
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 28 May 2024
Sec. Plant Breeding
Volume 15 - 2024 | https://doi.org/10.3389/fpls.2024.1402709
Wheat is a major food crop that plays a crucial role in the human diet. Various breeding technologies have been developed and refined to meet the increasing global wheat demand. Several studies have suggested breeding strategies that combine generation acceleration systems and molecular breeding methods to maximize breeding efficiency. However, real-world examples demonstrating the effective utilization of these strategies in breeding programs are lacking. In this study, we designed and demonstrated a synergized breeding strategy (SBS) that combines rapid and efficient breeding techniques, including speed breeding, speed vernalization, phenotypic selection, backcrossing, and marker-assisted selection. These breeding techniques were tailored to the specific characteristics of the breeding materials and objectives. Using the SBS approach, from artificial crossing to the initial observed yield trial under field conditions only took 3.5 years, resulting in a 53% reduction in the time required to develop a BC2 near-isogenic line (NIL) and achieving a higher recurrent genome recovery of 91.5% compared to traditional field conditions. We developed a new wheat NIL derived from cv. Jokyoung, a leading cultivar in Korea. Milyang56 exhibited improved protein content, sodium dodecyl sulfate-sedimentation value, and loaf volume compared to Jokyoung, which were attributed to introgression of the Glu-B1i allele from the donor parent, cv. Garnet. SBS represents a flexible breeding model that can be applied by breeders for developing breeding materials and mapping populations, as well as analyzing the environmental effects of specific genes or loci and for trait stacking.
Wheat (Triticum aestivum L.) serves as the primary ingredient in diverse products, including breads, noodles, and cookies. It also contributes to the human diet as the third most important cereal crop in terms of total production (Shewry and Hey, 2015; Kiszonas and Morris, 2018; Igrejas and Branlard, 2020). Numerous breeding efforts have been conducted to enhance productivity, pest and disease resistance, and end-use quality. However, breeders face persistent pressure to expedite the development of new cultivars in response to challenges such as climate change and escalating global demand owing to population growth (Hickey et al., 2019). Consequently, various breeding strategies, including rapid generational advancement, phenotypic selection (PS), and molecular breeding, have been developed and deployed to enhance breeding efficiency (Pandey et al., 2022).
The speed breeding (SB) system, which utilizes an extended 22-h photoperiod, has enabled the rapid advancement of wheat generation, allowing for the production of up to six generations of spring wheat (Ghosh et al., 2018; Watson et al., 2018). The speed vernalization (SV) system, with a higher vernalization temperature of 8–10°C and a 22-h photoperiod, has facilitated up to five generations for both spring and winter wheat (Cha et al., 2022b). Subsequently, a modified speed breeding system (mSB), combined with SV, was confirmed to be suitable for practical use in large breeding populations (Cha et al., 2023b). The mSB system reduces the energy input by 80% and maximizes space utilization by harnessing natural sunlight in conventional glasses instead of relying solely on artificial lights. PS for agronomic traits and multiple disease resistance can also be effectively conducted while accelerating generations under SB or mSB conditions (Hickey et al., 2012; Dinglasan et al., 2016; Riaz et al., 2016; Cha et al., 2023b). These series of SB systems are believed to accelerate the development of breeding materials, as well as the overall wheat research by facilitating the creation of mapping populations.
Although SB systems contribute to reduced breeding times, molecular breeding — including marker-assisted selection (MAS) and marker-assisted backcrossing (MABC) — has emerged as a powerful tool for the introgression of targeted genes or major effect Quantitative trait loci (QTLs) into elite varieties (Varshney et al., 2020). Widely adopted in global wheat breeding programs (Gupta et al., 2010), molecular breeding minimizes the population size and breeding efforts by enabling the selection of lines carrying target genes or loci before harvesting (Varshney et al., 2021). Several successful applications of MAS and MABC have been observed, particularly in introgressing disease resistance, such as powdery mildew, stripe, and stem rust, and improving quality traits, such as high-molecular-weight glutenin subunits (HMW-GS) and grain protein content (Randhawa et al., 2019; Jia et al., 2020; Zheng et al., 2020; Gupta et al., 2022). Among the many types of molecular markers, the kompetitive allele-specific polymerase chain reaction (KASP) genotyping technology has gained prominence in crop breeding programs because of its high throughput, time-saving attributes, and cost-effectiveness (Rasheed et al., 2016; Bohar et al., 2020; Kaur et al., 2020; Maccaferri et al., 2022). KASP assays are useful in routine breeding applications and aid in background screening during MABC, thereby reducing the need for excessive backcrossing by selecting candidate lines with the highest recurrent genome recovery (Kang et al., 2019).
When rapid generation advancement systems are combined with molecular breeding, both breeding speed and efficiency evidently increase (Ahmar et al., 2020; Cha et al., 2023a). While numerous theoretical models have proposed combinations of SB and molecular breeding methods (Hickey et al., 2019; Voss-Fels et al., 2019; Varshney et al., 2021; Samantara et al., 2022), real-world cases demonstrating the advantages and disadvantages as well as the efficiency and improvement points of applying this theory in practical wheat breeding systems are lacking. In this study, we demonstrated a synergized breeding method (SBS) that integrates SB, PS, and MABC to introgress the Glu-B1i allele into one of Korea’s leading bread wheat cultivars, Jokyoung. The Glu-B1i allele, which belongs to the HMW-GS group, has beneficial effects on bread-making quality, including higher protein content and gluten strength, good extensibility, and increased bread loaf volume (Guo et al., 2019; Guzmán et al., 2022; Cha et al., 2022a). This study comprised the entire process, from artificial crossing to field trial evaluation. We aimed to illustrate how SB systems can effectively synergize with modern breeding technologies, thereby making them more accessible to breeders worldwide.
Two wheat cultivars, sourced from the Gene Bank of the National Institute of Agricultural Sciences, Rural Development Administration (RDA), Republic of Korea (ROK), were employed to implement the synergized breeding strategy (SBS). The Korean hard white spring wheat cv. Jokyoung (IT 213249) served as the recurrent parent (Kang et al., 2006), whereas Canadian hard red spring wheat cv. Garnet (IT 205495) was used as the donor parent (McCallum and DePauw, 2008). The target gene of Garnet was Glu-B1i, located on chromosome 1B, which enhances bread quality by encoding the high-molecular-weight glutenin subunit (HMW-GS) 17 + 18 in wheat (Payne et al., 1987; Jang et al., 2017; Guo et al., 2019; Shin et al., 2020; Guzmán et al., 2022). Jokyoung is a leading bread wheat cultivar in Korea that harbors Glu-B1b, which encodes the HMW-GS 7 + 8 (Jang et al., 2017).
