
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 24 April 2024
Sec. Plant Nutrition
Volume 15 - 2024 | https://doi.org/10.3389/fpls.2024.1392355
 Qing Liao1,2,3
Qing Liao1,2,3 Ao-Mei Li1,2,3
Ao-Mei Li1,2,3 Ying Xing1,2,3
Ying Xing1,2,3 Pan-Xia Liang1,2,3
Pan-Xia Liang1,2,3 Ze-Pu Jiang1,2,3
Ze-Pu Jiang1,2,3 Yong-Xian Liu1,2,3*
Yong-Xian Liu1,2,3* Dong-Liang Huang1,2,3*
Dong-Liang Huang1,2,3*Selenium (Se) is a crucial micronutrient for human health. Plants are the primary source of Se for humans. Selenium in the soil serves as the primary source of Se for plants. The soil contains high total Se content in large areas in Guangxi, China. However, the available Se is low, hindering Se uptake by plants. Microorganisms play a pivotal role in the activation of Se in the soil, thereby enhancing its uptake by plants. In this study, selenobacteria were isolated from Se-rich soils in Guangxi. Then two selenobacteria strains, YLB1-6 and YLB2-1, representing the highest (30,000 μg/mL) and lowest (10,000 μg/mL) Se tolerance levels among the Se-tolerant bacteria, were selected for subsequent analysis. Although the two selenobacteria exhibited distinct effects, they can significantly transform Se species, resulting in a decrease in the soil residual Se (RES-Se) content while concurrently increasing the available Se (AVA-Se) content. Selenobacteria also enhance the transformation of Se valencies, with a significant increase observed in soluble Se6+ (SOL-Se6+). Additionally, selenobacteria can elevate the pH of acidic soil. Selenobacteria also promote the uptake of Se into plants. After treatment with YLB1-6 and YLB2-1, the Se content in the aboveground part of Chinese flowering cabbage increased by 1.96 times and 1.77 times, respectively, while the Se accumulation in the aboveground part of the plant significantly increased by 104.36% and 81.69%, respectively, compared to the control. Further whole-genome sequencing revealed the genetic difference between the two selenobacteria. Additionally, 46 and 38 candidate genes related to selenium utilization were identified from YLB1-6 and YLB2-1, respectively. This work accelerates our understanding of the potential molecular mechanism of Se biofortification by selenobacteria. It also provides microorganisms and gene targets for improving crop varieties or microorganisms to exploit the rich Se source in soil.
Selenium (Se) is one of the 15 essential trace elements in the human body, regulating numerous physiological processes, earning the reputation of “element of miraculous life”. Se in the soil serves as the primary source of Se for humans. The pathway “soil-plant-human” is currently the most efficient and common Se biofortification way to supply Se for humans (Chilimba et al., 2012; Premarathna et al., 2012; Schiavon et al., 2020; Hossain et al., 2021).
Guangxi has a Se-rich soil area of 75,700 km2, making it the largest designated Se-rich soil region in China. Our team collected and analyzed 1,500 soil samples across Guangxi, revealing an average Se content in the soil of 0.55 mg/kg, with the highest recorded value being 2.29 mg/kg (Liu et al., 2015). This level far exceeds the average soil Se content in China, which is 0.29 mg/kg (China National Environmental Monitoring Station, 1990), signifying that the soil Se content in Guangxi is rich and high. However, in Guangxi, the majority of soils are acidic, and Se in such soils mainly forms insoluble oxides and hydroxides with iron, resulting in low available Se content (Geng, 2010). This hinders the utilization of the rich soil Se resources in Guangxi. Therefore, improving the available Se content in soil has become an urgent requirement to exploit soil Se resources not only in Guangxi but also globally.
The fractions and species of selenium (Se) in the soil are pivotal factors influencing Se availability (Shardendu et al., 2003). Microorganisms play a significant role in influencing the fractions, species, migration, and bioavailability of soil Se (Stolz et al., 2002, 2006; Valdez Barillas et al., 2011; Xu et al., 2017). They facilitate the conversion of Se fractions and species through processes such as oxidation, reduction, assimilation, and methylation (Nancharaiah and Lens, 2015), thereby enhancing the uptake of soil Se by plants (Ye et al., 2020; Yang et al., 2021). Consequently, Se biofortification by microorganisms emerges as a crucial approach for utilizing Se sources in the soil. Co-inoculation with selenobacteria strains, such as Enterobacter sp. B16, and the arbuscular mycorrhizal fungus Glomus claroideum, has been shown to significantly increase Se levels in wheat grains (Durán et al., 2013). Three bacterial strains isolated from corn rhizosphere soil exhibited a robust activating effect on Se in the soil, enhancing the soil’s water-soluble Se content (Long et al., 2017). A Se-tolerant endophyte, Bacillus methylotrophicus CSN-1, isolated from the leaves of Cardamine hupingshanensis, a plant accumulating super-high levels of Se, can convert selenite into SeCys2, enhancing plant uptake and transport of Se (Zhang et al., 2018). Inoculation of Arbuscular mycorrhizal fungi has been demonstrated to significantly promote the uptake of selenate or selenite by winter wheat (Luo et al., 2019).
In addition, bacteria harbor extensive genetic resources. The identification of genes associated with Se transformation in bacteria can offer potential targets for the improvement of bacteria or plants through molecular technologies, thereby enhancing Se utilization. Whole-genome sequencing facilitates the decryption of the genetic code of microorganisms, providing insights into candidate genes involved in Se utilization and contributing to a deeper understanding of the mechanisms underlying Se biofortification (Peng et al., 2016).
To enhance the genetic and microbial resources available for the biofortification of selenium (Se) in Se-rich soil, this study focused on the isolation and identification of highly Se-tolerant microorganisms from Se-rich soils in Guangxi. These microorganisms exhibit the capability to alter Se species and valencies in the soil, thereby facilitating Se uptake by plants. Additionally, the genomes of two distinct selenobacteria, characterized by divergent Se utilization capacities, were sequenced. This genomic information serves as a valuable resource for understanding the genetic basis of Se utilization. This work provides new microorganisms and genetic sources for the exploitation of rich Se depository in soil, and accelerate our understanding on the mechanism regarding Se biofortification.
Solid Media: Nutrient agar, Gause’s synthetic agar, potato dextrose agar were procured from Guangdong Huankai Microbial Sci&Tech Co., Ltd. (China).
Liquid Media: Prepared by following the formulations of the aforementioned solid culture media, excluding the agar component. Autoclaving was performed at 121°C, 15 PSI for 20 minutes before use.
PBS Buffer: Purchased from Shenggong Biotechnology (Shanghai) Co., Ltd. (China).
Na2SeO3 (AR 98%): Procured from Shandong Xiyachem Chemical Industry Co., Ltd. (China).
Se Solution (100 mg/mL): Weighed 22.35 g of Na2SeO3 (AR 98%), dissolved it in deionized water to make up 100 mL. The solution was filtered through a 0.22 μM sterile filter, stored in the dark for further use.
All other chemical reagents are domestically produced and of analytical grade.
Soil samples were collected from Se-rich areas in Guangxi, including Yongfu, Bama, Yulin, Guiping, and Teng county. Ten grams of soil sample were added to 90 mL of sterilized PBS buffer containing 10 glass beads, followed by shaking at 30°C and 200 rpm for 30 minutes, standing for 5 minutes, and collecting the suspension. Subsequently, the suspension was centrifuged at 5000 rpm for 15 minutes, and the sediment was resuspended in 10 mL of PBS buffer and stored at 4°C. Two mL of the above sample was added to 100 mL of liquid medium with a Se concentration of 100 μg/mL. Pure cultures of strains were obtained using gradient dilution and the streak plate method. The purified strains were then cultured in a liquid medium. Next, 5 μL of the 24-48 hour culture was inoculated into solid culture media with different gradient Se concentrations. Strains with poor Se tolerance were systematically eliminated to obtain strains with stronger Se tolerance.
The 16S rDNA sequence of the highly Se-tolerant bacteria was determined by Nanning Guotuo Biotechnology Co., Ltd. (China). The acquired sequence was submitted to NCBI GenBank and compared to known sequences using the BLAST software. The phylogenetic tree was constructed using MEGA 5.0 software.
Bacteria: two bacterial strains, Bacillus cereus YLB1-6 (Registration number in China Center for Type Culture Collection: M2020342, CCTCC NO: M2020342) and Bacillus altitudinis YLB2-1 (Registration number in Guangdong Microbial Culture Collection Center: 62524, GDMCC NO: 62524), were utilized for pot inoculation experiments. These strains were inoculated into a liquid medium and incubated at 37°C, 200 rpm for 24 hours. Following centrifugation at 5000 rpm for 10 minutes, bacterial cells were collected, washed twice with sterile 0.85% sodium chloride (NaCl), and then resuspended in sterile water to achieve a final bacterial suspension of approximately 1×108 CFU/mL.
Soil: the soil selected for the pot experiment was obtained from Guiping City, Guangxi (109°56′59″ E, 23°18′36″ N). Characterized as lateritic red soil derived from granite, the basic physicochemical properties of the soil are as follows: total selenium content is 0.95 mg/kg, total nitrogen is 1.30 g/kg, total phosphorus is 0.736 g/kg, total potassium is 4.76 g/kg, alkali-soluble nitrogen is 129 mg/kg, readily available phosphorus is 46.0 mg/kg, readily available potassium is 82.6 mg/kg, organic matter is 16.0 g/kg, and the pH is 6.30. The soil underwent natural air-drying, grinding, and sieving through a 5 mm sieve. Before use, the soil was autoclaved at 121°C, 15 PSI for 30 minutes.
Plant: Chinese flowering cabbage was chosen as the plant for the experiment. Seeds were subjected to disinfection by immersion in 75% ethanol for 1 minute and 0.4% sodium hypochlorite for 2 minutes, followed by rinsing three times with sterile distilled water. Subsequently, the seeds were air-dried under aseptic conditions.
