
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 12 July 2024
Sec. Plant Breeding
Volume 15 - 2024 | https://doi.org/10.3389/fpls.2024.1402368
 Vignesh Mohanavel1
Vignesh Mohanavel1 Valarmathi Muthu1
Valarmathi Muthu1 Rohit Kambale1
Rohit Kambale1 Rakshana Palaniswamy1
Rakshana Palaniswamy1 Prisca Seeli2
Prisca Seeli2 Bharathi Ayyenar1
Bharathi Ayyenar1 Veeraranjani Rajagopalan1
Veeraranjani Rajagopalan1 Sudha Manickam1
Sudha Manickam1 Raghu Rajasekaran1
Raghu Rajasekaran1 Hifzur Rahman1,3
Hifzur Rahman1,3 Jagadeeshselvam Nallathambi1
Jagadeeshselvam Nallathambi1 Manonmani Swaminathan2
Manonmani Swaminathan2 Gopalakrishnan Chellappan2
Gopalakrishnan Chellappan2 Geethalakshmi Vellingiri4*
Geethalakshmi Vellingiri4* Raveendran Muthurajan1*
Raveendran Muthurajan1*Introduction: Rice, one of the major staple food crops is frequently affected by various biotic/abiotic stresses including drought, salinity, submergence, heat, Bacterial leaf blight, Brown plant hopper, Gall midge, Stem borer, Leaf folder etc. Sustained increase of yield growth is highly necessary to meet the projected demand in rice production during the year 2050. Hence, development of high yielding and multiple stress tolerant rice varieties adapted to wider environments will serve the need.
Methods: A systematic MAB approach was followed to pyramid eight major QTLs/genes controlling tolerance to major abiotic/biotic stresses viz., drought (qDTY1.1 and qDTY2.1), salinity (Saltol), submergence (Sub1), bacterial leaf blight (xa13 and Xa21), blast (Pi9) and gall midge (Gm4) in the genetic background of an elite rice culture CBMAS 14065 possessing high yield and desirable grain quality traits. Two advanced backcross derivatives of CBMAS 14065 possessing different combinations of target QTLs namely #27-1-39 (qDTY1.1+qDTY2.1+Sub1+xa13+Xa21+Gm4+Pi9) and #29-2-2 (qDTY1.1+qDTY2.1+Saltol+Xa21+Gm4+Pi9) were inter-mated.
Results: Inter-mated F1 progenies harboring all the eight target QTLs/genes were identified through foreground selection. Genotyping of the inter-mated F4 population identified 14 progenies possessing all eight target QTLs/genes under homozygous conditions. All the fourteen progenies were forwarded up to F8 generation and evaluated for their yield and tolerance to dehydration, salinity, submergence, blast and bacterial leaf blight. All the 14 progenies exhibited enhanced tolerance to dehydration and salinity stresses by registering lesser reduction in their chlorophyll content, relative water content, root length, root biomass etc., against their recurrent parent Improved White Ponni/CBMAS 14065. All the 14 progenies harboring Sub1 loci from FR13A exhibited enhanced survival (90 - 95%) under 2 weeks of submergence /flooding when compared to their recurrent parent CBMAS 14065 which showed 100% susceptibility The inter-mated population showed a enhanced level of resistance to bacterial leaf blight (Score = 0 to 2) against blast (Score – 0) whereas the susceptible check CO 39 and the recurrent parent CBMAS 14065 recorded high level of susceptibility (Score = 7 to 9).
Conclusion or discussion: Our study demonstrated the accelerated development of multiple stress tolerant rice genotypes through marker assisted pyramiding of target QTLs/genes using tightly linked markers. These multiple stress tolerant rice lines will serve as excellent genetic stocks for field testing/variety release and also as parental lines in future breeding programs for developing climate resilient super rice varieties.
Rice is a major cereal food crop to more than half of the global population (Khush and Jena, 2009; Roser et al., 2013), and rice production must be doubled by 2050 to meet the demand of the ever-growing global population (Yamori, 2013; Pandey et al., 2017). Current yield trends are insufficient to meet this demand, and declining land, water, labor, and nutrient resources pose further threats on global food security (Ray et al., 2013; Pradhan et al., 2019). Hence, increasing rice productivity is a key for achieving global food security.
Rice is cultivated in a wider range of environments and exposed to various abiotic and biotic stresses affecting yield (Pandey et al., 2017; Yugander et al., 2018). The changing climate aggravates the situation by increasing the occurrence of extreme weather events and changing the dynamics of pest and diseases. Abiotic stresses frequently affecting rice yields include drought, salinity, and submergence (Joneydi, 2012). Biotic stresses affecting rice yields include pests (brown plant hopper, leaf folder, stem borer, gall midge, etc.), pathogens (blast, bacterial leaf blight, false smut, etc.), and weeds. Crop losses due to major biotic stressors such as bacterial leaf blight, blast, and brown plant hopper are quite high (Hasan et al., 2015). In India, the area vulnerable to drought has increased by 57% since 1997, and the frequency of heavy rainfall and flash floods has increased by more than 80% since 2012 (The World Bank. 2023). Hence, a sustained increase in rice production depends on the development of resilient rice varieties tolerant to these biotic/abiotic stresses as well as requiring less labor, water, and nutrient resources.
Rice scientists persist in integrating crucial yield trait characteristics, including the 2Gs for the yield ceiling, by employing advanced breeding techniques, such as marker-assisted selection, haplotype-based breeding, and allele mining. The close proximity of the GW7 gene for grain weight and the DEP2 gene for grain number on chromosome 7 indicates the potential for simultaneous introgression of these two genes, provided that advantageous allelic variants exist in a single donor parent (Singh et al., 2024). Developing a multiple-stress-tolerant rice cultivar through conventional breeding approaches is practically very difficult. In conventional breeding, linkage drag continues to several generations and thus requires several backcross generations to eliminate undesirable traits. The discovery of major effect QTLs/genes controlling tolerance to major biotic/abiotic stresses, viz., drought, salinity, submergence, high temperature, bacterial leaf blight, blast, brown plant hopper, gall midge, low phosphorus, etc. (Supplementary Table S1) and advances in marker genotyping have paved the way for pyramiding of multiple QTLs/genes in a single genetic background through marker-assisted breeding (Janaki Ramayya et al., 2021).
In case of drought tolerance, qDTY1.1 and qDTY2.1 from a Philippine upland rice, Apo, have been used by several researchers to develop drought-tolerant versions of popular rice varieties (Venuprasad et al., 2009; Vikram et al., 2011; Muthu et al., 2020). Despite the existence of limited genetic variation for salinity tolerance traits in rice, intensive research efforts have identified a rice genotype, Pokkali, exhibiting a high level of tolerance to coastal salinity in India. Using Pokkali, a consistent QTL “Saltol” regulating Na/K uptake was identified and used by several researchers for the genetic improvement of salinity tolerance in rice (Singh et al., 2018). The discovery and utilization of a flood-tolerant rice “FR13A” discovered a mega effect of QTL “Sub1” regulating submergence tolerance in rice, which paved the way for deployment of MABB in rice (Neeraja et al., 2007).
Among the biotic stresses, blast is one of the most serious diseases causing yield reduction by up to 50% (Qu et al., 2006). Among >500 QTLs linked to blast resistance, only ~30 genes have been cloned and characterized (Das and Rao, 2015; Wu et al., 2016; Zheng et al., 2016). Out of this, Pi9 and Pi54 are the major genes offering broad-spectrum resistance against diverse isolates of M. oryzae (Qu et al., 2006). Against bacterial leaf blight (BLB) capable of causing 20%–40% yield reduction (Yasmin et al., 2017), 43 resistant genes have been identified (Supplementary Table S1). The combination of two or more resistant genes was found to be effective against prevailing strains of bacterial leaf blight in different locations of India. Pyramiding of xa13+ Xa21, either alone or in combination with other genes, viz., Xa2, Xa5, or Xa4, was proven effective in providing durable resistance to bacterial leaf blight. Gall midge is fast becoming a serious pest of rice in India, causing huge yield reduction. Several gall midge resistance genes have been identified (Gm1, Gm2, gm3, Gm4, Gm5, Gm6, Gm7, Gm8, Gm9, Gm10, and Gm11) and used in breeding rice against different biotypes of gall midge. Out of >10 Gm resistance genes, Gm4 was reported to be effective against major biotypes of Gm (Divya et al., 2018).
Developing “climate-smart rice” involves the development of rice varieties that can endure increasingly severe and harsh circumstances in the future (Reay and Reay, 2019). It is anticipated that climate-smart rice will contribute to a sustainable rise in rice output under environmental constraints that regularly arise in a single growing season (Bin and Zhang, 2023). Molecular marker techniques are more effective for transferring desired gene(s) in any combination(s) into any desired genetic backgrounds. Hence, molecular techniques offer novel prospects for the enhancement of rice yields amid changing pest/disease/abiotic stress conditions. Molecular breeders have developed many high-yielding rice varieties with resistance against several abiotic and biotic stress tolerances (Supplementary Table S2). Multiple-stress-tolerant rice varieties can provide yield security in areas subjected to multiple stresses in a single season. However, there are only limited varieties with more than two tolerance/resistance traits (Dixit et al., 2020).
In this study, we attempted to pyramid eight different QTLs/genes conferring tolerance/resistance against drought (qDTY1.1 and qDTY2.1), salinity (Saltol), submergence (Sub1), blast (Pi9), bacterial blight (xa13 and Xa21), and gall midge (Gm4) in an elite genetic stock CBMAS 14065.
A high-yielding, semi-dwarf, medium-duration, and medium slender grain rice culture CBMAS 14065 (derivative of a cross between the mega variety of South India, namely, Improved White Ponni and a drought-tolerant indica genotype Apo) was selected as the recurrent parent (Muthu et al., 2020). CBMAS 14065 harbors two major drought-tolerant QTLs (qDTY1.1 and qDTY2.1) of Apo but exhibited susceptibility to salinity, submergence, bacterial leaf blight, and blast. Two intermittent genetic stocks (BC2F1s) of CBMAS 14065—namely: (1) CBMAS 14065 (27–1-39) harboring qDTY1.1, qDTY2.1, Sub1, xa13, Xa21, Pi9, and Gm4 and (2) CBMAS 14065 (29–2-2) harboring qDTY1.1, qDTY2.1, Saltol, Xa21, Pi9, and Gm4—were used as parents for marker-assisted pyramiding strategy.
