
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 08 May 2024
Sec. Plant Metabolism and Chemodiversity
Volume 15 - 2024 | https://doi.org/10.3389/fpls.2024.1403060
This article is part of the Research TopicIn-Depth Interpretation of Critical Genomic Information Related to the Biosynthesis of Key Specialized (Secondary) Metabolism in Medicinal PlantsView all 10 articles
Paclitaxel (trade name Taxol) is a rare diterpenoid with anticancer activity isolated from Taxus. At present, paclitaxel is mainly produced by the semi-synthetic method using extract of Taxus tissues as raw materials. The studies of regulatory mechanisms in paclitaxel biosynthesis would promote the production of paclitaxel through tissue/cell culture approaches. Here, we systematically identified 990 transcription factors (TFs), 460 microRNAs (miRNAs), and 160 phased small interfering RNAs (phasiRNAs) in Taxus chinensis to explore their interactions and potential roles in regulation of paclitaxel synthesis. The expression levels of enzyme genes in cone and root were higher than those in leaf and bark. Nearly all enzyme genes in the paclitaxel synthesis pathway were significantly up-regulated after jasmonate treatment, except for GGPPS and CoA Ligase. The expression level of enzyme genes located in the latter steps of the synthesis pathway was significantly higher in female barks than in male. Regulatory TFs were inferred through co-expression network analysis, resulting in the identification of TFs from diverse families including MYB and AP2. Genes with ADP binding and copper ion binding functions were overrepresented in targets of miRNA genes. The miRNA targets were mainly enriched with genes in plant hormone signal transduction, mRNA surveillance pathway, cell cycle and DNA replication. Genes in oxidoreductase activity, protein-disulfide reductase activity were enriched in targets of phasiRNAs. Regulatory networks were further constructed including components of enzyme genes, TFs, miRNAs, and phasiRNAs. The hierarchical regulation of paclitaxel production by miRNAs and phasiRNAs indicates a robust regulation at post-transcriptional level. Our study on transcriptional and posttranscriptional regulation of paclitaxel synthesis provides clues for enhancing paclitaxel production using synthetic biology technology.
Paclitaxel is a terpenoid compound isolated from Taxus, which can promote the formation and stabilization of microtubules, prevent their depolymerization and inhibit cell division. Widely recognized as a first-line clinical drug, paclitaxel demonstrates curative effects on the treatments of breast cancer, ovarian cancer, and melanoma (De Furia, 1997; Yu et al., 2021). The massive extraction of paclitaxel has posed a serious threat to the growth of Taxus, resulting in the disastrous reduction of the natural Taxus population. Various approaches, including chemical synthesis, semi-chemical synthesis, plant tissue/cell culture, endophytic fungal synthesis and others, have been explored to develop sustainable methods for paclitaxel production (Fett-Neto et al., 1992; Balogu and Kinston, 1999). Many genes in the paclitaxel biosynthetic pathway were also identified, which can be used to screen bottleneck enzymes to optimize metabolic engineering for paclitaxel production (Howat et al., 2014; Kuang et al., 2019; Perez-Matas et al., 2024). The biosynthesis of paclitaxel involves at least 19 steps, from diterpene precursor geranylgeranyl diphosphate (GGPP) to the final product (Hefner et al., 1998; Jarchow-Choy et al., 2014; Liao et al., 2016). The initial step is the cyclization of GGPP to taxane by the key enzyme TS (taxane synthase) (Ansbacher et al., 2018). Subsequently, the tricyclic taxane backbone undergoes extremely complex modifications mediated by many oxygenases, acyltransferases, and benzoyltransferases, including the 2α-, 5α-, 7β-, 9α-, 10β-, and 13α-hydroxylases, and TAT (taxadienol 5α-O-acetyl transferase) and DBAT (10-deacetylbaccatin III 10-O-acetyltransferase) (Walker and Croteau, 2000; Walker et al., 2000; Jennewein and Croteau, 2001; Jennewein et al., 2001; Walker et al., 2002; Chau and Croteau, 2004; Kaspera and Croteau, 2006; Long et al., 2008). Recently, two key enzymes have been identified for artificial construction of the baccatin III biosynthetic pathway, including Taxane oxetanase 1 (TOT1) representing a previously unknown enzyme mechanism for oxetane ring formation and T9αH for the taxane oxidation of the C9- position (Jiang et al., 2024).
Transcription factors (TFs) play critical roles in the regulation of plant growth, development, and responses to diverse environmental stresses (Singh et al., 2002; Kahle et al., 2005). In Taxus, many TFs have been reported to be involved in the regulations of key genes in the paclitaxel biosynthesis pathway (Kuang et al., 2019; Mutanda et al., 2021). TcMYC2a (bHLH member) was considered to play an important role in the jasmonate-responsive expression of TASY, TAT, DBTNBT, T13αOH, and T5αOH genes (Zhang et al., 2018). Members of the MYB family, known for their roles in various secondary metabolite biosynthesis, also contribute to paclitaxel production (Cao et al., 2020; Yu et al., 2022). In the ERF family, a repressor TcERF12 and an activator TcERF15 affected paclitaxel biosynthesis by recognizing the GCC-box on the promoter region of the TS gene (Zhang et al., 2015b). TcWRKY1 significantly enhanced the transcription level of DBAT (Li et al., 2013). Engineering of single and/or a combination of TFs would tune the expression of multiple enzyme genes symmetrically for paclitaxel generation.
MicroRNAs (miRNAs) are a class of small non-coding single-stranded RNA molecules of approximately 20-24 nucleotides, which mediate the degradation or inhibition of target genes with diverse functions by sequence complementation (Jones-Rhoades, 2012; Shivaprasad et al., 2012; Deng et al., 2018). In T. chinensis and T. media, miRNAs have been reported to significantly correlated with genes in paclitaxel biosynthesis, such as T5H, TAT and T10H (Zhang et al., 2015a; Chen et al., 2020). Phased small interfering RNAs (phasiRNAs) are generated through DCL-catalyzed processing of dsRNA (double-stranded RNA) precursors, they are 21- or 24-nucleotide (nt) in length and start from a precisely defined 5' terminus by trigger miRNAs (Johnson et al., 2009; Dukowic-Schulze et al., 2016). The target genes of phasiRNAs play important roles in various transcriptional regulation processes, such as cell formation, meristem formation, cell cycle, anthocyanin synthesis, response to biotic and abiotic stresses, and so on (Fei et al., 2016; Xia et al., 2019; Liu et al., 2020). The phasiRNAs can also target the transcripts of other phasiRNAs, generating self-enhancing regulatory networks. Unraveling the miRNAs and phasiRNAs regulating paclitaxel biosynthetic genes holds the potential to overcome metabolic bottlenecks.
In this study, we systematically identified enzyme genes in the paclitaxel synthesis pathway and characterized their expression patterns in tissues and treatments. TFs, miRNAs, and phasiRNAs were screened for key regulatory genes in paclitaxel synthesis. The constructed regulatory networks would contribute to our understanding of the regulatory mechanisms in paclitaxel production. The interactions between miRNA/phasiRNA and their targeted protein-coding genes provide clues to promote paclitaxel production through the engineering of genes in posttranscriptional regulations.
Fresh young leaves of two female and two male individuals were collected from the natural distribution range of T. chinensis (109° 52′19′′N, 30° 60′03′′E) in Taiyanghe, Enshi city, Hubei Province, China. The leaf samples were collected in May, 2021. Published transcriptome and small RNA datasets were also downloaded for analysis including forty-two RNAseq samples (PRJNA730337 and PRJNA251671) and three sRNA samples (PRJNA173133 and PRJNA251671).
RNA-seq library was constructed using fresh young leaves of T. chinensis. Total RNA was extracted using the RNA prep Pure Plant Plus Kit according to the manufacturer’s instructions (LC-BIO TECHNOLOGIES (HANGZHOU) CO., LTD., China). Sequencing was performed using the Illumina NovaSeq platform (Illumina, San Diego, CA, USA) and paired-end raw reads were generated. To obtain high-quality reads, adapters and low-quality reads of the raw data were removed using Trimmomatic (version 0.39) (Bolger et al., 2014). RNA-seq reads were mapped onto the reference genome assembly using STAR (version 2.7.9; parameters: -two pass Mode) (Dobin et al., 2013) and the TPM was calculated to evaluate the expression level of each gene using the RSEM (Li and Dewey, 2011) pipeline after averaging some replicated samples. The numbers of sample replicates in tissue, treatment, and sex experiments in the differential expression analyses are six, one, and three, respectively. Significantly differentially expressed genes were evaluated using edgeR (Robinson et al., 2010) with |logFC| > 1 and FDR < 0.05.
