- School of Biological Sciences, Jining Medical University, Rizhao, Shandong, China
Small heat shock proteins (sHSPs) are molecular chaperones that are widely present in plants and play a vital role in the response of plants to various environmental stimuli. This study employed transgenic Arabidopsis to investigate the impact of the new tomato (Solanum lycopersicum) sHSP protein (SlHSP17.3) on salt stress tolerance. Transient conversion analysis of Arabidopsis protoplasts revealed that SlHSP17.3 localized to the cytoplasm. Furthermore, as suggested by expression analysis, salt stress stimulated SlHSP17.3 expression, suggesting that SlHSP17.3 is involved in the salt stress response of plants. SlHSP17.3-overexpressing plants presented greater germination rates, fresh weights, chlorophyll contents, and Fv/Fm ratios, as well as longer root lengths, lower reactive oxygen species (ROS) levels, and lighter cell membrane injury under salt stress. Furthermore, certain stress-related genes (AtCOR15, AtDREB1B, and AtHSFA2) were up-regulated in salt-stressed transgenic plants. Overall, SlHSP17.3 overexpression improved the salt stress resistance of transgenic plants, mainly through increasing AtCOR15, AtDREB1B, and AtHSFA2 expression.
1 Introduction
Salt stress is induced by elevated levels of soluble salts in the soil; it presents considerable challenges to plant growth and global agricultural productivity and constitutes a critical concern for global resources and ecology (Guo et al., 2022). Soil salinity, often exacerbated by irrigation practices and environmental factors such as arid climates and proximity to saltwater bodies, poses a threat to the sustainable cultivation of crops. Salt stress adversely affects various physio-biochemical events crucial for plant development.
Plants engage various complex adaptive mechanisms in response to salt stress to counteract harmful effects and preserve the cellular balance. This includes the activation of small heat shock proteins (sHSPs), the molecular chaperones instrumental for cellular responses to stress, which are especially useful in shielding cells against the negative impacts of thermal stress (Waters, 2013; Papsdorf and Richter, 2014; He et al., 2021). These proteins, typically ranging from 12–42 kDa, are recognized for their capacity to bind to and stabilize unfolded or partially folded proteins, thus preventing protein aggregation while assisting in later refolding (Sun et al., 2016). sHSPs, which are ancient and diverse, share one highly conserved α-crystallin domain (ACD), one variable N-terminal region, and one short C-terminal sequence. In most sHSPs, the ACD or HSP20 region includes nine highly conserved β-sheets (Hibshman et al., 2023). In accordance with their molecular weights, such HSPs can be classified into six categories: sHSPs, HSP40 (DnaJ family), HSP60, HSP70, HSP90, and HSP100 (Sun et al., 2012). sHSPs are currently recognized as the most extensively studied and largest HSP family (Basha et al., 2012). They are subclassified into cytosolic classes I (CI-type) and II (CII-type), mitochondria, chloroplasts, peroxisomes, and endoplasmic reticulum categories on the basis of sequence similarity and intracellular location (Siddique et al., 2008; Jiang et al., 2009). sHSP compartmentalization in plants not only highlights their role in maintaining cellular balance but also indicates functional specialization depending on their location, enhancing the overall stress resilience of plants.
sHSP expression within plants is induced by stresses such as heat, salt, cold, drought, heavy metals, and oxidation, underscoring their versatility and essential role in cellular defense mechanisms (Wu et al., 2022). These proteins are inducible, indicating a highly regulated transcriptional process, with heat shock factors (HSFs) activating the expression of HSPs, including sHSPs, upon stress (Wang et al., 2019a; Iqbal et al., 2022). This regulation plays a vital role in cell survival under unfavorable conditions, demonstrating the integral role of sHSPs in the cellular stress response. Research on plant sHSPs indicates that they are also involved in stress responses and development, not just functioning as molecular chaperones. For example, the expression of CI-type sHSPs increases during embryo formation and seed maturation (Wang et al., 2014). Additionally, sHSP proteins decrease light dependence during tomato seed germination (Koo et al., 2015) and protect specific cell types in plant embryo sacs under heat stress (Ambastha and Leshem, 2020a). Yang et al. (2022) reported that overexpression of GmHSP17.9 in transgenic soybeans results in significant increases in fresh weight, nodule number, poly-β-hydroxybutyrate (PHB) content, nitrogenase activity, urea, and total nitrogen level, along with a notable increase in seed yield. Feng et al. (2019) reported that CaHSP25.9 overexpression in Arabidopsis thaliana enhances the germination rate and length of roots under drought stress. Tomato SlHSP17.7 has been identified as a cofactor for SlCCX1-like, which targets endoplasmic reticulum (ER) membrane proteins to maintain intracellular Ca2+ homeostasis and reduce cold stress sensitivity (Zhang et al., 2020). As discovered by Qin et al. (2022), transgenic Arabidopsis plants overexpressing TaHSP17.6 grow more lateral roots to adapt to salt stress. According to Tian et al. (2022), compared with their wild-type (WT) counterparts, transgenic tobacco plants harboring the PtsHSP17.2 gene presented less variation in chlorophyll levels, malondialdehyde (MDA) levels, and relative electrolyte leakage upon heat stress. Hence, although the effects of stress-responsive sHSP genes have been documented, further extensive characterization of such abiotic stress responses is warranted to fully understand stress resistance in plants.
Numerous sHSP genes in plants respond to various signals; however, the specific roles of these genes in tomatoes remain unclear. This study isolated the stress-related sHSP gene SlHSP17.3 from tomatoes and characterized it to elucidate its functions. The expression of SlHSP17.3 can be triggered upon salt stress. Additionally, phylogenetic tree analysis together with subcellular localization studies revealed SlHSP17.3 as the CII-type sHSP. The overexpression of SlHSP17.3 in Arabidopsis enhances salt stress resistance.
2 Materials and methods
2.1 Plants and treatments
Wide-type Arabidopsis (WT, ecotype Columbia-0, retained by our laboratory) and T3 transgenic Arabidopsis plants (genetic transformation obtained by our laboratory) were cultivated in quartz sand at 22°C (day)/20°C (night), with a 14-h/10-h light/dark cycle and 200 μmol m-2s-1 photon flux density. The WT tomato variety (S. lycopersicum cv. L-402, retained by our laboratory) was cultivated in quartz sand at 25°C (day)/22°C (night), with a 16-h/8-h light/dark cycle and a 200 μmol m-2s-1 photon flux density. Each plant material was subjected to irrigation via Hoagland’s nutrient solution once/weekly.
To investigate the SlHSP17.3 expression profile upon salt treatment, Hoagland nutrient solution supplemented with 250 mM NaCl was added to the roots collected from six-week-old WT tomato plants, whereas Hoagland nutrient solution treatment alone was used for the control group. Moreover, 100 µM ABA was sprayed on plant leaves, while water was sprayed on control plant leaves. To analyze salt resistance, 200 mM NaCl solution was added to two-week-old WT and T3 transgenic Arabidopsis plants for a 14-day period.
2.2 Cloning and bioinformatics analysis of SlHSP17.3
The SlHSP17.3 coding sequence (CDS) was subjected to amplification based on WT tomato leaf cDNA via polymerase chain reaction (PCR) with specific primers (5′-CCATGGATGGATTTGAGGTTGATGGGT-3′ (forward), 5′-CACGTGAGCAACTTTGACCTGAATGG-3′ (reverse)). Then, we ligated the PCR products into the pMD19-T vector (TaKaRa, Beijing, China) prior to sequencing. Multiple sequence alignment was conducted with DNAMAN version 6.0 software (Lynnon Biosoft, USA), and phylogenetic analysis of the SlHSP17.3 protein was performed via MEGA version 5.05 software (Sudhir Kumar, Temple University, USA). Additionally, we acquired the amino acid sequences of sHSP proteins from various plants from GenBank (https://www.ncbi.nlm.nih.gov/genbank/) for comparison. The accession numbers are as follows: AtHSP21 (Arabidopsis thaliana, NM_118906); AtHSP25.3 (DQ446875); NtHSP26 (Nicotiana tabacum, D88584); OsHSP26 (Oryza sativa, AB020973); PhHSP21 (Petunia×hybrida, X54103); PsHSP21 (Pisum sativum, X07187); SlHSP21 (Solanum lycopersicum, LEU66300); TaHSP26.6 (Triticum aestivum, AF097659); TtHSP26.8 (Triticum turgidum ssp. Dicoccon, AJ971372; ZmHSP (Zea mays, EU966283); ZmHSP26 (L28712); AtHSP17.6 (NM_121240); AtHSP17.7 (O81822); CpHSP17.7 (Carica papaya, AY242075); GmHSP17.9 (Glycine max, P05477); LpHSP17.4 (Lycopersicon peruvianum, AY608694); MsHSP17 (Medicago sativa, X98617); PdHSP17.5 (Prunus dulcis, AF159562); PsHSP17.1 (P19242); SlHSP17.6 (LEU72396); ZmHSP17.5 (EU970990); AtHSP22.0 (Q38806); GmHSP22 (X63198); PsHSP22.7 (M33898); AcHSP (Ananas comosus, AY098528); AtHSP17.4 (NM_114492); AtHSP17.6A (NM_104679); AtHSP17.8 (NM_100614); AtHSP18.2 (NM_125364); CfHSPI (Capsicum frutescens, AY284925); CpHSP17.5 (AY387588); CsHSP17.5 (Castanea sativa, AJ582679); CsHSP (Camellia sinensis, EU727315); FaHSP (Fragaria×ananassa, U63631); HaHSP17.6 (Helianthus annuus, X59701); HaHSP17.9 (AJ237596); HvHSP17 (Hordeum vulgare ssp. vulgare) (Y07844); HvHSP18 (X64561); LpHSP19.9 (AJ225047); LpHSP20 (AJ225048); LpHSP20.1 (AJ225046); MdHSP1 (Malus×domestica, AF161179); MdHSP17.5 (EU636239); MsHSP18.2 (X58711); NtHSP18 (X70688); OsHSP16.9 (X60820); OsHSP17.8 (X75616); OsHSP18 (FJ383169); OsHSP20 (EU325986); RcHSP17.8 (Rosa chinensis, EF053229); SlHSP17.7 (AF123255); SlHSP17.8 (AF123256); SlHSP20 (SLU59917); TtHSP16.9 (AM709752); VuHSP17.7 (Vigna unguiculata, EF514500); ZmHSP17.2 (X65725); AtHSP15.7 (DQ403190); GmHSP (B0M1A7); OsHSP16.0 (Q652V8); AtHSP23.5 (NM_124523); AtHSP23.6 (NM_118652); PsHSP22 (X86222); SlHSP (AB017134); and ZmHSP22 (AY758275).
2.3 SlHSP17.3 subcellular localization
The full-length SlHSP17.3 CDS was amplified via the corresponding primers (5′-CTCGAGATGGATTTGAGGTTGATGGGT-3′ (forward), 5′-GGTACCGTAGCAACTTTGACCTGAATGG-3′ (reverse)). The amplified SlHSP17.3 CDS was subsequently inserted into the vector pEZS-NL after XhoI/KpnI digestion, resulting in the generation of a SlHSP17.3::EGFP (enhanced green fluorescent protein) fusion construct regulated via the cauliflower mosaic virus (CaMV) 35S promoter. We subsequently transfected Arabidopsis mesophyll protoplasts with both EGFP and SlHSP17.3::EGFP recombinant plasmids, and fluorescence was observed using laser confocal microscopy (Leica TCS SP8, Leica, Wetzlar, Germany).
2.4 Transgenic Arabidopsis plant transformation and identification
We cloned the SlHSP17.3 CDS in the CaMV 35S promoter-controlled pCAMBIA1302 binary expression vector and subsequently inserted this resulting recombinant plastid into Agrobacterium tumefaciens strain GV3101, which was then verified via PCR. Arabidopsis (Col-0) transformation was accomplished via the floral dip approach. After the transgenic plants were cultivated on half-strength MS agar plates supplemented with 25 µg/mL hygromycin for the identification of positive lines, DNA was extracted from WT plants and hygromycin-resistant T1 seedlings. The presence of target genes was confirmed by a PCR assay via the following primers: 35S promoter TACGCAGCAGGTCTCTCAAGACGAT (forward) and SlHSP17.3 CACGTGAGCAACTTTGACCTGAATGG (reverse). Subsequently, ten separate homozygous transgenic T3 lines were obtained. To ensure uniform viability, all the seeds utilized in each assay were collected at identical stages and preserved under identical conditions.