Three treatments were established, including a control group without bacteria (CK), YLB1-6 treatment, and YLB2-1 treatment, with three replicates for each treatment. Circular plastic pots, with an inner diameter of 18 cm and a height of 12 cm, were filled with 1.75 kg of soil. Following standard fertilization treatment, 0.2 g of urea and 0.4 g of potassium dihydrogen phosphate per kg of soil were added into each pot. Subsequently, 300 mL of distilled water was added to moisten the soil before sowing. In each pot, 10 Chinese flowering cabbage seeds were sown. The seedlings were thinned at the 4-leaves stage. and 6 seedlings, exhibiting similar growth vigor, were retained for each pot. Following this, 10 mL of bacterial suspension was inoculated into the rhizosphere of the plants, with an equal amount of sterile water serving as the control (CK). Sterile water was consistently employed throughout the pot experiment. The plants were cultivated in a glasshouse with natural light for a duration of 50 days before harvest. During the pot experiment, the temperature ranged between 20~35°C in the greenhouse, and soil moisture was maintained at approximately 70% of field capacity.
The entire plants were harvested after 50 days of growth, washed with distilled water, and dried with absorbent paper. The aboveground and belowground parts of the plants were separated, and the fresh weight of each part was measured. Subsequently, the samples were heated at 105 °C for 30 minutes and dried at 60°C until a constant weight was achieved, then weighed. The dried samples were ground into fine powder, digested with HNO3-HClO4 (V:V=4:1), and reduced with 6 mol/L HCl. The Se content was measured using the hydride generation-atomic fluorescence spectroscopy method, following the “National Food Safety Standard-Determination of Selenium in Foods (GB 5009.93-2017)”.
Se Translocation factor (TF) and total Se amount were calculated based on the formulas:
Total Se amount (µg/pot) = [Se content in aboveground parts (mg/kg) × the fresh weight of aboveground parts (g/pot) + Se content in underground parts (mg/kg) × the fresh weight of underground parts (g/pot)] ×1000. Here, 1000 is the conversion factor from mg to µg.
After removing debris, the soil was air-dried. A portion was sieved through a 1mm mesh for pH determination, and another portion was sieved through a 100-mesh sieve for analyses of soil Se species and Se valencies. The pH was measured using the potentiometric method. Various Se species in the soil, including soluble Se (SOL-Se), exchangeable Se and carbonate-bound Se (EX-Se), iron (Fe)/manganese (Mn) oxide-bound Se (FMO-Se), organic matter-bound Se and elemental Se (OM-Se), and residual Se (RES-Se), were extracted using the continuous leaching method (Qu et al., 1997; Xing et al., 2018). The Se valencies, including Se4+, Se6+, and Se2- in SOL-Se and EX-Se, were extracted following the method proposed by Wang et al. (2012). The Se species and valencies were measured using the hydride generation-atomic fluorescence spectroscopy method. The available Se in the soil (AVA-Se) is the sum of SOL-Se and EX-Se.
Genomic DNA was isolated from the cell suspension of the YLB1-6 and YLB 2-1 strains using the Wizard Genomic DNA Kit (Promega). DNA purification and concentration were estimated using Nanodrop and Qubit. DNA with high quality (OD260/280 = 1.8~2.0) was employed for further experiments. The genome was sequenced using PacBio SequelII platforms. The data obtained from the PacBio SequelII platform were utilized for bioinformatics analysis. The Flye software was employed for genome assembly, and Pilon was used to correct errors.
Glimmer version 3.02 was utilized for coding sequence (CDS) prediction, and the predicted CDSs were annotated from NR, Swiss-Prot, Pfam, Gene Ontology (GO), Clusters of Orthologous Groups (COG), and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases using the NCBI-BLAST software, BLAST. For tRNA prediction and rRNA prediction, tRNA-scan-SE and Barrnap were employed, respectively. The prediction of SSR, Islands, and CRISPR sites was conducted using the trf and CRT software.
Data were analyzed by a one-way analysis of variance, and paired Duncan’s new multiple range test was performed using SPSS 17.0. All experiments were conducted in triplicate, and the values are presented as means ± standard errors. The level of significance was set at 0.05, and extreme significance was set at 0.01.
A total of 62 strains with distinct morphologies were isolated from soil samples collected across various Se-rich areas in Guangxi, encompassing 32 bacterial strains, 9 actinomycete strains, 10 fungal strains, and 11 yeast strains (Figure 1A). Through gradient Se concentration screening, bacteria exhibited the highest Se tolerance, reaching a concentration of 30,000 μg/mL, followed by fungi at 3,000 μg/mL, yeast at 1,500 μg/mL, and actinomycetes at 900 μg/mL. Among the 32 bacterial strains, 8 demonstrated tolerance to Se concentrations exceeding 10,000 μg/mL. Specifically, one strain could withstand 30,000 μg/mL (YLB1-6), two strains tolerated 29,000 μg/mL (YLB1-33, YLB1-26), two strains endured 20,000 μg/mL (YLB1-2, YFB1-8), two strains survived at 11,000 μg/mL (BMB2-1, TXB1-8), and one strain at 10,000 μg/mL (YLB2-1). The 16S rDNA sequencing identified these 8 bacterial strains as belonging to 5 species: Bacillus licheniformis, Bacillus pumilus, Bacillus cereus, Bacillus altitudinis, and Serratia marcescens (Figure 1B). These bacterial strains are classified as selenobacteria, which were designated in previous works (Acuña et al., 2013; Durán et al., 2015, 2018).
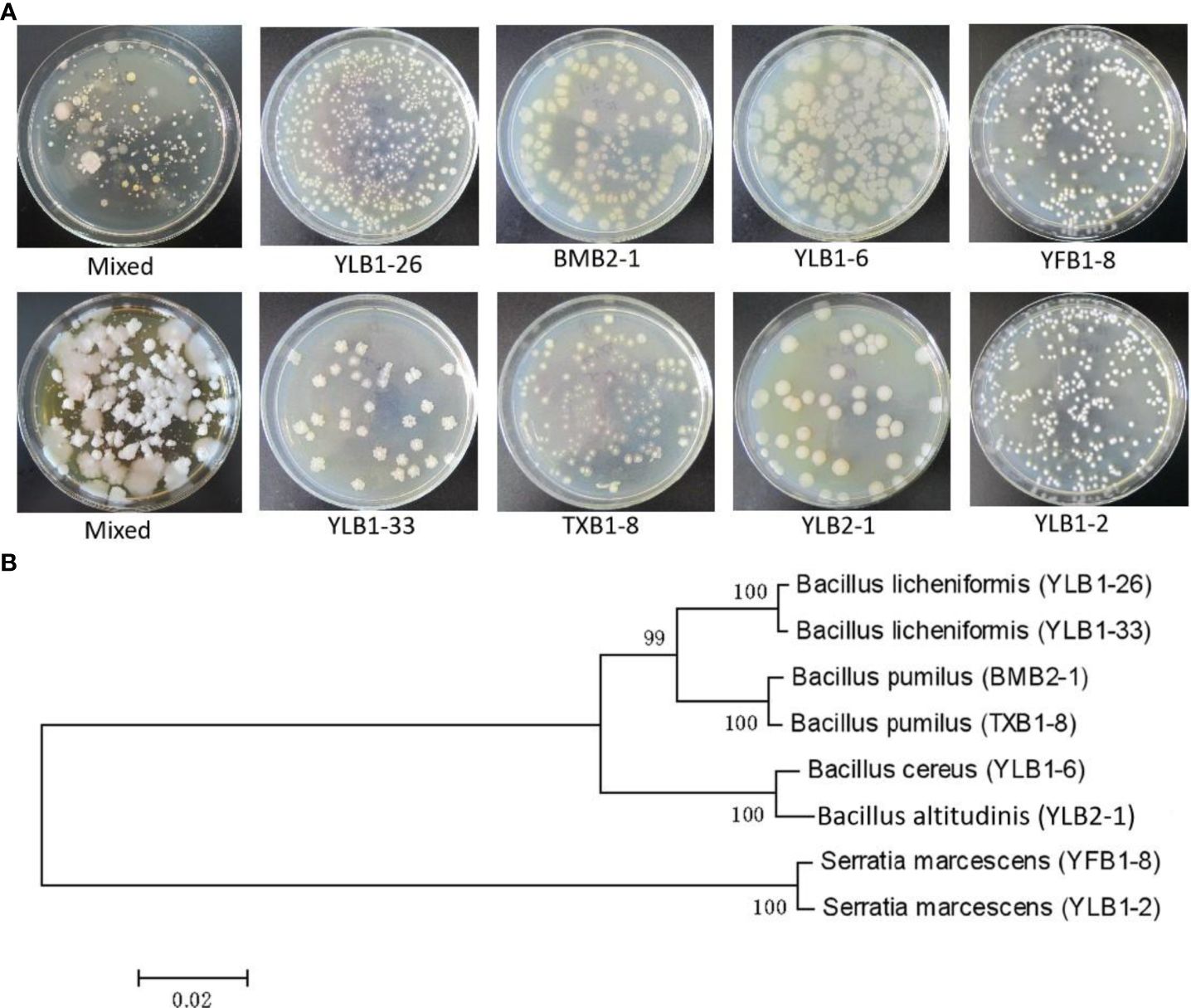
Figure 1 Isolation of Se-tolerant microorganisms. (A) Colony morphology of strains with high Se tolerance. (B) Phytogenetic analysis of Se-tolerant bacteria.
To assess the impact of selenobacteria on Se biofortification, two selenobacteria strains, YLB1-6 and YLB2-1, representing the highest (30,000 μg/mL) and lowest (10,000 μg/mL) Se tolerance levels among the 8 Se-tolerant bacteria, were selected for a pot experiment. Inoculation with selenobacteria significantly reduced soil RES-Se content while increasing AVA-Se content. However, different selenobacteria exhibited varying effects on Se transformation. In comparison to CK, YLB1-6 decreased soil RES-Se by 13.18% while significantly increasing FMO-Se, SOL-Se, and EX-Se content. The latter two species resulted in a 4.22% increase in AVA-Se. YLB2-1 treatment led to a 9.80% reduction in RES-Se but a significant increase in OM-Se and SOL-Se content, as well as a 3.17% increase in AVA-Se, mainly contributed by SOL-Se (Table 1).
In the CK treatment, the soil Se content in different valencies exhibited the trend: EX-Se4+ > EX-Se2- > SOL-Se4+ > SOL-Se6+. However, after selenobacteria inoculation, the soil Se content in different valencies showed the following trend: EX-Se4+ > EX-Se2- > SOL-Se6+ > SOL-Se4+. These results indicate that after inoculation with selenobacteria, SOL-Se6+ significantly increased, but SOL-Se4+ did not show a significant change. Among the EX-Se species, EX-Se4+ did not change significantly, while EX-Se2- increased extremely significantly (Figure 2).
