- 1College of Life and Environmental Sciences, Hangzhou Normal University, Hangzhou, China
- 2Key Laboratory of Vegetable Biology, College of Landscape and Horticulture, Yunnan Agricultural University, Kunming, China
The epigenetic machinery has received extensive attention due to its involvement in plant growth, development, and adaptation to environmental changes. Recent studies often highlight the epigenetic regulatory network by discussing various epigenetic mutants across various plant species. However, a systemic understanding of essential epigenetic regulatory mechanisms remains limited due to a lack of representative mutants involved in multiple biological processes. Colorless Non-ripening (Cnr), a spontaneous epimutant isolated from a commercial population, was initially characterized for its role in fruit ripening regulation. Cnr fruits exhibit an immature phenotype with yellow skin, attributed to hypermethylation of the SQUAMOSA PROMOTER BINDING PROTEIN-LIKE-CNR (SlSPL-CNR) promoter, resulting in the repression of gene expression. In addition to DNA methylation, this process also involves histone modification and microRNA, integrating multiple epigenetic regulatory factors. Interestingly, knockout mutants of SlSPL-CNR display phenotypical distinctions from Cnr in fruit ripening, indicating complex genetic and epigenetic control over the non-ripening phenotype in Cnr fruits. Accumulating evidence suggests that Cnr epimutation is pleiotropic, participating in various biological processes such as Cd stress, Fe deficiency, vivipary, and cell death. Therefore, the Cnr epimutant serve as an excellent model for unveiling how epigenetic mechanisms are involved in diverse biological processes. This review paper focuses on recent research advances regarding the Cnr epimutant, delving into its complex genetic and epigenetic regulatory mechanisms, with the aim of enhancing our understanding and facilitating the development of high-quality, high-yield crops through epigenetic modification.
Introduction
Epigenetic modification refers to alterations in chromatin structure rather than changes in the DNA sequence, encompassing DNA methylation, histones modification, chromatin remodeling and siRNAs (small interfering RNAs). Increasing evidence highlights the vital role of epigenetic regulation in plant growth, development, and stress resilience, as these modifications coordinate with transcription factors to regulate gene expression. For example, the quintuple mutant mddcc, lacking DNA methyltransferases such as MET1 (METHYLTRANSFERASE1), DRM1 (DOMAINS REARRANGED METHYLTRANSFERASE1), DRM2, CMT2 (CHROMOMETHYLASE2), and CMT3, displays extreme growth defects including reduced size and failure to transition to the floral stage due to widespread changes in DNA methylation (He et al., 2022). In Arabidopsis, mutations in REF6 (RELATIVE OF EARLY FLOWERING6), encoding a histone H3 lysine 27 demethylase, lead to pleiotropic development phenotypes such as early flowering and upward curling leaves upon overexpression (Lu et al., 2011). Additionally, under salinity stress, the DNA methylation reader OsSUVH7 regulates OsHKT1;5 (HIGH-AFFINITY POTASSIUM TRANSPORTER1;5) gene expression by mediating DNA methylation in the upstream transcriptional factors (Wang J. et al., 2020). Clearly, plants rely on epigenetic modifications for normal growth and adaptation to environmental changes.
Epigenetic modifications, such as DNA methylation, histones modification, and siRNAs, are interconnected rather than independent. For instance, the establishment of DNA methylation through the RNA-directed DNA methylation (RdDM) pathway involves siRNAs, which are associated with SUVH2/9 (SUPPRESSOR OF VARIEGATION 3–9 HOMOLOG PROTEIN2/9)-mediated H3K9me2 and the SWITCH/SUCROSE NONFERMENTING (SWI/SNF) chromatin-remodeling complex (Zhang et al., 2018). Thus, DNA methylation emerges as a major epigenetic modification by coordinating with and influencing other epigenetic modifications. However, our current understanding of how DNA methylation regulates plant growth, development, and stress responses primarily stems from studies on various DNA methylation-related mutants across different plant species. Therefore, integrating knowledge of DNA methylation with other genetic and epigenetic components to orchestrate growth and stress responses remains an ongoing challenge.
