
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 30 July 2024
Sec. Plant Pathogen Interactions
Volume 15 - 2024 | https://doi.org/10.3389/fpls.2024.1427916
An integral part of plant immunity is transcription reprogramming by concerted action of specific transcription factors that activate or repress genes through recruitment or release of RNA polymerase II (Pol II). Pol II is assembled into Pol II holoenzyme at the promoters through association with a group of general transcription factors including transcription factor IIB (TFIIB) to activate transcription. Unlike other eukaryotic organisms, plants have a large family of TFIIB-related proteins with 15 members in Arabidopsis including several plant-specific TFIIB-related proteins (BRPs). Molecular genetic analysis has revealed important roles of some BRPs in plant reproductive processes. In this study, we report that Arabidopsis knockout mutants for BRP1, the founding member of the BRP protein family, were normal in growth and development, but were hypersusceptible to the bacterial pathogen Psuedomonas syringae. The enhanced susceptibility of the brp1 mutants was associated with reduced expression of salicylic acid (SA) biosynthetic gene ISOCHORISMATE SYNTHASE 1 (ICS1) and SA-responsive PATHOGENESIS-RELATED (PR) genes. Pathogen-induced SA accumulation was reduced in the brp1 mutants and exogenous SA rescued the brp1 mutants for resistance to the bacterial pathogen. In uninfected plants, BRP1 was primarily associated with the plastids but pathogen infection induced its accumulation in the nucleus. BRP1 acted as a transcription activator in plant cells and binded to the promoter of ICS1. These results collectively indicate that BRP1 is a functionally specialized transcription factor that increasingly accumulates in the nucleus in response to pathogen infection to promote defense gene expression.
In most eukaryotes, three multi-subunit RNA polymerases (Pol I, II and III) are responsible for the transcription of nuclear genome (Vannini and Cramer, 2012). Pol II, which transcribes genes encoding mRNAs, small nuclear RNAs (snRNAs) and microRNA (miRNAs), has been most extensively studied for understanding transcription and transcriptional regulation. Pol II requires up to seven different general transcription factors (TATA box-binding protein or TBP, transcription factor IIA or TFIIA, TFIIB, TFIID, TFIIE, TFIIF and TFIIH) for transcription initiation (Cox et al., 2012; Vannini and Cramer, 2012). These general transcription factors recognize promoter elements, recruit and assist Pol II in DNA opening and initial RNA synthesis (Archuleta et al., 2024). Pol I and III, which synthesize 25S ribosomal RNA (rRNA) and small untranslated RNAs (tRNA and 5S rRNA), respectively, also require the same or similar general transcription factors for transcription initiation (Vannini and Cramer, 2012). Thus, each of these general transcription factors often has 2 to 4 paralogs. In yeast (Saccharomyces cerevisiae), TFIIB, a Pol II general transcription factor, has two paralogs, Rrn7 and Brf-1, as general transcription factors for Pol I and III, respectively (Knutson, 2013). On the other hand, there are thousands of gene-specific transcription factors in a typical eukaryotic organism that control complex tissue- and cell-specific gene expression. Unlike general transcription factors, many gene-specific transcription factors belong to large families of many members with both shared and distinct functions (Shiu et al., 2005; Qu and Zhu, 2006; Charoensawan et al., 2010; Moore and Goldberg, 2011; Catarino et al., 2016).
In addition to the three conserved Pols, plants contain Pol IV and V, which are required for small interfering RNAs (siRNA) biogenesis, siRNA targeting and RNA-directed DNA methylation (Ream et al., 2009; Huang et al., 2015). Plants also contain multiple copies of TBP with two in Arabidopsis (Heard et al., 1993). The largest number of plant general transcription factors belong to the TFIIB-related protein (BRP) family with 15 in Arabidopsis (Knutson, 2013; Ning et al., 2021). Phylogenetic analysis indicates that the expanded TFIIB-related protein family in plants can be grouped into five distinct subfamilies, three of which correspond to the TFIIB, Rrn7, and Brf clades conserved in all eukaryotes (Knutson, 2013). There are two additional TFIIB-related protein subfamilies, named BRP1 and BRP5, in plants that are not present in other eukaryotes (Knutson, 2013). Genetic studies have shown that mutants for Arabidopsis TFIIB1 (Zhou et al., 2013b), TFIIB2 (Zhou et al., 2013b), BRP2 (Cavel et al., 2011), BRP4 (Qin et al., 2014), BRP5 (Niu et al., 2013) and Maternal Effect Embryo Arrest 12 (MEE12) (Chen et al., 2007) are defective in pollen and/or endosperm development, indicating that they are regulators of plant reproductive processes. In addition, Arabidopsis MEE65 is a Rrn7 homolog for Pol I (Burton and Burton, 2014), while Arabidopsis MEE12 is a close Brf homolog for Pol III. The embryo arrest phenotype of mee12 and mee65 could be due to defective Pol I and III transcription, respectively.
Arabidopsis BRP1 was first described more than 20 years ago as a plant-specific TFIIB-related protein (originally named pBRP) based on its conserved TFIIB structural features and the ability to bind TBP (Lagrange et al., 2003). Intriguingly, Arabidopsis BRP1 was primarily localized to the cytoplasmic surface of the plastid envelope and accumulated in the nucleus only after chemical inhibition of the proteasome activity or in the fusca 6 (fus6) mutant deficient in the CONSTITUTIVE PHOTOMORPHOGENESIS 9 (COP9) signalosome, which targets proteasome-mediated degradation of transcription factors (Lagrange et al., 2003). Thus, Arabidopsis BRP1 is subject to rapid turnover in the nucleus by proteasome-mediated protein degradation. It has been proposed that plant BRP1 is a general transcription factor for Pol I but not for Pol II or III based on the types of promoters recognized by BRP1 from red algae Cyanidioschyzon merolae and Arabidopsis (Imamura et al., 2008). Chromatin immunoprecipitation (ChIP) analysis revealed that CmpBRP1 specifically recognized the rDNA promoter region in vivo, and the occupancy was correlated to de novo 18S rRNA synthesis (Imamura et al., 2008). On the other hand, BRP1 did not recognize the Pol II-dependent promoters of five light-responsive protein-coding genes or the Pol III-dependent 5S rDNA promoter (Imamura et al., 2008). Pol I-dependent transcription from the rDNA promoter in crude cell lysate was inhibited by the CmpBRP1 antibody or when the CmpBRP1–CmTBP binding site in the rDNA promoter was mutated (Imamura et al., 2008). It was also shown that CmpBRP1 co-immunoprecipitated and co-localized with the Pol I subunit, CmRPA190, in the cell (Imamura et al., 2008).
Other studies, however, have provided strong evidence for a role of Arabidopsis BRP1 in the transcription of protein-coding genes by Pol II. Arabidopsis BRP1 interacted with Agrobacterium transcription activator Virulence E3 (VirE3) and had a strong effect on VirE3-activated expression of protein-coding genes in plants (Garcia-Rodriguez et al., 2006; Niu et al., 2015; Li et al., 2021a). One of the strongly activated genes by VirE3 encodes VirE2-interacting Protein 1(VIP1)-binding F-box Protein (VBF; At1G56250), which affected the levels of VirE2 and VIP1 (Niu et al., 2015; Li et al., 2021a). In Arabidopsis cells, co-expression of VirE3 induced accumulation of BRP1 in the nucleus and co-expression of BRP1 enhanced VirE3-stimulated transcription of VBF (Niu et al., 2015). These results indicate that VirE3 targets the transcriptional machinery of plant cells to promote plant transformation by Agrobacterium (Niu et al., 2015). More importantly, BRP1 promoted VirE3-mediated transcription of a large number of protein-coding genes in Arabidopsis (Niu et al., 2015; Li et al., 2021a). These findings strongly indicate that plant-specific BRP1 functions in the transcription of protein-coding genes by Pol II.
We have been studying the roles of important protein quality control pathways including autophagy in plant responses to both biotic and abiotic stresses. We became interested in BRP1 because proteomic profiling revealed that it was elevated in the double mutants for the selective autophagy receptor Neighbor of BRCA1 (Breast Cancer Gene 1) Gene 1 (NBR1) and the chaperone-dependent ubiquitin E3 ligase Carboxy-terminal Heat Shock Protein 70-interacting Protein (CHIP) (Zhou et al., 2013a, 2014). To analyze the biological functions of BRP1, we generated brp1 knockout mutants and found them to be normal in growth and development. However, these brp1 mutants were hypersusceptible to the bacterial pathogen Psuedomonas syringae. Thus, unlike other characterized TFIIB-related proteins with critical roles in plant growth and development, BRP1 has an important role in plant immunity. To understand the molecular basis for the critical role of BRP1 in plant immunity, we compared wild-type (WT) and brp1 mutant plants for pathogen-induced expression of salicylic acid (SA) biosynthetic gene ISOCHORISMATE SYNTHASE 1 (ICS1) and SA-responsive PATHOGENESIS-RELATED (PR) genes. We also analyzed pathogen-induced accumulation of SA and the effects of exogenous SA on the disease resistance of the brp1 mutants. We further investigated pathogen-induced nuclear accumulation and the transcription regulatory activity of BRP1, as well as the direct binding to defense-related gene promoters by BRP1 in plant cells. These results collectively indicate that BRP1 plays a critical role in plant immunity through increased nuclear accumulation upon pathogen infection to promote gene expression associated with SA-mediated defense responses.
Arabidopsis (Arabidopsis thaliana) WT and mutant plants used in the study are all in the Col-0 background. Homozygous T-DNA insertion mutants brp1-1 (WiscDsLoxHs064_04H) and sa induction deficient2 (sid2; Salk_133146) were identified by PCR using primers flanking the T-DNA insertions listed in Supplementary Table 1. Arabidopsis were grown in growth chambers or rooms at 24°C, 120 µmol m-2s-1 light on a photoperiod of 12-hour light and 12-hour dark.
