
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Plant Sci., 04 June 2024
Sec. Plant Pathogen Interactions
Volume 15 - 2024 | https://doi.org/10.3389/fpls.2024.1385164
Biotic stresses caused by bacterial and fungal pathogens damage crops; identifying treatments that enhance disease resistance provides important information for understanding plant defenses and sustainable agriculture. Salt stress affects crop yields worldwide; however, studies have focused on the toxic sodium ion, leaving the effects of the chloride ion unclear. In this study, we found that irrigation with a combination of chloride salts (MgCl2, CaCl2, and KCl) suppressed the cell death phenotype of the ceramide kinase mutant acd5. Chloride salt pre-irrigation also significantly limited the cell death caused by Pseudomonas syringae pv maculicola infection and inhibited the multiplication of this bacterial pathogen in a mechanism partially dependent on the salicylic acid pathway. Moreover, chloride salt pre-irrigation improved plant defenses against the fungal pathogen challenge, confining the lesion area caused by Botrytis cinerea infection. Furthermore, the growth of herbivorous larvae of Spodoptera exigua was retarded by feeding on chloride salt irrigated plants. Thus, our data suggest that treatment with Cl- increases broad spectrum resistance to biotic challenges.
Plants have evolved many ways to defend against abiotic and biotic stresses. Different stressors provoke different defense response pathways and affect plant responses to subsequent stressors via synergistic or antagonistic crosstalk. Since the toxic threshold for Na+ is much lower than that of Cl-, researchers have focused on the mechanisms that respond to Na+ stress, rather than the effect of Cl- on plants. Under high-salt conditions, plants exclude ions such as Na+ from root cells to reduce uptake or unload ions from the xylem to the phloem to decrease ion transport to the shoot (Davenport et al., 2007; Zhu, 2016). The accumulation of osmotic substances helps to balance water potential and inhibit the influx of toxic ions from the apoplast into the cytosol (Hasegawa et al., 2000). In addition, plant cells compartmentalize high levels of Na+ in vacuoles to keep ion concentrations below the toxic level in the cytosol and other organelles; some plants can also secrete ions through salt glands (Munns and Tester, 2008).
Cl- accumulation can also dampen plant growth and reducing Cl- levels increases plant resistance to salinity; however, several studies have demonstrated that a suitable concentration of Cl- is vital for plant cell biology. For example, Cl- working as an osmotic regulator affects stomata behavior and participates in plant responses to drought and salinity (Wang et al., 2013; Geilfus et al., 2015; Zhang et al., 2017). Increasing Cl- contents in plants by treating the plants with Cl- salts can enhance the osmotic regulatory function of NO3- and increase the usage of NO3- (Wege et al., 2017; Rosales et al., 2020). Franco-Navarro et al. (2019) showed that Cl- can promote water use efficiency and CO2 diffusion in the intercellular space, thus increasing photosynthetic efficiency. Taken together, these observations suggest that a certain concentration of Cl- plays a positive role in plant resistance to abiotic stress and promotes plant growth.
In addition to its effect on abiotic stress, some studies suggest that Cl- affects biotic stress responses. For example, the chloride channel family protein CLCd can negatively regulate microbe pattern-triggered immunity (PTI), possibly via altered pH in the Golgi complex (Guo et al., 2014b). Pharmacological treatments showed that anion channels differentially participated in PTI and effector-triggered immunity (Guo et al., 2014a), but whether changing Cl- levels by irrigation would affect plant biotic responses remains unclear.
Yang et al. (2019) found that pre-irrigation with 300 mM NaCl drastically inhibited the cell-death phenotype of acd5 mutants, which lack ceramide kinase and exhibit accelerated cell death (Liang et al., 2003). To further investigate the molecular mechanism of the inhibitory effect of salt on cell death in acd5, we tried several kinds of salts to discover the main ions that affect cell death and found that Cl- is the major ion suppressing cell death in acd5. Cl- pre-irrigation also enhanced plant resistance to a broad range of biotic stresses, including hemibiotrophic and necrotrophic pathogen infection, and phytophagous insect attack. These results showed that treatment with a certain concentration of chloride salts enhanced plant resistance to a broad spectrum of biotic stresses. Further research to decipher the molecular relationships between Cl- and plant cell death, plant responses to biotic stresses, will deepen our understanding of the functions of inorganic ions in cellular processes. Our data provide new insight into the biological significance of Cl- as beneficial ion.
Arabidopsis wild-type (Col-0) and acd5 mutant plants were used in this study. Seeds were surface sterilized with 70% ethanol for 7 min, washed three times with sterile water, and sown on half-strength Murashige and Skoog (MS) medium plates supplemented with 1% sucrose and 0.6% agar. After 2 days of stratification at 4°C, plates were kept in the greenhouse at 22°C and a 16-h light/8-h dark cycle for 8 days. Then the seedlings were transplanted onto soil and grown in the same greenhouse.
Three-week-old soil-grown plants were irrigated with water, 300 mM chloride salt solution (composed of 60 mM MgCl2, 60 mM CaCl2, and 60 mM KCl), or other salt solutions as specified in the figure legends once to soil capacity, typically, 15 mL of salt solution was irrigated into one well of the 32-well seedling-planting tray. Thereafter, the plants were irrigated with water as required (Yang et al., 2019). For salt spraying treatment, chloride salt solutions of different concentrations containing 0.02% Silwet-77 were sprayed onto plants. The phenotype of plants was recorded as indicated in each figure legend.
Bacterial infection was conducted as previously described (Yang et al., 2019). Briefly, after 24 h of Cl- treatment, the 3rd and 4th leaves of three-week–old plants were infiltrated with 10 mM MgSO4 (mock), Pseudomonas syringae pv maculicola (Pma) DG3, DG6, or Pseudomonas syringae pv tomato (Pst) DC3000-GFP at OD600 as described in the corresponding figure legend. The virulent Pma DG3 is a recA derivative of Pma ES4326, and the avirulent Pma DG6 is a recA derivative of Pma ES4326 expressing the type-III effector avrRpt2 (Yao and Greenberg, 2006). These two pathogens have been maintained in Dr. Yao’s lab for years after being gifted by Dr. Greenberg. For the bacterial growth assay, leaf discs were collected at indicated time points to determine the population of bacteria as colony forming units. The colonization of DC3000-GFP was visualized under a fluorescence microscope (DM5000B, Leica).
