- Crop Science Centre, University of Cambridge, Lawrence Weaver Road, Cambridge, United Kingdom
Arbuscular mycorrhizal (AM) fungi engage with land plants in a widespread, mutualistic endosymbiosis which provides their hosts with increased access to nutrients and enhanced biotic and abiotic stress resistance. The potential for reducing fertiliser use and improving crop resilience has resulted in rapidly increasing scientific interest. Microscopic quantification of the level of AM colonization is of fundamental importance to this research, however the methods for recording and processing these data are time-consuming and tedious. In order to streamline these processes, we have developed AMScorer, an easy-to-use Excel spreadsheet, which enables the user to record data rapidly during from microscopy-based assays, and instantly performs the subsequent data processing steps. In our hands, AMScorer has more than halved the time required for data collection compared to paper-based methods. Subsequently, we developed AMReader, a user-friendly R package, which enables easy visualization and statistical analyses of data from AMScorer. These tools require only limited skills in Excel and R, and can accelerate research into AM symbioses, help researchers with variable resources to conduct research, and facilitate the storage and sharing of data from AM colonization assays. They are available for download at https://github.com/EJarrattBarnham/AMReader, along with an extensive user manual.
Introduction
The evolution of land plants has been shaped by their interactions with a diverse microbial community. Amongst these interactions, the mutually beneficial symbiosis with arbuscular mycorrhizal (AM) fungi is one of the oldest and most widespread (Remy et al., 1994; Treseder and Cross, 2006). At the start of this symbiosis, fungal spores germinate and extend extraradical hyphae. Then, upon contact with a potential host, AM fungi develop hyphopodia on the surface of epidermal cells, enter the root, and produce intraradical hyphae. Subsequently, AM fungi develop arbuscules in the cortex, and establish vesicles and new spores (Figure 1) (Smith and Read, 2008; van Creij et al., 2020). Arbuscules are the characteristic, highly branched structures which AM fungi produce beneath the walls of plant cells, surrounded by a plant-derived membrane. Across this membrane, plants provide the fungus with sugars and fatty acids, in return for inorganic nutrients such as phosphorous and nitrogen (Luginbuehl and Oldroyd, 2017; Paries and Gutjahr, 2023). In vascular plants, fungal colonization is restricted to root tissue, while in non-vascular plants AM fungi colonise other tissues, such as the thalli of liverworts and hornworts (Edwards et al., 2000; Wang and Qiu, 2006).
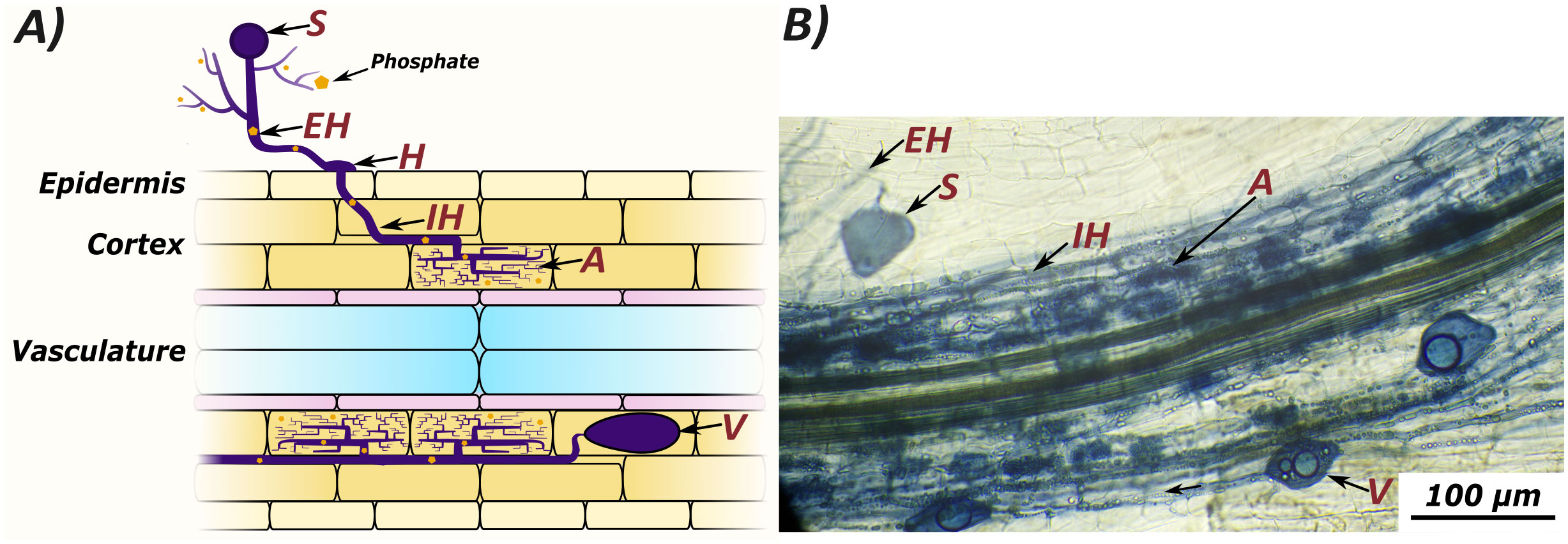
Figure 1 Arbuscular mycorrhizal colonization can be quantified by counting several microscopically visible structures: Spores (S), extraradical hyphae (EH), hyphopodia (H), intraradical hyphae (IH), arbuscules (A), and vesicles (V). Extraradical hyphae scavenge nutrients from the growth medium, which are transferred to the host plant at arbuscules. A graphic representation of fungal structures (A) compared to an image of colonised root tissue from Lotus japonicus stained with ink and vinegar (B).
AM fungi and their host plants are globally distributed (Treseder and Cross, 2006; Öpik et al., 2010), and AM fungi play important roles in both natural ecosystems and agricultural environments, offering plants both improved access to nutrients and increased abiotic and biotic stress resistance (Begum et al., 2019; Samuel and Veeramani, 2021; Bennett and Groten, 2022). Given their ecological importance, their significance for plant biology, and the potential for agricultural benefit, AM fungi have received increasing scientific and industrial interest.
