- 1Research Institute of Molecular Alchemy (RIMA), Gyeongsang National University, Jinju, Republic of Korea
- 2School of Biological Sciences and Technology, Chonnam National University, Gwangju, Republic of Korea
- 3Division of Applied Life Science (BK21 Four), Anti-aging Bio Cell factory Regional Leading Research Center (ABC-RLRC), Gyeongsang National University, Jinju, Republic of Korea
- 4Division of Applied Life Science (BK21 Four), ABC-RLRC, RIMA, Gyeongsang National University, Jinju, Republic of Korea
Floral transition from the vegetative to the reproductive stages is precisely regulated by both environmental and endogenous signals. Among these signals, photoperiod is one of the most important environmental factors for onset of flowering. A florigen, FLOWERING LOCUS T (FT) in Arabidopsis, has thought to be a major hub in the photoperiod-dependent flowering time regulation. Expression levels of FT likely correlates with potence of flowering. Under long days (LD), FT is mainly synthesized in leaves, and FT protein moves to shoot apical meristem (SAM) where it functions and in turns induces flowering. Recently, it has been reported that Arabidopsis grown under natural LD condition flowers earlier than that grown under laboratory LD condition, in which a red (R)/far-red (FR) ratio of light sources determines FT expression levels. Additionally, FT expression profile changes in response to combinatorial effects of FR light and photoperiod. FT orthologs exist in most of plants and functions are thought to be conserved. Although molecular mechanisms underlying photoperiodic transcriptional regulation of FT orthologs have been studied in several plants, such as rice, however, dynamics in expression profiles of FT orthologs have been less spotlighted. This review aims to revisit previously reported but overlooked expression information of FT orthologs from various plant species and classify these genes depending on the expression profiles. Plants, in general, could be classified into three groups depending on their photoperiodic flowering responses. Thus, we discuss relationship between photoperiodic responsiveness and expression of FT orthologs. Additionally, we also highlight the expression profiles of FT orthologs depending on their activities in flowering. Comparative analyses of diverse plant species will help to gain insight into molecular mechanisms for flowering in nature, and this can be utilized in the future for crop engineering to improve yield by controlling flowering time.
1 Introduction
Plants have evolved their flowering strategies to maximize reproductive success. Both environmental and endogenous signals influence the timing of flowering via integration in photoperiod, temperature, vernalization, aging, phytohormone, autonomous pathways, and carbohydrate metabolism (Teotia and Tang, 2015; Rehman et al., 2023). Among these signals, photoperiod is considered as the most crucial environmental cue for flowering time regulation, as it is a stable indicator of the seasonal changes. Indeed, photoperiod-dependent flowering regulation is likely conserved across angiosperms. Flowering plants can be categorized into three major groups based on photoperiodic responses for flowering induction: short day plants (SDPs), long day plants (LDPs), and day-neutral plants (DNPs). SDPs flower when the daylength is at or shorter than a certain length, while flowering in LDPs is induced as daylength increases. Flowering in DNPs is not dependent on photoperiod. Photoperiodic flowering regulation has been extensively studied in a model plant, Arabidopsis, which is a representative LDP. The onset of flowering is determined by the expression levels of FT (Krzymuski et al., 2015). FT mRNA is almost undetectable in unfavored SD conditions. However, FT expression gradually increases as the daylength increases. FT is mainly synthesized in a subset of phloem companion cells in the distal part of leaves (Takada and Goto, 2003; Chen et al., 2018). Arabidopsis recognizes the different photoperiods and rapidly determines the expression of FT. When SD-grown plants are transferred to LD, FT expression is promoted on the first day of transfer (Krzymuski et al., 2015). In line with this, Arabidopsis flowering can be promoted with exposure to a single LD light regime. It should be noted that, despite the positive correlation between FT expression and the onset of flowering time, FT-mediated flowering is not solely dependent on FT expression in leaves. Since FT is mainly transcribed and synthesized in leaves and then FT protein moves from leaves to the shoot apical meristem (SAM), where it functions to induce flowering, the amount of FT protein translocated into SAM could be determined by several regulatory mechanisms including stability of FT mRNA (Seo et al., 2011; Wu et al., 2013), the success of FT transport processes (such as uploading to and unloading from the phloem) (Liu et al., 2012; Zhu et al., 2016; Endo et al., 2018; Susila et al., 2021), the stability and activity of FT protein (Qin et al., 2017; Xia et al., 2020), etc.
In addition to the photoperiodic regulation, FT expression levels are regulated in a time-of-day-dependent manner. FT expression peaks at the end of daytime (dusk) under laboratory LD conditions (Figure 1A). A B-box (BBX) transcription factor, CONSTANS (CO), is a major transcription factor activating FT expression under LDs. Overall expression profiles of CO are similarly maintained regardless of photoperiod, as CO expression is controlled by the circadian clock (Shim et al., 2017). In contrast, the stability of CO protein is regulated by light conditions. Combined with the circadian clock-mediated transcriptional regulation of CO expression and light-dependent CO protein stability, CO can accumulate only in LD conditions, consequently inducing FT expression and flowering. This regulatory mechanism of photoperiodic flowering fits well with the external coincidence model (Pittendrigh and Minis, 1964). Although CO is one of the most important factors in this photoperiodic regulation of FT expression, numerous factors have been identified as regulators of FT expression. A recent review has summarized the complicated regulatory networks involved in photoperiod-dependent FT expression (Takagi et al., 2023).
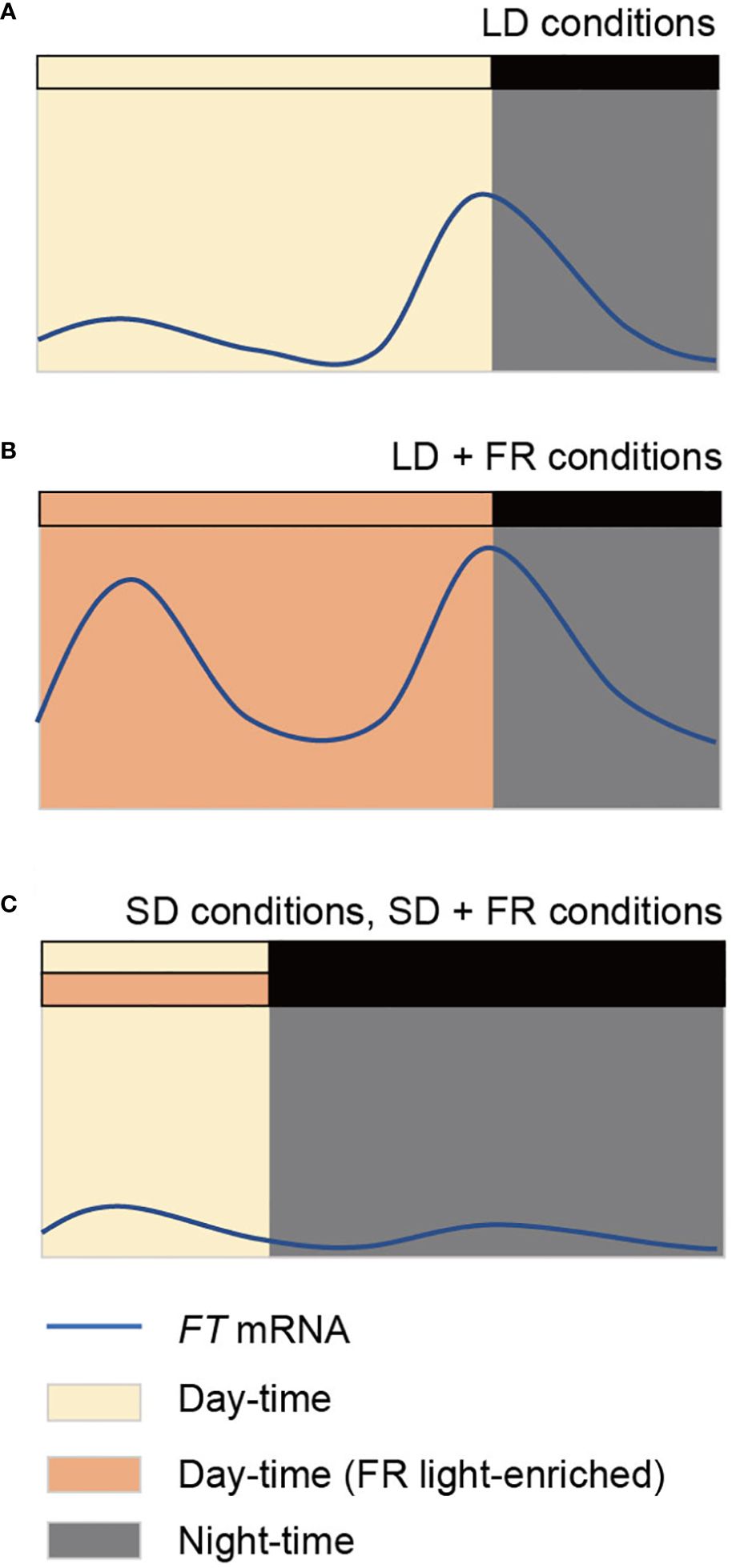
Figure 1 Diurnal expression profiles of FLOWERING LOCUS T (FT) in Arabidopsis seedlings. FT expression peaks once in a day at the end of day under laboratory long day (LD) conditions (A). Under natural LDs or when far-red (FR) light is supplemented in LD conditions (LD + FR), expression of FT peaks twice in a day in the morning and the evening (B). FT expression is low in short day (SD) conditions, both in the absence and presence of supplementary FR light (SD+FR) (C). Yellow and orange boxes indicate day period without and with supplementary FR light, respectively. Night period is indicated as dark or grey boxes.
Interestingly, a recent study has demonstrated that the FT expression profile differs when Arabidopsis grows in natural LD conditions compared in laboratory LDs, without altering the vasculature-specific expression pattern (Song et al., 2018) (Figure 1B). The expression of FT peaks in the morning as well as in the evening. The ratio of R/FR light is responsible for the variation in FT expression profiles between natural LDs and laboratory LDs. Typical laboratory conditions with fluorescent light sources have higher R/FR ratios than those of sunlight which has a R/FR ratio of approximately 1 (Holmes and Smith, 1977; Song et al., 2018). Laboratory LD conditions with supplementary FR light, mimicking the sunlight R/FR ratio, can create the bimodal FT expression profile and accelerate flowering (Song et al., 2018). In contrast, laboratory SD conditions with a R/FR ratio of 1 fail to induce FT expression (Figure 1C), suggesting that the regulation of FT expression in nature is still photoperiod-dependent. Among the previously identified FT-regulating factors, a R/FR light photoreceptor, phytochrome A (phy A), is necessary for FT expression especially in the morning. phyA induces FT expression partially via the stabilization of CO protein and increasing the activity of phytochrome interacting factors (PIFs) (Song et al., 2018; Lee et al., 2023). Although the supplementary FR light (adjusted R/FR ratio signal) in LD induces FT expression levels both in the morning and at dusk, they are likely regulated by different molecular mechanisms. Exposure to supplementary FR light does not immediately induce FT expression in the morning, while it rapidly induces the evening FT expression, suggesting that long-term transmission of light signal is required for the morning FT induction (Lee et al., 2023). This may explain the photoperiodic regulation of FT expression under FR light-enriched conditions. Nevertheless, detailed molecular mechanisms need to be elucidated.
