
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 30 May 2024
Sec. Plant Abiotic Stress
Volume 15 - 2024 | https://doi.org/10.3389/fpls.2024.1401414
The AT-hook motif nuclear localized (AHL) gene family is a highly conserved transcription factors involved in plant growth, development, and stress responses. However, AHLs have not been systematically analyzed in radish (Raphanus sativus). Therefore, we performed genome-wide identification and expression pattern, gene structure, and function verifications of radish AHLs. We identified 52 radish AHLs (RsAHL1–RsAHL52), which were unevenly distributed across nine chromosomes. Phylogenetic analysis showed that the RsAHLs were divided into two clades (A and B) and subdivided into three types (I, II, and III). Collinearity analysis revealed that the 52 RsAHLs produced 49 repeat events. Tissue expression profiles revealed differential expression of RsAHLs across different tissues, with higher expression observed in flower organs, particularly petals and anthers. qRT-PCR results indicated that RsAHLs responded to abscisic acid, methyl jasmonate, and abiotic stress (low and high temperatures and drought). Additionally, RsAHL14 induced a dwarf phenotype in tomato plants, and RsAHL14-overexpression tomato plants presented significantly decreased expression levels of the gibberellin (GA) synthetic genes ent-Copalyl diphosphatase, GA3ox-3/-4/-5, and GA20ox-1/-2/-3, but significantly increased expression of the degradation gene GA2ox-1/-3. Thus, RsAHL14 might affect plant growth by regulating GA content. Collectively, our study comprehensively identified RsAHLs in radish and provided a reference for further research on these genes.
Plants are subjected to various biotic and abiotic stresses during growth and development and resist these stresses through their own defense mechanisms (Alves et al., 2014). Transcription factors are activated under stress and play a defensive role by regulating the expression of defense-related genes (Chen et al., 2002). In Arabidopsis, AP2/EREBP, bZIP/HD-ZIP, Myb, and several zinc finger (ZF) transcription factors are either activated or suppressed under stress conditions (Shinozaki and Yamaguchi-Shinozaki, 2000). Notably, altering the expression levels of certain transcription factors can significantly alter plant stress resistance. For example, the overexpression of VvNAC17 can improve the sensitivity of Arabidopsis to abscisic acid (ABA) and enhance the ability of Arabidopsis to resist drought, salt, and freezing stress (Ju et al., 2020).
AT-hook motif nuclear localized (AHL) genes are transcription factors that were first discovered in the high-mobility group proteins (HMG) of mammals, and they play an important role in chromosome structure assembly and target gene transcription regulation (Goodwin et al., 1973). AHL genes are widely present in terrestrial plants. For example, 29 AHL genes were identified in Arabidopsis (Zhao et al., 2014), 37 in maize (Bishop et al., 2020), 63 in soybean (Wang et al., 2021b), 26 in rice (Kumar et al., 2023), 47 in carrot (Machaj and Grzebelus, 2021), 42 in Brassica rapa (Zhang et al., 2023), and 48, 51, and 99 in the three different cotton genomes (Zhao et al., 2020), respectively. AHL proteins contain two conserved domains: the AT-hook motif and the plants and prokaryotes conserved (PPC) domain (Zhao et al., 2013). The AT-hook motif is a small DNA-binding motif that can be divided into two types (I and II). Type I AT-hook contains the conserved sequence Arg-Gly-Arg-Pro, followed by Gly-Ser-Lys-Asn-Lys, while Type II AT-hook contains the conserved sequence Arg-Gly-Arg-Pro-Arg-Lys-Tyr. Both types have a characteristic structure centered on Arg-Gly-Arg-Pro (RGRP) residues (Zhang et al., 2022). This conserved amino acid sequence is necessary for protein-DNA interactions and nuclear localization (Do et al., 2006). The AT-hook motif specifically binds to AT-rich sequences in double-stranded DNA furrows to complete interactions with target genes (Aravind and Landsman, 1998). The PPC domain, also known as DUF296, contains approximately 120 amino acids and is located in the carboxylic acids relative to the AT-hooks (Preston and Hileman, 2012). PPC domains can be divided into Type A (without introns) and Type B (with introns), both of which contain a Gly-Arg-Phe-Glu-Ile-Leu core conserved sequence. Upstream of the conserved sequence, the Type A PPC domain has the Leu-Arg-Ser-His core conserved sequence, while the Type B PPC domain has the Phe-Thr-Pro-His core conserved sequence (Zhang et al., 2022). In plants, the main roles of PPC domains are nuclear localization and protein-protein interactions, suggesting that AHL proteins may be involved in regulating transcriptional activity (Fujimoto et al., 2004).
Based on the AT-hook motif sequence characteristics, sequence similarity, AT-hook motif combination with PPC, and affinity for DNA, AHL proteins can be divided into three categories: Types I, II, and III. Type I AHL proteins have polar amino acids at the C-end of the core sequence, and the second amino acid at the C-end is usually glycine, which has strong affinity. Type I AHL proteins contains one Type I AT-hook motif and Type A PPC domain. The second amino acid at the C-terminus of the Type II AHL proteins core sequence is usually lysine, which has a weak affinity. Type II AHL proteins contain two AT-hook motifs (Type I and Type II) and Type B PPC domain. The fourth conserved amino acid downstream of the C terminal of the Type III AHL proteins is lysine, and its affinity lies between those of Types I and II. Type III AHL proteins contains one Type II AT-hook motif and Type B PPC domain. Furthermore, the AHL proteins could be divided into two clades (clade A and clade B), in which Type I AHL proteins belonged to clade A, while Types II and III AHL proteins belonged to clade B (Zhao et al., 2014). Analysis of exon and intron numbers of maize AHL genes shows that Types II and III evolved from Type I (Bishop et al., 2020). AHL proteins can not only directly bind DNA but also affect the binding of other transcription factors to DNA, thereby indirectly regulating the transcriptional activity of target genes (Strick and Laemmli, 1995).
AHL genes are crucial for plant growth and development, organ building, and stress responses. In Arabidopsis thaliana, AHL22 can act as a chromatin remodeling factor to modify the structure of FLOWERING LOCUS T (FT) chromatin and regulate flowering time (Yun et al., 2012). Overexpression of AHL20 and AHL22 decreases the transcription levels of FT and delays flowering (Xiao et al., 2009; Yun et al., 2012; Tayengwa et al., 2020). Furthermore, AHL22, AHL27, and AHL29 negatively regulate hypocotyl elongation (Street et al., 2008; Zhao et al., 2013). AHL transcription factors and phytochrome-interacting factors (PIF) competitively bind to PIF target sites, reduce PIF binding to growth-promoting genes, and inhibit the transcriptional activation of these genes, thus repressing leaf petiole elongation (Favero et al., 2020). TEK/AHL16 could negatively regulate the flowering inhibitors MAF4 and MAF5 (Xu et al., 2013). During the aging process, AHL27 could delay the aging of Arabidopsis leaves, while AHL9 showed the opposite phenotype (Lim et al., 2007; Zhou et al., 2022). In plant defense and stress responses, overexpression of OsAHL1 could improve the drought resistance of rice (Zhou et al., 2016). Further studies showed that the OsAHL10, OsAHL13 and OsAHL20 were involved in the signaling of drought stress and salt stress (Ambadas et al., 2023). However, overexpression of AHL20 inhibited the expression of NHO1 and FRK1 induced by pathogen-associated molecular patterns (PAMPs) and was sensitive to toxic Pseudomonas syringae bacteria, indicating that AHL20 negatively regulates the defense ability of Arabidopsis (Lu et al., 2010). In addition, several AHL genes have been reported to regulate plant hormone balance, such as gibberellin (Matsushita et al., 2007), cytokinin (Rashotte et al., 2003), and jasmonic acid (Vom Endt et al., 2007).
Radish (Raphanus sativus) is an important cruciferous vegetable and one of the most widely cultivated root vegetables, and studies have suggested that radish can be used as an ideal model plant for root crops (Hoang et al., 2020). During radish growth, biotic and abiotic stresses can affect the growth and expansion of taproots, thus affecting radish yield (Hoang et al., 2020; Li et al., 2023). To date, studies on AHL genes have mainly focused on Arabidopsis, whereas studies on AHL genes in radishes are relatively scarce. Therefore, to gain a more comprehensive understanding of the important functions of AHL genes in plants, we identified and analyzed the RsAHL family in radishes in this study. The gene structure, phylogenetic tree, chromosome location, gene collinearity, conserved motifs, and promoter cis-elements of radish RsAHL family members were analyzed. Specific expression patterns of RsAHL family were also identified in various tissues, and their response to abiotic and hormone stress was observed. Furthermore, we identified the biological functions of RsAHL14 overexpression in tomatoes. Therefore, this study lays the foundation for further analysis of the role of RsAHL genes in radish growth, development, and stress responses.
The radish genome “NAU-LB” (BioProject number: PRJCA011486.) (Xu et al., 2023) was used to identify RsAHL family members in radish. Twenty-nine Arabidopsis and twenty-six rice AHL family protein sequences were used to construct a Hidden Markov Model (HMM) of known AHL protein family sequences using HMMER 3.0 software. The HMM was generated by computational analysis of known homologous gene sequences and used to predict whether the homologous gene sequences exist in other species. All potential RsAHL family sequences were identified in the radish protein sequences. In contrast, BLASTP (Altschul et al., 1990) (v2.10.1+) was used to compare all radish protein sequences with the obtained AHL family reference sequences (e-value=1*10–5), and the matched sequences were used as potential RsAHL family sequences. After synthesizing these candidate sequences, the PfamScan (Bateman et al., 2004) (v1.6) and Pfam A (Finn et al., 2014) (v33.1) databases were used to annotate the domain of the target sequence, and the sequence containing the PF03479 domain was determined as the final RsAHL protein sequence.
The RsAHL protein family sequences of radish, Arabidopsis, and rice were used to construct a neighbor-joining tree. First, MAFFT (v7.427) was used to perform multiple sequence comparisons, and MEGA10 software (Kumar et al., 2008) was used to build the neighbor-joining tree (the bootstrap was set to 1000), and then iTOL v6 (https://itol.embl.de/) was used to annotate the evolutionary tree.
“NAU-LB” genome sequence and general feature format (GFF) files were used to extract chromosome length and location information of RsAHL family members through the TBtools software. Subsequently, further visualization analysis was performed (Chen et al., 2020a).
Collinearity analysis was performed using MCScanX software (match score, 50; match size, 5; gap penalty, -1; overlap window, 5; e-value, 1e-5; max gaps, 25). Segment and tandem duplications caused by gene duplication events were analyzed.
