- 1Institute of Neurobiology, Jining Medical University, Jining, China
- 2Institute of Biotechnology and Health, Beijing Academy of Science and Technology, Beijing, China
- 3Rehabilitation Department, Traditional Chinese Medicine Hospital of Yanzhou District of Jining City, Jining, China
Introduction: The bZIP genes (bZIPs) are essential in numerous biological processes, including development and stress responses. Despite extensive research on bZIPs in many plants, a comprehensive genome-wide analysis of bZIPs in garlic has yet to be undertaken.
Methods: In this study, we identified and classified 64 AsbZIP genes (AsbZIPs) into 10 subfamilies. A systematic analysis of the evolutionary characteristics of these AsbZIPs, including chromosome location, gene structure, conserved motifs, and gene duplication, was conducted. Furthermore, we also examined the nucleotide diversity, cis-acting elements, and expression profiles of AsbZIPs in various tissues and under different abiotic stresses and hormone treatments.
Results and Discussion: Our findings revealed that gene replication plays a crucial role in the expansion of AsbZIPs, with a minor genetic bottleneck observed during domestication. Moreover, the identification of cis-acting elements suggested potential associations of AsbZIPs with garlic development, hormone, and stress responses. Several AsbZIPs exhibited tissue-preferential and stress/hormone-responsive expression patterns. Additionally, Asa7G01972 and Asa7G01379 were notably differentially expressed under various stresses and hormone treatments. Subsequent yeast two-hybridization and yeast induction experiments validated their interactions with Asa1G01577, a homologue of ABI5, reinforcing their importance in hormone and abiotic stress responses. This study unveiled the characteristics of the AsbZIP superfamily and lays a solid foundation for further functional analysis of AsbZIP in garlic.
Introduction
Transcription factors (TFs) play pivotal roles in regulating gene expression by recognizing and binding to promoters, which is essential for plant development and stress responses (Singh et al., 2002; Li et al., 2013). The basic leucine zipper (bZIP) family is one of the TF families and is widely present in eukaryotes (Rodriguez-Uribe and O’Connell, 2006; Nijhawan et al., 2008). The bZIP family is characterized by a conserved bZIP domain, usually comprising 60 to 80 amino acids, which contains a basic region and a leucine zipper region (Nijhawan et al., 2008). The basic region, positioned at the N-terminal end of the region, contains a conserved N-x7-R/K motif that is related to nuclear localization and binds to target DNA (Suckow et al., 1994; Correa et al., 2008). The leucine zipper region exhibits a relatively lower level of conservation, consisting of a repetitive sequence comprising leucine or other hydrophobic amino acids (Ile, Val, Phe, or Met). Specifically, 9 of these amino acids are positioned at the C-terminus, forming an amphipathic helix (Jakoby et al., 2002; Wei et al., 2012; Liu and Chu, 2015; Hu et al., 2016b).
The bZIP gene family has been comprehensively characterized and investigated at the genomic level across various plant species, including Arabidopsis thaliana (Jakoby et al., 2002), Oryza sativa (Nijhawan et al., 2008), Glycine max (Liao et al., 2008b), Sorghum bicolor (Wang et al., 2011), Zea mays (Wei et al., 2012), Vitis vinifera (Liu et al., 2014b), Cucumis sativus (Baloglu et al., 2014), Maninot esculenta (Hu et al., 2016b), Malus domestica (Zhao et al., 2016), Brassica napus (Zhou et al., 2017), Fragaria ananassa (Wang et al., 2017), Daucus carota (Que et al., 2015) and Hordeum vulgare (Pourabed et al., 2015). Previous studies have demonstrated the important role of bZIPs in diverse crucial biological processes, such as organ and tissue differentiation (Gangappa et al., 2013; Lozano-Sotomayor et al., 2016; Zhang et al., 2016; Tan et al., 2020), seed storage (Bombarely et al., 2012; Edwards et al., 2017), metabolic activity (Baena-Gonzalez et al., 2007), photomorphogenesis and light signal regulation (Joo et al., 2014; Babitha et al., 2015), salt and drought tolerance (Ying et al., 2012; Liu et al., 2014a), and hormone and sugar signaling processes (Fan et al., 2019; Liu et al., 2020). Specifically, HY5, a key transcription factor in light signal transduction, encodes a bZIP protein that regulates root and hypocotyl development in Arabidopsis thaliana (Oyama et al., 1997). AtZIP1 is involved in sugar signaling and influences seed growth and development (Wigge et al., 2005). Additionally, AtbZIP53 can form heterodimers with AtbZIP1, 10 or 25 to promote the transcriptional activation of seed maturation genes (Alonso et al., 2009). AtZIP17 and AtZIP24 are crucial in the salt stress response (Liu et al., 2008; Yang et al., 2009). In rice, OsbZIP46 is strongly upregulated under drought, heat, and abscisic acid (ABA) stresses (Tang et al., 2012). OsbZIP72 is induced by drought and ABA treatments (Lu et al., 2009). In addition, SlbZIP33 is involved in stress-induced response and plays a vital role in fruit ripening (Orellana et al., 2010; Bastias et al., 2014). GmbZIP15 increases the sensitivity of soybean to salt and drought stresses by negatively regulating the gene expression levels of GmWRKY12 and GmABF1 (Zhang et al., 2020a). IbbZIP1 strongly responds to ABA and is related to salt and drought tolerances in sweet potato (Kang et al., 2019). TabZIP6 is involved in cold tolerance by forming a dimer with two other bZIP proteins belonging to the S subfamily (Cai et al., 2018).
Garlic, originating from Central Asia, the Mediterranean and the Caucasus, has been cultivated for over 5,000 years. Like onion, which is the largest crop in the Allium genus, garlic is not only an economically important vegetable and spice, but is also widely applied in the pharmaceutical and nutritional industries (ETOH et al., 2001; Martin and Ernst, 2003; Kamenetsky et al., 2015). At present, due to global warming and the shrinking availability of cultivated land, enhancing the quality of key traits is particularly imperative for garlic breeding. bZIP genes play vital roles in numerous physiological processes; thus, comprehensive identification and analysis of bZIP gene family members are essential. However, a notable research gap exists in the investigation of bZIP genes in garlic. The successful assembly of the garlic chromosome genome provides a solid foundation for these endeavors (Sun et al., 2020).
In this work, 64 AsbZIP genes (AsbZIPs) were identified and separated into 10 groups on the basis of phylogenetic relationships; these groups were compared with those of Arabidopsis thaliana and Oryza sativa, which are representative species of dicotyledonous and monocotyledonous plants, respectively. Further comprehensive analyses of AsbZIPs, including gene structure, motif analysis, chromosome distribution, evolutionary characteristics and cis−acting elements, were conducted. Additionally, we investigated the variation atlas of single-nucleotide polymorphisms (SNPs) in AsbZIPs. Finally, we explored the expression profiles of AsbZIPs in different garlic tissues and under multiple stresses via quantitative real-time polymerase chain reaction (qRT-PCR). Our study provides a solid foundation for further functional investigations of AsbZIPs.
Materials and methods
Identification of bZIP genes in the garlic genome
Whole garlic genome data were downloaded from https://doi.org/10.6084/m9.Figshare.12570947.v1, and those of Arabidopsis thaliana and Oryza sativa were retrieved from the TAIR database (https://www.Arabidopsis.org/) and the RGAP database (https://rice.plantbiology.msu.edu/), respectively. The bZIP domains (PF00170 and PF07716) were retrieved from the PFAM database (http://pfam.xfam.org) (El-Gebali et al., 2019) and used to perform an HMM (hidden Markov model) search via the HMMER 3.0 program. NCBI-CDD (https://www.ncbi.nlm.nih.gov/cdd/) and SMART (http://smart.embl.de/) were used to further confirm the bZIP domain of potential bZIP gene family members. The ExPASy proteomics server (https://web.expasy.org/computepi/) was used to calculate the molecular weight (MW), isoelectric point (pI), instability index (I.I.), aliphatic index (A.I.), total number of negatively charged residues (Asp + Glu, n.c.r, %), total number of positively charged residues (Arg + Lys, p.c.r, %), and grand average hydropathicity (GRAVY) of the bZIP proteins in garlic. The subcellular localizations of the bZIP proteins were assessed via the Cell-PLoc 2.0 web server (http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/).
Phylogenetic analysis
The bZIP protein sequences from Allium sativum, Arabidopsis thaliana, and Oryza sativa were aligned via MUSCLE with default parameters. A phylogenetic tree for these proteins was established via the neighbor-joining (NJ) method in MEGA 7.0 software (Kumar et al., 2018), with 1000 iterations. The iTOL online software tool (https://itol.embl.de/) was used to output visual images.
Gene structure and conserved motif analysis
The Gene Structure Display Server (GSDS) (http://gsds.gao-lab.org/) (Hu et al., 2015) was employed to identify the exon and intron structures of all AsbZIPs. The MEME program (https://meme-suite.org/meme/) (Bailey et al., 2009) was used to investigate conserved motifs. The minimum and maximum lengths of the conserved motifs were set to 6 and 50, respectively, with a maximum of 10 conserved motifs.
Chromosomal localization and gene duplication analysis of AsbZIPs
The chromosomal positions of the AsbZIP genes were visualized via MapChart software (Stajich et al., 2002) according to the garlic genome annotation (https://doi.org/10.6084/m9.Figshare.12570947.v1). Using the Multiple Collinear Scan Toolkit (MCScanX), we investigated the gene duplication events of the AsbZIP genes (Wang et al., 2012). A tandem duplication event is a chromosome region with two or more adjacent genes within a 200 kb range, and duplicated pairs positioned on different chromosomes are defined as segmental duplication events (Cannon et al., 2004). Syntenic analysis of bZIP genes between garlic and four plant species (Arabidopsis thaliana, Oryza sativa, Zea mays, and Theobroma cacao) was conducted via Dual Synteny Plotter software (https://github.com/CJChen/TBtools) (Chen et al., 2020). The calculation of synonymous (Ks) and nonsynonymous (Ka) substitution rates and Ka/Ks ratios of each duplicated gene pair were carried out based on the coding sequences (CDS) alignments of bZIP genes via Ka/Ks calculator 2.0 software, and a Ka/Ks ratio > 1 was interpreted as positive selection, < 1 as purified selection, and = 1 as neutral evolution (Nei and Gojobori, 1986; Wang et al., 2010).
Analysis of cis-acting elements
Cis-acting elements within the 2000 bp region upstream of the transcriptional start site of each AsbZIP gene were identified via the PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html) (Chow et al., 2019).
Nucleotide variation and population structure
The resequencing data of 233 garlic samples were retrieved from the Genome Variation Map (project accession: PRJCA006629). Supplementary Table 1 presents the geographic distribution and detailed material information. SnpEff v4.3 was utilized for the annotation of SNPs (Cingolani et al., 2012). Additionally, the population structure was analyzed via ADMIXTURE v1.3.0, with K values ranging from 2 to 5. The phylogenetic tree was established by Treebest v1.9.2, and the Smartpca within EIGENSOFT v4.2 was used for the analysis of principal component analysis (PCA). Nucleotide diversity (π) and Wright’s F statistic (Fst) were calculated using VCFtools v0.1.16.
Plant materials and various stresses and hormone treatments
Garlic cloves (cv. Ershuizao) were planted in pots and cultivated in a chamber (16 h/8 h of light/dark, 30°/22° day/night). The treatment experiments were performed as described previously (Zhang et al., 2022b; Munim Twaij et al., 2023; Yao et al., 2023; Shi et al., 2024) with some modifications. The salt, cold and heat stress experiments were simulated with a 200 mM NaCl solution, at 4° and 30°, respectively. Five hormone treatments, including ABA (1 mg/L), GA3 (gibberellic acid, 200mg/L), MeJA (methyl jasmonic acid, 100 µM), IAA (indoleacetic acid, 100 µM), and SA (salicylic acid, 100 µM)), were conducted. Root and leaf samples were collected at 0, 6, 12, 24 and 48 hours after each treatment and promptly frozen in liquid nitrogen. For the drought treatment, irrigation was ceased for the seedlings in the treatment group, and those in the control group were irrigated normally. Roots and leaves were harvested at 0, 7 and 14 days after treatment. Additionally, freshly harvested garlic cloves were stored at 4°C and collected at 0, 10, 15, and 40 days after treatment. To investigate the tissue-specific expression profiles of the AsbZIPs, leaves, stems, pseudostems, roots, buds, bulbs and flowers were harvested at 192 days after planting. All samples were promptly frozen in liquid nitrogen and then preserved at -80°C for subsequent mRNA extraction.
RNA isolation and qRT-PCR analysis
RNA extraction kits (Vazyme, Nanjing, China) were used to extract total RNA according to the manufacturer’s instructions. Subsequently, reverse transcription of two micrograms of RNA was conducted via HiScript III RT SuperMix for qPCR (Vazyme, Nanjing, China). The qRT-PCR assay was conducted as previously described (Xu et al., 2013). Supplementary Table 2 shows the primers designed with Primer Express software (v3.0). The tubulin gene was used as the internal control gene, and the expression level of each gene was determined via the 2−ΔΔCt method (Livak and Schmittgen, 2001).
Gene regulatory network analysis
The co-expression network was constructed via 185 RNA-Seq datasets retrieved from the Gene Expression Omnibus (GEO) database (accession codes GSE211495, GSE186042, and GSE145455) and the Sequence Read Archive (SRA) database (accession codes PRJNA682570, PRJNA472416, and PRJNA683607). The high-quality reads were filtered via the NGS QC Toolkit (v2.3). TopHat (v2.0.0) was used to align these filtered reads to the garlic genome with default parameters. Then, the FPKM values and read counts of each garlic gene were calculated via Cufflinks (v2.0.2). To identify genes co-expressed with AsbZIP genes, weighted gene co-expression network analysis (WGCNA) was used to construct a co-expression network. For further investigation, genes within the top 5% highest weighted values associated with AsbZIP genes were selected.
Furthermore, to determine directional interactions in the transcriptional regulatory network related to AsbZIP genes, 2-kb upstream sequences of co-expressed genes of each AsbZIP gene were analyzed, and FIMO (Grant et al., 2011) was utilized to screen genes whose promoters included a significantly enriched motif of corresponding AsbZIP gene according to the high-quality TF binding motifs retrieved from the PlantTFDB database. A motif was regarded as present in a promoter if it had at least one match at a P value ≤ 10-4. The clusterProfiler package in R was used to perform GO enrichment analysis of the filtered co-expressed genes of each AsbZIP gene.
Prediction of the AsbZIP protein interaction network
STRING (https://cn.string-db.org/) was used to construct an interaction network for AsbZIP proteins according to their orthologous proteins in Arabidopsis thaliana.
Heterologous expression in yeast and yeast two-hybrid
To investigate the functions of Asa7G01972 and Asa7G01379 in salt stress, a specific primer pair for each gene (Supplementary Table 2) was used to amplify the DNA fragment, which was cloned in frame into the pYES2 recombinant vector. The recombinant and empty vector plasmids were separately transformed into yeast cells (BY4741) following the manufacturer’s protocol for the Yeastmaker™ Yeast Transformation System 2 (Clontech Laboratories, Inc., Palo Alto, CA, USA). Yeast transformants were cultivated in SD/-Ura liquid medium at 30° until the OD600 reached 0.5. The preculture was transferred to SG (-Ura, 2% galactose) and diluted to an OD600 of 0.4, followed by incubation with shaking for an additional 24 hours at 30°C to induce gene expression. Subsequently, the yeast cells were harvested for subsequent stress treatments (Ibrahim et al., 2001).