The synergized breeding strategy (SBS) incorporated three breeding tools: speed breeding (SB) system, phenotypic selection (PS), and marker-assisted backcrossing (MABC). The SBS scheme is illustrated in Figure 1. An initial artificial cross between Jokyoung and Garnet was conducted under field conditions. Two rounds of backcrossing with Jokyoung were performed under modified speed breeding combined with speed vernalization (SV+mSB), as described by Cha et al (Cha et al., 2023b). The BC2F2 population was evaluated under mSB conditions, which enabled visual phenotypic selection based on the difference in days to heading (DTH) between the two parental lines. BC2F3 lines were planted in the field during the spring season, and a second visual phenotypic selection was conducted considering DTH and grain color. Molecular markers were applied to the BC1F1, BC2F1, BC2F2, and BC2F4 generations to select lines closely resembling Jokyoug genetically and harboring the Glu-B1i gene from the donor parent. The foreground selection marker, cauBx642, was used in BC1F1, BC2F1, and BC2F2 to identify lines carrying Glu-B1i as heterozygous (BC1F1 and BC2F1) or homozygous (BC2F2). Background selection at BC2F4 was aimed at assessing the recurrent genomic background recovery. All BCnF1 plants were cultivated in 12-cm pots, which provide a larger number of spikes and grains (Cha et al., 2020), while plants from the BC2F3 generations were sown in 72-cell trays.
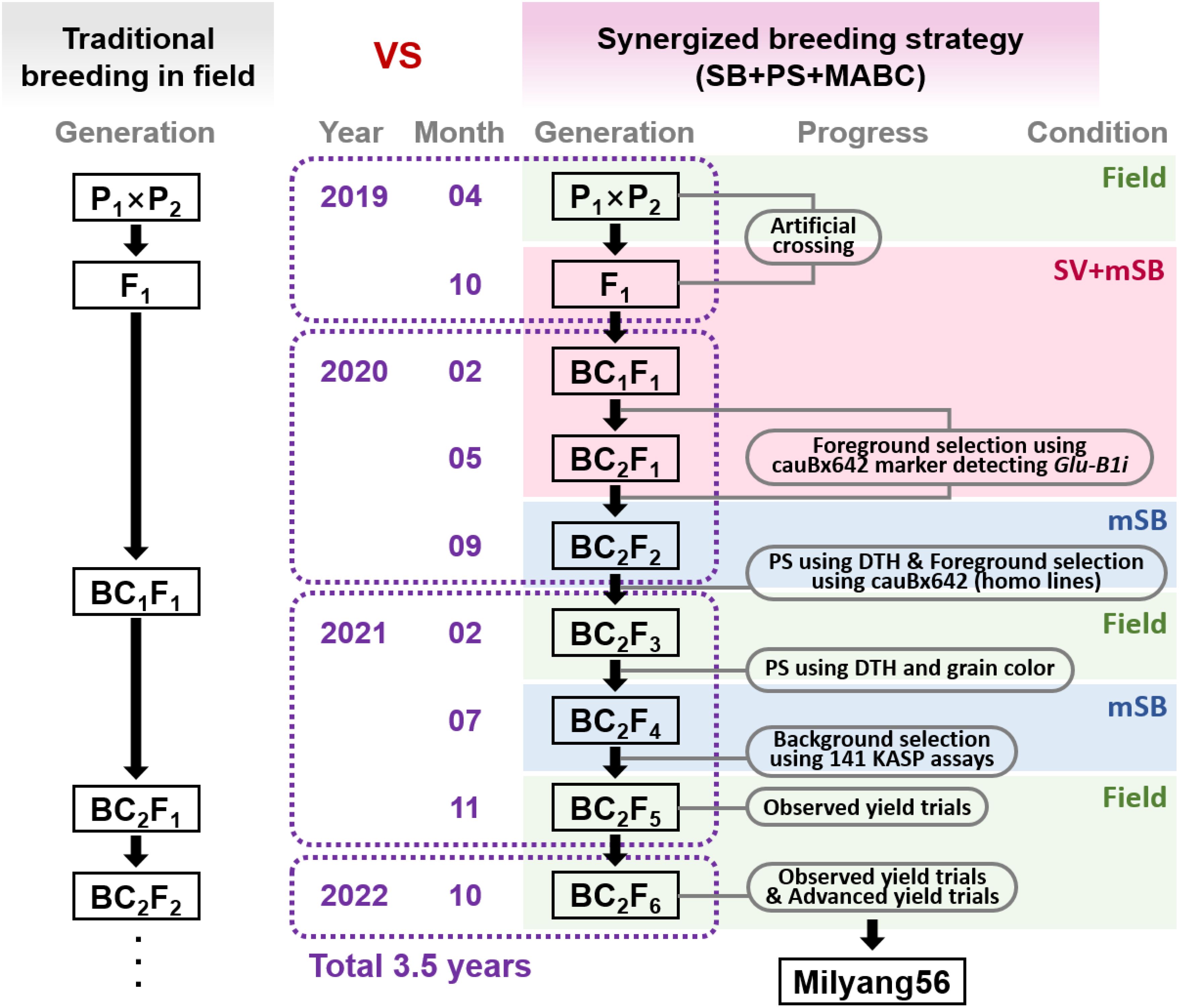
Figure 1 Synergized breeding strategy compared to traditional breeding method. The diagram represents the development of Milyang56 (Jokyoung*3/Garnet) by combining the speed breeding system (SB), phenotypic selection (PS), and marker-assisted backcrossing (MABC). mSB: modified speed breeding condition (22 h light, 2 h dark, 25 ± 3 °C in glasshouse); SV+mSB: speed vernalization (22 h light, 2 h dark in 8 °C for 4 weeks), then moved to the mSB condition; DTH: days to heading.
Genomic DNA was extracted from fresh seedling leaves of each plant using the MagMax DNA Multi-Sample Kit (Applied Biosystems, Woburn, MA, USA) and Kingfisher DNA extraction machine (Thermo Fisher Scientific, Waltham, MA, USA) following the manufacturer’s instructions. For foreground selection, the cauBx642 marker was used, as reported by Xu et al (Xu et al., 2008). The polymerase chain reaction (PCR) was carried out under the following conditions: initial denaturation at 95°C for 5 min, followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 59°C for 30 s, and extension at 72°C for 30; and final extension at 72°C for 5 min. PCR products were separated by electrophoresis on a 3% agarose gel at 200 V for 45 min.
For background selection, a total of 731 kompetitive allele-specific polymerase chain reaction (KASP) assays were used to identify the genetic background of the developed backcrossed lines. Among these 731 assays, 155 assays referred to CIMMIYT wheat molecular genetics (Dreisigacker et al., 2016), 70 assays to Rasheed et al (Rasheed et al., 2016), and the rest to the CerealsDB core marker set (Wilkinson et al., 2012). The PCR conditions were as follows: denaturation at 95°C for 15 min; 10 touchdown cycles at 95°C for 20 s, annealing starting at 65°C and decreasing by 1°C per cycle for 25 s; additional 30 annealing cycles at 95°C for 10 s and 57°C for 1 min; and final extension at 72°C for 5 min. Fluorescence detection and data analysis were performed using a QuantStudio3 real-time PCR instrument and QuantStudio Design & Analysis Software v1.5.1 (Applied Biosystems, Carlsbad, CA, USA).
All glasshouse-based and field tests were conducted at the National Institute of Crop Science (NICS), RDA, Miryang, ROK (35°29’32.9” N, 128°44’33.4” E). Agronomic traits, including days to heading (DTH), days to maturity (DTM), culm length (CL), spike length (SPL), number of tillers per plant (TN), number of grains per spike (GN), thousand-grain weight (TGW), test weight (TW), and grain yield (GY), were assessed following the RDA Standard Evaluation Manual for Agricultural Experiments and Research (RDA, 2012).
