- Key Laboratory of Superior Forage Germplasm in the Qinghai-Tibetan Plateau, Qinghai Academy of Animal Husbandry and Veterinary Sciences, Qinghai University, Xining, China
The TCP gene family members play multiple functions in plant growth and development and were named after the first three family members found in this family, TB1 (TEOSINTE BRANCHED 1), CYCLOIDEA (CYC), and Proliferating Cell Factor 1/2 (PCF1/2). Nitrogen (N) is a crucial element for forage yield; however, over-application of N fertilizer can increase agricultural production costs and environmental stress. Therefore, the discovery of low N tolerance genes is essential for the genetic improvement of superior oat germplasm and ecological protection. Oat (Avena sativa L.), is one of the world’s staple grass forages, but no genome-wide analysis of TCP genes and their roles in low-nitrogen stress has been performed. This study identified the oat TCP gene family members using bioinformatics techniques. It analyzed their phylogeny, gene structure analysis, and expression patterns. The results showed that the AsTCP gene family includes 49 members, and most of the AsTCP-encoded proteins are neutral or acidic proteins; the phylogenetic tree classified the AsTCP gene family members into three subfamilies, and each subfamily has different conserved structural domains and functions. In addition, multiple cis-acting elements were detected in the promoter of the AsTCP genes, which were associated with abiotic stress, light response, and hormone response. The 49 AsTCP genes identified from oat were unevenly distributed on 18 oat chromosomes. The results of real-time quantitative polymerase chain reaction (qRT-PCR) showed that the AsTCP genes had different expression levels in various tissues under low nitrogen stress, which indicated that these genes (such as AsTCP01, AsTCP03, AsTCP22, and AsTCP38) played multiple roles in the growth and development of oat. In conclusion, this study analyzed the AsTCP gene family and their potential functions in low nitrogen stress at the genome-wide level, which lays a foundation for further analysis of the functions of AsTCP genes in oat and provides a theoretical basis for the exploration of excellent stress tolerance genes in oat. This study provides an essential basis for future in-depth studies of the TCP gene family in other oat genera and reveals new research ideas to improve gene utilization.
1 Introduction
Transcription factors (TF), also known as trans-acting factors, are a class of protein factors that can bind to specific DNA sequences (cis-acting elements) in the upstream promoter region of target genes through their DNA binding domain (DBD) and have essential roles in regulating plant growth and development (Li et al., 2019). The TCP protein, as a group of plant-specific transcription factors, evolved from algae and bryophytes and is named after the earliest identified members of the family: TB1 (TEOSINTE BRANCHED 1) in maize, CYCLOIDEA (CYC) in Cynoglossum, and Proliferating Cell Factor 1/2 (PCF1/2) in rice (Cubas et al., 1999). All members of the TCP protein contain a highly conserved basic helix-loop-helix (bHLH) structure consisting of 59 amino acid residues and can be divided into two subclasses based on the structural differences of TCP, including Class I (TCP-P/PCF) and Class II (TCP-C/CIN, TCP-C/CYC and TB1) (Viola et al., 2023). The TCP gene family is widespread in the plant kingdom, and its family members can directly or indirectly bind to regulatory target genes, which regulate multiple processes of plant growth and development (Zhou et al., 2022). In recent years, with the completion of whole genome sequencing of various species, the identification and analysis of TCP family genes have been carried out one after another on some species, and some related studies on the functions of the TCP gene family have been done (Cubas et al., 1999). ZmTCP42 in maize (Zea mays) regulates some ABA or drought stress-regulated genes and is an essential positive regulator of drought tolerance (Ding et al., 2019). MaPCF10 and MaPCF13 of banana (Musa acuminata) may be functionally important in fruit development and ripening (Wang et al., 2022). SmTCP40 of passion fruit (Solanum melongena) was stimulated by the jasmonic acid analog methyl jasmonate and is involved in its pigment signaling (Si et al., 2023). DchTCP2 and DchTCP13 in Dendrobium chrysotoxum significantly affect lateral organ development (Huang et al., 2023). Negative regulation of the growth of BpTCP8, BpTCP14, and BpTCP19 meristems of Broussonetia papyrifera (Zhao et al., 2020). CsTCP14, CsTCP16, CsTCP18, CsTCP19, and CsTCP20 were identified as TCP transcription factors associated with drought stress response in Citrus sinensis (Liu D. H. et al., 2022). OsTCP19, identified in Oryza sativa, responds to drought and salt stress and is an essential node in cell signaling that cross-links stress and developmental pathways (Mukhopadhyay and Tyagi, 2015). Thus, the TCP family of related genes is extensively involved in plant growth and development processes and is essential in response to abiotic stress regulation.
Oat (Avena sativa L.), as an annual grain-feeding crop, belongs to the grass family and is grown in about 42 countries around the world due to their excellent traits such as coolness, high nutritional value, high-stress resistance, and good palatability (Singh et al., 2013). Oat germplasm is mainly grown on marginal lands to avoid competition with grain crops for arable land. As a result, oat are often exposed to unfavorable growing conditions such as high salinity, drought, extreme temperatures, and low nitrogen stress. Nitrogen, as one of the essential nutrients in plants, is widely used in current agricultural production because its application significantly promotes plant growth and increases crop yields (Liang et al., 2021). Statistics show that the total amount of nitrogen fertilizers produced globally to increase crop yields exceeds 1 million tons annually. Although the production of N fertilizers consumes only 1-2% of global energy consumption, the utilization rate of N fertilizers is less than 50% (Dobermann and Cassman, 2005). As the global population continues to increase, overuse of nitrogen fertilizers has triggered severe environmental problems such as environmental pollution and global warming. Therefore, without increasing the use of nitrogen fertilizers, improving the nitrogen utilization rate of pasture grasses and cultivating pasture grass species with high nitrogen utilization rates or low nitrogen tolerance are of great significance for the sustainable development of the pasture industry.
The publication of the whole genome information of oat provides theoretical primary data for identifying family genes and gene function-related studies at the whole genome level. In recent years, functional genomics-related studies of oats have become a hot spot. Currently, in the available analysis of transcription factors in oat, the gene families of GRAS (Pan et al., 2023), WRKY (Liu K. et al., 2022), and PYL (Mi et al., 2023) have been characterized. Many genes have been unearthed in resistance to abiotic stresses, such as salinity, drought, cold, and high temperature. Although the TCP gene family members were widely used for abiotic stress response in other plants, the AsTCP gene family has not been characterized, and its function in response to low nitrogen stress remains unclear.
In this study, we identified and analyzed the AsTCP gene family members at the whole genome level and systematically analyzed the physical and chemical properties, conserved motif analysis, phylogeny, chromosomal localization, covariance, and cis-acting elements of the TCP gene family members. Meanwhile, this study analyzed the expression patterns of TCP gene family members in different tissues based on the genetic validation of qRT-PCR under low nitrogen stress, which provides a theoretical basis for further in-depth research on the molecular mechanism of abiotic stress resistance of AsTCP genes and the mining of excellent oat genes.
2 Materials and methods
2.1 Identification and sequence analysis of AsTCP genes
The oat genome was obtained from the results of joint sequencing of units studied by PepsiCo and Corteva Agriscience (https://wheat.pw.usdaa.gov/jb/?data=/ggds/oat-ot3098-pepsico). Searching for TCP candidate genes from the oat genome, this study first explored and compared the structural domains in the oat genome using the HMMER software (Version 3.0, http://hmmer.org/). The TCP protein sequence of Arabidopsis was obtained from EnsemblPlants (http://ensemblgenomes.org/). Hidden Markov model (HMM) mapping of the structural domains of the Arabidopsis TCP gene family was searched based on the oat genome, and information about the structural domains of oat was extracted using the HMMER software. To demonstrate the existence of oat TCP proteins, TCP structural domains were identified online using NCBI’s Conserved Domain Database database (CDD, https://www.ncbi.nlm.nih.gov/) and Plam values to obtain oat TCP candidate sequences, and candidate sequences whose TCP structural domains were not conserved were deleted.
2.2 Phylogenetic analysis of AsTCP genes
To further explore the evolutionary relationship between Arabidopsis and oat, phylogenetic analyses were constructed in this study. Multiple sequence comparisons of TCP proteins from both species were performed using MEGA software (Kumar et al., 2018) (Version X, http://megasoftware.net/). We first explored the best model for tree building based on the “Parameter” coefficients in the “Model” column, and the DNA model predictions indicated that “WAG+G+F” was the best choice. Phylogenetic tree using maximum likelihood (ML) method (calibration parameter Bootstrap, repeated 1000 times). In the parameter setting, we execute the ML method (Where “Model/Method” selects “WAG with Freqs. (+F) model”; “Rates among Sites” select “Gamma Distributed (G)”; “Test of Phylogeny” select “Bootstrap method”; “Substitutions Type” select “Amino acid”; “No of Discrete Gamma Categories” set to 5; “No. of Bootstrap Replications” set to 1000; “Gaps/Missing Data Treatment” select “Partial deletion”; “Site Coverage Cutoff (%)” set to 95, and the rest of the parameters are set to default values), phylogenetic tree visualization using iTOL (https://itol.embl.de/).
2.3 Conserved motifs analysis of oat
To study the TCP characteristic structural domains in oat TCP sequences, in this study, multiple sequence comparison of AsTCP proteins was carried out using Jalview software (Version 2.10.5), with the parameters of the comparison process set to the default values, and the results of the comparison were colorized using BioEdit software (Hall et al., 2011). AsTCP conserved protein sequences were identified using the online tool MEME Suit (Bailey et al., 2009) (https://meme-suite.org/meme/tools/meme) and visualized using TBtools software. Physicochemical properties of oat TCP proteins were predicted using the ExPASy proteomics server, and their subcellular localization was predicted using Cell-PLoc online software (http://www.csbio.sjtu.edu.cn/bioinf/Cell-PLoc-2/).