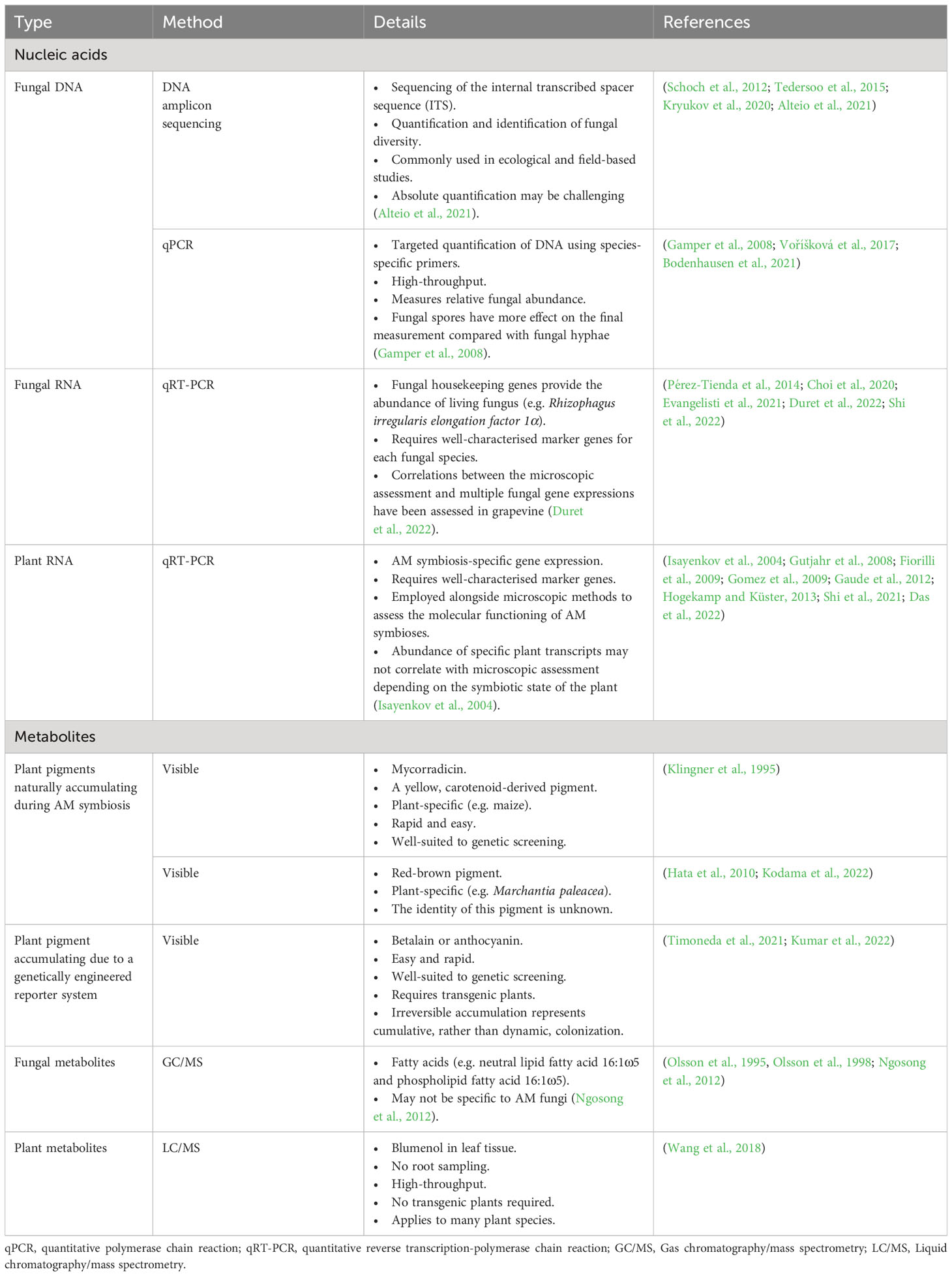
Most research into AM symbioses requires a means of assessing the abundance of AM fungi within the roots and rhizosphere. The earliest studies of AM fungi were dependent on microscopic analysis of stained plant tissue. While many alternative methods for quantifying AM colonization have since been developed using molecular proxies for the presence of AM fungi (Table 1), microscopy remains the gold-standard. This is because microscopy provides a direct measurement of fungal abundance without the need for homogenization of plant tissue. Consequently, microscopy enables the simultaneous assessment of the abundance, spatial distribution, and morphology of specific fungal structures. Microscopy, however, can be extremely time-intensive, and so represents a bottleneck for research into AM symbioses. Therefore, ways to efficiently record and process data from microscopic observations is of significant value.
To ease data collection by microscopic techniques, we have developed an Excel spreadsheet (AMScorer) which enables a user to easily record the presence or absence of all six fungal structures (extraradical hyphae, hyphopodia, intraradical hyphae, arbuscules, vesicles, and spores) during microscopic assays. An accompanying R package, AMReader, can read the data in AMScorer to rapidly produce graphical outputs with statistical analyses. These tools are user-friendly, compatible with many established methods of quantifying AM fungi, and are available at https://github.com/EJarrattBarnham/AMReader along with a comprehensive manual. Here, we present their key features.
Method
AMScorer has been developed to aid the recording and processing of data from AM root colonization assays. It is paired with AMReader to enable quick statistical analysis and data visualization. They are primarily designed to be compatible with the McGonigle magnified intersection method (McGonigle et al., 1990), or modifications thereof (Torabi et al., 2021). They can also be utilized for methods which quantify AM colonization in similar ways, such as the gridline intersect method (Giovannetti and Mosse, 1980). The specific details of these methods have been reviewed (Ho-Plágaro et al., 2020; Torabi et al., 2021). An example workflow for preparation of this material is outlined in Figure 2. Following preparation of the root material, AMScorer can be used to record AM colonization levels in either 100 fields-of-view or 100 intersections, recording the presence/absence of each fungal structure. For large root systems, it is critical to collect representative root fragments by thoroughly randomizing their selection. It is also possible to quantify multiple slides of root fragments, and subsequently average the results.