2.5 Amplification of the SlHSP17.3 promoter and GUS staining analysis
Using the cetyl-trimethylammonium bromide (CTAB) approach, the SlHSP17.3 promoter region was amplified from genomic DNA isolated from leaf samples of WT tomato plants. This genomic DNA served as the template for amplification. For GUS staining analysis, T3 transgenic Arabidopsis seedlings expressing SlHSP17.3pro::GUS were subjected to both natural conditions and salt stress treatment. GUS staining was conducted following the methods of Sundaresan et al. (1995). The quantitative analysis of GUS staining was mainly based on the description by Ambastha and Leshem (2020b).
2.6 Salt stress assay
In the salt assay, we sprayed surface sterile T3 and WT progeny seeds on half-strength MS media supplemented with NaCl at varying concentrations (0, 100, and 150 mM); following two days of vernalization in the dark at 4°C, we placed the plates in the chamber and measured the germination rate daily thereafter. To analyze root growth after germination, we first sowed the seeds onto half-strength MS media for a three-day period and subsequently added the seeds with emerged radicles to half-strength MS media containing various NaCl concentrations (0, 100, and 150 mM) for fivedays. We subsequently assessed root elongation and fresh weight. For the germination experiments, under different salt concentrations, each genotype had 49 seeds per plate with three plates per treatment, resulting in 147 seeds per genotype per experiment. For the root length experiments, under different salt concentrations, each genotype had five seedlings per plate with three plates per treatment, for a total of 15 seedlings per genotype per experiment. To analyze salt resistance in mature plants, 200 mM NaCl was applied for continuous treatment of four-week-old WT and T3 adult plants over a two-week duration, after which the phenotype was observed. The chlorophyll contents were quantified according to the approach by Kong et al. (2014). Each experiment was conducted three times.
2.7 Chlorophyll fluorescence analysis
Chlorophyll fluorescence imaging was conducted using a FluorCam multispectral fluorescence imaging system (PSI, Norfolk, USA) following the procedures outlined by Baker (2008). Chlorophyll fluorescence was measured using the Handy PEA instrument (Hansatech Instruments, Norfolk, UK) in accordance with the method from Ma et al. (2013). We subsequently determined the Fv/Fm ratio of PSII using the following equation: Fv/Fm = (Fm-Fo)/Fm.
2.8 Histochemical analysis and H2O2 and O2•− determinations
For hydrogen peroxide (H2O2) and superoxide radical (O2•-) staining, the leaves were immersed in 3,3’-diaminobenzidine (DAB) staining solution (DAB dissolved in 50 mM Tris-acetate buffer, pH 5.0, with a final concentration of 0.1 mg/mL) and nitro blue tetrazolium (NBT) staining solution (NBT dissolved in 25 mM phosphate buffer, pH 7.6, with a final concentration of 0.1 mg/mL), respectively. The leaves were incubated at room temperature in the dark overnight. Afterward, the leaves were removed and immersed in a bleaching solution (ethanol:acetic acid:glycerol at 4:1:1), boiled for 10 min for decolorization, and then photographed. Trypan blue staining was carried out as described by Choi et al. (2007). H2O2 and O2•− levels in both transgenic and WT leaf samples were subsequently analyzed via the method from Kong et al. (2014). For H2O2 and O2•- staining, 200 mM NaCl solution was added to four-week-old WT and T3 transgenic Arabidopsis plants for a two-day period. For trypan blue staining, 200 mM NaCl solution was added to four-week-old WT and T3 transgenic Arabidopsis plants for five days.
2.9 Physiological parameter measurements
For physiological parameter measurements, 200 mM NaCl solution was added to four-week-old WT and T3 transgenic Arabidopsis plants for two days. A total of 1.0 g of leaf tissue was rapidly ground in 5 mL of cold extraction buffer (50 mM potassium phosphate buffer, pH 7.0, containing 1 mM EDTA and 1% polyvinylpyrrolidone). After homogenization, the mixture was centrifuged at 12,000 ×g for 20 min at 4°C. The resulting supernatant was immediately used as the crude enzyme extract for assessing antioxidant enzyme activity. The determination of superoxide dismutase (SOD, EC 1.15.1.1) activity primarily followed the method of Giannopolitis and Ries (1977) with slight modifications. 0.1 mL aliquot of the crude enzyme solution (the control group using potassium phosphate buffer at 50 mM) was immediately added to the reaction mixture (1.5 mL of 50 mM potassium phosphate buffer, 0.3 mL of 65 mM methotrexate solution, 0.3 mL of 0.5 mM NBT solution, 0.3 mL of 0.1 mM EDTA-Na2, and 0.3 mL of 0.2 mM riboflavin) and placed in a glass tube. The control group was incubated in the dark, while the experimental group was exposed to light at 4000 lx for 30 minutes. Subsequently, the absorbance at 560 nm was measured. The determination of catalase (CAT, EC 1.11.1.6) activity primarily followed the method of Aebi (1984) with slight modifications. The following solutions in sequence (0.2 mL of crude enzyme extract, 1.5 mL of phosphate buffer at pH 7.8, and 1.0 mL of distilled water) were added in a 10 mL test tube. Then 0.3 mL of 0.1 mol L-1 H2O2 was added to the test tube, the timer started immediately, and the solution quickly transferred into a quartz cuvette. The absorbance at 240 nm was measured and a reading was taken every 1 minute for a total of 3 minutes. After the measurement, the enzyme activity was calculated according to the following formula: Catalase activity (U g-1 FW min-1) =△A240×VT/0.1×V×t×FW. VT represented total volume of the crude enzyme extract (mL). 0.1 refers to one enzyme activity unit (U) for every 0.1 decrease in A240. V represented volume of crude enzyme used for the determination (mL). t represented time from adding hydrogen peroxide to the last reading (min). FW represented fresh weight of the sample (g). The MDA level and relative electrical conductivity (REC) of the leaves were subsequently analyzed via the methods described by Kong et al. (2014). The ABA content of the leaves was assessed as described by Wang et al. (2019b).
2.10 Real-time quantitative PCR assay
The extraction of total leaf RNA was performed according to the method provided with the MolPure® Plant RNA Kit (Yeasen, Shanghai, China). Reverse transcription was carried out with the TaKaRa Reverse Transcription Kit to obtain cDNA. The relative gene expression was assessed via RT-qPCR, with cDNA used as the template and EF-1α (GenBank Accession No. LOC544055) and AtUbiquitin (At4g05320) as control genes. RT-qPCR conditions were 30 s at 95°C, 15 s at 95°C, 15 s at 55°C, and 15 s at 72°C for 42 cycles. Each treatment was repeated three times. Supplementary Table S1 displays the RT-qPCR primers used. For the RT-qPCR assay, 200 mM NaCl solution was added to four-week-old WT and T3 transgenic Arabidopsis plants for a two-day period.
2.11 Statistical analysis
SigmaPlot version 12.5 (Systat Software, San Jose, CA, USA) and SPSS version 18.0 (Chicago, IL, USA) were used for statistical analysis. The results are presented as the means ± standard deviations of at least three replicates. Significance levels are shown as *p < 0.05 and **p < 0.01 compared with the control.
3 Results
3.1 Identification and bioinformatics analysis of SlHSP17.3
The SlHSP17.3 gene is 773 base pairs (bp) long, and its open reading frame (ORF) is 468 bp. Its start codon (ATG) is located at the 148th nucleotide position, whereas the stop codon (TAG) is located at the 613th nucleotide position. The ORF is responsible for encoding one protein comprising 155 amino acids, and the estimated molecular weight is 17.3 kDa, whereas the isoelectric point is 6.75 (http://web.expasy.org/compute_pi/). According to a BLAST search of the tomato database (https://solgenomics.net/), this gene is located on chromosome 8 of the tomato genome. Phylogenetic tree analysis of reported sHSP proteins revealed that SlHSP17.3 is a member of the cytosolic class II sHSP protein group (Figure 1A). The cytosolic class II sHSP sequences revealed a distinctive N-terminal conserved domain (RDAKAMAATPADV) (Figure 1B). Moreover, the conserved C-terminal domain known as the ACD, comprising about 90 amino acids and containing consensus regions II/III, was identified. In addition, a polyproline motif (PPPEP) was detected in the C-terminus (Bondino et al., 2012; Li et al., 2016).
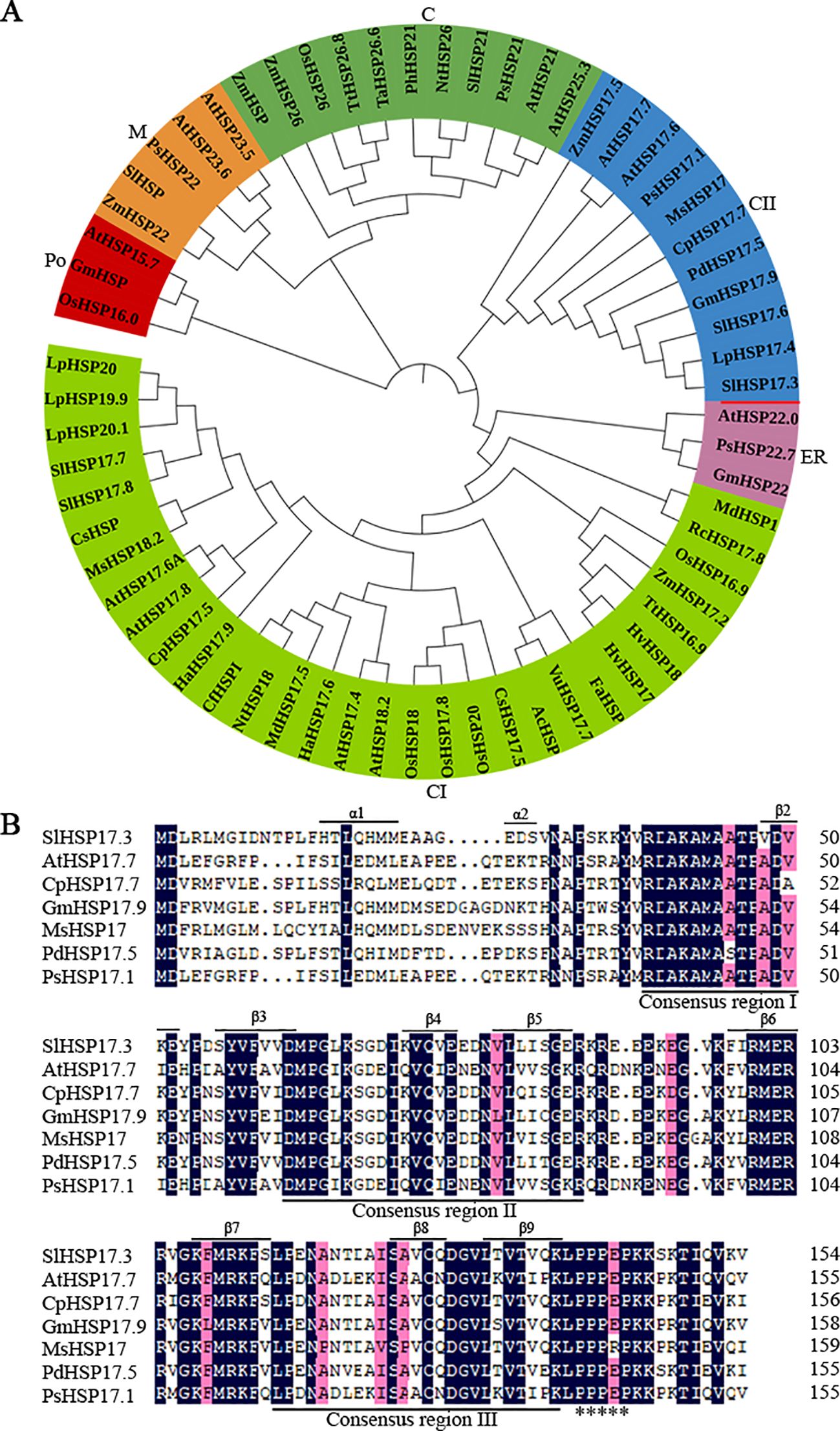
Figure 1. Phylogenetic tree and homologous sequence alignment analysis of sHSP proteins. (A) Phylogenetic tree analysis of SlHSP17.3 with additional sHSP proteins, which divided the sHSP protein family into six clades (C, chloroplast; CII, cytosolic II; ER, endoplasmic reticulum; CI, cytosolic I; Po, peroxisome; M, mitochondria). The unrooted phylogenetic tree of sHSP proteins was generated using the neighbor-joining approach in MEGA (version 5.05). The accession numbers are as follows: AtHSP21 (Arabidopsis thaliana, NM_118906); AtHSP25.3 (DQ446875); NtHSP26 (Nicotiana tabacum, D88584); OsHSP26 (Oryza sativa, AB020973); PhHSP21 (Petunia×hybrida, X54103); PsHSP21 (Pisum sativum, X07187); SlHSP21 (Solanum lycopersicum, LEU66300); TaHSP26.6 (Triticum aestivum, AF097659); TtHSP26.8 (Triticum turgidum ssp. Dicoccon, AJ971372; ZmHSP (Zea mays, EU966283); ZmHSP26 (L28712); AtHSP17.6 (NM_121240); AtHSP17.7 (O81822); CpHSP17.7 (Carica papaya, AY242075); GmHSP17.9 (Glycine max, P05477); LpHSP17.4 (Lycopersicon peruvianum, AY608694); MsHSP17 (Medicago sativa, X98617); PdHSP17.5 (Prunus dulcis, AF159562); PsHSP17.1 (P19242); SlHSP17.6 (LEU72396); ZmHSP17.5 (EU970990); AtHSP22.0 (Q38806); GmHSP22 (X63198); PsHSP22.7 (M33898); AcHSP (Ananas comosus, AY098528); AtHSP17.4 (NM_114492); AtHSP17.6A (NM_104679); AtHSP17.8 (NM_100614); AtHSP18.2 (NM_125364); CfHSPI (Capsicum frutescens, AY284925); CpHSP17.5 (AY387588); CsHSP17.5 (Castanea sativa, AJ582679); CsHSP (Camellia sinensis, EU727315); FaHSP (Fragaria×ananassa, U63631); HaHSP17.6 (Helianthus annuus, X59701); HaHSP17.9 (AJ237596); HvHSP17 (Hordeum vulgare ssp. vulgare) (Y07844); HvHSP18 (X64561); LpHSP19.9 (AJ225047); LpHSP20 (AJ225048); LpHSP20.1 (AJ225046); MdHSP1 (Malus×domestica, AF161179); MdHSP17.5 (EU636239); MsHSP18.2 (X58711); NtHSP18 (X70688); OsHSP16.9 (X60820); OsHSP17.8 (X75616); OsHSP18 (FJ383169); OsHSP20 (EU325986); RcHSP17.8 (Rosa chinensis, EF053229); SlHSP17.7 (AF123255); SlHSP17.8 (AF123256); SlHSP20 (SLU59917); TtHSP16.9 (AM709752); VuHSP17.7 (Vigna unguiculata, EF514500); ZmHSP17.2 (X65725); AtHSP15.7 (DQ403190); GmHSP (B0M1A7); OsHSP16.0 (Q652V8); AtHSP23.5 (NM_124523); AtHSP23.6 (NM_118652); PsHSP22 (X86222); SlHSP (AB017134); and ZmHSP22 (AY758275). (B) Multiple sequence alignment of SlHSP17.3 with additional sHSP proteins. Conserved motifs are underlined. The asterisk indicates one polyproline motif. Secondary structure prediction shows α-helix at the N-terminus and β-sheets in the C-terminal ACD (β2-β9) underlined.