2.4 Chromosomal location and homology analysis of AsTCP genes
The CDS sequences and gene sequences corresponding to all oat TCP genes were extracted from the oat genome files using TBtools, and the intron and exon structures of the AsTCP genes were predicted using the GSDS 2.0 (gene structure display server, http://gsds.gao-lab.org/) online web site Analysis. The position of the AsTCP gene corresponding to the chromosome was obtained from the GFF3 data of the oat genome (Chen H. et al., 2020). Gene localization on chromosomes was visualized using TBtools software (Chen C. et al., 2020). To explore the homology of TCP genes between oat and other plant species, we downloaded genome-wide data and gene annotation files from the online website EnsemblPlants (http://ensemblgenomes.org/) for Arabidopsis (Arabidopsis thaliana), rice (Oryza sativa), maize (Zea mays), wheat (Triticum aestivum), and barley (Hordeum vulgare) genome-wide data and gene annotation files. In this study, we analyzed the duplication events of the AsTCP genes using the One Step MCscanX function of TBtools software.
2.5 Cis-acting elements analysis of AsTCP genes
The cis-acting elements of the AsTCP gene family were predicted and analyzed using PLACE (http://www.dna.affrc.go.jp/PLACE/signalscan) online software using the bases 2000 bp upstream of the start codon of the gene family as the sequence, which was extracted by Tbtools software (Chen C. et al., 2020). The cis-acting elements of the AsTCP genes were visualized by the”Simple BioSequence Viewer” function of TBtools software.
2.6 Plant material and stress treatments
The oat variety selected in this experiment was “Jiayan No.2”, and the seeds were provided by the Qinghai Academy of Animal Science and Veterinary Medicine, Qinghai University. This experiment began on November 20, 2023, with full-grained seeds soaked in 75% ethanol for 1 minute for disinfection and then rinsed well with distilled water for germination. The oat seedlings with consistent growth were transferred to Hoagland nutrient solution (PH= 5.8) in 16 h light/8 h darkness with a light intensity of about 8000 lx. The seedlings were then grown and cultured at 28°C until the two-leaf stage, and the nutrient solution was replaced every two days. After seedlings grew to 3 weeks of age, we divided seedlings into two groups. One group was treated with Hoagland nutrient solution containing 1.25 mM NH4NO3 for low nitrogen stress (LN- Low nitrogen), and the other group was incubated with Hoagland nutrient solution holding 10 mM NH4NO3 (NN-normal nitrogen) (Wang Y. et al., 2023). The low N treatment time points were 7 and 14 days, with a control group at normal N levels. The second leaf from the top of each plant and a root sample from each plant were then taken, immersed in liquid nitrogen for quick freezing, and stored in a freezer at -80°C. Three independent biological replicates were performed, each with 3-6 individual plants.
2.7 Expression analysis of the AsTCP genes by qRT-PCR
Total RNA was extracted from oat root and leaf tissues under different treatment times using the MiniBEST Plant RNA Extraction Kit (Takara, China). Genomic DNA was removed by reverse transcription using PrimeScriptTM RT reagent Kit with gDNA Eraser (Takara), and first-strand cDNA was synthesized. cDNA concentration was determined using a Multiskan FC zymography (Thermo Fisher, USA) and uniformly diluted to 50 ng·µL-1 for qRT-PCR reactions. Specific primers were designed by Primer software (Version 5.0). The qRT-PCR analysis was performed using a LightCycler Fast Real-Time PCR system (Roche, Switzerland) with three technical replicates. The amplification reaction system consisted of 2 µL of diluted cDNA solution, 1 µL of forward and reverse primers, 12.5 µL of SYBR Premix Ex Taq II (RR047A, Takara), and 8.5 µL of sterilized water in a total volume of 20 µL. The reaction program consisted of 40 cycles of denaturation at 95°C for 30 s, denaturation at 95°C for 5 s, and annealing at 60°C for 30 s; 95°C for 15 s, 60°C for 30 s, and 95°C for 15 s. This study used the GAPDH gene reported by Tajti et al. as an internal reference gene (Tajti et al., 2021). This inner reference gene is identical to the studied species and showed largely stable expression levels in different tissues. To ensure the accuracy of the experimental results, three replications were performed for each sample (Table 1). The relative expression was calculated using the 2−(ΔΔCt) method (Livak and Schmittgen, 2001).
2.8 Statistical analysis
The data were organized using Microsoft Excel software, analyzed by ANOVA using SPSS 17.0 statistical software, and the least significant difference (LSD) method was used to compare the differences between groups of different data, with the level of significance set at α = 0.05. Graphing was performed using Origin Pro 2019b (Origin Lab Corporation, Inc. Northampton, Massachusetts, USA) software for graphing.
3 Results
3.1 Identification of AsTCP gene family members in oat
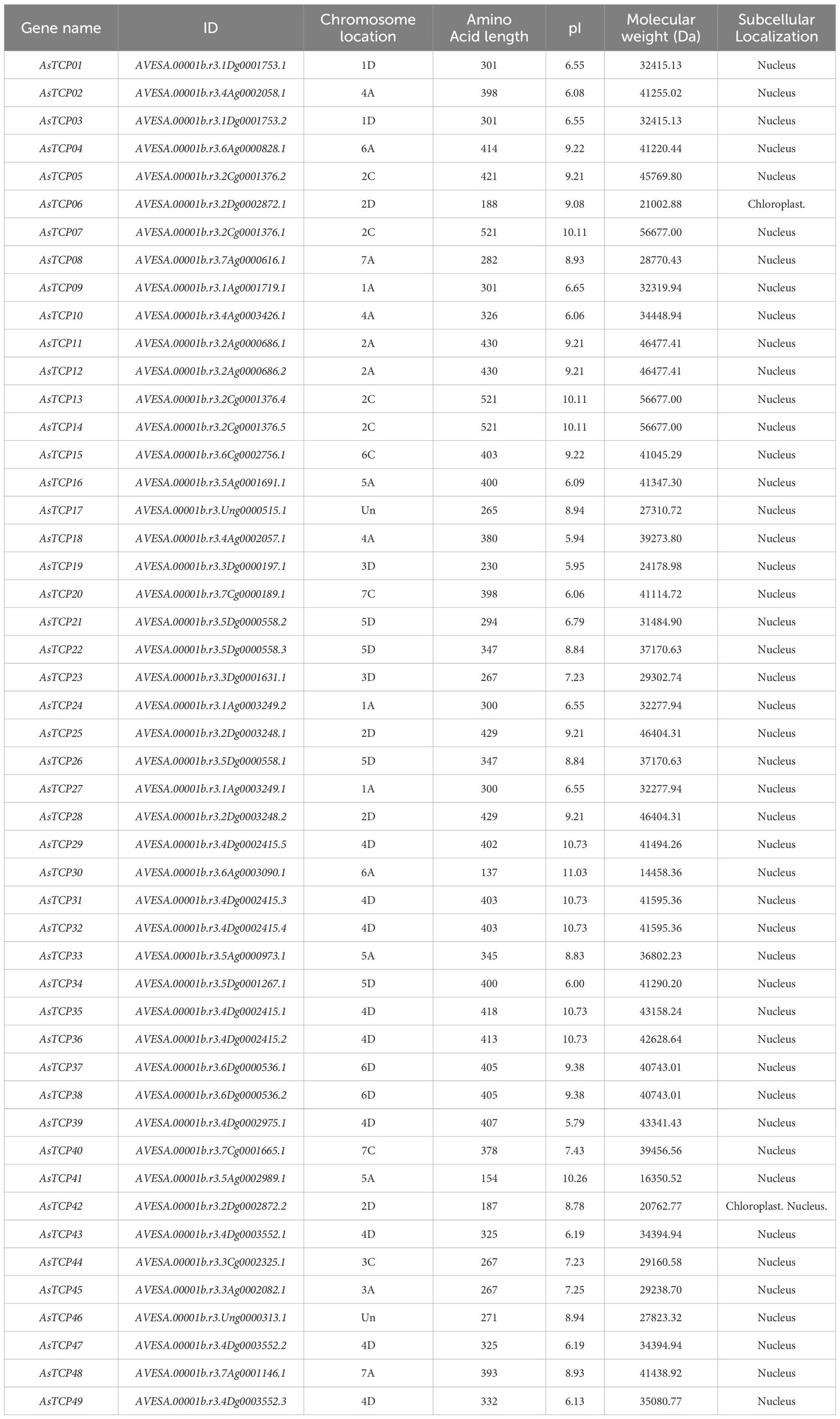
49 TCP proteins were identified in oat, named AsTCP01 to AsTCP49 based on their physical location on the chromosome (Supplementary Table S2), and analyzed for coding sequence length (CDS), protein molecular weight (Mw), isoelectric point (pI), and chromosomal location of the gene (Location) (Table 1). Among the 49 AsTCP proteins, the AsTCP41 protein was the smallest, with 154 amino acids, and the AsTCP07, AsTCP13, and AsTCP14 proteins were the largest, with 521 amino acids. The molecular weights of the proteins ranged from 14458.36 Da (AsTCP30) to 56677 Da (AsTCP07, AsTCP13, and AsTCP14), the pI values ranged from 5.79 (AsTCP39) to 10.73 (AsTCP29, AsTCP31, AsTCP32, AsTCP35, and AsTCP36) The mean pI value was 8.24, indicating that oat TCP protein was acidic.
