
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 18 June 2024
Sec. Plant Abiotic Stress
Volume 15 - 2024 | https://doi.org/10.3389/fpls.2024.1431835
This article is part of the Research Topic Advanced Breeding for Abiotic Stress Tolerance in Crops, Volume II View all 18 articles
Cotton fiber, the mainstay of the world’s textile industry, is formed by the differentiation of epidermal cells on the outer peridium of the ovule. The TBL gene family is involved in the regulation of epidermal hair development as well as response to abiotic stress. However, the function of TBL genes in cotton has not been systematically studied yet. Here, we identified 131 and 130 TBL genes in TM-1 (Gossypium hirsutum) and Hai7124 (Gossypium barbadense), respectively. Phylogenetic, gene structure, expression pattern and cis-element of promoter analysis were performed and compared. Single gene association analysis indicated that more TBL genes related to fiber quality traits were found in G. barbadense, whereas more genes associated with yield traits were found in G. hirsutum. One gene, GhTBL84 (GH_D04G0930), was induced by treatment at 4°C for 12 and 24 h in G. hirsutum and silencing of the GhTBL84 gene by VIGS technology in TM-1 can significantly improve the resistance of cotton seedlings to low temperature stress. In sum, our study conducted a genome-wide identification and comparative analysis of TBL family genes in G. hirsutum and G. barbadense and demonstrated a group of TBL genes significantly associated with fiber quality and excavated cold stress responsive gene, such as GhTBL84, providing a theoretical basis for further improving cotton agronomic traits.
Cotton is a warm-season crop, originating from tropical and subtropical regions, with an optimal growth temperature range of (28 ± 3°C), which still retains its sensitivity to low temperatures after a long period of domestication and selection by humans (Pham et al., 2018). Cotton fiber is a unicellular hair-like structure formed by a single cell protrusion from the ovule epidermis, which in turn differentiates into a highly elongated and thickened single cell of seed trichomes (Pham et al., 2018; Zhai et al., 2023). The fiber development can be broadly divided into four sequential and overlapping stages, mainly in terms of days post anthesis (DPA): cell initiation (-3 to 3 DPA), cell elongation (3 to 20 DPA), secondary cell wall (SCW) synthesis and thickening (20 to 45 DPA), and dehydration maturation (45 to 50 DPA) (Haigler et al., 2012). The number of fiber cell initiations determines cotton fiber yield, while cell elongation and cell wall thickening determine final fiber quality. G. hirsutum bolls are large, high yielding and adaptable, but their fiber quality is poor, while G. barbadense bolls are small, low yielding and late maturing, but their fiber quality is excellent and they are a very good raw material for spinning high count yarn (Avci et al., 2013).
The DUF231 (Domin of unknown function) family are proteins that contain a conserved TBL (trichome birefringence like) structural domain that is unique to plants. Studies have shown that TBL proteins can act as polysaccharide O-acetyltransferases and catalyse the O-acetylation of specific cell wall polymers (Yuan et al., 2016a; Stranne et al., 2018, b). TBL3 and CESA genes are transcriptionally coordinated, and knockdown of TBL3 reduces crystalline secondary wall cellulose in trichomes and stems in Arabidopsis (Bischoff et al., 2010). The Arabidopsis esk1/tbl32/tbl33 triple mutant shows a significant reduction in cellulose and xylan content, disorganization of secondary cell wall structures and severe collapse of xylem cell structures (Yuan et al., 2016a). In addition, The TBL family of proteins is also involved in the regulation of plant responses to abiotic stresses. The Arabidopsis esk1 mutant increased plant tolerance to cold stress, with a 5.5°C increase in freezing tolerance (Xin et al., 2007). The cotton GhTBL34 gene was located in the confidence interval of a major quantitative trait locus for Verticillium wilt (VW) resistance, and the VIGS test showed that silencing TBL34 reduced VW resistance in cotton (Zhao et al., 2021).
Cold stress is an environmental factor that has a significant impact on plant growth, productivity and survival, and changes the fluidity of plant cell membranes when the external ambient temperature is altered (Sangwan et al., 2002). This change may be sensed by membrane-localized proteins, possibly including receptor-like protein kinases and histidine kinases (Ding et al., 2019). The receptor-like cytoplasmic kinase cold-responsive protein kinase 1 (CRPK1) transduces cold signals from the plasma membrane to the nucleus via 14–3-3 and C-REPEAT binding factor (CBF) proteins, affecting freezing tolerance in Arabidopsis (Liu et al., 2017). Under cold stress, CBFs (also known as dehydration response element-binding proteins or DREB1) transcription factors from the DREB subfamily A-1 of the ERF/APETALA2 transcription factor family are essential for acquired cold tolerance (Guo et al., 2018; Kidokoro et al., 2022).
In this study, we demonstrated a group of TBL genes in the cotton genome and performed phylogenetic, gene structure, expression pattern and promoter element analyses. The TBL genes related to fiber quality were identified by single gene association analysis in G. hirsutum and G. barbadense. In addition, the cold stress responsive TBL gene was excavated, such as GhTBL84, and silencing the GhTBL84 gene can improve the cold resistance of cotton. It provides a theoretical basis for further improvement of cotton agronomic traits and abiotic stress resistance in cotton.
G. hirsutum cv. Texas Marker-1 (TM-1), the genetic standard line for G. hirsutum cotton, was used as the plant material in this study. The plants were cultured in a growth chamber with conditions set to 22°C day (16 h)/night (8 h), 65% humidity and 120 μmol m−2 s−1 of light intensity. For cold treatment, four-leaf cotton seedlings were transferred to a thermostat incubator and grown at 4°C for 72 hours for phenotypic observations.
The G. hirsutum, G. barbadense, G. arboreum and G. raimondii genome sequences were downloaded from COTTONOMICS (http://cotton.zju.edu.cn/index.htm) and CottonMD (https://yanglab.hzau.edu.cn/CottonMD.1). All Arabidopsis TBL protein sequences were obtained from TAIR database (https://www.arabidopsis.org/) were used as queries for BLASTP (https://blast.ncbi.nlm.nih.gov/Blast.cgi). The obtained protein sequences of TBLs were submitted to the NCBI conserved domain database (https://www.ncbi.nlm.nih.gov/cdd) for further confirmation.
The TBL members of Arabidopsis and four Gossypium species were aligned by multiple sequence alignment using MUSCLE (Edgar, 2004). MEGA (v.11.0) software was used to construct a phylogenetic tree using the maximum likelihood estimation (ML) method with a bootstrap set to 1000 times. The phylogenetic tree was further embellished using the online software iTOL (https://itol.embl.de/).
The molecular weight (MW) and isoelectric point (pI) of TBLs were obtained by ExPASy (https://web.expasy.org/protparam/). The subcellular location was predicted with Cell-PLoc 2.0 (http://www.csbio.sjtu.edu.cn/bioinf/Cell-PLoc-2/). The gene structures and conserved motifs of TBLs were analyzed using TBtools software (v 2.0) (Chen et al., 2023). For cis-elements of promoter analysis, upstream 2,000 bp sequences of the start codon was extracted and submitted to PlantCARE database (Lescot et al., 2002).