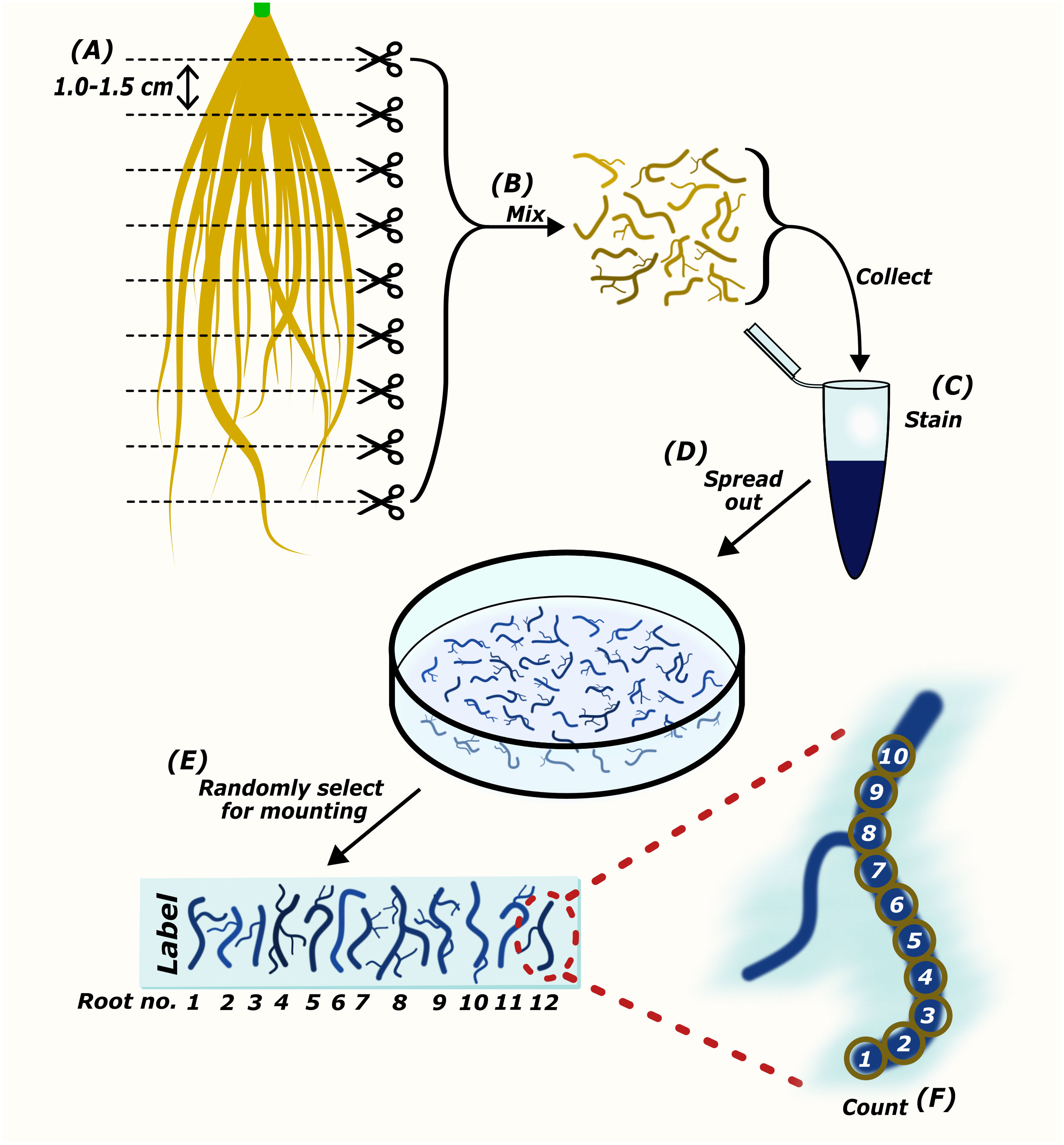
Figure 2 The preparation of root samples for microscopic quantification of AM colonization. Harvested roots are cut into sections approximately 1.0-1.5 cm in length (A). Root pieces are mixed thoroughly to collect representatives of the whole root system for staining (B, C). Root pieces are spread out in mounting medium (D). Randomly selected root pieces are mounted onto a microscopy slide (E). The abundance of AM fungi is counted either per field-of-view (Torabi et al., 2021), or using the McGonigle magnified intersection method (McGonigle et al., 1990) (F).
AMScorer
AMScorer (Excel) consists of 26 sheets (A to Z), each of which contains 15 counting tables (Figure 3A) (Supplementary File 1). Each table is intended to collect data from a biological replicate, often a single plant. In total, AMScorer can collect data from 390 plants per experiment, which we expect to be sufficient for large experimental designs. The counting tables exploit the Excel ‘Named Range’ sheet feature to enable all data inputs using just two buttons: ‘1’ and ‘TAB’ (Figure 4). This allows the user to operate the spreadsheet with one hand, while adjusting the microscope’s field-of-view with their other hand. Consequently, data can be collected simultaneously to microscopic observations. This greatly reduces the time needed for data collection. The data from these counting tables are then automatically fed into the main output, the ‘AM Results’ sheet, which displays percentage root length colonization of each structure per replicate. An ‘Information + Warnings’ sheet collects information about possible errors in data input to help ensure data accuracy (Figure 3B).
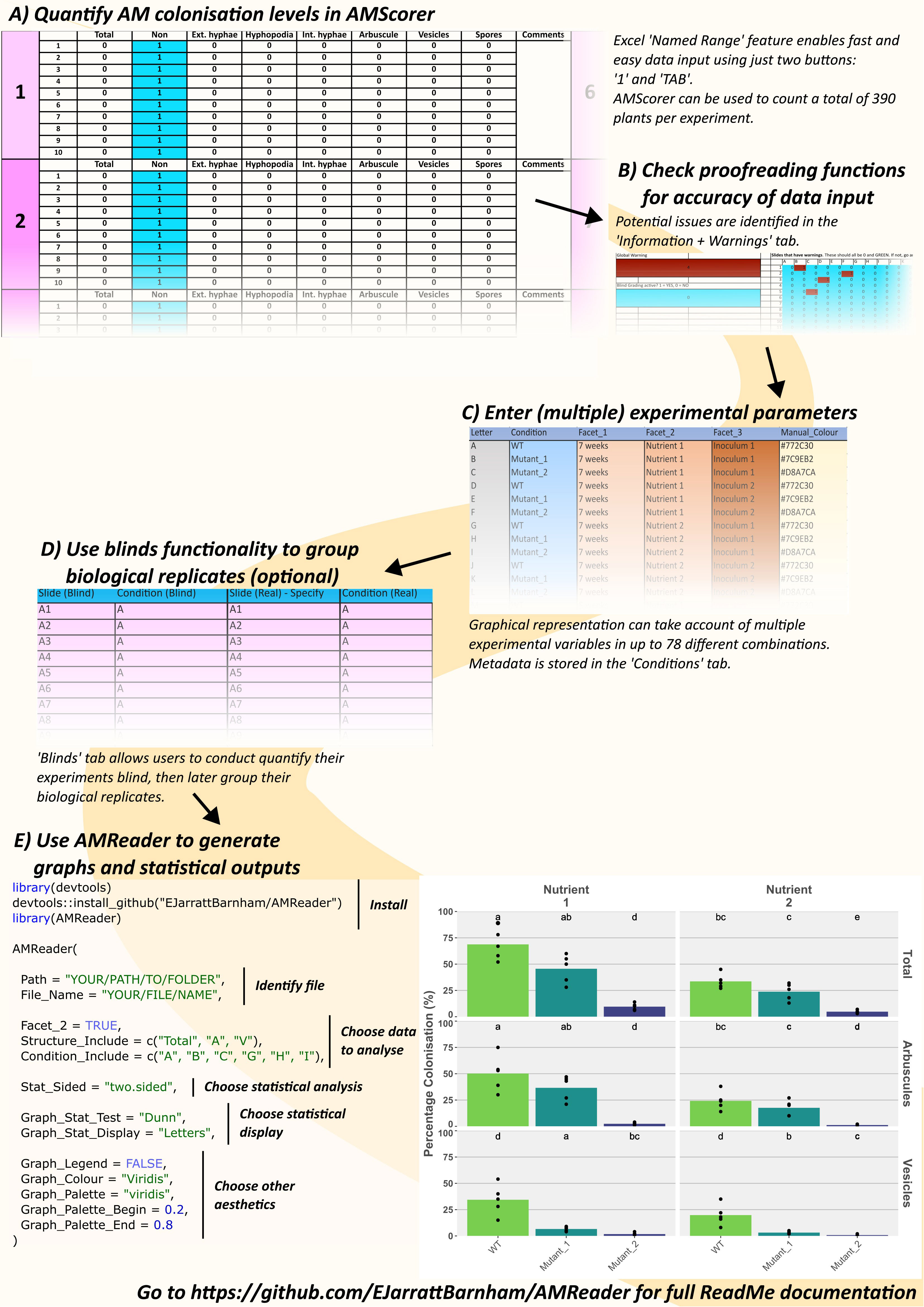
Figure 3 An example workflow using AMScorer and AMReader. One ‘Counting Table’ per plant allows the quantification of colonization in 390 plants per experiment (A). Proofreading methods ensure accurate data input (B). The ‘Conditions’ sheet can account for multiple experimental parameters with up to 78 different combinations (C). AMScorer enables easy blind counting and data to