
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 10 July 2024
Sec. Functional and Applied Plant Genomics
Volume 15 - 2024 | https://doi.org/10.3389/fpls.2024.1436998
This article is part of the Research Topic Improving Yield and Quality of Cereal Crops: Exploring and Utilizing Genes for Green and Efficient Traits View all 10 articles
 Bo Peng1*
Bo Peng1* Yan Liu1
Yan Liu1 Jing Qiu1
Jing Qiu1 Jing Peng2
Jing Peng2 Xiaoyu Sun1
Xiaoyu Sun1 Xiayu Tian1
Xiayu Tian1 Zhiguo Zhang3
Zhiguo Zhang3 Yaqin Huang4
Yaqin Huang4 Ruihua Pang1
Ruihua Pang1 Wei Zhou1
Wei Zhou1 Jinhui Zhao1
Jinhui Zhao1 Yanfang Sun1*
Yanfang Sun1* Quanxiu Wang1*
Quanxiu Wang1*Cytoplasmic 6-phosphogluconate dehydrogenase (G6PGH) is a key enzyme in the pentose phosphate pathway that is involved in regulating various biological processes such as material metabolism, and growth and development in plants. However, it was unclear if OsG6PGH1 affected rice grain quality traits. We perform yeast one-hybrid experiments and reveal that OsG6PGH1 may interact with OsAAP6. Subsequently, yeast in vivo point-to-point experiments and local surface plasmon resonance experiments verified that OsG6PGH1 can bind to OsAAP6. OsG6PGH1 in rice is a constitutive expressed gene that may be localized in the cytoplasm. OsAAP6 and protein-synthesis metabolism-related genes are significantly upregulated in OsG6PGH1 overexpressing transgenic positive endosperm, corresponding to a significant increase in the number of protein bodies II, promoting accumulation of related storage proteins, a significant increase in grain protein content (GPC), and improved rice nutritional quality. OsG6PGH1 positively regulates amylose content, negatively regulates chalkiness rate and taste value, significantly affects grain quality traits such as appearance, cooking, and eating qualities of rice, and is involved in regulating the expression of salt stress related genes, thereby enhancing the salt-stress tolerance of rice. Therefore, OsG6PGH1 represents an important genetic resource to assist in the design of high-quality and multi-resistant rice varieties.
Rice (Oryza sativa L.) is a globally important food crop, upon which more than half of the world’s and more than two-thirds of China’s populations rely as a dietary staple (Wang et al., 2018; Zhao et al., 2020; Li et al., 2022a). With continued improvement in societal living standards, the demand for high-quality rice is increasing (Zhou et al., 2019). Rice grain quality is measured in terms of its physical, chemical, and nutritional characteristics during growth, processing, storage, and consumption. These characteristics affect the nutritional value, taste, and processing performance of rice, and its appearance, nutritional, milling, processing, cooking, and eating qualities (Li et al., 2018; Custodio et al., 2019; Zhou et al., 2020; Zhao et al., 2022a).
Rice appearance is mainly affected by grain size and shape, chalkiness, and translucency. Nutritional quality is mostly affected by protein, starch, fatty and amino acid, vitamin, and mineral contents. The protein in rice is high in quality, and easily digested and absorbed in the gut (Peng et al., 2021; Li et al., 2022b; Gopika et al., 2022; Riaz et al., 2018). Grain protein is the second largest nutritional component after carbohydrates, and its content largely affects nutritional quality. An increase in grain protein content (GPC) improves the nutritional quality of rice. Based on solubility, grain proteins can be divided into glutelins, albumins, prolamins, and globulins (He et al., 2013; Peng et al., 2014; Chen et al., 2018). Milling and processing qualities mainly affect the performance of rice during processing, and are based on percentages of brown, milled, and head rice (Balindong et al., 2018). The brown rice rate directly affects processing quality, and indirectly its edible quality (Tong et al., 2019). The head rice rate is a grading indicator for grain quality. Cooking and eating qualities are the sensory and physicochemical properties of rice during cooking (e.g., extensibility, water absorption, swelling, gelatinization, color, form, aroma, and palatability) (Zhou et al., 2019; Shi et al., 2022), and are measured using amylose content, gelatinization temperature, and gel consistency (Zhou et al., 2019; Bao et al., 2023). Aspects of the cooking and eating quality are of greatest concern to consumers.
Amino acid permease (AAP) belongs to a family of amino acid transporters. AAPs play important roles in amino acid transport throughout plant tissues (Tegeder and Hammes et al., 2018). A total of 19 OsAAPs have been identified from rice, and most of them have been cloned and functionally identified. Of them, OsAAP1 and OsAAP4 regulate absorption and redistribution of neutral amino acids, thereby regulating rice growth, development, and yield (Ji et al., 2020; Fang et al., 2021; Erinaldo et al., 2022); OsAAP3 and OsAAP5 mainly transport alkaline amino acids, and have a negative effect on rice tillering and yield (Lu et al., 2018; Wang et al., 2019); OsAAP6 positively regulates GPC, thereby affecting the nutritional quality of rice (Peng et al., 2014); OsAAP7 and OsAAP16 can transport most amino acids (excepting aspartic acid and β-alanine) (Taylor et al., 2015); OsAAP8 mutation significantly increases GPC and amino acid contents (Peng et al., 2024); OsAAP10 and OsAAP11 regulate starch and protein contents in rice grains by affecting the transport of acidic and neutral amino acids, respectively (Wang et al., 2020; Yang et al., 2023a); OsAAP13 is mainly involved in amino acid transportation in above-ground parts of rice, and plays an important role in rice growth, development, and stress regulation (Nie et al., 2023); OsAAP14 is involved in nitrogen metabolism and positively regulates rice axillary bud growth, tillering, and yield (Yang et al., 2022); OsAAP15 affects rice yield by regulating panicle branch development (Yang et al., 2023b); and OsAAP17-related alleles positively regulate yield per plant in indica rice (Itishree et al., 2023). Thus, OsAAPs play important roles in regulating rice yield and quality traits.
The pentose phosphate pathway (PPP) is important for sugar metabolism in plants; its main physiological function is to produce reduced NADPH and generate pentose phosphate to participate in nucleic acid metabolism. PPP is the main source of NADPH in eukaryotic cells, and is involved in regulating metabolic intermediates in the biosynthesis process, which can participate in the synthesis of substances such as fatty and amino acids (Wang et al., 2017; Li et al., 2022c). 6-phosphogluconate dehydrogenase (G6PGH/6PGDH) is a key enzyme in the PPP, catalyzing the production of 5-phosphoribose and NADPH from 6-phosphogluconate. G6PGH has been successfully isolated and cloned from plants such as soybeans, corn, tomatoes, apples, Arabidopsis, rice, and wheat (Hanks et al., 1983; Tanksley and Kuehn, 1985; Bailey-Serres and Nguyen, 1992; Huang et al., 2003; Hölscher et al., 2016; Su et al., 2016). G6PGH participates in the plant’s response to various abiotic stresses, and plays an important role in responses to temperature extremes, salt, heavy metals, and osmotic and other stresses. In maize, 6PGDH can significantly increase yield under high temperature stress. After a double mutation of 6pgd1 and 6pgd2 in corn, the homozygous mutant showed delayed growth (Bailey-Serres and Nguyen, 1992); after the PGD3 mutation, the mutant exhibited severe granule filling and defective embryonic phenotype (Ribeiro et al., 2020). Overexpression of 6PGDH in Arabidopsis significantly enhances cold tolerance (Tian et al., 2021), and can accelerate growth of callus tissue in apples (Zhu et al., 2019). Transcription levels of two types of OsG6PGH isolated and cloned in rice (cytoplasmic 6-phosphogluconate dehydrogenase (OsG6PGH1) and plastid 6-phosphogluconate dehydrogenase (OsG6PGH2)) were significantly upregulated in rice seedlings under drought, high salt, cold, and abscisic acid treatment, indicating that OsG6PGH plays an important role in the response of rice to abiotic stress (Huang et al., 2003; Hou et al., 2007). However, it remained unclear if OsG6PGH was involved in biological processes such as rice growth, development, and substance metabolism, or if it affected grain quality.
We identified a major quantitative trait locus gene OsAAP6 that positively regulated GPC and affected rice nutritional quality (Peng et al., 2014, 2018). To gain a deeper understanding of the molecular mechanism by which OsAAP6 regulated GPC, we determined OsG6PGH1 to possibly interact with it (Peng et al., 2023). We further validated the ability of OsG6PGH1 to bind to OsAAP6 through in vivo point-to-point experiments in yeast and localized surface plasmon resonance experiments. In overexpressing OsG6PGH1 plants, we report OsG6PGH1 as capable of regulating the expression of OsAAP6 to affect the contents of the main storage substances in rice grains, and to have important effects on grain quality traits. Simultaneously, OsG6PGH1 participates in biological processes such as the salt-stress response. These results are important for molecular design and breeding of high-quality and multi-resistant rice varieties.
We first simultaneously transformed the plasmid pGADT7-Rec2-OsG6PGH1 carrying the target gene OsG6PGH1 and the bait plasmid pHIS2-OsAAP6 into yeast receptive cell Y187. Transformed yeast cells were cultured on SD-Trp-Leu solid medium at 30°C for 3–5 d (Peng et al., 2023). Select yeast monoclonal antibodies from this medium were inoculated into SD-Trp-Leu liquid medium. Cultivation was continued at 30°C for 12 h. The OD600 value of the yeast solution was adjusted for all samples to 0.4 by adding ddH2O to ensure consistency in experimental conditions. Next, the standardized yeast solution onto SD-Leu-Trp-His+150 mmol·L-1 3AT medium, and cultured at 30°C for 3–4 d to further validate the interaction between yeast clones and bait proteins (Peng et al., 2023; Zhang et al., 2024).
Binding analysis of OsG6PGH1 and OsAAP6 was performed using the OpenSPR surface plasmon resonance biosensor (Nicoya Life Science Inc., Kitchener, Canada). Using a standard coupling reaction of 1-ethyl-3-(3-dimethylpropyl)-carbodiimide (EDC) and N-hydroxysuccinimide (NHS), the OsAAP6 was fixed at a concentration of 50 μg·ml-1 onto the activated COOH sensor chip (Peng et al., 2023). A blocking solution was then injected into the sensor chip to reduce the possibility of non-specific binding. Then, prokaryotic total cell lysate protein solutions at concentrations of 15, 22.5, and 30 μg·ml-1 were introduced to the sensor chip. As the sample flows through the chip, OsAAP6 contained in it binds specifically to previously fixed antibodies, and is captured by the sensor (Liu et al., 2019). To detect the interaction between OsAAP6 and OsG6PGH1, we injected 50 μg·ml-1 IgG as a negative control and OsG6PGH1 antibody as a target protein into the sensor chip. By comparing the response signals of IgG (non-specific binding) and OsG6PGH1 antibody (with potential specific binding), it can be determined if OsG6PGH1 has interacted with OsAAP6.
