
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 30 May 2024
Sec. Plant Breeding
Volume 15 - 2024 | https://doi.org/10.3389/fpls.2024.1397018
The continuously refined genome assembly of the Chinese cabbage accession Chiifu is widely recognized as the reference for Brassica rapa. However, the high self-incompatibility of Chiifu limits its broader utilization. In this study, we report the development of self-compatible Chiifu lines through a meticulous marker-assisted selection (MAS) strategy, involving the substitution of the Chiifu allele of MLPK (M-locus protein kinase) with that from the self-compatible Yellow Sarson (YS). A YS-based marker (SC-MLPK) was employed to screen 841 B. rapa accessions, confirming that all eight accessions with the mlpk/mlpk (mm) genotype exhibited self-compatibility. Additionally, we designed 131 High-Resolution Melting (HRM) markers evenly distributed across the B. rapa genome as genomic background selection (GBS) markers to facilitate the introgression of self-compatibility from YS into Chiifu along with SC-MLPK. Genome background screening revealed that the BC3S1 population had a proportion of the recurrent parent genome (PR) ranging from 93.9% to 98.5%. From this population, we identified self-compatible individuals exhibiting a high number of pollen tubes penetrating stigmas (NPT) (>25) and a maximum compatibility index (CI) value of 7.5. Furthermore, we selected two individuals demonstrating significant similarity to Chiifu in both genetic background and morphological appearance, alongside self-compatibility. These selected individuals were self-pollinated to generate two novel lines designated as SC-Chiifu Lines. The development of these self-compatible Chiifu lines, together with the SC-MLPK marker and the set of HRM markers, represents valuable tools for B. rapa genetics and breeding.
Chinese cabbage (Brassica rapa ssp. pekinensis) accession Chiifu exhibits high self-incompatibility. Chiifu is the first Brassica accession to be sequenced and assembled as reference genome (Chiifu v1.5) (Wang et al., 2011). Advancements in sequencing technologies have led to the successive development of three updated genome assemblies (Chiifu v2.5, v3.0, and v4.0) (Cai et al., 2017; Zhang et al., 2018, 2023). The Chiifu genome serves as a prominent reference in the study of Brassica species, necessitating the mass selfed seeds. Additionally, the construction of a Chiifu mutant library assumes great significance, facilitating investigations into essential agronomic traits within B. rapa. However, the labor-intensive process of seed production in the highly self-incompatible Chiifu, reliant upon artificial pollination, poses challenges for its widespread application. In addressing this limitation, the development of a self-compatible Chiifu line with a high homozygosity of the genetic background (HGB) plays a crucial role in promoting genomic and genetic breeding studies of Brassica species.
The self-incompatibility (SI) in Chinese cabbage is regulated by a multi-allelic S locus, encompassing three genes associated with pollen-pistil recognition: SRK (S-locus receptor kinase), SCR/SP11 (S-locus cysteine-rich protein/S-locus protein 11), and SLG (S-locus glycoprotein) (Yamamoto et al., 2023). The genetic set of alleles of these genes at the same S locus is designated as the ‘S haplotype’ (Nasrallah and Nasrallah, 1993). Incompatible reactions occur when the pistil and pollen share the same S haplotype. Additionally, ARC1 (arm repeat-containing 1), THL1 (thioredoxin-h-like-1), MLPK, and FER-Rac/Rop signaling (FERONIA receptor kinase homolog or Rac/Rop guanosine triphosphatase signaling), not associated with the S locus, are involved in the pathways of the SI response (Stone et al., 1999; Cabrillac et al., 2001; Murase et al., 2004; Zhang et al., 2021).
Yellow Sarson (YS) (B. rapa ssp. tricolaris) carries a recessive mutation of the modifier (m) gene, eliminating the SI response in the stigma (Murase et al., 2004). The M gene, responsible for this trait, encodes a membrane-anchored protein kinase (MLPK). A SNP induces a specific amino acid substitution (Gly to Arg) at position 194 (G194R) (Murase et al., 2004). The dysfunctional G194R form of MLPK protein in mm plants can be restored through the transient expression of MLPK, thereby reinstating the ability of mm papilla cells to reject self-pollen (Murase et al., 2004). Two distinct forms of MLPK, MLPK form 1 (MLPKf1) and MLPK form 2 (MLPKf2), localize to the papilla cell membrane and directly interact with SRK to transduce SI signaling (Kakita et al., 2007). The Chiifu accession carries the S60 haplotype on chromosome A07 (Yamamoto et al., 2023), characterized by MLPK-dependent self-incompatibility (Ohata et al., 2023). However, the practical challenge of breeding self-compatible Chinese cabbage lines based on MLPK remains unresolved.
The fusion of genomics and breeding has accelerated the development of cultivars with enhanced traits (Dou et al., 2022; Liu et al., 2022; Cai et al., 2023; Li et al., 2023). Utilizing trait-specific markers in conjunction with genomic background analysis has proven to be a potent breeding strategy for overcoming SI. Notably, in Brassica oleracea, the construction of a linkage map has facilitated the identification of major quantitative trait loci (QTLs) linked to self-compatibility (SC). Moreover, SC genes have been rapidly introduced by marker-assisted backcrossing (MABC) using self-compatibility-specific markers and 36 genomic background selection (GBS) markers (Xiao et al., 2019). The silencing of SP11 gene within the S locus has led to the development of a self-compatible transgenic line in B. rapa (Jung et al., 2012). Leveraging the S locus in conjunction with 111 SSR markers distributed evenly on a physical map facilitated MABC, yielding a self-compatible line with a compatibility index (CI) of approximately 1.15 in Chinese cabbage (Zheng et al., 2022). Therefore, it is feasible to identify self-compatible accessions through screening B. rapa germplasm and subsequently develop self-compatible Chiifu lines via MABC.
HRM technology, capitalizing on the identification of distinct melting curves of PCR products, has emerged as a high throughput, accurate, and cost-effective tool for detecting genetic variations, including single nucleotide polymorphisms (SNPs), insertions or deletions (INDELs), and methylation (Herrmann et al., 2007; White et al., 2007; Distefano et al., 2012). In our previous study, we developed 148 INDEL markers for HRM genotyping in B. rapa, thereby expanding the repertoire of HRM markers in B. rapa (Chen et al., 2021). However, these HRM markers were solely designed based on a physical map as GBS markers. This result highlighted the advantage of HRM markers as genome background markers for marker-assisted selection.
This study aims to overcome the SI present in the B. rapa reference line Chiifu. We developed a functional-related marker (SC-MLPK) for MLPK. The accessions with mlpk/mlpk (mm) genotype identified by SC-MLPK across B. rapa subspecies proved all being self-compatible. Leveraging the constructed physical and genetic maps of B. rapa, we developed genetic background HRM markers evenly distributed across the genome. Through the combination of genotyping by the SC-MLPK marker and genetic background analysis for MABC, we successfully developed two self-compatible lines, designated as SC-Chiifu Lines by transferring the mm allele into Chiifu. Thus, this study provides valuable insights into the practical application of molecular markers for overcoming the SI in B. rapa.