The Y2H experiment was conducted according to the manufacturer’s instructions (Clontech Laboratories, Inc., Palo Alto, CA, USA). The CDS of Asa7G01972 or Asa7G01379 was inserted into the pGBKT7 vector to form the bait construct, whereas that of AsaABI5 (geneID: Asa1G01577) was cloned and inserted into the pGADT7 vector to generate the prey construct. The primers used for the Y2H assay are presented in Supplementary Table 2. These plasmids were then transformed into yeast cells (strain AH109), and the resulting yeast transformants were cultured on SD medium supplemented with 3-aminotriazole and X-α-gal but lacking tryptophan, leucine, histidine, and adenine at 30°C for 2−3 days.
Statistical analysis
The samples were harvested from three independent plants. Data from at least three replicates are shown as the means ± SDs. The statistical analysis, including Student’s t test, was conducted via SPSS software (version 17, SPSS Inc., Chicago, IL, USA). A significance criterion of P < 0.05 indicated statistical significance.
Results
Identification and characterization of bZIP genes in garlic
A total of 64 AsbZIP genes were identified via hmm search and confirmed via the NCBI-CDD and SMART databases (Supplementary Table 3). Apart from 3 genes (Asa0G04894, Asa0G01277, and Asa0G02642) located on unassigned scaffoldings, the remaining 61 genes were randomly distributed across 8 chromosomes. Specifically, chromosome 4 had the greatest number of genes, with 12, whereas chromosome 3 had the lowest number of genes, with only 4.
Gene characteristics were further analyzed. The CDS lengths of the AsbZIP genes ranged from 240 bp (Asa2G04074) to 1758 bp (Asa8G00330), with predicted molecular weights (MWs) ranging from 9.24256 to 64.00282 kDa. Asa7G01972 and Asa7G01871 presented the extremes in terms of isoelectric point (pI), with values of 4.89 and 11.24, respectively, indicating the lowest and highest pIs within the set. All the AsbZIP proteins had consistently negative GRAVY values, indicating their hydrophilic nature, which was coincident with their presumed roles as transcription factors. Apart from 8 genes (Asa3G03771, Asa4G00594, Asa4G00875, Asa4G04645, Asa5G05838, Asa7G02467, Asa7G05774, and Asa8G00330), the majority of proteins exhibited instability. Furthermore, all the AsbZIP proteins were localized in the nucleus.
Phylogenetic analysis and classification of AsbZIPs
bZIP protein sequences from garlic (64), Arabidopsis thaliana (78), and Oryza sativa (88) were used to construct a phylogenetic tree via the NJ method, delineating their evolutionary relationships (Figure 1). A total of 230 bZIPs from these species were divided into 10 subfamilies, labelled as Groups A, B, C, D, E, F, G, H, I, and S, on the basis of their classification in Arabidopsis thaliana (Jakoby et al., 2002). The subfamilies varied in size, with the largest encompassing 47 members (S group) and the smallest containing 9 members (B group). Notably, all the species contributed members to each identified subfamily.
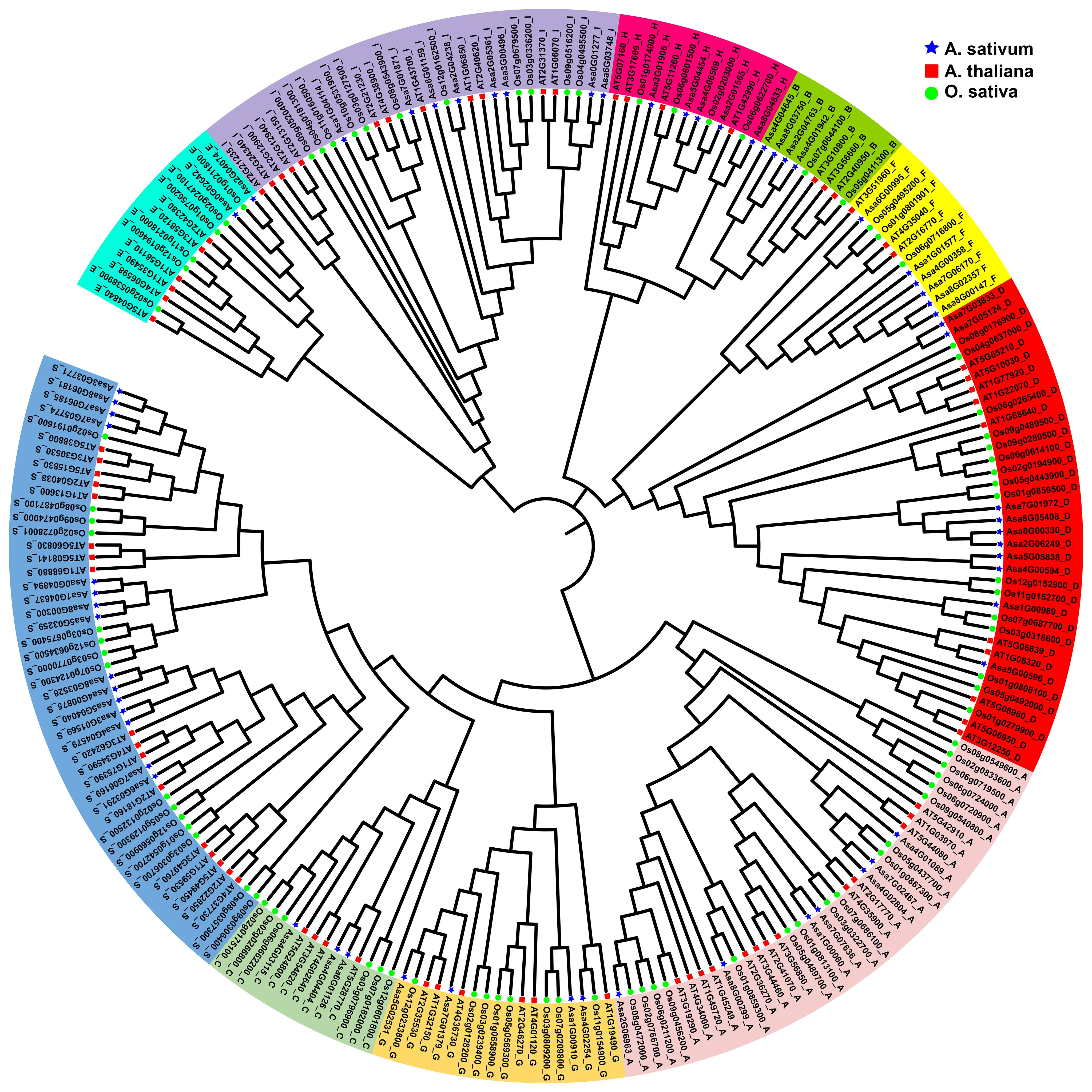
Figure 1 Phylogenetic relationships of the identified bZIP proteins in Allium sativum, Arabidopsis thaliana, and Oryza sativa. The colored regions represent bZIP genes of different subfamilies A to I, S. The blue stars indicate the AsbZIP proteins, the red squares represent the AtbZIP proteins, the green circles represent the OsbZIP proteins.
Gene structure and conserved motif analysis
The structure of exons and introns provides pivotal evidence for discerning phylogenetic relationships within gene families (Li et al., 2006). The number and distribution of exons and introns of AsbZIPs were investigated (Figures 2A, B). The results revealed that 17 AsbZIPs, constituting 27% of the total, lacked introns, and Groups D and I had the most abundant intron-lacking AsbZIP genes, with four. Among AsbZIPs with introns, the number of introns ranged from 1 to 11. The greatest number of introns among the AsbZIP genes was 11, and these genes belonged to Groups A, D, H, S and I. The AsbZIPs in Groups C and G contained either 0 or 3 introns, whereas those in Group E had either 0 or 1 introns. Generally, AsbZIP genes within the same subfamily presented similar gene structures.
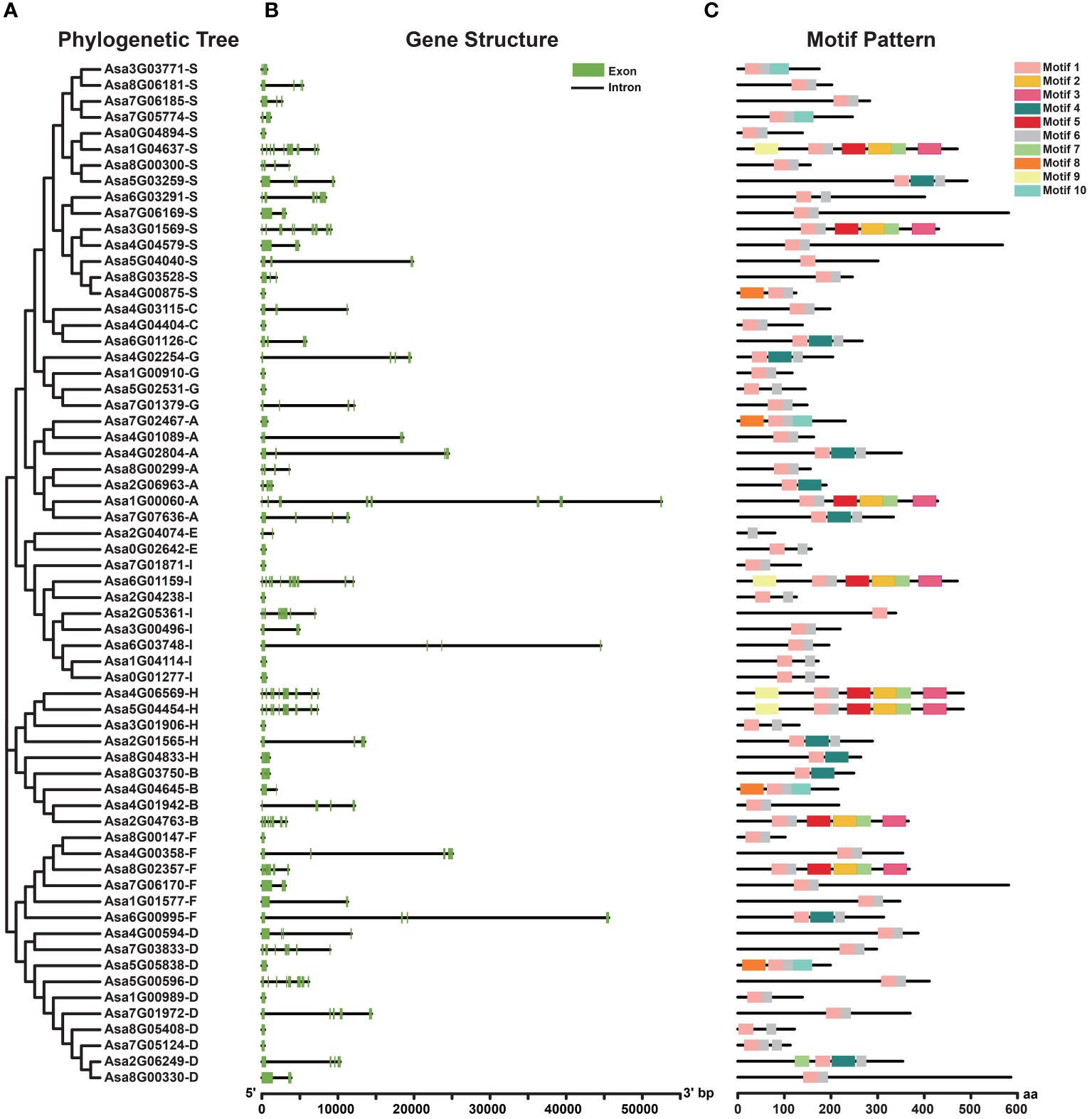
Figure 2 Phylogenetic relationships, gene structures and conserved protein motifs of the AsbZIP superfamily in garlic. (A) Phylogenetic relationships of 64 AsbZIP proteins. (B) Exon-intron structure of AsbZIP genes. Green boxes indicate exons, and black lines indicate introns. (C) The motif profile of AsbZIP proteins. These motifs are presented in differently colored boxes numbered 1-10. The sequence information of each motif is provided in Supplementary Table 4. The protein length can be estimated using the scale at the bottom.
Additionally, to gain insight into the divergence and characterization of AsbZIP proteins, 10 conserved motifs were identified (Figures 2A, C). These motifs varied in length, ranging from 21 (motif 6) to 50 (motifs 2, 3, 4, 5, 8 and 9) (Supplementary Table 4). The majority (89.06%) of the AsbZIP proteins presented the prevalent presence of motifs 1 and 6. Motif 8 was present in Groups A, B, D and S; motif 9 was found in Groups H, I and S; and motif 10 occurred in Groups A, B and S. Numerous motifs were found in particular groups, suggesting potential associations with distinct biological functions.
Chromosomal distribution, gene duplication events and synteny analysis of bZIPs in garlic
A total of 61 AsbZIP genes were unevenly distributed on 8 chromosomes, and three AsbZIP genes (Asa0G01277, Asa0G02642, and Asa0G04894) were present on the scaffolds (Figure 3). Chromosome 4 had the most genes, with 12 genes, followed by chromosome 7 (11 genes), chromosome 8 (10 genes) and chromosome 2 (7 genes). Chromosome 1 and chromosome 5 each featured an identical number of 6 AsbZIP genes. Chromosome 3 had the lowest number of AsbZIP TFs (4 genes).
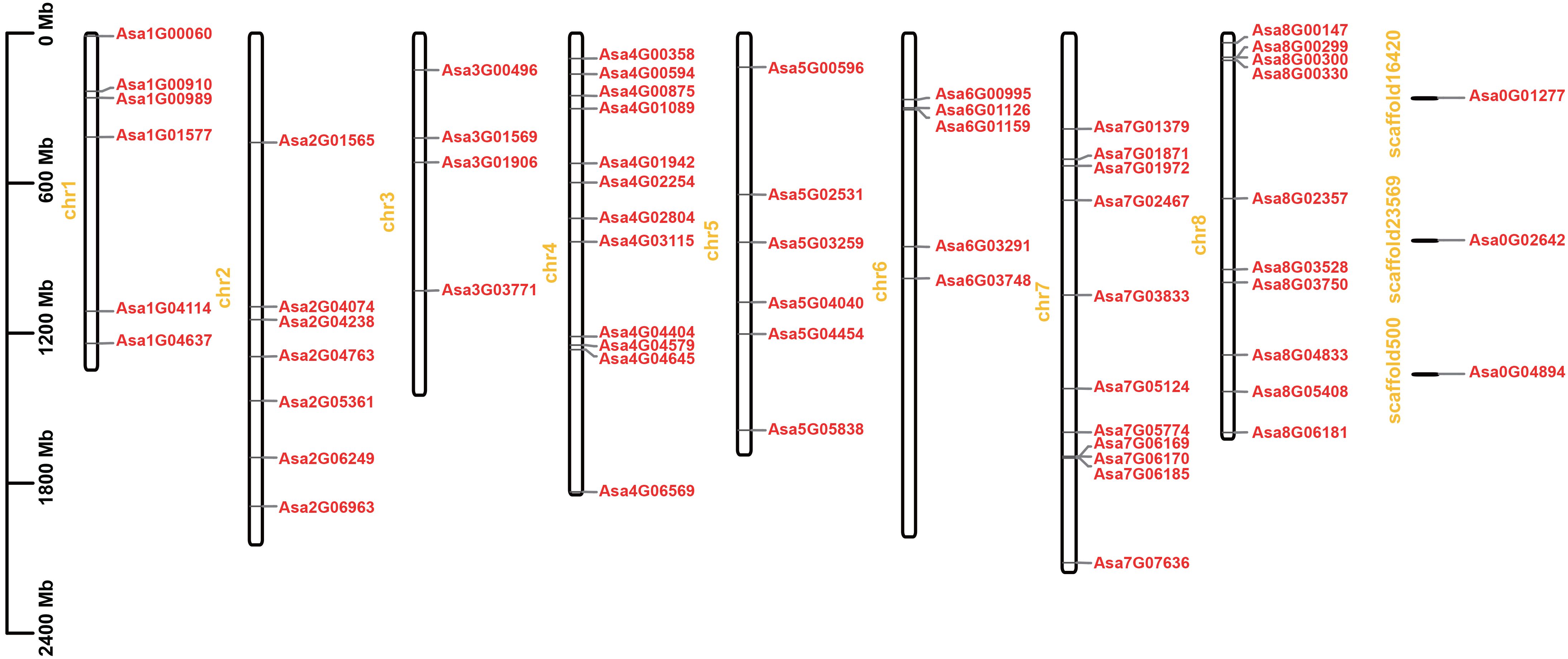
Figure 3 Chromosomal distribution of bZIP genes in garlic. The chromosome number is marked on the left of each chromosome in yellow. Chromosome lengths and gene positions can be inferred from the scale on the left.
