- 1Roy J. Carver Department of Biochemistry, Biophysics and Molecular Biology, Iowa State University, Ames, IA, United States
- 2Center for Metabolic Biology, Iowa State University, Ames, IA, United States
- 3Department of Genetics, Development, and Cell Biology, Iowa State University, Ames, IA, United States
The maize glossy2 and glossy2-like genes are homologs, which encode proteins that belong to the BAHD family of acyltransferases. In planta genetic studies have demonstrated that these genes may be involved in the elongation of very long chain fatty acids (VLCFAs) that are precursors of the cuticular wax fraction of the plant cuticle. VLCFAs are synthesized by a fatty acyl-CoA elongase complex (FAE) that consists of four component enzymes. Previously, we functionally identified the maize FAE component enzymes by their ability to complement haploid Saccharomyces cerevisiae strains that carry lethal deletion alleles for each FAE component enzyme. In this study we used these complemented haploid strains and wild-type diploid strains to evaluate whether the co-expression of either GLOSSY2 or GLOSSY2-LIKE with individual maize FAE component enzymes affects the VLCFA product-profile of the FAE system. Wild-type diploid strains produced VLCFAs of up to 28-carbon chain length. Co-expression of GLOSSY2 or GLOSSY2-LIKE with a combination of maize 3-ketoacyl-CoA synthases stimulated the synthesis of longer VLCFAs, up to 30-carbon chain lengths. However, such results could not be recapitulated when these co-expression experiments were conducted in the yeast haploid mutant strains that lacked individual components of the endogenous FAE system. Specifically, lethal yeast mutant strains that are genetically complemented by the expression of maize FAE-component enzymes produce VLCFAs that range between 20- and 26-carbon chain lengths. However, expressing either GLOSSY2 or GLOSSY2-LIKE in these complemented strains does not enable the synthesis of longer chain VLCFAs. These results indicate that the apparent stimulatory role of GLOSSY2 or GLOSSY2-LIKE to enable the synthesis of longer chain VLCFAs in diploid yeast cells may be associated with mixing plant enzyme components with the endogenous FAE complex.
1 Introduction
The plant cuticle provides the organism with the first physical barrier from deleterious agents in the environment. It consists of the polyester cutin, which is embedded and coated with a complex mixture of unique, solvent-extractable lipids, commonly referred to as the cuticular waxes. These cuticular waxes are primarily a mixture of linear very long chain fatty acids (VLCFAs) and derivatives (e.g., aldehydes, alcohols, hydrocarbons, ketones, and wax esters), predominantly of 24-carbon atoms and longer (Samuels et al., 2008). Because the cuticle is produced by a single layer of epidermal cells and secreted to the plant surface, the molecular mechanisms that determine this process have been difficult to characterize by classical biochemical strategies. However, mutant alleles that affect its deposition have been a powerful research tool to deciphering the metabolic processes that underlie the deposition of cuticular waxes. These include the glossy mutants of maize (Bianchi et al., 1985; Schnable et al., 1994), and eceriferum mutants of Arabidopsis (Koornneef et al., 1989).
The maize glossy2 (gl2) gene was initially identified in a mutant stock that failed to accumulate normal levels of cuticular waxes on seedling leaves (Hayes and Brewbaker, 1928). Subsequently, the molecular isolation of gl2 (Tacke et al., 1995) led to the realization that it is a homolog of the Arabidopsis CER2 gene (Xia et al., 1996). Both gl2 and cer2 mutations reduce the normal accumulation of cuticular waxes in maize and Arabidopsis, respectively, and both these mutations appear to affect the ability to elongate a specific chain-length of fatty acids. In maize, the gl2 mutation appears to block the ability to elongate fatty acids beyond the 30-carbon chain length (Bianchi et al., 1975), whereas in Arabidopsis the cer2 mutation affects the ability to elongate fatty acids beyond 26- or 28-carbon chain lengths (McNevin et al., 1993; Jenks et al., 1995; Negruk et al., 1996).
At the time of these initial molecular characterizations, the GL2 (Tacke et al., 1995) and CER2 (Xia et al., 1996) proteins were novel, and their biochemical functionality was unclear. However, upon subsequent parallel molecular and biochemical characterization of a larger collection of proteins (St-Pierre and Luca, 2000), it became obvious that these two proteins are members of the BAHD family of enzymes that share a characteristic active site motif, the HXXXDX-motif, which catalyze acyltransferase reactions required in the biosynthesis of a variety of specialized metabolites (D’Auria, 2006; Moghe et al., 2023). BAHD acyltransferases were apparently important during the terrestrial colonization by plants, evidenced by the evolutionary expansion of the number of BAHD genes encoded by angiosperm genomes (~50-200 genes per genome). This compares to fungal genomes, which appear to be the evolutionary origin of the BAHD protein as they contain fewer than five BAHD genes per genome (Kruse et al., 2022). This evolutionary expansion and diversification of BAHD proteins appears to have supported the establishment of a wide range of biosynthetic machinery that generates the rich array of specialized metabolites that occur in the plant kingdom.
BAHD proteins have been phylogenetically classified into eight major clades, each of which display unique abilities to catalyze the acylation of different types of substrates (D’Auria, 2006; Kruse et al., 2022; Moghe et al., 2023). The common feature among these biochemically characterized proteins is the ability to acylate either an alcohol or amine functional group, generating either ester or amide products, respectively. The exception to this generalization is the clade that contains the Arabidopsis CER2 and CER2-LIKE family of proteins (Xia et al., 1996; Haslam et al., 2015, 2017; Haslam and Kunst, 2020) and the maize GL2 and GL2-LIKE proteins (Tacke et al., 1995; Alexander et al., 2020). Although heterologous expression studies in yeast have indicated that the CER2 and CER2-LIKE proteins interact with one of the enzyme components of the fatty acid elongase (FAE) system, and thereby affect the fatty acid product profile of the system (Haslam et al., 2012; Haslam and Kunst, 2013; Haslam et al., 2015, 2017; Wang et al., 2017; Haslam and Kunst, 2020; Gonzales-Vigil et al., 2021), the exact biochemical mechanism of how this is achieved remains unclear.
The FAE complex carries out the elongation of preexisting 16- and 18-carbon fatty acyl-CoAs to acyl chains of 20 carbons and longer, using malonyl-CoA as the elongating substrate. This complex is composed of four enzymatic components that iteratively catalyze cycles of Claisen condensation-reduction-dehydration and reduction reactions, resulting in the elongation of the acyl chain by 2-carbon atoms per cycle (Leonard et al., 2004). The FAE system is metabolically significant because it generates very long chain fatty acyl-CoAs, which are used as substrates to assemble such complex lipids as membrane phospholipids, storage lipids, sphingolipids (Harrison et al., 2018; Haslam and Feussner, 2022), eicosanoids (Smith, 1989; Thulasingam and Haeggström, 2020), and the plant cuticle (Samuels et al., 2008; Yeats and Rose, 2013; Barbero, 2016).
Underlying the phylogenetic diversity of these VLCFA-derived lipids, the plant FAE system displays genetic and biochemical diversity among the enzymatic components that constitute the complex (Campbell et al., 2019). Specifically, two distinct types of enzymes catalyze the Claisen condensation reactions of FAE; these are 3-ketoacyl-CoA synthases (KCSs), initially identified as the product of the Arabidopsis FAE1 gene (James et al., 1995), and Elongation-defective proteins (ELOs), which were initially identified in yeast (Toke and Martin, 1996; Oh et al., 1997). The KCS enzymes occur exclusively in plants, whereas ELOs are present in plants, fungi, and animals (Leonard et al., 2004). The three other component-enzymes of the FAE complex are: i) 3-ketoacyl-CoA reductase (KCR), ii) 3-hydroxyacyl-CoA dehydratase (HCD) and iii) enoyl-CoA reductase (ECR). Apart from the KCR component, which was initially identified as the product of the maize glossy8a gene (Xu et al., 1997, 2002), the plant homologs of HCD and ECR have been identified by the genetic complementation of yeast strains that carry null allele mutations in genes that encode these functions (Gable et al., 2004; Bach et al., 2008; Campbell et al., 2019), namely the yeast PHS1 (Denic and Weissman, 2007) and TSC13 (Kohlwein et al., 2001) genes, respectively.
The maize genome encompasses considerable genetic redundancy among these enzymatic components, particularly in the enzymes that catalyze the first two reactions of the FAE cycle; there are 26 genes encoding for the KCS-type enzyme, possibly up to six genes encoding the ELO-type enzyme (Campbell et al., 2019; Stenback et al., 2022), and two genes encoding the KCR enzyme (Dietrich et al., 2005). In contrast, only single copy genes of the HCD and ECR component enzymes have been characterized to date (Campbell et al., 2019). The molecular genetic characterizations of the Arabidopsis CER2 and four CER2-LIKE proteins indicate that they interact with the KCS enzymatic component of FAE to affect the chain-length products of the FAE system (Haslam and Kunst, 2013; Haslam et al., 2015, 2017; Haslam and Kunst, 2020). The current study is premised on prior experiments, which indicated that one of the maize paralogs, GL2-LIKE, is a functional homolog of CER2, whereas the other, GL2, appears not to be a functional homolog. Both GL2 and GL2-LIKE share a high degree of sequence homology to CER2, including the canonical BAHD HXXXDX-catalytic motif (Alexander et al., 2020).
This study is focused on understanding the functionality of the GL2 and GL2-LIKE proteins in the context of the functionality model described above for the Arabidopsis homologs (i.e., CER2 and CER2-LIKE proteins) (reviewed by Haslam et al., 2017). The experiments described herein take advantage of the in vivo yeast-based system that has previously been used to functionally identify the individual components of the maize FAE complex (Campbell et al., 2019; Stenback et al., 2022), which made it possible to mix and match individual maize FAE enzyme components with either GL2 or GL2-LIKE proteins, and thereby test whether these maize BAHD proteins modify the in vivo generated VLCFA product-profile of the resulting yeast strains. The results of these experiments suggest that the apparent stimulatory role of GL2 or GL2-LIKE to enable the synthesis of longer chain VLCFAs may be associated with mixing plant components with the endogenous FAE enzyme complex.
2 Materials and methods
2.1 Molecular cloning
The ORFs coding for the GL2 (GRMZM2G098239; Zm00001d002353) and GL2-LIKE (GRMZM2G315767; Zm00001d024317) proteins were codon-optimized for expression in yeast using GeneOptimizer (GeneArt, LifeTechnologies) and in Arabidopsis (OptimumGene (GenScript; www.genscript.com), respectively. These sequences were chemically synthesized (GenScript; GeneArt, LifeTechnologies). These DNA fragments were cloned into high-copy episomal yeast plasmids, pAG426 (URA3) or pAG423 (HIS3) (Invitrogen, Carlsbad, CA) (Alberti et al., 2007) using either the Gateway® cloning system (Invitrogen, Carlsbad, CA) or In-Fusion® cloning system (Takara Bio USA, Inc., Mountain View, CA). Depending on the experiment, expression from the episomal plasmids was under the control of a galactose-inducible promoter (GAL1) or the constitutive glyceraldehyde-3-P-dehydrogenase (GPD) promoter. All recombinant yeast shuttle vectors were confirmed by DNA sequencing and were maintained in E. coli TOP10 cells (Invitrogen, Carlsbad, CA), using Luria Bertani (LB) media supplemented with the appropriate antibiotics.
