
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 30 July 2024
Sec. Plant Bioinformatics
Volume 15 - 2024 | https://doi.org/10.3389/fpls.2024.1425651
This article is part of the Research TopicOmics in Seed Development: Challenges and Opportunities for Improving of Seed Quality and Yield in Model and Crop PlantsView all 13 articles
 Rui Chen1
Rui Chen1 Gang Gu2
Gang Gu2 Binghui Zhang2
Binghui Zhang2 Chaofan Du3
Chaofan Du3 Xiaolu Lin3
Xiaolu Lin3 Weiwei Cai3
Weiwei Cai3 Yan Zheng1
Yan Zheng1 Tong Li1
Tong Li1 Ruiqi Wang1
Ruiqi Wang1 Xiaofang Xie1,4*
Xiaofang Xie1,4*The E3 enzyme in the UPS pathway is a crucial factor for inhibiting substrate specificity. In Solanaceae, the U-box E3 ubiquitin ligase has a complex relationship with plant growth and development, and plays a pivotal role in responding to various biotic and abiotic stresses. The analysis of the U-box gene family in Solanaceae and its expression profile under different stresses holds significant implications. A total of 116 tobacco NtU-boxs and 56 eggplant SmU-boxs were identified based on their respective genome sequences. Phylogenetic analysis of U-box genes in tobacco, eggplant, tomato, Arabidopsis, pepper, and potato revealed five distinct subgroups (I-V). Gene structure and protein motifs analysis found a high degree of conservation in both exon/intron organization and protein motifs among tobacco and eggplant U-box genes especially the members within the same subfamily. A total of 15 pairs of segmental duplication and 1 gene pair of tandem duplication were identified in tobacco based on the analysis of gene duplication events, while 10 pairs of segmental duplication in eggplant. It is speculated that segmental duplication events are the primary driver for the expansion of the U-box gene family in both tobacco and eggplant. The promoters of NtU-box and SmU-box genes contained cis-regulatory elements associated with cellular development, phytohormones, environment stress, and photoresponsive elements. Transcriptomic data analysis shows that the expression levels of the tobacco and eggplant U-box genes in different tissues and various abiotic stress conditions. Using cultivar Hongda of tobacco and cultivar Yanzhi of eggplant as materials, qRT-PCR analysis has revealed that 15 selected NtU-box genes and 8 SmU-box may play important roles in response to pathogen Ras invasion both in tobacco and eggplant.
The Ubiquitin-Proteasome System (UPS) is considered to be a major pathway of protein-specific degradation and plays an important role in the signal pathway of regulating environmental stresses in the post-translation stage of protein (Serrano et al., 2018; Stone, 2019; Sharma and Taganna, 2020). The system involves the coordinated catalytic activities of three types of enzymes, namely a large number of ubiquitin ligases (E3), together with a few ubiquitin activating enzymes (E1) and ubiquitin-conjugating enzymes (E2) (Yan et al., 2003; March and Farrona, 2018). In the process of ubiquitination, when ATP supplies energy, E1 activates the ubiquitin molecules and transmits them to E2, and E3 connects the ubiquitin binding E2 to the target protein, and finally achieves ubiquitination of the target protein (Finley and Chau, 1991; Pickart, 2001; Trenner et al., 2022). Ubiquitin ligases are crucial in the ubiquitin pathway as they specifically recognize the target proteins for ubiquitination, and it is also the most abundant enzyme in terms of quantity. Studies have shown that the ubiquitin ligases are classified into different families according to their structure, function, and substrate specificity (Kim et al., 2021a), but functional domains of four families (HECT, RING, U-box, and cullin) are common. U-box protein contains a 70-amino acid U-box domain, a single protein widely distributed in yeast, plants, and animals (Shu and Yang, 2017). The first plant elucidated U-box protein (PUB) family was in Arabidopsis, which contains 64 members (Azevedo et al., 2001; Trujillo, 2018). Subsequent studies have successively identified 82 members in wild emmer wheat (Yang et al., 2021a), 70 in Salvia miltiorrhiza (Pan et al., 2022), 77 in rice (Kim et al., 2021b), 121 in Phyllostachys edulis (Zhou et al., 2021), 67 in barley (Ryu et al., 2019) and 62 in tomato (Sharma and Taganna, 2020).
Numerous studies have shown that U-box proteins are involved in the regulation of plant hormone signal transduction, abiotic and biotic stress responses (Zeng et al., 2008). For example, AtU-box18 and AtU-box19 were found to coordinately function as regulatory components in development and stress response in Arabidopsis (Bergler and Hoth, 2011). Furthermore, NtACRE276 was confirmed to have the E3 ligase activity and involved in cell death and defense signaling, and its ortholog in Arabidopsis (AtU-box17) and canola (BnARC1) showed similar biological function (Yang et al., 2006). In wheat, TaPUB1 plays a key role in regulating the antioxidant capacity of diploid wheat under drought stress (Zhang et al., 2017), enhancing its resistance to powdery mildew fungi by controlling U-box proteins of CMPG1-V (Zhu et al., 2015). Similarly, in tomato, SIU-box13 and SIU-box40 were found to confer resistance against tomato yellow leaf curl virus (Sharma and Taganna, 2020).
Tobacco and eggplant are important crops. Extensive studies have demonstrated that U-box genes play vital roles in regulating diverse developmental processes and stress signaling in plants (Azevedo et al., 2001). Current research on members of the U-box gene family in tobacco and eggplant is limited, and their exact function is still unknown. Therefore, it is of great significance to systematically analyze the U-box gene family in tobacco and eggplant. The purpose of this study was to comprehensively analyze the U-box gene family by integrating transcriptome data of tobacco and eggplant and study the expression patterns of U-box gene family members under pathogen Ralstonia solanacearum L. (Ras) infection conditions. The results of this study lay an important foundation for further analysis of the function and trait improvement of the U-box gene family in tobacco and eggplant.
The genome sequence and annotation data of Solanaceae species, including tomato (ITAG2.4), eggplant (SME-HQ) and tobacco (Nitab-v4.5) were downloaded from the Sol Genomics Network (https://solgenomics.net/) (Fernandez-Pozo et al., 2015; Edwards et al., 2017). The local protein database of tobacco and eggplant was constructed by command ‘makeblastdb’ of the local BLAST tool (BLAST+ 2.13.0) and a total of 62 known tomato U-box protein sequences were used as seed sequences to align with the tobacco and eggplant protein sequences by BLASTP program (Sharma and Taganna, 2020) U-box domain (PF04564) was obtained from the Pfam database (version 37.0) (http://pfam.xfam.org/) (Mistry et al., 2021).Sequences with the similarity ≥30% and E≤ 1e−10 were considered as the candidate proteins. Subsequently, the candidate protein sequences were further analyzed for the presence of a U-box structure domain (PF04564) using the CDD program of NCBI (https://www.ncbi.nlm.nih.gov/cdd/) and SMART tool (http://smart.embl-heidelberg.de/). The candidate protein containing the U-box conserved domain was confirmed as the final U-box protein. These U-box genes of tobacco and eggplant were renamed as NtU-boxs and SmU-boxs, respectively. The physicochemical properties of the tobacco and eggplant U-box proteins were predicted and analyzed using the ExPASy software (https://www.expasy.org) (Wilkins et al., 1999) and the subcellular location of each U-box protein analysis was based on the Cell PLoc 2.0 (Chou and Shen, 2008). The gene structure, conserved motif, phylogenetic tree, chromosomal localization, and synteny were analyzed, and corresponding flow chart was provided in Supplementary Figure S1.
The GFF format file of gene structure for tobacco and eggplant was obtained from the Solanaceae genome database (https://solgenomics.net/) (Fernandez-Pozo et al., 2015; Edwards et al., 2017).The intron-exon gene structures of NtU-box and SmU-box genes were displayed using TBtools (version 2.097) (Chen et al., 2020) based on the gff3 files of the tobacco and eggplant genome. The conserved motifs of NtU-box and SmU-box proteins were analyzed using the online program MEME (https://meme-suite.org/meme/tools/meme) (Ma et al., 2014; Bailey et al., 2015) and the parameters were as follows: the number of motifs was set to 20, and the width range of motifs was set to be 5-200 amino acids respectively. Motif annotation was identified using the Pfam online tool (http://pfam-legacy.xfam.org/). U-box genes were submitted to the PlantCARE online program (Lescot et al., 2002) (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) for cis-acting elements prediction.
To explore the evolutionary relationship of the U-box gene family in plants, the full protein sequences of U-box from tobacco, eggplant, pepper, potato, tomato, and Arabidopsis were analyzed. The Clustal X software (Thompson et al., 1997) was used to perform multiple sequence alignment. The phylogenetic tree was constructed by the MEGA-11 (Tamura et al., 2021) tool using the maximum likelihood method (ML) with a bootstrap of 1000 replications. The ML is a significant statistical method for parameter estimation. The ITOL (version 6.0) tool (https://itol.embl.de/) was used to edit the phylogenetic tree of NtU-box proteins and SmU-box proteins.
Based on the annotation information and the full genome protein sequences of tobacco (Nitab-v4.5), eggplant (SME-HQ), tomato (ITAG2.4) and Arabidopsis (TAIR10), the MCScanX software (Wang et al., 2012) with default parameters was employed to analyze the possible segmental duplication, tandem duplication events, intra-genomic syntenic and inter-genomic collinearity blocks (Edwards et al., 2017) and the TBtools software (version 2.097) (Chen et al., 2020) was used for visualization.
To investigate the expression patterns U-box genes across different tissues and under various abiotic stresses, we analyzed the FPKM (Fragments Per Kilobase of transcript per Million mapped reads) values of NtU-box genes in different tissues (GSE233199) (Mo et al., 2023) under drought conditions (GSE214048) (Hu et al., 2022), as well as SmU-box genes in different tissues (PRJNA328564) (Barchi et al., 2019) and under high temperature conditions (Liu et al., 2023), which were download from NCBI(https://www.ncbi.nlm.nih.gov/geo/). A map was generated using the heatmap function of the R gplots package (Walter et al., 2015).