To explore the evolutionary patterns of the AsbZIP genes, tandem and segmental duplication events were analyzed. Two tandem duplication regions, Asa7G06170 and Asa7G06169, Asa8G00299 and Asa8G00300, are located on chromosomes 7 and 8, respectively (Figure 3; Supplementary Table 5). Furthermore, seven pairs of segmental duplicated genes (Asa4G00594 and Asa7G01379, Asa5G00596 and Asa7G01871, Asa6G03748 and Asa7G01871, Asa4G03115 and Asa8G04833, Asa5G00596 and Asa8G04833, Asa5G04040 and Asa8G00330, Asa8G03528 and Asa8G00330) were identified; these pairs were associated with chromosomes 4 and 7, chromosomes 5 and 7, chromosomes 6 and 7, chromosomes 4 and 8, chromosomes 5 and 8, chromosomes 5 and 8, and chromosome 8, respectively (Figure 4; Supplementary Table 5). These observations strongly suggest that tandem and segmental duplication played a significant role in the expansion of AsbZIP genes.
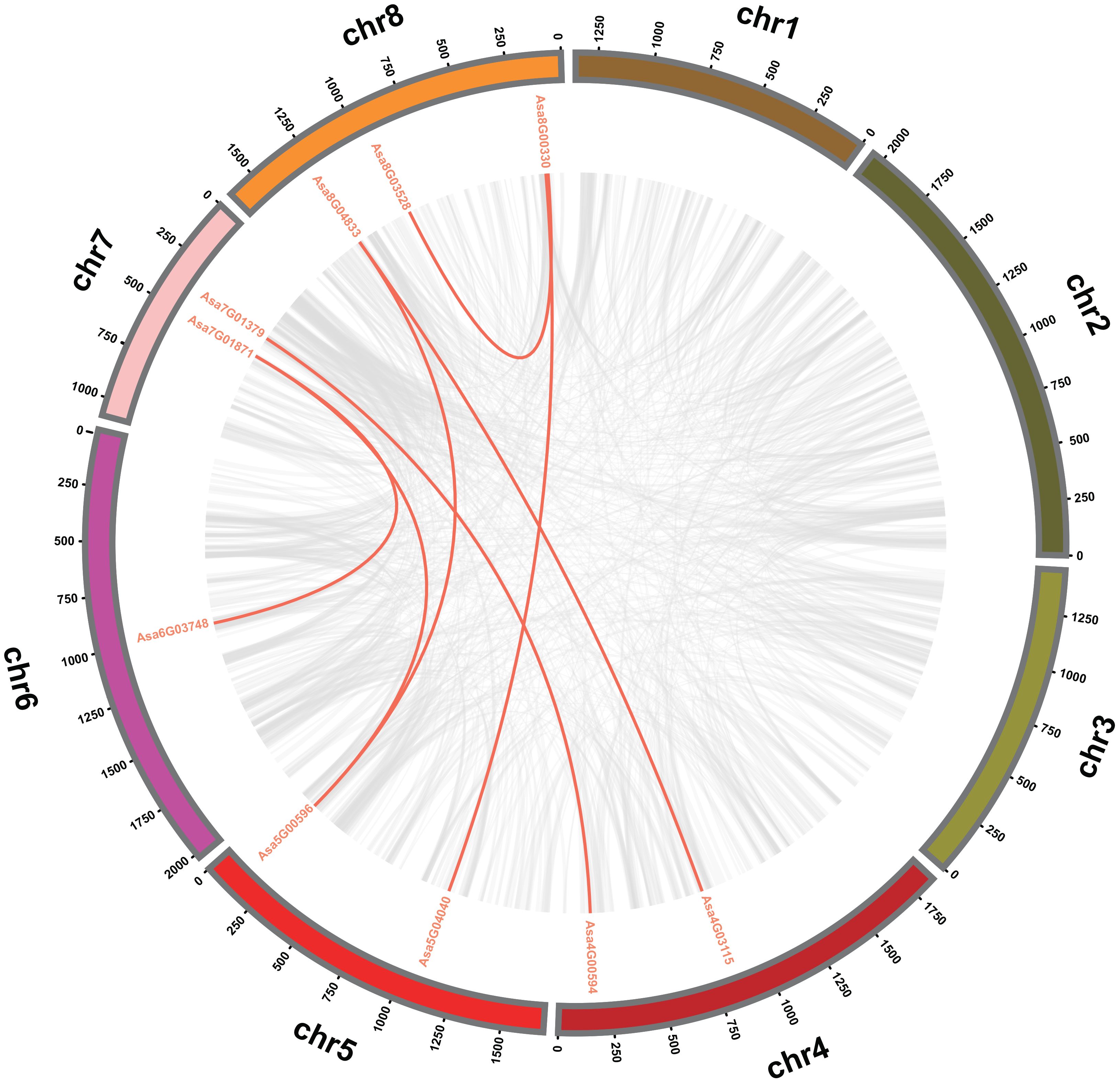
Figure 4 Interchromosomal relationships of the bZIP genes in garlic. Different-colored circles represent distinct chromosomes, with grey lines within the circles representing all syntenic blocks in the garlic genome and red lines within the circles indicating collinear blocks among AsbZIP genes.
To delve into the evolutionary restrictions of AsbZIP genes, the Ka vs. Ks substitution ratios were analyzed (Supplementary Table 5). The Ka/Ks ratios between Asa7G06170 and Asa7G06169, Asa8G00299 and Asa8G00300, Asa8G00330 and Asa8G03528, Asa8G00330 and Asa5G04040, Asa7G01871 and Asa6G03748, and Asa7G01871 and Asa5G00596 were less than 1, implying purifying selection, whereas those between Asa8G04833 and Asa5G00596, Asa8G04833 and Asa4G03115, and Asa7G01379 and Asa4G00594 were greater than 1, suggesting positive selection.
Evolutionary analysis of AsbZIP genes and several other species
To further elucidate the phylogenetic mechanisms of the AsbZIP family, we established syntenic maps of garlic compared with those of four other representative species: two monocotyledonous plants (Oryza sativa and Zea mays) and two dicotyledonous plants (Arabidopsis thaliana and Theobroma cacao) (Figure 5). Twenty-two bZIP genes in garlic exhibited collinearity associations with those in Oryza sativa (19 pairs), Zea mays (12 pairs), Arabidopsis thaliana (15 pairs), and Theobroma cacao (2 pairs). Additionally, 19 gene pairs consisting of only 6 AsbZIPs were detected between garlic and Oryza sativa, and Zea mays, indicating that these orthologous pairs appeared after the divergence between monocotyledonous and dicotyledonous plants.
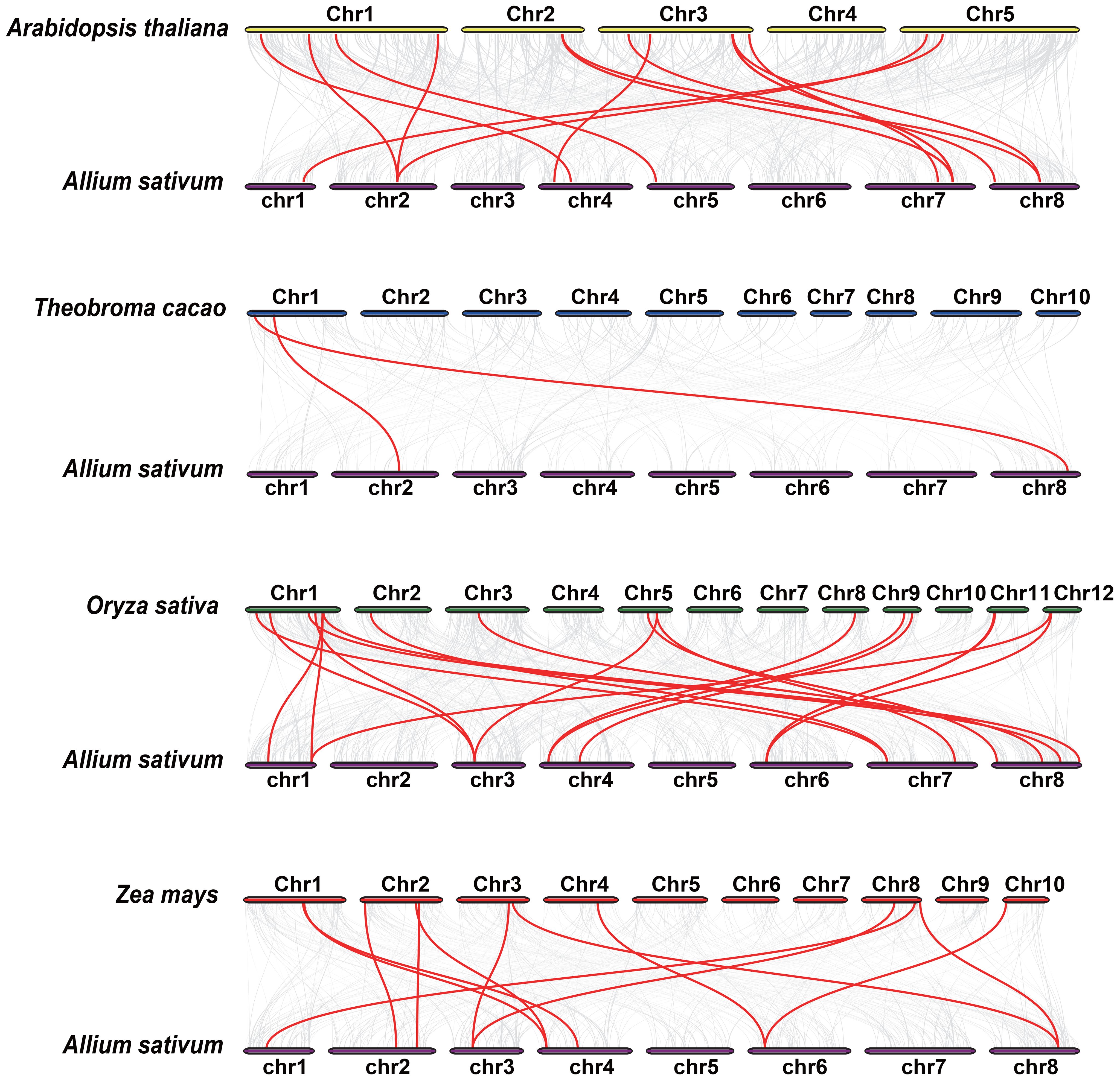
Figure 5 Syntenic analysis of bZIP genes between garlic and four representative plant species. Gray lines in the background represent collinear blocks between garlic and other plant genomes, whereas red lines highlight syntenic bZIP gene pairs.
Nucleotide variation and population structure of AsbZIP genes
Publicly available garlic resequencing data were used to explore AsbZIP-related SNPs, revealing the intricate sequence diversity of AsbZIP genes. The dedicated SNP calling pipeline successfully generated 2941 SNPs, each with high confidence (Supplementary Table 6). A predominant portion of AsbZIP-associated SNPs were enriched in intergenic regions, whereas others occurred within genic regions, including 4 missense, 40 intron, 1 synonymous, 43 downstream, and 70 upstream variants. The overall transition/transversion (Ts/Tv) ratio was 1.938, with G/A (21.73%) and C/T (19.25%) as the predominant allelic substitution patterns, indicating a higher frequency of mutations involving purine-to-purine or pyrimidine-to-pyrimidine transitions than mutations that switch pyrimidines to purines or purines to pyrimidines.
PCA was performed using AsbZIP-related SNPs to explore the interrelationship among the origin group and three distinct garlic cultivars (Figure 6A). The first eigenvector, which explained 54.12% of the total genetic variance, indicated divergence within these populations. The second and third eigenvectors, contributing 17.32% and 9.06% to genetic variation, respectively, facilitated the differentiation among these groups. The phylogenetic tree displayed consistent population affinities (Figure 6B). In accordance with the phylogenetic tree, ADMIXTURE analysis also validated the consistent group relatedness. At K=4, a clear demarcation according to geographical origin was observed. The existence of a genetic mixture between wild and landrace garlic indicated a possible domestication of cultivated garlic and the ongoing gene flow between wild and landrace garlic.
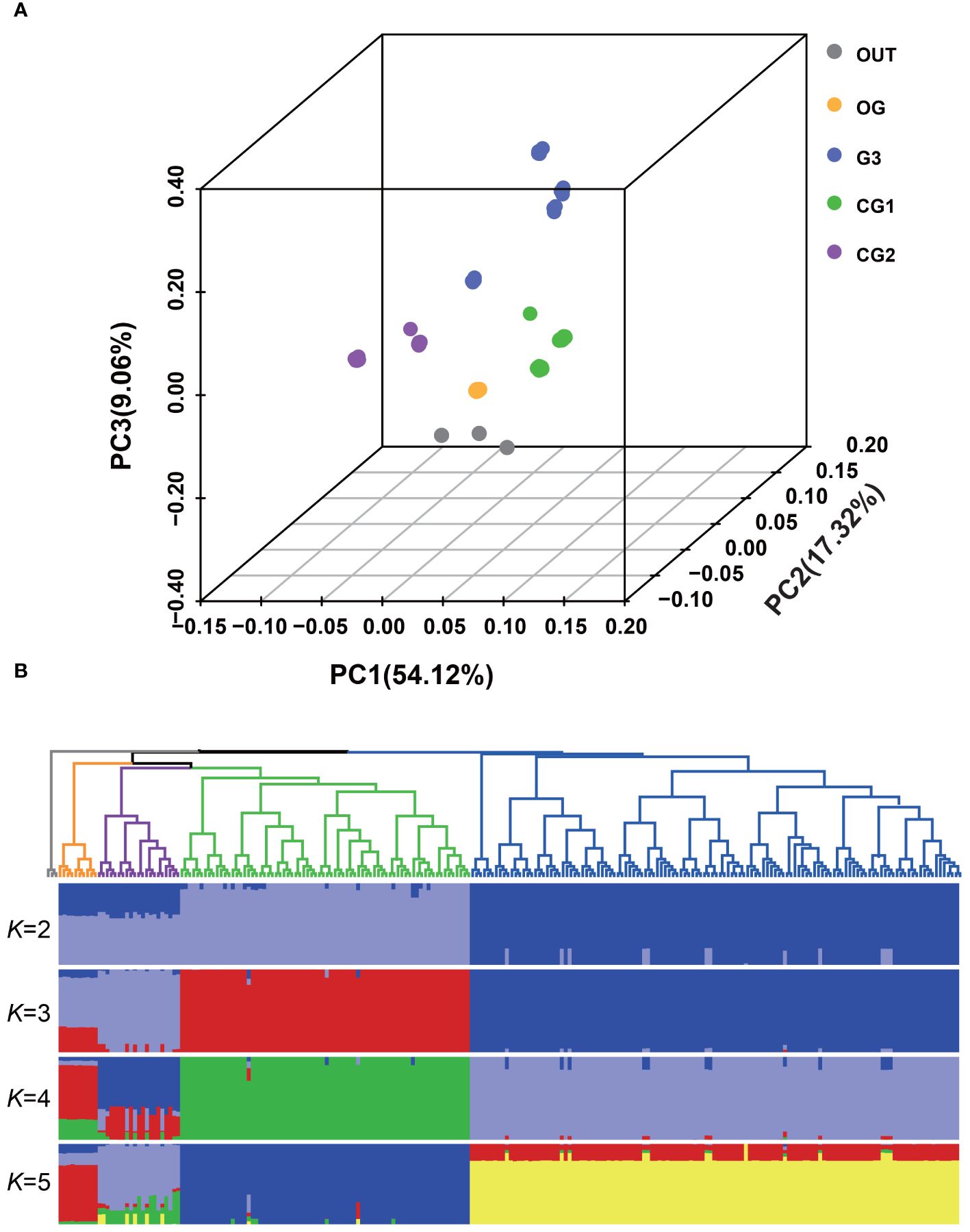
Figure 6 Population structure of wild and local garlic based on AsbZIP-related SNPs. (A) Plots of principal component analysis for the first (PC1), second (PC2), and third (PC3) components. Dot color indicates population and location. (B) Phylogenetic tree and population structure with K ranging from 2 to 5. The rooted tree was constructed using neighbor-joining method. The orders and positions of sample accessions on the x-axis are consistent with those in the neighbor-joining tree.