The chromosomal positions of TBL genes were obtained via a gff3 annotation file from COTTONOMICS (http://cotton.zju.edu.cn/index.htm) and displayed by MapChart (v.2.2) (Voorrips, 2002). MCScanX software was used to identify gene duplication and syntenic regions, and the results were visualized by TBtools software (v 2.0) (Chen et al., 2023). The protein interaction network prediction of GhTBLs and GbTBLs was analyzed by the STRING online database (https://cn.string-db.org/) (Szklarczyk et al., 2021).
The genetic variation information and phenotypic data of G. hirsutum and G. barbadense populations (Fang et al., 2017; Hu et al., 2019; Fang et al., 2021) were utilized to annotate variant loci using ANNOVAR (Wang et al., 2010). Subsequently, haplotype analysis of TBL gene family was conducted based on the typing of nonsynonymous mutations, and t-tests were performed on different haplotypes (P < 0.05), thereby eliminating TBL genes associated with agronomic traits. The results were visualized with R software.
The qRT-PCR reaction was performed following the instructions of HiScript II One Step qRT-PCR SYBR Green Kit (Q221–01, Nanjing Novozymes Bioscience and Tech-nology Co., Ltd., China). The reaction system consisted of 2 μL of cDNA, 0.4 μL of each upstream and downstream primer, 7.2 μL of ddH2O, and 10 μL of 2 × SYBR Premix Ex-TaqTM. The PCR reaction program was set as follows: 95°C for 3 min, followed by 40 cycles of 95°C for 10 s and 60°C for 30 s, and 95°C for 15 s and 60°C for 1 min. Each reaction was repeated three times, and the relative expression was calculated using the 2-ΔΔCT method (Livak and Schmittgen, 2001). The GhHis3 gene was selected as the internal reference gene (Mao et al., 2023), and all primer sequences were listed in Supplementary Table S8.
Silencing fragments of the GhTBL84 gene were selected using the SGN VIGS Tool (https://vigs.solgenomics.net/) and amplified by specific primers (Supplementary Table S8) from cDNA of TM-1 and cloned into a pTRV2 vector. The fusion vector TRV2::GhTBL84, TRV2:CLA1 (positive control) and empty vector TRV2::00 (negative control), TRV1::00 were transformed into Agrobacterium tumefaciens GV3101. The final construct was mixed with the helper vector (TRV1::00) in the ratio of 1:1 with OD600 = 2.0. Finally, the mixture was injected into the cotyledons of cotton TM-1 (Li J. et al., 2023).
Amino acid sequence similarity and conserved structural domains were identified in TBLs from four different cotton species: G. hirsutum (131), G. barbadense (130), G. arboreum (66), and G. raimondii (62). The predicted protein length and molecular weight (MW) of the cotton TBL genes ranged from 221 aa/40.11 kDa to 1,422 aa/99.13 kDa, while the theoretical isoelectric point (pI) ranged from 5.18 to 9.51. Subcellular localization predictions indicated that TBLs are distributed in various tissues of both G. hirsutum and G. barbadense. GhTBLs are mainly located in chloroplasts, indicating their potential involvement in regulating various cellular functions and playing a crucial role in photoregulation. It is worth noting that GbTBLs are primarily situated in the cell wall, while GbTBLs are believed to be associated with the development of cotton fibers. Detailed information about the predicted protein length, pI, MW, chromosomal location, and other related information is shown in Supplementary Tables S1 and S2.
To determine the evolutionary relationships of the cotton TBL gene family, a phylogenetic evolutionary tree was constructed using the Arabidopsis thaliana TBL gene family as a reference (Figure 1, Supplementary Table S3). The TBL gene family was classified into six subgroups (Groups I to VI) based on their affinities, which is consistent with previous reports (Stranne et al., 2018). Group IV has the largest of members, with 99 members, including 9 in A. thaliana, 30 in G. hirsutum, 29 in G. barbadense, 15 in G. arboreum, and 16 in G. raimondii. In contrast, Group VI has the lowest number of members, with only 21 members, including 2 in A. thaliana, 7 in G. hirsutum, 6 in G. barbadense, 3 in G. arboreum, and 3 in G. raimondii. The number of members in each subgroup were varied. However, each group contained an approximately equal number of G. hirsutum and G. barbadense members, which appeared in corresponding pairs. In addition, the number of G. hirsutum and G. barbadense members was nearly twice that of the diploid ancestral species, G. arboreum, and G. raimondii, consistent with the evolutionary relationship of the species. Five genes from G. hirsutum (GH_A01G1358, GH_A11G3750, GH_D04G1469, GH_D04G1899, GH_D09G0883) and four genes from G. barbadense (GB_A04G1681, GB_A09G1180, GB_D01G0463 and GB_D09G1031) were identified as lacking corresponding homologous genes, suggesting gene loss in some TBLs during the evolution (Supplementary Table S4).
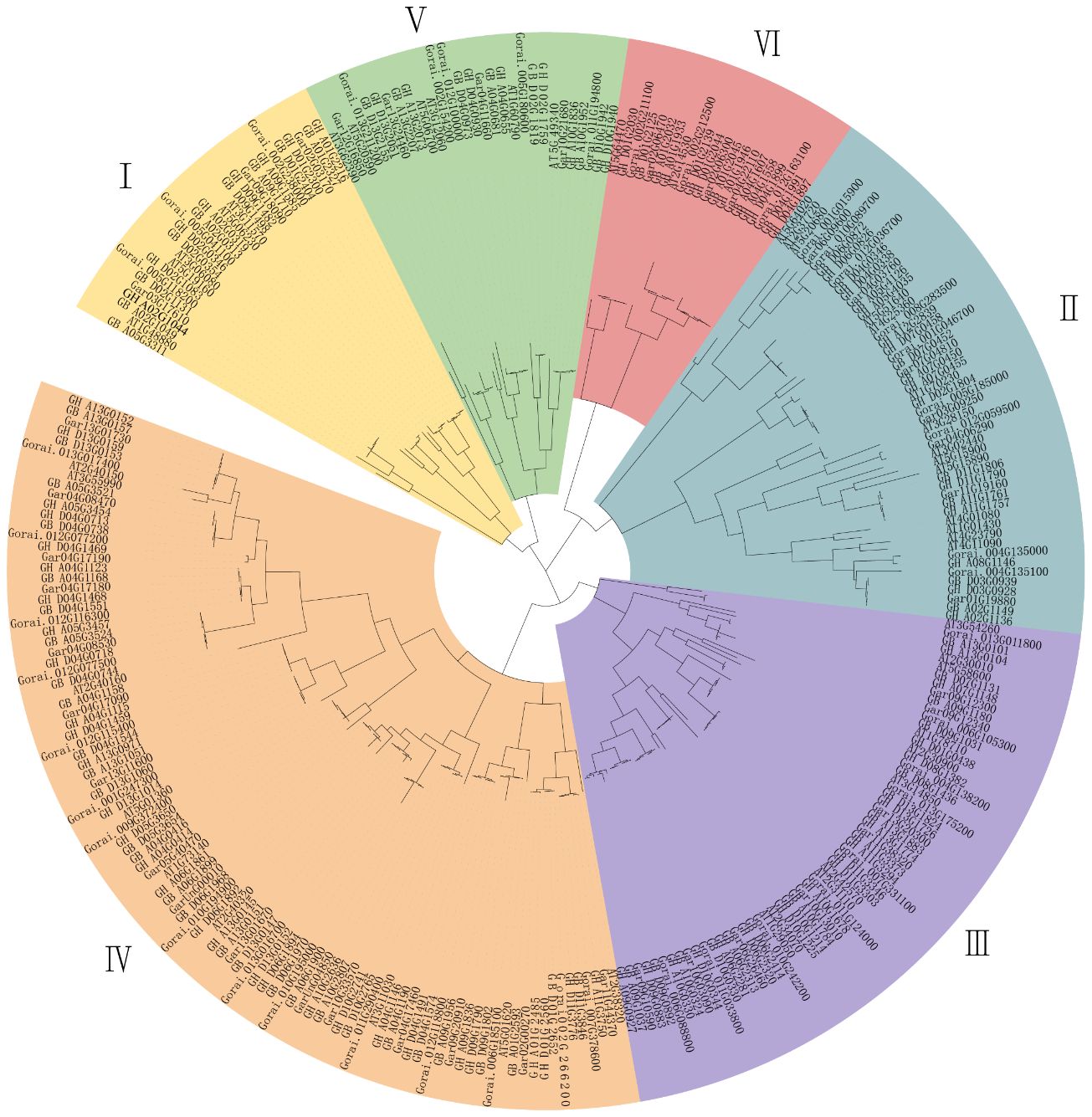
Figure 1 Phylogentic analysis of TBL gene family from Arabidopsis thaliana (AT), G. hirsutum (GH) and G. barbadense (GB), G. arboreum (Ga), G. raimondii (Gr). The phylogenetic tree was constructed using the maximum likelihood estimation (ML) method by MEGA 7.0 software.