To create mutants targeting the deletion of OsG6PGH1 based on CRISPR/Cas9 gene-editing tool, we commissioned Wuhan Astronomical Biotechnology Co., Ltd. to conduct genetic transformation experiments. PCR and Sanger sequencing techniques were used to detect and analyze the mutant sequence (Peng et al., 2024), ensuring the successful knockout of OsG6PGH1.
The cDNA fragment of OsG6PGH1 was cloned onto the intermediate vector PMD-18T to obtain the PMD-18T-OsG6PGH1 recombinant plasmid. Sequencing validation was then performed on the recombinant plasmid to ensure the accuracy of the OsG6PGH1 sequence. After verifying the accuracy, the target fragment was cut from PMD-18T-OsG6PGH1 and connected to the pCMBIA1301s overexpression vector to construct the overexpression plasmid pCMBIA1301s-OsG6PGH1. Wuhan Astronomical Biotechnology Co., Ltd. was again entrusted to use Agrobacterium-mediated genetic transformation to introduce this overexpression plasmid into rice cells, to overexpress the OsG6PGH1.
Total RNA was extracted using TRIzol reagent according to manufacturer instructions (TAKARA) (Peng et al., 2024). A reverse transcription kit (R202-02 HyperScript III RT SuperMix for qPCR with gDNA Remover) was used to reverse transcribe RNA into cDNA. qRT-PCR was performed using 2 SYBR Green Fast qPCR Master Mix (YiFeiXue Bio Technology) on quantitative analyzer (ABI7300), with a reaction program involving a predenaturation step at 95°C for 30 s, and an amplification reaction of 40 cycles, each including 95°C denaturation for 15 s, and a 60°C annealing extension for 30 s. To accurately evaluate changes in expression of target genes, β-actin was used as an internal reference gene, and relative expression levels were standardized using the 2−ΔΔCT calculation method (Wang et al., 2022). Three biological replicates were performed for each reaction using primers detailed in Supplementary Tables 1–4.
Based on the cDNA sequence of OsG6PGH1, we designed and detected primers using the NCBI website. Total RNA was extracted from root, stem, leaf, sword leaf, glume, stem node, and endosperm tissues, and reverse transcribed into cDNA. Expression levels of OsG6PGH1 in tissues were detected by qRT-PCR (Li et al., 2023). β-Actin was used as an internal reference gene in each replicated experiment.
Protoplasts were first extracted from rice seedlings cultured in darkness for 14 d. The subcellular localization vector OsG6PGH1-GFP and plasma membrane (PAT2-RFP) and nuclear (Ghd7-RFP) markers were co-transformed into the protoplasts of rice seedlings. Fluorescence signals in protoplasts were detected using confocal fluorescence microscopy (Peng et al., 2023); all fluorescence experiments were independently observed at least three times.
Field experiments were performed on rice plants at Xinyang Normal University, Henan Province. Planting density was 16.5 × 26.0 cm, with the field managed in accordance with local agricultural standards. Before measurement, harvested grains were air dried, and then stored at room temperature for at least 3 months. Fully grouted grains were used to detect rice quality and yield traits (Li et al., 2014). A near-infrared grain analyzer (INFRAEC Nova) was used to detect protein, amylose, and free fatty acid contents, and taste value (Peng et al., 2023). The gelatinization temperature of grain was measured using the alkaline digestion method (Peng et al., 2024), and polished rice gel consistency was determined following People’s Republic of China (GB/T17891-1999, high-quality rice) national standards.
The endosperm of 10 DAF rice was fixed in 2.5% glutaraldehyde with 0.2 mol L−1 phosphate buffer (pH 7.2) for at least 12 h. Fixed endosperm tissue was cut into 1 mm sections using an ultra-thin slicer (Power Tome XL, RMC). Sections were examined by TEM (H-7650, Hitachi). ImageJ software (NIH) was used to calculate the quantity and area of PBs in each TEM sample image (Peng et al., 2014).
The middle part of the grain was gently tapped with the back of a knife to allow natural breakage; a sharp blade was then used to cut the broken part to form a thin sheet of ~2–3 mm thickness. Slices were then divided into two: one part to observe the chalky area beneath a regular optical microscope; the other to be fixed on a copper sample stage with conductive adhesive, sprayed with platinum using a HUS-5GB vacuum coating machine, and examined by scanning electron microscope (Hitachi S-4800) (Xu et al., 2022).
Both OsG6PGH1(OX) positive and negative plants grew in the same environment. Endosperms were simultaneously collected, and then ground for proteomic sequencing (replicated three times per sample). Total protein was first extracted, and its concentration, purity, and integrity were examined to ensure sample usability. Obtained data were filtered to obtain clean readings aligned with the reference proteome sequence. Statistical tests were performed to obtain DEPs based on the annotation file of the proteome and reading position (i.e., expression level) compared with the proteome. Finally, Clusters of Orthologous Genes functional annotation, Gene Ontology, Kyoto Encyclopedia of Genes and Genomes, and Pfam enrichment analyses were performed on DEPs to identify biological processes and metabolic pathways of protein enrichment (Xiong et al., 2019).
After peeling and disinfecting OsG6PGH1 transgenic seeds, they were inoculated into 1/2 MS solid culture medium. After 7 d, rice seedlings with consistent growth were selected and placed into a hydroponic medium for acclimation for 7 d. Seedlings cultured for 14 d were inoculated in 50 mmol L−1 and 100 mmol L−1 concentrations of NaCl medium, and placed into a 28°C constant temperature light incubator. Nutrient solutions were changed every 2 d to prevent bacterial contamination (Zhao et al., 2022b). Seedling growth was monitored daily.
A total of 92 regulatory factors that may interact with OsAAP6 were screened through yeast one- hybrid experiments (Peng et al., 2023). One regulatory factor, the plasmid pGADT7-Rec2-G6PGH1, along with the bait plasmid pHIS2-OsAAP6, were transferred into yeast receptive cell Y187. On solid culture media, with the addition of 3AT inhibitors, the OsG6PGH1 yeast colony and positive control yeast colony (+) both grew well, but there were significant differences between them and the negative control (−) (Figure 1A). This indicates that OsG6PGH1 can interact with OsAAP6 in yeast.
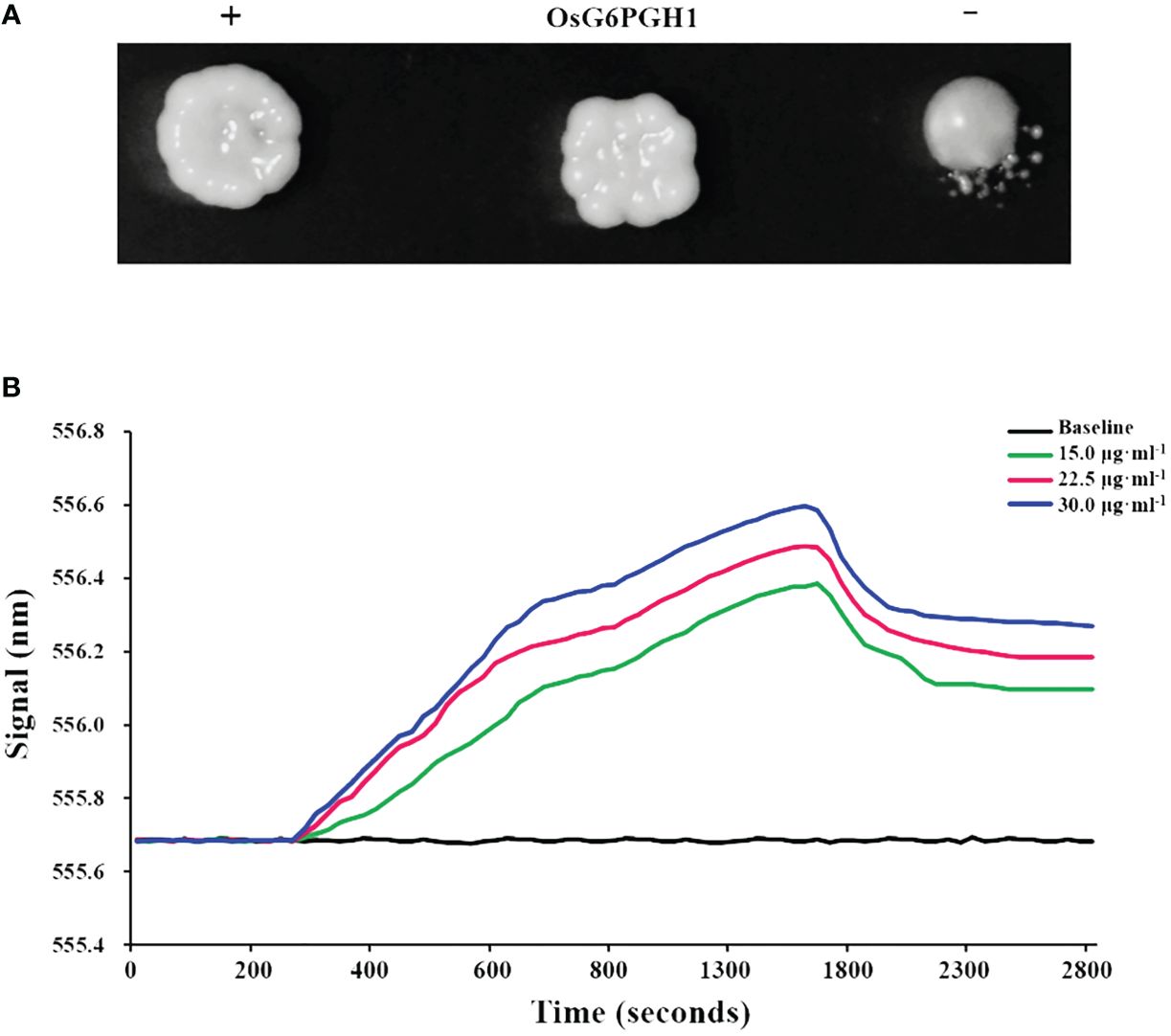
Figure 1 Verification of interactions between OsG6PGH1 and OsAAP6. (A) Yeast in vivo point-to-point experiment: +, positive control; −, negative control. (B) In vitro validation using localized surface plasmon resonance experiments.
To verify the interaction between OsG6PGH1 and OsAAP6, prokaryotic expression vectors pET-28a-OsG6PGH1 and pET-28a were transfected into Escherichia coli competent cells BL21 (DE3). In local surface plasmon resonance experiments, the curves for three concentrations all significantly trended upward at 280 s. This change indicated that an interaction occurred between OsG6PGH1 and OsAAP6, forming a complex (OsG6PGH1–OsAAP6). At 1650 s, there was a significant downward trend in curves for each concentration, indicating that the OsG6PGH1–OsAAP6 complex had undergone dissociation (Figure 1B). These dynamic changes indicate that OsG6PGH1 can bind to OsAAP6 in vitro.