be grouped in multiple ways (D). An example R code to read AMScorer sheet into AMReader (E). AMReader provides an easy way to (1) read data from AMScorer, (2) select which data to present, (3) conduct various statistical analyses, (4) display data alongside statistical information, and (5) adjust color schemes and other aesthetics of graphical outputs.

Figure 4 The main features of each counting table in AMScorer. The Excel “Named Range” feature enables selection of cells for data entry. Movement to the next cell in the counting table is achieved with the keyboard TAB button. Pressing SHIFT and TAB will allow movement to the previous cell. Cells where the presence of a fungal structure have been recorded change color to blue, whilst cells where there is no data entered will turn red and activate a corresponding warning. If AMScorer believes a data entry may have been accidental, it will highlight the “Too few counts?” warning. To record that no fungal structures were present in a sample, activate the “No fungus override” button by inputting a “1”. Counts of fungal structures are summed up below each table, and results transferred automatically to the “AM Results” sheet.
AMScorer also contains a ‘Conditions’ sheet where the user can provide metadata detailing the experimental parameters (Figure 3C). This may include, for example, genotypes, time points, and growth conditions. AMScorer can handle complex experimental designs with up to 4 different variables, and a total of 78 unique combinations of these variables.
Additionally, a ‘Blinds’ sheet enables users to perform colonization assays blind, and for collected data to be readily reorganized into sets of biological replicates (Figure 3D). A ‘Test’ sheet contains 15 counting tables which may be used for collecting data that is not intended to contribute to the main results, but which may be useful to the user. For example, it may be used to keep a record of colonization from plants sampled during an experiment to inform the timing of a full harvest.