3.2 Phylogenetic analysis and classification of AsTCP genes
To explore the phylogenetic relationships of oat TCP proteins, we constructed a phylogenetic tree based on the amino acid sequences of 49 AsTCP proteins and 24 AtTCP proteins (Supplementary Table S3) using the maximum likelihood (ML) method (Figure 1). Based on their homology with Arabidopsis TCP proteins, the 49 AsTCP genes were classified into two major branches and three subfamilies, of which the PCF subfamily included 14 AtTCP genes and 25 AsTCP genes; the CIN subfamily contained 8 AtTCP genes and 22 AsTCP genes; and the CYC/TB1 subfamily contained 3 AtTCP genes and 1 AsTCP genes (Figure 1). The PCF subfamily contained the most AsTCP genes, accounting for 51% of all AsTCP genes. The CYC/TB1 subfamily contained the minor AsTCP genes, accounting for only 2% of all AsTCP genes.
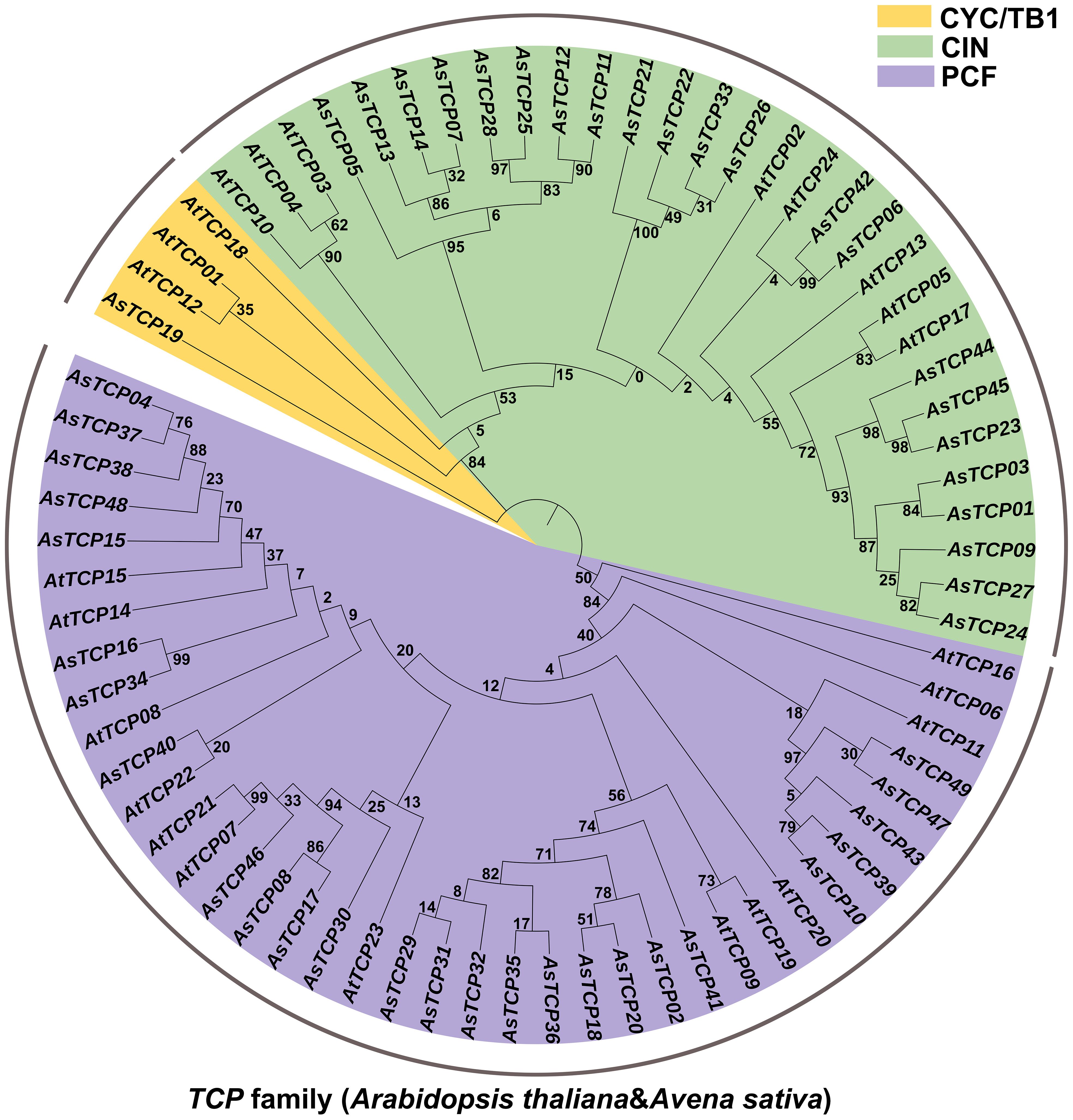
Figure 1 Phylogenetic tree of TCP proteins in Avena sativa and Arabidopsis thaliana. The TCP protein sequences of the two species were aligned by MEGA X with the MUS-CLE method, and the tree was built with the maximum likelihood (ML) method. The tree was further categorized into three distinct subfamilies in different colors.
3.3 Motif and gene structure analysis
The protein-conserved sequences and sequence logos of the oat TCP gene family members were analyzed using MEME online analysis software. A total of 10 conserved motifs (named motif 1~10) were identified, and more motifs were located at the C-terminal than at the N-terminal (Figure 2A). Most AsTCP proteins contained motif 1, motif 2, and motif 7. AsTCP18 did not have motif 3, while AsTCP06 and AsTCP44 only included motifs 1 and 2. Meanwhile, comparing and analyzing the results of the phylogenetic classification of the AsTCP genes, we found that the types, numbers, and order of protein motifs of the members of the same subfamily were very conserved. The closely related members mostly have similar motifs: The Class I group includes 1,2 and 4, while the Class II group includes 1,2,4, and 5.
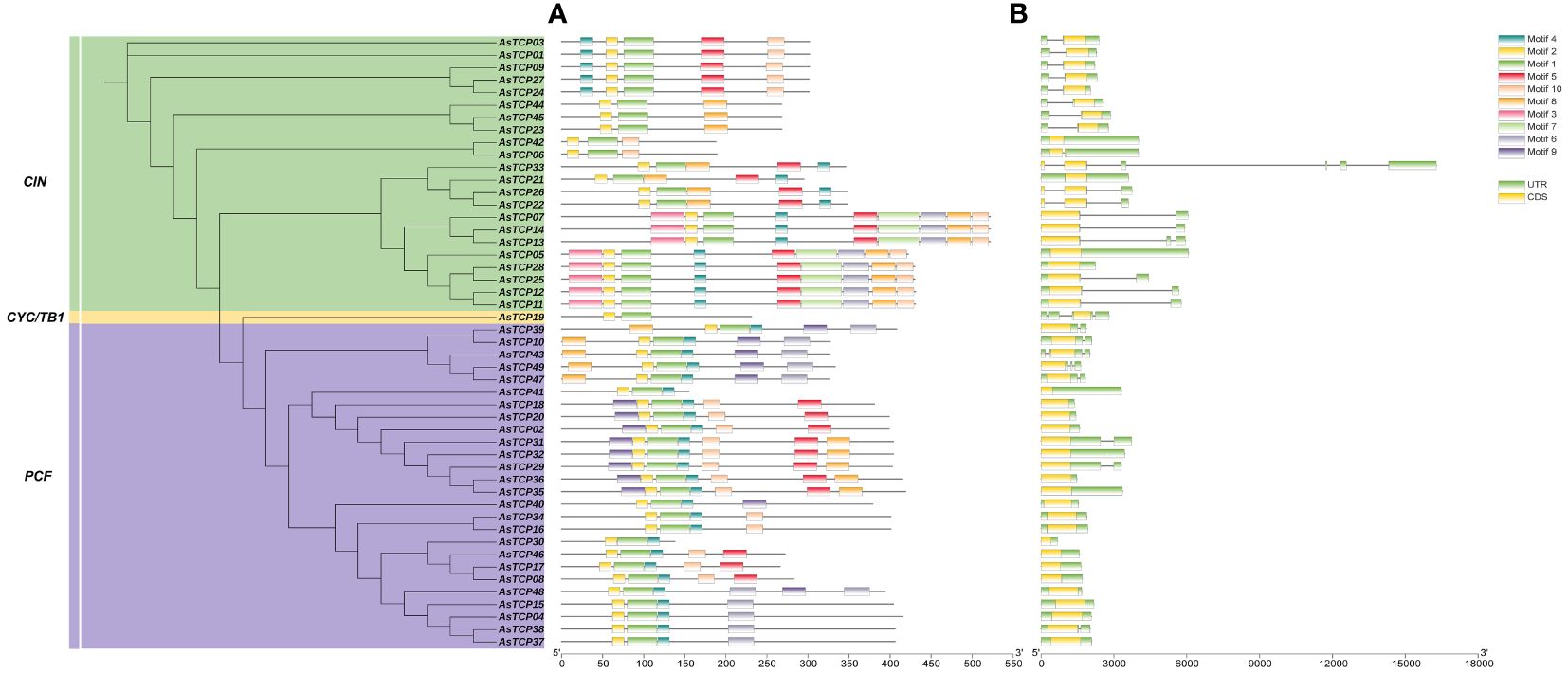
Figure 2 Structural analysis of AsTCP genes in oat. (A) The distribution of motifs in TCP proteins. (B) The exon-intron struc-ture of TCP gene.