To investigate expression levels of OsG6PGH1 in different rice tissues, RNA was extracted from root, stem, stem node, leaf, sword leaf, endosperm, and glume tissues. Expression patterns were analyzed using qRT-PCR. OsG6PGH1 was expressed in each tissue type (Figure 2A), and this result was in accordance with the distinct tissue expressions of OsG6PGH1 (LOC_Os06g02144) on CREP (Supplementary Figure 1), indicating it is a constitutive expression gene. Expression levels of OsG6PGH1 were higher in roots, and lower in the endosperm and glume (Figure 2A).
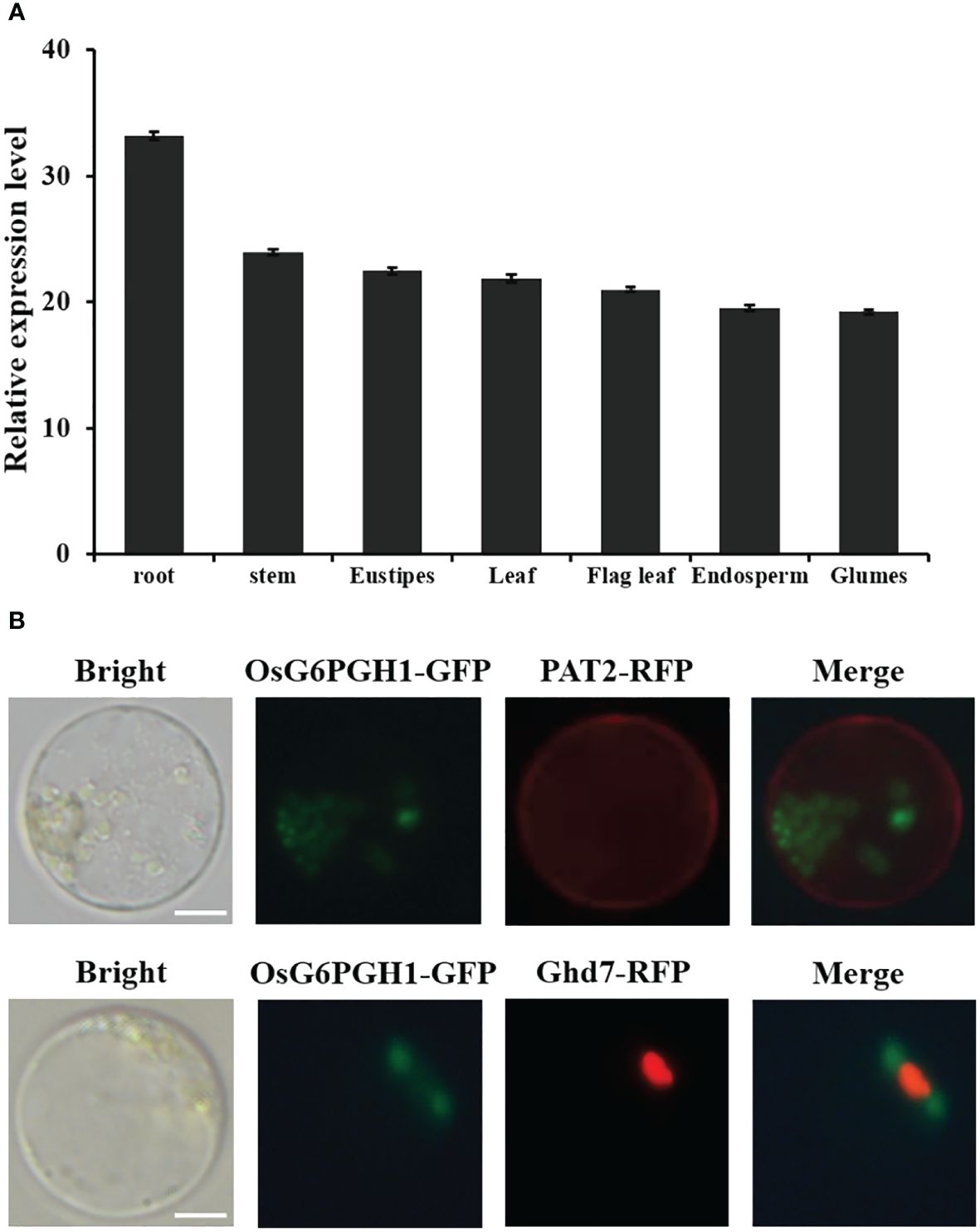
Figure 2 Analysis of OsG6PGH1 expression patterns and subcellular localization in rice. (A) Detection and analysis of expression levels in different tissues. (B) OsG6PGH1-GFP is not co-localized with either the membrane marker protein PAT2-RFP or nuclear marker protein Ghd7-RFP. Scale bars, 5 μm; error bars, standard error of the mean (SEM).
To investigate localization of OsG6PGH1 in cells, OsG6PGH1 was fused with the green fluorescent protein gene (GFP) (OsG6PGH1-GFP) and simultaneously co-expressed with the cell-membrane-localization marker gene PAT2-RFP and nuclear marker gene Ghd7-RFP in rice protoplasts, and observed using laser confocal electron microscopy. OsG6PGH1-GFP was not co-localized with membrane marker protein PAT2-RFP or nuclear marker protein Ghd7-RFP (Figure 2B), indicating that OsG6PGH1 was not localized in either the cell membrane or nucleus, but that it may be localized in the cytoplasm.
To investigate the biological function of OsG6PGH1 in rice, we created CRISPR/Cas9 knockout and overexpressing transgenic lines of OsG6PGH1. Based on the wild-type Zhonghua 11 (ZH11), a target site for OsG6PGH1 was designed. CRISPR/Cas9 gene editing tool was used to target and induce the fusion of sgRNA and Cas9 protein, and specific splicing was performed on the two PAM sites of OsG6PGH1. A gene edited expression vector was constructed, and 20 CRISPR/Cas9 gene edited plants were obtained through Agrobacterium-mediated genetic transformation (including 16 OsG6PGH1 positive mutants) (Supplementary Figure 2A). DNA was extracted from the leaves of the OsG6PGH1 mutants, and PCR amplification and DNA sequencing were performed; two types of mutations were found in the cDNA target sequence of OsG6PGH1: Osg6pgh1-1 (+1 bp) and Osg6pgh1-2 (+2 bp) (Supplementary Figure 2B). The target sequence of Osg6pgh1-1 has a 1 bp insertion and 3 bp substitution in its second exon, resulting in a frameshift mutation, and Osg6pgh1-2 has a 2 bp insertion and 4 bp substitution in its first and second exons, which also causes a frameshift mutation.
To obtain transgenic plants with OsG6PGH1 overexpression, we connected the cDNA of OsG6PGH1 to the intermediate vector PMD-18T to obtain PMD-18T-OsG6PGH1. After sequencing verification, the target fragment was connected to the pCMBIA1301s overexpression vector to obtain the OsG6PGH1 overexpression plasmid pCMBIA1301s-OsG6PGH1. Through Agrobacterium- mediated genetic transformation, 20 transgenic plants with OsG6PGH1 overexpression (OsG6PGH1(OX)) were obtained (including 15 positive plants) (Supplementary Figure 2C).
We performed qRT-PCR analysis on expression levels of OsAAP6 in the endosperm of OsG6PGH1 mutants (M2) and overexpressing transgenic F2 lines. There was no significant change in expression levels of OsAAP6 in the endosperm of OsG6PGH1 homozygous mutants, while expression levels significantly increased in the OsG6PGH1 overexpressing positive endosperm (Figure 3A).
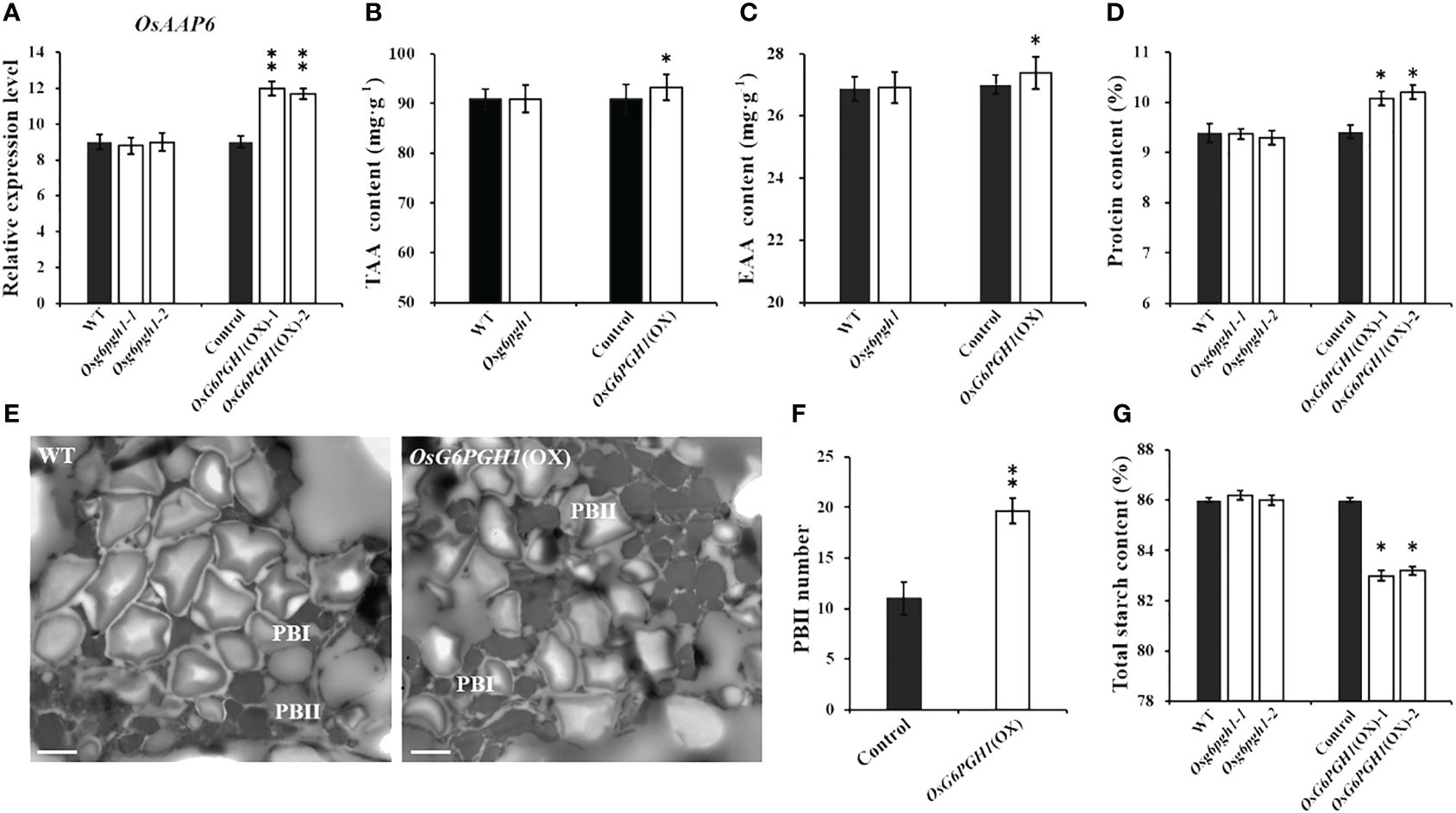
Figure 3 Detection and analysis of major nutrients in transgenic rice grains with OsG6PGH1. (A) Detection and analysis of expression levels of OsAAP6 in OsG6PGH1 transgenic endosperm. OsG6PGH1 transgenic plant: (B) total amino acids, (C) essential amino acids, and (D) grain protein content. (E) transmission electron microscopy image of endosperm of OsG6PGH1(OX) 10 days after fertilization; and analysis of (F) protein body II, and (G) total starch content. Significant differences are based on two tailed t-tests, where **P ≤ 0.01, *P ≤ 0.05. WT, Wild type; Control, OsG6PGH1(OX) negative control. Scale bars, 2 μm; error bars, standard error of the mean (SEM), n ≥ 50.