The epimutant Cnr (Colorless Non-ripening) in tomato initially demonstrated DNA hypermethylation in the promoter of SlSPL-CNR (SQUAMOSA PROMOTER BINDING PROTEIN-LIKE-CNR), resulting in the characteristics non-ripening phenotype of fruits (Manning et al., 2006). Subsequent investigations identified that SlCMT3 is responsible for mediating DNA hypermethylation in the SlSPL-CNR promoter, along with microRNA SlymiR157 (Chen et al., 2015a, 2015b). Furthermore, the ripening-related transcription factor FRUITFULL1 (FUL1, also known as TDR4), interacting with RIN (RIPENING-INHIBITOR) to regulate fruit ripening (Leseberg et al., 2008), undergoes H3K27me3 modification in the Cnr epimutant (Gao et al., 2019). These findings suggest that the Cnr epimutant integrates multiple epigenetic regulatory factors, including DNA methylation, microRNAs, and histone modifications, to regulate fruit ripening. Beyond fruit ripening, the Cnr epimutant has been implicated in various biological processes, such as Cd stress, Fe deficiency, vivipary, and cell death (Chen et al., 2018, 2022; Lai et al., 2020; Yao et al., 2020). Therefore, summarizing the pleiotropic effects of the Cnr epimutant contributes to a comprehensive understanding of epigenetic regulation across different biological processes in plants. This review aims to consolidate recent advancements concerning the Cnr epimutant as a representative case study, enhancing our comprehension of the multifaceted roles of epigenetic modifications in diverse biological processes. Ultimately, this understanding may illuminate new avenues for crop breeding by manipulating epigenetic modifications in the future.
Colorless non-ripening epimutant: its past and present
Tomato (Solanum lycopersicum) serves as an exceptional model crop for understanding the intricate process of fresh fruit ripening process, involving notable transformations in color, flavor, and texture. Being a climacteric fruit, tomato ripening is marked by a surge in respiration following an increase in ethylene biosynthesis (Biale, 1964; Alexander and Grierson, 2002). Among the naturally occurring mutants in tomato, Cnr stands out as an epimutant, initially identified in a commercial population of F1 hybrid cv Liberto plants in 1993. The distinct characteristics of Cnr fruit include non-ripening with yellow-colored pericarp tissues, along with reduced ethylene production and suppressed softening (Thompson et al., 1999). Furthermore, two additional spontaneous mutants, ripening inhibitor (rin) and non-ripening (nor), have been identified, each exhibiting similar rare ripening phenotypes to Cnr. These mutations, along with the Cnr epimutation, are regarded as major regulators functioning upstream of ethylene-mediated ripening pathways, thereby modulating the expression of numerous ripening-related genes (Vrebalov et al., 2002; Manning et al., 2006; Giovannoni, 2007). It is worth noting that Cnr epimutant represents an epigenetic variation, distinct from the gain-of-function genetic variation observed in rin and nor mutants. Notably, studies have shown that the fruit phenotype in heterozygous lines of hybrid progeny between Cnr and rin or nor does not significantly differ from that of Cnr alone, indicating the dominance of the Cnr allele among these three spontaneous mutations (Thompson et al., 1999; Wang R. et al., 2020). These findings highlight the pivotal role of Cnr epimutation in fruit ripening, positioning it as a master regulator.
Following the genetic investigation, biochemical analysis was conducted to elucidate the implications of Cnr epimutation-mediated non-ripening. The pericarp tissue of Cnr mutants displayed a depletion in carotenoid pigments and diminished cell-to-cell contacts (Thompson et al., 1999). Further cellular analysis revealed modifications in the middle lamella homogalacturonan within Cnr fruits, which result in impaired adhesion to calcium and a thicker cell wall throughout the pericarp compared to wild-type Ailsa Craig (AC) fruits (Orfila et al., 2001). Moreover, ripening-associated soluble pectic polysaccharides in Cnr fruits were found to be reduced in comparison with wild-type AC fruits at various ripeness stages (Orfila et al., 2002). These findings reveal a close link between the non-ripening phenotype of Cnr fruits with their cell wall properties. Moreover, changes in enzyme activity and gene expression associated with the cell wall were observed during the ripening process of Cnr fruits (Eriksson et al., 2004). Specifically, chitinase and peroxidase activity were enhanced in Cnr fruits compared to wild-type AC plants. However, ripening-related gene expression was profoundly inhibited in Cnr fruits. While these studies elucidated the non-ripening phenotype of Cnr fruits from physiological and biochemical aspects, the molecular understanding behind the nature of the Cnr epimutant remains lacking.
To delineate the molecular framework underlying the Cnr-mediated ripening process, the Cnr gene was subsequently characterized. Through positional cloning, the SBP-box (SQUAMOSA promoter binding protein-like) gene was identified at the Cnr locus, revealing hypermethylation in the 286-bp promoter region located 2.4 kb upstream from the initiation codon of SlSPL-CNR (Manning et al., 2006). This discovery strengthened our comprehension of the key role played by the SlSPL-CNR gene and epigenetic variation in tomato fruit ripening. Subsequently, whole-genome bisulfite sequencing was performed on AC and Cnr fruit, spanning from the immature stage to the ripe stage, elucidating the dynamic epigenome throughout the fruit ripening process (Zhong et al., 2013). Meanwhile, the promoter region binding transcription factors in fruit ripening regulatory process, such as RIN, were found to undergo demethylation, indicating that DNA methylation, in cooperation with transcription factor binding, is essential for the fruit ripening process. Surprisingly, the CRISP/Cas9-induced knockout mutant of SlSPL-CNR failed to replicate the non-ripening phenocopy of Cnr, implying that SlSPL-CNR contributed little in controlling Cnr fruit ripening (Gao et al., 2019). Overall, the traits of the Cnr epimutant may be attributed not only to SlSPL-CNR itself but also to DNA hypermethylation in other loci, indicating the epigenetic diversity of the Cnr epimutant during the fruit ripening process. Therefore, effectively harnessing the diverse epigenetic mechanisms of Cnr non-ripening phenotype to facilitate fruit breeding for postharvest quality poses a significant challenge in the future.