In the field, the BC2F3 lines were planted in late February 2021, whereas the BC2F5 lines were sown in early November 2021, representing the spring and winter growing seasons, respectively. BC2F5 lines underwent observed yield trials (OYT) in 2021, and BC2F6 lines were evaluated in both OYT and advanced yield trials (AYT) in 2022. For the OYT tests, lines were hill-seeded in the field with a spacing of 30 × 12 cm between and within the lines (a seed per hill). For the AYT tests, a rate of 14 kg/10a for each line was sown in the field as drill seeding, with 5 m rows and 30 cm spacing between rows. Fertilizer consisting of 9.1 kg N, 7.4 kg P2O5, and 3.9 kg K2O per 10 a was applied in all field tests.
To analyze flour quality, the OYT and AYT lines were milled using a Brabender Quadurmat Junior mill (CW Brabender Instruments, Inc., South Hackensack, NJ, USA) and a Buhler MLU 202 laboratory mill (Bühler AG, Uzwil, Switzerland), respectively.
HMW-Gs were extracted from the flour and identified using a lab-on-a-chip (Agilent 2100 bioanalyzer & protein 230 kit, Agilent Technologies, Waldbronn, Germany), following the series of processes proposed by Shin et al (Shin et al., 2020).
Flour protein content was determined using LECO FP628 (Laboratory Equipment Co., St. Joseph, Mich., USA) with a nitrogen factor of 5.7. The gluten content and index were measured using a Glutomatic 2200 (Perten Instruments AB, Sweden). The SDS sedimentation value was determined following the method proposed by Axford et al. (1979), with a modified sample amount of 3 g. All experiments, including bread-making quality, were conducted in accordance with the American Association of Cereal Chemists (AACC) methods (AACC, 2010), and the experimental results were standardized based on 14% moisture content.
RStudio (version 1.4.1717; RStudio, PBC, Boston, MA, USA) was used for all statistical analyses. Chi-square tests, analysis of variance, Duncan’s multiple range tests, and t-tests were performed using the ggplot2 and agricolae packages.
SBS was executed according to the scheme shown in Figure 1. The F1 seeds were obtained from a cross between cv. Jokyoung and cv. Garnet under field conditions were planted under SV+mSB conditions in October 2019. Jokyoung was subjected to two rounds of backcrossing under SV+mSB conditions, followed by two rounds of foreground selection using the cauBx642 marker to detect the Glu-B1i allele.
The first backcross commenced in December 2019, and BC1F1 seeds were planted in February 2020. Of the six BC1F1 plants, three that exhibited the heterozygous Glu-B1i allele were selected for the second backcrossing (Figure 1, Table 1). A total of 31 BC2F1 seeds were sown in May 2020. Among these, 13 plants carried the Glu-B1i allele heterozygously and were harvested by the family for the next generation (Figure 2A, Table 1).
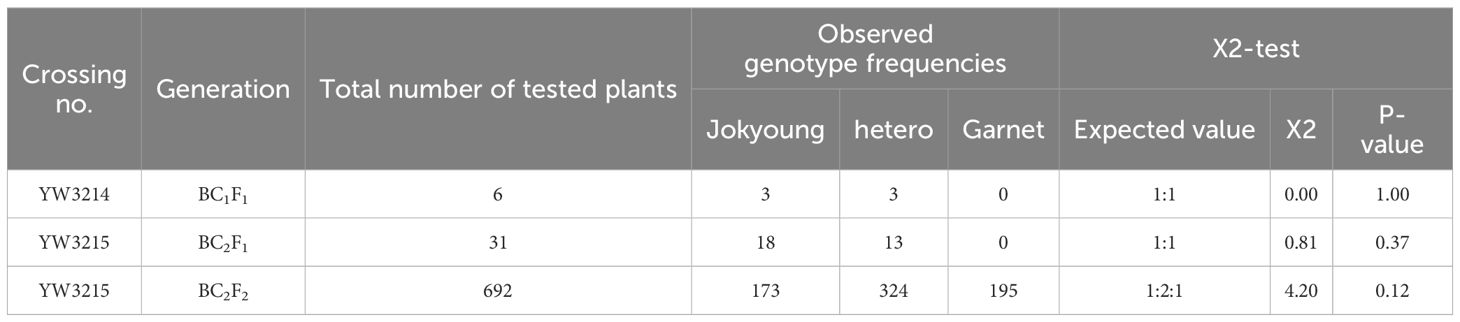
Table 1 Chi-square tests of the backcrossed populations for introducing the Glu-B1i allele into cv. Jokyoung.
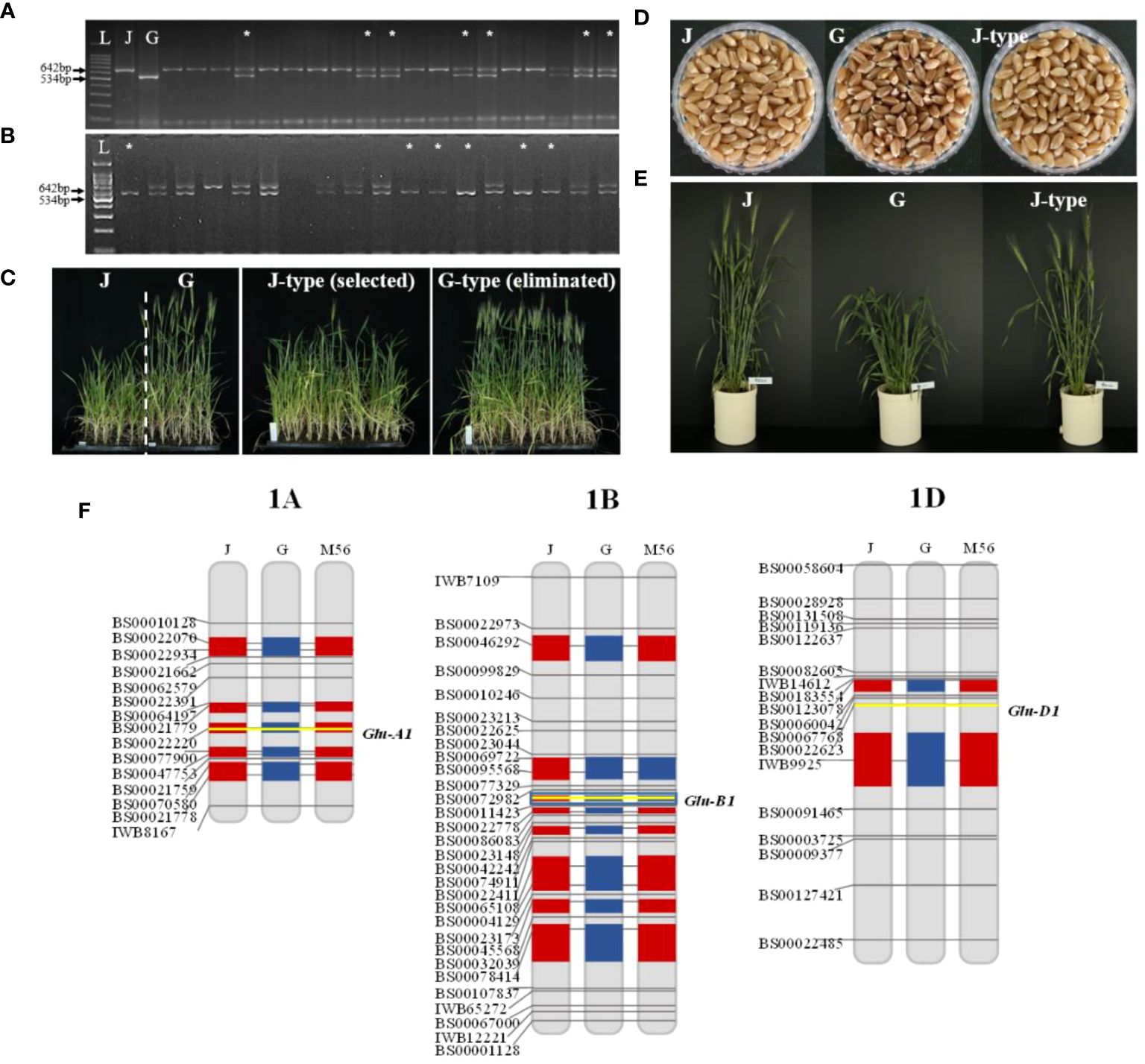
Figure 2 Genotypic and phenotypic selection in each generation. Foreground selection of Glu-B1i with cauBx642 marker in (A) BC2F1 and (B) BC2F2. Phenotypic selection in (C) BC2F2 families and (D, E) BC2F3 individuals. Background selection in (F) BC2F4. L: 100bp ladder marker, J: Jokyung, G: Garnet, *: Selected individuals.