Botrytis cinerea infection was performed as previously described (Zeng et al., 2021) with a minor modification. B. cinerea was cultured on V8 medium according to Zhang et al. (2015b) for 7 days. B. cinerea spores were suspended in potato dextrose broth (PDB) and adjusted to 1 × 105 spores mL-1. Five μL spore suspension was dropped onto the 3rd and 4th leaf of the plants. Then plants were covered with a dome and kept in the dark. The Spodoptera exigua larvae (2nd instar) were purchased from Henan Jiyuan Baiyun Industry (China). The feeding assay was performed as previously described (Huang et al., 2021).
Detection of the ROS induced by flg22 was performed according to Yang et al. (2019). Leaf discs were incubated with distilled water overnight in the dark. Then distilled water was substituted with 34 μg/mL L-012 (Wako), 20 μg/mL peroxidase, and 100 nM Flg22 or a Mock treatment without Flg22. Flg22-induced ROS was detected with a microplate detector (Spark, Tecan) by recording the relative light units (RLU) of luminescence.
The expression of FRK1 (At2g19190) was detected as described previously (Yang et al., 2019). Three hours post Flg22 infiltration, the samples for total RNA isolation were collected. Total RNA was extracted using the E.Z.N.A. plant RNA Kit (R6827-01, Omega) according to the manufacturer’s instructions. One µg RNA was reverse-transcribed into cDNA using the PrimeScript RT kit with genomic DNA Eraser (Takara). Ten µL reaction system containing 1 µL of diluted cDNA sample (1:20), 5 µL SYBR Premix Ex Taq II (Takara), and 0.4 µL gene-specific primers was mixed in each 96-well plate well and then quantitatively analyzed by quantitative PCR (Step One plus, ABI). ACTIN2 (At3g18780) was analyzed as the internal reference gene to normalize values for transcript abundance. The 2-△△CT method (Livak and Schmittgen, 2001) was used to calculate the relative expression level of FRK1. Three technical replicates were performed for each template and primer combination. The primers for amplification are as follows: FRK1-F, 5′-GCCAACGGAGACATTAGAG-3′; FRK1-R, 5′-CCATAACGACCTGACTCATC-3′; ACTIN2-F, 5′-GGTAACATTGTGCTCAGTGGTGG-3′; ACTIN2-R, 5′-GGTGCAACGACCTTAATCTTCAT-3′.
Transcriptome sequencing was conducted on the Illumina PE150 platform by Verygenome Technology (Guangzhou, China). The clean reads were aligned to the reference genome using HISAT2 (Sirén et al., 2014). Differential expression analysis was performed using the ‘DESeq2’ R package (1.18.0) (Wang et al., 2010). Differentially expressed genes were defined by log2(fold change) ≥ 1 and false discovery rate (FDR) < 0.05. Gene Ontology analysis was performed using the clusterProfiler R package (Yu et al., 2012) with FDR < 0.05. We have deposited the full RNA sequencing data in NCBI (PRJNA1073023, https://www.ncbi.nlm.nih.gov/sra/PRJNA1073023).
Cell death was visualized by trypan blue staining as previously described (Yang et al., 2019; Zeng et al., 2021). Plant leaves collected at the indicated time points were submerged in staining buffer (lactic acid: water-saturated phenol: glycerol: distilled water = 1:1:1:1, V:V:V:V) containing 0.25 mg/mL trypan blue, and incubated in boiling water for 2 min, then decolorized in chloral hydrate solution. The blue cell death lesions were recorded by a stereo microscope with the ×20 objective (SteREO Lumar.V12, Carl Zeiss), and analyzed by ImageJ software (ImageJ 1.48v).
Callose deposition was detected by aniline blue staining (Bi et al., 2014). At 12 hours post infiltration with 100 nM Flg22 or Mock (10 mM MgSO4), leaf samples were fixed in FAA (formaldehyde: acetic acid: ethanol: water=2:1:9:8, V:V:V:V) for 5 min. Then the samples were transferred into alcoholic lactophenol (95% ethanol:lactophenol=2:1, V:V), and incubated in boiling water for 2 min. The leaves were washed twice in 50% ethanol for 2 min. After being rinsed in distilled water, the samples were transferred into aniline blue staining buffer (0.01% aniline blue in 150 mM K2HPO4, pH 9.5) overnight, and visualized under a fluorescence microscope with the ×5 objective (DM5000B, Leica) and analyzed by ImageJ software.
Data are presented as means ± SE. Significant differences were determined by ANOVA post hoc tests (P < 0.05) using different letters or determined by Student’s t-test with P-values presented in each figure legend. Numbers of biological replicates or technical repeats are also given in each figure legend.
To decipher the mechanism by which 300 mM NaCl inhibits cell death in acd5 mutants, we tested salts with different anions or cations. As shown in Supplementary Figure S1A, the cell death lesions on acd5 were thoroughly suppressed by irrigation with 300 mM NaCl, 300 mM KCl, and 150 mM CaCl2, but were not suppressed by 300 mM NaNO3, 300 mM KNO3, 150 mM Na2SO4, or 150 mM K2SO4. This result suggests that the major ion suppressing cell death in acd5 is chloride (Cl-).
Furthermore, we tested a compound salt containing MgCl2, CaCl2, and KCl (Zhang et al., 2015a) to confirm the effect of Cl- on cell death. Indeed, 300 mM Cl- (60 mM MgCl2, 60 mM CaCl2, 60 mM KCl) also suppressed cell death in acd5 (Supplementary Figure S1B). Together, these results show that a certain concentration of Cl- can suppress the cell death caused by interrupted sphingolipid metabolism. To limit the influence of individual cations, we used the MgCl2–CaCl2–KCl compound solution for Cl- treatment in subsequent experiments described below.
Since NaCl also suppresses cell death lesions caused by pathogenic bacteria and enhances plant resistance (Singh et al., 2014; Yang et al., 2019), we next tested whether Cl- could recapitulate this effect. To this end, plants pretreated with Cl- were challenged with virulent Pseudomonas syringae pv. maculicola (Pma) DG3 and avirulent Pma DG6 (carrying the effector protein avrRpt2). Pma DG3 produces cell death induced by infection and Pma DG6 produces cell death as part of the hypersensitive response and disease resistance. As shown Figure 1, Cl- irrigation suppressed the cell death induced by Pma DG3 and Pma DG6, indicating Cl- could suppress cell death that resulted from susceptibility in response to virulent pathogen infection, and to resistance (hypersensitive response-related cell-death) in response to avirulent pathogen infection. We also tested whether the cations introduced by the chloride salt altered plant response to pathogenic bacteria. As shown in Supplementary Figure S2, although Cl- salt could effectively suppress the cell death induced by Pma DG3, the solution containing equivalent cations, such as 60 mM K+, 60 mM Mg2+, and 60 mM Ca2+ released by KNO3, 60 mM MgSO4, and 60 mM Ca(NO3)2, respectively, was ineffective to confine the cell death caused by DG3 infection. This result further confirmed that chloride ion irrigation could substantially alter plant cell fate confronting pathogen challenge. Moreover, spraying with Cl- suppressed the lesion development caused by spray infection with DG3 (Supplementaryray Figure S3). Thus, these results suggested that Cl- treatment could not only suppress the cell death in the lesion mimic mutant acd5, but could also suppress cell death induced by bacterial pathogens.