2.2 Yeast Strains and Media
The yeast strains, INVSc1, BY4743, BY4741, and BY4742 were obtained from Open Biosystems (ThermoFisher Scientific, Rockford, IL) and maintained in YPD (yeast peptone dextrose) or synthetic complete (SC) dropout media. Yeast strains carrying mutations in the endogenous FAE component genes and complemented by maize FAE component genes were previously described (Campbell et al., 2019; Stenback et al., 2022). All yeast strains expressing the maize GL2 or GL2-LIKE proteins were selected by their ability to grow on minimal medium (SD) lacking the appropriate amino acid or nucleobase (e.g., uracil). The induction of expression from the GAL1 promoter was accomplished by replacing glucose with 2% (w/v) galactose in SC medium. Yeast cultures were grown according to standard procedures in appropriate media at 30°C (Adams and Kaiser, 1998). For fatty acid analysis experiments, all strains were grown for 72 hours with exception of the INVSc1 strain that expressed maize proteins from the GAL1 promoter; these strains were grown for 48 hours.
2.3 Yeast transformation
Plasmids were transformed into yeast using a standard lithium acetate transformation protocol (Gietz and Woods, 2002). Briefly, 3 µL of salmon sperm DNA, 1 µg of plasmid DNA, and 100 µL transformation mix (200 µL of 2 M lithium acetate; 800 µL of 50% (w/v) PEG; 7.7 µL of β-mercaptoethanol) were added to a 1.5 mL microcentrifuge tube and mixed by vortexing. Yeast cells from a large colony were added to the mix, vortexed, and incubated at 37°C for 30 min. Samples were then centrifuged at 900 g for 5 min, and the cell pellet was resuspended in 200 µL of sterile water and plated on selective solid media. The plates were incubated at 30°C for 2-3 days. The non-recombinant vector was transformed into each strain and used as a control.
2.4 Fatty acid analysis
Cell pellets were lyophilized, and after the addition of 10 μg of nonadecanoic acid as an internal standard, fatty acids were extracted and converted to methyl esters by the addition of 1 mL 5% sulfuric acid in methanol, followed by incubation of the mixture at 80°C for 1 h. After cooling, 2 mL of 0.9% (w/v) aqueous NaCl was added, and fatty acid methyl esters were extracted thrice with 1 mL of 4:1 hexane: chloroform. After each extraction, phases were separated and the organic phase was removed and pooled, and samples were dried under a stream of nitrogen gas to a final volume of ~250 µL. One microliter of each sample was analyzed by gas chromatography-flame ionization detection (GC-FID) or gas chromatography-mass spectrometry (GC-MS).
GC-FID analysis was conducted with an Agilent 6890 GC, equipped with a DB-1 MS capillary column (15 m x 0.25 mm x 0.25 μm, Agilent 122-0112). Chromatography was conducted with helium gas, at a flow-rate of 1.2 mL/min, and an inlet temperature at 280 °C. The column oven temperature was initially held at 80°C for 1 min, then ramped at 15°C/min to 230°C and held for 2 min, and then ramped at 15°C/min to 340°C and held there for 2 min. For peak identification purposes, chromatograms were compared with fatty acid methyl ester standards (8:0-30:0) and parallel GC-MS analyses were queried against the NIST 14 Mass Spectral Library. Samples that were analyzed by GC-MS were first silylated with 50 µL N,O-Bis(trimethylsilyl)trifluoroacetamide (BSTFA) with 1% trimethylchlorosilane (TMCS) (Sigma-Aldrich), prior to chromatography.
2.5 Statistical analysis
Statistical significance of the differences in fatty acid profiles were determined by analyzing 3-6 replicates of each yeast strain, as defined in the captions of each figure. In all cases, statistical comparisons are made only from cultures that were grown in parallel in the same incubator. Fatty acid abundance data are reported in Supplementary Table 1, and statistical significance between genotypes was determined by Tukey’s Honest Significant Difference (HSD) test following ANOVA. The p-values for all Tukey’s HSD tests for each experiment are included in Supplementary Table 2.
2.6 Western blot analysis
Protein extracts were prepared from yeast cell pellets using the YeastBuster Protein Extraction Reagent, (Novagen Sigma-Aldrich, Inc., St. Louis, MO). In brief, weighed cell pellets were suspended in YeastBuster Reagent (5 ml/g cell pellet) and 0.5 M Tris(hydroxypropyl) phosphine (50 µl/g cell pellet). The cell suspensions were agitated for 15-20 min at room temperature and then centrifuged at 16,000 g for 20 min at 4°C. The supernatant was collected and subjected to SDS-PAGE, and proteins were electrophoretically transferred from the polyacrylamide gel to a nitrocellulose membrane using a Bio-Rad Criterion™ blotter (Bio-Rad, Hercules, CA). Membranes were first incubated in a solution of 5% (w/v) non-fat milk powder, dissolved in TBST buffer (0.1% (v/v) Tween 20, 0.15 M NaCl, 2 mM KCl, 20 mM Tris-HCl, pH 7.5) that contained GL2-specific antiserum (1:1000 dilution) (Alexander et al., 2020), and then with a solution containing horseradish peroxidase-linked to goat anti-mouse IgG antibody (Bio-Rad) (1:1000 dilution). After extensive washing with TBST buffer, the antigen-antibody complexes were detected using the Pierce ECL chemi-luminescent detection system (ThermoFischer Scientific) and visualized on the ChemiDoc XRS+ gel documentation system (Bio-Rad).
2.7 Reverse transcription-PCR analysis
Yeast strains expressing maize proteins were grown in liquid cultures and cells were pelleted by centrifugation. RNA was extracted from the resultant cell pellets utilizing the RNeasy Mini Kit (Qiagen, Germantown, MD) according to manufacturer’s instructions. Contaminating DNA was removed from the RNA samples by treatment with RQ1 RNase-Free DNase (Promega, Madison, WI), and 1 µg of RNA was used for cDNA synthesis using either the Superscript IV Synthesis Kit (ThermoFisher Scientific) or PrimeScript Synthesis Kit (Takara Bio USA, San Jose, CA) according to manufacturer’s instructions. PCR was conducted using the resultant cDNA as template, GoTaq Green Master Mix (Promega) and gene-specific primer pairs to assess the expression of ZmKCS6 (Primer 1: 5’-GTGAACCTCAAGCACGTCAA-3’ and Primer 2: 5’-CTCTTGTCGTCGTCGCTGAT-3’), GL2 (Primer 1: 5’-ATGGTTTTCGAACAACACGAAG-3’ and Primer 2: 5’-TTAAGCAACATGTAAAGCAGAACCC-3’), and GL2-LIKE (Primer 1: 5’-ATGGTTGTTGAGGCTAACTCTG-3’ and Primer 2: 5’-TCAAGCAACTCTAAGTGCATCC-3’). ZmKCS5 expression was assessed by utilizing Q5 High-Fidelity Polymerase with GC enhancer (NEB, UK) with gene-specific primer pairs (Primer 1: 5’- CAGAAGAACCTGCAGCTGTC-3’ and Primer 2: 5’- GCCGCTGCCGAAGCCGATCTGCCAG-3’). Each cDNA sample was also subjected to PCR analysis by using primers specific for the yeast GLC7 gene (Primer 1: 5’-CCAGATCTATATTCATAAAGCAACCC-3’ and Primer 2: 5’-GATAATTAGATTCTGGCGGGAATC-3’). This test PCR assay established that all cDNA samples lacked the GLC7 intron, indicating that the template samples were not contaminated by genomic DNA. The thermal cycling program for PCR was initiated at 95°C for 3 min, and then 35 cycles of incubations at 95°C for 30 s, 56°C for 45 s, and 72°C for 1.5 min; followed by a final extension step at 72°C for 5 min.
2.8 Accession numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers: GRMZM2G098239 and Zm00001d002353 (Gl2); GRMZM2G315767 and Zm00001d024317 (Gl2-like); GRMZM2G393897 and Zm00001d009608 (ZmKCS4); AC233893.1_FG003 and Zm00001d048061 (ZmKCS5); GRMZM2G164974 and Zm00001d028241(ZmKCS6); GRMZM2G160417 and Zm00001d039053 (ZmKCS15); AC205703.4_FG006 and Zm00001d017111(Gl8a; ZmKCR1); GRMZM2G087323 and Zm00001d050992 (Gl8b; ZmKCR2); GRMZM2G151087 and Zm00001d039856 (ZmHCD).
3 Results
3.1 Effect of co-expressing maize GL2 or GL2-LIKE with maize KCS homologs in wild-type diploid yeast strains
Based on the carbon chain lengths of the alkyl derivatives that occur in the cuticular waxes of maize (Bianchi et al., 1985), the maize FAE system should have the ability to produce VLCFAs as long as 34-carbon atoms. Moreover, in planta characterizations of mutations in the maize gl2 gene indicate that it may be involved in the ability of plant cells to produce VLCFAs beyond 26:0 and 28:0, and up to 34:0 (Bianchi et al., 1975; Alexander et al., 2020). Therefore, analogous to the Arabidopsis FAE system, where interactions between the Arabidopsis CER2-LIKE proteins and KCS paralogs stimulate the terminal elongation cycle(s) of the FAE system to generate longer chain VLCFAs (Haslam et al., 2017), yeast expression experiments evaluated whether GL2 or GL2-LIKE could affect the VLCFA profiles produced by co-expression with ZmKCS homologs. Initial experiments were conducted with ZmKCS5 or ZmKCS6, which are the closest homologs to the Arabidopsis CER6-encoded KCS (Campbell et al., 2019) that specifically interact with CER2 in the Arabidopsis FAE system (Haslam et al., 2015, 2017).
Figure 1 shows the results of such experiments using the diploid INVSc1 strain as the expression host. The wild-type INVSc1 strain yields a VLCFA titer of ~7.5 µmol/g dry weight, and these VLCFAs range from 20:0 to 28:0, with the most abundant VLCFA being 26:0. The individual expression of ZmKCS5, ZmKCS6, GL2 or GL2-LIKE does not significantly affect the VLCFA titers, as compared to the wild-type host strain (Figures 1A, B). However, the co-expression of ZmKCS5 (Figures 1A, C) or ZmKCS6 (Figures 1B, D) with either GL2 or GL2-LIKE induced an approximately 2-fold increase in the VLCFA titer (Figures 1A, B), and this is particularly associated with increased accumulation of the 24:0, 26:0 and 28:0 VLCFAs (Figures 1C, D). The fact that these increased titers occur only upon the co-expression of ZmKCS and GL2 homologs, and not when these maize proteins were individually expressed, is indicative that the GL2 homologs affect the activity of the ZmKCS proteins; possibly via interactions between the co-expressed proteins, analogous to the CER2-KCS interactions in Arabidopsis (Haslam et al., 2017).
