
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 25 June 2024
Sec. Plant Symbiotic Interactions
Volume 15 - 2024 | https://doi.org/10.3389/fpls.2024.1414193
This article is part of the Research Topic Use of Biostimulants in Beneficial Plant-Microbe Interactions View all 9 articles
Parts of this article's content have been modified or rectified in:
Erratum: The growth-promoting and disease-suppressing mechanisms of Trichoderma inoculation on peanut seedlings
 Xingqiang Wang1,2†
Xingqiang Wang1,2† Zhongjuan Zhao1,2†
Zhongjuan Zhao1,2† Hongmei Li1,2
Hongmei Li1,2 Yanli Wei1,2
Yanli Wei1,2 Jindong Hu1,2
Jindong Hu1,2 Han Yang1,2
Han Yang1,2 Yi Zhou2,3*
Yi Zhou2,3* Jishun Li1,2*
Jishun Li1,2*Trichoderma spp. is known for its ability to enhance plant growth and suppress disease, but the mechanisms for its interaction with host plants and pathogens remain unclear. This study investigated the transcriptomics and metabolomics of peanut plants (Arachis hypogaea L.) inoculated with Trichoderma harzianum QT20045, in the absence and presence of the stem rot pathogen Sclerotium rolfsii JN3011. Under the condition without pathogen stress, the peanut seedlings inoculated with QT20045 showed improved root length and plant weight, increased indole acetic acid (IAA) production, and reduced ethylene level, with more active 1-aminocyclopropane-1-carboxylate acid (ACC) synthase (ACS) and ACC oxidase (ACO), compared with the non-inoculated control. Under the pathogen stress, the biocontrol efficacy of QT20045 against S. rolfsii was 78.51%, with a similar effect on plant growth, and IAA and ethylene metabolisms to the condition with no biotic stress. Transcriptomic analysis of peanut root revealed that Trichoderma inoculation upregulated the expression of certain genes in the IAA family but downregulated the genes in the ACO family (AhACO1 and AhACO) and ACS family (AhACS3 and AhACS1) consistently in the absence and presence of pathogens. During pathogen stress, QT20045 inoculation leads to the downregulation of the genes in the pectinesterase family to keep the host plant’s cell wall stable, along with upregulation of the AhSUMM2 gene to activate plant defense responses. In vitro antagonistic test confirmed that QT20045 suppressed S. rolfsii growth through mechanisms of mycelial entanglement, papillary protrusions, and decomposition. Our findings highlight that Trichoderma inoculation is a promising tool for sustainable agriculture, offering multiple benefits from pathogen control to enhanced plant growth and soil health.
Stem rot disease in peanuts (Arachis hypogaea L.), caused by a soil-borne necrotrophic fungus Sclerotium rolfsii Sacc, is prevalent in the world’s major peanut-producing regions (Subedi et al., 2024), including China, India, US, and Argentina (Bot et al., 2011), and causes yield loss of up to 80% (Yan et al., 2021). S. rolfsii secretes an exceptionally large quantity of polygalacturonases, cutinase, and cellulase enzymes that break down the primary and secondary cell wall components of host cells (Ayyandurai et al., 2023). The disease shows up as the yellowing and dropping of the lower branches on the main stem as well as poor root growth, stem rot, and early death (Aydoğdu, 2023 ). This pathogen can also form sclerotia in the soil or in host plant debris for a long period, leading to continued infections (Schmid et al., 2010); hence, agricultural practices such as crop rotation, deep plowing, and residue removal were ineffective to control the stem rot disease (Jia et al., 2023). Chemical control including the use of fungicides such as tebuconazole, difenoconazole, and chlorothalonil (Cilliers et al., 2003) serves as the primary method to control this disease, but the extensive use of pesticides leads to soil pollution and pathogen resistance—for example, the United States Environmental Protection Agency has labeled tebuconazole as a potential carcinogen for humans (Cui et al., 2018). Tebuconazole has been observed to move from soil into aquatic systems via runoff and frequently detected at 0.010–0.115 μg/L in US streams (Bradley et al., 2017). Therefore, using novel biocontrol agents with effective antagonistic properties is considered as a sustainable approach for the management of peanut stem rot disease (Whipps, 2004).
Trichoderma spp. has been widely used as a beneficial fungus, which exhibits a strong capacity to promote host plant growth and suppress disease infection (López-Bucio et al., 2015; Zhou et al., 2020; Liu et al., 2022). Trichoderma strains stimulate plant growth, including seed germination, root elongation, flowering time, and fruit formation by modulating the plant hormone metabolism and signaling (Viterbo et al., 2010). Firstly, auxin-related compounds produced by T. virens played a great role in modulating the root architecture and activation of auxin-regulated gene expression in Arabidopsis plants (Contreras-Cornejo et al., 2009). In addition, the presence of the 1-aminocyclopropane-1-carboxylate acid (ACC) deaminase gene in Trichoderma species genomes leads to the regulation of ethylene levels in plants, and ethylene biosynthesis acts as a key regulator between environmental shifts and plant adaptive responses (Pieterse et al., 2012)—for example, the study on wheat seedling under salt stress revealed that the ACC deaminase activity from T. longibrachiatum strain increased the plant IAA concentration by 11%, decreased the ACC content by 22%, and reduced the ethylene level by 12% (Zhang et al., 2019).
Under biotic stress, Trichoderma is also an important biocontrol agent against pathogenic fungus. The antagonistic mechanism of Trichoderma against pathogens mainly includes mycoparasitism and inhibitory effect from secreting the antimicrobial compounds. In the mycoparasitism process, Trichoderma firstly recognizes the oligochitins in the pathogen’s cell wall and then grows toward the pathogen by chemotropism. Upon physical contact, the hyphae of Trichoderma wrap around those of the pathogen, triggering the release of enzymes that degrade the cell walls, such as chitinases and β-1,3-glucanases (Sood et al., 2020). Meanwhile, Trichoderma can also release the volatile organic compounds (VOCs) acting as antagonistic agents (Kanchiswamy et al., 2015). The study by which Trichoderma spp. (T. longibrachiatum and T. asperellum) was co-cultured with S. rolfsii found that the specific antimicrobial VOCs of isolongifolan-7-ol and transsesquisabinene hydrate showed antifungal activity against S. rolfsii (Ayyandurai et al., 2023). However, there is insufficient understanding regarding how Trichoderma influences plant hormone metabolism during its interaction with pathogens.
Trichoderma spp. not only directly interacts with phytopathogens but also reprograms the gene expression of the host plants, thereby stimulating defense mechanisms to suppress the pathogen (Lorito et al., 2010; Druzhinina et al., 2011; Ray et al., 2016). The plant response induced by Trichoderma inoculation was stronger than that of immunity triggered by pathogens (Cornejo et al., 2011). Even during the early stages of plant growth, there exists a substantial communication between the host plant and Trichoderma. Plant seedlings subjected to Trichoderma treatment, either with crude mycelium or purified proteins such as chitinase and cellulase, displayed various cellular alterations linked to defense mechanisms (Sriram et al., 2009), including rapid ion fluxes, the production of reactive oxygen compounds, the accumulation of phytoalexins, and the synthesis of pathogenesis-related proteins (Abbas et al., 2022).
For a legume plant which is able to form symbiosis with the rhizobia, there is an interaction between disease infection and symbiosis on the transcriptional response of the host plant, e.g., metabolism and signaling pathways (Maheshwari et al., 2021; Zhou et al., 2022). However, the impact of Trichoderma inoculation on the metabolism of legume plants in the presence and the absence of pathogen infections remained largely unknown. This study focuses on the hormone (IAA and ethylene) metabolism pathways of peanut host responding to the T. harzianum inoculation with and without the stress of S. rolfsii pathogen. The research objective is to evaluate the mechanisms of T. harzianum inoculation that promoted peanut growth under non-stress conditions and the mechanisms by which T. harzianum inoculation suppressed the S. rolfsii pathogen and modulated host plant metabolisms to resist disease.
Trichoderma harzianum strain QT20045 was used in this study. This strain was obtained from our Trichoderma Resource Bank in the Ecological Institute of Shandong Academy of Sciences, NCBI accession no. MH284507 (Hu et al., 2020). Sclerotium rolfsii JN3011 was isolated from diseased peanuts with stem rot collected from the peanut fields in Jinan City, China. The CAS number and brand of the key chemicals used in this article are listed in Supplementary Table S3.
The antagonistic effect of T. harzianum QT20045 against S. rolfsii was evaluated under in vitro condition using the dual culture method. Three treatments including T. harzianum QT20045 (+T-S), S. rolfsii (-T+S), and QT20045 and S. rolfsii dual culture (+T+S) were studied as a completely randomized design with three experimental replicates. T. harzianum QT20045 and S. rolfsii were purely cultured in potato dextrose agar (PDA) medium at 25°C for 4 days. The mycelium disc from each culture was co-inoculated on the edges of the PDA plate, which was then incubated at 25°C. The mycelium disc of individual fungus was also inoculated in separate PDA plates as the control. The percentage of inhibition was calculated as the relative increase for the diameter of pathogenic fungus without Trichoderma to the diameter of pathogenic fungus in the treatment with Trichoderma.