A total of 75 markers located in the vicinity of the eight different target QTLs/genes, viz., qDTY1.1, qDTY2.1, Saltol, Sub1, xa13, Xa21, Pi9, and Gm4, were surveyed for genetic polymorphism between the respective donor parents and recurrent parent CBMAS 14065 (Table 1). Genomic DNA was extracted from leaf samples of all of the inter-mated progenies following the modified CTAB protocol (Tiwari et al., 2017). DNA quality was checked using agarose gel electrophoresis and quantified by using a Nanodrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, USA). PCR reaction was carried out in a total volume of 10 μL containing 100 ng of DNA template, 6 μL of master mix (10x Taq Buffer, dNTPs, and Taq polymerase), and 0.5 μL each of 10 μM forward primer and reverse primers. All PCR reactions were amplified following the temperature profiles of initial denaturation at 94°C for 4 min, followed by 37 cycles of 94°C for 1 min, annealing at 55°C–60°C for 30 s, extension at 72°C for 1 min, and followed by a final extension at 72°C for 10 min using a thermal cycler (C1000 TOUCH PCR, Bio-Rad Inc., USA). PCR products were resolved in a 3% agarose gel stained with ethidium bromide, and the gel images were captured in a gel documentation system (Bio-Rad, USA).
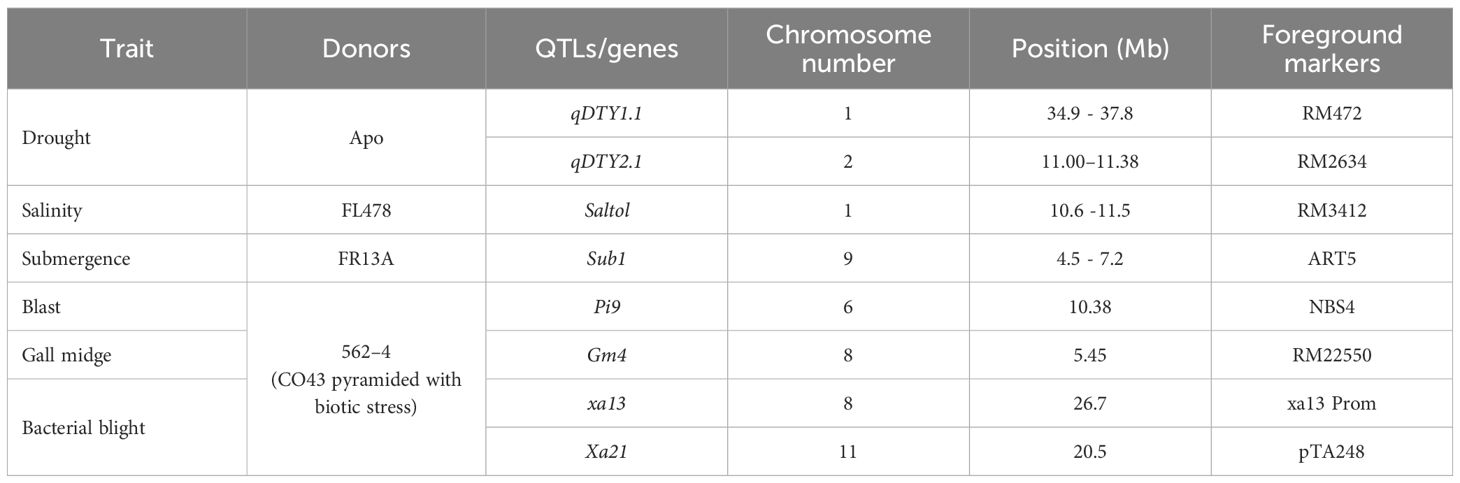
Table 1 Details on the target traits, donors, QTLs/genes, linked markers, and chromosomal location(s).
Two intermittent genetic stocks were inter-mated, and F1 progenies harboring the entire eight target QTLs/genes were identified using foreground selection (Figure 1). True F1s were forwarded to further generations and positive progenies were identified through foreground selection.
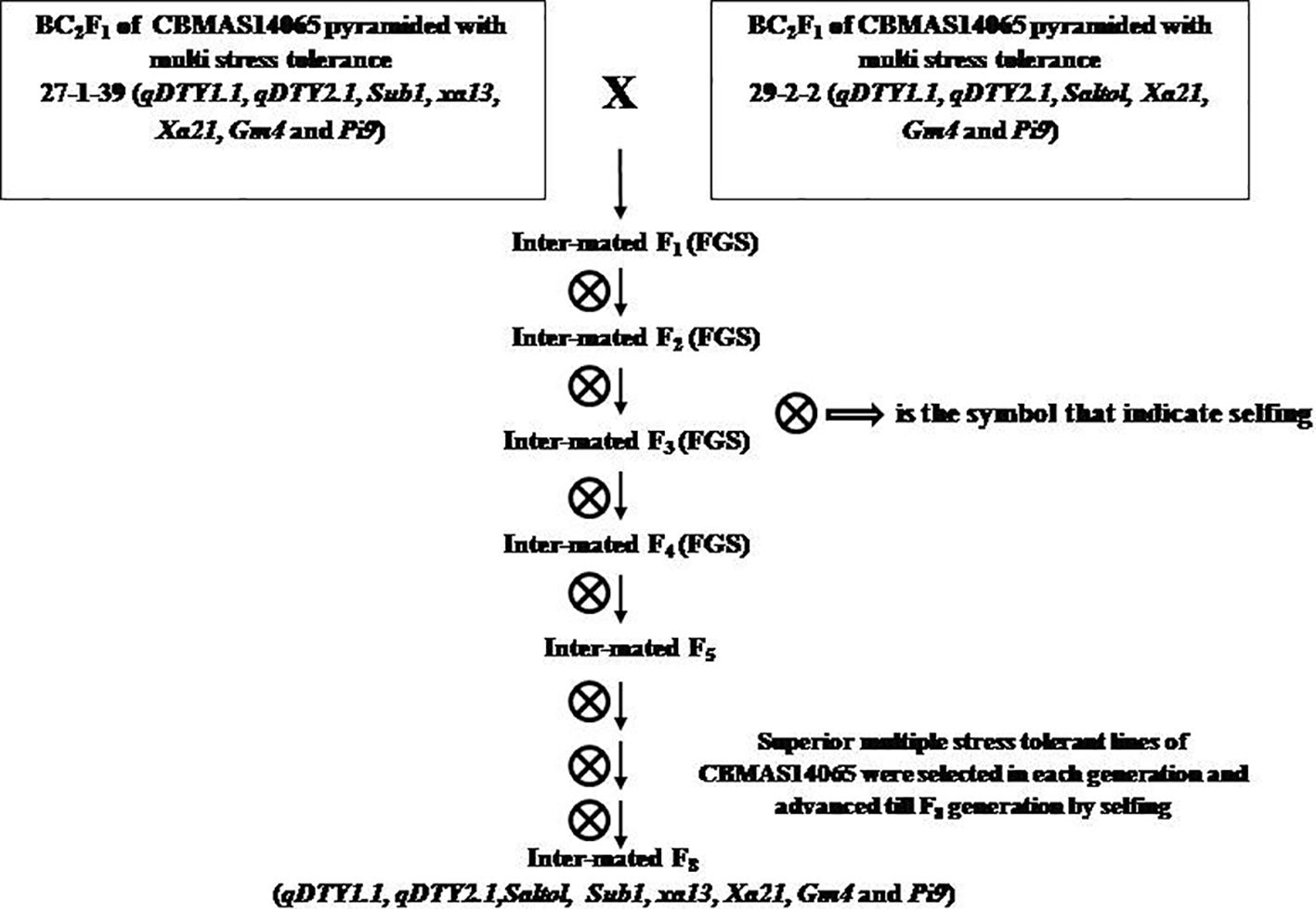
Figure 1 Breeding strategy followed for pyramiding 8 different QTLs/genes controlling tolerance against drought, salinity, submergence, blast, Bacterial leaf Blight and Gall midge in the genetic background of CBMAS14065.
The 14 IMF7 lines were evaluated for their responses to blast pathogen along with the recurrent parent, donor parent (# 562–4), and susceptible check CO 39 under uniform blast nursery at Department of Rice, Tamil Nadu Agricultural University, Coimbatore, India. Each entry was raised on a nursery bed with a row length of 50 cm and row spacing of 10 cm. For uniform transmission of disease after inoculation, one row of the susceptible check (CO 39) was planted between every four entries and along the borders of the nursery bed. Young seedlings at the four-leaf stage were inoculated with fungal spores at a concentration of 1 × 105 spores per milliliter using a fine nozzle sprayer, and high relative humidity was maintained in the nursery to favor disease development. The disease reaction in each progeny was recorded 35 days after sowing and periodically every 7 days until the susceptible check showed more than 80% of infection. Scoring of the disease reaction was done visually on a 0–9 SES scale according to the IRRI’s Standard Evaluation (SES) System (Supplementary Table S3).
Inter-mated F7 progenies (IMF7 progenies) carrying bacterial blight resistance genes (xa13 and Xa21) were grown in pots along with the donor and recurrent parents and subjected to artificial clip inoculation as described earlier (Kauffman, 1973). The bacterial leaf blight pathogen, Xanthomonas oryzae pv. oryzae (Xoo), was inoculated at a concentration of 109 CFU/mL onto the rice plants at the maximum tillering stage (50–55 days). A total of 12–15 fully developed leaves from four plants in each progeny were selected and inoculated by artificial clipping procedure. The disease reaction was assessed at 21 days after inoculation as outlined in a previous study (Ramalingam et al., 2017). Lesion length was measured from the cut tip of the inoculated leaves and scored on a 0–9 SES scale (Supplementary Table S4). The scores of five infected leaves from each progeny were averaged to get the mean score.
A total of 14 progenies were raised under hydroponic conditions (Yoshida, 1976) along with the recurrent parent CBMAS 14065 and donor FL 478. A total of five plants were maintained for each entry. The pH of the Yoshida solution was monitored every day and maintained between 4 and 4.5. Then, 21-day-old seedlings were subjected to salinity stress by adding NaCl into the Yoshida solution at two different concentrations (75 and 100 mM), and a set of respective control plants was maintained without any stress. Visual symptoms of salt injury were measured at 21 days after salinization following the modified Standard Evaluation Scoring (SES, 1996) system (Sultana et al., 2022) (Supplementary Table S5). Phenotypic observations, such as leaf rolling, shoot length, root length, biomass, relative water content (RWC), and drying symptoms, were scored. The scores of five plants were averaged to get the mean score for each progeny.