Ten motifs were identified in the TBL gene family of G. hirsutum and G. barbadense using the online tool MEME to predict conserved motifs. The conserved motifs of homologous genes in subgroups A and D of G. hirsutum and G. barbadense are similar in terms of their types, numbers, and distribution locations (Supplementary Figure S1). Motifs 4, 5, and 9 are present in both G. hirsutum and G. barbadense, indicating a high degree of conservation during evolutionary. Motif 1 is present in all family members except for GH_A08G1146. Furthermore, all contained motif 2 except for GH_A05G3406, GH_A06G1862, GB_A08G1294, and GB_D09G1498, suggesting that some of the TBL genes have lost motifs during evolution, which may have led to functional changes. Analysis of the exon and intron structures of the TBL genes showed that the genes within the same group had similar gene structures, with comparable exon lengths and distributions, indicating the reliability of the phylogenetic classification. However, individual genes, GH_D11G1790/GB_D11G1806 and GH_A11G1271/GB_A11G1277, have different gene lengths but encode amino acid sequences of the same length (Supplementary Table S4), suggesting that introns have undergone selective splicing during evolution, leading to gene functional diversity.
According to the genome annotation information of G. hirsutum and G. barbadense, we generated chromosomal distribution maps for the TBL gene family (Figure 2). The results showed that the 131 GhTBLs and 130 GbTBLs are unevenly distributed across all 26 chromosomes. For instance, there are 15 GhTBLs and 13 GbTBLs on chromosome D04, and one TBL is found on chromosome A03. The TBL family genes have been retained in homologous chromosomes during chromosome doubling events in heterotetraploid cotton and may have evolved to favor the D subgenome. There are certainly other genes that have undergone tandem duplication events, such as GH_A01G0442/GH_A01G0443 and GB_A01G0433 GB_A01G0434 on chromosome A01; GH_A06G1861/GH_A06G1862 and GB_A06G1899/GB_A06G1900 on chromosome A06; and GH_D04G0717/GH_D04G0718 and GB_D04G0345/GB_D04G0346 on chromosome D04. Tandem and segmental duplication are crucial for the generation of new gene family members during the evolutionary process, and the duplicated genes are basically one-to-one correspondence (Figure 3; Supplementary Tables S5, S6).
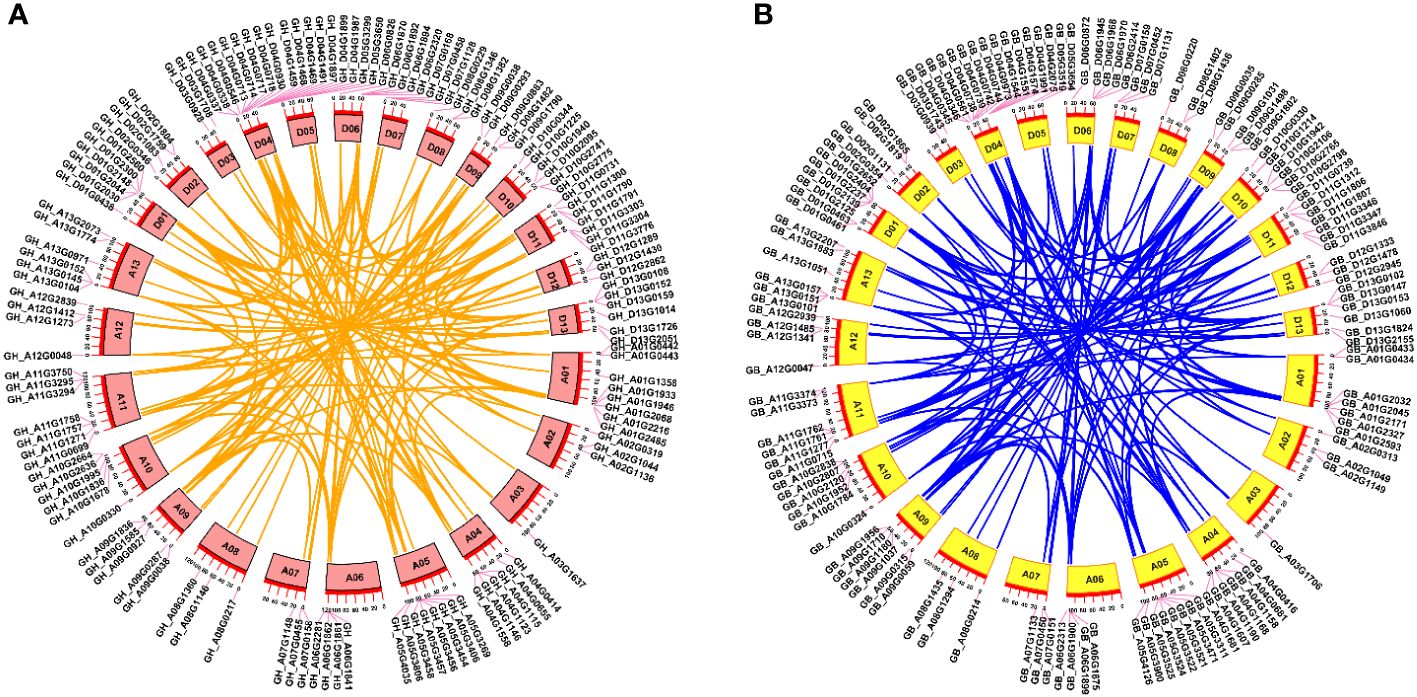
Figure 3 Collinearity analysis of TBL gene family from G. hirsutum (A) and G. barbadense (B). Orange and blue lines exhibited the link between paralogous gene pairs.