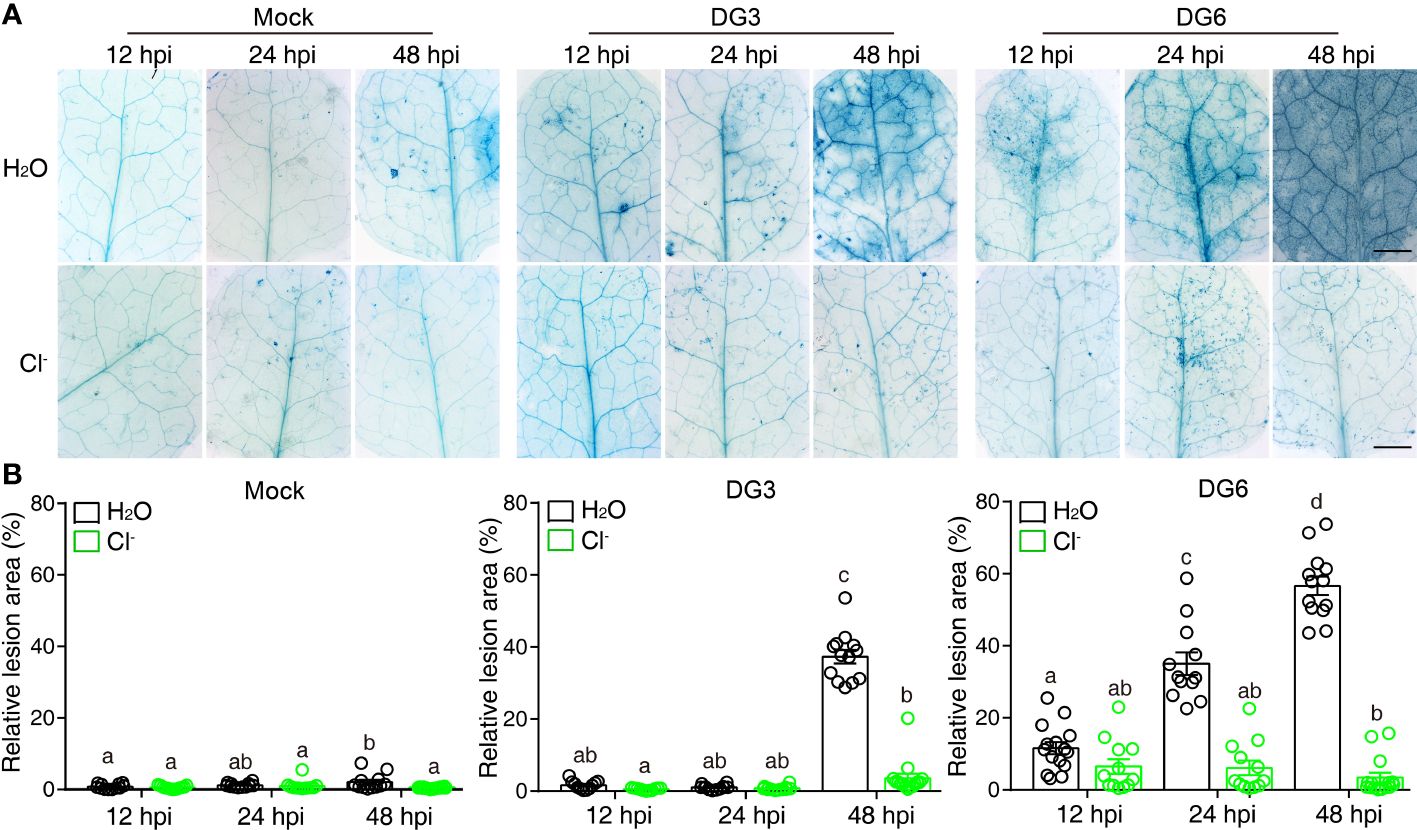
Figure 1 Cl- treatment hinders cell death lesions induced by bacterial pathogen. Three-week-old soil-grown plants were irrigated with H2O or 300 mM Cl-. 24 h later, the 3rd and the 4th leaf were infiltrated with Mock (10 mM MgSO4), Pseudomonas syringae pv maculicola (Pma) DG3 (OD = 0.001), or Pma DG6 (OD = 0.001). (A) Representative phenotype of cell death visualized by trypan blue straining. Samples were detached and stained at the indicated time points, bar = 2 mm. (B) Statistical analysis of cell-death lesions in (A). The relative cell death lesion area per leaf was quantified lesion area as a percentage of the leaf area in leaves photographed under a stereomicroscope and measured by Image J software (n ≥ 10). Data are presented as mean values ± SEM. Statistical differences were analyzed by ANOVA post hoc tests (P < 0.05) and significantly different samples are indicated with different letters. This experiment was repeated at least twice with similar results using independent samples.
Plant cell death is tightly related to resistance to pathogens. Therefore, we analyzed the interactions between plants and different pathogens to decipher the impact of Cl- treatment on pathogenesis. GFP-tagged Pseudomonas syringae pv. tomato (Pst) DC3000 (DC3000-GFP) was inoculated to visualize Pst colonization of the phyllosphere in situ (Kong et al., 2020). As shown in Figures 2A, B, DC3000-GFP remained pathogenic to Arabidopsis, and multiplied in plants irrigated with H2O, but not in plants irrigated with Cl-.