Fresh young leaves from the individuals described above were collected for small RNA extraction. sRNA sequencing libraries were prepared using TruSeq Small RNA Sample Prep Kits (USA). Single end reads of 50 bp was obtained using Illumina Hiseq2500 platform (LC-BIO TECHNOLOGIES (HANGZHOU) CO., LTD., China). Reads containing adapters and low-quality reads were trimmed using Cutadapt (version 2.10) (Martin, 2011). The reads were firstly aligned to the Rfam database (version 11.0) (Griffiths-Jones et al., 2005) using Bowtie (version 1.3.0) (Langmead, 2010) to remove non-coding RNAs (rRNA, tRNA, snRNA, scRNA, and snoRNA). The processed sRNA data was submitted to the ShortStack program to identify potential miRNA loci (foldsize = 500; mincov = 2; ranmax = 35) (Axtell, 2013). Loci that meet the N15 criteria or the Y criteria are retained. To identify known miRNAs, we compared candidate mature miRNAs with records in miRBase (version 22.1) (Kozomara et al., 2019) using PatMaN (version 1.2) (Mismatch ≤ 4) (Prüfer et al., 2008). The remaining sequences were then aligned to known miRNA precursor sequences to identify potential miRNA*s. All miRNA loci that meet the Y criteria are considered as novel miRNAs.
To identify potential phasiRNAs loci, processed sRNA data was submitted to the PHASIS (version 3.3) pipeline (Kakrana et al., 2017). Combining the previously identified miRNA sequence with phastrigs (the third module in the PHASIS pipeline), all the miRNA triggers of phasiRNAs were identified. The target genes of miRNAs and phasiRNAs were predicted using the online tool psRNATarget with the following criteria: maximum cutoff of score = 3; penalty for G:U pair = 0.5; penalty for other mismatched = 1; extra penalty weight for mismatched in seed region; HSP size = 19; penalty for opening gap = 2; and penalty for extending gap = 0.5 (Dai et al., 2018). TPTM (transcripts per 10 million reads) was calculated to evaluate the expression level of each miRNA and phasiRNA. GO (Harris et al., 2004) and KEGG (Ogata et al., 1999) enrichment analysis of target genes (FDR ≤ 0.05) was performed through the R package clusterProfiler (version 3.18.1) (Yu et al., 2012). The background gene sets were annotated with Swiss-prot database using Diamond software (E-value ≤ 10-5) (version 0.9.19) (Buchfink et al., 2021).
Protein coding genes in the paclitaxel synthesis pathway in T. chinensis were identified using Diamond Blastp (version 0.9.19) search (E-value ≤ 10-5) (Buchfink et al., 2021). In co-expression analysis, the quantitative results of transcriptome were applied for Spearman correlation analysis and weighted gene co-expression network analysis (WGCNA) (Langfelder and Horvath, 2008) was performed to identify co-expressed gene pairs (|ρ| ≥ 0.7). TFs of T. chinensis were identified at genome wide using iTAK software (Zheng et al., 2016). The regulatory networks of enzyme genes, TFs, miRNAs, and phasiRNAs were constructed by Cytoscape (version 3.8.2) (Shannon et al., 2003).
The expression patterns of genes involved in paclitaxel synthesis were investigated to gain insights into their transcriptional regulatory mechanisms (Song et al., 2021). 71 genes of diverse gene families were identified in the whole genome of T. chinensis (Supplementary Tables 1-3). These genes can be categorized into GGPPS (geranyl geranyl diphosphate synthase), T5αOH, T13αOH, TAT, T2αOH, T7βOH, T10βOH, TBT (taxane-2α-O-benzoyl transferase), BAPT (C-13-phenylpropanoyl-CoA transferase), DBTNBT (3’-N-debenzoyl-2’-deoxytaxol N-benzoyl transferase), CoA Ligase, PAM (phenylalanine aminomutase), T14βOH, TB506, and TXS (Figure 1). Previously, two gene clusters on chromosome 9 have been reported (Xiong et al., 2021). In analysis, extra T10βOH_like genes and T5αOH_like genes were identified in the gene cluster located on a 141.69-Mb region of chromosome 9 (616,470,670 - 758,158,182 bp; Supplementary Table 3; Xiong et al., 2021).
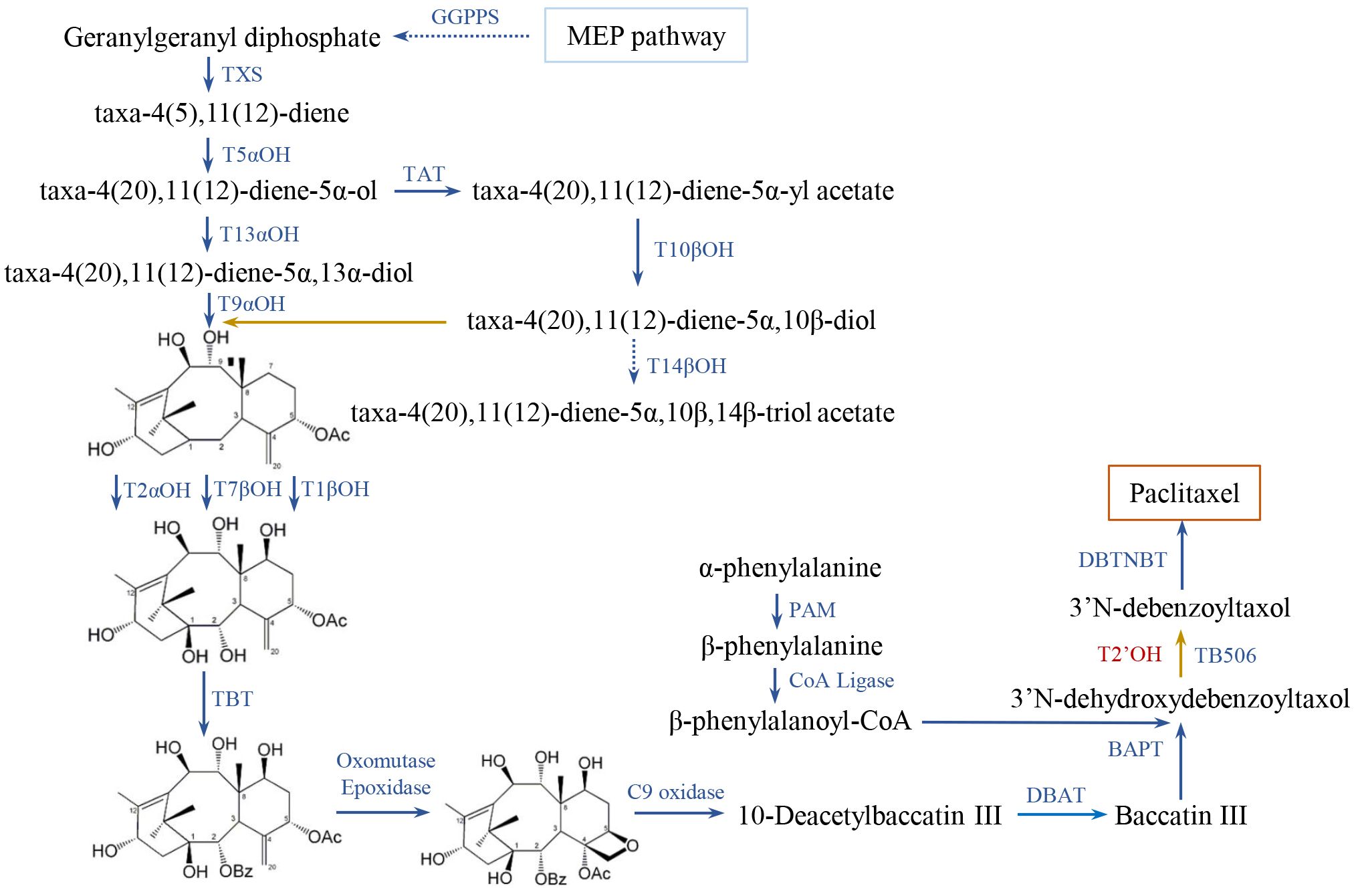
Figure 1 Biosynthetic pathway of paclitaxel in Taxus. Identified and unknown enzymes in the pathway are shown in blue and red, respectively. The full names of abbreviations are listed as: GGPPS, geranylgeranyl diphosphate synthase; TXS, Taxadiene synthase; T5αOH, taxane 5α-hydroxylase; TAT, taxadien-5α-ol-O-acetyl-transferase; T13αOH, taxane 13α-hydroxylase; T10βOH, taxane 10β-hydroxylase; T14βOH, taxane 14β-hydroxylase; T2αOH, taxane 2α-hydroxylase; T7βOH, taxane 7β-hydroxylase; TBT, taxane 2α-O-benzoyl transferase; DBAT, 10-deacetyl-baccatin III-10-Oacetyltransferase; PAM, phenylalanine aminomutase; BAPT, 13-O-(3-amino-3-phenylpropanoyl) transferase; DBTNBT, 30-N-debenzoyl-20-deoxytaxol-Nbenzoyltransferase. The metabolic processes were adapted from previous reports (Croteau et al., 2006; Cheng et al., 2021; Zhang et al., 2023; Jiang et al., 2024).