The determination of when to flower is crucial for reproductive success, impacting crop yield including grain yield and biomass (Blümel et al., 2015). Therefore, understanding the photoperiodic flowering of various plant species is essential. It has been widely accepted that FT orthologs are evolutionally and functionally conserved in flowering plants, and the expression of FT orthologs is required to promote flowering, regardless of whether a plant is a SDP, LDP, or DNP. Given the observed differences in FT expression profiles in Arabidopsis depending on light quality, especially R/FR ratios, it prompts us to investigate FT orthologs in diverse plant species to understand the significance of expression profiles on flowering in nature. Additionally, while FT orthologs in many plant species share similar gene structures in general (Figure 2), their functions are, in fact, diversified, playing roles either activators or repressors in floral regulation. Hence, we also focus on relationship between floral activities of FT orthologs and their expression profiles.
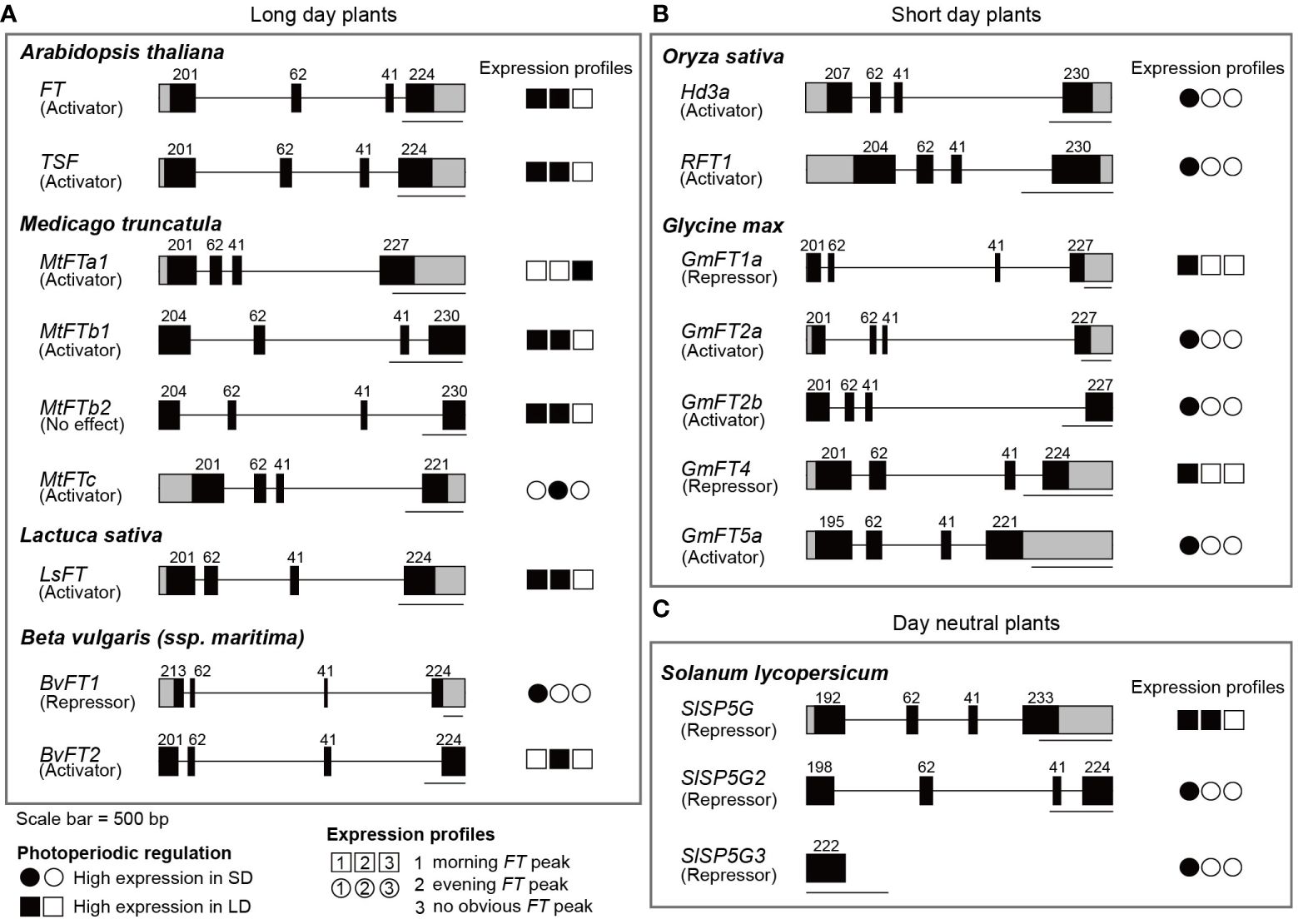
Figure 2 Gene structures and photoperiod-dependent expression profiles of FT and its ortholog genes. The gene structures of FT and FT orthologs in LDPs (A), SDPs (B), and a DNP (C) are indicated with their respective photoperiod-dependent expression profiles. Floral activities are denoted below the FT orthologs in parenthesis. In the gene structures, black and grey boxes represent exons and UTRs, respectively, and numbers above the exons indicate size of each exon in base pairs. A set of three circles or rectangles indicates expression patterns of FT ortholog in either SD or LD, depending on the photoperiod in which expression levels are high. The photoperiod-dependent expression profiles are denoted by the shape type; circles mean higher expression in SD than in LD, while rectangles mean higher expression in LD than in SD. Time-of-day expression profiles are indicated by the position of filled shapes; filled circles or rectangles denote expression peaks observed in the morning (position 1) or in the evening (position 2). A filled circle or rectangle at position 3 indicates that a gene is expressed irrespective of the time of day. Scale bar = 500 bp.
2 Conservation of expression profiles of FT orthologs across flowering plants
FT belongs to Phosphatidyl ethanolamine-binding protein (PEBP) family. A subfamily containing FT, named the FT-like clade, has been identified not only in angiosperms but also in gymnosperms (Jin et al., 2021). In flowering plants, the role of FT in flowering induction is evolutionally and functionally conserved. Besides its involvement in flowering time regulation, diverse roles of FT-like PEBP have been characterized, which has been summarized and reviewed (Jin et al., 2021). Although FT and its orthologs have distinct as well as shared roles, their gene structures are well-conserved (Almeida de Jesus et al., 2022; Liu et al., 2023) (Figure 2). Most FT orthologs consist of four exons, with few exceptions reported in banana and macadamia (Chaurasia et al., 2017; Yang et al., 2023). The size of exons is similar among FT orthologs, with the 2nd and 3rd exons having a consistent length (Liu et al., 2023). Conversely, the length of introns is variable, with an intron of over 3000bp present in sugar beet (BvFT1) and soybean (GmFT2a) among FT orthologs investigated in this review (Figure 2). The first intron of Arabidopsis FT, along with the promoter region, has been shown to be a regulatory region controlling FT expression in response to various environmental signals including light and temperature (Bratzel and Turck, 2015; Deng and Chua, 2015). Given that most FT orthologs have at least one intron longer than the average intron length of 165 bp in Arabidopsis (Feldmann and Goff, 2014), it is possible that mode of actions for regulation of FT expression by environmental changes is also conserved in various plant species.
Since a concept of florigen first arose in 1936 (Chailakhyan, 1936), extensive physiological analyses combined with molecular-genetic approaches in various plants have revealed that functionally conserved FT orthologs play key roles in flowering regulation. Regulatory mechanisms governing the expression of FT orthologs under diverse environmental conditions have been elucidated, particularly in model plants such as Arabidopsis and rice. In other plants, however, detailed regulatory mechanisms remain unclear. In this section, we compare expression profiles of FT orthologs in three plant groups, each exhibiting distinct responses to daylength for flowering: SDPs, LDPs, and DNPs. Additionally, we provide a summary of the effects of FR light on flowering regulation, accompanied by the expression of FT.
2.1 FT orthologs in LDPs
2.1.1 Light quality shapes FT expression profiles of Arabidopsis
The photoperiodic regulation of FT in LDPs has been extensively investigated by using Arabidopsis. The external coincidence model well fits into the photoperiodic regulation of flowering time in Arabidopsis, wherein flowering is induced only when a clock-controlled internal rhythm coincides with an external factor. The expression of CO follows a rhythmic pattern controlled by the circadian clock, gradually increasing from the middle of the day in LD conditions (Imaizumi, 2010). This increase is facilitated by the action of GIGANTEA (GI) and FLAVIN-BINDING, KELCH REPEAT, F-BOX (FKF1). GI expression, also regulated by the circadian clock, peaks in the middle of the day (Fowler et al., 1999). In LD conditions, the blue light receptor FKF1 accumulates and activates in the midday, forming a complex with abundant GI protein, subsequently activating CO expression. The stability of CO protein is light-regulated. HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENE1 (HOS1) and CONSTITUTIVE PHOTOMORPHOGENIC 1 (COP1)-SUPRESSOR OF PHYA-105 1 (SPA1)/-SPA3-SPA4 complex destabilize CO protein in the morning and in the night, respectively (Shim et al., 2017). In LD conditions, high CO expression coincides with light period, which in turn induces FT expression. In contrast, in SD conditions, when CO expression level is high, the dark period prevents CO protein stabilization and subsequent FT activation. This mechanistic model is based on the observed FT expression profile in laboratory LD/SD conditions. However, in nature, a morning FT peak as well as the evening FT peak appears. It has been shown that different R/FR ratios between sunlight (R/FR = 1) and laboratory conditions (R/FR > 2) determines the expression profiles (Song et al., 2018). Indeed, supplementary FR light in laboratory LD conditions mimics the bimodal FT expression profile observed in nature. Although the supplementary FR light induces the FT expression levels both in the morning and at dusk, the mechanisms underlying FR light-mediated FT induction likely differ between morning and evening. Firstly, the evening FT expression is induced in response to the supplementary FR light exposure as short as 2 hours (Lee et al., 2023). However, unlike induction of PIL1 expression, a marker of shade responses enriched by FR light, the evening FT expression could be induced when the 2 hr supplementary FR light was irradiated 6 hr ahead of dusk. This suggests that the evening FT expression is influenced by light quality (R/FR ratio) and the circadian clock, which is consistent with the classical external coincidence model. Secondly, the morning FT induction requires prolonged FR light exposure. Neither a 2hr, 4hr, or 8hr FR light exposure is sufficient for full morning FT expression (Lee et al., 2023). phyA, but not phyB, has thought to be crucial for the morning FT induction under natural LD or FR light-enriched LD conditions. Mutation on phyA abolishes the morning FT induction under FR light supplemented LD conditions, while mutation on phyB does not affect the morning FT expression level in laboratory LD conditions (Song et al., 2018). Furthermore, end-of-day FR light treatment accelerates flowering, mediated by phyB but not phyA (Cerdán and Chory, 2003). These findings, combined with the requirement of prolonged FR light exposure, support the notion that the morning FT expression is a FR high-irradiance response (HIR) via phyA. As part of phyA-mediated FT expression regulation, PIF7, together with other PIFs including PIF4 and PIF5, directly activates FT expression (Lee et al., 2023). EARLY FLOWEIRING 3 (ELF3) has shown to antagonize phyA in the regulation of morning FT expression. elf3 mutant significantly increased FT expression in the morning under both FR light-enriched LD conditions and laboratory LD conditions. ELF3 affects FT expression by regulating the stability of CO protein, in which ELF3 interacts with the COP1/SPA1 complex (Laubinger et al., 2006; Jang et al., 2008; Yu et al., 2008; Huang et al., 2016). Under simulated natural LD conditions, the abundance of ELF3 protein is lower compared to laboratory LD conditions, and the stability of CO protein increases throughout the day when ELF3 is mutated (Song et al., 2018). This suggests that supplementary FR light reduces ELF3 protein stability, thereby stabilizing CO protein and resulting in morning FT expression. Based on previously reported FT regulators and their molecular mechanisms, phyA and ELF3 have been identified as regulators of morning FT expression. However, considering observations that none of mutants except phyA and fhy1fhl examined by Song et al., 2018 specifically affected morning FT expression, it is possible that the regulation of morning FT expression is complex. Indeed, in addition to the prolonged FR light exposure, the induction of morning FT expression appears to be controlled by the circadian clock, as indicated by the presence of time windows most effective for FT induction by the supplementary FR light (Lee et al., 2023). FR light treatment from the late afternoon in the previous day to the early morning is sufficient to fully induce morning FT expression. Thus, the regulation of morning FT expression is tightly governed by both the circadian clock and prolonged FR light signals. Further investigations are required to elucidate the molecular mechanisms underlying FT expression under FR light enriched LD conditions.
