biol. 66, 897–908. doi: 10.1111/jipb.13648
Li, H., Wen, X., Huang, X., Wei, M., Chen, H., Yu, Y., et al. (2022). Genome-Wide identification and characterization of TCP gene family members in Melastoma candidum. Molecules 27, 9036. doi: 10.3390/molecules27249036
Lin, Y. F., Chen, Y. Y., Hsiao, Y. Y., Shen, C. Y., Hsu, J. L., Yeh, C. M., et al. (2016). Genome-wide identification and characterization of TCP genes involved in ovule development of Phalaenopsis equestris. J. Exp. Bot. 67, 5051–5066. doi: 10.1093/jxb/erw273
Ma, J., Wang, Q., Sun, R., Xie, F., Jones, D. C., Zhang, B. (2014). Genome-wide identification and expression analysis of TCP transcription factors in Gossypium raimondii. Sci. Rep. 4, 6645. doi: 10.1038/srep06645
Marrone, G., Di Lauro, M., Izzo, F., Cornali, K., Masci, C., Vita, C., et al. (2023). Possible beneficial effects of hydrolyzable tannins deriving from Castanea sativa L. @ in internal medicine. Nutrients 16, 45. doi: 10.3390/nu16010045
Melo, L. F. M. D., Aquino-Martins, V. G. D. Q., Silva, A. P. D., Oliveira Rocha, H. A., Scortecci, K. C. (2023). Biological and pharmacological aspects of tannins and potential biotechnological applications. Food Chem. 414, 135645. doi: 10.1016/j.foodchem.2023.135645
Mittasch, J., Bottcher, C., Frolova, N., Bonn, M., Milkowski, C. (2014). Identification of UGT84A13 as a candidate enzyme for the first committed step of gallotannin biosynthesis in pedunculate oak (Quercus robur). Phytochemistry 99, 44–51. doi: 10.1016/j.phytochem.2013.11.023
Niemetz, R., Gross, G. G. (2005). Enzymology of gallotannin and ellagitannin biosynthesis. Phytochemistry 17, 2001–2011. doi: 10.1016/j.phytochem.2005.01.009
Parapunova, V., Busscher, M., Busscher-Lange, J., Lammers, M., Karlova, R., Bovy, A. G., et al. (2014). Identification, cloning and characterization of the tomato TCP transcription factor family. BMC Plant Biol. 14, 157. doi: 10.1186/1471-2229-14-157
Rauluseviciute, I., Riudavets-Puig, R., Blanc-Mathieu, R., Castro-Mondragon, J. A., Ferenc, K., Kumar, V., et al. (2024). JASPAR 2024: 20th anniversary of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 52, D174–D182. doi: 10.1093/nar/gkad1059
Rohilla, M., Rishabh, Bansal, S., Garg, A., Dhiman, S., Dhankhar, S., et al. (2023). Discussing pathologic mechanisms of Diabetic retinopathy & therapeutic potentials of curcumin and beta-glucogallin in the management of Diabetic retinopathy. Biomed. pharmacother. 169, 115881. doi: 10.1016/j.biopha.2023.115881
Saigo, T., Wang, T., Watanabe, M., Tohge, T. (2020). Diversity of anthocyanin and proanthocyanin biosynthesis in land plants. Curr. Opin. Plant Biol. 55, 93–99. doi: 10.1016/j.pbi.2020.04.001
Sandhu, A. K., Gray, D. J., Lu, J., Gu, L. (2011). Effects of exogenous abscisic acid on antioxidant capacities, anthocyanins, and flavonol contents of muscadine grape (Vitis rotundifolia) skins. Food Chem. 126, 982–988. doi: 10.1016/j.foodchem.2010.11.105
Schommer, C., Palatnik, J. F., Aggarwal, P., Chételat, A., Cubas, P., Farmer, E. E., et al. (2008). Control of jasmonate biosynthesis and senescence by miR319 targets. PloS Biol. 6, e230. doi: 10.1371/journal.pbio.0060230
Shirmohammadli, Y., Efhamisisi, D., Pizzi, A. (2018). Tannins as a sustainable raw material for green chemistry: A review. Ind. Crops Prod. 126, 316–332. doi: 10.1016/j.indcrop.2018.10.034
Smeriglio, A., Barreca, D., Bellocco, E., Trombetta, D. (2017). Proanthocyanidins and hydrolysable tannins: occurrence, dietary intake and pharmacological effects. Br. J. Pharmacol. 174, 1244–1262. doi: 10.1111/bph.13630
