- 1Ministry of Education Key Laboratory of Xinjiang Phytomedicine Resource Utilization, Xinjiang Production and Construction Corps Key Laboratory of Oasis Town and Mountain-basin System Ecology, College of Life Sciences, Shihezi University, Shihezi, Xinjiang, China
- 2Xinjiang Second Medical College, Karamay, Xinjiang, China
Calcium-Dependent Protein Kinases (CDPKs) are a class of serine/threonine protein kinases encoded by several gene families that play key roles in biotic and abiotic stresses response and plant growth and development. However, snow lotus (Saussurea involucrata kar L.) CDPKs has rarely been reported. In this study, 20 CDPK genes in snow lotus were identified based on transcriptome data and classified into four groups (I-IV) based on their structural features and phylogenetic analyses. Among them, the transcript levels of SikCDPK1 were significantly induced by low temperature and multiple hormone treatments, and SikCDPK1 gene was found to have different expression in snow lotus seeds, leaves, stems and roots. The full-length promoter activity of SikCDPK1 gene was higher than that of the 5’ end deletion fragment, and the promoter fragment containing the low temperature inducing element had increased activation after low temperature treatment. The promoter activity of SikCDPK1 gene was mainly expressed in roots and rosette leaves. In addition, overexpressing plants of SikCDPK1 were more tolerant compared to the wild type after being subjected to low temperature stress. Physiological analyses indicated that SikCDPK1 improved plant tolerance to low temperature stress by maintaining cell membrane stability and reducing the accumulation of reactive oxygen species (ROS). These findings provided insights into CDPK gene families in snow lotus and broaden our understanding of the biological role of SikCDPK1 and the mechanism of low temperature stress tolerance in snow lotus.
1 Introduction
In plants, low temperature is a common environmental stress that has important effects on plant growth and development. To adapt to the complex and changing external environment, plants have evolved a series of regulatory mechanisms, in which the kinase signaling pathway plays an important role (Zhang et al., 2022). In past studies, an important class of plant kinase family, the CDPK family, has been identified, and members of the CDPK family play an important role in the regulation of a variety of biological processes, including low temperature response, through Ca2+ signaling (Yip Delormel and Boudsocq, 2019).
Ca2+ plays an irreplaceable role as an important and ubiquitous second messenger in plant growth and development as well as in biotic and abiotic stress response (Aldon et al., 2018). Indeed, calcium is both an essential plant mineral element and a secondary signaling molecule. It can keep calcium concentrations in the cytoplasm in the hundreds of nanomolar levels and outside the cell membrane in the 1-10 millimolar range (Luan and Wang, 2021). This natural concentration gap puts it in a Ca2+ transient state that can be used as a signal to rapidly sense external stimuli, which is also known as Ca2+ signatures (Allan et al., 2022).
CDPK is a specific calcium sensor in plants and typically contains four distinct structural domains connecting a Ser/Thr protein kinase structural domain to an EF-hands motif containing a Ca2+ binding structural domain flanked by N-terminal and C-terminal variable regions (Klimecka and Muszyńska, 2007). The VNTD structural domain is located at the N-terminal end of the CDPK and has a significantly different amino acid sequence and length. The amino acid sequences and lengths vary significantly, and most CDPKs contain N-myristoylation sites and S-palmitoylation sites, which have been linked not only to the subcellular localization of CDPK proteins but also to the specificity of substrate recognition (Hrabak et al., 2003; Asano et al., 2005; Stael et al., 2012; Asai et al., 2013). Upon binding Ca2+, CDPKs are activated by autophosphorylation and phosphorylation, and the active kinases phosphorylate downstream targets such as ions channel, transcription factors, and metabolic enzymes (Wernimont et al., 2010; Liese and Romeis, 2013).
Numerous studies have shown that members of the CDPK gene family are extensively involved in growth and development, biotic and abiotic stress response, and hormone metabolism in plants. CDPKs have been implicated in mediating Ca2+-regulated pollen tube growth as an important node in Ca2+ signaling pathways (Estruch et al., 1994; Moutinho et al., 1998). IbCDPK28 expression levels increased significantly in sweet potato tuberous roots during tuberization, and may play a functional role in regulating tuberous root growth (Li et al., 2022). Cold-induced calcium signal is perceived by AtCPK28 which in turn phosphorylates its downstream target NLP7 thereby enhancing cold tolerance in plants (Ding et al., 2022). VpCDPK9 and VpCDPK13 enhance powdery mildew resistance by regulating SA and ethylene signaling in vivo (Hu et al., 2021). AtCPK4 and AtCPK11 are two important positive regulators in the ABA signaling pathway (Zhu et al., 2007). Maize ZmCDPK1 plays a negative role in cold stress signaling (Mittal et al., 2017). Whereas ZmCDPK4 positively regulates ABA signaling and enhances drought stress tolerance (Jiang et al., 2013). CDPKs are also involved in the synthesis as well as signaling of ethylene, auxin, jasmonic acid, gibberellin and ABA hormones (Li et al., 2017). Together, these studies suggest that CDPK-mediated abiotic stress and hormone responses are complex and conserved in plants.
Given their importance, the CDPK gene family has been widely identified and characterized in many plant species. For example, 34, 31, 30, 28, 29, 31, and 29 CDPKs were found in Arabidopsis thaliana, Oryza sativa (rice), Brachypodium distachyon, Hordeum vulgare (barley), Setaria italica (Foxtail millet), Capsicum annuum (pepper) and Solanum lycopersicum (tomato) (Cheng et al., 2002; Ray et al., 2007; Wen et al., 2020; Yang et al., 2017; Yu et al., 2018; Cai et al., 2015; Hu et al., 2016). Although a number of CDPK family genes involved in low temperature response have been identified, the understanding of the specific functions of these genes in low temperature adversity is still limited. Snow lotus is an excellent plant that tolerates extreme climates, and can grow in rock crevice in mountains and cliffs with perennial snow, rarefied air and strong ultraviolet radiation. The study of its CDPKs gene is undoubtedly of great significance for elucidating the low temperature tolerance mechanism of snow lotus.
However, up to now, there have been no reports of snow lotus CDPKs, and the functional studies of snow lotus CDPK genes are also rarely reported. In this study, we first identified 20 SikCDPK genes from the low temperature transcriptome database of snow lotus. The sequence, structure and evolution of SikCDPKs were analyzed by bioinformatics methods. The representative gene SikCDPK1 was screened for cloning and expression analysis. The mechanism of SikCDPK1 in low temperature response was also explored, which provided new clues to further reveal the mechanism of low temperature tolerance in snow lotus adversity.
2 Materials and methods
2.1 Screening and identification of SikCDPK gene family members involved in low temperature response
The snow lotus low temperature transcriptome database was obtained from (http://www.shengtingbiology.com/SaussureaKBase/index.jsp), meanwhile, the Arabidopsis CDPK family member protein sequences from the Arabidopsis Information Resource (TAIR, https://www.arabidopsis.org/) database. The seed files (PF00036 and PF07714) of the CDPK gene family were downloaded by the Pfam online database (http://pfam.xfam.org/), and the protein sequences containing the conserved domains of CDPK were analyzed by hmmsearch. All identified CDPK proteins of snow lotus were submitted to Pfam and NCBI website’s (Conserved Domain Database, CDD) for conserved structural domain validation to confirm the snow lotus CDPK gene family members.
2.2 Sequence analysis and subcellular localization of SikCDPK gene family members involved in low temperature response
The molecular weight, isoelectric point (pI), hydrophilicity index and other physicochemical properties of the snow lotus CDPK gene family were queried by the ProtParam tool (http://web.ExPasy.orP/ProtParam/) of the ExPASy online software; the online tool WoLF PSORT (https://wolfpsort.hgc.jp/) was used to analyze the sequence and subcellular localization of the SikCDPK gene family members. The online tool Myristoylaton (https://web.expasy.org/myristoylator/) was used to predict N-myristoylation, and CSS-Plam was used to predict palmitoylation. TMHMMServerv2.0 was used to predict the transmembrane structural domains, and Wolf POSRT tool (https://www.genscript.com/psort.html) was used for subcellular localization.
2.3 Phylogenetic tree construction of SikCDPK gene family involved in low temperature response
The protein sequences of snow lotus CDPK and Arabidopsis CDPK were compared and analyzed using MEGA 7.0 software and ClustalX, and the phylogenetic tree was constructed using the Neighbor-Joining algorithm with the Bootstrap value set to 1000.
2.4 Conserved motifs of the SikCDPK gene family involved in low temperature response
The conserved motifs of the CDPK family of snow lotus were analyzed using Multiple Em for Motif Elicitation (MEME, https://meme-suite.org/meme/tools/meme) online software, with the parameters set to a maximum discovery number of motifs of 10 and a maximum length of motifs of 100 nt (Nucleotide).
2.5 Cloning and bioinformatics analysis of SikCDPK1 gene
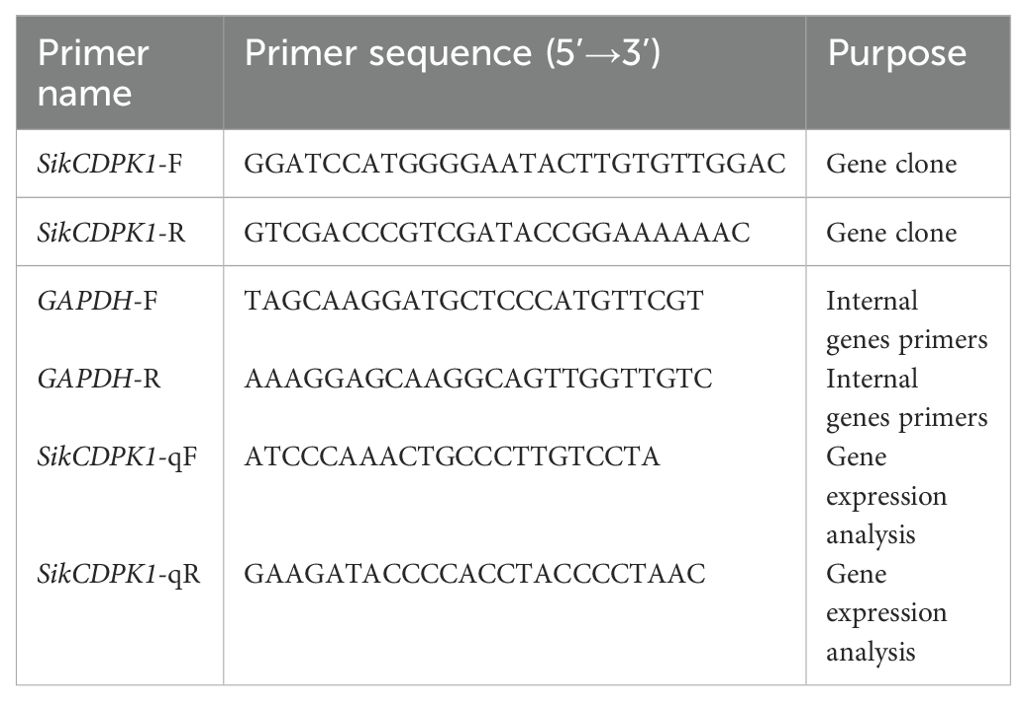
According to the low temperature transcriptome sequencing data of snow lotus, the GhCDPK1 gene sequence was used as a probe search, and after removing the duplicate sequences by multiple sequence comparison, the complete open reading frames (ORFs) of the sequences were analyzed one by one by using the ORF Finder tool (https://www.ncbi.nlm.nih.gov/orffinder) in NCBI. The ORF primers were designed in Table 1. The total RNA was extracted and the first strand of cDNA was synthesized according to the TaKaRa kit. The primers for the target genes, SikCDPK1-F and SikCDPK1-R, were used in Table 1 for RT-PCR amplification, and the PCR program was as follows: pre-denaturation at 94°C for 5 min; denaturation at 94°C for 30 s; replication at 63°C for 40 s; extension at 72°C for 1 min and 30 s; 30 cycles. The positive product was named as SikCDPK1, sequenced by Beijing Huada Genetics, and the conserved domains of SikCDPK1 gene were analyzed by SMART online analysis software (http://smart.embl-heidelberg.de/). The phylogenetic tree was constructed using ClustalX and MEGA7.0 software.