To assess the occurrence of genetic bottlenecks in AsbZIPs in garlic during acclimation, the population-based nucleotide diversity of AsbZIP genes was calculated. AsbZIP genes presented a slight decrease of only 0.00003% in nucleotide diversity from wild garlic to local garlic (Supplementary Figure 1), suggesting a weak genetic bottleneck during domestication. Additionally, we measured the degree of population differentiation via Wright’s F statistic. The calculated Fst index of 0.2235 between wild and landrace garlic within the AsbZIPs indicated that this gene family experienced relatively mild selective pressures during the domestication of garlic.
Analysis of Cis−acting elements in AsbZIP promoters
Regulatory mechanisms potentially regulating AsbZIP genes were investigated by analyzing the cis-elements present in their promoters. The results revealed that these cis-elements could be classified into three groups: development, hormone and stress-associated elements (Figure 7; Supplementary Figure 2). Several development-related cis-elements, such as the CAT-box (associated with meristem development), O2-site (associated with zein metabolism regulation), circadian (associated with circadian control), GCN4_motif (associated with endosperm development), and MSA-like (associated with cell cycle regulation), were located on the promoters of AsbZIPs. Ten hormone-responsive regulatory elements associated with ABA, IAA, GA, MeJA, and SA responses, including ABRE (29.48%), TGA-element (8.25%), AuxRR-core (2.59%), TGA-box (0.71%), P-box (5.19%), GARE-motif (3.54%), TATC-box (2.83%), TGACG-motif (19.81%), CGTCA-motif (19.81%) and TCA-element (7.78%), were discovered in the promoters of AsbZIPs. Additionally, seven stress-responsive regulatory elements that respond to various stress conditions, such as anaerobic induction, anoxic-specific inducibility, defense and stress responsiveness, drought inducibility, low-temperature responsiveness, salt responsiveness and wound-responsiveness, were identified, including ARE (47.74%), GC-motif (3.01%), TC-rich repeats (8.65%), MBS (20.3%), LTR (19.17%), DRE (0.38%) and WUN-motif (0.75%). These findings indicate that the transcriptional regulation of AsbZIPs is associated with development, hormone, and stress.
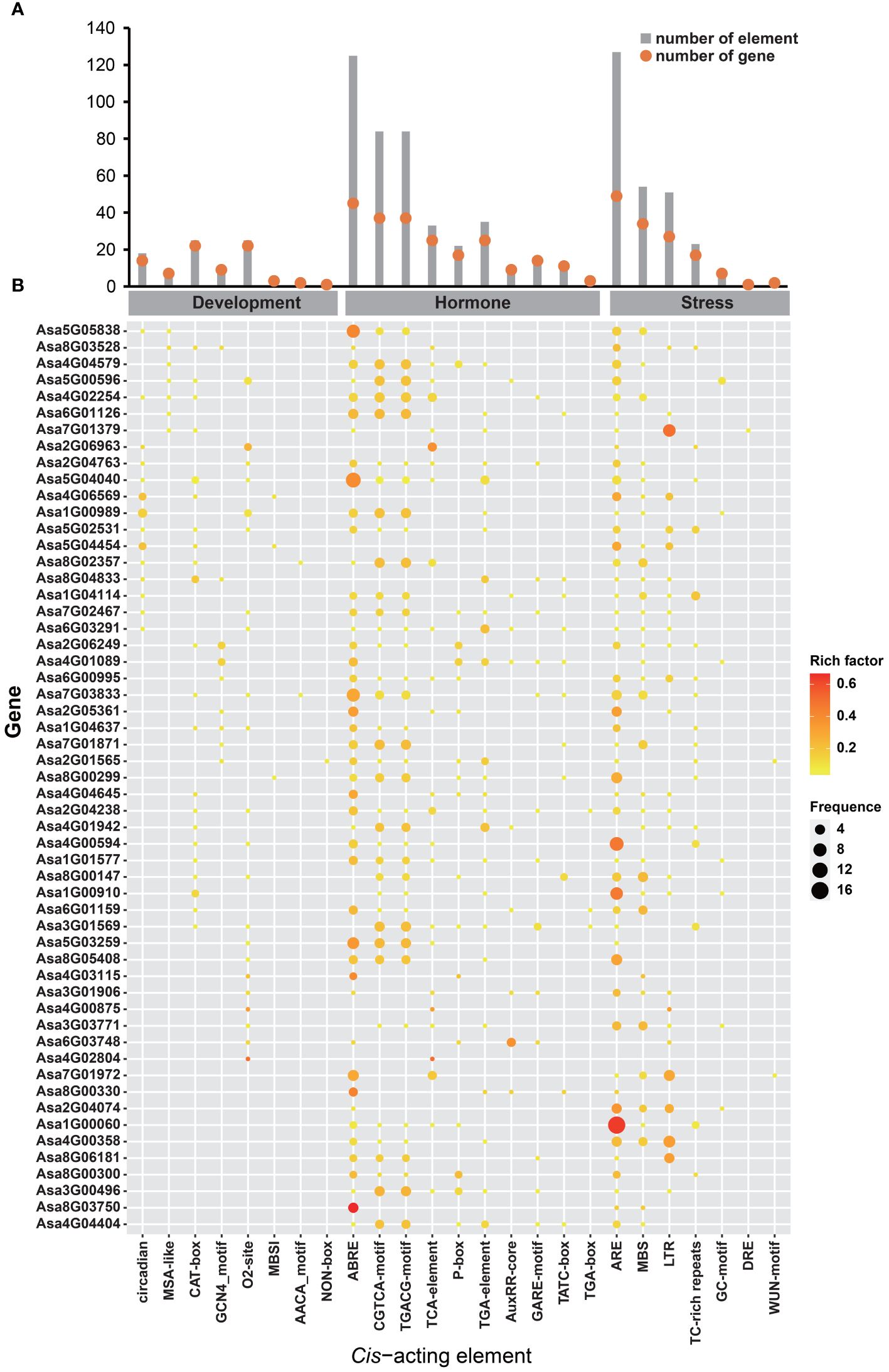
Figure 7 Cis-acting elements in the promoter of AsbZIP genes. (A) Number of each cis-acting element in the promoter regions of the AsbZIP genes. Red dots indicate the number of genes containing the corresponding cis-acting element, while gray boxes represent the total number of cis-acting elements. (B) Frequency of each cis-acting element in the promoters of AsbZIP genes. Cis-acting elements were categorized into development-related, hormone-related, and stress-related classes.
Expression patterns of AsbZIPs in multiple tissues of garlic
To investigate the function of the AsbZIP genes in growth and development, 14 AsbZIP genes were randomly selected, and their expression levels in various tissues (bulb, bud, leaf, pseudostem, sprout, root and flower) were determined. As shown in the Supplementary Figures 3A, 4 (Asa6G00995, Asa8G02357, Asa2G06963 and Asa4G03115), and 2 (Asa8G06181 and Asa7G01379) AsbZIP genes were relatively highly expressed in roots and leaves, respectively. Asa6G00995 presented high expression levels in roots, whereas its expression was relatively low in buds. The expression trend of Asa3G01906 was opposite to that of Asa6G00995; Asa3G01906 presented elevated expression levels in buds, with lower expression levels observed in roots. Additionally, Asa8G04833 exhibited high expression in flowers, whereas the expression level of Asa4G00594 was relatively high in bulbs. These distinct expression patterns suggest various roles of these genes in garlic development.
Expression patterns of AsbZIPs under abiotic stress
Given the crucial regulatory roles of bZIP transcription factors in response to adverse conditions, it is imperative to study the expression of AsbZIPs under various abiotic stress conditions. Figures 8A–D illustrate the expression profiles of AsbZIP genes under salt, drought, heat and cold stresses, respectively. Under salt stress, Asa4G00594, Asa7G01379, Asa2G06963 and Asa7G01972 were significantly upregulated in both leaves and roots at different time points. Asa6G00995, Asa8G02357, Asa2G04763, Asa7G02467 and Asa5G05838 presented induced expression and peaked at 12 h in leaves but presented suppressed expression in roots under salt stress. In contrast, Asa3G01906, Asa8G04833 and Asa4G03115 were downregulated in leaves and upregulated in roots at different time points. Additionally, Asa7G01972 presented the greatest change in gene expression level in both leaves (peaked at 12 h) and roots (peaked at 6 h) under salt stress, followed by Asa7G01379, which peaked at 12 h in both leaves and roots. Under drought stress, Asa4G00594, Asa3G01906, Asa7G01379, Asa7G01972 and Asa5G05838 were upregulated and peaked at 14 d, whereas Asa6G00995, Asa8G06181, Asa2G06963, Asa7G02467, Asa4G03115 and Asa8G05408 were downregulated at distinct time points in both roots and leaves, implying their pivotal roles in response to drought stress in distinct tissues. Furthermore, Asa7G01972 and Asa7G01379 were the genes whose expression was most predominantly induced in leaves and roots, respectively. Under heat stress, with the exception of Asa8G04833 and Asa2G06963, whose expression decreased, the other genes were induced in the leaves. Moreover, Asa8G06181 and Asa7G01379 presented the greatest induction at 24 h in the leaves and roots, respectively. Under cold treatment, Asa4G00594, Asa6G00995, Asa3G01906, Asa7G01379, Asa2G04763, Asa8G04833, Asa7G02467, Asa7G01972 and Asa8G05408 presented increased expression in the leaves. Among these genes, the expression of Asa4G00594, Asa6G00995, Asa7G01379, Asa8G04833, Asa7G02467 and Asa7G01972 peaked at 6 h. Seven genes (Asa4G00594, Asa6G00995, Asa3G01906, Asa7G01379, Asa8G04833, Asa7G02467 and Asa8G05408) were upregulated in the roots at all time points after cold treatment, with Asa3G01906 exhibiting the highest expression levels. In addition, under cold stress, the majority of AsbZIPs were induced in garlic cloves, except for Asa7G01379, Asa8G04833, and Asa5G05838 (Supplementary Figure 3B). Among these genes, the highest induction level was found in Asa2G06963 at 40 d, while Asa5G05838 was the most significantly inhibited gene. Overall, the predominant and distinct expression profiles of these genes suggest their potential roles in the response to abiotic stress.
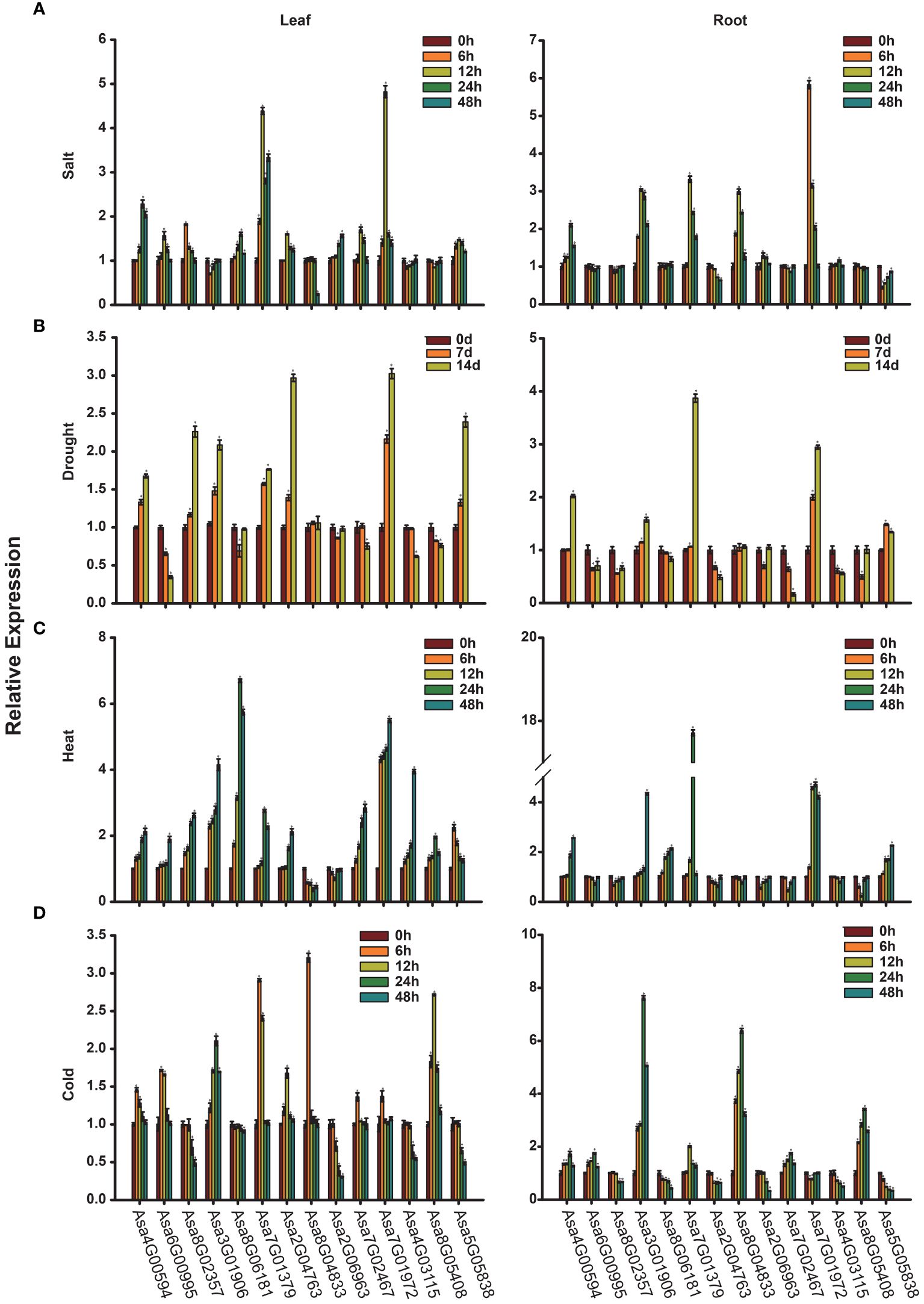
Figure 8 Expression analysis of AsbZIP genes. (A-D) The expression levels of AsbZIP genes under salt (A), drought (B), heat (C) and cold (D) detected by qRT-PCR. Error bars represent standard deviations from biological replicates. One asterisk (*) denotes a significant difference at P < 0.05, determined using Student’s t-test.