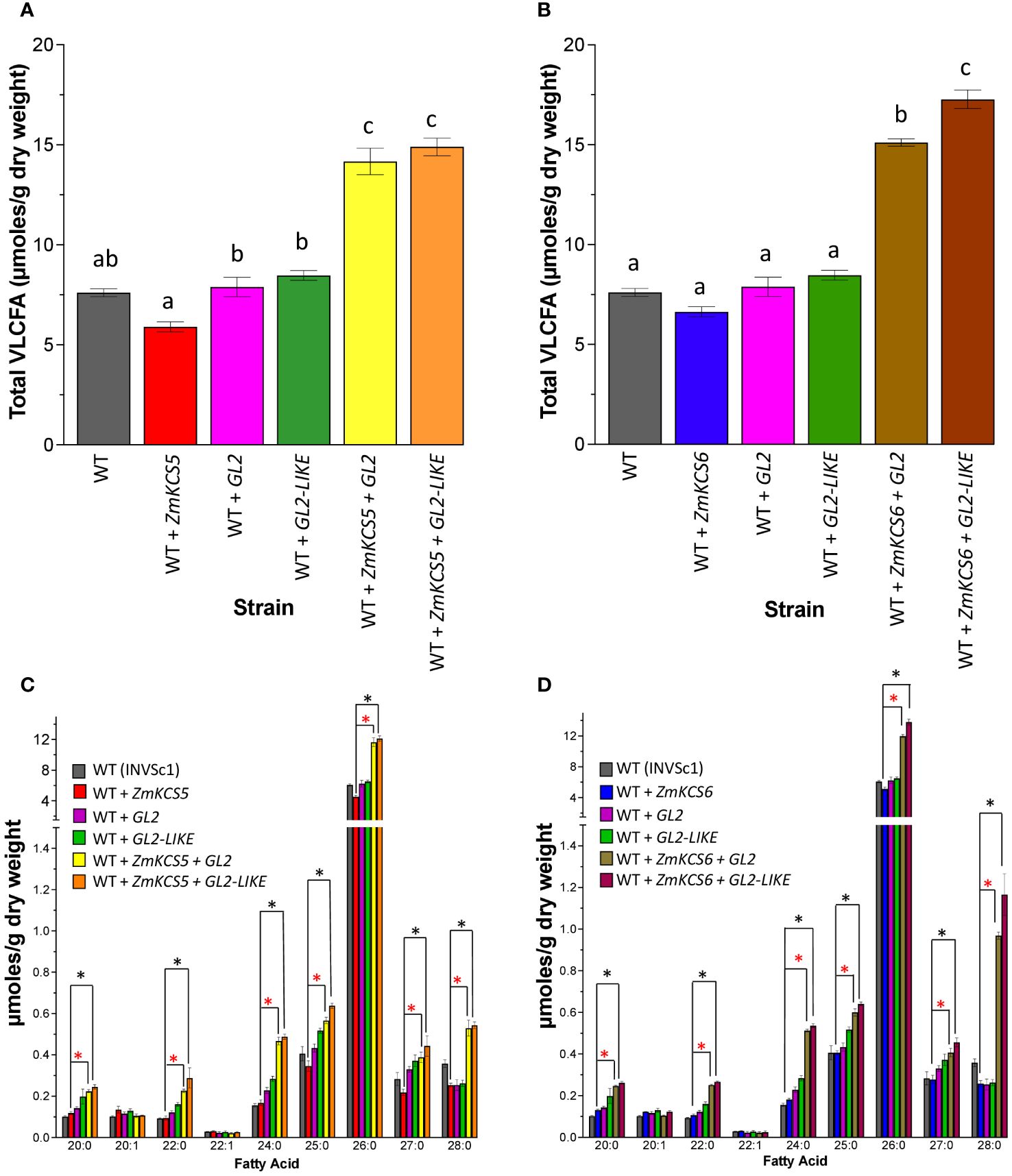
Figure 1 VLCFA accumulation in the INVSc1 yeast diploid strain co-expressing GL2 or GL2-LIKE with ZmKCS isozymes. (A) VLCFA accumulation in WT (INVSc1) and the isogenic yeast strains expressing individually or in combination ZmKCS5 and GL2 or GL2-LIKE. (B) VLCFA accumulation in WT (INVSc1) and the isogenic yeast strains expressing individually or in combination ZmKCS6 and GL2 or GL2-LIKE. (C) VLCFA composition in WT (INVSc1) and the isogenic yeast strains expressing individually or in combination ZmKCS5 and GL2 or GL2-LIKE. (D) VLCFA composition in WT (INVSc1) and the isogenic yeast strains expressing individually or in combination ZmKCS6 and GL2 or GL2-LIKE. All maize genes were episomally expressed under the transcriptional control of the galactose-inducible GAL1 promoter. All data are the average ± standard error (n = 6). Statistical significance of differences among all yeast strains was determined by Tukey’s HSD test (Supplementary Table 2). In (A, B), different letters above each data bar indicate statistically significant differences among the strains (p-value ≤ 0.05). In (C, D), asterisks identify statistically significant differences (p-value ≤ 0.05) between the strain expressing only a ZmKCS isozyme and the strain co-expressing the ZmKCS isozyme with GL2 (red asterisk) or with GL2-LIKE (black asterisk).
However, these results do not recapitulate the in planta expectation based on the gl2 mutant phenotype, which indicates that this gene may be involved in the ability of plant cells to produce VLCFAs of beyond 28:0 and up to 34:0 (Bianchi et al., 1975; Alexander et al., 2020). Therefore, we considered whether this may be associated with either the promoter that was used in the co-expression experiments (i.e., the galactose inducible GAL1 promoter) or by the particular yeast strain that was used, INVSc1, whose provenance is proprietary, and unknown. Hence, both the GL2 and GL2-LIKE proteins were individually co-expressed with either ZmKCS5 or ZmKCS6 under the control of the constitutive, glyceraldehyde-3-phosphate dehydrogenase (GPD) promoter (Bitter et al., 1987) in the diploid strain, BY4743, which has a well-defined pedigree (Brachmann et al., 1998).
In contrast to the results obtained with the GAL1 promoter, these experiments with the GPD promoter in either the INVSc1 or BY4743 diploid strains generated qualitatively different VLCFA profiles (Figure 2) that more accurately recapitulate the in planta chemotypes expected for the GL2 protein (Bianchi et al., 1975). Specifically, in both the INVSc1 (Figures 2A, B) and BY4743 (Figures 2C, D) strains, the co-expression of GL2 or GL2-LIKE with either ZmKCS5 or ZmKCS6 induced the accumulation of 30:0 VLCFA, a product that was undetectable in the other strains that singularly expressed ZmKCS5, ZmKCS6, GL2 or GL2-LIKE (compare Figures 1, 2). These distinct observations between the use of the GPD and GAL1 promoters in different diploid strains indicate that additional insights are required to dissect the complexity of co-expressing FAE components in heterologous biological systems, such as plant FAE components in yeast strains that also carry an endogenous FAE system.
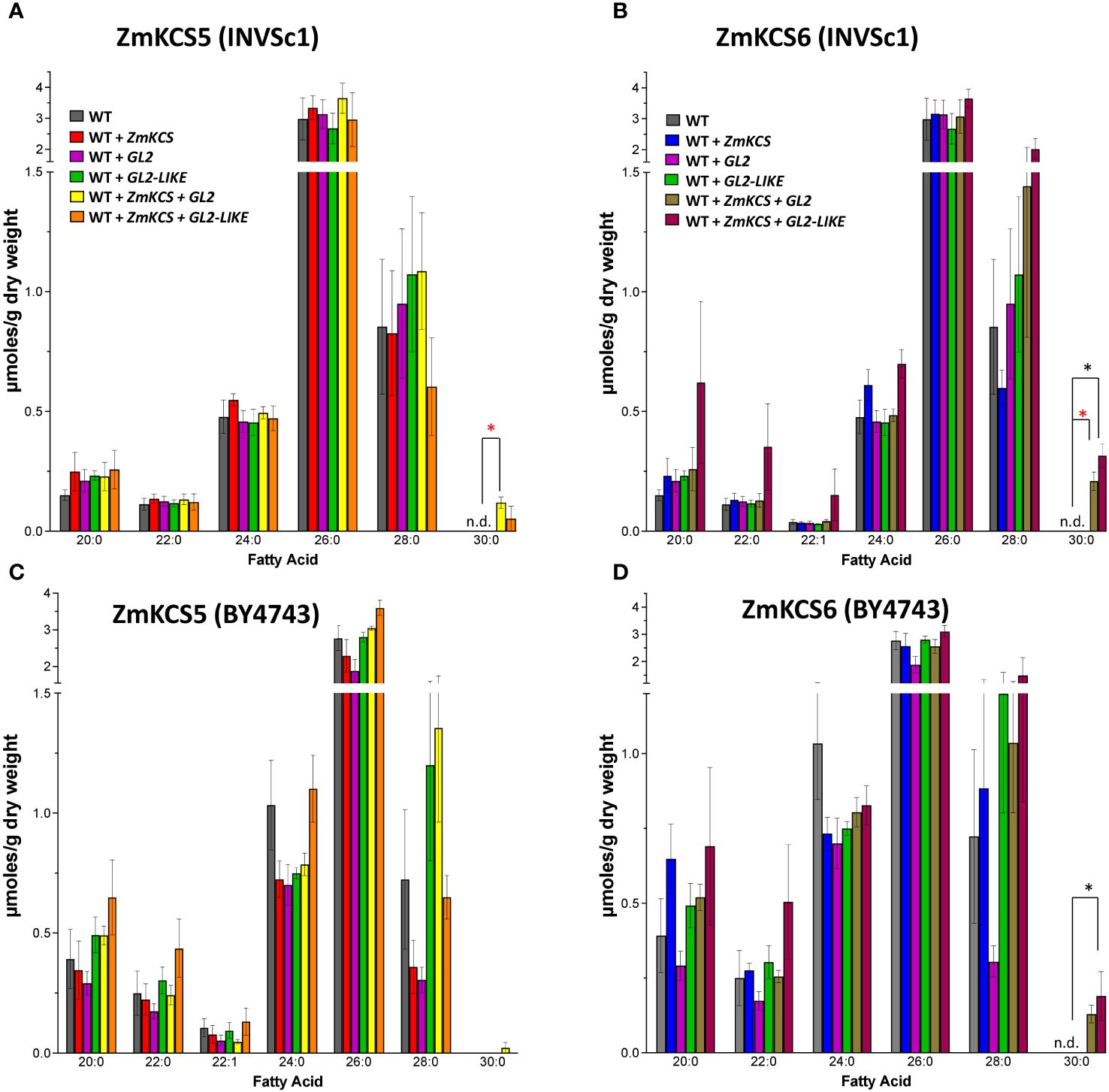
Figure 2 VLCFA accumulation in the yeast diploid strains, INVSc1 and BY4743, co-expressing GL2 or GL2-LIKE with ZmKCS isozymes.(A) VLCFA composition in WT (INVSc1), and the isogenic yeast strains expressing individually or in combination ZmKCS5 and GL2 or GL2-LIKE. (B) VLCFA composition in WT (INVSc1), and the isogenic yeast strains expressing individually or in combination ZmKCS6 and GL2 or GL2-LIKE. (C) VLCFA composition in WT (BY4743) and the isogenic yeast strains expressing individually or in combination ZmKCS5 and GL2 or GL2-LIKE. (D) VLCFA composition in WT (BY4743) and the isogenic yeast strains expressing individually or in combination ZmKCS6 and GL2 or GL2-LIKE. All maize genes were episomally expressed under the transcriptional control of the constitutive GPD promoter. All data are the average ± standard error (n = 3). Statistical significance of differences among all yeast strains was determined by Tukey’s HSD test (Supplementary Table 2). Asterisks identify statistically significant differences (p-value ≤ 0.05) between the strain expressing only a ZmKCS isozyme and the strain co-expressing the ZmKCS isozyme with GL2 (red asterisk) or with GL2-LIKE (black asterisk).