2.6 Analysis of spatio-temporal expression pattern of SikCDPK1 gene
The total RNA of snow lotus was extracted from the young stems and leaves of snow lotus grown for 45 days. Three biological repeats in each sample were performed. The first strand of cDNA was synthesized in accordance with the steps in the TaKaRa kit, and assayed by qRT-PCR. Amplification by qRT-PCR was performed using TB Green® Premix Ex Taq™ II (Takara). The qRT-PCR reaction was carried out with the primers in Table 1 for the endogenous genes of GAPDH-F and GAPDH-R, as well as the primer for the target gene, SikCDPK1. Reaction system: cDNA (1 µL), GAPDH-F/SikCDPK1-qF (0.25 µL), GAPDH-R/SikCDPK1-qR (0.25 µL), SYBR Green PCR Master Mix (5 µL), ddH2O (3.5 µL), and the reaction program: 95°C pre-denaturation for 5 min; 95°C 10 s, 62°C 30 s, 72°C 30 s 40 cycles. Three replicates were set up for each sample, and the average value was taken. The expression of snow lotus SikCDPK1 gene in different tissues and organs was calculated using the 2-ΔΔCt method (Livak and Schmittgen, 2001), and the experimental data were subjected to LSD (least significance difference) multiple ANOVA using the SPSS 18.0 software, with the test of significance set at P<0.05 (significant level) and P<0.01 (highly significant level), and graphs were made in using Excel software.
2.7 Analysis of SikCDPK1 gene involved in cold and hormone
The aseptic seedlings of snow lotus were placed in -4°C light incubator for cold stress treatment, and the leaf samples were taken from treatment 0, 1, 3, 6, 12, 24 h. Three replicates were set up for each sample, and qRT-PCR reaction was carried out in accordance with the above method, and the expression of SikCDPK1 gene was calculated at different times of the low temperature treatment.
Snow lotus seedlings grown for 3 months with stable and healthy growth were selected, rinsed and placed in MS liquid medium separately. After 48 hours of recovery, they were placed in MS medium supplemented with 100 µM ABA, 100 µM GA3, 1 mM SA and treated for 0, 1, 3, 6, 12 and 24 hours, respectively. Samples were collected, and three replicates were set up for each sample. qRT-PCR reactions were performed according to the above methods. The expression of SikCDPK1 gene after induction by low temperature, ABA, GA3, and SA was calculated by the 2-ΔΔCt method (Livak and Schmittgen, 2001), and the experimental data were subjected to LSD using SPSS 18.0 software. Multiple ANOVA analysis was performed, and GraphPad Prism 9.1 was used for graphing.
2.8 Cloning and activity analysis of SikCDPK1 gene promoter
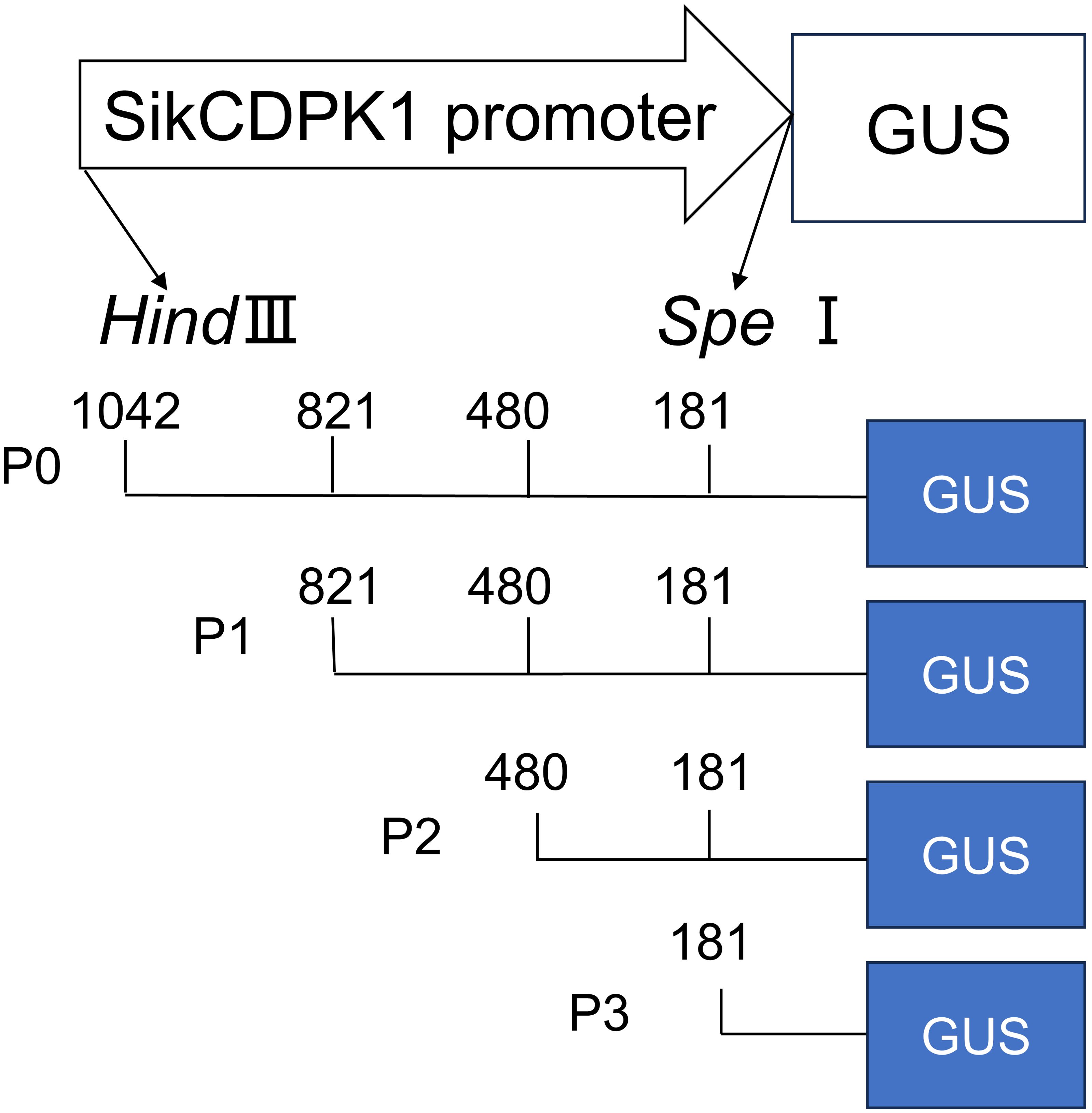
The SikCDPK1 promoter was cloned from snow lotus by TAIL-PCR, and the cis-acting elements of the promoter region were analyzed using the promoter online analysis software PlantCare (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/). The full-length promoter or 5’-end deletion promoter-driven GUS recombinant expression vector pCAMBIA1304-SikCDPK1-P0-GUS, P1-GUS, P2-GUS, and P3-GUS was constructed (Figure 1). Arabidopsis thaliana Col- 0 seeds were sterilized with 70% alcohol for 2 min and washed twice with sterile water; then sterilized with 2% NaClO for 7 min and washed with sterile water. The seeds were sterilized by 70% alcohol for 2 min and washed with sterile water for 2 times, then sterilized by adding 2% NaClO for 7 min and washed with sterile water for 5 times, and then spring-cleaned at 4°C for 3-4 d. The seeds were evenly sown on 1/2 of MS medium. They were incubated in a light incubator at 25°C for 16 h of light and 8 h of darkness, and then transplanted into pots containing vermiculite and nutrient soil (1:1 by volume) at the stage of 2-3 true leaves, and then cultured in an artificial climatic chamber under the same environmental conditions. Arabidopsis thaliana was genetically transformed by the flower drop method (Clough and Bent, 1998). After the transgenic Arabidopsis thaliana was screened to the purity, putting it in the low temperature (4°C). The P0-GUS pure transgenic Arabidopsis thaliana was used as a positive control and wild-type Arabidopsis thaliana (Col- 0) as a negative control, respectively. Mature leaves on the day of flowering were taken and immersed in 0.5 mg/L X- Gluc (Sigma) solution, and then decolorized with anhydrous ethanol to remove chlorophyll after being sheltered from light at 37°C overnight to observe whether blue color appeared or not, and photographed with a dissecting microscope (Vitha et al., 1995). The experiment was repeated three times independently. The promoter activity was analyzed in different tissues of Arabidopsis thaliana by GUS histochemical staining and low temperature treatment was analyzed.
2.9 Response of overexpressed SikCDPK1 gene to cold stress
Using Agrobacterium-mediated method, pCAMBIA2300-GV3101-SikCDPK1 was used to infest transformed tobacco leaf discs, and the screened positive trans-SikCDPK1 gene tobacco was put into the artificial climate chamber at -4°C, and subjected to cold stress treatment for 0, 3, 6, 9 and 12h respectively, and then the plants were put into the incubator at 25°C to recover for 24h, and the morphological changes of the tobacco in the various treatments were observed. The plants were then put into the incubator at 25°C for 24h to observe the morphological changes of tobacco in different treatments, and the samples were taken at different treatment stages and stored in the freezer at -80°C. Three biological repeats in each treatment were performed.
2.10 Physiological indices measurements
To check the physiological indices changes, the leaves were used to identify relative electricity conductivity (REC), contents of MDA, after low temperatures treatments, whereas some plants were kept untreated as controls. Half a gram of fresh leaf sample (chopped into small pieces) was dipped in distilled water for half an hour, and REC1 was measured by using an REC meter. REC2 was recorded by heating the samples in a water bath at 90°C for 50 min. The final value of relative electricity conductivity (REC; %) was determined with this formula: REC%=(REC1÷REC2) ×100 (Chattha et al., 2022). Malondialdehyde (MDA) content was measured according to (Schmedes and Hølmer, 1989) with slight modifications. Powdered samples (0.2 g) were homogenized in 10 mL of 10% (w/v) trichloroacetic acid (TCA). Homogenate was centrifuged at 9660× g for 10 min. Then, 2 mL of the supernatant was mixed with 2 mL of 10% (w/v) TCA containing 0.6% (w/v) of thiobarbituric acid (TBA) and incubated at 100°C for 20 min, and then quickly cooled on ice followed by centrifugation at 9660× g for 10 min. Absorbance at 532, 600, and 450 nm was measured using Jenway 7305 UV/Visible Spectrometer (Jenway, London, UK). The MDA content was calculated according to the formula MDA content (µM/g) = 6.45 (OD 532 − OD 600) − 0.56 OD 450. For the O2- detection, detached leaves were immersed in 100 mL staining solution containing 0.1% (w/v) nitroblue tetrazolium (NBT), 10 mM sodium azide, 50 mM potassium phosphate, pH 6.4 for 15 min. After stopping the reaction with 95% ethanol, the samples were decolorized in 96% ethanol under heating at 40°C (Jambunathan, 2010). Superoxide ions react with NBT and appear as blue. These stained leaves can be photographed by light microscope.