Sun, J., Shi, W., Wu, Y., Ji, J., Feng, J., Zhao, J., et al. (2021). Variations in Acorn Traits in Two Oak Species: Quercus mongolica Fisch. ex Ledeb. and Quercus variabilis Blume. Forests 12, 1755. doi: 10.3390/f12121755
Sun, L., Zou, X., Jiang, M., Wu, X., Chen, Y., Wang, Q., et al. (2020). The crystal structure of the TCP domain of PCF6 in Oryza sativa L. reveals an RHH-like fold. FEBS Lett. 594, 1296–1306. doi: 10.1002/1873-3468.13727
Tabarelli, M., Malnoy, M., Janik, K. (2022). Chasing consistency: an update of the TCP gene family of malus x domestica. Genes (Basel) 13, 1696. doi: 10.3390/genes13101696
Tahara, K., Nishiguchi, M., Funke, E., Miyazawa, S., Miyama, T., Milkowski, C. (2021). Dehydroquinate dehydratase/shikimate dehydrogenases involved in gallate biosynthesis of the aluminum-tolerant tree species Eucalyptus camaldulensis. Planta 253, 3. doi: 10.1007/s00425-020-03516-w
Takeda, T., Suwa, Y., Suzuki, M., Kitano, H., Ueguchi-Tanaka, M., Ashikari, M., et al. (2003). The OsTB1 gene negatively regulates lateral branching in rice. Plant Journal: Cell Mol. Biol. 33, 513–520. doi: 10.1046/j.1365-313X.2003.01648.x
Tatematsu, K., Nakabayashi, K., Kamiya, Y., Nambara, E. (2008). Transcription factor AtTCP14 regulates embryonic growth potential during seed germination in Arabidopsis thaliana. Plant J. 53, 42–52. doi: 10.1111/j.1365-313X.2007.03308.x
Tondi, G., Thevenon, M. F., Mies, B., Standfest, G., Petutschnigg, A., Wieland, S. (2013). Impregnation of Scots pine and beech with tannin solutions: effect of viscosity and wood anatomy in wood infiltration. Wood Sci. Technol. 47, 615–626. doi: 10.1007/s00226-012-0524-5
Wang, Q., Xu, G., Zhao, X., Zhang, Z., Wang, X., Liu, X., et al. (2020). Transcription factor TCP20 regulates peach bud endodormancy by inhibiting DAM5/DAM6 and interacting with ABF2. J. Exp. Bot. 4, 1585–1597. doi: 10.1093/jxb/erz516
Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40, e49. doi: 10.1093/nar/gkr1293
Xie, J., Chen, Y., Cai, G., Cai, R., Hu, Z., Wang, H. (2023). Tree Visualization By One Table (tvBOT): a web application for visualizing, modifying and annotating evolutionary trees. Nucleic Acids Res. 51, W587–W592. doi: 10.1093/nar/gkad359
Xie, P., Shi, J., Tang, S., Chen, C., Khan, A., Zhang, F., et al. (2019). Control of bird feeding behavior by tannin1 through modulating the biosynthesis of polyphenols and fatty acid-derived volatiles in sorghum. Mol. Plant 12, 1315–1324. doi: 10.1016/j.molp.2019.08.004
Xie, Y. G., Ma, Y. Y., Bi, P. P., Wei, W., Liu, J., Hu, Y., et al. (2020). Transcription factor FvTCP9 promotes strawberry fruit ripening by regulating the biosynthesis of abscisic acid and anthocyanins. Plant Physiol. Biochem. 146, 374–383. doi: 10.1016/j.plaphy.2019.11.004
Xu, Y., Liu, H., Gao, Y., Xiong, R., Wu, M., Zhang, K., et al. (2021). The TCP transcription factor PeTCP10 modulates salt tolerance in transgenic Arabidopsis. Plant Cell Rep. 40, 1971–1987. doi: 10.1007/s00299-021-02765-7
Xu, Y. M., Xiao, X. M., Zeng, Z. X., Tan, X. L., Liu, Z. L., Chen, J. W., et al. (2019). BrTCP7 transcription factor is associated with meJA-promoted leaf senescence by activating the expression of brOPR3 and brrccr. Int. J. Mol. Sci. 20 (16), 3963. doi: 10.3390/ijms20163963