3.2 Subcellular localization of SlHSP17.3
According to the ProtComp 9.0 database (http://www.softberry.com/berry.phtml) prediction, SlHSP17.3 is a potential cytoplasmic protein (Supplementary Table S2). For validation, transient transformation in vivo was conducted with Arabidopsis protoplasts isolated from leaves expressing 35S::EGFP and 35S::SlHSP17.3-EGFP fusion proteins (Figure 2A). 35S::EGFP exhibited dispersed green fluorescence throughout the protoplasts, with the exception of vacuoles (upper panels in Figure 2B). Conversely, transfection with the 35S::SlHSP17.3-EGFP fusion protein resulted in a clear concentration of green fluorescence within the cytoplasm (Figure 2B, lower panels). The results strongly support that SlHSP17.3 is a cytoplasmic protein.
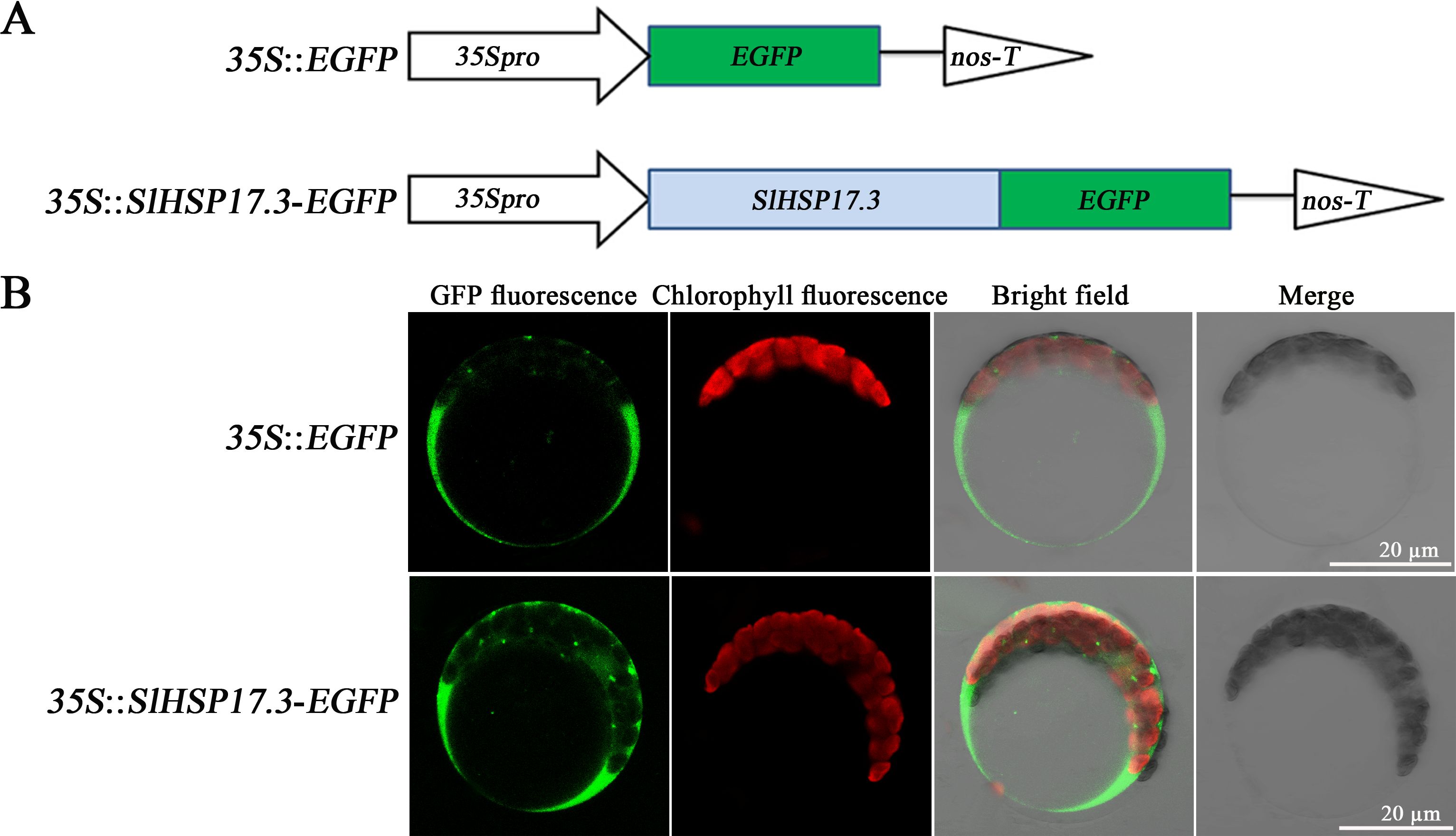
Figure 2. Subcellular localization of SlHSP17.3. (A) Structural pattern of the EGFP fusion protein. (B) 35S::EGFP (upper panel) and 35S::SlHSP17.3-EGFP (lower panel) transient expression in Arabidopsis protoplasts. Protoplasts were observed using laser confocal microscopy.
3.3 Expression of SlHSP17.3 induced by salt stress
To explore SlHSP17.3 tissue-specific expression, two methods were employed. First, we conducted RT-qPCR to analyze SlHSP17.3 expression patterns in various tomato organs. As shown in Figure 3A, SlHSP17.3 exhibited consistent expression across different organs, with a preference for expression in the leaves. Second, we used GUS staining to examine SlHSP17.3 expression in diverse tissues (Figure 3B). The results of the GUS staining analysis aligned with the RT-qPCR findings. Following treatment with 200 mM NaCl, the SlHSP17.3 transcription level first increased but then decreased, peaking on day 5 (Figure 4A). Additionally, GUS staining revealed that the color intensity of the cotyledonary leaves and stems of three-day-old seedlings under salt stress was darker than that of untreated seedlings (Supplementary Figure S1). The same trend was observed for the cotyledonary leaves and stems of seven-day-old seedlings; although there was little difference in color intensity among the true leaves of seven-day-old seedlings, quantitative analysis of GUS staining also supported this result (Figure 4B; Supplementary Figure S2). We speculated that the expression of SlHSP17.3 might be induced more prominently as the leaves mature. In summary, these experimental results indicate that the expression of SlHSP17.3 is significantly upregulated under salt stress.
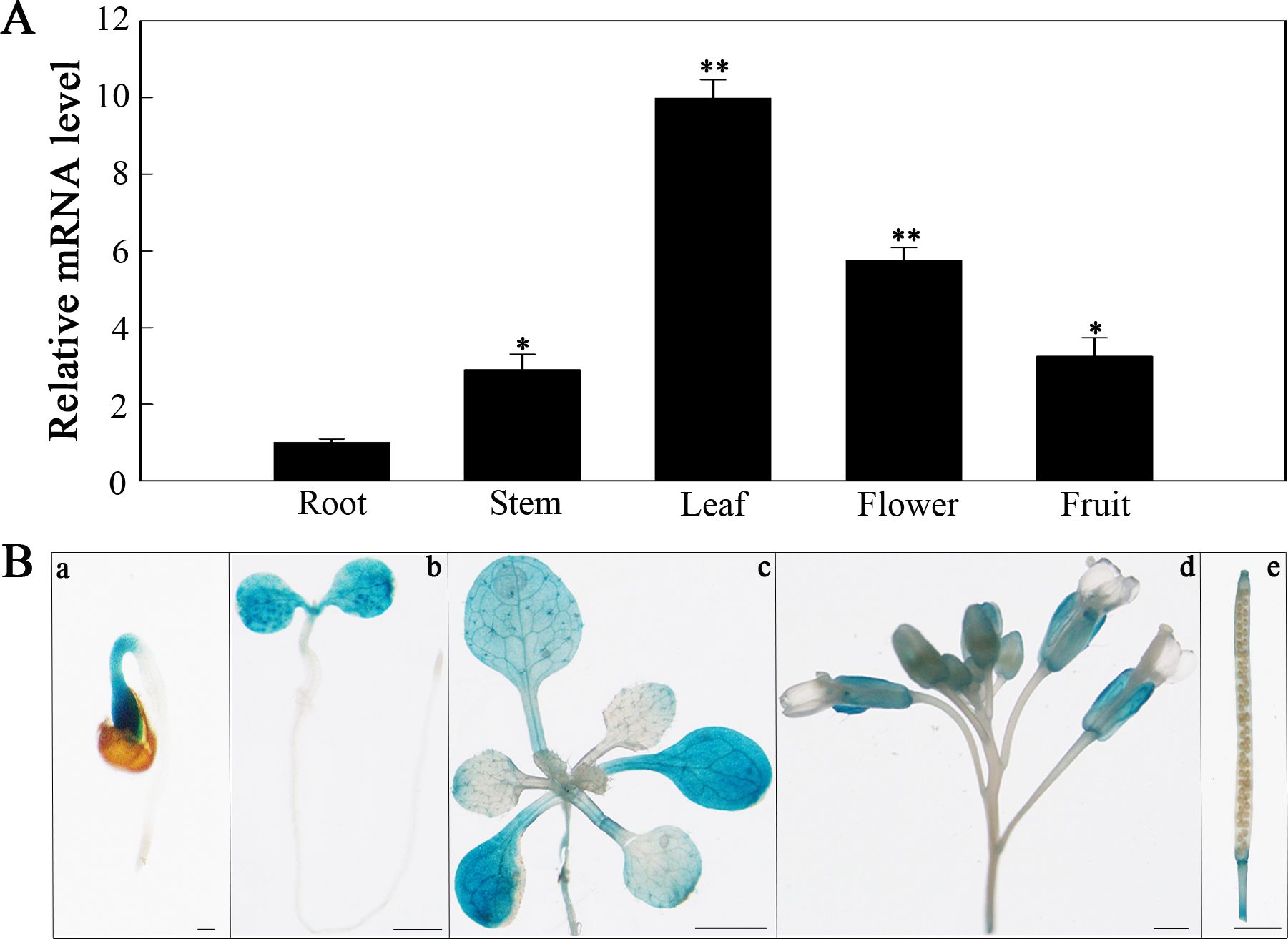
Figure 3. Tissue-specific expression analysis of SlHSP17.3. (A) RT-qPCR analysis of SlHSP17.3 expression patterns in tomato. Significance levels are shown to be * for p < 0.05 and ** for p < 0.01. (B) GUS staining analysis of SlHSP17.3pro::GUS in different tissues of transgenic Arabidopsis plants grown under natural conditions. a, 1 day (bars = 200 µm); b, 3 days (bars = 1 mm); c, 12 days (bars = 1 mm); d, inflorescence (bars = 100 µm); e, silique (bars = 1 mm).