The CDS and genome sequences of radish RsAHL family members were used to analyze the intron-exon structure using the GSDS website (http://gsds.cbi.pku.edu.cn/) for visualization. The MEME website was used to predict the conserved motif of RsAHLs, and TBtools software was used to visualize it.
The upstream 2 kb sequences of the RsAHLs promoter were extracted from the “NAU-LB” genome and then submitted to the PlantCARE (https://bioinformatics.psb.ugent.be/webtools/plantcare/html/) database to predict the distribution of cis-elements on the promoter. Subsequently, all cis-elements were classified, and those related to stress and hormone responses were extracted for visual analysis using the TBtools software.
Radish (Raphanus sativus) was cultivated at 25°C with 16 h of light and 8 h of darkness per day. Radish materials with consistent growth were selected for the stress treatment. Radish leaf samples were collected after cold treatment (4°C), high temperature treatment (42°C), PEG6000 treatment and hormone treatment (methyl jasmonate, abscisic acid) at 0 h, 1 h, 3 h, 6 h, 9 h, 12 h and 24 h.
The CDS of RsAHL14 was amplified using radish cDNA with specific amplification primers (Supplementary Table 4) and then cloned into pHELLSGATE8 (Chen et al., 2020b) to generate CaMV35S::AHL14 overexpression vector. This vector was transferred into Agrobacterium strain GV3101. Subsequently, genetic transformation was performed using MicroTom tomatoes as the background material. Transgenic plants were detected using PCR, and the primer sequences are shown in Supplementary Table 4.
Total RNA was extracted using TRIzol reagent (Vazyme, Nanjing, China) and reverse-transcribed into cDNA using the HiScript II 1st Strand cDNA Synthesis Kit (+ gDNA wiper) (Vazyme). Real-time fluorescence quantitative PCR (RT-qPCR) was performed using CFX384 Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA) in conjunction with ChamQ Universal SYBR® qPCR Master Mix (Vazyme). The relative expression levels of RsAHLs in radish tissues, abiotic stress samples, and overexpression plants were measured. The primer sequences are listed in Supplementary Table 4. Three biological replicates were used for each experiment. The radish PRII (RNA polymerase-II transcription factor) gene was used as the internal control (Xu et al., 2012) and the results were calculated using the 2−ΔΔCt method.
International Business Machine-Statistical Package for Social Sciences (IBM-SPSS) and Microsoft excel software were used for statistical analyses. All data were based on three independent biological replicates and expressed as the mean ± sd. Student’s t-test was used to calculate significant differences.
To identify AHLs in radish, 29 AtAHL and 26 OsAHL protein sequences were used as queries for BlastP and HMM searches against the radish genome. A total of 52 AHL proteins were identified and named RsAHL1–RsAHL52 (Supplementary Table 1), and then their physicochemical properties were analyzed. The RsAHLs ranged from 195 to 852 amino acids in length, with a relative molecular weight (MV) spanning between 20.86 and 94.62 kDa. Their isoelectric point (pI) was between 4.81and 9.83, while their aliphatic index ranged from 49.59 to 88.53. The grand average hydropathy (GRAVY) values of all RsAHLs were less than zero, indicating that RsAHL are strongly hydrophilic. Subcellular localization prediction showed that most of the RsAHL genes were located in the nucleus, followed by the chloroplast (Supplementary Table 1).
To further infer the evolutionary relationships between RsAHL and other AHL proteins, we constructed phylogenetic trees using multiple sequence alignments of AHL proteins from radish (Raphanus sativus), Arabidopsis, and rice (Oryza sativa L.). The results showed that the AHL genes of these three species were divided into two clades: clades A and B. Within clade A, radish, Arabidopsis, and rice accounted for 25, 15, and 15 proteins, respectively, whereas within clade B, they accounted for 27, 14, and 11 proteins, respectively. Furthermore, the two independent clades A and B could be further divided into three types, with Type I belonging to clade A and Types II and III belonging to clade B (Figure 1). This result implies a high degree of phylogenetic consistency among AHL genes across different species and confirms the homology between the different species, suggesting that family members of the same branch may have similar functions.
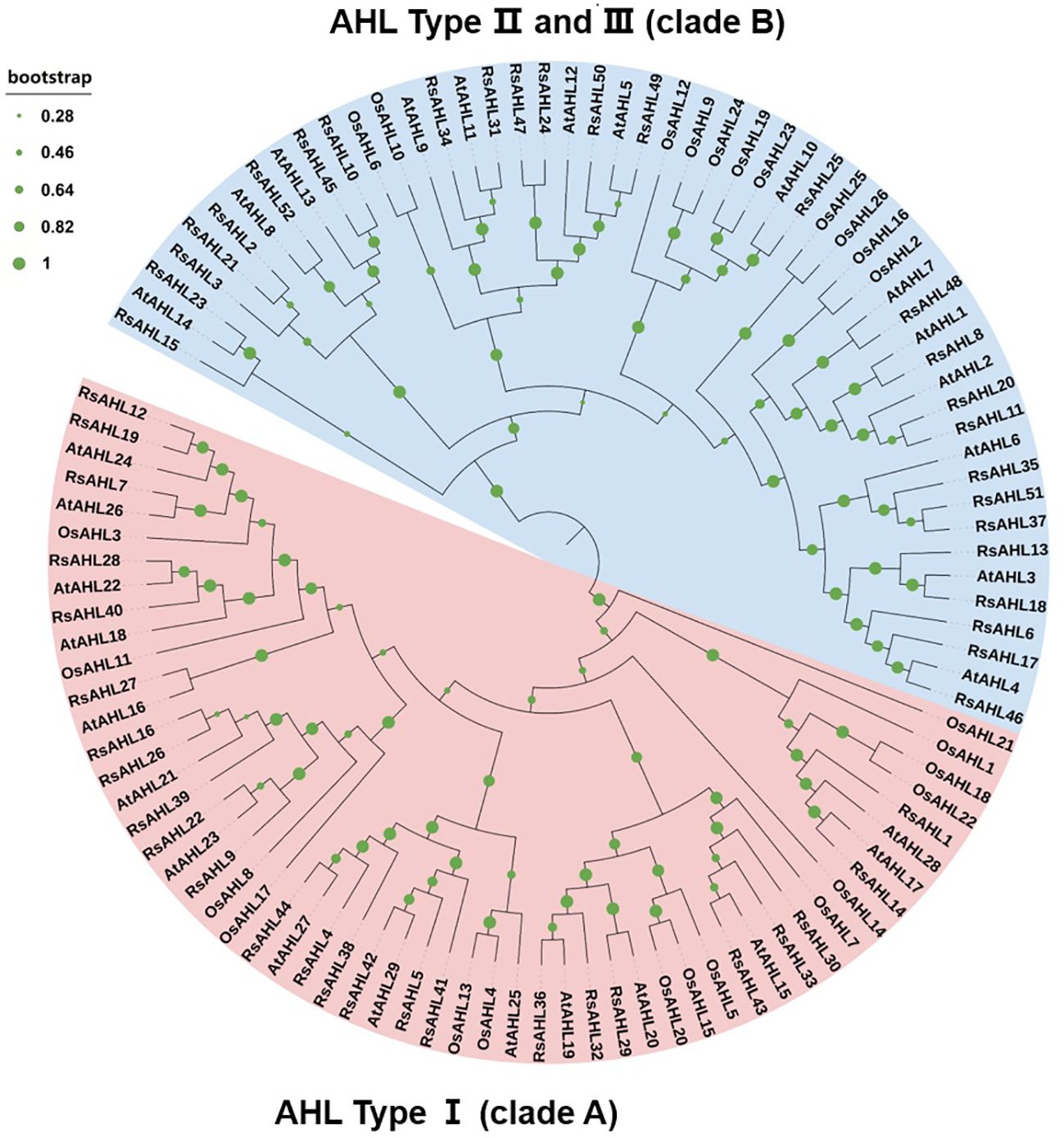
Figure 1 Phylogenetic tree of the AT-hook motif nuclear localized (AHL) family in radish, Arabidopsis, and rice. Different branch colors represent different subfamilies. The Rs- represents radish, the AT- represents Arabidopsis, and the LOC Os- represents rice.
Based on the location information of each RsAHL gene, we arranged the positions of the 52 RsAHL genes across nine chromosomes of the radish genome, and chromosome mapping indicated that they were 52 RsAHL genes were unevenly distributed across the chromosomes (Figure 2). RsAHLs were mainly located on RsChr2, RsChr4, and RsChr9, which contained 9, 14, and 7 genes, respectively. Notably, only two RsAHL genes were found on RsChr8, namely RsAHL44 and RsAHL45.
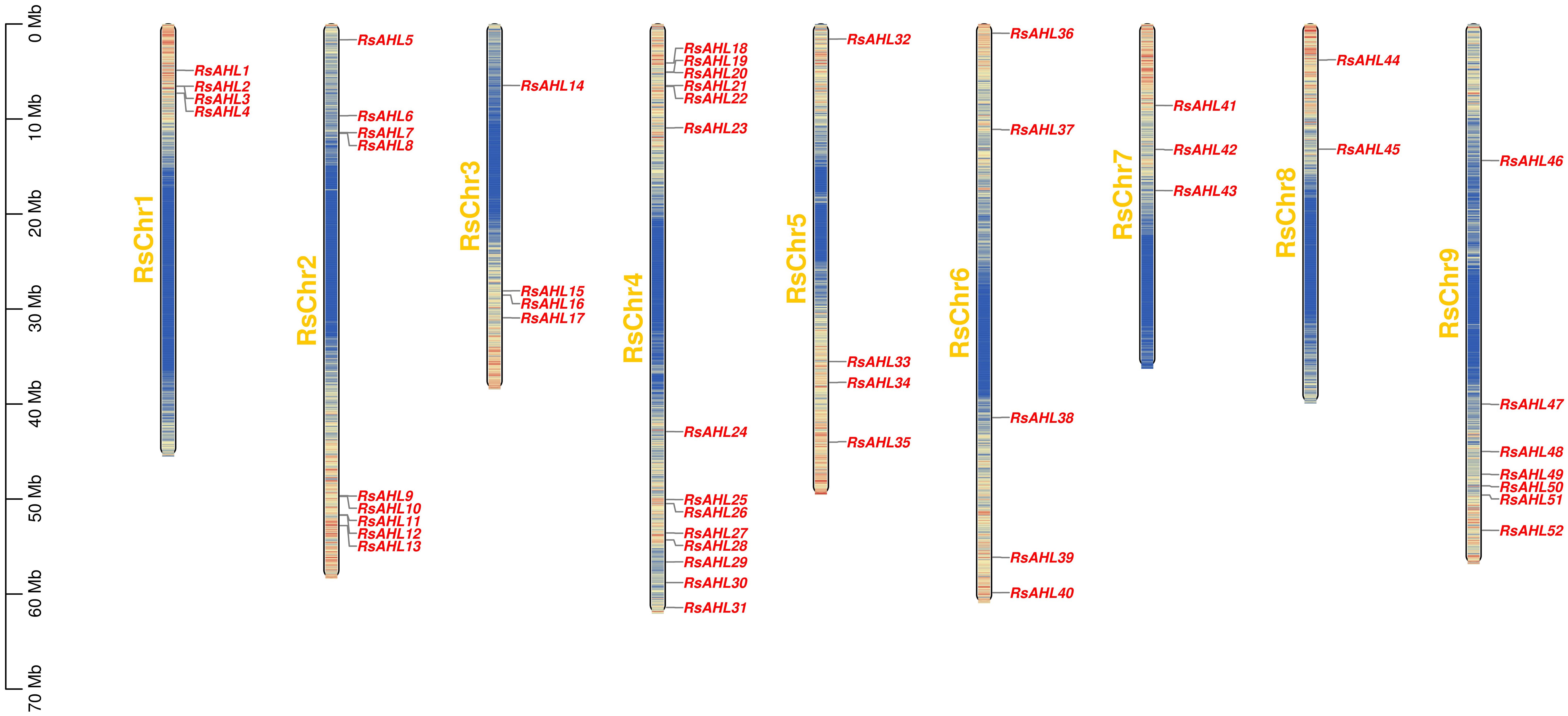
Figure 2 Arrangement of Raphanus sativus AHLs (RsAHLs) across nine chromosomes within radish. The name of each chromosome is on the left of the bar, while the gene name is on the right.