The conidia suspension of T. harzianum QT20045 was obtained by spores from the individual culture and dual culture with S. rolfsii, respectively. The procedure began by pouring 3 mL of sterilized distilled water onto the dishes, and the suspension was collected. The concentration of conidia was evaluated using a hemocytometer and then adjusted to reach a concentration of 1 × 108 CFU/mL. The Trichoderma suspension was used for the analysis of IAA production and ACC deaminase activity according to the method from Penrose and Glick (2010), respectively. The concentration of IAA was determined by colorimetric method with OD530 and calculated by comparison with the IAA standard curve. The total protein concentration in extract was determined by a dye-binding method using bovine serum albumin as the protein standard (Kruger, 2009). The activity of ACC deaminase was quantitatively evaluated by measuring the amount of α-ketobutyrate produced, expressed as µmol α-ketobutyrate produced per milligram of protein per hour.
The biocontrol efficacy of QT20045 against S. rolfsii was in vivo evaluated in peanut plants growing under greenhouse conditions. The experiment was carried out with four treatments and five replicates using a completely randomized design. Four treatments included inoculating with T. harzianum QT20045 and S. rolfsii and the non-inoculated controls (+T-S, -T-S, +T+S, and -T+S). The peanut seeds (cultivar “Luhua 19” provided by Shandong Luhua Agricultural Science and Technology Promotion Co. Ltd.) were sterilized in 70% ethanol for 2 min and 2.0% NaClO for 2 min and then washed with sterile water three times. The disinfected seeds were soaked in sterilized water or QT20045 conidia suspension (1 × 108 CFU/mL) for 1 h and then incubated at 25°C for 2 days. The germinated seeds were planted in pots (115-mm high × 92-mm wide × 114-mm long) containing 1 kg of sterilized soil with three seedlings in each pot and cultivated in the greenhouse at 28°C in the daytime and 18°C at night. The pathogenic fungal solution was prepared by culturing in PDB at 180 rpm for 7 days, and 20 mL solution was watered to the peanut seedlings’ root. The plants were harvested at 20 days after pathogen infection when the vegetation growth reached the maximum. Plant height was measured, and roots were washed with sterilized water for the further analysis. The disease incidence (percentage of plants showing disease symptoms) of peanut plants was assessed, and the severity of stem rot disease was scored visually on a scale of 0–4 based on the percentage of affected leaves and twigs: 0 indicated no symptoms, 1 represented 1%–33% affected, 2 indicated 34%–66% affected, 3 represented 67%–100% affected, and 4 indicated a dead plant (Carrero-Carron et al., 2016). The disease index (DI) of each treatment was calculated using the Townsend–Heuberger formula (Townsend and Heubergeb, 1943) as below:
where r is the rating value (0–4), Nr is the number of disease plants with a rating of r, and N is the total number of peanut plants.
The control efficacy was calculated using Abbott’s formula (Abbott, 1925): the relative increase for the disease severity without QT20045 (infected with S. rolfsii) to the disease severity in QT20045 treatment (infected with S. rolfsii).
The endogenous IAA was extracted from plant roots and purified using the modified method from Alemneh et al. (2021). Root samples (1 g) were frozen immediately after harvest and then homogenized with a mortar and pestle using 80% methanol for IAA extraction. High-performance liquid chromatography (HPLC; model Agilent 1100, Agilent Technologies, Waldbronn, Germany), utilizing a C18 column with a 5-µm particle size and an ultraviolet (UV) detector with a wavelength of 221 nm, was used for IAA detection. In the mobile phase, methanol/acetonitrile/acetic acid (0.1%), in a 60:5:35 ratio, was applied at a flow rate of 0.8 mL/min and column temperature of 30°C. The ethylene synthesis in peanut seedlings was measured following the method of Yamauchi et al. (2014). Fresh roots (1 g) were placed into a 20-mL airtight glass bottle with 10 mL water to maintain root moisture. After incubating, 1 mL gas was collected from the bottle and analyzed through gas chromatography–mass spectrometry (GC–MS, Agilent 7890B; Agilent, Foster City, CA, USA). The concentration of ethylene in a standard sample was determined to make the standard curve. Ethylene production was expressed in µmol per gram of root fresh weight per hour.
The activity of ACS was detected by modifying the method of Kato et al. (2000). Peanut root samples (0.3 g) were ground in liquid nitrogen, then transferred to a 2-mL centrifuge tube with 1.5 mL of a mixed solution containing 100 mM HEPPS-KOH extraction buffer (pH 8.5), 10 mM β-sulfhydryl ethanol, 10 µM pyridoxal phosphate, and 1 mM EDTA, and centrifuged at 12,000 rpm and 4°C for 20 min. The supernatant, as the crude enzyme extract, was determined by a dye-binding method using bovine serum albumin as the protein standard (Kruger, 2009). After that, 0.7 mL of the supernatant was transferred into a headspace vial with 2 mL of a mixed solution containing 50 mM HEPPS-KOH reaction buffer (pH 8.5), 250 µM S-adenosyl-L-methionine (SAM), and 10 µM pyridoxal phosphate and sealed with a silicone stopper. After incubation, 1 mL of gas was collected from the headspace vial to quantify the ethylene content using GC–MS. ACS activity was determined as the amount of ethylene converted from SAM during the reaction period and expressed as µmoles ACC per gram protein per hour.
The activity of ACC oxidase (ACO) was detected following the method described by Nguyen et al. (2023). Firstly, 0.3 g of peanut roots was ground in liquid nitrogen, then transferred to a headspace vial containing 4 mL of a solution containing 100 mM MOPS buffer (pH 7.2), 1 mM ACC, 1 mM ascorbic acid, and 200 µM FeSO4. To provide the CO2 for reaction, 50 mL of 30 mM NaHCO3 was injected into the bottle. The ethylene content in the gas was analyzed using GC–MS. The ACO activity was determined by quantifying the ethylene converted from ACC during the reaction, expressed as µmoles ACC per gram of protein per hour.
Total RNA was isolated from 0.1 g root samples (ground into powder in liquid nitrogen) using RNA prep Pure Plant Plus Kit (Norgen Biotek, Canada) according to the manufacturer’s instructions. RNA concentration and quality were determined using a NanoPhotometer spectrophotometer (Thermo Fisher Scientific, USA). mRNA was isolated from total RNA using magnetic beads attached to poly-T oligos. Fragmentation was conducted at high temperature with divalent cations in NEBNext’s First Strand Synthesis Reaction Buffer (5X). Random hexamer primers and M-MuLV Reverse Transcriptase (RNase H-) were employed to synthesize first-strand cDNA. Subsequently, second-strand cDNA synthesis was carried out using DNA Polymerase I and RNase H. The sequencing libraries were generated using NEBNext® UltraTM RNA Library Prep Kit for Illumina® (Illumina, USA) following the manufacturer’s recommendations. Sequencing was performed using the Illumina Novaseq PE150 platform. The raw data were quality-controlled using Fastp (version 0.23.1) (Chen et al., 2018) to produce clean data. The reference genome was built using Hisat2 (version 2.2.1) (Kim et al., 2015), and paired-end clean reads were aligned to the reference genome. Validated data were compared to the assembled transcriptome sequence, and FeatureCounts (version 2.0.1) (Liao et al., 2013) was used to determine the expression levels. DESeq2 (version 1.26.0) (Love et al., 2014) in R package was used for differential gene expression analysis, and genes with log fold change ≥1 and adjusted P-value<0.05 (the resulting P-values were adjusted using Benjamini and Hochberg’s approach to control the false discovery rate) were considered significantly differentially expressed genes (DEGs) (Jung et al., 2011). We used ClusterProfiler (version 3.4.4) (Yu et al., 2012) in R package for GO enrichment analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis (Gao et al., 2024).
We selected two of the most representative genes, meeting the criteria of log fold change >1 and adjusted P-value<0.05, from each of the IAA, ACO, and ACS families and one gene from each of the PR1, PME, and DFR families related to plant disease resistance. In total, the expression of nine selected DEGs was analyzed by RT-qPCR to validate the RNA-seq data. Primers with lengths that ranged from 200 to 300 bp were designed using Primer 5.0 software (Bai and Wong, 2004) (the primer sequences are listed in Supplementary Table S1). First-strand cDNA was reverse-transcribed from the extracted total RNA of the peanut roots for RNA-seq using the Evo M-MLV RT Premix kit (Promega, USA) according to the manufacturer’s instructions. RT-qPCR was performed using a CFX96 optical real-time detection system (Bio-Rad Laboratories Inc, USA). The 20-µL reactions contained 10 µL 2xPower SYBR® Green PCR Master Mix (Thermo Fisher Scientific, USA), 0.4 µL each of 10 µM forward and reverse primers, 2.0 µL cDNA samples, and 7.2 µL sterile and DNA-free water. The PCR program was as follows: pre-denaturation at 95°C for 30 s, denaturation at 95°C for 5 s, annealing at 54°C for 30 s, and extension at 72°C for 20 s, for a total of 40 cycles. Amplicon dissociation curves were recorded after cycle 40 by heating from 60°C to 95°C with a ramp speed of 1.0°C per min. All reactions were performed with three technical replicates for each of the biological replicates. Actin-7 was used as the internal reference gene, and the relative expression was calculated using the method from Schmittgen and Livak (2008).