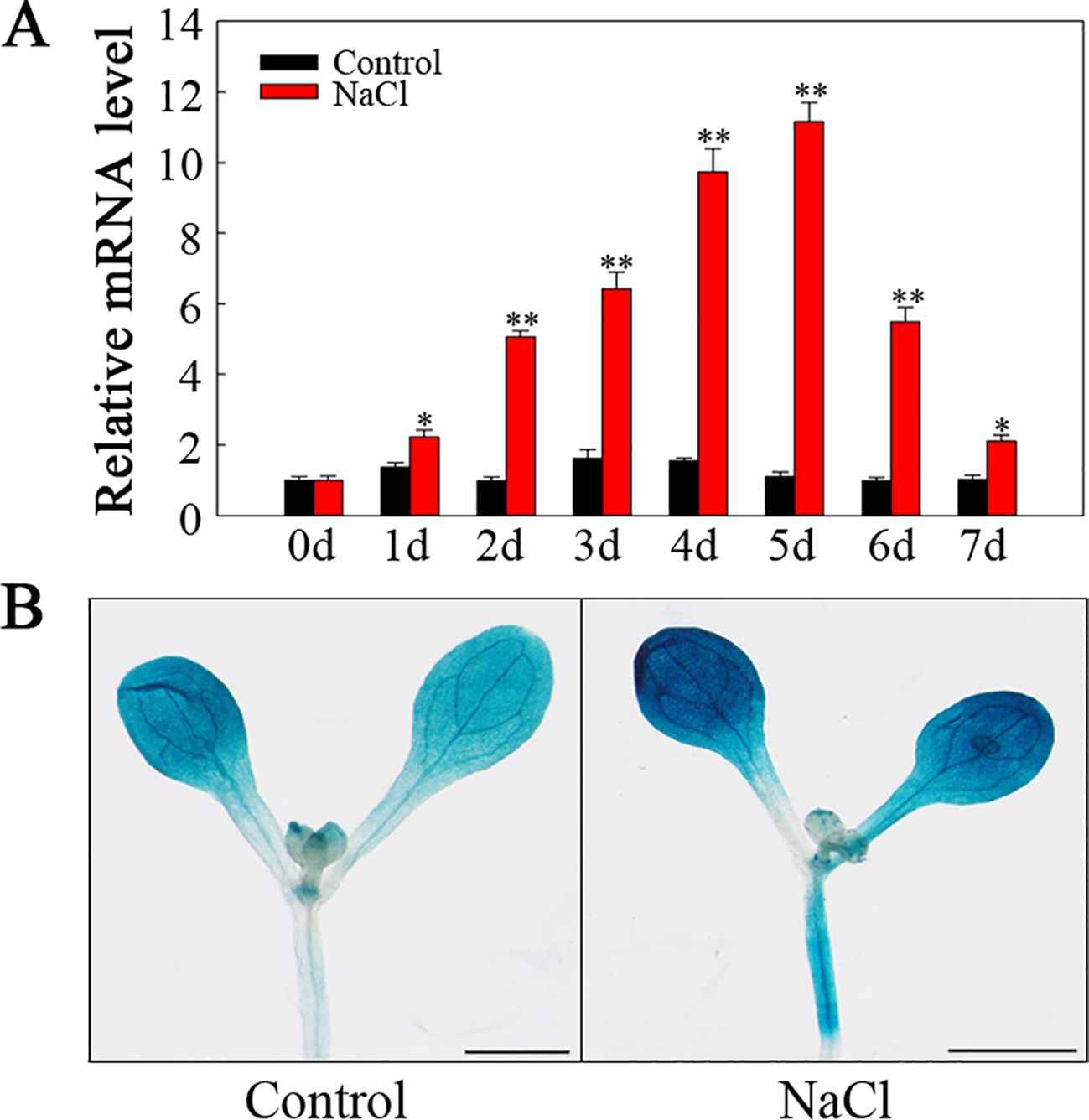
Figure 4. SlHSP17.3 expression levels and GUS staining upon salt stress. (A) SlHSP17.3 expression under salt stress according to RT-qPCR. (B) GUS staining analysis of seven-day-old leaves under normal conditions and after exposure to salt stress for one day. Scale bars = 1 mm. Significance levels are shown to be * for p < 0.05 and ** for p < 0.01.
3.4 Screening of transgenic plants
Ten T3 transgenic lines resistant to hygromycin were evaluated using RT-qPCR. Compared with the WT, all ten transgenic lines presented increased expression of SlHSP17.3 (Figure 5). From these lines, three were selected on the basis of their varying levels of relative expression: OE2 with low expression (113.6-fold), OE3 with moderate expression (225.6-fold), and OE4 with high expression (302.8-fold). These lines were selected for further experimentation.
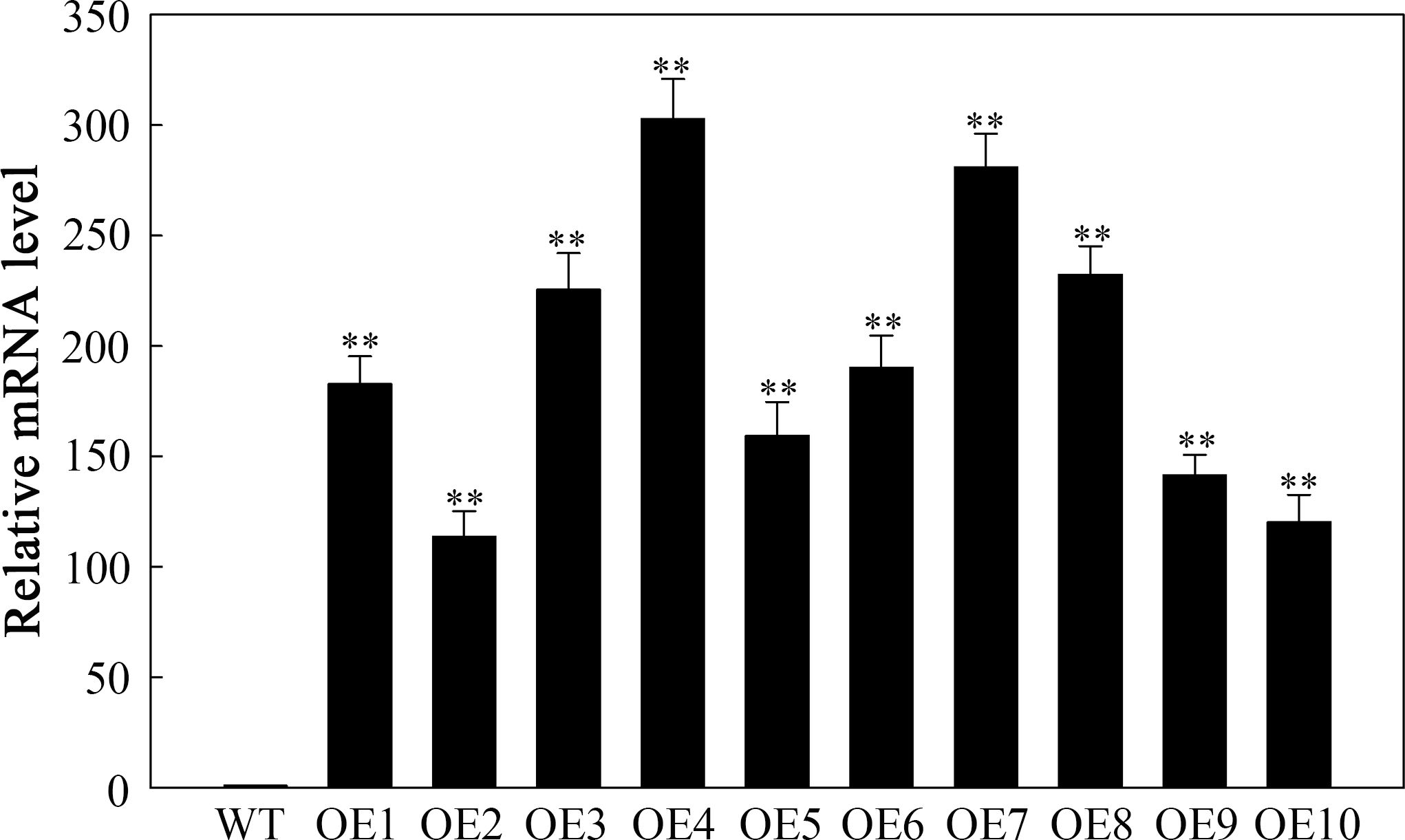
Figure 5. RT-qPCR detection of SlHSP17.3-overexpressing plants under natural conditions. Four-week-old WT and T3 generation transgenic Arabidopsis plants were used to detect the expression level of SlHSP17.3. Significance levels are shown as ** for p < 0.01.
3.5 SlHSP17.3 overexpression improves salt tolerance
To assess the potential role of SlHSP17.3 in the response of transgenic Arabidopsis plants to salt stress, we sowed seeds from both the transgenic and WT plants onto half-strength MS media supplemented with varying concentrations of NaCl. The germination rate did not markedly differ between the transgenic and WT plants on half-strength MS plates devoid of NaCl. Nonetheless, compared with WT plants, transgenic plants overexpressing SlHSP17.3 presented significantly greater germination rates under 100 and 150 mM NaCl conditions (Figures 6A, B). Additionally, a seedling growth assay was performed under salt stress. The control plants presented a phenotype consistent with that of all the transgenic plants, whereas the WT plants presented a decreased cotyledon size, shortened root length, and lower fresh weight under salt stress (Figures 6C, D).