A site in the second exon of the BRP1 gene, which is about 200 nucleotides from the 5’-end of its coding sequence, was selected as a target for genome editing. The target sequences (ATTGAAGGCGGTAATGAATCCGGT and AAACACCGGATTCATTACCGCCTT) were inserted into a plant CRISPR/Cas9 vector containing the Cas9 gene driven by the promoter of the YAO gene, which is preferentially expressed in the tissues undergoing cell division (Yan et al., 2015). Arabidopsis transformation was performed using the floral dipping method (Clough and Bent, 1998). For identification of mutations in the target site, the region was PCR-amplified using PCR primers flanking the target site (Supplementary Table 1) at T2 generation and directly sequenced.
Assays of Arabidopsis plant resistance to a virulent strain of Pseudomonas syringae pv tomato DC3000 (PstDC3000) were performed as previously described (Wang et al., 2014, 2015).
Total RNA was isolated from WT and mutant leaves using Trizol reagent (Invitrogen) and treated by DNase with Turbo DNA-free (Thermo Fisher Scientific) to remove contaminated DNA. cDNA was synthesized from 2.5 μg total RNA using SuperScript III reverse transcriptase (Invitrogen). Transcript levels were determined by RT-qPCR with ACTIN2 as an internal control using gene-specific primers (Supplementary Table 2) as previously described (Li et al., 2021b).
Free and total SA content was determined with a biosensor strain Acinetobacter species, ADPWH_lux, as described previously (Defraia et al., 2008). SA concentrations in the leaf samples were calculated based on the SA standard curve, which was constructed using the sid2 mutant leaf extract (Defraia et al., 2008).
The genomic sequence of BRP1 including its ~2.0 kb promoter was PCR-amplified using gene-specific primers (agcggcgcgccAGCGTTTGGGGTTTCTCACT and agcttaattaaGAAGTCTCCATGGGGATTATCAG). The amplified BRP1 genomic sequence was fused with a 4x myc epitope tag in a plant transformation vector as previously described (Wang et al., 2019). The construct was introduced into Arabidopsis plants using floral dipping method (Clough and Bent, 1998).
Chloroplasts were isolated from leaves of transgenic plants expressing myc-tagged BRP1 using a chloroplast isolation kit (Sigma-Aldrich, USA). Procedures for homogenization, removal of cell debris and leaf tissue by filtration, collection of total cell chloroplast fraction by centrifugation, and separation of intact chloroplasts using Percoll gradient were performed following the manufacturer’s protocol.
Nuclei were isolated from Arabidopsis leaves using a plant nuclei isolation kit CelLytic™ PN (Sigma-Aldrich, USA). Nuclei were first prepared from leaves with the nuclei isolation buffer provided in the kit. After mesh filtering, the cell lysate was centrifuged with 2.3 M sucrose at 12,000xg for 10 min and the nuclei pellet was collected by following the manufacturer’s protocol.
Total proteins from Arabidopsis leaves, isolated chloroplast and nuclei were isolated in a protein extraction buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM EDTA, 0.1% Triton X-100, 0.2% Nonidet P-40, 0.6 mM phenylmethylsulfonyl fluoride, 80 μM MG115, 80 μM MG132) and complete protease inhibitor cocktail tablet (Roche, USA). Protein isolation, electrophoretic separation, blotting and detection of BRP1-myc proteins using anti-myc antibodies were performed as previously described (Li et al., 2021b). Proteins isolated from chloroplast and unclei were also analyzed by protein blotting using antibodies against chloroplast-specific PsbH and nucleus-specific histone H4 proteins to assess the extent of cross-contamination. Anti-PsbH and histone H4 antibobies were obtained from Agrisera and Abcam, respectively.
Transgenic Arabidopsis plants containing a β-glucuronidase (GUS) reporter gene driven by a synthetic promoter consisting of the −100 minimal Cauliflower Mosaic Virus (CaMV) 35S promoter and eight copies of the LexA operator sequence have been described previously (Kim et al., 2006). To generate effector genes, the DNA fragment for the LexA DBD was digested from the plasmid pEG202 (Clontech) using HindIII and EcoRI and cloned into the same sites in pBluescript. The full-length BRP1 coding sequence was subsequently subcloned behind the LexA DBD to generate a translational fusion. The LexA DBD-BRP1 fusion genes were cloned into the XhoI and SpeI sites of pTA2002 behind the steroid-inducible promoter (Aoyama and Chua, 1997). As controls, the unfused LexA DBD and BRP1 genes were also cloned into the same sites of pTA7002. These effector constructs were directly transformed into the transgenic GUS reporter plants, and double transformants were identified through screening for antibiotic (hygromycin) resistance. Determination of the activation or repression of GUS reporter gene expression by the effector proteins was performed as described previously (Kim et al., 2006).
Six-week-old transgenic plants expressing myc-tagged BRP1 under its native promoter were inoculated with PstDC3000. Leaf samples were collected at 0 and 24 hours post inoculation (hpi) and processed as previously described (Birkenbihl et al., 2012). After cross-linking by vacuum infiltration of 1% formaldehyde solution, nuclei were isolated and sonicated. The sheared chromatin was incubated with anti-myc antibodies (ChIP grade; ABCAM). Immuncomplexes were collected with protein A-agarose and DNA was extracted and precipitated after reversing crosslinking. qPCR was performed using gene-specific primers (Supplementary Table 3).
To determine the role of Arabidopsis BRP1 directly, we isolated a brp1 T-DNA mutant (brp1-1) that contains a T-DNA insertion in the third exon (Figure 1A). RT-qPCR showed that the pbrp-1 mutant had little detectable full-length BRP1 transcripts (Supplementary Figure 1A). We also generated a second brp mutant (brp1-2) by targeting a site at the N-terminal domain of the BRP protein using CRIPR/cas9-mediated genome editing. The brp1-2 mutant contains a single A base insertion between nucleotides 232 and 233 of the BRP1 coding sequence (Figure 1A). This single nucleotide insertion in the brp1-2 mutant causes a reading frame shift and introduces a premature termination codon that is expected to produce a protein of 106 amino acid residues (Supplementary Figure 1B). Both brp1-1 and brp1-2 mutants grew and developed normally. In addition, the levels of rRNAs in the brp1 mutants based on the stained band intensities after electrophoretic separation of total RNA were not significantly reduced as compared to those in WT (data not shown).
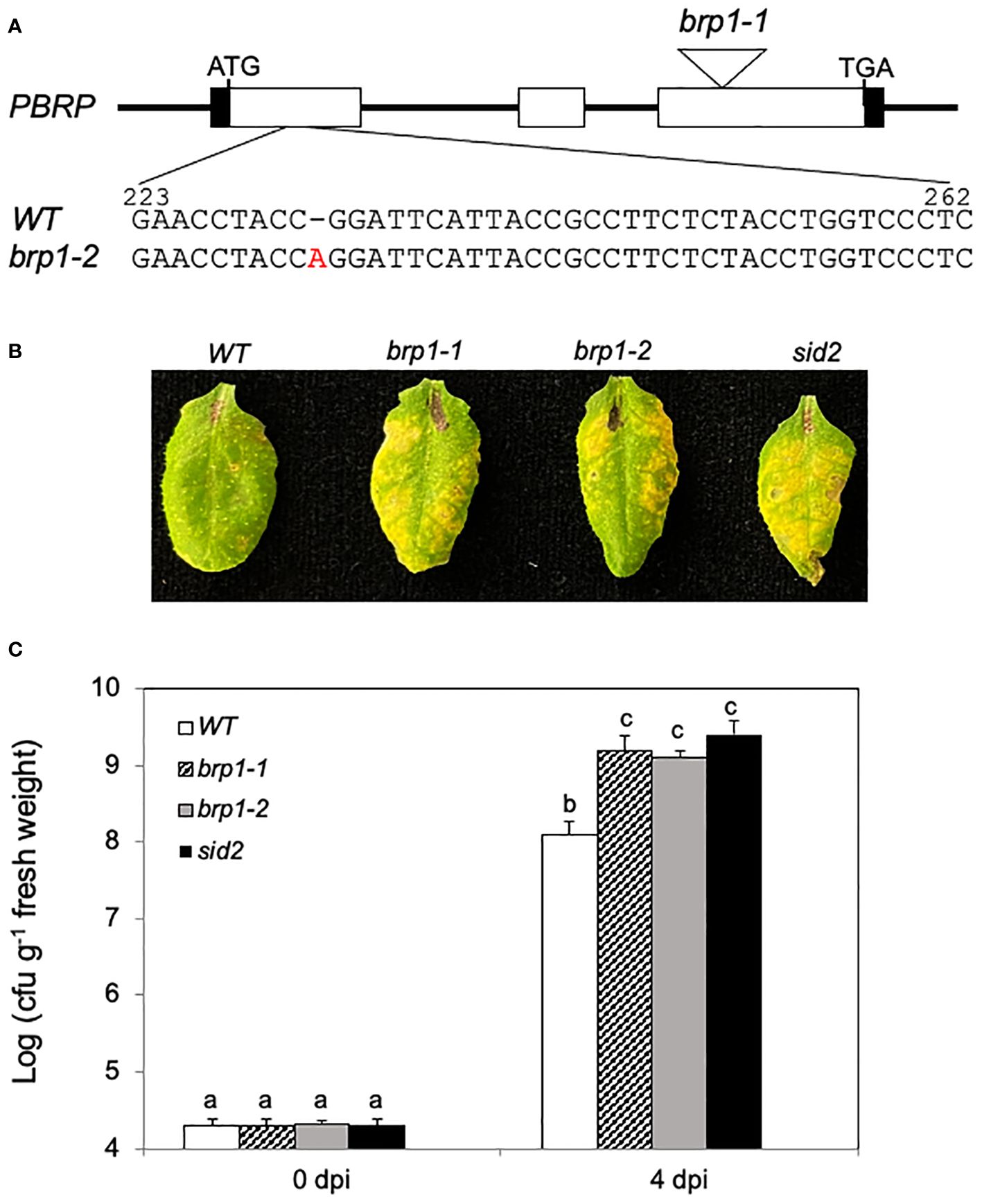
Figure 1 Compromised disease resistance of brp1 mutants to PstDC3000. (A) Arabidopsis BRP1 gene structure and mutants. The brp1-1 T-DNA mutant contains a T-DNA insertion in the third exon of BRP1. The brp1-2 mutant contains a single A base insertion between nucleotides 232 and 233 of the BRP1 coding sequence. (B) Disease symptom development after infection by the virulent PstDC3000. Leaves of 6 weeks old Arabidopsis Col-0 WT, brp1 and sid2 mutant plants were infiltrated with PstDC3000 (OD600 = 0.0002 in 10 mM MgCl2). Pictures of representative leaves were taken at 4 dpi. (C) Leaf samples were taken at 0 or 4 dpi to determine the bacterial growth. The means and standard errors were calculated from 10 plants (n=10) for each genotype. According to Duncan’s multiple range test (P=0.01), means of the values do not differ if they are indicated with the same letter. The experiment was repeated three times with similar results.
