
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 03 September 2024
Sec. Plant Systematics and Evolution
Volume 15 - 2024 | https://doi.org/10.3389/fpls.2024.1439206
A correction has been applied to this article in:
Corrigendum: Comparative plastome analyses and evolutionary relationships of 25 East Asian species within the medicinal plant genus Scrophularia (Scrophulariaceae)
Backgroud: Scrophularia L., a genus of the Scrophulariaceae, is a group of important medicinal plants used for eliminating heat and detoxifying. East Asia has an abundance of potentially medicinal Scrophularia species, and it serves as a secondary diversity center of the genus. However, the genomic resources available for germplasm identification and pharmaceutical exploration of East Asian Scrophularia are insufficient, hindering its commercial and industrial development. Additionally, the interspecific relationships of most East Asian Scrophularia species remain unclear.
Methods: In this study, we sequenced the leaves of 25 East Asian species of the genus Scrophularia, assembled and annotated the complete chloroplast genomes, and subsequently performed comparative and phylogenetic analyses on these genomes.
Results and discussion: The conserved plastome length of these 25 species ranged from 151,582 bp to 153,239 bp, containing a total of 132 coding genes, including 18 duplicated genes and 114 unique genes. Through genome alignment of these 25 species, 38-53 repeated sequences and 7 shared SSRs were identified, along with regions with high nucleotide polymorphism (Pi), which could potentially serve as molecular markers for species identification. The genome structure, gene content, and arrangement showed conservation, while variations were observed in the IR boundary regions and IGS. Phylogenetic inferences based on whole plastomes or on coding sequences (CDS) only yielded congruent results. We categorized the 25 East Asian Scrophularia species into six distinct clades and further explored their interspecies relationships using morphological characteristics, such as flower color, the relative position of stamens and corolla, and plant height. This could lay a genetic basis for future resource development of Scrophularia in East Asia.
Scrophularia L., a species-rich and complicated genus of Scrophulariaceae, comprises nearly 300 species across the Northern temperate zone (Hong et al., 1998; Wang, 2015). Southern Europe and the Mediterranean are the main center of Scrophularia diversity (Hong, 1983), while East Asia is the secondary center with a total of 42 documented species, 36 of which are recorded in China (Li et al., 1999; Wang, 2015). Many species of this genus possess high therapeutic properties and are extensively employed as herbal medications to treat fever, constipation, rheumatism, and inflammatory affections (Huang, 2018; Lee et al., 2021; Cui, 2023). It is notable that Scrophularia plants possess bioactive iridoids, such as harpagide and harpagoside, which are found in most species of this genus (Li et al., 1999; de Santos Galíndez et al., 2002; Pasdaran and Hamedi, 2017). Scrophularia ningpoensis Hemsl., which has a wider distribution in China and was officially listed in the Chinese Pharmacopoeia as the sole medicinal source of Scrophulariae Radix (SR, also called “Xuan Shen”), has been used over two thousand years (Chinese Pharmacopoeia Commission, 2020; Lee et al., 2021). While the remaining species in China tend to exhibit distinct regional characteristics (Supplementary Table 1) and are usually utilized as local folk remedies or ethnic medicines, such as S. spicata Franch., S. fargesii Franch., S. henryi Hemsl. and S. moellendorffii Maxim (Li et al., 1999). S. incisa Weinm. is a traditional Mongolian medicine, and its entire plant is used for treating measles and rash diseases (Editorial Board of the Chinese Materia Medica, 2003). S. dentata Royle ex Benth. is employed as an ethnic medicine known as “Alpine Tibetan herb” for the treatment of exanthema and fever (Zhang et al., 2013; Ni et al., 2016). S. buergeriana Miq., S. kakudensis Franch. and S. yoshimurae T. Yamaz., as the common substitution and adulterants of SR, have been employed in Korea and in Taiwan Province for over 20 years (Sagare et al., 2001; Nam et al., 2020; Manivannan et al., 2021; Guo et al., 2023b). However, due to restricted wild distribution and indiscriminate harvesting and exploitation, wild strains of S. ningpoensis are facing a diminishing genetic diversity (Wang and Wang, 2007; Zhao, 2008; Chen, 2014). Furthermore, S. ningpoensis is well-known as one of the “Zhe Ba Wei” (eight traditional Chinese medicines from Zhejiang Province), and it is grown in many other provinces across China by introduction breeding (Chen, 2011). Due to the decreasing cultivation area of authentic S.ningpoensis in Zhejiang (He et al., 2020), extensive cultivation in other provinces has intensified market competition (Zhang et al., 2022a). The intensification of market competition has led to an increase in artificial cultivation. However, owing to intensive artificial selection (Chen, 2011) and the unsustainable practices adopted by farmers, like long-term asexual reproduction, the genetic diversity of cultivated varieties and the quality of medicinal materials continue to decline (Yang, 2011). Therefore, it is crucial to clarify the species relationship of Scrophularia in East Asia and develop its medicinal resources reasonably. Efficient universal molecular markers are also essential to promote contemporary breeding projects in order to explore and conserve the germplasm of this medicinally and economically significant genus.
Despite the considerable medicinal worth of Scrophularia rhizomes, differentiating among species presents a challenge because of their analogous therapeutic properties and physical characteristics (Guo et al., 2023b). To understand species’ genetic background, since the 1990s, a multitude of approaches have been applied to explore the origin, genetic diversity and evolutionary relationships of Scrophularia (Ortega Olivencia and Devesa Alcaraz, 1993), including pollination system (Navarro-Pérez et al., 2013), plastid DNA datasets (e.g. trnL-trnF, psbA-trnH, trnQ-rps16 and trnS-trnG), and nuclear ribosomal DNA sequences (nuclear ITS) (Attar et al., 2011; Scheunert and Heubl, 2011; Navarro-Pérez et al., 2013; Scheunert and Heubl, 2014; 2017). Among these efforts, the most extensive sampling of phylogenetic relationships within the Scrophularia genus to date has been constructed using sequences from the nuclear ITS region and two plastid DNA regions, encompassing 147 species worldwide, but only 13 East Asian species included (Scheunert and Heubl, 2017). In addition, several subclades and infrageneric relationships had weak support, especially within the East-Asian lineage of S. sect. Scrophularia. Considering this, further investigation of East Asian Scrophularia using more comprehensive genomic information, including the plastome, would be of great interest and significance for advancing our understanding in this field.
East Asia is a natural plant floristic region and one of the most diverse and complex regions in terms of plant biodiversity worldwide (Boufford and Ōba, 1998; Li et al., 2015). Once a vital ice age refuge, it’s now seen as a hub for angiosperm diversification and possesses one of the world’s oldest and most complete series of plant diversity evolution (Li, 2008). East Asiatic Floristic Kingdom reflects the evolutionary history and interrelationships of species, revealing the impact of environmental and ecological interactions (Deng, 2015), from the uplift of Qinghai-Tibetan Plateau (An and Harrison, 2000; Liu et al., 2006) to monsoonal climates (Azani et al., 2019). Therefore, East Asia is a critical region for the origin and evolution of angiosperms worldwide, as well as an important area and natural laboratory for studying distribution processes. Most of China is part of the East Asiatic Floristic Kingdom (Chen et al., 2022b), a young biome from the Early Miocene (Tang and Li, 1996; Milne and Abbott, 2002; Yang et al., 2023), rich in both relict plants (Milne and Abbott, 2002) and young species. Investigating the inter-species relationships within East Asian Scrophularia could provide theoretical support for applications such as molecular plant breeding and the analysis of medicinal components, while also offering a useful perspective on the evolution of East Asian Scrophularia species.
Genomic data provide us with a convenient tool to explore inter-specific phylogeny and mechanisms of intra-specific differentiation (Lin, 2020; Chen et al., 2021; Zhang et al., 2022b). Despite some limitations, such as challenges in addressing incomplete lineage sorting (ILS) and hybridization, plastids with their conserved structure and low recombination are still valuable for sequencing and phylogenetic analysis in angiosperms (Daniell et al., 2016; Feng et al., 2022; Xiang et al., 2024). Overall, plastome sequences have been commonly utilized to build phylogenies for plants (Raubeson et al., 2007; Gao et al., 2010; Lin, 2020).
In this study, we compared and characterized the complete plastomes of 25 species of East Asian Scrophularia, with respect to checking the contraction and expansion of the IR regions, identifying rapidly evolving regions of plastid DNA (i.e. SSRs and differentiation hotspots such as repeat sequence) and calculating the protein-coding genes selective pressure. Another goal was to build a strongly supported phylogenetic trees of 44 plastomes, elucidating their evolutionary relationships. These research efforts are expected to be helpful for future research on medicinal resource development, cultivation and breeding, species identification, systematics and conservation of Scrophularia.
Leaves of 25 East Asian Scrophularia species were collected for sequencing (Supplementary Table 2). Voucher specimens were preserved in the Zhejiang University Herbarium (HZU). The CTAB method with modifications was employed to extract DNA from silica gel-dried leaves (Zhou et al., 2021). The lysis reagent Plant DNAzol (Invitrogen Corp. CTAB lysis solution) was used to extract the total genomic DNA from the leaf material of Scrophularia species. The MGIEasy Universal DNA Library Prep Set (96 RXN, Item No.: 1000006986) was used for the preparation of total libraries, which were then sequenced on the DNBSeq platform using the PE100 strategy at China National GenBank (CNGB) in Shenzhen, China. After obtaining the raw sequencing data, low-quality reads and adapters were filtered out using Trimmomatic v0.39 (Bolger et al., 2014). Through read mapping and gap-filling steps, GetOrganelle software was utilized iteratively de novo to assemble the complete plastome. Geneious software (Geneious Biologics 2023 (https://www.geneious.com/biopharma/, accessed on 10 May 2023)) was used to annotate the assembled plastomes and the annotations of rps12 gene and ycf1 gene was inspected with CPGview (Liu et al., 2023). Eventually, the complete plastomes of 25 East Asian Scrophularia species were all uploaded to NCBI Genbank database.
We used the online tool REPuter (https://bibiserv.cebitec.uni-bielefeld.de/reputer/) to annotate repeat sequences in the plastomes of the 25 Scrophularia species (Kurtz and Schleiermacher, 1999). This analysis included four types of repeats: forward repeats, reverse repeats, palindromic repeats, and complementary repeats. The parameter settings were as follows: a Hamming distance of 3, a minimum repeat size of 30 bp, and a maximum repeat count of 80. For the analysis of simple sequence repeats (SSRs) in the Scrophularia plastomes, we employed MISA (https://pgrc.ipk-gatersleben.de/misa/) with the following parameter settings for the minimum repeat unit sizes: mononucleotide repeats of 10, dinucleotide repeats of 6, trinucleotide repeats of 4, tetranucleotide repeats of 3, pentanucleotide repeats of 3, and hexanucleotide repeats of 3 (Beier et al., 2017).