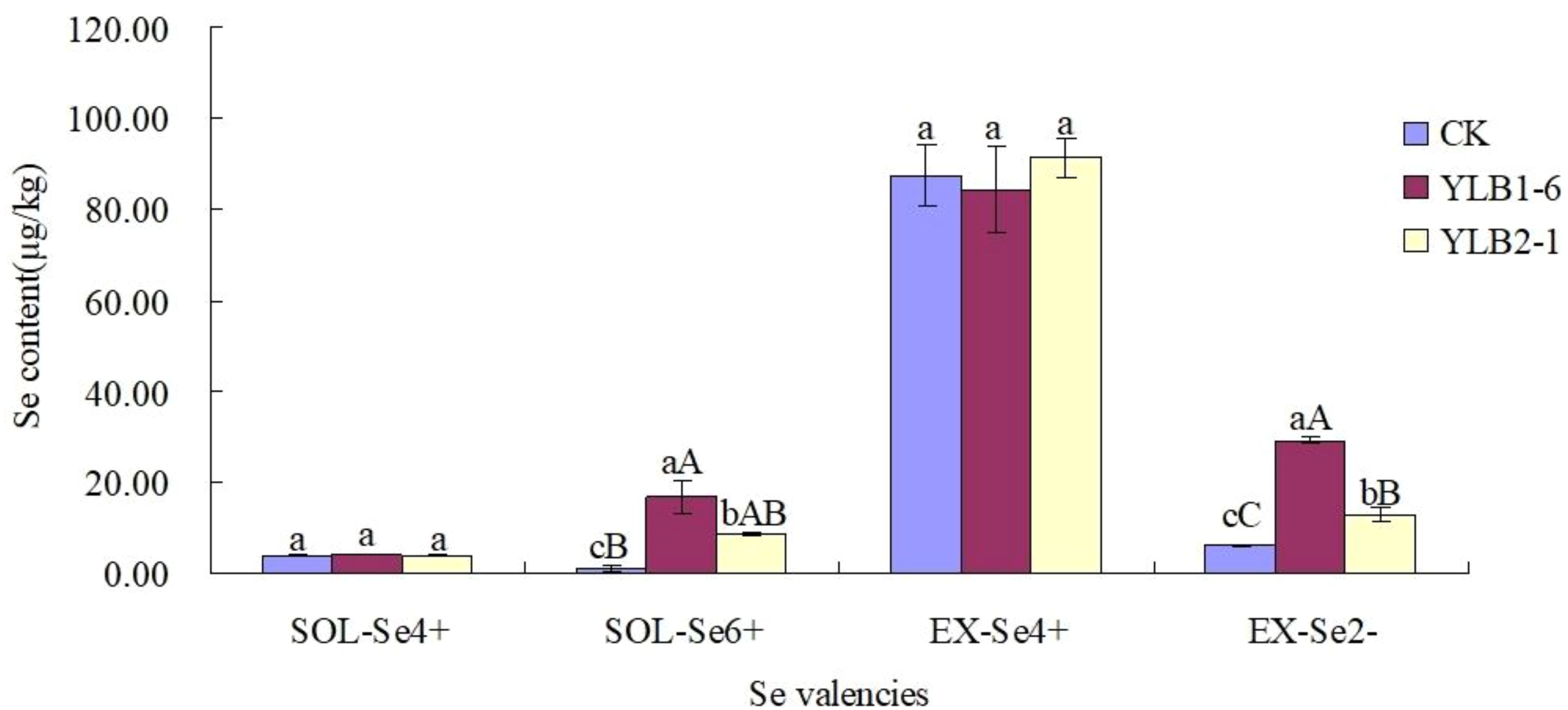
Figure 2 Effect of selenobacteria on Se valencies in Se-riched soil. Uppercase and lowercase letters in the same column indicate significant difference at 0.01 and 0.05 levels, respectively.
pH is a critical factor influencing Se availability. In pot experiments, in comparison to CK, the pH increased by 4.20% in the YLB1-6 treatment and 1.59% in the YLB2-1 treatment. The pH elevation in the YLB1-6 treatment reached a highly significant level (Table 2).
Following selenobacteria inoculation, the Se content in the aboveground part of Chinese flowering cabbage significantly exceeded that in the CK group. The Se content in the aboveground part after treatment with YLB1-6 and YLB2-1 was 1.96 times and 1.77 times that of the CK group, respectively, demonstrating that selenobacteria inoculation significantly enhances Se uptake in the aboveground part of the plant. However, a notable difference in the Se content in the underground part of the plant was observed between the two selenobacteria treatments. The Se content in the underground part of the YLB1-6 treatment did not differ significantly from that in the CK group, whereas the YLB2-1 treatment was significantly higher than that in the CK group, being 1.36 times that of the CK group. Additionally, the Se content in the underground part was higher than that in the aboveground part in all treatments. The Se content in the underground part of the CK group was 2.71 times that in the aboveground part, while in the YLB1-6 and YLB2-1 treatments, the Se content in the underground part was 1.22 times and 2.09 times that in the aboveground part, respectively (Figure 3A).
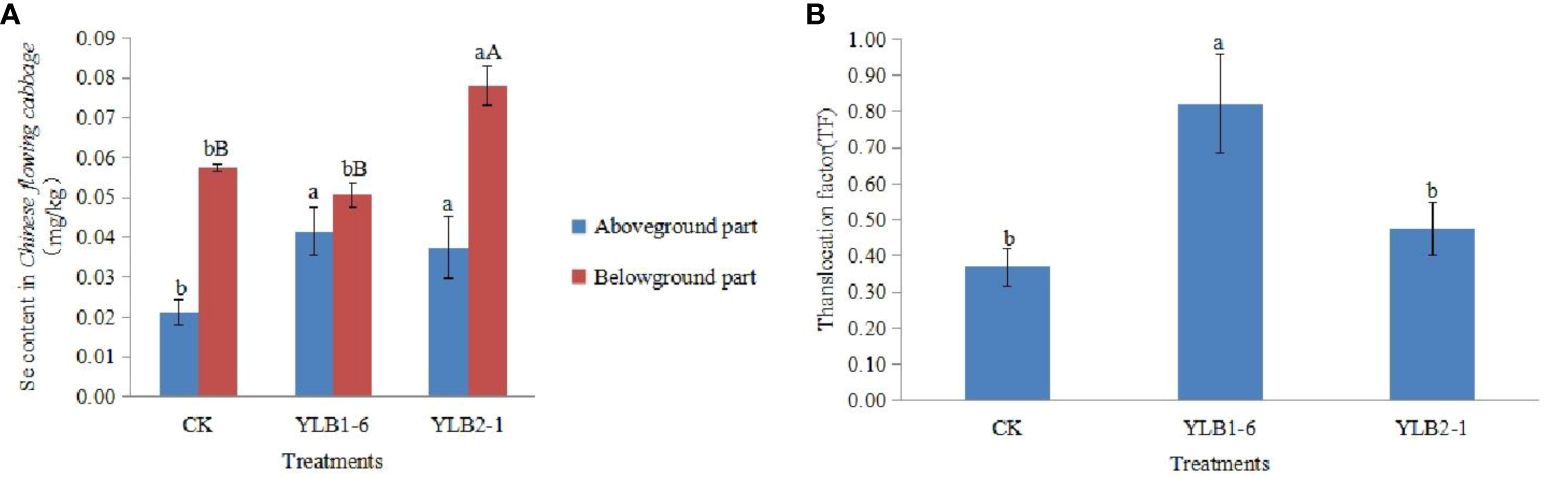
Figure 3 Effect of selenobacteria on Se uptake in Chinese Flowering Cabbage. (A) Se content in different parts of plants. (B) Se translocation factor in Chinese Flowering Cabbage. The uppercase and lowercase letters on the bar represent significant difference at 0.01 and 0.05 levels, respectively.
The Se Translocation Factor (TF) represents the ability of plants to transport Se from the underground part to the aboveground part. The TF for the CK treatment is 0.37, for the YLB1-6 treatment is 0.82, and for the YLB2-1 treatment is 0.48. Compared to CK, the TF of the YLB1-6 treatment increased by 123.14%, while the YLB2-1 treatment only increased by 29.29% (Figure 3B).
Even though the fresh weight of both the aboveground part and the underground part of the plant slightly increased with the bacteria treatments compared to CK, the differences, whether considering individual plant parts or the total fresh biomass, were not significant (Figure 4A). This suggests that inoculating selenobacteria does not improve the biomass of Chinese flowering cabbage.
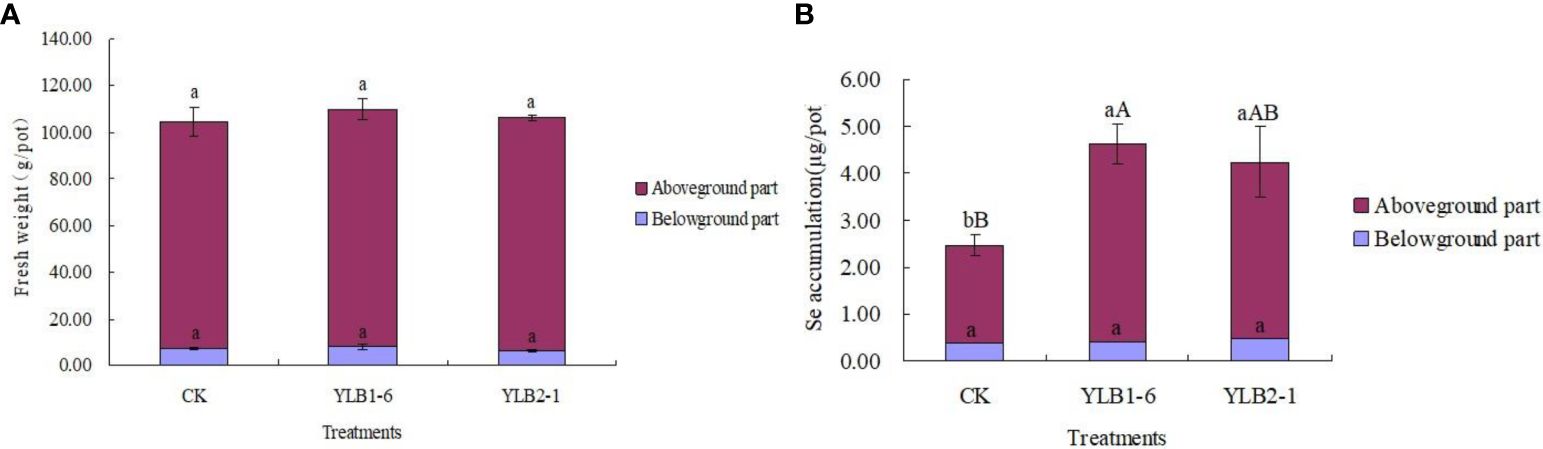
Figure 4 Effect of selenobacteria on the biomass (A) and Se accumulation (B) of Chinese flowering cabbage. The upcase and lowercase letters on the bar represent significant difference at 0.01 and 0.05 levels, respectively.