Through these features, AMScorer enables rapid and easy collection of data, which can subsequently be maintained and shared alongside information concerning the experimental design. Full details on the use of AMScorer are detailed in the documentation found in the GitHub repository.
AMReader
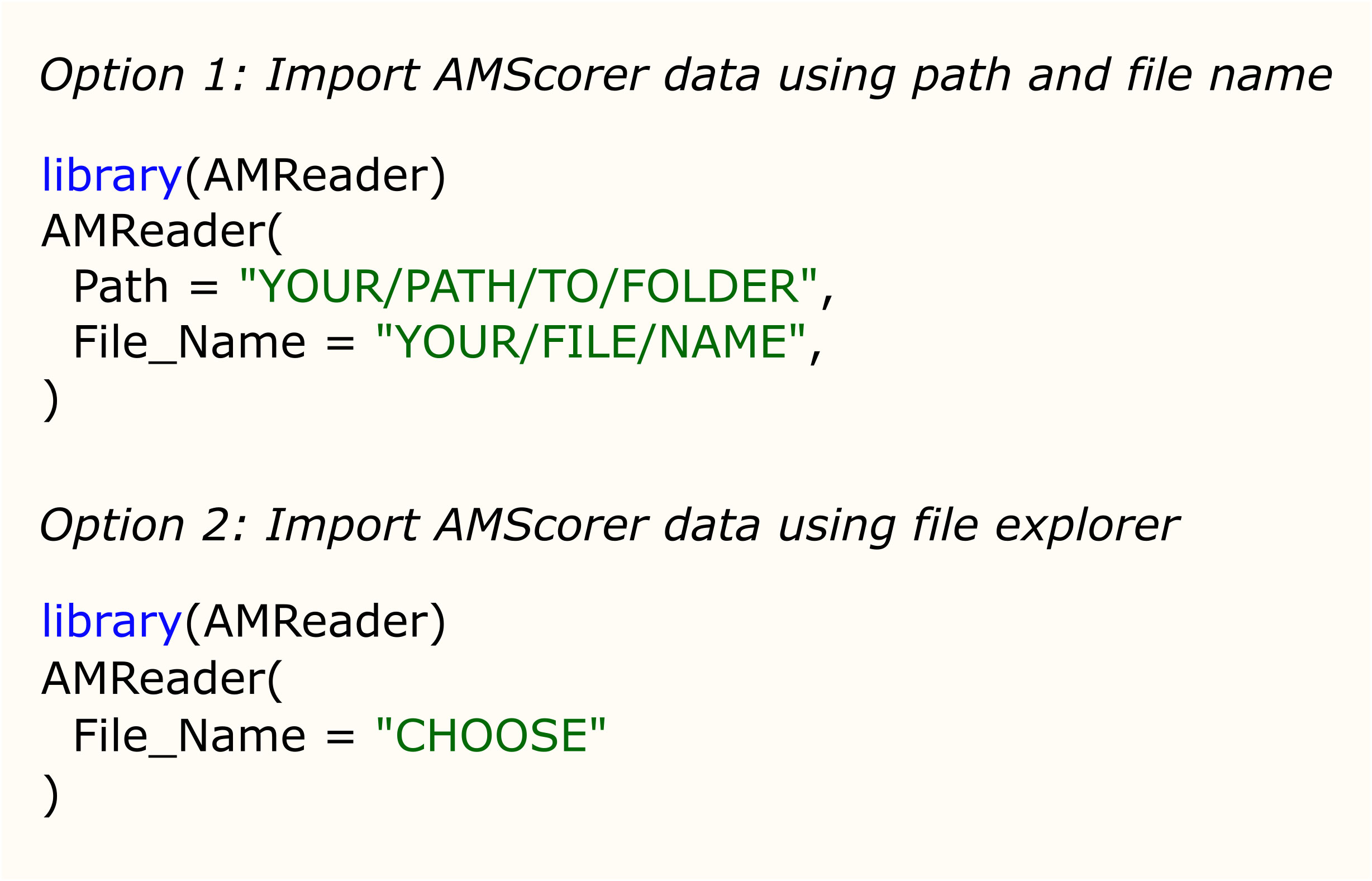
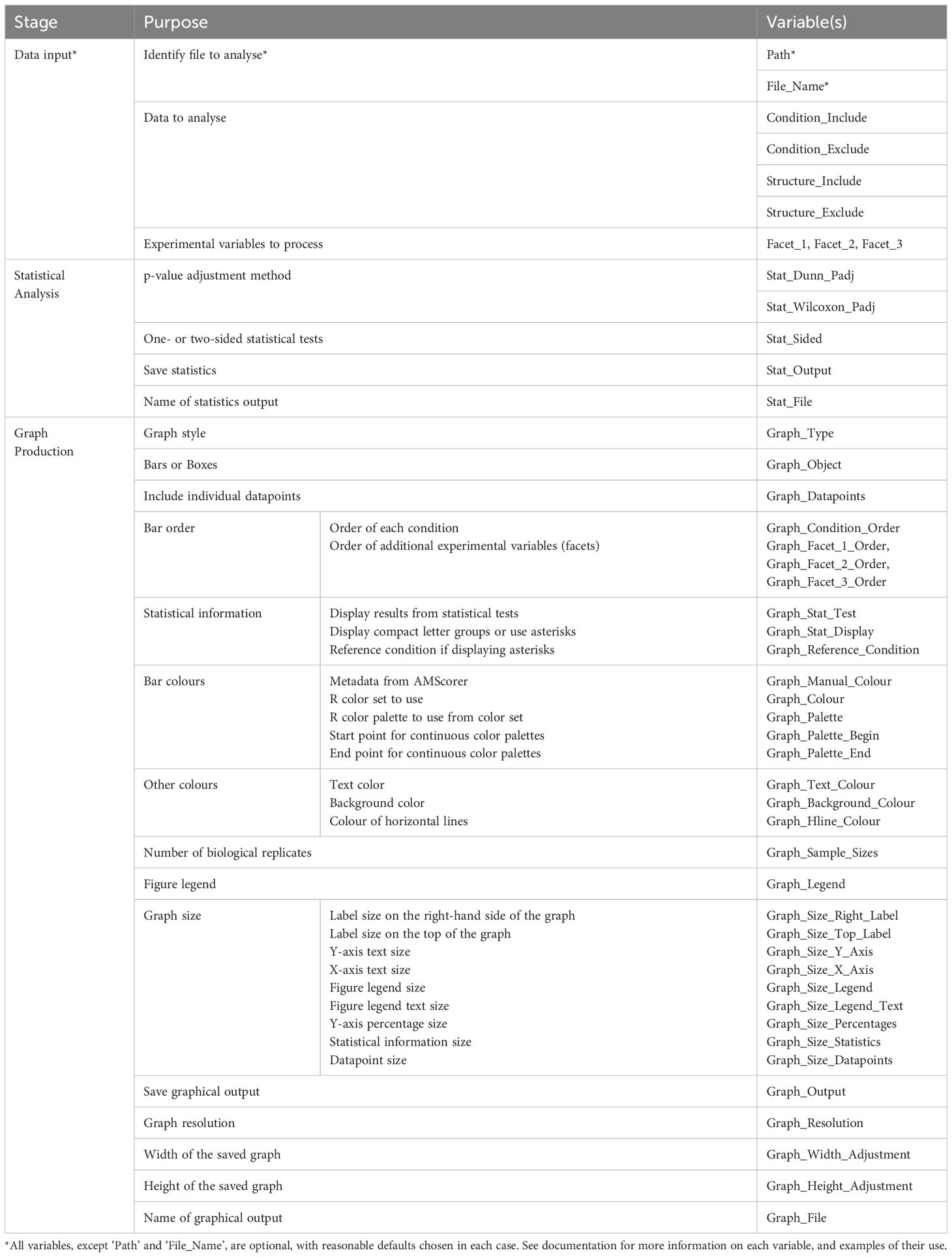
Following data collection, AMScorer can be read by the R package AMReader, which has been designed to: (1) Import data from AMScorer and process these data, (2) conduct statistical analyses, and (3) generate graphical displays (Figure 3E). Requiring only basic skills in R software, AMReader enables rapid and simple data analysis with just a single function. Its simplest input requires only the identification of the AMScorer file. This can be achieved either by defining its path and file name, or opening file explorer to locate the file (Figure 5). AMReader also contains extensive options for selecting data, choosing statistical tests, and adjusting the aesthetics of graphical outputs (Table 2). All these operations are controlled by the AMReader function, creating a user-friendly method to personalize and edit the output. A full set of examples, working through an example experimental dataset, are detailed in the documentation on the GitHub repository. For more experienced R users, the key outputs of AMReader are saved to the R environment, enabling user customization at any stage.
Results
Once microscopic quantification has been performed using AMScorer, the data will be automatically collected within the ‘AM Results’ sheet. In order to demonstrate the downstream functions of AMReader, a simulated dataset was generated (Supplementary File 2). This dataset is accessible within the GitHub repository under ‘man/Data’. Figure 6 demonstrates an example workflow for data analysis, which is also described below.

Figure 6 Example usage of AMReader. A simple workflow would involve loading data by identification of the AMScorer file (A), filtering of data to choose only those conditions and fungal structures of interest (B), statistical analysis (C), and later aesthetic changes to the graphical representation (D, E).