The B. rapa reference genome line Chiifu, characterized by a CI value < 1.0 at the anthesis stage, served as the receptor parent. Its S-haplotype is S60 (class II), as detailed in a recent study (Yamamoto et al., 2023). A Yellow Sarson line, R-o-18, exhibiting a CI value > 10, was employed as the donor parent. The initial cross involved mating the donor parent R-o-18 with the recurrent parent Chiifu, resulting in the generation of F1 plants. Subsequent backcrossing was systematically performed with Chiifu, leading to the development of backcross populations denoted as BC1, BC2, and BC3. From the BC3 generation, the BC3-1, BC3-6, and BC3-7 plants with the Mm genotype and high proportion of recurrent parent genome (PR) were self-pollinated to yield BC3S1 populations. Individuals with both phenotypic and genetic background similarity to Chiifu while demonstrating self-compatibility were selected from the BC3S1 generation to be self-pollinated to produce self-compatible Chiifu lines (Figure 1). Additionally, this study included 841 accessions of B. rapa from our laboratory to identify individuals with mm genotypes. These accessions encompassed 207 Chinese cabbage, 189 Pak choi (ssp. chinensis), 150 Caixin (ssp. parachinensis), 119 European turnip (ssp. rapa), 76 Oil seeds (ssp. oleifera), 41 Yellow Sarson (ssp. tricolaris), 24 Japanese komatsuna (ssp. perviridis), 23 Japanese mizuna (ssp. nipposinica), and 12 Taicai (ssp. chinensis var. tai-tsai) (Cheng et al., 2016).
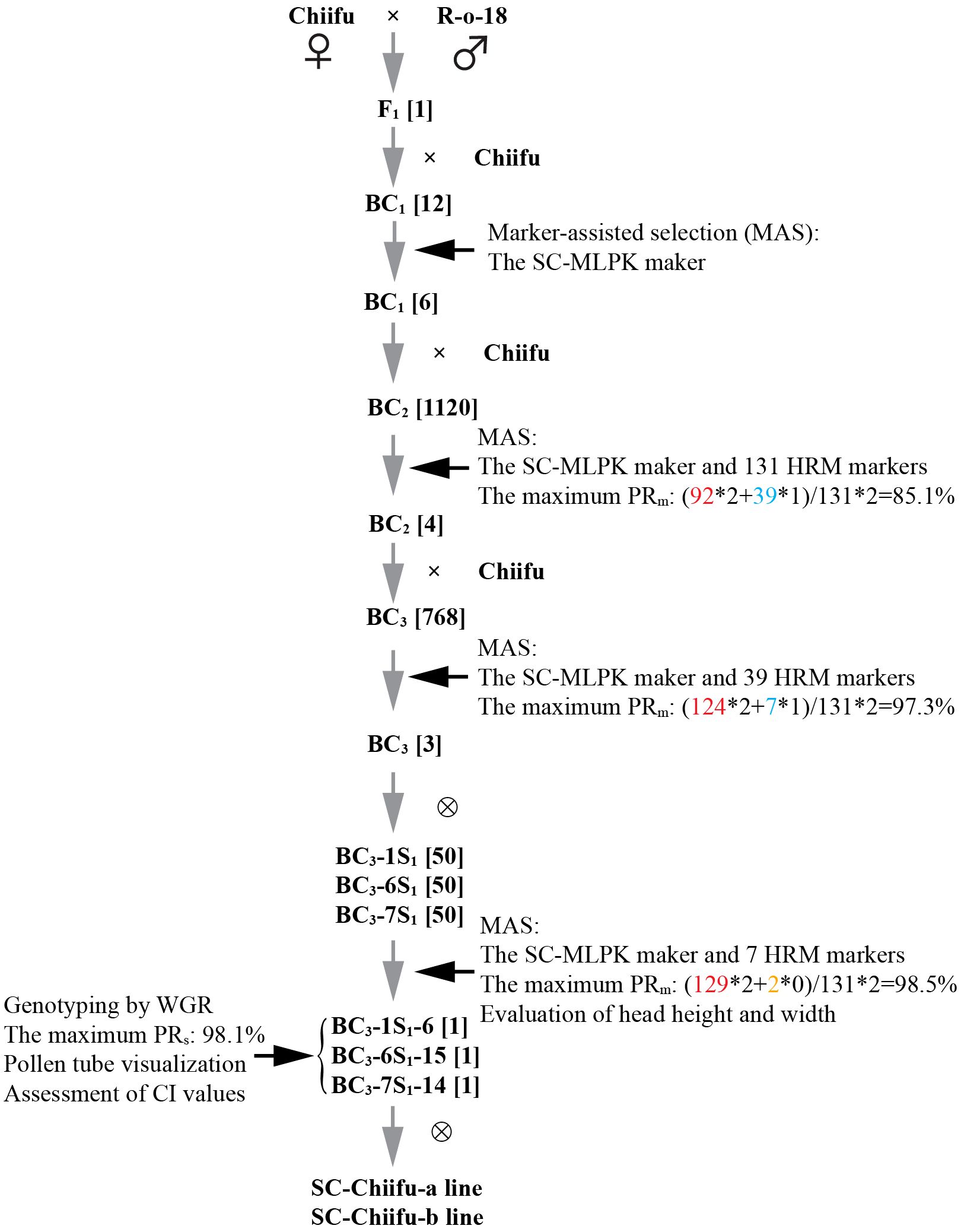
Figure 1 Overcoming high self-incompatibility in Chinese cabbage Chiifu through MABC. The numbers in square brackets indicate the number of plants. Gray arrows represent the process of constructing populations. Black arrows represent the selection by genotyping and phenotyping. The numbers in red, orange and cyan represent the number of markers with Chiifu, R-o-18 or heterozygote genotypes, respectively.
Genomic DNA was extracted from all samples by the modified CTAB method (Mavrodiev et al., 2021). Plant materials selected for DNA library construction included R-o-18, BC3-1S1-6, BC3-6S1-15, and BC3-7S1-14. These samples were chosen to represent key stages in the breeding program. For WGR, DNA libraries were prepared and subsequently sequenced on a BGISEQ-T7 platform employing paired-end 150 bp (PE150) reads. To ensure the production of high-quality data, the raw sequencing reads were filtered using the fastp software, adhering to default parameters (Chen et al., 2018).
High-quality reads were aligned to the B. rapa Chiifu reference genome version 3.0 using Burrows-Wheeler-Alignment (BWA) software with the “mem” method (Li and Durbin, 2009; Zhang et al., 2018). PCR-duplicated reads were filtered out using Samtools software (Li et al., 2009). Variant datasets (SNPs/INDELs) were obtained by applying stringent filters to exclude variants with low quality or multi-allelic features (QUAL < 60, QD < 20, MQ < 30, FS > 13) using GATK software (McKenna et al., 2010; Chang et al., 2023). Additionally, heterozygous loci in the variant datasets of R-o-18 were excluded. Sequence variants of the MLPK gene between the reference genome Chiifu v3.0 and R-o-18 aligned reads were visualized using the Integrated Genome Viewer (IGV) software (Robinson et al., 2011). The analysis was conducted with an updated version of genome annotation (Chiifu v3.5) to ensure precision in interpreting the genomic variations (Zhang et al., 2022b).
KASP genotyping was employed to discern specific SNPs associated with the MLPK gene. Prior to designing the MLPK-specific marker, an extensive investigation into SNPs affecting MLPK functionality was conducted. Initially, SNPs within MLPK were extracted from the SNP dataset of 524 B. rapa genomes, as reported in a previous study (Cai et al., 2021). Subsequently, the precise SNP responsible for the G194R mutation within MLPK, which has been reported by Murase et al. (2004), was identified. Utilizing this SNP information, an MLPK-specific marker was designed using Primer3 software (Untergasser et al., 2012). Furthermore, accessions carrying the mm genotype were identified from the SNP dataset of 524 B. rapa genomes, with one such accession serving as a positive control for subsequent KASP genotyping.
Amplification and allelic discrimination for the KASP marker were performed using the Scientific QuantStudio™ 12K Flex Real-Time PCR System (Thermo Fisher Scientific, USA). The KASP genotyping protocol followed the methodology outlined in a previous study (Zhang et al., 2022a). Details regarding the KASP reaction volume and thermal cycling conditions for SC-MLPK are provided in Supplementary Tables S1 and S2, respectively.