To analyze the response of the brp1 mutants to pathogen infection, we compared them with Col-0 WT and a SA-deficient sid2 mutant for response to the virulent bacterial pathogen PstDC3000. As shown in Figure 1B, at 4 days post inoculation (dpi), WT plants developed very mild symptoms of chlorosis. On the other hand, the two brp1 mutants developed more severe disease symptoms at 4 dpi than WT plants (Figure 1B). The enhanced disease symptoms in the brp1 mutants after PstDC3000 infection were similar to those in the sid2 mutant (Figure 1B), which accumulated greatly reduced levels of SA (Wildermuth et al., 2001; Garcion et al., 2008). The levels of bacterial growth in the brp1 and sid2 mutants were also 10 to 20 times higher than those in the WT plants (Figure 1C). Thus, the brp1 mutants were as susceptible to the bacterial pathogen as SA-deficient sid2 mutant plants (Figure 1).
SA-mediated defense signaling is important for resistance to P. syringae in Arabidopsis (Glazebrook, 2005). To analyze the molecular basis for enhanced susceptibility of the brp1 mutants to the bacterial pathogen, we compared WT, brp1 and sid2 mutants for the expression of SID2, which codes for SA biosynthetic enzyme ICS1 (Wildermuth et al., 2001), before and after PstDC3000 infection. At 0 dpi, no significant difference was observed in the transcript levels of SID2 between WT and the two brp1 mutants (Figure 2). After inoculation with PstDC3000, the levels of SID2 transcripts were increased by more than 6-fold in WT but only about 1.5-fold in the brp1 mutants by 1 dpi (Figure 2). Thus, pathogen-induced SID2 expression was compromised in the brp1 mutants. As expected, little expression of SID2 was detected either prior to or after PstDC3000 infection in the sid2 mutant due to disruption of the analyzed gene by a T-DNA insertion (Figure 2).
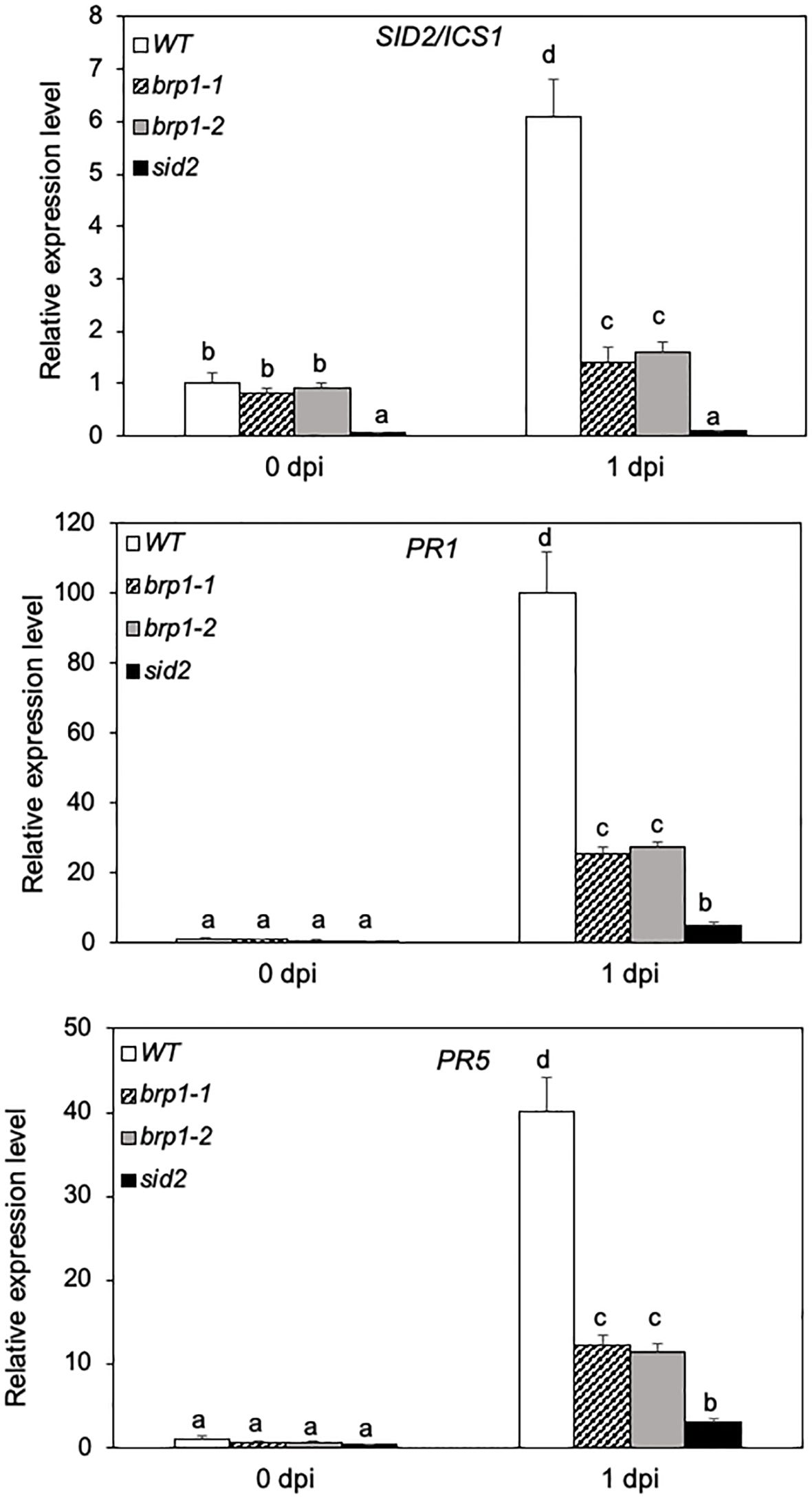
Figure 2 Reduced defense gene expression in pathogen-infected brp1 mutants. Leaves of 6 weeks-old Arabidopsis Col-0 WT, brp1 and sid2 mutant plants were infiltrated with PstDC3000 (OD600 = 0.0002 in 10 mM MgCl2). Total RNA was isolated from leaf samples collected at indicated dpi. Transcript levels of ICS1/SID2, PR1 and PR5 were determined using RT-qPCR. Error bars indicate SE (n = 3). According to Duncan’s multiple range test (P=0.01), means of the values do not differ if they are indicated with the same letter. The experiment was repeated twice with similar results.
We also compared the expression of SA-regulated PR genes in WT, brp1 and sid2 mutants. In WT, PR1 and PR5 transcripts were elevated by more than 100- and 40-fold, respectively, during the first dpi (Figure 2). In the brp1 mutants, however, there was only about 25- and 10-fold increase in the PR1 and PR5 transcripts, respectively, during the first dpi (Figure 2). Thus, pathogen-induced PR gene expression was also compromised in the brp1 mutants. The levels of transcripts for pathogen-induced PR1 and PR5 genes were even further reduced in the sid2 mutant when compared to those in WT and brp1 mutants (Figure 2).
The compromised phenotypes of the brp1 mutants in disease resistance (Figure 1) was correlated with reduced expression of SA biosynthetic gene ICS1/SID2 and SA-regulated PR gene expression (Figure 2). This correlation suggests that the brp1 mutants may be defective in SA production. Therefore, we compared the free SA and conjugated SA-glucoside (SAG) levels in WT, brp1 and sid2 mutants before and after PstDC3000 infection. At 0 dpi, basal levels of free SA were similar in WT and brp1 mutants (Figure 3A). At 1 dpi, free SA increased by more than 12-fold in WT, but only 4- to 5-fold in the brp1 mutants (Figure 3A). The levels of total SA (free SA and SAG) were already about 4 times lower in the brp1 mutants than in WT at 0 dpi (Figure 4B). At 1 dpi, the levels of total SA in WT were about 6 times higher in WT than in the brp1 mutants (Figure 3B). As previously reported, basal and pathogen-induced free SA and total SA levels in the sid2 mutant were only about 5 to 10% of those in WT (Figure 3).