The multifaceted role of SlSPL-CNR during fruit development
Initially, the Cnr locus was identified within SlSPL-CNR, where DNA hypermethylation was observed in its 286-bp promoter region. Notably, virus-induced gene silencing (VIGS) experiments illustrated that this hypermethylation correlated with the suppressed expression of SlSPL-CNR from the mature green stage to the ripening stage (Manning et al., 2006). Therefore, SlSPL-CNR was considered to contribute to the dominant Cnr non-ripening phenotype and was also believed to be necessary for ripening in last decades. SlSPL-CNR, a member of the SQUAMOSA PROMOTER BINDING PROTEIN-LIKE family transcription factors (TFs) first identified in Antirrhinum majus (Klein et al., 1996), harbors zinc-finger motifs and a monopartite nuclear localization signal deemed crucial for fruit ripening (Lai et al., 2020). The conservation of the canonical SPL core motif (GTAC) for binding the target gene promoter underscores its functional importance (Birkenbihl et al., 2005). However, SlSPL-CNR knockout lines generated by CRISPR/Cas9 technology replicated a less pronounced Cnr non-ripening phenotype (Gao et al., 2019), challenging previous assumptions about the function of Cnr epimutation. Additionally, proteomic analysis unveiled the complexity of SlSPL-CNR’s regulatory mechanism during Cnr fruit ripening (Zhou et al., 2022).
The involvement of SlSPL-CNR in fruit development extends beyond ripening. It modulates the ripening process by binding to the promoter of SlTCP18, a gene named after the three first characterized family members: TEOSINTE BRANCHED (TB) 1 from Zea mays, CYCLOIDEA (CYC) from Antirrhinum majus, and PROLIFERATING CELL FACTORS (PCFs) from Oryza sativa. This interaction affects the expression of genes crucial for fruit maturation (Parapunova et al., 2014). Knockout of SlSPL-CNR leads to increased flavonoid content within fruits, achieved by repressing SlMYB12, a key regulator of flavonoid biosynthesis (Zhou et al., 2023). Moreover, it directly controls genes associated with wax biosynthesis, notably SlCER1–2 (ECERIFERUM1) and SlCER6 (β-KETOACYL-COENZYME A SYNTHASE6), with the aim of reducing postharvest water loss and enhancing fruit quality (Chen et al., 2024). SlSPL-CNR’s upstream regulatory network involves intricate interactions with other genes like RIN and SlBL4 (BELL-LIKE HOMEODOMAIN4), influencing fruit ripening and chloroplast development (Zhou et al., 2012; Yan et al., 2020). Besides, homolog genes in Arabidopsis, such as AtSPL3, play pivotal roles in the floral transition (Cardon et al., 2002), expanding the understanding of SlSPL-CNR’s functionality. Moreover, recent research reveals that SlSPL-CNR impacts cellular homeostasis and stress responses during fruit development by interacting with SlSnRK1 (SUCROSE NON-FERMENTING 1-RELATED PROTEIN KINASE1), thereby implicating it in the regulation of cell death processes (Lai et al., 2020). These findings collectively emphasize the multifaceted regulatory roles of SlSPL-CNR in tomato fruit biology.
The upstream regulatory network in colorless non-ripening fruit ripening process
The regulatory mechanism underlying the Cnr non-ripening phenotype represent a fascinating intersection of genetic and epigenetic control. Especially, epigenetic modifications, including microRNAs (miRNAs) and DNA methylation, act as upstream regulatory factors and play a significant role in Cnr fruit ripening process. MiRNAs, known for their regulatory role in multiple biological processes, including growth, development, and stress responses, emerge as key players in the intricate network governing fruit ripening. Research has highlighted the involvement of miRNAs in controlling the expression of ripening transcription factors through mRNA cleavage during fruit ripening (Moxon et al., 2008; Mohorianu et al., 2011; Karlova et al., 2013). Specifically, SlymiR157 has been identified as directly targeting SlSPL-CNR, thereby influencing its expression by inducing mRNA degradation and translational repression, ultimately impacting the ripening process in Cnr fruits (Chen et al., 2015a).
