
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 24 February 2025
Sec. Plant Biotechnology
Volume 15 - 2024 | https://doi.org/10.3389/fpls.2024.1442324
 Bertrand Bachaumond Hankoua1*
Bertrand Bachaumond Hankoua1* Marieme Diao2
Marieme Diao2 Ayalew Ligaba-Osena3
Ayalew Ligaba-Osena3 Rafael A. Garcia4
Rafael A. Garcia4 Sarahani Harun5
Sarahani Harun5 Yogesh K. Ahlawat6,7
Yogesh K. Ahlawat6,7Cassava is a crucial source of daily calorie intake for millions of people in sub-Saharan Africa (SSA) but has an inferior protein content. Despite numerous attempts utilizing both traditional and biotechnological methods, efforts to address protein deficiency in cassava have yet to meet with much success. We aim to leverage modern biotechnologies to enhance cassava's nutritional value by creating bioengineered cassava cultivars with increased protein and starch content. In this study, we utilized Qui-Quine Starch (QQS), a novel orphan gene unique to Arabidopsis thaliana, to develop transgenic cassava plants with increased protein and starch accumulation in their tissues. A total of 10 independent transgenic cassava lines expressing QQS were successfully regenerated in this study, among which line R7 (F) demonstrated superior growth vigor. Quantitative RT-PCR verified the expression of the QQS gene in the transgenic lines. Data showed that QQS expression in cassava plants increased leaf protein content by 36% in line R’’’ (LA) L2 and root protein by 17% for the same line compared to their wild-type and empty vector (NPTII) control plants. Moreover, leaf-soluble total carbohydrates increased by 51.76% in line R (G) L2, and root-soluble total carbohydrates increased by 46.75% in line R7 (F). The novel function of QQS in increasing the starch content in the transgenic biomass is demonstrated. No significant change in the content of specific amino acids was observed among the lines and various plant parts. In addition, QQS expression revealed increased biomass, plant vigor, and early In vitro mini-tubers production for line R7 (F). Gene interaction study between AtQQS and 59 interacting partners generated 184 interactions or edges. These gene networks comprised several functional categories regulating the starch metabolic and auxin biosynthetic processes. The role of QQS in imparting starch and protein content of transgenic cassava plants is validated. The next logical step is the evaluation of biochemical profiles of cassava lines expressing QQS that reach maturity and the transferability of these findings to consumer-preferred cassava cultivars and local landraces grown in SSA. This study represents the first biotechnological report demonstrating a simultaneous increase of protein and starch content in bioengineered cassava.
More than 270 million people suffer from chronic malnutrition and 400 million from micronutrient deficiencies in Sub-Sahara Africa (SSA), where subsistence farmers and their families are most at risk from the implications of undernutrition (Graham and Welch, 1996; Harika et al., 2017; Fraval et al., 2019). Many of these farmers, 250 million and more of the poorest people in the world, rely on cassava as their staple food (Narayanan et al., 2019). Cassava roots’ starch provides over 25% of dietary energy for more than 250 million Africans (Sayre et al., 2011). Children consuming cassava as a staple food are at risk for inadequate zinc, iron, and vitamin A intake (Gegios et al., 2010; Narayanan et al., 2019). Moreover, a typical cassava-based diet provides less than 30% of the minimum daily requirement for protein and only 10%–20% for iron, zinc, and vitamin A (Sayre et al., 2011). Although cassava is regarded as a food security crop in many developing countries, it has crucial deficiencies in essential minerals, vitamins, and protein. If one’s diet primarily relies on staple foods like cassava, there may be adverse health consequences unless the diet is enriched with essential nutrients. Many individuals who rely solely on cassava for their dietary energy are poor and cannot afford supplementary foods to meet their protein needs. Consequently, enhancing the nutritional profile of cassava crops has emerged as a pressing concern for scientists, particularly cassava breeders, and biotechnologists, to ensure that those dependent on it obtain their daily nutrient intake.
To effectively address the issue of agricultural development in developing countries, it is critical to recognize the consumer-preferred crops and, most importantly, the staple crops that provide most of the calories. In (SSA), the starchy root crop cassava ranks first in total productivity and number two behind maize as a source of calories (FAO/WHO, 2002; FAO, 2012; FAO, 2013; Narayanan et al., 2019). For cassava, starch content represents 64-72 percent of its total carbohydrates, depending on the cultivars or genotypes. Classical and advanced molecular breeding approaches of cassava for increasing the overall biomass and enhanced starch content of root tuber have represented, over the years, the fundamental objective of various cassava improvement programs. Through these breeding objectives, cassava breeders in various cassava-producing countries worldwide have developed and adopted novel cassava cultivars with increased starch yield, quality, and overall biomass (Hillocks et al., 2002; Jennings and Iglesias, 2002; Malik et al., 2020; Amelework and Bairu, 2022). The application of biotechnological approaches for improving starch content and overall biomass in cassava has been attempted with some success. Starch production was increased by enhancing the activity of ADP-glucose pyrophosphorylase (AGPase) activity in transgenic cassava tuberous roots (Ihemere et al., 2006). To achieve this result, a modified form of the bacterial glgC gene by site-directed mutagenesis (G336D) was expressed under the control of a Class I patatin promoter in transgenic cassava plants (Ihemere et al., 2006). Data demonstrated that modified cassava plants exhibiting the highest level of AGPase activities had a 2.6-fold increase in total tuberous root biomass under greenhouse growing conditions. Additionally, these modified cassava plants with the highest AGPase activity in their tuberous root showed significant increases in their above-ground biomass (Ihemere et al., 2006). Additionally, cassava is known for its high level of cyanogen, which may render the crop poisonous for daily consumption (Chavarriaga-Aguirre et al., 2016). Breeding programs have attempted to overcome the high levels of cyanogen, viral diseases, and nutrient deficiencies in cassava by trying conventional and transgenic breeding methods (Chavarriaga-Aguirre et al., 2016). The amino acid content of cassava reveals a shallow composition of sulfur amino acids, which are essential for human health (Omole, 1977; Nassar and Sousa, 2007). Traditional breeding methods have met little success in increasing the protein content of cassava, mainly due to the absence of cultivars with high protein content in the available local landrace germplasms for use as parents in classical and advanced breeding approaches targeting protein enhancement. This setback makes transgenic technology more attractive in achieving protein enhancement in cassava. Biotechnology has been proven successful in developing low cyanogen content (Siritunga and Sayre, 2003, 2004), insect-resistant (Ladino et al., 2002), virus-resistant (Lin et al., 2019; Narayanan et al., 2021), and herbicide-resistant (Sarria et al., 2000) cassava cultivars, and many nutritional traits such as vitamin A (Welsch et al., 2010; Failla et al., 2012), iron (Ihemere et al., 2012; Narayanan et al., 2015); and vitamin B (Li K. T. et al., 2015) have been enhanced. However, the use of biotechnological tools to increase protein content in cassava has not been conducted intensively, and a few attempts over the years have met with little success. A global initiative addresses iron, zinc, and selenium deficiencies by identifying cassava genotypes naturally rich in these nutrients. This study evaluates 20 South American cassava genotypes for their potential in biofortification, finding promising candidates like MS018, DG014, and DG839, which exhibit elevated levels of Fe, Zn, and Se alongside superior photosynthetic capacity and increased yield potential. The research contributes to developing cassava genotypes with enhanced agronomic biofortification and improved yield prospects (Inacio et al., 2024).
The first attempt was to create a strong protein sink in roots, which was shown to be sufficient to elevate total root protein. A class I patatin promoter-driven cassava hydroxy nitrile lyase (HNL) construct was used to produce transgenic plants with a three-fold increase in total root protein and an 80% reduction in root linamarin levels (Narayanan et al., 2011). Cassava hydroxynitrile lyase (HNL) is an enzyme that catalyzes the conversion of acetone cyanohydrin to cyanide. Because this enzyme is localized in the apoplastic space around leaf cells, it could presumably be induced to accumulate, thereby overexpression without turning over. The second attempt consisted of accumulating proteins in roots expressing artificial storage proteins ASP1, designed to be rich in essential amino acids (Zhang et al., 2003).
These previous attempts suggest that increasing protein content in staple crops such as cassava, a diet of the malnourished, is still an extremely challenging task (Narayanan et al., 2011). No cassava cultivar with high protein content has been developed to meet the minimum daily requirements (MDR). We developed a novel bioengineered cassava cultivar and identified the best transgenic plants expressing a high protein-enhancing trait. Previous studies have demonstrated that the ectopic expression of QQS (Li L. et al., 2015) increased soybean protein independently of the genetic background and the original protein content of the cultivar. Furthermore, QQS is known to regulate the metabolic processes that affect the partitioning of carbon and nitrogen among proteins and carbohydrates, thereby resulting in a variation in leaf and seed composition (Li L. et al., 2015). Molecular and biochemical tools were employed to validate the transgenic nature of the cassava lines expressing QQS, and their nutritional profile was examined for protein and soluble total carbohydrate enhancement. QQS gene was expressed in the model cassava cultivar (cv 60444), which is not a preferred cultivar for farmers in Africa (Chavarriaga-Aguirre et al., 2016) however it was chosen because it is among the African germplasm for which genetic transformability has been demonstrated, and the time from Agrobacterium co-culture to the regeneration of whole plants is 4.5–5 months with escape rates of 5% or less (Chavarriaga-Aguirre et al., 2016). The subsequent integration of protein and starch nutritional traits into the genetic backgrounds of cassava favored by farmers in SSA (Hankoua et al., 2005, 2007; Nyaboga et al., 2013) should be attempted. This proof-of-concept demonstrated in this study using cv 60444 will guarantee critical success for delivering starch and, more importantly, protein-biofortified cassava products to millions of poor smallholder cassava farmers and consumers.
The study aimed to investigate the effects of introducing the Qui-Quine starch gene into cassava plants through genetic modification. Specifically, the study aims to assess how the constitutive overexpression of this gene influences the levels of both starch and protein content in transgenic cassava plants. Furthermore, the studies seek to elucidate the potential of this genetic modification approach to enhance cassava’s nutritional quality and yield, thereby contributing to efforts to improve food security and address malnutrition in regions where cassava is a staple crop. The orphan gene AtQQS (Qua-Quine Starch) and its interacting partner, a transcription factor NF-YC4 (Nuclear Factor Y, subunit C4), have been proven to elevate the levels of protein in leaves and seeds without influencing the growth and yield of agronomically significant species (Li L. et al., 2015). Other studies, unrelated to nutritional traits enhancement, demonstrated that the increased expression of AtQQS and NF-YC4 in Arabidopsis and soybean results in novel functionalities such as heightened resistance or decreased susceptibility to various pathogens, including viruses, bacteria, fungi, aphids, and soybean cyst nematodes (Qi et al., 2019). Furthermore, investigating the gene network involving AtQQS and its interactor, NF-YC4 proves crucial in unravelling the intricate mechanisms underlying the interaction between AtQQS and starch metabolic processes and its influence on auxin metabolic processes. This insight contributes to a more comprehensive understanding of the genetic regulatory pathways involved in these essential physiological processes.
The availability of the complete nucleotide sequence of the cassava genome (Prochnik et al., 2012) prompts us to optimize the sequence of the QQS gene for better expression in cassava by taking advantage of the preferred codon usage of the gene expression machinery of cassava (Dai et al., 2000). This codon optimization of QQS could positively impact the expression of QQS in transgenic cassava plants. The QQS gene sequence (GenBank# NM_113975) was sent for synthesis and QQS codon optimization at Integrated DNA Technologies, Inc. (Coralville, Iowa, USA). The synthesized and codon-optimized sequence was cloned at the Sac1 and Kpn1 restriction sites in the shuttle vector pCU57, a 2.7 Kb Genscript vector having a bacteria ampicillin-resistant marker (Supplementary Figure S1A). The sequence alignment of the original and optimized QQS sequences is shown (Supplementary Figure S2). In Supplementary Figure S2, nucleotides in “red” in the optimized sequence are codons preferred by the cassava expression machinery. The sequences of the restriction sites for SacI and KpnI restriction enzymes were added at both 3’ and 5’ of primers designed to amplify the QQS optimized sequence (GenBank# NM_113975) from the donor plasmid pCU57. These primer modifications facilitate restriction cloning of the synthesized QQS gene in a pSAT1shuttle vector (Tzfira et al., 2005), as shown in (Supplementary Figure S1B). PCR amplification using QQS’s specific primers and double restriction digestion of the pCU57-QQS vector with restriction enzymes SacI and Kpn I New England Biolabs (MA, USA) restriction enzymes were performed to confirm the presence of the optimized QQS insert before cloning into shuttle vector pSAT1 and binary vector pPZP-RC2-npt-II for plant expression as shown in (Supplementary Figure S1C).