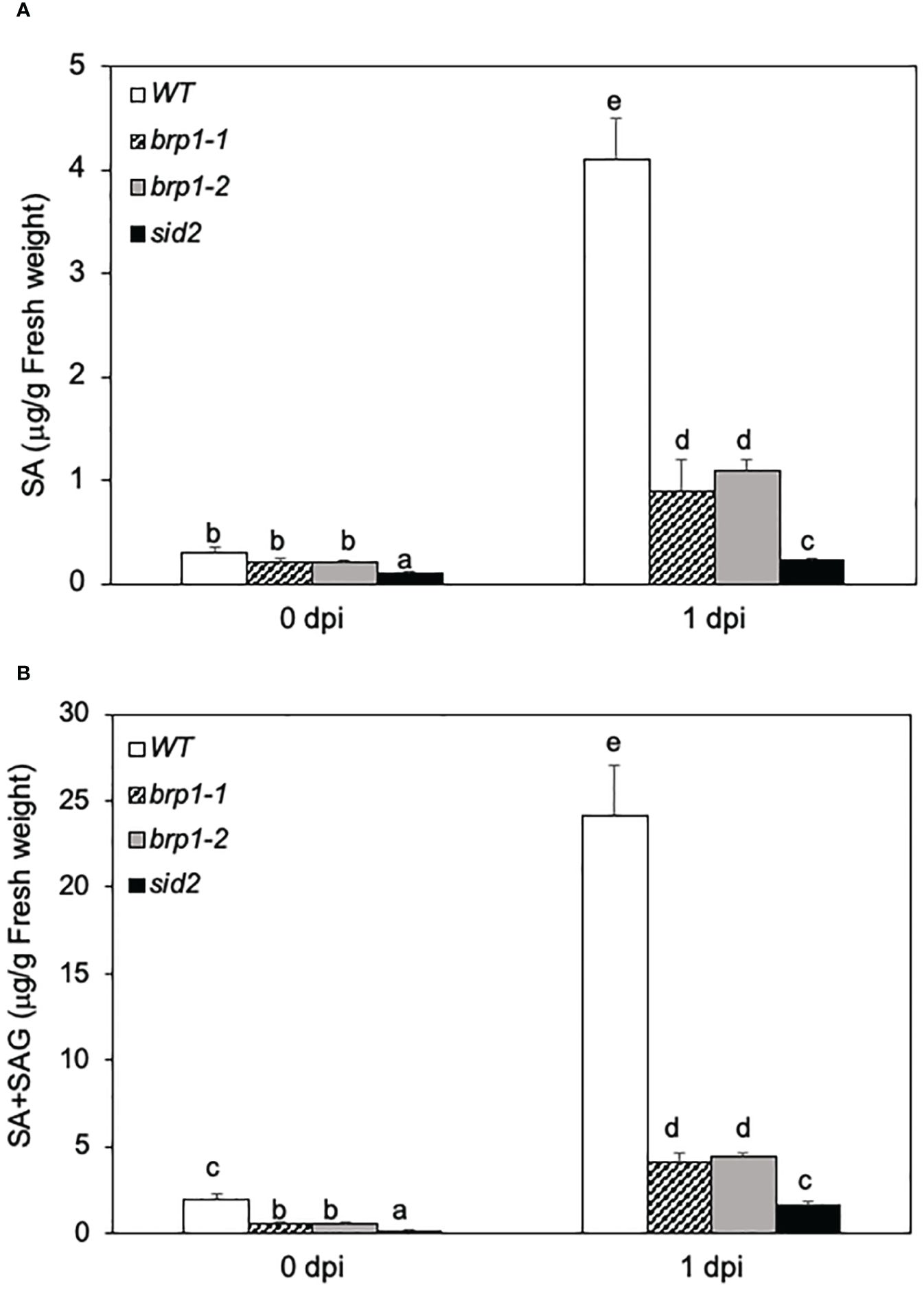
Figure 3 Reduced SA levels in the brp1 mutants.Leaves of 6 weeks-old Arabidopsis Col-0 WT, brp1 and sid2 mutant plants were infiltrated with PstDC3000 (OD600 = 0.0002 in 10 mM MgCl2). Inoculated leaves were sampled at indicated dpi and determined for both free SA (A) and free SA plus SAG (B) content using a bacterial SA biosensor. Error bars indicate SE (n = 5). According to Duncan’s multiple range test (P=0.01), means of the values do not differ if they are indicated with the same letter.
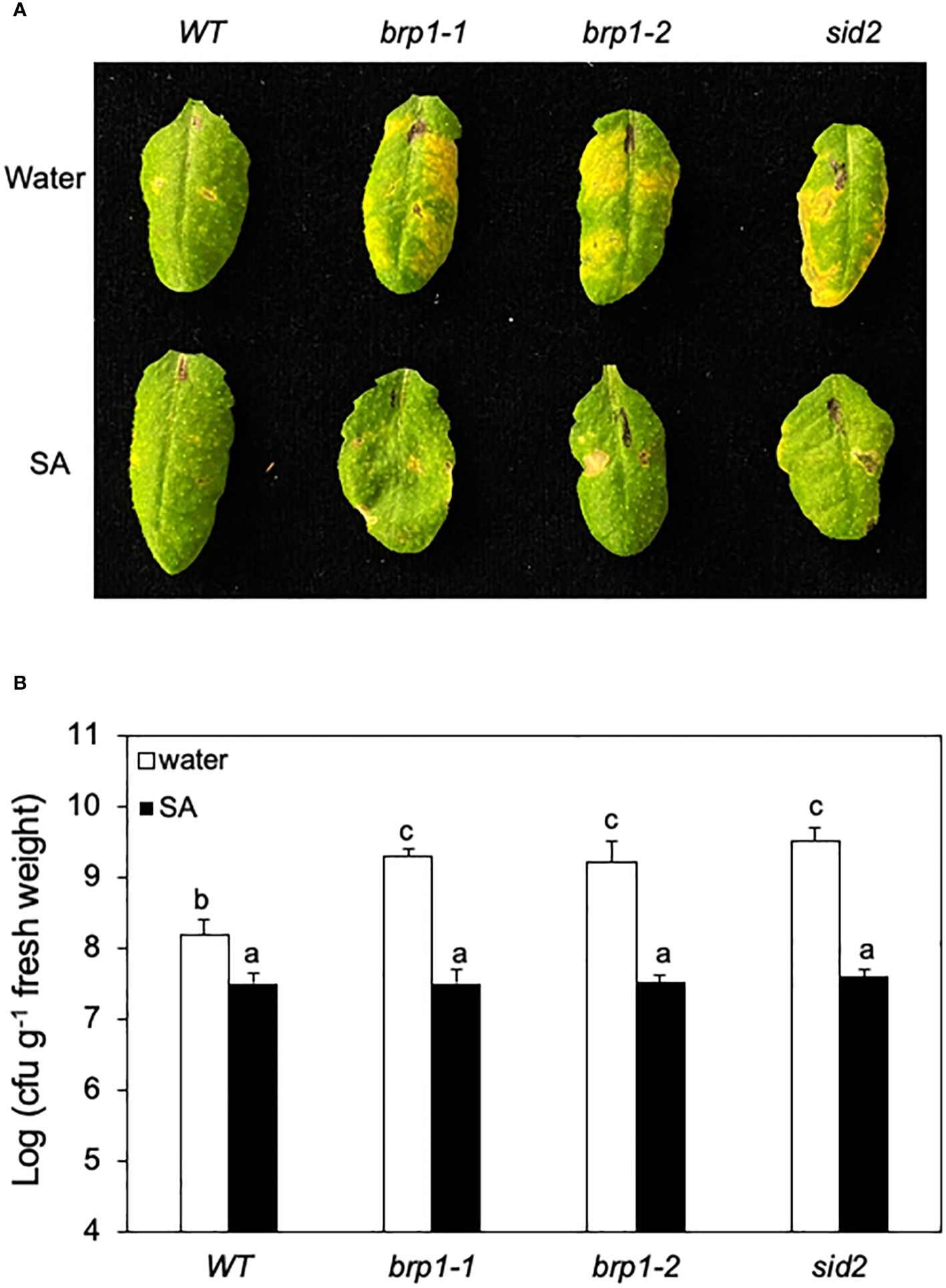
Figure 4 Rescue of brp1 by SA in disease resistance. (A) Six-week-old WT, brp1 and sid2 mutants were sprayed with water or SA (1 mM). The plants were infiltrated one day later with a suspension of PstDC3000 (OD600 = 0.0002 in 10 mM MgCl2). Images are of representative inoculated leaves taken at 4 dpi. (B) Effect on bacterial growth. Six-week-old WT and mutants were sprayed with water or SA (1 mM). Pathogen inoculation of WT and mutant plants was performed 1 day later. Samples were taken at 4 dpi to determine the growth of the bacterial pathogen. The means and standard errors were calculated from 6 plants for each treatment. According to Duncan’s multiple range test (P=0.01), means of the values do not differ if they are indicated with the same letter. These experiments were repeated three times with similar results.
To determine whether reduced SA accumulation caused disease susceptibility in the brp1 mutants, we tested whether exogenous SA could restore their disease resistance. We first sprayed WT, brp1 and sid2 mutant plants with water or SA, and one day later inoculated the sprayed plants with PstDC3000. After spraying with water, both the brp1 and sid2 mutants developed more severe disease symptoms (Figure 4A) and supported higher bacterial growth than WT plants did (Figure 4B). After SA treatment, however, both the brp1 and sid2 mutant had levels of disease symptom development (Figure 4A) and bacterial growth similar to those of SA-treated WT plants (Figure 4B). These results support that the compromised phenotypes of the brp1 mutants in disease resistance are caused by reduced SA accumulation after pathogen infection.
It has been previously shown that BRP1 is primarily localized to the plastid envelope and its accumulation in the nucleus was detected only after proteasome inhibition (Lagrange et al., 2003). Other published studies reported that co-expression of BRP1-interacting VirE3 transcription activator from Agrobacterium promoted nuclear accumulation of BRP1 (Garcia-Rodriguez et al., 2006; Niu et al., 2015). Given the critical role of BRP1 in plant responses to PstDC3000, we analyzed the effect of pathogen infection on its nuclear accumulation. Initially, we generated a BRP1-GFP fusion construct under control of its native promoter and attempted to use confocal microscopy to examine its accumulation and subcellular localization in response to pathogen infection. However, we observed only very low levels of fluorescent signals in the nucleus that were difficult for quantification. Therefore, we generated a 4xmyc-tagged BRP1 gene under control of its native promoter and investigated the change in both the levels and subcellular localization using subcellular fractionation and protein blotting. The construct was first transformed into the brp1-2 mutant and was found to fully complement the mutant for resistance to PstDC3000 and, therefore, is fully functional (Supplementary Figure 2). Protein blotting using an anti-myc antibody detected very low levels of PBRP-myc in plants at 0 hpi (Figure 5). At 12 and 24 hpi, increased levels of BRP1-myc were detected in the inoculated plants (Figure 5). RT-qPCR showed that this increase in BRP1-myc proteins was not associated with significant increase in BRP1 gene transcripts, indicating that pathogen-induced BRP1 protein accumulation involves a post-translational mechanism. We also analyzed the changes of BRP1 protein levels associated with isolated chloroplasts and nuclei from PstDC3000-infected Arabidopsis leaves. Protein blotting of isolated chloroplast and nuclear proteins using antibodies against chloroplast-specific PsbH and nuclear histone H4 proteins revealed little protein cross-contamination in isolated chloroplast and nuclear fractions (Supplementary Figure 3). As shown in Figure 5, myc-tagged BRP1 in the chloroplast fraction was detected at 0 hpi but significantly reduced at 12 and 24 hpi. By contrast, nuclear BRP1 was barely detectable at 0 hpi but substantially increased at 12 and 24 hpi (Figure 5). Thus, pathogen infection increased both the total protein level and nuclear accumulation of BRP1 in plant cells.