3.2 Effect of co-expressing maize GL2 or GL2-LIKE with maize KCS isozymes as replacements of the yeast ELO3 component-enzyme of FAE
To overcome the complexity of genetically adding heterologous maize FAE components to an intact FAE system (i.e., the yeast FAE), we co-expressed ZmKCS paralogs with either GL2 or GL2-LIKE in a yeast strain that lacked the analogous endogenous yeast FAE component enzyme. These experiments were conducted in a haploid strain (i.e., BY4742, which is one of the haploid progenitors of the diploid strain, BY4743 (Brachmann et al., 1998)) that carries mutations in genes coding for yeast FAE component genes. The yeast FAE system does not utilize KCS-type condensing enzymes, but rather utilizes ELO-type condensing enzymes, encoded by the ELO1, ELO2 or ELO3 genes (Toke and Martin, 1996; Oh et al., 1997). Because ELO3 has the ability to catalyze the terminal cycles of fatty acid elongation using preexisting 24:0 and 26:0 acyl-CoAs (Oh et al., 1997; Denic and Weissman, 2007), we first recapitulated the above experiments in the BY4742 haploid strain that carried an elo3 null allele (note that this strain carries functional ELO1 and ELO2 genes). In these experiments we individually expressed ZmKCS4, ZmKCS5, ZmKCS6 and ZmKCS15 in the elo3 mutant yeast strain, and compared the VLCFA profiles when these ZmKCS isozymes were co-expressed with either GL2 or GL2-LIKE (Figure 3). The rationale for including ZmKCS4 and ZmKCS15 isozymes is based on the fact that these two plant isozymes have been shown to be functional in yeast (Stenback et al., 2022).
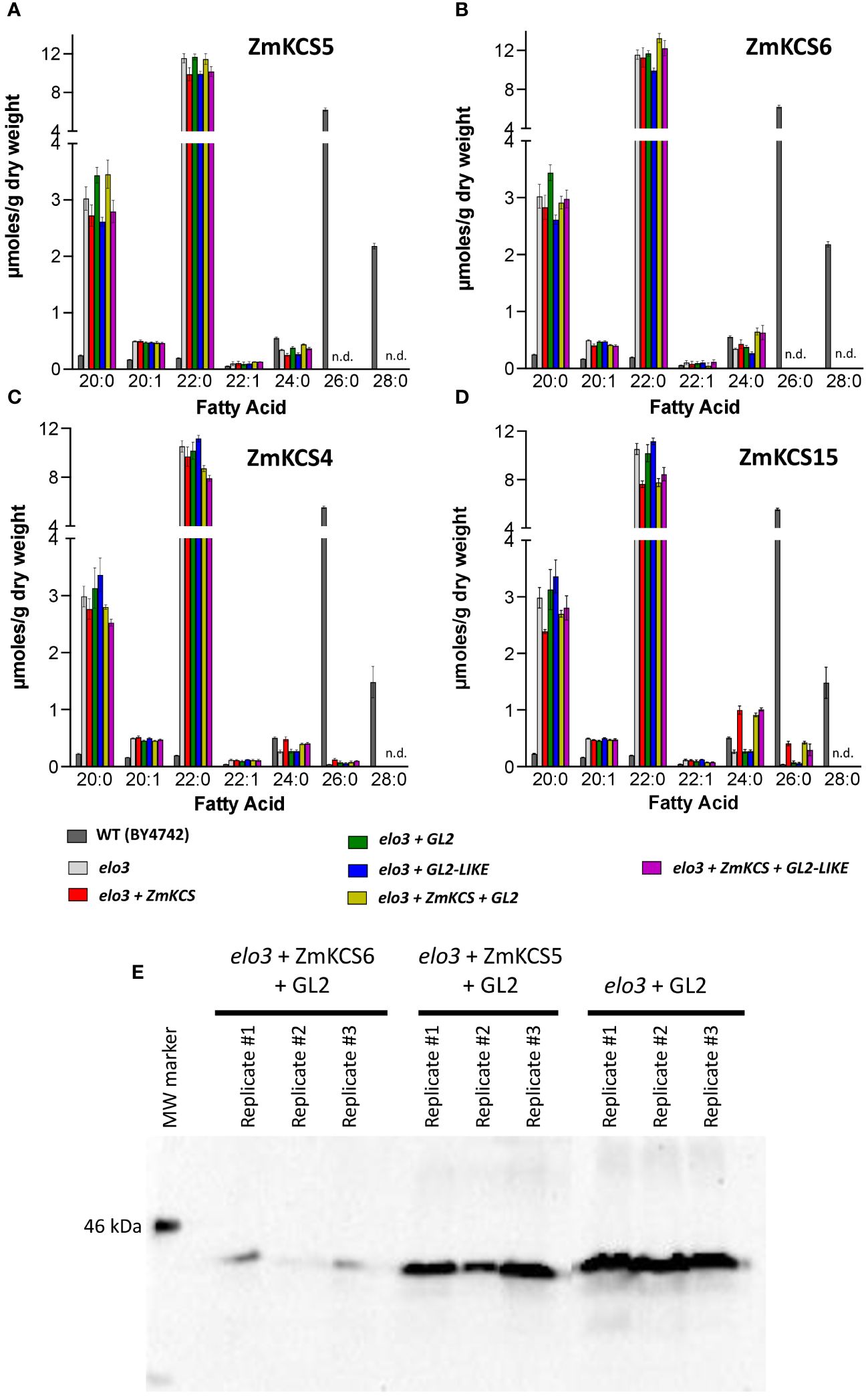
Figure 3 VLCFA accumulation in the yeast haploid strain (BY4742) carrying an elo3 null allele co-expressing GL2 or GL2-LIKE with ZmKCS isozymes. (A) VLCFA composition of yeast elo3 mutant strains expressing individually or in combination ZmKCS5 and GL2 or GL2-LIKE. (B) VLCFA composition of yeast elo3 mutant strains expressing individually or in combination ZmKCS6 and GL2 or GL2-LIKE. (C) VLCFA composition of yeast elo3 mutant strains expressing individually or in combination ZmKCS4 and GL2 or GL2-LIKE. (D) VLCFA composition of yeast elo3 mutant strains expressing individually or in combination ZmKCS15 and GL2 or GL2-LIKE. (E) GL2 protein expression demonstrated by Western blot analyses of the indicated strains (n=3). All maize genes were episomally expressed under the transcriptional control of the constitutive GPD promoter. All fatty acid composition data are the average ± standard error (n = 4). Statistical significance of differences among all yeast strains was determined by Tukey’s HSD test (Supplementary Table 2). There were no statistically significant differences (p-value ≤ 0.05) in VLCFA profiles between the strain expressing only a ZmKCS isozyme and the strain co-expressing the ZmKCS isozyme with GL2 or with GL2-LIKE.
As with the diploid strains, the wild-type BY4742 haploid strain produced VLCFAs that range between 20 and 28 carbon chain lengths, with 26:0 accounting for 80% of the VLCFAs. The effect of the elo3 mutation was to eliminate the accumulation of 26:0 and 28:0, and thus the predominant VLCFA in this mutant yeast strain was 24:0. The expression of ZmKCS5 or ZmKCS6 in the elo3 mutant strain had no effect on the fatty acid profiles (Figures 3A, B). However, the expression of ZmKCS4 or ZmKCS15 in the elo3 mutant strain resulted in partially restoring the ability of the strain to produce the longer VLCFAs; specifically these strains produced small but statistically significant quantities of 26:0 (Figures 3C, D). Thus, in contrast to ZmKCS4 or ZmKCS15, neither ZmKCS5 or ZmKCS6 were capable of replacing the elo3 function. Moreover, the co-expression of either GL2 or GL2-LIKE with each of these four ZmKCS isozymes was incapable of modifying the abilities of the resulting strains to produce larger quantities of the longer chain VLCFAs (i.e., 26:0, 28:0, or 30:0) (Figure 3).
3.3 Effect of co-expressing maize GL2 or GL2-LIKE with maize KCS isozymes that functionally replace the yeast ELO function
As demonstrated by the synthetic lethality associated with the elo2 elo3 double mutant, the ability to produce VLCFAs is essential for yeast viability (Toke and Martin, 1996; Oh et al., 1997). Moreover, prior characterizations have established that five ZmKCS isozymes (i.e., ZmKCS2, ZmKCS4, ZmKCS11, ZmKCS15, and ZmKCS20) are capable of genetically rescuing this lethality of the elo2 elo3 double mutant (Stenback et al., 2022). Therefore, we used these rescued strains to evaluate whether the co-expression of GL2 or GL2-LIKE could modify the product profile of the FAE system, indicative of interactions with the ZmKCS isozymes.
As previously reported (Stenback et al., 2022), in the absence of either GL2 or GL2-LIKE, the expression of ZmKCS2 enables the elongation of fatty acids to 22-carbon chain length, whereas ZmKCS4, ZmKCS11, and ZmKCS20 enables the elongation of fatty acids to 24-carbon chain length, and ZmKCS15 can elongate fatty acids up to 26-carbon atoms (Figure 4). These acyl-products include VLCFAs and 2-hydroxy-VLCFAs, which are hydroxylated post-synthesis in the assembly of ceramide-based lipids (Erdbrügger and Fröhlich, 2021). The co-expression of either GL2 or GL2-LIKE with each of these ZmKCS isozymes did not cause any qualitative change in the VLCFA profiles; namely there were no new VLCFA products generated that were absent from the strains that only expressed the ZmKCS isozyme. Rather, there were statistically significant quantitative changes in the accumulation of some of the VLCFA products. For example, GL2-LIKE caused increased accumulation of 2-hydroxy-22:0 or 2-hydroxy-24:0 when it was co-expressed with ZmKCS11 (Figure 4C) or ZmKCS15 (Figure 4D), respectively; the latter change was accompanied by the decreased accumulation of 26:0 (Figure 4D).