Nevertheless, Se accumulation in the aboveground part of the plant significantly increased when treated with YLB1-6 and YLB2-1, with increases of 104.36% and 81.69% compared to CK, respectively. Although there were no significant differences in Se accumulation in the underground part of the plant between the different treatments, the total Se accumulation in the plant of YLB1-6 and YLB2-1 treatments showed increases of 87.35% and 71.83%, respectively, compared to CK. Additionally, YLB1-6 exhibited a better performance in Se accumulation than YLB2-1 did (Figure 4B).
To unravel the genetic code of selenobacteria for Se biofortification, we conducted whole-genome sequencing of two selenobacteria strains with differing Se utilization abilities. For the YLB1-6 strain, characterized by higher Se utilization ability, the average genome depth was 35.32× (Figure 5A), ensuring complete genome coverage of 100%. The total gene length was 4,682,520 bp, with most genes falling within the 200 bp to 1400 bp range (Figure 5B), indicating the coding regions were fully sequenced, and the coding region occupied most proportion in the whole genome. The GC content of most CDS ranges from 30% to 40% (Figure 5C). Conversely, for the YLB2-5 strain with lower Se utilization ability, the average genome depth was 65.68×, also achieving complete genome coverage of 100%. The total gene length was 3,320,223 bp, with gene lengths mostly ranging from 200 bp to 1400 bp (Supplementary Figure S1), indicating comprehensive sequencing of coding regions dominating the genome. The GC content of most CDS ranged from 35% to 45% (Supplementary Figure S1).
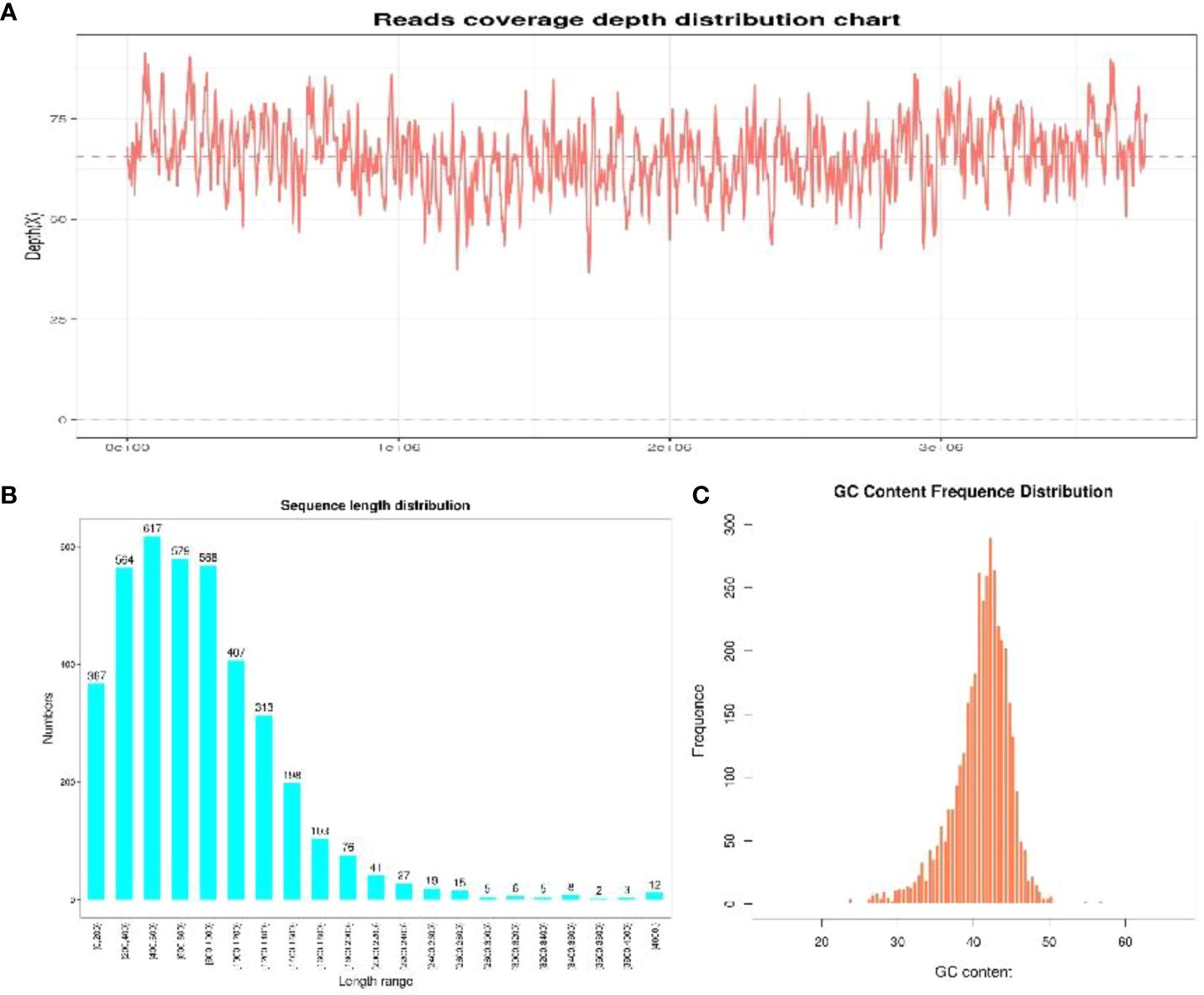
Figure 5 The sequencing quality of YLB1-6. (A) Reads coverage depth distribution chart. (B) Sequence length distribution. (C) GC content frequence distribution.
The genome of YLB1-6 comprises a 5,204,081 bp circular chromosome with an average G+C content of 35.39%, and a 425,392 bp circular plasmid DNA with a G+C content of 32.51%. Additionally, the genome encompasses 106 tRNA and 42 rRNA genes (Table 3). The chromosome houses 5,271 predicted genes (Figure 6A), while the plasmid genome contains 402 genes involved in RNA processing and modification, nucleotide transport and metabolism, amino acid transport and metabolism, signal transduction mechanisms, etc (Supplementary Figure S2).
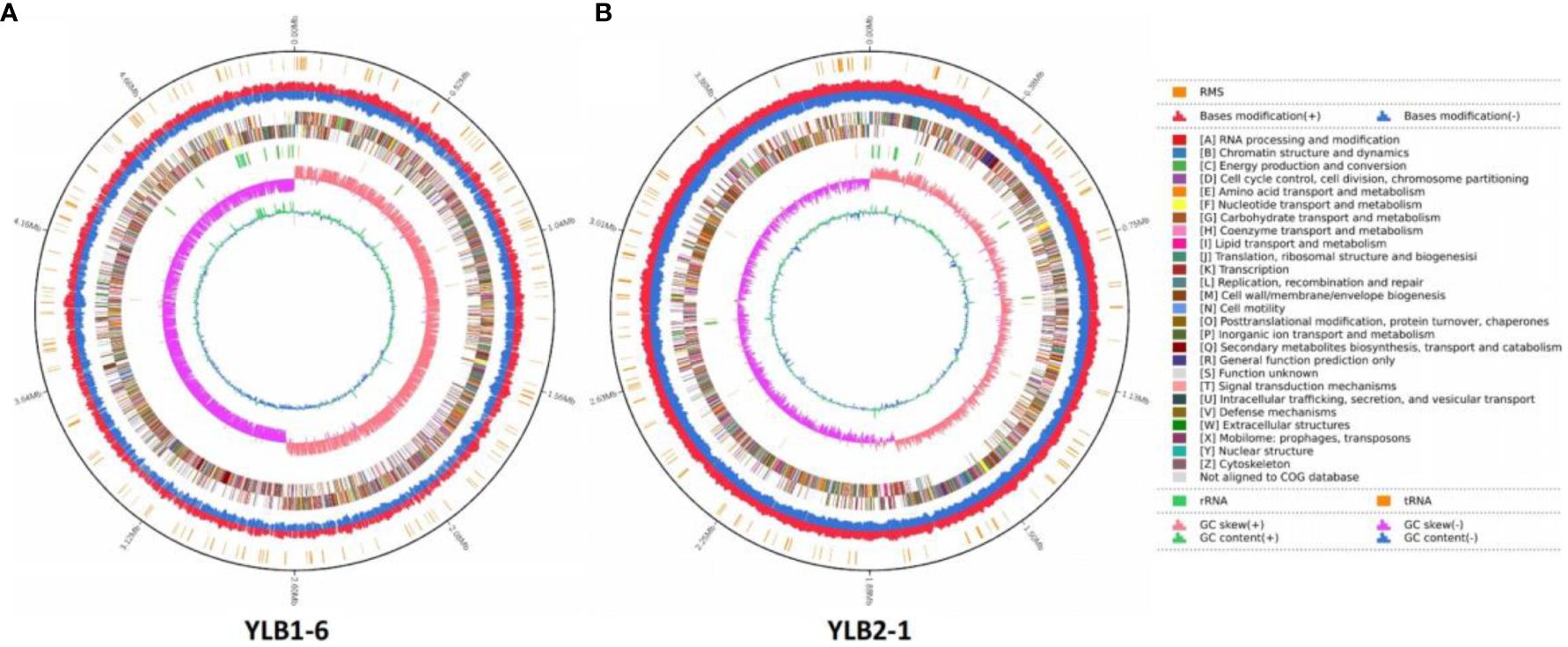
Figure 6 Landscape of the assembled YLB1-6 and YLB2-1 genome. From the outside, the first circle shows the RMS. The second circle shows the bases modification (+/-). The third circle shows the gene function classification. The fourth circle shows the GC content. The fifth circle shows the GC skew. (A) YLB1-6. (B) YLB2-1.
In contrast, the genome of YLB2-1 consists of a 3,757,945 bp circular chromosome without a plasmid, featuring an average G+C content of 41.36%. The genome also includes 81 tRNA and 24 rRNA genes (Table 3). The chromosome contains 3,935 predicted genes, also engaged in RNA processing and modification, nucleotide transport and metabolism, amino acid transport and metabolism, signal transduction mechanisms (Figure 6B).