An analysis of relative synonymous codon usage (RSCU) and effective number of codon (ENC) was conducted using CodonW V1.4.2 (https://codonw.sourceforge.net/). RSCU was employed to assess variations in the usage patterns of synonymous codons across the entire genome. It reflects the ratio of the observed frequency of a particular synonymous codon in the actual gene sample to its expected average frequency based on theoretical calculations (Sharp and Li, 1987; Chen et al., 2022a). The cusp program from the EMBOSS (https://emboss.toulouse.inra.fr/) website was used to compute the GC content associated with the three positions of codons (first, second, and third) in the entire plastome and within 25 plastomes (Rice et al., 2000).
To elucidate the intergenic and intra-species variations and gene structural composition in 25 species of East Asian Scrophularia, with S. alaschanica as the reference sequence, we employed mVISTA (https://genome.lbl.gov/vista/mvista/submit.shtml/) for genome-wide multiple sequence alignment. We utilized the shuffle-LAGAN mode, which enables global alignment and is the only mode capable of identifying gene rearrangements and inversions (Brudno et al., 2003; Frazer et al., 2004). Additionally, we conducted collinearity analysis using Mauve for multi-genome alignment of the Scrophularia species, aiming to detect rearrangements and inversions (Darling et al., 2004).
Variation in the size of the molecule is typically due to the expansion or contraction of the inverted repeat (IR) into or out of adjacent single-copy regions, as well as changes in sequence complexity caused by insertions or deletions of unique sequences (Plunkett and Downie, 2000). CPJSdraw online software (https://cloud.genepioneer.com:9929/#/tool/alltool/detail/335) was used to compare IR border expansion or contraction of the twenty-five sequences by directly uploading their gb format files (Li et al., 2023a).
We used a Perl script to extract protein-coding genes from each Scrophularia plastome. These sequences were then visualized and examined for divisibility by three using Geneious software. Using S. takesimensis (KP718628) as the reference sequence, we calculated the Ka/Ks values for each CDS using TBtools V1.113 (Chen et al., 2020).
CDS and intergenic spacers (IGS) were extracted with a Perl script (https://github.com/quxiaojian/Bioinformatic_Scripts/tree/master/extract_sequences_from_gb_files) and organized using Geneious software. The CDS and IGS were separately aligned using MAFFT v7.0 to construct matrices (Katoh and Standley, 2013). Using S. takesimensis (KP718628) as the reference sequence, nucleotide diversity analysis was performed in DnaSP v6.0 to determine the total number of mutations (Eta) and nucleotide diversity (Pi) in the 25 Scrophularia plastomes (Rozas et al., 2017).
A set of 44 plastomes from six genera in Scrophulariaceae, which included 34 individuals in Scrophularia, 2 in Verbascum, 2 in Buddleja, 2 in Eremophila, 2 in Myoporum, and 1 in Leucophyllum, as well as one in Plantaginaceae, were used. Aside from the Scrophularia species, the remaining 10 species are outgroups, with Digitalis lanata used as the root of the phylogenetic tree (Supplementary Table 3). These were chosen to construct two phylogenetic trees inferred from whole plastome data and from CDS data only, respectively. MAFFT V7 was used to align 44 plastome sequences under default parameters. IQ-TREE V1.6.8 was used to construct phylogenetic trees using the maximum likelihood method (Nguyen et al., 2015; Yang et al., 2022a). When running IQ-TREE, it will be executed twice: the first run is to select the best model, and the second run is to construct the tree using the best model. The preferred model for the whole genome was TVM+F+R2, while UNREST+FO+R2 was the optimal model for constructing phylogenetic trees using CDS. Based on Bayesian Information Criterion (BIC), the best models for both whole genome and CDS trees were confirmed as TVM+I+G substitution models using jModelTest v2.1.10 (Darriba et al., 2012). MrBayes V3.2.7 was employed for Bayesian inference phylogenetic tree construction (Ronquist and Huelsenbeck, 2003). The analyses were conducted with 2 million generations using the Markov Chain Monte Carlo (MCMC) algorithm. Trees were sampled every 100 iterations. The first 1/4 of the calculated trees were discarded as burn-in, and a consensus tree was constructed from the remaining trees to compute posterior probabilities (PPs).
The quality metrics of raw reads (Q20 and Q30) and clean reads (reads after quality trimming, reads assembled and coverage of assemblies) indicated good sequencing quality and high depth coverage, demonstrating that the sequencing depth was sufficient to support the assembly of the plastid genome (Supplementary Table 4). After assembly and annotation, we obtained the following structural information of the plastid genome. In the 25 East Asian Scrophularia species, plastomes had a total length ranging from 151,582bp to 153,239 bp. It consisted of a large single-copy region (LSC) spanning from 82,790 bp to 84,386 bp, a small single-copy region (SSC) ranging from 17,321 bp to 17,942 bp, and two IR regions with lengths between 25,392 bp and 25,570 bp. The Scrophularia plastomes encoded a total of 132 genes, comprising 18 duplicated genes and 114 unique genes, with 80 protein-coding genes, 4 ribosomal RNA (rRNA) genes and 30 transfer RNA (tRNA) genes. Among 114 unique genes, 10 protein-coding genes (petB, petD, atpE, ndhB (x2), rpl16, rpl2 (x2), rps16, rpoC1) and 5 tRNA genes (trnA-UGC, trnG-GCC, trnI-GAU, trnL-UAA, trnV-UAC) contained a single intron, while 3 genes (clpP, rps12, and ycf3) had 2 introns (Figure 1; Supplementary Table 5). These characteristics align with previous findings in Scrophularia plastomes (Wang et al., 2022; Guo et al., 2023b). The overall GC content ranged from 37.87% to 38.09%. The GC content in the LSC ranged from 35.93% to 36.20%, while in the SSC it ranged from 32.04% to 32.35%. The IR regions exhibited a GC content ranging from 43.08% to 43.22%, which was higher than that of both the LSC and SSC regions (Table 1).
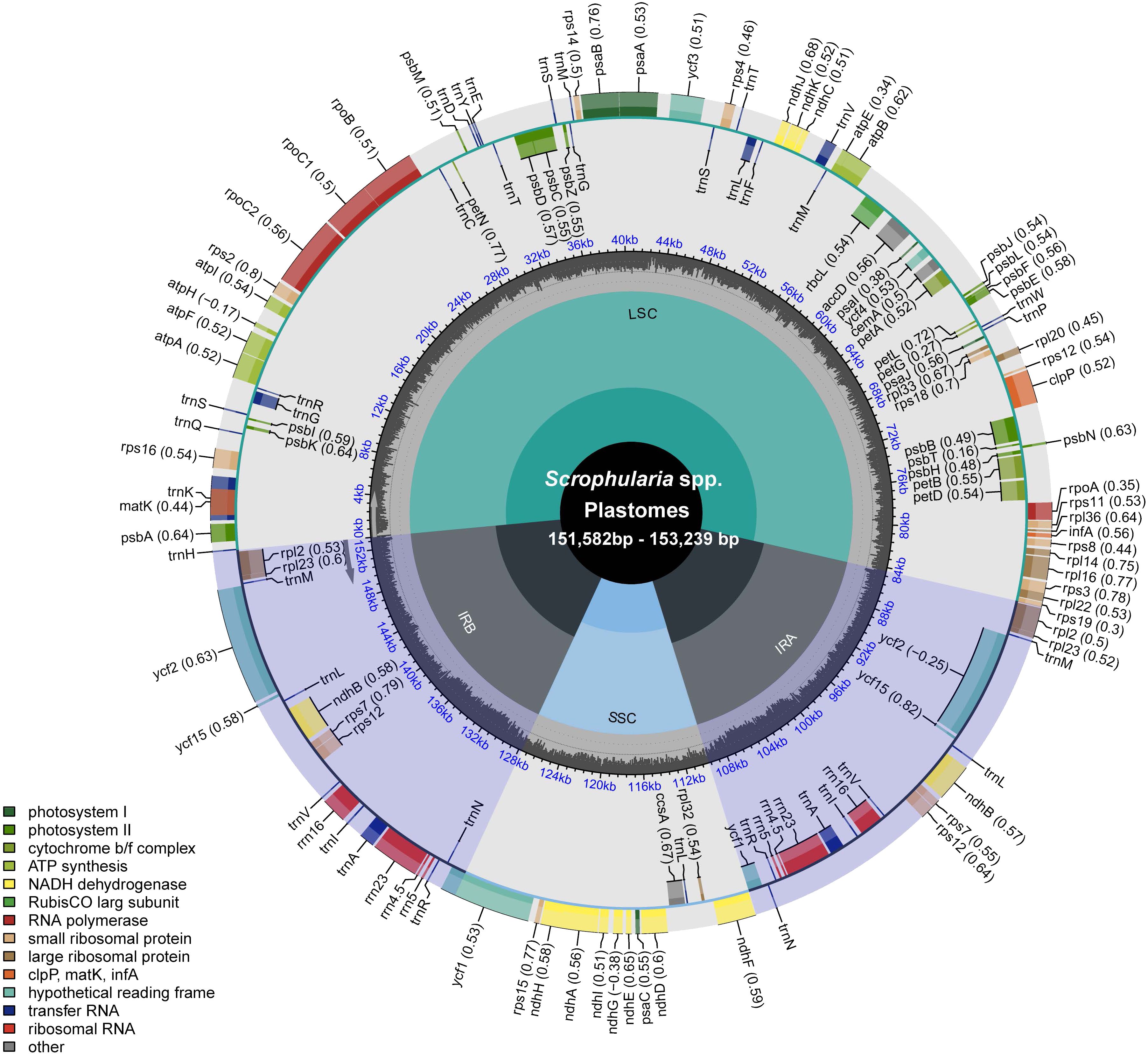
Figure 1. Circular gene map of Scrophularia plastomes. The map illustrates the characteristic quadruple structure of the plastome, where the blue regions represent the IR regions, and the gray regions represent the LSC and SSC regions. The transcription directions for the inner and outer genes are clockwise and anticlockwise, respectively. The black, uneven circle in the middle represents the GC content along the genome. The functional classification of the genes is shown in the left bottom corner.