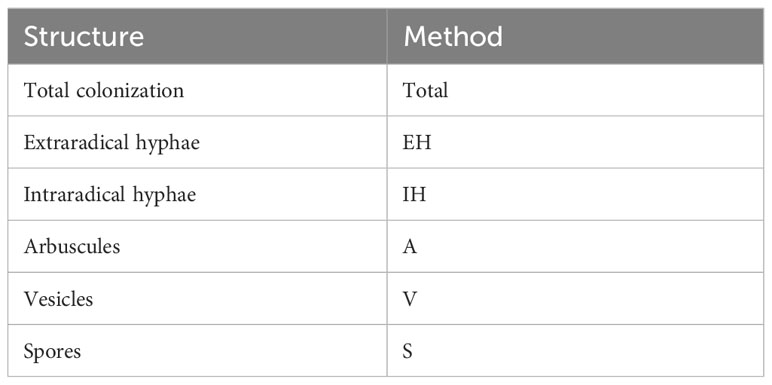
Firstly, the experimental data was loaded and displayed in full by activating all three faceting variables, corresponding to the three different experimental variables contained in the simulated data (Figure 6A). Subsequently, the data was filtered using the ‘Structure_Include’ and ‘Condition_Include’ variables (Figure 6B). Conditions were selected according to the letter codes stored in the ‘Conditions’ sheet (Figure 3C), and structures were selected according to their own codes (Table 3). Subsequently, statistical analysis was conducted by defining the statistical test using the ‘Graph_Stat_Test’ variable (Figure 6C). The user can choose from a range of parametric and non-parametric statistical tests, including the TukeyHSD and Wilcoxon pairwise tests. Using ‘Stat_Output’ and Stat_File’ would also allow the user to save an excel file containing reports from any statistical tests conducted on the given data.
AMReader currently supports multiple ways of displaying colonization data. For instance, a box plot may be preferable to bar plots, and this option can be chosen using the Graph_Object variable (Figure 6D). In addition, it is possible to produce a graph where the quantification of fungal structures is either separated into distinct facets (Graph_Type = “Facets”), or displayed alongside each other (Graph_Type = “Single”) (Figure 6E). Further personalization of the graph can be achieved by a great range of variables (Table 2), as illustrated here by changing the color of bars (Figure 6E).
We will seek to expand the range of graphical representations available in the near future, and would also welcome contributions from the community. Alternatively, all datafiles generated during the operation of AMReader are saved to the R environment so that more experienced R users can use AMReader for initial analyses, and then generate figures and results according to their own preferences.
Discussion
Quantification of AM colonization by microscopy represents a significant bottleneck for the study of AM symbioses. Many alternative, high-throughput methods have been developed to quantify AM colonization through molecular and metabolic changes induced by AM colonization. However, these rarely capture information on the spatial distribution, relative abundance, and morphological features of fungal structures. Consequently, microscopy remains a key requirement for many studies into AM symbioses. To reduce the time required for AM colonization assays, we have developed an Excel spreadsheet, AMScorer, which enables the user to record data during microscopy-based assays, and instantly performs the subsequent data processing steps. Over the course of an experiment, this greatly reduces the time required for quantification of AM colonization. In addition, AMScorer enables all data relevant to the experiment to be stored within a single spreadsheet, allowing easier preservation and dissemination of AM colonization data.
Recently, a novel machine learning approach has also been developed to aid the collection of AM colonization data (Evangelisti et al., 2021). Such methods hold promise for enabling full automation of AM colonization assays. Currently, however, they require high-resolution images with good contrast, which can be difficult to obtain in some plant species and are also time-intensive to acquire. Additionally, these tools require significant expertise in the computational methods. Here we provide tools which require limited technical knowledge, and are compatible with currently established methods for conducting colonization assays, and are compatible with downstream analysis pipelines, such as Ramf (Chiapello et al., 2019).
To facilitate data analysis, we have also developed AMReader, an R package which can transform the data collected by AMScorer into graphical representations quickly and easily. We have sought to develop AMReader so that it is usable by those with little experience in R software, creating a user-friendly system in which the user can control the R pipeline, without the need to write any additional layers of code. This distinguishes AMReader from currently available pipelines. We have, however, also ensured that more experienced R users can employ AMScorer and AMReader within their own, personalized, scripts.
Microscopic quantification of AM colonization represents a major bottleneck for research into AM colonization. AMScorer and AMReader represent new tools which can more than halve the time necessary for assays. We hope this will greatly accelerate research and facilitate sharing of data within the community.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
EJ-B: Conceptualization, Software, Validation, Writing – original draft, Writing – review & editing. GO: Funding acquisition, Writing – review & editing, Writing – original draft. JC: Funding acquisition, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. EJ-B. and GO were supported by the Bill and Melinda Gates Foundation and the Foreign, Commonwealth, and Development Office as OPP1172165, Engineering the Nitrogen Symbiosis for Africa. JC was supported by the Royal Society University Research Fellowship (URF/R1/211134) in the UK.
