- 1Bako National Maize Research Center, Bako, Ethiopia
- 2EthioAgri-CEFT, Addis Ababa, Ethiopia
- 3College of Agriculture and Veterinary Medicine, Jima University, Jima, Ethiopia
- 4Maize Improvement Unit, International Institute of Tropical Agriculture, Ibadan, Nigeria
Biofortification of provitamin A in maize is an attractive and sustainable remedy to the problem of vitamin A deficiency in developing countries. The utilization of molecular markers represents a promising avenue to facilitate the development of provitamin A (PVA)-enriched maize varieties. We screened 752 diverse tropical yellow/orange maize lines using kompetitive allele-specific PCR (KASP) makers to validate the use of KASP markers in PVA maize breeding. To this end, a total of 161 yellow/orange inbred lines, selected from among the 752 lines, were evaluated for their endosperm PVA and other carotenoid compounds levels in two separate trials composed of 63 and 98 inbred lines in 2020 and 2021, respectively. Significant differences (p < 0.001) were observed among the yellow maize inbred lines studied for all carotenoid profiles. An inbred line TZMI1017, introduced by the International Institute of Tropical Agriculture (IITA) showed the highest level of PVA (12.99 µg/g) and β-carotene (12.08 µg/g). The molecular screening showed 43 yellow maize inbred lines carrying at least three of the favorable alleles of the KASP markers. TZMI1017 inbred line also carried the favorable alleles of almost all markers. In addition, nine locally developed inbred lines had medium to high PVA concentrations varying from 5.11 µg/g to 10.76 µg/g and harbored the favorable alleles of all the KASP PVA markers. Association analysis between molecular markers and PVA content variation in the yellow/orange maize inbred lines did not reveal a significant, predictable correlation. Further investigation is warranted to elucidate the underlying genetic architecture of the PVA content in this germplasm. However, we recommend strategic utilization of the maize-inbred lines with higher PVA content to enhance the PVA profile of the breeding program’s germplasm.
1 Introduction
Maize is the most widely cultivated staple crop in Sub-Saharan Africa and provides over 20% of total calories in human diets (Badu-Apraku et al., 2020). In Ethiopia, maize is the second most widely cultivated staple crop after tef (Eragrostis tef), but it ranks first in terms of productivity and total production, with more than nine million households depending on it for their food and livelihoods (Abate et al., 2015). However, heavy dependence on the consumption of staple food crops like maize can contribute to micronutrient malnutrition such as vitamin A deficiency (VAD) (Menkir et al., 2018). VAD is prevalent in developing countries and is the main cause of preventable blindness and growth retardation in children and increased risk of maternal mortality in pregnant women (Olivia Pecukonis, 2017).
Biofortification of staple crops with essential micronutrients stands out as a cost-efficient and sustainable intervention against VAD (Gupta et al., 2015). Biofortification is the process of increasing the content of vitamins and minerals in the edible parts of staple crops through plant breeding techniques (Garg et al., 2018).
Maize biofortification efforts in Sub-Saharan Africa tackle the challenge of low provitamin A (PVA) content in local inbred lines (Parasanna et al., 2020). Breeders employed backcrossing, using high β-carotene temperate maize with elite white tropical inbred lines (Pixley et al., 2013); the inbred lines showed variation in carotenoid profiles (Babu et al., 2013; Sayadi Maazou et al., 2021). Conventionally, breeders relied on screening large numbers of genotypes for carotenoid profiles using high-performance liquid chromatography (HPLC) or ultra-performance liquid chromatography (UPLC) (Gupta et al., 2019). However, these techniques are expensive for routine PVA breeding programs. This is where molecular screening comes in—it allows researchers to detect alleles associated with high PVA content (Babu and Prasanna, 2014), offering a more cost-effective approach. Genetic markers, particularly for the crtRB1 gene in the carotenoid biosynthesis pathway (Muthusamy et al., 2015), facilitated breeding for high-PVA maize. Particularly, favorable alleles at crtRB1-3’TE showed promise for increasing PVA content (Azmach et al., 2013; Babu et al., 2013). These findings suggest PVA-enriched inbred lines and associated markers are valuable tools. Recently developed kompetitive allele-specific PCR (KASP) markers targeting the gene crtRB1 were reported to offer a reliable, high-throughput screening method for PVA content (Gowda et al., 2017). However, the genetic background in which these favorable alleles developed, population size, and marker–trait relationships may influence the effectiveness of such marker-assisted selection for main routine breeding programs (Babu et al., 2013).
In Ethiopia, yellow maize improvement was started in 2004 to meet the demand of the poultry industry, and subsequently, breeding for provitamin A enrichment for human nutrition began in 2008 through the introduction and evaluation of maize hybrids and inbred lines (Girum et al., 2012). Currently, the public maize breeding program in Ethiopia has developed a large number of yellow/orange maize inbred lines with diverse genetic backgrounds. In addition, the program has been introducing PVA inbred lines from IITA and CIMMYT and extensively evaluating them for various agronomic traits in multilocation field trials. Most of the locally developed yellow/orange inbred lines were derived from bi-parental crosses and backcrosses using a few temperate PVA trait donor inbred lines and locally adapted elite white inbred lines. These inbred lines have not been assayed for their provitamin A and other carotenoid contents in relation to their haplotype for the alleles of the crtRB1 gene. Therefore, the objectives of our study were to (i) assess the PVA and other carotenoid profiles of the adapted yellow/orange maize inbred lines for use as parents to develop hybrids with high PVA content and source populations of new inbred lines; (ii) evaluate the inbred lines using a set of KASP markers associated with allelic variants of a major crtRB1 gene; and (iii) select inbred lines carrying favorable alleles of high provitamin A content for subsequent use as parents to accelerate new PVA inbred line development using molecular markers.