A total of 1084 repeats were detected, consisting of 523 forward repeats, 536 palindromic repeats, 15 reverse repeats, and 10 complementary repeats (Supplementary Table 6). The lengths of the repeat sequences in the 25 Scrophularia plastomes varied from 38 (S. yunnanensis) to 53 (S. mapienensis). Among them, only 12 exhibited reverse repeats, while 9 had complementary repeats. S. buergeriana displayed the highest number of reverse repeats (3) and complementary repeats (2) (Figure 2). Regarding the base size, the majority of the dispersed repeat sequences were 30-39 bp in length, accounting for 79.61% of the total. A minority of repeats were 50 bp or longer (1.75%), with S. lijiangensis, S. spicata, and S. mandarinorum having the highest number (4) of dispersed repeats longer than 50 bp (Supplementary Table 6).
We observed a range of SSRs quantities in the plastomes of 25 East Asian Scrophularia species, with counts varying from 38 (S. musashiensis & S. fargesii) to 61 (S. yunnanensis). Among these SSRs, 1040 (93.86%) were comprised of A/T bp, the frequency of C and G nucleotides was low (0.45%). Besides single nucleotide repeats, we identified 10 different types of SSRs shared among the 25 Scrophularia plastomes, namely AT/TA, ATA, CAT, TTA, AAGA, AATA, ATCA, GAAA, and GTCT (Figure 3A, Supplementary Table 7). It is worth noting that the SSRs exhibited nucleotide variations, with each type of repeat (di-, tri-, tetra-, penta-, and hexanucleotide) corresponding to 7.9%, 2.8%, 7.6%, 0.7% and 0.2%, respectively (Supplementary Table 7). The SSRs were unevenly distributed in the plastome, with 79.9% located in the LSC region, 9.5% in the SSC region, and 10.6% in the IRb and IRa regions, indicating increased polymorphic variation in LSC region (Figure 3B).
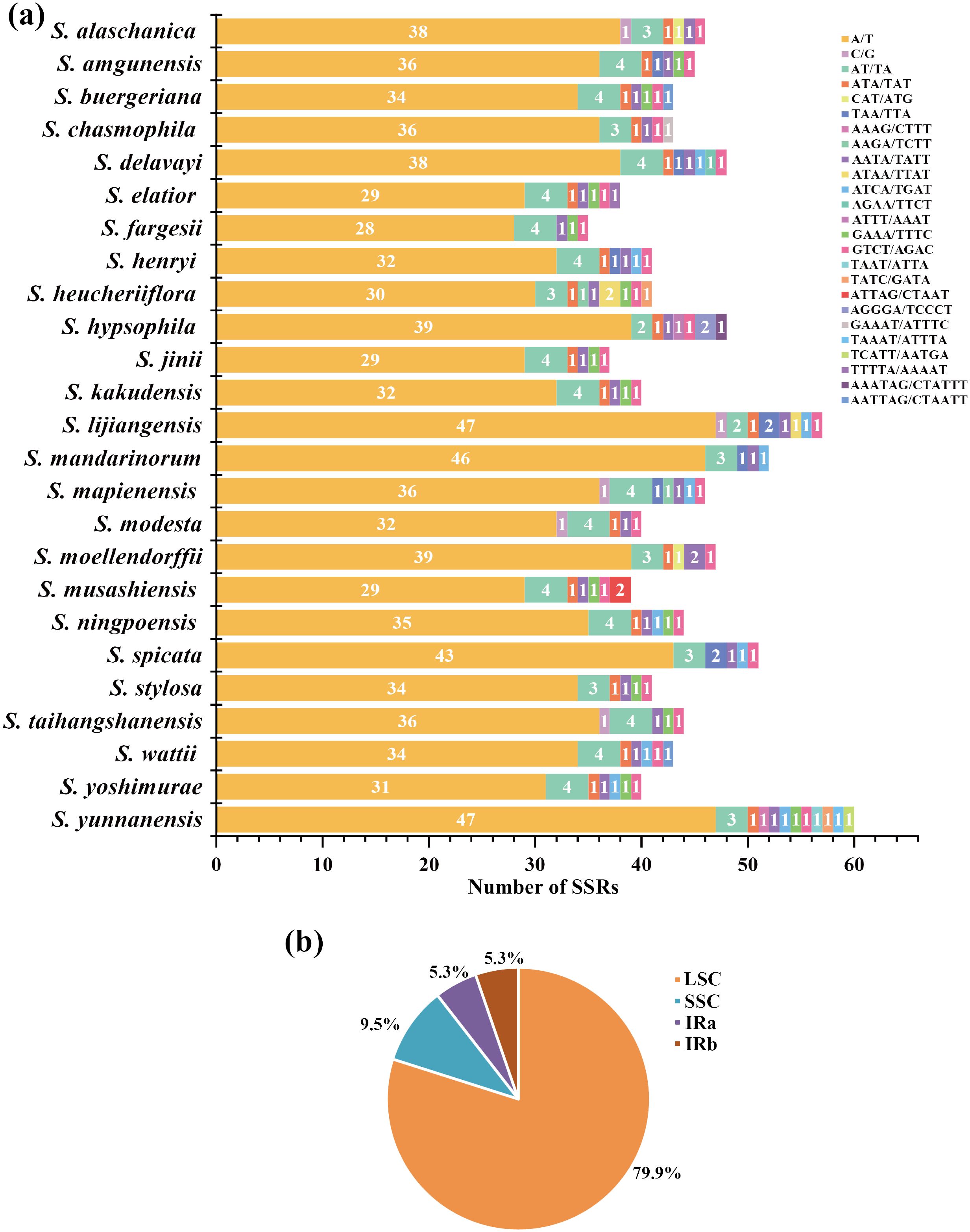
Figure 3. (A) Type and number of SSRs in 25 East Asian Scrophularia plastomes. (B) SSRs locus distribution (including LSC, SSC, IRa and IRb) of East Asian Scrophularia plastomes.
We conducted a statistical analysis of relative synonymous codon usage (RSCU) in 25 East Asian Scrophularia species, and the results were consistent across different species. The most and least frequently used amino acids were Leucine (Leu) (126,428) and Tryptophan (Trp) (17,404), respectively. Among the 30 codons analyzed, the RSCU values for each Scrophularia individual were greater than 1, indicating a preference for these codons. Among these preferential codons, the codon for Arginine (AGA) exhibited the highest preference, with an average RSCU value of 1.95. The codons UGG and AUG did not show any preference (RSCU = 1) (Figure 4; Supplementary Table 8). The effective number of codons (ENC) value typically ranges from 20 to 61, with lower values indicating stronger bias in codon usage away from random selection (Wu et al., 2007; Li et al., 2023b). Our research results revealed an ENC range of 55.36 (S. elatior) to 56.19 (S. hypsophila) across the 25 Scrophularia plastomes, with an average of 55.78.
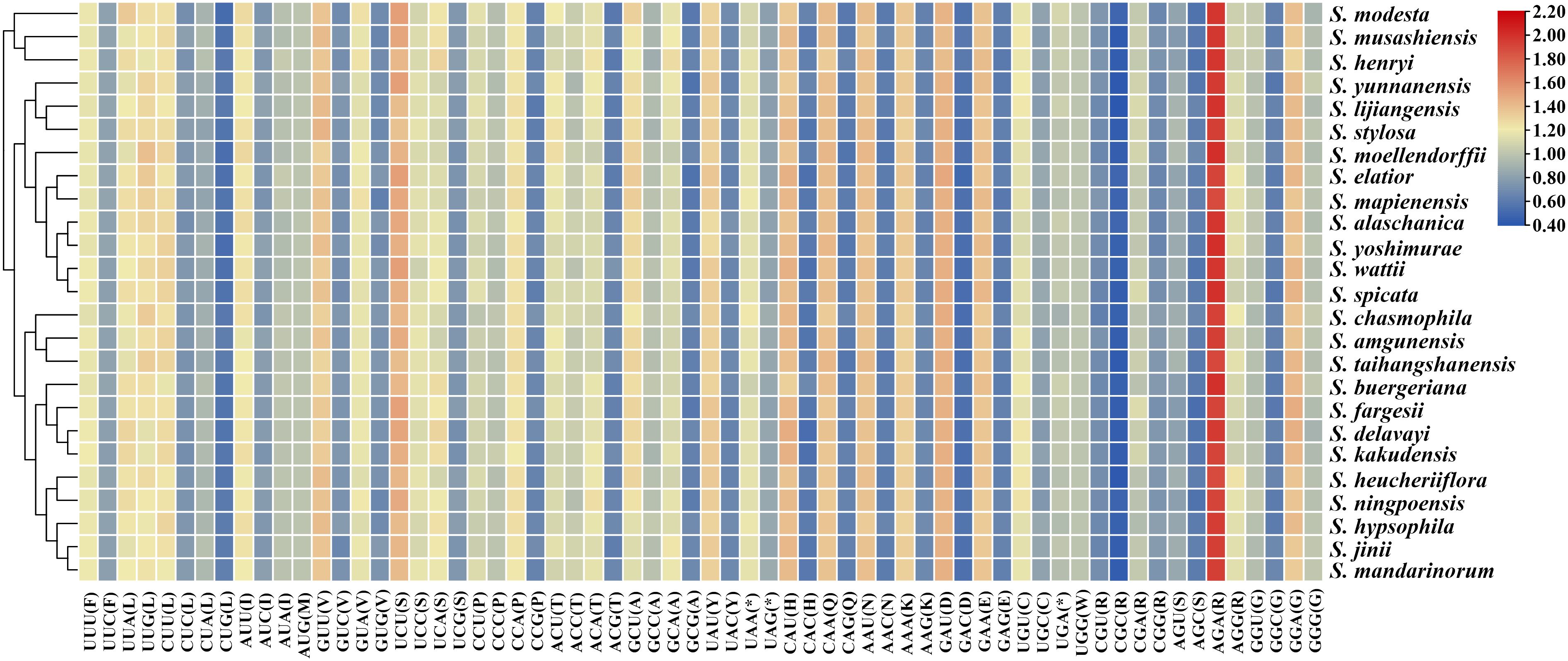
Figure 4. Heatmap of Relative Synonymous Codon Usage (RSCU) values for 25 East Asian Scrophularia plastomes.
Through the whole-genome alignment, we found minimal differences between the intra-genic and intergenic regions across the global view (Supplementary Figure 1). To analyze this further, we used Mauve for collinearity analysis, where homologous regions were shown in the same color. The plastome sequences of the 25 Scrophularia species displayed normal and single blocks. The homologous blocks were aligned in a linear manner, indicating a lack of rearrangements and inversions, resulting in good collinearity (Supplementary Figure 2).