A total of 14 inter-mated F7 progenies were raised under hydroponic conditions along with CBMAS 14065, Improved White Ponni, and Apo as described earlier. Dehydration stress was imposed on 21-day-old seedlings using PEG 6000 at two different concentrations, viz., 15% and 20% (Nahar et al., 2018), and a set of respective control plants was maintained without any stress. Visual symptoms of drought stress were measured at 21 days after the treatment following the modified Standard Evaluation Scoring (SES) system. Phenotypic observations such as leaf tip drying/rolling, shoot length, root length, relative water content (RWC), plant biomass, and dry symptoms were scored for both stressed and control plants. The scores from the five replications were averaged to get the mean score for each progeny.
Seedlings of IMF7 progenies, recurrent parent, and donor parent were raised in pots (three replications). Then, 21-day old seedlings were subjected to submergence stress by immersing the pots in a cement tank (the water level was maintained at a depth of 65 cm) containing water for 14 days. One pot was maintained under non-submerged conditions. Water depth was monitored every day to keep the plants under completely submerged conditions. After 14 days of submergence, the tank was de-submerged, and the plants were allowed to recover under natural conditions. At 14 days after de-submergence, the % plant survival (PPS), relative plant elongation under submergence (RPE), and chlorophyll reduction percentage of the plants were recorded using a SPAD meter (SPAD-502, Konica, Minolta, Germany). Data analysis and submergence scoring were assessed as per the earlier studies (Kaushal et al., 2018; Pradhan et al., 2019).
A total of 14 superior IMF7 progenies were evaluated for their agronomic performance along with the recurrent parent CBMAS 14065 and donor parents. A total of 13 plants per row and six rows per entry in 30 × 30-cm spacing were maintained. Different yield component traits, viz., plant height, days to first flowering, days to 50% flowering, number of tillers, flag leaf length and width, panicle length, 100-seeds weight, and single plant yield were recorded. All statistical analyses were carried out using Microsoft Excel 2021.
Two BC2F1 progenies of CBMAS 14065—namely, #27–1-39 harboring seven different target QTLs/genes [namely, qDTY1.1 and qDTY2.1 (drought tolerance), Sub1 (submergence tolerance), xa13 and Xa21 (bacterial leaf blight resistance), Pi9 (blast resistance), and Gm4 (gall midge resistance)] were crossed with another BC2F1 genetic stock of CBMAS 14065—namely #29–2-2 harboring six different QTLs/genes [namely qDTY1.1 and qDTY2.1 (drought tolerance), Saltol (salinity tolerance), Xa21 (bacterial leaf blight resistance), Pi9 (blast resistance), and Gm4 (gall midge resistance)]. Foreground selection among 51 inter-mated F1s resulted in the identification of one plant #13 having all eight QTLs (qDTY1.1 + qDTY2.1 + saltol + Sub1 + xa13 + Xa21 + Pi9 + Gm4) (Table 2) which was selfed and forwarded further. The foreground selection of 43 F2 progenies identified one progeny—namely, #13-31—harboring five loci under homozygous conditions and three loci under heterozygous conditions and three other progenies harboring four loci under homozygous conditions and with remaining four loci under heterozygous conditions (Table 2).
IMF2 progeny #13–31 harboring five loci under homozygous conditions was forwarded to the next generation. The foreground genotyping of 134 IMF3 progenies identified a total of five IMF3 progenies, viz., 13–31-11, 13–31-24, 13–31-26, 13–31-56, and 13–31-122, harboring seven loci under homozygous conditions and one loci under heterozygous condition in different combinations (Table 2).
An evaluation of 600 IMF4 progenies identified a total of 14 progenies harboring all of the eight target loci under homozygous conditions (Table 3; Figure 2). Upon evaluation of 14 IMF5, a total of 70 superior plants (five progenies from each of the 14 families) were selected based on their agronomic traits and forwarded to IMF6–IMF7 generations.
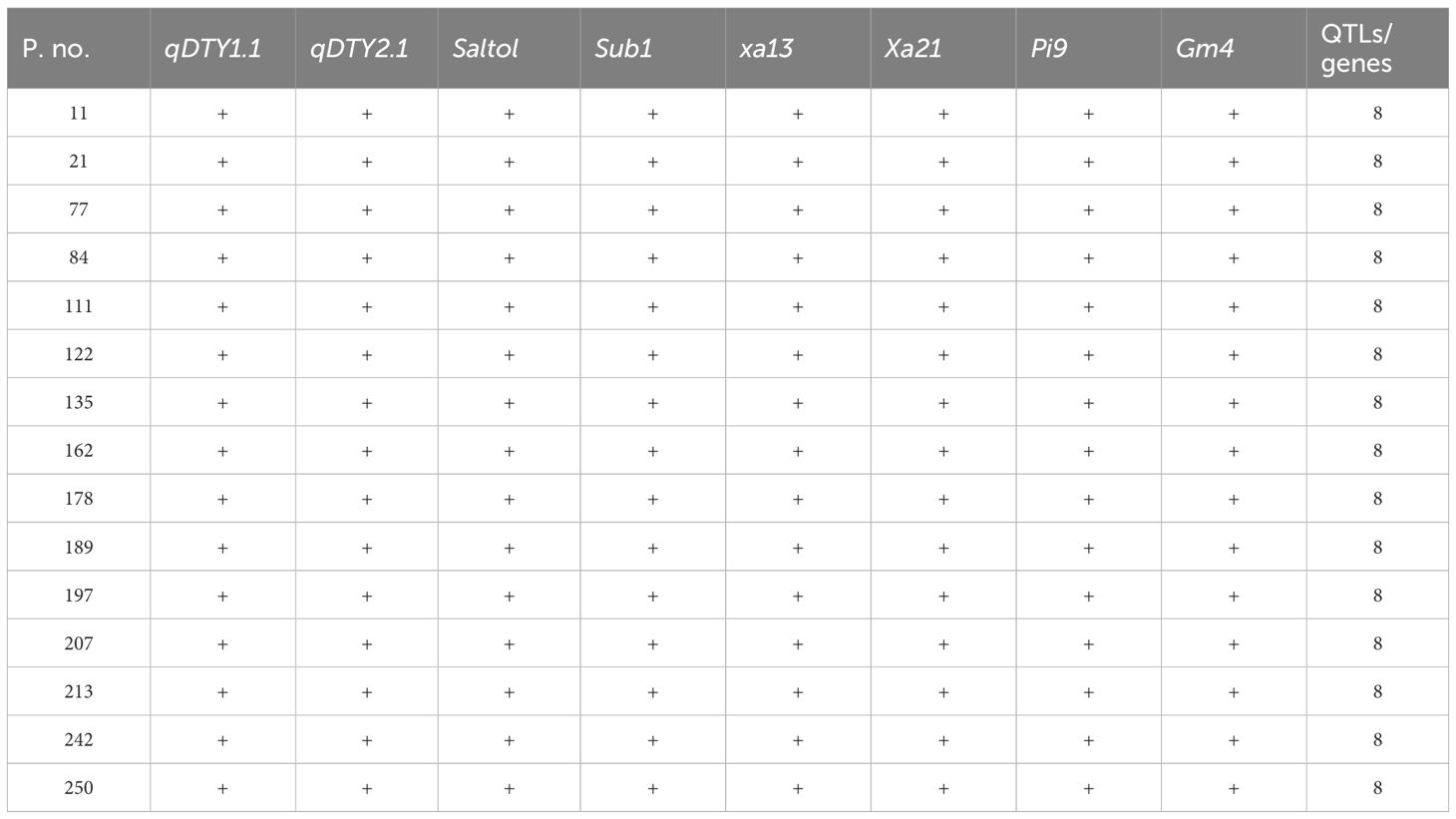
Table 3 Foreground selection of 14 IMF4 progenies of CBMAS 14065 showing the homozygosity of eight different target loci.
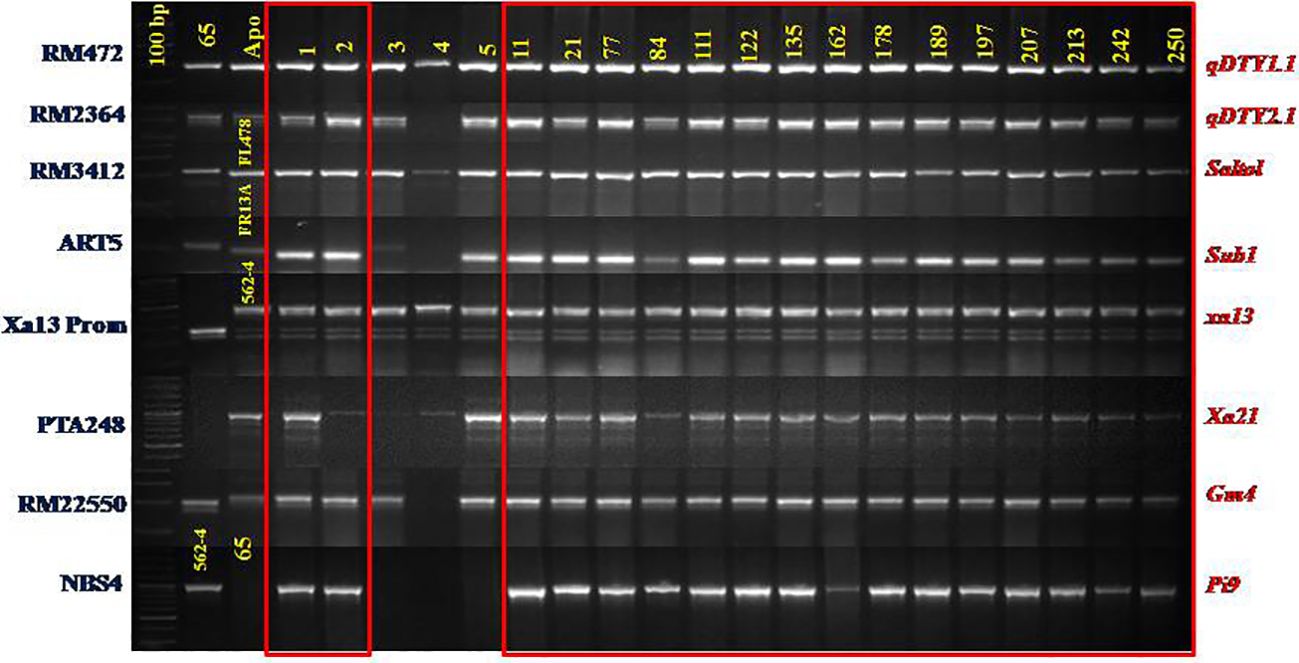
Figure 2 Foreground selection of IMF4 progenies of CBMAS 14065 using markers liked to 8 target QTLs/Genes.