In addition to miRNAs, epigenetic modifications, particularly DNA methylation, also play crucial roles in fruit ripening. Screening of genes encoding DNA methyltransferases has revealed the significance of SlCMT3 in the Cnr epimutant phenotype. Silencing of SlCMT3 in this context alleviates DNA hypermethylation observed at the SlSPL-CNR promoter and RIN binding sites, resulting in increased SlSPL-CNR expression and subsequent restoration of normal ripening in Cnr fruits (Chen et al., 2015b). Interestingly, the maintenance of DNA methylation dynamics in the Cnr epimutant primarily involves methylated cytosines in CG and CHG contexts. While the role of SlCMT3 in maintaining DNA methylation balance is evident (Chen et al., 2015b), further exploration suggests that SlMET1 might also be critical for sustaining DNA methylation alongside SlCMT3 in the Cnr epimutation (Yao et al., 2020). This aspect necessitates comprehensive investigation to elucidate the full scope of the epigenetic regulatory mechanisms governing the Cnr non-ripening phenotype.
Colorless non-ripening epimutation is involved in multiple biological process
Cnr epimutation participates in various biological processes, including phytohormones production and signaling, cell wall property modification, and extensive modification of the DNA methylome (Thompson et al., 1999; Orfila et al., 2001, 2002; Manning et al., 2006; Zhong et al., 2013). Such diverse involvements suggest a broader role for Cnr epimutation in various biological processes and stress responses. In the Cnr epimutant, DNA hypermethylation is observed in genes related to abscisic acid biosynthesis, such as SlNCED (9-CIS-EPOXYCAROTENOID-DIOXYGENASE), resulting in reduced expression of SlNCED and consequently diminished abscisic acid biosynthesis in seeds, which coincides with the vivipary phenotype observed in Cnr fruits (Yao et al., 2020). In addition, studies have revealed the role of Cnr epimutation under abiotic stress conditions. Under Cd stress, the Cnr epimutant exhibits increased sensitivity to Cd due to the repression of SlSPL-CNR expression, which directly impacts the SlNR (NITRATE REDUCTASE) promoter, influencing NO production and resulting in the accumulation of more Cd compared to wild-type AC plants (Chen et al., 2018). Considering the essential role of NO in Fe deficiency responses, the Cnr epimutant has been implicated in constitutive Fe deficiency responses even under Fe-sufficient conditions, as revealed through comparative physiological and transcriptomic analyses (Chen et al., 2022). Although initial evidence pointed to the importance of SlSPL-CNR in Fe-deficient responses in Cnr roots, subsequent studies have provided a comprehensive understanding of the epigenetic mechanisms regulating Fe homeostasis by Cnr epimutation. Hypermethylation of the SlSPL-CNR promoter in Cnr roots inhibits its expression (Chen et al., 2022), while SlSPL-CNR protein directly binds to SlbHLH101 (BASIC HELIX-LOOP-HELIX101) promoter, inhibiting its transcriptional expression (Zhu et al., 2022). Additionally, SlymiR157 targets SlSPL-CNR expression for mRNA degradation, consequently releasing SlbHLH101 expression to facilitate Fe-deficient responses (Zhu et al., 2022). Furthermore, SlMET1-dependent CG hypermethylation of the SlPME53 (PECTIN METHYLESTERASE53) intron induces its expression, regulating apoplastic Fe reutilization in the Cnr epimutant (Zhu et al., 2024). In summary, the retention of Fe in the apoplast and its uptake into the cytoplasm contribute to the constitutive Fe-deficient response of Cnr under Fe-sufficient conditions. Upon perceiving the Fe deficiency signals, retained Fe in the pectin enters the cell for reutilization. By dissecting both genetic and epigenetic factors, these findings systematically unveil the Cnr-mediated regulatory mechanisms in response to Fe deficiency in tomato, underscoring the importance of harmonizing the pleiotropy of Cnr epimutation for systematic functional exploration in Cnr research.
Prospectives
The pleiotropic effects of Cnr epimutation on various developmental processes and stress responses highlight extensive DNA methylome modification within the Cnr genome, giving rise to distinct regulatory mechanisms (Figure 1). Bisulfite sequencing has revealed that nearly half of the cytosine positions in the Cnr fruit genome are methylated (Zhong et al., 2013). This finding suggests the contribution of not only SlSPL-CNR but also other potential epigenetic loci to the non-ripening phenotype of Cnr. Therefore, comprehensive big data analyses are warranted to elucidate the genome-wide effects of Cnr epimutation in future studies.