Comparison of 25 plastomes of East Asian Scrophularia species revealed minor variations in the expansion and contraction of the IR regions. The IRa/SSC and IRb/LSC boundaries extended into the ycf1 and rps19 genes, leading to the generation of pseudogenes. In IRa, the length of the pseudogene ycf1 ranged from 209 to 978 bp, with the majority being around 870 bp. Specifically, S. chasmophila had a pseudogene ycf1 length of 978 bp, S. delavayi had 900 bp, and S. kakudensis had 231 bp. While the ycf1 gene, in those species, had a length ranging from 4,386 to 4,493 bp in the SSC region. Except for S. kakudensis, S. buergeriana, and S. ningpoensis, the pseudogene rps19 genes in the IRa region were at a distance of 0 bp from the LSC/IRa junction (JLA). The rps19 gene located at the JLB extended into the LSC region with a length of 238-252 bp (252 bp in S. wattii). In S. moellendorffii, the rpl2 gene completely situated in the IRb region experienced a slight contraction. It had a length of 1,483 bp, distinguishing it from the 1,492 bp length found in other species. Notably, in S. chasmophila, the ndhF gene expanded from the SSC region into the IRb region with an expansion length of 28 bp, while its length in the SSC region was 2,210 bp. In contrast, in the remaining species, the ndhF genes were entirely within the SSC region, with lengths mostly around 2,232 bp (Figure 5).

Figure 5. The contraction and expansion diagram of the IR region in the plastomes. The positions of LSC, IR, and SSC junctions were compared among 25 Scrophularia species. JLB stands for the junction between the long single copy and reverse repeat (LSC/IRb), JSB stands for the junction between reverse repeat and short single copy (IRb/SSC), JSA stands for the junction between short single copy and forward repeat (SSC/IRa), and JLA stands for the junction between forward repeat and long single copy (IRa/LSC).
A selection pressure analysis was performed on the protein-coding genes of 25 East Asian Scrophularia plastomes. Among the 80 analyzed protein-coding genes, the average Ka/Ks ratio was found to be 0.1172. The most conserved genes showed an average Ka/Ks value of 0 (excluding NA, where NA indicates Ks = 0, meaning no synonymous mutations), indicating strong purifying selection pressure. These genes include petB, petG, petN, psaC, psbA, psbD, psbE, psbI, psbJ, psbL, psbM, psbN, psbT, psbZ, and rps7. The top three Ka/Ks values were 2.6677 for ndhF gene in S. chasmophila, 2.5567 for ycf2 gene in S. fargesii, and 2.3657 for ndhF gene in S. musashiensis (Figure 6; Supplementary Table 9).
Using a Perl script, we extracted 80 CDS and 101 IGS from the plastomes of 25 East Asian Scrophularia species. Subsequently, nucleotide diversity (Pi) analysis was conducted using DnaSP. The Pi values ranged from 0 to 0.00694 in CDS and from 0 to 0.005691 in IGS. The IGS exhibited higher levels of polymorphism compared to the CDS. Higher Pi values were observed in CDS such as ycf1 (0.00694), matK (0.00645), rpl32 (0.00643), ndhF (0.00638), psbK (0.00592), and rps8 (0.00569) (Figure 7A; Supplementary Table 10). Similarly, IGS including trnH-GUG-psbA (0.05691), ndhD-psaC (0.03392), psbT-psbN (0.02508), ndhK-ndhC (0.01976), rpl32-trnL-UAG (0.01798), psbB-psbT (0.01778), petD-rpoA (0.01726), and rps18-rpl20 (0.01709) exhibited higher Pi values (Figure 7B; Supplementary Table 10).
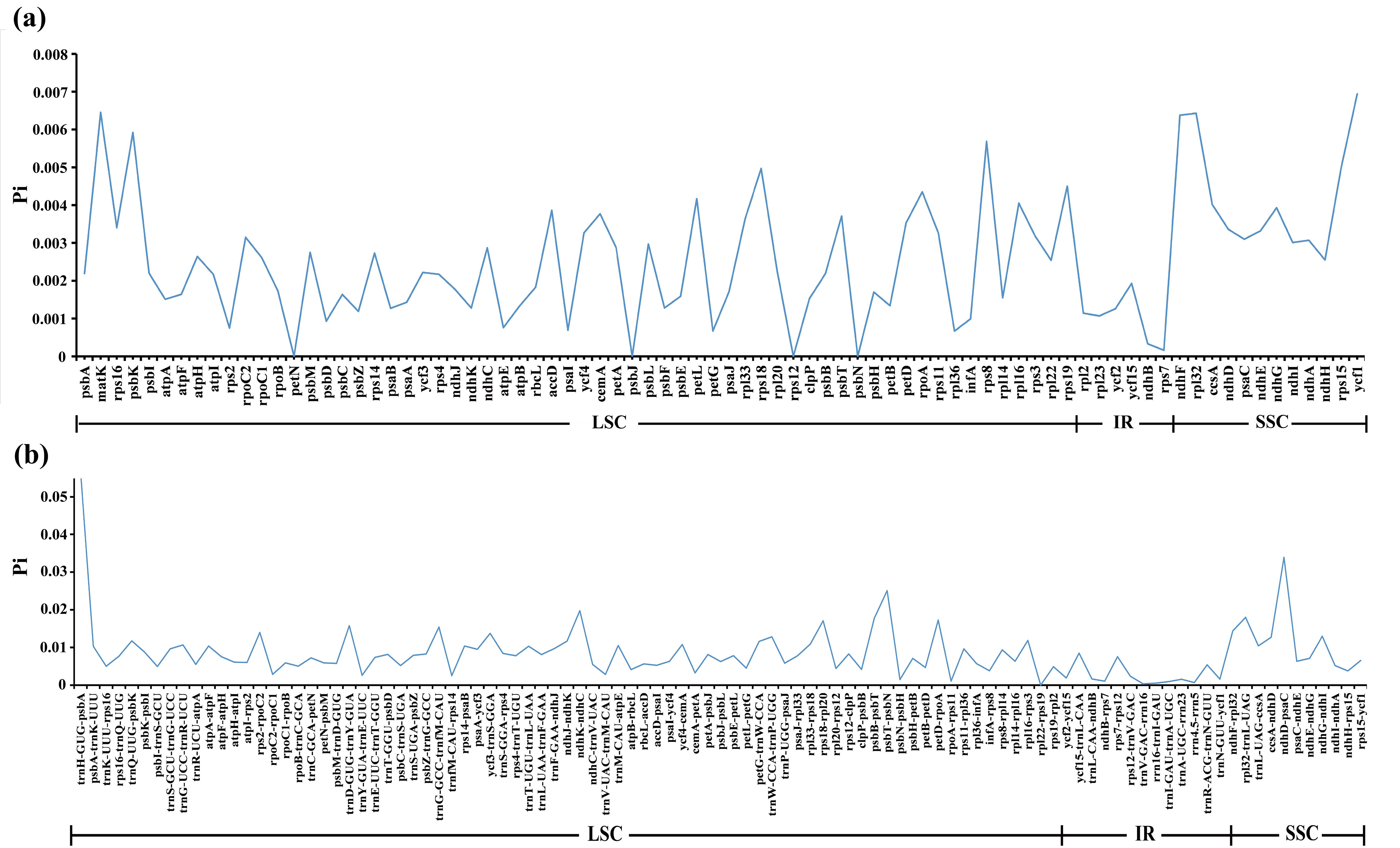
Figure 7. Comparison of nucleotide diversity (Pi) values. (A) among 80 CDS of 25 East Asian Scrophularia species. (B) among 101 IGS of 25 East Asian Scrophularia species.
Based on the analysis of complete plastome sequences and CDS data from 44 species, we constructed four phylogenetic trees using Bayesian inference (BI) and maximum likelihood (ML) methods. The topologies of these trees were so similar that we decided to show the tree inferred from the Maximum Likelihood method (Figure 8; Supplementary Figure 3). The majority of nodes received strong support (BI-PP/ML-BS = 1/100). Scrophulariaceae species throughout the tree could be categorized into three distinct monophyletic groups, including tribe Scrophularieae, tribe Buddlejeae, and tribes Myoporeae + Leucophylleae. The genus Scrophularia could be further divided into two sections: Scrophularia sect. Caninae and S. sect. Scrophularia; the 25 newly studied species belong to the latter section. The phylogenetic tree robustly supported that the 25 species fell into six monophyletic clades (A-F). Clade F was sister group to clades A-E. Within the core of the phylogenetic tree, the other clades formed a topology of [clade C + (clade A + B)].