3 Results
3.1 Identification and analysis of SikCDPK gene family involved in low temperature response
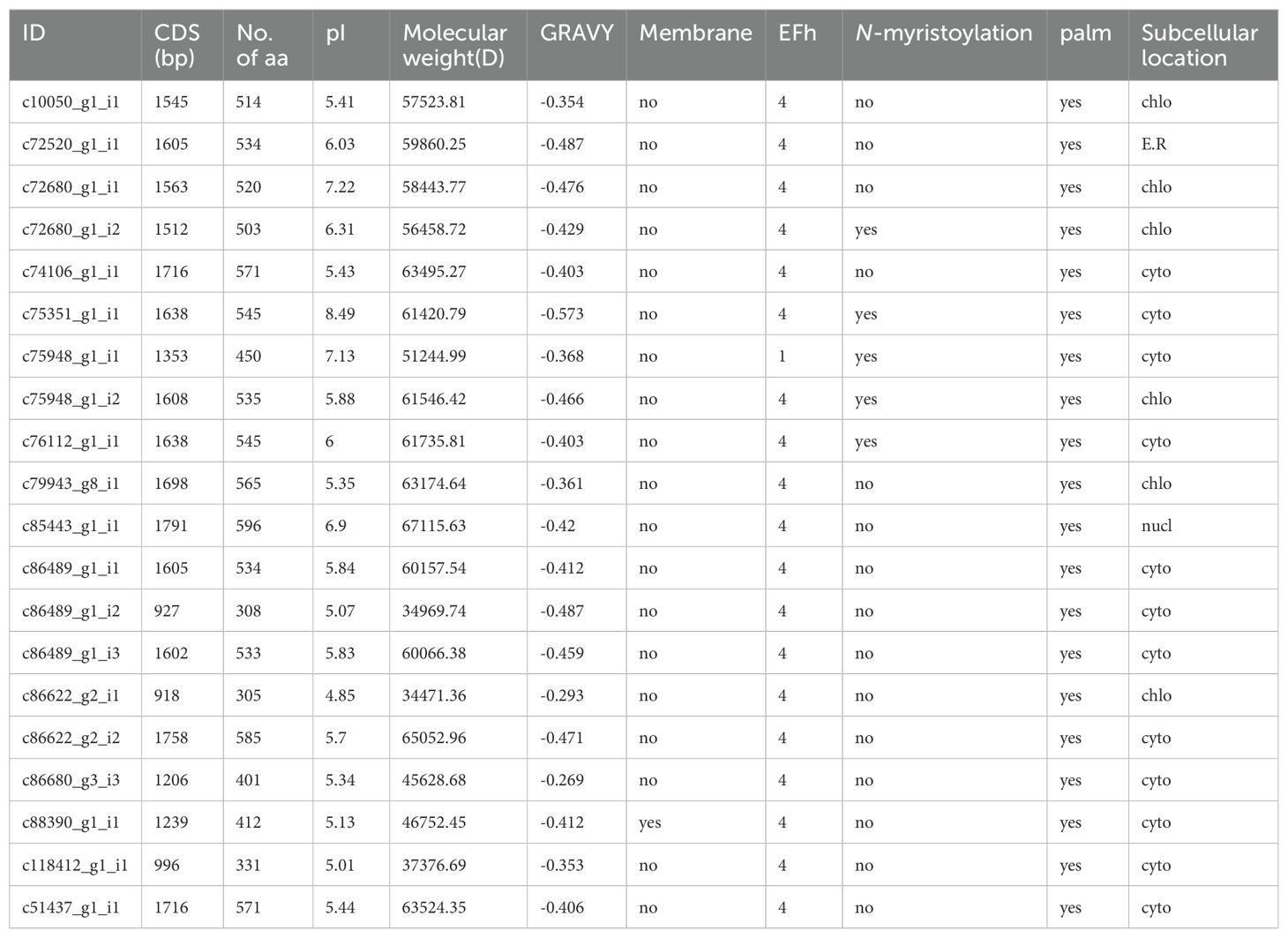
Using the snow lotus transcriptome as the reference sequence and HMMER 3.0 software to search for protein sequences containing the PF07714 model, and further analyzing the conserved structural domains by CDD and SMART software, a total of 20 SikCDPK family members were identified in the snow lotus transcriptome database (Table 2).
The sequence lengths of the snow lotus CDPK family members ranged from 918 bp (c86622_g2_i1) to 1791 bp (c85443_g1_i1), encoding 305 ~ 596 amino acids, respectively, and the relative molecular weights of the proteins ranged from 34,471.36 D to 67,115.63 D. PI analyses revealed that the theoretical isoelectric point of 20 SikCDPK proteins were between 4.85 (c86622_g2_i1) to 8.49 (c75351_g1_i1), of which 17 SikCDPK proteins with pI < 7.0 (mean 5.62) were acidic proteins, and 3 SikCDPK proteins with pI > 7.0 (mean 7.61) were basic proteins. Analysis of GRAVY revealed that the average coefficient of hydrophilicity of all 20 SikCDPK proteins was less than 0, indicating that all snow lotus CDPK proteins are hydrophilic proteins. Analysis with TMHMM Server software revealed that only one SikCDPK protein (c88390_g1_i1) had a transmembrane structure, while the others did not have a transmembrane structural domain. EFh analysis of the calcium-binding region revealed that 20 SikCDPK family members have CDPK-typical, Ca2+-binding EF-hands structural domains, of which 19 family members contain four EF-hands structures like most CDPKs in other species, whereas one family member protein (c75948_g1_i1) has only one EF-hands structure. Analysis of N-myristoylation and palmitoylation revealed that 5 genes in the SikCDPKs family have N-myristoylation sites and all 20 have palmitoylation sites. Analysis of prediction subcellular localization revealed that 12 SikCDPK proteins were located in the cytoplasm, 6 SikCDPK proteins were localized on chloroplasts, 1 SikCDPK protein was localized in the endoplasmic reticulum, and 1 SikCDPK protein was localized in the nucleus (Table 2).
3.2 Phylogenetic analysis of snow lotus SikCDPK families involved in low temperature response
In order to analyze the phylogenetic relationship of the SikCDPK gene family of snow lotus, in this study, the full-length protein sequences of 20 SikCDPKs and 34 AtCDPKs of Arabidopsis thaliana were imported into the MEGA 7.0 software, and the neighbor-joining method was used to construct a phylogenetic tree (Figure 2). With reference to the classification of Arabidopsis thaliana, the 20 SikCDPK proteins were classified into four subgroups, which were called subgroups of Group I, Group ll, Group III, and Group IV. Group I and Group III had the most members with 7 SikCDPK proteins, while Group IV subgroup had the least members with only 1 SikCDPK protein. An in-depth study of the clustering results of CDPK members revealed the presence of genes with similar evolutionary relationships in the same group, suggesting their homology.
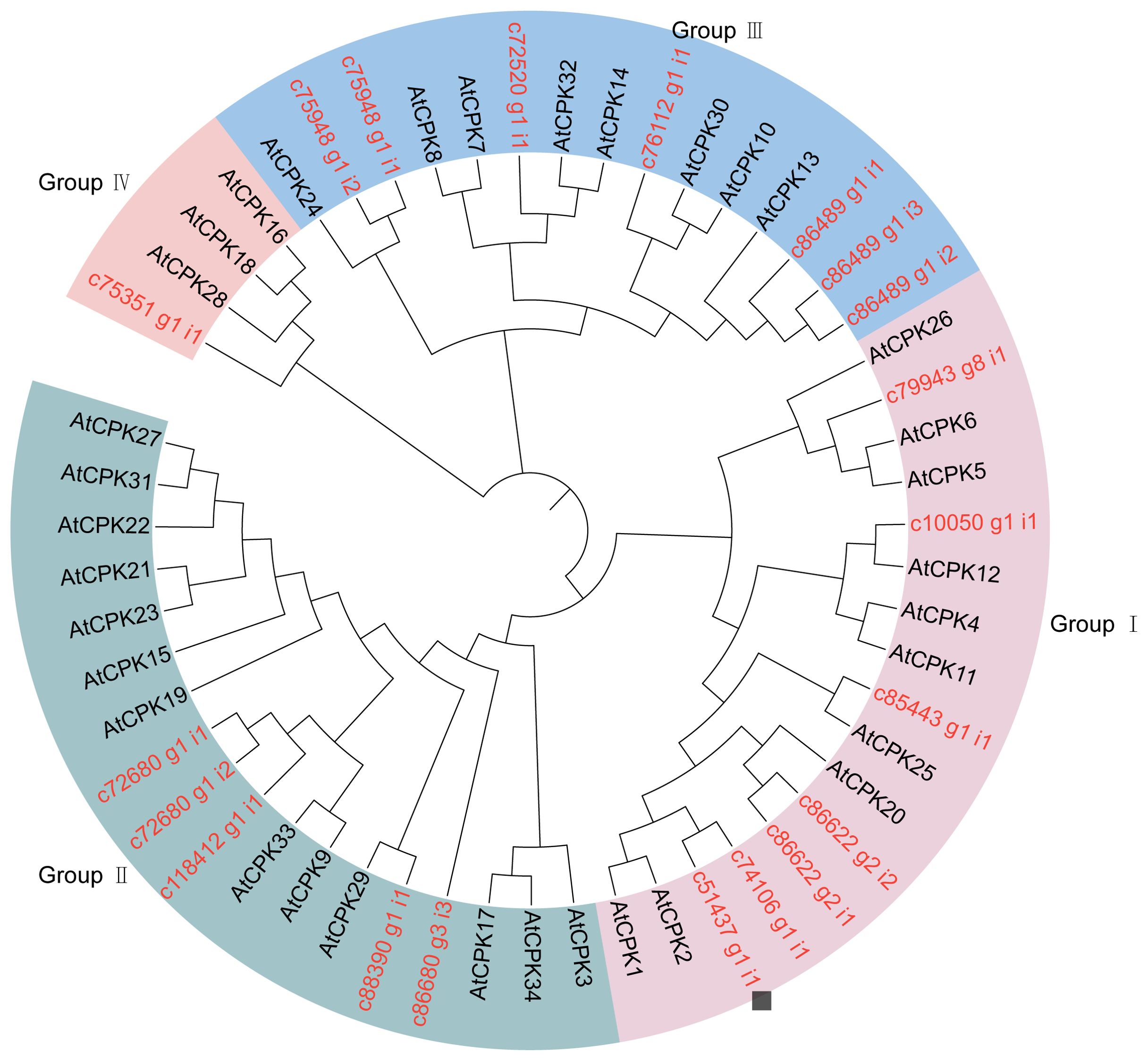
Figure 2. The evolutionary tree is composed of CDPKs from Arabidopsis and Saussurea involucrate The tree was generated using Mega 7.0 software by the Neighbor-joining method and bootstrap analysis (1,000 replicates) expressed in percentages. Members in the same group are clustered under the same color with a chromosomal location number ranging from; 20 SikCDPKs in snow lotus, and 34 AtCPKs in Arabidopsis.
3.3 Conserved motif analysis of snow lotus SikCDPK family involved in low temperature response
The conserved motifs of snow lotus SikCDPK protein sequences were analyzed by MEME online software, and a total of 10 conserved motifs were found (Figure 3), of which motif 1, motif 2, motif 7, and motif 10 were present in all SikCDPK protein sequences. Except for c75948-g1-i1, which does not have motif 4 and motif 8, c86489-g1-i2 and c86622- g2-i1, which do not have motif 3, motif 5, motif 6, and motif 9, c88390-g1-i1, which does not have motif 6, and c86680-g3-i3, which does not have motif 6 and motif8, but has 2 motif7; c118412-g1-i1 does not have motif6, motif9, and motif3. Except for c118412-g1-i1, which does not have motif6, motif9, and motif3, the other 14 SikCDPK protein sequences have the same 10 conserved motifs, suggesting that the genes of the CDPK family of the snow lotus may be functionally similar.