The purpose of cloning QQS optimized sequence into pSAT1 shuttle vector was to assign our gene of interest with genetic elements available in pSAT1 modular vectors such as a promoter (MAS and enhanced 35S in most cases), a multiple cloning site, a homing endonuclease or rare restriction enzyme site, a translation enhancer (5’-UTR from tobacco etch virus), and a terminator sequence as shown in the (Supplementary Figure S1B). Gibson’s assembly method was used for this cloning after multiple unsuccessful attempts using the ligation technique. Gibson assembly method is a ligation-free cloning technique developed to increase the efficiency and accuracy of DNA assembly to create expression plasmid (Gibson et al., 2009). The optimized QQS sequence was redesigned using Snap gene software to assign it with 5’ and 3’ overlap sequences (30 nucleotides were added at each 5’ and 3’ end). The software designed the corresponding QQS gene-specific Gibson primers and QQS fragment primers (Supplementary Table S1) to assign to the QQS gene 5’ and 3’ overlapping regions via PCR amplification. Gibson QQS fragment was generated through a PCR reaction involving the synthesized QQS fragment primers with overlapping regions and the plasmid pCU57, comporting the optimized QQS gene sequence as a template. Q5 High-fidelity DNA polymerase (New England Biolabs, USA) was used for the PCR reaction following the manufacturer’s protocol at an annealing temperature of 72°C (New England Biolabs Tm calculator tools). The outcome of this PCR reaction was an amplicon of a linearized QQS gene comporting overlapping sequences to its 5’ and 3’ ends. The pSAT1 vector, our initial entry shuttle vector, was also linearized using a double restriction enzymatic reaction with Sac1 and Kpn1 to generate a linearized vector for the Gibson reaction. Then, the linearized QQS gene comporting overlapping regions, the linearized vector was purified using gel, and purity and concentration were determined. The fragments (linearized QQS gene and PSAT1 vector) were combined in a single Gibson reaction following NEBuilder® HiFi DNA Assembly Master Mix/NEBuilder HiFi DNA Assembly Cloning Kit protocol. An aliquot from the Gibson reaction, supposed to contain the correct assembled vector pSAT1-QQS, as shown in the Supplementary Materials S1 (A), was transformed by heat shock into E. coli 5α competent cells (New England Biolabs, USA) for assembled pSAT1-QQS propagation and insert verification. Colony PCR and double restriction digestion of the assembled pSAT1-QQS vector with SacI and KpnI were performed to confirm the proper insertion of the QQS-optimized gene in this vector.
pPZP-RC2-npt-II, an 8783 bp plasmid , is a plant expression binary vector as described by Tzfira et al. (2005); Dafny-Yelin and Tzfira (2007); Zeevi et al. (2012) as shown in (Supplementary Figure S1C). The confirmed single-gene expression cassette originating from the pSAT1-QQS assembled vector underwent restriction digestion with the restriction homing enzyme AscI (New England Biolabs, Ipswich, MA, USA) to enable the 1,433 bp QQS expression cassette to be pulled out from the vector and to facilitate its insertion into the pPZP-RC2-npt-II final vector. The pPZP-RC2-npt-II binary vector (Goderis et al., 2002) was linearized through restriction digestion with AscI homing enzyme (New England Biolabs, Ipswich, MA, USA). Following the manufacturer’s protocol, the resultant digestion products were purified using the QIAGEN MiniElute Gel Extraction Kit (QIAGEN, USA). The product of the reaction, a linearized pPZP-RC2-npt-II, and the QQS expression cassette were dephosphorylated with the shrimp alkaline phosphatase (NEB, USA) following instructions from the manufacturers. The linearized and dephosphorylated QQS expression cassette and pPZP-RC2-npt-II vector were ligated using T4 DNA ligase (New England Biolabs, Ipswich, MA, USA). An aliquot of the ligation product was transformed into E. coli DH5α competent cells for assembled plasmid propagation and insert verification. Colony PCR was performed on positive colonies of E. coli DH5α, and five positive colonies were sequenced at Macrogen, USA, to confirm the integrity of the inserted optimized QQS sequence into pPZP-RC2-npt-II. In addition to colony PCR, the proper cloning of the QQS expression cassette into the pPZP-RC2-npt-II-QQS final expression vector was confirmed through restriction digestion with ASCI homing enzyme and expression cassette fragment confirmed on the 1% agarose gel electrophoresis. The correct QQS expression construct (pPZP-RC2-nptII-QQS) (Figure 1) was mobilized into disarmed A. tumefaciens strain LBA4404 (Lin et al., 2005) by heat shock. Colony PCR was performed on positive A. tumefaciens strain LBA4404 colonies to assert the conformity of the QQS expression construct into the disarmed A. tumefaciens strain LBA4404. Furthermore, A. tumefaciens strain LBA4404 trans-conjugant was used as a vir helper strain and vector system for cassava transformation (Taylor et al., 2001; Hankoua et al., 2006; Taylor N. et al., 2012; Ligaba-Osena et al., 2018).
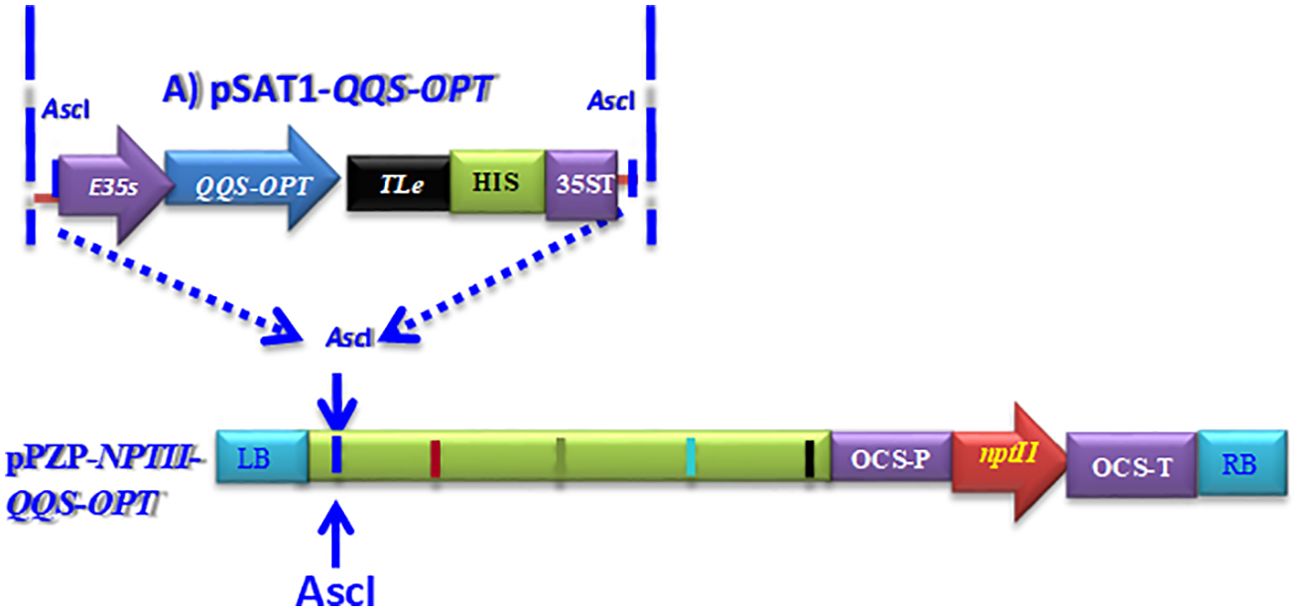
Figure 1. Schematic depiction of the pPZP-QQS-NPTII-OPT expression construct designed for the constitutive expression of optimized QQS in genetically modified cassava plants. Building the QQS expression construct involves cloning the optimized QQS into the pSAT1 shuttle vector, designated as pSAT1-QQS-OPT. The process of deriving the plant expression vector includes the use of the Ascl as a homing restriction enzyme and DNA cleavage point for targeted insertion of the QQS expression construct into the Ascl restriction site located within the T-DNA borders, right-border (RB), and left border (LB) of the pPZP-RC2-nptII binary vector to generate the pPZP-nptII-QQS-OPT expression vector for cassava transformation. Key elements of the expression construct include the Enhanced 35S promoter (E35S) to drive the expression of the optimized QQS in bioengineered cassava plants, the Translation Enhancer (TLE) derived from the potato etch virus, the 35S terminator sequence, and histidine tag for transgenic protein detection, nptII for plant selectable marker gene. Within the T-DNA borders of the pPZP-RC2-nptII binary vector, the nptII expression for plant-selectable marker selection will be controlled by the octopine synthase gene's promoter (OCS-P) and terminator (OCS-T).
The friable embryogenic callus (FEC) is routinely used as a target tissue for the genetic transformation of cassava. To induce and proliferate high-quality FEC tissues, we followed protocols described by (Taylor et al., 2001; Hankoua et al., 2006; Taylor N. et al., 2012) with limited modifications. Cassava cultivar TMS60444, a preferred genotype for gene insertion due to its performance in responding to the existing in vitro regeneration and genetic transformation systems of cassava (Ladino et al., 2002; Hankoua et al., 2006), was maintained as in vitro plantlets in a controlled growth chamber under the following conditions: 27°C, 80% relative humidity, and 16-h/8-h day/night cycles. Under these growing conditions, in vitro, plantlets are generated using stem segments comporting at least one axillary, apical, or both shoot bud cultured on Cassava Basic Medium with the following nutrients composition [4.43 g/L MS (Murashige and Skoog, 1962) salts with vitamins, 2% (w/v) sucrose, 2mM CuSO4, Supplementary Table S2]. According to Hankoua et al. (2006), Taylor et al. (2001) and Taylor N. et al. (2012), friable embryogenic calli were initiated from cyclic somatic embryos from cassava immature leaf lobes. Induced FEC was proliferated, cycled, and maintained as described by Taylor et al. (2001; Hankoua et al., 2006, 2012) in the MS2-P50 medium (Supplementary Table S2). To initiate FEC, high-quality and proliferating cyclic somatic embryos were fragmented into small pieces and incubated on GD250P [GD (Gresshoff and Doy, 1972) 2-50P, Supplementary Table S2] medium for FEC formation. The pure FEC was continuously selected from GD250P plates and subcultured every four weeks until homogenous and highly proliferating FEC clusters were obtained. Homogeneous and highly proliferating friable embryogenic calli (HPFEC) were continuously maintained on GD6-50P supplemented with 6% sucrose [GD (Gresshoff and Doy, 1972) 6-50P, Supplementary Table S2] before their utilization for genetic transformation.
Agrobacterium-mediated DNA delivery and recovery of in vitro regenerated plantlets were performed following the cassava transformation and regeneration techniques described by Hankoua et al., 2006; Taylor N. et al., 2012; Osena Ligaba et al., 2018. Agrobacterium tumefaciens strain LBA4404 vir helper strains, and transconjugants harboring the control binary plant transformation vector pPZP-RCS2-nptII without the optimized QQS gene of interest and comporting the nptII gene coding for the enzyme neomycin phosphotransferase (NPT), an aminoglycoside phosphotransferase conferring resistance to antibiotics such as kanamycin, neomycin, paromomycin and used as the plant selectable marker gene, the expression cassette for driving the expression of QQS optimized gene in cassava tissue were used for genetic transformation. Single colonies of Agrobacterium transconjugants LBA4404 carrying each expression construct were cultured in a 2 mL LB medium containing appropriate antibiotics for overnight growth for 10-12 hours under shaking at 220 rpm at 28°C. Approximately 0.5–1.5 mL of these overnight Agrobacterium suspensions initiated 25 mL of Yeas Mold (YM) medium (composition for 1 L of media: yeast extract 3 g, malt Extract 3 g, peptone 5 g, dextrose 10 g) supplemented with appropriate antibiotics and 100 μM acetosyringone.
The Agrobacterium suspension cultures grew and were monitored to reach an OD600 of 0.5–0.75. The bacteria suspensions with optimal OD600 were prepared according to Hankoua et al., 2006; and Osena Ligaba et al., 2018 before the inoculation of HPFEC. This suspension was washed and induced in a GD2 50P medium containing 200 µm of acetosyringone and Pluronic F-68, a non-ionic surfactant, at a concentration of 0.03%. HPFEC obtained from GD2-50P agar media (Supplementary Table S2) was inoculated with 1 mL of induced bacterial suspensions. Three independent genetic transformation events were performed for each expression construct. After inoculating HPFEC with these bacteria suspensions, tissues were vacuum infiltrated for at least 15 min with two pauses every 5 min. These inoculations and infiltrations were incubated at room temperature for approximately 30 min.
Excess Agrobacterium suspensions were removed after inoculation steps, and coculturing of the inoculated HPFEC tissues was performed according to protocols described in Hankoua et al., 2006; Taylor N. et al., 2012; Osena Ligaba et al., 2018. Inoculated HPFEC tissues were placed on a co-culture plate (GD2- 50P+100 µM acetosyringone) and incubated at 22°C with full lighting for two days. Two days after cocultivation, the inoculated HPFEC were washed twice into GD2 50P containing 500 mg of carbenicillin and then placed on a recovery media plate (GD2 50P+500 mg/l carbenicillin) for seven days to initiate cell division. After seven days of recovery, HPFEC tissues were transferred to Stage I selection medium GD2-50P containing 27.5 μM paromomycin and 500 mg/L carbenicillin. PCR validated transgenes insertions from genomic DNA samples isolated from proliferating putative transgenic homogeneous and proliferating friable embryogenic calli.
Putative transgenic HPFEC tissues, which are those that survived and proliferated on 27.5 µM paromomycin, were placed onto embryo maturation media (EMM) with the following composition [4.43 g/L MS salt with vitamins, 2% (w/v) sucrose, 45 µM paromomycin, 0.5 mg/L NAA, pH5.8, Supplementary Table S1] for the emergence of the putative transgenic embryo. Putative transformed maturing embryos were monitored from antibiotic selection media, then picked regularly, and transferred onto embryo germination media (EGM) [4.43 g/L MS salt with vitamins, 0.4 mg/L BAP, 2% (w/v) sucrose, Supplementary Table S2] for 3 to 4 weeks for putative transgenic shoots emergence. Germinated, putative transgenic shoots were transferred to CBM without antibiotics for rooting (Figure 2).