The Hongda variety of tobacco and Yanzhi variety of eggplant were cultivated using conventional cultivation methods. The seedlings were managed until the 3-5 leaf stage. A total of 100 tobacco seedlings and 100 eggplant seedlings were selected and inoculated with a highly efficient strain of Ralstonia solanacearum L. (Ras) that had been isolated and maintained by our laboratory (Gao et al., 2019). These plants were cultured in a greenhouse with high-humidity and high-temperature (approximately 80% humidity, 28~30°C, 14 h light exposure; 10 h dark environment). The seedlings of tobacco and eggplant were collected at 0 h, 12 h, 24 h, 48 h and 96 h after inoculation, with each biological sample consisting of 5 plants and a total of 3 replicates. For sampling, seedlings were uprooted and their roots were quickly washed with sterile water to remove any attached soil and pathogens. The cDNA synthesis was carried out using the SMART Kit (Takara). To evaluate the expression levels of the NtU-box and SmU-box genes, real-time quantitative PCR (qRT-PCR) was conducted using SYBR Green qPCR Premix (Universal), and the relative expression levels were calculated using the 2−ΔΔt method (Livak and Schmittgen, 2001). Three technical replicates were performed for each sample. The actin genes of both tobacco and eggplant were used as the internal reference gene, and the primers of NtU-box and SmU-box genes (Supplementary Table S1) were designed using primer3 software (https://bioinfo.ut.ee/primer3-0.4.0/).
In this study, a total of 116 U-box genes were identified in tobacco, while 56 U-box genes were in eggplant. These genes were renamed from NtU-box1 to NtU-box116 and SmU-box1 to SmU-box56, respectively. To characterize these NtU-box and SmU-box genes, gene ID and protein molecular weight (MW), theoretical isoelectric point (pI), subcellular localization, number of exon, and CDS sequences were analyzed (Supplementary Table S2). In tobacco, NtU-box genes contained 1 to 19 exons, and the relative molecular weight of their corresponding proteins varied greatly from 20890.72 Da (NtU-box112) to 194534.46 Da (NtU-box31), and the theoretical isoelectric point ranged from 5.12 (NtU-box47) to 9.47 (NtU-box110). In eggplant, SmU-box genes contained 1 to 18 exons. The predicted molecular weight ranged from 31784.29 Da (SmU-box53) to 166473.85 Da (SmU-box43), while the theoretical isoelectric point varied from 4.92 (SmU-box51) to 9.23 (SmU-box9). Subcellular localization analysis of NtU-box and SmU-box proteins showed that these proteins were mainly present in the nucleus. Among the 116 NtU-box proteins, 5 were located on the cytoplasm, 4 on the cell membrane, and 1 on the chloroplast, while other NtU-box proteins were located on the nucleus. Among the 56 SmU-box proteins, 3 were located in the cytoplasm, 1 was located in the chloroplast, and the other SmU-box proteins were located in the nucleus (Supplementary Table S2).
The analysis of chromosomal localization showed that some NtU-box genes could not acquire the precise location information due to the incomplete sequencing of the tobacco genome. Among the 116 NtU-box genes, a total of 64 genes were unevenly distributed on 24 chromosomes of tobacco, while the remaining 52 NtU-box genes were mapped to unassigned scaffolds (Figure 1A). Notably, chromosome 19 contained the largest number of NtU-boxs (8 genes), followed by chromosome 04 with 7 NtU-box genes, and chromosomes 13 and 14 with 6 NtU-box genes each. In contrast, chromosomes 03, 05, 11, 20, 21, and 24 each contained only one NtU-box gene, while chromosomes 01, 02, 08 and 23 had no NtU-box genes detected (Figure 1A). In addition, 1 pair of tandem duplication genes on chromosome 19 (NtU-box14/10) and 15 pairs of segmental duplication genes were identified in the tobacco NtU-box gene family (Figure 1A, Supplementary Table S3). The result of chromosomal location analysis revealed that 55 out of 56 SmU-box genes were unevenly distributed among the 12 chromosomes of eggplant, with only 1 SmU-box gene located on an unattributed scaffold. Chromosome 01 contained the largest number of SmU-boxs (12 genes), followed by chromosomes 03 and 11 with 7 SmU-box genes, and 5 SmU-box genes for chromosomes 04, 05, 09 and 12. However, chromosomes 02, 06, 07, and 10 contained 4, 3, 1, and 1 SmU-box genes, respectively, while none of the SmU-box genes were detected on chromosome 08 (Figure 1B). In the eggplant SmU-box gene family, 10 pairs of fragment duplication genes were identified, but no tandem repeats were identified (Figure 1B; Supplementary Table S3).
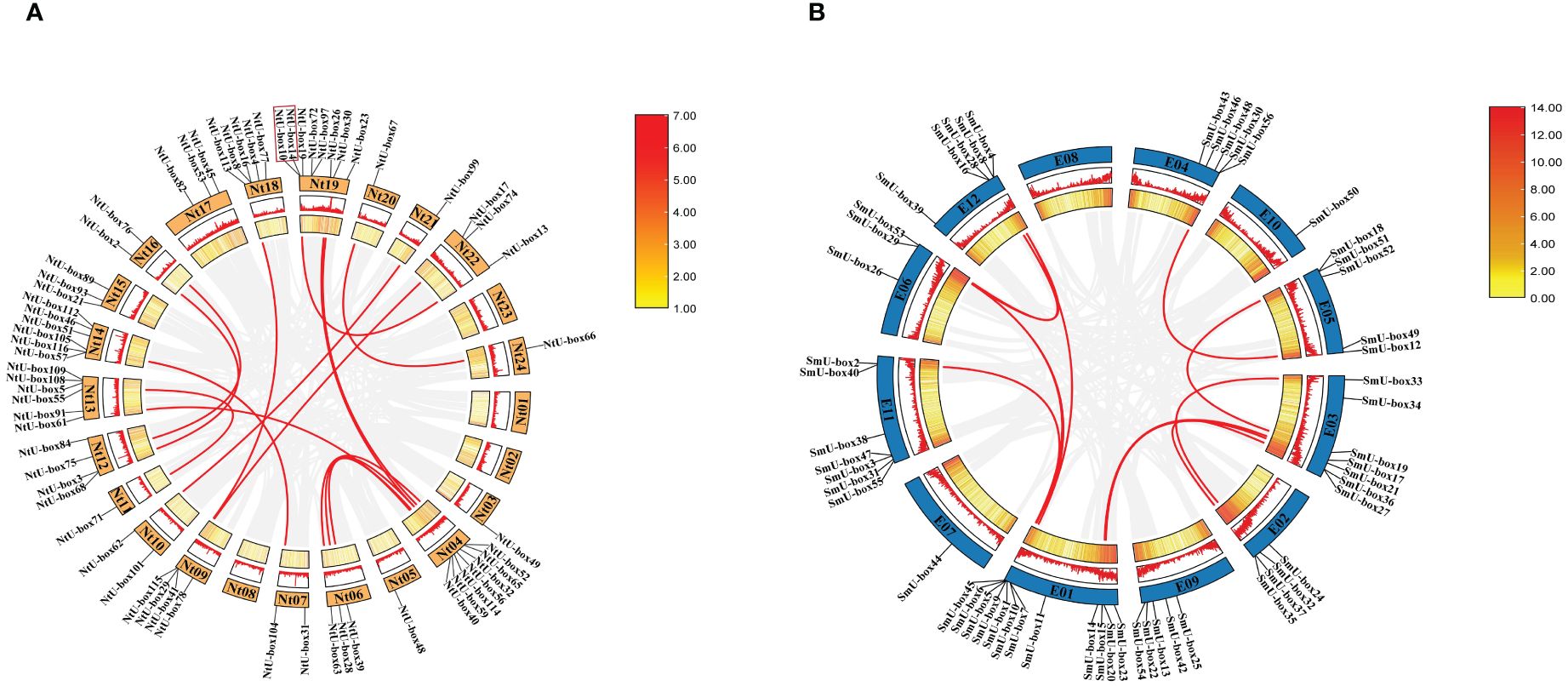
Figure 1 Gene distribution and duplication (A) NtU-box genes on the 24 chromosomes in tobacco (B) SmU-box genes on the 12 chromosomes in eggplant. Tandem-duplicated gene pairs of Nt19 are marked with red box, and segmental duplication genes are connected by red line.
A total of 22 orthologous genes were identified between tobacco and Arabidopsis based on the interspecies syntenic analysis, while there were 65 syntenic counterparts between tobacco and tomato (Figures 2A, B). A total of 68 orthologous genes were identified between eggplant and Arabidopsis based on the interspecies syntenic analysis, while there are 41 syntenic counterparts between eggplant and tomato (Figures 2A, B). The genomic regions around NtU-box41/61/62/74, SmU-box9/15/29/39/40 showed strong syntenic relationships with their counterparts in both Arabidopsis and tomato (Supplementary Table S4). Notably, good collinearity was detected among the U-box genes of four distinct species, even after undergoing speciation and long-term evolution, and the result suggested that these genes might have originated before Solanaceae species diversification and retained conserved functional roles.
