- 1Key Laboratory of Plant Molecular & Developmental Biology, College of Life Sciences, Yantai University, Yantai, Shandong, China
- 2State Key Laboratory of Crop Gene Resources and Breeding, Institute of Crop Sciences, Chinese Academy of Agricultural Sciences (CAAS), Beijing, China
- 3Nanfan Research Institute, Chinese Academy of Agricultural Sciences (CAAS), Sanya, Hainan, China
Introduction: The WD40 gene family, prevalent in eukaryotes, assumes diverse roles in cellular processes. Spartina alterniflora, a halophyte with exceptional salt tolerance, flood tolerance, reproduction, and diffusion ability, offers great potential for industrial applications and crop breeding analysis. The exploration of growth and development-related genes in this species offers immense potential for enhancing crop yield and environmental adaptability, particularly in industrialized plantations. However, the understanding of their role in regulating plant growth and development remains limited.
Methods: In this study, we conducted a comprehensive analysis of WD40 genes in S. alterniflora at the whole-genome level, delving into their characteristics such as physicochemical properties, phylogenetic relationships, gene architecture, and expression patterns. Additionally, we cloned the TTG1 gene, a gene in plant growth and development across diverse species.
Results: We identified a total of 582 WD40 proteins in the S. alterniflora genome, exhibiting an uneven distribution across chromosomes. Through phylogenetic analysis, we categorized the 582 SaWD40 proteins into 12 distinct clades. Examining the duplication patterns of SaWD40 genes, we observed a predominant role of segmental duplication in their expansion. A substantial proportion of SaWD40 gene duplication pairs underwent purifying selection through evolution. To explore the functional aspects, we selected SaTTG1, a homolog of Arabidopsis TTG1, for overexpression in Arabidopsis. Subcellular localization analysis revealed that the SaTTG1 protein localized in the nucleus and plasma membrane, exhibiting transcriptional activation in yeast cells. The overexpression of SaTTG1 in Arabidopsis resulted in early flowering and increased seed size.
Discussion: These outcomes significantly contribute to our understanding of WD40 gene functions in halophyte species. The findings not only serve as a valuable foundation for further investigations into WD40 genes in halophyte but also offer insights into the molecular mechanisms governing plant development, offering potential avenues in molecular breeding.
1 Introduction
The WD40 protein, alternatively known as the WD repeat protein, comprises a regulatory superfamily that is ubiquitously found in eukaryotes (Jain and Pandey, 2018). This protein features multiple WD repeat motifs, each consisting of 44–60 amino acid residues (Mishra et al., 2012). These WD repeat motifs are characterized by a glycine histidine dipeptide at the N-terminus and a tryptophan aspartate dipeptide at the C-terminus (Smith et al., 1999). Folding of WD repeat motifs creates a four-stranded antiparallel β-sheet, stabilizing the WD40 protein fold through a robust hydrogen bonding network between N- and C-termini (Xu and Min, 2011). WD40 domains typically contain four to eight WD repeats, forming β-propeller structures (Stirnimann et al., 2010). WD40 proteins have been systematically identified across various plant genomes, with 237 in Arabidopsis (Van Nocker and Ludwig, 2003), 743 in wheat (Hu et al., 2018), and 191 in Cucumis sativus (Chen et al., 2023).
Accumulating evidence underscores the diverse roles played by WD40 proteins in various physiological and biochemical processes in plants. TRANSPARENT TESTA GLABRA 1 (TTG1), an initially identified WD40 protein in Arabidopsis, serves as a component of the MYB-bHLH-WD40 complex, regulating anthocyanin biosynthesis in plants. This complex has been observed in diverse species such as rice (Sun et al., 2022), blueberry fruits (Zhao et al., 2019), Ficus carica (Fan et al., 2022), Punica granatum (Ben-Simhon et al., 2011), and Camellia sinensis (Liu et al., 2018). Furthermore, the overexpression of TTG-like gene CsWD40 in tobacco significantly increased anthocyanin content in the transgenic plant’s petals (Liu et al., 2018). Additionally, Ehd5 acts as a positive regulator of rice flowering, providing insights into the molecular mechanisms underlying heading date (Zhang et al., 2023). Moreover, Fvcpc2 plays a crucial role in regulating mushroom development and yield in Flammulina velutipes (Wu et al., 2020). These findings underscore the significance of WD40 proteins in plant growth and development.
WD40 genes emerge as pivotal regulators in abiotic stress responses and hormonal signaling cascades. One notable instance is GbLWD1-like, a WD40 gene from Ginkgo biloba, which enhances salt tolerance in transgenic Populus (Xin et al., 2021). XIW1 (XPO1-interacting WD40 protein 1) positively influences the abscisic acid response in Arabidopsis (Xu et al., 2019). A mutation in XIW1 results in the reduced induction of ABI5 and ABA-responsive genes under salt treatment (Cai et al., 2020). Heterologous overexpression of TaWD40D in Arabidopsis significantly enhances tolerance to hormonal responses during seed germination and abiotic stresses during seedling development (Kong et al., 2015). OsRACK1A, regulated by the circadian rhythm, plays a role in regulating salt stress responses (Zhang et al., 2018). These findings underscore the pivotal role of WD40 genes in plant stress tolerance and hormonal responses.
Spartina alterniflora, a halophyte thriving in coastal salt marshes, holds considerable economic value in coastal natural wetlands. Despite being an invasive species, this species produces more aboveground and underground biomass than local populations, making it a promising source of biochar (Liu et al., 2022), and a raw material for anaerobic digestion processes to produce biogas (Yang et al., 2009). Additionally, it has multiple applications such as livestock feed, fertilizer, and production of bio mineral liquids with various health benefits (Qin et al., 1998; Qiu et al., 2020; Zhou et al., 2023). The ideal ecological characteristics of biomass energy crops include C4 photosynthesis, long canopy duration, permanence, absence of pests or diseases, rapid spring growth, rigidity, and efficient water use (Raghu et al., 2006). S. alterniflora possesses most of these characteristics, making it an excellent candidate for industrial and energy development (Buhle et al., 2012). Therefore, exploring the excellent energy development genes of S. alterniflora is crucial for addressing energy security challenges.
S. alterniflora has potential for as biofuel feedstock, enhancing crop yield and environmental adaptability through the exploration of growth and development-related genes. However, limited research has focused on gene functional analysis in this species. Presently, reported studies on S. alterniflora genes have primarily focused on salt stress tolerance, with scarcely any research exploring the functions of its genome in plant growth and development. This gap in knowledge presents a significant opportunity for further exploration. In this investigation, we conducted a comprehensive analysis of WD40 genes at the whole-genome level in S. alterniflora, exploring their characteristics, including physical and chemical properties, phylogenetic tree, gene structure, and expression patterns. Furthermore, we cloned the TTG1 gene, which plays a pivotal role in plant growth and development across various species. Our aim is to decipher the functions of the WD40 gene family by studying the functional significance of TTG1, as it may provide insights into the regulation of flowering and seed size, crucial processes that directly impact crop yield and quality. This study lies in its comprehensive analysis of WD40 genes in S. alterniflora, particularly the cloning and functional exploration of the TTG1 gene, which bridges the knowledge gap on the role of this species’ genome in plant growth and development.
2 Materials and methods
2.1 Identification of WD40 genes in S. alterniflora
The genomic data, encompassing DNA, complete coding sequence (CDS), protein sequence, and positional annotations of S. alterniflora, were procured from our compiled genome. To identify WD40 proteins in S. alterniflora, we initially searched for sequences exhibiting the characteristic WD40 motif (PF00400) within the S. alterniflora protein sequences using HMMER 3.0. The putative SaWD40 proteins were validated using SMART (Letunic et al., 2021) and NCBI’s conserved domain database (Marchler-Bauer et al., 2015). Finally, only the proteins containing the WD40 repeat were retained for subsequent analysis.
To examine their physical attributes, all SaWD40 sequences were assessed using EXPASY to record the number of amino acids and molecular.