Because of the influence of OsAAP6 on the distribution of amino acids in grains and its positive regulation of grain protein content, we detected and analyzed amino acid contents and GPC in OsG6PGH1 mutants (M2) and overexpressing transgenic F2 grains. OsG6PGH1 affected the content of some amino acids (e.g., alanine, glutamic acid, leucine, lysine, methionine) (Supplementary Figure 3A). Total and essential amino acid contents in grains of OsG6PGH1(OX) positive plants significantly increased, while there was no difference in grains of OsG6PGH1 homozygous mutants (Figures 3B, C). Additionally, the GPC in OsG6PGH1(OX) positive plants increased significantly, while there was no significant difference in OsG6PGH1 mutants (Figure 3D). To further investigate if OsG6PGH1(OX) affected the protein bodies in rice endosperm, we observed the endosperm of OsG6PGH1(OX) 10 d after fertilization (DAF) using transmission electron microscopy (TEM). Protein bodies I (PB I) and protein bodies II (PB II) were clearly visible under TEM: PB I had relatively regular circular or elliptical shapes, and PB II had irregular shapes and stained uniformly (Figure 3E). Average cross-sectional areas of PB I and II in OsG6PGH1(OX) positive endosperms did not differ significantly from those in transgenic negative endosperms, while numbers of PB II significantly increased (Figure 3F; Supplementary Figures 3B–D). This indicates that overexpression of the OsG6PGH1 significantly increased the number of PB II in rice endosperm, leading to increased GPC.
To further investigate if OsG6PGH1 affected the accumulation of nutrients such as starch and free fatty acids in rice, starch and free fatty acid content was measured in OsG6PGH1 mutants (M2) and overexpressing transgenic F2 plants. Total starch contents in OsG6PGH1(OX) positive grains were significantly reduced, while total starch contents in OsG6PGH1 homozygous mutants did not change significantly (Figure 3G). There was no significant change in free fatty acid content in homozygous mutants of OsG6PGH1 or OsG6PGH1(OX) positive grains (Supplementary Figure 3E). Therefore, overexpression of OsG6PGH1 helps to increase GPC, but not the accumulation of starch in grains.
Because of the significant impact of OsG6PGH1(OX) on accumulation of protein and starch in grain, we examined expression levels of protein-and starch-metabolism-related genes in OsG6PGH1 mutants and overexpressed transgenic F2 endosperm. Gene expression levels correlated positively with grain protein synthesis and accumulation metabolism, with 10KD Prolamine, GluA1, 11S Globulin, Glutelin 4, GluB1, AlaAT, GluA3, GluA2, GluB4 significantly upregulated in the positive endosperm of OsG6PGH1(OX), but not in OsG6PGH1 homozygous mutants (Figures 4A–J). Expression levels of starch-synthesis-related genes (Susy2, Susy3) and starch-degradation metabolism-related genes (AMY3A, AMY3B, ISA1) were significantly downregulated in OsG6PGH1 homozygous mutants, and significantly upregulated in OsG6PGH1(OX) positive plants. Expression levels of genes related to starch synthesis (SS1, SSIVa, SBE) were significantly upregulated in OsG6PGH1 homozygous mutants, while expression levels were the opposite in OsG6PGH1(OX) positive plants (Figures 4K–S). The granule-bound starch synthase gene (GBSSI) related to amylose synthesis was significantly downregulated in OsG6PGH1 homozygous mutants and significantly upregulated in OsG6PGH1(OX) positive plants (Figure 4T). These results are consistent with a phenotype with significant increases in GPC and a significant decrease in starch content in rice grains with OsG6PGH1(OX).
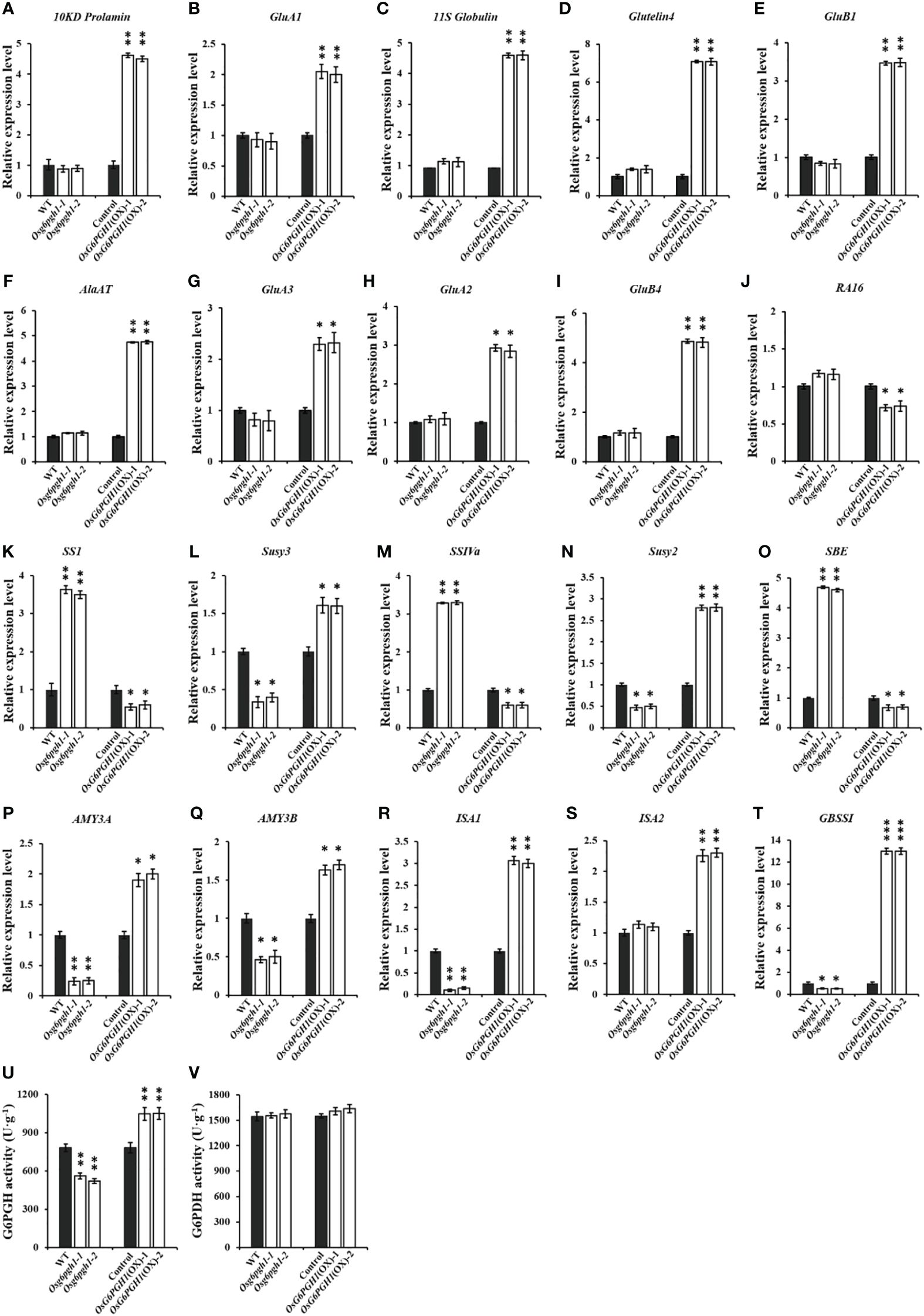
Figure 4 Expression and enzyme activities of related genes in OsG6PGH1 transgenic rice endosperm. (A–J) qRT-PCR analysis of protein-metabolism-related genes; (K–T) qRT-PCR analysis of starch-metabolism-related genes; and detection and analysis of enzyme activity of endosperm (U) 6PGDH and (V) G6PDH in OsG6PGH1 transgenic rice lines 10 DAF. Significant differences are based on two tailed t-tests, where ***P ≤ 0.001, **P ≤ 0.01, *P ≤ 0.05. WT, Wild type; Control, OsG6PGH1(OX) negative control. Error bars, standard error of the mean (SEM).
To investigate the effect of changes in OsG6PGH1 on the activity of upstream and downstream enzymes in the PPP, we measured the activity of 6-phosphogluconate dehydrogenase (G6PGH) and its upstream rate-limiting enzyme glucose-6-phosphate dehydrogenase (G6PDH) in the endosperm of rice 10 DAF. Activity of G6PGH in the endosperms of OsG6PGH1 transgenic F2 plants changed significantly: the activity of G6PGH in endosperm of OsG6PGH1 homozygous mutants was significantly reduced, and significantly enhanced in OsG6PGH1(OX) positive endosperms (Figure 4U); the activity of the upstream rate-limiting enzyme G6PDH did not change significantly in OsG6PGH1 transgenic plants (Figure 4V).
To investigate if OsG6PGH1 affects the cooking and eating quality of rice, we tested amylose content, taste value, gel consistency, and gelatinization temperature of OsG6PGH1 mutants and overexpressing transgenic F2 plants. Compared with wild type rice, the amylose content of OsG6PGH1 homozygous mutants was significantly lower, and taste value and gelatinization temperature were significantly higher. Amylose contents of OsG6PGH1(OX) positive grains were significantly higher, the taste value was significantly lower, and the gelatinization temperature did not change significantly (Figures 5A–C). Gel consistency of OsG6PGH1 mutants and overexpressing transgenic rice showed no significant changes (Supplementary Figure 4A).