At 21 days after imposing dehydration stress, CBMAS 14065 and all of the 14 inter-mated F7 progenies harboring qDTY1.1 and qDTY2.1 exhibited enhanced tolerance compared to the susceptible check Improved White Ponni. Four progenies (21–2, 122–2, 197–1, and 242–1) exhibited a lesser reduction of root (length and biomass) and shoot (length and biomass) growth-related traits under dehydration (Table 4; Figures 3A, B). The susceptible parent Improved White Ponni showed >25% reduction in its chlorophyll content and relative water content, whereas the tolerant parent Apo, recurrent parent CBMAS 14065, and IMF7 progenies showed a maximum of 15% reduction in their chlorophyll content and relative water content (Figure 3A).
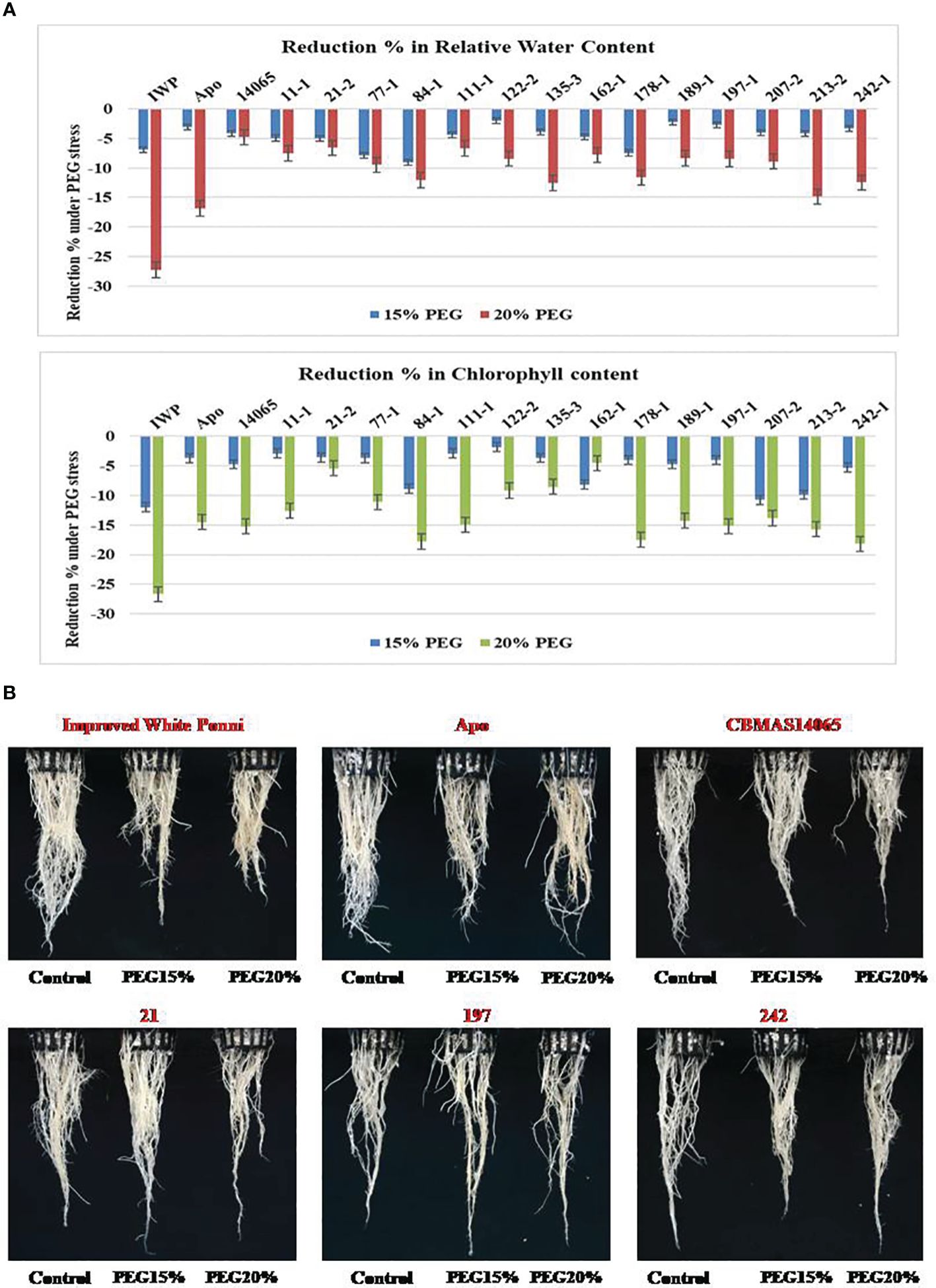
Figure 3 (A) Graphical representation of intermated F7 progenies of CBMAS14065 against drought tolerance using PEG. (B) Root growth behaviour of intermated F7 progenies of CBMAS14065 under dehydration induced by PEG6000 treatment.
Inter-mated F7 progenies of CBMAS 14065 harboring eight target QTLs, including the salinity tolerance QTL, namely, “Saltol”, were grown under hydroponic conditions and subjected to NaCl stress (75 and 100 mM). After 21 days of salinity, scoring was done based on the symptoms of salt injury using a modified standard evaluation system (SES) developed at IRRI, Philippines. The susceptible check IWP and the recurrent parent CBMAS 14065 exhibited more than 40% reduction in their relative water content and >50% reduction in the shoot–root dry biomass (Table 5; Figures 4A–C). Four progenies, viz., #21–2, #122–2, #197–1, and #242–1), exhibited a high level of tolerance to salinity by recording lesser % reduction in terms of chlorophyll content and relative water content.
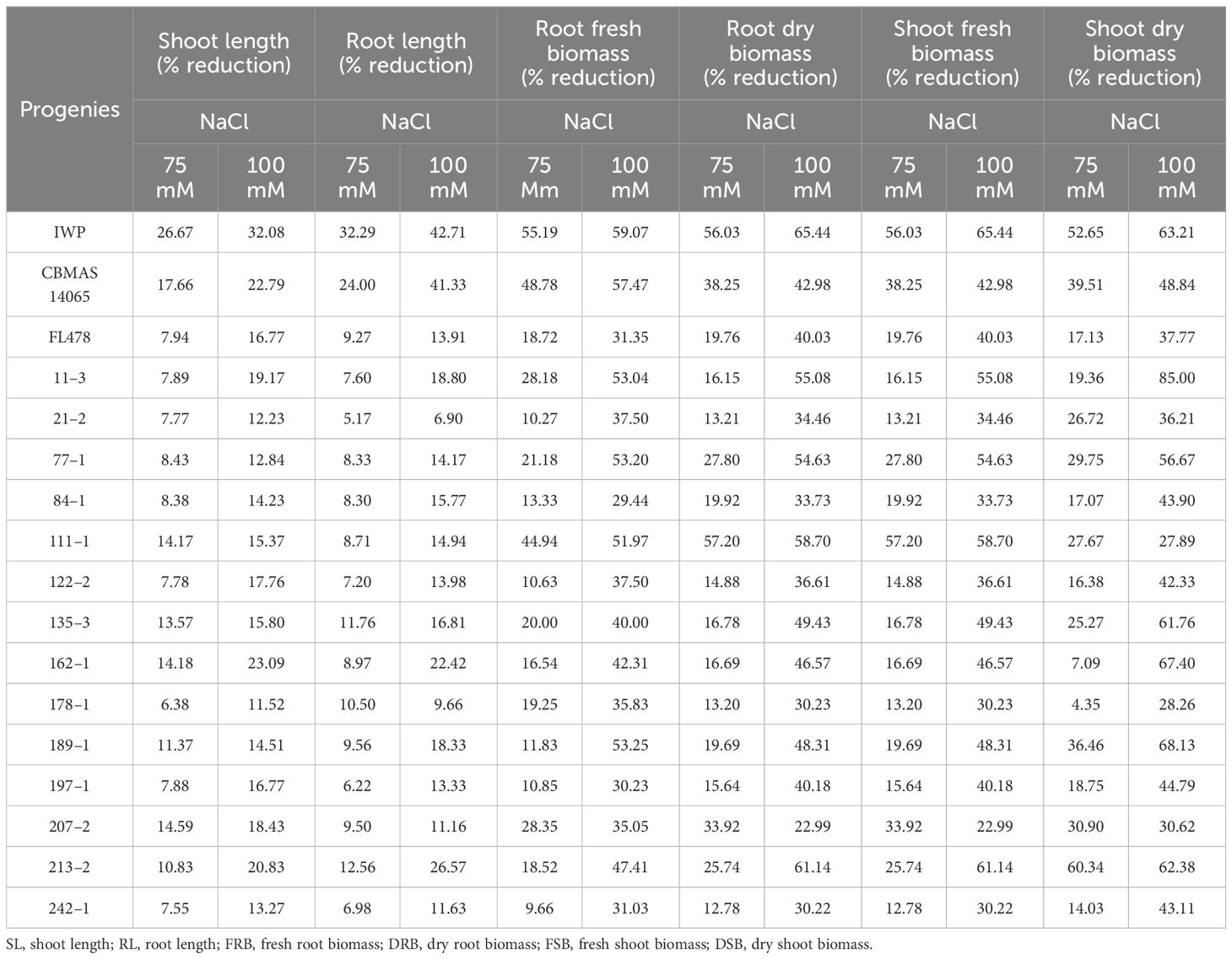
Table 5 Salinity-responsive changes in the shoot/root growth pattern among the inter-mated F7 progenies CBMAS 14065.