Cis-elements located in promoters play an important role in the regulation of gene expression. To gain more insight into the functions of TBL genes, the cis-regulatory elements in the 2000 bp upstream of the transcription start sites of cotton TBL genes (Figure S2). The study revealed the presence of various response elements, such as light responsive element, defense and stress responsive element involved in drought and low temperature, and hormone responsive element associated with auxin, gibberellin, salicylic acid, abscisic acid and MeJA. It is noteworthy that all TBL genes, except GH_D07G0458 and GB_A11G1277, contained multiple light responsive elements. This indicates that the cotton TBL genes might play an important role in light-regulated growth and development, and is involved in multiple regulatory pathways when cotton is exposed to environmental stress.
The interaction properties of GhTBL family proteins in G. hirsutum were analyzed using the STRING database. The results are presented in Supplementary Figure S3A, which shows 131 amino acid sequences of TBL family members in G. hirsutum in the protein interaction network, with 24 members, including 5 members with node interactions. Among them, GhTBL93 (GH_D05G3650) had the most interaction nodes, indicating that the protein requires interaction with other proteins to function properly. The protein nodes and interaction relationships of the GbTBL gene family in G. barbadense correspond to homologous genes in G. hirsutum, which is consistent with those in G. hirsutum (Supplementary Figure S3B). Among them, GbTBL92 (GB_D05G3654) has the most interaction nodes, indicating that this protein needs to interact with other proteins to function. There is no significant difference between the protein network of the interaction between G. hirsutum and G. barbadense.
To clarify the expression pattern of the cotton TBL genes, we analyzed their expression in different tissues: cotton root, stem, leaf, ovule and fiber (Figure 4). The TBL genes were all expressed in a tissue-specifical manner, indicating their potential involvement in various regulatory pathways during cotton growth and development. In the same group, genes exhibit similar expression patterns, while genes in different groups dis-play distinct expression patterns. During fiber development, GhTBL genes in Groups IV and V of G. hirsutum are significantly expressed, whereas GbTBL genes in Groups II and IV of G. barbadense are significantly expressed. In addition, the number of gene expressions in G. barbadense is higher than that in G. hirsutum. It is worth noting that there is a difference in the peak expression period of homologous genes between G. hirsutum and G. barbadense. Specifically, G. barbadense exhibits a delayed peak expression period during fiber development compared to G. hirsutum. To compare the expression trends during fiber development, we selected three pairs of homologous gene pairs in the G. hirsutum and G. barbadense lineage, the results suggests that G. barbadense fibers have a longer elongation and development period, potentially resulting in higher quality fibers (Supplementary Figure S4).
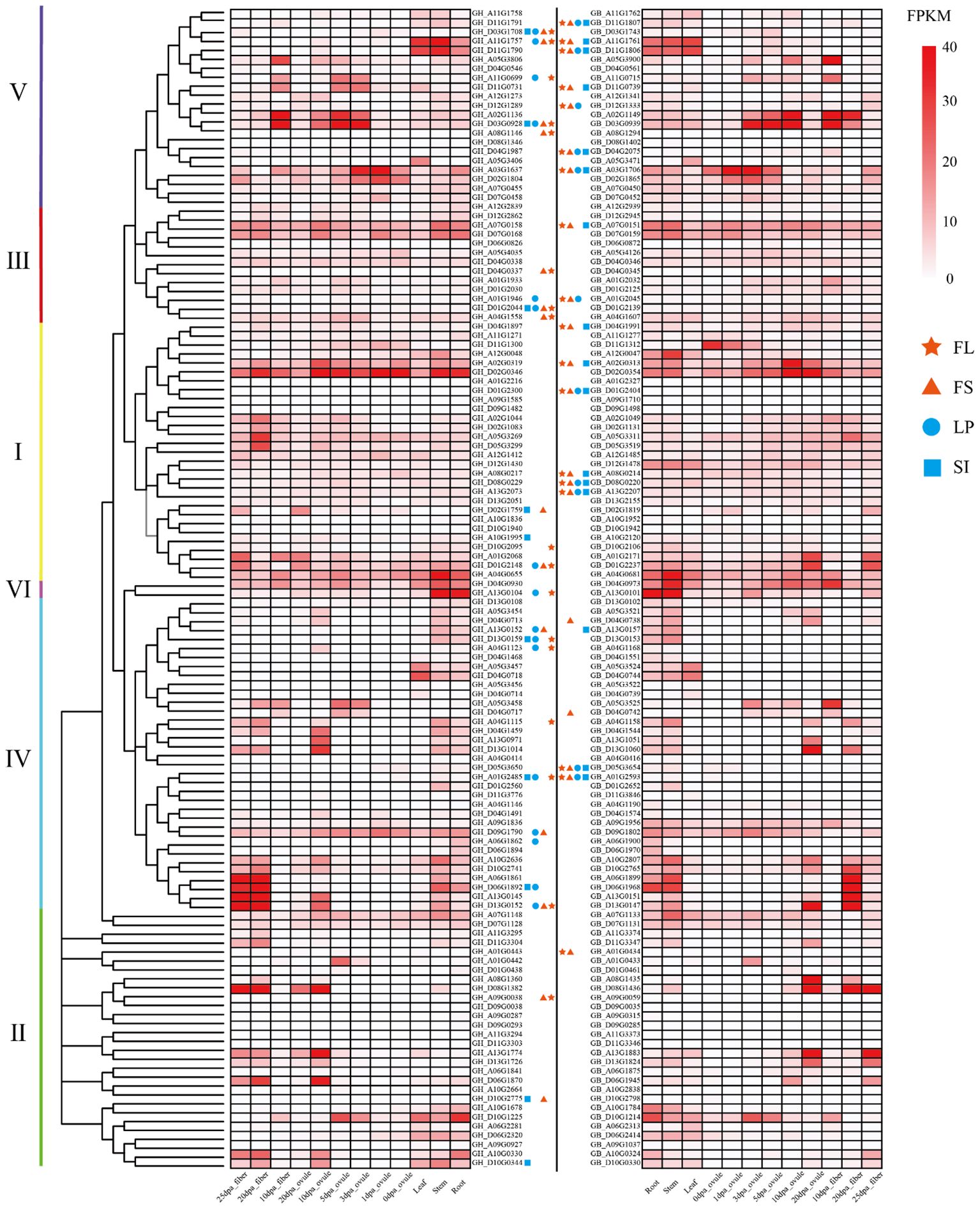
Figure 4 Correlation analysis between TBL genes of G. hirsutum and G. barbadense and fiber length (FL), fiber strength (FS), lint percentage (LP), and seed index (SI). DPA: Days post anthesis. The phylogenetic tree was constructed using the maximum likelihood estimation (ML) method by MEGA 7.0 software. The darker the color, the higher the expression.