Moreover, utilizing Island Path-DIMOB, PHAST, and Minced software, the genome of YLB1-6 revealed 7 gene islands, 44 CRISPR (clustered regularly interspaced short palindromic repeats), and 3 prophages (Table 3). In parallel, the genome of YLB2-1 displayed 9 gene islands, 6 CRISPR, and 2 prophages (Table 3).
Among the 5,673 annotated genes from YLB1-6, 5,644 genes exhibited the best hit in the Non-redundant protein sequence database (Nr database). Additionally, 4,017, 4,283, 3,734, and 2,561 genes/proteins were annotated in Swissprot, COG, GO, and KEGG databases, respectively, with 2,418 genes common to all these databases (Figure 7A). Furthermore, 2,616 genes were annotated in special function databases, with 530 (9.34%) in CARD, 88 (1.55%) in CAZY, 1,707 (30.09%) in PHI, 920 (16.22%) in VFDB, 1,439 (25.37%) in TCDB, and 199 (3.51%) in RMA (Figure 7B). Of the 5,644 genes annotated in the Nr database, 2,931 (51.93%) were common genes in the genus Bacillus, with 1,688 (29.90%) showing significant similarity to those in Bacillus cereus (Figure 7C), as which YLB1-6 was confirmed by 16S rDNA sequencing.
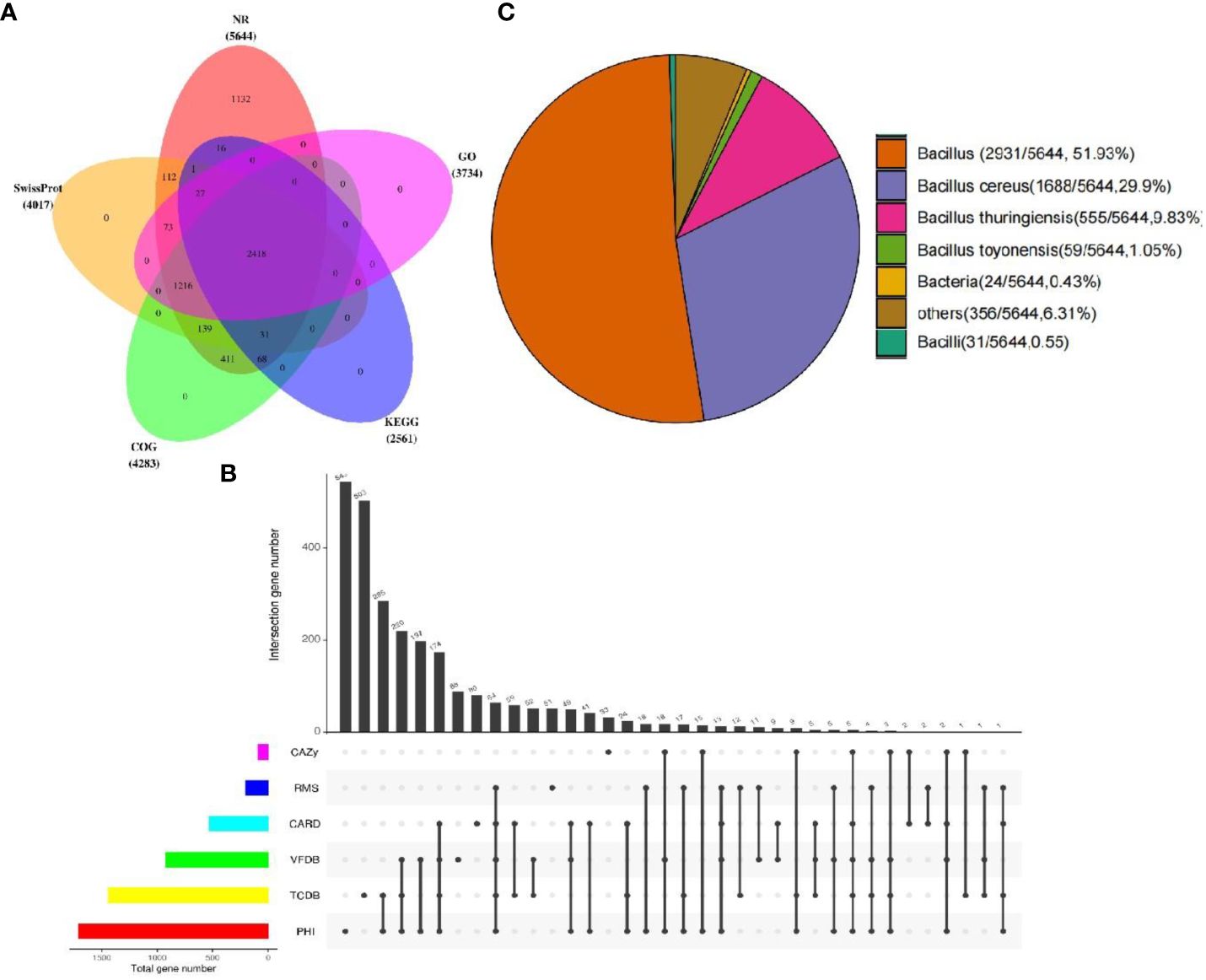
Figure 7 Annotated genes from YLB1-6. (A) The Venn diagram illustrates the distribution of genes across multiple databases. (B) Genes were annotated using five databases (CAZy, RMS, CARD, VFDB, TCDB, PHI), and common and unique genes were analyzed using UpSet plots (black indicates the presence of data at the point, grey indicates the absence of data at the point, and different points are connected to indicate intersection). (C) The distribution of species-annotated genes.
For YLB2-1, out of the 3,935 annotated genes, 3,872 genes had the best hit in the Nr database. Furthermore, 3,296, 3,084, 2,948, and 2,163 genes were identified in Swissprot, COG, GO, and KEGG, respectively, and 2,076 genes were common to all these databases (Supplementary Figure S3A). In addition, 1,905 genes were annotated in special function databases, including 304 (7.73%) in CARD, 82 (2.08%) in CAZY, 1,309 (33.27%) in PHI, 655 (16.65%) in VFDB, 1,016 (25.82%) in TCDB, and 119 (3.02%) in RMA (Supplementary Figure S3B). Of the 3,872 genes mapped in the Nr database, 2,475 (63.92%) were common genes in the genus Bacillus, while 830 (21.43%) showed significant similarity to those in Bacillus altitudinis (Supplementary Figure S3C), as which YLB2-1 was identified by 16S rDNA sequencing.
Presently, numerous Se utilization-related genes have been either experimentally validated or computationally identified in diverse bacteria and archaea. Through a comprehensive examination of the YLB1-6 genome for known Se utilization-related genes, a total of 46 genes were discerned. These include peroxiredoxin (11), thioredoxin (15), thioredoxin reductase (5), methionine sulfoxide reductase A (1), seryl-tRNA synthetase (1), yqeB (1), yqeC (2), nitrite reductase (3), formate dehydrogenase (3), and arsenate reductase (4), with peroxiredoxin and thioredoxin being the most prevalent. These genes participate in diverse activities such as thioredoxin peroxidase activity, oxidoreductase activity, peroxiredoxin activity, glutathione peroxidase activity, peptide-methionine (S)-S-oxide reductase activity, serine-tRNA ligase activity, and formate dehydrogenase (NAD+) activity (Table 4). Their potential significance in Se utilization by the selenobacterial strain YLB1-6 is underscored.
Conversely, in strain YLB2-1 (Table 5), 38 Se utilization-related genes were identified, encompassing peroxiredoxin (12), thioredoxin (10), thioredoxin reductase (5), arsenate reductase (4), formate dehydrogenase assembly factor FdhD (1), glutaredoxin (1), methionine sulfoxide reductase (1), nitrite reductase (3), and seryl-tRNA synthetase (1). The distinct repertoire of Se-related genes in these two strains sheds light on the genetic underpinnings of their disparate Se-utilization abilities.
Selenium, a crucial trace element, plays a significant role in maintaining the health of both humans and animals (Rayman, 2012). Consequently, there is a growing concern regarding Se-enriched foods and the techniques employed in their preparation. In many areas, soil serves as a Se source with high total Se content. Harnessing the Se reservoir in soil presents a cost-effective and efficient means of developing natural food with elevated Se levels. However, while many Se-rich soils boast high total Se content, the available Se content tends to be low. Microorganisms are pivotal in Se bioactivation (Stolz et al., 2006; Valdez Barillas et al., 2011), making the screening of microorganisms with Se activation ability a crucial approach for effectively utilizing the abundant Se in soil (Jong et al., 2015; Zare et al., 2017; Ullah et al., 2020).
The screening of Se-activating bacteria, also referred to as selenobacteria, commonly involves exposure to high-Se concentrations, as documented in various studies (Prakash et al., 2010; Acuña et al., 2013; Durán et al., 2015, 2018). Under these elevated Se conditions, these bacteria demonstrate the capacity to adapt and accumulate higher levels of Se (Pusztahelyi et al., 2015). This screening method not only enhances the efficiency of Se enrichment but also leads to a significant increase in the production of biological Se. For instance, the Se-tolerant selenobacteria strain BSN313 demonstrated the ability to enrich Se up to 2,123 µg/g of dry weight (Ullah et al., 2020). In a study conducted in Enshi, China, three bacteria, Acinetobacter baumannii, Bacillus licheniformis, and Bacillus subtilis were isolated from the Se ore zone. Among these, Acinetobacter exhibited a high Se tolerance of up to 25,000 µg/mL, while the other two strains of Bacillus demonstrated the ability to tolerate Se concentrations exceeding 33,000 µg/mL (Peng et al., 2012).
In this study, liquid media containing a gradient of high Se content were employed for the initial screening of Se-tolerant microorganisms sourced from Se-rich soils in Guangxi, China. Through the utilization of gradient dilution spread plates, a total of 62 microorganisms with distinctive colony characteristics were isolated from diverse Se-rich soil samples. Among these, 32 were bacteria, 9 were actinomycetes, 10 were fungi, and 11 were yeast. A more detailed analysis of Se tolerance revealed that bacteria exhibited the highest Se tolerance, while actinomycetes displayed the lowest. Among the identified selenobacteria, Bacillus cereus YLB1-6 exhibited exceptional Se tolerance, with the ability to withstand a Se content of 30,000 µg/mL. This finding is particularly rare in Se-rich areas of Guangxi. Even the least tolerant strain, Bacillus altitudinis YLB 2-1, demonstrated a significant tolerance level of 10,000 µg/mL Se content. This work contributes valuable microbial resources for the exploration of abundant Se sources in soil.