Expression analysis of AsbZIP genes under hormonal treatment
Numerous cis-elements associated with hormone response in the promoters of AsbZIP genes indicate pivotal roles of these genes in the plant hormone response. Therefore, the expression profiles of AsbZIP genes under hormonal stress conditions (ABA, GA3, IAA, MeJA, and SA) were investigated. The expression levels of 14 AsbZIPs in garlic leaves and roots at 6 h, 12 h, 24 h and 48 h after treatment were determined via qRT-PCR (Figures 9A–E). Under ABA stress, the increased expression patterns of Asa7G01379, Asa8G04833, Asa7G01972, and Asa5G05838 were detected at all time points, whereas other genes presented reduced expression in leaves. Among these genes, Asa7G01379 had the highest induction level and peaked at 24 h, followed by Asa7G01972, which peaked at 12 h; the most significantly inhibited level was obtained in Asa8G02357 at 12 h. In roots, Asa4G00594, Asa6G00995, Asa8G06181, Asa7G01379, Asa8G04833, Asa7G01972 and Asa4G03115 were predominantly induced by ABA at all time points, whereas other genes were inhibited. Furthermore, Asa7G01972 was most significantly induced and peaked at 12 h. Under GA3 treatment, most AsbZIP genes presented increased expression profiles, except for Asa8G02357, whose expression was most significantly induced in Asa7G01379 at 6 h in leaves. In roots, the expression levels of 9 AsbZIP genes, namely, Asa4G00594, Asa6G00995, Asa8G06181, Asa7G01379, Asa2G04763, Asa8G04833, Asa7G02467, Asa7G01972, and Asa4G03115, were induced by GA3 at different time points. Furthermore, 9 genes (Asa8G02357, Asa8G06181, Asa2G04763, Asa2G06963, Asa7G02467, Asa7G01972, Asa4G03115, Asa8G05408, and Asa5G05838) were upregulated in leaves after IAA treatment. Notably, Asa7G01972 was strongly induced by IAA treatment, with its expression peaking at 24 h. In roots treated with IAA, only Asa2G04763 was upregulated, with its expression peaking at 12 h. MeJA stress led to the upregulation of Asa8G02357, Asa7G01379, Asa2G06963, and Asa5G05838 in leaves, with Asa8G02357 peaking at 48 h and the other genes peaking at 24 h. After MeJA treatment, the expression of six genes (Asa8G02357, Asa8G06181, Asa2G04763, Asa8G04833, Asa7G01972, and Asa5G05838) in roots increased, and the expression of Asa2G04763 was notably increased at 6 h. After SA treatment, the expression of Asa8G02357, Asa7G01379, Asa2G04763, and Asa5G05838 was upregulated in leaves, and only Asa2G04763 presented increased expression and peaked at 6 h in roots. In summary, these results provide useful information for further functional investigations of AsbZIPs.
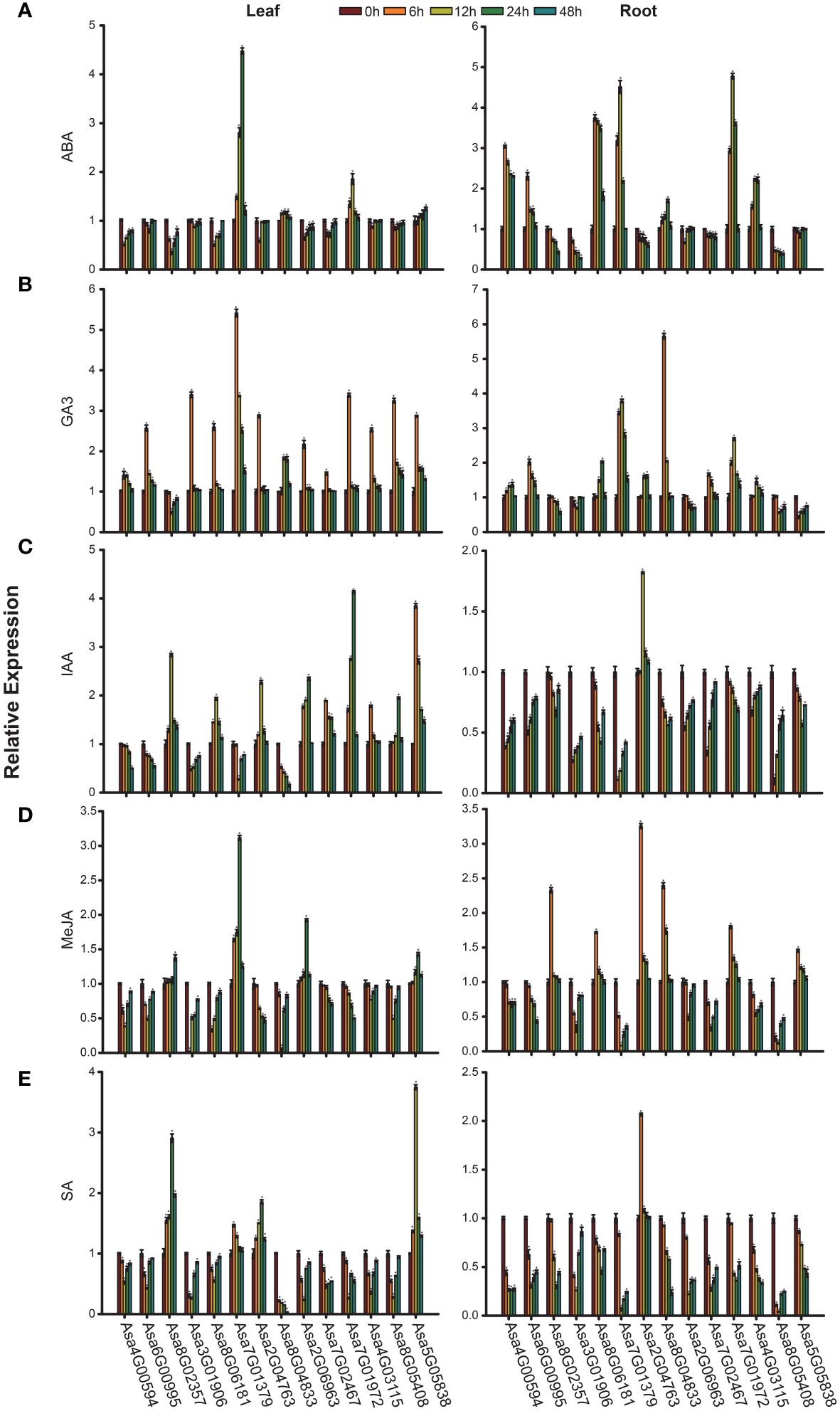
Figure 9 Expression patterns of AsbZIP genes under hormone treatment were examined by qRT-PCR. (A) Expression profiles of AsbZIP genes after exogenous ABA treatment. (B) Expression profiles of AsbZIP genes after exogenous GA3 treatment. (C) Expression profiles of AsbZIP genes after exogenous IAA treatment. (D) Expression profiles of AsbZIP genes after exogenous MeJA treatment. (E) Expression profiles of AsbZIP genes after exogenous SA treatment. The error bars represent standard deviations from biological replicates. A significance level of P < 0.05, as determined by Student’s t-test, is indicated by an asterisk (*).
Gene regulatory network analysis
Gene regulatory network analysis serves as an effective method for annotating gene function. The regulatory network of AsbZIP genes was constructed via WGCNA on the basis of 185 RNA-Seq samples (Supplementary Table 7) and the binding motifs of bZIP genes in Arabidopsis thaliana, resulting in 31 gene regulatory networks (Supplementary Table 8). The largest network was centered on Asa7G01972, which included 201 genes, whereas the smallest network, was centered on Asa0G04894, which included 8 genes.
To delve into the potential biological processes in which these genes might be involved, we conducted GO enrichment analysis. GO terms related to stress responses, such as the cellular response to abiotic stimuli, the cellular response to environmental stimuli and heat acclimation, were present in the majority of the AsbZIP genes, indicating their vital roles in the stress response (Figure 10). In addition to Asa3G01906, the gene regulatory networks of AsbZIP genes were enriched in the regulation of development and growth, indicating crucial roles of these genes in plant growth and development. Additionally, some GO terms associated with the hormone response, including abscisic acid-activated signaling pathway, gibberellin mediated signaling pathway and response to jasmonic acid, were enriched in the gene regulatory network of several AsbZIP genes (Asa0G02642, Asa1G00989, Asa3G00496, Asa3G01569, Asa4G1089, Asa4G02254, Asa4G02804, Asa4G04579, Asa5G03259, Asa6G00995, Asa6G01126, Asa6G01159, Asa6G03748, Asa7G01972, Asa7G02467, Asa7G05774, Asa8G00299 and Asa3G01906), indicating that these genes might play critical roles in hormone response regulation.
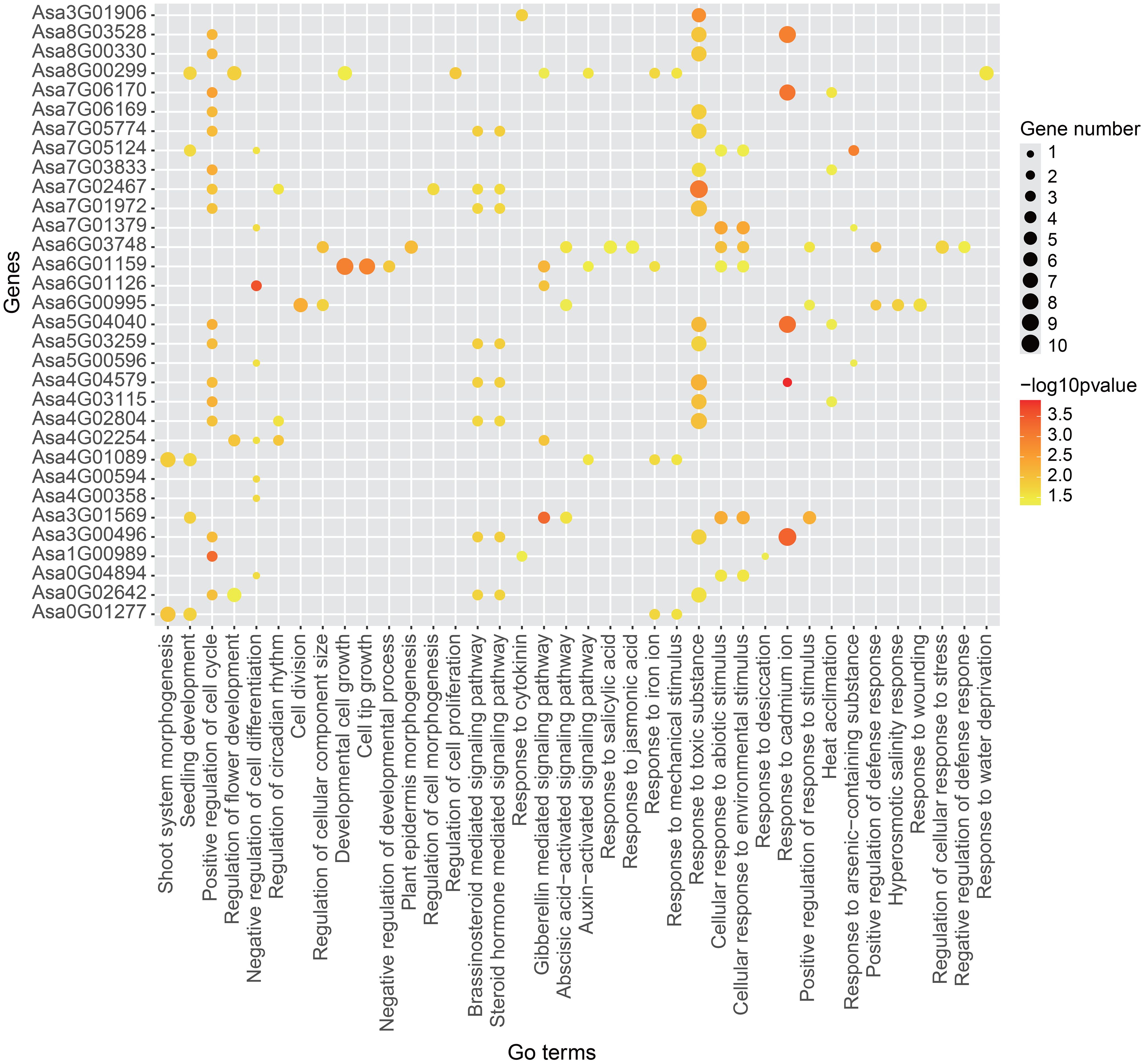
Figure 10 GO enrichment analysis of 31 gene sets of gene regulatory network. The size of each spot represents the number of genes enriched in specific term. The color of each spot indicates the enrichment significance level.
AsbZIP protein-protein interaction network prediction
Analyzing the functional associations among AsbZIP proteins is crucial for understanding the regulatory pathways within this protein family. To further elucidate the functions of the AsbZIP proteins, an interaction network of the AsbZIP proteins was constructed (Supplementary Figure 4). Asa1G01577 (homologue of ABI5), Asa4G04645 (homologue of ABF3), Asa5G05838 (homologue of ABF4), Asa4G00594 (homologue of ABF2), Asa7G06169 (homologue of ABF1), Asa8G03528 (homologue of DPBF4), Asa7G06185 (homologue of DPBF3) and Asa4G03115 (homologue of DPBF2) are associated with an abscisic acid-activated signaling pathway. Asa6G03748 (GBF4), Asa5G00596 (BZIP68), Asa7G03833 (GBF1), Asa2G05361 (BZIP16) and AT2G46270 (GBF3) are transcription factors regulated by diverse stimuli, such as light-induction or hormone control. Furthermore, Asa7G01379 (HY5) and Asa4G01089 (HYH) are transcription factors that promote photomorphogenesis in light.
Functional analysis of Asa7G01379 and Asa7G01972
Owing to their highly significant alterations in expression under salt stress and ABA treatment, Asa7G01379 and Asa7G01972 were selected for investigation of their biological functions via heterogeneous expression in yeast (Figure 11A). Compared with the yeast carrying the empty pYES2 vector, the growth of the yeast (BY4741) strains containing pYES2-Asa7G01379 and pYES2-Asa7G01972 was not significantly different under the control conditions but was greater under the salt treatment. These results suggest the important roles of Asa7G01379 and Asa7G01972 in salt stress.