After confirming the normal distribution of the measured variables, one-way analysis of variance (ANOVA) for a completely randomized design, with three replicates for the variables in the in vitro experiment and five replicates for the variables in the pot experiment, was performed using SPSS (version 26.0), and Duncan’s multiple-range test was applied to compare the different treatments at p<0.05.
In vitro test on PDA medium showed that T. harzianum QT20045 suppressed the growth of S. rolfsii (Figure 1A), and the percentage of inhibition was 97.60% ± 2.92%. Morphological changes of the S. rolfsii colony, when cultured with T. harzianum QT20045, were observed using an optical microscope (Figure 1B). The interactions between S. rolfsii and T. harzianum QT20045 include mycelial entanglement (Figure 1C), papillary protrusions (Figure 1D), and decomposition (Figure 1E).

Figure 1. Trichoderma harzianum QT20045 inhibits the mycelial growth of Sclerotium rolfsii. Two fungi were cultured in potato dextrose agar medium (A) with the treatments of -T+S (S. rolfsii individually), +T+S (S. rolfsii dual-cultured with T. harzianum QT20045), and +T-S (T. harzianum QT20045 individually). Morphological characteristics of S. rolfsii mycelia cultured without T. harzianum QT20045 (B) and interacted with T. harzianum QT20045 including entanglement (C), papillary protrusions (D), and decomposition (E).
The IAA concentration and ACC deaminase activity of T. harzianum QT20045 were measured under individual culture and dual culture with S. rolfsii (Figure 2). Compared with the individual culture, T. harzianum QT20045 under the dual culture decreased its IAA production by 27.46% (Figure 2A, p< 0.05) and ACC deaminase activity by 12.34% (Figure 2B, p< 0.05).
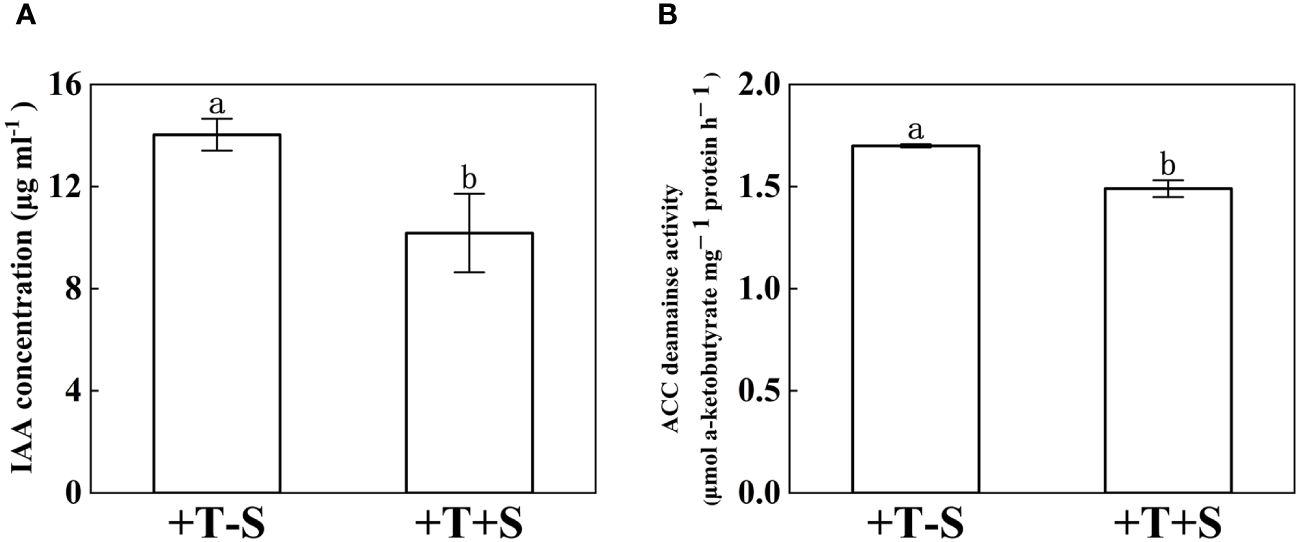
Figure 2. Indole acetic acid concentration (A) and 1-aminocyclopropane-1-carboxylate acid-deaminase activity (B) of T. harzianum QT20045 under individual culturing (+T-S) and dual-culturing with S. rolfsii (+T+S). The bars represent the average of the biological replicates (n = 3) with standard errors. Different letters denote significant differences at p< 0.05 by Duncan’s new multiple-range test.
The biocontrol efficacy of T. harzianum QT20045 against S. rolfsii was investigated under pot condition in the soil (Figure 3A). The result showed that the peanut seedlings treated with QT20045 displayed promoted growth when no pathogen was added and less disease infection when the pathogen was inoculated (Figure 3, Supplementary Table S1).
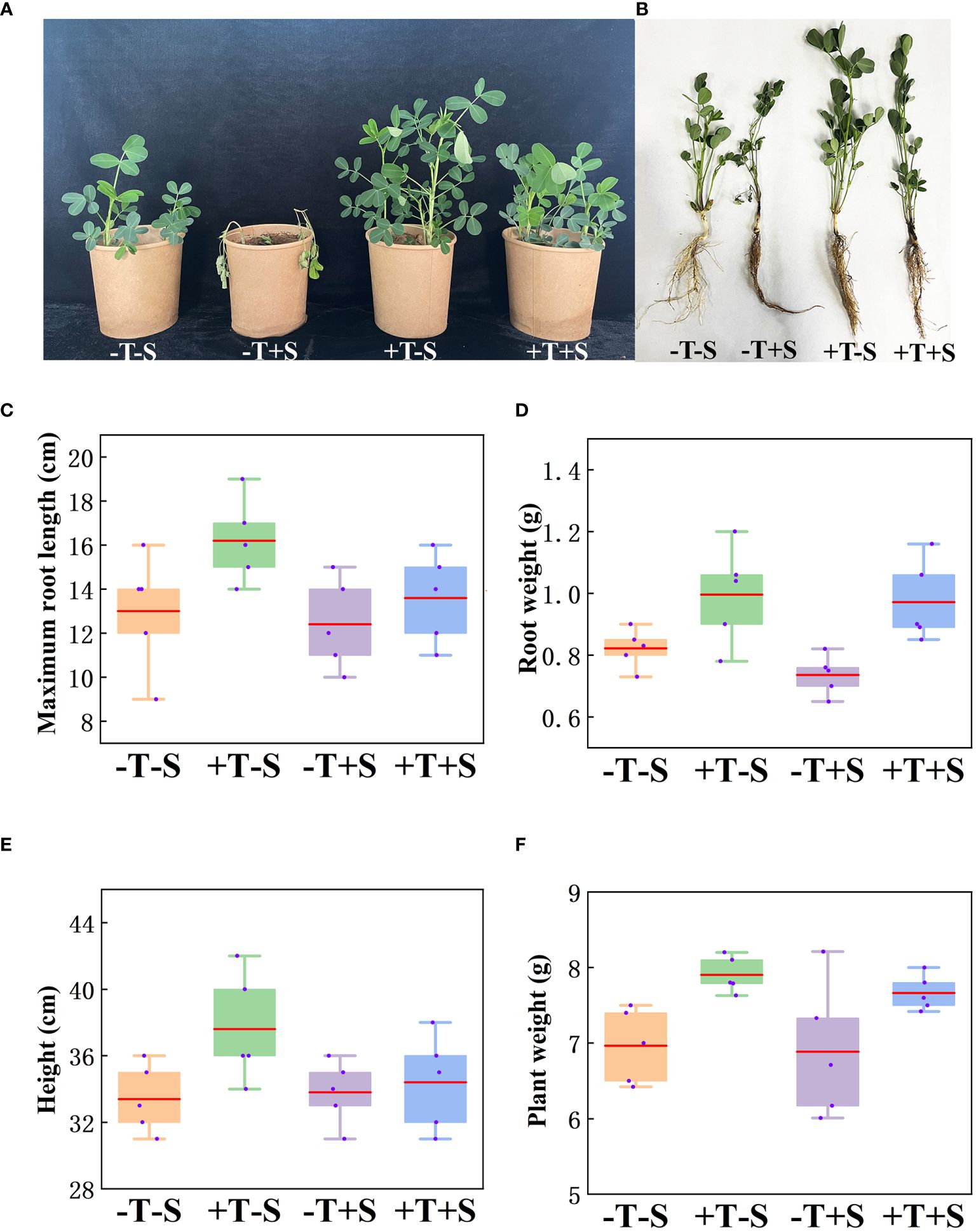
Figure 3. Growth-promoting and biocontrol effect of T. harzianum QT20045 on peanut seedlings after 20 days of growth, including the observed plant shoots (A) and roots (B) and the measured maximum root length (C), root weight (D), plant height (E), and plant weight (F). The experimental treatments included the combinations of inoculation with T. harzianum QT20045 (+T and -T) and the pathogen S. rolfsii (+S and -S).