The expression levels of identified genes in T. chinensis were further explored in diverse tissues and treatments. The results from tissues indicated most of these genes are expressed at a high level, and the expression of these genes is relatively higher in cone and root in comparison to in leaf and bark (Figure 2A). According to previous research results, the application of jasmonate can significantly induce the biosynthesis of paclitaxel and the expression levels of CYP725A subfamily genes in paclitaxel biosynthesis (Xiong et al., 2021). The transcriptional data of T. chinensis cell line treated with jasmonate were also included in our analysis. The data were collected at five time points (0h, 2h, 4h, 8h, 24h) with alcohol treatment as the control. The experimental results showed that, except for GGPPS and CoA Ligase, the expression of almost all enzyme genes in the entire synthesis pathway was significantly up-regulated by jasmonate treatment. The most significant effect was observed at the stage of treatment after four hours (Figure 2B). The differential expression patterns of genes in paclitaxel synthesis were also tested between female and male Taxus trees. The analysis results showed that the gene expression patterns varied by tissue. In barks, the expression level of enzyme genes located in the latter steps of the synthesis pathway is significantly higher in female plants than in male plants. In cones, the overall expression levels of enzyme genes were lower in male plants, however, the expression patterns were reverse in roots. In leaves, there was only a small portion of genes differentially expressed between female and male plants (Figure 2C). Notably, there was no significant expression difference between male leaves and female leaves from Enshi. It is speculated that this may be related to the leaf sampling stage and the physiological state.

Figure 2 Expression patterns of genes in paclitaxel synthesis in diverse conditions and treatments. (A) Expression patterns of differentially expressed genes in leaf, root, cone, and bark tissues of T. chinensis. The visualized data has been normalized through log2(TPM+1). (B) Differential expression patterns of genes in response to methyl jasmonate (MJ) treatment. The samples were from MJ treatment after 0h, 2h, 4h, 8h, and 24h using alcohol as control (CK). (C) Differential expression patterns of genes between female and male plants in the tested five samples. FB, female bark; MB, male bark; FC, female cone; MC, male cone; FR, female root; MR, male root; FL, female leaf; ML, male leaf. The differential expression analysis was performed using edgeR with cutoffs |logFC| > 1 and FDR < 0.05. “*” indicates significant up-regulation or significant down-regulation. The order of genes in the heatmap is based on the sequence of chemical reactions in paclitaxel synthesis pathway.
To characterize TFs involved in the regulation of paclitaxel synthesis, all members of TF families were identified at the whole genome level based on protein domains. A total of 990 TFs from 60 families were identified. The regulatory pairs between TFs and the target genes were inferred from correlation calculation and WGCNA. We performed Spearman correlation analysis on quantitative results of transcriptome sequencing data to screen and identify the expression associated genes of 71 enzyme genes (|ρ| ≥ 0.7). Most of the TFs were from MYB, AP2/ERF, bHLH, HB, LOB, MADS, and WRKY families (Figure 3A). In WGCNA analysis, 29 modules (height > 0.25) were generated based on expression similarity and clustering of gene trees. The enzyme genes in paclitaxel synthesis were mostly present in three modules (Green-yellow, Lightcyan1, and Lightcyan). Finally, 10 enzyme genes and 28 TFs were included in the regulatory networks (Figure 3B). These 10 enzyme genes are GGPPS-1, T5αOH_like_4, T13αOH-3, TAT-1, T2αOH-1, T10βOH_like_5, T10βOH_like_8, TBT-2, TBT-4 and BAPT-2, which are distributed in various steps of paclitaxel biosynthesis. The identified TFs showing co-expressed patterns with enzyme genes can serve as candidate genes to tune the expression of targets at transcription level.
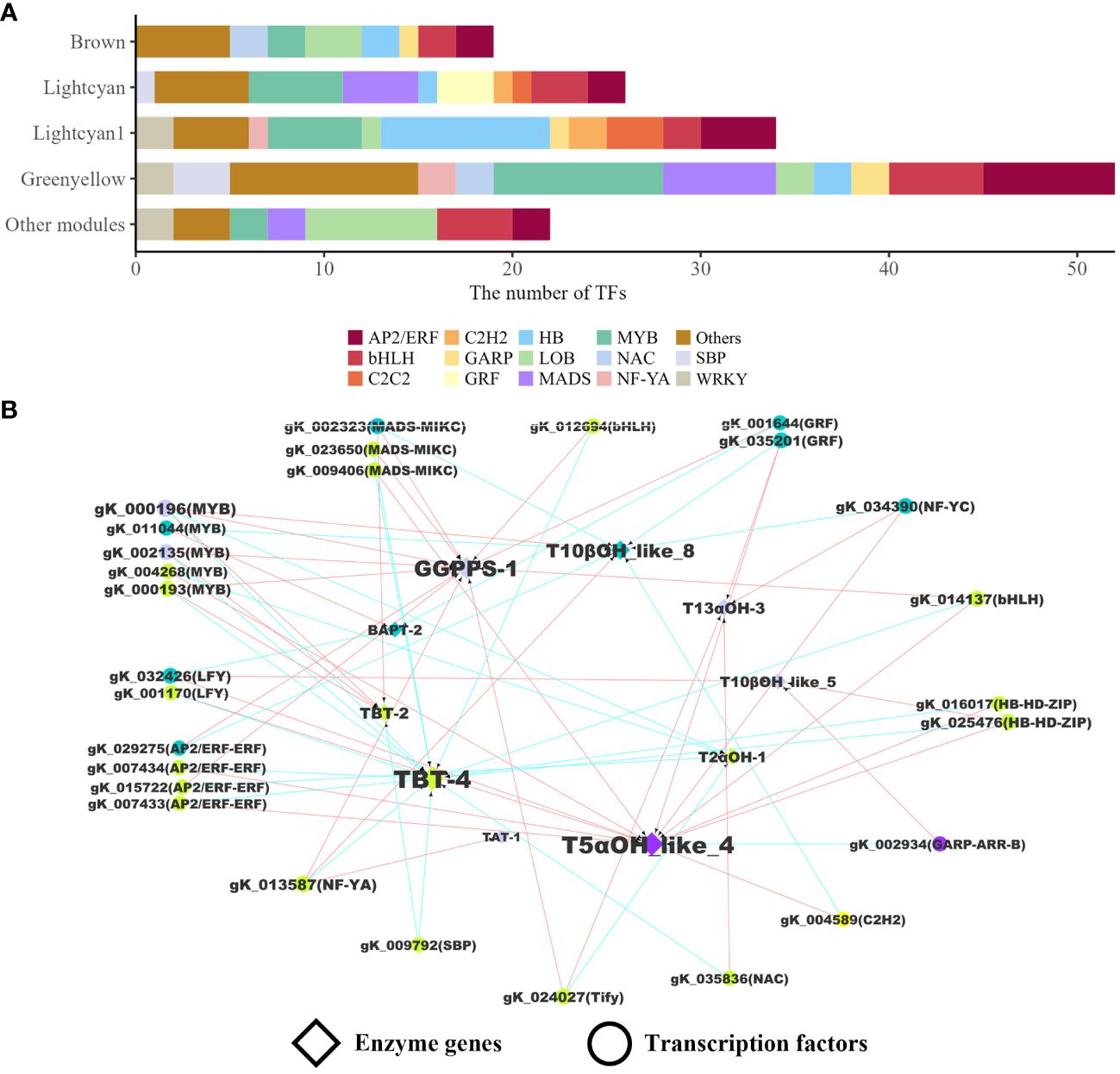
Figure 3 Transcriptional regulatory network in paclitaxel synthesis. (A) The number of TFs in WGCNA modules. (B) Regulatory network of enzyme genes and TFs. The colors represent WGCNA modules. The pairs of TFs and their targets were linked by arrows. The blue arrows indicate that the weight value between the two genes in WGCNA is greater than 0.3. The red arrows indicate that the absolute value of the Spearman correlation coefficient of two gene expression levels is greater than or equal to 0.7. Each TF in the co-expression network regulates two or more paclitaxel synthesis-related enzyme genes.