2.2 Classification and phylogenetic analysis of SaWD40s
To construct a phylogenetic tree for the SaWD40s, we first performed multiple sequence alignment of the full-length SaWD40 protein sequences using the T-COFFEE program (Magis et al., 2014). Subsequently, we employed the Neighbor-Joining (NJ) method in MEGA 7 to build the phylogenetic tree, with a Bootstrap value of 1000 and other default parameters (Kumar et al., 2016). Finally, we utilized Evolview v3 (Subramanian et al., 2019) to visualize the generated phylogenetic tree.
2.3 Gene duplication analysis
To analyze gene duplication events in SaWD40 genes, we employed MCScanX (Wang et al., 2012) with default settings to identify various gene duplicates, including whole-genome and tandem duplications. The chromosomal locations and duplicated WD40 gene pairs were visualized using TBtools software (Chen et al., 2020). Evolutionary dynamics of these gene pairs were assessed by calculating Ka/Ks values using KaKs_Calculator v 2.0 (Zhang, 2022). The divergence time was determined using the formula T = Ks/2λ, where Ks signifies the synonymous substitutions per site, and λ represents the rate of divergence for nuclear genes in plants. For monocot plants, the adopted λ value fell within the range of 5.1–7.1×10–9 synonymous substitutions per site per year.
2.4 Plant materials, RNA isolation, cDNA synthesis, and qRT-PCR
To conduct tissue-specific expression analysis, S. alterniflora seedlings were grown in an artificial chamber with a temperature of 24/22°C (day/night) and a photoperiod of 16 hours/8 hours (day/night). During the flowering stage, roots, stems, leaves, and inflorescences were collected for analysis. Immediately after sampling, the samples were rapidly frozen in liquid nitrogen and stored at -80°C for RNA extraction.
Total RNA was extracted from plants using the RNAsimple Total RNA Kit (Tiangen, Beijing) following the manufacturer’s instructions. The synthesis of the first-strand cDNA was performed using the FastKing RT Kit (Tiangen, Beijing), and the experimental steps from the kit’s manual were followed. qRT-PCR was conducted using the SYBR Green Pro Taq HS Kit (Tiangen, Beijing), following the manufacturer’s protocol. Data was normalized by the expression of GAPDH in S. alterniflora and AtEF1α4 in Arabidopsis. Each group of experiments was repeated in three biological replicates, and the relative expression levels were determined using the 2–ΔΔCt analysis method (Livak and Schmittgen, 2001). DNA extraction was carried out using the CTAB method. The primer sequences used in this study are listed in Supplementary Table 1.
2.5 Arabidopsis transformation and phenotypic analysis
The SaTTG1 CDS was efficiently amplified by RT-PCR and cloned into the pCAMBIA-1300 vector. Transgenic Arabidopsis Columbia-0 lines were obtained using the floral dip method (Clough and Bent, 1998). Positive lines were screened on 1/2 MS solid medium with 40mg/L Hygromycin B and genotype verified by PCR. Mature seeds from wild-type and transgenic lines were imaged using an SMZ25 microscope (Nikon).
2.6 Subcellular localization and transcriptional activity assays
To ascertain the subcellular localization of SaTTG1, the CDS of SaTTG1 was cloned into PAN580-GFP and introduced into rice (Oryza sativa) protoplasts. Subsequently, subcellular localization was observed using a confocal laser scanning microscope (Zeiss LSM 980 with Airysca, Germany). To assess transcriptional activity, the SaTTG1 CDS was ligated into the pGBKT7 vector for fusion expression. The resulting pGBKT7-SaTTG1 was introduced into Y2H, which were then serially diluted 10-fold and spotted on SD/-Trp plates. Transcriptional activity was evaluated by monitoring yeast growth on SD/-Trp-Ade-His plates. The pGBKT7 vector served as a negative control, with pGBKT7-P53 as a positive control.
2.7 Statistical analysis
Root, seed length, and width of Arabidopsis were quantified with ImageJ (Rueden et al., 2017) and analyzed in Microsoft Excel 2013. Error bars represent standard deviation, and significant differences are marked with numbers or letters.
3 Results
3.1 The S. alterniflora WD40 gene family
To unravel the functional aspects of the WD40 gene family in S. alterniflora, we employed the conserved domain of WD40 proteins (PF00400) as a query in the Pfam database, conducting a comprehensive search against the S. alterniflora genome protein sequences. This exploration led to the identification of a total of 582 SaWD40 members distributed across 31 chromosomes. Chromosome 7 exhibited the highest density of SaWD40 genes, while chromosomes 19 and 31 displayed the lowest density (Figure 1A). The nomenclature of SaWD40 genes was assigned as SaWD40–1 to SaWD40–582 based on their respective chromosome locations (Supplementary Table 2). The deduced SaWD40 DNA lengths ranged from 456 (SaWD40–276) to 52,866 bp (SaWD40–311), and protein lengths spanned from 70 (SaWD40–276) to 3,581 (SaWD40–30) amino acids (aa). The molecular weight varied between 7.44 (SaWD40–276) and 398.31 (SaWD40–30) kDa. Detailed physicochemical properties of S. alterniflora are comprehensively listed in Supplementary Table 2.
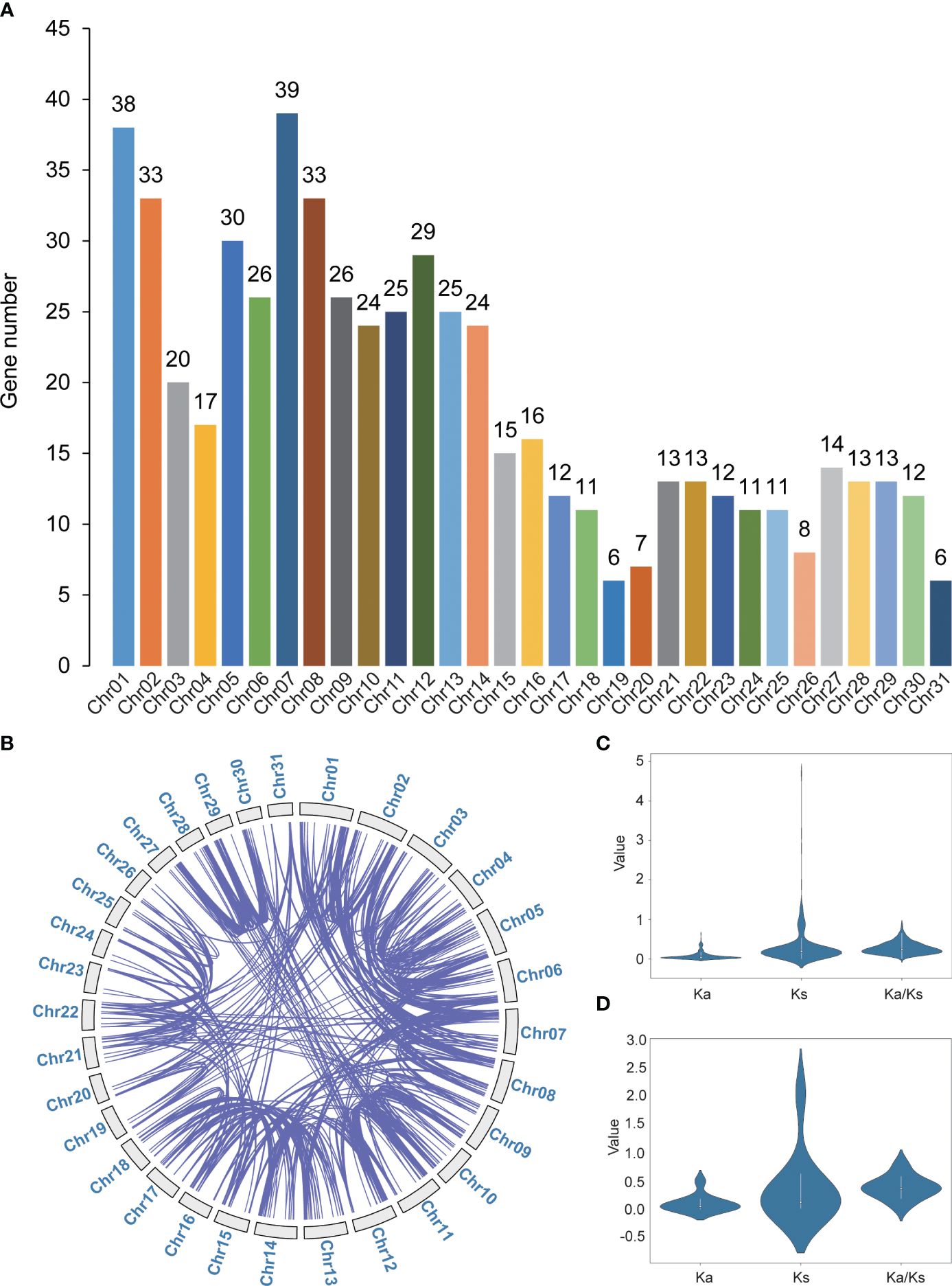
Figure 1 Identification and duplication analysis of the S. alterniflora WD40 gene family. (A) Chromosomal distribution and (B) segmental duplication events of SaWD40 genes. Connecting lines depict duplicated genes that together form pairs of duplicated genes. (C) The distribution values of ka, Ks, and Ka/Ks for segmental duplicate gene pairs. (D) The distribution values of ka, Ks, and Ka/Ks for tandem duplicate gene pairs. These values offer crucial insights into the selection pressures and evolutionary trends associated with these gene duplicates.