Under no pathogen inoculation, compared with the control (-T-S), the peanut plants treated with QT20045 (+T-S) displayed an obvious increase in maximum root length by 24.62%, root weight by 21.17%, plant weight by 13.50%, and plant height by 12.57% (Figures 3C–F). When a pathogen was present, compared with the control (-T+S), the peanut plants treated with QT20045 (+T+S) also significantly improved the maximum root length by 9.68%, root weight by 29.44%, plant weight by 11.30%, and plant height by 5.52% (Figures 3C–F). The disease incidence and disease index of peanut plants infected with S. rolfsii reached 61.27% and 36.19%, respectively, and T. harzianum QT20045 remarkably reduced them, resulting in the control efficiency of 78.51% (Supplementary Table S1).
Under no pathogen inoculation, the peanut seedlings treated with T. harzianum QT20045 (+T-S) produced more IAA (Figure 4A) and less ethylene (Figure 4B) compared with the non-Trichoderma-inoculated control (-T-S). Under the biotic stress of S. rolfsii, the application of T. harzianum QT20045 (+T+S) also increased the IAA production by 48.08% (Figure 4A) and decreased ethylene release by 25.18% (Figure 4B) in the roots of peanut plants compared with the control (-T+S).
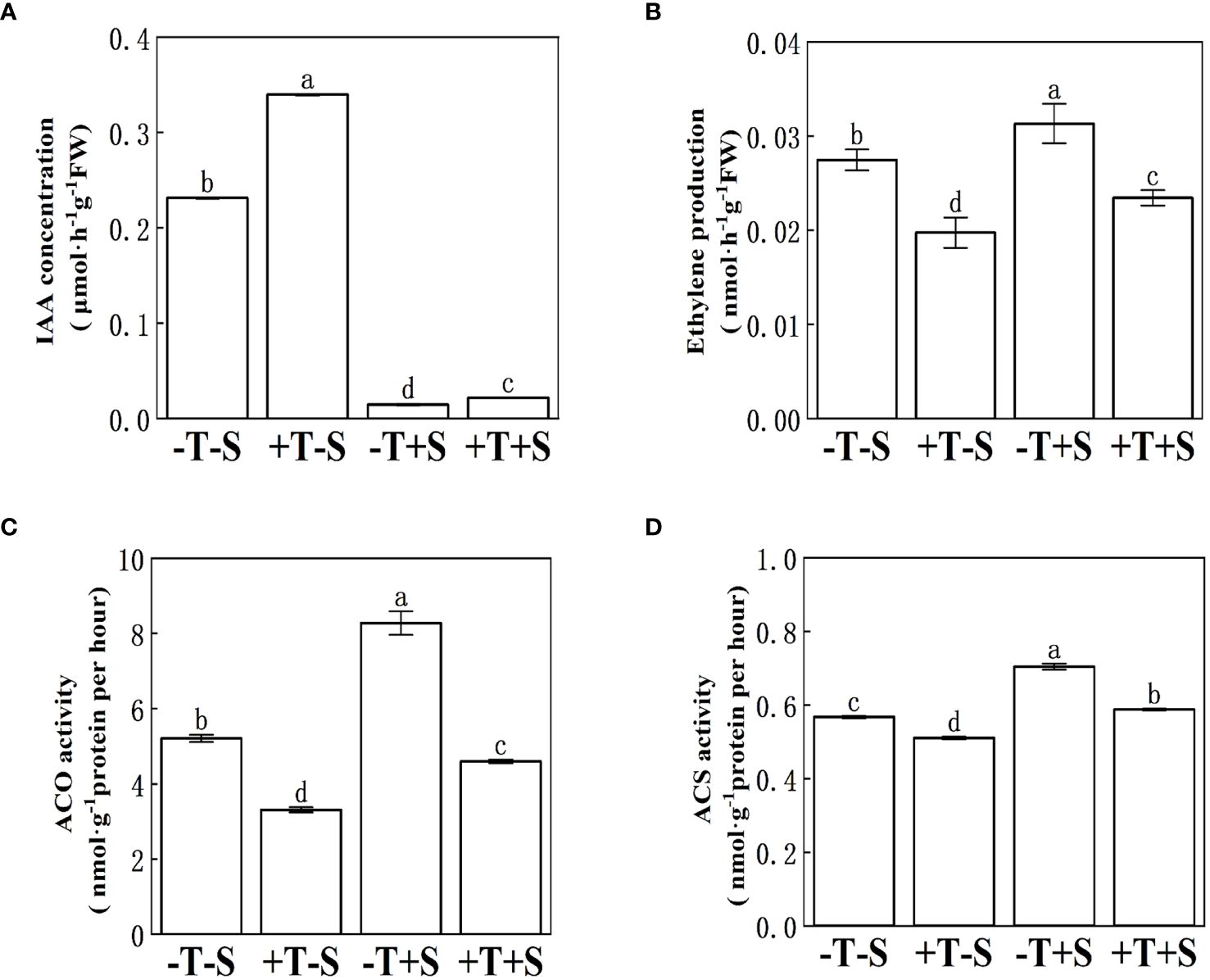
Figure 4. Effect of T. harzianum QT20045 inoculation on indole acetic acid content (A), ethylene production (B), 1-aminocyclopropane-1-carboxylate acid oxidase activity (C), and 1-aminocyclopropane-1-carboxylate acid synthase activity (D) in the roots of peanut seedlings. The experimental treatments included the combinations of inoculation with T. harzianum QT20045 (+T and -T) and the pathogen S. rolfsii (+S and -S). The bars represent the average of the biological replicates (n = 5) with standard errors. Different letters denote significant difference at p< 0.05 by Duncan’s new multiple-range test.
The effect of T. harzianum QT20045 inoculation on the ACS activity and ACO activity in the roots of peanut seedlings (Figures 4C, D) was consistent with its influence in ethylene release (Figure 4B). Compared with the non-inoculated control, T. harzianum QT20045 significantly reduced the ACS activity and ACO activity of the peanut plants when the pathogen was not (Figures 4C, D).
In order to analyze the genes related to the response of peanut plants to T. harzianum QT20045 (+T and -T) and S. rolfsii (+S and -S), RNA-seq was carried out using the total RNA of peanut seedlings. In total, 42.7, 43.1, 42.2, and 40.7 million reads with a length of 143–150 bp were obtained from -T-S, +T-S, -T+S, and +T+S treatments, respectively.
To focus on the effect of T. harzianum QT20045 on the transcriptomes of peanut plants, we selected two crucial comparison groups: +T-S vs. -T-S and +T+S vs. -T+S. Gene expression analysis showed that 336 DEGs were identified in +T-S vs. -T-S comparison, including 237 DEGs being upregulated and 99 DEGs being downregulated, and 4,317 DEGs were identified in +T+S vs. -T+S comparison, including 1,420 up-regulated DEGs and 2,897 downregulated DEGs (Figure 5A).