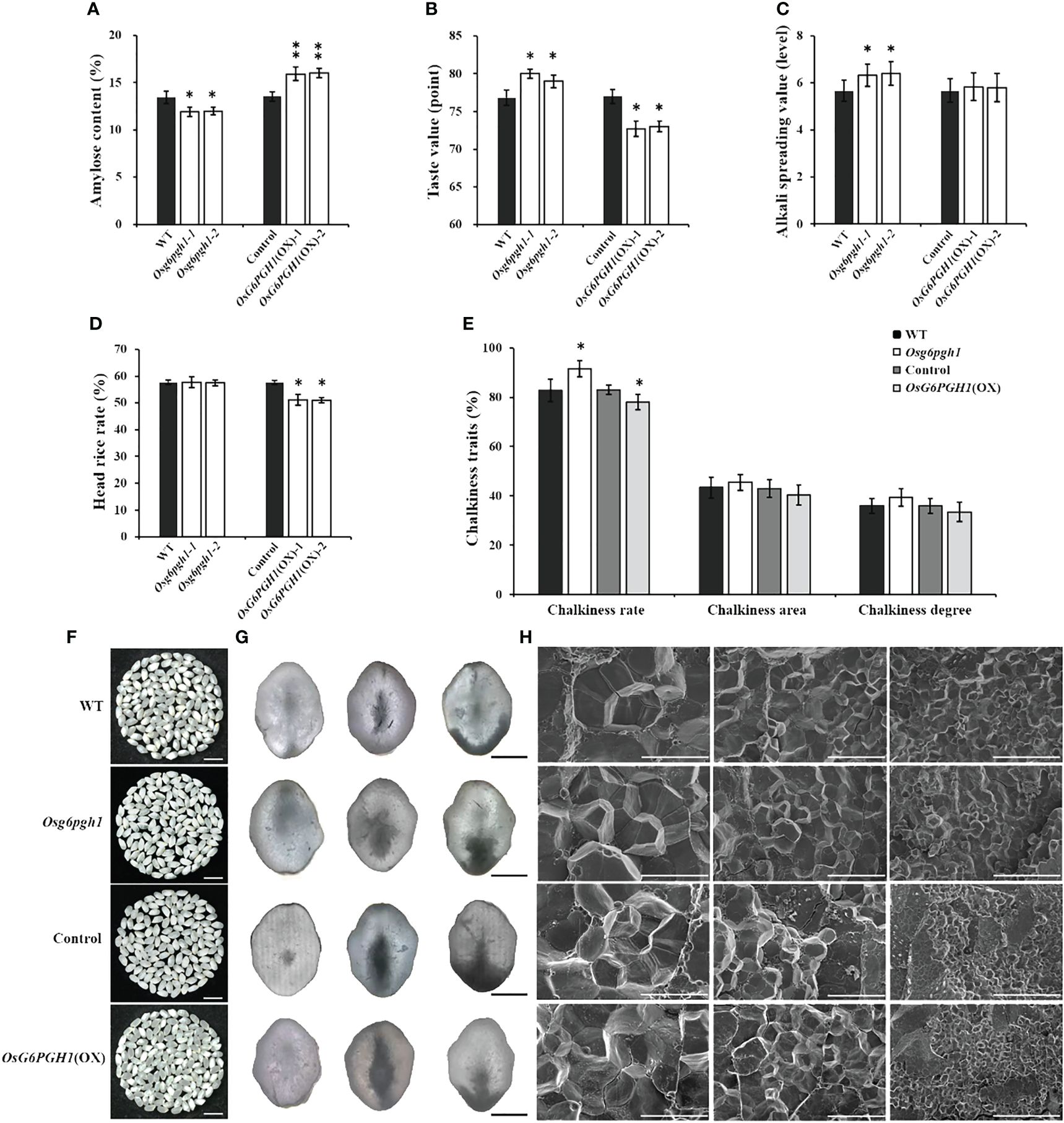
Figure 5 Analysis of cooking and eating, processing, and appearance quality traits of OsG6PGH1 transgenic rice. Detection and analysis of: (A) amylose content, (B) taste value, (C) gelatinization temperature; (D) head rice rate, (E) chalkiness traits, and (F) endosperm chalkiness characteristics. Scale bars, 1 cm. Images of: (G) chalkiness under optical microscope; columns from left (transparent), central (heart white), and right (belly white); scale bars, 1 mm; (H) Scanning electron microscopy of endosperm; scale bars, left to right: 1 mm, 10 μm, 20 μm, and 100 μm. Significant differences are based on two-tailed t-tests, where **P ≤ 0.01, *P ≤ 0.05. WT, Wild type; Control, OsG6PGH1 overexpression transgenic negative control. Error bars, standard error of the mean (SEM).
Because amylose contents of OsG6PGH1 mutants and overexpressing transgenic rice changed significantly, we analyzed their milling quality traits. The head rice rate of OsG6PGH1(OX) positive F2 rice significantly decreased (Figure 5D), while the brown and milled rice rates showed no significant change (Supplementary Figures 4B, C). Compared with wild type rice, there was no significant change in milling quality traits of OsG6PGH1 homozygous mutant rice (Figure 5D; Supplementary Figures 4B, C). These results indicate that upregulation of OsG6PGH1 expression does not improve rice cooking or eating quality, decreases the head rice rate, and affects the grain processing quality.
Chalkiness rates of OsG6PGH1 homozygous mutants increased significantly, while the chalkiness rate of OsG6PGH1(OX) rice was significantly reduced (Figures 5E, F). There were no significant changes in chalkiness area or degree in OsG6PGH1 mutants or overexpressing transgenic rice. The morphology and arrangement of starch granules in the endosperm of OsG6PGH1 mutants and overexpressing transgenic positive rice showed no significant change; both presented a densely arranged polygonal structure (Figures 5G, H). Therefore, OsG6PGH1 negatively regulated the chalkiness rate of grain, but did not affect chalkiness area or degree.
To investigate if OsG6PGH1 affected rice yield, we tested yield-related traits of OsG6PGH1 mutants (M2) and their overexpressing transgenic F2 plants. OsG6PGH1 homozygous mutant plant height was significantly reduced (Supplementary Figures 5A, B); there were no significant changes in the number of tillers per plant, number of spikelets per panicle, seed setting rate, thousand grain weight, or yield per plant (Supplementary Figures 5C–G). Numbers of spikelets per panicle of OsG6PGH1(OX) positive plants significantly decreased (Supplementary Figure 5D), and the setting rate significantly increased (Supplementary Figure 5E); other yield-related traits showed no significant changes, leading to no overall significant change in rice yield per plant (Supplementary Figure 5G). Because OsG6PGH1(OX) significantly affected grain protein and starch contents, we examined the grain type of OsG6PGH1 mutants and overexpressed transgenic seeds. There were no significant changes in grain length, width, or thickness of OsG6PGH1 homozygous mutants and overexpressing transgenic positive grains (Supplementary Figure 6). Therefore, although OsG6PGH1 affected plant height, number of spikelets per panicle, and seed setting rate, it did not affect rice yield per plant.
The GPC in OsG6PGH1(OX) positive grains significantly increased. To evaluate the accumulation of different storage proteins in grains, we performed proteomic analysis on OsG6PGH1(OX) endosperm. Upregulated proteins were screened using two methods: differential multiple (|log2 (Fold change)| ≥ 1.50) and significance level (P-value ≤ 0.05). Downregulated proteins were screened through two methods: differential multiple (|log2 (Fold change)| ≤ 1/1.50) and significance level (P-value ≤ 0.05). A total of 156 differentially expressed proteins (DEPs) were screened (Supplementary Figure 7A).
To understand the structure and function of these DEPs, functional annotation was performed using the COG database. Functions of DEPs were mainly related to amino acid transport and metabolism, carbohydrate transport and metabolism, and protein post-translational modification (Supplementary Figure 7B). GO functional enrichment analysis of these DEPs revealed significant differential enrichment in OsG6PGH1(OX) positive endosperms to occur mainly in cellular components and organelles, participation in metabolic processes, cellular processes, and other biological processes, with molecular functions such as binding and catalytic activity (Supplementary Figure 7C). KEGG pathway analysis on these DEPs revealed carbon metabolism, glycolysis/gluconeogenesis, starch and sucrose metabolism, and carbon fixation by photosynthetic organisms, to be pathways where DEPs were more concentrated in OsG6PGH1(OX) positive endosperms (Supplementary Figure 7D). Domain annotation and enrichment were performed on these DEPs through the Pfam database; protease inhibitors, seed storage, and LTP family were mostly enriched in OsG6PGH1(OX) positive endosperms (Supplementary Figure 7E). Results of proteomic analysis are consistent with upregulation of protein- and amylose-synthesis-related genes, and downregulation of amylopectin-metabolism-related genes in the positive endosperm of OsG6PGH1(OX)(Figure 4; Supplementary Table 5). This suggests that overexpression of OsG6PGH1 may promote protein synthesis and accumulation in the endosperm, while inhibiting corresponding starch synthesis and metabolic pathways.
Because OsG6PGH1, as a key gene in the PPP, may be involved in the salt stress response of rice, we performed salt stress treatment experiments on OsG6PGH1 mutants (M2) and OsG6PGH1(OX) transgenic F2 seedlings. Rice seedlings grown for 14 d in culture media were exposed to concentrations of 50 mmol·L−1 and 100 mmol·L−1 NaCl for 7 d. The survival rate of OsG6PGH1 homozygous mutant seedlings was significantly reduced compared with wild-type rice at 100 mmol·L−1 NaCl, but that of OsG6PGH1(OX) positive seedlings increased significantly in both salt stress treatments. This indicates that OsG6PGH1 improves salt stress resistance of rice seedlings (Figures 6A–C).