3.2 Gene duplication and phylogenetic analyses
Within the cohort of 582 SaWD40 genes, 490 genes participated in the formation of 605 segmentally duplicated gene pairs, while 20 genes contributed to 10 tandemly duplicated gene pairs (Figure 1B; Supplementary Table 3). This implies a significant role of segmental duplication in expanding the SaWD40 gene family. Further scrutiny of the synonymous (Ks) and non-synonymous (Ka) mutations in these gene pairs (Figures 1C, D; Supplementary Table 3) provided insights into the evolutionary dynamics. Moreover, the substitution rate (Ka/Ks) effectively determines positive selection pressure post-duplication, revealing evolutionary direction and selective strength in coding sequences. A Ka/Ks ratio of 1 indicates neutral selection, <1 purifying selection, and >1 positive selection. We calculated Ka, Ks, and Ka/Ks for each gene pair. The average Ka/Ks for tandem duplication SaWD40 genes (0.3875) was significantly higher than for segmental duplication genes (0.2735). Additionally, tandem and segmental duplication events were estimated to have occurred ~256.85–357.58 and ~205.48–286.06 Mya, respectively. This observation suggests the prevalence of purifying selection, indicating that these genes have encountered functional constraints and selective pressure in maintaining their essential functions.
To depict the evolutionary relationships among these 582 SaWD40 proteins, we constructed an unrooted phylogenetic tree utilizing the NJ method (Figure 2A; Supplementary Figure 1). The bootstrap values within the tree facilitated the division of these WD40 proteins into 12 clades. Clade IV boasted the largest contingent of WD40 members, encompassing 145 members, followed by 118 in Clade VII and 87 in Clade II, whereas Clade XI exhibited a more modest presence with only five members.
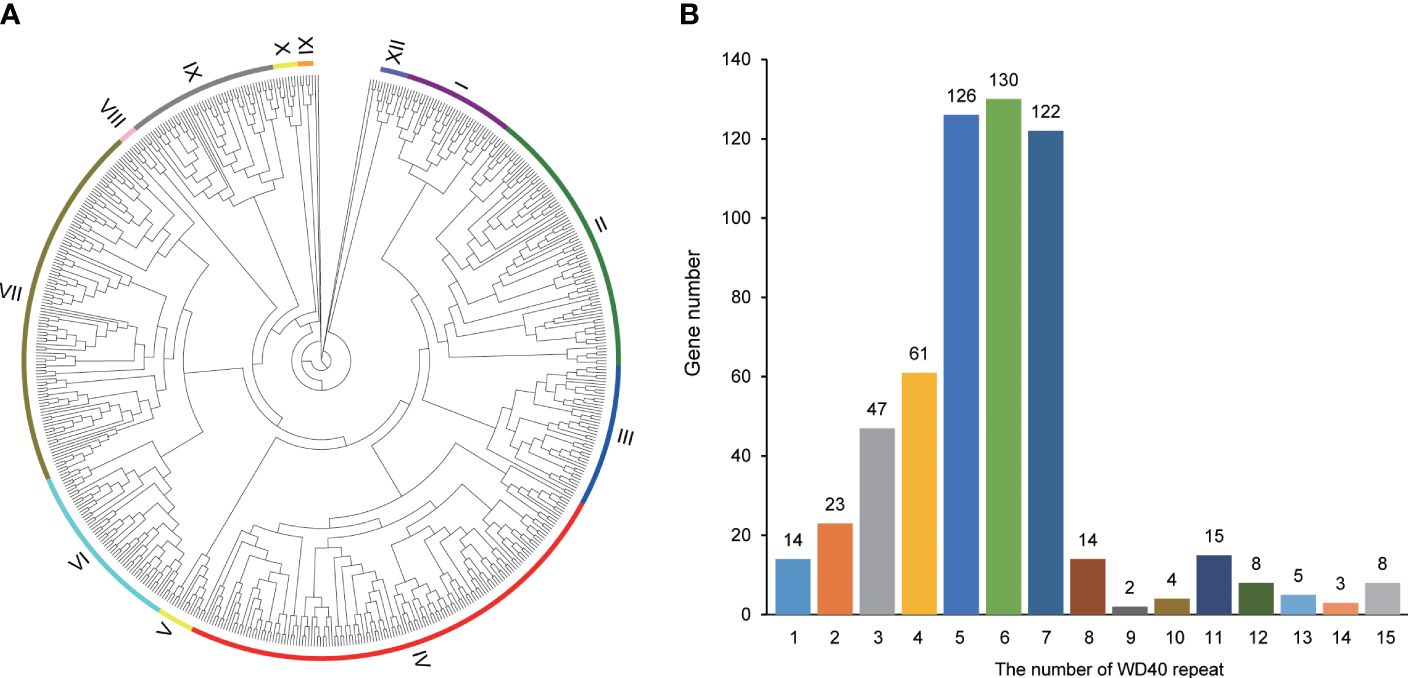
Figure 2 Phylogenetic and WD40 repeat analyses in S. alterniflora. (A) Phylogenetic analysis of WD40 proteins in S. alterniflora, constructed using the neighbor-joining method in MEGA 7. The tree exhibits 12 distinct phylogenetic clades with robust bootstrap support. (B) Summary of the number of WD40 repeats in S. alterniflora.
3.3 Gene structure and protein composition analyses
The SaWD40 genes exhibit a wide spectrum of WD40 repeats, ranging from 1 to 15 WD40 repeats (Figure 2B). Previous studies have indicated that proteins containing WD repeats typically possess 4–10 WD repeats (Van Nocker and Ludwig, 2003). In our SaWD40 protein repertoire, 459 members were identified to contain 4–10 WD repeats, while 39 members displayed more than 10 WD repeats.
We delved into the exon/intron structures of the SaWD40 genes. The analysis unveiled a variation in the number of exons within the SaWD40 gene structure, spanning from 1 to 41 (Supplementary Table 2), with an average of 10.83 exons. Remarkably, SaWD40–311 boasted the largest number of exons (41), followed by SaWD40–340 with 39 exons. In contrast, 21 SaWD40 (3.6%) genes were observed to possess only one exon. The prevalence of genes with seven exons stood out, constituting the highest percentage at 8.8% (51/582) among the total gene count.