2 Materials and methods
2.1 Plant materials
A total of 161 yellow/orange maize inbred lines along with checks (a locally developed white inbred line, BKL004, and a HarvestPluss inbred line, CLHP00003) were evaluated in two separate trials during the 2020 and 2021 main cropping seasons. The inbred lines had diverse genetic backgrounds, which include introductions from CIMMYT, IITA, and locally developed yellow/orange inbred lines. The local yellow/orange inbred lines at the F5 inbreeding stage were developed from crosses of adapted elite white maize inbred lines with introduced provitamin A donor lines through backcrossing and pedigree breeding methods.
2.2 Experimental site
The trials for carotenoid analysis and genotyping were planted at Bako Agricultural Research Center, Ethiopia. Bako represents the mid-altitude subhumid maize agroecology of Ethiopia. Its elevation is about 1,650 m above sea level and lies between 9°06′ north latitude and 37°09′ east longitude. Bako receives 1,200–1,500 mm of rainfall annually.
2.3 Analyses of carotenoids
Each of the two trials was composed of two sets of yellow/orange maize inbred lines with two checks; each trial was arranged in an alpha lattice design and blocks nested under two replications. Each inbred line was planted in one row with a row length of 5 m, a spacing of 75 cm, and a 25-cm distance between plants within a row. Trial I consisted of 63 inbred lines and was evaluated in the 2020 main cropping season, whereas trial II consisted of 98 inbred lines and was evaluated in the 2021 main cropping season.
Agronomic practices, including fertilizer application (NPS 150 kg/ha and UREA 250 kg/ha) and weed management, were carried out uniformly as per recommendation for the testing site.
Two plants were self-pollinated in each line to produce pure seeds for carotenoid analysis. Each ear was harvested separately, air-dried at ambient temperature, shelled, and stored in a cold room at 10°C for carotenoid analysis.
A hundred seed samples were drawn randomly from each plot and sent to CIMMYT, Mexico for carotenoid analysis. The carotenoid extraction and measurement were carried out as per the procedure CIMMYT’s laboratory manual (Muzhingi et al., 2017; Azmach et al., 2018).
Briefly, 600 mg of fine powder from each sample was transferred into a 15-mL glass tube container, to which 6 mL of ethanol with 0.1% of butylated hydroxyl toluene was added. Next, the tubes were shacked and vortexed for 15 s and incubated at 85°C for 5 min in a water bath. The tubes, which contain samples taken out from incubation and 500 μL of 80% potassium hydroxide added for saponification, were then shacked and vortexed.
Each tube sample was mixed with 3 mL of cold deionized water, 200 µL of internal standard β-Apo-80-carotenal, and 3 mL of hexane. The mixture was vortexed for 15 s, followed by a 3-min centrifugation at 3000 rpm. The supernatant (upper solvent phase) was pipetted into a 15-mL new test tube, placed on ice, and tightly covered. Again, 3 mL of hexane was added to each tube of sample and centrifuged. The supernatant (upper phase) was transferred to a new tube placed on ice and tightly covered.
Each extracted sample was dried under nitrogen gas using a Turbovap LV concentrator, reconstituted in 1 mL of 50:50 methanol:dichloroethane, and vortexed for 10 s. Using a mobile phase gradient from methanol:tert-butyl methyl ether (80:20 v/v) and a C30 Column (4.6 mm × 250 mm; 3 µm) to separate carotenoids, 50 µL of each sample was injected into the Acquity UPLC system. The solvent flow rate was 1 mL/min. The water acuity photodiode array detector is used to measure carotenoid absorbance at 450 nm wavelength (Muzhingi et al., 2017) and is connected to the software to view carotenoid profiles. Provitamin A was calculated as the sum of β-carotene and half of each of β-cryptoxanthin and α-carotene. Total carotenoid was calculated as the sum of PVA, lutein, and zeaxanthin. The concentration measurements were expressed in micrograms per gram of dry weight (DW).
2.4 Genotyping
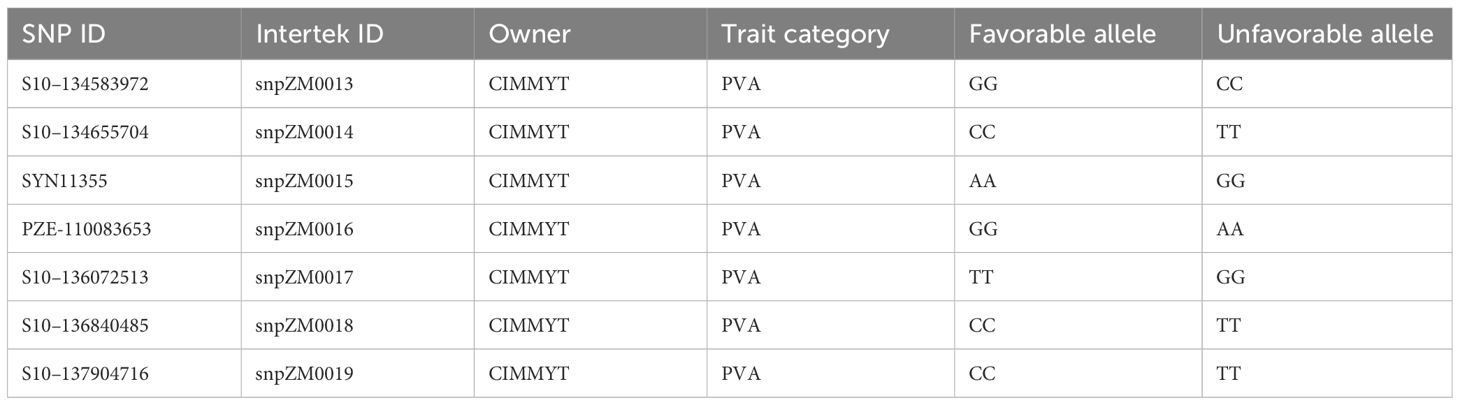
The 752 inbred lines (Supplementary Table S5), which also contain the 161 inbred lines analyzed for their carotenoid content, were genotyped using seven KASP PCR markers with key allelic variants of the crtRB1 gene. Each inbred line was planted on a single row of 5 m long at Bako Maize Research Center in the 2021 off-season. Young leaf samples were collected from five typical plants in each plot using labeled 96-well plates following the leaf sample collection procedure of the genotyping service provider. The leaf samples were dried in an oven at 50°C for 48 h, sealed, placed in a bucket containing silica gels, and stored in a cold room till samples were shipped to Sweden. DNA extraction protocol followed the CTAB method (Obi et al., 2020), and KASP PCR genotyping was carried out by Intertek AgriTech Laboratory, Sweden (https://www.intertek.com/agriculture/agritech/). The isolated DNA samples were genotyped using seven KASP markers presented in Table 1. PCR amplification and thermal cycling were performed according to the standard protocol for the KASP genotyping chemistry manual (https://www.biosearchtech.com/, accessed in 2020).