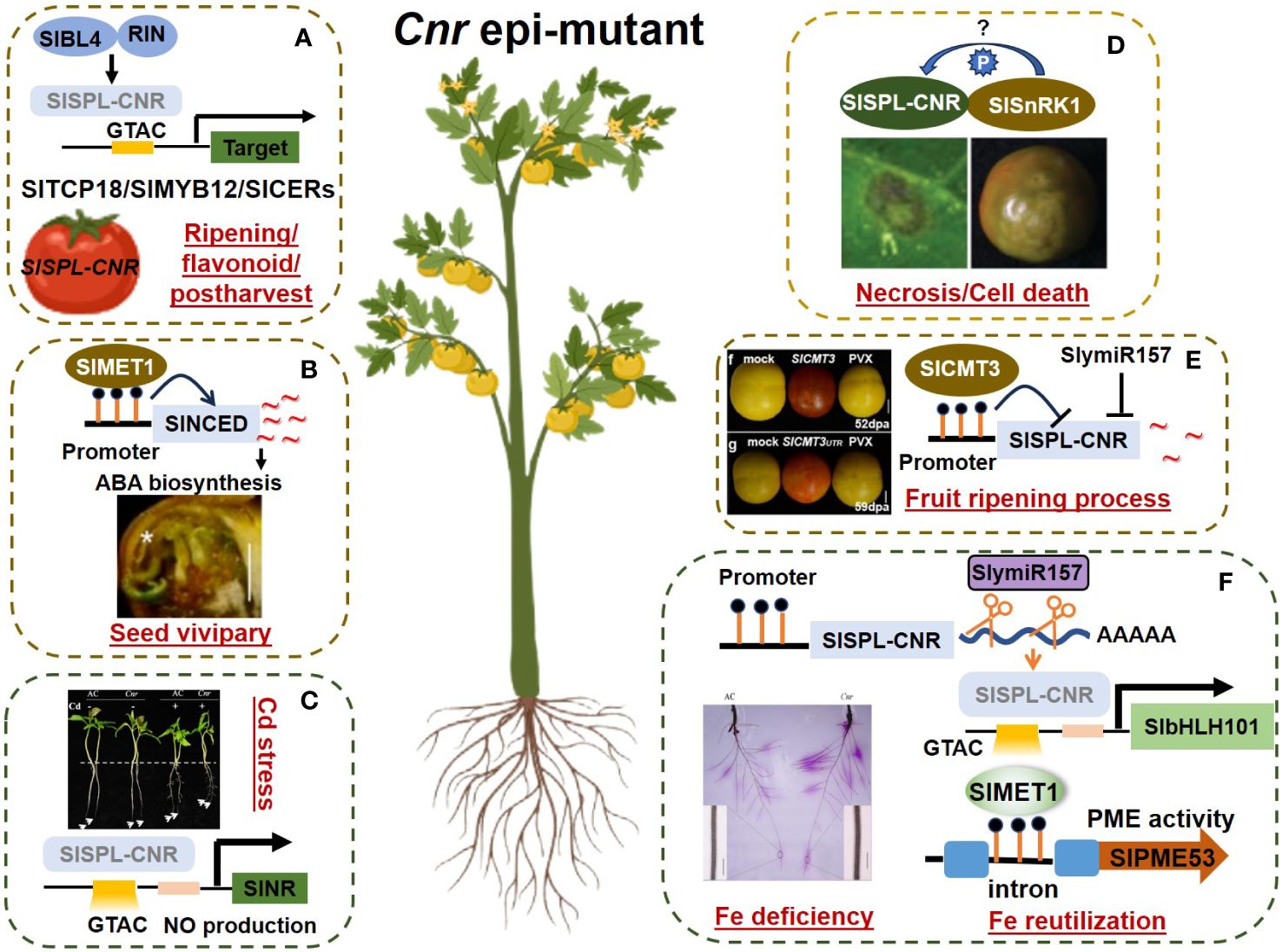
Figure 1 The pleiotropic role of Cnr epimutation in various processes. The regulatory network of Cnr epimutation in tomato is intricate and multifaceted, contributing to various crucial processes. Here’s an overview of each module and its associated references. Different modules (separated by dashed box) represent different processes. (A) The regulatory network of SlSPL-CNR in fruits involves ripening, flavonoid synthesis, and postharvest process. This module is supported by studies derived from Parapunova et al.(2014), Zhou et al. (2023) and Chen et al. (2024). SlBL4: BELL-LIKE HOMEODOMAIN4; SlSPL-CNR: SQUAMOSA PROMOTER BINDING PROTEIN-LIKE-CNR; RIN: RIPENING-INHIBITOR; SlTCP18: TEOSINTE BRANCHED & CYCLOIDEA & PROLIFERATING CELL FACTORS18; SlMYB12: R2R3-MYB TRANSCRIPTION FACTOR12; SlCERs: SlCER1–2 (ECERIFERUM1) and SlCER6 (β-KETOACYL-COENZYME A SYNTHASE6). (B) Cnr mediates vivipary tthrough METHYLTRANSFERASE1, as domonstrated by Yao et al. (2020). SlNCED: 9-CIS-EPOXYCAROTENOID-DIOXYGENASE. (C) SlSPL-CNR modulates Cd acquisition by targeting and repressing SlNR expression, thereby regulating NO production, as reported by Chen et al. (2018). SlNR: NITRATE REDUCTASE. (D) The interaction between SlSPL-CNR and SlSnRK1 mediates ripening and cell death, elucidated by Lai et al. (2020). SlSnPK1: SUCROSE NON-FERMENTING 1-RELATED PROTEIN KINASE1. (E) Upstream regulators of Cnr fruit ripening process, including Manning et al. (2006), Zhong et al. (2013), and Chen et al. (2015a, 2015b), provide insights into the regulatory cascade governing fruit ripening. SlCMT3: CHROMOMETHYLASE3; SlymiR157: microRNA miR157. (F) The SlymiR157−SlSPL-CNR−SlbHLH101 model and SlMET1−SlPME53 model are involved in Fe uptake and Fe reutilization, respectively, as described by Chen et al. (2022), and Zhu et al. (2022, 2024). SlMET1: METHYLTRANSFERASE1; SlbHLH101: BASIC HELIX-LOOP-HELIX101; SlPME53: PECTIN METHYLESTERASE53.
