- 1Department of Plant Sciences, Faculty of Biosciences (BIOVIT), Norwegian University of Life Sciences (NMBU), Ås, Norway
- 2Division of Biotechnology and Plant Health, Norwegian Institute of Bioeconomy Research (NIBIO), Ås, Norway
Phytophthora cactorum is a plant pathogenic oomycete that causes crown rot in strawberry leading to significant economic losses every year. To invade the host, P. cactorum secretes an arsenal of effectors that can manipulate host physiology and impair its defense system promoting infection. A transcriptome analysis was conducted on a susceptible wild strawberry genotype (Fragaria vesca) 48 hours post inoculation with P. cactorum to identify effectors expressed during the early infection stage. The analysis revealed 4,668 P. cactorum genes expressed during infection of F. vesca. A total of 539 secreted proteins encoded by transcripts were identified, including 120 carbohydrate-active enzymes, 40 RXLRs, 23 proteolytic enzymes, nine elicitins, seven cysteine rich proteins, seven necrosis inducing proteins and 216 hypothetical proteins with unknown function. Twenty of the 40 RXLR effector candidates were transiently expressed in Nicotiana benthamiana using agroinfiltration and five previously unreported RXLR effector genes (Pc741, Pc8318, Pc10890, Pc20813, and Pc22290) triggered cell death when transiently expressed. The identified cell death inducing RXLR effectors showed 31–66% identity to known RXLR effectors in different Phytophthora species having roles in pathogenicity including both activation and suppression of defense response in the host. Furthermore, homology analysis revealed that these cell death inducing RXLR effectors were highly conserved (82 - 100% identity) across 23 different strains of P. cactorum originating from apple or strawberry.
1 Introduction
Phytophthora cactorum is a devastating soil-borne oomycete pathogen that infects more than 200 plant species from 154 genera, including some of the most valuable horticultural plants such as apple, pear, and strawberry (Erwin and Ribeiro, 1996; Hantula et al., 2000; Sanchez et al., 2019; Chen et al., 2023). The pathogen can persist as resting oospores in soil for many years, even without a host plant, and during extreme environmental conditions. Phytophthora cactorum is challenging to control even with the use of chemicals due to oomyceticide (fungicide) resistance and the inefficiency of chemicals to different life stages of the pathogen (Marin et al., 2021; Ali et al., 2022). In the strawberry host, P. cactorum causes crown rot and leather rot, limiting the plant growth and quality of strawberry fruits, respectively, resulting in significant economic losses worldwide (Ellis and Grove, 1983; Stensvand et al., 1999). Crown rot symptoms include brown necrotic lesions in the rhizome (crown), which in severe cases result in wilting of the whole plant, whereas the leather rot affects the strawberry fruits, imparting an off-flavor taste and a pungent smell (Maas, 1998).
To establish an infection in the host plant, P. cactorum secretes an arsenal of effector proteins with diverse functions (Armitage et al., 2018; Gogoi et al., 2023b). These effectors are localized in the plant apoplastic spaces (apoplastic effectors) or translocate into the plant cell cytoplasm and diverse subcellular locations (cytoplasmic effectors) to enhance host colonization (Wang S. et al., 2019; Boevink et al., 2020). Apoplastic effectors include cysteine rich proteins, different cell wall degrading enzymes, elicitins, enzyme inhibitors, lipases, necrosis inducing proteins, phytotoxins, and proteolytic enzymes (proteases, peptidases). The most well studied group of cytoplasmic effectors from plant pathogenic oomycetes are the RXLRs (Arginine-any amino acid-Leucine-Arginine) and Crinklers (CRNs for crinkling and necrosis) (Kamoun, 2006; Hardham and Cahill, 2010; Wang Y. et al., 2019). The RXLR effectors have a conserved N-terminal RXLR amino acid motif often linked with an EER motif that mediates the translocation of the effector proteins into the host plant cells (Rehmany et al., 2005; Bhattacharjee et al., 2006; Whisson et al., 2007; Liu et al., 2019). Once inside the host cell, RXLRs can function both as activators of defense or suppressors of plant immunity (Birch et al., 2008; Oh et al., 2010; Anderson et al., 2015).
Plants possess a two layered immune system that responds to invading pathogens and impedes their growth. The first layer of immunity, known as pattern-triggered immunity (PTI), is activated when specific epitopes, called microbe- or pathogen associated molecular patterns (MAMPs/PAMPs), are recognized by the plant’s pattern recognition receptors (PRRs) (Medzhitov and Janeway, 1997; Jones and Dangl, 2006). The second layer of immunity, referred to as effector-triggered immunity (ETI), involves host resistance proteins that interact either directly or indirectly with specific effectors secreted by the pathogen, activating several defense signaling pathways (Jones and Dangl, 2006; Dodds and Rathjen, 2010; Dou and Zhou, 2012; Giraldo and Valent, 2013). Both PTI and ETI activation result in transcriptional reprogramming of the defense-related genes, which lead to the production of reactive oxygen species (ROS), secondary metabolites, hydrolytic enzymes, phytohormones and pathogenesis-related proteins (Tsuda and Katagiri, 2010; Naveed et al., 2020). The activation of defense responses in the host plant via PTI and ETI can also initiate a hypersensitive response or localized programmed cell death that can restrict growth of biotrophic and hemibiotrophic pathogens including Phytophthora species (Naveed et al., 2020).
Identifying and studying effector genes is crucial for understanding their roles in the interaction with host plants. Previous transcriptomic studies of different life stages of P. cactorum (mycelium, sporangia, zoospores, cysts, germinating cysts), and during infection of Nicotiana benthamiana and strawberry have identified several candidate effector genes (Chen et al., 2014, 2018; Nellist et al., 2021). Furthermore, in silico analyses of the sequenced genomes of P. cactorum have predicted hundreds of effector genes including RXLRs and CRNs (Armitage et al., 2018; Yang et al., 2018; Nellist et al., 2021; Gogoi et al., 2023b). To uncover the effectors produced by P. cactorum during the early and important phase of strawberry infection, the transcriptome was studied 48-hours post-inoculation of the rhizome (crown) of the susceptible Fragaria vesca genotype NCGR1218. Twenty candidate RXLR effector genes identified in the transcriptome study were transiently expressed in N. benthamiana leaves to examine potential cell death inducing responses.
2 Materials and methods
2.1 Plant material and Phytophthora cactorum
Diploid strawberry Fragaria vesca genotype NCGR1218 (susceptible to Phytophthora cactorum) was clonally propagated from runners and maintained in a greenhouse with a 16-hour photoperiod at 18°C. Nicotiana benthamiana plants were grown from seeds and were kept in a growth chamber at 21°C with a 16-hour photoperiod. Phytophthora cactorum strain 10300 previously isolated from a crown rot infected strawberry plant (Fragaria × ananassa) (Armitage et al., 2018) was routinely cultured on vegetable juice (V8) agar plates (Ferguson and Jeffers, 1999) at room temperature (~21°C) in the dark. A zoospore suspension was prepared from one-week old culture plates as described by Eikemo et al. (2000). The zoospores released from sporangia were counted using a hemocytometer and the concentration was adjusted to 2 × 105 zoospores/ml for inoculation of strawberry plants. The plants were gently wounded in the rhizome (crown) with a sterile scalpel and inoculated with 2 ml of the zoospore suspension or water (mock/control). Four biological replicates, with each replicate consisting of four individual plants were used for the inoculation experiment as well as the control treatment with water. The rhizome samples were harvested 48 hours after inoculation, flash frozen in liquid nitrogen and stored at -80°C until RNA isolation. The 48 hour time point represents the early infection stage of P. cactorum based on a temporal expression study of defense related genes in the resistant and susceptible F. vesca genotypes as previously described (Toljamo et al., 2016; Chen et al., 2016a; Gogoi et al., 2023a). No visible symptoms were observed at the time of harvest. Some additional plants, both zoospore inoculated and water control, were kept up to four weeks post inoculation to study the disease progression. Wilting and necrotic lesions were observed in the P. cactorum inoculated plants after two weeks while no symptoms were observed in the control plants (Gogoi et al., 2023a).
2.2 RNA extraction, transcriptome sequencing and analysis
Total RNA was extracted from the inoculated strawberry rhizomes using the Spectrum™ Plant Total RNA Kit (Sigma-Aldrich, USA) according to the manufacturer’s instructions. On-column DNase digestion was performed for 30 minutes on the isolated RNA to remove traces of genomic DNA contamination (Sigma-Aldrich, USA), as described by Gogoi et al. (2023a). Sequencing libraries were prepared using the TruSeq™ stranded total RNA library prep Kit (Illumina), and sequencing was performed using four lanes on an Illumina HiSeq 3/4000 System (2 × 150 bp) at the Norwegian Sequencing Centre, Oslo, Norway. An average of 54.8 million high-quality trimmed reads (SD 5.3 million) were obtained from the four replicates in this study. Transcriptome assembly and expression analysis were carried out as previously described (Gogoi et al., 2023a). Briefly, the transcripts were de novo assembled instead of mapping to the reference genome (P. cactorum, assembly ASM1686465v1), to recover both F. vesca and P. cactorum transcripts. Transcripts were quantified using the pseudo-alignment method Kallisto (Bray et al., 2016), and the normalization of the transcript counts was performed in the CLC genomic workbench v11.01 (Qiagen, Aarhus, Denmark) using the transcripts per million (TPM) method (Wagner et al., 2012). The longest transcript isoforms obtained from the de novo assembly were assigned a P. cactorum gene ID using BLASTN against the sequenced genome of the P. cactorum strain 10300 (GCA_003287315.1_Pcac_10300_v1_cds_from_genomic), with an expectation value e < 10–10 as a threshold. Transcripts having ≥ 99% identity to the P. cactorum 10300 genome were selected for the downstream analysis. The transcripts were annotated using Blast2GO v5.0 (Götz et al., 2008). The full-length sequences of the proteins encoded by the genes were retrieved using NCBI protein sequences (GCA_003287315.1_Pcac_10300_v1_protein), and these were used for further analysis. The full-length protein sequences encoded by transcripts were analyzed using the STRING database V11.5 (Szklarczyk et al., 2021), with gene ontology (GO) classification for functional annotation and the Kyoto encyclopedia of genes and genomes (KEGG) to predict biological pathways.
2.3 Secretome prediction and in-silico functional analysis of effector proteins
Effector proteins must be secreted in order to reach their cellular targets at the intercellular interface between the plant and pathogen or inside the host cell (Torto et al., 2003), and therefore the proteins encoded by transcripts were analyzed for signal peptides using SignalP5 (Armenteros et al., 2019b). The proteins predicted to have a signal peptide were further analyzed for transmembrane domains using Phobius (Käll et al., 2004), mitochondrial transit peptides using TargetP2.0 (Armenteros et al., 2019a), and endoplasmic reticulum retention signals (KDEL/HDEL motif) (Stornaiuolo et al., 2003) using PROSITE-Scan (de Castro et al., 2006). All proteins with a signal peptide in their N-termini with no more than one transmembrane domain and no mitochondrial transit peptide or endoplasmic reticulum retention motif were considered to be secreted proteins of P. cactorum.
The predicted secreted proteins were examined for carbohydrate active enzymes (CAZymes) using HMMER: dbCAN3-sub tool (e-value < 1e-15; coverage > 0.35) in the dbCAN3 meta server (Huang et al., 2018), while CRN and RXLR effectors were identified using the effectR package in R v4.2.0 (Tabima and Grünwald, 2019).