Meanwhile, we found that motif 1 and 2 appeared in all AsTCP genes, indicating that these motifs are strongly conserved and have important evolutionary significance for AsTCP gene family members. To understand the structural composition of AsTCP genes, the exon and intron structure maps of AsTCP genes were obtained by comparing the genomic DNA sequences of AsTCP genes (Figure 2B). The results showed that some AsTCP genes (19 genes, 39%) did not contain introns; AsTCP05 had five introns; the number of exons in oat TCP genes ranged from one to two, with most of the AsTCP genes containing only one exon, and only AsTCP06, AsTCP22, AsTCP26, and AsTCP33 contained two exons. All 49 AsTCP genes included an uncompiled region (UTR). From the results, it is clear that members of the same subfamily have similar gene structures.
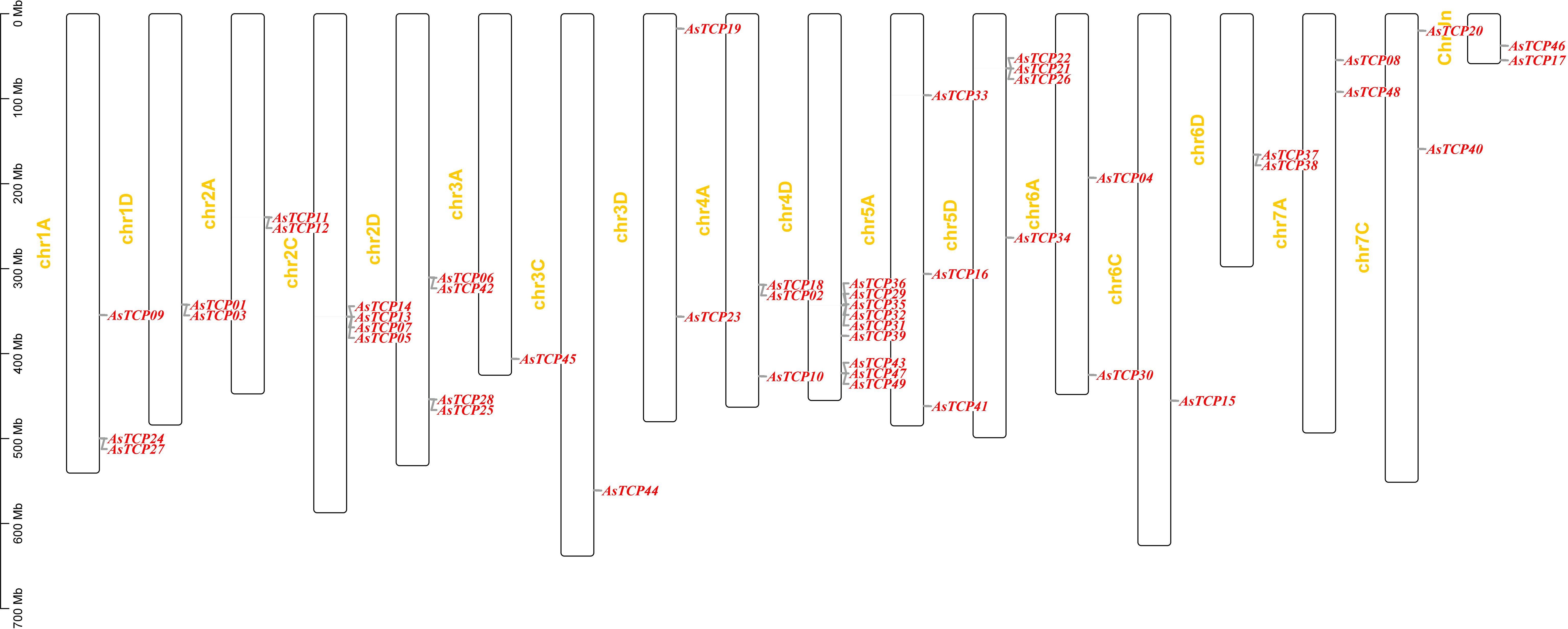
3.4 Chromosomal distribution and collinearity analysis of AsTCP genes
Based on the oat genome information, the distribution of all AsTCP genes on chromosomes was identified and analyzed (Figure 3). The results showed that 49 AsTCP genes were unevenly distributed on the 18 chromosomes of oat, among which nine genes were localized on chromosome 5A, which accounted for the most significant number of genes in the total number of genes, about 18.4%, followed by those on chromosome 2C (4 genes, about 8.2%), chromosome 2D (5 genes, about 8.2%) and chromosome 5D (4 genes, about 8.2%),. Only one gene was localized on chromosomes 3C, 3D, and 6C. In chromosomes, only one gene was localized (1 gene, about 2%). Tandem duplication events are essential for gene evolution and expansion. The results showed 23 regions of tandem duplication events in the oat genome (Figure 4, connected by red lines), indicating that these regions are hotspots for the distribution of AsTCP genes. The results suggest an evolutionary relationship between these AsTCP members, and their origins are related to chromosome duplication, implying that these genes may have similar functions. The results of collinearity analysis of AsTCP genes showed that genes with collinearity relationships are located within the same subfamily. To further explore the phylogenetic mechanisms of the oat TCP gene family, four representative comparative linear maps were constructed in this study to investigate the homology of TCP gene family members between oat and four representative species (Figure 5). The 4 species included 1 dicotyledon (Arabidopsis thaliana) and 3 monocotyledons (Hordeum vulgare, Triticum aestivum, and Oryza sativa). The results of covariance analysis showed that there were 7, 42, 39, and 94 homologous gene pairs between oat and Arabidopsis thaliana, Hordeum vulgare, Oryza sativa, and Triticum aestivum, respectively. Overall, the AsTCP genes consisted of more monocotyledonous covariant gene pairs, and the TCP genes were highly conserved evolutionarily between oat and Triticum aestivum, and the covariance analysis between oat and other species was critical for elucidating the evolution of the TCP genes.
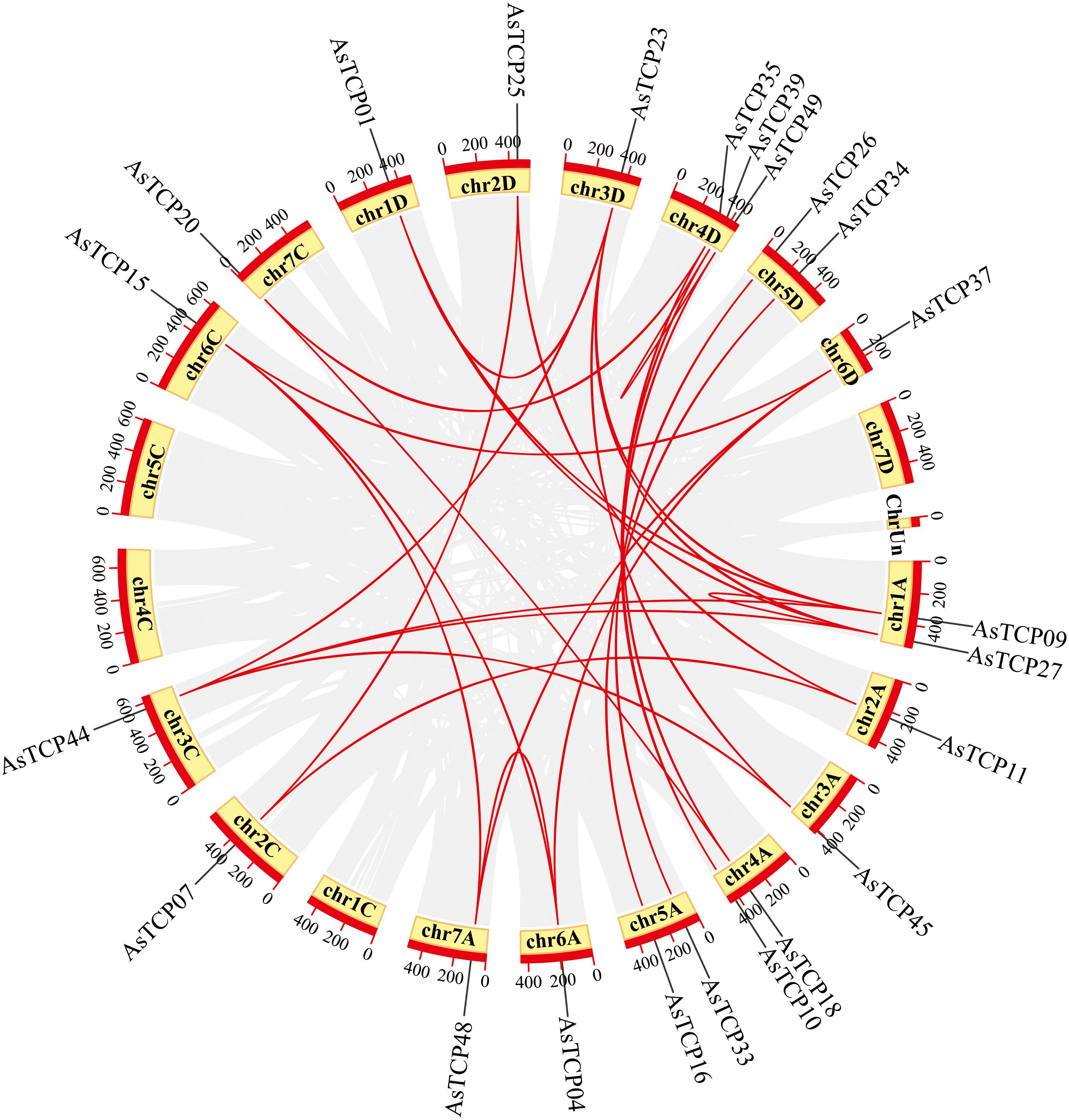
Figure 4 Chromosome localization of AsTCP duplicated genes in oat. The red lines represent the segmentally duplicated genes and the black bands represent the collinear block.