Gene duplication was the most important cause of gene family expansion during plant evolution. To further study the evolutionary relationship of RsAHL in radish, gene duplication events were analyzed. And 49 duplications events were identified in 41 RsAHL genes. Among them, RsAHL4 had the most collinearity relationship with other RsAHLs, including RsAHL5, RsAHL38, RsAHL41, RsAHL42 and RsAHL44. However, 10 RsAHLs (RsAHL1, RsAHL2, RsAHL3, RsAHL14, RsAHL15, RsAHL23, RsAHL25, RsAHL27, RsAHL29 and RsAHL48) had no collinearity relationship with the other RsAHLs (Figure 3; Supplementary Table 2). These results suggest that segment repeats play a crucial role in the expansion of the RsAHL family.
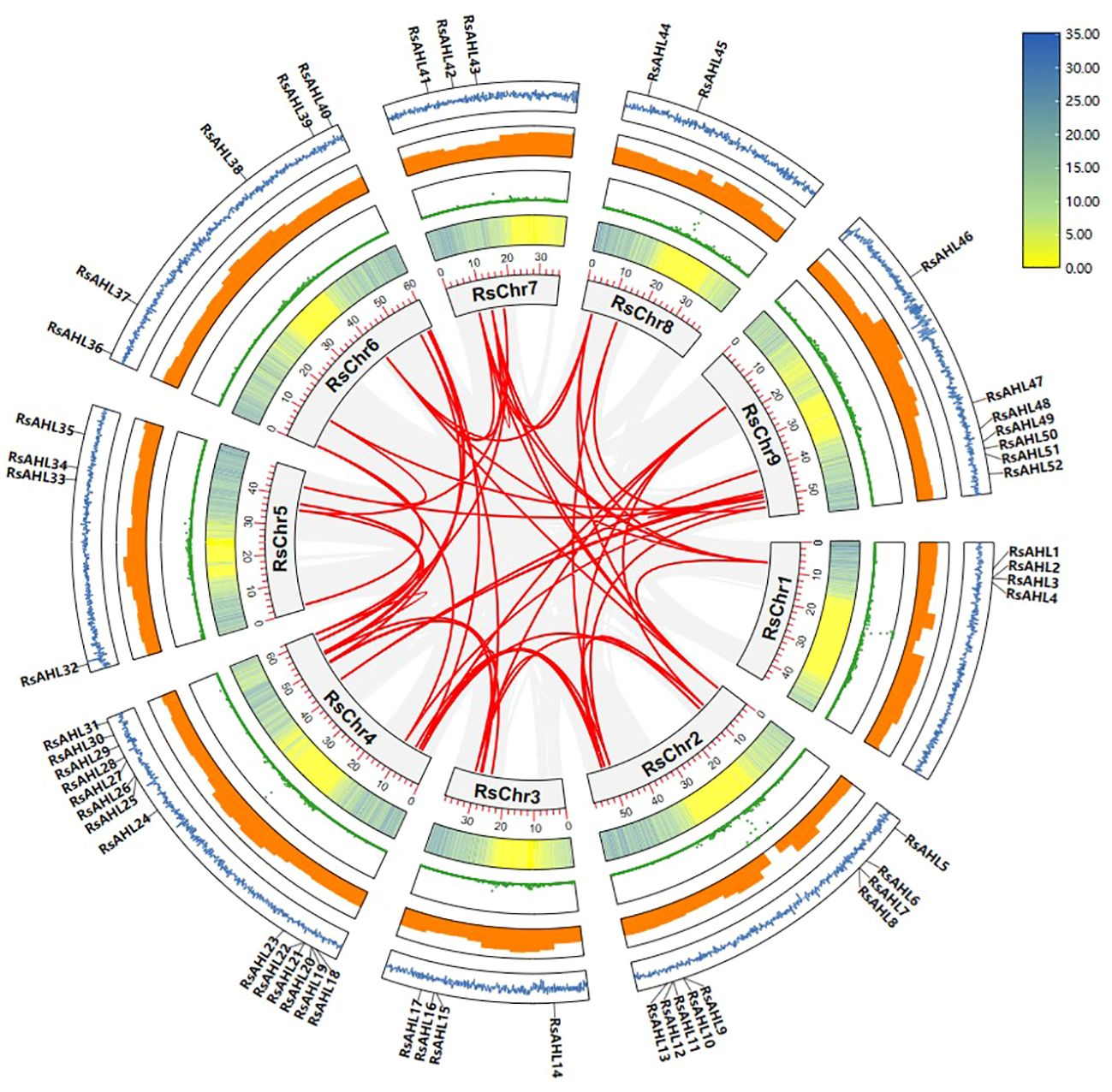
Figure 3 Collinearity analysis of the RsAHL gene family. The gray lines indicate all synteny blocks between each chromosome, and the red lines in the circle indicated duplicated AHL pairs. The circles from inside to outside represent gene density, N-ratio, GC-ratio and GC-skew.
To infer the evolutionary relationship between RsAHL genes, MEGA 10 was used to construct a phylogenetic tree of the 52 RsAHL protein sequences in radish (Figure 4A). We predicted the conserved protein motifs using the Multiple Expectation maximizations for Motif Elicitation (MEME) website (Figure 4B). We identified a total of 10 conserved motifs among the RsAHL proteins (Supplementary Figure 1; Table 1). These motifs ranged in length from 15 to 38 amino acids, with sites ranging from 5 to 51. Motifs 5 and 6 contained a conserved Arg-Gly-Arg core and belonged to the AT-hook motif family. In addition, motifs 5 and 6 were identified as Type I and Type II AT-hook motifs, respectively. Notably, we also identified the PPC domain (motif 2), which contains conserved Gly-Arg-Phe-Glu-Ile-Leu residues (Zhao et al., 2014). This motif has also been identified in maize, but not in soybean (Bishop et al., 2020; Wang et al., 2021b). Interestingly, almost all of the RsAHL genes contained motifs 1, 2, and 3, suggesting the consistency of the RsAHL protein sequences. Overall, the gene structure and motif prediction results indicated the consistency and evolutionary diversity of AHL genes in terrestrial plants.
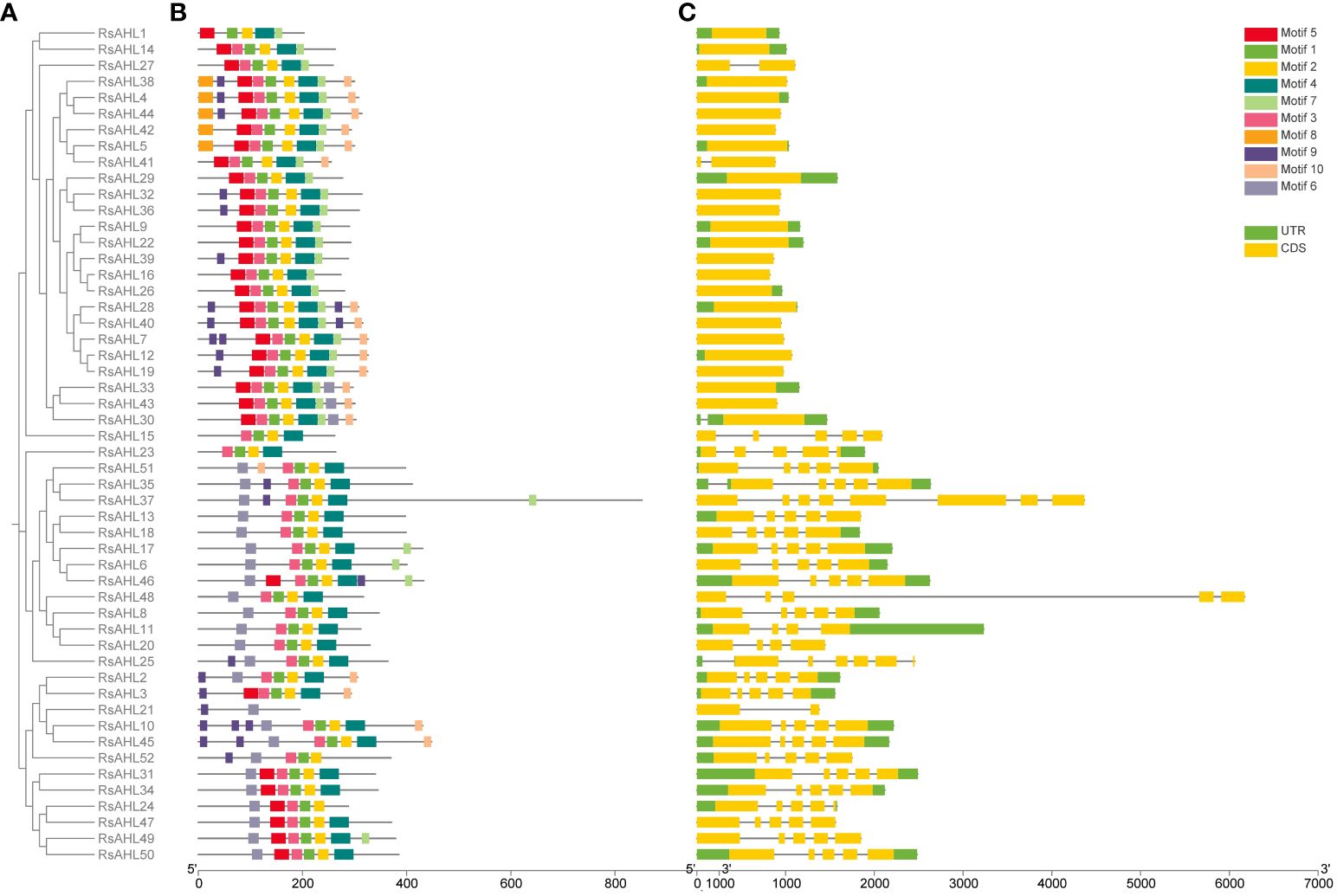
Figure 4 Phylogenetic tree, gene structure, and conserved motifs of RsAHLs in radish. (A) Phylogenetic tree was constructed using MEGA 10 in radish. (B) Motif structure diagram of RsAHL proteins. Different colors indicate different motif structures. (C) Gene structure diagram. The yellow box represents the coding sequence (CDS) region, the black thin line represents the intron, and the green represents the untranslated region (UTR).