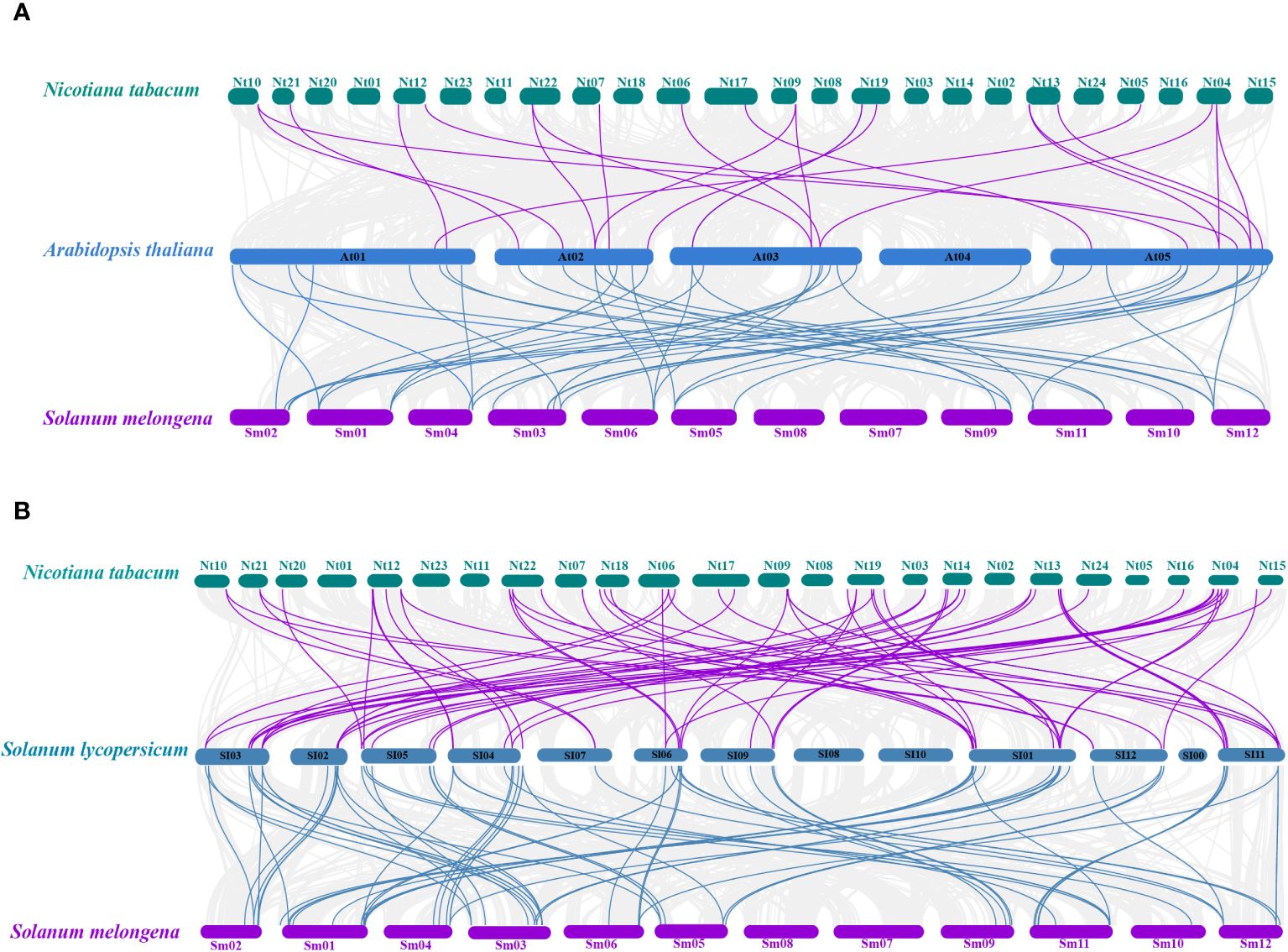
Figure 2 Collinearity analyses of U-box genes among tobacco, eggplant tomato and Arabidopsis. (A) Collinearity analyses among Arabidopsis thaliana, Nicotiana tabacum and Solanum melongena. (B) Collinearity analyses among Solanum Lycopersicon, Nicotiana tabacum and Solanum melongena. The gray line represents the co-collinearity of all genes among the three species, and the purple and blue line represents the collinearity among members of the U-box gene family.
To investigate the evolutionary relationship between NtU-box and SmU-box genes, a phylogenetic tree was constructed (Figures 3A, 4A). In tobacco, NtU-box genes were divided into 5 subgroups (I ~ V), with the largest members (41 members) in subgroup I. The subgroup represented more than 35.3% of the total NtU-box members. In contrast, subgroups II, IV, and V had only 6, 9 and 21 members, respectively. In eggplant, the largest members of eggplant (18 members) found in the subgroup I and this subgroup represented more than 32.1% of the total SmU-box members. In contrast, subgroups II, III, and V had only contained 3, 5 and 10 members, respectively. Gene structure of NtU-boxs were found that the number of exons varied from 1 (NtU-box1) to 19 (NtU-box105) (Figure 3B).

Figure 3 Gene structure and evolution of U-box family in tobacco (A) Phylogenetic relationships of NtU-boxs. Different subgroups were marked with different colors. (B) Intron-exon structure of NtU-boxs. UTR and CDS are represented by different colors. The scale bar of bottom demonstrates the length of exons and introns.
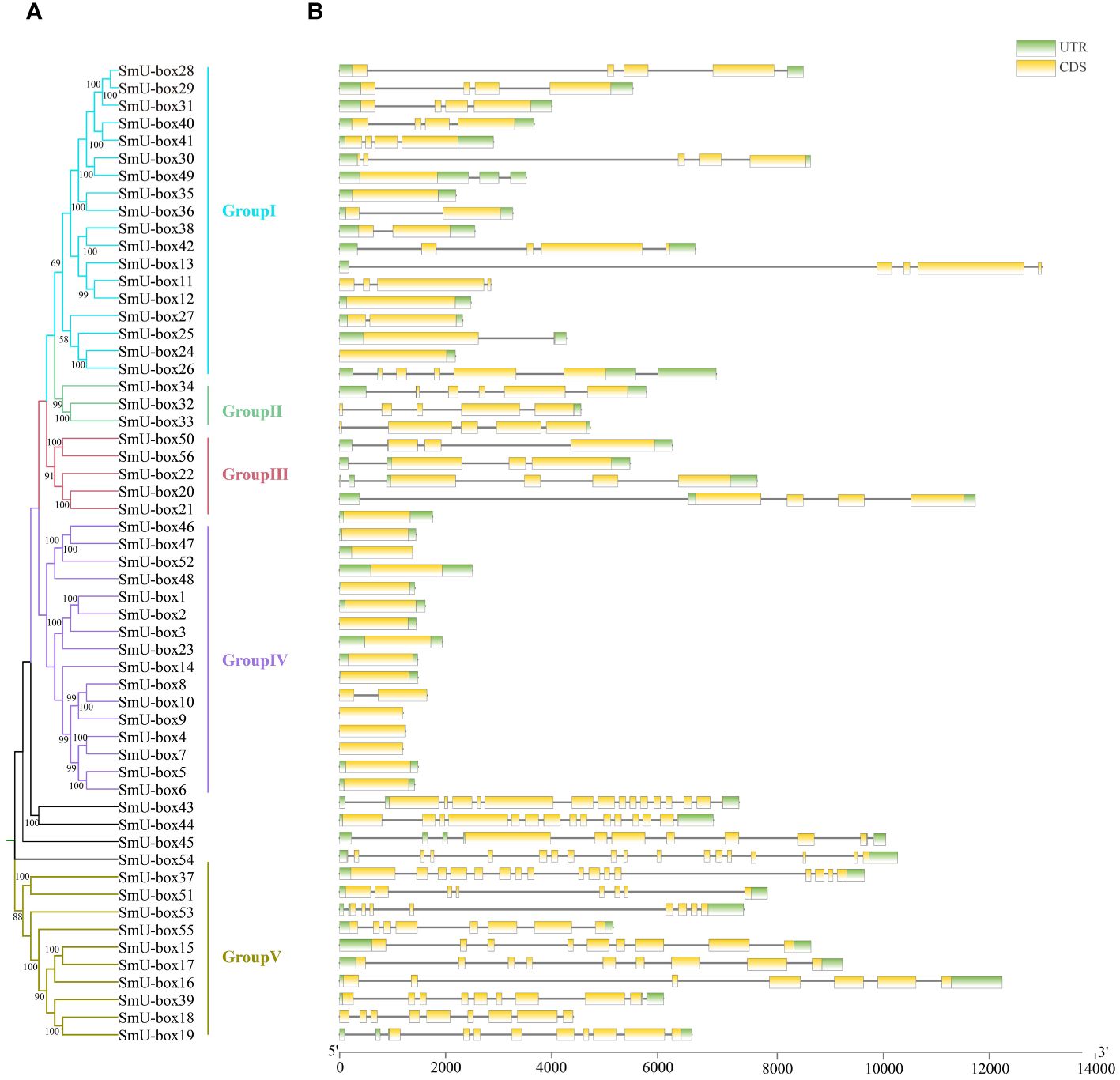
Figure 4 Gene structure and evolution of U-box family in eggplant. (A) Phylogenetic relationships of SmU-boxs. Different subgroups were marked with different colors. (B) Intron-exon structure of SmU-boxs. UTR and CDS are represented by different colors. The scale bar of bottom demonstrates the length of exons and introns.
The number of exons SmU-box gene ranging from 1 to 18. Among 116 U-box genes in tobacco, NtU-box105 and NtU-box106 contained the greatest number of exons (19), while 40 NtU-box genes (34.5%) only contained one exon (Figure 3B). Among 56 U-box genes in eggplant, 21 SmU-box genes (37.5%) contained one exon (Figure 4B). In addition, the U-box genes with similar gene structures were clustered into the same sub-clade. For example, most tobacco members of Group II only housed five exons. This result indicated that the members of the same groups exhibited similar gene structures.
A total of 20 conserved motifs have been identified in the 116 NtU-box and 56 SmU-box genes. The lengths and conserved sequences of each motif are listed in Supplementary Table S5. Among them, Motif 7 and Motif 1 were prevalent across most genes in all five groups of tobacco NtU-box proteins (Figure 5). Similarly, in eggplant SmU-box proteins Motif 1, Motif 4, Motif 6, and Motif 5 were found in most genes of the five groups, indicating their high conservation in U-box proteins (Figure 6). The protein sequences of the 20 motifs were uploaded to the CDD program for further domain analysis. Motif 1, Motif 2, and Motif 7 were annotated as components of the conserved U-box domain sequences, essential for maintaining the structural integrity of the U-box and facilitating ubiquitin linkage activity. Additionally, Motif 3 was annotated as part of the ARM conserved domain, which was the most common type in the U-box family. Moreover, genes in the same group on the phylogenetic tree exhibited similar conserved motifs. For example, all NtU-box genes in Group II had the same 7 motifs (Motifs 1, 2, 7, 9, 11, 15, and 20), suggesting potential functional similarities.