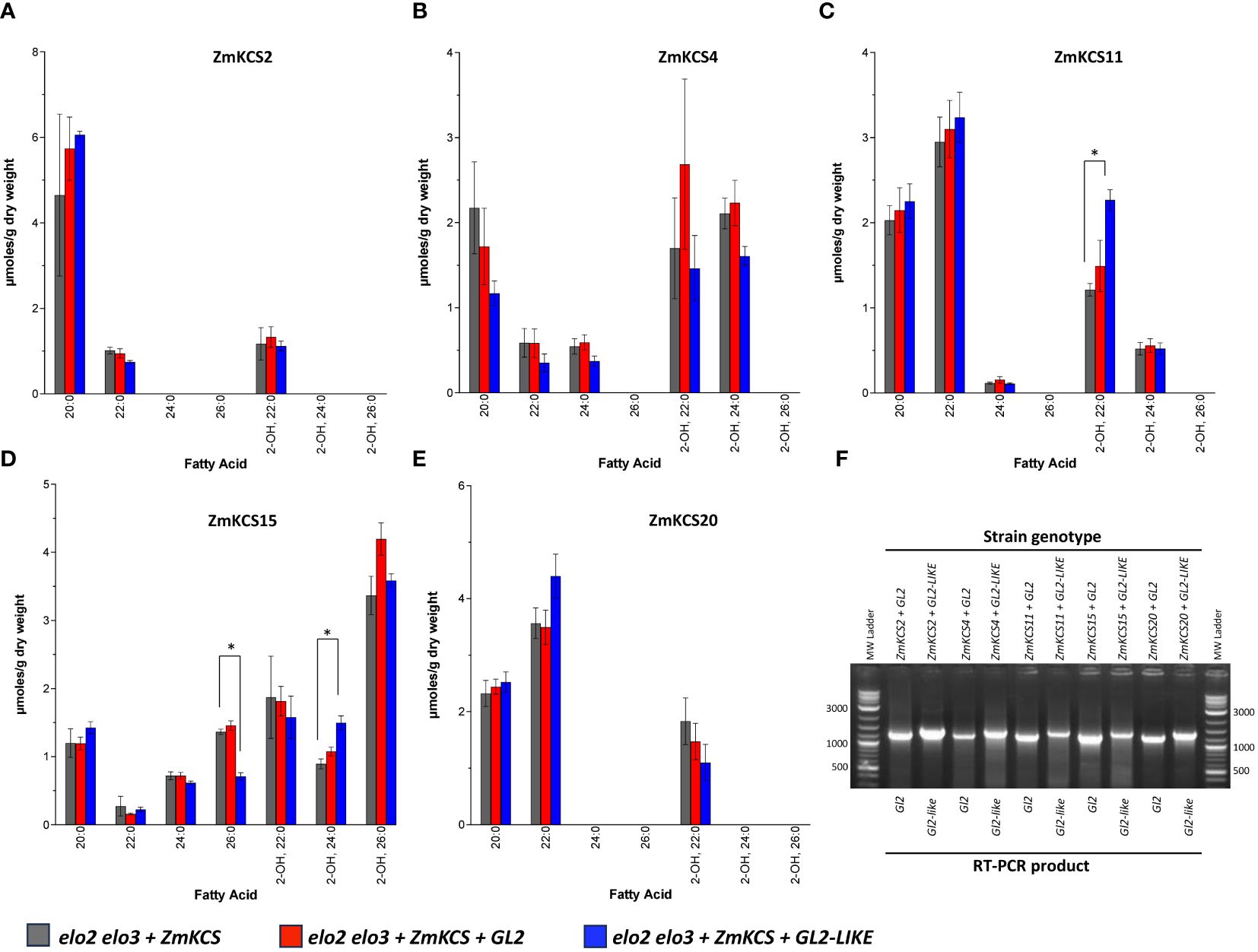
Figure 4 VLCFA accumulation upon co-expressing GL2 or GL2-LIKE with ZmKCS isozymes that genetically complement the lethality of the elo2 elo3 double mutant yeast strain. VLCFA composition of yeast elo2 elo3 double mutant strains expressing individually or in combination GL2 or GL2-LIKE and ZmKCS2 (A), ZmKCS4, (B) ZmKCS11 (C), ZmKCS15 (D), and ZmKCS20 (E). (F) RT-PCR analysis of the expression of the Gl2 (1300-bp product) and Gl2-like (1400-bp product) mRNAs in the indicated yeast strains. All maize genes were episomally expressed under the transcriptional control of the GPD constitutive promoter. Numerical data in panels (A–E) are the average ± standard error (n=3). Statistical significance of differences among all yeast strains was determined by Tukey’s HSD test (Supplementary Table 2). Asterisks identify statistically significant differences (p-value ≤ 0.05) between the strain expressing only a ZmKCS isozyme and the strain co-expressing that ZmKCS isozyme with GL2 (red asterisk) or with GL2-LIKE (black asterisk).
3.4 Effect of co-expressing maize GL2 or GL2-LIKE in combination with pairs of maize KCS isozymes
The findings observed with the GL2 and GL2-LIKE proteins (Figure 4) are not consistent with the model developed with the Arabidopsis homologs of these proteins (i.e., the CER2 and CER2-LIKE proteins) (Haslam et al., 2017). This difference between the maize and Arabidopsis homologs may be associated with the fact that the CER2 and CER2-LIKE proteins mediate this effect by specifically interacting with the CER6- or CER60-encoded KCS isozymes (Haslam et al., 2015). The difference between the two systems (i.e., maize versus Arabidopsis) may be due to the fact that the ZmKCS isozymes used in the current study are in a phylogenetic clade that is distinct from the Arabidopsis CER6- or CER60-encoding KCS isozymes. In maize, the phylogenetic homologs to the CER6-and CER60-encoding KCSs are ZmKCS5 and ZmKCS6 (Campbell et al., 2019). However, because ZmKCS5 and ZmKCS6 are incapable of complementing the yeast elo2 elo3 double mutant strain (Stenback et al., 2022), it was not possible to co-express them individually with GL2 or GL2-LIKE in the yeast system lacking the native condensing enzymes. Therefore, we co-expressed either ZmKCS5 or ZmKCS6 in the yeast elo2 elo3 double mutant strain that was rescued by genetic complementation by the expression of ZmKCS15. This complemented strain was chosen because it was capable of producing the longest VLCFAs (i.e., up to 26:0) (Stenback et al., 2022). Figure 5 shows that the co-expression of GL2 or GL2-LIKE with either ZmKCS5 or ZmKCS6 in the ZmKCS15-rescued elo2 elo3 double mutant had no effect on the VLCFA profiles that were generated.
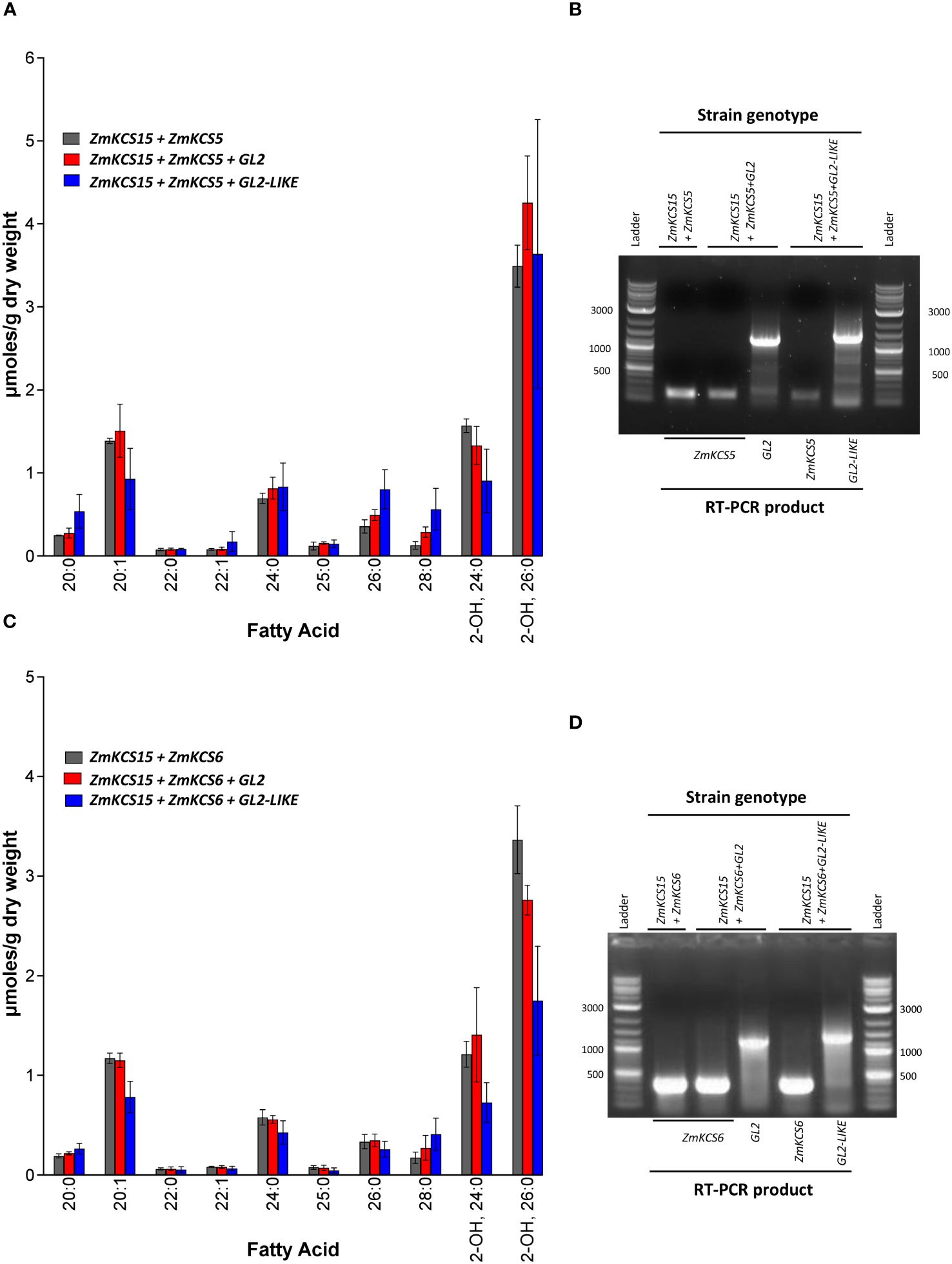
Figure 5 VLCFA accumulation upon co-expressing GL2 or GL2-LIKE with different combinations of ZmKCS isozymes that rescue the lethality of the elo2 elo3 double mutant yeast strain. (A) VLCFA composition of yeast elo2 elo3 double mutant strains genetically complemented by the combined co-expression of the ZmKCS15 and ZmKCS5 isozymes, in the absence or presence of either GL2 or GL2-LIKE. (B) RT-PCR analysis of the expression of the Gl2 (1300-bp product), Gl2-like (1400-bp product) and ZmKCS5 (168-bp product) mRNAs in the indicated yeast strains. (C) VLCFA composition of yeast elo2 elo3 double mutant strains genetically complemented by the combined co-expression of the ZmKCS15 and ZmKCS6 isozymes, in the absence or presence of either GL2 or GL2-LIKE. (D) RT-PCR analysis of the expression of the Gl2 (1300-bp product), Gl2-like (1400-bp product) and ZmKCS6 (330-bp product) mRNAs in the indicated yeast strains. All maize genes were episomally expressed under the transcriptional control of the GPD constitutive promoter. Numerical data in panels (A, C) are the average ± standard error (n=3). Statistical significance of differences among all yeast strains was determined by Tukey’s HSD test (Supplementary Table 2). There were no statistically significant differences (p-value ≤ 0.05) in VLCFA profiles between the strain co-expressing combinations of ZmKCS isozymes in the absence or presence of either GL2 or GL2-LIKE.
Comparing the VLCFA profiles in Figures 4D and 5A, we found that the co-expression of ZmKCS15 with ZmKCS5 in the elo2 elo3 double mutant is no different than solely expressing ZmKCS15 in this strain, and the co-expression of GL2 or GL2-LIKE with both ZmKCS15 and ZmKCS5 had little effect on the profile (Figure 5A). Similar results were obtained when ZmKCS15 expression was combined with ZmKCS6 expression; the most pronounced apparent effect, which is not statistically significant, was a 50% reduction in the accumulation of 2-hydroxy-26:0 upon the co-expression with GL2-LIKE (Figure 5B).
3.5 Effect of co-expressing GL2 and GL2-LIKE with other FAE components
Prior studies have indicated that protein-protein interactions among the FAE component enzymes are important for VLCFA biosynthesis; in the Arabidopsis system this includes interactions between CER2-LIKE proteins and the KCR, HCD and ECR components (Kim et al., 2022). Therefore, using the yeast strains that we previously developed to functionally characterize these additional components of the maize FAE system (Campbell et al., 2019), we evaluated whether the GL2 or GL2-LIKE proteins can interact with additional FAE components (i.e., ZmKCR1, ZmKCR2 and ZmHCD) and thereby affect the VLCFA product-profile of the FAE system.