Subsequently, we further analyzed the distribution of introns and exons in the RsAHL genes to explore the gene structure in radish (Figure 4C). The length of the RsAHL genes was between 824 and 6172 bp, among which the Type I genes were generally shorter than the Type II and III genes (Supplementary Table 1). Moreover, the numbers of introns and exons were diverse. We found that the genetic structure was similar for each type of gene. For example, Type I genes contained only one or two exons, and Types II and III contained more exons and introns than Type I, suggesting a diverse genetic structure of this subgroup. Therefore, we believe that Types II and III evolved from Type I. This result is consistent with that of the AHL gene family reported in maize (Bishop et al., 2020).
Cis-elements in promoters can be bound by transcription factors to regulate plant growth, development, defense, and stress responses by regulating gene expression. To infer the function of the RsAHL gene family, we used the PlantCARE database to perform cis-element analysis with a 2 kb promoter region of RsAHL. The functions of these cis-elements have also been identified (Supplementary Table 3). As shown in Figure 5, we found that all RsAHL promoters contained light response elements, including Box4, GT1-motif, G-box, TCT-motif, GATA-motif, I-box, AE-box, MRE, chs-CMA1a/2a, Sp1, ATCT-motif, AT1-motif, GA-motif, TCCC-motif, Box II, ACE, ATC-motif, LAMP-element, Gap-box, GTGGC-motif, chs-Unit 1, 3-AF1 binding site, 4 cl-CMA2b, ACA-motif, and LS7. The Box4 cis element exists in almost all RsAHL genes, with RsAHL46 containing the highest number. This suggests that RsAHL genes are likely regulated by light. Approximately 86.5% of RsAHL family members contained anaerobic induction elements (ARE), with RsAHL6 containing the highest number. Seven cis-elements were associated with hormonal responses: abscisic acid (ABRE and TCA-element), methyl jasmonate or MeJA (CGTCA-motif, TGACG-motif), and auxin (AuxRR-core, TGA-element, and AuxRE). Additionally, we found that 26.9%, 44.2%, and 48.1% of RsAHL family members contained low-temperature elements (LTR), drought induction elements (MBS), and defense and stress responsiveness (TC-rich repeats), respectively. These results indicate that the RsAHL gene family plays an important role in regulating plant growth, hormone responses, and abiotic stress responses.
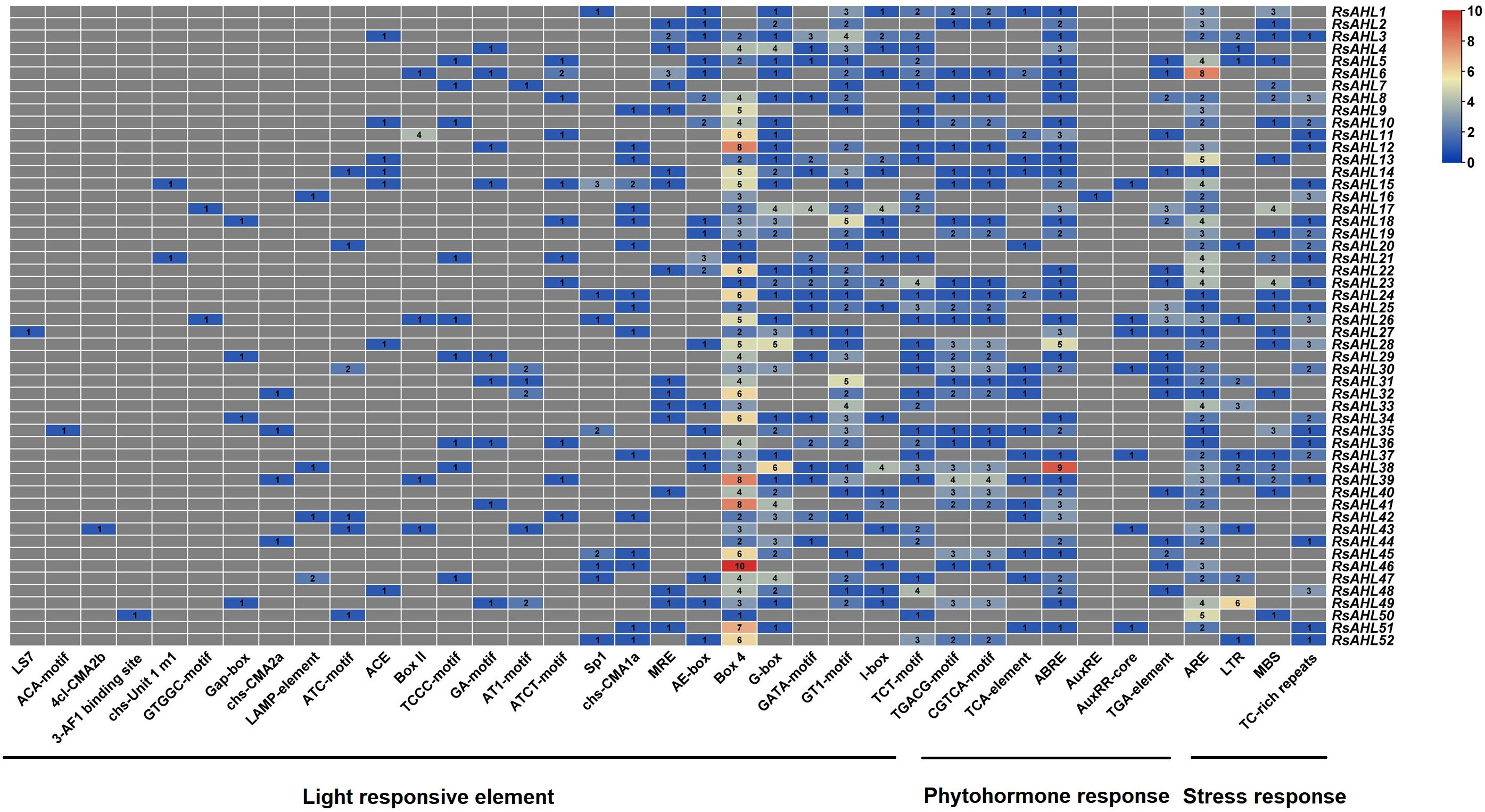
Figure 5 Number of cis-elements on the promoter of RsAHL genes in radish. Different colors represent the number of different cis-acting elements.
To study the expression of RsAHLs in various radish tissues, we extracted RNA from the taproots, stems, leaves, petals, calyx, filaments, and anthers of radish and detected the relative expression levels of RsAHLs among them (Figure 6). The results showed that RsAHLs had different expression patterns in different tissues of the radish plant. The relatively highest expression of RsAHL genes was observed in the anthers, petals, and taproots. In particular, RsAHL44 expression was the highest in petals, RsAHL33 expression was the highest in anthers, and RsAHL25 expression was the highest in taproots and calyxes. On the contrary, RsAHL11, RsAHL17, RsAHL18, RsAHL20, RsAHL32, RsAHL34, RsAHL46 and RsAHL49 exhibited relatively low expression across various organs. In addition, the expression levels of the RsAHL family in filaments were lower than those in other flower organs. These results indicate that RsAHL family is mainly expressed in the taproots and floral organs, suggesting the potential function of RsAHL genes in taproots and flowers.

Figure 6 Relative expression level of the RsAHL family in various tissues of radish. Different colored boxes indicate different levels of expression.
To further explore the expression pattern of the RsAHL family in radish under hormonal stress, qRT-PCR was performed to analyze the 52 RsAHL genes under different stress treatments. Based on the analysis of cis-acting elements in the promoter, MeJA and ABA were predicted to induce the strongest hormone responses. Subsequently, MeJA (200 μM) and ABA (10 μM) were further used to treat radish seedlings (Figure 7). These results showed that RsAHL genes presented varying responses to the MeJA and ABA treatments. Under ABA treatment, 14 RsAHL genes were significantly upregulated after 6 h, with RsAHL4, RsAHL8, and RsAHL24 showing a gradual increase in expression. RsAHL5, RsAHL17 and RsAHL22 were upregulated at 3 h and reached their highest expression levels at 12, 24, and 6 h, respectively. In contrast, RsAHL21, RsAHL31, RsAHL33 and RsAHL39 were down-regulated after ABA treatment, whereas RsAHL15 and RsAHL43 were down-regulated after 6 h (Figure 7A). Under MeJA treatment, the expression of RsAHL13, RsAHL21, RsAHL28, RsAHL32 and RsAHL45 were continuously upregulated at 1 h. RsAHL22 and RsAHL52 were upregulated at 3 h, RsAHL22 reached its highest level at 6 h and then decreased, while RsAHL52 reached its highest value at 12 h and then decreased. RsAHL24 expression was upregulated at 6 h, reached its highest level, and then decreased thereafter. In contrast, the expression of RsAHL4 was downregulated immediately after treatment, whereas that of RsAHL10, RsAHL15 and RsAHL46 began to decline after 3 h. Notably, RsAHL51 did not change significantly after treatment, indicating that it did not respond to MeJA induction (Figure 7B). Therefore, the expression of RsAHLs in most radish samples were significantly upregulated by the ABA and MeJA treatments. Particularly, the expression of RsAHLs was significantly upregulated 6 and 1 h after ABA and MeJA induction, respectively.