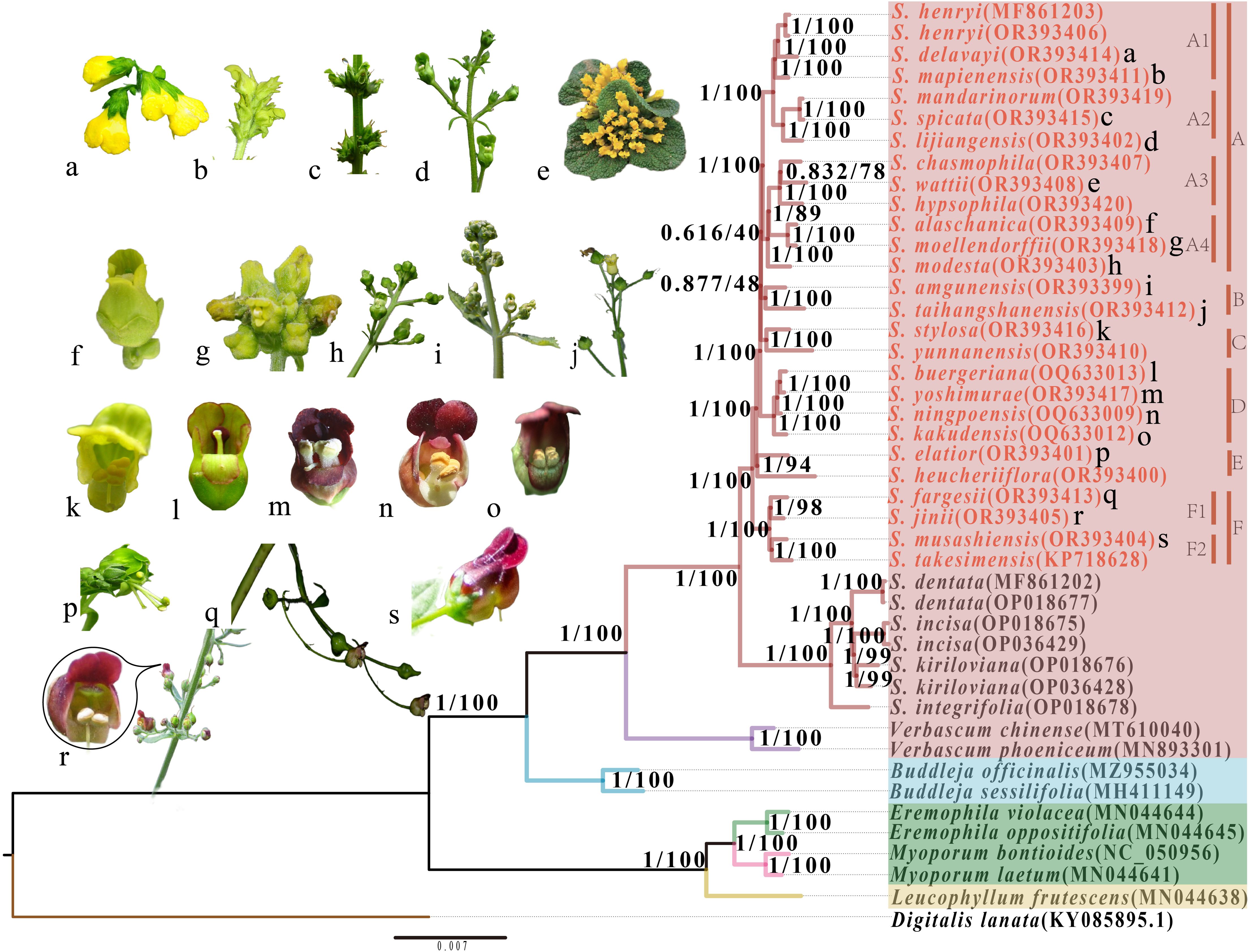
Figure 8. A phylogenetic tree of 30 Scrophularia species inferred from maximum likelihood based on the plastome sequence dataset. Support values above the branches, assessed by two methods (ML, BI), are listed as the order PP (posterior probability)/BS (bootstrap support). Genbank accession numbers of all species are given within the parentheses. Rectangular blocks of red, blue, green, and yellow represent the tribes Scrophularieae, Buddlejeae, Myoporeae and Leucophylleae, respectively. The four tribes all belong to the Scrophulariaceae, while Digitalis lanata belongs to the Plantaginaceae. The red branches represent Scrophularia, with the remaining species being outgroups. Within the red block, red font indicates Scrophularia sect. Scrophularia, while black font indicates Scrophularia sect. Caninae. The capital letters A-F represent different clades within S. sect. Scrophularia studied. Lower case letters a-s correspond to images of some species given on the left.
The plastome structure is generally conserved. This conservation indicates the presence of widespread evolutionary selective pressures associated with photosynthesis, which is the primary function of chloroplasts (Bungard, 2004). The majority of nonsynonymous substitutions are under purifying selection, implying that in most cases, natural selection eliminates harmful mutations and maintains amino acids unchanged (Hughes et al., 2008). In our study, the Ka/Ks values of most of genes were less than 1 (98.45%) in these Scrophularia species, while a few genes were greater than 1 (1.55%). It suggests that the majority of genes in the Scrophularia plastome are under purifying selection, with a few under significant positive selection, such as ycf2 gene (S. modesta, S. chasmophila, S. alaschanica, S. stylosa, S. yoshimurae, S. moellendorffii, S. hypsophila, S. buergeriana, S. ningpoensis) and ndhF gene (S. elatior, S. musashiensis, S. chasmophila, S. fargesii, S. delavayi, S. stylosa, S. yoshimurae, S. buergeriana, S. ningpoensis) (Figure 6).
Repetitive plastome sequences are crucial for genome rearrangement and genetic variation (Qi et al., 2017; Wei et al., 2020; Yang et al., 2022b). Within our study, a total of 1,084 repeat sequences were detected, the loci containing these repetitive sequences are critical hotspots for genomic reconfiguration. They provide information for understanding the evolutionary history and sequence divergence of plant species (Zong et al., 2019; Sun et al., 2020; Chong et al., 2022). Furthermore, they may provide abundant information for population studies and phylogenetic analysis (Gao et al., 2009; Nie et al., 2012; Zong et al., 2019). Of SSRs, 1040 (93.86%) consisted of A/T bp, with a notably low frequency of C and G nucleotides (0.45%), a pattern also observed in other medicinal plants such as Gentiana (Ni et al., 2017), Alpinia (Li et al., 2020), and Aconitum (Niu et al., 2023). SSRs in the plastome are frequently used as genetic markers in population genetics and evolutionary studies (Yang et al., 2016; Guo et al., 2020; Chen et al., 2023). Among the 25 samples of Scrophularia, a total of 7 common SSRs loci were identified. Analyzing similar SSRs in comparable genetic regions could be a potential approach for marking East Asian Scrophularia species for future population genetics studies, germplasm evaluation and resource conservation.
DNA sequence data from diverse organisms clearly show that synonymous codons for any given amino acid are not used with equal frequency, even though the choices among codons should be equivalent in terms of protein structure (Ikemura, 1985). The genomic GC content emerged as the strongest single determinant of codon usage variation across species (Plotkin and Kudla, 2011). The average GC content of the first, second, and third codon positions in the plastomes was 37.93%, 37.89%, and 38.15%, respectively. The overall GC3 content ranged from 37.87% (S. taihangshanensis) to 38.09% (S. jinii), all below 50%. There are a total of 28 codons with RSCU values greater than 1, of which 25 end with A/U, indicating a preference for codons ending in A/U in the plastomes of the 25 East Asian Scrophularia species. (Supplementary Table 8). The result is consistent with our previous research on the plastomes of Scrophularia (Wang et al., 2022; Guo et al., 2023b). Although the number and order of genes in the plastome are generally conserved, the IR regions expansion and contraction, a phenomenon known as “ebb and flow in plastomes”, is the main mechanism responsible for the variation in the plastome length of higher plant (Goulding et al., 1996; Kim and Lee, 2004; Zhu et al., 2016; Yin et al., 2018). Similar to many medicinal plants, the high GC content in the IR region may be caused by the elevated GC content of rRNA and tRNA in this region (Deng et al., 2021; Lu et al., 2022), as observed in various species such as Salvia (Liang et al., 2019), Polygonum (Guo et al., 2022), and Atractylodes (Xu et al., 2023). Additionally, the GC content in the IR regions is higher than in the LSC and SSC regions. Moreover, among 14 regions with higher Pi values, the majority were located in the LSC (64.28%), while a smaller portion was found in the SSC (35.72%). Notably, none of them were found in the IR regions. Overall, the IR displayed a lower level of polymorphism. This is consistent with previous reports, indicating that the IR regions are more conserved compared to the LSC and SSC regions (Wang et al., 2022; Guo et al., 2023b).
Comparative genomic analysis can contribute to gaining a comprehensive understanding of a genus (Sivashankari and Shanmughavel, 2007; Tonti-Filippini et al., 2017). Compared to protein-coding regions, the non-coding regions exhibited higher diversity and variability. The regions with overall significant differences, which are trnH-GUG-psbA, rps16-trnQ-UUG, and trnT-UGU-trnL-UAA, are in good agreement with the Pi calculation results. Research has shown that polymorphic indels and nucleotides of trnH-psbA could be used to authenticate most Kaempferia species (Techaprasan et al., 2010). The trnH-GUG-psbA has been supported by studies as a DNA barcode sequence for distinguishing the medicinal plant Datura metel and its adulterants (Bi et al., 2022). Therefore, the divergent hotspot regions could serve as candidate markers for future identification analyses within the Scrophularia genus. Developing universal primers targeting these hotspot regions would be of importance in revealing the molecular phylogenetics and conservation genetics of other Scrophularia species (Jia et al., 2017; Kong et al., 2017; Liu et al., 2021).
According to phylogenetic tree, tribe Scrophularieae was sister to tribe Buddlejeae, and together they formed a sister group to tribes Myoporeae + Leucophylleae. The interspecies relationships among these tribes were reasonably resolved and have also been confirmed in previous literature (Wang, 2015; Gao, 2021). We subdivided Scrophularia sect. Scrophularia into six clades (A-F) and discussed them based on morphological characteristics in the following. Subclades A1 and A2 formed a clade. In subclade A1, the stamens of S. mapienensis are about the same length as the lower lip of the corolla, which is yellow-white or purple (Figure 8b). The stamens of the other two species are half the length of the lower lip, and their bell-shaped or spherical corollas are yellow-green or yellow (Figure 8a). In subclade A2, S. spicata and S. mandarinorum, as sister taxa, can grow to over 1 meter in height, and the corolla color of species within subclade A2 is green or yellow-green (Figures 8c, d). Subclades A3 (S. chasmophila, S. wattii, and S. hypsophila) and A4 (S. alaschanica and S. moellendorffii) were sister groups and together they formed a sister relationship with S. modesta, which has green or yellowish-green corolla (Figure 8h). With the exception of S. chasmophila, which features a yellow-green corolla, the species belonging to subclades A3 and A4 are characterized by their bright yellow corollas (Figures 8e-g). The species of Scrophularia within subclade A3 are all perennial small herbaceous plants, especially S. wattii with scalelike and rosette leaves appressed to ground (Figure 8e). Clade B included S. taihangshanensis and S. amgunensis, both of which have yellow-green corollas (Figures 8i, j). S. stylosa with light yellow corolla and obcordate staminodes (Figure 8k) was sister to S. yunnanensis with green corolla, the two species constituting clade C. S. ningpoensis and its common medicinal substitutes or adulterants of S. buergeriana, S. kakudensis, and S. yoshimurae formed a monophyletic clade D. The corolla of S. buergeriana is green and S. kakudensis has outside green and inside purplish brown corolla, while the corollas of the other two species are purple (Figures 8l-o). Clade E included S. elatior and S. heucheriiflora, with their stamens extending beyond the green corolla (Figure 8p). The endemic Chinese species S. fargesii and S. jinii are sister taxa (subclade F1), while the Japanese endemics S. musashiensis and Korean endemics S. takesimensis clustered as subclade F2. These two subclades shared a common ancestor, with purple-red corollas and slender flower stalks (Figures 8q-s). The cyme inflorescence of S. fargesii consists of 1-3 or 5 flowers, while that of S. jinii comprises 1-7 flowers. As for leaf margin, S. jinii is deeply double serrate but S. fargesii is unequally double serrate (Wang et al., 2018).