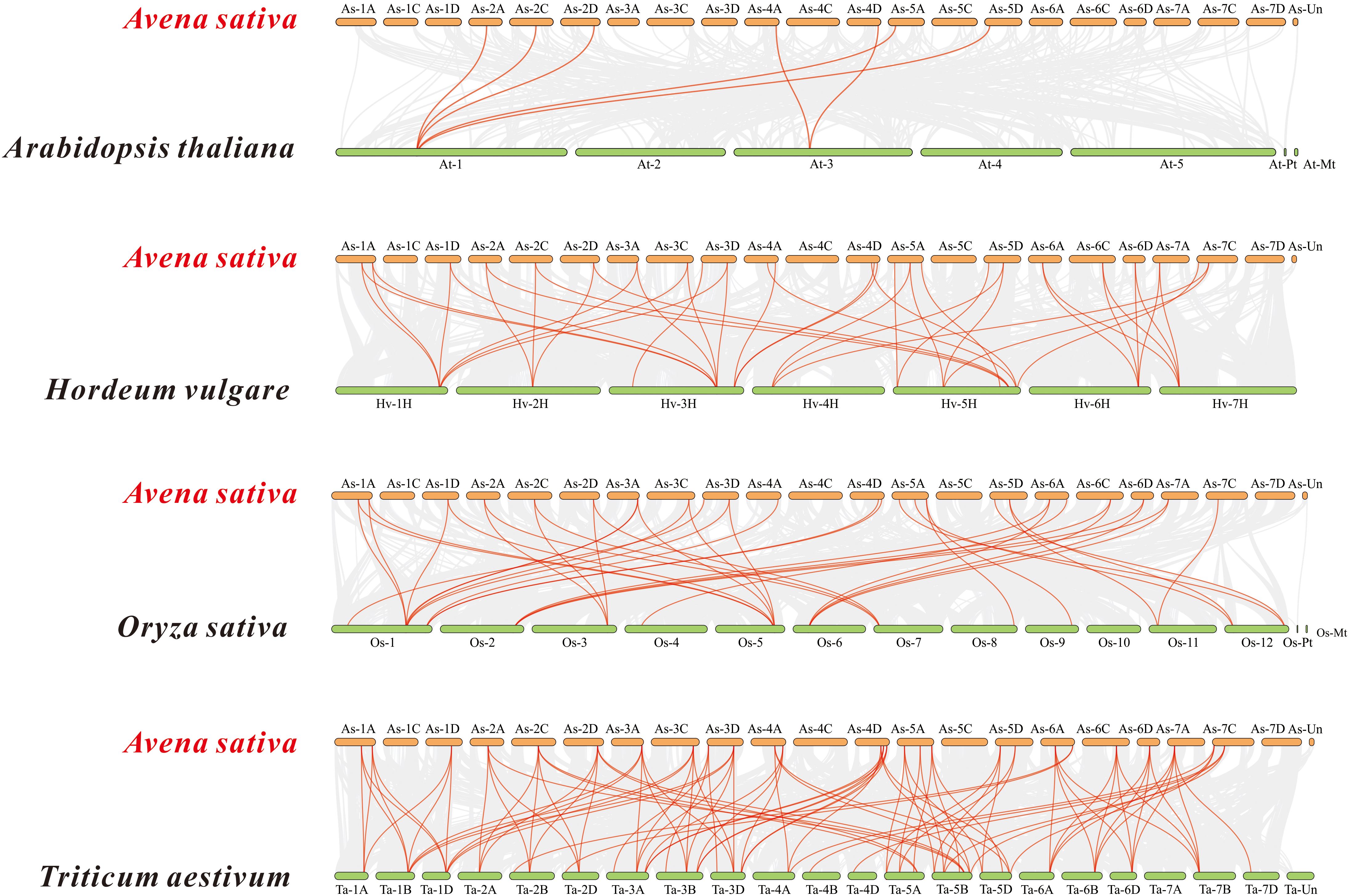
Figure 5 Synteny analyses of the TCP genes between oat and four representa-tive species. The collinear blocks within A. sativa and other specie genomes were displayed by the gray lines. The syntenic TCP gene pairs between oat and other species were highlighted with the red lines.
3.5 Cis-acting element analysis of AsTCP genes
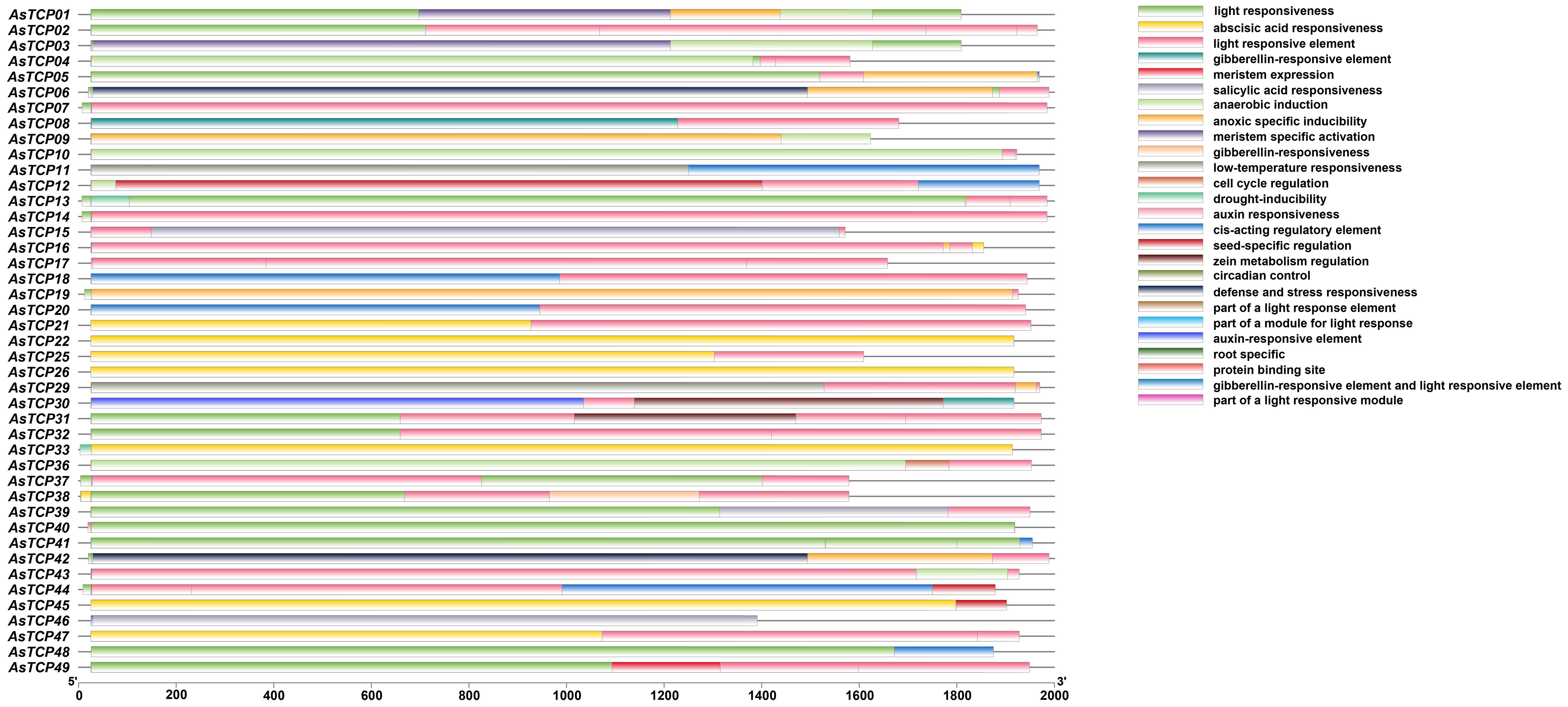
Cis-acting elements play essential roles in the transcriptional regulation of plant genes; they are specific binding sites involved in transcription initiation and regulation of protein binding. To predict the cis-acting elements contained in the promoter region of the AsTCP genes, the types of cis-elements and the potential functions of the contained elements of the AsTCP gene family members were analyzed using PLACE online software. The results showed (Figure 6, Supplementary Table S4) that the promoter regions of AsTCP gene family members contained a large number of cis-acting regulatory elements related to plant growth and development, hormone responses, and resistance, including light-responsive element (Sp1), abscisic acid-responsive element (ABRE), gibberellin-responsive element (TATC-box), low-temperature-stress-responsive element (LRE), and drought stress-responsive element (DRE/CRT) cis-acting elements. Different members of the AsTCP gene family contain similar cis-acting elements in their promoter regions regarding type and number. However, they also have some differences. For example, there are most AsTCP genes contain light-responsive elements (43 genes) and abscisic acid-responsive elements (41 genes), and some AsTCP genes contain drought-responsive elements (18 genes) and low-temperature stress-responsive elements (11 genes). The results showed that AsTCP19 of the CYC/TB1 subfamily contained a higher number of cis-acting elements, and the CIN subfamily’s AsTCP22 and AsTCP25 genes of the CIN subfamily contained fewer cis-acting elements. The cis-acting element analyses indicated that AsTCP genes play critical regulatory roles against various abiotic stresses in plants.