We evaluated if either GL2 or GL2-LIKE could affect changes in VLCFA profiles upon co-expression with either ZmKCR1 (gl8a) or ZmKCR2 (gl8b) (Xu et al., 1997, 2002; Dietrich et al., 2005). These co-expression experiments were conducted in yeast strains that lacked the endogenous yeast KCR (i.e., the ybr159Δ mutant strain) (Han et al., 2002). The yeast ybr159Δ mutation is lethal, but can be rescued by the expression of either ZmKCR1 or ZmKCR2 (Campbell et al., 2019). The resulting rescued yeast strains produced VLCFA profiles that are similar to those that occur in the wild-type strain, with 26:0 being the most abundant VLCFA (Figure 6). The co-expression of either GL2 or GL2-LIKE with the two maize KCR isozymes, ZmKCR1 (Figure 6A) or ZmKCR2 (Figure 6B), produced minor, but statistically significant quantitative changes in the VLCFA profiles, affecting only 22:0, 24:0, and 26:0.
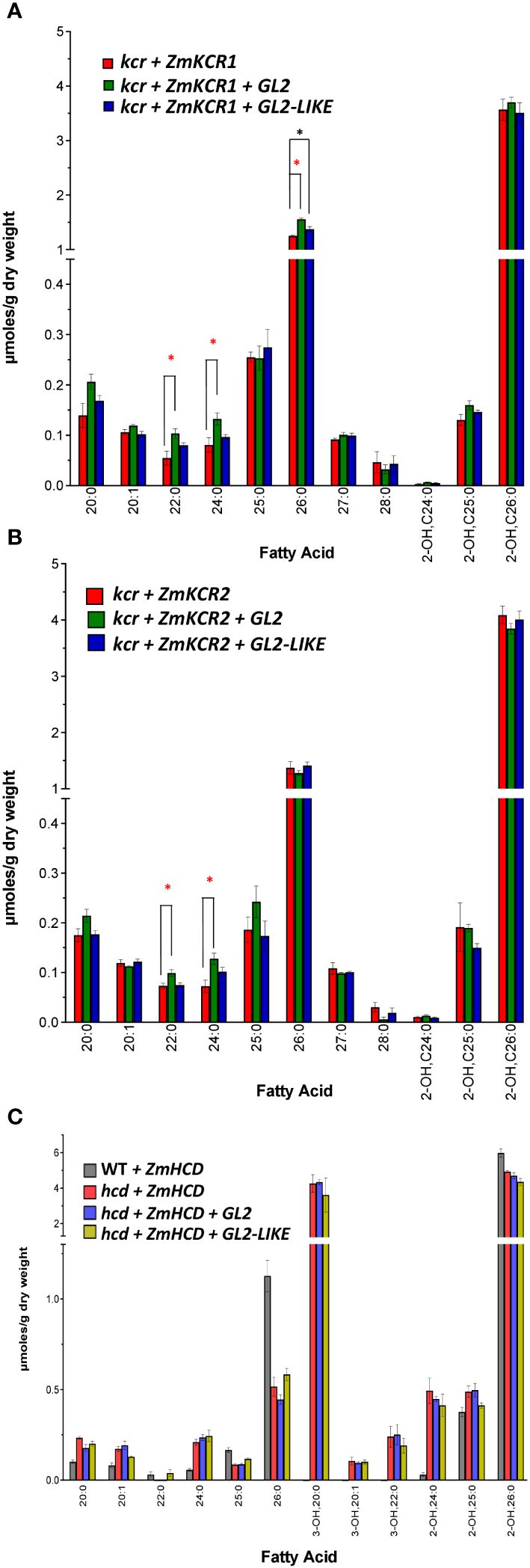
Figure 6 VLCFA accumulation upon co-expressing GL2 or GL2-LIKE with ZmKCR1, ZmKCR2 or ZmHCD that rescue the lethality of the yeast kcr or hcd null alleles, respectively. (A) VLCFA composition of the yeast kcr mutant strain genetically rescued by the expression of ZmKCR1, in the absence or presence of either GL2 or GL2-LIKE. (B) VLCFA composition of the yeast kcr mutant strain genetically rescued by the expression of ZmKCR2, in the absence or presence of either GL2 or GL2-LIKE. (C) VLCFA composition of the yeast hcd mutant strain genetically rescued by the expression of ZmHCD, in the absence or presence of either GL2 or GL2-LIKE. All maize genes were episomally expressed under the transcriptional control of the galactose-inducible GAL1 inducible promoter. Data are the average ± standard error (n=4). Statistical significance of differences among all yeast strains was determined by Tukey’s HSD test (Supplementary Table 2). Asterisks identify statistically significant differences (p-value ≤ 0.05) between the strain expressing GL2 (red asterisk) or with GL2-LIKE (black asterisk), relative to their absence.
Finally, using the same strategy we investigated if GL2 or GL2-LIKE interacts with ZmHCD to affect VLCFA profiles, by co-expressing the two combinations in the yeast hcd mutant strain (Campbell et al., 2019). Figure 6C compares the VLCFA profiles of the ZmHCD-rescued hcd mutant yeast strains, with and without the co-expression of either GL2 or GL2-LIKE. Although the VLCFA profile of the yeast strain is not altered when ZmHCD is expressed in the wild-type background, the profile is strikingly different when ZmHCD is expressed in the hcd mutant background. Specifically, ZmHCD complementation induces the appearance of 3-hydroxy-VLCFAs of 20 and 22 carbon chain lengths and a 50% reduction in 26:0. We suggest that this is because the maize HCD is not as efficient in replacing the endogenous yeast HCD in converting 3-hydroxyacyl-CoA to the enoyl-CoA, and thus 3-hydroxy-VLCFAs accumulate. Even so, the co-expression of ZmHCD with either GL2 or GL2-LIKE did not further alter the VLCFA profiles of the rescued strains (Figure 6C).
3.6 Molecular confirmation of the expression of maize genes in yeast
The expression of ZmKCS2, ZmKCS4, ZmKCS11, ZmKCS15 and ZmKCS20 proteins in yeast cells was genetically confirmed because their presence rescued the lethality associated with the yeast elo2 elo3 double mutant strain (e.g., Figure 4). In addition, changes in VLCFA profiles in a number of yeast strains indicate that the GL2 and GL2-LIKE proteins were successfully expressed (e.g., Figures 1, 2). However, in some strains that were engineered to express these proteins, the resultant VLCFA profiles were very similar to those generated by the host strain (e.g., Figures 3–5). Therefore, we sought molecular evidence to confirm successful expression of the GL2 and GL2-LIKE proteins, and ZmKCS isozymes. Such confirmation was obtained by either Western blot or RT-PCR analyses.
For example, Figure 3E shows Western blot analyses of protein extracts from a subset of the yeast elo3-mutant strains, which confirms that the GL2 protein was successfully expressed in these experiments, even though its expression did not generate a change in the VLCFA profile. Analogously, RT-PCR analysis confirmed the expression of the Gl2 and Gl2-like mRNA in the yeast elo2 elo3 double mutant strains that were genetically complemented by the expression of the ZmKCS2, ZmKCS4, ZmKCS11, ZmKCS15 or ZmKCS20 proteins (Figure 4F), even though the expression of Gl2 and Gl2-like did not qualitatively change the VLCFA profiles (Figures 4A–E).
Similarly, we evaluated the expression of mRNAs encoding for GL2 or GL2-LIKE proteins and either ZmKCS5 (Figure 5B) or ZmKCS6 (Figure 5D), which were co-expressed with ZmKCS15 in the yeast elo2 elo3 double mutant strain. This double mutant strain is only viable because of the expression of the ZmKCS15 protein (Figure 4D). Thus, in these strains ZmKCS15 was co-expressed with either ZmKCS5 or ZmKCS6, in the presence or absence of either GL2 or GL2-LIKE. RT-PCR analyses confirmed the expression of Gl2 and Gl2-like mRNAs and either ZmKCS5 (Figure 5B) or ZmKCS6 (Figure 5D), even though these genetic modifications did not affect changes in the VLCFA profiles (Figures 5A, C).
4 Discussion
The BAHD class of acyltransferases was initially identified by the biochemical characterization of four enzymes that are involved in the biosynthesis of plant secondary metabolites (St-Pierre and Luca, 2000). Subsequently, hundreds of these enzymes have been characterized from diverse phylogenetic sources, and they have been classified into eight different sequence-based clades, which also segregate these enzymes according to the chemical nature of the substrate that is acylated (Kruse et al., 2022; Moghe et al., 2023). One of these clades (i.e., Clade 2) contains proteins (i.e., GL2 and CER2) whose acyltransferase activity is uncharacterized. These two proteins were genetically defined by mutant alleles that affect the normal accumulation of cuticular waxes (Hayes and Brewbaker, 1928; Koornneef et al., 1989). Following the molecular characterization of these genetic loci (Tacke et al., 1995; Negruk et al., 1996; Xia et al., 1996), the homologous Gl2-like locus (Alexander et al., 2020) and four homologous CER2-LIKE loci (Pascal et al., 2013; Haslam et al., 2015; Haslam and Kunst, 2020) were identified in maize and Arabidopsis, respectively. Additionally, homologs of these proteins have been characterized from a number of different plant species, including rice (Wang et al., 2017), onion (Liu et al., 2023), broccoli (Han et al., 2021), cabbage (Ji et al., 2021), and poplar (Gonzales-Vigil et al., 2021).
The functionality of these proteins has been based on a model developed from extensive characterizations of the Arabidopsis CER2 and CER2-LIKE proteins (reviewed by Haslam et al. (2017)). These characterizations indicate that the CER2 and CER2-LIKE proteins are modulators of the product-profile of the FAE system, mediated by physical interactions with the KCS component of the FAE system (Haslam and Kunst, 2020; Kim et al., 2022). Hence, in the presence of CER2 or CER2-LIKE, the FAE system can produce longer chain VLCFAs than in their absence (Haslam et al., 2012, 2015; Haslam and Kunst, 2020). This functionality model was primarily developed from the analyses of yeast strains that co-express Arabidopsis KCS isozymes with CER2 or CER2-LIKE proteins. Specifically, the singular expression of an Arabidopsis KCS isozyme in yeast produces VLCFAs of only 28-carbon chain length. However, co-expression of CER2 or CER2-LIKE proteins with Arabidopsis KCS isozymes enables the yeast strain to produce VLCFAs of longer chain lengths, up to 34-carbon atoms (Haslam et al., 2012, 2015; Haslam and Kunst, 2020). Moreover, this modification of the product profile of the FAE system by CER2 and CER2-LIKE proteins displays specificity, occurring only with the Arabidopsis KCS6 (CER6; AT1G68530) and KCS5 (CER60; AT1G25450) proteins, and does not occur with KCS1 (AT1G01120), KCS10 (FIDDLEHEAD; AT2G26250), KCS9 (AT2G16280) or KCS20 (AT5G43760) (Haslam et al., 2015). This model provides an explanation for the observed in planta change in the cuticular wax phenotypes expressed by the loss-of-function cer2 or cer2-like mutants. Specifically, Arabidopsis cer2 mutants do not accumulate cuticular VLCFAs and derivative products that are greater than 28-carbon chain length, whereas in the wild-type state these cuticular components are derived from 30- to 34-carbon VLCFAs (Negruk et al., 1996; Haslam et al., 2012, 2015; Haslam and Kunst, 2020).