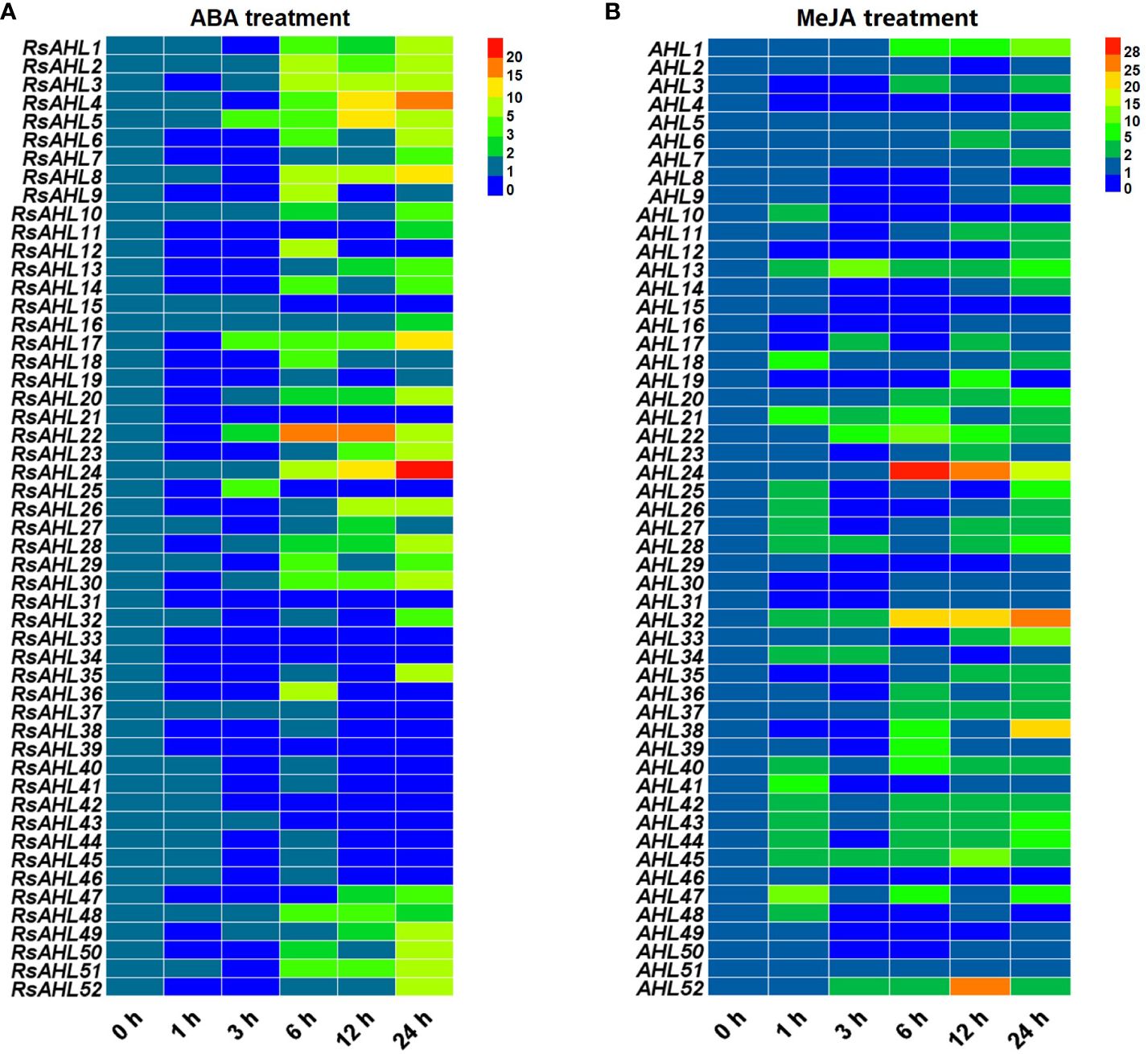
Figure 7 Relative expression level of the RsAHL family under hormone treatment. (A) Relative expression level of RsAHLs in radish seedlings after abscisic acid (ABA) treatment. (B) Relative expression level of RsAHLs in radish seedlings after methyl jasmonate (MeJA) treatment. Here, 0 h, 1 h, 3 h, 6 h, 12 h, and 24 h represent the time points of sampling after treatment. Different colored boxes indicate different levels of expression.
To investigate the roles of the RsAHL family under abiotic stress, the expression levels of RsAHLs were determined after cold (4°C), hot (42°C) and polyethylene glycol PEG6000 (20%) treatment. Under the 4°C treatment, the expression of most RsAHL genes was significantly upregulated after 3 h, and reached the highest level at 24 h. The expression of RsAHL20 reached its highest level at 3 h, while that of RsAHL52 reached its highest level at 12 h (Figure 8A). At 42 °C treatment, most of the RsAHL genes were significantly upregulated and reached their highest expression at 12 h. The expression levels of RsAHL11, RsAHL16, and RsAHL51 gradually increased after 6 h of treatment, whereas the expression levels of RsAHL21 and RsAHL35 increased after 1 h. In contrast, the expression levels of RsAHL18, RsAHL31 and RsAHL32 were downregulated after treatment (Figure 8B). Most RsAHLs were significantly upregulated after PEG6000 treatment. The expression levels of RsAHL8, RsAHL11, RsAHL21, RsAHL33, RsAHL40, RsAHL41, and RsAHL50 were consistently upregulated after treatment. Notably, 10 RsAHL genes were consistently upregulated after 3 h of treatment and RsAHL35 and RsAHL36 were gradually upregulated after 6 h of treatment. However, RsAHL3, RsAHL17, and RsAHL23 were downregulated after treatment (Figure 8C). These results suggest that RsAHL plays an important role in abiotic stress.
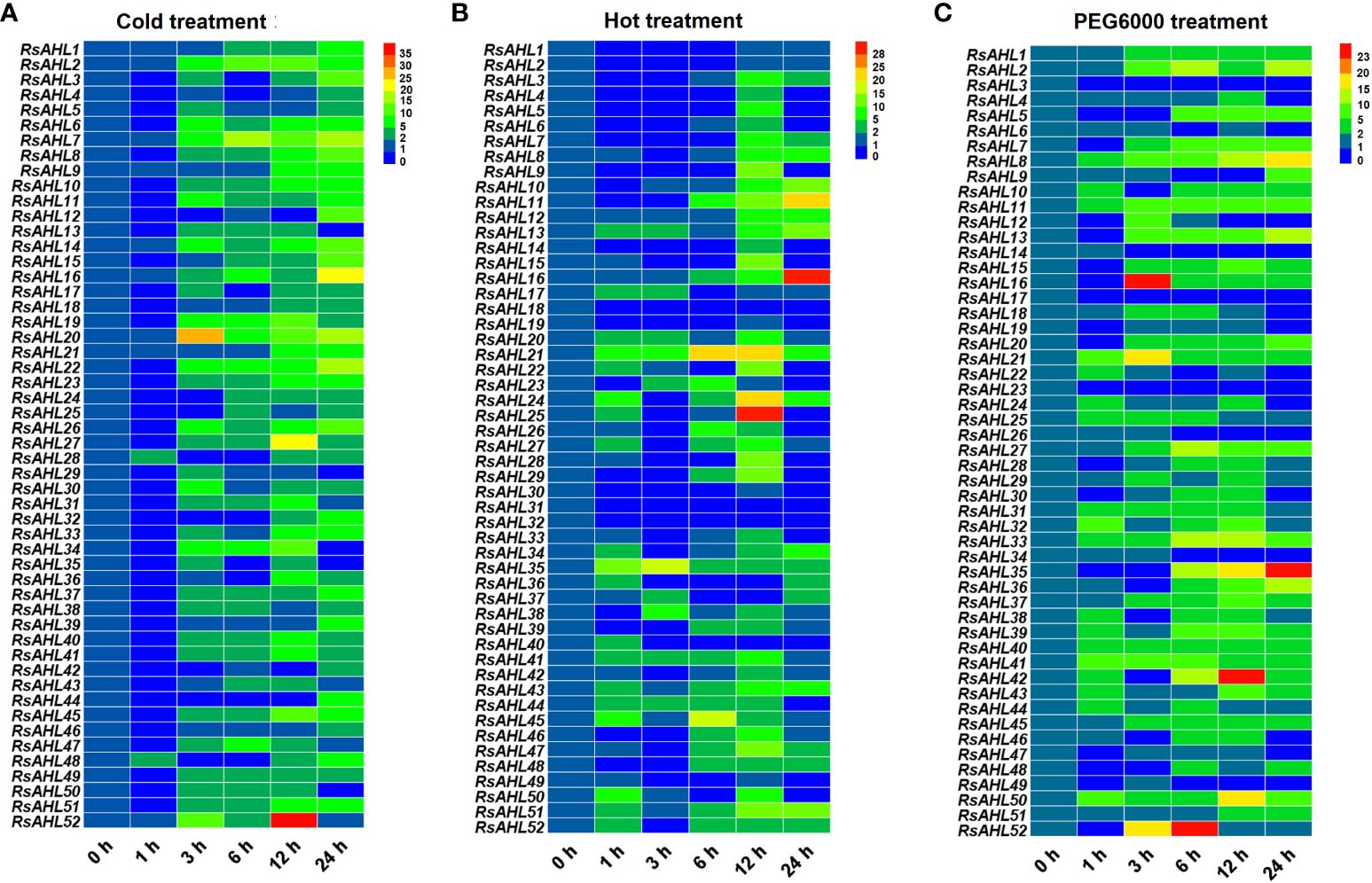
Figure 8 Expression patterns of RsAHLs under various abiotic stress. (A) Relative expression level of RsAHLs in radish seedlings after cold treatment (4°C). (B) Relative expression level of RsAHLs in radish seedlings after hot treatment (42°C). (C) Relative expression level of RsAHLs in radish seedlings after polyethylene glycol PEG6000 treatment. Here, 0 h, 1 h, 3 h, 6 h, 12 h, and 24 h represent the time points of sampling after treatment. Different colored boxes indicate different levels of expression.
To identify the biological function of the RsAHL family, we constructed overexpression vectors of several RsAHL genes and used MicroTom as the background material for genetic transformation. Using vector forward and gene reverse primers to detect the positive of transgenic materials, we obtained RsAHL14-overexpression positive plants (Supplementary Figure 2), with the RsAHL14-OE-1/-2/-5/-6/-12 lines used for further experiments. In the observation of the phenotype of the positive transgenic tomato plants, it was found that plants with RsAHL14 overexpression were dwarfed during the developmental process compared to the control (CK) (Figure 9A). Further, the plant height was significantly lower in RsAHL14-overexpression lines compared with CK (Figure 9C). To analyze the relative expression of RsAHL14 in the overexpression lines, RsAHL14-OE-4 (which showed low expression of RsAHL14 in positive materials) was used as the control. The results showed that the relative expression levels of RsAHL14 in the leaves of the RsAHL14-OE-1/-2/-5/-6/-12 lines were 5.0-, 4.4-, 5.9-, 23.6-, and 48.0-fold higher than that in the level of RsAHL14-OE-4, respectively (Figure 9B). Notably, previous studies have shown that changes in the gibberellin (GA) content in plants can cause dwarfing.