Additionally, all proteins characterized as secreted were searched against the pathogen-host interaction database (PHI-base) 4.14 using BLASTP (e-value <1e-5; ≥50% query coverage; bit score ≥50; identity ≥30%). The PHI-base comprises experimentally validated genes associated with pathogen virulence, which can aid in uncovering the role of secreted proteins in pathogenicity or disease development (Urban et al., 2022). The PHI-phenotypes ‘increased virulence’, ‘lethal’, ‘loss of pathogenicity’, and ‘reduced virulence’ are produced as a result of a mutation or altered expression of a specific gene in the pathogen, while the PHI-phenotype ‘plant avirulence determinant’ represents the effector gene required for the recognition of a pathogen in resistant hosts (Urban et al., 2022).
2.4 Protein homology and substitution rates analyses of the RXLR effectors
The predicted protein structures of 39 of the 40 RXLR effectors detected in this study (PC110_g6139 was unavailable) were downloaded as PDB files from the AlphaFold Protein Structure Database (Varadi et al., 2022). The predicted structure of the RXLR effectors were aligned using template modeling (TM)-align (Zhang and Skolnick, 2005) and a TM-score matrix was constructed for topological structural similarity assessment of the protein structures. Protein pairs with a TM-score > 0.5 are considered to share the same global fold, while those with a TM-score < 0.5 do not (Xu and Zhang, 2010). Additionally, homologs of the five RXLR effectors that induced cell death in N. benthamiana were identified from 23 different strains (Supplementary Material S5) of P. cactorum using OrthoFinder v2.4.0 (Emms and Kelly, 2019). The protein sequences of the identified homologs for each RXLR were aligned using Clustal Omega (Madeira et al., 2022) and phylograms were constructed using the neighbor-joining method and visualized using PRESTO (http://www.atgc-montpellier.fr/presto).
The rate of synonymous (α) and nonsynonymous (β) substitutions in the homologs of cell death inducing RXLR effectors were calculated for the four RXLRs that had variation within available sequences using FUBAR (Fast, Unconstrained Bayesian AppRoximation) from the HyPhy software v2.5.29 (MP) (Murrell et al., 2013; Kosakovsky Pond et al., 2020). Amino acids sites with β > α and posterior probability > 0.9 were considered under positive (diversifying) selection, whereas β < α (with posterior probability > 0.9) were considered under negative (purifying) selection.
2.5 Cloning of P. cactorum RXLR effector genes and their transient expression in Nicotiana benthamiana
To understand the role of RXLR effectors in inducing plant immunity, RXLR candidate genes were cloned in a plant expression vector and were transiently expressed in N. benthamiana leaves. For cloning, cDNA was used as a template for amplification of the target RXLR genes. Briefly, one microgram of total RNA isolated from the infected rhizome samples of the susceptible strawberry genotype NCGR1218 was used for cDNA synthesis using the iScript™ cDNA Synthesis Kit (Bio-Rad, USA). The full-length genes without the signal peptide region were amplified in a 50 µl PCR reaction mix with Phusion® High-Fidelity DNA Polymerase (5 U/µl) (New England Biolabs, USA) using gene-specific primers (Supplementary Table S1). PCR was performed in the T100 Thermal Cycler (BioRad, USA) with an initial denaturation at 95°C for five minutes followed by 35 cycles of denaturation at 95°C for 30 seconds, annealing at 60°C for 30 seconds, and polymerization at 72°C for 1 minute with a final extension of 12 minutes at 72°C. The amplified products were gel purified using QIAquick Gel Extraction Kit (Qiagen, USA), cloned into the Gateway entry vector pDONR™/Zeo (Thermo Fisher Scientific, USA) by BP recombination, and subsequently moved into the plant expression vector pK7WG2 (Karimi et al., 2002) through an LR recombination reaction (Thermo Fisher Scientific, USA). The constructs were transformed into chemically competent Escherichia coli DH5α cells (Library Efficiency™ DH5α competent cells, Invitrogen, USA) using the heat-shock transformation method (Froger and Hall, 2007). Transformants were selected using Luria-Bertani agar (LA) plates containing 50 µg/ml of zeocin and 100 µg/ml of spectinomycin for BP and LR transformants, respectively, followed by colony PCR in a 20 µl reaction mix with AmpliTaq DNA Polymerase (5 U/µl) (Applied Biosciences, USA) and PCR conditions as described above with primer specific annealing temperature (Supplementary Table S1). The recombinant plasmids were isolated using the Qiagen™ Plasmid Mini Kit (Qiagen, Germany), and the target insert sequence was verified using Sanger sequencing at Eurofins Genomics (Germany). The freeze-thaw transformation method was used to introduce the recombinant plasmid constructs into chemically competent cells of Agrobacterium tumefaciens strain AGL1 (Weigel and Glazebrook, 2006). The transformed bacterial colonies were selected on LA plates containing spectinomycin (100 µg/ml), carbenicillin (50 µg/ml) and rifampicin (15 µg/ml) and confirmed by colony PCR using gene-specific primers (Supplementary Table S1). A suspension of A. tumefaciens with an OD600 of 0.8 prepared as described by Du et al. (2014) was infiltrated into the top 3rd to 5th leaf of 3–4 weeks old N. benthamiana plants. The two P. cactorum genes, Pc16451 (PC110_g16451) and Pc22254 (PC110_g22254), previously reported to induce cell death in N. benthamiana, as RXLR6 and RXLR27, respectively (Chen et al., 2014) were included as positive controls. In addition INF1, which is a known elicitor of cell death from P. infestans (Kamoun et al., 1997, 1998), was used as a positive control, while the empty vector pK7WG2 was used as a negative control. The expression of the recombinant RXLR genes in N. benthamiana were confirmed three days after the agroinfiltration by reverse transcription PCR (RT-PCR). Briefly, the cDNA was synthesized from the total RNA isolated from agroinfiltrated leaves and PCR was performed using 2 µl cDNA as a template, and primers targeting short fragments of the RXLR genes (Supplementary Table S1). RNA was used as a template for a negative control (-RT control).
2.6 Histochemical assays
Agroinfiltrated N. benthamiana leaves showing visible cell death were stained using trypan blue to confirm the cell death response, as trypan blue is readily taken up by dead cells but not by living cells (Keogh et al., 1980). Briefly, leaves were boiled for 5–10 minutes in the trypan blue staining solution [10 ml DL-lactic acid (90%), 10 ml phenol (equilibrated with 10 mM Tris-HCL, pH 8.0), 1 mM EDTA, 10 ml glycerol (98%), 10 ml distilled water, 20 mg trypan blue] diluted with 96% ethanol (1:1 v/v), until the green color of the leaf disappeared (Bach-Pages and Preston, 2018). The leaves were further incubated in the solution at room temperature for 30 minutes, and destained overnight in chloral hydrate solution (2.5 g/ml) on a rotary shaker (40 rpm). Additionally, 3, 3´- diaminobenzidine (DAB) and aniline blue staining (Bach-Pages and Preston, 2018) were performed to assess oxidative burst/H2O2 accumulation and callose deposition, respectively, 3 days after agroinfiltration. The insoluble brown precipitate formed after DAB staining is a result of a peroxidase dependent polymerization of DAB with H2O2, indicating H2O2 accumulation, which is connected to cell death response (Alvarez et al., 1998; Steffens et al., 2013). Callose depositions are formed by plants upon recognition of non-self components and these fluoresce under UV light after binding with aniline blue (Jin and Mackey, 2017).
3 Results
3.1 Transcriptome profile and functional classification
From our previous defense transcriptome study of Fragaria vesca genotypes inoculated with Phytophthora cactorum (Gogoi et al., 2023a), a total of 412,970 de novo assembled transcript isoforms were obtained. Blasting the longest isoforms of de novo assembled transcripts (308,070) to the P. cactorum strain 10300 genome (GCA_003287315.1) resulted in 4,808 transcripts, which represent ca 1.6% of the total transcripts, of which 4,665 had more than 99% identity to P. cactorum genes (Supplementary Material S1). Three candidate RXLR effector genes, Pc741 (PC110_g741), Pc12148 (PC110_g12148), and Pc19826 (PC110_g19826), with less than 99% nucleotide identity to the genome of P. cactorum strain 10300, were manually curated based on the RXLR motif and their expression during infection. These genes were included in the final list of P. cactorum genes, amounting to a total of 4,668 genes (Supplementary Material S1).
The 4,668 proteins encoded by the P. cactorum transcripts were classified using gene ontology (GO) categories, and 3,365 of them were represented in 848 ontologies (517 biological process, 188 molecular function, and 143 cellular component) while KEGG analysis identified 2,009 functional transcripts assigned to 78 pathways (Figure 1; Supplementary Material S2).
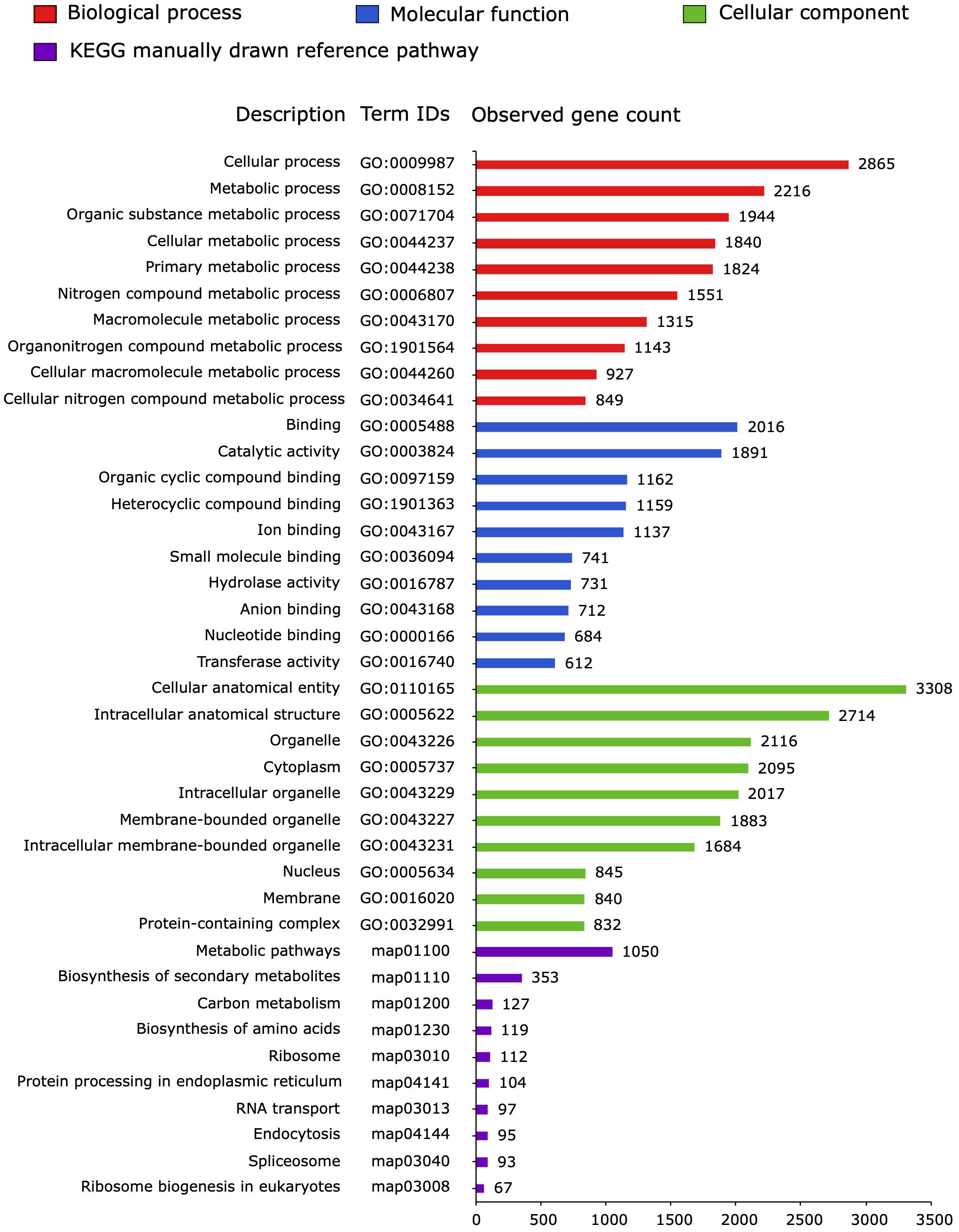
Figure 1 Gene ontology (GO) classification and Kyoto encyclopedia of genes and genome (KEGG) analysis of proteins encoded by transcripts of Phytophthora cactorum (accession GCA_003287315.1) detected in the rhizome (crown) of the susceptible Fragaria vesca NCGR1218, 48 hours after inoculation. The GO terms and KEGG pathways were categorized using the STRING V11.5 database (Szklarczyk et al., 2021). The top 10 most represented terms in each of the GO categories cellular component, molecular function, and biological process, as well as KEGG pathways are shown.