The CI value was determined following a previously published protocol (Xiao et al., 2019). Approximately 20 flowers per individual were manually self-pollinated one or two days after reaching full bloom. At the podding stage, CI was calculated using the formula: CI = number of seeds/number of pollinated flowers. Levels of SI, moderate self-compatibility (MSC), or SC were categorized as follows: SI (CI < 1); MSC (1 ≤ CI ≤ 4); SC (CI > 4). B. rapa accessions with the mm genotype, Chiifu, R-o-18, BC3S1 individuals with the mm genotype, were assessed of CI values. Additionally, from the BC2 generation, plants with Mm genotypes were self-pollinated to generate BC2S1 populations. BC2S1 individuals with the mm genotype were assessed of CI values. All pollinations were conducted in the experimental greenhouse of IVF-CAAS, Beijing, China, at around 10:00 a.m. on sunny days.
Genome-wide INDELs were selected from variant datasets derived between Chiifu and R-o-18. The Primer3 software was employed to design INDEL markers, utilizing 150 bp sequences flanking each INDEL locus (Untergasser et al., 2012; Chen et al., 2021). The designed INDEL markers produced amplicons ranging from 130 bp to 260 bp, with primer lengths between 18 bp and 23 bp. Melting temperature (Tm) predictions for each amplicon were carried out using the nearest-neighbor model (Breslauer et al., 1986; SantaLucia, 1998), setting parameters such as the concentration of sodium ions and reaction temperature to 1 M and 37 °C, respectively. ΔTm for each INDEL amplicon was predicted, and INDEL markers with a ΔTm greater than 0.5°C were retained as HRM markers.
To address the limitations of physical distance in reflecting genetic recombination rates, HRM markers were anchored to the constructed genetic linkage map in B. rapa based on their physical positions (Liu et al., 2016). Genome-wide HRM markers, evenly distributed across 10 chromosomes of B. rapa, were chosen for HRM genotyping, guided by both physical and genetic maps. Amplification and allelic discrimination for HRM genotyping were performed following a previously reported method (Chen et al., 2021).
WGR data of representative individuals (BC3-1S1-6, BC3-6S1-15, BC3-7S1-14, and R-o-18) were mapped to assembly of Chiifu. Firstly, whole-genome SNP datasets were generated using the Variant Calls method. Loci with missing genotypes were filtered out from the SNP datasets. Genotypes obtained from the SNP datasets were anchored to the physical map based on their respective physical positions. Graphical genotyping analysis was conducted using our Python script available at https://github.com/clc-CAAS/Genetic_map_construction.
PR was calculated after genotyping using the formula: PR = (Recurrent parent allele count * 2 + Heterozygous allele count * 1 + Donor parent allele count * 0)/Total allele count * 2. HGB was calculated after genotyping using the formula: HGB = Homozygous allele count/Total allele count. To distinguish genotyping by HRM markers and WGR data, PR and HGB were designated as PRm and HGBm, respectively, when calculated after genotyping by HRM markers, and as PRs and HGBs, respectively, when calculated after genotyping by WGR data.
Chinese cabbage flowers from Chiifu, R-o-18, BC3-1S1-6, BC3-6S1-15, and BC3-7S1-14 were manually pollinated with a consistent amount of self-pollen grains one or two days after reaching full bloom. Ten flowers were selected for each individual. After 6 hours, stigmas were cut 2 mm away from the stigmatic surface for aniline blue staining to visualize pollen tubes following a published approach (Zhang et al., 2021). Stigmas were fixed in Canoy’s fixative (methanol: acetic acid = 3:1), softened in 10 M NaOH, and stained with 0.1% aniline blue. Pollen tubes were visualized under UV fluorescence using a Leica MSV269 Stereo Microscope, and images were captured with a Leica DFC450 digital camera. SI, MSC or SC was assessed by the number of pollen tubes penetrating stigmas (NPT). The levels of SI, MSC or SC were classified as follows: SI (NPT < 10), MSC (10 ≤ NPT ≤ 25), SC (NPT > 25).
To identify the SNP responsible for inducing G194R within MLPK, we examined SNP datasets from 524 B. rapa genomes (Cai et al., 2021). We identified a total of 26 SNPs within MLPK (Supplementary Table S3). A single nucleotide substitution (G to C) at position 1277 bp (G1277C) in the fourth exon of MLPK was found to induce the G194R substitution (Figure 2A; Supplementary Table S4). This G1277C mutation was present in only five out of 524 B. rapa genomes, while the MLPK/MLPK (MM) genotype observed in 509 genomes and missing genotype data in 10 genomes. Additionally, sequence variants of MLPK between Chiifu and R-o-18 were visualized, as illustrated in Supplementary Figure S1. We developed a specific KASP marker named SC-MLPK based on the G1277C mutation (Figure 2B; Supplementary Table S5). We optimized the thermal cycling conditions of SC-MLPK for KASP genotyping, determining that 32 cycles after touchdown in the PCR program were superior to the native 26 cycles (Supplementary Figure S2; Supplementary Table S2).
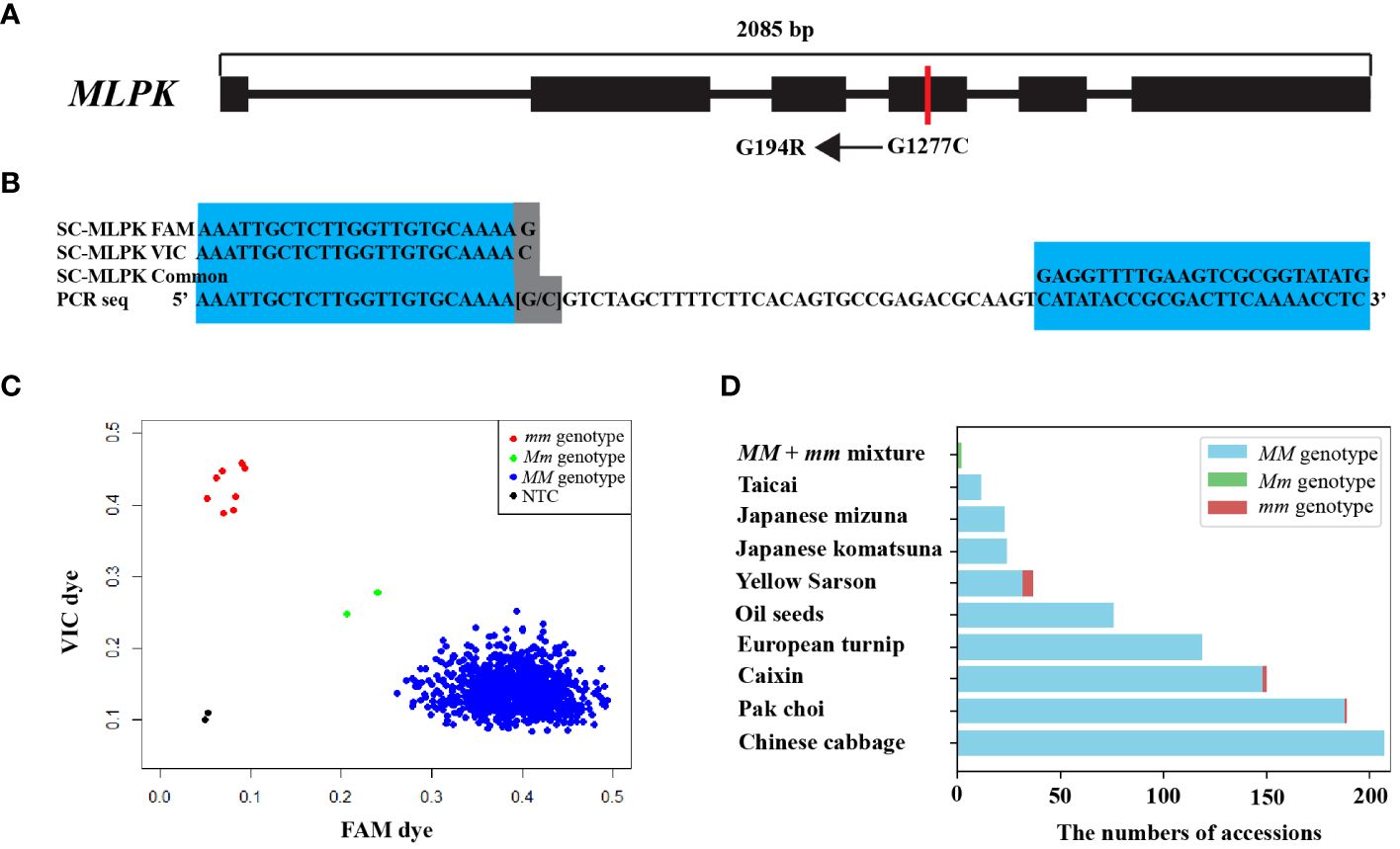
Figure 2 Structural organization of MLPK and KASP genotyping by the SC-MLPK marker. (A) Structural organization of the MLPK gene in B. rapa, highlighting “G1277C” in the coding sequence (CDS) between Chiifu and R-o-18. “G1277C” represents a single nucleotide substitution (G to C) at position 1277 bp of the MLPK gene. “G194R” represents a specific amino acid substitution (Gly to Arg) at position 194. Exons are denoted by black rectangles. (B) Sequences of the MLPK-specific marker SC-MLPK. (C) KASP genotyping in 841 B. rapa accessions using the SC-MLPK marker. Scattered plots showed clustering of alleles on the X- (FAM) and Y- (VIC) axes. Blue represents FAM-labeled allele (MM genotype); red represents VIC labeled allele (mm genotype); green represents the MLPK/mlpk (Mm) genotype, pesudo-heterogenetic sample by manually mixing equal amounts of DNA of R-o-18 and Chiifu; black represents the NTC (no-template control). (D) Distribution of mm plants across B. rapa subspecies.