By performing association analysis between TBL genes and major agronomic traits of G. hirsutum and G. barbadense, it was found that there is a correlation between TBL genes and agronomic traits related to fiber quality and yield (Supplementary Table S7). As shown in Figure 4, there were 17, 14, 16 and 10 genes related to fiber length (FL), fiber strength (FS), lint percentage (LP) and seed index (SI) in G. hirsutum, respectively. On the other hand, 18 genes were found to be associated with FL, 20 genes with FS, 11 genes with LP and 16 genes with SI in G. barbadense. Regarding the agronomic traits in G. hirsutum and G. barbadense, 26 and 21 genes respectively were found to be significantly related. The number of genes in G. hirsutum was significantly higher than that in G. barbadense. Analysis of the homologous genes revealed two pairs of homologous genes associated with FL (GH_A11G1757/GB_A11G1761 and GH_A01G2485/GB_A01G2593) and LP (GH_A01G1946/GB_A01G2045 and GH_A01G2485/GB_A01G2593) traits in both species, and only one pair of homologous genes associated with FS (GH_A11G1757/GB_A11G1761) and SI (GH_A01G2485/GB_A01G2593) traits. Differences in homologous genes between G. hirsutum and G. barbadense, based on the gene structure of the TBL genes, result in differences in trait association. For instance, GH_D11G1790 and GB_D11G1806 exhibit coding sequence differences in Group V (Supplementary Table S7). They differ in gene length by 123 bp, and GB_D11G1806 is expressed in fibers, while GH_D11G1790 is not. The association between GB_D11G1806 and the four agronomic traits mentioned is significant, whereas GH_D11G1790 shows no significant association with these traits.
The RNA-seq data downloaded from the public database was used to further analyze the expression patterns of TBL genes were in G. hirsutum and G. barbadense under different durations of cold, heat, salt, and drought stresses (Figure 5). The results showed that most of the TBL genes responded to the different stress treatments. In G. hirsutum, 59 TBL genes were induced by stress under four abiotic stress conditions, while only 29 genes were induced in G. barbadense. The induction genes of G. hirsutum and G. barbadense under four abiotic stress conditions differs among different evolutionary groups, with each having certain preferences. In G. hirsutum, the induced genes are mainly distributed in Groups II and IV, while in G. barbadense, they are mainly distributed in Groups I, III, and V. The induction of homologous genes in G. hirsutum and G. barbadense varies under different stress conditions for the same group. For example, GhTBL119 (GH_D11G1791) is induced under both high and low temperature conditions, whereas the homologous gene GbTBL118 (GB_D11G1807) is only induced under low temperature conditions. We also found that GhTBL84 (GH_D04G0930) in group I is induced under both 4°C and salt stress conditions, whereas the homologous gene GbTBL85 (GB_D04G0973) is not induced under all four abiotic stress conditions. Currently, compared with high temperature, salt and drought stresses, low-temperature stress has a greater impact on cotton during spring emergence, seedling stage and bolling stage, resulting in lower yield and poor quality. Therefore, we verified the role of GhTBL84 in cotton response to cold stress.
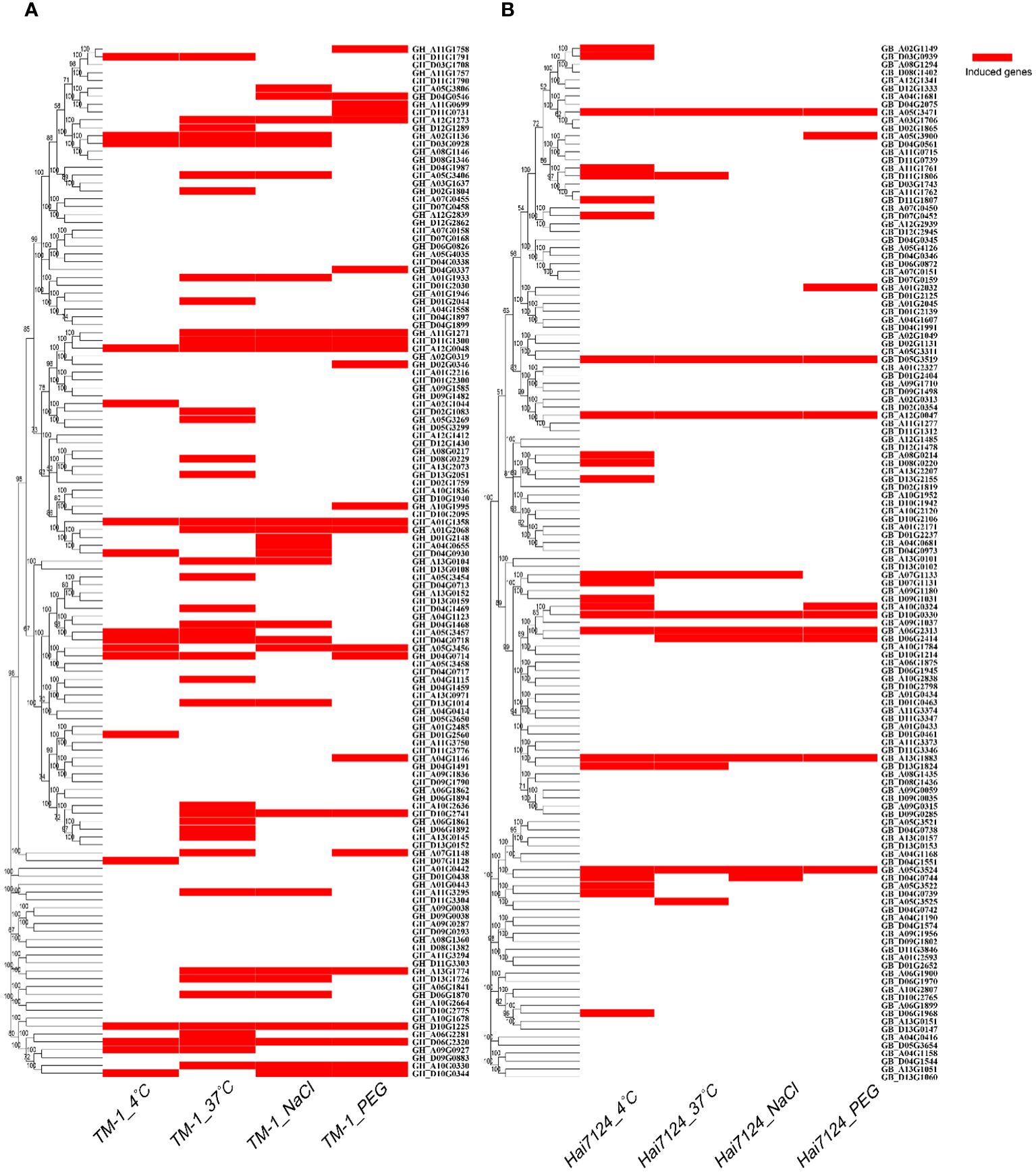
Figure 5 Induced genes of TBL gene family from G hirsutum (A) and G barbadense (B) under different abiotic stress. After 1 h, 3 h, 6 h, 12 h and 24 h treatment with stress (Low temperature: 4°C; High temperature: 37°C; NaCl: salt stress; PEG: drought stress), up-regulated or down-regulated gene expression was considered to be regulated by abiotic stress.