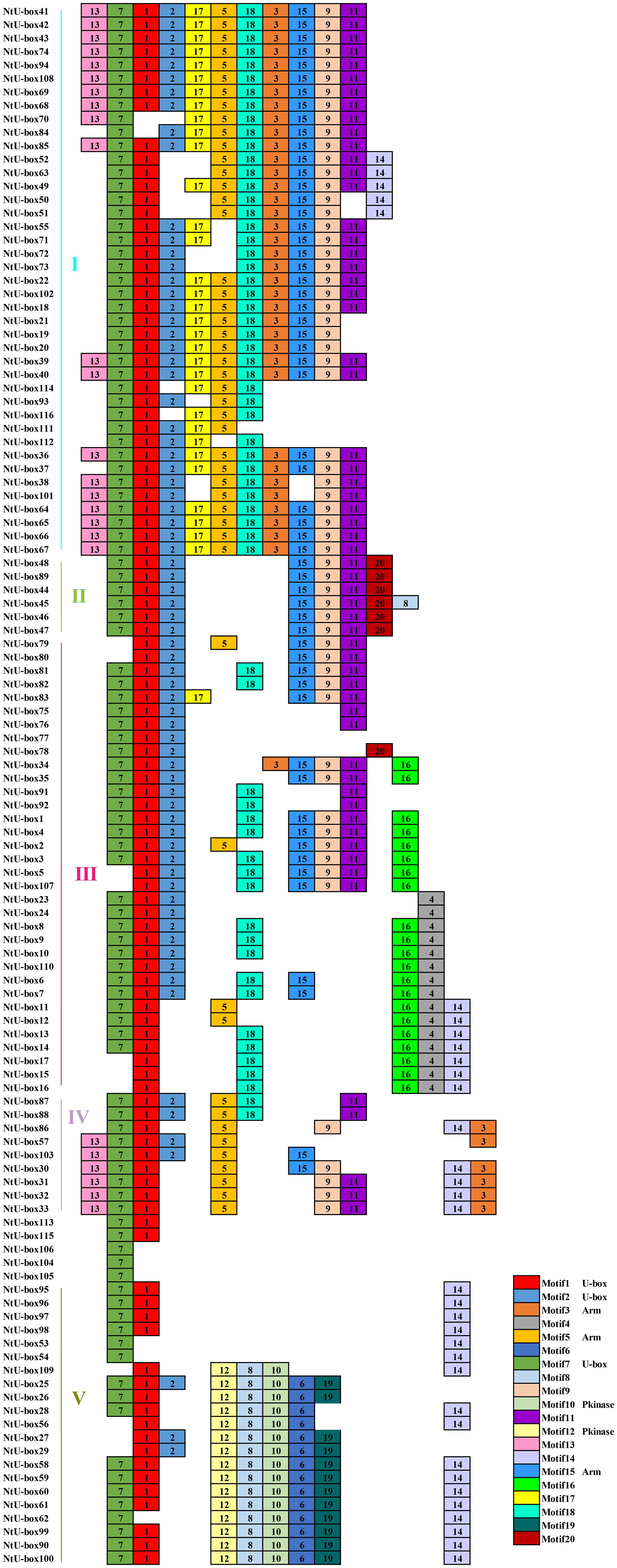
Figure 5 Conserved motifs for U-box proteins in tobacco. Different motifs are showed with different colored boxes and numbers (1-20).
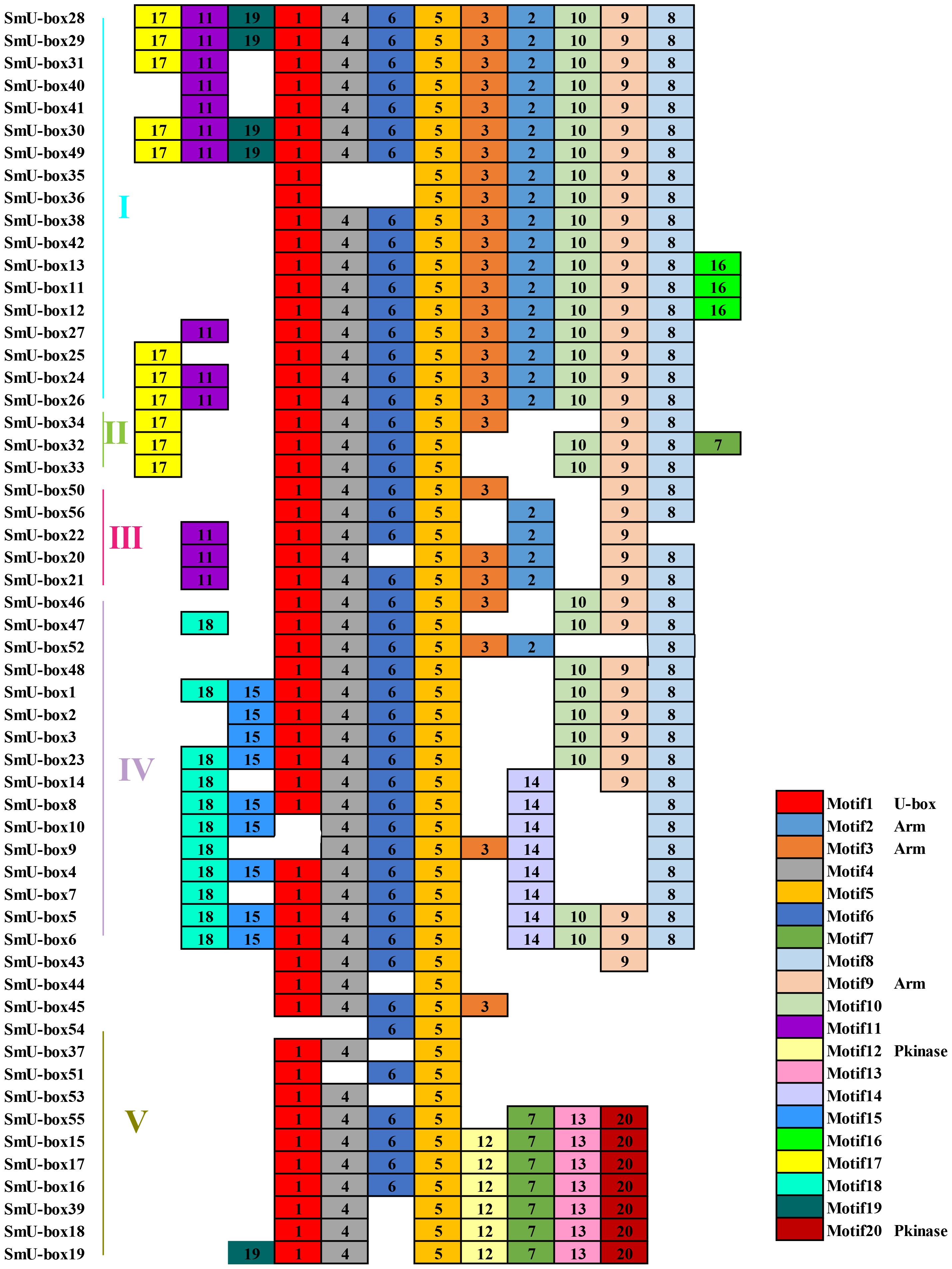
Figure 6 Conserved motifs for U-box proteins in eggplant. Different motifs are showed with different colored boxes and numbers (1-20).
To explore the evolution of the U-box gene family in plant, 428 U-box gene members from 6 species were selected to construct a phylogenetic tree (Figure 7), including tobacco (116), eggplant (56), potato (66), tomato (62), Arabidopsis thaliana (64), and pepper (64) (Supplementary Table S6). Based on previous study, 428 U-box genes were divided into 5 different subfamilies (Wang et al., 2021). Based on the phylogenetic tree, a total of 138 sister pairs of homologous proteins were identified, including 71 pairs of orthologous genes and 67 pairs of paralogous genes (Supplementary Table S7). Specifically, there were 41 paralogous pairs from tobacco, 17 pairs from Arabidopsis thaliana, 6 pairs from pepper, 2 pairs from potato, and 1 pair from eggplant.
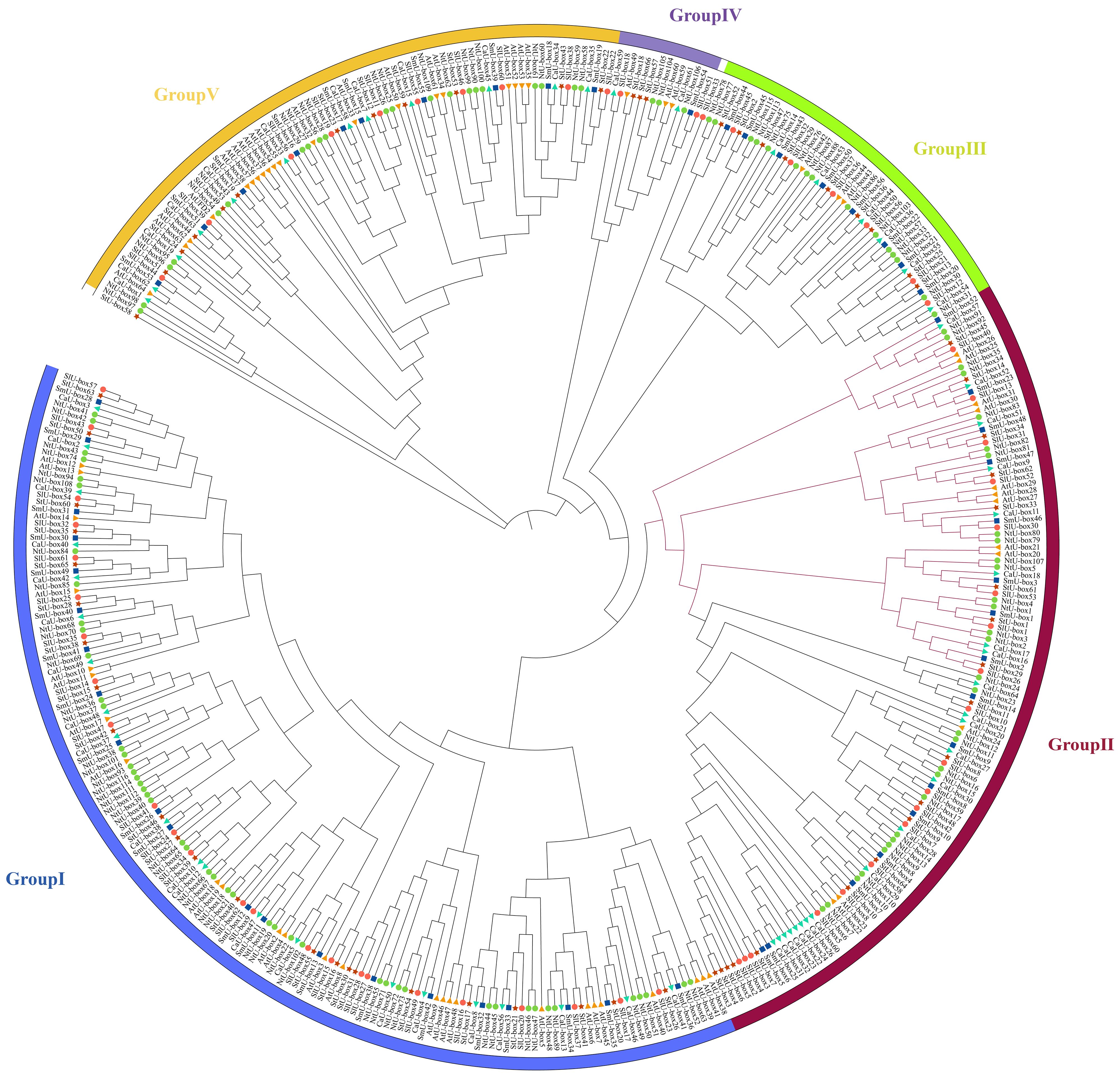
Figure 7 Phylogenetic relationship of 116 NtU-box and 56 SmU-box proteins, along with another 256 published U-box proteins. The phylogenetic relationships were generated by using MEGA-11 using the Maximum Likelihood (ML) method (1000 bootstrap replicates), and visualized with ITOL software. U-box proteins were classified into five distinct groups, as indicated by the different colors.