To identity accessions with mm genotypes, we genotyped 841 B. rapa accessions by SC-MLPK marker assay. Eight accessions with the mm genotype were identified, including five yellow sarson, one pak choi, two caixin. Notably, the mm genotype was not detected in any accessions of Chinese cabbage (Figures 2C, D). To further confirm the self-compatibility of the mm plants, we accessed CI values for these eight accessions (Table 1), revealing CI values ranged from 4.25 to 16.3 among them. These results demonstrated that all of the eight accessions with mm genotypes were self-compatible. Thus, the mm genotype emerged as a potential factor for overcoming self-incompatibility in B. rapa.
To facilitate the introgression of mm allele into Chiifu through MABC, we developed HRM markers which discriminate genome backgrounds between Chiifu and R-o-18. By aligning 10 Gb WGR data of R-o-18 to the reference genome Chiifu v3.0, a total of 665,095 INDELs were identified. Among these, 188,289 INDELs meeting stringent criteria of homozygosity and a length of ≥ 3 bp were selected for marker design. Furthermore, 131 co-dominant HRM markers (Supplementary Table S6) evenly distributed across 10 chromosomes of the B. rapa (Table 2), were successfully developed for MABC. The ΔTm (melting temperature difference) values of INDEL amplicons between Chiifu and R-o-18 ranged from 0.8 °C to 2 °C for these HRM markers. The number of HRM markers ranged from 10 on chromosomes A07 and A08 to 17 on chromosome A03. The average genome-wide density of HRM markers was 2.28 Mb, ranging from 1.48 Mb on A10 to 3.01 Mb on A09. Furthermore, the average genetic interval across the genome was 7.29 cM, with a range from 5.15 cM on A10 to 9.04 cM on A07 (Table 2). The HRM genotyping for a representative HRM marker on each chromosome is depicted in Supplementary Figure S3. The 131 evenly distributed HRM markers across 10 chromosomes were used for further MABC of mm allele introgression from R-o-18 into Chiifu.
To facilitate the selection of individuals harboring the Mm genotype in successive backcrossing populations, the KASP assay for the SC-MLPK marker was applied to the BC1, BC2, and BC3 populations. The distribution of individuals harboring the Mm genotype in each backcrossing population was as follows: BC1 (6/12), BC2 (n=534/1120), and BC3 (n=390/768). An extensive population comprising 1120 individuals was utilized in the BC2 population to expedite the selection process for plants with the Mm genotype and a high PRm. The segregation ratio of MM: Mm in each backcrossing population aligned with the anticipated Mendelian ratio of 1: 1 (P > 0.05).
To streamline the genetic background analysis in the extensive BC2 population, a two-step MAS strategy was implemented for individuals with the Mm genotype. In the first step, three HRM markers on chromosomes A01, A02, and A09 were used for preliminary selection (Supplementary Figures S4A-D). Subsequently, a total of 96 individuals selected from the first round screening were upon the second round selection using the left 128 HRM markers. Based on the result from assays by SC-MLPK and 131 HRM markers, the maximum PRm in the BC2 population was determined to be 85.1%. For background analysis of individuals with the Mm genotype in the BC3 population, 39 markers with heterozygous genotypes from the BC2 population were employed. The highest PRm was 97.3% among the BC3 population (Supplementary Figure S4D). These results elucidated that the genetic background of each backcrossing population progressively approached to that of Chiifu.
Three BC3 individuals with Mm genotypes as well as high PRm, including BC3-1 (PRm = 97.3%), BC3-6 (PRm = 95.1%), and BC3-7 (PRm = 94.7%), were retained for self-pollination to generate BC3S1 populations. From each BC3S1 population, ten mm genotype plants were selected to be genotyped using background HRM markers (Figure 3A). Genotyping revealed that PRm for the BC3S1 population ranged from 93.9% to 98.5%. Three mm individuals with high PRm and high HGBm, namely BC3-1S1-6, BC3-6S1-15, and BC3-7S1-14, were retained from BC3S1 population. Genotyping indicated that the PRm of BC3-1S1-6, BC3-6S1-15, and BC3-7S1-14 was 98.5%, 96.6%, and 95.8%, respectively. The HGBm of BC3-1S1-6, BC3-6S1-15, and BC3-7S1-14 was 100%, 96.2%, and 99.2%, respectively.