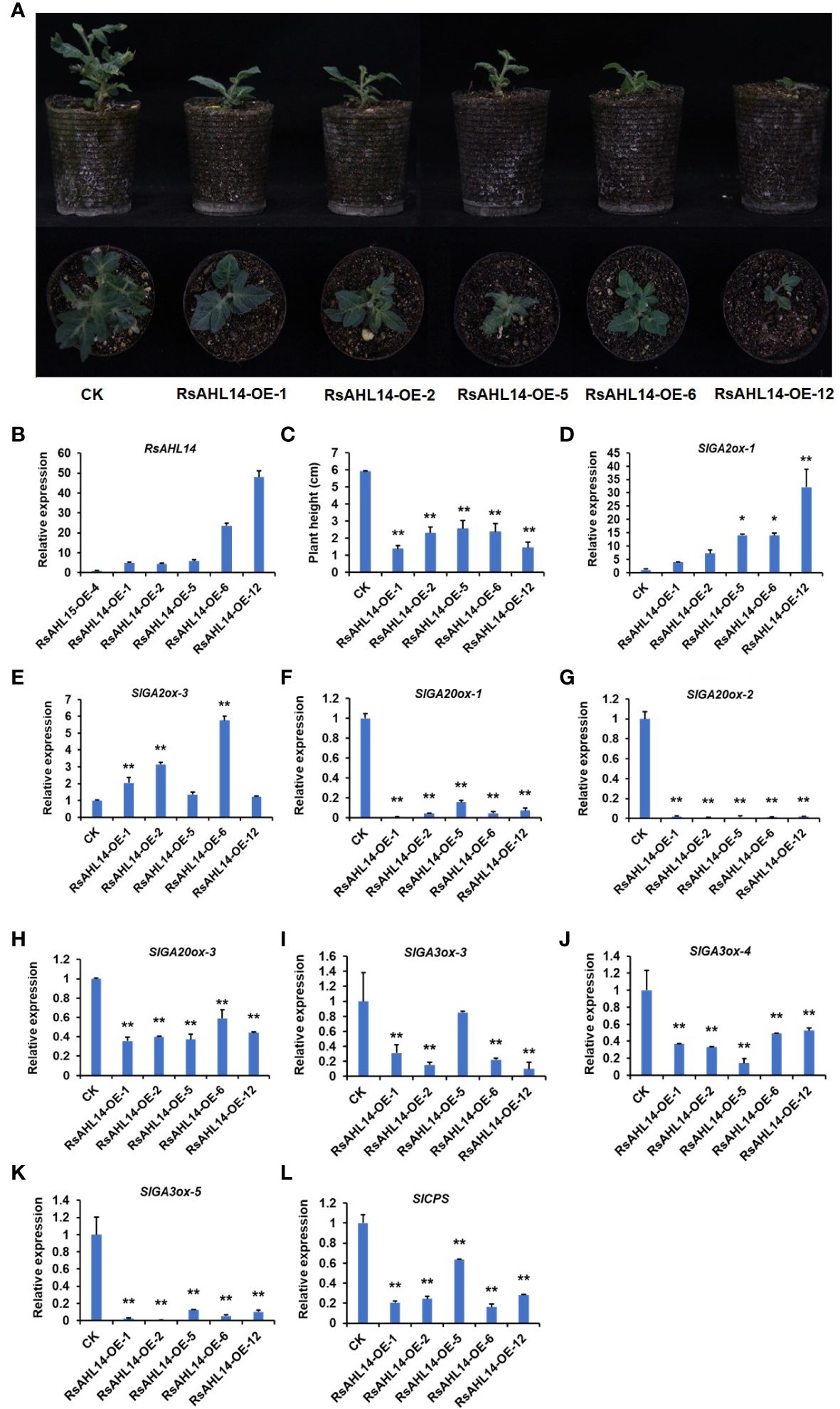
Figure 9 Overexpression of RsAHL14 causes tomato dwarfing by regulating the expression of GA synthesis and degradation genes. (A) Phenotypes of the RsAHL14-overexpression lines and CK in tomato. (B) Relative expression level of RsAHL14 in tomato leaves of the overexpression lines and CK. (C) Plant height of the RsAHL14-overexpression lines and CK. Relative expression level of SlGA2ox-1 (D), SlGA2ox-3 (E), SlGA20ox-1 (F), SlGA20ox-2 (G), SlGA20ox-3 (H), SlGA3ox-3 (I), SlGA3ox-4 (J), SlGA3ox-5 (K) and SlCPS (L) in tomato leaves of the overexpression lines and CK. The asterisks indicate significant difference (**P ≤ 0.01, *P ≤ 0.05).
To determine whether changes in GA lead to dwarfing, the expression of genes involved in GA biosynthesis and degradation pathways was detected in RsAHL14 overexpression plants. Among these, ent-Copalyl diphosphatase (CPS), ent-Kaurene synthase (KS), ent-Kaurene oxidase (KO), GA20-oxidase (GA20ox), and GA3-oxidase (GA3ox) are key enzymes in the GA biosynthetic pathway, while GA2-oxidase (GA2ox) is a key enzyme in the degradation pathway. As expected, compared with the CK, the relative expression levels of GA2ox-1 and GA2ox-3 were significantly increased in the RsAHL14-OE plants (Figures 9D, E), while those of GA20ox-1, GA20ox-2, GA20ox-3, GA3ox-3, GA3ox-4, GA3ox-5 and CPS were significantly decreased (Figures 9F-L). However, the expression levels of other GA-related genes were irregular in the overexpression lines (Supplementary Figure 3). This suggests that GA synthesis was hindered in RsAHL14-overexpression lines, the degradation rate was accelerated, and the resulting plants were dwarfed.
Radish is an important root crop and one of the main autumn and winter vegetables. AHLs play an important role in plant growth, development, and stress resistance in Arabidopsis, maize, rice, cotton, and soybeans (Kim et al., 2011; Zhao et al., 2014; Bishop et al., 2020; Zhao et al., 2020; Wang et al., 2021b). However, the identification of AHL proteins in radish has not yet been reported. In this study, we performed a genome-wide analysis of AHLs in radish and identified 52 RsAHLs. The phylogenetic analysis revealed that RsAHLs could be divided into two clades (A and B) and three types (I, II, and III) (Figure 1), which is consistent with the results obtained for other land plants. Notably, we identified a PPC domain (motif 2) in RsAHL proteins (Figure 4; Supplementary Figure 1; Table 1); however, no such domain was found in soybeans (Wang et al., 2021b). The PPC domains could interact with each other or with other transcription factors to regulate transcriptional activation (Zhao et al., 2013; Seo and Lee, 2021), suggesting a diverse biological function for RsAHLs.
In soybean and maize, AHL proteins are present in multiple organelles, including the nucleus, cytoplasm, and chloroplasts (Bishop et al., 2020; Wang et al., 2021b). Similar results were obtained for radish in this study (Supplementary Table 1), suggesting that the subcellular localization of AHLs is conserved across various species. All RsAHL gene families were unevenly distributed across nine chromosomes (Figure 2). Collinearity analysis showed that multiple gene duplication events occurred in the RsAHL genes within the radish genome (Figure 3; Supplementary Table 2), suggesting that RsAHLs expanded through gene duplication and gained and lost their functions.
Several studies have demonstrated the involvement of AHL genes in various stress responses (Yadeta et al., 2011; Zhou et al., 2016; Wong et al., 2019). Cis-elements in promoters are believed to influence plant growth, development, and stress responses (Yamaguchi-Shinozaki and Shinozaki, 2006; Himmelbach et al., 2010). Analysis of the cis element of the RsAHL promoter is conducive to a comprehensive understanding of its potential function. Most RsAHLs contained anaerobic induction elements and responded to ABA, MeJA, and auxins (Figure 5). In an anaerobic environment, plant root development is blocked, and epidermal cells are damaged or killed, leading to pathogen infection (Kuan and Erwin, 1980). Notably, all RsAHL genes can be photoinduced because their promoters contain light-responsive elements. In addition, some RsAHLs respond to low temperature and drought stress. In the study of grape and soybean, all grape and soybean AHL gene promoters contained light response elements, hormone response elements, and stress response elements (Li et al., 2021; Wang et al., 2021b). This suggests that the AHL genes in radish and other plant species affect plant growth, development, and stress response.
The AHL family is widely distributed in plants and is vital in the regulation of flower, hypocotyl, root, and leaf development (Street et al., 2008; Xiao et al., 2009; Yun et al., 2012; Tayengwa et al., 2020; Seo and Lee, 2021; Zhou et al., 2022). To better understand the specific expression patterns of RsAHL in radish, we analyzed the relative expression levels of 52 RsAHLs in different tissues. We found that RsAHLs were highly expressed in flower organs and taproots compared to that in other tissues (Figure 6). AHL3, AHL4, AHL18, and OsAHL1 regulate the development of root (Zhou et al., 2013, 2016; Sirl et al., 2020). Whereas, DP1, AHL16/TEK, AHL20, AHL21/GIK, AHL22, AHL23, AHL27, Baf1 regulates the development of various flower organs (Ng et al., 2009; Xiao et al., 2009; Gallavotti et al., 2011; Jin et al., 2011; Yun et al., 2012; Jia et al., 2015; Tayengwa et al., 2020). These results indicate that RsAHLs play a crucial role in the development of flowers and taproots in radish plants. In particular, DcAHLc1 plays an important role in the development of storage root in carrot (Macko-Podgórni et al., 2017).
Under biotic and abiotic stresses, the transcription level of AHL is significantly affected. For example, the overexpression of OsAHL1 can improve rice drought tolerance and resistance, participate in the oxidative stress response, and regulate leaf chlorophyll content (Zhou et al., 2016). Similarly, under the PEG treatment, the expression of grape AHL genes was induced by different degrees of stress (Li et al., 2021). We further found that radish RsAHL genes were also induced to different degrees under PEG treatment. Similar results were found after the 42°C heat treatment and 4°C cold treatment (Figure 8). A similar study induced change in PtrAHL12 and PtrAHL17 expression through ABA and drought treatments, respectively (Wang et al., 2021a). Our results showed that RsAHL gene expression was significantly upregulated after 6 h of ABA treatment, and after 6 and 12 h of MeJA treatment (Figure 7). These results imply a multifaceted role of RsAHL genes in plant stress responses and reveal that further examination of the biological functions of these genes is warranted to help improve the adaptability of radish under various abiotic stress conditions.