BC2F2 plants were cultivated in September 2020 under mSB conditions for phenotypic selection (PS) based on days to heading (DTH). As DTH varied between Jokyoung and Garnet under mSB conditions, PS was conducted within the BC2F2 populations (Figure 2C, Supplementary Table S1). Of the 31 BC2F2 families derived from each BC2F1 individual plant, 12 BC2F2 families with comparable DTH to Jokyoung and confirmed to carry Glu-B1i through bulked segregant analysis were chosen (Supplementary Table S1). Individuals from each family were assessed using the cauBx642 marker. Among the 692 individuals evaluated, 195 lines harboring homozygous Glu-B1i were selected for subsequent generations (Figure 2B, Table 1 and Supplementary Table S1).
A total of 133 BC2F3 lines, yielding a sufficient quantity of seeds for field trials, were planted in the field in February 2021. PS was conducted based on the differences in DTH and grain color between the two parental lines. Consequently, 27 lines showing a heading date similar to Jokyoung (May 6) and white grains were selected (Figures 2D, E, Supplementary Figure S1). Subsequently, the 27 selected BC3F4 lines were grown under mSB conditions for background selection.
Of the 731 KASP assays collected, 141 exhibited polymorphisms between the two parents (Supplementary Table S2). Initially, 27 BC3F4 lines were assessed using 101 KASP assays. Sixteen lines, showing more than 85% recurrent genome recovery and more than 95% homozygosity, were identified. Among these, eight lines with the highest recurrent genome recovery and homozygosity underwent further testing with an additional 40 KASP assays (Table 2, Supplementary Figure S2). As a result of testing a total of 141 polymorphic KASP assays, these eight lines showed an average recurrent parent genome recovery of 83.9% (79.4–91.5%). These eight lines were evaluated as observed yield trial (OYT) in November 2021, and NIL-1 demonstrated the highest background genome recovery (91.5%) and similar agronomic traits, including heading date, maturity date, culm length, and other agronomic characteristics, compared to Jokyoung while carrying the introduced Glu-B1i allele from Garnet (Figure 2F, Supplementary Table S3, Supplementary Figure S3, and Supplementary Figure S4). Implementing SBS, from the initial crossing to the planting of the BC2F6 generation as OYT, took only 3.5 years, whereas it would have taken 7.5 years with conventional breeding methods, consequently reducing the time by 53%.
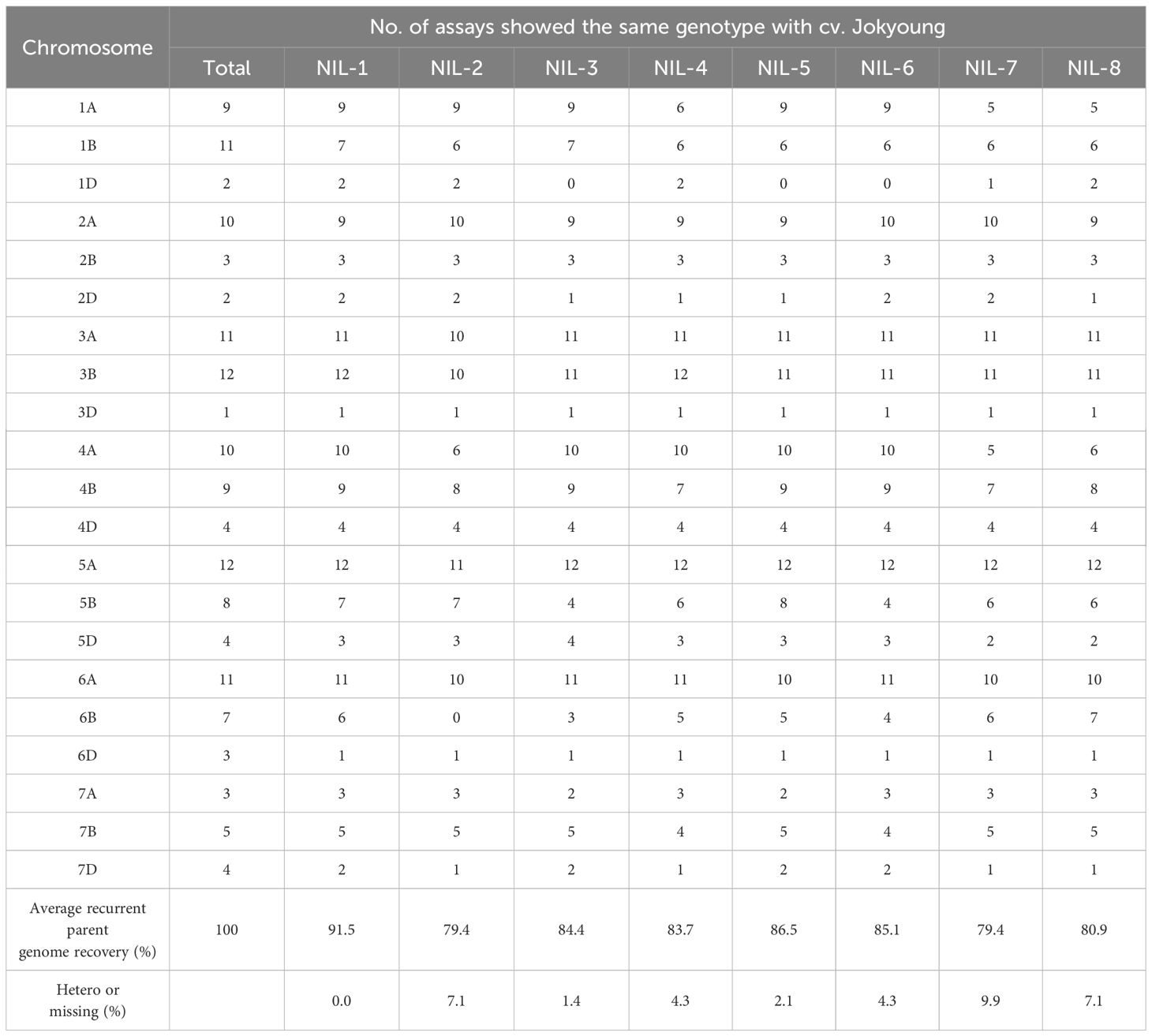
Table 2 Average recurrent genomic background recovery of eight Jokyoung-NIL-candidate lines in observed yield trials.