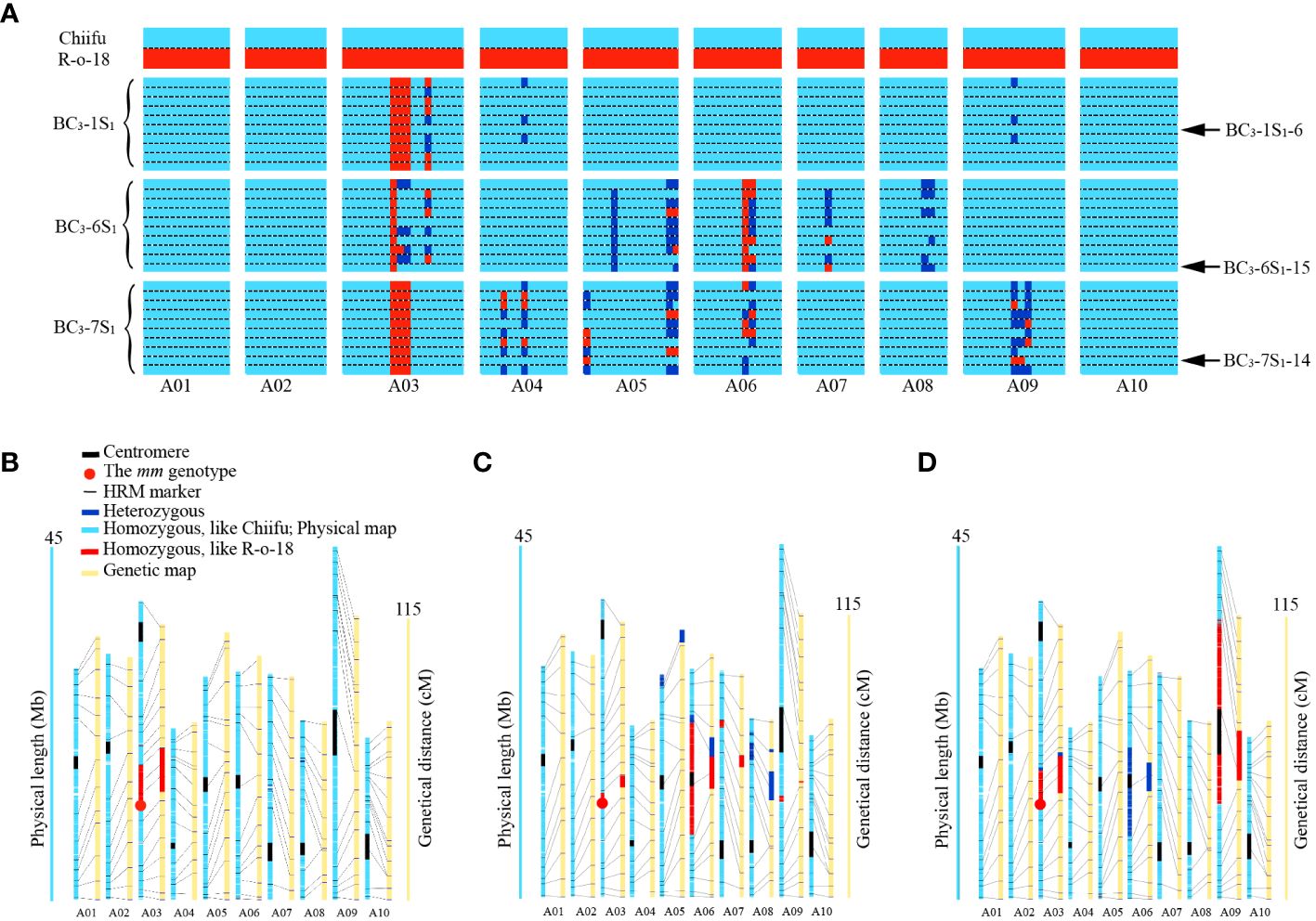
Figure 3 Genetic background analysis using markers and WGR. (A) Graphical genotyping based on polymorphism for SC-MLPK and 131 HRM markers. The vertical axis represents parental materials and ten introgression individuals of each BC3S1 population, while the horizontal axis represents SC-MLPK and 131 HRM markers (totaling 132 markers). Introgression individuals are grouped by different BC3S1 populations. The markers were organized according to their physical positions on the chromosomes. Arrow represents the selected individuals from each BC3S1 population. (B-D) Genotypes, determined by WGR, anchored to physical and genetic maps in BC3-1S1-6, BC3-6S1-15 and BC3-7S1-14, respectively. The yellow columns represent the genetic linkage groups corresponding to each chromosome. The centromere region is delineated by the black box. The red dot represents the mm genotype. Light blue segment indicated the homozygous genotype identical to Chiifu, red segment denoted the homozygous genotype identical to R-o-18, and navy blue segment represented the heterozygous genotype.
To further confirm the high similarity of genetic backgrounds between the three BC3S1 individuals to that of Chiifu, we conducted WGR for them. Genome-wide SNPs were extracted from aligned reads, utilizing the B. rapa genome Chiifu v3.0 as a reference. Loci with missing genotypes for each sample were filtered, and SNPs with homozygous genotypes in R-o-18 were retained, resulting in the identification of 1.6 million SNPs spanning chromosomes A01 to A10. These SNPs were anchored to both physical and genetic maps (Figures 3B-D). Based on the SNPs, boundaries of introgressed segments were delineated in BC3-1S1-6, BC3-6S1-15, and BC3-7S1-14 (Supplementary Table S7). Genotyping revealed that the PRs of BC3-1S1-6, BC3-6S1-15, and BC3-7S1-14 was 98.1%, 92.6%, and 88.5%, respectively. The HGBs of BC3-1S1-6, BC3-6S1-15, and BC3-7S1-14 was 99.6%, 97%, and 96%, respectively (Table 3). The individual plant (BC3-1S1-6) with the highest PRs of 98.1% in the BC3S1 population displayed a near-identical match with the recurrent parent Chiifu genotype, except for a specific region (approximately 5.2 Mb, from 12059865 to 17244940) of the MLPK gene linkage on chromosome A03 retaining the donor parent R-o-18 genotype (Figure 3B). This result indicated successful introgression of mm allele from R-o-18 into Chiifu.
To evaluate the phenotypic traits of individual plants within the BC3-1S1, BC3-6S1, and BC3-7S1 populations relative to Chiifu, we conducted a detailed analysis encompassing agronomic traits, pollen tube visualization, and assessment of CI value (Figure 4). For agronomic trait analysis, 10 plants with mm genotype were selected from each BC3S1 population. Before leafy heads were ready for harvest, head height and width were evaluated and compared with Chiifu (Table 4). The head height and width ranged from 23.5 to 25.1 cm and 10.7 to 13.0 cm, respectively. Notably, both the average head height and width in BC3-6S1 population were not significantly different from those of Chiifu. In each BC3S1 population, all individuals exhibited heading, hairy, and crinkle traits similar to Chiifu, while the R-o-18 line displayed non-heading, hairless, and smooth traits. Three individuals including BC3-6S1-15, BC3-1S1-6, and BC3-7S1-14 exhibiting both genotypic and phenotypic similarity to Chiifu were selected from each BC3S1 population (Figures 4A-C; Supplementary Figure S5).