2.5 Statistical analysis
The carotenoid data were analyzed using PROC GLM in SAS software version 9.3 (SAS institute, 2011), in which inbred lines were treated as fixed effects while blocks and replications were treated as random effects. A mean separation was performed to compare treatment means using an LSD test at a 5% level of significance. Similarly, the repeatability (R2) values for PVA and other carotenoids were estimated using the PROC GLM model using SAS software (version 9.3). The distribution of carotenoid traits was analyzed using R software version 3.1.4. Genotyping data of inbred lines were sorted and aligned in an Excel sheet to identify the presence of favorable alleles associated with provitamin A and other carotenoid profiles. The general linear model (GLM) was used to analyze the association of favorable alleles with carotenoid traits in the TASSEL 5.0 version (Bradbury et al., 2007).
3 Results
3.1 Carotenoid profile
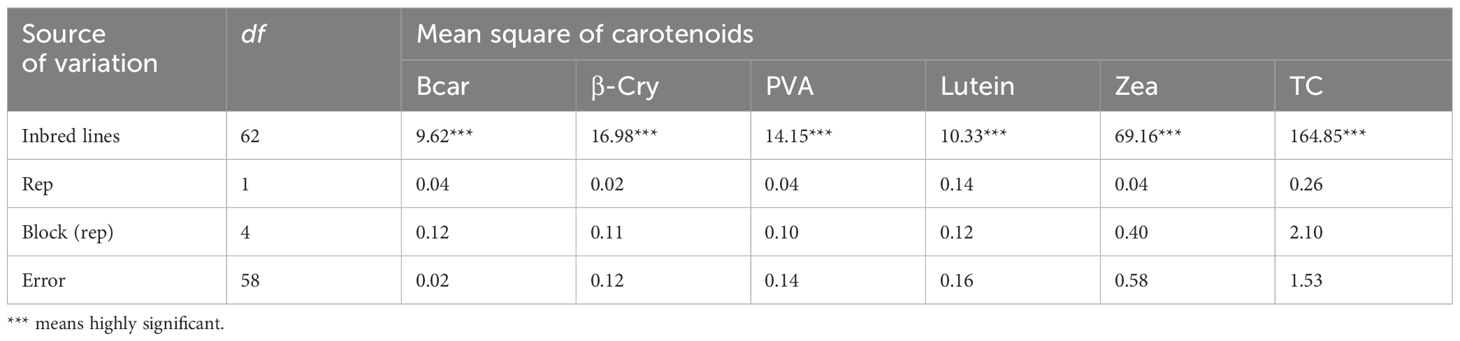
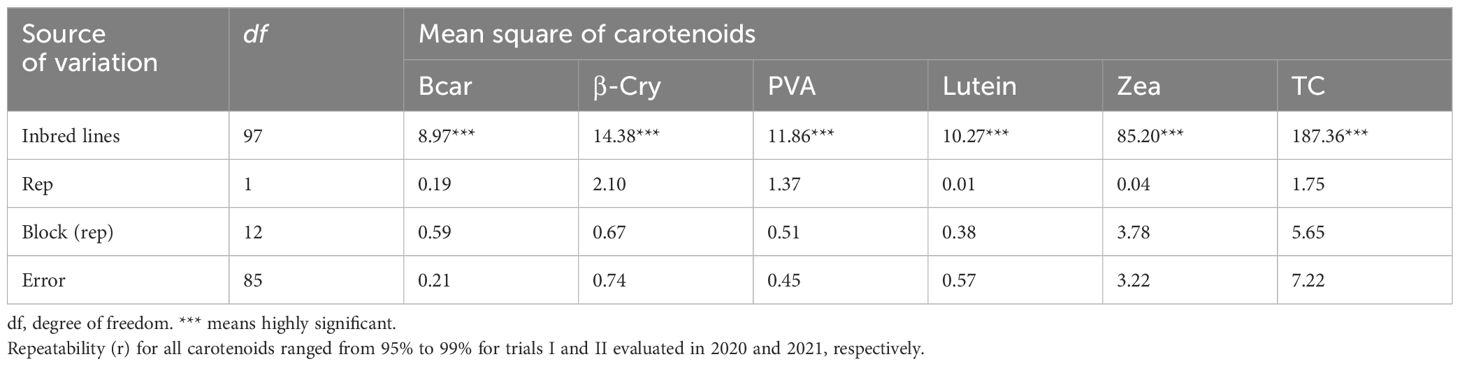
The analysis of variance showed highly significant differences (p < 0.001) among the yellow maize inbred lines in both trials I and II for PVA, β-carotene, β-cryptoxanthin, zeaxanthin, lutein, and total carotenoid concentrations (Tables 2, 3). The distributions of PVA and other carotenoids (µg/g) for the two trials (trial I, 63 inbred lines, and trial II, 98 inbred lines) are presented in boxplots (Figures 1A, B, respectively). The two trials displayed a similar distribution of carotenoids among the inbred lines. A few out-layers were observed for β-carotene and β-cryptoxanthin in both trials.