MicroRNAs (miRNAs) genes in T. chinensis were identified based on small RNA sequencing and secondary structure prediction. 460 miRNAs from 311 miRNA families were identified (Supplementary Table 4), among which 92 and 219 are known and novel families, respectively (Supplementary Table 5). The length of miRNAs ranged from 20 nt to 24 nt. The 21-nt miRNAs were most dominant. A significant bias toward U was observed at the first nucleotides of mature miRNA sequences (Supplementary Table 6). The distribution patterns of miRNA genes on the chromosomes indicated that they are abundant at arm regions of chromosomes and few of them distribute at the centromere regions (Figure 4A).
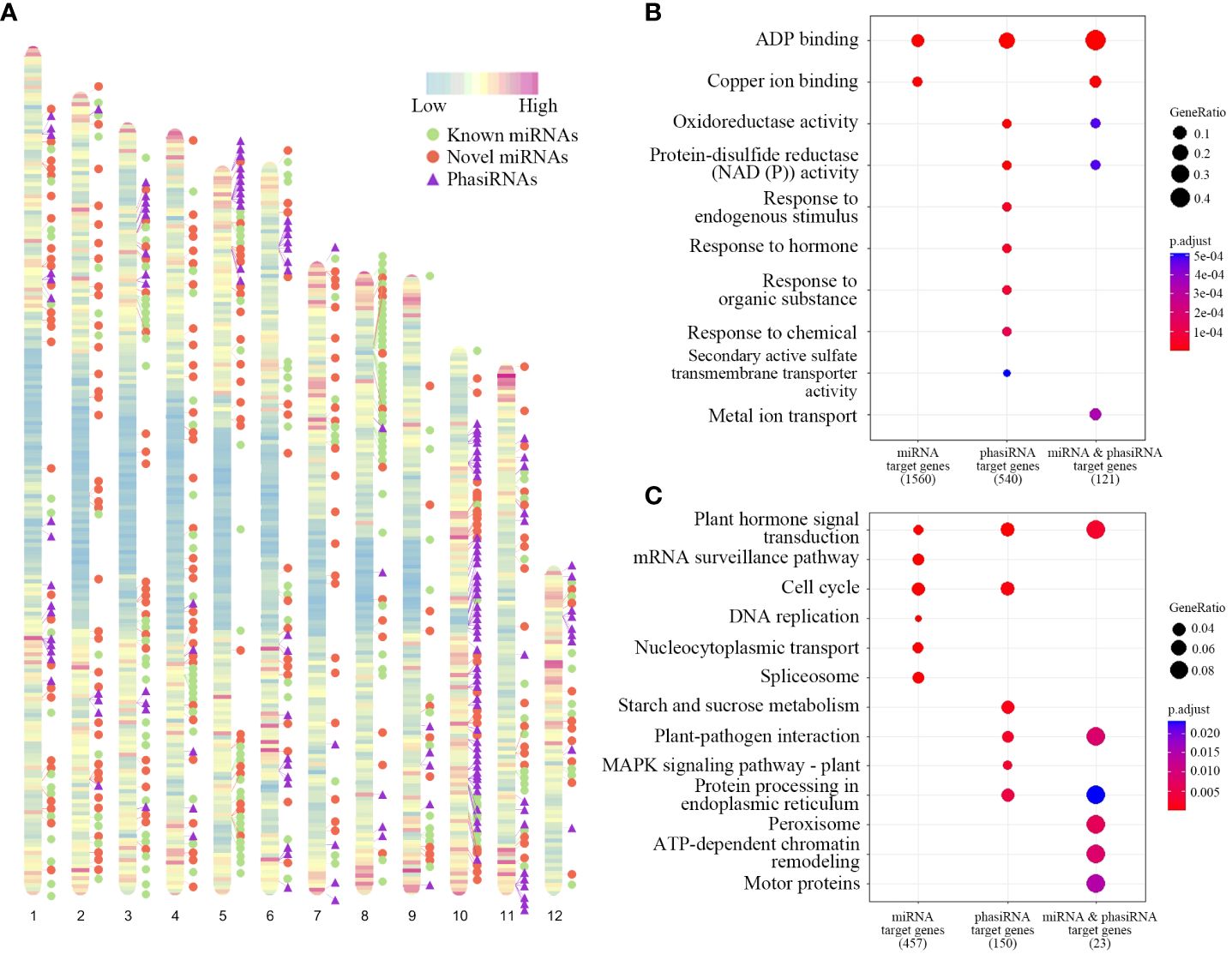
Figure 4 Chromosomal distributions of miRNA and phasiRNA genes and functional enrichment of their targets. (A) Heatmap represents the distribution of gene density on chromosomes. The locations of known miRNAs, novel miRNAs, and phasiRNAs were indicated as green dots, red dots, and purple triangles, respectively. (B, C) GO (B) and KEGG (C) enrichment analysis of genes targeted by miRNAs, phasiRNAs, and both of them.
Target genes were further predicted for all identified miRNAs, among which 49 genes were involved in paclitaxel synthesis (Supplementary Table 8). We performed GO enrichment and KEGG enrichment for the predicted target genes (Figures 4B, C). GO enrichment results indicated that genes with ADP binding and copper ion binding functions were overrepresented in targets of miRNA genes (Figure 4C). KEGG enrichment results show that the target genes of miRNA were mainly enriched with genes in plant hormone signal transduction, mRNA surveillance pathway, cell cycle and DNA replication (Figure 4C).
PhasiRNAs were also identified at genome level using small RNA reads. A total of 160 21-nt phasiRNAs were obtained, whereas no phasiRNAs with other read sizes were found in the analysis (Supplementary Table 7). Among all the phasiRNA genes, 48 are located on chromosome 10, accounting for more than a quarter of the total number. The distribution of phasiRNAs exhibited an enrichment at chromosome regions with high gene density (Figure 4A). The target genes of all the phasiRNAs were predicted, among which 18 genes were involved in paclitaxel synthesis (Supplementary Table 9). GO analysis results show that genes in ADP binding, oxidoreductase activity, protein-disulfide reductase activity were enriched in targets of phasiRNAs. KEGG enrichment results show that target genes of phasiRNA were mainly enriched in pathways including plant hormone signal transduction, cell cycle, starch and sucrose metabolism, and plant-pathogen interaction (Figure 4C). We also performed functional enrichment analysis for the intersection of target genes of miRNAs and phasiRNAs. The miRNA triggers of phasiRNAs were further predicted, resulting the identification of 280 miRNAs targeting all 160 phasiRNAs.