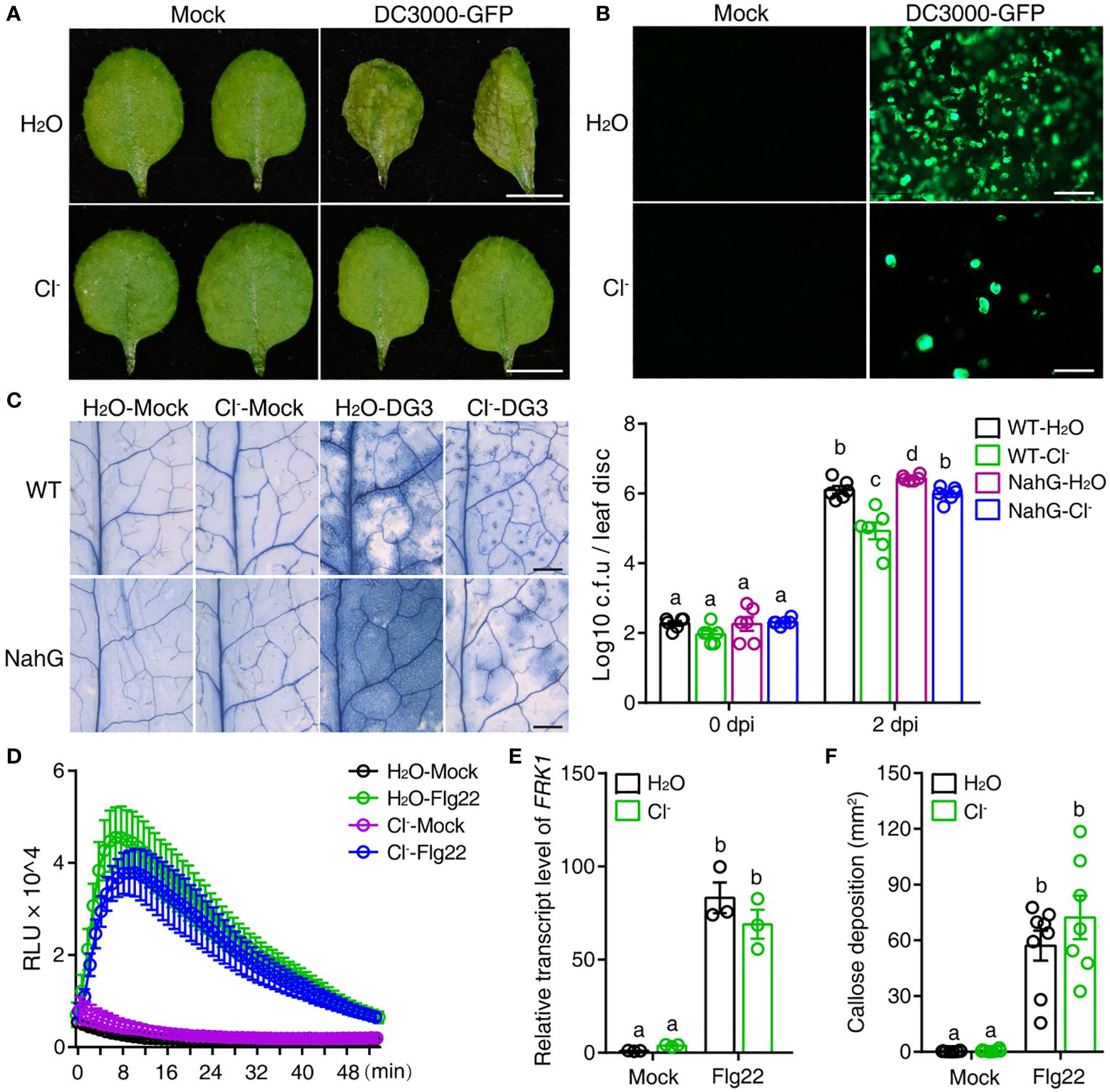
Figure 2 Cl- treatment increases plant resistance to bacterial pathogens. Three-week-old soil-grown plants were irrigated with H2O or 300 mM Cl-. 24 h later, the 3rd and the 4th leaves were infiltrated with Mock (10 mM MgSO4), or pathogenic agents, specifically DG3 (OD = 0.0005) or Pseudomonas syringae pv tomato (Pst) DC3000-GFP (OD = 0.01). (A) Representative phenotypes of leaves recorded 2 dpi with Mock or Pst DC3000-GFP infiltration, bar = 5 mm. (B) The proliferation of DC3000-GFP in leaves observed by fluorescence microscopy at 2 dpi, bar = 100 μm. (C) Representative phenotype of cell death visualized by trypan blue straining at 2 days post DG3 infection (dpi), bar = 1 mm. DG3 growth in inoculated leaves shown in the right panel. Leaf discs were taken at the indicated time (n = 6). (D) The reactive oxygen species (ROS) burst induced with 100 nM Flg22 or without Flg22 (Mock). At 24 h after chloride salt irrigation, leaf discs from the 3rd and 4th leaves were collected and prepared for Flg22-induced ROS detection (for Flg22 induction, n = 5; for Mock, n = 3). (E) Relative transcript level of FRK1 (At2g19190) upon Flg22 induction. RNA samples were collected 3 h post infiltration with 100 nM Flg22. (F) Quantification of callose deposition upon Flg22 or Mock (10 mM MgSO4) induction post salt treatment. Leaves were collected 12 h post infiltration and stained with aniline blue (n ≥ 7). Callose deposition per visual field was counted via Image J software. Data are presented as mean values ± SEM. Statistical differences were analyzed by ANOVA post hoc tests (P < 0.05) and significantly different samples are indicated with different letters. These experiments were repeated at least twice using independent samples.
Salicylic acid (SA) is the major phytohormone supporting plant resistance to bacterial pathogens. The Arabidopsis transgenic line NahG, which overexpresses a bacterial SA hydroxylase, accumulates little or no SA, leading to plant susceptibility to pathogens. In the present study, consistent with previous reports, NahG increased plant susceptibility to DG3, as indicated by much more severe cell death and more robust bacterial proliferation compared with control plants (Figure 2C). Intriguingly, Cl- pretreatment also suppressed the cell death induced by DG3 and dampened bacterial growth in NahG plants (Figure 2C). However, the multiplication of DG3 in NahG-Cl- was still significantly higher than that in WT-Cl- treatment, indicating that the SA pathway played a role in Cl–mediated enhancement of plant resistance to DG3.
Previously, Singh et al. (2014) discovered that certain abiotic stresses prime PTI, thus enhancing plant resistance to a subsequent biotic challenge. To check whether Cl- treatment also changed the properties of PTI, we examined responses to a molecular pattern, the Flg22 epitope of flagellin. As shown in Figure 2D, Flg22 induced a rapid reactive oxygen species (ROS) burst in plants treated with Cl- and control plants treated with H2O. Cl- treatment did not significantly alter the relative expression level of the PTI marker gene FRK1 and Flg22-induced callose deposition (Figures 2E, F). These results suggested that, in the present study, the enhancement of plant resistance to bacterial infection was partially dependent on the SA pathway, but not PTI responses.