Similarity in gene expression patterns implies similar functions, especially for homologous genes (Yang et al., 2021b). Based on published transcriptomic data, the expression patterns of U-box genes in major tissues were compared in tobacco (116), eggplant (56), and Arabidopsis thaliana (64). These NtU-box and SmU-box genes were classified into 7 categories according to their normalized expression levels (Supplementary Figure S2A). Among them, the U-box genes of group I specifically expressed in leaf with a high level; the U-box genes of group II expressed specifically high in all tissue; U-box genes in Cluster III mostly expressed in stem; U-box in group IV and V highly expressed in roots. Based on the tissue expression clustering characteristics, phylogenetic relationship of multispecies U-box genes were predicted (Supplementary Table S8).
In general, the functions of some NtU-box and SmU-box genes were mainly divided into the following four categories: 1. Play a role in formation of organs and regulator of flowering time. 2. Play a role in regulating root development, pollen tapetum development and ROS induced chloroplast degradation. 3. Play combinatory roles in response to drought stress. 4. Encodes a U-box domain-containing E3 ubiquitin ligase with central Ser/Thr protein kinase domain, and its expression is responsive to both phosphate (Pi) and phosphite (Phi) in both roots and shoots (Supplementary Table S8). Only those NtU-box and SmU-box genes that were homologous to the reported AtU-box genes, had highly similar expression patterns, and shared similar functions. Fox example, AtPUB18 and AtPUB19 function as regulators in the drought stress response (Liu et al., 2011) and their homologous genes NtU-box65/66/67 genes and SmU-box27 genes, which had similar expression patterns, were predicted to have similar functions (Supplementary Figure S2B). Transcriptome data showed that NtU-box65/66/67 genes and SmU-box27 genes showed a positive regulatory expression pattern under heat stress (Supplementary Table S9). AtPUB14 plays a role in organ formation and flowering time regulation (Feke et al., 2020), and its homologous gene SmU-box28/29/31 has a similar expression pattern in eggplant (Supplementary Figure S2C). Transcriptome data showed that SmU-box28/29/31 has a high expression level in eggplant flowers, which is expected to have similar functions with AtPUB14.
To investigate the potential function of U-box genes during plant development and upon exposure to various stresses, the cis-elements within the promoter regions of U-box genes were analyzed. A total of 36 cis-regulatory genes were identified in the promoter region of NtU-box genes, while 34 cis-regulatory genes were identified in the promoter region of SmU-box genes (Figure 8, Supplementary Table S10). These cis-regulatory genes can be divided into four categories, specifically cell development, plant hormones, environmental stress, and photoresponse elements (Supplementary Table S10). The 5 cis-acting elements related to cell development include CAT-box, MSA-like, CCAAT-box, MBSI, and HD-Zip 1. For phytohormone-responsive elements, the cis-acting elements include TGACG-motif, ABRE, P-box, TGA-element, TCA-element, AuxRR-core, TATC-box, GARE-motif, AuxRE, A-box, and O2-site. Additionally, ten light responsive elements were identified, including GT1-motif, G-Box, Box4, MRE, ATC-motif, Sp1, ATCT-motif, ACE, 3-AF1 binding site, and AAAC-motif. The expression of these genes might be regulated by phytohormones, various light-responsiveness cis-elements, defense signaling transduction, and abiotic stresses during the growth of tobacco and eggplant.

Figure 8 (A) Predicted cis-elements in NtU-box promoters. (B) Predicted cis-elements in SmU-box promoters. Different shapes and colors represent the different types of cis-elements. Annotations of cis-elements were listed in Supplementary Table S10.
Transcriptome analysis revealed diverse expression patterns of NtU-box and SmU-box genes in different tissues, which were clustered into three groups (Figure 9) (Yang et al., 2022). In tobacco (Figure 9A), group I comprised 41 NtU-box genes, with the majority showing high levels of expression in vegetative tissue, particularly in root. Conversely, NtU-box genes in groups II and III exhibited low levels of expression. A total of 5 SmU-box genes (SmU-box 54/42/53/34/41) in group II showed high levels of expression in all investigated tissues, including stem, leaf, radicle, cotyledons, root, and flower (Figure 9B), and 31SmU-box genes in group I exhibited moderate levels of expression. Conversely, SmU-box genes in group III showed nearly negligible expression, exhibited lower expression levels.
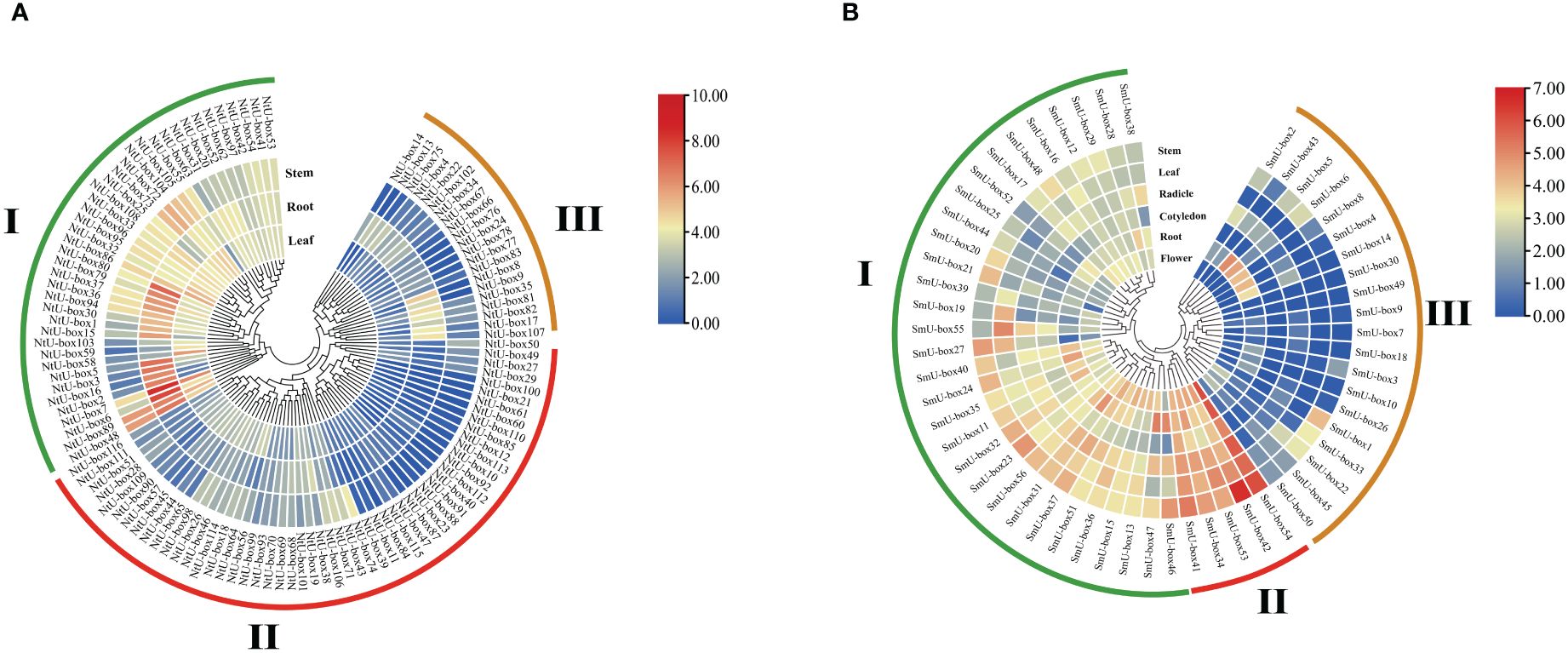
Figure 9 The expression patterns of tobacco and eggplant U-box genes in different tissues. (A) The expression patterns of NtU-box genes (B) The expression patterns of SmU-box genes. FPKM values for U-box genes were transformed by log2 (n+1).
To determine the expression profiles of U-box family genes under abiotic stress, the FPKM data of tobacco NtU-box genes under drought stress (Hu et al., 2022) and eggplant SmU-box genes under high temperature (Liu et al., 2023) were downloaded. The expression profiles of 62 NtU-box genes and 26 SmU-box genes were analyzed (Supplementary Table S9). The results revealed distinct expression patterns among the NtU-box and SmU-box gene members under various stress conditions (Figure 10). Specifically, the 62 NtU-box genes and 56 SmU-box genes were each classified into three groups (I ~ III). In tobacco, a total of 15 NtU-box genes were included in group I, and the high expression levels of these genes in the five drought stress stages mean that these genes can play an important role in the drought stress process, while the 32 NtU-box genes clustered in group II showed low or no expression in the whole drought stress process. In eggplant, a total of 14 SmU-box genes were included in group III, and the expression profiles of these genes exhibited a pattern of initial decrease followed by an increase across the three high-temperature stress stages, indicating their potential significance in responding to high-temperature stress. Conversely, the 6 SmU-box genes in group I showed either minimal or no expression throughout the high-temperature stress process. It is worth noting that the expression levels of NtU-box59 and NtU-box46 genes gradually decreased with the increase of drought time. In terms of high-temperature stress (Figure 10B), the expression level of the SmU-box27 gene in group II gradually increased with the increase of high-temperature time, while the majority of SmU-box genes (SmU-box54/32/22/38/39) reached the peak expression level at the 12 h of high-temperature stress. These results indicated the functional diversity of U-box members among different species.