Arabidopsis plants under canopy exhibit earlier flowering compared to those fully exposed to light. They perceive shaded conditions by sensing R/FR ratios, which is mediated by photoreceptors including phys. In shade, neighboring plants absorb R light but reflect FR light, leading to a decreased amount of R and an increased FR light. As a result, the R/FR ratio varies depending on the degree of shade. In response to the reduced R/FR ratio under shade, active phys turns into inactive state, triggering shade avoidance responses including acceleration of flowering (Casal, 2012). The early flowering phenotype in shade is predominantly dependent on phyB with contributions from phyD and phyE (Devlin et al., 1999, 1998). On the other hand, phyA has thought to be less effective in flowering regulation under shade, even though it mediates other shade avoidance responses (Franklin et al., 2003; Roig-Villanova and Martínez-García, 2016). It has been shown that both morning and evening FT expression gradually increase as the R/FR ratio decreases from normal laboratory white light (R/FR ratio = 2.0) to shade conditions (R/FR = 0.25) (Song et al., 2018). Given that the induction of FT expression requires functional phyA, these observations suggest that the phyA-mediated regulatory mechanism for FT induction under FR-enriched LD conditions may also partially contribute to early flowering under shade conditions.
2.1.2 Expression profiles of FT orthologs in other LDPs
Medicago truncatula (Medicago), belonging to the rosidae subclass along with Arabidopsis, is a model plant of legumes that can mediate nitrogen fixation. Its flowering is promoted by long days (Clarkson and Russell, 1975). Medicago possesses five FT ortholog genes, MtFTa1, MtFTa2, MtFTb1, MtFTb2, and MtFTc. Among these, MtFTa1, MtFTb1, and MtFTc exhibit flowering activation activity, as demonstrated by the complementation of the late flowering phenotype in Arabidopsis ft mutants upon overexpression of each FT ortholog gene (Laurie et al., 2011). The role of MtFTa1 in floral induction has been further confirmed in Medicago, however, mtftc mutants did not affect flowering in Medicago (Laurie et al., 2011). Except for MtFTa2, the expression of the four MtFT genes is dependent on photoperiod (Table 1). In LD conditions, MtFTa1, MtFTb1, and MtFTb2 are expressed, while their expression is either absent or marginal in SD conditions (Laurie et al., 2011). This implies that MtFTa1 and MtFTb1 are responsible for photoperiodic flowering regulation, while MtFTb2 may contribute to photoperiodic responses other than flowering. MtFTb1 and MtFTb2 show bimodal expression profiles in LD conditions with peaks in the morning (Zeitgeber Time, ZT4) and at dusk (ZT16), reminiscent of the Arabidopsis FT expression profile observed in FR-enriched LD conditions. In contrast, MtFTa1 expresses consistently throughout the day in LD. The expression of MtFTc also appears to be regulated by photoperiod, but unlike MtFTa1, MtFTb1, and MtFTb2, its expression level is higher in SD than in LD conditions (Laurie et al., 2011)(Table 1). In SD conditions, MtFTc expression levels start to increase in the morning and peak after dusk (during the night-time period), which is a unique pattern among the FT orthologs examined in this review (Laurie et al., 2011) (Table 1). Moreover, MtFTc expresses at a low level in leaves in LD conditions. This expression profile suggests that MtFTc may have little or weak contribution to flowering time regulation in LD, and its high expression in SD conditions implies a potential role in regulating flowering time in SD. Conversely, MtFTa2 is expressed in both LD and SD conditions. Its expression level in LD remains relatively constant throughout the day, while it peaks after dusk in SD conditions, similar to MtFTc. The inability of ectopic expression of MtFTa2 in Arabidopsis to rescue the late flowering of the ft mutant, combined with its high expression in tissues other than leaves such as roots, suggests diversified role(s) for MtFTa2 beyond flowering time regulation in Medicago. In addition to FT orthologs, orthologs in Medicago of photoperiodic flowering time-regulating factors such as CO and CO-like (COL), GI, and FKF1 genes have been identified, though their functions in flowering remain to be resolved or appear to be diversified (Hecht et al., 2005; Wong et al., 2014). Medicago possesses a single copy of PHYA gene. MtPHYA positively regulates flowering in LD, while its effect becomes weaker in SD, suggesting a photoperiod-dependent contribution of phyA to flowering (Jaudal et al., 2020). Despite the functional conservation of phyA between Medicago and Arabidopsis in flowering, it has been shown that flowering in Medicago sativa (M. Sativa), a close relative Medicago truncatula, is delayed under shade conditions. This shade-induced delay of flowering was also observed under a R/FR ratio of 0.8 (Lorenzo et al., 2019). Given the sunlight R/FR ratio close to 1, M. Sativa in its natural habitat may flower later than in laboratory LD conditions. Under shade with a R/FR ratio of 0.4–0.6, the expression levels of MsFTa1 and MsFTb1 were reduced around dusk compared to normal LD conditions, which is consistent with the reduction of expression of SQUAMOSA PROMOTER BINDING LIKE 3 (SPL3), a floral activator (Lorenzo et al., 2019). However, it should be noted that floral activators such as PIF3 and HB2 are up-regulated in the same condition. Further analysis is needed to understand how the regulatory pathway of flowering responds to supplementary FR light. The delay in flowering under shade in M. Sativa is likely a diverged strategy to increase survival rates by choosing the accumulation of resources instead of flowering under unfavorable conditions, as a characteristic of a perennial plant. However, Medicago truncatula, an annual plant, also exhibits similar responses to shade or supplementary FR light; flowering is delayed in shaded conditions (Mauro et al., 2014), and HB2 expression is up-regulated by supplementary FR light (Zhang et al., 2020). The delay in flowering due to shade has also been reported in another legume plant, soybean. Flowering of soybean is delayed under low R/FR light quality (Cober and Voldeng, 2001) (Discussed in Section 2.2.2). Thus, the determination of flowering time in response to different R/FR ratios may be governed by legume-specific mechanisms rather than whether it is an annual or a perennial plant.
Lactuca Sativa (Lettuce), a dicot like Arabidopsis, belongs to the asteridae subclass, and its flowering is induced with longer day lengths. Lettuce has been shown to have one copy of the FT ortholog gene, LsFT, which functions as a floral activator (Fukuda et al., 2011) (Table 1). While it remains unclear whether photoperiod affects LsFT expression levels, the rates of its accumulation during development in different cultivars positively correlate with floral induction (Fukuda et al., 2017), further supporting the functional conservation of FT ortholog in Lettuce. Under LD conditions, LsFT exhibits high expression in the morning and around dusk (Sgamma et al., 2016), resembling the expression pattern observed in Arabidopsis grown under FR light-enriched LD conditions. However, this result was obtained from lettuce grown in a greenhouse, thus it needs to be elucidated if the expression profile of LsFT is also controlled by FR light that is enriched in natural light conditions. Nevertheless, the sole FT-like gene in lettuce, LsFT, behaves similarly to Arabidopsis FT. FR light likely accelerates flowering in lettuce, as supplementary FR light in the presence of R/Blue/Green light increased stalk heights, indicative of a premature flowering response (Alrajhi et al., 2023). However, little is known whether LsFT is responsible for floral induction by the supplementary FR light. Being one of the most popular leafy vegetables, delayed flowering in lettuce is preferred to prevent stem elongation and bitterness (Assefa et al., 2019). Therefore, understanding the flowering time pathway of lettuce is crucial.
Beta vulgaris (Beet) belongs to Caryophyllidae, the third-largest subclass of the class Magnoliopsida (dicotyledons). Beet possesses two FT-like genes, BvFT1 and BvFT2, which interestingly exhibit opposing functions in flowering regulation. BvFT2 promotes flowering, while BvFT1 represses flowering in both beet and Arabidopsis when ectopically expressed (Pin et al., 2010) (Table 1). The antagonistic roles of BvFTs appear to be conferred by sequence variations in segment B of the fourth exon, encoding an external loop of PEBP proteins. In this segment, both BvFT proteins share a conserved key amino acid, Glutamine (Q), at residue 144 and 140 in BvFT1 and BvFT2, respectively, that differentiates FT from its close PEBP protein TFL1, a floral repressor (Ahn et al., 2006). However, only BvFT2 has sequence conservation throughout the entire segment B. Indeed, swapping the segment B sequences in BvFTs altered their inherent activity in opposite ways (Pin et al., 2010). In addition to their opposing roles, expression profiles of the two BvFTs differ. Similar to Arabidopsis FT in laboratory conditions, BvFT2 expresses high in LD but not in SD conditions with a peak around dusk in annual beets (Pin et al., 2010). On the contrary, BvFT1 is expressed highly in SD with a peak after dawn (Pin et al., 2010). In biennial beets without vernalization, the expression of BvFTs is photoperiod-independently regulated. BvFT1 expresses in both SD and LD with the same diurnal profile, while BvFT2 does not express, which is consistent with essential role of vernalization for floral induction in biennial plants. After vernalization, the expression profiles of each BvFTs resemble those in annual beets. As a root crop, delaying flowering in the first year of the biennial plant’s life cycle is necessary for increasing leaf and root mass for sugar and bioethanol production. Several flowering-regulating genes have been identified and characterized in the regulation of beet flowering, such as BOLTING TIME CONTROL 1 (BvBTC1), a key determinant of vernalization requirements between annual and biennial plants (Pin et al., 2012). As a positive regulator of flowering, BvBTC1 is likely involved in the responsiveness to LD, as shown that reduced BvBTC1 expression by RNAi caused induction of BvFT1 expression and reduction of BvFT2 expression (Pin et al., 2012). Additionally, several reports have demonstrated the negative effect of shade on leaf and root biomass of beet (Artru et al., 2018; Schambow et al., 2019). In 1965, Lane et al. showed that by using different types of lamps, flowering time varied depending on R/FR ratios. In general, supplementary FR light promotes flowering in beets in the presence of R light. Thus, beet determines flowering time in response to the light quality, likely via mechanisms similar to phyA-mediated flowering regulation in Arabidopsis.