3.2 Secretome analysis of P. cactorum genes expressed during strawberry infection
Of the 4,668 proteins encoded by the detected transcripts, 539 (~12%) were predicted to have signal peptides for secretion and are thus referred to as secreted proteins (Supplementary Material S1). The 539 secreted proteins belonged to several effector families including 120 carbohydrate active enzymes (CAZymes), 40 RXLRs, 23 proteolytic enzymes, nine elicitins, seven cysteine rich proteins, seven necrosis inducing proteins and three crinklers (CRN) (Table 1; Supplementary Table S1). The 120 identified CAZymes belonged to 52 sub-families potentially targeting different substrates of the plant cell (Table 2). The majority of the sub-families belonged to glycoside hydrolases (GHs). Of the 40 RXLR effectors, 31 had complete RXLR and EER motifs while nine had only the RXLR motif (Table 3).
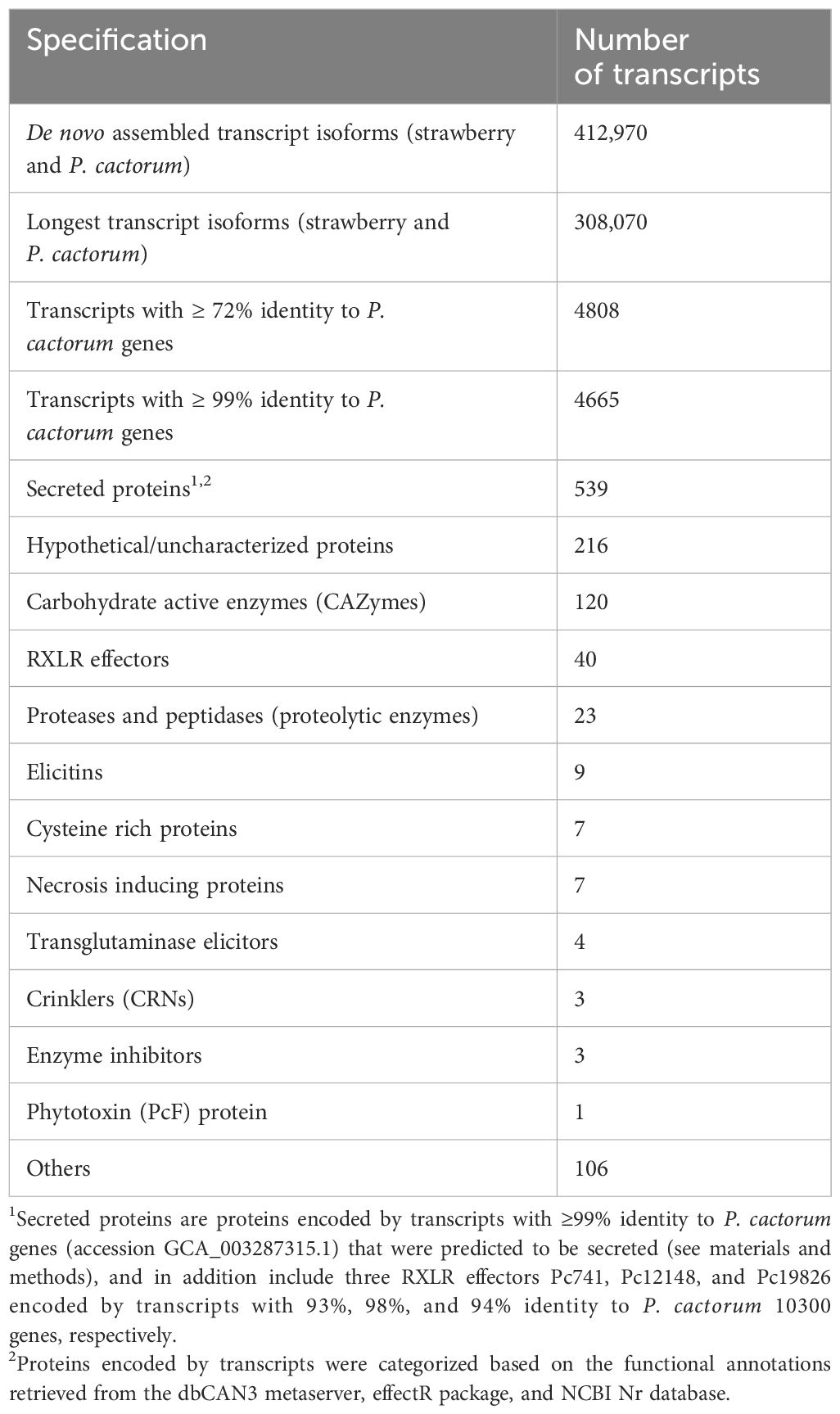
Table 1 Number and encoding properties of transcripts from Phytophthora cactorum detected 48 hours after inoculation into the crown (rhizome) of the susceptible Fragaria vesca NCGR1218.
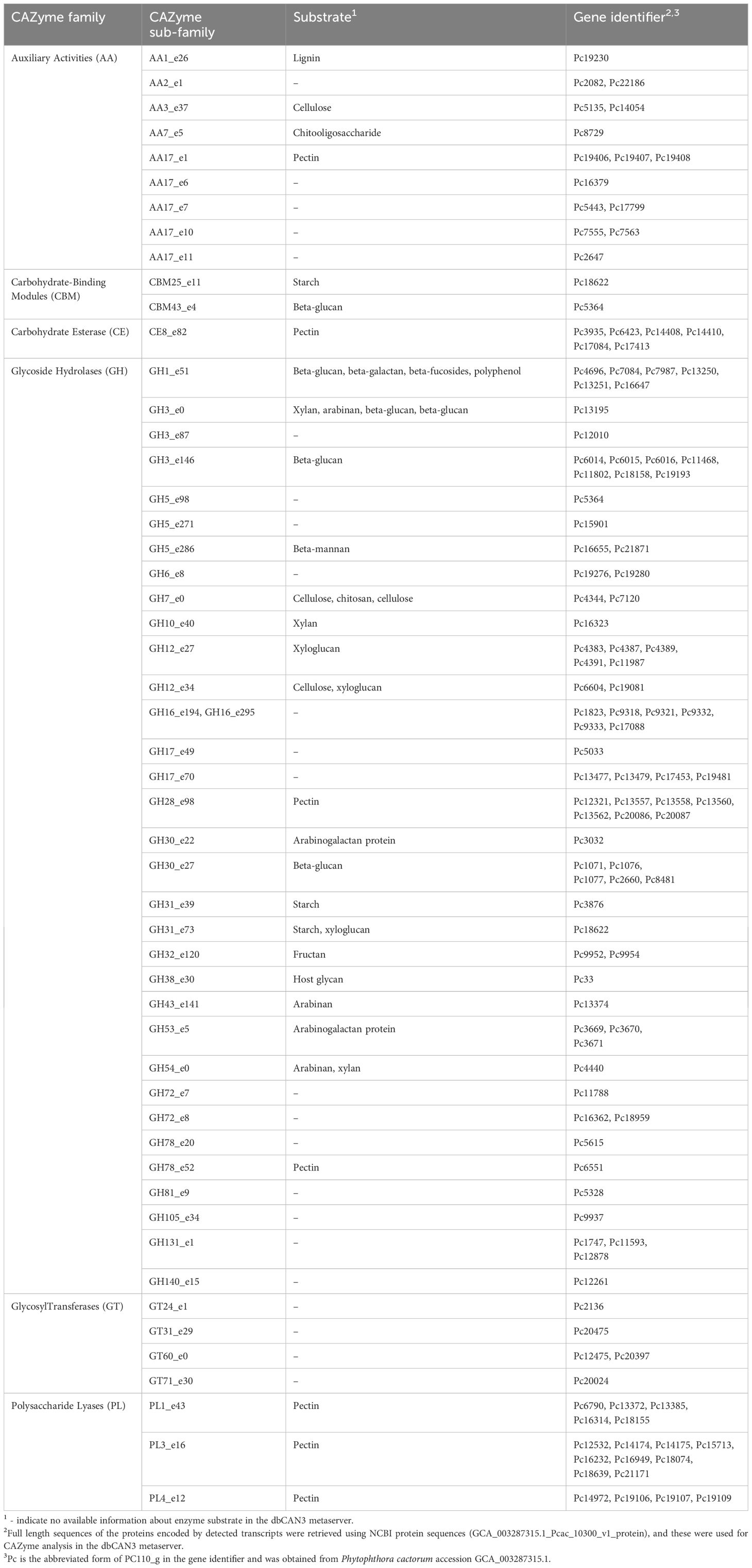
Table 2 Carbohydrate active enzymes (CAZymes) encoded by the transcripts detected 48 hours after inoculation with Phytophthora cactorum in the rhizome (crown) of the susceptible Fragaria vesca NCGR1218.
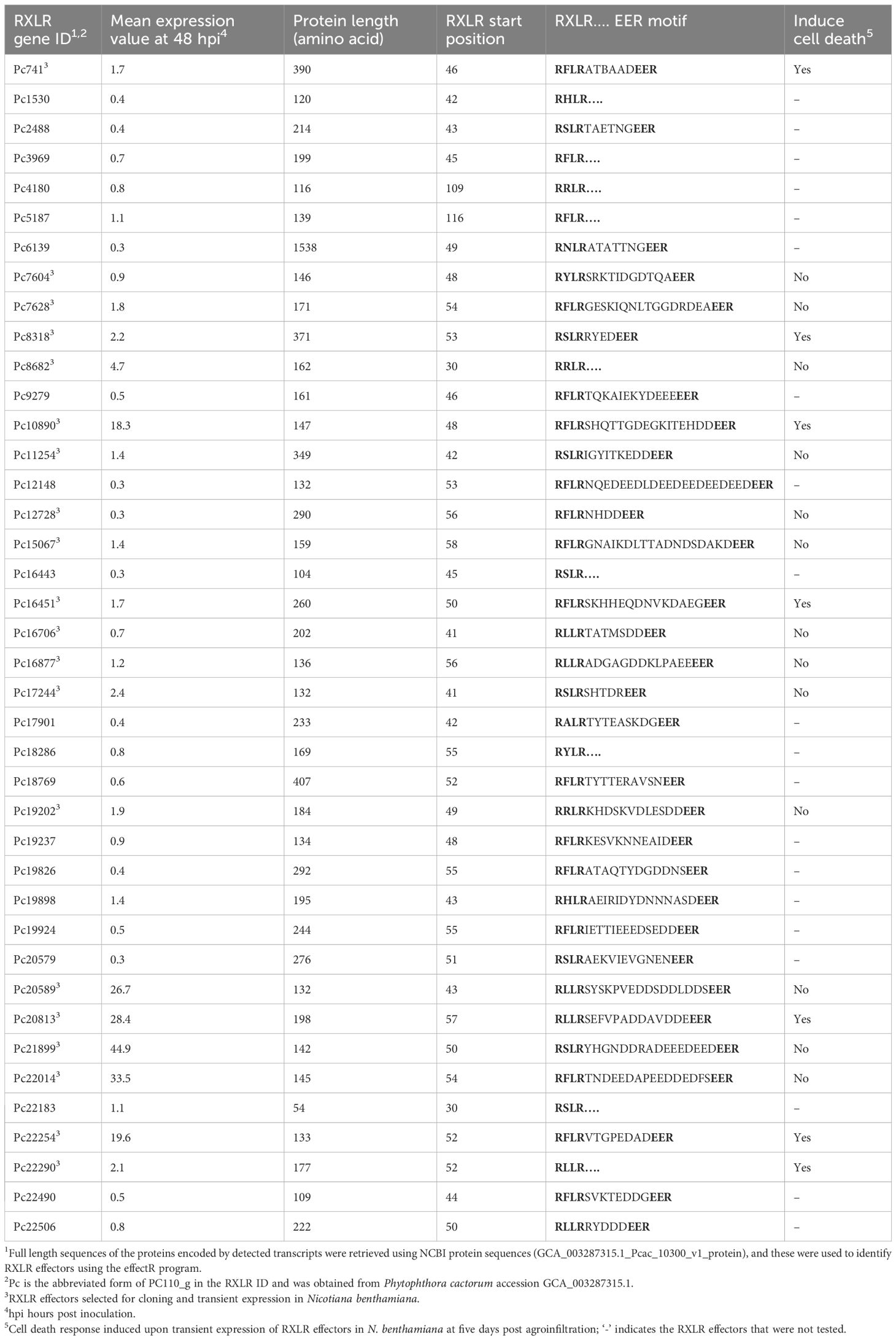
Table 3 RXLR effector candidates encoded by transcripts detected 48 hours after inoculation with Phytophthora cactorum in the rhizome (crown) of the susceptible Fragaria vesca NCGR1218.