3.6 Expression profiles of AsTCP family members under low nitrogen stress
To further investigate the expression levels of AsTCP genes in different tissues under low-nitrogen environmental conditions and to elucidate the exact functions of AsTCP genes, we validated 9 AsTCP genes among the identified AsTCP genes in qRT-PCR experiments, and the genes selected covered each AsTCP gene subfamily (Figure 7). Gene primers for qRT-PCR experiments were shown in Supplementary Table S1. According to the results of qRT-PCR experiments, some AsTCP genes were significantly expressed in both leaf and root tissues. The results showed differences in the expression patterns of AsTCP genes in different subfamilies, and the expression patterns of AsTCP genes in the same subfamily were similar. Under low-nitrogen environmental conditions, AsTCP32 and AsTCP38, members of the PCF subfamily, reached the highest level of expression in leaf and root tissues on the seventh day of stress, followed by a gradual decrease in expression. In addition, we observed different expression patterns in members of the same subfamily. For example, in the CIN subfamily, the expression of AsTCP03 tended to be up-regulated from 0 days to 7 days and declined at 14 days; conversely, the expression of AsTCP05 was consistently up-regulated from 0 days to 14 days. qRT-PCR results indicated differences in the expression of some AsTCP genes in leaf and root tissues. For example, the expression of the AsTCP01 gene in the CIN subfamily was higher in leaf tissues than in root tissues. In summary, by analyzing the expression patterns of different AsTCP genes under low-nitrogen stress, we can select some representative AsTCP genes to further study their importance in resisting low-nitrogen stress.
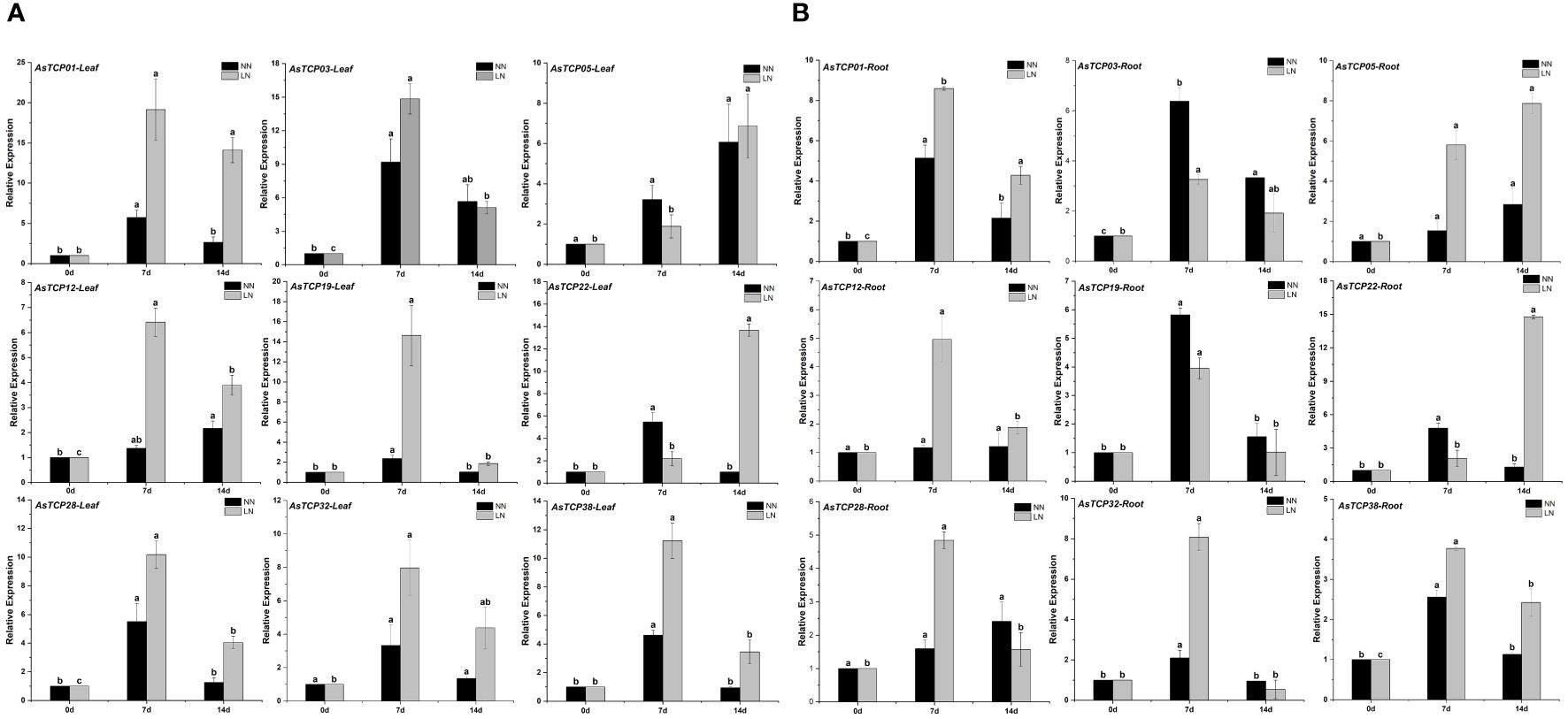
Figure 7 (A) The expression patterns of 9 AsTCP genes under low nitrogen stress in leaf were analyzed by real-time quantitative RT-PCR. (B) The expression patterns of 9 AsTCP genes under low nitrogen stress in root were analyzed by real-time quantitative RT-PCR. The relative expression levels which were normalized to GAPDH were determined by the comparative CT method (2−ΔΔCT) Three biological replicates were conducted for each experiment. This experiment uses the significant difference letter marking method, all the averages in order from large to small, and then in the largest average marked with the letter a, encountered with the difference between the average of the significant difference, marked with the letter b, and so on, where there is a the same marking of the letter that is, the difference is not significant, and where there is a different marking of the letter that is, the difference is significant.
4 Discussion
Recently, TCP gene family members have begun to be identified in different plant species with the publication of genome sequencing data from other plant species. In forages, the identification of TCP gene families in Medicago sativa (Zhang et al., 2023) and Dactylis glomerata (Wang C. et al., 2023) has also been completed. Oat is an annual crop in the grass family, with the advantages of stress resistance and high nutritional value. It is widely used in the grass and pasture industry (Fu and Yang, 2017). However, there are no reports on AsTCP gene family members at the genome-wide level. Therefore, the AsTCP genes were systematically analyzed using published oat genome-wide information and bioinformatics techniques in this study.
This study identified 49 AsTCP gene family members from the oat genome. Compared with plants such as Zea mays (46 genes), Triticum aestivum (66 genes), Hordeum vulgare (20 genes), and Cucumis sativus (27 genes), the number of AsTCP gene family members was on the high side which demonstrated that the number of TCP genes was closely related to the genome size and varied among species. Species with similar gene sequences to the AsTCP genes are mainly from grasses such as wheat and rice, suggesting that the AsTCP gene family members have similar evolutionary characteristics and affinities with the genes of these closely related species. Based on the tree-like branching structure of TCP proteins between Arabidopsis and oat, we further classified these AsTCP genes into three subfamilies: PCF, CIN, and CYC/TB1. In this study, we found that the AsTCP genes were unevenly distributed across 18 chromosomes in the oat genome (Figure 3), and it is noteworthy that chromosome 5A contained the most AsTCP genes (9 genes), which are mainly from the PCF and CIN subfamilies. Many studies have been conducted to review the functions of plant TCP genes, revealing their diversity: PCF subfamily-related genes promote the proliferation of young internodal cells and activate seed embryo growth potential (Tatematsu et al., 2008). The CIN subfamily regulates plant leaf, root, and petal development (Palatnik et al., 2003). Therefore, some critical AsTCP genes may play a role in regulating plant growth and development.
Earlier findings showed that gene duplication transferred TCP gene family members from prokaryotes to land plants. Our study found some AsTCP genes (19 genes, about 39%) did not contain introns. This structural feature is similar to that found in Camellia sinensis (87%) (Shang et al., 2022), Cymbidium goeringii (57%) (Liu D. et al., 2022), and Dactylis glomerata (18%) (Wang C. et al., 2023). Most AsTCP genes do not have introns, meaning their transcription times are shorter than those containing introns. Evidence shows these intronless genes can produce more rapid protein (Ma et al., 2020). This study explains why the AsTCP genes can respond quickly and develop a response mechanism under abiotic stress. With the increase in gene sequencing data, there is growing evidence that the ancestors of all eukaryotes had genes containing introns but that most eukaryotes experienced intron loss as their species evolved. The high proportion of genes that have lost introns in these plants (e.g., Avena sativa, Camellia sinensis, and Cymbidium goeringii) suggests that an intron loss event may have occurred during their evolution. Most transcription factors perform functional regulation by modulating protein interactions and DNA binding activity. These regulations mainly depend on modifying structural domains and motifs of transcription factors (Liu et al., 1999). In this study, 10 conserved motifs were identified in oat, and most AsTCP proteins contained all the conserved motifs, and the types, numbers, and order of conserved motifs of proteins encoded by members of the same subfamily were relatively consistent. Notably, there are only 2 motifs (motif 1 and motif 2) in AsTCP42, suggesting that these motifs may have indispensable functions in the CIN subfamily. This study indicates that these 2 motifs may have essential functions for the CIN subfamily. When 2 or more genes are sequentially arranged within 200 kb of each other on a chromosome, we call this a tandem duplication event (Holub, 2001). Compared with other species, the results for AsTCP genes showed no significant correlation between chromosome length and the number of AsTCP genes.