2.2 FT orthologs in SDPs
2.2.1 Two FT orthologs, Hd3a and RFT1, in rice
The mechanisms governing the photoperiodic regulation of flowering in SDPs have been demonstrated mostly in rice, one of the world’s major crops. Rice, a monocotyledonous flowering plant in the grass family Poaceae, initiates flowering in response to daylengths shorter than 13.5h (Itoh et al., 2010). In rice, two FT-like genes, Heading date 3a (Hd3a) and RICE FT1 (RFT1), play crucial roles as mobile florigens synthesized in leaves and essential for flowering (Kojima et al., 2002; Tamaki et al., 2007; Komiya et al., 2008) (Table 1). The expression of these florigens is responsive to the photoperiod, with higher expression levels observed in SD conditions. Under SD conditions (10 h light, 30°C/14 h dark, 25°C), the expression of Hd3a and RFT1 peaks at or before dawn, and then gradually decrease during the day (Izawa et al., 2002; Komiya et al., 2008). The diurnal expression profiles of Hd3a and RFT1 are regulated by the activity of OsGI, whose expression is under the control of the circadian clock with a peak at the end of day (Hayama et al., 2003, 2002; Lee and An, 2015). OsGI is necessary for the diurnal expression of HEADING DATE 1 (Hd1), a rice ortholog of CO, which activates the expression of Hd3a and RFT1 in the morning (Hayama et al., 2003). This pathway resembles the CO-FT pathway in Arabidopsis. Additionally, a rice-specific pathway has been identified. EARLY HEADING DATE 1 (Ehd1) encoding a B-type response regulator is upregulated by OsGI when blue light coincides with the morning phase, and activates Hd3a and RFT1 expression (Doi et al., 2004; Itoh et al., 2010). Ehd1-mediated flowering regulation pathway has thought to be absent in Arabidopsis, although its homologs are found in Arabidopsis and other plants (Bar et al., 2008). It plays a central role in photoperiod-mediated flowering in rice, which is recently extensively reviewed in Vicentini et al., 2023. Conversely, in LD conditions, Ehd1 expression is repressed by GRAIN NUMBER, PLANT HEIGHT AND HEADING DATE 7 (Ghd7), encoding a CCT-domain protein highly expressed in LD (Xue et al., 2008; Itoh et al., 2010). Despite lower expression levels of Hd3a and RFT1 in LD conditions compared to SD conditions, RFT1 has shown to be a major floral activator under LD conditions (Komiya et al., 2009). Indeed, RFT1 shows diurnal expression profile with a peak in the early morning and gradually increasing over the night (Komiya et al., 2009). The current model of photoperiodic flowering regulation mediated by Hd3a and RFT1 relies primarily on their on-off gene expression profiles between SD and LD, respectively. As the photoperiod lengthens in LD conditions, high expression of Hd1 coincides with the daytime phase. However, in contrast to the role of CO in Arabidopsis, Hd1 acts as a repressor for the expression of Hd3a and RFT1 in LD conditions (Hayama et al., 2003). Consequently, the accumulation of Hd1 suppresses the expression of Hd3a and RFT1, leading to reduced expression around dusk. Nevertheless, this model cannot explain the circadian clock-regulated RFT1 expression in LD, which is important for flowering in LD. The similar diurnal expression profiles of RFT1 between SD and LD suggest that photoperiodic regulation in rice differs from that in Arabidopsis. Ehd1 expression in LD showed similar expression patterns with RFT1 (Komiya et al., 2009), thus release of suppression of Ehd1 expression in LD conditions may accomplish with developmental processes.
FR light has been identified as a positive regulator flowering in rice, as demonstrated by experiments such as 2-hr FR light irradiation before a 10-hr dark period in LD conditions (14-hr light/10-hr dark) promoting flowering (Ikeda, 1985). In Arabidopsis, end-of-day (EOD)-FR light treatment counteracts the negative impact of R light by inactivating phyB (Smith, 1982; Franklin and Whitelam, 2005). Although the induction of flowering in Arabidopsis by EOD-FR light treatment is not solely dependent on phyB (Bagnall et al., 1995; Xie et al., 2020), it is possible that the 2-hr FR light irradiation before the dark period affects phyB activity in rice for floral induction. This hypothesis is supported by evidence that EOD-FR in SD conditions (10-hr light/14-hr dark) accelerated flowering in rice in a phyB-dependent manner (Takano et al., 2005). Another possibility is that the FR light signal itself, perceived by photoreceptors like phyA, plays a role in mediating flowering promotion, similar to Arabidopsis. Several observations support this idea: 1) 3-hr FR light was more effective at inducing flowering than 1-hr or 2-hr light treatment before the dark period in LD, and this FR light irradiation was more effective than extending the dark-period (Ikeda, 1985), 2) phyA mutant consistently exhibited delayed flowering in SD conditions (10-hr light/14-hr dark) (Takano et al., 2005; Ishikawa et al., 2009), which is different from the action of phyB, 3) simulated shade conditions accelerated flowering time, inducing RFT1 but not Hd3a expression (Cui et al., 2023). Given that phyA’s function in FR high-irradiance response (HIR) has been reported in photomorphogenesis in rice (Takano et al., 2005), similarly to Arabidopsis, these findings suggest a role of phyA in responding to prolonged FR light in flowering time regulation through the modulation of RFT1 (and Hd3a) expression.
2.2.2 Expression profiles of FT orthologs in other SDPs
Soybean (Glycine max) belongs to the rosidae subclass together with Arabidopsis and Medicago, however, unlike Arabidopsis and Medicago, its flowering is induced under short days. Soybean possesses ten FT-like genes. GmFT2a/2b, GmFT3a/3b, GmFT5a/5b are known to activate flowering, while GmFT1a/1b, GmFT4, and GmFT6 function as floral repressors (Lee et al., 2021) (Table 1). Their expression is controlled by photoperiod (Lee et al., 2021). The floral activators, GmFT2a/2b and GmFT5a, exhibit high expression levels in SD, peaking in the morning. On the other hand, GmFT1a and GmFT4, the floral repressors, are suppressed in SD but highly induced in LD. Interestingly, despite GmFT1a and GmFT4 showing high expression in LD conditions, their expression peaks are observed only in the morning. This is in contrast to FT orthologs of LDPs, where expression peaks either once dusk (e.g., Arabidopsis FT in the laboratory LD conditions) or twice in the morning as well as in the dusk (e.g., Arabidopsis FT in natural LD conditions). Moreover, in contrast to the role of phyA in flowering in Arabidopsis, soybean phyAs (GmPHYA2 and GmPHYA3) are known to negatively regulate flowering time. They achieve this by increasing the expression of the legume-specific regulator E1, which, in turn, represses the expression of the floral activators GmFT2a and GmFT5a while activating the expression of the floral repressors GmFT1a and GmFT4 (Zhai et al., 2014; Cao et al., 2017; Liu et al., 2018). E1 is specifically expressed in LD, but not in SD, with two peaks at both dawn and dusk in LD conditions (Xia et al., 2012), resembling the expression pattern of FT in Arabidopsis plants grown under FR light enriched-LD conditions. GmPHYAs promote E1 expression, partially through the regulation of circadian clock genes, the GmPRR3a/GmPRR3b(Tof11/Tof12)-GmLHY module. GmPHYs upregulate the expression of GmPRR3a/GmPRR3b, which then directly represses the expression of GmLHY genes. Subsequently, this relieves GmLHY-mediated suppression of E1, inducing its expression (Lu et al., 2020). Notably, mutations in GmLHY genes (lhy1a lhy1b lhy2a lhy2b) induces E1 expression at dusk in LD, while the morning E1 expression remains unaffected in the mutant (Lu et al., 2020), implying that E1 expression in the morning and at dusk may be regulated by at least two different modes of action, similar to the regulation of Arabidopsis FT expression. The soybean case study suggests that although the activity of FT orthologs has diversified as either activators or repressors, regulatory mechanisms for bimodal expression in flowering may have been conserved through the combined action of photoperiod and circadian clock. In addition to the negative effect of phyA on soybean flowering, low R/FR ratios differentially affect soybean flowering compared to Arabidopsis (Table 1). Although shade avoidance responses such as elongation of seedlings and stem, and reduction of leaf area and size are observed in soybeans under various growth conditions similar to those in Arabidopsis (Green-Tracewicz et al., 2012; Yang et al., 2014; Wu et al., 2017), onset of soybean flowering in LD (20 h Light/4 h Dark) is delayed as the R/FR ratio decreases, in which E7 is required (Cober and Voldeng, 2001). Under LD (20 h Light/4 h Dark) with a R/FR ratio of 2.7, flowering time in near-isogenic lines with E7 allele and e7 allele is indistinguishable. However, when the R/FR ratio is similar to or lower than the sunlight R/FR ratio, soybean lines with the E7 allele significantly delay flowering compared to those with the e7 allele. Although a low R/FR ratio in SD (12 h Light/12 h Dark) slightly delays flowering, the responsiveness is much weaker compared to that in LD. As phyA is thought to mainly function in LD to repress flowering in soybean, the different responsiveness to R/FR ratios between LD and SD may be due to activity of phyA. Not only soybeans but also other legume plants, including winged bean (Psophocarpus tetragonolobus) (Raai et al., 2020) and Medicago (as described in Section 2.1.2), have shown to delay flowering in response to shade. Lentil (Lens culinaris) have been reported to flower earlier in low R/FR conditions than in high R/FR conditions (Yuan et al., 2017), however, it should be noted that the R/FR ratio in this low R/FR condition is 1.6, even higher than the sunlight R/FR ratio. Thus, it is possible that legumes can induce a series of typical shade avoidance responses but not early flowering. Instead, they may have evolved to delay flowering under shade in a legume-specific manner.
2.3 FT orthologs in tomato, a day neutral plant
Solanum lycopersicum (tomato) belongs to the Asteridae subclass, along with lettuce. Tomato is typically classified as a DNP (Mizoguchi et al., 2007). Six FT-like genes in tomato have been identified (Table 1). Among them, SINGLE-FLOWER TRUSS/SELF-PRUNING 3D (SFT/SP3D) functions to induce flowering, while SP5G, SP5G2, SP5G3 function as floral repressors (Cao et al., 2018, 2016). SFT expresses photoperiod-independently, which is consistent of tomato being a DNP. However, its expression profiles differ between SD and LD conditions. Expression levels of SFT are relatively stable in SD, while it peaks in the morning in LD (Cao et al., 2016). Unlike SFT, the expression of three floral repressors, SP5G, SP5G2, and SP5G3, is photoperiod-dependently regulated. Expression of SP5G is highly expressed in LD, but not in SD, with two peaks in the morning and the evening (Cao et al., 2016), similar to the expression profiles of Arabidopsis FT and FT orthologs acting as floral activators in LDPs. Conversely, SP5G2 and SP5G3 are highly expressed in the morning in SD conditions. The diurnal expression profiles of SP5G2 and SP5G3 resemble those of the FT-like genes in SDPs and the floral repressor of beet, BvFT1 (Figure 2, Table 1). Although tomato flowering is regulated photoperiod-independently, the diurnal expression profiles of SP5G, SP5G2, and SP5G3 imply that photoperiodic regulation of gene expression exists in tomato.