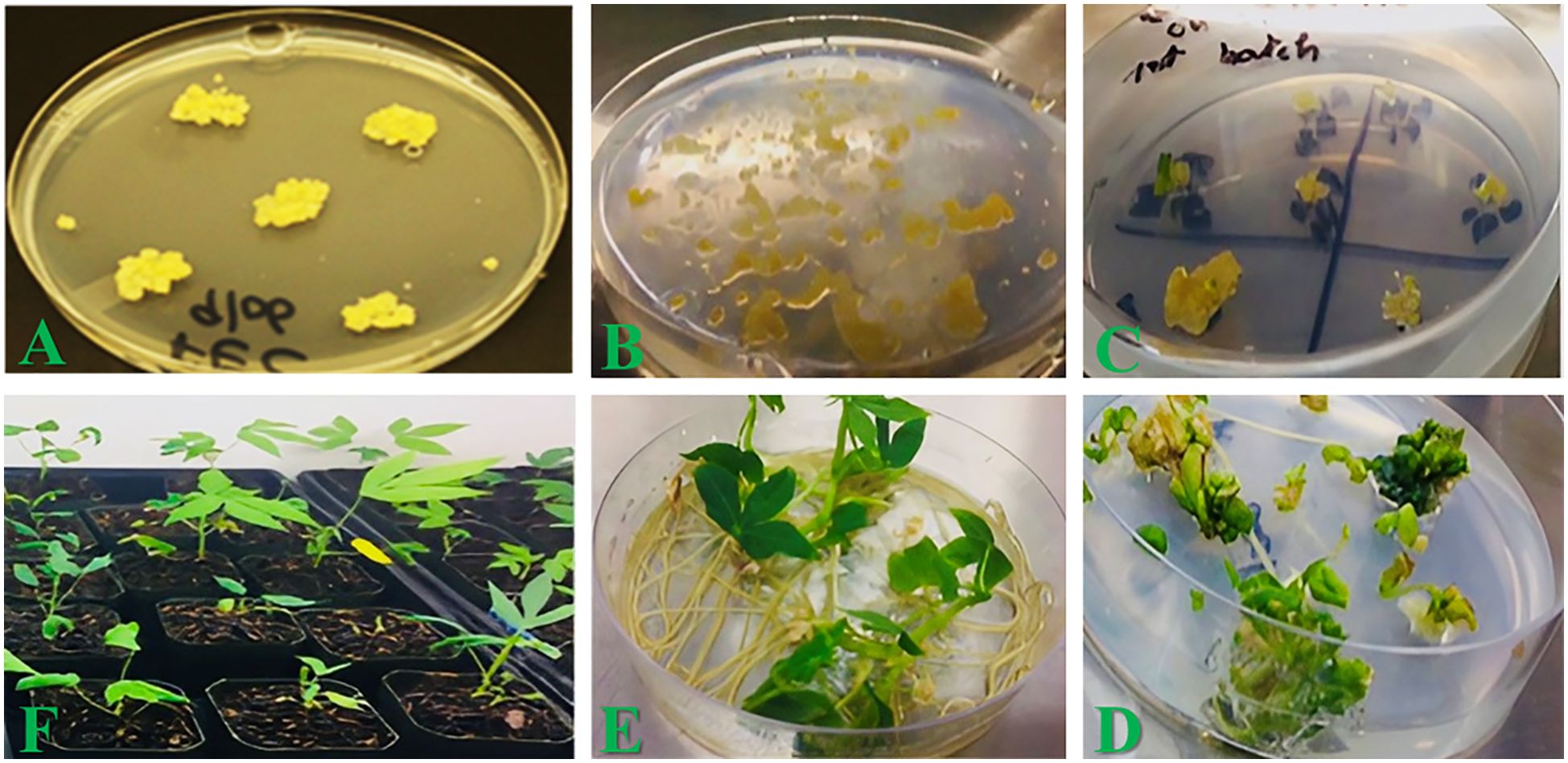
Figure 2. Tissue culture steps for transforming Highly Proliferating Friable Embryogenic Calli (HPFEC) of cassava, recovery, and hardening of transgenic plants. (A) HPFEC of cassava were generated as described in the Section "Materials and Methods." (B) Paromomycin selection of inoculated HPFEC with Agrobacterium strain LBA4404 containing an expression cassette of optimized QQS gene as in Figure 1 and production of putative transgenic calli. (C) Recovery of putative transgenic immature and cotyledonary stage somatic embryos selected under paromomycin pressure. (D) Germination and regeneration of 1st putative transgenic cotyledonary stage somatic embryos to recover putative transgenic shoots.). (E) Elongation and rooting of putative transgenic shoots to generate putative transgenic plantlets. (F) Transplantation of putative transgenic cassava plantlets into soil transgenic for acclimatization.
Three-week-old micro-propagated plantlets thriving on CBM (Supplementary Table S2) were transplanted in Kord 3.0 square pots filled with 230 mL Fafard 52 soil (Maryland Plants & supplies INC., MD, USA), then treated with Gnatrol to control fungal gnats for hardening. This acclimatization protocol was slightly modified (Zidenga et al., 2012), which used the Fafard 51 mix to acclimate in vitro cassava plantlets and was shown to promote mini-tuber development. Nitrogen-Phosphate-Potassium (NPK) fertilizer and all-purpose plant fertilizer, Miracle-Gro, USA, were added, by alternation, once a week to the Fafard 52 soil for the fertilization of the greenhouse acclimatized in vitro plantlets (Figure 2) until established plants were morphologically assessed and biomass harvest for downstream characterization.
Preliminary evidence of the insertion of the QQS gene into the genome of transgenic lines was validated through PCR reactions conducted at various stages of cassava plant regeneration and development. Biomass samples were collected from putative transgenic sources, including 1) maturing embryos, 2) three-week-old cassava plantlets cultivated on MS2 cassava maintenance medium, and 3) six-week-old soil-grown cassava plants that were well-established. The transgenic biomass derived from the six-week-old well-established soil-grown cassava plants was utilized for quantitative RT-PCR analysis to assess the expression level of the QQS gene across different transgenic lines and to conduct their biochemical characterization. Furthermore, to evaluate the expression or transcript levels of QQS in root and leaf tissues of various transgenic lines, total RNA was extracted from newly fully expanded leaves and roots of each selected transgenic line and wild-type non-transformed plants. The tissues were ground in liquid nitrogen using a mortar and pestle. Total RNA was extracted using Spectrum Plant Total RNA kit (Sigma, Aldrich, USA). For each transgenic line, 4 μg of the extracted RNA was treated with DNase using amplification-grade deoxyribonuclease I in a kit (Invitrogen Life Technology, USA). After DNase treatment, 16 μL of the resulting reaction was used to synthesize cDNA using superscript III first-strand cDNA synthesis system for RT-PCR (In vitro gen life technology, USA). The cDNA concentration was checked on Nanodrop, and then samples were normalized to 1.25 µg/μL for their use in quantitative real-time (QRT) PCR reaction. QRT-PCR QQS primers (Supplementary Table S3) were synthesized by Integrated DNA Technologies (IDT, Coralville, IA, USA). QRT-PCR was carried out using Power SYBR Green Master Mix (Applied Biosystems, USA). The Cassava tubulin gene was used as an internal control. Three technical replicates were used for each biological replicate (transgenic line), and the wild-type non-transgenic line was used as the reference sample. Relative gene expression was calculated using the ΔΔCT method available on the 7500 and 7500 fast real-time PCR system version 2.0.1 software. The mean and standard error of the RQ values collected from the real-time PCR data were used to compare gene expression levels between lines.
Leaves, stems, and roots biomasses were harvested from 6 weeks in-vitro and soil-grown of five selected QQS stable transgenic lines (R’’’(G) L1, R’’’(G) L2, R’’’(G) L3, R’’’ (LA) L2, R7 (F)), lines expressing only NPTII gene known as empty vector expressers, and wild-type plants expressing no gene for biochemical characterization. These biomasses were ground in liquid nitrogen and then stored at -80°C in 50 mL sterile centrifuge tubes. Later, these biomass samples were utilized for biochemical analysis to compare the total protein content, amino acid composition, and soluble carbohydrate content between wild-type non-transformed, empty vector expressers and stable transgenic lines expressing QQS.
For the total soluble protein extraction and quantification, 100 mg of ground tissues of various biomass were taken from a 50 mL sterile centrifuge previously stored at -80°C. Protein extraction was performed in triplicate for each cassava line and biomass type. Total Protein extraction was done using 1 mL of cold extraction buffer (200 mM NaCl, 1 mM Tris-HCl pH 7.8, 4% of 2- mercaptoethanol, and 1% complete protease inhibitor). After extraction of the total protein, the protein samples were vortexed and centrifuged to obtain protein pellets. These protein pellets were further suspended in 100 μL resuspension buffer, and the resulting aqueous solution was used for total protein quantification. Following the manufacturer protocol, total protein content was determined using a protein quantification CB-X kit (G Biosciences, Maryland Heights, MO, USA). The absorbance of the protein samples at 595 nm was recorded from a Thermofisher US-Vis 260 evolution spectrophotometer to determine the protein concentration (μg/μL) according to the best line of fit equation obtained by generating a standard curve using Bovine Serum Albumin (BSA) 2 mg/ml and according to the CB-X kit protocol.
Total carbohydrate extraction and quantification were performed on ground tissues of various biomass taken from a 50 mL sterile centrifuge previously stored at -80°C. For each cassava line and tissue type, 50 mg of ground tissues were used with two replicates per sample. Soluble total carbohydrate was extracted using 200 μL of an ice-cold assay buffer supplied by the soluble total carbohydrate Assay kit (Sigma-Aldrich, USA) used for the analysis. After carbohydrate extraction, aqueous samples were centrifuged (13,000g/5min) to obtain carbohydrate pellets. These carbohydrate pellets were further re-suspended in 30 μL buffer, and the resulting aqueous solution was used for total carbohydrate quantification following the manufacturer’s protocol. Water was used for samples with less than 30 μL of pellet resuspension solution to adjust the volume to a final volume of 30 μL as required by the protocol. The absorbance of the samples at 490 nm was used to determine the soluble total carbohydrate concentration (μg/50 mg) according to an equation obtained by generating a standard curve using a D-glucose standard 2 mg/ml and according to the soluble total carbohydrate Assay kit protocol.
Amino acid analysis was performed according to the method described in Cohen and De Antonis, 1994. Briefly, oven-dry samples were hydrolyzed in 6.1 N HCl containing a small amount of phenol; the hydrolysis flasks were extensively purged of oxygen using a PicoTag workstation (Waters Corp., Milford, MA) and then incubated at 110°C for 20h. Hydrolyzed samples were then evaporated to dryness under vacuum and derivatized using AccQFluor reagent (Waters) according to the manufacturer’s directions. Chromatography was performed using procedures described as ‘System 1’ in Cohen and De Antonis (1994), with α-aminobutyric acid as an internal standard. Separation was achieved using an AccQTag C18 reverse phase column (Waters) and detection by fluorescence using excitation with 250 nm light and measured emission at 395 nm. Hydrolysis, derivatization, and analysis of each sample were performed in triplicate.
This analysis constructed a gene network using the Gene IDs of the AtQQS gene in Arabidopsis: AtQQS (UniProt ID- Q3E7K4). First, the Gene ID (QQS) was used as a query against the STRING database via Cytoscape and StringApp (Doncheva et al., 2019). STRING is a biological network database that stores known and predicted interactions obtained from primary databases and computational approaches, respectively (Szklarczyk et al., 2023). Next, we evaluated the biological role of the generated network using ClueGO/CluePedia (Bindea et al., 2009) apps in Cytoscape. Finally, a right-sided hypergeometric test (enrichment) with Bonferroni correction was used to compute the false discovery rate of each pathway and biological process to assess its significance.
Statistical Analysis System (SAS) Enterprise Guide 5.1 was used for statistical analysis. Experiments were done in duplicates for soluble total carbohydrate content, and sampling was performed using independent biological triplicates for total protein content. One-way ANOVA was used to check differences between the mean values. Significant probability was set at p ≤ 0.05. In addition, Excel’s Microsoft version 2016 was used to check correlations between QQS transcript levels in transgenic root and leaf, the soluble total carbohydrates, and the soluble total protein content of these plant parts. Correlations analyses were also performed to determine any relationship between carbohydrate and protein content in leaves and roots.