Yang, Q., Chen, X., Li, J., Pei, Z., Chen, Y., Liu, P., et al. (2023). HB26, a member of HD-Zip I subfamily, is involved in the regulation of hydrolysable tannin biosynthesis in the cupules of Quercus variabilis by transactivation of UGT84A13. Ind. Crops Prod. 200, 116866. doi: 10.1016/j.indcrop.2023.116866
Yang, Q., Li, J., Wang, Y., Wang, Z., Pei, Z., Street, N. R., et al. (2024). Genomic basis of the distinct biosynthesis of β-glucogallin, a biochemical marker for hydrolyzable tannin production, in three oak species. New Phytol. 242, 2702–2718. doi: 10.1111/nph.19711
Yang, Q., Yang, B., Li, J., Wang, Y., Tao, R., Yang, F., et al. (2020). ABA-responsive ABRE-BINDING FACTOR3 activates DAM3 expression to promote bud dormancy in Asian pear. Plant Cell Environ. 43, 1360–1375. doi: 10.1111/pce.13744
Yao, X., Ma, H., Wang, J., Zhang, D. (2007). Genome-Wide comparative analysis and expression pattern of TCP gene families in Arabidopsis thaliana and Oryza sativa. J. Integr. Plant Biol. 6, 885–897. doi: 10.1111/j.1744-7909.2007.00509.x
Ye, Q., Zhang, S., Qiu, N., Liu, L., Wang, W., Xie, Q., et al. (2021). Identification and characterization of glucosyltransferase that forms 1-galloyl-β-d-glucogallin in Canarium album L., a functional fruit rich in hydrolysable tannins. Molecules 26, 4650. doi: 10.3390/molecules26154650
Yin, P., Yang, L., Li, K., Fan, H., Xue, Q., Li, X., et al. (2019). Bioactive components and antioxidant activities of oak cup crude extract and its four partially purified fractions by HPD-100 macroporous resin chromatography. Arab. J. Chem. 12, 249–261. doi: 10.1016/j.arabjc.2016.09.018
Yu, L., Chen, Q., Zheng, J., Xu, F., Ye, J., Zhang, W., et al. (2022). Genome-wide identification and expression pattern analysis of the TCP transcription factor family in Ginkgo biloba. Plant Signaling Behav. 17, 1994248–1994248. doi: 10.1080/15592324.2021.1994248
Yuan, Z., Fang, Y., Zhang, T., Fei, Z., Han, F., Liu, C., et al. (2018). The pomegranate (Punica granatum L.) genome provides insights into fruit quality and ovule developmental biology. Plant Biotechnol. J. 16, 1363–1374. doi: 10.1111/pbi.12875
Zhang, X., Bao, Y., Shan, D., Wang, Z., Song, X., Wang, Z., et al. (2018). Magnaporthe oryzae induces the expression of a microRNA to suppress the immune response in rice. Plant Physiol. 177, 352–368. doi: 10.1104/pp.17.01665
Zhou, D. (2016). Research on antibacterial activity and mechanism of extracts from Quercus variabilis Blume against foodborne pathogens. Xianyang (Shannxi): Northwest A&F University.
Zhou, D., Liu, Z. H., Wang, D. M., Li, D. W., Yang, L. N., Wang, W. (2019). Chemical composition, antibacterial activity and related mechanism of valonia and shell from Quercus variabilis Blume (Fagaceae) against Salmonella paratyphi a and Staphylococcus aureus. BMC Complement Altern. Med. 19, 271. doi: 10.1186/s12906-019-2690-6
Keywords: Quercus variabilis, traditional medicine, hydrolyzable tannins, TCP gene family, UGT84A13
Citation: Wang Y, Li J, Chen Y, Yu Z, Liu P, Li G and Yang Q (2024) Genome-wide identification of TCP transcription factors and their potential roles in hydrolyzable tannin production in Quercus variabilis cupule. Front. Plant Sci. 15:1444081. doi: 10.3389/fpls.2024.1444081
Received: 05 June 2024; Accepted: 18 July 2024;
Published: 06 August 2024.
Edited by:
Xinyi Guo, Central European Institute of Technology (CEITEC), CzechiaCopyright © 2024 Wang, Li, Chen, Yu, Liu, Li and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qinsong Yang, cXN5YW5nQGJqZnUuZWR1LmNu
 Yaochen Wang1,2,3
Yaochen Wang1,2,3 Guolei Li
Guolei Li Qinsong Yang
Qinsong Yang