NIL-1 was designated Milyang56 and compared with the recurrent parent Jokyoung across two tested years (2021 and 2022) and trial methods (OYT and advanced yield trial(AYT)). A two-year OYT test was conducted to assess the annual variation. Overall, no significant differences were observed between Jokyoung and Milyang56 in most major agronomic traits, including DTH, days to maturity (DTM), culm length (CL), number of grains per spike (GN), and thousand-grain weight (TGW), across tested years and trial methods (Figures 3A, B, Table 3, Supplementary Tables S4–S6). While most agronomic traits exhibited significant differences due to environmental variations, such as the test year and trial method, there were no significant differences between Jokyoung and Milyang56. Although Milyang56 showed a shorter spike length than Jokyoung across both tested years and trial methods, no significant differences were observed in TGW and grain yield between Jokyoung and Milyang56 (Figure 3C, Table 3 and Supplementary Table S5).
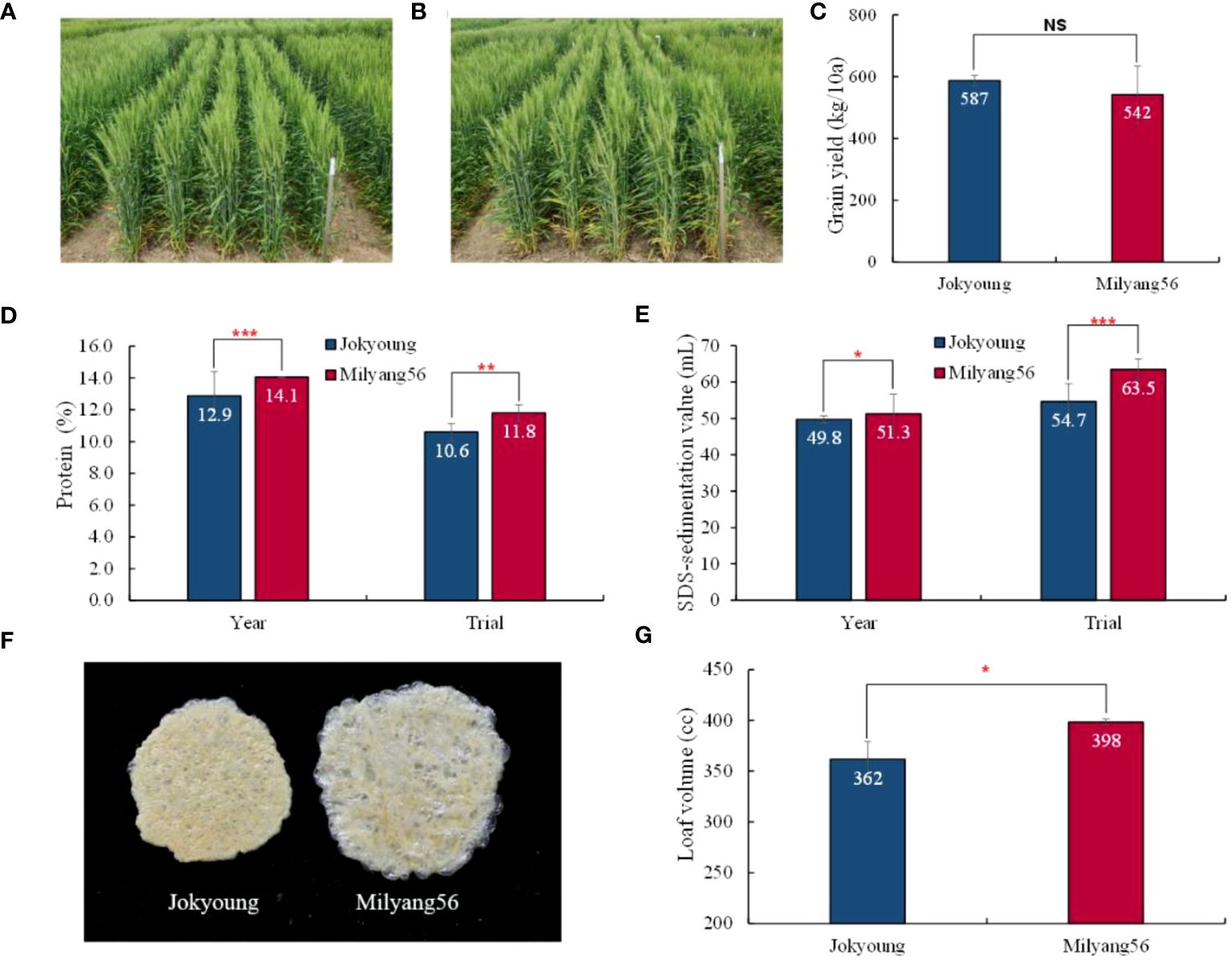
Figure 3 Comparison of plant phenotype and flour quality between Jokyoung and Milyang56 (Jokyoung-Glu-B1i-NIL) in field conditions. Phenotype of (A) Jokyoung and (B) Milyang56; Comparison of (C) grain yield in advanced yield test (AYT). For the comparison of (D) protein content, (E) SDS-sedimentation value, ‘Year’ represents the average performance in observed yield trials (OYT) over two years (2022–2023), while the ‘Trial’ signifies the average scores in the OYT and AYT in 2023. (F) dry gluten extracted from each 10 g-flour and (G) loaf volume tested in AYT. NS: no significant difference, * P<0.05, ** P<0.01, and *** P<0.001 significant difference according to t-test.
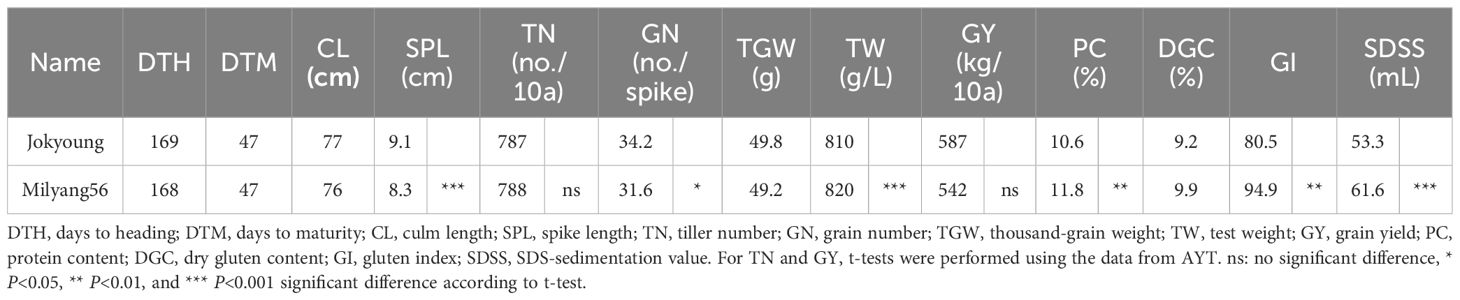
Table 3 Comparison of Jokyoung and Milyang56 with the average performance in observed yield trial and advanced yield trial (AYT) in 2023.