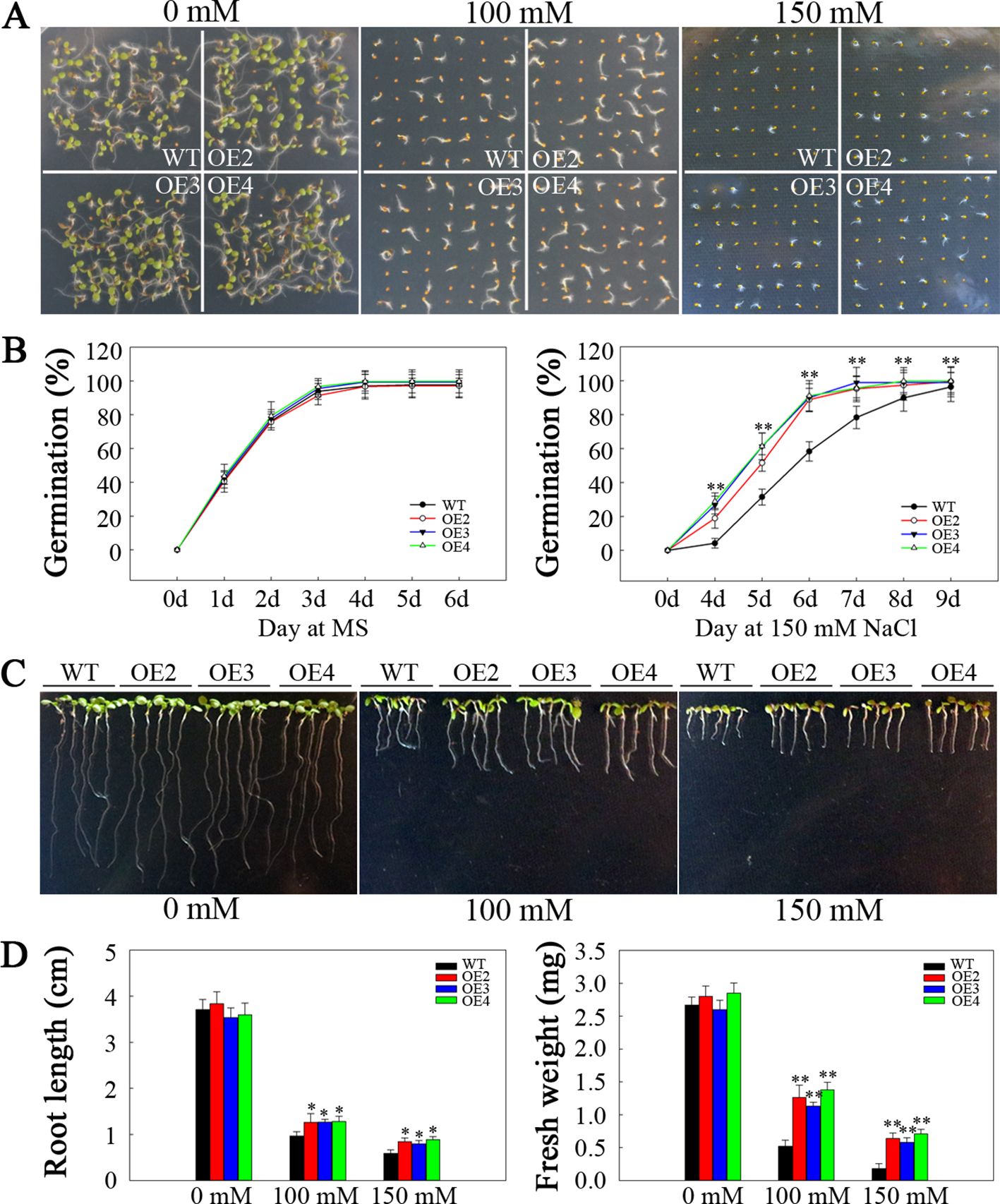
Figure 6. Analysis of the germination and seedling emergence of salt-stressed SlHSP17.3-overexpressing plants. (A) Seed germination under NaCl stress and normal conditions. (B) Germination rates of transgenic and WT plants under 150 mM NaCl stress and under normal conditions. (C) Seedling root phenotypes under NaCl stress and normal conditions. After sowing onto half-strength MS media for a three-day period, the seeds that had emerged from the radicle were placed onto half-strength MS media that contained varying concentrations of NaCl. The roots were photographed five days after transfer. (D) Seedling root length and fresh weight were measured five days after the plants were transferred to NaCl-containing plates. n = 147 per treatment for (A) and n = 15 per treatment for (C). Significance levels are shown to be * for p < 0.05 and ** for p < 0.01.
This study aimed to determine whether the improved salt resistance resulting from SlHSP17.3 overexpression extends to mature plants. For this purpose, two-week-old transgenic and WT lines were subjected to 200 mM NaCl treatment for two weeks. As shown in Figure 7A, the control lines exhibited healthy growth with no discernible differences in growth phenotype. However, following salt stress, both the transgenic and WT plants presented obvious growth inhibition to differing extents. Nevertheless, the degree of inhibition in the WT plants was notably greater than that in the transgenic plants. Compared with the transgenic plants, the WT plants presented significantly smaller leaves, lower chlorophyll contents (Figure 7B), and lower Fv/Fm values (Figure 7C). These findings were further corroborated by the chlorophyll fluorescence imaging results (Figure 7A). These experimental results indicate that SlHSP17.3 contributes to enhance the salt resistance of transgenic Arabidopsis plants.
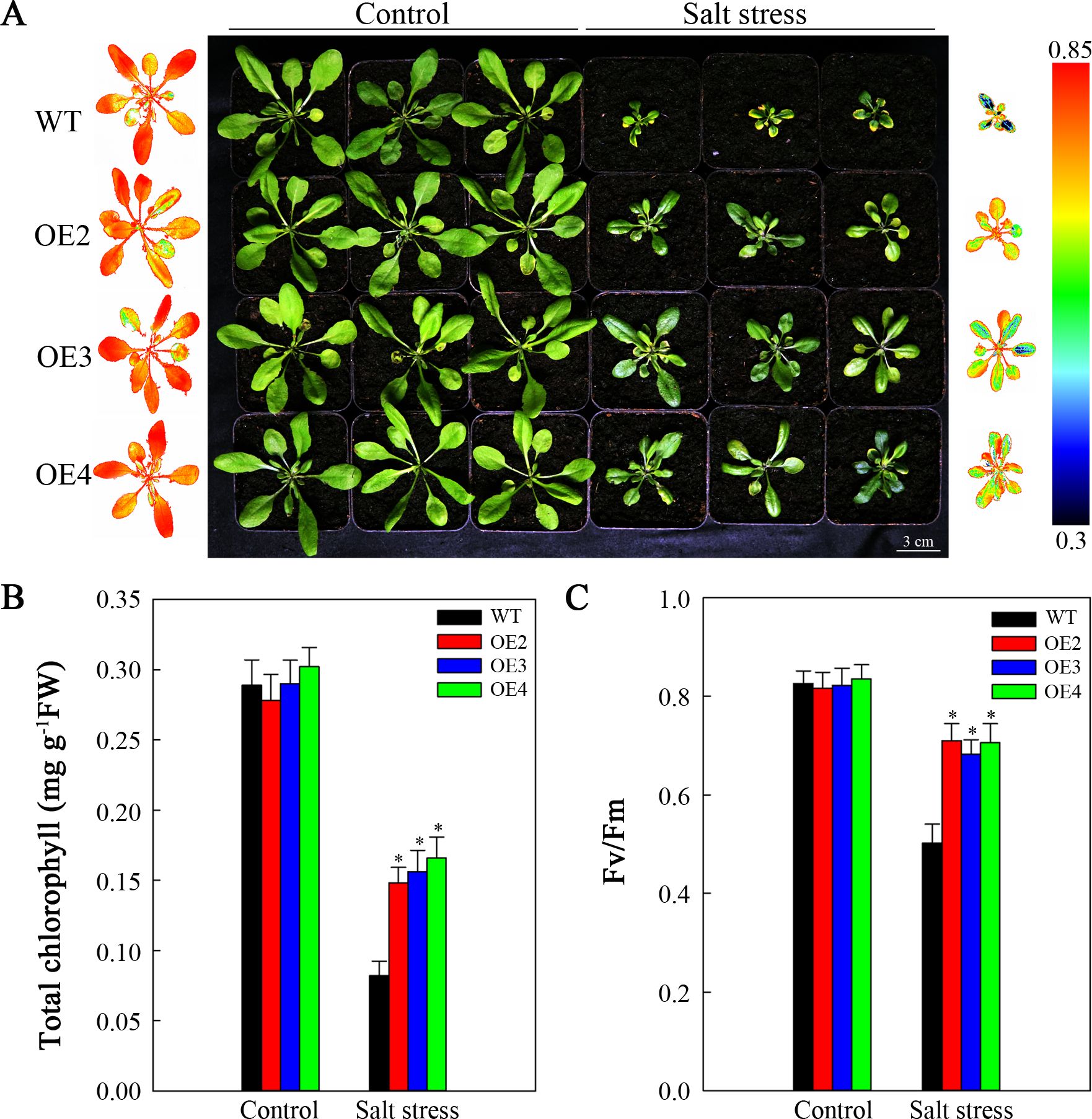
Figure 7. Analysis of the growth of mature salt-stressed SlHSP17.3-overexpressing plants. (A) Four-week-old WT and transgenic plants under 200 mM NaCl stress and normal conditions for two weeks. The chlorophyll fluorescence images on the left represent the leftmost column of plants in the control group, whereas the chlorophyll fluorescence images on the right represent the rightmost column of plants in the treatment group. (B) Total chlorophyll content. (C) Fv/Fm. Significance levels are shown as * for p < 0.05.
3.6 SlHSP17.3 overexpression decreases the ROS content in transgenic Arabidopsis
Salt stress typically leads to the generation of ROS. DAB and NBT staining were conducted to assess two primary ROS species, namely, H2O2 and O2•−, respectively. As shown in Figure 8, prior to treatment, the levels of H2O2 and O2•− were relatively low, especially H2O2, and the differences in WT compared with transgenic plants were not significant. Nonetheless, after two-day salt stress, brown polymerization product (DAB staining) accumulation noticeably elevated, particularly in the WT (Figure 8A). Similar trends were noted for the O2•− level (Figure 8B). The H2O2 and O2•− levels were consistent (Figures 8C, D). Therefore, SlHSP17.3 overexpression mitigates H2O2 and O2•− accumulation.
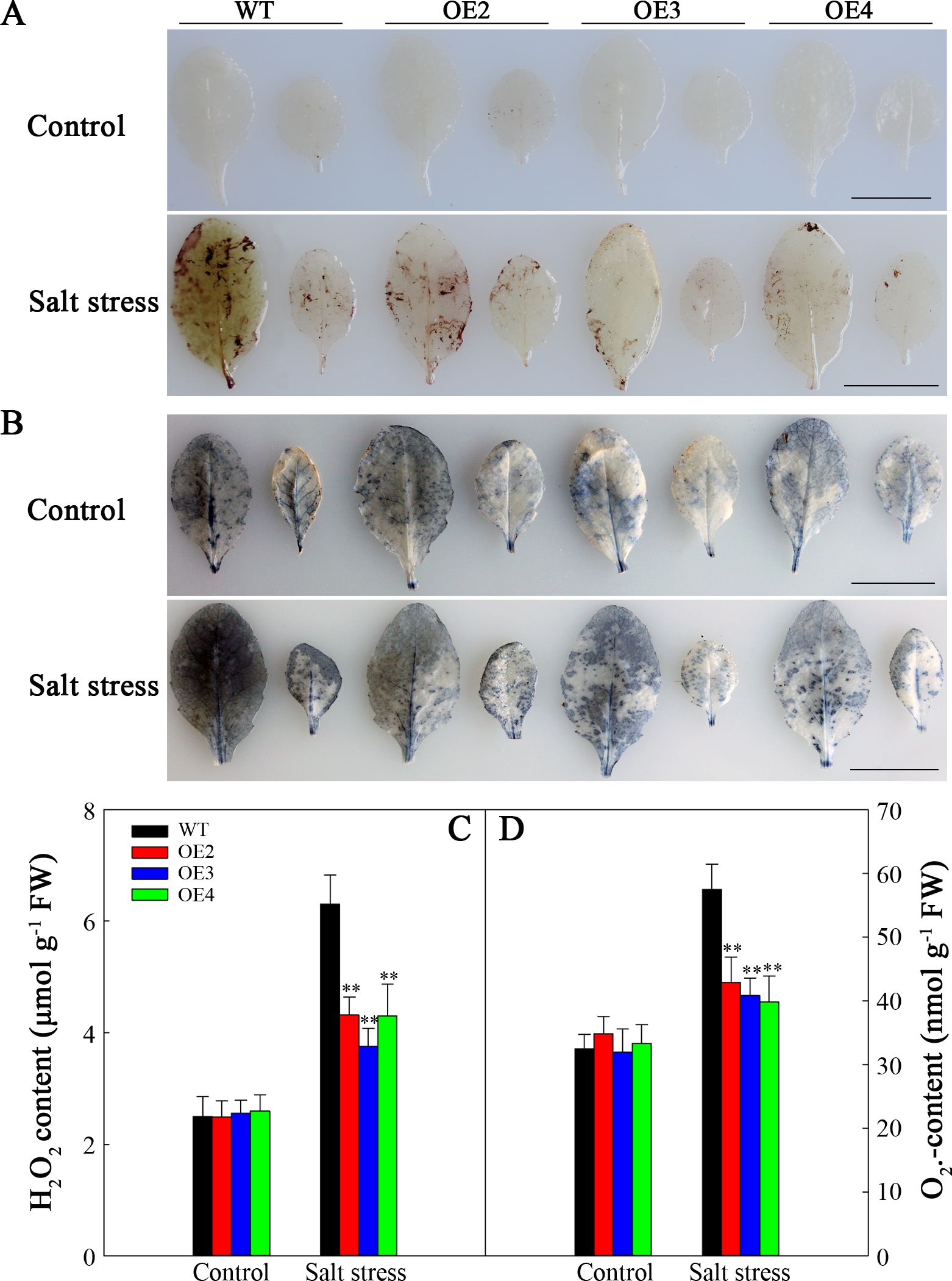
Figure 8. ROS correlation analysis of the WT and transgenic lines under salt stress. (A) DAB staining for H2O2. (B) NBT staining for O2•−. (C) H2O2 content. (D) O2•− content. Scale bars = 1 cm. Significance levels are shown as ** for p < 0.01.