Transcriptome data were used to identify the GhTBL84 gene, which showed a significant change in expression under cold stress (Supplementary Figure S5). To investigate the role of GhTBL84 in the response to cold stress, we utilized the VIGS assay to suppress the expression of GhTBL84 in cotton TM-1 seedlings. As shown in Figure 6A, the TRV2::GhCLA1 plants exhibited albino phenotypes. The leaf wilting and drooping amplitude of plants inoculated with TRV2::00 was significantly higher than that of TRV2::GhTBL84 plants under cold treatment. And the expression level of GhTBL84 in TRV2::GhTBL84 plants was significantly lower than TRV2::00 plants (Figure 6B), indicating that GhTBL84 may as a negative regulator response to cold stress.
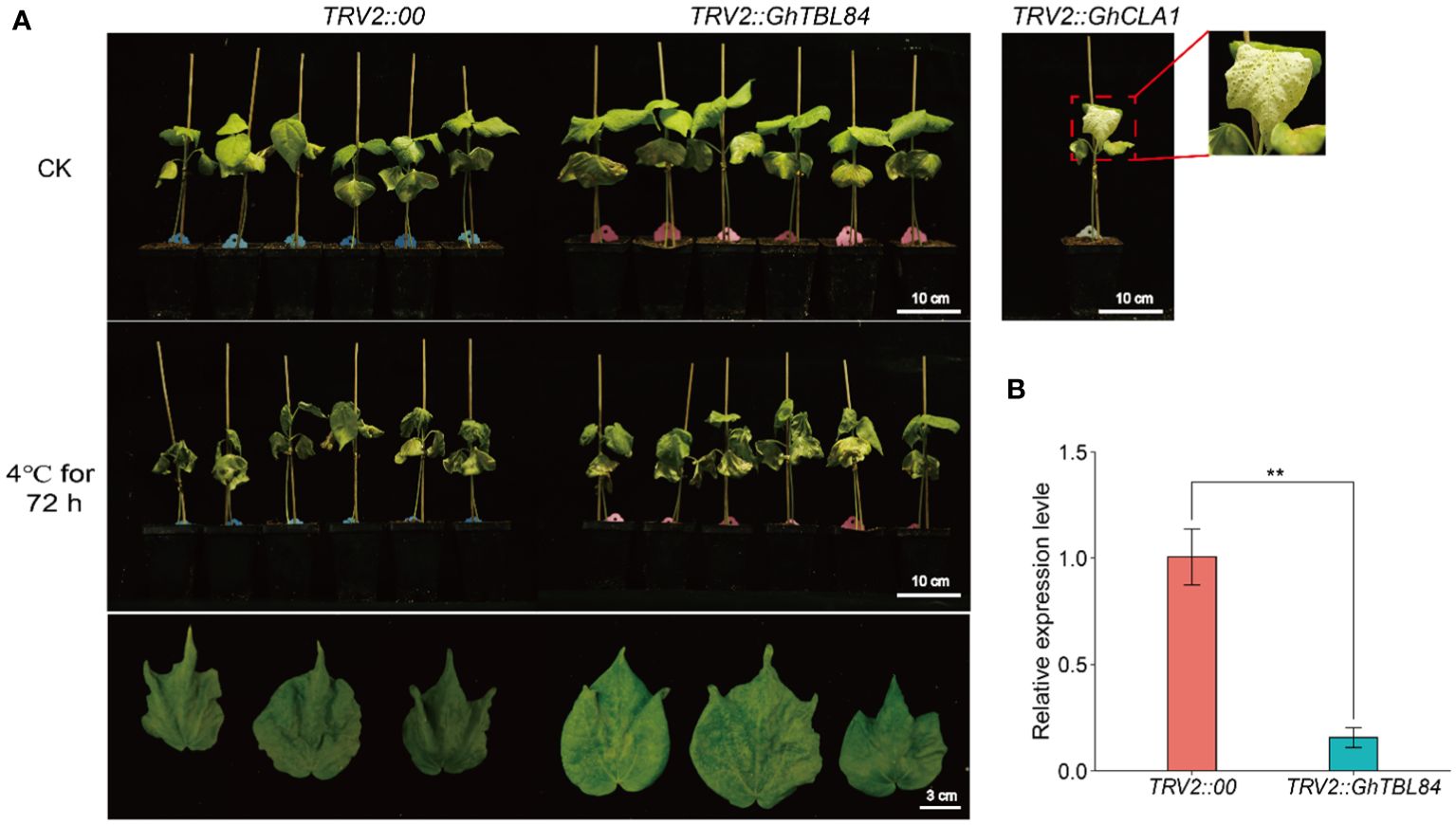
Figure 6 GhTBL84 responses to cold stress by VIGS validation. (A) Representative images showing the phenotypes of TRV2:00 and TRV2:GhTBL84 seedlings after cold treatment. (B) The relative expression of GhTBL84 in TRV2:00 and TRV2:GhTBL84 seedlings. Values represent means ± SD of three biological replicates. ** indicates significant differences at p < 0.01.
Cotton is a major cash crop, and its fibers hold an unassailable position in the textile industry. Fibers are formed by the protrusion of a single cell of the ovule epidermis and then differentiates, similar to the formation of epidermal hairs in A. thaliana. Therefore, many researchers have used the study of Arabidopsis trichomes as a substitute for investigating cotton fibers (Wang et al., 2004; Yang and Ye, 2013). The TBL gene families in Arabidopsis (Xiong et al., 2013), rice (Gao et al., 2017), quinoa (Ma et al., 2021), and rose (Tian et al., 2021) have been previously identified. However, the function of TBL genes in abiotic stress in cotton has not been extensively studied. In this study, we objectively explore the potential function of the cotton TBL gene family in cotton fiber development and response to abiotic stress.
In this study, 131 and 130 TBL genes were identified from two tetraploid cotton species, G. barbadense (Hai 7124) and G. hirsutum (TM-1), respectively. This result is similar to studies reported in cotton, although the number of gene family members is not the same, which may be related to the genomic data and sequence com-parison methods used by the researchers (Kabir et al., 2023; Li Z. et al., 2023). In comparison, there are 45 TBL family genes in A. thaliana and the number of genes in cotton is 2.8 times higher, suggesting that the TBL gene family may have undergone replication events such as whole genome duplication during evolution, resulting in significant gene expansion.
Collinearity analysis showed some occurrence of synonymous mutations. The Ka/Ks values of all gene pairs were less than 1 in G. hirsutum. In G. barbadense, only one gene pair (GB_D06G1968 and GB_D10G2765) had a Ka/Ks value of 1. 05, while the rest of the genes had a Ka/Ks value of less than 1. This result indicates that this one pair of genes had been under positive selection in the two cotton species and may be the key genes in the evolutionary process, and the rest of the genes are relatively conserved during evolution (Hurst, 2002).
The developmental stages of cotton fiber include five stages: cell initiation and differentiation, rapid elongation or primary cell wall synthesis, primary to secondary cell wall transition, secondary cell wall synthesis, and fiber maturation (Haigler et al., 2012). Secondary cellulose deposition is a part of the cotton fiber elongation (Hinchliffe et al., 2010).The TBL gene is a member of the plant-specific DUF231 structural domain gene family, which is responsible for the initiation of plant trichomes and the O-acetylation pathway in the cell wall. In rice mutant ostbl1 and ostbl2, not only was the plant height significantly reduced, but also the level of cell wall acetylation was affected (Gao et al., 2017). However, the role of TBLs in cotton has not been systematically studied.