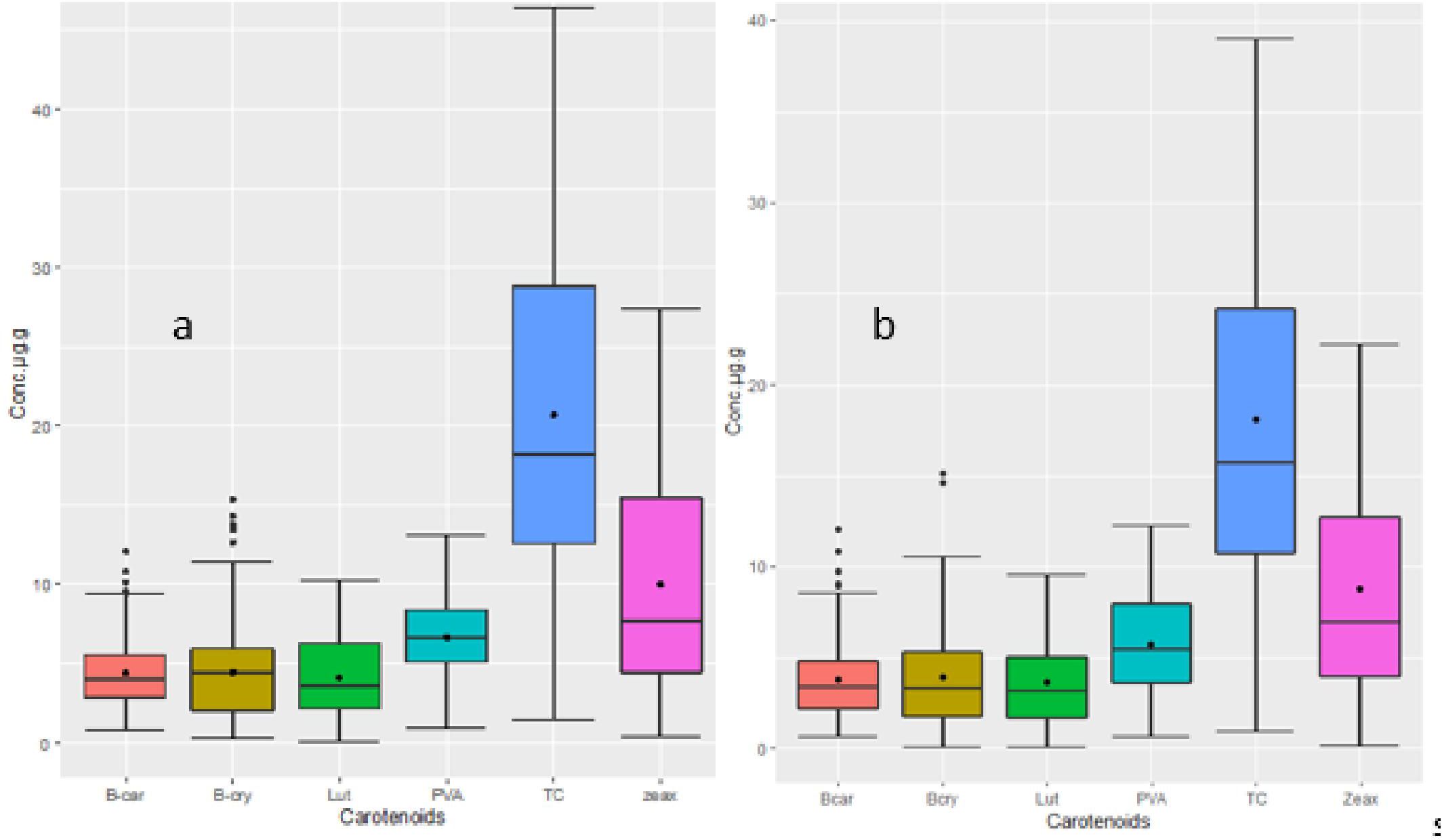
Figure 1 (A) Distribution of carotenoid concentrations for 98 inbred lines (Trial II). (B) Distribution of carotenoid concentrations for 63 inbred lines (Trial I). The upper and lower whisker (outside of the box plot) endpoints represent maximum and minimum concentrations, respectively. In the box plot, the lower and upper edges represent the first and third quartiles, the line inside the box represents the median, and the black point represents the mean. Carotenoids are abbreviated as follows: β-car, β-carotene; β-cry, cryptoxanthine; Lut, lutein; PVA, provitamin A; TC, total carotene; zeax, zeaxantine.
The PVA content varied from 0.7 µg/g to 12.99 µg/g with a mean value of 6.64 µg/g; the β-carotene value varied from 0.65 to 12.07 with a mean of 4.41 µg/g and that of β-cryptoxanthin varied from 0.10 to 14.85 with a mean of 4.45 µg/g. Zeaxanthin showed the highest level ranging from 8.76 µg/g to 24.75 μg/g whereas lutein had the lowest level, ranging from 0.05 µg/g to 8.91 µg/g (Supplementary Tables S1, S2).
The inbred line TZMI1017, introduced from IITA, had the highest level of provitamin A (12.99 µg/g), followed by CML 297 (11.33 µg/g) and a locally developed inbred line (IL00’E-9–1-1–1-1–1/(KUI carotenoid syn-FS17–3-2-B-B-B/(KU1409/DE3/KU1409) S2–18-2-B)-B-1(MAS: L4H1)-5-B-B-B)-B-11–1-2-B (11.08 µg/g) (Table 4) and also supplementary (Supplementary Table S6) for all carotenoids. On the other hand, the two checks (the white seeded inbred line (BKL004) and HarvestPlus inbred line (CLHP00003) had the lowest provitamin A content, 0.7 µg/g and 1.08 µg/g, respectively. Inbred lines with mean PVA contents of 6.64 µg/g to 12.99 µg/g can be used as donor parents in PVA maize breeding.