Furthermore, the methylome pattern of Cnr appears to vary depending on organ type or developmental stages, as evidenced by disparities between Cnr fruits and roots in bisulfite sequencing data (Zhu et al., 2024). Considering these observations alongside the diverse effects of Cnr on fruit ripening, leaf cell death, and root responses to abiotic stress, it is imperative to account for tissue-specific effects when evaluating the pleiotropic effects of Cnr. Moreover, the involvement of MicroRNA SlymiR157 in Cnr fruit ripening and the association of the ripening-related transcription factor TDR4/FUL1 with hyperH3K27me3 marks in the Cnr epimutant underscore the importance of exploring cooperative mechanisms beyond DNA methylation alone in the Cnr-mediated regulatory network across various biological processes. Future research efforts should delve deeper into the crosstalk between genetic regulation and epigenetic regulation, including DNA methylation modification, histone modification, and microRNAs, to enhance our understanding of the intricate regulatory network governed by Cnr epimutation.
Lastly, the potential application of Cnr epimutant, as representative epigenetic variations, in the development of novel strategies and technologies for crop improvement warrants consideration. For example, an ideal crop could be constructed in concordance with breeding stress-tolerant crop and fruit breeding for postharvest quality after deciphering the epigenetic codes of Cnr in stress responses and fruit ripening process, respectively. In summary, a systematic approach integrating multi-omics, tissue-specific, and the exploration of multiple genetic and epigenetic modifications is essential for constructing Cnr-mediated regulatory network, ultimately facilitating advancements in improving tomatoes.
Author contributions
HZ: Conceptualization, Writing – original draft, Writing – review & editing. JY: Supervision, Writing – original draft, Writing – review & editing, Funding acquisition. WC: Funding acquisition, Resources, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Natural Science Foundation of Zhejiang Province (LY24C150007 and LZ22C150001), China Postdoctoral Science Foundation (2019M652064), and of Hangzhou Normal University Dengfeng Project.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Alexander, L., Grierson, D. (2002). Ethylene biosynthesis and action in tomato: a model for climacteric fruit ripening. J. Exp. Bot. 53, 2039–2055. doi: 10.1093/jxb/erf072
Biale, J. B. (1964). Growth, Maturation, and Senescence in Fruits: Recent knowledge on growth regulation and on biological oxidations has been applied to studies with fruits. Science 146, 880–888. doi: 10.1126/science.146.3646.880
Birkenbihl, R. P., Jach, G., Saedler, H., Huijser, P. (2005). Functional dissection of the plant-specific SBP-domain: overlap of the DNA-binding and nuclear localization domains. J. Mol. Biol. 352, 585–596. doi: 10.1016/j.jmb.2005.07.013
Cardon, G. H., Höhmann, S., Nettesheim, K., Saedler, H., Huijser, P. (2002). Functional analysis of the Arabidopsis thaliana SBP-box gene SPL3: a novel gene involved in the floral transition. Plant J. 12, 367–377. doi: 10.1046/j.1365-313x.1997.12020367.x
Chen, D., Wang, T., Huang, H., Zhang, Q., Chen, X., Sun, Z., et al. (2024). SlCNR regulates postharvest water loss and wax accumulation in tomato fruit and directly represses the transcription of very-long-chain (VLC) alkane biosynthesis-related genes SlCER1-2 and SlCER6. Postharvest Biol. Tec. 208, 112641. doi: 10.1016/j.postharvbio.2023.112641
Chen, W., Jin, J., Lou, H., Liu, L., Kochian, L. V., Yang, J. (2018). LeSPL-CNR negatively regulates Cd acquisition through repressing nitrate reductase-mediated nitric oxide production in tomato. Planta 248, 893–907. doi: 10.1007/s00425-018-2949-z