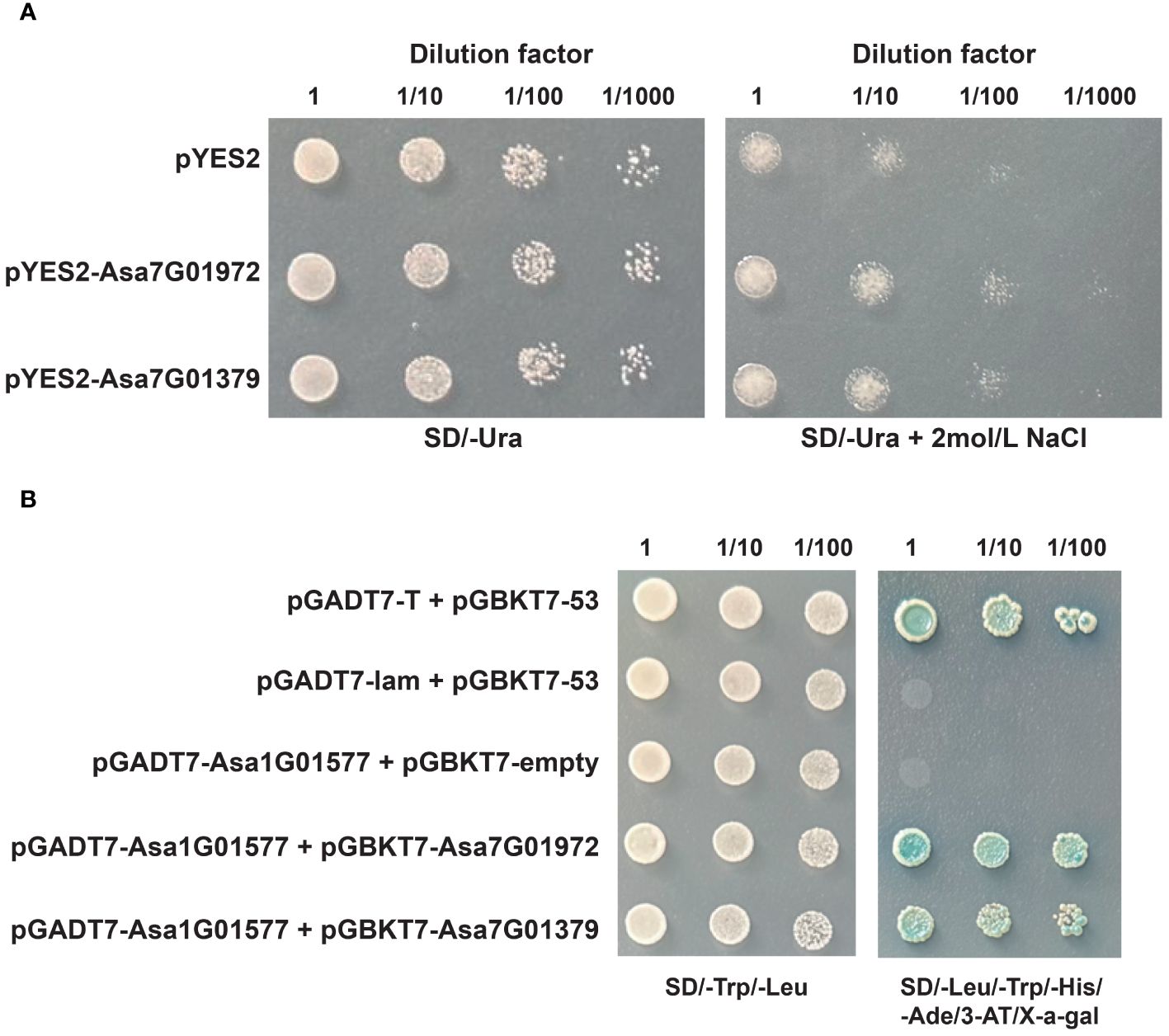
Figure 11 The functional analysis of Asa7G01972 and Asa7G01379. (A) The growth conditions of recombinant yeast BY4741 (pYES2-Asa7G01972/pYES2-Asa7G01979) and control yeast BY4741 (pYES2) were assessed under 2 mol/L NaCl treatment for 24 h Photographs of the plates were taken after 72 h of incubation at 30°. (B) Yeast two-hybrid analysis. 53-pGBKT7/T-pGADT7 was used as the positive control, and 53-pGBKT7/lam-pGADT7 was used as the negative control.
To confirm the potential protein-protein interaction profiles predicted by STRING, we performed Y2H assays. The results validated the protein interactions of Asa7G01379-Asa1G01577 and Asa7G01972-Asa1G01577 (Figure 11B), demonstrating the reliability of the predicted protein interactions. Owing to the vital role of Asa1G01577 (a homologue of ABI5) in the ABA signal transduction pathway, we presume that Asa7G01379 and Asa7G01972 might regulate the response to salt stress through the ABA pathway.
Discussion
Garlic, with its plump bulbs and high allicin content, is not only used as a vegetable and flavoring ingredient, but is also widely used in the pharmaceutical industry. The bZIP gene family is one of the most critical families in plants and plays pivotal roles in regulating plant growth, development, and response to both biotic and abiotic stresses (Rodriguez-Uribe and O’Connell, 2006; Prasad et al., 2012; Gangappa et al., 2013). Previous studies on bZIP transcription factors have focused mainly on Arabidopsis thaliana, crops and fruits (Jakoby et al., 2002; Nijhawan et al., 2008; Liao et al., 2008a; Liu et al., 2020; Kumar et al., 2021). However, a comprehensive study of AsbZIP genes at the genome-wide level in garlic has not been conducted thus far. Here, a comprehensive study of the bZIP gene family in garlic was performed, and 64 bZIP genes were identified in garlic (genome size: 16 GB). The number of bZIP genes in garlic was less than that in Arabidopsis thaliana (78, genome size: 117 Mb) (Jakoby et al., 2002), rice (89, genome size: 466 Mb) (Nijhawan et al., 2008), maize (125, genome size: 2182 Mb) (Bolle, 2004), soybean (160, genome size: 915 Mb) (Zhang et al., 2018), and Tartary buckwheat (96, genome size: 489 Mb) (Liu et al., 2019) but greater than that in grape (55, genome size: 490 Mb) (Liu et al., 2014b), indicating that there is no positive correlation between the number of bZIP genes and genome size. Phylogenetic analysis revealed that AsbZIPs can be divided into 10 subfamilies (Figure 1), which was consistent with the findings in Arabidopsis thaliana (Jakoby et al., 2002) and grape (Liu et al., 2014b), but lower than those in tartary buckwheat (11) (Liu et al., 2019), Chinese jujube (14) (Zhang et al., 2020b), poplar (12) (Zhao et al., 2021) and potato (11) (Kumar et al., 2021).
Furthermore, gene intron/exon structural analysis revealed variation in the number of introns within AsbZIP genes, ranging from 0 to 11 (Figure 2), which is similar to that of bZIP genes in other plants, such as rice (0-12) (Nijhawan et al., 2008), wheat (0-14) (Liang et al., 2022) and moso bamboo (0-17) (Pan et al., 2019). However, members within the same subfamily shared similar gene structures, supporting the results of the phylogenetic tree. The bZIP genes in subfamilies C/E/G have no more than three introns, supporting the hypothesis that genes with fewer introns may facilitate rapid environmental responsiveness (Shabalina et al., 2010). In addition, although there are variations in the motif components of distinct subfamilies, the motifs encoding the bZIP domain are highly conserved, and the majority of members of the same subfamily present similar motif components, which is coincident with the findings of previous studies (Zhou et al., 2017). For example, most AsbZIP proteins in subfamily S share motifs 1 and 6, with the exception of Asa5G04040, which only possesses motif 1. Several motifs (such as motifs 8, 9, and 10) found exclusively in specific subfamilies might contribute to the diverse biological functions of AsbZIP genes, which is consistent with previous reports (Wang et al., 2018; Liu et al., 2019).
The expansion of gene families through tandem or segmental duplication is crucial for functional diversity in evolution (Cannon et al., 2004). Furthermore, another study indicated that garlic has experienced three whole genome duplication events (Sun et al., 2020). Here, two tandem duplications and seven segmental duplications of AsbZIPs were identified in the garlic genome (Figures 3, 4), suggesting the vital role of gene replication in AsbZIP gene family expansion, which is consistent with previous reports (Nijhawan et al., 2008; Liu et al., 2014b, 2019). The number of paralogous pairs in garlic (9) was less than that in poplar (31) (Zhao et al., 2021), rice (34) (Nijhawan et al., 2008) and pear (68) (Ma et al., 2021). Additionally, the results of evolutionary analysis between garlic bZIP genes and their counterparts in two eudicots and two monocots revealed that there were more collinear gene pairs between garlic and Oryza sativa (Figure 5), indicating relatively close evolutionary relationships between these species.
The exposure of wild species to novel selection environments driven by human demands gradually induces different morphological and physiological alterations, eventually leading to their divergence from wild ancestors. This process represents domestication, a form of coevolution between plants and animals (Purugganan and Fuller, 2009). Cultivated garlic, which originates from the wild Allium longicuspis and Allium tuncelianum, has undergone genetic alterations leading to divergence in plant architecture and growth habits, referred to as the domestication syndrome (Li et al., 2022). Nevertheless, the alterations in AsbZIPs resulting from garlic domestication are poorly understood. Here, we identified 2941 AsbZIP-associated SNPs and observed their uneven distribution across the genome sequence (Supplementary Table 6), including a total of 2783 intergenic and 158 intragenic variations, which was consistent with the report presenting lower polymorphism of SNPs in intragenic regions than in intergenic regions (Hao and He, 2024). PCA, admixture and phylogenetic analysis successfully segregated all the accessions into two groups: wild garlic and local garlic (Figures 6A, B), which was consistent with the phylogenetic classification between wild and domesticated garlic of AsHSFs (Hao and He, 2024). In addition, domestication of garlic led to a bottleneck that decreased nucleotide diversity in alleles. The nucleotide diversity of AsbZIPs in wild garlic was slightly higher than that of domesticated garlic, with a decrease of only 0.00003%, which is lower than the average reduction of nucleotide diversity from wild garlic to landraces (Li et al., 2022), indicating a minor genetic bottleneck in AsbZIP genes during domestication. This result was confirmed by the Fst result, implying that AsbZIP genes did not experience strong selection pressure during domestication.
bZIP genes are related to plant development, and analyzing tissue-specific gene expression profiles facilitates to further comprehension of the biological functions of these genes. In Arabidopsis thaliana, HY5 (AtbZIP56) interacts with BBX25 to inhibit seedling photomorphogenesis through regulating the gene expression of BBX22 (Gangappa et al., 2013). In Oryza sativa, overexpression of OsbZIP49 resulted in the reduction of internode length and plant height, showcasing a tiller-spreading phenotype (Gangappa and Botto, 2016; Ma et al., 2018; Lorenzo, 2019; Ding et al., 2021; Han et al., 2023; Liu et al., 2023). Moreover, suppressed plant growth and decreased number of petals were observed in the Arabidopsis thaliana overexpressing the Capsicum annuum CabZIP1 gene (Lee et al., 2006). Here, we systematically investigated the expression profiles of AsbZIPs in several tissues of garlic. As shown in Supplementary Figure 3A, Asa2G04763, Asa7G02467, Asa8G05408 and Asa3G01906 were highly expressed in floral buds, and Asa6G00995, Asa8G02357, Asa4G03115 and Asa2G06963 presented relatively high expression levels in roots, whereas relatively high expression levels in leaves were observed for Asa8G06181 and Asa7G01379, suggesting potential specific functions of these genes in particular organs during the growth and development of garlic, which was comparable with tissue-specific expression profiles of bZIP genes in other species including Malus halliana (Wang et al., 2021b), Musa nana (Hu et al., 2016a) and Citrullus lanatus (Yang et al., 2019).
In addition, numerous studies have revealed that bZIP TFs are involved in responding to stresses. In wheat, TabZIP96 played a crucial role in the regulation of cold stress (Liang et al., 2022). In potato, the gene expression of StbZIP25 was induced under salt stress and could improve salt tolerance (Wang et al., 2021a). In pepper, CabZIP25-overexpression resulted in increased fresh weight and root length under salt stress (Gai et al., 2020). In Oryza sativa, OsbZIP71 could regulate the expression of OsNHX1 and COR413-TM1 to improve the drought and salinity tolerance (Liu et al., 2014a). In Vitis vinifera, the expression of VvbZIP23 was upregulated by several abiotic stresses, such as cold and drought stresses (Tak and Mhatre, 2013). Our results revealed that most AsbZIP genes were specifically induced or repressed under multiple types of stress (Figures 8A, D). In particular, Asa4G00594 is upregulated under salt and drought stresses, and its homologous gene AtABF2 is known to contribute to the tolerance to salinity and drought stresses (Fujita et al., 2005; Du et al., 2023). Moreover, Asa7G01379, encoding the AsHY5 protein, was upregulated significantly under salt stress, which is coincident with the findings of a previous report (Yang et al., 2023). The most of AsbZIP genes were induced under heat stress, except for Asa8G04833 and Asa2G06963. Under cold stress, the expression levels of Asa7G01379 and Asa2G06963 increased and decreased, respectively, which is consistent with the key roles of their homologues in cold acclimation in Arabidopsis thaliana (Oono et al., 2006; Perea-Resa et al., 2017). The expression profiles of AsbZIP genes under abiotic stresses provide potential avenues for garlic breeding to increase their ability to tolerance stresses.
An increasing number of studies have revealed that bZIP genes are involved in phytohormone signaling. In rice, OsbZIP72 and OsbZIP23 were identified as regulators of ABA response to affect drought tolerance (Xiang et al., 2008; Lu et al., 2009). In Arabidopsis thaliana, AtbZIP39, AtbZIP36, AtbZIP38, AtbZIP35 and AtbZIP37 have been found to play vital roles in the ABA signaling pathway (Choi et al., 2000; Uno et al., 2000; Lopez-Molina et al., 2001; Hossain et al., 2010; Liang et al., 2022). In Artemisia annua, the bZIP gene AaTGA6 was demonstrated as a key regulator in the SA signaling pathway (Lv et al., 2019), and AabZIP1 played a vital role in the regulation of ABA signaling (Zhang et al., 2022a). Here, the identification of cis-acting elements revealed the potential roles of AsbZIPs in phytohormone signal transduction (Figures 7A, B; Supplementary Figure 2). As shown in Figures 9A-E, the majority of AsbZIP genes showed significant alteration under several hormone stresses. For example, after ABA treatment, Asa4G00594 and Asa5G05838 were induced in roots and leaves, respectively, which is consistent with previous investigations (Choi et al., 2000; Kim et al., 2004). Particularly, Asa7G01379 and Asa7G01972 were the most significantly induced AsbZIP genes under salt stress, implying crucial functions of these genes for salt stress response. Furthermore, GO enrichment terms of gene regulatory network analysis showed that Asa7G01379 and Asa7G01972 might be involved in the stress response (Figure 10), which was confirmed by yeast-induced expression assay (Figure 11A). Besides, the Y2H experiment demonstrated the interactions of Asa7G01379-Asa1G01577 and Asa7G01972-Asa1G01577, as predicted by STRING (Figures 11B; Supplementary Figure 4), which is useful for revealing the biological functions of these AsbZIP proteins involved in the stress response through ABA signaling. According to the above results, Asa7G01379 and Asa7G01972 might be potential target locus for breeding stress-resistant garlic.