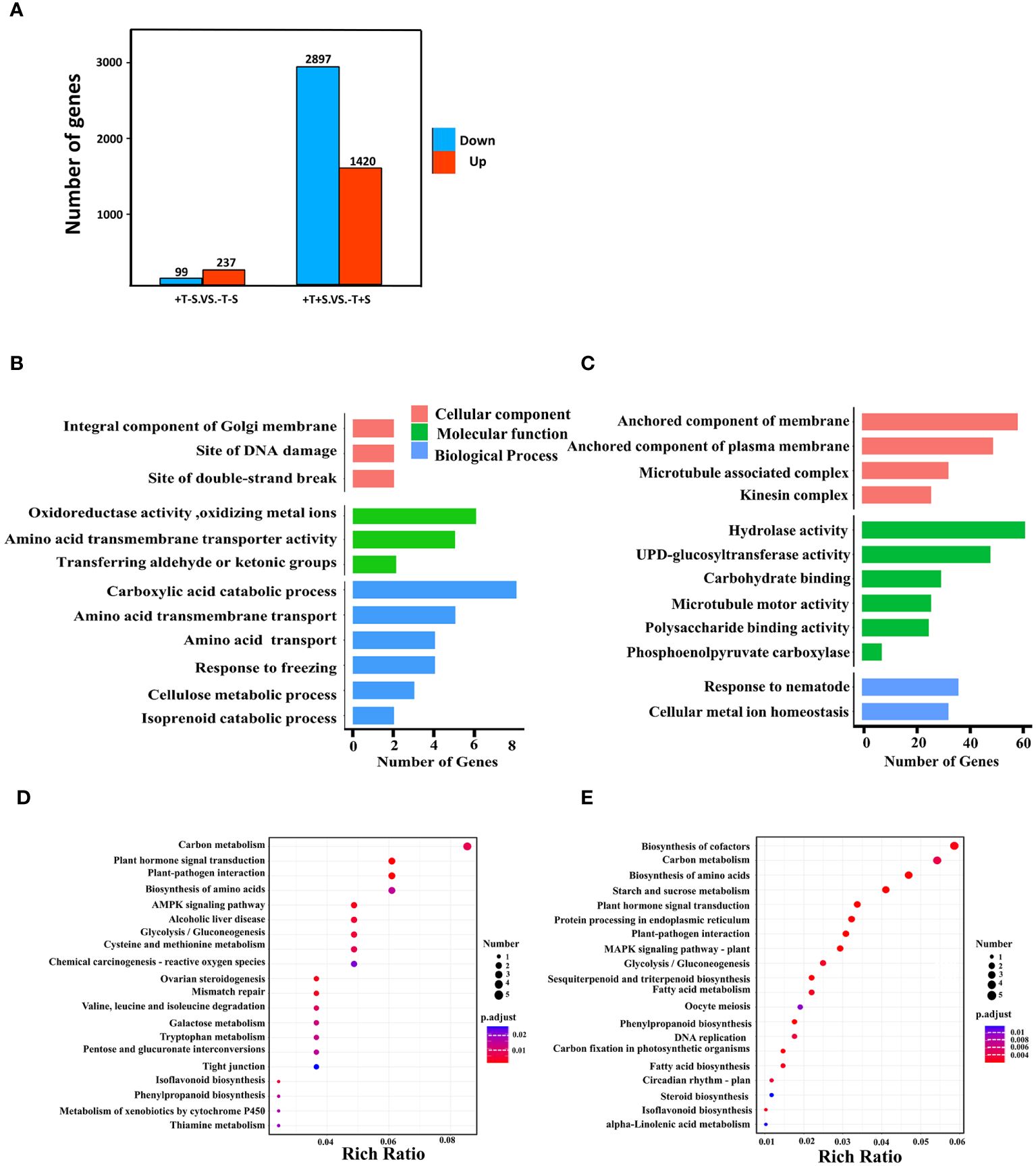
Figure 5. Effect of T. harzianum QT20045 inoculation (+T and -T) on root transcriptomes of peanut seedling in the absence and presence of the pathogen S. rolfsii (+S and -S) with focus on two comparisons +T-S vs. -T-S and +T+S vs. -T+S. Number of upregulated and downregulated differentially expressed genes (DEGs) in two comparisons (A). GO terms significantly enriched in biological processes, cellular components, and molecular functions for +T-S vs. -T-S comparison (B) and +T+S vs. -T+S comparison (C). KEGG pathway enrichment scatter map for +T-S vs. -T-S comparison (D) and +T+S vs. -T+S comparison (E). Rich ratio is the ratio of the DEGs number to all the gene numbers annotated in the pathway.
GO analysis revealed that all DEGs fell into three functional categories: biological process, cellular component, and molecular function. In the comparison of +T-S vs. -T-S, DEGs were significantly enriched in the functional groups of biological processes (e.g., carboxylic acid catabolism and amino acid transmembrane transport), molecular functions (e.g., oxidoreductase activity and amino acid transmembrane transporter activity), and cellular components (e.g., integral components of the Golgi membrane, DNA damage sites, and double-strand break sites; Figure 5B). In the +T+S vs. -T+S comparison, DEGs were significantly enriched in the pathways of biological processes including responses to nematodes and cellular metal ion homeostasis, molecular functions such as hydrolase activity and UDP-glucosyltransferase activity, and cellular components related to anchored membrane (Figure 5C).
To further explore DEG functions, we conducted a KEGG enrichment analysis. We identified 284 DEGs across 178 pathways from the +T-S vs. -T-S comparison. The top pathways included carbon metabolism (seven DEGs; two upregulated and five downregulated), plant hormone signal transduction (five DEGs; four upregulated and one downregulated), and plant–pathogen interaction (five DEGs; three up regulated and two down regulated) (Figure 5D). For the +T+S vs. -T+S comparison, 2,200 DEGs distributed across 378 pathways were identified, including the key pathways for biosynthesis of cofactors (40 DEGs; seven upregulated and 33 downregulated), carbon metabolism (37 DEGs; six upregulated and 31 downregulated), and biosynthesis of amino acids (32 DEGs; five upregulated and 27 downregulated) (Figure 5E).
When annotating the DEGs, we found that most of them were enriched in functions related to the modulation of plant growth and resistance to plant diseases. For the effect of QT20045 on plant growth in the absence of the pathogen, the IAA-related family (including the genes AhIAGLU: LOC_20541, AhIAA9: LOC_62526, AhIAGLU: LOC_21136, AhAUX28: LOC_71520) was significantly upregulated by Trichoderma inoculation. In contrast, the expression of the ACO family (AhACO1: LOC_02019, AhACO: LOC_64097), which has ACO activity and catalyzes the formation of ethylene from pyruvic acid, was significantly downregulated. Moreover, the ACS family (AhACS3: LOC_20599, AhACS1: LOC_97527) and the GRF family (AhUGT76F1: LOC_15453, AhUGT76B1: LOC_65278) were also downregulated by Trichoderma inoculation (Figure 6A).
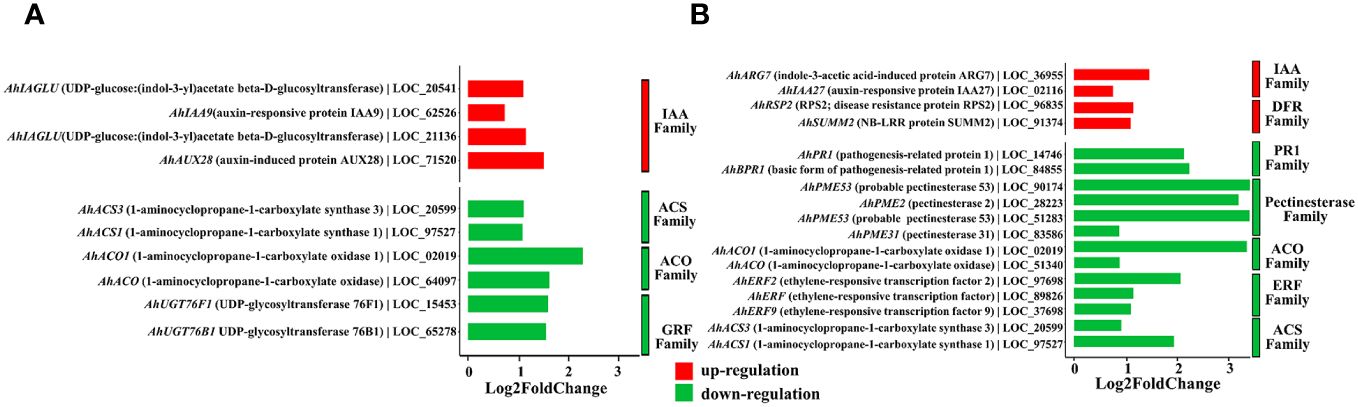
Figure 6. Differentially expressed genes of peanut seedling induced by T. harzianum QT20045 inoculation in the absence (A) (T-S vs. -T-S) and presence (B) (+T+S vs. -T+S) of the pathogen S. rolfsii.
Under the biotic stress of stem rot disease, significant upregulation by Trichoderma inoculation was observed in the IAA family (AhARG7: LOC_36955, AhIAA27: LOC_020016) and DFR family (AhSUMM2: LOC_91374, AhRSP2: LOC_96835) in the transcriptomes of peanut seedlings. Conversely, the pectinesterase family (AhPME31: LOC_83586, AhPME53: LOC_51283, AhPME2: LOC_28223, AhPME53: LOC_90174), another set of ACO-associated genes (AhACO1: LOC_02019, AhACO: LOC_64097), and the genes in the PR1 family (AhPR1: LOC_14746, AhBPR1: LOC_84855), the ERF family (AhERF2: LOC_97698, AhERF: LOC_89826, AhERF9: LOC_37698), and the ACS family (AhACS3: LOC_20599, AhACS1: LOC_97527) were significantly downregulated (Figure 6B).
To validate the RNA-seq data, nine DEGs were chosen to quantify the expression level using RT-qPCR (Figure 7, Supplementary Table S2). The RT-qPCR analysis confirmed that the expression patterns of all nine DEGs were consistent with the RNA-seq data (Figure 7). These findings validate the accuracy of the transcriptome data and confirm the association of these genes with the plant growth promotion and disease suppression effects from the strain QT20045.