Overall, 16 independent putative transgenic in vitro cassava plants were recovered from all Agrobacterium-co-cultures and plant recovery. Seven of these transgenic plants were lost due to recurrent in vitro microbial contaminations. In vitro, putative transgenic plantlets at the onset of soil transplantation look normal with no detectable morphological anomalies as compared with their non-transformed counterparts (WT) and NPTII (empty vector) expressers (Figure 3). All In vitro putative transgenic plantlets showed a 100% survival rate after transplanting in soil (Figure 3). Visual comparison of plant height and biomass production at 6-7 weeks after soil transplanting between wild-type, NPTII, and each transgenic cassava is shown in Figure 4. No marked variation in these agronomic parameters was observed between these lines, except that line R7 (B) showed slow growth with lower biomass production than the wild type and NPTII expresser (Figure 4). Symptoms of fascinated leaf, tiny stem, and slow growth were observed in two soil-established cassava expressing QQS lines (R7 (E), R7 (D) (Figure 5 Section A and Section B). Among all these transgenic cassava lines, line R7 (F) exhibited a particular phenotype in its growth habit and capability to produce more biomass (Figure 5A). It is evident in Figure 5A that Line R7 (F) grows more profusely and produces more biomass than the other transgenic lines, such as R’’’ (G) L2 and the WT. Furthermore, morphological differences between individual plants of line R7, including variation in growth rate, leaf shape, and size, were observed (Figure 5).

Figure 3. Phenotypes of soil established in vitro putative transgenic cassava lines expressing QQS gene compared to the wild type (WT) non-bioengineered and NPTII expressing transgenic lines. All In vitro plantlets showed a 100% survival rate after transplantation in soil, and transgenic plants in soil look normal with no detectable morphological anomalies compared to their non-transformed counterparts (WT) and NPTII (empty vector) expressers.

Figure 4. Comparative morphological analysis of 6-week-old in vitro putative transgenic cassava plants, established in soil, which illustrates distinctions between wild-type non-transformed plantlets and NPTД-expressing transgenic cassava, and QQS expressing lines R""(G)L1 (A), R""(G)L2 (B), R""(G)L3 (C), R""(LA)L1 (D), R""(LA)L2 (E), Old FEC (F), R7(F) (G), and R7(B) (H).

Figure 5. (A-D): QQS expressing transgenic cassava line R7(F) exhibits a remarkable ability to make more biomasses, including larger leaves (A, B) and root system (C) and the ability to initiate mini tubers faster in- vitro (D) (6-7 weeks) and in soil (3-4 weeks) compared to other QQS expressers and WT non-transformed cassava.
Moreover, Figure 5 section C shows the disparities in plant height, leaf production, and root size of these transgenic cassava lines. Line R7 (F) grows taller and produces more shoot and root biomass than line R’’’ (G) L3 and wild-type non-transformed plants. Similarly, line R7 (F) plant size and root size were compared with the ones obtained with lines R’’’ (LA) L2, R’’’ (G) L3, and R’’’ (G) L1. These morphological data demonstrate a higher and unique growth ability for line R7 (F) compared to all other transgenic lines and control non-transformed plants. All regenerated plants were transferred from In vitro culture to soil under the same growing conditions and evaluated at the same age (6-7 weeks). The final section, D in Figure 5, illustrates the higher capability of R7 (F) to induce mini tubers quicker after its In vitro plantlets were transferred to the soil between 6-7 weeks compared with other transgenic lines and wild-type plants, which did not exhibit this early tuberization capability after more weeks of growth in soil.
Genomic DNA was extracted from eleven randomly selected putative transgenic lines. Insertion of the NPTII and QQS was validated (data not shown) using the genomic DNA and gene-specific PCR primers for QQS (230 bp) and the plant selectable marker (NPTII, 790 bp) (Supplementary Table S1). The level of QQS gene expression in the leaf and root biomass of the wild-type and five putative transgenic lines was performed using SYBR green master mix and qRT-PCR primers (Supplementary Table S2), where the QQS was used as a target gene, and the Manihot esculenta Crantz) α-tubulin gene was used as an internal control. The results obtained from the expression analysis of QQS in leaves and roots are shown in Figure 6. The data showed that the QQS gene is differentially expressed in the leaf and root tissues of all the transgenic lines tested. Transcript levels varied widely between lines and tissue types. Furthermore, the putative transgenic line QQSL2 produced the highest transcript level for the leaf. In contrast, its transcript abundance was lower in the roots than in the other transgenic lines and the non-transgenic control. QQS expression in roots of lines QQSL3 and QQS L4 was higher than the non-transgenic control and other transgenic lines. In line QQSL5, the QQS transcript level was much lower than the other lines in leaf and root tissues. Moreover, the expression of QQS showed a very sharp decrease in the root compared to the leaf tissue in cassava line QQSL2. For root tissue, lines QQSL5 and QQSL2 generated the lowest level of QQS transcript, even below the transcript of the reference wild-type non-transformed plants.
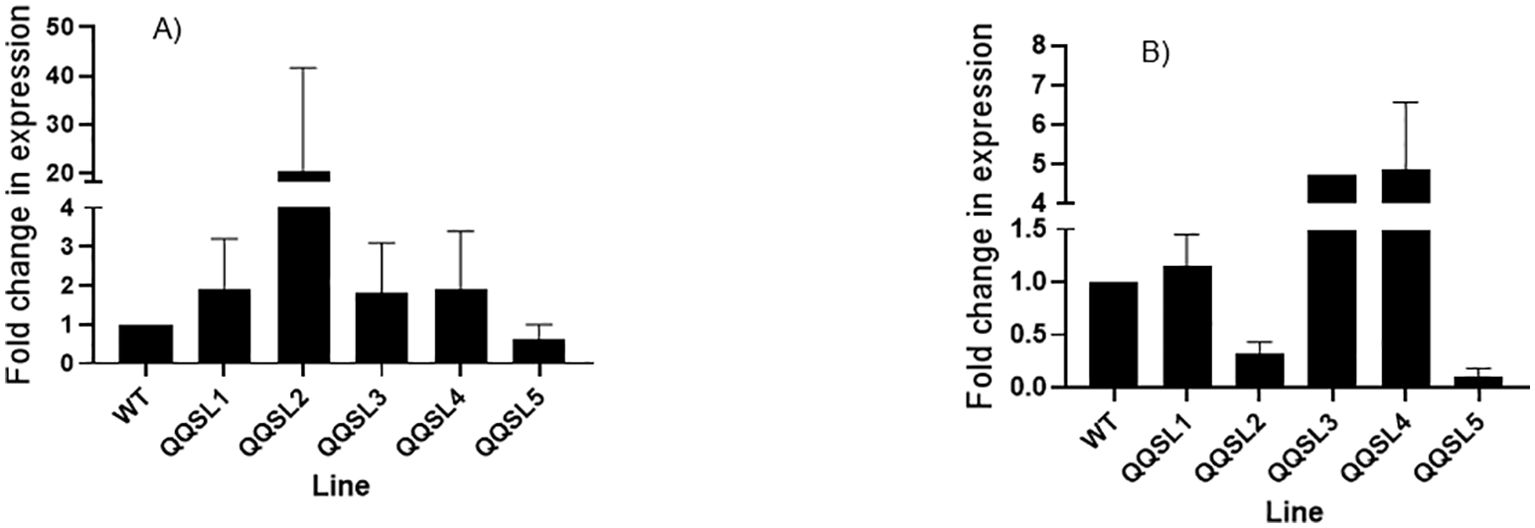
Figure 6. Quantitative real-time PCR analysis of QQS gene from leaf (A) and root tissues (B) of six-week- old soil-grown putative transgenic cassava plants. The graphs were plotted using the mean RQ of samples from each line and the standard error (SE) obtained after the real-time PCR. QQSL1: R"" (G) L1; QQSL2: R"" (G) L2; QQSL3: R”” (G) L3; QQSL4: R”” (LA) L1; QQSL5: R7 (F); (A) QQS transcript expression in different transgenic lines in roots, (B) QQS expression in different transgenic lines in cassava leaves. The samples were measured in triplicates. Data represent means Standard deviation; n=3 biological replicates. Error bars show the standard deviation from the mean. One-way ANOVA determined statistically significant differences from WT plants.
Biochemical analyses were performed on a wild-type non-transformed cassava line and an NPTII-expressing line, and five confirmed transgenic lines were named R’’’ (G) L1, R’’’ (G) L2, R’’’ (G) L3, R’’’ (LA) L2, and R7 (F). Positive quantitative Real-time PCR data in Figure 6 demonstrated that the QQS gene is well integrated into the genome of these transgenic lines. This positive data prompted us to select these lines for further biochemical analyses.
Biochemical analyses, including protein and soluble carbohydrate contents, were performed on wild-type non-transformed cassava line, NPTII expressing line, and five transgenic lines R’’’ (G) L1, R’’’ (G) L2, R’’’ (G) L3, R’’’ (LA) L2 and R7 (F). Total soluble protein was analyzed using the Coomassie Bradford protein assay kit following the manufacturer’s procedure using BSA as the standard (Thermi Scientific). Figure 7 shows that the samples’ soluble total protein concentration varied depending on the tissue types analyzed. All the stable transgenic lines and the two control non-QQS expressers (WT and NPTII) make more protein in the lead and stem biomass, with root biomass making less protein for all the lines. For leaf tissues, no significant differences were observed in total protein concentration between all the putative transgenic lines except biomass of line R7 (F), showing a slight reduction in soluble total protein concentration (1.1µg/µL) compared to other lines and the two controls non-QQS expressers (1.3 µg/µL for WT and NPTII). More variabilities in the total protein concentration were observed for stem and root biomass, with lines R’’’ (G) L1 (0.6 µg/µL) and R’’’ (G) L3 (0.5µg/µL) showing a significant protein decrease in total protein concentration compared to other lines, especially WT (0.75 µg/µL) and NPTII (0.70 µg/µL) biomass. Stem biomass from other lines exhibits total protein levels comparable to control plants. Variabilities in protein concentration of root biomass were observed in all the lines, with lines R’’’ (G) (LA) L2 (0.53 µg/µL) and R7 (F) (0.5µg/µL) showing a higher protein concentration compared to other lines, especially the WT non-transformed (0.42 µg/µL) and the NPTII expressing line (0.40 µg/µL). Root biomass from lines R’’’ (G) L1 (0.35 µg/µl) and R’’’ (G) L2 (0.55µg/µL) showed lower protein concentration.
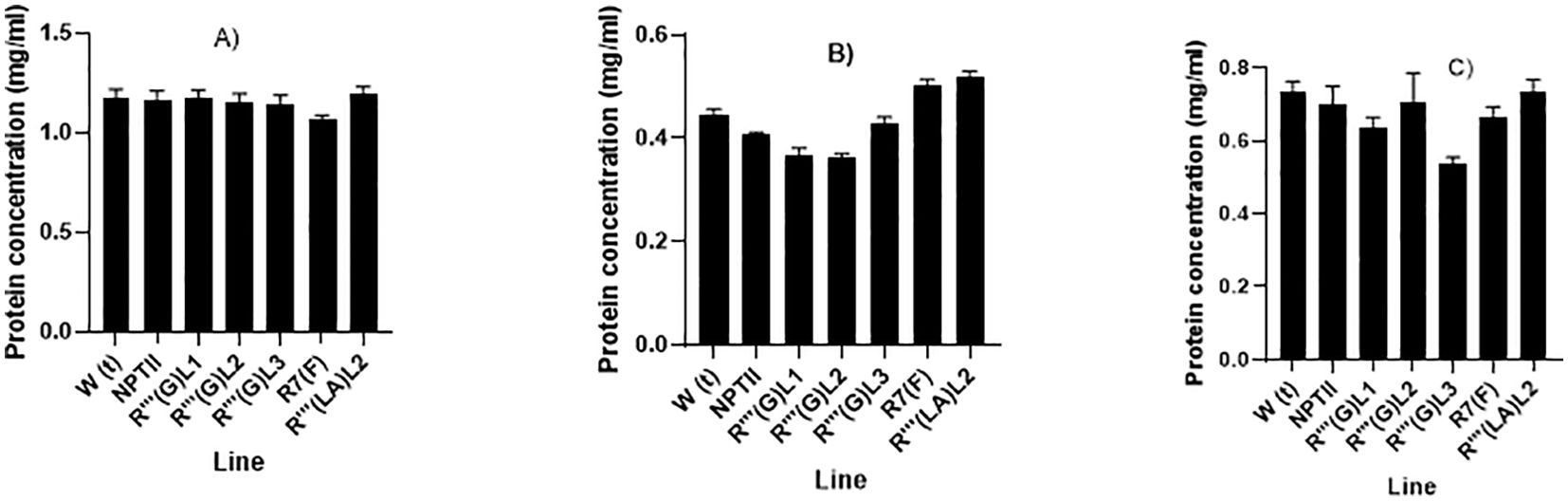
Figure 7. (A-C): Quantitative evaluation of total soluble protein concentration in leaves (A), stem (B) and root (C) tissues of six weeks in-vitro soil grown transgenic cassava plants. The graphs were plotted using the mean protein concentration of samples from each line and the standard error (SE) of the mean of three replicates.
Figure 8 depicts the amino acid composition data gathered from biomass samples obtained from leaves, stems, and roots of stably transformed cassava plants. Across all samples, a total of 18 amino acids were identified. The data summary shown in Figure 8 is displaced in (Supplementary Table S4). Data analysis suggests no significant variation in the levels of individual amino acids between the leaves, stems, and roots biomass of stably transformed cassava plants compared to two control groups, namely non-QQS expressers (WT and NPTII). Glycine, glutamic acid, aspartic acid, asparagine, and leucine are abundant across all lines and plant parts, with glycine and glutamic acid particularly abundant in stem biomass across all lines. Conversely, histidine and methionine are the least abundant amino acids in all lines, with methionine showing a relatively higher abundance in root biomass across all tested transgenic lines. Notably, cysteine, an essential amino acid, was undetected in the analyzed samples. However, it is plausible that cysteine may be present but undetected due to the technical limitations of the employed methodology.