In silico analysis of the secreted proteins of P. cactorum using the pathogen host interaction database (PHI-base) showed 129 proteins having 30–99% sequence identity with proteins known to influence host-pathogen interaction in oomycete, fungal, bacterial, or nematode species. These 129 proteins were categorized in different PHI-phenotypes, of which 70 were assigned to the ‘reduced virulence’ phenotype, 45 to ‘plant avirulence determinant’, 29 to ‘increased virulence’, and nine to ‘loss of pathogenicity’ (Figure 2) (Supplementary Material S3). Twenty-four of the proteins were assigned to more than one PHI-phenotype.
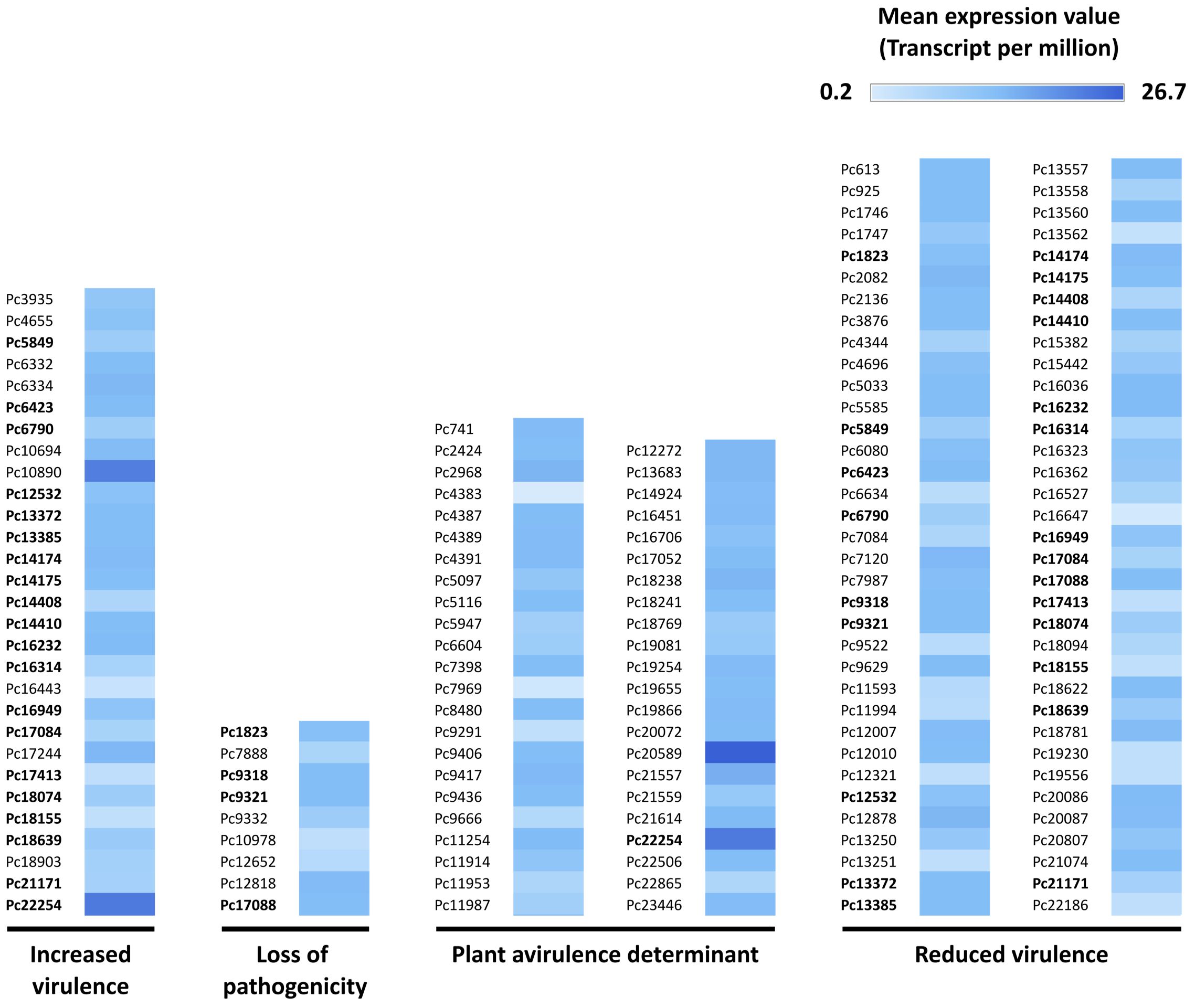
Figure 2 Secreted Phytophthora cactorum proteins with similarity to proteins that influence pathogen host interaction (PHI), based on the PHI-base (Urban et al., 2022). The proteins are deduced from transcripts detected in the rhizome (crown) of a susceptible Fragaria vesca genotype (NCGR1218), 48 hours after inoculation with P. cactorum. The PHI-phenotypes are resulting from a mutation or altered expression of the specific gene in the pathogen while ‘plant avirulence determinant’ represents an effector required for recognition of a pathogen by the resistant host. Pc is the abbreviated form of PC110_g in the gene identifier and is obtained from P. cactorum accession GCA_003287315.1. The gene IDs in bold are assigned to more than one PHI-phenotype.
3.3 RXLR effectors that trigger cell death in Nicotiana benthamiana
Twenty of the 40 candidate RXLR effector genes with high expression values during infection of strawberry or with high similarity to known Phytophthora effectors were selected for transient expression in N. benthamiana (Table 3). The expression of the agroinfiltrated RXLR effector genes in N. benthamiana were confirmed three days post agroinfiltration using RT-PCR (Supplementary Figure S1), while the phenotypic responses of the expressed RXLR genes were recorded five days post agroinfiltration. Five previously unreported RXLR effector genes, Pc741 (PC110_g741), Pc8318 (PC110_g8318), Pc10890 (PC110_g10890), Pc20813 (PC110_g20813), and Pc22290 (PC110_g22290) induced cell death when expressed in N. benthamiana leaves, which was confirmed with trypan blue staining (Figure 3). The cell death induced by the five RXLR effectors were further confirmed by 3, 3’- diaminobenzidine (DAB) staining, which showed a strong brown color precipitation in the RXLR agroinfiltrated regions, indicating an accumulation of H2O2 compared to the regions agroinfiltrated with the empty vector control (Supplementary Figure S2). Furthermore, aniline blue staining showed fluorescence in the RXLR agroinfiltrated regions with induced cell death, demonstrating accumulation of callose, in contrast to the regions agroinfiltrated with empty vector control (Supplementary Figure S3). These results indicate that the five RXLR effectors could activate the immune system of N. benthamiana.
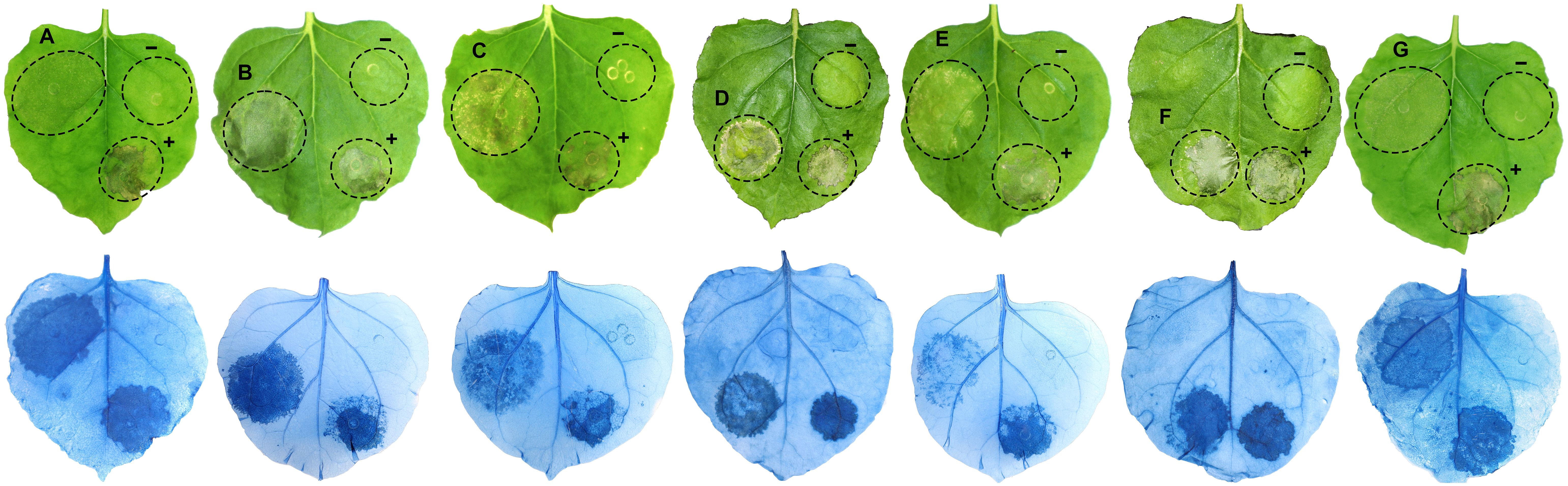
Figure 3 Cell death induced by Phytophthora cactorum RXLR effector genes (A) Pc741, (B) Pc8318, (C) Pc10890, (D) Pc16451, (E) Pc20813, (F) Pc22254, and (G) Pc22290 after transient expression in Nicotiana benthamiana leaves. The lower panel confirms cell death response after trypan blue staining. INF1, an elicitor gene of cell death from Phytophthora infestans was used as a positive control (+), and the empty vector pK7WG2 was used as a negative control (-) in each of the leaves. In addition, two previously reported cell death inducing RXLR genes (D) Pc16451 and (F) Pc22254 from P. cactorum (Chen et al., 2014) were included as positive controls. Images were taken five days after agroinfiltration of the gene constructs and after trypan blue staining of the same leaf. Pc is the abbreviated form of PC110_g from the gene identifier and is obtained from P. cactorum accession GCA_003287315.1.