Chen, W., Kong, J., Lai, T., Manning, K., Wu, C., Wang, Y., et al. (2015a). Tuning LeSPL-CNR expression by SlymiR157 affects tomato fruit ripening. Sci. Rep. 5, 7852. doi: 10.1038/srep07852
Chen, W., Kong, J., Qin, C., Yu, S., Tan, J., Chen, Y., et al. (2015b). Requirement of CHROMOMETHYLASE3 for somatic inheritance of the spontaneous tomato epimutation Colourless non-ripening. Sci. Rep. 5. doi: 10.1038/srep07852
Chen, W., Zhu, H., Wang, J., Han, G., Huang, R., Hong, Y., et al. (2022). Comparative physiological and transcriptomic analyses reveal altered Fe-deficiency responses in tomato epimutant Colorless Non-ripening. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.796893
Eriksson, E. M., Bovy, A., Manning, K., Harrison, L., Andrews, J., De Silva, J., et al. (2004). Effect of the Colorless non-ripening mutation on cell wall biochemistry and gene expression during tomato fruit development and ripening. Plant Physiol. 136, 4184–4197. doi: 10.1104/pp.104.045765
Gao, Y., Zhu, N., Zhu, X., Wu, M., Jiang, C., Grierson, D., et al. (2019). Diversity and redundancy of the ripening regulatory networks revealed by the fruitENCODE and the new CRISPR/Cas9 CNR and NOR mutants. Hortic. Res. 6, 39. doi: 10.1038/s41438-019-0122-x
Giovannoni, J. J. (2007). Fruit ripening mutants yield insights into ripening control. Curr. Opin. Plant Biol. 10, 283–289. doi: 10.1016/j.pbi.2007.04.008
He, L., Huang, H., Bradai, M., Zhao, C., You, Y., Ma, J., et al. (2022). DNA methylation-free Arabidopsis reveals crucial roles of DNA methylation in regulating gene expression and development. Nat. Commun. 13, 1335. doi: 10.1038/s41467-022-28940-2
Karlova, R., van Haarst, J. C., Maliepaard, C., van de Geest, H., Bovy, A. G., Lammers, M., et al. (2013). Identification of microRNA targets in tomato fruit development using high-throughput sequencing and degradome analysis. J. Exp. Bot. 64, 1863–1878. doi: 10.1093/jxb/ert049
Klein, J., Saedler, H., Huijser, P. (1996). A new family of DNA binding proteins includes putative transcriptional regulators of the Antirrhinum majus floral meristem identity gene SQUAMOSA. Mol. Gen. Genet. 250, 7–16. doi: 10.1007/BF02191820
Lai, T., Wang, X., Ye, B., Jin, M., Chen, W., Wang, Y., et al. (2020). Molecular and functional characterization of the SBP-box transcription factor SPL-CNR in tomato fruit ripening and cell death. J. Exp. Bot. 71, 2995–3011. doi: 10.1093/jxb/eraa067
Leseberg, C. H., Eissler, C. L., Wang, X., Johns, M. A., Duvall, M. R., Mao, L. (2008). Interaction study of MADS-domain proteins in tomato. J. Exp. Bot. 5, 2253–2265. doi: 10.1093/jxb/ern094
Lu, F., Cui, X., Zhang, S., Jenuwein, T., Cao, X. (2011). Arabidopsis REF6 is a histone H3 lysine 27 demethylase. Nat. Genet. 43, 715–719. doi: 10.1038/ng.854
Manning, K., Tor, M., Poole, M., Hong, Y., Thompson, A. J., King, G. J., et al. (2006). A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat. Genet. 38, 948–952. doi: 10.1038/ng1841
Mohorianu, I., Schwach, F., Jing, R., Lopez-Gomollon, S., Moxon, S., Szittya, G., et al. (2011). Profiling of short RNAs during fleshy fruit development reveals stage-specific sRNAome expression patterns. Plant J. 67, 232–246. doi: 10.1111/j.1365-313X.2011.04586.x
Moxon, S., Jing, R., Szittya, G., Schwach, F., Rusholme Pilcher, R. L., Moulton, V., et al. (2008). Deep sequencing of tomato short RNAs identifies microRNAs targeting genes involved in fruit ripening. Genome Res. 18, 1602–1609. doi: 10.1101/gr.080127.108
Orfila, C., Huisman, M. M. H., Willats, W. G. T., van Alebeek, G., Schols, H. A., Seymour, G. B., et al. (2002). Altered cell wall disassembly during ripening of Cnr tomato fruit: implications for cell adhesion and fruit softening. Planta 215, 440–447. doi: 10.1007/s00425-002-0753-1
Orfila, C., Seymour, G. B., Willats, W. G. T., Huxham, I. M., Jarvis, M. C., Dover, C. J., et al. (2001). Altered middle lamella homogalacturonan and disrupted deposition of (1-> 5)-alpha-L-arabinan in the pericarp of Cnr, a ripening mutant of tomato. Plant Physiol. 126, 210–221. doi: 10.1104/pp.126.1.210