Figure 8. Amino acid compositions of biomass harvested from the leaves, stem, and roots of stably transformed cassava plants. It provides a summary or description of the data related to the distribution and abundance of various amino acids in the different plant parts (leaves, stem, and roots) of stably transgenic cassava plants. For each amino acid, the mean value of each sample was plotted to obtain the figure above. BF-1 :R""(LA)L2 Leaves; BF-4:R""(G)L3 Leaves; BF-5:NPTII Leaves; BF-8:W(t) Leaves; BF-11: Pure FEC Leaves; BF-14: R""(G)L1 Leaves; BF-17: R7 (F) Leaves; BF-20: R""(LA)L2 Leaves; BF-3:R""(LA)L2 Roots; BF-7:NPTII Roots; BF-10:W(t) Roots; BF-13: Pure FEC Roots; BF-16:R""(G)L1 Roots; BF-19: R7 (F) Roots; BF-22:R""(G) L2 Roots; BF-2: R""(LA)L2 Stem; BF-6:NPTII Stem; BF-9: W(t) Stem; BF-12: Pure FEC Stem; BF-15: R""(G)L1 Stem; BF-18: R7(F)Stem; BF-21: R""(G)L2 Stem;.
The total soluble carbohydrate is determined by a kit purchased from Sigma according to the recommended procedure. Figure 9 shows large leaf and root carbohydrate concentration variabilities for all the lines analyzed. In leaf biomass, line R’’’(G)L1 and R’’’(G) L2 produced the highest soluble total carbohydrate content, reaching approximately 13µg/50mg compared to the control non-QQS expressers WT (9µg/50mg) and NPTII (11 µg/50 mg). For root sample, line R’’’(G) L1 (6 µg/50mg) and R’’’(G) L2 (5µg/50mg) produced the lowest carbohydrate content reaching approximately 13µg/50mg, and R7 (F) the highest (18µg/50mg) compared to the controls non-QQS expressers WT (11µg/50mg) and NPTII (10 µg/50mg). The root biomass of line R(7)(F) produces the highest carbohydrate content among all biomasses and lines tested.
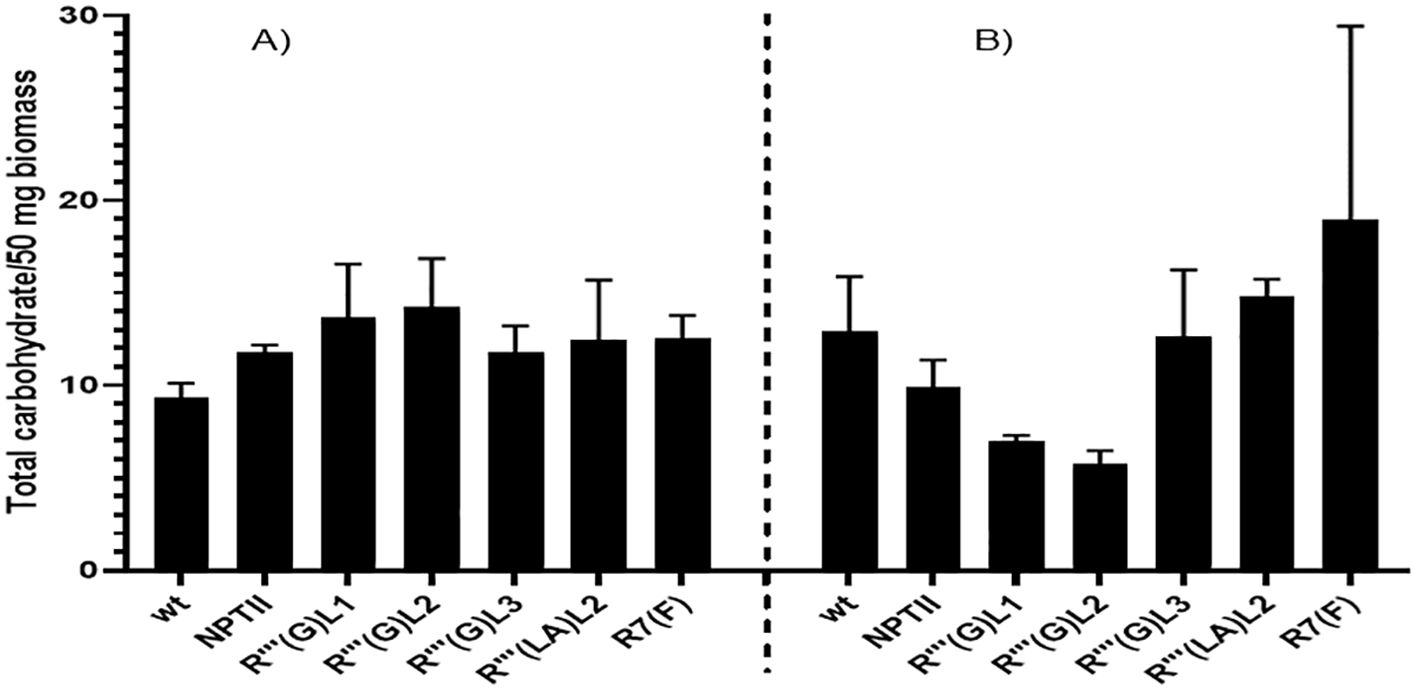
Figure 9. Quantitative evaluation of soluble total carbohydrate in leaves (A) and root tissues of wild-type, NPTII, and QQS expressing lines (B) of six weeks in vitro soil- grown cassava plants. The graphs were plotted using the mean total carbohydrate content of samples from each line and the standard error (SE) obtained after carbohydrate analysis. The samples were measured in triplicates. Data represents the standard error of two biological replicates. Bars represent the means and standard error of at least two independent experiments.
Figure 10 shows the relationship results between QQS transcript levels in leaf and root tissues and their corresponding soluble total carbohydrate or protein content (Figures 10A, B, C, G) as well as the relationship between carbohydrate and protein content between and within leaf and root biomass (D, E, F, H) for all transgenic lines tested. This correlation analysis refers to statistical relationships between the expression levels of QQS transcripts and the amounts of carbohydrates or proteins in a sample. These correlations can provide insights into how gene expression might influence the production or regulation of carbohydrates and proteins within a biological system. Based on the various equations of best fit and the value of the coefficient of determination R2 of all the correlation data, it is evident that no strong correlation is observed between the protein and carbohydrate content of the leaf and root biomasses and the level of QQS transcripts produced from these plant parts in all the lines. No relationship was also depicted between the leaf carbohydrate and leaf protein for all the lines. The only positive correlation observed was between root carbohydrates and root protein. A very poor correlation was observed between the content of leaf carbohydrate and root carbohydrate, as well as the content of leaf protein and root protein.
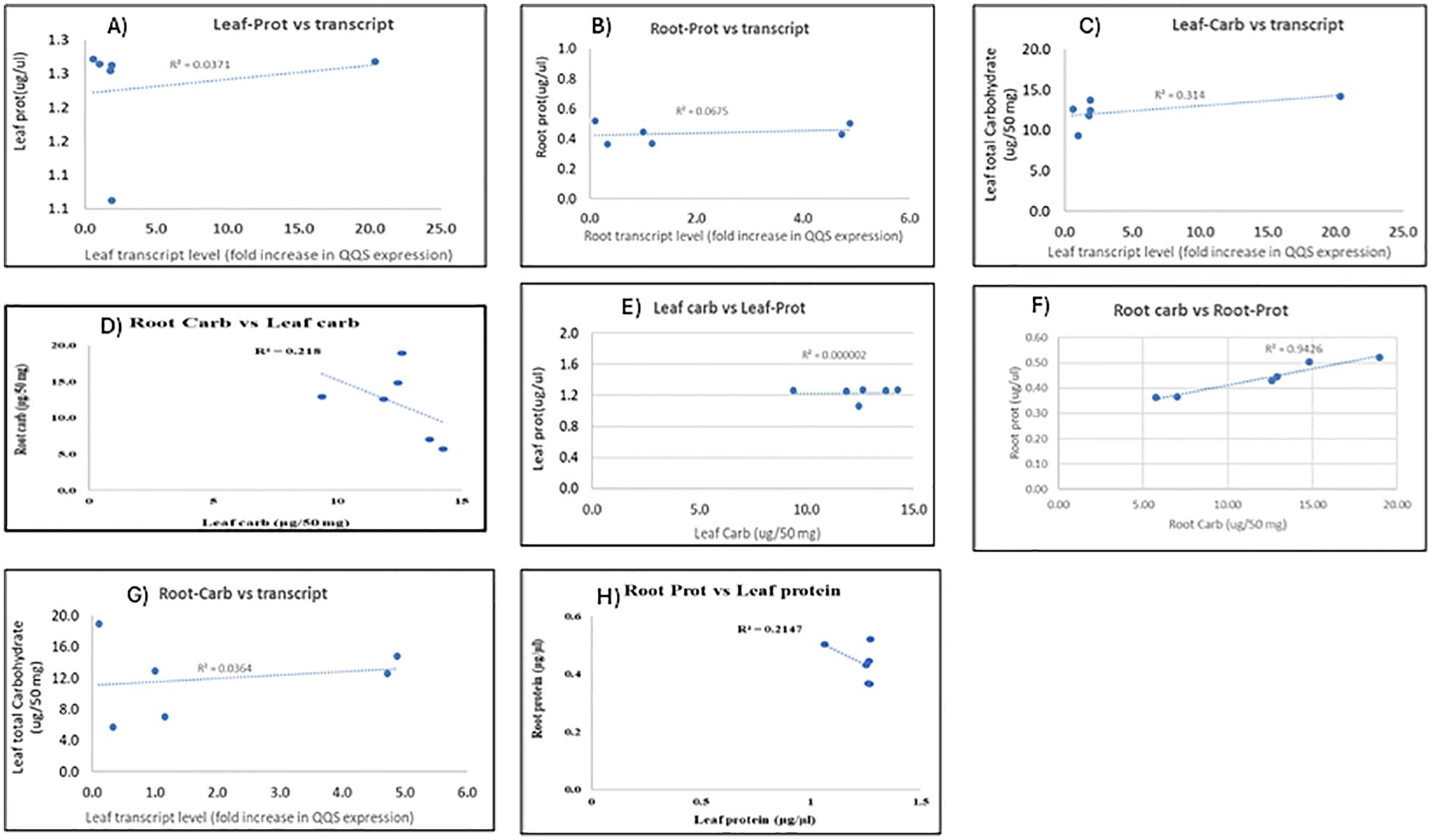
Figure 10. The correlation analysis between transcript expression level and carbohydrate or protein content, between carbohydrate and protein content of various plant parts (roots and leaves) of stably transformed cassava lines. The correlations between leaf; root protein content, and transcript level are depicted in Panels (A–C, G) show the correlations between leaf, root carbohydrate content, and transcript level. Panels (D–F, H) showed correlations between root and leaf carbohydrate content, leaf carbohydrate and leaf protein content, root carbohydrate and root protein content, and root and leaf protein content, respectively.
Using the STRING database, AtQQS was used as a bait gene to establish gene interaction. Figure 11 shows the interaction between the query gene with 59 interacting partners, generating 184 interactions or edges. The gene network consisted of several functional categories related to the regulation of the starch metabolic process and the regulation of the auxin biosynthetic process analyzed by ClueGO/CluePedia (Figure 12). Detailed information on the genes and biological processes are listed in (Supplementary Table S5).
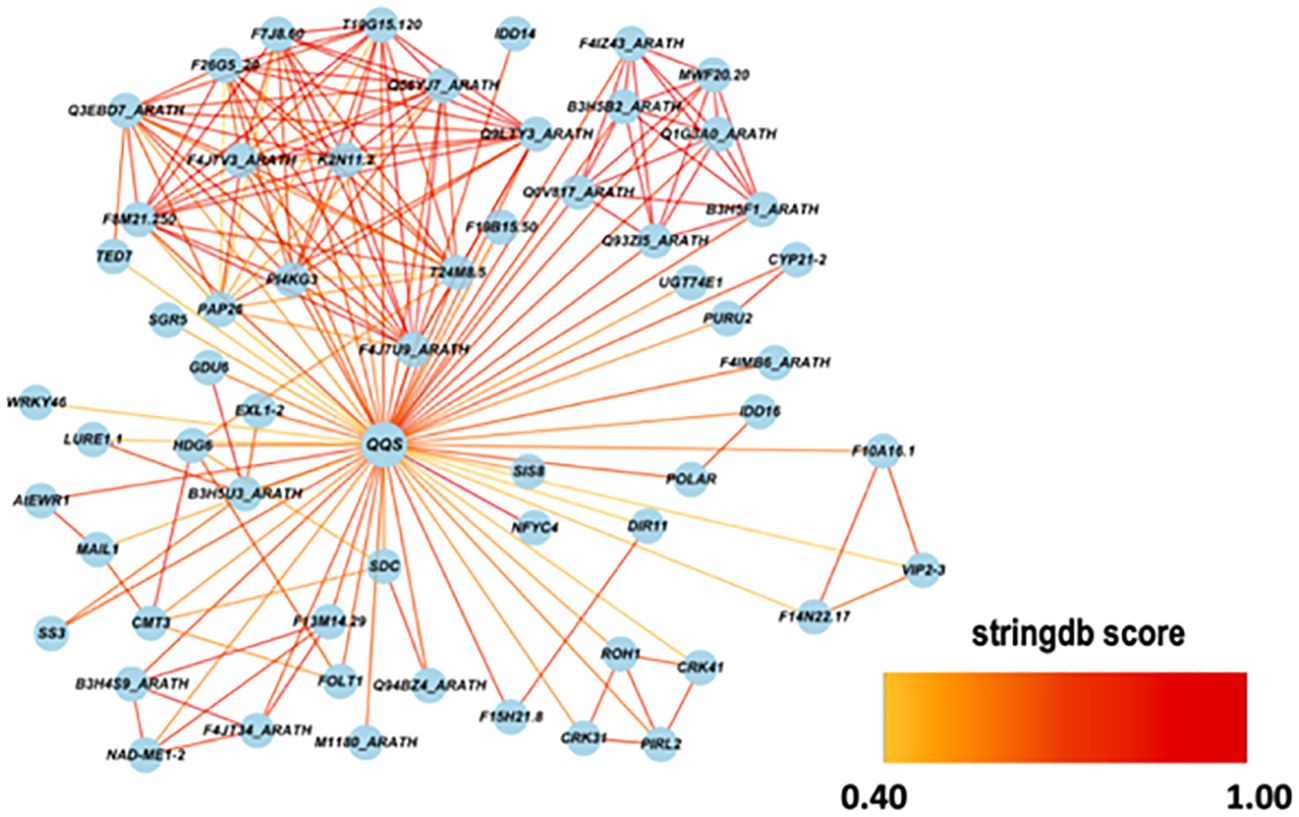
Figure 11. A gene network between AtQQS with 59 related genes and 184 interactions were obtained from the STRING database. The selected string score is 0.40 and above.