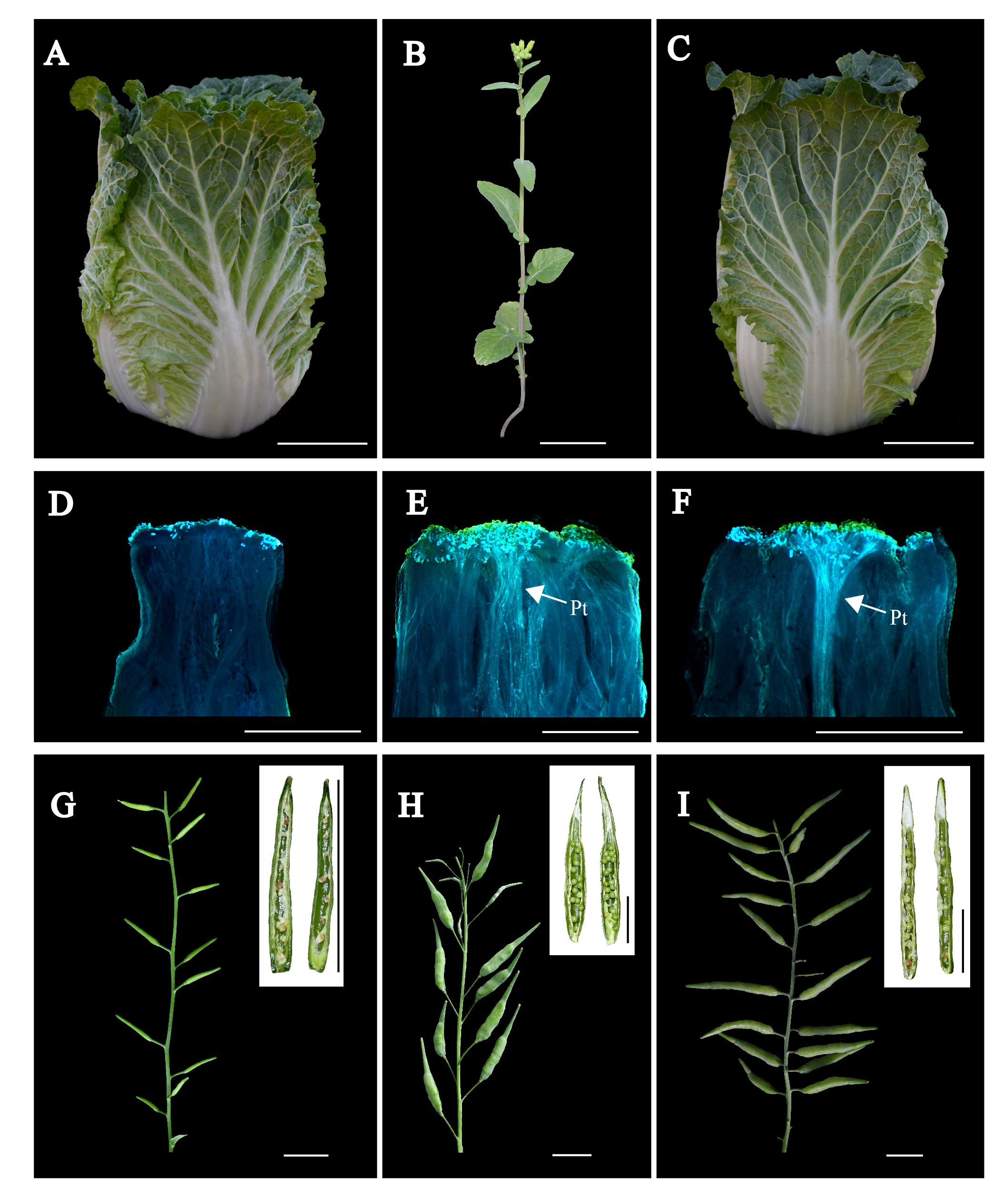
Figure 4 Comprehensive assessment of field and reproductive performance in Chiifu, R-o-18 and BC3-6S1-15 individual plants. (A–C) Field performance comparison of Chiifu, R-o-18, and BC3-6S1-15 plants. Scale bars in (A–C), 5 cm. (D–F) Fluorescence microscopic images depicting pollen tube germination at the stigmas of Chiifu, R-o-18, and BC3-6S1-15 individual plants, with “Pt” indicating the pollen tubes. Scale bars in (D–F), 1 mm. (G-I) Seed setting of Chiifu, R-o-18, and BC3-6S1-15 individual plants. Scale bars in (G–I), 2 cm.
Upon flowering, BC3-6S1-15, BC3-1S1-6, and BC3-7S1-14, together with Chiifu, R-o-18, were investigated for pollen tube elongation and CI. Fluorescence microscopy revealed that R-o-18, BC3-6S1-15, and BC3-7S1-14 plants had more than 25 pollen tubes, whereas no pollen tubes were observed in BC3-1S1-6 and Chiifu (Figures 4D-F; Supplementary Figure S6). During the seed-filling stage, seed setting was evaluated (Figures 4G-I). Notably, BC3-6S1-15 and BC3-7S1-14 plants exhibited significant SC, achieving CI values of 7.5 and 7.1, respectively. In contrast, BC3-1S1-6 exhibited SI with a CI of 0.1, similar to that of Chiifu (CI = 0.1). Consequently, the two newly developed plants (BC3-6S1-15, BC3-7S1-14), which exhibited phenotypic and genetic similarities to Chiifu while demonstrating SC, were self-pollinated to generate two novel lines designated as SC-Chiifu Lines (SC-Chiifu-a, SC-Chiifu-b, respectively). The CI values of the SC-Chiifu Lines compared to the CI value of Chiifu (CI = 0.1) increased tenfold. However, the CI values of the SC-Chiifu Lines remained lower than that observed for R-o-18 (CI = 12). These results highlight the successful transfer of self-compatibility from R-o-18 into Chiifu, leading to the development of two novel SC-Chiifu Lines.
In the BC2S1 population, 63 individuals with the mm genotype were selected for the assessment of CI values. The CI values varied significantly among these individuals, spanning from 0.2 to 13 (Supplementary Table S8). Subsequent CI analysis delineated 29 individuals exhibiting SC (CI > 4), 22 individuals displaying MSC (1 ≤ CI ≤ 4), and 12 individuals manifesting SI (CI < 1). This observation indicated a pronounced segregation of SC within the mm plants of the BC2S1 population.
Furthermore, variations in SC were also observed among the three selected individuals (BC3-6S1-15, BC3-7S1-14, and BC3-1S1-6) with the mm genotype. Given that all three plants in the BC3S1 population possessed the same mm genotype and similar genetic backgrounds, the observed segregation suggested the involvement of gene(s) beyond the MLPK locus in influencing self-compatibility in B. rapa. We postulated that the gene(s) responsible being located at the segments shared by BC3-6S1-15 and BC3-7S1-14 individuals are distinct from those in BC3-1S1-6. We examined S haplotypes in BC3-1S1-6, BC3-6S1-15, BC3-7S1-14, Chiifu and R-o-18 using S-specific primers (Oikawa et al., 2011). We found that BC3-1S1-6, BC3-6S1-15, and BC3-7S1-14, along with Chiifu, had S60 homozygotes, while R-o-18 exhibited an S54 homozygote. There was no correlation between CI value and S haplotype in BC3-1S1-6, BC3-6S1-15 and BC3-7S1-14. Based on our analysis on the introgressed segment boundaries in the three selected individuals, we identified four such segments (Figure 5). Specifically, four shared segments were located on chromosomes A03 (approximately 0.8 Mb, from 16451403 to 17244940), A06 (approximately 11.3 Mb, from 8101576 to 19449508), A07 (approximately 0.8 Mb, from 14002870 to 14798378), and A09 (approximately 0.7 Mb, from 12255709 to 12997755). These findings suggested that these shared segments emerged as candidate segments potentially influencing SC.
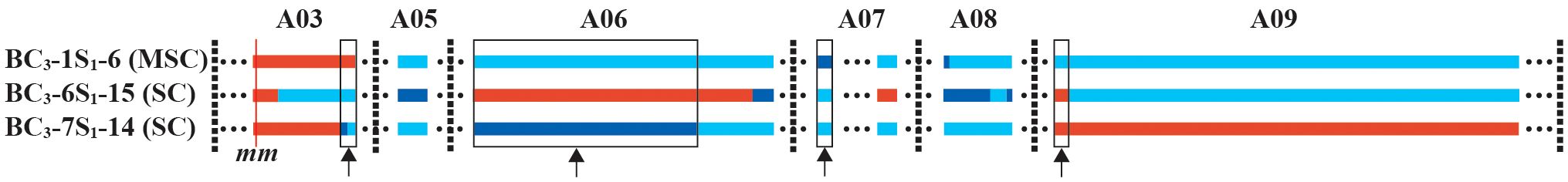
Figure 5 Identification of the shared segments in BC3-6S1-15 and BC3-7S1-14 but not in BC3-1S1-6. Light blue segments indicated the homozygous genotype identical to Chiifu; navy blue segments indicate the heterozygous genotype; red segments indicate the homozygous R-o-18 genotype. Arrows indicate the shared segments between BC3-6S1-15 and BC3-7S1-14.
The production of self-pollinated seeds is difficult for Chiifu due to its high self-incompatibility, hampering it being widely used in genetic studies although the reference genome assemblies have been continuously updated. The small bud size of Chiifu adds to the complexity, making it challenging to manually open for self-pollination attempts. Despite efforts to self-pollinate during the bud stage, success rates are often low due to its small bud size and high self-incompatibility. Furthermore, the combination of high self-incompatibility and strong crossing compatibility exacerbates the issue by frequently leading to presence of contaminated hybrid seeds in the limited seed yield obtained from self-pollination attempts. In this study, we addressed the self-incompatibility issue in Chiifu by introducing the mm allele into this line. Through this genetic modification, we successfully developed two SC-Chiifu lines that not only retained genetic backgrounds closely resembling Chiifu but also exhibited self-compatibility phenotypes. The development of these SC-Chiifu lines enabled efficient seed setting without the need for manual pollination. This not only alleviates the challenges associated with self-pollination but also mitigates the risk of cross-pollen contamination.