Conclusions
Here, we systematically investigated the genome-wide AsbZIP TFs in garlic. We identified 64 AsbZIP genes and conducted a comprehensive analysis of their physical characteristics, evolutionary associations, gene structures, conserved motifs, gene duplication events, nucleotide variation, population structure, expression patterns, and responses to abiotic stress and hormone treatment. On the basis of the above analysis, we speculate that AsbZIP genes play vital roles in the development and response of garlic to stress. Furthermore, our study revealed that 2 ABA-responsive genes, Asa7G01972 and Asa7G01379, are closely related to the response to salt stress in garlic. This study could inform subsequent functional analysis of AsbZIP genes, contributing to a more in-depth understanding of the molecular mechanisms underlying developmental processes and stress responses in garlic.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
SH: Conceptualization, Data curation, Funding acquisition, Investigation, Writing – original draft, Writing – review & editing. SX: Data curation, Investigation, Writing – review & editing. ZH: Data curation, Investigation, Writing – review & editing. XH: Conceptualization, Data curation, Funding acquisition, Investigation, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by Natural Science Foundation of Shandong Province (Grant No. ZR2023QC312), research initiation project of Jining Medical University (Grant No. 600993001) and research initiation project of Beijing Academy of Science and Technology (Grant No. 0420239352KF001-05).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1391248/full#supplementary-material
References
Alonso, R., Onate-Sanchez, L., Weltmeier, F., Ehlert, A., Diaz, I., Dietrich, K., et al. (2009). A pivotal role of the basic leucine zipper transcription factor bZIP53 in the regulation of Arabidopsis seed maturation gene expression based on heterodimerization and protein complex formation. Plant Cell 21, 1747–1761. doi: 10.1105/tpc.108.062968
Babitha, K. C., Ramu, S. V., Nataraja, K. N., Sheshshayee, M. S., Udayakumar, M. (2015). EcbZIP60, a basic leucine zipper transcription factor from Eleusine coracana L. improves abiotic stress tolerance in tobacco by activating unfolded protein response pathway. Mol. Breed. 35, 1–17. doi: 10.1007/s11032-015-0374-6
Baena-Gonzalez, E., Rolland, F., Thevelein, J. M., Sheen, J. (2007). A central integrator of transcription networks in plant stress and energy signalling. Nature 448, 938–942. doi: 10.1038/nature06069
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Baloglu, M. C., Eldem, V., Hajyzadeh, M., Unver, T. (2014). Genome-wide analysis of the bZIP transcription factors in cucumber. PloS One 9, e96014. doi: 10.1371/journal.pone.0096014
Bastias, A., Yanez, M., Osorio, S., Arbona, V., Gomez-Cadenas, A., Fernie, A. R., et al. (2014). The transcription factor AREB1 regulates primary metabolic pathways in tomato fruits. J. Exp. Bot. 65, 2351–2363. doi: 10.1093/jxb/eru114
Bolle, C. (2004). The role of GRAS proteins in plant signal transduction and development. Planta 218, 683–692. doi: 10.1007/s00425-004-1203-z
Bombarely, A., Rosli, H. G., Vrebalov, J., Moffett, P., Mueller, L. A., Martin, G. B. (2012). A draft genome sequence of Nicotiana benthamiana to enhance molecular plant-microbe biology researchMol. Plant Microbe Interact 25, 1523–1530. doi: 10.1094/MPMI-06-12-0148-TA
Cai, W., Yang, Y., Wang, W., Guo, G., Liu, W., Bi, C. (2018). Overexpression of a wheat (Triticum aestivum L.) bZIP transcription factor gene, TabZIP6, decreased the freezing tolerance of transgenic Arabidopsis seedlings by down-regulating the expression of CBFs. Plant Physiol. Biochem. 124, 100–111. doi: 10.1016/j.plaphy.2018.01.008
Cannon, S. B., Mitra, A., Baumgarten, A., Young, N. D., May, G. (2004). The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 4, 10. doi: 10.1186/1471-2229-4-10
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Choi, H., Hong, J., Ha, J., Kang, J., Kim, S. Y. (2000). ABFs, a family of ABA-responsive element binding factors. J. Biol. Chem. 275, 1723–1730. doi: 10.1074/jbc.275.3.1723
Chow, C. N., Lee, T. Y., Hung, Y. C., Li, G. Z., Tseng, K. C., Liu, Y. H., et al. (2019). PlantPAN3.0: a new and updated resource for reconstructing transcriptional regulatory networks from ChIP-seq experiments in plants. Nucleic Acids Res. 47, D1155–D1163. doi: 10.1093/nar/gky1081
Cingolani, P., Platts, A., Wang le, L., Coon, M., Nguyen, T., Wang, L., et al. (2012). A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6, 80–92. doi: 10.4161/fly.19695
Correa, L. G., Riano-Pachon, D. M., Schrago, C. G., dos Santos, R. V., Mueller-Roeber, B., Vincentz, M. (2008). The role of bZIP transcription factors in green plant evolution: adaptive features emerging from four founder genes. PloS One 3, e2944. doi: 10.1371/journal.pone.0002944
Ding, C., Lin, X., Zuo, Y., Yu, Z., Baerson, S. R., Pan, Z., et al. (2021). Transcription factor OsbZIP49 controls tiller angle and plant architecture through the induction of indole-3-acetic acid-amido synthetases in rice. Plant J. 108, 1346–1364. doi: 10.1111/tpj.15515
Du, J., Zhu, X., He, K., Kui, M., Zhang, J., Han, X., et al. (2023). CONSTANS interacts with and antagonizes ABF transcription factors during salt stress under long-day conditions. Plant Physiol. 193, 1675–1694. doi: 10.1093/plphys/kiad370
Edwards, K. D., Fernandez-Pozo, N., Drake-Stowe, K., Humphry, M., Evans, A. D., Bombarely, A., et al. (2017). A reference genome for Nicotiana tabacum enables map-based cloning of homeologous loci implicated in nitrogen utilization efficiency. BMC Genomics 18, 448. doi: 10.1186/s12864-017-3791-6
El-Gebali, S., Mistry, J., Bateman, A., Eddy, S. R., Luciani, A., Potter, S. C., et al. (2019). The Pfam protein families database in 2019. Nucleic Acids Res. 47, D427–D432. doi: 10.1093/nar/gky995
ETOH, T., WATANABE, H., IWAI, S. (2001). RAPD variation of garlic clones in the center of origin and the westernmost area of distribution. Memoirs of the Faculty of Agriculture Kagoshima University. 37, 21–27. Available at: http://hdl.handle.net/10232/2819.
Fan, L. X., Xu, L., Wang, Y., Tang, M. J., Liu, L. W. (2019). Genome- and Transcriptome-Wide Characterization of bZIP Gene Family Identifies Potential Members Involved in Abiotic Stress Response and Anthocyanin Biosynthesis in Radish (Raphanus sativus L.). Int. J. Mol. Sci. 20, 6334. doi: 10.3390/ijms20246334
Fujita, Y., Fujita, M., Satoh, R., Maruyama, K., Parvez, M. M., Seki, M., et al. (2005). AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell 17, 3470–3488. doi: 10.1105/tpc.105.035659
Gai, W., Ma, X., Qiao, Y., Shi, B., Ul Haq, S., Li, Q., et al. (2020). Characterization of the bZIP transcription factor family in pepper (L.): positively modulates the salt tolerance. Front. Plant Sci. 11, 139. doi: 10.3389/fpls.2020.00139
Gangappa, S. N., Botto, J. F. (2016). The multifaceted roles of HY5 in plant growth and development. Mol. Plant 9, 1353–1365. doi: 10.1016/j.molp.2016.07.002
Gangappa, S. N., Crocco, C. D., Johansson, H., Datta, S., Hettiarachchi, C., Holm, M., et al. (2013). The Arabidopsis B-BOX protein BBX25 interacts with HY5, negatively regulating BBX22 expression to suppress seedling photomorphogenesis. Plant Cell 25, 1243–1257. doi: 10.1105/tpc.113.109751
Grant, C. E., Bailey, T. L., Noble, W. S. (2011). FIMO: scanning for occurrences of a given motif. Bioinformatics 27, 1017–1018. doi: 10.1093/bioinformatics/btr064
Han, H., Wang, C., Yang, X., Wang, L., Ye, J., Xu, F., et al. (2023). Role of bZIP transcription factors in the regulation of plant secondary metabolism. Planta 258, 13. doi: 10.1007/s00425-023-04174-4
Hao, X., He, S. (2024). Genome-wide identification, classification and expression analysis of the heat shock transcription factor family in Garlic (Allium sativum L.). BMC Plant Biol. 24, 421. doi: 10.1186/s12870-024-05018-3
Hossain, M. A., Cho, J. I., Han, M., Ahn, C. H., Jeon, J. S., An, G., et al. (2010). The ABRE-binding bZIP transcription factor OsABF2 is a positive regulator of abiotic stress and ABA signaling in rice. J. Plant Physiol. 167, 1512–1520. doi: 10.1016/j.jplph.2010.05.008
Hu, B., Jin, J., Guo, A. Y., Zhang, H., Luo, J., Gao, G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Hu, W., Wang, L., Tie, W., Yan, Y., Ding, Z., Liu, J., et al. (2016a). Genome-wide analyses of the bZIP family reveal their involvement in the development, ripening and abiotic stress response in banana. Sci. Rep. 6, 30203. doi: 10.1038/srep30203
Hu, W., Yang, H., Yan, Y., Wei, Y., Tie, W., Ding, Z., et al. (2016b). Genome-wide characterization and analysis of bZIP transcription factor gene family related to abiotic stress in cassava. Sci. Rep. 6, 22783. doi: 10.1038/srep22783
Ibrahim, H. R., Matsuzaki, T., Aoki, T. (2001). Genetic evidence that antibacterial activity of lysozyme is independent of its catalytic function. FEBS Lett. 506, 27–32. doi: 10.1016/S0014-5793(01)02872-1
Jakoby, M., Weisshaar, B., Droge-Laser, W., Vicente-Carbajosa, J., Tiedemann, J., Kroj, T., et al. (2002). bZIP transcription factors in Arabidopsis. Trends Plant Sci. 7, 106–111. doi: 10.1016/S1360-1385(01)02223-3
Joo, J., Lee, Y. H., Song, S. I. (2014). Overexpression of the rice basic leucine zipper transcription factor OsbZIP12 confers drought tolerance to rice and makes seedlings hypersensitive to ABA. Plant Biotechnol. Rep. 8, 431–441. doi: 10.1007/s11816-014-0335-2
Kamenetsky, R., Faigenboim, A., Shemesh Mayer, E., Ben Michael, T., Gershberg, C., Kimhi, S., et al. (2015). Integrated transcriptome catalogue and organ-specific profiling of gene expression in fertile garlic (Allium sativum L.). BMC Genomics 16, 12. doi: 10.1186/s12864-015-1212-2
Kang, C., Zhai, H., He, S. Z., Zhao, N., Liu, Q. C. (2019). A novel sweetpotato bZIP transcription factor gene, IbbZIP1, is involved in salt and drought tolerance in transgenic Arabidopsis. Plant Cell Rep. 38, 1373–1382. doi: 10.1007/s00299-019-02441-x
Kim, S., Kang, J. Y., Cho, D. I., Park, J. H., Kim, S. Y. (2004). ABF2, an ABRE-binding bZIP factor, is an essential component of glucose signaling and its overexpression affects multiple stress tolerance. Plant J. 40, 75–87. doi: 10.1111/j.1365-313X.2004.02192.x
Kumar, P., Kumar, P., Sharma, D., Verma, S. K., Halterman, D., Kumar, A. (2021). Genome-wide identification and expression profiling of basic leucine zipper transcription factors following abiotic stresses in potato (Solanum tuberosum L.). PloS One 16, e0247864. doi: 10.1371/journal.pone.0247864
Kumar, S., Stecher, G., Li, M., Knyaz, C., Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Lee, S. C., Choi, H. W., Hwang, I. S., Choi, D. S., Hwang, B. K. (2006). Functional roles of the pepper pathogen-induced bZIP transcription factor, CAbZIP1, in enhanced resistance to pathogen infection and environmental stresses. Planta 224, 1209–1225. doi: 10.1007/s00425-006-0302-4
Li, J., Besseau, S., Toronen, P., Sipari, N., Kollist, H., Holm, L., et al. (2013). Defense-related transcription factors WRKY70 and WRKY54 modulate osmotic stress tolerance by regulating stomatal aperture in Arabidopsis. New Phytol. 200, 457–472. doi: 10.1111/nph.12378
Li, X., Duan, X., Jiang, H., Sun, Y., Tang, Y., Yuan, Z., et al. (2006). Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis. Plant Physiol. 141, 1167–1184. doi: 10.1104/pp.106.080580
Li, N., Zhang, X., Sun, X., Zhu, S., Cheng, Y., Liu, M., et al. (2022). Genomic insights into the evolutionary history and diversification of bulb traits in garlic. Genome Biol. 23, 188. doi: 10.1186/s13059-022-02756-1
Liang, Y., Xia, J., Jiang, Y., Bao, Y., Chen, H., Wang, D., et al. (2022). Genome-Wide Identification and Analysis of bZIP Gene Family and Resistance of TaABI5 (TabZIP96) under Freezing Stress in Wheat (Triticum aestivum). Int. J. Mol. Sci. 23, 2351. doi: 10.3390/ijms23042351
Liao, Y., Zou, H. F., Wang, H. W., Zhang, W. K., Ma, B., Zhang, J. S., et al. (2008a). Soybean GmMYB76, GmMYB92, and GmMYB177 genes confer stress tolerance in transgenic Arabidopsis plants. Cell Res. 18, 1047–1060. doi: 10.1038/cr.2008.280
Liao, Y., Zou, H. F., Wei, W., Hao, Y. J., Tian, A. G., Huang, J., et al. (2008b). Soybean GmbZIP44, GmbZIP62 and GmbZIP78 genes function as negative regulator of ABA signaling and confer salt and freezing tolerance in transgenic Arabidopsis. Planta 228, 225–240. doi: 10.1007/s00425-008-0731-3
Liu, Y., Chai, M., Zhang, M., He, Q., Su, Z., Priyadarshani, S., et al. (2020). Genome-wide analysis, characterization, and expression profile of the basic leucine zipper transcription factor family in pineapple. Int. J. Genomics 2020, 3165958. doi: 10.1155/2020/3165958
Liu, J., Chen, N., Chen, F., Cai, B., Dal Santo, S., Tornielli, G. B., et al. (2014b). Genome-wide analysis and expression profile of the bZIP transcription factor gene family in grapevine (Vitis vinifera). BMC Genomics 15, 281. doi: 10.1186/1471-2164-15-281
Liu, X., Chu, Z. Q. (2015). Genome-wide evolutionary characterization and analysis of bZIP transcription factors and their expression profiles in response to multiple abiotic stresses in Brachypodium distachyon. BMC Genomics 16, 227. doi: 10.1186/s12864-015-1457-9
Liu, C., Mao, B., Ou, S., Wang, W., Liu, L., Wu, Y., et al. (2014a). OsbZIP71, a bZIP transcription factor, confers salinity and drought tolerance in rice. Plant Mol. Biol. 84, 19–36. doi: 10.1007/s11103-013-0115-3
Liu, J. X., Srivastava, R., Howell, S. H. (2008). Stress-induced expression of an activated form of AtbZIP17 provides protection from salt stress in Arabidopsis. Plant Cell Environ. 31, 1735–1743. doi: 10.1111/j.1365-3040.2008.01873.x
Liu, H., Tang, X., Zhang, N., Li, S., Si, H. (2023). Role of bZIP transcription factors in plant salt stress. Int. J. Mol. Sci. 24, 7893. doi: 10.3390/ijms24097893