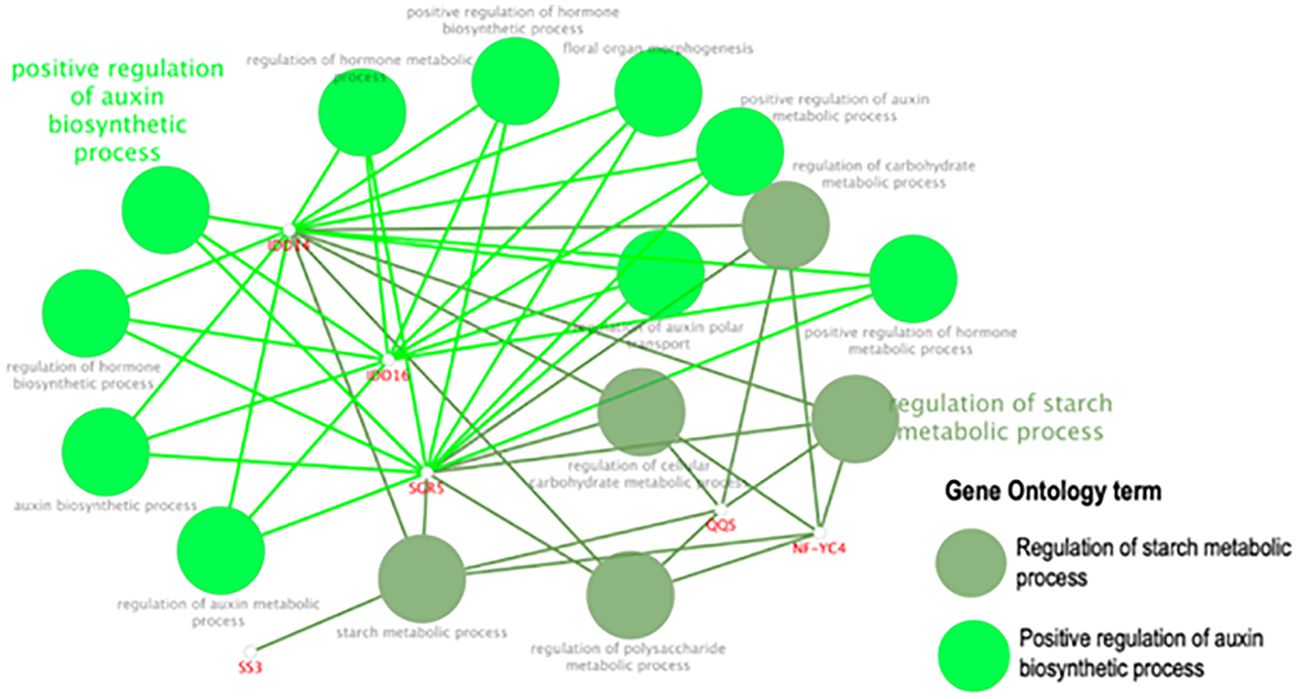
Figure 12. The GO enrichment using ClueGO/CluePedia apps from Cytoscape. The related genes are highlighted Supplementary Table S7 according to the biological processes involved.
SAS Enterprise Guide 5.1 was used for statistical analysis. Lines were used as the independent variable, and protein or carbohydrate content in leaf, root, and stem were used as the dependent variables, with a significant level of 5%.
In this study, we assembled the expression cassette of optimized QQS sequence into the plant transformation binary vector pPZP-RCS2-ocs-nptII (Goderis et al., 2002), one of the modular plant transformation vectors that allow flexible insertion of multiple expression units was achieved through the combination of ligation-free (Gibson et al., 2009) and ligation-dependent cloning system. Out of the remaining eight stably transgenic lines, only 5 were selected for molecular and biochemical analysis because root, leaf, and stem biomasses were available. Morphological characterization of the established and soil-grown transgenic cassava lines identified four independent lines (R7(D), R7 (B), R’’’ (LA)L1, R7 (E) out of the eight successfully regenerated transgenic lines (50%) presented some form of morphological abnormalities, but R7 (B), R7 (E) showed a pronounced abnormality characterized with tiny and fasciated leaves, very tiny stem, and an overall slow growth rate. Because of these physiological abnormalities, lines R7 (B) and R7 (E) were not used for the downstream biochemical analysis because of their inability to produce enough biomass from all plant parts. The low growth rate can be related to low photosynthetic capability due to the small leaf surface area of the malformed plants. The cassava target tissues, friable embryogenic callus (FEC) used for inserting QQS optimized sequence into cassava genome, were 24 months old and were established and maintained from organized embryogenic tissues (Taylor et al., 2001; Hankoua et al., 2006; Taylor N. et al., 2012). The observed growth abnormality of these malformed lines could be due to the age of the FEC used in this study, which was about twenty-four months old. This observation is consistent with Taylor N. et al. (2012), who suggested that transformation of FEC at an optimum stage (about nine weeks after induction from organized embryogenic tissue) and timely subculturing of transgenic tissues could reduce the formation of abnormal plants. These abnormalities could also be due to the adverse physiological and morphological effects of the recombinant QQS protein in transgenic cassava plants. The observed growth defect could be genetic or epigenetic variation (Machczyńska et al., 2015). The growth variability may also be due to the high-level expression of the optimized QQS gene, but this is contrary to the observation that the expression of QQS in soybean (Li L. et al., 2015) has no impact on the morphology of the field-grown transgenic soybean. However, 69.2% of the regenerated line presented enhanced morphological characteristics compared to their wild-type sibling plants with large leaves, very stick stems, high biomass production, and an early mini-tuber induction, particularly for line R7 (F) at 6-7 weeks after its in vitro plantlets were transferred in soil. In contrast to the observed outcomes in Arabidopsis thaliana and soybean, where the introduction of QQS expression resulted in regenerated lines that exhibited no discernible differences in morphology compared to their wild-type counterparts throughout their growth stages (Li et al., 2009; Li L. et al., 2015), the case of QQS expression in cassava plants, exemplified by line R7 (F), showcases a notable departure. Here, there is a notable enhancement in the physiological attributes of the plant, leading to an overall increase in plant vigor, a novel characteristic that may be attributed to the expression of QQS in cassava.
Data from the gene network analysis between AtQQS and its interacting partners generated by the STRING database and the GO enrichment using ClueGO/CluePedia showed that the gene network revealed several functional categories with AtQQS association to some transcription factors such as Zinc finger protein SHOOT GRAVITROPISM 5 (SGR5), a member of the SGR gene family known as regulating starch metabolism by supporting cellular gravitropism, a physiological process known to be very crucial for plant growth and crop productivity (Cho et al., 2024), and the protein indeterminate-domain 16 (IDD-16), a transcription factor regulating lateral organ morphogenesis and gravitropic responses as well as involved in the establishment of auxin gradients through the regulation of auxin biosynthesis and transport (Cui et al., 2013). There is a possibility that the expression of QQS in putative-transformed cassava line R7 (F) might positively regulate the expression of SGR5 and IDD-16 or their orthologues in cassava, enabling novel QQS expressing cassava lines to display excellent plant vigor. However, further detailed genomic studies such as global transcriptome on transgenic phenotype such as line R7 (F) will provide molecular and physiological cues on the nature of these genetic relationships, among others, between QQS, SGR5, and IDD-16. While biomass traits such as plant height, root, stem, and leaf biomass production are quantitative traits controlled by many genes, studies aimed at uncovering the molecular mechanism behind such traits discovered many regulators, such as transcription factors, which positively or negatively regulate the expression of these quantitative traits. Identifying such regulators (negative or positive) opened many avenues for targeting heterologous expressions or knockouts aimed to enhance the expression of quantitative traits. Studies recently showed that the overexpression of a single Dof transcription factor COG1 in Arabidopsis is a critical regulator of plant biomass by promoting photosynthesis and starch accumulation (Wei et al., 2023).
Initially identified as an orphan gene in Arabidopsis, QQS was initially characterized as a transcription factor that modulates metabolite allocation in soybean seeds (Li L. et al., 2015). The transgenic introduction of this transcription factor into the cassava genome, a crop known for its high starch production compared to the high protein content of soybean, may reveal additional interactions of QQS with other transcription factors, such as microRNA, known to play crucial roles in plant development, potentially resulting in enhanced biomass production (Zhang et al., 2015; Raza et al., 2023). Insertion of QQS and NPTII genes in the cassava genome was confirmed through gene-specific PCR using genomic DNA isolated from various tissues of different independent putative transgenic lines at an early stage of regeneration (3 months after transformation). Real-time PCR results revealed differentially expressed QQS between different lines first, within and between leaves and roots of these lines. The variability in the transcript level of a QQS sequence may be due to the combination of various critical genetic factors shown intensively to affect transcript levels, such as tissue type, promoter sequence, enhancer and codon usage, and transgene insertion location within the chromosome (Streatfield, 2007). We used a powerful constitutive promoter, the enhanced 35S promoter (Kay et al., 1987; Zhou et al., 2013), to drive QQS expression in all tissues of these putatively transformed lines and QQS expressing lines, but many studies have demonstrated that significant variability in the transcript levels of a transgene in various tissues is achievable even with a relatively less constitutive solid promoter and this could be due to transgenic position effect (Liu, 2013) and the physiological environment of the cell and tissue types where transgene is expressed. This position effect could explain why higher QQS expression was noticed in leaves, especially for the R’’’ (G) L2 line compared with other transgenic lines and the wild-type control. Furthermore, QQS expression in R’’’ (LA) L2 was low in leaves while very high in roots. In addition, QQS is highly expressed in R’’’ (G) L2 leaves while very low in the roots. Moreover, QQS was expressed at a low level in R7(F) both in leaves and roots, and the transgene position effect could account for these observed variabilities in QQS expression.