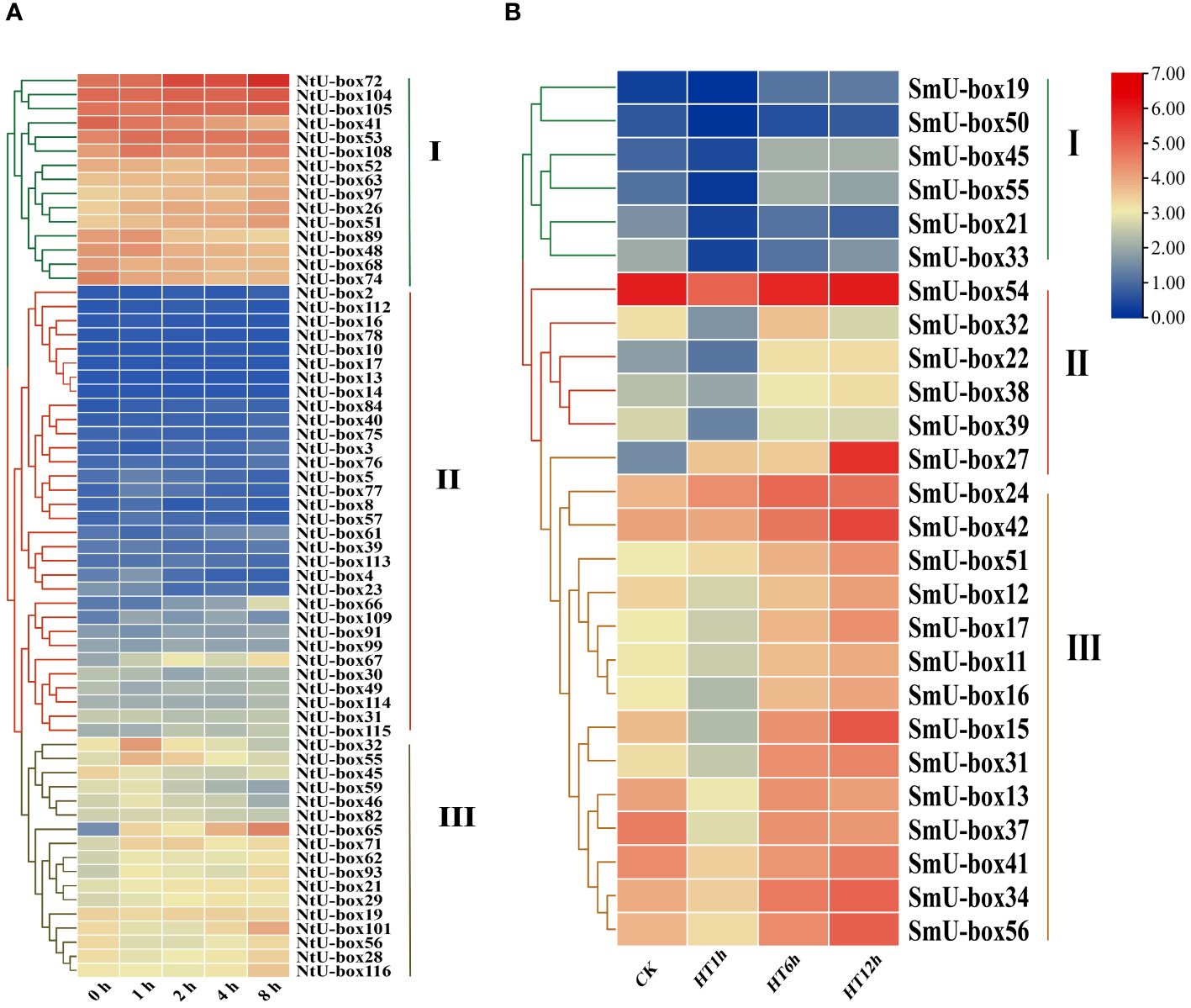
Figure 10 The expression profiles of tobacco and eggplant U-box genes under abiotic stresses. (A) The expression patterns of NtU-box genes under drought stress. (B) The expression patterns of SmU-box genes under high temperature. FPKM values for U-box genes were transformed by log2 (n+1).
No significant changes were observed in the seedlings of both tobacco and eggplant at the initial stage after being infected by Ralstonia solanacearum L. (Ras). In tobacco, the primary symptoms induced by Ras infection manifested in the seedling at 96 h (Figure 11A). During this stage, the seedling exhibiting leaf wilting and stem necrosis, with the roots turning yellow and exhibiting necrosis. In contrast, symptoms in the eggplant appeared later than those in tobacco, with notable symptoms appeared at 120 h (Figure 11B). The leaves appeared withered, and the basal part of the stem showed severe necrosis and turned black. It was reported that SIU-box13 and SIU-box40 in tomato, play a crucial role in the regulation of pathogen invasion (Sharma and Taganna, 2020). To further explore the potential functions of the U-box genes in tobacco and eggplant, a total of 15 NtU-box and 8 SmU-box genes that clustered with SIU-box13 and SIU-box40 in II subgroups of the phylogenetic tree (Figure 7) were selected for qRT-PCR analysis under Ras infection. The qRT-PCR analysis found that the majority of the selected genes showed significant response to Ras infection. In tobacco (Figure 12A), the expression levels of 7 NtU-box genes (NtU-box1/3/79/81/82/83/91) exhibited a trend of initial increased followed by decreased with the extension of time after inoculation. Among these genes, the expression levels of 5 genes (NtU-box3/79/81/82/83) were significantly up-regulated at 12 h after inoculation, while 2 genes (NtU-box2/4) displayed significant down-regulated and 6 genes (NtU-box5/34/35/80/92/107) showed a pattern of decrease followed by a slight increase. In eggplant (Figure 12B), all the 8 selected genes displayed up-regulated expression in response to the infection compared to the initial stage (0 h), and 2 genes (SmU-box1/2) exhibited significant up-regulation exceeding a 4-fold increase at 24 h post-inoculation.

Figure 11 Disease symptoms of tobacco and eggplant seedlings. (A) Tobacco seedlings at 0 h and 96 h post-inoculation with Ras. (B) Eggplant seedlings at 0 h and 120 h post inoculation with Ras. The basal parts of stems were magnified and shown in the circles.
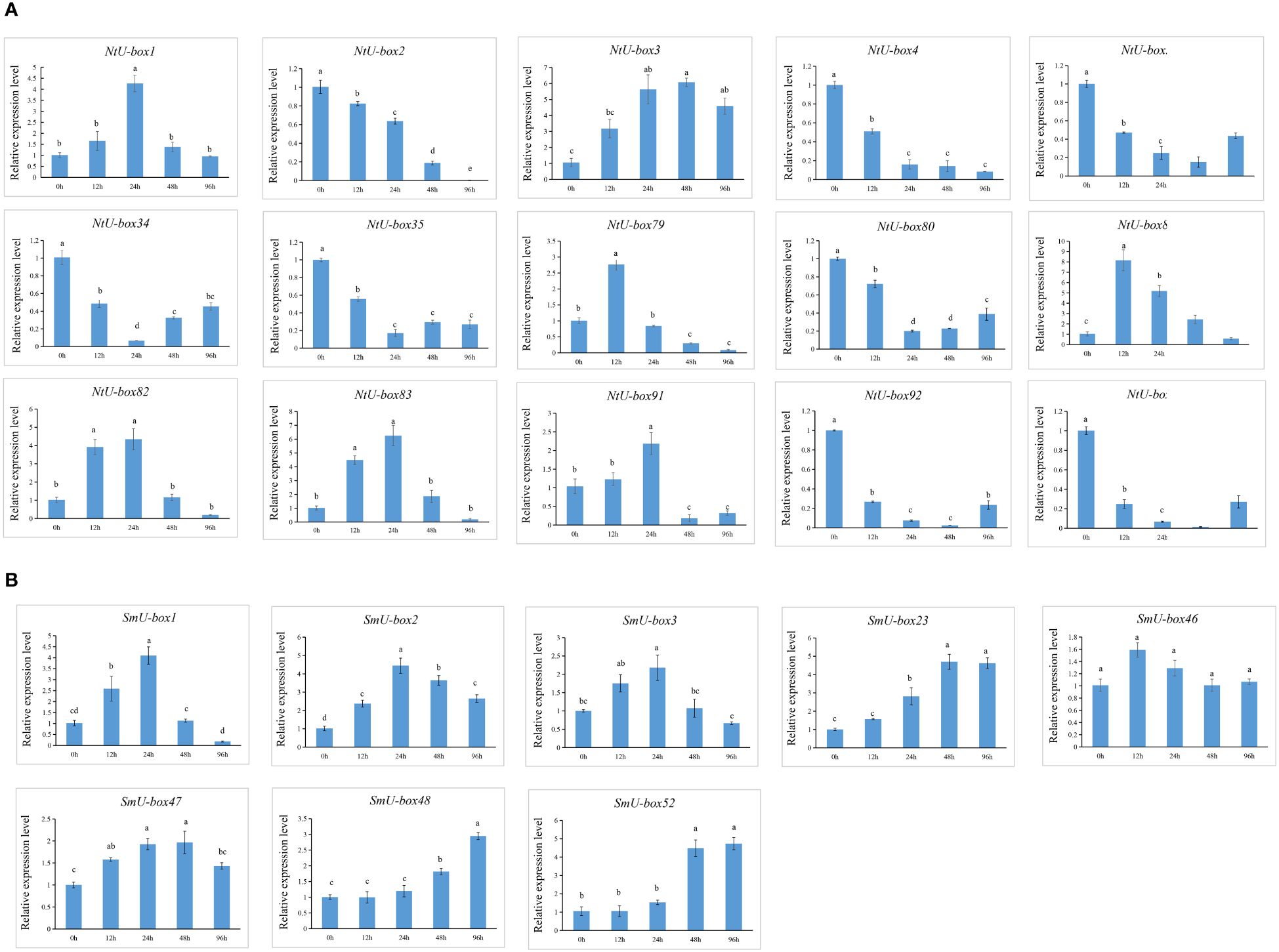
Figure 12 Expression profiles of U-box genes in response to Ras infection. (A) Relative expression level of 15 NtU-boxs in response to Ras infection. (B) Relative expression level of 8 SmU-boxs in response to Ras infection. Error bars are standard deviations of three biological replicates. Diverse letters on top of the bars indicate significant differences at P<0.05.