The differences in SOD and CAT activities were of no statistical significance between the WT plants and the transgenic lines under normal conditions. Nevertheless, following two-day salt stress, the SOD and CAT activities increased to varying degrees. Nonetheless, compared to those in the WT lines, the increasing magnitudes of their activities within transgenic plants increased (Figures 9A, B). To investigate the reasons behind these alterations in enzyme activity, AtSOD1 and AtCAT1 expression was assessed via RT-qPCR. As shown in Figures 9C, D, AtSOD1, and AtCAT1 expression was not markedly difference in transgenic plants compared with WT under normal conditions. At two days post salt stress, the expression of AtSOD1 and AtCAT1 significantly increased; however, there were no significant differences of the AtSOD1 and AtCAT1 expression level in WT and the transgenic lines. Therefore, the decreased ROS contents within the transgenic lines are associated with increased SOD and CAT activities attributed to SlHSP17.3 overexpression.
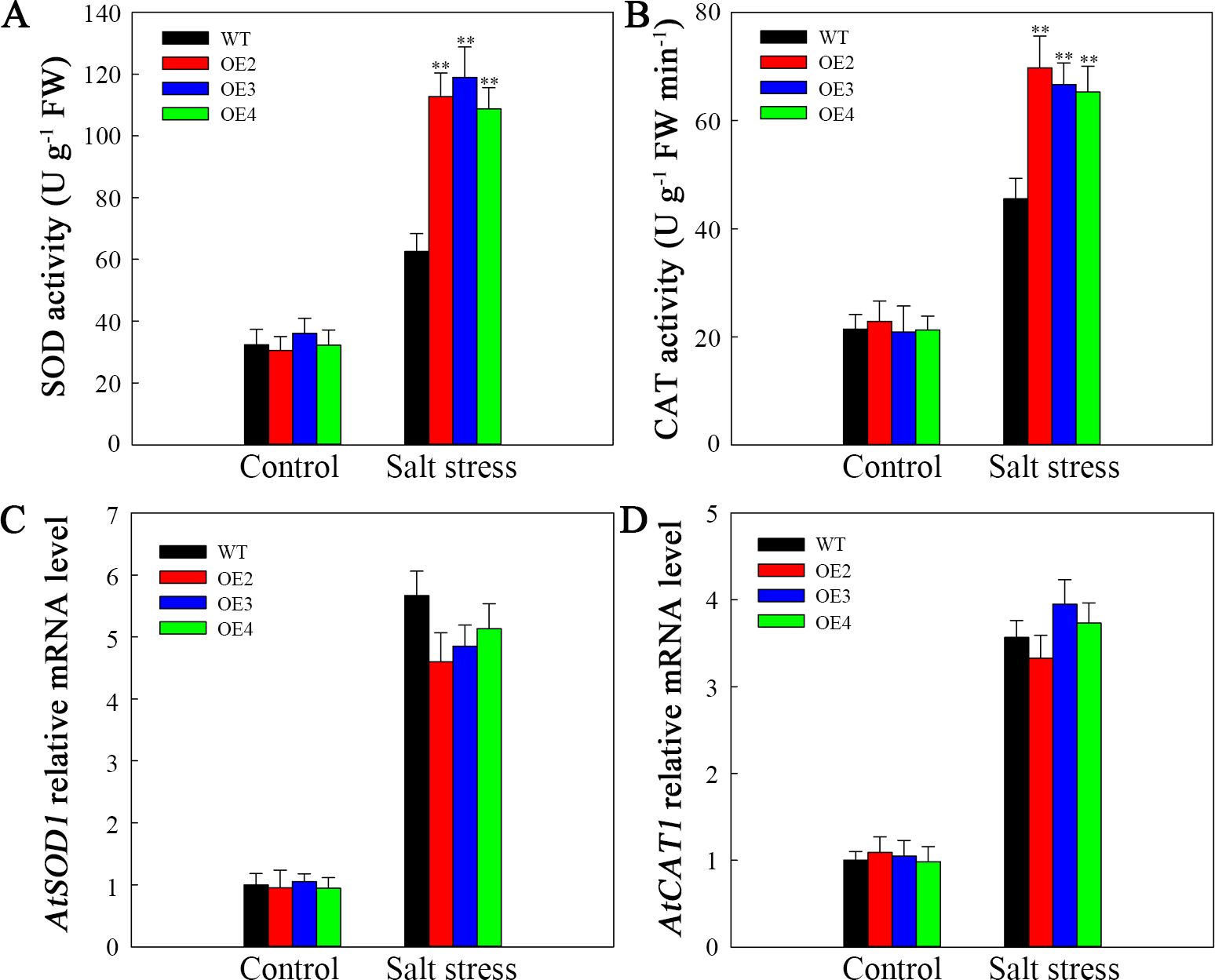
Figure 9. Antioxidase activities and antioxidase gene levels upon salt stress. (A) SOD activity. (B) CAT activity. (C) AtSOD1 expression. Database accession number of AtSOD1: AT1G08830. (D) AtCAT1 expression. Database accession number of AtCAT1: AT1G20630. Significance levels are shown as ** for p < 0.01.
3.7 SlHSP17.3 overexpression alleviated salt-induced damage to cells
The membranes of plant cells exhibit extremely high susceptibility to ROS-mediated lipid peroxidation. Trypan blue staining was used to analyze this susceptibility. After fiev days of salt stress, the WT plants presented darker blue coloration than did the transgenic plants (Figure 10A). Nonetheless, each plant had a similar degree of blue staining in the natural growth situations. To demonstrate the above findings, we assessed the MDA level and REC, which are known indicators of cell injury. Without salt stress, these indicators were not noticeably different between transgenic plants and WT. However, after salt stress, both indicators increased, with a more significant increase observed in WT than in those in the transgenic lines (Figures 10B, C). Consequently, SlHSP17.3 overexpression provides protection against salt stress-induced cell injury.
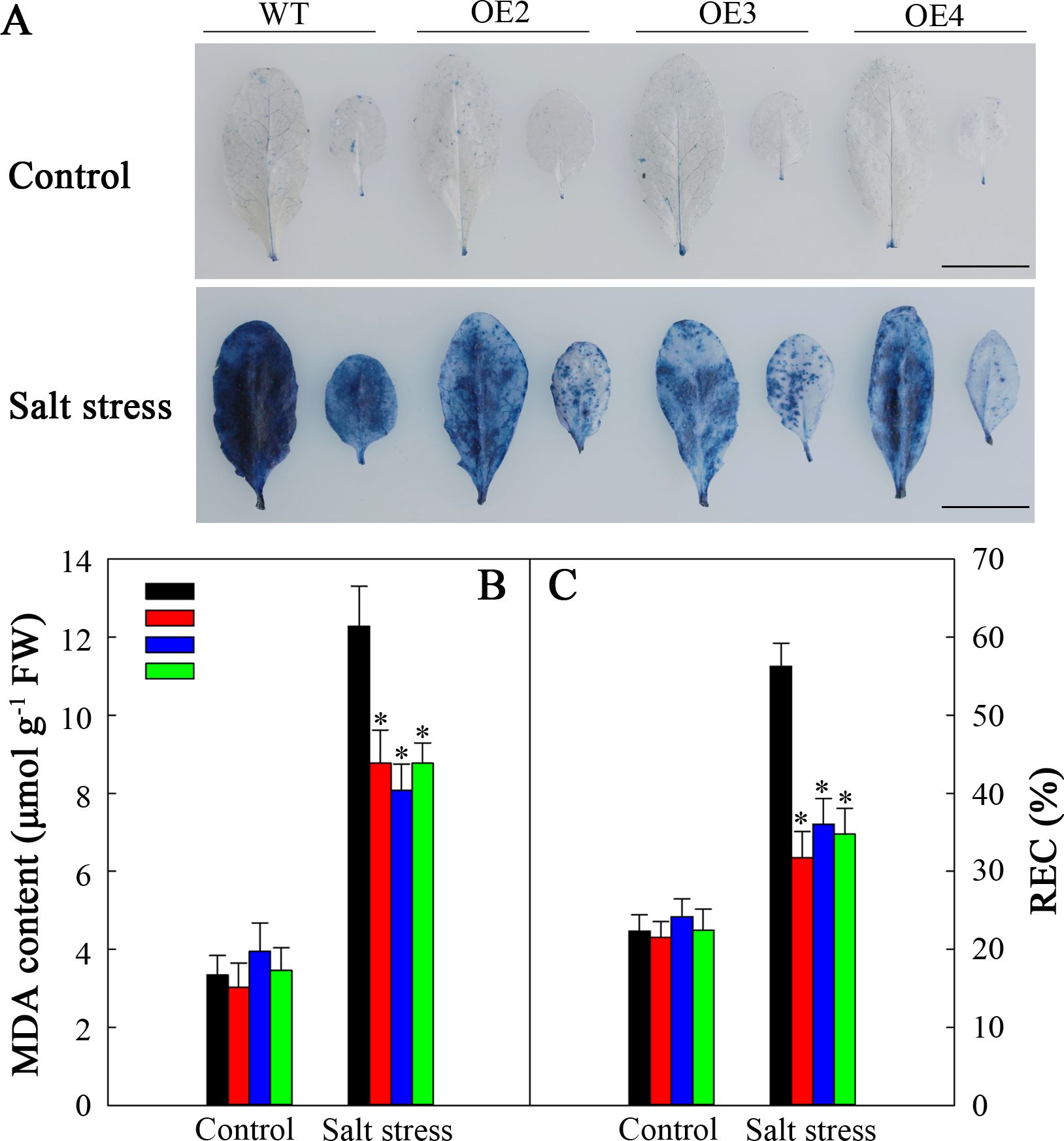
Figure 10. Cell injury in the WT and transgenic lines. (A) Trypan blue staining. The top panel shows plants under normal conditions, whereas the bottom panel shows plants under salt stress treatment for five days. (B) MDA levels of the WT and transgenic lines. (C) REC of the WT and transgenic lines. Scale bars = 1 cm. Significance levels are shown as * for p < 0.05.
3.8 SlHSP17.3 overexpression promotes the expression of certain stress-related genes upon salt stress
To explore how SlHSP17.3 promotes the salt resistance of transgenic Arabidopsis, we carried out RT-qPCR to assess various factors related to ABA biosynthesis, signaling, and stress response. Before or after salt stress, the expression levels of genes related to ABA biosynthesis (AtNCED3 and AtABI4) and ABA signaling (AtRAB18, AtRD29A, and AtMYB44) were not markedly different in WT and transgenic plants. However, following salt stress treatment, the expression of these genes increased in both the transgenic and WT plants (Figure 11A). Additionally, among the stress-related genes analyzed (AtCOR15, AtAPX2, AtDREB1B, and AtERF05), the expression of AtCOR15 and AtDREB1B significantly increased among transgenic plants compared with WT following salt stress (Figure 11B). Moreover, considering that heat shock transcription factors respond to heat stress and are related to other stress responses, we examined the expression of several of these factors (AtHSFA1, AtHSFA2, AtHSFB1, AtHSFB2, and AtHSFC). After salt stress, AtHSFA2 expression markedly increased in the transgenic plants compared with WT lines (Figure 11C). Consequently, such increased salt resistance in SlHSP17.3-overexpressing transgenic Arabidopsis plants could be associated with the upregulation of certain stress-related genes and certain HSFs.