Biochemical analysis of stably QQS expressing lines revealed either an increase or a decrease in protein content depending on lines and a general increase in leaf carbohydrates. In contrast, a decrease in root carbohydrate was recorded for R’’’(G) L1 and R’’’ (G) L2 putative transgenic lines. In general, four tendencies have been identified in the biochemical results. For some cases where the QQS was expressed at a low level in leaves (R7 (F)), protein content in leaves decreased compared to the wild-type non-transgenic, while soluble total carbohydrate content was very high. This tendency of plants expressing QQS to decrease protein content while increasing carbohydrate production when expressed at a deficient level was also found in some transgenic Arabidopsis thaliana and soybean lines expressing QQS (Li et al., 2009). In contrast, a high expression of QQS in the leaves of the line (R(G) L2) reveals no variation in the protein content compared to the wild type, while carbohydrate accumulation in this line (R(G) L2 increased considerably. Li et al. (2009) demonstrated that a high expression level of QQS in transgenic soybean led to an increased protein accumulation while carbohydrate accumulation was decreased. This observation does not align with the observed effect of high expression of QQS in cassava leaves of line (R (G) L2), possibly pointing to another molecular mechanism of this transcription factor in plant development in a related soybean plant, cassava. Moreover, in cassava roots, a shallow expression of QQS in line R7 (F) led to the modulation of protein and soluble total carbohydrates, increasing their content. In contrast, a very high QQS expression in stably transformed cassava line (R (LA) L2) revealed a high accumulation of protein and soluble total carbohydrates. However, a high expression of QQS in R’’’(G) L3 roots revealed no noticeable variation in protein and soluble total carbohydrate accumulation. No correlations were observed between the QQS transcript levels, carbohydrate, and protein content for leaf and root biomasses of all the transgenic lines. This lack of relationship demonstrates that the level of QQS transcripts in a specific plant part of an independent and stable transgenic line does not influence the ability of QQS protein to exert the ability to partition the allocation of metabolites during plant development. This phenomenon was also observed in soybean lines expressing QQS (Li L. et al., 2015). It was also demonstrated that the effect of transcription factors such as QQS on the starch and protein accumulation in lines from the same transformation event may vary as the expression level of the transgene in different lines of the same transformation event may be different (Shou et al., 2004). QQS is a regulatory protein, and it is not unusual for regulatory proteins to have shallow expression levels (Nagaraj et al., 2011). Thus, this could be explained by the fact that only a minimal concentration of QQS saturates the QQS receptor, and any increases over this specific concentration or level do not affect their ability to impact protein and starch contents in a specific QQS expressing lines through the ability of this transgenic QQS protein to exert metabolic partitioning. No correlation was found between the content of leaf carbohydrate and leaf protein, leaf carbohydrate and root protein, leaf carbohydrate and root carbohydrate, but root carbohydrate and root protein content were found to be highly correlated (R²=O.9). It will be interesting to see if this ability to accumulate starch and protein in young roots is translated to mature tuberous root which is known to accumulate high starch and low protein (Chavarriaga-Aguirre et al., 2016). The lack of a linear relationship between the content of carbohydrates and protein from leaf, root, and stem biomass of transgenic lines versus QQS transcript was also observed in QQS transcript accumulation in Arabidopsis and soybean (Li L. et al., 2015). However, in the case of Arabidopsis and soybean, the elevation of QQS RNA accumulation was related to an increase in protein accumulation and a decrease in starch accumulation (Li L. et al., 2015), therefore validating one of the unveiled molecular mechanisms of QQS (Li et al., 2009). For cassava, line R”‘ (G) L1 and line R”‘(G) L2 leaf protein did not increase significantly compared to the control plants but have significantly reduced carbohydrate content, and this could be attributed to the QQS expression and modulation effect in metabolites distribution as demonstrated in Arabidopsis and soybean QQS expressers. Transgenic cassava line R7(F) produced the lowest level of QQS transcript in both root and leave tissues but produced the highest soluble protein increase in the root tissue compared to all the other root and leaf samples for all the lines tested. This phenomenon was also observed in some soybean QQS expressers. This phenomenon is not unusual for regulatory proteins to correlate with shallow expression levels (Nagaraj et al., 2011). Therefore, only a minimal concentration of QQS saturates the QQS receptor (Li et al., 2009), and any increases over this concentration do not affect metabolite content. There might be other possible explanations for a lack of strong linear correlation between levels of QQS transcript and metabolite composition, such as QQS translational efficiency or stability or the effectiveness of the QQS protein to biochemically express its function or post-translational modification is limiting. Indeed, various post-translational regulatory mechanisms can come into play, as have been described for other transgenes (Lillo et al., 2004).
The expression of the QQS gene into cassava plants increased leaf protein until 1.36% in line R (LA) L2 and root protein until 17.02% for the same line. Moreover, leaf-soluble total carbohydrate increased to 51.76% in line R’’’(G) L2, and root-soluble total carbohydrate increased to 46.75% in line R(F). For line R7 (F), a low expression in leaves caused a decrease in leaf protein content by 9% while root protein, leaf, and root carbohydrate increased respectively by 13.03%, 34.29%, and 46.75%. A strong correlation (0.9) was found between root protein and total root carbohydrate accumulation. No correlation was found between the level of expression and chemical compound accumulation. Morphological observation has revealed a positive impact of QQS expression in cassava, giving a more vigorous plant with larger leaves, a stick stem, and enhanced biomass production. Our findings will be interesting for farmers and agriculture if the biochemical profiling results are confirmed and recapitulated in tuberous roots, stems, and leaves of mature transgenic cassava when grown in the field. Therefore, the novel cassava developed in this project will contribute to ensuring a well-balanced protein diet for the 800 million people in Asia, Africa, and Latin America who rely on the starch crop cassava as their staple food. The capability of QQS to increase the soluble total carbohydrate content and, consequently, starch accumulation, if validated in these transgenic mature plants, can be explored by starch and bioethanol producers. QQS has also been proven to decrease plant oil accumulation. To validate the molecular mechanism of QQS in transgenic cassava plants, the two-yeast hybrid system could be used in further experimentation to see if QQS will bind to the conserved transcription factor, nuclear factor Y, subunit C4 (NF-YC4), which is a ubiquitous factor modulating plant development. In terms of safety regarding genetically modified food (GMO), to this day, no concrete identification of toxicity was found in GMOs. Nevertheless, investigations should be done on transgenic QQS plants to ensure the safety of crops. It will be interesting to study how QQS impacts carbon reparation in plants and, therefore, causes variations in plant chemical composition and vigor. Western blot can be done for protein detection, and Southern blot can be used to check the integration of QQS into the cassava genome and evaluate transcript copy number. In addition, the model cassava cultivar (cv 60444), which is not a preferred cultivar for farmers in Africa (Chavarriaga-Aguirre et al., 2016) was used in this study; if the results observed are confirmed on tissues from matured plants, the new traits should be transferred to the consumers and farmers preferred cultivar such as local African cassava landrace variety TME 204 used for nutritional trait enhancement through biotechnology (Narayanan et al., 2019). Finally, a more robust biotechnological approach could be designed to simultaneously combine the expression of the QQS and AmA1 (Amaranth Albumin) gene to drive a significant increase in the total protein content in transgenic cassava tubers. Studies demonstrated that tuber-specific expression of AmA1 (Amaranth Albumin 1), a seed storage protein in transgenic potato tubers, revealed up to 60% increase in total protein content (Chakraborty et al., 2010). Furthermore, a comparative protein profiling of this transgenic potato stably expressing the AmA1 gene unveils proteome rebalancing as a molecular mechanism driving increased protein content in these transgenic potato tubers (Chakraborty et al., 2010). In vitro and In vivo studies on experimental animals should also be attempted to demonstrate that QQS transgenic proteins in transgenic cassava tubers are safe for human consumption.
The original contributions presented in the study are publicly available. This data can be found here: NCBI, PQ790632, PQ790633 and PQ790634.
BH: Writing – review & editing, Writing – original draft, Supervision, Resources, Methodology, Funding acquisition, Conceptualization. MD: Writing – review & editing, Validation, Methodology, Investigation, Conceptualization. AL-O: Validation, Methodology, Writing – review & editing. RG: Writing – review & editing, Validation, Methodology. SH: Writing – review & editing, Validation, Methodology. YA: Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. Cassava-related research was sponsored by USDA NIFA CBG Grants (# 2014-38821-22417, 2008-35504-04557) and Evans-Allen Grant (#6011-220017-21) to BH. USDA NIFA CBG Grant # 2024-38821-42144 provides funds to cover the publication cost of this manuscript. MD’s Graduate research thesis was supported by the USDA NIFA Evans-Allen Grant (#6011-220017-21).
We thank Dr. Alberta Aryee for their technical assistance in preparing the cassava tissue samples to analyze amino acid composition. We also thank members of the USDA-ARS Lab, including Dr. Garry Richards, for allowing MD to use their lab facilities as needed to complete some aspects of this research.
Author MD was employed by Reckitt Benckiser Inc.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1442324/full#supplementary-material
Amelework, A. B., Bairu, M. W. (2022). Advances in genetic analysis and breeding of cassava (Manihot esculenta crantz): A review. Plants 11, 1617. doi: 10.3390/plants11121617
Bindea, G., Mlecnik, B., Hackl, H., Charoentong, P., Tosolini, M., Kirilovsky, A., et al. (2009). ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25, 1091–1093. doi: 10.1093/bioinformatics/btp101
Chakraborty, S., Chakraborty, N., Agrawal, L., Ghosh, S., Narula, K., Shekhar, S., et al. (2010). Next-generation protein-rich potato expressing the seed protein gene AmA1 results from proteome rebalancing in the transgenic tuber. Proc. Natl. Acad. Sci. United States America 107, 17533–17538. doi: 10.1073/pnas.1006265107
Chavarriaga-Aguirre, P., Brand, A., Medina, A., Prías, M., Escobar, R., Martinez, J., et al. (2016). The potential of using biotechnology to improve cassava: a review. In Vitro Cell. Dev. Biology-Plant 52, 461–478. doi: 10.1007/s11627-016-9776-3
Cho, Y., Kim, Y., Lee, H., Sundong, K., Kang, J., Kadam, U. S., et al. (2024). Cellular and physiological functions of SGR family in gravitropic response in higher plants. J. Adv. Res. 1:S2090-1232(24)00039-0. doi: 10.1016/j.jare.2024.01.026
Cohen, S. A., De Antonis, K. M. (1994). Applications of amino acid derivatization with 6-aminoquinolyl-n-hydroxysuccinimidyl carbamate. Analysis of feed grains, intravenous solutions, and glycoproteins. J. Chromatogr. A 661, 25–34. doi: 10.1016/0021-9673(93)E0821-B
Cui, D., Zhao, J., Jing, Y., Fan, M., Liu, J., Wang, Z., et al. (2013). The arabidopsis IDD14, IDD15, and IDD16 cooperatively regulate lateral organ morphogenesis and gravitropism by promoting auxin biosynthesis and transport. PloS Genet. 9, e1003759. doi: 10.1371/journal.pgen.1003759
Dafny-Yelin, M., Tzfira, T. (2007). Delivery of multiple transgenes to plant cells. Plant Physiol. 145, 1118–1128. doi: 10.1104/pp.107.106104
Dai, Z., Hooker, B. S., Daniel, B., Anderson, D. B., Thomas, S. R. (2000). Improved plant-based production of E1 endoglucanase using potato: expression optimization and tissue targeting. Mol. Breed. 6, 277–285. doi: 10.1023/A:1009653011948
Doncheva, N. T., Morris, J. H., Gorodkin, J., Jensen, L. J. (2019). Cytoscape stringApp: network analysis and visualization of proteomics data. J. Proteome Res. 18, 623–632. doi: 10.1021/acs.jproteome.8b00702
Failla, M. L., Chitchumroonchokchai, C., Siritunga, D., De Moura, F. F., Fregene, M., Manary, M. J., et al. (2012). Retention during processing and bioaccessibility of β-carotene in high β-carotene transgenic cassava root. J. Agric. Food Chem. 60, 3861–3866. doi: 10.1021/jf204958w
FAO. (2012). Food Outlook Global Market. Global Information and Early Warning System (GIEWS), FAO, Rome: FAO, page 129.
Food and Agriculture Organization [FAO] (2002). “Vitamin A,” in FAO/WHO Expert Consultation on Human Vitamin and Mineral Requirements Chapter 7 (World Health Organization/Food and Agriculture Organization of the United Nations, Rome), 87–107.
Fraval, S., Hammond, J., Bogard, J. R., Van Etten, J., Herrero, M., Oosting, S. J., et al. (2019). Food access deficiencies in sub-saharan africa: prevalence and implications for agricultural interventions. Front. Sustain. Food Syst. 3. doi: 10.3389/fsufs.2019.00104
Gegios, A., Amthor, R., Maziya-Dixon, B., Egesi, C., Mallowa, S., Nungo, R., et al. (2010). Children consuming cassava as a staple food are at risk for inadequate zinc, iron, and vitamin A intake. Plant Foods Hum. Nutr. 65, 64–70. doi: 10.1007/s11130-010-0157-5
Gibson, D. G., Young, L., Chuang, R. Y., Venter, C., Hutchison, C. A., Smith, H. O. (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345. doi: 10.1038/nmeth.1318
Goderis, I. J. W. M., Miguel, F. C., Bolle, D., Francois, I. E. J. A., Wouters, P. F. J. W., Broekaert, W. F., et al. (2002). A set of modular plant transformation vectors allowing flexible insertion of up to six expression units. Plant Mol. Biol. 50, 17–27. doi: 10.1023/A:1016052416053
Graham, R. D., Welch, R. M. (1996). Breeding for staple food crops with high micronutrient density. Agricultural strategies for micronutrients 3. Washington, DC: International Food Policy Research Institute. Available online at: https://hdl.handle.net/10568/157196.
Gresshoff, P. M., Doy, C. H. (1972). Development and differentiation of haploid lycopersicum esculentum (Tomato). Planta 107, 161–170. doi: 10.1007/BF00387721
Hankoua, B., Mbanaso, A., Banda, R., Fauquet, C., Taylor, N. (2007). Enhancement of friable embryogenic callus production and plant recovery by tyrosine in a range of agronomically important West African cassava (Manihot esculenta Crantz) landraces. In Vitro Cell. Dev. Biol. - Animal. 43, S48–S49. doi: 10.1007/s11626-007-9028-2
Hankoua, B. B., Ng, S. Y. C., Fawole, I., Puonti-Kaerlas, J., Pillay, M., Dixon, A. G. O. (2005). Regeneration of a wide range of African cassava genotypes via shoot organogenesis from cotyledons of maturing somatic embryos and conformity of the field-established regenerants. Plant Cell Tiss Organ Cult. 82, 221–231. doi: 10.1007/s11240-005-0514-5
Hankoua, B. B., Taylor, N. J., Ng, S. Y. C., Fawole, I., Puonti-Kaerlas, J., Chellappan, P., et al. (2006). Production of the first transgenic cassava (Manihot esculenta Crantz) plants in Africa via direct shoot organogenesis of friable embryoids and germination of maturing somatic embryos. Afr. J. Biotechnol. 5, 1700–1712. doi: 10.5897/AJB06.248
Harika, R., Faber, M., Samuel, F., Mulugeta, A., Kimiywe, J., Eilander, A. (2017). Are low intakes and deficiencies in iron, vitamin A, zinc, and iodine of public health concern in Ethiopian, Kenyan, Nigerian, and South African children and adolescents? Food Nutr. Bull. 38, 405–427. doi: 10.1177/0379572117715818
Hillocks, R. J., Thresh, J. M., Bellotti, A. C. (Eds.) (2002). Cassava Biology, Production and Utilization (CABI). Available online at: https://www.cabidigitallibrary.org/.