Acknowledgments
We thank our colleagues at the Crop Science Centre, Cambridge, UK for feedback and suggestions during the development of AMScorer and AMReader. Especially we thank Doris Albinsky, Sarabeth Buckley, Raphaella Hull, Thiago Alexandre Moraes, Emily Servanté, Mara Sgroi, Eirini Vlachaki and Darius Zarrabian. We thank Uta Paszkowski for feedback on the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1405598/full#supplementary-material
References
Alteio, L. V., Séneca, J., Canarini, A., Angel, R., Jansa, J., Guseva, K., et al. (2021). A critical perspective on interpreting amplicon sequencing data in soil ecological research. Soil Biol. Biochem. 160, 1–3. doi: 10.1016/j.soilbio.2021.108357
Begum, N., Qin, C., Ahanger, M. A., Raza, S., Khan, M. I., Ashraf, M., et al. (2019). Role of arbuscular mycorrhizal fungi in plant growth regulation: implications in abiotic stress tolerance. Front. Plant Sci. 10, 1068. doi: 10.3389/fpls.2019.01068
Bennett, A. E., Groten, K. (2022). The costs and benefits of plant–arbuscular mycorrhizal fungal interactions. Annu. Rev. Plant Biol. 73, 649–672. doi: 10.1146/annurev-arplant-102820-124504
Bodenhausen, N., Deslandes-Hérold, G., Waelchli, J., Held, A., van der Heijden, M. G. A., Schlaeppi, K. (2021). Relative qPCR to quantify colonization of plant roots by arbuscular mycorrhizal fungi. Mycorrhiza 31, 137–148. doi: 10.1007/s00572-020-01014-1
Chiapello, M., Das, D., Gutjahr, C. (2019). Ramf: an open-source R package for statistical analysis and display of quantitative root colonization by arbuscular mycorrhiza fungi. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.01184
Choi, J., Lee, T., Cho, J., Servanté, E. K., Pucker, B., Summers, W., et al. (2020). The negative regulator SMAX1 controls mycorrhizal symbiosis and strigolactone biosynthesis in rice. Nat. Commun. 11, 2114. doi: 10.1038/s41467-020-16021-1
Das, D., Paries, M., Hobecker, K., Gigl, M., Dawid, C., Lam, H.-M., et al. (2022). PHOSPHATE STARVATION RESPONSE transcription factors enable arbuscular mycorrhiza symbiosis. Nat. Commun. 13, 477. doi: 10.1038/s41467-022-27976-8
Duret, M., Zhan, X., Belval, L., Le Jeune, C., Hussenet, R., Laloue, H., et al. (2022). Use of a RT-qPCR method to estimate mycorrhization intensity and symbiosis vitality in grapevine plants inoculated with Rhizophagus irregularis. Plants 11, 3237. doi: 10.3390/plants11233237
Edwards, D., Read, D. J., Duckett, J. G., Read, D. J., Duckett, J. G., Francis, R., et al. (2000). Symbiotic fungal associations in ‘lower’ land plants. Philos. Trans. R. Soc Lond. B. Biol. Sci. 355, 815–831. doi: 10.1098/rstb.2000.0617
Evangelisti, E., Turner, C., McDowell, A., Shenhav, L., Yunusov, T., Gavrin, A., et al. (2021). Deep learning-based quantification of arbuscular mycorrhizal fungi in plant roots. New Phytol. 232, 2207–2219. doi: 10.1111/nph.17697
Fiorilli, V., Catoni, M., Miozzi, L., Novero, M., Accotto, G. P., Lanfranco, L. (2009). Global and cell-type gene expression profiles in tomato plants colonized by an arbuscular mycorrhizal fungus. New Phytol. 184, 975–987. doi: 10.1111/j.1469-8137.2009.03031.x
Gamper, H. A., Young, J. P. W., Jones, D. L., Hodge, A. (2008). Real-time PCR and microscopy: Are the two methods measuring the same unit of arbuscular mycorrhizal fungal abundance? Fungal Genet. Biol. 45, 581–596. doi: 10.1016/j.fgb.2007.09.007
Gaude, N., Bortfeld, S., Duensing, N., Lohse, M., Krajinski, F. (2012). Arbuscule-containing and non-colonized cortical cells of mycorrhizal roots undergo extensive and specific reprogramming during arbuscular mycorrhizal development. Plant J. 69, 510–528. doi: 10.1111/j.1365-313X.2011.04810.x
Giovannetti, M., Mosse, B. (1980). An evaluation of techniques for measuring vesicular arbuscular mycorrhizal infection in roots. New Phytol. 84, 489–500. doi: 10.1111/j.1469-8137.1980.tb04556.x
Gomez, S. K., Javot, H., Deewatthanawong, P., Torres-Jerez, I., Tang, Y., Blancaflor, E. B., et al. (2009). Medicago truncatula and Glomus intraradices gene expression in cortical cells harboring arbuscules in the arbuscular mycorrhizal symbiosis. BMC Plant Biol. 9, 10. doi: 10.1186/1471-2229-9-10
Gutjahr, C., Banba, M., Croset, V., An, K., Miyao, A., An, G., et al. (2008). Arbuscular mycorrhiza–specific signaling in rice transcends the common symbiosis signaling pathway. Plant Cell 20, 2989–3005. doi: 10.1105/tpc.108.062414
Hata, S., Kobae, Y., Banba, M. (2010). “Interactions between plants and arbuscular mycorrhizal fungi,” in International Review of Cell and Molecular Biology. Ed. Jeon, K. W. (San Diego, USA: Academic Press), 1–48. doi: 10.1016/S1937-6448(10)81001-9
Hogekamp, C., Küster, H. (2013). A roadmap of cell-type specific gene expression during sequential stages of the arbuscular mycorrhiza symbiosis. BMC Genomics 14, 306. doi: 10.1186/1471-2164-14-306
Ho-Plágaro, T., Tamayo-Navarrete, M. I., García-Garrido, J. M. (2020). “Histochemical staining and quantification of arbuscular mycorrhizal fungal colonization,” in Arbuscular Mycorrhizal Fungi: Methods and Protocols. Eds. Ferrol, N., Lanfranco, L. (Springer US, New York, NY), 43–52. doi: 10.1007/978-1-0716-0603-2_4
Isayenkov, S., Fester, T., Hause, B. (2004). Rapid determination of fungal colonization and arbuscule formation in roots of Medicago truncatula using real-time (RT) PCR. J. Plant Physiol. 161, 1379–1383. doi: 10.1016/j.jplph.2004.04.012
Klingner, A., Bothe, H., Wray, V., Marner, F.-J. (1995). Identification of a yellow pigment formed in maize roots upon mycorrhizal colonization. Phytochemistry 38, 53–55. doi: 10.1016/0031-9422(94)00538-5
Kodama, K., Rich, M. K., Yoda, A., Shimazaki, S., Xie, X., Akiyama, K., et al. (2022). An ancestral function of strigolactones as symbiotic rhizosphere signals. Nat. Commun. 13, 3974. doi: 10.1038/s41467-022-31708-3
Kryukov, A. A., Gorbunova, A. O., Machs, E. M., Mikhaylova, Y. V., Rodionov, A. V., Zhurbenko, P. M., et al. (2020). Perspectives of using Illumina MiSeq for identification of arbuscular mycorrhizal fungi. Vavilov J. Genet. Breed. 24, 158–167. doi: 10.18699/VJ20.605