The regulatory connections among miRNAs, phasiRAs, TFs, and enzyme genes in paclitaxel synthesis were combined to construct a regulatory network, which exhibits hierarchical structures in gene regulation (Figure 5). In the network, 60 miRNAs, 9 phasiRNAs and 14 TFs were inferred to regulated 10 key enzyme genes. Among the miRNAs in the network, 15 miRNAs are present in T. chinensis leaf with high abundance (Table 1). Further analysis indicated that six miRNAs regulated enzyme genes directly, and the other six miRNA genes regulated the enzyme genes through targeting TFs. Three miRNAs were predicted to trigger the production of phasiRNAs targeting TFs, which in turn regulate the transcriptions of enzymes genes. The hierarchical regulation of paclitaxel production by miRNAs and phasiRNAs indicates a robust regulation at post-transcriptional level.
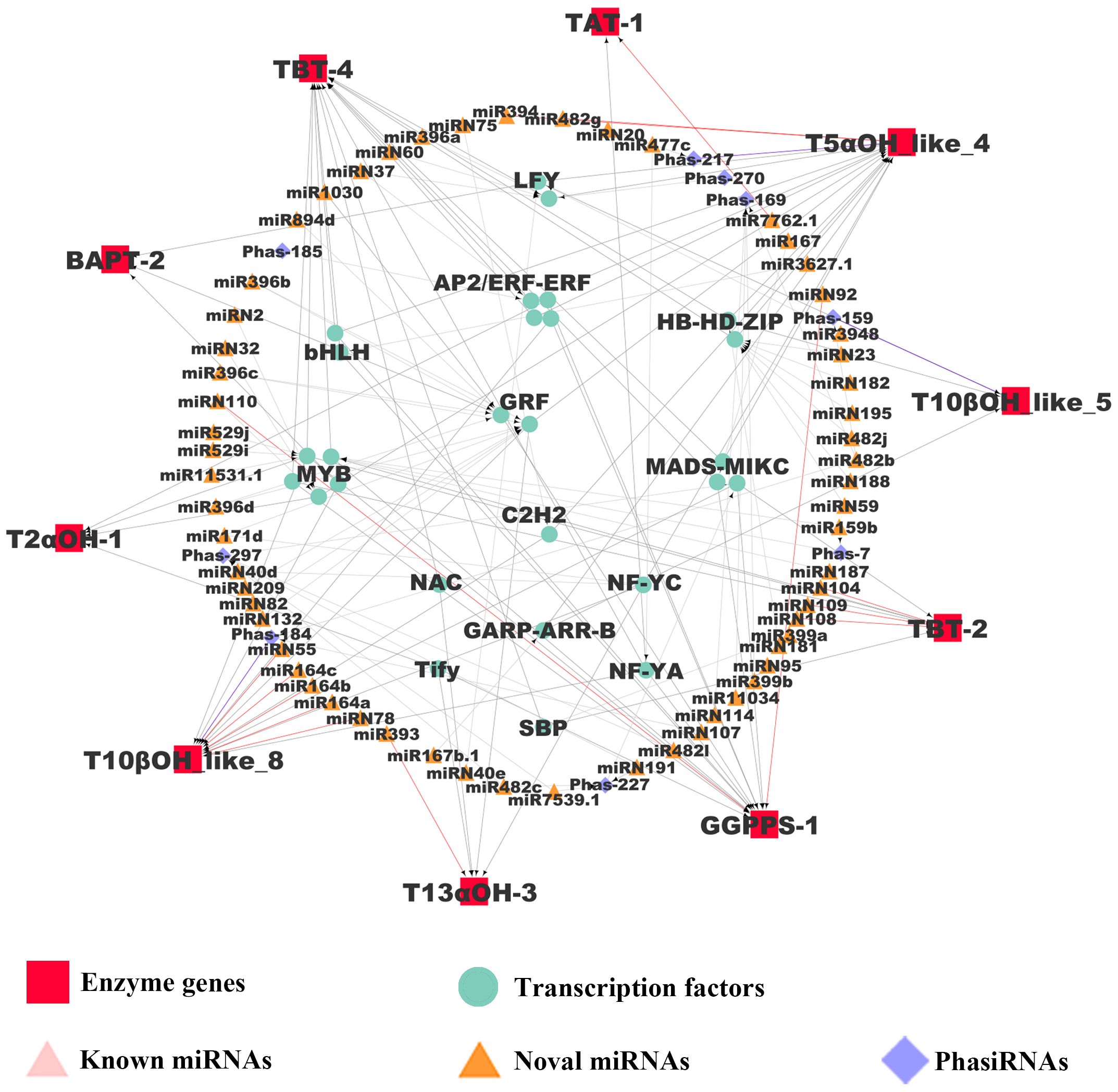
Figure 5 Regulatory network mediated by miRNAs and phasiRNAs in paclitaxel synthesis. The pairs of regulatory genes (miRNAs, phasiRNAs, and TFs) and their target genes were visualized with arrows. Red and purple lines represent miRNA/enzyme gene pairs and phasiRNA/enzyme gene pairs relationships, respectively. Gray lines represent miRNA/TF, phasiRNA/TF and TF/enzyme gene pairs.
As an anticancer drug, the molecular structure of paclitaxel is complex, with 11 stereocenters and a 17-carbon tetracyclic skeleton structure (Jennewein and Croteau, 2001). Despite many attempts to study its chemical synthesis, the intricate route, challenging reaction conditions, and low synthesis rate have posed significant hurdles for researchers. Consequently, attentions in the community have shifted toward the semi-synthesis method. The intermediate products like 10-deacetyl baccatin III (10-DAB) and baccatin III from taxanes were first extracted and applied for paclitaxel chemical synthesis. The semi-synthesis method is a high-purity, cost-effective approach, which has become the primary method for industrial production (Li et al., 2015). While the synthesis technology has been well studied, the production of paclitaxel remains constrained by limited Taxus resources. The large-scale culture of Taxus cells and the fermentation of endophytic fungi to produce paclitaxel have been avenues for breakthroughs. In the realm of metabolic engineering, especially with advancements in synthetic biology, successful large-scale synthesis of key natural products like artemisinin and ginsenosides from heterologous sources has been achieved (Ro et al., 2006; Yan et al., 2014; Sanchez-Muñoz et al., 2020; Schneider et al., 2020; Su et al., 2022).
The limiting factor affecting efficiency of paclitaxel semi-synthetic is the full picture of genes in the synthesis pathway. Six previously characterized Taxus genes can coordinatively produce key paclitaxel intermediates and serves as a crucial platform for the discovery of the remaining biosynthetic genes (Liu et al., 2024). The screening strategy for the biosynthesis pathway of paclitaxel is constantly being updated, new enzyme genes like TOT1 and T9αH1 were innovatively proposed and precisely located in the synthesis pathway (Jiang et al., 2024). In our study, most of the known genes involved in paclitaxel biosynthesis are located on an 80.46-Mb region and a 141.69-Mb region on chromosome 9, which were consistent with previous studies, and the number of identified enzyme genes in the clusters also increased (Supplementary Table 3; Xiong et al., 2021). The extra T10βOH_like genes and T5αOH_like genes identified in the 141.69-Mb region may have unknown novel functions, which requires further exploration and verification to provide insights and candidate genes for paclitaxel semi-synthesis.
The biosynthesis pathway of paclitaxel is fine-tuned at transcription level (Li et al., 2020; Li et al., 2022). More and more TFs are recognized as pivotal regulators in the paclitaxel pathway (Kuang et al., 2019; Mutanda et al., 2021). Previous reports have shown that some transcription factor, such as ERF, can bind the promoter of multiple genes in the pathway (Zhang et al., 2015b). At the same time, a single enzyme gene can be regulated by several TFs (Zhang et al., 2015b; Cao et al., 2020). Depending on different metabolic regulation purposes in paclitaxel biosynthesis, different transcription factors are used to construct various transcriptional regulation tools. Researchers are actively exploring diverse TFs through gene function validation to modulate each enzyme gene in the paclitaxel synthesis pathway, paving the way for enhanced transcriptional expression and improved synthesis efficiency. The TFs targeting to genes in paclitaxel pathway can also involve the regulation genes in the growth and development of T. chinensis (Li et al., 2022).