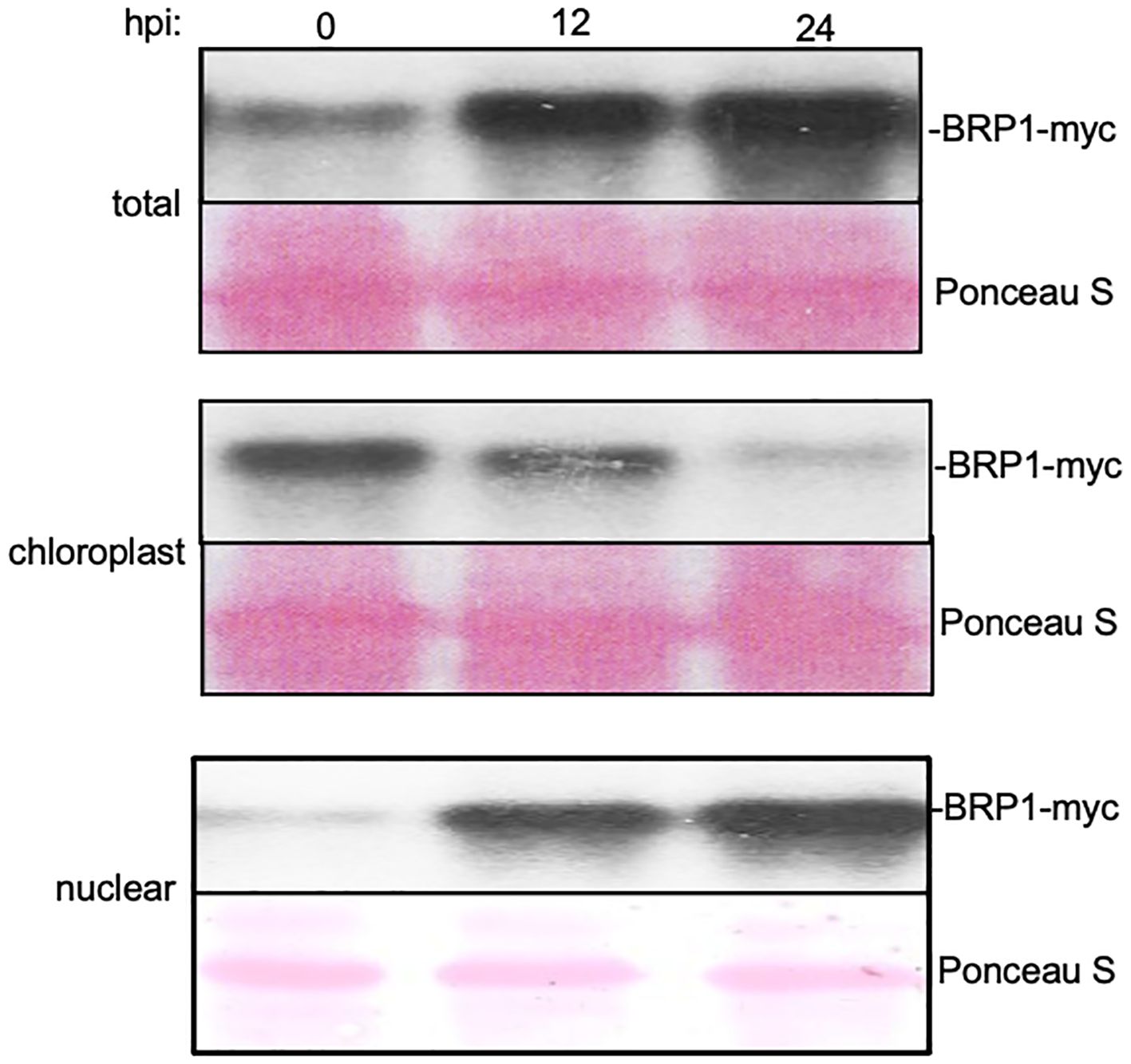
Figure 5 Pathogen-induced nuclear accumulation of BRP1 proteins. Transgenic brp1-2 mutant plants harboring a genomic BRP1-myc gene was inoculated with PstDC3000. Inoculated leaves were sampled at indicated hpi for isolation of chloroplasts and nuclei. BRP1-myc proteins in total (top), chloroplast (middle) and nuclear (bottom) protein extracts were determined by protein blotting using an anti-myc antibody. Ponceau S-stained blots are shown for loading controls. The experiment was repeated twice with similar results.
It has been proposed that BRP1 is a general transcription factor for Pol I but not for Pol II primarily based on promoter binding assays of BRP1 from red algae Cyanidioschyzon merolae and Arabidopsis (Imamura et al., 2008). However, these reported promoter binding ChIP assays only tested promoters of five light-responsive protein-coding genes as Pol II-dependent promoters, in addition to the promoters of rDNA and 5S rDNA as Pol I- and Pol III-dependent promoters, respectively (Imamura et al., 2008). On the other hand, BRP1 had strong effects on VirE3-depednent expression of plant protein-coding genes (Garcia-Rodriguez et al., 2006; Niu et al., 2015). Furthermore, the normal growth, development and accumulation of rRNAs but compromised phenotypes in disease resistance (Figure 1) and defense gene expression (Figure 2) in the brp1 mutants argue against BRP1 as a critical general transcription factor for Pol I.
To determine the role of BRP1 in transcription of protein-coding genes by Pol II in plant cells, we used a previously developed reporter-effector system to analyze the transcriptional regulatory activity of BRP1 through assays of its effects on the protein-coding GUS reporter gene in stably transformed plants. In this system, a synthetic promoter consisting of the -100 minimal CaMV 35S promoter sequence and eight copies of the LexA operator sequence was fused with the GUS reporter gene, subcloned into a plant transformation vector, and transformed into Arabidopsis plants (Kim et al., 2006) (Figure 6A). These transgenic plants contained low levels of expression of the GUS reporter gene due to the minimal CaMV 35S promoter, thereby making them possible for assays of transcription activation or repression by determining increase or decrease in GUS activities following coexpression of an effector protein (Kim et al., 2006). To generate the BRP1 effector, we fused its coding sequence with that of the DBD of LexA, subcloned the fusion effector behind the steroid-inducible Gal4 promoter in pTA7002 (Aoyama and Chua, 1997) and transformed into the transgenic GUS reporter lines (Figure 6A). Unfused BRP1 and LexADBD genes were also subcloned into pTA7002 and transformed into transgenic GUS reporter lines as controls (Figure 6A). Transgenic plants containing both the reporter and effector constructs were identified through antibiotic resistance screens and the effects of the effectors on the GUS reporter gene expression were determined by assays of the changes of GUS activities following DEX-induced effector gene expression. In the transgenic plants that expressed unfused BRP1 or LexA DBD effector, there was little change in the GUS activities after DEX treatment (Figure 6B). These results indicated that induced expression of BRP1 or LexA DBD alone had no significant effect on expression of the GUS reporter gene. In the transgenic plants harboring the LexA DBD-BRP1 effector gene, induction of the fusion effector gene after DEX treatment resulted in approximately 5-fold reduction in GUS activity (Figure 6B). These results strongly suggest that BRP1 is a transcriptional activator in plant cells.
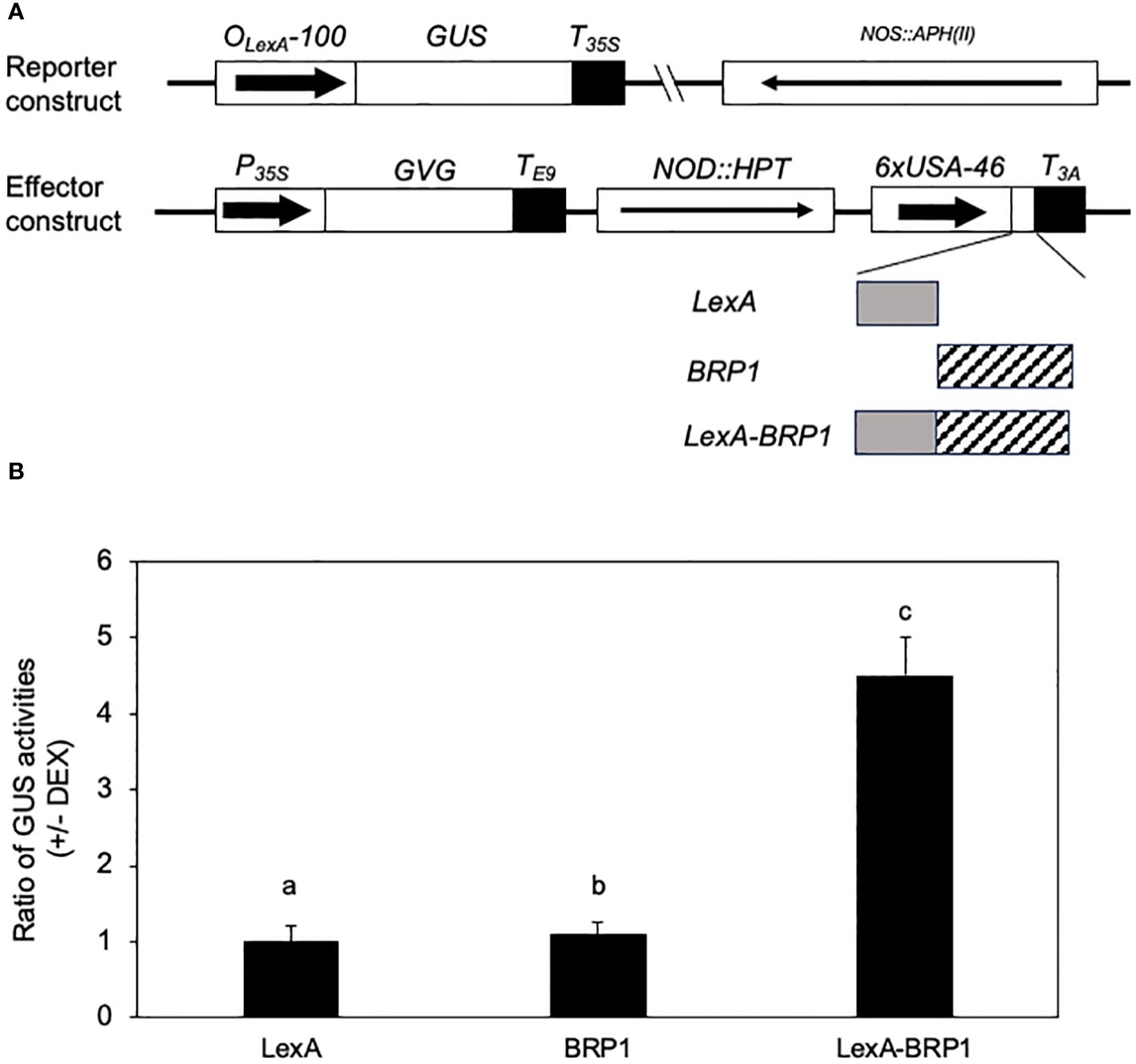
Figure 6 Transcription-activating activity of BRP1 in plant cells. (A) Constructs of reporter and effector genes. The GUS reporter gene is driven by a synthetic promoter consisting of the -100 minimal CaMV 35S promoter and eight copies of the LexA operator sequence. The effector genes were clone into pTA7002 behind the steroid-inducible promoter. The three effector genes encode LexADBA-BRP1 fusion protein (LexA-BRP1), LexA DBD (LexA), and BRP1, respectively. (B) Effects on the GUS reporter gene expression by induced expression of effector genes. The ratios of GUS activities were calculated from the GUS activities in the leaves harvested prior to DEX treatment over those determined in the leaves harvested 18 hours after DEX treatment. The means and errors were calculated from at least 10 positive transformants. According to Duncan’s multiple range test (P=0.01), means of the values do not differ if they are indicated with the same letter. The experiment was repeated twice with similar results.