U-box gene family members are present in almost all eukaryotes. The advent of genome sequencing has facilitated comprehensive analyses of the U-box gene family in numerous species, such as Arabidopsis thaliana (64), rice (77), soybeans (125), and tomato (62) (Wiborg et al., 2008; Zeng et al., 2008; Wang et al., 2016; Sharma and Taganna, 2020). In this study, a total of 116 U-box genes were identified in tobacco, which were larger than those in Arabidopsis thaliana and rice, but less than in soybean. As an allotetraploid, the genome size of Nicotiana tabacum is 4.5Gb, while that of Arabidopsis, rice and soybean is 125Mb, 430Mb and 1.025Gb, respectively (Kaul S et al., 2000; Burr, 2002; Schnable et al., 2009; Schmutz et al., 2010). In this case, it seems that there is no direct correlation between the number of U-box genes and genome size in these plants. The number of exons observed between NtU-box genes varies significantly, of which 64 genes contain two or more exons, 13 genes have more than 10 exons, and 39 genes possess only one exon. Similarly, for SmU-box genes, differences are notable: 32 genes contain two or more exons and 4 genes have more than 10 exons, and however, 20 genes possess only one exon. This suggested that distinct RNA splicing processes that could modulate the proportion of isoforms (active and non-active), crucial for stress regulation (Tang et al., 2021). Many SmU-box genes are either intron-less or have two introns, consistent with reports in tomato (Sharma and Taganna, 2020). U-box genes with multiple introns may act as a mutational buffer, protecting coding sequences from deleterious mutations. The presence of the intron-less genes underscores structural integrity within the U-box gene family, suggesting irregular distribution of U-box protein in different species (Zeng et al., 2008). The phylogenetic analysis of the U-box gene family showed a great similarity among all the five classes due to the presence of the core U-box domain in all the members (Figure 7), suggesting a shared ancestor predating the divergence of these species. Moreover, a single tandem duplication gene pair and 15 segmental duplication gene pairs were detected in tobacco (Figure 1A), while 10 segmental duplication gene pairs were identified in eggplant (Figure 1B), highlighting segmental duplication events as the primary driver for the expansion of the U-box gene family in both tobacco and eggplant. In addition, a good collinearity was detected among the U-box genes of four distinct species, even after undergoing speciation and long-term evolution. Syntenic analysis revealed a higher number of orthologous gene pairs between tobacco and tomato compared to those between tobacco and Arabidopsis (Figures 2A, B). Conversely, the count of orthologous gene pairs between eggplant and Arabidopsis exceeded those between eggplant and tomato (Figures 2A, B). The results suggested that species with close evolutionary relationships tend to exhibit greater similarity, higher homology, and increased conservation of the U-box genes.
In general, the evolution of gene family is predominantly determined by the organization of gene structures, whereas within a gene family, members of the same subfamily typically exhibit high conservation in both structure and function, reflecting their evolutionary relatedness. In this study, a total of 20 conserved motifs were identified in the U-box gene families of both tobacco and eggplant. Interestingly, despite belonging to the same or different subfamilies, U-box members exhibited variations in motif types and quantities. However, the differences in the same subfamily were notably smaller, indicating a higher level of conservation in motif composition within closely related members. These observations highlight the complexity of the tobacco and eggplant genomes and the differentiation and diversity of the function within the U-box gene family.
U-box genes play important roles in plant responses to both abiotic and biotic stresses (Adler et al., 2017; Sharma and Taganna, 2020; Tang et al., 2021). Functional analysis has revealed the molecular mechanisms involving U-box proteins involving U-box proteins in theses stress response (Cho et al., 2017). However, the role of the U-box gene family in tobacco and eggplant remains unclear. Stress refers to the phenomenon that plants are exposed to adverse conditions in their environment, causing their physiological processes to be negatively affected. Stress is usually divided into two categories: abiotic stress and biotic stress. Abiotic stress mainly refers to the unfavorable conditions caused by environmental factors, such as temperature, light, humidity, drought, salt and alkali. Biotic stresses include a range of biological factors that are harmful to plant survival and development, often stemming from infections and competition, including diseases, pests, and weeds (Atkinson and Urwin, 2012). These stress factors significantly affect crop growth and production. Therefore, mining excellent resistance genes in plants has become one of the main strategies to cope with various stress challenges. In this study, 116 NtU-boxs were identified in tobacco and 56 SmU-boxs were identified in eggplant, and their expressions under abiotic and biotic stresses were further analyzed. Transcriptomic data analysis revealed the different responses of plant U-box gene family members to abiotic stress, indicating functional differences within the family. In addition, SIU-box13 and SIU-box40 of tomato have been shown to play a key role in regulating pathogen invasion (Sharma and Taganna, 2020). Therefore, it can be inferred that the NtU-box and SmU-box genes clustered in the same subfamily with SIU-box13 and SIU-box40 may have similar functions. In this study, the expression patterns of 15 selected NtU-box genes and 8 SmU-box genes under Ras infection highlighting their important role in Ras resistance. Among the 15 NtU-box genes, the expression of 7 NtU-box genes (NtU-box1/3/79/81/82/83/91) was significantly up-regulated, and the NtU-box81 gene showed a strong response to Ras invasion, which increased more than 8 times at 12 h post-inoculation, while the remaining 8 NtU-box genes showed down-regulated expression patterns under Ras infection (Figure 12A). It is worth noting that certain NtU-box genes exhibit contrasting responses to bacterial and drought stresses. For instance, three genes, NtU-box3, NtU-box82 and NtU-box91 were up-regulated in response to Ras infection; whereas they showed down-regulated under drought conditions (Figures 10A, 12A; Supplementary Table S9). In eggplant, a similar phenomenon was observed. For example, all 8 selected SmU-box genes (SmU-box1/2/3/23/46/47/48/52) exhibited up-regulated in response to Ras infection (Figure 12B), while no significant changes were detected under high temperature stress condition (Figures 10B, 12B; Supplementary Table S9). This result suggested a functional differentiation in the tobacco NtU-box gene family in their responses to different stresses. Further study of these genes would greatly enhance the understanding of their functions in both tobacco and eggplant.
In this study, a total of 116 NtU-box genes and 56 SmU-box genes were identified in the genome of tobacco and eggplant, which were categorized into 5 subfamilies, respectively. These NtU-box genes and SmU-box genes were randomly distributed on the 24 chromosomes of tobacco and the 12 chromosomes of eggplant. Phylogenetic analysis suggested a shared ancestor predating the divergence of six species (tobacco, eggplant, potato, tomato, Arabidopsis thaliana, and pepper), and segmental duplication event was the primary driver for the expansion of the U-box gene family in both tobacco and eggplant. The promoters of NtU-box and SmU-box genes contained cis-regulatory elements associated with cell development, plant hormone response, photoresponsive elements, and stress response. The expression levels of the tobacco and eggplant U-box genes varied under various abiotic stress conditions. qRT-PCR analysis revealed that 15 selected NtU-box and 8 SmU-box genes play important roles in response to pathogen Ras invasion in tobacco and eggplant. Our results provided valuable information for further functional study of U-box genes in both tobacco and eggplant.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
RC: Writing – original draft. BZ: Writing – original draft, Data curation. GG: Writing – original draft, Resources. CD: Writing – review & editing, Resources. XL: Writing – review & editing, Resources. WC: Writing – review & editing, Resources. YZ: Writing – review & editing, Resources. TL: Writing – review & editing, Data curation. RW: Writing – review & editing, Data curation. XX: Writing – review & editing, Validation.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was financially supported by China Tobacco Company (110202201028 (LS-12) and Longyan Tobacco Company (LK-2022Y02; LK-2022Y06). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1425651/full#supplementary-material
Ras, Ralstonia solanacearum L.; MW, Molecular weight; pI, Isoelectric points; ML, maximum likelihood; NtU-box, U-box genes of Nicotiana tabacum; SmU-box, U-box genes of Solanum melongena; FPKM, Fragments Per Kilobase of transcript sequence per Millions base pairs sequenced; qRT-PCR, Quantitative real-time PCR.
Adler, G., Konrad, Z., Zamir, L., Mishra, A. K., Raveh, D., Bar-Zvi, D. (2017). The Arabidopsis paralogs, PUB46 and PUB48, encoding U-box E3 ubiquitin ligases, are essential for plant response to drought stress. BMC Plant Biol. 17, 8. doi: 10.1186/s12870-016-0963-5
Atkinson, N. J., Urwin, P. E. (2012). The interaction of plant biotic and abiotic stresses: from genes to the field. J. Exp. Bot. 63, 3523–3543. doi: 10.1093/jxb/ers100
Azevedo, C., Santos-Rosa, M. J., Shirasu, K. (2001). The U-box protein family in plants. Trends Plant Sci. 6, 354–358. doi: 10.1016/S1360-1385(01)01960-4
Bailey, T. L., Johnson, J., Grant, C. E., Noble, W. S. (2015). The MEME suite. Nucleic Acids Res. 43, W39–W49. doi: 10.1093/nar/gkv416
Barchi, L., Pietrella, M., Venturini, L., Minio, A., Toppino, L., Acquadro, A., et al. (2019). A chromosome-anchored eggplant genome sequence reveals key events in Solanaceae evolution. Sci. Rep. 9(1), 11769. doi: 10.1038/s41598-019-47985-w
Bergler, J., Hoth, S. (2011). Plant U-box armadillo repeat proteins AtPUB18 and AtPUB19 are involved in salt inhibition of germination in Arabidopsis. Plant Biol. (Stuttg) 13, 725–730. doi: 10.1111/j.1438-8677.2010.00431.x
Burr, B. (2002). Mapping and sequencing the rice genome. Plant Cell 14, 521–523. doi: 10.1105/tpc.140310