To decipher the effect of Cl- treatment on plant resistance, we next conducted transcriptome analysis. To this end, samples collected 9 hours post bacterial inoculation (hpi) were sequenced and analyzed (Supplementary Figure S4). Compared with H2O-Mock, C1–Mock treatment downregulated biotic resistance–related genes (Supplementary Figures S4B, D). This is consistent with a report that salt treatment can suppress the SA pathway (Yang et al., 2019). Consistent with the observed course of DG3 infection, the genes related to immune response, such as SA, jasmonate, and ethylene pathways, were strongly induced in plants infected with DG3 after H2O and Cl- pretreatment compared to H2O-Mock, Cl–Mock, respectively (Supplementary Figures S4B, D). Genes related to response to toxic substance and benzene-containing compound metabolic process were upregulated in Cl–DG3 compared with H2O-DG3, suggesting these two processes might take part in the mechanism by which Cl- enhances plant disease resistance.
Next, we investigated whether Cl- irrigation could enhance plant resistance to a necrotrophic pathogen, the fungi Botrytis cinerea. At 24 h after Cl- irrigation, a suspension of B. cinerea spores was dropped onto the leaves. As shown in Figures 3A, B, Cl- irrigation greatly suppressed the lesions induced by B. cinerea infection. These results show that Cl- irrigation can efficiently enhance plant resistance to necrotrophic pathogen infection.
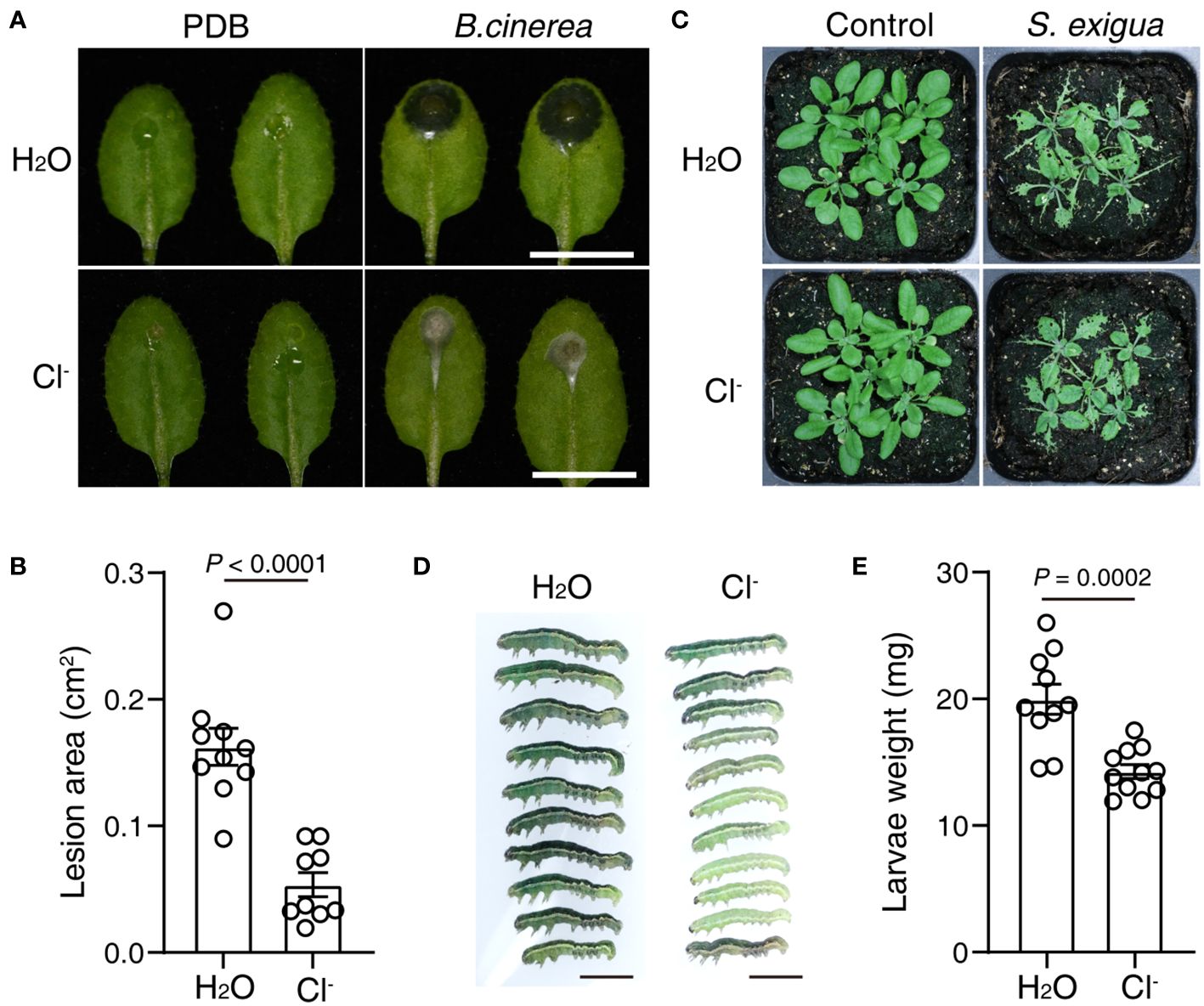
Figure 3 Cl- treatment promotes plant resistance to B. cinerea and an herbivorous insect. Three-week-old soil-grown plants were irrigated with H2O or 300 mM Cl-. 24 h later, the leaves were drop-inoculated with 5 μL Mock (PDB), or 5 μL 1×105 suspension of B. cinerea spores. (A) Photographs of representative leaves recorded 2 days post mock or spore infection, bar = 1 cm. (B) The quantification of lesion area in (A, n≥ 9). At 24 h after Cl- treatment, five plants were fed with or without five second instar larvae of S. exigua. (C) Representative photos of plants taken after 4 days of larvae feeding, bar = 5 mm. (D) The growth of larvae after 4 days of feeding, bar = 5 mm. (E) Quantification of the larvae weight. Statistical differences were analyzed by Student’s t-test. Data are presented as mean values ± SEM. These experiments were repeated twice using independent samples.