As a TFIIB-related protein, BRP1 does not necessarily bind DNA directly but could recognize the core promoter elements of its target genes such as TATA boxes through associated TBPs as a general transcription factor of Pol II. Since BRP1 positively regulates pathogen-induced expression of ICS1, PR1 and PR5, we performed ChIP using the myc-tagged BRP1 complementation lines in combination with qPCR to determine direct BRP1 binding to the core promoter regions of these defense-related genes. By using primer sets representing the core promoter regions (~100 nucleotides upstream of the putative transcript start sites) of the defense-related genes (Supplementary Table 3), we detected significant pathogen-dependent binding of BRP1 to the promoter of ICS1 (Figure 7). We also observed weak but significant binding of BRP1 to the core promoter region of PR1 (Figure 7). On the other hand, no significant binding of BRP1 to the core promoter region of PR5 was detected (Figure 7). These results indicated that BRP1 directly regulated expression of specific defense-related genes during plant defense responses.
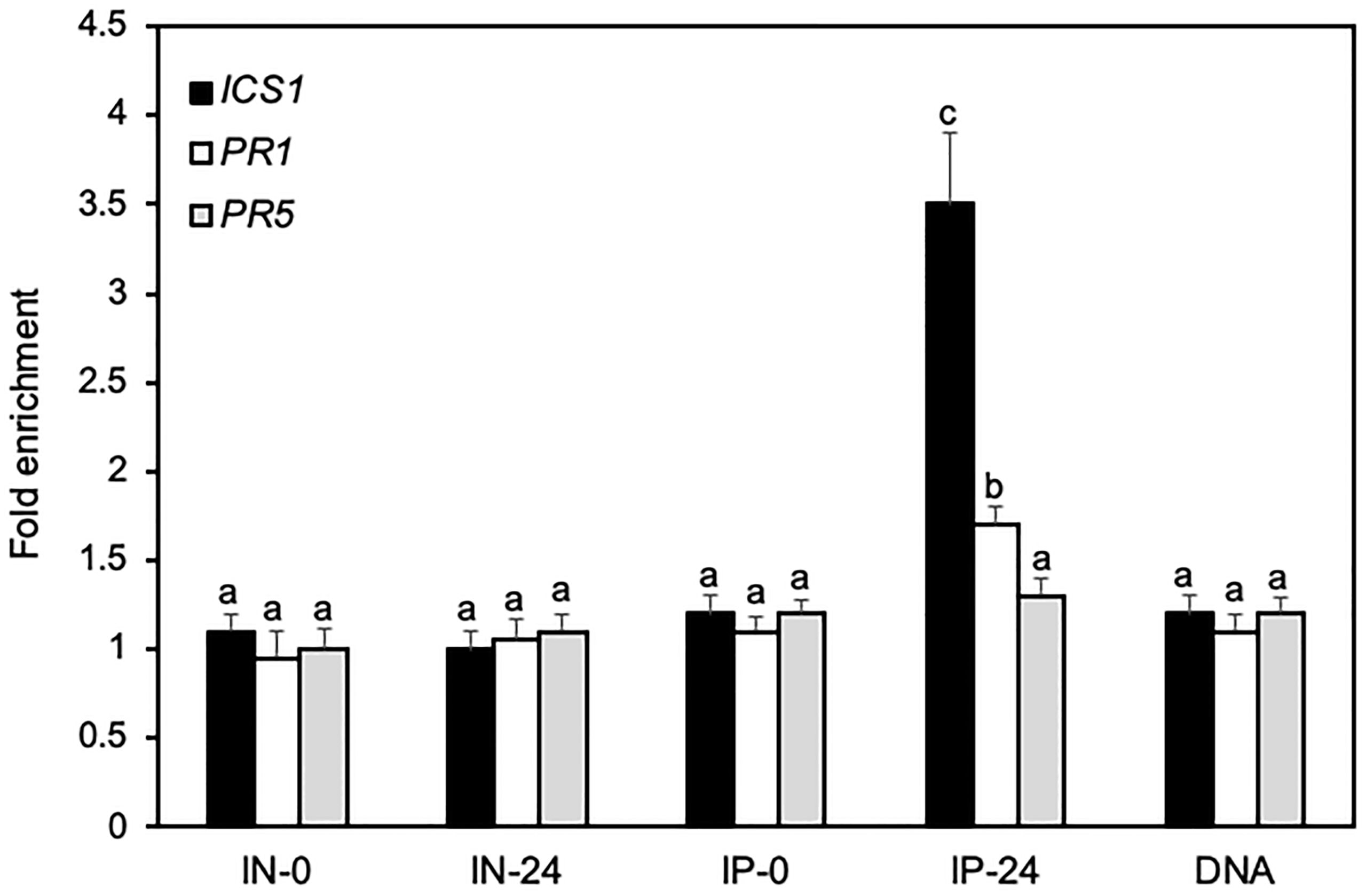
Figure 7 ChIP assays of direct binding of BRP1 to the core promoter elements of defense related genes. Transgenic myc-tagged complementation plants were inoculated with PstDC3000 and inoculated leaves were collected at 0 and 24 hpi and processed for ChIP assays. Input DNA before immunoprecipitation from plant leaves collected at 0 (IN-0) and 24 hpi (IN-24) and coimmunoprecipitated DNA using an anti-myc antibody (IP-0 and IP-24) were analyzed by qPCR using primers specific for the core promoter elements of ICS1, PR1 and PR5. The data are expressed as fold enrichment relative to a DNA fragment from RHIP (At4G26410) as a reference gene. Purified genomic DNA (DNA) was also included in the analysis for primer efficiency control. Error bars indicate SE (n = 3). According to Duncan’s multiple range test (P=0.01), means of the values do not differ if they are indicated with the same letter. The experiment was repeated twice with similar results.
There are 15 genes encoding TFIIB-like factors in the Arabidopsis genome (Knutson, 2013; Ning et al., 2021). Two of them (BRF4CTD and MEE12CTD) encode proteins that lack the conserved N-terminal zinc finger and the two cyclin fold repeats of TFIIB and are unlikely to act as TFIIB-like factors (Ning et al., 2021). Of the remaining 13 Arabidopsis TFIIB-like factors, at least 10 have been characterized through molecular genetic approaches and found to play important roles in gametogenesis, pollen tube growth guidance, embryogenesis, and endosperm development (Ning et al., 2021). Thus, the expansion and functional diversification of the TFIIB-related proteins in plants may contribute to the evolution of novel functions associated with plant-specific sexual reproductive processes. As sessile organisms, plants are also constantly exposed to a wide spectrum of biotic and abiotic stress conditions and have also evolved many unique stress- and defense-response mechanisms. Previously, Arabidopsis BRP1 has been implicated in the interaction between plants and Agrobacterium (Garcia-Rodriguez et al., 2006; Niu et al., 2015; Li et al., 2021a). In the present study, we have provided direct genetic evidence that the plant-specific TFIIB-related protein plays a critical role in plant defense responses.
Using T-DNA insertion and CRISPR/cas9 genome editing, we have generated two independent knockout mutants for Arabidopsis BRP1 (Figure 1A). Unlike knockout mutants for other characterized Arabidopsis TFIIB-related proteins, which are either lethal or severely compromised in important reproductive processes, the brp1 mutants displayed no significant phenotypes in growth and development. However, the brp1 mutants are highly susceptible to the bacterial pathogen P. syringae based on both enhanced symptoms and increased pathogen growth (Figure 1). Further analysis indicated that the hyper-susceptibility of the brp1 mutants to the bacterial pathogen was caused by compromised SA accumulation in association with reduced expression of SA biosynthetic gene SID2/ICS1 (Figures 2, 3). ChIP assays further indicated that BRP1 directly regulates pathogen-induced expression of SID2ICS1 (Figure 7). SA is an important defense signal with a critical role in both basal disease resistance and systemic acquired resistance mostly through transcriptional regulation of transcription program of plant defense genes (Seyfferth and Tsuda, 2014; Yan and Dong, 2014). Indeed, compromised disease resistance of the brp1 mutants was also correlated with defects in SA-regulated PR1 and PR5 gene expression (Figure 2). Thus, the plant-specific TFIIB-related protein has an important and specific role in plant immunity by promoting SA-mediated defense responses.