The complexity of metabolic pathways often leads to unexpected phenotypes in metabolic engineering. The overexpression of enzyme genes may result in the accumulation of toxic intermediate metabolites, while downregulation and knockout of key genes may lead to the shortage of metabolites required for cell growth (Wu et al., 2020). In addition, the modifying of a single gene may disrupt cell homeostasis, causing destructive effects on cell stress, growth, and division, leading to the decrease in synthesis efficiency of final products (Xu et al., 2020). Simultaneously modification of multiple genes could be ideal for perturbation of metabolic pathways. The metabolic regulatory system composed of TFs has been widely applied in metabolic engineering and synthetic biology due to its ability to globally and dynamically regulate target pathways (Deng et al., 2022). Based on the TFs regulatory network in our study, single and/or a combination of TFs can be knock-outed through genome editing or overexpressed by transgenic approaches to optimize transcription of enzyme genes.
MiRNAs have been reported to play significant roles in plant secondary metabolism (Zhai et al., 2011; Owusu Adjei et al., 2021). In our analysis, many enzyme genes in paclitaxel synthesis are predicted as targets of miRNAs and phasiRNAs, indicating the roles of posttranscriptional regulation in paclitaxel synthesis. Notably, key genes like GGPPS-1, BAPT-2, T13αOH-3, TBT-4, T10βOH_like_5, and T2αOH-1 are directly targeted by 15 miRNAs with high abundance in T. chinensis leaf. These miRNAs also target to TFs with functions in regulation of plant growth and development. The miRNAs could function as intermediate signals controlling the balance of paclitaxel synthesis and plant development (Ha and Kim, 2014).
The identification of miRNAs and phasiRNAs in paclitaxel synthesis could also provide novel strategies to elevate enzyme genes expression, thereby boosting paclitaxel production. The miRNA sponges and competing endogenous RNAs (ceRNAs) can bind to miRNAs and phasiRNAs to inhibit their functions (Panda, 2018; Smillie et al., 2018). Specific miRNA sponges and ceRNAs can be designed and overexpressed in tissues and cell lines of taxus to block key miRNAs and phaisRNAs, which could release the expression of enzyme genes. Genome editing techniques, such as CRISPR/Cas9 could also be applied to know out the expression of miRNAs and phasiRNAs to improve paclitaxel production (Deng et al., 2022).
In summary, our comprehensive investigation systematically identified enzyme-encoding genes involved in paclitaxel biosynthesis in T. chinensis and their transcriptional expression at diverse tissues. TFs, miRNAs and phasiRNAs were identified, followed by the construction of regulatory networks encompassing enzyme genes, and their upstream regulators. The hierarchical regulation of paclitaxel production by miRNAs and phasiRNAs indicates a robust regulation at post-transcriptional level. This study contributes valuable insights into the regulatory expression patterns of paclitaxel synthesis-related enzyme genes and provide guidance to elevate paclitaxel production.
Raw data of RNAseq and sRNAseq of studied samples from Enshi have been deposited in the NCBI SRA under the accession PRJNA1031429.
M-SS: Data curation, Formal analysis, Investigation, Visualization, Writing – original draft, Writing – review & editing. YJ: Formal analysis, Investigation, Writing – original draft. X-YC: Investigation, Writing – original draft. J-SC: Investigation, Writing – original draft. YG: Investigation, Writing – original draft. F-FF: Methodology, Supervision, Writing – original draft, Writing – review & editing. L-JX: Methodology, Supervision, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was supported by the National Key Research and Development Program of China (2022YFD2200601), the National Natural Science Foundation of China (32101559) and Natural Science Foundation of Jiangsu Province (BK20220411).
We thank the reviewers and editors for their constructive suggestions on our manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1403060/full#supplementary-material
Ansbacher, T., Freud, Y., Major, D. T. (2018). Slow-starter enzymes: role of active-site architecture in the catalytic control of the biosynthesis of taxadiene by taxadiene synthase. Biochemistry 57, 3773–3779. doi: 10.1021/acs.biochem.8b00452
Axtell, J. M. (2013). ShortStack: Comprehensive annotation and quantification of small RNA genes. RNA (New. York. N.Y.) 19, 740–751. doi: 10.1261/rna.035279.112
Balogu, E., Kinston, D. G. (1999). A new semisynthesis of paclitaxel from baccatin III. J. Nat. Prod. 62, 1068–1071. doi: 10.1021/np990040k
Bolger, A. M., Lohse, M., Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Buchfink, B., Reuter, K., Drost, H. G. (2021). Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat. Methods 18, 366–368. doi: 10.1038/s41592-021-01101-x
Cao, Y. P., Li, K., Li, Y. L., Zhao, X. P., Wang, L. H. (2020). MYB transcription factors as regulators of secondary metabolism in plants. Biol. (Basel). 9, 61. doi: 10.3390/biology9030061
Chau, M., Croteau, R. (2004). Molecular cloning and characterization of a cytochrome P450 taxoid 2alpha-hydroxylase involved in Taxol biosynthesis. Arch. Biochem. Biophys. 427, 48–57. doi: 10.1016/j.abb.2004.04.016
Chen, Y., Zhang, M., Jin, X. F., Tao, H. R., Wang, Y. M., Peng, B., et al. (2020). Transcriptional reprogramming strategies and miRNA-mediated regulation networks of Taxus media induced into callus cells from tissues. BMC Genomics 21, 168. doi: 10.1186/s12864-020-6576-2
Cheng, J., Wang, X., Liu, X. N., Zhu, X. X., Li, Z. H., Chu, H. Y., et al. (2021). Chromosome-level genome of Himalayan yew provides insights into the origin and evolution of the paclitaxel biosynthetic pathway. Mol. Plant 14, 11. doi: 10.1016/j.molp.2021.04.015
Croteau, R., Ketchum, R. E., Long, R. M., Kaspera, R., Wildung, M. R. (2006). Taxol biosynthesis and molecular genetics. Phytochem. Rev. 5, 75–97. doi: 10.1007/s11101-005-3748-2
Dai, X. B., Zhuang, Z. H., Zhao, P. X. (2018). psRNATarget: a plant small RNA target analysis server, (2017 release). Nucleic Acids Res. 46, W49–W54. doi: 10.1093/nar/gky316
De Furia, M. D. (1997). Paclitaxel (Taxol®): A new natural product with major anticancer activity. Phytomedicine 4, 273–282. doi: 10.1016/S0944-7113(97)80081-5
Deng, C., Wu, Y. K., Lv, X. Q., Li, J. H., Liu, Y. F., Du, G. C., et al. (2022). Refactoring transcription factors for metabolic engineering. Biotechnol. Adv. 57, 107935. doi: 10.1016/j.bioteChadv.2022.107935
Deng, P. C., Muhammad, S., Cao, M., Wu, L. (2018). Biogenesis and regulatory hierarchy of phased small interfering RNAs in plants. Plant Biotechnol. J. 16, 965–975. doi: 10.1111/pbi.12882
Deng, F. L., Zeng, F. R., Shen, Q. F., Abbas, A., Cheng, J. H., Jiang, W., et al. (2022). Molecular evolution and functional modification of plant miRNAs with CRISPR. Trends Plant Sci. 27, 890–907. doi: 10.1016/j.tplants.2022.01.009