Howeler, R. H., Lutaladio, N. B., Thomas, G. (2013). Save and grow cassava: A guide to sustainable production and identification. (Rome: Food and Agriculture Organization of the United Nations (FAO)), 142.
Ihemere, U. E., Narayanan, N. N., Sayre, R. T. (2012). Iron biofortification and homeostasis in transgenic cassava roots expressing the algal iron assimilatory gene, FEA1. Front. Plant Sci. 3, 171. doi: 10.3389/fpls.2012.00171
Ihemere, U., Arias-Garzon, D., Lawrence, S., Sayre, R. (2006). Genetic modification of cassava for enhanced starch production. Plant Biotechnology Journal 4, 453–465. doi: 10.1111/j.1467-7652.2006.00195.x
Inacio, K. A. M., Farfan, N. C., Carlos Eduardo, X. A., Gomes Polatto, M. A., Carrion, N. S., Santos Mendes, P. V., et al. (2024). Potential of cassava clones for iron, zinc, and selenium biofortification. Agriculture 14, 268. doi: 10.3390/agriculture14020268
Jennings, D. L., Iglesias, C. (2002). Breeding for crop improvement. In Cassava: Biology. Eds. Hillocks, R., Thresh, J. M., Bellotti, A. C. (CAB International), 149–166. Available online at: https://www.cabidigitallibrary.org/.
Kay, R., Chan, A., Daly, M., McPherson, J. (1987). Duplication of CaMV 35S promoter sequences creates a strong enhancer for plant genes. Science 236, 1299–1302. doi: 10.1126/science.236.4806.1299
Ladino, J. J., Echeverry, M., Mancilla, L. I., Lopez, D., Chavarriaga, P., Tohme, J., et al. (2002). Genetic transformation of cassava: confirmation of transgenesis in clone 60444 and analysis of CRY1Ab protein in transgenic lines. Preliminary data on the transformation of farmer-preferred cultivars SM1219-9 and CM3306-4 (Cali, Colombia: CIAT).
Li, K. T., Moulin, M., Mangel, N., Albersen, M., Verhoeven-Duif, N. M., Ma, Q., et al. (2015). Increased bioavailable vitamin B6 in field-grown transgenic cassava for dietary sufficiency. Nat. Biotechnol. 33, 1029–1032. doi: 10.1038/nbt.3318
Li, L., Foster, C. M., Gan, Q., Nettleton, D., James, M. G., Myers, A. M., et al. (2009). Identification of the novel protein QQS as a component of the starch metabolic network in Arabidopsis leaves. Plant J. 58, 485–498. doi: 10.1111/j.1365-313X.2009.03793.x
Li, L., Zheng, W., Zhu, Y., Ye, H., Tang, B., Arendsee, Z. W., et al. (2015). QQS orphan gene regulates carbon and nitrogen partitioning across species via NF-YC interactions. Proc. Natl. Acad. Sci. 112, 14734–14739. doi: 10.1073/pnas.1514670112
Ligaba-Osena, A., Jones, J., Donkor, E., Chandrayan, S., Pole, F., Wu, C.-H., et al. (2018). Novel bioengineered cassava expressing an archaeal starch degradation system and a bacterial ADP-glucose pyrophosphorylase for starch self-digestibility and yield increase. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00192
Lillo, C., Meyer, C., Lea, U. S., Provan, F., Oltedal, S. (2004). Mechanism and importance of post-translational regulation of nitrate reductase. J. Exp. Bot. 55, 1275–1282. doi: 10.1093/jxb/erh132
Lin, Z. J. D., Taylor, N. J., Bart, R. (2019). Engineering disease-resistant cassava. Cold Spring Harb. Perspect. Biol. 11. doi: 10.1101/cshperspect.a034595
Liu, C. (2013). Strategies for designing transgenic DNA constructs. Methods Mol. Biol. 1027, 183–201. doi: 10.1007/978-1-)60327-369-5_8
Machczyńska, J., Zimny, J., Bednarek, P. T. (2015). Tissue culture-induced genetic and epigenetic variation in triticale (× Triticosecale spp. Wittmack ex A. Camus 1927) regenerants. Plant Mol. Biol. 89, 279–292. doi: 10.1007/s11103-015-0368-0
Malik, A. I., Kongsil, P., Nguyễn, V. A., Ou, W., Sholihin, Srean, P., et al. (2020). Cassava breeding and agronomy in Asia: 50 years of history and future directions. Breed Sci. 70, 145–166. doi: 10.1270/jsbbs.18180
Murashige, T., Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 15, 473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x
Nagaraj, N., Wisniewski, J. R., Geiger, T., Cox, J., Kircher, M., Kelso, J., et al. (2011). Deep proteome and transcriptome mapping of a human cancer cell line. Mol. Syst. Biol. 7, 548. doi: 10.1038/msb.2011.81
Narayanan, N., Beyene, G., Chauhan, R. D., Gaitán-Solís, E., Gehan, J., Butts, P., et al. (2019). Biofortification of field-grown cassava by engineering expression of an iron transporter and ferritin. Nat. Biotechnol. 37, 144–151. doi: 10.1038/s41587-018-0002-1
Narayanan, N., Beyene, G., Chauhan, R. D., Gaitán-Solis, E., Grusak, M. A., Taylor, N., et al. (2015). Overexpression of Arabidopsis VIT1 increases the accumulation of iron in cassava roots and stems. Plant Sci. 240, 170–181. doi: 10.1016/j.plantsci.2015.09.007
Narayanan, N., Beyene, G., Chauhan, R. D., Grusak, M. A., Taylor, N. J. (2021). Stacking disease resistance and mineral biofortification in cassava varieties to enhance yields and consumer health. Plant Biotechnol. J. 19, 844–854. doi: 10.1111/pbi.13511
Narayanan, N. N., Ihemere, U., Ellery, C., Sayre, R. T. (2011). Overexpression of hydroxynitrile lyase in cassava roots elevates protein and free amino acids while reducing residual cyanogen levels. PloS One 6, e21996. doi: 10.1371/journal.pone.0021996
Nassar, N. M., Sousa, M. V. (2007). Amino acid profile in cassava and its interspecific hybrid. Genet. Mol. Res. 6, 292–297.
Nyaboga, E., Njiru, J., Nguu, E., Gruissem, W., Vanderschuren, H., Tripathi, L. (2013). Unlocking the potential of tropical root crop biotechnology in East Africa by establishing a genetic transformation platform for local farmer-preferred cassava cultivars. Front. Plant Sci. 4. doi: 10.3389/fpls.2013.00526
Omole, T. A. (1977). Cassava in the nutrition of layers: Cassava as Animal Feed Workshop, 18-20 Apr. 1977, Guelph, Ont., CA. Available online at: http://hdl.handle.net/10625/20145.
Prochnik, S., Marri, P. R., Desany, B., Rabinowicz, P. D., Kodira, C., Mohiuddin, M., et al. (2012). The cassava genome: current progress, future directions. Trop. Plant Biol. 5, 88–94. doi: 10.1007/s12042-011-9088-z
Qi, M., Zheng, W., Zhao, X., Hohenstein, J. D., Kandel, Y., Wang, Y., et al. (2019). QQS orphan gene and its interactor NF-YC4 reduce susceptibility to pathogens and pests. Plant Biotechnol. J. 17, 252–263. doi: 10.1111/pbi.12961
Raza, A., Charagh, S., Karikari, B., Sharif, R., Yadav, V., Mubarik, M. S., et al. (2023). miRNAs for crop improvement. Plant Physiol. Biochem. 201, 107857. doi: 10.1016/j.plaphy.2023.107857
Sarria, R., Torres, E., Angel, F., Chavarriaga, P., Roca, W. M. (2000). Transgenic plants of cassava (Manihot esculenta) with resistance to Basta obtained by Agrobacterium-mediated transformation. Plant Cell Rep. 19, 339–344. doi: 10.1007/s002990050737
Sayre, R., Beeching, J. R., Cahoon, E. B., Egesi, C., Fauquet, C., Fellman, J., et al. (2011). The BioCassava plus program: Biofortification of cassava for sub-Saharan Africa. Annu. Rev. Plant Biol. 62, 251–272. doi: 10.1146/annurev-arplant-042110-103751
Shou, H., Frame, B. R., Whitham, S. A., Wang, K. (2004). Assessment of transgenic maize events produced by particle bombardment or Agrobacterium-mediated transformation. Molecular Breeding 13, 201–208. doi: 10.1023/B:MOLB.0000018767.64586.53
Siritunga, D., Sayre, R. T. (2003). Engineering cyanogen synthesis and turnover in cassava (Manihot esculenta). Plant Mol. Biol. 53, 891–903. doi: 10.1007/s00425-003-1005-8
Siritunga, D., Sayre, R. T. (2004). Transgenic approaches for cyanogen reduction in cassava. World J. Microbiol. Biotechnol. 20, 769–780. doi: 10.1007/s11103-004-3415-9
Streatfield, S. J. (2007). Approaches to achieve high-level heterologous protein production in plants. Plant Biotechnol. J. 5, 2–15. doi: 10.1111/j.1467-7652.2006.00216.x
Szklarczyk, D., Kirsch, R., Koutrouli, M., Nastou, K., Mehryary, F., Hachilif, R., et al. (2023). The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 51, D638–D646.
Taylor, N., Gaitán-Solís, E., Moll, T., Trauterman, B., Jones, T., Pranjal, A., et al. (2012). A high-throughput platform for the production and analysis of transgenic cassava (Manihot esculenta) plants. Trop. Plant Biol. 5, 127–139. doi: 10.1007/s12042-012-9099-4
Taylor, N., Masona, M., Carcamo, R., Ho, T., Schöpke, C., Fauquet, C. M. (2001). Production of embryogenic tissues and regeneration of transgenic plants in cassava (Manihot esculenta Crantz). Euphytica 120, 25–34. doi: 10.1023/A:1017526814789
Tzfira, T., Tian, G. W., Croix, B., Vyas, S., Li, J., Leitner-Dagan, Y., et al. (2005). pSAT vectors: a modular series of plasmids for autofluorescent protein tagging and expression of multiple genes in plants. Plant Mol. Biol. 57, 503–516. doi: 10.1007/s11103-005-0340-5
Wei, Z., Zhang, H., Fang, M., Lin, S., Zhu, M., Li, Y., et al. (2023). The Dof transcription factor COG1 acts as a key regulator of plant biomass by promoting photosynthesis and starch accumulation. Mol. Plant 16, 1759–1772. doi: 10.1016/j.molp.2023.09.011
Welsch, R., Arango, J., Bär, C., Salazar, B., Al-Babili, S., Beltrán, J., et al. (2010). Provitamin A accumulation in cassava (Manihot esculenta Crantz) roots driven by a single nucleotide polymorphism in a phytoene synthase gene. Plant Cell 22, 3348–3356. doi: 10.1105/tpc.110.077560
Zeevi, V., Liang, Z., Arieli, U., Tzfira, T. (2012). Zinc finger nuclease and homing endonuclease-mediated assembly of multigene plant transformation vectors. Plant Physiol. 158, 132–144. doi: 10.1104/pp.111.184374
Zhang, P., Jaynes, J. M., Potrykus, I., Gruissem, W., Puonti-Kaerlas, J. (2003). Transfer and expression of an artificial storage protein (ASP1) gene in cassava (Manihot esculenta crantz). Transgenic Res. 12, 243–250. doi: 10.1023/A:1022918925882
Zhang, J., Li, S., Li, L., Li, M., Guo, C., Yao, J., et al. (2015). Exosome and exosomal microRNA: trafficking, sorting, and function. Genomics Proteomics Bioinf. 13, 17–24. doi: 10.1016/j.gpb.2015.02.001
Zhou, J., Yang, Y., Wang, X., Yu, F., Yu, C., Chen, J., et al. (2013). Enhanced transgene expression in rice following selection controlled by weak promoters. BMC Biotechnol. 13, 29. doi: 10.1186/1472-6750-13-29
Keywords: cassava, protein biofortification, protein deficiency, overexpression, QQS, starch improvement, food security, malnutrition
Citation: Hankoua BB, Diao M, Ligaba-Osena A, Garcia RA, Harun S and Ahlawat YK (2025) Constitutive overexpression of Qui-Quine Starch gene simultaneously improves starch and protein content in bioengineered cassava (Manihot esculenta Crantz). Front. Plant Sci. 15:1442324. doi: 10.3389/fpls.2024.1442324
Received: 01 June 2024; Accepted: 22 October 2024;
Published: 24 February 2025.
Edited by:
David W. M. Leung, University of Canterbury, New ZealandReviewed by:
Rambod Abiri, University of Guelph, CanadaCopyright © 2025 Hankoua, Diao, Ligaba-Osena, Garcia, Harun and Ahlawat. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bertrand Bachaumond Hankoua, YmhhbmtvdWFAZGVzdS5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.