Compared to the significant contribution of phytochromes and other photoreceptors in Arabidopsis to flowering regulation (Takagi et al., 2023), the effects of tomato photoreceptors on flowering time have been considered much weaker, with no obvious flowering phenotype observed in mutants of the photoreceptors, except for the blue light photoreceptors, cryptochrome 1a (cry1a)/cry2 mutant (Fantini et al., 2019). Indeed, the effect of supplementary FR light on tomato flowering varies depending on experimental conditions. Under a 12 h Light/12 h Dark cycle photoperiod, tomatoes flower later in low R/FR (0.6) ratio conditions than in conditions with R/FR ratios of 7.4 or 1.2 (Cao et al., 2018). In line with this, the expression of FT-like floral repressors, SP5G and SP5G2, is up-regulated. However, the delay in tomato flowering in this condition is mainly dependent on phyB1 rather than phyA. On the other hand, tomatoes grown in LD (16 h Light/8 h Dark) conditions with supplementary FR light in addition to broad-spectrum lighting in a greenhouse flower earlier than those without FR light (Meijer et al., 2023). Similarly, under R light-abundant LD conditions (16 h Light/8 h Dark, a mixture of 95% R light and 5% B light), the onset of flowering is accelerated in response to an increase in supplementary FR light (Kalaitzoglou et al., 2019). Given that the small change in R/FR ratios from the laboratory (>2.5) to sunlight (c. 1) results in different FT expression profiles in Arabidopsis, the different response of flowering may be attributed to the different experimental conditions with or without exposure to sunlight.
3 Future perspectives
The extensive study of FT orthologs has primarily focused on their functional conservation in flowering time regulation. However, the significance of their expression profiles has received less attention. The investigation of FT orthologs across various plants indicates that there may be conserved regulatory mechanisms underlying their expression. To advance our understanding in this area, future research should aim to establish rules governing the expression profiles of FT orthologs. This could involve:
3.1 Balance between activators and repressors in FT orthologs
FT-like PEBP proteins behave either as floral activators or repressors, with a conserved amino acid distinguishing FT orthologs from the floral repressor TFL1. Apart from sequence disparities, their expression profiles and regulatory mechanisms differ significantly. FT is expressed in leaves, and FT protein moves to the SAM. Conversely, TFL1 is expressed in the center of the SAM to repress flowering (Conti and Bradley, 2007; Baumann et al., 2015). In addition to spatial regulation, FT expression is temporally controlled in response to photoperiod as described above, while little is known about photoperiod dependency of TFL1 expression (Higuchi et al., 2013; Wickland and Hanzawa, 2015). The expression of FT-like floral repressors, such as BvFT1 in beet, Gm1a and GmFT4 in soybean, and SP5G, SP5G2, and SP5G3 in tomato, is photoperiod-dependent. Under non-facultative photoperiod conditions, the expression of these floral repressors, except for GmFT6 in soybean, tends to increase. This tendency is consistent with their function to repress flowering. Conversely, the expression of FT-like floral activators, except for MtFTc in Medicago, increased only under facultative photoperiod conditions. This implies that expression of both FT-like floral activators and repressors is under photoperiodic control, contributing the balance to determine flowering. The FT-like floral repressor in tobacco, NtFT2, interacts with Arabidopsis FD (Harig et al., 2012), suggesting that FT-like floral repressors, in addition to the floral activators, likely bind to FD orthologs. The floral repressor TFL1, which competes with FT in the SAM to repress flowering in Arabidopsis, also binds to FD protein (Abe et al., 2005; Wigge et al., 2005; Zhu et al., 2020). However, it is noted that the expression of TFL1 is restricted to the SAM. The FT-like floral repressors are thought to be expressed in leaves like the FT-like floral activators. In soybean, GmFT1a and GmFT4 are specifically expressed in cotyledons and trifoliate and unifoliate leaves (Liu et al., 2018; Lee et al., 2021). BvFT1 is also mainly expressed in leaves (Pin et al., 2010), and SP5G, SP5G2 and SP5G3 in tomato are expressed high in leaves and cotyledons (Cao et al., 2016). However, translocation of these FT-like floral repressors from leaves to the SAM through transporters similar to those responsible for FT-like floral activators, such as FT-INTERACTING PROTEIN 1 (FTIP1) and SODIUM POTASSIUM ROOT DEFECTIVE 1 (NaKR1) in Arabidopsis (Liu et al., 2012; Zhu et al., 2016), remains unexplored. Considering the similarity in expression profiles among the FT orthologs in the same plant, competition between activators and repressors may begin in the cells where they express. Thus, comprehensive investigations into the transcriptional regulatory mechanisms of multiple FT-like floral regulators are essential for understanding flowering in various plants.
3.2 Similar expression profiles of FT orthologs in different photoperiods
Due to the limited information available for SDPs, we summarized FT orthologs from two SDPs in this review. Therefore, we cannot generalize the expression profiles of FT orthologs of SDPs. However, we observed that all FT orthologs of SDPs examined in this review are expressed only in the morning, even under LD conditions, which differs from the expression profiles of FT orthologs in LDPs (Table 1, Figure 2). This observation can be further categorized into two cases; 1) different FT orthologs are expressed with similar profiles under different photoperiod, as seen in the case of soybean. 2) the same FT orthologs are expressed with the same profiles under different photoperiods, although the expression levels differ, as seen in the case of rice. In soybean, the peak expression time of floral activators (GmFT2a and GmFT5a) under SD conditions coincides with that of floral repressors (GmFT1a and GmFT4) under LD conditions. Rice FT orthologs, Hd3a and RFT1, peak only once in a day in the morning, both in SD and in LD. According to the prevailing model, photoperiod-dependent on-off expression could be explained by the functionality of major regulators, E1 in soybean and Hd1 and Ehd1 in rice. However, the time-of-day-dependent expression profiles of FT orthologs remain elusive. Interestingly, both plant species possess dual-function regulators: E1 in soybean and Hd1 in rice. In rice, the activity of Hd1 on Hd3a expression in LD is mediated by its interaction with DAYS TO HEADING 8 (DTH8) (Du et al., 2017). In SD, the Hd1-DTH8 complex is likely absent due to low expression of DTH8, resulting in the activation of Hd3a expression. Considering that E1 has been shown to possess transcriptional repression ability (Zhai et al., 2022), the activity of E1 is likely regulated by its binding partner(s). Additionally, both plant species have their unique regulatory factors: the legume-specific E1 and the rice-specific Ehd1. These pieces of evidence suggest that regulatory factors involved in the photoperiod-dependent expression of FT orthologs in SDPs remain undiscovered. Investigating whether SDPs have conserved regulatory mechanisms that confer the unique expression profiles of FT orthologs in SDPs would help expand our knowledge about the photoperiodic flowering regulation.
3.3 Multilayered regulatory mechanisms of FT transcript levels
Transcript levels are determined not only by de novo synthesis of mRNA but also by mRNA stability. However, quantitative RT-PCR, widely used for quantification of transcript levels, measures endpoint transcript levels, thus having a limitation in distinguishing between mRNA synthesis and its stability regulation. In Arabidopsis, FT mRNA stability is non-cell autonomously regulated by WEREWOLF (WER), an R2R3 MYB transcription factor (Seo et al., 2011). WER likely regulates the abundance of FT mRNA by stabilizing the transcripts rather than by increasing transcription. The wer mutant plants exhibited lower FT transcript levels compared to wild type, consequently displaying a late-flowering phenotype in a photoperiod-dependent manner (Seo et al., 2011). However, WER is expressed in both LD and SD conditions, and both WER mRNA and WER protein are mainly observed in the epidermis. These findings indicate that other factors are likely involved in the WER-mediated stabilization of FT mRNA. Therefore, further investigation is necessary to determine whether the photoperiod-dependent expression profiles of FT and its orthologs are shaped by de novo transcription and/or mRNA stability.
In addition to Arabidopsis FT, FT orthologs in Brachypodium distachyon and Cocos nucifera have been shown to be regulated at the post-transcriptional level (Wu et al., 2013; Qin et al., 2017; Xia et al., 2020). mRNAs of two FT orthologs in Brachypodium, BdFT1 and BdFT2, are targeted by miR5200 for mRNA cleavage (Wu et al., 2013). miR5200 expression is higher in SD conditions than in LD conditions, thus it inhibits the accumulation of BdFT1 and BdFT2 specifically in SD. Additionally, BdFT2 generates two different splicing isoforms, named BdFT2α and BdFT2β. BdFT2β lost its ability to bind to FD but retained the ability to form a heterodimer with BdFT2α or BdFT1, thereby resulting in attenuated formation of flowering inductive complexes (Qin et al., 2017). The mode of action derived by alternative splicing of BdFT2 is affected by an endogenous cue, as shown by gradual decrease in the BdFT2β/BdFT2α ratio as development progresses (Qin et al., 2017). Alternative splicing of FT orthologs has also been observed in wheat, barley and coconut (Qin et al., 2017; Xia et al., 2020). However, it remains unclear whether the alternative splicing of FT orthologs is photoperiod dependently regulated. Nevertheless, the accumulating evidence suggests that the mechanisms underlying the post-transcriptional regulation of FT transcripts may be conserved and regulated not only by endogenous cues but also environmental cues, such as photoperiod.
4 Conclusions
Since the identification of FT as a long-sought florigen in Arabidopsis, numerous studies have contributed to establishing the molecular mechanisms underlying how FT expression is elaborately regulated to determine flowering time. However, expression profiles of Arabidopsis FT appear to be differentially determined depending on light quality (Song et al., 2018; Lee et al., 2023). The current model hardly explains the bimodal FT expression profile in nature, particularly the morning FT expression. In this review, we aimed to gather and explore expression profiles of FT orthologs to gain insight into underlying regulatory mechanisms. Despite limitations in publicly available information for diurnal expression of FT orthologs in diverse plants, expression profiles of FT orthologs combined with their functional activity in flowering time regulation suggest that plants might share conserved regulatory mechanisms for the expression of FT orthologs. Firstly, FT orthologs tend to highly express in facultative photoperiodic conditions, as expected; high expression of FT orthologs of LDPs in LD conditions, while high expression of FT orthologs of SDPs in SD condition. Secondly, plants classified in the same type of photoperiodic flowering responses likely show similar expression profiles of FT orthologs; FT orthologs of LDPs peak either once in the late afternoon or twice in the morning and the evening in LD conditions, while FT orthologs of SDPs peak in the morning in SD conditions. Thus, not only FT orthologs per se but also regulatory mechanisms for the expression of FT orthologs may be conserved among plant species showing the same photoperiodic responses in flowering. However, it should be noted that, although FT orthologs in many plants are believed to function as florigens to promote flowering, several FT orthologs have shown to function as a repressor, which diversifies expression profiles of FT orthologs. The FT-like floral repressors examined in this review could be classified into two groups, based on their expression profiles. One consists of FT-like floral repressors that antagonistically express against the FT-like floral activators. These opposite expression patterns match with their role in flowering regulation to induce flowering only under favorable conditions. Another group contains FT-like floral repressors whose expression coincides with expression of FT-like floral activators in the same photoperiod and/or at the time of day. It remains unclear whether these FT-like floral repressors counteract the FT-like floral activators for flowering in the same conditions. Given the functional diversification of FT orthologs beyond floral regulation (Wang et al., 2022), it is possible that co-expression of FT-like floral activators and FT-like floral repressors may differentially contribute to processes other than flowering. Further exploration of redundancy among FT orthologs may give us clues to the significance of co-expression of FT orthologs.