3.4 Expression patterns of WD40 in different tissues
To elucidate the functional roles of WD40 genes, we conducted an analysis of their expression patterns across various tissues in the S. alterniflora transcriptome. As depicted in Figure 3 and detailed in Supplementary Table 4, with the exception of 11 genes (FPKM < 0.1) that exhibited undetectable expression levels across different tissues – namely SaWD40–29, -74, -103, -152, -194, -220, -271, -345, -374, -392, -580 – the remaining 565 genes can be detected exhibiting expression (FPKM ≥ 0.1) in certain tissues. Three hundred sixty genes displayed ubiquitous expression across all tissues. There were nine genes that exhibit distinct tissue-specific expression patterns. For instance, SaWD40–194 exhibited exclusive expression in roots, while SaWD40–346, SaWD40–379, SaWD40–391, and SaWD40–502 were specifically expressed in developing inflorescences; SaWD40–85, SaWD40–193, SaWD40–376, and SaWD40–500 displayed specific expression in mature seeds. Furthermore, we observed that certain genes are expressed at higher levels in specific tissues compared to other tissues. For instance, eight genes (SaWD40–73, SaWD40–252, SaWD40–256, SaWD40–293, SaWD40–258, SaWD40–283, SaWD40–444, SaWD40–477) exhibited higher expression levels in maturing seeds, while eight genes (SaWD40–19, SaWD40–102, SaWD40–107, SaWD40–125, SaWD40–219, SaWD40–272, SaWD40–488, SaWD40–514) demonstrated higher expression in developing inflorescences. These findings underscore the relatively widespread expression of WD40 genes across diverse tissues.
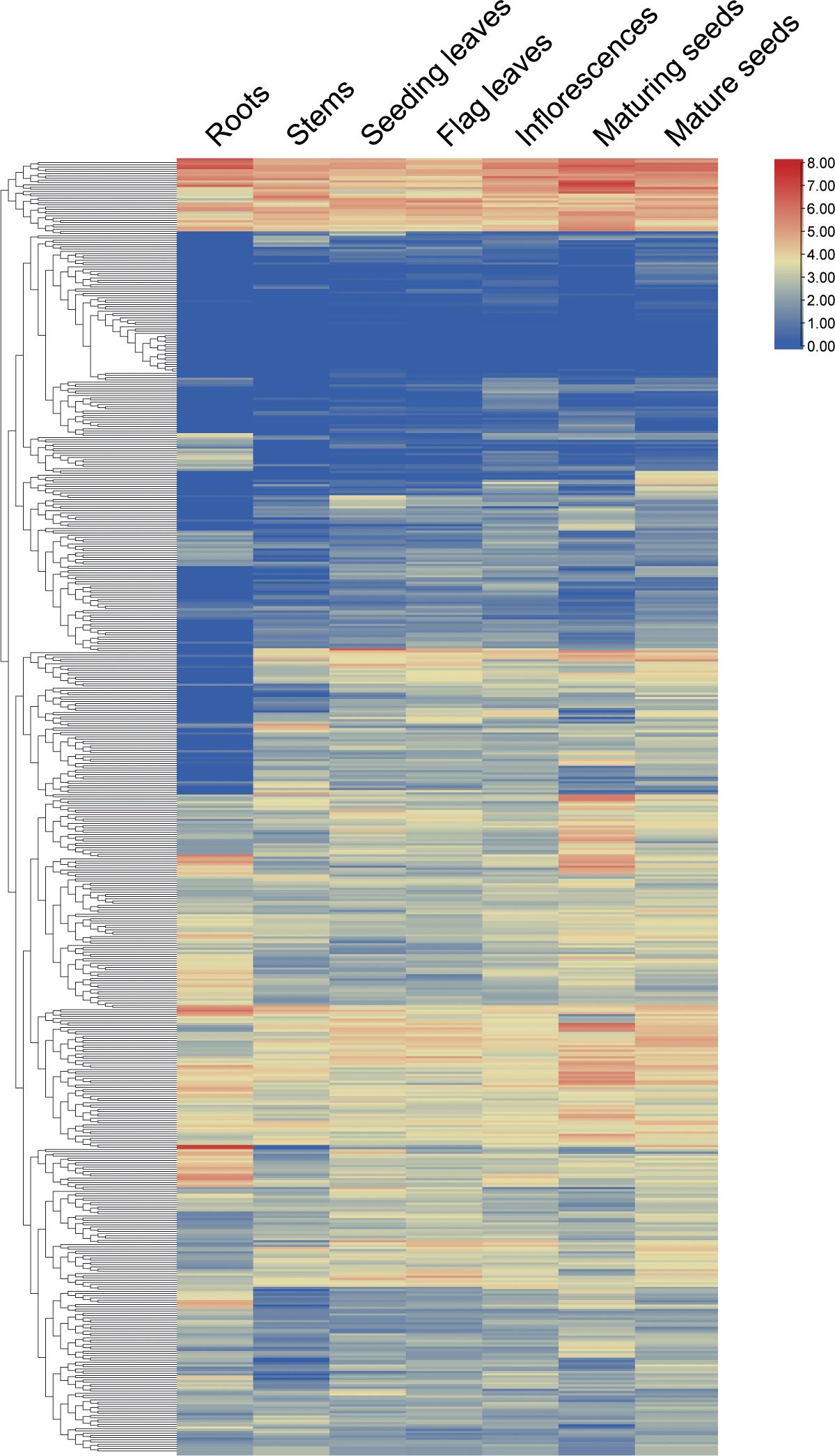
Figure 3 Expression patterns of SaWD40 genes in various tissues using RNA-seq data. The color intensity reflects expression levels, with red indicating higher expression and blue indicating lower expression.
3.5 Protein structure and expression patterns of SaTTG1
Analysis of expression patterns revealed that WD40 genes can be detected in various tissues of S. alterniflora, indicating their involvement in diverse growth and development stages of the plant. To validate the role of WD40 genes in plant growth and development, we aimed to functionally characterize them through heterologous expression in A. thaliana. TTG1, a gene encoding a WD40 protein, has been shown to play a pivotal role in multiple aspects of plant growth and development. Its involvement spans processes such as the accumulation of seed storage reserves (Chen et al., 2015), biosynthesis of anthocyanin and proanthocyanidin (Shan et al., 2019), regulation of circadian activity, epidermal cell fate, and pigmentation (Airoldi et al., 2019). However, to date, there have been no reports on the functional roles of WD40 genes in S. alterniflora. Expression pattern analysis revealed that SaWD40–256, a homolog of A. thaliana’s TTG1 gene, is expressed at its highest level in maturing seeds. Therefore, we aimed to functionally characterize the S. alterniflora TTG1 gene through heterologous expression in A. thaliana, with the goal of gaining deeper insights into the conservation of its functional properties.
Seeking a comprehensive understanding of the function of SaTTG1 (SaWD40–256) in S. alterniflora, we isolated the gene from young seedlings of this species. As depicted in Figure 4A, SaTTG1 exhibits a high sequence similarity with TTG1 proteins from Arabidopsis, Zea mays, and Oryza sativa. These proteins share a highly conserved WD40 repeat regulatory domain. To unravel the potential functions of the SaTTG1 protein, a phylogenetic analysis was conducted using SaTTG1 and TTG1 proteins from other plants. The results suggest that the functions of SaTTG1 more closely resemble those of monocot proteins than dicot proteins (Figure 4B; Supplementary Figure 2).