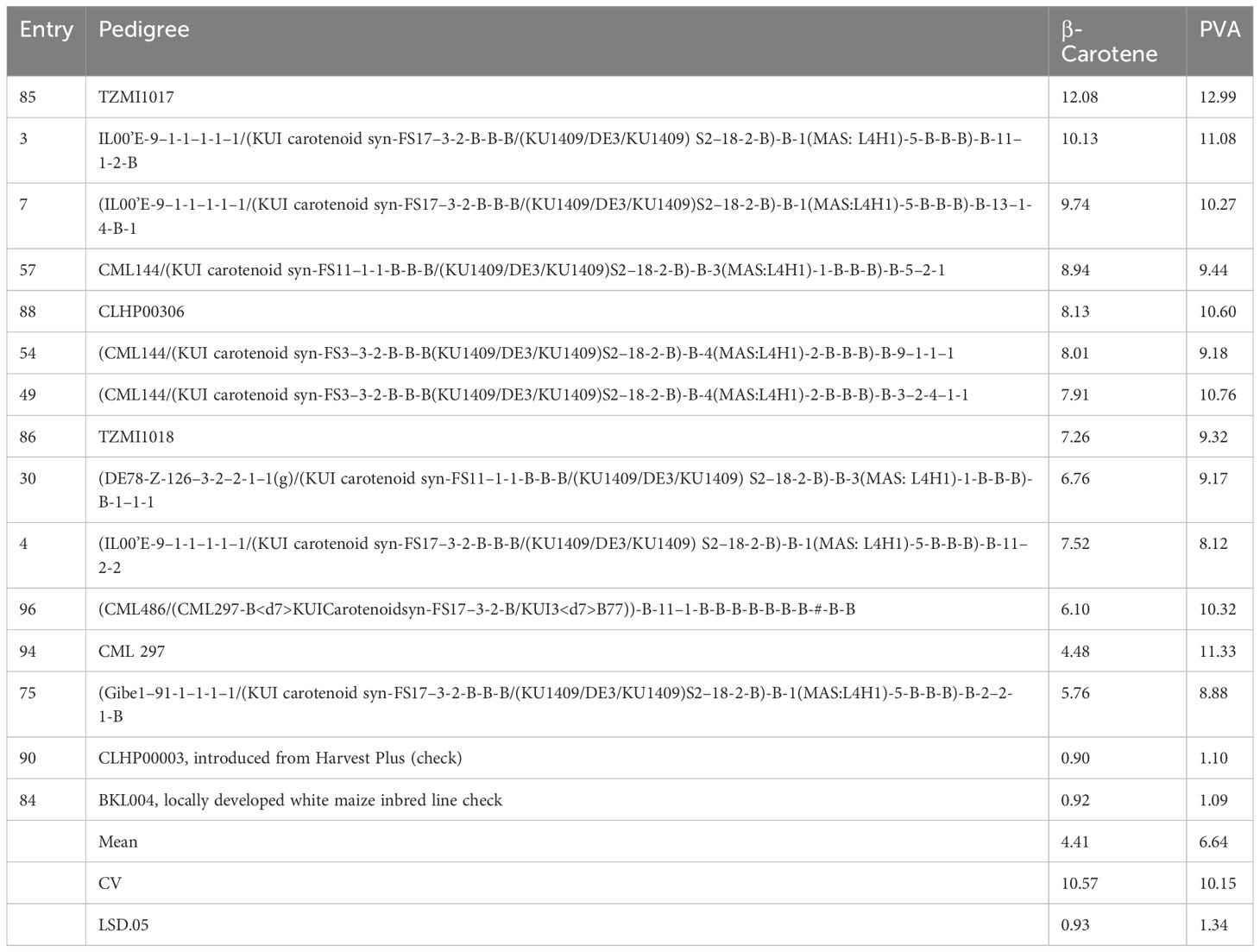
Table 4 Selected inbred lines among 98 inbred lines based on their mean value of β-carotene and provitamin A content (µg/g).
3.2 Molecular screening
Out of the 752 diverse yellow maize inbred lines screened with the seven KASP markers, about 113 inbred lines were found to be carrying one favorable allele by one of the KASP markers, whereas 16 inbred lines carried two favorable alleles at least by two KASP PCR markers. Moreover, 43 inbred lines carried three or more favorable alleles of the crtRB1 gene (Supplementary Table S3). Among 43 lines, 14 locally developed inbred lines carried favorable alleles of all markers of the crtRB1 gene, suggesting that introgression of favorable alleles into adapted elite lines was successful. Inbred lines TZMI1017 and CML297, introduced and adapted to local conditions carried five and six favorable alleles of KASP PVA markers, respectively.
Inbred lines carrying the favorable alleles of the KASP markers and having high PVA content were identified. Inbred line TZMI1017 showed the highest levels of PVA (12.99 µg/g) and β-carotene (12.08 µg/g) and carried the favorable alleles of almost all the markers evaluated (Table 5). Inbred line CML297 had high PVA (11.32 µg/g) with high β-cryptoxanthine concentration and carried the favorable alleles by five KASP markers used. In addition, nine locally developed inbred lines had medium to high PVA concentrations (5.11 µg/g to 10.76 µg/g) and carried favorable alleles of the seven KASP markers (Table 5). However, the general linear model association analysis result showed that the KASP markers were not significantly associated with variation in PVA carotenoids, β-carotene, and β-cryptoxan (Supplementary Table 4).