Dobin, A., Davis, C. A., Schlesinger, F., Drenkow, J., Zaleski, C., Jha, S., et al. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. doi: 10.1093/bioinformatics/bts635
Dukowic-Schulze, S., Sundararajan, A., Ramaraj, T., Kianian, S., Pawlowski, W. P., Mudge, J., et al. (2016). Novel meiotic miRNAs and indications for a role of phasiRNAs in meiosis. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00762
Fei, Q. L., Yang, L., Liang, W. Q., Zhang, D. B., Meyers, B. C. (2016). Dynamic changes of small RNAs in rice spikelet development reveal specialized reproductive phasiRNA pathways. J. Exp. Bot. 67, 6037–6049. doi: 10.1093/jxb/erw361
Fett-Neto, A. G., DiCosmo, F., Reynolds, W. F., Sakata, K. (1992). Cell culture of Taxus as a source of the antineoplastic drug taxol and related taxanes. Biotechnol. (N. Y). 10, 1572–1575. doi: 10.1038/nbt1292-1572
Griffiths-Jones, S., Moxon, S., Marshall, M., Khanna, A., Eddy, S. R., Bateman, A. (2005). Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 33, D121–D124. doi: 10.1093/nar/gki081
Ha, M., Kim, V. N. (2014). Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Bio 15, 509–524. doi: 10.1038/nrm3838
Harris, M. A., Clark, J., Ireland, A., Lomax, J., Ashburner, M., Foulger, R., et al. (2004). The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 32, D258–D261. doi: 10.1093/nar/gkh036
Hefner, J., Ketchum, R. E., Croteau, R. (1998). Cloning and functional expression of a cDNA encoding geranylgeranyl diphosphate synthase from Taxus canadensis and assessment of the role of this prenyltransferase in cells induced for taxol production. Arch. Biochem. Biophys. 360, 62–74. doi: 10.1006/abbi.1998.0926
Howat, S., Park, B., Oh, I. S., Jin, Y. W., Lee, E. K., Loake, G. J. (2014). Paclitaxel: biosynthesis, production and future prospects. N. Biotechnol. 31, 242–245. doi: 10.1016/j.nbt.2014.02.010
Jarchow-Choy, S. K., Koppisch, A. T., Fox, D. T. (2014). Synthetic routes to methylerythritol phosphate pathway intermediates and downstream isoprenoids. Curr. Org. Chem. 18, 1050–1072. doi: 10.2174/1385272819666140501001101
Jennewein, S., Croteau, R. (2001). Taxol: biosynthesis, molecular genetics, and biotechnological applications. Appl. Microbiol. Biotechnol. 57, 13–19. doi: 10.1007/s002530100757
Jennewein, S., Rithner, C. D., Williams, R. M., Croteau, R. B. (2001). Taxol biosynthesis: taxane 13 alpha-hydroxylase is a cytochrome P450-dependent monooxygenase. Proc. Natl. Acad. Sci. U. S. A. 98, 13595–13600. doi: 10.1073/pnas.251539398
Jiang, B., Gao, L., Wang, H. J., Sun, Y. P., Zhang, X. L., Ke, H., et al. (2024). Characterization and heterologous reconstitution of Taxus biosynthetic enzymes leading to baccatin III. Science 383, 622–629. doi: 10.1126/science.adj3484
Johnson, C., Kasprzewska, A., Tennessen, K., Fernandes, J., Nan, G. L., Walbot, V., et al. (2009). Clusters and superclusters of phased small RNAs in the developing inflorescence of rice. Genome Res. 19, 1429–1440. doi: 10.1101/gr.089854.108
Jones-Rhoades, M. W. (2012). Conservation and divergence in plant microRNAs. Plant Mol. Biol. 80, 3–16. doi: 10.1007/s11103-011-9829-2
Kahle, J., Baake, M., Doenecke, D., Albig, W. (2005). Subunits of the heterotrimeric transcription factor NF-Y are imported into the nucleus by distinct pathways involving importin beta and importin 13. Mol. Cell Biol. 25, 5339–5354. doi: 10.1128/mcb.25.13.5339-5354.2005
Kakrana, A., Li, P. C., Patel, P. R., Meyers, B. C. (2017). PHASIS: A computational suite for de novo discovery and characterization of phased, siRNA-generating loci and their miRNA triggers. bioRxiv, 158832. doi: 10.1101/158832
Kaspera, R., Croteau, R. (2006). Cytochrome P450 oxygenases of Taxol biosynthesis. Phytochem. Rev. 5, 433–444. doi: 10.1007/s11101-006-9006-4
Kozomara, A., Birgaoanu, M., Griffiths-Jones, S. (2019). miRBase: from microRNA sequences to function. Nucleic Acids Res. 47, D155–D162. doi: 10.1093/nar/gky1141
Kuang, X. J., Sun, S. J., Wei, J. H., Li, Y., Sun, C. (2019). Iso-Seq analysis of the Taxus cuspidata transcriptome reveals the complexity of Taxol biosynthesis. BMC Plant Biol. 19, 210. doi: 10.1186/s12870-019-1809-8
Langfelder, P., Horvath, S. (2008). WGCNA: an R package for weighted correlation network analysis. BMC Bioinf. 9, 559. doi: 10.1186/1471-2105-9-559
Langmead, B. (2010). Aligning short sequencing reads with Bowtie. Curr. Protoc. Bioinf. 32, 11.7.1–11.7.14. doi: 10.1002/0471250953.bi1107s32
Li, B., Dewey, C. N. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323. doi: 10.1186/1471-2105-12-323
Li, T. T., Li, B. B., Liao, C. L., Zhang, H. M., Wang, L. Z., Fu, T. T., et al. (2022). Transcriptome analysis provides insights into light condition effect on paclitaxel biosynthesis in yew saplings. BMC Plant Biol. 22, 577. doi: 10.1186/s12870-022-03958-2
Li, Y., Zhang, G. J., Pfeifer, B. A. (2015). Current and emerging options for taxol production. Adv. Biochem. Eng. Biotechnol. 148, 405–425. doi: 10.1007/10_2014_292
Li, J. W., Zhang, X. Y., Wu, H., Bai, Y. P. (2020). Transcription factor engineering for highthroughput strain evolution and organic acid bioproduction: a review. Front. Bioeng. Biotechnol. 8. doi: 10.3389/fbioe.2020.00098
Li, S., Zhang, P., Zhang, M., Fu, C., Yu, L. (2013). Functional analysis of a WRKY transcription factor involved in transcriptional activation of the DBAT gene in Taxus chinensis. Plant Biol. (Stuttg). 15, 19–26. doi: 10.1111/j.1438-8677.2012.00611.x
Liao, P., Hemmerlin, A., Bach, T. J., Chye, M. L. (2016). The potential of the mevalonate pathway for enhanced isoprenoid production. Biotechnol. Adv. 34, 697–713. doi: 10.1016/j.bioteChadv.2016.03.005
Liu, J. C., de la Peña, R., Tocol, C., Sattely, E. S. (2024). Reconstitution of early paclitaxel biosynthetic network. Nat. Commun. 15, 1419. doi: 10.1038/s41467-024-45574-8
Liu, Y. L., Teng, C., Xia, R., Meyers, B. C. (2020). PhasiRNAs in plants: their biogenesis, genic sources, and roles in stress responses, development, and reproduction. Plant Cell. 32, 3059–3080. doi: 10.1105/tpc.20.00335
Long, R. M., Lagisetti, C., Coates, R. M., Croteau, R. B. (2008). Specificity of the N-benzoyl transferase responsible for the last step of Taxol biosynthesis. Arch. Biochem. Biophys. 477, 384–389. doi: 10.1016/j.abb.2008.06.021
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17, 10–12. doi: 10.14806/ej.17.1.200
Mutanda, I., Li, J. H., Xu, F. L., Wang, Y. (2021). Recent advances in metabolic engineering, protein engineering, and transcriptome-guided insights toward synthetic production of Taxol. Front. Bioeng. Biotechnol. 9. doi: 10.3389/fbioe.2021.632269
Ogata, H., Goto, S., Sato, K., Fujibuchi, W., Bono, H., Kanehisa, M. (1999). KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 27, 29–34. doi: 10.1093/nar/27.1.29
Owusu Adjei, M., Zhou, X. Z. X., Mao, M. Q., Rafique, F., Ma, J. (2021). MicroRNAs roles in plants secondary metabolism. Plant Signal Behav. 16, 1915590. doi: 10.1080/15592324.2021.1915590
Panda, A. C. (2018). Circular RNAs act as miRNA sponges. Adv. Exp. Med. Biol. 1087, 67–79. doi: 10.1007/978-981-13-1426-1_6