Se in soil is commonly classified into five species: SOL-Se, EX-Se, FMO-Se, OM-Se, and RES-Se (Wang et al., 2012). SOL-Se and EX-Se are readily absorbed and utilized by plants, representing available soil Se (Hu et al., 2014). The valencies of Se in soil include Se2-, Se0, Se4+, and Se6+ (Fernández-Martínez and Charlet, 2009). Se valencies that plants can take up and utilize include Se4+ and Se6+ (Qin et al., 2017), as well as some Se2- forms, such as organic Se compounds like selenomethionine and selenocysteine (Guignardi and Schiavon, 2017).
Microorganisms play a pivotal role in transforming soil Se species and forming organic Se through metabolic processes involving Se transport, reduction, assimilation, oxidation, and methylation (Sarathchandra and Watkinson, 1981; Dowdle and Oremland, 1998; Winkel et al., 2012). The predominantly acidic red soils and lateritic soils in Se-rich areas of Guangxi, China, contain primarily selenite, which is prone to adsorption by iron oxides and clay minerals. However, microorganisms can transform adsorbed selenite into soluble Se for plant uptake (Shen and Zhou, 2011).
In this study, after inoculating selenobacteria, changes in Se species were observed, characterized by a decrease in RES-Se content and an increase in AVA-Se content in the soil. This indicates that selenobacteria can disrupt the equilibrium of Se species in the soil, promoting the transformation of residual Se into other Se species and thereby increasing the soil’s available Se content. Additionally, selenobacteria influence Se valency. After inoculation with selenobacteria, the soluble forms of SOL-Se6+ and EX-Se2- significantly increased. This suggests that the elevated levels of SOL-Se6+ and EX-Se2-, following the inoculation of selenobacteria, contribute to the increased available Se content in Se-rich red soils.
Soil pH is a crucial factor influencing the available Se content in soil, typically increasing with rising soil pH of acidic soil. In our study, pH measurements were conducted on the soil after pot experiments, revealing a significant increase in soil pH with the application of YLB1-6. Changes in soil pH can impact the transformation of Se species, thereby influencing Se availability. After inoculation with selenobacteria, acidic soil pH and soluble Se content significantly increased.
The pH of soil is known to mediate the adsorption and desorption processes of Se with soil components such as Fe and Al ions (Goh and Lim, 2004). Our findings indicate that both selenobacteria could transform RES-Se, although the resulting Se forms differed. This variation may be attributed to differences in the metabolic by-products produced by distinct selenobacteria, leading to the formation of different Se species. Additionally, pH influences soil redox potential, microbial species, and activity, thereby affecting Se valency (Goh and Lim, 2004). This explains the observed increase in SOL-Se6+ and EX-Se2- in the soil following the inoculation of selenobacteria.
Inoculating microorganisms has been shown to enhance the uptake of soil Se by plants (Yang et al., 2021). Bacterial inoculation has increased Se content in the leaves of various crops such as wheat (Acuña et al., 2013), tea (Xu et al., 2020), and rice (Huang et al., 2020), as well as in the leaves, shoots, and seeds of Brassica juncea (Lampis et al., 2009; Yasin et al., 2015). Our study affirms that the inoculation of selenobacteria significantly increased Se content in the above-ground part of Chinese flowering cabbage compared to the control. Considering the Se transformation profile in soil, this increase is attributed to the conversion of residual Se into available Se by selenobacteria, leading to an augmentation in soluble Se6+ and exchangeable Se2-. These forms are readily absorbed by plants, a phenomenon also supported by Kikkert and Berkelaar (2013). Moreover, Se transport rates follow the order Se6+ > Se2- > Se4+ (Chen, 2004; Jiang et al., 2015). Consequently, the Se6+ and Se2- taken up by plants can be easily transported to the above-ground parts, resulting in Se accumulation in the aboveground part rather than the roots.
Furthermore, the YLB1-6 treatment exhibited higher Se accumulation in Chinese flowering cabbage than the YLB2-1 treatment, indicating that the YLB1-6 strain possesses a stronger capacity for activating soil Se. Consequently, more Se was taken up and transferred into the aboveground parts of plants in the YLB1-6 treatment, leading to a higher Se Translocation Factor (TF) than that of the YLB2-1 treatment. Currently, many studies aimed at enhancing Se uptake in plants involve the addition of exogenous Se (Huang et al., 2018; Xiong et al., 2019; Guo et al., 2020). Our work demonstrates that selenobacteria can effectively activate soil Se for plant uptake, providing alternative solutions for the exploitation of rich soil Se resources.
Deciphering the molecular mechanisms and identifying key genes involved in Se biofortification in selenobacteria is crucial for laying theoretical foundations and providing genetic resources for enhancing Se utilization in crops and microbial species through molecular technology. Whole-genome sequencing of microorganisms offers a comprehensive understanding of their genetic information, serving as a molecular basis for exploring the Se transformation mechanisms of selenobacteria and identifying critical genes for Se biofortification (Zhang et al., 2022). In this study, the genomes of two selenobacteria strains, YLB1-6 and YLB2-5, representing the highest and lowest Se tolerance within the bacteria group isolated, were extensively analyzed.
The genome of YLB1-6 revealed 5,673 genes, including 47 related to Se utilization, such as peroxiredoxin, thioredoxin, thioredoxin reductase, methionine sulfoxide reductase A, seryl-tRNA synthetase, yqeB, yqeC, nitrite reductase, formate dehydrogenase, and arsenate reductase. Meanwhile, YLB2-5, with lower Se utilization ability, exhibited 3,935 genes, including 38 associated with Se utilization. Notably, YLB2-5 lacked the key genes yqeB and yqeC, and the overall number of Se-related genes was lower compared to YLB1-6. This discrepancy likely contributes to the distinct Se utilization abilities of these two strains.
Peroxiredoxin (Prx) and methionine-S-sulfoxide reductase A (MsrA) are vital members of the selenoprotein family, playing crucial roles in Se transformation and utilization (Speckmann et al., 2016; Brigelius-Flohé and Flohé, 2017). Thioredoxin reductase is essential for selenite reduction in certain bacteria, and the selenite reduction via the Trx system is a pivotal early step for bacterial selenoprotein biosynthesis (Tamura et al., 2011; Shimizu et al., 2021). The tRNASec, charged with serine to yield seryl-tRNASec by canonical seryl-tRNA synthetase (SerRS), is involved in the production of selenoproteins. YqeB and YqeC are predicted to be associated with the utilization of Se-cofactor. Nitrite reductase enzymatically reduces selenite to elemental Se, a crucial step in Se metabolism (Kessi and Hanselmann, 2004; Tobe and Mihara, 2018; Huang et al., 2021).
The identification of 46 and 38 genes related to Se utilization in the genomes of YLB1-6 and YLB2-5, respectively, underscores their importance in Se biofortification. These genes represent potential targets for crop or microorganism variety improvement to enhance Se utilization.
This study identified selenobacteria with a high Se-tolerant ability. Subsequent pot experiments demonstrated that these selenobacteria could transform the species and valencies of Se in the soil, facilitating Se uptake by plants. Whole-genome analysis unveiled the genetic elements of the two selenobacteria with contrasting Se utilization profiles and identified candidate genes related to Se utilization. This work significantly advances our understanding of the potential molecular mechanisms underlying Se biofortification by selenobacteria. Furthermore, it provides valuable microorganisms and gene targets for enhancing the Se utilization efficiency of crops or microorganisms, particularly for exploiting the rich Se sources in soil (Figure 8). This study also represents the first comprehensive genome dissection of selenobacteria.
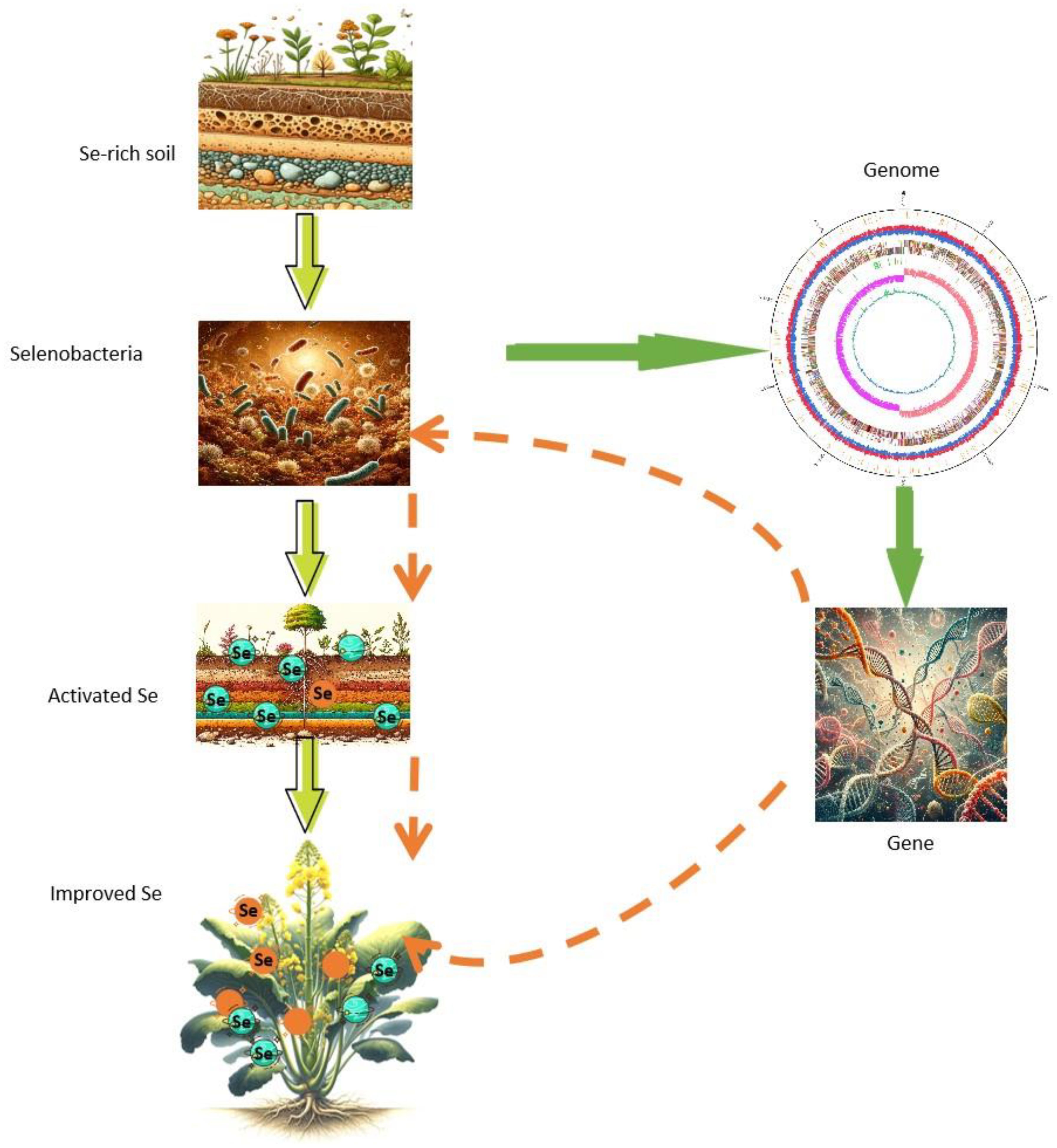
Figure 8 Isolation of selenobacteria and identification of candidate genes for Se fortification. Selenobacteria were isolated from selenium-rich soil, exhibiting the capability to activate selenium in the soil and facilitate its uptake into plants. The genomic analysis of selenobacteria unveiled candidate genes associated with selenium biofortification. These candidate genes can be leveraged to augment the selenobacteria’s proficiency in selenium activation, thereby enhancing the transformation of selenium into plants. Alternatively, these genes can be directly employed to enhance the plants’ capacity to uptake more selenium from the soil.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.cncb.ac.cn/, PRJCA021960.