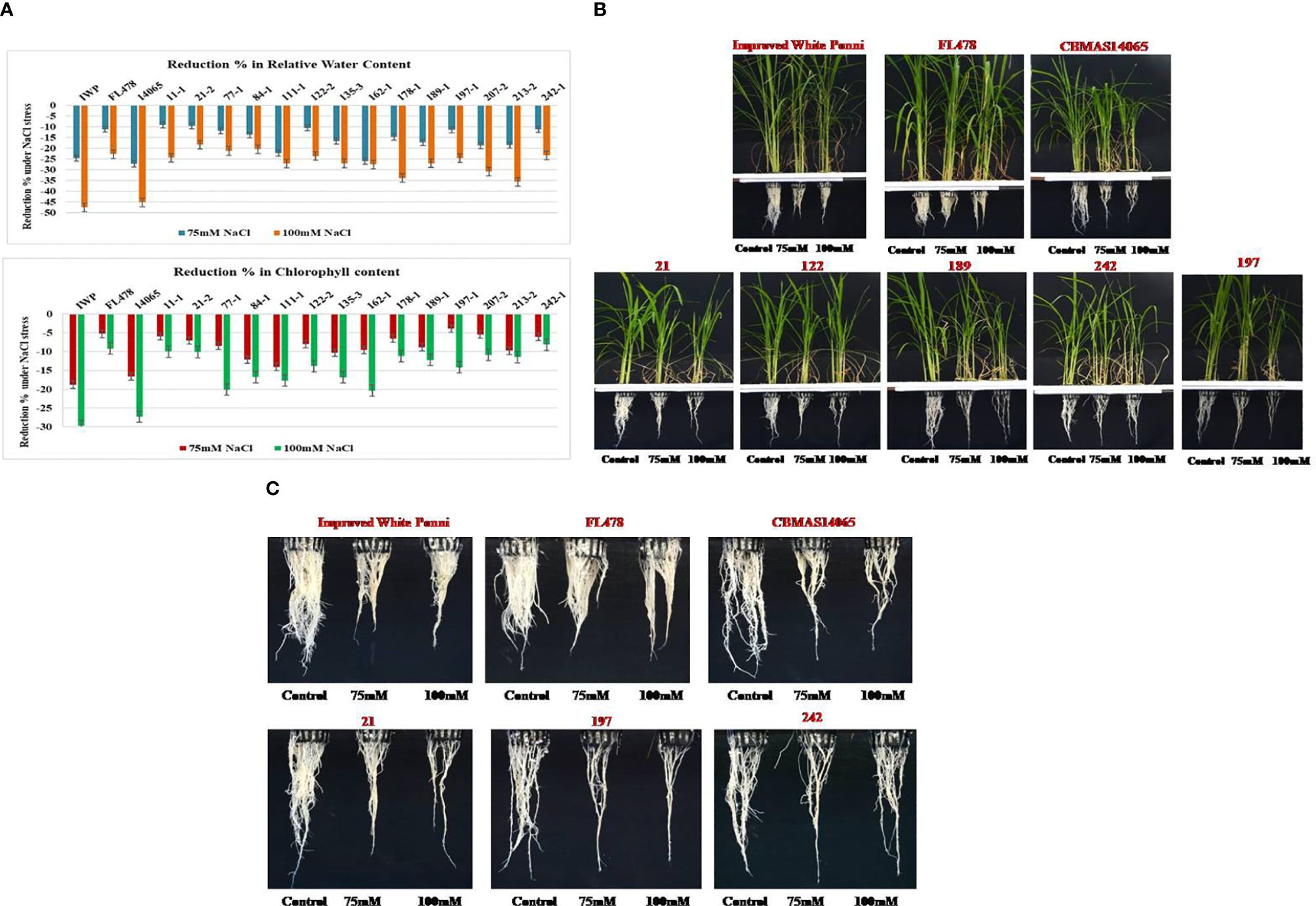
Figure 4 (A) Percent reduction in chlorophyll content and relative water content in the leaves of intermated F7 progenies of CBMAS14065 against NaCI stress. (B) Responses of rice genotypes against salinity stress induced by NaCI (Control-without stress; 75mM and 100mM refers to NaC1 contrentration). (C) Salinity responsive changes in the root growth pattern of inter-mated progenies of CBMAS14065.
The 21-day-old seedlings of 14 inter-mated F7 progenies of CBMAS 14065 grown in pots were exposed to submergence stress for 14 days in cement tanks along with their parents and appropriate checks and then de-submerged. After a stress period of another 21 days, parameters like plant height, number of plants, number of leaves, and chlorophyll content were measured before and after submergence to calculate the regeneration capacity of the progenies. Submergence-stress-tolerant progenies were identified based on their recovery/regeneration percentage. After 14 days of de-submergence, the recurrent parent CBMAS 14065 showed 100% mortality, whereas all of the 14 inter-mated F7 progenies showed enhanced tolerance (90%–100%) against 14 days of submergence, which was on par with the donor parent FR13A (Table 6; Figure 5). Relative plant elongation (RPE) during submergence was high in the recurrent parent CBMAS 14065 (123.42%), whereas the progenies exhibited significantly lesser elongation (80.69% to 113.64%) equal to the donor parent FR13A (Table 6).

Figure 5 Responses of inter-mated F7 progenies against 14 days of submergence stress. Control, no submerge; DAS refers to Days After Submergence; DADS refers to Days after De-Submergence.
All of the 14 inter-mated F7 progenies harboring a broad-spectrum blast resistance gene, Pi9, were screened under uniform blast nursery along with the recurrent parent (CBMAS 14065), donor parent (562–4), and susceptible check (CO 39) during the winter of 2021. All of the 14 inter-mated F7 progenies of CBMAS 14065 exhibited a high level of resistance to blast with a score of “0” comparable to the donor, #562–4. Recurrent parent CBMAS 14065 exhibited a high level of susceptibility with a score of 5 to 7 (Table 7; Figure 6).

Figure 6 Responses of recurrent parent (CBMAS14065), donor (#562-4) and inter-mated F7 progenies against blast pathogen.
The inter-mated F7 progenies of CBMAS 14065 pyramided with eight different target loci, including two bacterial leaf blight resistance loci—namely, xa13 and Xa21—exhibited a high level of resistance (lesion length of 0.3–1.0 cm) similar to that of the donor parent #562–4, whereas the recurrent parent showed high susceptibility with a leaf lesion length of 1.1–5.5 cm (Figures 7A, B).
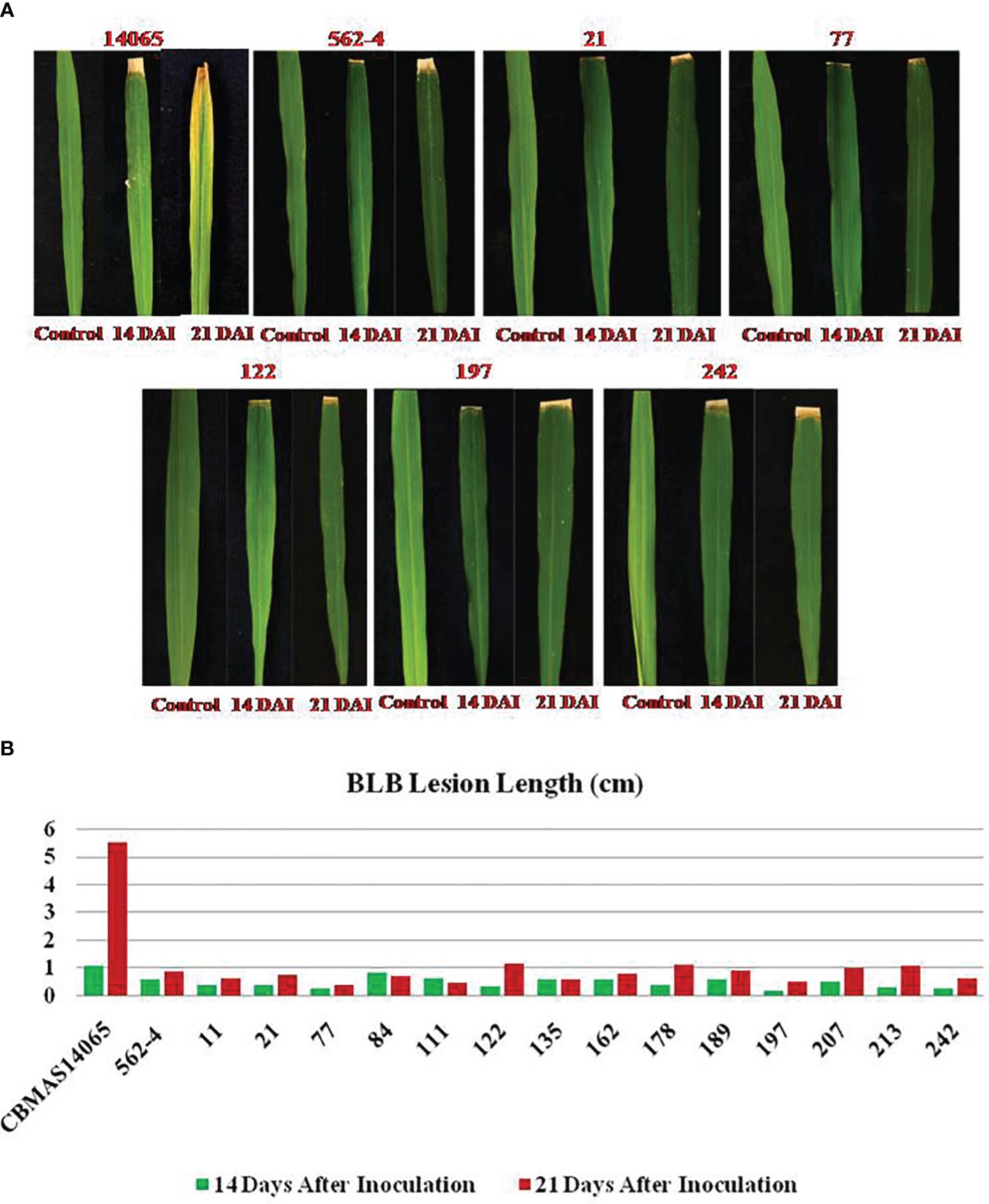
Figure 7 (A) Response of inter-mated of CBMAS14065 harboring xx13 and Xa21 and the parents against BLB infection. (B) Responses of inter-mated F7 progenies of CBMAS14065 harboring xx13 and Xa21 and the parents against BLB infection; Graphical representation of lesion of lesion at 14DAI and 21 DALI.