When the grains were milled, and the flour was tested, the protein content and SDS sedimentation value of Milyang56 were significantly higher than those of Jokyoung, irrespective of the tested years or trial methods (Figures 3D, E). Additionally, the gluten index tended to be higher in Milyang56, although no statistically significant difference was observed in the annual test (Figure 3F, Supplementary Table S5 and Supplementary Table S6). As a result of introgressing the Glu-B1i allele without altering the agronomic traits, the bread loaf volume of Milyang56 increased by 9.9% compared to that of Jokyoung (Figure 3G).
Our study demonstrated how a series of SB methods could be combined with other breeding techniques to form an upgraded and realized model within breeding programs. It showed breeders how to choose and apply proper generation acceleration systems according to the breeding purpose and characteristics of the breeding materials. Both the mSB and SV+mSB systems were utilized to develop a new wheat line, “Milyang56”. The mSB system notably reduced the DTH of spring wheat cultivars; however, differences in DTH between different cultivars still existed (Ghosh et al., 2018; Watson et al., 2018). Therefore, phenotypic selection was conducted using this difference point in the BC2F2 populations. In contrast, the SV+mSB condition was used in two rounds of backcrossing to exploit its ability to ensure a uniform DTH, enabling artificial crossing between parents with diverse genetic backgrounds (Cha et al., 2022b). The series of SB systems was also naturally combined with field tests, along with breeding purposes and schedules.
Molecular breeding methods, such as MAS and MABC, were also applied under mSB and SV+mSB conditions to enhance both the speed and efficiency of breeding. We demonstrated a real-world case of the SB + PS + molecular breeding model named SBS. Compared to traditional breeding methods under field conditions, SBS reduced breeding time by 53% for developing the BC2F6 NIL. Foreground selection was conducted for BC1F1 and BC2F1, and background selection was conducted for the BC2F4 generation. Milyang56 showed 91.5% recurrent parent genome recovery with Glu-B1i introgression from the donor parent despite only two rounds of backcrossing. The theoretical average percentages of recurrent parent genome recovery in the BC2 and BC3 generations were 87.5 and 93.8%, respectively. However, these percentages are only achieved with large populations and are usually lower in the smaller population sizes typically used in actual breeding programs (Collard et al., 2005). For example, the average genome recovery was reported to be 92.2% despite both foreground and background selection being conducted in the BC1F1 and BC2F1 generations of maize (Babu et al., 2005; Hasan et al., 2015). Although only one background selection was conducted in our study, a higher genome recovery than the theoretical average was retained because of the complementary PS.
However, opportunities exist to conduct MABC more efficiently. The accuracy of individual selection was improved when background selection was conducted in the lower generations, BC2F1 and BC2F2, rather than BC2F4. A NIL with a 96.2% recurrent genome recovery was developed as a result of background selection, with 71 assays for BC2F1 and six assays for the BC2F2 generation in rice (Kang et al., 2019). In practice, most MABC studies have conducted background selection for BC1F1, BC2F1, and BC2F2 (Vishwakarma et al., 2014; Rai et al., 2018; Bellundagi et al., 2022).
Background selection in early generations combined with PS may significantly increase recurrent genome recovery with fewer backcrossings compared to traditional methods. To conduct genotyping in a limited time (1–2 months before harvesting) in a series of SB conditions, breeders must be able to analyze the genotype of a large amount of breeding material simply and quickly. Therefore, we utilized KASP assays, which are highly accurate, inexpensive, and flexible (Yang et al., 2019). Different types of molecular markers can be utilized for both foreground and background selection, according to the breeder’s conditions.
HMW-GS Glu-B1i is known to enhance bread quality by increasing protein content, gluten strength, and gluten extensibility (Guo et al., 2019; Guzmán et al., 2022). However, environmental conditions affect the allelic performance of HMW-GSs in terms of the end-use quality of wheat (Kroupina et al., 2023). In Korea, Glu-B1i has been reported to have higher protein content and SDS sedimentation values than other alleles in 180 wheat varieties carrying various Glu-1 alleles (Cha et al., 2022a). However, important agronomic traits, especially DTH and DTM, were diverse in the varieties analyzed in this study, and no previous study has investigated the effects of Glu-B1i on similar genetic backgrounds in Korea. Therefore, we developed Jokyoung-NIL, Milyang56 and compared the effect of the Glu-B1i allele on end-use quality. Our study confirmed the positive effect of Glu-B1i on bread-making quality, along with previous studies using the SBS method and an extremely shortened breeding period. This method would facilitate the comparison of the effects of specific genes or loci in various environments by quickly developing NILs, and it can be utilized in gene stacking.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
J-KC: Conceptualization, Data curation, Investigation, Writing – original draft, Writing – review & editing. HP: Formal analysis, Investigation, Writing – review & editing. YK: Formal analysis, Investigation, Writing – review & editing. S-ML: Formal analysis, Investigation, Writing – review & editing. S-GJ: Data curation, Investigation, Writing – review & editing. SK: Conceptualization, Validation, Writing – review & editing. J-HL: Conceptualization, Funding acquisition, Methodology, Project administration, Supervision, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was carried out with the support of the “Research Program for Agriculture Science and Technology Development (Project Number: PJ017389012024)” Rural Development Administration, Republic of Korea.