QL: Conceptualization, Formal analysis, Funding acquisition, Investigation, Methodology, Writing – original draft. A-ML: Conceptualization, Data curation, Investigation, Methodology, Software, Visualization, Writing – original draft. YX: Formal analysis, Investigation, Methodology, Writing – review & editing. P-XL: Formal analysis, Investigation, Methodology, Writing – review & editing. Z-PJ: Formal analysis, Methodology, Project administration, Supervision, Writing – review & editing. Y-XL: Conceptualization, Project administration, Supervision, Writing – review & editing. D-LH: Conceptualization, Supervision, Writing – review & editing, Project administration.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (32160762), the Natural Science Foundation of Guangxi (2020GXNSFAA2970867), the Science and Technology Major Project of Guangxi (AA17202019), Guangxi Key Research and Development Programme (AB19245005), and National Natural Science Foundation of China Joint Fund Program (U23A2040).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The handling editor MS declared a past collaboration with the author Y-XL.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1392355/full#supplementary-material
Supplementary Figure 1 | The sequencing quality of YLB2-1. (A) Reads coverage depth distribution chart. (B) Sequence length distribution. (C) GC content frequence distribution.
Supplementary Figure 2 | The plasmic genome in YLB 1-6. From the outside, the first circle shows the RMS. The second circle shows the bases modification (+/-). The third circle shows the gene function classification. The fourth circle shows the GC content. The fifth circle shows the GC skew.
Supplementary Figure 3 | Annotated genes from YLB 2-1. (A) The Venn diagram of genes using multi databases. (B) Genes were annotated using five databases (CAZy, RMS, CARD, VFDB, TCDB, PHI), and the common and unique genes were analyzed using UpSet plots (black indicates the presence of data at the point, grey indicates the absence of data at the point, and different points are connected to indicate intersection). (C) The distribution of species annotated genes.
Acuña, J. J., Jorquera, M. A., Barra, P. J., Crowley, D. E., Mora, M. L. (2013). Selenobacteria selected from the rhizosphere as a potential tool for Se biofortification of wheat crops. Biol. Fertil Soils. 49, 175–185. doi: 10.1007/s00374-012-0705-2
Brigelius-Flohé, R., Flohé, L. (2017). Selenium and redox signaling. Arch. Biochem. Biophys. 617, 48–59. doi: 10.1016/j.abb.2016.08.003
Chen, D. Q. (2004). Progress of plant selenium assimilation and screening of resistant selenium mutant. Amino Acids Biotic resour. 26, 65–70. doi: 10.14188/j.ajsh.2004.02.023
Chilimba, A. D. C., Young, S. D., Black, C. R., Meacham, M. C., Lammel, J., Broadley, M. R. (2012). Assessing residual availability of selenium applied to maize crops in Malawi. Field Crop Res. 134, 11–18. doi: 10.1016/j.fcr.2012.04.010
China National Environmental Monitoring Station (1990). Background value of soil elements in China Vol. 87 (Beijing: CESP).
Dowdle, P. R., Oremland, R. S. (1998). Microbial oxidation of elemental selenium in soil slurries and bacterial cultures. EST 32, 3749–3755. doi: 10.1021/es970940s
Durán, P., Acuña, J. J., Gianfreda, L., Azcón, R., Funes-Collado, V., Mora, M. L. (2015). Endophytic selenobacteria as new inocula for selenium biofortification. Appl. Soil Ecol. 96, 319–326. doi: 10.1016/j.apsoil.2015.08.016
Durán, P., Acuna, J. J., Jorquera, M. A., Azcon, R., Borie, F., Cornejo, P., et al. (2013). Enhanced selenium content in wheat grain by co-inoculation of selenobacteria and arbuscular mycorrhizal fungi: a preliminary study as a potential Se biofortification strategy. J. Cereal Sci. 57, 275–280. doi: 10.1016/j.jcs.2012.11.012
Durán, P., Viscardi, S., Acuña, J. J., Cornejo, P., Azcón, R., Mora, M. L. (2018). Endophytic selenobacteria and arbuscular mycorrhizal fungus for Selenium biofortification and Gaeumannomyces graminis biocontrol. J. Soil Sci. Plant Nutr. 18, 1021–1035. doi: 10.4067/S0718-95162018005002902
Fernández-Martínez, A., Charlet, L. (2009). Selenium environmental cycling and bioavailability: A structural chemist point of view. Rev. Environ. Sci. Bio. 8, 81–110. doi: 10.1007/s11157-009-9145-3
Geng, J. M. (2010). Chemical Characteristics of Selenium in Paddy Soils and Genotypic Differences and Mechanism of Selenium Absorption and Accumulation of Rice in Hainan Province Vol. 7 (Haikou: Hainan University).
Goh, K. H., Lim, T. T. (2004). Geochemistry of inorganic arsenic and selenium in tropical soil: effect of reaction time, pH, and competitive anions on arsenic and selenium adsorption. Chemosphere 55, 849–859. doi: 10.1016/j.chemosphere.2003.11.041
Guignardi, Z., Schiavon, M. (2017). “Biochemistry of plant selenium uptake and metabolism,” in Selenium in Plants. Eds. Elizabeth, A. H. P., Lenny, H. E. W., Lin, Z. Q. (Springer International Publishing, New York), 21–34.
Guo, J. X., Luo, W. L., Fu, M., Li, G. H. (2020). Effects of basal application of selenium-enriched fertilizer on growth and nutrient elements’ content of Chinese flowering cabbage. Chin. Agric. Sci. Bull. 36, 52–56. doi: 10.11924/j.issn.1000-6850.casb2020-0022
Hossain, A., Skalicky, M., Brestic, M., Maitra, S., Sarkar, S., Ahmad, Z., et al. (2021). Selenium biofortification: roles, mechanisms, responses and prospects. Molecules 26, 881. doi: 10.3390/molecules26040881
Hu, B., Liang, D. L., Lei, L. M., Yu, D. S. (2014). Transformation of heavy metal fractions on soil urease and nitrate reductase activities in copper and selenium co-contaminated soil. Ecotoxicol. Environ. Saf. 110, 41–48. doi: 10.1016/j.ecoenv.2014.08.007
Huang, T. Q., Jiang, Z. P., Liang, P. X., Xing, Y., Liao, Q., Liu, Y. X., et al. (2018). Screening of enriched-selenium peanut cultivars and study on selenium-enriched production technology of peanut by fertilizing exogenous selenium. Soils 50, 1198–1202. doi: 10.13758/j.cnki.tr.2018.06.022
Huang, C. L., Wang, H. L., Shi, X. Y., Wang, Y., Shao, Y. H., Li, P., et al. (2020). Two new selenite reducing bacterial isolates from paddy soil and the potential Se biofortification of paddy rice. Ecotoxicology 30, 1465–1475. doi: 10.1007/s10646-020-02273-6
Huang, S., Wang, Y., Tang, C., Jia, H., Wu, L. (2021). Speeding up selenite bioremediation using the highly selenite-tolerant strain Providencia rettgeri HF16-A novel mechanism of selenite reduction based on proteomic analysis. J. Hazard Mater. 406, 124690. doi: 10.1016/j.jhazmat.2020.124690
Jiang, Y., Zeng, Z. H., Bu, Y., Ren, C. Z., Hu, Y. G. (2015). Effects of selenium fertilizer on grain yield, Se uptake and distribution in common buckwheat (Fagopyrum esculentum Moench). Plant Soil Environ. 61, 371–377. doi: 10.17221/284/2015-PSE
Jong, M. S., Reynolds, R. J. B., Richterova, K., Musilova, L., Staicu, L. C., Chocholata, I., et al. (2015). Selenium hyperaccumulators harbor a diverse endophytic bacterial community characterized by high selenium resistance and plant growth promoting properties. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00113
Kessi, J., Hanselmann, K. W. (2004). Similarities between the abiotic reduction of selenite with glutathione and the dissimilatory reaction mediated by Rhodospirillum rubrum and Escherichia coli. J. Biol. Chem. 279, 50662–50669. doi: 10.1074/jbc.M405887200
Kikkert, J., Berkelaar, E. (2013). Plant uptake and translocation of inorganic and organic forms of selenium. Arch. Environ. Con Tox. 65, 458–465. doi: 10.1007/s00244-013-9926-0
Lampis, S., Ferrari, A., Cunha-Queda, A. C. F., Alvarenga, P., Gregorio, S. D., Vallini, G. (2009). Selenite resistant rhizobacteria stimulate SeO32– phytoextraction by Brassica juncea in bioaugmented water-filtering artificial beds. Environ. Sci. pollut. R. 16, 663–670. doi: 10.1007/s11356-008-0088-y
Liu, Y. X., Yang, J. H., Shi, M. L., Jiang, Z. P., Xing, Y., Wu, Z. L. (2015). Analysis on the development prospect of selenium-rich functional agricultural products in Guangxi. Agri Tech. 35, 176–178. doi: 10.3969/j.issn.1671-962X.2015.01.084