The protein template modeling (TM)-score matrix for RXLR effectors revealed that the TM-scores for 99.6% of the pairwise comparisons of effectors were 0.5 or below (Supplementary Material S4), suggesting that they have different structures. For the five RXLRs that induced cell death in N. benthamiana (Pc741, Pc8318, Pc10890, Pc20813, and Pc22290), the TM scores were less than 0.4 in pairwise comparison with each other. Three RXLR effector pairs gave TM scores slightly above 0.5, which could indicate some structural similarity. However, the scores were all below 0.6, so not very confident. Phylogenetic analysis of these five RXLRs with homologs from 23 different P. cactorum strains revealed that most of the homologs from strawberry strains were highly similar. Some of the homologs of Pc741, Pc8318, and Pc22290 from the apple strains were phylogenetically distinct from particularly the crown rot strains (Supplementary Figure S4). Homologs of Pc10890 were detected only in 16 of the P. cactorum genomes available, and these were all identical (Supplementary Material S5).
In the cell death inducing RXLR effectors, from zero to four sites under positive selection were detected and from zero to six sites under negative selection (Supplementary Material S6). The RXLR effector Pc20813, which was among the P. cactorum genes with highest expression (13 to 17 fold higher than Pc741, Pc8318 and 22290), had exclusively positive selection sites compared to its homologs.
4 Discussion
In this study, the transcriptome of the strain P. cactorum 10300 was investigated during infection of the susceptible Fragaria vesca genotype NCGR1218 at 48 hours post inoculation, which is an early and presumably important phase of the infection (Toljamo et al., 2016; Gogoi et al., 2023a). No visible necrotic lesions were observed at this time point, which is consistent with the previous report on the Hawaii4 genotype, where only hyphal growth was visible on the root surfaces after inoculation with P. cactorum (Toljamo et al., 2016). Our study focused on the transcriptome profile of P. cactorum during infection of its natural host, while previous transcriptomics studies were either based on different life stages of P. cactorum, infection of model hosts like Nicotiana benthamiana, N. tabacum, and Solanum lycopersicum, or strawberry in vitro (Chen et al., 2014, 2018; Nellist et al., 2021). In total 4668 P. cactorum transcripts encoding proteins were detected. GO and KEGG analysis indicated that most of the proteins encoded by the transcripts had binding and catalytic activity and were involved in the metabolism of the pathogen (Figure 1), but 539 (˜12%) were predicted to encode secreted effector proteins belonging to different apoplastic and cytoplasmic effector groups (Table 1; Supplementary Material S1). Based on analysis of the secreted proteins encoded by the detected P. cactorum transcripts in the pathogen-host interaction (PHI)-database (Urban et al., 2022), 129 of 539 secreted proteins were 30–99% identical to proteins previously demonstrated to have influence on virulence of different pathogens, including Phytophthora spp. (Figure 2; Supplementary Material S3).
As much as 40% of the predicted secreted proteins encoded by P. cactorum transcripts detected during strawberry infection were hypothetical proteins with unknown function. Of the predicted secreted proteins with a putative function, most belonged to typical apoplastic effector groups of which the largest was carbohydrate active enzymes (CAZymes) that are known to participate in host cell wall degradation and metabolism to facilitate infection (Ospina-Giraldo et al., 2010). Of the 120 proteins belonging to the CAZymes, the majority were glycoside hydrolases (GHs). The GHs secreted by plant associated fungi and oomycetes represent the largest class of CAZymes (Bradley et al., 2022). Numerous CAZymes belonging to different families including AA17, CBM25, GH3, GH5, GH6, GH7, GH10, GH12, GH16, GH17, GH28 detected in this study have previously been described as virulence factors in different pathogens (Hardham and Blackman, 2018; Rafiei et al., 2021; Sabbadin et al., 2021; Bradley et al., 2022). For instance, the xylanases PpXYn1 and PpXyn2, belonging to the GH10 family, are virulence factors that degrade xylan and are upregulated during Phytophthora parasitica infection (Lai and Liou, 2018). However, some of the CAZymes that act as virulence factors may also elicit defense responses in the host. For example, the cellobiohydrolase PsGH7 and the xyloglucanase PsXEG1 (GH12 family) from Phytophthora sojae are virulence factors that promote infection, while they also elicit hypersensitive response in the soybean host, due to recognition as pathogen-associated molecular patterns (PAMPs) (Ma et al., 2015; Tan et al., 2020).
In addition to CAZymes, twenty-three proteolytic enzymes including proteases and peptidases were identified in our study. Proteolytic enzymes are important virulence factors in plant-pathogen interactions and may disrupt the host defense for successful invasion (Jashni et al., 2015). For instance, the cysteine protease genes PpCys44 and PpCys45 of P. parasitica have been shown to act as virulence factors during infection in N. benthamiana (Zhang et al., 2020). Three enzyme inhibitors, Kazal-type serine protease inhibitor, protease inhibitor epic4 and elastase-like inhibitors, were also identified. This group of proteins, particularly the protease inhibitors are known to bind and inhibit the function of host apoplastic proteases during plant colonization (Jashni et al., 2015). For instance, an extracellular kazal-like serine protease inhibitor EP1 from P. infestans has been shown to inhibit and interact with the pathogenesis-related P69B subtilisin-like serine protease of tomato (Tian et al., 2004). Similarly, protein inhibitors like EPIC genes were shown to be upregulated during plant infection and their products are known to inhibit papain-like cysteine proteases (Tian et al., 2007; Kaschani et al., 2010).
Furthermore, nine elicitins, seven cysteine rich proteins, seven necrosis inducing proteins (NPPs), four transglutaminase elicitors, and one phytotoxin protein were identified. Cysteine rich proteins, elicitins, and NPPs in general are considered PAMPs that may trigger immunity in host plants leading to cell death (Orsomando et al., 2001; Naveed et al., 2020; Midgley et al., 2022). However, some of these proteins also function as virulence factors in different pathogens. For instance, cysteine rich protein (SCR96) in P. cactorum (Chen et al., 2016b), elicitin protein (β-cinnamomin) in P. cinnamomi (Horta et al., 2010; Islam et al., 2019), cell wall transglutaminase elicitor in P. infestans (Brus-Szkalej et al., 2021), and phytotoxic protein SCR82 in P. capsici (Zhang et al., 2021) function as pathogen virulence factors in addition to triggering cell death as a result of PAMP recognition.
Of cytoplasmic effector groups, only three CRNs were identified in our study, of which PC110_g7969 showed 81% identity (86% similarity) to CRN1 from P. infestans that trigger necrotic responses in N. benthamiana and the host plant tomato (Torto et al., 2003). CRNs are known to result in cell death and chlorosis when expressed in the plant, however some of the CRNs can also suppress cell death, thereby promoting virulence. For instance, P. parasitica CRN effector PpCRN7 enhances INF1 induced cell death in N. benthamiana, while PpCRN20 suppresses it. Despite their contrasting functions, both PpCRN7 and PpCRN20 increase plant susceptibility to P. parasitica (Maximo et al., 2019).
Forty RXLR candidates were detected in the transcripts, which constitutes only 20% of the predicted RXLRs in the genome of P. cactorum (Armitage et al., 2018). This was 10% less than detected by Nellist et al. (2021) at 48h after inoculation, but they used tissue culture plants which are much more fragile than the plants used in our study thus promoting a more rapid infection process. Twenty of the 40 detected RXLR candidates were transiently expressed in N. benthamiana to investigate their cell death inducing properties and five previously unreported RXLR effector genes, Pc741, Pc8318, Pc10890, Pc20813, and Pc22290, induced cell death. Interestingly, Pc22290, an RXLR candidate without the EER motif induced cell death. The EER motif supposedly helps in translocation of RXLR effectors into the host cell, but an exact RXLR-EER sequence is not a requirement for its translocation (Dou et al., 2008; Van West et al., 2010; Chen et al., 2020). RXLR effectors that lack the EER motif have also previously been shown to induce cell death, e.g., the avirulence protein ATR13 from the oomycete pathogen Hyaloperonospora parasitica induced cell death in Arabidopsis (Allen et al., 2004).
The cell death region induced by RXLR effectors in N. benthamiana leaves showed accumulation of reactive oxygen species (ROS) such as H2O2 and callose deposits as confirmed by 3, 3’- diaminobenzidine (DAB) and aniline blue staining, respectively. Similar responses have been observed for RXLR effectors from other Phytophthora species (Yin et al., 2019; Situ et al., 2020). ROS like H2O2 strengthen the plant cell walls through oxidative cross-linking and also act as signaling molecules that induce defense responses (Alvarez et al., 1998; Steffens et al., 2013; Redza-Dutordoir and Averill-Bates, 2016). Furthermore, callose deposition is one of the immune responses deployed by plants upon recognition of ‘non-self’ components and is induced during PTI (Du et al., 2015). It is not yet clear whether the cell death induced is the direct effect of the RXLR effectors or due to recognition of the effector by resistance proteins in N. benthamiana. However, deposition of callose in the RXLR expressing leaves suggests that cell death is a result of host defense response (Jin and Mackey, 2017). It should be noted that these RXLR effectors not necessarily induce similar effects in strawberry, but the response in N. benthamiana is an indication of their relevance.
Since the RXLR effectors are known to target a wide range of cellular processes, the protein sequences of the identified cell death inducing RXLR effectors from P. cactorum were blasted against the UniProt database to look for similarities with functionally studied proteins from other pathogens. The RXLR effector Pc741 showed 44% sequence identity (54% similarity) to the P. capsici RXLR207 that induced ROS-mediated cell death in transgenic Arabidopsis and reduced pathogen colonization. However, a mutation in RXLR207 resulted in decreased pathogen virulence, suggesting it is crucial for infection (Li et al., 2019). The Pc8318 RXLR effector showed 54% identity (68% similarity) to the RXLR effector CRE5 (PITG_06308) of P. infestans, which did not induce cell death but in contrast suppressed cell death induced by Avh238, Avh241, BAX, INF1, thus indicating its potential roles in the virulence of Phytophthora spp (Yin et al., 2017). The Pc10890 RXLR effector showed 66% identity (77% similarity) with RXLR effector SFI6 from P. infestans, which like CRE5 did not induce cell death, but suppressed PAMP-triggered immunity in Arabidopsis and tomato, and increased virulence of P. infestans in N. benthamiana (Zheng et al., 2014). The Pc20813 RXLR effector showed 31% identity (47% similarity) with P. infestans RXLR effector SFI2 that induces cell death in N. benthamiana but attenuates flg22-induced immune response in tomato and Arabidopsis (Zheng et al., 2014). Furthermore, the Pc22290 RXLR effector showed 36% identity (53% similarity) with P. parasitica effector RXLR3 that did not induce cell death but significantly increased Phytophthora infection, and moderately suppressed INF1 induced cell death in N. benthamiana (Dalio et al., 2018). Although the five RXLR effectors induced cell death in N. benthamiana, it has yet to be confirmed if the induced cell death can inhibit P. cactorum infection. Thirteen of the 20 cloned RXLR effectors did not induce cell death in N. benthamiana indicating that these RXLRs do not directly activate the plant’s immune system. Some RXLR effectors that do not trigger cell death, instead can suppress PTI or ETI induced cell death thereby promoting pathogen virulence (Wang et al., 2023). The cell death suppression effect of RXLRs can be studied by co-infiltration with known effectors or elicitins that induce cell death in N. benthamiana (Zhan et al., 2022).
Two of the RXLR effectors that induced cell death in N. benthamiana, Pc741 and Pc22254, were assigned to the PHI-phenotype ‘plant avirulence determinant’, hence confirming the prediction from the PHI database and demonstrating that it can be a useful tool for selection of effector candidates for further characterization and functional studies.