The TBL gene family exhibited tissue-specific expression in cotton, indicating that it may be involved in multiple processes of cotton growth and development. Tissue expression pattern analysis revealed that 87 and 91 TBL genes were expressed at different stages of fiber development stage of fiber in G. barbadense and G. hirsutum, respectively. It suggested that TBL family genes may be associated with fiber in tetraploid cultivated cotton species (Kabir et al., 2023; Li J. et al., 2023). The expression peak of TBL genes in G. hirsutum during fiber development was obviously earlier than that in G. barbadense, which indicated that the genes involved in fiber development may be different in two cotton species, and ultimately caused the difference in fiber yield and quality.
Association analysis of TBL family genes with important agronomic traits such as FL, FS, SI, and LP in G. barbadense and G. hirsutum, showed that there were 18 and 20 genes related to yield and quality traits in G. barbadense, and 20 and 21 genes related to G. hirsutum, respectively. This suggests a higher correlation between the TBL family and fiber quality in G. barbadense. Previous studies have reported that GhTBL87 and GhTBL58 are relevant for fiber length (Kabir et al., 2023), which further confirmed the importance of TBL family genes in improving cotton fiber yield and quality. This provided the genetic resources for creating excellent cotton germplasm.
Abiotic stresses can also lead to differences in fiber quality. Promoter element perdition showed that cis-elements, including light-responsive, hormone-responsive, and low temperature responsive elements, differ in these genes. This suggests TBL genes may play a role in response to abiotic stress. For instance, in quinoa, TBL is involved in ethylene and salt stress responses (Ma et al., 2021). Similarly, TBL genes have been implicated in resistance to phytopathogenic bacteria in Arabidopsis, and RcTBL16 has been shown to be associated with gray mold in rose (Tian et al., 2021). Analysis of the expression patterns of GhTBL and GbTBL genes under abiotic stress showed that most the genes could be involved in the response to abiotic stress. In addition, more genes were significantly expressed in upland cotton than in island cotton, suggesting their response to abiotic stress may be more pronounced.
Cold stress disrupts normal physiological and metabolic processes in plant, alters cell morphology, and regulates respiration. This results in slow growth, wilting, drooping leaves, and wilting to death, ultimately lead to yield and quality loss (Kidokoro et al., 2022). Seedling emergence and fiber quality in cotton are significantly affected by cold stress. Several studies have been conducted on the physiological and molecular mechanisms of cotton in response to cold stress. For instance, research has shown that exposure to cold temperatures can result in changes to malate metabolism, soluble sugar metabolism, and cellulose synthesis. These changes ultimately lead to a significant reduction in the length of cotton fibers (Wang et al., 2024; Zheng et al., 2012). Similarly, transcriptome sequencing of stress-treated in TM-1 revealed a significant up-regulation of genes related to the ABA signaling pathway under cold stress (Hu et al., 2019). In this study, we found that the GhTBL84 gene responds to cold stress by characterizing the TBL gene family in cotton. Using VIGS technology, it was demonstrated that GhTBL84 was shown to negatively regulates the cold tolerance in cotton. However, the specific molecular mechanism was not explored in depth. Next, the molecular mechanism of the GhTBL84 gene involved in cold stress will be further analyzed by yeast two-hybrid (Y2H) and other methods. This will provide excellent genes for the development of cold-tolerant germplasm in cotton.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
LF: Writing – original draft, Writing – review & editing, Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization. XZ: Data curation, Conceptualization, Writing – review & editing. XM: Data curation, Writing – review & editing. WH: Validation, Writing – review & editing. YX: Formal analysis, Writing – review & editing. SH: Formal Analysis, Writing – review & editing. ZC: Data curation, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was supported by Ministry of Agriculture and Rural Affairs (2023ZD04039-01), the National Science Foundation of China (32172008), the Leading Innovative and Entrepreneur Team Introduction Program of Zhejiang (2019R01002).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1431835/full#supplementary-material
Avci, U., Pattathil, S., Singh, B., Brown, V. L., Hahn, M. G., Haigler, C. H. (2013). Cotton fiber cell walls of Gossypium hirsutum and Gossypium barbadense have differences related to loosely-bound xyloglucan. PloS One 8, e56315. doi: 10.1371/journal.pone.0056315
Bischoff, V., Nita, S., Neumetzler, L., Schindelasch, D., Urbain, A., Eshed, R., et al. (2010). TRICHOME BIREFRINGENCE and its homolog AT5G01360 encode plant-specific DUF231 proteins required for cellulose biosynthesis in Arabidopsis. Plant Physiol. 153, 590–602. doi: 10.1104/pp.110.153320
Chen, C., Wu, Y., Li, J., Wang, X., Zeng, Z., Xu, J., et al. (2023). TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 16, 1733–1742. doi: 10.1016/j.molp.2023.09.010
Ding, Y., Shi, Y., Yang, S. (2019). Advances and challenges in uncovering cold tolerance regulatory mechanisms in plants. New Phytol. 222, 1690–1704. doi: 10.1111/nph.15696
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Fang, L., Gong, H., Hu, Y., Liu, C., Zhou, B., Huang, T., et al. (2017). Genomic insights into divergence and dual domestication of cultivated allotetraploid cottons. Genome Biol. 18, 33. doi: 10.1186/s13059-017-1167-5
Fang, L., Zhao, T., Hu, Y., Si, Z., Zhu, X., Han, Z., et al. (2021). Divergent improvement of two cultivated allotetraploid cotton species. Plant Biotechnol. J. 19, 1325–1336. doi: 10.1111/pbi.13547
Gao, Y., He, C., Zhang, D., Liu, X., Xu, Z., Tian, Y., et al. (2017). Two trichome birefringence-like proteins mediate xylan acetylation, which is essential for leaf blight resistance in rice. Plant Physiol. 173, 470–481. doi: 10.1104/pp.16.01618
Guo, X., Liu, D., Chong, K. (2018). Cold signaling in plants: Insights into mechanisms and regulation. J. Integr. Plant Biol. 60, 745–756. doi: 10.1111/jipb.12706