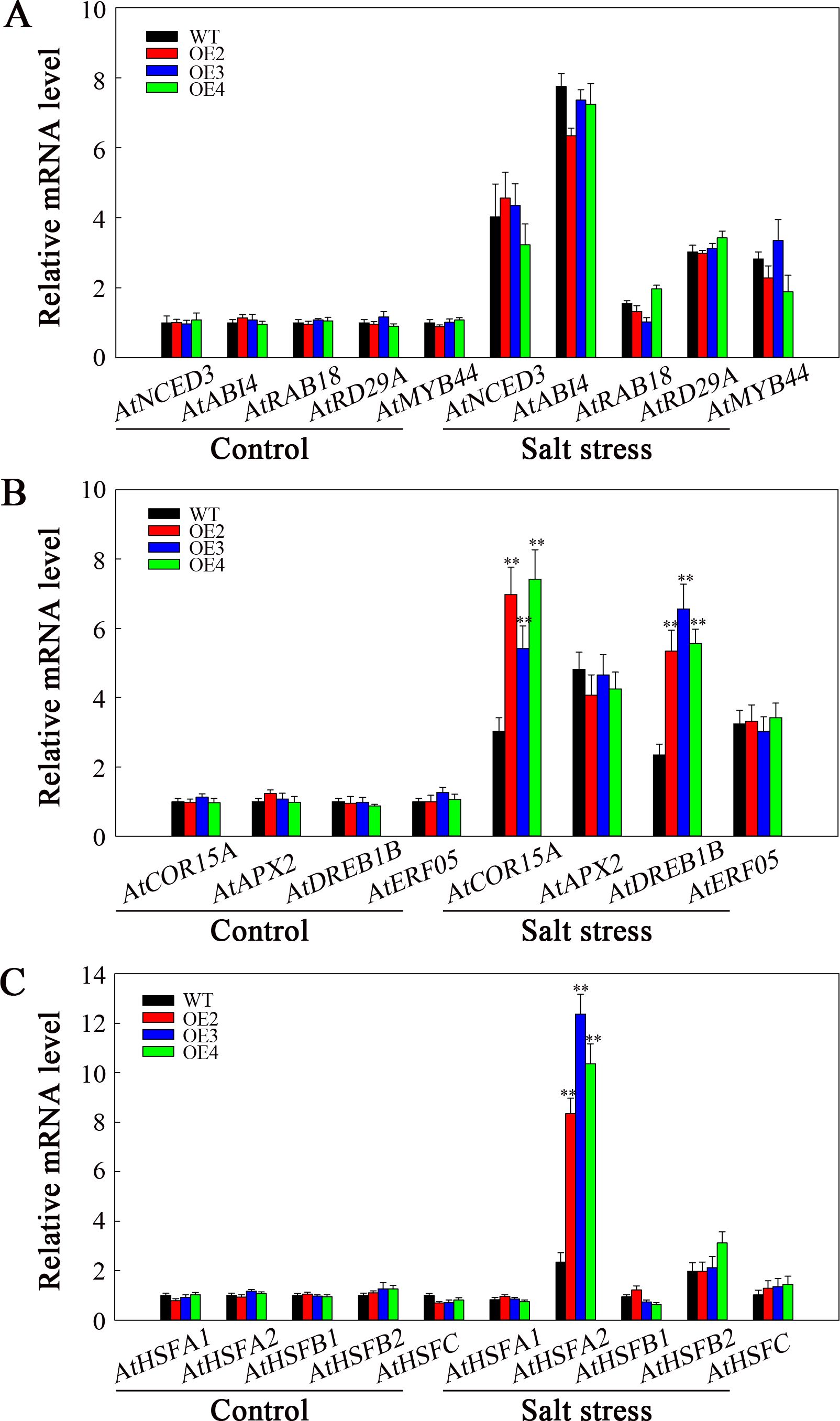
Figure 11. Analysis of ABA- and stress-related gene expression under salt stress. (A) ABA biosynthesis and signaling-related gene expression. (B) Stress-related gene expression. (C) AtHSF expression. Significance levels are shown as ** for p < 0.01.
4 Discussion
sHSPs constitute a group of molecular chaperones widely distributed in plants that provide protection against various environmental stresses. Many sHSPs are detected across various plants (Lopes-Caitar et al., 2013; Pandey et al., 2015; Tao et al., 2015; Guo et al., 2015). We have identified at least 42 sHSP proteins in tomato (unpublished). While recent studies on tomato sHSPs have focused mainly on their response to temperature stress, little attention has been given to understanding their effects on salt stress. For example, MasHSP24.4 overexpression enhances the high-temperature tolerance of transgenic tomato plants (Mahesh et al., 2013), and specific sHSPs, such as SlHsp17.4-CII and SlHsp23.8-M, play direct roles in chilling tolerance mechanisms in tomato genotypes (Ré et al., 2017). Additionally, SlHSP17.7 has been suggested to act as a cofactor for SlCCX1-like proteins, maintaining intracellular Ca2+ homeostasis and reducing cold stress sensitivity (Zhang et al., 2020). We isolated a novel tomato sHSP gene (SlHSP17.3) and found that its expression was associated with salt stress (Figure 4). Consistent with previous research (Port et al., 2004), our analysis revealed that SlHSP17.3 contains a characteristic α-crystallin domain (ACD) with a β-folded structure (Supplementary Figure S3). Evolutionary analysis indicated that SlHSP17.3 belongs to the cytosolic class II sHSP protein group (Figure 1). Subcellular localization experiments confirmed its cytoplasmic localization (Figure 2). Furthermore, functional studies demonstrated the positive influence of SlHSP17.3 on salt stress in transgenic Arabidopsis.
Salt stress poses a significant challenge to modern agriculture, profoundly impacting plant development and crop production. Many studies have indicated that plant sHSP proteins can exert both positive and negative regulatory effects upon plants when responding to salt stress (Jiang et al., 2009; Sun et al., 2016; Wang et al., 2020; He et al., 2021; Qin et al., 2022). Our findings revealed that overexpressing SlHSP17.3 promoted the salt resistance of transgenic Arabidopsis plants. After salt treatment, observations of the germination rate, seedling growth, membrane damage, mature transgenic plant phenotype, and associated physiological indices suggest that overexpressing SlHSP17.3 enhances the salt resistance of transgenic Arabidopsis (Figure 6, Figure 7, Figure 10). Taken together, our findings indicate the significant impact of SlHSP17.3 on plant responses to salt stress.
Furthermore, our research highlights the crucial role of SlHSP17.3 in alleviating salt-induced oxidative stress. ROS accumulation is a common consequence of salt stress, leading to cell injury and dysfunction (Napieraj et al., 2020). Both NBT and DAB staining, along with quantification, clearly demonstrated that overexpressing SlHSP17.3 substantially reduced ROS accumulation in salt-stressed transgenic Arabidopsis (Figure 8). This decrease in ROS levels is associated with increased antioxidase activities in SlHSP17.3-overexpressing plants, as shown in Figure 9A. Notably, this increased antioxidant enzyme activity does not correspond to elevated expression of antioxidant enzyme-encoding genes (Figure 9B). Instead, this finding suggests that SlHSP17.3 likely exerts its protective function as a molecular chaperone directly against oxidase activity. The observed upregulation of antioxidant enzymes and simultaneous reduction in ROS levels in SlHSP17.3-overexpressing plants upon salt stress underscore the pivotal role of SlHSP17.3 in scavenging ROS and mitigating oxidative stress. The above results provide insights into the intricate interplay between SlHSP17.3-mediated protein protection and ROS detoxification pathways, thereby enhancing overall stress resistance in plants.
sHSP protein expression in plants can positively or negatively impact salt stress by engaging in ABA-dependent/independent pathways, plant photosynthesis, and various stress response mechanisms (Sun et al., 2016; Li et al., 2018; Wang et al., 2020; He et al., 2021). Central to this regulatory network are the ABA responsiveness element (ABRE) and anaerobic response element (ARE) detected within sHSP gene promoters, which can be recognized by transcription factors, including AREB/ABF and MYB (Fujita et al., 2011; Banerjee and Roychoudhury, 2017). To explore the regulatory pathways of SlHSP17.3 under salt stress in greater depth, this study initially examined cis-acting elements within the SlHSP17.3 promoter and identified both ABRE and ARE (Supplementary Table S3). Consequently, we treated plants with ABA to analyze SlHSP17.3 gene expression, and our results revealed that the induction of SlHSP17.3 gene expression by ABA was not significant (Supplementary Figure S4). Additionally, the ABA content in the leaves of four-week-old WT and transgenic plants was measured with and without salt stress treatment. Although the ABA content increased in all lines after salt stress treatment, there was no significant difference in the ABA content between the WT and transgenic plants (Supplementary Figure S5). On the basis of our results, SlHSP17.3 may have different functions than those reported in previous studies. Then, we used RT-qPCR to identify ABA biosynthesis-, signaling-, and stress-related gene levels under salt stress. Following salt stress treatment, genes related to ABA biosynthesis (AtNCED3 and AtABI4) and ABA signaling (AtRAB18, AtRD29A, and AtMYB44) were upregulated to different extents. However, gene expression was not significantly different in transgenic plants compared with WT (Figure 11A). After salt stress, AtCOR15 and AtDREB1B expression in transgenic plants markedly increased compared with that in WT (Figure 11B). Based on these results, SlHSP17.3 overexpression enhances salt tolerance of transgenic Arabidopsis in an ABA-independent pathway through the up-regulation of certain stress-related genes. Furthermore, HSFs are related to heat shock responses and salt stress, where they collaborate with HSPs to improve plant resilience (Wang et al., 2019a; Iqbal et al., 2022). Therefore, selected HSF levels were examined, revealing that AtHSFA2 was markedly up-regulated in transgenic plants relative to WT lines following salt stress (Figure 11C). According to the above findings, overexpression of SlHSP17.3 contributes to the upregulation of specific HSFs, thereby increasing salt stress resistance in transgenic plants.