We would like to thank Dr. Jong-Min Ko (Rural Development Administration, Republic of Korea) for his valuable advice and support throughout the research project.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1402709/full#supplementary-material
AACC (2010). “Approved methods of analysis,” in Method 46-30.01. Crude protein-combustion method; 26-50.01. Brabender Quadrumat Jr.(Quadruplex) method; 54-40.02. Mixograph method; 38-12.02. Wet gluten, dry gluten, water-binding capacity, and gluten index; 10–10.03. Optimized straight-dough bread-making method, 11th Ed (Cereal & Grains Association, St. Paul. MN, USA). http://dx.doi.org/10.1094/AACCIntMethod-xx-xx.xx (each method number)
Ahmar, S., Gill, R. A., Jung, K.-H., Faheem, A., Qasim, M. U., Mubeen, M., et al. (2020). Conventional and molecular techniques from simple breeding to speed breeding in crop plants: recent advances and future outlook. Int. J. Mol. Sci. 21, 2590. doi: 10.3390/ijms21072590
Axford, D. W., McDermott, E. E., Redman, D. G.. (1979). Note on the sodium dodecyl sulfate test of bread making quality: comparision with pelschenke and Zeleny test. Cereal Chem. 56, 582–584. doi: 0009-0352/79/000095$3.00/0
Babu, R., Nair, S., Kumar, A., Venkatesh, S., Sekhar, J., Singh, N., et al. (2005). Two-generation marker-aided backcrossing for rapid conversion of normal maize lines to quality protein maize (QPM). Theor. Appl. Genet. 111, 888–897. doi: 10.1007/s00122-005-0011-6
Bellundagi, A., Ramya, K., Krishna, H., Jain, N., Shashikumara, P., Singh, P. K., et al. (2022). Marker-assisted backcross breeding for heat tolerance in bread wheat (Triticum aestivum L.). Front. Genet. 13, 1056783. doi: 10.3389/fgene.2022.1056783
Bohar, R., Chitkineni, A., Varshney, R. K. (2020). Genetic molecular markers to accelerate genetic gains in crops. Future Sci. 69, 158–160. doi: 10.2144/btn-2020-0066
Cha, J.-K., Lee, J.-H., Lee, S.-M., Ko, J.-M., Shin, D. (2020). Heading date and growth character of Korean wheat cultivars by controlling photoperiod for rapid generation advancement. Korean Soc. Breed. Sci. 52, 20–24. doi: 10.9787/KJBS.2020.52.1.20
Cha, J.-K., O’Connor, K., Alahmad, S., Lee, J.-H., Dinglasan, E., Park, H., et al. (2022b). Speed vernalization to accelerate generation advance in winter cereal crops. Mol. Plant 15, 1300–1309. doi: 10.1016/j.molp.2022.06.012
Cha, J.-K., Park, H., Choi, C., Kwon, Y., Lee, S.-M., Oh, K.-W., et al. (2023b). Acceleration of wheat breeding: enhancing efficiency and practical application of the speed breeding system. Plant Methods 19, 118. doi: 10.1186/s13007-023-01083-1
Cha, J.-K., Park, H., Kwon, Y., Lee, S.-M., Oh, K.-W., Lee, J.-H. (2023a). Genotyping the high protein content gene NAM-B1 in wheat (Triticum aestivum L.) and the development of a KASP marker to identify a functional haplotype. Agronomy 13, 1977. doi: 10.3390/agronomy13081977
Cha, J.-K., Shin, D., Park, H., Kwon, Y., Lee, S.-M., Ko, J.-M., et al. (2022a). Effects of high-molecular-weight glutenin subunits and agronomic traits on bread wheat quality parameters. Korean J. Crop Sci. 67, 111–120. doi: 10.7740/kjcs.2022.67.2.111
Collard, B. C., Jahufer, M., Brouwer, J., Pang, E. C. K. (2005). An introduction to markers, quantitative trait loci (QTL) mapping and marker-assisted selection for crop improvement: the basic concepts. Euphytica 142, 169–196. doi: 10.1007/s10681-005-1681-5
Dinglasan, E., Godwin, I. D., Mortlock, M. Y., Hickey, L. T. (2016). Resistance to yellow spot in wheat grown under accelerated growth conditions. Euphytica 209, 693–707. doi: 10.1007/s10681-016-1660-z
Dreisigacker, S., Sehgal, D., Reyes Jaimez, A., Luna Garrido, B., Muñoz Zavala, S., Núñez Ríos, C., et al. (2016). Laboratory protocols and applications to wheat breeding (Mexico City, Mexico: CIMMYT)
Ghosh, S., Watson, A., Gonzalez-Navarro, O. E., Ramirez-Gonzalez, R. H., Yanes, L., Mendoza-Suárez, M., et al. (2018). Speed breeding in growth chambers and glasshouses for crop breeding and model plant research. Nat. Protoc. 13, 2944–2963. doi: 10.1038/s41596-018-0072-z
Guo, H., Wu, J., Lu, Y., Yan, Y. (2019). High-molecular-weight glutenin 1Bx17 and 1By18 subunits encoded by Glu-B1i enhance rheological properties and breadmaking quality of wheat dough. J. Food Qual doi: 10.1155/2019/1958747
Gupta, P. K., Balyan, H. S., Chhuneja, P., Jaiswal, J. P., Tamhankar, S., Mishra, V. K., et al. (2022). Pyramiding of genes for grain protein content, grain quality, and rust resistance in eleven Indian bread wheat cultivars: A multi-institutional effort. Mol. Breed. 42, 21. doi: 10.1007/s11032-022-01277-w
Gupta, P., Langridge, P., Mir, R. (2010). Marker-assisted wheat breeding: present status and future possibilities. Mol. Breed. 26, 145–161. doi: 10.1007/s11032-009-9359-7
Guzmán, C., Crossa, J., Mondal, S., Govindan, V., Huerta, J., Crespo-Herrera, L., et al. (2022). Effects of glutenins (Glu-1 and Glu-3) allelic variation on dough properties and bread-making quality of CIMMYT bread wheat breeding lines. Field Crops Res. 284, 108585. doi: 10.1016/j.fcr.2022.108585
Hasan, M. M., Rafii, M. Y., Ismail, M. R., Mahmood, M., Rahim, H. A., Alam, M. A., et al. (2015). Marker-assisted backcrossing: a useful method for rice improvement. Biotechnol. Biotechnol. Equip. 29, 237–254. doi: 10.1080/13102818.2014.995920
Hickey, L. T., Hafeez, A. N., Robinson, H., Jackson, S. A., Leal-Bertioli, S. C., Tester, M., et al. (2019). Breeding crops to feed 10 billion. Nat. Biotechnol. 37, 744–754. doi: 10.1038/s41587-019-0152-9
Hickey, L. T., Wilkinson, P. M., Knight, C. R., Godwin, I. D., Kravchuk, O. Y., Aitken, E. A., et al. (2012). Rapid phenotyping for adult-plant resistance to stripe rust in wheat. Plant Breed. 131, 54–61. doi: 10.1111/j.1439-0523.2011.01925.x
Igrejas, G., Branlard, G. (2020). “The importance of wheat,” in Wheat quality for improving processing and human health, Springer, Cham. 1–7. doi: 10.1007/978-3-030-34163-3
Jang, Y.-R., Beom, H.-R., Altenbach, S. B., Lee, M.-K., Lim, S.-H., Lee, J.-Y. (2017). Improved method for reliable HMW-GS identification by RP-HPLC and SDS-PAGE in common wheat cultivars. Molecules 22, 1055. doi: 10.3390/molecules22071055
Jia, M., Xu, H., Liu, C., Mao, R., Li, H., Liu, J., et al. (2020). Characterization of the powdery mildew resistance gene in the elite wheat cultivar Jimai 23 and its application in marker-assisted selection. Front. Genet. 11, 241. doi: 10.3389/fgene.2020.00241
Kang, M., Lee, C., Park, D., Ku, B., Park, K., Ko, J., et al. (2006). A new white wheat cultivar, “Jokyoung” for bread making. Korean J. Breed. 38, 139–140.