Previously, it has been claimed that plant-specific BRP1 functions as a general transcription factor for Pol I, but not for Pol II or III (Imamura et al., 2008). This role of plant-specific BRP1 was primarily based on the binding of BRP1 to the Pol I-dependent rDNA promoters both in vitro and in vivo (Imamura et al., 2008). The reported study argued against a role of BRP1 in Pol II-dependent transcription because it failed to detect its binding to the promoters of five light-responsive protein-coding genes as Pol II-dependent promoters (Imamura et al., 2008). If BRP1 functions as a critical general transcription factor for Pol I responsible for transcription of rDNA, which accounts for over 50% of total cellular RNA, we expect that the Arabidopsis brp1 knockout mutants would display severe or even lethal phenotypes with greatly reduced levels of rRNAs. The lack of significant phenotypes in growth and development as well as the normal accumulation of rRNAs in the Arabidopsis brp1 knockout mutants strongly argues against a critical role of the plant-specific TFIIB-related protein in Pol I-dependent transcription. It is possible that BRP1 is involved in transcription by Pol I and the lack of effects from the loss of BRP1 on rRNA accumulation is resulted from the presence of additional TFIIB-related proteins as general transcription factor of Pol I.
On the other hand, Arabidopsis brp1 mutants were compromised in plant immunity due to defects in expression of genes involved in SA biosynthesis and other defense-related processes (Figures 1, 2). Furthermore, BRP1 is associated with the core promoter elements of ICS1 and, to a less extent, PR1 (Figure 7). Previously, it has been shown that plant BRP1 interacts with the virE3 transcription factor from Agrobacterium and promotes transcription of a large number of virE3-activated host protein-coding genes to promote Agrobacterium-mediated transformation (Garcia-Rodriguez et al., 2006; Niu et al., 2015; Li et al., 2021a). In addition, we have shown that BRP1 directly activated transcription of the protein-coding GUS reporter gene in plant cells (Figure 6). Therefore, plant-specific BRP1 functions as a critical transcription activator of plant genes involved in plant-microbe interactions and is targeted by plant pathogens such as Agrobacterium during the infection process to modulate plant host gene expression.
The critical role of BRP1 in plant-microbe interaction is consistent with the highly regulated nature of its subcellular localization. As previously reported (Lagrange et al., 2003), Arabidopsis BRP1 is primarily associated with chloroplasts in the absence of pathogen infection (Figure 5). Increased nuclear accumulation of BRP1 has been observed in plant cells after inhibition of proteasome or in COP9 mutants (Lagrange et al., 2003). Under normal growth condition, its nuclear accumulation is very limited probably through inhibition of nuclear translocation and the degradation of nuclear BRP1. The limited nuclear accumulation of BRP1 may be necessary to prevent unnecessary activation of defense-related genes. After infection by PstDC3000 infection, however, there was increased accumulation of BRP1 in the nucleus, concomitant with its decreased association in chloroplasts (Figure 5). Increased nuclear accumulation of BRP1 has also been observed in virE3-coexpressed cells (Garcia-Rodriguez et al., 2006). Bioinformatics analysis finds no nuclear localization signal in the protein, suggesting that the nuclear localization of BRP1 may be mediated by a piggyback mechanism through interaction with a nuclear protein such as Agrobacterium virE3, effector proteins from P. syringae and other plant host proteins. Further investigation of the mechanisms by which the subcellular localization of BRP1 is dynamically regulated under both normal and stress conditions could provide important new insights into the complex network of molecular events that balance plant growth with plant stress/defense responses.
Establishment of a TFIIB-related general transcription factor such as BRP1 in specific biological processes is highly significant as the findings challenge the paradigm of general transcription factors as universal regulators of class-specific gene expression. Structural and evolutionary analysis has shown that bacterial σ factors, archaeal transcription factor B (TFB) and eukaryotic TFIIB are homologs (Burton and Burton, 2014). Bacteria often contain a primary σ factor and many alternative σ factors for regulation of discrete sets of genes (Burton and Burton, 2014; Feklistov et al., 2014). Archaea also have multiple TFB factors that potentially mediate environmental responses, which may explain their extraordinary niche adaptation capability. In Halobacterium salinarum, a halophilic (salt-loving) member of the Archaea that grows in concentrations of NaCl near or at saturation, there are at least seven TFBs that direct environment-specific gene expression programs (Turkarslan et al., 2011). In eukaryotes, the functions of general transcription factors have been analyzed almost exclusively in the context of basal transcription and their possible roles in the regulation of physiology may have been under-appreciated. In yeast, ethanol production could be enhanced through the mutagenesis of TFIIB, suggesting that altering the function of a general transcription factor can have significant phenotypic consequences (D'Alessio et al., 2009). Furthermore, several studies have discovered regulatory roles of general transcription factors in cell-specific differentiation and development in eukaryotes (Lagrange et al., 2003; Juven-Gershon and Kadonaga, 2010). The large number of TFIIB-related proteins and their distinct roles in a broad spectrum of plant sexual reproduction and disease resistance present a new paradigm for the transcription and transcriptional regulation of genes, which can provide novel insights into the transcriptional programs that govern plant growth, development and responses to both biotic and abiotic stresses.
The identifiers for the Arabidopsis genes described in this article are as follows: BRP1 (AT4G36650), ICS1/SID2 (AT1G74710), PR1 (AT2G14610), PR5 (AT1G75040), Actin2 (AT3G18780), PsbH (ATCG00710), Histine H4 (AT2G28740 and RHIP (AT4G26410).
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
BX: Writing – review & editing, Data curation, Investigation. BF: Investigation, Writing – review & editing. ZC: Conceptualization, Funding acquisition, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by US National Science Foundation (grants no. IOS1758767 and MCB2241515).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1427916/full#supplementary-material
Supplementary Figure 1 | Characterization of Arabidopsis brp1 mutants. (A) Expression of BRP1 in WT and brp1-1 mutant. Total RNA was isolated from leaf samples collected from six-week-old plants. Transcript levels of BRP1 were determined using RT-qPCR. Error bars indicate SE (n = 3). A Student’s t-test was used for statistical analysis of the BRP1 transcript levels in WT versus in brp1-1 mutant (***indicates p-value < 0.001). (B) Predicted effect of the brp1-2 mutation on the translated product of BRP1. A single A base insertion between nucleotides 232 and 233 of the BRP1 coding sequence would cause a reading frame shift after amino acid residue 77 and introduce a premature termination codon after addition of 29 wrong amino acid residues (in red).
Supplementary Figure 2 | Complementation of brp1-2 by a myc-tagged BRP1 gene. (A) Six-week-old WT, brp1-2 mutant and two independent lines (L1 and L2) of brp1-2 mutant expressing a myc-tagged BRP1 gene under its native promoter were infiltrated with a suspension of PstDC3000 (OD600 = 0.0002 in 10 mM MgCl2). Images are of representative inoculated leaves taken at 4 dpi. (B) Effect on bacterial growth. Six-week-old WT, brp1-2 mutant and two independent lines (L1 and L2) of brp1-2 mutant expressing a myc-tagged BRP1 gene under its native promoter were infiltrated with a suspension of PstDC3000 (OD600 = 0.0002 in 10 mM MgCl2). Samples were taken at 4 dpi to determine the growth of the bacterial pathogen. The means and standard errors were calculated from 6 plants for each treatment. According to Duncan’s multiple range test (P=0.01), means of the values do not differ if they are indicated with the same letter. These experiments were repeated twice times with similar results.
Supplementary Figure 3 | Protein blotting for detection of potential cross-contamination of isolated chloroplast and nuclear fractions. Transgenic brp1-2 mutant plants harboring a genomic BRP1-myc gene was inoculated with PstDC3000. Inoculated leaves were sampled at indicated hpi for isolation of chloroplasts and nuclei. The same amount of proteins from each chloroplast and nuclear fraction was fractionated by electrophoresis and analyzed by protein blotting using an anti-PsbH or anti-histone H4 (H4) antibody. The experiment was repeated twice with similar results.
Aoyama, T., Chua, N. H. (1997). A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 11, 605–612. doi: 10.1046/j.1365-313X.1997.11030605.x
Archuleta, S. R., Goodrich, J. A., Kugel, J. F. (2024). Mechanisms and functions of the RNA polymerase II general transcription machinery during the transcription cycle. Biomolecules 14, 176. doi: 10.3390/biom14020176
Birkenbihl, R. P., Diezel, C., Somssich, I. E. (2012). Arabidopsis WRKY33 is a key transcriptional regulator of hormonal and metabolic responses toward infection. Plant Physiol. 159, 266–285. doi: 10.1104/pp.111.192641
Burton, S. P., Burton, Z. F. (2014). The sigma enigma: bacterial sigma factors, archaeal TFB and eukaryotic TFIIB are homologs. Transcription 5, e967599. doi: 10.4161/21541264.2014.967599
Catarino, B., Hetherington, A. J., Emms, D. M., Kelly, S., Dolan, L. (2016). The stepwise increase in the number of transcription factor families in the precambrian predated the diversification of plants on land. Mol. Biol. Evol. 33, 2815–2819. doi: 10.1093/molbev/msw155
Cavel, E., Pillot, M., Pontier, D., Lahmy, S., Bies-Etheve, N., Vega, D., et al. (2011). A plant-specific transcription factor IIB-related protein, pBRP2, is involved in endosperm growth control. PloS One 6, e17216. doi: 10.1371/journal.pone.0017216
Charoensawan, V., Wilson, D., Teichmann, S. A. (2010). Lineage-specific expansion of DNA-binding transcription factor families. Trends Genet. 26, 388–393. doi: 10.1016/j.tig.2010.06.004
Chen, Y. H., Li, H. J., Shi, D. Q., Yuan, L., Liu, J., Sreenivasan, R., et al. (2007). The central cell plays a critical role in pollen tube guidance in Arabidopsis. Plant Cell 19, 3563–3577. doi: 10.1105/tpc.107.053967
Clough, S. J., Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Cox, M. M., Doudna, J., O'donnell, M. (2012). Molecular biology: principle and practice (New York City, New York, USA: Freenman and Company).