Overall, we separated and identified a novel sHSP protein (SlHSP17.3) in tomato that belongs to the cytosolic class II sHSP family. Overexpressing SlHSP17.3 can alleviate ROS accumulation in salt-stressed transgenic Arabidopsis plants, and it can improve the salt stress resistance of transgenic Arabidopsis plants through up-regulating AtCOR15, AtDREB1B, and AtHSFA2.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author contributions
GC: Supervision, Writing – original draft. MN: Investigation, Writing – original draft. ZS: Investigation, Writing – original draft. HW: Investigation, Writing – original draft. SZ: Formal Analysis, Writing – original draft. FL: Formal Analysis, Software, Writing – review & editing. YW: Formal Analysis, Software, Writing – review & editing. GW: Conceptualization, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was supported by the Natural Science Foundation of Shandong (ZR2022QC240), the Natural Science Foundation of Rizhao (RZ2021ZR19), the Shandong Provincial University Youth Innovation Team, China (2022KJ102), the Shandong Province University Curriculum Ideological and Political Teaching Reform Research Project (SZ2023042), and the Innovation Training Project of University Student (S202410443271).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1443625/full#supplementary-material
References
Aebi, H. (1984). Catalase in vitro. Methods Enzymol. 105, 121–126. doi: 10.1016/s0076-6879(84)05016-3
Ambastha, V., Leshem, Y. (2020a). Differential cell persistence is observed in the Arabidopsis female gametophyte during heat stress. Plant Reprod. 33, 111–116. doi: 10.1007/s00497-020-00390-0
Ambastha, V., Leshem, Y. (2020b). Cyclin B1;1 activity is observed in lateral roots but not in the primary root during lethal salinity and salt stress recovery. Plant Signal Behav. 15, e1776026. doi: 10.1080/15592324.2020.1776026
Baker, N. R. (2008). Chlorophyll fluorescence: a probe of photosynthesis in vivo. Annu. Rev. Plant Biol. 59, 89–113. doi: 10.1146/annurev.arplant.59.032607.092759
Banerjee, A., Roychoudhury, A. (2017). Abscisic-acid-dependent basic leucine zipper (bZIP) transcription factors in plant abiotic stress. Protoplasma 254, 3–16. doi: 10.1007/s00709-015-0920-4
Basha, E., O'Neill, H., Vierling, E. (2012). Small heat shock proteins and α-crystallins: dynamic proteins with flexible functions. Trends Biochem. Sci. 37, 106–117. doi: 10.1016/j.tibs.2011.11.005
Bondino, H. G., Valle, E. M., Ten Have, A. (2012). Evolution and functional diversification of the small heat shock protein/α-crystallin family in higher plants. Planta 235, 1299–1313. doi: 10.1007/s00425-011-1575-9
Choi, H. W., Kim, Y. J., Lee, S. C., Hong, J. K., Hwang, B. K. (2007). Hydrogen peroxide generation by the pepper extracellular peroxidase CaPO2 activates local and systemic cell death and defense response to bacterial pathogens. Plant Physiol. 145, 890–904. doi: 10.1104/pp.107.103325
Feng, X. H., Zhang, H. X., Ali, M., Gai, W. X., Cheng, G. X., Yu, Q. H., et al. (2019). A small heat shock protein CaHsp25.9 positively regulates heat, salt, and drought stress tolerance in pepper (Capsicum annuum L.). Plant Physiol. Biochem. 142, 151–162. doi: 10.1016/j.plaphy.2019.07.001
Fujita, Y., Fujita, M., Shinozaki, K., Yamaguchi-Shinozaki, K. (2011). ABA-mediated transcriptional regulation in response to osmotic stress in plants. J. Plant Res. 124, 509–525. doi: 10.1007/s10265-011-0412-3
Giannopolitis, C. N., Ries, S. K. (1977). Superoxide dismutases: I. Occurrence in higher plants. Plant Physiol. 59, 309–314. doi: 10.1104/pp.59.2.309
Guo, M., Liu, J. H., Lu, J. P., Zhai, Y. F., Wang, H., Gong, Z. H., et al. (2015). Genome-wide analysis of the CaHsp20 gene family in pepper: comprehensive sequence and expression profile analysis under heat stress. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00806
Guo, M., Wang, X. S., Guo, H. D., Bai, S. Y., Khan, A., Wang, X. M., et al. (2022). Tomato salt tolerance mechanisms and their potential applications for fighting salinity: A review. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.949541
He, Y., Yao, Y., Li, L., Li, Y., Gao, J., Fan, M. (2021). A heat-shock 20 protein isolated from watermelon (ClHSP22.8) negatively regulates the response of Arabidopsis to salt stress via multiple signaling pathways. PeerJ 9, e10524. doi: 10.7717/peerj.10524
Hibshman, J. D., Carra, S., Goldstein, B. (2023). Tardigrade small heat shock proteins can limit desiccation-induced protein aggregation. Commun. Biol. 6, 121. doi: 10.1038/s42003-023-04512-y
Iqbal, M. Z., Jia, T., Tang, T., Anwar, M., Ali, A., Hassan, M. J., et al. (2022). A heat shock transcription factor TrHSFB2a of white clover negatively regulates drought, heat and salt stress tolerance in transgenic Arabidopsis. Int. J. Mol. Sci. 23, 12769. doi: 10.3390/ijms232112769
Jiang, C., Xu, J., Zhang, H., Zhang, X., Shi, J., Li, M., et al. (2009). A cytosolic class I small heat shock protein, RcHSP17.8, of Rosa chinensis confers resistance to a variety of stresses to Escherichia coli, yeast and Arabidopsis thaliana. Plant Cell Environ. 32, 1046–1059. doi: 10.1111/j.1365-3040.2009.01987.x
Kong, F., Deng, Y., Zhou, B., Wang, G., Wang, Y., Meng, Q. (2014). A chloroplast-targeted DnaJ protein contributes to maintenance of photosystem II under chilling stress. J. Exp. Bot. 65, 143–158. doi: 10.1093/jxb/ert357
Koo, H. J., Park, S. M., Kim, K. P., Suh, M. C., Lee, M. O., Lee, S. K., et al. (2015). Small heat shock proteins can release light dependence of tobacco seed during germination. Plant Physiol. 167, 1030–1038. doi: 10.1104/pp.114.252841
Li, M., Ji, L., Jia, Z., Yang, X., Meng, Q., Guo, S. (2018). Constitutive expression of CaHSP22.5 enhances chilling tolerance in transgenic tobacco by promoting the activity of antioxidative enzymes. Funct. Plant Biol. 45, 575–585. doi: 10.1071/fp17226
Li, Z. Y., Long, R. C., Zhang, T. J., Yang, Q. C., Kang, J. M. (2016). Molecular cloning and characterization of the MsHSP17.7 gene from Medicago sativa L. Mol. Biol. Rep. 43, 815–826. doi: 10.1007/s11033-016-4008-9
Lopes-Caitar, V. S., de Carvalho, M. C., Darben, L. M., Kuwahara, M. K., Nepomuceno, A. L., Dias, W. P., et al. (2013). Genome-wide analysis of the Hsp20 gene family in soybean: comprehensive sequence, genomic organization and expression profile analysis under abiotic and biotic stresses. BMC Genomics 14, 577. doi: 10.1186/1471-2164-14-577
Ma, N. N., Zuo, Y. Q., Liang, X. Q., Yin, B., Wang, G. D., Meng, Q. W. (2013). The multiple stress-responsive transcription factor SlNAC1 improves the chilling tolerance of tomato. Physiol. Plant 149, 474–486. doi: 10.1111/ppl.12049
Mahesh, U., Mamidala, P., Rapolu, S., Aragao, F., Souza, M., Rao, P., et al. (2013). Constitutive overexpression of small HSP24.4 gene in transgenic tomato conferring tolerance to high-temperature stress. Mol. Breed. 32, 687–697. doi: 10.1007/s11032-013-9901-5
Napieraj, N., Reda, M. G., Janicka, M. G. (2020). The role of NO in plant response to salt stress: interactions with polyamines. Funct. Plant Biol. 47, 865–879. doi: 10.1071/fp19047
Pandey, B., Kaur, A., Gupta, O. P., Sharma, I., Sharma, P. (2015). Identification of HSP20 gene family in wheat and barley and their differential expression profiling under heat stress. Appl. Biochem. Biotechnol. 175, 2427–2446. doi: 10.1007/s12010-014-1420-2
Papsdorf, K., Richter, K. (2014). Protein folding, misfolding and quality control: the role of molecular chaperones. Essays Biochem. 56, 53–68. doi: 10.1042/bse0560053
Port, M., Tripp, J., Zielinski, D., Weber, C., Heerklotz, D., Winkelhaus, S., et al. (2004). Role of Hsp17.4-CII as coregulator and cytoplasmic retention factor of tomato heat stress transcription factor HsfA2. Plant Physiol. 135, 1457–1470. doi: 10.1104/pp.104.042820
Qin, Y., Liu, X., Quan, X., Chen, J., Wang, Z. (2022). Heterologously expressing a wheat CI Small Heat Shock Protein gene enhances the salinity tolerance of Arabidopsis thaliana. J. Plant Growth Regul. 41, 236–243. doi: 10.1007/s00344-021-10296-4
Ré, M. D., Gonzalez, C., Escobar, M. R., Sossi, M. L., Valle, E. M., Boggio, S. B. (2017). Small heat shock proteins and the postharvest chilling tolerance of tomato fruit. Physiol. Plant 159, 148–160. doi: 10.1111/ppl.12491
Siddique, M., Gernhard, S., von Koskull-Döring, P., Vierling, E., Scharf, K. D. (2008). The plant sHSP superfamily: five new members in Arabidopsis thaliana with unexpected properties. Cell Stress Chaperones 13, 183–197. doi: 10.1007/s12192-008-0032-6
Sun, L., Liu, Y., Kong, X., Zhang, D., Pan, J., Zhou, Y., et al. (2012). ZmHSP16.9, a cytosolic class I small heat shock protein in maize (Zea mays), confers heat tolerance in transgenic tobacco. Plant Cell Rep. 31, 1473–1484. doi: 10.1007/s00299-012-1262-8
Sun, X., Sun, C., Li, Z., Hu, Q., Han, L., Luo, H. (2016). AsHSP17, a creeping bentgrass small heat shock protein modulates plant photosynthesis and ABA-dependent and independent signalling to attenuate plant response to abiotic stress. Plant Cell Environ. 39, 1320–1337. doi: 10.1111/pce.12683
Sundaresan, V., Springer, P., Volpe, T., Haward, S., Jones, J. D., Dean, C., et al. (1995). Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes Dev. 9, 1797–1810. doi: 10.1101/gad.9.14.1797
Tao, P., Guo, W. L., Li, B. Y., Wang, W. H., Yue, Z. C., Lei, J. L., et al. (2015). Genome-wide identification, classification, and expression analysis of sHSP genes in Chinese cabbage (Brassica rapa ssp pekinensis). Genet. Mol. Res. 14, 11975–11993. doi: 10.4238/2015.October.5.11
Tian, C., Zhang, Z., Huang, Y., Xu, J., Liu, Z., Xiang, Z., et al. (2022). Functional characterization of the Pinellia ternata cytoplasmic class II small heat shock protein gene PtsHSP17.2 via promoter analysis and overexpression in tobacco. Plant Physiol. Biochem. 177, 1–9. doi: 10.1016/j.plaphy.2022.02.017
Wang, G., Cai, G., Xu, N., Zhang, L., Sun, X., Guan, J., et al. (2019a). Novel DnaJ protein facilitates thermotolerance of transgenic tomatoes. Int. J. Mol. Sci. 20, 367. doi: 10.3390/ijms20020367
Wang, G., Xu, X., Wang, H., Liu, Q., Yang, X., Liao, L., et al. (2019b). A tomato transcription factor, SlDREB3 enhances the tolerance to chilling in transgenic tomato. Plant Physiol. Biochem. 142, 254–262. doi: 10.1016/j.plaphy.2019.07.017
Wang, J., Gao, X., Dong, J., Tian, X., Wang, J., Palta, J. A., et al. (2020). Over-expression of the heat-responsive wheat gene TaHSP23.9 in transgenic Arabidopsis conferred tolerance to heat and salt stress. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00243
Wang, W. Q., Ye, J. Q., Rogowska-Wrzesinska, A., Wojdyla, K. I., Jensen, O. N., Møller, I. M., et al. (2014). Proteomic comparison between maturation drying and prematurely imposed drying of Zea mays seeds reveals a potential role of maturation drying in preparing proteins for seed germination, seedling vigor, and pathogen resistance. J. Proteome Res. 13, 606–626. doi: 10.1021/pr4007574
Waters, E. R. (2013). The evolution, function, structure, and expression of the plant sHSPs. J. Exp. Bot. 64, 391–403. doi: 10.1093/jxb/ers355
Wu, J., Gao, T., Hu, J., Zhao, L., Yu, C., Ma, F. (2022). Research advances in function and regulation mechanisms of plant small heat shock proteins (sHSPs) under environmental stresses. Sci. Total Environ. 825, 154054. doi: 10.1016/j.scitotenv.2022.154054
Yang, Z., Du, H., Xing, X., Li, W., Kong, Y., Li, X., et al. (2022). A small heat shock protein, GmHSP17.9, from nodule confers symbiotic nitrogen fixation and seed yield in soybean. Plant Biotechnol. J. 20, 103–115. doi: 10.1111/pbi.13698
Keywords: salt stress, sHSP, SlHSP17.3, stress-related genes, transgenic Arabidopsis
Citation: Cai G, Niu M, Sun Z, Wang H, Zhang S, Liu F, Wu Y and Wang G (2024) A small heat shock protein (SlHSP17.3) in tomato plays a positive role in salt stress. Front. Plant Sci. 15:1443625. doi: 10.3389/fpls.2024.1443625
Received: 04 June 2024; Accepted: 25 September 2024;
Published: 11 October 2024.
Edited by:
Shalini Tiwari, South Dakota State University, United StatesReviewed by:
Sanchita Singh, Guru Gobind Singh Indraprastha University, IndiaYanping Jing, Jiangsu University, China
Copyright © 2024 Cai, Niu, Sun, Wang, Zhang, Liu, Wu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guodong Wang, Z2R3YW5nQG1haWwuam5tYy5lZHUuY24=
†ORCID: Guohua Cai, https://orcid.org/0009-0000-2126-6412
 Guohua Cai
Guohua Cai Mingyu Niu
Mingyu Niu Zhihao Sun
Zhihao Sun Huakun Wang
Huakun Wang Fei Liu
Fei Liu Guodong Wang
Guodong Wang