We also tested that if Cl- treatment could change plant resistance to herbivorous insect attack. At 24 h after Cl- irrigation, second-instar larvae of Spodoptera exigua were placed on WT plants. Intriguingly, larval growth was greatly stunted in WT plants irrigated with Cl- (Figures 3C, D). These results indicated that Cl- treatment also enhanced plant resistance to an herbivorous insect.
The crosstalk between abiotic and biotic stress responses is complicated and far from being understood. In the present study, our collective data show that an abiotic treatment, irrigation with a Cl- salt solution (MgCl2, KCl, and CaCl2), can remarkably enhance plant resistance to biotic stress, such as pathogen infection and herbivore attack. Furthermore, the enhanced plant resistance to a hemibiotrophic pathogen is only partially dependent on the SA pathway. We also demonstrated that Cl- is the major ion suppressing the accelerated cell death in the acd5 mutant.
As the main ions of salinity, Na+ and Cl- have been long considered toxic ions that impair plant growth and development in the field (Munns and Tester, 2008; Tavakkoli et al., 2011). However, exploration of the crosstalk between abiotic and biotic stresses reveals that certain abiotic stresses or inorganic ions can improve plant growth, nutrient utilization, and plant responses to biotic stresses (Wang et al., 2013; Singh et al., 2014; Geilfus et al., 2015; Wege et al., 2017; Zhang et al., 2017; Franco-Navarro et al., 2019; Yang et al., 2019; Rosales et al., 2020). Cl- salt treatment can change the morphology of leaf cells and reduce stomatal density, thus increasing water use efficiency, and improve photosynthetic performance, possibly by stimulating chloroplast biogenesis (Franco-Navarro et al., 2019). Cl- also facilitates the utilization of NO3- in plants, which provides a new guideline for developing agricultural fertilizers with an optimal NO3-/Cl- ratio to reduce excessive NO3- use (Rosales et al., 2020). Here, we discovered that Cl- can also suppress the accelerated cell death in acd5 (Supplementary Figure S1). This indicates that, besides promoting plant growth and nitrogen use, Cl- also participates in cell death regulation in plants, possibly by provoking antagonistic crosstalk between the phytohormone abscisic acid and the SA pathway, as observed for NaCl treatment (Yang et al., 2019). Since Cl- channels and Cl- itself as a signaling ion are tightly related to cell proliferation and innate immunity in animal cells (Valdivieso and Santa-Coloma, 2019; Lüscher et al., 2020; Chen et al., 2022), we also suspect that Cl- may inhibit cell death in acd5 in a phytohormone-independent manner. Therefore, further research to decipher the molecular mechanism, for example by screening for suppressors of Cl–mediated suppression of cell death in acd5, should provide interesting insight.
Compatible or incompatible plant–microbe interactions show differing extents of host cell death. We found that Cl- salt treatment effectively suppressed the cell death caused by not only the hemibiotrophic pathogen DG3, which undergoes a compatible interaction with Arabidopsis, but also DG6, in which recognition of the effector protein avrRpt2 by plant defenses induces an incompatible interaction (Figure 1; Yao and Greenberg, 2006). Cl- also enhanced plant resistance to pathogen infection by suppressing bacterial multiplication in intercellular spaces, and this enhancement is partially dependent on the SA pathway but not PTI responses, especially those mediated by FLS2 (Figure 2). This result indicated that the mechanism by which Cl- treatment enhanced disease resistance differed from that resulting from anion channel mutants or stimulated by channel inhibitors (Guo et al., 2014a, 2014b). Moreover, in the absence the pathogen infection, the genes related to immune responses and defense metabolites were downregulated (Supplementary Figure S4D), which is consistent with the observation that salt treatment suppresses the SA pathway. In addition, this inhibitory effect ceased when plants encountered a pathogen infection. These data suggested that the effect of chloride salt on biotic stress-responsive genes can also be regulated by upstream signaling pathways that are triggered by other environmental clues. RNA sequencing data showed the genes related to responses to toxic substance and benzene-containing compound metabolic process were enhanced in Cl–DG3 compared with H2O-DG3 treatment (Supplementary Figure S4), indicating these two processes play a role in Cl- salt–induced plant biotic resistance response, although the molecular mechanism needs further elucidation.
Cl- treatment also protected plants from the necrotrophic fungi B. cinerea and the herbivore S. exigua, which involve jasmonic acid (JA) signaling (Figure 3). This broad-spectrum resistance and the observation that the SA and JA pathways undergo antagonistic crosstalk (Pieterse et al., 2012), suggest that Cl- treatment does not merely exploit phytohormone pathways to enhance plant biotic resistance. We suspect Cl- could stimulate the accumulation of defense metabolites that are toxic to necrotrophic fungi and insects, a hypothesis that will need further validation.
Screening for suppressors of Cl–suppressed cell death in acd5 may enable us to uncover the effect of Cl- on plant cell death, and we will also learn from experiments in mammalian and yeast systems. Identifying the defense metabolites that are differentially abundant in plants under the combined treatment of Cl- irrigation and biotic stress may also be helpful and intriguing. Taken together, this research provides new clues to study the crosstalk between abiotic and biotic responses and underscores the influences and biological functions of the inorganic ion Cl- on plant cell death and defensive mechanisms.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: BioProject, PRJNA1073023.
YY: Conceptualization, Data curation, Methodology, Writing – original draft, Formal analysis, Funding acquisition. CY: Data curation, Methodology, Writing – original draft. JZ: Methodology, Writing – original draft. LX: Supervision, Writing – original draft. NY: Conceptualization, Funding acquisition, Supervision, Writing – review & editing, Resources.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Guangdong Provincial Key Laboratory of Plant Resources (2020PlantKF05), the start-up funding from Jiaying University (2023RC49) to YY, and the National Natural Science Foundation of China (32070196) to NY.
We thank Dr. Jun Liu (Sun Yat-sen University) for helping the B. cinerea infection experiment. We thank Prof. Zhaojun Ding (Shandong University) for gifting the DC3000-GFP strain.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1385164/full#supplementary-material
Supplementary Figure S1 | Salts containing chloride ions suppress the cell death phenotype in acd5 plants. (A) Three-week-old soil-grown plants were irrigated with H2O or 300 mM NaCl, 300 mM KCl, 300 mM NaNO3, 300 mM KNO3, 150 mM Na2SO4, 150 mM K2SO4, or 150 mM CaCl2 once to soil capacity. The phenotypes were recorded 2 weeks later, bar = 1 cm, inner bar = 2 mm. This experiment was repeated twice with similar results using independent samples (n= 12). (B) Cl- pretreatment suppresses the cell death phenotype of the acd5 mutant. Three-week-old soil-grown plants were irrigated with H2O or 300 mM Cl- (60 mM KCl, 60 mM MgCl2, 60 mM CaCl2) once to soil capacity. The phenotypes were recorded 2 weeks later, bar = 1 cm. This experiment was repeated at least twice with similar results using independent samples (n= 12).