We had previously characterized the functionality of the maize GL2 and GL2-LIKE proteins relative to CER2 function by in planta genetic complementation experiments, which indicated that the two maize homologs have overlapping and distinct functions. Specifically, the transgenic expression of either the GL2 or GL2-LIKE protein in Arabidopsis can complement the cer2 mutation by restoring the production of VLCFAs of 26:0 and greater chain lengths. However, whereas the GL2-LIKE protein requires an intact HXXXDX catalytic-motif to fully complement the cer2 function, GL2 protein can accomplish this complementation without an intact HXXXDX motif (Alexander et al., 2020).
In this study we explored the potential interactions between the GL2 or GL2-LIKE proteins with the maize FAE components, using yeast as the co-expression platform. Extrapolating from the functionality model developed for the homologous CER2 and CER2-LIKE proteins, we initially focused on the ability of GL2 or GL2-LIKE to affect the capability of ZmKCS isozymes to produce longer chain VLCFAs. We found that the effects of GL2 or GL2-LIKE were dependent on the yeast strain that was used and on the promoter that was used to drive the co-expression of these proteins. Specifically, when co-expression was controlled by the GAL1 promoter, in the diploid INVSc1 yeast strain, both GL2 or GL2-LIKE affected the ability of ZmKCSs to produce larger quantities of the longer chain VLCFAs, without enabling additional elongation cycles that would produce even longer chain products (i.e., only producing VLCFAs of up to 28-carbon atoms). However, when co-expression was controlled by the constitutive GPD promoter, in two diploid strains (i.e., INVSc1 or BY4743), both GL2 or GL2-LIKE stimulated the ability of ZmKCSs to produce the longer chain VLCFAs (i.e., 30:0) that could not be produced in the absence of GL2 or GL2-LIKE.
Although the latter results with the GPD promoters are consistent with the functionality model developed with the Arabidopsis CER2 and CER2-LIKE proteins (Haslam et al., 2017), we considered the possibility that the inconsistent results obtained with the GAL1 promoter in the INVSc1 strain may be associated with the complexity of co-expressing the ZmKCS isozymes with GL2 or GL2-LIKE in a host that has an intact endogenous FAE system. Adding to this complexity is the fact that the endogenous yeast FAE system does not utilize a KCS-type condensing enzyme, but rather utilizes a combination of three ELO-type condensing enzymes (ELO1, ELO2 and ELO3) (Toke and Martin, 1996; Oh et al., 1997). This complexity was partially overcome by taking advantage of yeast strains that we had previously developed, which were viable only because of the expression of maize FAE components (Campbell et al., 2019; Stenback et al., 2022). These strains carry null alleles in the endogenous FAE component genes (i.e., elo2 elo3 double mutant, kcr and hcd mutants), which is a lethal condition. However, the lethality associated with these mutants was rescued by the expression of individual maize FAE components. The co-expression of GL2 or GL2-LIKE in these strains did not significantly affect the VLCFA profiles produced by the resulting strains, in particular there was no induction of the synthesis of longer chain VLCFAs (i.e., of 30-carbons or longer), as was expected based on the functionality model developed with the Arabidopsis CER2 and CER2-LIKE genes (Haslam et al., 2017).
There are a number of potential explanations for these observations. For example, the GL2 and GL2-LIKE proteins may have functions that are distinct from the CER2 or CER2-LIKE proteins. However, our prior study, which demonstrated that the in planta expression of the two maize proteins can genetically complement the cer2 mutation, is inconsistent with this explanation (Alexander et al., 2020). Another possibility is that the effect of the GL2 and GL2-LIKE proteins on FAE is masked by the fact that the yeast strains that we developed expressed a hybrid FAE system that mixed maize and yeast components. This is a very viable explanation because bimolecular fluorescence complementation and yeast two-hybrid assays have indicated that protein–protein interactions are important in the assembly of a functional FAE system (Kim et al., 2022). This is of particular significance because the yeast FAE system uses an ELO-type enzyme to catalyze the Claisen condensation reaction to generate the new carbon-carbon bond that enables the elongation of the substrate (Toke and Martin, 1996; Oh et al., 1997), whereas the interaction between CER2 or CER2-LIKE proteins with FAE is mediated through interactions with the KCS component (Haslam et al., 2017). Indeed, with the many recent synthetic biological reagents that have been developed for yeast, it is becoming possible to evaluate this last possibility by reconstituting the entire plant FAE system. Most recently, the effect of the CER2 and CER2-LIKE proteins have been demonstrated with yeast strains that comprehensively express the Arabidopsis FAE system (Batsale et al., 2023), although these strains are also co-expressing the endogenous yeast FAE system. Based upon our prior reconstitution experiments (Campbell et al., 2019; Stenback et al., 2022) it should be possible to develop viable yeast strains that are solely expressing a maize FAE system, which can be used to specifically evaluate the functionality of GL2 or GL2-LIKE in affecting VLCFA production.
Data availability statement
The original contributions presented in the study are included in the article and Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author contributions
LEA: Data curation, Formal analysis, Investigation, Methodology, Supervision, Visualization, Writing – original draft. DW: Data curation, Formal analysis, Investigation, Methodology, Supervision, Visualization, Writing – original draft. KES: Investigation, Methodology, Writing – review & editing. KRC: Investigation, Methodology, Writing – review & editing. ET: Investigation, Methodology, Writing – review & editing. KF: Investigation, Methodology, Writing – review & editing. MAS: Investigation, Methodology, Writing – review & editing. LR: Investigation, Methodology, Writing – review & editing. MDY-N: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Writing – review & editing. BJN: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Visualization, Writing – review & editing. ML: Investigation, Visualization, Writing – original draft.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the State of Iowa, through Iowa State University’s Center for Metabolic Biology, by the National Science Foundation through grant MCB-2212799 to BJN and MDY-N, and the East Asia and Pacific Summer Institutes for U.S. Graduate Students (EAPSI) program of the National Science Foundation (grant OISE–1614020 to LEA). Additional support was provided by USDA-NIFA Hatch projects IOW0382 (BJN) and IOW03649 (MDY-N).
Acknowledgments
The authors acknowledge Dr. Alexis A. Campbell for providing the maize FAE component yeast replacement strains, and the W. M. Keck Metabolomics Research Laboratory, Office of Biotechnology, Iowa State University, Ames IA, for providing access to analytical instrumentation and Drs. Ann Perera and Lucas J. Showman for their assistance and support regarding metabolite profiling.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1403779/full#supplementary-material
References
Adams, A., Kaiser, C. (1998). Methods in yeast genetics: A Cold Spring Harbor Laboratory Course Manual (Plainview, NY: Cold Spring Harbor Laboratory Press).
Alberti, S., Gitler, A. D., Lindquist, S. (2007). A suite of Gateway cloning vectors for high-throughput genetic analysis in Saccharomyces cerevisiae. Yeast 24, 913–919. doi: 10.1002/yea.1502
Alexander, L. E., Okazaki, Y., Schelling, M. A., Davis, A., Zheng, X., Rizhsky, L., et al. (2020). Maize Glossy2 and Glossy2-like genes have overlapping and distinct functions in cuticular lipid deposition. Plant Physiol. 183, 840–853. doi: 10.1104/pp.20.00241
Bach, L., Michaelson, L. V., Haslam, R., Bellec, Y., Gissot, L., Marion, J., et al. (2008). The very-long-chain hydroxy fatty acyl-CoA dehydratase PASTICCINO2 is essential and limiting for plant development. Proc. Natl. Acad. Sci. U.S.A. 105, 14727–14731. doi: 10.1073/pnas.0805089105
Barbero, F. (2016). Cuticular lipids as a cross-talk among ants, plants and butterflies. Int. J. Mol. Sci. 17, 1966. doi: 10.3390/ijms17121966
Batsale, M., Alonso, M., Pascal, S., Thoraval, D., Haslam, R. P., Beaudoin, F., et al. (2023). Tackling functional redundancy of Arabidopsis fatty acid elongase complexes. Front. Plant Sci. 14, 1107333. doi: 10.3389/fpls.2023.1107333
Bianchi, G., Avato, P., Salamini, F. (1975). Glossy mutants of maize, VI chemical constituents of glossy-2 epicuticular waxes. Maydica 20, 165–173.
Bianchi, A., Bianchi, G., Avato, P., Salamini, F. (1985). Biosynthetic pathways of epicuticular wax of maize as assessed by mutation, light, plant age and inhibitor studies. Maydica 30, 179–198.