Kumar, A., Lin, H., Li, Q., Ruan, Y., Cousins, D., Li, F., et al. (2022). Anthocyanin pigmentation as a quantitative visual marker for arbuscular mycorrhizal fungal colonization of Medicago truncatula roots. New Phytol. 236, 1988–1998. doi: 10.1111/nph.18504
Luginbuehl, L. H., Oldroyd, G. E. D. (2017). Understanding the arbuscule at the heart of endomycorrhizal symbioses in plants. Curr. Biol. 27, R952–R963. doi: 10.1016/j.cub.2017.06.042
McGonigle, T. P., Miller, M. H., Evans, D. G., Fairchild, G. L., Swan, J. A. (1990). A new method which gives an objective measure of colonization of roots by vesicular—arbuscular mycorrhizal fungi. New Phytol. 115, 495–501. doi: 10.1111/j.1469-8137.1990.tb00476.x
Ngosong, C., Gabriel, E., Ruess, L. (2012). Use of the signature fatty acid 16:1ω5 as a tool to determine the distribution of arbuscular mycorrhizal fungi in soil. J. Lipids 2012, 1–8. doi: 10.1155/2012/236807
Olsson, P. A., Bååth, E., Jakobsen, I., Söderström, B. (1995). The use of phospholipid and neutral lipid fatty acids to estimate biomass of arbuscular mycorrhizal fungi in soil. Mycol. Res. 99, 623–629. doi: 10.1016/S0953-7562(09)80723-5
Olsson, P. A., Francis, R., Read, D. J., Söderström, B. (1998). Growth of arbuscular mycorrhizal mycelium in calcareous dune sand and its interaction with other soil microorganisms as estimated by measurement of specific fatty acids. Plant Soil 201, 9–16. doi: 10.1023/A:1004379404220
Öpik, M., Vanatoa, A., Vanatoa, E., Moora, M., Davison, J., Kalwij, J. M., et al. (2010). The online database MaarjAM reveals global and ecosystemic distribution patterns in arbuscular mycorrhizal fungi (Glomeromycota). New Phytol. 188, 223–241. doi: 10.1111/j.1469-8137.2010.03334.x
Paries, M., Gutjahr, C. (2023). The good, the bad, and the phosphate: regulation of beneficial and detrimental plant–microbe interactions by the plant phosphate status. New Phytol. 239, 29–46. doi: 10.1111/nph.18933
Pérez-Tienda, J., Corrêa, A., Azcón-Aguilar, C., Ferrol, N. (2014). Transcriptional regulation of host NH4+ transporters and GS/GOGAT pathway in arbuscular mycorrhizal rice roots. Plant Physiol. Biochem. 75, 1–8. doi: 10.1016/j.plaphy.2013.11.029
Remy, W., Taylor, T. N., Hass, H., Kerp, H. (1994). Four hundred-million-year-old vesicular arbuscular mycorrhizae. Proc. Natl. Acad. Sci. U. S. A. 91, 11841–11843. doi: 10.1073/pnas.91.25.11841
Samuel, S. S., Veeramani, A. (2021). ““Advantages of arbuscular mycorrhizal fungi (AMF) production for the profitability of agriculture and biofertilizer industry,”,” in Mycorrhizal Fungi. Ed. Radhakrishnan, R. (IntechOpen, Rijeka). doi: 10.5772/intechopen.95458
Schoch, C. L., Seifert, K. A., Huhndorf, S., Robert, V., Spouge, J. L., Levesque, C. A., et al. (2012). Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl. Acad. Sci. 109, 6241–6246. doi: 10.1073/pnas.1117018109
Shi, J., Zhao, B., Jin, R., Hou, L., Zhang, X., Dai, H., et al. (2022). A phosphate starvation response-regulated receptor-like kinase, OsADK1, is required for mycorrhizal symbiosis and phosphate starvation responses. New Phytol. 236, 2282–2293. doi: 10.1111/nph.18546
Shi, J., Zhao, B., Zheng, S., Zhang, X., Wang, X., Dong, W., et al. (2021). A phosphate starvation response-centered network regulates mycorrhizal symbiosis. Cell 184, 5527–5540. doi: 10.1016/j.cell.2021.09.030
Tedersoo, L., Anslan, S., Bahram, M., Põlme, S., Riit, T., Liiv, I., et al. (2015). Shotgun metagenomes and multiple primer pair-barcode combinations of amplicons reveal biases in metabarcoding analyses of fungi. MycoKeys 10, 1–43. doi: 10.3897/mycokeys.10.4852
Timoneda, A., Yunusov, T., Quan, C., Gavrin, A., Brockington, S. F., Schornack, S. (2021). MycoRed: Betalain pigments enable in vivo real-time visualization of arbuscular mycorrhizal colonization. PLoS Biol. 19, 1–27. doi: 10.1371/journal.pbio.3001326
Torabi, S., Varshney, K., Villaécija-Aguilar, J. A., Keymer, A., Gutjahr, C. (2021). Controlled assays for phenotyping the effects of strigolactone-like molecules on arbuscular mycorrhiza development. Methods Mol. Biol. Clifton NJ 2309, 157–177. doi: 10.1007/978-1-0716-1429-7_13
Treseder, K. K., Cross, A. (2006). Global distributions of arbuscular mycorrhizal fungi. Ecosystems 9, 305–316. doi: 10.1007/s10021-005-0110-x
van Creij, J., Wang, P., Limpens, E. (2020). “Chapter 20 - Arbuscular mycorrhiza, a fungal perspective,” in Molecular Aspects of Plant Beneficial Microbes in Agriculture. Eds. Sharma, V., Salwan, R., Al-Ani, L. K. T. (London, UK: Academic Press), 241–258. doi: 10.1016/B978-0-12-818469-1.00021-3
Voříšková, A., Jansa, J., Püschel, D., Krüger, M., Cajthaml, T., Vosátka, M., et al. (2017). Real-time PCR quantification of arbuscular mycorrhizal fungi: does the use of nuclear or mitochondrial markers make a difference? Mycorrhiza 27, 577–585. doi: 10.1007/s00572-017-0777-9
Wang, B., Qiu, Y.-L. (2006). Phylogenetic distribution and evolution of mycorrhizas in land plants. Mycorrhiza 16, 299–363. doi: 10.1007/s00572-005-0033-6
Keywords: AMReader, AMScorer, arbuscular mycorrhizal fungi, microscopy, root length colonization
Citation: Jarratt-Barnham E, Oldroyd GED and Choi J (2024) Efficiently recording and processing data from arbuscular mycorrhizal colonization assays using AMScorer and AMReader. Front. Plant Sci. 15:1405598. doi: 10.3389/fpls.2024.1405598
Received: 23 March 2024; Accepted: 25 April 2024;
Published: 17 May 2024.
Edited by:
Andrea Genre, University of Turin, ItalyReviewed by:
Mara Novero, University of Turin, ItalyLuca Giovannini, National Research Council (CNR), Italy
Copyright © 2024 Jarratt-Barnham, Oldroyd and Choi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Edwin Jarratt-Barnham, ZWNqMzlAY2FtLmFjLnVr; Jeongmin Choi, amM5MTNAY2FtLmFjLnVr
‡ORCID: Edwin Jarratt-Barnham, orcid.org/0000-0002-8796-2266
Giles E.D. Oldroyd, orcid.org/0000-0002-5245-6355
Jeongmin Choi, orcid.org/0000-0001-7753-3348
 Edwin Jarratt-Barnham
Edwin Jarratt-Barnham Giles E. D. Oldroyd‡
Giles E. D. Oldroyd‡ Jeongmin Choi
Jeongmin Choi