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Cho, S. K., Ryu, M. Y., Kim, J. H., Hong, J. S., Oh, T. R., Kim, W. T., et al. (2017). RING E3 ligases: key regulatory elements are involved in abiotic stress responses in plants. BMB Rep. 50, 393–400. doi: 10.5483/BMBRep.2017.50.8.128
Chou, K. C., Shen, H. B. (2008). Cell-PLoc: a package of Web servers for predicting subcellular localization of proteins in various organisms. Nat. Protoc. 3, 153–162. doi: 10.1038/nprot.2007.494
Edwards, K. D., Fernandez-Pozo, N., Drake-Stowe, K., Humphry, M., Evans, A. D., Bombarely, A., et al. (2017). A reference genome for Nicotiana tabacum enables map-based cloning of homeologous loci implicated in nitrogen utilization efficiency. BMC Genomics 18, 448. doi: 10.1186/s12864-017-3791-6
Feke, A. M., Hong, J., Liu, W., Gendron, J. M. (2020). A decoy library uncovers U-box E3 ubiquitin ligases that regulate flowering time in arabidopsis. Genetics 215, 699–712. doi: 10.1534/genetics.120.303199
Fernandez-Pozo, N., Menda, N., Edwards, J. D., Saha, S., Tecle, I. Y., Strickler, S. R., et al. (2015). The Sol Genomics Network (SGN)–from genotype to phenotype to breeding. Nucleic Acids Res. 43, D1036–D1041. doi: 10.1093/nar/gku1195
Finley, D., Chau, V. (1991). Ubiquitination. Annu. Rev. Cell Biol. 7, 25–69. doi: 10.1146/annurev.cb.07.110191.000325
Gao, W., Chen, R., Pan, M., Tang, W., Lan, T., Huang, L., et al. (2019). Early transcriptional response of seedling roots to Ralstonia solanacearum in tobacco (Nicotiana tabacum L.) European J. Plant Pathol 155, 527–536. doi: 10.1007/s10658-019-01788-x
Hu, Z., He, Z., Li, Y., Wang, Q., Yi, P., Yang, J., et al. (2022). Transcriptomic and metabolic regulatory network characterization of drought responses in tobacco. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1067076
Kaul, S., Koo, H. L., Jenkins, J., Rizzo, M, Rooney, T., Tallon, L. J., et al. (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. doi: 10.1038/35048692
Kim, M.-S., Kang, K.-K., Cho, Y.-G. (2021b). Molecular and functional analysis of U-box E3 ubiquitin ligase gene family in rice (Oryza sativa). Int. J. Mol. Sci. 22 (5), 12088. doi: 10.3390/ijms222112088
Kim, D. Y., Lee, Y. J., Hong, M. J., Kim, J. H., Seo, Y. W. (2021a). Genome wide analysis of U-box E3 ubiquitin ligases in wheat (Triticum aestivum L.). Int. J. Mol. Sci. 22, 2699. doi: 10.3390/ijms22052699
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Liu, R., Shu, B., Wang, Y., Yu, B., Wang, Y., Gan, Y., et al. (2023). Transcriptome analysis reveals key genes involved in the eggplant response to high-temperature stress. Environ. Exp. Bot. 211, 105369. doi: 10.1016/j.envexpbot.2023.105369
Liu, Y. C., Wu, Y. R., Huang, X. H., Sun, J., Xie, Q. (2011). AtPUB19, a U-box E3 ubiquitin ligase, negatively regulates abscisic acid and drought responses in Arabidopsis thaliana. Mol. Plant 4, 938–946. doi: 10.1093/mp/ssr030
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Ma, W., Noble, W. S., Bailey, T. L. (2014). Motif-based analysis of large nucleotide data sets using MEME-ChIP. Nat. Protoc. 9, 1428–1450. doi: 10.1038/nprot.2014.083
March, E., Farrona, S. (2018). Plant deubiquitinases and their role in the control of gene expression through modification of histones. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.02274
Mistry, J., Chuguransky, S., Williams, L., Qureshi, M., Salazar, G. A., Sonnhammer, E. L. L., et al. (2021). Pfam: The protein families database in 2021. Nucleic Acids Res. 49, D412–d419. doi: 10.1093/nar/gkaa913
Mo, Z., Huang, Y., Pu, T., Duan, L., Pi, K., Luo, J., et al. (2023). Genome-wide identification and characterization of Glutathione S-Transferases (GSTs) and their expression profile under abiotic stresses in tobacco (Nicotiana tabacum L.). BMC Genomics 24, 341. doi: 10.1186/s12864-023-09450-x
Pan, J., Li, X. I. N., Li, J., Liu, H. U. I., Hou, Z., Yang, Z., et al. (2022). Genome-wide identification of U-box gene family and expression analysis under abiotic stresses in Salvia miltiorrhiza. Biocell 46, 1751–1762. doi: 10.32604/biocell.2022.019455
Pickart, C. M. (2001). Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70, 503–533. doi: 10.1146/annurev.biochem.70.1.503
Ryu, M. Y., Cho, S. K., Hong, Y., Kim, J., Kim, J. H., Kim, G. M., et al. (2019). Classification of barley U-box E3 ligases and their expression patterns in response to drought and pathogen stresses. BMC Genomics 20, 326. doi: 10.1186/s12864-019-5696-z
Schmutz, J., Cannon, S. B., Schlueter, J., Ma, J., Mitros, T., Nelson, W., et al. (2010). Genome sequence of the palaeopolyploid soybean. Nature 463, 178–183. doi: 10.1038/nature08670
Schnable, P. S., Ware, D., Fulton, R. S., Stein, J. C., Wei, F., Pasternak, S., et al. (2009). The B73 maize genome: complexity, diversity, and dynamics. Science 326, 1112–1115. doi: 10.1126/science.1178534
Serrano, I., Campos, L., Rivas, S. (2018). Roles of E3 ubiquitin-ligases in nuclear protein homeostasis during plant stress responses. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00139
Sharma, B., Taganna, J. (2020). Genome-wide analysis of the U-box E3 ubiquitin ligase enzyme gene family in tomato. Sci. Rep. 10, 9581. doi: 10.1038/s41598-020-66553-1
Shu, K., Yang, W. (2017). E3 ubiquitin ligases: ubiquitous actors in plant development and abiotic stress responses. Plant Cell Physiol. 58, 1461–1476. doi: 10.1093/pcp/pcx071
Stone, S. L. (2019). Role of the ubiquitin proteasome system in plant response to abiotic stress. Int. Rev. Cell Mol. Biol. 343, 65–110. doi: 10.1016/bs.ircmb.2018.05.012
Tamura, K., Stecher, G., Kumar, S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Tang, X., Ghimire, S., Liu, W., Fu, X., Zhang, H., Sun, F., et al. (2021). Genome-wide identification of U-box genes and protein ubiquitination under PEG-induced drought stress in potato. Physiologia Plantarum 174 (1), e13475. doi: 10.1111/ppl.13475
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., Higgins, D. G. (1997). The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. doi: 10.1093/nar/25.24.4876
Trenner, J., Monaghan, J., Saeed, B., Quint, M., Shabek, N., Trujillo, M. (2022). Evolution and functions of plant U-box proteins: from protein quality control to signaling. Annu. Rev. Plant Biol. 73, 93–121. doi: 10.1146/annurev-arplant-102720-012310
Trujillo, M. (2018). News from the PUB: plant U-box type E3 ubiquitin ligases. J. Exp. Bot. 69, 371–384. doi: 10.1093/jxb/erx411
Walter, W., Sánchez-Cabo, F., Ricote, M. (2015). GOplot: an R package for visually combining expression data with functional analysis. Bioinformatics 31, 2912–2914. doi: 10.1093/bioinformatics/btv300
Wang, N., Liu, Y., Cong, Y., Wang, T., Zhong, X., Yang, S., et al. (2016). Genome-wide identification of soybean U-box E3 ubiquitin ligases and roles of gmPUB8 in negative regulation of drought stress response in arabidopsis. Plant Cell Physiol. 57, 1189–1209. doi: 10.1093/pcp/pcw068
Wang, C., Song, B., Dai, Y., Zhang, S., Huang, X. (2021). Genome-wide identification and functional analysis of U-box E3 ubiquitin ligases gene family related to drought stress response in Chinese white pear (Pyrus bretschneideri). BMC Plant Biol. 21 (1), 235. doi: 10.1186/s12870-021-03024-3
Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40, e49. doi: 10.1093/nar/gkr1293
Wiborg, J., O'Shea, C., Skriver, K. (2008). Biochemical function of typical and variant Arabidopsis thaliana U-box E3 ubiquitin-protein ligases. Biochem. J. 413, 447–457. doi: 10.1042/BJ20071568
Wilkins, M. R., Gasteiger, E., Bairoch, A., Sanchez, J. C., Williams, K. L., Appel, R. D., et al. (1999). Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 112, 531–552. doi: 10.1385/1-59259-584-7:531
Yan, J., Wang, J., Li, Q., Hwang, J. R., Patterson, C., Zhang, H. (2003). AtCHIP, a U-box-containing E3 ubiquitin ligase, plays a critical role in temperature stress tolerance in Arabidopsis. Plant Physiol. 132, 861–869. doi: 10.1104/pp.103.020800
Yang, Y., Chai, Y., Liu, J., Zheng, J., Zhao, Z., Amo, A., et al. (2021b). Amino acid transporter (AAT) gene family in foxtail millet (Setaria italica L.): widespread family expansion, functional differentiation, roles in quality formation and response to abiotic stresses. BMC Genomics 22 (1), 519. doi: 10.1186/s12864-021-07779-9
Yang, C. W., González-Lamothe, R., Ewan, R. A., Rowland, O., Yoshioka, H., Shenton, M., et al. (2006). The E3 ubiquitin ligase activity of arabidopsis PLANT U-BOX17 and its functional tobacco homolog ACRE276 are required for cell death and defense. Plant Cell 18, 1084–1098. doi: 10.1105/tpc.105.039198
Yang, Y., Wang, X., Zheng, J., Men, Y., Zhang, Y., Liu, L., et al. (2022). Amino acid transporter (AAT) gene family in Tartary buckwheat (Fagopyrum tataricum L. Gaertn.): Characterization, expression analysis and functional prediction. Int. J. Biol. Macromol 217, 330–344. doi: 10.1016/j.ijbiomac.2022.07.059
Yang, G., Ying, G., Wang, Z., Pan, W., Linghu, B., Pan, Y., et al. (2021a). Genome-wide identification and expression analysis of U-box gene family in wild emmer wheat (Triticum turgidum L. ssp. dicoccoides). Gene 799, 145840. doi: 10.1016/j.gene.2021.145840
Zeng, L. R., Park, C. H., Venu, R. C., Gough, J., Wang, G. L. (2008). Classification, expression pattern, and E3 ligase activity assay of rice U-box-containing proteins. Mol. Plant 1, 800–815. doi: 10.1093/mp/ssn044
Zhang, G., Zhang, M., Zhao, Z., Ren, Y., Li, Q., Wang, W. (2017). Wheat TaPUB1 modulates plant drought stress resistance by improving antioxidant capability. Sci. Rep. 7, 7549. doi: 10.1038/s41598-017-08181-w
Zhou, J., Hu, Y., Li, J., Yu, Z., Guo, Q. (2021). Genome-wide identification and expression analysis of the plant U-box protein gene family in phyllostachys edulis. Front. Genet. 12. doi: 10.3389/fgene.2021.710113
Keywords: U-box, biotic stress, expression analysis, phylogenetic analysis, Nicotiana tabacum L., Solanum melongena L.
Citation: Chen R, Gu G, Zhang B, Du C, Lin X, Cai W, Zheng Y, Li T, Wang R and Xie X (2024) Genome-wide identification and expression analysis of the U-box E3 ubiquitin ligase gene family related to bacterial wilt resistance in tobacco (Nicotiana tabacum L.) and eggplant (Solanum melongena L.). Front. Plant Sci. 15:1425651. doi: 10.3389/fpls.2024.1425651
Received: 30 April 2024; Accepted: 12 July 2024;
Published: 30 July 2024.
Edited by:
Zhaorong Hu, China Agricultural University, ChinaReviewed by:
Qibin Wu, Chinese Academy of Tropical Agricultural Sciences, ChinaCopyright © 2024 Chen, Gu, Zhang, Du, Lin, Cai, Zheng, Li, Wang and Xie. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaofang Xie, eHhmMzE3QGZhZnUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.