A total of 14 inter-mated progenies pyramided with eight different target loci were evaluated for their agronomic performance over two seasons (Kharif 2020 and Kharif 2021) along with the recurrent parent CBMAS 14065, Improved White Ponni, and relevant donors. Days to flowering among the 14 inter-mated progenies ranged between 96 to 103 days, while the recurrent parent CBMAS 14065 flowered in 113 days. The inter-mated progenies were found to possess a higher number of productive tillers (32 to 48 tillers) when compared to CBMAS 14065 (30 tillers). Overall, the inter-mated progenies of CBMAS 14065 were found to possess a higher yield potential (60 to 116 g/plant) when compared to CBMAS 14065 (58 g/plant). The 100-grain weight of inter-mated progenies ranged between 1.124 to 1.570 g when compared to 1.249 g recorded by CBMAS 14065. Overall, the pyramided IMF7 lines were found to be 10 days earlier than CBMAS 14065 in duration and 22–42 cm more dwarf than the popular variety Improved White Ponni (Table 8; Figure 8).
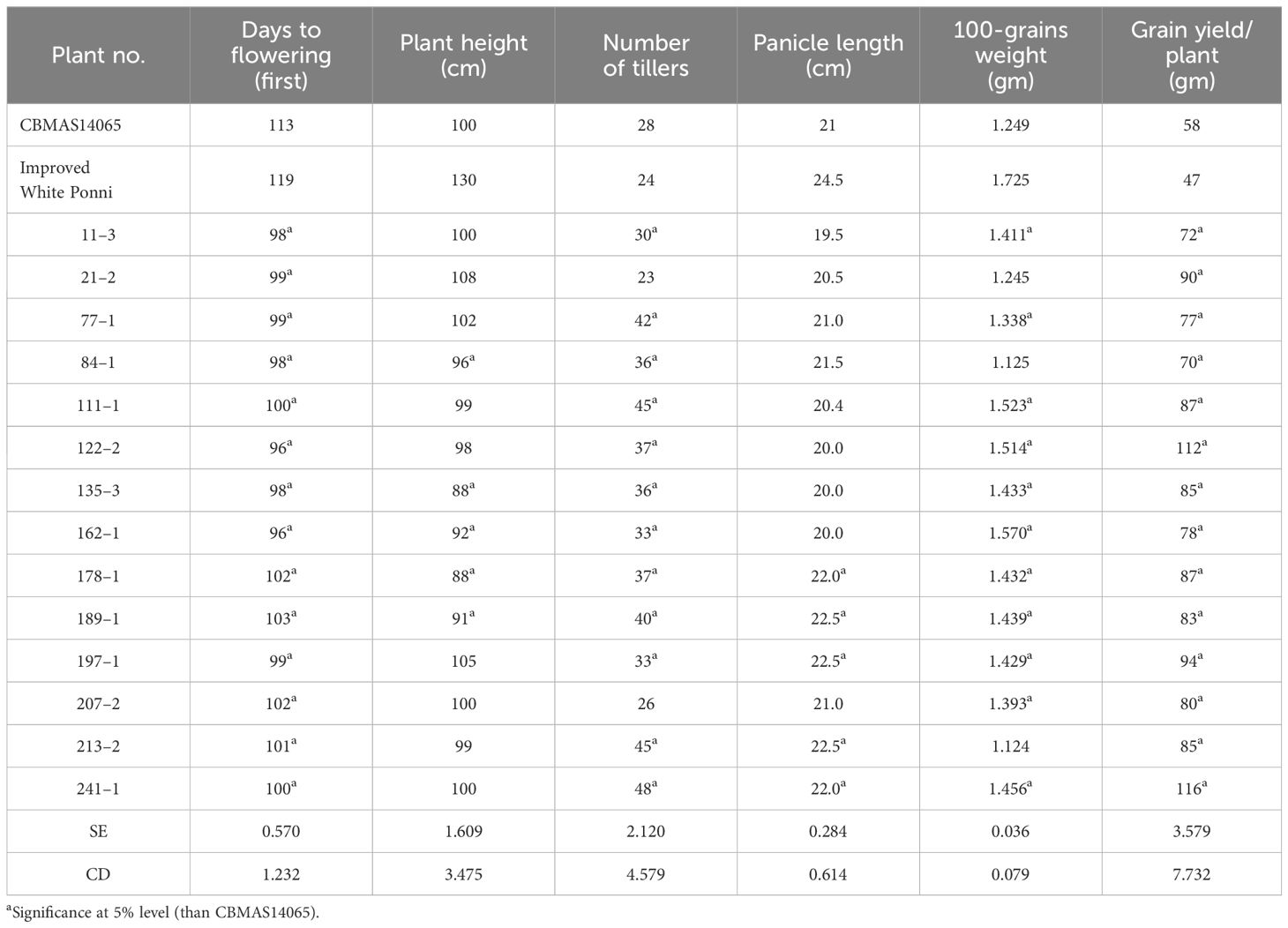
Table 8 Agronomic performance of the 14 inter-mated F7 progenies of CBMAS14065 pyramided with multiple-stress-tolerant QTLs/genes.

Figure 8 Photograph showing the planet morphology and grain characteristics of an IMF7 progeny along with recurrent Parent CBMAS14065.
Most of the rice varieties with high yields are susceptible to both biotic and abiotic stressors. Conventional breeding methods have been utilized effectively to produce numerous crop varieties with improved yield, but conventional breeding faces challenges (time-consuming and labor-intensive process) in pyramiding multiple-component traits/genomic loci conferring tolerance to various biotic/abiotic stresses. Recent advances in genotyping platforms and marker-assisted breeding have provided opportunities to pyramid or mobilize multiple QTLs/genes from several donors simultaneously into a single genetic background (Ramalingam et al., 2017; Das et al., 2018; Kumar et al., 2018; Rana et al., 2019; Valarmathi et al., 2019; Dixit et al., 2020; Muthu et al., 2020; Pancaldi and Trindade, 2020). Similar approaches have been successfully demonstrated in mobilizing QTLs/genes for various biotic (blast, bacterial leaf blight, and gall midge) and abiotic (salinity and submergence) stresses in the genetic background of popular rice varieties, viz., Naveen, Lalat, and Tapaswini (Das and Rao, 2015; Kumar et al., 2018; Janaki Ramayya et al., 2021).
CBMAS 14065 was developed as a semi-dwarf version of Improved White Ponni, and during field testing, this culture was found to exhibit a high level of susceptibility to salinity, submergence, blast, and bacterial leaf blight. Hence, attempts were made to pyramid tolerance to salinity (Saltol), submergence (Sub1), bacterial leaf blight (xa13 and Xa21), blast (Pi9), and gall midge (Gm4) along with the drought tolerance loci qDTY1.1 and qDTY2.1 already present in it. Except for drought and salinity tolerance, gene-based markers were used to monitor the introgression of genetic loci controlling tolerance to submergence, bacterial blight, and blast. Adequate numbers of progenies (~800 to 1,000) were maintained in each generation for the effective selection of targeted gene combinations. In the IMF4 generation, elite lines harboring all of the target genes/QTLs under homozygous conditions were identified and forwarded further through selfing. Under each generation, the presence of target loci was confirmed through foreground selection using trait-linked markers and yield potential.
Drought stress is a major problem affecting rice cultivation under both irrigated and rain-fed conditions, causing serious yield losses to the farmers. The discovery of major-effect QTLs, viz., qDTY1.1, qDTY2.1, qDTY12.1, etc., in Apo, Nagina 22, Adaysel, Way rarem, etc., enabled the deployment of marker-assisted breeding to improve drought tolerance or yield under drought in rice (Vikram et al., 2011; Ghimire et al., 2012; Mishra et al., 2013). In the present study, CBMAS 14065 and the inter-mated F7 population of CBMAS 14065 harboring qDTY1.1 and qDTY2.1 were screened for their tolerance to dehydration using polyethylene glycol (6000). CBMAS 14065 and the inter-mated F7 progenies pyramided with qDTY1.1 and qDTY2.1 recorded lesser reduction in their shoot length, root length, relative water content, root fresh biomass, root dry biomass, shoot fresh biomass, and shoot dry biomass during dehydration as compared to Improved White Ponni. This study has demonstrated the importance of drought-tolerant QTLs (qDTY1.1 and qDTY2.1) in alleviating drought-induced dehydration and other negative impacts similar to earlier reports (Venuprasad et al., 2009; Ghimire et al., 2012; Vikram et al., 2015).
Rice exhibits high sensitivity to salt stress at both vegetative and reproductive phases (Das et al., 2020). Intensive efforts in characterizing the salinity-tolerant Pokkali identified a QTL “Saltol” as regulating the uptake of cationic salts into the roots (Das et al., 2020). Since salinity is a major abiotic stress for rice, in the present study, a Pokkalai-derived RIL—namely, FL478—harboring the salinity-tolerant loci “Saltol” was used as a donor, and the introgression of “Saltol” loci conferred a comparable level of tolerance to salinity in the inter-mated F7 progenies of CBMAS 14065. The inter-mated F7 progenies of CBMAS 14065 and FL 478 recorded a relatively lesser reduction in their plant height and total dry matter accumulation when compared to that of the recurrent parent CBMAS 14065 and Improved White Ponni (Table 5) and the total dry matter production. The above-mentioned results were similar to that of the earlier reports where the Saltol-locus-introgressed lines of popular rice varieties exhibited enhanced tolerance to salinity than their recurrent parents (Valarmathi et al., 2019; Singh et al., 2021).