Liu, M., Wen, Y., Sun, W., Ma, Z., Huang, L., Wu, Q., et al. (2019). Genome-wide identification, phylogeny, evolutionary expansion and expression analyses of bZIP transcription factor family in tartaty buckwheat. BMC Genomics 20, 483. doi: 10.1186/s12864-019-5882-z
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lopez-Molina, L., Mongrand, S., Chua, N. H. (2001). A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 98, 4782–4787. doi: 10.1073/pnas.081594298
Lorenzo, O. (2019). bZIP edgetic mutations: at the frontier of plant metabolism, development and stress trade-off. J. Exp. Bot. 70, 5517–5520. doi: 10.1093/jxb/erz298
Lozano-Sotomayor, P., Chavez Montes, R. A., Silvestre-Vano, M., Herrera-Ubaldo, H., Greco, R., Pablo-Villa, J., et al. (2016). Altered expression of the bZIP transcription factor DRINK ME affects growth and reproductive development in Arabidopsis thaliana. Plant J. 88, 437–451. doi: 10.1111/tpj.13264
Lu, G., Gao, C. X., Zheng, X. N., Han, B. (2009). Identification of OsbZIP72 as a positive regulator of ABA response and drought tolerance in rice. Planta 229, 605–615. doi: 10.1007/s00425-008-0857-3
Lv, Z., Guo, Z., Zhang, L., Zhang, F., Jiang, W., Shen, Q., et al. (2019). Interaction of bZIP transcription factor TGA6 with salicylic acid signaling modulates artemisinin biosynthesis in Artemisia annua. J. Exp. Bot. 70, 3969–3979. doi: 10.1093/jxb/erz166
Ma, M., Chen, Q., Dong, H., Zhang, S., Huang, X. (2021). Genome-wide identification and expression analysis of the bZIP transcription factors, and functional analysis in response to drought and cold stresses in pear (Pyrus breschneideri). BMC Plant Biol. 21, 583. doi: 10.1186/s12870-021-03356-0
Ma, H., Liu, C., Li, Z., Ran, Q., Xie, G., Wang, B., et al. (2018). ZmbZIP4 contributes to stress resistance in maize by regulating ABA synthesis and root development. Plant Physiol. 178, 753–770. doi: 10.1104/pp.18.00436
Martin, K. W., Ernst, E. (2003). Herbal medicines for treatment of bacterial infections: a review of controlled clinical trials. J. Antimicrob. Chemother. 51, 241–246. doi: 10.1093/jac/dkg087
Munim Twaij, B., Jameel Ibraheem, L., Al-Shammari, R. H. H., Hasan, M., Akter Khoko, R., Sunzid Ahomed, M., et al. (2023). Identification and characterization of aldehyde dehydrogenase (ALDH) gene superfamily in garlic and expression profiling in response to drought, salinity, and ABA. Gene 860, 147215. doi: 10.1016/j.gene.2023.147215
Nei, M., Gojobori, T. (1986). Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3, 418–426. doi: 10.1093/oxfordjournals.molbev.a040410
Nijhawan, A., Jain, M., Tyagi, A. K., Khurana, J. P. (2008). Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice. Plant Physiol. 146, 333–350. doi: 10.1104/pp.107.112821
Oono, Y., Seki, M., Satou, M., Iida, K., Akiyama, K., Sakurai, T., et al. (2006). Monitoring expression profiles of Arabidopsis genes during cold acclimation and deacclimation using DNA microarrays. Funct. Integr. Genomics 6, 212–234. doi: 10.1007/s10142-005-0014-z
Orellana, S., Yanez, M., Espinoza, A., Verdugo, I., Gonzalez, E., Ruiz-Lara, S., et al. (2010). The transcription factor SlAREB1 confers drought, salt stress tolerance and regulates biotic and abiotic stress-related genes in tomato. Plant Cell Environ. 33, 2191–2208. doi: 10.1111/j.1365-3040.2010.02220.x
Oyama, T., Shimura, Y., Okada, K. (1997). The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev. 11, 2983–2995. doi: 10.1101/gad.11.22.2983
Pan, F., Wu, M., Hu, W., Liu, R., Yan, H., Xiang, Y. (2019). Genome-Wide Identification and Expression Analyses of the bZIP Transcription Factor Genes in moso bamboo (Phyllostachys edulis). Int. J. Mol. Sci. 20, 2203. doi: 10.3390/ijms20092203
Perea-Resa, C., Rodriguez-Milla, M. A., Iniesto, E., Rubio, V., Salinas, J. (2017). Prefoldins negatively regulate cold acclimation in arabidopsis thaliana by promoting nuclear proteasome-mediated HY5 degradation. Mol. Plant 10, 791–804. doi: 10.1016/j.molp.2017.03.012
Pourabed, E., Ghane Golmohamadi, F., Soleymani Monfared, P., Razavi, S. M., Shobbar, Z. S. (2015). Basic leucine zipper family in barley: genome-wide characterization of members and expression analysis. Mol. Biotechnol. 57, 12–26. doi: 10.1007/s12033-014-9797-2
Prasad, V. B., Gupta, N., Nandi, A., Chattopadhyay, S. (2012). HY1 genetically interacts with GBF1 and regulates the activity of the Z-box containing promoters in light signaling pathways in Arabidopsis thaliana. Mech. Dev. 129, 298–307. doi: 10.1016/j.mod.2012.06.004
Purugganan, M. D., Fuller, D. Q. (2009). The nature of selection during plant domestication. Nature 457, 843–848. doi: 10.1038/nature07895
Que, F., Wang, G. L., Huang, Y., Xu, Z. S., Wang, F., Xiong, A. S. (2015). Genomic identification of group A bZIP transcription factors and their responses to abiotic stress in carrot. Genet. Mol. Res. 14, 13274–13288. doi: 10.4238/2015.October.26.24
Rodriguez-Uribe, L., O’Connell, M. A. (2006). A root-specific bZIP transcription factor is responsive to water deficit stress in tepary bean (Phaseolus acutifolius) and common bean (P-vulgaris). J. Exp. Bot. 57, 1391–1398. doi: 10.1093/jxb/erj118
Shabalina, S. A., Ogurtsov, A. Y., Spiridonov, A. N., Novichkov, P. S., Spiridonov, N. A., Koonin, E. V. (2010). Distinct patterns of expression and evolution of intronless and intron-containing mammalian genes. Mol. Biol. Evol. 27, 1745–1749. doi: 10.1093/molbev/msq086
Shi, Y. P., Ding, G. H., Shen, H. T., Li, Z. H., Li, H. B., Xiao, G. H. (2024). Genome-wide identification and expression profiles analysis of the authentic response regulator gene family in licorice. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1309802
Singh, K., Foley, R. C., Onate-Sanchez, L. (2002). Transcription factors in plant defense and stress responses. Curr. Opin. Plant Biol. 5, 430–436. doi: 10.1016/S1369-5266(02)00289-3
Stajich, J. E., Block, D., Boulez, K., Brenner, S. E., Chervitz, S. A., Dagdigian, C., et al. (2002). The Bioperl toolkit: Perl modules for the life sciences. Genome Res. 12, 1611–1618. doi: 10.1101/gr.361602
Suckow, M., Schwamborn, K., Kisters-Woike, B., von Wilcken-Bergmann, B., Muller-Hill, B. (1994). Replacement of invariant bZip residues within the basic region of the yeast transcriptional activator GCN4 can change its DNA binding specificity. Nucleic Acids Res. 22, 4395–4404. doi: 10.1093/nar/22.21.4395
Sun, X. D., Zhu, S. Y., Li, N. Y., Cheng, Y., Zhao, J., Qiao, X. G., et al. (2020). A Chromosome-Level Genome Assembly of Garlic (Allium sativum) Provides Insights into Genome Evolution and Allicin Biosynthesis. Mol. Plant 13, 1328–1339. doi: 10.1016/j.molp.2020.07.019
Tak, H., Mhatre, M. (2013). Cloning and molecular characterization of a putative bZIP transcription factor VvbZIP23 from Vitis vinifera. Protoplasma 250, 333–345. doi: 10.1007/s00709-012-0417-3
Tan, S. L., Yang, Y. J., Liu, T., Zhang, S. B., Huang, W. (2020). Responses of photosystem I compared with photosystem II to combination of heat stress and fluctuating light in tobacco leaves. Plant Sci. 292, 110371. doi: 10.1016/j.plantsci.2019.110371
Tang, N., Zhang, H., Li, X., Xiao, J., Xiong, L. (2012). Constitutive activation of transcription factor OsbZIP46 improves drought tolerance in rice. Plant Physiol. 158, 1755–1768. doi: 10.1104/pp.111.190389
Uno, Y., Furihata, T., Abe, H., Yoshida, R., Shinozaki, K., Yamaguchi-Shinozaki, K. (2000). Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc. Natl. Acad. Sci. U.S.A. 97, 11632–11637. doi: 10.1073/pnas.190309197
Wang, X. L., Chen, X., Yang, T. B., Cheng, Q., Cheng, Z. M. (2017). Genome-Wide Identification of bZIP Family Genes Involved in Drought and Heat Stresses in Strawberry (Fragaria vesca). Int. J. Genomics 2017, 3981031. doi: 10.1155/2017/3981031
Wang, Q., Guo, C., Li, Z. Y., Sun, J. H., Wang, D., Xu, L. T., et al. (2021a). Identification and analysis of bZIP family genes in potato and their potential roles in stress responses. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.637343
Wang, Y. P., Tang, H. B., DeBarry, J. D., Tan, X., Li, J. P., Wang, X. Y., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40, e49. doi: 10.1093/nar/gkr1293
Wang, S., Zhang, R., Zhang, Z., Zhao, T., Zhang, D., Sofkova, S., et al. (2021b). Genome-wide analysis of the bZIP gene lineage in apple and functional analysis of MhABF in Malus halliana. Planta 254, 78. doi: 10.1007/s00425-021-03724-y
Wang, D., Zhang, Y., Zhang, Z., Zhu, J., Yu, J. (2010). KaKs_Calculator 2.0: a toolkit incorporating gamma-series methods and sliding window strategies. Genomics Proteomics Bioinf. 8, 77–80. doi: 10.1016/S1672-0229(10)60008-3
Wang, Y., Zhang, Y., Zhou, R., Dossa, K., Yu, J., Li, D., et al. (2018). Identification and characterization of the bZIP transcription factor family and its expression in response to abiotic stresses in sesame. PloS One 13, e0200850. doi: 10.1371/journal.pone.0200850
Wang, J., Zhou, J., Zhang, B., Vanitha, J., Ramachandran, S., Jiang, S. Y. (2011). Genome-wide expansion and expression divergence of the basic leucine zipper transcription factors in higher plants with an emphasis on sorghum. J. Integr. Plant Biol. 53, 212–231. doi: 10.1111/j.1744-7909.2010.01017.x
Wei, K., Chen, J., Wang, Y., Chen, Y., Chen, S., Lin, Y., et al. (2012). Genome-wide analysis of bZIP-encoding genes in maize. DNA Res. 19, 463–476. doi: 10.1093/dnares/dss026
Wigge, P. A., Kim, M. C., Jaeger, K. E., Busch, W., Schmid, M., Lohmann, J. U., et al. (2005). Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309, 1056–1059. doi: 10.1126/science.1114358
Xiang, Y., Tang, N., Du, H., Ye, H., Xiong, L. (2008). Characterization of OsbZIP23 as a key player of the basic leucine zipper transcription factor family for conferring abscisic acid sensitivity and salinity and drought tolerance in rice. Plant Physiol. 148, 1938–1952. doi: 10.1104/pp.108.128199
Xu, J., Duan, X., Yang, J., Beeching, J. R., Zhang, P. (2013). Enhanced reactive oxygen species scavenging by overproduction of superoxide dismutase and catalase delays postharvest physiological deterioration of cassava storage roots. Plant Physiol. 161, 1517–1528. doi: 10.1104/pp.112.212803
Yang, Y., Li, J., Li, H., Yang, Y., Guang, Y., Zhou, Y. (2019). The bZIP gene family in watermelon: genome-wide identification and expression analysis under cold stress and root-knot nematode infection. PeerJ 7, e7878. doi: 10.7717/peerj.7878
Yang, O., Popova, O. V., Suthoff, U., Luking, I., Dietz, K. J., Golldack, D. (2009). The Arabidopsis basic leucine zipper transcription factor AtbZIP24 regulates complex transcriptional networks involved in abiotic stress resistance. Gene 436, 45–55. doi: 10.1016/j.gene.2009.02.010
Yang, J., Qu, X., Li, T., Gao, Y., Du, H., Zheng, L., et al. (2023). HY5-HDA9 orchestrates the transcription of HsfA2 to modulate salt stress response in Arabidopsis. J. Integr. Plant Biol. 65, 45–63. doi: 10.1111/jipb.13372
Yao, X., Lai, D., Zhou, M., Ruan, J., Ma, C., Wu, W., et al. (2023). Genome-wide identification, evolution and expression pattern analysis of the GATA gene family in Sorghum bicolor. Front. Plant Sci. 14, 1163357. doi: 10.3389/fpls.2023.1163357
Ying, S., Zhang, D. F., Fu, J., Shi, Y. S., Song, Y. C., Wang, T. Y., et al. (2012). Cloning and characterization of a maize bZIP transcription factor, ZmbZIP72, confers drought and salt tolerance in transgenic Arabidopsis. Planta 235, 253–266. doi: 10.1007/s00425-011-1496-7
Zhang, Y., Gao, W., Li, H., Wang, Y., Li, D., Xue, C., et al. (2020b). Genome-wide analysis of the bZIP gene family in Chinese jujube (Ziziphus jujuba Mill.). BMC Genomics 21, 483. doi: 10.1186/s12864-020-06890-7
Zhang, X., Lin, H. M., Hu, H., Hu, X., Hu, L. (2016). Gamma-aminobutyric acid mediates nicotine biosynthesis in tobacco under flooding stress. Plant Divers. 38, 53–58. doi: 10.1016/j.pld.2016.05.004
Zhang, M., Liu, Y., Cai, H., Guo, M., Chai, M., She, Z., et al. (2020a). The bZIP transcription factor gmbZIP15 negatively regulates salt- and drought-stress responses in soybean. Int. J. Mol. Sci. 21, 7778. doi: 10.3390/ijms21207778
Zhang, M., Liu, Y., Shi, H., Guo, M., Chai, M., He, Q., et al. (2018). Evolutionary and expression analyses of soybean basic Leucine zipper transcription factor family. BMC Genomics 19, 159. doi: 10.1186/s12864-018-4511-6
Zhang, X., Yang, X., He, Q., Wang, Y., Liang, G., Liu, T. (2022b). Genome-wide identification and characterization of the GRAS transcription factors in garlic (Allium sativum L.). Front. Plant Sci. 13, 890052. doi: 10.3389/fpls.2022.890052
Zhang, H., Zhu, J., Gong, Z., Zhu, J. K. (2022a). Abiotic stress responses in plants. Nat. Rev. Genet. 23, 104–119. doi: 10.1038/s41576-021-00413-0
Zhao, K., Chen, S., Yao, W. J., Cheng, Z. H., Zhou, B. R., Jiang, T. B. (2021). Genome-wide analysis and expression profile of the bZIP gene family in poplar. BMC Plant Biol. 21, 122. doi: 10.1186/s12870-021-02879-w
Zhao, J., Guo, R., Guo, C., Hou, H., Wang, X., Gao, H. (2016). Evolutionary and expression analyses of the apple basic leucine zipper transcription factor family. Front. Plant Sci. 7, 376. doi: 10.3389/fpls.2016.00376
Keywords: genome-wide, garlic, bZIP, abiotic stress, expression profiles
Citation: He S, Xu S, He Z and Hao X (2024) Genome-wide identification, characterization and expression analysis of the bZIP transcription factors in garlic (Allium sativum L.). Front. Plant Sci. 15:1391248. doi: 10.3389/fpls.2024.1391248
Received: 25 February 2024; Accepted: 15 July 2024;
Published: 01 August 2024.
Edited by:
Umesh K. Reddy, West Virginia State University, United StatesReviewed by:
Wang Huasen, Qingdao Agricultural University, ChinaZilhas Ahmed Jewel, Sejong University, Republic of Korea
Copyright © 2024 He, Xu, He and Hao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaomeng Hao, aGFveGlhb21lbmdAbWFpbC5qbm1jLmVkdS5jbg==
 Shutao He
Shutao He Sen Xu1
Sen Xu1 Xiaomeng Hao
Xiaomeng Hao