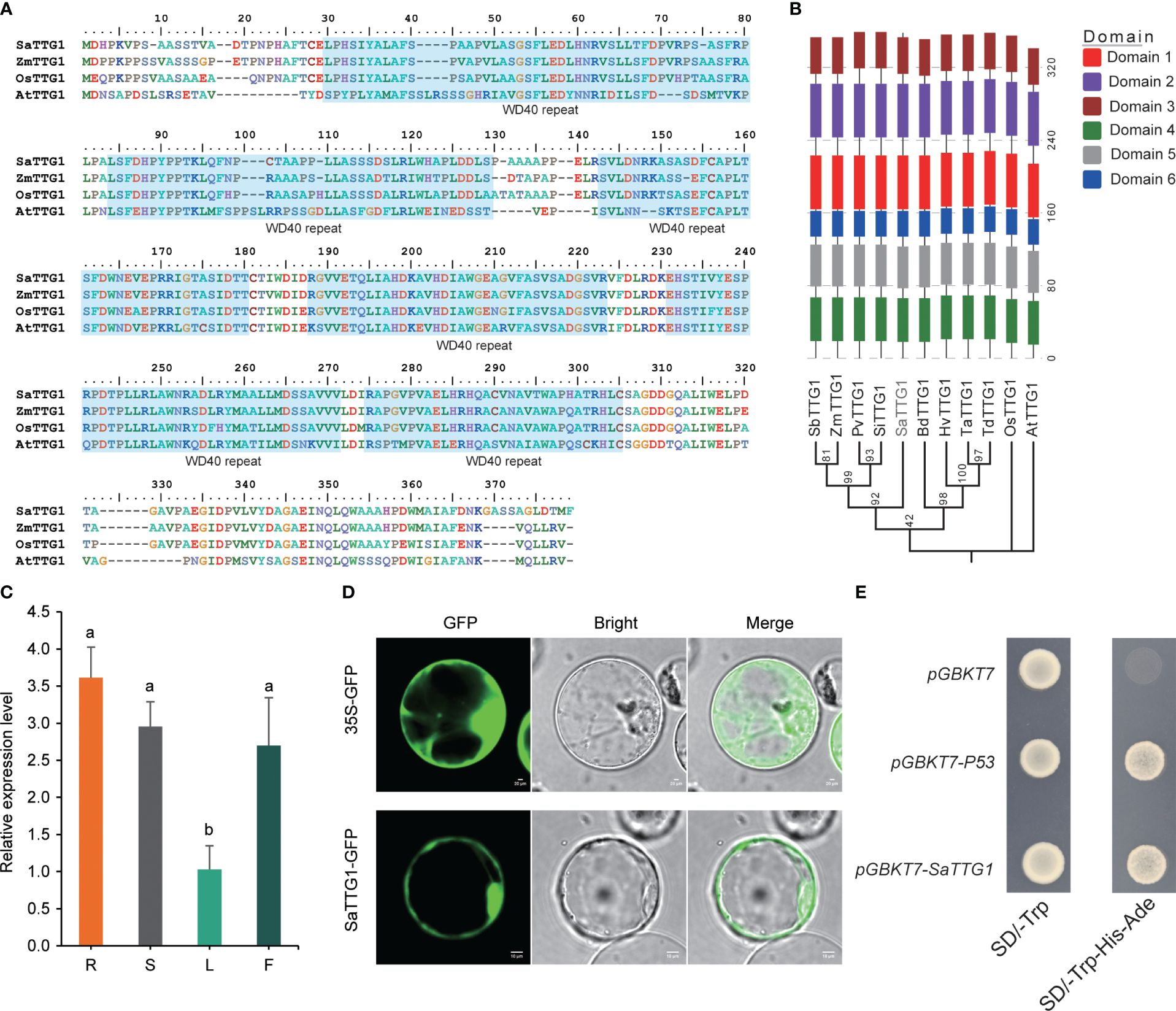
Figure 4 Protein structure and expression patterns of SaTTG1. (A) Sequence alignment of TTG1 proteins from S. alterniflora, Arabidopsis, rice, and maize. The blue shaded area indicates WD40 repeats. (B) Phylogenetic and conserved domain analyses of TTG1 proteins from monocot and dicot plants. Sorghum bicolor SbTTG1, AFN17366.1; Zea mays ZmTTG1, NP_001310302.1; Panicum virgatum PvTTG1, XP_039854481.1; Setaria italica SiTTG1, XP_004953461.1; Brachypodium distachyon BdTTG1, XP_003570109.1; Hordeum vulgare HvTTG1, XP_044952062.1; Triticum aestivum TaTTG1, XP_044403847.1; Triticum dicoccoides TdTTG1, XP_037447801.1; Oryza sativa OsTTG1, NP_001403759.1; Arabidopsis thaliana AtTTG1, CAC10524.1. (C) qRT-PCR analysis of SaTTG1 in various tissues. R, S, L, and F represent the root, stem, leaf, and inflorescence tissues of S. alterniflora, respectively. The values presented are expressed as means ± SD, with three biological replicates (n=3). Significant differences at P < 0.05 were determined using Duncan’s multiple range test, and are indicated by different letters. (D) Subcellular localization of the SaTTG1 protein fused with eGFP in rice protoplasts. Scale bar = 20 μm. (E) Transcriptional activity of SaTTG1 in yeast cells. Yeast cells containing pGBKT7-P53 were utilized as a positive control, while yeast cells containing pGBKT7 empty vector served as a negative control.
Figure 4C illustrates the presence of SaTTG1 transcript in various tissues, with the lowest expression level detected in leaves. This indicates that the gene plays a widespread role in the growth and developmental processes of plants. Subcellular localization studies revealed that SaTTG1, when transiently expressed as 2×35S::SaTTG1-eGFP in rice protoplasts, exhibited a fluorescent signal exclusively in the nucleus and plasma membrane (Figure 4D). Furthermore, yeast cells transformed with the pGBDT7-SaTTG1 fusion construct demonstrated activation of reporter genes and survival in the selective medium SD/-Trp-Ade-His (Figure 4E), affirming the transcriptional activation activity of SaTTG1 in yeast.
3.6 Overexpression of SaTTG1 in Arabidopsis modulates plant development
To examine the function of SaTTG1, we introduced the overexpression construct of 35S::SaTTG1 into the Arabidopsis wild-type Col-0. Through hygromycin selection, a total of 20 independent T1 transgenic plants were acquired. Subsequently, two independent T3 homozygous Col-0 35S::SaTTG1 transgenic lines (#1 and #2) were selected and validated by PCR (Figure 5A). qRT-PCR analysis corroborated the substantial expression of SaTTG1 in the respective transgenic lines, with no detection in the wild-type Col-0 (Figure 5B). These results demonstrate that the overexpression of SaTTG1 induces early flowering in Arabidopsis, and the number of rosette leaves in overexpressing plants is reduced (Figures 5C–F). The seed size of the transgenic lines surpassed that of Col-0, displaying augmented seed length and width (Figures 5G–I). Correspondingly, the 1000-seed weight of the transgenic lines exhibited a notable increase by 9.29% and 8.10% compared to Col-0 (Figure 5J). These findings underscore the significant role of SaTTG1 in the regulation of plant development.