Genetic background analysis within the SC-Chiifu Lines revealed that approximately 90% of the genetic segments originate from the recurrent parent, Chiifu, with the remaining 10% comprising exogenetic segments from the donor parent, R-o-18. Through the utilization of WGR data, we precisely delineated the boundaries of these exogenetic segments. This delineation offers valuable insights for future research endeavors involving the SC-Chiifu Lines. If the SC-Chiifu Lines are employed for updating the Chiifu genome, it is imperative to purify these exogenetic segments through targeted selection. Alternatively, for functional gene research, it may be feasible to disregard these exogenetic segments if they do not significantly impact the traits of interest, such as heading and leaf development. The SC-Chiifu Lines represent crucial plant materials for various applications, including the construction of a Chiifu mutant library and conducting Chiifu-based transgenic experiments. These lines are particularly valuable for breeding self-compatible heading Chinese cabbage, serving as ideal donor parents to provide self-compatibility and heading traits. In the study of self-compatibility mechanisms, both self-compatible SC-Chiifu lines and the self-incompatible BC3-1S1-6 individual have proven to be valuable resources. Furthermore, the seeds from the SC-Chiifu Lines described in this study will be made accessible to researchers for further functional genomics studies, thereby facilitating broader exploration and utilization of these valuable genetic resources.
B. rapa typically exhibits SI, overcoming this trait is not only crucial for utilizing the Chiifu line but also plays a pivotal role in enhancing B. rapa hybrid seed production using self-compatible lines within the male-sterile system. The conversion of male parental lines and maintainer lines into self-compatible lines is essential for the hybrid seed production process. Self-compatible lines streamline the reproduction process through honeybee pollination and circumvent crossing incompatibility during hybrid seed production. The G1277C mutation within MLPK represents a rare mutation. Among 841 accessions of B. rapa analyzed, only a minority, specifically eight accessions, exhibited the G1277C mutation. Assessment of CI values revealed that these eight accessions all demonstrated SC. In response to the G1277C, we developed a self-compatibility-specific marker, SC-MLPK. Following experimental optimization, this marker exhibited stable and accurate genotyping characteristics, making it widely applicable for the creation of self-compatible B. rapa lines. The successful utilization of SC-MLPK for overcoming SI serves as a valuable reference for the cultivation of self-compatible B. rapa lines.
The success of MABC critically depends on the development of molecular markers that are not only evenly distributed, operationally straightforward, and cost-effective but also high in density for precise genetic background analysis. HRM genotyping technology offers distinct advantages in meeting these criteria. In a previous study, we developed 148 HRM markers evenly distributed in the genome based on a physical map to differentiate between four heading and four non-heading B. rapa (Chen et al., 2021). In this study, we expanded upon this approach by developing 131 HRM markers evenly distributed across the B. rapa genome, guided by both physical and genetic maps, to differentiate between Chinese cabbage line Chiifu and yellow sarson line R-o-18. These 279 HRM markers represent a powerful tool in B. rapa genetics and breeding.
Notably we observed segregation in SI and SC among plants with mm genotypes in the both BC2S1 and BC3S1 generations. This is different from the observation in germplasm screening that all the eight accessions with G1277C mutation were SC. These observations suggested the potential for overcoming SI barriers or reinforcing SC through the incorporation of gene(s) beyond MLPK. Put differently, under a genetic background akin to Chiifu, individuals carrying solely mm genotypes prove inadequate in overcoming its self-incompatibility. To obtain self-compatible Chiifu lines, it is imperative to consider not only the MLPK locus but also other loci influencing self-compatibility. This necessity compromises our ability to achieve a PR of 100% in obtaining the SC-Chiifu Lines. Through the identification of boundaries of the three selected individuals in the BC3S1 generation, we detected four candidate segments potentially regulating SI. Additionally, lines SC-Chiifu-a and SC-Chiifu-b (CI > 7) overcame SI and reached SC levels, albeit not reaching the SC level of R-o-18 (CI = 12). This suggested that besides the gene(s) in the four candidate segments, S haplotype and MLPK, other gene(s) may influence SC. Further exploration through the construction of genetic populations can elucidate the genetic mechanisms involved in overcoming SI barriers or enhancing SC. Such investigations hold promise for advancing our understanding of SC mechanisms in Brassica and for developing more efficient breeding strategies.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
LC: Data curation, Formal analysis, Investigation, Methodology, Resources, Software, Writing – original draft, Writing – review & editing. JL: Resources, Writing – review & editing. XC: Formal analysis, Writing – review & editing. LZ: Resources, Writing – review & editing. YL: Methodology, Writing – review & editing. JW: Funding acquisition, Project administration, Writing – review & editing. XW: Funding acquisition, Project administration, Resources, Supervision, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Key Research and Development Program of China (2022YFF1003000), the Agricultural Science and Technology Innovation Program (ASTIP), and the Central Public-interest Scientific Institution Basal Research Fund (No. Y2023PT16).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1397018/full#supplementary-material
Breslauer, K. J., Frank, R., Blöcker, H., Marky, L. A. (1986). Predicting DNA duplex stability from the base sequence. Proc. Natl. Acad. Sci. U.S.A. 83, 3746–3750. doi: 10.1073/pnas.83.11.3746
Cabrillac, D., Cock, J. M., Dumas, C., Gaude, T. (2001). The S-locus receptor kinase is inhibited by thioredoxins and activated by pollen coat proteins. Nature 410, 220–223. doi: 10.1038/35065626
Cai, C., Wang, X., Liu, B., Wu, J., Liang, J., Cui, Y., et al. (2017). Brassica rapa Genome 2.0: A Reference Upgrade through Sequence Re-assembly and Gene Re-annotation. Mol. Plant 10, 649–651. doi: 10.1016/j.molp.2016.11.008
Cai, X., Chang, L., Zhang, T., Chen, H., Zhang, L., Lin, R., et al. (2021). Impacts of allopolyploidization and structural variation on intraspecific diversification in Brassica rapa. Genome Biol. 22, 166. doi: 10.1186/s13059-021-02383-2
Cai, X., Wu, J., Wang, X. (2023). Harvesting knowledge: illuminating advances in Brassica crops genomics and breeding. Horticulturae 9, 1332. doi: 10.3390/horticulturae9121332
Chang, L., Liang, J., Zhang, L., Zhang, Z., Cai, X., Wu, J., et al. (2023). A complex locus regulates highly lobed-leaf formation in Brassica juncea. Theor. Appl. Genet. 136, 224. doi: 10.1007/s00122-023-04473-0
Chen, R., Chang, L., Cai, X., Wu, J., Liang, J., Lin, R., et al. (2021). Development of inDel markers for Brassica rapa based on a high-resolution melting curve. Hortic. Plant J. 7, 31–37. doi: 10.1016/j.hpj.2020.05.003