Parapunova, V., Busscher, M., Busscher-Lange, J., Lammers, M., Karlova, R., Bovy, A. G., et al. (2014). Identification, cloning and characterization of the tomato TCP transcription factor family. Bmc. Plant Biol. 14, 157. doi: 10.1186/1471-2229-14-157
Thompson, A. J., Tor, M., Barry, C. S., Vrebalov, J., Orfila, C., Jarvis, M. C., et al. (1999). Molecular and genetic characterization of a novel pleiotropic tomato-ripening mutant. Plant Physiol. 120, 383–389. doi: 10.1104/pp.120.2.383
Vrebalov, J., Ruezinsky, D., Padmanabhan, V., White, R., Medrano, D., Drake, R., et al. (2002). A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (rin) locus. Science 296, 343–346. doi: 10.1126/science.1068181
Wang, J., Nan, N., Li, N., Liu, Y., Wang, T., Hwang, I., et al. (2020). A DNA methylation reader-chaperone regulator-transcription factor complex activates OsHKT1;5 expression during salinity stress. Plant Cell 32, 3535–3558. doi: 10.1105/tpc.20.00301
Wang, R., Lammers, M., Tikunov, Y., Bovy, A. G., Angenent, G. C., de Maagd, R. A. (2020). The rin, nor and Cnr spontaneous mutations inhibit tomato fruit ripening in additive and epistatic manners. Plant Sci. 294, 110436. doi: 10.1016/j.plantsci.2020
Yan, F., Gao, Y. S., Pang, X., Xu, X., Zhu, N., Chan, H., et al. (2020). BEL1-LIKE HOMEODOMAIN4 regulates chlorophyll accumulation, chloroplast development, and cell wall metabolism in tomato fruit. J. Exp. Bot. 71, 5549–5561. doi: 10.1093/jxb/eraa272
Yao, M., Chen, W., Kong, J., Zhang, X., Shi, N., Zhong, S., et al. (2020). METHYLTRANSFERASE1 and ripening modulate vivipary during tomato fruit development. Plant Physiol. 183, 1883–1897. doi: 10.1104/pp.20.00499
Zhang, H., Lang, Z., Zhu, J. (2018). Dynamics and function of DNA methylation in plants. Nat. Rev. Mol. Cell Biol. 19, 489–506. doi: 10.1038/s41580-018-0016-z
Zhong, S., Fei, Z., Chen, Y., Zheng, Y., Huang, M., Vrebalov, J., et al. (2013). Single-base resolution methylomes of tomato fruit development reveal epigenome modifications associated with ripening. Nat. Biotechnol. 31, 154–159. doi: 10.1038/nbt.2462
Zhou, L., Sun, Z., Hu, T., Chen, D., Chen, X., Zhang, Q., et al. (2023). Increasing flavonoid contents of tomato fruits through disruption of the SlSPL-CNR, a suppressor of SlMYB12 transcription activity. Plant Biotechnol. J. 22, 290–292. doi: 10.1111/pbi.14214
Zhou, T., Li, R., Yu, Q., Wang, J., Pan, J., Lai, T. (2022). Proteomic Changes in Response to Colorless nonripening Mutation during Tomato Fruit Ripening. Plants(Basel) 11. doi: 10.3390/plants11243570
Zhou, T., Zhang, H., Lai, T., Qin, C., Shi, N., Wang, H., et al. (2012). Virus-induced gene complementation reveals a transcription factor network in modulation of tomato fruit ripening. Sci. Rep. 2, 836. doi: 10.1038/srep00836
Zhu, H., Wang, J., Huang, R., Yang, Z., Fan, W., Huang, L., et al. (2024). Epigenetic modification of a pectin methylesterase gene activates apoplastic iron reutilization in tomato roots. Plant Physiol., kiae167. doi: 10.1093/plphys/kiae167
Keywords: colorless non-ripening (Cnr), crop breeding, epigenetic regulation, tomato, fruit ripening
Citation: Zhu H, Yang JL and Chen W (2024) Epigenetic insights into an epimutant colorless non-ripening: from fruit ripening to stress responses. Front. Plant Sci. 15:1440120. doi: 10.3389/fpls.2024.1440120
Received: 29 May 2024; Accepted: 12 June 2024;
Published: 02 July 2024.
Edited by:
Qi Chen, Kunming University of Science and Technology, ChinaReviewed by:
Shuai Li, Qingdao Agricultural University, ChinaTanveer Ahmad Khan, Independent Researcher, Aligarh, India
Copyright © 2024 Zhu, Yang and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weiwei Chen, 15858223807@163.com
 Huihui Zhu
Huihui Zhu Jian Li Yang
Jian Li Yang Weiwei Chen
Weiwei Chen