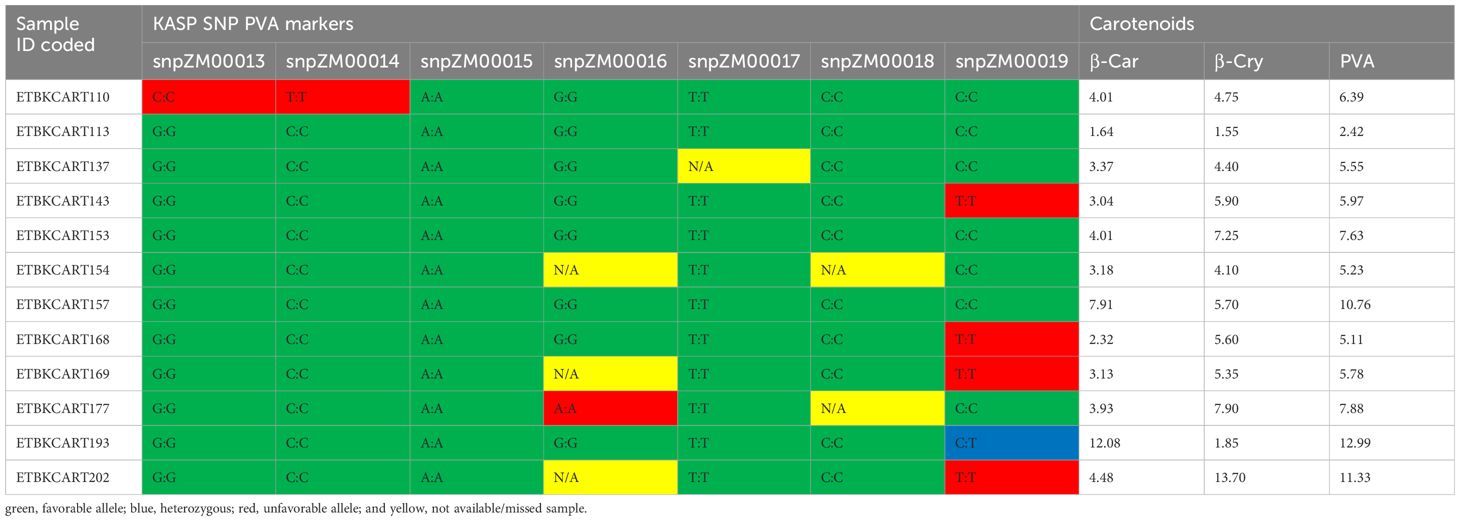
Table 5 Selected inbred lines carrying favorable alleles of the crtRB1 gene combined with high PVA content (µg g−1).
4 Discussion
Our result demonstrated high genetic variation among yellow maize inbred lines for all carotenoids, which was consistent with results in other studies (Muthusamy et al., 2015; Garg et al., 2018). Inbred lines with high β-carotene and PVA content were identified (Table 4). For example, inbred lines introduced from IITA and locally developed that contain PVA > 8 µg/g can be used as donor parents in hybrid and open variety development (Sayadi Maazou et al., 2021). The higher repeatability estimates, ranging from 95% to 99% for all carotenoid concentrations in both trials, indicate that the measured carotenoid values were reliable and the variation was mainly due to the genetic factors.
Different studies reported that genotype by environment interaction showed a minor and significant effect on PVA expression in maize. For instance, Menkir et al. (2008) and Muthusamy et al. (2015) reported that location and inbred lines by location interaction had a small fraction effect on lutein, α-carotene, and PVA. Similarly, there is a minor effect of genotype × environment interaction on lutein and zeaxanthin (Goswami et al., 2019a; Duo et al., 2021), whereas there is a nonsignificant effect on the PVA and β-carotene carotenoids of maize, indicating that β-carotene and PVA carotenoids are strongly influenced by genotypes. In the current study, we did not conduct line × year interaction, but the success of PVA breeding relies on the availability of genetic variation in yellow maize inbred lines (Menkir et al., 2008) and allelic diversity for PVA and other carotenoid content (Duo et al., 2021). On the other hand, variety × environment interaction showed effects on PVA content (Mengesha et al., 2019), and inbred lines × years of interaction showed significant effects on carotenoids except on β-carotene and PVA content (Menkir et al., 2015). Such inconsistent reports highlight that further investigation of genotype × environment interaction and other stress effects is required on PVA and other carotenoid content using a wide array of enriched yellow maize genetic backgrounds.
Marker-assisted selection is a useful tool to shorten the breeding cycle through the introgression of favorable alleles into agronomically superior elite lines to improve the nutritional quality of maize. We tested kompetitive allele-specific PCR (KASP) markers of the functional variations in the crtRB1 gene of the carotenoid biosynthesis pathway in maize endosperm to screen diverse maize inbred lines adapted to the mid-altitude subhumid agro-ecology of Ethiopia. Association analysis using the GLM model showed a nonsignificant association between the KASP markers and variations in PVA contents, β-carotene, and β-cryptoxanthin in the set of maize inbred lines studied. This is contrary to the results of Sayadi Maazou et al. (2021), who reported that KASP markers had a significant association with β-carotene and PVA accumulation and a negative, nonsignificant association with β-cryptoxanthin in maize. This could be due to the genetic backgrounds of the local inbred lines than those used in developing KASP markers, as that can affect marker–trait association (Babu et al., 2013). Likewise, the functional markers’ predictive accuracy seemed variable depending on the source of genotype and the combination of favorable alleles (Gedil and Menkir, 2019). Our results from similar studies (Esuma et al., 2022; Codjia et al., 2023) reported only one marker showed a significant association with PVA accumulation in cassava. Similarly, Vignesh et al. (2012) and Gebremeskel et al. (2018) reported that some lines carrying unfavorable alleles of the crtRB1 gene expressed high levels of PVA content, which might be due to ZmBCH1 and other genes. Such marker-to-trait association inconsistencies within a set of genotypes require further study, including phenotyping a larger sample of inbred lines and also checking for additional marker systems.