Furthermore, we found that RsAHL14-overexpression in tomato caused plant dwarfing, and numerous studies have shown that changes in the plant GA content can also cause dwarfing. For example, the dwarfing phenotypes of Arabidopsis ga3, maize dwarf3, and rice dwarf35 mutants are due to the transcription of GA synthetase P450 monooxygenase (P450) (Winkler and Helentjaris, 1995; Helliwell et al., 1998; Itoh et al., 2004). The GA content in plants is determined by synthesis and degradation pathways. In the present study, the relative expression levels of seven GA biosynthetic genes were significantly decreased, whereas the expression levels of SlGA2ox-1/-3 in the degradation pathway were significantly increased in RsAHL14-overexpression tomato plants (Figure 9). AGF1 (an AT-hook protein) regulates the GA balance by negatively regulating AtGA3ox-1 (Matsushita et al., 2007), suggesting that AHL genes play an important role in maintaining GA homeostasis. According to Yamaguchi’s study (Yamaguchi, 2008), a simple model of RsAHL14 expression in tomato plants was developed, and it was shown to cause changes in the expression of GA synthesis and degradation genes, which may lead to an imbalance of active and inactive GA content and ultimately lead to a tomato dwarf phenotype (Figure 10). However, the specific regulatory mechanism of RsAHL14 on GA needs to be further studied.

Figure 10 Model for the effects of RsAHL14 heterologous overexpression on the regulation of GA and dwarf phenotypes in tomato. GGPP, geranylgeranylpyrophosphate; CPP, copalyl pyrophosphate. Blue arrows represent a decrease, and red arrows represent an increase.
In this study, a total of 52 RsAHL genes were identified in radish, and they unevenly distributed on 9 chromosomes. The phylogenetic tree divided these genes into two clades and three types based on the AT-hook motif and PPC domain. Furthermore, the cis-acting elements of 2 kb promoter regions of RsAHL genes and their expression in different tissue were investigated, and their response to abiotic stress and hormones was also determined. Meanwhile, heterologous expression of RsAHL14 can induce dwarfing in tomato plants by regulating the expression of GA-related genes, thus suggesting the important relationship between RsAHL genes and plant hormones, especially GA. These results will provide a basis for further exploring the biological functions of the RsAHL family in growth regulation and stress responses.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author/s.
WC: Conceptualization, Data curation, Funding acquisition, Software, Visualization, Writing – original draft. LCh: Data curation, Investigation, Writing – review & editing. LCu: Resources, Supervision, Writing – review & editing. ZL: Investigation, Software, Validation, Writing – original draft. WY: Conceptualization, Funding acquisition, Supervision, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by grants from National Natural Science Foundation of China (3220181270); Natural Science Foundation of Hubei Province (2023AFB694); The fourth batch of modern agricultural industrial technology system high quality vegetable industrial technology system project in Hubei Province (2023HBSTX4-06); Wuhan Biological Breeding Major Project (2022021302024852); Youth Science Fund of Hubei Academy of Agricultural Sciences (2023NKYJJ07).
We would like to thank Editage (www.editage.cn) for English language editing.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1401414/full#supplementary-material
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Alves, M. S., Dadalto, S. P., Goncalves, A. B., de Souza, G. B., Barros, V. A., Fietto, L. G. (2014). Transcription factor functional protein-protein interactions in plant defense responses. Proteomes 2, 85–106. doi: 10.3390/proteomes2010085
Ambadas, D. A., Singh, A., Jha, R. K., Chauhan, D., Santhosh, B., Sharma, V. K. (2023). Genome-wide dissection of AT-hook motif nuclear-localized gene family and their expression profiling for drought and salt stress in rice (Oryza sativa). Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1283555
Aravind, L., Landsman, D. (1998). AT-hook motifs identified in a wide variety of DNA-binding proteins. Nucleic Acids Res. 26, 4413–4421. doi: 10.1093/nar/26.19.4413
Bateman, A., Coin, L., Durbin, R., Finn, R. D., Hollich, V., Griffiths-Jones, S., et al. (2004). The Pfam protein families database. Nucleic Acids Res. 32, D138–D141. doi: 10.1093/nar/gkh121
Bishop, E. H., Kumar, R., Luo, F., Saski, C., Sekhon, R. S. (2020). Genome-wide identification, expression profiling, and network analysis of AT-hook gene family in maize. Genomics 112, 1233–1244. doi: 10.1016/j.ygeno.2019.07.009440
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020a). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, W., Hu, T., Ye, J., Wang, B., Liu, G., Wang, Y., et al. (2020b). A CCAAT-binding factor, SlNFYA10, negatively regulates ascorbate accumulation by modulating the D-mannose/L-galactose pathway in tomato. Hortic. Res. 7, 200. doi: 10.1038/s41438-020-00418-6
Chen, W., Provart, N. J., Glazebrook, J., Katagiri, F., Chang, H. S., Eulgem, T., et al. (2002). Expression profile matrix of arabidopsis transcription factor genes suggests their putative functions in response to environmental stresses. Plant Cell. 14, 559–574. doi: 10.1105/tpc.010410
Do, H. J., Song, H., Yang, H. M., Kim, D. K., Kim, N. H., Kim, J. H. (2006). Identification of multiple nuclear localization signals in murine Elf3, an ETS transcription factor. FEBS Lett. 580, 1865–1871. doi: 10.1016/j.febslet.2006.02.049
Favero, D. S., Kawamura, A., Shibata, M., Takebayashi, A., Jung, J. H., Suzuki, T., et al. (2020). AT-Hook transcription factors restrict petiole growth by antagonizing PIFs. Curr. Biol. 30, 1454–1466. doi: 10.1016/j.cub.2020.02.017
Finn, R. D., Bateman, A., Clements, J., Coggill, P., Eberhardt, R. Y., Eddy, S. R., et al. (2014). Pfam: the protein families database. Nucleic Acids Res. 42, D222–D230. doi: 10.1093/nar/gkt1223
Fujimoto, S., Matsunaga, S., Yonemura, M., Uchiyama, S., Azuma, T., Fukui, K. (2004). Identification of a novel plant MAR DNA binding protein localized on chromosomal surfaces. Plant Mol. Biol. 56, 225–239. doi: 10.1007/s11103-004-3249-5
Gallavotti, A., Malcomber, S., Gaines, C., Stanfield, S., Whipple, C., Kellogg, E., et al. (2011). BARREN STALK FASTIGIATE1 is an AT-hook protein required for the formation of maize ears. Plant Cell. 23, 1756–1771. doi: 10.1105/tpc.111.084590
Goodwin, G. H., Sanders, C., Johns, E. W. (1973). A new group of chromatin-associated proteins with a high content of acidic and basic amino acids. Eur. J. Biochem. 38, 14–19. doi: 10.1111/j.1432-1033.1973.tb03026.x
Helliwell, C. A., Sheldon, C. C., Olive, M. R., Walker, A. R., Zeevaart, J. A., Peacock, W. J., et al. (1998). Cloning of the Arabidopsis ent-kaurene oxidase gene GA3. Proc. Natl. Acad. Sci. U.S.A. 95, 9019–9024. doi: 10.1073/pnas.95.15.9019
Himmelbach, A., Liu, L., Zierold, U., Altschmied, L., Maucher, H., Beier, F., et al. (2010). Promoters of the barley germin-like GER4 gene cluster enable strong transgene expression in response to pathogen attack. Plant Cell. 22, 937–952. doi: 10.1105/tpc.109.067934
Hoang, N. V., Choe, G., Zheng, Y., Aliaga Fandino, A. C., Sung, I., Hur, J., et al. (2020). Identification of conserved gene-regulatory networks that integrate environmental sensing and growth in the root cambium. Curr. Biol. 30, 2887–2900. doi: 10.1016/j.cub.2020.05.046
Itoh, H., Tatsumi, T., Sakamoto, T., Otomo, K., Toyomasu, T., Kitano, H., et al. (2004). A rice semi-dwarf gene, Tan-Ginbozu (D35), encodes the gibberellin biosynthesis enzyme, ent-kaurene oxidase. Plant Mol. Biol. 54, 533–547. doi: 10.1023/B:PLAN.0000038261.21060.47
Jia, Q. S., Zhu, J., Xu, X. F., Lou, Y., Zhang, Z. L., Zhang, Z. P., et al. (2015). Arabidopsis AT-hook protein TEK positively regulates the expression of arabinogalactan proteins for Nexine formation. Mol. Plant 8, 251–260. doi: 10.1016/j.molp.2014.10.001
Jin, Y., Luo, Q., Tong, H., Wang, A., Cheng, Z., Tang, J., et al. (2011). An AT-hook gene is required for palea formation and floral organ number control in rice. Dev. Biol. 359, 277–288. doi: 10.1016/j.ydbio.2011.08.023
Ju, Y., Yue, X., Min, Z., Wang, X., Fang, Y., Zhang, J. (2020). VvNAC17, a novel stress-responsive grapevine (Vitis vinifera L.) NAC transcription factor, increases sensitivity to abscisic acid and enhances salinity, freezing, and drought tolerance in transgenic Arabidopsis. Plant Physiol. Biochem. 146, 98–111. doi: 10.1016/j.plaphy.2019.11.002
Kim, H. B., Oh, C. J., Park, Y. C., Lee, Y., Choe, S., An, C. S., et al. (2011). Comprehensive analysis of AHL homologous genes encoding AT-hook motif nuclear localized protein in rice. BMB Rep. 44, 680–685. doi: 10.5483/BMBRep.2011.44.10.680
Kuan, T. L., Erwin, D. C. (1980). Predisposition effect of water saturation of soil on Phytophthora root rot of alfalfa. Eco. Epidemiol. 70, 981–986. doi: 10.1094/Phyto-70-981
Kumar, A., Singh, S., Mishra, A. (2023). Genome-wide identification and analyses of the AHL gene family in rice (Oryza sativa). 3 Biotech. 13, 248. doi: 10.1007/s13205-023-03666-0