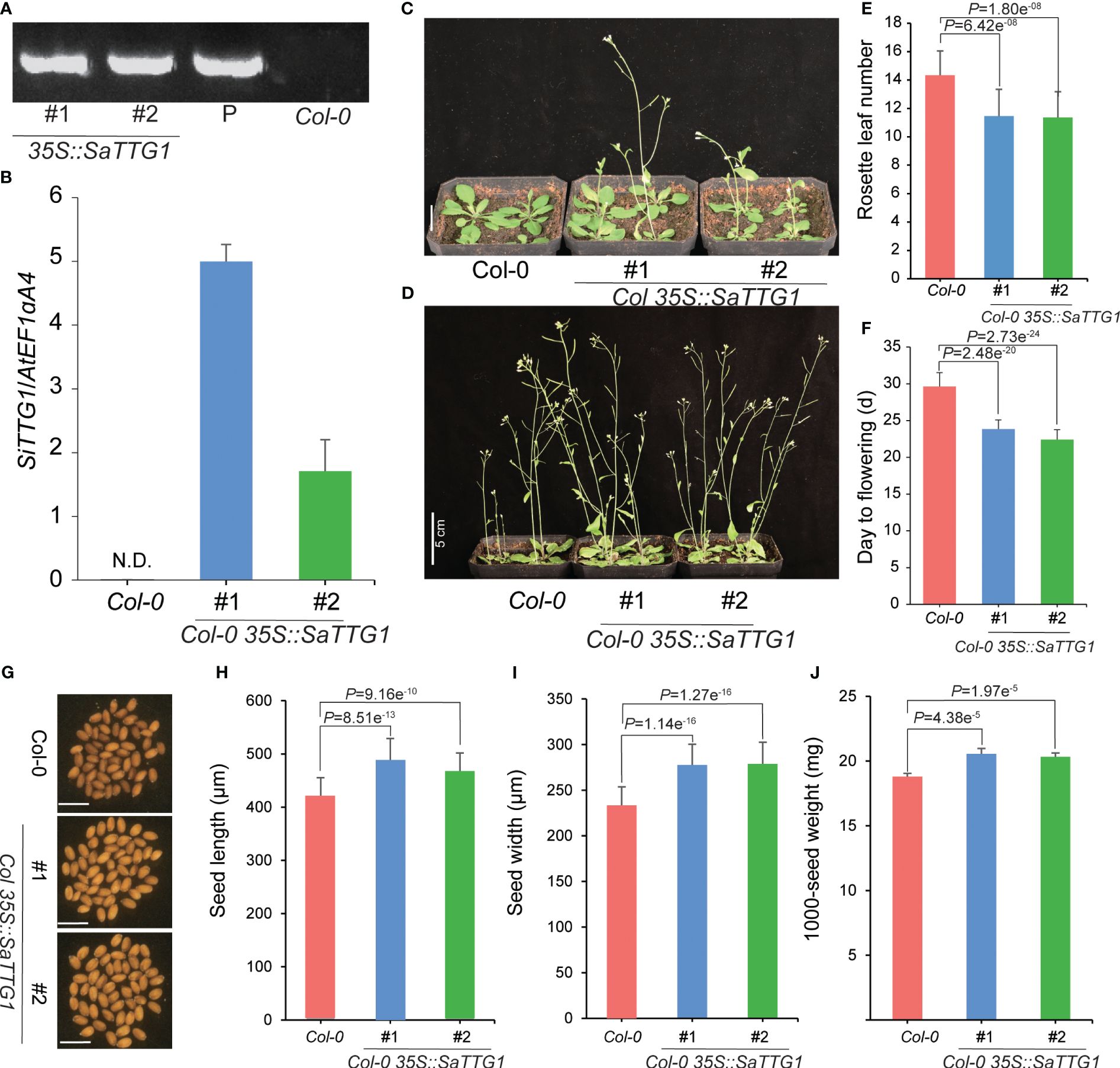
Figure 5 Overexpression of SaTTG1 in Arabidopsis regulates plant development. (A) PCR-based DNA genotyping of the transgenic lines using specific primers. (B) Relative expression levels of the transgenic lines assessed by qRT-PCR, normalized against the expression of the internal control AtEF1α;A4. (C, D) Phenotypes of Col-0 and transgenic lines at 21 (C), Scale bar = 1 cm) and 28 days (D), Scale bar = 5 cm), respectively. (E) Statistical analysis of the number of rosette leaves on the 28th day. (F) Flowering time statistics. (G–J) Seed phenotypes of Col-0 and transgenic lines, including (G) seed phenotype photo (Scale bar = 1 mm), (H) Seed length statistics (n = 30), (I) Seed width statistics (n = 30), and (J) 1000-seed weight statistics (n = 6). Error bars represent the standard deviation, and significance tests were conducted using Student’s t-tests.
4 Discussion
WD40 constitutes a substantial gene family identified in various plant species, encompassing Arabidopsis, wheat, Flammulina velutipes, Cucurbita maxima, and others (Van Nocker and Ludwig, 2003; Hu et al., 2018; Chen et al., 2023; Ji et al., 2023). Our investigation discerned a higher count of WD40 genes in S. alterniflora compared to other species, implying potential gene duplication events during evolution. In S. alterniflora, 490 SaWD40 genes formed 605 segmentally duplicated gene pairs, and 20 SaWD40 genes arranged into 10 tandemly duplicated gene pairs, reinforcing the notion that gene duplication has substantially contributed to the proliferation of WD40 genes. A meticulous analysis of Ka/Ks values for all duplicated gene pairs disclosed that the Ka/Ks ratio predominantly remained at less than 1, indicating of the prevailing influence of purifying selection on these genes.
Diversity in gene structure, denoting variations in gene sequences within a species, resulting in genetic polymorphism, was evident in SaWD40. The gene structure composition displayed considerable diversity, with the number of exons ranging from 1 to 41. Comparable diversity is witnessed in the gene structures of other plants. For instance, wheat WD40 genes exhibit a range of 1 to 39 exons (Hu et al., 2018), Cerasus humilis genes vary from 1 to 51 (Ji et al., 2023), and Cucurbita maxima genes showcase between 1 and 30 exons (Chen et al., 2023). This observation underscores the widespread distribution of WD40 across different species, indicative of the functional diversification of WD40 genes. Moreover, the uneven distribution of WD40 domains in the SaWD40 protein contributes to further nuances in the functional diversification of SaWD40 genes.
The WD40 gene stands as a pivotal player in plant growth and development, with TTG1 being a focal point in numerous studies, highlighting its predominant expression in tissues with anthocyanin accumulation. In this study, we observed the presence of SaTTG1 across different tissues, with the lowest expression level detected in leaves. The relatively low expression level of SaTTG1 in leaves may suggest that its function in this tissue is not dominant or may differ from its roles in other tissues. Diverse plant species exhibit tissue-specific expression patterns of TTG1, such as apple TTG1 primarily expressed in the peel (An et al., 2012), Chinese bayberry TTG1 gene highly expressed in the fruit (Liu et al., 2013), and Freesia×hybrida TTG1 mainly expressed in the petals (Shan et al., 2019). In Arabidopsis, TTG1 plays a crucial role in flowering time regulation (Paffendorf et al., 2020). Our observations align with these findings, demonstrating that overexpression of the SaTTG1 gene in Arabidopsis induces early flowering. The tissue-specific expression patterns observed across different plant species also highlight the specificity of TTG1’s functions in plant development. Overall, our results contribute to the understanding of the diverse roles of TTG1 in plant growth and development, particularly in relation to flowering time regulation.
Moreover, TTG1 emerges as a critical player in seed development, occupying a strategic position in the regulatory hierarchy governing seed filling. Directly targeted by FUS3, TTG1 regulates the accumulation of seed storage proteins and fatty acids during seed maturation (Chen et al., 2015). Overexpression of Setaria italica SiTTG1 has been shown to rectify reduced expression of mucilage biosynthetic genes, including genes involved in seed fatty acid and storage protein accumulation in ttg1–13 plants (Liu et al., 2017). Our research substantiates these roles, indicating that overexpression of SaTTG1 in Arabidopsis significantly influences seed size, underscoring TTG1 as a critical determinant in plant seed development.
Genes from industrial crops, such as switchgrass (Panicum virgatum), sugarcane (Saccharum officinarum), and cassava (Manihot esculenta), possess the potential to enhance the energy yield of plants through modification of growth and development processes. Overexpression of specific genes, such as switchgrass PvBiP2 and PvWOX3a in switchgrass, can increase biomass yield and enhance stem development, respectively (Yang et al., 2021; Song et al., 2023). In sugarcane, SNF4 and its sorghum ortholog SNF4 impact biomass and sugar yield (Upadhyaya et al., 2022), while overexpression of the SoSPS1 gene enhances sucrose content (Anur et al., 2020). Similarly, in cassava, overexpression of MeSLAH4 enhances nitrogen assimilation, growth, and yield (Song et al., 2022). These findings highlight the potential of industrial crops for enhancing energy production through genetic manipulation. As a halophyte, S. alterniflora has the characteristics of industrial and energy crops (Buhle et al., 2012). In this study, analysis of gene expression patterns revealed that a total of 360 WD40 genes are expressed in various tissues, with many genes exhibiting tissue-specific expression patterns. Overexpression of the SaTTG1 gene can alter seed size and weight, indicating its potential for improving seed yield. This finding suggests that the gene can also be used for genetic improvement in industrial crops.