Kang, J.-W., Shin, D., Cho, J.-H., Lee, J.-Y., Kwon, Y., Park, D.-S., et al. (2019). Accelerated development of rice stripe virus-resistant, near-isogenic rice lines through marker-assisted backcrossing. PloS One 14, e0225974. doi: 10.1371/journal.pone.0225974
Kaur, B., Mavi, G., Gill, M. S., Saini, D. K. (2020). Utilization of KASP technology for wheat improvement. Cereal Res. Commun. 48, 409–421. doi: 10.1007/s42976-020-00057-6
Kiszonas, A. M., Morris, C. F. (2018). Wheat breeding for quality: A historical review. Cereal Chem. 95, 17–34. doi: 10.1094/CCHEM-05-17-0103-FI
Kroupina, A. Y., Yanovsky, A. S., Korobkova, V. A., Bespalova, L. A., Arkhipov, A. V., Bukreeva, G. I., et al. (2023). Allelic variation of Glu-A1 and Glu-B1 genes in winter durum wheat and its effect on quality parameters. Foods 12, 1436. doi: 10.3390/foods12071436
Maccaferri, M., Martina, B., Roberto, T. (2022). “Sequence-based marekr assisted selection in wheat,” in Wheat improvement: food security in a changing climate (Cham: Springer International Publishing), 513–538. doi: 10.1007/978-3-030-90673-3
McCallum, B. D., DePauw, R. M. (2008). A review of wheat cultivars grown in the Canadian prairies. Can. J. Plant Sci. 88, 649–677. doi: 10.4141/CJPS07159
Pandey, S., Singh, A., Parida, S. K., Prasad, M. (2022). Combining speed breeding with traditional and genomics-assisted breeding for crop improvement. Plant Breed. 141, 301–313. doi: 10.1111/pbr.13012
Payne, P. I., Nightingale, M. A., Krattiger, A. F., Holt, L. M. (1987). The relationship between HMW glutenin subunit composition and the bread-making quality of British-grown wheat varieties. J. Sci. Food Agric. 40, 51–65. doi: 10.1002/jsfa.2740400108
Rai, N., Bellundagi, A., Kumar, P. K., Kalasapura Thimmappa, R., Rani, S., Sinha, N., et al. (2018). Marker-assisted backcross breeding for improvement of drought tolerance in bread wheat (Triticum aestivum L. em Thell). Plant Breed. 137, 514–526. doi: 10.1111/pbr.12605
Randhawa, M. S., Bains, N. S., Sohu, V. S., Chhuneja, P., Trethowan, R. M., Bariana, H. S., et al. (2019). Marker assisted transfer of stripe rust and stem rust resistance genes into four wheat cultivars. Agronomy 9, 497. doi: 10.3390/agronomy9090497
Rasheed, A., Wen, W., Gao, F., Zhai, S., Jin, H., Liu, J., et al. (2016). Development and validation of KASP assays for genes underpinning key economic traits in bread wheat. Theor. Appl. Genet. 129, 1843–1860. doi: 10.1007/s00122-016-2743-x
RDA (2012). Manual for standard evaluation method in agricultural experiment and research (Suwon: RDA Press)
Riaz, A., Periyannan, S., Aitken, E., Hickey, L. (2016). A rapid phenotyping method for adult plant resistance to leaf rust in wheat. Plant Methods 12, 1–10. doi: 10.1186/s13007-016-0117-7
Samantara, K., Bohra, A., Mohapatra, S. R., Prihatini, R., Asibe, F., Singh, L., et al. (2022). Breeding more crops in less time: A perspective on speed breeding. Biology 11, 275. doi: 10.3390/biology11020275
Shewry, P., Hey, S. (2015). The contribution of wheat to human diet and health. Food Energy Secur 4, 178–202. doi: 10.1002/fes3.64
Shin, D., Cha, J.-K., Lee, S.-M., Kabange, N. R., Lee, J.-H. (2020). Rapid and easy high-molecular-weight glutenin subunit identification system by lab-on-a-chip in wheat (Triticum aestivum L.). Plants 9, 1517. doi: 10.3390/plants9111517
Varshney, R. K., Barmukh, R., Roorkiwal, M., Qi, Y., Kholova, J., Tuberosa, R., et al. (2021). Breeding custom-designed crops for improved drought adaptation. Advanced Genet. 2, e202100017. doi: 10.1002/ggn2.202100017
Varshney, R. K., Sinha, P., Singh, V. K., Kumar, A., Zhang, Q., Bennetzen, J. L. (2020). 5Gs for crop genetic improvement. Curr. Opin. Plant Biol. 56, 190–196. doi: 10.1016/j.pbi.2019.12.004
Vishwakarma, M. K., Mishra, V., Gupta, P., Yadav, P., Kumar, H., Joshi, A. K. (2014). Introgression of the high grain protein gene Gpc-B1 in an elite wheat variety of Indo-Gangetic Plains through marker assisted backcross breeding. Curr. Plant Biol. 1, 60–67. doi: 10.1016/j.cpb.2014.09.003
Voss-Fels, K. P., Herzog, E., Dreisigacker, S., Sukumaran, S., Watson, A., Frisch, M., et al. (2019). ““SpeedGS” to accelerate genetic gain in spring wheat,” in Applications of genetic and genomic research in cereals (Elsevier), Woolhead publishing, 303–327
Watson, A., Ghosh, S., Williams, M. J., Cuddy, W. S., Simmonds, J., Rey, M.-D., et al. (2018). Speed breeding is a powerful tool to accelerate crop research and breeding. Nat. Plants 4, 23–29. doi: 10.1038/s41477-017-0083-8
Wilkinson, P. A., Winfield, M. O., Barker, G. L., Allen, A. M., Burridge, A., Coghill, J. A., et al. (2012). CerealsDB 2.0: an integrated resource for plant breeders and scientists. BMC Bioinf. 13, 1–6. doi: 10.1186/1471-2105-13-219
Xu, Q., Xu, J., Liu, C., Chang, C., Wang, C., You, M., et al. (2008). PCR-based markers for identification of HMW-GS at Glu-B1x loci in common wheat. J. Cereal Sci. 47, 394–398. doi: 10.1016/j.jcs.2007.05.002
Yang, G., Chen, S., Chen, L., Sun, K., Huang, C., Zhou, D., et al. (2019). Development of a core SNP arrays based on the KASP method for molecular breeding of rice. Rice 12, 1–18. doi: 10.1186/s12284-019-0272-3
Keywords: wheat, speed breeding, speed vernalization, marker-assisted backcrossing, molecular breeding, Glu-B1i
Citation: Cha J-K, Park H, Kwon Y, Lee S-M, Jang S-G, Kwon S-W and Lee J-H (2024) Synergizing breeding strategies via combining speed breeding, phenotypic selection, and marker-assisted backcrossing for the introgression of Glu-B1i in wheat. Front. Plant Sci. 15:1402709. doi: 10.3389/fpls.2024.1402709
Received: 18 March 2024; Accepted: 16 May 2024;
Published: 28 May 2024.
Edited by:
Sergio J. Ochatt, INRA UMR1347 Agroécologie, FranceReviewed by:
Roberto Tuberosa, University of Bologna, ItalyCopyright © 2024 Cha, Park, Kwon, Lee, Jang, Kwon and Lee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jong-Hee Lee, Y2NyaWxqaEBrb3JlYS5rcg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.