5 Conclusion
The genetic variation observed in PVA and other carotenoids among the inbred lines suggests their potential value for use as parents to enhance the PVA content through crossing, recycling, and strategic selection. The KASP markers examined in this study did not show a clear, predictable association with the observed PVA variation. However, it is crucial to continue investigating the underlying cause and test a more comprehensive marker system to streamline the regular PVA maize breeding program.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
BG: Data curation, Formal analysis, Software, Writing – original draft, Methodology. GA: Conceptualization, Methodology, Visualization, Writing – review & editing, Project administration. KB: Supervision, Visualization, Validation, Writing – review & editing. AM: Funding acquisition, Project administration, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was funded by the Harvestplus challenge program (OPP1019962).
Acknowledgments
The authors highly acknowledge the Harvest Plus ChallengeProgram for funding this work, particularly for carotenoid analysesand genotyping of the samples. Heartfelt thanks go to Ms AnaOliveira (EIB CIMMYT-Kenya) for providing plates, usefulcomments, and guidance on sample collection and facilitating thegenotyping by InterTek. The authors also express appreciation to all BNMRC staff who participated in planting, managing, and processing samples at Bako. The authors would like to extend their appreciation to the editor Emmanuel Oladeji Alamu, the reviewers Maryke T. Labuschagne and Edmore Gasura, and the Frontiers Production team for their work on this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The handling editor EA declared a past co-authorship with the author AM.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1406550/full#supplementary-material
Supplementary Table 1 | Mean, minimum, and maximum distribution of carotenoids (µg/g) in2021.
Supplementary Table 2 | Mean, minimum, and maximum distribution of carotenoids (µg/g) in 2020.
Supplementary Table 3 | Genotypes of 43 provitamin A maize inbred lines carrying three or more favorable alleles of the seven crtRB1-KASP PCR markers.
Supplementary Table 4 | Association of the presence of favorable alleles of KASP markers with mean of β-carotene, β-cryptoxanthine.
Supplementary Table 5 | Provitamin A in inbred lines. A list of inbred lines used for genotyping.
Supplementary Table 6 | Mean value of PVA and other carotenoid content (µg/g) among 98 yellow inbred lines.
References
Abate, T., Shiferaw, B., Menkir, A., Wegary, D., Kebede, Y., Tesfaye, K., et al. (2015). Factors that transformed maize productivity in Ethiopia. Food Secur. 7, 965–981. doi: 10.1007/s12571-015-0488-z
Azmach, G., Gedi, M., Mendkir, A., Spillane, C. (2013). Marker-trait association analysis of functional gene markers for provitamin A levels across diverse tropical yellow maize inbred lines. BMC Plant Bio 13, 227–242. doi: 10.1186/1471-2229-13-227
Azmach, G., Menkir, A., Spillane, C., Gedil, M. (2018). Genetic loci controlling carotenoid biosynthesis in diverse tropical maize lines. Genes Genomes Genet. 8, 1049–1065. doi: 10.1534/g3.117.300
Babu, R., Prasanna, B. M. (2014). “Molecular breeding for quality protein maize (QPM),” in Genomics of plant genetic resources. Eds. Tuberosa, R., Graner, A., Frison, E. (Springer Dordrecht Heidelberg, New York, London), 489–505.
Babu, R., Rojas, N. P., Gao, S., Yan, J., Pixley, K. (20132013). Validation of the effects of molecular marker polymorphisms in LcyE and CrtRB1 on provitamin A concentrations for 26 tropical maize populations. Theor. Appl. Genet. 126, 389–399. doi: 10.1007/s00122-012-1987-3
Badu-Apraku, B., Fakorede, B., Talabi, A. O., Obeng-Bio, E., Tchala, N., Oyekale, S. A. (2020). “Quantitative genetics, molecular techniques and agronomic performance of provitamin a maize in sub-Saharan Africa,” in Quantitative genetics, genomics and plant breeding (CABI, Wallingford UK), 276–324.
Bradbury, P. J., Zhang, Z., Kroon, D. E., Casstevens, T. M., Ramdoss, Y., Buckler, E. S. (2007). TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23, 2633–2635. doi: 10.1093/bioinformatics/btm308
Codjia, E. D., Olasanmi, B., Ugoji, C. E., Rabbi, L. Y. (2023). SNP-based marker-assisted selection for high provitamin A content in African cassava genetic background. South Afr. J. Sci. 119, 1–10. doi: 10.17159/sajs.2023/15115
Duo, H., Hossain, F., Muthusamy, V., Zanjeer, R. U., Goswami, R., Chand, G., et al. (2021). Development of sub-tropically adapted diverse provitamin-A rich maize inbreds through marker-assisted pedigree selection, their characterization and utilization in hybrid breeding. PLoS One 16, e0245497. doi: 10.1371/journal.pone.0245497
Esuma, W., Eyoo, O., Gwandu, F., Mukasa, S., Alicai, T., Ozimati, A., et al. (2022). Validation of KASP markers associated with cassava mosaic disease resistance, storage root dry matter and provitamin A carotenoid contents in Ugandan cassava germplasm. Front. Plant Sci. 13, 1017275. doi: 10.3389/fpls.2022.1017275
Garg, M., Sharma, N., Sharma, S., Kapoor, P., Kumar, A., Chunduri, V., et al. (2018). Biofortified crops generated by breeding, agronomy, and transgenic approaches are improving lives of millions of people around the world. Front. Nutr. 5, 12. doi: 10.3389/fnut.2018.00012
Gebremeskel, S., Garcia-Oliveira, A. L., Menkir, A., Adetimirin, V., Gedil, M. (2018). Effectiveness of predictive markers for marker assisted selection of pro-vitamin A carotenoids in medium-late maturing maize (Zea mays L.) inbred lines. J. Cereal Sci. 79, 27–34. doi: 10.1016/j.jcs.2017.09.001
Gedil, M., Menkir, A. (2019). An integrated molecular and conventional breeding scheme for enhancing genetic gain in maize in Africa. Front. Plant Sci. 10, 1430. doi: 10.3389/fpls.2019.01430
Girum, A., Mosisa, W., Legesse, W., Wende, A., Berhanu, T. (2012). Development of improved yellow maize germplasm in Ethiopia. Eds. Worku, M., Twumasi-Afriyie, S., Wolde, L., Tadesse, B., Demisie, G., et al (Addis Ababa, Ethiopia: EIAR/CIMMYT), 58–65. Meeting the Challenges of Global Climate Change and Food Security through Innovative Maize Research. Proceedings of the Third National Maize Workshop of Ethiopia April 18–20, 2011.