Advanced molecular biology techniques, coupled with bioinformatics, has facilitated in-depth exploration of flowering across diverse plant species. Moreover, sophisticated analyses using Arabidopsis has opened a new chapter for regulatory mechanisms of FT expression in nature. The FT orthologs across plant species could be classified according to the similarity of expression profiles. Thus, comparative analyses within and between plant species might offer insights into molecular mechanisms. The effect of phyA or FR light on expression profiles of FT orthologs in plants beyond Arabidopsis has received less attention. Most plants examined in this review respond to the supplementary FR light to promote flowering and phyA functions as a positive regulator, although soybean has shown the opposite effect of phyA and FR light on flowering. Investigation into the interplay between FR light (and phyA) and the expression of individual FT orthologs may be helpful to understand flowering in diverse plant species in their natural environments.
Author contributions
NL: Funding acquisition, Writing – original draft, Writing – review & editing. JS: Writing – review & editing, Funding acquisition. M-KK: Writing – review & editing. MK: Funding acquisition, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Research Foundation of Korea (Grant NRF 2021R1A5A8029490, 2022M3A9I3018121 and RS-2023-00301974); The Cooperative Research Program for Agriculture Science and Technology Development (Project No. PJ01577902), Rural Development Administration, Republic of Korea; The Technology Development Program (grant number, 20014582) funded by the Ministry of Trade, Industry & Energy (MOTIE, Korea).
Acknowledgments
We apologize for not being able to cite all exciting studies due to space limitations.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abe, M., Kobayashi, Y., Yamamoto, S., Daimon, Y., Yamaguchi, A., Ikeda, Y., et al. (2005). FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309 (5737), 1052–1056. doi: 10.1126/science.1115983
Ahn, J. H., Miller, D., Winter, V. J., Banfield, M. J., Jeong, H. L., So, Y. Y., et al. (2006). A divergent external loop confers antagonistic activity on floral regulators FT and TFL1. EMBO J. 25 (3), 605–614. doi: 10.1038/sj.emboj.7600950
Almeida de Jesus, D., Batista, D. M., Monteiro, E. F., Salzman, S., Carvalho, L. M., Santana, K., et al. (2022). Structural changes and adaptative evolutionary constraints in FLOWERING LOCUS T and TERMINAL FLOWER1-like genes of flowering plants. Front. Genet. 13. doi: 10.3389/fgene.2022.954015
Alrajhi, A. A., Alsahli, A. S., Alhelal, I. M., Rihan, H. Z., Fuller, M. P., Alsadon, A. A., et al. (2023). The effect of LED light spectra on the growth, yield and nutritional value of red and green lettuce (Lactuca sativa). Plants 12 (3), 463. doi: 10.3390/plants12030463
Artru, S., Lassois, L., Vancutsem, F., Reubens, B., Garré, S. (2018). Sugar beet development under dynamic shade environments in temperate conditions. Eur. J. Agron. 97, 38–47. doi: 10.1016/j.eja.2018.04.011
Assefa, A. D., Choi, S., Lee, J. E., Sung, J. S., Hur, O. S., Ro, N. Y., et al. (2019). Identification and quantification of selected metabolites in differently pigmented leaves of lettuce (Lactuca sativa L.) cultivars harvested at mature and bolting stages. BMC Chem. 13 (1), 56. doi: 10.1186/s13065–019-0570–2
Bagnall, D. J., King, R. W., Whitelam, G. C., Boylan, M. T., Wagner, D., Quail, P. H. (1995). Flowering responses to altered expression of phytochrome in mutants and transgenic lines of Arabidopsis thaliana (L.) Heynh. Plant Physiol. 108 (4), 1495–1503. doi: 10.1104/pp.108.4.1495
Bar, M., Aharon, M., Benjamin, S., Rotblat, B., Horowitz, M., Avni, A. (2008). AtEHDs, novel Arabidopsis EH-domain-containing proteins involved in endocytosis. Plant J. 55 (6), 1025–1038. doi: 10.1111/j.1365-313X.2008.03571.x
Baumann, K., Venail, J., Berbel, A., Domenech, M. J., Money, T., Conti, L., et al. (2015). Changing the spatial pattern of TFL1 expression reveals its key role in the shoot meristem in controlling Arabidopsis flowering architecture. J. Exp. Bot. 66 (15), 4769–4780. doi: 10.1093/jxb/erv247
Blümel, M., Dally, N., Jung, C. (2015). Flowering time regulation in crops-what did we learn from Arabidopsis? Curr. Opin. Biotechnol 32, 121–129. doi: 10.1016/j.copbio.2014.11.023
Bratzel, F., Turck, F. (2015). Molecular memories in the regulation of seasonal flowering: From competence to cessation. Genome Biol. 16, 192. doi: 10.1186/s13059-015-0770-6
Cao, K., Cui, L., Zhou, X., Ye, L., Zou, Z., Deng, S. (2016). Four tomato FLOWERING LOCUS T-like proteins act Antagonistically to regulate floral initiation. Front. Plant Sci. 6, 1213. doi: 10.3389/fpls.2015.01213
Cao, D., Takeshima, R., Zhao, C., Liu, B., Jun, A., Kong, F. (2017). Molecular mechanisms of flowering under long days and stem growth habit in soybean. J. Exp. Bot. 6 (8), 1873–1884. doi: 10.1093/jxb/erw394
Cao, K., Yan, F., Xu, D., Ai, K., Yu, J., Bao, E., et al. (2018). Phytochrome B1-dependent control of SP5G transcription is the basis of the night break and red to far-red light ratio effects in tomato flowering. BMC Plant Biol. 18 (1), 158. doi: 10.1186/s12870-018-1380-8
Cerdán, P. D., Chory, J. (2003). Regulation of flowering time by light quality. Nature 423 (6942), 881–885. doi: 10.1038/nature01636
Chailakhyan, M. K. (1936). New facts in support of the hormonal theory of plant development. Dokl Akad Nauk SSSR 13, 79–83.
Chaurasia, A. K., Patil, H. B., Krishna, B., Subramaniam, V. R., Sane, P. V., Sane, A. P. (2017). Flowering time in banana (Musa spp.), a day neutral plant, is controlled by at least three FLOWERING LOCUS T homologues. Sci. Rep. 7 (1), 5935. doi: 10.1038/s41598–017-06118-x
Chen, Q., Payyavula, R. S., Chen, L., Zhang, J., Zhang, C., Turgeon, R. (2018). FLOWERING LOCUS T mRNA is synthesized in specialized companion cells in Arabidopsis and Maryland Mammoth tobacco leaf veins. Proc. Natl. Acad. Sci. U.S.A. 115 (11), 2830–2835. doi: 10.1073/pnas.1719455115
Clarkson, N. M., Russell, J. S. (1975). Flowering responses to vernalization and photoperiod in annual medics (Medicago spp.). Aust. J. Agric. Res. 26 (5), 831–838. doi: 10.1071/AR9750831
Cober, E. R., Tanner, J. W., Voldeng, H. D. (1996). Soybean photoperiod-sensitivity loci respond differentially to light quality. Crop Sci. 36, 606–610. doi: 10.2135/cropsci1996.0011183x003600030014x
Cober, E. R., Voldeng, H. D. (2001). Low R:FR light quality delays flowering of E7E7 soybean lines. Crop Sci. 41, 1823–182. doi: 10.2135/cropsci2001.1823
Conti, L., Bradley, D. (2007). TERMINAL FLOWER1 is a mobile signal controlling Arabidopsis architecture. Plant Cell 19 (3), 767–778. doi: 10.1105/tpc.106.049767
Cui, Y., Zhu, M., Song, J., Fan, H., Xu, X., Wu, J., et al. (2023). Expression dynamics of phytochrome genes for the shade-avoidance response in densely direct-seeding rice. Front. Plant Sci. 13, 1105882. doi: 10.3389/fpls.2022.1105882
Deng, S., Chua, N. H. (2015). Inverted-repeat RNAs targeting FT intronic regions promote FT expression in arabidopsis. Plant Cell Physiol. 56 (8), 1667–1678. doi: 10.1093/pcp/pcv091
Devlin, P. F., Patel, S. R., Whitelam, G. C. (1998). Phytochrome E influences internode elongation and flowering time in Arabidopsis. Plant Cell 10 (9), 1479–1487. doi: 10.1105/tpc.10.9.1479
Devlin, P. F., Robson, P. R. H., Patel, S. R., Goosey, L., Sharrock, R. A., Whitelam, G. C. (1999). Phytochrome D acts in the shade-avoidance syndrome in Arabidopsis by controlling elongation growth and flowering time. Plant Physiol. 119 (3), 909–915. doi: 10.1104/pp.119.3.909
Doi, K., Izawa, T., Fuse, T., Yamanouchi, U., Kubo, T., Shimatani, Z., et al. (2004). Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes Dev. 18 (8), 926–936. doi: 10.1101/gad.1189604
Du, A., Tian, W., Wei, M., Yan, W., He, H., Zhou, D., et al. (2017). The DTH8-hd1 module mediates day-length-dependent regulation of rice flowering. Mol. Plant 10 (7), 948–961. doi: 10.1016/j.molp.2017.05.006
Endo, M., Yoshida, M., Sasaki, Y., Negishi, K., Horikawa, K., Daimon, Y., et al. (2018). Re-evaluation of florigen transport kinetics with separation of functions by mutations that uncouple flowering initiation and long-distance transport. Plant Cell Physiol. 59 (8), 1621–1629. doi: 10.1093/pcp/pcy063
Fantini, E., Sulli, M., Zhang, L., Aprea, G., Jiménez-Gómez, J. M., Bendahmane, A., et al. (2019). Pivotal roles of cryptochromes 1a and 2 in tomato development and physiology. Plant Physiol. 179 (2), 732–748. doi: 10.1104/pp.18.00793
Feldmann, K. A., Goff, S. A. (2014). The First Plant Genome Sequence-Arabidopsis thaliana. Adv. Bot. Res. 69, 91–117. doi: 10.1016/B978–0-12–417163–3.00004–4
Fowler, S., Lee, K., Onouchi, H., Samach, A., Richardson, K., Morris, B., et al. (1999). GIGANTEA: A circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J. 18 (17), 4679–4688. doi: 10.1093/emboj/18.17.4679
Franklin, K. A., Praekelt, U., Stoddart, W. M., Billingham, O. E., Halliday, K. J., Whitelam, G. C. (2003). Phytochromes B, D, and E act redundantly to control multiple physiological responses in Arabidopsis. Plant Physiol. 131 (3), 1340–1346. doi: 10.1104/pp.102.015487