Haigler, C. H., Betancur, L., Stiff, M. R., Tuttle, J. R. (2012). Cotton fiber: a powerful single-cell model for cell wall and cellulose research. Front. Plant Sci. 3. doi: 10.3389/fpls.2012.00104
Hinchliffe, D. J., Meredith, W. R., Yeater, K. M., Kim, H. J., Woodward, A. W., Chen, Z. J., et al. (2010). Near-isogenic cotton germplasm lines that differ in fiber-bundle strength have temporal differences in fiber gene expression patterns as revealed by comparative high-throughput profiling. Theor. Appl. Genet. 120, 1347–1366. doi: 10.1007/s00122-010-1260-6
Hu, Y., Chen, J., Fang, L., Zhang, Z., Ma, W., Niu, Y., et al. (2019). Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton. Nat. Genet. 51, 739–748. doi: 10.1038/s41588-019-0371-5
Hurst, L. D. (2002). The Ka/Ks ratio: diagnosing the form of sequence evolution. Trends Genet. 18, 486. doi: 10.1016/S0168-9525(02)02722-1
Kabir, N., Wang, X., Lu, L., Qanmber, G., Liu, L., Si, A., et al. (2023). Functional characterization of TBL genes revealed the role of GhTBL7 and GhTBL58 in cotton fiber elongation. Int. J. Biol. Macromol. 241, 124571. doi: 10.1016/j.ijbiomac.2023.124571
Kidokoro, S., Shinozaki, K., Yamaguchi-Shinozaki, K. (2022). Transcriptional regulatory network of plant cold-stress responses. Trends Plant Sci. 27, 922–935. doi: 10.1016/j.tplants.2022.01.008
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Li, J., Nie, K., Wang, L., Zhao, Y., Qu, M., Yang, D., et al. (2023). The molecular mechanism of GhbHLH121 in response to iron deficiency in cotton seedlings. Plants (Basel) 12. doi: 10.3390/plants12101955
Li, Z., Shi, Y., Xiao, X., Song, J., Li, P., Gong, J., et al. (2023). Genome-wide characterization of trichome birefringence-like genes provides insights into fiber yield improvement. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1127760
Liu, Z., Jia, Y., Ding, Y., Shi, Y., Li, Z., Guo, Y., et al. (2017). Plasma membrane crpk1-mediated phosphorylation of 14–3-3 proteins induces their nuclear import to fine-tune CBF signaling during cold response. Mol. Cell 66, 117–128.e115. doi: 10.1016/j.molcel.2017.02.016
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Ma, Q., Su, C., Dong, C. H. (2021). Genome-wide transcriptomic and proteomic exploration of molecular regulations in quinoa responses to ethylene and salt stress. Plants (Basel) 10. doi: 10.3390/plants10112281
Mao, Y., Dai, F., Si, Z., Fang, L., Zhang, T. (2023). Duplicate mutations of GhCYP450 lead to the production of ms5m6 male sterile line in cotton. Theor. Appl. Genet. 136, 2. doi: 10.1007/s00122-023-04296-z
Pham, H. M., Kebede, H., Ritchie, G., Trolinder, N., Wright, R. J. (2018). Alternative oxidase (AOX) over-expression improves cell expansion and elongation in cotton seedling exposed to cool temperatures. Theor. Appl. Genet. 131, 2287–2298. doi: 10.1007/s00122-018-3151-1
Sangwan, V., Orvar, B. L., Beyerly, J., Hirt, H., Dhindsa, R. S. (2002). Opposite changes in membrane fluidity mimic cold and heat stress activation of distinct plant MAP kinase pathways. Plant J. 31, 629–638. doi: 10.1046/j.1365-313X.2002.01384.x
Stranne, M., Ren, Y., Fimognari, L., Birdseye, D., Yan, J., Bardor, M., et al. (2018). TBL10 is required for O-acetylation of pectic rhamnogalacturonan-I in Arabidopsis thaliana. Plant J. 96, 772–785. doi: 10.1111/tpj.14067
Szklarczyk, D., Gable, A. L., Nastou, K. C., Lyon, D., Kirsch, R., Pyysalo, S., et al. (2021). The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 49, D605–d612. doi: 10.1093/nar/gkab835
Tian, Y., Zhang, S., Liu, X., Zhang, Z. (2021). Global investigation of TBL gene family in rose (Rosa chinensis) unveils RcTBL16 is a susceptibility gene in gray mold resistance. Front. Plant Sci. 12, 738880. doi: 10.3389/fpls.2021.738880
Voorrips, R. E. (2002). MapChart: software for the graphical presentation of linkage maps and QTLs. J. Hered. 93, 77–78. doi: 10.1093/jhered/93.1.77
Wang, K., Li, M., Hakonarson, H. (2010). ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164. doi: 10.1093/nar/gkq603
Wang, J., Liang, Y., Gong, Z., Zheng, J., Li, Z., Zhou, G., et al. (2024). Genomic and epigenomic insights into the mechanism of cold response in upland cotton (Gossypium hirsutum). Plant Physiol. Biochem. 206, 108206. doi: 10.1016/j.plaphy.2023.108206
Wang, S., Wang, J. W., Yu, N., Li, C. H., Luo, B., Gou, J. Y., et al. (2004). Control of plant trichome development by a cotton fiber MYB gene. Plant Cell 16, 2323–2334. doi: 10.1105/tpc.104.024844
Xin, Z., Mandaokar, A., Chen, J., Last, R. L., Browse, J. (2007). Arabidopsis ESK1 encodes a novel regulator of freezing tolerance. Plant J. 49, 786–799. doi: 10.1111/j.1365-313X.2006.02994.x
Xiong, G., Cheng, K., Pauly, M. (2013). Xylan O-acetylation impacts xylem development and enzymatic recalcitrance as indicated by the Arabidopsis mutant tbl29. Mol. Plant 6, 1373–1375. doi: 10.1093/mp/sst014
Yang, C., Ye, Z. (2013). Trichomes as models for studying plant cell differentiation. Cell Mol. Life Sci. 70, 1937–1948. doi: 10.1007/s00018-012-1147-6
Yuan, Y., Teng, Q., Zhong, R., Ye, Z. H. (2016a). Roles of Arabidopsis TBL34 and TBL35 in xylan acetylation and plant growth. Plant Sci. 243, 120–130. doi: 10.1016/j.plantsci.2015.12.007
Yuan, Y., Teng, Q., Zhong, R., Ye, Z. H. (2016b). TBL3 and TBL31, two arabidopsis DUF231 domain proteins, are required for 3-O-monoacetylation of xylan. Plant Cell Physiol. 57, 35–45. doi: 10.1093/pcp/pcv172
Zhai, Z., Zhang, K., Fang, Y., Yang, Y., Cao, X., Liu, L., et al. (2023). Systematically and comprehensively understanding the regulation of cotton fiber initiation: a review. Plants (Basel) 12. doi: 10.3390/plants12213771
Zhao, Y., Jing, H., Zhao, P., Chen, W., Li, X., Sang, X., et al. (2021). GhTBL34 is associated with verticillium wilt resistance in cotton. Int. J. Mol. Sci. 22. doi: 10.3390/ijms22179115
Keywords: cotton, TBL gene family, fiber quality, GhTBL84, VIGS, cold stress
Citation: Zhu X, Ma X, Hu W, Xing Y, Huang S, Chen Z and Fang L (2024) Genome-wide identification of TBL gene family and functional analysis of GhTBL84 under cold stress in cotton. Front. Plant Sci. 15:1431835. doi: 10.3389/fpls.2024.1431835
Received: 13 May 2024; Accepted: 03 June 2024;
Published: 18 June 2024.
Edited by:
Xinyang Wu, China Jiliang University, ChinaReviewed by:
Jianyong Wu, Chinese Academy of Agricultural Sciences, ChinaCopyright © 2024 Zhu, Ma, Hu, Xing, Huang, Chen and Fang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lei Fang, ZmFuZ2xAemp1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.