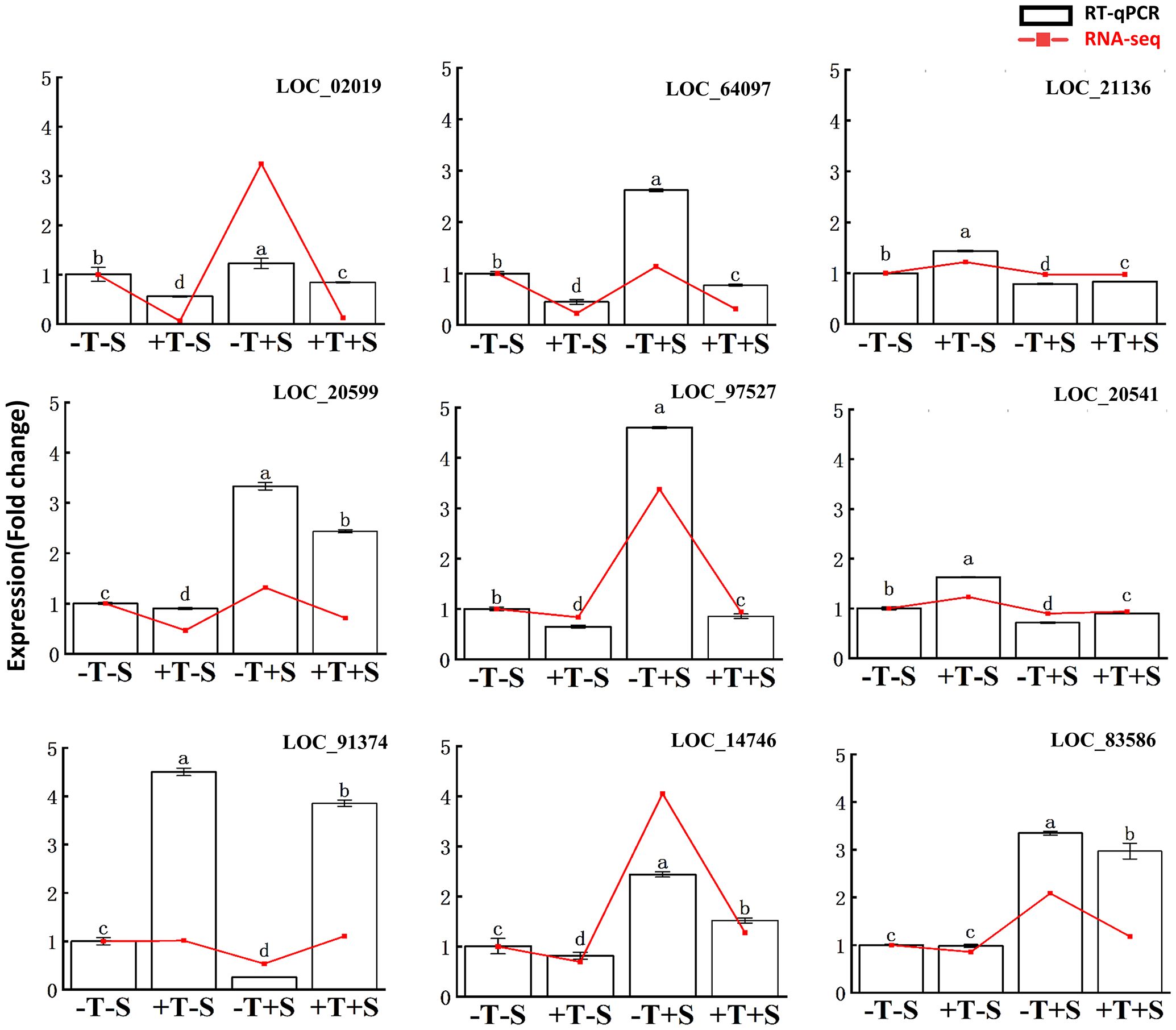
Figure 7. Reverse transcription-quantitative PCR validation of differentially expressed genes in the transcriptomes of peanut root. The relative change of gene expression in two methods was calculated for comparison. The bars represented the average of the biological replicates (n = 5) with standard errors. Different letters denote significant differences at p< 0.05 by Duncan’s new multiple-range test.
The study investigates the mechanisms underlying plant growth promotion and disease suppression by T. harzianum QT20045 in peanut seedlings. Under conditions without biotic stress, QT20045 significantly enhances root development and height in peanut seedlings by modulating hormone metabolism, particularly increasing IAA content and decreasing ethylene production. This modulation is attributed to the upregulation of genes involved in IAA biosynthesis and downregulation of genes in ethylene biosynthesis, especially both the ACS- and ACO-associated genes. Additionally, QT20045 exhibits antagonistic effects on the pathogen Sclerotium rolfsii, employing mechanisms such as entanglement, parasitism, and dissociation. Under the biotic stress of S. rolfsii, QT20045 upregulates genes related to pectinesterase, contributing to plant cell wall stability and resistance to pathogens, as well as the genes associated with immune receptors. Overall, T. harzianum QT20045 shows potential for enhancing plant growth and suppressing disease in peanut seedlings through hormone modulation, antagonistic activity against pathogens, and enhancement of plant defense mechanisms. The occurrence, prevalence, and severity of plant diseases are influenced by a complex interplay of climate, crop rotation, and agronomic practices (Singh et al., 2023), which can elevate the risks associated with applying (bio)pesticides if the disease does not develop. Our findings indicate that T. harzianum QT20045 has both the biofertilizer and biopesticide potential to promote peanut productivity irrespective of stem rot disease occurrence.
Under the condition with no biotic stress, T. harzianum QT20045 considerably enhanced the growth of peanut seedlings by fostering increased root development and height. This enhancement was attributed to its modulation of hormone metabolism within the host plants, including IAA and ethylene. Firstly, T. harzianum QT20045 considerably increased the IAA content in the roots of peanut seedlings, possibly because it could activate the expression of the genes related to IAA biosynthesis of the host plants, e.g., the genes involved in IAA biosynthesis AhIAGLU were upregulated by QT20045 (Figures 4, 7).
In addition, under no biotic stress, T. harzianum QT20045 was able to decrease ethylene production to promote plant growth. Ethylene biosynthesis consists of two primary continuous enzymatic reactions: (i) ACS converts S-adenosylmethionine to ACC and (ii) ACO transforms ACC into ethylene in various plant organs (Bleecker and Kende, 2000). Our study confirmed that T. harzianum QT20045 reduced ethylene biosynthesis by regulating both ACS and ACO enzymes. Firstly, the enzyme activity of ACS and ACO was decreased by Trichoderma inoculation (Figures 4C, D); then, we found that QT20045 suppresses the expression of both ACS-associated genes such as AhACS3 and AhACS1 and ACO-associated genes such as AhACO1 and AhACO, while the previous reports only confirmed that Trichoderma inoculation enhanced the ACC deaminase activity of the host plant, but not the specific reactions of ethylene biosynthesis (Qi and Zhao, 2013; Zhang et al., 2019).
In our study, T. harzianum QT20045 exhibited a significant antagonistic effect on S. rolfsii, and the pot experiment further confirmed the biocontrol efficacy of T. harzianum QT20045 for peanut stem rot disease. The antagonistic mechanisms Trichoderma employed included entanglement, parasitism, and dissociation for the mycelia of S. rolfsii (Figure 1). The results of this study are in accordance with the previous findings that Trichoderma species such as T. harzianum and T. longibrachiatum were able to inhibit the growth of S. rolfsii based on in vitro tests (Hirpara et al., 2017; Ayyandurai et al., 2023). Interestingly, PR1 gene is considered as an indicator showing a plant’s responses to fungal attack (Anisimova et al., 2021), while our results showed that the application of Trichoderma reduced the expression of genes in the PR1 family. This suggests that Trichoderma’s presence inhibits the growth of S. rolfsii in the soil, consequently weakening the host plant’s immune response.
Under the biotic stress of S. rolfsii, inoculation with T. harzianum QT20045 significantly improved the growth of peanut seedlings by promoting greater root growth and height while also decreasing disease infection. This improvement was linked to the elevated production of IAA by upregulating the genes of IAA biosynthesis AhIAGLU and the reduced release of ethylene by downregulating the genes of AhACO in the host plant. Therefore, it is interesting that the presence of peanut pathogens did not affect the plant-growth promoting effect of Trichoderma inoculation, even if the same genes were regulated by T. harzianum QT20045. Our results indicated the conserved effect of Trichoderma on the IAA and ethylene metabolisms of the host plants.
T. harzianum QT20045 inoculation leads to the downregulation of the genes in the pectinesterase family of the peanut seedlings, which indicated that Trichoderma was able to keep the host plant’s cell wall stable, reduce the degradation of the key cell wall components such as pectin, and enhance the plant’s resistance to pathogens during the infection. Plant cell walls, comprising polymers like cellulose, hemicellulose, pectin, and lignin, are vital not only for maintaining and supporting cells but also for resisting pathogen invasion. Ke et al. (2013) observed that cell-wall-degrading enzymes, including pectinesterase, were significantly upregulated during pathogen infection, leading to a marked reduction in the pectin content of infected plant cells compared to the control. The genes associated with pectinesterase of tomato plants was downregulated by all of the tested Trichoderma strains under biotic stress (Lorito et al., 2010), while another study showed that the pectinesterase protein was identified from maize root during the inoculation of Trichoderma under hydroponic conditions without pathogens (Guillermo et al., 2018), which indicated that the regulation of pectinesterase genes results from the combined effects of both Trichoderma and pathogens. Trichoderma inoculation also modulated certain responses of the plant’s immune system—for example, in plant responses to biotic stress, MAPK cascades are crucial for signal transmission, while SUMM2 acts as an immune receptor, responsible for monitoring the integrity of diverse elements within the MAPK cascade (Zhang et al., 2017). Our finding demonstrated that, under the biotic stress of S. rolfsii, T. harzianum QT20045 was able to upregulate the AhSUMM2 gene in the peanut seedling but did not affect the MAPK genes.