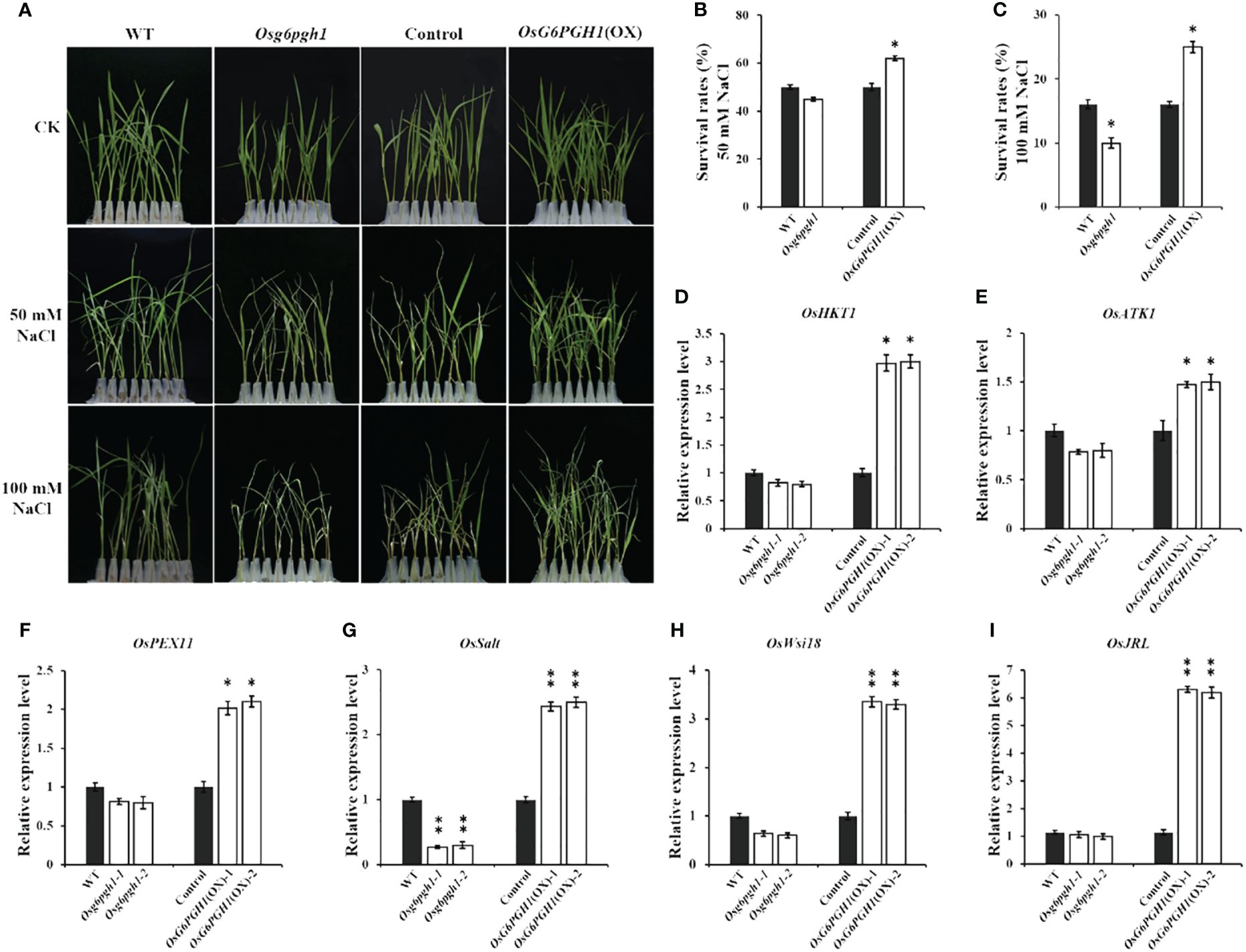
Figure 6 Salt stress tests and expression analysis of related genes in OsG6PGH1 transgenic rice seedlings. (A) Salt stress test. Survival rate of rice seedlings at (B) 50 mmol·L−1 NaCl, and (C) 100 mmol·L−1 NaCl. (D–I) qRT-PCR detection and analysis of salt stress-related genes. Significant differences are based on two-tailed t-tests, where **P ≤ 0.01, *P ≤ 0.05. WT, Wild type; Control, OsG6PGH1 overexpression transgenic negative control. Error bars, standard error of the mean (SEM).
To further explore the molecular mechanism of OsG6PGH1 regulating salt stress resistance in rice seedlings, we examined expression levels of six salt stress-related genes (OsPEX11, OsSalt, OsWsi18, OsJRL, OsHKT1, OsATK1) in OsG6PGH1 mutants and OsG6PGH1(OX) seedlings. qRT-PCR results revealed expression levels of these genes to be significantly increased in OsG6PGH1(OX) positive seedlings, while OsSalt was significantly reduced in OsG6PGH1 homozygous mutant plants (Figures 6D–I). These results are consistent with observations that salt stress resistance of OsG6PGH1 homozygous mutant seedlings is significantly reduced, while the salt stress resistance of transgenic positive rice seedlings is significantly enhanced.
AAPs are involved in various physiological metabolic processes in plants, and affect rice growth and development (Fang et al., 2021). In the rice genome, 19 OsAAPs (OsAAP1–19) have been identified, with most of them isolated, cloned, and identified (Peng et al., 2014; Taylor et al., 2015; Lu et al., 2018; Wang et al., 2019; Ji et al., 2020; Wang et al., 2020; Fang et al., 2021; Erinaldo et al., 2022; Yang et al., 2022; Yang et al., 2023a, b; Peng et al., 2024). Among them, OsAAP6 is a major quantitative trait locus gene that positively regulates GPC and nutritional quality of rice (Peng et al., 2014, 2019). To further elucidate the regulatory mechanism of OsAAP6, a total of 92 potential regulatory factors that may interact with it were screened through yeast one-hybrid experiments (Peng et al., 2023). The ability of OsG6PGH1 to bind to OsAAP6 was then validated through yeast in vivo point-to-point rotation experiments and local surface plasmon resonance experiments (Figure 1); it was involved in regulating expression of OsAAP6 in OsG6PGH1(OX). These results provide key clues for the in-depth analysis of OsAAP6 regulation of GPC, but also open up new possibilities for using OsAAP6 as a gene resource in molecular design breeding.
G6PGH is a key gene in the PPP, with multiple biological functions. After the double mutation of 6pgd1 and 6pgd2 in corn, the growth of homozygous individuals with the mutation was delayed (Bailey-Serres and Nguyen, 1992); overexpression of G6PGH in apples can accelerate growth of callus tissue (Zhu et al., 2019). We obtained OsG6PGH1 mutants using CRISPR/Cas9 gene-editing tool and created OsG6PGH1(OX) transgenic plants. OsG6PGH1 homozygous mutant plant height was significantly reduced, as were the number of spikelets per panicle in OsG6PGH1(OX) transgenic positive plants, while the seed setting rate increased significantly. Notably, there was no significant change in rice yield per plant in OsG6PGH1 homozygous mutants or overexpressing transgenic positive rice. Therefore. OsG6PGH1 may be involved in regulating rice growth and development, but not yield per plant.
Rice grains are rich in amino acids, with a relatively balanced composition (20 types of amino acids, including 8 essential amino acids) (Yang et al., 2022). Grain protein is also high-quality, and easily digestible and absorbed (Peng et al., 2023). GPC is a key factor in the nutritional quality of rice (Xia et al., 2022). Therefore, exploring genes related to GPC and analyzing their functions is important for molecular genetic breeding of high-quality, genetically improved rice. We report the positive regulatory factor OsAAP6 for GPC to be significantly upregulated in OsG6PGH1(OX) transgenic positive plants, resulting in a significant increase in GPC and a significant decrease in starch content. This indicates that OsG6PGH1 is involved in regulating accumulation of major storage substances in rice grains. Further observation of protein bodies in the endosperm of OsG6PGH1(OX) revealed no significant changes in the area and quantity of protein body I, and area of protein body II, but the quantity of protein body II increased significantly. This indicates that overexpression of OsG6PGH1 significantly increases the number of protein bodies II, leading to a significant increase in GPC (Figure 3). Detection and analysis of expression levels of protein- and starch-metabolism-related genes in OsG6PGH1 mutants and overexpressed transgenic endosperm (Figure 4) revealed expression levels of protein- and starch-synthesis metabolism-related genes in rice grains to be consistent with their corresponding main storage substances (protein and starch) contents. Proteomic analysis of OsG6PGH1(OX) endosperm revealed DEPs to mainly contain structural domains related to seed storage, have biological functions in metabolic processes, and participate in biological pathways such as carbon metabolism, and starch and sucrose metabolism (Supplementary Figure 6). Therefore, genes related to protein synthesis and metabolism are upregulated in OsG6PGH1(OX) transgenic positive endosperm, which can significantly increase numbers of protein bodies II, promote accumulation of related storage proteins in the endosperm, and increase GPC and nutritional quality.
Starch, an important component in rice grains, accounts for approximately 80%–90% of rice dry weight. This starch is composed of amylose and amylopectin (Zhao et al., 2022a), the composition and proportion of which directly affect grain quality. In rice endosperm, amylose is synthesized by the Waxy (Wx) gene encoding GBSSI (Zhang et al., 2019), and amylose content plays a decisive role in the eating quality of rice (Pang et al., 2016; Zhang et al., 2022). Inhibition of the branched starch synthesis related gene SSIIa leads to an increase in chalkiness characters, significantly affecting the appearance of rice (Zhang et al., 2011; Zhao et al., 2022b). The PPP is important for sugar metabolism in plants, and G6PGH is involved in biosynthesis and metabolism of plant starch. The cytoplasmic form of G6PGH is necessary for normal starch biosynthesis in maize endosperm. The PGD3 mutation of the maize exhibits severe granule filling and defective embryonic phenotype (Ribeiro et al., 2020). We report the amylose content in rice grains of OsG6PGH1 mutants to be significantly reduced, while the chalkiness rate, taste value, and gelatinization temperature all increase significantly. However, the amylose content of OsG6PGH1(OX) positive grains significantly increased, while the chalkiness rate, taste value, and head rice rate all decreased significantly (Figure 5). Therefore, OsG6PGH1 can affect rice quality traits such as appearance, cooking, eating, and processing qualities by regulating contents of main storage substances (protein and starch).
G6PGH plays an important role in the response of plants to abiotic stress (Chen et al., 2020). Transgenic G6PGH in maize can significantly improve heat tolerance, thereby increasing maize yield under high temperature stress (Ribeiro et al., 2020). Overexpression of 6PGDH in Arabidopsis significantly enhances cold tolerance (Tian et al., 2021). In rice seedlings treated with drought, cold, high salt, and abscisic acid, the transcription levels of OsG6PGH1 and OsG6PGH2 were upregulated (Hou et al., 2007). Under salt stress, expression of OsG6PGH in rice stems is upregulated (Huang et al., 2003), indicating that OsG6PGH actively participates in the response of rice to abiotic stress. We report OsG6PGH1 to be neither localized on the cell membrane nor in the rice nucleus (Figure 2), but possibly in the cytoplasm, consistent with previous results. Analysis of expression patterns reveals OsG6PGH1 to be a constitutive expression gene that is expressed in all rice tissues, consistent with expression of OsG6PGH in root, stem, and leaf tissues of cucumber (Wei et al., 2010). We report salt resistance in rice seedlings with OsG6PGH1 homozygous mutants to be significantly reduced, but for salt resistance in OsG6PGH1(OX) positive seedlings to be significantly enhanced (Figure 6), consistent with expression levels in corresponding salt stress-related genes. Therefore, under salt stress conditions, OsG6PGH1 can positively regulate expression of genes related to the salt stress response, thereby improving rice salt stress resistance.