Long, Y. C., Chen, X., Zhou, S. Q. (2017). Isolation, identification and assessment on selenium biofortification of Siderophore-producing rhizobacteria. Curr. Bio. 7, 402–408. doi: 10.19586/j.2095-2341.2017.0053
Luo, W. Q., Li, J., Ma, X. N., Niu, H., Hou, S. W., Wu, F. Y. (2019). Effect of arbuscular mycorrhizal fungi on uptake of selenate, selenite, and selenomethionine by roots of winter wheat. Plant Soil. 438, 71–83. doi: 10.1007/s11104-019-04001-4
Nancharaiah, Y. V., Lens, P. N. L. (2015). Ecology and biotechnology of selenium-respiring bacteria. Microbiol. Mol. Bio R. 79, 61–80. doi: 10.1128/MMBR.00037-14
Peng, Z. Q., Fan, J., Xiang, D. E., Shi, H. L., Xiang, B. K., Zuo, M. (2012). Isolation, screening, identification of three strains with high- Se tolerance. Stud. Trace Elem. Health 29, 4–6. doi: 10.1038/ismej.2015.246
Peng, T., Lin, J., Xu, Y. Z., Zhang, Y. (2016). Comparative genomics reveals new evolutionary and ecological patterns of selenium utilization in bacteria. Isme J. 10, 2048–2059. doi: 10.1038/ismej.2015.246
Prakash, D., Pandey, J., Tiwary, B. N., Jain, R. K. (2010). Physiological adaptations and tolerance towards higher concentration of selenite (Se+4) in Enterobacter sp. AR-4, Bacillus sp. AR-6 and Delftia tsuruhatensis AR-7. Extremophiles 14, 261–272. doi: 10.1007/s00792-010-0305-8
Premarathna, H. M. P. L., McLaughlin, M. J., Kirby, J. K., Hettiarachchi, G. M., Stacey, S. (2012). Influence of submergence and subsequent drainage on the partitioning and lability of added selenium fertilizers in a sulphur-containing Fluvisol. Eur. J. Soil Sci. 63, 514–522. doi: 10.1111/j.1365-2389.2012.01462.x
Pusztahelyi, T., Kovács, S., Pócsi, I., Prokisch, J. (2015). Selenite-stress selected mutant strains of probiotic bacteria for Se source production. J. Trace Elem. Med. Bio. 30, 96–101. doi: 10.1016/j.jtemb.2014.11.003
Qin, H. B., Zhu, J. M., Lin, Z. Q., Xu, W. P., Tan, D. C., Zheng, L. R., et al. (2017). Selenium speciation in seleniferous agricultural soils under different cropping systems using sequential extraction and x-ray absorption spectroscopy. Environ. pollut. 225, 361–369. doi: 10.1016/j.envpol.2017.02.062
Qu, J. G., Xu, B. X., Gong, S. C. (1997). Sequential extraction techniques for determination of selenium speciation in soils and sediments. Environ. Chem. 16, 277–283.
Rayman, M. P. (2012). Selenium and human health. Lancet 379, 1256–1268. doi: 10.1016/S0140-6736(11)61452-9
Sarathchandra, S. U., Watkinson, J. H. (1981). Oxidation of elemental selenium to selenite by Bacillus megaterium. Science 211, 600–601. doi: 10.1126/science.6779378
Schiavon, M., Nardi, S., Dalla Vecchia, F., Ertani, A. (2020). Selenium biofortification in the 21st century: status and challenges for healthy human nutrition. Plant Soil. 453, 245–270. doi: 10.1007/s11104-020-04635-9
Shardendu, U., Salhani, N., Boulyga, S. F., Stengel, E. (2003). Phytoremediation of selenium by two helophyte species in subsurface flow constructed wetland. Chemosphere 50, 967–973. doi: 10.1016/S0045-6535(02)00607-0
Shen, Y. C., Zhou, J. (2011). Occurrences, migration and transformation of selenium in soil. Geo Anhui. 21, 186–191. doi: 10.3969/j.issn.1005-6157.2011.03.006
Shimizu, A., Tobe, R., Aono, R., Inoue, M., Hagita, S., Kiriyama, K., et al. (2021). Initial step of selenite reduction via thioredoxin for bacterial selenoprotein biosynthesis. Int. J. Mol. Sci. 22, 10965. doi: 10.3390/ijms222010965
Speckmann, B., Steinbrenner, H., Grune, T., Klotz, L. O. (2016). Peroxynitrite: From interception to signaling. Arch. Biochem. Biophys. 595, 153–160. doi: 10.1016/j.abb.2015.06.022
Stolz, J. F., Basu, P., Oremland, R. S. (2002). Microbial transformation of elements: The case of arsenic and selenium. Int. Microbiol. 5, 201–207. doi: 10.1007/s10123-002-0091-y
Stolz, J. F., Basu, P., Santini, J. M., Oremland, R. S. (2006). Arsenic and selenium in microbial metabolism. Annu. Rev. Microbiol. 60, 107–130. doi: 10.1146/annurev.micro.60.080805.142053
Tamura, T., Sato, K., Komori, K., Imai, T., Kuwahara, M., Okugochi, T., et al. (2011). Selenite reduction by the thioredoxin system: Kinetics and identification of protein-bound selenide. Biosci. Biotechnol. Biochem. 75, 1184–1187. doi: 10.1271/bbb.100847
Tobe, R., Mihara, H. (2018). Delivery of selenium to selenophosphate synthetase for selenoprotein biosynthesis. Biochim. Biophys. Acta Gen. Subj. 1862, 2433–2440. doi: 10.1016/j.bbagen.2018.05.023
Ullah, A., Sun, B., Wang, F., Yin, X., Xu, B., Ali, N., et al. (2020). Isolation of selenium-resistant bacteria and advancement under enrichment conditions for selected probiotic Bacillus subtilis (BSN313). J. Food Biochem. 44, e13227. doi: 10.1111/jfbc.13227
Valdez Barillas, J. R., Quinn, C. F., Pilon-Smits, E. A. H. (2011). Selenium accumulation in plants–phytotechnological applications and ecological implications. Int. J. Phytoremed. 13, 166–178. doi: 10.1080/15226514.2011.568542
Wang, S. S., Liang, D. L., Wang, D., Wei, W., Dong, D. F., Zhi, Q. L. (2012). Selenium fractionation and speciation in agriculture soils and accumulation in corn(Zea mays L.) under field conditions in Shaanxi Province, China. Sci. Total Environ. 427-428, 159–164. doi: 10.1016/j.scitotenv.2012.03.091
Winkel, L. H., Johnson, C. A., Lenz, M., Grundl, T., Leupin, O. X., Amini, M., et al. (2012). Environmental selenium research: from microscopic processes to global understanding. Environ. Sci. Technol. 46, 571–579. doi: 10.1021/es203434d
Xing, Y., Liu, Y. X., Liang, P. X., Liao, Q., Pan, L. P., Chen, J. P., et al. (2018). Effects of phosphorus on selenium uptake of pakchoi and soil selenium morphology in Se-rich latosolic red soil and red soil. Soils 50, 1170–1175. doi: 10.13758/j.cnki.tr.2018.06.018
Xiong, J., Li, W. L., Pan, W. X., Wei, M. Z., Tang, X. H., Yan, H. F., et al. (2019). Effects of exogenous selenium application on selenium content and yield of purple sweet potato under different soil conditions. J. South. Agricul. 50, 1211–1218. doi: 10.3969/j.issn.2095-1191.2019.06.08
Xu, X., Cheng, W., Liu, X., You, H., Wu, G., Ding, K. M., et al. (2020). Selenate reduction and selenium enrichment of tea by the endophytic herbaspirillum sp. strain WT00C. Curr. Microbiol. 77, 588–601. doi: 10.1007/s00284-019-01682-z
Xu, Q. L., Wu, W. L., Zhao, G. S., Zhu, Y. Y., Guo, Y. B. (2017). Selenium metabolism in microorganisms. Microbiol. China 44, 207–216. doi: 10.13344/j.microbiol.China.160380
Yang, D., Hu, C., Wang, X., Shi, G., Li, Y., Fei, Y., et al. (2021). Microbes: a potential tool for selenium biofortification. Metallomics 13, mfab054. doi: 10.1093/mtomcs/mfab054
Yasin, M., Mehdawi, A. F. E. I., Jahn, C. E., Anwar, A., Turner, M. F. S., Faisal, M., et al. (2015). Seleniferous soils as a source for production of selenium-enriched foods and potential of bacteria to enhance plant selenium uptake. Plant Soil. 386, 385–394. doi: 10.1007/s11104-014-2270-y
Ye, Y., Qu, J., Pu, Y., Rao, S., Xu, F., Wu, C. (2020). Selenium biofortification of crop food by beneficial microorganisms. J. Fungi. 6, 59. doi: 10.3390/jof6020059
Zare, H., Vahidi, H., Owlia, P., Khujin, M. H., Khamisabadi, A. (2017). Yeast enriched with Selenium: A promising source of Selenomethionine and Seleno-Proteins. TPPS 1, 130–134. doi: 10.22037/tpps.v1i3.16778
Zhang, R., Fan, T., Li, M., Yuan, L. X. (2018). Isolation, identification and selenite metabolism of a selenium-tolerant endophyte from Cardamine hupingshanensis. Microbiol. China 45, 314–321. doi: 10.13344/j.microbiol.China.170208
Keywords: selenobacteria, Se, biofortification, genome, Se activation
Citation: Liao Q, Li A-M, Xing Y, Liang P-X, Jiang Z-P, Liu Y-X and Huang D-L (2024) Selenobacteria-mediated Se transformation and uptake involving the unique genetic code. Front. Plant Sci. 15:1392355. doi: 10.3389/fpls.2024.1392355
Received: 27 February 2024; Accepted: 11 April 2024;
Published: 24 April 2024.
Edited by:
M. J. I. Shohag, University of Florida, United StatesReviewed by:
Surjit Singh, Sister Nivedita University, IndiaCopyright © 2024 Liao, Li, Xing, Liang, Jiang, Liu and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yong-Xian Liu, bGl1eXgyN0AxNjMuY29t; Dong-Liang Huang, aGRsNjY2QDE2My5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.