5 Conclusion
In this investigation, we systematically identified 582 WD40 genes within the genome of S. alterniflora, showcasing an uneven distribution across the species’ 31 chromosomes. The expansion of SaWD40 genes is attributed to gene duplication events, introducing diversity in the composition of gene structures. SaTTG1, a member of the WD40 family, is localized in both the nucleus and plasma membrane, demonstrating transcriptional activation activity. When introduced into Arabidopsis, SaTTG1 significantly modifies the flowering time and seed size of the transgenic plants. Our study sheds light on the multifaceted functions of WD40 genes, laying the groundwork for further exploration of their intricate roles in the realm of plant biology.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
MY: Formal analysis, Investigation, Visualization, Writing – original draft. SKC: Formal analysis, Investigation, Visualization, Writing – original draft. JG: Data curation, Investigation, Validation, Writing – review & editing. SG: Data curation, Investigation, Validation, Writing – review & editing. SHC: Funding acquisition, Supervision, Validation, Writing – review & editing. HL: Conceptualization, Funding acquisition, Resources, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by STI 2030-Major Projects (2023ZD0407501), Nanfan special project, CAAS, Grant No. YBXM2407, Innovation Program of Chinese Academy of Agricultural Sciences, the Agricultural Fine Seed Project of Shandong Province, grant number 2023LZGC011, the Yantai City School and Local Integration Development Project (SM21JY05), and Shandong Province Key Research and Development Program (2021SFGC0303).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer ZL declared a shared affiliation with the authors MY, SC, and HL to the handling editor at the time of review.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1390461/full#supplementary-material
References
Airoldi, C.A., Hearn, T.J., Brockington, S.F., Webb, A.A.R., Glover, B.J. (2019). TTG1 proteins regulate circadian activity as well as epidermal cell fate and pigmentation. Nat Plants. 5, 1145–1153. doi: 10.1038/s41477-019-0544-3
An, X. H., Tian, Y., Chen, K. Q., Wang, X. F., Hao, Y. J. (2012). The apple WD40 protein MdTTG1 interacts with bHLH but not MYB proteins to regulate anthocyanin accumulation. J. Plant Physiol. 169, 710–717. doi: 10.1016/j.jplph.2012.01.015
Anur, R. M., Mufithah, N., Sawitri, W. D., Sakakibara, H., Sugiharto, B. (2020). Overexpression of sucrose phosphate synthase enhanced sucrose content and biomass production in transgenic sugarcane. Plants (Basel) 9, 200. doi: 10.3390/plants9020200
Ben-Simhon, Z., Judeinstein, S., Nadler-Hassar, T., Trainin, T., Bar-Ya'akov, I., Borochov-Neori, H., et al. (2011). A pomegranate (Punica granatum L.) WD40-repeat gene is a functional homologue of Arabidopsis TTG1 and is involved in the regulation of anthocyanin biosynthesis during pomegranate fruit development. Planta 234, 865–881. doi: 10.1007/s00425-011-1438-4
Buhle, E. R., Feist, B. E., Hilborn, R. (2012). Population dynamics and control of invasive Spartina alterniflora: inference and forecasting under uncertainty. Ecol. Appl. 22, 880–893. doi: 10.1890/11-0593.1
Cai, J., Huang, H., Xu, X., Zhu, G. (2020). An Arabidopsis WD40 repeat-containing protein XIW1 promotes salt inhibition of seed germination. Plant Signal. Behav. 15, 1712542. doi: 10.1080/15592324.2020.1712542
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, C., Yang, Y., Pan, L., Xia, W., Xu, L., Hua, B., et al. (2023). Genome-wide identification of WD40 proteins in Cucurbita maxima reveals its potential functions in fruit development. Genes (Basel) 14, 220. doi: 10.3390/genes14010220
Chen, M., Zhang, B., Li, C., Kulaveerasingam, H., Chew, F. T., Yu, H. (2015). TRANSPARENT TESTA GLABRA1 regulates the accumulation of seed storage reserves in Arabidopsis. Plant Physiol. 169, 391–402. doi: 10.1104/pp.15.00943
Clough, S. J., Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Fan, Z., Zhai, Y., Wang, Y., Zhang, L., Song, M., Flaishman, M. A., et al. (2022). Genome-wide analysis of anthocyanin biosynthesis regulatory WD40 gene FcTTG1 and related family in Ficus carica L. Front. Plant Sci. 13, 948084. doi: 10.3389/fpls.2022.948084
Hu, R., Xiao, J., Gu, T., Yu, X., Zhang, Y., Chang, J., et al. (2018). Genome-wide identification and analysis of WD40 proteins in wheat (Triticum aestivum L.). BMC Genomics 19, 803. doi: 10.1186/s12864-018-5157-0
Jain, B. P., Pandey, S. (2018). WD40 repeat proteins: signalling scaffold with diverse functions. Protein J. 37, 391–406. doi: 10.1007/s10930-018-9785-7
Ji, X. L., Zhang, M., Wang, D., Li, Z., Lang, S., Song, X. S. (2023). Genome-wide identification of WD40 superfamily in Cerasus humilis and functional characteristics of ChTTG1. Int. J. Biol. Macromol. 225, 376–388. doi: 10.1016/j.ijbiomac.2022.11.074
Kong, D., Li, M., Dong, Z., Ji, H., Li, X. (2015). Identification of TaWD40D, a wheat WD40 repeat-containing protein that is associated with plant tolerance to abiotic stresses. Plant Cell Rep. 34, 395–410. doi: 10.1007/s00299-014-1717-1
Kumar, S., Stecher, G., Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Letunic, I., Khedkar, S., Bork, P. (2021). SMART: recent updates, new developments and status in 2020. Nucleic. Acids Res. 49, D458–d460. doi: 10.1093/nar/gkaa937
Liu, X., Feng, C., Zhang, M., Yin, X., Xu, C., Chen, K. (2013). The MrWD40–1 gene of chinese bayberry (Myrica rubra) interacts with MYB and bHLH to enhance anthocyanin accumulation. Plant Mol. Biol. Rep. 31, 1474–1484. doi: 10.1007/s11105-013-0621-0
Liu, K., Qi, S., Li, D., Jin, C., Gao, C., Duan, S., et al. (2017). TRANSPARENT TESTA GLABRA 1 ubiquitously regulates plant growth and development from Arabidopsis to foxtail millet (Setaria italica). Plant Sci. 254, 60–69. doi: 10.1016/j.plantsci.2016.10.010
Liu, Y., Hou, H., Jiang, X., Wang, P., Dai, X., Chen, W., et al. (2018). A WD40 repeat protein from Camellia sinensis regulates anthocyanin and proanthocyanidin accumulation through the formation of MYB⁻bHLH⁻WD40 ternary complexes. Int. J. Mol. Sci. 19, 1686. doi: 10.3390/ijms19061686
Liu, W., Chen, X., Wang, J., Zhang, Y. (2022). Does the effect of flowering time on biomass allocation across latitudes differ between invasive and native salt marsh grass Spartina alterniflora? Ecol. Evol. 12, e8681. doi: 10.1002/ece3.8681
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Magis, C., Taly, J. F., Bussotti, G., Chang, J. M., Di Tommaso, P., Erb, I., et al. (2014). T-Coffee: Tree-based consistency objective function for alignment evaluation. Methods Mol. Biol. 1079, 117–129. doi: 10.1007/978-1-62703-646-7_7