D'Alessio, J. A., Wright, K. J., Tjian, R. (2009). Shifting players and paradigms in cell-specific transcription. Mol. Cell 36, 924–931. doi: 10.1016/j.molcel.2009.12.011
Defraia, C. T., Schmelz, E. A., Mou, Z. (2008). A rapid biosensor-based method for quantification of free and glucose-conjugated salicylic acid. Plant Methods 4, 28. doi: 10.1186/1746-4811-4-28
Feklistov, A., Sharon, B. D., Darst, S. A., Gross, C. A. (2014). Bacterial sigma factors: a historical, structural, and genomic perspective. Annu. Rev. Microbiol. 68, 357–376. doi: 10.1146/annurev-micro-092412-155737
Garcia-Rodriguez, F. M., Schrammeijer, B., Hooykaas, P. J. (2006). The Agrobacterium VirE3 effector protein: a potential plant transcriptional activator. Nucleic Acids Res. 34, 6496–6504. doi: 10.1093/nar/gkl877
Garcion, C., Lohmann, A., Lamodiere, E., Catinot, J., Buchala, A., Doermann, P., et al. (2008). Characterization and biological function of the ISOCHORISMATE SYNTHASE2 gene of Arabidopsis. Plant Physiol. 147, 1279–1287. doi: 10.1104/pp.108.119420
Glazebrook, J. (2005). Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43, 205–227. doi: 10.1146/annurev.phyto.43.040204.135923
Heard, D. J., Kiss, T., Filipowicz, W. (1993). Both Arabidopsis TATA binding protein (TBP) isoforms are functionally identical in RNA polymerase II and III transcription in plant cells: evidence for gene-specific changes in DNA binding specificity of TBP. EMBO J. 12, 3519–3528. doi: 10.1002/embj.1993.12.issue-9
Huang, Y., Kendall, T., Forsythe, E. S., Dorantes-Acosta, A., Li, S., Caballero-Perez, J., et al. (2015). Ancient origin and recent innovations of RNA polymerase IV and V. Mol. Biol. Evol. 32, 1788–1799. doi: 10.1093/molbev/msv060
Imamura, S., Hanaoka, M., Tanaka, K. (2008). The plant-specific TFIIB-related protein, pBrp, is a general transcription factor for RNA polymerase I. EMBO J. 27, 2317–2327. doi: 10.1038/emboj.2008.151
Juven-Gershon, T., Kadonaga, J. T. (2010). Regulation of gene expression via the core promoter and the basal transcriptional machinery. Dev. Biol. 339, 225–229. doi: 10.1016/j.ydbio.2009.08.009
Kim, K. C., Fan, B., Chen, Z. (2006). Pathogen-induced Arabidopsis WRKY7 is a transcriptional repressor and enhances plant susceptibility to Pseudomonas syringae. Plant Physiol. 142, 1180–1192. doi: 10.1104/pp.106.082487
Knutson, B. A. (2013). Emergence and expansion of TFIIB-like factors in the plant kingdom. Gene 526, 30–38. doi: 10.1016/j.gene.2013.04.022
Lagrange, T., Hakimi, M. A., Pontier, D., Courtois, F., Alcaraz, J. P., Grunwald, D., et al. (2003). Transcription factor IIB (TFIIB)-related protein (pBrp), a plant-specific member of the TFIIB-related protein family. Mol. Cell Biol. 23, 3274–3286. doi: 10.1128/MCB.23.9.3274-3286.2003
Li, X., Wang, Z., Fu, Y., Cheng, X., Zhang, Y., Fan, B., et al. (2021b). Two ubiquitin-associated ER proteins interact with COPT copper transporters and modulate their accumulation. Plant Physiol. 187, 2469–2484. doi: 10.1093/plphys/kiab381
Li, S., Xu, B., Niu, X., Lu, X., Cheng, J., Zhou, M., et al. (2021a). JAZ8 interacts with virE3 attenuating agrobacterium mediated root tumorigenesis. Front. Plant Sci. 12, 685533. doi: 10.3389/fpls.2021.685533
Moore, D. L., Goldberg, J. L. (2011). Multiple transcription factor families regulate axon growth and regeneration. Dev. Neurobiol. 71, 1186–1211. doi: 10.1002/dneu.20934
Ning, H., Yang, S., Fan, B., Zhu, C., Chen, Z. (2021). Expansion and functional diversification of TFIIB-like factors in plants. Int. J. Mol. Sci. 22, 1078. doi: 10.3390/ijms22031078
Niu, Q. K., Liang, Y., Zhou, J. J., Dou, X. Y., Gao, S. C., Chen, L. Q., et al. (2013). Pollen-expressed transcription factor 2 encodes a novel plant-specific TFIIB-related protein that is required for pollen germination and embryogenesis in Arabidopsis. Mol. Plant 6, 1091–1108. doi: 10.1093/mp/sst083
Niu, X., Zhou, M., Henkel, C. V., Van Heusden, G. P., Hooykaas, P. J. (2015). The Agrobacterium tumefaciens virulence protein VirE3 is a transcriptional activator of the F-box gene VBF. Plant J. 84, 914–924. doi: 10.1111/tpj.13048
Qin, Z., Zhang, X., Zhang, X., Xin, W., Li, J., Hu, Y. (2014). The Arabidopsis transcription factor IIB-related protein BRP4 is involved in the regulation of mitotic cell-cycle progression during male gametogenesis. J. Exp. Bot. 65, 2521–2531. doi: 10.1093/jxb/eru140
Qu, L. J., Zhu, Y. X. (2006). Transcription factor families in Arabidopsis: major progress and outstanding issues for future research. Curr. Opin. Plant Biol. 9, 544–549. doi: 10.1016/j.pbi.2006.07.005
Ream, T. S., Haag, J. R., Wierzbicki, A. T., Nicora, C. D., Norbeck, A. D., Zhu, J. K., et al. (2009). Subunit compositions of the RNA-silencing enzymes Pol IV and Pol V reveal their origins as specialized forms of RNA polymerase II. Mol. Cell 33, 192–203. doi: 10.1016/j.molcel.2008.12.015
Seyfferth, C., Tsuda, K. (2014). Salicylic acid signal transduction: the initiation of biosynthesis, perception and transcriptional reprogramming. Front. Plant Sci. 5, 697. doi: 10.3389/fpls.2014.00697
Shiu, S. H., Shih, M. C., Li, W. H. (2005). Transcription factor families have much higher expansion rates in plants than in animals. Plant Physiol. 139, 18–26. doi: 10.1104/pp.105.065110
Turkarslan, S., Reiss, D. J., Gibbins, G., Su, W. L., Pan, M., Bare, J. C., et al. (2011). Niche adaptation by expansion and reprogramming of general transcription factors. Mol. Syst. Biol. 7, 554. doi: 10.1038/msb.2011.87
Vannini, A., Cramer, P. (2012). Conservation between the RNA polymerase I, II, and III transcription initiation machineries. Mol. Cell 45, 439–446. doi: 10.1016/j.molcel.2012.01.023
Wang, Z., Li, X., Liu, N., Peng, Q., Wang, Y., Fan, B., et al. (2019). A family of NAI2-interacting proteins in the biogenesis of the ER body and related structures. Plant Physiol. 180, 212–227. doi: 10.1104/pp.18.01500
Wang, F., Shang, Y., Fan, B., Yu, J. Q., Chen, Z. (2014). Arabidopsis LIP5, a positive regulator of multivesicular body biogenesis, is a critical target of pathogen-responsive MAPK cascade in plant basal defense. PloS Pathog. 10, e1004243. doi: 10.1371/journal.ppat.1004243
Wang, F., Yang, Y., Wang, Z., Zhou, J., Fan, B., Chen, Z. (2015). A critical role of lyst-interacting protein5, a positive regulator of multivesicular body biogenesis, in plant responses to heat and salt stresses. Plant Physiol. 169, 497–511. doi: 10.1104/pp.15.00518
Wildermuth, M. C., Dewdney, J., Wu, G., Ausubel, F. M. (2001). Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414, 562–565. doi: 10.1038/35107108
Yan, S., Dong, X. (2014). Perception of the plant immune signal salicylic acid. Curr. Opin. Plant Biol. 20, 64–68. doi: 10.1016/j.pbi.2014.04.006
Yan, L., Wei, S., Wu, Y., Hu, R., Li, H., Yang, W., et al. (2015). High-efficiency genome editing in arabidopsis using YAO promoter-driven CRISPR/Cas9 system. Mol. Plant 8, 1820–1823. doi: 10.1016/j.molp.2015.10.004
Zhou, J. J., Liang, Y., Niu, Q. K., Chen, L. Q., Zhang, X. Q., Ye, D. (2013b). The Arabidopsis general transcription factor TFIIB1 (AtTFIIB1) is required for pollen tube growth and endosperm development. J. Exp. Bot. 64, 2205–2218. doi: 10.1093/jxb/ert078
Zhou, J., Wang, J., Cheng, Y., Chi, Y. J., Fan, B., Yu, J. Q., et al. (2013a). NBR1-mediated selective autophagy targets insoluble ubiquitinated protein aggregates in plant stress responses. PloS Genet. 9, e1003196. doi: 10.1371/journal.pgen.1003196
Keywords: TFIIB-related proteins, salicylic acid, plant immunity, isochorismate synthase 1, transcription factors, pathogenesis-related (PR) genes
Citation: Xu B, Fan B and Chen Z (2024) A critical role of a plant-specific TFIIB-related protein, BRP1, in salicylic acid-mediated immune response. Front. Plant Sci. 15:1427916. doi: 10.3389/fpls.2024.1427916
Received: 05 May 2024; Accepted: 15 July 2024;
Published: 30 July 2024.
Edited by:
Walter Gassmann, University of Missouri, United StatesReviewed by:
Shubho Chaudhuri, Bose Institute, IndiaCopyright © 2024 Xu, Fan and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhixiang Chen, emhpeGlhbmdAcHVyZHVlLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.