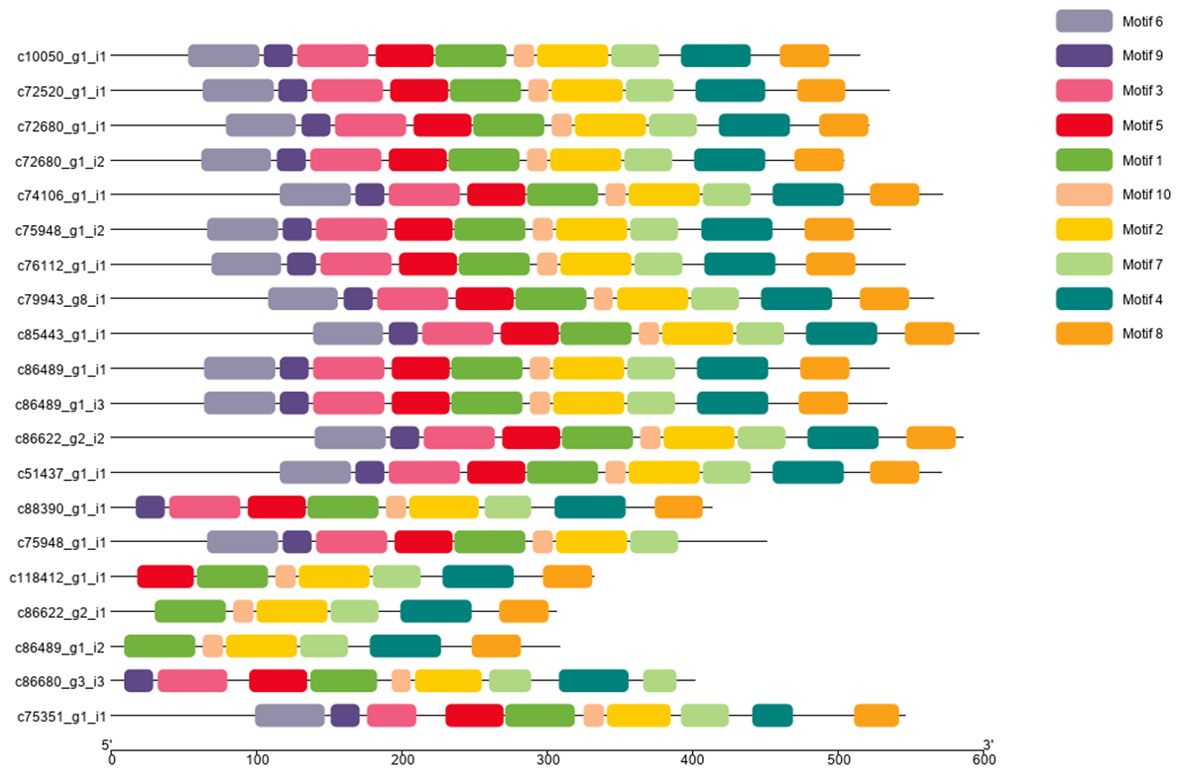
Figure 3. Conserved motif analysis of the CDPK protein sequence of Saussurea involucrate. The conserved motifs of SikCDPK proteins were discovered using the online searching tool MEME (http://memesuite.org/tools/meme). Different colors were used to indicate the conserved motifs.
3.4 Cloning and bioinformatics analysis of SikCDPK1 gene
After sequencing, we obtained the gene SikCDPK1 with a coding region length of 1716 bp, and the GenBank accession number is KU133953. The conserved domains of SikCDPK1 were analyzed by SMART online analysis tool, and the results showed that SikCDPK1 has typical CDPK conserved domains, i.e., Ser/Thr protein kinase domains at the N-terminal end, a linker region in the middle, and a regulatory region at the C-terminal end (which is also a calcium-binding region), and there are four EF-hands structural domains, and it belongs to the CDPK gene family (Figure 4).
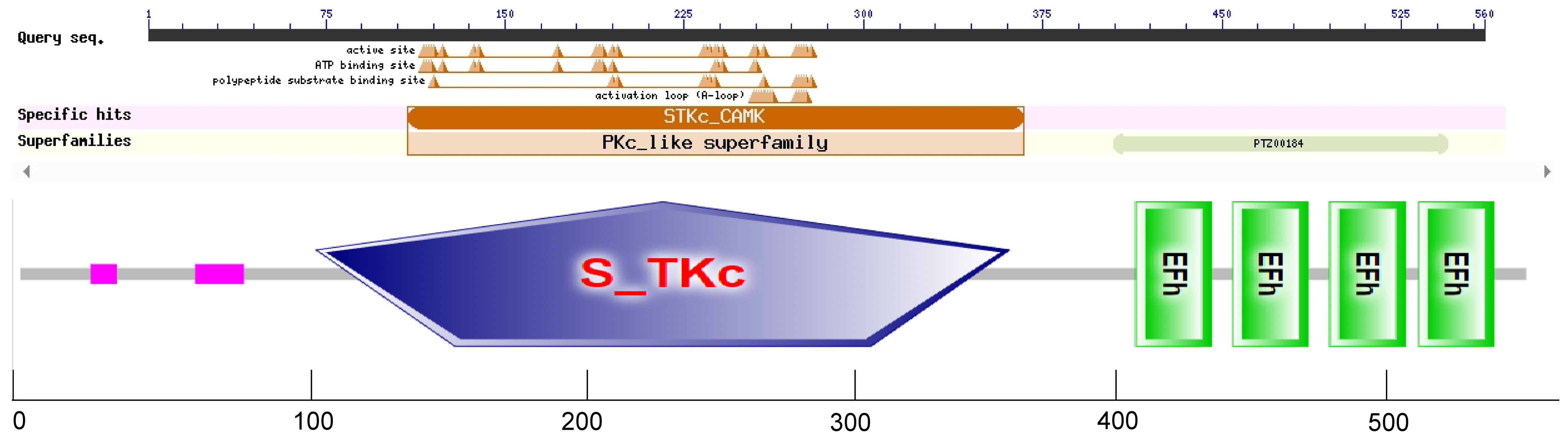
Figure 4. Domain organization of the SikCDPK1. The logos of domain organization were obtained from EMBL-EBI and SMART websites and were amended with Adobe Illustrator. The domains: V represents variable domain; K represents catalytic domain; I represents auto-inhibitory domain; C represents the region of calcium binding motifs: EF-hands.
3.5 Tissue differential expression
The expression of SikCDPK1 gene in roots, stems, leaves and seeds of snow lotus was explored by qRT-PCR. As shown in Figure 5, SikCDPK1 gene had the highest expression in seeds, others were leaves and stems in order, and the lowest expression was in roots. The gene expression in seeds, leaves compared with that in stems and roots reached significant differences. It indicates that SikCDPK1 gene is present in snow lotus seeds, leaves, stems and roots, but there are differences in expression in different organs.
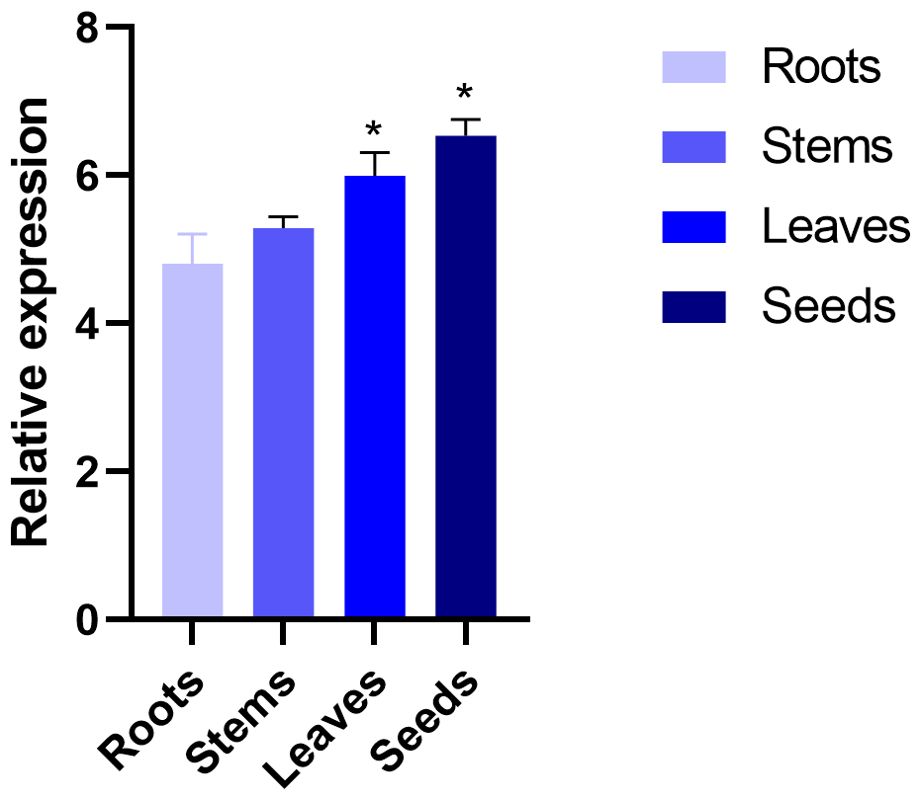
Figure 5. Tissue specific expression of SikCDPK1 gene. Quantitative PCR data are represented as mean ± SEM. *p < 0.05. All date represent mean values for three independent biological replicates.
3.6 Analysis of gene expression induced by low temperature, ABA, GA3 and SA
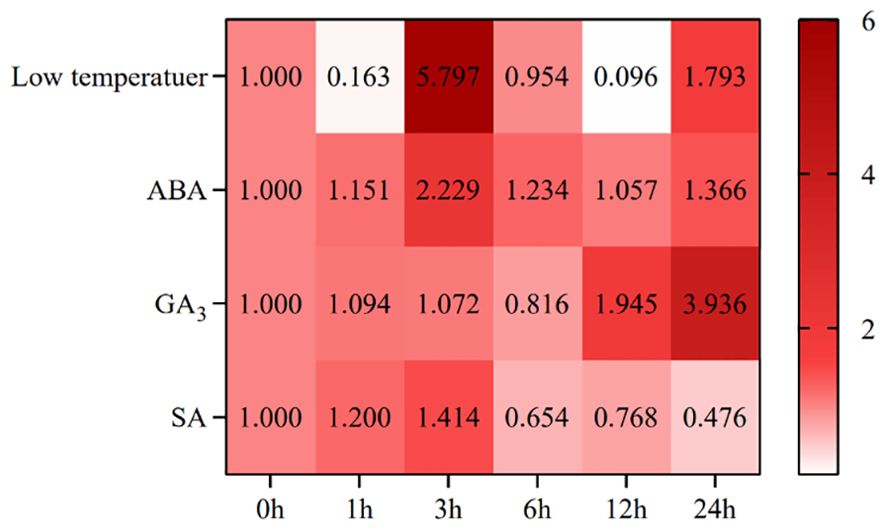
After low temperature, the expression of SikCDPK1 showed the trend of “decline-rise-decline-rise” (Figure 6), which was 0.16 times of CK at 1h, and then reached the maximum at 3h, with the expression amount 5.8 times of CK, which reached the level of highly significant difference compared with CK. At 6h, it reached the initial level, and then dropped to the minimum expression amount at 12h. At 24h, the expression level started to rise again, which was 1.79 times of CK.
After ABA, the expression of SikCDPK1 showed a “rising-declining-slowly rising” trend (Figure 6), reaching a peak at 3h, with an expression amount 2.23 times of CK, and reaching a highly significant difference compared with CK. At 12h, it reached the bottom, and the expression amount at the bottom was 1.057 times of CK, and then increased again at 24h, and the expression amount was 1.366 times of CK.
After GA3, the expression of SikCDPK1 showed a trend of “slightly increasing-decreasing-increasing” (Figure 6), reaching the peak at 6 h, and the expression at the bottom of the peak was 0.816 times of CK, and then continued to increase. At 12h, the expression was 1.945 times of CK. It reached the peak at 24h, the expression amount was 3.9 times of CK, and reached a highly significant difference level compared with CK.
After SA, the expression pattern of SikCDPK1 showed an overall “rising-declining” trend (Figure 6), reaching a peak at 3h, with an expression amount 1.4 times that of CK, which was significantly different from that of CK. After that, the expression continued to decline, and the lowest expression was at 24h, which was 0.476 times that of CK, which was significantly different from that of CK.
The above indicates that the expression of SikCDPK1 gene was not only affected by low temperature, but also by ABA, GA3 and SA.