About 12–14 Mha of rice crop in India is frequently affected by flash-flooding submergence, and the discovery of a major effect QTL “Sub 1” from flood-tolerant FR 13A paved the way to develop submergence-tolerant versions of popular rice varieties (Neeraja et al., 2007; Rahman et al., 2018). In the present study, efforts were made to pyramid the submergence-tolerant loci “Sub 1” from FR13A into CBMAS 14065 along with other biotic/abiotic-stress-tolerant QTLs/genes. All of the inter-mated F7 progenies exhibited 90%–96% of recovery after 14 days of submergence stress, whereas the recurrent parent CBMAS 14065 showed 100% susceptibility (Figure 5). Relative plant elongation percentage (RPE) was found to be high in the recurrent parent (123.42%) and found to be low in the donor parent and the inter-mated F7 progenies (80.69% to 113.84%). The marker-assisted introgression of Sub1 loci was followed to develop several flood-tolerant rice varieties—namely, CR Dhan 801, Bahuguni dhan-1 and Bahugunidhan-2, Swarna, Samba Mahsuri, TDK Sub1, BR 11 Sub1, CR 1009 Sub1, and CO 43 Sub1, which are now extensively cultivated in different parts of India (Septiningsih et al., 2015; Rahman et al., 2018; Sandhu et al., 2019). In a similar manner, the present study has paved the way for the development of submergence-tolerant rice in the genetic background of CBMAS 14065, a derivative of a popular rice variety of South India—namely, Improved White Ponni.
Out of the several blast resistance genes, Pi9 is reported to exhibit broad-spectrum resistance to different pathotypes of the blast fungus (Liu et al., 2002; Luo et al., 2017). Large-scale testing of rice genotypes harboring different blast resistance genes demonstrated that the efficacy of Pi9 was sufficient enough to confer a high level of resistance to the most virulent Eastern Indian pathotype of blast fungus (Imam et al., 2014; Variar et al, 2019). Hence, efforts were made to pyramid Pi9 into CBMAS 14065 along with target QTLs for other biotic/abiotic stresses. The inter-mated progenies of CBMAS 14065 harboring Pi9 locus exhibited a high level of resistance to a local isolate of the blast pathogen (score = 0) as compared to CBMAS 14065 and the susceptible check CO 39 (Score = 7 to 9) (Figure 6). Khanna et al. (2015) had successfully introgressed seven blast genes into Pusa Basmati 1 (PB1), and they discovered that, among the seven genes, the Pi9-carrying inter-mated population showed the highest resistance against the virulent races of blast diseases, particularly in hotspot areas. Furthermore, this study has opened up avenues for pyramiding other blast resistance like Pi54 which will further broaden the genetic base against blast.
The most realistic/practical way of enhancing resistance to bacterial leaf blight pathogen is to combine two or more R genes reported (Mundt et al., 1999). Among the numerous bacterial leaf blight genes reported, the broad-spectrum Xa21, in combination with xa13, demonstrated broad-spectrum resistance to diverse races of bacterial blight pathogen (Pradhan et al., 2016). In this study, IMF7 progenies harboring xa13 and Xa21 showed a high level of resistance to local isolate(s) of bacterial blight infection when compared to the recurrent parent CBMAS 14065 (Figures 7A, B). The pyramided lines showed significantly lesser lesion length (<0.3 cm) when compared to the recurrent parent. Pyramiding of multiple QTLs/genes conferring resistance to various biotic and abiotic stresses through conventional breeding is difficult and consumes a longer time. Recent advances in marker technology, genotyping, and mating designs provided an opportunity to accelerate the development of multiple-stress-tolerant rice with significant reduction in time, space, labor, and money. In the present study, the development of early generation backcross progenies of CBMAS 14065 harboring different stress-tolerant QTLs and the planned inter-mating of backcross progenies combined with precision genotyping and phenotyping demonstrated the accelerated development of climate-resilient rice requiring less chemicals. Further evaluation of these progenies across environments will identify elite lines exhibiting a consistent performance across environments. These rice genotypes will also serve as genetic stocks for further pyramiding of new traits to enhance their performance and climate resilience.
The present study clearly demonstrated the success of MAS in combining tolerance to multiple biotic and abiotic stresses while maintaining a higher yield potential and the preferred grain quality. The developed population containing eight distinct QTLs/genes in the current study demonstrated superiority over recurrent parents for multiple abiotic and biotic stresses (drought, submergence, salinity, blast, bacterial leaf blight, and gall midge) as well as for use as genetic stock in future breeding programs.
The original contributions presented in the study are included in the article / Supplementary Material, Further enquires can be directed to the corresponding Authors.
RM: Conceptualization, Funding acquisition, Project administration, Supervision, Writing – original draft, Writing – review & editing. VMo: Formal analysis, Investigation, Methodology, Writing – original draft. VMu: Investigation, Methodology, Writing – original draft. RK: Data curation, Software, Writing – original draft. RP: Data curation, Investigation, Writing – original draft. PS: Investigation, Validation, Writing – original draft. BA: Resources, Validation, Writing – original draft. VR: Resources, Supervision, Writing – original draft. SM: Project administration, Resources, Supervision, Writing – review & editing. RR: Methodology, Project administration, Resources, Writing – review & editing. HR: Methodology, Writing – review & editing. JN: Software, Validation, Writing – review & editing. MS: Project administration, Resources, Supervision, Writing – review & editing. GC: Resources, Validation, Writing – review & editing. GV: Project administration, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The authors thank the Department of Biotechnology, Government of India, New Delhi, India, for the funding support (BT/PR 13454/COE/34/43/2015).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1402368/full#supplementary-material
Bin, A. R., Zhang, J. (2023). Trends in rice research: 2030 and beyond. Food Energy Secur. 12, e390. doi: 10.1002/fes3.390
Das, G., Rao, G. (2015). Molecular marker assisted gene stacking for biotic and abiotic stress resistance genes in an elite rice cultivar. Front. Plant Sci. 6, 698. doi: 10.3389/fpls.2015.00698
Das, G., Rao, G. J., Varier, M., Prakash, A., Prasad, D. (2018). Improved Tapaswini having four BB resistance genes pyramided with six genes/QTLs, resistance/tolerance to biotic and abiotic stresses in rice. Sci. Rep. 8.
Divya, D., Nair, S., Bentur, J. (2018). Expression profiles of key genes involved in rice gall midge interactions reveal diversity in resistance pathways. Curr. Sci. 115, 74–82. doi: 10.18520/cs/v115/i1/74-82
Das, S., Kumar, A., Barman, M., Pal, S., Bandopadhyay, P. (2020). Impact of climate variability on phenology of rice. Agronomic Crops: Volume 3: Stress Responses Tolerance 13–28. doi: 10.1007/978-981-15-0025-1_2
Dixit, S., Singh, U. M., Singh, A. K., Alam, S., Venkateshwarlu, C., Nachimuthu, V. V., et al. (2020). Marker assisted forward breeding to combine multiple biotic-abiotic stress resistance/tolerance in rice. Rice 13, 1–15. doi: 10.1186/s12284-020-00391-7
Ghimire, K. H., Quiatchon, L. A., Vikram, P., Swamy, B. M., Dixit, S., Ahmed, H., et al. (2012). Identification and mapping of a QTL (qDTY1. 1) with a consistent effect on grain yield under drought. Field Crops Res. 131, 88–96. doi: 10.1016/j.fcr.2012.02.028
Hasan, M. M., Rafii, M. Y., Ismail, M. R., Mahmood, M., Rahim, H. A., Alam, M. A., et al. (2015). Marker-assisted backcrossing: a useful method for rice improvement. Biotechnol. Biotechnol. Equip. 29, 237–254. doi: 10.1080/13102818.2014.995920
Imam, J., Alam, S., Mandal, N. P., Variar, M., Shukla, P. (2014). Molecular screening for identification of blast resistance genes in North East and Eastern Indian rice germplasm (Oryza sativa L.) with PCR based makers. Euphytica 196, 199–211. doi: 10.1007/s10681-013-1024-x
Janaki Ramayya, P., Vinukonda, V. P., Singh, U. M., Alam, S., Venkateshwarlu, C., Vipparla, A. K., et al. (2021). Marker-assisted forward and backcross breeding for improvement of elite Indian rice variety Naveen for multiple biotic and abiotic stress tolerance. PloS One 16, e0256721. doi: 10.1371/journal.pone.0256721
Joneydi, M. S. (2012). Factors affecting in sustainability of agricultural production systems in Iran. Ann. Biol. Res. 3, 4578–4583.
Kauffman, H. (1973). An improved technique for evaluating resistance of rice varieties to Xanthomonas oryzae. Plant Dis. Rep. 57, 537–541.
Kaushal, L., Ulaganathan, K., Shenoy, V., Balachandran, S. (2018). Geno-and phenotyping of submergence tolerance and elongated uppermost internode traits in doubled haploids of rice. Euphytica 214, 1–16. doi: 10.1007/s10681-018-2305-1
Khanna, A., Sharma, V., Ellur, R. K., Shikari, A. B., Gopala Krishnan, S., Singh, U., et al. (2015). Development and evaluation of near-isogenic lines for major blast resistance gene (s) in Basmati rice. Theor. Appl. Genet. 128, 1243–1259. doi: 10.1007/s00122-015-2502-4
Khush, G. S., Jena, K. (2009). Current status and future prospects for research on blast resistance in rice (Oryza sativa L.). Adv. genetics Genomics control Rice blast Dis., 1–10.