To explore the evolutionary relationship between AsTCP gene family members and different plant species. We selected three monocotyledonous and one dicotyledonous plant species and analyzed the homology of their TCP genes. The results showed an excellent homology correlation between oat and wheat. Overall, monocotyledonous plants’ TCP gene family members showed better homology concordance than the identified AsTCP genes. Therefore, it is reasonable to speculate that the covariance between TCP genes correlates with the evolutionary divergence trends of the plant species in which they were found. The AsTCP genes originated from the same ancestor as the TCP gene in wheat. The results of phylogenetic clustering in angiosperms may be nuanced among species but are generally consistent. This study suggests a high degree of diversity and complexity of TCP genes in angiosperms (Zhao et al., 2018). Our phylogenetic tree is consistent with the phylogeny of TCP proteins from a wide range of Gramineae species (Chai et al., 2017). Oat and wheat are similar in the number of TCP proteins, but the genomic data for oat is much larger, indicating many repetitive DNA sequences in their genomes (Zhao et al., 2018). From the chromosome location map of oat (Figure 4), we can see the tandem duplication of multiple AsTCP genes, which suggests a high rate of gene duplication in oat.
The promoter region of the AsTCP gene family members contains many action elements, which can be classified into three major categories according to their mechanisms of action: light-responsive, stress-responsive, and hormone-responsive action elements. The results of cis-acting element analysis showed that there were six types of light-responsive elements in the promoter regions of AsTCP gene family members. However, different subfamily members’ types and numbers of elements varied greatly. This result indicates that various subfamily members respond differently under other light conditions. The analyses in this study showed that the AsTCP gene family promoters contain a variety of adversity stress-acting elements and hormone response-acting elements. Hormones were also shown to act as a trans-acting factor capable of binding to hormone-responsive elements on promoters to regulate the transcriptional expression of target genes (Gunes et al., 2007). Hormones are not only directly involved in the growth and development of organisms but also act as signal transducers (e.g., ABA) in response to adversities such as low temperature, high temperature, and salt stress (Mundy et al., 1990). Therefore, it is reasonable to speculate that the expression of AsTCP gene family members may be regulated by hormones, which in turn are involved in scavenging excessive reactive oxygen species from their cells in vivo, increasing their resistance to abiotic stresses.
With the publication of plant whole genome sequencing results, TCP genes involved in abiotic stress response have been identified from a wide range of plants, including species such as Zingiber officinale (Jiang et al., 2023), Panicum virgatum (Zheng et al., 2019), Camellia sinensis (Shang et al., 2022) and Solanum melongena (Li et al., 2022). This study investigated the expression patterns of AsTCP genes in different tissues under low nitrogen stress conditions. By performing qRT-PCR experiments, we analyzed the expression patterns of AsTCP gene family members under low-nitrogen stress conditions. The experimental results showed that all AsTCP gene family subfamilies have members that can respond to low nitrogen stress. The AsTCP gene family plays an essential regulatory role in the response of plants to abiotic stresses (Parapunova et al., 2014). With the publication of plant whole genome sequencing results, TCP genes involved in abiotic stress response have been identified from a wide range of plants, including species such as Zingiber officinale (Jiang et al., 2023), Panicum virgatum (Zheng et al., 2019), Camellia sinensis (Shang et al., 2022) and Solanum melongena (Li et al., 2022). This study investigated the expression patterns of AsTCP genes in different tissues under low nitrogen stress conditions. The TaTCP9 identified were expressed throughout the development of young and immature spikes, and most TaTCP genes were expressed in multiple tissues and developmental stages. Expression in various tissues and developmental stages suggests that these TCP genes play an essential role in wheat development (Zhao et al., 2018), ZmTCP42 identified from maize is a critical TCP gene in maize and plays a crucial role in response to drought stress (Ding et al., 2019), so in-depth studies on the members of the AsTCP gene family members are necessary for mining their superior traits and implementing molecular breeding. Therefore, in-depth research on the members of the AsTCP gene family is of great significance for mining its particular characteristics and implementing molecular breeding. To tap the AsTCP genes responsive under low nitrogen stress, we conducted a preliminary screening and found some AsTCP genes with significantly up-regulated expression under NH4NO3 treatment. The results showed that the expression levels of AsTCP01, AsTCP03, AsTCP22, and AsTCP38 genes tended to increase during the gradual extension of low nitrogen stress. In leaf tissues, AsTCP01 and AsTCP03 showed higher expression levels after seven days of stress. On the other hand, AsTCP22 remained at relatively high expression levels after 14 days of stress. In root tissues, the expression patterns were similar except for AsTCP05 and AsTCP22, which showed the highest expression levels after 14 days of stress. The results of this study can further confirm that the AsTCP gene family has an essential role in response to low nitrogen stress. Therefore, a genome-wide survey of the AsTCP gene family will provide a necessary reference for the search for high-quality stress-resistant AsTCP genes, further deepen the understanding of the molecular mechanism of stress resistance in oat, and provide a theoretical basis for the development of excellent stress-resistant oat materials through genetic engineering technology.
In summary, in this study, members of the AsTCP gene family were identified at the genome-wide level using a bioinformatics approach, and their gene structures, conserved motifs, and evolutionary relationships were analyzed. Meanwhile, the potential regulatory mechanisms of the identified AsTCP genes in response to low nitrogen stress were revealed by determining the chromosomal distribution of the genes and their expression patterns in different tissues. In addition, the results of cis-acting element and gene expression analyses provide a basis for further investigation of AsTCP gene family members in abiotic stress response and other biological functions. However, the current study has only touched upon the preliminary characterization of AsTCP genes, and further functional validation studies are still needed to explore the roles of AsTCP genes in different biological processes in depth. The analysis of gene families relies on the quality of species genomes, and the quality of species genomic information affects the results of gene family analysis. As our research progresses, we will synthesize and analyze future updates of oat genomic information to sincerely resolve the theory of oat response to abiotic stress and lay the foundation for creating new highly resilient oat germplasm through genetic engineering technology.
5 Conclusion
In this study, we identified 49 AsTCP genes distributed unevenly on different chromosomes of the oat genome. Further categorizing these AsTCP genes, we classified them into three subfamilies: CIN, PCF, and CYC/TB1. We analyzed the gene structures and found that most AsTCP genes lacked introns, indicating their gene structures were very conserved. By analysis of conserved structural domains and motif patterns, we discovered that TCP gene family members of the same subfamily or branch have similar features, which may imply that they have identical gene functions. By qRT-PCR analysis, we confirmed that some AsTCP genes (AsTCP01, AsTCP03, AsTCP22, and AsTCP38) may play regulatory roles under low-nitrogen stress conditions, and these genes may become the focus of future research. In conclusion, this study comprehensively investigated the characterization of AsTCP genes. It provided a theoretical basis for further understanding the biological functions of AsTCP genes through the in-depth analysis of the screened particular AsTCP genes.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
JP: Data curation, Writing – original draft, Writing – review & editing. ZLJ: Methodology, Software, Writing – review & editing. XM: Writing – review & editing. LD: Data curation, Writing – review & editing. ZFJ: Project administration, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by the Qinghai Natural Science Foundation Program -Innovation Team(2022-ZJ-902).
Acknowledgments
We thank Dr. Yue Wang for his valuable help in sample collection.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1382790/full#supplementary-material
References
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Chai, W., Jiang, P., Huang, G., Jiang, H., Li, Y. (2017). Identification and expression profiling analysis of TCP family genes involved in growth and development in maize. Physiol. Mol. Biol. Plants. 23, 779–791. doi: 10.1007/s12298-017-0476-1
Chen, C., Chen, H., Zhang, Y., Thormas, H. R., Frank, M. H., He, Y. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, H., Zeng, Y., Yang, Y., Huang, L., Tang, B., Zhang, H. (2020). Allele-aware chromosome-level genome assembly and efficient transgene-free genome editing for the autotetraploid cultivated alfalfa. Nat. Commun. 11, 2494. doi: 10.1038/s41467-020-16338-x
Cubas, P., Lauter, N., Doebley, J., Coen, E. (1999). The TCP domain: a motif found in proteins regulating plant growth and development. Plant J. 18, 215–222. doi: 10.1046/j.1365-313X.1999.00444.x
Ding, S., Cai, Z., Du, H., Wang, H. (2019). Genome-wide analysis of TCP family genes in Zea mays L. identified a role for ZmTCP42 in drought tolerance. Int. J. Mol. Sci. 20, 2762. doi: 10.3390/ijms20112762
Dobermann, A., Cassman, K. G. (2005). Cereal area and nitrogen use efficiency are drivers of future nitrogen fertilizer consumption. Sci. China Ser. C: Life Sci. 48:745–758. doi: 10.1007/BF03187115
Fu, Y. B., Yang, M. H. (2017). “Genotyping-by-sequencing and its application to oat genomic research,” in Oat. Ed. Gasparis, S., Methods in Molecular Biology 1536, 169–187. doi: 10.1007/978-1-4939-6682-0_13
Gunes, A., Inal, A., Alpaslan, M. (2007). Salicylic acid induced changes on some physiological parameters symptomatic for oxidative stress and mineral nutrition in maize (Zea mays L.) grown under salinity. J. Plant Physiol. 164, 728–736. doi: 10.1016/j.jplph.2005.12.009
Hall, T., Biosciences, I., Carlsbad, C. (2011). BioEdit: an important software for molecular biology. GERF Bull. Biosci. 2, 60–61.