Stress from S. rolfsii impeded peanut seedlings’ growth, but inoculation with the beneficial microorganism T. harzianum QT20045 significantly mitigated this negative impact. QT20045 exhibits beneficial effects on the growth of the host plants by enhancing ACC deaminase activity and IAA production. The key genes sensitive to Trichoderma inoculation were identified in the present study, including AhIAGLU for IAA metabolisms, AhACS and AhACO for ethylene modulation, and AhSUMM2 for enhancing the plant’s resistance to pathogens. These genes can be the focus in future research to investigate the mechanisms by which T. harzianum QT20045 regulates the genetic responses of the host plant. The use of Trichoderma can significantly diminish the reliance on chemical fertilizers and pesticides, promoting more sustainable and environmentally friendly agricultural practices and securing long-term soil fertility and health.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
XW: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. ZZ: Conceptualization, Data curation, Formal analysis, Validation, Visualization, Writing – original draft, Writing – review & editing. HL: Conceptualization, Investigation, Methodology, Validation, Writing – review & editing. YW: Conceptualization, Formal analysis, Supervision, Validation, Writing – review & editing. JH: Conceptualization, Data curation, Formal analysis, Software, Visualization, Writing – review & editing. HY: Conceptualization, Methodology, Writing – review & editing. YZ: Conceptualization, Formal analysis, Supervision, Validation, Visualization, Writing – review & editing. JL: Conceptualization, Formal analysis, Funding acquisition, Investigation, Project administration, Resources, Supervision, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was supported by the Natural Science Foundation of Shandong Province (Project ID: ZR2021MC085, ZR2022MC215, ZR2023MC045), and Independent training innovation team project from Jinan city (project ID: 2020GXRC003).
We gratefully acknowledge all the colleagues who have worked on this article and governments for their financial support.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1414193/full#supplementary-material
Abbas, A., Mubeen, M., Zheng, H., Sohail, M. A., Shakeel, Q., Solanki, M. K., et al. (2022). Trichoderma spp. genes involved in the biocontrol activity against Rhizoctonia solani. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.884469
Abbott, W. S. (1925). A method of computing the effectiveness of an insecticide. J. Economic Entomology 18, 265–267. doi: 10.1093/jee/18.2.265a
Alemneh, A. A., Cawthray, G. R., Zhou, Y., Ryder, M. H., Denton, M. D. (2021). Ability to produce indole acetic acid is associated with improved phosphate solubilising activity of rhizobacteria. Arch. Microbiol. 203, 3825–3837. doi: 10.1007/s00203-021-02364-w
Anisimova, O. K., Shchennikova, A. V., Kochieva, E. Z., Filyushin, M. A. (2021). Pathogenesis-related genes of pr1, pr2, pr4, and pr5 families are involved in the response to Fusarium infection in garlic (Allium sativum L.). Int. J. Mol. Sci. 22, 6688. doi: 10.3390/ijms22136688
Aydoğdu, M. (2023). Biological control of groundnut stem rot using native isolates of Trichoderma harzianum and T. aggressivum f. aggressivum. J. Plant Pathology. 105, 269–281. doi: 10.1007/s42161-022-01279-9
Ayyandurai, M., Akila, R., Manonmani, K., Harish, S., Mini, M. L., Vellaikumar, S. (2023). Deciphering the mechanism of Trichoderma spp. consortia possessing volatile organic compounds and antifungal metabolites in the suppression of Sclerotium rolfsii in groundnut. Physiol. Mol. Plant Pathology. 125, 102005. doi: 10.1016/j.pmpp.2023.102005
Bai, R. K., Wong, L. C. (2004). Detection and quantification of heteroplasmic mutant mitochondrial DNA by real-time amplification refractory mutation system quantitative PCR analysis: A single-step approach. Clin. Chem. 50, 996. doi: 10.1373/clinchem.2004.031153
Bleecker, A. B., Kende, H. (2000). Ethylene: a gaseous signal molecule in plants. Annu. Rev. Cell Dev. Biol. 16, 1–18. doi: 10.1146/annurev.cellbio.16.1.1
Bot, P. J., Akgul, D. S., Ozgonen, H., Erkilic, A. (2011). The effects of seed treatments with fungicides on stem rot caused by Sclerotium rolfsii sacc. in peanut. Pakistan J. Botany. 43, 2991–2996. doi: 10.1007/s12231-011-9174-z
Bradley, P. M., Journey, C. A., Romanok, K. M., Barber, L. B., Buxton, H. T., Foreman, W. T., et al. (2017). Expanded target-chemical analysis reveals extensive mixed-organic-contaminant exposure in US streams. Environ. Sci. Technology. 51, 4792–4802. doi: 10.1021/acs.est.7b00012
Carrero-Carron, I., Trapero-Casas, J. L., Olivares-Garcia, C., Monte, E., Hermosa, R., Jiménez-Díaz, R. M. (2016). Trichoderma asperellum is effective for biocontrol of Verticillium wilt in olive caused by the defoliating pathotype of Verticillium dahliae. Crop Protection. 88, 45–52. doi: 10.1016/j.cropro.2016.05.009
Chen, S., Zhou, Y., Chen, Y., Gu, J. (2018). Fastp: an ultra-fast all-in-one fastq preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Cilliers, A. J., Pretorius, Z. A., Van Wyk, P. S. (2003). Integrated control of Sclerotium rolfsii on groundnut in South Africa. J. Phytopathology. 151, 249–258. doi: 10.1046/j.1439-0434.2003.00715.x
Contreras-Cornejo, H. A., Macías-Rodríguez, L., Cortés-Penagos, C., López-Bucio, J. (2009). Trichoderma virens, a plant beneficial fungus, enhances biomass production and promotes lateral root growth through an auxin-dependent mechanism in Arabidopsis. Plant Physiol. 149, 1579–1592. doi: 10.1104/pp.108.130369
Cornejo, H. A. C., Rodriguez, L. M., Pena, E. B., Bucio, J. L. (2011). Trichoderma-induced plant immunity likely involves both hormonal-and camalexin-dependent mechanisms in Arabidopsis thaliana and confers resistance against necrotrophic fungi Botrytis cinerea. Plant Signaling Behavior. 6, 1554–1563. doi: 10.4161/psb.6.10.17443
Cui, N., Xu, H., Yao, S., He, Y., Zhang, H., Yu, Y. (2018). Chiral triazole fungicide tebuconazole: enantioselective bioaccumulation, bioactivity, acute toxicity, and dissipation in soils. Environ. Sci. pollut. Res. 25, 25468–25475. doi: 10.1007/s11356-018-2587-9
Druzhinina, I. S., Seidl-Seiboth, V., Herrera-Estrella, A., Horwitz, B. A., Kenerley, C. M., Monte, E., et al. (2011). Trichoderma: the genomics of opportunistic success. Nat. Rev. Microbiol. 9, 749–759. doi: 10.1038/nrmicro2637
Gao, Y., Zhang, G., Jiang, S., Liu, Y. X. (2024). Wekemo bioincloud: a user-friendly platform for meta-omics data analyses. iMeta 3, e175. doi: 10.1002/imt2.175
Guillermo, N. L., Greenwood, D. R., Martin, M., Christopher, W., Carla, E., Steyaert, J. M., et al. (2018). The apoplastic secretome of Trichoderma virens during interaction with maize roots shows an inhibition of plant defence and scavenging oxidative stress secreted proteins. Front. Plant Science. 9. doi: 10.3389/fpls.2018.00409
Hirpara, D. G., Gajera, H. P., Hirpara, H. Z., Golakiya, B. A. (2017). Antipathy of Trichoderma against Sclerotium rolfsii Sacc. evaluation of cell wall-degrading enzymatic activities and molecular diversity analysis of antagonists. J. Mol. Microbiol. Biotechnol. 27, 22–28. doi: 10.1159/000452997
Hu, J., Zhou, Y., Chen, K., Li, J., Wei, Y., Wang, Y., et al. (2020). Large-scale Trichoderma diversity was associated with ecosystem, climate and geographic location. Environ. Microbiol. 22, 1011–1024. doi: 10.1111/1462-2920.14798
Jia, S., Song, C., Dong, H., Yang, X., Li, X., Ji, M., et al. (2023). Evaluation of efficacy and mechanism of Bacillus velezensis CB13 for controlling peanut stem rot caused by Sclerotium rolfsii. Front. Microbiol. 14. doi: 10.3389/fmicb.2023.1111965