Marchler-Bauer, A., Derbyshire, M. K., Gonzales, N. R., Lu, S., Chitsaz, F., Geer, L. Y., et al. (2015). CDD: NCBI's conserved domain database. Nucleic. Acids Res. 43, D222–D226. doi: 10.1093/nar/gku1221
Mishra, A. K., Puranik, S., Prasad, M. (2012). Structure and regulatory networks of WD40 protein in plants. J. Plant Biochem. Biotechnol. 21, 32–39. doi: 10.1007/s13562-012-0134-1
Paffendorf, B., Qassrawi, R., Meys, A. M., Trimborn, L., Schrader, A. (2020). TRANSPARENT TESTA GLABRA 1 participates in flowering time regulation in Arabidopsis thaliana. PeerJ 8, e8303. doi: 10.7717/peerj.8303
Qin, P., Xie, M., Jiang, Y. (1998). Spartina green food ecological engineering1Paper presented at ICEE 96—International Conference on Ecological Engineering, Beijing, China, 7–11 October 1996.1. Ecol. Eng. 11, 147–156. doi: 10.1016/S0925-8574(98)00030-5
Qiu, Z., Tang, J., Chen, J., Zhang, Q. (2020). Remediation of cadmium-contaminated soil with biochar simultaneously improves biochar's recalcitrance. Environ. pollut. 256, 113436. doi: 10.1016/j.envpol.2019.113436
Raghu, S., Anderson, R. C., Daehler, C. C., Davis, A. S., Wiedenmann, R. N., Simberloff, D., et al. (2006). Ecology. Adding biofuels to the invasive species fire? Science 313, 1742. doi: 10.1126/science.1129313
Rueden, C. T., Schindelin, J., Hiner, M. C., DeZonia, B. E., Walter, A. E., Arena, E. T., et al. (2017). ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinf. 18, 529. doi: 10.1186/s12859-017-1934-z
Shan, X., Li, Y., Yang, S., Gao, R., Zhou, L., Bao, T., et al. (2019). A functional homologue of Arabidopsis TTG1 from Freesia interacts with bHLH proteins to regulate anthocyanin and proanthocyanidin biosynthesis in both Freesia hybrida and Arabidopsis thaliana. Plant Physiol. Biochem. 141, 60–72. doi: 10.1016/j.plaphy.2019.05.015
Smith, T. F., Gaitatzes, C., Saxena, K., Neer, E. J. (1999). The WD repeat: a common architecture for diverse functions. Trends Biochem. Sci. 24, 181–185. doi: 10.1016/S0968-0004(99)01384-5
Song, L., Wang, X., Zou, L., Prodhan, Z., Yang, J., Yang, J., et al. (2022). Cassava (Manihot esculenta) slow anion channel (MeSLAH4) gene overexpression enhances nitrogen assimilation, growth, and yield in rice. Front. Plant Sci. 13, 932947. doi: 10.3389/fpls.2022.932947
Song, G., Zhang, J., Wang, Y., Ji, Y., Fang, Z., Cai, Q., et al. (2023). Overexpression of PvBiP2 improved biomass yield and cadmium tolerance in switchgrass (Panicum virgatum L.). J. Hazard. Mater. 446, 130648. doi: 10.1016/j.jhazmat.2022.130648
Stirnimann, C. U., Petsalaki, E., Russell, R. B., Müller, C. W. (2010). WD40 proteins propel cellular networks. Trends Biochem. Sci. 35, 565–574. doi: 10.1016/j.tibs.2010.04.003
Subramanian, B., Gao, S., Lercher, M. J., Hu, S., Chen, W. H. (2019). Evolview v3: a webserver for visualization, annotation, and management of phylogenetic trees. Nucleic. Acids Res. 47, W270–w275. doi: 10.1093/nar/gkz357
Sun, X., Zhang, Z., Li, J., Zhang, H., Peng, Y., Li, Z. (2022). Uncovering hierarchical regulation among MYB-bHLH-WD40 proteins and manipulating anthocyanin pigmentation in rice. Int. J. Mol. Sci. 23, 8203. doi: 10.3390/ijms23158203
Upadhyaya, H. D., Wang, L., Prakash, C. S., Liu, Y., Gao, L., Meng, R., et al. (2022). Genome-wide association mapping identifies an SNF4 ortholog that impacts biomass and sugar yield in sorghum and sugarcane. J. Exp. Bot. 73, 3584–3596. doi: 10.1093/jxb/erac110
Van Nocker, S., Ludwig, P. (2003). The WD-repeat protein superfamily in Arabidopsis: conservation and divergence in structure and function. BMC Genomics 4, 50. doi: 10.1186/1471-2164-4-50
Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic. Acids Res. 40, e49. doi: 10.1093/nar/gkr1293
Wu, T., Zhang, Z., Hu, C., Zhang, L., Wei, S., Li, S. (2020). A WD40 protein encoding gene Fvcpc2 positively regulates mushroom development and yield in Flammulina velutipes. Front. Microbiol. 11, 498. doi: 10.3389/fmicb.2020.00498
Xin, Y., Wu, Y., Han, X., Xu, L. A. (2021). Overexpression of the Ginkgo biloba WD40 gene GbLWD1-like improves salt tolerance in transgenic Populus. Plant Sci. 313, 111092. doi: 10.1016/j.plantsci.2021.111092
Xu, C., Min, J. (2011). Structure and function of WD40 domain proteins. Protein Cell 2, 202–214. doi: 10.1007/s13238-011-1018-1
Xu, X., Wan, W., Jiang, G., Xi, Y., Huang, H., Cai, J., et al. (2019). Nucleocytoplasmic trafficking of the Arabidopsis WD40 repeat protein XIW1 regulates ABI5 stability and abscisic acid responses. Mol. Plant 12, 1598–1611. doi: 10.1016/j.molp.2019.07.001
Yang, S., Li, J., Zheng, Z., Meng, Z. (2009). Characterization of Spartina alterniflora as feedstock for anaerobic digestion. Biomass Bioenergy 33, 597–602. doi: 10.1016/j.biombioe.2008.09.007
Yang, R., Wu, Z., Bai, C., Sun, Z., Wang, M., Huo, Y., et al. (2021). Overexpression of PvWOX3a in switchgrass promotes stem development and increases plant height. Hortic. Res. 8, 252. doi: 10.1038/s41438-021-00678-w
Zhang, Z. (2022). KaKs_Calculator 3.0: calculating selective pressure on coding and non-coding sequences. Genomics Proteomics Bioinf. 20, 536–540. doi: 10.1016/j.gpb.2021.12.002
Zhang, X., Feng, Q., Miao, J., Zhu, J., Zhou, C., Fan, D., et al. (2023). The WD40 domain-containing protein Ehd5 positively regulates flowering in rice (Oryza sativa). Plant Cell 35, 4002–4019. doi: 10.1093/plcell/koad223
Zhang, D., Wang, Y., Shen, J., Yin, J., Li, D., Gao, Y., et al. (2018). OsRACK1A, encodes a circadian clock-regulated WD40 protein, negatively affect salt tolerance in rice. Rice (N Y) 11, 45. doi: 10.1186/s12284-018-0232-3
Zhao, M., Li, J., Zhu, L., Chang, P., Li, L., Zhang, L. (2019). Identification and characterization of MYB-bHLH-WD40 regulatory complex members controlling anthocyanidin biosynthesis in blueberry fruits development. Genes (Basel) 10, 496. doi: 10.3390/genes10070496
Keywords: Spartina alterniflora, WD40, TTG1, flowering time, seed size
Citation: Yang M, Chen S, Geng J, Gao S, Chen S and Li H (2024) Comprehensive analysis of the Spartina alterniflora WD40 gene family reveals the regulatory role of SaTTG1 in plant development. Front. Plant Sci. 15:1390461. doi: 10.3389/fpls.2024.1390461
Received: 23 February 2024; Accepted: 29 April 2024;
Published: 28 May 2024.
Edited by:
Yi-Hong Wang, University of Louisiana at Lafayette, United StatesReviewed by:
Minmin Miao, Yangzhou University, ChinaZhe Liang, Chinese Academy of Agricultural Sciences, China
Copyright © 2024 Yang, Chen, Geng, Gao, Chen and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huihui Li, bGlodWlodWlAY2Fhcy5jbg==; Shihua Chen, Y2hlbnNoQHl0dS5lZHUuY24=
†These authors have contributed equally to this work
 Maogeng Yang
Maogeng Yang Shoukun Chen
Shoukun Chen Jiahui Geng1,2,3
Jiahui Geng1,2,3 Huihui Li
Huihui Li