Only a few P. cactorum effectors have been functionally characterized so far: four RXLRs, one NPP, two elicitins, and four cysteine rich proteins (Chen et al., 2023). The five additional cell death inducing RXLRs identified in this study can in principle play important roles in pathogenicity, despite being recognized by N. benthamiana when transiently expressed. However, the direct roles of the RXLR effectors identified in this study, in the virulence of P. cactorum remain to be elucidated. The protein structural similarity analysis of the identified RXLR effectors indicated that 99.6% of them were structurally unrelated with TM scores 0.5 or below. This included the RXLRs that induced cell death in N. benthamiana. Homology analysis revealed that all cell death inducing effectors were conserved across different strains of P. cactorum, but that some of the homologs of Pc741, Pc8318, and Pc22290 from the apple strains were phylogenetically distinct from particularly the crown rot strains (Supplementary Figure S4, Supplementary Material S5). The RXLR effector Pc20813 exhibited only sites under positive selection, indicating that these amino acid residues have an important role in the host-pathogen interaction (Sironi et al., 2015). The Pc20813 gene was among the genes with highest expression during the infection and displayed greater divergence among its homologs in P. cactorum strains than the other cell death inducing effectors. This contrasts with previous findings that suggest genes with higher expression levels tend to diverge less than those with lower expression (Pál et al., 2001; Yang et al., 2012; Williamson et al., 2014). In three other RXLRs (Pc741, Pc8318 and Pc22290), more negative selection sites were observed suggesting that these genes might be under strong negative selection favoring amino acid substitutions that can affect pathogen’s fitness.
Previous studies have reported that pathogens drive effector evolution through mutation and sequence substitution in RXLR effector genes, thus promoting their virulence (Win and Kamoun, 2007; Jiang et al., 2008; Dong et al., 2009; Qutob et al., 2009; Cui et al., 2012). Further analysis of polymorphisms in the equivalent RXLR effector genes from other strains of P. cactorum could therefore provide more insights about their important domains and evolution of virulence mechanisms. In addition, identification of the host receptors or protein interacting partners of the identified cell death inducing RXLR effectors could help to understand signaling pathways involved in cell death and provide insights into resistance mechanism involved. These insights can further help in screening of resistant strawberry genotypes and development of future disease control strategies.
5 Conclusion
The present transcriptome study provides comprehensive insights of the Phytophthora cactorum genes expressed during the early stage of infection of the rhizome of the model plant, Fragaria vesca. A total of 4668 P. cactorum transcripts were identified, and 539 of these were predicted to encode secreted proteins belonging to different effector families including CAZymes, elicitins, cysteine rich proteins, necrosis inducing proteins, proteolytic enzymes and RXLRs. Twenty of the 40 RXLR effectors identified were transiently expressed in Nicotiana benthamiana, and five previously unreported RXLR effectors triggered cell death response. The functional roles of these RXLR effectors in strawberry infection is not yet known and need further investigation. Further research on the subcellular localization of these RXLR proteins in the plant cell, and interactions of host proteins and the RXLR effectors of P. cactorum can help to develop new strategies for breeding resistance in strawberry.
Data availability statement
The data presented in the study are deposited in the ArrayExpress repository, accession no. E-MTAB-12152, released on 1 January 2023. https://www.ebi.ac.uk/biostudies/arrayexpress/studies/E-MTAB-12152.
Author contributions
BG: Methodology, Formal Analysis, Writing – original draft, Investigation. AG: Writing – original draft, Methodology, Investigation, Formal Analysis. MP: Writing – review & editing, Investigation. AS: Writing – review & editing. MB: Writing – original draft, Supervision, Project administration, Investigation, Funding acquisition, Conceptualization.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. BG was funded by the European Union’s Horizon 2020 (H2020) Marie Skłodowska-Curie Actions Innovative Training Networks PROTECTA H2020-MSCA-ITN-2017 call, under the grant agreement number 766048. The work was also supported by NIBIO (basic funding) and the Research Council of Norway, grant number 326212.
Acknowledgments
Dr. Jahn Davik (NIBIO) is acknowledged for providing strawberry genotypes, Dr. Erik Lysøe (NIBIO) for helping in the initial assembling of the transcriptomics data, the RHIZO team at the Laboratory of Plant Science (LRSV), Toulouse, France for providing the AGL1 Agrobacterium strain, and Prof. Dr. Vivianne Vleeshouwers, Department of Plant Sciences, Wageningen University, The Netherlands for providing the INF1 construct.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1379970/full#supplementary-material
References
Ali, A., Kumar, R., Mazákova, J., Maňasová, M., Zouhar, M., Pánek, M. (2022). Evaluation of the ability of seven active ingredients of fungicides to suppress Phytophthora cactorum at diverse life stages, and variability in resistance found among isolates. J. Fungi 8, Article 1039. doi: 10.3390/jof8101039
Allen, R. L., Bittner-Eddy, P. D., Grenville-Briggs, L. J., Meitz, J. C., Rehmany, A. P., Rose, L. E., et al. (2004). Host-parasite coevolutionary conflict between Arabidopsis and downy mildew. Sci. (80-.) 306, 1957–1960. doi: 10.1126/science.1104022
Alvarez, M. E., Pennell, R. I., Meijer, P. J., Ishikawa, A., Dixon, R. A., Lamb, C. (1998). Reactive oxygen intermediates mediate a systemic signal network in the establishment of plant immunity. Cell 92, 773–784. doi: 10.1016/S0092-8674(00)81405-1
Anderson, R. G., Deb, D., Fedkenheuer, K., McDowell, J. M. (2015). Recent progress in RXLR effector research. Mol. Plant-Microbe Interact. 28, 1063–1072. doi: 10.1094/MPMI-01-15-0022-CR
Armenteros, J. J. A., Salvatore, M., Emanuelsson, O., Winther, O., Von Heijne, G., Elofsson, A., et al. (2019a). Detecting sequence signals in targeting peptides using deep learning. Life Sci. Alliance 2, e201900429. doi: 10.26508/lsa.201900429
Armenteros, J. J. A., Tsirigos, K. D., Sønderby, C. K., Petersen, T. N., Winther, O., Brunak, S., et al. (2019b). SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 37, 420–423. doi: 10.1038/s41587-019-0036-z
Armitage, A. D., Lysøe, E., Nellist, C. F., Lewis, L. A., Cano, L. M., Harrison, R. J., et al. (2018). Bioinformatic characterization of the effector repertoire of the strawberry pathogen Phytophthora cactorum. PloS One 13, e0202305. doi: 10.1371/journal.pone.0202305
Bach-Pages, M., Preston, G. M. (2018). Methods to quantify biotic-induced stress in plants. Methods Mol. Biol. 1734, 241–255. doi: 10.1007/978–1-4939–7604-1_19
Bhattacharjee, S., Hiller, N. L., Liolios, K., Win, J., Kanneganti, T. D., Young, C., et al. (2006). The malarial host-targeting signal is conserved in the irish potato famine pathogen. PloS Pathog. 2, e50. doi: 10.1371/journal.ppat.0020050
Birch, P. R. J., Boevink, P. C., Gilroy, E. M., Hein, I., Pritchard, L., Whisson, S. C. (2008). Oomycete RXLR effectors: delivery, functional redundancy and durable disease resistance. Curr. Opin. Plant Biol. 11, 373–379. doi: 10.1016/j.pbi.2008.04.005
Boevink, P. C., Birch, P. R. J., Turnbull, D., Whisson, S. C. (2020). Devastating intimacy: the cell biology of plant–Phytophthora interactions. New Phytol. 228, 445–458. doi: 10.1111/nph.16650
Bradley, E. L., Ökmen, B., Doehlemann, G., Henrissat, B., Bradshaw, R. E., Mesarich, C. H. (2022). Secreted glycoside hydrolase proteins as effectors and invasion patterns of plant-associated fungi and oomycetes. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.853106
Bray, N. L., Pimentel, H., Melsted, P., Pachter, L. (2016). Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527. doi: 10.1038/nbt.3519
Brus-Szkalej, M., Andersen, C. B., Vetukuri, R. R., Grenville-Briggs, L. J. (2021). A family of cell wall transglutaminases is essential for appressorium development and pathogenicity in Phytophthora infestans. bioRxiv. doi: 10.1101/2021.11.23.469665
Chen, X. R., Zhang, B. Y., Xing, Y. P., Li, Q. Y., Li, Y. P., Tong, Y. H., et al. (2014). Transcriptomic analysis of the phytopathogenic oomycete Phytophthora cactorum provides insights into infection-related effectors. BMC Genomics 15, Article 980. doi: 10.1186/1471-2164-15-980
Chen, X. R., Brurberg, M. B., Elameen, A., Klemsdal, S. S., Martinussen, I. (2016a). Expression of resistance gene analogs in woodland strawberry (Fragaria vesca) during infection with Phytophthora cactorum. Mol. Genet. Genomics 291, 1967–1978. doi: 10.1007/s00438-016-1232-x
Chen, X. R., Li, Y. P., Li, Q. Y., Xing, Y. P., Liu, B. B., Tong, Y. H., et al. (2016b). SCR96, a small cysteine-rich secretory protein of Phytophthora cactorum, can trigger cell death in the Solanaceae and is important for pathogenicity and oxidative stress tolerance. Mol. Plant Pathol. 17, 577–587. doi: 10.1111/mpp.12303
Chen, X. R., Huang, S. X., Zhang, Y., Sheng, G. L., Zhang, B. Y., Li, Q. Y., et al. (2018). Transcription profiling and identification of infection-related genes in Phytophthora cactorum. Mol. Genet. Genomics 293, 541–555. doi: 10.1007/s00438-017-1400-7
Chen, T., Liu, R., Dou, M., Li, M., Li, M., Yin, X., et al. (2020). Insight into function and subcellular localization of Plasmopara viticola putative RXLR effectors. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.00692
Chen, X. R., Wen, K., Zhou, X., Zhu, M. Y., Liu, Y., Jin, J. H., et al. (2023). The devastating oomycete phytopathogen Phytophthora cactorum: insights into its biology and molecular features. Mol. Plant Pathol. 24, 1017–1032. doi: 10.1111/mpp.13345
Cui, L., Yin, W., Dong, S., Wang, Y. (2012). Analysis of polymorphism and transcription of the effector gene Avr1b in Phytophthora sojae isolates from China virulent to Rps1b. Mol. Plant Pathol. 13, 114–122. doi: 10.1111/j.1364-3703.2011.00733.x
Dalio, R. J. D., Maximo, H. J., Oliveira, T. S., Dias, R. O., Breton, M. C., Felizatti, H., et al. (2018). Phytophthora parasitica effector PpRXLR2 suppresses Nicotiana benthamiana immunity. Mol. Plant-Microbe Interact. 31, 481–493. doi: 10.1094/MPMI-07-17-0158-FI
de Castro, E., Sigrist, C. J. A., Gattiker, A., Bulliard, V., Langendijk-Genevaux, P. S., Gasteiger, E., et al. (2006). ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 34, W362–W365. doi: 10.1093/nar/gkl124
Dodds, P. N., Rathjen, J. P. (2010). Plant immunity: towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 11, 539–548. doi: 10.1038/nrg2812
Dong, S., Qutob, D., Tedman-Jones, J., Kuflu, K., Yuanchao, W., Tyler, B. M., et al. (2009). The Phytophthora sojae avirulence locus Avr3c encodes a multi-copy RXLR effector with sequence polymorphisms among pathogen strains. PloS One 4, e5556. doi: 10.1371/journal.pone.0005556
Dou, D., Kale, S. D., Wang, X., Jiang, R. H. Y., Bruce, N. A., Arredondo, F. D., et al. (2008). RXLR-mediated entry of Phytophthora sojae effector Avr1b into soybean cells does not require pathogen-encoded machinery. Plant Cell 20, 1930–1947. doi: 10.1105/tpc.107.056093
Dou, D., Zhou, J. M. (2012). Phytopathogen effectors subverting host immunity: different foes, similar battleground. Cell Host Microbe 12, 484–495. doi: 10.1016/j.chom.2012.09.003
Du, Y., Mpina, M. H., Birch, P. R. J., Bouwmeester, K., Govers, F. (2015). Phytophthora infestans RXLR effector AVR1 interacts with exocyst component Sec5 to manipulate plant immunity. Plant Physiol. 169, 1975–1990. doi: 10.1104/pp.15.01169
Du, J., Rietman, H., Vleeshouwers, V. G. A. A. (2014). Agroinfiltration and PVX agroinfection in potato and Nicotiana benthamiana. J. Vis. Exp. 83, e50971. doi: 10.3791/50971
Eikemo, H., Stensvand, A., Tronsmo, A. M. (2000). Evaluation of methods of screening strawberry cultivars for resistance to crown rot caused by Phytophthora cactorum. Ann. Appl. Biol. 137, 237–244. doi: 10.1111/j.1744-7348.2000.tb00064.x
Ellis, M. A., Grove, G. G. (1983). Leather rot in Ohio strawberries. Plant Dis. 67, 549. doi: 10.1094/PD-67-549
Emms, D. M., Kelly, S. (2019). OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20, 1–14. doi: 10.1186/s13059-019-1832-y
Erwin, D. C., Ribeiro, O. K. (1996). Phytophthora diseases worldwide (St. Paul, MN: American Phytopathological Society (APS Press).