Kumar, S., Nei, M., Dudley, J., Tamura, K. (2008). MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform. 9, 299–306. doi: 10.1093/bib/bbn017
Li, C., Mao, B., Wang, K., Xu, L., Fan, L., Wang, Y., et al. (2023). RsERF40 contributes to cold stress tolerance and cell expansion of taproot in radish (Raphanus sativus L.). Hortic. Res. 10, uhad013. doi: 10.1093/hr/uhad013
Li, X., He, H., Wang, H., Wu, X., Wang, H., Mao, J. (2021). Identification and expression analysis of the AHL gene family in grape (Vitis vinifera). Plant Gene. 26, 100285. doi: 10.1016/j.plgene.2021.100285
Lim, P. O., Kim, Y., Breeze, E., Koo, J. C., Woo, H. R., Ryu, J. S., et al. (2007). Overexpression of a chromatin architecture-controlling AT-hook protein extends leaf longevity and increases the post-harvest storage life of plants. Plant J. 52, 1140–1153. doi: 10.1111/j.1365-313X.2007.03317.x
Lu, H., Zou, Y., Feng, N. (2010). Overexpression of AHL20 negatively regulates defenses in Arabidopsis. J. Integr. Plant Biol. 52, 801–808. doi: 10.1111/j.1744-7909.2010.00969.x
Machaj, G., Grzebelus, D. (2021). Characteristics of the AT-Hook Motif Containing Nuclear Localized (AHL) genes in Carrot provides insight into their role in plant growth and storage root development. Genes. (Basel). 12, 764. doi: 10.3390/genes12050764
Macko-Podgórni, A., Machaj, G., Stelmach, K., Senalik, D., Grzebelus, E., Iorizzo, M., et al. (2017). Characterization of a genomic region under selection in cultivated Carrot (Daucus carota subsp. sativus) reveals a candidate domestication gene. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00012
Matsushita, A., Furumoto, T., Ishida, S., Takahashi, Y. (2007). AGF1, an AT-hook protein, is necessary for the negative feedback of AtGA3ox1 encoding GA 3-oxidase. Plant Physiol. 143, 1152–1162. doi: 10.1104/pp.106.093542
Ng, K. H., Yu, H., Ito, T. (2009). AGAMOUS controls GIANT KILLER, a multifunctional chromatin modifier in reproductive organ patterning and differentiation. PloS Biol. 7, e1000251. doi: 10.1371/journal.pbio.1000251
Preston, J. C., Hileman, L. C. (2012). Parallel evolution of TCP and B-class genes in Commelinaceae flower bilateral symmetry. EvoDevo 3 (1), 6. doi: 10.1186/2041-9139-3-6
Rashotte, A. M., Carson, S. D., To, J. P., Kieber, J. J. (2003). Expression profiling of cytokinin action in Arabidopsis. Plant Physiol. 132, 1998–2011. doi: 10.1104/pp.103.021436
Seo, M., Lee, J. Y. (2021). Dissection of functional modules of AT-HOOK MOTIF NUCLEAR LOCALIZED PROTEIN 4 in the development of the root xylem. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.632078
Shinozaki, K., Yamaguchi-Shinozaki, K. (2000). Molecular responses to dehydration and low temperature: differences and cross-talk between two stress signaling pathways. Curr. Opin. Plant Biol. 3, 217–223. doi: 10.1016/S1369-5266(00)80068-0
Sirl, M., Snajdrova, T., Gutierrez-Alanis, D., Dubrovsky, J. G., Vielle-Calzada, J. P., Kulich, I., et al. (2020). At-Hook Motif Nuclear Localised Protein 18 as a novel modulator of root system architecture. Int. J. Mol. Sci. 21 (5), 1886. doi: 10.3390/ijms21051886
Street, I. H., Shah, P. K., Smith, A. M., Avery, N., Neff, M. M. (2008). The AT-hook-containing proteins SOB3/AHL29 and ESC/AHL27 are negative modulators of hypocotyl growth in Arabidopsis. Plant J. 54, 1–14. doi: 10.1111/j.1365-313X.2007.03393.x
Strick, R., Laemmli, U. K. (1995). SARs are cis DNA elements of chromosome dynamics: synthesis of a sar repressor protein. Cell 83, 1137–1148. doi: 10.1016/0092-8674(95)90140-X
Tayengwa, R., Sharma Koirala, P., Pierce, C. F., Werner, B. E., Neff, M. M. (2020). Overexpression of AtAHL20 causes delayed flowering in Arabidopsis via repression of FT expression. BMC Plant Biol. 20, 559. doi: 10.1186/s12870-020-02733-5
Vom Endt, D., Soares e Silva, M., Kijne, J. W., Pasquali, G., Memelink, J. (2007). Identification of a bipartite jasmonate-responsive promoter element in the Catharanthus roseus ORCA3 transcription factor gene that interacts specifically with AT-Hook DNA-binding proteins. Plant Physiol. 144, 1680–1689. doi: 10.1104/pp.107.096115
Wang, M., Chen, B., Zhou, W., Xie, L., Wang, L., Zhang, Y., et al. (2021b). Genome-wide identification and expression analysis of the AT-hook Motif Nuclear Localized gene family in soybean. BMC Genomics 22, 361. doi: 10.1186/s12864-021-07687-y
Wang, H., Leng, X., Yang, J., Zhang, M., Zeng, M., Xu, X., et al. (2021a). Comprehensive analysis of AHL gene family and their expression under drought stress and ABA treatment in Populus trichocarpa. Peer J. 9, e10932. doi: 10.7717/peerj.10932
Winkler, R. G., Helentjaris, T. (1995). The maize Dwarf3 gene encodes a cytochrome P450-mediated early step in Gibberellin biosynthesis. Plant Cell. 7, 1307–1317. doi: 10.1105/tpc.7.8.1307
Wong, M. M., Bhaskara, G. B., Wen, T. N., Lin, W. D., Nguyen, T. T., Chong, G. L., et al. (2019). Phosphoproteomics of Arabidopsis Highly ABA-induced1 identifies AT-Hook-Like10 phosphorylation required for stress growth regulation. Proc. Natl. Acad. Sci. U S A. 116, 2354–2363. doi: 10.1073/pnas.1819971116
Xiao, C., Chen, F., Yu, X., Lin, C., Fu, Y. (2009). Over-expression of an AT-hook gene, AHL22, delays flowering and inhibits the elongation of the hypocotyl in Arabidopsis thaliana. Plant Mol. Biol. 71, 39–50. doi: 10.1007/s11103-009-9507-9
Xu, Y., Gan, E. S., Ito, T. (2013). The AT-hook/PPC domain protein TEK negatively regulates floral repressors including MAF4 and MAF5. Plant Signal Behav. 8, e25006. doi: 10.4161/psb.25006
Xu, L., Wang, Y., Dong, J., Zhang, W., Tang, M., Zhang, W., et al. (2023). A chromosome-level genome assembly of radish (Raphanus sativus L.) reveals insights into genome adaptation and differential bolting regulation. Plant Biotechnol. J. 21, 990–1004. doi: 10.1111/pbi.14011
Xu, Y., Zhu, X., Gong, Y., Xu, L., Wang, Y., Liu, L. (2012). Evaluation of reference genes for gene expression studies in radish (Raphanus sativus L.) using quantitative real-time PCR. Biochem. Biophys. Res. Commun. 424, 398–403. doi: 10.1016/j.bbrc.2012.06.119
Yadeta, K. A., Hanemian, M., Smit, P., Hiemstra, J. A., Pereira, A., Marco, Y., et al. (2011). The Arabidopsis thaliana DNA-binding protein AHL19 mediates verticillium wilt resistance. Mol. Plant Microbe Interact. 24, 1582–1591. doi: 10.1094/MPMI-04-11-0090
Yamaguchi, S. (2008). Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 59, 225–251. doi: 10.1146/annurev.arplant.59.032607.092804
Yamaguchi-Shinozaki, K., Shinozaki, K. (2006). Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Plant Biol. 57, 781–803. doi: 10.1146/annurev.arplant.57.032905.105444
Yun, J., Kim, Y. S., Jung, J. H., Seo, P. J., Park, C. M. (2012). The AT-hook Motif-containing Protein AHL22 regulates flowering initiation by modifying FLOWERING LOCUS T chromatin in Arabidopsis. J. Biol. Chem. 287, 15307–15316. doi: 10.1074/jbc.M111.318477
Zhang, W., Cheng, X., Fang, D., Cao, J. (2022). AT-HOOK MOTIF NUCLEAR LOCALIZED (AHL) proteins of ancient origin radiate new functions. Int. J. Biol. Macromol. 214, 290–300. doi: 10.1016/j.ijbiomac.2022.06.100
Zhang, X., Li, J., Cao, Y., Huang, J., Duan, Q. (2023). Genome-wide identification and expression analysis under abiotic stress of BrAHL genes in Brassica rapa. Int. J. Mol. Sci. 24, 12447. doi: 10.3390/ijms241512447
Zhao, J., Favero, D. S., Peng, H., Neff, M. M. (2013). Arabidopsis thaliana AHL family modulates hypocotyl growth redundantly by interacting with each other via the PPC/DUF296 domain. Proc. Natl. Acad. Sci. U S A. 110, E4688–E4697. doi: 10.1073/pnas.1219277110577
Zhao, J., Favero, D. S., Qiu, J., Roalson, E. H., Neff, M. M. (2014). Insights into the evolution and diversification of the AT-hook Motif Nuclear Localized gene family in land plants. BMC Plant Biol. 14, 266. doi: 10.1186/s12870-014-0266-7
Zhao, L., Lü, Y., Chen, W., Yao, J., Li, Y., Li, Q., et al. (2020). Genome-wide identification and analyses of the AHL gene family in cotton (Gossypium). BMC Genomics 21, 69. doi: 10.1186/s12864-019-6406-6
Zhou, J., Wang, X., Lee, J., Lee, J. (2013). Cell-to-cell movement of two interacting AT-hook factors in Arabidopsis root vascular tissue patterning. Plant Cell. 25, 187–201. doi: 10.1105/tpc.112.102210
Zhou, L., Liu, Z., Liu, Y., Kong, D., Li, T., Yu, S., et al. (2016). A novel gene OsAHL1 improves both drought avoidance and drought tolerance in rice. Sci. Rep. 6, 30264. doi: 10.1038/srep30264
Keywords: Radish (Raphanus sativus), AHL genes, genome-wide analysis, gene expression, stress response
Citation: Chen W, Chen L, Cui L, Liu Z and Yuan W (2024) Genome-wide analysis of radish AHL gene family and functional verification of RsAHL14 in tomato. Front. Plant Sci. 15:1401414. doi: 10.3389/fpls.2024.1401414
Received: 15 March 2024; Accepted: 15 May 2024;
Published: 30 May 2024.
Edited by:
Sudhakar Reddy Palakolanu, International Crops Research Institute for the Semi-Arid Tropics (ICRISAT), IndiaReviewed by:
Dariusz Grzebelus, University of Agriculture in Krakow, PolandCopyright © 2024 Chen, Chen, Cui, Liu and Yuan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weiling Yuan, eXdpaW5nMjAyMUBoYmFhcy5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.