Supplementary Figure S2 | Cl- but not the cations is vital for the cell death suppression. Three-week-old soil-grown plants were irrigated with H2O, 300 mM Cl-, or a salt mixture (60 mM KNO3, 60 mM MgSO4, and 60 mM Ca(NO3)2). 24 h later, the 3rd and the 4th leaf were infiltrated with Mock (10 mM MgSO4), or Pma DG3 (OD = 0.001). (A) Representative leaves recorded 2 dpi with Mock or Pma DG3 infiltration (bar = 5 mm). (B) Representative pictures of cell death visualized by trypan blue straining. Samples were detached and stained at the indicated time points, bar = 1 mm. (C) Statistical analysis of cell-death lesions in (B, n ≥ 12). The relative cell death lesion area per leaf was quantified as a percentage of lesion area in leaf area photographed under a stereomicroscope and measured by Image J software. Data are presented as mean values ± SEM. Statistical differences were analyzed by ANOVA post hoc tests (P < 0.05) and significantly different samples are indicated with different letters. This experiment was repeated twice using independent samples.
Supplementary Figure S3 | Spray treatment with Cl- enhances plant resistance to DG3. Three-week-old soil-grown plants were sprayed with H2O, 100 mM, 200 mM, or 300 mM Cl- containing 0.02% Silwet-77. At 24 h later, the plants were sprayed with Mock (10 mM MgSO4 and 0.02% Silwet-77) treatment or Pseudomonas syringae pv maculicola (Pma) DG3 (OD = 0.05, 0.02% Silwet-77). Five plants were tested for each treatment. (A) Representative plant showing cell death visualized by trypan blue staining. Samples were harvested 2 days post DG3 spraying, bar = 1 cm. (B) The representative cell death phenotype of the 3rd and 4th leaves detached from plants in (A), bar = 5 mm. (C) The cell death lesions recorded by stereo microscopy, bar = 1 mm. (D) Statistical analysis of cell death lesions in (C). The cell death area per leaf was quantified as the ratio of lesion area under a stereomicroscope, and analyzed using Image J software. Data are presented as mean values ± SEM. Statistical differences were analyzed by ANOVA post hoc tests (P < 0.05) and significantly different samples are indicated with different letters. This experiment was repeated twice with similar results using independent samples.
Supplementary Figure S4 | Transcriptome analysis upon treatment with Cl- and DG3. Three-week-old soil-grown plants were irrigated with H2O or 300 mM Cl-. 24 h later, the 3rd and the 4th leaf were infiltrated with Mock (10 mM MgSO4) treatment, or DG3 (OD = 0.001). Samples were collected 9 h post DG3 infection for total RNA isolation and sequencing. Data analysis was based on Log FC ≥ 1 and P < 0.05. (A, C) Venn diagrams showing the numbers of genes upregulated (A) or downregulated (C) upon combined treatment with salt and infection with DG3. (B, D) Gene ontology and cluster analysis for the biological processes upregulated (B) or downregulated (D) upon treatment with salt and infection with DG3.
Bi, F. C., Liu, Z., Wu, J. X., Liang, H., Xi, X. L., Fang, C., et al. (2014). Loss of ceramide kinase in Arabidopsis impairs defenses and promotes ceramide accumulation and mitochondrial H2O2 bursts. Plant Cell 26, 3449–3467. doi: 10.1105/tpc.114.127050
Chen, L., Guan, W. J., Qiu, Z. E., Xu, J. B., Bai, X., Hou, X. C., et al. (2022). SARS-CoV-2 nucleocapsid protein triggers hyperinflammation via protein-protein interaction-mediated intracellular Cl- accumulation in respiratory epithelium. Signal Transduction Targeted Ther. 7, 255. doi: 10.1038/s41392-022-01048-1
Davenport, R. J., Munoz-Mayor, A., Jha, D., Essah, P. A., Rus, A., Tester, M. (2007). The Na+ transporter AtHKT1;1 controls retrieval of Na+ from the xylem in Arabidopsis. Plant Cell Environ. 30, 497–507. doi: 10.1111/j.1365-3040.2007.01637.x
Franco-Navarro, J. D., Rosales, M. A., Cubero-Font, P., Calvo, P., Alvarez, R., Diaz-Espejo, A., et al. (2019). Chloride as a macronutrient increases water-use efficiency by anatomically driven reduced stomatal conductance and increased mesophyll diffusion to CO2. Plant J. 99, 815–831. doi: 10.1111/tpj.14423
Geilfus, C. M., Mithofer, A., Ludwig-Muller, J., Zorb, C., Muehling, K. H. (2015). Chloride- inducible transient apoplastic alkalinizations induce stomata closure by controlling abscisic acid distribution between leaf apoplast and guard cells in salt-stressed Vicia faba. New Phytol. 208, 803–816. doi: 10.1111/nph.13507
Guo, W., Wang, C., Zuo, Z., Qiu, J. L. (2014a). The roles of anion channels in Arabidopsis immunity. Plant Signaling Behav. 9, e29230. doi: 10.4161/psb.29230
Guo, W., Zuo, Z., Cheng, X., Sun, J., Li, H., Li, L., et al. (2014b). The chloride channel family gene CLCd negatively regulates pathogen-associated molecular pattern (PAMP)-triggered immunity in Arabidopsis. J. Exp. Bot. 65, 1205–1215. doi: 10.1093/jxb/ert484
Hasegawa, P. M., Bressan, R. A., Zhu, J. K., Bohnert, H. J. (2000). Plant cellular and molecular responses to high salinity. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51, 463–499. doi: 10.1146/annurev.arplant.51.1.463
Huang, L. Q., Chen, D. K., Li, P. P., Bao, H. N., Liu, H. Z., Yin, J., et al. (2021). Jasmonates modulate sphingolipid metabolism and accelerate cell death in the ceramide kinase mutant acd5. Plant Physiol. 187, 1713–1727. doi: 10.1093/plphys/kiab362