3.7 Promoter cloning and activity analysis of SikCDPK1 gene
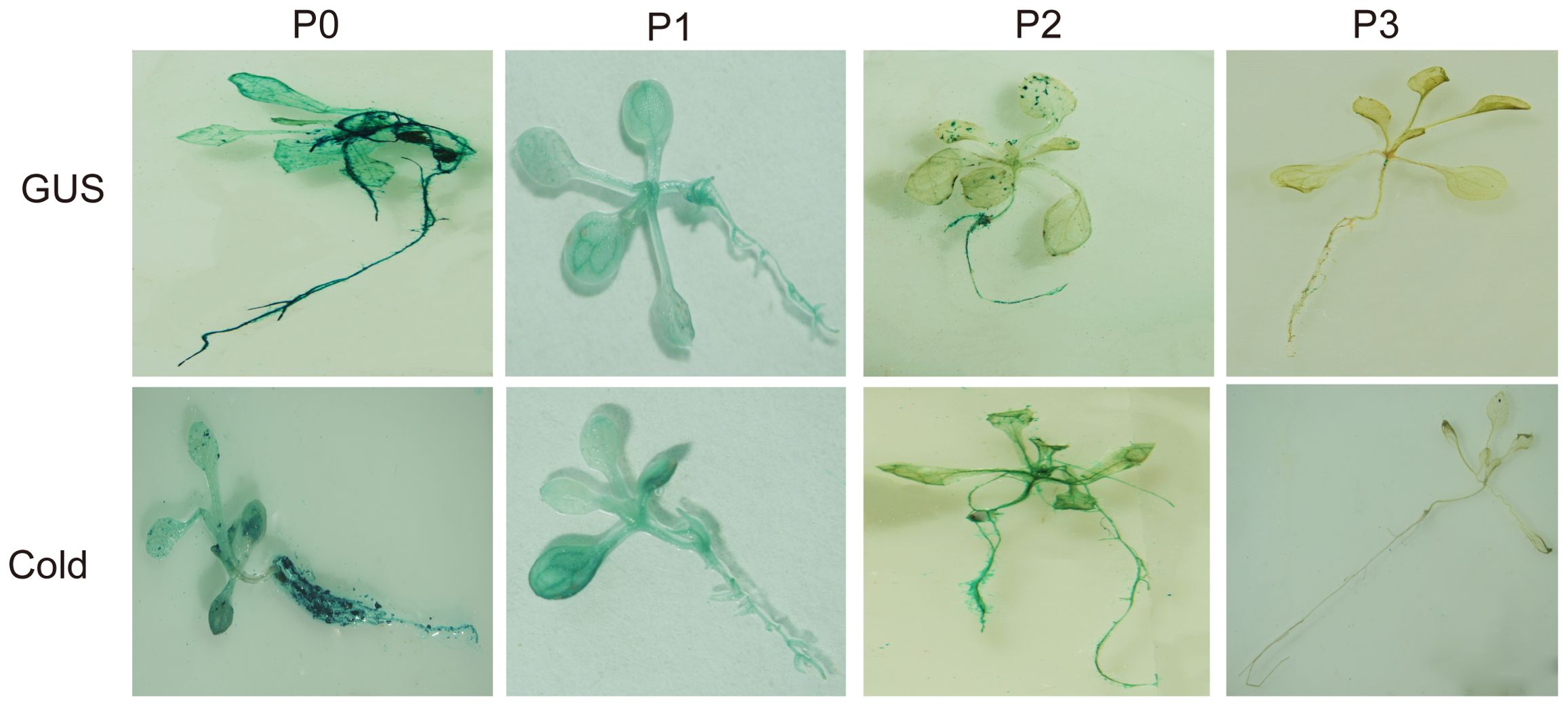
Analysis of the cloned SikCDPK1 gene promoter obtained using PlantCARE revealed that the sequence contained several CAAT box and TATA box conserved elements, and also included abiotic stress-responsive homeopathic action elements: response to dehydration-responsive element MYB-Core, CBFHV with ACGTATERD1, salt-induced response element GT1GMSCAM4, cold-induced response element LTRECOREATCOR15, and light-responsive element G-box; cis-acting elements in response to hormone-induced stresses: methyl jasmonate-responsive elements CGTCA-motif and TGACG-motif, abscisic acid-responsive element DPBFCOREDCDC3, ABA-responsive element ABRE, gibberellin-responsive element TATC-box, etc. (Figure 7). In order to deeply investigate the function of SikCDPK1 promoter, based on the full-length promoter P0, three deletion fragments responding to different stresses were designed according to the distribution of cis-regulatory elements, named P1, P2, and P3, respectively. Seedlings of GUS transgenic homozygous plants driven by the P0 promoter and its different 5’ end deletion fragments were stained with GUS histochemistry (Figure 8), and it was found that Arabidopsis thaliana with the transversion of the full-length sequence P0 was the most darkly colored, and the strongest coloring was found in the roots, stems, and leaves of the seedlings. The trans-promoter P1 fragment was relatively weakly colored, the trans-promoter P2 fragment was even more inferior, and the Arabidopsis seedlings with the trans-promoter P3 fragment did not show any coloration, indicating that the promoter activity of pSikCDPK1 was from strong to weak as P0> P1> P2> P3. The color of promoter P2 was darkened and GUS activity was enhanced after low-temperature treatment. This is because the promoter sequence contains low-temperature inducible elements such as LTRECOREATCOR15 and CBFHV.
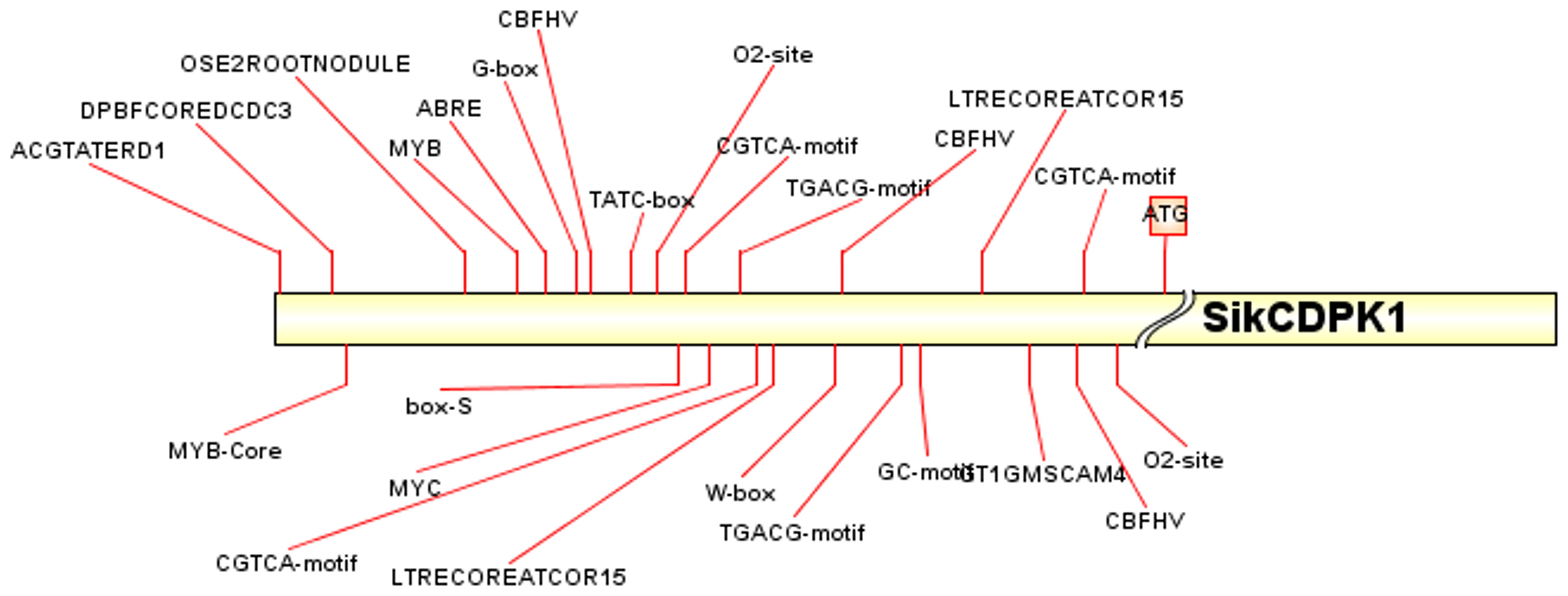
Figure 7. The main cis-regulatory element distribution of the promoter of SiKCDPK1 gene. ATG is the start of the gene; Promoter elements to the left of the ATG are labeled with a red line.
GUS histochemical staining of GUS transgenic homozygous plants driven by the full-length fragment of the trans-P0 promoter revealed that GUS expression was detected mainly in roots (Figure 9A), rosette leaves (Figure 9B) floral pods (Figure 9D), and fruit stalks at the attachment of the angiosperm (Figure 9F), with higher expression in roots and rosette leaves, but not in suprachordate leaves (Figure 9C), petals (Figure 9E), young fruits, mature angiosperms, and seeds (Figure 9G), suggesting that the promoter of the SikCDPK1 gene has a clear tissue expression specificity and exerts its expression-regulatory role mainly in roots and rosette leaves.
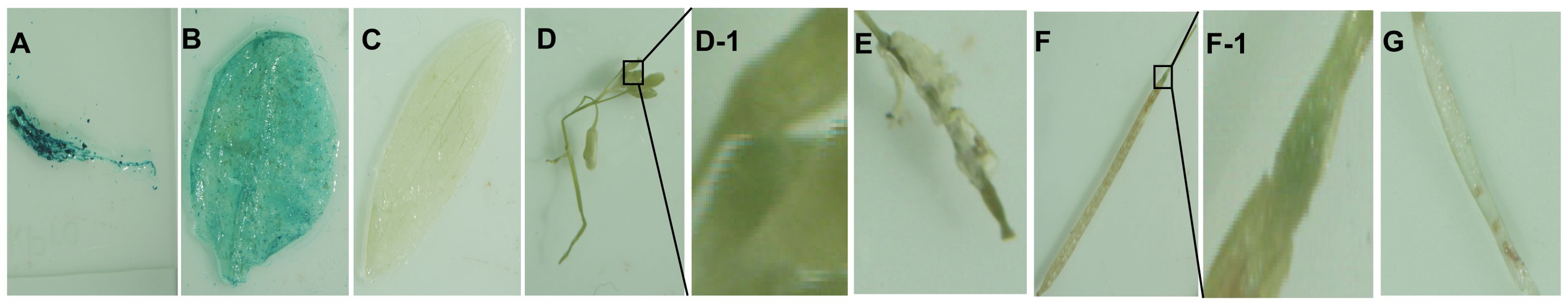
Figure 9. GUS assay results of different tissues of A. thaliana harboring SikCDPK1 promoter. (A) Root, (B) Rosette leaf, (C) Stem leaf, (D) Flower and stem, (D-1). Magnification of (D, E) Petal, (F) Silique, (F-1). Magnification of (F, G) Seed.
3.8 The overexpression of SikCDPK1 improves low temperature tolerance of tobacco
The wild type and transgenic tobacco lines were put into the -4°C artificial climate chamber for low temperature stress treatment. As shown in Figure 10A, there was no obvious phenotypic difference between the wild type and transgenic strains at room temperature; after 3 h in the incubator at -4°C, the leaves of wild type tobacco slightly drooped, which showed the characteristics of mild cold injury, and there was no obvious phenotypic change in the transgenic tobacco; after 6h of low temperature stress, the symptoms of cold injury of the wild type tobacco were more and more obvious, with the leaves moderately wilting and appearing as spots, while the transgenic tobacco did not have any obvious change. After 9h of low temperature stress, the wild type tobacco was basically completely wilted and waterlogged, with only the top leaflet buds showing signs of life, while the leaves of the transgenic tobacco were slightly weakened at this time; after 12h of low temperature stress, the wild type tobacco was completely wilted, and the leaves of the transgenic tobacco were curled up and drooping, which was a severe form of wilting. After 24h of room temperature recovery, there was no change in the wild-type tobacco, while the leaves of the transgenic tobacco began to stretch. High-level expression of SikCDPK1 gene in transgenic tobacco detected by real time fluorescence quantitative PCR (qRT-PCR) assay (Figure 10B).