However, there were also branches with relatively low support rate and short length in the phylogenetic trees, such as the relationships between Clade C and Clade (A+B), as well as between Clade B and Clade A. We speculate that there may have been a rapid radiation. Given the potential for minimal genetic disparities among species during rapid differentiation, the swift evolutionary radiation of species in a condensed timeframe may contribute to diminished support rates within the phylogenetic tree (Guo et al., 2023a; Liu et al., 2024). Consequently, these branches with low supports may stem from the accumulation of multiple lineages over a short period. To validate this phenomenon, additional studies will be needed increasing the number of individuals and delving into the population genomics of the East Asian Scrophularia. By examining variation information, we can discuss the genetic structure, gene flow, speciation mechanisms, and evolutionary dynamics of East Asian Scrophularia populations.
It is also important to consider that, while plastid genomes are helpful for constructing phylogenetic trees, they still have certain limitations. For example, they do not adequately address issues such as ILS, hybridization, and whole genome duplication (McLay et al., 2023; Brown et al., 2024; Session, 2024; Wang et al., 2024). As we know, introgression and ILS are common mechanisms that lead to cytonuclear discordance. Although cytonuclear discordance is widespread and often considered an obstacle in phylogenetic and taxonomic studies, it can also provide valuable information (Duan et al., 2023). Currently, S. ningpoensis is the primary species cultivated on a large scale. Through genomic research on the East Asian Scrophularia, we anticipate gaining insights into the genus, which will aid in molecular breeding and unlock the medicinal potential of a broader range of Scrophularia species. It will contribute to the rational exploration, full utilization, and sustainable cultivation and harvesting of medicinal plant resources, ensuring their conservation and sustainable use for future generations.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
XW: Formal analysis, Methodology, Software, Writing – original draft, Writing – review & editing. LG: Formal analysis, Methodology, Software, Writing – original draft, Writing – review & editing. LD: Formal analysis, Visualization, Writing – review & editing. LM: Data curation, Resources, Writing – review & editing. RW: Conceptualization, Funding acquisition, Supervision, Writing – review & editing. PL: Conceptualization, Investigation, Resources, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was financially supported by the Zhejiang Province Basic Public Welfare Research Program Project (LGN22H280005), Key Research and Development Program of Zhejiang Province (grant no. 2023C02017), the Fundamental Research Funds of Zhejiang Sci-Tech University (24042128-Y), General Scientific Research Projects of the Department of Education of Zhejiang Province (No. Y202353132).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1439206/full#supplementary-material
An, Y., Harrison, T. M. (2000). Geologic evolution of the Himalayan-Tibetan orogen. Annu. Rev. Earth lanetary Sci. 28, 211–280. doi: 10.1146/annurev.earth.28.1.211
Attar, F., Riahi, M., Daemi, F., Aghabeigi, F. (2011). Preliminary molecular phylogeny of Eurasian Scrophularia (Scrophulariaceae) based on DNA sequence data from trnS-trnG and ITS regions. Plant Biosyst. - Plant Biosyst. 145, 1–9. doi: 10.1080/11263504.2011.590826
Azani, N., Bruneau, A., Wojciechowski, M. F., Zarre, S. (2019). Miocene climate change as a driving force for multiple origins of annual species in Astragalus (Fabaceae, Papilionoideae). Mol. Phylogenet Evol. 137, 210–221. doi: 10.1016/j.ympev.2019.05.008
Beier, S., Thiel, T., Münch, T., Scholz, U., Mascher, M. (2017). MISA-web: a web server for microsatellite prediction. Bioinformatics 33, 2583–2585. doi: 10.1093/bioinformatics/btx198
Bi, G., Ding, Y., Wang, L., Hu, S., Li, H., Lei, M., et al. (2022). Characterization and phylogenetic analysis of complete chloroplast genome of Datura metel and Brugmansia arborea. Chin. Traditional Herbal Drugs 53, 7191–7200. doi: 10.7501/j.issn.0253-2670.2022.22.023
Bolger, A. M., Lohse, M., Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Boufford, D. E., Ōba, H. (Eds.) (1998). Sino-Japanese flora: its characteristics and diversification (Tokyo: University Museum, University of Tokyo).
Brown, M. R., Abbott, R. J., Twyford, A. D. (2024). The emerging importance of cross-ploidy hybridisation and introgression. Mol. Ecol. 33, e17315. doi: 10.1111/mec.17315
Brudno, M., Malde, S., Poliakov, A., Do, C. B., Couronne, O., Dubchak, I., et al. (2003). Glocal alignment: finding rearrangements during alignment. Bioinformatics 19, i54–i62. doi: 10.1093/bioinformatics/btg1005
Bungard, R. A. (2004). Photosynthetic evolution in parasitic plants: insight from the chloroplast genome. BioEssays 26, 235–247. doi: 10.1002/bies.10405
Chen, C. (2011). Studies on domestication & phylogeography of Scrophularia ningpoensis Hemsley and phylogeny of Eastern Asia Scrophularia. [Doctoral dissertation]. (Zhejiang (China): Zhejiang University).
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, L. (2014). Studies on dynamics of characteristic secondary metabolites in Scrophularia ningpoensis in relation to environmental conditions and genetic variation. [Master's dissertation]. (Zhejiang (China): Zhejiang University).
Chen, X., Mao, Y., Chai, W., Yan, K., Liang, Z., Xia, P. (2023). Genome-wide identification and expression analysis of MYB gene family under nitrogen stress in Panax notoginseng. Protoplasma 260, 189–205. doi: 10.1007/s00709-022-01770-1
Chen, Y., Gong, M., Nie, X., Qi, Z., Liu, X., Jin, Q., et al. (2021). Characterization of botanical origin of selected popular purple Eleutherococcus tea grown in Yunnan province of China and quantification of Its anthocyanins using spectrophotometric method. Food Sci. Technol. 42, e91121. doi: 10.1590/fst.91121
Chen, Z., Zhang, X., Hu, H., Niu, Y., Ye, J., Zhang, Q. (2022b). Progress and prospect of phytogeography in China. Acta Geographica Sin. 77, 120–132. doi: 10.11821/dlxb202201009
Chen, M., Zhang, M., Liang, Z., He, Q. (2022a). Characterization and comparative analysis of chloroplast genomes in five Uncaria species endemic to China. Int. J. Mol. Sci. 23, 11617. doi: 10.3390/ijms231911617
Chinese Pharmacopoeia Commission (2020). Pharmacopoeia of the People's Republic of China (Peijing: China Medical Science and Technology Press).
Chong, X., Li, Y., Yan, M., Wang, Y., Li, M., Zhou, Y., et al. (2022). Comparative chloroplast genome analysis of 10 Ilex species and the development of species-specific identification markers. Ind. Crops Products 187, 115408. doi: 10.1016/j.indcrop.2022.115408
Cui, C. (2023). Standardization Research on English Translation of Core Terms and Main Components in TCM. [Doctoral dissertation]. (Beijing (China): China Academy of Chinese Medical Sciences).
Daniell, H., Lin, C., Yu, M., Chang, W. (2016). Chloroplast genomes: diversity, evolution, and applications in genetic engineering. Genome Biol. 17, 134. doi: 10.1186/s13059-016-1004-2
Darling, A. C., Mau, B., Blattner, F. R., Perna, N. T. (2004). Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14, 1394–1403. doi: 10.1101/gr.2289704
Darriba, D., Taboada, G. L., Doallo, R., Posada, D. (2012). jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9, 772–772. doi: 10.1038/nmeth.2109
Deng, T. (2015). Origin and diversification of important character taxa in the East Asian flora. [Doctoral dissertation]. (Yunnan (China): YunNan university).
Deng, Y., Liu, X., Zhuo, D., Zeng, F. (2021). Structural characteristics and phylogenetic relationship of chloroplast genome of Syzygium acuminatissimum. Guangdong Agric. Sci. 48, 38–46. doi: 10.16768/j.issn.1004-874X.2021.06.006
de Santos Galíndez, J., Díaz Lanza, A. M., Fernández Matellano, L. (2002). Biologically active substances from the genus Scrophularia. Pharm. Biol. 40, 45–59. doi: 10.1076/phbi.40.1.45.5864
Duan, L., Fu, L., Chen, H. (2023). Phylogenomic cytonuclear discordance and evolutionary histories of plants and animals. Sci. China Life Sci. 66, 2946–2948. doi: 10.1007/s11427-023-2456-6
Feng, J., Jin, X., Zhang, S., Yang, J., Fei, S., Huang, Y., et al. (2022). Smilax weniae, a new species of Smilacaceae from limestone areas bordering Guizhou and Guangxi, China. Plants 11, 1032. doi: 10.3390/plants11081032
Editorial Board of the Chinese Materia Medica (2003). Chinese Materia Medica: Mongolian Medicine Volume. Shanghai (China): Shanghai Scientific & Technical Publishers.
Frazer, K. A., Pachter, L., Poliakov, A., Rubin, E. M., Dubchak, I. (2004). VISTA: computational tools for comparative genomics. Nucleic Acids Res. 32, W273–W279. doi: 10.1093/nar/gkh458
Gao, J. (2021). Phylogenetic and biogeographical research of Scrophularia incisa complex. [Master's dissertation]. (Zhejiang (China): Zhejiang Sci-Tech University).
Gao, L., Su, Y., Wang, T. (2010). Plastid genome sequencing, comparative genomics, and phylogenomics: current status and prospects. J. Systematics Evol. 48, 77–93. doi: 10.1111/j.1759-6831.2010.00071.x
Gao, L., Yi, X., Yang, Y., Su, Y., Wang, T. (2009). Complete chloroplast genome sequence of a tree fern Alsophila spinulosa: insights into evolutionary changes in fern chloroplast genomes. BMC evolutionary Biol. 9, 1–14. doi: 10.1186/1471-2148-9-130
Goulding, S. E., Wolfe, K., Olmstead, R., Morden, C. (1996). Ebb and flow of the chloroplast inverted repeat. Mol. Gen. Genet. MGG 252, 195–206. doi: 10.1007/BF02173220
Guo, L., Guo, S., Xu, J., He, L., Carlson, J. E., Hou, X. (2020). Phylogenetic analysis based on chloroplast genome uncover evolutionary relationship of all the nine species and six cultivars of tree peony. Ind. Crops Products 153, 112567. doi: 10.1016/j.indcrop.2020.112567
Guo, S., Liao, X., Chen, S., Liao, B., Guo, Y., Cheng, R., et al. (2022). A comparative analysis of the chloroplast genomes of four Polygonum medicinal plants. Front. Genet. 13. doi: 10.3389/fgene.2022.764534
Guo, C., Luo, Y., Gao, L., Yi, T., Li, H., Yang, J., et al. (2023a). Phylogenomics and the flowering plant tree of life. J. Integr. Plant Biol. 65, 299–323. doi: 10.1111/jipb.13415
Guo, L., Wang, X., Wang, R., Li, P. (2023b). Characterization and comparative analysis of chloroplast genomes of medicinal herb Scrophularia ningpoensis and its common adulterants (Scrophulariaceae). Int. J. Mol. Sci. 24, 10034. doi: 10.3390/ijms241210034
He, B., Jiang, J., Xu, D., Wang, S. (2020). Production and sale status, development countermeasures of Chinese medicinal materials industry in Zhejiang Province in 2019. J. Zhejiang Agric. Sci. 61, 1776–1779+1800. doi: 10.16178/j.issn.0528-9017.20200922
Hong, D. (1983). The distribution of Scrophulariaceae in the holarctic with special reference to the floristic relationships between eastern Asia and eastern north America. Ann. Missouri Botanical Garden 70, 701–712. doi: 10.2307/2398985
Hong, D., Yang, H., Jin, C., Holmgren, N. H., Fischer, M. A., Mill, R. R. (1998). “Scrophulariaceae,” in Flora of China. Ed. Wu, Z.-Y. (Missouri Botanical Garden Press, Science Press, St. Louis, MI, US, Beijing, China).