Goswami, R., Zunjare, R. U., Khan, S., Muthusamy, V., Baveja, A., Das, A. K. (2019a). Genetic variability of kernel provitamin-A in sub-tropically adapted maize hybrids possessing rare allele of β-carotene hydroxylase. Cereal Res. Commun. 47, 205–215. doi: 10.1556/0806.47.2019.12
Gowda, M., Worku, M., Nair, S. K., Palacios-Rojas, N., Prasanna, B. M. (2017). “Quality assurance/quality control (QA/QC) in maize breeding and seed production,” in Theory and Practice (CIMMYT, Nairobi, Kenya).
Gupta, H. S., Hossain, F., Muthusam, V. (2015). Biofortification of maize: An Indian perspective. Indian J.Genet. 75, 1–22. doi: 10.5958/0975-6906.2015.00001.2
Gupta, H. S., Hossain, F., Muthusamy, V., Zunjare, R. U. (2019). “Marker-assisted breeding for enrichment of provitaminA in maize,” in Quality breeding in field crops (Springer, Cham), 139–157.
Mengesha, W., Menkir, A., Meseka, S., Bossey, B., Afolabi, A., Burgueno, J., et al. (2019). Factor analysis to investigate genotype and genotype× environment interaction effects on pro-vitamin A content and yield in maize synthetics. Euphytica 215, 1–15. doi: 10.1007/s10681-019-2505-3
Menkir, A., Liu, W., White, W. S., Maziya-Dixon, B., Rocheford, T. (2008). Carotenoid diversity in tropical-adapted yellow maize inbred lines. Food Chem. 109, 521–529. doi: 10.1016/j.foodchem.2008.01.002
Menkir, A., Natalia, P., Oladeji, A., Maria Cristina, D., Thanda, D., Bussie, M., et al. (2018). VitaminA-Biofortified Maize: Exploiting Native Genetic Variation for Nutrient Enrichment. Science Brief: Biofortification No. 2 (HarvestPlus, and Crop Trust (Bonn, Germany: Science Brief: Biofortification).
Menkir, A., Rocheford, T., Maziya-Dixon, B., Tanumihardjo, S. (2015). Exploiting natural variation in exotic germplasm for increasing provitamin-A carotenoids in tropical maize. Euphytica 205, 203–217. doi: 10.1007/s10681-015-1426-z
Muthusamy, V., Hossain, F., Thirunavukkarasu, N., Saha, S., Gupta, H. S. (2015). Allelic variations for lycopene ϵ-cyclaseand β-carotene hydroxylase genes in maize inbreds and their utilization in β-carotene enrichment programme. Cogent Food Agric. 1, 1. doi: 10.1080/23311932.2015.1033141
Muzhingi, T., Palacios, N., Miranda, A., Cabrera, M. L., Yeum, K. J., Tang, G. (2017). Genetic variation of carotenoids, vitamin E and phenolic compounds in biofortified maize. J. Sci. Food Agric. 1–9. doi: 10.1002/jsfa.7798
Obi, Q. N., Menkir, A., Babalola, D. F., Gedil, M. (2020). Development of Efficient Genotyping Workflow for Accelerating Maize Improvement in Developing Countries. (Research square) 1–22. doi: 10.21203/rs.3.rs-129326/v1
Olivia Pecukonis (2017). Global data on vitamin A status is critical for program decision but often outdated, reveals new study. Available online at: https://www.nutritionintl.org/news/.
Parasanna, B. M., Palacios-Rojas, N., Hossain, F., Muthusamy, V., Menkir, A., Dhliwayo, T., et al. (2020). Molecular breeding for nutritionally enriched maize: status and prospects. Front. Genet. 10. doi: 10.3389/fgene.2019.013924
Pixley, K., Palacios, N. R., Babu, R., Mutale, R., Surles, R., Simpungwe, E. (2013). “Biofortification of maize with provitamin A carotenoids,” in Carotenoids in Human Health. Ed. Tanumihardo, S. A. (inger Science and Business Media, New York), 271–292. doi: 10.1007/978-1-62703-203-2
Sayadi Maazou, A. R., Gedil, M., Adetimirin, V. O., Meseka, S., Mengesha, W., Babalola, D., et al. (2014). Comparative assessment of effectiveness of alternative genotyping assays for characterizing carotenoids accumulation in tropical maize inbred lines. Agronomy 11 (10), 2022.
Keywords: biofortification, carotenoids profile, provitamin A, favorable allele, marker-assisted selection (MAS), yellow/orange maize inbred lines
Citation: Garoma B, Azimach G, Bante K and Menkir A (2024) Assessing the carotenoid profiles and allelic diversity of yellow maize inbred lines adapted to mid-altitude subhumid maize agroecology in Ethiopia. Front. Plant Sci. 15:1406550. doi: 10.3389/fpls.2024.1406550
Received: 25 March 2024; Accepted: 19 June 2024;
Published: 23 July 2024.
Edited by:
Emmanuel Oladeji Alamu, International Institute of Tropical Agriculture, ZambiaReviewed by:
Maryke T. Labuschagne, University of the Free State, South AfricaEdmore Gasura, University of Zimbabwe, Zimbabwe
Copyright © 2024 Garoma, Azimach, Bante and Menkir. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Belay Garoma, YmVsYXlnYXJvbWFAZ21haWwuY29t
 Belay Garoma
Belay Garoma Girum Azimach
Girum Azimach Kassahun Bante
Kassahun Bante Abebe Menkir
Abebe Menkir