Chen, S., Zhou, Y., Chen, Y., Gu, J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Cheng, F., Wu, J., Cai, C., Fu, L., Liang, J., Borm, T., et al. (2016). Genome resequencing and comparative variome analysis in a Brassica rapa and Brassica oleracea collection. Sci. Data 3, 160119. doi: 10.1038/sdata.2016.119
Distefano, G., Caruso, M., La Malfa, S., Gentile, A., Wu, S. B. (2012). High resolution melting analysis is a more sensitive and effective alternative to gel-based platforms in analysis of SSR–an example in citrus. PLoS One 7, e44202. doi: 10.1371/journal.pone.0044202
Dou, J., Wang, Y., Yang, H., Niu, H., Liu, D., Yang, S., et al. (2022). Development of branchless watermelon near isogenic lines by marker assisted selection. Hortic. Plant J. 8, 627–636. doi: 10.1016/j.hpj.2022.07.003
Herrmann, M. G., Durtschi, J. D., Wittwer, C. T., Voelkerding, K. V. (2007). Expanded instrument comparison of amplicon DNA melting analysis for mutation scanning and genotyping. Clin. Chem. 53, 1544–1548. doi: 10.1373/clinchem.2007.088120
Jung, H. J., Jung, H. J., Ahmed, N. U., Park, J. I., Kang, K. K., Hur, Y., et al. (2012). Development of self-compatible B. rapa by RNAi-mediated S locus gene silencing. PLoS One 7, e49497. doi: 10.1371/journal.pone.0049497
Kakita, M., Murase, K., Iwano, M., Matsumoto, T., Watanabe, M., Shiba, H., et al. (2007). Two distinct forms of M-locus protein kinase localize to the plasma membrane and interact directly with S-locus receptor kinase to transduce self-incompatibility signaling in Brassica rapa. Plant Cell 19, 3961–3973. doi: 10.1105/tpc.106.049999
Li, H., Durbin, R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
Li, H., Gu, A., Feng, D., Li, N., Yang, R., Zhang, X., et al. (2023). Fast tracking alien gene discovery by molecular markers in a late flowering Chinese cabbage-cabbage translocation line ‘AT7–4’. Hortic. Plant J. 9, 89–97. doi: 10.1016/j.hpj.2021.11.009
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Liu, X., Dong, S., Miao, H., Bo, K., Li, C., Yang, Y., et al. (2022). Genome-wide analysis of expansins and their role in fruit spine development in cucumber (Cucumis sativus L.). Hortic. Plant J. 8, 757–768. doi: 10.1016/j.hpj.2021.11.004
Liu, J., Liu, B., Cheng, F., Liang, J., Wang, X., Wu, J. (2016). A high density linkage map facilitates QTL mapping of flowering time in Brassica rapa. Hortic. Plant J. 2, 217–223. doi: 10.1016/j.hpj.2016.11.003
Mavrodiev, E. V., Dervinis, C., Whitten, W. M., Gitzendanner, M. A., Kirst, M., Kim, S., et al. (2021). A new, simple, highly scalable, and efficient protocol for genomic DNA extraction from diverse plant taxa. Appl. Plant Sci. 9, e11413. doi: 10.1002/aps3.11413
McKenna, A., Hanna, M., Banks, E., Sivachenko, A., Cibulskis, K., Kernytsky, A., et al. (2010). The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303. doi: 10.1101/gr.107524.110
Murase, K., Shiba, H., Iwano, M., Che, F. S., Watanabe, M., Isogai, A., et al. (2004). A membrane-anchored protein kinase involved in Brassica self-incompatibility signaling. Science 303, 1516–1519. doi: 10.1126/science.1093586
Nasrallah, J. B., Nasrallah, M. E. (1993). Pollen[mdash]Stigma signaling in the sporophytic self-incompatibility response. Plant Cell 5, 1325–1335. doi: 10.2307/3869785
Ohata, M., Takada, Y., Sato, Y., Okamoto, T., Murase, K., Takayama, S., et al. (2023). MLPK function is not required for self-incompatibility in the S29 haplotype of Brassica rapa L. Plant Reprod. 36, 255–262. doi: 10.1007/s00497-023-00463-w
Oikawa, E., Takuno, S., Izumita, A., Sakamoto, K., Hanzawa, H., Kitashiba, H., et al. (2011). Simple and efficient methods for S genotyping and S screening in genus Brassica by dot-blot analysis. Mol. Breed. 28, 1–12. doi: 10.1007/s11032-010-9455-8
Robinson, J. T., Thorvaldsdóttir, H., Winckler, W., Guttman, M., Lander, E. S., Getz, G., et al. (2011). Integrative genomics viewer. Nat. Biotechnol. 29, 24–26. doi: 10.1038/nbt.1754
SantaLucia, J., Jr. (1998). A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl. Acad. Sci. U.S.A. 95, 1460–1465. doi: 10.1073/pnas.95.4.1460
Stone, S. L., Arnoldo, M., Goring, D. R. (1999). A breakdown of Brassica self-incompatibility in ARC1 antisense transgenic plants. Science 286, 1729–1731. doi: 10.1126/science.286.5445.1729
Untergasser, A., Cutcutache, I., Koressaar, T., Ye, J., Faircloth, B. C., Remm, M., et al. (2012). Primer3–new capabilities and interfaces. Nucleic Acids Res. 40, e115. doi: 10.1093/nar/gks596
Wang, X., Wang, H., Wang, J., Sun, R., Wu, J., Liu, S., et al. (2011). The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 43, 1035–1039. doi: 10.1038/ng.919
White, H. E., Hall, V. J., Cross, N. C. (2007). Methylation-sensitive high-resolution melting-curve analysis of the SNRPN gene as a diagnostic screen for Prader-Willi and Angelman syndromes. Clin. Chem. 53, 1960–1962. doi: 10.1373/clinchem.2007.093351
Xiao, Z., Han, F., Hu, Y., Xue, Y., Fang, Z., Yang, L., et al. (2019). Overcoming cabbage crossing incompatibility by the development and application of self-compatibility-QTL- specific markers and genome-wide background analysis. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00189
Yamamoto, M., Ishii, T., Ogura, M., Akanuma, T., Zhu, X. Y., Kitashiba, H. (2023). S haplotype collection in Brassicaceae crops-an updated list of S haplotypes. Breed Sci. 73, 132–145. doi: 10.1270/jsbbs.22091
Zhang, L., Cai, X., Wu, J., Liu, M., Grob, S., Cheng, F., et al. (2018). Improved Brassica rapa reference genome by single-molecule sequencing and chromosome conformation capture technologies. Hortic. Res. 5, 50. doi: 10.1038/s41438-018-0071-9
Zhang, Z., Guo, J., Cai, X., Li, Y., Xi, X., Lin, R., et al. (2022b). Improved reference genome annotation of Brassica rapa by pacific biosciences RNA sequencing. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.841618
Zhang, L., Huang, J., Su, S., Wei, X., Yang, L., Zhao, H., et al. (2021). FERONIA receptor kinase-regulated reactive oxygen species mediate self-incompatibility in Brassica rapa. Curr. Biol. 31, 3004–3016.e3004. doi: 10.1016/j.cub.2021.04.060
Zhang, L., Li, X., Chang, L., Wang, T., Liang, J., Lin, R., et al. (2022a). Expanding the genetic variation of Brassica juncea by introgression of the Brassica rapa Genome. Hortic. Res. 9. doi: 10.1093/hr/uhab054
Zhang, L., Liang, J., Chen, H., Zhang, Z., Wu, J., Wang, X. (2023). A near-complete genome assembly of Brassica rapa provides new insights into the evolution of centromeres. Plant Biotechnol. J. 21, 1022–1032. doi: 10.1111/pbi.14015
Keywords: Chinese cabbage, self-incompatibility, MLPK, HRM (High-Resolution Melting), MAS (Marker-Assisted Selection)
Citation: Chang L, Liang J, Cai X, Zhang L, Li Y, Wu J and Wang X (2024) Development of self-compatible Chinese cabbage lines of Chiifu through marker-assisted selection. Front. Plant Sci. 15:1397018. doi: 10.3389/fpls.2024.1397018
Received: 06 March 2024; Accepted: 20 May 2024;
Published: 30 May 2024.
Edited by:
Ryo Fujimoto, Kobe University, JapanReviewed by:
Chaobo Tong, Chinese Academy of Agricultural Sciences, ChinaCopyright © 2024 Chang, Liang, Cai, Zhang, Li, Wu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaowu Wang, d2FuZ3hpYW93dUBjYWFzLmNu; Jian Wu, d3VqaWFuQGNhYXMuY24=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.