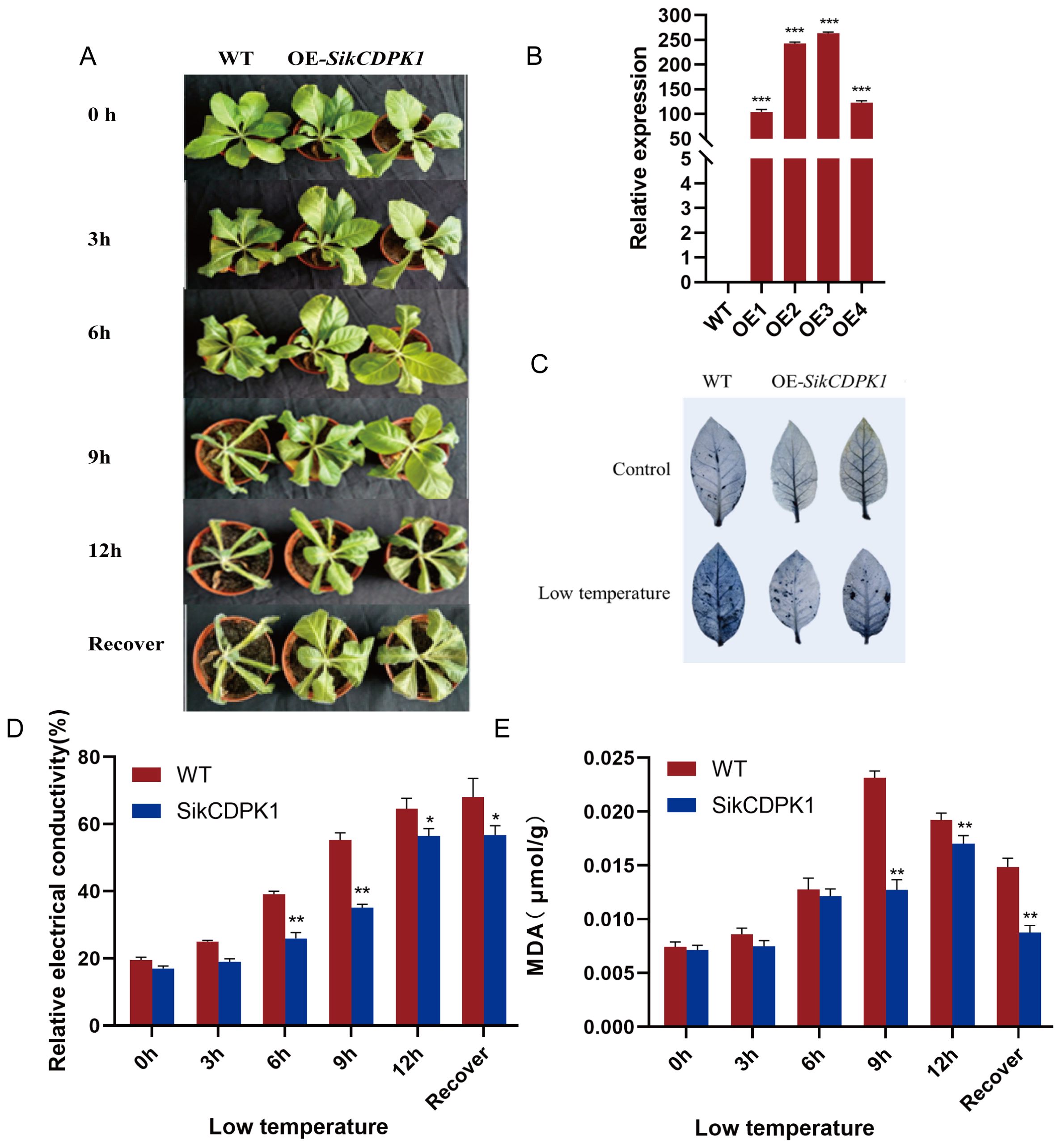
Figure 10. Overexpression of SikCDPK1 improves cold tolerance of Tobacco. (A) The phenotypes of wild type tobacco and transgenic lines after low temperature for 0h、3h、6 h、9 h、12 h and recover. Bars =1 cm. (B) The expression level of overexpressed-SikCDPK1 in Tobacco. wild type Tobacco; OE1, OE2, OE3 and OE4. (C) NBT staining of wild type tobacco and OE-SikCDPK1 tobacco under low temperature. (D) Relative electricity conductivity. (E) MDA concentrations. Statistically significant differences compared to wild type. Values represent the mean ± SE from three biological replicates. *P< 0.05, **P< 0.01, ***p<0.001.
Under normal growth conditions, there was no obvious difference between the leaves of wild type tobacco and transgenic tobacco, but the transgenic tobacco formed blue-black compound compared with the wild type tobacco plants after low temperature stress treatment, indicating that the transgenic tobacco lines accumulated relatively less ROS under low temperature stress conditions (Figure 10C).
The relative electricity conductivity and MDA activity of wild type and transgenic tobacco were measured at different treatment times under low temperature stress, and it was found that the relative electricity conductivity of both wild type and transgenic tobacco showed an obvious increasing trend with the extension of the stress time, and the relative conductivity of transgenic tobacco began to decrease after 1 day of recovery at room temperature, while the wild type still maintained an increasing trend; the relative electricity conductivity of transgenic plants was significantly lower than that of wild type during the whole process. The relative electricity conductivity of the transgenic plants was significantly lower than that of the wild type throughout the whole process, and reached a highly significant level at the 6h and 9h. This indicated that ion leakage caused by low temperature is related to injury of the cell membrane and its dysfunction. Thus lower ROS accumulation in transgenic tobacco causes lower accumulation of MDA, which suggests less damage to the cell membrane, so its integrity is better preserved. Hence the lower value of relative electrical conductivity (Figure 10D).
Before low temperature treatment, there was little difference between the MDA contents of wild type and transgenic tobacco, and with the extension of low temperature stress, the MDA contents of both wild type and transgenic tobacco showed an increasing trend, and the MDA contents of transgenic plants were lower than those of wild type, and reached a highly significant level at 9h. The MDA contents began to decrease at the recovery stage of room temperature. It indicated that the cell membrane of transgenic plants was less damaged under low temperature stress (Figure 10E).
4 Discussion
4.1 The identification and analysis of CDPKs in Saussurea involucrata
CDPK genes have been found in several plant genomes, such as Arabidopsis (Cheng et al., 2002), rice (Ray et al., 2007), and barley (Yang et al., 2017). In this study, we searched the transcriptome database of snow lotus based on conserved structural domains and identified 20 CDPKs in snow lotus. 34, 31, and 28 CDPKs were found in Arabidopsis thaliana, rice, and barley, respectively, which were much higher than those found in snow lotus (Cheng et al., 2002). A phylogenetic tree was constructed to categorize snow lotus CDPKs into four major subfamilies, which were consistent with Arabidopsis subfamilies. However, each subfamily in snow lotus has fewer members than in Arabidopsis.
The conduction of Ca2+ in CDPK genes is mainly through EF-hands, as well as the activation threshold of Ca2+ may be related to the differences in the number and location of EF-hands (Klimecka and Muszyńska, 2007). Generally, CDPK genes in plants have four EF-hands; however, there are exceptions for some CDPK genes in Arabidopsis and maize, which contain two or three EF-hands (Cheng et al., 2002; Kong et al., 2013). In our study, all SikCDPKs (except c75948_g1_i1) contained four EF-hands (Table 2). Therefore, the biological functional differences between c75948_g1_i1 (one EF-hand) and other SikCDPKs (four EF-hands) can be further explored.
CDPKs are widely distributed in plants (Simeunovic et al., 2016). Many members of the CDPK family play important roles in plant response to abiotic stress. OsCDPK1 regulates rice salt and drought tolerance (Ho et al., 2013). The CDPK family is involved in drought or salt stress adaptation by inducing ABA responsive genes and adjusting the ABA-induced anionic channel (SLAC1, SLAH3), leading to stomata adjustment in Arabidopsis (Edel and Kudla, 2016; Zhang et al., 2020). Overexpression of MDCPK1A in tobacco removes ROS accumulation and regulates the expression of stress-related genes, thereby significantly increasing cold and salt resistance (Dong et al., 2020). Excessive expression of ZMCPK1 in corn leaf inhibits the expression of cold inducing marker gene ZMERF3. The ectopic expression of ZMCPK1 in Arabidopsis reduced the adaptation of the plant in terms of cold resistance, indicating that ZMCPK1 acts as a negative regulatory factor under cold stress (Weckwerth et al., 2015). Arabidopsis CPK28 functions as a negative regulator of an immune signal which responds to immune responses by regulating BIK1 (multi-mode identification receptor (PRR)) (Simeunovic et al., 2016). CDPK can also be used as a negative regulator of the stress response. Transgenic plants overexpressing CDPK are more sensitive to abiotic stress and biotic stress. Arabidopsis cpk23 mutant increases tolerance to drought and salt stress, but resistance to drought and salt stress is reduced in AtCPK23 overexpressing plants (Ma and Wu, 2007). Taken together, it is apparent that CDPKs participate in plant abiotic and biotic stress in the form of positive and negative regulation. Virus-induced silencing of CDPK2 and CDPK3 showed that CDPK1 and CDPK2 are involved in the regulation of AVR9/CF-9 genes, and CDPK is a necessary condition for plant AVR9/CF-9 induced hypersensitivity during pathogen infection (Romeis et al., 2001). CDPK10 in maize is also involved in defense signaling pathways (Murillo et al., 2001).
The specificity of signaling can be determined by the subcellular location of CDPK proteins, which can be regulated at the translational and post-translational levels. Predictions indicate that SikCDPKs are mainly located in several cellular compartments, namely chloroplasts, nucleus, and cytoplasm. The distribution of SikCDPKs in different cellular compartments suggests that there are also differences in signaling specificity. It was also predicted that most SikCDPKs contain N-myristoylation sites, and CDPK genes with N-myristoylation motifs can promote protein-membrane and protein-protein interactions (Lu and Hrabak, 2013; Martín and Busconi, 2000). Myristoylation may be part of a major signaling process that directs these proteins to cell membrane binding sites of the 19 SikCDPKs identified in snow lotus, all contain palmitoylation sites. Palmitoylation sites may provide important clues for subcellular localization (Martín and Busconi, 2000).
Sequence motifs are becoming increasingly important in gene regulation analysis. In this study, we found the similarity in the distribution of motifs in the SikCDPKs family by analyzing motifs. By resolving sequence motifs, functions in biological processes can be resolved. When a large number of repetitive sequence structures appear in a certain large class of sequences, it is valuable to explore its significance (Bailey et al., 2015). Therefore, in the subsequent process of exploring the functions of SikCDPKs, the biological significance involved in each biological process can be further understood based on motifs.
4.2 SikCDPK1 plays a positive role in conferring low temperature tolerance
A new calcium-dependent protein kinase gene, SikCDPK1, was cloned from snow lotus, and the real-time quantitative PCR results showed that the expression of the SikCDPK1 gene was induced by low temperature stress, which was similar to the results of the previous studies on PeCPK10 and OsCPK13 (Chen et al., 2013; Wan et al., 2007).