In summary, OsG6PGH1 was screened, identified, and validated to interact with OsAAP6 through yeast one-hybrid technology, and in vivo point-to-point rotation and local surface plasmon resonance tests. OsG6PGH1 is a constitutive expressed gene, and its encoded OsG6PGH1 may be located in the cytoplasm. Expression levels of OsAAP6 were significantly upregulated in OsG6PGH1(OX) transgenic positive plants, resulting in a significant increase in grain protein content (GPC), and improved nutritional quality of rice. OsG6PGH1 positively regulates amylose content, and negatively regulates rice chalkiness rates and taste values, significantly affecting various quality traits such as appearance, cooking, and eating qualities of rice. Additionally, OsG6PGH1 is involved in regulating the expression of salt-stress-related genes, enhancing the salt-stress tolerance of rice. Therefore, OsG6PGH1 affects multiple quality traits, participates in the salt stress response of rice, and provides important genetic resources for molecular design and breeding of high-quality and multi-resistant rice varieties.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
BP: Writing – original draft, Writing – review & editing, Data curation, Funding acquisition, Investigation, Methodology, Project administration. YL: Conceptualization, Data curation, Formal analysis, Methodology, Writing – original draft. JQ: Data curation, Formal analysis, Methodology, Writing – original draft. JP: Conceptualization, Data curation, Formal analysis, Visualization, Writing – original draft. XS: Conceptualization, Data curation, Methodology, Writing – original draft. XT: Data curation, Methodology, Resources, Writing – original draft. ZZ: Formal analysis, Methodology, Resources, Writing – original draft. YH: Funding acquisition, Investigation, Methodology, Writing – original draft. RP: Formal analysis, Investigation, Supervision, Writing – original draft. WZ: Conceptualization, Data curation, Methodology, Writing – original draft. JZ: Data curation, Formal analysis, Investigation, Methodology, Writing – original draft. YS: Data curation, Formal analysis, Investigation, Writing – review & editing. QW: Data curation, Investigation, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was financially supported by National Natural Science Foundation of China (U2004141; 12305399), Key Project of Science and Technology in Henan Province (222102110141; 242102110266; 242102110270), Key R&D and Promotion Project of Xinyang-Science and Technology Research Project (20230025), Nanhu Scholars Program for Young Scholars of XYNU, and Student Research Fund Project of XYNU (202410477040).
We thank EditSprings (https://www.editsprings.cn) for providing the expert linguistic services.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1436998/full#supplementary-material
Bailey-Serres, J., Nguyen, M. T. (1992). Purification and characterization of cytosolic 6-phosphogluconate dehydrogenase isozymes from maize. Plant Physiol. 100, 1580–1583. doi: 10.1104/pp.100.3.1580
Balindong, L. J., Ward, M. R., Rose, J. T., Liu, L., Raymond, C. A., Snell, P. J., et al. (2018). Rice grain protein composition influences head rice yield. Cereal Chem. 95, 253–263. doi: 10.1002/cche.10031
Bao, J. S., Deng, B. W., Zhang, B. L. (2023). Molecular and genetic bases of rice cooking and eating quality: an updated review. Cereal Chem. 100, 1220–1233. doi: 10.1002/cche.10715
Chen, L., Kuai, P., Ye, M. F., Zhou, S. X., Lu, J., Lou, Y. G. (2020). Overexpression of a cytosolic 6-phosphogluconate dehydrogenase gene enhances the resistance of rice to Nilaparvata lugens. Plants 9, 1529. doi: 10.3390/plants9111529
Chen, P. L., Shen, Z. K., Ming, L. C., Li, Y. B., Dan, W. H., Lou, G. M., et al. (2018). Genetic basis of variation in rice seed storage protein (albumin, globulin, prolamin, and glutelin) content revealed by genome-wide association analysis. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00612
Custodio, M. C., Cuevas, R. P., Ynion, J., Laborte, A. G., Velasco, M. L., Demont, M. (2019). Rice quality: How is it defined by consumers, industry, food scientists and geneticists? Trends Food Sci. Technol. 92, 122–137. doi: 10.1016/j.tifs.2019.07.039
Erinaldo, G. P., Cassia, P. C. B., Carlos, A. B., Leandro, A. S., Joviana, L., Claudete, S. C., et al. (2022). The amino acid transporter OsAAP1 regulates the fertility of spikelets and the efficient use of n in rice. Plant Soil. 480, 507–521. doi: 10.1007/S11104-022-05598-9
Fang, Z. M., Wu, B. W., Ji, Y. Y. (2021). The amino acid transporter OSAAP4 contributes to rice tillering and grain yield by regulating neutral amino acid allocation through two splicing variants. Rice 14, 2. doi: 10.1186/s12284-020-00446-9
Gopika, J., Aarti, B., Prince, C., Melinda, F., Fogarasi, S. (2022). A narrative review on rice proteins: current scenario and food industrial application. Polymers 14, 3003. doi: 10.3390/polym14153003
Hanks, J. F., Schubert, K., Tolbert, N. E. (1983). Isolation and characterization of infected and uninfected cells from soybean nodules role of uninfected cells in ureide synthesis. Plant Physiol. 71, 869–873. doi: 10.1104/pp.71.4.869
He, Y., Wang, S. Z., Ding, Y. (2013). Identification of novel glutelin subunits and a comparison of glutelin composition between japonica and indica rice (Oryza sativa L.). J. Cereal Sci. 57, 362–371. doi: 10.1016/j.jcs.2012.12.009
Hou, F. Y., Ji, H., Yu, S. L., Zhang, H. S. (2007). The 6-phosphogluconate dehydrogenase genes are responsive to abiotic stresses in rice. J. Integr. Plant Biol. 49, 655–663. doi: 10.1111/j.1744-7909.2007.00460.x
Hölscher, C., Lutterbey, M. C., Lansing, H., Meyer, T., Schaewen, A. V. (2016). Defects in peroxisomal 6-phosphogluconate dehydrogenase isoform PGD2 prevent gametophytic interaction in Arabidopsis thaliana. Plant Physiol. 171, 192–205. doi: 10.1104/pp.15.01301
Huang, J., Zhang, H., Wang, J., Yang., J. (2003). Molecular cloning and characterization of rice 6-phosphogluconate dehydrogenase gene that is up-regulated by salt stress. Mol. Biol. Rep. 30, 223–227. doi: 10.1023/A:1026392422995
Itishree, N., Bijayalaxmi, S., Chinmay, P., Cayalvizhi, B., Seenichamy, R. P., Jawahar, L. K., et al. (2023). Genome-wide analysis of amino acid transporter gene family revealed that the allele unique to the aus variety is associated with amino acid permease 17 (OsAAP17) amplifies both the tiller count and yield in indica rice (Oryza sativa l.). Agronomy 13. doi: 10.3390/AGRONOMY13102629
Ji, Y. Y., Huang, W. T., Wu, B. W., Fang, Z. M., Wang, X. L. (2020). The amino acid transporter AAP1 mediates growth and grain yield by regulating neutral amino acid uptake and reallocation in Oryza sativa. J. Exp. Bot. 71, 4763–4777. doi: 10.1093/jxb/eraa256
Li, P., Chen, Y. H., Lu, J., Zhang, C. Q., Liu, Q. Q., Li, Q. F. (2022a). Genes and their molecular functions determining seed structure, components, and quality of rice. Rice 15, 18. doi: 10.1186/s12284-022-00562-8
Li, Y. B., Fan, C. C., Xing, Y. Z., Yun, P., Luo, L. J., Yan, B., et al. (2014). Chalk5 encodes a vacuolar h(+)-translocating pyrophosphatase influencing grain chalkiness in rice. Nat. Genet. 46, 398–404. doi: 10.1038/ng.2923
Li, Q. P., Zhang, C. L., Wen, J. C., Chen, L. J., Shi, Y. T., Yang, Q. H., et al. (2023). Transcriptome analyses show changes in gene expression triggered by a 31-bp indel within OsSUT3 5’UTR in rice panicle. Int. J. Mol. Sci. 24, 10640. doi: 10.3390/ijms241310640
Li, Y. S., Peng, K. K., Tian, Y., Ren, Z. P., Cang, J., Xu, Q. H., et al. (2022c). Study on lncRNA1808 regulating cold resistance response of PPP rate-limiting enzyme (Ta6PGDH) in winter wheat. J. Triticeae Crops 42, 1047–1058. doi: 10.7606/j.issn.1009-1041.2022.09.01
Li, Y., Xiao, J., Chen, L., Huang, X., Cheng, Z., Han, B., et al. (2018). Rice functional genomics research: past decade and future. Mol. Plant 11, 359–380. doi: 10.1016/j.molp.2018.01.007
Li, Y. F., Yang, Z., Yang, C. K., Liu, Z. H., Shen, S. Q., Zhan, C. S., et al. (2022b). The NET locus determines the food taste, cooking and nutrition quality of rice. Sci. Bull. 67, 2045–2049. doi: 10.1016/j.scib.2022.09.023
Liu, Y., Zhang, N., Li, P., Yu, L., Chen, S. M., Zhang, Y., et al. (2019). Low-cost localized surface plasmon resonance biosensing platform with a response enhancement for protein detection. Nanomaterials 9, 1019. doi: 10.3390/nano9071019
Lu, K., Wu, B. W., Wang, J., Zhu, W., Nie, H. P., Qian, J. J., et al. (2018). Blocking amino acid transporter OsAAP3 improves grain yield by promoting outgrowth buds and increasing tiller number in rice. Plant Biotechnol. J. 16, 1710–1722. doi: 10.1111/pbi.12907
Nie, S. S., Li, H. Y., Zhang, L., Yang, G., Hang, J. N., Huang, W. T., et al. (2023). Analysis of spatiotemporal expression characteristics of rice amino acid permeability enzyme gene OsAAP13. Mol. Plant Breed. 21, 7667–7676. doi: 10.13271/j.mpb.021.007667
Pang, Y. L., Ali, J., Wang, X. Q., Franje, N. J., Revilleza, J. E., Xu, J. L., et al. (2016). Relationship of rice grain amylose, gelatinization temperature and pasting properties for breeding better eating and cooking quality of rice varieties. PloS One 11, e0168483. doi: 10.1371/journal.pone.0168483