Perez-Matas, E., Hidalgo-Martinez, D., Moyano, E., Palazon, J., Bonfill, M. (2024). Overexpression of BAPT and DBTNBT genes in Taxus baccata in vitro cultures to enhance the biotechnological production of paclitaxel. Plant Biotechnol. J. 22, 233–247. doi: 10.1111/pbi.14182
Prüfer, K., Stenzel, U., Dannemann, M., Green, R. E., Lachmann, M., Kelso, J. (2008). PatMaN: rapid alignment of short sequences to large databases. Bioinformatics 24, 1530–1531. doi: 10.1093/bioinformatics/btn223
Ro, D. K., Paradise, E. M., Ouellet, M., Fisher, K. J., Newman, K. L., Ndungu, J. M., et al. (2006). Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature 440, 940–943. doi: 10.1038/nature04640
Robinson, M. D., McCarthy, D. J., Smyth, G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139. doi: 10.1093/bioinformatics/btp616
Sanchez-Muñoz, R., Perez-Mata, E., Almagro, L., Cusido, R. M., Bonfill, M., Palazon, J., et al. (2020). A novel hydroxylation step in the taxane biosynthetic pathway: a new approach to paclitaxel production by synthetic biology. Front. Bioeng. Biotechnol. 8. doi: 10.3389/fbioe.2020.00410
Schneider, F., Samarin, K., Zanella, S., Gaich, T. (2020). Total synthesis of the complex taxane diterpene canataxpropellane. Science 367, 676–681. doi: 10.1126/science.aay9173
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Shivaprasad, P. V., Chen, H. M., Patel, K., Bond, D. M., Santos, B. A., Baulcombe, D. C. (2012). A microRNA superfamily regulates nucleotide binding site-leucine-rich repeats and other mRNAs. Plant Cell. 24, 859–874. doi: 10.1105/tpc.111.095380
Singh, K., Foley, R. C., Oñate-Sánchez, L. (2002). Transcription factors in plant defense and stress responses. Curr. Opin. Plant Biol. 5, 430–436. doi: 10.1016/s1369-5266(02)00289-3
Smillie, C. L., Sirey, T., Ponting, C. P. (2018). Complexities of post-transcriptional regulation and the modeling of ceRNA crosstalk. Crit. Rev. Biochem. Mol. Biol. 53, 231–245. doi: 10.1080/10409238.2018.1447542
Song, C., Fu, F. F., Yang, L. L., Niu, Y., Tian, Z. Y., He, X. X., et al. (2021). Taxus yunnanensis genome offers insights into gymnosperm phylogeny and taxol production. Commun. Biol. 4, 1203. doi: 10.1038/s42003-021-02697-8
Su, X. J., Yang, L. L., Wang, D. L., Shu, Z. Q., Yang, Y. C., Chen, S. L., et al. (2022). 1 K Medicinal Plant Genome Database: an integrated database combining genomes and metabolites of medicinal plants. Hortic. Res. 9, uhac075. doi: 10.1093/hr/uhac075
Walker, K., Croteau, R. (2000). Taxol biosynthesis: molecular cloning of a benzoyl-CoA:taxane 2alpha-O-benzoyltransferase cDNA from taxus and functional expression in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 97, 13591–13596. doi: 10.1073/pnas.250491997
Walker, K., Fujisaki, S., Long, R., Croteau., R. (2002). Molecular cloning and heterologous expression of the C-13 phenylpropanoid side chain-CoA acyltransferase that functions in Taxol biosynthesis. Proc. Natl. Acad. Sci. U. S. A. 99, 12715–12720. doi: 10.1073/pnas.192463699
Walker, K., Schoendorf, A., Croteau, R. (2000). Molecular cloning of a taxa-4(20),11(12)-dien-5alpha-ol-O-acetyl transferase cDNA from Taxus and functional expression in Escherichia coli. Arch. Biochem. Biophys. 374, 371–380. doi: 10.1006/abbi.1999.1609
Wu, Y. K., Chen, T. C., Liu, Y. F., Tian, R. Z., Lv, X. Q., Li, J. H., et al. (2020). Design of a programmable biosensor-CRISPRi genetic circuits for dynamic and autonomous dual-control of metabolic flux in Bacillus subtilis. Nucleic Acids Res. 48, 996–1009. doi: 10.1093/nar/gkz1123
Xia, R., Chen, C. J., Pokhrel, S., Ma, W. Q., Huang, K., Patel, P., et al. (2019). 24-nt reproductive phasiRNAs are broadly present in angiosperms. Nat. Commun. 10, 627. doi: 10.1038/s41467-019-08543-0
Xiong, X. Y., Gou, J. B., Liao, Q. G., Li, Y. L., Zhou, Q., Bi, G. Q., et al. (2021). The Taxus genome provides insights into paclitaxel biosynthesis. Nat. Plants. 7, 1026–1036. doi: 10.1038/s41477-021-00963-5
Xu, X. H., Liu, Y. F., Du, G. C., Ledesma-Amaro, R., Liu, L. (2020). Microbial chassis development for natural product biosynthesis. Trends Biotechnol. 38, 779–796. doi: 10.1016/j.tibtech.2020.01.002
Yan, X., Fan, Y., Wei, W., Wang, P. P., Liu, Q. F., Wei, Y. J., et al. (2014). Production of bioactive ginsenoside compound K in metabolically engineered yeast. Cell Res. 24, 770–773. doi: 10.1038/cr.2014.28
Yu, C. N., Huang, J. F., Wu, Q. C., Zhang, C. C., Li, X. L., Xu, X. Y., et al. (2022). Role of female-predominant MYB39-bHLH13 complex in sexually dimorphic accumulation of taxol in Taxus media. Hortic. Res. 9, uhac062. doi: 10.1093/hr/uhac062
Yu, G. C., Wang, L. G., Han, Y. Y., He, Q. Y. (2012). clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287. doi: 10.1089/omi.2011.0118
Yu, C. N., Zhang, C. C., Xu, X. Y., Huang, J. F., Chen, Y. Y., Luo, X. J., et al. (2021). Omic analysis of the endangered Taxaceae species Pseudotaxus chienii revealed the differences in taxol biosynthesis pathway between Pseudotaxus and Taxus yunnanensis trees. BMC Plant Biol. 21, 104. doi: 10.1186/s12870-021-02883-0
Zhai, J. X., Jeong, D. H., De Paoli, E., Park, S., Rosen, B. D., Li, Y. P., et al. (2011). MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev. 25, 2540–2553. doi: 10.1101/gad.177527.111
Zhang, M., Dong, Y. S., Nie, L., Lu, M. B., Fu, C. H., Yu, L. J. (2015a). High-throughput sequencing reveals miRNA effects on the primary and secondary production properties in long-term subcultured Taxus cells. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00604
Zhang, M., Jin, X. F., Chen, Y., Wei, M., Liao, W. F., Zhao, S. Y., et al. (2018). TcMYC2a, a basic Helix-Loop-Helix transcription factor, transduces JA-signals and regulates taxol biosynthesis in Taxus chinensis. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00863
Zhang, M., Li, S. T., Nie, L., Chen, Q. P., Xu, X. P., Yu, L. J., et al. (2015b). Two jasmonate-responsive factors, TcERF12 and TcERF15, respectively act as repressor and activator of tasy gene of taxol biosynthesis in Taxus chinensis. Plant Mol. Biol. 89, 463–473. doi: 10.1007/s11103-015-0382-2
Zhang, Y. J., Wiese, L., Fang, H., Alseekh, S., Perez de Souza, L., Scossa, F., et al. (2023). Synthetic biology identifies the minimal gene set required for paclitaxel biosynthesis in a plant chassis. Mol. Plant 16, 1951–1961. doi: 10.1016/j.molp.2023.10.016
Keywords: paclitaxel, Taxus chinensis, transcription factors, microRNA, phasiRNA, gene regulatory network
Citation: Sun M-S, Jia Y, Chen X-Y, Chen J-S, Guo Y, Fu F-F and Xue L-J (2024) Regulatory microRNAs and phasiRNAs of paclitaxel biosynthesis in Taxus chinensis. Front. Plant Sci. 15:1403060. doi: 10.3389/fpls.2024.1403060
Received: 18 March 2024; Accepted: 22 April 2024;
Published: 08 May 2024.
Edited by:
Xinyi Guo, Central European Institute of Technology (CEITEC), CzechiaReviewed by:
Jianchao Ma, Henan University, ChinaCopyright © 2024 Sun, Jia, Chen, Chen, Guo, Fu and Xue. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fang-Fang Fu, ZmZmdUBuamZ1LmVkdS5jbg==; Liang-Jiao Xue, bHh1ZUBuamZ1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.