To further investigate the function of SikCDPK1, overexpressing SikCDPK1 was found to have no significant difference between the transgenic plants and the wild-type phenotype. This is similar to the findings of OsCPK13, suggesting that the SikCDPK1 gene has little effect on the growth status of tobacco. Low-temperature stress was applied to tobacco trans-SikCDPK1 gene, and the transgenic plants were significantly better than the wild type in terms of low temperature tolerance. This was similar to the results of overexpression of AtCDPK1, PeCPK10, OsCPK13 and VaCPK20 genes (Atif et al., 2019; Chen et al., 2013; Wan et al., 2007; Dubrovina et al., 2015), and the transgenic plants significantly improved the plant resistance to low temperature. Measurement of physiological indexes in SikCDPK1-transgenic tobacco under low temperature stress revealed that the MDA content and conductivity were lower than those of the wild type plants. Plant cells produce excess of reactive oxygen species (ROS), such as H2O2, O2- and -OH, and the accumulation of MDA were subsequently found (Upadhyaya and Khan, 2007). The high expression of ROS could result in the deep damage of plants, such as lipid peroxidation and electrolyte leakage (Mishra et al., 2009). Notably, MDA produced in membranes could destroy membrane structure and cause protein degradation, and then it aggravated by the damage of low temperature (Martinez et al., 2018; Petrov et al., 2015). For keeping the balance of ROS numbers, plants had evolved some enzymatic and non-enzymatic antioxidant systems to eliminate or reduce the ROS damage induced by low temperature stress (Wani et al., 2018; Wang et al., 2016). Thus it can reduce the damage of the plasma membrane caused by low temperature, and make the transgenic plants significantly increase the ability to tolerate low temperature.
Ca2+ is an important second messenger in the low-temperature signaling pathway (Monroy and Dhindsa, 1995). When Arabidopsis thaliana or Medicago sativa (alfalfa) face low temperature stress, Ca2+ from extracellular stores flows into the cells and causes a rapid increase in the cytoplasmic Ca2+ content. This transient increase in Ca2+ concentration is necessary to induce the expression of cold-adapted genes and cold tolerance in cold-resistant plants (Cao et al., 2023). In Arabidopsis, the full expression of some cold-regulated genes, such as the CRT/DRE-controlled cor6.6, is dependent on an increase in Ca2+ levels (Thomashow, 1999). When Ca2+ chelating agents - BAPTA, or Ca2+ channel blockers -La3+, inhibited the low-temperature-induced Ca2+ input, the expression of low-temperature induced cas15 gene was attenuated, and the cold-adapted genes were not expressed at the same time (Cao et al., 2023). Gene expression was reduced, and the cold acclimatization ability of alfalfa was reduced. Therefore, it is speculated that it may be that the inhibition of Ca2+ input with the prolongation of low-temperature treatment affected the expression of the SikCDPK1 gene, resulting in the incomplete up-regulation of SikCDPK1.
4.3 Demonstration of SikCDPK1 promoter activity in transgenic plants
Promoters are important regulatory elements that determine the temporal and spatial order of gene expression to a certain extent, and the study of promoter activity and function can help to investigate the function and expression regulation mechanism of genes in depth. In order to understand the expression regulation of snow lotus SikCDPK1 gene in response to adversity, our group cloned the promoter sequence of the SikCDPK1 gene by TAIL-PCR (Shi et al., 2022). The promoter sequence prediction revealed that the promoter region contains not only the TATA-box and CAAT-box (Alok et al., 2020), but also MYB-core、ACGTATERD1、LTRECOREATCOR15、CBFHV and other cis-acting elements related to low temperature. Transient expression of GUS or other marker genes has been used in assessing activity and tissue specificity of plant promoters (Basu et al., 2003). SikCDPK1 promoter P0 can promote the expression of GUS gene under low-temperature stress, and it has strong expression specificity in different tissues. The SikCDPK1 promoter has a promoter activity center more than 480 bp upstream of the ATG start codon, and the promoter P0 and P2 is able to be induced by low temperature. Interestingly, the SikCDPK1 gene had the lowest expression in roots, while the SikCDPK1 promoter had the highest activity in roots. This may be the receptors are different and the gene regulatory mechanisms have changed, resulting in inconsistent expression (Hernando et al., 2017).
Although the results of this experiment showed that the SikCDPK1 gene significantly improved the low temperature tolerance of transgenic tobacco, the transcriptome database of snow lotus was still incomplete, and the chromosomal mapping results were not clear, so it was difficult to find and name more SikCDPKs gene family members. In the future, the SikCDPKs family genes related to stress resistance can be mined from the complete snow lotus transcriptome database, and the function, gene regulation of the genes can be studied in depth. These studies will provide a theoretical basis for exploring the upstream regulatory sequences of the SikCDPK1 gene and analyzing the regulatory mechanism of the SikCDPK1 gene in response to adversity.
5 Conclusions
In this study, 20 CDPK genes with conserved motifs that were significantly similar to those of Arabidopsis thaliana and rice were identified from snow lotus, and these SikCDPKs could be divided into four subgroups. Among them, SikCDPK1 responds to low temperature and hormonal treatment, and is expressed differently in different organs of snow lotus. The promoter of SikCDPK1 gene contains a cold induction element, and the promoter activity is increased after low temperature. The degree of damage in overexpressing SikCDPK1 plants was lower after low temperature treatment, the ROS accumulation was less, and the relative conductivity and MDA content changed slightly. The results indicated that SikCDPK1 could positively regulate the low temperature tolerance of plants by activating the cold-inducible element in the promoter, improving gene expression, maintaining cell membrane stability, and reducing the accumulation of reactive oxygen species. Our study demonstrated that SikCDPK1 positively regulates cold tolerance, providing a new genetic resource for genetic improvement of low temperature tolerance in snow lotus. Future studies should continue to improve the construction of a transcriptome database for the CDPK family in snow lotus. Investigating the signaling network and biochemical functions of SikCDPK1 to gain a deeper understanding of its molecular mechanism in regulating low temperature stress tolerance.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
GS: Conceptualization, Data curation, Formal analysis, Writing – original draft, Writing – review & editing. YL: Investigation, Methodology, Visualization, Writing – original draft. XT: Methodology, Software, Supervision, Writing – original draft. JG: Formal analysis, Methodology, Validation, Writing – original draft. XZ: Funding acquisition, Project administration, Resources, Writing – review & editing, Conceptualization.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (Grant No.31760066, 32360457).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1436651/full#supplementary-material
Abbreviations
CK, Control check; CDPKs, Calcium-Dependent Protein Kinases; ROS, Reactive oxygen species; REC, Relative electrical conductivity; MDA, Malondialdehyde.
References
Aldon, D., Mbengue, M., Mazars, C., Galaud, J. P. (2018). Calcium signalling in plant biotic interactions. Int. J. Mol. Sci. 19, 665. doi: 10.3390/ijms19030665
Allan, C., Morris, R. J., Meisrimler, C. N. (2022). Encoding, transmission, decoding, and specificity of calcium signals in plants. J. Exp. Bot. 73, 3372–3385. doi: 10.1093/jxb/erac105
Alok, A., Kaur, J., Tiwari, S. (2020). Functional characterization of wheat myoinositol oxygenase promoter under different abiotic stress conditions in Arabidopsis thaliana. Biotechnol. Lett. 42, 2035–2047. doi: 10.1007/s10529-020-02967-1
Asai, S., Ichikawa, T., Nomura, H., Kobayashi, M., Kamiyoshihara, Y., Mori, H. (2013). The variable domain of a plant calcium-dependent protein kinase (CDPK) confers subcellular localization and substrate recognition for NADPH oxidase. J. Biol. Chem. 288, 14332–14340. doi: 10.1074/jbc.M112.448910
Asano, T., Tanaka, N., Yang, G., Hayashi, N., Komatsu, S. (2005). Genome-wide identification of the rice calcium-dependent protein kinase and its closely related kinase gene families: comprehensive analysis of the CDPKs gene family in rice. Plant Cell Physiol. 46, 356–366. doi: 10.1093/pcp/pci035
Atif, R. M., Shahid, L., Waqas, M., Ali, B., Rashid, M. A.R., Azeem, F. (2019). Insights on calcium-dependent protein kinases (CPKs) signaling for abiotic stress tolerance in plants. Int. J. Mol. Sci. 20, 5298. doi: 10.3390/ijms20215298
Bailey, T. L., Johnson, J., Grant, C. E., Noble, W. S. (2015). The MEME suite. Nucleic Acids Res. 43, W39–W49. doi: 10.1093/nar/gkv416
Basu, C., Kausch, A. P., Luo, H., Chandlee, J. M. (2003). Promoter analysis in transient assays using a GUS reporter gene construct in creeping bentgrass (Agrostis palustris). Journal of plant physiology 160, 1233–1239.
Cai, H., Cheng, J., Yan, Y., Xiao, Z., Li, J., Mou, S. (2015). Genome-wide identification and expression analysis of calcium-dependent protein kinase and its closely related kinase genes in Capsicum annuum. Front. Plant Sci. 6, 737. doi: 10.3389/fpls.2015.00737
Cao, Y., Hwarari, D., Radani, Y., Guan, Y., Yang, L. (2023). Molecular mechanism underlying plant response to cold stress. Phyton 92, 03-04. doi: 10.32604/phyton.2023.024929
Chattha, M. U., Amjad, T., Khan, I., Nawaz, M., Ali, M., Chattha, M. B. (2022). Mulberry based zinc nano-particles mitigate salinity induced toxic effects and improve the grain yield and zinc bio-fortification of wheat by improving antioxidant activities, photosynthetic performance, and accumulation of osmolytes and hormones. Front. Plant Sci. 13, 920570. doi: 10.3389/fpls.2022.920570
Chen, J., Xue, B., Xia, X., Yin, W. (2013). A novel calcium-dependent protein kinase gene from Populus euphratica, confers both drought and cold stress tolerance. Biochem. Biophys. Res. Commun. 441, 630–636. doi: 10.1016/j.bbrc.2013.10.103
Cheng, S. H., Willmann, M. R., Chen, H. C., Sheen, J. (2002). Calcium signaling through protein kinases. The Arabidopsis calcium-dependent protein kinase gene family. Plant Physiol. 129, 469–485. doi: 10.1104/pp.005645
Clough, S. J., Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium- mediated transformation of Arabidopsis thaliana. Plant J. Cell Mol. Biol. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Ding, Y., Yang, H., Wu, S., Fu, D., Li, M., Gong, Z., et al. (2022). CPK28-NLP7 module integrates cold-induced Ca2+ signal and transcriptional reprogramming in Arabidopsis. Sci. Adv. 8, eabn7901. doi: 10.1126/sciadv.abn7901
Dong, H., Wu, C., Luo, C., Wei, M., Qu, S., Wang, S. (2020). Overexpression of MdCPK1a gene, a calcium dependent protein kinase in apple, increase tobacco cold tolerance via scavenging ROS accumulation. PloS One 15, e0242139. doi: 10.1371/journal.pone.0242139