Ferguson, A. J., Jeffers, S. N. (1999). Detecting multiple species of Phytophthora in container mixes from ornamental crop nurseries. Plant Dis. 83, 1129–1136. doi: 10.1094/PDIS.1999.83.12.1129
Froger, A., Hall, J. E. (2007). Transformation of plasmid DNA into E. coli using the heat shock method. J. Vis. Exp. 6, Article 253. doi: 10.3791/253
Giraldo, M. C., Valent, B. (2013). Filamentous plant pathogen effectors in action. Nat. Rev. Microbiol. 11, 800–814. doi: 10.1038/nrmicro3119
Gogoi, A., Lysøe, E., Eikemo, H., Stensvand, A., Davik, J., Brurberg, M. B. (2023a). Comparative transcriptome analysis reveals novel candidate resistance genes involved in defense against Phytophthora cactorum in strawberry. Int. J. Mol. Sci. 24, Article 10851. doi: 10.3390/ijms241310851
Gogoi, A., Rossmann, S. L., Lysøe, E., Stensvand, A., Brurberg, M. B. (2023b). Genome analysis of Phytophthora cactorum strains associated with crown- and leather-rot in strawberry. Front. Microbiol. 14. doi: 10.3389/fmicb.2023.1214924
Götz, S., García-Gómez, J. M., Terol, J., Williams, T. D., Nagaraj, S. H., Nueda, M. J., et al. (2008). High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 36, 3420–3435. doi: 10.1093/nar/gkn176
Hantula, J., Lilja, A., Nuorteva, H., Parikka, P., Werres, S. (2000). Pathogenicity, morphology and genetic variation of Phytophthora cactorum from strawberry, apple, rhododendron, and silver birch. Mycol. Res. 104, 1062–1068. doi: 10.1017/S0953756200002999
Hardham, A. R., Blackman, L. M. (2018). Phytophthora cinnamomi. Mol. Plant Pathol. 19, 260–285. doi: 10.1111/mpp.12568
Hardham, A. R., Cahill, D. M. (2010). The role of oomycete effectors in plant-pathogen interactions. Funct. Plant Biol. 37, 919–925. doi: 10.1071/FP10073
Horta, M., Caetano, P., Medeira, C., Maia, I., Cravador, A. (2010). Involvement of the β-cinnamomin elicitin in infection and colonization of cork oak roots by Phytophthora cinnamomi. Eur. J. Plant Pathol. 127, 427–436. doi: 10.1007/s10658-010-9609-x
Huang, L., Zhang, H., Wu, P., Entwistle, S., Li, X., Yohe, T., et al. (2018). DbCAN-seq: a database of carbohydrate-active enzyme (CAZyme) sequence and annotation. Nucleic Acids Res. 46, D516–D521. doi: 10.1093/nar/gkx894
Islam, M. T., Hussain, H. I., Russo, R., Chambery, A., Amoresano, A., Schallmey, A., et al. (2019). Functional analysis of elicitins and identification of cell wall proteins in Phytophthora cinnamomi. Physiol. Mol. Plant Pathol. 107, 21–32. doi: 10.1016/j.pmpp.2019.04.003
Jashni, M. K., Mehrabi, R., Collemare, J., Mesarich, C. H., de Wit, P. J. G. M. (2015). The battle in the apoplast: further insights into the roles of proteases and their inhibitors in plant–pathogen interactions. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00584
Jiang, R. H. Y., Tripathy, S., Govers, F., Tyler, B. M. (2008). RXLR effector reservoir in two Phytophthora species is dominated by a single rapidly evolving superfamily with more than 700 members. Proc. Natl. Acad. Sci. U. S. A. 105, 4874–4879. doi: 10.1073/pnas.0709303105
Jin, L., Mackey, D. M. (2017). Measuring callose deposition, an indicator of cell wall reinforcement, during bacterial infection in Arabidopsis. Methods Mol. Biol. 1578, 195–205. doi: 10.1007/978–1-4939–6859-6_16
Jones, J. D. G., Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Kaschani, F., Shabab, M., Bozkurt, T., Shindo, T., Schornack, S., Gu, C., et al. (2010). An effector-targeted protease contributes to defense against Phytophthora infestans and is under diversifying selection in natural hosts. Plant Physiol. 154 (4), 1794–1804. doi: 10.1104/pp.110.158030
Käll, L., Krogh, A., Sonnhammer, E. L. L. (2004). A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 338, 1027–1036. doi: 10.1016/j.jmb.2004.03.016
Kamoun, S. (2006). A catalogue of the effector secretome of plant pathogenic ooomycetes. Annu. Rev. Phytopathol. 44, 41–60. doi: 10.1146/annurev.phyto.44.070505.143436
Kamoun, S., Van West, P., De Jong, A. J., De Groot, K. E., Vleeshouwers, V. G. A. A., Govers, F. (1997). A gene encoding a protein elicitor of Phytophthora infestans is down- regulated during infection of potato. Mol. Plant-Microbe Interact. 10, 13–20. doi: 10.1094/MPMI.1997.10.1.13
Kamoun, S., Van West, P., Vleeshouwers, V. G. A. A., De Groot, K. E., Govers, F. (1998). Resistance of Nicotiana benthamiana to Phytophthora infestans is mediated by the recognition of the elicitor protein INF1. Plant Cell 10, 1413–1425. doi: 10.1105/tpc.10.9.1413
Karimi, M., Inzé, D., Depicker, A. (2002). GATEWAY™ vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7, 193–195. doi: 10.1016/S1360-1385(02)02251-3
Keogh, R. C., Deverall, B. J., McLeod, S. (1980). Comparison of histological and physiological responses to Phakopsora pachyrhizi in resistant and susceptible soybean. Trans. Br. Mycol. Soc 74, 329–333. doi: 10.1016/S0007-1536(80)80163-X
Kosakovsky Pond, S. L., Poon, A. F. Y., Velazquez, R., Weaver, S., Hepler, N. L., Murrell, B., et al. (2020). HyPhy 2.5 - a customizable platform for evolutionary hypothesis testing using phylogenies. Mol. Biol. Evol. 37, 295–299. doi: 10.1093/molbev/msz197
Lai, M. W., Liou, R. F. (2018). Two genes encoding GH10 xylanases are essential for the virulence of the oomycete plant pathogen Phytophthora parasitica. Curr. Genet. 64, 931–943. doi: 10.1007/s00294-018-0814-z
Li, Q., Ai, G., Shen, D., Zou, F., Wang, J., Bai, T., et al. (2019). A Phytophthora capsici effector targets ACD11 binding partners that regulate ROS-mediated defense response in Arabidopsis. Mol. Plant 12, 565–581. doi: 10.1016/j.molp.2019.01.018
Liu, L., Xu, L., Jia, Q., Pan, R., Oelmüller, R., Zhang, W., et al. (2019). Arms race: diverse effector proteins with conserved motifs. Plant Signal. Behav. 14, Article 1557008. doi: 10.1080/15592324.2018.1557008
Ma, Z., Song, T., Zhu, L., Ye, W., Wang, Y., Shao, Y., et al. (2015). A Phytophthora sojae glycoside hydrolase 12 protein is a major virulence factor during soybean infection and is recognized as a PAMP. Plant Cell 27, 2057–2072. doi: 10.1105/tpc.15.00390
Maas, J. L. (1998). “Compendium of strawberry diseases, second edition,” in Your Expert Diagnostic Guide for Strawberry Diseases (St. Paul, MN: APS Publications), 16–81. doi: 10.1094/9780890546178.003
Madeira, F., Pearce, M., Tivey, A. R. N., Basutkar, P., Lee, J., Edbali, O., et al. (2022). Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic Acids Res. 50, W276–W279. doi: 10.1093/nar/gkac240
Marin, M. V., Seijo, T. E., Zuchelli, E., Peres, N. A. (2021). Resistance to mefenoxam of Phytophthora cactorum and Phytophthora nicotianae causing crown and leather rot in Florida strawberry. Plant Dis. 105, 3490–3495. doi: 10.1094/PDIS-11-20-2474-RE
Maximo, H. J., Dalio, R. J. D., Dias, R. O., Litholdo, C. G., Felizatti, H. L., MaChado, M. A. (2019). PpCRN7 and PpCRN20 of Phythophthora parasitica regulate plant cell death leading to enhancement of host susceptibility. BMC Plant Biol. 19, Article 544. doi: 10.1186/s12870-019-2129-8
Medzhitov, R., Janeway, C. A. (1997). Innate immunity: the virtues of a non-clonal system of recognition. Cell 91, 295–298. doi: 10.1016/S0092-8674(00)80412-2
Midgley, K. A., van den Berg, N., Swart, V. (2022). Unraveling plant cell death during Phytophthora infection. Microorganisms 10, Article 1139. doi: 10.3390/microorganisms10061139
Murrell, B., Moola, S., Mabona, A., Weighill, T., Sheward, D., Kosakovsky Pond, S. L., et al. (2013). FUBAR: a fast, unconstrained Bayesian approximation for inferring selection. Mol. Biol. Evol. 30, 1196–1205. doi: 10.1093/molbev/mst030
Naveed, Z. A., Wei, X., Chen, J., Mubeen, H., Ali, G. S. (2020). The PTI to ETI continuum in Phytophthora-plant interactions. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.593905
Nellist, C. F., Armitage, A. D., Bates, H. J., Sobczyk, M. K., Luberti, M., Lewis, L. A., et al. (2021). Comparative analysis of host-associated variation in Phytophthora cactorum. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.679936
Oh, S. K., Kamoun, S., Choi, D. (2010). Oomycetes RXLR effectors function as both activator and suppressor of plant immunity. Plant Pathol. J. 26, 209–215. doi: 10.5423/PPJ.2010.26.3.209
Orsomando, G., Lorenzi, M., Raffaelli, N., Dalla Rizza, M., Mezzetti, B., Ruggieri, S. (2001). Phytotoxic protein PcF, purification, characterization, and cDNA sequencing of a novel hydroxyproline-containing factor secreted by the strawberry pathogen Phytophthora cactorum. J. Biol. Chem. 276, 21578–21584. doi: 10.1074/jbc.M101377200
Ospina-Giraldo, M. D., Griffith, J. G., Laird, E. W., Mingora, C. (2010). The CAZyome of Phytophthora spp.: a comprehensive analysis of the gene complement coding for carbohydrate-active enzymes in species of the genus Phytophthora. BMC Genomics 11, 1–16. doi: 10.1186/1471-2164-11-525
Pál, C., Papp, B., Hurst, L. D. (2001). Highly expressed genes in yeast evolve slowly. Genetics 158, 927–931. doi: 10.1093/genetics/158.2.927
Qutob, D., Tedman-Jones, J., Dong, S., Kuflu, K., Pham, H., Wang, Y., et al. (2009). Copy number variation and transcriptional polymorphisms of Phytophthora sojae RXLR effector genes Avr1a and Avr3a. PloS One 4, e5066. doi: 10.1371/annotation/2a2adcf8-afbc-4d46-92c6-d543d6b29182
Rafiei, V., Vélëz, H., Tzelepis, G. (2021). The role of glycoside hydrolases in phytopathogenic fungi and oomycetes virulence. Int. J. Mol. Sci. 22, Article 9359. doi: 10.3390/ijms22179359