Huang, L. (2018). Shennongjia Chinese Medicinal Resources Illustrated Manual, Vol. 4. Fuzhou (China): Fujian Science and Technology Publishing House.
Hughes, A. L., Friedman, R., Rivailler, P., French, J. O. (2008). Synonymous and nonsynonymous polymorphisms versus divergences in bacterial genomes. Mol. Biol. Evol. 25, 2199–2209. doi: 10.1093/molbev/msn166
Ikemura, T. (1985). Codon usage and tRNA content in unicellular and multicellular organisms. Mol. Biol. Evol. 2, 13–34. doi: 10.1093/oxfordjournals.molbev.a040335
Jia, Q., Wang, J., Zhu, J., Hua, W., Shang, Y., Yang, J., et al. (2017). Toward identification of black lemma and pericarp gene Blp1 in barley combining bulked segregant analysis and specific-locus amplified fragment sequencing. Front. Plant Sci. 8, 1414. doi: 10.3389/fpls.2017.01414
Katoh, K., Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kim, K., Lee, H. (2004). Complete chloroplast genome sequences from Korean ginseng (Panax schinseng Nees) and comparative analysis of sequence evolution among 17 vascular plants. DNA Res. 11, 247–261. doi: 10.1093/dnares/11.4.247
Kong, H., Liu, W., Yao, G., Gong, W. (2017). A comparison of chloroplast genome sequences in Aconitum (Ranunculaceae): a traditional herbal medicinal genus. PeerJ 5, e4018. doi: 10.7717/peerj.4018
Kurtz, S., Schleiermacher, C. (1999). REPuter: fast computation of maximal repeats in complete genomes. Bioinf. (Oxford England) 15, 426–427. doi: 10.1093/bioinformatics/15.5.426
Lee, H., Kim, H., Lee, D., Choi, B., Yang, S. (2021). Scrophulariae radix: an overview of its biological activities and nutraceutical and pharmaceutical applications. Molecules 26, 5250. doi: 10.3390/molecules26175250
Li, D. (2008). Floristics and plant biogeography in China. J. Integr. Plant Biol. 50, 771–777. doi: 10.1111/j.1744-7909.2008.00711.x
Li, H., Guo, Q., Xu, L., Gao, H., Liu, L., Zhou, X. (2023a). CPJSdraw: analysis and visualization of junction sites of chloroplast genomes. PeerJ 11, e15326. doi: 10.7717/peerj.15326
Li, Y., Jiang, S., Zhu, D. (1999). Research progress on the chemical constituents and pharmacological activities of Scrophularia species. Chin. Traditional Herbal Drugs 04), 307–310.
Li, R., Kraft, N. J., Yu, H., Li, H. (2015). Seed plant phylogenetic diversity and species richness in conservation planning within a global biodiversity hotspot in eastern Asia. Conserv. Biol. 29, 1552–1562. doi: 10.1111/cobi.12586
Li, Q., Luo, Y., Sha, A., Xiao, W., Xiong, Z., Chen, X., et al. (2023b). Analysis of synonymous codon usage patterns in mitochondrial genomes of nine Amanita species. Front. Microbiol. 14. doi: 10.3389/fmicb.2023.1134228
Li, D., Zhu, G., Xu, Y., Ye, Y., Liu, J. (2020). Complete chloroplast genomes of three medicinal Alpinia species: genome organization, comparative analyses and phylogenetic relationships in family Zingiberaceae. Plants 9, 286. doi: 10.3390/plants9020286
Liang, C., Wang, L., Lei, J., Duan, B., Ma, W., Xiao, S., et al. (2019). A comparative analysis of the chloroplast genomes of four Salvia medicinal plants. Engineering 5, 907–915. doi: 10.1016/j.eng.2019.01.017
Lin, N. (2020). Study on phylogenetic evolution and phylogeoregion of featured taxa in the East Asian flora. [Doctoral dissertation]. (Wuhan (China): Wuhan Botanical Garden, Chinese Academy of Sciences).
Liu, S., Ni, Y., Li, J., Zhang, X., Yang, H., Chen, H., et al. (2023). CPGView: A package for visualizing detailed chloroplast genome structures. Mol. Ecol. Resour. 23, 694–704. doi: 10.1111/1755-0998.13729
Liu, J., Wang, Y., Wang, A., Hideaki, O., Abbott, R. J. (2006). Radiation and diversification within the Ligularia-Cremanthodium-Parasenecio complex (Asteraceae) triggered by uplift of the Qinghai-Tibetan Plateau. Mol. Phylogenet Evol. 38, 31–49. doi: 10.1016/j.ympev.2005.09.010
Liu, L., Zhang, Y., Li, P. (2021). Development of genomic resources for the genus Celtis (Cannabaceae) based on genome skimming data. Plant Diversity 43, 43–53. doi: 10.1016/j.pld.2020.09.005
Liu, J., Zhou, S., Liu, Y., Zhao, B., Yu, D., Zhong, M., et al. (2024). Genomes of Meniocus linifolius and Tetracme quadricornis reveal the ancestral karyotype and genomic features of core Brassicaceae. Plant Commun. 5, 100878. doi: 10.1016/j.xplc.2024.100878
Lu, R., Chen, M., Feng, Y., Yuan, N., Zhang, Y., Cao, M., et al. (2022). Comparative plastome analyses and genomic resource development in wild rice (Zizania spp., Poaceae) using genome skimming data. Ind. Crops Products 186, 115244. doi: 10.1016/j.indcrop.2022.115244
Manivannan, A., Soundararajan, P., Park, Y. G., Jeong, B. R. (2021). Physiological and proteomic insights into red and blue light-mediated enhancement of in vitro growth in Scrophularia kakudensis—A potential medicinal plant. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.607007
McLay, T. G. B., Fowler, R. M., Fahey, P. S., Murphy, D. J., Udovicic, F., Cantrill, D. J., et al. (2023). Phylogenomics reveals extreme gene tree discordance in a lineage of dominant trees: hybridization, introgression, and incomplete lineage sorting blur deep evolutionary relationships despite clear species groupings in Eucalyptus subgenus Eudesmia. Mol. Phylogenet. Evol. 187, 107869. doi: 10.1016/j.ympev.2023.107869
Milne, R. I., Abbott, R. J. (2002). “The origin and evolution of tertiary relict floras,” in Advances in Botanical Research (United States: Academic Press), 281–314.
Nam, H., Lee, A. Y., Seo, Y., Park, I., Yang, S., Chun, J., et al. (2020). Three Scrophularia Species (Scrophularia buergeriana, S. koraiensis, and S. takesimensis) Inhibit RANKL-Induced Osteoclast Differentiation in Bone Marrow-Derived Macrophages. Plants 9, 1656. doi: 10.3390/plants9121656
Navarro-Pérez, M. L., López, J., Fernández-Mazuecos, M., Rodríguez-Riaño, T., Vargas, P., Ortega-Olivencia, A. (2013). The role of birds and insects in pollination shifts of Scrophularia (Scrophulariaceae). Mol. Phylogenet. Evol. 69, 239–254. doi: 10.1016/j.ympev.2013.05.027
Nguyen, L.-T., Schmidt, H. A., Von Haeseler, A., Minh, B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Ni, L., Zhao, Z., Dorje, G., Ma, M. (2016). The complete chloroplast genome of Ye-Xing-Ba (Scrophularia dentata; Scrophulariaceae), an alpine Tibetan herb. PLoS One 11, e0158488. doi: 10.1371/journal.pone.0158488
Ni, L., Zhao, Z., Xu, H., Chen, S., Dorje, G. (2017). Chloroplast genome structures in Gentiana (Gentianaceae), based on three medicinal alpine plants used in Tibetan herbal medicine. Curr. Genet. 63, 241–252. doi: 10.1007/s00294-016-0631-1
Nie, X., Lv, S., Zhang, Y., Du, X., Wang, L., Biradar, S. S., et al. (2012). Complete chloroplast genome sequence of a major invasive species, crofton weed (Ageratina adenophora). PloS One 7, e36869. doi: 10.1371/journal.pone.0036869
Niu, Y., Su, T., Wu, C., Deng, J., Yang, F. (2023). Complete chloroplast genome sequences of the medicinal plant Aconitum transsectum (Ranunculaceae): comparative analysis and phylogenetic relationships. BMC Genomics 24, 1–15. doi: 10.1186/s12864-023-09180-0
Ortega Olivencia, A., Devesa Alcaraz, J. (1993). "Revisión del género Scrophularia L. (Scrophulariaceae) en la península Ibérica e Islas Baleares." in Ruizia (Madrid: Real Jardín Botánico Consejo Superior de Investigaciones Científicas), 11, 5–157.