Kong, X., Zhang, C., Zheng, H., Sun, M., Zhang, F., Zhang, M., et al. (2020). Antagonistic interaction between Auxin and SA signaling pathways regulates bacterial infection through lateral root in Arabidopsis. Cell Rep. 32, 108060. doi: 10.1016/j.celrep.2020.108060
Liang, H., Yao, N., Song, J. T., Luo, S., Lu, H., Greenberg, J. T. (2003). Ceramides modulate programmed cell death in plants. Genes Dev. 17, 2636–2641. doi: 10.1101/gad.1140503
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-△△CT) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lüscher, B. P., Vachel, L., Ohana, E., Muallem, S. (2020). Cl- as a bona fide signaling ion. Am. J. Physiol. Cell Physiol. 318, C125–C136. doi: 10.1152/ajpcell.00354.2019
Munns, R., Tester, M. (2008). Mechanisms of salinity tolerance. Annu. Rev. Plant Physiol. Plant Mol. Biol. 59, 651–681. doi: 10.1146/annurev.arplant.59.032607.092911
Pieterse, C. M., van der Does, D., Zamioudis, C., Leon-Reyes, A., Van Wees, S. C. (2012). Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 28, 489–521. doi: 10.1146/annurev-cellbio-092910-154055
Rosales, M. A., Franco-Navarro, J. D., Peinado-Torrubia, P., Díaz-Rueda, P., Alvarez, R., Colmenero-Flores, J. M. (2020). Chloride improves nitrate utilization and NUE in plants. Front. Plant Sci. 11, 442. doi: 10.3389/fpls.2020.00442
Singh, P., Yekondi, S., Chen, P. W., Tsai, C. H., Yu, C. W., Wu, K., et al. (2014). Environmental history modulates Arabidopsis pattern-triggered immunity in a HISTONE ACETYLTRANSFERASE1-dependent manner. Plant Cell 26, 2676–2688. doi: 10.1105/tpc.114.123356
Sirén, J., Välimäki, N., Mäkinen, V. (2014). Indexing graphs for path queries with applications in genome research. IEEE/ACM Trans. Comput. Biol. Bioinform. 11, 375–388. doi: 10.1109/Tcbb.2013.2297101
Tavakkoli, E., Fatehi, F., Coventry, S., Rengasamy, P., McDonald, G. K. (2011). Additive effects of Na+ and Cl- ions on barley growth under salinity stress. J. Exp. Bot. 62, 2189–2203. doi: 10.1093/jxb/erq422
Valdivieso, A. G., Santa-Coloma, T. A. (2019). The chloride anion as a signalling effector. Biol. Rev. 94, 1839–1856. doi: 10.1111/brv.12536
Wang, L. K., Feng, Z. X., Wang, X., Wang, X. W., Zhang, X. G. (2010). DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26, 136–138. doi: 10.1093/bioinformatics/btp612
Wang, Y., Chen, Z. H., Zhang, B., Hills, A., Blatt, M. R. (2013). PYR/PYL/RCAR abscisic acid receptors regulate K+ and Cl- channels through reactive oxygen species- mediated activation of Ca2+ channels at the plasma membrane of intact Arabidopsis guard cells. Plant Physiol. 163, 566–577. doi: 10.1104/pp.113.219758
Wege, S., Gilliham, M., Henderson, S. W. (2017). Chloride: not simply a ‘cheap osmoticum’, but a beneficial plant macronutrient. J. Exp. Bot. 68, 3057–3069. doi: 10.1093/jxb/erx050
Yang, Y. B., Yin, J., Huang, L. Q., Li, J., Chen, D. K., Yao, N. (2019). Salt enhances disease resistance and suppresses cell death in ceramide kinase mutants. Plant Physiol. 181, 319–331. doi: 10.1104/pp.19.00613
Yao, N., Greenberg, J. T. (2006). Arabidopsis ACCELERATED CELL DEATH2 modulates programmed cell death. Plant Cell 18, 397–411. doi: 10.1105/tpc.105.036251
Yu, G. C., Wang, L. G., Han, Y. Y., He, Q. Y. (2012). clusterProfiler: an R Package for comparing biological themes among gene clusters. Omics. 16, 284–287. doi: 10.1089/omi.2011.0118
Zeng, H. Y., Liu, Y., Chen, D. K., Bao, H. N., Huang, L. Q., Yin, J., et al. (2021). The immune components ENHANCED DISEASE SUSCEPTIBILITY 1 and PHYTOALEXIN DEFICIENT 4 are required for cell death caused by overaccumulation of ceramides in Arabidopsis. Plant J. 107, 1447–1465. doi: 10.1111/tpj.15393
Zhang, H., Zhao, F. G., Tang, R. J., Yu, Y., Song, J., Wang, Y. (2017). Two tonoplast MATE proteins function as turgor-regulating chloride channels in Arabidopsis. Proc. Natl. Acad. Sci. United States America 114, E2036–E2045. doi: 10.1073/pnas.1616203114
Zhang, W. W., Yang, H. Q., You, S. Z., Fan, S. L., Ran, K. (2015a). MhNCED3, a gene encoding 9-cis-epoxycarotenoid dioxygenase in Malus hupehensis Rehd., enhances plant tolerance to Cl- stress by reducing Cl- accumulation. Plant Physiol. Biochem. 89, 85–91. doi: 10.1016/j.plaphy.2015.02.012
Zhang, Y., Jin, X., Ouyang, Z., Li, X., Liu, B., Huang, L., et al. (2015b). Vitamin B6 contributes to disease resistance against Pseudomonas syringae pv. tomato DC3000 and Botrytis cinerea in Arabidopsis thaliana. J. Plant Physiol. 175, 21–25. doi: 10.1016/j.jplph.2014.06.023
Keywords: salt stress, cell death, broad spectrum resistance, salicylic acid, Arabidopsis
Citation: Yang YB, Yang C, Zheng JR, Xu LZ and Yao N (2024) Chloride salt enhances plant resistance to biotic stresses. Front. Plant Sci. 15:1385164. doi: 10.3389/fpls.2024.1385164
Received: 12 February 2024; Accepted: 15 May 2024;
Published: 04 June 2024.
Edited by:
Hua Lu, University of Maryland, United StatesReviewed by:
Abhay K. Pandey, North Bengal Regional R & D Center, IndiaCopyright © 2024 Yang, Yang, Zheng, Xu and Yao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liang-Zheng Xu, eHVsaWFuZ3poZW5nanl1QDEyNi5jb20=; Nan Yao, eWFvbmFuQG1haWwuc3lzdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.