Dubrovina, A. S., Kiselev, K. V., Khristenko, V. S., Aleynova, O. A. (2015). VaCPK20, a calcium-dependent protein kinase gene of wild grapevine Vitis amurensis Rupr., mediates cold and drought stress tolerance. J. Plant Physiol. 185, 1–12. doi: 10.1016/j.jplph.2015.05.020
Edel, K. H., Kudla, J. (2016). Integration of calcium and ABA signaling. Curr. Opin. Plant Biol. 33, 83–91. doi: 10.1016/j.pbi.2016.06.010
Estruch, J. J., Kadwell, S., Merlin, E., Crossland, L. (1994). Cloning and characterization of a maize pollen-specific calcium-dependent calmodulin-independent protein kinase. Proc. Natl. Acad. Sci. U.S.A 91, 8837–8841. doi: 10.1073/pnas.91.19.8837
Hernando, C. E., Romanowski, A., Yanovsky, M. J. (2017). Transcriptional and post-transcriptional control of the plant circadian gene regulatory network. Biochim. Biophys. Acta Gene Regul. Mech. 1860, 84–94. doi: 10.1016/j.bbagrm.2016.07.001
Ho, S. L., Huang, L. F., Lu, C. A., He, S. L., Wang, C. C., Yu, S. P. (2013). Sugar starvation-and GA-inducible calcium-dependent protein kinase 1 feedback regulates GA biosynthesis and activates a 14-3-3 protein to confer drought tolerance in rice seedlings. Plant Mol. Biol. 81, 347–361. doi: 10.1007/s11103-012-0006-z
Hrabak, E. M., Chan, C. W. M., Gribskov, M., Harper, J. F., Choi, J. H., Halford, N., et al. (2003). The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol. 132, 666–680. doi: 10.1104/pp.102.011999
Hu, Y., Cheng, Y., Yu, X., Liu, J., Yang, L., Gao, Y., et al. (2021). Overexpression of two CDPKs from wild Chinese grapevine enhances powdery mildew resistance in Vitis vinifera and Arabidopsis. New Phytol. 230, 2029–2046. doi: 10.1111/nph.17285
Hu, Z., Lv, X., Xia, X., Zhou, J., Shi, K., Yu, J., et al. (2016). Genome-wide identification and expression analysis of calcium-dependent protein kinase in tomato. Front. Plant Sci. 7, 469. doi: 10.3389/fpls.2016.00469
Jambunathan, N. (2010). Determination and detection of reactive oxygen species (ROS), lipid peroxidation, and electrolyte leakage in plants. Plant Stress tolerance: Methods Protoc. 639, 291–297. doi: 10.1007/978-1-60761-702-0_18
Jiang, S., Zhang, D., Wang, L., Pan, J., Liu, Y., Kong, X., et al. (2013). A maize calcium-dependent protein kinase gene, ZmCPK4, positively regulated abscisic acid signaling and enhanced drought stress tolerance in transgenic Arabidopsis. Plant Physiol. Biochem. 71, 112–120. doi: 10.1016/j.plaphy.2013.07.004
Klimecka, M., Muszyńska, G. (2007). Structure and functions of plant calcium-dependent protein kinases. Acta Biochimica Polonica 54, 219–233. doi: 10.1186/1471-2164-14-433
Kong, X., Lv, W., Jiang, S., Zhang, D., Cai, G., Pan, J., et al. (2013). Genome-wide identification and expression analysis of calcium-dependent protein kinase in maize. BMC Genomics 14, 1–15. doi: 10.1186/1471-2164-14-433
Li, X., Zhao, L., Zhang, H., Liu, Q., Zhai, H., Zhao, N., et al. (2022). Genome-wide identification and characterization of CDPK family reveal their involvements in growth and development and abiotic stress in sweet potato and its two diploid relatives. Int. J. Mol. Sci. 23, 3088. doi: 10.3390/ijms23063088
Liese, A., Romeis, T. (2013). Biochemical regulation of in vivo function of plant calcium-dependent protein kinases (CDPK). Biochim. Biophys. Acta (BBA)-Molecular Cell Res. 1833, 1582–1589. doi: 10.1016/j.bbamcr.2012.10.024
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2– ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lu, S. X., Hrabak, E. M. (2013). The myristoylated amino-terminus of an Arabidopsis calcium dependent protein kinase mediates plasma membrane localization. Plant Mol. Biol. 82, 267–278. doi: 10.1007/s11103-013-0061-0
Luan, S., Wang, C. (2021). Calcium signaling mechanisms across kingdoms. Annu. Rev. Cell Dev. Biol. 37, 311–340. doi: 10.1146/annurev-cellbio-120219-035210
Ma, S. Y., Wu, W. H. (2007). AtCPK23 functions in Arabidopsis responses to drought and salt stresses. Plant Mol. Biol. 65, 511–518. doi: 10.1007/s11103-007-9187-2
Martín, M. L., Busconi, L. (2000). Membrane localization of a rice calcium-dependent protein kinase (CDPK) is mediated by myristoylation and palmitoylation. Plant J. 24, 429–435. doi: 10.1111/j.1365-313X.2000.00889.x
Martinez, V., Nieves-Cordones, M., Lopez-Delacalle, M., Rodenas, R., Mestre, T. C., Garcia-Sanchez, F., et al. (2018). Tolerance to stress combination in tomato plants: new insights in the protective role of melatonin. Molecules 23, 535. doi: 10.3390/molecules23030535
Mishra, V., Srivastava, G., Prasad, S. M. (2009). Antioxidant response of bitter gourd (Momordica charantia L.) seedlings to interactive effect of dimethoate and UV-B irradiation. Scientia Hortic. 120, 373–378. doi: 10.1016/j.scienta.2008.11.024
Mittal, S., Mallikarjuna, M. G., Rao, A. R., Jain, P. A., Dash, P. K., Thirunavukkarasu, N.. (2017). Comparative analysis of CDPK family in maize, Arabidopsis, rice, and sorghum revealed potential targets for drought tolerance improvement. Front. Chem. 5, 115. doi: 10.3389/fchem.2017.00115
Monroy, A. F., Dhindsa, R. S. (1995). Low-temperature signal transduction: induction of cold acclimation-specific genes of alfalfa by calcium at 25 degrees C. Plant Cell 7, 321–331. doi: 10.1105/tpc.7.3.321
Moutinho, A., Trewavas, A. J., Malho, R. (1998). Relocation of a Ca2+-dependent protein kinase activity during pollen tube reorientation. Plant Cell 10, 1499–1510. doi: 10.1105/tpc.10.9.1499
Murillo, I., Jaeck, E., Cordero, M. J., San Segundo, B. (2001). Transcriptional activation of a maize calcium-dependent protein kinase gene in response to fungal elicitors and infection. Plant Mol. Biol. 45, 145–158. doi: 10.1023/A:1006430707075
Petrov, V., Hille, J., Mueller-Roeber, B., Gechev, T. S. (2015). ROS-mediated abiotic stress-induced programmed cell death in plants. Front. Plant Sci. 6, 69. doi: 10.3389/fpls.2015.00069
Ray, S., Agarwal, P., Arora, R., Kapoor, S., Tyagi, A. K. (2007). Expression analysis of calcium-dependent protein kinase gene family during reproductive development and abiotic stress conditions in rice (Oryza sativa L. ssp. indica). Mol. Genet. Genomics 278, 493–505. doi: 10.1007/s00438-007-0267-4
Romeis, T., Ludwig, A. A., Martin, R., Jones, J. D. (2001). Calcium-dependent protein kinases play an essential role in a plant defence response. EMBO J 20, 5556–5567. doi: 10.1093/emboj/20.20.5556
Schmedes, A., Hølmer, G. (1989). A new thiobarbituric acid (TBA) method for determining free malondialdehyde (MDA) and hydroperoxides selectively as a measure of lipid peroxidation. J. Am. Oil Chemists’ Soc. 66, 813–817. doi: 10.1007/BF02653674
Shi, G. Z., Wang, Z. Y., Sun, Q., Zhu, X. X. (2022). Cloning and activity analysis of SikCDPK1 promoter from saussurea involucrate. Biotechnol. Bull. 38, 191–197. doi: 10.13560/j.cnki.biotech.bull.1985.2022-0570
Simeunovic, A., Mair, A., Wurzinger, B., Teige, M. (2016). Know where your clients are: subcellular localization and targets of calcium-dependent protein kinases. J. Exp. Bot. 67, 3855–3872. doi: 10.1093/jxb/erw157
Stael, S., Wurzinger, B., Mair, A., Mehlmer, N., Vothknecht, U. C., Teige, M.. (2012). Plant organellar calcium signalling: an emerging field. J. Exp. Bot. 63, 1525–1542. doi: 10.1093/jxb/err394
Thomashow, M. F. (1999). Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Biol. 50, 571–599. doi: 10.1146/annurev.arplant.50.1.571
Upadhyaya, H., Khan, M. P. (2007). Hydrogen peroxide induces oxidative stress in detached leaves of Oryza sativa L. Gen. Appl. Plant Physiol. 33, 83–95.
Vitha, S., Beneš, K., Phillips, J. P., Gartland, K. M. (1995). Histochemical GUS analysis. Agrobacterium Protoc. 44, 185–193. doi: 10.1385/0-89603-302-3:185
Wan, B., Lin, Y., Mou, T. (2007). Expression of rice Ca (2+)-dependent protein kinases (CDPKs) genes under different environmental stresses. FEBS Lett. 581, 1179–1189. doi: 10.1016/j.febslet.2007.02.030
Wang, F., Liu, J., Zhou, L., Andrabi, K. I., John, R. (2016). Senescence-specific change in ROS scavenging enzyme activities and regulation of various SOD isozymes to ROS levels in psf mutant rice leaves. Plant Physiol. Biochem. 109, 248–261. doi: 10.1016/j.plaphy.2016.10.005
Wani, M. A., Jan, N., Qazi, H. A., Pan, G., Li, Z., Cheng, F. (2018). Cold stress induces biochemical changes, fatty acid profile, antioxidant system and gene expression in Capsella bursa pastoris L[J]. Acta Physiologiae Plantarum 40, 1–14. doi: 10.1007/s11738-018-2747-z
Weckwerth, P., Ehlert, B., Romeis, T. (2015). ZmCPK1, a calcium-independent kinase member of the Zea mays CDPK gene family, functions as a negative regulator in cold stress signalling. Plant Cell Environ. 38, 544–558. doi: 10.1111/pce.12414
Wen, F., Ye, F., Zhao, Z., Liao, L., Li, T., Jia, M., et al. (2020). Genome-wide survey and expression analysis of calcium-dependent protein kinase (CDPK) in grass Brachypodium distachyon. BMC Genomics 21, 1–17. doi: 10.1186/s12864-020-6475-6
Wernimont, A. K., Artz, J. D., Finerty, P., Jr., Lin, Y. H., Amani, M., Allali-Hassani, A., et al. (2010). Structures of apicomplexan calcium-dependent protein kinases reveal mechanism of activation by calcium. Nat. Struct. Mol. Biol. 17, 596–601. doi: 10.1038/nsmb.1795
Yang, Y., Wang, Q., Chen, Q., Yin, X., Qian, M., Sun, X., et al. (2017). Genome-wide survey indicates diverse physiological roles of the barley (Hordeum vulgare L.) calcium-dependent protein kinase genes. Sci. Rep. 7, 5306. doi: 10.1038/s41598-017-05646-w
Yip Delormel, T., Boudsocq, M. (2019). Properties and functions of calcium-dependent protein kinases and their relatives in Arabidopsis thaliana. New Phytol. 224, 585–604. doi: 10.1111/nph.16088
Yu, T. F., Zhao, W. Y., Fu, J. D., Liu, Y. W., Chen, M., Zhou, Y. B., et al. (2018). Genome-wide analysis of CDPK family in foxtail millet and determination of SiCDPK24 functions in drought stress. Front. Plant Sci. 9, 651. doi: 10.3389/fpls.2018.00651
Zhang, H., Liu, D., Yang, B., Zhu, J. K. (2020). Arabidopsis CPK6 positively regulates ABA signaling and drought tolerance through phosphorylating ABA-responsive element-binding factors. J. Exp. Bot. 71, 188–203. doi: 10.1093/jxb/erz432
Zhang, H. M., Zhu, J. H., Gong, Z. Z., Liu, W. Z., Mu, B., Song, H., et al. (2022). Abiotic stress responses in plants. Nat. Rev. Genet. 23, 104–119. doi: 10.1038/s41576-021-00413-0
Keywords: low temperature, SikCDPK1, expression analysis, functional exploration, calcium-dependent protein kinases (CDPKs)
Citation: Shi G, Liu Y, Tian X, Guo J and Zhu X (2024) Identification of SikCDPK family genes to low-temperature by RNA-seq approaches and functional analysis of SikCDPK1 in Saussurea involucrata (Kar. & Kir.). Front. Plant Sci. 15:1436651. doi: 10.3389/fpls.2024.1436651
Received: 22 May 2024; Accepted: 15 August 2024;
Published: 30 September 2024.
Edited by:
László Szabados, Hungarian Academy of Sciences (MTA), HungaryReviewed by:
Anna Kulik, Polish Academy of Sciences, PolandSudhir Kumar, Iowa State University, United States
Xi Zhu, Chinese Academy of Tropical Agricultural Sciences, China
Copyright © 2024 Shi, Liu, Tian, Guo and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xinxia Zhu, MzAyNjQxMzE2QHFxLmNvbQ==
 Guangzhen Shi
Guangzhen Shi Yuling Liu1
Yuling Liu1