Bitter, G. A., Egan, K. M., Koski, R. A., Jones, M. O., Elliott, S. G., Giffin, J. C. (1987). Expression and secretion vectors for yeast. Methods Enzymol. 153, 516–544. doi: 10.1016/0076-6879(87)53076-2
Brachmann, C. B., Davies, A., Cost, G. J., Caputo, E., Li, J., Hieter, P., et al. (1998). Designer deletion strains derived from Saccharomyces cerevisiae S288C: A useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14, 115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2
Campbell, A. A., Stenback, K. E., Flyckt, K., Hoang, T., Perera, M. A. D. N., Nikolau, B. J. (2019). A single-cell platform for reconstituting and characterizing fatty acid elongase component enzymes. PloS One 14, e0213620. doi: 10.1371/journal.pone.0213620
D’Auria, J. C. (2006). Acyltransferases in plants: A good time to be BAHD. Curr. Op Plant Biol. 9, 331–340. doi: 10.1016/j.pbi.2006.03.016
Denic, V., Weissman, J. S. (2007). A molecular caliper mechanism for determining very long-chain fatty acid length. Cell 130, 663–677. doi: 10.1016/j.cell.2007.06.031
Dietrich, C. R., Perera, M. A. D. N., Yandeau-Nelson, M. D., Meeley, R. B., Nikolau, B. J., Schnable, P. S. (2005). Characterization of two GL8 paralogs reveals that the 3-ketoacyl reductase component of fatty acid elongase is essential for maize (Zea mays L.) development. Plant J. 42, 844–861. doi: 10.1111/j.1365-313X.2005.02418.x
Erdbrügger, P., Fröhlich, F. (2021). The role of very long chain fatty acids in yeast physiology and human diseases. Biol. Chem. 402, 25–38. doi: 10.1515/hsz-2020-0234
Gable, K., Garton, S., Napier, J. A., Dunn, T. M. (2004). Functional characterization of the Arabidopsis thaliana orthologue of Tsc13p, the enoyl reductase of the yeast microsomal fatty acid elongating system. J. Exp. Bot. 55, 543–545. doi: 10.1093/jxb/erh061
Gietz, R. D., Woods, R. A. (2002). Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 350, 87–96. doi: 10.1016/s0076-6879(02)50957-5
Gonzales-Vigil, E., von Loessl, M. E., Chen, J. Y., Li, S., Haslam, T. M., Kunst, L., et al. (2021). Understanding the role of Populus ECERIFERUM2-likes in the biosynthesis of very-long-chain fatty acids for cuticular waxes. Plant Cell Physiol. 62, 827–838. doi: 10.1093/pcp/pcab040
Han, G., Gable, K., Kohlwein, S. D., Beaudoin, F., Napier, J. A., Dunn, T. M. (2002). The Saccharomyces cerevisiae YBR159w gene encodes the 3-ketoreductase of the microsomal fatty acid elongase. J. Biol. Chem. 277, 35440–35449. doi: 10.1074/jbc.M205620200
Han, F., Huang, J., Xie, Q., Liu, Y., Fang, Z., Yang, L., et al. (2021). Genetic mapping and candidate gene identification of BoGL5, a gene essential for cuticular wax biosynthesis in broccoli. BMC Genomics 22, 811. doi: 10.1186/s12864-021-08143-7
Harrison, P. J., Dunn, T. M., Campopiano, D. J. (2018). Sphingolipid biosynthesis in man and microbes. Nat. Prod Rep. 35, 921–954. doi: 10.1039/C8NP00019K
Haslam, T. M., Feussner, I. (2022). Diversity in sphingolipid metabolism across land plants. J. Exp. Bot. 73, 2785–2798. doi: 10.1093/jxb/erab558
Haslam, T. M., Gerelle, W. K., Graham, S. W., Kunst, L. (2017). The unique role of the ECERIFERUM2-LIKE clade of the BAHD acyltransferase superfamily in cuticular wax metabolism. Plants 6, 23. doi: 10.3390/plants6020023
Haslam, T. M., Haslam, R., Thoraval, D., Pascal, S., Delude, C., Domergue, F., et al. (2015). ECERIFERUM2-LIKE proteins have unique biochemical and physiological functions in very-long-chain fatty acid elongation. Plant Physiol. 167, 682–692. doi: 10.1104/pp.114.253195
Haslam, T. M., Kunst, L. (2013). Extending the story of very-long-chain fatty acid elongation. Plant Sci. 210, 93–107. doi: 10.1016/j.plantsci.2013.05.008
Haslam, T. M., Kunst, L. (2020). Arabidopsis ECERIFERUM2-LIKEs Are mediators of condensing enzyme function. Plant Cell Physiol. 61, 2126–2138. doi: 10.1093/pcp/pcaa133
Haslam, T. M., Mañas-Fernández, A., Zhao, L., Kunst, L. (2012). Arabidopsis ECERIFERUM2 is a component of the fatty acid elongation machinery required for fatty acid extension to exceptional lengths. Plant Physiol. 160, 1164–1174. doi: 10.1104/pp.112.201640
Hayes, H. K., Brewbaker, H. E. (1928). Glossy seedlings in maize. Am. Nat. 62, 228–235. doi: 10.1086/280202
James, D. W., Jr., Lim, E., Keller, J., Plooy, I., Ralston, E., Dooner, H. K. (1995). Directed tagging of the Arabidopsis FATTY ACID ELONGATION1 (FAE1) gene with the maize transposon Activator. Plant Cell 7, 309–319. doi: 10.1105/tpc.7.3.309
Jenks, M. A., Tuttle, H. A., Eigenbrode, S. D., Feldmann, K. A. (1995). Leaf epicuticular waxes of the Eceriferum mutants in Arabidopsis. Plant Physiol. 108, 369–377. doi: 10.1104/pp.108.1.369
Ji, J., Cao, W., Tong, L., Fang, Z., Zhang, Y., Zhuang, M., et al. (2021). Identification and validation of an ECERIFERUM2- LIKE gene controlling cuticular wax biosynthesis in cabbage (Brassica oleracea L. var. capitata L.). Theor. Appl. Genet. 134, 4055–4066. doi: 10.1007/s00122-021-03947-3
Kim, J., Kim, R. J., Lee, S. B., Suh, M. C. (2022). Protein-protein interactions in fatty acid elongase complexes are important for very-long-chain fatty acid synthesis. J. Exp. Bot. 73, 3004–3017. doi: 10.1093/jxb/erab543
Kohlwein, S. D., Eder, S., Oh, C. S., Martin, C. E., Gable, K., Bacikova, D., et al. (2001). Tsc13p is required for fatty acid elongation and localizes to a novel structure at the nuclear-vacuolar interface in Saccharomyces cerevisiae. Mol. Cell Biol. 21, 109–125. doi: 10.1128/MCB.21.1.109-125.2001
Koornneef, M., Hanhart, C. J., Thiel, F. (1989). A genetic and phenotypic description of Eceriferum (Cer) mutants in Arabidopsis thaliana. J. Hered 80, 118–122. doi: 10.1093/oxfordjournals.jhered.a110808
Kruse, L. H., Weigle, A. T., Irfan, M., Martínez-Gómez, J., Chobirko, J. D., Schaffer, J. E., et al. (2022). Orthology-based analysis helps map evolutionary diversification and predict substrate class use of BAHD acyltransferases. Plant J. 111, 1453–1468. doi: 10.1111/tpj.15902
Leonard, A. E., Pereira, S. L., Sprecher, H., Huang, Y. S. (2004). Elongation of long-chain fatty acids. Prog. Lipid Res. 43, 36–54. doi: 10.1016/S0163-7827(03)00040-7
Liu, L., Xu, H., Zhang, W., Xing, J., Zhu, M., Zhang, Y., et al. (2023). Genome-wide analysis of the BAHD Family in elsh onion and CER2-LIKEs Involved in Wax Metabolism. Genes 14, 1286. doi: 10.3390/genes14061286
McNevin, J. P., Woodward, W., Hannoufa, A., Feldmann, K. A., Lemieux, B. (1993). Isolation and characterization of eceriferum (cer) mutants induced by T-DNA insertions in Arabidopsis thaliana. Genome 36, 610–618. doi: 10.1139/g93-082
Moghe, G., Kruse, L. H., Petersen, M., Scossa, F., Fernie, A. R., Gaquerel, E., et al. (2023). BAHD company: The ever-expanding roles of the BAHD acyltransferase gene family in plants. Ann. Rev. Plant Biol. 74, 165–194. doi: 10.1146/annurev-arplant-062922-050122
Negruk, V., Yang, P., Subramanian, M., McNevin, J. P., Lemieux, B. (1996). Molecular cloning and characterization of the CER2 gene of Arabidopsis thaliana. Plant J. 9, 137–145. doi: 10.1046/j.1365-313X.1996.09020137.x
Oh, C. S., Toke, D. A., Mandala, S., Martin, C. E. (1997). ELO2 and ELO3, homologues of the Saccharomyces cerevisiae ELO1 gene, function in fatty acid elongation and are required for sphingolipid formation. J. Biol. Chem. 272, 17376–17384. doi: 10.1074/jbc.272.28.17376
Pascal, S., Bernard, A., Sorel, M., Pervent, M., Vile, D., Haslam, R. P., et al. (2013). The Arabidopsis cer26 mutant, like the cer2 mutant, is specifically affected in the very long chain fatty acid elongation process. Plant J. 73, 733–746. doi: 10.1111/tpj.12060
Samuels, L., Kunst, L., Jetter, R. (2008). Sealing plant surfaces: Cuticular wax formation by epidermal cells. Annu. Rev. Plant Biol. 59, 683–707. doi: 10.1146/annurev.arplant.59.103006.093219
Schnable, P. S., Stinard, P. S., Wen, T.-J., Heinen, S., Weber, D., Zhang, L., et al. (1994). The genetics of cuticular wax biosynthesis. Maydica 39, 279–287.
Smith, W. L. (1989). The eicosanoids and their biochemical mechanisms of action. Biochem. J. 259, 315–324. doi: 10.1042/bj2590315
Stenback, K. E., Flyckt, K. S., Hoang, T., Campbell, A. A., Nikolau, B. J. (2022). Modifying the yeast very long chain fatty acid biosynthetic machinery by the expression of plant 3-ketoacyl CoA synthase isozymes. Sci. Rep. 12, 13235. doi: 10.1038/s41598-022-17080-8
St-Pierre, B., Luca, V. D. (2000). Evolution of acyltransferase genes: Origin and diversification of the BAHD superfamily of acyltransferases involved in secondary metabolism. Recent Adv. Phytochem. 34, 285–315. doi: 10.1016/S0079-9920(00)80010-6
Tacke, E., Korfhage, C., Michel, D., Maddaloni, M., Motto, M., Lanzini, S., et al. (1995). Transposon tagging of the maize Glossy2 locus with the transposable element En/Spm. Plant J. 8, 907–917. doi: 10.1046/j.1365-313X.1995.8060907.x
Thulasingam, M., Haeggström, J. Z. (2020). Integral membrane enzymes in eicosanoid metabolism: Structures, mechanisms and inhibitor design. J. Mol. Biol. 432, 4999–5022. doi: 10.1016/j.jmb.2020.07.020
Toke, D. A., Martin, C. E. (1996). Isolation and characterization of a gene affecting fatty acid elongation in Saccharomyces cerevisiae. J. Biol. Chem. 271, 18413–18422. doi: 10.1074/jbc.271.31.18413
Wang, X., Guan, Y., Zhang, D., Dong, X., Tian, L., Qu, L. Q. (2017). A β-ketoacyl-CoA synthase is involved in rice leaf cuticular wax synthesis and requires a CER2-LIKE protein as a cofactor. Plant Physiol. 173, 944–955. doi: 10.1104/pp.16.01527
Xia, Y., Nikolau, B. J., Schnable, P. S. (1996). Cloning and characterization of CER2, an Arabidopsis gene that affects cuticular wax accumulation. Plant Cell 8, 1291–1304. doi: 10.1105/tpc.8.8.1291
Xu, X., Dietrich, C. R., Delledonne, M., Xia, Y., Wen, T. J., Robertson, D. S., et al. (1997). Sequence analysis of the cloned glossy8 gene of maize suggests that it may code for a β-ketoacyl reductase required for the biosynthesis of cuticular waxes. Plant Physiol. 115, 501–510. doi: 10.1104/pp.115.2.501
Xu, X., Dietrich, C. R., Lessire, R., Nikolau, B. J., Schnable, P. S. (2002). The endoplasmic reticulum-associated maize GL8 protein is a component of the acyl-coenzyme A elongase involved in the production of cuticular waxes. Plant Physiol. 128, 924–934. doi: 10.1104/pp.010621
Keywords: fatty acid elongase, very long chain fatty acids, GLOSSY2, GLOSSY2-LIKE, BAHD proteins, maize, cuticular waxes, yeast
Citation: Alexander LE, Winkelman D, Stenback KE, Lane M, Campbell KR, Trost E, Flyckt K, Schelling MA, Rizhsky L, Yandeau-Nelson MD and Nikolau BJ (2024) The impact of the GLOSSY2 and GLOSSY2-LIKE BAHD-proteins in affecting the product profile of the maize fatty acid elongase. Front. Plant Sci. 15:1403779. doi: 10.3389/fpls.2024.1403779
Received: 19 March 2024; Accepted: 20 June 2024;
Published: 11 July 2024.
Edited by:
Lei Zhao, Academy of Sciences (CAS), ChinaReviewed by:
Xia Wan, Chinese Academy of Agricultural Sciences, ChinaHongyan Yao, Fudan University, China
Thomas J. Bach, Université de Strasbourg, France
Joaquín J. Salas, Spanish National Research Council (CSIC), Spain
Copyright © 2024 Alexander, Winkelman, Stenback, Lane, Campbell, Trost, Flyckt, Schelling, Rizhsky, Yandeau-Nelson and Nikolau. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Basil J. Nikolau, ZGltbWFzQGlhc3RhdGUuZWR1; Marna D. Yandeau-Nelson, bXluQGlhc3RhdGUuZWR1
†These authors share first authorship
‡These authors share senior authorship
 Liza Esther Alexander1,2†
Liza Esther Alexander1,2† Dirk Winkelman
Dirk Winkelman Kenna E. Stenback
Kenna E. Stenback Kayla Flyckt
Kayla Flyckt Marna D. Yandeau-Nelson
Marna D. Yandeau-Nelson Basil J. Nikolau
Basil J. Nikolau