Kumar, A., Sandhu, N., Dixit, S., Yadav, S., Swamy, B., Shamsudin, N. A. A. (2018). Marker-assisted selection strategy to pyramid two or more QTLs for quantitative trait-grain yield under drought. Rice 11, 1–16. doi: 10.1186/s12284-018-0227-0
Liu, G., Lu, G., Zeng, L., Wang, G.-L. (2002). Two broad-spectrum blast resistance genes, Pi9 (t) and Pi2 (t), are physically linked on rice chromosome 6. Mol. Genet. Genomics 267, 472–480. doi: 10.1007/s00438-002-0677-2
Luo, W., Huang, M., Guo, T., Xiao, W., Wang, J., Yang, G., et al. (2017). Marker-assisted selection for rice blast resistance genes Pi2 and Pi9 through high-resolution melting of a gene-targeted amplicon. Plant Breed. 136, 67–73. doi: 10.1111/pbr.12447
Mishra, K. K., Vikram, P., Yadaw, R. B., Swamy, B. M., Dixit, S., Cruz, M. T. S., et al. (2013). qDTY 12. 1: a locus with a consistent effect on grain yield under drought in rice. BMC Genet. 14, 1–10. doi: 10.1186/1471-2156-14-12
Mundt, C. C., Ahmed, H. U., Finckh, M. R., Nieva, L. P., Alfonso, R. F. (1999). Primary disease gradients of bacterial blight of rice. Phytopathology 89, 64–67. doi: 10.1094/PHYTO.1999.89.1.64
Muthu, V., Abbai, R., Nallathambi, J., Rahman, H., Ramasamy, S., Kambale, R., et al. (2020). Pyramiding QTLs controlling tolerance against drought, salinity, and submergence in rice through marker assisted breeding. PloS One 15, e0227421. doi: 10.1371/journal.pone.0227421
Nahar, S., Sahoo, L., Tanti, B. (2018). Screening of drought tolerant rice through morpho-physiological and biochemical approaches. Biocatalysis Agric. Biotechnol. 15, 150–159. doi: 10.1016/j.bcab.2018.06.002
Neeraja, C. N., Maghirang-Rodriguez, R., Pamplona, A., Heuer, S., Collard, B. C., Septiningsih, E. M., et al. (2007). A marker-assisted backcross approach for developing submergence-tolerant rice cultivars. Theor. Appl. Genet. 115, 767–776. doi: 10.1007/s00122-007-0607-0
Pancaldi, F., Trindade, L. M. (2020). Marginal lands to grow novel bio-based crops: a plant breeding perspective. Front. Plant Sci. 11, 227. doi: 10.3389/fpls.2020.00227
Pandey, P., Irulappan, V., Bagavathiannan, M. V., Senthil-Kumar, M. (2017). Impact of combined abiotic and biotic stresses on plant growth and avenues for crop improvement by exploiting physio-morphological traits. Front. Plant Sci. 8, 537. doi: 10.3389/fpls.2017.00537
Pradhan, S. K., Nayak, D. K., Pandit, E., Behera, L., Anandan, A., Mukherjee, A. K., et al. (2016). Incorporation of bacterial blight resistance genes into lowland rice cultivar through marker-assisted backcross breeding. Phytopathology 106, 710–718. doi: 10.1094/PHYTO-09-15-0226-R
Pradhan, S. K., Pandit, E., Pawar, S., Baksh, S. Y., Mukherjee, A. K., Mohanty, S. P. (2019). Development of flash-flood tolerant and durable bacterial blight resistant versions of mega rice variety ‘Swarna’through marker-assisted backcross breeding. Sci. Rep. 9, 12810. doi: 10.1038/s41598-019-49176-z
Qu, S., Liu, G., Zhou, B., Bellizzi, M., Zeng, L., Dai, L., et al. (2006). The broad-spectrum blast resistance gene Pi9 encodes a nucleotide-binding site–leucine-rich repeat protein and is a member of a multigene family in rice. Genetics 172, 1901–1914. doi: 10.1534/genetics.105.044891
Rahman, H., Dakshinamurthi, V., Ramasamy, S., Manickam, S., Kaliyaperumal, A. K., Raha, S., et al. (2018). Introgression of submergence tolerance into CO 43, a popular rice variety of India, through marker-assisted backcross breeding. Czech J. Genet. Plant Breed. 54, 101–108. doi: 10.17221/149/2017-CJGPB
Ramalingam, J., Savitha, P., Alagarasan, G., Saraswathi, R., Chandrababu, R. (2017). Functional marker assisted improvement of stable cytoplasmic male sterile lines of rice for bacterial blight resistance. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01131
Rana, M. M., Takamatsu, T., Baslam, M., Kaneko, K., Itoh, K., Harada, N., et al. (2019). Salt tolerance improvement in rice through efficient SNP marker-assisted selection coupled with speed-breeding. Int. J. Mol. Sci. 20. doi: 10.3390/ijms20102585
Ray, D. K., Mueller, N. D., West, P. C., Foley, J. A. (2013). Yield trends are insufficient to double global crop production by 2050. PloS One 8, e66428. doi: 10.1371/journal.pone.0066428
Reay, D., Reay, D. (2019). Climate-smart rice. Climate-Smart Food, 121–133. doi: 10.1007/978-3-030-18206-9
Roser, M., Ritchie, H., Ortiz-Ospina, E., Rodés-Guirao, L. (2013). World population growth. Our World data. Available at: https://ourworldindata.org/worldpopulation-growth.
Sandhu, N., Dixit, S., Swamy, B., Raman, A., Kumar, S., Singh, S., et al. (2019). Marker assisted breeding to develop multiple stress tolerant varieties for flood and drought prone areas. Rice 12, 1–16. doi: 10.1186/s12284-019-0269-y
Septiningsih, E. M., Hidayatun, N., Sanchez, D. L., Nugraha, Y., Carandang, J., Pamplona, A. M., et al. (2015). Accelerating the development of new submergence tolerant rice varieties: the case of Ciherang-Sub1 and PSB Rc18-Sub1. Euphytica 202, 259–268. doi: 10.1007/s10681-014-1287-x
Singh, G., Kaur, N., Khanna, R., Kaur, R., Gudi, S., Kaur, R., et al. (2024). 2Gs and plant architecture: breaking grain yield ceiling through breeding approaches for next wave of revolution in rice (Oryza sativa L.). Crit. Rev. Biotechnol. 44, 139–162. doi: 10.1080/07388551.2022.2112648
Singh, V. J., Vinod, K. K., Krishnan, S. G., Singh, A. K. (2021). Rice adaptation to climate change: opportunities and priorities in molecular breeding. Mol. Breed. Rice abiotic Stress tolerance Nutr. Qual., 1–25. doi: 10.1002/9781119633174.ch1
Singh, V. K., Singh, B. D., Kumar, A., Maurya, S., Krishnan, S. G., Vinod, K. K., et al. (2018). Marker-assisted introgression of Saltol QTL enhances seedling stage salt tolerance in the rice variety “Pusa Basmati 1. Int. J. Genomics 2018. doi: 10.1155/2018/8319879
Sultana, H., Somaddar, U., Samanta, S. C., Chowdhury, A. K., Saha, G. (2022). Diversity analysis of Bangladeshi coastal rice landraces (Oryza sativa) for morpho-physiological and molecular markers’ responses to seedling salinity tolerance. Plant Breed. Biotechnol. 10, 115–127. doi: 10.9787/PBB.2022.10.2.115
Tiwari, S., Tomar, R., Tripathi, M., Ahuja, A. (2017). Modified protocol for plant genomic DNA isolation. Indian Res. J. Genet. Biotechnol. 9, 478–485.
Valarmathi, M., Sasikala, R., Rahman, H., Jagadeeshselvam, N., Kambale, R., Raveendran, M. (2019). Development of salinity tolerant version of a popular rice variety improved white ponni through marker assisted back cross breeding. Plant Physiol. Rep. 24, 262–271. doi: 10.1007/s40502-019-0440-x
Variar, M., Cruz, C. V., Carrillo, M., Bhatt, J., Sangar, R. (2019). “Rice blast in India and strategies to develop durably resistant cultivars,” in Advances in genetics, genomics and control of rice blast disease (Netherlands: Springer), 359–374.
Venuprasad, R., Dalid, C., Del Valle, M., Zhao, D., Espiritu, M., Sta Cruz, M., et al. (2009). Identification and characterization of large-effect quantitative trait loci for grain yield under lowland drought stress in rice using bulk-segregant analysis. Theor. Appl. Genet. 120, 177–190. doi: 10.1007/s00122-009-1168-1
Vikram, P., Swamy, B. M., Dixit, S., Ahmed, H. U., Teresa Sta Cruz, M., Singh, A. K., et al. (2011). qDTY 1.1, a major QTL for rice grain yield under reproductive-stage drought stress with a consistent effect in multiple elite genetic backgrounds. BMC Genet. 12, 1–15. doi: 10.1186/1471-2156-12-89
Vikram, P., Swamy, B. M., Dixit, S., Singh, R., Singh, B. P., Miro, B., et al. (2015). Drought susceptibility of modern rice varieties: an effect of linkage of drought tolerance with undesirable traits. Sci. Rep. 5, 14799. doi: 10.1038/srep14799
Wu, J., Kim, S. G., Kang, K. Y., Kim, J.-G., Park, S.-R., Gupta, R., et al. (2016). Overexpression of a pathogenesis-related protein 10 enhances biotic and abiotic stress tolerance in rice. Plant Pathol. J. 32, 552. doi: 10.5423/PPJ.OA.06.2016.0141
Yamori, W. (2013). Improving photosynthesis to increase food and fuel production by biotechnological strategies in crops. J. Plant Biochem. Physiol. 1. doi: 10.4172/2329-9029.1000113
Yasmin, S., Hafeez, F. Y., Mirza, M. S., Rasul, M., Arshad, H. M., Zubair, M., et al. (2017)Biocontrol of bacterial leaf blight of rice and profiling of secondary metabolites produced by rhizospheric Pseudomonas aeruginosa BRp3. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.01895
Yoshida, S. (1976). Laboratory manual for physiological studies of rice. Int. Rice Res. Ins Philippines 23, 61–66.
Yugander, A., Sundaram, R. M., Singh, K., Ladhalakshmi, D., Subba Rao, L. V., Madhav, M. S., et al. (2018). Incorporation of the novel bacterial blight resistance gene Xa38 into the genetic background of elite rice variety Improved Samba Mahsuri. PloS One 13, e0198260. doi: 10.1371/journal.pone.0198260
Keywords: rice, multiple-stress tolerance, abiotic and biotic, marker-assisted backcrossing, QTL pyramiding
Citation: Mohanavel V, Muthu V, Kambale R, Palaniswamy R, Seeli P, Ayyenar B, Rajagopalan V, Manickam S, Rajasekaran R, Rahman H, Nallathambi J, Swaminathan M, Chellappan G, Vellingiri G and Muthurajan R (2024) Marker-assisted breeding accelerates the development of multiple-stress-tolerant rice genotypes adapted to wider environments. Front. Plant Sci. 15:1402368. doi: 10.3389/fpls.2024.1402368
Received: 17 March 2024; Accepted: 03 June 2024;
Published: 12 July 2024.
Edited by:
Sambasivam Periyannan, University of Southern Queensland, AustraliaReviewed by:
Gurjeet Singh, Texas A&M University, United StatesCopyright © 2024 Mohanavel, Muthu, Kambale, Palaniswamy, Seeli, Ayyenar, Rajagopalan, Manickam, Rajasekaran, Rahman, Nallathambi, Swaminathan, Chellappan, Vellingiri and Muthurajan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Raveendran Muthurajan, cmF2ZWVuZHJhbnRuYXVAZ21haWwuY29t; Geethalakshmi Vellingiri, Z2VldGhhQHRuYXUuYWMuaW4=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.