Redza-Dutordoir, M., Averill-Bates, D. A. (2016). Activation of apoptosis signaling pathways by reactive oxygen species. Biochim. Biophys. Acta - Mol. Cell Res. 1863, 2977–2992. doi: 10.1016/j.bbamcr.2016.09.012
Rehmany, A. P., Gordon, A., Rose, L. E., Allen, R. L., Armstrong, M. R., Whisson, S. C., et al. (2005). Differential recognition of highly divergent downy mildew avirulence gene alleles by RPP1 resistance genes from two Arabidopsis lines. Plant Cell 17, 1839–1850. doi: 10.1105/tpc.105.031807
Sabbadin, F., Urresti, S., Henrissat, B., Avrova, A. O., Welsh, L. R. J., Lindley, P. J., et al. (2021). Secreted pectin monooxygenases drive plant infection by pathogenic oomycetes. Sci. (80-.) 373, 774–779. doi: 10.1126/science.abj1342
Sanchez, A. D., Sosa, M. C., Lutz, M. C., Carreño, G. A., Ousset, M. J., Lucero, G. S. (2019). Identification and pathogenicity of Phytophthora species in pear commercial orchards in Argentina. Eur. J. Plant Pathol. 154, 811–822. doi: 10.1007/s10658-019-01705-2
Sironi, M., Cagliani, R., Forni, D., Clerici, M. (2015). Evolutionary insights into host–pathogen interactions from mammalian sequence data. Nat. Rev. Genet. 16, 224–236. doi: 10.1038/nrg3905
Situ, J., Jiang, L., Fan, X., Yang, W., Li, W., Xi, P., et al. (2020). An RXLR effector PlAvh142 from Peronophythora litchii triggers plant cell death and contributes to virulence. Mol. Plant Pathol. 21, 415–428. doi: 10.1111/mpp.12905
Steffens, B., Steffen-Heins, A., Sauter, M. (2013). Reactive oxygen species mediate growth and death in submerged plants. Front. Plant Sci. 4. doi: 10.3389/fpls.2013.00179
Stensvand, A., Herrero, M. L., Talgø, V. (1999). Crown rot caused by Phytophthora cactorum in Norwegian strawberry production. EPPO Bull. 29, 155–158. doi: 10.1111/j.1365-2338.1999.tb00809.x
Stornaiuolo, M., Lotti, L. V., Borgese, N., Torrisi, M. R., Mottola, G., Martire, G., et al. (2003). KDEL and KKXX retrieval signals appended to the same reporter protein determine different trafficking between endoplasmic reticulum, intermediate compartment, and golgi complex. Mol. Biol. Cell 14, 823–1294. doi: 10.1091/mbc.e02-08-0468
Szklarczyk, D., Gable, A. L., Nastou, K. C., Lyon, D., Kirsch, R., Pyysalo, S., et al. (2021). The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 49, D605–D612. doi: 10.1093/nar/gkab835
Tabima, J. F., Grünwald, N. J. (2019). EffectR: an expandable R package to predict candidate RXLR and CRN effectors in oomycetes using motif searches. Mol. Plant-Microbe Interact. 32, 1067–1076. doi: 10.1094/MPMI-10-18-0279-TA
Tan, X., Hu, Y., Jia, Y., Hou, X., Xu, Q., Han, C., et al. (2020). A conserved glycoside hydrolase family 7 cellobiohydrolase PsGH7a of Phytophthora sojae is required for full virulence on soybean. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.01285
Tian, M., Huitema, E., Da Cunha, L., Torto-Alalibo, T., Kamoun, S. (2004). A Kazal-like extracellular serine protease inhibitor from Phytophthora infestans targets the tomato pathogenesis-related protease P69B. J. Biol. Chem. 279 (25), 26370–26377. doi: 10.1074/jbc.M400941200
Tian, M., Win, J., Song, J., van der Hoorn, R., van der Knaap, E., Kamoun, S. (2007). A Phytophthora infestans cystatin-like protein targets a novel tomato papain-like apoplastic protease. Plant Physiol. 143 (1), 364–377. doi: 10.1104/pp.106.090050
Toljamo, A., Blande, D., Kärenlampi, S., Kokko, H. (2016). Reprogramming of strawberry (Fragaria vesca) root transcriptome in response to Phytophthora cactorum. PloS One 11, 1–21. doi: 10.1371/journal.pone.0161078
Torto, T. A., Li, S., Styer, A., Huitema, E., Testa, A., Gow, N. A. R., et al. (2003). EST mining and functional expression assays identify extracellular effector proteins from the plant pathogen Phytophthora. Genome Res. 13, 1675–1685. doi: 10.1101/gr.910003
Tsuda, K., Katagiri, F. (2010). Comparing signaling mechanisms engaged in pattern-triggered and effector-triggered immunity. Curr. Opin. Plant Biol. 13, 459–465. doi: 10.1016/j.pbi.2010.04.006
Urban, M., Cuzick, A., Seager, J., Wood, V., Rutherford, K., Venkatesh, S. Y., et al. (2022). PHI-base in 2022: a multi-species phenotype database for pathogen-host interactions. Nucleic Acids Res. 50, D837–D847. doi: 10.1093/nar/gkab1037
Van West, P., De Bruijn, I., Minor, K. L., Phillips, A. J., Robertson, E. J., Wawra, S., et al. (2010). The putative RXLR effector protein SpHtp1 from the fish pathogenic oomycete Saprolegnia parasitica is translocated into fish cells. FEMS Microbiol. Lett. 310, 127–137. doi: 10.1111/j.1574-6968.2010.02055.x
Varadi, M., Anyango, S., Deshpande, M., Nair, S., Natassia, C., Yordanova, G., et al. (2022). AlphaFold protein structure database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 50, D439–D444. doi: 10.1093/nar/gkab1061
Wagner, G. P., Kin, K., Lynch, V. J. (2012). Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci. 131, 281–285. doi: 10.1007/s12064-012-0162-3
Wang, S., McLellan, H., Boevink, P. C., Birch, P. R. J. (2023). RxLR effectors: Master modulators, modifiers and manipulators. Mol. Plant Microbe Interact. 36, 754–763. doi: 10.1094/MPMI-05-23-0054-CR
Wang, S., McLellan, H., Bukharova, T., He, Q., Murphy, F., Shi, J., et al. (2019). Phytophthora infestans RXLR effectors act in concert at diverse subcellular locations to enhance host colonization. J. Exp. Bot. 70, 343–356. doi: 10.1093/jxb/ery360
Wang, Y., Tyler, B. M., Wang, Y. (2019). Defense and counterdefense during plant-pathogenic oomycete infection. Annu. Rev. Microbiol. 73, 667–696. doi: 10.1146/annurev-micro-020518-120022
Weigel, D., Glazebrook, J. (2006). Transformation of Agrobacterium using the freeze-thaw method. Cold Spring Harb. Protoc. 2006. doi: 10.1101/pdb.prot4666
Whisson, S. C., Boevink, P. C., Moleleki, L., Avrova, A. O., Morales, J. G., Gilroy, E. M., et al. (2007). A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature 450, 115–118. doi: 10.1038/nature06203
Williamson, R. J., Josephs, E. B., Platts, A. E., Hazzouri, K. M., Haudry, A., Blanchette, M., et al. (2014). Evidence for widespread positive and negative selection in coding and conserved noncoding regions of Capsella grandiflora. PloS Genet. 10, e1004622. doi: 10.1371/journal.pgen.1004622
Win, J., Kamoun, S. (2007). Adaptive evolution has targeted the C-terminal domain of the RXLR effectors of plant pathogenic oomycetes. Plant Signal. Behav. 193, 2349–2369. doi: 10.1105/tpc.107.051037
Xu, J., Zhang, Y. (2010). How significant is a protein structure similarity with TM-score = 0.5? Bioinformatics 26, 889–895. doi: 10.1093/bioinformatics/btq066
Yang, M., Duan, S., Mei, X., Huang, H., Chen, W., Liu, Y., et al. (2018). The Phytophthora cactorum genome provides insights into the adaptation to host defense compounds and fungicides. Sci. Rep. 8, 1–11. doi: 10.1038/s41598-018-24939-2
Yang, J. R., Liao, B. Y., Zhuang, S. M., Zhang, J. (2012). Protein misinteraction avoidance causes highly expressed proteins to evolve slowly. Proc. Natl. Acad. Sci. U. S. A. 109, E831–E840. doi: 10.1073/pnas.1117408109
Yin, J., Gu, B., Huang, G., Tian, Y., Quan, J., Lindqvist-Kreuze, H., et al. (2017). Conserved RXLR effector genes of Phytophthora infestans expressed at the early stage of potato infection are suppressive to host defense. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.02155
Yin, X., Shang, B., Dou, M., Liu, R., Chen, T., Xiang, G., et al. (2019). The nuclear-localized RXLR effector PvAvh74 from Plasmopara viticola induces cell death and immunity responses in Nicotiana benthamiana. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.01531
Zhan, Z., Liu, H., Yang, Y., Liu, S., Li, X., Piao, Z. (2022). Identification and characterization of putative effectors from Plasmodiophora brassicae that suppress or induce cell death in Nicotiana benthamiana. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.881992
Zhang, Z. H., Jin, J. H., Sheng, G. L., Xing, Y. P., Liu, W., Zhou, X., et al. (2021). A small cysteine-rich phytotoxic protein of Phytophthora capsici functions as both plant defense elicitor and virulence factor. Mol. Plant-Microbe Interact. 34, 891–903. doi: 10.1094/MPMI-01-21-0025-R
Zhang, Q., Li, W., Yang, J., Xu, J., Meng, Y., Shan, W. (2020). Two Phytophthora parasitica cysteine protease genes, PpCys44 and PpCys45, trigger cell death in various Nicotiana spp. and act as virulence factors. Mol. Plant Pathol. 21, 541–554. doi: 10.1111/mpp.12915
Zhang, Y., Skolnick, J. (2005). TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 33, 2302–2309. doi: 10.1093/nar/gki524
Keywords: oomycete, RNA-Seq, agroinfiltration, disease resistance response, host-pathogen interaction
Citation: Ghimire B, Gogoi A, Poudel M, Stensvand A and Brurberg MB (2024) Transcriptome analysis of Phytophthora cactorum infecting strawberry identified RXLR effectors that induce cell death when transiently expressed in Nicotiana benthamiana. Front. Plant Sci. 15:1379970. doi: 10.3389/fpls.2024.1379970
Received: 31 January 2024; Accepted: 06 May 2024;
Published: 24 May 2024.
Edited by:
Manuel Avilés, Universidad de Sevilla, SpainReviewed by:
Peter Henry, United States Department of Agriculture, United StatesCarsten Pedersen, University of Copenhagen, Denmark
Copyright © 2024 Ghimire, Gogoi, Poudel, Stensvand and Brurberg. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: May Bente Brurberg, bWF5LmJydXJiZXJnQG5pYmlvLm5v
 Bikal Ghimire
Bikal Ghimire Anupam Gogoi
Anupam Gogoi Mandeep Poudel
Mandeep Poudel Arne Stensvand
Arne Stensvand May Bente Brurberg
May Bente Brurberg