Peng, B., He, L. L., Tian, X. Y., Xin, Q. Q., Sun, Y. F., Song, X. H., et al. (2019). Expression analysis of OsAAP6 gene under different nitrogen conditions and its effects on grain nutritional quality in rice. Southwest China J. Agric. Sci. 32, 1973–1979. doi: 10.16213/j.cnki.scjas.2019.9.002
Peng, B., Kong, D. Y., Huang, Y. J., Jia, Y. Y., Ma, C. X., Song, H., et al. (2018). Bioinformatics analysis of OsAAP6 gene structure and function in rice. Southwest China J. Agric. Sci. 31, 429–436. doi: 10.16213/j.cnki.scjas.2018.3.001
Peng, B., Sun, X. Y., Tian, X. Y., Kong, D. Y., He, L. L., Peng, J., et al. (2023). OsNAC74 affects grain protein content and various biological traits by regulating OsAAP6 expression in rice. Mol. Breed. 43, 87. doi: 10.1007/s11032-023-01433-w
Peng, B., Wang, L., Q. Fan, C. C., Jiang, G. H., Luo, L. J., Li, Y. B., et al. (2014). Comparative mapping of chalkiness components in rice using five populations across two environments. BMC Genet. 15, 49. doi: 10.1186/1471-2156-15-49
Peng, B., Zhang, Q. X., Liu, Y., Zhao, Q., Zhao, J. H., Zhang, Z. G., et al. (2024). OsAAP8 mutation leads to significant improvement in the nutritional quality and appearance of rice grains. Mol. Breed. 44, 34. doi: 10.1007/s11032-024-01473-w
Peng, B., Zhang, Q. X., Tian, X. Y., Sun, Y. F., Huang, X. H., Pang, R. H., et al. (2021). Influencing factors of grain nutritional quality and its genetic improvement strategy in rice. J. Biotechnol. Res. 7, 1–11. doi: 10.32861/jbr.16
Riaz, M., Akhter, M., Iqbal, M., Ali, S., Khan, R. A. R., Raza, M., et al. (2018). Estimation of amylose, protein and moisture content stability of rice in multi locations. Afr. J. Agric. Res. 13, 1213–1219. doi: 10.5897/AJAR
Ribeiro, C., Hennen-Bierwagen, T. A., Myers, A. M., Cline, K., Settles, A. M. (2020). Engineering 6-phosphogluconate dehydrogenase improves grain yield in heat-stressed maize. Proc. Natl. Acad. Sci. United States America 117, 33177–33185. doi: 10.1073/pnas.2010179117
Shi, S. J., Wang, E. T., Li, C. X., Cai, M. L., Cheng, B., Cao, C. G., et al. (2022). Use of protein content, amylose content, and RVA parameters to evaluate the taste quality of rice. Front. Nutr. 8, 758547. doi: 10.3389/fnut.2021.758547
Su, L., Liu, X., An, J. P., Li, H. H., Zhao, J., Hao, Y. J., et al. (2016). Molecular cloning and functional analysis of a 6-phosphogluconate dehydrogenase gene Md6PGDH1 in apple. Acta Hortic. Sin. 43, 1225–1235. doi: 10.16420/j.issn.0513-353x.2016-0202
Tanksley, S. D., Kuehn, G. D. (1985). Genetics, subcellular localization, and molecular characterization of 6-phosphogluconate dehydrogenase isozymes in tomato. Biochem. Genet. 23, 441–454. doi: 10.1007/BF00499085
Taylor, M. R., Reinders, A., Ward., J. M. (2015). Transport function of rice amino acid permeases (AAPs). Plant Cell Physiol. 56, 1355–1363. doi: 10.1093/pcp/pcv053
Tegeder, M., Hammes, U. Z. (2018). The way out and in: phloem loading and unloading of amino acids. Curr. Opin. Plant Biol. 43, 16–21. doi: 10.1016/j.pbi.2017.12.002
Tian, Y., Peng, K. K., Bao, Y. Z., Zhang, D., Meng, J., Wang, D. J., et al. (2021). Glucose-6-phosphate dehydrogenase and 6-phosphogluconate dehydrogenase genes of winter wheat enhance the cold tolerance of transgenic Arabidopsis. Plant Physiol. Biochem. 161, 86–97. doi: 10.1016/j.plaphy.2021.02.005
Tong, C., Gao, H., Luo, S., Liu, L., Bao, J. (2019). Impact of postharvest operations on rice grain quality: a review. Compr. Rev. Food Sci. Food Saf. 18, 626–640. doi: 10.1111/1541-4337.12439
Wang, J. W., Tian, H., Yu, X., Zheng, L. P. (2017). Glucose-6-phosphate dehydrogenase plays critical role in artemisinin production of Artemisia annua under salt stress. Biol. Plantarum 61, 529–539. doi: 10.1007/s10535-016-0674-7
Wang, J., Wu, B. W., Lu, K., Qian, J. J., Chen, Y. P., Fang, Z. M. (2019). The amino acid permease 5 (OsAAP5) regulates tiller number and grain yield in rice. Plant Physiol. 180, 1031–1045. doi: 10.1104/pp.19.00034
Wang, S. Y., Yang, Y. H., Guo, M., Zhong, C. Y., Yan, C. J., Sun, S. Y. (2020). Targeted mutagenesis of amino acid transporter genes for rice quality improvement using the CRISPR/Cas9 system. Crop J. 8, 457–464. doi: 10.1016/j.cj.2020.02.005
Wang, Z., Zhou, Y., Ren, X.-Y., Wei, K., Fan, X. L., Huang, L. C., et al. (2022). Co-overexpression of two key source genes, OsBMY4 and OsISA3, improves multiple key traits of rice seeds. J. Agric. Food Chem. 71, 615–625. doi: 10.1021/acs.jafc.2c06039
Wang, Y. P., Wei, Z. Y., Xing, S. C. (2018). Stable plastid transformation of rice, a monocot cereal crop. Biochem. Biophys. Res. Commun. 503, 2376–2379. doi: 10.1016/j.bbrc.2018.06.164
Wei, Y., Cheng, X. Y., Li, Z. L., Wang, Y. P., Shi, J. L., Wu, Z. M., et al. (2010). Cloning of 6-phosphogluconate dehydrogenase gene cDNA fragments from cucumber and expression analysis. J. Nanjing Agric. Univ. 33, 37–42.
Xia, D., Wang, Y. P., Shi, Q. Y., Wu, B., Yu, X. M., Zhang, C. Q., et al. (2022). Effects of Wx genotype, nitrogen fertilization, and temperature on rice grain quality. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.901541
Xiong, Q. X., Zhong, L., Shen, T. H., Cao, C. H., He, H. H., Chen, X. R. (2019). iTRAQ-based quantitative proteomic and physiological analysis of the response to N deficiency and the compensation effect in rice. BMC Genomics 20, 681. doi: 10.1186/s12864-019-6031-4
Xu, H. Y., Li, S. F., Kazeem, B. B., Ajadi, A. A., Luo, J. J., Yin, M., et al. (2022). Five rice seed-specific NF-YC genes redundantly regulate grain quality and seed germination via interfering gibberellin pathway. Int. J. Mol. Sci. 23, 8382. doi: 10.3390/ijms23158382
Yang, G., Wei, X. L., Fang, Z. M. (2022). Melatonin mediates axillary bud outgrowth by improving nitrogen assimilation and transport in rice. Front. Plant Sci. 13, 900262. doi: 10.3389/fpls.2022.900262
Yang, X. Y., Yang, G., Wei, X. L., Huang, W. T., Fang, Z. M. (2023a). OsAAP15, an amino acid transporter in response to nitrogen concentration, mediates panicle branching and grain yield in rice. Plant Sci. 330, 111640. doi: 10.1016/j.plantsci.2023.111640
Yang, Y. H., Zhang, Y., Sun, Z. X., Shen, Z. Y., Li, Y. G., Guo, Y. F., et al. (2023b). Knocking out OsAAP11 to improve rice grain quality using CRISPR/Cas9 system. Int. J. Mol. Sci. 24, 14360. doi: 10.3390/ijms241814360
Zhang, C. Q., Zhu, J. H., Chen, S. J., Fan, X. L., Li, Q. F., Lu, Y., et al. (2019). Wxlv, the ancestral allele of rice waxy gene. Mol. Plant 12, 1157–1166. doi: 10.1016/j.molp.2019.05.011
Zhang, G. Y., Cheng, Z. J., Zhang, X., Guo, X. P., Su, N., Jiang, L., et al. (2011). Double repression of soluble starch synthase genes SSIIa and SSIIIa in rice (Oryza sativa l.) uncovers interactive effects on the physicochemical properties of starch. Genome 54, 448–459. doi: 10.1139/g11-010
Zhang, W., Liu, Y. X., Luo, X. L., Zeng, X. F. (2022). Pasting, cooking, and digestible properties of; japonica; rice with different amylose contents. Int. J. Food Properties. 25, 936–947. doi: 10.1080/10942912.2022.2069806
Zhang, X. F., Zhu, Q., Hua, Y. Y., Jia, L. H. Y., Qiu, S. Y., Chen, Y. J., et al. (2024). Screening and validation of OsCYP22 interacting proteins in rice. Acta Agronomica Sin. 50, 1628–1634. doi: 10.3724/SP.J.1006.2024.32041
Zhao, H., Li, Z. X., Wang, Y. Y., Wang, J. Y., Xiao, M. G., Liu, H., et al. (2022b). Cellulose synthase-like protein OsCSLD4 plays an important role in the response of rice to salt stress by mediating abscisic acid biosynthesis to regulate osmotic stress tolerance. Plant Biotechnol. J. 20, 468–484. doi: 10.1111/pbi.13729
Zhao, M., Lin, Y., Chen, H. (2020). Improving nutritional quality of rice for human health. Theor. Appl. Genet. 133, 1397–1413. doi: 10.1007/s00122-019-03530-x
Zhao, D. S., Zhang, C. Q., Li, Q. F., Liu, Q. Q. (2022a). Genetic control of grain appearance quality in rice. Biotechnol. Adv. 60, 108014. doi: 10.1016/j.biotechadv.2022.108014
Zhou, H., Xia, D., He, Y. Q. (2019). Rice grain quality—traditional traits for high quality rice and health-plus substances. Mol. Breeding. 40, 201–204. doi: 10.1007/s11032-019-1080-6
Zhou, T. Y., Zhou, Q., Li, E. P., Yuan, L. M., Wang, W. L., Zhang, H., et al. (2020). Effects of nitrogen fertilizer on structure and physicochemical properties of ‘super’ rice starch. Carbohydr. Polymers. 239, 116237. doi: 10.1016/j.carbpol.2020.116237
Keywords: rice, OsG6PGH1, grain quality, proteome, salt stress
Citation: Peng B, Liu Y, Qiu J, Peng J, Sun X, Tian X, Zhang Z, Huang Y, Pang R, Zhou W, Zhao J, Sun Y and Wang Q (2024) OsG6PGH1 affects various grain quality traits and participates in the salt stress response of rice. Front. Plant Sci. 15:1436998. doi: 10.3389/fpls.2024.1436998
Received: 23 May 2024; Accepted: 25 June 2024;
Published: 10 July 2024.
Edited by:
Hao Zhou, Sichuan Agricultural University, ChinaCopyright © 2024 Peng, Liu, Qiu, Peng, Sun, Tian, Zhang, Huang, Pang, Zhou, Zhao, Sun and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bo Peng, cGVuZ2JvQHh5bnUuZWR1LmNu; Yanfang Sun, MzcwMzgwNDYwQHFxLmNvbQ==; Quanxiu Wang, d2FuZ3F1YW54aXVAeHludS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.