Jung, K., Friede, T., Beissbarth, T. (2011). Reporting FDR analogous confidence intervals for the log fold change of differentially expressed genes. BMC Bioinf. 12, 288. doi: 10.1186/1471-2105-12-288
Kanchiswamy, C. N., Malnoy, M., Maffei, M. E. (2015). Chemical diversity of microbial volatiles and their potential for plant growth and productivity. Front. Plant Science. 6. doi: 10.3389/fpls.2015.00151
Kato, M., Hayakawa, Y., Hyodo, H., Ikoma, Y., Yano, M. (2000). Wound-induced ethylene synthesis and expression and formation of 1-aminocyclopropane-1-carboxylate (ACC) synthase, ACC oxidase, phenylalanine ammonia-lyase, and peroxidase in wounded mesocarp tissue of Cucurbita maxima. Plant Cell Physiol. 41, 440–447. doi: 10.1093/pcp/41.4.440
Ke, X., Huang, L., Han, Q., Gao, X., Kang, Z. (2013). Histological and cytological investigations of the infection and colonization of apple bark by valsa Mali var. Mali. Australas. Plant Pathology. 42, 85–93. doi: 10.1007/s13313-012-0158-y
Kim, D., Langmead, B., Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Kruger, N. J. (2009). The Bradford method for protein quantitation. Protein Protoc. Handbook., 17–24. doi: 10.1007/978-1-59745-198-7_4
Liao, Y., Smyth, G. K., Shi, W. (2013). Featurecounts: an efficient general-purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. doi: 10.1093/bioinformatics/btt656
Liu, M., Philp, J., Wang, Y., Hu, J., Wei, Y., Li, J., et al. (2022). Plant growth-promoting rhizobacteria Burkholderia Vietnamiensis B418 inhibits root-knot nematode on watermelon by modifying the rhizosphere microbial community. Sci. Rep. 12, 8381. doi: 10.1038/s41598-022-12472-2
López-Bucio, J., Pelagio-Flores, R., Herrera-Estrella, A. (2015). Trichoderma as biostimulant: exploiting the multilevel properties of a plant beneficial fungus. Scientia Horticulturae. 196, 109–123. doi: 10.1016/j.scienta.2015.08.043
Lorito, M., Woo, S. L., Harman, G. E., Monte, E. (2010). Translational research on Trichoderma: from’omics to the field. Annu. Rev. Phytopathology. 48, 395. doi: 10.1146/annurev-phyto-073009-114314
Love, M. I., Huber, W., Anders, S. (2014). Moderated estimation of fold change and dispersion for rna-seq data with deseq2. Genome Biol. 15, 550. doi: 10.1186/s13059-014-0550-8
Maheshwari, R., Bhutani, N., Kumar, P., Suneja, P. (2021). Plant growth promoting potential of multifarious endophytic Pseudomonas lini strain isolated from Cicer arietinum L. Israel J. Plant Sci. 69, 50–60. doi: 10.1163/22238980-bja10047
Nguyen, T. M. V., Hertog, M. L., Van de Poel, B., Tran, D. T., Nicolaï, B. (2023). Targeted system approach to ethylene biosynthesis and signaling of a heat tolerant tomato cultivar; the impact of growing season on fruit ripening. Front. Plant Science. 14. doi: 10.3389/fpls.2023.1195020
Penrose, D. M., Glick, B. R. (2010). Methods for isolating and characterizing ACC deaminase-containing plant growth-promoting rhizobacteria. Physiologia Plantarum. 118, 10–15. doi: 10.1111/j.1399-3054.2003.00086.x
Pieterse, C. M., van der Does, D., Zamioudis, C., Leon-Reyes, A., Van Wees, S. C. (2012). Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 28, 489–521. doi: 10.1146/annurev-cellbio-092910-154055
Qi, W., Zhao, L. (2013). Study of the siderophore-producing Trichoderma asperellum Q1 on cucumber growth promotion under salt stress. J. Basic Microbiol. 53, 355–364. doi: 10.1002/jobm.201200031
Ray, S., Singh, V., Singh, S., Sarma, B. K., Singh, H. B. (2016). Biochemical and histochemical analyses revealing endophytic alcaligenes faecalis mediated suppression of oxidative stress in Abelmoschus esculentus challenged with Sclerotium rolfsii. Plant Physiol. Biochem. 109, 430–441. doi: 10.1016/j.plaphy.2016.10.019
Schmid, J., Müller-Hagen, D., Bekel, T., Funk, L., Stahl, U., Sieber, V., et al. (2010). Transcriptome sequencing and comparative transcriptome analysis of the scleroglucan producer Sclerotium rolfsii. BMC Genomics 11, 1–16. doi: 10.1186/1471-2164-11-329
Schmittgen, T. D., Livak, K. ,. J. (2008). Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3, 1101–1108. doi: 10.1038/nprot.2008.73
Singh, B. K., Delgado-Baquerizo, M., Egidi, E., Guirado, E., Leach, J. E., Liu, H., et al. (2023). Climate change impacts on plant pathogens, food security and paths forward. Nat. Rev. Microbiol. 21, 640–656. doi: 10.1038/s41579-023-00900-7
Sood, M., Kapoor, D., Kumar, V., Sheteiwy, M. S., Ramakrishnan, M., Landi, M., et al. (2020). Trichoderma: The “secrets” of a multitalented biocontrol agent. Plants 9, 762. doi: 10.3390/plants9060762
Sriram, S., Manasa, S. B., Savitha, M. J. (2009). Potential use of elicitors from Trichoderma in induced systemic resistance for the management of Phytophthora capsici in red pepper. J. Biol. Control. 23, 449–456.
Subedi, S., Ojha, M., Lozada, D. N., Madugula, S., Sanogo, S., Steiner, R., et al. (2024). Phenotypic evaluation of USDA peanut (Arachis hypogaea L.) mini-core collection for resistance against stem rot caused by Sclerotium rolfsii Sacc. Arch. Phytopathol. Plant Protection. 56, 1–16. doi: 10.1080/03235408.2024.2321632
Townsend, G. R., Heubergeb, J. W. (1943). Methods for estimating losses caused by diseases in fungicide experiments. Plant Dis. Reporter. 27 (17), 340–343. doi: 10.5555/19451100061
Viterbo, A., Landau, U., Kim, S., Chernin, L., Chet, I. (2010). Characterization of ACC deaminase from the biocontrol and plant growth-promoting agent Trichoderma asperellum T203. FEMS Microbiol. Letters. 305, 42–48. doi: 10.1111/fml.2010.305.issue-1
Whipps, J. M. (2004). Prospects and limitations for mycorrhizas in biocontrol of root pathogens. Can. J. Botany. 82, 1198–1227. doi: 10.1139/b04-082
Yamauchi, T., Watanabe, K., Fukazawa, A., Mori, H., Abe, F., Kawaguchi, K., et al. (2014). Ethylene and reactive oxygen species are involved in root aerenchyma formation and adaptation of wheat seedlings to oxygen-deficient conditions. J. Exp. Botany. 65, 261–273. doi: 10.1093/jxb/ert371
Yan, L., Wang, Z., Song, W., Fan, P., Kang, Y., Lei, Y., et al. (2021). Genome sequencing and comparative genomic analysis of highly and weakly aggressive strains of Sclerotium rolfsii, the causal agent of peanut stem rot. BMC Genomics 22, 1–15. doi: 10.1186/s12864-021-07534-0
Yu, G., Wang, L. G., Han, Y., He, Q. Y. (2012). Clusterprofiler: an r package for comparing biological themes among gene clusters. Omics-a J. Integr. Biol. 16, 284–287. doi: 10.1089/omi.2011.0118
Zhang, S., Gan, Y., Xu, B. (2019). Mechanisms of the iaa and acc-deaminase producing strain of Trichoderma longibrachiatum t6 in enhancing wheat seedling tolerance to Nacl stress. BMC Plant Biol. 19, 1–18. doi: 10.1186/s12870-018-1618-5
Zhang, Z., Liu, Y., Huang, H., Gao, M., Wu, D., Kong, Q., et al. (2017). The NLR protein SUMM2 senses the disruption of an immune signaling MAP kinase cascade via CRCK3. EMBO Rep. 18, 292–302. doi: 10.15252/embr.201642704
Zhou, Y., Wang, Y., Chen, K., Wu, Y., Hu, J., Wei, Y., et al. (2020). Near-complete genomes of two Trichoderma species: A resource for biological control of plant pathogens. Mol. Plant-Microbe Interact. 33, 1036–1039. doi: 10.1094/MPMI-03-20-0076-A
Keywords: soil health, microbiome, omics, ACCd, fungal, legume
Citation: Wang X, Zhao Z, Li H, Wei Y, Hu J, Yang H, Zhou Y and Li J (2024) The growth-promoting and disease-suppressing mechanisms of Trichoderma inoculation on peanut seedlings. Front. Plant Sci. 15:1414193. doi: 10.3389/fpls.2024.1414193
Received: 08 April 2024; Accepted: 05 June 2024;
Published: 25 June 2024.
Edited by:
Ricardo Aroca, Spanish National Research Council (CSIC), SpainReviewed by:
Elsherbiny A. Elsherbiny, Mansoura University, EgyptCopyright © 2024 Wang, Zhao, Li, Wei, Hu, Yang, Zhou and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yi Zhou, eWkuemhvdUBhZGVsYWlkZS5lZHUuYXU=; Jishun Li, eWV3dTJAc2Rhcy5vcmc=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.