Pasdaran, A., Hamedi, A. (2017). The genus Scrophularia: a source of iridoids and terpenoids with a diverse biological activity. Pharm. Biol. 55, 2211–2233. doi: 10.1080/13880209.2017.1397178
Plotkin, J. B., Kudla, G. (2011). Synonymous but not the same: the causes and consequences of codon bias. Nat. Rev. Genet. 12, 32–42. doi: 10.1038/nrg2899
Plunkett, G. M., Downie, S. R. (2000). Expansion and contraction of the chloroplast inverted repeat in Apiaceae subfamily Apioideae. Systematic Bot. 25, 648–667. doi: 10.2307/2666726
Qi, Z., Shen, C., Han, Y., Shen, W., Yang, M., Liu, J., et al. (2017). Development of microsatellite loci in Mediterranean sarsaparilla (Smilax aspera; Smilacaceae) using transcriptome data. Appl. Plant Sci. 5, 1700005. doi: 10.3732/apps.1700005
Raubeson, L. A., Peery, R., Chumley, T. W., Dziubek, C., Fourcade, H. M., Boore, J. L., et al. (2007). Comparative chloroplast genomics: analyses including new sequences from the angiosperms Nuphar advena and. Ranunculus macranthus. 8, 174. doi: 10.1186/1471-2164-8-174
Rice, P., Longden, I., Bleasby, A. (2000). EMBOSS: the European molecular biology open software suite. Trends Genet. 16, 276–277. doi: 10.1016/S0168-9525(00)02024-2
Ronquist, F., Huelsenbeck, J. P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574. doi: 10.1093/bioinformatics/btg180
Rozas, J., Ferrer-Mata, A., Sánchez-DelBarrio, J. C., Guirao-Rico, S., Librado, P., Ramos-Onsins, S. E., et al. (2017). DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 34, 3299–3302. doi: 10.1093/molbev/msx248
Sagare, A. P., Kuo, C., Chueh, F., Tsay, H. (2001). De novo regeneration of Scrophularia yoshimurae Yamazaki (Scrophulariaceae) and quantitative analysis of harpagoside, an iridoid glucoside, formed in aerial and underground parts of in vitro propagated and wild plants by HPLC. Biol. Pharm. Bull. 24, 1311–1315. doi: 10.1248/bpb.24.1311
Scheunert, A., Heubl, G. (2011). Phylogenetic relationships among New World Scrophularia L. (Scrophulariaceae): new insights inferred from DNA sequence data. Plant Systematics Evol. 291, 69–89. doi: 10.1007/s00606-010-0369-z
Scheunert, A., Heubl, G. (2014). Diversification of Scrophularia (Scrophulariaceae) in the Western Mediterranean and Macaronesia – Phylogenetic relationships, reticulate evolution and biogeographic patterns. Mol. Phylogenet. Evol. 70, 296–313. doi: 10.1016/j.ympev.2013.09.023
Scheunert, A., Heubl, G. (2017). Against all odds: reconstructing the evolutionary history of Scrophularia (Scrophulariaceae) despite high levels of incongruence and reticulate evolution. Organisms Diversity Evol. 17, 323–349. doi: 10.1007/s13127-016-0316-0
Session, A. M. (2024). Allopolyploid subgenome identification and implications for evolutionary analysis. Trends Genet. 40, 621–631. doi: 10.1016/j.tig.2024.03.008
Sharp, P. M., Li, W. (1987). The codon adaptation index-a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 15, 1281–1295. doi: 10.1093/nar/15.3.1281
Sivashankari, S., Shanmughavel, P. (2007). Comparative genomics-a perspective. Bioinformation 1, 376. doi: 10.6026/97320630001376
Sun, J., Wang, Y., Liu, Y., Xu, C., Yuan, Q., Guo, L., et al. (2020). Evolutionary and phylogenetic aspects of the chloroplast genome of Chaenomeles species. Sci. Rep. 10, 11466. doi: 10.1038/s41598-020-67943-1
Tang, Y., Li, L. (1996). On historical elements and Tertiary Precursors of eastern Asiatic angiospermous flora–based on phytogeography of Staphyleaceae, Morinaceae and Caprifoliaceae. J. Systematics Evol. 34, 453–478.
Techaprasan, J., Klinbunga, S., Ngamriabsakul, C., Jenjittikul, T. (2010). Genetic variation of Kaempferia (Zingiberaceae) in Thailand based on chloroplast DNA (psbA-trnH and petA-psbJ) sequences. Genet Mol Res. 9, 1957–1973. doi: 10.4238/vol9-4gmr873
Tonti-Filippini, J., Nevill, P. G., Dixon, K., Small, I. (2017). What can we do with 1000 plastid genomes? Plant J. 90, 808–818. doi: 10.1111/tpj.13491
Wang, R. (2015). Phylogeny and Biogeography of Scrophularia and Phylogeography of S. incisa Complex. [Doctoral dissertation]. (Zhejiang (China): Zhejiang University).
Wang, R., Gao, J., Feng, J., Yang, Z., Qi, Z., Li, P., et al. (2022). Comparative and phylogenetic analyses of complete chloroplast genomes of Scrophularia incisa complex (Scrophulariaceae). Genes 13, 1691. doi: 10.3390/genes13101691
Wang, J., Wang, J. (2007). The Evolution of Chinese Medicinal Material Varieties and Their Authenticity (Beijing: China Medical Science and Technology Press).
Wang, Y., Wu, X., Chen, Y., Xu, C., Wang, Y., Wang, Q. (2024). Phylogenomic analyses revealed widely occurring hybridization events across Elsholtzieae (Lamiaceae). Mol. Phylogenet. Evol. 198, 108112. doi: 10.1016/j.ympev.2024.108112
Wang, R., Xia, M., Tan, J., Chen, C., Jin, X., Li, P., et al. (2018). A new species of Scrophularia (Scrophulariaceae) from Hubei, China. Phytotaxa 350, 1. doi: 10.11646/phytotaxa.350.1.1
Wei, F., Tang, D., Wei, K., Qin, F., Li, L., Lin, Y., et al. (2020). The complete chloroplast genome sequence of the medicinal plant Sophora tonkinensis. Sci. Rep. 10, 12473. doi: 10.1038/s41598-020-69549-z
Wu, X., Wu, S., Ren, D., Zhu, Y., He, F. (2007). The analysis method and progress in the study of codon bias. Hereditas 29, 420–426. doi: 10.1360/yc-007-0420
Xiang, Y., Wang, X., Ding, L., Bai, X., Feng, Y., Qi, Z., et al. (2024). Deciphering the Plastomic Code of Chinese Hog-Peanut (Amphicarpaea edgeworthii Benth., Leguminosae): Comparative Genomics and Evolutionary Insights within the Phaseoleae Tribe. Genes 15, 88. doi: 10.3390/genes15010088
Xu, W., Lu, R., Li, J., Xia, M., Chen, G., Li, P. (2023). Comparative plastome analyses and evolutionary relationships of all species and cultivars within the medicinal plant genus Atractylodes. Ind. Crops Products 201, 116974. doi: 10.1016/j.indcrop.2023.116974
Yang, S. (2011). Studies on phytochemical diversity of Scrophularia ningpoensis Hemls. and its association with genetic and environmental variation. [Doctoral dissertation]. (Zhejiang (China): Zhejiang University).
Yang, L., Feng, Y., Ding, L., Tong, L., Qi, Z., Yan, X. (2022a). The complete chloroplast genome sequence of Elatostema stewardii Merr. (Urticaceae). Mitochondrial DNA B Resour. 7, 1494–1496. doi: 10.1080/23802359.2022.2108348
Yang, Q., Jiang, Y., Wang, Y., Han, R., Liang, Z., He, Q., et al. (2022b). SSR loci analysis in transcriptome and molecular Marker development in Polygonatum sibiricum. BioMed. Res. Int. 2022, 4237913. doi: 10.1155/2022/4237913
Yang, X., Zhang, X., Xue, T., Zhang, X., Yang, F., Yu, J., et al. (2023). Phylogenomics and historical biogeography of Hydrangeeae (Hydrangeaceae) elucidate the effects of geologic and climatic dynamics on diversification. Proc. Biol. Sci. 290, 20230659. doi: 10.1098/rspb.2023.0659
Yang, Y., Zhou, T., Duan, D., Yang, J., Feng, L., Zhao, G. (2016). Comparative analysis of the complete chloroplast genomes of five Quercus species. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00959
Yin, K., Zhang, Y., Li, Y., Du, F. K. (2018). Different natural selection pressures on the atpF gene in evergreen sclerophyllous and deciduous oak species: evidence from comparative analysis of the complete chloroplast genome of Quercus aquifolioides with other oak species. Int. J. Mol. Sci. 19, 1042. doi: 10.3390/ijms19041042
Zhang, M., Chen, K., Li, Y., Wang, P., Zhang, L. (2022a). Investigation of production status and prospect analysis of Zhejiang Dao-di herb:Scrophulariae Radix. China J. Traditional Chin. Med. Pharm. 37, 4845–4848.
Zhang, M., Chen, M., Zhang, X., Chen, S., Liang, Z. (2022b). The complete chloroplast genome sequence of traditional Chinese medicine Uncaria macrophylla (Rubiaceae). Mitochondrial DNA B Resour. 7, 694–695. doi: 10.1080/23802359.2022.2048215
Zhang, L., Yang, Z., Jia, Q., Dorje, G., Zhao, Z., Guo, F., et al. (2013). Two new phenylpropanoid glycosides with interesterification from Scrophularia dentata Royle ex Benth. J. Mol. structure 1049, 299–302. doi: 10.1016/j.molstruc.2013.05.039
Zhao, Z. (2008). The study on germplast resources diversity of Scrophularian ningpoensis Hemsl. [Master's dissertation]. (Shaanxi (China): Northwest A&F University).
Zhou, X., Zhang, Z., Huang, Y., Xiao, H., Wu, J., Qi, Z., et al. (2021). Conservation genomics of wild red sage (Salvia miltiorrhiza) and its endangered relatives in China: Population structure and interspecific relationships revealed from 2b-RAD data. Front. Genet. 12, 688323. doi: 10.3389/fgene.2021.688323
Zhu, A., Guo, W., Gupta, S., Fan, W., Mower, J. P. (2016). Evolutionary dynamics of the plastid inverted repeat: the effects of expansion, contraction, and loss on substitution rates. New Phytol. 209, 1747–1756. doi: 10.1111/nph.13743
Keywords: Scrophularia, medicinal plant, East Asia, chloroplast genome, comparative analysis, phylogenomics
Citation: Wang X, Guo L, Ding L, Medina L, Wang R and Li P (2024) Comparative plastome analyses and evolutionary relationships of 25 East Asian species within the medicinal plant genus Scrophularia (Scrophulariaceae). Front. Plant Sci. 15:1439206. doi: 10.3389/fpls.2024.1439206
Received: 27 May 2024; Accepted: 19 August 2024;
Published: 03 September 2024.
Edited by:
Gerald Matthias Schneeweiss, University of Vienna, AustriaReviewed by:
Massoud Ranjbar, Bu-Ali Sina University, IranCopyright © 2024 Wang, Guo, Ding, Medina, Wang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ruihong Wang, cnVpaG9uZzg5NzdAenN0dS5lZHUuY24=; Pan Li, cGFubGlAemp1LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.