Franklin, K. A., Whitelam, G. C. (2005). Phytochromes and shade-avoidance responses in plants. Ann. Bot. 96 (2), 169–175. doi: 10.1093/aob/mci165
Fukuda, M., Matsuo, S., Kikuchi, K., Kawazu, Y., Fujiyama, R., Honda, I. (2011). Isolation and functional characterization of the FLOWERING LOCUS T homolog, the LsFT gene, in lettuce. J. Plant Physiol. 168 (13), 1602–1607. doi: 10.1016/j.jplph.2011.02.004
Fukuda, M., Yanai, Y., Nakano, Y., Sasaki, H., Uragami, A., Okada, K. (2017). Isolation and gene expression analysis of flowering-related genes in lettuce (Lactuca sativa L.). Hort J. 86, 340–348. doi: 10.2503/hortj.OKD-036
Green-Tracewicz, E., Page, E. R., Swanton, C. J. (2012). Light quality and the critical period for weed control in soybean. Weed Sci. 60 (1), 86–91. doi: 10.1614/WS-D-11-00072.1
Harig, L., Beinecke, F. A., Oltmanns, J., Muth, J., Müller, O., Rüping, B., et al. (2012). Proteins from the FLOWERING LOCUS T-like subclade of the PEBP family act antagonistically to regulate floral initiation in tobacco. Plant J. 72 (6), 908–921. doi: 10.1111/j.1365-313X.2012.05125.x
Hayama, R., Izawa, T., Shimamoto, K. (2002). Isolation of rice genes possibly involved in the photoperiodic control of flowering by a fluorescent differential display method. Plant Cell Physiol. 43 (5), 494–504. doi: 10.1093/pcp/pcf059
Hayama, R., Yokoi, S., Tamaki, S., Yano, M., Shimamoto, K. (2003). Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 422 (6933), 719–722. doi: 10.1038/nature01549
Hecht, V., Foucher, F., Ferrándiz, C., Macknight, R., Navarro, C., Morin, J., et al. (2005). Conservation of Arabidopsis flowering genes in model legumes. Plant Physiol. 137 (4), 1420–1434. doi: 10.1104/pp.104.057018
Higuchi, Y., Narumi, T., Oda, A., Nakano, Y., Sumitomo, K., Fukai, S., et al. (2013). The gated induction system of a systemic floral inhibitor, antiflorigen, determines obligate short-day flowering in chrysanthemums. Proc. Natl. Acad. Sci. U.S.A. 110 (42), 17137–17142. doi: 10.1073/pnas.1307617110
Holmes, M. G., Smith, H. (1977). THE FUNCTION OF PHYTOCHROME IN THE NATURAL ENVIRONMENT—I. CHARACTERIZATION OF DAYLIGHT FOR STUDIES IN PHOTOMORPHOGENESIS AND PHOTOPERIODISM. Photochem. Photobiol. 25, 533–538. doi: 10.1111/j.1751–1097.1977.tb09124.x
Huang, H., Alvarez, S., Bindbeutel, R., Shen, Z., Naldrett, M. J., Evans, B. S., et al. (2016). Identification of evening complex associated proteins in Arabidopsis by affinity purification and mass spectrometry. Mol. Cell. Proteomics 15 (1), 201–217. doi: 10.1074/mcp.M115.054064
Ikeda, K. (1985). Photoperiodic flower induction in rice plants as influenced by light intensity and quality. Jpn Agric. Res. Q 18, 164–170
Imaizumi, T. (2010). Arabidopsis circadian clock and photoperiodism: time to think about location. Curr. Opin. Plant Biol. 13 (1), 83–89. doi: 10.1016/j.pbi.2009.09.007
Ishikawa, R., Shinomura, T., Takano, M., Shimamoto, K. (2009). Phytochrome dependent quantitative control of Hd3a transcription is the basis of the night breakeffect in rice flowering. Genes Genet. Syst. 84 (2), 179–184. doi: 10.1266/ggs.84.179
Itoh, H., Nonoue, Y., Yano, M., Izawa, T. (2010). A pair of floral regulators sets critical day length for Hd3a florigen expression in rice. Nat. Genet. 42 (7), 635–638. doi: 10.1038/ng.606
Izawa, T., Oikawa, T., Sugiyama, N., Tanisaka, T., Yano, M., Shimamoto, K. (2002). Phytochrome mediates the external light signal to repress FT orthologs in photoperiodic flowering of rice. Genes Dev. 16 (15), 2006–2020. doi: 10.1101/gad.999202
Jang, S., Marchal, V., Panigrahi, K. C. S., Wenkel, S., Soppe, W., Deng, X. W., et al. (2008). Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO J. 27 (8), 1277–1288. doi: 10.1038/emboj.2008.68
Jaudal, M., Wen, J., Mysore, K. S., Putterill, J. (2020). Medicago PHYA promotes flowering, primary stem elongation and expression of flowering time genes in long days. BMC Plant Biol. 20 (1), 329. doi: 10.1186/s12870-020-02540-y
Jin, S., Nasim, Z., Susila, H., Ahn, J. H. (2021). Evolution and functional diversification of FLOWERING LOCUS T/TERMINAL FLOWER 1 family genes in plants. Semin. Cell Dev. Biol. 109, 20–30. doi: 10.1016/j.semcdb.2020.05.007
Kalaitzoglou, P., van Ieperen, W., Harbinson, J., van der Meer, M., Martinakos, S., Weerheim, K., et al. (2019). Effects of continuous or end-of-day far-red light on tomato plant growth, morphology, light absorption, and fruit production. Front. Plant Sci. 10, 322. doi: 10.3389/fpls.2019.00322
Kojima, S., Takahashi, Y., Kobayashi, Y., Monna, L., Sasaki, T., Araki, T., et al. (2002). Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol. 43 (10), 1096–1105. doi: 10.1093/pcp/pcf156
Komiya, R., Ikegami, A., Tamaki, S., Yokoi, S., Shimamoto, K. (2008). Hd3a and RFT1 are essential for flowering in rice. Development 135 (4), 767–774. doi: 10.1242/dev.008631
Komiya, R., Yokoi, S., Shimamoto, K. (2009). A gene network for long-day flowering activates RFT1 encoding a mobile flowering signal in rice. Development 136 (20), 3443–3450. doi: 10.1242/dev.040170
Krzymuski, M., Andrés, F., Cagnola, J. I., Jang, S., Yanovsky, M. J., Coupland, G., et al. (2015). The dynamics of FLOWERING LOCUS T expression encodes long-day information. Plant J. 83 (6), 952–961. doi: 10.1111/tpj.12938
Lane, H. C., Cathey, H. M., Evans, L. T. (1965). The dependence of flowering in several long-day plants on the spectral composition of light extending the photoperiod. Am. J. Bot. 52 (10), 1006–1014. doi: 10.2307/2440130
Laubinger, S., Marchal, V., Gentillhomme, J., Wenkel, S., Adrian, J., Jang, S., et al. (2006). Arabidopsis SPA proteins regulate photoperiodic flowering and interact with the floral inducer CONSTANS to regulate its stability. Development 133 (16), 3213–3222. doi: 10.1242/dev.02481
Laurie, R. E., Diwadkar, P., Jaudal, M., Zhang, L., Hecht, V., Wen, J., et al. (2011). The Medicago Flowering locus T homolog, MtFTa1, is a key regulator of flowering time. Plant Physiol. 156 (4), 2207–2224. doi: 10.1104/pp.111.180182
Lee, Y. S., An, G. (2015). OsGI controls flowering time by modulating rhythmic flowering time regulators preferentially under short day in rice. J. Plant Biol. 58, 137–145. doi: 10.1007/s12374-015-0007-y
Lee, S. H., Choi, C. W., Park, K. M., Jung, W. H., Chun, H. J., Baek, D., et al. (2021). Diversification in functions and expressions of soybean FLOWERING LOCUS T genes fine-tunes seasonal flowering. Front. Plant Sci. 12, 613675. doi: 10.3389/fpls.2021.613675
Lee, N., Ozaki, Y., Hempton, A. K., Takagi, H., Purusuwashi, S., Song, Y. H., et al. (2023). The FLOWERING LOCUS T gene expression is controlled by high-irradiance response and external coincidence mechanism in long days in Arabidopsis. New Phytol. 239 (1), 208–221. doi: 10.1111/nph.18932
Liu, W., Jiang, B., Ma, L., Zhang, S., Zhai, H., Xu, X., et al. (2018). Functional diversification of Flowering Locus T homologs in soybean: GmFT1a and GmFT2a/5a have opposite roles in controlling flowering and maturation. New Phytol. 217 (3), 1335–1345. doi: 10.1111/nph.14884
Liu, H., Liu, X., Chang, X., Chen, F., Lin, Z., Zhang, L. (2023). Large-scale analyses of angiosperm Flowering Locus T genes reveal duplication and functional divergence in monocots. Front. Plant Sci. 13, 1039500. doi: 10.3389/fpls.2022.1039500
Liu, L., Liu, C., Hou, X., Xi, W., Shen, L., Tao, Z., et al. (2012). FTIP1 is an essential regulator required for florigen transport. PloS Biol. 10. doi: 10.1371/journal.pbio.1001313
Lorenzo, C. D., Alonso Iserte, J., Sanchez Lamas, M., Antonietti, M. S., Garcia Gagliardi, P., Hernando, C. E., et al. (2019). Shade delays flowering in Medicago sativa. Plant J. 99 (1), 7–22. doi: 10.1111/tpj.14333
Lu, S., Dong, L., Fang, C., Liu, S., Kong, L., Cheng, Q., et al. (2020). Stepwise selection on homeologous PRR genes controlling flowering and maturity during soybean domestication. Nat. Genet. 52 (4), 428–436. doi: 10.1038/s41588-020-0604-7
Mauro, R. P., Sortino, O., Dipasquale, M., Mauromicale, G. (2014). Phenological and growth response of legume cover crops to shading. J. Agric. Sci. 152 (6), 917–931. doi: 10.1017/S0021859613000592
Meijer, D., Doesburg, F.v., Jungerling, L., Weldegergis, B. T., Kappers, I. F., Oystaeyen, A.V., et al. (2023). Supplemental far-red light influences flowering traits and interactions with a pollinator in tomato crops. Environ. Exp. Bot. 213, 105438. doi: 10.1016/j.envexpbot.2023.105438
Mizoguchi, T., Niinuma, K., Yoshida, R. (2007). Day-neutral response of photoperiodic flowering in tomatoes: Possible implications based on recent molecular genetics of Arabidopsis and rice. Plant Biotechnol. 24 (1), 83–86. doi: 10.5511/plantbiotechnology.24.83
Mockler, T., Yang, H., Yu, X., Parikh, D., Cheng, Y. C., Dolan, S., et al. (2003). Regulation of photoperiodic flowering by Arabidopsis photoreceptors. Proc. Natl. Acad. Sci. USA. 100 (4), 2140–5. doi: 10.5511/plantbiotechnology.24.83
Pin, P. A., Benlloch, R., Bonnet, D., Wremerth-Weich, E., Kraft, T., Gielen, J. J. L., et al. (2010). An antagonistic pair of FT homologs mediates the control of flowering time in sugar beet. Science 330 (6009), 1397–1400. doi: 10.1126/science.1197004