Holub, E. B. (2001). The arms race is ancient history in Arabidopsis, the wildflower. Nat. Rev. Genet. 2, 516–527. doi: 10.1038/35080508
Huang, Y., Zhao, X., Zheng, Q., He, X., Zhang, M. M., Ke, S., et al. (2023). Genome-wide identification of TCP gene family in dendrobium and their expression patterns in dendrobium chrysotoxum. Int. J. Mol. Sci. 24, 14320. doi: 10.3390/ijms241814320
Jiang, Y., Jiang, D., Xia, M., Gong, M., Li, H., Xing, H., et al. (2023). Genome-wide identification and expression analysis of the TCP gene family related to developmental and abiotic stress in ginger. Plants 12, 3389. doi: 10.3390/plants12193389
Kumar, S., Stecher, G., Li, M., Knyaz, C., Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. evolution. 35, 1547. doi: 10.1093/molbev/msy096
Li, D., Tang, X., Dong, Y., Wang, Y., Shi, S., Li, S., et al. (2022). Comparative genomic investigation of TCP gene family in eggplant (Solanum melongena L.) and expression analysis under divergent treatments. Plant Cell Rep. 41, 2213–2228. doi: 10.1007/s00299-022-02918-2
Li, J., Han, G., Sun, C., Sui, N. (2019). Research advances of MYB transcription factors in plant stress resistance and breeding. Plant Signaling Behavior. 14, 1613131. doi: 10.1080/15592324.2019.1613131
Liang, H., Tao, D., Zhang, Q., Zhang, S., Wang, J., Liu, L., et al. (2021). Nitrogen fertilizer application rate impacts eating and cooking quality of rice after storage. PloS One 16, e0253189. doi: 10.1371/journal.pone.0253189
Liu, K., Ju, Z., Jia, Z., Liang, G., Ma, X., Liu, W. (2022). Genome-wide identification and characterization of the oat (Avena sativa L.) WRKY transcription factor family. Genes 13, 1918. doi: 10.3390/genes13101918
Liu, D. H., Luo, Y., Han, H., Liu, Y. Z., Alam, S., Zhao, H. X., et al. (2022). Genome-wide analysis of citrus TCP transcription factors and their responses to abiotic stresses. BMC Plant Biol. 22, 1–14. doi: 10.1186/s12870-022-03709-3
Liu, L., White, M. J., MacRae, T. H. (1999). Transcription factors and their genes in higher plants: functional domains, evolution and regulation. Eur. J. Biochem. 262, 247–257. doi: 10.1046/j.1432-1327.1999.00349.x
Liu, D., Zhang, C., Zhao, X., Ke, S., Li, Y., Zhang, D. (2022). Genome-wide analysis of the TCP gene family and their expression pattern in Cymbidium goeringii. Front. Plant Science. 13. doi: 10.3389/fpls.2022.1068969
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2– ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Ma, L. T., Zhu, T., Wang, H. R., Zhou, H., Shao, L., Ding, Q. (2020). Genome-wide identification, phylogenetic analysis and expression profiling of the late embryogenesis-abundant (LEA) gene family in Brachypodium distachyon. Funct. Plant Biol. 48, 386–401. doi: 10.1071/FP20143
Mi, W., Liu, K., Liang, G., Jia, Z., Ma, X., Ju, Z. (2023). Genome-wide identification and characterization of ABA receptor pyrabactin resistance 1-like protein (PYL) family in oat. PeerJ 11, e16181. doi: 10.7717/peerj.16181
Mukhopadhyay, P., Tyagi, A. K. (2015). OsTCP19 influences developmental and abiotic stress signaling by modulatingABI4-mediated pathways. Sci. Rep. 5, 9998. doi: 10.1038/srep09998
Mundy, J., Yamaguchi-Shinozaki, K., Chua, N. H. (1990). Nuclear proteins bind conserved elements in the abscisic acid-responsive promoter of a rice rab gene. Proc. Natl. Acad. Sci. 87, 1406–1410. doi: 10.1073/pnas.87.4.1406
Palatnik, J. F., Allen, E., Wu, X., Schommer, C., Schwab, R., Carrington, J. C., et al. (2003). Control of leaf morphogenesis by microRNAs. Nature 425, 257–263. doi: 10.1038/nature01958
Pan, J., Zhou, Q., Wang, H., Chen, Y., Wang, Z., Zhang, J. (2023). Genome-wide identification and characterization of abiotic stress responsive GRAS family genes in oat (Avena sativa). PeerJ 11, e15370. doi: 10.7717/peerj.15370
Parapunova, V., Busscher, M., Busscher-Lange, J., lammers, M., Karlova, R., Bovy, A. G., et al. (2014). Identification, cloning and characterization of the tomato TCP transcription factor family. BMC Plant Biol. 14, 1–17. doi: 10.1186/1471-2229-14-157
Shang, X., Han, Z., Zhang, D., Wang, Y., Qin, H., Zou, Z. (2022). Genome-wide analysis of the TCP gene family and their expression pattern analysis in tea plant (Camellia sinensis). Front. Plant Science. 13. doi: 10.3389/fpls.2022.840350
Si, C., Zhan, D., Wang, L., Sun, X., Zhong, Q., Yang, S. (2023). Systematic investigation of TCP gene family: genome-wide identification and light-regulated gene expression analysis in Pepino (Solanum muricatum). Cells 12, 1015. doi: 10.3390/cells12071015
Singh, R., De, S., Belkheir, A. (2013). Avena sativa (Oat), a potential neutraceutical and therapeutic agent: an overview. Crit. Rev. Food Sci. Nutr. 53, 126–144. doi: 10.1080/10408398.2010.526725
Tajti, J., Pál, M., Janda, T. (2021). Validation of reference genes for studying different abiotic stresses in oat (Avena sativa L.) by RT-qPCR. Plants 10, 1272. doi: 10.3390/plants10071272
Tatematsu, K., Nakabayashi, K., Kamiya, Y., Nambara, E. (2008). Transcription factor AtTCP14 regulates embryonic growth potential during seed germination in Arabidopsis thaliana. Plant J. 53, 42–52. doi: 10.1111/j.1365-313X.2007.03308.x
Viola, I. L., Alem, A. L., Jure, R. M., Gonzalez, D. H. (2023). Physiological roles and mechanisms of action of class I TCP transcription factors. Int. J. Mol. Sci. 24, 5437. doi: 10.3390/ijms24065437
Viola, I. L., Gonzalez, D. H. (2023). TCP transcription factors in plant reproductive development: juggling multiple roles. Biomolecules 13, 750. doi: 10.3390/biom13050750
Wang, C., Feng, G., Xu, X., Huang, L., Nie, G., Li, D. (2023). Genome-wide identification, characterization, and expression of TCP genes family in orchardgrass. Genes 14, 925. doi: 10.3390/genes14040925
Wang, Y., Liu, K., Liang, G., Jia, Z., Ju, Z., Ma, X. (2023). Comprehensive evaluation of low nitrogen tolerance in oat (Avena sativa L.) seedlings. Agronomy 13, 604. doi: 10.3390/agronomy13020604
Wang, J., Wang, Z., Jia, C., Miao, H., Zhang, J., Liu, J., et al. (2022). Genome-wide identification and transcript analysis of TCP gene family in Banana (Musa acuminata L.). Biochem. Genet. 60, 204–222. doi: 10.1007/s10528-021-10100-8
Zhang, M., Qin, S., Yan, J., Li, L., Xu, M., Liu, Y. (2023). Genome-wide identification and analysis of TCP family genes in Medicago sativa reveal their critical roles in Na+/K+ homeostasis. BMC Plant Biol. 23, 1–17. doi: 10.1186/s12870-023-04318-4
Zhao, M., Peng, X., Chen, N., Shen, S. (2020). Genome-wide identification of the TCP gene family in Broussonetia papyrifera and functional analysis of BpTCP8, 14 and 19 in shoot branching. Plants 9, 1301. doi: 10.3390/plants9101301
Zhao, J., Zhai, Z., Li, Y., Geng, S., Song, G., Guan, J. (2018). Genome-wide identification and expression profiling of the TCP family genes in spike and grain development of wheat (Triticum aestivum L.). Front. Plant science. 9. doi: 10.3389/fpls.2018.01282
Zheng, A., Sun, F., Cheng, T., Wang, Y., Xie, K., Zhang, C. (2019). Genome-wide identification of members of the TCP gene family in switchgrass (Panicum virgatum L.) and analysis of their expression. Gene 702, 89–98. doi: 10.1016/j.gene.2019.03.059
Keywords: oat, TCP, genome-wide, expression profile, low nitrogen stress
Citation: Pan J, Ju Z, Ma X, Duan L and Jia Z (2024) Genome-wide characterization of TCP family and their potential roles in abiotic stress resistance of oat (Avena sativa L.). Front. Plant Sci. 15:1382790. doi: 10.3389/fpls.2024.1382790
Received: 06 February 2024; Accepted: 26 March 2024;
Published: 08 April 2024.
Edited by:
Douglas S. Domingues, University of São Paulo, BrazilReviewed by:
Milind B. Ratnaparkhe, ICAR Indian Institute of Soybean Research, IndiaSadaruddin Chachar, Zhongkai University of Agriculture and Engineering, China
Mohammad Irfan, Jamia Millia Islamia, India
Copyright © 2024 Pan, Ju, Ma, Duan and Jia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhifeng Jia, anpoZmVuZ0AxNjMuY29t
 Jing Pan
Jing Pan Zeliang Ju
Zeliang Ju Xiang Ma
Xiang Ma