Pin, P. A., Zhang, W., Vogt, S. H., Dally, N., Büttner, B., Schulze-Buxloh, G., et al. (2012). The role of a pseudo-response regulator gene in life cycle adaptation and domestication of beet. Curr. Biol. 22 (12), 1095–1101. doi: 10.1016/j.cub.2012.04.007
Pittendrigh, C. S., Minis, D. H. (1964). The entrainment of circadian oscillations by light and their role as photoperiodic clocks. Am. Nat. 98, 261–294. doi: 10.1086/282327
Qin, Z., Wu, J., Geng, S., Feng, N., Chen, F., Kong, X., et al. (2017). Regulation of FT splicing by an endogenous cue in temperate grasses. Nat. Commun. 8, 14320. doi: 10.1038/ncomms14320
Raai, M. N., Zain, N. A. M., Osman, N., Rejab, N. A., Sahruzaini, N. A., Cheng, A. (2020). Effects of shading on the growth, development and yield of winged bean (Psophocarpus tetragonolobus). Ciencia Rural 50, p.e20190570. doi: 10.1590/0103-8478cr20190570
Rehman, S., Bahadur, S., Xia, W. (2023). An overview of floral regulatory genes in annual and perennial plants. Gene 147699. doi: 10.1016/j.gene.2023.147699
Roig-Villanova, I., Martínez-García, J. F. (2016). Plant responses to vegetation proximity: A whole life avoiding shade. Front. Plant Sci. 7, 180038. doi: 10.3389/fpls.2016.00236
Schambow, T. J., Adjesiwor, A. T., Lorent, L., Kniss, A. R. (2019). Shade avoidance cues reduce Beta vulgaris growth. Weed Sci. 67 (3), 311–317. doi: 10.1017/wsc.2019.2
Seo, E., Yu, J., Ryu, K. H., Lee, M. M., Lee, I. (2011). WEREWOLF, a regulator of root hair pattern formation, controls flowering time through the regulation of FT mRNA stability. Plant Physiol. 156 (4), 1867–1877. doi: 10.1104/pp.111.176685
Sgamma, T., Pape, J., Massiah, A., Jackson, S. (2016). Selection of reference genes for diurnal and developmental time-course real-time PCR expression analyses in lettuce. Plant Methods 12, 21. doi: 10.1186/s13007-016-0121-y
Shim, J. S., Kubota, A., Imaizumi, T. (2017). Circadian clock and photoperiodic flowering in Arabidopsis: CONSTANS is a Hub for Signal integration. Plant Physiol. 173 (1), 5–15. doi: 10.1104/pp.16.01327
Smith, H. (1982). Light quality, photoperception, and plant strategy. Annu. Rev. Plant Physiol. 33 (1), 481–518. doi: 10.1146/annurev.pp.33.060182.002405
Song, Y. H., Kubota, A., Kwon, M. S., Covington, M. F., Lee, N., Taagen, E. R., et al. (2018). Molecular basis of flowering under natural long-day conditions in Arabidopsis. Nat. Plants 4 (10), 824–835. doi: 10.1038/s41477-018-0253-3
Susila, H., Jurić, S., Liu, L., Gawarecka, K., Chung, K. S., Jin, S., et al. (2021). Florigen sequestration in cellular membranes modulates temperature-responsive flowering. Science 373 (6559), 1137–1142. doi: 10.1126/science.abh4054
Takada, S., Goto, K. (2003). Terminal flower2, an arabidopsis homolog of heterochromatin protein1, counteracts the activation of flowering locus T by constans in the vascular tissues of leaves to regulate flowering time. Plant Cell 15 (12), 2856–2865. doi: 10.1105/tpc.016345
Takagi, H., Hempton, A. K., Imaizumi, T. (2023). Photoperiodic flowering in Arabidopsis: Multilayered regulatory mechanisms of CONSTANS and the florigen FLOWERING LOCUS T. Plant Commun 4 (3), 100552. doi: 10.1016/j.xplc.2023.100552
Takano, M., Inagaki, N., Xie, X., Yuzurihara, N., Hihara, F., Ishizuka, T., et al. (2005). Distinct and cooperative functions of phytochromes A, B, and C in the control of deetiolation and flowering in rice. Plant Cell 17 (12), 3311–3325. doi: 10.1105/tpc.105.035899
Tamaki, S., Matsuo, S., Hann, L. W., Yokoi, S., Shimamoto, K. (2007). Hd3a protein is a mobile flowering signal in rice. Science 316 (5827), 1033–1036. doi: 10.1126/science.1141753
Teotia, S., Tang, G. (2015). To bloom or not to bloom: Role of micrornas in plant flowering. Mol. Plant 8 (3), 359–377. doi: 10.1016/j.molp.2014.12.018
Vicentini, G., Biancucci, M., Mineri, L., Chirivì, D., Giaume, F., Miao, Y., et al. (2023). Environmental control of rice flowering time. Plant Commun. 4 (5), 100610. doi: 10.1016/j.xplc.2023.100610
Wang, S., Yang, Y., Chen, F., Jiang, J. (2022). Functional diversification and molecular mechanisms of FLOWERING LOCUS T/TERMINAL FLOWER 1 family genes in horticultural plants. Mol. Horticulture 2 (1), 19. doi: 10.1186/s43897-022-00039-8
Wickland, D. P., Hanzawa, Y. (2015). The FLOWERING LOCUS T/TERMINAL FLOWER 1 gene family: functional evolution and molecular mechanisms. Mol. Plant 8 (7), 983–997. doi: 10.1016/j.molp.2015.01.007
Wigge, P. A., Kim, M. C., Jaeger, K. E., Busch, W., Schmid, M., Lohmann, J. U., et al. (2005). Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309 (5737), 1056–1059. doi: 10.1126/science.1114358
Wong, A. C. S., Hecht, V. F. G., Picard, K., Diwadkar, P., Laurie, R. E., Wen, J., et al. (2014). Isolation and functional analysis of CONSTANS-LIKE genes suggests that a central role for CONSTANS in flowering time control is not evolutionarily conserved in Medicago truncatula. Front. Plant Sci. 5, 486. doi: 10.3389/fpls.2014.00486
Wu, Y., Gong, W., Yang, W. (2017). Shade inhibits leaf size by controlling cell proliferation and enlargement in soybean. Sci. Rep. 7 (1), 9259. doi: 10.1038/s41598–017-10026–5
Wu, L., Liu, D., Wu, J., Zhang, R., Qin, Z., Liu, D., et al. (2013). Regulation of FLOWERING LOCUS t by a MicroRNA in Brachypodium distachyon. Plant Cell 25 (11), 4363–4377. doi: 10.1105/tpc.113.118620
Xia, W., Liu, R., Zhang, J., Mason, A. S., Li, Z., Gong, S., et al. (2020). Alternative splicing of flowering time gene FT is associated with halving of time to flowering in coconut. Sci. Rep. 10 (1), 11640. doi: 10.1038/s41598–020-68431–2
Xia, Z., Watanabe, S., Yamada, T., Tsubokura, Y., Nakashima, H., Zhai, H., et al. (2012). Positional cloning and characterization reveal the molecular basis for soybean maturity locus E1 that regulates photoperiodic flowering. Proc. Natl. Acad. Sci. U.S.A. 109 (32), E2155–E2164. doi: 10.1073/pnas.1117982109
Xie, Y., Zhou, Q., Zhao, Y., Li, Q., Liu, Y., Ma, M., et al. (2020). FHY3 and FAR1 integrate light signals with the miR156-SPL module-mediated aging pathway to regulate arabidopsis flowering. Mol. Plant 13 (3), 483–498. doi: 10.1016/j.molp.2020.01.013
Xue, W., Xing, Y., Weng, X., Zhao, Y., Tang, W., Wang, L., et al. (2008). Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 40 (6), 761–767. doi: 10.1038/ng.143
Yang, F., Huang, S., Gao, R., Liu, W., Yong, T., Wang, X., et al. (2014). Growth of soybean seedlings in relay strip intercropping systems in relation to light quantity and red: Far-red ratio. Field Crops Res. 155, 245–253. doi: 10.1016/j.fcr.2013.08.011
Yang, J., Ning, C., Liu, Z., Zheng, C., Mao, Y., Wu, Q., et al. (2023). Genome-wide characterization of PEBP gene family and functional analysis of TERMINAL FLOWER 1 homologs in macadamia integrifolia. Plants 12 (14), 2692. doi: 10.3390/plants12142692
Yu, J. W., Rubio, V., Lee, N. Y., Bai, S., Lee, S. Y., Kim, S. S., et al. (2008). COP1 and ELF3 control circadian function and photoperiodic flowering by regulating GI stability. Mol. Cell 32 (5), 617–630. doi: 10.1016/j.molcel.2008.09.026
Yuan, H. Y., Saha, S., Vandenberg, A., Bett, K. E. (2017). Flowering and growth responses of cultivated lentil and wild Lens germplasm toward the differences in red to far-red ratio and photosynthetically active radiation. Front. Plant Sci. 8, 386. doi: 10.3389/fpls.2017.00386
Zhai, H., Lü, S., Liang, S., Wu, H., Zhang, X., Liu, B., et al. (2014). GmFT4, a homolog of FLOWERING LOCUS T, is positively regulated by E1 and functions as a flowering repressor in soybean. PloS One 9 (2), e89030. doi: 10.1371/journal.pone.0089030
Zhai, H., Wan, Z., Jiao, S., Zhou, J., Xu, K., Nan, H., et al. (2022). GmMDE genes bridge the maturity gene E1 and florigens in photoperiodic regulation of flowering in soybean. Plant Physiol. 189 (2), 1021–1036. doi: 10.1093/plphys/kiac092
Zhang, X., Liu, L., Wang, H., Gu, Z., Liu, Y., Wang, M., et al. (2020). Mtpin1 and mtpin3 play dual roles in regulation of shade avoidance response under different environments in Medicago truncatula. Int. J. Mol. Sci. 21 (22), 8742. doi: 10.3390/ijms21228742
Zhu, Y., Klasfeld, S., Jeong, C. W., Jin, R., Goto, K., Yamaguchi, N., et al. (2020). TERMINAL FLOWER 1-FD complex target genes and competition with FLOWERING LOCUS T. Nat. Commun. 11 (1), 5118. doi: 10.1038/s41467-020-18782-1
Keywords: photoperiodic flowering, FLOWERING LOCUS T, florigen, expression profiles, flowering plants
Citation: Lee N, Shim JS, Kang M-K and Kwon M (2024) Insight from expression profiles of FT orthologs in plants: conserved photoperiodic transcriptional regulatory mechanisms. Front. Plant Sci. 15:1397714. doi: 10.3389/fpls.2024.1397714
Received: 08 March 2024; Accepted: 20 May 2024;
Published: 03 June 2024.
Edited by:
Amy Leigh Klocko, University of Colorado Springs, United StatesReviewed by:
Consuelo Guerrero Ruiz, University of Malaga, SpainHeng Sun, Chinese Academy of Sciences (CAS), China
Copyright © 2024 Lee, Shim, Kang and Kwon. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Moonhyuk Kwon, bWt3b25AZ251LmFjLmty; Nayoung Lee, bmF5b3VuZ0BnbnUuYWMua3I=
 Nayoung Lee
Nayoung Lee Jae Sung Shim
Jae Sung Shim Min-Kyoung Kang3
Min-Kyoung Kang3 Moonhyuk Kwon
Moonhyuk Kwon